Abstract
Spore formation in the budding yeast, Saccharomyces cerevisiae, involves de novo creation of four prospore membranes, each of which surrounds a haploid nucleus resulting from meiosis. The meiotic outer plaque (MOP) is a meiosis-specific protein complex associated with each meiosis II spindle pole body (SPB). Vesicle fusion on the MOP surface creates an initial prospore membrane anchored to the SPB. Ady4 is a meiosis-specific MOP component that stabilizes the MOP-prospore membrane interaction. We show that Ady4 recruits the lipid kinase, Mss4, to the MOP. MSS4 overexpression suppresses the ady4∆ spore formation defect, suggesting that a specific lipid environment provided by Mss4 promotes maintenance of prospore membrane attachment to MOPs. The meiosis-specific Spo21 protein is an essential structural MOP component. We show that the Spo21 N terminus contains an amphipathic helix that binds to prospore membranes. A mutant in SPO21 that removes positive charges from this helix shares phenotypic similarities to ady4∆. We propose that Mss4 generates negatively charged lipids in prospore membranes that enhance binding by the positively charged N terminus of Spo21, thereby providing a mechanism by which the MOP-prospore membrane interaction is stabilized.
INTRODUCTION
Sporulation is a specialized program of gametogenesis in Saccharomyces cerevisiae resulting in the packaging of nuclei containing haploid chromosome sets into spores (Neiman, 2011). Sporulation is triggered when diploid cells are cultured in a nonfermentable carbon source in the absence of nitrogen (Neiman, 2011). Sporulation is coupled with meiosis; cells undergo DNA replication and then two rounds of chromosomal segregation to give rise to four haploid chromosome sets that are then encapsulated within spores. The process of spore formation begins after anaphase II, when the haploid chromosome sets present at each of the four poles of the meiosis II spindle are captured within newly formed membranes, termed prospore membranes (Neiman, 1998). In yeast, spindle pole bodies (SPBs; equivalent to centrosomes in higher eukaryotes) are embedded in the nuclear envelope and required for prospore membrane formation (Moens and Rapport, 1971). Prospore membranes are double layered and generated de novo from Golgi-derived secretory vesicles (Neiman, 1998). At the onset of meiosis II, a novel structure called the meiosis II outer plaque (MOP) is formed on the cytoplasmic face of each SPB (Moens and Rapport, 1971). Prospore membrane formation initiates when vesicles dock onto a MOP and fuse to create the new membrane compartment, which then expands beyond the MOP to engulf the chromosomes (Moens and Rapport, 1971; Knop and Strasser, 2000; Bajgier et al., 2001). Throughout this expansion, each prospore membrane maintains contact with its MOP, and this connection is essential to successfully capture the chromosomes (Mathieson et al., 2010a).
There are three meiosis-specific proteins essential for the MOP structure: Spo21, Mpc54, and Spo74 (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). A deletion of any one of the genes encoding these proteins eliminates both MOPs and prospore membranes. As a result, no nuclei are packaged and no spores are formed (Knop and Strasser, 2000; Bajgier et al., 2001; Nickas et al., 2003). A fourth meiosis-specific MOP protein is Ady4 (Nickas et al., 2003). The role of Ady4 in spore formation is distinct from that of the other MOP proteins. ADY4 is dispensable for MOP and prospore membrane formation. However, an ady4Δ diploid exhibits premature disassembly of zero to four MOPs within a single cell, leading to asci with zero to four spores (Nickas et al., 2003; Mathieson et al., 2010b). In asci that contain two haploid spores (called dyads), the nucleus in each spore may be derived either from the same spindle (sister dyads) or from one pole of each meiosis II spindle (nonsister dyads; Davidow et al., 1980). The distribution of sister versus nonsister dyads in ady4Δ dyads is random. In contrast, diploids hemizygous or hypomorphic for the other MOP genes are biased toward nonsister dyads (Bajgier et al., 2001; Wesp et al., 2001; Nickas et al., 2003). Another difference between Ady4 and the other MOP proteins is that Ady4 shuttles on and off the MOP during prospore membrane formation while the other components remain stably associated to the outer plaque (Mathieson et al., 2010a).
The observation that ady4Δ asci contain fewer than four spores due to stochastic disassembly of MOPs suggests that Ady4 helps maintain the connection between MOPs and the growing prospore membranes (Nickas et al., 2003; Mathieson et al., 2010a). However, the mechanism by which Ady4 does this was unclear. This report shows that Ady4 recruits the phosphatidylinositol-4-phosphate-5-kinase (PI4P-5 kinase) Mss4 to the SPB. Furthermore, we demonstrate that the N terminus of the MOP component Spo21 contains a positively charged amphipathic helix that binds specifically to prospore membranes. A spo21 mutant lacking negative charges in this motif exhibits similar phenotypes as ady4Δ, including increased formation of random dyads. Our results suggest that Spo21 binding to acidic phospholipids generated by Mss4 in prospore membranes stabilizes the connection of MOPs to prospore membranes.
RESULTS
Ady4 and Mss4 interact in the two-hybrid assay
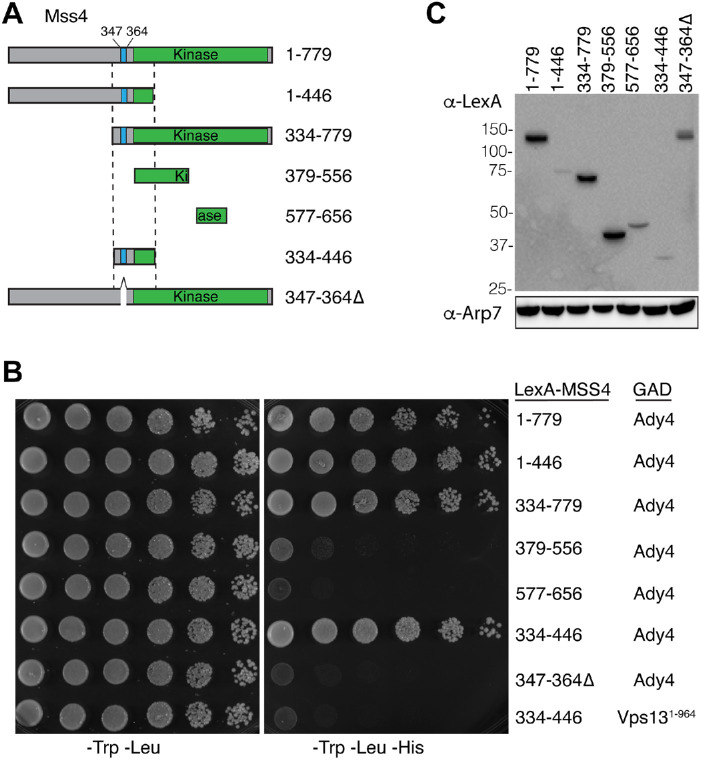
The variability in the number of spores due to the random disassembly of MOPs in ady4Δ suggests that Ady4 stabilizes the MOP-prospore membrane association either directly, or indirectly by interaction with another protein. In an earlier study, a high-throughput two-hybrid screen revealed an interaction between Ady4 and the lipid kinase Mss4 (Yu et al., 2008). MSS4 is an essential gene that encodes the sole PI4P-5 kinase in budding yeast, which localizes to the plasma membrane and the nucleus and is required for prospore membrane growth (Desrivieres et al., 1998; Homma et al., 1998; Audhya and Emr, 2003; Rudge et al., 2004; Mendonsa and Engebrecht, 2009). To map the Ady4 interaction domain on Mss4, various lexA-MSS4 fusions (Figure 1A) were tested for interaction with ADY4 fused to the GAL4 activation domain (GAD) using the two-hybrid system (Hollenberg et al., 1995). The assay strain contains HIS3 under control of a promoter containing lexA binding sites. Growth on solid medium that selects for both plasmids and lacks histidine (−Trp −Leu −His) is indicative of a protein–protein interaction (Figure 1, B and C). Consistent with the high-throughput study, full-length lexA-MSS4 (amino acids [aa] 1–779) supported growth on selective medium when combined with GAD-ADY4 (Figure 1B). lexA-MSS4 truncations encoding aa 1–446 and 334–779 also interacted with GAD-ADY4, suggesting that the Ady4 interaction domain lies in the overlap between these two fragments (aa 334–446). This region was sufficient for Ady4 binding as lexA-MSS4334–446 also supported growth on the medium lacking histidine when combined with GAD-ADY4 but not when paired with an unrelated GAD fusion, GAD-VPS131–994, as a specificity control (Figure 1B).
FIGURE 1:
Ady4 interacts with Mss4 in the yeast two-hybrid system. (A) Schematic of the lexA-Mss4 fusions used for the yeast two-hybrid assay in panel B. Numbers indicate amino acid positions. The blue bar indicates a lysine-rich sequence, while the green bar indicates the kinase domain. (B) Strain L40 (lexAop::HIS3) was transformed with plasmids carrying GAD-ADY4 or GAD-VPS131–964 (as a specificity control) and the indicated lexA-MSS4 fusions. Cells were grown to saturation and 10-fold dilutions were spotted onto solid medium lacking tryptophan and leucine (left panel) or lacking tryptophan, leucine, and histidine (right panel). Growth in the absence of histidine indicates an interaction between the LexA and GAD fusion proteins. (C) Western blot of the LexA-Mss4 fusions used in panel B. Extracts from the cells used in the spotting assay were probed with antibodies to either LexA or Arp7 as a loading control.
The 334–446 Ady4 interaction domain in Mss4 is located upstream of the lipid kinase domain (Figure 1A). Within this region is a lysine-rich sequence between aa 347 and 364 that has been previously shown to function as a nuclear localization sequence for Mss4 (Figure 1A; Audhya and Emr, 2003). Deletion of this region from full-length MSS4 (lexA-mss4347–364∆) abolished interaction with Ady4 (Figure 1B). The fusion with this deletion was expressed at higher levels than some fusions (e.g., LexA-Mss4344–446) that show interaction with Ady4, indicating that the loss of interaction is not due to changes in protein stability (Figure 1C). Thus, this 17-aa basic patch on Mss4 is required for interaction with Ady4.
Overexpression of MSS4 or STT4 suppresses the ady4∆ spore formation phenotype
To examine if the two-hybrid interaction between Mss4 and Ady4 is functionally relevant, MSS4 was overexpressed in an ady4Δ diploid to see if this affected the spore number phenotype. Partial suppression was observed, as the number of asci with monads was reduced from 20% in ady4Δ to 7% in ady4Δ/2μ MSS4, while the number of triads/tetrads increased from 15% to 49% (Figure 2, A and B). Mss4 converts phosphatidylinositol 4 phosphate (PI4P) to phosphatidylinositol 4,5, bisphosphate (PI4,5P2) and so acts downstream from a PI4-kinase. Increasing the amount of PI4P might, therefore, also suppress ady4Δ by providing more substrate for Mss4 to phosphorylate. There are three PI-4 kinases in Saccharomyces cerevisiae, LSB6, PIK1, and STT4. Lsb6 and Pik1 localize primarily to the vacuolar membrane and Golgi, respectively (Han et al., 2002; Strahl et al., 2005). STT4 encodes a PI-4 kinase that is present in both plasma and prospore membranes where it generates a pool of PI4P (Audhya and Emr, 2002; Nakamura et al., 2021). High copy expression of either LSB6 or PIK1 in ady4Δ did not increase the number of spores/tetrad (Figure 2B). In contrast, STT4 expressed from a low copy centromere vector suppressed the ady4Δ spore formation phenotype as well as high copy MSS4 (Figure 2B). These results suggest that increasing PI-phosphate levels in the prospore membrane are the basis for suppression of ady4∆ and that Ady4 might promote PI-phosphate levels through its interaction with Mss4.
FIGURE 2:
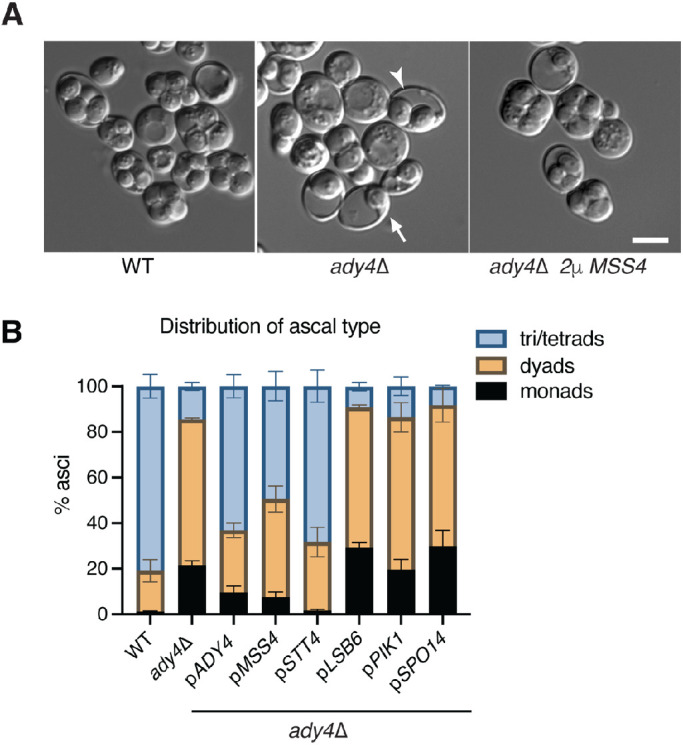
STT4 and MSS4 are dosage suppressors of the ady4∆ spore formation defect. (A) AN120 (WT) and KM6 (ady4∆) cells were sporulated and the cultures examined by light microscopy. Arrows highlight examples of asci with reduced spore numbers in ady4∆. KM6 (ady4∆) carrying pRS425-MSS4 displayed asci with four spores. Scale bar = 5 µm. (B) KM6 (ady4∆) was transformed with plasmids carrying the indicated gene (pRS426-ADY4-GST, pRS425 MSS4, pRS415 STT4, yEP351-LSB6, and yEP351-SPO14), sporulated for 3 d, and the cultures were examined by light microscopy. The distribution of ascal types in each culture is shown. Error bars indicate one SD; 400 cells were scored for each strain from each of three independent colonies.
SPO14 encodes a phospholipase D, which catalyzes the hydrolysis of phosphatidylcholine into phosphatidic acid and is essential for prospore membrane formation (Rose et al., 1995; Rudge et al., 1998). Spo14 localizes to prospore membranes and its activity is regulated by PI4,5P2 (Rudge et al., 1998; Sciorra et al., 2002). It was possible therefore, that the PI4,5P2 created by overexpression of MSS4 indirectly bypasses ady4Δ through activation of Spo14. If true, then overexpression of SPO14 might also suppress the distribution of different ascal types observed in ady4Δ. Such suppression was not observed, however (Figure 2B). While this result suggests that the MSS4 suppression ady4Δ is not occurring indirectly through activation of Spo14, the possibility that SPO14 was not sufficiently overexpressed to exhibit suppression has not been ruled out.
ADY4 is required for Mss4 localization to prospore membrane precursor vesicles
The ability of MSS4 overexpression to suppress the ady4Δ spore number defect, coupled with fact that MSS4 is required to make prospore membranes, suggests Mss4 generates negatively charged PI4,5P2 in prospore membranes that promotes association with MOPs. The Ady4-Mss4 two-hybrid interaction further suggests that the function of Ady4 may be to efficiently recruit Mss4 to the SPB where it can interact with prospore membrane lipids. In this case, overexpression of MSS4 in ady4Δ raises the Mss4 protein level sufficiently such that Mss4 localization to the spindle pole or prospore membrane occurs independently of ADY4. This hypothesis predicts that Mss4 should be present in the vicinity of MOPs. However, while Mss4-GFP (green fluorescent protein) was observed at plasma membranes in both vegetative and sporulating cells, it has not been detected at prospore membranes (Rudge et al., 2004; Nakamura et al., 2021). One possibility is that the pool of Mss4-GFP at prospore membranes is below the threshold of detection by fluorescence microscopy. The fluorescent signal for Mss4 was therefore amplified using a split GFP system (Chen et al., 2020). MSS4 was fused to five tandem repeats encoding the 11th β-strand of the GFP β-barrel (GFP β11) and this fusion was cotransformed into cells with a second plasmid constitutively expressing the remaining sequence of GFP (GFP β1–10). In vivo assembly of the separate parts of GFP results in multiple GFP moieties tagged to Mss4 (hereafter referred to simply as Mss4-5xGFP; Cabantous et al., 2005; Kamiyama et al., 2016). In vegetative cells, expression of Mss4-5xGFP produced brighter fluorescence than Mss4-GFP without changing the distribution of the protein (Supplemental Figure S1).
To determine whether the brighter signal of Mss4-5xGFP allowed detection of the protein at MOPs, cells from a sporulating culture containing Mss4-5xGFP were identified using Spc42-TagBFP, an SPB marker, to detect the four SPBs from the meiosis II spindles. In these cells, the bulk of Mss4-5xGFP still localized primarily on the plasma membrane (Figure 3A). It could be the case that Mss4 was too diffusely distributed throughout the prospore membrane to generate a discrete signal even with the brighter Mss4-5xGFP. SSO1 encodes a SNARE protein required for vesicle fusion at the MOP (Oyen et al., 2004; Nakanishi et al., 2006). In sso1∆ cells, fusion of prospore membrane precursor vesicles is blocked and clusters of these vesicles accumulate at MOPs (Nakanishi et al., 2006). Localization of Mss4-5xGFP was therefore examined in meiosis II cells from an sso1∆ diploid (Figure 3, A and B). In these cells, intracellular foci of Mss4 fluorescence that overlapped with the blue SPB signal were readily visible. More than 80% of meiosis II SPBs in the sso1∆ strain colocalized with Mss4-5xGFP (Figure 3, A and B). As association of the Mss4-5xGFP signal with the SPB requires sso1∆, these foci likely represent localization of Mss4-5xGFP to the SPB-associated vesicles that accumulate in this mutant. Furthermore, ADY4 is required for recruitment to these foci as meiosis II SPB-associated Mss4-5xGFP signal was not seen in sso1Δ ady4Δ cells (Figure 3B).
FIGURE 3:
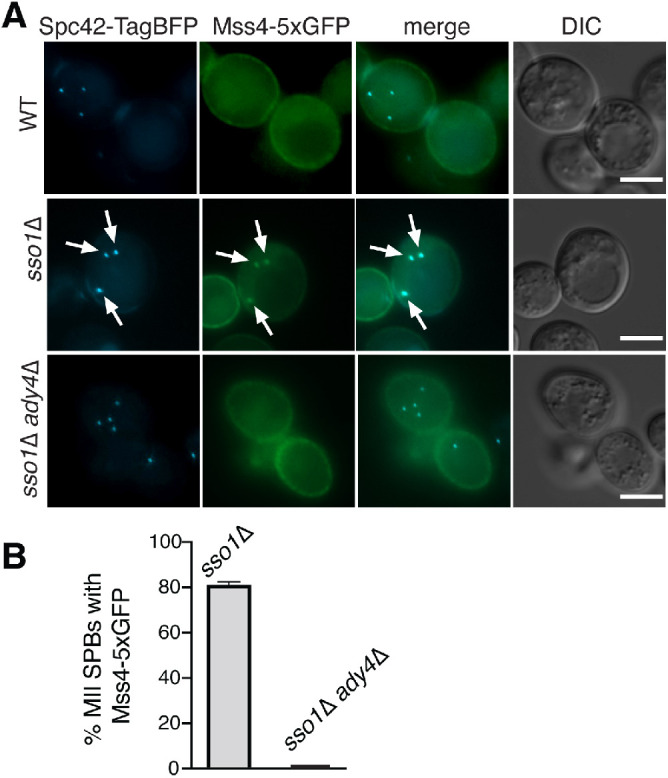
Ady4 is a targeting subunit for Mss4. (A) Wild-type (AN120) containing SPC42::TagBFP (pRS306-spc42c-TagBFP), sso1∆ SPC42::TagBFP (GND10), and sso1∆ ady4∆ SPC42::TagBFP (GND11) diploids were transformed with plasmids expressing GFP1-10 (pRS423-PTEF1-GFP1-10) and MSS4-5xGFP11 (pRS414-Mss4-5xGFP11). Images of cells in meiosis II are shown for each strain. Arrows indicate colocalization of Mss4-5xGFP with the Spc42 marker. Scale bars = 5 µm. (B) The frequency of colocalization of Mss4 and Spc42 was measured in the cultures shown in A. For sso1∆ SPC42::TagBFP (n = 358) and sso1∆ ady4∆ (n = 225), at least 50 meiosis II SPBs were scored in six and four independent experiments, respectively. Error bars indicate one SD.
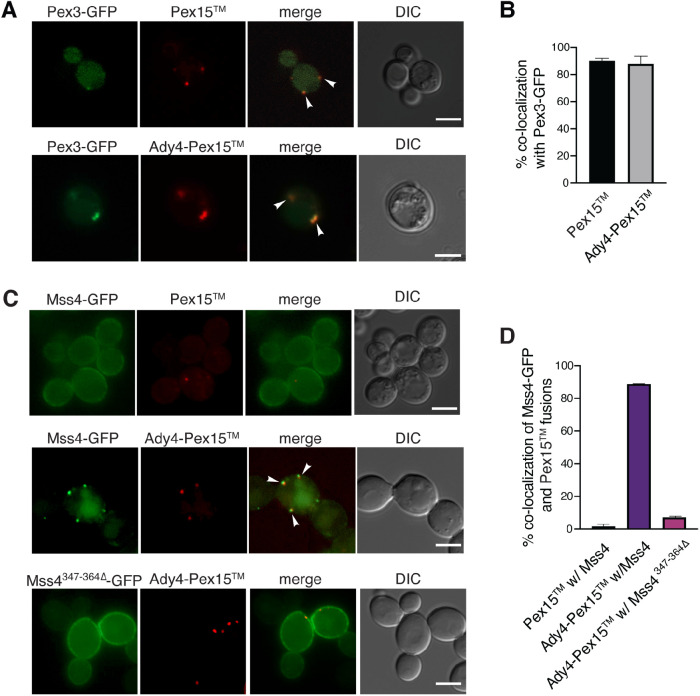
Ectopic localization of Ady4 to peroxisomes is sufficient to recruit Mss4
If the Ady4-Mss4 protein interaction is sufficient to target Mss4 to MOPs in sporulating cells, localizing Ady4 to a different organelle in vegetative cells should result in recruitment of Mss4 to that organelle as well. Ady4 was therefore fused to the C-terminal transmembrane domain of the peroxisomal protein, Pex15, which is sufficient to target heterologous proteins to the cytoplasmic surface of the peroxisome (Halbach et al., 2006). To visualize this protein fragment, Pex15 was tagged at its N terminus with the fluorescent mOrange protein to make mOrange-Pex15 (hereafter referred to simply as Pex15; Shaner et al., 2004). The fusion gene was constitutively expressed using the TEF1 promoter (Mumberg et al., 1995). Pex15 exhibited cytosolic foci that displayed frequent colocalization (87%) with the peroxisomal marker Pex3-GFP, confirming that Pex15 is an effective peroxisome targeting sequence (Figure 4, A and B; Hohfeld et al., 1991; Huh et al., 2003). Fusion of Ady4 to the N terminus of mOrangePex15 to generate Ady4-mOrange-Pex15 (hereafter Ady4-Pex15) was sufficient to target Ady4 to the peroxisome as well (Figure 4, A and B)
FIGURE 4:
Ady4 can target Mss4 to an ectopic location in vegetative WT cells. (A) A WT diploid (AN120) was transformed with plasmids expressing PEX3-eGFP (pRS424-PTEF1Pex3-eGFP) and PEX15 (pRS316-PTEF1-mOrangePex15) or ADY4- PEX15 (pRS426-PTEF1Ady4-mOrange-Pex15) and localization of the fusion proteins was examined by fluorescence microcopy. Arrowheads indicate colocalization of Pex3 and the Pex15 fusions. Scale bars = 5 µm. DIC indicates differential interference contrast microscopy. (B) Quantification of the fraction of Pex3-GFP foci displaying Pex15 or Ady4-Pex15 colocalization with Mss4-GFP fusions. More than 100 Pex3-eGFP foci were analyzed in three independent experiments. Error bars indicate one SD. (C) WT (AN120) expressing MSS4-GFP (pRS414 Mss4-GFP) with PEX15 or ADY4- PEX15 and WT expressing Mss4347–364∆-GFP (pRS414- Mss4347–364∆-GFP) with Ady4-Pex15 were examined for colocalization of the markers. Arrowheads indicate colocalization of Mss4 and Ady4-Pex15. Scale bars = 5 µm. (D) Quantification of the fraction of Pex15 foci associated with Mss4-GFP signal. For Pex15 with Mss4-GFP, about 2% colocalization was seen in a total of 190 Pex15 foci scored in five independent experiments. For Mss4-GFP and Ady4-Pex15, colocalization was ∼85% (153 Ady4-Pex15 foci examined in four independent experiments). For Mss4347–364∆-GFP with Ady4-mOrange-Pex15 colocalization was 7% (221 Ady4-Pex15 foci scored in five independent experiments). Error bars indicate one SD.
Pex15 or Ady4-Pex15 was then coexpressed with Mss4-GFP and localization of the fusion proteins examined by fluorescence microscopy. No colocalization was observed between Pex15 and Mss4-GFP (Figure 4, C and D). By contrast, Mss4-GFP was efficiently recruited to peroxisomes in the presence of Ady4-Pex15, as 88% of Mss4-GFP foci overlapped with Ady4-Pex15 foci (Figure 4, C and D). Colocalization was dependent upon the protein interaction between Mss4 and Ady4, as it was greatly reduced when Mss4-GFP contained a deletion of the Ady4 interaction region (aa 347–364; Figure 4, C and D). These data provide further evidence that Ady4 acts as a targeting factor for Mss4 and that it does so by binding to a patch of basic residues within Mss4.
An amphipathic helix in the N terminus of Spo21 binds to prospore membranes
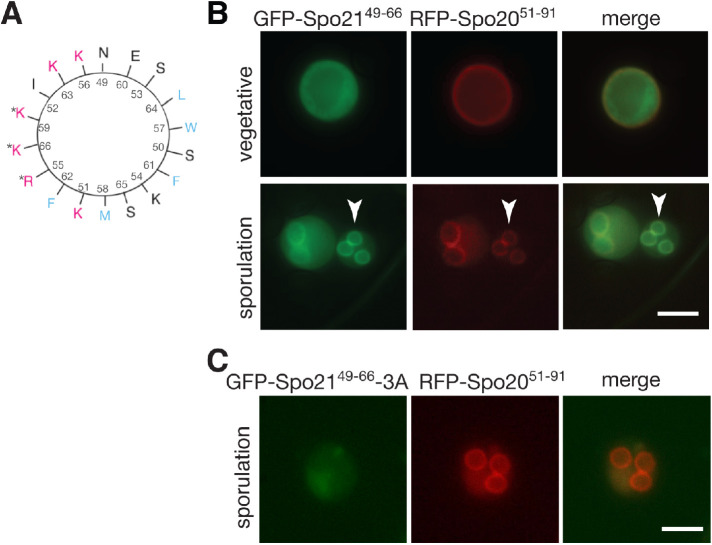
Ady4 recruits Mss4 to the SPB and the connection of the prospore membrane to the MOP is frequently lost in ady4∆ cells (Figure 3; Nickas et al., 2003; Mathieson et al., 2010a). These observations suggest that PI4,5P2 generated by Mss4 may be important for the connection of MOPs to prospore membranes. Two MOP proteins, Spo21 and Mpc54, have N termini located proximal to prospore membranes, raising the possibility that one of them binds to prospore membranes via interaction with PI4,5P2 (Mathieson et al., 2010a). Amphipathic helices with positively charged and hydrophobic faces are common membrane-binding motifs (Segrest et al., 1974; Epand et al., 1995). The N terminus of Spo21 includes a potential amphipathic helix containing aa 49–66 (Figure 5A). AlphaFold models of the Spo21 structure predict this region is helical as well (Jumper et al., 2021; Varadi et al., 2022).
FIGURE 5:
A predicted amphipathic helix in the N terminus of Spo21 targets GFP to prospore membranes. (A) A helical wheel depicting Spo21 amino acids 49–66. Arginine (R) and lysine (K) residues on the positively charged face are in red. Hydrophobic residues (F, phenylalanine; M, methionine; W, tryptophan) are in blue. Asterisks mark the residues replaced with alanine in the spo21-3A allele. (B) A WT strain (AN120) was transformed with plasmids expressing GFP-SPO2149–66 and the prospore membrane marker RFP-SPO2051–91 and imaged in both vegetative growth and sporulation. Arrowheads highlight an example of GFP-Spo2149–66 at the prospore membrane. (C) A WT diploid transformed with plasmids expressing GFP-SPO2149–66-3A and RFP-SPO2051–91 and imaged in sporulation. Scale bars = 5 µm.
The ability of the Spo21 helix to bind to membranes in vivo was tested by fusing the sequence encoding this region to GFP under the control of the TEF1 promoter (GFP-Spo2149–66). Localization of GFP-Spo2149–66 was compared with Spo2051–91-mRFP, which contains a similar amphipathic helix from the N-terminal region of the SNARE Spo20 (aa 51–91) fused to mRFP that binds to plasma membranes in vegetative cells and relocalizes to prospore membranes during sporulation (Nakanishi et al., 2004). When the two constructs were examined in vegetative yeast cells, Spo2051–91-mRFP was seen clearly at the plasma membrane while GFP-Spo2149–66 fluorescence was located throughout the cytoplasm (Figure 5B). By contrast, GFP-Spo2149–66 showed strong colocalization with Spo2051–91-mRFP at prospore membranes in sporulating cells (100% colocalization, 100 cells scored; Figure 5B), suggesting that this helix of Spo21 has specific affinity for prospore membranes.
Amphipathic helices bind membranes, in part, through electrostatic interactions between positively charged lysine or arginine side chains and negatively charged lipid head groups (Segrest et al., 1974). To determine if the positive residues in the Spo21 helix are required for membrane binding, three of these aa (R55, K59, and K66; Figure 5, A and C) were mutated to alanine in the context of the GFP fusion (GFP-Spo2149–66-3A). When GFP-Spo2149–66-3A was expressed in sporulating cells, it failed to localize to prospore membranes (0% of cells colocalized with Spo2051–91-mRFP; Figure 5C). These data demonstrate that Spo21 binds to prospore membranes through a positively charged region in its N terminus.
Interaction of Spo21 with prospore membranes promotes formation of four spored asci
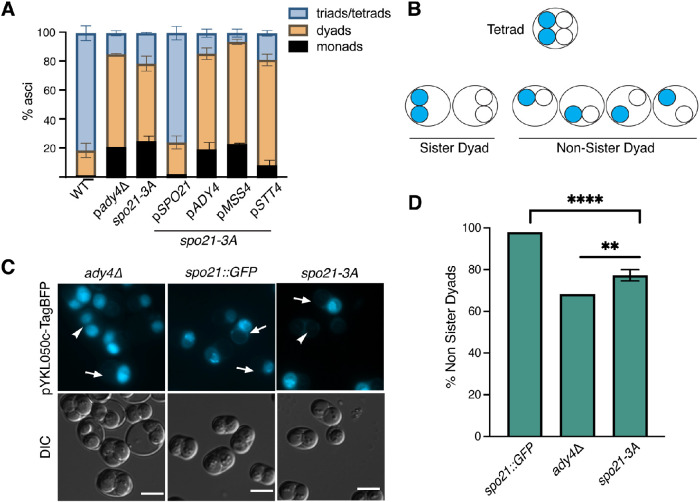
To examine the functional significance of Spo21’s ability to bind prospore membranes, a diploid homozygous for the R55A, K59A, and K66A mutations (spo21-3A) was analyzed for the number of spores in each ascus. In contrast to the spo21∆ that makes no spores (Knop and Strasser, 2000; Bajgier et al., 2001), the spo21-3A diploid produced asci with varying numbers of spores, similar to ady4∆ (Figure 6A; Nickas et al., 2003). This phenotype was complemented by the addition of SPO21 on a plasmid, confirming the sporulation defect is due to the recessive spo21-3A mutant (Figure 6A). Unlike ady4∆, however, the reduced spore phenotype of spo21-3A was not suppressed by additional copies of ADY4, MSS4, or STT4 (Figure 6A).
FIGURE 6:
Positive charges in the Spo21 N terminus promote tetrad formation. (A) Distribution of ascal types in WT (AN120) ady4∆ (AC18), spo21-3A (GND4), and GND4 transformed with plasmids expressing SPO21 (pRS426-Spo21), ADY4 (pRS426-Ady4-GST), MSS4 (pRS424-Mss4), or STT4 (pRS426-Stt4). Each strain was sporulated and the number of spores per ascus was scored by light microscopy. For each strain, 400 asci were scored in each of three independent experiments. (B) A schematic of nonsister dyad formation in cells heterozygous for the centromere-linked, spore autonomous, fluorescent marker PYKL050c::TagBFP::TRP1. For cells packaging dyads randomly, 2/3 of the dyads formed will be nonsister dyads. (C) Representative images of asci of ady4∆ (AC18-cen), spo21::GFP (AN230-cen), and spo21-3A (GND9) from the experiments in panel A. Arrows highlight examples of nonsister dyads; arrowheads indicate sister dyads. Scale bars = 5 µm. (D) Quantification of dyad types in the strains in C. Four asterisks indicate the frequency of nonsister dyads in spo21-3A is significantly different than in spo21::GFP (p < 0.0001; Fisher’s exact test). Two asterisks indicate that the frequency of nonsister dyads in spo21-3A is significantly different from ady4∆ (p < 0.003). For spo21::GFP, n = 257 dyads scored one experiment; ady4∆ n = 325 dyads scored one experiment; spo21-3A, at least 90 dyads scored in each of three independent experiments.
The majority of spo21-3A asci were dyads containing two haploid spores (Figure 6A). Dyads arise when only two of the four haploid chromosome sets are packaged into spores. Sister dyads contain spores derived from the two nuclei at opposite poles of the same meiosis II spindle, while nonsister dyads contain spores containing a nucleus from one pole of each meiosis II spindle (Figure 6B). A hypomorphic allele of SPO21, spo21::GFP, produces exclusively nonsister dyads, while ady4Δ asci are randomly distributed between sister and nonsister dyads (Bajgier et al., 2001; Nickas et al., 2003). To determine the type of dyads produced by spo21-3A, a gene encoding the blue fluorescent protein, TagBFP, under the control of a spore autonomous promoter was integrated at the tightly centromere-linked trp1 locus on one chromosome IV homologue to create a strain heterozygous for the reporter (Thacker et al., 2011). Sister dyads contained either two blue or two nonfluorescent spores, while nonsister dyads had only one blue spore (Figure 6, B and C; Thacker et al., 2011). As expected, the spo21::GFP strain produced almost exclusively nonsister dyads (98%), while the ady4Δ strain showed a random distribution of dyad types (66% nonsister dyads; Figure 6, C and D). Dyads produced by the spo21-3A mutant fell between these two standards (74% nonsister dyads). This value was significantly different from spo21::GFP (Fischer’s exact test; p value <0.0001). While the spo21-3A value was also significantly different from ady4∆ (p < 0.003), these data indicate that spo21-3A, unlike a hypomorphic allele of SPO21, does not produce exclusively nonsister dyads but leads to the appearance of significant numbers of random dyads as in ady4Δ. These results are consistent with the possibility that this helical region binds to lipids generated through Ady4-dependent recruitment of Mss4 and that loss of this binding produces a similar phenotype to ady4∆.
Binding of Spo21 to prospore membranes is necessary for proper prospore membrane number
Another expected distinction between spo21::GFP and ady4∆ is the number of prospore membranes formed per cell. Nonsister dyads result from initiation of only two prospore membranes, one on each daughter SPB at meiosis II (Nickas et al., 2003). In ady4Δ cells, dyads result from stochastic disassembly of MOPs during prospore membrane growth (Bajgier et al., 2001; Nickas et al., 2003; Mathieson et al., 2010b). Because disassembly occurs after prospore membranes initiate, the number of prospore membranes in individual ady4Δ cells should be close to four. spo21::GFP, ady4Δ, and spo21-3A strains were sporulated and the fluorescent prospore membrane marker Spo2051–91-TagBFP (Lin et al., 2013) was used to assess the number of prospore membranes present in meiosis II cells (Figure 7A). As expected, more than 85% of wild-type (WT) cells displayed three or four prospore membranes. By contrast, spo21::GFP cells exhibited almost exclusively one or two prospore membranes. Somewhat surprisingly, ady4∆ cells also showed significantly reduced numbers of prospore membranes, with less than 20% of cells displaying three or four membranes (Figure 7A). While morphological variability in prospore membranes has been previously reported in ady4∆ cells, reduced numbers of membranes was not previously noted (Nickas et al., 2003). This result suggests that the reduced number of spores in ady4Δ might result from prospore membrane initiation defects in addition to instability of the MOP.
FIGURE 7:
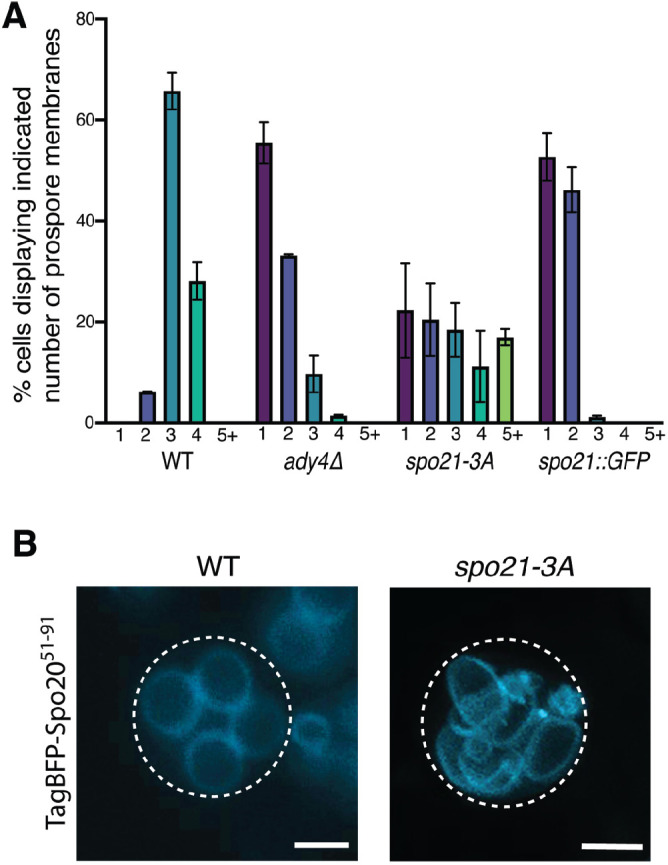
Positive charges in the Spo21 N terminus promote proper prospore membrane number. (A) WT (AN120), ady4∆ (AC18), spo21::GFP (AN230), and spo21-3A (GND4) cells were transformed with the prospore membrane marker TagBFP-SPO2051–91. Cells were sporulated and the number of prospore membranes in individual cells were scored. At least 80 cells were scored in each of three independent experiments for each strain. (B) A maximum intensity projection of a WT cell with four prospore membranes and a spo21-3A cell displaying more than four prospore membranes. Scale bars = 2 µm.
Once again, spo21-3A displayed a unique phenotype. While cells with one or two prospore membranes were seen, almost 20% of cells contained more than four prospore membranes (Figure 7, A and B). The source of these supernumerary prospore membranes is not clear, but this phenotype distinguishes spo21-3A from both ady4Δ and previously characterized spo21 alleles.
DISCUSSION
A mechanism for connecting MOPs to developing prospore membranes
Vesicle fusion at MOPs initiates prospore membrane formation and the association between a prospore membrane and its MOP must be maintained for a nucleus to become completely enclosed within the membrane (Moens and Rapport, 1971; Neiman, 1998; Mathieson et al., 2010a) The results reported here suggest a mechanism by which this association is created and maintained. Our model is that Ady4 promotes recruitment of Mss4 to a region of the prospore membrane near the MOP where Mss4 generates a localized pool of the negatively charged lipid, PI4,5P2 (Figure 8). This lipid environment in turn promotes binding of the Spo21 N-terminal helix to prospore membranes, which stabilizes the MOP structure. The connection between a MOP and a prospore membrane is weakened in ady4Δ due to reduced levels of PI4,5P2 and by reduced Spo21 binding to lipids in spo21-3A. As a result, prospore membranes separate from MOPs, leading to reduced numbers of spores in the ascus. At least in the case of ady4Δ, this release results in disassembly of the MOP structure (Nickas et al., 2003; Mathieson et al., 2010a).
FIGURE 8:
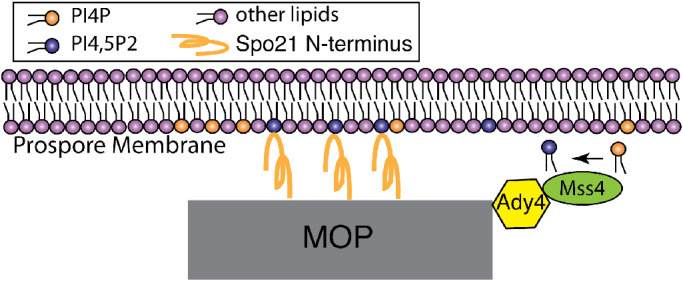
Model for Ady4 during prospore membrane formation. During early prospore membrane formation, Ady4 recruits Mss4 to the SPB. Mss4 generates a pool of PI4,5P2 in the prospore membrane near the SPB, thereby facilitating membrane binding of the amphipathic helix in the N terminus of Spo21.
The PI4,5P2 lipid is important for prospore membrane formation
The role of PI4,5P2 in prospore membrane formation has been unclear. Earlier work demonstrated that high copy MSS4 stimulates prospore membrane formation and that the t-SNARE Sso1 can bind to PI4,5P2, suggesting that PI4,5P2 might be important for Sso1 function mediating vesicle fusion at the prospore membrane (Mendonsa and Engebrecht, 2009). Moreover, Spo14, which is essential for prospore membrane formation, requires PI4,5P2 as a cofactor (Rose et al., 1995; Sciorra et al., 2002). However, neither the Mss4 protein nor its product PI4,5P2 are detected in growing prospore membranes (Nakamura et al., 2021). Our results indicate that there is ADY4-dependent recruitment of Mss4 to incipient prospore membranes and therefore, there is likely a pool of PI4,5P2 in these membranes as well. Thus, our data support the idea that PI4,5P2 is important for early prospore membrane formation events.
A role for Mss4 in Sso1 function and prospore membrane initiation is consistent with the reduced number of prospore membranes per cell observed in ady4Δ. The absence of ADY4-dependent Mss4 and PI4,5P2 at SPBs could lead to reduced efficiency of initiation, resulting in fewer prospore membranes. Cells lacking ADY4 also display heterogeneity in prospore membrane size (Nickas et al., 2003). Inefficient prospore membrane initiation leading to delayed formation at some SPBs could explain this phenotype of ady4Δ as well.
The suppression of ady4Δ by overexpression of STT4 or MSS4 implies that some part of the MOP can bind to the lipids produced by these kinases. We report that a predicted amphipathic helix near the N terminus of Spo21 binds specifically to prospore membranes. Both ady4Δ cells and cells with mutations in this helix (spo21-3A) make asci with reduced numbers of spores. However, in many ady4Δ cells the number of prospore membranes is reduced, but in spo21-3A the number is increased. Our model posits that both Ady4 and Spo21 help maintain MOP-prospore membrane association but in different ways. Ady4’s effect is indirect: it brings Mss4 to the incipient membrane to create a lipid environment conducive to Spo21 binding and perhaps other functions. Deletion of ady4∆ therefore changes the lipid composition of prospore membranes, affecting both initiation and attachment of prospore membranes and their association with MOPs. However, the effect of the spo21-3A mutant is more direct in that it reduces the affinity for the incipient prospore membrane even though the lipid environment is normal. In this case, the release of prospore membranes from MOPs, due to the weakened connection to the MOPs, might result in cycles of initiation and release, producing extra prospore membranes.
What is the ligand of the Spo21 amphipathic helix?
A reporter that binds specifically to PI4,5P2 localizes predominantly to the plasma membrane both in vegetative growth and throughout sporulation until the time of prospore membrane closure (Audhya and Emr, 2003; Nakamura et al., 2021). This is opposite of the behavior we report for GFP-Spo2148–66, which is predominantly cytosolic in vegetative cells but then localizes to growing prospore membranes. Thus, although Ady4 recruits Mss4, it is unlikely that the Spo21 N terminus only binds to PI4,5P2. Rather, we propose that this helix simply binds to negatively charged lipids and the recruitment of Mss4 raises the level of such lipids in the vicinity of the MOP. Although the cell uses PI4,5P2 in this context, other phospholipids with acidic head groups might work as well, which is consistent with our finding that increased expression of STT4 can also rescue ady4Δ. The Spo21 amphipathic helix, therefore, may function similarly to the lipid-binding amphipathic helix from the SNARE Spo20, which lacks lipid specificity in vitro, but shows specificity for phosphatidic acid–enriched membranes in vivo, likely by partitioning to the membrane of highest negative charge (Nakanishi et al., 2004; Horchani et al., 2014).
Ady4 may represent a conserved Mss4 targeting protein
Ady4 binds to a central region of Mss4 that is N-terminal to the kinase catalytic domain (Figure 1; Audhya and Emr, 2003). Although this central region is not conserved in mammalian Mss4 orthologues, it is well conserved in Mss4 homologues throughout the fungi. Unlike the structural proteins of the MOP, which are not well conserved at the primary sequence level in other yeasts, ADY4 has clear orthologues throughout the Saccharomycotina. The ADY4 homologue in Clavispora lusitaniae is also induced in sporulation, suggesting that the role of the gene product in spore formation may be conserved (Sherwood et al., 2014). However, ADY4 orthologues are also found in yeasts such as Candida albicans that do not form ascospores. The conservation of Ady4 may reflect its use as a targeting factor for Mss4 at other intracellular locations in these yeasts.
MATERIALS AND METHODS
Request a protocol through Bio-protocol.
Yeast strains
All strains used in this study are in the SK1 background and are listed in Table 1. Standard media and methods were used (Rose et al., 1990). To generate the homozygous spo21-3A diploid GND4, two-step gene replacement was used (Rothstein, 1991). The URA3 spo21-3A plasmid, pRS306-spo21-3A, was linearized using SwaI and integrated at the SPO21 promoter in the spo21∆::HIS3MX6 haploids AN178-3A and AN178-3B. Cells that had lost the URA3 gene by recombination were selected on medium containing 5-fluoro-orotic acid (5-FOA; Brachmann et al., 1998). 5-FOA–resistant colonies were then screened for histidine auxotrophy, indicating loss of spo21Δ::HIS3MX6. The resulting spo21-3A haploids GNH3 and GNH4 were crossed to generate GND4. To generate the diploid GND9 (spo21-3A, PYKL050c-TagBFP::TRP1) pRS404-PYKL050c-TagBFP was linearized with PmlI and integrated at the trp1 locus in GND4. GND10 (sso1∆, spc42::TagBFP) was made by integrating pRS306-spc42c-TagBFP digested with AflII at the spc42 locus in the sso1Δ haploids GN01 and GN02. 5-FOA–resistant colonies were selected and recombinants that contained spc42::TagBFP were identified by screening of colonies by microscopy for blue fluorescence. These haploids were mated to make GND10. GND11 (sso1∆ ady4∆ spc42::TagBFP) was made similarly by first introducing pRS306-spc42c-TagBFP into the sso1Δ ady4Δ haploids EMH3 and EMH4 (Mathieson et al., 2010a). To generate the sso1∆::kanMX6 diploid GN03, the sso1∆:HIS3MX6 allele in strains HI1 and HI2 (Nakanishi et al., 2006) was first converted to sso1∆::kanMX6. To do this, the primers HNO161 and HNO162 used to amplify the kanMX6 cassette form pFA6-Kan (Longtine et al., 1998) and the PCR product used to transform HI1 and HI2. G418-resistant transformants were screened for loss of the HIS3MX6 cassette by growth on –His medium. The resulting haploids, GN01 and GN02, were mated to generate GN03. The ady4Δ haploids KM3 and KM5 were made by disrupting the ADY4 ORF with the kanMX6 cassette (Longtine et al., 1998) in haploids SKY3574 and SKY3575 (Thacker et al., 2011) and crossed to form the ady4∆ diploid KM6. All disruptions were confirmed using the PCR.
TABLE 1:
List of strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| AC18 | MAT a /MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3∆SK/his3∆SK arg4-NspI/ARG4 lys2/lys2 ho∆::LYS2/ho∆:LYS2 rme1::LEU2/RME1 ady4∆::HIS3MX6/ady4∆::HIS3MX6 | This study |
| AC18-cen |
MAT
a
/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3∆SK/his3∆SK arg4-NspI/ARG4 lys2/lys2 ho∆::LYS2/ho∆:LYS2 rme1::LEU2/RME1 ady4∆::HIS3MX6/ady4∆::HIS3MX6
PrYKL050c::TagBFP::TRP1/trp1 |
This study |
| AN117-16D | MATa his3∆SK ho::LYS2 leu2 lys2 trp1::hisG ura3 | Neiman et al., 2000 |
| AN117-4B | MATa arg4-NspI his3∆SK ho∆::LYS2 leu2 lys2 rme1::LEU2 trp1::hisG ura3 | Neiman et al., 2000 |
| AN120 | MATa/MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3∆SK/his3∆SK arg4-NspI/ARG4 lys2/lys2 ho∆::LYS2/ho∆::LYS2 rme1::LEU2/RME1 | Neiman et al., 2000 |
| AN178-3A | MATa ura3 leu2 his3∆SK arg4 lys2 ho∆LYS2 rme1::LEU2 spo21∆::HIS3MX6 | Bajgier et al., 2001 |
| AN178-3B | MATα ura3 leu2 trp1::hisG his3∆SK lys2 ho∆LYS2 spo21∆::HIS3MX6 | Bajgier et al., 2001 |
| AN230 | MATa /MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3∆SK/his3∆SK arg4-NspI/ARG4 lys2/lys2 ho∆::LYS2/ho∆::LYS2 rme1::LEU2/RME1 spo21::GFP/spo21::GFP | Bajgier et al., 2001 |
| AN230-cen | MAT a /MATα ura3/ura3 leu2/leu2 trp1::hisG/trp1::hisG his3∆SK/his3∆SK arg4-NspI/ARG4 lys2/lys2 ho∆::LYS2/ho∆::LYS2 rme1::LEU2/RME1 spo21::GFP/spo21::GFP PrYKL050c::TagBFP::TRP1/trp1 | This study |
| EMD10 | MATa /MATα arg4-Nsp1/ARG4 his3∆SK/his3∆SK ura3/ura3 leu2/leu2 lys2/lys2 trp1::hisG/trp1::hisG RME1/rme1::LEU2 sso1∆::HIS3MX6/sso1∆::HIS3MX6 ady4∆::HIS3MX6/ady4∆::HIS3MX6 | Mathieson et al., 2010b |
| GN01 | MATa ura3 leu2 trp1::hisG his3∆SK lys2 ho::LYS2 sso1∆::kanMX6 | This study |
| GN02 | MATα ura3 leu2 trp1::hisG his3∆SK lys2 ho::LYS2 rme1::LEU2 sso1∆::kanMX6 | This study |
| GN03 | MATa /MATα ura3/ura3 trp1::hisG/trp1::hisG leu2/leu2 his3∆SK/his3∆SK lys2/lys2 arg4/ARG4 rme1::LEU2/RME1 ho∆LYS2/ho∆LYS2 sso1∆::kanMX6/sso1∆::kanMX6 | This study |
| GND10 | MATa /MATα arg4-Nsp1/ARG4 his3∆SK/his3∆SK ura3/ura3 leu2/leu2 lys2/lys2 trp1::hisG/trp1::hisG RME1/rme1::LEU2 sso1∆::HIS3MX6/sso1∆::HIS3MX6 SPC42::TagBFP/SPC42::TagBFP | This study |
| GND11 | MATa /MATα ura3/ura3 trp1::hisG/trp1::hisG leu2/leu2 his3∆SK/his3∆SK lys2/lys2 arg4/ARG4 rme1::LEU2/RME1 ho∆LYS2/ho∆LYS2 ady4∆HIS3MX6/ady4∆HIS3MX6 sso1∆HIS3MX6/sso1∆HIS3MX6 SPC42::TagBFP/SPC42::TagBFP | This study |
| GND4 | MATa /MATα ura3/ura3 leu2/leu2 his3∆SK/his3∆SK arg4-Nsp1/ARG4 lys2/lys2 ho∆LYS2/ho∆::LYS2 RME1/rme1::LEU2 spo21-3A/spo21-3A | This study |
| GND9 | MATa /MATα ura3/ura3 leu2/leu2 his3∆SK/his3∆SK arg4-Nsp1/ARG4 lys2/lys2 ho∆LYS2/ho∆::LYS2 RME1/rme1::LEU2 spo21-3A/spo21-3A PrYKL050c::TagBFP::TRP1/trp1 | This study |
| GNH3 | MATa ura3 leu2 his3∆SK arg4 lys2 ho∆LYS2 rme1::LEU2 spo21-3A | This study |
| GNH4 | MATα ura3 leu2 trp1::hisG his3∆SK lys2 ho∆LYS2 spo21-3A | This study |
| HI1 | MATα ura3 his3∆SK trp1::hisG arg4-Nsp1 lys2 ho::Lys2 rme1::LEU2 le2 sso1::his5+ | Nakanishi et al., 2006 |
| HI2 | Mat a ura3 leu2 trp1-hisG his3∆SK lys2 ho::LYS2 sso1::his5+ | Nakanishi et al., 2006 |
| HI3 | MATa /MATα ura3/ura3 trp1::hisG/trp1::hisG leu2/leu2 his3∆SK/his3∆SK lys2/lys2 arg4/ARG4 rme1::LEU2/RME1 ho∆LYS2/ho∆LYS2 sso1∆::HIS3MX6/sso1::HIS3MX6 | Nakanishi et al., 2006 |
| KM3 | MATa ura3 leu2::hisG trp1:hisG ho∆::LYS2 CEN8::mCerulean-TRP1 ady4∆::kanMX6 | This study |
| KM5 | MATα ura3 leu2::hisG trp1::hisG ho∆::LYS2 CEN8::tdtomato-URA3 ady4∆::KANMX6 | This study |
| KM6 | MATa /MATα ura3/ura3 leu2::hisG/leu2::hisG trp1:hisG/trp1:hisG lys2/ ho∆LYS2 CEN8::mCerulean-TRP1/CEN8::tdtomato-URA3 ady4∆::kanMX6/ady4∆::kanMX6 | This study |
| L40 | MATa leu2 ade2 his3 trp1 LYS::lexAop-HIS3 URA3::lexAop-lacZ | Hollenberg et al., 1995 |
Plasmids
The plasmids used in this study are listed in Table 2 and oligonucleotides are listed in Supplemental Table 1. The plasmid pRS425-PIK1 was constructed by a three-piece Gibson Assembly using a NEBuilder HiFi DNA Assembly kit (New England Biolabs; catalogue #E2621L). To generate the three fragments to assemble, the plasmid pRS425 was digested with BamHI and XhoI and the 5′ and 3′ halves of the PIK1 gene, including ∼500 base pairs of upstream or downstream sequence were amplified using the primer pairs GNO89/GNO92 and GNO90/GNO91, respectively. To build pRS414-Mss4 5xGFP11, primers GNO04 and GNO05 were used to amplify the five copies of the GFP 11th β strand with flanking PacI/AscI sites from pRS426-PGAL1-5xGFP11 (Chen et al., 2020). The fragment was digested with PacI and AscI and ligated with similarly digested pRS414-Mss4-GFP (Ling et al., 2012). To build pRS426-PTEF1-1-10GFP, primers OKZ189 and OKZ190 were used to amplify the GFP 1–10 fragment from pRS425-PTEF1-GFP1-10 (Chen et al., 2020). The resulting PCR product was digested with SacI and XhoI and ligated into pRS423-PTEF1 (Mumberg et al., 1995) digested with SacI and XhoI. To build pRS306-SPO21 (pKZ208), primers ANO477 (anneals 300 nucleotides upstream of the SPO21 open reading frame with homology to pRS306) and ANO478 (432 nucleotides downstream from the SPO21 stop codon with homology to pRS306), were used to amplify a fragment containing SPO21 from BY4741 genomic DNA. The product was inserted into KpnI and SacI digested pRS306 by Gibson Assembly. pRS306-spo21-3A was constructed by PCR using complementary primers GNO27 and GNO28, which introduce alanine codons in place of codons 55, 59, and 66 in the SPO21 coding region. The template used for the reaction was pRS306-SPO21. The PCR reaction was treated with DpnI to degrade the template plasmid and circularized by ligation with T4 ligase before transformation into Escherichia coli. To generate fusions of GFP to the SPO21 amphipathic helix, complementary oligos encoding a start codon and SPO21 codons 48–66 were annealed to make a duplex fragment with BamHI and ClaI sticky ends. This fragment was then ligated into pRS426-PTEF-GFP (Nakanishi et al., 2004) digested with BamHI and ClaI. Oligos ANO475 and ANO476 were used to construct pRS426 PTEF GFP-Spo2148–66 and GNO68 and GNO69 were used to construct pRS426 PTEF GFP-Spo2148–66-3A. To build 314-HTB1mOrange2 (pKZ10), first the HTB1-GFP fusion was amplified from the yeast GFP collection (Huh et al., 2003) using primers KZO49 and KZO50. The resulting PCR product was digested with ApaI and XhoI and ligated with similarly digested pRS314 (Sikorski and Hieter, 1989) to create pRS314-HTB1-GFP (pKZ2). Plasmids carrying S. cerevisiae codon-optimized mOrange2 or TagBFP cloned into pUC57 were purchased from Genewiz (Newark, NJ) and then the pRS314-HTB1-GFP and pUC57-mOrange2-ScOpt plasmids were both digested with PacI and AscI. The pRS314-HTB1 backbone and mOrange2 fragments were ligated using T4 ligase to generate pRS314-HTB1-mOrange2. To generate pRS404-PYKL050c-TagBFP (pKZ247), the CFP marker from psK692 (Thacker et al., 2011) was swapped with TagBFP in a three-fragment Gibson Assembly. First, oligos OKZ67 and OKZ501 were used to amplify a fragment of pRS404-PYKL050c-CFP (Thacker et al., 2011) containing the region of the YKL050c promoter to the ampicillin resistance gene. OKZ68 and OKZ504 were used to amplify a second fragment from the PGK1 terminator to an overlapping stretch of the ampicillin resistance gene in pRS404-PYKL050c-CFP. Lastly, oligos OKZ502 and OKZ503 were used to amplify TagBFP from pUC57-TagBFP-ScOpt with homology at the 5′ end to the YKL050c promoter and to the PGK1 terminator at the 3′ end. The three fragments were then assembled using Gibson Assembly. pRS306-spc42C-TagBFP(pKZ219) was assembled in two steps. First, oligos OKZ386 and OKZ387 were used to amplify the SPC42 C terminus and its 3′ untranslated region (nucleotide 566 of the SPC42 open reading frame to 386 nucleotides downstream from the stop codon) from BY4741 genomic DNA. The PCR product was then assembled with KpnI and SacI digested pRS306 by Gibson Assembly to make pRS306-spc42C (pKZ218). Next, primers OKZ390 and OKZ391 were used to amplify TagBFP from pUC57-TagBFP-ScOpt and the PCR product was then assembled with ClaI digested pRS306-spc42C (the ClaI site is just 5′ of the SPC42 stop codon) by Gibson Assembly to make pRS306-spc42C-TagBFP.
TABLE 2:
Plasmids used in this study.
| Plasmids | Description | Source |
|---|---|---|
| pBTM116-MSS4 FL | lexA fused to the N terminus of Mss4 | This study |
| pBTM116-MSS4I | lexA fused to the N terminus of Mss4 1–446 | This study |
| pBTM116-MSS4II | lexA fused to the N terminus of Mss4 334–779 | This study |
| pBTM116-MSS4III | lexA fused to the N terminus of Mss4 379–556 | This study |
| pBTM116-MSS4IV | lexA fused to the N terminus of Mss4 557–756 | This study |
| pBTM116-MSS4M | lexA fused to the N terminus of Mss4 334–446 | This study |
| pBTM116-MSS4N∆ | lexA fused to the N terminus of Mss4 with the internal deletion 347–364∆ | This study |
| pFA6-KanMX6 | Template for knockout PCR | Longtine et al., 1998 |
| pGADGH-ADY4 | Full-length GAD fusion | Nickas et al., 2003 |
| pGADGH-VPS13I | GAD fused to the N-terminal 964 aa of Vps13 | This study |
| pRS306-spc42C | C-terminal GFP for tagging | This study |
| pRS306-Spo21 | Integrating WT SPO21 | This study |
| pRS306-Spo21-3AR55A,K59A and K66A | Integrating mutant SPO21 | This study |
| pRS306-sSpc42C-TagBFP | Spc42 C terminus fused to TagBFP | This study |
| pRS314-HTB1-GFP | Histone H2B fused to GFP | This study |
| pRS314-HTB1-mOrange | Histone H2B fused to mOrange2 | This study |
| pRS316-HTB1-mOrange | Histone H2B fused to mOrange2 | This study |
| pRS404-PYKL050c-CFP | Spore-specific reporter | Thacker et al., 2011 |
| pRS404-PYKL050c-TagBFP | TagBFP under spore autonomous promoter | This study |
| pRS414-Mss4 GFP | Single GFP tag | Ling et al., 2012 |
| pRS414-Mss4-5xGFP11 | Mss4 fused to five copies of GFP β strand 11 | This study |
| pRS414-Mss4347–364∆GFP | GFP-tagged internal deletion | Ling et al., 2012 |
| pRS415-Stt4 GFP | C-terminal GFP tag | Audhya and Emr, 2002 |
| pRS423-PTEF1-1-10GFP | β strands 1–10 of GFP | This study |
| pRS424-PTEF1-Pex3-yEGFP | Pex3 GFP fusion | This study |
| pRS425-Mss4 | High copy WT gene | Coluccio et al., 2004 |
| pRS425-Pik1 | High copy WT gene | This study |
| pRS425-PTEF1-1-10GFP | β strands 1–10 of GFP | Chen et al., 2020 |
| pRS426-PGAL1-5xGFP11 | Tandem copies of 11th β strand | Chen et al., 2020 |
| pRS426-PTEF1pRS426-GFP Spo14 | Constitutive promoterInternal GFP-tagged WT gene | Mumberg et al., 1995 Rudge et al., 2001 |
| pRS426-PTEF1-1-10GFP | β strands 1–10 of GFP | This study |
| pRS426-PTEF1-Ady4-mOrange Pex15 | Peroxisome-targeted Ady4-mOrange | This study |
| pRS426-PTEF1-GFP-Spo2149–66 | Amphipathic helix of Spo21 | This study |
| pRS426-PTEF1-GFP-Spo2149–66-3A | Mutant amphipathic helix of Spo21 | This study |
| pRS426-PTEF1-mOrange2 Pex15 | Peroxisome-targeted mOrange | This study |
| pRS426-PTEF1-Spo20-51–91TagBFP | Prospore membrane marker | Jin et al., 2017 |
| pRS426-Stt4 | High copy WT gene | This study |
| pUC57-mOrange2-ScOpt | Saccharomyces codon-optimized mOrange2 | This study |
| pUC57-TagBFP-ScOptYEP351-GFP SPO14 | Saccharomyces codon-optimized TagBFPHigh copy GFP-tagged WT gene | This studyRudge et al., 1998 |
| YEP351-LSB6 | High copy WT gene | Audhya and Emr, 2002 |
The different lexA-MSS4 fusions were made by first amplifying different regions of the MSS4 coding region from genomic DNA from BY4741: primers OKZ253 and OKZ264 were used to amplify a fragment encoding Mss4 1-446, OKZ265 and OKZ254 for Mss4 334-779, OKZ265 and OKZ264 for Mss4 334-446, OKZ284 and OKZ285 for Mss4 379-556, and OKZ286 and OKZ287 for Mss4 557-756aa. All primer pairs carry homology to the LexA DNA binding domain and to the ADH1 terminator of pSTT91 (Hollenberg et al., 1995) and were introduced into EcoRI and BamHI digested pSTT91 by Gibson Assembly. The LexA-Mss4-347-364∆ plasmid was constructed by three-fragment Gibson Assembly using pSTT91 plasmid digested with EcoRI and BamHI, a fragment encoding aa 1–346 of Mss4 amplified from pSTT91-Mss4 using oligos OKZ253 and GNO52 and a fragment encoding aa 365–779 of Mss4 using oligos GNO51 and OKZ254. For GAD-VPS13 1–964 aa, the VPS13 segment was amplified from BY4741 genomic DNA using OKZ110 and OKZ115 and inserted into pGAD424 linearized with EcoRI and BamHI by Gibson Assembly.
To build pRS426-PTEF1mOrange2 (pKZ58), primers OKZ40 and OKZ41 were used to amplify mOrange from pUC57-mOrange2-ScOpt and introduced into pRS426-PTEF1 (Mumberg et al., 1995) digested with SpeI and XhoI by Gibson Assembly. pRS424-PTEF1-mOrange2 was then digested with BamHI and XhoI and the region encoding the transmembrane domain of Pex15 (aa 315–383) including the stop codon was amplified from genomic DNA using the primers KZO94 and KZO100 and these two fragments were combined by Gibson Assembly to generate pRS426-PTEF1-mOrange2-Pex15 (pKZ54). To build pRS426-PTEF1-Ady4-mOrange2-Pex15, primers GNO38 and GNO39 were used to amplify Ady4 with flanking homology to the TEF promoter and to mOrange2 and combined with SpeI linearized pRS426-PTEF1-mOrange2-Pex15 by Gibson Assembly. To build pRS426-STT4, the STT4 ORF as well as 400 base pairs upstream and 300 base pairs downstream were amplified from genomic DNA in two halves using the oligo pairs GNO70/GNO72 and GNO71/GNO73. The two resulting fragments were cloned into EcoRI and HindIII digested pRS426 using three-fragment Gibson Assembly.
Sporulation
For liquid sporulation, cells were inoculated in YPD or selective SD media and grown at 30°C overnight. The following day 3 ml of cells were diluted into 35 ml of YPA media and grown at 30°C to an OD660 of 0.9–1.2 for approximately 12–16 h. The cells were washed with distilled water, resuspended at an OD660 of ∼1.0 in 2% potassium acetate, and incubated at 30˚C with shaking. Cells were imaged 5 to 6 h after transfer to potassium acetate. For sporulation on solid medium, cells were first patched onto YPD or selective medium and incubated overnight at 30°C. The following day the cells were replica-plated onto sporulation plates (1% potassium acetate, 2% agar, 0.05% yeast extract, and 0.05% glucose) and left to grow at room temperature overnight for microscopy analysis or at 30°C for 2 d before sporulation and ascal types were assessed.
Yeast two-hybrid assay and spotting assay
The L40 strain was used for yeast two-hybrid experiments; it contains LexA operators upstream of two reporter genes, lacZ and HIS3 (Hollenberg et al., 1995). LexA and GAD fusions of interest were transformed into the L40 strain using a standard yeast transformation protocol. Spotting assays were performed as described (Chen et al., 2018).
Western blotting and antibodies
Proteins were extracted from cells for immunoblotting by resuspension in 5% trichloroacetic acid with gentle agitation at 4°C for 10 min. The proteins were precipitated by centrifugation at 1000 × g for 5 min and resuspended in 1 ml of acetone. The proteins were spun down at 16,000 × g for 7 min, the acetone was aspirated via vacuum, and the cell pellets were dried overnight. The cells were lysed using the following lysis buffer 50 mM Tris, pH 7.5, 1 mM EDTA, 13.75 μl of 1M dithiothreitol , 55 μl of 100 mM phenylmethylsulfonyl fluoride with one protease inhibitor cocktail added (Roche; 04693132001). Lysis buffer (200 μl) was added to the cell pellets along with 200 μl of glass beads and lysed using a Fast Prep 24 bead beater (MP Bio; 116004500). After bead beating, 150 μl of 2× protein sample buffer was added, then the cells were incubated at 95°C for 5 min. Lastly, the cell debris and glass beads were spun down at 16,000 × g for 5 min and the supernatant was transferred to a new tube. The supernatant (5 μl) was loaded onto a 10% SDS-polyacrylamide gel and transferred to a polyvinylidene membrane. All antibodies were obtained from Santa Cruz Biotechnologies; the primary antibodies were anti-Arp7 (sc-8961) and anti-Lex A (sc-365999) at 1:5000 dilution for each. Secondary antibodies were horseradish peroxidase (HRP)–conjugated mouse–anti-goat (sc-2354) and HRP-conjugated goat–anti-mouse (sc-2005) at 1:10,000 dilution for each.
Fluorescence microscopy
Live cells were imaged for fluorescence microscopy using a Zeiss Imager Z2 microscope (Carl Zeiss, Thorn-wood, NY) with a Zeiss Axiocam 702 mono digital camera. To acquire images, ZEN 3.0 (Blue edition) software was used. To prepare figures, Adobe Photoshop and Illustrator were used.
Supplementary Material
Acknowledgments
The authors thank Alison Coluccio for strains and members of the Neiman lab for helpful discussions. This work was supported by National Institutes of Health Grants no. R35 GM-140684 to N.M.H. and no. R01 GM-072540 to A.M.N., as well as a Revise and Resubmit seed grant from the Stony Brook Research Foundation to A.M.N.
Abbreviations used:
- aa
amino acids
- DIC
differential interference contrast
- 5-FOA
%-fluoroorotic acid
- GAD
Gal4 activation domain
- MOP
meosis II outer plaque
- PCR
polymerase chain reaction
- PI4P
phosphatidylinositol-4-phosphate
- PI4P-5K
phosphatidylinositol-4-phosphate-5-kinase
- PI4,5P2
phosphatidylinositol-4,5-bisphosphate
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-11-0515) on March 1, 2023.
REFERENCES
- Audhya A, Emr SD (2002). Stt4 PI 4-kinase localizes to the plasma membrane and functions in the Pkc1-mediated MAP kinase cascade. Dev Cell 2, 593–605. [DOI] [PubMed] [Google Scholar]
- Audhya A, Emr SD (2003). Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J 22, 4223–4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgier BK, Malzone M, Nickas M, Neiman AM (2001). SPO21 is required for meiosis-specific modification of the spindle pole body in yeast. Mol Biol Cell 12, 1611–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD (1998). Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132. [DOI] [PubMed] [Google Scholar]
- Cabantous S, Terwilliger TC, Waldo GS (2005). Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat Biotechnol 23, 102–107. [DOI] [PubMed] [Google Scholar]
- Chen X, Gaglione R, Leong T, Bednor L, de Los Santos T, Luk E, Airola M, Hollingsworth NM (2018). Mek1 coordinates meiotic progression with DNA break repair by directly phosphorylating and inhibiting the yeast pachytene exit regulator Ndt80. PLoS Genet 14, e1007832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhao G, Zahumensky J, Honey S, Futcher B (2020). Differential scaling of gene expression with cell size may explain size control in budding yeast. Mol Cell 78, 359–370.e356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio A, Malzone M, Neiman AM (2004). Genetic evidence of a role for membrane lipid composition in the regulation of soluble NEM-sensitive factor receptor function in Saccharomyces cerevisiae. Genetics 166, 89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidow LS, Goetsch L, Byers B (1980). Preferential occurrence of nonsister spores in two-spored Asci of Saccharomyces cerevisiae: evidence for regulation of spore-wall formation by the spindle pole body. Genetics 94, 581–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desrivieres S, Cooke FT, Parker PJ, Hall MN (1998). MSS4, a phosphatidylinositol-4-phosphate 5-kinase required for organization of the actin cytoskeleton in Saccharomyces cerevisiae. J Biol Chem 273, 15787–15793. [DOI] [PubMed] [Google Scholar]
- Epand RM, Shai Y, Segrest JP, Anantharamaiah GM (1995). Mechanisms for the modulation of membrane bilayer properties by amphipathic helical peptides. Biopolymers 37, 319–338. [DOI] [PubMed] [Google Scholar]
- Han GS, Audhya A, Markley DJ, Emr SD, Carman GM (2002). The Saccharomyces cerevisiaeLSB6 gene encodes phosphatidylinositol 4-kinase activity. J Biol Chem 277, 47709–47718. [DOI] [PubMed] [Google Scholar]
- Halbach A, Landgraf C, Lorenzen S, Rosenkranz K, Volkmer-Engert R, Erdmann R, Rottensteiner H (2006). Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J Cell Sci 119, 2508–2517. [DOI] [PubMed] [Google Scholar]
- Hohfeld J, Veenhuis M, Kunau WH (1991). PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol 114, 1167–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenberg SM, Sternglanz R, Cheng PF, Weintraub H (1995). Identification of a new family of tissue-specific basic helix-loop-helix proteins with a two-hybrid system. Mol Cell Biol 15, 3813–3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma K, Terui S, Minemura M, Qadota H, Anraku Y, Kanaho Y, Ohya Y (1998). Phosphatidylinositol-4-phosphate 5-kinase localized on the plasma membrane is essential for yeast cell morphogenesis. J Biol Chem 273, 15779–15786. [DOI] [PubMed] [Google Scholar]
- Horchani H, de Saint-Jean M, Barelli H, Antonny B (2014). Interaction of the Spo20 membrane-sensor motif with phosphatidic acid and other anionic lipids, and influence of the membrane environment. PLoS One 9, e113484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O’Shea EK (2003). Global analysis of protein localization in budding yeast. Nature 425, 686–691. [DOI] [PubMed] [Google Scholar]
- Jin L, Zhang K, Sternglanz R, Neiman AM (2017). Predicted RNA binding proteins Pes4 and Mip6 regulate mRNA levels, translation, and localization during sporulation in budding yeast. Mol Cell Biol 37, e00408–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiyama D, Sekine S, Barsi-Rhyne B, Hu J, Chen B, Gilbert LA, Ishikawa H, Leonetti MD, Marshall WF, Weissman JS, Huang B (2016). Versatile protein tagging in cells with split fluorescent protein. Nat Commun 7, 11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Strasser K (2000). Role of the spindle pole body of yeast in mediating assembly of the prospore membrane during meiosis. EMBO J 19, 3657–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CP, Kim C, Smith SO, Neiman AM (2013). A highly redundant gene network controls assembly of the outer spore wall in S.cerevisiae. PLoS Genet 9, e1003700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling Y, Stefan CJ, Macgurn JA, Audhya A, Emr SD (2012). The dual PH domain protein Opy1 functions as a sensor and modulator of PtdIns(4,5)P(2) synthesis. EMBO J 31, 2882–2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR (1998). Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Mathieson EM, Schwartz C, Neiman AM (2010a). Membrane assembly modulates the stability of the meiotic spindle-pole body. J Cell Sci 123, 2481–2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson EM, Suda Y, Nickas M, Snydsman B, Davis TN, Muller EG, Neiman AM (2010b). Vesicle docking to the spindle pole body is necessary to recruit the exocyst during membrane formation in Saccharomyces cerevisiae. Mol Biol Cell 21, 3693–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendonsa R, Engebrecht J (2009). Phosphatidylinositol-4,5-bisphosphate and phospholipase D-generated phosphatidic acid specify SNARE-mediated vesicle fusion for prospore membrane formation. Eukaryot Cell 8, 1094–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens PB, Rapport E (1971). Spindles, spindle plaques, and meiosis in the yeast Saccharomyces cerevisiae (Hansen). J Cell Biol 50, 344–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1995). Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Nakamura TS, Suda Y, Muneshige K, Fujieda Y, Okumura Y, Inoue I, Tanaka T, Takahashi T, Nakanishi H, Gao XD, et al. (2021). Suppression of Vps13 adaptor protein mutants reveals a central role for PI4P in regulating prospore membrane extension. PLoS Genet 17, e1009727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, de los Santos P, Neiman AM (2004). Positive and negative regulation of a SNARE protein by control of intracellular localization. Mol Biol Cell 15, 1802–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Morishita M, Schwartz CL, Coluccio A, Engebrecht J, Neiman AM (2006). Phospholipase D and the SNARE Sso1p are necessary for vesicle fusion during sporulation in yeast. J Cell Sci 119, 1406–1415. [DOI] [PubMed] [Google Scholar]
- Neiman AM (1998). Prospore membrane formation defines a developmentally regulated branch of the secretory pathway in yeast. J Cell Biol 140, 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM (2011). Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189, 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman AM, Katz L, Brennwald PJ (2000). Identification of domains required for developmentally regulated SNARE function in Saccharomyces cerevisiae. Genetics 155, 1643–1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickas ME, Schwartz C, Neiman AM (2003). Ady4p and Spo74p are components of the meiotic spindle pole body that promote growth of the prospore membrane in Saccharomyces cerevisiae. Eukaryot Cell 2, 431–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyen M, Jantti J, Keranen S, Ronne H (2004). Mapping of sporulation-specific functions in the yeast syntaxin gene SSO1. Curr Genet 45, 76–82. [DOI] [PubMed] [Google Scholar]
- Rose K, Rudge SA, Frohman MA, Morris AJ, Engebrecht J (1995). Phospholipase D signaling is essential for meiosis. Proc Natl Acad Sci USA 92, 12151–12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P (1990). Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press. [Google Scholar]
- Rothstein R (1991). Targeting, disruption, replacement, and allele rescue: integrative DNA transformation in yeast. Methods Enzymol 194, 281–301. [DOI] [PubMed] [Google Scholar]
- Rudge SA, Morris AJ, Engebrecht J (1998). Relocalization of phospholipase D activity mediates membrane formation during meiosis. J Cell Biol 140, 81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge SA, Pettitt TR, Zhou C, Wakelam MJ, Engebrecht JA (2001). SPO14 separation-of-function mutations define unique roles for phospholipase D in secretion and cellular differentiation in Saccharomyces cerevisiae. Genetics 158, 1431–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudge SA, Sciorra VA, Iwamoto M, Zhou C, Strahl T, Morris AJ, Thorner J, Engebrecht J (2004). Roles of phosphoinositides and of Spo14p (phospholipase D)-generated phosphatidic acid during yeast sporulation. Mol Biol Cell 15, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciorra VA, Rudge SA, Wang J, McLaughlin S, Engebrecht J, Morris AJ (2002). Dual role for phosphoinositides in regulation of yeast and mammalian phospholipase D enzymes. J Cell Biol 159, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segrest JP, Jackson RL, Morrisett JD, Gotto AM Jr (1974). A molecular theory of lipid-protein interactions in the plasma lipoproteins. FEBS Lett 38, 247–258. [DOI] [PubMed] [Google Scholar]
- Shaner NC, Campbell RE, Steinbach PA, Giepmans BN, Palmer AE, Tsien RY (2004). Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol 22, 1567–1572. [DOI] [PubMed] [Google Scholar]
- Sherwood RK, Scaduto CM, Torres SE, Bennett RJ (2014). Convergent evolution of a fused sexual cycle promotes the haploid lifestyle. Nature 506, 387–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P (1989). A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl T, Hama H, Dewald DB, Thorner J (2005). Yeast phosphatidylinositol-4-kinase, Pik1, has essential roles at the Golgi and in the nucleus. J. Cell Biol 171, 967–979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Lam I, Knop M, Keeney S (2011). Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae. Genetics 189, 423–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, Yuan D, Stroe O, Wood G, Laydon A, et al. (2022). AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res 50, D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesp A, Prinz S, Fink GR (2001). Conservative duplication of spindle poles during meiosis in Saccharomyces cerevisiae. J Bacteriol 183, 2372–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Braun P, Yildirim MA, Lemmens I, Venkatesan K, Sahalie J, Hirozane-Kishikawa T, Gebreab F, Li N, Simonis N, et al. (2008). High-quality binary protein interaction map of the yeast interactome network. Science 322, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.