Abstract
Microvilli are defining morphological features of the apical surfaces in diverse epithelial tissues. To develop our understanding of microvillus biogenesis, we used a biotin proximity–labeling approach to uncover new molecules enriched near EPS8, a well-studied marker of the microvillus distal tip compartment. Mass spectrometry of biotinylated hits identified KIAA1671, a large (∼200 kDa), disordered, and previously uncharacterized protein. Based on immunofluorescent staining and expression of fluorescent protein–tagged constructs, we found that KIAA1671 localizes to the base of the brush border in native intestinal tissue and polarized epithelial–cell culture models, as well as dynamic actin-rich structures in unpolarized, nonepithelial cell types. Live imaging also revealed that during the early stages of microvillar growth, KIAA1671 colocalizes with EPS8 in diffraction-limited puncta. However, once elongation of the core bundle begins, these two factors separate, with EPS8 tracking the distal end and KIAA1671 remaining behind at the base of the structure. These results suggest that KIAA1671 cooperates with EPS8 and potentially other assembly factors to initiate growth of microvilli on the apical surface. These findings offer new details on how transporting epithelial cells builds the brush border and may inform our understanding of how apical specializations are assembled in other epithelial contexts.
INTRODUCTION
Microvilli are actin bundle–supported protrusions that decorate the apical surfaces of diverse epithelial cell types, imparting these tissues with specialized functions ranging from solute uptake to mechanosensation. In the case of transporting epithelia, microvilli are organized into densely packed arrays that maximize apical plasma membrane holding capacity and thus the functional surface area available for interacting with the luminal space. Known as “brush borders,” these critical solute-transporting organelles drive nutrient uptake in the intestinal tract and filtrate absorption in the kidney tubule (Welling and Welling, 1975; Helander and Fandriks, 2014). Exaggerated microvilli known as stereocilia also play central roles in the mechanosensory function of the cochlea and of vestibular epithelial systems (McGrath et al., 2017). Though microvilli and similar fingerlike protrusions have evolved to function broadly in physiology, our understanding of mechanisms that control the assembly of these structures during differentiation remains limited. The goal of this study was to identity new proteins that are well positioned to drive the growth of microvilli during the maturation of transporting epithelial cells.
Ultrastructural studies dating back several decades were the first to provide key details on microvillus architecture (Granger and Baker, 1950). An individual microvillus extends a micrometer or more from the apical surface, supported by a core bundle of 20–40 actin filaments that are bundled in parallel (Mooseker and Tilney, 1975a; Ohta et al., 2012). All filaments within the core bundle are oriented with their barbed ends—the preferred site of new actin monomer incorporation—out in the distal tip, whereas the pointed ends extend down into the subapical cytoplasm and are anchored in a filamentous network known as the terminal web (Mooseker and Tilney, 1975b). Subsequent biochemical studies revealed that actin filaments within the core are bundled through the action of villin-1 (VIL1), plastin-1 (PLS1, also known as fimbrin), espin (ESPN), and, most recently characterized, mitotic spindle positioning (MISP) (Bretscher and Weber, 1979, 1980; Bartles et al., 1998; Morales et al., 2022). Additionally, core actin bundles are linked to the overlying plasma membrane by the ezrin–radixin–moesin (ERM) family proteins ezrin (EZR), myosin-1a (MYO1A), and myosin-6 (MYO6; Bretscher, 1983; Howe and Mooseker, 1983; Berryman et al., 1993; Hegan et al., 2012), which collectively provide long-term structural stability and prevent individual protrusions from fusing with their neighbors (Tyska et al., 2005; Casaletto et al., 2011; Hegan et al., 2012).
Long-standing hypotheses on how microvilli form during differentiation were inspired by TEM images that revealed electron–dense plaque at the distal ends of these protrusions (Mooseker and Tilney, 1975a); similar features were eventually found at the distal ends of stereocilia and filopodia as well (Svitkina et al., 2003; Rzadzinska et al., 2004). Situated between the plasma membrane and the barbed ends of core bundle actin filaments, factors in this complex would be well positioned to regulate actin monomer incorporation and thus protrusion growth (Tilney and Cardell, 1970; Mooseker and Tilney, 1975b). Consistent with this proposal, proteins that localize to the distal tip compartment have been implicated in protrusion elongation in several investigations. For microvilli, one example is brain-specific angiogenesis inhibitor 1-associated protein 2-like protein 1 (BAIAP2L1, also known as insulin receptor tyrosine kinase substrate; Postema et al., 2018). In addition to its N-terminal I-BAR domain, BAIAP2L1 contains a central SH3 domain and a WH2 domain that binds to actin (Yamagishi et al., 2004; Millard et al., 2007; Postema et al., 2018). In live cells, BAIAP2L1 tracks the ends of growing protrusions and knockdown of BAIAP2L1 in human intestinal epithelial cells results in shorter microvilli, suggesting a function in protrusion elongation under normal conditions (Postema et al., 2018).
Epidermal growth factor receptor pathway substrate 8 (EPS8) is another tip-localized factor that stands out, based on its remarkably specific enrichment at the distal ends of microvilli, stereocilia, and filopodia (Croce et al., 2004; Manor et al., 2011; Zampini et al., 2011; Zwaenepoel et al., 2012; Chou et al., 2014; Avenarius et al., 2017; Postema et al., 2018). EPS8 contains an N-terminal phosphotyrosine-binding domain (PTB), an src-homology 3 (SH3)-binding domain, proline–rich regions that interact with BAIAP2L1, and a C-terminal actin-binding “effector region” (Mongiovi et al., 1999; Disanza et al., 2004; Uhlik et al., 2005; Hertzog et al., 2010). Motifs in the actin-binding region of EPS8 exhibit both filament bundling and capping activity in vitro (Hertzog et al., 2010), and studies in live cells indicate that these motifs do hold direct barbed end–binding activity (Gaeta et al., 2021a). However, EPS8 loss-of-function experiments in a range of animal and cell culture models uniformly lead to shorter protrusions, suggesting that this factor promotes elongation rather than capping under normal conditions (Tocchetti et al., 2010; Manor et al., 2011; Zampini et al., 2011). Additional data in favor of this proposal come from recent studies of microvillus biogenesis, which showed that new core actin bundles grow de novo and as “daughters” from the sides of preexisting “mother” protrusions; in both cases, new bundles emerge from puncta marked by EPS8 and BAIAP2L1, which remain enriched on the distal ends of actively growing protrusions (Gaeta et al., 2021a). Finally, mutations in EPS8 that disrupt actin binding also impair the formation of EPS8 puncta, core bundle formation, and microvillar motility observed in these live imaging assays (Meenderink et al., 2019; Gaeta et al., 2021a).
Given the exquisitely specific tip localization of EPS8 and the mounting evidence of its involvement in protrusion elongation, we leveraged this factor to screen for new distal-tip residents that might also hold the potential for controlling microvillar growth. Proteomic studies of isolated brush borders have contributed rich datasets on the molecular composition of the apical actin cytoskeleton, although conventional multidimensional protein identification technology approaches lack spatial resolution (McConnell et al., 2011; Revenu et al., 2012; Yoshida et al., 2016). One promising method for defining local proteomes involves proximity labeling, which takes advantage of modified Escherichia coli biotin ligases that nonspecifically biotinylate surrounding proteins (Roux et al., 2012; Kim et al., 2016). By anchoring such a ligase to a protein of interest, neighboring proteins can be biotinylated, isolated, and then identified using mass spectrometry. As an example, biotin proximity labeling using the modified enzyme TurboID was recently used to identify proteins at the apical noncentrosomal microtubule–organizing center in the intestine of Caenorhabditis elegans (Sanchez et al., 2021).
In this study, we used EPS8 to spatially anchor the BioID2 biotin ligase (Kim et al., 2016) at the distal tips of microvilli in human intestinal epithelial-cell lines. This strategy allowed us to identify the uncharacterized protein KIAA1671 as a new resident of brush-border microvilli. KIAA1671 localizes to the base of the brush border in native intestinal tissue and polarized epithelial cell–culture models, and to dynamic actin-rich structures in nonepithelial-cell types. In time-lapse observations of microvillus biogenesis, we found that KIAA1671 localizes to sites of both de novo and daughter core-bundle growth. We also observed that KIAA1671 initially colocalizes with EPS8 in diffraction-limited puncta, although once growth of a core bundle begins, these two factors separate, with EPS8 tracking the elongating distal end and KIAA1671 remaining behind at the base of the structure. Collectively, these results suggest that KIAA1671 might cooperate with EPS8 and potentially other core-bundle assembly factors to initiate the growth of new microvilli on the apical surface. These results highlight the utility of proximity labeling for identifying features of local proteomes, even when spatial proximity is transient. These findings may also hold implications for understanding how apical specializations are assembled in other epithelial contexts.
RESULTS AND DISCUSSION
A BioID2 screen reveals new EPS8 proximal factors
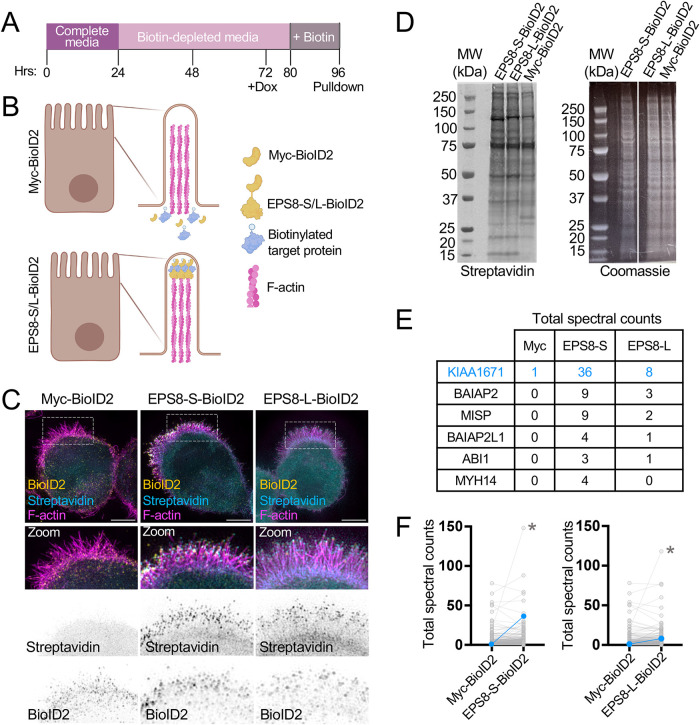
To search for new distal tip residents that might function in microvillus biogenesis, we expressed variants of EPS8 tagged with the biotin ligase BioID2 in the Ls174T-W4 (W4) human intestinal epithelial cell line to enable biotin proximity labeling of EPS8-labeled microvillus tips. W4 cells were engineered to activate the STRADα/LKB1 pathway when treated with doxycycline, which in turn leads to polarity establishment and brush border formation (Baas et al., 2004). W4 cells stably expressing EPS8-S-BioID2 (S = short linker, predicted labeling radius ∼10 nm), EPS8-L-BioID2 (L = long linker, predicted labeling radius ∼25 nm; Kim et al., 2014, 2016), or myc-BioID2 as an unanchored negative control were incubated in biotin-depleted media for ∼48 h before induction of brush border formation and supplementation with 50 μM biotin overnight. The next day, tagged proteins were collected by streptavidin pulldown and then subjected to mass spectrometry for identification (Figure 1, A and B). As EPS8 targets the distal ends of microvilli, we expected that cells expressing BioID2-tagged EPS8 constructs would demonstrate higher levels of biotin incorporation in the brush border than cells expressing the myc-BioID2 control. Indeed, cells expressing the EPS8-S-BioID2 and EPS8-L-BioID2 constructs revealed extensive biotinylation in the brush border compared with controls, as detected by streptavidin staining (Figure 1C). Western blotting for biotinylated proteins in whole-cell lysates using a fluorescent streptavidin conjugate also confirmed biotin incorporation in all samples (Figure 1D). Biotinylated proteins in these samples were subsequently captured by streptavidin pulldown and identified using mass spectrometry, which revealed over 280 hits across all three samples, 56 of which were unique to the EPS8-S-BioID2 and EPS8-L-BioID2 conditions. As expected, we identified multiple known EPS8 binding partners, including brain-specific angiogenesis inhibitor 1-associated protein 2 (BAIAP2), BAIAP2L1, ABI1 (Disanza et al., 2004, 2006; Postema et al., 2018), as well as other brush border–resident proteins such as MISP and MYH14 (Chinowsky et al., 2020; Morales et al., 2022). However, one of the most striking hits was the uncharacterized protein KIAA1671, which demonstrated elevated total spectral counts in EPS8-S-BioID2 and EPS8-L-BioID2 samples (Figure 1, E and F).
FIGURE 1:
Utilizing biotin proximity labeling to probe for molecules involved in microvilli biogenesis. (A) Schematic timeline of W4 induction and biotin supplementation before streptavidin pull down. (B) Schematic representation of a W4 cell expressing the myc-BioID2 construct and EPS8-S/L-BioID2 constructs. Upon addition of 50 µM biotin to cell culture medium overnight, the BioID2 construct will promiscuously biotinylate proteins within an ∼10–25 nm radius. (C) Representative cells expressing either the myc-BioID2 control, EPS8-BioID2-HA, or EPS8-13xL-BioID2-HA constructs (yellow) and stained with streptavin-488 (cyan) and phalloidin (magenta) to determine the extent of biotinylation in the brush border of W4 cells. (D) Biotinylated proteins from EPS8-S-BioID2-HA, EPS8-L-BioID2-HA, or myc-BioID2 samples separated by SDS page and detected with a fluorescent streptavidin conjugate (left). Coomassie staining of previously stated samples to confirm equal total protein input for pulldown (right). (E) Highlighted raw total spectra counts of known EPS8 interacting proteins or brush border resident proteins detected in myc-BioID2, EPS8-S-BioID2, and EPS8-L-BioID2 samples. (F) Plots comparing total spectral counts between proteins detected in myc-BioID2 vs. EPS8-s-BioID2 or EPS8-L-BioID2 samples. Connected blue data points indicate KIAA1671 spectral counts, and asterisks indicate spectral counts of EPS8-S/L-BioID2 protein in samples.
Identification of KIAA1671 as a new resident of the brush border actin cytoskeleton
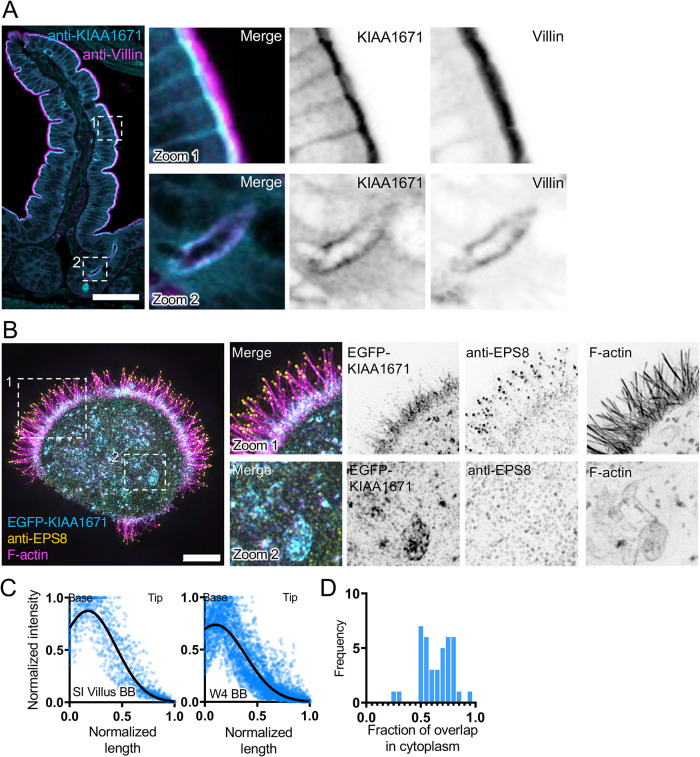
To further explore the utility of hits derived from our EPS8-anchored BioID2 screen, we decided to focus validation studies on the uncharacterized, KIAA1671 (1806 a.a., ∼196 kDa). Although KIAA1671 was previously identified in large-scale genomic and proteomic studies (Huttlin et al., 2017), its biological roles remain entirely unstudied. In silico domain analysis using IUPRED2A (Meszaros et al., 2018; Erdos and Dosztanyi, 2020) suggests that KIAA1671 is largely disordered (Supplemental Figure 1, A and B), although a tankyrase binding domain is annotated near the C-terminus. Further validation by immunofluorescence and construct expression revealed that KIAA1671 robustly targets F-actin–rich networks (Figure 2). In mouse small intestinal tissue, endogenous KIAA1671 localizes to the subapical compartment in villus enterocytes, where the rootlet ends of microvilli embed in the terminal web (Figure 2A, zoom 1, and 2C); apical staining was also observed in intestinal crypts (Figure 2A, zoom 2). Similarly to native tissue, W4 cells exhibited terminal web enrichment of EGFP-KIAA1671 (Figure 2B, zoom 1, and 2C), and in this context a subpopulation also localized to F-actin–rich structures in the cytoplasm (Figure 2B, zoom 2). Costaining these cells for endogenous EPS8 revealed that most of this factor targeted microvillar tips as expected, although we noted a faint band of signal that overlapped with KIAA1671 in the terminal web (Figure 2B, zoom 1). Quantifying the fraction of overlap between the EGFP-KIAA1671 and phalloidin-568 signals in the cytoplasm of W4 cells revealed that the majority (66%) of total cytoplasmic EGFP-KIAA1671 signal merged with F-actin (Figure 2D). Taken together, these results confirm that KIAA1671 is a bona fide resident of the brush-border actin cytoskeleton in polarized epithelial cells.
FIGURE 2:
KIAA1671 is an uncharacterized protein that targets the brush border cytoskeleton. (A) A paraffin-embedded mouse small-intestinal tissue section stained with anti-KIAA1671 (cyan) and anti-Villin (magenta) antibodies. Zoom 1 highlights apical staining on the villus, while Zoom 2 highlights apical staining in intestinal crypts. Scale bar = 50 µm. (B) Representative W4 cell expressing an EGFP-KIAA1671 construct (cyan), stained with an anti-EPS8 antibody (yellow) and phalloidin to mark F-actin (magenta). Zoom 1 highlights KIAA1671 at the base of the brush border, while Zoom 2 highlights KIAA1671 localization in the cytoplasm. (C) Linescans measuring anti-KIAA1671 (Left) or EGFP-KIAA1671 (Right) at the apical surfaces of intestinal tissue and W4 cells, respectively. n = 63 linescans of villus intestinal tissue from one independent staining; n = 40 line cans of W4 brush borders from three independent experiments. (D) Quantification of the fraction of overlap between KIAA1671 and F-actin. Bars indicate the fraction of total KIAA1671 signal that overlaps with F-actin signal. n = 40 cells from three independent experiments. All images are displayed as maximum-intensity projections.
KIAA1671 associates with dynamic F-actin networks in nonpolarized cells
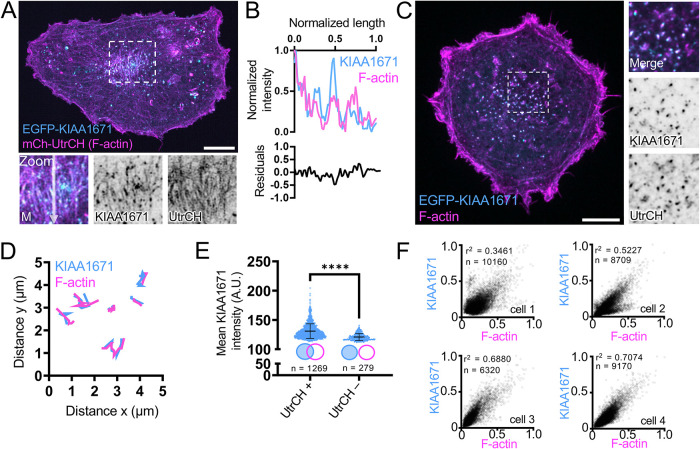
To determine if the localization of KIAA1671 to F-actin–rich structures was unique to the highly specialized actin cytoskeleton found in polarized intestinal epithelial cells, we also expressed EGFP-KIAA1671 in the nonpolarizing line B16F1. Transfection experiments revealed that EGFP-KIAA1671 localizes to F-actin positive linear features that labeled with a fluorescently tagged utrophin calponin homology (UtrCH) domain (mCherry-UtrCH; Figure 3, A and B). We also observed a prominent population of cytoplasmic puncta that were KIAA1671- and F-actin–positive. Time-lapse imaging of this population using spinning disk confocal microscopy showed that KIAA1671-/F-actin–containing puncta are motile, suggesting these are sites of active actin polymerization. Analysis of puncta motion over a 45-s window revealed that the trajectories of KIAA1671 and F-actin signals exhibited significant and sustained spatial overlap through time (Figure 3, C and D; Supplemental Video 1). Additionally, two-dimensional analysis of KIAA1671 puncta that were associated with F-actin signal (Figure 3E, UtrCH +, n = 5701 representative puncta from a single cell) demonstrated higher mean intensities than for puncta that were not F-actin–associated (Figure 3E, UtrCH -, n = 1563 representative puncta from a single cell). Consistent with these analyses, when we correlated the intensities of many KIAA1671/F-actin puncta, we found a significant positive relationship between these two signals across multiple cells (Figure 3F). Thus, higher levels of KIAA1671 are associated with higher levels of F-actin. KIAA1671’s general colocalization with F-actin–rich structures, comigration with F-actin–rich puncta, and positive correlation with F-actin content are suggestive of an actin-binding protein that may promote, either directly or indirectly, the accumulation of actin polymer.
FIGURE 3:
KIAA1671 overlaps with dynamic actin-rich structures in unpolarized cells. (A) A representative B16F1 cell expressing EGFP-KIAA1671 and mCherry-UtrCH to mark F-actin. Zooms highlight linear actin features. Scale bar = 10 µm. (B) Linescan of linear features as highlighted in A, zoom. (C) B16F1 cell displaying cytoplasmic puncta of EGFP-KIAA1671 and mCherry-UtrCH. Scale bar = 10 µm. (D) Representative trajectory analysis of 6 EGFP-KIAA1671 and mCherry-UtrCH puncta as represented in C. Traces measured at 5-s intervals sampled over a 45-s time period from a single imaging field. (E) 2D puncta analysis comparing mean EGFP-KIAA1671 intensities of puncta either associated (UtrCH +) or not associated (UtrCH-) with UtrCH puncta. Plots are representative from a single cell over a 15-min imaging window. **** p < 0.0001 using a two-tailed Mann–Whitney test, n = 1269 UtrCH + puncta; n = 279 UtrCH – puncta. Plots show mean ± SD. (F) Two-dimensional intensity correlations of EGFP-KIAA1671 puncta with mCherry-UtrCH from six cells over two independent experiments. p values were calculated from a two-tailed test. All images are displayed as maximum intensity projections.
Movie S1.
KIAA1671 and EPS8 transiently colocalize in puncta during microvillus biogenesis
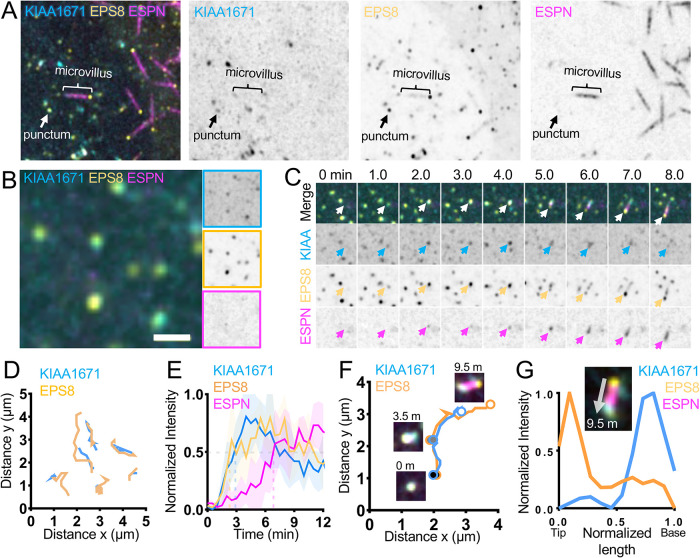
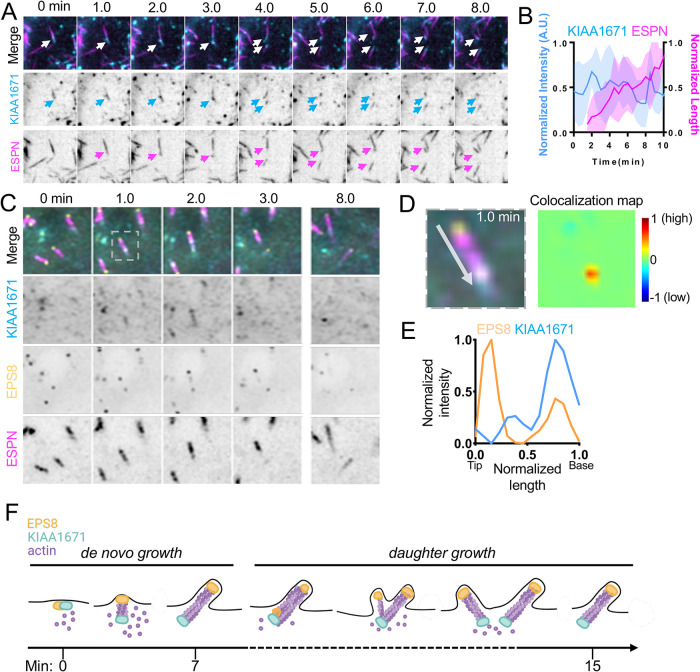
As alluded to, KIAA1671 localizes to F-actin networks in diverse contexts, although in the specific case of polarized epithelial cells, it localizes to the rootlet end of the microvillus, which is—paradoxically—the opposite end of the protrusion from where EPS8 resides. Given the significant spatial separation between these compartments (∼1 μm or more), how might an EPS8-anchored biotin ligase encounter a protein near the basal ends of microvilli? One potential answer derives from the fact that microvilli are actively growing on the surfaces of W4 cells during the biotinylation step in our labeling assay. Considering this point, EPS8 and KIAA1671 might only transiently colocalize at some point during the growth process. To explore this possibility, we turned to live imaging of microvillar biogenesis using the LLC-PK1-CL4 (CL4) cell culture model. We previously used this system to directly visualize the motility and growth of individual microvilli from an en face perspective (Meenderink et al., 2019; Gaeta et al., 2021a,b). CL4 cells stably expressing EGFP-EPS8 and mCherry-ESPN were transiently transfected with Halo-KIAA1671 (labeled with JFx646) and then subjected to spinning-disk confocal microscopy to enable visualization of microvillus biogenesis. Time-lapse datasets revealed a population of KIAA1671 and EPS8 that are microvillus–associated (Figure 4A) as well as a separate population of colocalized punctate structures that lacked ESPN signal (Figure 4B). Analyzing the motion of KIAA1671 and EPS8 puncta at 30-s intervals over a period of 30 min revealed overlapping trajectories, suggesting that these proteins are moving through the cytoplasm in complexes (Figure 4, B and D). We also observed that KIAA1671 and EPS8 coaccumulate at future sites of de novo microvillar growth; in many of these cases, minutes later, ESPN-positive actin bundles elongated from these puncta (Figure 4, C and E). Notably, as these core bundles elongate, EPS8 and KIAA1671 signals separate, with EPS8 marking the growing distal tip of the microvillus and KIAA1671 remaining behind at the proximal end (Figure 4, F and G; Supplemental Video 2).
FIGURE 4:
KIAA1671 and EPS8 puncta associate and then separate during microvillus growth. (A) CL4 cell displaying microvilli as well as cytoplasmic KIAA1671 and EPS8 puncta in cells expressing Halo-KIAA1671, EGFP-EPS8, and mCherry-ESPN. Box width = 15 µm. (B) Representative image of EGFP-EPS8 and Halo-KIAA1671 puncta in CL4 cells. Single-channel images are denoted by border outline in corresponding color (right). (C) Montage of a de novo microvillus growth event in a CL4 cell expressing EGFP-KIAA1671 (KIAA, cyan), EGFP-EPS8 (yellow), and mCherry-ESPN (magenta). Box width = 4 µm. (D) Trajectory analysis of Halo-KIAA1671 and EGFP-EPS8 puncta as represented in B. Traces are measured at 30-s intervals sampled from a 30-min time period from a single imaging field. (E) Averaged normalized intensity vs. time curves representing 5 de novo microvillus growth events from the imaging experiment shown in C. Solid line represents mean and shaded area represents SD. (F) Trajectory analysis of Halo-KIAA1671 (cyan) and EGFP-EPS8 (yellow) during a de novo microvillus growth event. Large, shaded circles represent indicated time points during the trajectory. (G) Image of the 9.5-min time point indicated in F (Top) with linescan demonstrating separation of the Halo-KIAA1671 and EGFP-EPS8 signals to opposite ends of the microvillus. All images are displayed as maximum-intensity projections.
Movie S2.
In addition to de novo growth, new “daughter” microvilli also grow from preexisting “mother” protrusions (Gaeta et al., 2021a), generating branched intermediates similar in appearance to the “forked” microvilli observed on the surfaces of diverse epithelial tissues (Tilney and Cardell, 1970; Zhu et al., 2022). Careful inspection of our time-lapse datasets revealed that EGFP-KIAA1671 localizes at the basal end of a mother microvillus (marked by mCherry-ESPN) in the minutes leading up to daughter formation (Figure 5, A and B). Previous studies established that EPS8 marks the site of new daughter growth from an established mother (Gaeta et al., 2021a). Consistent with this, KIAA1671 and EPS8 are both found at the base of nascent mother protrusion before elongation of a daughter microvillus begins (Figure 5C; Supplemental Video 3). Indeed, colocalization map analysis of the 1-min time point in Figure 5A shows that before daughter core bundle elongation, KIAA1671 and EPS8 signals exhibit strong colocalization at the base of the microvillus (Figure 5, D and E).
FIGURE 5:
KIAA1671 and EPS8 puncta overlap during daughter microvillus growth. (A) Representative montage of daughter microvillus growth event. Box width = 9 µm. (B) Quantification of EGFP-KIAA1671 intensity and microvillus length vs. time of daughter growth events. n = 12 daughter growth events from seven cells over two independent experiments. (C) Montage of a daughter microvillus growth event in CL4 cells expressing Halo-KIAA1671, EGFP-EPS8, and mCherry-ESPN. Box width = 5 µm. (D) Zoom of the 1-min timepoint in C (Left), with colocalization heat map analysis (Right). Warmer colors on heat map represent a higher degree of colocalization, while cooler colors represent a low degree of colocalization. (E) Linescan of zoom shown in D. (F) Schematic of both de novo and daughter microvillus growth illustrating KIAA1671 and EPS8 together at early stages of de novo growth, followed by separation as the microvillus matures. During daughter growth KIAA1671 and EPS8 mark spots of new bundle formation on the mother bundle. As the new daughter bundle grows, EPS8 remains at the distal tip while KIAA1671 stays at the base of the microvillus.
Movie S3.
Collectively, these time-lapse observations show that KIAA1671 and EPS8 transiently colocalize in puncta at the earliest stages of microvillar growth, and then separate as the microvillus elongates (Figure 5F), which provides a likely explanation for the biotinylation of KIAA1671 in our EPS8 anchored proximity labeling screen.
Using a proximity labeling approach, we identified the previously uncharacterized protein KIAA1671 as a bona fide resident of the brush border actin cytoskeleton. In native intestinal epithelial cells, KIAA1671 localizes to the base of microvilli in the subapical terminal web, and expression studies in culture models also indicate that this factor localizes to dynamic actin networks independent of the cellular context. However, we were presented with a paradox when we considered the localization of KIAA1671 relative to EPS8, which was used to anchor the BioID2 moiety in our screen. EPS8 localizes to the distal tips, while KIAA1671 is found at the bases of microvilli, separated by a distance of at least 1 µm, outside the predicted range of proximity labeling (∼25 nm, using the long linker). We employed live-cell imaging to investigate whether the interaction between EPS8 and KIAA1671 might be transient in nature. Indeed, time-lapse datasets revealed that EPS8 and KIAA1671 do associate in dynamic puncta with a subset giving rise to microvilli de novo. Remarkably, as the core actin bundle begins to elongate, the EPS8 and KIAA1671 signals separate, with EPS8 occupying the elongating distal end as expected, and KIAA1671 remaining behind at the base of the protrusion. The end result is a microvillus with EPS8 marking the distal end and KIAA1671 highlighting the rootlet, a distribution consistent with what we observe in fixed intestinal epithelial tissue and cell culture models at steady state. We also observed a similar pattern of initial transient colocalization followed by spatial separation in instances of mother–daughter growth. Thus, using a spatially targeted proteomic screen, we identified a novel factor that may cooperate with EPS8 during the early stages of microvillus biogenesis.
Do our results implicate KIAA1671 as a binding partner of EPS8? Biotin proximity labeling in conjunction with live-cell imaging of KIAA1671 and EPS8 during microvillus biogenesis suggests that these proteins transiently associate in a complex. However, neither of these techniques implies direct protein–protein interaction, and thus further biochemical characterization is necessary to determine whether EPS8 and KIAA1671 are binding partners. In terms of structural mechanisms that could mediate direct binding, EPS8 contains a central SH3 domain that mediates protein:protein interactions by binding to proline-rich sequences in interacting proteins. The SH3 domain of EPS8 binds to PxxDY motifs instead of canonical PxxP motifs (Mongiovi et al., 1999), and primary sequence analysis of KIAA1671 indicates the presence of multiple proline-rich stretches. Such regions are common features of intrinsically disordered proteins (Theillet et al., 2013), presenting a possibility for future studies aimed toward characterizing KIAA1671/EPS8 interactions. Independent of the details of association, our data do indicate that KIAA1671 and EPS8 cooperate early in this process of core bundle assembly. Interestingly, transmission electron micrographs of microvillus regrowth events in native tissue revealed electron-dense plaques at the apical surfaces of intestinal cells, which were hypothesized to be microvillus precursors (Tilney and Cardell, 1970). KIAA1671 and EPS8 do colocalize in cytoplasmic puncta that lack ESPN signal, and it is tempting to speculate that these features correspond to electron-dense plaques noted in those classic ultrastructural studies.
How might KIAA1671 impact the dynamics of the actin cytoskeleton? Bioinformatics analysis identified a tankyrase binding protein motif at the C-terminus of KIAA1671. Intriguingly, the related tankyrase binding domain containing protein TNKS1BP1 (also known as TAB182) has been shown to associate with actin, and through its C-terminus directly binds the CAPZA2 subunit of heterodimeric capping protein (CP) (Ohishi et al., 2017). Local alignment of KIAA1671 and TNKS1BP1 indicates that its CAPZA2-binding region a.a. 1543–1635 displays 32.4% identity and 51% similarity with KIAA1671 a.a. 1617–1711 (Ohishi et al., 2017; Supplemental Figure 2C), and previous large-scale affinity purification proteomic studies also detected an interaction between KIAA1671 and CAPZA2 (Huttlin et al., 2021). What are the functional implications of an interaction with CAPZA2? CP binds to the barbed ends of actin filaments with subnanomolar affinity, potently inhibiting incorporation of new monomers (Wear et al., 2003). However, several factors tune CP activity using steric or allosteric mechanisms. Notably, CARMIL family proteins (including CKIP-1, WASHCAP, CD2AP, CapZip, and Twinfilin) allosterically reduce CP affinity for the filament barbed end. Direct binding of these proteins to CP is mediated by a capping protein interaction (CPI) motif (Bruck et al., 2006; Hernandez-Valladares et al., 2010; McConnell et al., 2020). Interestingly, local alignment of the CARMIL1 CPI (a.a. 958–1082) indicates weak identity with KIAA1671 a.a. 1013–1137 (Supplemental Figure 2D), leading to the intriguing possibility that this factor might hold the potential to regulate CP activity and drive actin polymerization during microvillar growth.
Additional insight into the function of KIAA1671 might also be derived from its dynamic localization during microvillus growth. As alluded to above, KIAA1671 initially colocalizes with EPS8, but as the nascent core bundle begins to elongate, these two factors separate and become enriched at opposite ends. This dynamic pattern is reminiscent of the behavior of the actin nucleator APC and the formin mDia1, as observed in vitro (Breitsprecher et al., 2012). APC and mDia1 initially colocalize in puncta that represent a nucleation complex, but as the associated actin filament begins to elongate, APC remains behind at the pointed end, while mDia1 tracks the elongating barbed end (Breitsprecher et al., 2012). In this case, the persistent pointed-end localization of APC is a telltale sign of its activity as a templating filament nucleator. Although the current level of annotation on KIAA1671 does not point to canonical actin monomer binding motifs, which would be needed to underpin nucleation activity, this remains an open functional hypothesis that could be tested in future cell-biological studies.
MATERIALS AND METHODS
Cell culture
Ls174T-W4 cells (female human colon epithelial cells) were cultured in DMEM with high glucose and 2 mM l-glutamine supplemented with 10% tetracycline-free fetal bovine serum (FBS), 1% glutamine, G-418 (1 mg/ml), blasticidin (10 μg/ml), and phleomycin (20 μg/ml). LLC-PK1-CL4 and B16F1 melanoma cells were cultured in DMEM with high glucose and 2 mM l-glutamine supplemented with 10% FBS and 1% glutamine. All cultures were maintained at 37°C with 5% CO2.
Cloning and Constructs
An empty vector pLVX-MCS-puro construct was created by digesting the pLVX-AcGFP vector (Takara) with NdeI and XhoI. The region of the CMV promoter that was cut during this process was religated, with the region amplified via PCR using primers 5′→3′ forward CCCCCGCCCATTGACGTCAATAATGACG and 5′→3′ reverse TAAGCACTCGAGGGTGGCGACCGGTAGAT, creating the vector pLVX-MCS-puro. BioID2-HA and 13xLinker-BioID2-HA sequences were amplified via PCR flanked with XhoI and ApaI cut sites 5′→3′ forward CCCCCGCCCATTGACGTCAATAATGACG and 5′→3′ reverse TAAGCACTCGAGGGTGGCGACCGGTAGAT. These PCR products were digested and inserted into the pLVX-MCS-puro vector to create pLVX-MCS-BioID2 and pLVX-MCS-13xLinker-BioID2. The pLVX-myc-BioID2 vector was created via Gibson assembly, using PCR to amplify the insert sequence 5′→3′ forward GGATCTACCGGTCGCCACCCAGACCCAAGCTGGCTAGC, 5′→3′ reverse GATCCGGTGGATCCCGGGCCTCGAGGCTGATCAGCGGTTTAAAC, and inserted into the pLVX-MCS-puro backbone digested with XhoI and ApaI using the Gibson assembly master mix (NEB). Full-length human EPS8 was amplified via PCR using primers 5′→3′ forward TAAGCACTCGAGGCCACCATGAATGGTCATATTTCTAATCATCC and 5′→3′ reverse TAAGCAGAATTCGTGACTGCTTCCTTCATCAAAAGATTCC, inserted into the pLVX-MCS-BioID2-HA and pLVX-MCS-13xLinker-BioID2-HA via restriction digest with XhoI and EcoRI. The EGFP-KIAA1671 plasmid was purchased from Genscript, encoding ORF clone accession number NM_001145206.1, inserted into a pcDNA3.1(+)-N-eGFP backbone. pHalo-C1-KIAA1671 was generated using by PCR using primers 5′→3′ forward ACCATGGCCACGCGG and 5′→3′ reverse TCAAACTTGGTTCTCATAAAGACTTTGCCT, topo cloned into the pCR8 vector, and then shuttled into the pHalo-C1-GW vector. Full-length human EPS8 was generated via PCR using primers 5′→3′ forward ATGAATGGTCATATTTCTAATCATCCC and 5′→3′ reverse TTAGTGACTGCTTCCTTCATCAAAAGATTCC topo cloned into the EGFP-C1-GW vector. All constructs were verified by sequencing.
Stable cell line generation and transient transfection
Stable cell lines.
To create W4 BioID2 and CL4 mCherry-ESPN (encoding human espin 2B)–expressing stable cells, target cells were transduced with lentivirus expressing pLVX-EPS8-BioID2-HA, pLVX-EPS8-13xLinker-BioID2-HA, pLVX-myc-BioID2, or pLVX-mCherry-ESPN. Lentivirus was generated using HEK293FT cells cotransfected with pLVX-EPS8-BioID2-HA, pLVX-EPS8-13xLinker-BioID2-HA, pLVX-myc-BioID2, or pLVX-mCherry-ESPN and psPAX2 and pMD2.G using Lipofectamine2000 (ThermoFisher) according to the manufacturer’s instructions. Media were changed 16 h after transfection, and virus was collected 72 and 96 h after transfection and concentrated using Lenti-X concentrator (Clontech). Cells were then transduced with virus after growing to 75% confluency and supplemented with 10 μg/ml polybrene. Media were changed 1 day after transduction and cells were placed under selection with puromycin (10 μg/ml concentration) 72 h after transduction. To create double stable CL4 cell lines, cells either transiently transfected with EGFP tagged constructs and subsequently selected with 1 mg/ml G418 for approximately 2–4 wk (EGFP-EPS8, GFP-KIAA1671) and transduced with lentivirus expressing pLVX-mCherry-ESPN. Cells expressing EGFP-KIAA1671 and mCherry-ESPN were plated onto plasma-cleaned 35-mm glass-bottomed dishes (Cellvis, D35-20-1.5-N) and imaged at subconfluency.
Transient transfection.
B16F1 melanoma cells were transiently transfected with EGFP-KIAA1671 and mCherry-UtrCH using Lipofectamine2000 according to the manufacturer’s instructions. The next day, cells were plated onto plasma-cleaned 35-mm glass-bottomed dishes coated with 50 µg/ml laminin, allowed to adhere for at least 3 h, and then imaged. CL4 cells stably expressing EGFP-EPS8 and mCherry-ESPN were transiently transfected with Halo-KIAA1671 using Lipofectamine 2000 according to the manufacturer’s instructions. After 4 h, cells were plated onto plasma-cleaned 35-mm glass-bottomed dishes (Cellvis, D35-20-1.5-N) and imaged the following day at subconfluency. For labeling of Halo-tagged constructs, cells were incubated for 1 h in JFX-646 dye (Janelia Farms, diluted 1:1000 in CL4 medium), briefly rinsed with phosphate-buffered saline (PBS), and then imaged in CL4 medium.
Biotin proximity labeling pull down and mass spectrometry
W4 cells stably transduced with pLVX-EPS8-BioID2, pLVX-EPS8-13xLBioID2, or pLVX-myc-BioID2 were grown to ∼80% confluency in T-175 flasks and incubated for 48 h in biotin-depleted media before induction with doxycycline. Biotin (50 μM) was added to cell culture media after 8 h of induction with doxycycline, and induced cells were incubated overnight. The following pull-down protocol was drawn largely from Roux et al. (2018). Cells were rinsed 2× in DPBS and pelleted at 200 × g for 5 min. Pelleted cells were resuspended in room temperature Lysis Buffer (8 M urea, 50 mM Tris-HCl, 1 mM DTT, and 1× Halt Protease Inhibitor, pH 7.4). Triton X-100 was added to make a final vol/vol of 1%, and suspension was mixed by trituration and placed on ice. Samples were then lysed with a 27 ½ gauge needle attached to a 1-ml syringe 6× each and diluted with prechilled lysis buffer to favor affinity capture (Roux et al., 2018). Samples were then spun at 16,500 × g for 10 min at 4°C. Supernatant was added to equilibrated streptavidin sepharose high- performance beads (GE Healthcare) and rotated overnight at 4°C. Beads were then pelleted at 1000 × g and resuspended in 1 ml wash buffer (8 M urea, 50 mM Tris-HCl, pH 7.4) 4× and resuspended in 1 ml wash buffer. Ten percent of beads were saved for Western blot, with the other 90% resuspended in 1 mM biotin + 50 mM NH4HCO3 in preparation for mass spectrometry analysis. Data in Figure 1, E and F, show spectral data from a single experiment. Mass spectrometry was performed through the Vanderbilt Mass spectrometry core. Briefly, to the bead slurry, 50 µl of 8 M urea 100 mM tris-HCl pH 8.0 was added, and disulfide bonds were reduced using TCEP and alyklated to prevent reformation with iodoacetimde. Slurry was further diluted using 300 µl of 100 mM tris-HCL pH 8.0; 5 µg of sequencing-grade trypsin was added and allowed to digest overnight at 37°C. A 5-µl sample of the resulting peptides was analyzed by data-dependent LC-MS/MS. Briefly, peptides were autosampled onto a 200 mm by 0.1 mm (Jupiter 3 µm, 300 A) self-packed analytical column coupled directly to an LTQ linear ion trap mass spectrometer (ThermoFisher) using a nanoelectrospray source and resolved using an aqueous-to-organic gradient. Both the intact masses (MS) and fragmentation patterns (MS/MS) of the peptides were collected in a data-dependent manner utilizing dynamic exclusion to maximize depth of coverage. Resulting peptide MS/MS spectral data were searched against the canonical human protein database (Uniprot), to which common contaminants and reversed version of each protein, using SEQUEST (https://link.springer.com/article/10.1016/1044-0305(94)80016-2). Peptide spectral matches were collated, filtered, and compared using Scaffold (Proteome Software).
Immunofluorescence staining
Cultured cells.
Cells were rinsed 1× in prewarmed PBS and fixed by incubating in prewarmed 4% paraformaldehyde for 15 min at 37°C. Cells were then rinsed 3× for 5 min with PBS and permeabilized in 0.1% Triton X-100 for 15 min at RT. Cells were then rinsed with PBS 3× for 5 min and then blocked in 5% BSA for 1 h at 37°C. Following blocking, cells were briefly rinsed 1× with PBS and incubated with 1° antibodies for 1 h at 37°C (anti-HA rabbit, Invitrogen H6908; anti-myc rabbit; Abcam ab9106) or Alexa Fluor-488 conjugated streptavidin (Invitrogen S32354), both at 1:1000 dilution. Cells were then rinsed 4× for 5 min with PBS and incubated with 2° antibodies (Alexa Fluor 647 F(ab’)2-goat anti-rabbit IgG (H+L) A21246, 1:1000 and Alexa-Fluor 568 Phalloidin (Invitrogen A12380)), 1:200 for 30 min at RT. Following 2° antibody incubation, cells were rinsed 4× for 5 min in PBS and mounted on glass slides using ProLong Gold antifade reagent (Life Technologies, Invitrogen, P36930).
Tissue.
Paraffin-embedded tissue sections of mouse WT small intestinal Swiss rolls and kidney sections were deparaffinized using HistoClear solution (Fisher) and rehydrated in a descending graded ethanol series (100%, 95%, 90%, 70%, 50%) and rinsed 3× with PBS for 5 min. Slides were then subjected to an antigen retrieval step consisting of boiling for 1 h in a solution of 10 mM Tris (pH 9.0) and 0.5 mM ethylene glycol–bis(β-aminoethyl ether)–-N,N,N′,N′-tetraacetic acid (EGTA). Slides were then washed in PBS 3× and blocked with 5% normal goat serum (NGS) ON at 4°C. After blocking, slides were briefly rinsed with PBS and stained ON at 4°C with antibodies (rabbit anti-KIAA1671, Invitrogen, PA5-53737; mouse anti-Villin, Santa Cruz, sc-66022 1:50) in 1% NGS in PBS. After being washed with PBS 4×, samples were incubated with secondary antibodies (Alexa Fluor 488 F(ab’)2-goat anti-rabbit IgG (H+L), Invitrogen A11070, 1:1000; Alexa Fluor 568 F(ab’)2-goat anti-mouse IgG (H+L), Invitrogen A11019, 1:1000) in 1% NGS in PBS for 1–2 h at RT. Slides were then washed 4× with PBS, dehydrated in an ascending ethanol series (50%, 70%, 90%, 95%,100%), and mounted using ProLong Gold Antifade reagent (Life Technologies, Invitrogen, P36930).
Microscopy
Laser scanning confocal microscopy was performed using a Nikon A1 equipped with 488- and 561-nm excitation lines and 100×/1.49 NA TIRF, 40×/1.3 NA oil, and 25×/1.05 NA silicon immersion objectives. Structured illumination microscopy was performed using a Nikon N-SIM equipped with an Andor DU-897 EMCCD camera, four excitation lines (405, 488, 561, and 647 nm), and a 100×/1.49 NA TIRF objective. All images were reconstructed using Nikon Elements software with matching reconstruction parameters. Spinning disk confocal microscopy was performed using a Nikon Ti2 inverted light microscope equipped with a Yokogawa CSU-X1 spinning disk head, a Photometrics Prime 95B sCMOS or Hamamatsu Fusion BT sCMOS camera, 488- and 561-nm excitation lasers, and a 100×/1.49 NA TIRF objective. Cells were maintained in a stage top incubator at 37°C with 5% CO2 (Tokai Hit).
Quantification and statistical analysis
Line scan analysis.
To perform line scan analysis, lines were drawn along the axes of microvilli or linear actin features on maximum-intensity projected images in FIJI. Linewidth was set for the following: tissue, 1; W4 cells, 30; B16F1 cells, 5; CL4 microvilli, 4–5. For intestinal tissue, line scan analysis was conducted on 100× images, while a 40× image is represented in Figure 2A. The length and intensity axes were then normalized from 0 to 1, combined, and fitted to a Gaussian where appropriate (GraphPad Prism). Residual plots were calculated from the difference between raw EGFP-KIAA1671 intensity signals and plotted over the normalized length.
Fraction of overlap.
To calculate the fraction of overlap between the KIAA1671 and phalloidin signals in the cytoplasm of W4 cells, a 5 × 5 µm area was cropped and individual channels were manually thresholded in Nikon Elements. A binary layer “AND” was created to measure the pixel area of overlapping pixel intensities between the 488- and 561-nm channels. To calculate the fraction of overlap, the “AND” area fraction was divided by the total KIAA1671 area fraction.
Trajectory analysis.
Using the Nikon elements “spot detection” to detect puncta and “track binaries” to measure XY trajectories, a subset of puncta from a 5 × 5 µm area or a single representative de novo growth event was tracked over a period indicated in the figure legends. XY coordinates were then plotted in GraphPad Prism.
Comparison of UtrCH + and UtrCH - EGFP-KIAA1671 marked puncta.
Using Nikon Elements “spot detection” to detect puncta, binary layers of EGFP and mCherry channels were created. The binary “HAVING” function was used to segment EGFP (i.e., KIAA1671) puncta that were associated with mCherry (i.e., UtrCH) puncta. “Track binaries” was then used to measure KIAA puncta intensities over time from a 10 × 10 µm image area. Areas that contained a large “dropletlike” KIAA1671 signal were excluded from analysis. Values were plotted and statistical difference was evaluated using a Mann–Whitney test in GraphPad Prism.
Puncta intensity correlations.
Using Nikon Elements “spot detection” to detect EGFP-KIAA1671 puncta and mCherry-UtrCH puncta on maximum-intensity projected images, additional binary layers were then created using binary image operations. To detect KIAA1671 puncta associated with UtrCH puncta, the “HAVING” operation was used. To detect KIAA1671 puncta that were not associated with UtrCH puncta, the binary layer of KIAA1671 puncta “HAVING” UtrCH was subtracted from the total KIAA1671 binary layer. The same operations were performed to determine UtrCH puncta associated and not associated with KIAA1671 puncta. Mean intensities were measured using the “track binaries” function to track intensities of puncta over time throughout the movie.
Intensity correlations.
Using Nikon Elements “spot detection,” EGFP-KIAA1671 and mCherry-UtrCH puncta were detected using maximum-intensity projected images. Plots represent all thresholded puncta in all frames of time-lapse movies. Intensities from individual channels were normalized and plotted, and Pearson r2 and p values were calculated using GraphPad Prism.
Time series analysis of de novo microvillus growth.
Events were analyzed manually using tools in FIJI. All measurements were performed on z-projected images deconvolved using the Richardson–Lucy algorithm Nikon Elements Software. For EGFP and mCherry coexpressing cells, t = 0 was defined as three frames (1.5 min) before the appearance of the EGFP signal. A circular ROI was used to measure the mean EGFP signal, Halo-JF646, and mCherry signal for each microvillus. Events were excluded if they held less than three frames (1.5 min) before the appearance of the EGFP signal, if the microvillus collapsed before reaching steady state growth, if a single bundle immediately gave rise to multiple microvilli, or if significant overlap with adjacent microvilli confounded our analysis. Using Prism 10 (GraphPad), mean intensity values were first normalized to account for intensity differences between microvilli, with the smallest value equal to 0 and the largest value equal to 1. Normalized values were then averaged and plotted with SD.
Time series analysis of daughter microvillus growth.
Events were analyzed manually using tools in FIJI. All measurements were performed on z-projected images deconvolved using the Richardson–Lucy algorithm in Nikon Elements software. For EGFP and mCherry coexpressing cells, t = 0 was defined as three frames (1.5 min) before the appearance of the mCherry signal. A circular ROI was used to measure the mean EGFP signal for each daughter microvillus. The line tool was then used to measure the length of the daughter bundle over time. Events were excluded if they held less than three frames (1.5 min) before the appearance of the mCherry-ESPN + daughter bundle or if significant overlap with adjacent microvilli confounded our analysis. Using Prism 10 (GraphPad), mean intensity and length values were first normalized to account for intensity and length differences between microvilli, with the smallest value equal to 0 and the largest value equal to 1. Normalized values were then averaged and plotted with SD.
Colocalization colormap.
The colocalization heatmap was created using the colocalization colormap plugin in FIJI on cropped maximum-intensity projected images.
Local protein alignment.
Protein sequences were aligned using EMBOSS Water local alignment https://www.ebi.ac.uk/Tools/psa/ with the BLOSUM62 matrix.
Supplementary Material
Acknowledgments
The authors would like to thank all members of the Tyska laboratory, the members of the Vanderbilt Microtubules and Motors Club, the Epithelial Biology Center, and the Cellular, Biochemical, and Molecular Sciences Training program for feedback and guidance. Microscopy was performed in part by the Vanderbilt Cell Imaging Shared Resource. Mass spectrometry analysis was performed through the Vanderbilt Mass Spectrometry Core lab. This work was supported by Vanderbilt Cellular, Biochemical and Molecular Sciences Training Grant 5T32GM 008554-25 (I.M.G.), the NIH NIDDK National Research Service Award F31 DK122692 (I.M.G.), and NIH Grants R01 DK111949, R01 DK125546, and R01 DK095811 (M.J.T.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations used:
- I-BAR
inverse bin-amphiphysin-rvs
- JFx646
janelia fluor x646
- PTB
phosphotyrosine binding
- SH3
src-homology 3
- TEM
transmission electron microscopy
- TNKS1BP1
tankyrase 1 binding protein 1
- WH2
WASP homology domain-2.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E22-11-0498) on February 15, 2023.
REFERENCES
- Avenarius MR, Krey JF, Dumont RA, Morgan CP, Benson CB, Vijayakumar S, Cunningham CL, Scheffer DI, Corey DP, Muller U, et al. (2017). Heterodimeric capping protein is required for stereocilia length and width regulation. J Cell Biol 216, 3861–3881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas AF, Kuipers J, van der Wel NN, Batlle E, Koerten HK, Peters PJ, Clevers HC (2004). Complete polarization of single intestinal epithelial cells upon activation of LKB1 by STRAD. Cell 116, 457–466. [DOI] [PubMed] [Google Scholar]
- Bartles JR, Zheng L, Li A, Wierda A, Chen B (1998). Small espin: a third actin-bundling protein and potential forked protein ortholog in brush border microvilli. J Cell Biol 143, 107–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berryman M, Franck Z, Bretscher A (1993). Ezrin is concentrated in the apical microvilli of a wide variety of epithelial cells whereas moesin is found primarily in endothelial cells. J Cell Sci 105, 1025–1043. [DOI] [PubMed] [Google Scholar]
- Breitsprecher D, Jaiswal R, Bombardier JP, Gould CJ, Gelles J, Goode BL (2012). Rocket launcher mechanism of collaborative actin assembly defined by single-molecule imaging. Science 336, 1164–1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A (1983). Purification of an 80,000-dalton protein that is a component of the isolated microvillus cytoskeleton, and its localization in nonmuscle cells. J Cell Biol 97, 425–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K (1979). Villin: the major microfilament-associated protein of the intestinal microvillus. Proc Natl Acad Sci USA 76, 2321–2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A, Weber K (1980). Fimbrin, a new microfilament-associated protein present in microvilli and other cell surface structures. J Cell Biol 86, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruck S, Huber TB, Ingham RJ, Kim K, Niederstrasser H, Allen PM, Pawson T, Cooper JA, Shaw AS (2006). Identification of a novel inhibitory actin-capping protein binding motif in CD2-associated protein. J Biol Chem 281, 19196–19203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casaletto JB, Saotome I, Curto M, McClatchey AI (2011). Ezrin-mediated apical integrity is required for intestinal homeostasis. Proc Natl Acad Sci USA 108, 11924–11929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinowsky CR, Pinette JA, Meenderink LM, Lau KS, Tyska MJ (2020). Nonmuscle myosin-2 contractility-dependent actin turnover limits the length of epithelial microvilli. Mol Biol Cell 31, 2803–2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou AM, Sem KP, Wright GD, Sudhaharan T, Ahmed S (2014). Dynamin1 is a novel target for IRSp53 protein and works with mammalian enabled (Mena) protein and Eps8 to regulate filopodial dynamics. J Biol Chem 289, 24383–24396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croce A, Cassata G, Disanza A, Gagliani MC, Tacchetti C, Malabarba MG, Carlier MF, Scita G, Baumeister R, Di Fiore PP (2004). A novel actin barbed-end-capping activity in EPS-8 regulates apical morphogenesis in intestinal cells of Caenorhabditis elegans. Nat Cell Biol 6, 1173–1179. [DOI] [PubMed] [Google Scholar]
- Disanza A, Carlier MF, Stradal TE, Didry D, Frittoli E, Confalonieri S, Croce A, Wehland J, Di Fiore PP, Scita G (2004). Eps8 controls actin-based motility by capping the barbed ends of actin filaments. Nat Cell Biol 6, 1180–1188. [DOI] [PubMed] [Google Scholar]
- Disanza A, Mantoani S, Hertzog M, Gerboth S, Frittoli E, Steffen A, Berhoerster K, Kreienkamp HJ, Milanesi F, Di Fiore PP, et al. (2006). Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat Cell Biol 8, 1337–1347. [DOI] [PubMed] [Google Scholar]
- Erdos G, Dosztanyi Z (2020). Analyzing protein disorder with IUPred2A. Curr Protoc Bioinformatics 70, e99. [DOI] [PubMed] [Google Scholar]
- Gaeta IM, Meenderink LM, Postema MM, Cencer CS, Tyska MJ (2021a). Direct visualization of epithelial microvilli biogenesis. Curr Biol 31, 2561–2575.e2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaeta IM, Meenderink LM, Tyska MJ (2021b). A protocol for imaging microvilli biogenesis on the surface of cultured porcine kidney epithelial cell monolayers. STAR Protoc 2, 100998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger B, Baker RF (1950). Electron microscope investigation of the striated border of intestinal epithelium. Anat Rec 107, 423–441. [DOI] [PubMed] [Google Scholar]
- Hegan PS, Giral H, Levi M, Mooseker MS (2012). Myosin VI is required for maintenance of brush border structure, composition, and membrane trafficking functions in the intestinal epithelial cell. Cytoskeleton (Hoboken) 69, 235–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helander HF, Fandriks L (2014). Surface area of the digestive tract - revisited. Scand J Gastroenterol 49, 681–689. [DOI] [PubMed] [Google Scholar]
- Hernandez-Valladares M, Kim T, Kannan B, Tung A, Aguda AH, Larsson M, Cooper JA, Robinson RC (2010). Structural characterization of a capping protein interaction motif defines a family of actin filament regulators. Nat Struct Mol Biol 17, 497–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertzog M, Milanesi F, Hazelwood L, Disanza A, Liu H, Perlade E, Malabarba MG, Pasqualato S, Maiolica A, Confalonieri S, et al. (2010). Molecular basis for the dual function of Eps8 on actin dynamics: bundling and capping. PLoS Biol 8, e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe CL, Mooseker MS (1983). Characterization of the 110-kdalton actin–calmodulin-, and membrane-binding protein from microvilli of intestinal epithelial cells. J Cell Biol 97, 974–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Navarrete-Perea J, Cannon JR, Baltier K, Gebreab F, Gygi MP, Thornock A, Zarraga G, Tam S, et al. (2021). Dual proteome-scale networks reveal cell-specific remodeling of the human interactome. Cell 184, 3022–3040.e3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttlin EL, Bruckner RJ, Paulo JA, Cannon JR, Ting L, Baltier K, Colby G, Gebreab F, Gygi MP, Parzen H, et al. (2017). Architecture of the human interactome defines protein communities and disease networks. Nature 545, 505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Birendra KC, Zhu W, Motamedchaboki K, Doye V, Roux KJ (2014). Probing nuclear pore complex architecture with proximity-dependent biotinylation. Proc Natl Acad Sci USA 111, E2453–2461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Jensen SC, Noble KA, Kc B, Roux KH, Motamedchaboki K, Roux KJ (2016). An improved smaller biotin ligase for BioID proximity labeling. Mol Biol Cell 27, 1188–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Disanza A, Grati M, Andrade L, Lin H, Di Fiore PP, Scita G, Kachar B (2011). Regulation of stereocilia length by myosin XVa and whirlin depends on the actin-regulatory protein Eps8. Curr Biol 21, 167–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell P, Mekel M, Kozlov AG, Mooren OL, Lohman TM, Cooper JA (2020). Comparative analysis of CPI-motif regulation of biochemical functions of actin capping protein. Biochemistry 59, 1202–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Benesh AE, Mao S, Tabb DL, Tyska MJ (2011). Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol 300, G914–G926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Roy P, Perrin BJ (2017). Stereocilia morphogenesis and maintenance through regulation of actin stability. Semin Cell Dev Biol 65, 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenderink LM, Gaeta IM, Postema MM, Cencer CS, Chinowsky CR, Krystofiak ES, Millis BA, Tyska MJ (2019). Actin dynamics drive microvillar motility and clustering during brush border assembly. Dev Cell 50, 545–556.e544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meszaros B, Erdos G, Dosztanyi Z (2018). IUPred2A: context-dependent prediction of protein disorder as a function of redox state and protein binding. Nucleic Acids Res 46, W329–W337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millard TH, Dawson J, Machesky LM (2007). Characterisation of IRTKS, a novel IRSp53/MIM family actin regulator with distinct filament bundling properties. Journal of Cell Science 120, 1663–1672. [DOI] [PubMed] [Google Scholar]
- Mongiovi AM, Romano PR, Panni S, Mendoza M, Wong WT, Musacchio A, Cesareni G, Di Fiore PP (1999). A novel peptide-SH3 interaction. EMBO J 18, 5300–5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, Tilney LG (1975a). Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. J Cell Biol 67, 725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooseker MS, Tilney LG (1975b). Organization of an actin filament-membrane complex. Filament polarity and membrane attachment in the microvilli of intestinal epithelial cells. The Journal of Cell Biology 67, 725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales EA, Arnaiz C, Krystofiak ES, Zanic M, Tyska MJ (2022). Mitotic spindle positioning (MISP) is an actin bundler that selectively stabilizes the rootlets of epithelial microvilli. Cell Rep 39, 110692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi T, Yoshida H, Katori M, Migita T, Muramatsu Y, Miyake M, Ishikawa Y, Saiura A, Iemura SI, Natsume T, Seimiya H (2017). Tankyrase-binding protein TNKS1BP1 regulates actin cytoskeleton rearrangement and cancer cell invasion. Cancer Res 77, 2328–2338. [DOI] [PubMed] [Google Scholar]
- Ohta K, Higashi R, Sawaguchi A, Nakamura K (2012). Helical arrangement of filaments in microvillar actin bundles. J Struct Biol 177, 513–519. [DOI] [PubMed] [Google Scholar]
- Postema MM, Grega-Larson NE, Neininger AC, Tyska MJ (2018). IRTKS (BAIAP2L1) elongates epithelial microvilli using EPS8-dependent and independent mechanisms. Curr Biol 28, 2876–2888.e2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revenu C, Ubelmann F, Hurbain I, El-Marjou F, Dingli F, Loew D, Delacour D, Gilet J, Brot-Laroche E, Rivero F, et al. (2012). A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol Biol Cell 23, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Burke B, May DG (2018). BioID: A screen for protein–protein interactions. Curr Protoc Protein Sci 91, 19 23 11-19 23 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux KJ, Kim DI, Raida M, Burke B (2012). A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J Cell Biol 196, 801–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzadzinska AK, Schneider ME, Davies C, Riordan GP, Kachar B (2004). An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. J Cell Biol 164, 887–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez AD, Branon TC, Cote LE, Papagiannakis A, Liang X, Pickett MA, Shen K, Jacobs-Wagner C, Ting AY, Feldman JL (2021). Proximity labeling reveals non-centrosomal microtubule-organizing center components required for microtubule growth and localization. Curr Biol 31, 3586–3600.e3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Bulanova EA, Chaga OY, Vignjevic DM, Kojima S, Vasiliev JM, Borisy GG (2003). Mechanism of filopodia initiation by reorganization of a dendritic network. J Cell Biol 160, 409–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theillet FX, Kalmar L, Tompa P, Han KH, Selenko P, Dunker AK, Daughdrill GW, Uversky VN (2013). The alphabet of intrinsic disorder: I. Act like a Pro: on the abundance and roles of proline residues in intrinsically disordered proteins. Intrinsically Disord Proteins 1, e24360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Cardell RR (1970). Factors controlling the reassembly of the microvillous border of the small intestine of the salamander. J Cell Biol 47, 408–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tocchetti A, Soppo CB, Zani F, Bianchi F, Gagliani MC, Pozzi B, Rozman J, Elvert R, Ehrhardt N, Rathkolb B, et al. (2010). Loss of the actin remodeler Eps8 causes intestinal defects and improved metabolic status in mice. PLoS One 5, e9468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyska MJ, Mackey AT, Huang JD, Copeland NG, Jenkins NA, Mooseker MS (2005). Myosin-1a is critical for normal brush border structure and composition. Mol Biol Cell 16, 2443–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlik MT, Temple B, Bencharit S, Kimple AJ, Siderovski DP, Johnson GL (2005). Structural and evolutionary division of phosphotyrosine binding (PTB) domains. J Mol Biol 345, 1–20. [DOI] [PubMed] [Google Scholar]
- Wear MA, Yamashita A, Kim K, Maeda Y, Cooper JA (2003). How capping protein binds the barbed end of the actin filament. Curr Biol 13, 1531–1537. [DOI] [PubMed] [Google Scholar]
- Welling LW, Welling DJ (1975). Surface areas of brush border and lateral cell walls in the rabbit proximal nephron. Kidney Int 8, 343–348. [DOI] [PubMed] [Google Scholar]
- Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N (2004). A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J Biol Chem 279, 14929–14936. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Fukutomi T, Kimura T, Sakurai H, Hatano R, Yamamoto H, Mukaisho K, Hattori T, Sugihara H, Asano S (2016). Comprehensive proteome analysis of brush border membrane fraction of ileum of ezrin knockdown mice. Biomed Res 37, 127–139. [DOI] [PubMed] [Google Scholar]
- Zampini V, Ruttiger L, Johnson SL, Franz C, Furness DN, Waldhaus J, Xiong H, Hackney CM, Holley MC, Offenhauser N, et al. (2011). Eps8 regulates hair bundle length and functional maturation of mammalian auditory hair cells. PLoS Biol 9, e1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Li M, Zhao R, Li M, Chai Y, Zhu Z, Yang Y, Li W, Xie Z, Li X, et al. (2022). In situ structure of intestinal apical surface reveals nanobristles on microvilli. Proc Natl Acad Sci USA 119, e2122249119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel I, Naba A, Da Cunha MM, Del Maestro L, Formstecher E, Louvard D, Arpin M (2012). Ezrin regulates microvillus morphogenesis by promoting distinct activities of Eps8 proteins. Mol Biol Cell 23, 1080–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.