Abstract
Background and Purpose
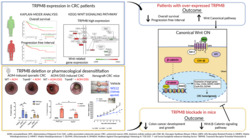
Transient receptor potential melastatin type‐8 (TRPM8) is a cold‐sensitive cation channel protein belonging to the TRP superfamily of ion channels. Here, we reveal the molecular mechanism of TRPM8 and its clinical relevance in colorectal cancer (CRC).
Experimental Approach
TRPM8 expression and its correlation with the survival rate of CRC patients was analysed. To identify the key pathways and genes related to TRPM8 high expression, Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were conducted in CRC patients. TRPM8 functional role was assessed by using Trpm8 −/− mice in models of sporadic and colitis‐associated colon cancer. TRPM8 pharmacological targeting by WS12 was evaluated in murine models of CRC.
Key Results
TRPM8 is overexpressed in colon primary tumours and in CD326+ tumour cell fraction. TRPM8 high expression was related to lower survival rate of CRC patients, Wnt–Frizzled signalling hyperactivation and adenomatous polyposis coli down‐regulation. In sporadic and colitis‐associated models of colon cancer, either absence or pharmacological desensitization of TRPM8 reduced tumour development via inhibition of the oncogenic Wnt/β‐catenin signalling. TRPM8 pharmacological blockade reduced tumour growth in CRC xenograft mice by reducing the transcription of Wnt signalling regulators and the activation of β‐catenin and its target oncogenes such as C‐Myc and Cyclin D1.
Conclusion and Implications
Human data provide valuable insights to propose TRPM8 as a prognostic marker with a negative predictive value for CRC patient survival. Animal experiments demonstrate TRPM8 involvement in colon cancer pathophysiology and its potential as a drug target for CRC.
Keywords: colon cancer, pharmacology, transient receptor potential channels, TRPM8, Wnt/β‐catenin
Abbreviations
- AOM
azoxymethane
- APC
adenomatous polyposis coli
- Axin 2
axis inhibition protein 2
- CAC
colitis‐associated colon cancer
- CRC
colorectal cancer
- DAPI
4′,6‐diamidino‐2‐phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- DSS
dextran sulfate sodium
- EpCAM
epithelial cell adhesion molecule
- FZD
Frizzled
- GAPDH
glyceraldehyde 3‐phosphate dehydrogenase
- GSA
gene set analysis
- GSEA
gene set enrichment analysis
- HEK
human embryonic kidney
- IHC
immunohistochemistry
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- LRP5
low‐density lipoprotein receptor‐related protein 5
- LRP6
low‐density lipoprotein receptor‐related protein 6
- MEICS
murine endoscopic index of colitis severity
- MEM
minimum essential media
- OLFM4
olfactomedin protein 4
- PFI
progression‐free interval
- Sox‐9
SRY‐box transcription factor 9
- TBS‐T
tris‐buffered saline‐Tween‐20
- TCGA
The Cancer Genome Atlas
- TRP
transient receptor potential
- TRPM8
transient receptor potential melastatin type‐8
- Wnt
wingless‐related integration site
What is already known
TRPM8 is a cold‐sensitive protein belonging to the TRP superfamily of ion channels.
TRPM8 is dysregulated in tumour tissues including prostate and breast cancer.
What does this study add
TRPM8 up‐regulation in CRC patients is correlated with a poor survival and Wnt/Frizzled signalling hyperactivation.
TRPM8 genetic or pharmacological blockade inhibits murine colon carcinogenesis by regulating Wnt/β‐catenin signalling pathway.
What is the clinical significance
TRPM8 may be a prognostic marker in CRC with negative predictive value for patient survival.
TRPM8 may be considered as targeted therapy for CRC treatment.
1. INTRODUCTION
Colorectal cancer (CRC) represents one of the leading causes of cancer‐related deaths in men and women with a significant health burden worldwide (Miller et al., 2022). The overall incidence and prevalence of CRC are rising due to the ageing of the population as well as to the increase in established risk factors such as sedentary lifestyles, smoking, obesity and chronic inflammatory disorders (e.g. inflammatory bowel diseases and primary sclerosing cholangitis) (Douaiher et al., 2017; Nunez et al., 2019). It is estimated that by the year 2035, the total number of deaths due to colon cancer will increase by 60% (Sawicki et al., 2021). Sporadic CRC develops mostly via a multistep process involving a sequence of genetic, morphological and histological alterations that accumulate over 10 to 15 years (Rawla et al., 2019). CRC development is mainly symptomless until the metastatic stage of the disease (McQuade et al., 2017). As the 5‐year survival rate for CRC patients diagnosed in the early stages is significantly higher than in the advanced stages (about 60% vs. 10%, respectively) (McQuade et al., 2017), there is an urgent and demanding need to made the population aware of prevention therapies as well as to identify new prognostic markers useful in early diagnosis. Furthermore, the diversity of human tumour cells, in both molecular heterogeneity and plasticity, represents a new challenge for current research and personalized precision medicine. Discovering new targets that are unique to CRC can be more therapeutically beneficial.
The transient receptor potential (TRP) cation channel, subfamily melastatin (M), member 8 (TRPM8), is a cold‐sensitive six‐pass transmembrane Ca2+ protein belonging to the TRP superfamily of ion channels, which is mainly detected in cold‐sensitive peripheral sensory neurons (Bautista et al., 2007; Moran, 2018). Although epithelial cells commonly express low amounts of TRPM8, its expression is much higher in tumour cells (Tsavaler et al., 2001). TRPM8 is strongly up‐regulated in several cancers such as prostate, breast, pancreas and skin, whereas it is dramatically reduced during metastasis in the androgen‐independent prostate cancer (Henshall et al., 2003; Tsavaler et al., 2001; Yee, 2016). Concerning the gastrointestinal tract, studies have shown that TRPM8 is expressed in CRC‐immortalized cells (Borrelli et al., 2014) and, more recently, this protein has been found to be up‐regulated in gastric cancer patients with metastasis as well as in CRC patients with liver metastasis (Liu et al., 2021; Xu et al., 2021). Nonetheless, the TRPM8 mechanism of action and, importantly, pathophysiological and clinical significance in colon carcinogenesis are still largely fragmented and undefined. Thus, our study aimed to mechanistically investigate the possible prognostic and pathophysiological role of TRPM8 in colon cancer.
We report here, for the first time, that high TRPM8 expression in colon cancer specimens predicts low survival in CRC patients and that TRPM8 aberrant expression is specifically detected in tumour cells isolated from bulk tumours of CRC patients. We also provide evidence that genetic deletion of Trpm8 protects mice from chemically induced sporadic colon cancer and colitis‐associated colon cancer (CAC), suggesting its key function in tumour initiation and growth. Finally, we show that pharmacological desensitization of TRPM8 reduces tumour progression in chemically induced colon cancer as well as in CRC‐xenografted mice, with a mechanism involving the Wnt/β‐catenin pathway.
2. METHODS
2.1. Patient samples
Frozen tissue biopsies of 34 primary adenocarcinomas and 34 normal tissues were obtained from CRC patients following the tumour resection at the Department of Public Health (University of Naples Federico II) and were processed for mRNA and protein extraction. Healthy specimens were collected at a distance of at least 10–15 cm from the tumour lesion of the same patients. Patients with a well‐established diagnosis of CRC were included (see Table S1 for tumour node metastasis classification). Exclusion criteria were age less than 35 or more than 85 years. Human studies were approved by the Ethics Committee of the University of Federico II (Naples, Italy; Protocol Number 350/19). All patients gave their written and informed consent.
2.2. Isolation of primary tumour cells from CRC biopsies and flow cytometric analysis
Fresh biopsy tissues were dissociated using the human tumour dissociation kit (RRID:SCR_020276, Cat. 130‐095‐929, Miltenyi Biotec, Auburn, CA, USA). Briefly, the GentleMACS dissociator (RRID:SCR_020267), which combines enzymatic disaggregation with a mechanical disruption process, was used to obtain a single‐cell suspension from the bulk tumour. Thus, primary tumour cells were isolated from single‐cell suspensions by using a tumour cell isolation kit (130‐108‐339, Miltenyi Biotec) in conformity with the manufacturer's protocols. Single‐cell suspensions were analysed by flow cytometry before and after tumour cell isolation. In brief, primary cells were pelleted, washed and resuspended in phosphate‐buffered saline (PBS) containing 0.5% bovine serum albumin (BSA) and 2 mM of ethylenediamine tetraacetic acid (EDTA). For surface staining, cells (5 × 105) were incubated for 10 min at 4°C with the following fluorochrome‐labelled anti‐human antibodies: CD326 (EpCAM)‐APC (clone REA764) (RRID:AB_871664), CD45‐PerCP‐Vio 700 (clone REA747) (RRID:AB_2889583) and CD31‐PE (clone REA1028) (RRID:AB_2660561) (Miltenyi Biotec). Cells were gated for positivity to CD326 and negativity to CD31 and CD45. The flow cytometry analysis was carried out with BriCyte E6 (Mindray, P.R. China), and data were analysed by FlowJo (Tree Star, Inc.). Cell suspension was also proceeded for the mRNA extraction by using miRNeasy Mini Kit (74104, Qiagen, Hilden, Germany), according to the manufacturer's protocol.
2.3. Data collection and bioinformatics analyses
The Cancer Genome Atlas (TCGA) (https://www.cancer.gov/tcga, RRID:SCR_003193) repository collected high‐throughput and clinical data of many tumours and normal tissues from many organs. To evaluate differences in prognosis between high and low TRPM8 expressions, survival curves were estimated by the Kaplan–Meier estimator and compared using log‐rank tests. To visualize multiple genomic alteration events in a single overview together with some clinical parameters, cellular localization and immunohistochemistry (IHC), from Human Atlas tumour and normal tissues, we produced an enriched oncoprint heatmap (additional information is reported in the supporting information).
2.4. Animals
Mice were housed at the animal house of the Department of Pharmacy, University of Naples Federico II (Italy), under controlled temperature (23 ± 2°C), humidity (60%) and with a 12‐h light and 12‐h dark cycle. For experimental models of CRC induced by azoxymethane (AOM) and/or AOM/dextran sulfate sodium (DSS), male wild‐type (WT; C57BL/6J mice, Stock No. 000664, RRID:IMSR_JAX:000664) and Trpm8 −/− mice (B6.129P2‐Trpm8tm1Jul/J, Stock No. 008198, RRID: IMSR_JAX:008198) were obtained from the Jackson Laboratories, fed ad libitum with standard food (Mucedola srl, Settimo Milanese, Italy) and housed in standard polycarbonate cages. Trpm8 −/− mice have been previously described (Bautista et al., 2007) and they develop normally without any spontaneous disease emerging under the breeding conditions. Female BALB/c nude mice, used in the xenograft model of CRC, were obtained from Charles River (Sant'Angelo Lodigiano, Italy), fed with sterile mouse food and maintained in pathogen‐free conditions. Six‐ to 8‐week‐old mice were used for all experiments unless otherwise stated. Mice were randomly allocated to different experimental groups, and outcome assessments were performed single blind. All animals were anaesthetized with enflurane inhalation before being humanely killed by carbon dioxide. Death was further assessed by confirmation of rigour mortis. All efforts were made to minimize the number of animals used and their suffering. Experimental procedures and protocols were in compliance with National (Direttiva 2010/63/UE) laws and policies and approved by the Italian Ministry of Health (Protocol Numbers 1101/2015‐PR and 481/2020‐PR). Animal studies are reported in compliance with the ARRIVE guidelines (Percie du Sert et al., 2020) and with the recommendations made by the British Journal of Pharmacology (Lilley et al., 2020).
2.5. Materials
WS12 was purchased from Tocris (Bio‐Techne, Milan, Italy). All the reagents for in vitro cell cultures and ex vivo analysis were provided by Merck Sigma (Merck Life Science, Milan, Italy), Aurogene (Rome, Italy), Thermo Fisher Scientific (Monza, Italy), Corning (Milan, Italy) and Bio‐Rad (Milan, Italy). For in vivo studies, WS12 was dissolved in ethanol/Tween‐20/saline (2:1:7), 2 ml·kg−1, and for in vitro studies in 0.1% ethanol. Vehicle had no effect on the responses under the study. WS12 dose was selected based on previous studies reporting its specificity for TRPM8 in vivo (Liu et al., 2013).
2.6. Cell culture
Cells derived from human colon cancer (i.e., HCT116, RRID:CVCL_0291) were purchased from ATCC (LGC Standards, Milan, Italy) and used between 18 and 27 passages. Immortalized healthy human colonic epithelial cells (HCEC), derived from human colon biopsies, were kindly gifted from Fondazione Callerio Onlus (Trieste, Italy). CRC and HCEC cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Euroclone) supplemented with 10% (v/v) fetal bovine serum (FBS), 1 mmol·L−1 l‐glutamine, 1 mmol·L−1 sodium pyruvate, 0.1 mmol·L−1 non‐essential amino acids and 100 U·ml−1 antibiotics (penicillin and streptomycin) at 37°C in 5% CO2. Human embryonic kidney 293 (HEK‐293) cells were grown on 100‐mm‐diameter Petri dishes as monolayers in minimum essential media (MEM; Life Technology, Milan, Italy) supplemented with non‐essential amino acids, 10% FBS and 2 mM l‐glutamine and were maintained under 5% CO2 at 37°C. Cell viability was evaluated by trypan blue exclusion, and cell lines were confirmed to be Mycoplasma free (by using Mycoplasma PCR Detection Kit, Cat. G238, ABM). The medium was changed every 48 h in conformity with the manufacturer's protocols.
2.7. Induction of tumours
-
a
Azoxymethane (AOM) model of sporadic CRC
AOM (40 mg·kg−1 in total) (Cat. A5486, Sigma‐Aldrich, Italy) was administered intraperitoneally in WT and Trpm8 −/− mice at the single dose of 10 mg·kg−1, once per week for 4 weeks. WS12 (10 mg·kg−1) or vehicle was given intraperitoneally every 3 days, starting 1 week before the first administration of AOM. All animals were anaesthetized by enflurane inhalation before being killed by asphyxiation with CO2 12 weeks (84 days) after the first injection of AOM (Pagano et al., 2017). Death was further assessed by confirmation of rigour mortis. Tumour count was determined at Day 91 microscopically in longitudinally cut specimens.
-
b
Azoxymethane (AOM)/dextran sulfate sodium (DSS) model of CAC
WT and Trpm8 −/− mice were injected with AOM (12.5 mg·kg−1, i.p). Colitis was induced by two cycles of 2.5% DSS (molecular weight 36–50 kDa; 02160110‐CF, MP Biomedicals, Canada) in drinking water for 5 days, followed by a 16‐day tap water period and a final cycle of 2% DSS followed by a 5‐day tap water period (Pagano et al., 2021). The disease activity index (DAI) score was assessed considering stool consistency and presence of blood. All animals were anaesthetized as described above at day 63 and the tumour count was determined microscopically in longitudinally cut specimens. Inflammation grade was evaluated by measuring colon weight/colon length ratio and spleen weight.
-
c
Xenograft model of CRC
HCT116 cells (2 × 106 cells per mouse) were subcutaneously injected into the right flanks of each athymic mouse in a total volume of 200 μl (50% cell suspension in PBS and 50% Matrigel™) (Cat. 356237, Corning). Tumours were allowed to grow to 200–300 mm3 in volume (designated Day 1), and mice were randomly assigned to the treatment groups. Then mice were intraperitoneally injected every day with WS12 (10 mg·kg−1) or vehicle. The tumour volume was measured every day by digital callipers and calculated using the formula volume = π/6 × length × width2. Mice were humanely killed as described before when the human endpoint tumour volume was 1500 mm3. Thus, tumours were excised, photographed, weighed and processed as due.
2.8. Exclusion criteria
For in vivo studies, the starting group sizes were identical but changed finally by losses. In the AOM/DSS model, all groups were initially designed to contain the equal number of mice, but one mouse of the Trpm8 −/− vehicle group died from fighting in the cage. In the AOM/WS12 model, all groups were initially designed to contain the equal number of mice, but three mice of the Trpm8 −/− AOM + WS12 group died from fighting in the cage. Dead animals, whose data were not recorded, were excluded.
2.9. Endoscopy for evaluating colon pathology
High‐resolution endoscopy in live mice was assessed. Briefly, mice were anaesthetized by subcutaneous injection of alfaxolone (5 mg·kg−1) and xylazine (5 mg·kg−1) and examined with a mini‐endoscope (Mainz COLOVIEW® System, Karl Storz). High‐quality endoscopy video and photos were used for monitoring and grading tumours and inflammation. Scoring of colitis activity was expressed as a modified murine endoscopic index of colitis severity (MEICS) based on the observed signs of inflammation. The MEICS consisted of the following four parameters: (i) thickening of the colon, (ii) changes of the vascular pattern, (iii) fibrin visible and (iv) stool consistency (Becker et al., 2005). Tumour development was scored by the total number as well as the size of tumours. Tumours were counted to obtain the overall number. Tumour sizes were graded as follows: Grade 1 (small but detectable), Grade 2 (tumour covering up to one eighth of the colonic circumference), Grade 3 (tumour covering up to one fourth of the colonic circumference), Grade 4 (tumour covering up to half of the colonic circumference) and Grade 5 (tumour covering more than half of the colonic circumference). Endoscopic grading was performed for each parameter (scores 0–3), leading to a cumulative score between 0 and 15 (Becker et al., 2005).
2.10. Confocal immunofluorescence analysis
Slides of colon tissue (5 μm) were deparaffinized with xylene and rehydrated with decreasing gradient alcohol. Antigen retrieval was carried out by pressure‐cooking slides for 3 min in 0.01 M citrate buffer (pH 6.0). To avoid non‐specific interactions of antibodies, the slides were treated for 2 h in 5% BSA in PBS. Immunostaining was performed by incubation overnight with anti‐β‐catenin (1:100, Alexa Fluor® 488, Cat. 562505, BD Pharmingen™) at 4°C. The slides were mounted on microscope slides using Mowiol + 4′,6‐diamidino‐2‐phenylindole (DAPI) for nuclear staining. The images were acquired at room temperature and detected under 63× magnification (numerical aperture of the objective lens: 1,4), by using a laser‐scanning confocal microscope with AiryScan2 module (Zeiss, LSM 900 associated) (Vanacore et al., 2018). Three fields for each colon were analysed by Zeiss Zen blue edition software.
2.11. Western blot
The Immuno‐related procedures used comply with the recommendations made by the British Journal of Pharmacology (Alexander et al., 2018). Xenograft tumours were homogenized using FastPrep‐24 homogenizer (Cat. 116004500, MP Biomedicals); human biopsies were homogenized using the program Protein_1 on a gentleMACS tissue Dissociator (Miltenyi Biotec). Total cellular proteins were extracted by solubilizing the homogenized human and murine tumours in lysis buffer containing 50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1% NP‐40, 1 mM EDTA, 0.25% sodium deoxycholate, 10 mM NaF, 10 μM Na3VO4, 1 mM phenylmethanesulfonyl fluoride (PMSF) and protease inhibitor cocktail (10 g·mg−1 aprotinin, 10 g·ml−1 pepstatin and 10 g·ml−1 leupeptin); and 50 mM β‐glycerophosphate, 100 μM sodium orthovanadate, 2 mM MgCl2, 1 mM ethylene glycol tetraacetic acid (EGTA), 1 mM DTT, 1 mM PMSF, 1 mM leupeptin and 0.5 mM aprotinin, respectively. Protein concentration was determined with the DC Protein Assay Kit (Cat. 5000112, Bio‐Rad): 40 μg of the xenograft extract and 50 μg of the human biopsy extracts were fractionated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE, Bio‐Rad) and transferred onto polyvinylidene difluoride (PVDF) membranes. After incubation with 5% BSA (Cat. A9418, Sigma‐Aldrich) in tris‐buffered saline‐Tween‐20 (TBS‐T) (10 mM Tris, pH 8.0, 150 mM NaCl, 0.5% [v/v] Tween‐20), membranes were incubated overnight with the following fresh primary antibodies (all from Cell Signaling Technology, except where indicated): anti‐β‐catenin (Cat. 9582, 1:1000), anti‐α tubulin (Cat. 2125, 1:1000), anti‐TRPM8 (Thermo Fisher Scientific, Cat. MA5‐35474) and anti‐β‐actin (Santa Cruz Biotechnology Inc., Cat. SC‐47778). Therefore, secondary fresh antibodies anti‐rabbit IgG (Cat. 7074, 1:2000) and anti‐mouse IgG (Bioss, Woburn, MA, USA, 0296G‐HRP), horseradish peroxidase (HRP) linked, were added and detected. Western blot detection was performed with enhanced chemiluminescence using Chemidoc XRS (RRID:SCR_019690, Bio‐Rad) and analysed using Image Lab Version 6.10.7 (12012931, Bio‐Rad).
2.12. RNA extraction and gene expression profiling
Xenograft tumour and CRC biopsies were homogenized using FastPrep‐24 homogenizer (MP Biomedicals); single‐cell suspension of primary tumours was obtained as described above. RNA was extracted using RNeasy Mini Kit (Qiagen), according to the manufacturer's protocol. The quantification and quality analysis of RNA was performed on NanoDrop OneC spectrophotometer (Cat. 701‐058112, Thermo Fisher Scientific). cDNA was synthesized by reverse transcription of 1 μg of RNA using the High‐Capacity cDNA Reverse Transcription Kit (Cat. 43‐688‐14, Applied Biosystems, Italy), and reverse transcription polymerase chain reaction (RT–PCR) was completed using Fast SYBR Green Master Mix (Cat. 43‐856‐12, Applied Biosystems) as the reporter dye, glyceraldehyde 3‐phosphate dehydrogenase (GAPDH) as the reference gene and gene‐specific primers completely listed in Table S2. The relative abundance of the genes was expressed by using the 2−ΔΔCt formula.
2.13. RNA sequencing
High‐quality RNA was extracted from mouse colon, using the PureLink™ RNA Mini Kit (Cat. 12183020, Thermo Fisher Scientific, Waltham, MA, USA), according to the manufacturer's instructions. RNA quality and quantity were assessed with the Qubit® RNA HS Assay Kit (Cat. Q32852, Invitrogen, Carlsbad, CA, USA) and a Qubit 3.0 fluorometer (Cat. Q33216, Invitrogen) followed by agarose gel electrophoresis. Libraries were generated using the Ion AmpliSeq™ Transcriptome Mouse Gene Expression Core Panel and Chef‐Ready Kit (comprehensive evaluation of AmpliSeq transcriptome, a whole transcriptome RNA sequencing methodology) (Cat. A36554, Thermo Fisher Scientific). Briefly, 10 ng of RNA was reverse transcribed with SuperScript™ Vilo™ cDNA Synthesis Kit (11754050, Thermo Fisher Scientific) before library preparation on the Ion Chef™ instrument (Cat. A30070, Thermo Fisher Scientific). The resulting cDNA was amplified to prepare barcoded libraries using the Ion Code™ PCR Plate and the Ion AmpliSeq™ Transcriptome Mouse Gene Expression Core Panel (Thermo Fisher Scientific), Chef‐Ready Kit, according to the manufacturer's instructions. Barcoded libraries were combined to a final concentration of 100 pM and used to prepare Template‐Positive Ion Sphere™ (Cat. 4468656, Thermo Fisher Scientific) Particles to load on Ion 540™ Chips, using the Ion 540™ Kit‐Chef (Cat. A30011, Thermo Fisher Scientific). Sequencing was performed on an Ion S5™ Sequencer with Torrent Suite™ Software Version 6 (Thermo Fisher Scientific). The analyses were performed with a range of fold < −2 and > +2 and a P value < 0.05, using Transcriptome Analysis Console Software (Version 4.0.1), certified for AmpliSeq analysis (Thermo Fisher Scientific).
2.14. Stable clone generation and TRPM8 intracellular calcium assay
Selection of stable TRPM8 expression in HEK‐293 cell clones was achieved by incubating the cells in the presence of 1000 μg·ml−1 of geneticin (G418; Cat. 10131035, Thermo Fisher, Milan, Italy) for 1 month (De Petrocellis et al., 2007). Quantitative real‐time PCR was also periodically carried out to measure rat TRPM8 expression following published procedures (Iannotti et al., 2014). Briefly, to determine the effect of WS12, HEK‐TRPM8 cells were loaded with the methyl ester Fluo4‐AM (Cat. F14201, Thermo Fisher) 4 μM in dimethyl sulfoxide (DMSO) containing 0.02% Pluronic F‐127 (Cat. P3000MP) for 1 h in the dark at room temperature in MEM not containing FBS. After the incubation with Fluo4‐AM, cells were washed twice in Tyrode's buffer (145 mM NaCl, 2.5 mM KCl, 1.5 mM CaCl2, 1.2 mM MgCl2, 10 mM d‐glucose and 10 mM HEPES, pH 7.4), resuspended in Tyrode's buffer and transferred into quartz cuvettes of the spectrofluorimeter (Perkin‐Elmer LS50B; PerkinElmer Life and Analytical Sciences, Waltham, MA, USA) under continuous stirring. All media and reagents were purchased from Life Technology. Potency was expressed as the concentration of test substances exerting a half‐maximal agonist effect (i.e., half‐maximal increases in [Ca2+]i)(EC50) calculated by using GraphPad Prism. The efficacy of the agonists was determined by normalizing effects relative to the maximum Ca2+ influx on [Ca2+]i observed with 4 μM ionomycin (Cayman, USA). Non‐transfected HEK‐293 cells were used as a control. Measurement of [Ca2+]i was performed at 22°C with a Fluorescence Peltier System (PTP‐1, Perkin‐Elmer). To test the effect of WS12 on TRPM8 desensitization, cells were stimulated with a triple stimulation of WS12 (0.2 μM), with stimulus repeated every 5 min. Change in the intracellular calcium concentration [Ca2+]i was determined in the presence of crescent concentrations of WS12 by measuring cell fluorescence at 25°C (λex = 488 nm, λem = 516 nm). Curve fitting (sigmoidal concentration–response variable slope) and parameter estimation were performed with GraphPad Prism® 8 (GraphPad Software Inc., San Diego, CA, USA).
2.15. Data and statistical analysis
The data and statistical analysis comply with the recommendations of the British Journal of Pharmacology on experimental design and analysis in pharmacology (Curtis et al., 2018). The study was designed to generate groups of equal size, using randomization and blinded analysis. All data are expressed as mean ± SEM, and statistical analysis was performed using GraphPad Prism 8 software. Outliers (if any) were identified by ROUT test. As all data were normally distributed (normality was tested using the Anderson–Darling method), statistical analysis was performed using Student's paired and unpaired t test (for comparing a single treatment mean with a control mean) and Student's multiple t test, one‐way analysis of variance (ANOVA) followed by Dunnett's multiple comparisons test or two‐way ANOVA followed by Tukey–Kramer's, Sidak's and Tukey's multiple comparisons test (for analysis of multiple treatment means). Kaplan–Meier survival analysis was evaluated using the Mantel–Cox test. All flow cytometry data were analysed using FlowJo Versions 9–10 (FlowJo LLC). P value < 0.05 was considered to be significant. G Power was used for sample size calculation. In experimental models of cancer, six to nine mice were randomly assigned to each experimental group; where not indicated, ex vivo analysis was performed on five mice of each experimental group. Post hoc tests were run only when F achieved P < 0.05 and there was no variance inhomogeneity. All measurements were undertaken only for n ≥ 5.
2.16. Nomenclature of targets and ligands
Key protein targets and ligands in this article are hyperlinked to corresponding entries in http://www.guidetopharmacology.org and are permanently archived in the Concise Guide to PHARMACOLOGY 2021/22 (Alexander, Christopoulos et al., 2021; Alexander, Fabbro et al., 2021; Alexander, Mathie et al., 2021).
3. RESULTS
3.1. High expression of TRPM8 predicts low survival in patients with CRC
In order to understand the impact of TRPM8 in CRC, we investigated the expression profile of 283 TCGA samples. Although TRPM8 expression is widely ranged (Tukey's five‐number summary: 0, 0.2, 0.86, 1.65 and 8.75), the expression of the 3rd (upper) quartile of the gene (1.65) is below the expression of the 1st (lower) quartile (3.7) of all the genes, resulting in a very low expression of Trpm8. To verify if such variation in TRPM8 expression could affect CRC prognosis, we investigated the disease‐specific survival and progression‐free interval (PFI) of patients by the log‐rank test (Figure 1a,b). Interestingly, by dividing samples according to the tertile of the TRPM8 expression, the results show a difference in survival of CRC patients. Specifically, the patients belonging to the tertile with a low or a complete absence of TRPM8 show a significant increase in disease‐specific survival and PFI compared with the patients with higher TRPM8 expression (Figure 1a,b). This result paired with the overall low expression of the gene, suggesting a mechanism present/absent linked to the TRPM8 gene.
FIGURE 1.
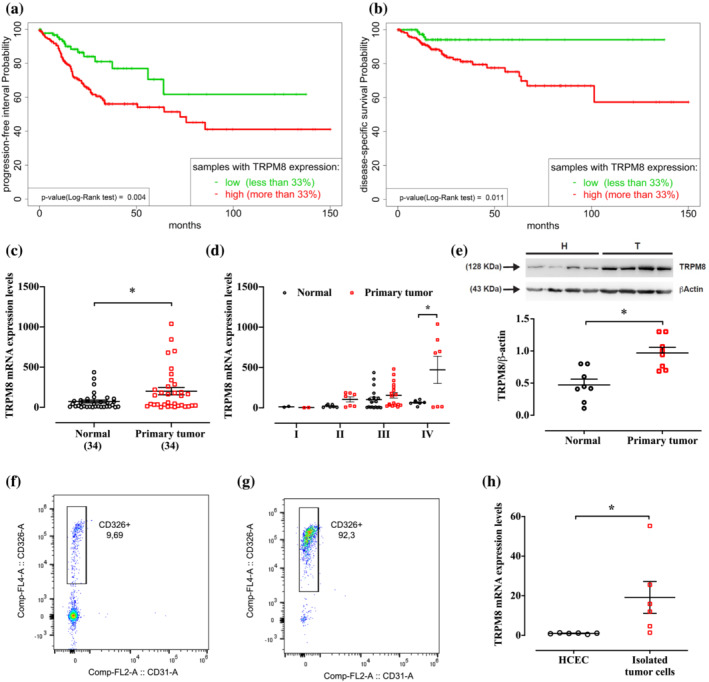
Transient receptor potential melastatin type‐8 (TRPM8) channel expression in patients with colorectal cancer (CRC). (a, b) Kaplan–Meier survival analysis of CRC patients with low TRPM8 expression (green) and high TRPM8 expression (red) was used to analyse the (a) progression‐free survival probability and (b) disease‐specific survival probability. P values were determined using the log‐rank test, and the exact values are reported at the left bottom of the figures. (c, d) mRNA expression of TRPM8 was analysed by reverse transcription quantitative polymerase chain reaction (RT–qPCR) and calculated by using the 2−ΔΔCt formula (n = 34 different biological samples) in normal tissues (collected at least 10 cm from the tumour lesion) and primary tumours removed from patients with CRC diagnosis. Error bars represent ±SEM. P values were determined using paired t test. *P < 0.05 versus normal. (e) Representative immunoblots of TRPM8 protein levels in human CRC specimens (primary tumours) and the surrounding nontumorous tissues (normal). Lower panel shows densitometric analysis of western blot analysis. Results show mean ± SEM of eight different biological samples. P value were determined using paired t tests. *P < 0.05 versus normal. (f, g) Representative flow cytometric analysis showing the enrichment of CD326+ human tumour cells before (f) and after (g) cells purification from primary specimens. CD31 was used to exclude the infiltration of leukocytes and endothelial cells. (h) mRNA expression of TRPM8 was analysed by RT–qPCR and calculated by using the 2−ΔΔCt formula (n = 6 independent experiments) in immortalized human colonic epithelial cells (HCEC) and primary tumour cells isolated from the bulk tumour by magnetic separation. Error bars represent ±SEM. P values were determined using unpaired t tests. *P < 0.05 versus HCEC.
3.2. TRPM8 is significantly up‐regulated in human CRC biopsies
In order to unravel the role of TRPM8 in human CRC, we additionally measured the expression profile of TRPM8 in human CRC specimens as well as in the surrounding normal colon tissues. RT–PCR analysis showed a significant up‐regulation of TRPM8 in tumour specimens compared with normal tissues (n = 34 CRC patients) (Figure 1c). A sub‐analysis carried out according to the tumour node metastasis staging system (i.e., pT1, pT2, pT3 and pT4; see Table S1) showed that TRPM8 expression was significantly up‐regulated only in stage pT4, with no significant changes in pT1–pT2–pT3 stages (Figure 1d). Immunoblot analysis showed that TRPM8 was poorly expressed in normal colonic tissue, whereas it was significantly overexpressed in the adjacent adenocarcinoma lesions collected from CRC patients (Figure 1e).
To clarify whether the TRPM8 overexpression in the bulk tumour was linked to CRC cells rather than to cells of non‐tumour origin (e.g. endothelial cells, fibroblasts and lymphocyte subpopulation) contained in the tumour mass, we dissociated human CRC biopsies so as to obtain a single‐cell suspension containing about 10% of CD326+ tumour cells (Figure 1f). CD326 (epithelial cell adhesion molecule [EpCAM]) is a pleiotropic molecule described as a dominant surface antigen on human colon carcinoma (Patriarca et al., 2012). As shown in Figure 1g, the single‐cell suspension was then strongly enriched (about 92%) in tumour CD326+ cells by using the magnetic separation of non‐tumour cells (Figure 1f,g). Comparison of sorted CD326+ tumour cells and healthy HCEC revealed that TRPM8 was up‐regulated in isolated primary CRC cells (Figure 1h).
Overall, these data highlight TRPM8 overexpression in human CRC tissues, and its aberrant expression is selectively associated with the tumoral epithelial compartment.
3.3. Trpm8 deletion reduces sporadic colon cancer and CAC development in mice
To find out if Trpm8 deletion influenced experimental tumorigenesis, we used a genetic approach in models of sporadic (induced by AOM) and colitis‐associated cancer (as reported in the schematic illustrations in Figures 2a and 3a).
FIGURE 2.
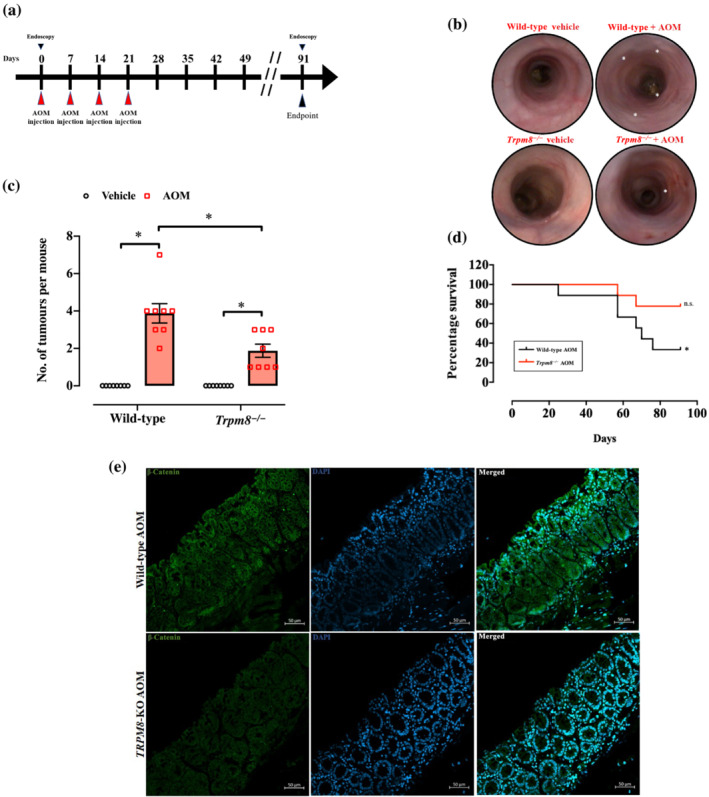
Loss of Trpm8 reduces azoxymethane (AOM)‐induced sporadic colon tumours. (a) Schematic representation of AOM protocol and timeline. Mice were injected with 10 mg·kg−1 of AOM at the indicated time. (b) Representative colonoscopy images of wild‐type (WT) and Trpm8 −/− mice carried out on the day of killing. All mice were subjected to endoscopy (n = 8). (c) Number of tumours per mouse in WT and Trpm8 −/− mouse colon treated with vehicle (black) or AOM (red) (n = 8 mice). Error bars represent ±SEM. P value was determined using two‐way ANOVA followed by Sidak's multiple comparisons test. *P < 0.05 Trpm8 −/− AOM versus vehicle, *P < 0.05 Trpm8 −/− AOM versus WT AOM and *P < 0.05 WT AOM versus vehicle. (d) Kaplan–Meier survival analysis of WT and Trpm8 −/− mice treated with AOM. P values were determined using the Mantel–Cox test. n.s. = non‐significant and *P < 0.05 versus Day 0. (e) Representative confocal images of β‐catenin (green) and 4′,6‐diamidino‐2‐phenylindole (DAPI) (blue) immunostaining in WT and Trpm8 −/− tumours collected at Day 91. Scale bars = 50 μm. Analyses were carried out from three mice for each experimental group.
FIGURE 3.
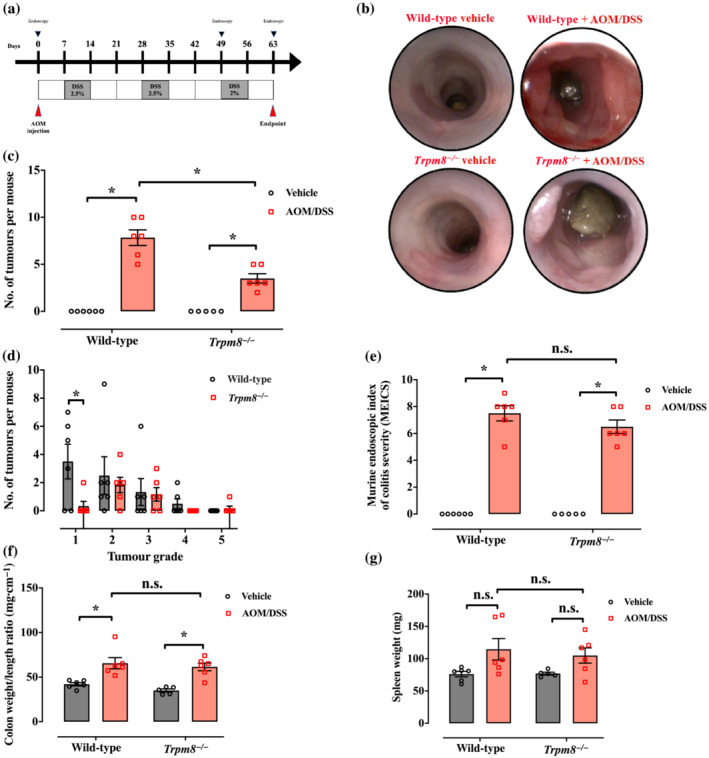
Loss of Trpm8 reduces azoxymethane (AOM)/dextran sodium sulfate (DSS)‐induced colitis‐associated colon cancer. (a) Schematic representation of AOM/DSS protocol and timeline. Mice were injected intraperitoneally with 12.5 mg·kg−1 AOM on the first day of the experiment. DSS was administered in drinking water at the indicated doses at the indicated time. (b) Representative colonoscopy images taken at the end of the carcinogenic protocol (Day 63). All mice were subjected to endoscopy (n = 6). (c) Number of tumours per mouse counted microscopically in wild‐type and Trpm8 −/− mice, treated with vehicle (black) or AOM/DSS (red); n = 6 mice for wild‐type vehicle and wild‐type and Trpm8 −/− AOM; n = 5 mice for Trpm8 −/− vehicle. Error bars represent ±SEM. P values were determined using two‐way ANOVA followed by Sidak's multiple comparisons test. *P < 0.05 Trpm8 −/− AOM/DSS versus vehicle, *P < 0.05 Trpm8 −/− AOM/DSS versus wild‐type AOM/DSS and *P < 0.05 wild‐type AOM/DSS versus vehicle. (d) Tumour grade of wild‐type (black) and Trpm8 −/− (red) mouse tumours, evaluated by colonoscopy (tumours counted in n = 6 mice). Error bars represent ±SEM. P value was determined using two‐way ANOVA followed by Sidak's multiple comparisons test. *P < 0.05 Grade 1 Trpm8 −/− versus wild‐type mice. (e) Analysis of the inflammatory parameters by the murine endoscopic index of colitis severity (MEICS) in wild‐type and Trpm8 −/− mice treated with vehicle (black) or AOM/DSS (red); n = 6 mice for wild‐type vehicle and wild‐type and Trpm8 −/− AOM; n = 5 mice for Trpm8 −/− vehicle. Error bars represent ±SEM. P values were determined using two‐way ANOVA followed by Sidak's multiple comparisons test. n.s. = non‐significant Trpm8 −/− AOM/DSS versus wild‐type AOM/DSS and *P < 0.05 wild‐type and Trpm8 −/− AOM/DSS versus vehicle. (f, g) Analysis of the macroscopic inflammatory parameters colon weight/colon length ratio (f) and spleen weight (g) in wild‐type and Trpm8 −/− mice treated with vehicle (black) or AOM/DSS (red); n = 6 mice for wild‐type vehicle and wild‐type and Trpm8 −/− AOM; n = 5 mice for Trpm8 −/− vehicle. Error bars represent ±SEM. P values were determined using two‐way ANOVA followed by Sidak's multiple comparisons test. n.s. = non‐significant and *P < 0.05 (f) wild‐type and Trpm8 −/− AOM/DSS versus vehicle.
As shown in Figure 2b,c, the endoscopy was carried out on Day 91, then mice were killed, and their colons were analysed for tumour numbers. Intestinal mucosa from mice under control conditions (both WT and Trpm8 −/− mice) did not show any phenotypical difference. The endoscopic evaluation and the count of tumour numbers showed decreased tumour frequency in AOM‐treated Trpm8 −/− mice compared with WT mice (Figure 2b). Neither WT nor Trpm8 −/− mice that underwent control treatment developed tumours (Figure 2c). Instead, after AOM administration, Trpm8 −/− mice presented a significant lower number of colonic tumours (Figure 2c) compared with WT mice. Additionally, Trpm8 −/− mice did not lose body weight after AOM treatment (Figure S1A). Of relevance, the survival rate was significantly reduced in WT, but not in Trpm8 −/− , mice Figure 2d).
As β‐catenin activation plays a critical role in colon tumorigenesis (Arnold et al., 2020), we qualitatively observed its expression by confocal immunofluorescence in the AOM‐treated mice. As shown in Figure 2e, a clearly higher β‐catenin localization was detected in WT tumours compared with Trpm8 −/− tumours, indicating that the Wnt/β‐catenin pathway could be clearly linked to TRPM8 function in colon carcinogenesis.
In the CAC model of colon carcinogenesis, we compared WT and Trpm8 −/− mice subjected to the AOM/DSS protocol (Figure 3a). After Cycle 1 of DSS, Trpm8 −/− mice lost a greater fraction of body weight compared with WT mice (Figure S1B). The hyper‐susceptibility of Trpm8 −/− mice to DSS‐induced intestinal inflammation has been previously documented (de Jong et al., 2015).
To explore CAC progression, the first endoscopic evaluation was assessed before the last cycle of DSS (Day 49). Endoscopic evaluation showed a reduced tumour development in Trpm8 −/− mice, with a trend towards a reduction in a tumour multiplicity (Figure S1C,D). At the end of the treatment (Day 63), in AOM/DSS‐treated animals, Trpm8 −/− mice had significantly lower total tumour numbers than WT mice (Figure 3b,c). Notably, AOM/DSS‐treated Trpm8 −/− mice displayed a significant lower frequency of Grade 1 tumours compared with AOM/DSS‐treated WT mice (Figure 3d), suggesting a possible function of Trpm8 in tumour initiation rather than progression. Taken together, these results suggest that Trpm8 deletion functionally reduces colon carcinogenesis triggered by inflammation. However, Trpm8 −/− mice did not show significant differences in all of the investigated inflammatory parameters (i.e., MEICS, colon weight/colon length ratio and spleen weight) compared with WT mice (Figure 3e–g).
Altogether, these data were consistent with a decisive role of TRPM8 in colon carcinogenesis in both sporadic colon cancer and CAC.
3.4. TRPM8 expression affects the Wnt network in colon cancer
As we found differences in the expression of β‐catenin (a key indicator of hyperactive Wnt signalling, Nusse & Clevers, 2017) between AOM‐treated WT and Trpm8 −/− mice (Figure 2e), and considering that such signalling crucially drives colonic regeneration and proliferation, we looked up Wnt pathway overrepresentation in the TCGA database of 268 CRC patients. Interestingly, both gene set analysis (GSA) (by hypergeometric test) and gene set enrichment analysis (GSEA) showed Kyoto Encyclopedia of Genes and Genomes (KEGG) Wnt signalling pathway gene set overrepresentation.
In the canonical Wnt pathway, Wnt initiates the signal transduction through two types of cell‐surface receptors: the low‐density lipoprotein receptor‐related proteins 5 and 6 (LRP5/LRP6) and the Frizzled (FZD) family of serpentine proteins (Clevers & Nusse, 2012; MacDonald & He, 2012). We found that a high expression of TRPM8 correlated with hyperactivation of the Wnt–Frizzled signalling in CRC patients (Figure 4). The proteins directly activated by frizzled receptors are unknown, but it has been repeatedly reported that mutation or deletion of the adenomatous polyposis coli (APC) protein occurs early during colorectal tumorigenesis (Powell et al., 1992). TRPM8 high expression was correlated with a down‐regulation of the APC gene and, unexpectedly, with an up‐regulation of NOTUM in CRC patients of our dataset (Figure 4a–c). NOTUM is a highly conserved secreted feedback antagonist of Wnt signalling (Kakugawa et al., 2015), recently identified as the primary mediator driving fixation of Apc‐mutant clones of intestinal stem cells (Flanagan et al., 2021).
FIGURE 4.
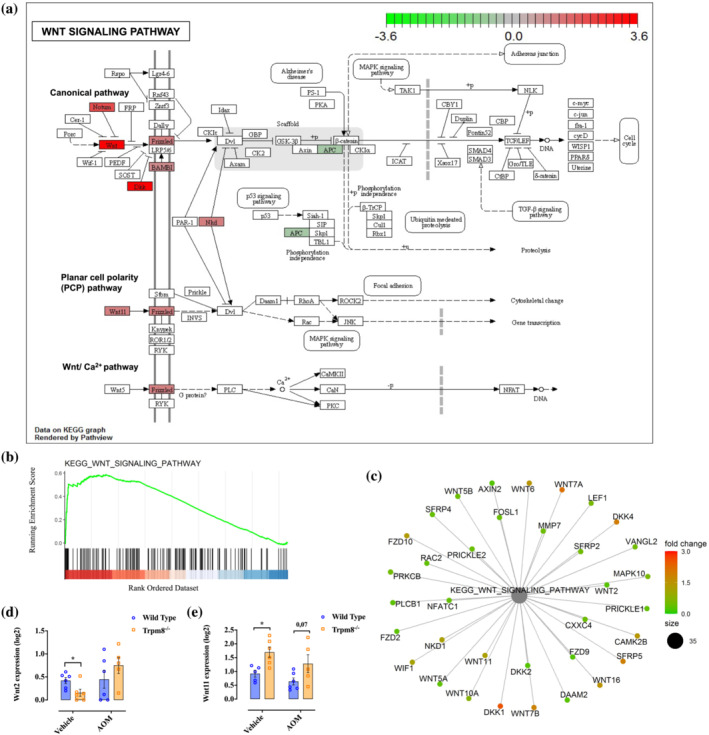
Kyoto Encyclopedia of Genes and Genomes (KEGG) Wnt signalling pathway. (a) Overlay of the differentially expressed genes (DEGs) between high and low Trpm8 expression groups on the KEGG pathway. The overexpressed and underexpressed genes are represented in red and green, respectively. (b) Gene set enrichment analysis (GSEA) enrichment plot. In green, the enrichment profile; in black on the X‐axis, the hits. Colour scale on the X‐axis goes from high (red) to low (blue) expression of genes in the high Trpm8 group. (c) Cnetplot showing the linkages between genes and biological concepts. The overexpressed and underexpressed genes are represented in red and green, respectively. (d, e) Expression of Wnt2 (d) and Wnt11 (e) by RNA sequencing analysis on the colon of wild‐type (blue) and Trpm8 −/− (orange) mice, treated with vehicle or azoxymethane (AOM) (n = 6 mice). P values were determined using multiple Student's t test. *P < 0.05 wild‐type vehicle versus Trpm8 −/− vehicle.
To recapitulate the genetic features, the protein abundance, the localization and the susceptibility to drugs of the Wnt pathway, we have depicted the oncoprint heatmap of this pathway (Figure S2). Although there are no differences between the high and low TRPM8‐expressing samples, such a heatmap allowed us to investigate each gene of the KEGG pathway by identifying the aberrations, both punctually and per sample or gene, using side histograms (Figure S2).
Because these results suggest hyperactivation of Wnt signalling in CRC patients with high levels of TRPM8, we analysed the Wnt/β‐catenin pathway in colons from AOM‐treated mice by RNA sequencing analysis. In physiological states, Trpm8 −/− mice showed a significant reduction in Wnt2 levels compared with WT mice (Figure 4d). Interestingly, in physiological states, we also observed that Trpm8 −/− mice have higher levels of Wnt11 (believed to activate the Wnt planar cell polarity signalling, Krishnamurthy & Kurzrock, 2018) than WT mice (Figure 4e).
3.5. TRPM8 pharmacological modulation by WS12 reduces experimental colon cancer growth
The data described above prompted us to ask whether pharmacological modulation of TRPM8 could be an effective strategy to reduce colon cancer development and growth.
For this purpose, we used WS12, one of the most potent available TRPM8 ligands (Bodding et al., 2007; Melanaphy et al., 2016). In our experiments, we confirmed the ability of this compound to activate and rapidly desensitize TRPM8 channels in HEK‐293 cells overexpressing the rat TRPM8 (Figure S3). The efficacy of using agonists of TRPM8 to induce its rapid desensitization (hence inactivation) has been an intriguing pharmacological approach (Diver et al., 2019) that has already been demonstrated to counteract intestinal inflammation (Ramachandran et al., 2013).
In the AOM model of sporadic colon cancer (Figure 5a), WS12 treatment (10 mg·kg−1, i.p.), starting 1 week before the carcinogenic agent, significantly reduced AOM‐induced tumours in WT, but not in Trpm8 −/− , mice (Figure 5b). These results strongly suggest that WS12 acts via TRPM8 desensitization.
FIGURE 5.
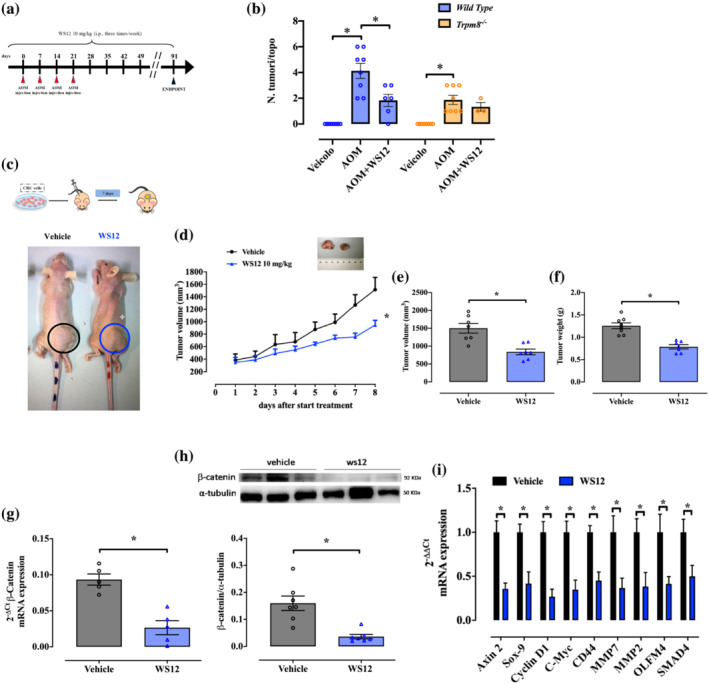
Pharmacological modulation of Trpm8 reduces tumour growth in azoxymethane (AOM) and xenograft models of colon cancer by impairing the Wnt/β‐catenin pathway. (a) Schematic representation of the AOM protocol and timeline. Mice were injected with 10 mg·kg−1 of AOM at the indicated time. WS12 (10 mg·kg−1) was administered intraperitoneally every other day. (b) Number of tumours per mouse counted on wild‐type (blue) and Trpm8 −/− (orange) mouse colon, treated with vehicle (n = 8 mice), AOM (n = 8 mice) or AOM + WS12 (n = 6 wild‐type mice and n = 3 Trpm8 −/− mice, three dead mice values were excluded, and this experimental group was not subjected to statistical analysis). P values were determined using two‐way ANOVA followed by Tukey's multiple comparisons test. *P < 0.05 Trpm8 −/− AOM versus vehicle, *P < 0.05 wild‐type AOM + WS12 versus AOM and *P < 0.05 wild‐type AOM versus vehicle. (c) Upper panel: Schematic representation of the xenograft protocol. WS12 (10 mg·kg−1) was administered intraperitoneally every day. Lower panel: Pictures of athymic mice bearing xenograft tumours on the day of killing. Tumors are indicated with black (vehicle) and blue (WS12) circles (d) Analysis of tumour volume of mice receiving vehicle (black) or WS12 (blue) in an 8‐day time course. Tumour size was measured every day and the volume was calculated. Each dot represents the mean ± SEM of seven mice. P values were determined using the Tukey–Kramer test; *P < 0.05 versus vehicle. (e, f) Analysis of tumour volume (e) and weight (f) of the explanted tumours from mice treated with vehicle (black) or WS12 (blue), at the end of the experiment (n = 7). P values were determined using unpaired Student's t tests; *P < 0.05 versus vehicle. (g) β‐Catenin mRNA expression was evaluated by reverse transcription quantitative polymerase chain reaction (RT–qPCR) and calculated by using the 2−ΔΔCt formula in xenografted tumours explanted from mice treated with vehicle (black) or WS12 (blue) (n = 5 different biological samples). P values were determined using unpaired Student's t tests; *P < 0.05 versus vehicle. (h) Representative immunoblots of β‐catenin protein levels in xenografted tumours explanted from mice receiving vehicle (black) or WS12 (blue). Lower panel shows densitometric analysis of western blot analysis. Results show mean ± SEM of seven different biological samples for each experimental group (i.e., vehicle and WS12) (n = 1 outlier has been removed from WS12 group with ROUT test). P values were determined using unpaired Student's t tests; *P < 0.05 versus vehicle. (i) Wnt signalling pathway‐associated genes (Axin 2 [axis inhibition protein 2], Sox‐9 [SRY‐box transcription factor 9], Cyclin D1, C‐Myc, CD44, MMP7 [matrix metalloproteinase 7], MMP2 [matrix metalloproteinase 2], OLFM4 [olfactomedin protein 4] and SMAD4) mRNA expression was evaluated by RT–qPCR and calculated by using the 2−ΔΔCt formula in xenografted tumours explanted from mice treated with vehicle (black) or WS12 (blue) (n = 5 different biological samples). P values were determined using multiple Student's t tests; *P < 0.05 versus vehicle.
Next, we evaluated the effect of WS12 in the HCT116 (adenocarcinoma cells expressing TRPM8) xenograft model of tumour (Figure 5c, upper panel). WS12 treatment (10 mg·kg−1, i.p.) caused a significant reduction in tumour growth compared with vehicle‐treated mice (Figure 5c, lower panel, d). As shown in Figure 5e,f, collected tumours were significantly smaller in size and weighed less in WS12‐treated mice (Figure 5e,f).
3.6. WS12 impairs the canonical Wnt/β‐catenin pathway in CRC xenografted tumours
To identify WS12 molecular mechanisms, we examined the Wnt pathway in xenografted tumours. On the basis of our results in patients (Figure 4a–c) and in the AOM model (Figures 2 and 4d,e) of colon carcinogenesis, we investigated the Wnt/β‐catenin pathway in tumours derived from WS12‐treated mice. Both mRNA and protein expressions of β‐catenin were significantly reduced in tumours from WS12‐treated mice (Figures 5g,h). Accordingly, WS12 reduced the expression of Axin 2 (axis inhibition protein 2), Sox‐9 (SRY‐box transcription factor 9), Cyclin D1, C‐Myc, CD44, MMP7 (matrix metalloproteinase 7), MMP2 (matrix metalloproteinase 2), OLFM4 (olfactomedin protein 4) and SMAD4, which are important regulatory downstream genes of the Wnt/β‐catenin signalling pathway (Figure 5i).
Collectively, these results suggest that WS12 could specifically perturbate the Wnt signalling by reducing the transcription of regulatory genes of that pathway, which are involved in CRC initiation, persistence and invasion.
4. DISCUSSION
TRPM8 is a member within the subset of temperature‐sensitive TRP channels and it is the main receptor involved in cold sensation (Clapham, 2003). TRPM8 was firstly detected in the prostate, but it is widely expressed by sensory neurons (Clapham, 2003). In prostate cancer, TRPM8 androgen‐dependent overexpression seems to be required for cancer cell survival (Lunardi et al., 2021; Zhang & Barritt, 2004), and TRPM8 has been proposed as a prognostic marker in such disease (Zhang & Barritt, 2004, 2006). The aberrant expression of TRPM8 has been also found in many other cancers (Hemida et al., 2021; Kijpornyongpan et al., 2014; Ouadid‐Ahidouch et al., 2013; Xu et al., 2021; Yee et al., 2012). Either activation (by increasing cytosolic Ca2+ levels) or antagonism/desensitization (by decreasing cytosolic Ca2+ levels) of TRPM8 (Zhang & Barritt, 2006) has been shown to affect in vitro cell growth and/or survival (Huang et al., 2020; Li et al., 2009; Okamoto et al., 2012; Yee, 2015). Furthermore, we have shown that the Cannabis component, cannabigerol, which nonselectively binds and antagonizes TRPM8, reduces Caco‐2 cell viability (Borrelli et al., 2014). Finally, a very recent study observed an aberrant TRPM8 expression in CRC patients with liver metastasis (Liu et al., 2021).
In the current study, we report that TRPM8 expression is significantly increased in tumour biopsies of stage pT4 CRC patients. We show for the first time that TRPM8 overexpression in human colon tumours is significantly associated with poor disease prognosis of CRC. Specifically, TRPM8 high expression in colon tumours (more than 33% of samples) predicts a significant lower PFI as well as disease‐specific survival. Also, an activated Wnt pathway seems to characterize the biology of these ‘high expressing TRPM8/low survival’ colon tumours. Additionally, we have shown an aberrant and selective TRPM8 expression in primary tumour cells isolated from CRC patients.
By using experimental models of sporadic CRC and CAC, we found that genetic deletion of TRPM8 reduced colon tumorigenesis. Hence, the loss of Trpm8 was sufficient to significantly reduce tumour incidence in both sporadic and inflammation‐induced intestinal tumour models, thus clearly featuring the critical role of TRPM8 in tumour development. Although TRPM8 has been defined as an ‘anti‐inflammatory’ target in gastrointestinal tract (de Jong et al., 2015; Hosoya et al., 2014; Khalil et al., 2016; Ramachandran et al., 2013), in our experiments TRPM8 did not affect the inflamed parameters related to tumour development in the AOM/DSS model. Similarly, Ramachandran and colleagues (2013) reported that Trpm8 −/− mice with colitis, although presenting elevated colonic levels of the inflammatory neuropeptide calcitonin‐gene‐related peptide compared with WT mice, did not differ from WT mice when inflammatory parameters were measured.
Mutations in β‐catenin, first described in colon cancer and melanoma (Rubinfeld et al., 1997), occur in a wide variety of solid tumours (Reya & Clevers, 2005). It has been broadly demonstrated that Wnt signalling controls the levels of the key effector β‐catenin for signal transduction (Clevers & Nusse, 2012). Therefore, we explored the Wnt signalling pathway in CRC patients with high TRPM8 expression in our dataset. Accordingly, TRPM8 high expression (more than 33% of our population) was correlated with hyperactivation of Wnt and Frizzled and down‐regulation of APC. APC negatively regulates Wnt signalling by directing the phosphorylation and degradation of β‐catenin (Morin et al., 1997). Consequently, we looked at the genetic profile of WT and Trpm8 −/− mice treated or not treated with the carcinogenic agent AOM by using RNA sequencing. Remarkably, the loss of Trpm8 in mice caused up‐regulation of Wnt11, which is only implicated in non‐canonical Wnt pathways. Such signalling is independent of β‐catenin and involves protein kinase C and Jun amino‐terminal kinase (Djiane et al., 2000; Miller et al., 1999; Pandur et al., 2002). Collectively, these findings suggest that TRPM8 could induce colon tumorigenesis possibly by perturbating the Wnt canonical pathway. In agreement with this hypothesis, Wnt2, whose overexpression promotes CRC growth, invasion and angiogenesis (Unterleuthner et al., 2020; Zhang et al., 2020), was significantly down‐regulated in healthy Trpm8 −/− mice compared with WT mice in our experiments. Taken all together, these results leads us to hypothesize that Trpm8 deletion might reduce Wnt/β‐catenin signalling. Overall, we hypothesize that tumour cells take advantage of up‐regulating TRPM8 by causing an aberrant activation of the β‐catenin signalling pathway that, in turns, fuels and further sustains tumour development. However, more in‐depth studies are necessary to depict the functional relevance of such aberrant activation in terms of effects on one or more hallmarks of cancer (e.g., proliferation and tumour spread).
It is well established that prostaglandin E2 (PGE2 ) promotes colon cancer cell growth and development through the activation of the β‐catenin signalling pathway (Aizawa et al., 2019; Castellone et al., 2005; Park et al., 2013). In addition, TRPM8 activation is associated with PGE2 release in pathophysiological states (Aizawa et al., 2019; Park et al., 2013). As we have shown here that TRPM8 modulation has a protective role in colon cancer, possibly due to the inactivation of the β‐catenin pathway, we cannot exclude the possibility that the final effector of TRPM8‐mediated effects could be PGE2. This aspect is relevant and deserved further ad hoc investigations.
In the light of our data about the role of TRPM8 in colon cancer pathophysiology and its prognostic value in cancer diagnosis (discussed above), we ultimately explored the possibility to consider TRPM8 as a target for drug discovery. For this purpose, we modulated TRPM8 by using WS12, previously reported as a selective and potent TRPM8 ligand (Bodding et al., 2007; Melanaphy et al., 2016). First, we found that WS12 was able to activate (and desensitize after subsequent stimulations) TRPM8 overexpressed in HEK‐293 cells. Next, WS12 reduced AOM‐induced tumours in WT, but not in Trpm8 −/−, mice, thus suggesting that its antitumour effect was TRPM8 mediated. Finally, pharmacological treatment with WS12 reduced tumour growth in CRC‐xenografted mice. Interestingly, in xenografted mice, the antitumor action of WS12 was associated with the reduction of the canonical Wnt pathway, in line with our results obtained with Trpm8 −/− mice in the AOM model of colon carcinogenesis. Specifically, WS12 reduced TRPM8‐mediated activation of β‐catenin and subsequently the induction of its main target genes, such as C‐Myc, Cyclin D1 and CD44. Overall, from a translational point of view, our data demonstrated that pharmacological modulation of TRPM8 might be effective for both preventive and therapeutic strategies. Future experiments with APCMin mice, which are genetically predisposed to intestinal adenoma formation, will firmly establish if TRPM8 modulation is potentially useful in the prevention in familiar CRC/polyposis.
In conclusion, we have provided new evidence pointing to a pivotal role of TRPM8 in experimental colon carcinogenesis. This is based on the observation that TRPM8 overexpression correlated well with poor prognosis in CRC patients and that TRPM8 deletion or pharmacological targeting protected mice from tumour development. Mechanistically, the carcinogenetic effect of TRPM8 seems to be associated with activation of the Wnt2/β‐catenin pathway. From a translational point of view, our human data suggest the possibility of using TRPM8 as a new prognostic marker for CRC and our animal data indicate the existence of a new agent in CRC pathophysiology representing an innovative molecular target for CRC treatment/prevention.
AUTHOR CONTRIBUTIONS
Conceptualization and project administration: EP, BR and AAI. Methodology: EP, DC, FI, FC, GL, MFN, TV, PD, SL, PS and RC. Formal analysis and interpretation of data: EP, BR and FB. Collection and stadiation of CRC human biopsies: MDA, RL and MDL. RNA sequencing data: SM and SF. Bioinformatic analysis: PZ and GF. Writing—original draft: EP and AAI; writing—review and editing: BR, FB and VD. Funding acquisition: AAI, VD and FB.
CONFLICTS OF INTEREST
The authors declare no conflicts of interest.
DECLARATION OF TRANSPARENCY AND SCIENTIFIC RIGOUR
This Declaration acknowledges that this paper adheres to the principles for transparent reporting and scientific rigour of preclinical research as stated in the BJP guidelines for Design & Analysis, Immunoblotting and Immunochemistry, and Animal Experimentation and as recommended by funding agencies, publishers and other organizations engaged with supporting research.
Supporting information
Figure S1. Investigated parameters in AOM and AOM/DSS models.
(A). Weight changes in the AOM model. Weight was monitored once a week at the beginning of every week. Weights are expressed as percentage of the initial weight, and each dot represents the mean ± SEM of 8 mice. P values were assessed by Two‐Way ANOVA followed by Tukey's multiple comparisons test. * p < 0.05
(B). Weight changes in the AOM/DSS model. Weight was monitored every other day for the entire duration of the experiment. Weights are expressed as percentage of the initial weight and each dot represents the mean ± SEM of 6 mice. P value was assessed by Two‐Way ANOVA followed by Tukey's multiple comparisons test. *p < 0.05
(C). Representative colonoscopy images of wild type (WT) and Trpm8−/− mice carried out at day 49 of AOM protocol.
(D). Number of tumors/mouse in WT (black) and Trpm8 −/− (red) mice treated with AOM counted during the endoscopy at day 49 (n = 3). Error bars represent ± SEM. Exploratory data not subjected to statistical analysis.
(E). Tumor area evaluation by endoscopic measuring of WT (black) and Trpm8−/− (red) mice treated with AOM/DSS (n = 6). Error bars represent ± SEM. P values were determined using unpaired Student's t test. n.s. non significant.
(F). Disease Activity Index (DAI) score of WT (red) and Trpm8 −/− (blue) mice treated with AOM/DSS. DAI score was measured every other day during DSS cycles, and once a week during washout periods. DAI was assessed considering stool consistency and presence of blood as reported in the text. Each dot represents the mean ± SEM of 6 mice.
Figure S2. Oncoprint visualization of clinical and genomic features of the KEGG Wnt pathway in the 268 TCGA CRC patients.
Protein abundance in both CRC and normal colo‐rectal tissues, as well as localization and druggability are reported. In the HM, red represents amplification, blue represents deletion and green dots mutations of gene. On the left the cellular localization together with the druggability are reported for each gene. On the right, a barplot recapitulating the frequency of the aberrations for each gene and the immunohistochemistry status of the tumors and the healthy controls according to the Tumor Atlas. On top, TRPM8 subgroups microsatellite status and barplot recapitulating the frequency of the aberrations for each sample.
Figure S3. Effect of WS12 on HEK‐293 cells stably expressing TRPM8.
(A). WS12 effect on calcium influx induced by ionomycin (4μM). The concentration of WS12 is expressed as log10[M]. Error bars represent ± SEM
(B). Effect of triple WS12 (0.2 μM) stimulus on induced TRPM8 gating of Ca2 + influx into TRPM8‐HEK‐293 cells. Data are expressed as a normalized response to the first stimulus (n = 5 different experiments). Error bars represent ± SEM. P‐values were determined using one way ANOVA followed by Dunnett's multiple comparisons test. * p < 0.05
TRPM8‐HEK‐293 cells response to triple WS12 stimulation.
Figure S4. RNA sequencing of Wild‐Type and Trpm8 −/− mice.
Expression of Wnt2b, Wnt3, Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7b, Wnt9a, Wnt10b, Apc, Apc2 by RNA sequencing analysis on the colon of wild Type (blue) and Trpm8 −/− (orange) mice, treated with vehicle or AOM (n = 6). P values were determined using multiple Student's t‐test. * p < 0.05.
Table S1. Patients data and TNM classification.
Table S2. Sequences for Homo sapiens primers of the genes screened by Quantitative Real‐Time Polymerase Chain Reaction.
ACKNOWLEDGEMENTS
We are grateful to Dr. Aniello Schiano Moriello for technical help with HEK‐293 stably overexpressing recombinant rat TRPM8 with Ca2+ measurement. Open Access Funding provided by Universita degli Studi di Napoli Federico II within the CRUI‐CARE Agreement.
Pagano, E. , Romano, B. , Cicia, D. , Iannotti, F. A. , Venneri, T. , Lucariello, G. , Nanì, M. F. , Cattaneo, F. , De Cicco, P. , D'Armiento, M. , De Luca, M. , Lionetti, R. , Lama, S. , Stiuso, P. , Zoppoli, P. , Falco, G. , Marchianò, S. , Fiorucci, S. , Capasso, R. , … Izzo, A. A. (2023). TRPM8 indicates poor prognosis in colorectal cancer patients and its pharmacological targeting reduces tumour growth in mice by inhibiting Wnt/β‐catenin signalling. British Journal of Pharmacology, 180(2), 235–251. 10.1111/bph.15960
Funding information This work was supported by the National Grant Progetti di Rilevante Interesse Nazionale (PRIN) 2017 (project numbers 2017XC73BW and 2017E84AA4) from the Ministero dell'Università e della Ricerca (MIUR) and the Regione Campania‐POR Campania FESR 2014/2020 (project number B61G18000470007).
Ester Pagano and Barbara Romano equally contributed to this paper.
Contributor Information
Francesca Borrelli, Email: franborr@unina.it.
Angelo A. Izzo, Email: aaizzo@unina.it.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request.
REFERENCES
- Aizawa, N. , Ohshiro, H. , Watanabe, S. , Kume, H. , Homma, Y. , & Igawa, Y. (2019). RQ‐00434739, a novel TRPM8 antagonist, inhibits prostaglandin E2‐induced hyperactivity of the primary bladder afferent nerves in rats. Life Sciences, 218, 89–95. 10.1016/j.lfs.2018.12.031 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. , Christopoulos, A. , Davenport, A. P. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Southan, C. , Davies, J. A. , Abbracchio, M. P. , & CGTP Collaborators Alexander . (2021). THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: G protein‐coupled receptors. British Journal of Pharmacology, 178(S1), S27–S156. 10.1111/bph.15538 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. , Fabbro, D. , Kelly, E. , Mathie, A. , Peters, J. A. , Veale, E. L. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Southan, C. , Davies, J. A. , Boison, D. , Burns, K. E. , Dessauer, C. , Gertsch, J. , Helsby, N. A. , Izzo, A. A. , Koesling, D. , … Wong, S. S. (2021). THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: Enzymes. British Journal of Pharmacology, 178(S1), S313–S411. 10.1111/bph.15542 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. , Mathie, A. , Peters, J. A. , Veale, E. L. , Striessnig, J. , Kelly, E. , Armstrong, J. F. , Faccenda, E. , Harding, S. D. , Pawson, A. J. , Southan, C. , Davies, J. A. , Aldrich, R. W. , Attali, B. , Baggetta, A. M. , Becirovic, E. , Biel, M. , Bill, R. M. , Catterall, W. A. , … Zhu, M. (2021). THE CONCISE GUIDE TO PHARMACOLOGY 2021/22: Ion channels. British Journal of Pharmacology, 178(S1), S157–S245. 10.1111/bph.15539 [DOI] [PubMed] [Google Scholar]
- Alexander, S. P. H. , Roberts, R. E. , Broughton, B. R. S. , Sobey, C. G. , George, C. H. , Stanford, S. C. , Cirino, G. , Docherty, J. R. , Giembycz, M. A. , Hoyer, D. , Insel, P. A. , Izzo, A. A. , Ji, Y. , MacEwan, D. J. , Mangum, J. , Wonnacott, S. , & Ahluwalia, A. (2018). Goals and practicalities of immunoblotting and immunohistochemistry: A guide for submission to the British Journal of Pharmacology . British Journal of Pharmacology, 175, 407–411. 10.1111/bph.14112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold, A. , Tronser, M. , Sers, C. , Ahadova, A. , Endris, V. , Mamlouk, S. , Horst, D. , Möbs, M. , Bischoff, P. , Kloor, M. , & Bläker, H. (2020). The majority of β‐catenin mutations in colorectal cancer is homozygous. BMC Cancer, 20, 1038. 10.1186/s12885-020-07537-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bautista, D. M. , Siemens, J. , Glazer, J. M. , Tsuruda, P. R. , Basbaum, A. I. , Stucky, C. L. , Jordt, S. E. , & Julius, D. (2007). The menthol receptor TRPM8 is the principal detector of environmental cold. Nature, 448, 204–208. 10.1038/nature05910 [DOI] [PubMed] [Google Scholar]
- Becker, C. , Fantini, M. C. , Wirtz, S. , Nikolaev, A. , Kiesslich, R. , Lehr, H. A. , Galle, P. R. , & Neurath, M. F. (2005). In vivo imaging of colitis and colon cancer development in mice using high resolution chromoendoscopy. Gut, 54, 950–954. 10.1136/gut.2004.061283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodding, M. , Wissenbach, U. , & Flockerzi, V. (2007). Characterisation of TRPM8 as a pharmacophore receptor. Cell Calcium, 42, 618–628. 10.1016/j.ceca.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Borrelli, F. , Pagano, E. , Romano, B. , Panzera, S. , Maiello, F. , Coppola, D. , de Petrocellis, L. , Buono, L. , Orlando, P. , & Izzo, A. A. (2014). Colon carcinogenesis is inhibited by the TRPM8 antagonist cannabigerol, a Cannabis‐derived non‐psychotropic cannabinoid. Carcinogenesis, 35, 2787–2797. 10.1093/carcin/bgu205 [DOI] [PubMed] [Google Scholar]
- Castellone, M. D. , Teramoto, H. , Williams, B. O. , Druey, K. M. , & Gutkind, J. S. (2005). Prostaglandin E2 promotes colon cancer cell growth through a Gs‐axin‐ß‐catenin signaling axis. Science, 310, 1504–1510. 10.1126/science.1116221 [DOI] [PubMed] [Google Scholar]
- Clapham, D. E. (2003). TRP channels as cellular sensors. Nature, 426, 517–524. 10.1038/nature02196 [DOI] [PubMed] [Google Scholar]
- Clevers, H. , & Nusse, R. (2012). Wnt/β‐catenin signaling and disease. Cell, 149, 1192–1205. 10.1016/j.cell.2012.05.012 [DOI] [PubMed] [Google Scholar]
- Curtis, M. J. , Alexander, S. , Cirino, G. , Docherty, J. R. , George, C. H. , Giembycz, M. A. , Hoyer, D. , Insel, P. A. , Izzo, A. A. , Ji, Y. , MacEwan, D. J. , Sobey, C. G. , Stanford, S. C. , Teixeira, M. M. , Wonnacott, S. , & Ahluwalia, A. (2018). Experimental design and analysis and their reporting II: Updated and simplified guidance for authors and peer reviewers. British Journal of Pharmacology, 175, 987–993. 10.1111/bph.14153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong, P. R. , Takahashi, N. , Peiris, M. , Bertin, S. , Lee, J. , Gareau, M. G. , Paniagua, A. , Harris, A. R. , Herdman, D. S. , Corr, M. , Blackshaw, L. A. , & Raz, E. (2015). TRPM8 on mucosal sensory nerves regulates colitogenic responses by innate immune cells via CGRP. Mucosal Immunology, 8, 491–504. 10.1038/mi.2014.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Petrocellis, L. , Starowicz, K. , Moriello, A. S. , Vivese, M. , Orlando, P. , & Di Marzo, V. (2007). Regulation of transient receptor potential channels of melastatin type 8 (TRPM8): Effect of cAMP, cannabinoid CB1 receptors and endovanilloids. Experimental Cell Research, 313, 1911–1920. 10.1016/j.yexcr.2007.01.008 [DOI] [PubMed] [Google Scholar]
- Diver, M. M. , Cheng, Y. , & Julius, D. (2019). Structural insights into TRPM8 inhibition and desensitization. Science, 365, 1434–1440. 10.1126/science.aax6672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djiane, A. , Riou, J. , Umbhauer, M. , Boucaut, J. , & Shi, D. (2000). Role of frizzled 7 in the regulation of convergent extension movements during gastrulation in Xenopus laevis. Development, 127, 3091–3100. 10.1242/dev.127.14.3091 [DOI] [PubMed] [Google Scholar]
- Douaiher, J. , Ravipati, A. , Grams, B. , Chowdhury, S. , Alatise, O. , & Are, C. (2017). Colorectal cancer‐global burden, trends, and geographical variations. Journal of Surgical Oncology, 115, 619–630. 10.1002/jso.24578 [DOI] [PubMed] [Google Scholar]
- Flanagan, D. J. , Pentinmikko, N. , Luopajärvi, K. , Willis, N. J. , Gilroy, K. , Raven, A. P. , Mcgarry, L. , Englund, J. I. , Webb, A. T. , Scharaw, S. , Nasreddin, N. , Hodder, M. C. , Ridgway, R. A. , Minnee, E. , Sphyris, N. , Gilchrist, E. , Najumudeen, A. K. , Romagnolo, B. , Perret, C. , … Sansom, O. J. (2021). NOTUM from Apc‐mutant cells biases clonal competition to initiate cancer. Nature, 594, 430–435. 10.1038/s41586-021-03525-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemida, A. S. , Hammam, M. A. , Heriz, N. , & Shehata, W. A. (2021). Expression of transient receptor potential channel of melastatin number 8 (TRPM8) in non‐melanoma skin cancer: A clinical and immunohistochemical study. Journal of Immunoassay & Immunochemistry, 42(6), 620–632. 10.1080/15321819.2021.1918709 [DOI] [PubMed] [Google Scholar]
- Henshall, S. M. , Afar, D. E. , Hiller, J. , Horvath, L. G. , Quinn, D. I. , Rasiah, K. K. , Gish, K. , Willhite, D. , Kench, J. G. , Gardiner‐Garden, M. , Stricker, P. D. , Scher, H. I. , Grygiel, J. J. , Agus, D. B. , Mack, D. H. , & Sutherland, R. L. (2003). Survival analysis of genome‐wide gene expression profiles of prostate cancers identifies new prognostic targets of disease relapse. Cancer Research, 63, 4196–4203. [PubMed] [Google Scholar]
- Hosoya, T. , Matsumoto, K. , Tashima, K. , Nakamura, H. , Fujino, H. , Murayama, T. , & Horie, S. (2014). TRPM8 has a key role in experimental colitis‐induced visceral hyperalgesia in mice. Neurogastroenterology and Motility, 26, 1112–1121. 10.1111/nmo.12368 [DOI] [PubMed] [Google Scholar]
- Huang, Y. , Li, S. , Jia, Z. , Zhao, W. , Zhou, C. , Zhang, R. , Ali, D. W. , Michalak, M. , Chen, X. Z. , & Tang, J. (2020). Transient receptor potential melastatin 8 (TRPM8) channel regulates proliferation and migration of breast cancer cells by activating the AMPK‐ULK1 pathway to enhance basal autophagy. Frontiers in Oncology, 10, 573127. 10.3389/fonc.2020.573127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iannotti, F. A. , Hill, C. L. , Leo, A. , Alhusaini, A. , Soubrane, C. , Mazzarella, E. , Russo, E. , Whalley, B. J. , di Marzo, V. , & Stephens, G. J. (2014). Nonpsychotropic plant cannabinoids, cannabidivarin (CBDV) and cannabidiol (CBD), activate and desensitize transient receptor potential vanilloid 1 (TRPV1) channels in vitro: Potential for the treatment of neuronal hyperexcitability. ACS Chemical Neuroscience, 5, 1131–1141. 10.1021/cn5000524 [DOI] [PubMed] [Google Scholar]
- Kakugawa, S. , Langton, P. F. , Zebisch, M. , Howell, S. , Chang, T. H. , Liu, Y. , Feizi, T. , Bineva, G. , O'Reilly, N. , Snijders, A. P. , Jones, E. Y. , & Vincent, J. P. (2015). Notum deacylates Wnt proteins to suppress signalling activity. Nature, 519, 187–192. 10.1038/nature14259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil, M. , Babes, A. , Lakra, R. , Försch, S. , Reeh, P. W. , Wirtz, S. , Becker, C. , Neurath, M. F. , & Engel, M. A. (2016). Transient receptor potential melastatin 8 ion channel in macrophages modulates colitis through a balance‐shift in TNF‐alpha and interleukin‐10 production. Mucosal Immunology, 9, 1500–1513. 10.1038/mi.2016.16 [DOI] [PubMed] [Google Scholar]
- Kijpornyongpan, T. , Sereemaspun, A. , & Chanchao, C. (2014). Dose‐dependent cytotoxic effects of menthol on human malignant melanoma A‐375 cells: Correlation with TRPM8 transcript expression. Asian Pacific Journal of Cancer Prevention, 15, 1551–1556. 10.7314/APJCP.2014.15.4.1551 [DOI] [PubMed] [Google Scholar]
- Krishnamurthy, N. , & Kurzrock, R. (2018). Targeting the Wnt/beta‐catenin pathway in cancer: Update on effectors and inhibitors. Cancer Treatment Reviews, 62, 50–60. 10.1016/j.ctrv.2017.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Q. , Wang, X. , Yang, Z. , Wang, B. , & Li, S. (2009). Menthol induces cell death via the TRPM8 channel in the human bladder cancer cell line T24. Oncology, 77, 335–341. 10.1159/000264627 [DOI] [PubMed] [Google Scholar]
- Lilley, E. , Stanford, S. C. , Kendall, D. E. , Alexander, S. P. H. , Cirino, G. , Docherty, J. R. , George, C. H. , Insel, P. A. , Izzo, A. A. , Ji, Y. , Panettieri, R. A. , Sobey, C. G. , Stefanska, B. , Stephens, G. , Teixeira, M. , & Ahluwalia, A. (2020). ARRIVE 2.0 and the British Journal of Pharmacology: Updated guidance for 2020. British Journal of Pharmacology, 177, 3611–3616. 10.1111/bph.15178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, B. , Fan, L. , Balakrishna, S. , Sui, A. , Morris, J. B. , & Jordt, S. E. (2013). TRPM8 is the principal mediator of menthol‐induced analgesia of acute and inflammatory pain. Pain, 154, 2169–2177. 10.1016/j.pain.2013.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. J. , Li, L. Z. , & Xu, P. (2021). Upregulation of TRPM8 can promote the colon cancer liver metastasis through mediating Akt/GSK‐3 signal pathway. Biotechnology and Applied Biochemistry, 69, 230–239. 10.1002/bab.2102 [DOI] [PubMed] [Google Scholar]
- Lunardi, A. , Barbareschi, M. , Carbone, F. G. , Morelli, L. , Brunelli, M. , Fortuna, N. , Genovesi, S. , & Alaimo, A. (2021). TRPM8 protein expression in hormone naive local and lymph node metastatic prostate cancer. Pathologica, 113, 95–101. 10.32074/1591-951X-262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald, B. T. , & He, X. (2012). Frizzled and LRP5/6 receptors for Wnt/β‐catenin signaling. Cold Spring Harbor Perspectives in Biology, 4, a007880. 10.1101/cshperspect.a007880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuade, R. M. , Stojanovska, V. , Bornstein, J. C. , & Nurgali, K. (2017). Colorectal cancer chemotherapy: The evolution of treatment and new approaches. Current Medicinal Chemistry, 24, 1537–1557. 10.2174/0929867324666170111152436 [DOI] [PubMed] [Google Scholar]
- Melanaphy, D. , Johnson, C. D. , Kustov, M. V. , Watson, C. A. , Borysova, L. , Burdyga, T. V. , & Zholos, A. V. (2016). Ion channel mechanisms of rat tail artery contraction‐relaxation by menthol involving, respectively, TRPM8 activation and L‐type Ca2+ channel inhibition. American Journal of Physiology. Heart and Circulatory Physiology, 311, H1416–H1430. 10.1152/ajpheart.00222.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. R. , Hocking, A. M. , Brown, J. D. , & Moon, R. T. (1999). Mechanism and function of signal transduction by the Wnt/β‐catenin and Wnt/Ca2+ pathways. Oncogene, 18, 7860–7872. 10.1038/sj.onc.1203245 [DOI] [PubMed] [Google Scholar]
- Miller, K. D. , Nogueira, L. , Devasia, T. , Mariotto, A. B. , Yabroff, K. R. , Jemal, A. , Kramer, J. , & Siegel, R. L. (2022). Cancer treatment and survivorship statistics, 2022. CA: A Cancer Journal for Clinicians, 72, 409–436. 10.3322/caac.21731 [DOI] [PubMed] [Google Scholar]
- Moran, M. M. (2018). TRP channels as potential drug targets. Annual Review of Pharmacology and Toxicology, 58, 309–330. 10.1146/annurev-pharmtox-010617-052832 [DOI] [PubMed] [Google Scholar]
- Morin, P. J. , Sparks, A. B. , Korinek, V. , Barker, N. , Clevers, H. , Vogelstein, B. , & Kinzler, K. W. (1997). Activation of β‐catenin‐Tcf signaling in colon cancer by mutations in β‐catenin or APC. Science, 275, 1787–1790. 10.1126/science.275.5307.1787 [DOI] [PubMed] [Google Scholar]
- Nunez, F. P. , Quera, P. R. , & Gomollon, F. (2019). Primary sclerosing cholangitis and inflammatory bowel disease: Intestine‐liver interrelation. Gastroenterología Y Hepatología, 42, 316–325. [DOI] [PubMed] [Google Scholar]
- Nusse, R. , & Clevers, H. (2017). Wnt/β‐catenin signaling, disease, and emerging therapeutic modalities. Cell, 169, 985–999. 10.1016/j.cell.2017.05.016 [DOI] [PubMed] [Google Scholar]
- Okamoto, Y. , Ohkubo, T. , Ikebe, T. , & Yamazaki, J. (2012). Blockade of TRPM8 activity reduces the invasion potential of oral squamous carcinoma cell lines. International Journal of Oncology, 40, 1431–1440. 10.3892/ijo.2012.1340 [DOI] [PubMed] [Google Scholar]
- Ouadid‐Ahidouch, H. , Dhennin‐Duthille, I. , Gautier, M. , Sevestre, H. , & Ahidouch, A. (2013). TRP channels: Diagnostic markers and therapeutic targets for breast cancer? Trends in Molecular Medicine, 19, 117–124. 10.1016/j.molmed.2012.11.004 [DOI] [PubMed] [Google Scholar]
- Pagano, E. , Borrelli, F. , Orlando, P. , Romano, B. , Monti, M. , Morbidelli, L. , Aviello, G. , Imperatore, R. , Capasso, R. , Piscitelli, F. , Buono, L. , di Marzo, V. , & Izzo, A. A. (2017). Pharmacological inhibition of MAGL attenuates experimental colon carcinogenesis. Pharmacological Research, 119, 227–236. 10.1016/j.phrs.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Pagano, E. , Elias, J. E. , Schneditz, G. , Saveljeva, S. , Holland, L. M. , Borrelli, F. , Karlsen, T. H. , Kaser, A. , & Kaneider, N. C. (2021). Activation of the GPR35 pathway drives angiogenesis in the tumour microenvironment. Gut, 71, 509–520. 10.1136/gutjnl-2020-323363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandur, P. , Lasche, M. , Eisenberg, L. M. , & Kuhl, M. (2002). Wnt‐11 activation of a non‐canonical Wnt signalling pathway is required for cardiogenesis. Nature, 418, 636–641. 10.1038/nature00921 [DOI] [PubMed] [Google Scholar]
- Park, N. H. , Na, Y. J. , Choi, H. T. , Cho, J. C. , & Lee, H. K. (2013). Activation of transient receptor potential melastatin 8 reduces ultraviolet B‐induced prostaglandin E2 production in keratinocytes. The Journal of Dermatology, 40, 919–922. 10.1111/1346-8138.12288 [DOI] [PubMed] [Google Scholar]
- Patriarca, C. , Macchi, R. M. , Marschner, A. K. , & Mellstedt, H. (2012). Epithelial cell adhesion molecule expression (CD326) in cancer: A short review. Cancer Treatment Reviews, 38, 68–75. 10.1016/j.ctrv.2011.04.002 [DOI] [PubMed] [Google Scholar]
- Percie du Sert, N. , Hurst, V. , Ahluwalia, A. , Alam, S. , Avey, M. T. , Baker, M. , Browne, W. J. , Clark, A. , Cuthill, I. C. , Dirnagl, U. , Emerson, M. , Garner, P. , Holgate, S. T. , Howells, D. W. , Karp, N. A. , Lazic, S. E. , Lidster, K. , MacCallum, C. J. , Macleod, M. , … Würbel, H. (2020). The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. British Journal of Pharmacology, 177, 3617–3624. 10.1111/bph.15193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell, S. M. , Zilz, N. , Beazer‐Barclay, Y. , Bryan, T. M. , Hamilton, S. R. , Thibodeau, S. N. , Vogelstein, B. , & Kinzler, K. W. (1992). APC mutations occur early during colorectal tumorigenesis. Nature, 359, 235–237. 10.1038/359235a0 [DOI] [PubMed] [Google Scholar]
- Ramachandran, R. , Hyun, E. , Zhao, L. , Lapointe, T. K. , Chapman, K. , Hirota, C. L. , Ghosh, S. , McKemy, D. D. , Vergnolle, N. , Beck, P. L. , Altier, C. , & Hollenberg, M. D. (2013). TRPM8 activation attenuates inflammatory responses in mouse models of colitis. Proceedings of the National Academy of Sciences of the United States of America, 110, 7476–7481. 10.1073/pnas.1217431110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawla, P. , Sunkara, T. , & Barsouk, A. (2019). Epidemiology of colorectal cancer: Incidence, mortality, survival, and risk factors. Przeglad Gastroenterologiczny, 14, 89–103. 10.5114/pg.2018.81072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya, T. , & Clevers, H. (2005). Wnt signalling in stem cells and cancer. Nature, 434, 843–850. 10.1038/nature03319 [DOI] [PubMed] [Google Scholar]
- Rubinfeld, B. , Robbins, P. , El‐Gamil, M. , Albert, I. , Porfiri, E. , & Polakis, P. (1997). Stabilization of β‐catenin by genetic defects in melanoma cell lines. Science, 275, 1790–1792. 10.1126/science.275.5307.1790 [DOI] [PubMed] [Google Scholar]
- Sawicki, T. , Ruszkowska, M. , Danielewicz, A. , Niedzwiedzka, E. , Arlukowicz, T. , & Przybylowicz, K. E. (2021). A review of colorectal cancer in terms of epidemiology, risk factors, development, symptoms and diagnosis. Cancers (Basel), 13, 2025. 10.3390/cancers13092025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsavaler, L. , Shapero, M. H. , Morkowski, S. , & Laus, R. (2001). Trp‐p8, a novel prostate‐specific gene, is up‐regulated in prostate cancer and other malignancies and shares high homology with transient receptor potential calcium channel proteins. Cancer Research, 61, 3760–3769. [PubMed] [Google Scholar]
- Unterleuthner, D. , Neuhold, P. , Schwarz, K. , Janker, L. , Neuditschko, B. , Nivarthi, H. , Crncec, I. , Kramer, N. , Unger, C. , Hengstschläger, M. , Eferl, R. , Moriggl, R. , Sommergruber, W. , Gerner, C. , & Dolznig, H. (2020). Cancer‐associated fibroblast‐derived WNT2 increases tumor angiogenesis in colon cancer. Angiogenesis, 23, 159–177. 10.1007/s10456-019-09688-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanacore, D. , Messina, G. , Lama, S. , Bitti, G. , Ambrosio, P. , Tenore, G. , Messina, A. , Monda, V. , Zappavigna, S. , Boccellino, M. , Novellino, E. , Monda, M. , & Stiuso, P. (2018). Effect of restriction vegan diet's on muscle mass, oxidative status, and myocytes differentiation: A pilot study. Journal of Cellular Physiology, 233, 9345–9353. 10.1002/jcp.26427 [DOI] [PubMed] [Google Scholar]
- Xu, Q. , Kong, N. , Zhang, J. , Bai, N. , Bi, J. , & Li, W. (2021). Expression of transient receptor potential cation channel subfamily M member 8 in gastric cancer and its clinical significance. Experimental and Therapeutic Medicine, 21, 377. 10.3892/etm.2021.9808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee, N. S. (2015). Roles of TRPM8 ion channels in cancer: Proliferation, survival, and invasion. Cancers (Basel), 7, 2134–2146. 10.3390/cancers7040882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee, N. S. (2016). TRPM8 ion channels as potential cancer biomarker and target in pancreatic cancer. Advances in Protein Chemistry and Structural Biology, 104, 127–155. 10.1016/bs.apcsb.2016.01.001 [DOI] [PubMed] [Google Scholar]
- Yee, N. S. , Chan, A. S. , Yee, J. D. , & Yee, R. K. (2012). TRPM7 and TRPM8 ion channels in pancreatic adenocarcinoma: Potential roles as cancer biomarkers and targets. Scientifica (Cairo), 2012, 415158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L. , & Barritt, G. J. (2004). Evidence that TRPM8 is an androgen‐dependent Ca2+ channel required for the survival of prostate cancer cells. Cancer Research, 64, 8365–8373. 10.1158/0008-5472.CAN-04-2146 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , & Barritt, G. J. (2006). TRPM8 in prostate cancer cells: A potential diagnostic and prognostic marker with a secretory function? Endocrine‐Related Cancer, 13, 27–38. 10.1677/erc.1.01093 [DOI] [PubMed] [Google Scholar]
- Zhang, L. , Jiang, X. , Li, Y. , Fan, Q. , Li, H. , Jin, L. , Li, L. , Jin, Y. , Zhang, T. , Mao, Y. , & Hua, D. (2020). Clinical correlation of Wnt2 and COL8A1 with colon adenocarcinoma prognosis. Frontiers in Oncology, 10, 1504. 10.3389/fonc.2020.01504 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Investigated parameters in AOM and AOM/DSS models.
(A). Weight changes in the AOM model. Weight was monitored once a week at the beginning of every week. Weights are expressed as percentage of the initial weight, and each dot represents the mean ± SEM of 8 mice. P values were assessed by Two‐Way ANOVA followed by Tukey's multiple comparisons test. * p < 0.05
(B). Weight changes in the AOM/DSS model. Weight was monitored every other day for the entire duration of the experiment. Weights are expressed as percentage of the initial weight and each dot represents the mean ± SEM of 6 mice. P value was assessed by Two‐Way ANOVA followed by Tukey's multiple comparisons test. *p < 0.05
(C). Representative colonoscopy images of wild type (WT) and Trpm8−/− mice carried out at day 49 of AOM protocol.
(D). Number of tumors/mouse in WT (black) and Trpm8 −/− (red) mice treated with AOM counted during the endoscopy at day 49 (n = 3). Error bars represent ± SEM. Exploratory data not subjected to statistical analysis.
(E). Tumor area evaluation by endoscopic measuring of WT (black) and Trpm8−/− (red) mice treated with AOM/DSS (n = 6). Error bars represent ± SEM. P values were determined using unpaired Student's t test. n.s. non significant.
(F). Disease Activity Index (DAI) score of WT (red) and Trpm8 −/− (blue) mice treated with AOM/DSS. DAI score was measured every other day during DSS cycles, and once a week during washout periods. DAI was assessed considering stool consistency and presence of blood as reported in the text. Each dot represents the mean ± SEM of 6 mice.
Figure S2. Oncoprint visualization of clinical and genomic features of the KEGG Wnt pathway in the 268 TCGA CRC patients.
Protein abundance in both CRC and normal colo‐rectal tissues, as well as localization and druggability are reported. In the HM, red represents amplification, blue represents deletion and green dots mutations of gene. On the left the cellular localization together with the druggability are reported for each gene. On the right, a barplot recapitulating the frequency of the aberrations for each gene and the immunohistochemistry status of the tumors and the healthy controls according to the Tumor Atlas. On top, TRPM8 subgroups microsatellite status and barplot recapitulating the frequency of the aberrations for each sample.
Figure S3. Effect of WS12 on HEK‐293 cells stably expressing TRPM8.
(A). WS12 effect on calcium influx induced by ionomycin (4μM). The concentration of WS12 is expressed as log10[M]. Error bars represent ± SEM
(B). Effect of triple WS12 (0.2 μM) stimulus on induced TRPM8 gating of Ca2 + influx into TRPM8‐HEK‐293 cells. Data are expressed as a normalized response to the first stimulus (n = 5 different experiments). Error bars represent ± SEM. P‐values were determined using one way ANOVA followed by Dunnett's multiple comparisons test. * p < 0.05
TRPM8‐HEK‐293 cells response to triple WS12 stimulation.
Figure S4. RNA sequencing of Wild‐Type and Trpm8 −/− mice.
Expression of Wnt2b, Wnt3, Wnt4, Wnt5a, Wnt5b, Wnt6, Wnt7b, Wnt9a, Wnt10b, Apc, Apc2 by RNA sequencing analysis on the colon of wild Type (blue) and Trpm8 −/− (orange) mice, treated with vehicle or AOM (n = 6). P values were determined using multiple Student's t‐test. * p < 0.05.
Table S1. Patients data and TNM classification.
Table S2. Sequences for Homo sapiens primers of the genes screened by Quantitative Real‐Time Polymerase Chain Reaction.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


