Abstract
The metabolism of malignant cells differs significantly from that of healthy cells and thus, it is possible to perform metabolic imaging to reveal not only the exact location of a tumor, but also intratumoral areas of high metabolic activity. Herein, we demonstrate the feasibility of metabolic tumor imaging using signal‐enhanced 1‐13C‐pyruvate‐d3, which is rapidly enhanced via para‐hydrogen, and thus, the signal is amplified by several orders of magnitudes in less than a minute. Using as a model, human melanoma xenografts injected with signal‐enhanced 1‐13C‐pyruvate‐d3, we show that the conversion of pyruvate into lactate can be monitored along with its kinetics, which could pave the way for rapidly detecting and monitoring changes in tumor metabolism.
Keywords: hyperpolarization, NMR, metabolites, tumor, cancer
The feasibility to image cancer with MRI by rapidly enhanced 1‐13C pyruvate‐d3 using para‐hydrogen is introduced.
Introduction
Based upon the principle of nuclear magnetic resonance (NMR), magnetic resonance imaging (MRI) is a powerful imaging tool in clinical applications and medical diagnoses. [1] Although MRI is used widely, it has relatively low sensitivity. To overcome this limitation, signal enhancement – or hyperpolarization – strategies have been introduced,[ 2 , 3 , 4 , 5 , 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 , 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 ] which allow for boosting NMR signals by over 10 000‐fold compared to the normal, thermally polarized signal. [2] To investigate metabolism, the enhancement of 13C‐enriched metabolites is particularly valuable because the hyperpolarized state can typically be tracked for 2–3 minutes. [4] Regarding cancer imaging, the main and major focus over the past years has been the use of 13C‐enriched pyruvate.[ 4 , 5 , 6 , 7 ] This has been pursued exclusively with dissolution dynamic nuclear polarization (dDNP) and recently been applied to and used in clinical studies. [6] However, dDNP is a rather slow approach that requires tens of minutes to an hour to produce a dose of signal‐enhanced pyruvate as the contrast agent.[ 3 , 9 ] Thereby, the molecule of interest, e. g. 13C‐pyruvate, is cooled, together with radicals, to temperatures below 2 K using liquid helium and irradiation with microwaves. After the respective time, the contrast agent solution is rapidly thawed and used for studies. These rather long procedures can be a significant translational burden for application and use in preclinical studies and clinical studies of large patient cohorts.
A rapid method, with promise to solve the challenges of the state‐of‐the‐art technology for metabolic imaging, are para‐hydrogen‐based hyperpolarization approaches. With these, the singlet spin order of para‐hydrogen is used in a rapid reaction (seconds) to obtain hyperpolarized signals. There are two approaches to using para‐hydrogen: 1) Para‐Hydrogen Induced Polarization (PHIP) in which an unsaturated bond is hydrogenated[ 10 , 11 , 12 , 13 , 14 , 15 , 16 , 17 , 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 ] and 2) Signal Amplification by Reversible Exchange (SABRE) wherein a substrate is hyperpolarized via a temporarily stable complex comprising a catalyst and para‐hydrogen.[ 38 , 39 , 40 , 41 , 42 , 43 , 44 , 45 , 46 , 47 , 48 ] In the case of SABRE, the structure of the signal‐enhanced metabolite is not altered and pyruvate can be enhanced. However, thus far, SABRE is limited to organic solvents and has not been used yet to conduct biological experiments. [48] The idea to use PHIP for biomedical studies has been around since the early 2000s, but because of the lack of metabolically active compounds that can be reacted with para‐hydrogen, it has mainly been used for angiography experiments.[ 16 , 17 , 18 , 19 , 20 ] Given the development of catalysts, which allows trans‐hydrogenation reactions, it has recently become possible to enhance the molecule fumarate and to conduct metabolic imaging experiments in animals such as mouse.[ 20 , 21 ] To obtain a substantial amount of metabolites in a biocompatible solution, the PHIP‐SAH (PHIP by means of Side Arm Hydrogenation) approach was developed.[ 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 ] Introduced in 2015, it requires a suitable precursor of a metabolite that is hydrogenated with para‐hydrogen, followed by a subsequent spin order transfer (SOT) of the hydrogen spin order to a 13C spin of interest in the metabolite. [23] Although cleavage of the metabolite yields the signal‐enhanced metabolites, polarization in the context of the first studies was relatively low (single digit percentages) and thus lagged an order of magnitude behind dDNP.[ 23 , 49 ] However, in vivo studies of models of heart disease showed that this approach can be used to enhance the pyruvate signal and to monitor metabolism in living mice. [24] Thus far, pulsed magnetic resonance approaches have proven to yield the highest levels of signal‐enhancement with PHIP‐SAH.[ 28 , 29 , 30 , 31 , 32 , 33 , 34 , 35 , 36 , 37 ] We have recently introduced the MINERVA protocol (Maximizing Insensitive Nuclear Enhancement Reached Via para‐hydrogen Amplification) to maximize obtainable signal enhancement and demonstrated that large levels of 13C polarization similar to dDNP can be achieved (60 % 1‐13C‐ethyl pyruvate‐d6 polarization on the precursor and 28 % of 1‐13C‐pyruvate‐d3 in solution).[ 32 , 37 ] The entire MINERVA protocol includes an optimized metabolic precursor, a pulsed NMR experiment and purification as depicted in Figure 1. The approach was first tested by us in cellular models for Parkinson's disease and evidence was provided that pyruvate‐d3 can serve as a biomarker for the neurodegenerative disease. [32]
Figure 1.
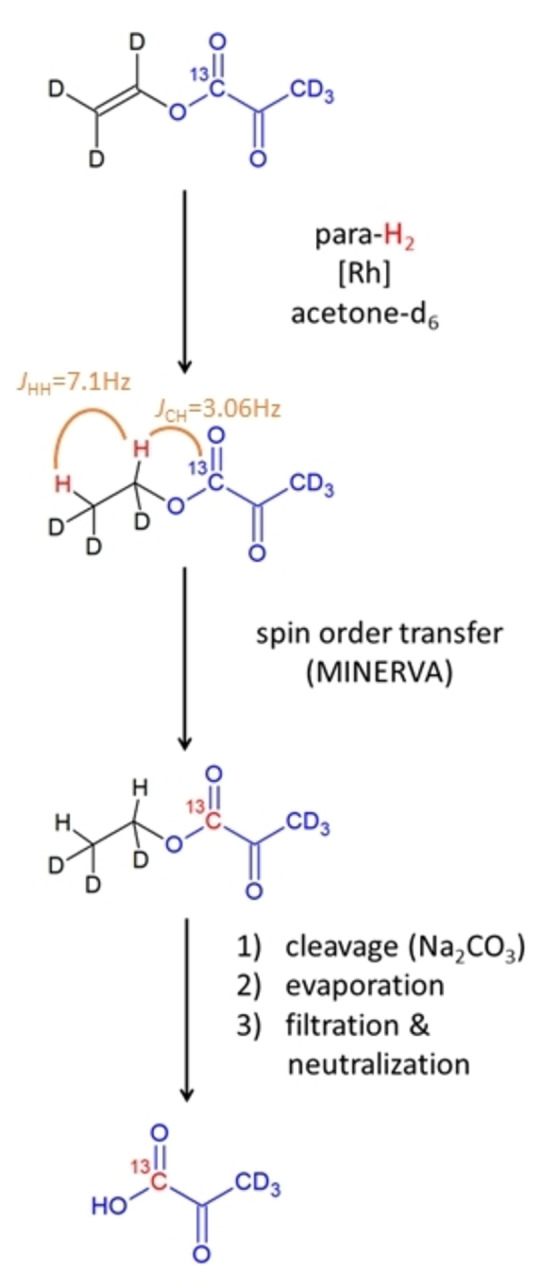
Procedure to rapidly enhance 13C‐pyruvate for metabolic tumor imaging.
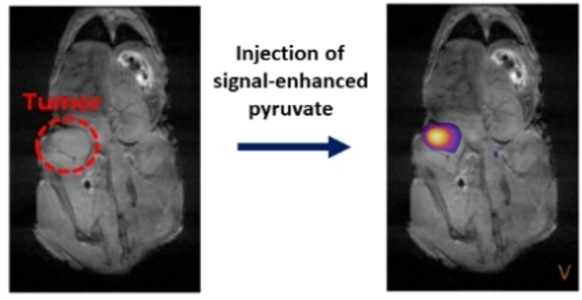
In this communication we show that pyruvate‐d3 rapidly signal enhanced via para‐hydrogen in large concentrations (50 mM) can be used for metabolic tumor imaging. In particular, we show in melanoma mouse models that pyruvate‐d3 is converted into lactate specifically in tumors and that a metabolic map of the tumor can be obtained. Moreover, we show that the flux of the pyruvate‐lactate conversion can be monitored which could act as a relevant parameter to monitor future treatments. [4] Overall, the development, described here, represent a first step towards the clinical applicability of a fast hyperpolarization approach.
Results and Discussion
To establish the enhancement level that can be obtained for 1‐13C pyruvate, we have used the pulse sequence with the respective timings displayed in Figure 2A and quantified polarization of 50 mM precursor (2B) and of pyruvate at the site after injection (2 C). The latter thereby represents the polarization that enters into the bloodstream with an injection volume of 70 μL. To this end, 50 mM vinyl pyruvate‐d6 and 13 mM commercially available catalyst ([1,4‐Bis(diphenylphosphino)butane](1,5‐cyclooctadiene) rhodium(I) tetrafluoroborate) were dissolved in acetone‐d6 and 0.2 mL transferred to a 5 mm NMR tube. The samples were degassed by bubbling N2 gas for 1 minute and transferred into the bore of the magnet. Para‐hydrogen was supplied to the solution for 20 s at a pressure of 7 bar at a temperature of 330 K. After hydrogenation and a settling time of the solution of 1 s, the 1‐13C‐carbon was hyperpolarized via the MINERVA sequence as displayed in Figure 2A. At this stage, the polarization of the precursor can be measured as in Figure 2B, and for three samples, an average polarization of 53±2 % was obtained.
Figure 2.
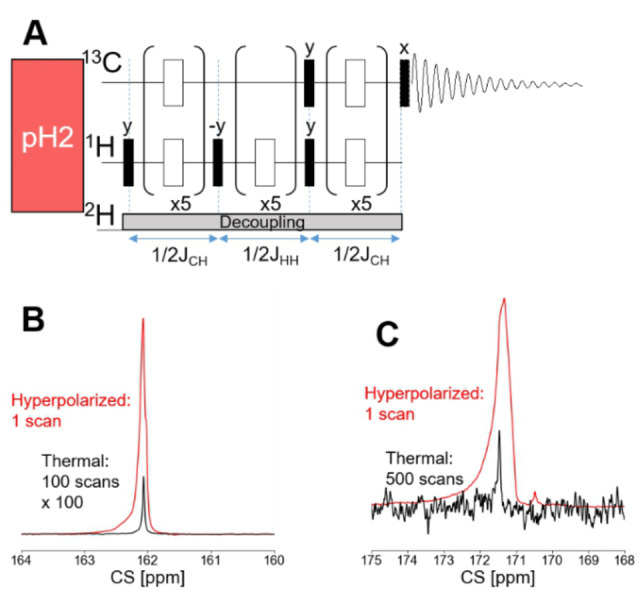
A) MINERVA pulse sequence for the SOT. The delays were calculated using the coupling values JCH=3.06 Hz and JHH=7.1 Hz. A constant z‐gradient of 0.2 G/cm during para‐hydrogen supply and 0.04 % during the pulse sequence as well as 2H decoupling were used. Black bars: 90° pulses, white bars: 180° pulses. B) Overlay of the hyperpolarized spectrum (single scan) of vinyl pyruvate (red) immediately after the pulse sequence and the thermal spectrum of the same sample (black) with 100 averages and scaled up by a factor of 100. The average 13C polarization is 53+/−2 % (standard deviation). C) Overlay of the hyperpolarized pyruvate after injection into the imaging magnet (red), acquired with an 18° pulse in one scan and the thermal spectrum of the same sample (black). The thermal spectrum was acquired after addition of 5 mM Gadobutrol, which reduced the T1 to below 1 s, using a pulse acquire sequence with 1H‐decoupling and 500 scans with 90° pulses.
We performed three separate experiments, which encompassed the procedure of hyperpolarizing, and purifying and detecting the signal enhancement following injection through a catheter into the imaging system. After the MINERVA transfer, a last 90° pulse returned the polarization into z‐axis. Cleavage of the side arm was performed using 0.075 mL of 150 mM Na2CO3 in H2O. For mixing, gaseous N2 was guided through the solution for 2 s and after a settling time of 0.5 s, the acetone was evaporated by vacuum pumping for 10 s at 323 K in a heated water bath outside of the magnet, where the catalyst precipitates. Following evaporation, the pH was adjusted to physiological conditions ∼7.4 using 0.075 mL 100 mM HEPES with a pH ∼3. The remaining solution was drawn from the NMR tube into a syringe through a filter with a pore size of 1.2 um and thereafter, transferred immediately to the imaging magnet. During this transfer, the syringe was attached to a magnetic plate to reduce relaxation losses. The obtained average polarization was 12±4 % (Figure 2C) for an injection volume of 70 μL into an NMR tube. The entire procedure, i. e. after the precursor is hyperpolarized and until the contrast agents was injected, took 67 seconds. Given a T 1 of 1‐13C‐pyruvate‐d3 of 59 s (see Figure S1), the polarization losses can be attributed mainly to relaxation effects.
Following establishment of this procedure, we used it to investigate, in vivo, subcutaneous human melanoma xenografts above the flanks in Balb/c nu/nu mice (n=3). To ensure correct positioning of each animal and with reference to anatomical details, a high‐resolution proton image was taken first. For this, a FISP sequence with a field of view of 42×30 mm2 and an image size of 140×100 mm2 were used. Fifty sections/images, with a slice thickness of 1.7 mm, were captured along the coronal plane. First, we analyzed the conversion kinetics of hyperpolarized pyruvate in the tumor‐bearing mice. For this, we conducted spectroscopic experiments with low‐flip angle excitations to obtain the pyruvate‐to‐lactate conversion rates. Fifteen seconds after the injection, a train of 18° flip angle pulses was initiated and a spectrum acquired every 4 seconds. A spectrum, depicted in Figure 3, shows that the tumor cells rapidly take up the injected pyruvate. As previously reported in the scientific literature, changes in the slopes of the curves are an indicator of treatments and thus, the obtained pyruvate to lactate conversion rate kPL=0.068+/−0.008 1/s is a valuable parameter. [4] Details on how the parameter was obtained are described in the SI supplement. We would like to note that although derivatization of pyruvate can happen easily, leading to different impurities that can be misguiding for the peak assignment, the chemical shift value of the peak at 176.5 ppm however indicates the formation of alanine. [50]
Figure 3.
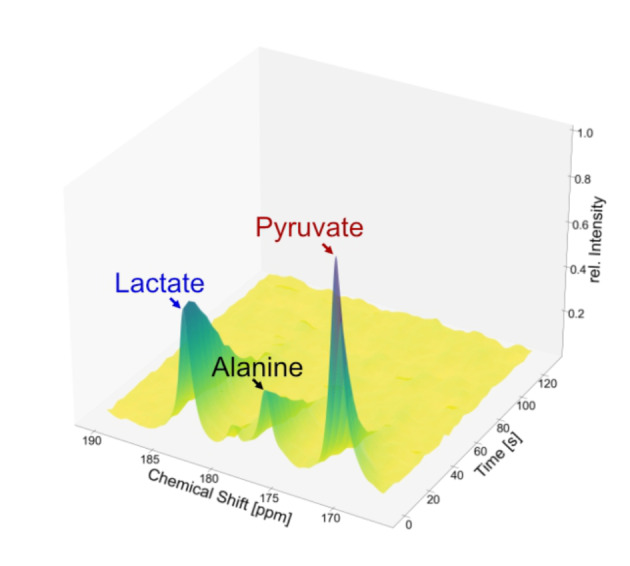
13C NMR spectra upon injection of hyperpolarized 1‐13C‐pyruvate‐d3 into a mouse. The pyruvate signal is visible at 170.9 ppm, the alanine signal at 176.5 ppm, and the lactate signal at 183.1 ppm. Between the lactate and the alanine signal, a pyruvate‐hydrate peak is detected at 179.2 ppm. Acquisition was started after 15 s, and the spectra were recorded with 18° pulses 4 s after one another. Intensities were normalized to the highest signal. The conversion rate kPL=0.068+/−0.008 1/s.
In separate experiments, 13C MRSI (magnetic resonance spectroscopic imaging) of the melanoma tumor‐bearing mice were acquired using an EPSI (echo planar spectroscopic imaging) sequence, again with coronal slice orientation. To ensure that the injection was successful, a spectrum with 10° flip angle was acquired before taking the EPSI (Figure 4A). For the EPSI, an excitation pulse with a 22° flip angle, an echo time of 2.58 ms, and a repetition time of 100 ms were used. The thickness of the slice was chosen to be 30 mm, the image size 14×10 mm2, and a field of view of 42×30 mm2 to obtain a resolution of 3×3 mm2. The spectra were acquired with the offset at 173 ppm, a spectral range of 47.9 ppm, and 100 spectral points leading to a spectral resolution of 0.47 ppm. The images of the tumor‐ bearing mice are shown in Figures 4B, 4 C and 4D. Images were captured 20 seconds after injection. According to kinetic experiments, a maximum in lactate was observed after 20 seconds. The proton image (Figure 4b) shows the location of the tumors and a spectroscopic image of pyruvate (Figure 4C) and lactate (Figure 4D) the metabolic activity of the tumors. Shown in Figure 4E is a tissue section of the tumors stained with hematoxylin and eosin (H&E) and in Figure 4F, a tissue section of the tumors that was probed with an anti‐human antibody cocktail to the three melanoma cell markers MART‐1, tyrosinase, gp100 and counterstained with fluorescent DAPI.
Figure 4.
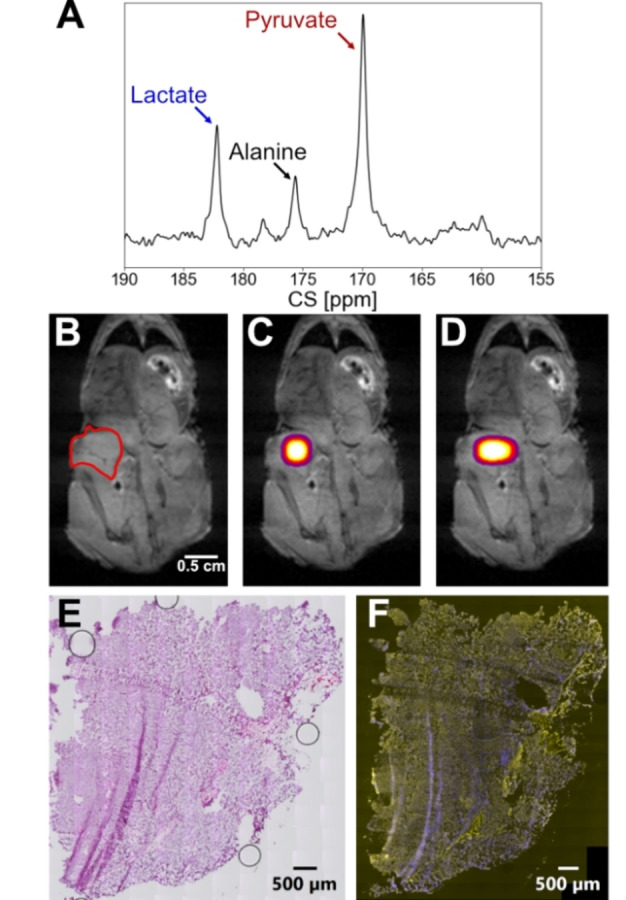
A) Single scan 13C spectrum acquired with a 10° pulse 10 seconds after the injection of hyperpolarized pyruvate. B) Anatomical 1H image with the tumor region indicated in read. C) 13C pyruvate image and D) lactate image in the tumor region. The image is acquired 20 seconds after the injection. Only data points with SNR≥3 are shown. E) Human melanoma‐xenograft tissue section stained with H&E. F) Human melanoma‐xenograft tissue section probed with a human melanoma cell marker antibody cocktail to MART‐1, tyrosinase, gp100 (pseudocolored yellow) and counterstained with fluorescent DAPI (pseudocolored blue).
Conclusion
In summary, using our rapid signal‐enhancement approach with para‐hydrogen, we have demonstrated its capacity for metabolic imaging of tumors such as human melanoma, which is the most aggressive type of skin cancer. Specifically, using spectroscopic imaging and magnetic resonance spectroscopy, we show that signal‐enhanced 1‐13C pyruvate‐d3 can be used to monitor the metabolism in tumors and to record the conversion kinetics over time. To improve the polarization procedure, automation of the process and construction of sophisticated reactors would now be needed. As we have shown, polarization losses are almost exclusively attributable to T 1 relaxation, hence a reduction of the purification and transportation time to e. g. only 30 seconds, i. e. half of T 1 of 13C‐pyruvate‐d3, could result in an overall 13C polarization of 32 % based on our reported initial polarization. In addition, we expect that an optimization of the hydrogenation procedure itself can result in even higher degrees of signal enhancement. Based upon our experimental approach and the data shown here, we believe that future investigations using this rapid approach can now be conducted to study tumors and other diseases. The rapid approach with para‐hydrogen is attractive because in vivo studies can be conducted quickly with a promising approach of quasi‐continuous contrast agent delivery and devices for polarization can also be implemented in already existing scanners for enhancing metabolites.[ 20 , 51 ] It can also be envisioned that in the future, tumor diagnostics in the clinic can be conducted, which will require upscaling of the devices to enhance signals of metabolic contrast agents and efforts to obtain regulatory approval. With respect to clinical translation it should be of note that the overall pyruvate concentration circulating through the blood stream of the mouse and assuming equal distribution is about 2 mM which is similar to estimated values in clinical trials using DNP. [52] We therefore expect that upon upscaling of the process, a high chance to demonstrate successful patient studies is given.
Experimental Section
Materials and Instruments
All chemicals were purchased from commercial suppliers. The hyperpolarization as well as the transfer experiments were carried out in a Bruker ultrashield 300 MHz (7.05 T) spectrometer. The in vivo experiments were carried out in a Bruker ultrashield 300 MHz (7.05 T) spectrometer (wide bore) equipped with a MicWB40 rf probe and Micro 2.5 WB gradient system. Para‐enriched hydrogen was obtained at 20 K with a He‐cooled para‐hydrogen generator provided by a Cryocooler system (Sumimoto HC‐4A helium compressor, Sumimoto Cold Head CH‐204 with ph2 reaction chamber by ColdEdge Technologies, temperature controller Lake Shore Cryotronics, Inc.) and delivered by a home‐built valve and tubing system.
Animal Housing
Animals used in this study were housed in a Zentrale Tierexperimentelle Einrichtung (ZTE) of the Universitätsmedizin Göttingen (UMG) Göttingen (Lower Saxony, Germany) under standard laboratory conditions in a 12 h light cycle with food and water available ad libitum.
Human Melanoma Xenografts
Carried out under approved Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit (LAVES) protocol (20/3563), the metastatic human melanoma cells WM983‐B, suspended in 0.1 ml sterile phosphate‐buffered saline, were injected subcutaneously over the right as well as the left flank of 4–5 weeks old female nude mice (CAnN.Cg‐Foxn1nu/Crl) (Charles River Laboratories Charles River GmbH & Co. KG, Sulzfeld, Germany). When the tumors had reached a size of 5 mm in any direction, the animals were anaesthetized and scanned by MRI.
Narcosis and Monitoring
For initial narcosis, a solution consisting of 4 ml 0,9 % sterile NaCl solution (B. Braun, Melsungen, Germany) 0,4 ml Medetomidine (1 mg/ml; Zoetis Deutschland GmbH, Berlin, Germany) and 0,6 ml Ketamine (50 mg/ml; Inresa Arzneimittel GmbH, Freiburg, Germany) was prepared. Animals were weighed, followed by intraperitoneal (i. p.) injection of 5 μl narcosis solution per gram body weight. After ensuring that the animal was anaesthetized, a catheter was placed into its lateral tail vein. During the time the animal remained in the NMR machine, narcosis was maintained using Isoflurane (Baxter Deutschland GmbH, Heidelberg), which was administered via a breathing mask with a Multi Delivery System (Rothacher Medical GmbH, Heitenried, Switzerland). Breathing and temperature were monitored during the procedure using a Model 1030 MR‐compatible Small Animal Monitoring and Gating System (SA Instruments Inc. New York, USA). The isoflurane level was adjusted (between 0.5 & 1.5 %) every five minutes to keep the breathing frequency of the animals between 80 and 140 Hz.
Immunohistochemistry
Following monitoring, the mice were sacrificed and the tumors resected and cryopreserved. Five‐μm tissue sections, prepared from the human melanoma xenografts, were fixed with 4 % PFA in PBS, treated with a 1X solution of TrueBlack lipofuscin autofluorescence quencher (Biotium), blocked with 10 % goat serum in PBS and probed with a mouse anti‐human antibody cocktail to the three melanoma cell markers MART‐1, tyrosinase, gp100 (Novus Biologicals). Thereafter, the tissue sections were probed with a goat anti‐mouse DyLight 755 secondary antibody (Novus Biologicals), counterstained with fluorescent DAPI, followed by addition of Fluoromount‐G mounting medium. In addition, a five‐μm tumor xenograft tissue section was stained with hematoxylin and eosin (H&E) and mounted with Entellan (Merck). Images of the tissue sections were acquired with a 20x/0.80 air objective on a SLIDEVIEW VS200 research slide scanner (Evident) and thereafter, processed with OlyVIA‐Software (Evident).
Conflict of interest
The authors declare no conflict of interest.
1.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Acknowledgments
The following funding sources are acknowledged: S. G. thanks the Max Planck Society, the Deutsche Forschungsgemeinschaft (DFG) for funding (grants 418416679, 426677227 and 450146057).This project has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (Grant agreement No. 949180). A. B. S. acknowledges funding support from the German Cancer Consortium (DKTK), and the DFG (grant numbers SCHM 3694/1‐1, SCHM 3694/2‐1, SFB 1479). We thank Dr. Belov and the chemical synthesis facility team for support in the scale up of the deuterated pyruvate precursor. The authors thank Prof. Susann Boretius and Dr. Jessica König supporting this work. We also thank Robin Paschke and Dominik Peix from the electronic workshop of the MPI‐NAT for their support with the monitoring system. Open Access funding enabled and organized by Projekt DEAL.
Hune T., Mamone S., Schroeder H., Jagtap A. P., Sternkopf S., Stevanato G., Korchak S., Fokken C., Müller C. A., Schmidt A. B., Becker D., Glöggler S., ChemPhysChem 2023, 24, e202200615.
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.
References
- 1. Brown R. W., Cheng Y. C. N., Haake E. M., Thompson M. R., Venkatesan R., Magnetic resonance imaging: physical principles and sequence design, Wiley, 2014. [Google Scholar]
- 2. Ardenkjaer-Larsen J. H., Fridlund B., Gram A., Hansson G., Hansson L., Lerche M. H., Servin R., Thanning M., Golman K., Proc. Natl. Acad. Sci. USA 2003, 100, 10158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Golman K., in't Zandt R., Thaning M., Proc. Natl. Acad. Sci. USA 2006, 103, 11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Day S. E., Kettunen M. I., Gallagher F. A., Hu D. E., Lerche M., Wolber J., Golman K., Ardenkjaer-Larsen J. H., Brindle K. M., Nat. Med. 2007, 13, 1382. [DOI] [PubMed] [Google Scholar]
- 5. Gallagher F. A., Kettunen M. I., Day S. E., Hu D. E., Ardenkjaer-Larsen J. H., in't Zandt R., Jensen P. R., Karlsson M., Golman K., Lerche M. H., Brindle K. M., Nature 2008, 453, 940. [DOI] [PubMed] [Google Scholar]
- 6. Nelson S. J., Kurhanewicz J., Vigneron D. B., Larson P. E. Z., Harzstark A. L., Ferrone M., van Criekinge M., Chang J. W., Bok R., Park I., Reed G., Carvajal L., Small E. J., Munster P., Weinberg V. K., Ardenkjaer-Larsen J. H., Chen A. P., Hurd R. E., Odegardstuen L.-I., Robb F. J., Tropp J., Murray J. A., Sci. Transl. Med. 2013, 198, 198ra108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kurhanewicz J., Vigneron D. B., Brindle K., Chekmenev E. Y., Comment A., Cunningham C. H., DeBerardinis R. J., Green G. G., Leach M. O., Rajan S. S., Rizi R. R., Ross B. D., Warren W. S., Malloy C. R., Neoplasia 2011, 13, 81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Jannin S., Dumez J.-N., Giraudeau P., Kurzbach D., J. Magn. Reson. 2019, 305, 41–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jannin S., Bornet A., Melzi R., Bodenhausen G., Chem. Phys. Lett. 2012, 549, 99–102. [Google Scholar]
- 10. Bowers C. R., Weitekamp D. P., Phys. Rev. Lett. 1986, 57, 2645. [DOI] [PubMed] [Google Scholar]
- 11. Trantzschel T., Bernarding J., Plaumann M., Lego D., Gutmann T., Ratajczyk T., Dillenberger S., Buntkowsky G., Bargon J., Bommerich U., Phys. Chem. Chem. Phys. 2012, 14, 5601. [DOI] [PubMed] [Google Scholar]
- 12. Salnikov O. G., Kovtunov K. V., Koptyug I. V., Sci. Rep. 2015, 5, 13930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shchepin R. V., Barskiy D. A., Coffey A. M., Manzanera Esteve I. V., Chekmenev E. Y., Angew. Chem. Int. Ed. 2016, 55, 6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kovtunov K. V., Barskiy D. A., Shchepin R. V., Salnikov O. G., Prosvirin I. P., Bukhtiyarov A. V., Kovtunova L. M., Bukhtiyarov V. I., Koptyug I. V., Chekmenev E. Y., Chem. Eur. J. 2016, 22, 16446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haake M., Natterer J., Bargon J., J. Am. Chem. Soc. 1996, 118, 8688. [Google Scholar]
- 16. Goldman M., Johannesson H., Axelsson O., Karlsson M., C. R. Chim. 2006, 9, 357. [Google Scholar]
- 17. Bhattacharya P., Chekmenev E. Y., Perman W. H., Harris K. C., Lin A. P., Norton V. A., Tan C. T., Ross B. D., Weitekamp D. P., J. Magn. Reson. 2007, 186, 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bhattacharya P., Chekmenev E. Y., Reynolds W. F., Wagner S., Zacharias N., Chan H. R., Bünger R., Ross B. D., NMR Biomed. 2011, 24, 1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmidt A. B., Berner S., Schimpf W., Müller C., Lickert T., Schwader-lapp N., Knecht S., Skinner J. G., Dost A., Rovedo P., Hennig J., von Elverfeldt D., Hövener J.-B., Nat. Commun. 2017, 8, 14535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmidt A. B., Berner S., Braig M., Zimmermann M., Hennig J., von Everfeldt D., Hövener J.-B., PLoS One 2018, 13, e0200141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knecht S., Blanchard J. W., Barskiy D., Cavallari E., Dagys L., Van Dyke E., Tsukanov M., Bliemel B., Münnemann K., Aime S., Reineri F., Levitt M. H., Buntkowsky G., Pines A., Blümler P., Budker D., Eills J., Proc. Natl. Acad. Sci. USA 2021, 118, e2025383118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stewart N. J., Nakano H., Sugai S., Tomohiro M., Kase Y., Uchio Y., Yamaguchi T., Matsuo Y., Naganuma T., Takeda N., Nishimura I., Hirata H., Hashimoto T., Matsumoto S., ChemPhysChem. 2021, 22, 915–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reineri F., Boi T., Aime S., Nat. Commun. 2015, 6, 5858. [DOI] [PubMed] [Google Scholar]
- 24. Cavallari E., Carrera C., Sorge M., Bonne G., Muchir A., Aime S., Reineri F., Sci. Rep. 2018, 8, 8366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cavallari E., Carrera C., Aime S., Reineri F., J. Magn. Reson. 2018, 289, 12. [DOI] [PubMed] [Google Scholar]
- 26. Cavallari E., Carrera C., Aime S., Reineri F., ChemPhysChem. 2018, 20, 318. [DOI] [PubMed] [Google Scholar]
- 27. Hövener J.-B., Pravdivtsev A. N., Kidd B., Bowers C. R., Glöggler S., Kovtunov K. V., Plaumann M., Katz-Brull R., Buckenmaier K., Jerschow A., Reineri F., Theis T., Shchepin R. V., Wagner S., Bhattacharya P., Zacharias N. M., Chekmenev E. Y., Angew. Chem. Int. Ed. 2018, 57, 11140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Korchak S., Yang S., Mamone S., Glöggler S., ChemistryOpen 2018, 7, 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Korchak S., Mamone S., Glöggler S., ChemistryOpen 2018, 7, 672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Korchak S., Emondts M., Mamone S., Blümich B., Glöggler S., Phys. Chem. Chem. Phys. 2019, 23, 22849–22856. [DOI] [PubMed] [Google Scholar]
- 31. Korchak S., Jagtap A. P., Glöggler S., Chem. Sci. 2021, 12, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ding Y., Korchak S., Mamone S., Jagtap A. P., Stevanato G., Sternkopf S., Moll D., Schroeder H., Becker S., Fischer A., Gerhardt E., Outeiro T. F., Opazo F., Griesinger C., Glöggler S., Chemistry-Methods 2022, 2, e202200023. [Google Scholar]
- 33. Dagys L., Jagtap A. P., Korchak S., Mamone S., Saul P., Levitt M. H., Glöggler S., Analyst 2021, 146, 1772–1778. [DOI] [PubMed] [Google Scholar]
- 34. Kaltschnee L., Jagtap A. P., McCormick J., Wagner S., Bouchard L.-S., Utz M., Griesinger C., Glöggler S., Chem. Eur. J. 2019, 25, 11031–11035. [DOI] [PubMed] [Google Scholar]
- 35. McCormick J., Korchak S., Mamone S., Ertas Y. N., Liu Z., Verlinsky L., Wagner S., Glöggler S., Bouchard L.-S., Angew. Chem. Int. Ed. 2018, 130, 10852–10856. [DOI] [PubMed] [Google Scholar]
- 36. Schmidt A. B., Brahms A., Ellermann F., Knecht S., Berner S., Hennig J., von Elverfeldt D., Herges R., Hövener J.-B., Pravdivtsev A. N., Phys. Chem. Chem. Phys. 2021, 23, 26645–26652. [DOI] [PubMed] [Google Scholar]
- 37. Mamone S., Jagtap A. P., Korchak S., Ding Y., Sternkopf S., Glöggler S., Angew. Chem. Int. Ed. 2022, 61, e202206298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Adams R. W., Aguilar J. A., Atkinson K. D., Cowley M. J., Elliott P. I. P., Duckett S. B., Green G. G. R., Khazal I. G., Lopez-Serrano J., Williamson D. C., Science 2009, 323, 1708. [DOI] [PubMed] [Google Scholar]
- 39. Rayner P. J., Burns M. J., Olaru A. M., Norcott P., Fekete M., Green G. G. R., Highton L. A. R., Mewis R. E., Duckett S. B., Proc. Natl. Acad. Sci. USA 2017, 114, E3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barskiy D. A., Kovtunov K. V., Koptyug I. V., He P., Groome K. A., Best Q. A., Shi F., Goodson B. M., Shchepin R. V., Coffey A. M., Waddell K. W., Chekmenev E. Y., J. Am. Chem. Soc. 2014, 136, 3322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suefke M., Lehmkuhl S., Liebisch S. A., Blümich B., Appelt S., Nat. Phys. 2017, 13, 568. [Google Scholar]
- 42. Eshuis N., Hermkens N., van Weerdenburg B. J. A., Feiters M. C., Rutjes F. P. J., Wijmenga S. S., Tessari M., J. Am. Chem. Soc. 2014, 136, 2695. [DOI] [PubMed] [Google Scholar]
- 43. Theis T., Truong M. L., Coffey A. M., Shchepin R. V., Waddell K. W., Shi F., Goodson B. M., Warren W. S., Chekmenev E. Y., J. Am. Chem. Soc. 2015, 137, 1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Spannring P., Reile I., Emondts M., Schleker P. P. M., Hermkens N. K. J., van der Zwaluw N. G. J., van Weerdenburg B. J. A., Tinnemans P., Tessari M., Blümich B., Rutjes F. P. J. T., Feiters M. C., Chem. Eur. J. 2016, 22, 9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Truong M. L., Shi F., He P., Yuan B., Plunkett K. N., Coffey A. M., Shchepin R. V., Barskiy D. A., Kovtunov K. V., Koptyug I. V., Waddell K. W., Goodson B. M., Chekmenev E. Y., J. Phys. Chem. B 2014, 118, 13882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Colell J. F. P., Emondts M., Logan A. W. J., Shen K., Bae J., Shchepin R. V., G. X. Ortiz, Jr. , Spannring P., Wange Q., Malcolmson S. J., Chekmenev E. Y., Feiters M. C., Rutjes F. P. J. T., Blümich B., Theis T., Warren W. S., J. Am. Chem. Soc. 2017, 139, 7761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Iali W., Roy S. S., Tickner B. J., Ahwal F., Kennerley A. J., Duckett S. B., Angew. Chem. Int. Ed. 2019, 58, 10271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. TomHon P., Abdulmojeed M., Adelabu I., Nantogma S., Kabir M. S. H., Lehmkuhl S., Chekmenev E. Y., Theis T., J. Am. Chem. Soc. 2022, 144, 282–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Carrera C., Cavallari E., Digilio G., Bondar O., Aime S., Reineri F., ChemPhysChem 2021, 22, 1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harris T., Gamliel A., Sosna J., Gomori J. M., Katz-Brull R., Appl. Magn. Reson. 2018, 49, 1085. [Google Scholar]
- 51. Schmidt A. B., Zimmermann M., Berner S., de Massin H., Müller C. A., Ivantaev V., Hennig J., Elverfeldt D. v., Hövener J.-B., Comm. Chem. 2022, 5, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tran M., Latifoltojar A., Neves J. B., Papoutsaki M.-V., Gong F., Comment A., Costa A. S. H., Glaser M., Tran-Dang M.-A., El Sheikh S., Piga W., Bainbridge A., Barnes A., Young T., Jeraj H., Awais R., Adeleke S., Holt C., O'Callaghan J., Twyman F., Atkinson D., Frezza C., Arstad E., Gadian D., Emberton M., Punwani S., BJR Case Rep. 2019, 5, 2019003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Supporting Information
Data Availability Statement
The data that support the findings of this study are available in the supplementary material of this article.


