Abstract
Background
High-level BK polyomavirus (BKPyV) replication in allogeneic hematopoietic cell transplantation (HCT) predicts failing immune control and BKPyV-associated hemorrhagic cystitis.
Methods
To identify molecular markers of BKPyV replication and disease, we scrutinized BKPyV DNA-loads in longitudinal urine and plasma pairs from 20 HCT patients using quantitative nucleic acid testing (QNAT), DNase-I treatment prior to QNAT, next-generation sequencing (NGS), and tested cell-mediated immunity.
Results
We found that larger QNAT amplicons led to under-quantification and false-negatives results (P < .001). DNase-I reduced urine and plasma BKPyV-loads by >90% (P < .001), indicating non-encapsidated BKPyV genomes. DNase-resistant urine BKPyV-loads remained infectious in cell culture. BKPyV genome fragmentation of ≤250 bp impaired NGS coverage of genetic variation using 1000-bp and 5000-bp amplicons. Conversely, 250-bp amplicons captured viral minority variants. We identified genotype-specific and genotype-independent changes in capsid Vp1 or T-antigen predicted to escape from antibody neutralization or cytotoxic CD8 T-cells, respectively. Genotype-specific changes in immunodominant 9mers were associated with reduced or absent CD8 T-cell responses. Thus, failure to control BKPyV replication in HCT Patients may involve insufficient genotype-specific cytotoxic CD8 T-cell responses, potentially predictable by low neutralizing antibodies as well as genotype-independent immune escape.
Conclusions
Our results provide new insights for patient evaluation and for designing immune protection through neutralizing antibodies, adoptive T-cell therapy, or vaccines.
Keywords: BK polyomavirus, BKPyV, hemorrhagic cystitis, hematopoietic cell transplantation, HCT, T cell, CD8, epitope, LTag, Vp1, neutralizing antibody, immune escape
BKPyV genome loads in allogeneic hematopoietic cell transplantation are DNase-I sensitive, nonencapsidated DNA fragments of ≤250 bp. This can impair detection of BKPyV diversity by next-generation sequencing and specifically genotype-associated changes mediating BKPyV immune escape from cytotoxic CD8 T-cell killing.
BK polyomavirus (BKPyV) complicates 5%–25% of adult allogeneic hematopoietic cell transplantations (HCT) [1–3]. While asymptomatic, low-level BKPyV replication with urinary shedding of <10 000 genome equivalent copies (c)/mL is common in healthy blood donors [4], frequencies increase from 10% to 80% in patients undergoing HCT [5, 6]. In parallel, urine BKPyV-loads increase dramatically from <1000 c/mL to >10 million c/mL [7], in line with conditioning and immunosuppression ablating local and systemic immune control [8, 9]. In HCT patients exposed to bladder-urotoxic conditioning, sustained high-level BKPyV replication in urothelial cells can lead to denudation of the bladder epithelium with urine leakage and painful inflammation, the hallmarks of BKPyV-associated hemorrhagic cystitis (BKPyV-HC) [10, 11]. Despite the pathophysiology of BKPyV-HC being still incompletely understood and partly confounded by different case definitions, diagnostic procedures, and spontaneous resolution, repeatedly reported risk factors include conditioning with cyclophosphamide, unrelated or HLA-mismatched donors, and graft-versus-host disease (GVHD) [2, 3]. Low or absent T-cell immunity is thought to increase the risk and prolong the course of BKPyV disease [9, 12, 13]. Viral virulence determinants include mutations and rearrangements of the viral genome, including those in the noncoding control region (NCCR) increasing replicative fitness [14–16], as well as amino acid exchanges permitting escape from neutralizing antibodies (nAbs) and cytotoxic CD8 T-cells. BKPyV-specific nAbs target the serotype-specific outer loop domains of the major capsid protein Vp1 encoded in the late viral gene region (LVGR) [17]. BKPyV-specific cytotoxic CD8 T-cells are preferentially directed against the small tumor antigen (sTag) and large tumor antigen (LTag) encoded in the early viral gene region (EVGR) [18, 19], and recognize closely spaced clusters of immunodominant 9mer epitopes presented across common HLA class I types [20, 21]. 9mer-specific CD8 T-cell responses correlate with clearance of BKPyV replication [21]. Single immunodominant 9mers are presented by several HLA types such as HLA-B7, HLA-B8, and HLA-B51 linked to protection from BKPyV replication [22, 23]. Analyzing publicly available BKPyV genome sequences has revealed non-synonymous amino acid exchanges in immunodominant LTag 9mers, which significantly reduced HLA-A/HLA-B binding and reduced or abrogated 9mer-specific CD8 T-cell responses [24]. Based on these findings, we conducted a detailed molecular characterization of urine and plasma BKPyV loads in our allogeneic HCT patients [25].
METHODS
Patients and Eligibility Criteria
We searched the laboratory database of Clinical Virology, University Hospital Basel, for patients with urine BKPyV-QNATs between January 2019 and July 2021 (Figure 1A ). Eligible patients were (1) aged ≥ 18 years, (2) HCT, and (3) had ≥2 consecutive BKPyV-positive urine samples. Exclusion criteria were incomplete information about patient history and other important findings or leading diagnoses such as viral infections (eg, herpes simplex virus, cytomegalovirus, adenovirus), bacterial infection, GVHD, or metastatic neoplasia [2]. We excluded patients with GVHD as its onset and treatment may potentially confound associations assessed in this study.
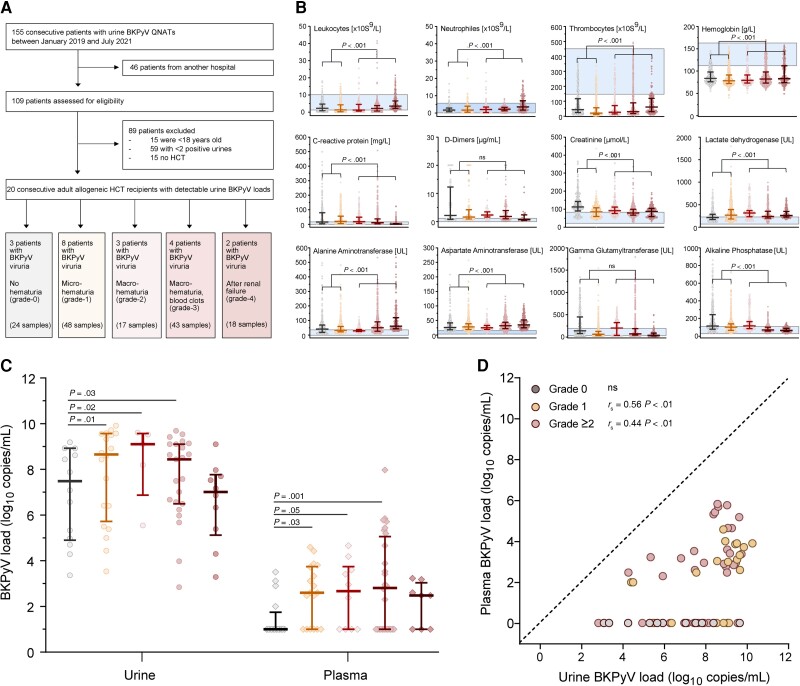
Figure 1.
Patients' characteristics. The epidemiology and patients' demographics are displayed in Supplementary Table 1. Patients provided longitudinal plasma (total n = 73) and urine samples (total n = 77) after HCT. Samples from patients without hematuria are displayed in grey, with microscopic (grade 1) hematuria in yellow, and hematuria grade ≥2 in shades of red according to severity. A, Flow diagram of the study. B, Cellular and blood chemistry parameters of the 20 HCT patients at the onset of BKPyV replication (± 7 d; median, 25th and 75th percentiles, minimum, maximum). Blue area indicates the reference values of the parameters in the general healthy population (P value by Mann-Whitney U test). C, Urine and plasma BKPyV loads of the 20 HCT patients by hematuria grade (median, 25th and 75th percentiles, minimum, maximum; P value by Mann-Whitney U test). D, Time-matched urine and plasma samples of the 20 HCT patients (n = 72; dashed line indicates 100% agreement level, P value by Spearman rank correlation). Abbreviations: BKPyV, BK polyomavirus; HCT, hematopoietic cell transplantation; ns, not significant; QNAT, quantitative nucleic acid testing.
We performed a detailed chart review to collect demographic, clinical, and laboratory data from eligible patients (Supplementary Table 1). Among the 155 patients assessed for eligibility, 46 (30%) were patients from another affiliated hospital. Of the remaining 109 patients, 89 (57%) met 1 or more exclusion criteria (Supplementary Table 2), leaving 20 for the in-depth study (Supplementary Table 1). Patients provided longitudinal plasma (total n = 73) and urine samples (total n = 77) after HCT (range, 1 to 495 days), with a median of 3 urine and 3 plasma samples per patient (Supplementary Table 1).
Diagnosis of BKPyV-HC required the triad of (1) clinical signs of cystitis such as dysuria, lower abdominal pain, urge, and/or frequency; (2) macroscopic hematuria (≥grade 2); and (3) high-level BKPyV viruria defined as >7 log10 c/mL [2]. The severity of hematuria was graded as microscopic (grade 1), macroscopic (grade 2), macroscopic with clots (grade 3), or macroscopic with clots and after renal failure secondary to urinary tract obstruction (grade 4) [2, 26].
The study was conducted according to good laboratory practice and in accordance with the Declaration of Helsinki and national and institutional standards for laboratory quality control and was approved by the Ethical Committee of North-Western and Central Switzerland (EKNZ 2020-00769).
DNase Digestion, QNAT Assays, and Amplicon-Based Next-Generation Sequencing
DNase-I treatment was done prior to DNA extraction as described previously [27, 28]. Following DNA extraction, the pangenotype BKPyV-QNATs LTag-(3.1) 88 bp, 133 bp, and 239 bp were performed to quantify BKPyV DNA-loads. Cell-free human genomic DNA was quantified using aspartoacylase gene QNAT [27]. Details on the amplicon-based next-generation sequencing (NGS) approach are provided in the Supplementary Methods.
BKPyV Culture From Urine and Plasma
Urine and plasma were either treated with DNase-I as described [27] or left untreated, and then compared for BKPyV isolation using COS-7 cell culture (details are provided in the Supplementary Methods). Supernatant BKPyV-loads and immunofluorescence staining was done at 6 days postinfection as described previously [29].
Characterization of BKPyV-Specific CD8 T-Cell Responses
The Immune Epitope Database analysis resource tool was used to predict the binding score of genotype-specific or variant immunodominant LTag-9mer T-cell epitopes [20–22].
For functional characterization of BKPyV-specific CD8 T-cell responses, CD14+ cells were isolated from peripheral blood mononuclear cells (PBMCs) of 2 healthy BKPyV IgG-seropositive blood donors (blood donor No. 1, HLA-A1/24, HLA-B8/B51; blood donor No. 2, HLA-A11/23, HLA-B8/B49) and differentiated into mature monocyte-derived dendritic cells (mMo-DCs) [25]. mMo-DCs were pulsed with 1 µg/mL of overlapping LTag-27mer pools [25], reflecting genotype I (27mP-gI) and genotype IV (27mP-gIV), for 4 hours and cocultured with autologous CD14− cells for 9 days. On day 4, interleukin 2 (IL-2; 10 ng/mL; PeproTech) and IL-7 (5 ng/mL; PeproTech) were added. After expansion, cells were restimulated with a pool of immunodominant BKPyV genotype I or genotype IV LTag-9mers (9 mP-gI and 9 mP-gIV); or with single-genotype I LTag-9mers (TTKEKAQIL and LTRDPYHTI) or genotype IV LTag-9mers (TTKEKALIL and LTRDPYYII). Intracellular interferon-γ (IFN-γ) production by CD4 and CD8 T cells was measured by flow cytometry.
Flow Cytometry Analysis
Expanded cells were rechallenged with LTag-derived peptides for 6 hours in the presence of Golgi-Stop (Becton Dickinson). Medium alone served as negative control and Staphylococcus enterotoxin B (3 µg/mL; Sigma Aldrich) as positive control. CD4 (Becton Dickinson) and CD8 (BioLegend) antibodies were used for extracellular staining. For intracellular staining, IFN-y antibody (Becton Dickinson) was used. Read-out was done on a Fortessa cytometer (Becton Dickinson) and analyzed using FlowJo version 10.8.1.
BKPyV and JC Polyomavirus Serology Analysis
BKPyV genotype I and IV as well as JC polyomavirus (JCPyV) IgG antibodies levels were determined in plasma samples of 2 healthy blood donors by a standard enzyme-linked immunosorbent assay (ELISA) using Vp1 virus-like particles purified from baculovirus expression systems [30]. Plasma samples were diluted 1:200, and a cutoff of OD492 > 0.100 defined a reactive serologic response.
Statistical Analysis
All statistical data analysis was done in R version 3.6.1 (https://cran.r-project.org), and Prism version 8 (Graphpad Software) was used for data visualization. Details on the bioinformatics approach are provided in the Supplementary Material.
RESULTS
Among 109 consecutive patients with BKPyV diagnosed between January 2019 and July 2021, we identified 20 (18%) matching the study criteria (Figure 1A and Supplementary Table 1). The HCT patients presented with high-level BKPyV viruria defined as >10 million c/mL (>7 log10) at a median of 38 days after HCT (interquartile range [IQR], 28–49 days) and median 21 days after engraftment (IQR, 13–28 days). High-level BKPyV viruria lasted for median 65 days after onset (IQR, 53–106 days). Three out of 20 (15%) patients had urine BKPyV-loads without cystitis and hematuria (grade 0), 8 (40%) patients had clinical signs of cystitis and high-level BKPyV replication with microhematuria only (grade 1), and 9 (45%) patients were diagnosed with BKPyV-HC with grade ≥ 2 hematuria (Figure 1A ). Cidofovir was not administered. In BKPyV-HC patients, routine laboratory analyses were notable for low platelets and hemoglobin levels, higher neutrophils (Figure 1B), elevated C-reactive protein and D-dimer concentrations, and borderline or elevated serum-creatinine and enzyme levels such as lactate dehydrogenase (LDH), aspartate aminotransferase (ASAT), and alanine transaminase (ALAT).
While all 20 patients showed peak urine BKPyV-loads >7 log10 c/mL, 15 out of 17 (88%) patients with hematuria showed detectable BKPyV in plasma with a median viral load of 3.2 log10 c/mL (IQR, 2.8–3.9 log10 c/mL). Urine and plasma BKPyV-loads tended to be higher in patients with BKPyV-HC (Figure 1C ). When analyzing 72 time-matched urine and plasma pairs from these 20 HCT patients, BKPyV was detectable in all urine samples at median 5.0 log10 c/mL higher levels (IQR, 3.7–6.2 log10 c/mL; Figure 1D ). Plasma BKPyV-loads were undetectable in 33 pairs (46%) and detectable in 39 pairs (56%; > 1000 c/mL in 25, 35%). The results were in line with the reported lower sensitivity, but higher specificity of plasma BKPyV-loads, while the oppositive was true for urine BKPyV-loads of >10 million c/mL.
To assess the BKPyV load in longitudinal urine (n = 77) and plasma (n = 73) samples, we used 3 previously validated BKPyV-QNATs with different amplicon size (88 bp, 133 bp, and 239 bp) [27]. Significantly higher BKPyV-loads were obtained with the 88-bp and 133-bp QNATs compared to the 239-bp QNAT (P < .001; Figure 2A ). The impact of amplicon length was consistent and led to substantial underquantification of 2.5 log10 c/mL with the 239-bp compared to the 88-bp QNAT in both urine and plasma samples, and almost doubled the fraction of undetectable plasma samples to 49 (67%).
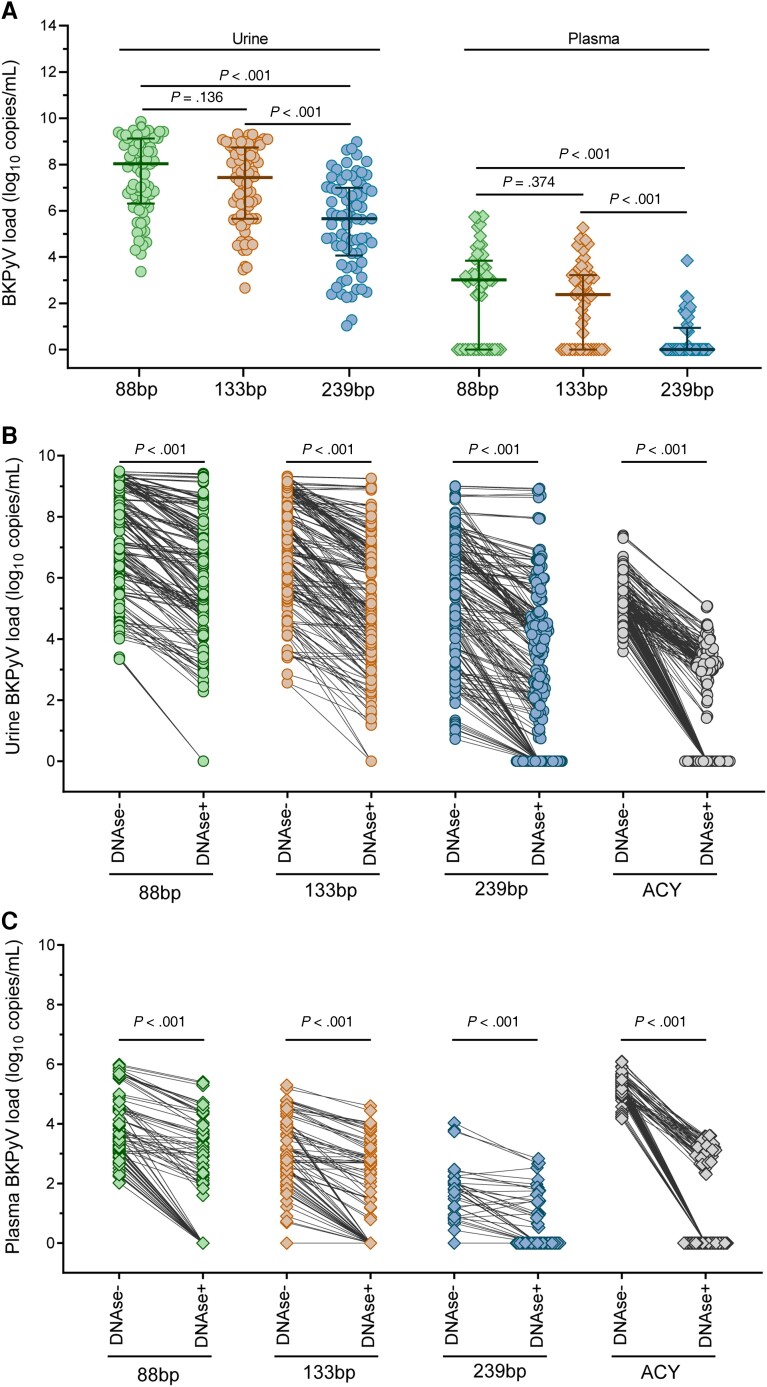
Figure 2.
Assessment of BKPyV genome load, DNase protection, and amplicon length. BKPyV loads were retrospectively analyzed in longitudinal urine (n = 77) and plasma (n = 73) samples from 20 HCT patients by 3 QNATs with different amplicon size (88 bp, 133 bp, and 239 bp). DNase I sensitivity of urine and plasma BKPyV genome loads as well as of cell-free human genomic DNA was assessed by DNAse I digestion prior to nucleic acid extraction, followed by BKPyV and ACY QNAT. Statistical comparison of nonparametric data was done using Mann-Whitney U test. A, Impact of BKPyV DNA fragment size on retrospective urine and plasma BKPyV load quantification (median, 25th and 75th percentiles). B, DNase I sensitivity of urine BKPyV loads and cell-free human genomic DNA. C, DNase I sensitivity of plasma BKPyV loads and cell-free human genomic DNA. Abbreviations: ACY, aspartoacylase; BKPyV, BK polyomavirus; HCT, hematopoietic cell transplantation; QNAT, quantitative nucleic acid testing.
Because nonencapsidated BKPyV genome fragmentation has been demonstrated as a major reason for aggravating the impact of amplicon size on BKPyV-loads [27], we compared BKPyV-loads without and with DNase treatment prior to nucleic acid extraction (Figure 2). The results indicated that DNase digestion significantly reduced urine BKPyV loads by approximately 90% in all QNATs (P < .001). Specifically, DNase digestion decreased urine BKPyV loads by >2.0 log10 c/mL in 26 (33%) urine samples as determined by the 88-bp QNAT (Figure 3A ). Likewise, larger amplicon sizes and DNase digestion decreased plasma BKPyV-load quantification and led to a higher proportion of undetectable BKPyV (88 bp, n = 27, 37%; 133 bp, n = 29, 40%; 239 bp, n = 52, 71%; Figure 2A–2C and Figure 3B ).
Figure 3.
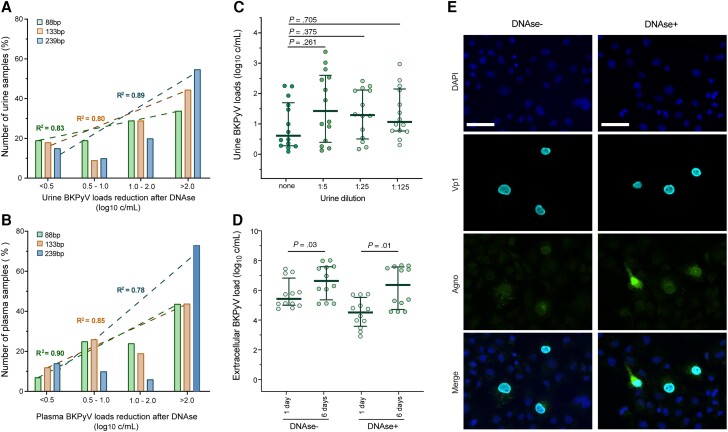
Assessment of urine and plasma BKPyV load reduction after DNase I digestion. A, Reduction in urine BKPyV load after DNase I digestion. Linear regression analysis for 88-bp, 133-bp, and 239-bp QNATs. B, Reduction in plasma BKPyV load after DNase I digestion. Linear regression analysis for 88-bp, 133-bp, and 239-bp QNATs. C, Serial dilution and DNase I treatment of urines without significant reduction in urine BKPyV loads as determined by 88-bp QNAT in 15 urines with <0.5 log10 c/mL from (A); median, 25th and 75th percentiles; Mann-Whitney U test. D, BKPyV culture from untreated (DNase−) or DNase I treated (DNase+) urine samples in COS-7 cells. BKPyV loads were determined at 1 day and 6 days postinfection with the 88-bp QNAT (median, 25th and 75th percentiles; Mann-Whitney U test). E, Immunofluorescence staining of BKPyV culture from untreated (DNase−) or DNase I treated (DNase+) urine samples in COS-7 cells at 6 days postinfection. DAPI staining of the COS-7 cell nucleus (blue), BKPyV-specific viral Vp1 capsid (cyan), and agno protein (green). Scale bars represent 50 µm. Abbreviations: BKPyV, BK polyomavirus; DAPI, 4′,6-diamidino-2-phenylindole; QNAT, quantitative nucleic acid testing.
However, 15 (19%) urine samples with median BKPyV-loads of 9.2 log10 c/mL (IQR, 8.5–9.7 log10 c/mL) showed no significant decrease in BKPyV-load (<0.5 log10 c/mL) even though cell-free human DNA loads declined. To exclude saturation of DNase activity by high BKPyV-loads, serial dilution of the 15 urine samples was performed prior to DNase digestion, nucleic acid extraction, and QNAT (Figure 3C ). The results showed no significant decrease suggesting that BKPyV-DNA may be largely protected inside virions in line with early electron microscopy studies [31]. We therefore assessed the infectiousness of urine BKPyV-loads without and with prior DNase treatment using cell culture [16]. Urine BKPyV-loads were significantly reduced upon DNase treatment at a median of 0.9 log10 c/mL at 1 day postinfection (P < .01; Figure 3D ). After 6 days of cell culture, a significant BKPyV-load increase of 1.0 log10 c/mL for DNase-untreated and 1.6 log10 c/mL for DNase-treated urine samples was observed (P < .05; Figure 3D ), indicating that DNase-protected urine BKPyV genomes corresponded to infectious BKPyV. This finding was further strengthened by immunofluorescence staining of the BKPyV-specific Vp1 viral capsid and agno protein (Figure 3E ). In contrast, BKPyV isolation by cell culture from plasma samples failed despite high viral loads of 6.0 log10 c/mL. Taken together, the results identify genomic DNA fragments as the major form of urine and plasma BKPyV-loads, and recognize DNase-resistant BKPyV-loads in urine as infectious units.
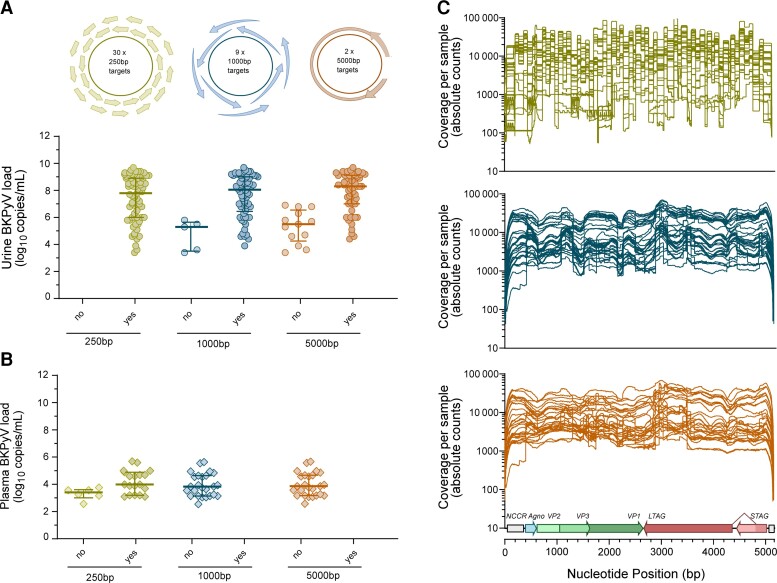
To investigate the impact of these findings on the molecular assessment of BKPyV genome diversity by NGS, we compared the yield of 30 amplicon targets of 250 bp, 9 amplicons of 1000 bp, and 2 amplicons of 5000 bp covering the BKPyV 5.1-kb genome (Figure 4A ). For urine, 25-bp NGS yielded complete BKPyV genome sequences for all 77 samples, while 1000-bp and 5000-bp nucleic acid testing (NAT) yielded sequences in 74 (96%) and 63 (82%) samples, when BKPyV loads were above 10 million c/mL (Figure 4A ). For plasma, only 29 of 73 (40%) samples had detectable BKPyV-loads, and 15 of 29 (52%) samples yielded complete BKPyV genome sequence with 250-bp NGS, while 1000-bp and 5000-bp NGS yielded no amplicons (Figure 4B ). Taken together, this corresponds well with the documented presence of both unprotected DNA genome fragments and protected full-length DNA genomes in urine, and highly fragmented BKPyV DNA in plasma, allowing assessment of BKPyV sequence diversity only with small 250-bp amplicons.
Figure 4.
NGS coverage in urine and plasma samples. NGS was performed from longitudinal urine (n = 77) and plasma samples (n = 29) from the 20 hematopoietic cell transplantation patients using amplicons with different lengths (250 bp, 1000 bp, and 5000 bp). A, Targeted NGS from urine using different amplicon targets. B, Targeted NGS from plasma using different amplicon targets. C, NGS coverage of full BKPyV genome sequences from urine samples using 250-bp, 1000-bp, and 5000-bp targets (n = 63). Abbreviations: BKPyV, BK polyomavirus; NGS, next-generation sequencing; no, no target amplicon present; yes, target amplicon present.
NGS coverage of full BKPyV genome sequences was high (mean coverage >1000) with increasing tendency toward smaller 250-bp amplicons (Figure 4C ). As expected, full-genome NGS coverage showed intersample variation, and was higher for gene sequences in the EVGR and LVGR compared to the nonstructural NCCR known to display higher sequence variability (Figure 4C ).
The NCCRs of urine genomes showed no rearrangements but only the linear archetype architecture of O-P-Q-R-S in all 20 patients. No nucleotide exchanges were found in the group 1 DNA binding sites of SP1-4 and ETS2 known to activate EVGR expression of archetype NCCRs similar to NCCR rearrangements.
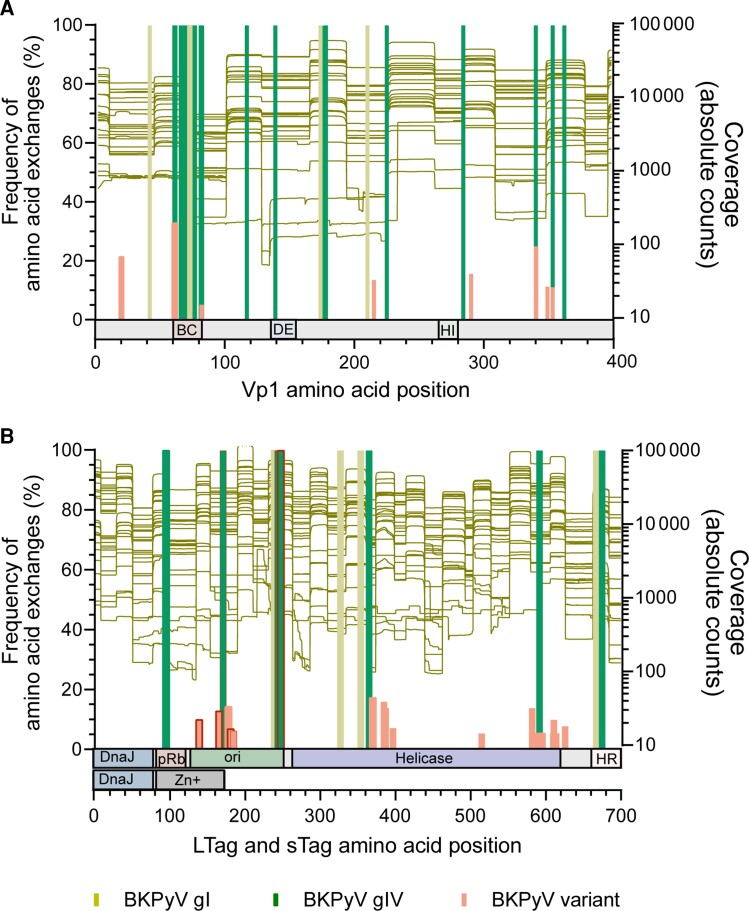
BKPyV genome diversity was identified as genotype-specific amino acid exchanges in all 20 HCT patients with BKPyV subtype Ib-2 (n = 8; 40%), Ic (n = 8; 40%), Ia (n = 2; 10%), and IVc-2 (n = 2; 10%) and was consistent in all 3 amplicon sizes where available. Importantly, genotypes determined by amino acid exchanges in Vp1 matched the genotype sequences in LTag in all urine and plasma samples from the same patient and did not change over the study period. However, minority species were identified solely with 250-bp NGS and represented genotype-independent BKPyV variants. Seven Vp1 minority variants were present in both urine and plasma, and 1 case of R215S was only identified in plasma. Moreover, we identified a hotspot of BKPyV genotype I- and IV-specific as well as variant-associated amino acid exchanges in the BC loop of Vp1 at positions 61 to 82, previously associated with escape from nAbs (Figure 5A ). For LTag, we identified hotspots of genotype-specific and variant-associated changes in the C- and N-terminus of the DNA replication binding domain (ori), the C-terminus of the helicase, and in the N-terminus of the host range domain where amino acid exchanges clustered (Figure 5B ). BKPyV variants were located in the ori and the helicase domain, while none were found in the DnaJ homology region shared between LTag and sTag.
Figure 5.
Variation in immunogenic Vp1 and LTag epitopes determined by NGS. Proportion of sequences with BKPyV genotype-specific and variant (genotype-independent) amino acid exchanges in viral capsid Vp1 and the LTag protein compared to the archetype WW [32] reference sequence (BKPyV subtype Ib-1; accession No. AB211371.1) as determined by 250-bp NGS (left y-axis). Vp1 and LTag target region coverage of 250-bp NGS (right y-axis). A, Frequency of amino acid exchanges in the Vp1 protein. The external BC, DE, and HI loops in Vp1 are indicated below the diagram. B, Frequency of amino acid exchanges in the LTag and sTag proteins as determined by 250-bp NGS. The respective domains in LTag and sTag (according to DeCaprio et al [33]) are indicated below the diagram. Of note, the amino-terminal part including the DnaJ homology domain is identical for sTag and LTag, whereas the carboxyterminal domains indicated are specific for sTag or LTag. Changes in immunodominant 9mer T-cell epitopes with significant impact on HLA-A/HLA-B-binding scores are indicated by a red frame. Abbreviations: BKPyV, BK polyomavirus; DnaJ, DnaJ homology region; gI, genotype I; gIV, genotype IV; HR, host range domain; LTag, large tumor antigen; NGS, next-generation sequencing; ori, origin of DNA replication binding domain; pRb, retinoblastoma protein binding domain.
To investigate potential immunological consequences of the amino acid exchanges on the LTag-specific cytotoxic CD8 T-cell responses, we applied the Immune Epitope Database tool to predict changes in the binding score of LTag-9mer epitopes [21]. Using wild-type BKPyV genotype Ib-1 as reference, we found genotype-specific amino acid exchanges at 11 individual LTag positions, of which 5 (45%) occurred in previously identified immunodominant 9mer epitopes. Genotype-independent amino acid exchanges were identified at 18 LTag positions of BKPyV variants, of which 7 (39%) affected immunodominant 9mer epitopes. Several of these amino acid exchanges were associated with a significant change in HLA-A and HLA-B binding (Table 1).
Table 1.
Amino Acid Changes in Immunodominant 9mer Epitopes of BKPyV Wild-Type Reference (Genotype Ib-1) in Genotype-Specific and Genotype-Independent Large T Antigen and the Predicted Effect on HLA-A/HLA-B Binding Scores
| BKPyV Genotype Ib-1 Reference 9mer Amino Acida | 9mer Amino Acid Starting Positionb | HLA Type | HLA-A/HLA-B Binding Scorec | BKPyV 9mer Amino Acid Sequence | Type | HLA Type | HLA-A/HLA-B Binding Scorec | Change HLA-A/HLA-B Binding Score (n, %) | |
|---|---|---|---|---|---|---|---|---|---|
| DPKDFPSDL | 136 | HLA-B*51:01 | 0.3471 | DPK N FPSDL | Variant (genotype independent) | HLA-B*51:01 | 0.2536↓ | −0.0935 | (−27%) |
| HLA-B*08:01 | 0.3197 | HLA-B*08:01 | 0.3492 | 0.0295 | (9%) | ||||
| HLA-B*07:02 | 0.2099 | HLA-B*07:02 | 0.1908 | −0.0191 | (−9%) | ||||
| HLA-A*24:02 | 0.0003 | HLA-A*24:02 | 0.0002 | ||||||
| TTKEKAQIL | 165 | HLA-B*51:01 | 0.0280 | TTKEKA L IL | Genotype IV specific | HLA-B*51:01 | 0.0350 | 0.007 | (25%) |
| HLA-B*08:01 | 0.7030 | HLA-B*08:01 | 0.6460↓ | −0.057 | (−8%) | ||||
| HLA-B*07:02 | 0.0350 | HLA-B*07:02 | 0.0490 | 0.014 | (40%) | ||||
| HLA-A*24:02 | 0.0050 | HLA-A*24:02 | 0.0080 | 0.003 | (60%) | ||||
| TTKEKAQIL | 165 | HLA-B*51:01 | 0.0321 | TT Q EKAQIL | Variant (genotype independent) | HLA-B*51:01 | 0.0278 | −0.0043 | (−13%) |
| HLA-B*08:01 | 0.7233 | HLA-B*08:01 | 0.2580↓ | −0.4653 | (−64%) | ||||
| HLA-B*07:02 | 0.0570 | HLA-B*07:02 | 0.0237 | −0.0333 | (−58%) | ||||
| HLA-A*24:02 | 0.0039 | HLA-A*24:02 | 0.0046 | 0.0007 | (18%) | ||||
| LMEKYSVTF | 176 | HLA-B*51:01 | 0.0126 | LMEKYSVT I | Variant (genotype independent) | HLA-B*51:01 | 0.0333 | 0.0207 | (164%) |
| HLA-B*08:01 | 0.1209 | HLA-B*08:01 | 0.0438↓ | −0.0771 | (−64%) | ||||
| HLA-B*07:02 | 0.0073 | HLA-B*07:02 | 0.0012 | −0.0061 | (−84%) | ||||
| HLA-A*24:02 | 0.1870 | HLA-A*24:02 | 0.0049 | −0.1821 | (−97%) | ||||
| LMEKYSVTF | 177 | HLA-B*51:01 | 0.0130 | LMEK C SVTF | Variant (genotype independent) | HLA-B*51:01 | 0.0042 | −0.0088 | (−68%) |
| HLA-B*08:01 | 0.1210 | HLA-B*08:01 | 0.0201↓ | −0.1009 | (−83%) | ||||
| HLA-B*07:02 | 0.0070 | HLA-B*07:02 | 0.0032 | −0.0038 | (−54%) | ||||
| HLA-A*24:02 | 0.1870 | HLA-A*24:02 | 0.0547↓ | −0.1323 | (−71%) | ||||
| LTRDPYHTI | 238 | HLA-B*51:01 | 0.4160 | LTRDPY YI I | Genotype IV specific | HLA-B*51:01 | 0.2040↓ | −0.212 | (−51%) |
| HLA-B*08:01 | 0.2080 | HLA-B*08:01 | 0.0750↓ | −0.133 | (−64%) | ||||
| HLA-B*07:02 | 0.1280 | HLA-B*07:02 | 0.0380↓ | −0.09 | (−70%) | ||||
| HLA-A*24:02 | 0.0390 | HLA-A*24:02 | 0.0197 | −0.0193 | (−49%) | ||||
Abbreviations: aa, amino acid; BKPyV, BK polyomavirus; HLA, human leukocyte antigen.
BKPyV subtype Ib-1 (accession No. AB211371.1).
According to BKPyV subtype Ib-1 (accession No. AB211371.1).
HLA binding was predicted with the Immune Epitope Database and Analysis Resource tool (http://tools.iedb.org/main/). A difference of ≥0.05 in the HLA-binding score was interpreted as a significant change in binding (bold).
In detail, a genotype IV-specific Q171L amino acid exchange in the 9mer165 TTKEKAQIL decreased the predicted HLA-B*08:01 binding score by 8% (Table 1). Similar genotype IV-specific changes were found for 9mer238 LTRDPYHTI, for which the combined H244Y and T245I change resulted in a dramatic decrease of the HLA-B*07:02, B*08:01, and HLA-B*51:01 binding score by >50% (Table 1). Other overlapping LTag-9mers such as TKEKALILY, KEKALILYK, KALILYKKL, and PYYIIEESI appeared to be less affected (Supplementary Table 3).
Genotype-independent changes were found for 9mers such as K171Q changing 9mer165 (TTKEKALIL), which decreased the predicted binding for HLA-B*08:01 by 64% (Table 1). Other BKPyV variants were found at LTag starting positions 136, 176, and 177 (Table 1). Interestingly, the F185I exchange only affected the binding score for HLA-B*08:01 when it occurred at the anchor position of the LTag-9mer epitope (9mer176; LMEKYSVTF; Table 1), while it did not have an effect on HLA-B binding at other positions (eg, MEKYSVTII; Supplementary Table 3). Taken together, the data indicate that the highly conserved LTag carried amino acid exchanges in immunodominant 9mer T-cell epitopes that were either BKPyV genotype-specific or genotype-independent variants that significantly impair HLA class I binding and hence presentation to cytotoxic CD8 T cells.
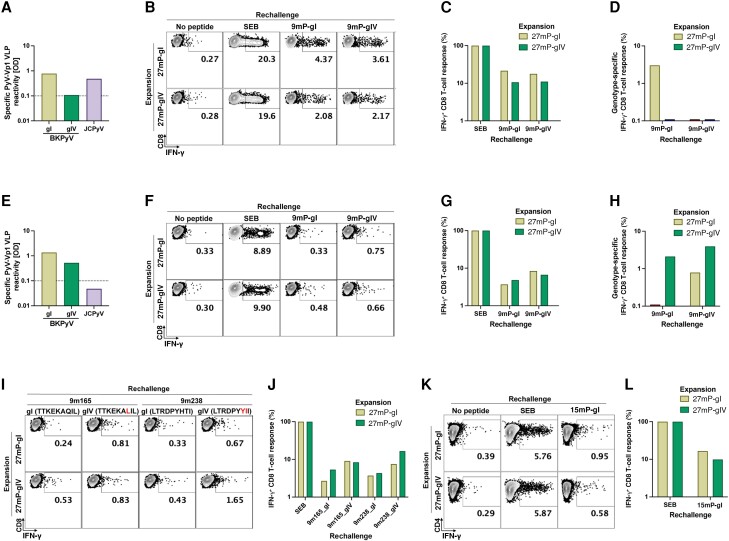
Given the principal implications of these predictions on CD8 T-cell control of BKPyV replication in immunocompromised patients, we explored the functional impact by comparing CD8 T-cell responses to genotype I- and IV-specific LTag-9mers in healthy blood donors. Blood donor No. 1 (HLA-A1/24, HLA-B8/B51) was BKPyV genotype I seropositive but seronegative for BKPyV genotype IV (Figure 6A ). Upon rechallenge, CD8 T-cell responses to the immunodominant 9mer pool containing genotype IV-specific LTag (9mP-gIV) was significantly lower than those to the genotype I 9mP-gI (Figure 6B and 6C ). This effect was most pronounced when common (genotype-independent) 9mers were excluded, using single genotype I- or genotype IV-specific 9mers for CD8 T-cell stimulation (Figure 6D ). Blood donor No. 2 (HLA-A11/23, HLA-B8/B49) was seropositive for BKPyV genotype I and genotype IV (Figure 6E ). Genotype I- and IV-specific LTag-9mers stimulated IFN-γ production by CD8 T-cells (Figure 6F and 6G ), whereby the responses were higher for genotype IV-specific LTag-9mers (Figure 6E ). To examine the contribution of single genotype-specific 9mers, we compared 9mer167 and 9mer238 (Table 1). The results revealed CD8 T-cell responses to both genotype I- and genotype IV-specific peptides, whereby the responses were highest following genotype IV expansion with 27mP-gIV expansion and matching restimulation with 9mer238-gIV (Figure 6I and 6J ). Our previous work suggested a role for CD4 T-cell help during 27mP expansion. Indeed, CD4 T-cell responses were found for both, but slightly higher responses for genotype I following homologous expansion and restimulation (Figure 6K and 6L ).
Figure 6.
BKPyV gI- and gIV-specific CD8 T-cell response. PBMCs of 2 healthy donors were used to expand BKPyV genotype-specific T-cells in vitro. After expansion with gI- or gIV-specific LTag overlapping 27mer pools (27 mP-gI and 27 mP-gIV), cells were restimulated with a pool of immunodominant BKPyV gI or gIV 9mers (9 mP-gI and 9 mP-gIV), with gI 9mers (TTKEKAQIL and LTRDPYHTI) or gIV 9mers (TTKEKALIL and LTRDPYYII), or a genotype-independent 9mer (9m27). Stimulation by SEB was used to determine the proportion of IFN-γ–producing T-cells. IFN-γ production was measured by intracellular staining using FACS. BKPyV gI- and gIV-specific IgG antibodies and JCPyV IgG specific antibodies from donor 1 (A) and donor 2 (E). Assessment of functional T-cell responses by rechallenge with BKPyV gI- and gIV-specific LTag-9mers and measurement of IFN-γ production using FACS in donor 1 (B) and donor 2 (F). Normalization of CD8 T-cells responding to BKPyV gI- and gIV-specific LTag-9mers to the total number of T-cells that could be stimulated by SEB from donor 11 (C) and donor 2 (G). Exclusion of T-cells stimulated by the genotype-independent LTag-9mer (9m27; corresponding to 3.75% and 0.68% after 27mP-gI expansion and 2.2% and 0.27% after 27mP-gIV expansion for donor 1 and donor 2; respectively) leaving solely BKPyV gI- and gIV-specific CD8 T-cell responses from donor 1 (D) and donor 2 (H). Rechallenge of CD8 T-cells from donor 2 using single BKPyV gI- or gIV-specific LTag-9mers, 9m165 and 9m238 (I). Normalization of CD8 T-cells responding to BKPyV gI- and gIV-specific LTag-9mers to the total number of T-cells that could be stimulated by SEB (J). Rechallenge of CD4 T cells from donor 2 using a pool of BKPyV gI-specific LTag 15mers (K). Normalization of CD4 T cells responding to BKPyV gI-specific LTag 15mers to the total number of T cells that could be stimulated by SEB (L). Abbreviations: BKPyV, BK polyomavirus; FACS, fluorescence-activated cell sorting; gI, genotype I; gIV, genotype IV; IFN-γ, interferon-γ; IgG, immunoglobulin G; JCPyV, JC polyomavirus; LTag, large tumor antigen; OD, optical density; SEB, Staphylococcus enterotoxin B; VLP, virus-like particle.
DISCUSSION
Given the uncertainties regarding pathogenesis, diagnosis, and outcome of BKPyV-HC, we conducted an in-depth molecular characterization of BKPyV replication in 20 allogeneic HCT recipients. We observed that, first, urine and plasma BKPyV loads are largely derived from unprotected viral genome DNA fragments. Thus, DNase digestion prior to nucleic acid extraction and QNAT resulted in >90% decline of BKPyV loads. In line with the fragmented nature of the viral genomes, the decline in BKPyV loads by DNase digestion was most pronounced for the largest QNAT amplicon of 239 bp, which was 100-fold lower in 50% and 70% of urine and plasma samples, respectively, and more undetectable results compared to the 88-bp reference. Thus, unprotected BKPyV genome fragments susceptible to enzymatic degradation may contribute to underquantification of BKPyV loads, especially by assays having larger amplicons. These results extend previous observations on BKPyV and cytomegalovirus loads in plasma of solid organ transplant patients [27, 28, 34]. We conclude that clinical and laboratory studies relying on BKPyV DNA-loads need to ensure that targets are not degraded. Moreover, international calibrators should consider standardizing genome equivalents by QNAT amplicons of around 100 bp rather than averaging results from different assays [28]. Second, urine specimens differ from plasma by containing significant amounts of DNase-protected BKPyV genomes, reflecting infectious virions. As a control, cell-free human DNA loads of aspartoacylase ASPA gene remained DNase sensitive in the same samples. Indeed, pre-PCR studies relied on directly visualizing cell-free polyomavirus particles in urine of immunocompromised patients by negative staining and electron microscopy [6, 31, 35, 36]. Thus, high-level BKPyV replication can produce urine viral loads >10 million c/mL containing a significant fraction of infectious units and hence a source of virus transmission, especially among HCT patients [12, 37, 38]. Third, BKPyV genome fragmentation leads to underestimating the genetic variation by NGS in an amplicon-size–dependent manner. Thus, NGS of plasma BKPyV DNA failed in all samples when using amplicons of 1000 bp or 5000 bp, even at high plasma viral loads of up to 1 million c/mL. NGS coverage remained successful in urine across a range of urine viral loads although the failure rate tended to be higher for the 5000-bp amplicons. Using the 250-bp NGS, we identified minority variants in both urine and plasma, indicating that BKPyV replication generated variants by intrahost evolution. Our analysis revealed sequence changes in the EVGR and LVGR, but not in the NCCR. We did not find alterations of SP1-4 or ETS1-2 known to increase EVGR expression from linear archetype NCCRs [16, 39]. Because NCCR changes emerge first in plasma of kidney transplant patients with prolonged BKPyV replication [14], further study of allogeneic HCT recipients with protracted BKPyV-HC may be warranted.
Given the central role of failing virus-specific immunity in the pathogenesis of BKPyV diseases in transplant patients [1, 9], we focused on BKPyV genotypes having the capsid protein Vp1 and LTag as the main target of nAbs and cytotoxic CD8 T cells, respectively. We found that only 10 out of the 20 HCT patients showed BKPyV genotypes commonly associated with Europe (Ib-2 n = 8 and IVc-2 n = 2), while BKPyV genotypes Ia and Ic prevalent in Africa and Asia were identified in the remaining patients [40]. The genotype-specific amino acid exchanges in Vp1 were mostly found in the BC loop and are the major serotype determinants. Although certain monoclonal nAbs reportedly cross-neutralize different BKPyV serotypes [41], the highest nAb titers are serotype specific and target the BC loop [42]. Thus, serotype-specific immunity in allogeneic HCT donors may be important predictors of duration and clearance of BKPyV replication and HC posttransplant.
Indeed, we obtained novel insights regarding BKPyV diversity and cytotoxic CD8 T-cell control [20, 21, 25]. First, the BKPyV genotypes identified in the LTAG sequences perfectly matched the serotypes defined by the VP1 sequences. This validates the 250-bp NGS coverage because the LTAG and VP1 sequences are approximately 2000 bp apart. Second, we identified BKPyV genotype-specific and genotype-independent amino acid changes in immunodominant LTag-9mer epitopes, which were predicted to decrease HLA-class I binding to CD8 T cells. Third, we could demonstrate that genotype-specific 9mers elicit genotype-specific CD8 T-cell responses. As predicted from the decrease in HLA class I binding, this was most dramatic for the immunodominant BKPyV-genotype-IV encoded LTag-9mer238 epitope LTRDPYYII, impairing presentation by HLA-B*07:02, B*08:01, and HLA-B*51:01, and genotype I-specific CD8 T-cell activation.
Thus, BKPyV replication after allogeneic HCT may result from (1) reactivation and high-level viruria of the recipient BKPyV during the allogeneic HCT immune gap; (2) exposure to immunosuppressive drugs for GVHD prophylaxis [9, 43]; (3) impaired serotype Vp1 antibody and corresponding LTag genotype-specific T-cell control; and (4) viral variants escaping T-cell control [24]. Based on the findings of this study, prospective clinical studies can be designed to investigate the diagnostic and functional role as well as the prevalence and magnitude of these steps and may help to optimize antiviral strategies as well as vaccination and adoptive transfer of nAbs and T cells [41, 44, 45].
Limitations of our study are the retrospective nature of patient inclusion and requiring longitudinal samples for analysis. As the cohort size of 20 (17%) of 155 consecutive patients with high-level BKPyV viruria may appear low, we provide the detailed flow chart for appreciating potential biases. However, a well-designed HCT study from 2013 to 2018 [46] identified 70 of 923 patients with high-level viruria in ≥2 samples. Thus, our study of 20 adult HCT patients from 2019 to 2021 seems proportional and, together with the high data density and the in-depth molecular analysis, may support a cautious interpretation.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Supplementary Material
Contributor Information
Karoline Leuzinger, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland; Clinical Virology, University Hospital Basel, Basel, Switzerland.
Amandeep Kaur, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland.
Maud Wilhelm, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland.
Konstantin Frank, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland.
Caroline A Hillenbrand, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland.
Fabian H Weissbach, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland.
Hans H Hirsch, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, Basel, Switzerland; Clinical Virology, University Hospital Basel, Basel, Switzerland; Infectious Diseases and Hospital Epidemiology, University Hospital Basel, Basel, Switzerland.
Notes
Acknowledgments. We thank the biomedical analysts Vroni Del Zenero and Marion Wernli, Transplantation and Clinical Virology, Department Biomedicine, University of Basel, and Bettina Oettli, Nadine Doppler, and Katia Bir, Clinical Virology, University Hospital Basel, in Basel, Switzerland for expert help and assistance.
Author contributions. K. L. and H. H. H. designed the study, validated results, extracted data, performed analyses, reviewed data, and wrote the manuscript. A. K., M. W., K. F., C. A. H., and F. W. performed analyses, reviewed data, and contributed to writing the manuscript.
Financial support. This work was supported by the University of Basel (grant number MM2109 personal appointment grant to H. H. H.); and in part by the Swiss Transplant Cohort Study (grant number FUP148 to K. L. and H. H. H.).
References
- 1. Graf FE, Hirsch HH. BK polyomavirus after solid organ and hematopoietic cell transplantation: one virus–three diseases. In: Morris MI, Kotton CN, Wolfe C, eds. Emerging transplant infections. Cham, Switzerland: Springer Nature, 2020:1–26. [Google Scholar]
- 2. Cesaro S, Dalianis T, Hanssen Rinaldo C, et al. . ECIL guidelines for the prevention, diagnosis and treatment of BK polyomavirus-associated haemorrhagic cystitis in haematopoietic stem cell transplant recipients. J Antimicrob Chemother 2018; 73:12–21. [DOI] [PubMed] [Google Scholar]
- 3. Imlay H, Xie H, Leisenring WM, et al. . Presentation of BK polyomavirus-associated hemorrhagic cystitis after allogeneic hematopoietic cell transplantation. Blood Adv 2020; 4:617–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Egli A, Infanti L, Dumoulin A, et al. . Prevalence of polyomavirus BK and JC infection and replication in 400 healthy blood donors. J Infect Dis 2009; 199:837–46. [DOI] [PubMed] [Google Scholar]
- 5. Arthur RR, Shah KV, Baust SJ, Santos GW, Saral R. Association of BK viruria with hemorrhagic cystitis in recipients of bone marrow transplants. N Engl J Med 1986; 315:230–4. [DOI] [PubMed] [Google Scholar]
- 6. Apperley JF, Rice SJ, Bishop JA, et al. . Late-onset hemorrhagic cystitis associated with urinary excretion of polyomaviruses after bone marrow transplantation. Transplantation 1987; 43:108–12. [DOI] [PubMed] [Google Scholar]
- 7. Leung AY, Suen CK, Lie AK, Liang RH, Yuen KY, Kwong YL. Quantification of polyoma BK viruria in hemorrhagic cystitis complicating bone marrow transplantation. Blood 2001; 98:1971–8. [DOI] [PubMed] [Google Scholar]
- 8. Hirsch HH. Human polyomavirus and papillomavirus infection and disease posttransplant. In: Ljungman P, Snydman D, eds. Transplant infections. 4th ed. Cham: Springer, 2016:631–52. [Google Scholar]
- 9. Kaur A, Wilhelm M, Wilk S, Hirsch HH. BK polyomavirus-specific antibody and T-cell responses in kidney transplantation: update. Curr Opin Infect Dis 2019; 32:575–83. [DOI] [PubMed] [Google Scholar]
- 10. Binet I, Nickeleit V, Hirsch HH. Polyomavirus infections in transplant recipients. Curr Opin Organ Transplant 2000; 5:210–6. [Google Scholar]
- 11. Hirsch HH, Pergam SA. Human adenovirus, polyomavirus, and parvovirus infections in patients undergoing hematopoietic stem-cell transplantation. In: Forman SJ, Negrin RS, Antin H, Appelbaum FR, eds. Thomas' hematopoietic cell transplantation. 5th ed. John Wiley & Sons Ltd, 2016:1090–104. [Google Scholar]
- 12. Koskenvuo M, Dumoulin A, Lautenschlager I, et al. . BK polyomavirus-associated hemorrhagic cystitis among pediatric allogeneic bone marrow transplant recipients: treatment response and evidence for nosocomial transmission. J Clin Virol 2013; 56:77–81. [DOI] [PubMed] [Google Scholar]
- 13. Rubert L, Quartuccio G, Giudo I, et al. . Role of BK virus-specific immunity in BKV-related haemorrhagic cystitis after paediatric allogeneic HSCT. 39th Annual Meeting of the European Group for Blood and Marrow Transplantation. London, 2013. [Google Scholar]
- 14. Gosert R, Rinaldo CH, Funk GA, et al. . Polyomavirus BK with rearranged noncoding control region emerge in vivo in renal transplant patients and increase viral replication and cytopathology. J Exp Med 2008; 205:841–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Priftakis P, Bogdanovic G, Kalantari M, Dalianis T. Overrepresentation of point mutations in the Sp1 site of the non-coding control region of BK virus in bone marrow transplanted patients with haemorrhagic cystitis. J Clin Virol 2001; 21:1–7. [DOI] [PubMed] [Google Scholar]
- 16. Bethge T, Hachemi HA, Manzetti J, Gosert R, Schaffner W, Hirsch HH. Sp1 sites in the noncoding control region of BK polyomavirus are key regulators of bidirectional viral early and late gene expression. J Virol 2015; 89:3396–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Solis M, Velay A, Porcher R, et al. . Neutralizing antibody-mediated response and risk of BK virus-associated nephropathy. J Am Soc Nephrol 2018; 29:326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Binggeli S, Egli A, Dickenmann M, Binet I, Steiger J, Hirsch HH. BKV replication and cellular immune responses in renal transplant recipients. Am J Transplant 2006; 6:2218–9. [DOI] [PubMed] [Google Scholar]
- 19. Binggeli S, Egli A, Schaub S, et al. . Polyomavirus BK-specific cellular immune response to VP1 and large T-antigen in kidney transplant recipients. Am J Transplant 2007; 7:1131–9. [DOI] [PubMed] [Google Scholar]
- 20. Cioni M, Leboeuf C, Comoli P, Ginevri F, Hirsch HH. Characterization of immunodominant BK polyomavirus 9mer epitope T cell responses. Am J Transplant 2016; 16:1193–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leboeuf C, Wilk S, Achermann R, et al. . BK polyomavirus-specific 9mer CD8 T cell responses correlate with clearance of BK viremia in kidney transplant recipients: first report from the Swiss transplant cohort study. Am J Transplant 2017; 17:2591–600. [DOI] [PubMed] [Google Scholar]
- 22. Willhelm M, Wilk S, Kaur A, Hirsch HH. Swiss transplant cohort S. Can HLA-B51 protect against BKPyV-DNAemia? Transplantation 2019; 103:e384–e5. [DOI] [PubMed] [Google Scholar]
- 23. Wunderink HF, Haasnoot GW, de Brouwer CS, et al. . Reduced risk of BK polyomavirus infection in HLA-B51-positive kidney transplant recipients. Transplantation 2019; 103:604–12. [DOI] [PubMed] [Google Scholar]
- 24. Leuzinger K, Kaur A, Wilhelm M, Hirsch HH. Variations in BK polyomavirus immunodominant large tumor antigen-specific 9mer CD8 T-cell epitopes predict altered HLA-presentation and immune failure. Viruses 2020; 12:1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wilhelm M, Kaur A, Wernli M, Hirsch HH. BK Polyomavirus (BKPyV)-specific CD8 T-cell expansion in vitro using 27mer peptide antigens for developing adoptive T-cell transfer and vaccination. J Infect Dis 2021; 223:1410–22. [DOI] [PubMed] [Google Scholar]
- 26. Bedi A, Miller CB, Hanson JL, et al. . Association of BK virus with failure of prophylaxis against hemorrhagic cystitis following bone marrow transplantation. J Clin Oncol 1995; 13:1103–9. [DOI] [PubMed] [Google Scholar]
- 27. Leuzinger K, Naegele K, Schaub S, Hirsch HH. Quantification of plasma BK polyomavirus loads is affected by sequence variability, amplicon length, and non-encapsidated viral DNA genome fragments. J Clin Virol 2019; 121:104210. [DOI] [PubMed] [Google Scholar]
- 28. Naegele K, Lautenschlager I, Gosert R, et al. . Cytomegalovirus sequence variability, amplicon length, and DNase-sensitive non-encapsidated genomes are obstacles to standardization and commutability of plasma viral load results. J Clin Virol 2018; 104:39–47. [DOI] [PubMed] [Google Scholar]
- 29. Manzetti J, Weissbach FH, Graf FE, et al. . BK Polyomavirus evades innate immune sensing by disrupting the mitochondrial network and promotes mitophagy. iScience 2020; 23:101257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kardas P, Leboeuf C, Hirsch HH. Optimizing JC and BK polyomavirus IgG testing for seroepidemiology and patient counseling. J Clin Virol 2015; 71:28–33. [DOI] [PubMed] [Google Scholar]
- 31. Hogan TF, Padgett BL, Walker DL, Borden EC, McBain JA. Rapid detection and identification of JC virus and BK virus in human urine by using immunofluorescence microscopy. J Clin Microbiol 1980; 11:178–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chauhan S, Lecatsas G, EH Harley. Genome analysis of BK(WW) viral DNA cloned directly from human urine. Intervirology 1984; 22:170–6. [DOI] [PubMed] [Google Scholar]
- 33. DeCaprio JA, Imperiale MJ, Hirsch HH. Polyomaviridae. In: Howley PM, Knipe DM, eds. Fields virology, DNA viruses. 7th ed. Vol. 2. Chapter 1. Philadelphia: Lippincott Williams & Wilkins, 2021. [Google Scholar]
- 34. Tong Y, Pang XL, Mabilangan C, Preiksaitis JK. Determination of the biological form of human cytomegalovirus DNA in the plasma of solid-organ transplant recipients. J Infect Dis 2017; 215:1094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Coleman DV, Gardner SD, Field AM. Human polyomavirus infection in renal allograft recipients. Br Med J 1973; 3:371–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reese JM, Reissing M, Daniel RW, Shah KV. Occurrence of BK virus and BK virus-specific antibodies in the urine of patients receiving chemotherapy for malignancy. Infect Immun 1975; 11:1375–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hosoi H, Murata S, Suzuki T, et al. . A cluster of BK polyomavirus-associated hemorrhagic cystitis after allogeneic hematopoietic stem cell transplantation. Transpl Infect Dis 2021; 23:e13736. [DOI] [PubMed] [Google Scholar]
- 38. Onda Y, Kanda J, Hanaoka N, et al. . Possible nosocomial transmission of virus-associated hemorrhagic cystitis after allogeneic hematopoietic stem cell transplantation. Ann Hematol 2021; 100:753–61. [DOI] [PubMed] [Google Scholar]
- 39. Bethge T, Ajuh E, Hirsch HH. Imperfect symmetry of Sp1 and core promoter sequences regulates early and late virus gene expression of the bidirectional BK polyomavirus noncoding control region. J Virol 2016; 90:10083–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torres C. Evolution and molecular epidemiology of polyomaviruses. Infect Genet Evol 2020; 79:104150. [DOI] [PubMed] [Google Scholar]
- 41. Jordan S, Limaye AP, Fischbach B, et al. . A randomized phase 2 study of Mau868 Vs placebo to treat Bk viremia in kidney transplant recipients. Am J Transplant 2022; 22:1–3. [Google Scholar]
- 42. Jin L, Gibson PE, Knowles WA, Clewley JP. BK virus antigenic variants: sequence analysis within the capsid VP1 epitope. J Med Virol 1993; 39:50–6. [DOI] [PubMed] [Google Scholar]
- 43. Hirsch HH, Yakhontova K, Lu M, Manzetti J. BK polyomavirus replication in renal tubular epithelial cells is inhibited by sirolimus, but activated by tacrolimus through a pathway involving FKBP-12. Am J Transplant 2016; 16:821–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Nelson AS, Heyenbruch D, Rubinstein JD, et al. . Virus-specific T-cell therapy to treat BK polyomavirus infection in bone marrow and solid organ transplant recipients. Blood Adv 2020; 4:5745–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wali RK, Singh M, Wojciechowski D, et al. . Posoleucel as preemptive therapy for BKV infection in kidney transplant recipients: safety and tolerability in a phase 2 trial. Am J Transplant 2022; 22:1–3. [Google Scholar]
- 46. Laskin BL, Denburg MR, Furth SL, et al. . The natural history of BK polyomavirus and the host immune response after stem cell transplantation. Clin Infect Dis 2020; 71:3044–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.