Abstract
Background
B‐Cell CLL/Lymphoma 11B (BCL11B) is a C2H2 zinc finger transcription factor that has broad biological functions and is essential for the development of the immune system, neural system, cardiovascular system, dermis, and dentition. Variants of BCL11B have been found in patients with neurodevelopmental disorders and immunodeficiency.
Materials and Methods
Whole‐exome sequencing (WES) and clinical examinations were performed to identify the etiology of our patient. A variant in the BCL11B gene, NM_138576.4: c.1206delG (p.Phe403Serfs*2) was found and led to frameshift truncation.
Results
We reported a male patient with developmental delay and cerebral palsy who carried the BCL11B variant. The detailed clinical features, such as brain structure and immune detection, were described and reviewed in comparison to previous patients.
Conclusions
The BCL11B‐related neurodevelopmental disorders are rare, and only 17 variants in 25 patients have been found to date. Our report expands the variants spectrum of BCL11B and increases the case of neurodevelopmental abnormalities.
Keywords: BCL11B, cerebral palsy, developmental delay, whole‐exome sequencing
The BCL11B‐related neurodevelopmental disorder was rare, only 14 variants were found to date. Our report expands the variants spectrum of BCL11B and increases the case of neurodevelopmental abnormalities.
1. INTRODUCTION
BCL11B (B‐Cell CLL/Lymphoma 11B) encodes a C2H2‐type zinc finger protein; it is a homolog gene for BCL11A that was first identified in 2001 (Satterwhite et al., 2001). BCL11B is a zinc finger transcription factor, and its functions are most widely studied in the immune and nervous systems. Bcl11b is widely expressed in various subsets of T‐cell and plays a crucial role in the regulation of T‐cell development, function, and survival (Avram & Califano, 2014). In neuronal networks, Bcl11b has also been shown to be expressed in corticospinal motor neurons (CSMNs). Based on a study of Bcl11b‐deficient mice, Bcl11b is required to establish cortical connections to the spinal cord (Arlotta et al., 2005). In addition, conditional deletion of Bcl11b in the mouse hippocampus leads to the loss of excitatory synapses and abnormal synaptic transmission (De Bruyckere et al., 2018).
The first patient with the BCL11B variant, who presented with “leaky” SCID, was reported in 2016. Zebrafish functional experiments revealed that a BCL11B variant caused human multisystem anomalies with SCID and that BCL11B was crucial in hematopoietic progenitors (Punwani et al., 2016). Subsequently, successive reports demonstrated the pathogenicity of BCL11B variants (Alfei et al., 2021; Lessel et al., 2018; Lu et al., 2021; Prasad et al., 2020; Qiao et al., 2019; Yang et al., 2020). All patients presented with developmental delay, most of whom presented with intellectual disability, mild facial dysmorphisms, and impaired T‐cell development. Variants in BCL11B are now associated with “Immunodeficiency 49 (OMIM: 617237)” and “Intellectual developmental disorder with speech delay, dysmorphic facies, and T‐cell abnormalities (OMIM: 618092)”. However, the reported patients and variants are limited. Further expansion of variant sites and phenotypes is beneficial for a better understanding of this disease.
Here, we report a four‐year‐ and five‐month‐old boy who presented with cerebral palsy and global developmental delay and was identified with a de novo variant in BCL11B. The frameshift variant NM_138576.4: c.1206delG (p.Phe403Serfs*2) was predicted to cause protein truncation (p.Phe403Serfs*2) and possibly disrupt the protein function.
2. MATERIALS AND METHODS
2.1. Patient
Written informed consent was obtained from the legal guardians of the patient to participate in this study. The study was approved by the Human Ethics Committees of the Children's Hospital affiliated with Zhejiang University. The clinical course, cerebral magnetic resonance imaging (MRI), and genetic testing of our patient were analyzed. The immunophenotype of our patient was determined by peripheral blood and serum.
2.2. Whole‐exome sequencing
Genetic analysis was performed using WES. The source of the sample was the peripheral blood of the patient and his parents. The genomic DNA library was captured using the IDT XGen Exome Research Panel. WES was then performed on the NovaSeq 6000 Sequencing platform using paired‐end reads and compared to the human reference genome (GRCh38/hg38). The variants were annotated by ANNOVAR (McKenna et al., 2010). The pathogenicity of the identified variants was predicted by multiple software programs, such as Mutation Taster, Polyphen2, and Sorting Intolerant from Tolerant (SIFT). All variants were filtered by inherent patterns, frequency, clinical characteristics, and public databases. Pathogenic variants were finally screened according to the American College of Medical Genetics and Genomics (ACMG) (Richards et al., 2015). Sanger sequencing was performed in the trio family. Sanger sequencing was performed to verify the variants we detected in WES.
3. RESULTS
3.1. Case description
A 4‐year‐ and 5‐month‐old boy, with an older brother and nonconsanguineous parents in good health, initially presented with a developmental delay in motor and language when he was 9 months old. He could grab, but not pinch. He was unable to sit without support and unable to crawl, and he had an abnormal posture of standing on his tiptoes. Gesell Developmental Scale results (2019.4.25) showed cognitive and motor delays. MRI results (2019.11.23) showed widened extracerebral space and less signal reduction in the internal capsule (Figure 1a,b). The patient was diagnosed with mixed cerebral palsy and was followed by rehabilitation (2020.12.25‐2021.1.15; 2021.3.1‐2021.3.9). He made some cognitive and motor progress. A repeat MRI (2021.01.12) showed widened extracerebral space and a lucid septum cyst (Figure 1c,d). Immune detection (2021.7.21) showed high CD8 and low NK cells and IgG2 (Table 1). However, the patient had no obvious immunodeficiency phenotype. He had not experienced infections or allergies. After various rehabilitation therapy, such as sling exercise training, physiotherapy, occupational, and speech therapy, his development improved markedly. When he was 3 years and 3 months old, he could climb stairs, stand, and walk with support, and he could grasp objects with both hands, but with poor accuracy and coordination. He could name a few animals, and his vocabulary was more than 20 words, but he could not speak phrases. He had poor separation of the lower extremities. Currently, he has poor overall development, with the language area slightly stronger than the other areas, and the motor area slightly behind.
FIGURE 1.
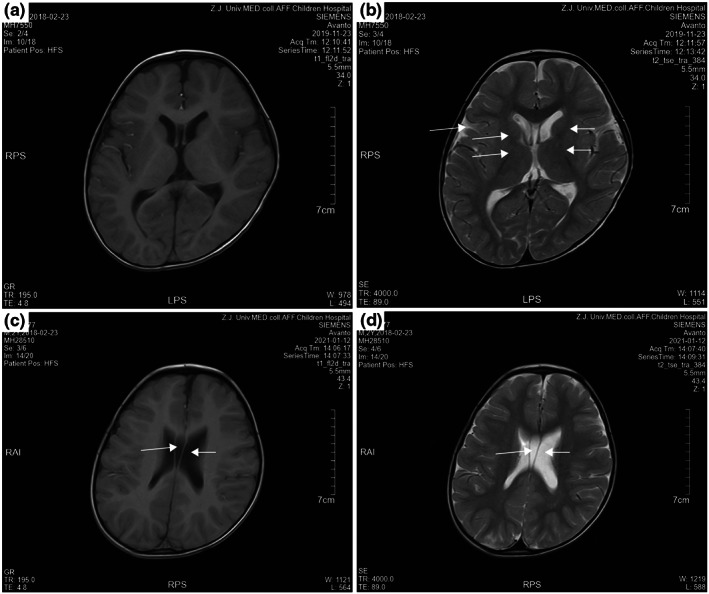
MRI results in our patient in 2019 (a, b) and 2021 (c, d). (a) T1‐weighted axial image. (b) T2‐weighted axial images. The widened extracerebral space and the internal capsule showed less signal reduction. (c, d) T1‐ and T2‐weighted axial images showed a widened extracerebral space and lucid septum cysts.
TABLE 1.
The immune results in our patient (2021.7.21)
| Cluster of differentiation | Value | Reference |
|---|---|---|
| CD19 | 24.4 | 18.5% ~ 28.0% |
| CD3 | 67.3 | 56.0% ~ 68.0% |
| CD4 | 29.7 | 29.0% ~ 40.0% |
| CD8 | 27.2↑ | 19.0% ~ 25.0% |
| CD3−CD16+CD56+ | 4.7↓ | 9.0% ~ 19.0% |
| CD4/CD8− | 1.09↓ | 1.1–2.0 |
| Immunoglobulin subclass | ||
| IgG1 | 5.74 | 4.90 ~ 11.4 g/L |
| IgG2 | 1.16↓ | 1.50 ~ 6.40 g/L |
| IgG3 | 0.08↓ | 0.20 ~ 1.10 g/L |
| IgG4 | 0.80 | 0.08 ~ 1.40 g/L |
3.2. Identification of gene variation
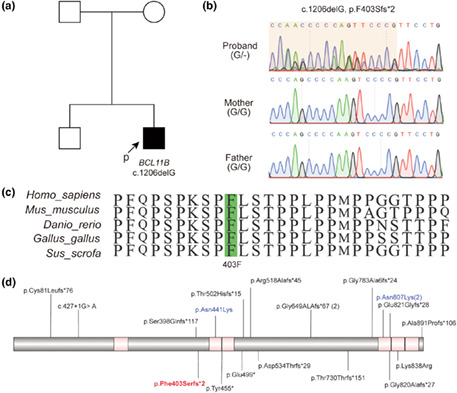
WES was performed to further identify the etiology of our patient. A heterozygous BCL11B variant in exon 4, NM_138576.4: c.1206delG (p.Phe403Serfs*2) was identified, resulting in a frameshift variation p.Phe403Serfs*2, which could be expected to result in protein truncation (Figure 2). The de novo variant in our patient was verified by the Sanger sequencing and his parents did not carry the variant. This variant was not reported or included in the gnomAD, ExAC, or ClinVar databases, indicating its rarity. The probability of being loss‐of‐function intolerant (pLI) in BCL11B was 0.99 and was considered an extremely intolerant transcript. Finally, the BCL11B variant in our patient was classified as likely pathogenic (PVS1_Strong+PS2_Moderate+PM2_Supporting) according to ACMG guidelines.
FIGURE 2.
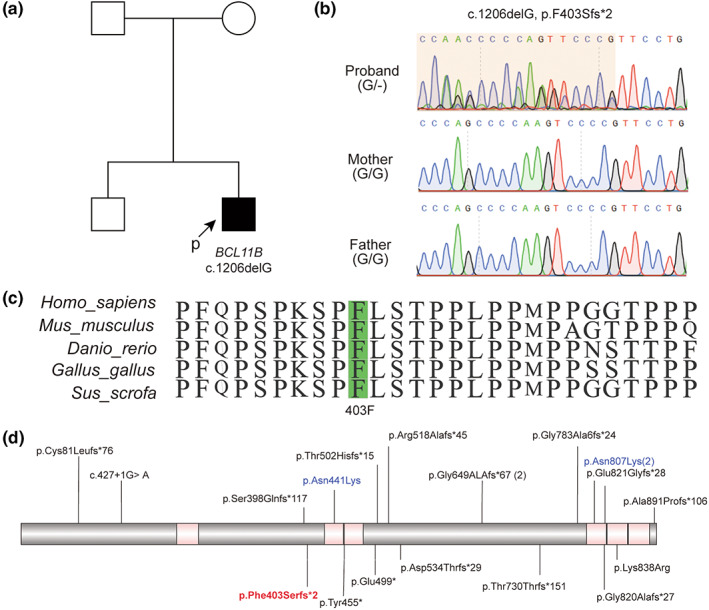
Identification of the variant in BCL11B. (a) The pedigree of our patient. The proband is highlighted with a filled square with an arrow. (b) Sanger sequencing in the trio family. A de novo 1206C deletion was identified by sequencing. (c) The changed 403F amino acid was conserved in multiple species. (d) Schematic of variants in the BCL11B gene in neurodevelopmental disease patients. The variant in our patient is shown in red text. Missense variants in the BCL11B gene are shown in blue text.
4. DISCUSSION
BCL11B is a zinc finger protein transcription factor with multiple functions in the development of the immune and nervous cutaneous systems. BCL11B plays an important role in the development of deep‐layer corticospinal projection neurons and striatal middle spiny neurons (Nikouei et al., 2016). Additionally, in mice, Bcl11b is highly expressed in medium spiny neurons (MSNs) and the hippocampus, in which Bcl11b (KO/KO) mouse MSNs fail to differentiate (Arlotta et al., 2008), and selective loss of Bcl11b expression in the adult hippocampus results in impaired spatial working memory (Simon et al., 2016). In addition, Bcl11b (KO/KO) mouse exhibit a defect in embryonic tooth development (Katsuragi et al., 2013).
Recent studies in BCL11B variant patients have presented an immunophenotype, a developmental delay, and other clinical features, such as abnormal facial appearance and dental anomalies, which are reviewed in Table 2. A total of 10 studies have reported 17 BCL11B variants and 25 patients, including 3 missense variants, 1 splice variant, and 13 truncated variants. These findings show an association between BCL11B variants and “Immunodeficiency 49” and “Intellectual developmental disorder with dysmorphic facies, speech delay, and T‐cell abnormalities.” Interestingly, patients with missense variants tend to have a more severe immunodeficiency, which may be due to the loss of DNA binding (Lessel et al., 2018). Our patient with BCL11B frameshift variants had no history of allergies or infections. A hematological evaluation of his immune system was performed (Table 1) and found some unusual changes were found. However, no immunodeficiency was found in our patient, which was also the same as previously reported cases (Lessel et al., 2018).
TABLE 2.
BCL11B variants (NM_138576.4) and associated phenotypes in individuals with developmental delay
| BCL11B variants | Ref.(PMID) | Age (year) | Gender | Autistic features | Speech impairment | Motor delay | Intellectual disability | Abnormal facial appearance | Refractive error | Dental anomalies | Immune response | MRI abnormal |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| p.N441KAsn441Lys | 27959755 | 0 | M | NA | + | + | + | + | NA | NA | + | Callosal agenesis |
| p.Gly820Alafs*27 | 29985992 | 3 11/12 | F | + | + | + | + | + | + | + | − | − |
| p.Gly649Alafs*67 | 15 1/12 | M | − | + | + | + | + | − | + | + | − | |
| p.Ala891Profs*106 | 1 2/3 | M | + | + | + | + | + | + | − | + | − | |
| p.Thr502Hisfs*15 | 1 5/6 | F | − | + | + | + | + | − | − | − | − | |
| p.N807KAsn807Lys | 2 1/4 | M | − | + | + | + | + | − | + | + | − | |
| p.Cys81Leufs*76 | 17 7/12 | M | − | + | + | + | + | − | − | − | − | |
| p.D534TfsAsp534Thrfs*29 | 13 | F | + | + | + | + | + | − | − | − | − | |
| 46,XY,t(4;14)(p15;q32.1) | 11 | M | + | + | + | + | + | − | − | − | − | |
| 46,XY,t(4;14)(q31.1;q32.2) | 29 | M | − | + | − | + | + | − | − | − | Moderate ectopia of amygdala | |
| p.GluE499* | 6 1/2 | F | − | + | + | + | + | + | + | − | − | |
| p.TyrY455* | 9 11/12 | M | − | + | + | + | + | + | − | − | Hypoplasia of the globus pallidus | |
| p.ArgR518Alafs*45 | 9 3/4 | M | − | + | + | + | + | − | − | + | − | |
| p.Thr730Thrfs*151 | 31347296 | 5 | F | NA | + | + | + | + | NA | NA | + | − |
| p.Ser398GlnQfs*117 | 33194885 | 1 5/12 | F | NA | + | + | + | + | NA | NA | + | Abnormal myelination of the white matter |
| p.Gly649Alafs*67 | 32659295 | 4 | F | NA | + | + | + | + | NA | + | NA | Dysgenesis of the corpus callosum and enlargement of the frontal horns of the lateral ventricles bilaterally |
| p.N807KAsn807Lys | 9 | F | NA | + | + | + | + | + | + | − | − | |
| p.N441KAsn441Lys | 34844266 | 0 | M | NA | + | + | + | + | + | − | NA | Corpus callosum dysgenesis, mildly enlarged frontal horns of lateral ventricles |
| p.Cys826TyrY | 34887873 | 14 | F | NA | − | − | + | − | − | − | + | − |
| p.Gly783Ala6fs*24 | 36275064 | 2 1/12 | M | − | + | + | NA | + | − | − | + | Widening of the extracerebral interval, abnormal signals of the cornu occipitale and a reduced right‐brain cerebral volume |
| p.Lys838Arg | 36202297 | 5 | F | − | + | + | NA | NA | NA | NA | NA | Frontotemporal substance reduction with dilated CSF spaces |
| c.427+1G>A | 36176959 | 11 | F | + | + | + | + | + | NA | + | + | − |
| 3 10/12 | M | − | + | + | + | + | NA | + | + | − | ||
| 35 | F | − | + | + | + | + | NA | − | − | NA | ||
| p.Glu821Glyfs*28 | 3 8/12 | M | − | + | + | + | + | NA | + | + | − | |
| p.PheF403Serfs*2 | Our report | 4 5/12 | M | − | + | + | + | − | − | − | − | Widened extracerebral space and the internal capsule showed less signal reduction |
Abbreviation: NA, not known available.
The clinical features of patients with BCL11B gene variants are heterogeneous, and recent studies have suggested that craniosynostosis may be associated with this gene variant(Gaillard et al., 2021). Further research is required to clarify the relationship between craniosynostosis and BCL11B variation. Additionally, whether the brain structure of patients is abnormal also varies. Only five of the 19 patients reported thus far had abnormal MRI findings (Table 2). Our patient showed abnormal MRI results, with a widened extracerebral space in 2019 and lateral ventricle enlargement in 2021 (Figure 1). In addition to extending the abnormal phenotype on MRI, our patient was the second to be diagnosed with cerebral palsy among BCL11B variant patients. He presented with poor upper extremity control, bilateral foot eversion, standing on tiptoe, and toe grip. A previous case diagnosis of choreoathetotic cerebral palsy manifested as an extension of the arms with rotation of the wrist, the extension of the neck, flexion of the spine, and continuous dyskinetic movements of the tongue (Prasad et al., 2020). Richer clinical features are presented due to the patient carrying a missense variant (p.N807K), such as abnormal facial appearance, dental anomalies, dysgenesis of the corpus callosum, and bilateral enlargement of the frontal horns of the lateral ventricles bilaterally.
5. CONCLUSIONS
In summary, variants in the BCL11B gene showed high phenotypic heterogeneity, but the mechanisms that affect neurodevelopment remain limited. Our report expands the spectrum of DNA variants of the BCL11B gene and neurodevelopmental abnormalities of BCL11B‐related disorders. The clinical phenotype and gene variation spectrum need to be expanded through additional cases, and more detailed functions need to be studied in the future.
AUTHOR CONTRIBUTIONS
Haifeng Li and Yonglin Yu conceived and designed the experiments. Xiaoyi Jia and Hongwei Yin did the patient recruitment and clinical analysis. Fan Yang and Zuozhen Yang did the WES and molecular analysis. Yonglin Yu and Hongfang Jiang wrote the first draft of the manuscript. All authors reviewed and approved the final manuscript.
CONFLICT OF INTEREST
The authors declare that they have no competing interests.
ETHICAL COMPLIANCE
The parents of the proband were informed by the consultant in the Department of Pediatric Neurology about the purpose of the DNA analysis. The samples were investigated after obtaining informed consent from the parents. This work was approved by the Children's Hospital affiliated with Zhejiang University.
ACKNOWLEDGMENTS
We wish to gratefully thank the patient's parents for allowing us to publish this clinical report. And also thank Cipher Gene for their sequencing knowledge support. This work was supported by the Technological Research Program of Zhejiang (LBY21H170002 2021).
Yu, Y. , Jia, X. , Yin, H. , Jiang, H. , Du, Y. , Yang, F. , Yang, Z. , & Li, H. (2023). A novel variant in BCL11B in an individual with neurodevelopmental delay: A case report. Molecular Genetics & Genomic Medicine, 11, e2132. 10.1002/mgg3.2132
REFERENCES
- Alfei, E. , Cattaneo, E. , Spaccini, L. , Iascone, M. , Veggiotti, P. , & Doneda, C. (2021). Progressive clinical and neuroradiological findings in a child with BCL11B missense mutation: Expanding the phenotypic spectrum of related disorder. Neuropediatrics, 53, 283–286. 10.1055/s-0041-1736193 [DOI] [PubMed] [Google Scholar]
- Arlotta, P. , Molyneaux, B. J. , Chen, J. , Inoue, J. , Kominami, R. , & Macklis, J. D. (2005). Neuronal subtype‐specific genes that control corticospinal motor neuron development in vivo. Neuron, 45(2), 207–221. 10.1016/j.neuron.2004.12.036 [DOI] [PubMed] [Google Scholar]
- Arlotta, P. , Molyneaux, B. J. , Jabaudon, D. , Yoshida, Y. , & Macklis, J. D. (2008). Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum. The Journal of Neuroscience, 28(3), 622–632. 10.1523/jneurosci.2986-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avram, D. , & Califano, D. (2014). The multifaceted roles of Bcl11b in thymic and peripheral T cells: Impact on immune diseases. Journal of Immunology, 193(5), 2059–2065. 10.4049/jimmunol.1400930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bruyckere, E. , Simon, R. , Nestel, S. , Heimrich, B. , Kätzel, D. , Egorov, A. V. , Liu, P. , Jenkins, N. A. , Copeland, N. G. , Schwegler, H. , Draguhn, A. , & Britsch, S. (2018). Stability and function of hippocampal mossy fiber synapses depend on Bcl11b/Ctip2. Frontiers in Molecular Neuroscience, 11, 103. 10.3389/fnmol.2018.00103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillard, L. , Goverde, A. , van den Bosch, Q. C. C. , Jehee, F. S. , Brosens, E. , Veenma, D. , Magielsen, F. , de Klein, A. , IMJ, M. , & van Dooren, M. F. (2021). Case report and review of the literature: Congenital diaphragmatic hernia and Craniosynostosis, a coincidence or common cause? Frontiers in Pediatrics, 9, 772800. 10.3389/fped.2021.772800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsuragi, Y. , Anraku, J. , Nakatomi, M. , Ida‐Yonemochi, H. , Obata, M. , Mishima, Y. , Sakuraba, Y. , Gondo, Y. , Kodama, Y. , Nishikawa, A. , Takagi, R. , Ohshima, H. , & Kominami, R. (2013). Bcl11b transcription factor plays a role in the maintenance of the ameloblast‐progenitors in mouse adult maxillary incisors. Mechanisms of Development, 130(9–10), 482–492. 10.1016/j.mod.2013.05.002 [DOI] [PubMed] [Google Scholar]
- Lessel, D. , Gehbauer, C. , Bramswig, N. C. , Schluth‐Bolard, C. , Venkataramanappa, S. , van Gassen, K. L. I. , Hempel, M. , Haack, T. B. , Baresic, A. , Genetti, C. A. , Funari, M. F. A. , Lessel, I. , Kuhlmann, L. , Simon, R. , Liu, P. , Denecke, J. , Kuechler, A. , de Kruijff, I. , … Kubisch, C. (2018). BCL11B mutations in patients affected by a neurodevelopmental disorder with reduced type 2 innate lymphoid cells. Brain, 141(8), 2299–2311. 10.1093/brain/awy173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, H. Y. , Sertori, R. , Contreras, A. V. , Hamer, M. , Messing, M. , Del Bel, K. L. , Lopez‐Rangel, E. , Chan, E. S. , Rehmus, W. , Milner, J. D. , KM, M. N. , Lehman, A. , Wiest, D. L. , & Turvey, S. E. (2021). A novel germline heterozygous BCL11B variant causing severe atopic disease and immune dysregulation. Frontiers in Immunology, 12, 788278. 10.3389/fimmu.2021.788278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna, A. , Hanna, M. , Banks, E. , Sivachenko, A. , Cibulskis, K. , Kernytsky, A. , Garimella, K. , Altshuler, D. , Gabriel, S. , Daly, M. , & DePristo, M. A. (2010). The genome analysis toolkit: A MapReduce framework for analyzing next‐generation DNA sequencing data. Genome Research, 20(9), 1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikouei, K. , Muñoz‐Manchado, A. B. , & Hjerling‐Leffler, J. (2016). BCL11B/CTIP2 is highly expressed in GABAergic interneurons of the mouse somatosensory cortex. Journal of Chemical Neuroanatomy, 71, 1–5. 10.1016/j.jchemneu.2015.12.004 [DOI] [PubMed] [Google Scholar]
- Prasad, M. , Balci, T. B. , Prasad, C. , Andrews, J. D. , Lee, R. , Jurkiewicz, M. T. , Napier, M. P. , Colaiacovo, S. , Guillen Sacoto, M. J. , & Karp, N. (2020). BCL11B‐related disorder in two Canadian children: Expanding the clinical phenotype. European Journal of Medical Genetics, 63(9), 104007. 10.1016/j.ejmg.2020.104007 [DOI] [PubMed] [Google Scholar]
- Punwani, D. , Zhang, Y. , Yu, J. , Cowan, M. J. , Rana, S. , Kwan, A. , Adhikari, A. N. , Lizama, C. O. , Mendelsohn, B. A. , Fahl, S. P. , Chellappan, A. , Srinivasan, R. , Brenner, S. E. , Wiest, D. L. , & Puck, J. M. (2016). Multisystem anomalies in severe combined immunodeficiency with mutant BCL11B. The New England Journal of Medicine, 375(22), 2165–2176. 10.1056/NEJMoa1509164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao, F. , Wang, C. , Luo, C. , Wang, Y. , Shao, B. , Tan, J. , Hu, P. , & Xu, Z. (2019). A De novo heterozygous frameshift mutation identified in BCL11B causes neurodevelopmental disorder by whole exome sequencing. Molecular Genetics & Genomic Medicine, 7(9), e897. 10.1002/mgg3.897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S. , Aziz, N. , Bale, S. , Bick, D. , Das, S. , Gastier‐Foster, J. , Grody, W. W. , Hegde, M. , Lyon, E. , Spector, E. , Voelkerding, K. , & Rehm, H. L. (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterwhite, E. , Sonoki, T. , Willis, T. G. , Harder, L. , Nowak, R. , Arriola, E. L. , Liu, H. , Price, H. P. , Gesk, S. , Steinemann, D. , Schlegelberger, B. , Oscier, D. G. , Siebert, R. , Tucker, P. W. , & Dyer, M. J. (2001). The BCL11 gene family: Involvement of BCL11A in lymphoid malignancies. Blood, 98(12), 3413–3420. 10.1182/blood.v98.12.3413 [DOI] [PubMed] [Google Scholar]
- Simon, R. , Baumann, L. , Fischer, J. , Seigfried, F. A. , De Bruyckere, E. , Liu, P. , Jenkins, N. A. , Copeland, N. G. , Schwegler, H. , & Britsch, S. (2016). Structure‐function integrity of the adult hippocampus depends on the transcription factor Bcl11b/Ctip2. Genes, Brain, and Behavior, 15(4), 405–419. 10.1111/gbb.12287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, S. , Kang, Q. , Hou, Y. , Wang, L. , Li, L. , Liu, S. , Liao, H. , Cao, Z. , Yang, L. , & Xiao, Z. (2020). Mutant BCL11B in a patient with a neurodevelopmental disorder and T‐cell abnormalities. Frontiers in Pediatrics, 8, 544894. 10.3389/fped.2020.544894 [DOI] [PMC free article] [PubMed] [Google Scholar]


