Abstract
Photosystem II repair in chloroplasts is a critical process involved in maintaining a plant’s photosynthetic activity under cold stress. FtsH (filamentation temperature-sensitive H) is an essential metalloprotease that is required for chloroplast photosystem II repair. However, the role of FtsH in tea plants and its regulatory mechanism under cold stress remains elusive. In this study, we cloned a FtsH homolog gene in tea plants, named CsFtsH5, and found that CsFtsH5 was located in the chloroplast and cytomembrane. RT-qPCR showed that the expression of CsFtsH5 was increased with leaf maturity and was significantly induced by light and cold stress. Transient knockdown CsFtsH5 expression in tea leaves using antisense oligonucleotides resulted in hypersensitivity to cold stress, along with higher relative electrolyte leakage and lower Fv/Fm values. To investigate the molecular mechanism underlying CsFtsH5 involvement in the cold stress, we focused on the calcineurin B-like-interacting protein kinase 11 (CsCIPK11), which had a tissue expression pattern similar to that of CsFtsH5 and was also upregulated by light and cold stress. Yeast two-hybrid and dual luciferase (Luc) complementation assays revealed that CsFtsH5 interacted with CsCIPK11. Furthermore, the Dual-Luc assay showed that CsCIPK11-CsFtsH5 interaction might enhance CsFtsH5 stability. Altogether, our study demonstrates that CsFtsH5 is associated with CsCIPK11 and plays a positive role in maintaining the photosynthetic activity of tea plants in response to low temperatures.
Keywords: cold, CsCIPK11, CsFtsH5, photosynthetic activity, tea plant
1. Introduction
Light and temperature are two critical environmental factors regulating physiological and developmental processes in plants. Exposure to adverse environmental conditions, including extreme light and temperature, can lead to oxidative stress in plants. The organelle that is most vulnerable to oxidative damage is the chloroplast, the proper function of which is a prerequisite for the normal growth and development of plants. Chloroplasts are semi-autonomous organelles assembled with functional multi-subunit photosynthetic complexes, including photosystem II (PSII) and photosystem I (PSI) [1,2]. Photodamage to the PSII complex is an unavoidable process under stress conditions. Thus, quality control of the PSII complex is critical for maintaining photosynthetic activity in the presence of excessive reactive oxygen species (ROS) [3,4]. Photodamage primarily affects the PSII reaction center protein D1. The rapid and selective removal of photodamaged D1 is a critical step in the reactivation and reassembly of PSII, thus ensuring the maintenance of photosynthetic activity. In fact, plants have developed effective mechanisms for degrading damaged D1 [5,6]. Research indicates that the thylakoid membrane-bound FtsH metalloprotease plays a central role in the degradation of the damaged D1 protein [7,8].
FtsH is a conserved membrane-anchored metalloprotease that contains an ATPase domain and a catalytic zinc-binding site [9]. FtsH was first identified in prokaryotes [10] and then demonstrated in the chloroplasts and mitochondria of eukaryotic cells [11,12]. To date, twelve FtsH genes have been identified in arabidopsis (Arabidopsis thaliana (L.) Heyn.) [13]. Functional redundancy and sequence homology analyses show that FtsH in thylakoid membranes are present in hexameric heterocomplexes, consisting of four major isomers divided into two major isoforms: Type A (FtsH2 and FtsH8) and Type B (FtsH1 and FtsH5) [13,14]. FtsH2 and FtsH5 are the dominant subunits, and mutations in either cause impaired chloroplast development and maintenance, resulting in leaf variegation phenotypes [13]. Additionally, arabidopsis ftsh5 and ftsh2 mutants exhibited susceptibility to photooxidative stress with concomitant excessive ROS accumulation [13,15], and ftsh2 mutants exhibited susceptibility to 10 °C low temperature and failure to control PSII quality [16].
Although chloroplast FtsH5 and FtsH2 are predominantly responsible for PSII repair and photoprotection, it remains unclear how their functions are modulated. Several reports have suggested that proteins associated with the thylakoid membranes are involved in modulating FtsH proteases. In arabidopsis, a null mutation in THYLAKOID FORMATION (thf1) exhibited a leaf variegation phenotype, and the thf1 mutant had reduced FtsH proteases [17,18,19]. GTPase AtEngA interacts with FtsH complexes and represses FtsH activities [20]. AtFIP, a small thylakoid-anchored protein, interacts with type A FtsH subunits and may negatively modulate FtsH activity [21]. In addition, studies have suggested that FtsH, especially FtsH5, can be phosphorylated in a calcium-dependent manner in the thylakoid, which may partially influence the stability, proteolytic activity, and development of the FtsH complex [22]. AtCIPK13, a calcineurin B-like (CBL)-interacting protein kinase located in the chloroplast, has been speculated to be a candidate kinase involved in calcium-dependent signal transduction in chloroplasts [23,24]. However, the specific kinase responsible for the calcium-dependent protein phosphorylation of FtsH has not been characterized in detail.
FtsH proteases have been widely studied in multiple plant species; however, they are not well studied in tea plants (Camellia sinensis (L.) O. Kuntze). Being a low temperature-sensitive economic crop, a tea plant’s ability to tolerate cold stress has become a vital research topic in the context of climate change. We have previously reported that the calcium signaling pathway and the family of CIPK kinases play a vital role in the cold response of tea plants [25,26]. In the current study, we aimed to determine if CsFtsH5, the ortholog of AtFtsH5, was upregulated by light and low temperature. Here, we report that CsFtsH5 was involved in the cold tolerance of tea plants and interacted with CsCIPK11, which enhanced the accumulation of CsFtsH5 protein. We surmise that the CsFtsH5 regulation by CsCIPK11 is a key process for adjusting chloroplast functions to adapt to environmental needs. These findings will serve as a foundation for further studies on calcium-dependent phosphorylation of thylakoid proteins.
2. Results
2.1. Sequence and Phylogenetic Analyses of CsFtsH5 in Tea Plants
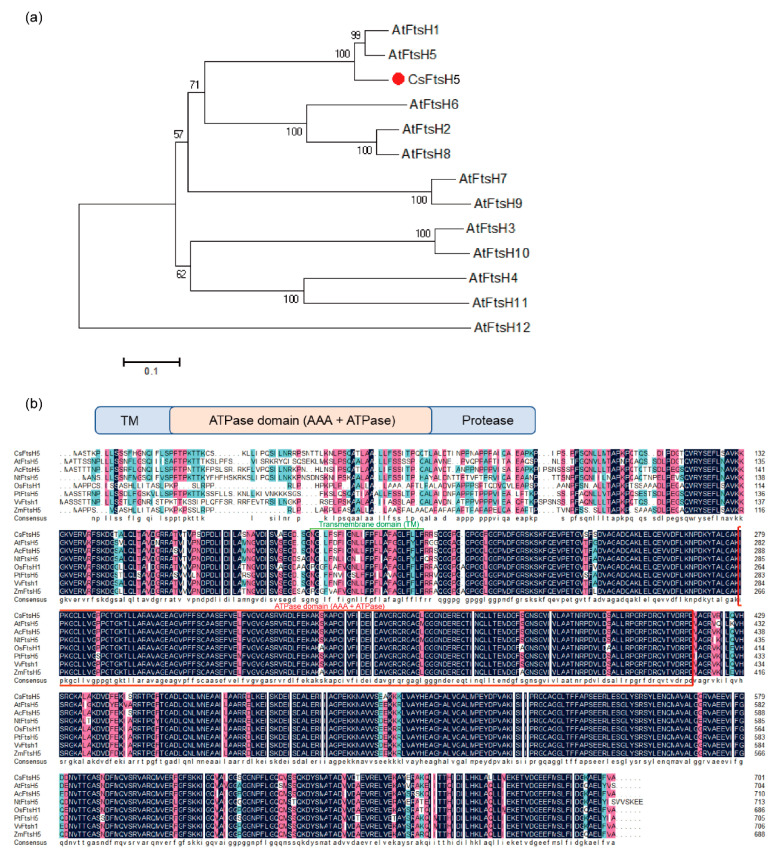
The gene CsFtsH5 was named according to its arabidopsis homolog and was cloned from the leaf cDNA of the ‘Longjing 43’ tea plant. CsFtsH5 encodes a protein of 701 amino acid residues and has a deduced molecular mass of 75.13 kDa. To investigate the evolutionary relationship of CsFtsH5, a phylogenetic tree was constructed using the amino acid sequences of arabidopsis FtsHs. As expected, AtFtsH5 and AtFtsH1 were classified into the same group, and CsFtsH5 shared the closest relationship with AtFtsH5 (85.2% homology) rather than AtFtsH1 (83.9% homology) (Figure 1a). Similar to the typical bioinformatic characteristics of FtsH [9,27], FtsH5 consists of a transmembrane domain at the N-terminus, followed by a highly conserved AAA+ ATPase domain, which is responsible for the binding and hydrolysis of ATP, and a protease domain (Figure 1b). According to the result of multiple sequence alignment, CsFtsH5 shared a high similarity in amino acid sequence with formerly reported FtsH5s in other species in the AAA+ ATPase and protease domains, yet they displayed variation at the N-terminal domain (Figure 1b), indicating that CsFtsH5 may undertake a similar role to other homologous proteins in mediating protein degradation.
Figure 1.
Phylogenetic analyses and protein sequence alignment of CsFtsH5. (a) Neighbor-joining phylogenetic tree of CsFtsH5 and AtFtsHs. The red dot indicates CsFtsH5. (b) Bioinformatic features of CsFtsH5 domains and multiple sequences alignment of CsFtsH5. At: Arabidopsis thaliana; Pt: Populus trichocarpa; Nt: Nicotiana tabacum; Zm: Zea mays L.; Vv: Vitis vinifera; Os: Oryza sativa; Ac: Actinidia Chinensis.
2.2. Expression Pattern and Sub-Cellular Localization of CsFtsH5
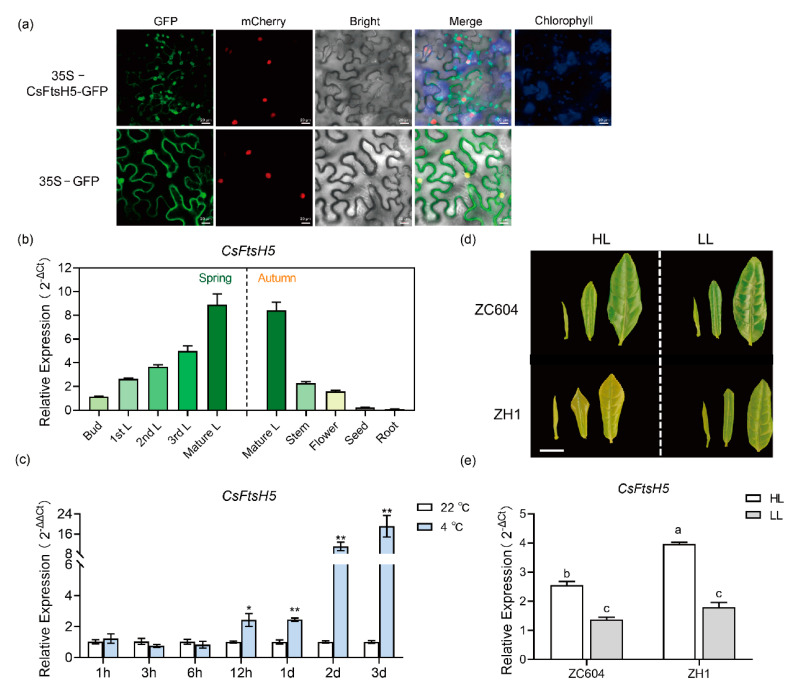
To explore the subcellular localization and potential function of the CsFtsH5 protein, the 35S-GFP (green fluorescent protein) construct fused with CsFtsH5 was generated and transiently expressed in H2B-RFP transgenic N. benthamiana leaves [28]. The vector control, which contained GFP alone, was found throughout the cell, whereas the CsFtsH5-GFP protein was visible in the chloroplast and membrane, which implied the potential role of CsFtsH5 in leaf photosynthesis (Figure 2a). The expression pattern of CsFtsH5 was analyzed in different organs of tea plants. RT-qPCR results showed that CsFtsH5 was detected in all selected tissues, including the first leaf (1st L), second leaf (2nd L), third leaf (3rd L), and mature leaf (mature L) sampled in the spring, and the mature L, stem, flower, seed, and root sampled in the autumn. The levels of CsFtsH5 expression differed among the tissues. CsFtsH5 was highly expressed in leaves, whereas lower expression was detected in roots, flowers, and seeds. Interestingly, the increased expression of CsFtsH5 occurred with leaf maturity (Figure 2b). These results suggest that CsFtsH5 is involved in tea plant growth and development, particularly important for the development of leaves in tea plants.
Figure 2.
Expression profiles and sub-cellular localization of CsFtsH5. (a) Sub-cellular localization of CsFtsH5 protein in H2B-RFP transgenic Nicotiana benthamiana leaves. The empty 35S-GFP was a positive control. Nuclei are indicated by the H2B-RFP (red), and the chloroplasts are shown in blue. Scale bar = 20 µm. (b)The expression patterns of CsFtsH5 in tea plant tissues, including bud, 1st L, 2nd L, 3rd L, mature L, stem, flower, seed, and root. Samples were collected on April, 2020, and October, 2020. (c) The CsFtsH5 expression in tea plants under 22 °C and 4 °C treatments for 1, 3, 6, and 12 h, as well as 1, 2, and 3 d. Data are shown as the mean ± SEM (n = 3). Asterisks indicate significant differences according to t-tests (* p < 0.05, ** p < 0.01). (d) The phenotype of 2 leaves and 1 bud of ZC604 and ZH1 under 100% and 10% sunlight conditions. Scale bar = 1 cm. (e) The expression patterns of CsFtsH5 in leaves of the green-leaf cultivar of ZC604 cultivar and the yellow-leaf cultivar of ZH1 under 100% and 10% sunlight conditions. Means of three replicates and standard errors are presented; different letters above the column indicate a significant difference at p < 0.05 using LSD’s test. Values in (b,e) are expressed relative to the expression levels of the reference gene using the formula 2−ΔCt. Values in (c) are expressed relative to the expression levels of the 22 °C group using the formula 2−ΔΔCt. CsPTB was used as a reference gene in expression analysis.
Given that FtsH is required for chloroplast photosystem II repair, which is a critical process in maintaining a plant’s photosynthetic activity under cold and light stress, we analyzed the response pattern of CsFtsH5 in tea plants to cold stress. RT-qPCR results showed that CsFtsH5 expression was gradually elevated after 12 h under the 4 °C treatment and peaked at the 3 d time point (18.8-fold; Figure 2c). Further, we analyzed the expression pattern of CsFtsH5 in tea leaves under different light intensity treatments (100% sunlight, HL (high light); 10% sunlight, LL (low light)). RT-qPCR results showed that CsFtsH5 was significantly induced by light in both tea cultivars, i.e., the green-leaf cultivar ‘ZC604’ (‘Zhongcha 604’) and the chlorophyll-deficient cultivar ‘ZH1’ (‘Zhonghuang 1’) [29], whose leaves exhibited yellow color under high-light conditions and green under low-light conditions (Figure 2d). Interestingly, the expression level of CsFtsH5 in ‘ZH1’ was higher than that in ‘ZC604’ under HL conditions (Figure 2e), indicating that CsFtsH5 expression is likely related to chloroplast function. These results suggest that CsFtsH5 is involved in light and low-temperature responses in tea plants.
2.3. CsFtsH5 Down-Regulation Causes Tea Leaf Hypersensitivity to Cold Stress
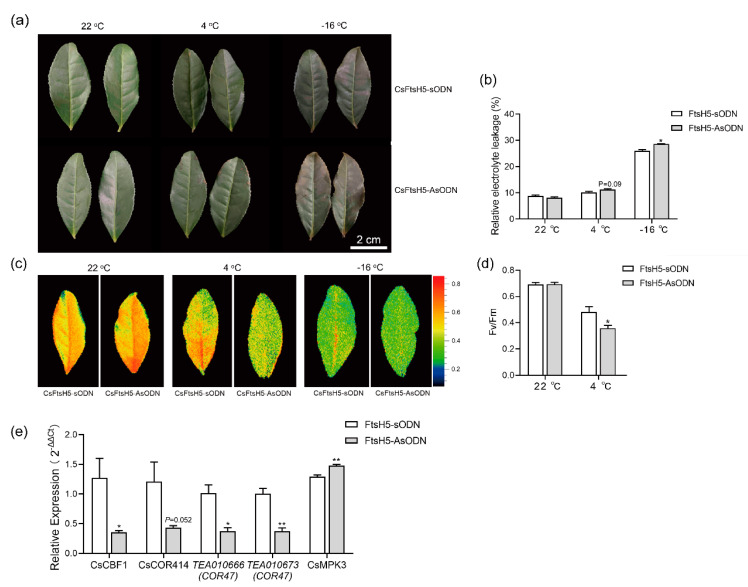
To examine the role of CsFtsH5 in the cold stress responses of the tea plant, we next effectively and transiently suppressed CsFtsH5 expression (Figure S1) by using the gene-specific antisense oligonucleotides (AsODN) method, as previously described by Zhao et al. [30]. There were no obvious differences in plant phenotypes between CsFtsH5-AsODN and CsFtsH5-sODN plants under normal conditions (Figure 3a). However, under cold and freezing stress, the blade edge of CsFtsH5-AsODN leaves was more damaged than that of CsFtsH5-sODN leaves (Figure 3a). We then measured the Fv/Fm values and the relative electrolyte leakage (REL) of the tea leaves before and after low-temperature stress. The CsFtsH5-AsODN plants exhibited significantly lower Fv/Fm values and relatively higher REL values (p = 0.09) than the CsFtsH5-sODN plants under cold stress (Figure 3b–d). Further, the REL value of CsFtsH5-AsODN plants was significantly higher than that of CsFtsH5-sODN plants under freezing stress (Figure 3b). These results suggest that down-regulation of CsFtsH5 leads to enhanced cold sensitivity in tea plants.
Figure 3.
CsFtsH5-AsODN tea plants showed hypersensitivity to cold stress. (a) Phenotypes of CsFtsH5-AsODN and CsFtsH5-sODN plants treated with cold stress (4 °C for 5 d followed by 22 °C recovery for 2 d) and freezing stress (22 °C for 4.5 d followed by −16 °C for 12 h, then 22 °C recovery for 2 d). Control group was grown at 22 °C for 7 d. Scale bar = 2 cm. (b–d). Relative electrolyte leakage, chlorophyll fluorescence images, and Fv/Fm value of CsFtsH5-AsODN and CsFtsH5-sODN plants treated with low temperature. (e) The relative expression of CsCBF and CsCOR in CsFtsH5-AsODN and CsFtsH5-sODN plants under 4 °C low temperature. Mean and standard deviation values were obtained from at least three independent experiments. The asterisks indicated significantly different values between CsFtsH5-AsODN and CsFtsH5-sODN plants according to t-tests (* p < 0.05, ** p < 0.01).
The CBF pathway plays a critical role in plant cold response. Thus, we next examined the expression pattern of the genes involved in the CBF pathway, including the positive regulators of CsCBF1, CsCOR414, and CsCOR47, and the negative regulator of CsMPK3 in CsFtsH5-AsODN and CsFtsH5-sODN plants under cold stress [31]. The results showed that compared with CsFtsH5-sODN leaves, the transcript abundance of CsCBF1, CsCOR414, and CsCOR47 was significantly repressed in CsFtsH5-AsODN tea leaves under cold stress, whereas CsMPK3 was highly expressed in CsFtsH5-AsODN (Figure 3e). Taken together, these data indicate that CsFtsH5 plays a positive role in regulating cold-regulated gene expression in tea plants.
2.4. Interaction between CsFtsH5 and CsCIPK11
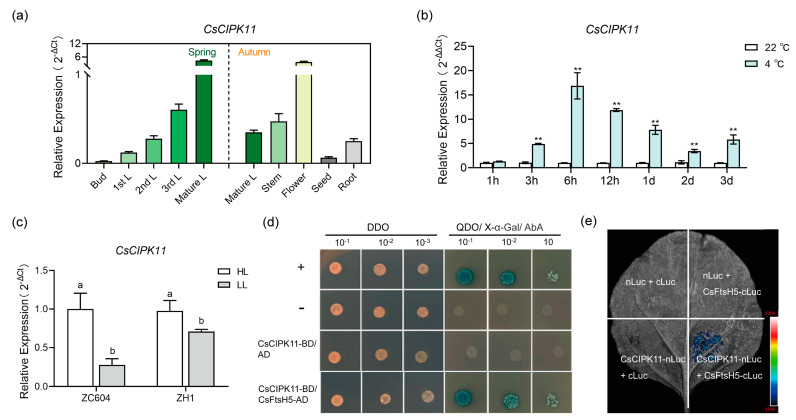
Next, we investigated the molecular mechanism underlying CsFtsH5 involvement in the cold stress response. Considering our previous research involving the CsCIPK family, we focused on CsCIPK11, which showed a tissue expression pattern similar to that of CsFtsH5 (Figure 4a). Expression analysis showed that CsCIPK11 was highly expressed in mature L and flower and exhibited lower expression in bud and seed. Meanwhile, consistent with CsFtsH5, the expression level of CsCIPK11 was positively related to the maturity of the tea leaf (Figure 4a). Further, the expression of CsCIPK11 was also induced by cold stress and light signals (Figure 4b,c). Exposed to 4 °C low temperature, CsCIPK11 in young leaf was rapidly and significantly induced at the 3 h point, and its highest expression appeared at 6 h (Figure 4b). In addition, CsCIPK11 was significantly induced by light in ‘ZC604’ and ‘ZH1’ (Figure 4c).
Figure 4.
CsFtsH5’s interaction with CsCIPK11. (a) The expression patterns of CsCIPK11 in tea plant tissues, including bud, 1st L, 2nd L, 3rd L, mature L, stem, flower, seed, and root. Samples were collected on April 2020 and October 2020. (b) The CsCIPK11 expression in tea plants under 22 °C and 4 °C treatments for 1, 3, 6, and 12 h, as well as 1, 2, and 3 d. Data are shown as the mean ± SEM (n = 3). Asterisks indicate significant differences according to t-tests (** p < 0.01). (c) The expression patterns of CsCIPK11 in leaves of ZC604 cultivar and ZH1 cultivar under 100% and 10% sunlight conditions. Means of three replicates and standard errors are presented; different letters above the column indicate significant difference at p < 0.05 using LSD’s test. Values in (a,c) are expressed relative to the expression levels of reference gene using formula 2−ΔCt. Values in (b) are expressed relative to the expression levels of 22 °C group using formula 2−ΔΔCt. CsPTB was used as a reference gene in expression analysis. (d) Y2H assay of pGBKT7-CsCIPK11 and pGADT7-CsFtsH5 in AH109 strain. DDO, SD/−Leu/−Trp; QDO, SD/−Leu/−Trp/−His/−Ade; X, X-α-gal. pGBKT7-53 and pGADT7-T acted as the positive control, while pGBKT7-lam and pGADT7-T served as the negative control. (e) Luciferase complementation imaging analysis of CsCIPK11-nLuc and CsFtsH5-cLuc in Nicotiana benthamiana leaves. CsCIPK11-nLuc with cLuc, nLuc with CsFtsH5-cLuc, and nLuc with cLuc served as controls.
Furthermore, the yeast two-hybrid (Y2H) assay showed that cells co-transformed with CsFtsH5 and CsCIPK11 grew and turned blue on QDO (SD/−Leu/−Trp/−His/−Ade; X-α-gal) medium (Figure 4d), indicating that the interaction between CsCIPK11 and CsFtsH5 occurred in yeast. Subsequently, the luciferase complementation imaging (LCI) assay showed that an obvious luminescence signal appeared only in the regions of leaves co-expressing CsFtsH5-cLuc and CsCIPK11-nLuc, whereas the Luc signal was not detected when the empty vector was expressed in the same proportions (Figure 4e). These data are strongly suggestive of an interaction between CsFtsH5 and CsCIPK11.
2.5. CsCIPK11 May Contribute to CsFtsH5 Protein Stability
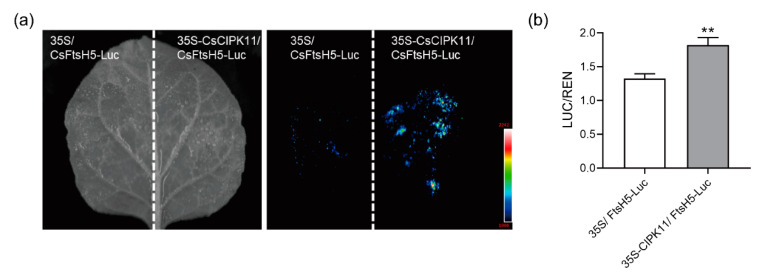
In order to explore the molecular mechanism of the interaction, we further investigated whether the interaction between CsCIPK11 and CsFtsH5 affected the stability of the latter. We co-expressed 35S-CsCIPK11 and an empty vector with 35S-CsFtsH5-Luc in N. benthamiana leaves. The fluorescence signal was stronger in leaves co-expressing 35S-CsCIPK11 and 35S-CsFtsH5-Luc than in leaves co-expressing the 35S-empty vector and 35S-CsFtsH5-Luc (Figure 5a). Further, the relative LUC/REN activity measurements also support the fluorescence signal results (Figure 5b). Thus, our data indicate that CsCIPK11 interacted with CsFtsH5 and increased its accumulation in vivo. CsCIPK11-CsFtsH5 interaction may enhance the stability of CsFtsH5.
Figure 5.
CsCIPK11 may stabilize the CsFtsH5 protein. (a) Fluorescence observations in Nicotiana benthamiana leaves that expressed 35S-CsCIPK11/35S-CsFtsH5-Luc and 35S-empty vector/35S-CsFtsH5-Luc. (b) Relative LUC/REN activity measurements in dual-Luciferase assays. The relative LUC/REN value is the average of six biological replicates. Error bars indicate the SEM of six biological replicates. Asterisks indicate significant differences according to t-tests (** p < 0.01).
3. Discussion
The chloroplast is the most susceptible organelle to low-temperature-induced oxidative damage. FtsH metalloproteases are identified as the master regulators in maintaining the photosynthetic activity of chloroplast [8,32]. Considering the growth and development of tea plants are limited by low temperatures, a better understanding of the role of CsFtsH in cold stress responses can provide meaningful information for adversity-resistance breeding in tea plants.
3.1. Metalloprotease CsFtsH5 of Tea Plant Is Homologous to FtsH5
FtsH proteases are membrane-bound metalloproteases that are present in eubacteria, animals, and plants. Here, we isolated and identified CsFtsH5 from tea plants. Sequence and phylogenetic analyses showed that CsFtsH5 is a protein homologous to FtsH5 and contains highly conserved ATPase and protease domains (Figure 1a,b), indicating that FtsH is an evolutionarily conserved protein across various species. The ATPase domain is involved in protein degradation, and its central pore allows substrate proteins to unfurl and enter the proteolytic chamber, where polypeptide chains are gradually broken down into short peptides [33]. Thus, we speculate that CsFtsH5 may be involved in protein degradation, especially D1 degradation, similar to AtFtsH2 in arabidopsis and PvFtsH2 in common beans [7,16,34,35]. The role of CsFtsH5 in D1 degradation requires further investigation. Interestingly, while AtFtsH5 is located only in the chloroplast [15], we found that CsFtsH5 was located both in the chloroplast and membrane (Figure 2a). This observation raises the possibility of different roles played by FtsH5 between arabidopsis and tea plants. Loss of AtFtsH5 caused leaf variegation in arabidopsis [15]. CsFtsH5 was primarily expressed in leaves, and the expression level was positively correlated with leaf maturity (Figure 2b), suggesting that CsFtsH5 is potentially involved in leaf development in tea plants. Further, we found that similar to AtFtsH5 [15], CsFtsH5 is also a light-responsive gene (Figure 2d,e). Noticeably, in response to light exposure, yellow leaves displayed greater CsFtsH5 expression compared to green leaves (Figure 2e). These results are consistent with those obtained by a previous study on chrysanthemums, which reported that CmFtsH expression is regulated by light and is higher expressed in the yellow leaf tissue [36]. Thus, such responses indicate a feedback regulation in which chrysanthemun and tea plants enter an altered physiological status followed by activation of CmFtsH and CsFtsH5, respectively.
3.2. Repressing CsFtsH5 Facilitated the Cold Sensitivity of Tea Plants
FtsH proteases are necessary for maintaining the quality of membrane protein and preventing chloroplast damage from cold and heat [16,37], which coincided with the location of CsFtsH5 in the chloroplast and membrane (Figure 2a). We noticed a significant induction of CsFtsH5 expression by low-temperature treatment (Figure 2c), indicating that CsFtsH5 is responsive to low temperatures in tea plants.
When arabidopsis is exposed to high temperatures, the photosynthetic capability of the ftsh11 mutant is greatly reduced. Further, the AtFtsH11 protease alleviates the thermotolerance damage of the photosynthesis apparatus [38]. Compared with the wild type, ftsh2 mutants exhibit a drastic decline in PSII activity and a significant cell death response under low-temperature and high-light-intensity conditions [16]. Those results showed that FtsH participated in temperature stress by regulating chloroplast activity. Our study also characterized the role of CsFtsH5 in response to low temperatures. Downregulation of CsFtsH5 enhanced cold sensitivity (Figure 3), suggesting that CsFtsH5 protected the tea plant from low-temperature stress, likely by maintaining the PSII activity, as indicated by the Fv/Fm results. In addition, the downregulating of CsFtsH5 in tea leaves suppressed the CBF-mediated cold response pathway (Figure 3e). We speculate that suppressing CsFtsH5 in tea leaves induced PSII damage and oxidative stress in the chloroplast, which may influence the CBF pathway and plant cold tolerance. However, the underlying mechanism needs to be further explored in future studies.
3.3. Regulation of CsFtsH5 by CsCIPK11 Likely Mediates the Cold Response of Tea Plants
Several studies have reported the molecular mechanisms underlying FtsH regulation in plants. For example, a soybean study reported that GmFtsH25 interacts with photosystem I light-harvesting complex 2 (GmLHCa2), which contributes to enhanced photosynthesis [39]. An arabidopsis study found that AtFtsH11 is a critical regulator of photosynthetic thermotolerance. AtFtsH11 interacted with the ATP synthase assembly factor BFA3, which degraded under high temperatures, leading to the adjustment of ATP synthase assembly in the chloroplast in response to heat stress [40]. Furthermore, AtTHF1 is necessary for AtFtsH to accumulate normally in the thylakoid membrane. AtFtsH proteases were reduced in the thf1 mutant, leading to a leaf variegation phenotype [19]. AtEngA interacts with AtFtsH, and overexpression of EngA enhances FtsH stability, which engenders the dysfunction of FtsH protease activity in chloroplasts [20]. Therefore, proper turnover of FtsH proteases is crucial for their activity [20]. However, the detailed molecular mechanisms underlying FtsH5 regulation in plants remain unclear.
Ca2+ is a vital secondary messenger in both plant development and early stress responses [41]. AtFtsH5 is phosphorylated in the thylakoid in a calcium-dependent manner, which possibly regulates FtsH stability in thylakoid membranes [22,24]. Extensive functional studies have demonstrated the broad involvement of CIPK family members in plant development and stress responses. For example, AtCIPK13 was found to be located in the chloroplast; therefore, AtCIPK13 was assumed to operate in chloroplasts and mediate calcium-dependent signal transduction pathways. However, detailed characterization has not yet been performed [22,23]. In agreement with this evidence, the current study found that the expression pattern of CsCIPK11 was congruent with CsFtsH5 in leaves with different maturities, as well as in response to cold and light (Figure 4a–c). Based on this result, we assessed the evidence for the interaction between CsFtsH5 and CsCIPK11 (Figure 4d,e) and found that CsCIPK11 may affect the stability of CsFtsH5 by interacting with it (Figure 5). Ca2+ is an important second messenger related to cold stimulus, and CIPK mediates the translation of Ca2+ information into downstream-specific responses. The response of CsCIPK11 was quicker than that of CsFtsH5 under cold stress (Figure 2c and Figure 4b). We speculated that CsCIPK11 might be the upstream regulator of CsFtsH5 under cold stress. Considering that the phosphorylation of thylakoid proteins is one of the most important regulatory mechanisms of photosynthesis, it is not yet clear if CsCIPK11 enhances the stability of CsFtsH5 protein through phosphorylation. Future studies should assess the role of CsCIPK11 and characterize whether phosphorylation of CsFtsH5 may be a means to alter its activity.
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
Tea plant tissues, including bud, 1st L, 2nd L, 3rd L, mature L, stem, flower, seed, and root, were collected for RNA extraction from the 5-year-old ‘Longjing 43’ tea plant cultivar grown in the field at the Tea Research Institute of the Chinese Academy of Agricultural Sciences, Hangzhou, People’s Republic of China. Samples were collected in the spring (April 2020) and autumn (October 2020). To investigate CsFtsH5 and CsCIPK11 expression in response to cold stress, ‘Longjing 43’ tea plants were treated at 22 °C (control treatment) and 4 °C (experimental treatment) and maintained under the same photoperiod (12 h/12 h, light/dark). The first mature leaves were harvested for RNA extraction at 1, 3, 6, and 12 h, as well as 1, 2, and 3 d after treatment. To investigate CsFtsH5 and CsCIPK11 expression in response to light, 10-year-old tea plant cultivars of ‘ZC604’ and ‘ZH1’ with the same phenological phase were transferred to 100% and 10% sunlight conditions, and treatment was started from March 22 to April 8, 2019. Young shoots with two leaves and one bud were randomly collected for RNA extraction. Three independent biological replicates were used for each analysis. All samples were stored at −80 °C after being flash-frozen in liquid nitrogen for further experiments.
4.2. RNA Extraction and RT-qPCR Reactions
Total RNA of tea plant leaves and other tissues was isolated using the TIANGEN RNAprep Pure Plant Kit (Beijing, China), according to the manufacturer’s instructions. Complementary DNA (cDNA) templates for quantitative RT-PCR (RT-qPCR) were prepared using a TaKaRa PrimeScript™ RT reagent kit (Kusatsu, Japan) with 1 μg of total RNA according to the manufacturer’s protocol. cDNA was amplified by PCR using a Roche Light Cycler 480 (Basel, Switzerland) with SYBR Green I Master Mix of Roche (Basel, Switzerland). Two technical replicates and three biological replicates were performed. CsPTB was used as a reference gene in the expression level analysis using the formula 2−ΔCt or 2−ΔΔCt [42]. All RT-qPCR primer sequences are listed in Table S1.
4.3. Sequence Alignment and Phylogenetic Tree Analysis
RT-PCR was used to amplify the CDS of CsFtsH5 from the cDNA of ‘Longjing 43’ leaves. Primers are listed in Table S2. The molecular weight of the CsFtsH5 protein was computed by ProtParam (https://web.expasy.org/protparam/, accessed on 1 May 2022). AtFtsH and CsFtsH5 protein sequences were used to construct a phylogenetic tree. Alignment and phylogenetic tree assembly were performed using default settings and the neighbor-joining algorithm of MEGA version 7; 1000 bootstrap trials were used. The amino acid sequences of AtFtsH and CsFtsH5 are shown in Table S3. The AAA+ ATPase domain was identified using the NCBI Conserved Domain Database (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 15 August 2022). The homologous sequence of CsFtsH5 was analyzed using BLASTp against the NCBI database. The alignment of FtsH5 protein sequences was performed using DNAMAN.
4.4. Subcellular Localization of CsFtsH5
To determine the subcellular localization of CsFtsH5, its open reading frame was cloned into a 35S-GFP vector which contained a C-terminal eGFP fragment [43]. The 35S-GFP and 35S-CsFtsH5-GFP vector plasmids were transformed into Agrobacterium strain GV3101 and transiently expressed in Nicotiana benthamiana expressing an H2B-RFP protein [28]. After 48 h, confocal microscopy images of the leaf cells were taken using a Zeiss LSM710 confocal laser scanning microscope (Oberkochen, Germany).
4.5. Down-Regulation of CsFtsH5 Gene in Tea Plant
According to a previously characterized procedure for gene suppression in tea leaves [30], candidate AsODNs were obtained using Soligo software 2.2 with CsFtsH5 as the input sequence. AsODNs and sODNs were synthesized by Youkang Biosystems Company (Zhejiang, China) (Table S4). To suppress the expression of CsFtsH5 in ‘Longjing 43’ tea plant leaves, tea plants were grown under dark conditions for 20 min before injection, 1 mL of 25 μM CsFtsH5-AsODN solution was injected fully into the lower epidermis of the first mature leaf, and leaves injected with the equivalent sODN were used as controls.
4.6. Low-Temperature Treatment and Measurement of Fv/Fm and REL
CsFtsH5-AsODN plants and CsFtsH5-sODN plants were treated with cold stress (4 °C for 5 d, followed by 22 °C recovery for 2 d) and freezing stress (22 °C for 4.5 d, followed by −16 °C for 12 h, and then 22 °C recovery for 2 d) immediately after injection with oligonucleotides. CsFtsH5-sODN plants were used as controls. The phenotypes of the CsFtsH5-AsODN and CsFtsH5-sODN plants were recorded after recovery. Injected leaves were immediately collected after low-temperature treatment to measure the maximum photochemical efficiency of PSII (Fv/Fm) and REL. For REL measurement, 10 leaf discs punched using a 1 cm diameter hole puncher were collected and placed in a 50 mL tube with 10 mL of distilled water. The tubes were shaken at 200 rpm at 25 °C for 2 h. The conductivity of the solutions was measured at 25 °C using an Orion 5 Star conductivity meter of Thermo Fisher Scientific (Waltham, Massachusetts, USA) at 25 °C, as previously described [44]. Three independent biological replicates were used for REL determination. To measure Fv/Fm, CsFtsH5-AsODN and CsFtsH5-sODN plants were placed in the dark for 20 min, and Fv/Fm values were measured using FluorCam 7 of Photon Systems Instruments (Drasov, Czech Republic). The fluorescence parameters were integrated over the entire leaf area. Eight independent biological replicates were used for Fv/Fm determination.
4.7. Y2H
The full-length CDS of CsFtsH5 and CsCIPK11 were amplified from the cDNA of ‘Longjing 43’ tea plant leaves and cloned into pGADT7 (AD) and pGBKT7 (BD), respectively. According to the manufacturer’s protocol, the recombinant AD and BD vectors were co-transformed into yeast AH109 chemically competent cells of Weidi (Shanghai, China) in specified combinations. The AH109 cells co-expressing pGADT7-T with pGBKT7-p53 were used as the positive control (+). The AH109 cells co-expressing pGADT7-T with pGBKT7-lam and CsCIPK11-BD with the AD empty vector were used as the negative control. Positive transformants grown on DDO:SD/−Leu/−Trp medium (without tryptophan and leucine) were plated on the screening medium QDO: SD/−Ade/−His/−Leu/−Trp (without tryptophan, leucine, adenine, and histidine) to investigate the interactions. Serial dilutions (OD600 = 0.2, 0.02, and 0.002) were plated on DDO and QDO medium. Then, the plates were incubated at 30 °C for 7 days and recorded.
4.8. LCI Analysis
To perform the LCI assays, CsFtsH5 and CsCIPK11 CDSs were cloned into the C-terminal half of luciferase (C-Luc) and into the N-terminal half of luciferase (N-Luc). The plasmids were transformed into Agrobacterium strain GV3101-P19 of Weidi (Shanghai, China) and transiently expressed in N. benthamiana. The leaves co-expressing CsCIPK11-nLuc with cLuc, nLuc with CsFtsH-cLuc, as well as nLuc with cLuc, were used as the negative control. After 48 h, the LCI assay was performed using a chemiluminescence image analysis system of Tanon (Shanghai, China).
4.9. Dual-Luc Assays
The Dual-Luc assay was performed to analyze the stability of CsFtsH5, as previously described [45]. The coding regions of CsFtsH5 were placed under the control of the 35S promoter, and the cassettes were inserted into pGreenII 0800-Luc, which contains the 35S-REN cassette, to generate pGreenII 0800-35S-CsFtsH5-Luc constructs. Subsequently, 35S-CsCIPK11 and 35S-empty vectors were co-expressed with 35S-CsFtsH5-Luc constructs in N. benthamiana. After 48 h, the LCI assay was performed using a chemiluminescence image analysis system of Tanon (Shanghai, China). These N. benthamiana specimens were then collected for the Dual-Luc assay using the Dual-Luciferase Reporter System of Promega (Madison, WI, USA) according to the manufacturer’s instructions, as previously described [46]. Six independent biological replicates were taken. The REN activity for each reaction was used as an internal control.
4.10. Statistical Analysis
All experiments were performed with at least three independent biological replicates. Data represent the mean ± SEM of biological replicates. Data were statistically analyzed using t-tests and LSD tests performed using SPSS. Images were handled using GraphPad Prism 9, Adobe Photoshop, and Adobe Illustrator.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24076288/s1.
Author Contributions
L.W. and D.N. conceived, designed, and supervised the experiment. T.D., Y.W., J.P., J.W., H.W., M.H., N.L. and X.H. performed the experiments. L.W., T.D., Y.Y. and X.W. analyzed and discussed the data. T.D. and L.W. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
All the authors in the manuscript have no conflicts of interest.
Funding Statement
This work was supported by the National Natural Science Foundation of China (U22A20499 and 31870685), the Central Public Interest Scientific Institution Basal Research Fund (1610212018007), the Project for Collaborative Promotion of Major Technology of Zhejiang province (2022XTTGCY01-02), and the China Agriculture Research System of MOF and MARA (CARS-19-01A).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Zimorski V., Ku C., Martin W.F., Gould S.B. Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 2014;22:38–48. doi: 10.1016/j.mib.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 2.Leister D. Chloroplast research in the genomic age. Trends Genet. 2003;19:47–56. doi: 10.1016/S0168-9525(02)00003-3. [DOI] [PubMed] [Google Scholar]
- 3.Mishra N.P., Francke C., van Gorkom H.J., Ghanotakis D.F. Destructive role of singlet oxygen during aerobic illumi-nation of the photosystem II core complex. Biochim. Biophys. Acta. 1994;1186:81–90. doi: 10.1016/0005-2728(94)90138-4. [DOI] [Google Scholar]
- 4.Nixon P.J., Michoux F., Yu J., Boehm M., Komenda J. Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 2010;106:1–16. doi: 10.1093/aob/mcq059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Järvi S., Suorsa M., Aro E.-M. Photosystem II repair in plant chloroplasts—Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta (BBA) Bioenerg. 2015;1847:900–909. doi: 10.1016/j.bbabio.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 6.Kato Y., Sakamoto W. Protein quality control in chloroplasts: A current model of D1 protein degradation in the photo-system II repair cycle. J. Biochem. 2009;146:463–469. doi: 10.1093/jb/mvp073. [DOI] [PubMed] [Google Scholar]
- 7.Bailey S., Thompson E., Nixon P.J., Horton P., Mullineaux C.W., Robinson C., Mann N.H. A Critical Role for the Var2 FtsH Homologue of Arabidopsis thaliana in the Photosystem II Repair Cycle in Vivo. J. Biol. Chem. 2002;277:2006–2011. doi: 10.1074/jbc.M105878200. [DOI] [PubMed] [Google Scholar]
- 8.Kato Y., Miura E., Ido K., Ifuku K., Sakamoto W. The Variegated Mutants Lacking Chloroplastic FtsHs Are Defective in D1 Degradation and Accumulate Reactive Oxygen Species. Plant Physiol. 2009;151:1790–1801. doi: 10.1104/pp.109.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suno R., Niwa H., Tsuchiya D., Zhang X., Yoshida M., Morikawa K. Structure of the Whole Cytosolic Region of ATP-Dependent Protease FtsH. Mol. Cell. 2006;22:575–585. doi: 10.1016/j.molcel.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 10.Santos D., Almeida D.F.D. Isolation and characterization of a new temperature-sensitive cell division mutant of esch-erichia coli K-12. J. Bacteriol. 1975;124:1502–1507. doi: 10.1128/jb.124.3.1502-1507.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adam Z., Zaltsman A., Sinvany-Villalobo G., Sakamoto W. FtsH proteases in chloroplasts and cyanobacteria. Physiol. Plant. 2005;123:386–390. doi: 10.1111/j.1399-3054.2004.00436.x. [DOI] [Google Scholar]
- 12.Tatsuta T., Langer T. AAA proteases in mitochondria: Diverse functions of membrane-bound proteolytic machines. Res. Microbiol. 2009;160:711–717. doi: 10.1016/j.resmic.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. Coordinated regulation and complex formation of yellow varie-gated1 and yellow variegated2, chloroplastic FtsH metalloproteases involved in the repair cycle of photosystem II in Ara-bidopsis thylakoid membranes. Plant Cell. 2003;15:2843–2855. doi: 10.1105/tpc.017319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaltsman A., Ori N., Adam Z. Two types of FtsH protease subunits are required for chloroplast biogenesis and pho-tosystem II repair in Arabidopsis. Plant Cell. 2005;17:2782–2790. doi: 10.1105/tpc.105.035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakamoto W., Tamura T., Hanba-Tomita Y., Murata M. The VAR1 locus of Arabidopsis encodes a chloroplastic FtsH and is responsible for leaf variegation in the mutant alleles. Genes Cells Devoted Mol. Cell. Mech. 2002;7:769–780. doi: 10.1046/j.1365-2443.2002.00558.x. [DOI] [PubMed] [Google Scholar]
- 16.Duan J., Lee K.P., Dogra V., Zhang S., Liu K., Caceres-Moreno C., Lv S., Xing W., Kato Y., Sakamoto W., et al. Impaired PSII Proteostasis Promotes Retrograde Signaling via Salicylic Acid. Plant Physiol. 2019;180:2182–2197. doi: 10.1104/pp.19.00483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q., Sullivan R.W., Kight A., Henry R.L., Huang J., Jones A.M., Korth K.L. Deletion of the Chloroplast-Localized Thylakoid Formation1 Gene Product in Arabidopsis Leads to Deficient Thylakoid Formation and Variegated Leaves. Plant Physiol. 2004;136:3594–3604. doi: 10.1104/pp.104.049841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang W., Chen Q., Zhu Y., Hu F., Zhang L., Ma Z., He Z., Huang J. Arabidopsis thylakoid formation 1 is a criti-cal regulator for dynamics of PSII–LHCII complexes in leaf senescence and excess light. Mol. Plant. 2013;6:1673–1691. doi: 10.1093/mp/sst069. [DOI] [PubMed] [Google Scholar]
- 19.Zhang L., Wei Q., Wu W., Cheng Y., Hu G., Hu F., Sun Y., Zhu Y., Sakamoto W., Huang J. Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidop-sis. Plant J. Cell Mol. Biol. 2009;58:1041–1053. doi: 10.1111/j.1365-313X.2009.03843.x. [DOI] [PubMed] [Google Scholar]
- 20.Kato Y., Hyodo K., Sakamoto W. The Photosystem II Repair Cycle Requires FtsH Turnover through the EngA GTPase. Plant Physiol. 2018;178:596–611. doi: 10.1104/pp.18.00652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes K.L., Rodrigues R.A.O., Silva M.C., Braga W.G.S., Silva-Filho M.C. The Zinc-Finger Thylakoid-Membrane Protein FIP Is Involved with Abiotic Stress Response in Arabidopsis thaliana. Front. Plant Sci. 2018;9:504. doi: 10.3389/fpls.2018.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stael S., Rocha A.G., Wimberger T., Anrather D., Vothknecht U.C., Teige M. Cross-talk between calcium signalling and protein phosphorylation at the thylakoid. J. Exp. Bot. 2011;63:1725–1733. doi: 10.1093/jxb/err403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schliebner I., Pribil M., Zühlke J., Dietzmann A., Leister D. A Survey of chloroplast protein kinases and phospha-tases in Arabidopsis thaliana. Curr. Genom. 2008;9:184–190. doi: 10.2174/138920208784340740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kato Y., Sakamoto W. Phosphorylation of the Chloroplastic Metalloprotease FtsH in Arabidopsis Characterized by Phos-Tag SDS-PAGE. Front. Plant Sci. 2019;10:1080. doi: 10.3389/fpls.2019.01080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X.-C., Zhao Q.-Y., Ma C.-L., Zhang Z.-H., Cao H.-L., Kong Y.-M., Yue C., Hao X.-Y., Chen L., Ma J.-Q., et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genom. 2013;14:415. doi: 10.1186/1471-2164-14-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang L., Feng X., Yao L., Ding C., Lei L., Hao X., Li N., Zeng J., Yang Y., Wang X. Characterization of CBL–CIPK signaling complexes and their involvement in cold response in tea plant. Plant Physiol. Biochem. 2020;154:195–203. doi: 10.1016/j.plaphy.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Tomoyasu T., Yuki T., Morimura S., Mori H., Yamanaka K., Niki H., Hiraga S., Ogura T. The Escherichia coli FtsH protein is a prokaryotic member of a protein family of putative ATPases involved in membrane functions, cell cycle control, and gene expression. J. Bacteriol. 1993;175:1344–1351. doi: 10.1128/jb.175.5.1344-1351.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li F., Zhao N., Li Z., Xu X., Wang Y., Yang X., Liu S.-S., Wang A., Zhou X. A calmodulin-like protein suppresses RNA silencing and promotes geminivirus infection by degrading SGS3 via the autophagy pathway in Nicotiana benthamiana. PLoS Pathog. 2017;13:e1006213. doi: 10.1371/journal.ppat.1006213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L., Cao H., Chen C., Yue C., Hao X., Yang Y., Wang X. Complementary transcriptomic and proteomic analyses of a chlorophyll-deficient tea plant cultivar reveal multiple metabolic pathway changes. J. Proteom. 2016;130:160–169. doi: 10.1016/j.jprot.2015.08.019. [DOI] [PubMed] [Google Scholar]
- 30.Zhao M., Zhang N., Gao T., Jin J., Jing T., Wang J., Wu Y., Wan X., Schwab W., Song C. Sesquiterpene glucosyla-tion mediated by glucosyltransferase UGT91Q2 is involved in the modulation of cold stress tolerance in tea plants. New Phytol. 2020;226:362–372. doi: 10.1111/nph.16364. [DOI] [PubMed] [Google Scholar]
- 31.Wang L., Di T., Peng J., Li Y., Li N., Hao X., Ding C., Huang J., Zeng J., Yang Y., et al. Comparative metabolomic analysis reveals the involvement of catechins in adaptation mechanism to cold stress in tea plant (Camellia sinensis var. sinensis) Environ. Exp. Bot. 2022;201:104978. doi: 10.1016/j.envexpbot.2022.104978. [DOI] [Google Scholar]
- 32.Komenda J., Tichy M., Prásil O., Knoppová J., Kuviková S., de Vries R., Nixon P.J. The exposed N-terminal tail of the D1 subunit is required for rapid D1 degradation during photosystem II repair in Synechocystis sp PCC 6803. Plant Cell. 2007;19:2839–2854. doi: 10.1105/tpc.107.053868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okuno T., Ogura T. FtsH protease-mediated regulation of various cellular functions. Regul. Proteolysis Microorg. 2013;66:53–69. doi: 10.1007/978-94-007-5940-4_3. [DOI] [PubMed] [Google Scholar]
- 34.Xu K., Zhu J., Zhai H., Wu H., Gao Y., Li Y., Zhu X., Xia Z. A critical role of PvFtsH2 in the degradation of pho-todamaged D1 protein in common bean. Hortic. Res. 2021;8:126. doi: 10.1038/s41438-021-00554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshioka-Nishimura M., Yamamoto Y. Quality control of Photosystem II: The molecular basis for the action of FtsH protease and the dynamics of the thylakoid membranes. J. Photochem. Photobiol. B Biol. 2014;137:100–106. doi: 10.1016/j.jphotobiol.2014.02.012. [DOI] [PubMed] [Google Scholar]
- 36.Chang Q., Chen S., Chen Y., Deng Y., Chen F., Zhang F., Wang S. Anatomical and Physiological Differences and Differentially Expressed Genes Between the Green and Yellow Leaf Tissue in a Variegated Chrysanthemum Variety. Mol. Biotechnol. 2012;54:393–411. doi: 10.1007/s12033-012-9578-8. [DOI] [PubMed] [Google Scholar]
- 37.Wagner R., Aigner H., Pružinská A., Jänkänpää H.J., Jansson S., Funk C. Fitness analyses of Arabidopsis thaliana mutants depleted of FtsH metalloproteases and characterization of three FtsH6 deletion mutants exposed to high light stress, senescence and chilling. New Phytol. 2011;191:449–458. doi: 10.1111/j.1469-8137.2011.03684.x. [DOI] [PubMed] [Google Scholar]
- 38.Chen J., Burke J.J., Velten J., Xin Z. FtsH11 protease plays a critical role in Arabidopsis thermotolerance. Plant J. 2006;48:73–84. doi: 10.1111/j.1365-313X.2006.02855.x. [DOI] [PubMed] [Google Scholar]
- 39.Wang L., Yang Y., Yang Z., Li W., Hu D., Yu H., Li X., Cheng H., Kan G., Che Z., et al. GmFtsH25 overexpression increases soybean seed yield by enhancing photosynthesis and photo-synthates. J. Integr. Plant Biol. 2022 doi: 10.1111/jipb.13405. ahead of print . [DOI] [PubMed] [Google Scholar]
- 40.Yue X., Ke X., Shi Y., Li Y., Zhang C., Wang Y., Hou X. Chloroplast inner envelope protein FtsH11 is involved in the adjustment of assembly of chloroplast ATP synthase under heat stress. Plant Cell Environ. 2023;46:850–864. doi: 10.1111/pce.14525. [DOI] [PubMed] [Google Scholar]
- 41.Hepler P.K. Calcium: A Central Regulator of Plant Growth and Development. Plant Cell. 2005;17:2142–2155. doi: 10.1105/tpc.105.032508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao X., Horvath D.P., Chao W.S., Yang Y., Wang X., Xiao B. Identification and Evaluation of Reliable Reference Genes for Quantitative Real-Time PCR Analysis in Tea Plant (Camellia sinensis (L.) O. Kuntze) Int. J. Mol. Sci. 2014;15:22155–22172. doi: 10.3390/ijms151222155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao L., Ding C., Hao X., Zeng J., Yang Y., Wang X., Wang L. CsSWEET1a and CsSWEET17 Mediate Growth and Freezing Tolerance by Promoting Sugar Transport across the Plasma Membrane. Plant Cell Physiol. 2020;61:1669–1682. doi: 10.1093/pcp/pcaa091. [DOI] [PubMed] [Google Scholar]
- 44.Wang L., Yao L., Hao X., Li N., Qian W., Yue C., Ding C., Zeng J., Yang Y., Wang X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018;96:577–592. doi: 10.1007/s11103-018-0716-y. [DOI] [PubMed] [Google Scholar]
- 45.Xu F., He S., Zhang J., Mao Z., Wang W., Li T., Hua J., Du S., Xu P., Li L., et al. Photoactivated CRY1 and phyB Interact Directly with AUX/IAA Proteins to Inhibit Auxin Signaling in Arabidopsis. Mol. Plant. 2017;11:523–541. doi: 10.1016/j.molp.2017.12.003. [DOI] [PubMed] [Google Scholar]
- 46.Peng J., Li N., Di T., Ding C., Li X., Wu Y., Hao X., Wang Y., Yang Y., Wang X., et al. The interaction of CsWRKY4 and CsOCP3 with CsICE1 regulates CsCBF1/3 and mediates stress response in tea plant (Camellia sinensis) Environ. Exp. Bot. 2022;199:104892. doi: 10.1016/j.envexpbot.2022.104892. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.