Abstract
The mechanistic target of rapamycin (mTOR) is a key regulator of cell growth, integrating growth factor signaling and nutrient availability, and is a downstream effector of oncogenic receptor tyrosine kinases (RTKs) and PI3K/Akt signaling. Thus, activating mTOR mutations would be expected to enhance growth in many tumor types. However, tumor sequencing data revealed that mTOR mutations are enriched only in Renal Clear Cell Carcinoma (RCCC) – a clinically hypervascular tumor unlikely to be constrained by nutrient availability. To further define this cancer type-specific restriction, activating mutations in mTOR were studied. All mTOR mutants tested enhanced growth in a cell type agnostic manner under nutrient-replete conditions, but were detrimental to cell survival in nutrient-poor conditions. Consistent with this, analysis of tumor data demonstrated that oncogenic mutations in the nutrient-sensing arm of the mTOR pathway display a similar phenotype and were found to be exceedingly rare in human cancers of all types. Together, these data suggest that maintaining the ability to turn off mTOR signaling in response to changing nutrient availability is retained in most naturally occurring tumors.
Keywords: mTOR, Renal Clear Cell Carcinoma, oncogenesis, nutrient-sensing
Introduction
Cellular metabolic reprogramming to promote growth and proliferation is one of the hallmarks of cancer. Normal cells, after being stimulated by growth factors, initiate a receptor tyrosine kinase (RTK)-PI3K-Akt-pathway that leads to the activation of the kinase mechanistic target of rapamycin (mTOR). mTOR is the catalytic subunit of both the mTOR complex 1 (mTORC1) and the mTOR complex 2 (mTORC2)(1) (2). mTORC2 contributes to growth factor signaling and the regulation of cytoskeletal rearrangements, while mTORC1 promotes cell growth by increasing nutrient uptake, de novo lipid, nucleotide, and protein synthesis, and regulating cell cycle progression (3–5). mTORC1 also inhibits catabolic processes such as autophagy and lysosomal biogenesis (Figure 1a) (2,6–8). In the absence of growth signals, mTORC1 remains inactive as a result of Tuberous sclerosis complex (TSC) inhibition of Rheb, the GTPase that activates mTOR kinase. Growth factor signaling through Akt inhibits the TSC complex and promotes mTORC1 activation. In addition to regulation by growth factor signaling, mTORC1 activity is also modulated by a nutrient sensing regulatory arm (9).
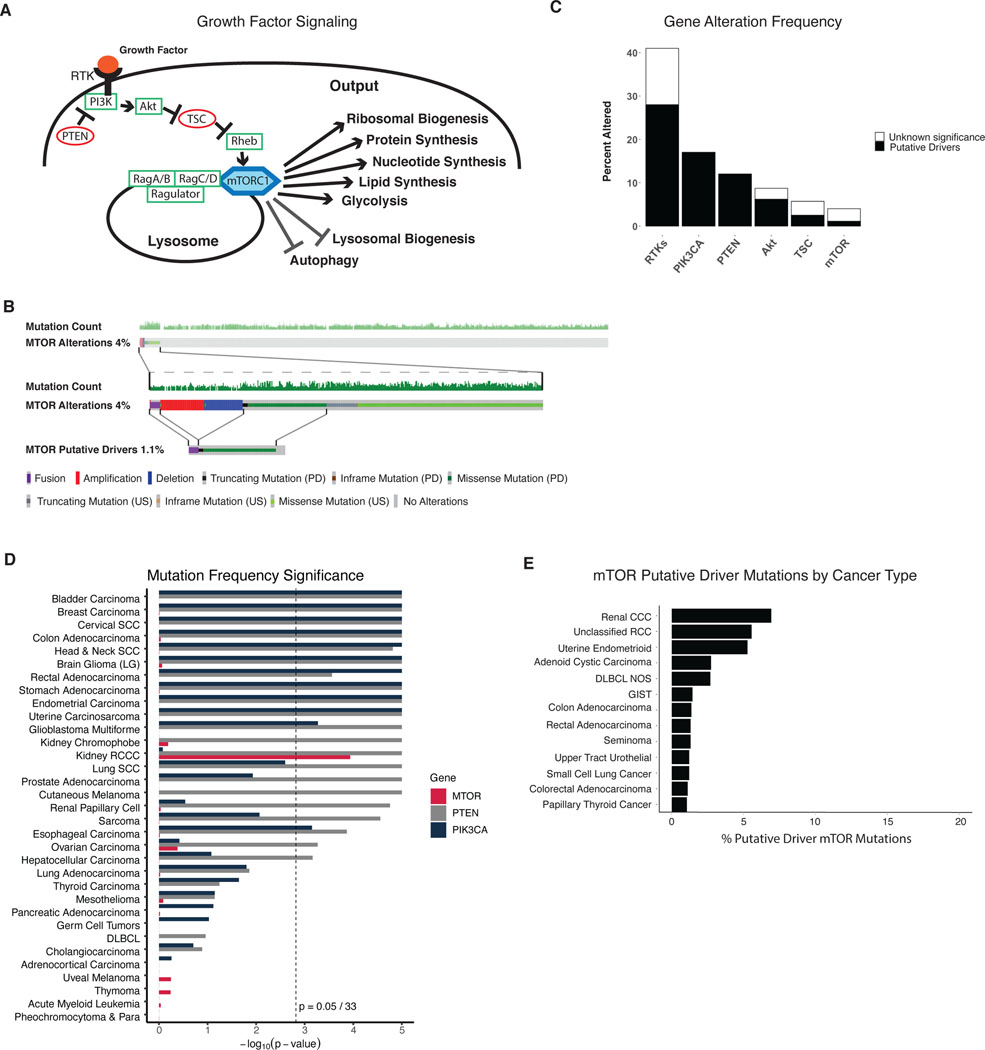
Figure 1: mTOR mutations are rare and significantly enriched only in Renal Clear Cell Carcinoma.
A) Model of Growth factor mediated signaling resulting in mTORC1 activation and cellular output of mTORC1 activity. B) Oncoprint of mTOR alterations within the TCGA pan-cancer dataset. Top panel indicates each sample within the TCGA, with color coded alterations of mTOR shown in 4% of samples. Middle panel shows samples with mTOR alterations at higher magnification. Bottom panel shows only putative driver mTOR alterations. PD = putative driver, US = unknown significance. C) Gene alteration frequency of different oncogenes within the TCGA pan-cancer dataset. RTKs include EGFR, ERBB2, ERBB3, ERBB4, MET, ALK, RET, ROS1, FGFR1, FGFR2, FGFR3, KDR, FLT1, FLT2, FLT3, FLT4, and PDGFRA. Akt includes Akt1, Akt2, and Akt3. TSC includes TSC1 and TSC2. (D) MutSigCV analysis of TCGA pan-cancer studies of mTOR (red), PTEN (grey), and PIK3CA (dark blue), showing the p-values for the significance of the mutational frequency of each gene within each TCGA cancer study. Dotted lines show p = 0.05/33. E) Enrichment of putative driver mTOR mutations by cancer type within MSK IMPACT dataset.
While some oncogenic drivers can indirectly activate mTOR, the canonical oncogenic pathway regulating mTOR is the RTK-PI3K-Akt-mTOR axis(10,11). This pathway is negatively regulated by two major tumor suppressors, the TSC1/TSC2 complex and PTEN. Mutations in each of these upstream components are thought to have the ultimate effect of activating mTOR to promote uncontrolled growth. Supporting a role of mTOR in oncogenesis, gain of function mutations in mTOR have been isolated from tumor cells and demonstrated to exhibit constitutive kinase activity (12–14). Yet clinically, mTOR inhibitors have shown limited activity (15). In this study, we show that while mutations in RTKs and PI3K are significantly enriched in multiple tumor types, activating mutations of mTOR are significantly enriched only in RCCC. Examining the properties of cancer cells in which activating mTOR mutants are ectopically introduced, we consistently found that such mutations were detrimental to cell growth whenever either amino acids, glucose, or oxygen became limiting. This suggests that the ability to modulate mTORC1 in response to variations of nutrient levels in the tumor microenvironment was maintained in most human tumors. To examine this possibility, we investigated whether mutations in the nutrient sensing arm of mTORC1 were ever observed in spontaneously arising human cancers. Analysis of TCGA Pan-cancer data base revealed that mutations in the components of the nutrient-sensing arm of mTORC1 regulation are exceedingly rare, despite the fact that such mutations lead to constitutive activation of mTORC1.
Materials and Methods:
Reagents.
Reagents Antibodies were from: Cell Signaling #4377 phospho-T389 S6K1, #2708 S6K1, #2920 Akt, #2983 mTOR, #4060 phospho-S473 Akt, #4377 phospho-T389 S6K1 #2855 phospho-T37/46 4E-BP1, #6888 phospho-S757 Ulk1, #8054 Ulk1. Secondary antibodies were from: GE Healthcare (HRP-linked anti-rabbit, HRP-linked anti-mouse). Inhibitors were from: Tocris Bioscience (PD98059, Torin 1). Amino acid starvation media were from Athena (DMEM/F12 0423) or prepared by the Memorial Sloan Kettering Cancer Center media core facility. Dialyzed FBS was from Gemini Biosystems and BSA (A1470) was from Sigma.
Cell Lines and gRNA Constructs.
K-RasG12D and WT MEFs were generated in the Thompson lab, as described previously (6). PTEN KO MEFs and MEFs expressing myr-Akt were generated in the Thompson lab, as described previously (16). Caki-1 cells were kindly provided by J. Hsieh. KPC (KrasLSL-G12D Trp53LSL-R272H Pdx1-Cre) murine pancreatic ductal adenocarcinoma cell lines were kindly provided by Scott Lowe. Generation of cell lines expressing tetracycline-inducible wild-type or mutant mTOR cells were made as described, with plasmids for TRE-Flag tagged WT or mutant mTOR kindly provided by J. Hsieh (17). Expression of mTOR WT and Mutants was induced by adding 1μg/mL doxycycline (Sigma) to culture medium.
gRNAs targeting DEPDC5 were cloned into LentiCRISPR v2. Plasmids were cotransfected with lentivirus packaging plasmids into HEK293T cells using Lipofectamine 2000 Transfection Reagent (Life Technologies), fresh media were added after 16 h, and viral supernatants were collected 2 d after infection. Target cells were infected by addition of viral supernatant and 8 μg/mL polybrene. Twenty-four hours after infection, cells were selected with appropriate antibiotics.
All cell lines were verified to be mycoplasma-negative by MycoAlert Mycoplasma Detection Kit (LT07–318, Lonza). Cell lines were tested bi-annually. All cells used for experiments were maintained for less than 20 passages, and the majority of experiments were performed in cells passaged less than 10 passages.
Cell Culture and Nutrient Starvation Experiments.
Cell culture experiments were performed at 37 °C and 5% CO2 in DMEM or DMEM/F12 with 10% dialyzed FBS (molecular weight cutoff 10,000), 100 units/mL penicillin, 100 μg/mL streptomycin, 17.5 mM glucose, and 4 mM glutamine with 1ug/mL doxycycline to induce mTOR WT or Mutant expression, unless otherwise specified.
For proliferation assays, MEFs were plated in complete medium, 5 h later or overnight briefly rinsed, and then cultured in starvation medium as indicated. For 5% amino acid (AA) experiments, AAs were supplemented to total 5% of DMEM/F12 amino acid solution. For hypoxia experiments, cells were cultured in a hypoxia chamber at 1% or 0.5% oxygen for the indicated length prior to counting. Cell numbers at the start of treatment (d0) and 1–4 days later were counted in triplicates using the Multisizer 3 Coulter Counter (Beckman) and normalized to the cell number at d0.
Data Analysis.
TCGA pan-cancer studies and MSK IMPACT data were accessed using cBioPortal version 3.6 (18,19). Putative driver mutations were identified by filtering variant annotation counts identified by OncoKB driver annotation, Hotspots, cBioPortal >= 10, and COSMIC >= 10 (the number of occurrences of mutations at the same amino acid position present in the COSMIC database) (20). Significance of mutual exclusion and co-occurrence of alterations of MTOR, PTEN, and PIK3CA in RCCC and endometrial cancer were also obtained from cBioPortal. Graphs were created using GraphPad Prism software. In vitro experimental data are representative of three independent experiments, and each independent experiment is done with samples in triplicate. Statistical significance is based on one-way ANOVA. p values are as indicated: ns = not significant; * , p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. R package maftools=v2.6.0 was used to format TCGA MC3 public access MAF files ahead of analysis with MutSigCV=v1.41 using default full-coverage and covariate files (21). Statistical significance reported on TCGA MC3 data is based on Bonferroni correction for the number of cancer types considered (n=33). Code used to generate figures, and reproducibility details are available at https://github.com/morrislab/mtor-rccc.
Data Availability
TCGA MC3 data is publicly available via the NIH Genomic Data Commons (https://gdc.cancer.gov/about-data/publications/mc3-2017). All PCAWG analysis results (Supplemental Figure 1d) were downloaded from the ICGC data portal (https://dcc.icgc.org/releases/PCAWG/drivers/p-values).
Western blotting.
Protein extracts were prepared by using 1 × RIPA buffer (20–188, Millipore) supplemented with protease (1860932, ThermoFisher) and phosphatase inhibitors (78428, ThermoFisher). Equal amounts of total protein were separated on NuPAGE Bis-Tris, transferred to nitrocellulose membranes and subjected to Western blotting with indicated primary antibodies.
Measurement of glucose and glutamine consumption and lactate secretion.
Cells were plated in 6-well cell culture plates at a concentration aimed to reach 0.5–1 × 106 cells at the time of harvest. Media were exchanged for an assay period of 16 h, then collected, centrifuged, and analyzed using a 2950 Biochemistry Analyzer (YSI Life Sciences) to determine glucose, glutamine, and lactate concentration. Absolute rates of consumption/secretion of these metabolites were calculated by subtracting the concentration in medium incubated for the same amount of time without cells and then normalizing to the cell number at the time of harvest, media volume, and hours of incubation.
Results:
Unlike upstream pathway components, mTOR is rarely mutated in human cancer.
Using the TCGA Pan-Cancer Atlas and Memorial Sloan Kettering IMPACT tumor sequencing data, the prevalence of mTOR alterations was examined. Despite mTOR’s central role in directing cellular growth (Figure 1a), mTOR alterations are rare; occurring in only 4 % of tumors sequenced in the TCGA Pan-Cancer Atlas dataset (Figure 1b), and 3 % of tumors sequenced within the MSK IMPACT dataset (Supplemental Figure 1a) (18,19,22). Many of these identified alterations are unlikely to be oncogenic (23,24). For example, the mTOR deletions found on sequencing analysis would not be predicted to aid oncogenesis. Overall, there were no excess amplifications over deletions of mTOR, suggesting an absence of selection for mTOR amplification as an oncogenic driver. When more stringent criteria were used to identify putative driver alterations, such as OncoKB annotation and hotspot analysis, potential activating alterations in mTOR only made up 1.1% of all samples within the TCGA pan-cancer atlas (Figure 1b), and 0.8% in IMPACT (Supplemental Figure 1a).
We then looked at the frequency of activating alterations in upstream effectors within this pathway including RTKs, PI3K and Akt; or deletions in PTEN, the main suppressor of pathway activity (Figure 1c and Supplemental Figure 1b). RTKs were found to be altered in >40% of all tumor samples within the TCGA pan-cancer dataset; with nearly 75% of these being identified as potential oncogenic drivers (Figure 1c). Thus, oncogenic alterations in RTKs occur nearly 30-fold more often than potentially activating mTOR alterations. The 16 most commonly altered RTKs were grouped in the above analysis as they are cell-type-specific and we analyzed a pan-cancer dataset. Recognizing that a comparison of multiple genes to one gene is imperfect, we performed additional comparison of alteration frequencies within the Breast Invasive Carcinoma TCGA tumor sequencing data. ERBB2 as a representative RTK was altered in 14% of samples, with nearly all of these alterations acting as putative drivers. However, only 0.4% of samples contained potentially activating mutations in mTOR, again demonstrating the rarity of driver mTOR mutations as compared to upstream activators of the mTOR pathway (Supplemental Figure 1c). Other upstream activators of the mTOR pathway are also frequently altered in cancers. PIK3CA, PTEN, and Akt are altered in 17%, 12%, and 8% of tumor samples, respectively; and the vast majority of these mutations have been classified by OncoKB and hotspot analysis as putative drivers. Thus, potentially activating alterations of mTOR are rare compared to oncogenic alterations within upstream effectors of the RTK/PI3K/Akt/mTOR pathway.
mTOR mutations are enriched only in Renal Clear Cell Carcinoma.
While the putative driver filters within cBioportal are an expedient way to differentiate passenger from driver alterations noted on sequencing datasets, it introduces bias as less-well studied genes may have true driver mutations that are not identified and included within the putative driver annotation. To further address the issue of passenger versus driver mutations, we performed a MutSigCV analysis on mTOR mutations as well as PIK3CA or PTEN, across the TCGA pan-cancer studies. MutSigCV tests whether mutations within genes occur at a rate higher than expected by chance, by taking into account covariates such as background mutation rate, gene expression level, replication timing, gene size, and chromatin state – all of which affect the expected mutational rate within a gene (23).
Using MutSigCV analysis on whole exome sequencing TCGA Pan-cancer Atlas data, we found that mTOR mutations were significantly enriched only in tumors reported in the Kidney RCCC TCGA cancer study (Figure 1d). A similar analysis on whole genome sequenced cancers also revealed significant enrichment in mTOR mutations exclusively in RCCC (Supplemental Figure 1d) (25,26). In contrast, PIK3CA and PTEN mutations were significantly enriched in many cancer types (Figure 1d, Supplemental Figure 1d). In fact, cancers not showing enrichment of PTEN or PI3K mutations were largely rare cancers known to display tissue-specific genetic mutational profiles such as in germ cell tumors and uveal melanoma, providing validation of this analysis (27,28). To further validate our MutSigCV analysis, we examined the mutational significance of other common oncogenes and tumor suppressors, and found that these were significantly mutated within cancer types where they are known to act as drivers. As a control we analyzed titin, which is commonly mutated in cancers due to its size, which was not significantly enriched in any cancer type (Supplemental Figure 1e).
We also examined clinically annotated mTOR driver mutations and found that RCCC and endometrial cancer had the highest proportion of clinically annotated putative driver mTOR mutations across MSK IMPACT and the TCGA studies (Figure 1e and Supplemental Figure 1f). The relatively high proportion of annotated mTOR mutations within Endometrial cancer was surprising, as mTOR mutations were not found to be enriched in endometrial cancers in our MutSigCV analysis. Additional analysis of the endometrial cancer TCGA dataset showed that mTOR mutations co-occur with PIK3CA or PTEN mutations in 90% of tumor samples. This was in contrast to RCCC, where only 5% of mTOR mutations had co-occurring PIK3CA or PTEN mutations (Supplemental Table 1 and 2 and Supplemental Figure 1g). Further analysis and study will be necessary to investigate why mTOR activating mutations in endometrial cancer are enriched in tumors with upstream PIK3CA or PTEN mutations.
Naturally occurring mutations in mTOR reduce cell survival and growth that depends on macropinocytosis.
One potential reason for the rarity of mTOR mutations could be that mTOR is not susceptible to activation through point mutations. However, examples of activating mTOR point mutations have been reported and validated as activating, with hotspot activating mutations mostly occurring in mTOR’s FAT, FRB, or kinase domains (Figure 2a) (12,13). Thus, hyperactive mTOR mutations exist but are not enriched as initiating mutations or selected during tumor progression and metastasis in most tumors, as seen by the lack of enrichment of these mutations across tumors other than RCCC.
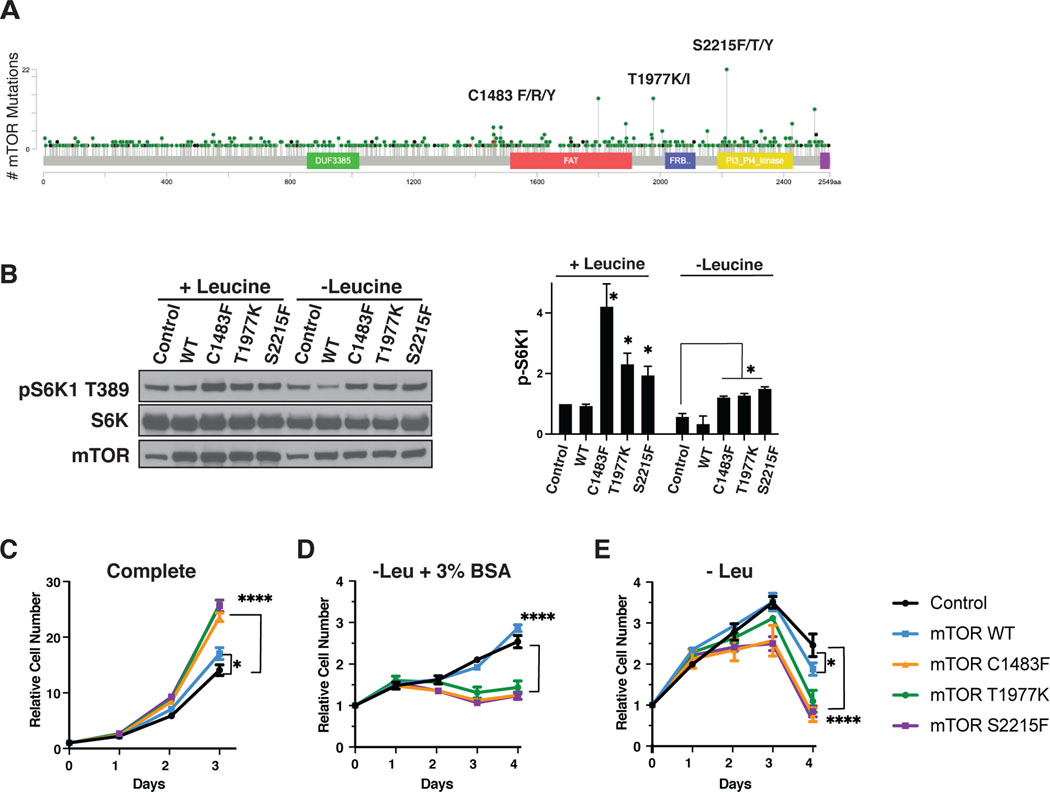
Figure 2: Activating mTOR mutations inhibit growth dependent on exogenous proteins.
A) Lollipop plot of mutations within the mTOR protein. S2215F/T/Y, T1977K/I, and C1483F/R/Y, three clinically validated activating mTOR hotspot mutations used in subsequent in vitro experiments are labeled. B) Western blot of KrasG12D MEFs with either control mTOR (empty vector), or expressing mTOR WT or hyperactivating mutants cultured in complete or leucine deficient medium for 16 hrs, paired with quantification of pS6K1 protein expression. This is a representative blot from three independent experiments. Growth curves with KrasG12D mutant MEFs expressing control (empty vector), mTOR WT or hyperactivating mutants in C) complete medium, D) medium without leucine supplemented with 3% bovine serum albumin (BSA), and E) in medium without leucine. The cell number at indicated days relative to the number at the start of the treatment (d0) is shown. Results are representative of three independent experiments performed in triplicate and are expressed as mean ± SD (error bars) of three replicates. Statistical significance is based on one-way ANOVA. p values are as indicated: ns = not significant; * , p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
The Hotspot mutations reported to date in mTOR have been associated with increased tumor cell growth when assayed in vitro (12). Despite their potential to occur, it is surprising that activating mTOR mutations appear to be significantly enriched only in RCCC. Recently it was found that wild type mTOR is a negative regulator of exogenous protein utilization initiated by RTK/Ras/PI3K stimulation of macropinocytosis (6). Macropinocytosis has been shown to be an important pathway to support tumor cell growth in vivo in KRas-driven pancreatic adenocarcinoma, where essential nutrients like amino acids become limiting, and inhibition of mTOR is beneficial for tumor growth in these conditions (29,30). To test whether activating mutations in mTOR can suppress growth dependent on exogenous protein utilization, we expressed several recurring gain-of-function mTOR mutants in KRas-transformed cells, known to be able to utilize exogenous proteins for growth when essential amino acids are limiting.
Three previously validated activating mTOR mutants were selected representing the most common naturally occurring variants within different functional domains of mTOR: C1483F in the F1 cluster of the FAT domain, T1977K in the F3 cluster of the FAT domain, and S2215F in the K1 cluster of the kinase domain (Figure 2a) (12,17). As expected, both KRas-transformed cells expressing empty vector (control) and those expressing mTOR WT retained the ability to sense leucine deprivation and inactivate mTORC1, as shown by decreased phosphorylation of mTOR target S6K1 (pS6K1) (Figure 2b). Cells expressing mutant mTOR had higher levels of pS6K1 at baseline in complete media, and while there was some decrease of this level under leucine starvation, the cells maintained a higher level of pS6K1 under leucine starvation compared to mTOR WT or control cells. Consistent with previous studies, these mTOR mutants had a growth advantage over control KRas-transformed cells or cells expressing wild type (WT) mTOR in complete medium (Figure 2c). However, KRas-transformed cells expressing activating mTOR mutants were unable to utilize exogenously supplied bovine serum albumin (BSA) as an amino acid source for growth in the setting of leucine deprivation – while control and mTOR WT Ras-transformed cells were able to grow under these same conditions (Figure 2d). Moreover, the mTOR mutants were detrimental in conditions of leucine deprivation without supplementation of exogenous BSA, leading to more cell death while Control and mTOR WT Ras-transformed cells remained viable (Figure 2e).
Activating mutations of mTOR compromise cell survival under amino acid-limiting conditions.
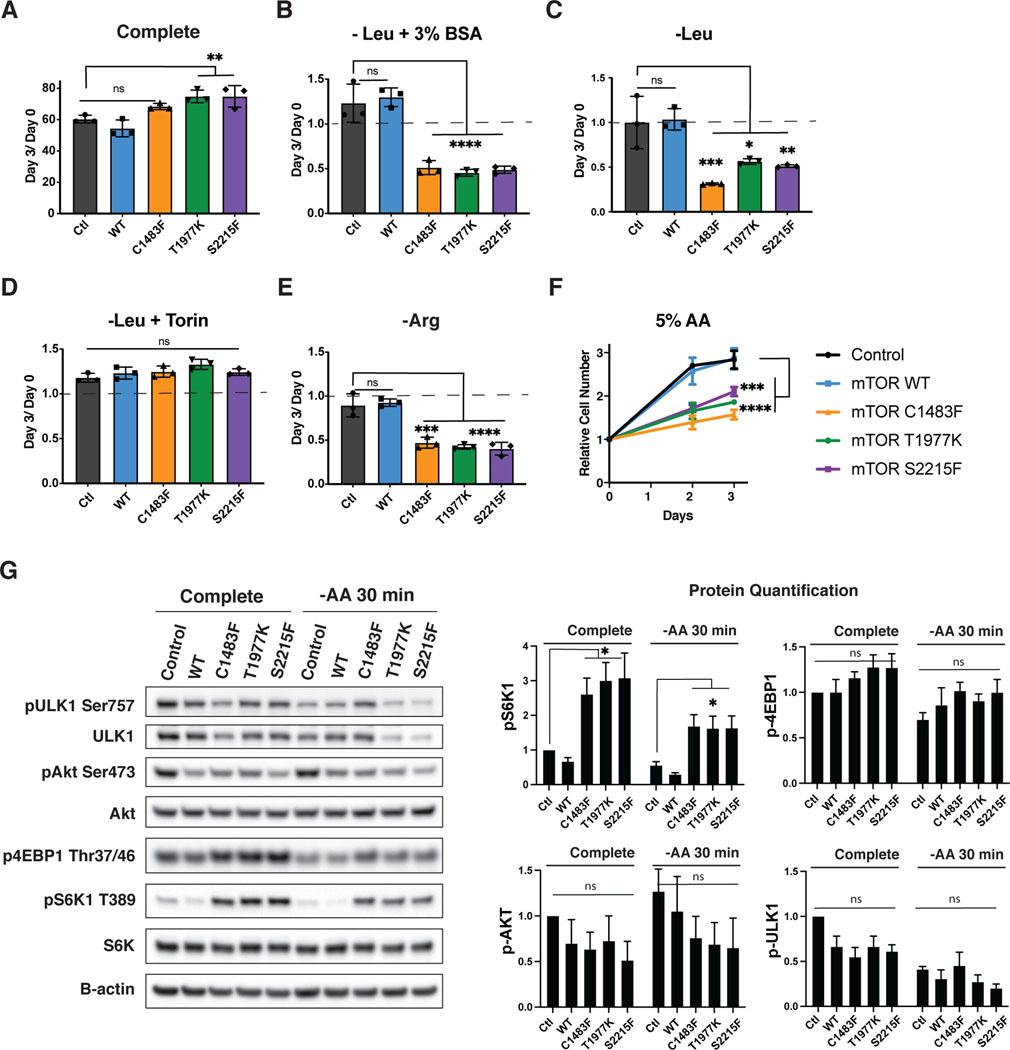
We then wanted to determine whether the growth detriment seen in Ras-transformed cells with activating mTOR mutants was dependent on Ras activation, or whether it was generalizable to cells which do not robustly activate macropinocytosis and exogenous protein utilization under periods of amino acid starvation. Thus, we did similar experiments in Ras-WT MEFs expressing empty vector (control), WT mTOR, or mutant mTOR. As expected, the mTOR mutants showed a growth advantage when nutrients were replete (Figure 3a). Unlike the KRasG12D cells, these Ras WT cells were unable to significantly grow in the setting of leucine starvation with BSA supplementation. However, the Control and mTOR WT cells remained viable under conditions of leucine starvation (with minimal growth with supplemental BSA), while the mTOR mutants had increased cell death under leucine starvation with or without BSA (Figures 3b and 3c). This survival-defect was dependent on mTOR activity – as mTOR inhibition with Torin rescued the death of leucine-starved mTOR-mutant cells (Fig 3d). Similar to leucine starvation, decreased survival was shown with arginine deprivation, where the mTOR mutants uniformly had more cell death while the control and mTOR WT cells maintained viability during this same period (Figure 3e). Glutamine starvation – which is not sensed by mTOR via the canonical RagGTPase signaling pathway, was equally deleterious to cells regardless of whether they expressed activating mTOR mutations (Supplemental Figure 2a).
Figure 3: Activating mTOR mutations decrease growth and survival under amino acid deficient conditions.
Relative cell number of WT MEFs expressing empty vector (control) or expressing mTOR WT or mTOR mutant treated with A) complete, B) leucine-deficient + 3% BSA, C) leucine-deficient, D) leucine-deficient + 250nM Torin, E) arginine-deficient, and F) 5% amino acids. This represents the cell number at d3 over d0, where d0 is the number of cells at the start of treatment, except in Figure F where the cell number at indicated days relative to the number at the start of treatment (d0) is shown. The dotted line in B–E represents cell number at the start of treatment. Results are representative of three independent experiments performed in triplicate and are expressed as mean ± SD (error bars) of three replicates. Statistical significance is based on one-way ANOVA. p values are as indicated: ns = not significant; * , p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. G) Western blot of WT MEFs with either control mTOR (empty vector), or expressing mTOR WT or hyperactivating mutants cultured in complete or amino acid deficient medium for 30 minutes, paired with quantification of phospho-protein expression. This is a representative blot from three independent experiments expressed as mean ± SE (error bars). Statistical significance is based on one-way ANOVA as indicated: ns = not significant; *, p ≤ 0.05.
In addition to starving cells of individual amino acids known to be sensed by mTOR, we also tested whether generally depleting all amino acids would similarly be detrimental in growth or survival of these mTOR mutants. Thus, we grew the mTOR mutant cells in complete medium, or medium containing only 5% of the normal amino acid composition. Similar to experiments with leucine or arginine deprivation, the mTOR mutants showed a significant disadvantage in growth in 5% amino acid media as compared to the control or mTOR WT transfected cells (Figure 3f). These findings are in contrast to expression of active Akt (myr-Akt) or deletion of PTEN in cells growing in 5% AA. Neither of these alterations were deleterious to cell growth or survival when grown in 5% amino acid media, with or without the supplementation of BSA. (Supplemental Figure 2b and 2c).
As expected, the mTOR mutants had increased mTORC1 activity under complete conditions, as seen by increased pS6K1 in cells expressing mTOR mutants. pS6K1 was retained under amino acid starvation only in cells expressing mTOR mutants, and was maintained at 24 hrs (Figure 3g and Supporting Figure 2e). The mTOR mutants also increased phosphorylation of 4EBP1 (p4EBP1), another mTORC1 target, in complete conditions. While p4EBP1 levels remained higher in cells expressing activating mTOR mutants compared to WT mTOR or control cells under amino acid starvation, this change was small, and did not persist at 24hrs of amino acid starvation (Figure 3g and Supporting Figure 2e). We did not see significant increases in Ulk1 phosphorylation (pUlk1) in cells expressing activating mTOR mutants in either complete or amino acid deficient conditions. We also did not see increased mTORC2 activity, as seen by phosphorylation of Akt at serine 473 (pAkt S473). In fact, cells expressing either WT mTOR or the activating mTOR mutants had decreased pAkt S473 compared to control cells expressing an empty vector.
Activating mutations in mTOR are detrimental when glucose is limiting.
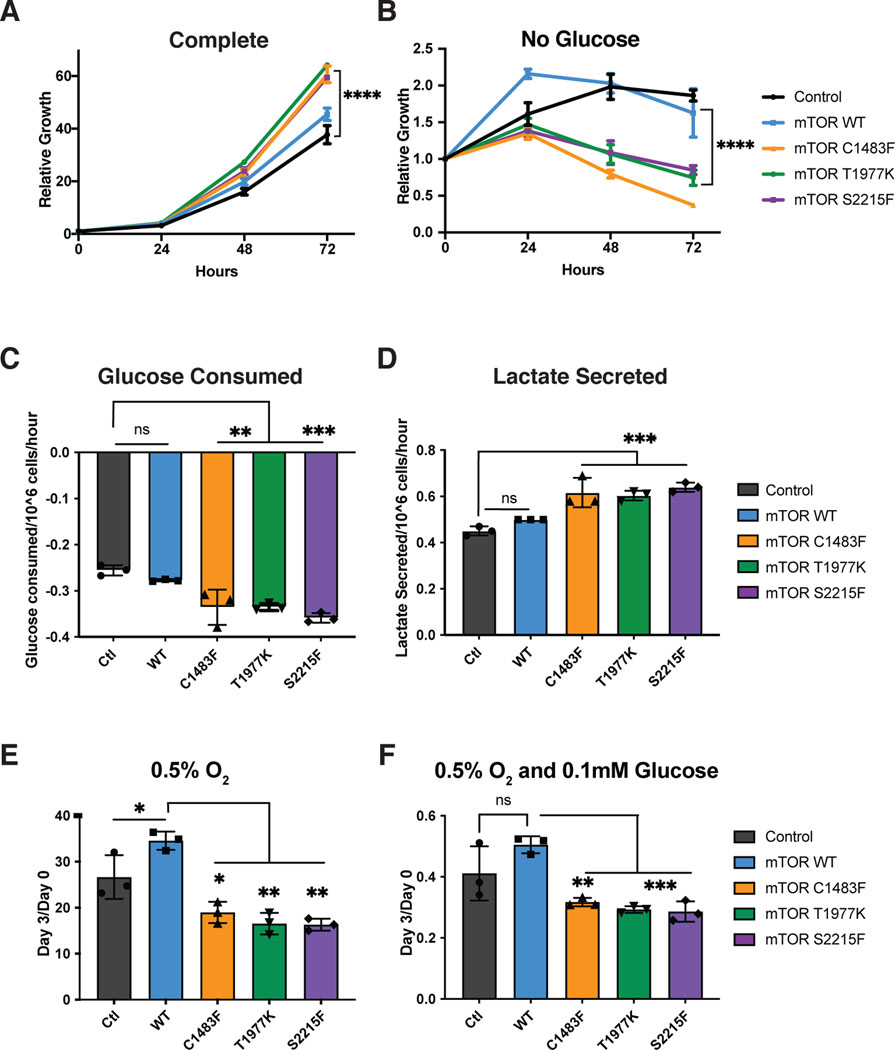
In addition to amino acids, another nutrient critical to cellular survival and growth is glucose – which has been shown to be depleted within tumors (30,31). mTOR senses glucose directly via its metabolite DHAP, as well as indirectly via AMPK (32). Cells with loss of TSC are ‘addicted’ to glucose, and have more cell death upon glucose withdrawal (33). We hypothesized that glucose limitation, similar to amino acid limitation, would be detrimental to cells with hyperactive mTOR mutations. In agreement with this hypothesis, we found that cells expressing hyperactive mTOR mutants, despite showing a growth advantage in complete medium, had significantly more cell death in conditions of glucose starvation as compared to control cells or cells expressing mTOR WT (Figure 4a and b). We examined glucose utilization of these cells and found that the mTOR mutants utilized significantly more glucose and secreted more of this as lactate, although the glucose consumption/lactate secretion ratio remained unchanged (Figure 4c and d). Given the increased utilization of glucose by the mTOR mutant cells, we examined cell growth in various glucose-limiting conditions by doing a glucose titration. We found a detrimental effect of mTOR mutants compared to Control or WT mTOR on growth and survival in the low glucose conditions of 1mM or 0.1mM as well as with complete glucose starvation (Supplemental Figure 3a-d).
Figure 4: Activating mTOR mutations are detrimental under glucose and O2 limiting conditions.
Relative cell number of WT MEFs expressing empty vector (control) or expressing mTOR WT or mTOR mutants treated with A) complete medium or B) medium without glucose. The cell number at indicated days relative to the number at the start of the treatment (d0) is shown. C) Glucose consumed and D) Lactate secreted per million cells per hour. Relative cell number of WT Mefs expressing empty vector (control) or expressing mTOR WT or mTOR mutants treated with E) 0.5% O2 or F) 0.5% O2 and 0.1mM Glucose. This represents the cell number at d3 over d0, where d0 is the number of cells at the start of treatment. Results are representative of three independent experiments performed in triplicate and are expressed as mean ± SD (error bars) of three replicates. Statistical significance is based on one-way ANOVA. p values are as indicated: ns = not significant; * , p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
Activating mutations in mTOR are detrimental when oxygen is limiting.
In addition to sensing metabolites such as amino acids and glucose, mTORC1 senses cellular oxygen availability, both as a result of changes to energy charge through AMPK, as well as sensing oxygen directly via REDD1 (34). Previous work showed that TSC null cells had increased growth when grown under hypoxic conditions with 1% oxygen (34). In agreement with these studies, we found that cells expressing activating mTOR mutants were able to grow better under hypoxic conditions with 1% O2 (Supplemental Figure 3e). However, this was not true when oxygen levels were lowered to 0.5% O2, a level potentially seen in a tumor environment that not only stabilizes HIF, but also interferes with respiration (35). Cells expressing activating mTOR mutants grew slower in 0.5% O2 compared to control cell or cells expressing WT mTOR (Figure 4e). Cells expressing mTOR activating mutants also had more cell death when cultured in low glucose at 0.5% O2 (Figure 4f) and low amino acids at 0.5% O2 (Supplemental Figure 3f).
The survival disadvantage caused by activating mTOR mutations in nutrient limiting conditions is cell type agnostic.
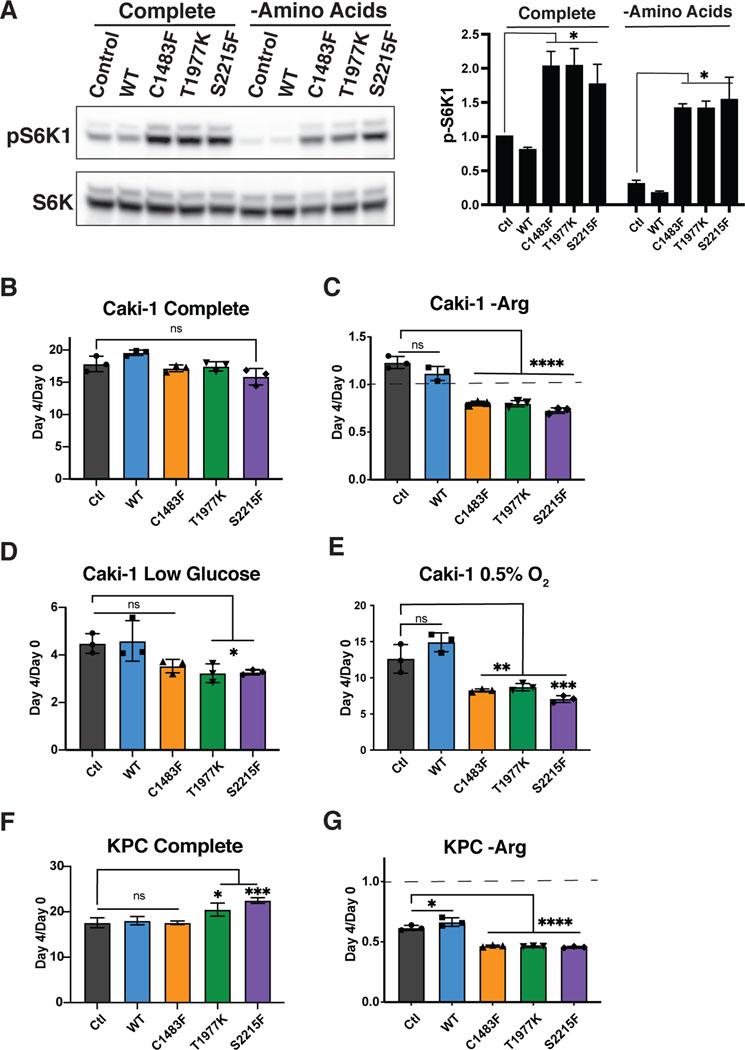
Despite the survival disadvantage conferred by activating mTOR mutations under nutrient-limited conditions, these mTOR mutations are significantly enriched in RCCC. We hypothesize that the enrichment of mTOR mutants in RCCC may occur because these cancers are hypervascular and thus rarely deprived of nutrients. However, an alternative possibility is that renal cell cancers are uniquely able to tolerate mTOR hyperactivation during periods of nutrient limitation due to some cell-type specific behavior. To further evaluate this possibility, we expressed mTOR WT and hyperactive mutants in Caki-1 cells, an RCCC cell line (Figure 5a and Supplemental Figure 4a). Similar to experiments in KRas mutant or WT MEFs, these mTOR mutants retained mTORC1 signaling despite amino acid deprivation (Figure 5a). Caki-1 cells expressing mutant or WT mTOR had similar growth properties (Figure 5b), yet a clear detriment with increased cell death was seen in the mTOR mutant-expressing cells starved of arginine or leucine (Figure 5c and Supplemental Figure 4b). We also tested the effect of glucose limitation and hypoxia, and similar to our result in MEFs, the Caki-1 mTOR mutant cells had a growth disadvantage under low glucose conditions (Figure 5d), as well as in hypoxic conditions with 0.5% O2 alone (Figure 5e), or in combination with low amino acids or low glucose conditions (Supplemental Figure 4c and d).
Figure 5: The detrimental effects of activating mTOR mutations are cell type agnostic.
A) Western blot of Caki-1 cells expressing control mTOR (empty vector), mTOR WT, or mTOR mutants grown in complete media or media deprived of all amino acids for 30 minutes, with graph representing protein quantification of pS6K1. Results are representative of at least three independent experiments. Relative cell number of Caki-1 cells expressing empty vector (control) or expressing mTOR WT or mTOR mutants treated with the following conditions: B) complete, C) arginine deficient D) 1mM Glucose, E) 0.5% O2, expressed as mean ± SE (error bars). Statistical significance is based on one-way ANOVA as indicated; *, p ≤ 0.05. Cell numbers at day 4 relative to the number at the start of treatment (d0) are shown. Relative cell number of KPC cells expressing empty vector (control) or expressing mTOR WT or mTOR mutants in F) complete or G) minus arginine medium. The cell numbers at day 4 relative to the number at the start of treatment (d0) are shown. The dotted line in Figures C and G represents cell number at the start of treatment. Results are representative of at least two independent experiments performed in triplicate and are expressed as mean ± SD (error bars) of three replicates. Statistical significance is based on one-way ANOVA. p values are as indicated: ns = not significant; * , p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
In addition to examining the effect of hyperactivating mTOR mutations within a hypervascular cancer such as RCCC, we also examined the effects of these mutations within hypovascular cancers such as pancreatic adenocarcinoma. We expressed WT and hyperactive mTOR mutants in KPC cells, a murine pancreatic cancer cell line derived from Kras and TP53 mutated pancreatic cells (Supplemental Figure 4e). Consistent with all other cell lines examined, we found increased cell death upon arginine starvation in KPC cells expressing mTOR mutants compared to those expressing WT mTOR (Figure 5f and g).
The nutrient-sensing components regulating mTORC1 activity are rarely mutated.
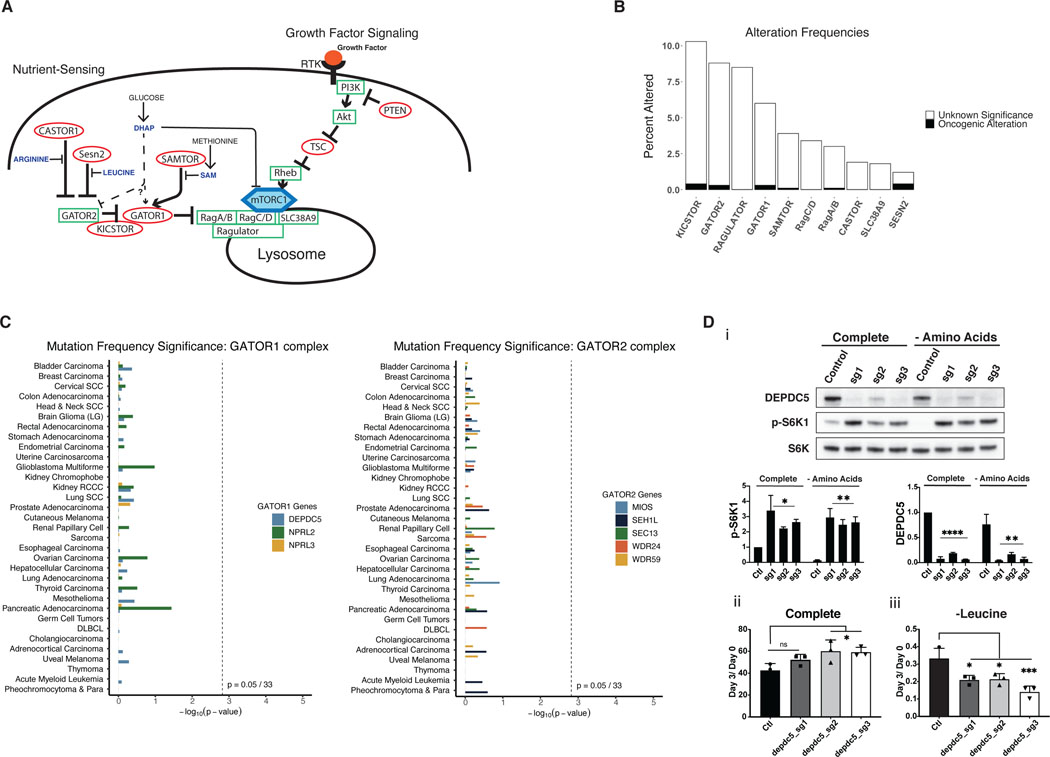
mTORC1 activity is regulated by two regulatory arms: a growth factor sensing arm, and a nutrient sensing arm. Rheb, the final effector of the growth factor sensing arm, can only activate mTORC1 if it is appropriately anchored at the lysosome by the Rag GTPases. The functions of the Rag GTPases are regulated by the GATOR1 complex, a component of the nutrient-sensing arm regulating mTOR (Figure 6a) (2). The localization of mTORC1 at the lysosome is disrupted if specific amino acid and other nutrient conditions are not met. Thus, a lack of these signals leads to incomplete activation of mTORC1 even in the presence of growth factors (9). Given the detrimental effects of hyperactive mTOR in the setting of amino acid or glucose limitation, we hypothesized that the ability of mTOR to sense nutrient availability – and inactivate in the setting of nutrient limitation – might be critical for cellular survival and continued growth during periods of nutrient stress.
Figure 6: Mutations in the nutrient-sensing arm of mTORC1 are rare and not significantly mutated in cancers.
A) Model of both growth factor signaling and nutrient sensing arms of the mTORC1 pathway. The nutrient sensing arm inactivates mTORC1 via GATOR1/2 if nutrients are limited, even in the presence of growth factor signaling. B) Mutational frequency of genes within the nutrient-sensing arm of the mTORC1 pathway in the TCGA pan-cancer dataset. KICSTOR includes C12ORF66, ITFG2, KPTN, and SZT2. GATOR2 includes MIOS, SEH1L, SEC13, WDR24, and WDR59. RAGULATOR includes LAMTOR1, LAMTOR2, LAMTOR3, LAMTOR4, LAMTOR5. GATOR1 includes DEPDC5, NPRL2, and NPRL3. CASTOR includes CASTOR1 and CASTOR2. C) MutSigCV analysis of GATOR1 and GATOR2 genes, shown as p-values for mutational frequency significance of each gene within each TCGA cancer study. D) i) Western blot of control MEFs or DEPDC5 CRISPR KO MEFs treated with complete or amino acid deficient medium for 30 minutes, paired with quantification of pS6K1 and DEPDC5 protein expression. This is a representative blot from three independent experiments. Relative cell number of MEFs, control or with DEPDC5 CRISPR KO in ii) complete medium and iii) amino acid deficient medium. The cell numbers at day 3 relative to the number at the start of treatment (d0) are shown. Results are representative of at least two independent experiments performed in triplicate and are expressed as mean ± SD (error bars) of three replicates. Statistical significance is based on one-way ANOVA. p values are as indicated: ns = not significant; * , p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001.
To test this hypothesis, we examined the prevalence of mutations within the nutrient-sensing arm of the mTORC1 pathway. Only rare alterations within each gene examined in this pathway were found in the TCGA pan-cancer dataset. Furthermore, when filtered for oncogenic alterations using OncoKB annotation and hotspot analysis, alterations within nutrient sensing genes were exceedingly rare (Figure 6b). To avoid bias arising from relying on putative driver annotation among these less-well studied genes, we performed a MutSigCV analysis of all GATOR1 complex and GATOR2 complex components across the TCGA pan-cancer studies. Of this set of 8 genes, none were significantly enriched in any cancer type (Figure 6c).
Given the rarity of driver mutations within the nutrient-sensing arm of mTORC1, we hypothesized that alterations in this pathway that would activate mTORC1 might behave similarly to activating mTOR mutations. To test this, we made DEPDC5 knockout (KO) MEFs, as DEPDC5 is an essential component of GATOR1. As expected, the KOs retained S6K1 phosphorylation under leucine deprivation compared to control cells (Figure 6d i). The DEPDC5 KOs also grew better under nutrient replete conditions; however, they exhibited increased cell death under leucine starvation (Figure 6d ii and iii). Thus, inactivating nutrient-sensing and thereby inappropriately retaining mTORC1 activity is detrimental in amino-acid deficient conditions.
Discussion.
In this study we analyzed existing tumor databases to determine the frequency with which activating mutations in mTOR are observed relative to other components in the RTK/PI3K/Akt/mTOR signaling pathway. Surprisingly, using MutSigCV analysis, we found that mTOR mutations are only significantly enriched in RCCC. This result was unexpected given mTOR’s central role in promoting cell growth. While the present findings lend support to previous work suggesting that mTOR driver mutations undergo positive selection in RCCC (24), the lack of a significant number of mTOR mutations across the rest of the cancer landscape was unexpected. However, the present findings are consistent with clinical data showing poor overall efficacy of mTOR-inhibitors as cancer therapy, with the notable exception of RCCC (15). In contrast, mutations in upstream activators of the mTOR pathway including RTKs and PI3K were found to be significantly enriched in most cancer types. These results confirm the importance of the RTK/PI3K/Akt/mTOR pathway to oncogenesis. However, the data suggest that RTKs or PI3K are the central genes within this pathway that contribute to oncogenesis.
One potential reason for the apparent advantage of RTK or PI3K driver mutants over mTOR driver mutants is that PI3K activates multiple simultaneous pathways in addition to mTOR that contribute to oncogenesis, including Akt-mediated inhibition of FoxO and GSK3 (36). In addition, we and others have shown that PI3K activates alternative nutrient scavenging via macropinocytosis (16). When nutrients are abundant, PI3K-mediated activation of mTORC1 inhibits the catabolism of these scavenged proteins in the lysosome. However, in the setting of nutrient stress, mTORC1 is inhibited and catabolism of PI3K-mediated uptake of exogenous proteins or necrotic cells provides an additional nutrient-source.
Another potential reason for the rarity of oncogenic mTOR mutations is that mTOR activation negatively feeds back to decrease signaling through PI3K (37). The impact of this negative regulation is highlighted by work investigating the different malignant potential of tumors arising in mice with deletion of PTEN vs TSC. Both of these alterations led to increased mTOR activity (38). However, PTEN-deleted tumors maintained high levels of PI3K-induced PIP3 as well as activated Akt and mTOR. In contrast, TSC null cells had high mTOR activity, but mTOR-mediated negative feedback ultimately led to decreased PI3K and Akt activity, contributing to the less malignant phenotype compared to PTEN-deleted tumors. Concomitant PTEN deletion with TSC deletion overcame the negative regulation of PI3K/Akt and allowed TSC deleted tumors to become more malignant similar to PTEN-deleted tumors (38). We found that a similar situation may be occurring in endometrial cancer where PTEN and mTOR are frequently mutated in the same tumor.
While the negative-regulatory effects of hyperactive mTOR on PI3K and Akt is a potential explanation for the more malignant phenotype of PTEN vs. TSC-deleted tumors, it does not fully explain the scarcity of mTOR driver mutations in cancer nor their restriction to RCCC. A growing body of evidence indicates that mTORC1 needs to be inactivated for cells to survive periods of nutrient limitation. Indeed, hyperactive mTORC1 signaling mediated by loss of TSC was shown to be a liability in tumor-like stress conditions, rendering cells dependent on desaturated lipids and glucose (39)(33). In addition, our lab has previously shown that mTORC1 inhibits degradation and utilization of exogenous proteins as a nutrient source, and that inhibition of mTORC1 is beneficial to cell growth and survival under nutrient-limiting conditions (6). Furthermore, mTORC1 inhibits autophagy, which is required to maintain nucleotide pools and prevent energy crisis during starvation in cancer cells (40). Thus in settings of nutrient-stress, hyperactivation of mTORC1 would be expected to be detrimental, as cells would be unable to inactivate mTOR to pause growth to balance ATP and nutrient supply with demand, nor activate autophagy and alternative nutrient scavenging strategies (29,30,33,40). Supporting this model, our in vitro results showed that activating mTOR mutations, which maintain mTOR activity despite nutrient restriction, were detrimental to cells under amino acid, glucose, or oxygen-limiting conditions. We found that activating mTOR mutants had sustained increases in mTORC1 activity, but not in mTORC2 activity, specifically implicating aberrant mTORC1 activity as the driver of these nutrient vulnerabilities. The role of mTORC1 is to integrate the sufficiency of various nutrient signals before committing the cell to growth. This explains how inappropriate activation of mTORC1 can be detrimental to cells under so many nutrient-limiting conditions, and implicates inhibition of mTORC1 as an important stress response in growing cells.
The differential effect of activating mTOR mutants in 1% and 0.5% O2 is particularly instructional. At 1% O2, cells begin adapting to lower oxygen conditions to limit ATP demand, despite the fact that oxygen is not yet limiting for respiration. In this circumstance, activating mTOR mutations can maintain ATP-demanding growth programs and give cells a growth advantage. However, these same mutations become deleterious as oxygen levels continue to fall and become limiting for respiration and other oxygen-dependent reactions critical for growth (35). Under these circumstances, inappropriately active mTOR becomes a liability as it maintains growth programs that the cell is unable to accommodate, leading to decreased growth or increased cell death.
RCCC is distinctively hypervascular both due to the physiology of the kidneys – which utilize fenestrated capillaries and allow the passage of nutrients through the endothelial barrier – as well as the VHL mutations that drive the majority of these cancers via angiogenesis (41,42). As such, RCCCs are much less likely to experience nutrient stress in comparison to other tumor types, as their growth is not limited by the compromised ability of existing vasculature to support the growing tumor. This may partially explain why RCCC is an exception, allowing for enrichment of mTOR mutations. The fact that the deleterious effects of mTOR mutants in nutrient-poor conditions is cell type agnostic further supports the microenvironment-centric view explaining the ability of RCCCs to tolerate activating mTOR mutations. However, there are likely other cell type and tissue-type specific contributions that allow RCCCs to uniquely tolerate activating mTOR mutations.
The present study also demonstrates that a functional nutrient-sensing arm of mTOR is maintained in nearly all human tumors and is critical for growth and survival in the setting of nutrient stress. A knockout of a component of GATOR1, the nutrient-sensor and negative regulator of mTORC1, phenocopied hyperactivating mTOR mutants by showing a growth advantage under nutrient-replete conditions, but a detriment under leucine-starved conditions. Consistent with this, we found that mutations within the nutrient sensing arm of the mTOR pathway are exceedingly rare. These results further demonstrate the importance of being able to inactivate mTOR when key nutrients are scarce, and help explain why mTOR driver mutations are rare outside RCCCs. Conceivably, retaining the ability to turn off mTOR signaling is essential for cancer cells to adapt to the fluctuating nutrient supply that is characteristic for poorly vascularized tumors.
These data potentially explain why, although the RTK/PI3K/Akt/mTOR pathway is critical for developmental cell growth and homeostatic proliferation, the oncogenic potential of the pathway components are not equal. Cell-autonomous survival and proliferation is supported more by proximal pathway components. In contrast, distal pathway components when constitutively activated can be deleterious to cell autonomous regulation of growth and survival. These findings provide insight into the vast differences in the observed mutation rates of pathway components and the potential of targeted inhibitors of the RTK/PI3K/Akt/mTOR pathway to disrupt tumor growth.
Supplementary Material
Significance.
The rarity of oncogenic mTOR mutations suggest that, despite the importance of mTOR in cell growth, cells need to inactivate mTOR to survive periods of nutrient stress. This may partially explain the limited clinical activity of mTOR inhibitors as anti-cancer therapeutics, and may inform future trials targeting the mTOR pathway.
Acknowledgements:
The results published here are in part based upon data generated by the TCGA Research Network: https://www.cancer.gov/tcga. We thank members of the C.B.T. laboratory for critical review of the data presented in this manuscript. A.A.B is the recipient of an American Cancer Society Postdoctoral Fellowship. This work was performed with assistance from the Donald B. and Catherine C. Marron Cancer Metabolism Center, Memorial Sloan Kettering. C.F.H was supported by Canadian Institute for Health Research Project Grant #388344, the Province of Ontario and the ACM SIGHPC Computational and Data Science Fellowship. This work was supported by the Cancer Center Support Grant (P30CA008748) to Memorial Sloan Kettering Cancer Center and grants from the National Cancer Institute (to C.B.T.) and from Sloan Kettering Institute (to Q.M.). Q.M. is a Canada CIFAR AI chair.
Financial Support:
A.A.B is the recipient of an American Cancer Society Postdoctoral Fellowship. C.F.H was supported by Canadian Institute for Health Research Project Grant #388344, the Province of Ontario and the ACM SIGHPC Computational and Data Science Fellowship. This work was supported by the Cancer Center Support Grant (P30CA008748) to Memorial Sloan Kettering Cancer Center and grants from the National Cancer Institute (to C.B.T.) and from Sloan Kettering Institute (to Q.M.). Q.M. is a Canada CIFAR AI chair.
Conflict of Interest:
C.B.T. is a founder of Agios Pharmaceuticals and a member of its scientific advisory board. He is also a former member of the Board of Directors and former stockholder of Merck and Charles River Laboratories. He holds patents related to cellular metabolism.
References
- 1.De Berardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci. Adv American Association for the Advancement of Science; 2016. page e1600200–e1600200. [DOI] [PMC free article] [PubMed]
- 2.Liu GY, Sabatini DM. mTOR at the nexus of nutrition, growth, ageing and disease. Nat Rev Mol Cell Biol. 2020;21:183–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ben-Sahra I, Manning BD. mTORC1 signaling and the metabolic control of cell growth. Curr Opin Cell Biol. NIH Public Access; 2017;45:72–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson TR, Sengupta SS, Harris TE, Carmack AE, Kang SA, Balderas E, et al. mTOR Complex 1 Regulates Lipin 1 Localization to Control the SREBP Pathway. Cell. 2011;146:408–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edinger AL, Thompson CB. Akt maintains cell size and survival by increasing mTOR-dependent nutrient uptake. Mol Biol Cell. American Society for Cell Biology; 2002;13:2276–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Palm W, Park Y, Wright K, Pavlova NN, Tuveson DA, Thompson CB. The Utilization of Extracellular Proteins as Nutrients Is Suppressed by mTORC1. Cell. 2015;162:259–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim J, Kundu M, Viollet B, Guan K-L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. Nature Publishing Group; 2011;13:132–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martina JA, Chen Y, Gucek M, Puertollano R. MTORC1 functions as a transcriptional regulator of autophagy by preventing nuclear transport of TFEB. Autophagy. Taylor & Francis; 2012;8:903–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag Complex Targets mTORC1 to the Lysosomal Surface and Is Necessary for Its Activation by Amino Acids. Cell. Cell Press; 2010;141:290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vander Broek R, Mohan S, Eytan D, Chen Z, Van Waes C. The PI3K/Akt/mTOR axis in head and neck cancer: functions, aberrations, cross-talk, and therapies. Oral Dis. 2015;21:815–25. [DOI] [PubMed] [Google Scholar]
- 11.Martelli AM, Evangelisti C, Chiarini F, McCubrey JA. The Phosphatidylinositol 3-kinase/AKT/mTOR signaling network as a therapeutic target in acute myelogenous leukemia patients. Oncotarget. 2010;1:89–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grabiner BC, Nardi V, Birsoy K, Possemato R, Shen K, Sinha S, et al. A diverse array of cancer-associated MTOR mutations are hyperactivating and can predict rapamycin sensitivity. Cancer Discov. American Association for Cancer Research Inc.; 2014;4:554–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sato T, Nakashima A, Guo L, Coffman K, Tamanoi F. Single amino-acid changes that confer constitutive activation of mTOR are discovered in human cancer. Oncogene. Nature Publishing Group; 2010;29:2746–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Urano J, Sato T, Matsuo T, Otsubo Y, Yamamoto M, Tamanoi F. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci U S A. National Academy of Sciences; 2007;104:3514–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hua H, Kong Q, Zhang H, Wang J, Luo T, Jiang Y. Targeting mTOR for cancer therapy. J. Hematol. Oncol BioMed Central Ltd.; 2019. page 1–19. [DOI] [PMC free article] [PubMed]
- 16.Palm W, Araki J, King B, DeMatteo RG, Thompson CB. Critical role for PI3-kinase in regulating the use of proteins as an amino acid source. Proc Natl Acad Sci U S A. 2017;114:E8628–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Pham CG, Albanese SK, Dong Y, Oyama T, Lee C-HH, et al. Mechanistically distinct cancer-associated mTOR activation clusters predict sensitivity to rapamycin. American Society for Clinical Investigation; 2016;126:3526–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. American Association for Cancer Research; 2012;2:401–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. NIH Public Access; 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chakravarty D, Gao J, Phillips S, Kundra R, Zhang H, Wang J, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. American Society of Clinical Oncology (ASCO); 2017;1–16. [DOI] [PMC free article] [PubMed]
- 21.Mayakonda A, Lin DC, Assenov Y, Plass C, Koeffler HP. Maftools: Efficient and comprehensive analysis of somatic variants in cancer. Genome Res. Cold Spring Harbor Laboratory Press; 2018;28:1747–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. Nature Publishing Group; 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lawrence MS, Stojanov P, Polak P, Kryukov GV., Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. Nature Publishing Group; 2013;499:214–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martincorena I, Raine KM, Gerstung M, Dawson KJ, Haase K, Van Loo P, et al. Universal Patterns of Selection in Cancer and Somatic Tissues. Cell. 2017;171:1029–1041.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ellrott K, Bailey MH, Saksena G, Covington KR, Kandoth C, Stewart C, et al. Scalable Open Science Approach for Mutation Calling of Tumor Exomes Using Multiple Genomic Pipelines. Cell Syst. Cell Press; 2018;6:271–281.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rheinbay E, Nielsen MM, Abascal F, Wala JA, Shapira O, Tiao G, et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature. Nature Research; 2020;578:102–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harbour JW. The genetics of uveal melanoma: An emerging framework for targeted therapy. Pigment Cell Melanoma Res. John Wiley & Sons, Ltd; 2012;25:171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheikine Y, Genega E, Melamed J, Lee P, Reuter VE, Ye H. Molecular genetics of testicular germ cell tumors. Am J Cancer Res. e-Century Publishing Corporation; 2012;2:153–67. [PMC free article] [PubMed] [Google Scholar]
- 29.Commisso C, Davidson SM, Soydaner-Azeloglu RG, Parker SJ, Kamphorst JJ, Hackett S, et al. Macropinocytosis of protein is an amino acid supply route in Ras-transformed cells. Nature. NIH Public Access; 2013;497:633–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamphorst JJ, Nofal M, Commisso C, Hackett SR, Lu W, Grabocka E, et al. Human pancreatic cancer tumors are nutrient poor and tumor cells actively scavenge extracellular protein. Cancer Res. NIH Public Access; 2015;75:544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan MR, Danai LV, Lewis CA, Chan SH, Gui DY, Kunchok T, et al. Quantification of microenvironmental metabolites in murine cancers reveals determinants of tumor nutrient availability. Elife. 2019;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orozco JM, Krawczyk PA, Scaria SM, Cangelosi AL, Chan SH, Kunchok T, et al. Dihydroxyacetone phosphate signals glucose availability to mTORC1. Nat Metab. Nature Research; 2020;2:893–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Choo AY, Kim SG, Vander Heiden MG, Mahoney SJ, Vu H, Yoon SO, et al. Glucose Addiction of TSC Null Cells Is Caused by Failed mTORC1-Dependent Balancing of Metabolic Demand with Supply. Mol Cell. 2010;38:487–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brugarolas J, Lei K, Hurley RL, Manning BD, Reiling JH, Hafen E, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. Cold Spring Harbor Laboratory Press; 2004;18:2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochachka PW, Buck LT, Doll CJ, Land SC. Unifying theory of hypoxia tolerance: Molecular/metabolic defense and rescue mechanisms for surviving oxygen lack. Proc Natl Acad Sci U S A. 1996;93:9493–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manning BD, Toker A. AKT/PKB Signaling: Navigating the Network. Cell. Cell Press; 2017. page 381–405. [DOI] [PMC free article] [PubMed]
- 37.Takano A, Usui I, Haruta T, Kawahara J, Uno T, Iwata M, et al. Mammalian Target of Rapamycin Pathway Regulates Insulin Signaling via Subcellular Redistribution of Insulin Receptor Substrate 1 and Integrates Nutritional Signals and Metabolic Signals of Insulin. Mol Cell Biol. American Society for Microbiology; 2001;21:5050–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Manning BD, Logsdon MN, Lipovsky AI, Abbott D, Kwiatkowski DJ, Cantley LC. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. Cold Spring Harbor Laboratory Press; 2005;19:1773–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Young RM, Ackerman D, Quinn ZL, Mancuso A, Gruber M, Liu L, et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. Cold Spring Harbor Laboratory Press; 2013;27:1115–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poillet-Perez L, White E. Role of tumor and host autophagy in cancer metabolism. Genes Dev. Cold Spring Harbor Laboratory Press; 2019. page 610–9. [DOI] [PMC free article] [PubMed]
- 41.Augustin HG, Koh GY. Organotypic vasculature: From descriptive heterogeneity to functional pathophysiology. Science (80-. ). 2017. [DOI] [PubMed]
- 42.Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. Nature Publishing Group; 2013;45:860–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
TCGA MC3 data is publicly available via the NIH Genomic Data Commons (https://gdc.cancer.gov/about-data/publications/mc3-2017). All PCAWG analysis results (Supplemental Figure 1d) were downloaded from the ICGC data portal (https://dcc.icgc.org/releases/PCAWG/drivers/p-values).