Abstract
Metastasis is the main cause of death in many cancers including colorectal cancer (CRC); however, the underlying mechanisms responsible for metastatic progression remain largely unknown. We found that nuclear TYRO3 receptor tyrosine kinase is a strong predictor of poor overall survival in patients with CRC. The metastasis-promoting function of nuclear TYRO3 requires its kinase activity and matrix metalloproteinase–2 (MMP-2)–mediated cleavage but is independent of ligand binding. Using proteomic analysis, we identified bromodomain-containing protein 3 (BRD3), an acetyl-lysine reading epigenetic regulator, as one of nuclear TYRO3’s substrates. Chromatin immunoprecipitation-sequencing data reveal that TYRO3-phosphorylated BRD3 regulates genes involved in anti-apoptosis and epithelial-mesenchymal transition. Inhibition of MMP-2 or BRD3 activity by selective inhibitors abrogates nuclear TYRO3-induced drug resistance and metastasis in organoid culture and in orthotopic mouse models. These data demonstrate that MMP-2/TYRO3/BRD3 axis promotes the metastasis of CRC, and blocking this signaling cascade is a promising approach to ameliorate CRC malignancy.
Matrix metalloproteinase-2-processed nuclear TYRO3 promotes colorectal cancer malignancy through BRD3.
INTRODUCTION
Activation of oncogenes and inactivation of tumor-suppressor genes are known to promote the initiation and progression of colorectal cancer (CRC) (1–5). However, the underlying mechanisms leading to metastasis, the main cause of death in patients with CRC, remain largely uncharacterized. Receptor tyrosine kinases (RTKs) are important plasma membrane receivers that perceive extracellular cues, convert them into chemical messages, and further transmit them into the nucleus to initiate cellular responses. The canonical function of the RTK is activated via a ligand-dependent manner (6). Aberrant expression and/or activation of RTKs have been implicated in cancer development and progression (7, 8). However, small-molecule inhibitors that disrupt RTK-mediated signaling transduction, such as regorafenib and axitinib, have demonstrated unsatisfied efficacy in blocking RTK-mediated cancer malignancy (9, 10), indicating that noncanonical pathways may exist to exert procancerous functions of RTKs.
Studies have shown that RTK endocytosis and trafficking to the nucleus, as a holoreceptor or truncated fragment, occur in several cell types (11, 12). At least 12 subfamilies of RTKs have noncanonical pathways that regulate transcription, DNA damage and repair, and cancer development (12–17). For example, epidermal growth factor receptor nuclear localization has been shown to increase chemotherapy resistance and reduce overall survival (14, 18).
The members of the TYRO3, AXL, and MER (TAM) family are the less studied RTKs, and their roles in cancer malignancy are relatively unclear. Classically, protein S and growth arrest–specific gene 6 bind to TAM to activate the downstream PI3K/AKT signaling pathway, thereby instigating diverse biological functions (19, 20). Aberrant expression of TAM has been shown to be associated with tumorigenesis, metastasis, and drug resistance (20, 21). A recent study also showed that TYRO3 is an oncogene that contributes to CRC progression and poor prognosis (22). Although administering human monoclonal TYRO3 antibody attenuates TYRO3-induced epithelial-mesenchymal transition (EMT) and tumor growth (22); like other cancer therapies, the treatment is limited because of the partial response. These findings suggest that there are some unknown mechanisms responsible for TYRO3-mediated CRC pathogenesis, which remain to be discovered.
Here, we present a mechanism for TYRO3 nuclear translocation in CRC through matrix metalloproteinase–2 (MMP-2). Nuclear TYRO3 phosphorylates bromodomain-containing protein 3 (BRD3), an acetyl-lysine reader, epigenetically regulating a group of genes to facilitate CRC progression and metastasis. Blocking nuclear translocation of TYRO3 by selective MMP-2 inhibitor or inhibiting nuclear TYRO3 function through ablating BRD3 effectively prevents CRC metastasis.
RESULTS
Nuclear translocation of TYRO3 C terminus promotes CRC malignancy
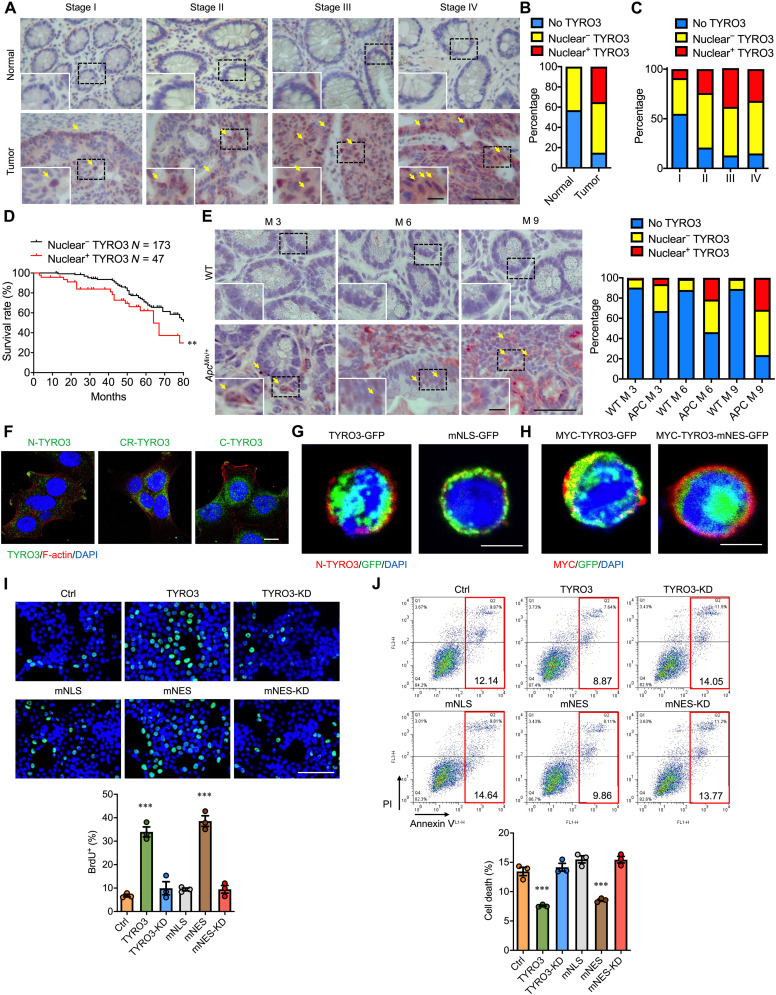
To investigate how TYRO3 promotes tumor progression in CRC, we performed immunohistochemical staining on 265 paired colon cancer and adjacent normal tissues and found a strong nuclear TYRO3-immunoreactive signal in about 35% of the cancer cells, but not in normal cells (Fig. 1, A to C). Moreover, patients with nuclear TYRO3 had worse prognosis than patients without nuclear TYRO3. The 6-year survival rate was lowered to 40% in nuclear TYRO3-positive patients (Fig. 1D).
Fig. 1. Nuclear TYRO3 expression correlates with malignancy of CRC and promotes procancerous functions.
(A) Tissue sections from paired normal and CRC were subjected to IHC staining of TYRO3 (red). Arrows indicate TYRO3+ in nucleus. Scale bar, 50 μm. Inset scale bar, 10 μm. (B) Accumulated percentage of TYRO3 staining in normal and cancer tissues. (C) Accumulated percentage of TYRO3 subcellular localization in different stages of cancer (stage I = 11, stage II = 29, stage III = 191, stage IV = 34). (D) Kaplan-Meier plot of survival rates in CRC patients with nuclear TYRO3 negative (n = 173) and nuclear TYRO3 positive (n = 47). (E) Colon tissue sections collected from wild-type (WT) and ApcMin/+ mice (n = 6 mice per group) were processed for TYRO3 IHC staining (red). Scale bar, 50 μm. Inset scale bar, 10 μm. Quantitative data are shown. Arrows indicate TYRO3+ in nucleus. (F) Subcellular distribution of TYRO3 was detected by immunofluorescence (IF) staining (N-TYRO3, anti–N-terminal TYRO3; CR-TYRO3, anti-central region TYRO3; C-TYRO3, anti–C-terminal TYRO3). Scale bar, 10 μm. (G) IF staining images showed location of N-terminal TYRO3 (red) and C-terminal TYRO3 (GFP) in HCT116 cells. Scale bar, 10 μm. (H) Images showed locations of N terminus (red, MYC) and C terminus (GFP) of TYRO3. Scale bar, 10 μm. (I) Representative images (top) and quantified result (bottom) of bromodeoxyuridine (BrdU) incorporation assay. Scale bar, 100 μm. (J) The percentages of cell death were analyzed from both annexin V–fluorescein isothiocyanate (FITC+)/propidium iodide (PI)− and annexin V-FITC+/PI+ by flow cytometry (n = 3). **P < 0.01 and ***P < 0.001.
The development of CRC may begin from noncancerous adenoma or polyp, which can be classified into hyperplastic polyp, tubular adenoma, villo-tubular adenoma (VTA), villous adenoma (VA), and serrated polyp (SP). Histologically, VTA, VA, and SP are highly associated with the development of CRC later in life (23, 24). To characterize the expression of nuclear TYRO3 in different types of polyps, we assessed the expression of nuclear TYRO3 in different polyps and found that levels of nuclear TYRO3 were increased in VTA, VA, and SP (fig. S1A), suggesting that nuclear TYRO3 may play some roles in polyp-carcinoma transformation.
We next investigated nuclear TYRO3 expression in two different mouse models of CRC. In the azoxymethane/dextran sodium sulfate (AOM/DSS) mouse model (colitis-associated CRC), cytosolic and nuclear TYRO3 expression was observed 2 weeks after AOM/DSS treatment, and the staining intensity and percentage of positive cells increased with time (fig. S1B). In the ApcMin/+ genetic mouse model of spontaneous CRC, nuclear TYRO3 was detected in as early as 3 months of age, and the percentage of nuclear TYRO3-positive cells increased along with the time (Fig. 1E). These results demonstrate that nuclear TYRO3 appears in the early stage of CRC and suggest it plays an important pathological function in tumor progression.
Immunofluorescence (IF) staining showed that the central region and C terminus of endogenous TYRO3 were detected in the nucleus, while the N terminus of TYRO3 was retained in the plasma membrane (Fig. 1F). Nuclear translocation of TYRO3 is confirmed in TYRO3–green fluorescent protein (GFP)–transfected cells (fig. S1C). Bioinformatic analysis identified a nuclear localization signal (NLS) sequence at amino acids 450 to 460 and a nuclear export signal (NES) sequence at 802 to 901 (fig. S1D). Mutation of the NLS (mNLS) prevented TYRO3 nuclear translocation (Fig. 1G and fig. S1E), while mutation of NES (mNES) showed nucleus only GFP signal (Fig. 1H and fig. S1F).
Functionally, wild-type TYRO3 or nucleus-retained TYRO3 (mNES-TYRO3) stimulated cell proliferation, while kinase dead (K550A) TYRO3 (TYRO3-KD and mNES-TYRO3-KD) or cytoplasm TYRO3 (mNLS-TYRO3) failed to induce cell proliferation (Fig. 1I and fig. S1G). Transfection with TYRO3 or mNES-TYRO3 but not TYRO3-KD or mNLS-TYRO3 significantly reduced the level of activated caspase-3 (fig. S1, H to J). Consistently, TYRO3 and mNES-TYRO3 decreased cell death compared to the control, while TYRO3-KD, mNLS-TYRO3, and mNES-TYRO3-KD failed to do so (Fig. 1J). Together, our data show the level of nuclear TYRO3 is positively correlated with CRC malignancy, and kinase activity is needed for nuclear TYRO3 to promote the malignant phenotype.
Proteolytic cleavage confers TYRO3 nuclear translocation
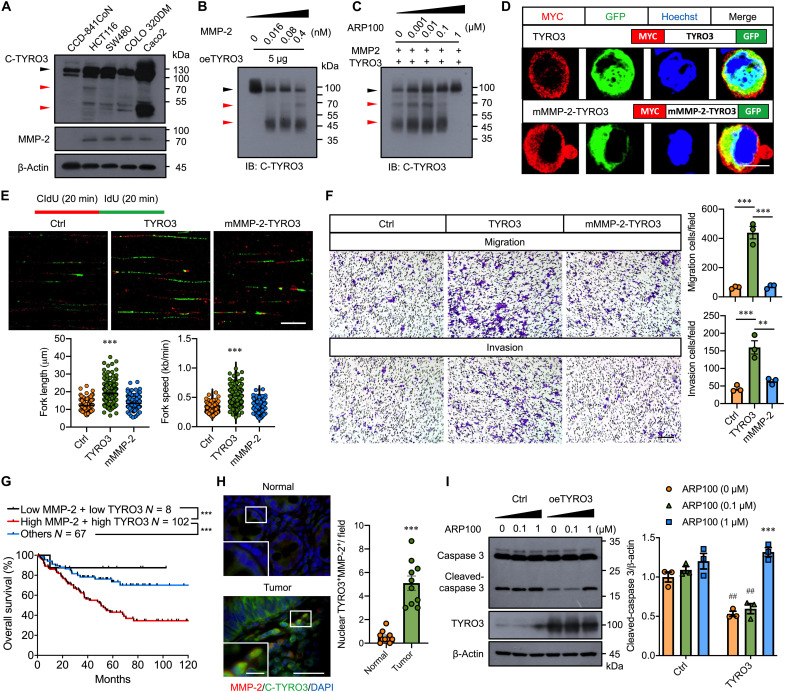
As we reasoned that TYRO3 is cleaved by proteases to cause intracellular domain (ICD)–TYRO3 nuclear translocation, we performed amino acid sequence analysis to predict potential proteases. Two MMP-2 cleavage sites were identified on TYRO3 at amino acid 247 to 251 and 442 to 446, which yield 26.4/70.6 kDa and 47.5/49.5 kDa fragments, respectively (fig. S2A). By immunoblotting, full-length TYRO3 was elevated in all CRC cell lines compared to that in the normal cell line (CCD-841CoN) (Fig. 2A, black arrowhead). Smaller bands around 50 and 70 kDa were also observed in cancer but not in normal cell lysates (Fig. 2A, red arrowhead). Consistent with this notion, the expression of MMP-2 was detected in CRC cell lines but not in normal colon cells (Fig. 2A). Digestion assay showed that the level of full length TYRO3 decreased while bands at 70 and 50 kDa increased along with the increase of MMP-2 (Fig. 2B). Administration of selective MMP-2 inhibitor, ARP100, dose-dependently inhibited the digestion of TYRO3 (Fig. 2C). We generated MMP-2 cleavage site mutated TYRO3 (mMMP-2–MYC–TYRO3–GFP) (fig. S2A) and found that MYC-tag and GFP signal were colocalized in the cytosol but not in the nucleus in mMMP-2–MYC–TYRO3–GFP–transfected cells (Fig. 2D). These data confirmed that TYRO3 traffics to the nucleus through MMP-2 cleavage. Functionally, overexpression of mMMP-2–TYRO3 failed to induce DNA replication and cell proliferation (Fig. 2E and fig. S2B). Cell migration and invasion abilities were also reduced in mMMP-2–TYRO3–overexpressed cells as compared to cells transfected with wild-type TYRO3 (Fig. 2F and fig. S2C).
Fig. 2. MMP-2 cleaves TYRO3 to promote nuclear translocation and cancer progression.
(A) Representative Western blots showed the expression of C-TYRO3, MMP-2, and β-actin in normal colon cell line (CCD-841CoN) and CRC cell lines. (B) Representative Western blots of TYRO3 in lysates of TYRO3-overexpressed HCT116 cells incubated with different amounts of recombinant MMP-2. Black arrowhead, full-length TYRO3; red arrowhead, cleaved TYRO3. (C) Representative Western blots showed TYRO3 in lysates and MMP-2 (0.4 nM) incubated with different amounts of MMP-2 inhibitor, APR100. Black arrowhead, full-length TYRO3; red arrowhead, cleaved TYRO3. IB, immunoblot. (D) Confocal images showed subcellular location of N-terminal TYRO3 (red) and C-terminal TYRO3 (GFP). Scale bar, 10 μm. (E) Representative images showed DNA fibers and quantification of replication forks (n = 150). Scale bar, 10 μm. (F) Transwell migration and invasion assays were performed after transfection. Representative images and quantification are shown. Scale bar, 200 μm. (G) The impact of MMP-2 and TYRO3 expression on overall survival (177 patients with CRC) in GSE17536 cohort (downloaded from Gene Expression Omnibus data). (H) Representative images and quantified show results of tissue sections from normal and tumor (n = 10) immunostained for MMP-2 (red) and C-TYRO3 (green). Scale bar, 200 μm. Inset scale bar, 50 μm. (I) Representative Western blots and quantified data showed the levels of caspase 3, TYRO3, and β-actin in HCT116 cells transfected with the indicated plasmids. Asterisks indicate statistical differences in intragroup comparison, and pound signs indicate statistical differences in intergroup comparison. ** and ##P < 0.01, ***P < 0.001.
To explore the clinical relevance of MMP-2 expression in CRC pathogenesis, we analyzed mRNA expression profiles and survival information in a colon cancer dataset (GSE17536) (25). Kaplan-Meier analysis showed that high MMP-2/high TYRO3 had the lowest overall survival rate (Fig. 2G). The immunostaining also showed that MMP-2 and nuclear TYRO3 were markedly up-regulated and coexpressed in the CRC tissues (Fig. 2H). Moreover, inhibition of MMP-2 activity abolished the anti-apoptotic effect exerted by TYRO3 overexpression (Fig. 2I).
Targeting MMP-2 activity prevents CRC progression in mice
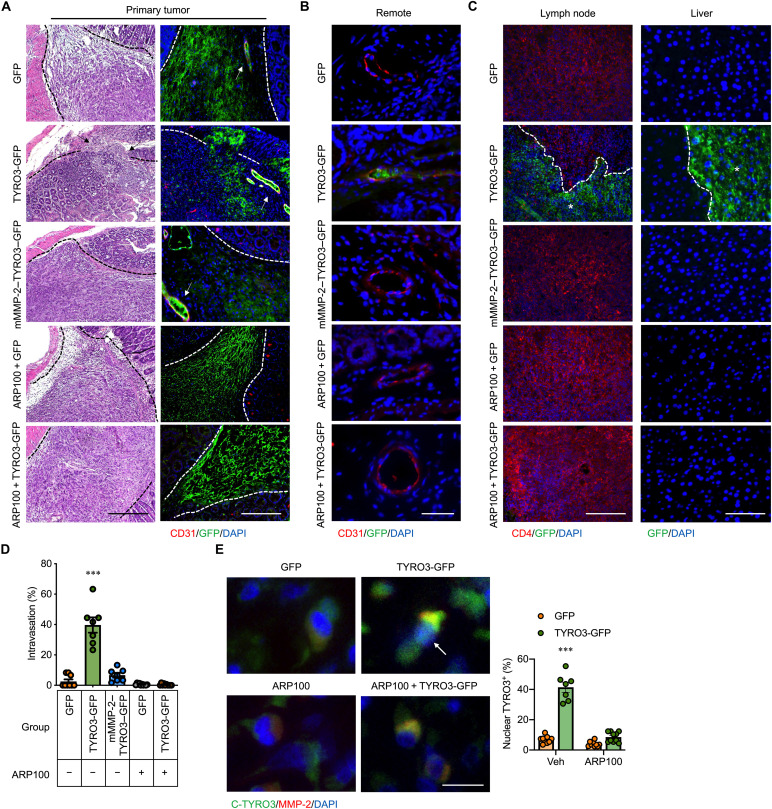
To test the metastasis-promoting ability of MMP-2–cleaved nuclear TYRO3, mice with orthotopic injection of TYRO3-GFP–overexpressed HCT116 cells were treated with or without ARP100. Treatment with ARP100 reduced CRC primary tumor size (fig. S3A). Histological analyses identified muscular invasion, remote (>100 μm away from the primary tumor) vascular invasion, lymph node invasion, and liver metastasis in the TYRO3-GFP group (Fig. 3, A to D, and fig. S3, B and C). In contrast, the primary tumor was restrained by the surrounding tissues as a clear boundary was seen in the mMMP-2–TYRO3–GFP group and the ARP100-treated TYRO3-GFP group (Fig. 3A). ARP100 treatment indeed prevented nuclear translocation of TYRO3 in vivo (Fig. 3E). No remote invasion or metastasis was observed in the mMMP-2–TYRO3–GFP group and the ARP100-treated TYRO3-GFP group (Fig. 3, A to D, and fig. S3C). These data demonstrate that disrupting MMP-2–mediated TYRO3 nuclear translocation is an effective approach for blocking CRC metastasis.
Fig. 3. TYRO3 promotes CRC metastasis through MMP-2–dependent nuclear translocation.
HCT116 cells stably express different constructs were injected into the cecal wall of NOD/SCID mice. ARP100 (5 mg/kg per day) or vehicle control (dimethyl sulfoxide) was delivered through osmotic pumps immediately after orthotopic injection for 1 month. (A) The representative pictures show the lesion in the cecum after 1 month by hematoxylin and eosin and IF staining (GFP, n = 9; TYRO3-GFP, n = 7; mMMP-2–GFP, n = 7; ARP100 + GFP, n = 10; ARP100 + TYRO3-GFP, n = 10). Arrows indicate the invasive cancer (left). Scale bar, 100 μm. Dashed lines delineate the boundaries segregating cancer and normal tissues. Arrows indicate cancer cells in the lumen of blood vessels (right). (B) Remote area (over 100 μm away from tumor) sections were stained with anti-GFP antibody (green) for cancer cells, anti-CD31 antibody (red) for endothelial cells, and DAPI for nucleus. Scale bar, 100 μm. (C) Lymph nodes were stained with anti-GFP antibody (green) for cancer cells, anti-CD4 antibody (red) for lymphocytes, and DAPI for nucleus. Scale bar, 100 μm. Liver tissue sections were stained with anti-GFP antibody (green) and counterstained with DAPI. Scale bar, 50 μm. Dashed lines delineate the boundaries segregating cancer and normal tissues. White stars indicate the cancers. (D) Percentage of mice with intravasated cancer cells at remote area. (E) Representative pictures show orthotopic tumor tissue sections stained with anti–C-TYRO3 antibody (green), anti–MMP-2 antibody (red), and DAPI (left). Arrow indicates the nuclear translocation of TYRO3. Scale bar, 10 μm. Right shows the percentage of mice with nuclear TYRO3 cancer cells. ***P < 0.001.
ICD-TYRO3 phenocopies the function of MMP-2–cleaved nuclear TYRO3
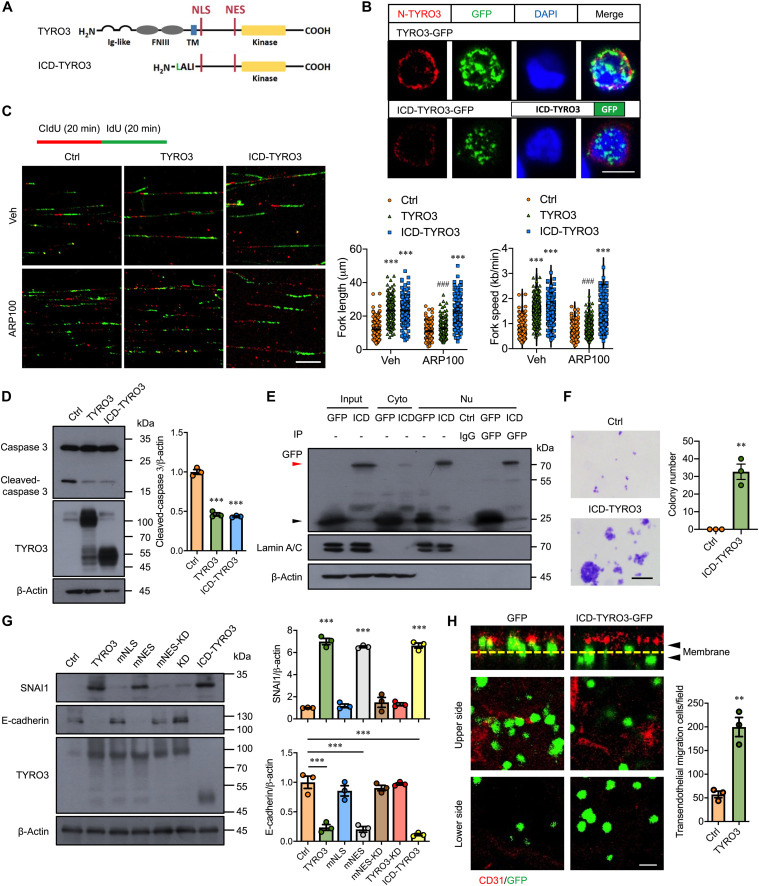
To verify the functional role of nuclear TYRO3, we generated an ICD-TYRO3 construct (Fig. 4A) and transfected it into HCT116 cells. ICD-TYRO3-GFP was mainly detected in the nuclei and much less in the cytosol (Fig. 4B). Expression of ICD-TYRO3 significantly increased nascent DNA biosynthesis (Fig. 4C) and cell proliferation (fig. S4A), inhibited caspase-3 activation (Fig. 4D), and reduced cell death percentage (fig. S4B). In contrast, treatment with ARP100 (1 μM) blunted the promotive effect in full-length TYRO3-transfected cells but not in ICD-TYRO3–overexpressed cells (Fig. 4C), indicating that ICD-TYRO3 is functionally equivalent to MMP-2–processed nuclear TYRO3.
Fig. 4. ICD-TYRO3 promotes normal colon cell transformation and CRC malignancy.
(A) Schematic drawing shows the sequence of TYRO3 and ICD-TYRO3. The ICD-TYRO3 starts from the MMP-2 cleavage site. Ig, immunoglobulin; TM, transmembrane domain. (B) Representative confocal images showed anti-N terminus of TYRO3 (red) and GFP signal detection in HCT116 cells after transfection. Scale bar, 10 μm. (C) Lengths and speed of nascent replication tracts labeled with CldU (red) and IdU (green) were measured (n = 150) after treatment. Scale bar, 10 μm. Asterisks indicate statistical differences in intragroup comparison and pound signs indicate statistical differences in intergroup comparison. (D) Representative Western blots and quantified data showed the levels of caspase 3, TYRO3, and β-actin in HCT116 cells transfected with the indicated plasmids. (E) Representative Western blots showed the expression of ICD-TYRO3 (ICD) in the nucleus of CCD-841CoN cells after transfection. Cyto, cytosolic fraction; Nu, nuclear fraction; Ctrl, control IgG; red arrowhead, ICD-TYRO3-GFP; black arrowhead, GFP. (F) Colony formation assay was performed for 21 days after transfection of CCD-841CoN with control (Ctrl) or ICD-TYRO3 plasmid. Left, representative images; right, quantification analysis. Scale bar, 100 μm. (G) Representative Western blots and quantified data showed the levels of SNAI1, E-cadherin, TYRO3, and β-actin in HCT116 cells transfected with the indicated plasmids. (H) Representative confocal images and quantified data showed the HCT116 transendothelial migration. CD31, endothelial cells (red); Yellow dashed lines, transwell filters; arrowheads, top and bottom sides of the filters. Scale bar, 10 μm. **P < 0.01; ### and ***P < 0.001.
Next, we tested whether forced expression of ICD-TYRO3 in normal colon cells, which has no detectable 50-kDa TYRO3 fragment (Fig. 2A), can cause cell transformation. Western blotting and GFP pull-down assay confirmed that ICD-TYRO3-GFP can be detected in the nucleus (Fig. 4E). Anchorage-independent colony formation assays demonstrated that transfection of ICD-TYRO3 was sufficient to drive normal colon cells growing on soft agar (Fig. 4F), indicating that forced expression of ICD-TYRO3 is sufficient to transform normal colon cells into cancerous cells.
To test whether the cancer-promoting effects result from TYRO3 nuclear translocation, we determined the molecular changes related to EMT after forced expression of nuclear TYRO3. The results demonstrated that overexpression of TYRO3, mNES-TYRO3, or ICD-TYRO3 induced the expression of Zinc Finger E-Box Binding Homeobox 1 (ZEB1), Snail Family Transcriptional Repressor 1 (SNAI1), and Snail Family Transcriptional Repressor 2 (SLUG) but decreased the expression of E-cadherin (Fig. 4G and fig. S4C). Moreover, overexpression of ICD-TYRO3 significantly facilitated transendothelial migration, a process equivalent to intravasation (Fig. 4H). Next, we evaluated the drug-resistant abilities of nuclear TYRO3 overexpressing cells with or without 5-fluouracil (5-FU) treatment. TYRO3, mNES-TYRO3, and ICD-TYRO3 reduced 5-FU–induced cell death compared with the control. However, the protective effect was not seen in mNLS-TYRO3–, mNES-TYRO3-KD–, and TYRO3-KD–transfected cells (fig. S4D). Together, these results indicated that nuclear TYRO3 is the key component to drive colon cancer malignancy.
Nuclear TYRO3 promotes CRC malignancy in vivo
We next evaluated nuclear TYRO3’s function in promoting CRC metastasis using the orthotopic mouse model. The tumor mass in the ICD-TYRO3-GFP group was larger than that in the GFP and mNLS-TYRO3-GFP groups (fig. S5A). An invasive CRC in the ICD-TYRO3-GFP group was observed upon histological analysis (fig. S5B). Moreover, the detection of GFP-positive cancer cells in the remote blood vessel confirms the dissemination of cancer cells promoted by the ICD-TYRO3-GFP (fig. S5C). Last, we observed peritoneal dissemination, lymph node invasion, and liver metastasis in the ICD-TYRO3-GFP–injected mice (fig. S5, D and E). These results indicated that ICD-TYRO3 drives CRC metastasis in mice.
BRD3 mediates nuclear TYRO3-induced malignant phenotypes
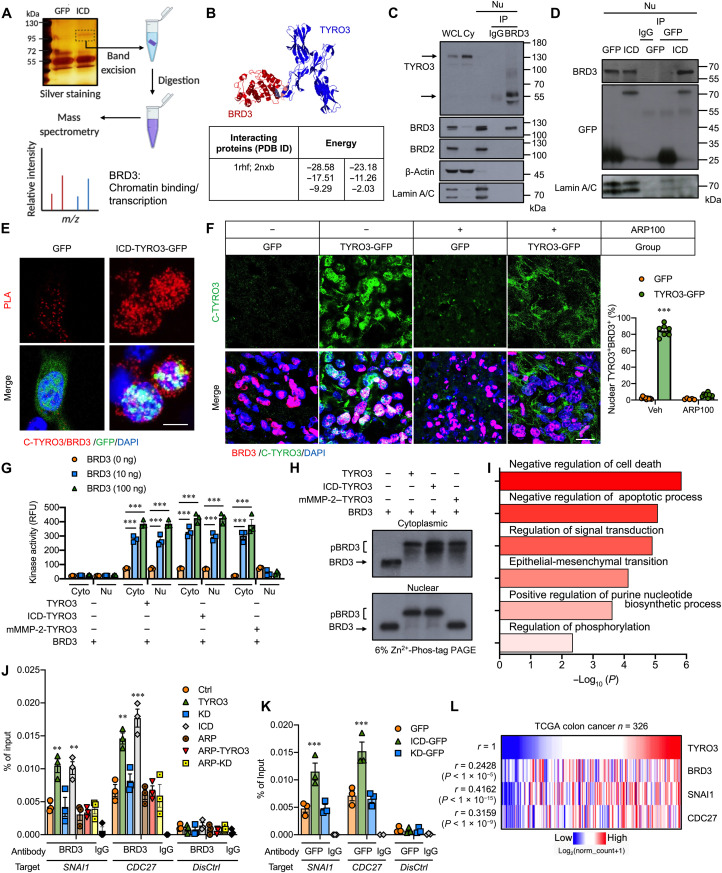
We next explored the molecular mechanism responsible for ICD-TYRO3–driven cancer development by proteomic analysis (Fig. 5A) with a focus on transcription factors or cofactors. Among the candidate proteins (fig. S6A), BRD3, an acetyl-lysine reader, was identified as a nuclear partner of TYRO3. The interaction between TYRO3 [Protein Data Bank (PDB) ID: 1rhf] and BRD3 (PDB ID: 2nxb) was validated by Protein Interactions using Structural Matching algorithm (Fig. 5B). Coimmunoprecipitation (co-IP) experiment (Fig. 5, C and D) and proximity ligation assay (PLA; Fig. 5E) confirmed that nuclear TYRO3 indeed binds BRD3. The in vitro data were further strengthened by in vivo observation that nuclear TYRO3 colocalized with BRD3 in the TYRO3-GFP group but not in the MMP-2 inhibitor–treated tumor (Fig. 5F). Moreover, protein kinase activity assay demonstrated that BRD3 was phosphorylated by recombinant TYRO3 and immunoprecipitants of nuclear and cytoplasmic fractions derived from TYRO3- and ICD-TYRO3–transfected HCT-116 cells in a time- and dose- dependent manner (Fig. 5G and fig. S6, B and C). In contrast, only cytosolic immunoprecipitant can phosphorylate BRD3 in mMMP-2–TYRO3–transfected cells (Fig. 5G). The phosphorylated BRD3 band was evidently separated from the unphosphorylated BRD3 when analyzed by Phos-tag SDS–polyacrylamide gel electrophoresis (PAGE) (Fig. 5H and fig. S6D).
Fig. 5. Nuclear TYRO3 phosphorylates BRD3 to promote CRC malignancy.
(A) Schematic drawing shows the flow chart of identifying BRD3 as ICD-TYRO3 nuclear partner by mass spectrometry from CCD-841CoN cells. (B) The interaction between TYRO3 (1rhf) and BRD3 (2nxb) and binding energies predicted by Structural Matching software. (C and D) Representative Western blots show the binding of endogenous TYRO3 and BRD3 in the nucleus of HCT116 cells (C) and ICD-TYRO3 and BRD3 in CCD-841CoN cells transiently transfected with GFP or ICD-TYRO3-GFP (D). Lamin A/C, loading control; WCL, whole cell lysate; Cy, cytosolic fraction; Nu, nuclear fraction; IgG, control. (E) Representative images show the binding of TYRO3 and BRD3 by in situ PLA. Scale bar, 5 μm. (F) Representative images and quantified data of nuclear TYRO3+/BRD3+ cells in orthotopic colon cancers (GFP, n = 9; TYRO3-GFP, n = 7; ARP100 + GFP, n = 10; ARP100 + TYRO3-GFP, n = 10). Scale bar, 10 μm. (G) The kinase activity of TYRO3 in phosphorylating BRD3. RFU, relative fluorescence units. (H) Representative Western blots show phosphorylated BRD3 (pBRD3) and unphosphorylated BRD3. The proteins were separated by 6% Zn2+-Phos-tag SDS–polyacrylamide gel electrophoresis (PAGE). (I) Biological processes associated with genes bound by BRD3 in nuclear TYRO3-overexpressing cells. (J) Results of ChIP-qPCR show the occupancies of BRD3 at the promoters of targeted genes. Distal primers were used as a quality control. (K) Results of ChIP-qPCR show the occupancies of TYRO3 at the promoters of SNAI1 and CDC27. Lysates from HCT-116 cells stably transfected with different constructs were immunoprecipitated using anti-GFP antibody and subjected for PCR amplification. (L) Heatmap shows the correlation of TYRO3 level with BRD3, SNAI1, and CDC27 by analyzing TCGA colon cancer dataset. **P < 0.01 and ***P < 0.001. m/z, mass/charge ratio.
Using chromatin immunoprecipitation sequencing (ChIP-seq) analysis, we showed that phosphorylated BRD3 binding is particularly enriched in genes involved in negative regulation of cell death, regulation of signal transduction, EMT, positive regulation of nucleotide biosynthesis, and regulation of phosphorylation (Fig. 5I). As a proof of concept, we checked whether CDC27 and SNAI1 were regulated by BRD3 since they are key regulators of TYRO3-induced CRC cell proliferation and metastasis. Integrative Genomic Viewer analysis identified BRD3 binding site on SNAI1 and CDC27 genes (fig. S6E). ChIP–quantitative polymerase chain reaction (qPCR) showed that BRD3 and nuclear TYRO3 occupancies at SNAI1 and CDC27 were enriched in TYRO3- and ICD-TYRO3–overexpressed cells but not the TYRO3-KD–overexpressed cells (Fig. 5, J and K). We then checked the coexpression of TYRO3/BRD3/SNAI1/CDC27 in The Cancer Genome Atlas (TCGA) database and confirmed that the expression of TYRO3 was positively correlated with BRD3, SNAI1, and CDC27 (Fig. 5L). Together, these data indicate that MMP-2–released nuclear TYRO3 phosphorylates BRD3 to confer increased oncogenic activity.
Ablating BRD3 eradicates TYRO3-induced CRC metastasis in mice
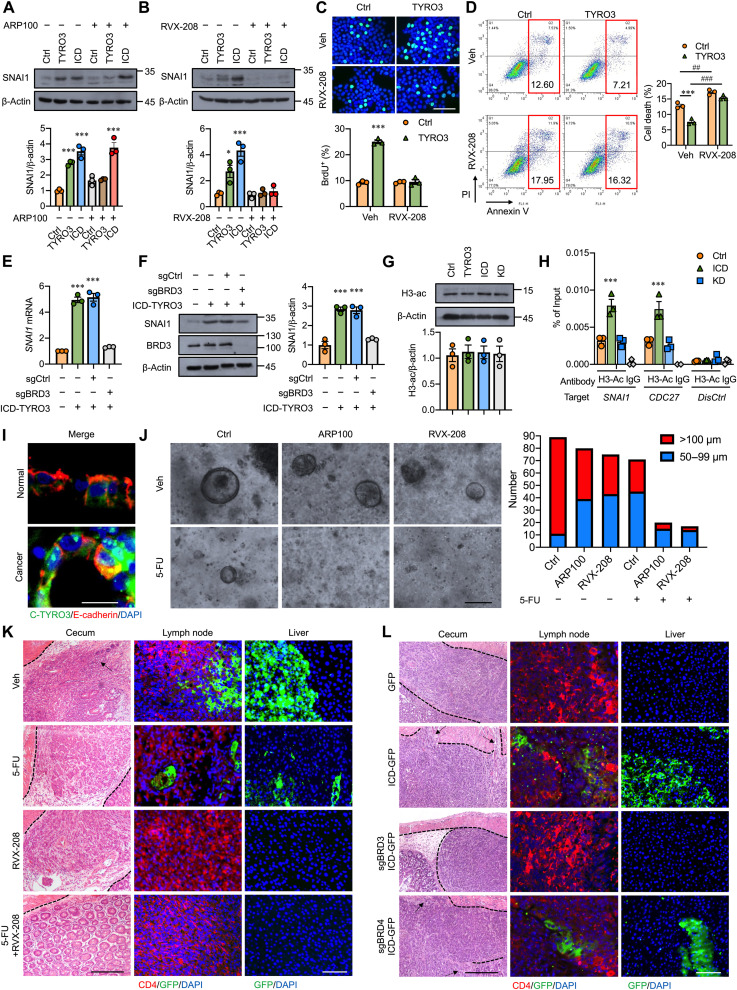
We then tested whether targeting BRD3 can abolish nuclear TYRO3-mediated CRC malignancy. First, inhibition of MMP-2 or BRD3 activity significantly reduced TYRO3-promoted SNAI1 expression; however, only BRD3 inhibitor (RVX-208) can block ICD-TYRO3–induced SNAI1 up-regulation (Fig. 6, A and B). Treatment with RVX-208 not only inhibited TYRO3-induced SNAI1 expression but also abolished TYRO3-induced cell growth and anti-apoptosis (Fig. 6, B to D). Knockdown of BRD3 but not BRD4 abolished ICD-TYRO3–induced SNAI1 expression (fig. S7, A-D). We then generated BRD3 or BRD4 knockout lines in ICD-TYRO3–overexpressed cells using the CRISPR-Cas9 system and confirmed the reduction of SNAI1 in the BRD3 but not the BRD4 knockout cells (Fig. 6, E and F, and fig. S7, E and F). Since BRD3 binds to acetylated histone, we aimed to test whether overexpression of TYRO3 causes any histone acetylation changes. Global acetylation of histone H3 are not different between control and TYRO3-overexpressed cells (Fig. 6G). However, ChIP-qPCR showed that histone H3 acetylation occupancies in SNAI1 and CDC27 promoters were enriched in ICD-TYRO3–overexpressed cells but not the TYRO3-KD–overexpressed cells (Fig. 6H), indicating that nuclear TYRO3 induces local histone H3 acetylation at the promoters of its targeted genes. Consistently, ICD-TYRO3–promoted transendothelial migration was blunted in the BRD3−/− but not BRD4−/− cells (fig. S7G).
Fig. 6. Inhibition of BRD3 eradicates TYRO3-induced CRC metastasis in mice.
(A) Representative Western blots and quantified result show SNAI1 in cells transfected with indicated plasmids in the presence (+) or absence (−) of MMP-2 inhibitor, ARP100 (1 μM). (B) Representative Western blots and quantified result show the levels of SNAI1 in HCT116 cells treated with (+) or without (−) BRD3 inhibitor, RVX-208 (10 μM). (C and D) Representative images and quantified results show BrdU+ (C) and annexin V+/PI+ (D) cells treated with or without BRD3 inhibitor. Scale bar, 50 μm. (E) Real-time reverse transcription–qRCR shows the expression of SNAI1. (F) Representative Western blot and quantified result show SNAI1 in ICD-TYRO3–overexpressed HCT116 cells with or without BRD3 knockout. (G) Representative Western blot and quantified result show global acetylation of histone H3 in HCT116 cells. (H) Potential target genes of BRD3 were analyzed by ChIP-qPCR. (I) Representative images show location of C-TYRO3, E-cadherin, and DAPI in colon organoids. Scale bar, 10 μm. (J) Representative images and quantified result of organoids (n = 3, 30 organoids per subject) treated with or without inhibitors were shown. Scale bar, 100 μm. (K) Representative images show local invasion and metastasis of cancer cells with or without different drug treatments. Arrows indicate the invasive cancer. Scale bar: 100 μm (left column); 50 μm (middle and right columns). (L) Representative images show local invasion and metastasis of cancer cells with or without BRD3 (sgBRD3) or BRD4 (sgBRD4) knockout. Arrows indicate the invasive cancer. Scale bars, 100 μm (left) and 50 μm (right). *P < 0.05, ##P < 0.01, ### and ***P < 0.001.
We then tested the therapeutic potential of BRD3 inhibitor in the organoid culture and orthotopic mouse model. Treatment with ARP100, RVX-208, or 5-FU alone reduced the size and number of cancer organoids. Combined treatment with ARP100 and 5-FU or RVX-208 and 5-FU markedly inhibited organoid development (Fig. 6, I and J). In the in vivo study, the sizes of tumors are smaller in 5-FU– and RVX-208–treated mice as compared to those in vehicle-treated mice (fig. S7H). The tumors are either very small or even invisible in the 5-FU + RVX-208 group (fig. S7H). Histological examination found that cancer cells had metastasized to the lymph node and liver in the vehicle group. Treatment with 5-FU reduced the incidence of cancer metastasis, but lymph node and liver metastatic cancer cells were still easily seen (Fig. 6K). Treatment with RVX-208 alone or combined treatment with 5-FU and RVX-208 completely abolished cancer metastasis (Fig. 6K). To validate the RVX-208’s effect is through inhibiting BRD3’s activity instead of potential off-target effect, we performed another in vivo study by orthotopically injected control (GFP only), ICD-TYRO3–overexpressed cells, BRD3−/− ICD-TYRO3-GFP cells, or BRD4−/− ICD-TYRO3-GFP cells and let the tumor grow for 1 month. The tumor mass was smaller in BRD3−/− group as compared to GFP, ICD-TYRO3-GFP, and BRD4−/− ICD-TYRO3-GFP groups (fig. S7I). An invasive CRC in the ICD-TYRO3-GFP and BRD4−/− ICD-TYRO3-GFP groups were observed upon histological analysis (Fig. 6L). Last, we observed lymph node invasion and liver metastasis in the ICD-TYRO3-GFP– and BRD4−/− ICD-TYRO3-GFP–injected mice but not BRD3−/− ICD-TYRO3-GFP–injected mice (Fig. 6L). Together, these results confirmed that nuclear TYRO3’s pathological effects were mediated by BRD3 but not BRD4.
DISCUSSION
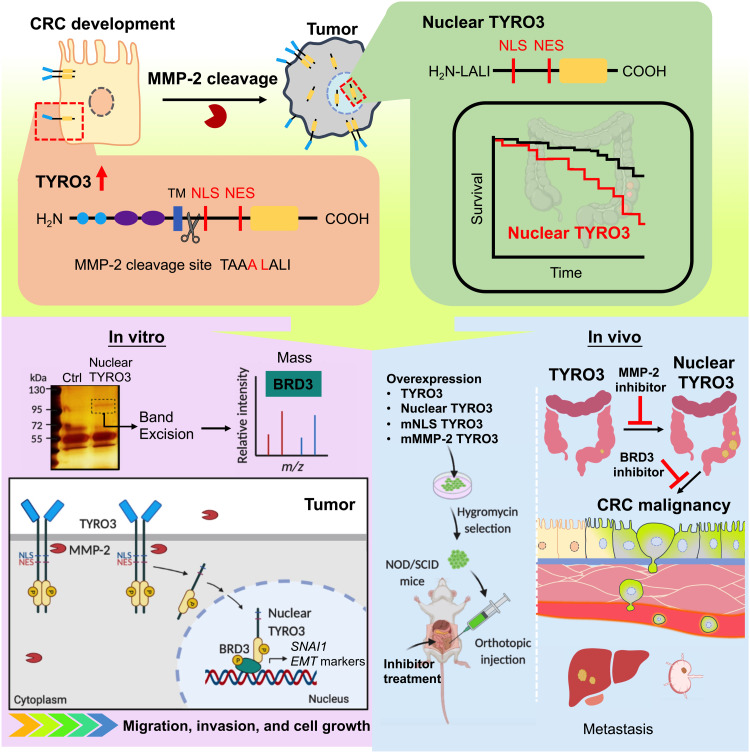
CRC is among the most common and deadliest neoplasms in the world. Current clinical treatments for CRC rely on surgery, chemotherapy, radiotherapy, and canonical signaling targeted therapy. However, the treatment effect is not satisfactory due, at least in part, to the uncharacterized mechanisms responsible for the pathological processes. Here, we identify that the ligand-independent activation of ICD-TYRO3 nuclear translocation is mediated by intracellular MMP-2–dependent proteolysis. Nuclear TYRO3 plays a protumor function by phosphorylating BRD3 to induce EMT, cell growth, and metastasis. The selective MMP-2 inhibitor prevents TYRO3 nuclear translocation while the BRD3 inhibitor and knockout blunt nuclear TYRO3’s function, thereby reducing the protumor phenotype of colon cancer cells and preventing CRC metastasis in mice (Fig. 7). These observations may lead to the development of potential therapeutic strategies for CRC by disrupting the translocation and/or function of nuclear TYRO3.
Fig. 7. Model of nuclear TYRO3 contributing to CRC malignancy.
We identified that MMP-2 releases ICD-TYRO3 to reduce survival rate and promote CRC migration, invasion, and cell growth through BRD3. TYRO3-induced CRC progression and metastasis are reversed by MMP-2 selective inhibitor in orthotopic mouse models.
Nuclear translocation of RTKs has been known for more than 20 years. Although the functions of most nuclear RTKs remain largely unknown, some nuclear RTKs have been shown to be associated with drug resistance and transcriptional regulation (16, 18, 26, 27). Compared to those well-known RTKs, the TAM family members are less studied. MER is the first reported TAM family member that can be detected in the nucleus in human leukemia cell lines under prolonged ligand stimulation (17). Here, we report TYRO3 as another TAM member functioning in the nucleus and show that nuclear translocation of TYRO3 is independent of its cognate ligands.
The translocation of RTKs to the nucleus can be mediated through different trafficking mechanisms (15, 28–30). Here, we found that TYRO3 nuclear translocation is mediated by MMP-2–dependent cleavage. TYRO3 has two MMP-2 cutting sites; one in the extracellular domain and the other locates immediately downstream of the transmembrane domain. Cleavage at the second MMP-2 cutting site generates a 50-kDa ICD-TYRO3, which contains nuclear localization sequence and kinase domain. Although MMP is widely considered as the protease of extracellular matrix proteins, increasing evidence suggested that MMP can also target intracellular substrates and regulate disease progression (31, 32). Mutating the MMP-2 cleaving sequence or administration of MMP-2 inhibitor blocks TYRO3 nuclear translocation and concomitantly abolishes TYRO-3–mediated pathological processes in vitro and in vivo. Collectively, these data demonstrate that MMP-2 is a critical regulator that controls TYRO3 nuclear translocation and subsequently CRC malignancy.
Although TYRO3 up-regulation has been detected in several cancers (22, 33–38), the cancer-promoting mechanism remains largely uncharacterized. Aberrant expression of MMP-2 triggers ligand-independent nuclear translocation of TYRO3 leading to drug resistance. This may explain why human anti-TYRO3 antibody (22) or U.S. Food and Drug Administration–approved small-molecule inhibitor (39) is only partially effective since they fail to target nuclear TYRO3. Therefore, preventing TYRO3 from translocating to the nucleus may be a better alternative to increase CRC drug sensitivity.
Our result that TYRO3 can serve as a transcription coactivator to enhance gene transcription by phosphorylating BRD3 demonstrates the noncanonical function of TYRO3. Bromodomain and extra-terminal domain (BET) proteins such as BRD3 can bind directly to the acetylated histones to promote the transcription of multiple oncogenic genes, including PI3K, AKT, and MYC (40, 41). In this study, we demonstrated that nuclear TYRO3 enhances histone H3 acetylation at the promoters of its target genes, which may increase the binding of BRD3 to these regions. Our data also showed that TYRO3 and BRD3 bind to the similar region of SNAI1 and CDC27 promoters. It has been reported that BRD4 phosphorylation in patient-derived CRC cell line promotes its chromatin binding ability (42). Here, we proved that BRD3 is phosphorylated by TYRO3 and is required for enhancing the expression of SNAI1, a multifunctional transcription factor that regulates EMT and drug resistance in many cancers. Preventing BRD3 phosphorylation by blocking TYRO3 nuclear translocation or forced expression of a kinase dead TYRO3 failed to enhance BRD3 binding to SNAI1 promoter and to induce SNAI1 expression. These data suggest that BRD3 phosphorylation is important for exerting its function.
To date, more than 10 BET inhibitors have advanced to early-stage clinical trials for patients with different types of cancer (43). BET inhibitors, such as A1874 and JQ1, have been demonstrated to inhibit CRC cell proliferation and migration (44, 45); however, the underlying mechanism is not clear. Here, we provide evidence to unravel the underlying mechanism of how BET inhibitors work in ameliorating cancer malignancy. BRD3 is an important player to transmit the TYRO3-mediated signaling in promoting CRC proliferation, anti-apoptosis, and metastasis. Treatment with RVX-208 or knockout of BRD3 to block BRD3’s binding activity not only reduces primary tumor size and prevents cancer metastasis but also synergizes the anticancer drug’s efficacy. Further study is warranted to characterize whether BET inhibitors are more pronounced in treating cancers with nuclear RTKs.
In summary, our findings strongly support that MMP-2–processed ICD-TYRO3 nuclear translocation is essential for CRC malignancy. Nuclear TYRO3 plays a protumor function to promote cell proliferation, drug resistance, and metastasis, which is likely mediated through BRD3 phosphorylation. Thus, targeting TYRO3 nuclear translocation or function may provide an unconventional therapeutic approach to eradicate CRC progression.
MATERIALS AND METHODS
Clinical samples
This study was reviewed and approved by the Institutional Reviewing Board of National Cheng Kung University Hospital (approved number: A-ER-109-065). All patients provided signed informed consent. CRC tissues (n = 265), polyps (n = 12 per type), colon tissue specimens for organoid (n = 3) were obtained from patients by surgical resection at the Department of Surgery in a university hospital. The protocol was approved by the Institutional Review Board, and informed consent was obtained from each patient.
Organoid culture
For establishing human colon organoids, colon tissue specimens from both genders were cut into 5-mm pieces and washed eight times with PBS supplemented with antibiotics (GeneDireX, CC501-011). Tissue fragments were digested with collagenase XI (100 U/ml; Sigma-Aldrich, C7657) and dispase II (0.125 mg/ml; Roche, 04942078001) (nontumor tissue) or liberase (0.28 WU/ml; Roche, 5401135001) (tumor tissue) at 37°C with shake for 40 min. After then, the reactions were stopped by fetal bovine serum (FBS; Corning, 35-010-CV) and washed with PBS supplemented containing 10% FBS for three times. Tissue fragments were filtered through a cell strainer (70 μm), centrifuged, and resuspended in Matrigel (BD Biosciences, 354230). Matrigel containing cell clusters were seeded into 24-well tissue culture plates and allowed to polymerize for 10 min at 37°C. The organoids were overlaid with 500-μl culture medium composed of advanced Dulbecco’s modified Eagle’s medium/F12 (Gibco,12500062) supplemented with 1 mM N-acetylcysteine (R&D Systems, 5619), 1 nM gastrin I (R&D Systems, 3006/1), 1× GlutaMAX (Gibco, 35050079), recombinant human epidermal growth factor (rhEGF) (50 ng/ml; Cell Guidance Systems, GFH26), recombinant human R-Spondin 1 (rhR-Spondin 1) (1 μg/ml; STEMCELL Technologies, 78213), rhNoggin (100 ng/ml; BioVision, 4675), 2 μM A83-01(STEMCELL Technologies, 72024), 10 μM SB202190 (BioVision, 2493), rhWnt-3a (60 ng/ml; Taiclone, tcup7973) (nontumor tissue), 1× N2 supplement (R&D Systems, AR009), 1× B-27 supplement (R&D Systems, AR008), 1× antibiotics, and 10 mM Hepes (Gibco, 15630130). Following the initial seeding of the cultures, 10 mM nicotinamide (R&D Systems, 4106/50), 2.5 μM CHIR 99021(R&D Systems, 4423/10), and 10 μM Y-27632 dihydrochloride kinase inhibitor (BioVision, 1596) were also added to the media for 2 to 3days. For organoid viability experiment, 200 colon organoids in 10-μl Matrigel were seeded per well on a prewarmed 96-well plate. Then, 50 μl of 5-FU (1.5 μg/ml) and/or inhibitor in the medium was added for 2 days. Organoid diameter was measured by ImageJ.
Animal studies
All animal experiments were performed following protocols approved by the Institutional Animal Care and Use Committee of National Cheng Kung University (approved number: 109214). C57BL/6, NOD/SCID, and ApcMin/+ male mice were maintained under standard conditions. The colon carcinogenesis mouse model was induced by AOM/DSS as described previously (22). In the orthotopic transplantation human colon tumor model, the 8-week-old NOD/SCID male mice were anesthetized and the hygromycin-resistant HCT116 (1 × 105) cells with stable overexpression of different constructs carrying GFP reporter in 50 μl of PBS (mixed with Matrigel at 1:1) were injected into the cecal wall. Selective MMP-2 inhibitor, ARP100 (5 mg/kg per day; Cayman Chemical, 13321), was delivered by an intraperitoneally implanted osmotic minipump (Alzet, model 2004). 5-FU (10 mg/kg, 5 days/week), BRD3 inhibitor, RVX-208 (10 mg/kg, 5 days/week, Selleckchem, S7295), or vehicle control was given by intraperitoneal injection. Tissues were harvested 1 month after surgery. All animal experiments were performed following protocols approved by the Institutional Animal Care and Use Committee.
Cell culture
CCD-841 CoN (CRL-1790, RRID:CVCL_2871), HCT116 (CCL-247, RRID:CVCL_0291), SW480 (CCL-228, RRID:CVCL_0546), COLO 320DM (CCL-220, RRID:CVCL_0219), Caco-2 (HTB-37, RRID:CVCL_0025), and HUVEC (CRL-1730, RRID:CVCL_2959) were purchased from American Type Culture Collection (Manassas) and maintained at 37°C and 5% CO2. CCD-841 CoN were cultured in Eagle’s minimum essential medium (Gibco, 61100061) supplemented with 10% FBS, HCT116 was cultured in McCoy’s 5A (Sigma-Aldrich, M4892) supplemented with 10% FBS, SW480 and COLO 320DM were cultured in RPMI 1640 (Gibco, 31800022) supplemented with 10% FBS, Caco-2 was cultured in Eagle’s minimum essential medium supplemented with 20% FBS, and HUVEC were cultured in medium 199 (Gibco, 31100-035) supplemented with heparin (25 U/ml; Sigma-Aldrich, H3149), endothelial cell growth supplement (30 μg/ml; Millipore, 02-102), and 10% FBS. Transient transfections were done using Lipofectamine 2000 (Invitrogen, 11668019), following the manufacturer’s protocol, and cultured for 1 day with or without ARP100 or RVX-208 treatment before samples were processed. For EMT marker analysis, cells were collected after 5 days of the transfection with or without ARP100 or RVX-208 treatment. For the generation of stable clones, transfected HCT116 cells were cultured for several weeks in media containing hygromycin B (100 μg/ml; Roche, 31282-04-9) selection.
Antibodies, plasmids, and reagents
Antibodies used in this study for Western blot (WB), immunohistochemistry (IHC) analysis, and
IF include anti-TYRO3 antibody (Cell Signaling Technology, 5585, RRID: AB_10706782, 1:200 for IF and 1:2000 for WB; OriGene, AP14424PU-N, RRID:AB_1771180, 1:200 for IHC; Abnova, H00007301-M05, RRID:AB_581805, 1:200 for IHC; Sigma-Aldrich, SAB1411375, 1:200 for IHC), Myc-Tag (Cell Signaling Technology, 2278S, RRID:AB_490778, 1:500 for IF), bromodeoxyuridine (BrdU; Millipore, MAB3510P, RRID:AB_94898, 1:200 for IF; BD Biosciences, BD347580, RRID:AB_10015219, 1:200 for IF; Abcam, Ab6326, 1:200 for IF), annexin V (BD Biosciences, BD 556419, RRID:AB_2665412, 1:50 for flow cytometry), GFP (GeneTex, GTX113617, RRID:AB_1950371, 1:500 for IF and1:5000 for WB), caspase-3 (GeneTex, GTX110543, RRID:AB_10722709, 1:5000 for WB), MMP-2 (GeneTex, GTX104577, RRID:AB_1950932, 1:5000 for WB; GeneTex, GTX30147, RRID:AB_369568, 1:200 for IF), β-actin (GeneTex, GTX109639, RRID:AB_1949572, 1:10,000 for WB), lamin A/C (GeneTex, GTX101127, RRID:AB_10729290, 1:10,000 for WB), CD31 (Bioss, BS-0468R, RRID:AB_10885355, 1:200 for IF; BD Biosciences, 550274, RRID:AB_393571, 1:100 for IF), CD4 (Abclonal, A0362, RRID:AB_2757146, 1:200 for IF), SNAI1 (Novus, NBP2-27293, catalog no. NBP2-27293, 1:1000 for WB), SLUG (GeneTex, GTX128796, RRID:AB_2885815, 1:2000 for WB), ZEB1 (GeneTex, GTX105278, RRID:AB_11162905, 1:2000 for WB), E-cadherin (Proteintech, 20874-1-AP, RRID:AB_10697811, 1:2000 for WB), BRD3 (Proteintech, 11859-1-AP, RRID:AB_206, 1:2000 for WB; Novus, H00008019-M01, RRID:AB_537205, 1:200 for IHC), BRD2 (Cell Signaling Technology, 5848, RRID:AB_10835146, 1:2000 for WB), BRD4 (Cell Signaling Technology, 13440, RRID:AB_2687578, 1:2000 for WB), and histone H3ac (Active Motif, 39139, RRID:AB_2687871, 1:2000 for WB). The human TYRO3 expression vector contains full-length cDNA (BC051756; GenDiscovery Biotenology Inc., Taipei, Taiwan, ROC) was purchased from OriGene and further ligated to pcDNA5/To vector (Invitrogen). The rhBRD3 was purchased from Abnova (H00008019-P01). The rhTYRO3 was purchased from Origene (TP308260).
Histology
Tissues were fixed with 4% paraformaldehyde, and 6-μm paraffin sections of tissue were subjected to hematoxylin and eosin staining following standard procedures. For immunohistochemical staining, after antigen retrieval, tissue sections were incubated with anti–C-terminal of TYRO3 antibodies (Origene) and then secondary antibodies and then visualized with aminoethyl carbazole. For immunofluorescent staining, sections of the cecum, lymph node, and liver tissue were incubated with the antibodies as indicated, including anti-TYRO3 (Cell Signaling Technology), anti-GFP (GeneTex), anti-CD31 (BD Biosciences, Bioss), anti-CD4 (Abclonal), and anti–MMP-2 (Proteintech).
Determination of TYRO3 subcellular distribution
HCT116 cells or transiently transfected HCT116 cells were cultured on μ-slides VI0.4 (ibidi) and fixed in 4% paraformaldehyde. After washing with PBS containing 0.3% Triton X-100, cells were blocked with 5% goat serum before the cells were stained with anti-TYRO3 (Cell Signaling Technology, Origene, Abnova, Sigma), anti–Myc-Tag (Cell Signaling Technology), F-actin (Cayman), and anti-GFP (GeneTex) and counterstained with 4′,6-diamidino-2-phenylindole (DAPI) or Hoechst. Fluorescence was captured using a laser confocal microscope (Olympus FV1000).
Site-directed mutagenesis
The human TYRO3 expression vector containing full-length cDNA (BC051756; GenDiscovery Biotenology Inc., Taipei, Taiwan, ROC) was ligated to pcDNA5/To vector (Invitrogen). For single-site mutation or deletion, the PCR reaction of 20 μl contained 200 ng of template, 0.4 μM primer pair (table S1), 200 μM deoxynucleotide triphosphates, and 3.75 U of Pfu DNA polymerase (Agilent). The PCR reactions were taken for transformation.
BrdU incorporation assay
After transfecting for 24 hours, HCT116 cells were serum-starved for 24 hours. Cells were grown for 18 hours in fresh media before labeling with 10 μM BrdU for 40 min, fixed with 4% paraformaldehyde for 10 min, and washed with 0.3% Triton X-100 in PBS. DNA was denatured with 2 N HCl for 30 min. Cells were incubated overnight with anti-BrdU antibody (Millipore) at 4°C and counterstained with DAPI for nuclei. ImageJ was used to analyze the results.
Western blotting
Western blotting was done following the standard protocols. Polyvinylidene difluoride membranes were labeled with anti–caspase-3 (Cell Signaling), anti-TYRO3 (Cell Signaling), anti-GFP (GeneTex), anti–MMP-2 (GeneTex), anti-lamin A/C (GeneTex), anti-BRD3 (Proteintech, Abnova), anti-BRD2 (Cell Signaling), anti-SNAI1 (Novus), anti-SLUG (GeneTex), anti-ZEB1 (GeneTex), anti–E-cadherin (Proteintech), or anti–β-actin (GeneTex) antibodies. Densitometry was done using ImageJ program from at least three independent experiments.
Flow cytometry
Treated cells were harvested and incubated with fluorescein isothiocyanate (FITC)–conjugated annexin V antibody (BD Biosciences) and propidium iodide (PI; Sigma-Aldrich) at room temperature for 20 min before fluorescence was measured by flow cytometry (FACS Can). FlowJo software was used to analyze the results.
MMP-2 in vitro digestion
TYRO3 was overexpressed in HCT116 cells, harvested by hypotonic buffer, and dispersed in different concentration of APMA (1 mM, Sigma-Aldrich, A9563)–activated human MMP-2 (0, 0.0016, 0.08, or 0.4 nM) in MMP digestion buffer [50 mM tris HCl, 150 mM NaCl, 5 mM CaCl2, 1 mM ZnCl2, and 0.01% Brij-35 (pH 7.5)]. MMP-2 inhibitor, ARP100 (0, 1, 10, 100, 1000 nM; Caymen Chemical), was mixed in the assay buffer with 0.4 nM MMP-2 and 5 μg of TYRO3. After 5 hours of incubation at 37°C, the mixture was used for Western blotting.
DNA fork assay
Cells were incubated with 10 μM 5-chloro-2′-deoxyuridine (CldU; Cayman Chemical, 18155) for 20 min followed by 100 μM idoxuridine (IdU; Cayman Chemical, 20222) for 20 min. Next, cells were harvested, and DNA fibers were spread on microslides to analysis of CldU (Abcam) and IdU tracts (BD Biosciences). Fluorescence was captured by using a laser confocal microscope (Olympus FV1000). The length of DNA fibers was measured by ImageJ.
Migration and invasion assay
For tumor cell transendothelial migration assay, HUVEC cells (5 × 104 cells per well) were added to the transwell cell culture chambers with polycarbonate membranes of 8.0-mm diameter pores (Millipore). Transfected HCT116 and SW480 cells were plated on their upper side for 24 hours and fixed in 4% paraformaldehyde. HUVEC were stained with anti-CD31 (Bioss) antibody. Transwell filters were examined by using a laser confocal microscope (Olympus FV1000). For migration and invasion assay, transfected HCT116 cells (1 × 105 cells per well) were seeded on the transwell cell culture chambers with polycarbonate membranes of 8.0-mm diameter pores (Millipore) for 24 hours, fixed in 4% paraformaldehyde for 10 min, and stained using crystal violet solution for 30 min. Stained cells were washed with water gently. Images were captured and quantified by ImageJ.
Colony formation assay
After transfection, CCD-841CoN cells (1 × 105 cell per well) were seeded on six-well plates for 3 weeks. The cells were fixed with 4% paraformaldehyde for 10 min and stained using crystal violet solution for 30 min. Stained cells were washed with water gently. Images were captured and the number of colonies (<50 μM) was counted.
Proteomic analysis
After transfection, the co-IP of nuclear protein complexes was performed using the nuclear complex co-IP (Active Motif) according to the manufacturer’s instructions. The co-IP samples were subjected to mass spectrometric analyses. Data acquisitions in positive mode was executed with ESI Compass for otofSeries 1.6 for Impact HD (TOFControl Version 3.3) (Bruker Daltonik). Analyses of the data were performed using the Compass DataAnalysis (Version 4.1 SR1) (Bruker Daltonik) and generated peak listed in mgf format. The protein identifications were executed with tandem mass spectrometry peak lists using Mascot software version 2.5 (Matrix Science, London, UK). The peptide sequences were searched against Homo sapiens (human) taxonomy (326053 sequences were obtained from NCBI on 15 Jan 2019) in UniProt protein database. Mass tolerances for peptide and product ions were set to 50 ppm and 0.5 Da; the instrument setting was specified as “ESI-QUAD-TOF.” Semi-trypsin was designated as the protease with two missing cleavages was allowed. Carboxymethylation on cysteine was set as the fixed modification, while oxidation of methionine was searched as variable modifications. Proteins were accepted if they had at least one rank 1 peptide with a Mascot ion score of more than 40.0 (P < 0.05).
Proximity ligation assay
The PLA was performed using the Duolink In Situ Red Starter Kit Mouse/Rabbit (Sigma-Aldrich, DUO92101) according to the manufacturer’s instructions. Briefly, nontransfected or transfected cells were followed by fixation, incubation with rabbit anti-TYRO3 and mouse anti-BRD3, incubation with the PLA probes, and ligation and amplification according to the manufacturer’s instructions. Imaging was performed with Olympus FV1000.
Protein kinase assay
The protein kinase assay was performed using the ADPsensor Universal Kinase Activity Assay Kit (BioVision, K212-100) according to the manufacturer’s instructions. Briefly, recombinant TYRO3 and BRD3 were diluted in kinase assay buffer and incubated in the reaction mix for 90 min. Signals were measured at excitation/emission = 535/587 nm. Samples were collected for phosphate-affinity SDS-PAGE. In a separated experiment, HCT-116 cells stably express TYRO3-GFP (TYRO3), ICD-TYRO3-GFP (ICD-TYRO3), or mMMP-2–TYRO3–GFP (mMMP-2–TYRO3) were lysed and immunoprecipitated with anti-GFP antibody from cytoplasmic fraction (Cyto) or nuclear fraction (Nu). The immunoprecipitants were incubated with BRD3 recombinant proteins as described above.
Chromatin immunoprecipitation sequencing
After treatment, ChIP using BRD3, GFP, or acetylation of H3 antibody was performed using the SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer’s instructions. Samples were collected for ChIP-seq or qPCR. All primer sequences used in individual reactions are shown in table S1. For ChIP-seq, DNA libraries from ICD-TYRO3– and KD-TYRO3–overexpressed cells were prepared for sequencing using the KAPA Hyper Prep Kit (KR0961-v6.17, Kapa Biosystems). Bcl2fastq version 2.19.1 (no lane splitting) software was used for base calling. ChIP-seq reads were aligned to the hg19 genome assembly using BWA-MEM software (version 0.7.17). Peaks were called using CLC Workbanch 8.5.1 with the ChIP-Seq Analysis (legacy) setting.
BRD3/BRD4 CRISPR-Cas9 knockout cell
To generate CRISPR-Cas9–mediated BRD3 and BRD4 knockout cells, the predesigned and validated guide RNA (gRNA) sequences targeting BRD3 and BRD4 (46) were synthesized and cloned into eSpCas9-2A-Neo vector that was customizedly designed and modified from eSpCas9-2A-Puro (PX459) V2.0 by GenScript (www.genscript.com/). All cloned vectors were verified by sanger sequencing. The ICD-TYRO3-GFP cells were transfected with gRNA-containing eSpCas9-2A-Neo plasmids using Lipofectamine 2000 and selected with G418 at the final concentration of 1.5 mg/ml. All oligonucleotides used to generate gRNA-containing constructs are shown in table S1.
Statistical analysis
All cell-related results were performed at least three times. Statistical analyses were performed by GraphPad Prism 6.0. Statistical significance was analyzed by Student’s t test, one-way analysis of variance (ANOVA), or two-way ANOVA followed by Tukey’s multiple comparisons test, respectively. The results were exhibited as mean ± SEM. Statistical significance was defined as *P < 0.05, **P < 0.01, or ***P < 0.001. The correlation of IHC with clinical prognostic factors were calculated and analyzed by χ2 test. For survival data, Kaplan-Meier curves were plotted and compared using a log-rank test.
Supplementary Material
Acknowledgments
We thank the technical supports provided by the “Bioimaging Core Facility of the National Core Facility for Biopharmaceuticals, Ministry of Science and Technology, Taiwan,” the “Center for Genomic Medicine, College of Medicine, National Cheng Kung University,” “Center for Bioinformatics and Digital Health,” and “Center for Research Resources and Development of Kaohsiung Medical University in protein identification and confocal image analysis.” We are also grateful to Y.-H. Yeh, Y.-T. Tang, Y.-Y. Lai, and M.-F. Huang for the technical support in clinical specimen handling, animal husbandry and surgery, immunohistochemical staining, and bioinformatic analysis.
Funding: This work was supported by grants from the Ministry of Science and Technology of Taiwan (MOST 109-2811-B-006-536, MOST 110-2811-B-006-541, MOST 109-2326-B-037-002-MY3, and MOST 108-2321-B-006-006) and from the National Health Research Institute, Taiwan (NHRI-EX110-11016BI).
Author contributions: Conceptualization: P.-L.H., C.-W.C., S.-Y.C., and S.-J.T. Methodology: P.-L.H., C.-W.C., Y.-A.T., B.-W.L., S.-C.L., Y.-S.L., S.-Y.C., H.S.S., and S.-J.T. Investigation: P.-L.H., C.-W.C., Y.-A.T., B.-W.L., S.-C.L., Y.-S.L., H.S.S., and S.-J.T. Visualization: P.-L.H., C.-W.C., S.-Y.C., and H.S.S. Supervision: S.-J.T. and H.S.S. Writing—original draft: P.-L.H. Writing—revision and editing: P.-L.H. and S.-J.T. All authors have read and commented on the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Mass spectrometry raw data and search results files have been deposited to the ProteomeXchange Consortium via the PRIDE (47) partner repository with the dataset identifier PXD029743 and 10.6019/PXD029743. The ChIP-seq data have been deposited in the NCBI GEO under accession number 188378.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Table S1
Erratum published 10 November 2023:
In the original files, figure S1F was a duplicate of the image used for Figure 1G. Figure S1F has been updated to include the correct image. The authors’ results and conclusions are not affected by this correction. The supplementary materials PDF has been updated. The original version is available here:
REFERENCES AND NOTES
- 1.Erreni M., Mantovani A., Allavena P., Tumor-associated macrophages (TAM) and inflammation in colorectal cancer. Cancer Microenviron. 4, 141–154 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huth E. J., Case K., Annals of internal medicine at age 75: Reflections on the past 25 years. Ann. Intern. Med. 137, 34–45 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Fearon E. R., Vogelstein B., A genetic model for colorectal tumorigenesis. Cell 61, 759–767 (1990). [DOI] [PubMed] [Google Scholar]
- 4.Lin S. C., Chien C. W., Lee J. C., Yeh Y. C., Hsu K. F., Lai Y. Y., Lin S. C., Tsai S. J., Suppression of dual-specificity phosphatase-2 by hypoxia increases chemoresistance and malignancy in human cancer cells. J. Clin. Invest. 121, 1905–1916 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang X., Li C. F., Zhang L., Wu C. Y., Han L., Jin G., Rezaeian A. H., Han F., Liu C., Xu C., Xu X., Huang C. Y., Tsai F. J., Tsai C. H., Watabe K., Lin H. K., TRAF6 restricts p53 mitochondrial translocation, apoptosis, and tumor suppression. Mol. Cell 64, 803–814 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schlessinger J., Cell signaling by receptor tyrosine kinases. Cell 103, 211–225 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Ghosh S., Marrocco I., Yarden Y., Roles for receptor tyrosine kinases in tumor progression and implications for cancer treatment. Adv. Cancer Res. 147, 1–57 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Du Z., Lovly C. M., Mechanisms of receptor tyrosine kinase activation in cancer. Mol. Cancer 17, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Argiles G., Saunders M. P., Rivera F., Sobrero A., Benson A., Ponce C. G., Cascinu S., Van Cutsem E., Macpherson I. R., Strumberg D., Kohne C. H., Zalcberg J., Wagner A., Garosi V. L., Grunert J., Tabernero J., Ciardiello F., Regorafenib plus modified FOLFOX6 as first-line treatment of metastatic colorectal cancer: A phase II trial. Eur. J. Cancer 51, 942–949 (2015). [DOI] [PubMed] [Google Scholar]
- 10.Kindler H. L., Ioka T., Richel D. J., Bennouna J., Letourneau R., Okusaka T., Funakoshi A., Furuse J., Park Y. S., Ohkawa S., Springett G. M., Wasan H. S., Trask P. C., Bycott P., Ricart A. D., Kim S., Van Cutsem E., Axitinib plus gemcitabine versus placebo plus gemcitabine in patients with advanced pancreatic adenocarcinoma: A double-blind randomised phase 3 study. Lancet Oncol. 12, 256–262 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Tomas A., Futter C. E., Eden E. R., EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 24, 26–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carpenter G., Liao H. J., Trafficking of receptor tyrosine kinases to the nucleus. Exp. Cell Res. 315, 1556–1566 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuo H. Y., Huang Y. S., Tseng C. H., Chen Y. C., Chang Y. W., Shih H. M., Wu C. W., PML represses lung cancer metastasis by suppressing the nuclear EGFR-mediated transcriptional activation of MMP2. Cell Cycle 13, 3132–3142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liccardi G., Hartley J. A., Hochhauser D., EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 71, 1103–1114 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin S. Y., Makino K., Xia W., Matin A., Wen Y., Kwong K. Y., Bourguignon L., Hung M. C., Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Ni C. Y., Murphy M. P., Golde T. E., Carpenter G., γ-secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Migdall-Wilson J., Bates C., Schlegel J., Brandao L., Linger R. M., DeRyckere D., Graham D. K., Prolonged exposure to a Mer ligand in leukemia: Gas6 favors expression of a partial Mer glycoform and reveals a novel role for Mer in the nucleus. PLOS ONE 7, e31635 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C., Iida M., Dunn E. F., Ghia A. J., Wheeler D. L., Nuclear EGFR contributes to acquired resistance to cetuximab. Oncogene 28, 3801–3813 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vouri M., Hafizi S., TAM receptor tyrosine kinases in cancer drug resistance. Cancer Res. 77, 2775–2778 (2017). [DOI] [PubMed] [Google Scholar]
- 20.Hsu P. L., Jou J., Tsai S. J., TYRO3: A potential therapeutic target in cancer. Exp. Biol. Med. 244, 83–99 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song X., Wang H., Logsdon C. D., Rashid A., Fleming J. B., Abbruzzese J. L., Gomez H. F., Evans D. B., Wang H., Overexpression of receptor tyrosine kinase Axl promotes tumor cell invasion and survival in pancreatic ductal adenocarcinoma. Cancer 117, 734–743 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chien C. W., Hou P. C., Wu H. C., Chang Y. L., Lin S. C., Lin S. C., Lin B. W., Lee J. C., Chang Y. J., Sun H. S., Tsai S. J., Targeting TYRO3 inhibits epithelial-mesenchymal transition and increases drug sensitivity in colon cancer. Oncogene 35, 5872–5881 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Spring K. J., Zhao Z. Z., Karamatic R., Walsh M. D., Whitehall V. L., Pike T., Simms L. A., Young J., James M., Montgomery G. W., Appleyard M., Hewett D., Togashi K., Jass J. R., Leggett B. A., High prevalence of sessile serrated adenomas with BRAF mutations: A prospective study of patients undergoing colonoscopy. Gastroenterology 131, 1400–1407 (2006). [DOI] [PubMed] [Google Scholar]
- 24.Selby J. V., Friedman G. D., Quesenberry C. P. Jr., Weiss N. S., A case-control study of screening sigmoidoscopy and mortality from colorectal cancer. N. Engl. J. Med. 326, 653–657 (1992). [DOI] [PubMed] [Google Scholar]
- 25.Smith J. J., Deane N. G., Wu F., Merchant N. B., Zhang B., Jiang A., Lu P., Johnson J. C., Schmidt C., Bailey C. E., Eschrich S., Kis C., Levy S., Washington M. K., Heslin M. J., Coffey R. J., Yeatman T. J., Shyr Y., Beauchamp R. D., Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 138, 958–968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wheeler D. L., Huang S., Kruser T. J., Nechrebecki M. M., Armstrong E. A., Benavente S., Gondi V., Hsu K. T., Harari P. M., Mechanisms of acquired resistance to cetuximab: Role of HER (ErbB) family members. Oncogene 27, 3944–3956 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chioni A. M., Grose R., FGFR1 cleavage and nuclear translocation regulates breast cancer cell behavior. J. Cell Biol. 197, 801–817 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen M. K., Hung M. C., Proteolytic cleavage, trafficking, and functions of nuclear receptor tyrosine kinases. FEBS J. 282, 3693–3721 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carpenter G., Liao H. J., Receptor tyrosine kinases in the nucleus. Cold Spring Harb. Perspect. Biol. 5, a008979 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Y. N., Hung M. C., Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell Biosci. 2, 13 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ali M. A., Chow A. K., Kandasamy A. D., Fan X., West L. J., Crawford B. D., Simmen T., Schulz R., Mechanisms of cytosolic targeting of matrix metalloproteinase-2. J. Cell. Physiol. 227, 3397–3404 (2012). [DOI] [PubMed] [Google Scholar]
- 32.Schulz R., Intracellular targets of matrix metalloproteinase-2 in cardiac disease: Rationale and therapeutic approaches. Annu. Rev. Pharmacol. Toxicol. 47, 211–242 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J., Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 (2007). [DOI] [PubMed] [Google Scholar]
- 34.Demarest S. J., Gardner J., Vendel M. C., Ailor E., Szak S., Huang F., Doern A., Tan X., Yang W., Grueneberg D. A., Richards E. J., Endege W. O., Harlow E., Koopman L. A., Evaluation of Tyro3 expression, Gas6-mediated Akt phosphorylation, and the impact of anti-Tyro3 antibodies in melanoma cell lines. Biochemistry 52, 3102–3118 (2013). [DOI] [PubMed] [Google Scholar]
- 35.Avilla E., Guarino V., Visciano C., Liotti F., Svelto M., Krishnamoorthy G., Franco R., Melillo R. M., Activation of TYRO3/AXL tyrosine kinase receptors in thyroid cancer. Cancer Res. 71, 1792–1804 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Kabir T. D., Ganda C., Brown R. M., Beveridge D. J., Richardson K. L., Chaturvedi V., Candy P., Epis M., Wintle L., Kalinowski F., Kopp C., Stuart L. M., Yeoh G. C., George J., Leedman P. J., A microRNA-7/growth arrest specific 6/TYRO3 axis regulates the growth and invasiveness of sorafenib-resistant cells in human hepatocellular carcinoma. Hepatology 67, 216–231 (2018). [DOI] [PubMed] [Google Scholar]
- 37.Qin A., Qian W., MicroRNA-7 inhibits colorectal cancer cell proliferation, migration and invasion via TYRO3 and phosphoinositide 3-kinase/protein B kinase/mammalian target of rapamycin pathway suppression. Int. J. Mol. Med. 42, 2503–2514 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee C., Overexpression of Tyro3 receptor tyrosine kinase leads to the acquisition of taxol resistance in ovarian cancer cells. Mol. Med. Rep. 12, 1485–1492 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Singh H., Brave M., Beaver J. A., Cheng J., Tang S., Zahalka E., Palmby T. R., Venugopal R., Song P., Liu Q., Liu C., Yu J., Chen X. H., Wang X., Wang Y., Kluetz P. G., Daniels S. R., Papadopoulos E. J., Sridhara R., McKee A. E., Ibrahim A., Kim G., Pazdur R., U.S. Food and Drug Administration approval: Cabozantinib for the treatment of advanced renal cell carcinoma. Clin. Cancer Res. 23, 330–335 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Grayson A. R., Walsh E. M., Cameron M. J., Godec J., Ashworth T., Ambrose J. M., Aserlind A. B., Wang H., Evan G., Kluk M. J., Bradner J. E., Aster J. C., French C. A., MYC, a downstream target of BRD-NUT, is necessary and sufficient for the blockade of differentiation in NUT midline carcinoma. Oncogene 33, 1736–1742 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stratikopoulos E. E., Dendy M., Szabolcs M., Khaykin A. J., Lefebvre C., Zhou M. M., Parsons R., Kinase and BET inhibitors together clamp inhibition of PI3K signaling and overcome resistance to therapy. Cancer Cell 27, 837–851 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang W., Tang Y. A., Xiao Q., Lee W. C., Cheng B., Niu Z., Oguz G., Feng M., Lee P. L., Li B., Yang Z. H., Chen Y. F., Lan P., Wu X. J., Yu Q., Stromal induction of BRD4 phosphorylation results in chromatin remodeling and BET inhibitor resistance in colorectal cancer. Nat. Commun. 12, 4441 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hajmirza A., Emadali A., Gauthier A., Casasnovas O., Gressin R., Callanan M. B., BET family protein BRD4: An emerging actor in NFκB signaling in inflammation and cancer. Biomedicine 6, 16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Qin A. C., Jin H., Song Y., Gao Y., Chen Y. F., Zhou L. N., Wang S. S., Lu X. S., The therapeutic effect of the BRD4-degrading PROTAC A1874 in human colon cancer cells. Cell Death Dis. 11, 805 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang Y., Tian S., Xiong J., Zhou Y., Song H., Liu C., JQ-1 inhibits colon cancer proliferation via suppressing Wnt/β-catenin signaling and miR-21. Chem. Res. Toxicol. 31, 302–307 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Perez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., Tiwary S., Cox J., Audain E., Walzer M., Jarnuczak A. F., Ternent T., Brazma A., Vizcaino J. A., The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 47, D442–D450 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Table S1
Erratum published 10 November 2023:
In the original files, figure S1F was a duplicate of the image used for Figure 1G. Figure S1F has been updated to include the correct image. The authors’ results and conclusions are not affected by this correction. The supplementary materials PDF has been updated. The original version is available here: