Abstract
Plant chitinases (EC 3.2.1.14) are pathogenesis-related (PR) proteins and are well studied in many plant species. However, little is known about the genomic organization and expression of chitinase genes in strawberries (Fragaria vesca). Here, 23 FvChi genes were identified in the genome of strawberry (F. vesca) and divided into GH18 and GH19 subfamilies based on phylogenetic relationships. A detailed bioinformatics analysis of the FvChi genes was performed, including gene physicochemical properties, chromosomal location, exon–intron distribution, domain arrangement, and subcellular localization. Twenty-two FvChi genes showed upregulation after Colletotrichum gloeosporioides infection. Following the exogenous application of SA, FvChi-3, 4, and 5 showed significant changes in expression. The ectopic expression of FvChi-14 in Arabidopsis thaliana increased resistance to C. higginsianum via controlling the SA and JA signaling pathway genes (AtPR1, AtICS1, AtPDF1.2, and AtLOX3). The FvChi-14 protein location was predicted in the cell wall or extracellular matrix. We speculate that FvChi-14 is involved in disease resistance by regulating the SA and JA signaling pathways. The findings of this study provide a theoretical reference for the functional studies of FvChi genes and new candidates for strawberry stress resistance breeding programs.
Keywords: biotic stress, pathogen, genome, evolutionary history, PR3
1. Introduction
Chitinases (EC.3.2.1.14) are glycosyl hydrolases widely found in various organisms in nature, and are thought to be one group of pathogenesis-related (PR) proteins [1]. Normally, these proteins are expressed at basal levels, but pathogen attacks or abiotic stress can significantly increase their expression, leading to systemic acquired resistance (SAR) in plants [2]. Chitinase mainly hydrolyzes β-1,4 glycosidic bonds in chitin polymers to generate N-acetyl-D-glucosamine (GlcNAc) oligomers, which interfere with or degrade chitin [3]. Chitin is a major structural component of fungal cell walls, and thus chitinase plays critical roles in plant defense against pathogens [4]. Viral/fungal infections induce plant chitinase activity, through which plants receive signals about the fungal attack. Apoplastic chitinase induces the release of fungal inducers, and vacuolar chitinase then acts to defend against pathogens [5]. Apoplastic chitinase produces disease-related signals that induce plant immunity, and vacuolar chitinase degrades newly synthesized chitin chains and inhibits fungal growth [6]. Plant chitinases can be divided into two subfamilies: 18 (GH18) and 19 (GH19) of glycosyl hydrolases, which can be further divided into five distinct classes (class I–V). Classes I, II, and IV are members of family GH19, while classes III and V belong to family GH18 [7]. Chitinases of family GH18 are ubiquitous among organisms, whereas family GH19 chitinases are restricted to plants and Streptomyces [8]. Members of the GH19 subfamily have an N-terminal chitin-binding domain (CBD) structure that can combine with chitin to enhance plant defense against pests and stress resistance [3].
The antifungal effects of chitinase in plants have been demonstrated in many studies. Chitinase genes have critical roles in multiple processes, including plant development and defense against stress. For example, after inoculating MnChi18 transgenic Arabidopsis with Botrytis cinerea, the activity of catalase (CAT) and the amount of malondialdehyde (MDA) decreased, showing that overexpression of MnChi18 could boost the resistance to B. cinerea [9]. The transient expression of CaChiIII7 significantly induced the expression of many defense-related genes and the high expression in pepper leaves along with H2O2 biosynthesis, and improved the basic resistance of pepper to C. acutatum [10]. When the resistant variety of cucumber ‘JIN5-508′ was inoculated with Podosphaera xanthii, the expression level of the chitinase genes increased significantly, which triggered a papillary reaction to stop the invasion of powdery mildew [11]. Studies on the molecular control of defense responses in pine trees have revealed that after being attacked by potential signaling molecules found in angiosperms and necrotizing diseases, plants produce several chitinase homologs [12]. Chitinase genes Chi23, Chi32, and Chi47 were silenced, and this significantly decreased cotton’s resistance to Verticillium dahliae, indicating their function in V. dahlia resistance [13]. Moreover, some studies have reported the role of chitinase genes in abiotic stress tolerance. The expression of the class III AtChiA gene was upregulated following salt and wound stresses [14]. ScChiI1 and ScChiIV1 showed upregulation under PEG, NaCl, and CuCl2 stress [15]. Strawberry (Fragaria ananassa Duch.) is a high-value fruit popular among the public for its delicious taste and rich nutrition [16]. However, strawberry fruit is vulnerable to mechanical damage and infection by plant pathogens, including viruses, bacteria, and fungi, during harvesting and storage due to the lack of shell protection [17]. Anthracnose is a serious fungal disease caused by the necrotizing fungus Colletotrichum spp., which mostly infects strawberry roots and stolons during development [18]. The main anthracnose species reported to infect strawberries are C. gloeosporioides and C. fragariae in China [19]. One of the most prevalent types of strawberry PR genes with hydrolytic activity has been found in the chitinase family [20]. There is little known about the roles of chitinase genes in antifungal disease resistance in strawberries [17,21,22]. Detailed information on the disease resistance mechanisms and pathways of chitinase genes in strawberries is lacking.
The chitinase gene family has been studied in a variety of plants, such as cotton (Gossypium) [13], cucumber (Cucumis sativus) [23], tomato (Solanum lycopersicum) [24], tea (Camellia sinensis) [5], cabbage (Brassica oleracea) [25], mulberry (Morus notabilis) [9], domesticated apple (Malus domestica), and wild apple (Malus sieversii) [26]. However, less information is available about the roles of chitinase genes in strawberry. The important roles of chitinase genes in different crops and lack of detailed information in strawberry justifies the need of identification, expression profiling, and functional characterization of chitinase genes in this crop. We executed detailed bioinformatics analysis of chitinase genes in F. vesca, including determining gene numbers, gene structures, motif distribution patterns, and evolutionary history analysis. Further, we performed expression profiling of chitinase genes in different plant parts and following C. gloeosporioides infection and hormone treatments. Based on the results of the initial study, FvChi-14 was isolated from strawberry and overexpressed in Arabidopsis for functional studies. Overexpression of FvChi-14 significantly increased the resistance of transgenic Arabidopsis to anthracnose. Our study provides new candidates for disease resistance breeding of strawberry and insights about the functions of FvChi.
2. Results
2.1. Identification and Physiochemical Properties of FvChi Proteins
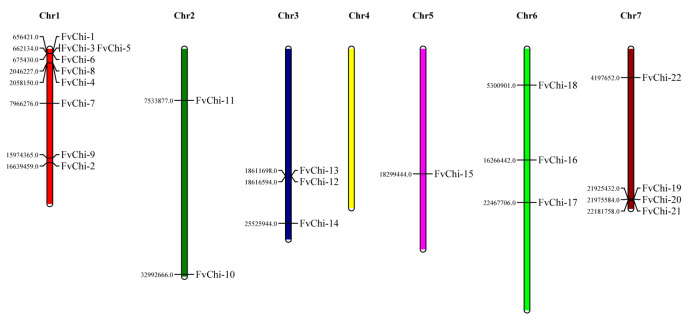
Twenty-three annotated FvChi members with chitin hydrolysis or chitinase-like functions were found in the F. vesca genomes (Table 1). The physicochemical properties and basic information are mentioned in Table 1. Sequence analysis showed that the amino acids of FvChi members ranged in length from 227 AA (FvChi-23) to 777 AA (FvChi-3), with protein molecular weights of 24.662 (FvChi-23) to 87.535 (FvChi-3) kDa and isoelectric points (pI) of 4.59 (FvChi-23) to 8.95 (FvChi-6). FvChi-1 to FvChi-22 were unevenly distributed on six chromosomes, and there was no member on chromosome four. Interestingly, FvChi-23 was present on an unknown chromosome (Figure 1). The highest and the lowest number of genes were present on chromosomes 1 (9 genes) and 5 (1 gene), respectively. Most of the FvChi members were predicted to be localized in the cell wall (39.1%) and vacuole (39.1%).
Table 1.
The details of chitinase genes identified in F. vesca.
| Name | Accession No. | Locus Name | Chr. | Location | CDS/bp | ORF/aa | Mw/kDa | pI | Predicted Location |
|---|---|---|---|---|---|---|---|---|---|
| FvChi-1 | XM_004288652.2 | XP_004288700.2 | 1 | 656,421–657,812 | 1248 | 415 | 46.867 | 8.91 | cell wall |
| FvChi-2 | XM_004288324.2 | XP_004288372.1 | 1 | 16,639,459–16,642,667 | 825 | 274 | 30.213 | 8.72 | extracell |
| FvChi-3 | XM_004288653.2 | XP_004288701.2 | 1 | 662,134–665,736 | 2334 | 777 | 87.535 | 7.59 | chloroplast |
| FvChi-4 | XM_011470234.1 | XP_011468536.1 | 1 | 2,058,150–2,059,443 | 1119 | 372 | 41.070 | 5.91 | cell wall |
| FvChi-5 | XM_011461675.1 | XP_011459977.1 | 1 | 662,134–665,736 | 2325 | 774 | 87.266 | 7.59 | cell membrane |
| FvChi-6 | XM_004287056.2 | XP_004287104.1 | 1 | 675,430–677,228 | 1134 | 377 | 41.072 | 8.95 | cell wall |
| FvChi-7 | XM_004287696.2 | XP_004287744.1 | 1 | 7,966,276–7,968,174 | 981 | 326 | 34.390 | 5.18 | vacuole |
| FvChi-8 | XM_011470322.1 | XP_011468624.1 | 1 | 2,046,227–2,050,302 | 2322 | 773 | 86.727 | 6.4 | cell membrane |
| FvChi-9 | XM_004289370.2 | XP_004289418.1 | 1 | 15,974,364–15,976,008 | 822 | 273 | 30.613 | 4.88 | cell wall |
| FvChi-10 | XM_004291897.2 | XP_004291945.1 | 2 | 32,992,664–32,994,601 | 1134 | 377 | 41.423 | 8.9 | cell wall |
| FvChi-11 | XM_004289994.2 | XP_004290042.1 | 2 | 7,533,877–7,535,599 | 1044 | 347 | 36.715 | 8.23 | vacuole |
| FvChi-12 | XM_004294591.2 | XP_004294639.1 | 3 | 18,616,594–18,618,119 | 954 | 317 | 35.684 | 7.17 | cell wall |
| FvChi-13 | XM_004295915.2 | XP_004295963.2 | 3 | 8,611,698–18,612,883 | 912 | 303 | 33.957 | 7.09 | cell wall |
| FvChi-14 | XM_004295058.2 | XP_004295106.1 | 3 | 25,525,945–25,527,885 | 972 | 323 | 35.775 | 6.2 | extracell |
| FvChi-15 | XM_004299998.2 | XP_004300046.2 | 5 | 18,299,444–18,301,125 | 918 | 305 | 33.816 | 8.56 | vacuole |
| FvChi-16 | XM_011468684.1 | XP_011466986.1 | 6 | 16,266,442–16,268,685 | 912 | 303 | 33.529 | 4.7 | cell wall |
| FvChi-17 | XM_011468993.1 | XP_011467295.1 | 6 | 22,467,706–22,470,336 | 888 | 295 | 31.370 | 4.64 | vacuole |
| FvChi-18 | XM_004304955.2 | XP_004305003.1 | 6 | 5,300,901–5,302,053 | 912 | 303 | 33.551 | 5.05 | cell wall |
| FvChi-19 | XM_004307979.2 | XP_004308027.1 | 7 | 21,925,431–21,926,910 | 927 | 308 | 33.000 | 8.62 | vacuole |
| FvChi-20 | XM_011471995.1 | XP_011470297.1 | 7 | 21,975,584–21,977,021 | 900 | 299 | 31.611 | 7.51 | vacuole |
| FvChi-21 | XM_004308005.2 | XP_004308053.1 | 7 | 22,181,758–22,183,063 | 900 | 299 | 31.562 | 5.28 | vacuole |
| FvChi-22 | XM_004306663.2 | XP_004306711.2 | 7 | 4,197,652–4,198,786 | 906 | 301 | 32.382 | 8.08 | vacuole |
| FvChi-23 | XM_004309786.2 | XP_004309834.1 | / | 355,466–356,526 | 684 | 227 | 24.662 | 4.59 | extracell |
Abbreviations: Chr.: chromosome; CDS: coding sequence; ORF: open reading frame; pI: isoelectric point.
Figure 1.
Chromosomal distribution of the FvChi genes. The numbers indicate where the genes start on the chromosomes. Based on their chromosomal order, the FvChi genes were given the names FvChi-1 to FvChi-23.
2.2. Phylogenetic Studies of FvChi Genes
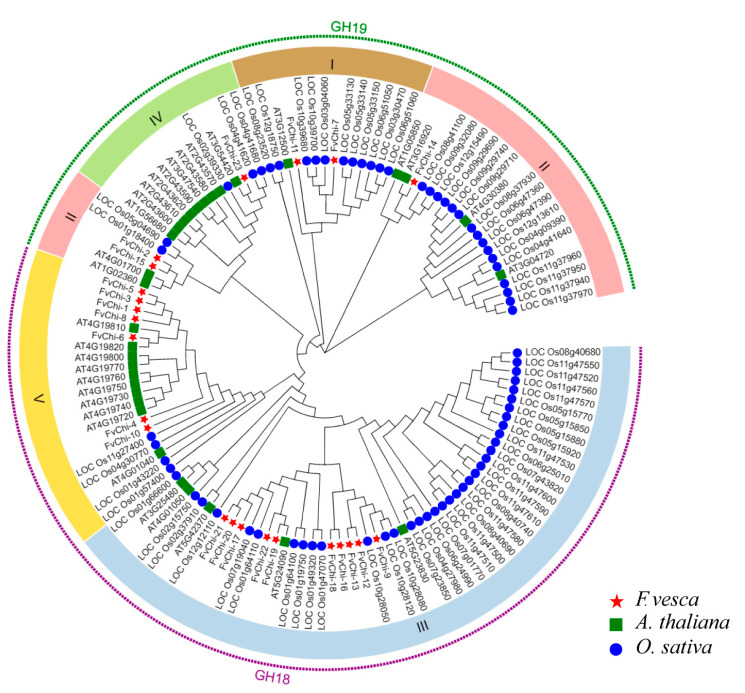
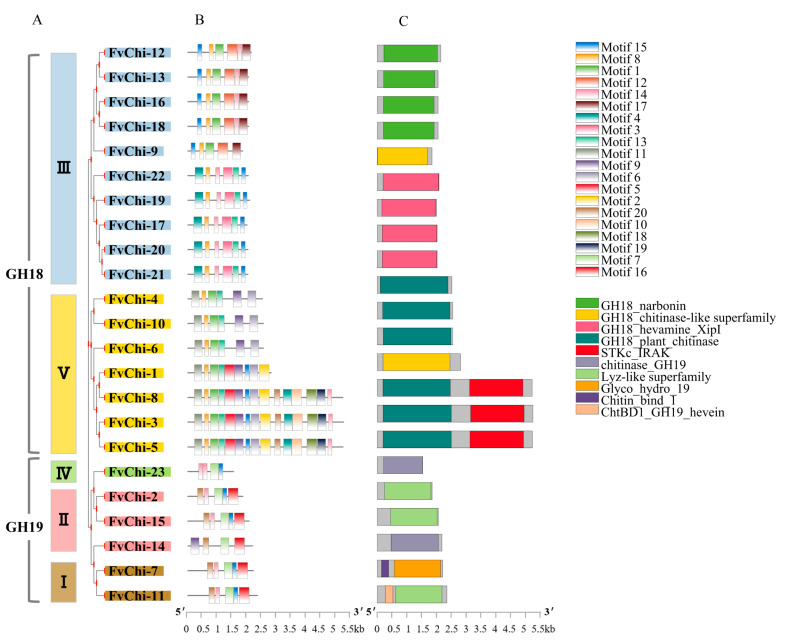
Based on the phylogenetic tree constructed among chitinase genes of strawberry, rice, and Arabidopsis (Figure 2), FvChi genes were divided into two subfamilies, GH18 and GH19. There were 17 and 6 genes in the GH18 and GH19 subfamilies, respectively. Moreover, GH18 and GH19 can be divided into subclasses (categories) having two and three classes, respectively (Figure 3A). According to motif and domain analysis, there were close relationships among members of the same subfamily (Figure 3B,C and Table S2). However, differences were found in the identified motifs of the two subfamilies GH18 and GH19. For example, three members (FvChi-3, FvChi-5 and FvChi-8) of class V were highly similar, and all have a catalytic domain of serine/threonine kinases (STK). Two members (FvChi-2 and FvChi-15) of class IV have a lysozyme-like domain whose family contains several members, including chitinase. Furthermore, two members (FvChi-7 and FvChi-11) of class I have a chitin-bind domain (CBD), which has a role in the recognition and binding of chitin subunits. The members of class III and class V apparently have a distinct glycosyl hydrolase family 18 (GH18) domain. Motif 8 was found in all members of the GH18 subfamily, indicating its importance for the strawberry chitinase GH18 subfamily, while motifs 7 and 16 were only found in GH19 subfamily proteins. However, motifs 9, 14, and 15 were found in both subfamilies.
Figure 2.
Phylogenetic analysis of chitinases proteins from A. thaliana, O. sativa, and F. vesca. Stars, squares, and circles represent F. vesca, A. thaliana, and O. sativa proteins, respectively.
Figure 3.
Gene structure analysis of FvChi. (A) Phylogenetic analysis and classification. Different boxes denote different subgroups. (B) Motif distribution patterns. (C) Domain distributions.
2.3. Gene Structure Architecture of FvChi Genes and Expansion Patterns
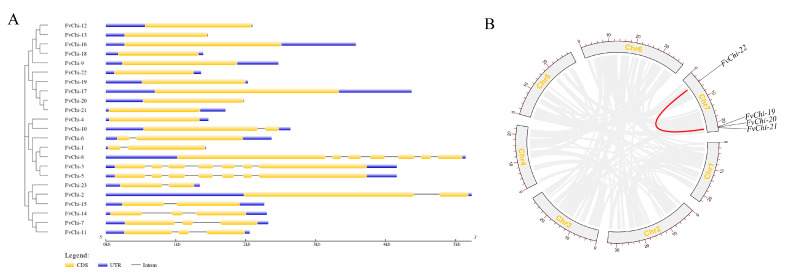
Exon–intron analysis revealed significant differences in gene structure and number of introns between the GH18 and GH19 subfamilies (Figure 4A). A total of 64.7% members of the GH18 subfamily do not have introns, while all members of the GH19 subfamily have introns. Further, members from the same class exhibited a closely linked gene structure based on a similar number or length of exons. Three genes (FvChi-3, FvChi-5, and FvChi-8) of class V have the most introns (six), while other members have one or no intron. Intraspecies collinearity analysis was executed to elaborate the evolution and origin dynamics of FvChi genes (Figure 4B). Three groups of genes with high homology showed clear genetic relationships, revealing that they evolved by tandem replication and formed gene clusters with similar sequences and functions. Only one set of collinearity of segmentally duplicated genes (FvChi-19, 20, 21, and FvChi-22) was identified on chromosome 7.
Figure 4.
Exon–intron structure and collinearity analysis of FvChi genes. (A) Exon–intron structure of FvChi genes. UTRs are shown as blue boxes, exons as yellow boxes, and introns as black lines. (B) Collinearity analysis of FvChi genes. The syntenic relationships of FvChi genes are connected by a red line.
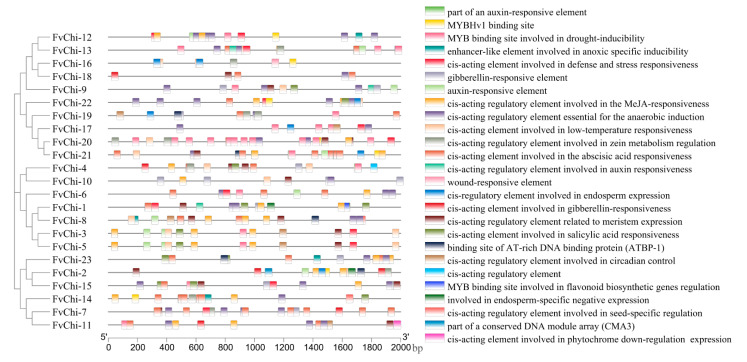
2.4. Analysis of Cis-Acting Elements in FvChi Gene Promoters
The cis-elements contained in the promoter region of the FvChi member have various roles in plant growth and development. Cis-elements were mainly sensitive to abscisic acid (ABA), salicylic acid (SA), methyl jasmonate (MeJA), anaerobic, and drought conditions (Figure 5 and Tables S3 and S4). MeJA-sensitive elements were identified in the promoter region of the 18 FvChi members, and the highest numbers were present in FvChi-3, FvChi-5, and FvChi-8. The promoters of nine genes contained SA-responsive elements, suggesting that most FvChi members might have roles in biotic stress tolerance. Abscisic acid-responsive elements (ABREs), ARE (anaerobic induction), and LTR (low-temperature) elements were identified in the promoter regions of 17, 17, and 18 genes, respectively. Elements associated with anoxia stress were found in 18 FvChi genes. Moreover, TC-rich repeats, ACE (light), MBS (drought), AuxRE (auxin), circadian, CAT-box (meristem), and GARE (gibberellin)-responsive cis-elements were also found in the promoter regions of some members.
Figure 5.
Promoter analysis of FvChi genes.
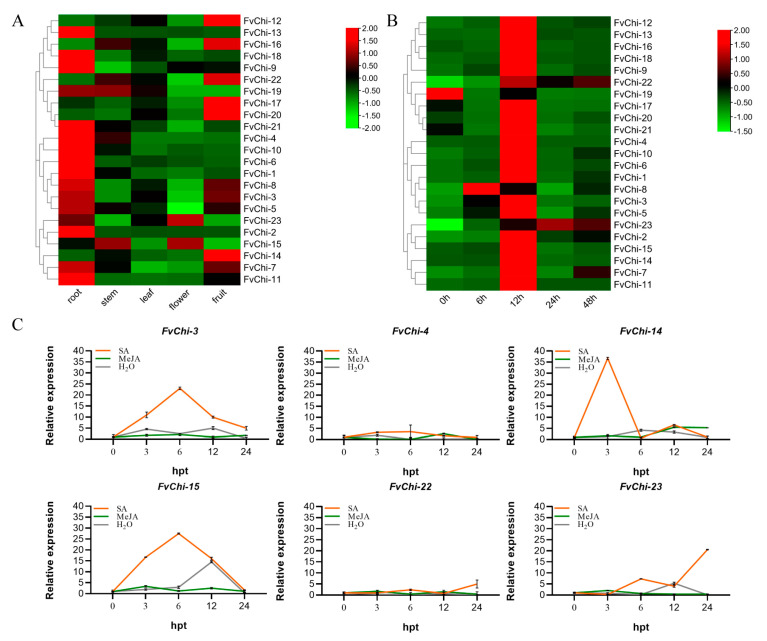
2.5. Expression Analysis of FvChi Gene in Tissue/Organs
Expression patterns of FvChi members were analyzed by quantitative reverse transcription PCR (qRT-PCR) (Figure 6A). Compared to other tissues/organs, most of the genes were highly expressed in roots such as FvChi-1, 2, 4, 6, 9, 10, 11, 13, and 18. On the other hand, the expression levels of FvChi members in flowers were very low. FvChi-15 and 19 showed stem specific expression and FvChi genes displayed ubiquitous expression in leaves. FvChi-12, 14, 16, 17, 20, and 22 showed high expression in fruits. As anticipated, closely related genes showed similar expression patterns; for example, members of class III (FvChi-12, 16, and 22) showed high expression in fruits and low expression in flowers. The results suggested that the FvChi genes have different roles in different strawberry organs, and homologs may have similar functions.
Figure 6.
FvChi gene expression profiles in different tissue/organs and response to C. gloeosporioides inoculation and hormones treatment by qRT-PCR. (A) FvChi gene expression in different tissue/organs including root, stem, leaf, flower, and fruit. (B) FvChi gene expression profiles in response to C. gloeosporioides inoculation. (C) FvChi gene expression profiles in response to SA, MeJA, and water treatments. Red and green color scale denotes high and low expression levels, respectively. Error bars indicate the standard deviation (SD) of three biological replicates.
2.6. Expression Analysis of FvChi Genes Following C. gloeosporioides Inoculation
The expression levels of FvChi genes were measured in strawberry leaves following C. gloeosporioides inoculation (Figure 6B). FvChi genes exhibited different expression patterns and can be divided into four groups based on expression levels. The Group I (FvChi-19) gene was downregulated over time after inoculation with C. gloeosporioides. The Group II (FvChi-8) gene initially showed upregulation and then was subsequently downregulated after 6 h. The members (FvChi-1, 2, 3, 4, 5, 6, 7, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 20, 21 and 22) of Group III also showed the same trend—initially upregulation and subsequent downregulation—but showed a peak expression at 12 hpi. Group IV contained only FvChi-23, which is upregulated at 24 hpi and subsequently downregulated. Overall, the relative expression levels of most FvChi genes increased significantly after C. gloeosporioides infection and reached a maximum at 12 hpi.
2.7. Expression Analysis of Selected FvChi Genes Following SA and MeJA Treatments
The responses of six FvChi genes (FvChi-3, 4, 14, 15, 22 and 23) were estimated against two hormone (SA and MeJA) applications (Figure 6C). Following SA treatment, FvChi-3, 14, and 15 showed the highest increase (23.00, 36.67, and 27.47, respectively), followed by a subsequent decrease. The other three genes showed a slight change in expression at 6 hpt. Overall, most of the genes showed peak expression at 6 hpt. As far as the response to MeJA application is concerned, there was no significant change in expression.
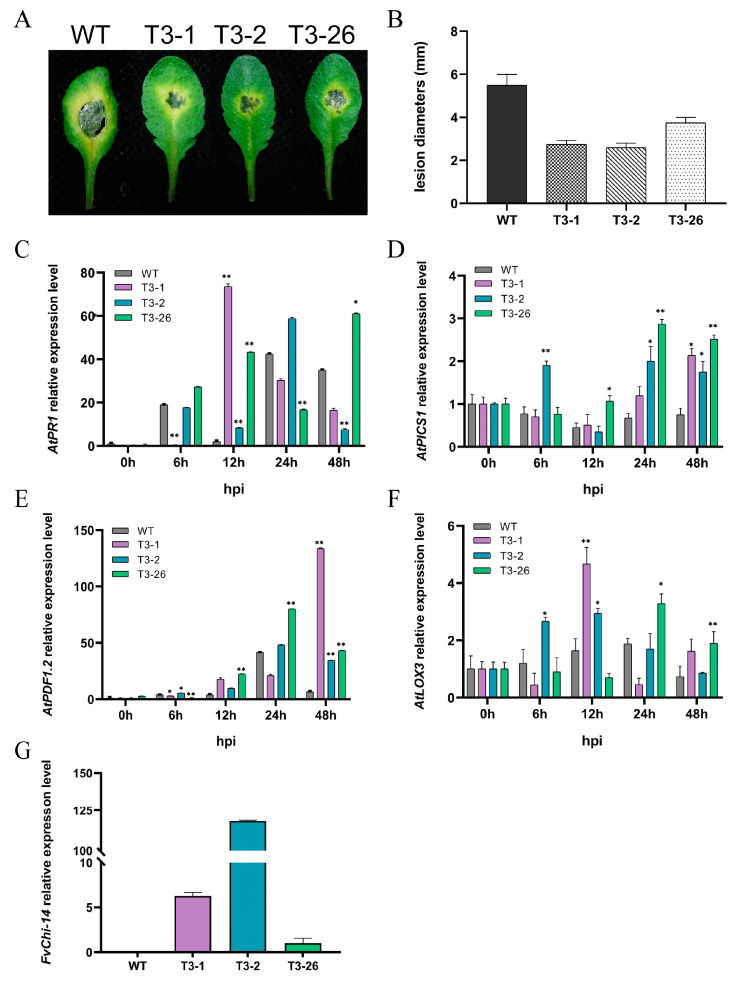
2.8. FvChi14 Overexpression in A. thaliana Enhances Resistance to C. higginsianum
Bioinformatics and expression profiling revealed that the FvChi14 gene was abundantly induced in strawberry when inoculated with C. gloeosporioides, and the expression was maximally upregulated with external hormone treatment (SA). Therefore, the FvChi14 gene was selected for more detailed analysis and overexpressed in Arabidopsis (Figure S1). A total of 43 T2 transgenic plants were obtained, and out of these, 3 independent T3 transgenic lines (T3-1, T3-2, and T3-26) having the strongest resistance to C. higginsianum were chosen for further studies. The FvChi-14 transcript was not shown in the WT, and the highest expression was noticed in the transgenic line T3-2 and the lowest expression in T3-26 (Figure 7G). After 48h of C. higginsianum inoculation with suspended drops, the leaves of the three transgenic lines and Col-0 plant showed obvious translucent necrotic spots surrounded by a yellow halo. However, the lesion diameter of the transgenic plant leaves was significantly smaller than that of the wild type (Figure 7A,B). To find the molecular basis of FvChi-14 resistance to C. higginsianum, the expression levels of some genes having presumed roles in SA and JA signaling pathways were detected using qRT-PCR (Figure 7C–F). PR1 is a SAR molecular marker downstream of SA accumulation; ICS1 is required for SA biosynthesis. At 12 hpi, AtPR1 showed 37.5- and 22.1-fold higher expression in the T3-1 and T3-26 transgenic lines, respectively, compared to WT plants (Figure 7C). AtPR1 was upregulated in T3-26 at 48 hpi but downregulated in T3-1 and T3-2 (Figure 7C). The relative expression of AtICS1 in transgenic strains showed a similar trend and was significantly higher than that of WT at 24–48 hpi (Figure 7D). In particular, the expressions of AtICS1 in T3-2 and T3-26 strains were 3.00- and 4.30-fold higher than that of WT at 24 hpi, respectively. AtICS1 was consistently upregulated in T3-1 at 48 hpi, while the other transgenic strains started to be downregulated. AtPDF1.2 and AtLOX3 have roles in JA signaling pathways. AtPDF1.2 showed higher expression levels at all time points in transgenic lines compared to WT. AtPDF1.2 in T3-1 was significantly upregulated between 24 and 48 hpi, while it showed slight downregulation in T3-2 and T3-26. Moreover, AtPDF1.2 was overexpressed 20.5-fold higher in T3-1 compared to WT at 48 hpi (Figure 7E). Similarly, the relative expression of AtLOX3 was 3.10-fold higher in T3-26 than in WT at 48 hpi (Figure 7F). The relative expression of AtLOX3 peaked at 12 hpi in T3-1 and 2, and at 24 hpi in T3-26. The expression patterns of defense-related genes were different in each transgenic line.
Figure 7.
Overexpression of FvChi-14 in Arabidopsis following C. higginsianum infection. Data are the mean values and SDs of three replications. (A) Symptoms of disease in Col-0 and transgenic A. thaliana leaves after 48 h of inoculation. (B) At 48 hpi, the average lesion diameter on leaves. (C) Expression analysis of AtPR1 gene. (D) Expression analysis of AtICS1 gene. (E) Expression analysis of AtPDF1.2 gene. (F) Expression analysis of AtLOX3 gene. (G) The transcript level of FvChi-14 in leaves of WT and transgenic lines. Asterisks represent a statistically significant difference (* p < 0.05, ** p < 0.01, Student’s t test) between wild-type and transgenic lines.
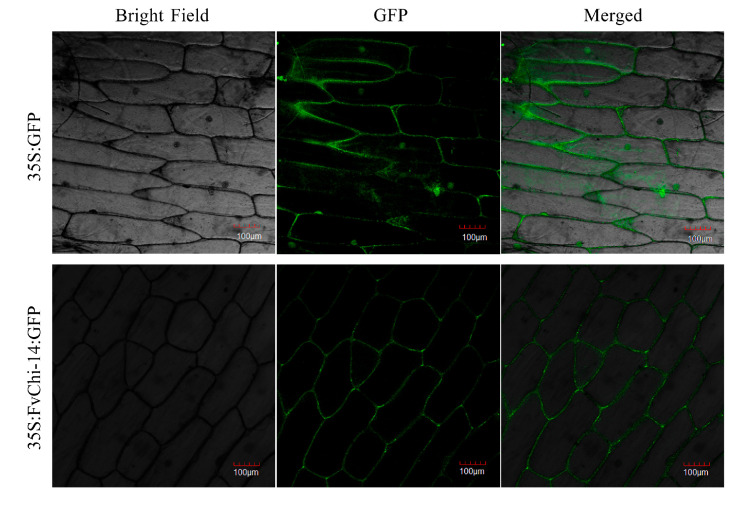
2.9. Subcellular Localization of FvChi-14 in Onion Cells
The subcellular location of the FvChi-14 was validated by transient expression, using GFP protein as a tag in onion cells. Onion epidermal cells were transfected with Agrobacterium containing 35S:FvChi-14:GFP fusion vector and 35S:GFP was used as control. Confocal microscopic examination of onion epidermal cells revealed that the FvChi-14 protein was predicted to be localized to the cell wall or extracellular matrix and exhibited green fluorescence signals (Figure 8).
Figure 8.
Subcellular localization of FvChi-14 in onion epidermal cells. Scale bar: 100 µm.
2.10. Prokaryotic Expression and Western Blot Analysis of FvChi-14
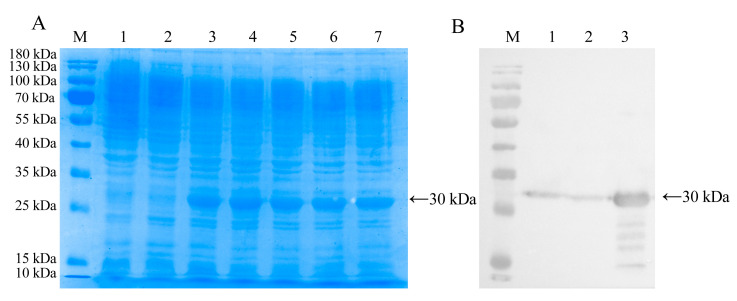
To investigate the potential resistance to abiotic stress, FvChi-14 was ectopically expressed in E. coli BL21 (DE3). The recombinant strain pGEX-4T-1-FvChi-14 showed a specific band at about 30kD in SDS-PAGE analysis after inducing protein expression (Figure 9A), which was similar to the protein size predicted by the ProtParam tool. Western blot analysis further verified that the specific band appeared at the corresponding position of the recombinant protein (Figure 9B), indicating that FvChi-14 protein was correctly expressed in E. coli.
Figure 9.
Prokaryotic expression of FvChi-14 in E. coli and immunoblotting analysis. Note: M: protein ladder; (A) 12% SDS-PAGE analysis of recombinant FvChi-14 protein. 1: pGEX-4T-1; 2: recombinant strain without IPTG induction; 3–7: recombinant strain induced by 0.1, 0.2, 0.4, 0.8, 1.6 mmol·L−1IPTG, respectively. (B) Immunoblotting analysis of the recombinant FvChi-14 protein with anti-GST antibody. 1: pGEX-4T-1; 2: recombinant strain without IPTG induction; 3: recombinant strain induced by 0.2 mmol·L−1IPTG.
3. Discussion
In response to developmental cues and pathogen attacks, higher plants have evolved several defense mechanisms. Different pathogenesis-related (PR) protein-coding genes are expressed in plants [27]. PR3/chitinases are the most important PRs, playing vital roles in resistance to pathogen infection. PR3 are lytic enzymes found in many higher plants and can directly attack the structural components of fungi and insects [28]. Nowadays, anthracnose caused by Colletotrichum spp. has seriously affected the development of the strawberry industry [29]. Chitinases are considered one of the key factors in plant resistance to fungal diseases. The transformation of chitinase genes in resistant strawberry varieties via transgenic technology has attracted the attention of researchers. Therefore, we conducted a detailed and systematic study to identify and characterize the chitinase gene family in the strawberry genome. Twenty-three chitinase genes were found in the diploid strawberry genome, and one gene (FvChi-14) was selected for functional studies. There are 24 chitinase genes in A. thaliana [7]. The close number of genes suggest that these species might have had a close relationship before speciation. However, the other studied species have different numbers of genes, for example, Brassica juncea, Capsicum annuum, and Glycine max have 47, 31, and 38, genes, respectively [2,3,30]. The different numbers of chitinase genes may be due to the differences in ploidy level and genome size between species [31].
The phylogenetic studies of chitinase sequences in F. vesca, A. thaliana, and O. sativa indicate that the proportion of class III genes in strawberries and rice is high (Figure 2). This may be related to the fact that class III chitinases are relatively conserved in different crops. The lysozyme activity of some class III chitinases suggests that plant class III chitinases may have biological effects different from those of other classes [32]. It has been shown that different class III chitinase isomers respond differently to Fusarium oxysporum infection, and melon resistance to F. oxysporum infection is linked with some subtypes of this enzyme [33]. Oschib1 and Oschib2 acted with extracellular chitinase to hydrolyze chitin oligosaccharides released from the cell wall of the pathogenic fungi [34]. In our experiment, the expression levels of the class III chitinase genes, FvChi-9 and FvChi-18, were upregulated by 55.70 and 54.20 times, respectively, after inoculation with C. gloeosporioides. Future research is needed to explore the roles of class III chitinases in disease resistance.
Gene intron density and the regulation of genes in response to stress are inversely correlated [35]. Because long and abundant introns extend transcription time, genes with few introns can be rapidly regulated following stress [30]. Stress-related gene families such as FvbZIP [36], SsLRR-RLK [37], and TaZF-HD [38] all have lower numbers of introns. Our results are also in line with these findings, and among the 23 chitinase genes in F. vesca, 11 genes did not contain introns and 9 genes had 1 to 2 introns (Figure 4A). Based on the previous findings and our results, we surmise that most of the FvChi genes can have roles in disease resistance. Moreover, the occurrence of stress-related cis-elements (Figure 5), changes in the expression of genes following C. gloeosporioide inoculation, and the response to SA treatments also support this hypothesis. However, functional analysis is required for confirmation.
Based on the bioinformatics and response against C. gloeosporioides, SA, and MeJA treatments, the FvChi-14 gene belonging to class II was selected for functional studies (Figure 6). Our findings are in line with earlier research showing that class II chitinases can be activated by ethylene, salicylic acid, or fungal spore treatment, and are extensively expressed in healthy plants [39]. Class II chitinases may perform various roles such as playing a role in plant defense and endogenous regulatory functions during plant development [40]. Overexpression of FvChi-14 in A. thaliana increased resistance to the fungus C. higginsianum. Following in vitro leaf inoculation with C. higginsianum, the lesion size on the leaves of transgenic lines (T3-1, T3-2 and T3-26) was significantly smaller than that of the wild plants. After this, the whole plant was sprayed with inoculated pathogen suspensions, and the relative expression levels of AtPR1, AtICS1, AtPDF1.2, and AtLOX3 were measured at 0, 3, 6, 12, 24, and 48 hpi. These resistance-related genes showed higher relative expression levels at 12-48 hpi in FvChi-14 transgenic plants than in control plants (Figure 7).
Our results are supported by previous findings; for example, overexpression of barley class II chitinase genes in transgenic wheat enhanced resistance to Fusarium graminearum in greenhouse and field conditions [41]. The Na2CO3-responsive chitinase gene LcCHI2 of class II improved pathogen resistance in transgenic maize and tobacco [42]. Compared to control cultivars, transgenic sugarcane lines treated with the barley chitinase class II gene exhibited better resistance to inoculation with C. falcatum [43]. Furthermore, the relative expression of AtPR1 peaked earlier at about 12hpi; AtPR1 expression levels were upregulated in T3-26 and downregulated in T3-1 and T3-2 at 48 hpi, while the relative expression of AtPDF1.2 peaked later at 48hpi, especially in the T3-1 line. This may be related to the fact that the genus Colletotrichum spp. followed the hemibiotrophic mode of infection [44]. For defense against biotrophic pathogens, SA is a main signaling molecule, while JA-mediated defense signaling is usually effective against necrotizing pathogens [45,46]. A potential explanation to these results could be as follows: FvChi-14 overexpression lines in A. thaliana first upregulate the relative expression level of the SA pathway-related gene AtPR1 and then upregulate the relative expression level of the JA defense pathway gene AtPDF1.2 after C. higginsianum infection. In conclusion, overexpression of the FvChi-14 gene can improve the resistance of A. thaliana to C. higginsianum. SA is a natural signaling molecule that activates the expression of defense genes and triggers changes in components of the redox signal transduction pathway [47]. SA scavenges hydroxyl radicals in plants and prevents the inactivation of catalase by H2O2, thus protecting plants from oxidative damage [48]. Chitinase genes are a class of defense response genes that are closely related to SAR in plants. SAR is often accompanied by the production of reactive oxygen species (ROS). Chitinase may induce the SA pathway by increasing ROS in response to disease resistance in plants. Chitinase has been shown to be induced by JA rather than SA, but the two pathways are not completely independent and cross each other at different sites [49]. Studies have shown that ROS may be involved in JA-induced chitinase accumulation in leaves and leaf sheaths in rice [50]. External application of MeJA enhanced the activity of inductive-type chitinase [51]. However, in this study, MeJA did not significantly induce the expression of some strawberry chitinase genes. (Figure 6C). Induction of SA and JA signaling patterns can alter gene expression and lead to specific defense responses to stress in plants [10]. The specific action mechanism of chitinase in these signaling modes needs further study. As anticipated, most chitinase proteins were predicted in the cell wall and vacuole in strawberry (Table 1). An N- or C-terminal targeting region on all plant chitinases sends the enzymes first to the endoplasmic reticulum and subsequently to either the vacuole or the apoplast [52]. Moreover, FvChi-14 protein was predicted to act in the extracellular matrix or vacuole, and the transient expression of tag protein GFP in onion epidermal cells confirmed that the subcellular location of FvChi-14 is the cell wall or the extracellular matrix (Figure 8).
4. Materials and Methods
4.1. Plant Materials and Seedlings Treatment
The strawberry (F. vesca ssp.) and A. thaliana (Col-0) plants grown in the plant incubators were used as experimental material. The growing conditions for strawberry were maintained at 23–25 °C and 85% humidity, while for A. thaliana, conditions were 21 °C, 70% relative humidity, and a 12 h light/12 h dark cycle.
Pathogen inoculation (C. gloeosporioides and C. higginsianum) and plant maintenance were carried out according to our previous experiment [53]. For the purpose of determining the biological efficacy of FvChi-14 against anthracnose, C. higginsianum was used to infect both A. thaliana transgenic lines and the Col-0 wild types [54]. For RNA extraction, leaf samples were collected at 0, 6, 12, 24, and 48 h postinoculation (hpi). Strawberry leaves were treated with 1.5 mL of either 50 mM methyl jasmonate (MeJA) or 5 mM salicylic acid (SA) solutions, and distilled water was used as a control. After spraying, the leaves were collected at 0, 3, 6, 12, and 24 h post-treatment (hpt). Strawberry leaves, stolons, flowers, fruits, and roots were collected and quickly immersed in liquid nitrogen for constitutive expression analysis. After this, samples were stored at −80 °C for further use and every experiment was performed with three biological and technical replicates.
4.2. Identification of Chitinase Genes in Strawberry
The genomes and gene annotation files of F. vesca were retrieved from NCBI (https://www.ncbi.nlm.nih.gov/genome/browse#!/overview/, accessed on 10 January 2021). The chitinase protein sequences of A. thaliana and O. sativa were downloaded from the Phytozome database (https://phytozome.jgi.doe.gov/pz/portal.html, accessed on 20 January 2021). CDS was extracted from strawberry genomes by TBtools and translated into protein sequences. A. thaliana chitinase protein sequences were used as queries against F. vesca protein sequences. Possible members were obtained after blast comparison. The obtained sequences of chitinase proteins were verified using Swiss-Prot database and Batch CD-Search (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, accessed on 30 January 2021). Twenty-three putative chitinase genes were screened and renamed FvChi-1- FvChi-23 based on their position on the chromosome. Basic information about selected proteins, such as the start and end sites, was retrieved from the NCBI.
4.3. Characterization Analysis of Strawberry Chitinase Genes
The accession number, chromosomal location, CDS length, and other information of FvChi genes were retrieved by NCBI. The physicochemical properties of FvChi genes were estimated using The ProtParam tool of ExPASy Server (https://web.expasy.org/protparam/, accessed on 10 February 2021), including protein molecular weight (MW) and isoelectric point (pI) values. The Plant-mPLoc Server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/, accessed on 10 February 2021) was used for the prediction of subcellular locations of proteins.
4.4. Gene Structure and Motif Distribution Patterns Analysis
The members of the chitinase gene family were examined using the online MEME application (http://meme-suite.org/, accessed on 20 February 2021). All the default parameters were used, and the motifs were searched up to 20. The conserved domain of the members of the chitinase gene family was examined using the web application Batch CD-Search. TBtools was used to visualize the results [18], and Splign (https://www.ncbi.nlm.nih.gov/sutils/splign/splign.cgi?textpage=online&level=form, accessed on 20 February 2021) was used to study the intron and exon distribution patterns. The results were displayed using Gene Structure Display Server 2.0 (gao-lab.org).
4.5. Phylogenetic Studies of FvChi Genes
Protein sequences were used for phylogenetic tree construction. The phylogenetic trees were generated using MEGA X and the maximum-likelihood (ML) method with the following conditions: 1000 bootstrap values, the WAG model, and 95% partial deletion. The phylogeny was determined among FvChi proteins, and another was generated among chitinase proteins of A. thaliana, O. sativa, and F. vesca.
4.6. Expansion Pattern and Promoter Analysis of FvChi Genes
The strawberry genomic database was compared with itself using the blast function of TBtools. The output results were imported into MCScan (http://chibba.pgml.uga.edu/mcscan2/, accessed on 22 February 2021) to determine collinear blocks under a default criterion [55]. The outcomes were shown using TBtools. The 2 Kb upstream nucleotide sequences of FvChi genes were downloaded and studied by PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 22 February 2021) for cis-elements.
4.7. Expression Analysis of FvChi Genes
Each sample was measured at 0.1 g, and total RNA was extracted from strawberry organs or treated leaves using RNAprep Pure Plant Kit (Tiangen, China). Sample total RNA concentration was measured using a Thermo Scientific NanoDrop 2000 instrument. Total RNA concentration was greater than 100 μg/μL, and OD260/280 values were 1.8–2.1. The first-strand cDNA was synthesized using the PrimeScript™RT kit (Takara) and contained 500ng total RNA per 10 μL cDNA reversal system. DNAMAN Version 10 software (https://www.lynnon.com/index.html, accessed on 27 February 2021) was used for designing primers, and Table S1 contains a list of the primers. The qRT-PCR was performed on Roche LightCycler96 machine for expression analysis. The PCR conditions were 30 s at 95 °C, 40 cycles of 5 s at 95 °C, 30 s at 60 °C, and a final step of 15 s at 72 °C. Each reaction mixture (20 μL) consisted of 0.5 μL of each primer (final concentration: 0.25 μM), 2 μL of diluted cDNA, 10 μL of TB Green Premix Ex Taq, 0.4 μL of ROX Reference Dye, and 6.6 μL of sterile water. The 2−∆∆CT method was used for the calculation of relative expression values [56], and the actin gene (GenBank accession number AB116565) was used as an internal reference. Each reaction was repeated three times biologically and technically.
4.8. Plasmid Construction and A. thaliana Transformation
Reverse transcription was used to create first strand of cDNA from 1 g of total RNA using the PrimerScriptTM-II 1st Strand cDNA Synthesis kit (TaKaRa Bio Inc. Dalian, China). The ORF sequence of the FvChi-14 (972bp) was amplified with LA Taq (TaKaRa Bio. Inc.). The PCR product was subsequently cloned into pMD20-T (TaKaRa Bio Inc.) and sequenced (FuZhou ShangYa BioInc., Fuzhou, China). The resulting sequence was cloned into the binary vector pCAMBIA1300-HA (CAMBIA company) using specific primers FvChi-14-clone-F and FvChi-14-clone-R containing BamH I and Spe I sites (Table S1). The sequence was also submitted to GenBank (Acc. NO. OQ211094). By using the freeze–thaw technique, the recombinant plasmid was transformed into Agrobacterium tumefaciens GV3101 [57], and then the recombinant was transferred into wild A. thaliana by floral dip method [58]. The solid MS medium containing 50 mg/L hygromycin was used for the screening of transgenic plants [59].
4.9. Subcellular Localization of FvChi-14 in Onion Cells
The ORF region of FvChi-14 was cloned into the pGFPc vector using specific primers FvChi-14-PE-F and FvChi-14-PE-R containing BamH I and X-bal sites (Table S1). The constructed fusion vector was verified by sequencing and transformed into A. tumefaciens GV3101 and was injected into onion epidermal cells for 48 h culture. The fluorescence reaction of GFP in the onion epidermis was observed by laser scanning confocal microscope (OLYMPUS IX83-FV3000).
4.10. Prokaryotic Expression and Western Blot Analysis of FvChi-14
The ORF region of FvChi-14 was cloned into the pGEX-4T-1 vector using specific primers FvChi-14-PE-F and FvChi-14-PE-R containing BamH I and Sma I sites (Table S1). The sequence of recombinant vector was verified by sequencing. The recombinant plasmid and original vector were introduced into E. coli BL21 (DE3) and cultivated in LB medium containing 100 mg·L−1 ampicillin.
The control strain (BL21: pGEX-4T-1) and recombinant strain (BL21: pGEX-4T-1- FvChi-14) were cultured at 200 rpm and 37 °C overnight. A total of 1% of the cultures were then inoculated to fresh LB medium containing 100 mg·L−1 ampicillin, and value was adjusted to 0.5 OD600. The recombinant strain was added with 0.2 mmol·L−1 isopropyl β-D-1-thiogalactopyranoside (IPTG) and cultured at 37 °C for 4 h to induce FvChi-14 protein expression. A total of 2 mL of the induced expression solution was centrifuged at 10,000× g for 1 min, and the bacteria were resuspended with 200 μL PBS buffer. The 60 μL bacterial suspension was added with 20 μL 4 × sample buffer (0.250 M Tris base, 0.28 M SDS, 40% glycerol, 20% 2-mercapto-ethanol, bromphenol blue), boiled at 100 °C for 10 min, and centrifuged at 12,000× g for 10 min. The supernatant was retained, and the target protein was resolved by 12% SDS-PAGE (SDS-PAGE Gel Kit, Solarbio, China). The gels were stained with InstaBlue protein stain solution (APE×BIO, Des Moines, IA, USA), and the results were observed with Gel Doc XR+ System (Bio-Rad, Hercules, CA, USA).
Proteins were electroblotted onto an NC membrane using semidry electrophoretic transfer cell (BIORAD, USA) following separation on 12% SDS-PAGE. The membrane was then blocked at room temperature for 3 h in blocking buffer (5% nonfat dried milk). The membrane was rinsed with PBST (1 × PBS, 1‰ tween 20) for 30 min after being treated with the anti-GST mouse monoclonal antibody (TransGen Biotech, Beijing, China) at 4 °C overnight. The membrane was then incubated with the goat anti-mouse IgG secondary antibody. Gel Doc XR+ System was used to analyze the data after staining the membrane with 2 mL of eECL chromogen (Cowin Bio., Beijing, China).
5. Conclusions
There are 23 chitinase genes in F. vesca, which can be divided into 5 subclasses. Most of the FvChi genes were differentially expressed in various organs of strawberry and showed upregulation following C. gloeosporioides inoculation. Further, some genes were strongly induced by hormone (SA and JA) treatment. In addition, overexpression of FvChi-14 increased the resistance of A. thaliana to C. higginsianum, via regulating the SA and JA signaling pathways. The FvChi-14 protein may be localized in the cell wall or extracellular matrix. This study offers insight into the potential roles of strawberry chitinase genes in biotic resistance and provides new candidates for stress resistance breeding programs. The study provides a basis for the functional characterization of FvChi genes.
Acknowledgments
C. gloeosporioides and C. higginsianum strain Ch-1 were provided by Han Yongchao and Zheng Lu, respectively.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/plants12071543/s1, Figure S1: The deduced amino acid and nucleotide sequence of FvChi-14. The underlined part denotes chitinase domain.; Table S1: Details on the primer sequences used; Table S2: Motif sequences identified found in FvChi genes; Table S3: Signaling hormones and stresses-related cis-acting elements in the promoter regions of FvChi genes; Table S4: Functions of the cis-elements found in the promoters of FvChi genes.
Author Contributions
Conceptualization, Z.W.; data curation, T.H.; investigation, T.H., J.F., G.J., Y.L., Q.Z. and N.L.; resources, Q.C. and Z.W.; writing—original draft, T.H.; writing—review and editing, T.H., B.A. and Z.W. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This research was funded by the National Natural Science Foundation of China (32072526).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Wen X., Tengsheng Z., Dandan L., Sufang A., Jinna H., Genyi L. Evolution and Expression Divergence of the Chitinase Gene Family against Leptosphaeria maculans and Sclerotinia sclerotiorum Infection in Brassica napus. J. Henan Agric. Sci. 2020;49:89–103. doi: 10.15933/j.cnki.1004-3268.2020.02.012. [DOI] [Google Scholar]
- 2.Lv P., Zhang C., Xie P., Yang X., El-Sheikh M.A., Hefft D.I., Ahmad P., Zhao T., Bhat J.A. Genome-Wide Identification and Expression Analyses of the Chitinase Gene Family in Response to White Mold and Drought Stress in Soybean (Glycine max) Life. 2022;12:1340. doi: 10.3390/life12091340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liu Z., Yu W., Cai W., Liu Z., Zang Y., Yuan Y., Wu B., Lv M. Genome-wide Identification and Expression Analysis of CTL Gene Family Memebers in Capsicum annuum L. Chin. J. Trop. Crops. 2021;42:3101–3110. [Google Scholar]
- 4.Tobias P.A., Christie N., Naidoo S., Guest D.I., Kulheim C. Identification of the Eucalyptus grandis chitinase gene family and expression characterization under different biotic stress challenges. Tree Physiol. 2017;37:565–582. doi: 10.1093/treephys/tpx010. [DOI] [PubMed] [Google Scholar]
- 5.Bordoloi K.S., Krishnatreya D.B., Baruah P.M., Borah A.K., Mondal T.K., Agarwala N. Genome-wide identification and expression profiling of chitinase genes in tea (Camellia sinensis (L.) O. Kuntze) under biotic stress conditions. Physiol. Mol. Biol. Plants. 2021;27:369–385. doi: 10.1007/s12298-021-00947-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandra S., Dutta A.K., Chandrashekara K.N., Acharya K. In silico characterization, homology modeling of Ca-mellia sinensis chitinase and its evolutionary analyses with other plant chitinases. Proc. Natl. Acad. Sci. India Sect. B Biol. Sci. 2015;87:685–695. doi: 10.1007/s40011-015-0634-6. [DOI] [Google Scholar]
- 7.Passarinho P.A., de Vries S.C. Arabidopsis Chitinases: A Genomic Survey. Arab. Book. 2002;1:e0023. doi: 10.1199/tab.0023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cletus J., Balasubramanian V., Vashisht D., Sakthivel N. Transgenic expression of plant chitinases to enhance disease resistance. Biotechnol. Lett. 2013;35:1719–1732. doi: 10.1007/s10529-013-1269-4. [DOI] [PubMed] [Google Scholar]
- 9.Xin Y., Wang D., Han S., Li S., Gong N., Fan Y., Ji X. Characterization of the Chitinase Gene Family in Mulberry (Morus notabilis) and MnChi18 Involved in Resistance to Botrytis cinerea. Genes. 2021;13:98. doi: 10.3390/genes13010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ali M., Li Q.-H., Zou T., Wei A.-M., Gombojab G., Lu G., Gong Z.-H. Chitinase Gene Positively Regulates Hypersensitive and Defense Responses of Pepper to Colletotrichum acutatum Infection. Int. J. Mol. Sci. 2020;21:6624. doi: 10.3390/ijms21186624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jingjing L., Renying Z., Xiaohua Q., Qiang X., Xuehao C. A Preliminary Study on Cloning of Chitinase Gene and Relationships to Resistance ofPowdery Mildew in Cucumber (Cucumis sativus L.) Mol. Plant Breed. 2015;13:1584–1591. doi: 10.13271/j.mpb.013.001584. [DOI] [Google Scholar]
- 12.Davis J.M., Wu H., Cooke J.E.K., Reed J.M., Luce K.S., Michler C.H. Pathogen Challenge, Salicylic Acid, and Jasmonic Acid Regulate Expression of Chitinase Gene Homologs in Pine. Am. Phytopathol. Soc. 2002;15:380–387. doi: 10.1094/MPMI.2002.15.4.380. [DOI] [PubMed] [Google Scholar]
- 13.Xu J., Xu X., Tian L., Wang G., Zhang X., Wang X., Guo W. Discovery and identification of candidate genes from the chitinase gene family for Verticillium dahliae resistance in cotton. Sci. Rep. 2016;6:29022. doi: 10.1038/srep29022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takenaka Y., Nakano S., Tamoi M., Sakuda S., Fukamizo T. Chitinase gene expression in response to environmental stresses in Arabidopsis thaliana: Chitinase inhibitor allosamidin enhances stress tolerance. Biosci. Biotechnol. Biochem. 2009;73:1066–1071. doi: 10.1271/bbb.80837. [DOI] [PubMed] [Google Scholar]
- 15.Su Y., Xu L., Wang S., Wang Z., Yang Y., Chen Y., Que Y. Identification, phylogeny, and transcript of chitinase family genes in sugarcane. Sci. Rep. 2015;5:10708. doi: 10.1038/srep10708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rasiukevičiūtė N., Rugienius R., Šikšnianienė J.B. Genetic diversity of Botrytis cinerea from strawberry in Lithuania. Zemdirb. Agric. 2018;105:265–270. doi: 10.13080/z-a.2018.105.034. [DOI] [Google Scholar]
- 17.Hassan E.A., Mostafa Y.S., Alamri S., Hashem M., Nafady N.A. Biosafe Management of Botrytis Grey Mold of Strawberry Fruit by Novel Bioagents. Plants. 2021;10:2737. doi: 10.3390/plants10122737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X.Y., Dai D.J., Zhao S.F., Shen Y., Wang H.D., Zhang C.Q. Genetic Diversity of Colletotrichum spp. Causing Strawberry Anthracnose in Zhejiang, China. Plant Dis. 2020;104:1351–1357. doi: 10.1094/PDIS-09-19-2026-RE. [DOI] [PubMed] [Google Scholar]
- 19.Han Y.C., Zeng X.G., Xiang F.Y., Ren L., Chen F.Y., Gu Y.C. Distribution and Characteristics of Colletotrichum spp. Associated with Anthracnose of Strawberry in Hubei, China. Plant Dis. 2016;100:996–1006. doi: 10.1094/PDIS-09-15-1016-RE. [DOI] [PubMed] [Google Scholar]
- 20.Amil-Ruiz F., Blanco-Portales R., Munoz-Blanco J., Caballero J.L. The strawberry plant defense mechanism: A molecular review. Plant Cell. Physiol. 2011;52:1873–1903. doi: 10.1093/pcp/pcr136. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto S., Shiraishi S., Suzuki S. Are cyclic lipopeptides produced by Bacillus amyloliquefaciens S13-3 responsible for the plant defence response in strawberry against Colletotrichum gloeosporioides? Lett. Appl. Microbiol. 2015;60:379–386. doi: 10.1111/lam.12382. [DOI] [PubMed] [Google Scholar]
- 22.Valenzuela-Riffo F., Zuniga P.E., Morales-Quintana L., Lolas M., Caceres M., Figueroa C.R. Priming of Defense Systems and Upregulation of MYC2 and JAZ1 Genes after Botrytis cinerea Inoculation in Methyl Jasmonate-Treated Strawberry Fruits. Plants. 2020;9:447. doi: 10.3390/plants9040447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartholomew E.S., Black K., Feng Z., Liu W., Shan N., Zhang X., Wu L., Bailey L., Zhu N., Qi C., et al. Comprehensive Analysis of the Chitinase Gene Family in Cucumber (Cucumis sativus L.): From Gene Identification and Evolution to Expression in Response to Fusarium oxysporum. Int. J. Mol. Sci. 2019;20:5309. doi: 10.3390/ijms20215309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cao J., Tan X. Comprehensive Analysis of the Chitinase Family Genes in Tomato (Solanum lycopersicum) Plants. 2019;8:52. doi: 10.3390/plants8030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu M., Lu S., Zhuang M., Zhang Y., Lv H., Ji J., Hou X., Fang Z., Wang Y., Yang L. Genome-wide identification and expression analysis of the Brassica oleracea L. chitin-binding genes and response to pathogens infections. Planta. 2021;253:80. doi: 10.1007/s00425-021-03596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haxim Y., Kahar G., Zhang X., Si Y., Waheed A., Liu X., Wen X., Li X., Zhang D. Genome-wide characterization of the chitinase gene family in wild apple (Malus sieversii) and domesticated apple (Malus domestica) reveals its role in resistance to Valsa mali. Front. Plant Sci. 2022;13:1007936. doi: 10.3389/fpls.2022.1007936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh A., Kirubakaran S.I., Sakthivel N. Heterologous expression of new antifungal chitinase from wheat. Protein Expr. Purif. 2007;56:100–109. doi: 10.1016/j.pep.2007.06.013. [DOI] [PubMed] [Google Scholar]
- 28.Ahmed N.U., Park J.I., Seo M.S., Kumar T.S., Lee I.H., Park B.S., Nou I.S. Identification and expression analysis of chitinase genes related to biotic stress resistance in Brassica. Mol. Biol. Rep. 2012;39:3649–3657. doi: 10.1007/s11033-011-1139-x. [DOI] [PubMed] [Google Scholar]
- 29.Chung P.C., Wu H.Y., Chen Y.C., Hung T.H., Chung C.L. Development of a nested PCR assay for detecting Colletotrichum siamense and Colletotrichum fructicola on symptomless strawberry plants. PLoS ONE. 2022;17:e0270687. doi: 10.1371/journal.pone.0270687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mir Z.A., Ali S., Shivaraj S.M., Bhat J.A., Singh A., Yadav P., Rawat S., Paplao P.K., Grover A. Genome-wide identification and characterization of Chitinase gene family in Brassica juncea and Camelina sativa in response to Alternaria brassicae. Genomics. 2020;112:749–763. doi: 10.1016/j.ygeno.2019.05.011. [DOI] [PubMed] [Google Scholar]
- 31.Kagale S., Koh C., Nixon J., Bollina V., Clarke W.E., Tuteja R., Spillane C., Robinson S.J., Links M.G., Clarke C., et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014;5:3706. doi: 10.1038/ncomms4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamel F., Boivin R., Tremblay C., Bellemare G. Structural and Evolutionary Relationships Among Chitinases of Flowering Plants. Mol. Evol. Vol. 1997;44:614–624. doi: 10.1007/PL00006184. [DOI] [PubMed] [Google Scholar]
- 33.Balde J.A., Francisco R., Queiroz A., Regalado A.P., Ricardo C.P., Veloso M.M. Immunolocalization of a class III chitinase in two muskmelon cultivars reacting differently to Fusarium oxysporum f. sp. melonis. J. Plant Physiol. 2006;163:19–25. doi: 10.1016/j.jplph.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka J., Takashima T., Abe N., Fukamizo T., Numata T., Ohnuma T. Characterization of two rice GH18 chitinases belonging to family 8 of plant pathogenesis-related proteins. Plant Sci. 2023;326:111524. doi: 10.1016/j.plantsci.2022.111524. [DOI] [PubMed] [Google Scholar]
- 35.Jeffares D.C., Penkett C.J., Bahler J. Rapidly regulated genes are intron poor. Trends Genet. 2008;24:375–378. doi: 10.1016/j.tig.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 36.Wang X.L., Chen X., Yang T.B., Cheng Q., Cheng Z.M. Genome-Wide Identification of bZIP Family Genes Involved in Drought and Heat Stresses in Strawberry (Fragaria vesca) Int. J. Genom. 2017;2017:3981031. doi: 10.1155/2017/3981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cheng W., Wang Z., Xu F., Ahmad W., Lu G., Su Y., Xu L. Genome-Wide Identification of LRR-RLK Family in Saccharum and Expression Analysis in Response to Biotic and Abiotic Stress. Curr. Issues Mol. Biol. 2021;43:1632–1651. doi: 10.3390/cimb43030116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu H., Xia P., Hu Y., Zhan C., Li Y., Gong S., Li Y., Ma D. Genome-wide identification of ZF-HD gene family in Triticum aestivum: Molecular evolution mechanism and function analysis. PLoS ONE. 2021;16:e0256579. doi: 10.1371/journal.pone.0256579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kellmann J.W., Kleinow T., Engelhardt K., Philipp C., Wegener D., Schell J., Schreier P.H. Characterization of two class II chitinase genes from peanut and expression studies in transgenic tobacco plants. Plant Mol. Biol. 1996;30:351–358. doi: 10.1007/BF00020121. [DOI] [PubMed] [Google Scholar]
- 40.Ponath Y., Vollberg H., Hahlbrock K., Kombrink E. Two differentially regulated class II chitinases from parsley. Biol. Chem. 2000;381:667–678. doi: 10.1515/BC.2000.087. [DOI] [PubMed] [Google Scholar]
- 41.Shin S., Mackintosh C.A., Lewis J., Heinen S.J., Radmer L., Dill-Macky R., Baldridge G.D., Zeyen R.J., Muehlbauer G.J. Transgenic wheat expressing a barley class II chitinase gene has enhanced resistance against Fusarium graminearum. J. Exp. Bot. 2008;59:2371–2378. doi: 10.1093/jxb/ern103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu X., Yu Y., Liu Q., Deng S., Jin X., Yin Y., Guo J., Li N., Liu Y., Han S., et al. A Na(2)CO(3)-Responsive Chitinase Gene From Leymus chinensis Improve Pathogen Resistance and Saline-Alkali Stress Tolerance in Transgenic Tobacco and Maize. Front. Plant Sci. 2020;11:504. doi: 10.3389/fpls.2020.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tariq M., Khan A., Tabassum B., Toufiq N., Bhatti M.U., Riaz S., Nasir I.A., Husnain T. Antifungal activity of chitinase II against Colletotrichum falcatum Went. causing red rot disease in transgenic sugarcane. Turk. J. Biol. 2018;42:45–53. doi: 10.3906/biy-1709-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dubouzet J.G., Maeda S., Sugano S., Ohtake M., Hayashi N., Ichikawa T., Kondou Y., Kuroda H., Horii Y., Matsui M., et al. Screening for resistance against Pseudomonas syringae in rice-FOX Arabidopsis lines identified a putative receptor-like cytoplasmic kinase gene that confers resistance to major bacterial and fungal pathogens in Arabidopsis and rice. Plant Biotechnol. J. 2011;9:466–485. doi: 10.1111/j.1467-7652.2010.00568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsuda K., Katagiri F. Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 2010;13:459–465. doi: 10.1016/j.pbi.2010.04.006. [DOI] [PubMed] [Google Scholar]
- 46.Glazebrook J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 2005;43:205–227. doi: 10.1146/annurev.phyto.43.040204.135923. [DOI] [PubMed] [Google Scholar]
- 47.Pulla R.K., Lee O.R., In J.G., Parvin S., Kim Y.J., Shim J.S., Sun H., Kim Y.J., Senthil K., Yang D.C. Identification and characterization of class I chitinase in Panax ginseng C. A. Meyer. Mol. Biol. Rep. 2011;38:95–102. doi: 10.1007/s11033-010-0082-6. [DOI] [PubMed] [Google Scholar]
- 48.Fobert P.R., Després C. Redox control of systemic acquired resistance. Curr. Opin. Plant Biol. 2005;8:378–382. doi: 10.1016/j.pbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 49.Rawat S., Ali S., Mittra B., Grover A. Expression analysis of chitinase upon challenge inoculation to Alternaria wounding and defense inducers in Brassica juncea. Biotechnol. Rep. 2017;13:72–79. doi: 10.1016/j.btre.2017.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rakwal R., Yang G., Komatsu S. Chitinase induced by jasmonic acid, methyl jasmonate, ethylene and protein phosphatase inhibitors in rice. Print. Neth. 2004;31:113–119. doi: 10.1023/B:MOLE.0000031407.18708.95. [DOI] [PubMed] [Google Scholar]
- 51.Buzi A., Chilosi G., Magro P. Induction of Resistance in Melon Seedlings Against Soil-borne Fungal Pathogens by Gaseous Treatments with Methyl Jasmonate and Ethylene. J. Phytopathol. 2004;152:491–497. doi: 10.1111/j.1439-0434.2004.00885.x. [DOI] [Google Scholar]
- 52.Sun Y.L., Hong S.K. Effect of chitinase on resistance to fungal pathogens in sea buckthorn, Hippophae rhamnoides, and cloning of Class I and III chitinase genes. Biochem. Genet. 2012;50:600–615. doi: 10.1007/s10528-012-9504-6. [DOI] [PubMed] [Google Scholar]
- 53.Shi Y., Shen Y., Ahmad B., Yao L., He T., Fan J., Liu Y., Chen Q., Wen Z. Genome-wide Identification and Expression Analysis of Dirigent Gene Family in Strawberry (Fragaria vesca) and Functional Characterization of FvDIR13. Sci. Hortic. 2022;297:110913. doi: 10.1016/j.scienta.2022.110913. [DOI] [Google Scholar]
- 54.Casado-Díaz A., Encinas-Villarejo S., Santos B.d.l., Schilirò E., Yubero-Serrano E.-M., Amil-Ruíz F., Pocovi M.I., Pliego-Alfaro F., Dorado G., Rey M., et al. Analysis of strawberry genes differentially expressed in response to Colletotrichum infection. Physiol. Plant. 2006;128:633–650. doi: 10.1111/j.1399-3054.2006.00798.x. [DOI] [Google Scholar]
- 55.Jiu S., Haider M.S., Kurjogi M.M., Zhang K., Zhu X., Fang J. Genome-wide Characterization and Expression Analysis of Sugar Transporter Family Genes in Woodland Strawberry. Plant Genome. 2018;11:170103. doi: 10.3835/plantgenome2017.11.0103. [DOI] [PubMed] [Google Scholar]
- 56.Mouhu K., Hytonen T., Folta K., Rantanen M., Paulin L., Auvinen P., Elomaa P. Identification of flowering genes in strawberry, a perennial SD plant. BMC Plant Biol. 2009;9:122. doi: 10.1186/1471-2229-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yun-zhou Y., Juan D., Gang W., Jing J. Studies on the Freeze-Thaw Method of Transforming Recombinant Plasmid DNA into Agrobacterium tumefaciens. J. Jilin Agric. Univ. 2003;25:257–259. doi: 10.13327/j.jjlau.2003.03.006. [DOI] [Google Scholar]
- 58.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 59.Jiang W., Yang B., Weeks D.P. Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE. 2014;9:e99225. doi: 10.1371/journal.pone.0099225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No new data were created or analyzed in this study. Data sharing is not applicable to this article.