Moore et al. identify a new role for the ubiquitin ligase RFWD3 in promoting replication fork reversal. They demonstrate that RFWD3 promotes recruitment of the DNA translocase ZRANB3 to stalled replication forks by inducing polyubiquitination of PCNA and interaction with ZRANB3.
Abstract
Replication fork reversal is an important mechanism to protect the stability of stalled forks and thereby preserve genomic integrity. While multiple enzymes have been identified that can remodel forks, their regulation remains poorly understood. Here, we demonstrate that the ubiquitin ligase RFWD3, whose mutation causes Fanconi Anemia, promotes recruitment of the DNA translocase ZRANB3 to stalled replication forks and ubiquitinated sites of DNA damage. Using electron microscopy, we show that RFWD3 stimulates fork remodeling in a ZRANB3-epistatic manner. Fork reversal is known to promote nascent DNA degradation in BRCA2-deficient cells. Consistent with a role for RFWD3 in fork reversal, inactivation of RFWD3 in these cells rescues fork degradation and collapse, analogous to ZRANB3 inactivation. RFWD3 loss impairs ZRANB3 localization to spontaneous nuclear foci induced by inhibition of the PCNA deubiquitinase USP1. We demonstrate that RFWD3 promotes PCNA ubiquitination and interaction with ZRANB3, providing a mechanism for RFWD3-dependent recruitment of ZRANB3. Together, these results uncover a new role for RFWD3 in regulating ZRANB3-dependent fork remodeling.
Introduction
DNA replication is continually threatened by obstacles arising from both endogenous and exogenous sources that stall replication forks. To maintain genomic integrity, cells have evolved pathways to preserve the stability of stalled forks and facilitate the accurate restart of replication. One such mechanism involves the reversal of stalled replication forks into four-way junctions, in which the two nascent DNA strands anneal to form a fourth regressed arm (Neelsen and Lopes, 2015; Quinet et al., 2017). Replication fork reversal can be triggered by numerous types of replication stress, including DNA lesions, nucleotide depletion, DNA secondary structures, and topological constraints (Neelsen and Lopes, 2015; Ray Chaudhuri et al., 2012; Zellweger et al., 2015). Reversal can protect forks by allowing them to pause in a stable structure while an obstructing lesion is removed or until a converging fork completes replication of the damaged region. Fork reversal can also facilitate bypass of the lesion through a template-switching mechanism involving use of the complementary nascent DNA strand as an undamaged template for DNA synthesis (Cortez, 2019; Neelsen and Lopes, 2015; Quinet et al., 2021).
Multiple remodeling enzymes can catalyze the reversal of stalled replication forks (Neelsen and Lopes, 2015). Among the best characterized are the SNF2 family translocases ZRANB3 (Ciccia et al., 2012; Vujanovic et al., 2017), SMARCAL1 (Bétous et al., 2013; Bétous et al., 2012; Ciccia et al., 2012; Kolinjivadi et al., 2017; Taglialatela et al., 2017), and HLTF (Bai et al., 2020; Blastyák et al., 2010; Kile et al., 2015). For each of these enzymes, fork remodeling activity has been demonstrated in both in vitro assays and in cells. Consistent with a protective role for these translocases during replication fork metabolism in wild-type cells, their depletion impairs the restart of stalled replication forks (Blastyák et al., 2010; Ciccia et al., 2009; Ciccia et al., 2012; Yuan et al., 2012). ZRANB3 is recruited to stalled replication forks through the interaction of its NZF domain with K63-linked polyubiquitin chains on the DNA sliding clamp PCNA (Ciccia et al., 2012; Weston et al., 2012; Yuan et al., 2012). Monoubiquitination of PCNA is performed by the E3 ubiquitin ligase RAD18, while polyubiquitin chain extension is mediated by the ligases HLTF and SHPRH (Mailand et al., 2013). Previous studies have suggested the involvement of additional ubiquitin ligases, as mouse embryonic fibroblasts (MEFs) with inactivation of both HLTF and SHPRH can still generate polyubiquitinated PCNA (Krijger et al., 2011), and residual monoubiquitinated PCNA has been detected in RAD18-deficient cells (Simpson et al., 2006; Leung et al., 2018).
In cells deficient for the tumor suppressors BRCA1 and BRCA2, replication fork reversal can be a vulnerability for fork stability, as reversed forks are targets for degradation in these cells. BRCA1 and BRCA2 promote RAD51 filament formation, which protects nascent DNA at stalled replication forks from nucleolytic resection (Hashimoto et al., 2010; Schlacher et al., 2011; Schlacher et al., 2012). The regressed arms of reversed forks resemble one-ended double-strand breaks (DSBs) and are the structural intermediates targeted for degradation in BRCA1/2-deficient cells (Kolinjivadi et al., 2017; Lemaçon et al., 2017; Mijic et al., 2017; Taglialatela et al., 2017). BRCA1/2-dependent coating of these arms with RAD51 protects against nucleolytic exposure. Importantly, depletion of ZRANB3 and other fork reversal enzymes such as SMARCAL1 from BRCA1/2-deficient cells prevents the degradation of nascent DNA at stalled forks (Kolinjivadi et al., 2017; Mijic et al., 2017; Taglialatela et al., 2017), reduces genomic instability (Taglialatela et al., 2017), and promotes resistance to fork stalling agents (Duan et al., 2020; Taglialatela et al., 2017).
Biallelic germline mutation of the ubiquitin ligase RFWD3, which is also known as FANCW, causes the genomic instability syndrome Fanconi Anemia (FA; Knies et al., 2017). We previously demonstrated that RFWD3 ubiquitinates the single-stranded DNA (ssDNA) binding factor RPA to promote homologous recombination (HR) at stalled replication forks and replication fork restart (Elia et al., 2015). Ubiquitination occurred on numerous sites within multiple RPA subunits, reminiscent of protein group sumoylation that promotes protein recruitment in yeast HR (Psakhye and Jentsch, 2012), and we proposed that RFWD3-dependent ubiquitination of RPA may promote the localization of repair factors. We also considered that ubiquitination of RPA may displace it from ssDNA via extraction by the ubiquitin segregase VCP/p97. Consistent with this model, RFWD3 loss was reported to cause the persistence of RPA foci (Feeney et al., 2017; Inano et al., 2017) and to inhibit DNA damage-induced interaction of RPA with VCP (Inano et al., 2017). RFWD3-dependent displacement of RPA from a postsynaptic intermediate may be necessary for late steps in HR (Inano et al., 2017). More recently, a role for RFWD3 in the recruitment of TLS polymerases and translesion synthesis (TLS) in Xenopus oocytes has been described (Gallina et al., 2021).
Here, we demonstrate that RFWD3 promotes the recruitment of ZRANB3 to stalled replication forks in a manner that depends on its ubiquitin ligase activity. Through the analysis of replication intermediates by electron microscopy (EM), we show that RFWD3 stimulates the reversal of stalled forks in a ZRANB3-epistatic fashion. Consistent with a role for RFWD3 in fork reversal, we find that RFWD3 depletion in U2OS cells or RFWD3 mutation in FA patient cells rescues nascent DNA degradation, fork collapse into DSBs, and sensitivity to hydroxyurea upon BRCA2 depletion. These phenotypes are similar to those of ZRANB3 loss in BRCA2-deficient cells. RFWD3 inactivation impairs ZRANB3 localization to spontaneous foci induced by depletion of USP1, a deubiquitinase that acts on PCNA. We find that RFWD3 promotes PCNA ubiquitination and interaction with ZRANB3, shedding light on why ZRANB3 recruitment is dependent on RFWD3. Collectively, these results uncover a new role for RFWD3 in replication fork remodeling by promoting PCNA ubiquitination and ZRANB3 recruitment.
Results
Depletion of RFWD3 rescues HU sensitivity in BRCA2-deficient cells
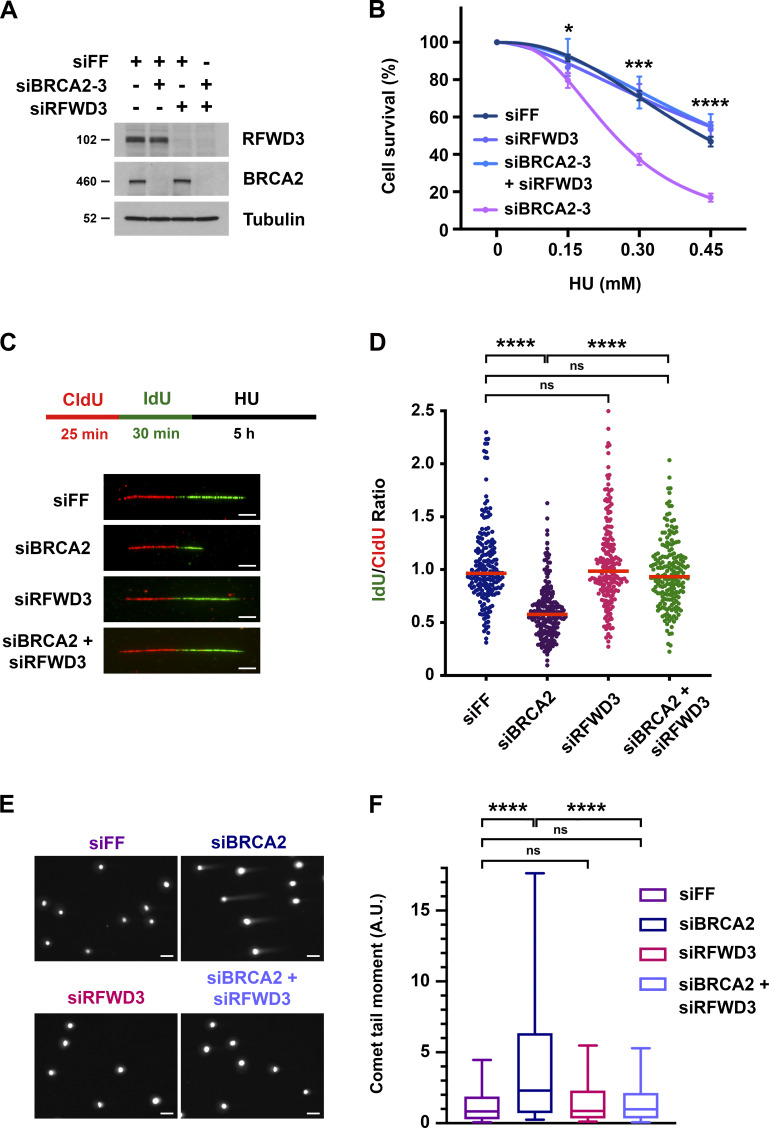
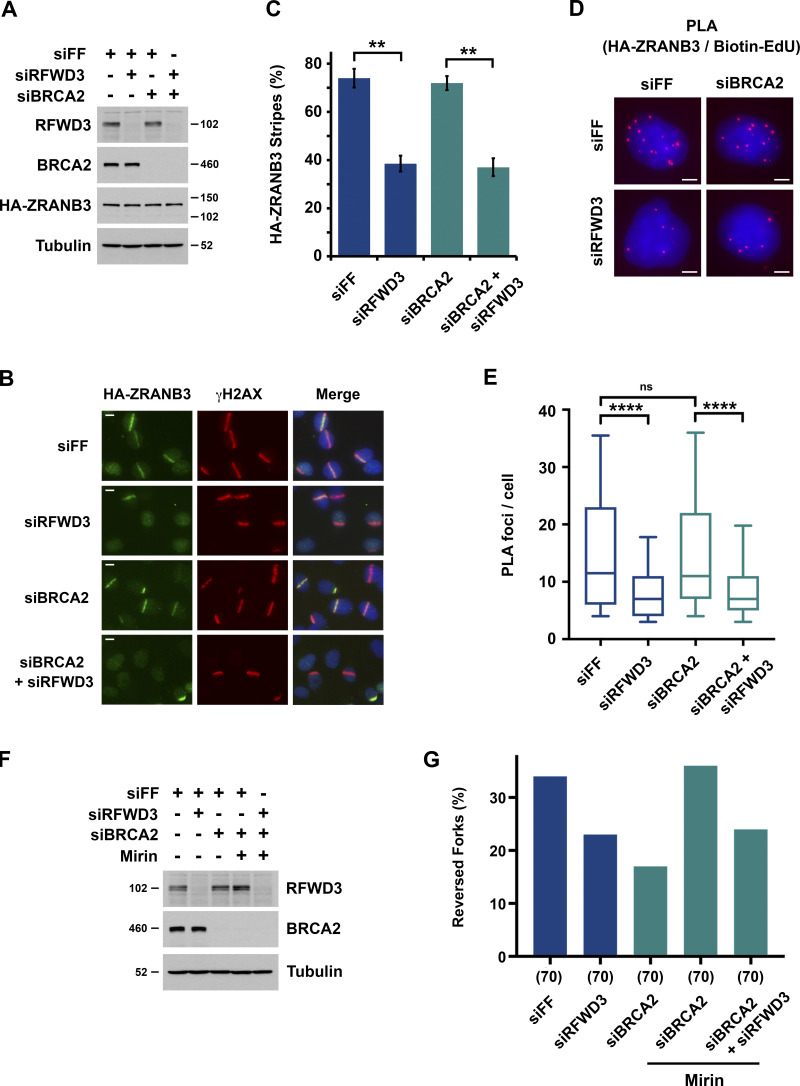
We previously demonstrated that the ubiquitin ligase RFWD3, which is mutated in FA, promotes homologous recombination (HR) at stalled replication forks (Elia et al., 2015). BRCA2 is another factor that functions in HR and whose biallelic mutation causes FA (Ceccaldi et al., 2016; Howlett et al., 2002; Kottemann and Smogorzewska, 2013). We wanted to better understand where RFWD3 functions in HR relative to BRCA2. We therefore performed epistasis experiments involving codepletion of BRCA2 and RFWD3 and examined sensitivity to the replication fork stalling agent hydroxyurea (HU). Depletion of BRCA2 in U2OS cells caused twofold sensitization to HU as determined by LC50 values (Fig. 1, A and B and Fig. S1, A and B) and consistent with prior results in BRCA2 mutant fibroblasts (Rickman et al., 2020). Remarkably, we saw that codepletion of RFWD3 was not epistatic with BRCA2 depletion nor amplified the sensitivity of BRCA2-depleted cells to HU, but rather reversed it (Fig. 1 B and Fig. S1, A and B), as observed recently (Duan et al., 2020). We validated this finding with independent siRNAs against BRCA2 and RFWD3 (Fig. S1, C and D). These results suggested that RFWD3 depletion reversed some functional defect caused by BRCA2 deficiency, and we next sought to identify that defect.
Figure 1.
Depletion of RFWD3 rescues HU sensitivity, nascent DNA degradation, and stalled fork collapse in BRCA2-deficient cells. (A) Detection of RFWD3 and BRCA2 levels in U2OS cells used in Fig. 1 B. (B) HU sensitivity of U2OS cells transfected with siBRCA2-3 and /or siRFWD3-4. Cell survival is normalized to the untreated control for each siRNA condition. Data represent the mean and SD of three replicates per HU dose and siRNA condition. Asterisks indicate P-values for RFWD3/BRCA2 versus BRCA2 depletion using an unpaired t test (*P < 0.05; ***P < 0.001; ****P < 0.0001). Data are representative of three independent experiments, for which mean LC50 values are provided in Fig. S1 B. (C) Schematic for single DNA fiber analysis to detect nascent DNA degradation at stalled forks. U2OS cells transfected with siBRCA2-3 and /or siRFWD3-4 were labeled with sequential CldU (25 min) and IdU (30 min) and then treated with 2 mM HU (5 h). Representative images are provided for replication tracks containing both CldU and IdU from cells transfected with the indicated siRNAs. Scale bars, 5 µm. (D) IdU/CldU replication track ratios in U2OS cells treated as in Fig. 1 C. Median values from >200 replication tracks are represented by red lines (n.s., not significant; ****P < 0.0001; Mann Whitney test). (E) Representative images of neutral comet tails in U2OS cells transfected with siBRCA2-3 and/or siRFWD3-4 and treated with HU for 24 h. Scale bars, 50 µm. (F) Box plot of neutral comet-tail moments in U2OS cells from Fig. 1 E. Whiskers represent the 10th and 90th percentiles. More than 300 cells were scored for each condition (n.s., not significant; ****P < 0.0001; Mann Whitney test).
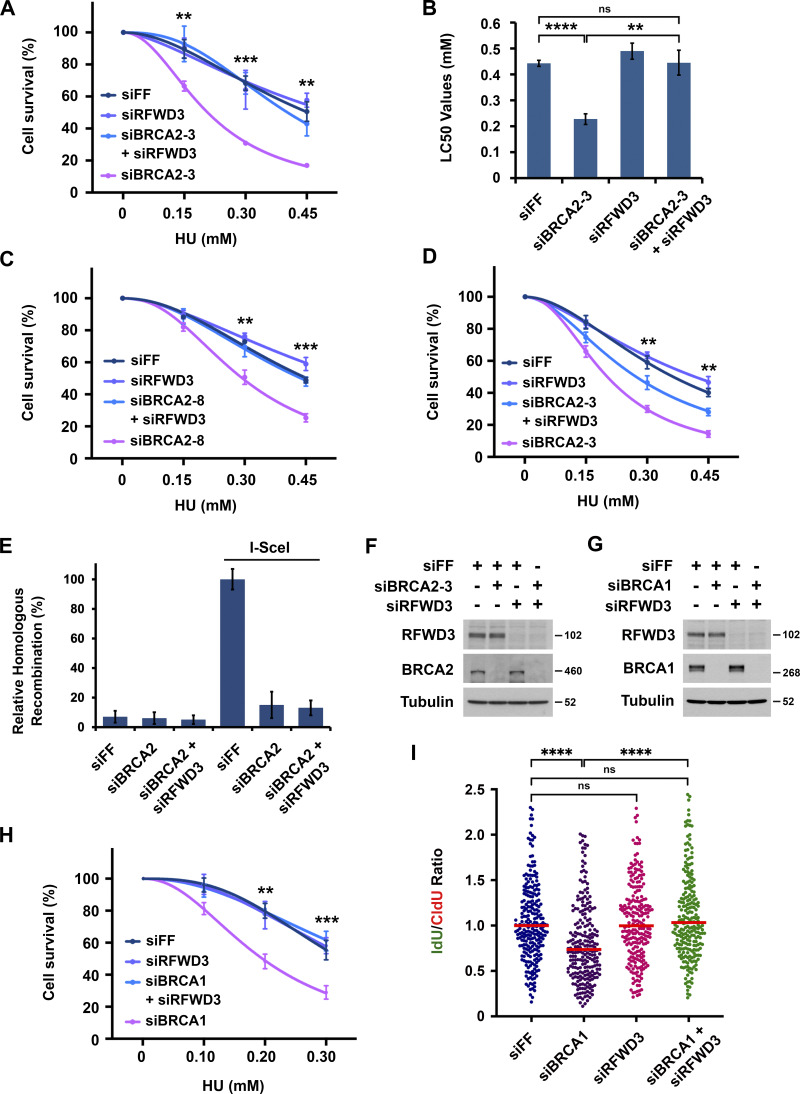
Figure S1.
Inactivation of RFWD3 reverses HU sensitivity and stalled fork degradation in BRCA1/2 -deficient cells. (A–D) Sensitivity of U2OS cells to HU upon transfection of the indicated siRNAs against BRCA2 and either siRFWD3-4 (Fig. S1, A–C) or siRFWD3-2 (Fig. S1 D). Cell survival is normalized to the untreated control for each siRNA condition. Data represent the mean and SD of three replicates per HU dose and siRNA condition. Asterisks on cell viability curves indicate P values for RFWD3/BRCA2 codepletion versus BRCA2 depletion using an unpaired t test (**P < 0.01; ***P < 0.001). LC50 values in Fig. S1 B are mean and SD from three independent experiments (**P < 0.01; ****P < 0.0001, unpaired t test). (E) U2OS cells with a stably integrated DR-GFP reporter were transfected with the indicated siRNAs and then infected with adenovirus expressing I-SceI to induce a DSB within the reporter. GFP-positive cells were detected by flow cytometry as a measure of homologous recombination efficiency. (F) Detection of BRCA2 and RFWD3 depletion in U2OS cells upon transfection of siBRCA2-3 and siRFWD3-4. Cells were used in Fig. 1 D. (G) Detection of BRCA1 and RFWD3 levels in U2OS cells upon transfection of siBRCA1 and siRFWD3-4. Cells were used in Fig. S1 H. (H) Sensitivity of U2OS cells to HU upon transfection of siBRCA1 and/or siRFWD3-4. Data are analyzed as in Fig. S1, A–D and represent the mean and SD of three replicates per HU dose and siRNA condition. Asterisks indicate P values for RFWD3/BRCA1 versus BRCA1 depletion using an unpaired t test (**P < 0.01; ***P < 0.001). (I) U2OS cells were transfected with siBRCA1 and/or siRFWD3-4. They were labeled with sequential CldU (25 min) and IdU (30 min) and then treated with 2 mM HU (5 h) as in Fig. 1 C. Median values for IdU/CldU track ratios (from >200 tracks) are represented by red lines (n.s., not significant; ****P < 0.0001; Mann Whitney test).
RFWD3 depletion rescues replication fork degradation and collapse in BRCA2-deficient cells
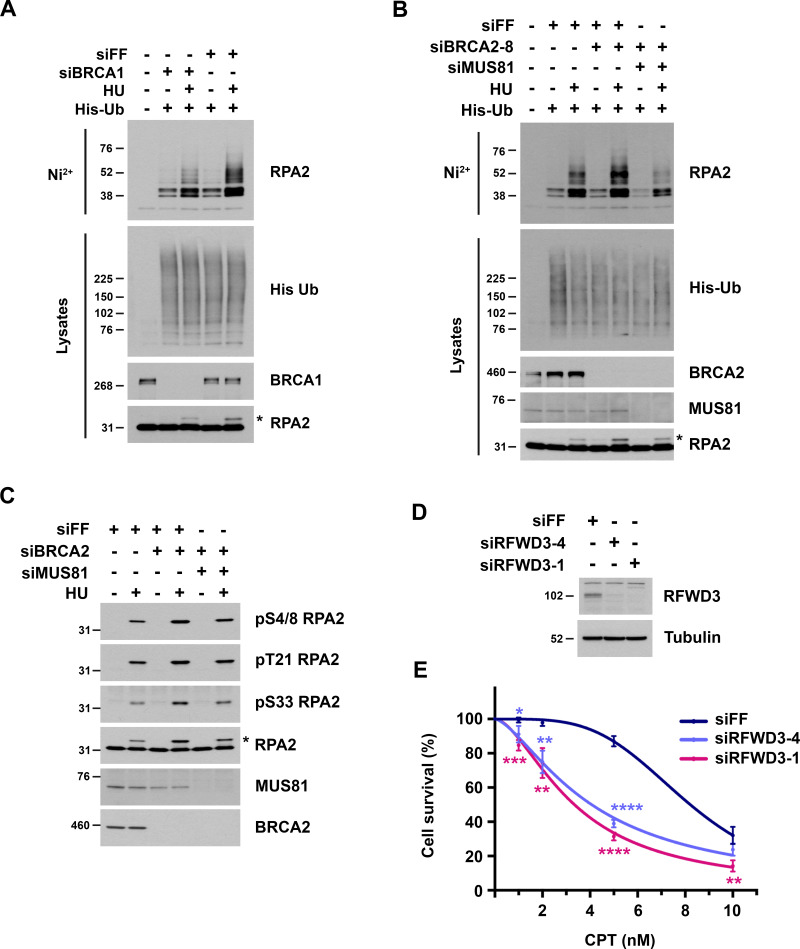
To determine the mechanism by which RFWD3 inactivation rescues HU sensitivity in BRCA2-deficient cells, we examined the effect of RFWD3 depletion on known functions of BRCA2, the first of which was homologous recombination. The DR-GFP reporter allows the measurement of HR at a DSB generated by the endonuclease I-SceI (Xia et al., 2006). Utilizing this reporter in U2OS cells, we found that BRCA2 depletion caused a decrease in HR as expected. This defect was not reversed by codepletion of RFWD3 (Fig. S1 E), which was also expected given that RFWD3 promotes HR itself. In addition to HR, BRCA2 is known to protect stalled replication forks from degradation by loading RAD51 onto the regressed DNA ends of reversed forks, which are targets for nucleases (Chen et al., 2018; Pasero and Vindigni, 2017; Rickman and Smogorzewska, 2019). To examine fork protection mediated by BRCA2, we therefore measured nascent DNA degradation at stalled forks using single DNA fiber analysis (Fig. 1 C). U2OS cells were sequentially pulsed with the nucleotide analogs chlorodeoxyuridine (CldU) and iododeoxyuridine (IdU) and then treated with HU for 5 h to stall forks. The lengths of DNA fiber tracks with continuous IdU and CldU staining were measured, and fork degradation was detected by a decrease in the IdU/CldU length ratio. As expected (Schlacher et al., 2011), the IdU/CldU ratio decreased nearly twofold upon depletion of BRCA2. It did not change appreciably upon depletion of RFWD3 alone but was completely reversed in BRCA2-deficient cells by RFWD3 codepletion (Fig. 1 D and Fig. S1 F). In addition to BRCA2, BRCA1 is known to protect against fork degradation (Schlacher et al., 2012) by promoting RAD51 loading onto reversed forks. We found that RFWD3 depletion also rescued nascent DNA degradation (Fig. S1 I) and HU sensitivity (Fig. S1 H) in BRCA1-deficient cells.
Degradation of stalled replication forks in BRCA2-deficient cells can lead to fork collapse and genomic instability. Prior studies have demonstrated that in the presence of HU, BRCA2 deficiency can cause the formation of double-strand breaks (DSBs) and chromosome aberrations, which can be rescued by inhibition of the nucleases MRE11 or EXO1 (Lemaçon et al., 2017; Ray Chaudhuri et al., 2016; Schlacher et al., 2011). Using neutral comet assay, we examined the generation of DSBs in BRCA2-deficient cells treated with HU to stall forks. Consistent with published data, we found that BRCA2 depletion increased the mean comet tail moment representative of DSB accumulation. Importantly, codepletion of RFWD3 completely reversed this increase in comet tail length (Fig. 1, E and F). These results demonstrate that inactivation of RFWD3 in BRCA2-deficient cells can rescue not only the degradation of stalled forks but their collapse into DSBs, consistent with our results showing the rescue of HU sensitivity upon RFWD3 depletion in these cells.
Mutations in the RFWD3 ubiquitin ligase and WD40 domains rescue nascent DNA degradation in BRCA2-deficient cells
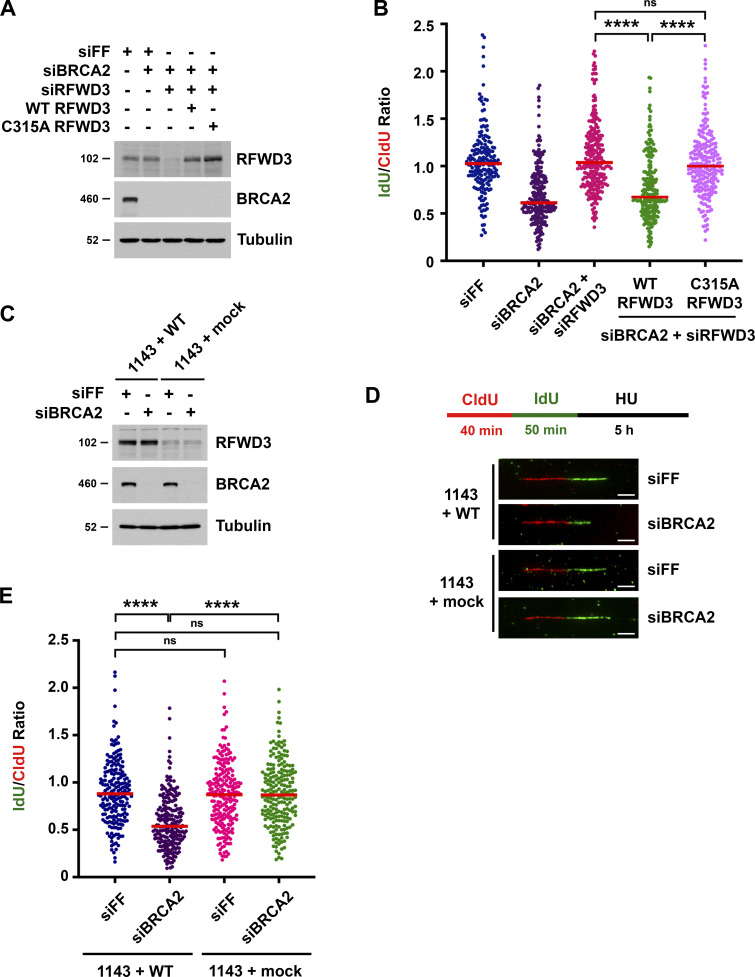
RFWD3 possesses an N-terminal RING domain that is responsible for its ubiquitin ligase activity (Fu et al., 2010) and a C-terminal WD40 domain that mediates its recruitment to nuclear foci in response to HU and mitomycin C (Feeney et al., 2017; Knies et al., 2017; Liu et al., 2011). To determine whether the ubiquitin ligase activity of RFWD3 is necessary for promoting nascent DNA degradation in BRCA2-deficient cells, we complemented RFWD3-depleted cells with siRNA-resistant WT and RING-mutant (C315A) RFWD3. WT RFWD3 restored fork degradation in BRCA2-deficient cells whereas C315A RFWD3 did not (Fig. 2, A and B). We also examined nascent DNA degradation upon BRCA2 depletion in immortalized skin fibroblasts (1143 cells) from an FA patient with RFWD3 mutation (Knies et al., 2017). One allele in these cells contains a missense mutation (I639K) in the WD40 domain of RFWD3, while the other allele does not express a protein product due to a frameshift resulting in premature termination prior to the RING domain. BRCA2 depletion induced nascent DNA degradation in 1143 cells complemented with WT RFWD3 (1143 + WT) but not in cells complemented with empty vector (1143 + mock; Fig. 2, C–E). Together, these findings demonstrate that the ubiquitin ligase and WD40 domains of RFWD3 are necessary for promoting fork degradation upon BRCA2 loss.
Figure 2.
Mutations in the RFWD3 ubiquitin ligase and WD40 domains rescue nascent DNA degradation in BRCA2-deficient cells. (A) Detection of RFWD3 and BRCA2 levels in U2OS cells for the experiment in Fig. 2 B. (B) U2OS cells expressing siRNA-resistant RFWD3 (WT or C315A) were transfected with siBRCA2-3 and siRFWD3-4, and they were compared with U2OS cells transfected with siBRCA2-3 with or without siRFWD3-4. Cells were labeled with sequential CldU (25 min) and IdU (30 min) and then treated with 2 mM HU (5 h) as in Fig. 1 C. Median values for IdU/CldU track ratios (from >200 tracks) are represented by red lines (n.s., not significant; ****P < 0.0001; Mann Whitney test). (C) Detection of RFWD3 and BRCA2 levels in FA patient fibroblasts for the experiment in Fig. 2 D. (D) Schematic for single DNA fiber analysis to detect nascent DNA degradation at stalled forks in FA patient fibroblasts complemented with WT RFWD3 (1143 + WT) or empty vector (1143 + mock). Cells were labeled with sequential CldU (40 min) and IdU (50 min) and then treated with 2 mM HU (5 h). Representative images are provided for CldU and IdU-containing replication tracks upon transfection with siFF or siBRCA2-3. Scale bars, 5 µm. (E) IdU/CldU replication length ratios in FA patient fibroblasts treated as in Fig. 2 D. Median values from >200 replication tracks are represented by red lines (n.s., not significant; ****P < 0.0001; Mann Whitney test).
RFWD3 promotes recruitment of the translocase ZRANB3 to ubiquitinated sites of DNA damage
We next sought to understand why RFWD3 inactivation rescued fork degradation and collapse in BRCA2-deficient cells. These phenotypes were reminiscent of those involving depletion of the fork remodeling enzyme ZRANB3 in BRCA2-deficient cells. ZRANB3 contains a SNF2 family translocase domain that catalyzes the reversal of stalled replication forks (Ciccia et al., 2012; Vujanovic et al., 2017) and thereby promotes fork degradation in BRCA2-deficient cells. Depletion of ZRANB3 rescues nascent DNA degradation (Mijic et al., 2017; Taglialatela et al., 2017) and replication fork collapse (Taglialatela et al., 2017) in these cells. ZRANB3 also plays an important role in replication fork remodeling in wild-type cells, where its fork reversal activity can facilitate the restart of stalled forks (Ciccia et al., 2012). We previously found that RFWD3 depletion inhibits replication fork restart in wild-type cells (Elia et al., 2015), representing an additional resemblance between phenotypes observed upon loss of RFWD3 or ZRANB3.
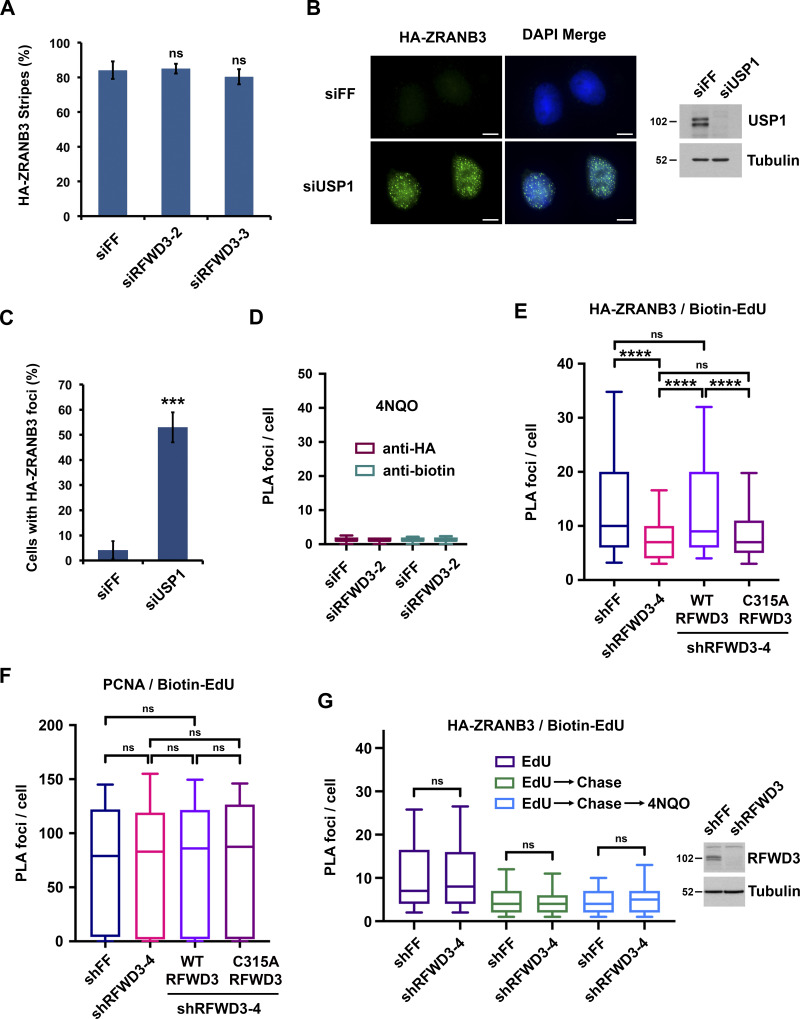
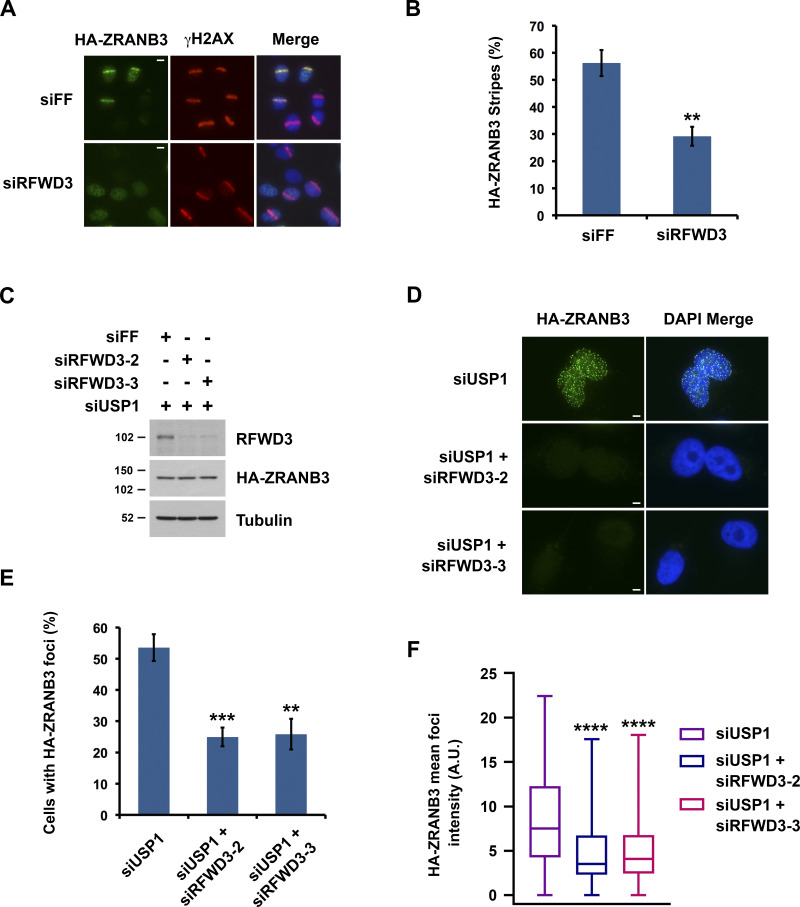
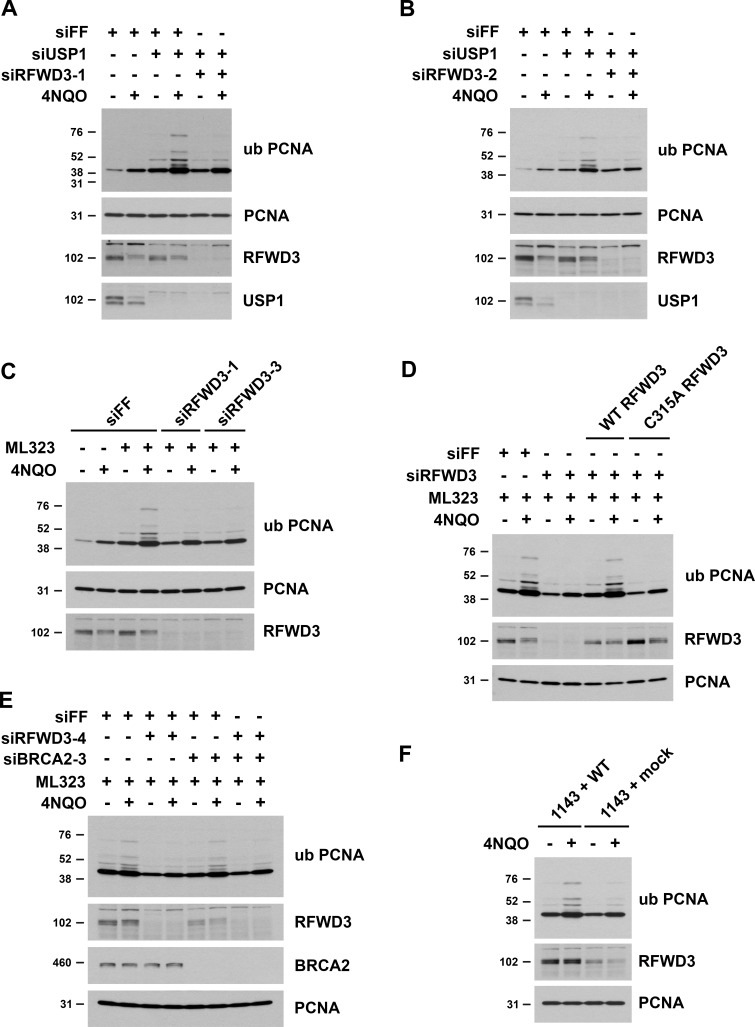
Similarity between RFWD3 and ZRANB3 inactivation phenotypes suggested that RFWD3 may regulate ZRANB3. Notably, ZRANB3 has a ubiquitin-binding NZF domain and is known to be recruited to stalled replication forks via interaction of this domain with ubiquitinated PCNA. We hypothesized that RFWD3 may promote the recruitment of ZRANB3 to stalled replication forks. To investigate this possibility, we initially examined localization of HA-tagged ZRANB3 to sites of UV laser microirradiation. Recruitment of ZRANB3 to such sites is known to depend on the binding of PCNA to multiple regions within ZRANB3. The PIP box and APIM motifs of ZRANB3 mediate ubiquitin-independent interactions, while the NZF domain of ZRANB3 associates with K63-linked polyubiquitinated PCNA (Ciccia et al., 2012; Weston et al., 2012). ZRANB3 is transiently recruited to laser-induced stripes with maximal localization at 5–15 min and reduced localization at 30 min. Depletion of the K63-specific E2 conjugating enzyme UBC13 reduces ZRANB3 recruitment at 30 min but not 5 min. This finding led to the model that initial ZRANB3 recruitment is ubiquitin-independent and relies on interaction of its PIP box and APIM motif with non-modified PCNA, while subsequent retention of ZRANB3 depends on interaction of its NZF domain with ubiquitinated PCNA (Ciccia et al., 2012). We found that depletion of RFWD3 from U2OS cells did not affect HA-ZRANB3 recruitment to laser stripes at 10 min (Fig. S2 A) but reduced its localization twofold at 30 min (Fig. 3, A and B). This level of reduction is consistent with that for UBC13 depletion (Ciccia et al., 2012) and supports the model that RFWD3 promotes the retention of ZRANB3 at DNA damage sites.
Figure S2.
Effect of RFWD3 depletion on ZRANB3 and PCNA recruitment. (A) U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs, fixed 10 min after UV laser irradiation, and stained with anti-HA and anti-γH2AX antibodies. The percentage of γH2AX-positive cells with HA-ZRANB3 colocalization at laser-generated stripes was quantitated. Data represent the mean and SD from three independent experiments (n.s., not significant; unpaired t test). (B) U2OS cells expressing HA-ZRANB3 were transfected with siUSP1 to induce ZRANB3 localization to nuclear foci in the absence of exogenous DNA damage. Foci were detected by staining with anti-HA antibody (green). Immunoblot shows the level of USP1 depletion. Scale bars, 10 µm. (C) Percentage of U2OS cells with more than ten HA-ZRANB3 foci upon transfection of siUSP1 as described in Fig. S2 B. Data represent the mean and SD from two independent experiments (***P < 0.001; unpaired t test). (D) U2OS cells expressing HA-ZRANB3 were treated as in Fig. 4 B except PLA was performed with either anti-HA or anti-biotin antibody alone as negative controls. All conditions were treated with 4NQO. Whiskers on box plots represent the 10th and 90th percentiles for >200 cells. (E) U2OS cells expressing HA-ZRANB3 were transduced with shRFWD3-4 or non-targeting shFF along with either shRNA-resistant WT or C315A RFWD3. They were pulsed with 10 µM EdU for 10 min, treated with 4 mM HU and 10 µM EdU for 4 h, and fixed. Biotin was conjugated to EdU by click chemistry, and PLA was performed with anti-HA and anti-biotin antibodies. Whiskers on box plots (>200 cells) represent the 10th and 90th percentiles (n.s., not significant; ****P < 0.0001; Mann Whitney test). (F) U2OS cells expressing HA-ZRANB3 were treated as in Fig. 4, D–F except PLA was performed with anti-PCNA and anti-biotin antibodies. Whiskers on box plots (>200 cells) represent the 10th and 90th percentiles (n.s., not significant; Mann Whitney test). RFWD3 depletion efficiency is provided in Fig. 4 D. (G) U2OS cells expressing HA-ZRANB3 were transduced with the indicated shRNAs, pulsed with 15 µM EdU for 10 min, chased with 15 µM thymidine for 2 h, treated with 1 µg/ml 4NQO for 4 h, and fixed. Biotin was conjugated to EdU, and PLA was performed with anti-HA and anti-biotin antibodies. Whiskers on box plots (>200 cells) represent the 10th and 90th percentiles (n.s., not significant; Mann Whitney test). Immunoblot shows level of RFWD3 depletion.
Figure 3.
RFWD3 promotes the recruitment of ZRANB3 to ubiquitinated sites of DNA damage. (A) Localization of HA-ZRANB3 to DNA damage sites generated by UV laser microirradiation. U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs, fixed 30 min after laser irradiation, and stained with anti-HA (green) and anti-γH2AX (red) antibodies. Images are representative of results quantitated in Fig. 3 B. Scale bars, 10 µm. (B) Graph showing the percentage of γH2AX-positive U2OS cells with HA-ZRANB3 colocalization at UV laser stripes. Data correspond to conditions in Fig. 3 A and represent the mean and SD (**P < 0.01, unpaired t test). (C) Detection of RFWD3 and HA-ZRANB3 levels in U2OS cells used in Fig. 3, E and F. Immunoblot was performed with antibodies against RFWD3 and HA. (D) U2OS cells expressing HA-ZRANB3 were transfected with siUSP1 to induce ZRANB3 localization to nuclear foci in the absence of exogenous DNA damage. Cells were also co-transfected with the indicated control or RFWD3 siRNAs and stained with anti-HA antibody (green). Scale bars, 5 µm. (E) Percentage of U2OS cells with more than ten HA-ZRANB3 foci upon co-transfection of siUSP1 and the indicated siRNAs, as described in Fig. 3 D. Data represent the mean and SD from three independent experiments (**P < 0.01; ***P < 0.001; unpaired t test). (F) U2OS cells expressing HA-ZRANB3 were transfected as in Fig. 3 D. The mean intensity of HA-ZRANB3 foci in each cell is depicted as a box plot for a population of more than 200 cells for each siRNA condition. Whiskers represent the minimum and maximum (n.s., not significant; ****P < 0.0001; Mann Whitney test).
We next set out to determine whether RFWD3 promotes the localization of ZRANB3 to ubiquitinated sites of DNA damage. It is known that ZRANB3 spontaneously localizes to nuclear foci upon depletion of the deubiquitinase USP1 (Ciccia et al., 2012), whose known substrates include PCNA (Huang et al., 2006; Kee and Huang, 2015). We confirmed that USP1 depletion induced the localization of HA-ZRANB3 to nuclear foci in the absence of exogenous DNA damage (Fig. S2, B and C). Importantly, codepletion of RFWD3 using two independent siRNAs reduced the percentage of cells with HA-ZRANB3 nuclear foci by a factor of two (Fig. 3, C–E). RFWD3 depletion with both siRNAs also caused a twofold decrease in the mean intensity of HA-ZRANB3 foci (Fig. 3 F). These results suggest that RFWD3 promotes the localization of ZRANB3 to ubiquitinated substrates of USP1 at DNA damage sites.
RFWD3 promotes the recruitment of ZRANB3 to stalled replication forks
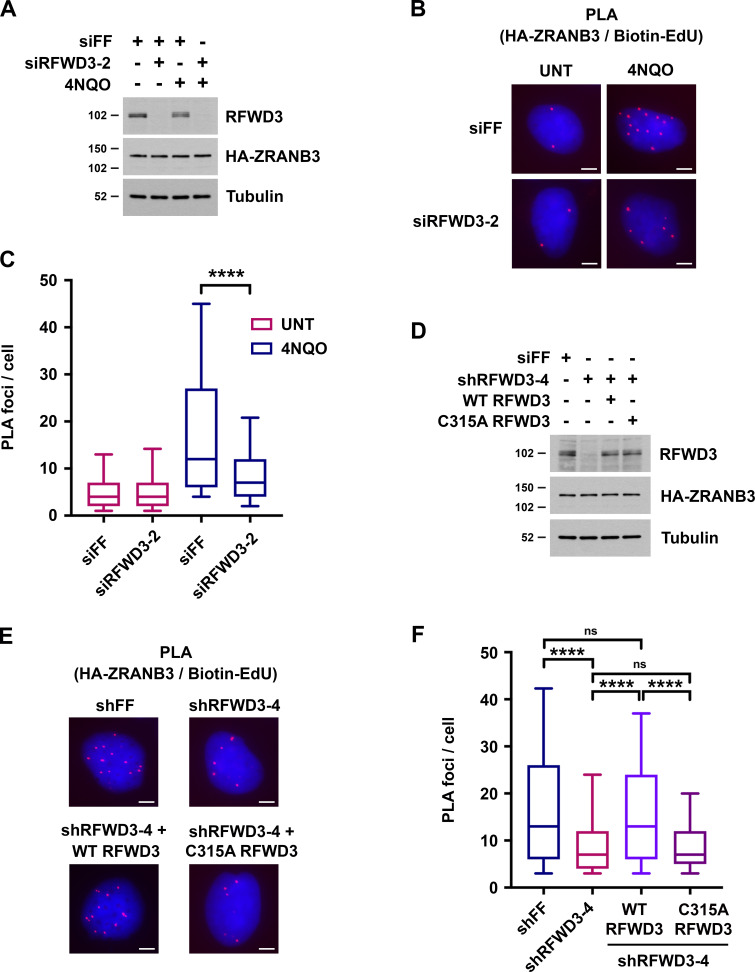
We next sought to determine whether RFWD3 promotes ZRANB3 recruitment to stalled replication forks. For this purpose, we used a proximity ligation assay (PLA) to measure colocalization of ZRANB3 with nascent DNA at stalled forks (Petruk et al., 2012; Roy et al., 2018; Taglialatela et al., 2017). U2OS cells expressing HA-ZRANB3 were treated with EdU for 10 min to label nascent DNA and then with the UV mimetic 4NQO for 4 h to stall forks. Biotin was conjugated to EdU by click chemistry, and PLA was performed with anti-HA and anti-biotin antibodies. We found that ZRANB3 was recruited to PLA foci representing biotin-labeled nascent DNA at stalled forks in the presence of 4NQO (Fig. 4, B and C). As a control, we confirmed that PLA foci were negligible in 4NQO-treated cells when either anti-HA or anti-biotin were used alone in the assay (Fig. S2 D). Importantly, RFWD3 depletion decreased PLA foci in 4NQO-treated cells by a factor of two (Fig. 4, A–C), consistent with the magnitude by which RFWD3 loss reduced ZRANB3 localization to laser stripes (Fig. 3 B) and ubiquitinated USP1 targets (Fig. 3, E and F). RFWD3 depletion with an independent shRNA also caused a twofold decrease in PLA foci (Fig. 4, D–F). Complementation of these cells with shRNA-resistant WT RFWD3 rescued ZRANB3 recruitment to 4NQO-stalled forks, whereas complementation with C315A RFWD3 did not (Fig. 4, D–F). RFWD3 depletion also caused a twofold decrease in ZRANB3 recruitment to HU-stalled forks, and this decrease was similarly rescued by complementation of RFWD3-depleted cells with WT but not C315A RFWD3 (Fig. S2 E). These data demonstrate that the ubiquitin ligase activity of RFWD3 is necessary for promoting ZRANB3 recruitment. As a control, we showed that RFWD3 depletion did not affect PCNA loading onto nascent DNA at stalled forks, as detected by PLA with anti-PCNA and anti-biotin antibodies (Fig. S2 F). We also showed that RFWD3 depletion did not affect ZRANB3 localization to chromatin when EdU labeling was chased with thymidine for 2 h (Fig. S2 G). Together, these findings demonstrate that RFWD3 promotes the recruitment of ZRANB3 to stalled replication forks in a ubiquitin-dependent manner.
Figure 4.
RFWD3 promotes ZRANB3 recruitment to nascent DNA at stalled replication forks. (A) Detection of RFWD3 and HA-ZRANB3 levels in U2OS cells used in Fig. 4, B and C. Immunoblot was performed with antibodies against RFWD3 and HA. (B) U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs, pulsed with 10 µM EdU for 10 min, treated with 1 µg/ml 4NQO for 4 h, and fixed. Biotin was conjugated to EdU by click chemistry, and proximity ligation assay (PLA) was performed with anti-HA and anti-biotin antibodies. Images are representative of results quantitated in Fig. 4 C. Scale bars, 5 µm. (C) Box plot depicting PLA foci per cell for each condition (>200 cells) in Fig. 4 B. Whiskers represent the 10th and 90th percentiles (****P < 0.0001, Mann Whitney test). (D) Detection of RFWD3 and HA-ZRANB3 levels in U2OS cells used in Fig. 4, E and F, and Fig. S2 F. Immunoblot was performed with antibodies against RFWD3 and HA. (E) U2OS cells expressing HA-ZRANB3 were transduced with shRFWD3-4 or non-targeting shFF along with either shRNA-resistant WT or C315A RFWD3. They were then pulsed with 10 µM EdU for 10 min, treated with 1 µg/ml 4NQO for 4 h, and fixed. Biotin was conjugated to EdU, and PLA was performed with antibodies against HA and biotin. Scale bars, 5 µm. (F) Box plot of PLA foci per cell for each condition (>200 cells) in Fig. 4 E. Whiskers represent the 10th and 90th percentiles (n.s., not significant; ****P < 0.0001; Mann Whitney test).
RFWD3 promotes the reversal of stalled replication forks
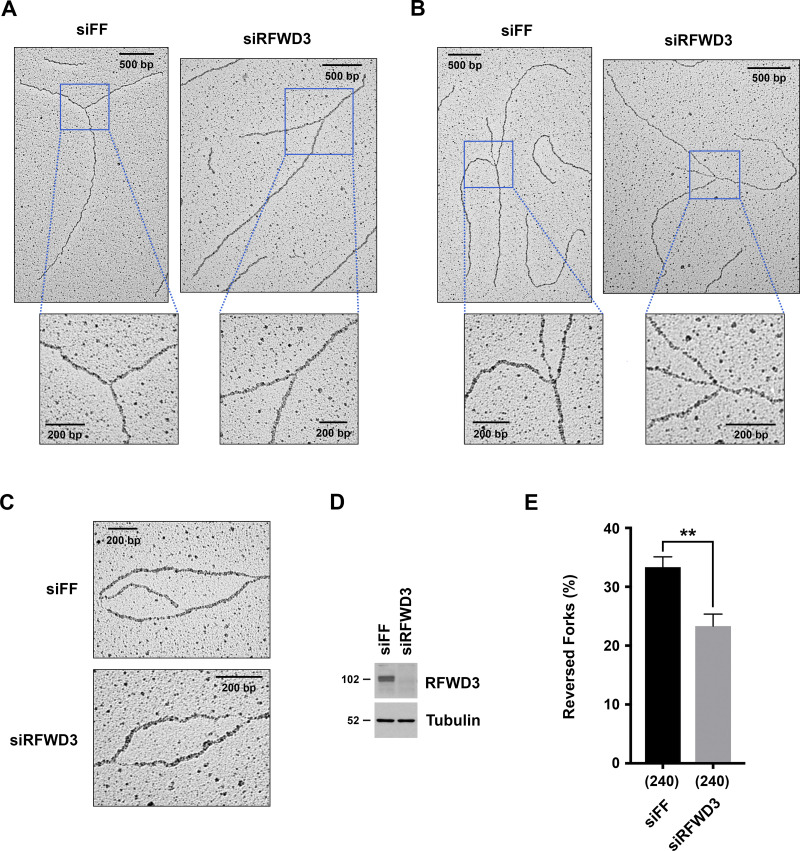
Decreased localization of ZRANB3 to stalled replication forks upon depletion of RFWD3 may explain our observation that replication fork degradation is rescued in BRCA2-deficient cells upon RFWD3 loss. This model predicts that RFWD3 should promote the reversal of stalled replication forks. To examine this possibility, we directly visualized replication intermediates at high resolution by electron microscopy and determined the percentage of reversed forks upon fork stalling. U2OS cells were treated with 2 mM HU for 5 h, and DNA structures preserved by psoralen crosslinking, including replication bubbles, were imaged (Fig. 5, A–C and Fig. S3, A and B). Treatment with HU yielded a high frequency of reversed forks as reported previously for U2OS cells (Krishnamoorthy et al., 2021; Lemaçon et al., 2017; Zellweger et al., 2015). Importantly, we found that RFWD3 depletion in three independent experiments significantly decreased this percentage (Fig. 5, D and E and Fig. S3 C). These results demonstrate that RFWD3 promotes the HU-induced reversal of stalled forks.
Figure 5.
RFWD3 promotes the reversal of stalled replication forks. (A and B) Representative images of normal (A) and reversed (B) replication forks detected by electron microscopy (EM) in control or RFWD3-depleted U2OS cells upon HU treatment. (C) Representative images of DNA replication bubbles detected by EM in HU-treated control and RFWD3-depleted U2OS cells. (D) Detection of RFWD3 levels in U2OS cells transfected with siRFWD3-4 for the experiment in Fig. 5 E. See Fig. S3 C for the depletion efficiency of other replicates used in Fig. 5 E. (E) Percentage of reversed replication forks detected by EM in control and RFWD3-depleted U2OS cells upon HU treatment (2 mM for 5 h). The mean and SEM from three independent experiments are shown (**P < 0.01, t test, see Fig. S3 C). The number of replication intermediates analyzed for each condition is indicated in parentheses.
Figure S3.
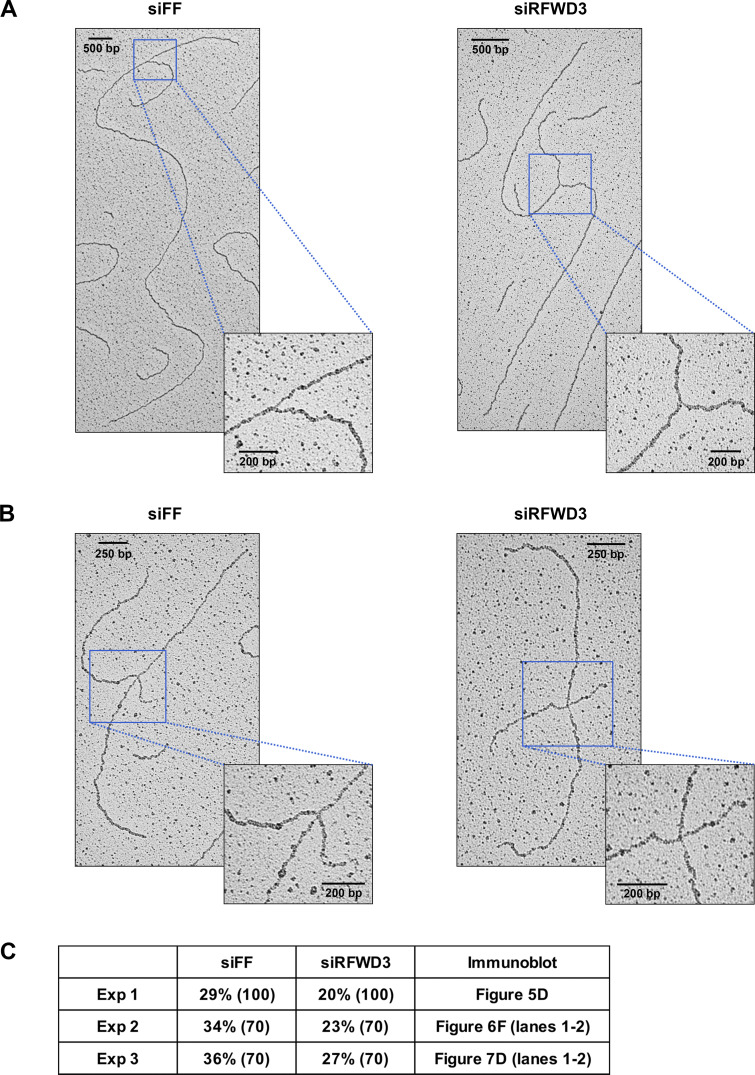
RFWD3 promotes the reversal of stalled replication forks. (A and B) Representative images of normal (A) and reversed (B) replication forks detected by electron microscopy (EM) in control or RFWD3-depleted U2OS upon HU treatment (2 mM for 5 h). Quantitation of reversed forks is provided in Fig. 5 E and Fig. S3 C. (C) Percentage of reversed replication forks identified by EM in the three independent experiments quantitated in Fig. 5 E. The number of replication intermediates is indicated in parentheses. Immunoblot figures showing the efficiency of RFWD3 depletion for each experiment are specified. Decrease in reversed forks correlates with the efficiency of RFWD3 depletion, which is weaker for the third experiment. Reversed fork values for the siFF and siRFWD3 conditions are effectively paired, as the first experiment was performed with a less potent preparation of HU (siFF vs. siRFWD3, P < 0.01 by paired t test, P < 0.05 by unpaired t test).
RFWD3 stimulates ZRANB3 recruitment and replication fork reversal in BRCA2-deficient cells
We propose that RFWD3 depletion rescues nascent DNA degradation in BRCA2-deficient cells by inhibiting ZRANB3-dependent fork reversal. This model relies on preservation of the functional relationship between RFWD3 and ZRANB3 in a BRCA2-deficient background. We therefore investigated whether RFWD3 promotes ZRANB3 recruitment and replication fork remodeling in not only BRCA2-proficient but also BRCA2-deficient cells. We found that RFWD3 depletion decreased HA-ZRANB3 localization to UV laser stripes to the same extent in both the presence and absence of BRCA2 codepletion (Fig. 6, A–C). We also performed PLA to examine ZRANB3 localization to EdU-labeled nascent DNA at stalled forks in BRCA2-depleted cells. Again, we found that RFWD3 depletion decreased ZRANB3 recruitment to 4NQO-stalled forks by the same magnitude in both BRCA2-deficient and proficient cells (Fig. 6, D and E).
Figure 6.
RFWD3 stimulates ZRANB3 recruitment and replication fork reversal in BRCA2-deficient cells. (A) Immunoblot showing RFWD3, BRCA2, and HA-ZRANB3 levels in U2OS cells used in Fig. 6, B–E. HA-ZRANB3 was detected with anti-HA antibody. (B) U2OS cells expressing HA-ZRANB3 were transfected with siRFWD3-2 and/or siBRCA2-3, fixed 30 min after UV laser irradiation, and stained with anti-HA (green) and anti-γH2AX (red) antibodies. Scale bars, 10 µm. (C) Graph showing the percentage of γH2AX-positive cells with HA-ZRANB3 colocalization at UV laser stripes corresponding to the experiment in Fig. 6 B. Data represent the mean and SD from two independent experiments (**P < 0.01, unpaired t test). (D) U2OS cells expressing HA-ZRANB3 were transfected with siRFWD3-2 and/or siBRCA2-3 and treated with 10 µM EdU for 10 min followed by 1 µg/ml 4NQO for 4 h. After cell fixation, biotin was conjugated to EdU by click chemistry, and proximity ligation assay (PLA) was performed with anti-HA and anti-biotin antibodies. Images are representative of results quantitated in Fig. 6 E. Scale bars, 5 µm. (E) Box plot showing the distribution of PLA foci per cell for each condition (>200 cells) in Fig. 6 D. Whiskers represent the 10th and 90th percentiles (****P < 0.0001, Mann Whitney test). (F) Detection of RFWD3 and BRCA2 levels in U2OS cells used in Fig. 6 G. (G) U2OS cells were transfected with siRFWD3-4 and/or siBRCA2-3 and treated with 2 mM HU for 5 h. Where indicated, 50 µM mirin was added concurrently with HU. Replication intermediates were detected by electron microscopy, and the percentage of reversed forks was measured in a single replicate. The number of replication intermediates analyzed for each condition is indicated in parentheses.
We next determined whether RFWD3 promotes replication fork remodeling in a BRCA2-deficient background. BRCA2 depletion is known to cause MRE11-dependent degradation of reversed replication forks detected by electron microscopy (Lemaçon et al., 2017; Mijic et al., 2017). To examine fork remodeling in BRCA2-depleted cells, we therefore protected reversed forks by adding the MRE11 inhibitor mirin during HU treatment. Consistent with prior reports (Lemaçon et al., 2017; Mijic et al., 2017), BRCA2 depletion reduced reversed forks twofold, and this decrease was fully rescued by the addition of mirin (Fig. 6, F and G). Importantly, we found that RFWD3 depletion decreased reversed forks to the same extent in both the presence and absence of BRCA2 codepletion (Fig. 6, F and G). In summary, these findings demonstrate that regulation of ZRANB3 recruitment and fork remodeling by RFWD3 is preserved in a BRCA2-deficient background.
RFWD3 and ZRANB3 epistasis in replication fork remodeling phenotypes
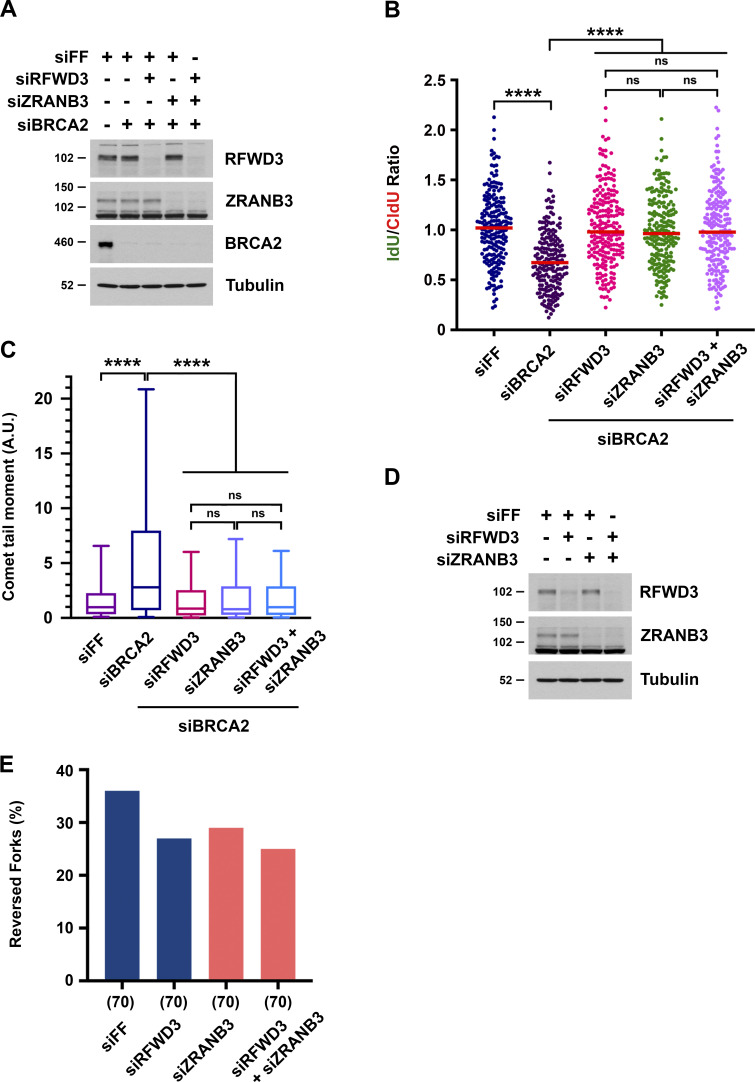
RFWD3 and ZRANB3 depletion lead to similar phenotypes, with loss of either protein rescuing both nascent DNA degradation and replication fork collapse in BRCA2-deficient cells. For ZRANB3 loss, these phenotypes have been attributed to suppression of the reversed fork substrate targeted by nucleases. For RFWD3 loss, we propose these phenotypes also result from impaired fork remodeling by ZRANB3. To further characterize whether RFWD3 and ZRANB3 function in the same pathway, we performed epistasis experiments involving their codepletion and examined fork degradation and collapse in BRCA2-deficient cells. Individual or combined depletion of RFWD3 and ZRANB3 suppressed both nascent DNA degradation (Fig. 7, A and B) and fork collapse into DSBs (Fig. 7 C) by the same magnitude. We also performed epistasis experiments examining replication fork reversal by electron microscopy. ZRANB3 depletion decreased the percentage of reversed forks observed upon treatment with HU. Importantly, we found that RFWD3 depletion decreased fork reversal to the same extent as either ZRANB3 depletion by itself or combined depletion of both RFWD3 and ZRANB3 (Fig. 7, D and E), consistent with an epistatic relationship between these factors. Together, these results suggest that RFWD3 functions in the same fork remodeling pathway as ZRANB3.
Figure 7.
RFWD3 and ZRANB3 epistasis in replication fork remodeling phenotypes. (A) Detection of RFWD3, ZRANB3, and BRCA2 levels in U2OS cells used in Fig. 7, B and C. (B) U2OS cells were transfected with siBRCA2-3, siRFWD3-4, and/or siZRANB3. They were labeled with sequential CldU (25 min) and IdU (30 min) and then treated with 2 mM HU (5 h) as in Fig. 1 C. Median values for IdU/CldU track ratios (from >200 tracks) are represented by red lines (n.s., not significant; ****P < 0.0001; Mann Whitney test). (C) Neutral comet-tail moments in U2OS cells transfected with siBRCA2-3, siRFWD3-4, and/or siZRANB3 and treated with 2 mM HU for 24 h. Whiskers represent the 10th and 90th percentiles. More than 200 cells were analyzed for each condition (****P < 0.0001, Mann Whitney test). (D) Detection of RFWD3 and ZRANB3 levels in U2OS cells used in Fig. 7 E. (E) U2OS cells transfected with siRFWD3-4 and/or siZRANB3 were treated with 2 mM HU for 5 h. Replication intermediates were detected by electron microscopy, and the percentage of reversed forks was measured in a single replicate. The number of replication intermediates analyzed for each condition is indicated in parentheses.
RFWD3 does not promote interaction of ZRANB3 with ubiquitinated RPA
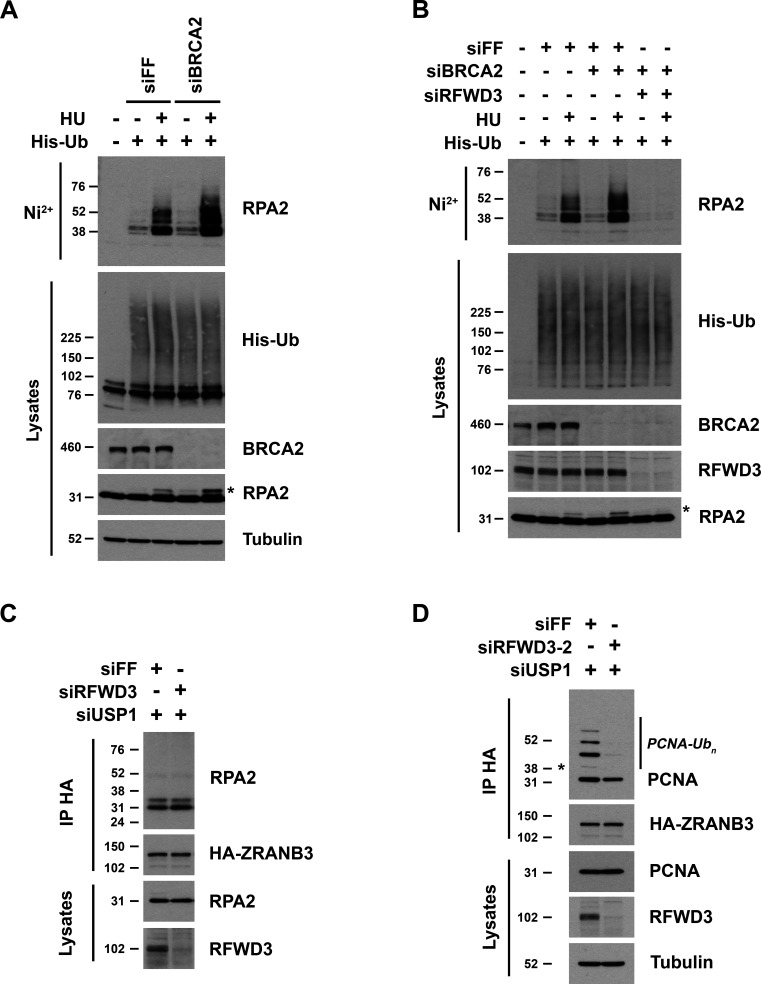
Having demonstrated that RFWD3 promotes the localization of ZRANB3 to stalled replication forks, we next sought to determine the mechanism through which RFWD3 regulates this recruitment. We showed previously that RFWD3 ubiquitinates the ssDNA-binding factor RPA in response to HU- or 4NQO-induced fork stalling (Elia et al., 2015). We wondered whether ubiquitinated RPA could serve as a scaffold for the recruitment of ZRANB3. To determine whether ubiquitinated RPA is available at stalled forks in BRCA2-deficient cells, we examined the effect of BRCA2 depletion on RPA ubiquitination. We expressed His-tagged ubiquitin in cells, purified ubiquitinated proteins by nickel pulldown under denaturing conditions, and immunoblotted for endogenous RPA2. We found that BRCA2 depletion actually increased both RPA ubiquitination and phosphorylation (Fig. 8 A), the latter indicated by a slower migrating form of RPA2 in lysates. RFWD3 depletion abolished the induction of both modifications (Fig. 8 B), consistent with its known roles in ubiquitinating RPA and thereby promoting RPA phosphorylation in BRCA2-proficient cells (Elia et al., 2015). These findings confirmed that RPA ubiquitination is available at stalled forks in BRCA2-deficient cells.
Figure 8.
RFWD3 promotes interaction of ZRANB3 with polyubiquitinated PCNA and not ubiquitinated RPA. (A and B) HeLa cells (A) or U2OS cells (B) were transfected with siFF, siBRCA2-8, and/or siRFWD3-4 and then transfected 1 d later with His-ubiquitin. After one additional day, they were treated with 2 mM HU for 2 h. Cells were lysed under denaturing conditions, and His-tagged ubiquitinated proteins were purified by nickel beads and immunoblotted for endogenous RPA2. His-ubiquitin in lysates was detected by anti-His immunoblot. Asterisk represents phosphorylated RPA2 detected as a slower migrating band in RPA2 lysate immunoblots. (C) U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs and treated with UV radiation (30 J/m2). After 3 h, protein complexes were crosslinked with formaldehyde, and anti-HA immunoprecipitated proteins were immunoblotted for endogenous RPA2 and HA. (D) U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs and treated with ATRi (10 µM) for 30 min followed by UV radiation (30 J/m2) and incubation in ATRi (10 µM) for another 3 h. Protein complexes were crosslinked with formaldehyde, and anti-HA immunoprecipitated proteins were immunoblotted for endogenous PCNA and HA. Asterisk represents monoubiquitinated band of PCNA.
We next sought to determine whether RFWD3 promotes interaction between ZRANB3 and ubiquitinated RPA2. The fraction of cellular RPA2 ubiquitinated in response to replication stress is small and cannot be observed by immunoblotting RPA2 in lysates (Elia et al., 2015). Detection of RPA ubiquitination by expression of exogenous ubiquitin as above and purification under denaturing conditions precludes interaction studies. If ZRANB3 interacts with ubiquitinated RPA, we reasoned that immunoprecipitation of HA-ZRANB3 may enrich for ubiquitinated RPA and allow observation by immunoblot. Given that USP1 depletion increased RFWD3-dependent localization of ZRANB3 to sites of DNA damage (Fig. S2. B and C and Fig. 3, D–F), we performed this experiment in the presence of USP1 depletion. Additionally, we performed formaldehyde crosslinking to preserve potentially transient interactions with ZRANB3. This approach, involving both USP1 depletion and formaldehyde crosslinking, was previously utilized to detect the interaction of ZRANB3 with polyubiquitinated PCNA (Ciccia et al., 2012). Using these conditions, we were unable to detect ZRANB3 binding to any ubiquitinated forms of RPA2. We did observe interactions with two RPA2 bands (Fig. 8 C), but comparing their sizes to ubiquitinated bands in Fig. 8 A suggested they were non-modified and phosphorylated RPA2, and these interactions were not affected by RFWD3 depletion (Fig. 8 C). Given potential difficulties in enriching for ubiquitinated RPA, these results do not completely rule out RPA-mediated recruitment of ZRANB3, but they do not support it as a likely mechanism.
Interestingly, we found that BRCA1 depletion decreased HU-induced RPA2 ubiquitination and phosphorylation (Fig. S4 A). We wondered why BRCA2 depletion increased both modifications (Fig. 8, A and B) while BRCA1 depletion did not. We considered the possibility that increased RPA ubiquitination in BRCA2-depleted cells results from ssDNA generation upon resection of collapsed replication forks. Nascent DNA degradation upon BRCA1 depletion may also be expected to increase fork collapse. However, given the known role of BRCA1 in promoting the resection of DSBs (Chen et al., 2018), BRCA1 depletion may also diminish resection at the DSB arising from fork collapse. Consistent with this model, we found that depletion of the endonuclease MUS81 reversed the induction of both RPA ubiquitination and phosphorylation upon BRCA2 loss (Fig. S4, B and C). These results suggest that fork breakage contributes to the increase in these modifications upon BRCA2 loss and are addressed further in the Discussion.
Figure S4.
Analysis of RPA2 ubiquitination and phosphorylation in BRCA1/2-deficient cells. (A and B) U2OS cells were transfected with the indicated siRNAs and then transfected 1 d later with His-ubiquitin. After one additional day, they were treated with 2 mM HU for 2 h and lysed under denaturing conditions. His-tagged ubiquitinated proteins were purified by nickel beads and immunoblotted for endogenous RPA2. His-ubiquitin in lysates was detected by anti-His immunoblot. Asterisks represent phosphorylated RPA2 detected as a slower migrating band in the RPA2 lysate immunoblots. (C) U2OS cells were transfected with siFF, siBRCA2-3, and/or siMUS81 and treated with 4 mM HU for 4 h. Phosphorylated RPA2 was detected in lysates using the indicated phospho-specific antibodies. Asterisk is the same as in Fig. S4, A and B. (D and E) Sensitivity of U2OS cells to camptothecin (CPT) upon RFWD3 depletion using the indicated siRNAs. Cell survival is normalized to the untreated control for each siRNA. Data represent the mean and SD of three replicates per CPT dose and siRNA condition. Asterisks indicate P values for siRFWD3 versus siFF using an unpaired t test (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001). Immunoblot shows RFWD3 depletion for each siRNA.
RFWD3 promotes PCNA polyubiquitination and interaction with ZRANB3
Recruitment of ZRANB3 to sites of DNA damage depends on interaction with polyubiquitinated PCNA. We next considered whether RFWD3 may regulate this interaction and perhaps even contribute to the ubiquitination of PCNA. Detection of PCNA polyubiquitination can be challenging, so we utilized conditions previously established to maximize its generation (Ciccia et al., 2012). These conditions involved UV irradiation in the presence of both ATR inhibition and the depletion of USP1, which regulates the levels of both mono- and polyubiquitinated PCNA (Brun et al., 2010; Huang et al., 2006; Yang and Zou, 2009), as well as formaldehyde crosslinking as described above. We immunoprecipitated HA-ZRANB3 from U2OS cells exposed to these conditions and successfully detected interaction with endogenous polyubiquitinated PCNA (Fig. 8 D). Consistent with prior results (Ciccia et al., 2012), ZRANB3 interacted preferentially with polyubiquitinated rather than monoubiquitinated PCNA. Importantly, RFWD3 depletion strongly reduced binding to multiple polyubiquitinated forms of PCNA. We also observed a small reduction in binding to non-modified PCNA, which likely arises from the incorporation of non-ubiquitinated PCNA monomers into homotrimers containing ubiquitinated PCNA. Together, these findings suggest that RFWD3 promotes ZRANB3 recruitment through the regulation of its interaction with polyubiquitinated PCNA.
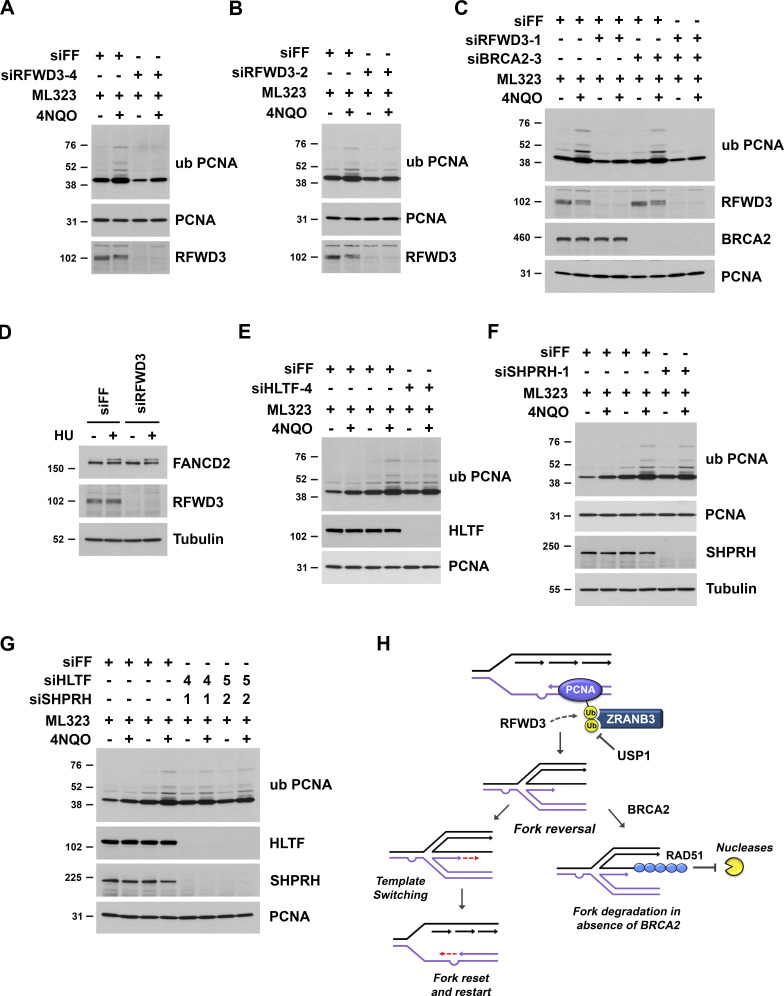
To investigate more directly whether RFWD3 promotes PCNA ubiquitination, we examined endogenous ubiquitinated PCNA in immunoblots of U2OS lysates. To generate polyubiquitinated PCNA, we again utilized USP1 depletion, which induced multiple slower-migrating bands detected by an antibody against PCNA ubiquitinated on K164. Importantly, we found that RFWD3 depletion strongly inhibited the induction of polyubiquitinated PCNA by the UV mimetic 4NQO in the presence of USP1 depletion (Fig. 9, A and B). We also observed a decrease in 4NQO-induced monoubiquitination of PCNA upon RFWD3 depletion. To generate polyubiquitinated PCNA, we additionally utilized the USP1 inhibitor ML323. We found that RFWD3 depletion with four independent siRNAs again inhibited the induction of polyubiquitinated PCNA by 4NQO in the presence of ML323 (Fig. 9 C and Fig. S5, A and B). These results demonstrate opposing roles for USP1 and RFWD3 in PCNA ubiquitination, which parallel their roles in ZRANB3 recruitment. We rescued the defect in PCNA ubiquitination by complementing RFWD3-depleted cells with siRNA-resistant WT RFWD3 but not with RING mutant (C315A) RFWD3, demonstrating the ubiquitin ligase activity of RFWD3 is necessary for promoting PCNA ubiquitination (Fig. 9 D).
Figure 9.
RFWD3 ubiquitin ligase activity promotes the polyubiquitination of PCNA. (A and B) U2OS cells were transfected with the indicated siRNAs, treated with 1 µg/ml 4NQO for 4 h, and endogenous PCNA ubiquitination was detected in whole cell lysates using an antibody against K164-ubiquitinated PCNA. (C and E) U2OS cells were transfected with the indicated siRNAs, treated with 30 µM ML323 for 4 h, and then treated with 1 µg/ml 4NQO in the presence of 30 µM ML323 for another 4 h. Endogenous PCNA ubiquitination was detected in whole cell lysates using an antibody against ubiquityl-K164 PCNA. (D) U2OS cells expressing siRNA-resistant RFWD3 (WT or C315A) were transfected with siRFWD3-4 and compared with U2OS cells transfected with siFF or siRFWD3-4. Cells were treated with 30 µM ML323 for 4 h and then with 1 µg/ml 4NQO in the presence of 30 µM ML323 for another 4 h. PCNA ubiquitination was detected using an antibody against K164-ubiquitinated PCNA. (F) FA patient fibroblasts complemented with WT RFWD3 (1143 + WT) or empty vector (1143 + mock) were treated with 1 µg/ml 4NQO for 4 h. PCNA ubiquitination was detected in whole cell lysates using an antibody against ubiquityl-K164 PCNA.
Figure S5.
RFWD3 promotes the polyubiquitination of PCNA. (A–C, and E–G) U2OS cells were transfected with the indicated siRNAs, treated with 30 µM ML323 for 4 h, and then treated with 1 µg/ml 4NQO in the presence of 30 µM ML323 for another 4 h. Endogenous PCNA ubiquitination was detected in whole cell lysates using an antibody against K164-ubiquitinated PCNA. (D) U2OS cells transfected with siFF or siRFWD3-4 were treated with 2 mM HU for 2 h. Whole-cell lysates were immunoblotted for FANCD2 and RFWD3. (H) Model showing that RFWD3 promotes PCNA polyubiquitination to recruit ZRANB3 and remodel stalled replication forks. Fork reversal allows template switching and replication fork restart in BRCA2-proficient cells. In BRCA2-deficient cells, however, reversal can lead to fork degradation.
We next investigated whether the regulation of PCNA ubiquitination by RFWD3 is preserved in a BRCA2-deficient background. RFWD3 depletion with two independent siRNAs decreased PCNA ubiquitination in U2OS cells to the same extent in both the presence and absence of BRCA2 codepletion (Fig. 9 E and Fig. S5 C). These findings are consistent with our results that RFWD3 promotes fork remodeling (Fig. 6 G) and ZRANB3 recruitment to both stalled forks (Fig. 6, D and E) and UV stripes (Fig. 6, B and C) to the same extent in BRCA2-deficient versus proficient cells. We also examined PCNA ubiquitination in FA patient fibroblasts (1143 cells) with RFWD3 mutation. PCNA polyubiquitination was significantly lower in 1143 cells complemented with empty vector (1143 + mock) than in 1143 cells complemented with WT RFWD3 (1143 + WT; Fig. 9 F), demonstrating RFWD3 promotes PCNA ubiquitination in this cell line too.
Lastly, RFWD3 and USP1 have opposing roles in ZRANB3 recruitment (Fig. 3, D–F and Fig. S2, B and C). Because USP1 is additionally known to deubiquitinate FANCD2, we also examined the dependency of FANCD2 monoubiquitination on RFWD3. ZRANB3 binds polyubiquitin chains, making FANCD2 an unlikely candidate for ZRANB3 recruitment, and we found that RFWD3 depletion did not affect FANCD2 ubiquitination (Fig. S5 D), consistent with prior reports (Feeney et al., 2017; Inano et al., 2017; Knies et al., 2017). In summary, these results suggest that RFWD3 stimulates ZRANB3 recruitment through ubiquitin-dependent interaction with PCNA and by promoting the ubiquitination of PCNA itself.
Discussion
RFWD3 depletion protects stalled replication forks from degradation and collapse in BRCA2-deficient cells
Prior reports have demonstrated that loss of multiple factors in the Fanconi Anemia (FA) pathway (FANCA, FANCB, FANCD2, and FANCJ) results in the degradation of nascent DNA at stalled replication forks (Bhat et al., 2018; Kim et al., 2015; Peng et al., 2018; Schlacher et al., 2012). We demonstrate here that depletion of the FA gene RFWD3 does not induce nascent DNA degradation but rather protects against it in BRCA2-deficient cells. Consistent with this finding, RFWD3 depletion in BRCA2 deficient cells also rescues the collapse of stalled forks into DSBs and sensitivity to HU. This phenotype may have therapeutic implications, as RFWD3 loss could promote resistance of BRCA2 mutant tumors to chemotherapeutic agents that induce replication stress. Future work will be necessary to determine the effect of RFWD3 inactivation on the sensitivity of BRCA2-deficient cells to fork-stalling agents other than HU.
During the preparation of our manuscript, another study reported that RFWD3 depletion rescues fork degradation and HU sensitivity in BRCA2-deficient cells (Duan et al., 2020). This study proposed that forks coated with ubiquitinated RPA are resistant to repair and that RFWD3 depletion facilitates fork repair by eliminating this intermediate. It suggested that the ubiquitination of RPA promotes its accumulation at stalled forks to inhibit RAD51 loading. Earlier reports have demonstrated that RFWD3-dependent RPA ubiquitination promotes repair via homologous recombination at stalled forks (Elia et al., 2015; Feeney et al., 2017; Inano et al., 2017; Knies et al., 2017) and that ubiquitination of RPA stimulates its displacement from DNA (Feeney et al., 2017; Inano et al., 2017). We have found that RFWD3 promotes ZRANB3 recruitment and replication fork remodeling. We propose that inhibition of RFWD3 in BRCA2-deficient cells suppresses generation of the reversed fork substrate necessary for nascent DNA degradation and thereby protects against fork degradation in these cells. We posit that increased RPA ubiquitination and phosphorylation in BRCA2-deficient cells result from ssDNA generation due to fork degradation and collapse (see next paragraph). Consequently, RFWD3 depletion in BRCA2-deficient cells decreases RPA ubiquitination and phosphorylation by not only removing the responsible ubiquitin ligase but by decreasing fork resection and ssDNA generation. This model does not rely on RPA ubiquitination playing a causative role but rather serving as a correlative marker for ssDNA generation in BRCA2-deficient cells.
We found that BRCA2 depletion increased RPA ubiquitination and phosphorylation whereas BRCA1 depletion in contrast decreased both modifications in response to replication fork stalling. Why these factors regulate RPA ubiquitination in opposite directions is an interesting question. We have found that the kinetics of RPA ubiquitination and phosphorylation mirror each other (Elia et al., 2015) and that genetic perturbations influence these modifications in usually the same direction, as observed for BRCA1/2 depletion. RPA phosphorylation is considered a surrogate for ssDNA generation (Zeman and Cimprich, 2014). RPA ubiquitination may be viewed similarly, as helicase–polymerase uncoupling by either HU or aphidicolin strongly induces it (Elia et al., 2015). Indeed, the direction by which these modifications change upon BRCA2 versus BRCA1 depletion parallels ssDNA levels measured by BrdU staining (Duan et al., 2020; Pathania et al., 2011). One explanation for increased RPA ubiquitination upon BRCA2 depletion is that it represents ssDNA generation at one-ended DSBs arising from fork collapse. In BRCA2-deficient cells, increased fork degradation may lead to collapse, and impaired RAD51 loading may cause the persistence of resected DSB ends coated with RPA. In the absence of BRCA1, fork degradation will also be increased, but resection at DSBs resulting from fork collapse may be decreased, given the role of BRCA1 in promoting DSB resection (Chen et al., 2018). The absence of increased RPA ubiquitination in BRCA1-deficient cells may thus reflect lower resection at collapsed forks. Consistent with fork breakage contributing to RPA ubiquitination, we found that MUS81 depletion reversed the induction of RPA ubiquitination and phosphorylation upon BRCA2 loss in U2OS cells. However, it remains unclear why BRCA1 depletion would cause a net decrease in ssDNA despite increased resection of regressed fork ends. BRCA1 may have novel roles in regulating ssDNA generation through alternative means, highlighting differences in stalled fork intermediates occurring in BRCA1- versus BRCA2-deficient cells (Duan et al., 2020).
RFWD3 promotes ZRANB3 recruitment and replication fork remodeling
We recognized that phenotypes of RFWD3 depletion in wild-type and BRCA2-deficient cells resembled those of the fork remodeling enzyme ZRANB3. Loss of either RFWD3 or ZRANB3 impairs the restart of stalled replication forks in wild-type cells (Ciccia et al., 2012; Elia et al., 2015) and rescues fork degradation and collapse in BRCA2-deficient cells (Mijic et al., 2017; Taglialatela et al., 2017). For ZRANB3, both of these phenotypes are due to its function in reversing stalled replication forks. As an additional similarity, RFWD3 depletion causes sensitivity to camptothecin (Fig. S4, D and E), as previously reported for ZRANB3 loss (Ciccia et al., 2012). We found that RFWD3 depletion impaired ZRANB3 recruitment to stalled replication forks and to sites of UV laser irradiation in both BRCA2-proficient and BRCA2-deficient cells. RFWD3 depletion also inhibited ZRANB3 localization to spontaneous nuclear foci induced by USP1 depletion. These results suggest that RFWD3 promotes the localization of ZRANB3 to ubiquitinated sites that are substrates of USP1, and they establish antagonistic roles for RFWD3 and USP1 in ZRANB3 recruitment. Opposing roles for these enzymes are consistent with RFWD3 function in the FA pathway and USP1 antagonism of other FA ubiquitin ligases.
Multiple SNF2 family translocases, including ZRANB3, SMARCAL1, and HLTF, demonstrate fork remodeling activity in both in vitro assays and in cells (Poole and Cortez, 2017). Loss of any one of these translocases in cells causes a partial reduction in reversed fork formation as detected by electron microscopy (Bai et al., 2020; Kolinjivadi et al., 2017; Taglialatela et al., 2017; Vujanovic et al., 2017). Incomplete abrogation of fork reversal upon loss of a single translocase suggests that these remodelers act in a nonredundant fashion, consistent with the finding that they recognize different fork structures in vitro (Bétous et al., 2013; Kile et al., 2015). Inactivation of only one translocase may not affect the reversal of fork structures targeted by other remodelers. Importantly, despite partially reducing fork reversal, loss of either ZRANB3, SMARCAL1, or HLTF alone completely rescues nascent DNA degradation (Mijic et al., 2017; Taglialatela et al., 2017). It has been proposed that the extensive fork degradation observed by DNA fiber analysis (thousands of bases) occurs through numerous rounds of fork reversal and resection, during which multiple fork structures may arise that require remodelers with distinct substrate specificities (Bhat and Cortez, 2018; Taglialatela et al., 2017). Inhibiting any one of these remodelers may disrupt this cyclic pathway and thereby completely abolish nascent DNA degradation detected by DNA fiber analysis. We found that RFWD3 depletion caused a partial decrease in fork reversal (Fig. 5 E, Fig. 6 G, and Fig. S3 C), which was comparable to the level of reduction for ZRANB3 depletion upon HU treatment (Fig. 7 E). Inhibition of ZRANB3-mediated fork reversal upon RFWD3 depletion may impair this cyclic pathway and similarly have a larger effect on fork degradation than expected from the magnitude of the fork reversal defect.
RFWD3 promotes PCNA polyubiquitination to recruit ZRANB3
We found that ZRANB3 recruitment to stalled forks depends on the ubiquitin ligase activity of RFWD3. In seeking to elucidate a ubiquitin-dependent mechanism for RFWD3-regulated ZRANB3 localization, we considered RPA and PCNA as potential platforms for ZRANB3 recruitment. We were unable to detect the binding of ZRANB3 to any ubiquitinated forms of RPA, suggesting that RPA is not involved in this process. These results are also consistent with RPA inhibiting the reversal activity of ZRANB3 toward damaged fork substrates in vitro (Bétous et al., 2013). We did detect the interaction of ZRANB3 with ubiquitinated PCNA, and consistent with prior results (Ciccia et al., 2012), this interaction was more pronounced for polyubiquitinated rather than monoubiquitinated forms of PCNA. Importantly, we found that RFWD3 depletion significantly reduced the binding of ZRANB3 to polyubiquitinated PCNA. These results suggest that RFWD3 promotes ZRANB3 recruitment through the regulation of PCNA polyubiquitination. However, it is also possible that RFWD3 promotes the ubiquitination of additional substrates that recruit ZRANB3 in certain genetic contexts.
The ubiquitination of PCNA in vertebrates involves the activity of multiple E3 ubiquitin ligases. Monoubiquitination of K164 is mediated by RAD18 (Kannouche et al., 2004; Watanabe et al., 2004), while K63-linked ubiquitin chain extension at this site is mediated by the ligases HLTF and SHPRH (Motegi et al., 2008; Motegi et al., 2006; Unk et al., 2008; Unk et al., 2006). Interestingly, prior reports have suggested the existence of other ubiquitin ligases, as MEFs lacking both HLTF and SHPRH can still produce polyubiquitinated PCNA (Krijger et al., 2011), and residual monoubiquitinated PCNA has been observed in RAD18-deficient cells (Leung et al., 2018; Simpson et al., 2006). To examine the ubiquitination of endogenous PCNA and its dependence on RFWD3, we utilized an antibody against K164-ubiquitinated PCNA. These experiments required USP1 depletion to adequately detect polyubiquitinated forms of PCNA by immunoblot. Importantly, we found that RFWD3 depletion inhibited the induction of both PCNA mono- and polyubiquitination, with a seemingly larger effect on PCNA polyubiquitination. Inhibition of polyubiquitination was greater than that observed for individual or combined depletion of HLTF and SHPRH (Fig. S5, E–G). PCNA polyubiquitination is known to involve K63 linkages, which are recognized by the NZF domain of ZRANB3 (Ciccia et al., 2012). Notably, we showed previously that RFWD3 is capable of promoting UBC13-dependent K63-linked polyubiquitination in cells (Elia et al., 2015). These results identify RFWD3 as another ubiquitin ligase that promotes the ubiquitination of PCNA.
PCNA ubiquitination plays a critical role in regulating two DNA damage tolerance (DDT) pathways that can bypass lesions encountered during DNA replication. The extent of PCNA ubiquitination dictates which of these pathways predominates when DDT is evoked (Branzei and Psakhye, 2016; Mailand et al., 2013). PCNA monoubiquitination promotes translesion synthesis (TLS) by directly recruiting error-prone TLS polymerases that can synthesize DNA across the lesion. Alternatively, PCNA polyubiquitination promotes template switching (TS), which is a recombination-based pathway that uses the nascent DNA strand of the undamaged sister chromatid as a template for DNA synthesis across the lesion. One version of template switching involves fork reversal (Conti and Smogorzewska, 2020; Cortez, 2019; Neelsen and Lopes, 2015; Quinet et al., 2021; Yeeles et al., 2013), which has led to the proposal that PCNA polyubiquitination may promote template switching through the recruitment of ZRANB3 to reverse replication forks (Ciccia et al., 2012; Quinet et al., 2021). During the preparation of our manuscript, another report demonstrated that RFWD3 promotes PCNA ubiquitination and translesion synthesis in Xenopus oocytes (Gallina et al., 2021). We demonstrate here that RFWD3 promotes replication fork reversal and PCNA polyubiquitination to recruit ZRANB3, which are events that have both been linked to template switching. Our results therefore provide functional evidence that lay the foundation for a role of RFWD3-dependent PCNA ubiquitination in not only translesion synthesis but importantly also in template switching (Fig. S5 H). In this regard, our work also helps to explain a recent study suggesting that RFWD3 participates in a DDT pathway that is independent of some TLS polymerases but for which no mechanism was provided (Kanao et al., 2022).
We proposed previously that RFWD3-dependent ubiquitination of RPA on multiple sites and subunits may stimulate the recruitment of repair factors (Elia et al., 2015), similar to multisite protein group sumoylation in yeast (Psakhye and Jentsch, 2012). We show here that RFWD3 promotes the ubiquitination of another substrate PCNA to stimulate ZRANB3 localization in human cells, while other reports have demonstrated that RFWD3 promotes recruitment of the TLS polymerases REV1-Poζ, Polη, and Polκ (Gallina et al., 2021), the helicase MCM8 (Inano et al., 2017), and the SNF2 family translocase RAD54 (Inano et al., 2017). Interestingly, RAD54 is known to have fork reversal activity in vitro (Bugreev et al., 2011). It is unclear whether RAD54 inactivation can rescue nascent DNA degradation in BRCA2-deficient cells, but precedence clearly exists for RFWD3-dependent recruitment of SNF2 family translocases. Our data elucidate a new role for RFWD3 in replication fork remodeling and expand the repertoire of ways in which this Fanconi gene can modulate fork dynamics.
Materials and methods
Cell culture
U2OS cells were cultured in McCoy's medium supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. HeLa cells were grown in DMEM with the same supplements. U2OS cells stably expressing HA-ZRANB3 were described previously (Ciccia et al., 2012). Immortalized patient fibroblasts stably transfected with a plasmid containing WT RFWD3 (1143 + WT) or empty vector (1143 + mock) were described previously (Knies et al., 2017) and cultured in DMEM with 15% fetal bovine serum and 1% penicillin/streptomycin. DR-U2OS cells with a stably integrated DR-GFP reporter were generously provided by Maria Jasin (Xia et al., 2006). siRNAs were reverse transfected into cells at 10–20 nM using Lipofectamine RNAiMAX reagent (13778150; Thermo Fisher Scientific). Plasmids were transfected into cells using FuGENE 6 (E2691; Promega) or using HeLa MONSTER and TransIt reagents (MIR 2900; MiRus).
siRNAs, plasmids, and antibodies
The following siRNAs were used: siBRCA1 (5′-CAACAUGCCCACAGAUCAA-3′), siBRCA2-3 (5′-GGUAUCAGAUGCUUCAUUA-3′), siBRCA2-8 (5′-UAAGGAACGUCAAGAGAUA-3′), siFF (5′-CGUACGCGGAAUACUUCGA-3′), siHLTF-4 (5′-CCAGAUGACUUUCUAACUA-3′), siHLTF-5 (5′-GGACUACGCUAUUACACGG-3′), siMUS81 (5′-CAGCCCUGGUGGAUCGAUA-3′), siRFWD3-1 (5′-CCAUUUGAGGUGAACCGUA-3′), siRFWD3-2 (5′-GGAAACAGGCCGAGUUAGA-3′), siRFWD3-3 (5′-GUUAAGAUGUUGAGUACUG-3′), siRFWD3-4 (5′-GGACCUACUUGCAAACUAU-3′), siSHPRH-1 (5′-GCACAAAUCAGUCGUGUUA-3′), siSHPRH-2 (5′-GAAGAAAUGUGGAACUGAA-3′), siUSP1 (5′-GAAAUACACAGCCAAGUAA-3′), and siZRANB3 (5′-GAUCAGACAUCACACGAUU-3′). The following shRNAs were used: shFF (5′-CCCGCCTGAAGTCTCTGATTAA-3′) and shRFWD3-4 (5′-AGAGGACCTACTTGCAAACTAT-3′). pCS2-His-Ub was generously provided by Michael Rape. The RFWD3 RNAi-resistant construct was generously provided by Junjie Chen (Gong and Chen, 2011) and is resistant to siRFWD3-4 and shRFWD3-4.
For Western blot, the following rabbit antibodies were used: BRCA2 (A300-005A; Bethyl), FANCD2 (NB100-182; Novus), HLTF (ab183042; Abcam), PCNA (ab18197; Abcam), PCNA ubiquityl-Lys164 (13439; Cell Signaling), RFWD3 (A301-397A; Bethyl), RPA32 (A300-244A; Bethyl), RPA32 pS4/8 (A300-245A; Bethyl), RPA32 pT21 (ab109394; Abcam), RPA32 pS33 (A300-246A; Bethyl, lot 3), SHPRH (ab80129; Abcam), β-Tubulin (2128; Cell Signaling), USP1 (A301-699A; Bethyl), and ZRANB3 (23111-1-AP; Proteintech). Mouse antibodies included BRCA1 (sc-6954; Santa Cruz), His (ab49781; Abcam), and MUS81 (ab14387; Abcam), while rat antibodies included HA (12013819001; Roche). Secondary antibodies were goat anti-rabbit IgG-HRP (111-035-003; Jackson ImmunoResearch) and goat anti-mouse IgG-HRP (115-035-003; Jackson ImmunoResearch).
Cell survival assay
U2OS cells were transfected with indicated siRNAs, harvested 24 h later, and seeded into 96-well plates at a density of 1.0 × 103 per well. After 24 h, cells were treated with HU at the indicated concentrations for 4 d, washed three times with media, and then incubated for another 2 d. Alternatively, cells were treated with camptothecin at the indicated doses for 24 h, washed three times with media, and then incubated for another 4 d. Fresh media followed by CellTiter-Glo 2.0 Reagent (G9242; Promega) was added to each well in 1:1 volume. The well contents were mixed for 2 min at 700 rpm in an orbital shaker to induce cell lysis, and plates were incubated at room temperature for 25 min to stabilize the luminescent signal. Luminescence was quantitated on a Perkin Elmer EnVision microplate reader, and cell viability was calculated by normalizing signals from treated samples to untreated controls for each siRNA condition. Data represent the mean and SD of three replicates per HU dose and siRNA condition.
DNA fiber analysis
Exponentially growing U2OS cells were incubated with 25 µM CldU (C6891; Sigma-Aldrich) for 25 min, washed with PBS, and incubated with 125 µM IdU (I7125; Sigma-Aldrich) for 30 min. 1143 + WT and 1143 + mock cells were incubated with 50 µM CldU for 40 min, washed with PBS, and incubated with 250 µM IdU for 50 min. Cells were washed with PBS and treated with 2 mM HU (H8627; Sigma-Aldrich) for 5 h. Labeled cells were subsequently trypsinized and resuspended in ice-cold PBS. This suspension (2.5 µl) was spotted onto glass slides and lysed with 7.5 µl of spreading buffer (0.5% SDS in 200 mM Tris-HCl, pH 7.4 and 50 mM EDTA) at room temperature. After 3 min, DNA fibers were spread by tilting slides at 15° relative to horizontal. Slides were air-dried, fixed in methanol and glacial acetic acid (3:1) for 4 min, and denatured with 2.5 M HCl for 30 min at room temperature. Slides were washed in PBS and blocked in 2% BSA in PBS-T (PBS, 0.05% Triton X-100) for 1 h at room temperature. To detect CldU and IdU, respectively, slides were stained with rat anti-BrdU (1:100, Abcam ab6326) and mouse anti-BrdU (1:20, Becton Dickinson 347580) in 2% BSA in PBS-T for 2 h at 37°C. Slides were subsequently washed in PBS-T and stained with Cy3 donkey anti-rat IgG (1:100, 712-165-153; Jackson ImmunoResearch) and Alexa Fluor 488 donkey anti-mouse IgG (1:100, 715-545-151; Jackson ImmunoResearch) in 2% BSA in PBS-T for 1 h at room temperature. Slides were washed in PBS-T and mounted in Prolong Gold Antifade (P36934; Thermo Fisher Scientific) and stored at 4°C. Replication tracks were imaged on a Nikon Eclipse 90i microscope with a Retiga 200R camera and measured using ImageJ software. In each experiment, 200 or more individual tracks were measured.
Neutral comet assay
DSBs resulting from replication fork collapse were evaluated by neutral comet assay. U2OS cells in six-well plates were treated with 2 mM HU (H8627; Sigma-Aldrich) for 24 h. Cells were harvested, resuspended in cold PBS, and combined at a 1:10 ratio (v/v) with 1% molten LMAgarose in PBS (4250-050-02; Trevigen). This solution was then added to glass Flare slides (3950-300-02; Trevigen) in the amount of 1,200 cells per well. Slides were incubated in cell lysis solution (4250-050-01; Trevigen) at 4°C for 1 h and then equilibrated in neutral electrophoresis buffer (100 mM Tris, 300 mM NaOAc, pH 9) for 30 min at 4°C. DNA migration was performed in neutral electrophoresis buffer at 1 V per cm (interelectrode distance) for 1 h at 4°C. Slides were subsequently immersed in DNA precipitation buffer (1 M NH4OAc, 87% EtOH) for 30 min, fixed with 70% ethanol for 30 min, and dried at room temperature. Comets were labeled with SYBR Gold Nucleic Acid Gel Stain (S11494; Thermo Fisher Scientific) for 30 min at room temperature. Slides were gently washed with water and mounted in Prolong Gold Antifade (P36934; Thermo Fisher Scientific). Images were acquired on a Nikon Eclipse 90i microscope with a Retiga 200R camera and analyzed using ImageJ (OpenComet software). More than 200 comets were analyzed for each condition.
Immunofluorescence microscopy
U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs. For UV laser microirradiation experiments, cells were treated with 10 µM BrdU for 24 h prior to irradiation using an Arcturus Veritas Laser Capture Microscope. At the indicated time points, cells were pre-extracted with 0.5% Triton X-100 in PBS at 4°C for 3 min, washed gently with PBS, fixed with 4% paraformaldehyde in PBS at room temperature for 10 min, washed again with PBS, and permeabilized with 0.5% NP-40 in PBS for 10 min. HA-ZRANB3 was detected using rabbit HA antibody (3724; Cell Signaling) followed by Alexa Fluor 488 anti-rabbit antibody (A11008; Thermo Fisher Scientific). γ-H2AX was detected using a mouse monoclonal antibody (05–636; Millipore) followed by Alexa Fluor 594 anti-mouse antibody (A11005; Thermo Fisher Scientific). Nuclei were stained with DAPI, and images were acquired on a Nikon Eclipse 90i microscope with a 60x 1.20 NA water immersion objective and a Retiga 200R camera using Nikon NIS-Elements AR software. Foci number, foci intensity, and stripe number were quantitated using Cell Profiler or ImageJ. More than 200 cells were analyzed for each condition.
Proximity ligation assay on nascent DNA at stalled replication forks
U2OS cells expressing HA-ZRANB3 were treated with 10 µM or 15 µM EdU (20518; Cayman Chemical) for 10 min, washed with PBS, and then treated with 1 µg/ml 4NQO or 4 mM HU for 4 h as specified in figure legends. In chase experiments, PBS washes were followed by treatment with 15 µM thymidine (T1895; Sigma-Aldrich) for 2 h and then with 1 µg/ml 4NQO for 4 h. Cells were pre-extracted with 0.5% Triton X-100 in PBS at 4°C for 3 min, washed gently with PBS, fixed with 4% paraformaldehyde in PBS at room temperature for 10 min, washed again with PBS, and permeabilized with 0.5% NP-40 in PBS for 10 min. After blocking in 3% BSA in PBS with 0.05% Tween for 30 min, Click-iT reaction was performed to conjugate biotin-azide (B10184; Thermo Fisher Scientific) to EdU. Cells were then incubated with mouse monoclonal anti-biotin (200-002-211; Jackson ImmunoResearch) along with either rabbit anti-HA (3724; Cell Signaling) or rabbit anti-PCNA (ab18197; Abcam). Negative controls were performed with each of these antibodies by themselves. PLA was performed with anti-Mouse MINUS (DUO92004; Sigma-Aldrich), anti-Rabbit PLUS (DUO92002; Sigma-Aldrich), and Duolink In Situ Detection Reagents Red (DUO92008; Sigma-Aldrich Duolink) according to manufacturer instructions. Nuclei were stained with DAPI, and images were acquired on a Nikon Eclipse 90i microscope with a 60x 1.20 NA water immersion objective and a Retiga 200R camera using Nikon NIS-Elements AR software. The number of foci in each cell was quantitated using ImageJ. More than 200 cells were analyzed for each condition.
Purification of His-tagged ubiquitinated proteins
U2OS or HeLa cells were transfected with pCS2 His-ubiquitin using FuGENE 6 (E2691; Promega) or using HeLa MONSTER and TransIt reagents (MIR 2900; MiRus). After 24 h, cells were treated with 2 mM HU (H8627; Sigma-Aldrich) or 1 µg/ml 4NQO (N8141; Sigma-Aldrich) for 2 h and harvested. Cell pellets were lysed by sonication in Buffer A (6 M Guanidine HCl, 100 mM NaH2PO4/Na2HPO4, 15 mM imidazole, pH 8) with 2 mM N-Ethylmaleimide (04259; Sigma-Aldrich). Lysates were incubated with Ni-NTA agarose and rotated at room temperature for 3 h. Beads were subsequently washed once in Buffer A, twice in 1:3 Buffer (1 volume of Buffer A/3 volumes of TI Buffer), and finally once with TI Buffer (25 mM Tris, 20 mM imidazole, pH 6.8). Bound complexes were eluted in Laemmli sample buffer with 250 mM imidazole and heated for 10 min at 95°C.
Protein immunoprecipitation
U2OS cells expressing HA-ZRANB3 were transfected with the indicated siRNAs. After 48 h, cells were treated with the ATR inhibitor VE-821 (S8007; Selleck) for 30 min, exposed to 30 J/m2 UV irradiation, and harvested 4 h later. Cells were fixed in 0.5% formaldehyde in PBS for 5 min at room temperature, and glycine was added to a final concentration of 125 mM to quench formaldehyde. Cells were washed twice with PBS, resuspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8) with 1% SDS, and washed twice with TE buffer. Each of these steps was performed at 4°C and involved centrifugation at 2,000 g for 5 min. Cells were then resuspended in cold TE buffer with 0.1% SDS and sonicated. Lysates were centrifuged at 14,000 rpm for 10 min, and the supernatant was supplemented with NaCl and NP-40 to final concentrations of 150 mM and 0.1%, respectively. Lysates were incubated with anti-HA agarose (A2095; Sigma-Aldrich) and rotated at 4°C overnight. Beads were subsequently washed 5 times with 10 mM Tris, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40. Protein crosslinks were reversed by incubation of beads in Laemmli sample buffer at 95°C for 25 min, and eluates were analyzed by immunoblot.
Electron microscopy
Visualization of replication intermediates by electron microscopy was performed as described previously (Neelsen et al., 2014) with some modifications. U2OS cells were treated with 2 mM HU in the presence or absence of 50 µM mirin for 5 h. They were subsequently harvested by trypsinization, washed with ice-cold PBS, resuspended in 10 ml cold PBS (5 × 106 cells), and transferred to 6-cm tissue culture plates. Trimethylpsoralen (T6137; Sigma-Aldrich) was added to a final concentration of 10 mg/ml, and the plates were incubated on a pre-cooled metal surface in the dark for 5 min. Cells were then UV-irradiated for 5 min with a monochromatic 365 nm lamp. This cycle of trimethylpsoralen addition, dark incubation, and irradiation was repeated three times. Cells were then washed twice with cold PBS, and genomic DNA was extracted using a QIAGEN cell culture DNA kit (13323; Qiagen) according to manufacturer instructions. Genomic DNA (30 µg) was resuspended in TE buffer (10 mM Tris, 1 mM EDTA, pH 8) and digested with 150 U of PvuII HF (R3151; New England Biolabs) for 4 h at 37°C. After purification using a QIAGEN Genomic-tip 20/G (10223; Qiagen), DNA was desalted and concentrated using an Amicon Ultra-0.5 Centrifugal Filter Unit with Ultracel-100 (UFC510024; Millipore). DNA was run on a 0.8% agarose gel to examine quality and concentration. It was then processed for rotary shadowing and platinum coating as previously described (Neelsen et al., 2014) using a Med20 evaporator (Leica). Images were acquired at the IFOM EM facility using a TECNAI12 electron microscope equipped with a GATAN camera run by Digital Micrograph software.
Homologous recombination assay
U2OS cells with a stably integrated DR-GFP reporter (Xia et al., 2006) were transfected with the indicated siRNAs 48 h prior to infection with adenovirus expressing I-SceI restriction enzyme. The percentage of GFP-positive cells was determined 48 h after adenoviral infection by flow cytometry using a BD-LSRII Flow Cytometer (Becton Dickinson). Data were collected using BD FACS Diva software (Becton Dickinson), and analysis was performed using FlowJo Software.
Statistical analysis
Statistical analyses were performed using GraphPad Prism 8. Statistical differences in DNA fiber length ratios, comet tail moments, and proximity ligation foci were determined by a two-tailed Mann Whitney test. For these experiments, whiskers in box plots represent the 10th and 90th percentiles unless otherwise noted. Statistical differences in cell survival, laser stripe localization, nuclear foci number, and fork reversal were determined by a two-tailed, unpaired t test as specified in figure legends. For these experiments, data represent the mean and SD unless otherwise noted. For t tests, data distributions were assumed to be normal but were not formally tested. For all experiments: n.s. P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001.
Online supplemental material
Fig. S1 (related to Fig. 1 and Fig. 2) shows that RFWD3 depletion reverses HU sensitivity in BRCA1- and BRCA2-deficient cells, rescues nascent DNA degradation in BRCA1-deficient cells, and does not restore homologous recombination in BRCA2-deficient cells. Fig. S2 (related to Fig. 3 and Fig. 4) demonstrates that RFWD3 depletion decreases ZRANB3 recruitment to HU-stalled replication forks but does not affect ZRANB3 localization to chromatin behind forks, PCNA recruitment to stalled forks, or ZRANB3 localization to UV laser stripes at 10 min. Fig. S3 (related to Fig. 5) provides fork reversal data for individual biological replicates and EM images of replication intermediates. Fig. S4 (related to Fig. 8) shows that BRCA1 depletion decreases RPA ubiquitination and phosphorylation, while BRCA2 depletion increases both modifications in a MUS81-dependent manner. Fig. S5 (related to Fig. 9) demonstrates that RFWD3 depletion decreases PCNA polyubiquitination to a greater extent than individual or combined depletion of HLTF and SHPRH. It also shows that RFWD3 promotes PCNA ubiquitination in BRCA2-deficient cells and provides a model for RFWD3 function in replication fork remodeling.
Acknowledgments
We thank L. Zou, S. Pathania, and members of the Elia and Zou laboratories for helpful discussions. We thank M. Jasin for U2OS cells with a stably integrated DR-GFP reporter.
This project was supported by funding to A.E.H. Elia from a Burroughs Wellcome Fund Career Award for Medical Scientists, a National Cancer Institute Federal Share Award, and a Breast Cancer Alliance Young Investigator Grant. V. Costanzo is funded by the Associazione Italiana per la Ricerca sul Cancro (AIRC-IG Ref 21824) and by the Fondazione Regionale per la Ricerca Biomedica (Ref IANG-CRC). Y. Wijesekara Hanthi is supported by an AIRC fellowship (Ref 26596). A. Ciccia is funded by the National Institutes of Health (R01CA197774). D. Schindler is funded by the Schroeder Kurth Fund and departmental support.
Author contributions: C.E. Moore and A.E.H. Elia designed the project. C.E. Moore, S.E. Yalcindag, H. Czeladko, R. Ravindranathan, Y. Wijesekara Hanthi, J.C. Levy, V. Sannino, and A.E.H. Elia performed experiments and analyzed data. Y. Wijesekara Hanthi and V. Sannino performed electron microscopy with supervision from V. Costanzo. D. Schindler provided Fanconi Anemia patient 1143 cells. A. Ciccia, V. Costanzo, and D. Schindler provided expertise and feedback. C.E. Moore and A.E.H. Elia wrote the paper with input from all the authors. A.E.H. Elia supervised the project.
References
- Bai, G., Kermi C., Stoy H., Schiltz C.J., Bacal J., Zaino A.M., Hadden M.K., Eichman B.F., Lopes M., and Cimprich K.A.. 2020. HLTF promotes fork reversal, limiting replication stress resistance and preventing multiple mechanisms of unrestrained DNA synthesis. Mol. Cell. 78:1237–1251.e7. 10.1016/j.molcel.2020.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous, R., Couch F.B., Mason A.C., Eichman B.F., Manosas M., and Cortez D.. 2013. Substrate-selective repair and restart of replication forks by DNA translocases. Cell Rep. 3:1958–1969. 10.1016/j.celrep.2013.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bétous, R., Mason A.C., Rambo R.P., Bansbach C.E., Badu-Nkansah A., Sirbu B.M., Eichman B.F., and Cortez D.. 2012. SMARCAL1 catalyzes fork regression and Holliday junction migration to maintain genome stability during DNA replication. Genes Dev. 26:151–162. 10.1101/gad.178459.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, K.P., and Cortez D.. 2018. RPA and RAD51: Fork reversal, fork protection, and genome stability. Nat. Struct. Mol. Biol. 25:446–453. 10.1038/s41594-018-0075-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhat, K.P., Krishnamoorthy A., Dungrawala H., Garcin E.B., Modesti M., and Cortez D.. 2018. RADX modulates RAD51 activity to control replication fork protection. Cell Rep. 24:538–545. 10.1016/j.celrep.2018.06.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blastyák, A., Hajdú I., Unk I., and Haracska L.. 2010. Role of double-stranded DNA translocase activity of human HLTF in replication of damaged DNA. Mol. Cell. Biol. 30:684–693. 10.1128/MCB.00863-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branzei, D., and Psakhye I.. 2016. DNA damage tolerance. Curr. Opin. Cell Biol. 40:137–144. 10.1016/j.ceb.2016.03.015 [DOI] [PubMed] [Google Scholar]
- Brun, J., Chiu R.K., Wouters B.G., and Gray D.A.. 2010. Regulation of PCNA polyubiquitination in human cells. BMC Res. Notes. 3:85. 10.1186/1756-0500-3-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugreev, D.V., Rossi M.J., and Mazin A.V.. 2011. Cooperation of RAD51 and RAD54 in regression of a model replication fork. Nucleic Acids Res. 39:2153–2164. 10.1093/nar/gkq1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccaldi, R., Sarangi P., and D’Andrea A.D.. 2016. The Fanconi anaemia pathway: New players and new functions. Nat. Rev. Mol. Cell Biol. 17:337–349. 10.1038/nrm.2016.48 [DOI] [PubMed] [Google Scholar]
- Chen, C.C., Feng W., Lim P.X., Kass E.M., and Jasin M.. 2018. Homology-directed repair and the role of BRCA1, BRCA2, and related proteins in genome integrity and cancer. Annu. Rev. Cancer Biol. 2:313–336. 10.1146/annurev-cancerbio-030617-050502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia, A., Bredemeyer A.L., Sowa M.E., Terret M.E., Jallepalli P.V., Harper J.W., and Elledge S.J.. 2009. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23:2415–2425. 10.1101/gad.1832309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia, A., Nimonkar A.V., Hu Y., Hajdu I., Achar Y.J., Izhar L., Petit S.A., Adamson B., Yoon J.C., Kowalczykowski S.C., et al. 2012. Polyubiquitinated PCNA recruits the ZRANB3 translocase to maintain genomic integrity after replication stress. Mol. Cell. 47:396–409. 10.1016/j.molcel.2012.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti, B.A., and Smogorzewska A.. 2020. Mechanisms of direct replication restart at stressed replisomes. DNA Repair. 95:102947. 10.1016/j.dnarep.2020.102947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortez, D. 2019. Replication-coupled DNA repair. Mol. Cell. 74:866–876. 10.1016/j.molcel.2019.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan, H., Mansour S., Reed R., Gillis M.K., Parent B., Liu B., Sztupinszki Z., Birkbak N., Szallasi Z., Elia A.E.H., et al. 2020. E3 ligase RFWD3 is a novel modulator of stalled fork stability in BRCA2-deficient cells. J. Cell Biol. 219:219. 10.1083/jcb.201908192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia, A.E., Wang D.C., Willis N.A., Boardman A.P., Hajdu I., Adeyemi R.O., Lowry E., Gygi S.P., Scully R., and Elledge S.J.. 2015. RFWD3-Dependent ubiquitination of RPA regulates repair at stalled replication forks. Mol. Cell. 60:280–293. 10.1016/j.molcel.2015.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feeney, L., Muñoz I.M., Lachaud C., Toth R., Appleton P.L., Schindler D., and Rouse J.. 2017. RPA-mediated recruitment of the E3 ligase RFWD3 is vital for interstrand crosslink repair and human health. Mol. Cell. 66:610–621.e4. 10.1016/j.molcel.2017.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, X., Yucer N., Liu S., Li M., Yi P., Mu J.J., Yang T., Chu J., Jung S.Y., O’Malley B.W., et al. 2010. RFWD3-Mdm2 ubiquitin ligase complex positively regulates p53 stability in response to DNA damage. Proc. Natl. Acad. Sci. USA. 107:4579–4584. 10.1073/pnas.0912094107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallina, I., Hendriks I.A., Hoffmann S., Larsen N.B., Johansen J., Colding-Christensen C.S., Schubert L., Sellés-Baiget S., Fábián Z., Kühbacher U., et al. 2021. The ubiquitin ligase RFWD3 is required for translesion DNA synthesis. Mol. Cell. 81:442–458.e9. 10.1016/j.molcel.2020.11.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong, Z., and Chen J.. 2011. E3 ligase RFWD3 participates in replication checkpoint control. J. Biol. Chem. 286:22308–22313. 10.1074/jbc.M111.222869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto, Y., Ray Chaudhuri A., Lopes M., and Costanzo V.. 2010. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 17:1305–1311. 10.1038/nsmb.1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howlett, N.G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G., et al. 2002. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 297:606–609. 10.1126/science.1073834 [DOI] [PubMed] [Google Scholar]
- Huang, T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., and D’Andrea A.D.. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8:339–347. 10.1038/ncb1378 [DOI] [PubMed] [Google Scholar]
- Inano, S., Sato K., Katsuki Y., Kobayashi W., Tanaka H., Nakajima K., Nakada S., Miyoshi H., Knies K., Takaori-Kondo A., et al. 2017. RFWD3-Mediated ubiquitination promotes timely removal of both RPA and RAD51 from DNA damage sites to facilitate homologous recombination. Mol. Cell. 66:622–634.e8. 10.1016/j.molcel.2017.04.022 [DOI] [PubMed] [Google Scholar]
- Kanao, R., Kawai H., Taniguchi T., Takata M., and Masutani C.. 2022. RFWD3 and translesion DNA polymerases contribute to PCNA modification-dependent DNA damage tolerance. Life Sci. Alliance. 5:5. 10.26508/lsa.202201584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche, P.L., Wing J., and Lehmann A.R.. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 14:491–500. 10.1016/S1097-2765(04)00259-X [DOI] [PubMed] [Google Scholar]
- Kee, Y., and Huang T.T.. 2015. Role of deubiquitinating enzymes in DNA repair. Mol. Cell. Biol. 36:524–544. 10.1128/MCB.00847-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kile, A.C., Chavez D.A., Bacal J., Eldirany S., Korzhnev D.M., Bezsonova I., Eichman B.F., and Cimprich K.A.. 2015. HLTF’s ancient HIRAN domain binds 3′ DNA ends to drive replication fork reversal. Mol. Cell. 58:1090–1100. 10.1016/j.molcel.2015.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, T.M., Son M.Y., Dodds S., Hu L., Luo G., and Hasty P.. 2015. RECQL5 and BLM exhibit divergent functions in cells defective for the Fanconi anemia pathway. Nucleic Acids Res. 43:893–903. 10.1093/nar/gku1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knies, K., Inano S., Ramírez M.J., Ishiai M., Surrallés J., Takata M., and Schindler D.. 2017. Biallelic mutations in the ubiquitin ligase RFWD3 cause Fanconi anemia. J. Clin. Invest. 127:3013–3027. 10.1172/JCI92069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolinjivadi, A.M., Sannino V., De Antoni A., Zadorozhny K., Kilkenny M., Técher H., Baldi G., Shen R., Ciccia A., Pellegrini L., et al. 2017. Smarcal1-Mediated fork reversal triggers mre11-dependent degradation of nascent DNA in the absence of Brca2 and stable Rad51 nucleofilaments. Mol. Cell. 67:867–881.e7. 10.1016/j.molcel.2017.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kottemann, M.C., and Smogorzewska A.. 2013. Fanconi anaemia and the repair of Watson and Crick DNA crosslinks. Nature. 493:356–363. 10.1038/nature11863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijger, P.H., Lee K.Y., Wit N., van den Berk P.C., Wu X., Roest H.P., Maas A., Ding H., Hoeijmakers J.H., Myung K., et al. 2011. HLTF and SHPRH are not essential for PCNA polyubiquitination, survival and somatic hypermutation: Existence of an alternative E3 ligase. DNA Repair (Amst). 10:438–444. 10.1016/j.dnarep.2010.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy, A., Jackson J., Mohamed T., Adolph M., Vindigni A., and Cortez D.. 2021. RADX prevents genome instability by confining replication fork reversal to stalled forks. Mol. Cell. 81:3007–3017.e5. 10.1016/j.molcel.2021.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaçon, D., Jackson J., Quinet A., Brickner J.R., Li S., Yazinski S., You Z., Ira G., Zou L., Mosammaparast N., et al. 2017. MRE11 and EXO1 nucleases degrade reversed forks and elicit MUS81-dependent fork rescue in BRCA2-deficient cells. Nat. Commun. 8:860. 10.1038/s41467-017-01180-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, W., Baxley R.M., Moldovan G.L., and Bielinsky A.K.. 2018. Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes (Basel). 10:10. 10.3390/genes10010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S., Chu J., Yucer N., Leng M., Wang S.Y., Chen B.P., Hittelman W.N., and Wang Y.. 2011. RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 286:22314–22322. 10.1074/jbc.M111.222802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand, N., Gibbs-Seymour I., and Bekker-Jensen S.. 2013. Regulation of PCNA-protein interactions for genome stability. Nat. Rev. Mol. Cell Biol. 14:269–282. 10.1038/nrm3562 [DOI] [PubMed] [Google Scholar]
- Mijic, S., Zellweger R., Chappidi N., Berti M., Jacobs K., Mutreja K., Ursich S., Ray Chaudhuri A., Nussenzweig A., Janscak P., et al. 2017. Replication fork reversal triggers fork degradation in BRCA2-defective cells. Nat. Commun. 8:859. 10.1038/s41467-017-01164-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi, A., Liaw H.J., Lee K.Y., Roest H.P., Maas A., Wu X., Moinova H., Markowitz S.D., Ding H., Hoeijmakers J.H., et al. 2008. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication forks. Proc. Natl. Acad. Sci. USA. 105:12411–12416. 10.1073/pnas.0805685105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motegi, A., Sood R., Moinova H., Markowitz S.D., Liu P.P., and Myung K.. 2006. Human SHPRH suppresses genomic instability through proliferating cell nuclear antigen polyubiquitination. J. Cell Biol. 175:703–708. 10.1083/jcb.200606145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen, K.J., Chaudhuri A.R., Follonier C., Herrador R., and Lopes M.. 2014. Visualization and interpretation of eukaryotic DNA replication intermediates in vivo by electron microscopy. Methods Mol. Biol. 1094:177–208. 10.1007/978-1-62703-706-8_15 [DOI] [PubMed] [Google Scholar]
- Neelsen, K.J., and Lopes M.. 2015. Replication fork reversal in eukaryotes: From dead end to dynamic response. Nat. Rev. Mol. Cell Biol. 16:207–220. 10.1038/nrm3935 [DOI] [PubMed] [Google Scholar]
- Pasero, P., and Vindigni A.. 2017. Nucleases acting at stalled forks: How to reboot the replication program with a few shortcuts. Annu. Rev. Genet. 51:477–499. 10.1146/annurev-genet-120116-024745 [DOI] [PubMed] [Google Scholar]
- Pathania, S., Nguyen J., Hill S.J., Scully R., Adelmant G.O., Marto J.A., Feunteun J., and Livingston D.M.. 2011. BRCA1 is required for postreplication repair after UV-induced DNA damage. Mol. Cell. 44:235–251. 10.1016/j.molcel.2011.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, M., Cong K., Panzarino N.J., Nayak S., Calvo J., Deng B., Zhu L.J., Morocz M., Hegedus L., Haracska L., et al. 2018. Opposing roles of FANCJ and HLTF protect forks and restrain replication during stress. Cell Rep. 24:3251–3261. 10.1016/j.celrep.2018.08.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk, S., Sedkov Y., Johnston D.M., Hodgson J.W., Black K.L., Kovermann S.K., Beck S., Canaani E., Brock H.W., and Mazo A.. 2012. TrxG and PcG proteins but not methylated histones remain associated with DNA through replication. Cell. 150:922–933. 10.1016/j.cell.2012.06.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole, L.A., and Cortez D.. 2017. Functions of SMARCAL1, ZRANB3, and HLTF in maintaining genome stability. Crit. Rev. Biochem. Mol. Biol. 52:696–714. 10.1080/10409238.2017.1380597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Psakhye, I., and Jentsch S.. 2012. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 151:807–820. 10.1016/j.cell.2012.10.021 [DOI] [PubMed] [Google Scholar]
- Quinet, A., Lemaçon D., and Vindigni A.. 2017. Replication fork reversal: Players and guardians. Mol. Cell. 68:830–833. 10.1016/j.molcel.2017.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinet, A., Tirman S., Cybulla E., Meroni A., and Vindigni A.. 2021. To skip or not to skip: Choosing repriming to tolerate DNA damage. Mol. Cell. 81:649–658. 10.1016/j.molcel.2021.01.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri, A., Callen E., Ding X., Gogola E., Duarte A.A., Lee J.E., Wong N., Lafarga V., Calvo J.A., Panzarino N.J., et al. 2016. Replication fork stability confers chemoresistance in BRCA-deficient cells. Nature. 535:382–387. 10.1038/nature18325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray Chaudhuri, A., Hashimoto Y., Herrador R., Neelsen K.J., Fachinetti D., Bermejo R., Cocito A., Costanzo V., and Lopes M.. 2012. Topoisomerase I poisoning results in PARP-mediated replication fork reversal. Nat. Struct. Mol. Biol. 19:417–423. 10.1038/nsmb.2258 [DOI] [PubMed] [Google Scholar]
- Rickman, K., and Smogorzewska A.. 2019. Advances in understanding DNA processing and protection at stalled replication forks. J. Cell Biol. 218:1096–1107. 10.1083/jcb.201809012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman, K.A., Noonan R.J., Lach F.P., Sridhar S., Wang A.T., Abhyankar A., Huang A., Kelly M., Auerbach A.D., and Smogorzewska A.. 2020. Distinct roles of BRCA2 in replication fork protection in response to hydroxyurea and DNA interstrand cross-links. Genes Dev. 34:832–846. 10.1101/gad.336446.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy, S., Luzwick J.W., and Schlacher K.. 2018. SIRF: Quantitative in situ analysis of protein interactions at DNA replication forks. J. Cell Biol. 217:1521–1536. 10.1083/jcb.201709121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher, K., Christ N., Siaud N., Egashira A., Wu H., and Jasin M.. 2011. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 145:529–542. 10.1016/j.cell.2011.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher, K., Wu H., and Jasin M.. 2012. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell. 22:106–116. 10.1016/j.ccr.2012.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson, L.J., Ross A.L., Szüts D., Alviani C.A., Oestergaard V.H., Patel K.J., and Sale J.E.. 2006. RAD18-independent ubiquitination of proliferating-cell nuclear antigen in the avian cell line DT40. EMBO Rep. 7:927–932. 10.1038/sj.embor.7400777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taglialatela, A., Alvarez S., Leuzzi G., Sannino V., Ranjha L., Huang J.W., Madubata C., Anand R., Levy B., Rabadan R., et al. 2017. Restoration of replication fork stability in BRCA1- and BRCA2-deficient cells by inactivation of SNF2-family fork remodelers. Mol. Cell. 68:414–430.e8. 10.1016/j.molcel.2017.09.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk, I., Hajdú I., Fátyol K., Hurwitz J., Yoon J.H., Prakash L., Prakash S., and Haracska L.. 2008. Human HLTF functions as a ubiquitin ligase for proliferating cell nuclear antigen polyubiquitination. Proc. Natl. Acad. Sci. USA. 105:3768–3773. 10.1073/pnas.0800563105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unk, I., Hajdú I., Fátyol K., Szakál B., Blastyák A., Bermudez V., Hurwitz J., Prakash L., Prakash S., and Haracska L.. 2006. Human SHPRH is a ubiquitin ligase for Mms2-Ubc13-dependent polyubiquitylation of proliferating cell nuclear antigen. Proc. Natl. Acad. Sci. USA. 103:18107–18112. 10.1073/pnas.0608595103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujanovic, M., Krietsch J., Raso M.C., Terraneo N., Zellweger R., Schmid J.A., Taglialatela A., Huang J.W., Holland C.L., Zwicky K., et al. 2017. Replication fork slowing and reversal upon DNA damage require PCNA polyubiquitination and ZRANB3 DNA translocase activity. Mol. Cell. 67:882–890.e5. 10.1016/j.molcel.2017.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., and Yamaizumi M.. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886–3896. 10.1038/sj.emboj.7600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weston, R., Peeters H., and Ahel D.. 2012. ZRANB3 is a structure-specific ATP-dependent endonuclease involved in replication stress response. Genes Dev. 26:1558–1572. 10.1101/gad.193516.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, B., Sheng Q., Nakanishi K., Ohashi A., Wu J., Christ N., Liu X., Jasin M., Couch F.J., and Livingston D.M.. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol. Cell. 22:719–729. 10.1016/j.molcel.2006.05.022 [DOI] [PubMed] [Google Scholar]
- Yang, X.H., and Zou L.. 2009. Dual functions of DNA replication forks in checkpoint signaling and PCNA ubiquitination. Cell Cycle. 8:191–194. 10.4161/cc.8.2.7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeeles, J.T., Poli J., Marians K.J., and Pasero P.. 2013. Rescuing stalled or damaged replication forks. Cold Spring Harb. Perspect. Biol. 5:a012815. 10.1101/cshperspect.a012815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, J., Ghosal G., and Chen J.. 2012. The HARP-like domain-containing protein AH2/ZRANB3 binds to PCNA and participates in cellular response to replication stress. Mol. Cell. 47:410–421. 10.1016/j.molcel.2012.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zellweger, R., Dalcher D., Mutreja K., Berti M., Schmid J.A., Herrador R., Vindigni A., and Lopes M.. 2015. Rad51-mediated replication fork reversal is a global response to genotoxic treatments in human cells. J. Cell Biol. 208:563–579. 10.1083/jcb.201406099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeman, M.K., and Cimprich K.A.. 2014. Causes and consequences of replication stress. Nat. Cell Biol. 16:2–9. 10.1038/ncb2897 [DOI] [PMC free article] [PubMed] [Google Scholar]