Abstract
Dormancy and heteromorphism are innate seed properties that control germination timing through adaptation to the prevailing environment. The degree of variation in dormancy depth within a seed population differs considerably depending on the genotype and maternal environment. Dormancy is therefore a key trait of annual weeds to time seedling emergence across seasons. Seed heteromorphism, the production of distinct seed morphs (in color, mass or other morphological characteristics) on the same individual plant, is considered to be a bet-hedging strategy in unpredictable environments. Heteromorphic species evolved independently in several plant families and the distinct seed morphs provide an additional degree of variation. Here we conducted a comparative morphological and molecular analysis of the dimorphic seeds (black and brown) of the Amaranthaceae weed Chenopodium album. Freshly harvested black and brown seeds differed in their dormancy and germination responses to ambient temperature. The black seed morph of seedlot #1 was dormant and 2/3rd of the seed population had non-deep physiological dormancy which was released by after-ripening (AR) or gibberellin (GA) treatment. The deeper dormancy of the remaining 1/3rd non-germinating seeds required in addition ethylene and nitrate for its release. The black seeds of seedlot #2 and the brown seed morphs of both seedlots were non-dormant with 2/3rd of the seeds germinating in the fresh mature state. The dimorphic seeds and seedlots differed in testa (outer seed coat) thickness in that thick testas of black seeds of seedlot #1 conferred coat-imposed dormancy. The dimorphic seeds and seedlots differed in their abscisic acid (ABA) and GA contents in the dry state and during imbibition in that GA biosynthesis was highest in brown seeds and ABA degradation was faster in seedlot #2. Chenopodium genes for GA and ABA metabolism were identified and their distinct transcript expression patterns were quantified in dry and imbibed C. album seeds. Phylogenetic analyses of the Amaranthaceae sequences revealed a high proportion of expanded gene families within the Chenopodium genus. The identified hormonal, molecular and morphological mechanisms and dormancy variation of the dimorphic seeds of C. album and other Amaranthaceae are compared and discussed as adaptations to variable and stressful environments.
Keywords: coat-imposed dormancy, gibberellins, hormone metabolism, seed coat properties, seed heteromorphism, thermal time modelling, abscisic acid, weed seed bank
1. Introduction
Predicting weed emergence in crop production systems is a global challenge that requires understanding mechanisms of weed ecology and trait evolution in response to climate change and altered agricultural practices (Neve et al., 2009; Westwood et al., 2018; Nakabayashi and Leubner-Metzger, 2021). The life-history strategy of weeds seems to be a blend of phenotypic plasticity and local adaptation for which within-species and within-seedlot variation in seed dormancy depth, as well as variation in dormancy release requirements are key to weed emergence timing (Baskin and Baskin, 2006; Walck et al., 2011; Batlla et al., 2020). The primary dormancy of freshly harvested mature seeds is established during seed maturation and its depth depends on the environmental conditions during the mother plant’s reproductive growth (Karssen, 1970; Penfield and MacGregor, 2017; Fernández Farnocchia et al., 2021). The control of germination by dormancy can be considered as block(s) to the completion of germination of an intact viable seed under otherwise favorable conditions, namely when the seed becomes non-dormant (Finch-Savage and Leubner-Metzger, 2006). The regulation of dormancy and germination in response to ambient environmental factors such as temperature is achieved, at least in part, by the balance and sensitivity to abscisic acid (ABA) and gibberellins (GA). ABA biosynthesis and signaling dominate the dormant state, whereas GA biosynthesis and signaling dominate the transition to germination. Morphological aspects of seed dormancy and germination include embryo shape and size, and the biophysical properties of the seed coat, endosperm and other tissues which may confer coat dormancy (Chahtane et al., 2017; Steinbrecher and Leubner-Metzger, 2017; Baskin and Baskin, 2019; Holloway et al., 2021; Walker et al., 2021).
Most plants produce one kind of seeds and fruits (diaspore monomorphism) which may exhibit variation in dormancy depth within the seedlot. Interestingly, some species evolved a heteromorphism strategy in which an individual plant produces two (dimorphism) or more (heteromorphism) kinds of diaspores (Imbert, 2002; Matilla et al., 2005; Baskin et al., 2014; Liu et al., 2018; Gianella et al., 2021). The heteromorphic seeds may differ in characteristics such as shape, color, size or mass, and this is often accompanied by different dispersal mechanisms, germination characteristics and dormancy levels. Plants with seed heteromorphism are commonly annuals of disturbed sites and stressful environments. Diaspore heteromorphism is considered a bet-hedging strategy in adaptation to unpredictable environments. Examples for dimorphic annuals include the Brassicaceae Aethionema arabicum and Diptychocarpus strictus (Baskin et al., 2014; Lenser et al., 2016; Arshad et al., 2021) and the Amaranthaceae Suaeda salsa (Wang et al., 2015; Li et al., 2016; Xu et al., 2017), other Suaeda and Atriplex species (Baskin et al., 2014; Liu et al., 2018), and Chenopodium album (Yao et al., 2010). The seed morphs of these Amaranthaceae species differ in size and seed coat color (black versus brown) and their distinct adaptive roles has been investigated in saline habitats (Yao et al., 2010; Baskin et al., 2014; Liu et al., 2018). In general, the brown seeds were found to be non-dormant and more salt-tolerant compared to the dormant black seeds which form a persistent soil seed bank. Amaranthaceae seed internal structure is characterized by a peripheral embryo and by variation of seed coat thickness which determines the coat-imposed physiological dormancy (Karssen, 1970; Baskin and Baskin, 2019; Fernández Farnocchia et al., 2021; Nakabayashi and Leubner-Metzger, 2021).
Chenopodium album L. (common lambsquarter, fat-hen) is a cosmopolitan, annual weed species of notable economic importance (Bajwa et al., 2019; Krak et al., 2019). It is among the most competitive and difficult-to-control weeds in several cropping systems and has developed resistance to multiple herbicide mode of actions. Using the C. album aggregate as a model, Krak et al. (2019) revealed the importance of Asia as one of the main centers of diversity for this weedy species. This work also showed that human-mediated dispersal of ruderal and weed plant species accompanied the westward spread of agriculture in the Neolithic period. The successful competitiveness of C. album as a weed derived predominantly from the ability to persist in the soil bank in the seed stage through complex regulation of physiological seed dormancy (Bouwmeester and Karssen, 1989; Bouwmeester and Karssen, 1993; Yao et al., 2010). Yao et al. (2010) comparatively investigated the seed morphology, dormancy and germination physiology of black and brown seeds of two C. album populations from semi-arid and light-saline environments in China which produced a similar proportion of the two seed morphs. In contrast to this, C. album weed populations from arable land in Europe produce only a small proportion (approx. 3%) of brown seeds. Most previous research on C. album was therefore focussed on the germination strategy of the black seeds to a better control of this harmful weed (Karssen, 1968; Karssen, 1976a; Karssen, 1976b; Saini et al., 1985; Bouwmeester and Karssen, 1993; Murdoch and Roberts, 1997; Bajwa et al., 2019). The underpinning molecular mechanisms of the C. album dimorphic seed’s dormancy and germination are largely unknown.
In the present study, we comparatively investigate the brown and black seed morphs of two C. album populations with different origin in Great Britain, seedlot #1 from an arable field followed by glasshouse propagation, and seedlot #2 from direct collection in an urban environment. The hormonal, molecular and morphological mechanisms of the seed morphs and seedlots differed considerably. This provided insight into the complex regulation of their dormancies and its variation in depth between and within seed morphs and seedlots. These findings are compared with results from other Amaranthaceae species and discussed in the wider context of seed dimorphism and dormancy mechanisms in weed seed biology.
2. Results
2.1. Dimorphic seeds of C. album differ in dormancy, seed coat morphology and germination responses to ambient temperatures
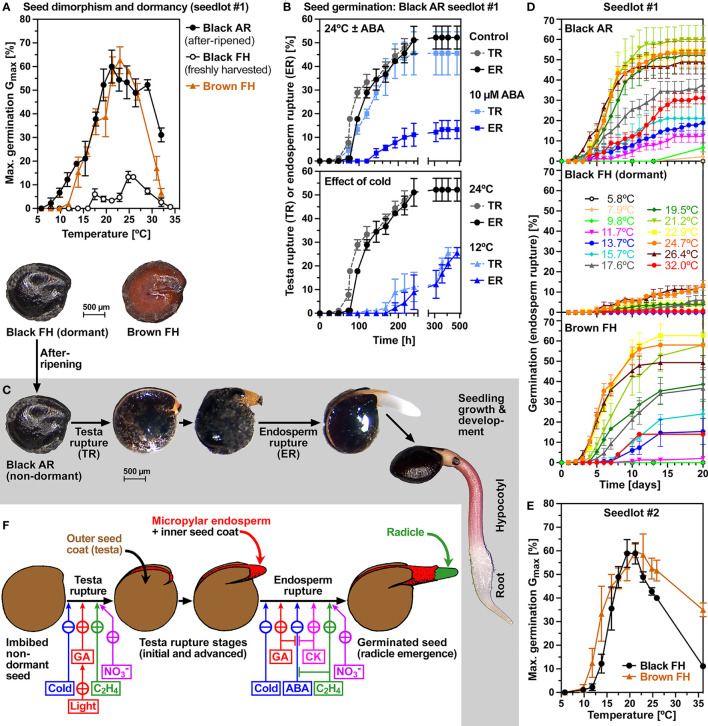
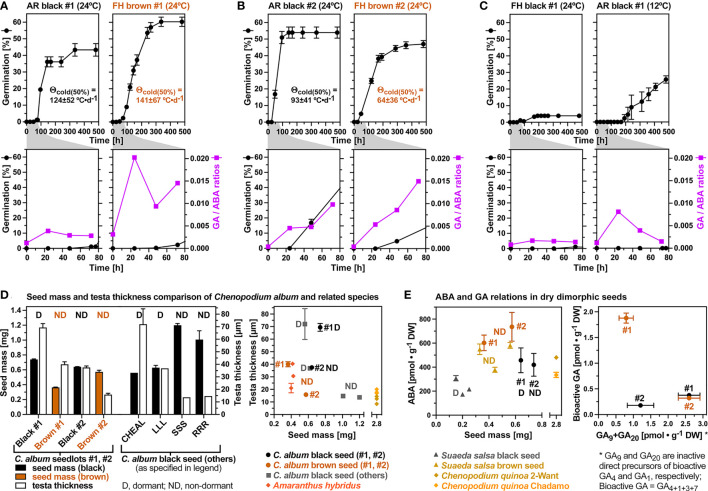
Figure 1 shows that the dimorphic seeds of C. album seedlots #1 and #2 differ in color (seed morphs: black and brown), dormancy and germination responses to ambient temperatures. Freshly harvested (FH) mature seeds of seedlot #1 (obtained by propagating accession #1 in the glasshouse) contained ca. 95% black seeds which were dormant and did not germinate over a wide range of temperatures ( Figures 1A, D ). The maximum germination percentage (Gmax) of these dormant populations of black seeds was ca. 10% at 25°C. Three months of seed after-ripening (AR, i.e. dry storage at room temperature) caused dormancy release resulting in ca. 60% Gmax and in widening of the permissive temperature window for germination of the black AR seeds ( Figures 1A, D ). In contrast to black seeds of seedlot #1 which were dormant in the FH state, black seeds of seedlot #2 (obtained by collecting seeds of accession #2 in the wild) were non-dormant upon harvest and germinated readily with ca. 60% Gmax ( Figure 1E ). In contrast to black seeds, brown seeds of accessions #1 and #2 were non-dormant in the FH state and germinated readily with ca. 60% Gmax without an AR storage requirement ( Figures 1A, D, E ). The distinct dimorphic seeds (black and brown) of these two distinct accessions (#1 and #2) were therefore highly suited to conduct a comparative analysis of the underpinning mechanisms.
Figure 1.
Germination physiology and temperature responses of Chenopodium album dimorphic seeds. (A) Seed dimorphism and temperature profile of C. album seedlot #1 maximum germination percentages (Gmax). The Gmax comparison at the temperatures indicated of mature freshly harvested (FH) black and brown seeds and after-ripened (AR) seeds imbibed at 24°C. (B) The effect of cis-S(+)-abscisic acid (ABA) at 24°C and of cold temperature (12°C) on the time courses of testa rupture (TR) and endosperm rupture (ER). (C) Physiological stages and visible events during the germination and subsequent seedling growth of C. album dimorphic seeds. (D) The kinetics of germination (scored as ER over time) of seedlot #1 seed morphs at different temperatures. (E) Seed dimorphism and temperature profile of C. album seedlot #2 Gmax. (F) Control of visible events (TR, ER) during C. album germination and the effects of light, cold, nitrate and hormonal (ABA; GA, gibberellins; C2H4, ethylene; CK, cytokinins) treatments (this work and Karssen, 1976a; Karssen, 1976b); drawing of seed stages from (Karssen, 1976b) reproduced with permission. (A-F) Mean ± SEM values are presented of at least 3 petri dishes each with 30 seeds imbibed in continuous white light.
Population-based thermal-time threshold modeling revealed that the non-dormant states of the black and brown seed morphs of seedlot #1 differed in their germination responses to ambient temperature ( Supplementary Figures 1A, B ). While the optimal (Topt) and base (Tbase) temperatures of black AR #1 seeds were 24°C and 8°C, respectively, the corresponding Topt and Tbase of brown seeds were ca. 2°C higher. In the sub-optimal (cold) temperature range the two morphs differed slightly in their thermal time constants (Θcold(50%)) which exhibited broad distributions (large SD) around their 50% values ( Supplementary Figures 1A, B ). In the supra-optimal (warm) temperature range the ceiling temperature (Tc(50%)) of the black seed population #1 was higher and more broadly distributed (35°C, large SD) compared to of the brown population (31°C, small SD) ( Supplementary Figures 1E, F ). In contrast to seedlot #1, the black and brown seed morphs of seedlot #2 were very similar in their Topt (ca. 22°C) and Tbase (ca. 10.5°C) temperatures, and their Θcold(50%) exhibited sharper distributions (small SDs) ( Supplementary Figures 1C, D ). The obtained Topt (22-24°C) were very similar to published values (22-25°C), while the obtained Tbase (8-10.5°C) temperatures were higher compared to the published values (3-6°C) for black C. album seeds (Karssen, 1976b; Murdoch et al., 1989; Roman et al., 1999; Guillemin et al., 2013). In these publications Gmax values of ca. 90% were observed, but others obtained much lower Gmax values of 50-60% for black C. album seeds even after 6 months of AR storage (Karssen, 1976a; Yao et al., 2010). The observed Gmax of ca. 60% in our C. album seedlots #1 and #2 ( Figure 1 ) was associated, even after 3 months of AR storage, with considerable fraction populations of non-germinating seeds ( Supplementary Figure 1 ).
The internal morphology of C. album seeds is characterized by a peripheral embryo which is curled around the perisperm ( Figure 2 ). The radicle and lower hypocotyl are covered with a living endosperm layer which is confined to the embryo’s radicle end and does not fully surround the entire embryo ( Figure 2C ). The embryo is however fully surrounded by a very thin inner seed coat (tegmen) and thicker hard outer seed coat (testa). Testa rupture (TR) and subsequent endosperm rupture (ER) are two visible events during the germination of black and brown C. album seeds ( Figure 1C ). At the TR stage with ruptured testa the expanding radicle remains covered by exposed micropylar endosperm and inner seed coat ( Figure 2A ). Subsequent ER rupture precedes the completion of germination by radicle emergence ( Figure 1C ). Light is known to be required for inducing TR (Karssen, 1976a) and abscisic acid (ABA) known to inhibit ER (Karssen, 1968) of black C. album seeds ( Figure 1F ). We demonstrate here that light is also required for the germination of brown C. album seeds ( Supplementary Figure 2A ), and that ABA delays germination by inhibiting ER without affecting the timing of TR of black ( Figure 1B ) and brown ( Supplementary Figure 2B ) seeds. Kinetin was shown to promote ER of black C. album seeds (Karssen, 1976a), but did not affect the seed germination of seedlots #1 and #2 ( Supplementary Figure 2C ). In contrast to light and ABA targeting either TR or ER, we found that cold temperature targets both processes and delayed C. album TR and ER ( Figure 1B ). The working model presented in Figure 1F summarizes how hormonal and environmental cues target C. album TR and ER.
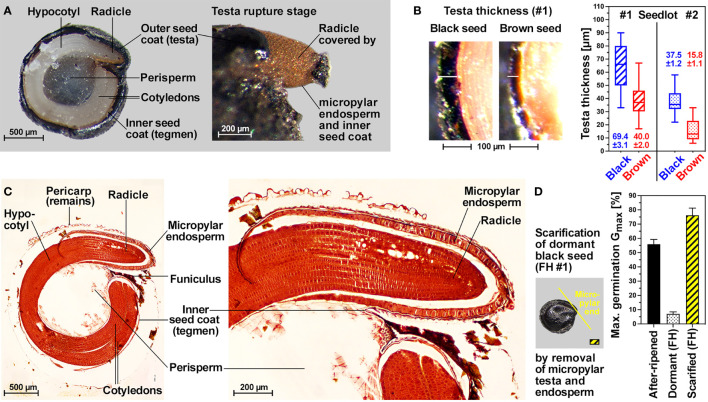
Figure 2.
Internal morphology, seed coat scarification and testa thickness of C. album dimorphic seeds. (A) Cross-section of dry black seed and visible endosperm protrusion after TR. (B) Images and box-and-whisker plot (min-max) of testa thickness of black and brown seed. Mean ± SEM values are presented of at least 50 seeds of each seed type. (C) Microscopic cross-section highlighting tissue features of C. album seeds after having undergone methyl methylacrylate Technovit fixation embedding protocol stained in safranin and counter stained in methylene blue. (D) Maximum germination (mean ± SEM values) of seedlot #1 dormant black, after-ripened black and mechanically scarified dormant black seeds.
Consistent with a role of testa thickness in C. album dormancy, black seeds of seedlot #1 (dormant) have a ca. 2-fold thicker testa compared to non-dormant brown seeds of seedlot #1 ( Figure 2B ). The testa thickness of black seeds of seedlot #2 (non-dormant) and brown seeds of seedlot #1 are roughly equal, and the testa of brown seeds of seedlot #2 is ca. 2-fold thinner ( Figure 2B ). It therefore seems that, at least for these two seed batches, that the dimorphic seeds differ in that the testas of brown seeds are thinner, and that the observed dormancy of black #1 seeds is associated with the thickest testa. Consistent with a role of the thick testa and the presence of coat dormancy in FH black #1 seeds, scarification of the dormant seed by seed coat removal at the micropylar end caused germination ( Figure 2D ). A scarification-induced Gmax of ca. 75% was observed for black #1 seeds which exceeds the dormancy-releasing effect of after-ripening (Gmax ca. 60% in AR black seeds, Figure 1 ). Together with the distinct responses to temperature and ABA ( Figure 1B ), the observed release of the coat-imposed dormancy suggests that it is controlled by the seed’s hormonal network.
2.2. Distinct hormonal regulation of dormancy and germination in dimorphic C. album seeds
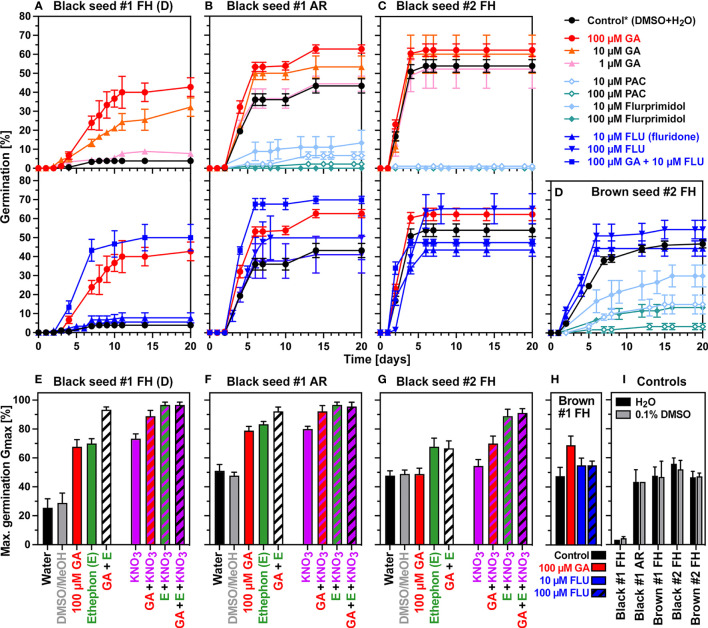
Figure 3 shows that gibberellins (GA) and ABA are key regulators of C. album seed dormancy and germination. Treatment experiments of FH black seeds of seedlot #1 with GA4+7 released their dormancy in a concentration-dependent manner with the 100 µM GA4+7 concentration exhibiting the highest dormancy-breaking activity ( Figure 3A ). While the carotenoid biosynthesis inhibitor fluridone (FLU, inhibits ABA biosynthesis) alone had no dormancy-releasing effect, the combined treatment of 100 µM GA4+7 plus FLU released the dormancy of FH black #1 seeds to the ca. 50-70% Gmax values observed for AR black #1 seeds ( Figures 3A, B ). Treatment combinations of GA, ethephon (E, an ethylene-releasing compound), and KNO3 resulted in Gmax values of >90% ( Figure 3E ). These results showed that GA is the predominant factor controlling dormancy release for the majority of dormant seeds, and that another fraction population of deeper dormant seeds require a more complex hormonal regulation which in addition to GA and ABA involves ethylene and nitrate signaling for the full dormancy release ( Figures 3E-G ). The black FH black seeds of seedlot #1 therefore exhibits two layers of physiological dormancy (PD): 2/3rd of the population has nondeep PD (nPD) which can be released by GA and AR, and 1/3rd of the population has a deeper PD (dPD) which cannot be released by GA and AR alone. This finding sheds new light on seed hetermorphism as a bet-hedging strategy to establish persistent seeds banks for this species.
Figure 3.
Germination responses of C. album dimorphic seeds to treatment with phytohormones and biosynthesis inhibitors. (A-D) Effects of gibberellins (GA4+7), flurprimidol, PAC, and fluridone (FLU), on the germination time courses of dormant black #1 (A), AR black #1 (B), black #2 (C), and brown #2 (D) seeds. (E-G) Effects of 100 µM GA4+7 (GA), 3.5 mM ethephon (E, ethylene release compound), and 10 mM KNO3 on the maximal germination percentages of dormant black #1 (E), AR black #1 (F), and black #2 (G) seed populations. (H) Effects of GA and FLU on brown #1 seeds. (I) Maximal germination percentages of water (H2O) and 0.1% (v/v) dimethyl sulfoxide (DMSO) controls. Mean ± SEM values are presented. AR, after-ripened; D, dormant; FLU, fluridone; MeOH, methanol; PAC, paclobutrazol; * indicates mean value between DMSO and H2O controls.
Pharmacological experiments with AR black seeds of seedlot #1 ( Figure 3B ) and FH black seeds of seedlot #2 ( Figure 3C ) demonstrated that GA4+7 promoted the germination of non-dormant seeds. The use of the GA biosynthesis inhibitors paclobutrazol (PAC) and flurprimidol further demonstrated that GA biosynthesis is essentially required for the germination of black seeds. Treatment with the carotenoid/ABA biosynthesis inhibitor fluridone (FLU) alone did not appreciably affect the germination of black seeds, but when combined with 100 µM GA4+7 it enhanced the GA effect ( Figures 3B, C ). As for the dormancy release, treatment combinations of GA, E and KNO3 caused maximum promotion and resulted in Gmax values of >90% ( Figures 3F, G ). The germination of FH brown seeds of seedlot #1 ( Figure 3H ) and seedlot #2 ( Supplementary Figure 2D ) was also promoted by GA4+7 treatment. Results with the GA biosynthesis inhibitors PAC and flurprimidol demonstrated that GA biosynthesis is also required for the germination of brown seeds ( Figure 3D ). From the different dose responses for PAC and flurprimidol we conclude that the germination of brown seeds may differ in their GA requirement or contents compared to black seeds ( Figure 3 ). These findings highlight the importance of GA biosynthesis and GA-ABA interactions for C. album seed germination and suggests that the two accessions (#1 and #2) and the seed morphs (black and brown) differ in their hormone metabolism and sensitivities.
2.3. Complex GA and ABA metabolism during dimorphic seed dormancy and germination
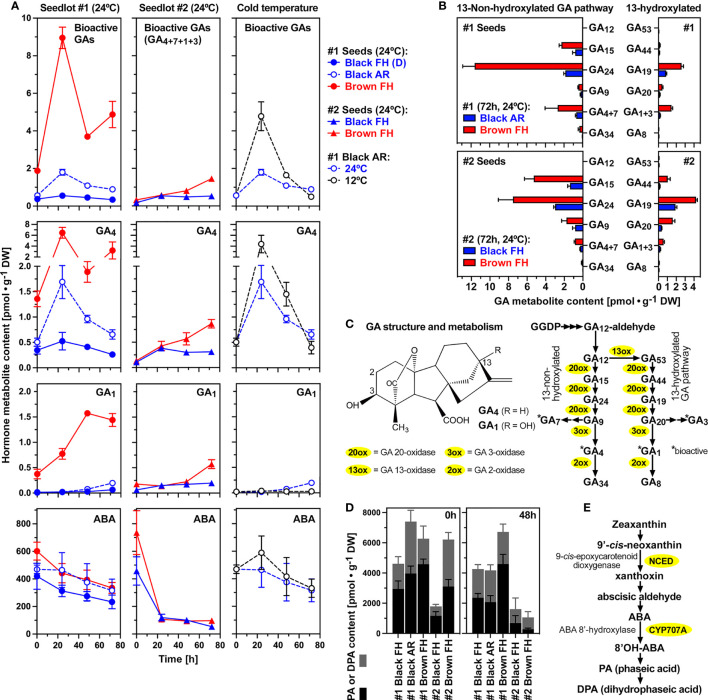
Figure 4 shows that the seed morphs (black and brown) and their dormancy state, as well as the accessions (#1 and #2) differ considerably in their GA and ABA metabolism. Bioactive GA (GA4, GA7, GA1, GA3) accumulated ca. 5-fold during the early imbibition of FH brown #1 seeds at 24°C ( Figure 4A ). A ca. 5-fold accumulation during the early imbibition was also observed in AR #1 black seeds, but the overall contents of bioactive GA accumulation in brown seeds were ca. 5-fold higher compared to AR black seeds. In contrast to brown and AR black seeds, no appreciable accumulation of bioactive GAs was observed in FH (D) black #1 seeds ( Figure 4A ). The contents of bioactive GAs in dry seeds of seedlot #1 were higher compared to dry seeds of seedlot #2. Starting from very low contents of bioactive GAs, accumulation during imbibition was also observed in brown #2 seeds, but the overall contents of bioactive GA accumulation remained much lower compared to brown #1 seeds ( Figure 4A ). Bioactive GA accumulation also occurred during the early imbibition of black #2 seeds, and remained lower compared to brown #2 seeds as it was the case for the #1 seed morphs ( Figure 4A ). These results highlight the importance of bioactive GA production in non-dormant seeds which is consistent with the results obtained with the GA biosynthesis inhibitors demonstrating that de novo GA biosynthesis was required for the completion of seed germination ( Figure 3 ). Brown seeds of seedlot #2 were more tolerant to GA biosynthesis inhibitors compared to black seeds ( Figure 3D ), which could be due to the observed increased de novo GA biosynthesis and bioactive GA accumulation in brown seeds compared to black seeds ( Figure 4 ).
Figure 4.
Gibberellin (GA) and abscisic acid (ABA) metabolite profiling during early imbibition of (C) album dimorphic seeds. (A) Temporal hormone metabolite content comparison of bioactive GAs and ABA in seedlots #1 and #2 at 24°C, and AR #1 black seeds at 12°C; Mean ± SEM values are presented. (B) GA biosynthesis pathway metabolite contents after 72h imbibition of dimorphic seeds from seedlots #1 and #2. (C) Molecular structure of bioactive C19-gibberellins GA4 and GA1 (left), GA metabolism and enzymes of the 13-non-hydroxylated and the 13-hydroxylated pathways (right). (D) Phaseic acid (PA) and dihydrophaseic acid (DPA) contents in dimorphic seeds of seedlots #1 and #2 in dry (left) and 48h imbibed (right) seeds. (E) ABA biosynthesis and catabolic pathway with key enzymes indicated.
The primary bioactive C19-GA accumulating during C. album seed germination was GA4 ( Figure 4A ), a product of the 13-non-hydroxylation GA biosynthesis pathway ( Figure 4C ). Accumulation of GA1, a C19-GA product of the 13-hydroxylation GA biosynthesis pathway, occurred to a lower extend, suggesting that the 13-non-hydroxylation GA biosynthesis pathway is dominant in C. album seeds ( Figure 4 ). Figure 4B demonstrates that this is indeed the case in both seed morphs (black and brown) and both accessions (#1 and #2). Especially in brown seeds much higher contents of the 13-non-hydroxylated precursors GA15 and GA24 were observed as compared to the corresponding 13-hydroxylated GA44 and GA19 metabolites ( Figure 4B ). Detail time course analysis of all major GA metabolites demonstrates the dominance of the 13-non-hydroxylation pathway during imbibition ( Supplementary Figure 3 ). In addition to GA4, also the 13-non-hydroxylated bioactive GA7 accumulated in seedlot #1, but not in seedlot #2. In contrast to this, the accumulation of 13-hydroxylated bioactive GA1 was not accompanied by appreciable GA3 accumulation ( Supplementary Figure 3 ).
In contrast to the accumulation of bioactive GA, the ABA contents of seeds declined during imbibition ( Figure 4A ). This decline did not appreciably differ among seed states within each seedlot, but was more rapid in black and brown seeds of seedlot #2 compared to seedlot #1. This raised the question of whether biosynthesis or catabolism of ABA is regulated differently between the black and brown seed morphs or between dormant vs non-dormant seeds. ABA degradation by ABA 8’-hydroxylases (CYP707A) and the subsequent accumulation of phaseic acid (PA) and dihydrophaseic acid (DPA) metabolites ( Figure 4E ) was evident in dry and imbibed black and brown seeds of both accessions ( Figure 4D ). PA and DPA contents were significantly higher in the dry seeds of AR #1 black and #1 brown seeds compared to FH #1 black seeds ( Figure 4D ), and subsequently declined during imbibition consistent with ABA declining across these seed states ( Figure 4A ). Imbibition of AR black #1 seeds at cold temperature (12°C) enhanced the accumulation of bioactive GAs without appreciably affecting the decline in ABA levels ( Figure 4A ).
2.4. Identification and phylogeny of Chenopodium GA and ABA metabolism genes
Key enzymes in ABA and GA metabolism include 9-cis-epoxycarotenoid dioxygenases (NCED), ABA 8’-hydroxylases (CYP707A), and GA-oxidases for GA biosynthesis (GA20ox, GA3ox) and inactivation (GA2ox) (Nambara et al., 2010; Urbanova et al., 2011; Hedden, 2020). They are encoded by multigene families (Huang et al., 2015; Lange and Lange, 2020; Arshad et al., 2021; Li et al., 2021; Sabir et al., 2022) and for Arabidopsis thaliana the members expressed during seed dormancy and germination are well known (Ogawa et al., 2003; Kushiro et al., 2004; Lefebvre et al., 2006; Okamoto et al., 2006; Nambara et al., 2010). To identify Chenopodium sequences of GA and ABA metabolism genes expressed in seeds we mined the Chenopodium quinoa genome (Jarvis et al., 2017) and conducted BLAST analyses with the A. thaliana and C. quinoa sequences via TAIR and Phytozome (Goodstein et al., 2012) as presented in detail in Supplementary Figure 4 . Using the obtained C. quinoa gene IDs we also identified differentially expressed genes (DEGs) during C. quinoa seed germination ( Supplementary Figure 5 ) from published transcriptomes (Wu et al., 2020; Hao et al., 2022). The combined information was used for the successful PCR cloning of 14 C. album cDNA sequences for GA and ABA metabolism genes ( Supplementary Figure 6 ).
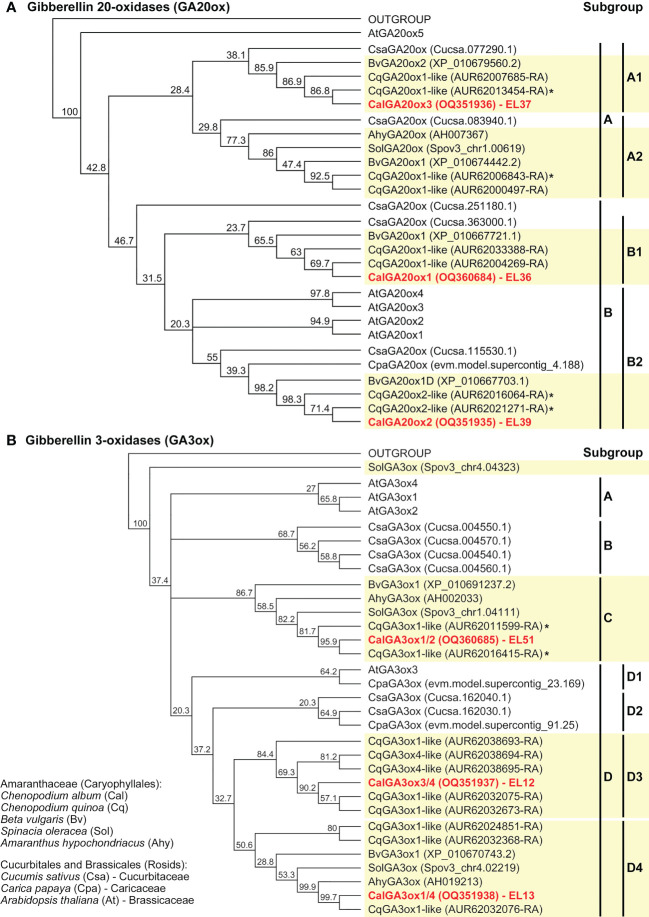
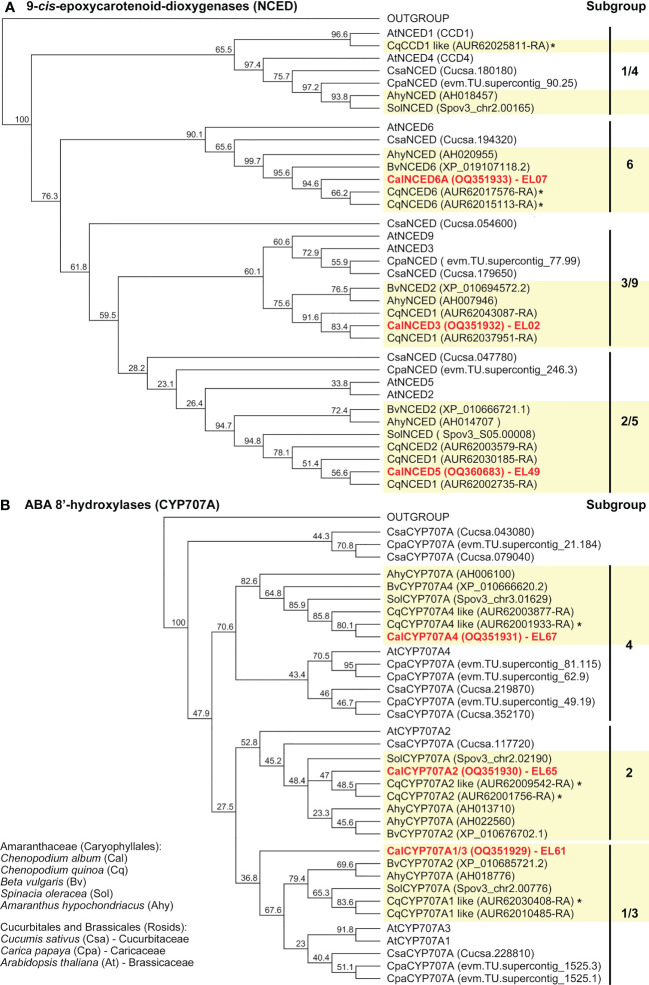
The phylogenetic trees for the GA ( Figure 5 ) and ABA ( Figure 6 and Supplementary Figure 7 ) metabolism genes support the observed gene family expansion and diversification within the Chenopodium lineage of the Amaranthaceae (Ma et al., 2021). The identified C. album sequences cover different Amaranthaceae groups ( Figures 5 , Figure 6 , and Supplementary Figure 7 ) and constitute putative orthologs to almost all the DEGs expressed in germinating C. quinoa seeds ( Supplementary Figure 5 ). The economically important C. quinoa is an allotetraploid species with a sequenced genome (Jarvis et al., 2017). For the ABA metabolism genes ( Figure 6 ) this expansion in Chenopodium was within each of the CYP707A1/3, CYP707A2, CYP707A4, NCED2/5, NCED3/9 and NCED6 subgroups known from A. thaliana. While each of these subgroups contained at least one A. thaliana type member, there was not in all subgroups contained a representative for Beta vulgaris, but each subgroup contained two to three C. quinoa representatives and C. album representative ( Figure 6 ). For the GA oxidase genes, the BLAST analyses ( Supplementary Figure 4 ) and phylogenetic trees suggest a more diversified gene expansion in the Chenopodium lineage. For the GA20ox ( Figure 5A ), GA3ox ( Figure 5B ) and GA2ox ( Supplementary Figure 7 ) genes the Amaranthaceae formed subgroups which were in many cases distinct from the other species and often did not contain Arabidopsis genes. An example for this among the GA3ox genes are the D3 and D4 subgroups which contain only Chenopodium and Amaranthaceae sequences, respectively ( Figure 5B ). Corresponding Brassicales (AtGA3ox3, Carica papaya) and Cucurbitaceae (Cucumis sativus) sequences were in the D1 and D2 subgroups. Other Arabidopsis (AtGA3ox1, AtGA3ox2, AtGA3ox4), Cucumis and Amaranthaceae sequences cluster separately in the A, B and C subgroups. In contrast to GA20ox and GA3ox where an abundance of Chenopodium genes has evolved and forms multiple subgroups, for the GA2ox genes A. thaliana has a similar number and diversity ( Supplementary Figure 7 ). The identified C. album GA2ox2 is a member of the C19-GA 2-oxidases which deactivate bioactive C19-GAs (Lange and Lange, 2020). The 14 identified C. album sequences ( Supplementary Figure 6 ) cover almost each major Chenopodium subgroup of GAox20, GAox3, NCED and CYP707A ( Figures 5 , Figure 6 ) genes and were used to analyze their expression in imbibed seeds with RT-qPCR.
Figure 5.
Phylogenetic tree of the predicted amino acid sequences of GA-oxidases. Known and putative amino acid GA20ox (A) and GA3ox (B) sequences of Chenopodium quinoa, Chenopodium album and other Amaranthaceae plus selected Brassicales and Cucurbitales species (as indicated) were aligned using ClustalW and Neighbor-Joining trees were built as described in methods. Note the expansion and diversification of the Chenopodium GA20ox and GA3ox gene subfamilies as contrasted to Beta vulgaris, C. sativum and A. thaliana. Chenopodium quinoa ( Supplementary Figure 4 ) and C. album ( Supplementary Figure 6 ) sequences (EL36, EL37, EL39, EL12, EL13, EL51) representing these subgroups were identified. An * indicates identified DEGs during C. quinoa seed germination ( Supplementary Figure 5 ).
Figure 6.
Phylogenetic tree of the predicted amino acid sequences of 9-cis-epoxycarotenoid dioxygenases (NCED) and ABA 8’-hydroxylases (CYP707A). Known and putative amino acid NCED (A) and CYP707A (B) sequences of Chenopodium quinoa, Chenopodium album and other Amaranthaceae plus selected Brassicales and Cucurbitales species (as indicated) were aligned using ClustalW and Neighbor-Joining trees were built as described in methods. Chenopodium quinoa ( Supplementary Figure 4 ) and C. album ( Supplementary Figure 6 ) sequences (EL02, EL07, EL49, EL61, EL65, EL67) representing the major subgroups were identified. An * indicates identified DEGs during C. quinoa seed germination ( Supplementary Figure 5 ).
2.5. Transcript expression patterns of GA and ABA metabolism genes in C. album seeds
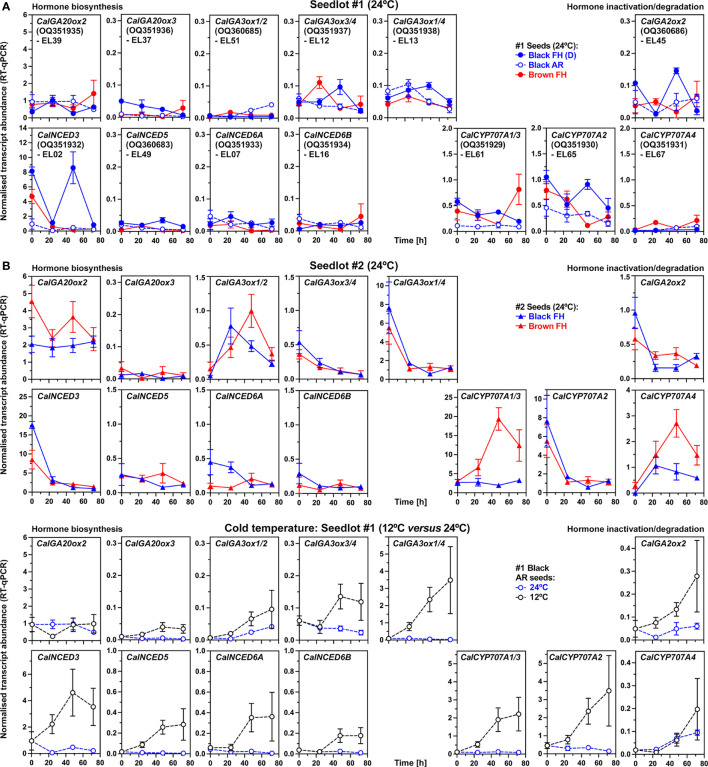
Gibberellin 20-oxidases (GA20ox) catalyze the formation of the inactive C19-precursors GA9 and GA20 of bioactive GA4 and GA1, respectively ( Figure 4C ). Among the three identified C. album GA20-oxidases CalGA20ox2 transcripts were most abundant in seeds, while CalGA20ox3 was only weakly expressed ( Figure 7 ). The CalGA20ox2 transcript abundances in seeds of seedlot #2 were 2-4 fold higher compared to seedlot #1 at 24°C ( Figures 7A, B ), as were the GA9 and GA20 contents ( Figure 4B ). Gibberellin 3-oxidases (GA3ox) catalyze the 3-hydroxylation which converts the inactive direct precursors GA9 and GA20 into bioactive GA4 and GA1, respectively ( Figure 4C ). CalGA3ox1/2 transcript abundances in seeds of seedlot #2 were higher compared to seedlot #1 at 24°C ( Figures 7A, B ). In seedlot #2 they also increased over time in parallel with bioactive GA accumulation, suggesting that the required GA3ox enzyme activity for the stronger accumulation of bioactive GA in seedlot #1 did not depend on additional transcript accumulation ( Figure 4A ). Transcript abundances of CalGA2ox2, encoding GA2ox which catalyzes GA inactivation by 2-hydroxylation ( Figure 4C ), were low and decreasing at 24°C except for dormant black #1 seeds ( Figure 7A ). Finally, the enhanced production of bioactive GA in cold-imbibed AR black #1 seeds ( Figure 4A ), was associated with the major accumulation of CalGA3ox1/4, and weak accumulation of CalGA3ox1/2, CalGA3ox3/4 and CalGA2ox2 transcripts ( Figure 7C ). Transcripts of their GA3ox and GA2ox orthologs accumulated during C. quinoa seed germination, while transcript abundances of GA20ox orthologs declined ( Supplementary Figure 5 ).
Figure 7.
RT-qPCR analysis of transcript abundance patterns in C. album dimorphic seeds for key genes in gibberellin (GA) and abscisic acid (ABA) metabolism. (A) RT-qPCR analysis of seedlot #1 dimorphic seeds imbibed at 24°C. The normalized transcript abundances of C. album GA-oxidase (GA20ox, GA3ox, GA2ox), 9-cis-epoxycarotenoid dioxygenase (NCED) and ABA 8’-hydroxylase (CYP707A) genes. For details about identified C. album genes see main text and Supplementary Figure 6 . (B) RT-qPCR analysis of seedlot #2 dimorphic seeds imbibed at 24°C. (A) RT-qPCR analysis of seedlot #1 AR black seeds imbibed at 12°C and 24°C. Mean ± SEM of at least 3 biological replicates.
In contrast to the accumulation of bioactive GA, the ABA contents of seeds declined during imbibition from contents in the dry state which were higher in brown as compared to black seeds ( Figure 4A ). This raised the question of whether biosynthesis or catabolism of ABA is regulated differently between the seed morphs and between different dormancy states. Four 9-cis-epoxycarotenoid dioxygenase (NCED) genes were expressed in C. album seeds, with transcript abundances of CalNCED3 declining in non-dormant seeds imbibed at 24°C ( Figures 7A, B ). In FH black #1 seeds the CalNCED3 transcript abundances increased in a transient manner ( Figure 7A ), suggesting a role in maintaining dormancy. In black and brown #2 seeds imbibed at 24°C the CalNCED5, CalNCED6A and CalNCED6B transcript abundances were low ( Figures 7A, B ). ABA degradation by ABA 8’-hydroxylases (CYP707A) and the PA/DPA pathway was more rapid in black and brown seeds of seedlot #2 compared to seedlot #1 ( Figure 4 ). While CalCYP707A2 transcript abundances in general declined upon imbibition at 24°C, those of CalCYP707A1/3 and CalCYP707A4 accumulated in the brown seeds of both seedlots ( Figures 7A, B ). Finally, the apparently similar pattern of ABA contents in cold-imbibed AR black #1 seeds ( Figure 4A ), was associated by the accumulation of transcripts of all four NCEDs and all three CYP707As ( Figure 7C ). A possible interpretation of these cold responses is that the CalNCED and CalGA2ox2 transcripts accumulate in the 1/3rd non-germinating seeds with deeper dormancy, while the CalCYP707A and CalGA3ox transcripts accumulate in the 2/3rd of germinating (non-dormant) seeds in the population. These findings also support that a tight interaction between GA and ABA, leading to distinct GA/ABA ratios ( Figure 8 ) are important which is referred to in the discussion.
Figure 8.
Chenopodium album dimorphic seed GA/ABA ratios and comparison of Amaranthaceae testa thicknesses and dry seed hormone contents. (A) Germination time courses and bioactive GA/ABA ratios of seedlot #1 black (after-ripened) and brown seeds imbibed at 24°C. (B) Germination time courses and bioactive GA/ABA ratios of seedlot #2 dimorphic seeds imbibed at 24°C. Mean ± SEM values are presented. The thermal-time constants Θcold(50%) ± SD from the population-based threshold modeling ( Supplementary Figure 1 ) are indicated and those of seedlot #1 are similar to Chenopodium quinoa (141-152 °C•d-1 from Ceccato et al., 2015). (C) Germination time courses and bioactive GA/ABA ratios of seedlot #1 black (after-ripened) imbibed at 12°C and 24°C. (D) Comparisons of C. album seed mass and testa thickness of the dimorphic seeds of seedlots #1 and #2 (this work) with published work: CHEAL (Gardarin et al., 2010), and LLL, SSS, RRR (Karssen, 1970) (left panel). Comparisons of C. album seed mass and testa thickness with Amaranthus hybridus (Gardarin et al., 2010; Nakabayashi and Leubner-Metzger, 2021) and C. quinoa (Ceccato et al., 2015) (right panel). Note that for C. album seedlots #1 and #2 the D (dormant) refers to the freshly harvested mature seed with full physiological dormancy (non-deep and deep PD; nPD+dPD) for seedlot #1, whereas the ND refers to the 2/3rd of non-dormant seed with nPD not induced during seed maturation. (E) Comparisons of ABA and GA relations in dry dimorphic seeds of C. album (this work), Suaeda salsa (Wang et al., 2015; Li et al., 2016; Xu et al., 2017), and C. quinoa (Ceccato et al., 2015).
3. Discussion
3.1. Roles for testa thickness and color in C. album dimorphic seed dormancy
Phylogenetic analyses of the internal seed morphology and associated ecophysiological seed traits led to the conclusion that heterodiaspory within the Amaranthaceae sensu lato evolved as a bet-hedging strategy in lineages with fast seed germination (Kadereit et al., 2017). Within the genus Chenopodium this however did not lead to seed heteromorphism for the very fast germinating C. quinoa (T50% 0-2 days), but it did for the annual weed C. album (T50% 2-7 days) with its black and brown seed morphs (this work and Yao et al., 2010). A peripheral embryo with annular shape surrounding the central perisperm (nutrient tissue) and with the outer embryo edge aligned against the inner surface of the bitegmic seed coat is considered as the ancestral morphological structure of Amaranthaceae seeds (Prego et al., 1998; Sukhorukov et al., 2015; Kadereit et al., 2017; Baskin and Baskin, 2019; Vandelook et al., 2021). We found in the two C. album seedlots (#1 and #2) the testa is much thicker compared to the very thin tegmen ( Figure 2 ), a finding which was also made for other Amaranthaceae (Prego et al., 1998; Ceccato et al., 2015; Sukhorukov et al., 2015). The C. album seed morphs share this ancestral internal seed morphology trait with other Chenopodium species and other Amaranthaceae. Seed coat thickness of weed species is positively related to their soil bank persistence (Gardarin et al., 2010). Together with seed coat pigments (color) and other biophysical properties such as permeability the thickness determines dormancy and longevity as key seed traits (Finch-Savage and Leubner-Metzger, 2006; MacGregor et al., 2015; Song et al., 2017; Steinbrecher and Leubner-Metzger, 2017; Nakabayashi and Leubner-Metzger, 2021).
Although the brown seed morph was consistently associated with non-dormancy (ND) in C. album (this work and Yao et al., 2010), Suaeda salsa (Wang et al., 2015; Li et al., 2016; Xu et al., 2017) and other Amaranthaceae with brown and black seeds (Wang et al., 2008; Baskin et al., 2014; Bhatt and Santo, 2016; Liu et al., 2018), the black seed morph was not always dormant ( Figure 8D ). The black seed coat pigment per se is therefore not sufficient to confer the coat-imposed dormancy. Seed mass differences between the morphs were also not associated with this trait as the brown seed morph can be either smaller ( Figure 8D ) or larger ( Figure 8E and Yao et al., 2010). Scarification of the dormant (D) black seeds of C. album seedlot #1 clearly demonstrated that this is indeed coat-imposed dormancy associated with distinct testa thickness of the two seedlots as well as the morphs within each seedlot ( Figure 8D ). A thin testa of < 30 µm was associated with the ND of black (SSS, RRR; i.e. two types of short-day conditions for mother plant growth used by Karssen, 1970) and brown (#2) C. album seeds, as well as with C. quinoa seeds which are larger compared to C. album ( Figure 8D ). A thick testa of up to ca. 70 µm was associated with dormant black C. album seeds (#1, CHEAL), but if the testa thickness is ca. 40 µm it was associated with either D black (LLL; long-day conditions for mother plant growth used by Karssen, 1970) or ND brown (#1) C. album seeds ( Figure 8D ). For C. album long-day conditions (LLL) during mother plant reproduction are known to lead to thicker testa and deeper dormancy, whereas short-day conditions (RRR) lead to thinner testa and low dormancy (Karssen, 1970). In agreement with this, the long-day conditions used for our glasshouse production of seedlot #1 led to D black seeds with thick testas of ca. 70 µm ( Figures 2B , 8D ), while a testa thickness ca. 40 µm was observed for the ND brown #1 seeds. In addition to day length also temperature during mother plant reproduction is known to affect coat-imposed dormancy for example of Amaranthus hybridus ( Figure 8D and Fernández Farnocchia et al., 2021) and of A. thaliana (MacGregor et al., 2015).
3.2. Complex hormonal regulation of Amaranthaceae seed dormancy and germination
Dormancy differences associated with distinct physicochemical seed and fruit coat properties are known to affect the flux of hormones and oxygen required for the control of germination (Hermann et al., 2007; Steinbrecher and Leubner-Metzger, 2017; Nakabayashi and Leubner-Metzger, 2021). Two C. quinoa accessions which differed in their dry seed ABA contents ( Figure 8E ) also differed in the amount of ABA leaching into the medium upon seed imbibition (Ceccato et al., 2015). Their dry seed ABA contents were very similar to those of black and brown C. album and S. salsa seeds ( Figure 8E ). In agreement with differences in testa permeability of the C. album seed morphs, imbibed black and brown seeds of seedlot #2 differed in the dose responses for GA biosynthesis inhibitors ( Figure 3A ). The dry black (ND) seeds of seedlot #2 also contained lower ABA degradation metabolites compared to the brown #2 (ND), the brown #1 (ND) and black #1 (D, AR) seeds ( Figure 4C ), suggesting that distinct oxygen permeability of the testa may have caused a distinct ABA metabolism during seed maturation. Interestingly, ABA contents in dry states were higher for the brown seed morphs compared to the black seed morphs of C. album and S. salsa ( Figure 8E and Wang et al., 2015; Li et al., 2016; Xu et al., 2017). Dormancy in these species was therefore also not associated with higher dry seed ABA contents, and ABA degradation occurred during seed imbibition in ND and D seeds of both morphs. The rate of ABA degradation during imbibition was higher in brown compared to black seeds of S. salsa (Li et al., 2016), but in C. album the rate did not appreciably differ between the black and brown seed morphs ( Figure 4A ). The rate of ABA degradation during imbibition was however higher in C. album seedlot #2 as compared to seedlot #1. This suggests that ratios and interaction of ABA with dormancy-releasing hormones and distinct hormone sensitivities may be more important than ABA contents itself for the seed responses to environmental cues such as ambient temperature.
Seedlots of C. album (black seeds) and C. quinoa are known to differ in their responses to ambient temperatures and water potentials (Roman et al., 1999; Soureshjani et al., 2022). In agreement with this, C. album seedlots #1 and #2 differed in their temperature responses and this was mainly obvious in the sub-optimal (colder) temperature range ( Figure 1 and Supplementary Figure S1 ). The release of the nondeep physiological dormancy (increase in Gmax) during the after-ripening storage of black #1 seeds was associated with widening of the permissive window mainly towards colder temperatures. Within a seedlot the dimorphic seeds (AR black #1 seeds) differed less in their responses to cold temperatures, but they differed more in the supra-optimal (warmer) temperature range. Chenopodium album seeds germinate with TR and ER as two subsequent visible events which allows to assign targets for hormones and environmental cues ( Figure 1 and Karssen, 1968; Karssen, 1976a; Karssen, 1976b). We found that while ABA has ER as its sole target, cold temperatures (12°C) close to the Tbase (8°C) delayed both TR and ER ( Figure 1B ). Light, GA, cytokinins, ethylene and nitrate were shown to act as ABA antagonists to fully release the dormancy and increase germination speed by targeting TR and ER, respectively ( Figure 1F ). Earlier work demonstrated that black seeds even in the after-ripened state required light for germination, light increased the Gmax in a seedlot-specific manner typically to 55-85% (Karssen, 1970; Karssen, 1976a; Machabée and Saini, 1991; Altenhofen and Dekker, 2013). In agreement with this earlier work, we found that the Gmax of AR black and brown seeds of seedlot #1 was very low in darkness and increased in the light ca. 60% ( Supplementary Figure S2 ). Light is known to act via increasing GA biosynthesis and signaling to release dormancy and promote germination (Finch-Savage and Leubner-Metzger, 2006). Treatment of imbibed seeds with GA biosynthesis inhibitors in the light demonstrated that both morphs (black and brown) required GA biosynthesis also for germination ( Figure 3 ). Furthermore, treatment with bioactive GA and fluridone caused dormancy release resulting in a Gmax of up to ca. 70% seed germination. We conclude from these findings that 2/3rd of the seed populations have either a layer of nondeep PD (nPD) which can be released by GA and AR in black seeds of seedlot #1, or have ND if this layer of nPD was not induced during the seed maturation as it was observed in brown seeds of both seedlots, and black seeds of seedlot #2. These triggered our detail investigation of the GA and ABA metabolism and raised the importance of the GA/ABA ratios in the dimorphic seeds.
3.3. Diversification and expression of Chenopodium GA and ABA metabolism genes
Comparative analysis of 5 sequenced Amaranthaceae and 9 other plant genomes (Ma et al., 2021) revealed a high proportion of expanded gene families associated with a recent whole genome duplication event in this lineage. In contrast to the gene family expansion and diversification observed in the C. quinoa genome, the genomes of Beta vulgaris and Amaranthus cruentus (Ma et al., 2021) were rather characterized by gene family contractions. The genus Chenopodium with its >150 species has a worldwide distribution. The economically important C. quinoa is an allotetraploid which resulted from hybridization of diploid ancestral lineages (Jarvis et al., 2017). For C. album the allohexaploid origin resulting from hybridization between diploid and tetraploid ancestral lineages was confirmed (Krak et al., 2016) and the evolutionary histories of Eurasian representatives reconstructed (Mandak et al., 2018). Our detailed sequence analyses of the identified C. album genes revealed their putative C. quinoa orthologs with numerous sequences ( Figures 5 , Figure 6 , and Supplementary Figure 7 ). The phylogenetic analyses of GA and ABA metabolism genes support an extended diversification of the Chenopodium GA20ox, GA3ox, NCED, and CYP707A genes within the Amaranthaceae in a manner distinct from the A. thaliana. Biochemical analyses not only with the Arabidopsis proteins, but also with Cucumis proteins from other subgroups ( Figure 5 ) demonstrated that they have GA-oxidase enzyme activity (Lange and Lange, 2020), suggesting that this is also the case for related diversified Chenopodium subgroup members. Divergence and subgroups without Arabidopsis genes were also observed for the GA-oxidase subfamilies in other species in phylogenetic analyses (Giacomelli et al., 2013; Huang et al., 2015; Sabir et al., 2022). The situation is similar for NCED (Arshad et al., 2021; Li et al., 2021) and CYP707A (Zheng et al., 2012). However, even the comprehensive work of Huang et al. (2015) into the divergence and adaptive evolution of the GA-oxidase genes in plants did not include any Amaranthaceae sequences. Inclusion of Amaranthaceae sequences in phylogenies is however relevant to also cover the Caryophyllales which is distinct from the Rosids and Asterids. Transcriptome analyses of C. quinoa seed germination (Wu et al., 2020; Hao et al., 2022) demonstrated that for several cases the corresponding orthologs were expressed during C. quinoa and C. album imbibition as detailed in Supplementary Figure 5 .
Taken together, distinct patterns of gene expression and ABA and GA metabolites were observed for C. album seeds. They differed between the dimorphic seeds (black versus brown) as well as between the two accessions (#1 versus #2). We found that the 13-non-hydroxylated GA biosynthesis pathway dominates in both dimorphs and both seedlots, with GA4 as the major bioactive GA ( Figure 4 ). The 13-non-hydroxylated GA biosynthesis pathway and GA4 as bioactive GA was also evident in dimorphic Sueada salsa seeds (Li et al., 2016), but the 13-hydroxylated pathway was not studied in this species. When dry C. album seeds were compared for their contents in bioactive GAs and their direct precursors (GA9+GA20) distinct strategies became evident ( Figure 8E ). Dry brown seeds of seedlot #1 had mainly bioactive GAs stored, whereas dry seeds of seedlot #2 mainly the direct precursors. Dry black seeds of seedlot #1 also mainly had the direct precursors stored, whereas dry black seeds of seedlot #2 were low in both. Together with ABA degradation this led to the observed differences in the GA/ABA ratios ( Figure 8 ). For seedlot #1 the GA/ABA ratio remained very low in FH (dormant) black seeds, increased >3-fold in AR black and >22-fold in brown seeds ( Figures 8A, C ).
For seedlot #1 this change in GA/ABA ratio was not associated without major changes in the transcript abundance patterns for the key GA biosynthesis enzymes during imbibition, which is consistent with stored bioactive GAs or direct precursors in the dry seeds ( Figure 8E ). In dormant black #1 seeds the ABA biosynthesis gene CalNCED3 was induced during imbibition suggesting a role in dormancy maintenance ( Figure 7A ). In agreement with this, for the B. vulgaris corresponding ABA metabolism gene upregulation in imbibed seeds during dormancy (NCEDs) and germination (CYP707As) has been reported (Hermann et al., 2007; Hourston et al., 2022). In contrast to C. album seedlot #1, the morphs of seedlot #2 did not appreciably differ in their GA/ABA ratio patterns which increased 5-10 fold ( Figure 8B ). This was associated with higher transcript abundance for CalGA20ox2 and up-regulation of the CalGA3ox1/2, CalCYP707A1/3 and CalCYP707A4 genes especially in brown seeds of seedlot #2 ( Figure 7B ). The up-regulation of the CalCYP707A1/3 and CalCYP707A4 genes explains the rapid ABA degradation and steep increase in the GA/ABA ratios in the dimorphic seeds of seedlot #2 ( Figure 8B ). The importance of GA-ABA interaction was also evident upon ABA treatment of imbibed C. quinoa seeds which inhibited the decline in the transcript abundances observed in the control for CqGA20ox2-like, CqNCED6, CqCYP707A2 and CqCYP707A2-like ( Supplementary Figure 5 ).
3.4. Hierachic layering of dormancy release and seed dimorphism as bet-hedging strategies
Interestingly, when AR black #1 seeds were imbibed at cold temperature (12°C) the GA/ABA ratios transiently increased ( Figure 8C ) and both GA and ABA biosynthesis and degradation genes were up-regulated ( Figure 7C ). A possible interpretation for this is that the GA biosynthesis (CalGA3ox1/2, CalGA3ox3/4, CalGA3ox1/4) and ABA degradation (CalCYP707A1/3, CalCYP707A2, CalCYP707A4) genes are induced in the germinating seeds of the population as a cold stratification response. These non-dormant seeds constitute 2/3rd of the population for which nondeep PD (nPD) was released by the AR storage. In contrast to this, the induction of the GA inactivation (CalGA2ox2) and ABA biosynthesis (CalNCED3, CalNCED5, CalNCED6A, CalNCED6B) genes could be associated with the non-germinating 1/3rd of the population which has a distinct layer of deeper PD (dPD). This dPD can be released by a combination of ethylene and nitrate ( Figures 3E-G ). Machabée and Saini (1991) discovered that imbibed C. album seeds produce ethylene, that dormancy release of some seedlots require ethylene action, and that this occurs in a temperature-dependent manner and is further enhanced by nitrate.
Our analysis sheds new light on seed hetermorhism as a bet-hedging strategy to establish persistent seeds banks for this species. The observation that the population has a fraction with nondeep physiological dormancy (nPD) and the other deep physiological dormancy (dPD) suggest a distinct bet-hedging strategy via the hierarchical layering of sensitivity to environmental signals. Similar findings were made for the dormancy release in A. thaliana Cvi (Finch-Savage et al., 2007) in that the light requirement persisted also in the AR state and that the sensitivities to cold and nitrate differed between seedlots. The results in both species are consistent with a role for the GA/ABA balance in integrating dormancy release signals. Arabidopsis thaliana seeds continually adjust their dormancy status by sensing a range of environmental signals related to temporal (e.g. temperature) and spatial (e.g. light and nitrate) change that indicate conditions suitable for germination (Finch-Savage and Footitt, 2017). This apparent hierarchy and layering of dormancy depth in the dimorphic seeds of C. album therefore provides a bet-hedging strategy in which a fraction of the population forms a persistent seed bank. Future research will reveal how the C. album seed morphs differ in dormancy cycling behaviour.
4. Conclusion
Using C. album as weed model system, we conducted a comparative morphological and molecular analysis of its dimorphic seeds (black and brown) with two seedlots (#1 and #2) of distinct origin. This analysis revealed distinct patterns of GA and ABA metabolism and related gene expression between the seedlots and morphs. In non-dormant brown seeds the testa thickness was half of the corresponding black seeds. Freshly harvested black seeds of seedlot #1 had a thick testa and testa-imposed dormancy which can be released by scarification at the micropylar end. After-ripening or GA treatment caused dormancy release in black #1 seeds and released the nondeep PD (nPD) of 2/3rd of the seeds in the population. Black seeds of seedlot #2 had thinner testas, their thickness was half of black #1 and equal to brown #2, and brown seeds of seedlot #2 had an even thinner testa. The 2/3rd of the seedlot #2 black seeds were non-dormant in the FH state, suggesting that nPD was not induced in this population. Earlier work with black C. album seeds concluded that depending on the photoperiod during reproduction two distinct dormancy layers can be induced (Karssen, 1970). We speculate that the first one is nPD, which is not induced in black seeds of seedlot #2, but is induced in black seed of seedlot #1 in associated with a thicker testa and AR and GA responsiveness to release the nPD. This first dormancy layer is mechanistically associated with the investigated GA and ABA as summarized in Figure 8 . It explains the germination behavior of 2/3rd of the seeds in the populations. We propose that the 1/3rd non-germinating black seeds in the populations have a second deeper dormancy layer (dPD) imposed which can be released by a combination of ethylene and nitrate ( Figures 3E-G and Machabée and Saini, 1991). Our future work with the dimorphic seeds of C. album seedlots will address this second layer of seed dormancy including with transcriptome analysis. Understanding and integrating complex mechanisms of seed dormancy dynamics with realistic ecophysiological simulations are essential for more sustainable weed management strategies (Baskin and Baskin, 2006; Batlla et al., 2020; Nakabayashi and Leubner-Metzger, 2021).
5. Materials and methods
5.1. Plant material
Seeds of Chenopodium album L. accession #1 were initially obtained from Syngenta Ltd. in 2015 (field sample PS-9522, Brechin, Angus, UK, 56.73 N -2.66 W) and stored in hermetically sealed containers containing silica gel at room temperature. These seeds were re-propagated at Royal Holloway’s in-house glasshouse and harvested at maturity (seedlot #1). Seeds were sown in 2/3rd Levington F2+S seed compost and 1/3rd perlite (Evergreen Garden Care Ltd., Frimley, UK) containing 0.4 g/L of Exemptor® insecticide (Bayer CropScience Limited, Cambridge, UK) and watered daily. Soil and plants were sprayed with Provado® Ultimate Bug Killer (Bayer) biweekly to limit pest growth. Growth conditions set in the glasshouse were a 16-h photoperiod, and day/night temperature 20°C/18°C ± 2°C. Chenopodium album accession #2 was obtained from direct collection of seeds during autumn 2020 outside Egham’s leisure center (51.43 N 0.54 W), Surrey, UK (seedlot #2). Freshly harvested (FH) seeds were dried for 1 week at 14% relative humidity (RH) and seedlots cleaned using a gravity separator. Brown seeds were manually separated from the mainly black seeds of the populations. After-ripened (AR) seeds were obtained from FH seeds by after-ripening dry storage (20°C, 33% RH) for 11 weeks. This duration of AR storage is known to be sufficient to fully release a first layer of C. album primary seed dormancy; a second dormancy layer (if present) is not released by the AR storage (Karssen, 1970; Machabée and Saini, 1991).
5.2. Seed germination assays
Seed germination assays were performed using 3 replicates of 30 seeds in 60-mm Petri dishes. Seeds were imbibed with 2 ml of autoclaved deionized water on two layers of filter papers (MN713; Macherey-Nagel, Dueren, Germany) and sealed using parafilm. The assays were performed in MLR-352 Versatile Environmental Test Chambers (Panasonic, Bracknell, UK) under constant white light (100 µmol m-2 s -1) at 24°C. Temperature response assays were performed on a GRD1-LH temperature gradient plate device (Grant Instruments Ltd., Cambridge, UK). Dose-response germination assays were performed using gibberellin A4+7 (GA4+7; Duchefa Biochemie, Haarlem, The Netherlands), cis,trans-S(+)-ABA (ABA; Duchefa), flurprimidol (Sigma, St Louis, MO, USA), fluridone (FLU; Duchefa). Compounds were supplied from liquid stocks dissolved in dimethyl sulfoxide (DMSO) stored at -20°C. All dilutions were adjusted to the same DMSO concentration of 0.1% (v/v) and all experiments included a 0.1% (v/v) DMSO control as well as a water control. Germination was recorded daily using a stereomicroscope and scored as completed upon visible radicle emergence through the testa and endosperm. In experiments where testa rupture (TR) and endosperm rupture (ER) were scored separately, multiple daily recordings were taken.
5.3. Microscopy and morphological analyses
Images of dry seed cross sections were taken using a DCF480 digital camera attached to a MZ 12.5 stereomicroscope (Leica, Wetzlar, Germany). Seed coat thickness represents testa thickness (the tegmen is very thin) and was measured using IMAGEJ (version 1.52i; National Institute of Health, Bethesda, MD, USA). For higher-resolution microscopy images, five seeds were punctured at the micropylar end using a sterile needle and 5 were left as controls. These seeds were imbibed for 24 hrs inside a 60-mm Ppetri dish. The seeds were then fixed in 4% paraformaldehyde in PEM buffer (0.1 M 1, 4-piperazinediethanesulfonic acid (pH 6.9), 2 mM triethylene glycol diamine tetraacetic acid and 1 mM magnesium sulfate) (Zeller, 2001) before being vacuumed for 5 min, sealed with vacuum off for 15 min, then slowly release before repeating the vacuum process 3 times. Seeds were then washed with PBS buffer (138 mM NaCl; 2.7 mM KCl, pH 7.4) for two 30 min periods before being subject to a dehydration through an ethanol gradient. Then the seeds undergo imbedding with methyl methylacrylate resin from the Technovit 7100 protocol (Kulzer Technik, Wehrheim, Germany), modified by adding an ethanol: resin gradient to improve uptake into starchy tissues. The polymerization blocking was next performed following the Technovit histology protocol (Matsushima, 2014). Sections of 5 µm thickness cuts were sliced from the blocks using a HM 355S microtome (Thermo Scientific, Loughborough, UK), mounted on slides, stained in 1% safranin solution for 5 minutes, counterstained in 0.1% methylene blue 15 minutes and analyzed using a Nikon ECLIPSE Ni-E microscope (Nikon, Amstelveen, The Netherlands).
5.4. Hormone extraction and quantification
Five biological replicates of 10 mg black and brown C. album seeds in the dry state (0h) and imbibed for 24h, 48h and 72h at either 24°C or 12°C were used. Seeds were then snap frozen in liquid nitrogen and lyophilized for 2 days using a freeze dryer LYOVAC GT 2 (SEIB Industrie, Gothenburg, Sweden) and metabolites were extracted. The levels of GA and ABA pathway metabolites, physiologically active or non-active, were quantified by UHPLC-MS/MS as described previously (Turečková et al., 2009; Urbanova et al., 2013; Walker et al., 2021; Hourston et al., 2022).
5.5. PCR cloning of C. album sequences and phylogenetic analyses
To identify Chenopodium sequences of GA and ABA metabolism genes expressed in seeds we mined the Chenopodium quinoa genome (Jarvis et al., 2017) and conducted BLAST analyses with the A. thaliana and C. quinoa sequences via TAIR and Phytozome (Goodstein et al., 2012) as presented in detail in Supplementary Figure 4 . The combined information of the BLAST analyses and subgroups in phylogenetic trees was used to design primers based on C. quinoa sequences ( Supplementary Table 1 ) targeting the sub-groups for the PCR cloning of C. album sequences expressed in seeds. PCR products were sent to Eurofins Genomics (Ebersberg, Germany) for Sanger sequencing. A total of 17 sequences of various lengths were obtained ( Supplementary Table 1 ), analyzed by BLAST ( Supplementary Figure 6 ) and deposited in the GenBank as BankIt submissions 2668556 and 2664064 which provided the allocated accession numbers ( Supplementary Table 1 ). Phylogenetic analysis of Amaranthaceae, Brassicales and Cucurbitales GA oxidases ( Figure 5 , Supplementary Figure 7 ), NCEDs and CYP707As ( Figure 6 ) with known and putative amino acid sequences aligned using ClustalW (BLOSUM cost matrix, Gap open cost 10, Gap extend cost 0.1) and Neighbor-Joining trees (Saitou and Nei, 1987) were built using Geneious 8.1.9 Tree Builder (Geneious, San Diego, CA, USA) using Jukes-Cantor distance. Consensus support (minimum 20%) was determined using bootstrap (1000).
5.6. Gene expression analysis via RT-qPCR
Four replicate samples of 20 mg black and brown C. album seeds were taken from seedlots #1 or #2 in the dry state (0h) or imbibed for 24h, 48h and 72h at either 24°C or 12°C, as indicated. Samples were homogenized in liquid nitrogen using a pestle and mortar, before addition of 1 ml extraction buffer containing 2% (w/v) hexadecyltrimethyl-ammonium bromide (CTAB), 2% (w/v) polyvinyl pyrrolidone MW 4000 (PVP), 100 mM Tris-HCl (pH 8), 25 mM ethylenediaminetetraacetic acid (EDTA, pH 8), 2 M NaCl and 2% (w/v) β-mercaptoethanol. Using this, the RNA extraction was performed as described in Graeber et al. (2011) with their listed modifications. RNA quantification was performed using a NanoDrop™ spectrophotometer (ND-1000, Thermo Scientific™, Delaware, USA) and agarose gel electrophoresis was used to confirm the presence of RNA. RNA integrity was assessed by using Aligent Bioanalyzer™ (Aligent Technologies), the three replicates with the highest RIN values for each treatment were selected for cDNA synthesis. One µg of RNA was used for cDNA synthesis using the Invitrogen™ SuperScript™ III First-Strand synthesis kit (Thermo Scientific, Loughborough, UK) following the manufacturer’s protocol.
The C. album GAox, NCED and CYP707A sequences plus reference genes (ACT7-like, CDC27B, PUB33-like) were used to design specific primers for the RT-qPCR analysis ( Supplementary Table 2 ). A cDNA pool template (combination of all samples, black, brown, dormant, non-dormant, dry and imbibed) was used to verify the primers ( Supplementary Table 2 ). Subsequently, cDNAs from each of the three replicates per sampling point were used to conduct the qPCR analysis. RT-qPCR reactions were performed using a CFX96 Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) and was conducted as detailed in Graeber et al. (2011). After baseline correction was removed, Cq values and PCR efficiencies were calculated using PCR miner of the raw fluorescence data values (Zhao and Fernald, 2005). Reference gene suitability was assessed by the geNorm software (https://genorm.cmgg.be/), which identified two reference genes (ACT7-like, CDC27B) for seedlot #1 samples and two reference genes (PUB33-like, CDC27B) for seedlot #2 samples as the most stable and uniformly expressed across all treatments. Candidate genes were normalized against the geometric mean of the expression levels of the two reference genes as described (Graeber et al., 2011). Ct and normalized relative gene expression levels were plotted on a logarithmic scale using a non-linear fit on GraphPad Prism v7 (GraphPad Software, San Diego, CA, USA).
5.7. Thermal-time modeling and statistical analysis
The cardinal temperatures permissible for germination including base temperature (Tbase), optimal temperature (Topt) and ceiling temperature (Tc), and the thermal time constants Θ were identified by population-based threshold modelling for thermal time (“heat sums”) (Roman et al., 1999; Bradford, 2002; Steckel et al., 2004; Finch-Savage and Leubner-Metzger, 2006; Batlla and Benech-Arnold, 2015; Soureshjani et al., 2022). To achieve this, the germination rates GR, i.e. the inverse of time to germination for a given percentage of the population (1/tg), this was plotted against temperature ( Supplementary Figure 1 ). Linear regression analysis with GraphPad Prism v7 was used to calculate regression lines in the sub-optimal (colder) and supra-optimal (warmer) temperature regions. Their intercepts were used to estimate Tb, To and Tc, respectively ( Supplementary Figure 1 ). Their slopes provide the thermal time constants to germination (Θcold(g) and Θwarm(g)). At sub-optimal temperatures, Θcold(g) varies among individual seed fractions in the population and is normal distributed around Θcold(50%). At supra-optimal temperatures, Tc(g) is normal distributed around Tc(50%) ( Supplementary Figure 1 ). Statistical analyses of obtained germination curves were by using a non-linear fit on GraphPad Prism v7 (GraphPad Software, San Diego, CA, USA). This program was also used to calculate mean ± SD and SEM values.
Data availability statement
The datasets presented in this study can be found on Royal Holloways figshare: https://doi.org/10.17637/rh.21829842.v1.
Author contributions
EL, AS, KN, MS and GL-M conceptualized and designed the research. EL, MP, VT, and DT performed experiments. EL, MP, VT, DT, KN and GL-M analyzed and interpreted data. EL and GL-M wrote the original manuscript draft. All authors complemented on the draft and were thereby involved in shaping the manuscript. All authors contributed to the article and approved the submitted version.
Acknowledgments
We thank Yan Li, Thomas Holloway and Nahema Venceslai for scientific discussions, and Diane Grant, Sarah Rabjohn, Mark Levy and Chongboi Haokip for their support with plant propagation and seed processing.
Funding Statement
This research was funded by the Biotechnology and Biological Sciences Research Council to GL-M (BBSRC-LINK grant no. BB/M02203X/1), the BBSRC DTP iCASE National Productivity Investment Fund (NPIF) training grant to EL (BB/R505730/1, project reference 10887496), and direct funding from Syngenta Ltd. to KN and GL-M. The work was additionally supported by the “Centre for Experimental Plant Biology” (No. CZ.02.1.01/0.0/0.0/16_019/0000738).
Conflict of interest
Author AS was employed by Syngenta.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This BBSRC-funded study received additional direct funding and in-kind support by Syngenta Ltd. The company funder had the following involvement with the study: general conceptualization and overall design the research, collection and provision of seedlot #1, extension of the PhD studentship stipend beyond BBSRC-funding.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1156794/full#supplementary-material
References
- Altenhofen L. M., Dekker J. (2013). Complex regulation of Chenopodium album seed germination. Appl. Ecol. Environ. Sci. 1, 133–142. doi: 10.12691/aees-1-6-6 [DOI] [Google Scholar]
- Arshad W., Lenser T., Wilhelmsson P. K. I., Chandler J. O., Steinbrecher T., Marone F., et al. (2021). A tale of two morphs: developmental patterns and mechanisms of seed coat differentiation in the dimorphic diaspore model Aethionema arabicum (Brassicaceae). Plant J. 107, 166–181. doi: 10.1111/tpj.15283 [DOI] [PubMed] [Google Scholar]
- Bajwa A. A., Zulfiqar U., Sadia S., Bhowmik P., Chauhan B. S. (2019). A global perspective on the biology, impact and management of Chenopodium album and Chenopodium murale: Two troublesome agricultural and environmental weeds. Environ. Sci. pollut. Res. 26, 5357–5371. doi: 10.1007/s11356-018-04104-y [DOI] [PubMed] [Google Scholar]
- Baskin C. C., Baskin J. M. (2006). The natural history of soil seed banks of arable land. Weed Sci. 54, 549–557. doi: 10.1614/WS-05-034R.1 [DOI] [Google Scholar]
- Baskin C. C., Baskin J. M. (2019). Martin's peripheral embryo - unique but not a phylogenetic 'orphan' at the base of his family tree: a tribute to the insight of a pioneer seed biologist. Seed Sci. Res. 29, 155–166. doi: 10.1017/S0960258519000175 [DOI] [Google Scholar]
- Baskin J. M., Lu J. J., Baskin C. C., Tan D. Y., Wang L. (2014). Diaspore dispersal ability and degree of dormancy in heteromorphic species of cold deserts of northwest China: A review. Perspect. Plant Ecology Evol. Systematics 16, 93–99. doi: 10.1016/j.ppees.2014.02.004 [DOI] [Google Scholar]
- Batlla D., Benech-Arnold R. L. (2015). A framework for the interpretation of temperature effects on dormancy and germination in seed populations showing dormancy. Seed Sci. Res. 25, 147–158. doi: 10.1017/S0960258514000452 [DOI] [Google Scholar]
- Batlla D., Ghersa C. M., Benech-Arnold R. L. (2020). Dormancy, a critical trait for weed success in crop production systems. Pest Manage. Sci. 76, 1189–1194. doi: 10.1002/ps.5707 [DOI] [PubMed] [Google Scholar]
- Bhatt A., Santo A. (2016). Germination and recovery of heteromorphic seeds of Atriplex canescens (Amaranthaceae) under increasing salinity. Plant Ecol. 217, 1069–1079. doi: 10.1007/s11258-016-0633-6 [DOI] [Google Scholar]
- Bouwmeester H. J., Karssen C. M. (1989). Environmental factors influencing the expression of dormancy patterns in weed seeds. Ann. Bot. 63, 113–120. doi: 10.1093/oxfordjournals.aob.a087713 [DOI] [Google Scholar]
- Bouwmeester H. J., Karssen C. M. (1993). Seasonal periodicity in germination of seeds of Chenopodium album l. Ann. Bot. 72, 463–473. doi: 10.1006/anbo.1993.1133 [DOI] [Google Scholar]
- Bradford K. J. (2002). Applications of hydrothermal time to quantifying and modeling seed germination and dormancy. Weed Sci. 50, 248–260. doi: 10.1614/0043-1745(2002)050[0248:AOHTTQ]2.0.CO;2 [DOI] [Google Scholar]
- Ceccato D., Bertero D., Batlla D., Galati B. (2015). Structural aspects of dormancy in quinoa (Chenopodium quinoa): importance and possible action mechanisms of the seed coat. Seed Sci. Res. 25, 267–275. doi: 10.1017/S096025851500015X [DOI] [Google Scholar]
- Chahtane H., Kim W., Lopez-Molina L. (2017). Primary seed dormancy: a temporally multilayered riddle waiting to be unlocked. J. Exp. Bot. 68, 857–869. doi: 10.1093/jxb/erw377 [DOI] [PubMed] [Google Scholar]
- Fernández Farnocchia R. B., Benech-Arnold R. L., Mantese A., Battla D. (2021). Optimization of next-generation emergence timing of Amaranthus hybridus is determined through seed dormancy modulation by the maternal environment. J. Exp. Bot. 72, 4283–4297. doi: 10.1093/jxb/erab141 [DOI] [PubMed] [Google Scholar]
- Finch-Savage W. E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x [DOI] [PubMed] [Google Scholar]
- Finch-Savage W. E., Cadman C. S. C., Toorop P. E., Lynn J. R., Hilhorst H. W. M. (2007). Seed dormancy release in arabidopsis cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51, 60–78. doi: 10.1111/j.1365-313X.2007.03118.x [DOI] [PubMed] [Google Scholar]
- Finch-Savage W. E., Footitt S. (2017). Seed dormancy cycling and the regulation of dormancy mechanisms to time germination in variable field environments. J. Exp. Bot. 68, 843–856. doi: 10.1093/jxb/erw477 [DOI] [PubMed] [Google Scholar]
- Gardarin A., Dürr C., Mannino M. R., Busset H., Colbach N. (2010). Seed mortality in the soil is related to seed coat thickness. Seed Sci. Res. 20, 243–256. doi: 10.1017/S0960258510000255 [DOI] [Google Scholar]
- Giacomelli L., Rota-Stabelli O., Masuero D., Acheampong A. K., Moretto M., Caputi L., et al. (2013). Gibberellin metabolism in Vitis vinifera l. during bloom and fruit-set: functional characterization and evolution of grapevine gibberellin oxidases. J. Exp. Bot. 64, 4403–4419. doi: 10.1093/jxb/ert251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gianella M., Bradford K. J., Guzzon F. (2021). Ecological, (epi)genetic and physiological aspects of bet-hedging in angiosperms. Plant Reprod. 34, 21–36. doi: 10.1007/s00497-020-00402-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein D. M., Shu S. Q., Howson R., Neupane R., Hayes R. D., Fazo J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K., Linkies A., Wood A. T., Leubner-Metzger G. (2011). A guideline to family-wide comparative state-of-the-art quantitative RT-PCR analysis exemplified with a brassicaceae cross-species seed germination case study. Plant Cell 23, 2045–2063. doi: 10.1105/tpc.111.084103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemin J. P., Gardarin A., Granger S., Reibel C., Munier-Jolain N., Colbach N. (2013). Assessing potential germination period of weeds with base temperatures and base water potentials. Weed Res. 53, 76–87. doi: 10.1111/wre.12000 [DOI] [Google Scholar]
- Hao Y. Q., Hong Y. C., Guo H. M., Qin P. Y., Huang A. N., Yang X. S., et al. (2022). Transcriptomic and metabolomic landscape of quinoa during seed germination. BMC Plant Biol. 22, 237. doi: 10.1186/s12870-022-03621-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. (2020). The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 61, 1832–1849. doi: 10.1093/pcp/pcaa092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann K., Meinhard J., Dobrev P., Linkies A., Pesek B., Heß B., et al. (2007). 1-Aminocyclopropane-1-carboxylic acid and abscisic acid during the germination of sugar beet (Beta vulgaris l.) - a comparative study of fruits and seeds. J. Exp. Bot. 58, 3047–3060. doi: 10.1093/jxb/erm162 [DOI] [PubMed] [Google Scholar]
- Holloway T., Steinbrecher T., Pérez M., Seville A., Stock D., Nakabayashi K., et al. (2021). Coleorhiza-enforced seed dormancy: A novel mechanism to control germination in grasses. New Phytol. 229, 2179–2191. doi: 10.1111/nph.16948 [DOI] [PubMed] [Google Scholar]
- Hourston J. E., Steinbrecher T., Chandler J. O., Pérez M., Dietrich K., Turečková V., et al. (2022). Cold-induced secondary dormancy and its regulatory mechanisms in beta vulgaris. Plant Cell Environ. 45, 1315–1332. doi: 10.1111/pce.14264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Wang X., Ge S., Rao G. Y. (2015). Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evolutionary Biol. 15, 207. doi: 10.1186/s12862-015-0490-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbert E. (2002). Ecological consequences and ontogeny of seed heteromorphism. Perspect. Plant Ecology Evol. Systematics 5, 13–36. doi: 10.1078/1433-8319-00021 [DOI] [Google Scholar]
- Jarvis D. E., Ho Y. S., Lightfoot D. J., Schmockel S. M., Li B., Borm T. J. A., et al. (2017). The genome of Chenopodium quinoa . Nature 542, 307–315. doi: 10.1038/nature21370 [DOI] [PubMed] [Google Scholar]
- Kadereit G., Newton R. J., Vandelook F. (2017). Evolutionary ecology of fast seed germination-a case study in Amaranthaceae/Chenopodiaceae. Perspect. Plant Ecol. Evol. Systematics 29, 1–11. doi: 10.1016/j.ppees.2017.09.007 [DOI] [Google Scholar]
- Karssen C. M. (1968). The light promoted germination of the seeds of Chenopodium album L. - II. Effects of (RS)-abscisic acid. Acta Botanica Neerlandica 17, 293–308. doi: 10.1111/J.1438-8677.1968.TB00129.X [DOI] [Google Scholar]
- Karssen C. M. (1970). Light promoted germination of seeds of Chenopodium album L . - III. Effect of photoperiod during growth and development of plants on dormancy of produced seeds. Acta Botanica Neerlandica 19, 81–94. [Google Scholar]
- Karssen C. M. (1976. a). Two sites of hormonal action during germination of Chenopodium album seeds. Physiologia Plantarum 36, 264–270. doi: 10.1111/j.1399-3054.1976.tb04426.x [DOI] [Google Scholar]
- Karssen C. M. (1976. b). Uptake and effect of abscisic acid during induction and progress of radicle growth in seeds of Chenopodium album . Physiologia Plantarum 36, 259–263. doi: 10.1111/j.1399-3054.1976.tb04425.x [DOI] [Google Scholar]
- Krak K., Habibi F., Douda J., Vit P., Lomonosova M. N., Wang L., et al. (2019). Human-mediated dispersal of weed species during the Holocene: A case study of Chenopodium album agg. J. Biogeography 46, 1007–1019. doi: 10.1111/jbi.13545 [DOI] [Google Scholar]
- Krak K., Vit P., Belyayev A., Douda J., Hreusova L., Mandak B. (2016). Allopolyploid origin of Chenopodium album s. str. (Chenopodiaceae): a molecular and cytogenetic insight. PLoS One 11, e0161063. doi: 10.1371/journal.pone.0161063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., et al. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. Eur. Mol. Biol. Organ. J. 23, 1647–1656. doi: 10.1038/sj.emboj.7600121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T., Lange M. J. P. (2020). The multifunctional dioxygenases of gibberellin synthesis. Plant Cell Physiol. 61, 1869–1879. doi: 10.1093/pcp/pcaa051 [DOI] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., et al. (2006). Functional analysis of arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. doi: 10.1111/j.1365-313X.2005.02622.x [DOI] [PubMed] [Google Scholar]
- Lenser T., Graeber K., Cevik O. S., Adiguzel N., Donmez A. A., Grosche C., et al. (2016). Developmental control and plasticity of fruit and seed dimorphism in Aethionema arabicum . Plant Physiol. 172, 1691–1707. doi: 10.1104/pp.16.00838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Yamaguchi S., Khan M. A., An P., Liu X., Tran L. S. (2016). Roles of gibberellins and abscisic acid in regulating germination of Suaeda salsa dimorphic seeds under salt stress. Front. Plant Sci. 6, 1235. doi: 10.3389/fpls.2015.01235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q. H., Yu X. T., Chen L., Zhao G., Li S. Z., Zhou H., et al. (2021). Genome-wide identification and expression analysis of the NCED family in cotton (Gossypium hirsutum l.). PLoS One 16, e0246021. doi: 10.1371/journal.pone.0246021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R. R., Wang L., Tanveer M., Song J. (2018). Seed heteromorphism: an important adaptation of halophytes for habitat heterogeneity. Front. Plant Sci. 9, 1515. doi: 10.3389/fpls.2018.01515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X., Vaistij F. E., Li Y., Van Rensburg W. S. J., Harvey S., Bairu M. W., et al. (2021). A chromosome-level Amaranthus cruentus genome assembly highlights gene family evolution and biosynthetic gene clusters that may underpin the nutritional value of this traditional crop. Plant J. 107, 613–628. doi: 10.1111/tpj.15298 [DOI] [PubMed] [Google Scholar]
- MacGregor D. R., Kendall S. L., Florance H., Fedi F., Moore K., Paszkiewicz K., et al. (2015). Seed production temperature regulation of primary dormancy occurs through control of seed coat phenylpropanoid metabolism. New Phytol. 205, 642–652. doi: 10.1111/nph.13090 [DOI] [PubMed] [Google Scholar]
- Machabée S., Saini H. S. (1991). Differences in the requirement for endogenous ethylene during germination of dormant and non-dormant seeds of Chenopodium album l. J. Plant Physiol. 138, 97–101. doi: 10.1016/S0176-1617(11)80737-6 [DOI] [Google Scholar]
- Mandak B., Krak K., Vit P., Lomonosova M. N., Belyayev A., Habibi F., et al. (2018). Hybridization and polyploidization within the Chenopodium album aggregate analysed by means of cytological and molecular markers. Mol. Phylogenet. Evol. 129, 189–201. doi: 10.1016/j.ympev.2018.08.016 [DOI] [PubMed] [Google Scholar]
- Matilla A., Gallardo M., Puga-Hermida M. I. (2005). Structural, physiological and molecular aspects of heterogeneity in seeds: a review. Seed Sci. Res. 15, 63–76. doi: 10.1079/SSR2005203 [DOI] [Google Scholar]
- Matsushima R. (2014). Thin sections of technovit 7100 resin of rice endosperm and staining. Bio-protocol J. 4, e1239. doi: 10.21769/BioProtoc.1239 [DOI] [Google Scholar]
- Murdoch A. J., Roberts E. H. (1997). Temperature and the rate of germination of dormant seeds of Chenopodium album . Basic Appl. Aspects Seed Biol. 30, 547–553. doi: 10.1007/978-94-011-5716-2_60 [DOI] [Google Scholar]
- Murdoch A. J., Roberts E. H., Goedert C. O. (1989). A model for germination responses to alternating temperatures. Ann. Bot. 63, 97–111. doi: 10.1093/oxfordjournals.aob.a087733 [DOI] [Google Scholar]
- Nakabayashi K., Leubner-Metzger G. (2021). Seed dormancy and weed emergence: from simulating environmental change to understanding trait plasticity, adaptive evolution, and population fitness. J. Exp. Bot. 72, 4181–4185. doi: 10.1093/jxb/erab150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E., Okamoto M., Tatematsu K., Yano R., Seo M., Kamiya Y. (2010). Abscisic acid and the control of seed dormany and germination. Seed Sci. Res. 20, 55–67. doi: 10.1017/S0960258510000012 [DOI] [Google Scholar]
- Neve P., Vila-Aiub M., Roux F. (2009). Evolutionary-thinking in agricultural weed management. New Phytol. 184, 783–793. doi: 10.1111/j.1469-8137.2009.03034.x [DOI] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15, 1591–1604. doi: 10.1105/tpc.011650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto M., Kuwahara A., Seo M., Kushiro T., Asami T., Hirai N., et al. (2006). CYP707A1 and CYP707A2, which encode ABA 8'-hydroxylases, are indispensable for a proper control of seed dormancy and germination in arabidopsis. Plant Physiol. 141, 97–107. doi: 10.1104/pp.106.079475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., MacGregor D. R. (2017). Effects of environmental variation during seed production on seed dormancy and germination. J. Exp. Bot. 68, 819–825. doi: 10.1093/jxb/erw436 [DOI] [PubMed] [Google Scholar]
- Prego I., Maldonado S., Otegui M. (1998). Seed structure and localization of reserves in Chenopodium quinoa . Ann. Bot. 82, 481–488. doi: 10.1006/anbo.1998.0704 [DOI] [Google Scholar]
- Roman E. S., Thomas A. G., Murphy S. D., Swanton C. J. (1999). Modeling germination and seedling elongation of common lambsquarters (Chenopodium album). Weed Sci. 47, 149–155. doi: 10.1017/S0043174500091554 [DOI] [Google Scholar]
- Sabir I. A., Manzoor M. A., Shah I. H., Abbas F., Liu X. J., Fiaz S., et al. (2022). Evolutionary and integrative analysis of gibberellin-dioxygenase gene family and their expression profile in three rosaceae genomes (F. vesca, P. mume, and P. avium) under phytohormone stress. Front. Plant Sci. 13, 942969. doi: 10.3389/fpls.2022.942969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini H. S., Bassi P. K., Spencer M. S. (1985). Seed germination in Chenopodium album l. - relationships between nitrate and the effects of plant hormones. Plant Physiol. 77, 940–943. doi: 10.1104/pp.77.4.940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N., Nei M. (1987). The neighbor-joining method - a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454 [DOI] [PubMed] [Google Scholar]
- Song J., Shi W. W., Liu R. R., Xu Y. G., Sui N., Zhou J. C., et al. (2017). The role of the seed coat in adaptation of dimorphic seeds of the euhalophyte Suaeda salsa to salinity. Plant Species Biol. 32, 107–114. doi: 10.1111/1442-1984.12132 [DOI] [Google Scholar]
- Soureshjani H. K., Bahador M., Tadayon M. R., Dehkordi A. G. (2022). Modeling seed germination of quinoa (Chenopodium quinoa willd.) at different temperatures and water potentials. Acta Physiologiae Plantarum 44, 102. doi: 10.1007/s11738-022-03425-3 [DOI] [Google Scholar]
- Steckel L. E., Sprague C. L., Stoller E. W., Wax L. M. (2004). Temperature effects on germination of nine amaranthus species. Weed Sci. 52, 217–221. doi: 10.1614/WS-03-012R [DOI] [Google Scholar]
- Steinbrecher T., Leubner-Metzger G. (2017). The biomechanics of seed germination. J. Exp. Bot. 68, 765–783. doi: 10.1093/jxb/erw428 [DOI] [PubMed] [Google Scholar]
- Sukhorukov A. P., Mavrodiev E. V., Struwig M., Nilova M. V., Dzhalilova K. K., Balandin S. A., et al. (2015). One-seeded fruits in the core caryophyllales: Their origin and structural diversity. PLoS One 10, e0117974. doi: 10.1371/journal.pone.0117974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turečková V., Novák O., Strnad M. (2009). Profiling ABA metabolites in Nicotiana tabacum l. leaves by ultra-performance liquid chromatography-electrospray tandem mass spectrometry. Talanta 80, 390–399. doi: 10.1016/j.talanta.2009.06.027 [DOI] [PubMed] [Google Scholar]
- Urbanova T., Tarkowská D., Novák O., Hedden P., Strnad M. (2013). Analysis of gibberellins as free acids by ultra performance liquid chromatography-tandem mass spectrometry. Talanta 112, 85–94. doi: 10.1016/j.talanta.2013.03.068 [DOI] [PubMed] [Google Scholar]
- Urbanova T., Tarkowska D., Strnad M., Hedden P. (2011). Gibberellins - terpenoid plant hormones: biological importance and chemical analysis. Collection Czechoslovak Chem. Commun. 76, 1669–1686. doi: 10.1135/cccc2011098 [DOI] [Google Scholar]
- Vandelook F., Newton R. J., Bobon N., Bohley K., Kadereit G. (2021). Evolution and ecology of seed internal morphology in relation to germination characteristics in amaranthaceae. Ann. Bot. 127, 799–811. doi: 10.1093/aob/mcab012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walck J. L., Hidayati S. N., Dixon K. W., Thompson K., Poschlod P. (2011). Climate change and plant regeneration from seed. Global Change Biol. 17, 2145–2161. doi: 10.1111/j.1365-2486.2010.02368.x [DOI] [Google Scholar]
- Walker M., Pérez M., Steinbrecher T., Gawthrop F., Pavlovic I., Novák O., et al. (2021). Molecular mechanisms and hormonal regulation underpinning morphological dormancy: a case study using Apium graveolens (Apiaceae). Plant J. 108, 1020–1036. doi: 10.1111/tpj.15489 [DOI] [PubMed] [Google Scholar]
- Wang L., Huang Z., Baskin C. C., Baskin J. M., Dong M. (2008). Germination of dimorphic seeds of the desert annual halophyte Suaeda aralocaspica (Chenopodiaceae), a C4 plant without kranz anatomy. Ann. Bot. 102, 757–769. doi: 10.1093/aob/mcn158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Xu Y. G., Wang S., Shi W., Liu R., Feng G., et al. (2015). Salinity affects production and salt tolerance of dimorphic seeds of Suaeda salsa . Plant Physiol. Biochem. 95, 41–48. doi: 10.1016/j.plaphy.2015.07.005 [DOI] [PubMed] [Google Scholar]
- Westwood J. H., Charudattan R., Duke S. O., Fennimore S. A., Marrone P., Slaughter D. C., et al. (2018). Weed management in 2050: Perspectives on the future of weed science. Weed Sci. 66, 275–285. doi: 10.1017/wsc.2017.78 [DOI] [Google Scholar]
- Wu Q., Bai X., Wu X. Y., Xiang D. B., Wan Y., Luo Y. M., et al. (2020). Transcriptome profiling identifies transcription factors and key homologs involved in seed dormancy and germination regulation of Chenopodium quinoa . Plant Physiol. Biochem. 151, 443–456. doi: 10.1016/j.plaphy.2020.03.050 [DOI] [PubMed] [Google Scholar]
- Xu Y., Zhao Y., Duan H., Sui N., Yuan F., Song J. (2017). Transcriptomic profiling of genes in matured dimorphic seeds of euhalophyte Suaeda salsa . BMC Genomics 18, 727. doi: 10.1186/s12864-017-4104-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S., Lan H., Zhang F. (2010). Variation of seed heteromorphism in Chenopodium album and the effect of salinity stress on the descendants. Ann. Bot. 105, 1015–1025. doi: 10.1093/aob/mcq060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller R. (2001). Fixation, embedding, and sectioning of tissues, embryos, and single cells. Curr. Protoc. Pharmacol. 7, A.3D.1–A.3D.9. doi: 10.1002/0471141755.pha03ds07 [DOI] [PubMed] [Google Scholar]
- Zhao S., Fernald R. D. (2005). Comprehensive algorithm for quantitative real-time polymerase chain reaction. J. Comput. Biol. 12, 1047–1064. doi: 10.1089/cmb.2005.12.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Huang Y. Y., Xian W. H., Wang J. X., Liao H. (2012). Identification and expression analysis of the Glycine max CYP707A gene family in response to drought and salt stresses. Ann. Bot. 110, 743–756. doi: 10.1093/aob/mcs133 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found on Royal Holloways figshare: https://doi.org/10.17637/rh.21829842.v1.