Abstract
Background
Cockayne syndrome (CS) is a DNA repair disorder primarily associated with pathogenic variants in ERCC6 and ERCC8. As in other Mendelian disorders, there are a number of genetically unsolved CS cases.
Methods
We ascertained five individuals with monoallelic pathogenic variants in MORC2, previously associated with three dominantly inherited phenotypes: an axonal form of Charcot-Marie-Tooth disease type 2Z; a syndrome of developmental delay, impaired growth, dysmorphic facies, and axonal neuropathy; and a rare form of spinal muscular atrophy.
Results
One of these individuals bore a strong phenotypic resemblance to CS. We then identified monoallelic pathogenic MORC2 variants in three of five genetically unsolved individuals with a clinical diagnosis of CS. In total, we identified eight individuals with MORC2-related disorder, four of whom had clinical features strongly suggestive of CS.
Conclusions
Our findings indicate that some forms of MORC2-related disorder have phenotypic similarities to CS, including features of accelerated aging. Unlike classic DNA repair disorders, MORC2-related disorder does not appear to be associated with a defect in transcription-coupled nucleotide excision repair and follows a dominant pattern of inheritance with variants typically arising de novo. Such de novo pathogenic variants present particular challenges with regard to both initial gene discovery and diagnostic evaluations. MORC2 should be included in diagnostic genetic test panels targeting the evaluation of microcephaly and/or suspected DNA repair disorders. Future studies of MORC2 and its protein product, coupled with further phenotypic characterization, will help to optimize the diagnosis, understanding, and therapy of the associated disorders.
Keywords: Cockayne syndrome, MORC2, DNA repair, Microcephaly
Introduction
Cockayne syndrome (CS), one of the classic inherited DNA repair disorders, is associated with multiorgan system complications, including neurodevelopmental/intellectual disabilities, microcephaly, poor growth, visual impairments (corneal opacification and/or cataracts), sensorineural hearing loss, demyelinating neuropathy, hepatic involvement, kidney dysfunction, skin photosensitivity, and dental anomalies.1, 2, 3, 4, 5 The two classic genes associated with CS are ERCC6 (CSB) and ERCC8 (CSA), whose protein products are key components of transcription-coupled nucleotide excision repair (TC-NER).6,7 Owing to phenotypic overlap with other DNA repair disorders such as subsets of xeroderma pigmentosum (XP) and trichothiodystrophy, individuals who present with clinical signs of CS may also have pathogenic variants in ERCC1,8 ERCC2 (XPD),9 ERCC3 (XPB),10 ERCC4 (XPF),11 ERCC5 (XPG),12 and XPA.13 A broad genetic test panel that includes both the two classic genes and other related genes such as these will yield the diagnosis in many cases. However, some individuals with features of CS remain genetically undiagnosed despite extensive testing.
The protein encoded by MORC2 (MIM:616688), named microrchidia CW-type zinc finger 2 (MORC2), is part of a superfamily of proteins involved in chromatin remodeling, epigenetic transcriptional regulation, DNA repair, and fatty acid biosynthesis.14, 15, 16, 17, 18 Characterization of germline de novo and rare inherited monoallelic dominant mutations has connected MORC2 with three overlapping phenotypes: Charcot-Marie-Tooth disease type 2Z; a syndrome of developmental delay, impaired growth, facial dysmorphia, and axonal neuropathy; and rarely an adult-onset form of non-5q spinal muscular atrophy.19, 20, 21
It has come to our attention that some individuals with pathogenic variants in MORC2 were initially given a clinical diagnosis of CS, leading us to explore this potential association further.
Methods
Ascertainment and enrollment
The cohort was ascertained via the Amy and Friends Cockayne Syndrome (CS) and Trichothiodystrophy (TTD) Support Group and the UK National CS/TTD Service. This was either through direct contact with families or indirectly through enrollment in collaborators' protocols, yielding an international cohort of individuals known to have pathogenic variants in MORC2 and individuals with CS phenotypes whose clinical testing did not yield a clear genetic diagnosis. All participants were enrolled through protocols approved by the University of Minnesota Institutional Review Board or by ethics committees at collaborating institutions, including a biorepository at Guy's and St. Thomas's Hospital (United Kingdom) of genetically unsolved CS cases, Imperial College and The Portland Hospital London (United Kingdom), the Université de Strasbourg (France), University Children's Hospital Zürich (Switzerland), and the Montreal Children's Hospital and McGill University Health Center Research Ethics Board.
Clinical data collection and analysis
Clinical and genetic test data were gathered from participating families and collaborators after enrollment. Data categories included gender/sex, age at onset, age at clinical diagnosis, birth history, developmental history, family history, growth parameters, visual complications, hearing loss, neurological complications, hepatic and/or renal involvement, skin photosensitivity, dental concerns, neuroimaging findings, electromyography findings, and clinical genetic test results. A CS diagnostic score that includes clinical and neuroradiological features, along with a CS severity score that includes anthropometric and neurological measures, was calculated for each participant.2
Genetic screening for MORC2 variants in undiagnosed participants
Sanger sequencing for MORC2 was performed on genomic DNA (gDNA) from five individuals with a clinical diagnosis of CS who did not have pathogenic variants in genes known to be associated with CS on clinical genetic testing. We designed 16 primer pairs to amplify all 26 exons. Potential primers were assessed for basic primer criteria including melting temperature, hairpins, self-complementarity, and specificity using OligoCalc (v.3.27) and NCBI Primerblast 8. PCR was performed on gDNA using Jumpstart Taq Polymerase (MilliporeSigma) or Q5 Hot Start High Fidelity Polymerase (NewEngland Biolabs). The PCR products were purified using ExoSAP-IT (ThermoFisher Scientific) before being sent to the University of Minnesota Genomics Center or Eurofins for Sanger sequencing. Chromatograms were analyzed using Chromas Pro (v.2.1.10); variants were annotated and assessed using UCSC Genome browser, dbSNP, ClinVar, CADD (V. GRCh38-1.6), PolyPhen2, gnomAD, and PROVEAN/SIFT.22, 23, 24, 25, 26, 27, 28, 29 Pathogenicity categories were assigned using American College of Medical Genetics (ACMG) standards and guidelines for variant interpretation.30 All variants were reported for transcript NM_001303256.3(MORC2).
Results
Demographic and genetic data
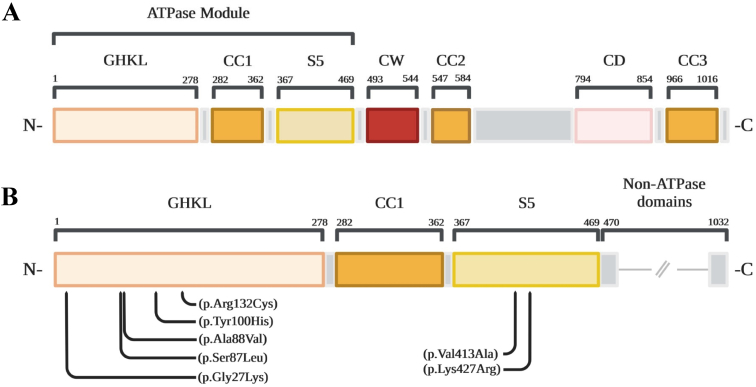
Participants were numbered in order of the pathogenic variant positions identified in MORC2, from 5′ to 3′. Ages of the participants ranged from three to 27 years. All participants were reported to have symptom onset in the first six to 18 months of life, most commonly presenting initially with developmental delay. There was some variability in their original clinical diagnosis: Participants 3 to 6 were all suspected of having a DNA repair disorder such as CS before the identification of pathogenic MORC2 variants, whereas the remaining participants were found to have pathogenic MORC2 variants during evaluations for other presentations. Five individuals with various clinical presentations located in the United States, Germany, Canada, France, and the United Kingdom (participants 1, 2, 6, 7, and 8, respectively) were previously confirmed to have pathogenic/likely pathogenic variants in MORC2. Participant 6 was recently published in a case report.31 A UK-based biorepository with CS phenotypes (Fig 1) was screened for MORC2 variants. These individuals did not have mutations in any of the 14 genes used to screen for mutations in nucleotide excision repair genes. Three of these five tested positive for monoallelic pathogenic MORC2 variants (participants 3, 4, and 5), including two siblings (participants 3 and 4), bringing the total cohort to eight individuals in seven families (Table 1). The pathogenic MORC2 variants were confirmed to be monoallelic in all affected individuals, as well as de novo by trio testing in all affected individuals. Participants 3 and 4 were siblings carrying the same variant, with parental testing indicating that neither parent carried the variant somatically, indicating a high likelihood of gonadal mosaicism in one of the parents. All pathogenic variants were located in the ATPase region (Fig 2). Participants 1 to 6 all carried disease-causing variants that affected the GHKL domain, whereas participants 7 and 8 had pathogenic variants in the S5 domain. Fibroblasts from the four participants with MORC2 variants that were tested showed a normal response in recovery of RNA synthesis after UV irradiation, the gold-standard test for cells deficient in genes involved in TC-NER (Table 1).
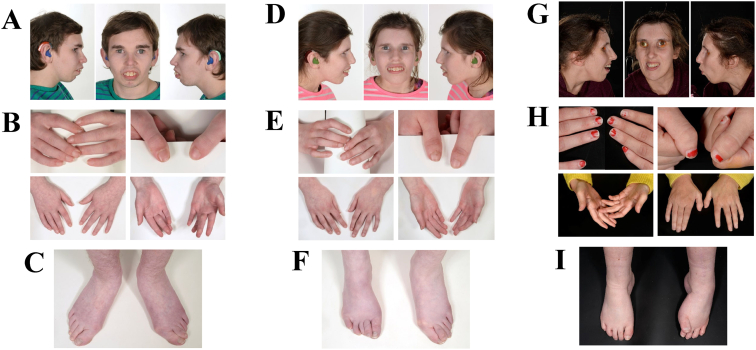
FIGURE 1.
(A-C) Images of the face, hands, and feet from participant 3 showing enophthalmos, mouth with incompetent lips, prominent teeth, emerging retrognathia, livedo reticularis, and poor peripheral circulation. (D-F) Images of the face, hands, and feet from participant 4, sibling of participant 3, showing similar features. (G-I) Images of the face, hands, and feet from participant 5 showed marked enophthalmos, retrognathia, fixed distal contractures in the feet with overlapping toes, and pes cavus suggestive of a peripheral neuropathy. Families consented for photography of all individuals shown. The color version of this figure is available in the online edition.
TABLE 1.
Summary of Participant Genetic and Clinical Information
| Case # | Genetic |
Clinical |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide Change | Amino Acid Change | Domain | Previously Reported | Classification | Inheritance | RRS Testing | Symptom Onset | Age When MORC2 Variant Was Identified | Current Age | Phenotype | Clinical CS Diagnostic Score2 | CS Severity Score2 | |
| 1 | c.79G>A | p.(Gly27Lys) | GHKL | + | Pathogenic | De novo | ND | 6 m | 16 y | 17 y | Spastic diplegic CP | 8 | 9 |
| 2 | c.260C>T | p.(Ser87Leu) | GHKL | + | Pathogenic | De novo | ND | 6 m | 4 y | 8 y | CMT2Z | 6 | 9 |
| 3 | c.263C>T | p.(Ala88Val) | GHKL | + | Pathogenic | De novo∗ | Normal | 18 m | 20 y | 20 y | CS-like | 8 | 5 |
| 4 | c.263C>T | p.(Ala88Val) | GHKL | + | Pathogenic | De novo∗ | ND | 12 m | 19 y | 19 y | CS-like | 10 | 5 |
| 5 | c.298T>C | p.(Tyr100His) | GHKL | – | Likely Pathogenic | De novo | Normal | 3 y | ≥24 y | 27 y | CS-like | 11 | 6 |
| 6 | c.394C>T | p.(Arg132Cys) | GHKL | + | Pathogenic | De novo | Normal | 17 m | 14 y | 15 y | CS-like | 10 | 6 |
| 7 | c.1238T>C | p.(Val413Ala) | S5 | + | Likely Pathogenic | De novo | Normal | 9 m | 3 y | 4 y | Global dev. delay | 6 | 7 |
| 8 | c.1280A>G | p.(Lys427Arg) | S5 | – | Likely Pathogenic | De novo | ND | <1 y | 3 y | 3 y | CMT2Z | 4 | 11 |
Abbreviations:
CMT = Charcot-Marie-Tooth disease
CP = Cerebral palsy
CS = Cockayne syndrome
dev. = Developmental
GHKL = Gyrase protein domain
ND = Not done
RRS = Recovery of RNA synthesis after UV irradiation
S5 = Ribosomal protein S5 domain
Note: All subjects were monoallelic for disease-causing MORC2 variants. Participant 6 was previously published.31 All variants are annotated for transcript NM_001303256.3(MORC2). CMT, Charcot-Marie-Tooth disease; CS, Cockayne syndrome; dev, developmental; GHKL, gyrase protein domain; S5, ribosomal protein S5 domain; ND, not done; RRS, recovery of RNA synthesis after UV-irradiation.; y, years; m, months; CP, cerebral palsy.
Participants 3 and 4 are full siblings; parental testing was negative for the MORC2 variant in question, indicating gonadal mosaicism.
FIGURE 2.
(A) A schematic diagram of the MORC2 protein domain structure. The ATPase module is at the N terminus including the GHKL, CC1, and S5 domains. (B) An inset of the diagram from (A) demonstrating the locations of the pathogenic variants in the current cohort. All pathogenic variants localize to the ATPase module. The color version of this figure is available in the online edition.
Neurological manifestations
All participants had neurological symptoms (Table 2). Of the eight participants, four had microcephaly (<5th percentile), whereas another had a head circumference at ∼10th percentile. Of the seven participants who achieved independent ambulation, all experienced gait disturbances of varying severity, most often related to balance. Other frequent signs and symptoms included abnormal tendon reflexes (either increased or decreased) in six of eight participants, as well as tremors in five of eight participants. Participants 1, 2, and 5 to 8 had at least one brain magnetic resonance imaging (MRI), and the reports were reviewed in all (original images were unavailable for review). Neuroimaging in participants 6 to 8 demonstrated cerebellar and vermian atrophy, as well as increased T2 signal in subjects 1, 6, and 7, localizing to the parietal region for participant 6 and to the basal ganglia in participant 7. Participant 2 had normal brain MRI results at age 2 years 4 months, but has not had a more recent imaging reported. Participant 5 had a brain MRI in early childhood, but no abnormal findings were reported; this is also the only participant who experienced seizures. Three participants had increased muscle tone, and five participants had decreased tone. No participants had muscle biopsy reports available at this time. Proximal and distal limb weakness was present in participants 1 to 4 and 7, but not in participants 6 or 8. Limb strength in participant 5 was not documented. Participant 1 experienced upper limb weakness, participants 3 and 4 had lower limb weakness, and participants 2 and 7 experienced upper and lower limb weakness. Participant 2 had severe weakness, never achieving independent ambulation.
TABLE 2.
Frequencies of Clinical Features
| Total | % | Case # |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| Neurological | ||||||||||
| Gait abnormalities | 7/7 | 100.0 | + | N/A | + | + | + | + | + | + |
| A/hypo/hyper-reflexia | 6/8 | 75.0 | – | + | + | + | – | + | + | + |
| Neuropathy, confirmed/suspected | 6/8 | 75.0 | – | + | + | + | + | + | – | + |
| Axonal | 2/5 | 40.0 | – | + | N/A | N/A | N/A | + | – | – |
| Abnormal brain MRI | 4/6 | 66.7 | + | – | N/A | N/A | – | + | + | + |
| T2 hyperintensive signal | 3/6 | 50.0 | + | – | N/A | N/A | – | + | + | – |
| Cerebellar/vermian atrophy | 3/6 | 50.0 | – | – | N/A | N/A | – | + | + | + |
| Tremors | 5/8 | 62.5 | – | + | – | + | + | + | – | + |
| Seizures | 1/8 | 12.5 | – | – | – | – | + | – | – | – |
| Musculoskeletal | ||||||||||
| Muscle tone abnormalities | 8/8 | 100.0 | + | + | + | + | + | + | + | + |
| Hypotonia | 5/8 | 62.5 | + | + | – | – | – | + | + | + |
| Hypertonia | 3/8 | 37.5 | – | – | + | + | + | – | – | – |
| Limb weakness | 5/7 | 71.4 | + | + | + | + | N/A | – | + | – |
| Proximal weakness | 5/7 | 71.4 | + | + | + | + | N/A | – | + | – |
| Distal weakness | 5/7 | 71.4 | + | + | + | + | N/A | – | + | – |
| Contractures | 4/8 | 50.0 | – | + | + | + | + | – | – | – |
| Scoliosis | 2/8 | 25.0 | – | + | – | – | – | + | – | – |
| Developmental | ||||||||||
| Short stature | 8/8∗ | 100.0 | + | +∗ | + | + | + | + | + | + |
| Motor developmental delay | 8/8 | 100.0 | + | + | + | + | + | + | + | + |
| Intellectual disability | 7/8 | 87.5 | + | + | + | + | + | + | + | – |
| Facial dysmorphism | 4/7 | 57.1 | N/A | – | + | + | + | + | – | – |
| Microcephaly | 4/8 | 50.0 | – | – | + | + | + | + | – | – |
| Other | ||||||||||
| Hearing loss | 6/8 | 75.0 | + | – | + | + | + | + | + | – |
| Vitamin D deficiency | 4/6 | 66.7 | + | + | + | + | – | N/A | – | N/A |
| Dental caries | 4/7 | 57.1 | + | – | NA | + | + | + | – | – |
| Retinopathy† | 3/7† | 42.9 | + | – | + | N/A | –† | + | – | – |
| Dermatologic symptoms | 2/8 | 25.0 | – | – | + | – | + | + | – | – |
| Constipation | 2/8 | 25.0 | – | + | – | – | – | + | – | – |
| Endocrine | 2/8 | 25.0 | – | – | – | – | + | + | – | – |
| Hyperthyroid | 1/8 | 12.5 | – | – | – | – | + | – | – | – |
| Precocious puberty | 1/8 | 12.5 | – | – | – | – | – | + | – | – |
| Cataracts | 1/8 | 12.5 | – | – | – | – | – | + | – | – |
Abbreviations:
MRI = Magnetic resonance imaging
N/A = Not applicable
Borderline for short stature.
Suspected to have retinopathy, confirmation pending.
Neurodevelopmental features
All participants except for individual 8 were reported to have intellectual disability and attended special needs programs at school. Language abilities were notable for limited vocabulary, short sentences, and the use of sign language and gestures.
Musculoskeletal manifestations
Participants 2, 3, 4, and 5 had contractures, and participants 2 and 6 had scoliosis, which was mild in the latter.
Other organ systems involvement
All participants had short stature. Hearing loss was noted in participants 1 and 3 to 7. Retinal dystrophy was also confirmed in participants 1, 3, and 6 and suspected in subject 5 with confirmation pending. Subject 4's family declined testing for retinal dystrophy. Participants 1, 3, 4, and 6 were also noted to have vision loss. Participant 3 had exotropia of the left eye and a concern about possible retinopathy was raised; however, this was not confirmed. Participant 2 exhibited glaucoma and intermittent strabismus but not retinal dystrophy. Participants 3, 5, and 6 had dermatologic issues, but were not confirmed to have the skin photosensitivity that is seen in CS. Participant 3 developed a possible photoaggravated eczema on his hands. Participant 5 has had mild sunburn reactions during the summer months that can be alleviated by the application of sunscreen alone. Participant 6 had a “polymorphous light eruption,” which lacked several features associated with CS photosensitivity. Participants 1 and 4 to 6 had frequent dental caries. Participants 2 and 6 had chronic constipation. Participants 5 and 6 had hyperthyroidism and precocious puberty, respectively. Participants 1 to 4 were noted to have vitamin D deficiency, whereas participants 5 and 7 did not, and participants 6 and 8 were not assessed for this at the time of data collection.
Comparison of CS versus MORC2 phenotypic classification
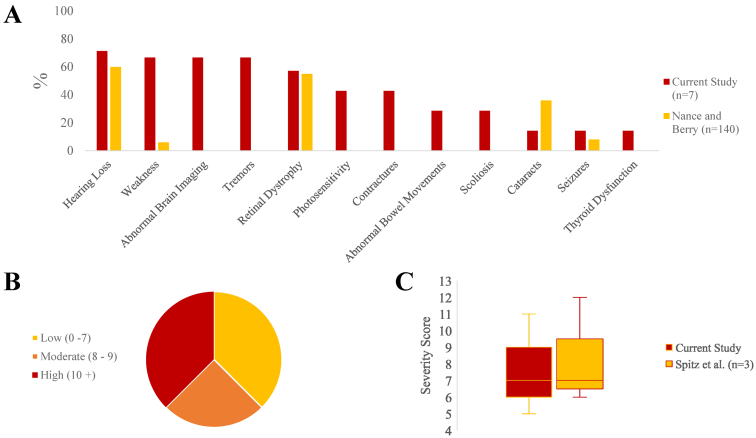
The frequency of CS-related symptoms, including hearing loss, weakness, retinal dystrophy, seizures, and thyroid dysfunction all occurred at similar rates in our cohort compared with prior reports of individuals with CS (Fig 3A).1,5 Each participant was assessed using recently published criteria for assessing diagnostic likelihood and clinical severity for CS.2 On this scale, participants 4 to 6 were in the high likelihood range for CS based on clinical scores, whereas participants 1 and 3 had scores that were in the moderate likelihood range (Fig 3B). Participants 2, 7, and 8 had scores associated with a lower probability of CS. Applying the severity scoring system, the participant score median was 6.5 and ranged from 5 to 11, resembling the patterns previously described the milder CS subgroups (Fig 3C).2
FIGURE 3.
(A) A graph indicating the frequencies of noted symptoms in our participants shows similar patterns to data from a classic prior study of CS.1 (B) A graph indicating the distribution of CS likelihood scores using a published scoring system.2 (C) A graph indicating the distribution of CS severity scores based on clinical criteria, using a published scoring system, with CS type III data from that publication shown for comparison.2 Note that a lower score indicates increased severity. The color version of this figure is available in the online edition.
Discussion
The patterns and severity of clinical presentations associated with pathogenic variants in MORC2 are variable, with a constellation of phenotypes described in the literature.18,19,21,32 Some of these studies have noted potential genotype-phenotype correlations, with symptoms of a syndrome of developmental delay, impaired growth, facial dysmorphia, and axonal neuropathy being somewhat more commonly diagnosed in individuals with mutations in the GHKL region of the ATP-binding domain of the protein, whereas Charcot-Marie-Tooth disease type 2Z diagnoses tended to be associated with variants in the S5 domain.18,32 These are general associations, however, and it is not possible to predict a phenotype reliably based on genotype information alone.33
We suspect that there are two main reasons that the connection between MORC2-related disorders and CS is only now coming to light: the lack of evidence for TC-NER defects in MORC2 deficiency in vitro and the autosomal dominant inheritance pattern. However, this phenotypic link suggests that MORC2 may contribute to a different aspect of gDNA homeostasis. Murine models with knock-in mutations in the homologous gene Morc2a have shown accumulations of DNA damage in nerve cells.34 Several hypotheses have been postulated on the disease mechanism associated with MORC2 pathogenic variants, including the potential for pathogenic variants to cause inappropriate responses to DNA damage. MORC2 contributes to the complex process of chromatin remodeling that is crucial at various steps of repairing double-stranded DNA breaks, potentially through its DNA-dependent ATPase activity, so that damaged DNA first becomes accessible for repair, and then returns to its protected chromatin structure.16 In particular, MORC2 can form a homodimer at its C-terminal coiled-coil domain that appears to facilitate the attenuation of histone-DNA interactions.35
As had been noted in prior studies, pathogenic variants in MORC2 exhibit considerable variability in clinical manifestations. No two participants had identical presentations even among siblings found to carry the same mutation, as was seen in the case of participants 3 and 4. Although all participants exhibited neurological and developmental symptoms, the specific symptoms varied in each category. This is not unlike CS, which has also been noted to exhibit phenotypic variability, making assessments of clinical severity and prognosis difficult early in the disease course based on initial presentation and the pathogenic variant. However, the common themes that emerge from our MORC2 cohort are strikingly similar in both form and frequency to those found in CS, including neurodevelopmental, dermatologic, and metabolic abnormalities. Poor growth and microcephaly also occurred with considerable frequency, mirroring findings in CS.1
Participants 2, 7, and 8 had the mildest phenotypes, exhibiting fewer symptoms that primarily involved the peripheral nervous system, and their CS diagnostic likelihood scores were consistently lower. These individuals would not likely meet clinical criteria for CS. It has been noted that certain variants are associated with peripheral versus central symptoms; pathogenic variants in the S5-fold region tend to manifest with peripheral symptoms,18 which is where the variants for participants 6 and 7 were identified. However, these three participants are also the youngest in the cohort, and it is possible that they will develop more CS-like symptoms over time.
One striking genetic aspect of our findings is the dominant pattern of inheritance, in contrast to the autosomal recessive pattern seen in classic DNA repair disorders. Classically, dominant pathogenic variants in severe inherited diseases were thought to be rare. However, it has become increasingly recognized that such dominant variants may occur, and that they tend to arise de novo, as has been described in some forms of congenital myopathy36 and as we have found in our cohort. Given that all of the pathogenic variants in our cohort are missense changes affecting the ATPase region, we suspect that the disease mechanism is more likely to be due to a toxic gain of function in a dominant negative manner, given that the protein homodimerizes, rather than due to a loss of function.
Understanding both the phenotypic spectrum of MORC2-related disorder and the genetic spectrum of CS is of critical importance. This knowledge has implications for genetic diagnostic evaluations, prognosis, ongoing clinical care, and genetic counseling. Our findings point to a commonality in phenotypes between classic CS and some individuals with pathogenic variants in MORC2. Further studies are needed to elucidate the specific molecular mechanisms by which these phenotypes arise. DNA repair failure, epigenetic silencing, and metabolic disruption have all been implicated as consequences of MORC2 dysfunction. It may be that more than one of these pathways is involved, and that a genotype-phenotype correlation may exist between which mutation is noted and what pathway is affected. This in turn would be a possible explanation for the wide spectrum of recognized phenotypes. In cases in which CS is a diagnostic consideration, MORC2 should be included in genetic diagnostic panels. Individuals confirmed to have pathogenic variants in MORC2 may benefit from multidisciplinary care akin to that needed for individuals with classical forms of CS.
Acknowledgments and Funding
We appreciate the participation of all the families included in this cohort. Amy & Friends Cockayne Syndrome and Trichothiodystrophy Support, and the clinical team at the National CS/TTD Service in London, United Kingdom, assisted with participant ascertainment. Jill Clayton-Smith, MD, FRCP (Manchester Centre for Genomic Medicine) contributed to the diagnostic evaluation of one of the participants. This study was supported by the National Initiative for Cockayne Syndrome (NICS), FDA CBER R01 FD007483, the Canadian Institutes of Health Research (Grant numbers 377869 and 426534), Compute Canada (www.computecanada.ca/), and the McGill University and Genome Quebec Innovation Center. G. Bernard has received the Clinical Research Scholar Junior 1 award from the Fonds de Recherche du Quebec – Santé (FRQS) (2012-2016), the New Investigator Salary Award from the CIHR (2017-2022), and the Clinical Research Scholar Senior award from the FRQS (2022-2025). A.F. Theil is supported by grants from European Research Council Advanced grant (340988) and Oncode Institute (partly financed by the Dutch Cancer Society).
Footnotes
Conflicts of interest declaration: G. Bernard would like to disclose that she is/was a consultant for Passage Bio Inc (2020-2022) and Ionis (2019). She is/was a site investigator for the Alexander's disease trial of Ionis (2021 to now), Metachromatic leukodystrophy of Shire/Takeda (2020-2021), Krabbe and GM1 gene therapy trials of Passage Bio (2021-now), Passage Bio GM1 natural history study (2021-now), and Adrenoleukodystrophy/Hematopoietic stem cell transplantation natural history study of Bluebird Bio (2019) and a site subinvestigator for the MPS II gene therapy trial of Regenxbio (2021 to now). She has received unrestricted educational grants from Takeda (2021-2022). She serves on the Scientific Advisory Board of the Pelizaeus-Merzbacher Foundation and the Yaya Foundation Scientific and Clinical Advisory Council and is the Chair of the Medical and Scientific Advisory Board of the United Leukodystrophy Foundation. She is a member of the Vanishing White Matter Consortium and the H-ABC Clinical Advisory Board and the Chair of the POLR3-related (4H) Leukodystrophy Consortium. She is on the editorial boards of Neurology Genetics, Frontiers in Neurology – Neurogenetics, and Journal of Medical Genetics. P. B. Kang reports financial support was provided by National Initiative for Cockayne Syndrome. C. A. Pacak reports financial support was provided by US Food and Drug Administration. G. Bernard reports financial support was provided by Canadian Institutes of Health Research. The remaining authors declare that there are no conflicts of interests regarding the publication of this article.
References
- 1.Nance M.A., Berry S.A. Cockayne syndrome: review of 140 cases. Am J Med Genet. 1992;42:68–84. doi: 10.1002/ajmg.1320420115. [DOI] [PubMed] [Google Scholar]
- 2.Spitz M.A., Severac F., Obringer C., et al. Diagnostic and severity scores for Cockayne syndrome. Orphanet J Rare Dis. 2021;16:63. doi: 10.1186/s13023-021-01686-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baer S., Tuzin N., Kang P.B., et al. Growth charts in Cockayne syndrome type 1 and type 2. Eur J Med Genet. 2021;64 doi: 10.1016/j.ejmg.2020.104105. [DOI] [PubMed] [Google Scholar]
- 4.Maguina M., Kang P.B., Tsai A., Pacak C.A. Peripheral neuropathies associated with DNA repair disorders. Muscle Nerve. 2023;67:101–110. doi: 10.1002/mus.27721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilson B.T., Stark Z., Sutton R.E., et al. The Cockayne Syndrome Natural History (CoSyNH) study: clinical findings in 102 individuals and recommendations for care. Genet Med. 2016;18:483–493. doi: 10.1038/gim.2015.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Laugel V., Dalloz C., Durand M., et al. Mutation update for the CSB/ERCC6 and CSA/ERCC8 genes involved in Cockayne syndrome. Hum Mutat. 2010;31:113–126. doi: 10.1002/humu.21154. [DOI] [PubMed] [Google Scholar]
- 7.Calmels N., Botta E., Jia N., et al. Functional and clinical relevance of novel mutations in a large cohort of patients with Cockayne syndrome. J Med Genet. 2018;55:329–343. doi: 10.1136/jmedgenet-2017-104877. [DOI] [PubMed] [Google Scholar]
- 8.Westerveld A., Hoeijmakers J.H., van Duin M., et al. Molecular cloning of a human DNA repair gene. Nature. 1984;310:425–429. doi: 10.1038/310425a0. [DOI] [PubMed] [Google Scholar]
- 9.Takayama K., Salazar E.P., Broughton B.C., et al. Defects in the DNA repair and transcription gene ERCC2(XPD) in trichothiodystrophy. Am J Hum Genet. 1996;58:263–270. [PMC free article] [PubMed] [Google Scholar]
- 10.Weeda G., Eveno E., Donker I., et al. A mutation in the XPB/ERCC3 DNA repair transcription gene, associated with trichothiodystrophy. Am J Hum Genet. 1997;60:320–329. [PMC free article] [PubMed] [Google Scholar]
- 11.Sijbers A.M., de Laat W.L., Ariza R.R., et al. Xeroderma pigmentosum group F caused by a defect in a structure-specific DNA repair endonuclease. Cell. 1996;86:811–822. doi: 10.1016/s0092-8674(00)80155-5. [DOI] [PubMed] [Google Scholar]
- 12.O’Donovan A., Wood R.D. Identical defects in DNA repair in xeroderma pigmentosum group G and rodent ERCC group 5. Nature. 1993;363:185–188. doi: 10.1038/363185a0. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K., Miura N., Satokata I., et al. Analysis of a human DNA excision repair gene involved in group A xeroderma pigmentosum and containing a zinc-finger domain. Nature. 1990;348:73–76. doi: 10.1038/348073a0. [DOI] [PubMed] [Google Scholar]
- 14.Inoue N. New gene family defined by MORC, a nuclear protein required for mouse spermatogenesis. Hum Mol Genet. 1999;8:1201–1207. doi: 10.1093/hmg/8.7.1201. [DOI] [PubMed] [Google Scholar]
- 15.Tchasovnikarova I.A., Timms R.T., Douse C.H., et al. Hyperactivation of HUSH complex function by Charcot–Marie–Tooth disease mutation in MORC2. Nat Genet. 2017;49:1035–1044. doi: 10.1038/ng.3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li D.-Q., Nair S.S., Ohshiro K., et al. MORC2 signaling integrates phosphorylation-dependent, ATPase-coupled chromatin remodeling during the DNA damage response. Cell Rep. 2012;2:1657–1669. doi: 10.1016/j.celrep.2012.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sánchez-Solana B., Li D.-Q., Kumar R. Cytosolic functions of MORC2 in lipogenesis and adipogenesis. Biochim Biophys Acta. 2014;1843:316–326. doi: 10.1016/j.bbamcr.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacquier A., Roubille S., Lomonte P., Schaeffer L. Microrchidia CW-type zinc finger 2, a chromatin modifier in a spectrum of peripheral neuropathies. Front Cell Neurosci. 2022;16 doi: 10.3389/fncel.2022.896854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevilla T., Lupo V., Martínez-Rubio D., et al. Mutations in the MORC2 gene cause axonal Charcot–Marie–Tooth disease. Brain. 2016;139:62–72. doi: 10.1093/brain/awv311. [DOI] [PubMed] [Google Scholar]
- 20.Karakaya M., Storbeck M., Strathmann E.A., et al. Targeted sequencing with expanded gene profile enables high diagnostic yield in non-5q-spinal muscular atrophies. Hum Mutat. 2018;39:1284–1298. doi: 10.1002/humu.23560. [DOI] [PubMed] [Google Scholar]
- 21.Hyun Y.S., Hong Y.B., Choi B.-O., Chung K.W. Clinico-genetics in Korean Charcot-Marie-Tooth disease type 2Z with MORC2 mutations. Brain. 2016;139:e40. doi: 10.1093/brain/aww082. [DOI] [PubMed] [Google Scholar]
- 22.Kent W.J., Sugnet C.W., Furey T.S., et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sherry S.T. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Landrum M.J., Lee J.M., Benson M., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rentzsch P., Schubach M., Shendure J., Kircher M. CADD-Splice—improving genome-wide variant effect prediction using deep learning-derived splice scores. Genome Med. 2021;13:31. doi: 10.1186/s13073-021-00835-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karczewski K.J., Francioli L.C., Tiao G., et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi Y. Proceedings of the ACM Conference on Bioinformatics, Computational Biology and Biomedicine - BCB ’12. ACM Press; Orlando, FL: 2012. A fast computation of pairwise sequence alignment scores between a protein and a set of single-locus variants of another protein [Internet] pp. 414–417. [Google Scholar]
- 28.Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7 doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Choi Y., Chan A.P. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richards S., Aziz N., Bale S., et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mirchi A., Derksen A., Tran L.T., et al. A Cockayne-like phenotype resulting from a de novo variant in MORC2: expanding the phenotype of MORC2-related disorders. Neurogenetics. 2022;23:271–274. doi: 10.1007/s10048-022-00697-2. [DOI] [PubMed] [Google Scholar]
- 32.Guillen Sacoto M.J., Tchasovnikarova I.A., Torti E., et al. De novo variants in the ATPase module of MORC2 cause a neurodevelopmental disorder with growth retardation and variable craniofacial dysmorphism. Am J Hum Genet. 2020;107:352–363. doi: 10.1016/j.ajhg.2020.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Semplicini C., Ollagnon-Roman E., Leonard-Louis S., et al. High intra-familiar clinical variability in MORC2 mutated CMT2 patients. Brain. 2017;140:e21. doi: 10.1093/brain/awx019. [DOI] [PubMed] [Google Scholar]
- 34.Lee G.S., Kwak G., Bae J.H., et al. Morc2a p.S87L mutant mice develop peripheral and central neuropathies associated with neuronal DNA damage and apoptosis. Dis Model Mech. 2021;14 doi: 10.1242/dmm.049123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie H.-Y., Zhang T.-M., Hu S.-Y., Shao Z.-M., Li D.-Q. Dimerization of MORC2 through its C-terminal coiled-coil domain enhances chromatin dynamics and promotes DNA repair. Cell Commun Signal. 2019;17:160. doi: 10.1186/s12964-019-0477-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nowak K.J., Wattanasirichaigoon D., Goebel H.H., et al. Mutations in the skeletal muscle α-actin gene in patients with actin myopathy and nemaline myopathy. Nat Genet. 1999;23:208–212. doi: 10.1038/13837. [DOI] [PubMed] [Google Scholar]