Abstract
The biliary system is a highly branched tubular network consisting of intrahepatic bile ducts (IHBDs) and extrahepatic bile ducts (EHBDs). IHBDs are derived from hepatic progenitor cells, while EHBDs originate directly from the endoderm through a separate branching morphogenetic process. Traits that are important for cancer are often found to overlap in developmental and other processes. Therefore, it has been suggested that intrahepatic cholangiocarcinomas (iCCAs) and extrahepatic cholangiocarcinomas (eCCAs) have different developmental mechanisms. While much evidence is being gathered on the mechanism of iCCAs, the evidence for eCCA is still very limited. The main reason for this is that there are very few appropriate animal models for eCCA. We can gain important insights from these animal models, particularly genetically engineered mouse models (GEMMs). GEMMs are immunocompetent and mimic human CCA subtypes with a specific mutational pattern, allowing the development of precancerous lesions, that is, biliary intraepithelial neoplasia (BilIN) and intraductal papillary neoplasm of the bile duct (IPNB). This review provides a summary of the pathogenesis and mechanisms of eCCA that can be revealed by GEMMs. Furthermore, we discuss several clinical questions, such as whether BilIN and IPNB really become malignant, whether the peribiliary gland is the origin of eCCAs, and others.
Keywords: animal model, bile duct cancer, biliary tree, BilIN, cholangiocarcinoma, extrahepatic duct, genetically engineered mouse model, IPNB, peribiliary gland, precancerous lesion
Abbreviations
- BilIN
biliary intraepithelial neoplasia
- BTSCs
biliary stem/progenitor cells
- eCCAs
extrahepatic cholangiocarcinomas
- EHBDs
extrahepatic bile ducts
- GEMMs
genetically engineered mouse models
- iCCAs
intrahepatic cholangiocarcinomas
- IHBDs
intrahepatic bile ducts
- IPNB
intraductal papillary neoplasm of the bile duct
- PBGs
peribiliary glands
INTRODUCTION
The mammalian biliary system is a network of highly branched tubular forms consisting of intrahepatic bile ducts (IHBDs) and extrahepatic bile ducts (EHBDs). 1 , 2 , 3 EHBDs are divided into hilar regional bile ducts and distal bile ducts, with the hilar regional bile ducts extending up to the confluence of the gallbladder ducts and the distal bile ducts extending from the confluence of the gallbladder ducts to the duodenal wall. 1 , 2 , 3
Bile duct formation requires cell–cell interactions, which in turn regulate cell differentiation and morphogenesis. 4 Despite the two origins of the bile ducts, IHBDs and EHBDs are lined by a common cell type, cholangiocytes, which forms a biliary tree that drains bile produced by hepatocytes to the duodenum through a network of IHBDs and EHBDs. 5 However, the intrahepatic and extrahepatic biliary systems occur separately, and it is still unclear how they are connected.
In addition, the bile duct epithelial cells, both intrahepatic and extrahepatic, are directly connected to the peribiliary glands (PBGs), which are thought to have regenerative capacity and progenitor cell properties. 6 , 7 , 8 , 9 Therefore, the development of the PBGs should also be mentioned.
Cholangiocarcinoma (CCA), also called bile duct cancer, is a malignant tumor arising from the epithelial cells of the biliary tree. 10 , 11 Recently, the importance of tumor localization of CCA has been emphasized as well its heterogeneity. 12 There are two main types of CCAs: intrahepatic CCA (iCCA) and extrahepatic CCA (eCCA), including both perihilar and distal CCA. These types have differences regarding their etiology, molecular alterations, pathogenesis, behavior, potential diagnostic or prognostic biomarkers, and treatment. 12 , 13 , 14 , 15 By introducing this knowledge of CCA into animal models, it may be possible to assess how various genetic mutations and changes are related to the pathophysiology of human disease. While much evidence regarding the mechanisms of cancer originating from the IHBDs has been accumulated from both animal models and humans, 14 , 15 , 16 , 17 , 18 those of EHBDs are not well understood.
Among animal models, genetically engineered mouse models (GEMMs) are an excellent tool in cancer research and are based on genetic alterations in human cancers. 19 , 20 Most tumors develop in sequence from early to late stage disease in the tumor microenvironment, which is composed of immune cells, endothelial cells and fibroblasts, mimicking human cancers. 18 , 21 , 22 There are precancerous lesions in CCAs, such as biliary intraepithelial neoplasia (BilIN) and intraductal papillary neoplasm of the bile duct (IPNB); 23 however, whether these precancerous lesions progress to cancer is controversial. Furthermore, other clinical and pathological questions remain.
It is well known that the developmental mechanisms of organs have similarities with the cells involved in tumor development and the mechanism of carcinogenesis. 24 Thus, in this review, we describe the processes that are relevant to the development of EHBDs and their embryology, and focus on the few but valuable GEMMs of eCCAs and what we can learn about the pathophysiology of human CCAs.
DEVELOPMENT OF EHBDS
The extrahepatic biliary duct system and intrahepatic biliary duct system, which have different origins, are interconnected to form a continuous biliary network, or bile duct tree. The extrahepatic biliary system consists of the hepatic duct, ductus deferens, gallbladder, and common bile duct.
The development of EHBDs is initiated by the formation of a diverticulum located ventral to the liver (Figure 1a). The gallbladder arises as a dilatation of the hepatic diverticulum and the common bile duct and grows by lengthening the caudal side of the hepatic diverticulum. Above the junction of the bladder and the common bile duct, the common hepatic duct progresses and widens in a funnel‐like fashion, reforming into individual hepatic ducts that collect bile from the hepatic lobe.
Figure 1.
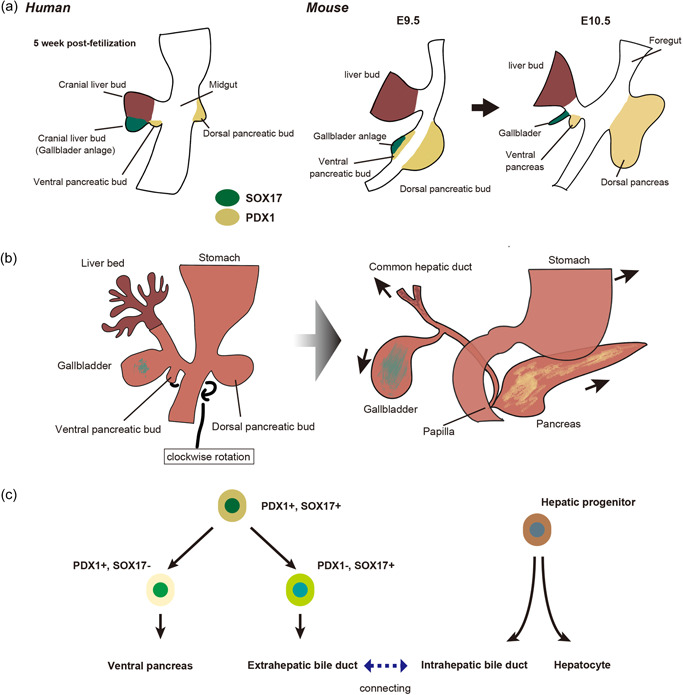
Development of the extrahepatic biliary tract. (a) Embryological development of the extrahepatic biliary tract. In humans, the extrahepatic duct originates from the caudal part of the hepatic bud, whereas in mice, the gallbladder primordium is associated with the ventral pancreas. E, embryonic day. (b) Development of the liver, pancreas, and biliary tree. (Left) The biliary system (gallbladder and cystic duct) initially arises as a tubular structure (pars cystica) formed by the elongation of the caudal portion of the hepatic diverticulum. The dorsal and ventral pancreas fuse after the ventral bud rotates clockwise around the caudal side of the foregut. (Right) During fusion of the dorsal and ventral pancreas, the attachments of the developing common bile duct and pancreatic duct are moved to their native positions in the dorsal duodenum. The pancreas drains mainly from the ventral pancreatic duct and joins the common bile duct at the level of the greater papilla. The main pancreatic duct arises from the merging of dorsal and ventral pancreatic ducts. (c) Molecular regulation of the extrahepatic and intrahepatic biliary tract. The transcription factors PDX1 and SOX17 regulate the development of the extrahepatic biliary tract.
In humans, the EHBDs are in contact with the liver at any point in the development of the liver and EHBDs. 25 , 26 In mice, there is a transient period when the EHBDs are not in contact with the liver, 27 but it is not well understood how the extrahepatic and intrahepatic systems anastomose. The intrahepatic portion of the hepatic duct originates from the intrahepatic ductal plate, while the extrahepatic portion originates from the parietal wavy portion of the common hepatic duct. 25
In mice, the origin of the EHBDs is somewhat different to that of humans in terms of molecular biology. EHBDs are not derived from hepatoblasts, but from the endodermal region located just caudal to the hepatic diverticulum (Figure 1a). This region develops into the ventral pancreatic tissue, common bile duct, and gallbladder, and is characterized by the expression of the transcription factors SRY‐box transcription factor 17 (SOX17) and pancreatic and duodenal homeobox factor‐1 (PDX1). 28
The biliary system, including the gallbladder and cystic duct, initially arises as a tubular structure (pars cystica) formed by the elongation of the caudal portion of the hepatic diverticulum (Figure 1b). The dorsal and ventral pancreas fuse after the ventral bud rotates clockwise around the caudal side of the foregut. 29 During fusion of the dorsal and ventral pancreas, the attachments of the developing common bile duct and pancreatic duct are moved to their native positions in the dorsal duodenum. The pancreas drains mainly from the ventral pancreatic duct and joins the common bile duct at the level of the greater papilla. The main pancreatic duct arises from merging of the dorsal and ventral pancreatic ducts.
SOX17 may be an important determinant of how cells within the PDX1 domain are allocated to the two distinct fates of the pancreas and EHBDs (Figure 1c). Expression analysis and fate mapping experiments have shown that EHBDs arise from a pool of pancreatic biliary progenitor cells co‐expressing SOX17 and PDX1 in the foregut endoderm of E8.5. 30 SOX17 is essential for determining the biliary fate of mice, and when SOX17 is lost, EHBDs do not form and are replaced by pancreatic tissue. 28 , 31 In addition, haploinsufficiency of SOX17 causes hypoplasia of gallbladder and the development of short ectopic EHBDs, while IHBDs are uninfluenced. 32
Proper separation of the EHBDs from the ventral pancreatic lineage is also regulated by Hes family BHLH transcription factor 1 (HES1), a transcriptional effector of NOTCH signaling; Hes1‐deficient mice develop gallbladder dysplasia and undergo ectopic pancreas formation, indicating that HES1 is required to repress the fate of the pancreas within the EHBD. 33 , 34 This phenotype is similar to the phenotype of Sox17 conditional knockout mice, indicating an interaction between HES1 and SOX17 during EHBD development. In fact, inactivation of SOX17 downregulates HES1 expression, while inactivation of HES1 paradoxically expands the SOX17 domain, 28 , 35 suggesting the existence of a SOX17–HES1 feedback loop. Although PDX1 is essential for pancreatic development, it is also expressed in EHBDs 36 and is required for the differentiation of mucin‐producing cells and PBGs. 37
That mesenchymal signaling contributes to the patterning of EHBDs is evident from the fact that fibroblast growth factor 10 (FGF10) mutants show shortening of hepatopancreatic ducts and their composition is unclear. 38 In addition, ectopic hepatocytes and pancreatic cells are observed in FGF10 mutants, indicating that hepatopancreatic cells and pancreatic cells are mis‐differentiated, which suggests that FGF10 broadly regulates the development of the hepatopancreatic endoderm region. Recently, a study of biliary atresia revealed that the bile ducts have special anatomical features that prevent bile acid‐mediated damage and other problems. 39 In particular, the special protective features of the cholangiocytes of EHBDs include an apical glycocalyx with a unique apical membrane and a protective bicarbonate layer. 40 , 41 , 42 , 43 The affinity of FGF10 binding to FGF receptors is increased by heparan sulfate, which are attached to proteoglycans. 44 The submucosal layer of EHBD may also have a protective function, and studies of the glycocalyx, which is mainly composed of heparan sulfate proteoglycans, may be necessary.
DEVELOPMENT AND TUMORIGENESIS OF PBGS
Development of peribiliary glands (PBGs)
Peribiliary glands are microscopic structures distributed mainly along the IHBDs, EHBDs, and gallbladder ducts (Figure 2). During development, the PBGs (especially the extramural glands) surrounding the IHBDs originate from the ductal plate. 45 During the seventh week of pregnancy, ductal plates are formed from immature hepatocyte progenitors surrounding the branches of the portal vein in the hilar region of the liver. At approximately 30 weeks of gestation, extrahepatic PBGs appear from the EHBDs, but not from the gallbladder duct. 46 At approximately 40 weeks of gestation, the ductal plate transforms into an immature PBG through a doubled cord and duct. After birth, the apexes of the immature PBGs keep increasing in number, finally forming a widespread network that connects each segment of the extrahepatic biliary system. 46 , 47 , 48 Subsequently, organization proceeds and maturation is completed at approximately 15 years of age. 45 The cells that form the network co‐express maturation markers such as cytokeratin 19 (CK19) and endodermal markers such as SOX17 and PDX1, whose function is postulated to be proliferation in response to injury to restore mucosal form and function. 48
Figure 2.
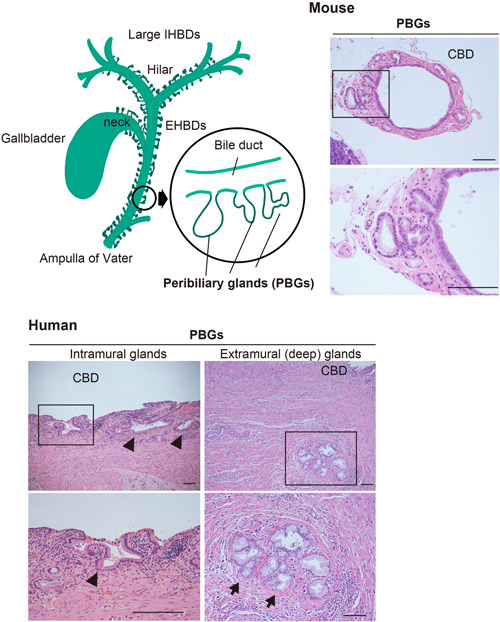
Schematic illustration with representative microphotographs (H&E staining) of common bile ducts with PBGs in humans and mice. (a) The lower image shows the area enclosed by the rectangle in the upper image. In humans, PBGs are known to consist of the intramural (arrowheads) and extramural glands (arrows). EHBD, extrahepatic bile duct; IHBD, intrahepatic bile duct; PBGs, peribiliary glands. Scale bars, 100 μm.
Peribiliary glands are categorized into two types according to their location within the biliary wall: intramural biliary glands and extramural biliary glands. 45 , 49 Substances secreted by both types of glands are discharged into the lumen of the bile duct through conduits from each gland. 45 , 50 Intramural glands are simple tubular mucous glands that are present randomly within the bile duct wall, and they drain directly into the lumen of the bile duct. 45 Extramural glands are branched tubular alveolar glands composed of serous, mucous, and mixed glands, which are arranged in a lobular fashion and are located in the periductal connective tissue. Furthermore, most of the endocrine cells expressing somatostatin are mixed with other serous glandular cells. 45 The ectopic pancreas resembles a pancreatic exocrine gland that does not contain islet of Langerhans cells, but rarely occurs alone or mixed with other serous acini of extramural biliary glands. 45 The PBGs, especially the extramural glands, are a complex of fine vasculature consisting of inflow arteries, capillary plexus (also called peribiliary vascular plexus) and outflow veins, lymphatic vessels, and nerve fibers. These anatomical complexes have been described as a PBG network 45 , 51 and are important structures in the spread of inflammatory and neoplastic lesions. Trophoblast surface protein 2 (TROP2) has been identified as a marker that distinguishes TROP2‐positive luminal cholangiocytes from TROP2‐negative peribiliary constituent epithelial cells, 52 but further elucidation is awaited.
PBGs as a niche for biliary stem/progenitor cells
The PBGs are believed to be a niche for biliary stem/progenitor cells (BTSCs) (Figure 2). In particular, BTSCs have been reported to differentiate into hepatocytes, cholangiocytes, and pancreatic cells. 53 In addition, PBGs are responsible for several physiological functions, such as the secretion of mucous glycoproteins, local immunity, enzyme secretion, regulation of bile concentration, and turnover, regeneration, and repair of the biliary epithelium. 45 , 54 The PBGs include the pancreatic exocrine glands, which have pancreatic exocrine enzymes. 45 Because of these functions and histological features, the PBGs are inextricably related to various biliary tract diseases. 8 , 55 , 56 , 57 Moreover, it has been speculated that cancer stem cells, 58 which are involved in the development and progression of primary biliary tract cancer, originate from the conversion of normal stem/progenitor cells, that is, BTSCs. 57
Although PBG cells consist almost exclusively of serous‐mucous epithelial cells, a few stem/progenitor cells are involved in the PBG, depending on the location of the PBG. 59 As a longitudinal or proximal–distal axis, we observe a high density of primitive stem cells in PBGs near the duodenum and more devoted progenitor cells near the liver and pancreas. 56 Stem cell niches with pluripotency (NANOG, OCT4, SOX2, and KLF4, but not MYC), self‐renewal (SALL4), and proliferation (MIB‐1) phenotypes are observed near the duodenum, and when proceeding to the pancreas, pancreatic/endoderm markers (SOX17, PDX1, and LGR5) and endocrine maturation markers (NGN3, MUC6, and insulin) have been found. 7 , 28 In the more proximal portion of the EHBD, closer to the hepatic portal, PBGs express biliary/endoderm markers (SOX17 and SOX9). The bile ducts become intrahepatic, and some PBG cells express albumin, suggesting hepatocyte maturation. 7 A radial axis can be identified in which cells with primitive stem cell characteristics are most abundant in the epidural PBGs located near the fibromuscular outer layer of the bile ducts. 56 This is interpreted as a maturing lineage from the deepest PBG to the bile duct epithelium on the luminal side. It has been demonstrated that EpCAM can be used as a transit lineage marker to identify intermediate cells between the deepest PBG cells and the lumen. 7 , 59 Cells expressing maturation markers (albumin, CK19, and CK7) gradually migrate to the surface epithelium and appear. Repopulation of the biliary epithelium from PBGs is similar to the natural turnover of the intestine, which is also derived from the embryonic endoderm. The most primitive PBG cells are present in the basal PBG, while transit‐amplifying cells are present in the intermediate compartment and mature cells are contiguous to the epithelium. 56 , 60 Similar to intestinal crypts, PBGs are stimulated to proliferate and repair the epithelium in pathological conditions of luminal epithelial injury or defects. 54 , 61 , 62 The intestinal epithelium is renewed every 4–5 days by the proliferation of stem cells in the crypts. 63 In contrast, the turnover of the epithelium of the bile ducts appears to be much slower.
PBGs as a site of tumor origin
Intraductal papillary neoplasm of the bile duct is a papillary tumor that arises in the bile ducts of intrahepatic and extrahepatic regions, and is centered on a fibrovascular core (Figure 3). 64 , 65 It was defined and listed in the World Health Organization (WHO) classification in 2010 66 as a precancerous and early cancerous lesion of CCA, together with mucinous cystic neoplasm (MCN) and BilIN (Figure 3). IPNB is prone to obstructive jaundice and cholangitis, and it has been reported that 40% of IPNBs are malignant at the time of diagnosis. 67 , 68 , 69 , 70 , 71 Nakanishi et al. 72 reported the first case of IPNB that may have originated from PBG histologically after surgical resection of IPNB. This lesion contained an in situ carcinoma at the base of the PBG. 72 Another case of IPNB that may have originated from a PBG has also been reported. Even in MUC6‐expressing gastric mucin of IPNB, MUC6 is not expressed in the epithelium lining the bile ducts 73 and this strong expression of MUC6 in IPNBs may be due to the PBG origin of IPNBs. This was also confirmed in other studies. 74 , 75 A case of IPNB reported by Miyata et al. showed that the cystic tumor did not communicate with the bile duct lumen and lacked the characteristics of MCN, indicating that the communication with the bile duct lumen may be lost during the development and progression of IPNB from a PBG. 74 Taken together, IPNB may originate, at least in part, from PBGs. In addition, it has been suggested that IPNBs derived from PBGs are preneoplastic lesions of mucin‐producing CCAs that morphologically resemble PBGs, 76 and our group has shown that some IPNBs progress in multistep carcinogenesis to become invasive carcinomas. 77
Figure 3.
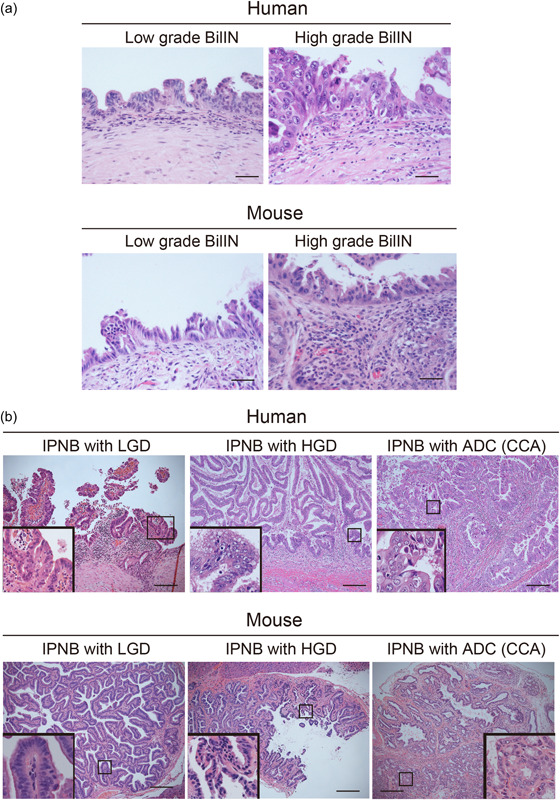
Representative microphotographs (H&E staining) of BilIN (a) and IPNB (b), including CCA, in the mouse model (iFGF10 model) and human patients. (a) Low‐ and high‐grade BilIN‐like lesions in mice are from liver‐specific iFGF10 (LSL‐rtTA;Alb‐Cre;Tet‐Fgf10) and iFGF10‐KrasG12D (LSL‐rtTA;Alb‐Cre;Tet‐Fgf10;LSL‐Kras G12D ) mice, respectively. (b) IPNB‐like lesions with LGD, HGD, and ADC in mice are from iFGF10 (Rosa‐rtTA; Tet‐Fgf10), iFGF10‐KrasG12D (Rosa‐rtTA; Tet‐Fgf10; LSL‐Kras G12D ; Krt19‐iCre), and iFGF10‐KrasG12D ‐p53R172H (Rosa‐rtTA; Tet‐Fgf10; LSL‐Kras G12D ; Krt19‐iCre; p53 R172H/+ mice, respectively. Please note that although mouse models usually show milder atypia than human lesions, biliary lesions in human patients are, in part, similar to those in mouse models. Each inset shows the higher magnification photo. ADC, adenocarcinoma; BilIN, biliary intraepithelial neoplasia; CCA, cholangiocarcinoma; IPNB, intraductal papillary neoplasm of the bile duct; HGD, high‐grade dysplasia; LGD, low‐grade dysplasia. The classification of the biliary lesions is based on the WHO Classification of Tumors of the Digestive System, 5th edn. Lyon: IARC Press, 2019. Scale bars, 50 μm (a), 200 μm (b).
MOUSE MODELS OF ECCAS
Importance of GEMMs
Understanding the histopathological classification of the various human CCAs is important for the future development and analysis of animal models, and that of human CCA from the progression of lesions from premalignant to malignant states is rapidly evolving. 12 , 13 , 15 , 23 , 78 , 79 In particular, the role of GEMMs in the study of CCA is to reproduce early‐stage lesions that are rarely detected in humans and to understand their mechanisms.
In the eCCA model established by our group, the development of biliary hyperplasia and low‐grade adenomatous lesions was observed, such as IPNB‐like 77 and BilIN‐like lesions (Figure 3). However, it is difficult to determine from human pathology whether these lesions remain hyperplastic or if they have the potential to become malignant. Animal models, particularly GEMMs, can provide some evidence for this issue by providing information on the surrounding tumor microenvironment.
The genetic mechanisms of tumorigenesis have recently been found to be very different between IHBDs and EHBDs in human patients. 15 , 80 , 81 , 82 However, compared with IHBD cancer, the mechanism of EHBD cancer is unknown. This is partly because of the lack of appropriate animal models. To elucidate the carcinogenic mechanism and identify the cellular origin of tumors and cancer, a good mouse model is essential. In addition, GEMMs are very effective tools because they can guide how normal cells become tumorous or malignant; that is, how they progress from a normal state to a precancerous state and an invasive or metastatic state, by recreating abnormalities in genes, inflammation, and the surrounding environment. 21 For these reasons, GEMMs of eCCA have an important role in understanding pathogenesis. 18
GEMMs of eCCA
What is lacking in most pre‐clinical approaches is the use of mouse models characterized by spontaneous cancer development in immunocompetent hosts. 83 , 84 An example of such a model is the GEMM, which spontaneously develops cancers. GEMMs have advantages and disadvantages. However, they are very effective tools because they can guide how normal cells become tumors or malignant; that is, how they progress from a normal state to a precancerous state and an invasive or metastatic state by recreating abnormalities in genes, inflammation, and the surrounding environment. There are very few reports of GEMMs of eCCA (Table 1). However, the histopathology illustrates the complexity and diversity of eCCA in humans.
Table 1.
GEMMs of extrahepatic tumorigenesis
| Name | Tumor location | Phenotype | Tumor latency | Reference | |
|---|---|---|---|---|---|
| KT‐K19 CreERT model: LSL‐Kras G12D; Tgfbr2 flox/flox; Krt19 CreERT | Kras G12D activation; Tgfbr2 loss; Cdh1 loss | EHBDs, PBGs | Biliary hyperplasia | 3 months∼ | Nakagawa et al. 85 |
| Under the control of Krt19 promoter | |||||
| KTC‐K19 CreERT model: LSL‐Kras G12D; Tgfbr2 flox/flox; Cdh1flox/flox; Krt19 CreERT | Kras G12D activation; Tgfbr2 loss; Cdh1 loss | EHBDs, PBGs | Invasive carcinoma (moderately/poorly differentiated adenocarcinoma) | 4 weeks∼ | Nakagawa et al. 85 |
| Under the control of Krt19 promoter | |||||
| S+ KPP model: Sox9‐CreERT2; LSL‐Kras G12D; PTEN flox/flox | ‐ | EHBDs, PBGs, GB neck, pancreas head | Biliary adenoma (IPNB with low/mediate dysplasia) In this model, tumor lesions are more malignant outside of EHBD | 3 months ∼ (after TM) | Lin et al. 86 |
| Under the control of Sox9 promoter | |||||
| iFGF10 model: Rosa26‐rtTA; Tet‐Fgf10‐ires‐tdTomato | Fgf10 overexpression | EHBDs, PBGs, large IHBDs, GB neck | BilIN, IPNB, IPNB with invasive carcinoma (with Kras G12D, p53 R172H, and/or p16 flox/flox mice) | 4 weeks ∼ (Dox continuous administration) | Tomita et al. 77 |
| Under the control of Rosa26 promoter | |||||
| Pdx1‐FGF10 model: LSL‐rtTA; Pdx1‐Cre; Tet‐Fgf10‐ires‐tdTomato | Fgf10 overexpression | EHBDs, PBGs, GB neck, pancreas | IPNB, IPNB with invasive carcinoma (with Kras G12D, p53 R172H, and/or p16 flox/flox mice) | 4 weeks ∼ (Dox continuous administration) | Tomita et al. 77 |
| Under the control of Pdx1 promoter | |||||
| Krt19‐FGF10 model: LSL‐rtTA; Krt19‐iCre; Tet‐Fgf10‐ires‐tdTomato | Fgf10 overexpression | EHBDs, PBGs, large IHBDs, GB neck | IPNB, IPNB with invasive carcinoma (with Kras G12D, p53 R172H, and/or p16 flox/flox mice) | 4 weeks ∼ (Dox continuous administration) | Tomita et al. 77 |
| Under the control of Krt19 promoter | |||||
| Sox9‐FGF10 model: LSL‐rtTA; Sox9‐CreERT2; Tet‐Fgf10‐ires‐tdTomato | Fgf10 overexpression | EXHBDS, PBGs, large IHBDs, GB neck | IPNB, IPNB with invasive carcinoma (with Kras G12D, p53 R172H, and/or p16 flox/flox mice) | 4 weeks ∼ (Dox continuous administration after TM) | Tomita et al. 77 |
| Under the control of Sox9 promoter |
Note: Alb‐FGF10 model (LSL‐rtTA; Alb‐Cre; Tet‐Fgf10‐ires‐tdTomato) develops BilIN, IPNB, and cholangiocarcinoma only in small and large IHBDs (Tomita et al. 77 ). Hepatocyte‐FGF10 model (LSL‐rtTA; Tet‐Fgf10‐ires‐tdTomato with AAV‐Ttr‐Cre injection) develops BilIN and IPNB only in small and large IHBDs (Tomita et al. 77 ).
Abbreviations: BilIN, biliary intraepithelial neoplasia; Dox, doxycyclin; EHBD, extrahepatic bile duct; GB, gallbladder; GEMMs, genetically engineered mouse models; IHBD, intrahepatic bile duct; IPNB, intraductal papillary neoplasms of the bile duct; PBG, peribiliary gland; TM, tamoxifen.
Krt19 CreERT Cdh1fox/flox, KrasG12D, and Tgfbr2flox/flox mice
Nakagawa et al. 87 focused on the abnormalities in the RAS and TGFβ/SMAD signaling pathways in human CCA, and crossed LSL (loxp‐stop‐loxp) ‐Kras G12D, Tgfbr2 flox/flox , and Krt19 CreERT mice to develop a tamoxifen‐induced bile duct epithelial cell‐specific mouse model. They generated a CCA mouse model (KT‐K19 CreERT) with tamoxifen‐induced bile duct epithelial cell‐specific activation of G12D mutant Kras and deletion of Tgfbr2. Furthermore, by crossing KT‐K19 CreERT mice with Cdh1 flox/flox mice (KTC‐K19 CreERT), further deleting the adhesion molecule CDH1 (E‐cadherin), they were able to develop early invasive lesions within 4 weeks. This mouse model showed that invasive eCCA developed at an early stage within 4 weeks. 87
Histopathological observation of the neoplastic lesions in the biliary tree revealed that adenocarcinoma cells with mild to moderate atypia expanded along the EHBD wall (so‐called periductal invasion) and extended into the intrahepatic hilar region, including the large IHBD. However, in the peripheral small IHBDs, only lesions with dysplastic changes were observed, and peripheral‐type iCCA did not develop, even though the genetic recombination rate between sites of the biliary tree was similar (approximately 40%). In addition, CCA extended to the gallbladder ducts, but the gallbladder was poorly altered. Consequently, the susceptibility of tumorigenesis to these mutations differed depending on the site of the biliary tree.
In these KTC‐K19 CreERT mice, deletion of E‐cadherin resulted in loss of intercellular adhesion of bile duct epithelial cells in the EHBDs and detachment of bile duct epithelial cells from the EHBD wall; loss of epithelium due to the deletion of E‐cadherin thereafter led to inflammation and a regenerative response by the PBGs. A time course of biliary epithelial cells of EHBD tissues showed that the PBGs incrementally enlarged and became morphologically atypical, accompanied by mitosis. It has been demonstrated that IL‐33 released by inflammation may also be involved in the unique distribution of CCA in KTC‐K19 CreERT mice. 85
In addition, cell fate mapping experiments conducted by crossing Rosa26‐LSL‐LacZ reporter mice (KTC‐LacZ‐K19 CreERT) suggested that the cellular origin of CCA involves PBGs. In fact, the distribution of CCA in KTC‐K19 CreERT mice was almost identical to the distribution of PBGs (i.e., EHBDs and surrounding large IHBDs, but not peripheral small IHBDs).
In humans, proliferation and hyperplastic changes, that is, cystic dilatation and papillary changes, of PBGs similar to those observed in this mouse model have been observed, 54 , 88 , 89 , 90 and early and precancerous lesions of CCA have been noted in PBGs. 88 Overall, this mouse model is an excellent tool that provides valuable information on PBGs and carcinogenesis, which are considered to harbor stem/progenitor cells in large IHBDs and EHBDs.
Sox9‐CreERT2; KrasG12D; Ptenflox/flox mice
Lin et al. 86 mainly generated and analyzed mouse models of iCCA, in which they reported the formation of tumors in EHBDs and the pancreas. To investigate whether KRAS activation and Pten deletion in the bile ducts induce CCA, they used tamoxifen‐inducible Cre‐transfected mice under the control of Sox9 promoter. Sox9 is transiently required for biliary tree development 91 and is expressed in intrahepatic and extrahepatic biliary epithelial cells, as well as in progenitor cells that maintain biliary duct epithelium. 92 , 93 They generated a Sox9‐CreERT2, LSL‐Kras G12D , Pten flox/flox (S+ KPP) mouse model. As a result, not only did iCCA and eCCA develop in these S+ KPP mice, but pancreatic cancer also appeared.
Specifically, in the S+ KPP mice, tumor development was examined 3 months after tamoxifen administration; analysis of the pathological tissues of the S+ KPP mice revealed the presence of various pathologies in the liver, biliary tree, and pancreas, which had not been previously reported. Focal hyperplastic lesions were identified in the epithelium of the interlobular bile ducts, EHBDs, and gallbladder, and epithelial growths with mitotic changes were observed. Scattered cystic collections of dilated bile ducts were evident. Lin et al. 86 referred to these two types of lesions as biliary adenomas.
They also showed loss of epithelial polarity, irregular edges, infiltration of inflammatory cells, and periductal fibrotic changes or desmoplastic changes. In the pancreatic lesions, the pancreatic ducts also showed malignant papillary ductal lesions similar to the bile ducts described above, and the peritumoral stroma showed desmoplastic changes similar to the bile ducts. Finally, the biliary adenoma in this model could be classified as an IPNB in humans. Histopathologically, the tumors grew not only in the hepatic bile duct but also in the pancreatic duct in S+ KPP mice. Although the tumors did not progress to cancer, they served as a mouse model for studying tumors that develop in the bile ducts from the IHBDs to the EHBDs to the pancreas, namely, the biliary tree.
From a molecular biology point of view, the loss of PTEN expression in biliary tract cancer is rare in humans, 12 , 78 , 79 , 94 but mutations in KRAS are some of the most common genetic abnormalities in human cancers. 95 Although such a combination of mutations is quite uncommon in human disease, the present model faithfully recapitulates many of the critical histopathological features of human disease. Therefore, considering the diversity of CCA, it is a very useful mouse model for histopathology.
iFGF10 mouse
The IPNB‐like lesions observed in the S+ KPP mice 86 described above are also noted in humans and are neoplastic lesions that can be classified and diagnosed on the basis of histopathological morphology. 66 IPNB is a papillary‐like lesion that can be observed grossly and microscopically in the bile ducts and is considered to be a precancerous lesion of CCA (Figure 3). 23 In Japan, it was reported in 2012 that young employees of a printing factory who had been using a large amount of chlorinated organic cleaning agents developed CCA at an extremely high rate, which became a major social problem. 67 , 68 , 69 Detailed analysis of the histopathology of this CCA revealed that the main tumors were iCCA of the mass‐forming or intrahepatic growth type and eCCA of the papillary type, most of which were located in relatively large bile ducts such as EHBDs and hilar bile ducts. 67 , 68 , 69 Chronic bile duct injury and characteristic morphological lesions such as BilIN‐ and IPNB‐like lesions were observed in a wide range of bile ducts, suggesting a multistep carcinogenesis mechanism, that is, precancerous lesions. 67 , 68 , 69 However, both the mechanism of developing distinct neoplastic lesions in the biliary tree, such as IPNB, and the fact that they are precancerous lesions remain controversial. One of the reasons for this is the lack of animal models that can faithfully reproduce this pathology.
Our group focused on the ligand fibroblast growth factor 10 (FGF10) and its receptor FGFR2, which are involved in the elongation and bifurcation of glandular ducts in the lung and pancreas, 96 , 97 and the downstream extracellular signal‐regulated kinase (ERK) signaling pathway. 98 We succeeded in creating a mouse model of intraductal papillary CCA with IPNB‐like features (Figure 3). 77 Furthermore, by analyzing the model in detail, we clarified part of the mechanism of the development, maintenance, and carcinogenesis of IPNBs, and showed the possibility of its therapeutic application.
Abnormal expression of FGF10 leads to the development of pancreatic epithelial hyperplasia, papillary adenoma of the lung, and prostatic intraepithelial neoplasia. 99 , 100 , 101 Therefore, we generated mice capable of inducing overexpression of the secretory protein FGF10 by doxycycline (Dox) treatment. In brief, we generated a Dox‐inducible transgenic mouse model (Col1a1:tetO:mouse Fgf10:ires:tdTomato:Rosa26:rtTA, hereafter referred to as “iFGF10”). The use of adult iFGF10 mice allows for the precise, reversible, and efficient spatiotemporal control of Fgf10 expression using the Tet system. 102 These iFGF10 mice overexpress FGF10 in Rosa26‐promoter dependent manner, indicating that this expression is broadly universal in the body.
Upon activation of FGF10–ERK signaling, papillary and cystic neoplastic lesions, which histopathologically mimic human IPNB, developed mainly in the EHBDs (Figure 3). Using cytokeratin 19 (Krt19)–iCre transgenic mice 103 and others that we generated in our previous study, the paracrine and autocrine actions of FGF10 caused widespread IPNB‐like lesions in the biliary tree.
In this study, we found that liver‐specific FGF10 overexpression leads to the development of bile duct cancers in the IHBDs but not EHBDs. We generated LSL‐rtTA;Alb‐Cre;Tet‐Fgf10 mice (Figure 3). In this mouse, FGF10 overexpression is induced in hepatic and biliary epithelial cells within the liver under the control of the Alb promoter. Furthermore, we found that, by crossing LSL‐Kras G12D mice, high‐grade BilIN‐like lesions in the peripheral IHBDs and IPNB in the hilar hepatic ducts develop in these liver‐specific FGF10‐inducible mice (Figure 3). Consequently, FGF10‐inducible mice developed papillary tumors at various locations in the biliary tree, depending on the location of their expression.
Using the cell lineage tracing method with the LSL‐LacZ reporter mouse, we showed that FGF10‐induced IPNBs originate from biliary epithelial cells including biliary stem/progenitor cells (SOX9 lineage) and PBGs (PDX1 lineage) (Figure 4). We also demonstrated that IPNB may indeed be a precancerous lesion. By crossing iFGF10 mice with LSL‐Kras G12D , p53 mutant, and p16 conditional knockout mice, a subset of FGF10‐induced IPNBs develop invasive carcinomas. In addition, the development and progression of FGF10‐induced IPNBs were suppressed by inhibition of the FGF10–FGFR2–RAS–ERK signaling pathway, indicating that this pathway could be a therapeutic target.
Figure 4.
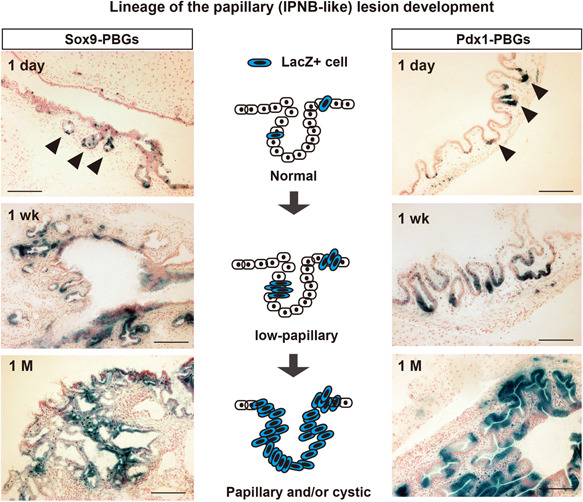
The PBG is a site of origin of papillary tumorigenesis in EHBDs of the iFGF10 mouse model. Cell lineage tracing of Sox9‐ and Pdx1‐expressing PBG cells during the progression from normal to papillary (IPNB‐like) lesions development in I FGF10 mice. Photos of X‐gal staining the in Sox9‐iFGF10 model (LSL‐rtTA; Sox9‐CreERT2; Tet‐Fgf10) and Pdx1‐iFGF10 model (LSL‐rtTA; Pdx1‐Cre; Tet‐Fgf10) crossed with LSL‐LacZ reporter mice. Dates indicate 1 day, 1 wks (weeks), and 1 M (month) since the start of Dox administration after marking the Sox9‐ or Pdx1‐expressing cells. Arrowhead, PBG. Bars, 200 µm. EHBD, extrahepatic bile duct; PBGs, peribiliary glands.
Furthermore, we verified that FGF10 and ERK are activated in human IPNB cases, indicating that FGF10–FGFR2–RAS–ERK signaling is involved in the development of CCA with papillary morphology, not only in mouse models but also in humans. What is unique about this mouse model is that it shows that papillary lesions occur in the biliary tree even in the absence of genetic mutations, and that inflammation increases the degree of atypia. The iFGF10 mouse model is extremely unique because a single secreted growth factor induces aberrant tubule formation in the biliary tree, thus leading to malignant transformation. Further analyses using this model are needed to clarify the pathogenesis of EHBD cancers.
LESSONS FROM GEMMS IN ECCAS
Are IPNB and BilIN precancerous lesions?
BilIN and IPNB in human CCA are considered to be precancerous lesions. 23 This has been demonstrated by the iFGF10 mouse model, 77 in which both IPNB‐ and BilIN‐like lesions can develop CCAs with invasive carcinoma (see Section 3.3.3). This is the first evidence obtained from a GEMM of eCCA. GEMMs can create not only genetic alterations that are found in humans in the biliary epithelium, but also assess changes in the surrounding microenvironment due to inflammation or chemical exposure, for example, inflammatory cell infiltration, fibrosis, and so on. 18 , 21
For example, in human diagnostic pathology, papillomatosis, which generates papillary and cystic dilated ducts, was classified as a benign lesion (currently, it is classified as IPNB with low‐grade dysplasia 23 ). iFGF10 mice develop this papillomatosis‐like lesion, which then progresses to a malignant lesion with more dysplasia resulting from the continuous severe inflammation present. This suggests that some cases of BiIIN and IPNB should be followed as premalignant and precancerous lesions in the clinical setting.
Are BiIIN and IPNB true neoplasms?
Papillary morphology in hepatobiliary pancreatic ducts, including low‐papillary, tubular, and cystic changes, is observed not only in premalignant lesions but also when inflammation is present, such as in hepatitis and cholangitis as reactive atypia. 104 The problem is differentiating between reactive atypia and neoplastic malignancy in diagnostic pathology. 104 For example, low‐grade BilIN involves reactive and metaplastic changes as well as mild atypia, leading to serious concerns for pathologists regarding the rapid histological diagnosis of resected bile duct margins during surgery.
The iFGF10 mouse, which has a Tet‐on inducible system, 102 can induce FGF10‐dependent papillary and cystic morphology of the biliary epithelium (see Section 3.3.3). Unlike conventional conditional knockout mice, such as those with the Cre‐loxP system, the iFGF10 mouse can control the on‐off expression FGF10 expression through Dox administration, leading to the development of IPNB‐like 77 and BilIN‐like lesions. Only FGF10 overexpresssion leads to develop IPNB‐like lesions. After that, the withdrawal of FGF10 induction leads to develop some flat or low‐papillary lesions, suggesting that reactive hyperplastic lesions. However, some IPNB‐like lesions keep still papillary morphology. Thus, we think that most of FGF10‐induced papillary lesions are reactive hyperplastic lesions and some are neoplasms. These results suggested that some FGF10‐induced papillary lesions are reactive hyperplastic lesions, and some are neoplasms. However, when genetic alterations, such as KrasG12D, p53, and/or p16 mutations were added to FGF10 induction in iFGF10 mice, papillary morphology and cellular atypia almost keeps even in the withdrawal of FGF10 induction, thus suggesting that the malignancy promoted by genetic alterations such as those in Kras, Tp53, and Cdkn2a(p16) is irreversible.
On the other hand, secreted FGF10 and inflammation cause not only papillary morphology but also hyperplastic and metaplastic changes. Intriguingly, papillary morphology is reversible, but metaplastic and mucin‐producing statuses are irreversible.
Taken together, these results provide some insights into clinical problems. For example, IPNB with low‐grade dysplasia with severe inflammation, which often causes bile duct obstruction, 105 , 106 would be treated by suppression of inflammation. However, genetic analysis will be more important for predicting malignant transformation.
Which genetic alterations contribute to CCA progression in mice and humans?
In‐depth sequencing analyses of human CCAs have accumulated evidence that molecular profiles vary between iCCA and eCCA. 107 , 108 , 109 , 110 , 111 , 112 , 113 , 114 In eCCA, most frequent genetic aberrations in targetable pathways of interest are KRAS mutation, TP53 mutation, SMAD4 mutation, CDKN2A/CDKN2B (including P16 INK4A ) loss, ERBB2/ERBB3 amplification, and ARID1A mutations (Figure 5a). 109 , 111 , 115 Among these alterations, KRAS mutation, TP53 mutation, SMAD4 mutation, CDKN2A/CDKN2B (including P16 INK4A ) loss, and ARID1A mutations are also frequent in iCCA. 109 , 111 , 115 Intriguingly, because KRAS mutation, TP53 mutation, and CDKN2A (P16 INK4A ) loss are strong oncogenic alterations in our iFGF10 GEMM, 77 these three genes may be oncogenic genes to induce eCCAs in humans (Figure 5b). Several gene alterations in the GEMMs of eCCAs are related to the progress the tumor development; however, these are not always consistent with the alterations of human CCAs.
Figure 5.
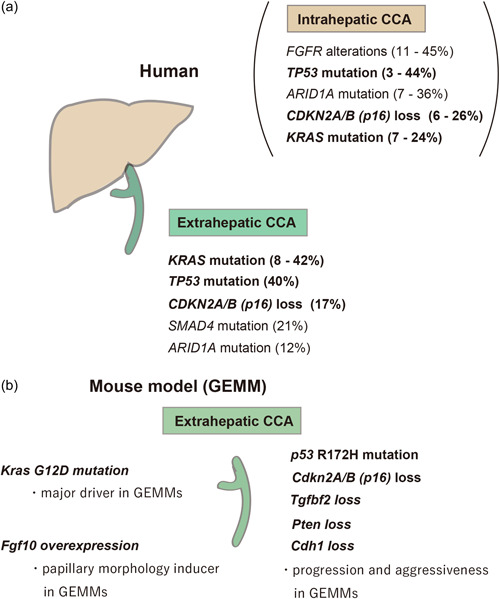
General overview of frequent gene alterations in eCCAs of human and mice. (a) The most frequent gene alterations of human intrahepatic and eCCAs. (b) Gene alterations of extrahepatic CCA mouse models. CCA, cholangiocarcinoma; eCCAs, extrahepatic CCAs.
The development of GEMMs with tissue‐specific conditionally expressed oncogenic changes has led to the development of gene‐driven, spontaneous tumor model mice. Current GEMMs of eCCA are very similar to those of pancreatic adenocarcinomas regarding the major genetic alterations that cause tumor initiation and progression. In pancreatic cancer models, the main driver gene that can initiate tumors is Kras G12D mutation. 116 , 117 , 118 In the eCCA models, Kras mutation also cause the initiation and progression of tumors. 77 , 85 , 86 , 119 Intriguingly, some IPNBs without dysplasia in iFGF10 mice develop malignant lesions, that is, high‐grade dysplasia and adenocarcinoma, through the addition of the Kras G12D mutation (Figure 5b). 77
The iFGF10 model harboring Tp53 R172H mutation or Cdkn2a(p16) Ink4a along with Kras G12D mutation develops more aggressive tumors such as adenocarcinoma, faithfully recapitulating most of the pathological features of human eCCA (Figures 3 and 5). 77 Tp53 and Cdkn2a (p16 Ink4a ) genetic alterations progress malignancy and invasiveness in the GEMMs of both pancreatic cancer and eCCA, 77 , 118 suggesting that multistep carcinogenesis is a mechanism of eCCAs. This also supports the similarity of the pathogenesis between pancreatic cancer and CCA. In human pancreatic ductal adenocarcinoma, activating KRAS mutations and p16Ink4a inactivation are near universal events. 120 In mouse models, Kras G12D initiates the formation of premalignant pancreatic ductal lesions, and loss of either p16Ink4a or p53 enables their malignant progression. 118 Furthermore, this similarity regarding Kras, Tp53, and Cdkn2a (p16 Ink4a ) genetic alterations is found in IPNB and intraductal papillary mucinous neoplasm, which is considered to be a counterpart of IPNB, in mice and humans. 118
By analyzing these GEMMs, we learned that p53 and p16 alterations along with mutated KRAS are strong oncogenic factors and can have therapeutic implications in eCCA as well as in pancreatic cancer.
Limitations of GEMMs in eCCA
The three GEMMs studied in this review represent the entire spectrum of human eCCAs, and some are also useful for studying the multistage pathogenesis of these tumors. However, in general, the difference in pathological features between mouse models and human specimens is a critically argued issue. 121 First, mouse lesions are milder cellular atypia rather than human lesions. Second, there are more human eCCAs with invasion and metastasis than the GEMMs. Third, most human eCCAs are solitary, whereas multifocality seems common in the GEMMs. The mouse is a mammal, yet it is not a human. There are several apparent distinctions between the two species. Our researchers must be aware of some of the critical biological differences between the two species in eCCAs and other cancers. A detailed understanding of the pathology and biology of the individual GEMMs for eCCAs is essential for selecting the most appropriate model for future studies.
CONCLUSIONS
The biliary tree, especially EHBDs, remains a mystery, not only in tumorigenesis but also in development and disease. The first clue to unraveling this mystery is the need for experimental analysis such as mouse models. Certainly, GEMMs have advantages and disadvantages. However, they are very effective tools because they can guide how normal cells become tumorous or malignant by recreating abnormalities in genes, epigenetic defects, inflammation, and the surrounding environment. Especially in EHBDs, histopathological analysis is necessary to understand the morphology in detail and to obtain a good understanding of the few available mouse models.
Future studies will focus on the molecular mechanisms that drive such oncogenesis and further progression to invasion and metastasis, while maintaining the relevance to human CCA as much as possible. In contrast to the progress made in the study of iCCA, eCCA is increasingly complex, and is both a challenging and highly ambitious task.
AUTHOR CONTRIBUTIONS
Hiroyuki Tomita drafted and made the manuscript and figures. Akira Hara supervised the draft and revision.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
ACKNOWLEDGMENTS
Hiroyuki Tomita has been announced as the winner of The Japanese Society of Pathology; Pathology Research Award in 2021. This work was supported by a grant from JSPS KAKENHI (grant JP 12891150) to Hiroyuki Tomita. We thank all members of the Tumor Pathology Department at Gifu University. We also thank H. Nikki March, PhD, from Edanz (https://jp.edanz.com/ac) for editing a draft of this manuscript.
Tomita H, Hara A. Development of extrahepatic bile ducts and mechanisms of tumorigenesis: Lessons from mouse models. Pathol. Int. 2022;72:589–605. 10.1111/pin.13287
REFERENCES
- 1. Roskams T, Desmet V. Embryology of extra‐ and intrahepatic bile ducts, the ductal plate. Anat Rec (Hoboken). 2008;291:628–35. [DOI] [PubMed] [Google Scholar]
- 2. Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43:257–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cardinale V, Wang Y, Carpino G, Mendel G, Alpini G, Gaudio E, et al. The biliary tree‐‐a reservoir of multipotent stem cells. Nat Rev Gastroenterol Hepatol. 2012;9:231–40. [DOI] [PubMed] [Google Scholar]
- 4. Raynaud P, Carpentier R, Antoniou A, Lemaigre FP. Biliary differentiation and bile duct morphogenesis in development and disease. Int J Biochem Cell Biol. 2011;43:245–56. [DOI] [PubMed] [Google Scholar]
- 5. Han Y, Glaser S, Meng F, Francis H, Marzioni M, McDaniel K, et al. Recent advances in the morphological and functional heterogeneity of the biliary epithelium. Exp Biol Med. 2013;238:549–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cardinale V, Wang Y, Carpino G, Cui CB, Gatto M, Rossi M, et al. Multipotent stem/progenitor cells in human biliary tree give rise to hepatocytes, cholangiocytes, and pancreatic islets. Hepatology. 2011;54:2159–72. [DOI] [PubMed] [Google Scholar]
- 7. Carpino G, Cardinale V, Onori P, Franchitto A, Berloco PB, Rossi M, et al. Biliary tree stem/progenitor cells in glands of extrahepatic and intraheptic bile ducts: an anatomical in situ study yielding evidence of maturational lineages. J Anat. 2012;220:186–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lanzoni G, Cardinale V, Carpino G. The hepatic, biliary, and pancreatic network of stem/progenitor cell niches in humans: a new reference frame for disease and regeneration. Hepatology. 2016;64:277–86. [DOI] [PubMed] [Google Scholar]
- 9. Lemaigre FP. Development of the intrahepatic and extrahepatic biliary tract: a framework for understanding congenital diseases. Annu Rev Pathol: Mech Dis. 2020;15:1–22. [DOI] [PubMed] [Google Scholar]
- 10. Alsaleh M, Leftley Z, Barbera TA, Sithithaworn P, Khuntikeo N, Loilome W, et al. Cholangiocarcinoma: a guide for the nonspecialist. Int J Gen Med. 2019;12:13–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kendall T, Verheij J, Gaudio E, Evert M, Guido M, Goeppert B, et al. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019;39(suppl 1):7–18. [DOI] [PubMed] [Google Scholar]
- 12. Banales JM, Marin JJG, Lamarca A, Rodrigues PM, Khan SA, Roberts LR, et al. Cholangiocarcinoma 2020: the next horizon in mechanisms and management. Nat Rev Gastroenterol Hepatol. 2020;17:557–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lendvai G, Szekerczés T, Illyés I, Dóra R, Kontsek E, Gógl A, et al. Cholangiocarcinoma: classification, histopathology and molecular carcinogenesis. Pathol Oncol Res. 2020;26:3–15. [DOI] [PubMed] [Google Scholar]
- 14. Brown ZJ, Hewitt DB, Pawlik TM. Biomarkers of intrahepatic cholangiocarcinoma: diagnosis and response to therapy. Front Biosci (Landmark Ed). 2022;27:85. [DOI] [PubMed] [Google Scholar]
- 15. Chung T, Park YN. Up‐to‐date pathologic classification and molecular characteristics of intrahepatic cholangiocarcinoma. Front Med. 2022;9:857140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Walter D, Döring C, Feldhahn M, Battke F, Hartmann S, Winkelmann R, et al. Intratumoral heterogeneity of intrahepatic cholangiocarcinoma. Oncotarget. 2017;8:14957–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loeuillard E, Fischbach SR, Gores GJ, Rizvi S. Animal models of cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2019;1865:982–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mohr R, Ozdirik B, Knorr J, Wree A, Demir M, Tacke F, et al. In vivo models for cholangiocarcinoma‐what can we learn for human disease? Int J Mol Sci. 2020;21:4993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Adams JM, Cory S. Transgenic models of tumor development. Science. 1991;254:1161–7. [DOI] [PubMed] [Google Scholar]
- 20. Frese KK, Tuveson DA. Maximizing mouse cancer models. Nat Rev Cancer. 2007;7:654–8. [DOI] [PubMed] [Google Scholar]
- 21. Kersten K, Visser KE, Miltenburg MH, Jonkers J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol Med. 2017;9:137–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pirenne S, Lemaigre FP. Genetically engineered animal models of biliary tract cancers. Curr Opin Gastroenterol. 2020;36:90–8. [DOI] [PubMed] [Google Scholar]
- 23. WHO Classification of Tumours Editorial Board. WHO classification of tumors of the digestive system. 5th ed. Lyon: IARC Press; 2019. [Google Scholar]
- 24. Kho AT, Zhao Q, Cai Z, Butte AJ, Kim JYH, Pomeroy SL, et al. Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 2004;18:629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tan CEL, Moscoso GJ. The developing human biliary system at the porta hepatis level between 11 and 25 weeks of gestation: a way to understanding biliary atresia. Pathol Int. 1994;44:600–10. [DOI] [PubMed] [Google Scholar]
- 26. Tan CEL, Moscoso GJ. The developing human biliary system at the porta hepatis level between 29 days and 8 weeks of gestation: a way to understanding biliary atresia. Pathol Int. 1994;44:587–99. [DOI] [PubMed] [Google Scholar]
- 27. Lemaigre FP. Molecular mechanisms of biliary development. Prog Mol Biol Transl Sci. 2010;97:103–26. [DOI] [PubMed] [Google Scholar]
- 28. Spence JR, Lange AW, Lin SCJ, Kaestner KH, Lowy AM, Kim I, et al. Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Dev Cell. 2009;17:62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Adkins RB, Jr. , Chapman WC, Reddy VS. Embryology, anatomy, and surgical applications of the extrahepatic biliary system. Surg Clin North Am. 2000;80:363–79. [DOI] [PubMed] [Google Scholar]
- 30. Woods A, Heslegrave AJ, Muckett PJ, Levene AP, Clements M, Mobberley M, et al. LKB1 is required for hepatic bile acid transport and canalicular membrane integrity in mice. Biochem J. 2011;434:49–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Uemura M, Hara K, Shitara H, Ishii R, Tsunekawa N, Miura Y, et al. Expression and function of mouse Sox17 gene in the specification of gallbladder/bile‐duct progenitors during early foregut morphogenesis. Biochem Biophys Res Commun. 2010;391:357–63. [DOI] [PubMed] [Google Scholar]
- 32. Uemura M, Ozawa A, Nagata T, Kurasawa K, Tsunekawa N, Nobuhisa I, et al. Sox17 haploinsufficiency results in perinatal biliary atresia and hepatitis in C57BL/6 background mice. Development. 2013;140:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sumazaki R, Shiojiri N, Isoyama S, Masu M, Keino‐Masu K, Osawa M, et al. Conversion of biliary system to pancreatic tissue in Hes1‐deficient mice. Nature Genet. 2004;36:83–7. [DOI] [PubMed] [Google Scholar]
- 34. Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Horiguchi M, Kuhara T, et al. Ectopic pancreas formation in Hes1‐knockout mice reveals plasticity of endodermal progenitors of the gut, bile duct, and pancreas. J Clin Invest. 2006;116:1484–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zaret KS, Grompe M. Generation and regeneration of cells of the liver and pancreas. Science. 2008;322:1490–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, et al. PDX‐1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–95. [DOI] [PubMed] [Google Scholar]
- 37. Fukuda A, Kawaguchi Y, Furuyama K, Kodama S, Kuhara T, Horiguchi M, et al. Loss of the major duodenal papilla results in brown pigment biliary stone formation in pdx1 null mice. Gastroenterology. 2006;130:855–67. [DOI] [PubMed] [Google Scholar]
- 38. Dong PDS, Munson CA, Norton W, Crosnier C, Pan X, Gong Z, et al. Fgf10 regulates hepatopancreatic ductal system patterning and differentiation. Nature Genet. 2007;39:397–402. [DOI] [PubMed] [Google Scholar]
- 39. Khandekar G, Llewellyn J, Kriegermeier A, Waisbourd‐Zinman O, Johnson N, Du Y, et al. Coordinated development of the mouse extrahepatic bile duct: implications for neonatal susceptibility to biliary injury. J Hepatol. 2020;72:135–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tietz PS, Holman RT, Miller LJ, LaRusso NF. Isolation and characterization of rat cholangiocyte vesicles enriched in apical or basolateral plasma membrane domains. Biochemistry. 1995;34:15436–43. [DOI] [PubMed] [Google Scholar]
- 41. Hohenester S, Maillette de Buy Wenniger L, Paulusma CC, van Vliet SJ, Jefferson DM, Oude Elferink RP, et al. A biliary HCO3‐ umbrella constitutes a protective mechanism against bile acid‐induced injury in human cholangiocytes. Hepatology. 2012;55:173–83. [DOI] [PubMed] [Google Scholar]
- 42. Maillette de Buy Wenniger LJ, Hohenester S, Maroni L, van Vliet SJ, Oude Elferink RP, Beuers U. The cholangiocyte glycocalyx stabilizes the ‘biliary HCO3 umbrella’: an integrated line of defense against toxic bile acids. Dig Dis. 2015;33:397–407. [DOI] [PubMed] [Google Scholar]
- 43. Tachi M, Okada H, Matsuhashi N, Takemura G, Suzuki K, Fukuda H, et al. Human colorectal cancer infrastructure constructed by the glycocalyx. J Clin Med. 2019;8:1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Patel VN, Likar KM, Zisman‐Rozen S, Cowherd SN, Lassiter KS, Sher I, et al. Specific heparan sulfate structures modulate FGF10‐mediated submandibular gland epithelial morphogenesis and differentiation. J Biol Chem. 2008;283:9308–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakanuma Y, Katayanagi K, Terada T, Saito K. Intrahepatic peribiliary glands of humans. I. Anatomy, development and presumed functions. J Gastroenterol Hepatol. 1994;9:75–9. [DOI] [PubMed] [Google Scholar]
- 46. Terada T. Development of extrahepatic bile duct excluding gall bladder in human fetuses: histological, histochemical, and immunohistochemical analysis. Microsc Res Tech. 2014;77:832–40. [DOI] [PubMed] [Google Scholar]
- 47. Spitz L, Petropoulos A. The development of the glands of the common bile duct. J Pathol. 1979;128:213–20. [DOI] [PubMed] [Google Scholar]
- 48. DiPaola F, Shivakumar P, Pfister J, Walters S, Sabla G, Bezerra JA. Identification of intramural epithelial networks linked to peribiliary glands that express progenitor cell markers and proliferate after injury in mice. Hepatology. 2013;58:1486–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nakanuma Y. A novel approach to biliary tract pathology based on similarities to pancreatic counterparts: is the biliary tract an incomplete pancreas? Pathol Int. 2010;60:419–29. [DOI] [PubMed] [Google Scholar]
- 50. Ishida F, Terada T, Nakanuma Y. Histologic and scanning electron microscopic observations of intrahepatic peribiliary glands in normal human livers. Lab Invest. 1989;60:260–5. [PubMed] [Google Scholar]
- 51. Sato H, Nakanuma Y, Kozaka K, Sato Y, Ikeda H. Spread of hilar cholangiocarcinomas via peribiliary gland network: a hither‐to‐unrecognized route of periductal infiltration. Int J Clin Exp Pathol. 2013;6:318–22. [PMC free article] [PubMed] [Google Scholar]
- 52. Matsui S, Harada K, Miyata N, Okochi H, Miyajima A, Tanaka M. Characterization of peribiliary gland‐constituting cells based on differential expression of trophoblast cell surface protein 2 in biliary tract. Am J Pathol. 2018;188:2059–73. [DOI] [PubMed] [Google Scholar]
- 53. Carpino G, Renzi A, Franchitto A, Cardinale V, Onori P, Reid L, et al. Stem/progenitor cell niches involved in hepatic and biliary regeneration. Stem Cells Int. 2016;2016:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Carpino G, Cardinale V, Renzi A, Hov JR, Berloco PB, Rossi M, et al. Activation of biliary tree stem cells within peribiliary glands in primary sclerosing cholangitis. J Hepatol. 2015;63:1220–8. [DOI] [PubMed] [Google Scholar]
- 55. Nakanuma Y, Harada K, Sasaki M, Sato Y. Proposal of a new disease concept “biliary diseases with pancreatic counterparts”. Anatomical and pathological bases. Histol. Histopathol. 2014;29:1–10. [DOI] [PubMed] [Google Scholar]
- 56. de Jong IEM, van Leeuwen OB, Lisman T, Gouw ASH, Porte RJ. Repopulating the biliary tree from the peribiliary glands. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1524–31. [DOI] [PubMed] [Google Scholar]
- 57. Matsubara T, Kozaka K, Matsui O, Nakanuma Y, Uesaka K, Inoue D, et al. Peribiliary glands: development, dysfunction, related conditions and imaging findings. Abdom Radiol. 2020;45:416–36. [DOI] [PubMed] [Google Scholar]
- 58. Oikawa T. Cancer stem cells and their cellular origins in primary liver and biliary tract cancers. Hepatology. 2016;64:645–51. [DOI] [PubMed] [Google Scholar]
- 59. Wang Y, Lanzoni G, Carpino G, Cui CB, Dominguez‐Bendala J, Wauthier E, et al. Biliary tree stem cells, precursors to pancreatic committed progenitors: evidence for possible life‐long pancreatic organogenesis. Stem Cells. 2013;31:1966–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Simons BD, Clevers H. Stem cell self‐renewal in intestinal crypt. Exp Cell Res. 2011;317:2719–24. [DOI] [PubMed] [Google Scholar]
- 61. Igarashi S. Participation of peribiliary glands in biliary tract pathophysiologies. World J Hepatol. 2013;5:425–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. op den Dries S, Westerkamp AC, Karimian N, Gouw ASH, Bruinsma BG, Markmann JF, et al. Injury to peribiliary glands and vascular plexus before liver transplantation predicts formation of non‐anastomotic biliary strictures. J Hepatol. 2014;60:1172–9. [DOI] [PubMed] [Google Scholar]
- 63. van der Flier LG, Clevers H. Stem cells, self‐renewal, and differentiation in the intestinal epithelium. Annu Rev Physiol. 2009;71:241–60. [DOI] [PubMed] [Google Scholar]
- 64. Nakanuma Y, Sasaki M, Ishikawa A, Tsui W, Chen TC, Huang SF. Biliary papillary neoplasm of the liver. Histol. Histopathol. 2002;17:851–61. [DOI] [PubMed] [Google Scholar]
- 65. Zen Y, Fujii T, Itatsu K, Nakamura K, Minato H, Kasashima S, et al. Biliary papillary tumors share pathological features with intraductal papillary mucinous neoplasm of the pancreas. Hepatology. 2006;44:1333–43. [DOI] [PubMed] [Google Scholar]
- 66. Bosman FT, Carneiro F, Hruban RH, Theise ND. WHO classification of tumors of the digestive system. 4th ed. Lyon: IARC Press; 2010. [Google Scholar]
- 67. Kubo S, Kinoshita M, Takemura S, Tanaka S, Shinkawa H, Nishioka T, et al. Characteristics of printing company workers newly diagnosed with occupational cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2014;21:809–17. [DOI] [PubMed] [Google Scholar]
- 68. Kubo S, Nakanuma Y, Takemura S, Sakata C, Urata Y, Nozawa A, et al. Case series of 17 patients with cholangiocarcinoma among young adult workers of a printing company in Japan. J Hepatobiliary Pancreat Sci. 2014;21:479–88. [DOI] [PubMed] [Google Scholar]
- 69. Sato Y, Kubo S, Takemura S, Sugawara Y, Tanaka S, Fujikawa M, et al. Different carcinogenic process in cholangiocarcinoma cases epidemically developing among workers of a printing company in Japan. Int J Clin Exp Pathol. 2014;7:4745–54. [PMC free article] [PubMed] [Google Scholar]
- 70. Hamano G, Kubo S, Takemura S, Tanaka S, Shinkawa H, Kinoshita M, et al. Comparison of clinicopathological characteristics between patients with occupational and non‐occupational intrahepatic cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2016;23:389–96. [DOI] [PubMed] [Google Scholar]
- 71. Kinoshita M, Kubo S, Nakanuma Y, Sato Y, Takemura S, Tanaka S, et al. Pathological spectrum of bile duct lesions from chronic bile duct injury to invasive cholangiocarcinoma corresponding to bile duct imaging findings of occupational cholangiocarcinoma. J Hepatobiliary Pancreat Sci. 2016;23:92–101. [DOI] [PubMed] [Google Scholar]
- 72. Nakanishi Y, Zen Y, Hirano S, Tanaka E, Takahashi O, Yonemori A, et al. Intraductal oncocytic papillary neoplasm of the bile duct: the first case of peribiliary gland origin. J Hepatobiliary Pancreat Surg. 2009;16:869–73. [DOI] [PubMed] [Google Scholar]
- 73. Nakanishi Y, Nakanuma Y, Ohara M, Iwao T, Kimura N, Ishidate T, et al. Intraductal papillary neoplasm arising from peribiliary glands connecting with the inferior branch of the bile duct of the anterior segment of the liver. Pathol Int. 2011;61:773–7. [DOI] [PubMed] [Google Scholar]
- 74. Miyata T, Uesaka K, Nakanuma Y. Cystic and papillary neoplasm at the hepatic hilum possibly originating in the peribiliary glands. Case Rep Pathol. 2016;2016:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Uchida T, Yamamoto Y, Ito T, Okamura Y, Sugiura T, Uesaka K, et al. Cystic micropapillary neoplasm of peribiliary glands with concomitant perihilar cholangiocarcinoma. World J Gastroenterol. 2016;22:2391–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Cardinale V, Wang Y, Carpino G, Reid LM, Gaudio E, Alvaro D. Mucin‐producing cholangiocarcinoma might derive from biliary tree stem/progenitor cells located in peribiliary glands. Hepatology. 2012;55:2041–2. [DOI] [PubMed] [Google Scholar]
- 77. Tomita H, Tanaka K, Hirata A, Okada H, Imai H, Shirakami Y, et al. Inhibition of FGF10‐ERK signal activation suppresses intraductal papillary neoplasm of the bile duct and its associated carcinomas. Cell Rep. 2021;34:108772. [DOI] [PubMed] [Google Scholar]
- 78. Louis C, Papoutsoglou P, Coulouarn C. Molecular classification of cholangiocarcinoma. Curr Opin Gastroenterol. 2020;36:57–62. [DOI] [PubMed] [Google Scholar]
- 79. Montal R, Sia D, Montironi C, Leow WQ, Esteban‐Fabró R, Pinyol R, et al. Molecular classification and therapeutic targets in extrahepatic cholangiocarcinoma. J Hepatol. 2020;73:315–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nature Genet. 2013;45:1470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, et al. Genomic spectra of biliary tract cancer. Nature Genet. 2015;47:1003–10. [DOI] [PubMed] [Google Scholar]
- 82. Bekaii‐Saab TS, Bridgewater J, Normanno N. Practical considerations in screening for genetic alterations in cholangiocarcinoma. Ann Oncol. 2021;32:1111–26. [DOI] [PubMed] [Google Scholar]
- 83. Hansen K, Khanna C. Spontaneous and genetically engineered animal models. Eur J Cancer. 2004;40:858–80. [DOI] [PubMed] [Google Scholar]
- 84. Onaciu A, Munteanu R, Munteanu VC, Gulei D, Raduly L, Feder RI, et al. Spontaneous and induced animal models for cancer research. Diagnostics. 2020;10:660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Nakagawa H, Suzuki N, Hirata Y, Hikiba Y, Hayakawa Y, Kinoshita H, et al. Biliary epithelial injury‐induced regenerative response by IL‐33 promotes cholangiocarcinogenesis from peribiliary glands. Proc Natl Acad Sci U S A. 2017;114:E3806–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Lin Y, Fang Z, Jiang T, Wan Z, Pan Y, Ma Y, et al. Combination of Kras activation and PTEN deletion contributes to murine hepatopancreatic ductal malignancy. Cancer Lett. 2018;421:161–69. [DOI] [PubMed] [Google Scholar]
- 87. Nakagawa H, Hikiba Y, Hirata Y, Font‐Burgada J, Sakamoto K, Hayakawa Y, et al. Loss of liver E‐cadherin induces sclerosing cholangitis and promotes carcinogenesis. Proc Natl Acad Sci U S A. 2014;111:1090–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Terada T, Nakanuma Y. Pathological observations of intrahepatic peribiliary glands in 1,000 consecutive autopsy livers. III. Hepatology. 1990;12:1229–33. [DOI] [PubMed] [Google Scholar]
- 89. Hughes NR, Pairojkul C, Royce SG, Clouston A, Bhathal PS. Liver fluke‐associated and sporadic cholangiocarcinoma: an immunohistochemical study of bile duct, peribiliary gland and tumour cell phenotypes. J Clin Pathol. 2006;59:1073–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sutton ME, Dries S, Koster MH, Lisman T, Gouw ASH, Porte RJ. Regeneration of human extrahepatic biliary epithelium: the peribiliary glands as progenitor cell compartment. Liver Int. 2012;32:554–9. [DOI] [PubMed] [Google Scholar]
- 91. Antoniou A, Raynaud P, Cordi S, Zong Y, Tronche F, Stanger BZ, et al. Intrahepatic bile ducts develop according to a new mode of tubulogenesis regulated by the transcription factor SOX9. Gastroenterology. 2009;136:2325–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, et al. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY‐related gene SOX9. Cell. 1994;79:1111–20. [DOI] [PubMed] [Google Scholar]
- 93. Mazur PK, Riener MO, Jochum W, Kristiansen G, Weber A, Schmid RM, et al. Expression and clinicopathological significance of notch signaling and cell‐fate genes in biliary tract cancer. Am J Gastroenterol. 2012;107:126–32. [DOI] [PubMed] [Google Scholar]
- 94. Rizvi S, Gores GJ. Pathogenesis, diagnosis, and management of cholangiocarcinoma. Gastroenterology. 2013;145:1215–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JSC, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. [DOI] [PubMed] [Google Scholar]
- 96. Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–78. [DOI] [PubMed] [Google Scholar]
- 97. Sekine K, Ohuchi H, Fujiwara M, Yamasaki M, Yoshizawa T, Sato T, et al. Fgf10 is essential for limb and lung formation. Nature Genet. 1999;21:138–41. [DOI] [PubMed] [Google Scholar]
- 98. Tang N, Marshall WF, McMahon M, Metzger RJ, Martin GR. Control of mitotic spindle angle by the RAS‐regulated ERK1/2 pathway determines lung tube shape. Science. 2011;333:342–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Clark JC, Tichelaar JW, Wert SE, Itoh N, Perl AKT, Stahlman MT, et al. FGF‐10 disrupts lung morphogenesis and causes pulmonary adenomas in vivo. Am J Physiol Lung Cell Mol Physiol. 2001;280:L705–15. [DOI] [PubMed] [Google Scholar]
- 100. Norgaard GA, Jensen JN, Jensen J. FGF10 signaling maintains the pancreatic progenitor cell state revealing a novel role of Notch in organ development. Dev Biol. 2003;264:323–38. [DOI] [PubMed] [Google Scholar]
- 101. Memarzadeh S, Xin L, Mulholland DJ, Mansukhani A, Wu H, Teitell MA, et al. Enhanced paracrine FGF10 expression promotes formation of multifocal prostate adenocarcinoma and an increase in epithelial androgen receptor. Cancer Cell. 2007;12:572–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Beard C, Hochedlinger K, Plath K, Wutz A, Jaenisch R. Efficient method to generate single‐copy transgenic mice by site‐specific integration in embryonic stem cells. Genesis. 2006;44:23–8. [DOI] [PubMed] [Google Scholar]
- 103. Kanayama T, Tomita H, Binh NH, Hatano Y, Aoki H, Okada H, et al. Characterization of a BAC transgenic mouse expressing Krt19‐driven iCre recombinase in its digestive organs. PLoS One. 2019;14:e0220818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zen Y, Adsay NV, Bardadin K, Colombari R, Ferrell L, Haga H, et al. Biliary intraepithelial neoplasia: an international interobserver agreement study and proposal for diagnostic criteria. Mod Pathol. 2007;20:701–9. [DOI] [PubMed] [Google Scholar]
- 105. Nanashima A, Imamura N, Sumida Y, Hiyoshi M, Hamada T, Nagayasu T. Clinicopathological aspects and diagnostic problems in patients with intraductal papillary neoplasm of the bile duct. Anticancer Res. 2018;38:2343–52. [DOI] [PubMed] [Google Scholar]
- 106. Luvira V. Progression of intraductal papillary neoplasm of the bile duct (IPNB): a proposed model through the observation of patients with non‐resected tumors. Ann Hepatol. 2021;23:100299. [DOI] [PubMed] [Google Scholar]
- 107. Borad MJ, Champion MD, Egan JB, Liang WS, Fonseca R, Bryce AH, et al. Integrated genomic characterization reveals novel, therapeutically relevant drug targets in FGFR and EGFR pathways in sporadic intrahepatic cholangiocarcinoma. PLoS Genet. 2014;10:e1004135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Ross JS, Wang K, Gay L, Al‐Rohil R, Rand JV, Jones DM, et al. New routes to targeted therapy of intrahepatic cholangiocarcinomas revealed by next‐generation sequencing. Oncologist. 2014;19:235–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Javle M, Bekaii‐Saab T, Jain A, Wang Y, Kelley RK, Wang K, et al. Biliary cancer: utility of next‐generation sequencing for clinical management. Cancer. 2016;122:3838–47. [DOI] [PubMed] [Google Scholar]
- 110. Farshidfar F, Zheng S, Gingras MC, Newton Y, Shih J, Robertson AG, et al Integrative genomic analysis of cholangiocarcinoma identifies distinct IDH‐mutant molecular profiles. Cell Rep. 2017;18:2780–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Valle JW, Lamarca A, Goyal L, Barriuso J, Zhu AX. New horizons for precision medicine in biliary tract cancers. Cancer Discovery. 2017;7:943–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu M, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res. 2018;24:4154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Bagante F, Ruzzenente A, Conci S, Rusev BC, Simbolo M, Campagnaro T, et al. Patterns of gene mutations in bile duct cancers: is it time to overcome the anatomical classification? HPB. 2019;21:1648–55. [DOI] [PubMed] [Google Scholar]
- 114. Boerner T, Drill E, Pak LM, Nguyen B, Sigel CS, Doussot A, et al. Genetic determinants of outcome in intrahepatic cholangiocarcinoma. Hepatology. 2021;74:1429–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Lamarca A, Barriuso J, McNamara MG, Valle JW. Molecular targeted therapies: ready for “prime time” in biliary tract cancer. J Hepatol. 2020;73:170–85. [DOI] [PubMed] [Google Scholar]
- 116. Mazur PK, Siveke JT. Genetically engineered mouse models of pancreatic cancer: unravelling tumour biology and progressing translational oncology. Gut. 2012;61:1488–500. [DOI] [PubMed] [Google Scholar]
- 117. Guerra C, Barbacid M. Genetically engineered mouse models of pancreatic adenocarcinoma. Mol Oncol. 2013;7:232–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Mallya K, Gautam SK, Aithal A, Batra SK, Jain M. Modeling pancreatic cancer in mice for experimental therapeutics. Biochim Biophys Acta Rev Cancer. 2021;1876:188554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Nakagawa H, Suzuki N, Koike K. Mouse model for cholangiocarcinoma from peribiliary glands. Methods Mol Biol. 2019;1905:237–45. [DOI] [PubMed] [Google Scholar]
- 120. Buscail L, Bournet B, Cordelier P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat Rev Gastroenterol Hepatol. 2020;17:153–68. [DOI] [PubMed] [Google Scholar]
- 121. Hruban RH, Adsay NV, Albores‐Saavedra J, Anver MR, Biankin AV, Boivin GP, et al. Pathology of genetically engineered mouse models of pancreatic exocrine cancer: consensus report and recommendations. Cancer Res. 2006;66:95–106. [DOI] [PubMed] [Google Scholar]


