Summary
Ectomycorrhizal exploration types are commonly assumed to denote spatial foraging patterns and resource‐related niches of extraradical mycelia. However, empirical evidence of the consistency of foraging strategies within exploration types is lacking.
Here, we analysed ectomycorrhizal foraging patterns by incubating root‐excluding ingrowth mesh bags filled with six different substrates in mature Picea abies forests. High‐throughput sequencing was used to characterise ectomycorrhizal fungal communities in the mesh bags and on adjacent fine roots after one growing season.
Contrary to expectations, many ectomycorrhizal genera of exploration types that are thought to produce little extraradical mycelium colonised ingrowth bags extensively, whereas genera commonly associated with ample mycelial production occurred sparsely in ingrowth bags relative to their abundance on roots.
Previous assumptions about soil foraging patterns of exploration types do not seem to hold. Instead, we propose that variation in the proliferation of extraradical mycelium is related to intergeneric differences in mycelial longevity and the mobility of targeted resources.
Keywords: boreal forest, cafeteria experiment, ectomycorrhizal exploration types, fungal networks, nutrient foraging, soil fungi
Introduction
Ectomycorrhizal fungi link tree roots to the soil environment by forming extraradical mycelium that extends into the soil from the mycorrhizal root tip (Smith & Read, 2008). The extent and morphology of the extraradical mycelium are important traits that may be linked to functional variation among species and genera (Agerer, 2001), which may relate further to ecosystem processes. For example, mycelium is an important precursor of soil organic matter (Clemmensen et al., 2013; Adamczyk et al., 2019), and morphological differences have been linked to decomposer capacity (Clemmensen et al., 2021; Argiroff et al., 2022). Ectomycorrhizal fungi can be classified into different ‘soil exploration types’ based on general morphological traits of the colonised root tips and emanating mycelium. These exploration types have been hypothesised to reflect the extent and manner of extraradical mycelial proliferation in the soil. The ‘contact’ type is described as having dense, smooth, hydrophilic mantles and only few emanating hyphae, while the ‘short‐distance’ type produces abundant short, nonaggregated hyphae in the near vicinity of the root tip. By contrast, the ‘medium‐distance smooth’ and ‘long‐distance’ types produce little extraradical mycelium close to the root but form cords, which vary in length and hydrophobicity. The ‘medium‐distance fringe’ and ‘mat’ types form extensive mycelia with many aggregated, hydrophobic cords (Agerer, 2001). The medium‐distance fringe, mat and long‐distance types have been associated with high mycelial biomass production (Hobbie & Agerer, 2010).
Exploration types have, to some extent, been found to reflect niche differentiation of ectomycorrhizal fungi. Those with none or few cords (contact, short‐distance and medium‐distance smooth) have been proposed to maximise the area of hydrophilic hyphae that extend into the soil and thereby promote rapid uptake of mobile N and decrease leaching (Hobbie & Agerer, 2010; Bahr et al., 2015). Types with hydrophobic mantles and cords (medium‐distance fringe and long‐distance) may instead display more directed growth towards discrete patches of immobile, organic resources (Finlay & Read, 1986; Cairney, 2005; Hobbie & Agerer, 2010). Furthermore, ectomycorrhizal fungi may respond to small‐scale variation in substrate quality by adapting local mycelial proliferation and spatial distribution (Rosling et al., 2004; Kluting et al., 2019). Exploration types have also been suggested to reflect patterns of community assembly via their different abilities to colonise new roots, with cord‐forming types being more successful in habitats with low root density (Peay et al., 2011).
It is important to point out that the exploration types originally were defined based on morphological investigations of root tips (Agerer, 2001) and hypotheses about the extent and foraging patterns of extraradical mycelia have largely been extrapolated from the amount and morphology of hyphae emanating from the mantle and often based on a few species (Weigt et al., 2011). Although exploration types are used as equivalents to foraging strategies (Tedersoo & Smith, 2013), whether they correspond to systematic and consistent differences in soil foraging remains uncertain. In comparisons between fungal communities on roots and in soil, species forming contact‐type mycorrhizas were underrepresented in the soil (Genney et al., 2006; Kjøller, 2006), implying a poor ability to forage for nutrients away from the roots. Kjøller (2006) found that species with exploration types assumed to form extraradical mycelia (short‐distance, medium‐distance and long‐distance) occurred abundantly in root‐free ingrowth mesh bags relative to roots. Similarly, Parrent & Vilgalys (2007) found that Tylospora (short‐distance) and Amanita (medium‐distance smooth) were prolific in ingrowth mesh bags and in bulk soil, although rarely observed as ectomycorrhizas, suggesting a high capacity to forage for nutrients away from the roots. Despite similarities in the results of these studies, divergent patterns have been observed for the long‐distance genus Suillus. Parrent & Vilgalys (2007) found Suillus to be an extensive soil coloniser despite being rare on roots, while Genney et al. (2006) observed the opposite relationship. Thus, more empirical evidence is needed to underpin a better understanding of how exploration types relate to proliferation of extraradical mycelium and selective colonisation of soil niches.
In this study, we conducted a ‘cafeteria experiment’ (Krebs, 1999), in which we incubated root‐excluding mycelial ingrowth mesh bags filled with different soil and sand substrates in mature, boreal Picea abies forests during one growing season. We investigated whether ectomycorrhizal fungi assigned to the same exploration type share common foraging patterns by comparing ectomycorrhizal communities in the ingrowth bags with those on adjacent fine roots, using DNA‐based metabarcoding. DNA has many drawbacks as a marker of mycelial biomass (Baldrian et al., 2013). For instance, copy numbers per hyphal material vary between taxa and most likely also between different tissue types within taxa. Nevertheless, DNA metabarcoding enables semiquantitative assessment (Castaño et al., 2020) of colonisation patterns of different fungal taxa and is particularly useful to evaluate relative differences among communities. Furthermore, we can provide information for taxa that are difficult to isolate, for which information from laboratory microcosms is scarce. The cafeteria experiment setup has been used commonly in animal ecology, where inference of ecology can be made based on the choices of animals when offered different food sources. Here, we used a similar methodology to investigate the extent and selectivity of extraradical mycelial foraging by different ectomycorrhizal fungi. The cafeterias offered a set of six different mesh bags, filled either with soil with varying pH and organic matter and nutrient contents, or with inert sand with or without apatite as a phosphorus (P) source. To capture a larger diversity of fungal species and foraging traits across different environments, cafeterias were incubated in 10 forests that varied in soil pH and inorganic N availability.
Based on the general morphological traits of their exploration types, we hypothesised that ectomycorrhizal fungi forage for soil resources in different ways. Contact type mycorrhizal fungi were expected to have low abundance in all bags due to their limited extension into the soil. The short‐distance and medium‐distance smooth exploration types, which form hydrophilic mycelia that absorb soluble, low‐molecular‐size nutrients from the surrounding soil, were expected to colonise all substrates (including inert sand) without preference, in a space‐filling manner. The hydrophobic medium‐distance fringe and long‐distance exploration types are thought to forage for immobile organic resources by forming cords and are expected to explore the ingrowth bags, particularly those with soil, with extensively proliferating extraradical mycelium (Finlay & Read, 1986; Leake et al., 2001) in a manner analogous to saprotrophic cord‐forming fungi (Boddy, 1999).
Furthermore, if fungal micro‐niches are determined by direct and local environmental filtering of mycelial colonisation (Rosling et al., 2004; Kluting et al., 2019), we expected fungal taxa to diverge in their colonisation of different soil substrates. Specifically, we expected species of short‐distance and medium‐distance smooth types, which are relatively more abundant in nutrient‐rich environments (Moeller et al., 2014; Sterkenburg et al., 2015; Defrenne et al., 2019; Pellitier & Zak, 2021), to predominantly colonise more nutrient‐rich soil substrates, also on the local ‘cafeteria scale’. By contrast, we expected cord‐forming species (mainly of medium‐fringe type), which are often abundant in nutrient‐poor environments, to be relatively more abundant in nutrient‐poor substrates due to lower competition from other ectomycorrhizal fungi.
Finally, we amended sand bags with apatite to investigate preferential colonisation by specific genera potentially involved in P mining by mineral weathering. Such a trait may be a competitive advantage in N‐rich but P‐limited forests, and increased ectomycorrhizal biomass production has been observed in apatite‐amended ingrowth bags in nemo‐boreal P. abies forests subjected to high rates of N deposition (14.5 kg ha−1 yr−1) (Wallander & Thelin, 2008; Almeida et al., 2019).
Materials and Methods
Ingrowth mesh bags
Aiming for a high variation in soil properties, organic topsoil or mull‐rich mineral soil (top 10 cm) was collected in autumn 2017 from four Swedish Picea abies (L.) H. Karst forests. Two soils were collected in central Sweden and two in southern Sweden. One of the southern soils had been subjected to P fertilisation. Some chemical characteristics of the soils are described in more detail in Table 1. Green parts of mosses and roots coarser than 2 mm in diameter were removed, and the soils were stored in bags at room temperature and in darkness for 17 months to reduce background levels of ectomycorrhizal DNA (Bååth et al., 2004). The soils remained moist throughout the storage period.
Table 1.
Characteristics of substrates used in the ingrowth mesh bags in the cafeteria experiment.
| Substrate | N deposition (kg ha−1 yr−1) | At collection | After 17 months preincubation | ||||
|---|---|---|---|---|---|---|---|
| pH | pH | Mineralised N (μg g−1 OM) | OM (%) | C/N | EMF DNA (%) | ||
| Organic, central Sweden | 5 | 4.2 | 4.4 | 2540 | 63 | 26 | 5.6 |
| Mull soil, central Sweden | 5 | 6.1 | 4.9 | 3220 | 8 | 16 | 4.5 |
| Organic, southern Sweden | 10–15 | 4.1 | 4.9 | 2090 | 66 | 25 | 0.6 |
| P‐fertilised organic, southern Sweden | 10–15 | 3.6 | 4.9 | 1340 | 71 | 29 | 3.1 |
| Sand | – | – | – | – | 0 | – | 0 |
| Sand with 1% apatite | – | – | – | – | 0 | – | 0 |
The soil was collected from mature Picea abies forests in two areas with contrasting atmospheric N deposition levels. pH was measured at the time of collection and after 17 months of preincubation. Mineralised N was calculated as the difference in inorganic N between un‐sieved, wet soils and sieved, dried soils after preincubation. Organic matter (OM) % and C/N were measured after preincubation. The proportion of ectomycorrhizal fungal (EMF) DNA was acquired through sequencing of ITS2 markers.
After 17 months, the soils were frozen, ground in a custom‐built freeze‐mill, sieved through 2 mm mesh, soaked in deionised water to wash out soluble nutrients and drained. The soils were dried at 40°C and filled into cylindrical mesh bags (8 cm long, 2.5 cm diameter; 50 μm mesh size; Sintab Product AB, Malmö, Sweden), which allowed ingrowth of fungal mycelium but excluded tree roots (Wallander et al., 2001); c. 7 g of organic topsoil and c. 21 g of mull soil were used to fill the bags. Additional bags were filled with either sand (c. 40 g; 0.36–2.0 mm; 99.6% SiO2; Silversand 90; Sibelco Nordic AB, Västerås, Sweden) or sand mixed with 1% apatite (Krantz, Bonn, Germany; sourced from Madagascar) with a grain size of 0.65–2.00 mm.
pH of the soil substrates was measured in a 1 : 5 v/v ratio of soil and deionised H2O with an 855 Robotic Titrosampler and an Aquatrode Plus combined pH electrode (Metrohm, Herisau, Switzerland) at collection and after 17 months of preincubation in room temperature. After sieving, washing and drying the preincubated soils, inorganic N concentrations (NH4 + and NO3 −) were measured by extraction in a 1 : 2.5 w/w ratio of soil and 2 M KCl and analysed on an autoanalyser (Bran + Luebbe XY‐2 Sampler; Seal Analytical Inc., Emu Plains, NSW, Australia). Organic matter concentration was determined by loss on ignition at 550°C for 5 h, and C/N was measured in a combustion elemental analyser (TruMac CN; Leco, St Joseph, MI, USA).
Site selection and field incubation
Ten mature (> 70 yr) P. abies‐dominated forests in central Sweden (latitude: 59.2–60.5°N) were selected for field incubation (Table 2). Forests with contrasting soil fertility were selected based on visual assessments (e.g. composition of understorey vegetation, contribution of deciduous trees and soil type). More productive sites had moder/mineral soils, some contribution of deciduous trees (Betula, Populus, Corylus) and an understorey consisting of mosses (Rhytidiadelphus sp., Hylocomium splendens, Ptilium crista‐castrensis and Pleurozium schreberi), grasses and ferns. Less productive sites had podzolised soil with a distinct organic layer (organic topsoil) overlying mineral soil, some contribution of Pinus sylvestris and understorey vegetation consisting of mosses (Hylocomium splendens and Pleurozium schreberi) and dwarf shrubs (Vaccinium myrtillus and Vaccinium vitis‐idaea).
Table 2.
Characteristics of the forests used for the incubation of the cafeterias.
| Soil type | Coordinates (WSG 84) | Basal area (m2 ha−1) | NH4 +–N (μg g−1 OM) | NO3 −–N (μg g−1 OM) | pH | |
|---|---|---|---|---|---|---|
| Latitude | Longitude | |||||
| Podzol | 59.27 | 14.84 | 42.9 | 24.8 (26.1) | 0.5 (0.4) | 4.1 (0.5) |
| Podzol | 59.69 | 14.88 | 39.3 | 24.9 (19.2) | 0.4 (0.3) | 4.2 (0.2) |
| Podzol | 60.29 | 17.05 | 36.0 | 19.2 (4.5) | 0.5 (0.2) | 4.4 (0.3) |
| Podzol | 60.28 | 17.74 | 34.0 | 18.4 (20.5) | 0.2 (0.1) | 4.5 (0.2) |
| Mull | 59.62 | 15.20 | 53.5 | 18.6 (9.4) | 0.1 (0.1) | 4.8 (0.2) |
| Podzol | 60.07 | 17.80 | 44.1 | 29.6 (9.6) | 0.3 (0.2) | 5.1 (0.8) |
| Mull | 59.88 | 17.35 | N/A | 23.8 (14.7) | 1.1 (2.5) | 5.2 (0.3) |
| Mull | 59.96 | 18.19 | 37.9 | 34.1 (8) | 0.1 (0.2) | 5.5 (0.3) |
| Mull | 60.18 | 17.86 | 28.3 | 20.6 (10.8) | 0 (0) | 5.6 (0.3) |
| Mull | 60.55 | 17.95 | 35.7 | 63.7 (21.1) | 0.5 (0.2) | 6.4 (0.8) |
Basal area was determined by measuring the diameter at breast height of all living trees within a 10 m radius from the central point of the plots. Soil characteristics were determined from one soil core taken in the centre of each cafeteria representing the same depth as the bags. The numbers in parentheses are within‐stand standard deviations of n = 5 cafeterias.
Bags were soaked in deionised H2O for a couple of minutes and placed in holes made by removing a soil core (2.5 cm in diameter) with a metal soil corer. At sites without a distinct organic layer, bags were placed vertically down to 8 cm depth from the soil surface and at sites with podzol soils, they were placed with the bottom of the bag at the organic–mineral soil interface. The six bags, containing different substrates (Table 1), were grouped in five replicate ‘cafeterias’ per site, spaced at least 5 m apart, each containing one bag of each substrate in a circle with 1 m diameter and even spacing of bags (Supporting Information Fig. S1). In total, 10 sites × 6 substrates × 5 replicates = 300 bags were incubated.
The incubation period lasted 153–160 d (May–November), after which the bags were retrieved from the soil (277 bags were recovered), placed individually in 50 ml tubes and frozen at −20°C within 8 h. At the time of bag collection, two soil cores (3 cm in diameter) were sampled from the middle of each cafeteria, spanning the same depth as the bags.
Sample preparation and soil chemical analysis
One of the two soil cores per cafeteria was used to measure soil chemical characteristics. The core was gently homogenised before a subsample (5 ml) was used to analyse pH as described previously. Another subsample was used to extract ammonium and nitrate as described previously. Picea abies roots (< 2 mm in diameter) were retrieved from the second central soil core and rinsed carefully. Cleaned roots and substrates from ingrowth mesh bags were freeze‐dried and finely ground; soils and roots in a Precellys homogenizer (Bertin Instruments, Montigny la Bretonneux, France); and sand in a ball mill (LMLW‐320/2; Laarmann, Roermond, the Netherlands).
Fungal community analysis
DNA was extracted from 20 to 50 mg (roots), 75 mg (organic soils: central, southern, southern P fertilised), 250 mg (mull soil) or 500 mg (sand and sand amended with apatite) of material with the NucleoSpin Soil kit (Macherey‐Nagel, Duren, Germany), and extracts were diluted to a DNA concentration of 0.5 ng μl−1. DNA was also extracted from samples of substrates that were not incubated in the field. Amplicons of the ITS2 region were produced by PCR using the forward primer fITS7 (Ihrmark et al., 2012) and the reverse primer ITS4 (White et al., 1990) with unique identification tags attached to both primers. Reactions (50 μl) were run with 12.5 ng of DNA template, 0.2 mM dNTP, 0.025 U μl−1 Dreamtaq polymerase (Thermo Fischer Scientific, Waltham, MA, USA) and 0.5 μM of each primer. PCR was performed with denaturation at 94°C, annealing at 56°C and extension at 72°C for 30 s each and cycle numbers (28–35) optimised to ensure that the reaction was in the exponential phase (Castaño et al., 2020). Negative controls with deionised H2O instead of DNA template were included. A total of 267 ingrowth mesh bag samples, 46 root samples and samples of nonincubated substrates (to characterise background communities) were successfully amplified and cleaned with Sera‐Mag (Cytiva, Marlborough, MA, USA) according to the manufacturer's instructions. Amplicon concentrations were measured fluorometrically on a Qubit (Invitrogen), and the PCR products were merged in equal amounts into four pools and cleaned with the EZNA Cycle Pure Kit (Omega Bio‐Tek., Norcross, GA, USA). Library preparation and sequencing were conducted by SciLifeLab (NGI, Uppsala, Sweden) on the PacBio Sequel I platform (Pacific Biosciences, Menlo Park, CA, USA) in one SMRT cell per pool. The PacBio platform was chosen to minimise biases due to ITS2 length variation (Castaño et al., 2020). Raw sequences were filtered and clustered in the bioinformatics pipeline Scata (https://scata.mykopat.slu.se/; Ihrmark et al., 2012), accepting sequences with length > 100 bp, mean quality > 20, single base quality > 3, primer sequence similarity > 90% and intact identification tags. Genotypes found only once in the whole data set were removed, and sequences were clustered into species hypotheses (Kõljalg et al., 2013) using single‐linkage clustering with 98.5% similarity to the closest neighbour required for sequences to join clusters.
Species were identified by comparing representative sequences to the UNITE database (Nilsson et al., 2019), and only ectomycorrhizal fungal species hypotheses (n = 342) were selected for further analyses. Relative abundances of any ectomycorrhizal species (in the total fungal community) found in soil substrates before field incubations (i.e. background levels, 0.6–5.6%; Table 1) were subtracted from their relative abundances measured after incubation (31–43%). After this background correction, relative abundances of ectomycorrhizal genera were calculated as their share of the ectomycorrhizal community. We selected 12 genera that were present on roots in at least 10 cafeterias (out of a total of 50) to be included in the statistical analyses. Exploration types were assigned according to Tedersoo & Smith (2013) and the DEEMY database (Agerer & Rambold, 2004; http://www.deemy.de/), and in cases of known intrageneric variation, the exploration type of the dominant species was used. The tested genera and their assigned exploration types, hydrophobicity and expected foraging patterns are listed in Table 3.
Table 3.
Functional traits of the 12 most frequent ectomycorrhizal genera.
| Genus | Exploration type | Hydrophobicity | Expectation |
|---|---|---|---|
| Amanita | Medium‐distance smooth | Hydrophobic | Uncertain |
| Amphinema | Medium‐distance fringe | Hydrophobic | Prolific in soil bags |
| Cenococcum | Short‐distance | Hydrophilic | Prolific in all bags |
| Cortinarius | Medium‐distance fringe | Hydrophobic | Prolific in soil bags |
| Hyaloscypha | Contact | Hydrophilic | Little colonisation of bags |
| Hygrophorus | Short‐distance | Prolific in all bags | |
| Lactarius | Contact | Hydrophilic | Little colonisation of bags |
| Piloderma | Medium‐distance fringe | Hydrophobic | Prolific in soil bags |
| Pseudotomentella | Medium‐distance smooth | Hydrophilic | Prolific in all bags |
| Russula | Contact | Hydrophilic | Little colonisation of bags |
| Tomentella | Medium‐distance smooth | Hydrophilic | Prolific in all bags |
| Tylospora | Short‐distance | Hydrophilic | Prolific in all bags |
Exploration types, hydrophobicity (based on Lilleskov et al., 2011) and expected extraradical mycelial proliferation of the 12 most frequent genera in the cafeteria study.
Statistical analysis
For each ectomycorrhizal genus in each cafeteria, we calculated a log ratio of the relative abundance in ingrowth bags relative to roots (Eqn 1) and in soil bags relative to sand bags (Eqn 2). Only cafeterias where the genus was present on roots were included in these calculations, with 11–39 cafeterias assessed depending on the genus.
| (Eqn 1) |
| (Eqn 2) |
where μ = 1/(mean sequencing depth × 6), that is the lowest expected relative abundance, was added to avoid zeros.
To evaluate whether exploration type was a good predictor for mycelial growth patterns, we used mixed‐effect linear models (the lmertest and lme4 packages) (Bates et al., 2015; Kuznetsova et al., 2017) in R (v.4.0.3; R Core Team, 2020) with the log ratios as response variables, exploration type as explanatory variable and genus, and cafeteria nested within site as random factors. Next, the log ratios for each genus were tested to investigate whether individual genera were characterised by little or prolific extraradical mycelial growth (i.e. low or high soil/root ratio) or a preference for soil over inert sand. This was done by testing whether the intercept of mixed‐effect linear models with log ratio as response variable and cafeteria nested within site as random factor was significantly different from zero. P‐values from all genera‐specific models were corrected for testing of multiple taxa by the Benjamini–Hochberg method (Benjamini & Hochberg, 1995). We also tested whether different types of substrates recruited different ectomycorrhizal communities, that is if the extraradical mycelia of different genera displayed preference for specific substrates. Substrate effects on community composition at the genus level among soil bags (humus and mull substrates) and among sand bags (sand and apatite amended sand) were evaluated by PERMANOVA (adonis2 function in the vegan package in R; Oksanen et al., 2022) with 1000 permutations constrained to samples within each cafeteria. Individual mixed‐effect linear models were applied for specific genera with square root transformed relative abundance (Hellinger transformation) as the response variable, substrate as a fixed factor and cafeteria nested within site as a random factor. P‐values were corrected for multiple testing as described previously, and genera with P ≤ 0.05 were subjected to Tukey's HSD post hoc tests with the emmeans package (Lenth et al., 2021). Graphs were produced with ggplot2 (Wickham et al., 2020).
Results
A total of 903 787 sequences passed the quality check, and after the removal of unique sequences, 466 080 sequences were clustered into 2668 species hypotheses, whereof 342 (272 499 sequences accounting for 0.6–95% of total reads from each sample; mean 46% and median 44%) were ectomycorrhizal and 235 (244 713 sequences accounting for 0.2–95% of total reads from each sample; mean 41% and median 38%) belonged to the 12 most frequently encountered genera (Table 3). These genera together accounted for on average 87% of the ectomycorrhizal reads in individual samples (range: 9–100%; median: 97%).
Exploration type was not a good predictor of relative colonisation of bags vs roots (P = 0.7) or soil bags vs sand bags (P = 0.3; Table S1). Amphinema, Tomentella and Tylospora had a high ratio of extraradical mycelium to root‐associated mycelium and were, thus, prolific bag colonisers, despite being of different exploration types. By contrast, Hyaloscypha, Hygrophorus, Cortinarius and Piloderma were more abundant on roots than as extraradical mycelium, although the two latter genera are of medium‐distance fringe type and, thus, expected to proliferate far into the soil (Tables S2, S3). Lactarius, Russula, Amanita and Piloderma showed a preference for soil bags, whereas Amphinema was more prolific in sand bags. Furthermore, Lactarius and Russula occurred as abundantly in ingrowth bags as on roots, despite being of the contact type (Fig. 1; Tables S2, S4).
Fig. 1.
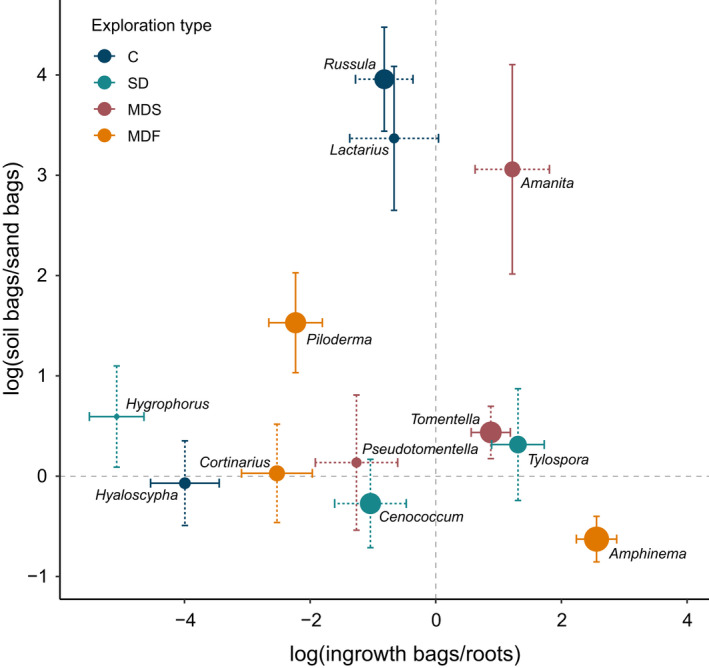
Model estimated log ratios of ectomycorrhizal genera. Log ratio was calculated as the log‐transformed ratio of the relative abundances of each genus in ingrowth bags and on roots (x‐axis) and in soil bags and sand bags (y‐axis). Point sizes correspond to the overall relative abundance of each genus in the ectomycorrhizal community. C, contact; MDF, medium‐distance fringe; MDS, medium‐distance smooth; SD, short‐distance. Dashed vertical and horizontal lines denote equal abundances in bags and on roots, and in soil bags and sand bags respectively. Error bars show SE, and solid bars denote a significant difference from zero lines (Benjamini–Hochberg‐adjusted P ≤ 0.05) based on mixed‐effect linear models. Model outputs are reported in Supporting Information Tables S1, S3, and S4, and relative abundances are reported in Table S2.
Community composition on the genus level differed between different types of soil bags (P = 0.001; Table S5), with three genera displaying a preference for some of the soil substrates; Amphinema and Tomentella were less abundant in the southern soils, where Cenococcum was more prolific (Fig. 2; Tables S6–S8). Apatite‐amended bags did not diverge in community composition from nonamended sand (P = 0.4; Tables S9, S10).
Fig. 2.
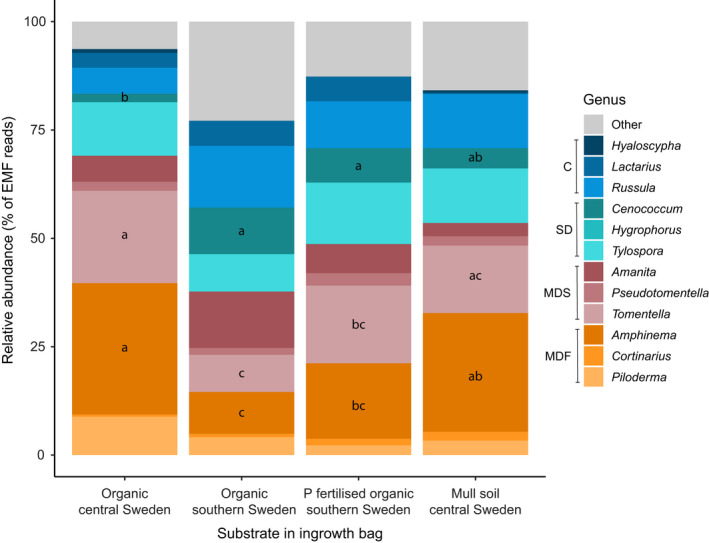
Ectomycorrhizal community composition in soil bags. Relative abundances of the 12 most frequent ectomycorrhizal genera colonising ingrowth bags filled with four different soil substrates. C, contact; MDF, medium‐distance fringe; MDS, medium‐distance smooth; SD, short‐distance. Different lowercase letters indicate significant effects (P ≤ 0.05) among substrate types for the respective genus according to post hoc Tukey HSD tests. Relative abundances are reported in Table S6, and model outputs are reported in Tables S7 and S8.
Discussion
All in all, we found little support for the utility of exploration types to predict patterns of extraradical mycelial foraging. Contrary to our hypothesis, contact‐type genera did not generally colonise bags to a lesser extent than others (Fig. 1); only the ascomycete genus Hyaloscypha behaved as expected for a contact type and colonised roots more extensively than ingrowth bags. The other two contact‐type genera, Russula and Lactarius, preferably colonised soil substrates over sand (Fig. 1), which may explain why Kjøller (2006), who used sandbags, concluded that these genera have limited mycelial proliferation. The detection of abundant DNA of some contact‐type genera in soil substrates suggests that they (Russulaceae) may colonise the soil matrix with extraradical mycelium more extensively than proposed by Agerer (2001), in line with the observation that Lactarius rufus was present in bulk soil without being detected on ectomycorrhizal root tips (Genney et al., 2006). Presumably, contact types may extend from the roots with fine emanating hyphae that are not readily visible and selectively target organic hotspots. Amanita (medium‐distance smooth) behaved similarly.
We hypothesised that the extraradical mycelium of the hydrophilic short‐distance and medium‐distance smooth exploration types would expand throughout the soil without preference for any particular substrate in a space‐filling manner, and, thus, be prolific in all types of ingrowth bags. Tomentella (medium‐distance smooth) and Tylospora (short‐distance) were indeed more abundant in bags than on roots. However, the short‐distance type Hygrophorus was, by contrast, mainly found on roots.
Contrary to our expectations, Piloderma and Cortinarius, which both mainly form medium‐distance fringe type mycorrhizas, did not proliferate extensively in the ingrowth bags, despite being widely considered to produce large amounts of extraradical mycelial biomass (Hobbie & Agerer, 2010). On the contrary, Amphinema (also medium‐distance fringe) colonised bags vigorously, despite its scarce representation on root tips. Furthermore, we did not find consistent support for the hypothesis that hydrophobic, cord‐forming genera would prefer organic substrates over sand bags; Piloderma was, as expected, more abundant in soil bags, but Cortinarius had no significant preference. Amphinema was even preferentially found in sand bags, potentially being outcompeted (or diluted) by more selective foragers in soil bags (Fig. 1).
Amphinema, Tomentella and Cenococcum differed significantly in relative abundance between ingrowth bags with different types of soil. Although the underlying mechanism is not clear, this observation suggests that some ectomycorrhizal fungi can detect and selectively direct extraradical mycelial growth towards specific substrates, and/or that mycelial colonisation may be confined to specific micro‐niches by local environmental filtering (Rosling et al., 2004). However, contrary to our hypothesis, short‐distance and medium‐distance smooth types, which are supposedly nitrophilic (Moeller et al., 2014; Sterkenburg et al., 2015; Defrenne et al., 2019; Pellitier & Zak, 2021), did not preferably colonise the mull soil, which had the lowest C : N ratio and highest inorganic N mineralisation (Table 2). The lack of community response to apatite amendment in sand bags is concordant with the results of Hedh et al. (2008), possibly indicating a low demand for mineral‐bound P or large functional redundancy in terms of weathering.
Although we conclude that exploration types are not consistent predictors of soil foraging, we observed systematic differences among genera regarding their extraradical mycelial proliferation in different substrates. Tedersoo et al. (2012) claimed that ectomycorrhizal lineage is a better predictor of functional attributes than exploration type. However, we also observed contrasting patterns within lineages (e.g. Piloderma vs Tylospora and Amphinema in the Athelioid lineage).
Piloderma and Cortinarius have been highlighted as having high extraradical biomass, but we rather observed low extraradical proliferation (DNA in the bags) of these genera. Still, these genera often dominate ectomycorrhizal fungal communities and attain a high biomass in old, nutrient‐limited boreal forests (Sterkenburg et al., 2015; Kyaschenko et al., 2017). Accumulation of extraradical mycelial biomass does not depend solely on growth rate, but also on biomass turnover (Clemmensen et al., 2013, 2015; Ekblad et al., 2013; Hagenbo et al., 2017). Species with slow turnover of extraradical hyphae, for example by forming long‐lived cords (Treseder et al., 2005), may attain high biomass over a long period of time in spite of slow growth. Here, we studied colonisation of bags during only one growing season while Hagenbo et al. (2018) studied successional colonisation of bags over a longer period and observed that Cortinarius progressively increased over multiple years, suggesting slow but persistent net accumulation of perennial mycelial biomass. Furthermore, Piloderma selectively colonised soil substrates with organic resources, while Cortinarius did not display such preference. Members of Cortinarius are known for their capacity to decompose and derive nutrients from complex organic substrates (Bödeker et al., 2014; Lindahl et al., 2021). These two genera are also recognised as nitrophobic (Lilleskov et al., 2011; van der Linde et al., 2018), as is Hygrophorus (Solly et al., 2017). By contrast, the genera that proliferated extensively in soil bags, relative to their more scarce representation on roots, that is Amphinema, Tylospora and Tomentella, have often been described as nitrophilic (Kranabetter et al., 2009; Sterkenburg et al., 2015; Hagenbo et al., 2018), at least in a boreal context with low nitrogen deposition. Amphinema was also more abundant in sand bags than in soil bags, suggesting a space‐filling growth strategy. Ample production of extraradical mycelium may be advantageous at high levels of mobile, inorganic nutrients, by minimising leaching and retaining nutrients in the mycorrhizal system (Hobbie & Agerer, 2010; Bahr et al., 2015). However, Amphinema has hydrophobic cords, implying that this growth strategy is not restricted to noncord‐forming, hydrophilic exploration types.
As exploration types do not seem to be consistent predictors of mycelial foraging, we see a need for alternative frameworks, for example based on nitrophobicity and/or hydrophobicity (Almeida et al., 2022). Most low‐proliferating taxa in our study are recognised as nitrophobic, hydrophobic and linked to exploitation of solid organic resources in nutrient‐limited environments. A long mycelial lifespan may enable them to accumulate high biomass over time (Hagenbo et al., 2017) in spite of scarce resource availability. The high‐proliferating taxa, on the contrary, are relatively nitrophilic/hydrophilic and may rather employ a space‐filling strategy to minimise losses of soluble inorganic nutrients under rich conditions. These coordinated traits are likely to be continuously distributed along a trait axis (van der Linde et al., 2018) similar to the leaf economics spectrum of plants (Wright et al., 2004). Further characterisation of trait axes will facilitate understanding of ecological niches, plasticity and adaptations of ectomycorrhizal fungal species along environmental gradients. To this end, more studies are needed to assess ectomycorrhizal foraging patterns in other types of environments, ideally also with more directly quantitative methods, as relative DNA abundances are only indirectly linked to mycelial biomass.
Author contributions
KJ, KC, HW and BL designed the study. KJ collected the data and performed the analyses. KJ wrote the first draft of the manuscript. All authors contributed to the interpretation and writing.
Supporting information
Fig. S1 Illustration of cafeteria setup.
Table S1 Model output from lme‐models of the log‐ratio of ectomycorrhizal exploration types in bags relative to roots, and in soil bags relative to sand bags.
Table S2 Relative abundance of ectomycorrhizal fungi on roots, and sand‐ or soil‐filled ingrowth bags.
Table S3 Model output from lme‐models of log‐ratio of ectomycorrhizal genera in bags relative to roots.
Table S4 Model output from lme‐models of log‐ratio of ectomycorrhizal genera in soil‐filled relative to sand‐filled ingrowth meshbags.
Table S5 Model output from PERMANOVA testing the effect of different soil substrates on ectomycorrhizal fungal community composition in ingrowth mesh bags.
Table S6 Relative abundance of ectomycorrhizal fungi in ingrowth bags filled with different organic substrates.
Table S7 Model output from models testing the effect of substrates in ingrowth meshbags on ectomycorrhizal fungal genera.
Table S8 Output from post hoc Tukey HSD test on ectomycorrhizal genera that displayed preference towards any soil substrate.
Table S9 Model output from PERMANOVA testing the effect of different sand substrates on ectomycorrhizal fungal community composition in ingrowth mesh bags.
Table S10 Relative abundance of ectomycorrhizal fungi in ingrowth bags filled with sand.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
This work was supported by the Swedish Research Council (VR) (project no. 2015‐04882).
Data availability
Data and code needed to reproduce analyses are available on Dryad Digital Repository (doi: 10.5061/dryad.08kprr55q; Jörgensen et al., 2022). Sequence data are published in NCBI‐SRA under project PRJNA796466.
References
- Adamczyk B, Sietiö O‐M, Biasi C, Heinonsalo J. 2019. Interaction between tannins and fungal necromass stabilizes fungal residues in boreal forest soils. New Phytologist 223: 16–21. [DOI] [PubMed] [Google Scholar]
- Agerer R. 2001. Exploration types of ectomycorrhizae. Mycorrhiza 11: 107–114. [Google Scholar]
- Agerer R, Rambold G. 2004. –2022. DEEMY – an information system for characterization and determination of ectomycorrhizae . München, Germany: www.deemy.de
- Almeida JP, Rosenstock NP, Forsmark B, Bergh J, Wallander H. 2019. Ectomycorrhizal community composition and function in a spruce forest transitioning between nitrogen and phosphorus limitation. Fungal Ecology 40: 20–31. [Google Scholar]
- Almeida JP, Rosenstock NP, Woche SK, Guggenberger G, Wallander H. 2022. Nitrophobic ectomycorrhizal fungi are associated with enhanced hydrophobicity of soil organic matter in a Norway spruce forest. Biogeosciences 19: 3713–3726. [Google Scholar]
- Argiroff WA, Zak DR, Pellitier PT, Upchurch RA, Belke JP. 2022. Decay by ectomycorrhizal fungi couples soil organic matter to nitrogen availability. Ecology Letters 25: 391–404. [DOI] [PubMed] [Google Scholar]
- Bååth E, Nilsson LO, Göransson H, Wallander H. 2004. Can the extent of degradation of soil fungal mycelium during soil incubation be used to estimate ectomycorrhizal biomass in soil? Soil Biology and Biochemistry 36: 2105–2109. [Google Scholar]
- Bahr A, Ellström M, Bergh J, Wallander H. 2015. Nitrogen leaching and ectomycorrhizal nitrogen retention capacity in a Norway spruce forest fertilized with nitrogen and phosphorus. Plant and Soil 390: 323–335. [Google Scholar]
- Baldrian P, Větrovský T, Cajthaml T, Dobiášová P, Petránková M, Šnajdr J, Eichlerová I. 2013. Estimation of fungal biomass in forest litter and soil. Fungal Ecology 6: 1–11. [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. 2015. Fitting linear mixed‐effects models using lme4. Journal of Statistical Software 67: 1–48. [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B (Methodological) 57: 289–300. [Google Scholar]
- Boddy L. 1999. Saprotrophic cord‐forming fungi: meeting the challenge of heterogeneous environments. Mycologia 91: 13–32. [Google Scholar]
- Bödeker ITM, Clemmensen KE, de Boer W, Martin F, Olson Å, Lindahl BD. 2014. Ectomycorrhizal Cortinarius species participate in enzymatic oxidation of humus in northern forest ecosystems. New Phytologist 203: 245–256. [DOI] [PubMed] [Google Scholar]
- Cairney JWG. 2005. Basidiomycete mycelia in forest soils: dimensions, dynamics and roles in nutrient distribution. Mycological Research 109: 7–20. [DOI] [PubMed] [Google Scholar]
- Castaño C, Berlin A, Durling MB, Ihrmark K, Lindahl BD, Stenlid J, Clemmensen KE, Olson Å. 2020. Optimized metabarcoding with Pacific biosciences enables semi‐quantitative analysis of fungal communities. New Phytologist 228: 1149–1158. [DOI] [PubMed] [Google Scholar]
- Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, Stenlid J, Finlay RD, Wardle DA, Lindahl BD. 2013. Roots and associated fungi drive long‐term carbon sequestration in boreal forest. Science 339: 1615–1618. [DOI] [PubMed] [Google Scholar]
- Clemmensen KE, Durling MB, Michelsen A, Hallin S, Finlay RD, Lindahl BD. 2021. A tipping point in carbon storage when forest expands into tundra is related to mycorrhizal recycling of nitrogen. Ecology Letters 24: 1193–1204. [DOI] [PubMed] [Google Scholar]
- Clemmensen KE, Finlay RD, Dahlberg A, Stenlid J, Wardle DA, Lindahl BD. 2015. Carbon sequestration is related to mycorrhizal fungal community shifts during long‐term succession in boreal forests. New Phytologist 205: 1525–1536. [DOI] [PubMed] [Google Scholar]
- Defrenne CE, Philpott TJ, Guichon SHA, Roach WJ, Pickles BJ, Simard SW. 2019. Shifts in ectomycorrhizal fungal communities and exploration types relate to the environment and fine‐root traits across interior Douglas‐fir forests of western Canada. Frontiers in Plant Science 10: 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblad A, Wallander H, Godbold DL, Cruz C, Johnson D, Baldrian P, Björk RG, Epron D, Kieliszewska‐Rokicka B, Kjøller R et al. 2013. The production and turnover of extramatrical mycelium of ectomycorrhizal fungi in forest soils: role in carbon cycling. Plant and Soil 366: 1–27. [Google Scholar]
- Finlay R, Read D. 1986. The structure and function of the vegetative mycelium of ectomycorrhizal plants 1. Translocation of C‐14‐labeled carbon between plants interconnected by a common mycelium. New Phytologist 103: 143–156. [Google Scholar]
- Genney DR, Anderson IC, Alexander IJ. 2006. Fine‐scale distribution of pine ectomycorrhizas and their extramatrical mycelium. New Phytologist 170: 381–390. [DOI] [PubMed] [Google Scholar]
- Hagenbo A, Clemmensen KE, Finlay RD, Kyaschenko J, Lindahl BD, Fransson P, Ekblad A. 2017. Changes in turnover rather than production regulate biomass of ectomycorrhizal fungal mycelium across a Pinus sylvestris chronosequence. New Phytologist 214: 424–431. [DOI] [PubMed] [Google Scholar]
- Hagenbo A, Kyaschenko J, Clemmensen KE, Lindahl BD, Fransson P. 2018. Fungal community shifts underpin declining mycelial production and turnover across a Pinus sylvestris chronosequence. Journal of Ecology 106: 490–501. [Google Scholar]
- Hedh J, Wallander H, Erland S. 2008. Ectomycorrhizal mycelial species composition in apatite amended and non‐amended mesh bags buried in a phosphorus‐poor spruce forest. Mycological Research 112: 681–688. [DOI] [PubMed] [Google Scholar]
- Hobbie EA, Agerer R. 2010. Nitrogen isotopes in ectomycorrhizal sporocarps correspond to belowground exploration types. Plant and Soil 327: 71–83. [Google Scholar]
- Ihrmark K, Bödeker ITM, Cruz‐Martinez K, Friberg H, Kubartova A, Schenck J, Strid Y, Stenlid J, Brandström‐Durling M, Clemmensen KE et al. 2012. New primers to amplify the fungal ITS2 region – evaluation by 454‐sequencing of artificial and natural communities. FEMS Microbiology Ecology 82: 666–677. [DOI] [PubMed] [Google Scholar]
- Jörgensen K, Clemmensen K, Wallander H, Lindahl BD. 2022. Data for: Do ectomycorrhizal exploration types reflect mycelial foraging? Dryad. doi: 10.5061/dryad.08kprr55q. [DOI] [PMC free article] [PubMed]
- Kjøller R. 2006. Disproportionate abundance between ectomycorrhizal root tips and their associated mycelia. FEMS Microbiology Ecology 58: 214–224. [DOI] [PubMed] [Google Scholar]
- Kluting K, Clemmensen K, Jonaitis S, Vasaitis R, Holmström S, Finlay R, Rosling A. 2019. Distribution patterns of fungal taxa and inferred functional traits reflect the non‐uniform vertical stratification of soil microhabitats in a coastal pine forest. FEMS Microbiology Ecology 95: fiz149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, Bates ST, Bruns TD, Bengtsson‐Palme J, Callaghan TM et al. 2013. Towards a unified paradigm for sequence‐based identification of fungi. Molecular Ecology 22: 5271–5277. [DOI] [PubMed] [Google Scholar]
- Kranabetter JM, Durall DM, MacKenzie WH. 2009. Diversity and species distribution of ectomycorrhizal fungi along productivity gradients of a southern boreal forest. Mycorrhiza 19: 99–111. [DOI] [PubMed] [Google Scholar]
- Krebs CJ. 1999. Ecological methodology, 2nd edn. Menlo Park, CA, USA: Benjamin/Cummings. [Google Scholar]
- Kuznetsova A, Brockhoff PB, Christensen RHB, Jensen SP. 2017. lmerTest: tests in linear mixed effects models. Journal of Statistical Software 82: 1–26. [Google Scholar]
- Kyaschenko J, Clemmensen KE, Hagenbo A, Karltun E, Lindahl BD. 2017. Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. The ISME Journal 11: 863–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leake JR, Donnelly DP, Saunders EM, Boddy L, Read DJ. 2001. Rates and quantities of carbon flux to ectomycorrhizal mycelium following C‐14 pulse labeling of Pinus sylvestris seedlings: effects of litter patches and interaction with a wood‐decomposer fungus. Tree Physiology 21: 71–82. [DOI] [PubMed] [Google Scholar]
- Lenth RV, Buerkner P, Herve M, Love J, Riebl H, Singmann H. 2021. emmeans: estimated marginal means, aka least‐squares means . [WWW document] URL https://cran.r‐project.org/package=emmeans [accessed 9 April 2021].
- Lilleskov EA, Hobbie EA, Horton TR. 2011. Conservation of ectomycorrhizal fungi: exploring the linkages between functional and taxonomic responses to anthropogenic N deposition. Fungal Ecology 4: 174–183. [Google Scholar]
- Lindahl BD, Kyaschenko J, Varenius K, Clemmensen KE, Dahlberg A, Karltun E, Stendahl J. 2021. A group of ectomycorrhizal fungi restricts organic matter accumulation in boreal forests. Ecology Letters 47: 1341–1351. [DOI] [PubMed] [Google Scholar]
- van der Linde S, Suz LM, Orme CDL, Cox F, Andreae H, Asi E, Atkinson B, Benham S, Carroll C, Cools N et al. 2018. Environment and host as large‐scale controls of ectomycorrhizal fungi. Nature 558: 243–248. [DOI] [PubMed] [Google Scholar]
- Moeller HV, Peay KG, Fukami T. 2014. Ectomycorrhizal fungal traits reflect environmental conditions along a coastal California edaphic gradient. FEMS Microbiology Ecology 87: 797–806. [DOI] [PubMed] [Google Scholar]
- Nilsson RH, Larsson K‐H, Taylor AFS, Bengtsson‐Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glöckner FO, Tedersoo L et al. 2019. The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Research 47: D259–D264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Simpson GL, Blanchet FG, Kindt R, Legendre P, Minchin PR, O'Hara RB, Solymos P, Stevens MHH, Szoecs E et al. 2022. vegan: community ecology package . [WWW document] URL https://github.com/vegandevs/vegan [accessed 7 June 2022].
- Parrent JL, Vilgalys R. 2007. Biomass and compositional responses of ectomycorrhizal fungal hyphae to elevated CO2 and nitrogen fertilization. New Phytologist 176: 164–174. [DOI] [PubMed] [Google Scholar]
- Peay KG, Kennedy PG, Bruns TD. 2011. Rethinking ectomycorrhizal succession: are root density and hyphal exploration types drivers of spatial and temporal zonation? Fungal Ecology 4: 233–240. [Google Scholar]
- Pellitier PT, Zak DR. 2021. Ectomycorrhizal fungal decay traits along a soil nitrogen gradient. New Phytologist 232: 2152–2164. [DOI] [PubMed] [Google Scholar]
- R Core Team . 2020. R: a language and environment for statistical computing , v.4.0.3. [WWW document] URL https://www.R‐project.org/ [accessed 27 November 2020].
- Rosling A, Lindahl BD, Finlay RD. 2004. Carbon allocation to ectomycorrhizal roots and mycelium colonising different mineral substrates. New Phytologist 162: 795–802. [DOI] [PubMed] [Google Scholar]
- Smith SE, Read DJ. 2008. Mycorrhizal symbiosis, 3rd edn. London, UK: Academic Press. [Google Scholar]
- Solly EF, Lindahl BD, Dawes MA, Peter M, Souza RC, Rixen C, Hagedorn F. 2017. Experimental soil warming shifts the fungal community composition at the alpine treeline. New Phytologist 215: 766–778. [DOI] [PubMed] [Google Scholar]
- Sterkenburg E, Bahr A, Brandström Durling M, Clemmensen KE, Lindahl BD. 2015. Changes in fungal communities along a boreal forest soil fertility gradient. New Phytologist 207: 1145–1158. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Naadel T, Bahram M, Pritsch K, Buegger F, Leal M, Kõljalg U, Põldmaa K. 2012. Enzymatic activities and stable isotope patterns of ectomycorrhizal fungi in relation to phylogeny and exploration types in an afrotropical rain forest. New Phytologist 195: 832–843. [DOI] [PubMed] [Google Scholar]
- Tedersoo L, Smith ME. 2013. Lineages of ectomycorrhizal fungi revisited: foraging strategies and novel lineages revealed by sequences from belowground. Fungal Biology Reviews 27: 83–99. [Google Scholar]
- Treseder KK, Allen MF, Ruess RW, Pregitzer KS, Hendrick RL. 2005. Lifespans of fungal rhizomorphs under nitrogen fertilization in a pinyon‐juniper woodland. Plant and Soil 270: 249–255. [Google Scholar]
- Wallander H, Nilsson LO, Hagerberg D, Baath E. 2001. Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytologist 151: 753–760. [DOI] [PubMed] [Google Scholar]
- Wallander H, Thelin G. 2008. The stimulating effect of apatite on ectomycorrhizal growth diminishes after PK fertilization. Soil Biology & Biochemistry 40: 2517–2522. [Google Scholar]
- Weigt RB, Raidl S, Verma R, Agerer R. 2011. Exploration type‐specific standard values of extramatrical mycelium – a step towards quantifying ectomycorrhizal space occupation and biomass in natural soil. Mycological Progress 11: 287–297. [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor J. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: PCR protocols. San Diego, CA, USA: Academic Press, 315–322. [Google Scholar]
- Wickham H, Chang W, Henry L, Pedersen TL, Takahashi K, Wilke C, Woo K, Yutani H, Dunnington D, RStudio . 2020. ggplot2: create elegant data visualisations using the grammar of graphics . [WWW document] URL https://github.com/tidyverse/ggplot2 [accessed 9 November 2020].
- Wright IJ, Reich PB, Westoby M, Ackerly DD, Baruch Z, Bongers F, Cavender‐Bares J, Chapin T, Cornelissen JHC, Diemer M et al. 2004. The worldwide leaf economics spectrum. Nature 428: 821–827. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Illustration of cafeteria setup.
Table S1 Model output from lme‐models of the log‐ratio of ectomycorrhizal exploration types in bags relative to roots, and in soil bags relative to sand bags.
Table S2 Relative abundance of ectomycorrhizal fungi on roots, and sand‐ or soil‐filled ingrowth bags.
Table S3 Model output from lme‐models of log‐ratio of ectomycorrhizal genera in bags relative to roots.
Table S4 Model output from lme‐models of log‐ratio of ectomycorrhizal genera in soil‐filled relative to sand‐filled ingrowth meshbags.
Table S5 Model output from PERMANOVA testing the effect of different soil substrates on ectomycorrhizal fungal community composition in ingrowth mesh bags.
Table S6 Relative abundance of ectomycorrhizal fungi in ingrowth bags filled with different organic substrates.
Table S7 Model output from models testing the effect of substrates in ingrowth meshbags on ectomycorrhizal fungal genera.
Table S8 Output from post hoc Tukey HSD test on ectomycorrhizal genera that displayed preference towards any soil substrate.
Table S9 Model output from PERMANOVA testing the effect of different sand substrates on ectomycorrhizal fungal community composition in ingrowth mesh bags.
Table S10 Relative abundance of ectomycorrhizal fungi in ingrowth bags filled with sand.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
Data and code needed to reproduce analyses are available on Dryad Digital Repository (doi: 10.5061/dryad.08kprr55q; Jörgensen et al., 2022). Sequence data are published in NCBI‐SRA under project PRJNA796466.


