Abstract
Natural killer (NK) cells with tissue‐residency features (trNK cells) are a new subpopulation of NK cells, which plays an important role in tissue homeostasis. However, the characteristics of trNK cells in the tumor microenvironment (TME) of human cancers remain unclear. Using multicolor flow cytometry, we investigated the quantity, phenotype, and function of trNK cells in biospecimens freshly resected from 60 non‐small cell lung cancer (NSCLC) patients. We successfully identified a new CD69+CXCR6+ trNK subset with an immunomodulatory‐like and exhausted phenotype, specifically accumulated in the TME of NSCLC. In vitro experiments showed that CD69+CXCR6+ trNK cells more readily secreted IFN‐γ and TNF‐α spontaneously. Furthermore, the production of IFN‐γ and TNF‐α by tumor‐infiltrating CD69+CXCR6+ trNK cells was not induced by their reactivation in vitro, which is analogous to T‐cell exhaustion. Finally, we demonstrated that the dysfunction of CD69+CXCR6+ trNK cells could be partly ameliorated by PD‐1 and CTLA‐4 blockade. In summary, we identified a new dysfunctional CD69+CXCR6+ trNK cell subset that specifically accumulates in the TME of NSCLC. Our findings suggest that CD69+CXCR6+ trNK cells are a promising target for immune checkpoint inhibitors in the treatment of NSCLC.
Keywords: CXCR6, exhaustion, immunotherapy, lung cancer, tissue‐resident NK cells
This study identified a CD69+CXCR6+ tissue‐resident NK (trNK) subset specifically accumulated in the tumor microenvironment of human non‐small cell lung cancer patients. CD69+CXCR6+ trNK cells showed an immunomodulatory‐like and exhaustion phenotype, and secreted IFN‐γ and TNF‐α rapidly.
Introduction
Lung cancer remains the leading cause of tumor‐related death and the second contributor to new cancer diagnoses worldwide [1], and various approaches have been developed to improve the prognosis of lung cancer patients. Immunotherapy based on immune checkpoint inhibitors (ICIs) is a promising treatment method that aims to block the immune inhibitory receptors on certain immune cells, unleashing the antitumor capacity of those cells [2, 3]. Unfortunately, although these immunotherapies have shown tremendous advances in their therapeutic effects, the populations of lung cancer patients that benefit from these therapies or suffer from treatment cytotoxicity are heterogeneous [4]. This dilemma is attributed to the diverse response of various immune cells to ICIs, but the details of how ICIs influence specific immune cell subgroups remain unclear.
Like well‐known cytotoxic CD8+ T cells, natural killer (NK) cells have attracted considerable interest in the field of cancer immunotherapy [5, 6]. Previous studies have shown that NK cells constitute a crucial member of the innate lymphoid cell (ILC) family, which exerts tremendous antitumor effects by both direct cytolytic functions and the secretion of proinflammatory chemokines [5, 7]. Increased levels of NK cells infiltrating the tumor microenvironment (TME) and gene expression that specifically supports NK cell functions have been associated with better prognoses in patients with hematological and solid neoplasms [8]. Therefore, enhancing the dysfunction of NK cells in solid tumors with ICIs improves the prognoses of these cancer patients. Moreover, several unique NK cell subgroups have been shown to be necessary in mediating the full therapeutic efficacy of ICI blockade [9, 10], implying a relationship between immunotherapy and NK cells in lung cancer.
The human lung offers one of the largest exchange surfaces with the exterior environment in the body, and the complexity of its local immune environment has been demonstrated in previous work [11, 12]. Accumulating evidence supports the important roles of tissue‐resident immune cells against diverse infections and malignancies [13, 14]. These subsets of nonrecirculating immune cells provide localized immunity in terms of both the innate and adaptive systems, but previous studies have more frequently investigated tissue‐resident memory T (Trm) cells than other tissue‐resident immune cells, including tissue‐resident NK (trNK) cells [11]. Furthermore, until now, most knowledge of human NK cells has been drawn from ex vivo experiments on conventional NK (cNK) cells derived from the peripheral blood [15], which differed from trNK cell from peripheral organs in function and development [16]. Nonetheless, recent publications have identified increased numbers of trNK cell subsets in different human tissues [11, 12] and verified several hallmark tissue‐resident surface molecules on NK cells, including CD69, CD49a (α1 integrin), CD103 (αE integrin), and members of the C‐X‐C motif chemokine receptor (CXCR) family [17]. Several reports have identified novel Trm cell subsets with potential specialized effector functions according to the expression of certain tissue‐resident markers on the cells [18, 19], and a corresponding investigation of trNK cells in lung cancer patients is urgently required.
Here, we identified a novel subset of CD69+CXCR6+ trNK cells that accumulate specifically in the TME of human non‐small cell lung cancer (NSCLC), the commonest subtype of lung cancer, accounting for 85% of total lung cancer cases [20]. We also demonstrate that CD69+CXCR6+ trNK cells express tissue‐resident, immunomodulatory‐like, and exhausted phenotypes. Interestingly, CD69+CXCR6+ trNK cells derived from both tumor and normal lung tissues secrete greater amounts of cytokines than unactivated cNK cells. However, this greater cytokine secretion by tumor‐infiltrating CD69+CXCR6+ trNK cells was not observed when they were reactivated in vitro, indicating the dysfunction of trNK cells in NSCLC. A further analysis showed that the in vitro blockade of PD‐1 and CTLA‐4 specifically promoted the secretion of cytokines by tumor‐infiltrating CD69+CXCR6+ trNK cells.
Overall, our study demonstrates the unique biological characteristic of tumor‐infiltrating CD69+CXCR6+ trNK cells, and these findings should improve the therapeutic efficacy of ICIs in NSCLC.
Results
CD69+CXCR6+ trNK cells accumulate in the microenvironment of human NSCLC
Among the various immune cells residing in lung tissues, increasing numbers of NK cell subsets have recently been precisely identified and studied [21]. In the present study, we collected tumor and paired normal lung tissue samples resected from 52 patients with NSCLC and normal lung tissue samples resected from eight patients, in the hope of detecting the infiltration and identifying the phenotypes of discrete NK cell subtypes in human NSCLC. The preoperative clinicopathological parameters of the 52 tumor tissue donors are presented in Table S1. NK cells were defined as CD3−CD14−CD19−CD45+CD56+ lymphocytes, and the gating strategy is shown in Fig. S1A.
The occurrence and cell counts (per 100,000 singlets) of NK cells were significantly lower in the tumor tissues than in the paired adjacent normal tissues, which implies the incompetence of the innate antitumor immune function during the early progression of NSCLC (Fig. 1A and B). NK cells expressing the traditional tissue residency marker CD69 and the regulator of lymphocyte homing patterns CXCR6 were analyzed with flow cytometry, which identified three main subpopulations: CD69+CXCR6+ double‐positive (DP) NK cells, CD69+CXCR6− single‐positive (SP) NK cells, and CD69−CXCR6− double‐negative (DN) NK cells (Fig. S1B). To investigate the reliability of CXCR6 staining, we first uploaded an unstained cell suspension directly to the flow cytometer, as the negative control (Fig. S1C–D). Because Trm cells were shown to strongly express CXCR6 [22], they were compared with the CD69+CXCR6+ trNK cells as a positive control (Fig. S1E–F). Finally, we uploaded samples stained with fluorescent antibodies directed against different CXCR6 clonotypes, and presented fluorescence minus one (FMO) control as well (Fig. S1G–H).
Figure 1.
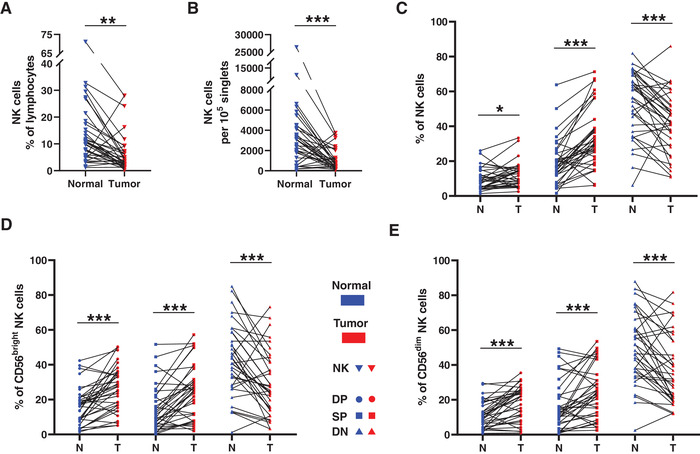
The distribution of the trNK cells in NSCLC. A−B. Line chart summarizes the proportion of NK cells in lymphocytes (A) and the counting of NK cells per 100,000 singlets (B) between the normal and tumor tissues. C−E. The comparisons of DP (Double positive, CD69+CXCR6+), SP (Single positive, CD69+CXCR6−), and DN (Double negative, CD69−CXCR6−) NK cell distribution in total NK cells (C), CD56bright NK cells (D), and CD56dim NK cells (E) between the normal (N) and tumor (T) tissues. Data are shown as the line chart. Data were obtained from the normal and paired tumor tissues resected from 35 of 52 operative NSCLC patients, pooled from three independent experiments. Symbols represent one individual sample. *p < 0.05, **p < 0.01, ***p < 0.001 (two‐tailed paired t‐test, and Wilcoxon matched‐pairs signed rank test for non‐parametric pairing t‐tests).
Consistent with the distribution of NK cells in the human lung, the levels of DN cNK cells were reduced in the tumor tissues, whereas those of CD69+CXCR6+ trNK cells and SP trNK cells were significantly elevated in the NK cells (Fig. 1C), confirming the specificity and predominance of trNK cells during in the antitumor immune response in NSCLC. The NK cells were further delineated into two major subsets: CD56bright NK cells and CD56dim NK cells. The DN expression of CD69 and CXCR6 or the SP expression of CD69 was upregulated on both CD56bright and CD56dim NK cells in the pulmonary tumor tissues (Fig. 1D and E), suggesting the accumulation of trNK cells in the intratumoral CD56bright and CD56dim cells and in the total NK cells of human NSCLC tissues.
The phenotype of tumor‐infiltrating CD69+CXCR6+ trNK cells in NSCLC
We then analyzed the phenotypes of the NK cells in NSCLC. Tissue residency was examined first (Fig. 2A), and the CD69+CXCR6+ trNK cells and SP trNK cells included more CD103+ cells than did the DN cNK cells. However, there was no difference in the frequency of CD103+ cells in the two trNK cell subsets (CD69+CXCR6+ and SP) (Fig. 2B and C). Furthermore, the CD69+CXCR6+ trNK cells infiltrating the tumors expressed more CD103 than did those in normal tissue, emphasizing the significance of trNK cells in regulating the progression of malignancy (Fig. 2D).
Figure 2.
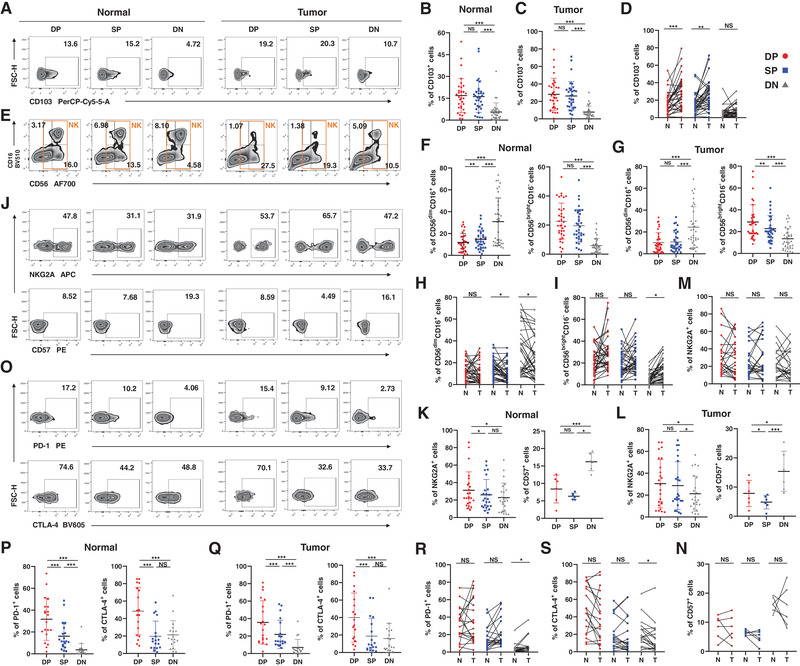
The phenotype of the trNK cells in NSCLC. (A−D) The percentages of CD103+ cells in DP, SP, and DN NK cells from the normal (B) and tumor tissues (C). The comparisons of the percentages of CD103+ cells in DP, SP, and DN NK cells between the normal (N) and tumor (T) tissues (D). Data are shown as the representative flow analysis (A), mean ± SEM (B−C), and line chart (D). (E−I) The percentages of CD56dimCD16+ and CD56brightCD16− cells in DP, SP, and DN NK cells from the normal (F) and tumor tissues (G). The comparisons of the percentages of CD56dimCD16+ (H) and CD56brightCD16− cells (I) in DP, SP, and DN NK cells between the normal (N) and tumor (T) tissues. Data are shown as the representative flow analysis (E), mean ± SEM (F−G), and line chart (H−I). (J−N) The percentages of NKG2A+ and CD57+ cells in DP, SP, and DN NK cells from the normal (K) and tumor tissues (L). The comparisons of the percentages of NKG2A+ (M) and CD57+ cells (N) in DP, SP, and DN NK cells between the normal (N) and tumor (T) tissues. Data are shown as the representative flow analysis (J), mean ± SEM (K−L), and line chart (M−N). (O−S) The percentages of PD‐1+ and CTLA‐4+ cells in DP, SP, and DN NK cells from the normal (P) and tumor tissues (Q). The comparisons of the percentages of PD‐1+ (R) and CTLA‐4+ cells (S) in DP, SP, and DN NK cells between the normal (N) and tumor (T) tissues. Data are shown as the representative flow analysis (O), mean ± SEM (P−Q), and line chart (R−S). Data were obtained from the normal and paired tumor tissues resected from 35 of 52 operative NSCLC patients, pooled from three independent experiments. 33 of 52 samples were stained by CD103 (A−D); 35 of 52 samples were stained by CD56 and CD16 (E−I); 24 of 52 samples were stained by NKG2A (J−N); six of 52 samples were stained by CD57 (J−N); 20 of 52 samples were stained by PD‐1 (J−N); 18 of 52 samples were stained by CTLA‐4 (O−S). Symbols represent one individual sample. *p < 0.05, **p < 0.01, ***p < 0.001 (two‐tailed t‐test and Wilcoxon signed rank test in B−C, F−G, K−L, P−Q; two‐tailed paired t‐test and Wilcoxon matched‐pairs signed rank test in D, H−I, M−N, R−S).
It is widely accepted that human NK cells from the peripheral blood can be divided into two major subsets: CD56brightCD16− and CD56dimCD16+ [23]. Our results confirm that CD69+CXCR6+ trNK cells from both normal and tumor tissues showed a lower frequency of the CD56dimCD16+ phenotype and an increased frequency of the CD56brightCD16− phenotype than did DN cNK cells. The CD56dimCD16+ phenotype remained predominant among the DN cNK cells (Fig. 2E–G). CD56/CD16 expression on the CD69+CXCR6+ trNK cells was similar in both the cells from normal tissues and those from tumor tissues (Fig. 2H and I).
The CD56bright subset of NK cells has been shown to be the precursor of the CD56dim subset in the linear maturation model of human NK cells [24]. Therefore, a larger proportion of the CD56brightCD16− subset implies a higher level of cell immaturity. The expression of NKG2A and CD57, both markers of NK cell maturation or differentiation [25], was then examined in the NK cell subgroups (Fig. 2J). The CD69+CXCR6+ trNK cells included more NKG2A+ cells and fewer CD57+ cells than the DN cNK cells, and the CD69+CXCR6+ trNK cells also included more NKG2A+ cells than the SP trNK cells in the normal tissues (Fig. 2K and L). No difference in NKG2A or CD57 expression on CD69+CXCR6+ trNK cells was observed between the normal and tumor tissues (Fig. 2M and N).
Because Trm cells have been shown to express increased immune checkpoint regulators [26], we analyzed the expression levels of PD‐1, CTLA‐4, TIGIT, LAG‐3, and TIM‐3 on human CD69+CXCR6+ trNK cells (Fig. 2O–S, Fig. S2). The validity of fluorescent staining was confirmed with FMO and negative controls (Fig. S1I‐J). According to our results, CD69+CXCR6+ trNK cells from both normal and tumor tissues showed greater expression of PD‐1, CTLA‐4, and TIM‐3 than the other NK cells, but only CD69+CXCR6+ trNK cells from normal tissues expressed more TIGIT and LAG‐3 than the other NK cells (Fig. 2P and Q, Fig. S2B–C). More TIM‐3+ cells were detected among the CD69+CXCR6+ trNK cells from tumor tissues than among their counterparts from normal tissues (Fig. S2D).
CD69+CXCR6+ trNK cells secrete elevated antitumor cytokines
NK cells exert antitumor effects through the secretion of cytokines and direct cytotoxicity [27]. Therefore, we analyzed the functions of CD69+CXCR6+ trNK cells in NSCLC from these two perspectives.
First, we cultured cell suspensions obtained from paired normal and tumor tissues, and analyzed the cytokine secretion profiles of the NK cells with flow cytometry (Fig. 3A). In both the normal and tumor tissues, the intracellular expression of IFN‐γ was significantly higher in CD69+CXCR6+ trNK cells than in SP trNK cells or DN cNK cells (Fig. 3B). CD69+CXCR6+ trNK cells from normal lung tissues also expressed more intracellular TNF‐α than SP trNK cells or DN cNK cells, but this difference between CD69+CXCR6+ trNK cells and DN cNK cells was not observed in tumor tissues (Fig. 3C). We then cultured the cell suspensions in the presence of brefeldin A (BFA) and monensin, to enhance the staining of intracellular cytokines [28]. The results were similar, showing that CD69+CXCR6+ trNK cells still produced more IFN‐γ and TNF‐α than SP trNK cells or DN cNK cells (Fig. S3A–C). No difference in IFN‐γ or TNF‐α expression on CD69+CXCR6+ trNK cells was observed between the normal and tumor tissues (Fig. S3D–E).
Figure 3.
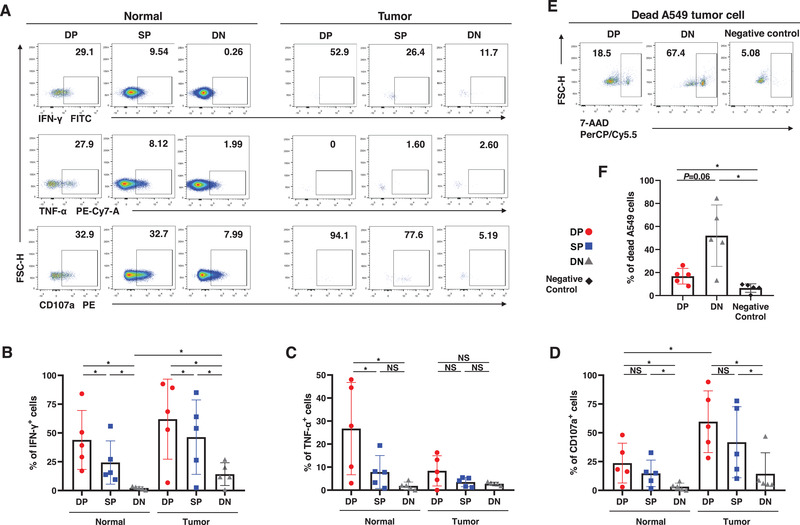
Functions of the trNK cells in NSCLC. (A−D) The percentages of IFN‐γ+ (B), TNF‐α+ (C), and CD107a+ cells (D) in DP, SP, and DN NK cells from the normal and tumor tissues. Data are shown as the representative flow analysis (A) and mean ± SEM (B−D). (E−F) The percentages of the dead A549 lung cancer cells after co‐culture with sorted DP and DN NK cells. Negative control refers to as the A549 cells cultured alone. Data are shown as the representative flow analysis (E) and mean ± SEM (F). Data were obtained from the normal and paired tumor tissues resected from ten of 52 operative NSCLC patients, pooled from three independent experiments. Symbols represent one individual sample. *p < 0.05, **p < 0.01, ***p < 0.001 (two‐tailed t‐test and Wilcoxon signed rank test in B−D and F).
To further clarify the cytokine secretion function of trNK cells, we individually sorted CD69+CXCR6+ trNK cells and DN cNK cells from normal tissues. The gating strategy and post‐sorting verification are shown in Fig. S1K–L. Our results proved that the concentration of IFN‐γ was significantly higher in the culture supernatant of the CD69+CXCR6+ trNK cells than in that of the DN cNK cells (Fig. S3F–G).
The cytotoxicity characteristics of NK cells in NSCLC were also evaluated. We first showed with flow cytometry that the proportion of CD107a+ cells was higher in CD69+CXCR6+ trNK cells than in cNK cells from both normal and tumor tissues (Fig. 3A and D). CD69+CXCR6+ trNK cells and DN cNK cells were then sorted from the cell suspensions, and cocultured with A549 lung cancer cells for 12 h. The gating strategy of A549 cells is shown in Fig. S1M. Unlike the overexpression of CD107a, CD69+CXCR6+ trNK cells induced lower proportion of dead A549 cells than did DN cNK cells after the coculture (Fig. 3E and F). Intracellular staining of granzyme B also showed that CD69+CXCR6+ trNK cells prepared from normal tissues produced less granzyme B than DN cNK cells, whereas the granzyme B secreted did not vary among the three subgroups in tumor‐derived cell suspensions of NK cells (Fig. S3H–J). The intracellular expression of perforin was also similar among the NK cell subgroups, regardless of whether the NK cells were obtained from normal or tumor tissues (Fig. S3H–J). CD69+CXCR6+ trNK cells from normal tissues expressed more granzyme‐B than did those from tumor tissues, but no difference in perforin expression on CD69+CXCR6+ trNK cells was observed between the normal and tumor tissues (Fig. S3K–L).
Taken together, these results indicate that the cytokine production of CD69+CXCR6+ trNK cells is more significant in NSCLC than their direct cytotoxicity. Since the secretion of IFN‐γ and TNF by NK cells has been shown to induce direct antitumor cytotoxicity and mediate tumor cell death [5, 27], CD69+CXCR6+ trNK cells are considered vital in controlling tumor progression.
Cytokine secretion by tumor‐infiltrating CD69+CXCR6+ trNK cells is restricted
The function of NK cells is influenced by various stimuli. To investigate whether this also applies to CD69+CXCR6+ trNK cells, we stimulated cell suspensions separately with lung cancer cells, IL‐15, and phorbol myristate acetate (PMA) and ionomycin. Then we analyzed the cytokine production by CD69+CXCR6+ trNK cells relative to that by other NK cell subsets.
First, CD69+CXCR6+ trNK cells and CD69−CXCR6− cNK cells were sorted from a cell suspension prepared from normal pulmonary tissue (Fig. 4A). The sorted CD69+CXCR6+ trNK cells included more IFN‐γ+ and TNF‐α+ cells than the cNK cells (Fig. 4B and C). During coculture with A549 cells, the CD69+CXCR6+ trNK cells also produced more IFN‐γ and TNF‐α than cNK cells cocultured with A549 cells (Fig. 4B and C). However, they produced less IFN‐γ than their unstimulated counterparts, which directly indicates that their capacity to secrete immunomodulatory cytokines is restricted by tumor cells (Fig. 4B).
Figure 4.
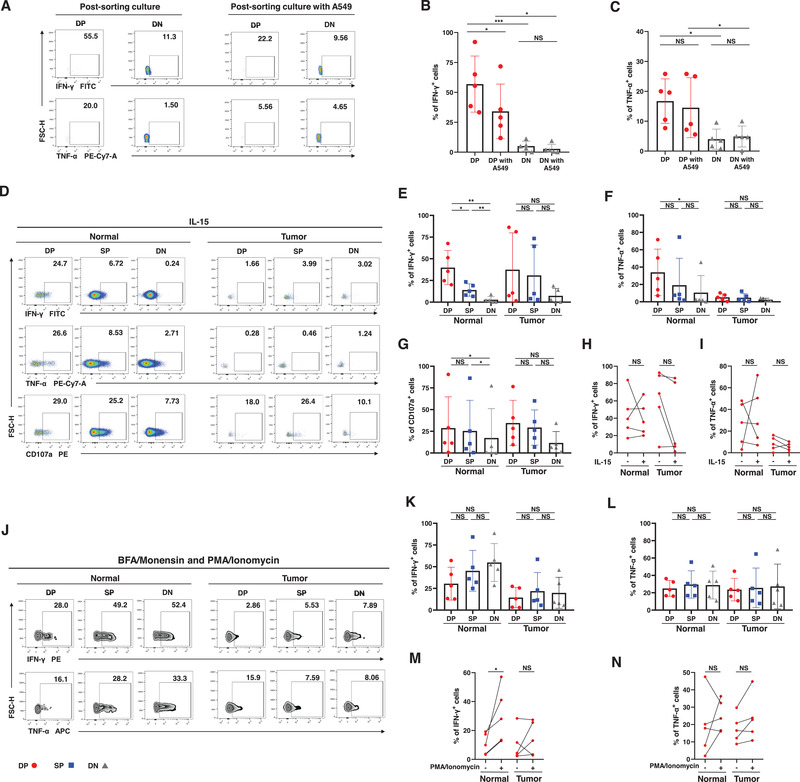
Functions of the stimulated trNK cells in NSCLC. (A−C) The percentages of IFN‐γ+ (B) and TNF‐α+ cells (C) in sorted DP and DN NK cells from the normal tissues during culture alone or coculture with A549 lung cancer cells. Data are shown as the representative flow analysis (A) and mean ± SEM (B−C). (D−G) The percentages of IFN‐γ+ (E), TNF‐α+ (F), and CD107a+ cells (G) in DP, SP, and DN NK cells from the normal and tumor tissues stimulated with IL‐15. Data are shown as the representative flow analysis (D) and mean ± SEM (E−G). (H−I) The comparisons of the percentage of IFN‐γ+ (H) and TNF‐α+ cells (I) in DP NK cells from the normal and tumor tissues between the unstimulated cells and the cells stimulated with IL‐15. Data are shown as line chart (H−I). (J−L) The percentages of IFN‐γ+ (K) and TNF‐α+ cells (L) in DP, SP, and DN NK cells from the normal and tumor tissues stimulated with PMA and Ionomycin. Data are shown as the representative flow analysis (J) and mean ± SEM (K−L). (M−N) The comparisons of the percentage of IFN‐γ+ (M) and TNF‐α+ cells (N) in DP NK cells from the normal and tumor tissues between the unstimulated cells and the cells stimulated with PMA and Ionomycin. Data are shown as line chart (M−N). Data were obtained from the normal tissues resected from five operative NSCLC patients (A−C), pooled from three independent experiments. Data were obtained from the normal and paired tumor tissues resected from 10 of 52 operative NSCLC patients (D−N), pooled from three independent experiments. Symbols represent one individual sample. *p < 0.05, **p < 0.01, ***p < 0.001 (two‐tailed t‐test and Wilcoxon signed rank test in [B,C], [E−G], and [K,L], two‐tailed paired t‐test and Wilcoxon matched‐pairs signed rank test in [H,I] and [M,N]).
NK cells were then stimulated with IL‐15, added to cell suspensions. CD69+CXCR6+ trNK cells obtained from normal pulmonary tissues included more IFN‐γ+ cells, TNF‐α+ cells, and CD107a+ cells than did DN cNK cells (Fig. 4D–G). In contrast, this variation was not observed between intratumoral CD69+CXCR6+ trNK cells and DN cNK cells (Fig. 4D–G). Notably, the function of NK cells was enhanced in the presence of IL‐15 [29], whereas the addition of IL‐15 did not increase the frequency of IFN‐γ+ cells or TNF‐α+ cells in the CD69+CXCR6+ trNK cells, indicating the dysfunction of trNK cells in human NSCLC (Fig. 4H–I).
We also stimulated pulmonary NK cells with PMA and Ionomycin, a nonspecific NK cell activator that increases the production of IFN‐γ and TNF‐α by NK cells [30]. However, activated CD69+CXCR6+ trNK cells failed to show greater IFN‐γ or TNF‐α production than DN cNK cells from either normal or tumor tissues (Fig. 4J–L), which was inconsistent with their inactivated counterparts. Direct comparisons further demonstrated that PMA and Ionomycin stimulation only effectively improved IFN‐γ production by CD69+CXCR6+ trNK cells from normal tissues (Fig. 4M). The addition of PMA and Ionomycin did not upregulate the production of TNF‐α or IFN‐γ by CD69+CXCR6+ trNK cells that had infiltrated tumor tissue (Fig. 4M and N).
In summary, using A549 lung cancer cells, IL‐15, and PMA and Ionomycin to stimulate cells, we demonstrated the dysfunction of CD69+CXCR6+ trNK cells and that CD69+CXCR6+ trNK cells infiltrating tumor tissues were significantly more restrained.
PD‐1 and CTLA‐4 inhibit cytokine secretion by CD69+CXCR6+ trNK cells
The inhibition of NK cell activities by immune checkpoint molecules has recently been reported [5, 31]. In this study, we confirmed the exhaustion phenotype of CD69+CXCR6+ trNK cells. Consequently, we hypothesized that the elevated expression of immune checkpoint markers, such as PD‐1 and CTLA‐4, on CD69+CXCR6+ trNK cells contributes to their dysfunction during stimulation, and thereby the in vitro blockade of PD‐1 and CTLA‐4 eliminates this dysfunction.
To design blockade strategies, we first evaluated the coexpression of inhibitory molecules. In 18 normal and tumor tissues from NSCLC patients, the proportion of PD‐1+ CD69+CXCR6+ trNK cells was positively associated with the proportion of CTLA‐4+ CD69+CXCR6+ trNK cells in normal tissues (p < 0.001, Spearman's r = 0.792; Fig. 5A), tumor tissues (p < 0.001, Spearman's r = 0.589; Fig. 5B), and both combined (p < 0.001, Spearman's r = 0.608: Fig. 5C). Recent clinical evidence has also shown that the combination of two or more ICIs enhances the prognosis of NSCLC [32], so used PD‐1 blockade alone, CTLA‐4 blockade alone, and PD‐1 and CTLA‐4 blockade combined to treat NK cells stimulated with PMA and ionomycin (Fig. 5D and E).
Figure 5.
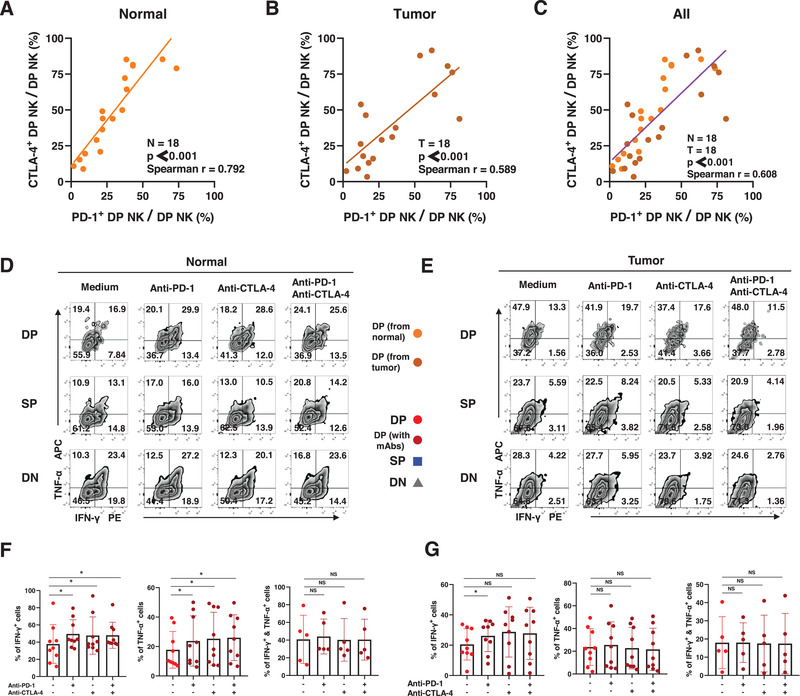
Cytokine secretion by trNK cells after the blockade(s) of PD‐1 and CTLA‐4 in NSCLC. (A−C) Spearman correlation analysis and linear regression analysis assess the correlation between the proportion of PD‐1+ DP NK cells and CTLA‐4+ DP NK cells from the normal tissues (A), the tumor tissues (B), and taken together (C). (D−E) The representative flow analysis of the intracellular expression of IFN‐γ and TNF‐α in DP, SP, and DN NK cells from the normal (D) and tumor tissues (E) when stimulated with BFA and monensin and PMA and Ionomycin. (F−G) The comparisons of intracellular expression of IFN‐γ and TNF‐α in DP NK cells from the normal (F) and tumor tissues (G) between spontaneous cells and cells after the blockade(s) of PD‐1 and CTLA‐4. Data are shown as mean ± SEM. Data were obtained from the normal and paired tumor tissues resected from 18 of 52 operative NSCLC patients (A−C), pooled from three independent experiments. Data were obtained from the normal and paired tumor tissues resected from nine of 52 operative NSCLC patients (D−G), pooled from three independent experiments. Symbols represent one individual sample. *p < 0.05, **p < 0.01, ***p < 0.001 (Spearman's correlation and linear regression tests in [A−C]; two‐tailed t‐test and Wilcoxon signed rank test in [F−G]).
Our results showed that the capacity of CD69+CXCR6+ trNK cells from normal tissue to secrete IFN‐γ and TNF‐α was enhanced by treatment with anti‐PD‐1 antibody or anti‐CTLA‐4 antibody alone or combined, which confirmed the hypothesis that PD‐1 and CTLA‐4 inhibit the secretion of cytokines by CD69+CXCR6+ trNK cells (Fig. 5F). The secretion of IFN‐γ by tumor‐infiltrating CD69+CXCR6+ trNK cells was enhanced by the anti‐PD‐1 antibody treatment (Fig. 5G). Neither the production of TNF‐α nor the coproduction of IFN‐γ and TNF‐α by tumors was altered by the blockade of PD‐1 or CTLA‐4 in CD69+CXCR6+ trNK cells (Fig. 5F and G). We also compared the secretion of cytokines by CD69+CXCR6+ trNK cells and DN cNK cells during stimulation. When treated with ICIs, the activated CD69+CXCR6+ trNK cells secreted more IFN‐γ and TNF‐α than SP trNK cells or DN cNK cells in most cases (Fig. S4), consistent with their unactivated counterparts.
Discussion
As a promising therapeutic treatment for different malignancies, immunotherapies aim to unleash the full efficacy of antitumor agents mediated by the human immune system, and consequently to limit tumor development [33]. For many decades, it has been widely believed that most lymphocytes involved in the immunosurveillance of tumors recirculate throughout the body constantly during their lifespans [22, 34]. However, with rapid advances in immunology, mounting evidence suggests that different tissue‐resident lymphocyte subsets occur in the peripheral nonlymphoid organs, contrary to the traditional opinion [34]. Further research into the development and functions of tissue‐resident lymphocytes has demonstrated their tremendous capacity to control tumor initiation, growth, and metastasis [13, 21, 22], Therefore, they are ideal targets for immunotherapy. However, compared with Trm cells, which mainly mediate the local adaptive immune response, our knowledge of trNK cells, which mediate the local innate immune response, remains limited, which limits the clinical application of immunotherapies.
In this study, we characterized the unique CD69+CXCR6+ trNK cell subset in human lung cancer, and showed the enrichment of these trNK cells in solid tumor tissues from patients with NSCLC compared with their levels in normal lung tissue. We also demonstrated the predominant role of CD69+CXCR6+ trNK cells in regulating antitumor activity via their extraordinary capacity to secrete IFN‐γ and TNF‐α. Whereas this capacity is impaired in activated CD69+CXCR6+ trNK cells, PD‐1‐ and CTLA‐4‐neutralizing antibodies significantly rescued this dysfunction. Because the role of trNK cells and their heterogeneous distribution among cell populations make them a promising target for ICIs [16], our results also provide a novel explanation of the varying efficiency of immunotherapy mediated by ICIs in human lung cancer.
We first analyzed the distribution of trNK cells in human lung cancer, and showed that the coexpression of CD69 and CXCR6 defines a novel subset of trNK cells in lung tissues resected from patients with operable NSCLC. The tumor tissues in these samples were enriched in trNK cells compared with the adjacent normal lung tissue (Fig. 1). Although CD69 is specifically expressed on trNK cells among the NK cells in the peripheral organs of humans and mice, CD49a and CD103 have also been used to identify trNK cells [17]. These have also been shown to promote the retention of immune cells under the regulation of transforming growth factor β (TGF‐β) [35, 36]. In contrast, the functional involvement of CXCR6 in the retention of lymphocytes is regulated by the interaction between CXCR6 and its ligand CXCL16 [37]. Although CD69+CXCR6+ trNK cells and SP trNK cells both show increased expression of the tissue‐resident marker CD103 compared with that on DN cNK cells (Fig. 2), our analysis showed that CD69+CXCR6+ trNK cells express several unique features, distinct from those expressed by SP trNK cells, in the NSCLC microenvironment. Therefore, the combination of CD69 and CXCR6 allows the discrimination of trNK cells and cNK cells, and implies that human solid tumors potentially mediate a specific pathway of tissue‐resident cell recruitment, in addition to the regulation of CD49a and CD103 expression by TGF‐β [36].
Previous reports have indicated that the dominant phenotypes and functions of human trNK cells vary from those of their conventional counterparts at various anatomical sites, including the lymph nodes, tonsils, and liver [38]. However, the diversity of the lung trNK cell populations has rarely been studied, especially in terms of pulmonary malignancies. According to our phenotypic analyses, CD69+CXCR6+ trNK cells and SP trNK cells showed an abundance of the CD56brightCD16− subtype and immature characteristics, similar to those of the immunomodulatory CD56bright NK cell subset, which has a capacity for the rapid secretion of cytokines including IFN‐γ and TNF‐α [29]. The distribution of the CD56brightCD16− phenotype was similar in the CD69+CXCR6+ trNK cells from normal and tumor tissues, which implies that the immunomodulatory function of trNK cells is not disrupted by the TME. Elevated proportions of PD‐1+ or CTLA‐4+ cells were also observed in CD69+CXCR6+ trNK cells from both normal and tumor tissues (Fig. 2). Other inhibitory receptors, including TIGIT, TIM‐3, and LAG‐3, were more abundantly expressed on CD69+CXCR6+ trNK cells as well, which confirms that trNK cells maintain the exhaustion phenotype in NSCLC (Fig. S2).
Because the phenotypic results were contradictory, we performed in vitro experiments to evaluate the functions of CD69+CXCR6+ trNK cells. We used flow cytometry and enzyme‐linked immunosorbent assays (ELISAs) to confirm the greater production of IFN‐γ and TNF‐α by CD69+CXCR6+ trNK cells than by cNK cells (Fig. 3, Fig. S3). The lower proportion of dead lung cancer cells after coculture with NK cells and the relatively low expression of granzyme B confirmed that the direct cytotoxicity of CD69+CXCR6+ trNK cells against tumor cells was not superior to that of DN cNK cells (Fig. 3, Fig. S3). Notably, the cytokine secretion capacity of CD69+CXCR6+ trNK cells was not affected by the TME, which is consistent with their stable immunomodulatory‐like phenotype shown in Fig. 2.
To further clarify the influence of elevated immune inhibitors on CD69+CXCR6+ trNK cells, we analyzed the functions of human NK cells under various stimuli (Fig. 4). First, the production of IFN‐γ and TNF‐α by CD69+CXCR6+ trNK cells decreased during coculture with A549 lung cancer cells. Moreover, IL‐15, a universal stimulant of multiple immune cells that has been tested in a clinical trial [39, 40], failed to increase the proportions of IFN‐γ+ and TNF‐α+ cells in the CD69+CXCR6+ trNK cells. Furthermore, when tumor‐infiltrating CD69+CXCR6+ trNK cells were stimulated with PMA and ionomycin, their intracellular expression of IFN‐γ and TNF‐α was not upregulated, which was inconsistent with other NK cells stimulated with PMA and ionomycin [30]. Thus, we demonstrated the dysfunction of CD69+CXCR6+ trNK cells by stimulating them with lung cancer cells, cytokines, and a nonspecific NK cell activator.
Therefore, we presume that the exhaustion phenotype impairs the function of activated CD69+CXCR6+ trNK cells, which consequently facilitates tumor immune escape under pathological conditions. Based on this hypothesis, we tested the effects of blocking PD‐1 and CTLA‐4, individually or together, on activated CD69+CXCR6+ trNK cells, and reevaluated their functionality after the blockade(s) (Fig. 5, Fig. S4). The activated CD69+CXCR6+ trNK cells from normal lung tissues expressed more IFN‐γ and TNF‐α after the in vitro blockade than before the blockade, whereas their counterpart cells from the tumor tissues did not. The failure to improve the function of activated CD69+CXCR6+ trNK cells derived from tumor tissues was attributable to the elevation of other inhibitory markers on them, such as TIM‐3 (Fig. S2D). The upregulation of TIM‐3 in tumor‐infiltrating NK cells was found capable of suppressing their cytokine secretion and cytotoxic activity [41], which was consistent with our current results.
These results convincingly clarify the controversy regarding the role of NK cells in therapies involving inhibitory checkpoint blockade [42]. For instance, some models maintain that PD‐1 and CTLA‐4 are expressed by tumor‐infiltrating cytotoxic T cells rather than by NK cells, and that blockade therapy with PD‐1‐ and CTLA‐4‐directed ICIs mainly affects T cells in the TME [43, 44]. Some published studies have contested this view, and have shown that melanoma patients who responded to PD‐1 or CTLA‐4 blockade therapy had increased MIP‐1β‐ and CD69‐expressing NK cells [45]. Moreover, an elevated frequency of PD‐1+ cells was associated with the highest functional activity in NK cells expressing CD69 when activated in vitro, although CD69 is commonly regarded as a signal that activates lymphocytes [46, 47]. Our results provide an innovative perspective on CD69+ NK cells in terms of their tissue residency, and demonstrate that trNK cells, especially the unique CD69+CXCR6+ trNK cell subtype, is the major contributor to the elevated proportions of PD‐1+ and CTLA‐4+ cells in the entire NK group. Therefore, trNK cells also participate in the clinical benefits of ICI‐based immunotherapies in NSCLC via their enhanced capacity to secrete cytokines IFN‐γ and TNF‐α.
In conclusion, our study has demonstrated the accumulation of CD69+CXCR6+ trNK cells in NSCLC. This cell subgroup shows an immunomodulatory‐like and exhausted phenotype, and a distinct capacity to secrete IFN‐γ and TNF‐α. In vitro experiments confirmed the dysfunction of CD69+CXCR6+ trNK cells during stimulation, and the blockade of ICIs could ameliorate this dysfunction. These results extend our understanding of the antitumor effects of trNK cells in human lung cancer.
Materials and methods
Tissue collection
The study included tissues from 60 patients diagnosed with a malignant pulmonary NSCLC tumor with an identified histological type, who underwent surgical resection at the Second Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou, China. 52 of the patients donated tumor and paired normal lung tissues, and eight of them donated normal lung tissue only. None of the patients had received chemotherapy or radiotherapy before surgery. Tumorous (homogeneous cellularity, without necrotic foci) and autologous normal lung tissues were obtained during surgery. Normal adjacent tissue was obtained from a macroscopically normal part of the excised pulmonary tissue at least 5 cm from the tumor tissue. The removed tissue was immersed in RPMI‐1640 medium (Sigma, RNBK7760) and immediately transported to the laboratory.
Human NSCLC sample preparation
To investigate the distribution and phenotypes of NK cells and their specific subsets in NSCLC, freshly excised paired normal and tumor tissues from 60 patients were uniformly cut into small pieces. The shredded tissue samples were then divided into at least three 5 mL tubes as experimental replicates.
We then added 5 mL of RPMI‐1640 containing 10% fetal bovine serum (FBS; Thermo Fisher Scientific, 10100147), 5 mg of type 1 collagenase (Cell Signaling Technology, 62648), and 5 mg of type 4 collagenase (Cell Signaling Technology, 44204) into the tubes containing 5 mL of shredded tissue samples, and digested them at 37°C (Miltenyi, gentleMACS™ Tissue Dissociator). After digestion, we filtered the mixtures to remove any tissue particles, and the cell suspensions were allowed to stand. Finally, after the cells were counted, we centrifuged them at 250×g for 10 min, and resuspended them in complete RPMI medium (RPMI‐1640 supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin), based on the protocols of subsequent experiments.
Distribution and phenotypic analysis with cell‐surface staining
In the distribution and phenotypic analyses, the cells were resuspended and divided into several centrifuge tubes. Each tube contained 50 μL of cell suspension, containing at least 1,000,000 cells. We then added 50 μL of cell staining buffer (CSB; Biolegend, 420201) containing 0.5 μL of fluorescence‐labeled antibody, and stored the samples in the dark for 15 min at 20–24 °C. All fluorescence‐labeled antibodies used in the present study are shown in Supplementary Table 2.
CSB (500 μL) was then added to the stained cell suspensions, which were centrifuged at 250×g for 5 min. Finally, we resuspended the cells in 300 μL of CSB, filtered them through a 40 μm filter, and analyzed the samples on a FACSCanto II system (BD Biosciences). The percentage of positive cells under each gate was calculated with the FlowJo software (Tree Star, Version 10).
Functional analyses with intracellular cytokine staining
In the functional analyses, each cell suspension was resuspended and divided into different numbers of subsamples according to the experimental plan. Each subsample contained at least 1,000,000 cells, and was cultured in a 12‐well cell culture plate (1 mL/well) for 12 h at 37°C. The interventions included the addition of IL‐15 (10 ng; Proteintech), BFA and monensin (3 mg and 1.4 mg; Multi Sciences, CS1002), BFA and monensin (3 mg and 1.4 mg) and PMA and ionomycin (50 μg and 1 mg; Multi Sciences, CS1001).
After incubation, the cell suspensions were transferred to centrifuge tubes and washed once with phosphate‐buffered saline (PBS) before subsequent experiments. We then added 250 μL of freshly diluted permeabilization buffer (Biolegend, 421402), and placed the samples at 4°C for 25 min. After adding 200 μL of intracellular staining permeabilization wash buffer (Biolegend, 421002), we centrifuged the cell suspensions at 550×g for 10 min and removed the supernatants. We then added 50 μL of CSB containing 1 μL of fluorescence‐labeled antibody, and incubated the samples in the dark for 30 min.
CSB (500 μL) was then added to the stained cell suspensions, which were centrifuged at 550×g for 10 min. The methods used to resuspend, detect the stained cells, and calculate their proportions were as described in the cell‐surface staining section.
Function analyses by direct cell cytotoxicity evaluation
To examine the functions of CD69+CXCR6+ trNK cells more accurately, cell suspensions from normal pulmonary tissues were first stained with fluorescence‐labeled antibody directed against CD69 or CXCR6, as described in the cell‐surface staining section. The cells were then sorted with the FACSCalibur system (BD Biosciences), according to the manufacturer's protocol, and post‐sorting tests were performed to ensure the validity of the results.
Single‐cell suspensions of CD69+CXCR6+ trNK cells or DN cNK cells were prepared and separated into aliquots, each containing 10,000 cells. Each aliquot was cocultured with 10,000 A549 lung cancer cells in a 96‐well culture plate (200 μL/well) at 37°C for 24 h.
The proportions of dead A549 cells were determined after staining with 7‐AAD Viability Staining Solution (Biolegend, 420403). Dead A549 cells were identified as 7‐AAD+ cells. After resuspending the samples into tubes each containing 50 μL of cell suspension, we then added 50 μL of CSB containing 1 μL of 7‐AAD solution. After storing the samples in the dark for 5 min, we detected the stained cells with the flow cytometry as described above as soon as possible.
Function analyses by ELISA
To confirm the secretion of IFN‐γ by CD69+CXCR6+ trNK cells, we first prepared single‐cell suspensions of CD69+CXCR6+ trNK cells and DN cNK cells, as described above. We then separated the suspensions into aliquots, each containing 100,000 cells, and cultured them in a 96‐well culture plate (200 μL/well) at 37°C for 24 h.
After incubation and centrifugation at 500 rpm for 5 min, the supernatants of the single‐cell suspensions were collected. The IFN‐γ concentrations were determined with an ELISA (Proteintech, KE00146), according to the protocol of the manufacturer.
Changes in the functionality of the NK cells after blockade with ICIs
To investigate how the blockade of immune checkpoints affects the functionality of NK cells in NSCLC, cell suspensions were prepared from normal lung tissues of eight patients with the method described above. The cells were resuspended and divided into different subsamples, each containing at least 1,000,000 cells. We cultured them in complete RPMI medium supplemented with BFA and monensin and PMA and ionomycin in 12‐well cell culture plates (1 mL/well) at 37°C for 12 h.
At the beginning of the culture period, we also added 10 μL of neutralizing anti‐human PD‐1 antibody (1 mg/mL; Biolegend, 329926) alone, 10 μL of neutralizing anti‐human CTLA‐4 antibody (1 mg/mL; Biolegend, 349932) alone, or 10 μL of anti‐human PD‐1 antibody and 10 μL of anti‐human CTLA‐4 antibody (combined in the well).
After incubation, intracellular cytokine staining, flow cytometric analysis, and calculations were performed as described above.
Statistical analysis
GraphPad Prism software version 8.0 was used in the statistical analyses. Each symbol in the results represented one sample, and the results in the form of a bar diagram were presented as mean ± SEM. Differences in the percentages of positively stained cells between the normal and tumor tissues were determined by paired t‐tests (Wilcoxon matched‐pairs signed rank tests for non‐parametric pairing t‐tests). Differences in the percentages of positively stained cells among NK cells were determined by t‐tests (Wilcoxon signed rank tests for non‐parametric t‐tests). Correlations between PD‐1+ DP NK cells and CTLA‐4+ DP NK cells were evaluated with Spearman's correlation and linear regression tests. The data were analyzed by two‐tailed tests unless otherwise specified while the p value < 0.05 was regarded as statistically significant.
Conflict of interest
The authors declare no commercial or financial conflict of interest.
Ethics approval
The Ethical Committee of the Second Affiliated Hospital of Zhejiang University School of Medicine has approved the studies, and the informed consent of all participating subjects was obtained.
Author contributions
X.K.C., Y.Y.C., Z.W.X., M.J.L., Z.X.H., D.C., L.F.Z., and T.H. performed experiments. X.K.C., Y.Y.C., and Z.W.X. analyzed the data. X.K.C., D.W., and P.W. designed experiments, interpreted the data, and wrote the manuscript. P.W. and Y.C. supervised the project.
Peer review
The peer review history for this article is available at https://publons.com/publon/10.1002/eji.202149608
Abbreviations
- NK
natural killer
- trNK
tissue‐resident NK
- TME
tumor microenvironment
- NSCLC
non‐small cell lung cancer
- ICI
immune checkpoint inhibitor
- ILC
innate lymphoid cell
- Trm
resident memory T
- CXCR
C‐X‐C motif chemokine receptor
- DP
CD69+CXCR6+ double positive
- SP
CD69+CXCR6− single positive
- DN
CD69−CXCR6− double negative
Supporting information
Supporting Information
Acknowledgments
The authors thank the Jian Huang lab for technical assistance and helpful support. This work was supported by National Natural Science Foundation of China (81572800 to P.W., 82073141 to P.W., 82073142 to D.W.), the Fundamental Research Funds for the Central Universities (2019QNA7025 to P.W.) and the Natural Science Foundation of Zhejiang Province (LR22H160006 and LY15H160041 to P.W., LY19H160050 to D.W.).
Contributor Information
Pin Wu, Email: pinwu@zju.edu.cn.
Ying Chai, Email: chaiy@zju.edu.cn.
Data availability statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- 1. Sung, H. , Ferlay, J. , Siegel, R. L. , Laversanne, M. , Soerjomataram, I. , Jemal, A. and Bray, F. , Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021. 71(3):209‐49. [DOI] [PubMed] [Google Scholar]
- 2. Sharma, P. and Allison, J. P. , The future of immune checkpoint therapy. Science 2015. 348(6230):56‐61. [DOI] [PubMed] [Google Scholar]
- 3. Sharma, P. and Allison, J. P. , Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 2015. 161(2):205‐14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. The Lancet Respiratory M . Lung cancer immunotherapy biomarkers: refine not reject. Lancet Respir. Med. 2018. 6(6):403. [DOI] [PubMed] [Google Scholar]
- 5. Shimasaki, N. , Jain, A. , Campana, D. , NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 2020. 19(3):200‐18. [DOI] [PubMed] [Google Scholar]
- 6. Sivori, S. , Pende, D. , Quatrini, L. , Pietra, G. , Della Chiesa, M. , Vacca, P. , Tumino, N. et al. NK cells and ILCs in tumor immunotherapy. Mol. Aspects Med. 2020. 100870. [DOI] [PubMed] [Google Scholar]
- 7. Demaria, O. , Gauthier, L. , Debroas, G. and Vivier, E. , Natural killer cell engagers in cancer immunotherapy: next generation of immuno‐oncology treatments. Eur. J. Immunol. 2021. 51(8):1934‐42. [DOI] [PubMed] [Google Scholar]
- 8. Melaiu, O. , Lucarini, V. , Cifaldi, L. and Fruci, D. , Influence of the tumor microenvironment on NK cell function in solid tumors. Front. Immunol. 2019. 10: 3038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Concha‐Benavente, F. , Kansy, B. , Moskovitz, J. , Moy, J. , Chandran, U. and Ferris, R. L. , PD‐L1 mediates dysfunction in activated PD‐1(+) NK cells in head and neck cancer patients. Cancer Immunol. Res. 2018. 6(12): 1548‐60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barry, K. C. , Hsu, J. , Broz, M. L. , Cueto, F. J. , Binnewies, M. , Combes, A. J. , Nelson, A. E. et al. A natural killer‐dendritic cell axis defines checkpoint therapy‐responsive tumor microenvironments. Nat. Med. 2018. 24(8): 1178‐91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ardain, A. , Marakalala, M. J. and Leslie, A. , Tissue‐resident innate immunity in the lung. Immunology 2020. 159(3): 245‐56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hervier, B. , Russick, J. , Cremer, I. and Vieillard, V. , NK cells in the human lungs. Front. Immunol. 2019. 10: 1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Amsen, D. , van Gisbergen, K. , Hombrink, P. and van Lier, R. A. W. , Tissue‐resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 2018. 19(6): 538‐46. [DOI] [PubMed] [Google Scholar]
- 14. Suarez‐Ramirez, J. E. , Chandiran, K. , Brocke, S. and Cauley, L. S. , Immunity to respiratory infection is reinforced through early proliferation of lymphoid TRM cells and prompt arrival of effect or CD8 T cells in the lungs. Front. Immunol. 2019. 10: 1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wu, S. Y. , Fu, T. , Jiang, Y. Z. and Shao, Z. M. , Natural killer cells in cancer biology and therapy. Mol. Cancer. 2020. 19(1): 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dogra, P. , Rancan, C. , Ma, W. , Toth, M. , Senda, T. , Carpenter, D. J. , Kubota, M. et al. Tissue Determinants of human NK cell development, function, and residence. Cell 2020. 180(4): 749‐63.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marquardt, N. , Kekalainen, E. , Chen, P. , Lourda, M. , Wilson, J. N. , Scharenberg, M. , Bergman, P. et al. Unique transcriptional and protein‐expression signature in human lung tissue‐resident NK cells. Nat. Commun. 2019. 10(1): 3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheuk, S. , Schlums, H. , Gallais Serezal, I. , Martini, E. , Chiang, S. C. , Marquardt, N. , Gibbs, A. et al. CD49a expression defines tissue‐resident CD8(+) T cells poised for cytotoxic function in human skin. Immunity 2017. 46(2): 287‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duhen, T. , Duhen, R. , Montler, R. , Moses, J. , Moudgil, T. , de Miranda, N. F. , Goodall, C. P. et al. Co‐expression of CD39 and CD103 identifies tumor‐reactive CD8 T cells in human solid tumors. Nat. Commun. 2018. 9(1): 2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bade, B. C. and Dela Cruz, C. S. , Lung cancer 2020: epidemiology, etiology, and prevention. Clin. Chest Med. 2020. 41(1): 1‐24. [DOI] [PubMed] [Google Scholar]
- 21. Wang, X. , Tian, Z. and Peng, H. , Tissue‐resident memory‐like ILCs: innate counterparts of TRM cells. Protein Cell 2020. 11(2): 85‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sun, H. , Sun, C. , Xiao, W. and Sun, R. , Tissue‐resident lymphocytes: from adaptive to innate immunity. Cell. Mol. Immunol. 2019. 16(3):205‐15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lanier, L. L. , Le, A. M. , Civin, C. I. , Loken, M. R. and Phillips, J. H. , The relationship of CD16 (Leu‐11) and Leu‐19 (NKH‐1) antigen expression on human peripheral blood NK cells and cytotoxic T lymphocytes. J. Immunol. 1986. 136(12): 4480‐6. [PubMed] [Google Scholar]
- 24. Yang, C. , Siebert, J. R. , Burns, R. , Gerbec, Z. J. , Bonacci, B. , Rymaszewski, A. , Rau, M. et al. Heterogeneity of human bone marrow and blood natural killer cells defined by single‐cell transcriptome. Nat. Commun. 2019. 10(1): 3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bjorkstrom, N. K. , Riese, P. , Heuts, F. , Andersson, S. , Fauriat, C. , Ivarsson, M. A. , Björklund, A. T. et al. Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK‐cell differentiation uncoupled from NK‐cell education. Blood 2010. 116(19): 3853‐64. [DOI] [PubMed] [Google Scholar]
- 26. Banchereau, R. , Chitre, A. S. , Scherl, A. , Wu, T. D. , Patil, N. S. , de Almeida, P. , Kadel Iii, E. E. et al. Intratumoral CD103+ CD8+ T cells predict response to PD‐L1 blockade. J. Immunother. Cancer 2021. 9(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wolf, N. K. , Kissiov, D. U. and Raulet, D. H. , Roles of natural killer cells in immunity to cancer, and applications to immunotherapy. Nat. Rev. Immunol. 2022. [DOI] [PubMed] [Google Scholar]
- 28. Morvan, M. G. and Lanier, L. L. , NK cells and cancer: you can teach innate cells new tricks. Nat. Rev. Cancer 2016. 16(1): 7‐19. [DOI] [PubMed] [Google Scholar]
- 29. Freud, A. G. , Mundy‐Bosse, B. L. , Yu, J. and Caligiuri, M. A. , The broad spectrum of human natural killer cell diversity. Immunity 2017. 47(5): 820‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen, S. , Yang, M. , Du, J. , Li, D. , Li, Z. , Cai, C. , Ma, Y. et al. The self‐specific activation receptor SLAM family is critical for NK cell education. Immunity 2016. 45(2): 292‐304. [DOI] [PubMed] [Google Scholar]
- 31. Sivori, S. , Vacca, P. , Del Zotto, G. , Munari, E. , Mingari, M. C. and Moretta, L. , Human NK cells: surface receptors, inhibitory checkpoints, and translational applications. Cell. Mol. Immunol. 2019. 16(5): 430‐41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chae, Y. K. , Arya, A. , Iams, W. , Cruz, M. R. , Chandra, S. , Choi, J. and Giles, F. , Current landscape and future of dual anti‐CTLA4 and PD‐1/PD‐L1 blockade immunotherapy in cancer; lessons learned from clinical trials with melanoma and non‐small cell lung cancer (NSCLC). J. Immunother. Cancer 2018. 6(1): 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Baumeister, S. H. , Freeman, G. J. , Dranoff, G. and Sharpe, A. H. , Coinhibitory pathways in immunotherapy for cancer. Annu. Rev. Immunol. 2016. 34: 539‐73. [DOI] [PubMed] [Google Scholar]
- 34. Mueller, S. N. and Mackay, L. K. , Tissue‐resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 2016. 16(2): 79‐89. [DOI] [PubMed] [Google Scholar]
- 35. Mackay, L. K. , Rahimpour, A. , Ma, J. Z. , Collins, N. , Stock, A. T. , Hafon, M. L. , Vega‐Ramos, J. et al. The developmental pathway for CD103(+)CD8+ tissue‐resident memory T cells of skin. Nat. Immunol. 2013. 14(12): 1294‐301. [DOI] [PubMed] [Google Scholar]
- 36. Zhang, N. and Bevan, M. J. , Transforming growth factor‐beta signaling controls the formation and maintenance of gut‐resident memory T cells by regulating migration and retention. Immunity 2013. 39(4): 687‐96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wein, A. N. , McMaster, S. R. , Takamura, S. , Dunbar, P. R. , Cartwright, E. K. , Hayward, S. L. , McManus, D. T. et al. CXCR6 regulates localization of tissue‐resident memory CD8 T cells to the airways. J. Exp. Med. 2019. 216(12): 2748‐62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Melsen, J. E. , Lugthart, G. , Lankester, A. C. and Schilham, M. W. , Human circulating and tissue‐resident CD56(bright) natural killer cell populations. Front. Immunol. 2016. 7: 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang, M. , Wen, B. , Anton, O. M. , Yao, Z. , Dubois, S. , Ju, W. , Sato, N. et al. IL‐15 enhanced antibody‐dependent cellular cytotoxicity mediated by NK cells and macrophages. Proc. Natl. Acad. Sci. U. S. A. 2018. 115(46): E10915‐E24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chauvin, J. M. , Ka, M. , Pagliano, O. , Menna, C. , Ding, Q. , DeBlasio, R. , Sanders, C. et al. IL‐15 Stimulation with TIGIT blockade reverses CD155‐mediated NK‐cell dysfunction in melanoma. Clin. Cancer Res. 2020. 26(20): 5520‐33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tan, S. , Xu, Y. , Wang, Z. , Wang, T. , Du, X. , Song, X. , Guo, X. et al. Tim‐3 hampers tumor surveillance of liver‐resident and conventional NK cells by disrupting PI3K signaling. Cancer Res. 2020. 80(5): 1130‐42. [DOI] [PubMed] [Google Scholar]
- 42. Myers, J. A. and Miller, J. S. , Exploring the NK cell platform for cancer immunotherapy. Nat. Rev. Clin. Oncol. 2021. 18(2): 85‐100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zhang, Q. , Bi, J. , Zheng, X. , Chen, Y. , Wang, H. , Wu, W. , Wang, Z. et al. Blockade of the checkpoint receptor TIGIT prevents NK cell exhaustion and elicits potent anti‐tumor immunity. Nat. Immunol. 2018. 19(7): 723‐32. [DOI] [PubMed] [Google Scholar]
- 44. Judge, S. J. , Dunai, C. , Aguilar, E. G. , Vick, S. C. , Sturgill, I. R. , Khuat, L. T. , Stoffel, K. M. et al. Minimal PD‐1 expression in mouse and human NK cells under diverse conditions. J. Clin. Invest. 2020. 130(6): 3051‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Subrahmanyam, P. B. , Dong, Z. , Gusenleitner, D. , Giobbie‐Hurder, A. , Severgnini, M. , Zhou, J. , Manos, M. et al. Distinct predictive biomarker candidates for response to anti‐CTLA‐4 and anti‐PD‐1 immunotherapy in melanoma patients. J. Immunother. Cancer 2018. 6(1): 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hsu, J. , Hodgins, J. J. , Marathe, M. , Nicolai, C. J. , Bourgeois‐Daigneault, M. C. , Trevino, T. N. , Azimi, C. S. et al. Contribution of NK cells to immunotherapy mediated by PD‐1/PD‐L1 blockade. J. Clin. Invest. 2018. 128(10): 4654‐68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu, Y. , Cheng, Y. , Xu, Y. , Wang, Z. , Du, X. , Li, C. , Peng, J. et al.Increased expression of programmed cell death protein 1 on NK cells inhibits NK‐cell‐mediated anti‐tumor function and indicates poor prognosis in digestive cancers. Oncogene 2017. 36(44): 6143‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


