Abstract
T cell responses play an important role in protection against beta-coronavirus infections, including SARS-CoV-2, where they associate with decreased COVID-19 disease severity and duration. To enhance T cell immunity across epitopes infrequently altered in SARS-CoV-2 variants, we designed BNT162b4, an mRNA vaccine component that is intended to be combined with BNT162b2, the spike-protein-encoding vaccine. BNT162b4 encodes variant-conserved, immunogenic segments of the SARS-CoV-2 nucleocapsid, membrane, and ORF1ab proteins, targeting diverse HLA alleles. BNT162b4 elicits polyfunctional CD4+ and CD8+ T cell responses to diverse epitopes in animal models, alone or when co-administered with BNT162b2 while preserving spike-specific immunity. Importantly, we demonstrate that BNT162b4 protects hamsters from severe disease and reduces viral titers following challenge with viral variants. These data suggest that a combination of BNT162b2 and BNT162b4 could reduce COVID-19 disease severity and duration caused by circulating or future variants. BNT162b4 is currently being clinically evaluated in combination with the BA.4/BA.5 Omicron-updated bivalent BNT162b2 (NCT05541861).
Keywords: SARS-CoV-2, COVID-19, viral variants, mRNA T cell vaccine, vaccine design, HLA ligandomics, T cell responses, immune protection
Graphical abstract

Adding non-spike targeting components to mRNA vaccination elicits promising T cell responses against SARS-CoV-2 variant strains in rodent models of COVID-19.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), a novel beta-coronavirus belonging to the sarbecovirus family, is a highly transmissible and pathogenic respiratory virus responsible for the COVID-19 pandemic. Vaccines encoding the SARS-Cov-2 spike (S) protein, which is responsible for viral entry into cells, have been used as a key intervention for the mitigation of the COVID-19 pandemic. Recently, highly infectious SARS-CoV-2 variants such as Omicron and its sublineages have emerged, some of which evade neutralizing antibodies raised by vaccination or by prior exposure.1 , 2 , 3 , 4 , 5 , 6 , 7 While currently approved S-protein-encoding vaccines remain effective at protecting from severe disease by both antibody and cell-mediated mechanisms, harnessing cellular immunity to conserved antigens could provide more robust protection from severe disease caused by circulating and future variants.8 , 9
Both CD4+ and CD8+ T cell responses have been shown to be important in the control and clearance of beta-coronaviruses.10 , 11 , 12 , 13 Generally, upon recognition of their cognate epitopes on major histocompatibility complex (MHC) class II molecules, CD4+ T cells help orchestrate the overall immune response to infection through cytokine and chemokine secretion, and provide support to antibody production by B cells, as well as cytotoxic functions of other immune cells. CD8+ T cells can recognize pathogen-infected cells through epitopes presented on the host cell surface by MHC class I molecules. Upon detection of infected cells, CD8+ T cells exert cytotoxic functions, leading to the elimination of the infected cells and limit intra-host pathogen spread. Following infection by SARS-CoV, which is closely related to SARS-CoV-2, neutralizing antibody levels decay over time,14 , 15 while virus-specific T cell memory persists in convalescent patients after a decade.16 A recent study reported that nucleocapsid (N)-specific T cells persisted for 17 years post-SARS-CoV infection, highlighting the potential for durable T cell responses raised against SARS-CoV-2.17 Functionally, depletion of CD8+ T cells in SARS-CoV-2 convalescent non-human primates reduced the protection conferred by natural immunity from re-challenge, demonstrating the importance of cellular immune responses, especially in the case of low baseline antibody titers.18 Depletion of CD8+ T cells following immunization against the SARS-CoV-2 S and/or N protein also abrogated protection against subsequent challenge in animal models showing their importance in a vaccination setting.19 , 20 In humans, the mRNA spike-encoding vaccine BNT162b2 induced long-lasting T cell responses.21 , 22 In breakthrough infections following vaccination, T cell activation correlated positively with the rate of viral clearance and negatively with the peak of the viral titer.23 Recently, it’s been reported that breakthrough cases following vaccination were associated with reduced S-specific T cell responses; in the absence of the variant-reactive T cells, chances for a breakthrough infection were higher.24 However, the exact role, mechanism, and contribution of vaccine-induced T cells in humans remain to be elucidated.25 In addition, vaccine-specific data focus on S-specific T cell responses, which may differ from T cell responses targeting other viral antigens.
Importantly, studies with human samples during the COVID-19 pandemic suggested that SARS-CoV-2-specific T cells may provide durable immunity against severe COVID-19 and were associated with a shortened course of disease.26 , 27 , 28 , 29 SARS-CoV-2-specific T cell responses following infection or vaccination are often detectable earlier than antibody responses. Rapid cellular responses correlate with reduced disease severity following infection, and the S-specific T cell responses induced by vaccination with the S-protein-encoding BNT162b2 are associated with the efficacy of this COVID-19 vaccine.29 , 30 , 31 Additionally, an increase in CD8+ T cells in hematological cancer patients with compromised B cell compartments correlated with increased survival after SARS-CoV-2 infection.32 Together, the evidence suggests the important and durable nature of the cell-mediated immune response in control of SARS-CoV-2, which could be further harnessed in vaccine development.
Recent studies have demonstrated that the N protein, membrane (M) protein, and Open Reading Frame 1ab (ORF1ab) polyprotein are among the most T cell immunogenic SARS-CoV-2 antigens.33 , 34 , 35 , 36 Additionally, the benefits of multi-antigen vaccines are being explored.19 , 37 , 38 One study suggested that memory T cell responses, which activate faster than antibody production, could provide significant protection via rapid viral clearance upon recognition of ORF1ab epitopes.39 A significant advantage T cell epitopes have over neutralizing antibody epitopes is their conservation across SARS-CoV-2 variants. While antibodies of different individuals exposed to a given SARS-CoV-2 variant converge on a small set of similar neutralizing epitopes, T cell epitopes are diversified between individuals due to their specific Human Leukocyte Antigen (HLA) haplotypes, providing a further obstacle for viral evasion of T cell immunity.40
To expand T cell responses beyond the S protein, we developed BNT162b4, an mRNA vaccine component designed to boost cellular immunity against SARS-CoV-2. BNT162b4 encodes segments of the SARS-CoV-2 N, M, and ORF1ab proteins and is intended to be administered with S-protein-encoding vaccines, such as BNT162b2.41 , 42 Here we show that BNT162b4 generates T cell epitopes presented on HLA-I complexes in human cell lines detectable by mass spectrometry (MS). Immunization of mice with BNT162b4 generates a robust T cell response against the N, M, and ORF1ab SARS-CoV-2 proteins when administered either alone or in combination with the S-protein-encoding BNT162b2. Finally, we demonstrate that BNT162b4 protects hamsters from severe COVID-19 following challenge with various SARS-CoV-2 variants and, when paired with BNT162b2, bolsters immunity against SARS-CoV-2. BNT162b4 is currently being clinically evaluated in combination with the BA.4/BA.5 Omicron-updated bivalent BNT162b2 (NCT05541861).
Results
BNT162b4 vaccine design and conservation of antigens across SARS-CoV-2 variants
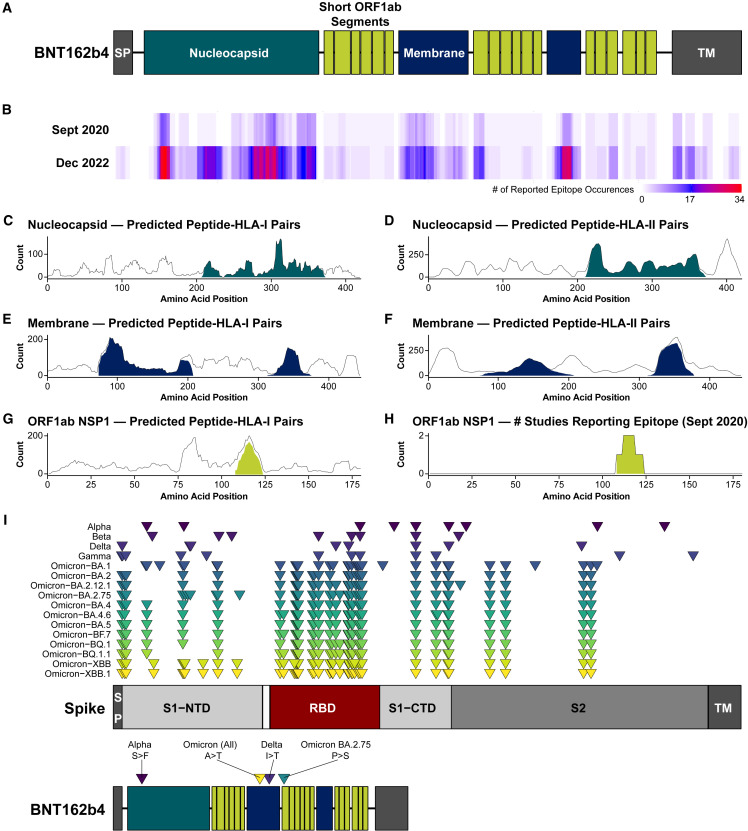
BNT162b4 was rationally designed in September 2020, before the emergence of significant SARS-CoV-2 variants, to maximize T cell immunogenicity across a diverse human population. Based on then-available reported immunogenicity from SARS-CoV-2 convalescent individuals combined with predicted immunogenicity33 , 35 , 36 , 43 , 44 , 45 , 46 , 47 , 48 , 49 and protein expression data from infected cells,48 , 50 , 51 , 52 we selected N, M, and non-structural proteins (NSP) 1–4 from ORF1ab for inclusion in the T cell-targeting vaccine component (Figure 1 A; Table S1). For the N and M proteins, segments that are enriched for both reported and predicted CD8+ and CD4+ T-cell epitopes were selected (Figures 1B–1F; Tables S2–S3). For ORF1ab, we employed a different approach, due to its length (>7,000 amino acids), using a greater number of shorter segments containing minimal HLA-I-targeting epitopes from throughout the genomic region encoding NSPs 1–4. NSPs 1–4 were specifically chosen because they are among the first proteins to be synthesized upon viral infection, and they are present in a greater abundance than other ORF1ab NSPs.53 , 54 , 55 From these four NSPs, considering only predicted epitopes that were also previously demonstrated as immunogenic (circa September 2020), 18 sequence segments of 9–16 amino acids were selected for inclusion in BNT162b4 (Figures 1G–1H; Figure S1 ; Table S1). Altogether, BNT162b4 contains 2,257 unique predicted peptide-HLA-I pairs across 105 HLA-I alleles and 1,992 unique predicted peptide-HLA-II pairs across 70 HLA-II alleles (Tables S2–S3).48 Each antigenic segment selected for inclusion in the vaccine is predicted to contain epitopes restricted by HLA-I alleles present in 99% or more of the global population.48
Figure 1.
Antigen selection and design of BNT162b4
(A) Schematic of BNT162b4 containing highly immunogenic segments of the SARS-CoV-2 N (green) and M proteins (blue), and short segments containing minimal epitopes from the ORF1ab NSPs 1–4 (lime-green).
(B) Epitope counts reported in contemporaneous literature circa September 2020 and December 2022. See STAR Methods section for references.
(C–G) Predicted epitope presentation from Poran et al., 2020.48.
(H) Regions of ORF1ab NSP1 reported as immunogenic in contemporaneous literature circa September 2020. For plots (C)–(H), lines represent the number of unique peptide-HLA pairs at each residue along the viral protein, and the shaded regions represent the number of unique peptide-HLA pairs contained entirely within the segment included in BNT162b4.
(I) Schematic summarizing non-synonymous mutations in the S protein (top) and along BNT162b4 (bottom) from past and currently circulating variants. Amino acid substitutions are indicated along BNT162b4. Length of proteins are to scale. SP, signal peptide; S1-NTD, N-terminal domain of S1 subunit of the S protein; RBD, receptor-binding domain; S1-CTD, C-terminal domain of S1 subunit of the S protein; S2, S2 subunit of the S protein; TM, transmembrane domain. Lists of variant mutations are retrieved from: WHO, CoVariants, Cov-lineages, or BV-BRC.
See also Figure S1.
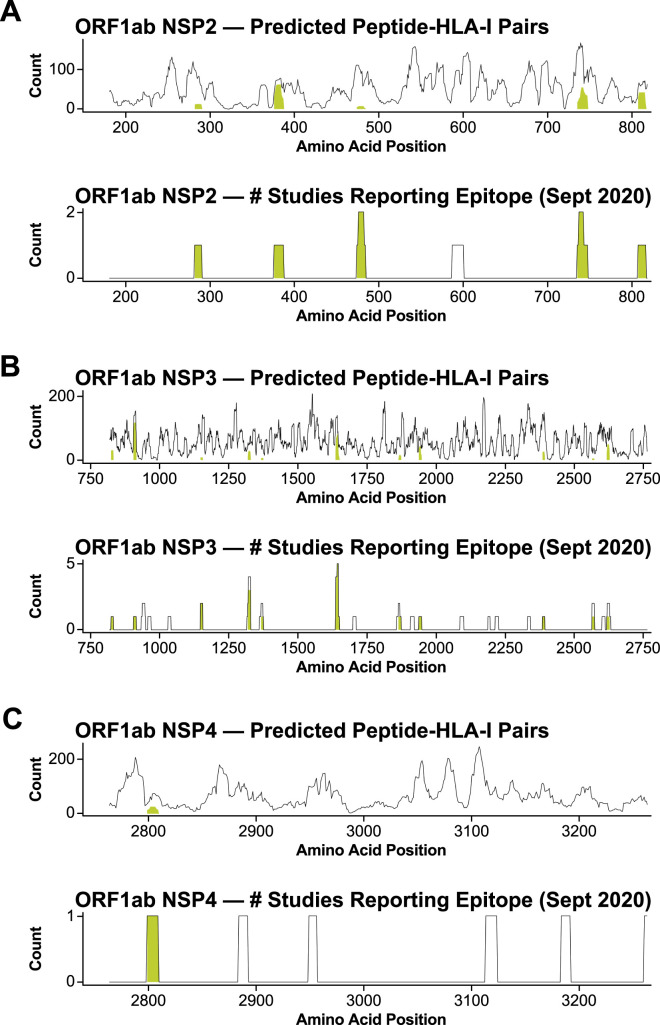
Figure S1.
Antigen selection and design of BNT162b4, related to Figure 1
Segments of (A) ORF1ab NSP2, (B) NSP3, and (C) NSP4 selected for inclusion in BNT162b4 based on predicted HLA-I epitope density (top; Poran et al., 202048) and regions of each antigen cited as immunogenic in the literature circa September 2020 (bottom). Lines represent the number of unique peptide-HLA pairs at each point along the viral protein, and the shaded regions represent the number of unique peptide-HLA pairs contained entirely within the selected segment. The x axis numbering is based on the full length ORF1ab polyprotein with numbers indicating amino acid position.
To investigate whether BNT162b4 may be impacted by variants that have replaced the ancestral SARS-CoV-2 strain, we mapped the mutations present in variants recognized by the World Health Organization (WHO) (as of December 2022) onto BNT162b4. Compared to the S protein, which contains over 40 non-synonymous mutations in some Omicron variants such as XBB, with most of them concentrated in the receptor-binding domain (RBD), BNT162b4 contains only four non-synonymous mutations across these variants, with only two non-synonymous mutations from circulating Omicron variants as of December 2022 (Figure 1I).
Mass spectrometry HLA ligandomics analysis identifies BNT162b4-derived HLA-presented epitopes
To examine whether BNT162b4 can generate T cell epitopes that are processed and presented onto HLA complexes, we used targeted MS to detect peptides from BNT162b4 bound to HLA-I molecules. To accurately determine to which HLA allele an epitope is bound, we used monoallelic profiling of tagged allele constructs (MAPTAC) to exogenously express and isolate a single HLA allele from the background of endogenous HLA alleles present in the A375 cell line56 , 57 as well as the BB7.2 antibody that targets endogenous A∗02:01 and the pan-HLA-I antibody W6/32 to profile epitopes bound to other A375 endogenous alleles (A∗01:01, B∗44:03, C∗16:01) in the A375 cell line. The peptide list for targeted MS analysis was generated from sequences in BNT162b4 predicted to bind the tested HLA alleles48 or observed by MS in Weingarten-Gabbay et al.54 Altogether, we identified 19 unique peptide epitopes across seven HLA alleles and observed 30 unique peptide-HLA pairs. Verified peptides spanned the entire length of BNT162b4 and peptides from all three encoded viral antigens were detected (Figure 2 ; Table 1 ). Notably, all 19 MS-verified epitopes were previously reported as immunogenic or were contained within a peptide previously reported as immunogenic, with nine of them being cited as “immunodominant” in previous studies.33 , 43 , 58 , 59 , 60 Additionally, the immunodominant ORF1ab epitopes PTDNYITTY and TTDPSFLGRY that were rationally designed for processing with cleavable linkers were presented on almost every HLA allele that was tested regardless of predicted presentation (Table 1; Figure S2 A). Several epitopes in this study appear to be “nested” as a subsequence of a longer epitope on the same HLA allele but have distinct chromatographic retention times indicating that they are not artifacts of MS analysis (Figure S2B). No epitopes were detected from the first M segment, likely due to peptides derived from this sequence being more hydrophobic61 and therefore more challenging to recover from sample processing and to detect by MS with the methods employed here (Figure S2C). These MS-detected epitopes represent only a subset of all the epitopes, which are expected to be presented by HLA alleles not tested in this study. In addition to targeted MS analysis, data-dependent analysis (DDA) was performed on the samples to look for potential junction-spanning epitopes presented on HLA complexes. While DDA analyses revealed 6 epitopes derived from BNT162b4, including 5 from the short ORF1ab segments, none of the observed epitopes spanned a junction between adjacent segments (Table 1; Table S4). Together, these data indicate that the BNT162b4 mRNA is translated in its entirety, generating epitopes that are processed and presented onto human HLA complexes for potential immune recognition by T cells.
Figure 2.
BNT162b4 is a source of HLA-presented epitopes as directly measured by mass spectrometry
(A) 19 unique BNT162b4-derived epitopes were detected by targeted mass spectrometry, with at least one epitope coming from each encoded SARS-CoV-2 antigen.
(B and C) (B) Isotopically encoded peptides were used to generate chromatographs with each color representing a unique fragment ion specific to the indicated peptide (arrows indicate apex of chromatographic peak), and (C) head-to-toe plots that confirm matching retention time, matching fragmentation pattern, and specificity to only BNT162b4 transfected samples (shown is epitope #18: ORF1ab PTDNYITTY on A∗01:01). Epitope sequence and allele restriction can be found in Table 1. Additional chromatographs, head-to-toe plots, and complete DDA results can be found in the raw data repository (see data and code availability).
Table 1.
Epitopes encoded in BNT162b4 that were detected by mass spectrometry
| Epitope Number | Epitope Sequence |
HLA Allele(s) | Antigen |
|---|---|---|---|
| 1 | LLLDRLNQL | A∗02:01 | Nucleocapsid |
| 2 | KTFPPTEPK | A∗11:01, A∗03:01 | Nucleocapsid |
| 3 | KTFPPTEPKK | A∗11:01, A∗03:01 | Nucleocapsid |
| 4 | KTIQPRVEKb | A∗11:01b, A∗03:01 | ORF1ab NSP2 |
| 5 | YLFDESGEF | A∗02:01 | ORF1ab NSP3 |
| 6 | YLFDESGEFKLb | A∗02:01b | ORF1ab NSP3 |
| 7 | SEVGPEHSL | B∗44:03a | ORF1ab NSP2 |
| 8 | SEVGPEHSLAEY | B∗44:03a | ORF1ab NSP2 |
| 9 | TTDPSFLG | A∗01:01 | ORF1ab NSP3 |
| 10 | TTDPSFLGR | A∗01:01, A∗11:01 | ORF1ab NSP3 |
| 11 | TTDPSFLGRYb | A∗01:01b, A∗02:01, A∗03:01, B∗08:01 | ORF1ab NSP3 |
| 12 | TTDPSFLGRYMb | A∗01:01b | ORF1ab NSP3 |
| 13 | QLMCQPILL | A∗02:01 | ORF1ab NSP3 |
| 14 | AEAELAKNV | B∗44:03a | ORF1ab NSP3 |
| 15 | LPKEITVATb | A∗11:01, B∗08:01b | Membrane |
| 16 | VATSRTLSY | C∗16:01a | Membrane |
| 17 | ATSRTLSYYK | A∗11:01 | Membrane |
| 18 | PTDNYITTYb | A∗01:01b, A∗02:01, A∗03:01, B∗08:01 | ORF1ab NSP3 |
| 19 | FGDDTVIEV | A∗02:01 | ORF1ab NSP3 |
Epitope observed only in pan-HLA-I W6/32 pulldown, allele inferred from prediction of sequence with A375 HLA alleles. Epitope numbers map to Figure 2A.
Epitopes were observed in both targeted and discovery mode MS analysis for the corresponding allele.
Figure S2.
BNT162b4 is a source of HLA-presented epitopes as directly measured by mass spectrometry, related to Figure 2
(A) Chromatographic traces for immunodominant ORF1ab epitopes PTDYNYITTY and TTDPSFLGRY, which were observed to be presented on all alleles tested (except A∗11:01) in this study. Each color represents a unique fragment ion specific to the indicated peptide. Arrows indicate apex of chromatographic peak. (B) Retention time comparison of epitope peptides with nesting sequences. (C) BNT162b4 overlaid with MS-observed epitopes and Kyte-Doolittle hydropathy scoring.61 Scores range from −4.5 (more hydrophilic) to 4.5 (more hydrophobic).
BNT162b4 induces CD4+ and CD8+ T cell responses in murine models
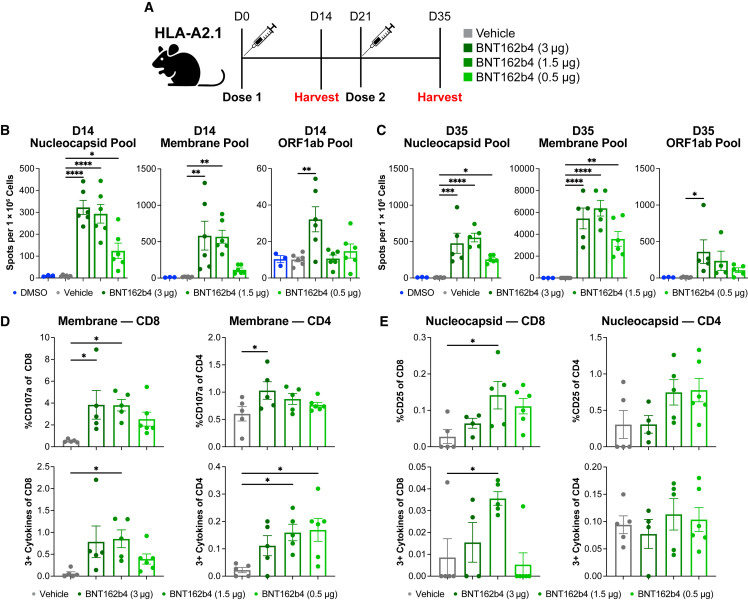
The immunogenicity of BNT162b4 was investigated in several different murine models in either one-dose or two-dose settings. HLA-A2.1 transgenic mice, which express a chimeric human HLA-A∗02:01 that is able to interact with mouse CD8+, were immunized with a dose titration of the lipid-nanoparticle encapsulated mRNA, and T cell responses were analyzed using interferon-ɣ (IFN-ɣ)-specific ELISpot and intracellular staining by flow cytometry (ICS) (Figure 3 ).62 After stimulation with either N or M peptide pools, splenocytes of the groups treated with either 3 μg or 1.5 μg of the BNT162b4 displayed significantly higher IFN-ɣ spot counts than vehicle control splenocytes. Mean spot counts for all treated groups given two vaccinations were higher than mice given one vaccination (Figures 3B and 3C). Of note, even though only three of the 18 included ORF1ab epitopes in BNT162b4 are predicted to bind HLA-A∗02:01, splenocytes stimulated with the ORF1ab peptide pools demonstrated significantly higher IFN-ɣ levels compared to the vehicle control at either timepoint in the highest dose cohort (Figures 3B and 3C). Both CD4+ and CD8+ T cells exhibited upregulation of activation markers, cytokine responses, and degranulation markers, indicating polyfunctionality and cytotoxic potential through expression of three or more of the following: IFN-ɣ, TNF-α, IL-2, or CD107a (Figures 3D and 3E). Meso scale discovery (MSD) analysis on supernatants from peptide-pulsed splenocytes indicate a strong Th1 cytokine response, as shown by low levels of IL-4 and elevated levels of IFN-ɣ, TNF-α, and IL-2 in immunized mice (Figures S3 A and S3B). The immunogenicity of BNT162b4 was also tested in the K18-hACE2 C57BL/6J (H2kb) transgenic model, as well as a BALB/c mouse model (H2kd), to highlight the robustness of the immune responses across different multiple MHC alleles (Figures S4 and S5 , respectively).63 Taken together, these data indicate that BNT162b4 can produce T cell responses against N, M, and ORF1ab in multiple animal models with differing MHC backgrounds, and the phenotype of these T cells is in line with previous reports of BNT162b2-induced T cell responses.21 , 41 , 42 , 64
Figure 3.
T-cell immunogenicity of BNT162b4
(A) Schematic of immunogenicity experiment (n = 6 animals/group).
(B) IFN-ɣ ELISpot data from peptide-pulsed splenocytes at day 14 after a prime immunization only, in triplicate. The N protein pool contained 39 peptides, the M pool contained 22 peptides, and the ORF1ab pool contained 19 peptides.
(C) IFN-ɣ ELISpot data from peptide-pulsed splenocytes at day 35 after two doses of BNT162b2, triplicate.
(D) Flow phenotyping and intracellular staining on CD8+ T cells and CD4+ T cells in day 35 splenocytes pulsed with M peptide pools.
(E) Flow phenotyping and intracellular staining on CD8+ T cells and CD4+ T cells inday 35 splenocytes pulsed with N peptide pools. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. Error bars indicate standard error of the mean (SEM).
See also Figures S3 and S4.
Figure S3.
Cytokine secretion profile of BNT162b4-immunized HLA-A2.1 Tg animals, related to Figure 3
(A) Th1 and Th2 cytokine levels in supernatants from M protein peptide-pulsed splenocytes analyzed via electrochemiluminescence immunoassay (ECLIA). (B) Th1 and Th2 cytokine levels in supernatants from N protein peptide-pulsed splenocytes analyzed via electrochemiluminescence immunoassay (ECLIA). Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. Error bars indicate standard error of the mean (SEM).
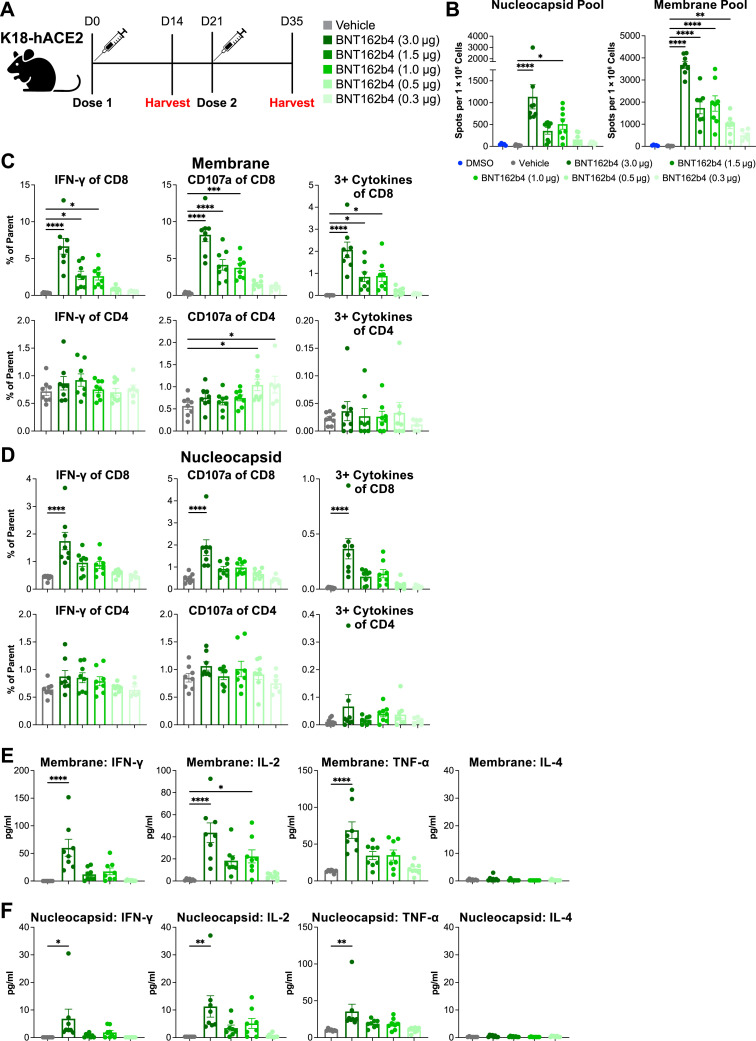
Figure S4.
Immunogenicity of BNT162b4 in K18-hACE2 C57BL/6 mouse model, related to Figure 3
(A) Schematic of the immunogenicity experiment (n = 7–8 animals per group). (B) IFN-ɣ ELISpot data from peptide-pulsed splenocytes at day 35 after two doses of BNT162b2, performed in triplicate. (C) Flow phenotyping and intracellular staining on CD8+ and CD4+ T cells of day 35 splenocytes pulsed with M peptide pools, performed in triplicate. (D) Flow phenotyping and intracellular staining on CD8+ and CD4+ T cells of day 35 splenocytes pulsed with N peptide pools. (E) Th1 and Th2 cytokine levels in supernatants from M protein peptide-pulsed splenocytes analyzed via electrochemiluminescence immunoassay (ECLIA). (F) Th1 and Th2 cytokine levels in supernatants from N protein peptide-pulsed splenocytes analyzed via ECLIA. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. Error bars indicate standard error of the mean (SEM).
Figure S5.
Immunogenicity of BNT162b4 combined with BNT162b2 in BALB/c mouse model, related to Figure 3
(A) Schematic of the combination immunogenicity experiment (n = 8 animals per group). (B) S protein S1-specific IgG titers from immunized animals, all dilutions performed in duplicate. (C) Kinetics of S1-specific IgG development. (D) Neutralizing antibody titers of immunized animals at day 14 and day 35. (E) IFN-ɣ ELISpot from peptide-pulsed splenocytes either pulsed with an N-terminal S protein peptide pool (right) or a C-terminal S protein peptide pool (left), performed in triplicate. (F) Polyfunctional CD8+ (left) or CD4+ (right) T cells reactive against either an S protein N-terminal (top) or C-terminal (bottom) peptide pool. (G) IFN-ɣ ELISpot from peptide-pulsed splenocytes either pulsed with an N peptide pool (left) or an M peptide pool (right). (H) Polyfunctional CD8+ (left) or CD4+ (right) T cells reactive against either an N (top) or M (bottom) peptide pool. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. LLOD: lower limit of detection. Error bars indicate standard error of the mean (SEM).
Immunization with BNT162b4 maintains the BNT162b2-driven immune response
As BNT162b4 is designed to complement the S-protein-encoding BNT162b2 vaccine, we sought to confirm that co-administration of 1 μg BNT162b4 and 1 μg BNT162b2 maintained the S-specific immune response induced by 1 μg BNT162b2 alone (Figure S5A). The combination of BNT162b4 with BNT162b2 maintained the high S1-specific antibody titers seen with BNT162b2 alone (Figures S5B and S5C). Neutralizing antibody titers followed a similar trend, where animals receiving a combination of BNT162b4 and BNT162b2 showed comparable neutralizing titers of animals receiving BNT16b2 alone (Figure S5D). Additionally, the levels of S-specific T cells generated in animals immunized with a combination of BNT162b2 and BNT162b4 were similar to those generated by immunization with BNT162b2 alone, further demonstrating that combination with BNT162b4 maintains the BNT162b2 immune response (Figures S5E–S5H). Notably, N and M specific responses were reduced in the group receiving both BNT162b2 and BNT162b4 compared to the group receiving BNT162b4 alone, though in both cases the responses detected were significantly stronger than the vehicle control (Figures S5G and S5H).
The combination of BNT162b4 and BNT162b2 demonstrates immunogenicity in animals with previous BNT162b2 vaccination
A large portion of the population worldwide has been previously exposed to the SARS-CoV-2 S protein either by vaccination or infection. To understand how vaccination with BNT162b4 would affect pre-existing immune responses to S protein in these individuals, we sought to assess the impact of introducing BNT162b4 following vaccination with the S-protein-encoding BNT162b2, similarly to how BNT162b4 is currently being clinically evaluated in combination with the BA.4/BA.5 Omicron-updated bivalent BNT162b2 (NCT05541861). HLA-A2.1 transgenic mice first immunized with two 1 μg doses of a primary series of BNT162b2 were subsequently given a third dose of vehicle, 1 μg BNT162b4, 1 μg BNT162b2, or separately formulated BNT162b2 and BNT162b4 at a 3:1 ratio of 1 μg BNT162b2 and 0.33 μg BNT162b4 (Figure 4 A).
Figure 4.
Impact of pre-existing BNT162b2 immunogenicity on vaccination with BNT162b4 combined with BNT162b2
(A) Schematic of in vivo experiment to assess impact of pre-existing S-specific immune responses (n = 4–8 mice/group).
(B) S protein S1-specific IgG titers at day 56 after vaccination, all dilutions performed in duplicate.
(C) S protein S1-IgG kinetics throughout treatment.
(D) 50% pseudovirus neutralization (pVN50) geometric mean titers (GMTs) against SARS-CoV-2 wild type strain in sera collected 14 days post-dose 3, showing lower limit of detection (LLOD).
(E) IFN-ɣ ELISpot T cell responses with peptide-pulsed splenocytes using an S protein peptide pool, triplicate.
(F) Polyfunctional CD8+ and CD4+ T cells from splenocytes peptide-pulsed using an S protein peptide pool.
(G) IFN-ɣ ELISpot T cell responses from splenocytes peptide-pulsed with BNT162b4-related peptide pools, triplicate. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. Error bars indicate standard error of the mean (SEM).
See also Figures S5 and S6.
After two doses of BNT162b2, immunization with BNT162b4 had no impact on S protein S1 IgG titers as compared to vehicle control (Figure 4B). Furthermore, total S protein S1 IgG levels showed that treatment with a third dose of co-administered BNT162b2 and BNT162b4 led to the generation of similar levels of antibodies against the S1 antigen as treatment with a third dose of BNT162b2 alone (Figure 4C). Compared to BNT162b2 alone, immunization with BNT162b2 co-administered with BNT162b4, either as primary vaccination series or as a third dose after BNT162b2 pre-immunization, showed similar levels of neutralizing antibodies (Figure 4D).
ELISpot assay analysis confirmed that T cell responses against the S protein were similar whether or not BNT162b4 was combined with BNT162b2 in the third immunization (Figure 4E, right). Animals receiving BNT162b4 alone in the third immunization exhibited higher S-specific T cell responses as compared with those receiving vehicle control for their third immunization (Figure 4E, left). ICS analysis further indicated that a third immunization of BNT162b4 alone or in combination with BNT162b2 leads to similar polyfunctional responses after peptide stimulation when compared with a third treatment of BNT162b2 alone (Figure 4F). Inclusion of BNT162b4 in the third dose led to strong T cell responses to the N, M, and ORF1ab segments, thus broadening the T cell response against additional SARS-CoV-2 target proteins (Figure 4G).
Additionally, in immunized K18-hACE2 mice, S-specific T cells seen in the peripheral blood of animals given BNT162b4 demonstrated similar frequencies as animals given BNT162b2 alone, indicating that the development of S-specific T cells were not impacted (Figure S6 ). S-specific T cells in animals given a third dose of either BNT162b4 alone or in combination with BNT162b2 showed a slightly higher prevalence of earlier memory phenotypes compared to animals given BNT162b2 alone (Figure S6H). Overall, immunization with a third dose of BNT162b4 alone or co-administered with BNT162b2 maintains S-specific immunity and induces a broader range of vaccine-induced T cells to include responses to the N, M, and ORF1ab antigens.
Figure S6.
Impact of pre-existing BNT162b2 immunity on vaccination with BNT162b4 co-administered with BNT162b2 in K18-hACE2 mice, related to Figure 4
(A) Schematic of the in vivo experiment to assess impact of pre-existing BNT162b2 immune responses (n = 6 animals per group). (B) S protein S1-specific IgG titers at day 56 after treatment, all dilutions performed in duplicate. (C) S protein S1-IgG kinetics throughout treatment. (D) Neutralizing titers of immunized animals at day 56 using a pVNT assay. (E) IFN-ɣ ELISpot T-cell responses with peptide-pulsed splenocytes using an S protein peptide pool, performed in triplicate. (F) Multimer staining for S-specific T cells in peripheral blood. (G) IFN-ɣ ELISpot T-cell responses from splenocytes peptide-pulsed with BNT162b4-related peptide pools. (H) Memory phenotype of S-specific T cells in the draining lymph nodes. The vehicle group was gated on total CD8+ T cells. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. Error bars indicate standard error of the mean (SEM).
T cell receptor repertoire and phenotype analysis for BNT162b4-vaccinated mice
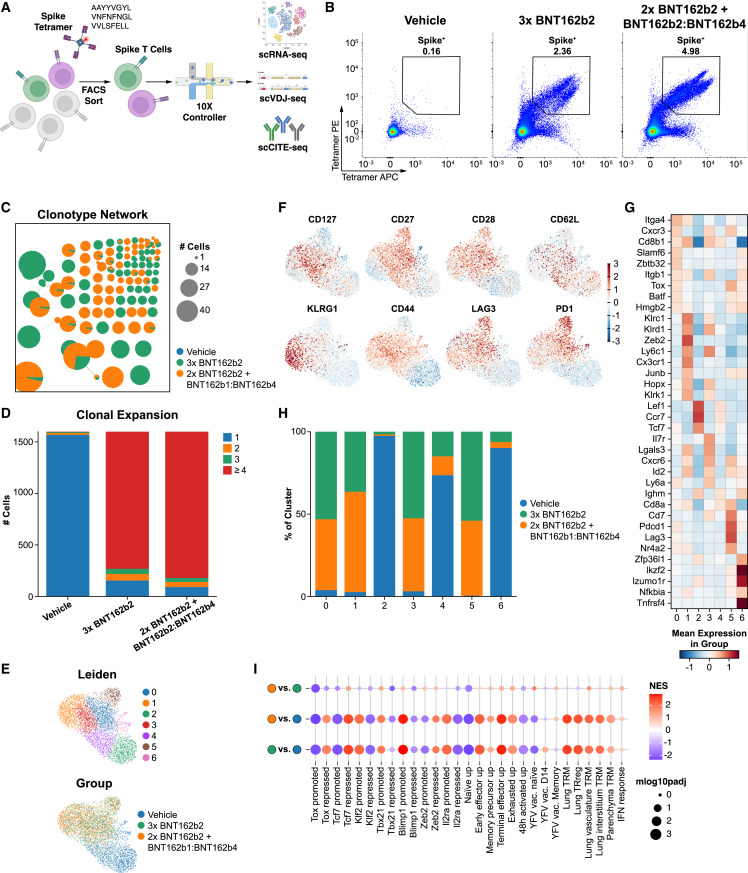
We sought to better understand the impact of the inclusion of BNT162b4 on the S-specific T cell phenotype and the T cell receptor (TCR) repertoire through single-cell RNA sequencing. HLA-A2.1 Tg animals were immunized with two doses of a primary series of BNT162b2 to generate S-specific immunity and then immunized with a subsequent dose of either BNT162b2 alone or a combination of BNT162b2 and BNT162b4, as described in Figure 4A. S-specific CD8+ T cells were enriched from splenocytes from multiple animals using MHC-tetramers loaded with three highly immunogenic S protein epitopes (Figure 5 A). Nucleic acids from T cells were isolated and used to generate VDJ Cellular Indexing of Transcriptomes and Epitopes (CITE)65 libraries to study the impact of the inclusion of BNT162b4 on T cell clonality, expansion, and functional phenotype (Figures 5A and 5B).
Figure 5.
Impact of BNT162b4 on the S-specific TCR repertoire and T cell phenotype
(A) Schematic of single-cell RNA sequencing experiment to generate GEX, CITE, and VDJ libraries of S-specific T cells of immunized animals (n = 4 animals/group).
(B) Representative plot of flow sorting gating for S-specific T cells for multimodal scRNA-seq using MHC tetramers.
(C) Clonotype network of largest clones (≥5 cells) from animals immunized with either three doses of BNT162b2 or 2 doses of BNT162b2 followed by a dose of combined BNT162b2 and BNT162b4. Each circle represents a unique CDR3 clone (both alpha/beta considered); circle size represents the number of cells with the corresponding TCR clone; connected circles represent clonotype clusters with similar CDR3 sequences.66
(D) Number of unique T cells in each treatment group belonging to clones containing 1, 2, 3, or ≥4 cells.
(E) UMAP representation of single-cell transcriptomes from sorted T cells, colored by either cluster assignment (top) or experimental group (bottom).
(F) Normalized and scaled cell surface expression of CITE-seq analysis of selected memory and activation markers.
(G) Heatmap of top differentially expressed (sorted by t test score) genes by cluster. Color scale indicates mean expression in the group.
(H) Cluster composition of cells from each treatment group.
(I) Gene set enrichment analysis of differentially enriched gene sets comparing immunized animals to control animals and between groups immunized with BNT162b2 alone versus a combination of BNT162b2 and BNT162b4. Color scale indicates normalized enrichment score (NES).
Analysis of VDJ sequences indicated that animals immunized with BNT162b4 in combination with BNT162b2 demonstrated a statistically significant increase in the number of clones undergoing clonal expansion of S-specific T cells when compared with animals immunized with BNT162b2 only (Figures 5C and 5D, as evaluated by chi-square test performed on an equal number of randomly subsampled cells, p value <4.83e-7 in 100 independent samplings). To examine the cell phenotypes, T cells were clustered applying the Leiden clustering algorithm to the gene expression profiles (Figure 5E). Gene expression data highlighted that immunization with both BNT162b4 and BNT162b2 led to both activated and memory clusters of T cells, similar to immunization with BNT162b2 alone (Figures 5E–5G). Additionally, CITE analysis showed both groups contained clusters of memory cells, as demonstrated by the presence of memory markers such as CCR7, CD62L, and CD27, and clusters of actively expanding T cells, as demonstrated by the upregulation of PD1, CD44, and KLRG1 (Figure 5F). We assessed the composition of each cluster with regards to the sample of origin: vehicle, a third immunization with BNT162b2 alone, or a third immunization with both BNT162b2 and BNT162b4. The cells from immunized animals were similarly distributed across clusters (especially clusters 0, 1, 3, and 5), clustering distinctly from unimmunized animals (clusters 2, 4, and 6; Figure 5H). Gene set enrichment analysis highlighted animals immunized with a combination of BNT162b4 and BNT162b2 showed a high degree of similarity to animals immunized with BNT162b2 alone (Figure 5I). Taken together, these data indicate that the phenotype and functionality of the S-specific T cells is maintained when BNT162b2 is administered alone or together with BNT162b4.
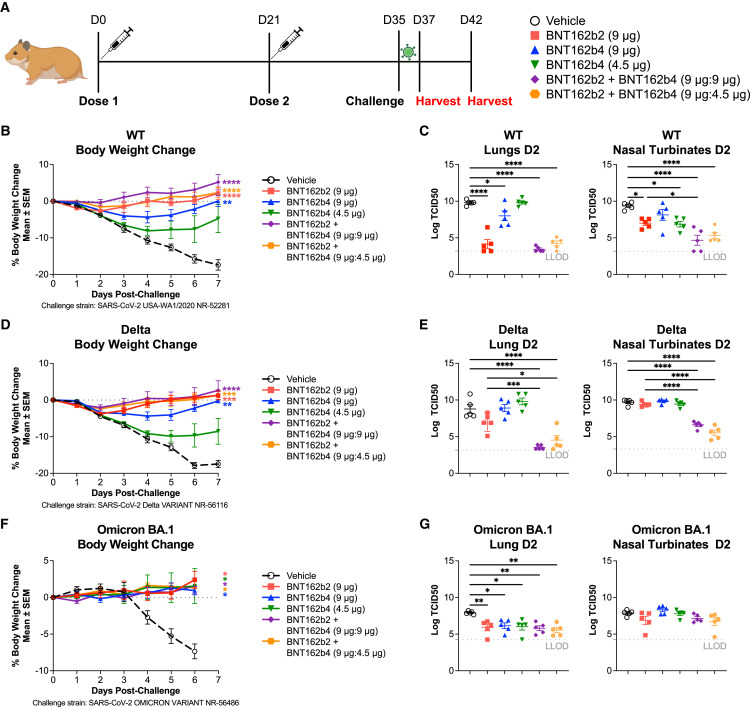
BNT162b4 confers protective immunity against viral challenge of golden Syrian hamsters
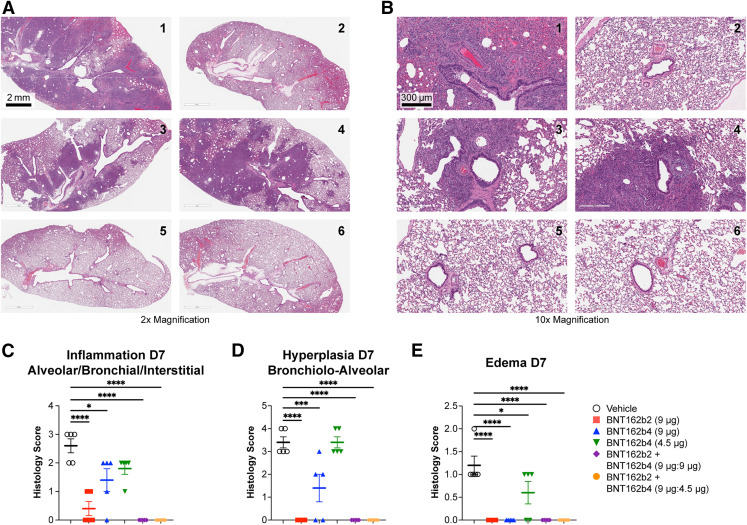
To determine whether T cell immunogenicity from BNT162b4 can lead to protection against severe SARS-CoV-2 infection, we immunized golden Syrian hamsters with two doses of 9 μg or 4.5 μg of BNT162b4 alone or in combination with BNT162b2 at various ratios. Animals were challenged with an intranasal inoculation of wild-type SARS-CoV-2 strain USA-WA1/2020 two weeks after the last immunization, and disease severity was measured by weight loss, viral titers, and tissue pathology in the lungs at harvest (Figures 6 A and S7 ). Immunization with BNT162b2 led to protection from the wild-type SARS-CoV-2 strain, as seen by lack of weight loss and negligible viral titers present in the lung and nasal turbinates (Figures 6B and 6C). Animals immunized with a combination of BNT162b2 and BNT162b4 were also protected against viral challenge, as seen by negligible viral titers in both the lungs as well as in nasal turbinates (Figures 6B and 6C). BNT162b4 immunization alone also led to significant protection against challenge in a dose-dependent manner, as indicated by a lower degree of weight loss compared to the vehicle control (Figure 6B). Viral titers in both the lungs and nasal turbinates from the animals immunized with BNT162b4 alone were also lower when compared to unimmunized animals, demonstrating that T cells play a role in limiting the severity of disease (Figure 6B). Animals immunized with the combination of BNT162b2 and BNT162b4 showed lower viral titers in nasal turbinates compared to animals immunized with BNT16b2 alone, indicating that a broader T cell response can improve the protection in the upper respiratory tract (Figure 6C). Analysis of lung pathology of animals seven days post-infection further supported the results from viral lung titer analysis. Animals immunized with BNT162b2 alone or in combination with BNT162b4 showed complete protection against SARS-CoV-2 infection associated inflammation, hyperplasia, and edema (Figure S7). Immunization with BNT162b4 alone at the higher dose led to a decreased severity of microscopic findings compared to unimmunized animals (Figure S7).
Figure 6.
BNT162b4 confers protective immunity in a golden Syrian hamster model
(A) Schematic of in vivo efficacy model in golden Syrian hamsters (n = 10 animals/group).
(B) Body weight change post challenge with WT SARS-CoV-2 in immunized animals.
(C) TCID50 viral titers from lung homogenates and nasal turbinates homogenates, day 2 post-infection, measurements in quadruplicate.
(D) Body weight change post challenge with Delta SARS-CoV-2 strain in immunized animals following the experimental schematic in (A).
(E) TCID50 viral titers from lung homogenates and nasal turbinates homogenates, day 2 post-infection from half of each cohort challenged with the Delta SARS-CoV-2 strain.
(F) Body weight change post challenge with Omicron BA.1 SARS-CoV-2 strain in immunized animals following the experimental schematic in (A).
(G) TCID50 viral titers from lung homogenates and nasal turbinates homogenates, day 2 post-infection from half of the cohort challenged with the Omicron BA.1 SARS-CoV-2 strain, measurements in quadruplicate.
Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. LLOD, lower limit of detection. Error bars indicate standard error of the mean (SEM). See also Figure S7.
Figure S7.
Lung pathology of golden Syrian hamsters post challenge with WT SARS-CoV-2, related to Figure 6
H&E staining from lungs of WT SARS-CoV2-infected animals 7 days post-infection: 1.3× magnification (A) and 10× magnification (B). SARS-CoV-2-related microscopic findings are characterized by dark staining consolidated areas; any areas perceived as dark in Groups 2 and 5 are considered artificial airway collapse due to poor formalin inflation. Histopathological score of lung tissue from infected animals 7 days post-infection for (C) inflammation, (D) hyperplasia, and (E) edema. Score was determined based on the percentage of findings in each area for each animal based on the following scoring: 0 – no pathological change; 1 – minimal <10% affected area; 2 – mild 10–25% area, 3 – moderate 25–50% area, 4 – marked 50–95% area. Statistical significance was assessed by one-way ANOVA with Dunnett’s multiple comparison post-test. Multiplicity-adjusted p values are shown. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p < 0.0001. Error bars indicate standard error of the mean (SEM).
To study the impact of BNT162b4 on protection against viral variants, hamsters were immunized with BNT162b2, BNT162b4, or their combination, and challenged with an intranasal inoculation of either the highly infectious Delta variant that demonstrates severe pathologies in humans67 and hamsters68 , 69 , 70 (Figures 6D and 6E) or the highly mutated Omicron BA.1 variant (Figures 6F and 6G). These data demonstrated protection against weight loss following challenge with either the Delta variant or the Omicron BA.1 variant. Viral titers in the lung and nasal turbinates following challenge with the Delta variant were significantly reduced in animals immunized with a combination of BNT162b2 and BNT162b4 (Figure 6E) as compared to animals immunized with BNT162b2 alone. The benefit conferred by immunization with a combination of both vaccines was more marked in the nasal turbinates compared to the lung, as similarly seen following the wild-type (WT) viral challenge. Compared to the Delta and WT challenges, challenge with the Omicron BA.1 variant demonstrated lower pathogenicity, as indicated by less severe weight loss and lower overall viral titers, as previously described.68 Following Omicron BA.1 challenge, all animals demonstrated reduced viral burden in the lungs, whether immunized with BNT162b2, BNT162b4, or their combination, and differences in nasal turbinate viral titers following Omicron challenge were less notable among groups (Figure 6G). Combined, these data suggest that immunity raised by BNT162b4 could be protective against symptoms caused by multiple viral variants of SARS-CoV-2 and, in combination with a variant-adapted BNT162b2, could reduce the likelihood that future variants cause severe disease in vaccinated individuals.
Discussion
Spike protein-encoding mRNA vaccines have been highly efficacious against disease caused by most dominant SARS-CoV-2 variants. However, the decrease in neutralizing antibody titers over time along with the rise of antigenically distinct S proteins resulted in increased breakthrough infections necessitating vaccines that could offer more durable immunity against emerging SARS-CoV-2 variants and associated disease. Recent studies have demonstrated that cellular immunity is far less impacted by viral variants, and strong, diverse cellular immune responses may counter-balance less effective neutralizing antibody titers.1 , 2 , 3 , 4 , 5 , 6 Additionally, several studies across various viral models have highlighted the importance of the T cell response in limiting viral replication and load18 , 19 , 20 , 38 through clearance of virally infected cells. Thus, inclusion of additional T cell antigens into vaccines has often been suggested to improve the efficacy and durability of the immune response in an ever-changing viral landscape.9 Unlike antibody targets, T cell antigens are not limited to extracellular proteins, and therefore many non-S antigens are a rich source of potential T cell epitopes. Aiming to develop an mRNA-encoded vaccine component with multiple, conserved T cell epitopes that could be used universally against all common SARS-CoV-2 variants, we focused our approach on conserved segments of immunogenic SARS-CoV-2 antigens. We designed and evaluated BNT162b4, an mRNA vaccine component encoding highly immunogenic segments of the N protein, M protein, and short segments containing minimal epitopes from the ORF1ab polyprotein.
Using targeted mass spectrometry, we showed that BNT162b4 generates T cell epitopes from all three encoded antigens that are presented on a broad set of HLA alleles. Concatenating segments from different viral antigens could lead to the generation of junctional epitopes that are not present in the SARS-CoV-2 proteome. By inclusion of a short stretch of amino acids that is expected to enhance cleavage,56 , 71 we aim to both increase the chances that the desired epitopes are processed from the polyprotein and reduce the likelihood of generation of junctional peptides.72 , 73 This is supported by the lack of detection of junctional epitopes by DDA MS and the frequent detection of junction-adjacent ORF1ab epitopes by our HLA ligandomics analysis, suggesting effective cleavage at these sites.
In vivo, BNT162b4 generates robust, multifunctional T cell responses against three antigens in multiple mouse models. The combination of BNT162b4 and BNT162b2 broadens the antigen-specific T cell repertoire but does not negatively impact the BNT162b2 induced neutralizing antibody response against the S protein. Similarly, co-administration of BNT162b4 with BNT162b2 does not impair the repertoire, frequency, and functional phenotype of S-specific T cells induced by BNT162b2. However, a decrease in cytokine secretion in N- and M-specific T cells is detected when BNT162b4 is given in combination with BNT162b2 compared with BNT162b4 alone. This may indicate some competition between the responses to the S protein versus the responses to N and M proteins. While the exact reason is unknown, it could potentially be caused due to differences in epitope abundance and presentation of BNT162b2 versus BNT162b4 epitopes or immunodominance of S-specific T cells. It is important to note that despite this potential interference, BNT162b4 broadens the total T cell responses to additional SARS-CoV-2 antigens.
Most importantly, we found that the immunity induced by BNT162b4 reduces disease severity in a SARS-CoV-2 hamster challenge model across various viral variants. Combining BNT162b2 with BNT162b4 enhances the protection against viral challenge, and this added benefit is most clearly demonstrated when hamsters are challenged with the Delta variant, which is characterized by a more severe disease pathology.74 The ability for viral-specific T cells to directly protect against severe disease has been suggested in several studies through correlations and preclinical experiments.18 , 19 , 20 , 38 Here, we directly demonstrate that the addition of the T cell-targeting vaccine component BNT162b4 further enhances immune protection to that conferred by BNT162b2 when the two are combined, as measured through lower viral titers, improved lung pathology, and a decrease of weight loss seen in animals challenged with multiple SARS-CoV-2 variants. Interestingly, when animals are challenged by the WT strain or the Delta variant, BNT162b4-driven immune responses confers protection that appears to be particularly influential in nasal turbinates, potentially suggesting a role for increased immunity in the mucosal membrane, where accessibility to IgG is limited and offers less protection.75 When animals are challenged with the Omicron BA.1 variant, the lack of detection of reduction in viral titers could potentially be due to the lower overall viral burden of the variant or the lower inflammatory response associated with the Omicron strain.68 , 76 The protection in the upper respiratory tract offered by the addition of BNT162b4, whether through the presence of tissue-resident memory T cells or T cell migration upon infection is particularly attractive, as it could point to a potential for a decreased transmission rate.77 , 78 , 79 , 80 This phenomenon is similar to a study that suggested that T cell responses to ORF1ab polyprotein could give rise to more abortive infections, potentially through targeting of early infected cells.39 Potent memory T cell responses activated rapidly following infection would likely mean less time for intra-host viral spread and activation of immune-evasion mechanisms.
While there is evidence for the importance of T cell responses in the control of SARS-CoV-2 infection, how a vaccine focused on raising T cell responses will help prevent COVID-19 in a clinical setting is unknown. In studies using mRNA S-protein-encoding vaccines, T cell responses against mutated variant epitopes stay largely intact; however, the ability of a vaccine to control infection depends on the subject’s neutralizing antibody levels to the infecting variant.5 , 81 , 82 This has also been reported in the macaque model where animals had decreased levels of neutralization titers against the beta variant and thus reduced control of infection, even when T cell responses were preserved.83 Additionally, studies monitoring the boosting of T cell responses after multiple vaccinations show that S-specific T cells do not expand more than after an individual’s primary vaccination series even though protection against infection is increased in correlation with neutralizing antibody levels.84 , 85 The extent to which T cells protect against infection is unknown; however, data demonstrating that T cells play a critical role in protective immunity against severe disease caused by SARS-CoV-2 in the hamster model warrant further testing of BNT162b4 in combination with an S-protein-encoding vaccine in humans. The beneficial effects of inclusion of the T cell-targeting vaccine BNT162b4 with the BA.4/BA.5 Omicron-adapted BNT162b2 is currently being evaluated in an ongoing clinical trial (NCT05541861).
Limitations of the study
All immunogenicity and challenge studies were performed in animal models, which may not translate to humans. In addition, BNT162b4 was optimized for antigen presentation on HLA complexes, not mouse or hamster MHCs. However, our mass spectrometry analysis confirms that epitopes from BNT162b4 are indeed processed and presented onto HLA complexes in human cell lines. We therefore expect that the immunogenicity and protection observed in these animal models will translate to humans.
For a SARS-CoV-2 challenge model, we have opted to use the golden Syrian hamster model due to its relative adherence to SARS-CoV-2 immunopathology in humans, as compared to well-described mouse models of efficacy.74 , 86 Although the dose we used in our SARS-CoV-2 challenge does not lead to lethality, virally challenged hamsters have marked weight loss and lung pathology, allowing it to serve as a useful model to study immune protection and disease severity, especially in adherence to human pathophysiology. The non-lethal hamster model is more reflective of natural infection and the protective role of the different immune components (compared with a very artificially high dose needed in lethal challenge). To further expand on this topic, we used three different variants of SARS-CoV-2 (Ancestral, Delta, and Omicron) in these hamster challenges to assess various disease severities and BNT162b4-induced protection. It was important to assess the ability of BNT162b4 to control severe disease with different SARS-CoV-2 variants, and thus the hamster model was preferable to a mouse adapted SARS-CoV-2 model. When using human-infecting SARS-CoV-2 strains in the K18-hACE2 mouse model, the expression of hACE2 in the mouse brain in the K18-hACE2 model87 leads to pathology and mortality not only due to respiratory pathology, but also due to neurotoxicity.73 , 85 Since we believe the residence of T cells within the appropriate tissues may play a role in control of disease, we chose an animal model that is physiologically relevant to human disease. Furthermore, the expected mechanism of action for T cell responses in viral clearance makes the non-lethal model suitable to study T-cell-targeting vaccines.
All in vivo animal studies were performed in SARS-CoV-2-naive models. This model may have been appropriate at the outset of the pandemic, but a significant portion of the human population has been infected by SARS-CoV-2 and may have pre-existing memory T cells to non-S viral antigens. In addition, many individuals have been vaccinated with inactivated virion vaccines, which also raise immunity that is not restricted to the S protein. The S-specific cellular immunity raised by BNT162b2 has been shown to be superior to that raised by an inactivated virus vaccine.88 , 89 This suggests that BNT162b4 could boost memory T cell responses or induce de novo responses in these subjects, which was not studied in our preclinical models. While we did not study the effect of BNT162b4 in SARS-CoV-2-experienced animal models, we expect that BNT162b4 would boost the existing T cell immunity acquired during infection, providing additional protection. Therefore, our studies in naive models likely underestimate the magnitude of the immunogenicity of BNT162b4, and administration to individuals with prior immunity may offer even further enhanced protection.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD45.2 BV421 | Biolegend | 109832; RRID:AB_2565511 |
| Anti-mouse CD3 PE-Cy7 | Biolegend | 100220; RRID:AB_173205 |
| Anti-mouse CD19 BV650 | Biolegend | 115540; RRID:AB_2563067 |
| Anti-mouse CD4 BV510 | Biolegend | 100559; RRID:AB_2562608 |
| Anti-mouse CD8 AF488 | Biolegend | 100723; RRID:AB_389304 |
| Anti-mouse CD69 APC | Biolegend | 104514; RRID:AB_492843 |
| Anti-mouse IFN-ɣ AF700 | Biolegend | 505823; RRID:AB_2561299 |
| Anti-mouse IL2 BV785 | Biolegend | 503843; RRID:AB_2832801 |
| Anti-mouse TNFα BV650 | Biolegend | 506333; RRID:AB_2562450 |
| Anti-mouse CD62L PE | Biolegend | 104408; RRID:AB_313095 |
| Anti-mouse CD44 BV711 | Biolegend | 103057; RRID:AB_2564214 |
| Anti-mouse PD1 PerCPCy5.5 | Biolegend | 109120; RRID:AB_2566641 |
| Anti-mouse CD25 BUV395 | BD Biosciences | 564022; RRID:AB_2722574 |
| Anti-mouse CD27 BUV805 | BD Biosciences | 741959; RRID:AB_2871267 |
| Anti-mouse CD107a PEDazzle594 | Biolegend | 121624; RRID:AB_2616690 |
| Anti-pan-HLA-I clone W6/32 | BioXCell | BE0079; RRID: AB_1107730 |
| Anti-HLA-A02 clone BB7.2 | BioXCell | Produced custom from ATCC hybridoma HB-82 |
| Anti-mouse CD8 TotalSeqC | Biolegend | 100785; RRID:AB_2819776 |
| Anti-mouse CD62L TotalSeqC | Biolegend | 104455; RRID:AB_2819800 |
| Anti-mouse CD44 TotalSeqC | Biolegend | 103063; RRID:AB_2813930 |
| Anti-mouse PD1 TotalSeqC | Biolegend | 109127; RRID:AB_2819819 |
| Anti-mouse CXCR5 TotalSeqC | Biolegend | 145537; RRID:AB_2814051 |
| Anti-mouse CD27 TotalSeqC | Biolegend | 124245; RRID:AB_2832448 |
| Anti-mouse CD39 TotalSeqC | Biolegend | 143815; RRID:AB_2819888 |
| Anti-mouse CD127 TotalSeqC | Biolegend | 135047; RRID:AB_2819874 |
| Anti-mouse TIM3 TotalSeqC | Biolegend | 119739; RRID:AB_2832424 |
| Anti-mouse LAG3 TotalSeqC | Biolegend | 125237; RRID:AB_2832450 |
| Anti-mouse CD28 TotalSeqC | Biolegend | 102133; RRID:AB_2832287 |
| Anti-mouse CD69 TotalSeqC | Biolegend | 104551; RRID:AB_2832333 |
| Anti-mouse CD38 TotalSeqC | Biolegend | 102735; RRID:AB_2819786 |
| Anti-mouse KLRG1 TotalSeqC | Biolegend | 138433; RRID:AB_2800649 |
| Anti-mouse CD95 TotalSeqC | Biolegend | 152616; RRID:AB_2860719 |
| Anti-mouse CD138 TotalSeqC | Biolegend | 142538; RRID:AB_2860696 |
| Anti-mouse CD273 TotalSeqC | Biolegend | 107229; RRID:AB_2860615 |
| Anti-mouse CD73 TotalSeqC | Biolegend | 127237; RRID:AB_2860665 |
| Anti-mouse CD80 TotalSeqC | Biolegend | 104755; RRID:AB_2819807 |
| Goat anti-mouse IgG HRP | 115-035-071 | Jackson ImmunoResearch; RRID: AB_2338506 |
| Goat anti-rabbit IgG HRP | A0545-1mL | Sigma-Aldrich; RRID: AB_257896 |
| Human SARS coronavirus spike S1subunit antibody | Sino Biological | 40150-RP01; RRID: AB_2827979 |
| Human SARS-CoV/SARS-CoV-2-Nucleocapsid Antibody | Sino Biological | 40143-MM05; RRID:AB_2827977 |
| Bacterial and virus strains | ||
| SARS-CoV-2 WT (USA-WA1/2020) | BEI Resources | NR-52281 |
| SARS-CoV-2 delta variant | BEI Resources | NR-56116 |
| SARS-CoV-2 omicron BA.1 variant | BEI Resources | NR-56486 |
| Chemicals, peptides, and recombinant proteins | ||
| Sodium bicarbonate | Sigma-Aldrich Chemie GmbH |
S5761 |
| Sodium carbonate | Sigma-Aldrich Chemie GmbH |
S7795 |
| Sodium pyruvate | Gibco | 11360–070 |
| Sulfuric acid 25% EMSURE® |
VWR International GmbH |
1007161000 |
| TMB One (3,3′,5,5′-Tetramethylbenzidine) ready-to-use-solution | Biotrend Chemikalien GmbH |
4380A |
| Tween 20 | Carl Roth GmbH & Co. KG | 9127.1 |
| Live/Dead fixable Near-IR | Thermo Fisher | L10119 |
| AOPI | Nexcelom | CS2-106-25mL |
| Monensin | eBioscience/invitrogen | 00-4505-51 |
| Brefeldin A | Biolegend | 420601 |
| BD Cytofix | BD biosciences | 554655 |
| BD Cytofix/Cytoperm Kit | BD biosciences | 554714 |
| Lipofectamine® LTX & PLUS | Thermo Fisher | 15338100 |
| Potassium bicarbonate | AppliChem GmbH | A2375,1000 |
| Casein blocking buffer | Sigma-Aldrich | B6429 |
| Concanavalin A | Sigma-Aldrich | C0412-5MG |
| Isoflurane | Piramal Critical Care | 9714675 |
| Heavy isotopically labeled peptides | BioNTech US (in-house) | N/A |
| TMTzero Label Reagent | Thermo Fisher | 90067 |
| TMT11-131C Label Reagent | Thermo Fisher | A37724 |
| BirA biotin ligase | BioNTech US (in-house) | N/A |
| PepMix™ SARS-CoV-2 (Spike Glycoprotein) | JPT | PM-WCPV-S-2 |
| Nucleocapsid peptide pool | BioNTech US (in-house) | N/A |
| Membrane peptide pool | BioNTech US (in-house) | N/A |
| ORF1ab peptide pool | BioNTech US (in-house) | N/A |
| Critical commercial assays | ||
| Mouse IFN-ɣ ELISpot kit | R&D Systems | EL485 |
| Chromium Next GEM Automated Single Cell 5′ cDNA Kit v2, 24 rxns | 10X Genomics | 1000425 |
| Chromium Next GEM Chip K Automated Single Cell Kit, | 10X Genomics | 1000297 |
| Automated Library Construction Kit, | 10X Genomics | 1000428 |
| Dual Index Kit TT Set A | 10X Genomics | 1000215 |
| Chromium Automated Single Cell Mouse TCR Amplification & Library Construction Kit, | 10X Genomics | 1000310 |
| Chromium Automated Single Cell 5′ Feature Barcode Library Construction Kit | 10X Genomics | 1000455 |
| Dual Index Kit TN Set A, | 10X Genomics | 1000250 |
| V-PLEX Plus Proinflammatory Panel1 Mouse Kit | Meso Scale Discovery | K15048G |
| Deposited data | ||
| Mass spectrometry RAW files | https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp | MassIVE repository: MSV000091008 |
| R code for epitope analysis and figures | Zenodo and GitHub | Zenodo https://doi.org/10.5281/zenodo.7789841; https://github.com/YolandaHuitianDiao/BNT162b4_epitopes.git |
| Python code for single cell sequencing analysis | Zenodo and GitHub | Zenodo https://doi.org/10.5281/zenodo.7789841; https://github.com/YolandaHuitianDiao/BNT162b4_scRNA.git |
| Single cell RNA-seq RAW files | NCBI GEO | GEO: GSE222403 |
| Experimental models: Cell lines | ||
| A375 | ATCC | CRL-1619 |
| Vero-76 cells | ATCC | CRL-1587 |
| Calu-3 cells | ATCC | HTB-55 |
| Experimental models: Organisms/strains | ||
| BALB/C | Janvier Labs | BALB/cJRj |
| HLA-A2.1 random transgenic CB6F1-Tg(HLA-A∗0201/H2-Kb)A∗0201 |
Taconic | HLA-A2.1 (CB6F1) |
| K18 h-ACE2 B6.Cg-Tg(K18-ACE2)2Prlmn/J |
Jackson Laboratories | 034860 |
| Golden Syrian Hamster | Envigo | HsdHAN-AURA |
| Recombinant DNA | ||
| VSV-ΔG-GFP vector | Kerafast | EH1004 |
| pcDNA3.1-derived expression plasmid | Invitrogen | V79020 |
| Software and algorithms | ||
| Excel | Microsoft | N/A |
| GraphPad Prism 8 | GraphPad Software Inc. | N/A |
| Gen5 software 3.0.9 | BioTek | N/A |
| ImmunoCapture 7.0.13.1 | ImmunoSpot | N/A |
| ImmunoSpot® analysis software version 7.0.26.0 | ImmunoSpot | N/A |
| IncuCyte Live Cell Analysis system | Satorius | N/A |
| BD FACSDiva software version 8.0.1 | BD Biosciences | N/A |
| FlowJo v 10.7.2 | BD Biosciences | N/A |
| cellranger-6.0.1 | 10x Genomics | N/A |
| R v 4.1.0 | The R Foundation | N/A |
| RStudio | Posit | N/A |
| Python 3.9.15 | Python Software Foundation | N/A |
| Scanpy | PyPI | N/A |
| Scirpy | PyPI | N/A |
| Muon | PyPI | N/A |
| Spectrum Mill v BI.07.04.210 | The Broad Institute of MIT and Harvard | N/A |
| Interactive Peptide Spectral Annotator tool | Brademan et al.90 | http://www.interactivepeptidespectralannotator.com/PeptideAnnotator.html |
| Skyline-daily v 21.2.1.424 or newer | University of Washington | N/A |
| Xcalibur v 4.6.67.17 or newer | Thermo Fisher | N/A |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Asaf Poran (asaf.poran@biontech.us).
Materials availability
There are restrictions to the availability of the DNA/mRNA constructs presented in the study, subject to completion of a material transfer agreement.
Experimental model and subject details
Animals and study design
Female HLA-A2.1 random transgenic animals (Taconic) were used at 5–10 weeks of age. HLA-A2.1 mice were housed under barrier and specific-pathogen-free (SPF) conditions in individually ventilated cages (Innovive individually ventilated cages, kept under positive pressure during the study) with a maximum of five animals per cage. The temperature and relative humidity in the cages and animal unit were kept at 20–24°C and 45–55%, respectively, and the air change (AC) rate in the cages was 75 AC/hr. Cages contained corn cob type bedding (The Anderson’s) and additional nesting material was changed weekly. Autoclaved Harlan Teklad 2918 irradiated diet and autoclaved acidified water from Innovive were provided ad libitum and changed at least once weekly. All materials were autoclaved prior to use. These studies were conducted in compliance with CRL IACUC under protocol No. I034.
Female BALB/c mice (Janvier labs) and K18-hACE2 (Jackson Laboratory) were used at 8–10 weeks of age. All experiments and protocols involving BALB/c mice were approved by the local authorities (local animal welfare committee), conducted according to the Federation of European Laboratory Animal Science Associations (FELASA) recommendations and in compliance with the German animal welfare act and Directive 2010/63/EU. Only animals with an unobjectionable health status were selected for testing procedures. Mice were housed at BioNTech SE’s animal facility under barrier and specific-pathogen-free (SPF) conditions in individually ventilated cages (Sealsafe GM500 IVC Green Line) with a maximum of five animals per cage. The temperature and relative humidity in the cages and animal unit were kept at 20–24°C and 45–55%, respectively, and the AC rate in the cages was 75 AC/hr. Cages contained dust-free bedding made of debarked chopped aspen wood (Abedd LAB & VET Service GmbH) and additional nesting material was changed weekly. Autoclaved ssniff M-Z food (sniff Spezialdiäten GmbH) and autoclaved tap water were provided ad libitum and changed at least once weekly. All materials were autoclaved prior to use.
Golden Syrian hamsters (Envigo) were used at 6–8 weeks of age. 50% of animals were male and 50% of animals used were female. Housing and handling of the animals were performed in accordance with the standards of the AAALAC International, the Animal Welfare Act as amended, and the Public Health Service Policy. Handling of animals was in compliance with the Biosafety in Microbiological and Biomedical Laboratories (BMBL), fifth edition (Centers for Disease Control). These studies were performed under IACUC-approved protocol no. 22–035. Animals were cared for and housed in ABSL-2/ABSL-3 containment with labeled cages, up to five hamsters per cage. Animal room temperatures were maintained at 20-26°C. The relative humidity of animal rooms were maintained at 30–70%. Lab Diet 5001 rodent diet (Ref # RHI 5021) was supplied to the animals and available ad libitum during the entire study period. Water was available ad libitum during the entire study. Samples of the water were analyzed by BIOQUAL for specified microbiological content and for chlorine level, pH hardness, and heavy metals. No contaminants that would affect the results of the study were known to be present in the water. BIOQUAL provided environmental enrichment in accordance with OLAW requirements, and in compliance with the Guide for the Care and Use of Laboratory Animals.
Cell lines and primary culture
A375 cell lines stably expressing biotin acceptor peptide (BAP)-tagged HLA alleles used for ligandomics experiments were cultured in DMEM +10% FBS +5% penicillin/streptomycin. For titration of VSV/SARS-CoV-2-S pseudovirus, Vero 76 cells (ATCC CRL-1587) were cultured in DMEM and 10% FBS. Primary splenocytes from animal experiments were cultured in X-VIVO +1% pen-Step +1% Glutamax. Calu-3 cells were cultured in EMEM +10% FBS.
SARS-CoV-2 virus stocks
The SARS-Cov-2 USA-WA1/2020 isolate (NR-52281), Delta (NR-56116) and Omicron BA.1 (NR-56486) were used in hamster challenge experiments (obtained from BEI resources). Virus was expanded in Calu-3 cells (ATCC HTB-55). All experiments with SARS-CoV-2 were performed in biosafety level 3 (BSL3) containment laboratories.
Method details
BNT162b4 vaccine design
In the design of BNT162b4, the M and N protein-encoding segments and the groups of sequences encoding ORF1ab proteins were concatenated using four amino acids linkers that are intended to maximize cleavability of minimal epitopes that are immediately following an N or M sequences. To promote cleavage and release of ORF1ab minimal epitope sequences themselves, the segments were arranged in an order promoting cleavage. A secretion signal from the SARS-CoV-2 S protein was added to the N-terminus of the string separated by a non-immunogenic linker,91 and an S protein transmembrane domain was added to the C-terminus of the string separated by another non-immunogenic linker to enhance trafficking to the endosomal compartment.92 BNT162b4 uses the same RNA platform including performance-optimizing backbone design, nucleoside-modified chemistry, and lipid nanoparticle (LNP) formulation as BNT162b2.41
Literature survey of experimentally-validated epitopes included in BNT162b4
A retrospective survey of literature-reported epitopes (as of December 2022) was done to compare to known epitopes circa September 2020 (Figure 1B). The data included in this figure are compiled from Agerer et al.93; Bilich et al.94; Ferretti et al.33; Francis et al.58; Gangaev et al.43; Gauttier et al.44; Habel et al.95; Hu et al.96; Joag et al.97; Kared et al.59; Lee et al.98; Lineburg et al.99; Liu et al.100; Mateus et al.45; Nagler et al.101; Nelde et al.46; Nguyen et al.102; Nielsen et al.103; Obermair et al.104; Peng et al.47; Poran et al.48; Rha et al.105; Saini et al.60; Schreibing et al.106; Schulien et al.30; Sekine et al.49; Shomuradova et al.107; Snyder et al.35; Tarke et al.36; Weingarten-Gabbay et al.54; and Zhao et al.13 A sequence may be counted more than one time per study if the epitope appears in overlapping test peptides.
Transfection of BNT162b4 mRNA into A375 MAPTAC cell lines
A375 cell lines stably expressing biotin acceptor peptide (BAP)-tagged HLA alleles56 , 57 were cultured in DMEM +10% FBS +5% penicillin/streptomycin. 50 million cells were plated in a T225 flask and allowed to adhere overnight. The next day, cells were transfected with 250 μg BNT162b4 RNA using Lipofectamine MessengerMAX reagent according to the manufacturer’s protocol. After 24 h of transfection, cells were harvested, washed with PBS, pelleted, and stored at −80°C until processing for MS analysis.
Isolation of HLA-I peptide complexes
Frozen cell pellets were thawed on ice for 20 min then gently lysed by hand pipetting in cold lysis buffer [20 mM Tris-Cl pH 8, 100 mM NaCl, 6 mM MgCl2, 1.5% (v/v) Triton X-100, 60 mM octyl B-D-glucopyranoside, 0.2 mM 2-iodoacetamide, 1 mM EDTA pH 8, 1 mM PMSF, 1X cOmplete EDTA-free protease inhibitor cocktail (Roche)] at a ratio of 1.2 mL lysis buffer per 50 x 106 cells. Lysates were incubated end-over-end at 4°C for 15 min with Benzonase nuclease (Millipore-Sigma) at a ratio of >250 units Benzonase per 50 x 106 cells to degrade DNA/RNA, then centrifuged at 15,000 x g at 4°C for 20 min and passed through a 0.2 μm filter to remove cellular debris and insoluble materials. For pan-class-I HLA and HLA-A∗02:01-specific pulldowns, cleared supernatants were transferred directly onto Gammabind Sepharose resin (Cytiva Life Sciences) charged with W6/32 (BioXCell BE0079) or BB7.2 (BioXCell produced custom from ATCC hybridoma HB-82) antibody, respectively. The BB7.2 and W6/32 beads were prepared by washing the Gammabind Sepharose resin three times with 1 mL cold PBS, charging overnight with antibody at a ratio of 100 μg antibody and 75 μL bead slurry per 50 x 106 cells, and washing three more times with 1 mL PBS prior to applying the lysate. For monoallelic affinity pulldowns, cleared supernatants were transferred to new 5 mL Protein LoBind Eppendorf tubes and BAP-tagged HLA molecules were biotinylated by incubating end-over-end at room temperature (RT) for 10 min in 5 mL tubes with 0.56 μM biotin, 1 mM ATP/1 mM magnesium acetate, and 3 μM BirA. The supernatants were incubated end-over-end at 4°C for at least 30 min with a volume corresponding to 200 μL of Pierce high-capacity NeutrAvidin beaded agarose resin (Thermo Fisher Scientific) slurry per 50 x 106 cells to affinity-enrich biotinylated-HLA-peptide complexes. The NeutrAvidin beaded agarose resin was washed three times with 1 mL cold PBS before use. The HLA-bound resins were transferred to a 96-well filter plate (Agilent) for all washing and elution steps. Prior to the transfer, the filter plate was conditioned twice with 1 mL methanol, and once each with 1 mL acetonitrile, 1 mL 1% (v/v) formic acid, and 1 mL wash buffer.
HLA-I peptide elution and filtration
The HLA-bound resins were washed four times with 1 mL wash buffer (20 mM Tris-HCl pH 8, 100 mM NaCl, 60 mM octyl-B-glucopyranoside, 0.2 mM 2-iodoacetamide, 1 mM EDTA pH 8), followed by four washes of 1 mL 10 mM Tris pH 8.0. All steps were drained to waste using nitrogen gas on a positive-pressure manifold at 3–6 psi. Spouts of the filter plate were wiped with an ethanol soaked Kimwipe to remove residual detergent prior to elution. Peptides were eluted directly into a pre-conditioned 96-well AcroPrep Advanced 10K molecular weight cut-off (MWCO) filter plate (Pall) stacked on top of a 96-well deep-well collection plate. Prior to use, the MWCO filter plate was passivated with 1% (v/v) of BSA in PBS for at least 1 h, rinsed with 2 x 400 μL of water, then acidified with 400 μL of 10% (v/v) acetic acid. The second water rinse and acidification steps were centrifuged through the MWCO filter at 2,000 x g for 15 min. The filter plate loaded with HLA-bound resin was stacked on top of the MWCO plate, and the resin was incubated twice for 5 min with 200 μL 10% (v/v) acetic acid to dissociate peptides from HLA molecules. The eluate was drained into the MWCO plate by applying nitrogen gas at ∼1 psi. During the first incubation, 100 fmol of Pierce peptide retention time calibration mix were added to the sample. Following the acetic acid elutions, the resin was washed with 200 μL 1.5 M Trizma base pH 8.5 (Sigma) and the elution was transferred to the MWCO plate by applying nitrogen gas at ∼1 psi. The MWCO plate was stacked on top of a 2 mL Protein LoBind 96-well collection plate (Eppendorf), and MWCO ultrafiltration was performed at 2,000 x g for 30 min. Following ultrafiltration, an additional 400 μL 1.5 M Trizma base pH 8.5 was added to the top of the MWCO plate, and the samples were centrifuged again at 2,000 x g for 30 min.
Reduction, alkylation, and on-column TMT modification
Following ultrafiltration, samples were tested to ensure the pH was between 6 and 8.5. For samples where the pH was too low, 1.5 M Trizma base pH 8.5 was added until proper pH was achieved. Cysteine sulfhydryls were reduced with the addition of TCEP to 5 mM for 30 min at 60°C and alkylated with the addition of iodoacetamide to 15 mM for 30 min at room temperature in the dark. Samples were acidified prior to desalting with the addition of neat formic acid (10% v/v final).
Desalting and on-column TMT labeling were performed on a Waters Sep-Pak C18 μElution plate stacked on top of a waste collection reservoir. Unless otherwise noted, all steps were drained to waste by applying 3–6 psi of nitrogen gas on a positive-pressure manifold. The C18 plate was equilibrated twice with 500 μL of methanol, followed by 250 μL 99.9% acetonitrile with 0.1% (v/v) formic acid, and four times with 1% (v/v) formic acid. Samples were then loaded onto the plate and pushed over the C18 resin at 1–2 psi. The deep-well sample collection plate was rinsed twice with 500 μL of 1% (v/v) formic acid to remove residual sample, and the rinse volume transferred to the C18 plate. TMTzero reagent (Thermo Fisher Scientific) was equilibrated to room temperature for at least 3 min prior to resuspension in 15 μL anhydrous acetonitrile with vigorous vortexing. The TMTzero reagent was further diluted in 500 μL 100 mM HEPES pH 8.5. The diluted TMTzero reagent was added to the C18 μElution plate and pushed over the C18 resin at ∼1 psi (about 5 min for the full volume to drain). Unreacted TMT was quenched with the addition of 500 μL 1% (v/v) hydroxylamine in 100 mM Tris at ∼1 psi. The C18 plate was washed four times with 500 μL 1% (v/v) formic acid. Peptides were eluted into a PCR receiver plate (Bio-Rad) using a stepwise gradient of 20 μL 15% acetonitrile with 1% (v/v) formic acid, 20 μL of 50% acetonitrile with 1% (v/v) formic acid, and 20 μL 80% acetonitrile with 1% (v/v) formic acid. Elution steps were performed on a tabletop centrifuge at 300 x g at room temperature for 30 s. Eluates were frozen, dried by vacuum centrifugation, and stored at −20°C until analysis.
HLA-I-peptide sequencing by MS
Samples were resuspended by adding 11 μL 3% (v/v) acetonitrile, 5% (v/v) formic acid supplemented with 145–200 fmol of each TMT-131C labeled heavy synthetic peptide per 8 μL injection. Samples were vortexed vigorously to resuspend the peptides and then briefly centrifuged to collect the sample in the bottom of each well. All nLC-MS/MS analyses employed the same liquid chromatography separation conditions described below. Peptides were chromatographically separated using an EASY-nLC 1200 System (Thermo Fisher Scientific) fitted with a PicoFrit 75 μm inner diameter and 10 μm emitter Nanospray column (New Objective) packed at 1,000 psi of pressure with helium to 30 cm with ReproSil-Pur 120 Å C18-AQ 1.9 μm packing material (Dr. Maisch GmbH) and heated at 60°C during separation. The column was equilibrated with 10X bed volume of solvent A [3% (v/v) acetonitrile/0.1% (v/v) formic acid], samples were loaded in 8 μL 3% (v/v) acetonitrile/5% (v/v) formic acid. Peptides were eluted into an Exploris 480 Orbitrap Mass Spectrometer (Thermo Fisher Scientific) equipped with a Nanospray Flex Ion source at an electrospray voltage of 2.0 kV.
For targeted MS analysis, 8 μL of the 11 μL sample was injected. Peptides were eluted with a linear gradient of 6–40% Solvent B [80% (v/v) acetonitrile/0.1% (v/v) formic acid] over 54 min, 40–100% Solvent B over 40 min, then held at 100% Solvent B for 5 min and 50% Solvent B for 9 min to wash the column. Linear gradients ran at a rate of 200 nL/min. Data were acquired using internal standard triggered parallel reaction monitoring.108 Fast, low-resolution precursor scans were used to look for m/z values in an inclusion list associated with the TMT-131C labeled heavy synthetic peptide internal standards. When an m/z value from the inclusion list was observed, a fast, low-resolution tandem mass spectrum (MS/MS) survey scan was performed, and characteristic fragment ions associated with the peptide were monitored. If five or more monitored ions were observed within a 15 ppm tolerance, a second MS/MS scan was performed with a mass offset equal to the difference between the TMT-131C labeled heavy synthetic peptide and the TMTzero labeled target HLA peptide. The second MS/MS scan was performed with isolation window set to 0.7 m/z, first mass set to 135 m/z, normalized HCD collision energy set to 38%, normalized AGC target set to 300%, maximum fill time set to 250 ms, Orbitrap resolution set to 60K, and data type set to centroided. The master cycle time was set to 3 s between successive precursor scans.
For unbiased discovery mode analysis, 2 μL of the 11 μL sample was injected. Peptides were eluted with a linear gradient of 6–30% Solvent B [80% (v/v) acetonitrile/0.1% (v/v) formic acid] over 84 min, 30–60% Solvent B over 9 min, 60–90% Solvent B over 1 min, then held at 90% Solvent B for 5 min and 50% Solvent B for 9 min to wash the column. Linear gradients ran at a rate of 200 nL/min. Precursor scans were acquired with a mass range of 350–1700 m/z, AGC set to 300%, maximum fill time set to auto, Orbitrap resolution set to 60K, and data type set to profile. Precursor ions with charges between 1 and 4 were selected for fragmentation, with precursors with a charge of 1 isolated only if the m/z was between 700 and 1700 m/z. MS/MS scans were performed with isolation window set to 0.7 m/z, scan range set to 120–1700, normalized HCD collision energy set to 32%, normalized AGC set to 100%, maximum fill time set to auto, Orbitrap resolution set to 45K, and data type set to centroid.
MS data analysis
Targeted data analysis was performed using Skyline-daily software (version 21.2.1.424 or newer). HLA peptides were identified based on matching retention time and fragmentation pattern with the TMT-131C heavy labeled synthetic peptides. Spectral visualizations were generated using the Interactive Peptide Spectral Annotator tool90 with extracted ion chromatogram (XIC) data exported from each RAW file using Qual Browser Thermo Xcalibur software. Skyline was used to identify peptide retention time, and the Peptide Spectral Annotator tool was used to confirm endogenous peptide detections by comparing endogenous and synthetic peptide spectra. Oftentimes, targeted analyses of low abundant peptides yield spectra where ion intensities associated with the targeted peptide are very low compared to the base peak of the spectrum. In these instances, to aid in the identification of a potential hit, a second “zoomed” spectrum was generated. In a zoomed spectrum, peaks from the endogenous peptide spectrum greater than the most intense annotated fragment ion in the corresponding synthetic peptide spectrum were deleted.
Tandem mass spectra (MS/MS) acquired with data-dependent acquisition (DDA) were interpreted using Spectrum Mill MS Proteomics software package v6.0 pre-release (Agilent Technologies). MS/MS spectra were excluded from searching if they did not have a precursor MH+ in the range of 600–2500, had a precursor charge >5, or had a minimum of <5 detected peaks. Merging of similar spectra with the same precursor m/z acquired in the same chromatographic peak was disabled. Protease specificity was set to none. Cysteine carbamidomethylation and an iterative TMTzero-TMT131C mix was set as a fixed modification. Methionine oxidation, cysteine cysteinylation, peptide N-terminal glutamine to pyroglutamic acid, 13C5 15N valine, 13C6 15N leucine, 13C6 15N isoleucine, 13C6 15N2 lysine, and 13C6 15N4 arginine were set as variable modifications. Datasets were searched against the BNT162b4 protein sequence concatenated to databases containing the human proteome (UCSC Genome Browser hg19 annotation; 63,691 entries) and a database of 264 common assay contaminants.109 A precursor mass tolerance of ±10 ppm, product mass tolerance of ±10 ppm, and a minimum scored peak intensity of 30% was used. Peptide spectrum matches (PSMs) for individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module to apply target-decoy based FDR estimation at the PSM rank to set scoring threshold criteria. An auto thresholds strategy using a minimum sequence length of 7, automatic variable range precursor mass filtering, and score and delta Rank1 – Rank2 score thresholds optimized across all LC-MS/MS runs for an HLA allele yielding a PSM FDR estimate of <1% for each precursor charge state. Endogenous HLA peptides were differentiated from spiked-in synthetic peptides by considering only peptides that were labeled exclusively with TMTzero and lacked any heavy-isotope-labeled amino acids.
Animal care and monitoring
Routine animal monitoring was carried out daily including inspection for dead mice and control of food and water supplies. The health of each mouse was closely assessed at least once weekly, and the results were documented in health monitoring sheets. The general physical condition of the mice was assessed according to the following parameters: mice were observed daily and weighed at least weekly for the duration of the study. Animals that lost more than 20% body weight were euthanized immediately. Mice were observed for the following clinical signs for the duration of the study: 1.) Moderate toxicity signs: intermittent hunching, ruffled coat, intermittent abnormal breathing, or intermittent tremors and convulsions; 2.) Substantial toxicity signs: prolonged immobility, hunched, dehydration, skin tenting, labored breathing, or prolonged prostration. Animals exhibiting signs of substantial toxicity will be euthanized immediately.
Immunization and endpoint collection
One to two days prior to injection, the injection site on the leg of the animal was shaved. The drug product was delivered by IM injection into the M. gastrocnemius (M. soleus was used if M. gastrocnemius was not available) in a volume of 20 μL per injection with a 30g needle insulin syringe. The injection site was observed every 24 h for 48 h post injection and at least once weekly throughout the study. At the endpoint timepoints, mice were euthanized with CO2 followed by a secondary method of euthanasia, cervical dislocation. Upon euthanasia, cardiac puncture was performed to collect whole blood, which was then placed in serum separator tubes for 30 min at RT and then centrifuged to collect serum. Spleens and both inguinal lymph nodes were collected, and these tissues were weighed and stored in ice-cold RPMI media for subsequent cell preparation.
Preparation of splenocytes and lymph nodes
Single-cell suspensions were prepared from spleens and lymph nodes by homogenizing them through 70 μm cell meshes using the plunger of a syringe. Splenocytes and lymph nodes were washed with an excess volume of DPBS followed by centrifugation at 300 × g for 6 min at RT and discarding the supernatants. Erythrocytes were then lysed with erythrocyte lysis buffer (154 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) for 5 min at RT. The reaction was stopped with an excess volume of media with 10% FBS. After another washing step, cells were resuspended in complete RPMI medium (10% FBS, 1% NEAA, 1% sodium pyruvate, 0.5% penicillin/streptomycin) and passed through a 70 μm cell mesh again. Cells were counted and stored short-term at 37°C for use on the same day or frozen in liquid nitrogen (i.e., cells were resuspended in 1 mL FBS/10% DMSO or in CryoStor 10 freezing media). For the use of frozen splenocytes in ELISpot analysis, immediately after thawing, pre-warmed FBS was added to splenocytes. Two washing steps using pre-warmed PBS were performed to remove DMSO before splenocytes were counted.
Serum collection
Blood collection took place via the vena facialis. Without prior anesthesia, mice were tightly held for blood collection, and the vena facialis was punctured using a lancet in a precise and short movement. Blood was collected into yellow-cap serum separator tubes from all mice, starting 14 days after intramuscular test article injection. After 30 min incubation at RT, blood was processed to collect serum by centrifugation at 2500 rpm, 4°C, for 10 min. Serum was stored at −70°C.
ELISpot assay
ELISpot assays with fresh or frozen splenocytes were performed according to the manufacturer’s protocol (with minor modifications as described below) using the R&D systems mouse IFN-ɣ ELISpot kit. Briefly, 96-well ELISpot plates were washed with PBS and blocked with serum-free medium for at least 1 h at 37°C. 100 μL of the splenocyte solution (fresh cells: 3 × 105 cells) were transferred to the respective wells of the 96-well ELISpot plate. Another 100 μL of overlapping peptide pools or controls were added in the following concentrations:
-
•
overlapping peptide mix against S protein pool: 0.3 μM final concentration per peptide [315 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
overlapping peptide mix against N protein pool: 0.3 μM final concentration per peptide [39 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
overlapping peptide mix against M protein pool: 0.3 μM final concentration per peptide [22 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
overlapping peptide mix against ORF1ab pool: 0.3 μM final concentration per peptide [19 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
Concanavalin A (ConA): 0.3 μM final concentration
For the positive control, the splenocytes were stimulated with ConA. For a non-stimulation control medium with DMSO was added. Plates were incubated overnight in a 37°C humidified incubator with 5% CO2. After approximately 18 h, cells were removed from the plates and the detection protocol of spots was initiated. To this end, the detection antibody, Streptavidin-ALP, and the ready-to-use substrate were added to the wells according to the manufacturer’s protocol. After plate drying for 2–3 h under the laminar flow or overnight, an ELISpot plate reader (ImmunoSpot S6 Core Analyzer, CTL) was used to count and analyze spot numbers per well.
Electrochemiluminescence immunoassay (ECLIA): Meso Scale Discovery (MSD) assay
MSD was performed in accordance with the Custom V-Plex manufacturer protocol. All reagents and samples were thawed to RT and 150 μL of a blocker was added to each well of the pre-coated plate. The plate was sealed and allowed to incubate for 1 h at RT with shaking. Next, the standards were prepared to generate the calibration curve. Plates were then washed after the incubation time and samples along with standards were added to the plate. 25 μL of sample were diluted once over with dilution agent 41 so that each well contained 50 μL. The plate was then allowed to incubate for 2 h at RT while shaking. The antibody detection solution was prepared as a master (1x in PBS) and distributed across the wells. The plate was then sealed and shaken at 800 rpm for 2 h at RT. The plate was then washed and the 2x read buffer solution was prepared and 150 μL of the buffer was distributed into each well before the plate was read on the Meso Sector S 600 plate reader according to manufacturer instructions.
Flow cytometry and intracellular cytokine staining
3 × 105 fresh splenocytes in 100 μL X-Vivo media were transferred to 96-well flat-bottom cell culture plates. 100 μL of an overlapping peptide pool or controls were added in the following concentrations:
-
•
overlapping peptide mix against S protein pool: 0.3 μM final concentration per peptide [315 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
overlapping peptide mix against N protein pool: 0.3 μM final concentration per peptide [39 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
overlapping peptide mix against M protein pool: 0.3 μM final concentration per peptide [22 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
overlapping peptide mix against ORF1ab pool: 0.3 μM final concentration per peptide [19 overlapping peptides – 15-mer long peptides with 11 amino acid overlap]
-
•
Concanavalin A (ConA): 0.3 μM final concentration
As a non-stimulation control, only medium with DMSO was added to detect unspecific background signals. Plates were incubated for 18 h in a 37°C humidified incubator with 5% CO2 before adding 5 μg/mL brefeldin A and 2 μM monensin. After another 4 h, cells were harvested and transferred to a 96-well, U-bottom plate for flow cytometry staining using the following antibodies from Biolegend: CD45.2 BV421 - 109832, CD3 PE-Cy7 - 100220, CD19 BV605 - 115540, CD4 BV510 - 100559, CD8 AF488 - 100723, CD62L PE - 104408, CD44 BV711 - 103057, PD1 PerCP-Cy5.5 - 109120, CD25 PUB395 - 564022, CD69 APC - 104514, CD27 BUV805 - 741959, CD107a PE Dazzle 594–121624, IFN-ɣ AF700 - 505823, IL2 BV785 – 503843, and TNFa BV650 - 506333. After the staining procedure, cells were resuspended in 100 μL FACS buffer (PBS +0.1% BSA) for flow cytometry analysis using a LSRFortessa- X20 (BD). Cytokine secretion and cytotoxic potential was measured through expression of three or more of the following: IFN-ɣ, TNF-α, IL-2 or CD107a.
ELISA
Serum samples were tested in 96-well plates for their antigen-specific antibody concentration. Each well of a MaxiSorp plate was coated with 100 ng/100 μL recombinant protein per well or isotype controls according to plate layout. Plates were incubated at 4°C overnight. Wells were washed and blocked with 1x blocking buffer and incubated at 37°C for 1 h on a shaker. Plates were washed and dilutions of the positive control antibody/serum samples were added and incubated at 37°C for 1 h on a shaker. Plates were washed three times with 300 μL/well PBS-T and the diluted secondary antibodies were incubated at 37°C for 45 min on shaker. After a wash, TMB substrate was added and incubated for 8 min at RT. The reaction was stopped with 100 μL 25% sulfuric acid and read on plate reader (450 nm, ref. 620 nm).
For concentration analysis, the signal of the specific samples was correlated with the positive control antibody. For reciprocal serum endpoint titer, the serum dilution that emitted the OD exceeding 4-fold background was used. The background was defined as the OD signal given by the recombinant protein incubated with the secondary detection anti-mouse IgG antibody only.
Pseudovirus-based neutralization assay
Replication-deficient vesicular stomatitis virus (VSV) that lacks the genetic information for the VSV envelope glycoprotein VSV-G but encodes green fluorescent protein (GFP) and luciferase was used for SARS-CoV-2-S pseudovirus generation. VSV pseudotypes were generated according to a published protocol.110 In brief, HEK293T/17 cells cultured in DMEM supplemented with 10% FBS were transfected with a pcDNA3.1-derived expression plasmid (Invitrogen) coding for the SARS-CoV-2 S protein (GenBank ID: QHD43416.1) with shortened cytoplasmic tail, i.e., pSARS-CoV-2-S-CΔ19, using Lipofectamine LTX & PLUS Reagent (Invitrogen) following the manufacturer’s instructions. The cytoplasmic tail was truncated for the 19 C-terminal amino acids to facilitate a more efficient integration of SARS-CoV-2-S into VSV virions analogous to SARS-CoV-S pseudotyped VSV.111 At 24 h post transfection, cells were inoculated with VSV-G trans-complemented VSV-ΔG-GFP/Luc vector (Indiana strain, de novo generated by reverse genetics from plasmid112 at a multiplicity of infection (MOI) of three and incubated for 2 h at 37°C and 7.5% CO2. Next, the inoculum was removed, cells were washed twice with PBS, and standard culture medium which contained 0.5 μg/mL anti-VSV-G antibody (clone 8G5F11) was added to neutralize residual input virus. 24 h after infection, VSV/SARS-CoV-2-S pseudovirus-containing supernatants were harvested, filtered (0.45 μm) and stored at −80°C in aliquots until further use.
For titration of VSV/SARS-CoV-2-S pseudovirus, 4 × 104 Vero 76 cells (ATCC CRL-1587) per 96-well were seeded in DMEM and 10% FBS. Cells were incubated for 4 to 6 h at 37°C and 7.5% CO2. Meanwhile, 2-fold, eight-step serial dilutions were prepared in 96-well V-bottom plates beginning with undiluted pseudovirus supernatant. Vero 76 wells were inoculated with 50 μL of the diluted pseudovirus supernatant and incubated for 16 to 24 h at 37°C and 7.5% CO2. Each dilution was tested in duplicate wells. After the incubation, the cell culture plates were removed from the incubator, the supernatant was discarded and 50 μL RPMI medium (Gibco) was added per well. In addition, 50 μL Bright-Glo substrate (Promega) was added per well and the relative luciferase units were recorded using a Tecan Spark microplate reader (Tecan). The relative luciferase units induced by a defined volume of a SARS-CoV-2 wild-type strain S glycoprotein pseudovirus reference batch previously described in Muik et al., 20216 that corresponds to an infectious titer of 200 transducing units (TU) per mL, was used as a comparator. Input volumes for the SARS-CoV-2 pseudovirus batches were calculated to normalize the infectious titer based on the relative luciferase units relative to the reference.
For the pVNT assay, Vero 76 cells were thawed, diluted to 2.67 × 105 cells/mL in assay medium (DMEM and 10% FBS) and seeded in 96-well flat-bottom plates at 4 × 104 cells per well. Cells were incubated for 4 to 6 h at 37°C and 7.5% CO2. Initial dilutions of mouse serum samples were prepared by adding 10 μL of serum to 50 μL assay medium in a 96-well V-bottom plate. Seven additional dilutions were subsequently prepared in 2-fold dilution steps, by iteratively transferring 30 μL of diluted sera to wells containing 30 μL assay medium. VSV/SARS-CoV-2 pseudovirus was thawed and diluted to obtain 200 TU per well. Pseudovirus/serum dilution mix was incubated for 5 min at RT on a microplate shaker at 750 rpm, and additional 5 min at RT without agitation. Pseudovirus/serum dilution mix was then added to the seeded Vero 76 cells (50 μL mix per well), followed by incubation for 16 to 24 h at 37°C and 5% CO2. Each dilution of serum samples was tested in duplicate wells. Vero 76 cells incubated with pseudovirus in the absence of mouse sera were used as positive controls. Vero 76 cells incubated without pseudovirus were used as negative controls. After the incubation, supernatants were removed, and RPMI and Bright-Glo substrate was added. Luminescence was recorded on a Tecan Spark microplate reader, and neutralization titers were calculated as the reciprocal of the highest serum dilution that still resulted in 50% reduction in luminescence. Results for all pseudovirus neutralization experiments were expressed as geometric mean titers (GMT) of duplicates. If no neutralization was observed, an arbitrary titer value of half of LLOD was reported.
10X single-cell RNA sequencing
Splenocytes from immunized animals were thawed and incubated with fluorescently-labeled S protein tetramers or peptide MHC tetramers presenting three different highly immunogenic T-cell epitopes AAYYVGYL, VNFNFNGL, and VVLSFELL. Cells were labeled with the following TotalSeqC antibodies from Biolegend: CD8 (100785), CD62L (104455), CD44 (103063), PD1 (109127), CXCR5 (145537), CD27 (124245), CD39 (143815), CD127 (135047), TIM3 (119739), LAG3 (125237), CD28 (102133), CD69 (104551), CD38 (102735), KLRG1 (138433), CD95 (152616), CD138 (142538), CD273 (107229), CD73 (127237), and CD80 (104755). The following hashing antibodies were used to differentiate between mice: Hash 1 (155861), Hash 2 (155863), Hash 3 (155865), Hash 4 (155867), Hash 5 (155869). Antigen-positive cells were sorted using the BD FACSAria Cell Sorter, and up to 8000 cells per animal were collected. Cells were pooled by treatment group, and each group was loaded into a channel on the 10X Chromium Connect using Chromium Next Automated GEM 5′ v2 kit. Gene expression libraries were generated using the automated library construction kit, CITE-seq libraries were generated using the Chromium Automated 5′ Feature Barcode kit and VDJ libraries were generated using Chromium Automated Mouse BCR/TCR Amplification and Library Construction kit. Sequencing was carried out on the Illumina NovaSeq 6000 platform with an S1 flow cell. Experimental schematics were created using BioRender.com.
ScRNAseq data processing and analysis
Fastq reads from gene expression, CITE and VDJ library generated by NovaSeq were aligned to GRCm38 reference genome (provided by cellranger: https://support.10xgenomics.com/single-cell-gene-expression/software/downloads/latest) using cellranger 6.0.1 with multi option. The resulted count matrices and V(D)J contig annotations were used for downstream analysis performed with Scanpy113 and Scirpy.66 Custom script was used for dehashing. Briefly, total reads of each hashtag were normalized to 10,000. Ratio of normalized hash counts were calculated for each cell. Cells with less than five total raw hash counts were labeled as negative; and cells with highest single category normalized hash count ratio of less than 0.8 were labeled as miscellaneous (misc). Cells with at least five counts and ≥ 0.8 single category normalized hash count were labeled by the highest ratio hash category.
For gene expression library, to filter low quality cells and doublets, cells with <500 raw counts or >5% reads from mitochondria genes and cells with >3000 genes based on raw read count were filtered out. Genes observed in less than three cells were filtered. Gene expression matrices were normalized to 10,000 per cell and log1p transformed with scanpy options (sc.pp.normalize_total, sc.pp.log1p). Highly variable genes were identified with scanpy option sc.pp.highly_variable_genes. Resulted normalized count matrices were regressed (sc.pp.regress_out) and scaled (sc.pp.scale).
For CITE measurements, cells with <50 raw counts were filtered out. CITE-seq count matrices were denoised with dsb (python interface with MUON)114 , 115 by using empty droplet count matrices as background. Then the matrices were scaled with sc.pp.scale to max_value = 10.
Normalized and scaled gene expression and CITE reads were merged by inner join to keep cells that pass quality cut-off in both libraries. V(D)J library annotations were also merged the data object based on cell barcodes. Cells with valid single pair of V(D)J annotation were kept for further analysis. For downstream analysis, cells from each group were randomly downsampled so that cells originated from animals immunized with BNT162b2 alone or BNT162b2 and BNT162b4 group are capped to the same cell number for distribution analysis (gene expression-based clusters or clonal expansion).
For gene expression-based clustering analysis, scaled count matrices (without CITE) were used and TCR/immunoglobulin genes were filtered out. Dimensionality reduction was performed with PCA (sc.tl.pca) and top 10–20 PCs were selected based on variance ratio for computing neighborhood graph (sc.pp.neighbors). UMAP and Leiden clustering were performed with scanpy functions sc.tl.umap and sc.tl.leiden to compute 2D projection and extract clusters.
V(D)J clonotype analysis was performed with scirpy.66 Cells with single pair of valid immune receptor pairing were used for CDR3 neighborhood graph and clonotype definition analysis. Scirpy function ir.dist was used to compute sequence-distance similarity CDR3 sequences. To calculate clonal expansion, tl.clonal_expansion function was used.
Viral challenge in a hamster model
Male and female golden Syrian hamsters between 6 and 8 weeks old were immunized intramuscularly on days 0 and 21. On day 35, animals were challenged through intranasal inoculation with either the WT (USA-WA1/2020), Delta, or Omicron BA.1 SARS-CoV-2 virus for virus challenge. Virus was expanded in Calu-3 cells (ATCC HTB-55) from a seed stock (BEI Resources Cat. #: WT: WAS Lot 1215020-1235, Delta:NR-56116, and Omicron:NR-56486). The intranasal inoculation (IN) was performed on Ketamine/Xylazine anesthetized hamsters. The animals were challenged with a total volume of 100 μL per animal (50 μL/nare). The infectious titers of the WT stock are 6 X 105 PFU/mL and used at a 1:10 dilution. The Delta challenge stock titer was at 1.4 X106 TCID50/mL and used at a 1:10 dilution. The Omicron BA.1 challenge stock titer was at 4.8 X106 TCID50/mL and used at a 1:10 dilution. After challenge, animals are monitored and weighed daily. Two days post challenge, D37, half of the cohort was euthanized, and lung and nasal turbinates were collected from infected animals. Seven days post challenge, D42, the remaining animals were euthanized, and tissues were collected for analysis.
TCID50 assay for determination of infectious viral load
Vero E6 cells were plated at 25,000 cells per well in DMEM + 10% FBS + Gentamicin and incubated overnight to be 80–100% confluent the following day. Media was aspirated out and replaced with fresh media and a serial dilution of live virus. The plates for the samples were incubated again at 37°C, 5.0% CO2 for 4 days. After 4 days, wells were visually inspected for cytopathic effect (CPE). Non-infected wells were identified by a clear confluent cell layer. Infected cells demonstrated cell rounding. The presence of CPE was recorded as a plus (+) and absence of CPE as minus (−). The TCID50 was calculated using the Read-Muench formula. If no infection was observed, an arbitrary titer value of the limit of detection [LLOD] was reported.
Histopathology
At necropsy, the left lung was collected and placed in 10% neutral buffered formalin for histopathologic analysis. Tissue sections were trimmed, blinded and processed to hematoxylin and eosin (H&E) stained slides and examined by a board-certified pathologist at Experimental Pathology Laboratories, Inc. (EPL) in Sterling, Virginia.
Quantification and statistical analysis
Datasets were assessed for significance using one-way analysis of variance (ANOVA) using GraphPad Prism 9 (GraphPad Software). The statistical details of comparisons can be found in the figures, figure legends, and results.
Acknowledgments
We are grateful to the members of BioNTech SE for their support and assistance with this study. We thank the many colleagues at BioNTech SE and Pfizer, Inc. who developed and produced the BNT162b2 vaccine. We especially thank Meghan Bushway, Alexander Muik, Joseph Murphy, Nadine Salisch, and Lynda Stuart for their expertise and insight; Jesse Dong, Jonathan McGee, Heidi Jackson, Madeline Worob, Yuting Huang, Daniel Kallin, and Alyssa Kruithof for peptide synthesis and manufacturing; Yiling Qiu for flow cytometry and cell-sorting support; Janani Sridar and Paul Turcott for tetramer reagent generation; Jaewon Choi and Brandon Conn for the A375 stable cell lines; Klara Dehler, Saskia Krapp, Helena Feist, and Jonathan Mottl for their support in the serological analyses; Su Hyun Lee, Huizhi Zhou, Hongling Liao, Taeyong Kim, and Andrey Tovchigrechko for their support sequencing the NGS libraries. We thank Andrew Finlayson of BioNTech SE for copyediting support. We thank our colleagues at BioModels, LLC. and BIOQUAL, Inc. for their support of the in vivo studies. We thank John Welle from Acumen Medical Communications for graphic design support. The graphical abstract was adapted from BioRender.com. The study was sponsored by BioNTech SE.
Author contributions
Conceptualization, C.M.A., R.B.G., and A.P.; Methodology, C.M.A., Y.J.X., D.A.R., H.D., D.H., J.R.S., T.Z., S.F., K.T., and A.P.; Investigation, C.M.A., Y.J.X., D.A.R., H.D., D.H., S.M., K.M., B.K., T.E.S., A.L., A.Z., S.A.K., C.I.C.C., S.H., A.P.H., Y.L.-L., K.W., and A.P.; Data Curation, C.M.A., Y.J.X., D.A.R., and A.P.; Writing – Original Draft, C.M.A., Y.J.X., D.A.R., H.D., R.B.G., A.P.; Writing – Review & Editing, C.M.A., Y.J.X., D.A.R., H.D., Ö.T., U.S., R.B.G., A.P.; Supervision, C.M.A., A.B.V., T.A.A., Ö.T., U.S., R.B.G., A.P.
Declaration of interests
U.S. and Ö.T. are management board members and employees at BioNTech SE. R.B.G., Board of Directors, Alkermes plc, Infinity Pharmaceuticals, and Zai Laboratory, and Scientific Advisory Board, Leap Therapeutics; consultant Third Rock Ventures, stockholder and employee of BioNTech US. C.M.A., Y.J.X., D.A.R., H.D., D.H., S.M., K.M., B.K., T.E.S., A.L., A.Z., S.A.K., C.I.C.C., S.H., A.P.H., T.Z., Y.L.-L., K.W., A.B.V., J.R.S., S.F., K.T., K.W., T.A.A., Ö.T., U.S., R.B.G., and A.P. are either current or past employees of BioNTech SE or BioNTech US, are stockholders, and/or are inventors on patents and patent applications related to RNA technology and COVID-19 vaccines.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Published: April 13, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.cell.2023.04.007.
Supplemental information
Data and code availability
-
•
The RAW MS data files and interpreted data were uploaded to the MassIVE repository: MSV000091008 (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp).
-
•
Single-cell RNA sequencing data are deposited on GEO: GSE222403 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•All codes for analysis and plots are available on Zenodo: https://doi.org/10.5281/zenodo.7789841 and GitHub:
- o Epitope analysis: https://github.com/YolandaHuitianDiao/BNT162b4_epitopes.git.
- o Single cell analysis: https://github.com/YolandaHuitianDiao/BNT162b4_scRNA.git.
References
- 1.Cao Y., Jian F., Wang J., Yu Y., Song W., Yisimayi A., Wang J., An R., Chen X., Zhang N., et al. Imprinted SARS-CoV-2 humoral immunity induces convergent Omicron RBD evolution. Nature. 2023;614:521–529. doi: 10.1038/s41586-022-05644-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao Y., Wang J., Jian F., Xiao T., Song W., Yisimayi A., Huang W., Li Q., Wang P., An R., et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. 2022;602:657–663. doi: 10.1038/s41586-021-04385-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jian F., Yu Y., Song W., Yisimayi A., Yu L., Gao Y., Zhang N., Wang Y., Shao F., Hao X., et al. Further humoral immunity evasion of emerging SARS-CoV-2 BA.4 and BA.5 subvariants. Lancet Infect. Dis. 2022;22:1535–1537. doi: 10.1016/S1473-3099(22)00642-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurhade C., Zou J., Xia H., Cai H., Yang Q., Cutler M., Cooper D., Muik A., Jansen K.U., Xie X., et al. Neutralization of Omicron BA.1, BA.2, and BA.3 SARS-CoV-2 by 3 doses of BNT162b2 vaccine. Nat. Commun. 2022;13:3602. doi: 10.1038/s41467-022-30681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muik, A., Lui, B.G., Diao, H., Fu, Y., Bacher, M., Toker, A., Grosser, J., Ozhelvaci, O., Grikscheit, K., Hoehl, S., et al. (2022). Progressive Loss of Conserved Spike Protein Neutralizing Antibody Sites in Omicron Sublineages Is Balanced by Preserved T-Cell Recognition Epitopes. bioRxiv, 2022.2012.2015.520569. 10.1101/2022.12.15.520569
- 6.Muik A., Wallisch A.K., Sanger B., Swanson K.A., Muhl J., Chen W., Cai H., Maurus D., Sarkar R., Tureci O., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021;371:1152–1153. doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q., Iketani S., Li Z., Liu L., Guo Y., Huang Y., Bowen A.D., Liu M., Wang M., Yu J., et al. Alarming antibody evasion properties of rising SARS-CoV-2 BQ and XBB subvariants. Cell. 2023;186:279–286.e8. doi: 10.1016/j.cell.2022.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sette A., Crotty S. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021;184:861–880. doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wherry E.J., Barouch D.H. T cell immunity to COVID-19 vaccines. Science. 2022;377:821–822. doi: 10.1126/science.add2897. [DOI] [PubMed] [Google Scholar]
- 10.Channappanavar R., Fett C., Zhao J., Meyerholz D.K., Perlman S. Virus-specific memory CD8 T cells provide substantial protection from lethal severe acute respiratory syndrome coronavirus infection. J. Virol. 2014;88:11034–11044. doi: 10.1128/JVI.01505-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Channappanavar R., Zhao J., Perlman S. T cell-mediated immune response to respiratory coronaviruses. Immunol. Res. 2014;59:118–128. doi: 10.1007/s12026-014-8534-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao J., Zhao J., Perlman S. T cell responses are required for protection from clinical disease and for virus clearance in severe acute respiratory syndrome coronavirus-infected mice. J. Virol. 2010;84:9318–9325. doi: 10.1128/JVI.01049-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao J., Zhao J., Mangalam A.K., Channappanavar R., Fett C., Meyerholz D.K., Agnihothram S., Baric R.S., David C.S., Perlman S. Airway Memory CD4(+) T Cells Mediate Protective Immunity against Emerging Respiratory Coronaviruses. Immunity. 2016;44:1379–1391. doi: 10.1016/j.immuni.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen K.W., Linderman S.L., Moodie Z., Czartoski J., Lai L., Mantus G., Norwood C., Nyhoff L.E., Edara V.V., Floyd K., et al. Longitudinal analysis shows durable and broad immune memory after SARS-CoV-2 infection with persisting antibody responses and memory B and T cells. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dan J.M., Mateus J., Kato Y., Hastie K.M., Yu E.D., Faliti C.E., Grifoni A., Ramirez S.I., Haupt S., Frazier A., et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371 doi: 10.1126/science.abf4063. eabf4063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ng O.W., Chia A., Tan A.T., Jadi R.S., Leong H.N., Bertoletti A., Tan Y.J. Memory T cell responses targeting the SARS coronavirus persist up to 11 years post-infection. Vaccine. 2016;34:2008–2014. doi: 10.1016/j.vaccine.2016.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M., et al. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 18.McMahan K., Yu J., Mercado N.B., Loos C., Tostanoski L.H., Chandrashekar A., Liu J., Peter L., Atyeo C., Zhu A., et al. Correlates of protection against SARS-CoV-2 in rhesus macaques. Nature. 2021;590:630–634. doi: 10.1038/s41586-020-03041-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hajnik R.L., Plante J.A., Liang Y., Alameh M.G., Tang J., Bonam S.R., Zhong C., Adam A., Scharton D., Rafael G.H., et al. Dual spike and nucleocapsid mRNA vaccination confer protection against SARS-CoV-2 Omicron and Delta variants in preclinical models. Sci. Transl. Med. 2022;14 doi: 10.1126/scitranslmed.abq1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu J., Yu J., McMahan K., Jacob-Dolan C., He X., Giffin V., Wu C., Sciacca M., Powers O., Nampanya F., et al. CD8 T cells contribute to vaccine protection against SARS-CoV-2 in macaques. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abq7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrera G., Picozza M., D'Orso S., Placido R., Pirronello M., Verdiani A., Termine A., Fabrizio C., Giannessi F., Sambucci M., et al. BNT162b2 vaccination induces durable SARS-CoV-2-specific T cells with a stem cell memory phenotype. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abl5344. [DOI] [PubMed] [Google Scholar]
- 22.Hurme A., Jalkanen P., Heroum J., Liedes O., Vara S., Melin M., Terasjarvi J., He Q., Poysti S., Hanninen A., et al. Long-Lasting T Cell Responses in BNT162b2 COVID-19 mRNA Vaccinees and COVID-19 Convalescent Patients. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.869990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koutsakos M., Reynaldi A., Lee W.S., Nguyen J., Amarasena T., Taiaroa G., Kinsella P., Liew K.C., Tran T., Kent H.E., Tan H.X., Rowntree L.C., Nguyen T.H.O., Thomas P.G., Kedzierska K., Petersen J., Rossjohn J., Williamson D.A., Khoury D., Davenport M.P., Kent S.J., Wheatley A.K., Juno J.A. SARS-CoV-2 breakthrough infection induces rapid memory and de novo T cell responses. Immunity. 2023;56:879–892.e4. doi: 10.1016/j.immuni.2023.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neale I., Ali M., Kronsteiner B., Longet S., Abraham P., Deeks A.S., Brown A., Moore S.C., Stafford L., Dobson S.L., et al. CD4+ and CD8+ T cell and antibody correlates of protection against Delta vaccine breakthrough infection: A nested case-control study within the PITCH study. medRxiv. 2023 doi: 10.1101/2023.02.16.23285748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent S.J., Khoury D.S., Reynaldi A., Juno J.A., Wheatley A.K., Stadler E., John Wherry E., Triccas J., Sasson S.C., Cromer D., Davenport M.P. Disentangling the relative importance of T cell responses in COVID-19: leading actors or supporting cast? Nat. Rev. Immunol. 2022;22:387–397. doi: 10.1038/s41577-022-00716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lafon E., Diem G., Witting C., Zaderer V., Bellmann-Weiler R.M., Reindl M., Bauer A., Griesmacher A., Fux V., Hoermann G., et al. Potent SARS-CoV-2-Specific T Cell Immunity and Low Anaphylatoxin Levels Correlate With Mild Disease Progression in COVID-19 Patients. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.684014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Bert N., Clapham H.E., Tan A.T., Chia W.N., Tham C.Y.L., Lim J.M., Kunasegaran K., Tan L.W.L., Dutertre C.A., Shankar N., et al. Highly functional virus-specific cellular immune response in asymptomatic SARS-CoV-2 infection. J. Exp. Med. 2021;218 doi: 10.1084/jem.20202617. e20202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rydyznski Moderbacher C., Ramirez S.I., Dan J.M., Grifoni A., Hastie K.M., Weiskopf D., Belanger S., Abbott R.K., Kim C., Choi J., et al. Antigen-Specific Adaptive Immunity to SARS-CoV-2 in Acute COVID-19 and Associations with Age and Disease Severity. Cell. 2020;183:996–1012.e19. doi: 10.1016/j.cell.2020.09.038. e1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan A.T., Linster M., Tan C.W., Le Bert N., Chia W.N., Kunasegaran K., Zhuang Y., Tham C.Y.L., Chia A., Smith G.J.D., et al. Early induction of functional SARS-CoV-2-specific T cells associates with rapid viral clearance and mild disease in COVID-19 patients. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulien I., Kemming J., Oberhardt V., Wild K., Seidel L.M., Killmer S., Sagar Daul, F., Daul F., Salvat Lago M., Decker A., et al. Characterization of pre-existing and induced SARS-CoV-2-specific CD8(+) T cells. Nat. Med. 2021;27:78–85. doi: 10.1038/s41591-020-01143-2. [DOI] [PubMed] [Google Scholar]
- 31.Kalimuddin S., Tham C.Y.L., Qui M., de Alwis R., Sim J.X.Y., Lim J.M.E., Tan H.C., Syenina A., Zhang S.L., Le Bert N., et al. Early T cell and binding antibody responses are associated with COVID-19 RNA vaccine efficacy onset. Med (N Y) 2021;2:682–688.e4. doi: 10.1016/j.medj.2021.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bange E.M., Han N.A., Wileyto P., Kim J.Y., Gouma S., Robinson J., Greenplate A.R., Hwee M.A., Porterfield F., Owoyemi O., et al. CD8(+) T cells contribute to survival in patients with COVID-19 and hematologic cancer. Nat. Med. 2021;27:1280–1289. doi: 10.1038/s41591-021-01386-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferretti A.P., Kula T., Wang Y., Nguyen D.M.V., Weinheimer A., Dunlap G.S., Xu Q., Nabilsi N., Perullo C.R., Cristofaro A.W., et al. Unbiased Screens Show CD8(+) T Cells of COVID-19 Patients Recognize Shared Epitopes in SARS-CoV-2 that Largely Reside outside the Spike Protein. Immunity. 2020;53:1095–1107.e3. doi: 10.1016/j.immuni.2020.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grifoni A., Weiskopf D., Ramirez S.I., Mateus J., Dan J.M., Moderbacher C.R., Rawlings S.A., Sutherland A., Premkumar L., Jadi R.S., et al. Targets of T Cell Responses to SARS-CoV-2 Coronavirus in Humans with COVID-19 Disease and Unexposed Individuals. Cell. 2020;181:1489–1501.e15. doi: 10.1016/j.cell.2020.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Snyder T.M., Gittelman R.M., Klinger M., May D.H., Osborne E.J., Taniguchi R., Zahid H.J., Kaplan I.M., Dines J.N., Noakes M.T., et al. Vol. 2007. 2020. Magnitude and Dynamics of the T-Cell Response to SARS-CoV-2 Infection at Both Individual and Population Levels. medRxiv. 2020. 2031.20165647. [DOI] [Google Scholar]
- 36.Tarke A., Sidney J., Kidd C.K., Dan J.M., Ramirez S.I., Yu E.D., Mateus J., da Silva Antunes R., Moore E., Rubiro P., et al. Comprehensive analysis of T cell immunodominance and immunoprevalence of SARS-CoV-2 epitopes in COVID-19 cases. Cell Rep. Med. 2021;2 doi: 10.1016/j.xcrm.2021.100204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matchett W.E., Joag V., Stolley J.M., Shepherd F.K., Quarnstrom C.F., Mickelson C.K., Wijeyesinghe S., Soerens A.G., Becker S., Thiede J.M., et al. Cutting Edge: Nucleocapsid Vaccine Elicits Spike-Independent SARS-CoV-2 Protective Immunity. J. Immunol. 2021;207:376–379. doi: 10.4049/jimmunol.2100421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carter B., Huang P., Liu G., Liang Y., Lin P.J.C., Peng B.-H., McKay L.G.A., Dimitrakakis A., Hsu J., Tat V., et al. A pan-variant mRNA-LNP T cell vaccine protects HLA transgenic mice from mortality after infection with SARS-CoV-2 Beta. Front. Immunol. 2023;14 doi: 10.3389/fimmu.2023.1135815. 1135815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swadling L., Diniz M.O., Schmidt N.M., Amin O.E., Chandran A., Shaw E., Pade C., Gibbons J.M., Le Bert N., Tan A.T., et al. Pre-existing polymerase-specific T cells expand in abortive seronegative SARS-CoV-2. Nature. 2022;601:110–117. doi: 10.1038/s41586-021-04186-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Foix A., Lopez D., Diez-Fuertes F., McConnell M.J., Martin-Galiano A.J. Predicted impact of the viral mutational landscape on the cytotoxic response against SARS-CoV-2. PLoS Comput. Biol. 2022;18 doi: 10.1371/journal.pcbi.1009726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., Kranz L.M., Walzer K.C., Hein S., Guler A., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021;592:283–289. doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 42.Sahin U., Muik A., Vogler I., Derhovanessian E., Kranz L.M., Vormehr M., Quandt J., Bidmon N., Ulges A., Baum A., et al. BNT162b2 vaccine induces neutralizing antibodies and poly-specific T cells in humans. Nature. 2021;595:572–577. doi: 10.1038/s41586-021-03653-6. [DOI] [PubMed] [Google Scholar]
- 43.Gangaev A., Ketelaars S.L.C., Isaeva O.I., Patiwael S., Dopler A., Hoefakker K., De Biasi S., Gibellini L., Mussini C., Guaraldi G., et al. Identification and characterization of a SARS-CoV-2 specific CD8(+) T cell response with immunodominant features. Nat. Commun. 2021;12:2593. doi: 10.1038/s41467-021-22811-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gauttier V., Morello A., Girault I., Mary C., Belarif L., Desselle A., Wilhelm E., Bourquard T., Pengam S., Teppaz G., et al. Tissue-resident memory CD8 T-cell responses elicited by a single injection of a multi-target COVID-19 vaccine. bioRxiv. 2020 doi: 10.1101/2020.08.14.240093. 2020.2008.2014. [DOI] [Google Scholar]
- 45.Mateus J., Grifoni A., Tarke A., Sidney J., Ramirez S.I., Dan J.M., Burger Z.C., Rawlings S.A., Smith D.M., Phillips E., et al. Selective and cross-reactive SARS-CoV-2 T cell epitopes in unexposed humans. Science. 2020;370:89–94. doi: 10.1126/science.abd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nelde A., Bilich T., Heitmann J.S., Maringer Y., Salih H.R., Roerden M., Lubke M., Bauer J., Rieth J., Wacker M., et al. SARS-CoV-2-derived peptides define heterologous and COVID-19-induced T cell recognition. Nat. Immunol. 2021;22:74–85. doi: 10.1038/s41590-020-00808-x. [DOI] [PubMed] [Google Scholar]
- 47.Peng Y., Mentzer A.J., Liu G., Yao X., Yin Z., Dong D., Dejnirattisai W., Rostron T., Supasa P., Liu C., et al. Broad and strong memory CD4(+) and CD8(+) T cells induced by SARS-CoV-2 in UK convalescent individuals following COVID-19. Nat. Immunol. 2020;21:1336–1345. doi: 10.1038/s41590-020-0782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Poran A., Harjanto D., Malloy M., Arieta C.M., Rothenberg D.A., Lenkala D., van Buuren M.M., Addona T.A., Rooney M.S., Srinivasan L., Gaynor R.B. Sequence-based prediction of SARS-CoV-2 vaccine targets using a mass spectrometry-based bioinformatics predictor identifies immunogenic T cell epitopes. Genome Med. 2020;12:70. doi: 10.1186/s13073-020-00767-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sekine T., Perez-Potti A., Rivera-Ballesteros O., Stralin K., Gorin J.B., Olsson A., Llewellyn-Lacey S., Kamal H., Bogdanovic G., Muschiol S., et al. Robust T Cell Immunity in Convalescent Individuals with Asymptomatic or Mild COVID-19. Cell. 2020;183:158–168.e14. doi: 10.1016/j.cell.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bezstarosti K., Lamers M.M., Doff W.A.S., Wever P.C., Thai K.T.D., van Kampen J.J.A., Haagmans B.L., Demmers J.A.A. Targeted proteomics as a tool to detect SARS-CoV-2 proteins in clinical specimens. PLoS One. 2021;16 doi: 10.1371/journal.pone.0259165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bojkova D., Klann K., Koch B., Widera M., Krause D., Ciesek S., Cinatl J., Munch C. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. 2020;583:469–472. doi: 10.1038/s41586-020-2332-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davidson A.D., Williamson M.K., Lewis S., Shoemark D., Carroll M.W., Heesom K.J., Zambon M., Ellis J., Lewis P.A., Hiscox J.A., Matthews D.A. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 2020;12:68. doi: 10.1186/s13073-020-00763-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouhaddou M., Memon D., Meyer B., White K.M., Rezelj V.V., Correa Marrero M., Polacco B.J., Melnyk J.E., Ulferts S., Kaake R.M., et al. The Global Phosphorylation Landscape of SARS-CoV-2 Infection. Cell. 2020;182:685–712.e19. doi: 10.1016/j.cell.2020.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weingarten-Gabbay S., Klaeger S., Sarkizova S., Pearlman L.R., Chen D.Y., Gallagher K.M.E., Bauer M.R., Taylor H.B., Dunn W.A., Tarr C., et al. Profiling SARS-CoV-2 HLA-I peptidome reveals T cell epitopes from out-of-frame ORFs. Cell. 2021;184:3962–3980.e17. doi: 10.1016/j.cell.2021.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Perlman S., Netland J. Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol. 2009;7:439–450. doi: 10.1038/nrmicro2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abelin J.G., Harjanto D., Malloy M., Suri P., Colson T., Goulding S.P., Creech A.L., Serrano L.R., Nasir G., Nasrullah Y., et al. Defining HLA-II Ligand Processing and Binding Rules with Mass Spectrometry Enhances Cancer Epitope Prediction. Immunity. 2019;51:766–779.e17. doi: 10.1016/j.immuni.2019.08.012. [DOI] [PubMed] [Google Scholar]
- 57.Choi J., Goulding S.P., Conn B.P., McGann C.D., Dietze J.L., Kohler J., Lenkala D., Boudot A., Rothenberg D.A., Turcott P.J., et al. Systematic discovery and validation of T cell targets directed against oncogenic KRAS mutations. Cell Rep. Methods. 2021;1 doi: 10.1016/j.crmeth.2021.100084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Francis J.M., Leistritz-Edwards D., Dunn A., Tarr C., Lehman J., Dempsey C., Hamel A., Rayon V., Liu G., Wang Y., et al. Allelic variation in class I HLA determines CD8(+) T cell repertoire shape and cross-reactive memory responses to SARS-CoV-2. Sci. Immunol. 2022;7 doi: 10.1126/sciimmunol.abk3070. eabk3070, , eabk3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kared H., Redd A.D., Bloch E.M., Bonny T.S., Sumatoh H., Kairi F., Carbajo D., Abel B., Newell E.W., Bettinotti M.P., et al. SARS-CoV-2-specific CD8+ T cell responses in convalescent COVID-19 individuals. J. Clin. Invest. 2021;131 doi: 10.1172/JCI145476. e145476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Saini S.K., Hersby D.S., Tamhane T., Povlsen H.R., Amaya Hernandez S.P., Nielsen M., Gang A.O., Hadrup S.R. SARS-CoV-2 genome-wide T cell epitope mapping reveals immunodominance and substantial CD8(+) T cell activation in COVID-19 patients. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abf7550. eabf7550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kyte J., Doolittle R.F. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 62.Vitiello A., Marchesini D., Furze J., Sherman L.A., Chesnut R.W. Analysis of the HLA-restricted influenza-specific cytotoxic T lymphocyte response in transgenic mice carrying a chimeric human-mouse class I major histocompatibility complex. J. Exp. Med. 1991;173:1007–1015. doi: 10.1084/jem.173.4.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McCray P.B., Jr., Pewe L., Wohlford-Lenane C., Hickey M., Manzel L., Shi L., Netland J., Jia H.P., Halabi C., Sigmund C.D., et al. Lethal infection of K18-hACE2 mice infected with severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:813–821. doi: 10.1128/JVI.02012-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., Baum A., Pascal K., Quandt J., Maurus D., et al. COVID-19 vaccine BNT162b1 elicits human antibody and T(H)1 T cell responses. Nature. 2020;586:594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 65.Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sturm G., Szabo T., Fotakis G., Haider M., Rieder D., Trajanoski Z., Finotello F. Scirpy: a Scanpy extension for analyzing single-cell T-cell receptor-sequencing data. Bioinformatics. 2020;36:4817–4818. doi: 10.1093/bioinformatics/btaa611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., Seaman S.R., Harris R.J., Hope R., Lopez-Bernal J., et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis. 2022;22:35–42. doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yuan S., Ye Z.W., Liang R., Tang K., Zhang A.J., Lu G., Ong C.P., Man Poon V.K., Chan C.C.S., Mok B.W.Y., et al. Pathogenicity, transmissibility, and fitness of SARS-CoV-2 Omicron in Syrian hamsters. Science. 2022;377:428–433. doi: 10.1126/science.abn8939. [DOI] [PubMed] [Google Scholar]
- 69.Storm, N., Crossland, N.A., McKay, L.G.A., and Griffiths, A. (2022). Comparative Infection and Pathogenesis of SARS-CoV-2 Omicron and Delta Variants in Aged and Young Syrian Hamsters. bioRxiv, 2022.2003.2002.482662. 10.1101/2022.03.02.482662
- 70.Li B., Deng A., Li K., Hu Y., Li Z., Shi Y., Xiong Q., Liu Z., Guo Q., Zou L., et al. Viral infection and transmission in a large, well-traced outbreak caused by the SARS-CoV-2 Delta variant. Nat. Commun. 2022;13:460. doi: 10.1038/s41467-022-28089-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Abelin J.G., Keskin D.B., Sarkizova S., Hartigan C.R., Zhang W., Sidney J., Stevens J., Lane W., Zhang G.L., Eisenhaure T.M., et al. Mass Spectrometry Profiling of HLA-Associated Peptidomes in Mono-allelic Cells Enables More Accurate Epitope Prediction. Immunity. 2017;46:315–326. doi: 10.1016/j.immuni.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schubert B., Kohlbacher O. Designing string-of-beads vaccines with optimal spacers. Genome Med. 2016;8:9. doi: 10.1186/s13073-016-0263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ali M., Foldvari Z., Giannakopoulou E., Boschen M.L., Stronen E., Yang W., Toebes M., Schubert B., Kohlbacher O., Schumacher T.N., Olweus J. Induction of neoantigen-reactive T cells from healthy donors. Nat. Protoc. 2019;14:1926–1943. doi: 10.1038/s41596-019-0170-6. [DOI] [PubMed] [Google Scholar]
- 74.Shou S., Liu M., Yang Y., Kang N., Song Y., Tan D., Liu N., Wang F., Liu J., Xie Y. Animal Models for COVID-19: Hamsters, Mouse, Ferret, Mink, Tree Shrew, and Non-human Primates. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.626553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wellford S.A., Moseman A.P., Dao K., Wright K.E., Chen A., Plevin J.E., Liao T.C., Mehta N., Moseman E.A. Mucosal plasma cells are required to protect the upper airway and brain from infection. Immunity. 2022;55:2118–2134.e6. doi: 10.1016/j.immuni.2022.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Suzuki R., Yamasoba D., Kimura I., Wang L., Kishimoto M., Ito J., Morioka Y., Nao N., Nasser H., Uriu K., et al. Attenuated fusogenicity and pathogenicity of SARS-CoV-2 Omicron variant. Nature. 2022;603:700–705. doi: 10.1038/s41586-022-04462-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hakki S., Zhou J., Jonnerby J., Singanayagam A., Barnett J.L., Madon K.J., Koycheva A., Kelly C., Houston H., Nevin S., et al. Onset and window of SARS-CoV-2 infectiousness and temporal correlation with symptom onset: a prospective, longitudinal, community cohort study. Lancet Respir. Med. 2022;10:1061–1073. doi: 10.1016/S2213-2600(22)00226-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Levine-Tiefenbrun M., Yelin I., Alapi H., Katz R., Herzel E., Kuint J., Chodick G., Gazit S., Patalon T., Kishony R. Viral loads of Delta-variant SARS-CoV-2 breakthrough infections after vaccination and booster with BNT162b2. Nat. Med. 2021;27:2108–2110. doi: 10.1038/s41591-021-01575-4. [DOI] [PubMed] [Google Scholar]
- 79.Wolfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Muller M.A., Niemeyer D., Jones T.C., Vollmar P., Rothe C., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 80.Jones T.C., Biele G., Muhlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., Bleicker T., Tesch J., Schmidt M.L., Sander L.E., et al. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373:eabi5273. doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kustin T., Harel N., Finkel U., Perchik S., Harari S., Tahor M., Caspi I., Levy R., Leshchinsky M., Ken Dror S., et al. Evidence for increased breakthrough rates of SARS-CoV-2 variants of concern in BNT162b2-mRNA-vaccinated individuals. Nat. Med. 2021;27:1379–1384. doi: 10.1038/s41591-021-01413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Tang P., Hasan M.R., Chemaitelly H., Yassine H.M., Benslimane F.M., Al Khatib H.A., AlMukdad S., Coyle P., Ayoub H.H., Al Kanaani Z., et al. BNT162b2 and mRNA-1273 COVID-19 vaccine effectiveness against the SARS-CoV-2 Delta variant in Qatar. Nat. Med. 2021;27:2136–2143. doi: 10.1038/s41591-021-01583-4. [DOI] [PubMed] [Google Scholar]
- 83.Chandrashekar A., Liu J., Yu J., McMahan K., Tostanoski L.H., Jacob-Dolan C., Mercado N.B., Anioke T., Chang A., Gardner S., et al. Prior infection with SARS-CoV-2 WA1/2020 partially protects rhesus macaques against reinfection with B.1.1.7 and B.1.351 variants. Sci. Transl. Med. 2021;13 doi: 10.1126/scitranslmed.abj2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Painter M.M., Mathew D., Goel R.R., Apostolidis S.A., Pattekar A., Kuthuru O., Baxter A.E., Herati R.S., Oldridge D.A., Gouma S., et al. Rapid induction of antigen-specific CD4(+) T cells is associated with coordinated humoral and cellular immunity to SARS-CoV-2 mRNA vaccination. Immunity. 2021;54:2133–2142.e3. doi: 10.1016/j.immuni.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Angyal A., Longet S., Moore S.C., Payne R.P., Harding A., Tipton T., Rongkard P., Ali M., Hering L.M., Meardon N., et al. T-cell and antibody responses to first BNT162b2 vaccine dose in previously infected and SARS-CoV-2-naive UK health-care workers: a multicentre prospective cohort study. Lancet. Microbe. 2022;3:e21–e31. doi: 10.1016/S2666-5247(21)00275-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Munoz-Fontela C., Widerspick L., Albrecht R.A., Beer M., Carroll M.W., de Wit E., Diamond M.S., Dowling W.E., Funnell S.G.P., Garcia-Sastre A., et al. Advances and gaps in SARS-CoV-2 infection models. PLoS Pathog. 2022;18 doi: 10.1371/journal.ppat.1010161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dong W., Mead H., Tian L., Park J.G., Garcia J.I., Jaramillo S., Barr T., Kollath D.S., Coyne V.K., Stone N.E., et al. The K18-Human ACE2 Transgenic Mouse Model Recapitulates Non-severe and Severe COVID-19 in Response to an Infectious Dose of the SARS-CoV-2 Virus. J. Virol. 2022;96 doi: 10.1128/JVI.00964-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Valyi-Nagy I., Matula Z., Gonczi M., Tasnady S., Bekő G., Reti M., Ajzner E., Uher F. Comparison of antibody and T cell responses elicited by BBIBP-CorV (Sinopharm) and BNT162b2 (Pfizer-BioNTech) vaccines against SARS-CoV-2 in healthy adult humans. Geroscience. 2021;43:2321–2331. doi: 10.1007/s11357-021-00471-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lim J.M.E., Hang S.K., Hariharaputran S., Chia A., Tan N., Lee E.S., Chng E., Lim P.L., Young B.E., Lye D.C., et al. A comparative characterization of SARS-CoV-2-specific T cells induced by mRNA or inactive virus COVID-19 vaccines. Cell Rep. Med. 2022;3 doi: 10.1016/j.xcrm.2022.100793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brademan D.R., Riley N.M., Kwiecien N.W., Coon J.J. Interactive Peptide Spectral Annotator: A Versatile Web-based Tool for Proteomic Applications. Mol. Cell. Proteomics. 2019;18 doi: 10.1074/mcp.TIR118.001209. S193-S201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sahin U., Derhovanessian E., Miller M., Kloke B.P., Simon P., Lower M., Bukur V., Tadmor A.D., Luxemburger U., Schrors B., et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature. 2017;547:222–226. doi: 10.1038/nature23003. [DOI] [PubMed] [Google Scholar]
- 92.Kreiter S., Selmi A., Diken M., Sebastian M., Osterloh P., Schild H., Huber C., Tureci O., Sahin U. Increased antigen presentation efficiency by coupling antigens to MHC class I trafficking signals. J. Immunol. 2008;180:309–318. doi: 10.4049/jimmunol.180.1.309. [DOI] [PubMed] [Google Scholar]
- 93.Agerer B., Koblischke M., Gudipati V., Montano-Gutierrez L.F., Smyth M., Popa A., Genger J.W., Endler L., Florian D.M., Muhlgrabner V., et al. SARS-CoV-2 mutations in MHC-I-restricted epitopes evade CD8(+) T cell responses. Sci. Immunol. 2021;6 doi: 10.1126/sciimmunol.abg6461. eabg6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bilich T., Nelde A., Heitmann J.S., Maringer Y., Roerden M., Bauer J., Rieth J., Wacker M., Peter A., Horber S., et al. T cell and antibody kinetics delineate SARS-CoV-2 peptides mediating long-term immune responses in COVID-19 convalescent individuals. Sci. Transl. Med. 2021;13:eabf7517. doi: 10.1126/scitranslmed.abf7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Habel J.R., Nguyen T.H.O., van de Sandt C.E., Juno J.A., Chaurasia P., Wragg K., Koutsakos M., Hensen L., Jia X., Chua B., et al. Suboptimal SARS-CoV-2-specific CD8(+) T cell response associated with the prominent HLA-A∗02:01 phenotype. Proc. Natl. Acad. Sci. USA. 2020;117:24384–24391. doi: 10.1073/pnas.2015486117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hu C., Shen M., Han X., Chen Q., Li L., Chen S., Zhang J., Gao F., Wang W., Wang Y., et al. Identification of cross-reactive CD8(+) T cell receptors with high functional avidity to a SARS-CoV-2 immunodominant epitope and its natural mutant variants. Genes Dis. 2022;9:216–229. doi: 10.1016/j.gendis.2021.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Joag V., Wijeyesinghe S., Stolley J.M., Quarnstrom C.F., Dileepan T., Soerens A.G., Sangala J.A., O'Flanagan S.D., Gavil N.V., Hong S.W., et al. Cutting Edge: Mouse SARS-CoV-2 Epitope Reveals Infection and Vaccine-Elicited CD8 T Cell Responses. J. Immunol. 2021;206:931–935. doi: 10.4049/jimmunol.2001400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lee E., Sandgren K., Duette G., Stylianou V.V., Khanna R., Eden J.S., Blyth E., Gottlieb D., Cunningham A.L., Palmer S. Identification of SARS-CoV-2 Nucleocapsid and Spike T-Cell Epitopes for Assessing T-Cell Immunity. J. Virol. 2021;95 doi: 10.1128/JVI.02002-20. e02002-e02020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lineburg K.E., Grant E.J., Swaminathan S., Chatzileontiadou D.S.M., Szeto C., Sloane H., Panikkar A., Raju J., Crooks P., Rehan S., et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses. Immunity. 2021;54:1055–1065.e5. doi: 10.1016/j.immuni.2021.04.006. e1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu J., Sun Y., Qi J., Chu F., Wu H., Gao F., Li T., Yan J., Gao G.F. The membrane protein of severe acute respiratory syndrome coronavirus acts as a dominant immunogen revealed by a clustering region of novel functionally and structurally defined cytotoxic T-lymphocyte epitopes. J. Infect. Dis. 2010;202:1171–1180. doi: 10.1086/656315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nagler A., Kalaora S., Barbolin C., Gangaev A., Ketelaars S.L.C., Alon M., Pai J., Benedek G., Yahalom-Ronen Y., Erez N., et al. Identification of presented SARS-CoV-2 HLA class I and HLA class II peptides using HLA peptidomics. Cell Rep. 2021;35 doi: 10.1016/j.celrep.2021.109305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Nguyen T.H.O., Rowntree L.C., Petersen J., Chua B.Y., Hensen L., Kedzierski L., van de Sandt C.E., Chaurasia P., Tan H.X., Habel J.R., et al. CD8(+) T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope display high naive precursor frequency and TCR promiscuity. Immunity. 2021;54:1066–1082.e5. doi: 10.1016/j.immuni.2021.04.009. e1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nielsen S.S., Vibholm L.K., Monrad I., Olesen R., Frattari G.S., Pahus M.H., Hojen J.F., Gunst J.D., Erikstrup C., Holleufer A., et al. SARS-CoV-2 elicits robust adaptive immune responses regardless of disease severity. EBioMedicine. 2021;68 doi: 10.1016/j.ebiom.2021.103410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Obermair F.J., Renoux F., Heer S., Lee C.H., Cereghetti N., Loi M., Maestri G., Haldner Y., Wuigk R., Iosefson O., et al. High-resolution profiling of MHC II peptide presentation capacity reveals SARS-CoV-2 CD4 T cell targets and mechanisms of immune escape. Sci. Adv. 2022;8 doi: 10.1126/sciadv.abl5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Rha M.S., Jeong H.W., Ko J.H., Choi S.J., Seo I.H., Lee J.S., Sa M., Kim A.R., Joo E.J., Ahn J.Y., et al. PD-1-Expressing SARS-CoV-2-Specific CD8(+) T Cells Are Not Exhausted, but Functional in Patients with COVID-19. Immunity. 2021;54:44–52.e3. doi: 10.1016/j.immuni.2020.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schreibing F., Hannani M.T., Kim H., Nagai J.S., Ticconi F., Fewings E., Bleckwehl T., Begemann M., Torow N., Kuppe C., et al. Dissecting CD8+ T cell pathology of severe SARS-CoV-2 infection by single-cell immunoprofiling. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1066176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Shomuradova A.S., Vagida M.S., Sheetikov S.A., Zornikova K.V., Kiryukhin D., Titov A., Peshkova I.O., Khmelevskaya A., Dianov D.V., Malasheva M., et al. SARS-CoV-2 Epitopes Are Recognized by a Public and Diverse Repertoire of Human T Cell Receptors. Immunity. 2020;53:1245–1257.e5. doi: 10.1016/j.immuni.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gallien S., Kim S.Y., Domon B. Large-Scale Targeted Proteomics Using Internal Standard Triggered-Parallel Reaction Monitoring (IS-PRM) Mol. Cell. Proteomics. 2015;14:1630–1644. doi: 10.1074/mcp.O114.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoffmann M., Kleine-Weber H., Schroeder S., Kruger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fukushi S., Mizutani T., Saijo M., Matsuyama S., Miyajima N., Taguchi F., Itamura S., Kurane I., Morikawa S. Vesicular stomatitis virus pseudotyped with severe acute respiratory syndrome coronavirus spike protein. J. Gen. Virol. 2005;86:2269–2274. doi: 10.1099/vir.0.80955-0. [DOI] [PubMed] [Google Scholar]
- 112.Lawson N.D., Stillman E.A., Whitt M.A., Rose J.K. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Wolf F.A., Angerer P., Theis F.J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 2018;19:15. doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bredikhin D., Kats I., Stegle O. MUON: multimodal omics analysis framework. Genome Biol. 2022;23:42. doi: 10.1186/s13059-021-02577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mule M.P., Martins A.J., Tsang J.S. Normalizing and denoising protein expression data from droplet-based single cell profiling. Nat. Commun. 2022;13:2099. doi: 10.1038/s41467-022-29356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RAW MS data files and interpreted data were uploaded to the MassIVE repository: MSV000091008 (https://massive.ucsd.edu/ProteoSAFe/static/massive.jsp).
-
•
Single-cell RNA sequencing data are deposited on GEO: GSE222403 and are publicly available as of the date of publication. Accession numbers are listed in the key resources table.
-
•All codes for analysis and plots are available on Zenodo: https://doi.org/10.5281/zenodo.7789841 and GitHub:
- o Epitope analysis: https://github.com/YolandaHuitianDiao/BNT162b4_epitopes.git.
- o Single cell analysis: https://github.com/YolandaHuitianDiao/BNT162b4_scRNA.git.