Abstract
The pedogenesis from the mineral substrate released upon glacier melting has been explained with the succession of consortia of pioneer microorganisms, whose structure and functionality are determined by the environmental conditions developing in the moraine. However, the microbiome variability that can be expected in the environmentally heterogeneous niches occurring in a moraine at a given successional stage is poorly investigated. In a 50 m2 area in the forefield of the Lobuche glacier (Himalayas, 5050 m above sea level), we studied six sites of primary colonization presenting different topographical features (orientation, elevation and slope) and harbouring greyish/dark biological soil crusts (BSCs). The spatial vicinity of the sites opposed to their topographical differences, allowed us to examine the effect of environmental conditions independently from the time of deglaciation. The bacterial microbiome diversity and their co‐occurrence network, the bacterial metabolisms predicted from 16S rRNA gene high‐throughput sequencing, and the microbiome intact polar lipids were investigated in the BSCs and the underlying sediment deep layers (DLs). Different bacterial microbiomes inhabited the BSCs and the DLs, and their composition varied among sites, indicating a niche‐specific role of the micro‐environmental conditions in the bacterial communities' assembly. In the heterogeneous sediments of glacier moraines, physico‐chemical and micro‐climatic variations at the site‐spatial scale are crucial in shaping the microbiome microvariability and structuring the pioneer bacterial communities during pedogenesis.
INTRODUCTION
The Himalayas has been defined as the ‘third pole’ because of a relatively large land surface coverage by ice glaciers and the harsh climatic conditions above 5000 m altitude. Such conditions, which include large diurnal temperature fluctuations, strong winds and high UV radiations, make the Himalayas one of the most extreme terrestrial environments on Earth (Yao et al., 2012). Unlike other similar ecosystems, such as the hyper‐arid Atacama Desert (Wierzchos et al., 2018) and the cold Antarctic desert (Orellana et al., 2018), the Himalayas region has a less extensive microbial ecology investigation (Dhakar & Pandey, 2020; Ezzat et al., 2022), because of the geographical remoteness and roughness that historically hampered its exploration (Lami et al., 2010; Matthews et al., 2020). As other mountain regions worldwide, also the Himalayas ecosystems are threatened by climate change (Hamid et al., 2020; Tak & Keshari, 2020; Zhao et al., 2020) that determines a progressive melting of glaciers and recession of their fronts (Adler et al., 2019; Maurer et al., 2019; Shean et al., 2020). Thus, scientific investigations focused on the changes in the hydrogeology of rivers, high‐altitude basins, ice melting, precipitations (Jury et al., 2020; Salerno et al., 2015), and their consequences on the vulnerability of local communities and ecosystems (Ashton & Zhu, 2020; Heath et al., 2020; Xu et al., 2009) are strongly demanded.
As an effect of glacier melting, a bare mineral substrate is exposed during ice retreat, increasing the size of ice‐free areas in the foreland (Diolaiuti et al., 2011; Nuth et al., 2010). During the time out of the ice cover, this oligotrophic substrate, poor in nutrients and with a small amount of organic carbon, nitrogen and other nutrients, undergoes physical, chemical and microbiological modifications that favour primary colonization and the associated biogeochemical transformations (Garrido‐Benavent et al., 2020) that pave the later stages of plants biocenosis establishment (Borin et al., 2010; Mapelli et al., 2011, 2018). In this context, glacier forefields provide unique opportunities to study the early stages of soil pedogenesis and biotic colonization, a poorly investigated aspect in high‐elevation ecosystems like the Third Pole (Sherpa et al., 2020).
In the early succession stages, pioneer microorganisms, including cyanobacteria, chemoheterotrophic and diazotrophic bacteria, fungi, algae, lichens and bryophytes, are the main players in colonizing the moraine barren debris (Warren et al., 2019; Wietrzyk‐Pełka et al., 2020) and glacier‐fed streams (Busi et al., 2022; Ezzat et al., 2022; Fodelianakis et al., 2022; Kohler et al., 2022). These pioneer microorganisms can form complex structures named biological soil crusts (BSCs) (Perera et al., 2018), contributing to key ecosystem services, such as biogeochemical cycles, water retention and stabilization/consolidation of the proto‐soil against erosion (Barger et al., 2006; Ghiloufi et al., 2019; Maestre et al., 2012; Mapelli et al., 2012; Rippin et al., 2018; Weber et al., 2015). Among prokaryotes forming the BSCs, the aerobic photoautotrophs cyanobacteria are the most studied (Pessi et al., 2019): they guarantee primary productivity, providing nitrogen and carbon to the substrates for the proliferation of the other BSC components, such as chemolithoautotrophic microorganisms—equally important in the primary succession processes. For instance, in the high Arctic moraine of the Midtre Lovénbreen glacier, the rock weathering mediated by the chemolithoautotrophic bacterium Acidithiobacillus ferrooxidans increased water retention, favouring the formation of a soil fertility island, which fostered plant establishment and growth (Borin et al., 2010; Mapelli et al., 2011).
The physical and chemical composition of the proto‐soil matrix, together with the climatic factors, affect the dynamics, structure and functionality of the BSCs‐associated communities in a complex interplay between the biotic and abiotic components of the ecosystem (Bourquin et al., 2022; Mallen‐Cooper et al., 2020; Schulz et al., 2013). The stages of succession can last several years in cold and high‐elevation environments (Schmidt et al., 2008) and BSCs from different points of moraine chronosequences differ remarkably in their development and microbial diversity (Mapelli et al., 2018). However, moraines are not topographically uniform and the terrain layout in each point of a chronosequence can be rather variable in terms of slope, spatial orientation, exposure to light and climatic factors (wind, precipitations, etc.), thus imposing variable selective pressures that determine different communities.
In the present study, we selected the foreland of the Lobuche glacier (5050 m above sea level [asl]) in the Khumbu Valley, located in the eastern area of the Sagarmatha National Park in Nepal, as a model site to investigate the involvement of BSCs in the process of primary colonization of the mineral substrate released in the glacier moraine. We test the hypotheses that: (i) bacterial communities associated with the BSCs differ from those of the underlying deep layers (DLs; i.e., mineral substrate between BSC and permafrost) and (ii) irrespective of the substrate types (BSC and DL), bacterial communities are specific for each site because of the niche‐specific microenvironmental conditions. The rationale behind the hypothesis is that the variable topography, microenvironment and substrate properties across the irregular spots in glacier moraines weigh in the shaping of microbial diversity in the rocky DLs and the overlying BSCs. A study area consisting of a 50 m2 stone pit has been identified in the ablation tongue of the glacier and it is featured by a mosaic of hills and depressions (Jones et al., 2019). Six sites presenting greyish/dark BSCs as signs of primary colonization were selected within such area (Figure 1). In each of the sites, BSCs have developed under different microenvironmental conditions. The close spatial proximity of these sites within the studied area abolishes the effect of time associated with points along chronosequences, thus allowing us to examine the effect of environmental variability. The BSCs and the corresponding belowground DLs were collected from each site to unveil the interactions among the micro‐climatic and physico‐chemical conditions and the microbial components in structuring the pioneer colonization of the barren substrate and driving the process of soil formation in the high‐elevation environment of the Himalayas. We combined membrane lipid analysis and high‐throughput sequencing of the bacterial 16S rRNA gene to unveil the community structure of the microbiome inhabiting the BSCs and the DLs. Through this approach, we have identified environmental factors driving a site‐dependent bacterial assembly of BSCs and DLs that add the microbiome microvariability perspective to the current knowledge on pedogenesis at the Third Pole.
FIGURE 1.
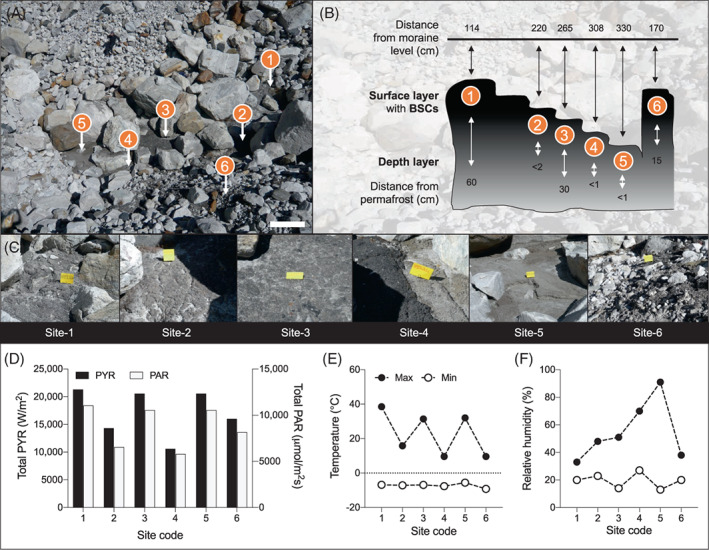
Location and environmental characteristics of the sampling microsites within the Lobuche glacier moraine. (A) Representative picture of the studied area. The six microsites selected for sampling are indicated. (B) Schematic representation of the sampling area; relative distances from BSCs to moraine level and the underlying permafrost are reported (values are expressed in centimetres). (C) Images of greyish/black biological soil crusts covering the mineral substrate in the six sites; yellow markers: 7 × 3 cm. (D) Solar radiation received by the sites reported as PYR solar radiation (W/m2; primary y‐axis) and photosynthetically active radiation photon flux (PAR, μmol/m2s; secondary y‐axis); results are expressed as the sum of the values measured in 24 h. (E) Air temperature and (F) air relative humidity, respectively; minimum and maximum values are reported as white and black circles, respectively.
EXPERIMENTAL PROCEDURES
Study sites and sample collection
In October 2009, an area showing evidence of primary colonization was identified in the forefield of the Lobuche glacier, Nepal, at 5050 m asl (27°57′24.92″ N, 86°48′35.81″ E; Figure 1A). The region is classified as a cold desert, experiencing long winter periods with temperatures below 0°C, while summer periods are short and arid, although snowmelt and occasionally rainfall pulses locally increase moisture content (Derin et al., 2019; Salerno et al., 2015). The study area was in a stone pit in the glacier forefield, located in the relic debris‐mantled ablation zone (~1 km in length) that is disconnected from the ice accumulation zone of the glacier. The topography of this ablation tongue is featured by a mosaic of hills and depressions (Jones et al., 2019). Samples were collected from six sites (named 1–6) located in a pit depression (from 1.14 to 3.30 m depth compared to the moraine level; schematic representation in Figure 1B), covering an overall area of 50 m2. The sites were selected based on the presence of greyish/dark biological patinas (thereafter defined as BSCs) positioned in different topographical conditions (hills/depressions) present in the moraine (Figure 1). The BSCs were poorly developed (thicknesses up to 5 mm; Table 1) without any presence of lichens, mosses and vascular plants (Figure 1C). For each of the six sites, BSCs and the relative below mineral substrates (hereafter defined as DL) were collected in triplicate ∼5 cm apart from each other and stored at −20°C for subsequent analysis. Samples for physico‐chemical characterization were air‐dried for 24 h and stored at room temperature until the analysis in the laboratory. Samples of BSC and DL were collected from each site and immediately soaked in methanol for further intact polar lipid (IPL) analysis.
TABLE 1.
Physico‐chemical characterization of biological soil crusts (BSCs) and depth layers (DLs) in the different sites (1–6) within the study area in the forefield of the Lobuche glacier.
| Layer | Site 1 | Site 2 | Site 3 | Site 4 | Site 5 | Site 6 |
|---|---|---|---|---|---|---|
| BSC | ||||||
| Colour | Grey | Brown‐grey | Brown‐grey | Black | Light grey | Black d |
| Thickness (mm) | 1 | 3 | 1 | 2 | 0.8 | 5 |
| RH% a | 5.2 ± 0.2 | 54.7 ± 4.1 | 26.8 ± 2.2 | 43.9 ± 5.3 | 16.7 ± 3.3 | 5.6 ± 1.5 |
| pH | 6.52 ± 0.01 | 5.44 ± 0.2 | 6.43 ± 0.17 | 6.17 ± 0.04 | NA | 4.78 ± 0.04 |
| C (g kg−1 DM) | 18 ± 1.9 | 56.6 ± 12.1 | 24.2 ± 7.8 | 46.1 ± 11.3 | NA | 70.5 ± 6.2 |
| N (g kg−1 DM) | 1.5 ± 0.7 | 3.5 ± 1.1 | 2.6 ± 0.6 | 3.9 ± 1.1 | NA | 3.1 ± 0.2 |
| DOC (mg kg−1 DM) | 1296 ± 6 | 2033 ± 15 | 2073 ± 21 | 3399 ± 28 | NA | 2430 ± 18 |
| IC50 b | 1.36 ± 0.08 | 3.45 ± 0.6 | 2.11 ± 0.17 | 3.65 ± 0.21 | NA | 3.55 ± 0.2 |
| DL | ||||||
| Thickness (cm) | 60 | 2 | 30 | 0.8 | 2 | 15 |
| WHC | 20.1 ± 0.4 | 18.1 ± 1.8 | 20.8 ± 2.2 | 19.3 ± 2.3 | 20.9 ± 0.5 | 23.3 ± 0.7 |
| pH | 5.76 ± 0.85 | 6.03 ± 0.06 | 6.24 ± 0.18 | 6.4 ± 0.03 | 6.66 ± 0.35 | 6.24 ± 0.46 |
| CEC | 1.89 ± 0.43 | 2.02 ± 0.34 | 1.36 ± 0.04 | 1.62 ± 0.04 | 2.11 ± 0.04 | 2.69 ± 1.05 |
| K exchangeable c | 389 ± 53 | 341 ± 23 | 380 ± 1 | 326 ± 4 | 366 ± 7 | 376 ± 32 |
| Ca exchangeable c | 340 ± 53 | 323 ± 69 | 423 ± 37 | 285 ± 48 | 378 ± 33 | 313 ± 46 |
| Al exchangeable c | 21.7 ± 0.6 | 7.7 ± 0.6 | 10.9 ± 1.2 | 5.4 ± 0.1 | 9.3 ± 0.2 | 4.1 ± 0.1 |
| Mg exchangeable c | 64.8 ± 13.9 | 52.5 ± 20.7 | 57 ± 21.4 | 69.2 ± 9.2 | 84.4 ± 9.2 | 73.5 ± 10.7 |
| Na exchangeable c | 53.6 ± 7.2 | 61.4 ± 8.5 | 68.3 ± 4.7 | 69 ± 7 | 236.7 ± 38.6 | 78.2 ± 2.9 |
| Fe exchangeable c | 33.5 ± 5.5 | 32.2 ± 8 | 27.3 ± 4.3 | 23.6 ± 0.6 | 69.2 ± 9.2 | 38.9 ± 0.8 |
| P | 5.6 ± 1.4 | 10.5 ± 0.9 | 13 ± 1.6 | 12.2 ± 4.2 | 7 ± 1.2 | 11.99 ± 0.27 |
| TKN | 74 ± 4 | 71.1 ± 3.2 | 70.5 ± 3.1 | 59.1 ± 2.6 | 48.2 ± 2.9 | 36.19 ± 2.06 |
| C | u.l.d. | u.l.d. | u.l.d. | u.l.d. | u.l.d. | u.l.d. |
| Microsurface | 4.81 ± 0.03 | 4.73 ± 0.28 | 4.37 ± 0.18 | 5.09 ± 0.62 | 4.87 ± 0.06 | 5.04 ± 0.76 |
Abbreviations: CEC, cation exchange capacity; DM, dry matter; DOC, dissolved organic carbon; NA, not available due to the limited amount of material available; RH, relative humidity; TKN, total Kjeldahl nitrogen; ULD, under the limit of detection; WHC, water holding capacity.
Measured in the field with the hydrometer.
Antioxidant activity measured as milligrams of DPPH per milligrams of dry BSC.
Exchangeable measured as milligrams per kilogram of DM.
Globular.
Measurement of environmental parameters in situ
Environmental parameters such as minimal and maximal air temperature (°C), air humidity (%), percentage of moisture in BSC (% relative humidity [RH]; hygrometer Sama Tolls, Italy), total solar radiation (PYR) and photosynthetically active radiation (PAR; PAR SENSOR QSO‐S PAR Photon Flux, METER Group, Inc., USA) were recorded to assess the micro‐environmental influence on bacterial community structure in each site.
Physico‐chemical and geological analysis
DLs were sieved after drying into fine (<2 mm) and coarse (>2 mm) particles. The pH of DLs was determined in an aqueous solution using a 1:2.5 soil/water ratio; total nitrogen by the Kjeldahl method (total Kjeldahl nitrogen [TKN]); available P using the Olsen method; and organic carbon by wet oxidation. For cation exchange capacity (CEC) and exchangeable Ca, Mg, K, Al, Fe and Na determinations, samples were saturated with BaCl2‐triethanolamine solution (pH 8.1) and exchangeable cations were determined by inductively coupled plasma (ICP‐MAS VARIAN, Liberty AX, Walnut Creek, CA, USA). All analytical data were determined on the fine particle fraction (<2 mm); all the above methods were described in Mapelli et al., 2018. Water holding capacity was determined by the Stackman box method (Mapelli et al., 2018).
In the case of BSC, samples were sieved after drying; pH, nitrogen and carbon content were determined as indicated above. Dissolved organic matter (DOM) was extracted from transect crust with deionized water, using 5 g of equivalent dry material (1:2 solid:liquid ratio, w/w) in a Dubnoff bath at 60 rpm for 30 min at room temperature. The obtained suspension was centrifuged for 15 min at 6500 rpm and then vacuum filtered twice: firstly by using a fast glass fibre filter (Whatman GF 6) and then by using a cellulose acetate membrane filter (Whatman OE 67) of 0.45 μm, then its organic carbon content (dissolved organic carbon [DOC]) was determined as reported before (Mapelli et al., 2018). BSCs antioxidant activity was assessed using the DPPH radical scavenging method (Gulcin, 2020). Briefly, an aliquot of DPPH (125 μM) (Prot. N. D9132, Sigma Aldrich, Dermstadt, Germany) solution in methanol was added to DOM. The decrease in absorbance at 517 nm was recorded by a spectrophotometer (Cary 60 UV–Vis, Agilent Technologies, Santa Clara, CA, USA). The results were expressed as IC50, that is, the DOC concentration that scavenges 50% of DPPH determined after 30 min of reaction. All the analyses were performed in triplicate. The biological material of BSC at Site 6 was not enough to perform physico‐chemical analyses.
The physico‐chemical tables containing the data from BSCs and DLs of the sites were squared transformed and used to create a resemblance matrix using the Euclidean distance in PRIMER (Anderson et al., 2008). Canonical analysis of principal components (CAP) was used to visualize site variation. Significant differences in physico‐chemical composition were investigated by permutational analysis of variance (PERMANOVA) in Primer (Anderson et al., 2008), considering the factor ‘Site’ as a fixed and orthogonal factor (5 and 6 levels in BSCs and DLs, respectively). PERMANOVA pair‐wise tests were also conducted to evaluate the effect of ‘Site’ in both sample categories, BSC and DLs. The contribution of the variables to the physico‐chemical differences among sites was assessed by the analysis of similarity percentages (SIMPER) in PRIMER (Anderson et al., 2008).
X‐ray powder diffraction
X‐ray powder diffraction (XRD) analyses were performed after micro‐sampling proto‐soil crusts. XRD measurements were carried out using an X'Pert Panalytical Diffractometer working in Bragg–Brentano geometry and equipped with an X'Celeretor Detector. Each sample was manually ground in an agate mortar and then analysed in the 3–80° 2 range (Cu‐wavelength, 40 kV, 40 mA) with a step size of 0.02° and a counting time of 20 s. Qualitative mineralogical phase analysis was performed based on peak position (Patterson, 1955), and then indications about their quantities were obtained based on the peak intensities.
Scanning electron microscope analysis
Ultramicroscopic analyses employed a Cambridge 360 scanning electron microscope (SEM), imaging both secondary and back‐scattered electrons. Some elemental analyses were performed to identify mineral constituents, with an energy dispersive x‐ray analysis (EDS Link Isis 300) requiring carbon‐coated samples: energy dispersive x‐ray spectroscopy with an accelerating voltage of 20 kV, filament intensity 1.70 A and probe intensity of 280 pA.
Intact polar lipid analysis
To analyse lipid biomarkers approximately, 2.5–8 g (dry weight) of soil material was collected and transferred with methanol (MeOH)‐cleaned spatulas into combusted glass vials and sealed with MeOH‐cleaned Teflon‐coated screw caps. The samples were immediately inundated with MeOH to prevent further microbial activity and stored at ambient temperatures in the dark. In the laboratory, 2 μg phospholipid standard di‐C21‐PC (Avanti Polar Lipids, USA) was added to all samples and lipids were extracted using a modified Bligh and Dyer procedure (Sturt et al., 2004). For the first two extraction steps, MeOH:dichloromethane (DCM):phosphate buffer (2:1:0.8, v/v) was used, followed by a third step using MeOH:DCM (1:3, v/v). For IPL analysis, an aliquot of the total lipid extract was measured in positive ion mode on a Bruker maXis Plus ultra‐high‐resolution quadrupole time‐of‐flight mass spectrometer coupled with an electrospray ionization source to a Dionex Ultimate 3000RS ultra‐high‐pressure liquid chromatograph. A Waters Acquity UHPLC BEH Amide column was used for HILIC (hydrophilic interaction liquid chromatography) separation of IPLs following the protocol described in Wörmer et al., 2013. For quantification, peak areas of individual IPLs were normalized to the internal di‐C21‐PC standard and response factors were corrected using commercially available standards as described in Schubotz et al., 2018. A second aliquot was separated into a sterol‐containing alcohol fraction and free fatty acid‐containing acid fraction using a solid‐phase extraction (Hinrichs et al., 2000) after derivatization with BSTFA and BF3, to form respective TMS (trimethylsilyl)‐derivatives and FAMEs (fatty acid methyl esters), the alcohol and fatty acid fractions were analysed on a ThermoFinnigan Trace GC coupled to a Finnigan DSQ for identification and a Finnigan FID for quantification.
The quantitative IPLs data from BSCs and DLs were log‐transformed to avoid overdispersion before creating a resemblance matrix using the Euclidean distance in PRIMER (Anderson et al., 2008). CAP was used to evaluate differences in IPLs composition (Anderson et al., 2008), considering the factor ‘Type’ as a fixed factor (two levels: BSCs and DLs). To explore and visualize the distribution of samples in the ordination space, the principal coordinates analysis (PCoA) was built in PRIMER.
DNA extraction and sequencing
Total DNA was extracted from 0.5 ± 0.1 g of dry BSC and DL samples with the PowerSoil DNA Isolation Kit (MoBio Inc., CA, USA), following the manufacturer's instructions. DNA quality was evaluated on 0.8% agarose gel, quantified using a NanoDrop Microvolume Spectrophotometer (Thermo Fisher Scientific) and stored at −20°C until further processing. High‐throughput Illumina sequencing was performed on the V3–V4 hypervariable regions of the 16S rRNA gene fragments by PCR amplification using 341F and 785R primers at Macrogen Inc., South Korea. Raw sequences were analysed with QIIME pipeline, including quality filtering, trimming, dereplication of the sequences and creation of operational taxonomic units (OTUs at 97% of similarity), as previously described (Booth et al., 2019). Representative sequences of each OTUs were aligned with the database in QIIME using uclust (Caporaso et al., 2010) and blast commands to search against the SILVA version 138 (Quast et al., 2012). After removing singletons, sequences non‐assigned to bacteria (i.e., plastids, archaea and unassigned), and sequences with relative abundance <0.001% in the entire dataset, a total of 3,477,016 reads with an average length of 300 bp were obtained (range of reads per sample: 37,725–133,904; Table S1). Rarefaction curves for the BSC and DL samples were reported in Figure S1. Sequences were deposited to the Sequence Read Archive of NCBI under the BioProject PRJNA698068. The bacterial OTU table was used to calculate alpha‐diversity indices (Shannon diversity and observed richness) in R using the Phyloseq package. Occupancy‐abundance curves were generated by calculating the number of samples in which a certain OTU was detected and its total relative abundance. The OTUs shared among different sites in BSCs and DLs were identified by a Venn‐diagram analysis in R. OTUs were tested for their differential abundance (enrichment and depletion; i.e., log2‐fold change) between BSC and DL using the DEseq2 package in R (Love et al., 2014). Considering BSC and DL separately, we computed the differential abundance of OTUs across sites and plotted the related taxonomic data through a differential heat tree using the package Metacoder (Foster et al., 2017). Linear discriminant analysis effect size (LEfSe) was further used to determine the bacterial discriminants between sites (Segata et al., 2011). The most important OTUs that contribute to the dissimilarity between bacterial communities among micro‐environments in BSC and DL were defined by SIMPER in PRIMER (Anderson et al., 2008). Beta‐diversity of bacterial communities was analysed using the compositional Bray–Curtis (BC) similarity matrix of the relative log‐transformed OTUs‐table in PRIMER (Anderson et al., 2008). The BC‐similarity matrix was used to perform the CAP and PERMANOVA to statistically test the impact of the categorical explanatory variable ‘Type’ (two levels: BSC and DL), along with the variable ‘Site’ nested in ‘Type’ (6 levels for both BSC and DL: 1, 2, 3, 4, 5 and 6). PERMANOVA pair‐wise tests were also conducted to evaluate the effect of ‘Site’ for BSC and DL. We computed the beta‐diversity components by using the beta.div.comp function of the package adespatial (Dray et al., 2018). The OTU table was used to infer the mechanisms driving the bacterial communities assembly in BSCs and DLs by applying a phylogenetic bin‐based null model (iCAMP) version 1.2.9 with recommended default settings (Ning et al., 2020). Sloan neutral community model was applied to predict the relationship between the frequency of occurrence of taxa in a community, that is, BSC and DL, and assessed the potential importance of the neutral or stochastic process in shaping bacterial community (Sloan et al., 2006). This model evaluates whether the microbial assembly from a metacommunity follows a neutral model or a niche‐based process as a function of the metacommunity log abundance. The fitting of the model was performed in the R environment using non‐linear least‐squares fitting and the minpack.lm, Hmisc and stats4 packages (Chen et al., 2019). The 95% confidence interval around all fitting statistics was calculated by bootstrapping with 1000 replicates. The taxa were subsequently separated into three fractions depending on whether they occurred more frequently than the neutral model predictions (over‐predicted fraction), less frequently than the neutral model predictions (under‐predicted fraction), or within the 95% confidence interval of the neutral model predictions (neutrality interval). The proportion of variability R 2 quantifies the fit level of detection frequency to the model, Nm is an estimate of dispersal between communities in which N is the metacommunity size and m is the rate of the individuals immigrating from the source community into the local community. We also applied this model by considering a taxon's abundance in a source metacommunity (DL or BSC) and its occurrence frequency in the target metacommunity (BSC or DL) to test the source‐sink hypothesis (Burns et al., 2016). Finally, to quantitatively estimate the potential contribution of DL bacterial communities (here given as source environment) to those of BSC (here given as sink environment), we used the Bayesian‐based SourceTracker (Knights et al., 2011) available as an R package at http://sourcetracker.sf.net.
The rate of decay of the bacterial community's similarity (BC) across the sites was evaluated in function of the site distance (m), and Euclidean distance matrices of physico‐chemical and micro‐climatic conditions of the sites and of the IPL diversity for both BSCs and DLs; linear regressions were computed in GraphPad Prism. Significant physico‐chemical variables explaining the bacterial communities structure (16S rRNA gene‐based) were assessed by using a distance‐based multivariate linear model (DistLM) in Primer (Anderson et al., 2008) and the overall best solutions (lowest corrected Akaike information criterion [AICc]) was indicated. The set of physico‐chemical variables measured in BSC and DL (explanatory variable) was used to assess the amount of variation of the bacterial community (multivariate response variables) by running the function Best.sq.r() from the mvabund package in R (Wang et al., 2012). The three most important physicochemical variables detected were further used to identify which bacterial members (relative abundance) were positively/negatively correlated with them in the two layers by running the Metacoder package in R (Foster et al., 2017).
Co‐occurrence network analysis
The bacterial phylotypes enriched in BSC and DL samples were used to build two correlation networks by calculating all pairwise Spearman correlation coefficients among these bacterial taxa in CoNet (Faust & Raes, 2016). We kept both negative and positive correlations with Spearman's correlation coefficient ρ > 0.5 and p < 0.01 to provide information on microbial taxa that may respond robustly to the environmental conditions of BSC and DL. The co‐occurrence network was visualized with Gephi (Bastian et al., 2009) and default parameters were used to identify their topological indices.
RESULTS
Environmental conditions of the studied area
The Lobuche glacier moraine is constituted by a randomly distributed patchwork of rocks with variable topographical features that is the norm across the debris‐mantled ablation zone of the glacier. In an area of 50 m2, we selected six sites in which greyish/dark BSCs develop on top of the mineral substrates DL (Figures 1A,C). The thickness of BSCs ranged from 0.8 to 5 mm, while DLs varied depending on the level of the underlying permafrost and ranged from <1 to 60 cm (Table 1). Evaluation of climatic parameters revealed that each site was characterized by unique micro‐environmental conditions in terms of time and intensity of exposition to light, temperature and RH (Figure 1D,F). Due to their position within the studied area (Figure 1A,B), the sites were differently exposed to the solar irradiation (Figures 1D and S2): sites 1, 3 and 5 received, on the day of sampling, about 8 h of sunlight per day (20,824 ± 425 PYR and 10,720 ± 291 PAR), while sites 2 and 6 received 6 h (15,181 ± 1204 PYR and 7360 ± 1154 PAR) and Site 4 received only 4 h (10,618 PYR and 5803 PAR) because partially shadowed by the surrounding rocks. Variable daily temperatures (25 ± 13°C) were also observed, reaching values over 30°C in sites 1, 3 and 5; while values below 20°C were recorded in sites 2, 4 and 6 (Figure 1E). All the sites had similar minimal temperatures of air (−7 ± 1°C) during the night. Air RH showed a specific pattern in which higher values (>50%) were observed in sites 3, 4 and 5, which were located more in‐depth compared to the average level of the moraine (Figure 1B), while sites 1, 2 and 6 showed values <50%; the minimum RH ranged between 13% and 27% across all sites (Figure 1F).
Physico‐chemical characterization of BSCs and DLs
Chemical microanalysis in conjunction with SEM (Figure S3a) revealed that BSCs presented on their surface mineral and organic constituents tightly connected. Calcium carbonate nodules were also observed in BSCs, along with nodules constituted by coalescent carbonate microtubules (Figures S3b,c). In addition, nuclear magnetic resonance (NMR) analysis revealed that the organic matter present in the BSCs was dominated by O‐CH3 or N‐alkyl, O‐alkyl C and di‐O‐alkyl C (from 60.91% to 72.65%), and aliphatic C bonded to other aliphatic chain or H (from 11.44% to 19.55%) but with different relative abundance across the six sites (Table S2). The physico‐chemical characteristics (Table 1) significantly differed across the sites (PERMANOVA: F 4,10 = 95.01, p = 0.001; Figure S4; pairwise comparison in Table S3). Among the chemical variables measured, DOC, RH and carbon (C) explained most of the observed chemical–physical diversity of BSCs across sites (up to 94%, Table S4a).
The DLs were consistently composed of thin grains of quartz, feldspar and mica (Table S5), while carbon was not detectable (Table 1). The DLs samples had high sand content (>90%) followed by a small proportion of loam (5%–6%) and clay (0.5%–5%), except for Site 5 where the latest represented up to 26% and 11% of the substrate material, respectively (Table S5). Despite similarity in term of texture, DLs had different compositions across the six sites (Table 1; PERMANOVA: F 5,12 = 31.71, p = 0.001; pairwise comparison in Table S3b). The most important variables that characterized such diversity among DLs in terms of physico‐chemical parameters were TKN and ion content (Fe, Na, TKN, Al and K; Table S4b).
Diversity of IPLs across BSCs and DLs
IPLs are major components of cells membrane and provide information about the living microbial community in the environmental samples (Schubotz et al., 2018). Based on their quantification, we found that the microbial biomass was mainly associated with BSCs, reaching up to 30‐fold higher levels than DLs (Figure 2A; average fold change, 8). The IPLs composition showed a separation between BSC and DL samples (CAP, choice of m = 2, trace statistic = 0.31, p = 0.034; PCoA in Figure 2B). Among IPLs, glycolipids (i.e., monoglycosyl diacylglycerod, diglycosyl diacylglycerol, triglycosyl diacylglycerol, sulfoquinovosyl diacylglycerol and glycoronic acid diacylglycerol) which are found in thylakoid membranes of all phototrophic organisms (Hölzl & Dörmann, 2007) were the most abundant lipid class: on average they represented 33% and 27% of total IPLs in BSCs and DLs, respectively (Figure 2C). Notably, the heterocyst glycolipids, a subgroup of glycolipids typical of nitrogen‐fixing filamentous cyanobacteria (Nichols & Wood, 1968), constituted 23% (range, 7%–42%) and 15% (6%–32%) of the total IPLs in BSCs and DLs, respectively. Betaine lipids, typical of lower plants and of some cyanobacteria (Klug & Benning, 2001; Řezanka et al., 2004), were abundant at all sites, representing up to 30% of total IPLs (Figure 2C). High relative levels of ornithine lipids, particularly trimethyl ornithine lipids that are exclusive bacterial markers (López‐Lara & Geiger, 2017; Moore, 2021), were observed in BSCs of sites 2, 3 and 6 (relative abundance range, 2%–3.8%). Even though eukaryotic biomass (e.g., mosses, lichens and bryophytes) was not visible in BSCs, the detection of sterols revealed its presence. The distribution of sterols (mainly ergosterol, stigmasterol and sitosterol) are comparable to what has been reported for Sphagnum species (Bryophyta) found in peat bogs (Baas et al., 2000) and were mainly detected in the BSCs of all sites, in varying relative abundances (Table S6). Sterols were below detection in the DLs Sites 1 and 6 (Table S6) and were mainly composed by phosphatidyl inositol (PI‐DAG), phosphatidyl choline, phosphatidyl ethanolamine, phosphatidyl glycerol and OH‐PC‐DAG (Figure 2C). Some of the IPLs detected presented multiple hydroxylations; these include betaine lipids, ornithine and trimethyl ornithine lipids and phosphatidyl choline lipids (Table S7). Betaine lipids were also observed with mixed ether and ester side chains and phosphatidyl ethanolamine as diether diacylglycerol, which suggests a bacterial origin for these lipids (Schubotz et al., 2009, 2018). It is important to note that while micro‐climatic conditions of the sites at the sampling time did not show any significant correlation with the content of IPLs groups (Pearson correlation p > 0.05), the RH of BSCs was significantly correlated with the content of glycolipids, phospholipids and betaine lipids (p < 0.05 and R 2, 0.89–0.91). No significant correlations were detected between the physico‐chemistry of DLs and the extracted IPLs. Major fatty acid carbon chain lengths (Table S6) and distributions of free fatty acids (Table S8) were also analysed. The most dominant combined carbon chain lengths of all IPL were C36:3, C36:4 and C34:3, C34:2, C34:1, C32:2 and C32:3, which matches with the distribution of the dominant free fatty acids C16:0, C18:1 and C18:2 (Figure S5 and Table S8).
FIGURE 2.
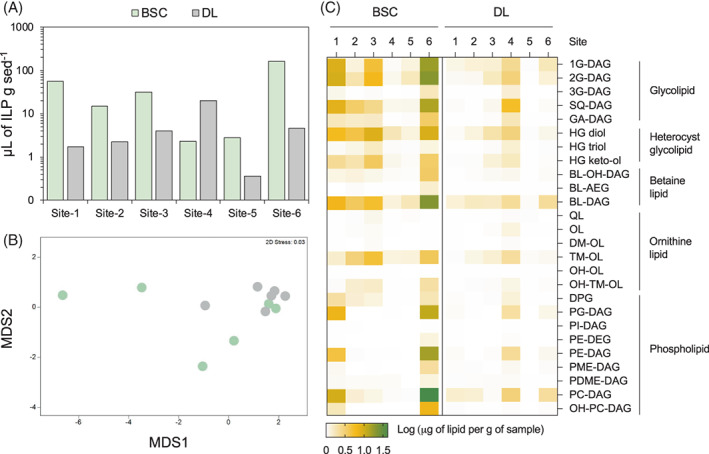
Intact polar lipid concentrations and compositions in BSCs and DLs across the six microsites. (A) Lipid concentration across the six sites in BSCs and DLs. Values are expressed as μL of IPL per gram of sediment. (B) Non‐metric multi‐dimensional scaling plot (based on Euclidean distances) of IPL composition for BSC and DL samples. (C) Relative abundance of the different IPL components across sites in BSC and DL samples; values are expressed as microlitres of IPL per gram of sediment and visualized as log‐transformed.
Structure and assembly of bacterial communities associated with BSCs and DLs
The bacterial communities associated with BSC and DL samples (3769 and 3886 OTUs, respectively; Table S1) were dominated by the presence of few abundant OTUs (9 and 11 OTUs, respectively, with relative abundance >1%) that accounted for up to 14% and 20% of the total number of reads, respectively (Figure S6). A long tail of rare OTUs with relative abundance ranging from 1% to 0.001% was detected in all the bacterial communities (3760 and 3875, respectively; Pareto‐like distribution in Figure S6). Most OTUs belonged to the phyla Proteobacteria (relative abundance, 26% and 24.1% in BSCs and DLs, respectively), Bacteroidetes (15.7% and 15.3%), Firmicutes (11.4% and 15.3%), Planctomycetes (8.7% and 7.7%), Acidobacteria (7.1% and 8.3%), Actinobacteria (5.9% and 9.4%), Cyanobacteria (6.9% and 3.3%), Chloroflexi (3.9% and 3.3%) and Armatimonadetes (3.8% and 2.2%; Figure S7). The bacterial communities associated with DL samples exhibited a higher diversity compared to those with BSCs in terms of species richness (number of OTUs; Mann–Whitney test, p = 0.0008), while Shannon's and Simpson's indices did not differ among the two types of samples (p = 0.51 and p = 0.48, respectively; Figure S8). The most abundant OTUs were conserved and consistently present in both BSCs and DLs (99% of shared OTUs and 91% of generalist OTUs, the latest defined as the OTUs equally distributed among BSCs and DLs; Figures 3A,B).
FIGURE 3.
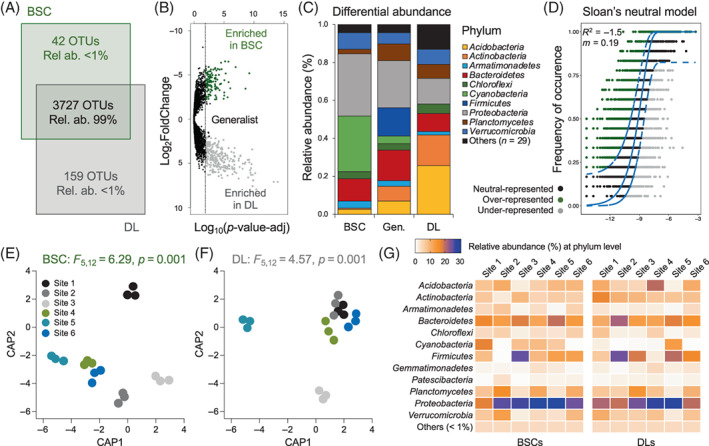
Distribution and assembly of bacterial communities associated with BSC and DL. (A) Venn diagram showing the total number of unique and shared OTUs between BSCs and DLs. (B) Volcano plot showing the OTUs that have a differential abundance (twofold changes, p < 0.05) between BSCs and DLs: OTUs enriched in BSCs are indicated by green dots and those enriched in DLs in green; generalist OTUs that do not show significantly different distribution between BSc and DL are indicated with black dots. (C) Taxonomy of BSC‐ and DL‐enriched OTUs and of generalist OTUs. (D) Fitting Sloan neutral community model with DL as the source bacterial community and BSC as the destination bacterial community (sink). The predicted neutral model (solid blue line) is surrounded by 95% confidence intervals (dashed blue lines). The frequency of detection of bacterial OTUs in BSC (sink community, y‐axis) is plotted against the relative abundance of bacterial OTUs in DL (source community, x‐axis) to indicate that the OTU is most likely neutrally dispersed into BSC (black points), over‐represented (selected for, green points), or under‐represented (selected against, grey points). (E, F) canonical analysis of principal coordinates (CAP) ordination based on Bray–Curtis similarity distances of (E) BSC and (F) DL. Colours indicate the different microsites analysed. (G) Relative abundance of most abundant phyla (average, >1%) across sites in BSC and DL samples; values are expressed as percentages calculated on the mean of the three replicates analysed.
Despite the high number of shared and generalist OTUs, the bacterial communities that inhabited BSCs and DLs differed significantly (PERMANOVA: F 1,34 = 2.94, p = 0.006; CAP cross‐validation 100%; variation explained, 25%). This specificity was explained by the differential distribution pattern (enrichment and depletion) between BSCs and DLs of 10% of OTUs (Figure 3B); 7% (n = 256) and 3% (n = 110) of OTUs were enriched in DLs and BSCs, respectively, and accounted for 8% and 10% of the total relative abundance in each type of samples. Among these, members of Alphaproteobacteria (27.7%), Oxyphotobacteria within cyanobacteria (29%) and Bacteroidia (12%) dominated the BSC‐enriched community, while Acidobacteria (26%; Blastocatellia 12.5% and Holophagae 7.4%), Actinobacteria (16%; Acidimicrobiia 11.8% and Thermoleophilia 3.7%) and Gammaproteobacteria (10.5%) were the main members of the DL‐enriched group (Figures 3C). We further evaluated the fitting of BSC and DL bacterial communities to the neutral community model (Sloan et al., 2006). While the neutral model failed to fit the frequency of occurrence of the BSC bacterial community (coefficient of the neutral fit, R 2 < 0 and migration rate, m = 0.0218; Figure S9a), it poorly fit that of DL (R 2 = 0.16 and m = 0.0435; Figure S9b). These results indicated that in both cases, a more substantial role of habitat (niche) filtering compared to stochastic processes drives the bacterial community assembly, that is, enrichment/exclusion of species with appropriate/inappropriate traits for given abiotic and biotic environmental conditions. In addition, when the source‐sink hypothesis was tested using DL bacterial communities as the source and BSC bacterial communities as the sink, the occurrence frequency of bacterial taxa did not fit the neutral community model (R 2 < 0 and m = 0.019; Figure 3D): the composition of BSCs is not driven by a source‐sink relationship with DL, and the probability that an individual who dies or leaves the BSC community is replaced by another individual immigrating from the DL community is limited. We ran the same analysis by considering BSC and DL bacterial communities as source and sink, respectively, and also in this case we rejected the source‐sink hypothesis (Figure S10). Even though the source‐sink hypotheses were rejected in both the combinations (DL vs. BSC and BSC vs. DL), we found that a limited part of the BSC bacteria community potentially originated/transferred from the below DL layer based on the community‐wide Bayesian model (SourceTracker; Figure S11). However, this portion decreased with the increasing thickness of the BSCs (regression in Figure S11b). The remaining BSC taxa could not be explained by DL and were identified by SourceTracker as ‘unknown’.
BSCs and DLs bacterial co‐occurrence
The network properties of the BSCs and DLs samples were largely comparable in size (number of nodes and total interactions) and had similar topological features (Table S9; Figure S12). For instance, OTUs engaging in significant associations represented 42% and 33% of the total OTUs present in the original community of BSC and DL, respectively. The associations (edges) were equally distributed among positive (co‐presence) and negative (mutual exclusion) interactions (Table S9). Both co‐occurrence networks followed a power‐law degree distribution in which most nodes had few connections (degree 1–5, 74% and 69% of nodes in BSCs and DLs, respectively) and only few nodes had numerous connections (Figure S13), suggesting the presence of a non‐random co‐occurrence pattern. Compared to the BSC network, the network of DLs showed higher degrees (Welch‐corrected test: t = 2.37, df = 661.1, p = 0.018) and higher density, along with lower path length and modularity (Table S9; Figure S12c). In addition, the DL nodes had lower betweenness (t = 4.57, df = 828.51, p < 0.0001), higher closeness (t = 2.016, df = 866, p = 0.044) and eccentricity (t = 3.40, df = 860.6, p = 0.0007) than those in BSCs samples (Figures S12d–f). These data indicate that the members of the DL‐associated bacterial communities established more complex, well‐connected and closer interactions compared to those occurring in BSCs.
The network interactions in BSCs and DLs showed a comparable taxonomic profile (proportion of node per phylum; Figure S12g) in agreement with the phylogenetic analysis previously described (Figure S7). The largest number of nodes was represented by members affiliated to Firmicutes, Bacteroidetes, Alphaproteobacteria, Acidobacteria and Actinobacteria, followed by Planctomycetes, Gammaproteobacteria, Chloroflexi, Cyanobacteria, Verrrucomicrobia and Armatimonadetes, and members of minor groups (i.e., Deltaproteobacteria, Deinococcus‐Thermus, FBP, Gemmatimonadetes, Omnitrophicaeota, Patescibacteria and WPS‐2). The phyla/classes detected with the higher node frequency were also responsible for the principal interactions within the networks, as revealed by the positive relationship between the number of nodes and degree per each taxonomic group (BSCs: p < 0.0001, R 2 = 0.76; DLs: p < 0.0001, R 2 = 0.86; Figure S14). A large cluster of densely connected nodes of Firmicutes, (classes of Bacilli, Clostridia, Erysipelotrichia and Negativicutes), Bacteroidetes, Actinobacteria and Acidobacteria was present in both BSCs and DLs networks (Figure S12g). Keystone species (here defined as nodes with >1% of total degree) belonged to Firmicutes (3 OTUs; Clostridia, Negativicutes and Erysipelotrichia), Patescibacteria (1 OTU; Saccharimonadia), Acidobacteria (1 OTUs; Acidobacteriia) and Bacteroidetes (1 OTU; Bacteroidia) in BSCs, while they were affiliated to Acidobacteria (3 OTUs; Acidobacteriia), Actinobacteria (1 OTUs; Actinobacteria and Coriobacteria), Bacteroidetes (2 OTUs; Bacteroidia), Firmicutes (12 OTUs; Clostridia and Negativicutes) and Proteobacteria (2 OTUs; Gammaproteobacteria) in DLs. Despite a large compositional similarity among the bacterial members interacting in the BSC and DL networks, substantial differences were detected in the proportion (twofold enrichment/depletion) of nodes that belong to the phylum Armatimonadetes and Verrucomicrobia, and in the degrees of Planctomycetes nodes (Figure S12g).
Micro‐environmental niche effect in BSCs and DLs associated bacterial communities
The components of beta‐diversity were analysed to identify the ecological processes that govern the assembly of bacterial communities in BSCs and DLs. Similar processes were identified in the aboveground BSCs and belowground DLs: the dominant processes are species replacement (i.e., mediated by the heterogeneity of the micro‐environments represented by each site) and similarity (i.e., due to the overall harsh environmental conditions), while richness difference was the less represented process (Figure S15). We inferred the mechanisms regulating the bacterial community assembly with a phylogenetic bin‐based null model (iCAMP) and found that it was consistently dominated by dispersal limitation and homogeneous selection in both BSCs (47% and 25%, respectively) and DLs (53% and 23%, respectively). Such mechanisms defined significantly different BSCs and DLs bacterial communities across the six sites (PERMANOVA: F 5,12 = 6.29, p = 0.001 and F 5,12 = 4.57, p = 0.001 in BSCs and DLs, respectively; Figures 3E,F; CAP cross‐validation 100%; pairwise comparison in Table S10). Biodiversity analysis performed separately on the six sites showed that the number of shared bacterial OTUs in BSCs and DLs was 62% and 74%, respectively. We consistently detected site‐specific OTUs (range of site‐specific OTUs: 0–122 and 0–107 in BSCs and DLs, respectively), along with site‐enriched and site‐co‐shared taxa (Figures 4G, S16 and S17). LEfSe analysis detected a total of 26 bacterial discriminants that changed according to the sites in BSCs‐associated bacterial communities (Figure S18). Members of Cyanobacteria were the bacterial discriminants in Site 1, those of Verrucomicrobia, Chloroflexi, Firmicutes, Acidobacteria and Alphaproteobacteria in Site 2, those of Chloroflexi, Armatimonadetes and Cyanobacteria in Site 3, Plantomycetes, Bacteroidetes, Chloroflexi and Cyanobacteria in Site 4, and those of Actinobacteria, Acidobacteria, Alphaproteobacteria and Firmicutes in Site 6 (details in Figure S18). Notably, no bacterial discriminants were detected in the BSC of Site 5 nor in all the six DL sites. SIMPER analysis was also performed to unveil the OTUs that contributed predominantly to the site diversity. In BSCs‐associated bacterial microbiomes, 43 OTUs (over 3769) played a crucial role, contributing up to 10% of dissimilarity. Among these, members of 22 families were identified, including Acetobacteraceae, Bacteroidaceae, Burkholderiaceae, Ruminococcaceae and Sphingomonadaceae as the dominants. On the other hand, the differences between sites in DL bacterial communities were mainly explained by the differential distribution of 54 OTUs (over 3886) belonging to 29 families (e.g., Bacteroidaceae, Bifidobacteriaceae, Burkholderiaceae, Chitinophagaceae, Ruminococcaceae and Lachnospiraceae) and contributing to 10% of the observed dissimilarity.
FIGURE 4.
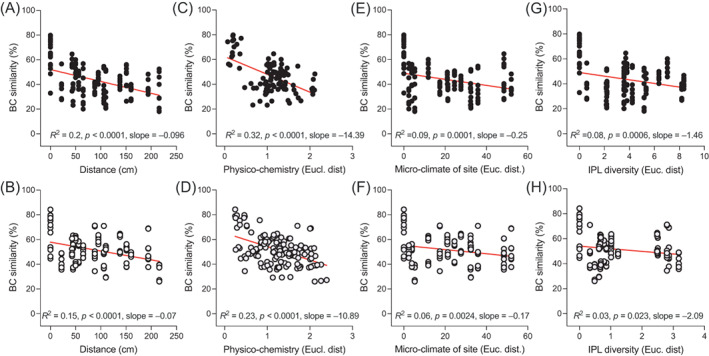
Relationship of beta‐diversity and environmental parameters characterizing the microsites. Bray–Curtis similarity distance decay in function of (A, B) the vertical distance from the above moraine level, (C, D) the overall physico‐chemical conditions (Euclidean distance of data reported in Table 1), (E, F) the micro‐climatic conditions of each site at the day of sampling (Euclidean distance of data reported in Figure 1), and (G, H) the intact lipid composition (Euclidean distance of data reported in Figure 2) in BSC and DL samples, respectively. Red lines indicate significant linear regressions; p‐values, R 2 and slopes are reported in the graphs.
FAPROTAX analysis provided insights into the predicted bacterial ecological functions (Figure S19). This showed a consistently widespread distribution among sites (both BSC and DL) of chemoheterotrophic metabolisms (i.e., aerobic chemoheterotrophy and chemoheterotrophy) and a less yet widespread distribution of fermentative metabolisms. Photosynthetic metabolisms were found in several but not in all BSC samples and in one of the DL samples. The overall picture indicates a general site specificity for the deduced metabolisms in the BSC and DL layers of the examined sediments.
Factor shaping diversity of bacterial communities associated with BSC and DL among sites
The role of the micro‐niches in shaping the associated‐bacterial communities was further shown by the linear decrease of bacterial similarity with the increasing differences among the topographical (i.e., distance among sites; BSC: p < 0.0001, R 2 = 0.2, slope = −0.096, DL: p < 0.0001, R 2 = 0.15, slope = −0.07; Figures 4A,B), physico‐chemical (BSC: p < 0.0001, R 2 = 0.32, slope = −14.39, DL: p < 0.0001, R 2 = 0.23, slope = −10.89; Figures 4C,D), micro‐climatic (BSC: p = 0.0001, R 2 = 0.09, slope = −0.25, DL: p = 0.0024, R 2 = 0.06, slope = −0.17; Figures 4E,F) and IPL signature (BSC: p = 0.0006, R 2 = 0.08, slope = −1.46, DL: p = 0.023, R 2 = 0.03, slope = −2.09; Figures 4G,H) characteristics of the different sites. These results indicate that the unique conditions defining the different sites are important drivers for structuring bacterial communities.
To further understand the forces that shape the BSC and DL bacterial community variance among sites, we analysed the contribution of the physico‐chemical variables. We focused on the physico‐chemical variables because they were the most important in explaining the observed diversity according to the slopes of the regressions (Figure 4). In BSCs, pH, DOC and crust RH (RH%) were the principal physico‐chemical variables (AICc = 105.79, R 2 = 0.63, Table S11a) that explained up to 40% of the variance (Figure 5A). Correlation analysis showed that (i) DOC had a significantly positive correlation with Proteobacteria phylum, Reyranellales and Anaerolineae RBG‐13‐54‐9 orders, Elsteraceae, Reyranellaceae and Myxococcales P3OB‐42 families, and Brevundimonas genus; (ii) pH was positively correlated with Cyanobacteria and Deinococcus‐Thermus phyla, Deinococci and Oxyphotobacteria classes, Blastocatellales and Deinococcales orders, Blastocatellaceae, Intrasporangiaceae, Deinococcaceae and Hymenobacteraceae families, and Nostoc PCC‐73102, Deinococcus, Hymenobacter and Gemmatirosa genera, while it was negatively correlated with Microgenomatia class, WD260 order, CPla‐3 termite group and KD3‐93 families, and Acidipila, Phenylobacterium, Rhizobacter and Telmatocola genera; (iii) RH was positively correlated with members of Omnitrophicaeota phylum, Dojkabacteria WS6 class, Steroidobacterales order, Steroidobacteraceae, Terrimicrobiaceae and Sandaracinaceae families, and Tahibacter, Terrimicrobium and Pelosinus genera.
FIGURE 5.
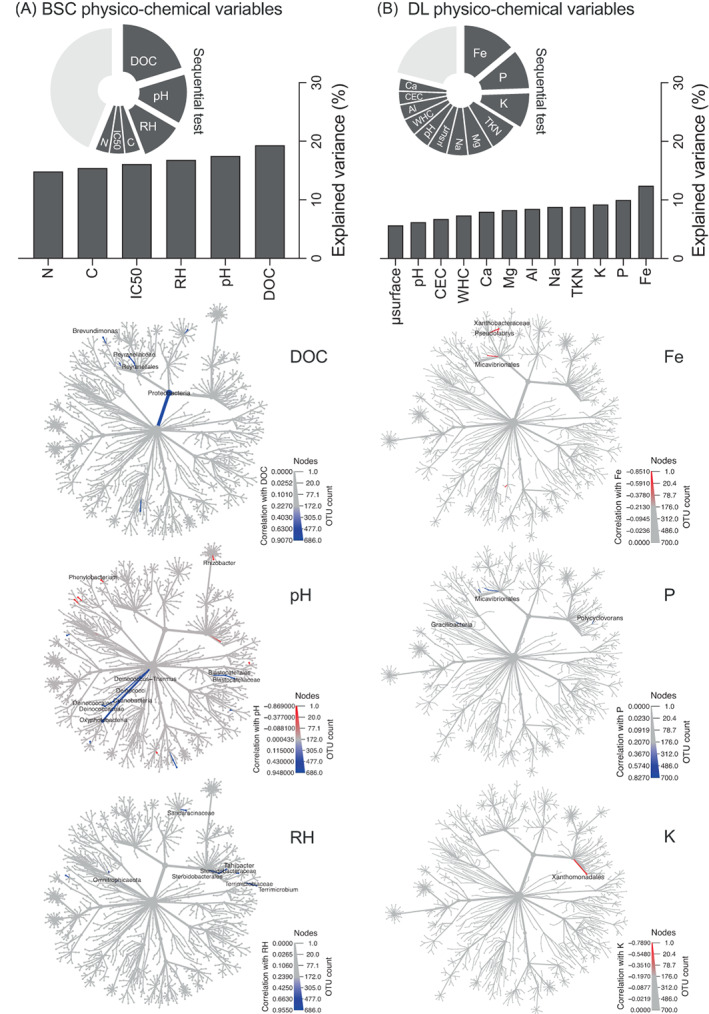
Effect of site‐environmental factors on bacterial communities' diversity in BSCs and DLs. Results of the variation partitioning model used to identify the effects of the physio‐chemical explanatory variables (Table 1) on the bacterial communities of (A) BSC and (B) DL are reported as bar plots. Results of sequential test run considering all the factors are reported as pie charts. The three most important variables explaining the variance of the bacteria communities in BSC (i.e., DOC, pH and RH) and DL (i.e., Fe, P and K) are used to evaluate their correlation with the bacterial taxa inhabiting the samples. Metacoder‐generated trees indicate the taxa that significantly (p < 0.05) correlated positively (shade of blue) or negatively (shade of red) with the selected physico‐chemical variables. Taxa non‐significantly correlated with such variables are indicated in grey.
In the case of DLs, the variability of bacterial communities was mainly explained by Fe, P and K, (AICc = 127.12, R 2 = 0.46, Table S11b) which account for almost 30% of the entire variance (Figure 5B): (i) Fe was negatively correlated with the presence of members of Micavibrionales order, Xanthobacteraceae and Simkaniaceae families, and Pseudolabrys genus; (ii) P was positively correlated with members of Gracilibacteria class, Micavibrionales order, Elsteraceae and Micavibrionaceae families, and Polycyclovorans genus; (iii) K was positively correlated with the members of Xanthomonadales order.
DISCUSSION
In this study, we investigated six sites of primary colonization with BSCs in a 50 m2 area of the Lobuche moraine. The study area was in the relic debris‐mantled ablation zone no anymore connected to the glacier dynamics (Jones et al., 2019). We chose this area because in a relatively small space it presents all hummock and depression sites with variable spatial orientation, altitude and slope that characterize the overall Lobuche moraine but minimizes the intrinsic variability associated with time in glacier‐moraine chronosequences. Thus, the sites were representative of the different topographical conditions occurring all over the moraine but did not reflect the intrinsic variability due to the time from deglaciation.
Due to their topographic position in the studied area, the six sites were exposed to distinct environmental conditions in terms of irradiation, air temperature and humidity. The spatial orientation and elevation of the sites determine the level of exposure to the environmental agents (wind, sunlight irradiation, dust deposition, humidity, etc.) and so influence the ecological dynamics of the living community (Austin et al., 2004; Fischer & Subbotina, 2014; Kidron et al., 2010), at least in the summer season when the moraine is not under a snow cover. However, we must consider that environmental measures over a very short time window, such as those used in this study, cannot be extended all over the different seasons and our environmental measures must be considered under the limited time window they were obtained. The six sites also presented a set of specific physico‐chemical properties (organic matter, pH and soil nutrient concentrations) that make them distinct niches in which unique bacterial communities are selected both at the BSC and DL levels. In many soil ecosystems, microbial community composition has been shown to be shaped by soil physico‐chemical conditions, such as pH in Antarctica (Chu et al., 2010), nutrients in North American ecosystems (Ramirez et al., 2012) and moisture in Chinese steppe (Zhao et al., 2016). Nevertheless, the presence of multiple niches in a relatively small area in the glacier forefield suggests that events ongoing in the early stages of primary succession are patchy and heterogeneous, rather than coordinated and/or strongly affected by macro‐climatic conditions of the overall moraine, and these differences are mirrored in the inhabiting microbiomes.
The BSCs and DLs of the Lobuche moraine were found to host different bacterial communities, which were composed of phototrophic taxa but also included a large component of non‐phototrophic microorganisms. The functionality of BSCs as pioneer colonizers in the extreme cold ecosystems of the Himalayas and elsewhere was mainly investigated for phototrophs (Janatková et al., 2013; Schmidt et al., 2008), but recent mounting evidence poses attention to the non‐photosynthetic microorganisms as contributors to ecosystem stability and multifunctionality (Ezzat et al., 2022; Jousset et al., 2017; Lynch & Neufeld, 2015; Wang et al., 2020). For instance, it has been recently shown that cryospheric ecosystems are inhabited by generalist taxa with Proteobacteria and Bacteroidota as key taxonomic groups (Bourquin et al., 2022; Gupta et al., 2015; Mapelli et al., 2011, 2018; Schmidt et al., 2008). The occurrence of the same bacterial phyla in geographically distinct areas is indicative of their ability to adopt similar metabolic strategies to survive in oligotrophic and cold environments (Srinivas et al., 2011), facilitating a multitude of phototrophic, photoheterotrophic and chemolithotrophic processes (Borin et al., 2010; Mapelli et al., 2018).
The BSCs and DLs of the Lobuche moraine were found to share a large number of generalist bacterial OTUs. Despite the dominance of such generalists, the consistent selective pressure of BSCs and DL affects the assembly of bacterial communities with site‐specific patterns even at short distances. For instance, among the taxa that discriminate between BSCs and DLs microbiomes, Alphaproteobacteria and Bacteroidetes were enriched in the BSC. Alphaproteobacteria prefer nutrient‐rich environments, and in deglaciating soil their abundance increase with soil age (Fernández‐Martínez et al., 2017), while Bacteriodetes are inhabitants of BSCs occurring in cold desert ecosystems like the Eastern Pamir (Khomutovska et al., 2021). Furthermore, BSCs were enriched in bacteria affiliated with Burkholderiales, typical inhabitants of ice‐dominated environments, that are seeded in the mineral substrate released by ice melting and colonize soils at initial and medium stages of development (Mapelli et al., 2011; Schmidt et al., 2008). On the contrary, DLs were enriched in Actinobacteria, Proteobacteria and Acidobacteria phyla which are among the most abundant taxa inhabiting bare soils of cold deserts worldwide (Bourquin et al., 2022; Leung et al., 2020) and were found in other Himalayan forefields (Srinivas et al., 2011). The limited dispersion and homogenous selection resulted in being the main ecological processes in the assembly of microbial communities in BSC and DL, suggesting that bacteria had a life–death dynamics that allowed community composition to drift apart due to spatial isolation between sites (Fodelianakis et al., 2020; 2021), as it was also observed in permafrost‐associated microbiomes in Alaskan forests (Bottos et al., 2018). The rejection of the neutral community model for both BSC and DL and of the source‐sink hypotheses support that bacterial community assembly is mainly deterministic rather than stochastic and it is driven by environmental factors and microbial species interactions. For instance, the distinct BSC habitat hosts bacterial communities predominantly trapped in EPS matrices to form complex structures/films covering the underlying substrate in the moraine, with consequent limitation of bacterial dispersal from DL to BSC and vice versa. An alternate interpretation of the data is that the environmental/ecological conditions of BSC and DL are not similar enough to experience comparable levels of selection (e.g., C content), as expected for those species identified as ‘neutrally distributed’.
Glacier ecosystems are low‐energy input environments (Schostag et al., 2019) where microorganisms have to survive under sub‐zero temperatures and are subjected to freeze–thaw cycles, standing as quasi‐closed systems (Graham et al., 2012). Under these conditions, dispersal is reduced, and nutrients supply is governed by local dynamics involving cellular dormancy (Choudoir & DeAngelis, 2022) and recycling of energy by exploiting the biomass of necrotic cells (Shoemaker et al., 2021). During dormancy, cells are in a viable but not‐cultivable state, a low‐energy condition accompanied by a reduction in cellular size. Upon perception of favourable conditions, cells exit dormancy and restore the vegetative growth (Su et al., 2013). At the same time, thawing cycles may lead to cellular lysis, so the dead microorganisms may release organic compounds to fuel living cells. By tracking population dynamics under energy‐limited conditions, it was observed that cells could use the necrotic biomass to sustain growth and evolve through the natural selection (Shoemaker et al., 2021). This bacterial survival strategy under energy limitation seems to be conserved across the bacterial kingdom, suggesting that this trend may be common to hot and cold deserts, where niche‐filtering by the climate and edaphic factors drive community assembly (Lee et al., 2016; Pointing et al., 2015).
IPL analysis deepens the knowledge of the resident microbiomes by providing information that helps in clarifying the adaptation and diversity of the entire community (Sturt et al., 2004). The presence in the study area of oxidized phosphatidyl choline, betaine and ornithine lipids having one to multiple hydroxylations and oxylipins indicates that microorganisms have to cope with high oxidative and photo‐oxidative stress, associated with low temperature and high irradiation (Amiraux, 2017). Furthermore, the predominant fatty acids had monounsaturated chains, a molecular component of the biochemical strategy used by psychrophiles to maintain cell membrane integrity in low‐temperature environments (Collins & Margesin, 2019). A higher abundance of glycolipids and heterocyst glycolipids in the BSCs compared to DLs is owed to the switch of glycolipid‐containing phototrophs on the surface to non‐phototroph organisms in the DLs (Kalisch et al., 2016). While little is known about the turnover of diacylglycerol glycolipids in soils after cell death, studies have shown that heterocyst glycolipids can be preserved in laminated sediments for millions of years (Bauersachs et al., 2010). Therefore, detecting heterocyst glycolipids in the deeper layers could be the trace of non‐degraded detrital material from the surface that has been buried over time. IPL analysis supports the bacterial metabolisms inferred by the 16S rRNA gene sequences dataset, which shows that phototrophic‐related functions are enriched in the BSCs compared to DLs. Besides phototrophs, representatives of the Firmicutes, Alphaproteobacteria and Gammaproteobacteria, are also capable of synthesizing glycolipids (Hölzl & Dörmann, 2007). While the abundant presence of trimethyl ornithine lipids can be most likely assigned to Planctomycetes in both the BSCs and DLs (Moore, 2021), the overall high levels of glycolipids and aminolipids suggest some nutrient limitation, especially of phosphorus (Schubotz et al., 2018; Van Mooy & Fredricks, 2010).
The results of IPL and physico‐chemical analyses corroborate that in the studied area the bacterial structure and nutritional properties of the DLs are poorly affected by the overlying BSCs. According to IPL analysis, most of the microbial biomass accumulates in the BSCs, implying a faster turnover of nutrients in the crust with intermediate metabolites bound to the microbial loop and not released in the underlying mineral layer. A similar trend of biomass and nutrients accumulation within the crust surface rather than in the bare soil was observed along an elevation gradient from 5300 to 5900 m asl in the Tibetan plateau (Chu et al., 2016; Janatková et al., 2013). NMR analysis of BSCs revealed the load of O‐alkyl, alkyl and N‐alkyl carbon, indicating that the input of carbon derived from the biocrust comprises polysaccharides and aliphatic biopolymers, in agreement with the BSC‐produced extracellular polymeric substances (Mugnai et al., 2020; Rossi et al., 2018). Carbon transfer from the biocrusts to the substrate below was mainly observed in temperate conditions for well‐developed BSC like those moss‐dominated (Dümig et al., 2012). In the case of the Lobuche BSCs studied here, we speculate that the higher summer temperatures they experience, as opposed to DLs that are in contact with the permafrost, might enhance the nutrient turnover and consumption, thus limiting the transfer to the lower layers. Indeed, temperature strongly influences soil organic carbon formation in glacier foreland, showing a faster increase in its accumulation in forefields that experience warmer periods (Khedim et al., 2021). Because of the low temperatures and limited N availability in Nepalese soil, organic carbon decomposes at relatively low rates and a gradient in temperature between the BSC surface and the below‐layers could further affect such decomposition rate (Chu et al., 2016). Due to the limited metabolic exchanges with the overlying BSCs, DL communities may experience extreme starvation conditions for nutrients and carbon availability. The DL bacterial communities were less abundant but with a higher richness than the overlying BSC, suggestive of a reduced enrichment process (Krauze et al., 2021). Such microbiomes tend to establish more connected networks that may contribute to increase niche adaptation and survival success under extreme oligotrophic conditions (Dong et al., 2022). Higher network topological features in DL than in BSC evoke synergistic strategies through associations that optimize the exchange of limited nutrients for growth and survival (Perera et al., 2018). Another important parameter that significantly affected the microbial diversity of BSCs was their RH. Water content and its availability are crucial factors controlling the distribution and growth of soil microbes (Stres et al., 2008; Van Horn et al., 2014). The influence of water availability on BSC microbiomes has been documented for cyanobacteria that promptly move toward the surface of wetted soils and resume photosynthesis (Garcia‐Pichel & Pringault, 2001). A suitable moisture content improves microbial activity, reinforces ecological functions, promotes the formation of soil fertility islands and accelerates the process of pedogenesis (Borin et al., 2010; Li et al., 2016). The abovementioned differences in carbon, DOC and moisture in the vertical soil, concomitantly with the temperature gradients occurring in the Lobuche forefield, may strongly affect the diversity of the soil microbiomes inhabiting the crusts and the DLs.
CONCLUSIONS
The study of primary colonization in the forefield of the Lobuche glacier in the Himalayas plateau indicates that the biogeochemical heterogeneity of BSCs and DLs recorded at the small spatial scale in the studied area contributes to the natural diversification of the moraine that, over time, can feed the formation of the soil. Our results point out that the irregular topography of the moraine governs such heterogeneity and steers the bipartite interaction between (i) the environmental conditions (such as physico‐chemical and micro‐climatic) of the sites, and (ii) the bacterial communities inhabiting the crust and the deeper substrate layers. We propose that, besides the study of time‐dependent chronosequences, an assessment of the microscale heterogeneity at each chronosequence point should be considered and acknowledged for a comprehensive learning of pedogenesis in moraines released by glacier ice.
AUTHOR CONTRIBUTIONS
Conceptualization: ER, RM, MF, SB and DD. Data curation: RM and MF. Formal analysis: RM, MF, ER,BS, FS. Funding aquisition: DD, SB. Investigation: ER, RM, MF, BS, FS, FM, SC, LB, LT,FT. Project administration: SB, FA, DD. Resources: SB, DD, FA, FS, LT. Software: RM, MF. Supervision: DD. Validation: ER, RM, MF, BS, FS, FM, SC, LB, LT,FT. Visualization: ER, RM, MF. Writing‐original draft: ER, RM, MF, BS, FS, FM, LT, FA, SB, DD. Writing‐review and editing: ER, RM, MF, DD.
CONFLICT OF INTEREST
Authors have no conflict of interest to declare.
Supporting information
Appendix S1: Supporting information
ACKNOWLEDGEMENTS
This study was carried out within the framework of the Ev‐K2‐CNR Project in collaboration with the Nepal Academy of Science and Technology as foreseen by the Memorandum of Understanding between Nepal and Italy and with the contributions from the Italian National Research Council and the Italian Ministry of Foreign Affairs. King Abdullah University of Science and Technology supported the study through baseline research funds to Daniele Daffonchio. Eleonora Rolli and Sara Borin thank the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska‐Curie grant agreement number 841317, project acronym ‘SENSE’.
Rolli, E. , Marasco, R. , Fusi, M. , Scaglia, B. , Schubotz, F. , Mapelli, F. et al. (2022) Environmental micro‐niche filtering shapes bacterial pioneer communities during primary colonization of a Himalayas' glacier forefield. Environmental Microbiology, 24(12), 5998–6016. Available from: 10.1111/1462-2920.16268
Eleonora Rolli and Ramona Marasco contributed equally.
Funding information King Abdullah University of Science and Technology
DATA AVAILABILITY STATEMENT
The data that supports the findings of this study are available in the table and supplementary material of this article. Sequences were deposited to the Sequence Read Archive of NCBI under the BioProject PRJNA698068.
REFERENCES
- Adler, C. , Huggel, C. , Orlove, B. & Nolin, A. (2019) Climate change in the mountain cryosphere: impacts and responses. Regional Environmental Change, 19, 1225–1228. [Google Scholar]
- Amiraux, R. (2017) Ice biota degradation in the Arctic environment: impact of bacterial stress state on this material's preservation and burial. Aix‐Marseille University.
- Anderson, M.M.J.J. , Gorley, R.N.R.N. & Clarke, K.R.R. (2008) PERMANOVA+ for PRIMER: guide to software and statistical methods. Plymouth: PRIMER‐E. [Google Scholar]
- Ashton, P. & Zhu, H. (2020) The tropical‐subtropical evergreen forest transition in East Asia: an exploration. Plant Divers, 42, 255–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin, A.T. , Yahdjian, L. , Stark, J.M. , Belnap, J. , Porporato, A. , Norton, U. et al. (2004) Water pulses and biogeochemical cycles in arid and semiarid ecosystems. Oecologia, 141, 221–235. [DOI] [PubMed] [Google Scholar]
- Baas, M., Pancost, R., van Geel, B. & SinningheDamstéa, J.A. (2000) A comparative study of lipids in Sphagnum species. Organicgeochemistry, 31, 535–541. [Google Scholar]
- Barger, N.N. , Herrick, J.E. , Van Zee, J. & Belnap, J. (2006) Impacts of biological soil crust disturbance and composition on C and N loss from water erosion. Biogeochemistry, 77, 247–263. [Google Scholar]
- Bastian, M. , Heymann, S. & Jacomy, M. (2009) Gephi: an open source software for exploring and manipulating networks. Third International AAAI Conference on Weblogs and Social Media, 8, 361–362. [Google Scholar]
- Bauersachs, T. , Speelman, E.N. , Hopmans, E.C. , Reichart, G.‐J. , Schouten, S. & Damste, J.S.S. (2010) Fossilized glycolipids reveal past oceanic N2 fixation by heterocystous cyanobacteria. Proceedings of the National Academy of Sciences, 107, 19190–19194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Booth, J.M.J.M. , Fusi, M. , Marasco, R. , Mbobo, T. & Daffonchio, D. (2019) Fiddler crab bioturbation determines consistent changes in bacterial communities across contrasting environmental conditions. Scientific Reports, 9, 3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borin, S. , Ventura, S. , Tambone, F. , Mapelli, F. , Schubotz, F. , Brusetti, L. et al. (2010) Rock weathering creates oases of life in a High Arctic desert. Environmental Microbiology, 12, 293–303. [DOI] [PubMed] [Google Scholar]
- Bottos, E.M. , Kennedy, D.W. , Romero, E.B. , Fansler, S.J. , Brown, J.M. , Bramer, L.M. et al. (2018) Dispersal limitation and thermodynamic constraints govern spatial structure of permafrost microbial communities. FEMS Microbiology Ecology, 94, 1–14. [DOI] [PubMed] [Google Scholar]
- Bourquin, M. , Busi, S.B. , Fodelianakis, S. , Peter, H. , Washburne, A. , Kohler, T.J. et al. (2022) The microbiome of cryospheric ecosystems. Nature Communications, 13, 3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, A.R. , Stephens, W.Z. , Stagaman, K. , Wong, S. , Rawls, J.F. , Guillemin, K. et al. (2016) Contribution of neutral processes to the assembly of gut microbial communities in the zebrafish over host development. The ISME Journal, 10, 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busi, S.B. , Bourquin, M. , Fodelianakis, S. , Michoud, G. , Kohler, T.J. , Peter, H. et al. (2022) Genomic and metabolic adaptations of biofilms to ecological windows of opportunity in glacier‐fed streams. Nature Communications, 13, 2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso, J.G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F.D. , Costello, E.K. et al. (2010) QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, W. , Ren, K. , Isabwe, A. , Chen, H. , Liu, M. & Yang, J. (2019) Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome, 7, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudoir, M.J. & DeAngelis, K.M. (2022) A framework for integrating microbial dispersal modes into soil ecosystem ecology. iScience, 25, 103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, H. , Fierer, N. , Lauber, C.L. , Caporaso, J.G. , Knight, R. & Grogan, P. (2010) Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environmental Microbiology, 12, 2998–3006. [DOI] [PubMed] [Google Scholar]
- Chu, H. , Sun, H. , Tripathi, B.M. , Adams, J.M. , Huang, R. , Zhang, Y. et al. (2016) Bacterial community dissimilarity between the surface and subsurface soils equals horizontal differences over several kilometers in the western Tibetan Plateau. Environmental Microbiology, 18, 1523–1533. [DOI] [PubMed] [Google Scholar]
- Collins, T. & Margesin, R. (2019) Psychrophilic lifestyles: mechanisms of adaptation and biotechnological tools. Applied Microbiology and Biotechnology, 103, 2857–2871. [DOI] [PubMed] [Google Scholar]
- Derin, Y. , Anagnostou, E. , Berne, A. , Borga, M. , Boudevillain, B. , Buytaert, W. et al. (2019) Evaluation of GPM‐era global satellite precipitation products over multiple complex terrain regions. Remote Sensing, 11, 2936. [Google Scholar]
- Dhakar, K. & Pandey, A. (2020) Microbial ecology from the Himalayan cryosphere perspective. Microorganisms, 8, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diolaiuti, G.A. , Maragno, D. , D'Agata, C. , Smiraglia, C. & Bocchiola, D. (2011) Glacier retreat and climate change: documenting the last 50 years of alpine glacier history from area and geometry changes of dosdè piazzi glaciers (Lombardy Alps, Italy). Progress in Physical Geography, 35, 161–182. [Google Scholar]
- Dong, K. , Yu, Z. , Kerfahi, D. , Lee, S.s. , Li, N. , Yang, T. et al. (2022) Soil microbial co‐occurrence networks become less connected with soil development in a high Arctic glacier foreland succession. Science of the Total Environment, 813, 152565. [DOI] [PubMed] [Google Scholar]
- Dray, S. , Blanchet, G. , Borcard, D. , Guenard, G. , Jombart, T. , Larocque, G. et al. (2018) Package “adespatial”. R Package, 2018, pp. 3–8.
- Dümig, A. , Veste, M. , Hagedorn, F. , Fischer, T. , Lange, P. , Spröte, R. et al. (2012) Biological soil crusts on initial soils: organic carbon dynamics and chemistry under temperate climatic conditions. Biogeosciences Discussions, 1, 851–894. [Google Scholar]
- Ezzat, L. , Fodelianakis, S. , Kohler, T.J. , Bourquin, M. , Brandani, J. , Busi, S.B. et al. (2022) Benthic biofilms in glacier‐fed streams from Scandinavia to the Himalayas host distinct bacterial communities compared with the streamwater. Applied and Environmental Microbiology, 88, e00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust, K. & Raes, J. (2016) CoNet app: inference of biological association networks using Cytoscape. F1000Research, 5, 1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Martínez, M.A. , Pérez‐Ortega, S. , Pointing, S.B. , Allan Green, T.G. , Pintado, A. , Rozzi, R. et al. (2017) Microbial succession dynamics along glacier forefield chronosequences in Tierra del Fuego (Chile). Polar Biology, 40, 1939–1957. [Google Scholar]
- Fischer, T. & Subbotina, M. (2014) Climatic and soil texture threshold values for cryptogamic cover development: a meta analysis. Biologia (Bratisl), 69, 1520–1530. [Google Scholar]
- Fodelianakis, S. , Valenzuela‐Cuevas, A. , Barozzi, A. & Daffonchio, D. (2021) Direct quantification of ecological drift at the population level in synthetic bacterial communities. The ISME Journal, 15, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodelianakis, S. , Washburne, A.D. , Bourquin, M. , Pramateftaki, P. , Kohler, T.J. , Styllas, M. et al. (2020) Homogeneous selection promotes microdiversity in the glacier‐fed stream microbiome 2. bioRxiv, 2020.12.03.409391.
- Fodelianakis, S. , Washburne, A.D. , Bourquin, M. , Pramateftaki, P. , Kohler, T.J. , Styllas, M. et al. (2022) Microdiversity characterizes prevalent phylogenetic clades in the glacier‐fed stream microbiome. The ISME Journal, 16, 666–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster, Z.S.L. , Sharpton, T.J. & Grünwald, N.J. (2017) Metacoder: an R package for visualization and manipulation of community taxonomic diversity data. PLoS Computational Biology, 13, e1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Pichel, F. & Pringault, O. (2001) Microbiology: cyanobacteria track water in desert soils. Nature, 413, 380–381. [DOI] [PubMed] [Google Scholar]
- Garrido‐Benavent, I. , Pérez‐Ortega, S. , Durán, J. , Ascaso, C. , Pointing, S.B. , Rodríguez‐Cielos, R. et al. (2020) Differential colonization and succession of microbial communities in rock and soil substrates on a maritime Antarctic glacier forefield. Frontiers in Microbiology, 11, 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghiloufi, W. , Seo, J. , Kim, J. , Chaieb, M. & Kang, H. (2019) Effects of biological soil crusts on enzyme activities and microbial community in soils of an arid ecosystem. Microbial Ecology, 77, 201–216. [DOI] [PubMed] [Google Scholar]
- Graham, D.E. , Wallenstein, M.D. , Vishnivetskaya, T.A. , Waldrop, M.P. , Phelps, T.J. , Pfiffner, S.M. et al. (2012) Microbes in thawing permafrost: the unknown variable in the climate change equation. The ISME Journal, 6, 709–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulcin, İ. (2020) Antioxidants and antioxidant methods: an updated overview. Archives of Toxicology, 94, 651–715. [DOI] [PubMed] [Google Scholar]
- Gupta, P. , Sangwan, N. , Lal, R. & Vakhlu, J. (2015) Bacterial diversity of Drass, cold desert in Western Himalaya, and its comparison with Antarctic and Arctic. Archives of Microbiology, 197, 851–860. [DOI] [PubMed] [Google Scholar]
- Hamid, M. , Khuroo, A.A. , Malik, A.H. , Ahmad, R. , Singh, C.P. , Dolezal, J. et al. (2020) Early evidence of shifts in Alpine summit vegetation: a case study from Kashmir Himalaya. Frontiers in Plant Science, 11, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, L.C. , Tiwari, P. , Sadhukhan, B. , Tiwari, S. , Chapagain, P. , Xu, T. et al. (2020) Building climate change resilience by using a versatile toolkit for local governments and communities in rural Himalaya. Environmental Research, 188, 109636. [DOI] [PubMed] [Google Scholar]
- Hinrichs, K.U. , Summons, R.E. , Orphan, V. , Sylva, S.P. & Hayes, J.M. (2000) Molecular and isotopic analysis of anaerobic methaneoxidizing communities in marine sediments. Organic Geochemistry, 31, 1685–1701. [Google Scholar]
- Hölzl, G. & Dörmann, P. (2007) Structure and function of glycoglycerolipids in plants and bacteria. Progress in Lipid Research, 46, 225–243. [DOI] [PubMed] [Google Scholar]
- Janatková, K. , Řeháková, K. , Doležal, J. , Šimek, M. , Chlumská, Z. , Dvorský, M. et al. (2013) Community structure of soil phototrophs along environmental gradients in arid Himalaya. Environmental Microbiology, 15, 2505–2516. [DOI] [PubMed] [Google Scholar]
- Jones, D.B. , Harrison, S. & Anderson, K. (2019) Mountain glacier‐to‐rock glacier transition. Global and Planetary Change, 181, 102999. [Google Scholar]
- Jousset, A. , Bienhold, C. , Chatzinotas, A. , Gallien, L. , Gobet, A. , Kurm, V. et al. (2017) Where less may be more: how the rare biosphere pulls ecosystems strings. The ISME Journal, 11, 853–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jury, M.W. , Mendlik, T. , Tani, S. , Truhetz, H. , Maraun, D. , Immerzeel, W.W. et al. (2020) Climate projections for glacier change modelling over the Himalayas. International Journal of Climatology, 40, 1738–1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalisch, B. , Dörmann, P. & Hölzl, G. (2016) DGDG and glycolipids in plants and algae. Sub‐Cellular Biochemistry, 86, 51–83. [DOI] [PubMed] [Google Scholar]
- Khedim, N. , Cécillon, L. , Poulenard, J. , Barré, P. , Baudin, F. , Marta, S. et al. (2021) Topsoil organic matter build‐up in glacier forelands around the world. Global Change Biology, 27, 1662–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khomutovska, N. , de los Ríos, A. , Syczewski, M.D. & Jasser, I. (2021) Connectivity of edaphic and endolithic microbial niches in cold mountain desert of eastern pamir (Tajikistan). Biology, 10, 1–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidron, G.J. , Vonshak, A. , Dor, I. , Barinova, S. & Abeliovich, A. (2010) Properties and spatial distribution of microbiotic crusts in the Negev Desert, Israel. Catena, 82, 92–101. [Google Scholar]
- Klug, R.M. & Benning, C. (2001) Two enzymes of diacylglyceryl‐O‐4′‐(N,N,N,‐trimethyl)‐homoserine biosynthesis are encoded by btaA and btaB in the purple bacterium Rhodobacter sphaeroides. Proceedings of the National Academy of Sciences of the United States of America, 98, 5910–5915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knights, D. , Kuczynski, J. , Charlson, E.S. , Zaneveld, J. , Mozer, M.C. , Collman, R.G. et al. (2011) Bayesian community‐wide culture‐independent microbial source tracking. Nature Methods, 8, 761–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler, T.J. , Fodelianakis, S. , Michoud, G. , Ezzat, L. , Bourquin, M. , Peter, H. et al. (2022) Glacier shrinkage will accelerate downstream decomposition of organic matter and alters microbiome structure and function. Global Change Biology, 28, 3846–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krauze, P. , Wagner, D. , Yang, S. , Spinola, D. & Kühn, P. (2021) Influence of prokaryotic microorganisms on initial soil formation along a glacier forefield on King George Island, maritime Antarctica. Scientific Reports, 11, 13135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lami, A. , Marchetto, A. , Musazzi, S. , Salerno, F. , Tartari, G. , Guilizzoni, P. et al. (2010) Chemical and biological response of two small lakes in the Khumbu Valley, Himalayas (Nepal) to short‐term variability and climatic change as detected by long‐term monitoring and paleolimnological methods. Hydrobiologia, 648, 189–205. [Google Scholar]
- Lee, K.C. , Archer, S.D.J. , Boyle, R.H. , Lacap‐Bugler, D.C. , Belnap, J. & Pointing, S.B. (2016) Niche filtering of bacteria in soil and rock habitats of the Colorado Plateau Desert, Utah, USA. Frontiers in Microbiology, 7, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, P.M. , Bay, S.K. , Meier, D.V. , Chiri, E. , Cowan, D.A. , Gillor, O. et al. (2020) Energetic basis of microbial growth and persistence in desert ecosystems. mSystems, 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H. , Li, R. , Rossi, F. , Li, D. , De Philippis, R. , Hu, C. et al. (2016) Differentiation of microbial activity and functional diversity between various biocrust elements in a heterogeneous crustal community. Catena, 147, 138–145. [Google Scholar]
- Love, M.I. , Huber, W. & Anders, S. (2014) Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M.D.J. & Neufeld, J.D. (2015) Ecology and exploration of the rare biosphere. Nature Reviews. Microbiology, 13, 217–229. [DOI] [PubMed] [Google Scholar]
- López‐Lara, I.M. & Geiger, O. (2017) Bacterial lipid diversity. Biochimica et Biophysica Acta ‐ Molecular and Cell Biology of Lipids, 1862, 1287–1299. [DOI] [PubMed] [Google Scholar]
- Maestre, F.T. , Quero, J.L. , Gotelli, N.J. , Escudero, A. , Ochoa, V. , Delgado‐Baquerizo, M. et al. (2012) Plant species richness and ecosystem multifunctionality in global drylands. Science, 335, 214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallen‐Cooper, M. , Bowker, M.A. , Antoninka, A.J. & Eldridge, D.J. (2020) A practical guide to measuring functional indicators and traits in biocrusts. Restoration Ecology, 28, S56–S66. [Google Scholar]
- Mapelli, F. , Marasco, R. , Balloi, A. , Rolli, E. , Cappitelli, F. , Daffonchio, D. et al. (2012) Mineral‐microbe interactions: biotechnological potential of bioweathering. Journal of Biotechnology, 157, 473–481. [DOI] [PubMed] [Google Scholar]
- Mapelli, F. , Marasco, R. , Fusi, M. , Scaglia, B. , Tsiamis, G. , Rolli, E. et al. (2018) The stage of soil development modulates rhizosphere effect along a High Arctic desert chronosequence. The ISME Journal, 12, 1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapelli, F. , Marasco, R. , Rizzi, A. , Baldi, F. , Ventura, S. , Daffonchio, D. et al. (2011) Bacterial communities involved in soil formation and plant establishment triggered by pyrite bioweathering on Arctic moraines. Microbial Ecology, 61, 438–447. [DOI] [PubMed] [Google Scholar]
- Matthews, T. , Perry, L.B. , Koch, I. , Aryal, D. , Khadka, A. , Shrestha, D. et al. (2020) Going to extremes. Bulletin of the American Meteorological Society, 101, 1870–1890. [Google Scholar]
- Maurer, J.M. , Schaefer, J.M. , Rupper, S. & Corley, A. (2019) Acceleration of ice loss across the Himalayas over the past 40 years. Science Advances, 5, eaav7266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, E.K. (2021) Trimethylornithine membrane lipids: discovered in planctomycetes and identified in diverse environments. Metabolites, 11, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mugnai, G. , Rossi, F. , Mascalchi, C. , Ventura, S. & De Philippis, R. (2020) High Arctic biocrusts: characterization of the exopolysaccharidic matrix. Polar Biology, 43, 1805–1815. [Google Scholar]
- Nichols, B.W. & Wood, B.J. (1968) The occurrence and biosynthesis of gamma‐linolenic acid in a blue‐green alga, Spirulina platensis . Lipids, 3, 46–50. [DOI] [PubMed] [Google Scholar]
- Ning, D. , Yuan, M. , Wu, L. , Zhang, Y. , Guo, X. , Zhou, X. et al. (2020) A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nature Communications, 11, 14717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuth, C. , Moholdt, G. , Kohler, J. , Hagen, J.O. & Kääb, A. (2010) Svalbard glacier elevation changes and contribution to sea level rise. Journal of Geophysical Research ‐ Earth Surface, 115, 1–16. [Google Scholar]
- Orellana, R. , Macaya, C. , Bravo, G. , Dorochesi, F. , Cumsille, A. , Valencia, R. et al. (2018) Living at the frontiers of life: extremophiles in Chile and their potential for bioremediation. Frontiers in Microbiology, 9, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson, A.L. (1955) X‐ray diffraction procedures for polycrystalline and amorphous materials. Journal of the American Chemical Society, 77, 2030–2031. [Google Scholar]
- Perera, I. , Subashchandrabose, S.R. , Venkateswarlu, K. , Naidu, R. & Megharaj, M. (2018) Consortia of cyanobacteria/microalgae and bacteria in desert soils: an underexplored microbiota. Applied Microbiology and Biotechnology, 102, 7351–7363. [DOI] [PubMed] [Google Scholar]
- Pessi, I.S. , Pushkareva, E. , Lara, Y. , Borderie, F. , Wilmotte, A. & Elster, J. (2019) Marked succession of cyanobacterial communities following glacier retreat in the High Arctic. Microbial Ecology, 77, 136–147. [DOI] [PubMed] [Google Scholar]
- Pointing, S.B. , Büdel, B. , Convey, P. , Gillman, L.N. , Körner, C. , Leuzinger, S. et al. (2015) Biogeography of photoautotrophs in the high polar biome. Frontiers in Plant Science, 6, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C. , Pruesse, E. , Yilmaz, P. , Gerken, J. , Schweer, T. , Yarza, P. et al. (2012) The SILVA ribosomal RNA gene database project: improved data processing and web‐based tools. Nucleic Acids Research, 41, D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez, K.S. , Craine, J.M. & Fierer, N. (2012) Consistent effects of nitrogen amendments on soil microbial communities and processes across biomes. Global Change Biology, 18, 1918–1927. [Google Scholar]
- Rippin, M. , Lange, S. , Sausen, N. & Becker, B. (2018) Biodiversity of biological soil crusts from the polar regions revealed by metabarcoding. FEMS Microbiology Ecology, 94, 1–15. [DOI] [PubMed] [Google Scholar]
- Rossi, F. , Mugnai, G. & De Philippis, R. (2018) Complex role of the polymeric matrix in biological soil crusts. Plant and Soil, 429, 19–34. [Google Scholar]
- Salerno, F. , Guyennon, N. , Thakuri, S. , Viviano, G. , Romano, E. , Vuillermoz, E. et al. (2015) Weak precipitation, warm winters and springs impact glaciers of south slopes of Mt. Everest (central Himalaya) in the last 2 decades (1994–2013). The Cryosphere, 9, 1229–1247. [Google Scholar]
- Schmidt, S.K. , Reed, S.C. , Nemergut, D.R. , Grandy, A.S. , Cleveland, C.C. , Weintraub, M.N. et al. (2008) The earliest stages of ecosystem succession in high‐elevation (5000 metres above sea level), recently deglaciated soils. Proceedings of the Royal Society B: Biological Sciences, 275, 2793–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schostag, M. , Priemé, A. , Jacquiod, S. , Russel, J. , Ekelund, F. & Jacobsen, C.S. (2019) Bacterial and protozoan dynamics upon thawing and freezing of an active layer permafrost soil. The ISME Journal, 13, 1345–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubotz, F. , Wakeham, S.G. , Lipp, J.S. , Fredricks, H.F. & Hinrichs, K.U. (2009) Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environmental Microbiology, 11, 2720–2734. [DOI] [PubMed] [Google Scholar]
- Schubotz, F. , Xie, S. , Lipp, J.S. , Hinrichs, K.U. & Wakeham, S.G. (2018) Intact polar lipids in the water column of the eastern tropical North Pacific: abundance and structural variety of non‐phosphorus lipids. Biogeosciences, 15, 6481–6501. [Google Scholar]
- Schulz, S. , Brankatschk, R. , Dümig, A. , Kögel‐Knabner, I. , Schloter, M. & Zeyer, J. (2013) The role of microorganisms at different stages of ecosystem development for soil formation. Biogeosciences, 10, 3983–3996. [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W.S. et al. (2011) Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shean, D.E. , Bhushan, S. , Montesano, P. , Rounce, D.R. , Arendt, A. & Osmanoglu, B. (2020) A systematic, regional assessment of high mountain Asia glacier mass balance. Frontiers in Earth Science, 7, 363. [Google Scholar]
- Sherpa, M.T. , Najar, I.N. , Das, S. & Thakur, N. (2020) Exploration of microbial diversity of Himalayan glacier moraine soil using 16S amplicon sequencing and phospholipid fatty acid analysis approaches. Current Microbiology, 78, 78–85. [DOI] [PubMed] [Google Scholar]
- Shoemaker, W.R. , Jones, S.E. , Muscarella, M.E. , Behringer, M.G. , Lehmkuhl, B.K. & Lennon, J.T. (2021) Microbial population dynamics and evolutionary outcomes under extreme energy limitation. Proceedings of the National Academy of Sciences, 118, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan, W.T. , Lunn, M. , Woodcock, S. , Head, I.M. , Nee, S. & Curtis, T.P. (2006) Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environmental Microbiology, 8, 732–740. [DOI] [PubMed] [Google Scholar]
- Srinivas, T.N.R. , Singh, S.M. , Pradhan, S. , Pratibha, M.S. , Kishore, K.H. , Singh, A.K. et al. (2011) Comparison of bacterial diversity in proglacial soil from Kafni Glacier, Himalayan Mountain ranges, India, with the bacterial diversity of other glaciers in the world. Extremophiles, 15, 673–690. [DOI] [PubMed] [Google Scholar]
- Stres, B. , Danevčič, T. , Pal, L. , Fuka, M.M. , Resman, L. , Leskovec, S. et al. (2008) Influence of temperature and soil water content on bacterial, archaeal and denitrifying microbial communities in drained fen grassland soil microcosms. FEMS Microbiology Ecology, 66, 110–122. [DOI] [PubMed] [Google Scholar]
- Sturt, H.F. , Summons, R.E. , Smith, K. , Elvert, M. & Hinrichs, K.U. (2004) Intact polar membrane lipids in prokaryotes and sediments deciphered by high‐performance liquid chromatography/electrospray ionization multistage mass spectrometry—new biomarkers for biogeochemistry and microbial ecology. Rapid Communications in Mass Spectrometry, 18, 617–628. [DOI] [PubMed] [Google Scholar]
- Su, X. , Chen, X. , Hu, J. , Shen, C. & Ding, L. (2013) Exploring the potential environmental functions of viable but non‐culturable bacteria. World Journal of Microbiology and Biotechnology, 29, 2213–2218. [DOI] [PubMed] [Google Scholar]
- Tak, S. & Keshari, A.K. (2020) Investigating mass balance of Parvati glacier in Himalaya using satellite imagery based model. Scientific Reports, 10, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Horn, D.J. , Okie, J.G. , Buelow, H.N. , Gooseff, M.N. , Barrett, J.E. & Takacs‐Vesbach, C.D. (2014) Soil microbial responses to increased moisture and organic resources along a salinity gradient in a polar desert. Applied and Environmental Microbiology, 80, 3034–3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mooy, B.A.S. & Fredricks, H.F. (2010) Bacterial and eukaryotic intact polar lipids in the eastern subtropical South Pacific: water‐column distribution, planktonic sources, and fatty acid composition. Geochimica et Cosmochimica Acta, 74, 6499–6516. [Google Scholar]
- Wang, X.C. , Wu, J. , Guan, M.L. , Zhao, C.H. , Geng, P. & Zhao, Q. (2020) Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. The Plant Journal, 101, 637–652. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Naumann, U. , Wright, S.T. & Warton, D.I. (2012) mvabund—an R package for model‐based analysis of multivariate abundance data. Methods in Ecology and Evolution, 3, 471–474. [Google Scholar]
- Warren, S.D. , Clair, L.L. , Stark, L.R. , Lewis, L.A. , Pombubpa, N. , Kurbessoian, T. et al. (2019) Reproduction and dispersal of biological soil crust organisms. Frontiers in Ecology and Evolution, 7, 1–17. [Google Scholar]
- Weber, B. , Wu, D. , Tamm, A. , Ruckteschler, N. , Rodríguez‐Caballero, E. , Steinkamp, J. et al. (2015) Biological soil crusts accelerate the nitrogen cycle through large NO and HONO emissions in drylands. Proceedings of the National Academy of Sciences of the United States of America, 112, 15384–15389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wierzchos, J. , Casero, M.C. , Artieda, O. & Ascaso, C. (2018) Endolithic microbial habitats as refuges for life in polyextreme environment of the Atacama Desert. Current Opinion in Microbiology, 43, 124–131. [DOI] [PubMed] [Google Scholar]
- Wietrzyk‐Pełka, P. , Rola, K. , Szymański, W. & Węgrzyn, M.H. (2020) Organic carbon accumulation in the glacier forelands with regard to variability of environmental conditions in different ecogenesis stages of high Arctic ecosystems. Science of the Total Environment, 717, 1–12. [DOI] [PubMed] [Google Scholar]
- Wörmer, L. , Lipp, J.S. , Schröder, J.M. & Hinrichs, K.U. (2013) Application of two new LC‐ESI‐MS methods for improved detection of intact polar lipids (IPLs) in environmental samples. Organic Geochemistry, 59, 10–21. [Google Scholar]
- Xu, J. , Grumbine, R.E. , Shrestha, A. , Eriksson, M. , Yang, X. , Wang, Y. et al. (2009) The melting Himalayas: cascading effects of climate change on water, biodiversity, and livelihoods. Conservation Biology, 23, 520–530. [DOI] [PubMed] [Google Scholar]
- Yao, T. , Thompson, L.G. , Mosbrugger, V. , Zhang, F. , Ma, Y. , Luo, T. et al. (2012) Third pole environment (TPE). Environment and Development, 3, 52–64. [Google Scholar]
- Zhao, C. , Miao, Y. , Yu, C. , Zhu, L. , Wang, F. , Jiang, L. et al. (2016) Soil microbial community composition and respiration along an experimental precipitation gradient in a semiarid steppe. Scientific Reports, 6, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, X. , Wang, X. , Wei, J. , Jiang, Z. , Zhang, Y. & Liu, S. (2020) Spatiotemporal variability of glacier changes and their controlling factors in the Kanchenjunga region, Himalaya based on multi‐source remote sensing data from 1975 to 2015. Science of the Total Environment, 745, 140995. [DOI] [PubMed] [Google Scholar]
- Řezanka, T. , Temina, M. , Tolstikov, A.G. & Dembitsky, V.M. (2004) Natural microbial UV radiation filters—mycosporine‐like amino acids. Folia Microbiologia (Praha), 49, 339–352. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1: Supporting information
Data Availability Statement
The data that supports the findings of this study are available in the table and supplementary material of this article. Sequences were deposited to the Sequence Read Archive of NCBI under the BioProject PRJNA698068.


