Summary
Ester‐linked p‐coumarate (pCA) is a hallmark feature of the secondary cell walls in commelinid monocot plants. It has been shown that pCA groups arise during lignin polymerisation from the participation of monolignol conjugates assembled by p‐coumaroyl‐CoA:monolignol transferase (PMT) enzymes, members of the BAHD superfamily of acyltransferases.
Herein, we report that a eudicot species, kenaf (Hibiscus cannabinus), naturally contains p‐coumaroylated lignin in the core tissues of the stems but not in the bast fibres. Moreover, we identified a novel acyltransferase, HcPMT, that shares <30% amino acid identity with known monocot PMT sequences.
Recombinant HcPMT showed a preference in enzyme assays for p‐coumaroyl‐CoA and benzoyl‐CoA as acyl donor substrates and sinapyl alcohol as an acyl acceptor. Heterologous expression of HcPMT in hybrid poplar trees led to the incorporation of pCA in lignin, but no improvement in the saccharification potential of the wood.
This work illustrates the value in mining diverse plant taxa for new monolignol acyltransferases. Furthermore, the occurrence of pCA outside monocot lineages may represent another example of convergent evolution in lignin structure. This discovery expands textbook views on cell wall biochemistry and provides a new molecular tool for engineering the lignin of biomass feedstock plants.
Keywords: acylated lignin, BAHD monolignol acyltransferase, cell‐wall‐bound phenolics, convergent evolution, designer lignins, p‐coumarate pendent groups, plant cell walls
Introduction
Lignin is a phenolic polymer found predominantly in the secondary cell walls of vascular plants. The deposition of lignin alters the physicochemical properties of cellulosic cell walls, thereby contributing to mechanical strength, water conduction and plant defence (Barros et al., 2015). At the same time, lignin presents a formidable challenge for the industrial use of plant biomass due to its considerable chemical recalcitrance (Yoo et al., 2020). An improved understanding of lignin biosynthesis and evolution could be harnessed to enhance feedstock species, thereby economising the sustainable use of plant biomass for the production of biochemicals, biofuels, biomaterials and an array of other bioproducts.
Lignin is assembled primarily from three monolignol precursors – p‐coumaryl, coniferyl and sinapyl alcohols – which, respectively, form p‐hydroxyphenyl (H), guaiacyl (G) and syringyl (S) lignin units once incorporated into a polymer (Boerjan et al., 2003). The biosynthesis of monolignols occurs in the cytosol via the phenylpropanoid pathway. Following the export of monolignols from the cell, lignin polymerisation proceeds by the coupling of monolignol radicals generated by peroxidase and laccase enzymes. As this assembly process is stochastic and dependent solely upon the supply and chemical propensity of monomers for radical coupling, the composition and structure of lignin varies widely across different cell types and plant lineages (Ralph et al., 2004; Novo‐Uzal et al., 2012).
The diversification of plants has given rise to a host of other lignin constituents in addition to the three canonical monomers. For example, the lignin of commelinid monocots features ester‐linked p‐coumarate pendent groups (pCA; Smith, 1955a; Higuchi et al., 1967). Similarly, p‐hydroxybenzoate moieties (pHB) decorate the lignins of poplar and willow (Smith, 1955b; Goacher et al., 2021). The lignin found in the bast fibres of kenaf is naturally acetylated (Ralph, 1996). And quite remarkably, the fronds of Canary Island date palms have lignin with ester‐linked pCA, pHB, benzoate (BA), ferulate (FA), vanillate (VA) and acetate (Ac) groups (Karlen et al., 2017). Although the biological role of these lignin acylations remains largely unknown, there is considerable interest in harnessing this biochemistry to create designer lignins (Mottiar et al., 2016).
Ester‐linked pendent groups arise from the incorporation of γ‐acylated monolignol conjugates that participate in radical coupling reactions (Lu & Ralph, 2002, 2008; Fig. S1A). Members of the BAHD superfamily of acyltransferases catalyse the assembly of these conjugates before their export and polymerisation (R. D. Hatfield et al., 2008); Withers et al., 2012; Marita et al., 2014; Petrik et al., 2014; Smith et al., 2022). Moreover, due to the inherent plasticity of lignification, acyltransferases have been successfully deployed for lignin engineering. For example, heterologous expression of p‐coumaroyl‐CoA:monolignol transferase (PMT) genes from rice, Brachypodium and maize led to the incorporation of novel pCA groups into the lignin of transgenic plants, including in Arabidopsis, poplar and alfalfa (Smith et al., 2015; Marita et al., 2016; Sibout et al., 2016).
Conjugates of pCA are predominantly assembled in monocots with sinapyl alcohol (Grabber et al., 1996), suggesting that PMT enzymes may have a bias in monolignol substrate preference. It has been proposed that one role of acylated monolignols may be to enhance the incorporation of S‐lignin units during lignification through radical transfer mechanisms (Takahama et al., 1996). Although pCA is a good substrate for peroxidase and laccase enzymes and it can become integrated into synthetic dehydrogenative polymers formed in vitro, pCA occurs primarily as free phenolic pendent groups in lignin (Ralph et al., 1994). This is because phenoxy radicals of pCA preferentially undergo radical transfer rather than coupling reactions during lignification (R. Hatfield et al., 2008); Gani et al., 2019), meaning that only the monolignol moieties of conjugates participate in polymerisation. By contrast, monolignol–FA conjugates engage in radical coupling efficiently at both ends, and may thereby become integrated into lignin polymer backbones (Wilkerson et al., 2014). In this way, the incorporation of backbone‐integrated ferulate esters introduces chemically labile linkages that readily improves the efficiency of lignin depolymerisation (Kim et al., 2017). On the contrary, the impact of pendent pCA groups on biomass recalcitrance remains less clear.
Cell‐wall‐bound pCA groups have traditionally been considered a unique feature in the cell walls of monocots (Harris & Hartley, 1980; Karlen et al., 2018). However, herein we report that the lignin of kenaf (Hibiscus cannabinus), a eudicot species, naturally contains ester‐linked pCA pendent groups in the core tissues of stems. This was unequivocally shown using NMR and derivatisation followed by reductive cleavage (DFRC) methods. We further characterised a BAHD acyltransferase from kenaf that showed activity as a PMT in assays of recombinant enzyme, and when expressed heterologously in transgenic poplar trees. These observations support a growing consensus that lignin acylation, including the incorporation of pCA groups, is not phylogenetically limited to a few select clades but rather occurs more broadly in diverse plant taxa.
Materials and Methods
Lignin analysis by NMR and DFRC
Two‐dimensional 1H–13C HSQC NMR spectra were collected using enzyme‐lignin samples prepared by enzymatic digestion with Cellulysin cellulase (from Trichoderma viride; Calbiochem, Sigma‐Aldrich, Burlington, MA, USA) following ball‐milling of extractive‐free cell wall material, as described previously (Kim & Ralph, 2010). Kenaf core and bast samples were collected from mature stems. HSQC spectra were obtained using a Biospin Avance 700 MHz spectrometer (Bruker Corp., Billerica, MA, USA) with an adiabatic‐pulse programme (hsqcetgpsisp2.2). The dimethyl sulphoxide peak at δH 2.5 ppm and δC 39.5 ppm was used for spectral calibration, and volume integrations were performed using TopSpin 4.08 (Bruker Corp.). Proportions were expressed on a basis where % pCA = ½ pCA2/6/100% aromatics, % pHB = ½ pHB2/6/100% aromatics, 100% aromatics = ½ (H2/6) + G2 + ½ (S2/6 + ) and 100% aliphatics = Aα + Bα + ½ (Cα) + ½ (). Peak identifications were based on previous reports (Kim & Ralph, 2010).
Ester‐linked conjugates in lignin were released by DFRC, as described previously (Regner et al., 2018). For kenaf samples, products corresponding to the peracetates of cis‐ and trans‐isomers of coniferyl 7,8‐dihydro‐p‐coumarate (G–DHpCA), sinapyl 7,8‐dihydro‐p‐coumarate (S–DHpCA) and coniferyl 7,8‐dihydro‐ferulate (G–DHFA) were detected using multiple reaction monitoring mass detection. For poplar samples, the peracetates of cis‐ and trans‐sinapyl p‐hydroxybenzoate (S–pHB) were also measured, although coniferyl p‐hydroxybenzoate (G–pHB) was not detected.
Gene identification
Total RNA was isolated from the fresh stems of actively growing kenaf plants using the TRIzol reagent (Invitrogen, ThermoFisher Scientific, Carlsbad, CA, USA). cDNA was synthesised using the Creator SMART cDNA library construction kit (Clontech, Mountain View, CA, USA) and was subjected to 454 sequencing using a 454 GSFLX Titanium Sequencer (Roche). The resulting RNA‐seq dataset was then analysed as described previously (Peers, 2012; Wilkerson et al., 2018).
Sequence reads were first trimmed and compiled by de novo assembly using clc genomics workbench (Qiagen Aarhus A/S, Aarhus, Denmark) with the default parameters (mismatch cost, 2; deletion cost, 3; insertion cost, 3; length fraction, 0.5; and similarity, 0.8). The resulting database was used as a reference for an RNA‐seq experiment in which the reads were mapped back onto the assembled contigs and tabulated. The amino acid sequences of three known monocot PMTs (OsPMT, Os01g18744; BdPMT, Bradi2g36910; and ZmPMT, GRMZM2G028104) were then used in tBlastn queries against the contigs database to generate a shortlist of candidate BAHDs expressed in kenaf stems (Table S1). Next, each contig was annotated wherever possible with the closest homologue from Arabidopsis, identified by Blastx using the TAIR10 genome assembly with Araport11 annotations. Many candidates were not highly expressed, while others were homologues of previously characterised proteins. However, five contigs, each with relatively high expression levels, matched to a single Arabidopsis locus that was annotated only as an HXXXD‐type acyltransferase. From this, a corresponding coding sequence was selected for further study and was denoted HcPMT. Finally, a synthetic version of this 1335‐bp sequence was codon‐optimised for ease of synthesis, and was procured from GeneArt (ThermoFisher, Waltham, MA, USA).
Recombinant protein expression
The HcPMT coding sequence was transferred into the pDONR221 plasmid and then, using the Gateway cloning system (Invitrogen), into the pDEST17 expression vector, which contains an N‐terminal His6 tag to enable immobilised metal affinity chromatography (see Table S2 for primers and PCR conditions). This plasmid was then transformed into chemically competent BL21 (DE3) pLysS Escherichia coli cells (Invitrogen) for protein expression.
A starter culture was inoculated in 10 ml of TB‐Amp media (12 g l−1 tryptone, 24 g l−1 yeast extract, 2.3 g l−1 KH2PO4, 12.5 g l−1 K2HPO4, 4 ml l−1 glycerol, 50 μg ml−1 ampicillin). After 16 h of incubation at 37°C on an orbital shaker at 200 rpm, the cells were transferred into 250 ml of fresh TB‐Amp media in a 1‐l baffled flask to a starting OD600 of 0.1. After 6 h of additional cultivation, the OD600 reached 0.8 and the temperature was decreased to 16°C. Protein expression was then induced by adding isopropyl‐β‐d‐thiogalactopyranoside to a concentration of 10 mM. After 16 h of further cultivation at 16°C, bacterial cells were harvested by centrifugation at 4000 g and 4°C.
The cell pellet was resuspended in an equal volume of Bugbuster protein extraction reagent (Sigma‐Aldrich) containing 1 U ml−1 benzonase nuclease and 0.5 mM phenylmethylsulphonyl fluoride and slowly agitated at room temperature for 20 min. Cell debris was then pelleted by centrifugation at 16 000 g and 4°C, and the supernatant was gradually concentrated and buffer‐exchanged 10 000‐fold with 20 mM sodium phosphate buffer (pH 7.4) using an Amicon Ultra 15 ml 10 K centrifugal concentrator unit (Sigma‐Aldrich). The 5‐ml volume of crude protein was then loaded onto a His GraviTrap flow‐through column (GE Healthcare, Chicago, IL, USA). After washing the column with 8 ml of 20 mM sodium phosphate buffer containing 500 mM sodium chloride, elution was performed using 2‐ml portions of the same buffer, but containing increasing concentrations of imidazole (50, 100, 150, 200, 250, 300, and 600 mM). SDS‐PAGE analysis revealed that the fraction eluted using 150 mM contained a protein with an apparent molecular weight close to the theoretical size of HcPMT (c. 50 kDa). This fraction was buffer‐exchanged 10 000‐fold with 25 mM sodium phosphate buffer (pH 5.8), and the protein concentration was determined using the Bio‐Rad protein assay (Bio‐Rad Laboratories, Hercules, CA, USA). The enzyme preparation was flash‐frozen by pipetting droplets into liquid nitrogen and then stored at −80°C until further use.
Enzyme characterisation
The purified enzyme was tested for activity with p‐coumaroyl‐CoA (pCA‐CoA), benzoyl‐CoA (BA‐CoA), p‐hydroxybenzoyl‐CoA (pHB‐CoA), feruloyl‐CoA (FA‐CoA), and acetyl‐CoA (Ac‐CoA) as acyl donors, and p‐coumaryl, coniferyl and sinapyl alcohols as monolignol acyl acceptors. BA‐CoA and Ac‐CoA were commercially available (Sigma‐Aldrich), whereas the other CoAs were enzymatically synthesised using Nt4CL1, as described previously (Beuerle & Pichersky, 2002). The monolignol conjugates used as authentic standards were produced following established chemical synthesis methodology (Y. Zhu et al., 2013).
Enzyme assays were set up as 50‐μl reactions containing 50 mM sodium phosphate buffer (pH 6), 1 mM dithiothreitol, 1 mM of each CoA thioester, 1 mM of each monolignol and 5 μg of purified HcPMT enzyme (1 μg μl−1). The reactions were incubated at room temperature for 60 min at which point they were terminated by adding 50 μl of 100 mM hydrochloric acid, solubilised by adding 900 μl methanol and then filtered through 0.2‐μm nylon syringe filters. Reaction products were analysed by liquid chromatography coupled with mass spectrometry (LC–MS, LCMS8040; Shimadzu Corp., Kyoto, Japan) using a Kinetex XB‐C18 column (250 × 4.60 mm, 100 Å; Phenomenex, Torrance, CA, USA) with water (A) and methanol (B) as eluents at a flow rate of 1 ml min−1. The binary gradient programme was ramped as follows: initial condition of 5% eluent B in eluent A, a linear increase to 100% eluent B over 30 min, hold for 4 min, then return to 5% eluent B over 1 min and hold for 10 min to re‐equilibrate the column. Reaction products were monitored using a photodiode array detector (λ = 250–400 nm) along with mass spectrometry in both positive and negative ion modes (Q3 scans between m/z 120 and m/z 600). Products were verified by comparing retention times and mass fragmentation patterns to authentic standards.
Transgenic hybrid poplar
The Gateway cloning system (Invitrogen) was used to transfer the synthetic HcPMT coding sequence from pDONR221 into the plant expression vector pMDC32, which contains a 35S promoter (Curtis & Grossniklaus, 2003). This binary vector was transferred into Agrobacterium tumefaciens EHA105 by freeze–thaw transformation of chemically competent cells. Agrobacterium‐mediated transformation of hybrid poplar (Populus alba × grandidentata; P39) was then performed as described previously (Mottiar et al., 2022).
Five transgenic lines were selected by PCR screening and multiplied in tissue culture. Before planting in the glasshouse, all plants were propagated from apical stem pieces to ensure even initial growth. For each transgenic line, five biological replicates were planted with a peat‐based soil in two‐gallon pots, grown in a glasshouse under an 18‐h daylight regime and watered as needed with fertigated water. Immediately upon planting and for the subsequent 2 wk, the plants were covered with transparent plastic cups to maintain humidity during acclimatisation. As they grew, the trees were supported with bamboo stakes and monitored regularly for any signs of disease or pests.
Poplar trees were harvested after 18 wk of growth. The plant heights and stem diameters were recorded, and the leaves and branches were removed. The bark was peeled from the stems, and the developing xylem was scraped from the wood and flash‐frozen in liquid nitrogen. Debarked stems were air‐dried and then later cut into small pieces and comminuted to a powder using a Wiley mill fitted with a 40‐mesh sieve. Extractive‐free samples were obtained by extracting wood powder for 24 h with hot acetone in a Soxhlet extraction apparatus.
Transgene expression
Xylem scrapings were ground to a fine powder in a mortar and pestle with liquid nitrogen, and total RNA was isolated using a CTAB‐based procedure (Kolosova et al., 2004). Following DNase treatment with the Turbo DNA‐free kit (Ambion, ThermoFisher), RNA was used in the synthesis of first‐strand cDNA with the OneScript Plus cDNA synthesis kit (Applied Biological Materials Inc., Richmond, BC, Canada). Next, the cDNA was diluted 16‐fold and used in qPCR reactions prepared with BrightGreen qPCR Master Mix (Applied Biological Materials Inc.). Gene‐specific primers validated over a wide range of template concentrations were used. Reference reactions were performed using primers designed to amplify the poplar actin and elongation factor 1β sequences. Thermal cycling, fluorescence detection, melt‐curve analysis and the determination of relative expression were performed using a CFX96 Touch Real‐Time PCR Detection System (Bio‐Rad Laboratories).
Lignin content, composition, molecular weight and histology
The lignin content and composition were evaluated using established methodology for Klason lignin determination and thioacidolysis, as described previously (Huntley et al., 2003; Robinson & Mansfield, 2009). Lignin molecular weight was analysed for enzyme‐lignin preparations using gel permeation chromatography, and lignin histology was examined by staining transverse stem cross sections with phloroglucinol‐HCl and toluidine blue, as described elsewhere (Mottiar et al., 2022).
Cell‐wall‐bound phenolic acids and acetyl
Cell‐wall‐bound phenolic acids were released by mild alkaline hydrolysis (saponification) and quantified using HPLC analysis, as described previously (Goacher et al., 2021). Cell‐wall‐bound acetyl was measured in a similar manner, as described previously (Mottiar et al., 2020).
Structural polysaccharides and saccharification potential
The composition of structural polysaccharides was measured by using high‐performance anion‐exchange liquid chromatography. Briefly, 15‐μl aliquots of 1 : 10 diluted acid hydrolysate from the Klason lignin hydrolysis were injected onto a CarboPac PA1 anion‐exchange column (Dionex Corp., Sunnyvale, CA, USA), and the six major neutral sugars were separated using 1 ml min−1 water as the eluent. Pulsed amperometric detection was enabled by the post‐column addition of 200 mM NaOH at 0.5 ml min−1.
Saccharification potential was evaluated by pretreating wood powder with either 2% sulphuric acid or 62.5 mM sodium hydroxide for 3 h at 90°C and then subjecting it to enzymatic hydrolysis using 0.2 FPU ml−1 of Cellic Ctec3 HS enzyme cocktail in 100 mM sodium acetate buffer at 50°C. Reactions were terminated after 0.5, 1, 2, 6, 12, 24 or 72 h by heating to 95°C for 10 min. Finally, the release of glucose and xylose was measured by anion‐exchange chromatography.
Analysis of PMT sequences
The amino acid sequence of HcPMT was aligned with three commelinid monocot PMT sequences – OsPMT (Os01g18744), BdPMT (Bradi2g36910) and ZmPMT (GRMZM2G028104) – using the clustal omega algorithm (Sievers et al., 2011). Sequence identity and similarity were calculated using the SMS Ident and Sim tool (Stothard, 2000). Next, putative homologues of HcPMT were identified from the following sequenced eudicot genomes using Blast with a cut‐off E‐value of 1 × 10−100: Populus trichocarpa (JGI v3.0), Arabidopsis thaliana (TAIR 10), Vitis vinifera (Genoscope 12×), Medicago truncatula (MTGD Mt4.0) and Solanum lycopersicum (iTAG 4.0). Amino acid sequences were aligned using MegaX with the Muscle algorithm and the default parameters (Kumar et al., 2018). This alignment was then subjected to a maximum likelihood analysis to construct a phylogenetic tree. The sequences for OsPMT, BdPMT and ZmPMT were included as an outgroup, and the phylogeny was resampled using 1000 bootstrap iterations.
Replicates and statistics
Enzyme assays were performed in triplicate. For the measurement of tree heights, stem diameters, transgene expression, lignin content, lignin composition, cell‐wall‐bound pCA, pHB and acetyl, structural polysaccharides and saccharification potential, five biological replicates were analysed per line and triplicate technical replicates were processed for each. The DFRC reactions for transgenic poplars were conducted with three biological replicates per line measured in triplicate. Three biological replicates were analysed by NMR and gel permeation chromatography for transgenic poplars. Representative images are provided for lignin histology. One sample each of mature kenaf stem bast and core tissue was used for NMR and DFRC. Statistically significant differences between the transgenic poplar lines and the WT control were evaluated by a one‐way ANOVA with a post hoc Dunnett's test performed using Spss Statistics 27 (IBM, Armonk, NY, USA), except for the transgene expression results where a Tukey–Kramer test was performed instead to compare the transgenic lines to one another.
Sequences
The nucleotide and amino acid sequences of HcPMT are provided in Fig. S2. The sequence of HcPMT was also reported as part of a recent genome sequence assembly and was assigned a gene identification number of GWHGACDB058585.1 (Zhang et al., 2020).
Results and Discussion
Kenaf lignin is naturally p‐coumaroylated
The bast fibres of kenaf contain an S‐rich lignin that is extensively acetylated (Ralph, 1996). It has been shown that acetyl groups predominantly occur on the γ‐OH of S‐lignin units and that they derive from the incorporation of preacylated monolignols during lignification (Lu & Ralph, 2002). In contrast to the bast fibres, kenaf stem core tissues inherently possess a lignin that is less acetylated and has lower levels of S units (Seca et al., 1998). Furthermore, alkaline hydrolysis indicated that mature kenaf stems may have cell‐wall‐bound pCA and FA (Geronikaki et al., 1979; Morrison III et al., 1999). However, pyrolysis GC–MS of kenaf uncovered only trace amounts of hydroxycinnamic acids (Gutiérrez et al., 2004).
To examine this further, we analysed kenaf bast and core tissues using solution‐state two‐dimensional 1H–13C heteronuclear single‐quantum coherence (HSQC) NMR. Spectra collected from enzyme‐lignin preparations showed well‐resolved aromatic signatures corresponding to H‐, G‐, and S‐lignin in both bast and core tissues, with bast fibres having a higher proportion of S units (Fig. 1a). This analysis also revealed the presence of pCA and FA groups in the core tissues, but not in bast fibres. DFRC reactions, which cleave β‐aryl ether bonds in lignin while leaving ester‐linkages intact, were then performed next (Fig. 1b). Diagnostic dihydro‐p‐coumarate (DHpCA) products derived from pCA conjugates with S‐lignin (S–DHpCA) and G‐lignin units (G–DHpCA) were detected in kenaf stem core tissues, but not in bast fibres. Products corresponding to monolignol–FA conjugates were also observed, as has been reported previously (Karlen et al., 2016). Notably, pCA was associated with both G and S units, whereas FA was exclusively conjugated with G units. We also used a modified version of DFRC to look for acetylated lignin units; however, these groups were only detected in the bast, as has been reported previously (Lu & Ralph, 2002).
Fig. 1.
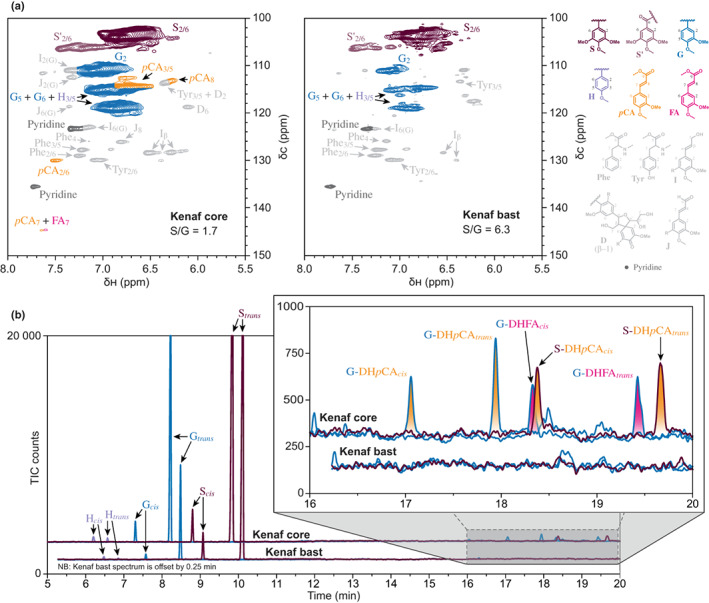
Evidence that kenaf core lignin naturally contains pCA groups. (a) 2D‐NMR 1H–13C HSQC spectra for enzyme‐lignin preparations of kenaf core and bast tissues, as labelled, showing the aromatics region. The colour coding and peak annotations for p‐hydroxyphenyl (H, mauve), guaiacyl (G, blue) and syringyl units (S and S′, dark and light purple), as well as p‐coumarate (pCA, orange) and ferulate groups (FA, pink), are elaborated with the structures shown. Also shown, in grey, are peaks corresponding to phenylalanine (Phe), tyrosine (Tyr), cinnamyl alcohol endgroups (I), cinnamaldehyde endgroups (J) and spirodienone units (D). (b) Multiple reaction monitoring traces from GC–MS analysis of derivatisation followed by reductive cleavage reaction products corresponding to kenaf core and bast tissues, as labelled. The spectrum for kenaf bast is delayed by 15 s. Peaks corresponding to cis‐ and trans‐ isomers of monolignols (H, mauve; G, blue; S, dark purple) and monolignols conjugated with p‐coumarate (coniferyl 7,8‐dihydro‐p‐coumarate, G–DHpCA and sinapyl 7,8‐dihydro‐p‐coumarate, S–DHpCA, yellow) and ferulate (coniferyl 7,8‐dihydro‐ferulate, G–DHFA, pink) are labelled and colour‐coded with reference to the structures shown.
The observation that pCA groups occur exclusively on the lignin of core tissues in kenaf stems whereas acetyl groups prevail in bast fibres illustrates that kenaf lignin acylation may be highly controlled in a spatiotemporal manner. This is certainly not the only example of such fine‐scale patterning of lignin composition and structure. For example, the outer tissues of Canary Island date palm fronds contain lignin with pCA and pHB groups, whereas the lignin found in the inner tissues contains pHB moieties as well as fewer pCA and BA groups (Karlen et al., 2017). At an even finer scale, pHB groups in poplar stems occur in the cell walls of xylem fibres but are largely absent from adjacent vessels (Goacher et al., 2021). Whether such variability in acylation substantially impacts the physicochemical properties of cell walls, and thereby plays any important biological role remains unclear (Mottiar & Mansfield, 2022). Differences in lignin acylation may be driven by spatially restricted expression of the corresponding monolignol acyltransferases; however, the supply of requisite substrates could also play an important role.
Our analysis proves the presence of pCA in the core tissues of kenaf stems and provides a more complete view of kenaf lignin structures (Fig. S1B,C). Recently, pCA was observed (but not remarked upon) in mulberry (Morus alba), another eudicot species but from the family Moraceae (Yamamoto et al., 2020). These two examples illustrate that p‐coumaroylation is not restricted to monocot lineages, as has long been the prevailing view. In an extensive survey of dicots, pCA was released from the cell walls of diverse plant taxa including members of the Bignoniaceae, Caryophyllaceae, Gesneriaceae, Meliaceae, Rubiaceae and Styracaceae families (Hartley & Harris, 1981). There have even been reports of small amounts of pCA in Populus spp., particularly in the bark tissues (Pearl & Beyer, 1960; Sun et al., 2001). However, it remains to be seen how widely pCA occurs within the eudicots and whether pCA represents a substantial fraction of the total lignin in any of these taxa.
Whether or not p‐coumaroylation, or any lignin acylation for that matter, has an important biological function remains an open question. However, it is becoming clear that acylated lignin is far more common than had been previously recognised (del Río et al., 2022). To date, five PMT enzymes have been identified and characterised, and all occur in commelinid monocots: rice (OsPMT; Withers et al., 2012), Brachypodium (BdPMT; Petrik et al., 2014), maize (ZmPMT; Marita et al., 2014), sorghum and switchgrass (SbPMT and PvPMT; Smith et al., 2022). In order to shed light on the occurrence of p‐coumaroylation within the eudicots and potentially develop a new tool for lignin engineering, we set out to characterise a novel BAHD acyltransferase from kenaf, HcPMT.
Recombinant HcPMT produces monolignol–pCA conjugates
HcPMT protein was expressed in E. coli and purified by immobilised metal affinity chromatography (Fig. S3). The recombinant enzyme was first tested in a series of pairwise assays with individual acyl donor substrates (pCA‐CoA, BA‐CoA, pHB‐CoA, FA‐CoA and Ac‐CoA) and monolignol acyl acceptors (p‐coumaryl, coniferyl and sinapyl alcohols). When provided with pCA‐CoA and any of the three monolignols, recombinant HcPMT produced monolignol–pCA conjugates (Fig. S4A–C). HcPMT also reacted with BA‐CoA to produce coniferyl–BA and sinapyl–BA, but not p‐coumaryl–BA (Fig. S4D–F). Similarly, coniferyl–pHB and sinapyl–pHB were produced with pHB‐CoA (Fig. S4G–I). However, feruloyl‐CoA yielded only sinapyl–FA as a product (Fig. S4J–L). As FA is linked only to G units in kenaf lignin, HcPMT is likely not responsible for feruloylation, consistent with a previous report that a distinct feruloyl‐CoA:monolignol transferase (FMT) occurs in kenaf (Wilkerson et al., 2018). And finally, HcPMT was able to produce sinapyl–Ac and coniferyl–Ac when provided with Ac‐CoA (Fig. S4M–O).
To determine the preferred acyl donor, HcPMT was subjected to a series of substrate competition assays with all five acyl‐CoA substrates along with each monolignol provided separately. In the presence of the five acyl donors and p‐coumaryl alcohol, recombinant HcPMT produced only p‐coumaryl–pCA (Fig. 2a). With coniferyl alcohol as the acyl acceptor, coniferyl–pCA was produced along with a lesser amount of coniferyl–BA (Fig. 2b). Similarly, with sinapyl alcohol and all five acyl donors provided, sinapyl–pCA and sinapyl–BA were produced (Fig. 2c). The identities of the conjugation products were confirmed by mass spectrometry and with reference to authentic standards. Thus, among the five acyl donor substrates tested, recombinant HcPMT was most active with pCA‐CoA and BA‐CoA. Since acetyl‐CoA was not a preferred substrate, it is very unlikely that HcPMT is a contributor to lignin acetylation in kenaf. In summary, the analysis of recombinant HcPMT supports its designation as a p‐coumaroyl‐CoA:monolignol transferase enzyme having a preference for sinapyl alcohol.
Fig. 2.
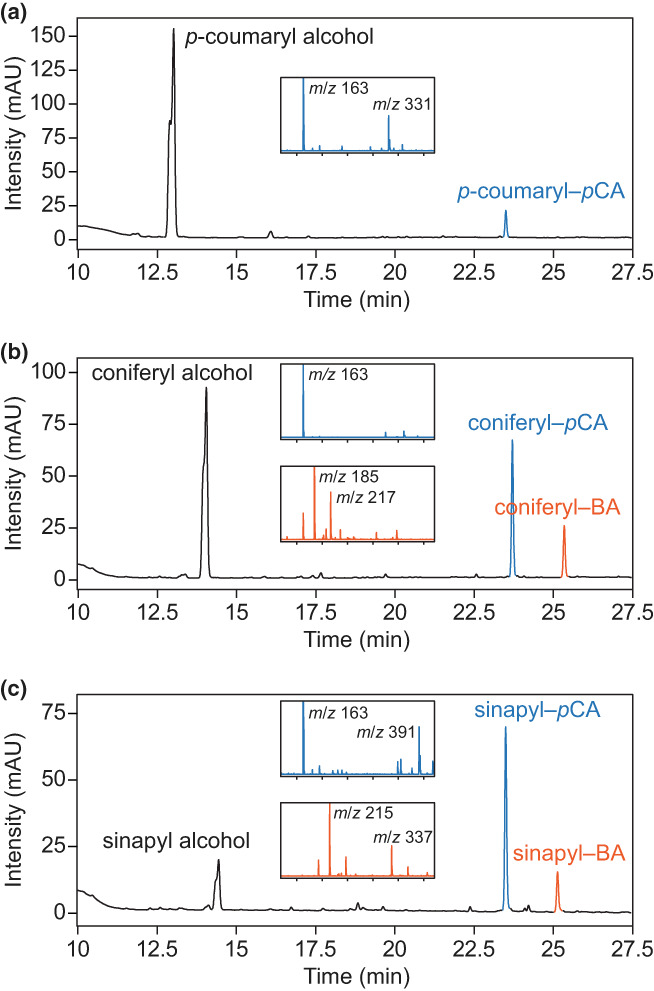
Competitive enzyme activity assays testing HcPMT substrate preferences. (a) In the presence of p‐coumaryl alcohol and five CoA thioesters (pCA‐CoA, BA‐CoA, pHBA‐CoA, FA‐CoA and Ac‐CoA), HcPMT produces only p‐coumaryl–pCA. (b) In the presence of coniferyl alcohol and all five CoA thioesters, HcPMT produces coniferyl–pCA and coniferyl–BA. (c) In the presence of sinapyl alcohol and all five CoA thioesters, HcPMT produces sinapyl–pCA and sinapyl–BA. Chromatographic traces show the absorbance at 273 nm. The insets show mass spectra for monolignol–pCA conjugates in blue and monolignol–BA conjugates in orange with m/z values labelled. All reactions were performed at room temperature for 60 min with 1 μg purified recombinant enzyme, 1 mM dithiothreitol, 1 mM of each CoA thioester and 1 mM of each monolignol in 50 mM phosphate buffer (pH 6).
Transgenic poplar trees expressing HcPMT have altered lignin
Although in vitro assays are invaluable for assessing substrate preference and catalytic potential, they do not necessarily reflect the true enzyme activity in vivo as enzymes that are apparently bifunctional may be catalytically limited by the availability of substrates. We therefore generated transgenic hybrid poplar trees to test the effects of HcPMT in planta. Five transgenic poplar lines representing independent stable transformation events were selected for in‐depth analysis and were grown under controlled conditions in a glasshouse (Fig. 3a). After 16 wk of cultivation, some of the transgenic lines appeared to be stunted compared with wild‐type (WT) control trees. Lines 4 and 5 were significantly shorter, and line 5 had a smaller average stem diameter (Fig. 3b). These lines also showed the highest levels of transgene expression, as measured by RT‐qPCR (Fig. S5). In addition, line 5 exhibited extensive sylleptic branching, which can be a sign of metabolic stress in hybrid poplars.
Fig. 3.
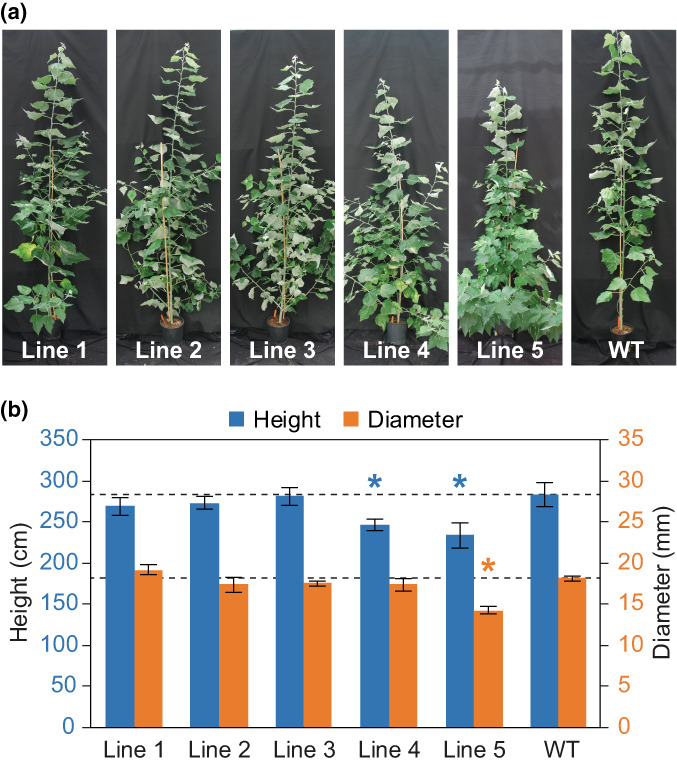
Transgenic hybrid poplar trees expressing HcPMT. (a) Representative photographs of each transgenic line and the wild‐type (WT) control. (b) Heights and stem diameters measured at the time of harvest are plotted on the left and right axes, respectively, in blue and orange. Values marked with an asterisk are significantly different from the WT control (one‐way ANOVA with Dunnett's test, n = 5 for each line using technical triplicates, P‐value < 0.05). Error bars correspond to SD.
To ascertain whether the expression of HcPMT affected the biosynthesis and assembly of cell wall components, the Klason lignin content was measured using samples of extractive‐free powdered wood from each line. Compared with WT control trees, there were no significant differences in total lignin content, nor in the proportions of acid‐insoluble or acid‐soluble lignin (Fig. 4a). Similarly, no consistent differences were noted in the composition of structural polysaccharides (Table S3). There were, however, significant differences in lignin composition with lines 1 and 5 having a greater proportion of S units compared with WT control trees (Fig. 4a). Microscopy of stem cross sections stained with phloroglucinol‐HCl and toluidine blue revealed that there were no obvious changes in lignin distribution, nor in the proportion or dimensions of vessels (Fig. S6). Next, the molecular weights of enzyme‐lignin preparations were assessed using gel permeation chromatography. Compared with WT control trees, lignin from lines 1, 3, 4 and 5 had greater weight‐averaged molecular weight, M W, as well as an increased dispersity index, Đ M (Fig. 4b).
Fig. 4.
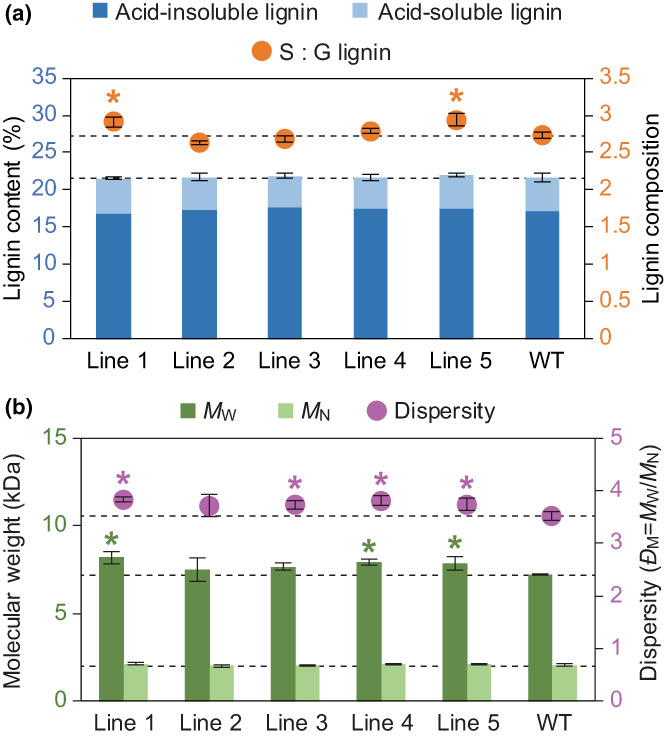
Lignin analysis of transgenic poplar expressing HcPMT. (a) The Klason lignin content is plotted on the left axis with the acid‐insoluble and acid‐soluble fractions shown in dark and light blue, respectively. The ratio of syringyl (S) : guaiacyl (G) lignin monomers is plotted on the right axis in orange. (b) The molecular weight of enzyme‐lignin samples is plotted on the left axis on a weight basis (M W) in dark green and on a number basis (M N) in light green. The dispersity (Đ M = M W/M N) is plotted on the right axis in magenta. Values marked with an asterisk are significantly different from the wild‐type (WT) control (one‐way ANOVA with Dunnett's test, n = 5 for each line using triplicates with P‐value < 0.05 in a and n = 3 for each line with P‐value < 0.1 in b). Error bars correspond to SD.
Mild alkaline hydrolysis reactions were then performed in attempts to selectively release ester‐linked cell‐wall‐bound components (Figs 5a, S7). Substantial quantities of pCA were released from extractive‐free wood powder for all five transgenic lines, whereas WT control trees yielded only trace amounts. Poplar lignin is naturally p‐hydroxybenzoylated and slight reductions in pHB groups were observed as a consequence of pCA incorporation. Interestingly, it was observed that the biosynthesis and the incorporation of pCA groups did not come at the expense of pHB when OsPMT was expressed in poplar (Smith et al., 2015). Although recombinant HcPMT also showed some activity in vitro on BA‐CoA, and it is known that BA is compatible with lignification in poplar (Kim et al., 2020), only trace amounts of BA groups were detected (Fig. S7B inset). There was also no change in cell‐wall‐bound Ac groups, further indicating that HcPMT does not produce acetylated monolignols in planta.
Fig. 5.
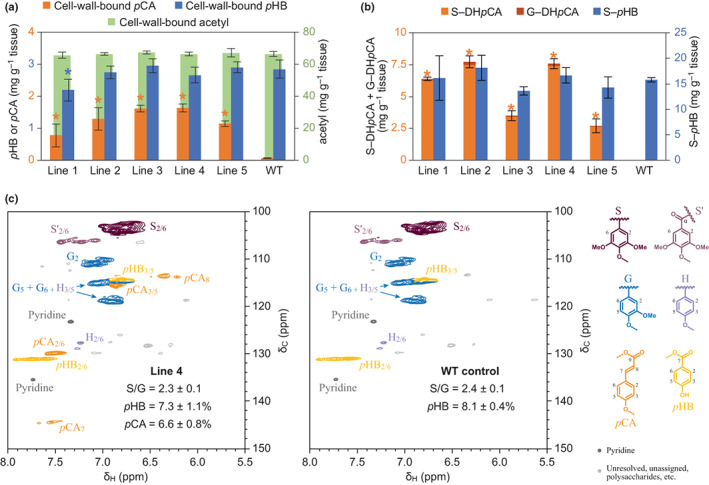
Analysis of lignin acylations in transgenic poplar expressing HcPMT. (a) The amounts of ester‐linked p‐coumarate (pCA) and p‐hydroxybenzoate (pHB) released by alkaline hydrolysis are plotted on the left axis in orange and blue, respectively. Cell‐wall‐bound acetyl is plotted on the right axis in green. (b) The amounts of the peracetates of sinapyl dihydro‐p‐coumarate (S–DHpCA) and coniferyl dihydro‐p‐coumarate (G–DHpCA) released by DFRC are plotted on the left axis in light orange and dark orange, respectively. Sinapyl p‐hydroxybenzoate (S–pHB) is plotted on the right axis in blue. Values marked with an asterisk are significantly different from the wild‐type (WT) control (one‐way ANOVA with Dunnett's test, n = 5 in (a) and n = 3 in (b) for each line using technical triplicates, P‐value < 0.05). Error bars correspond to SD. (c) Partial 2D‐NMR 1H–13C HSQC spectra for enzyme‐lignin samples of line 4 and the WT control, as labelled, showing the aromatic region. The peak annotations for p‐hydroxyphenyl (H, light purple), guaiacyl (G, blue) and syringyl units (S and S′, dark and light purple), as well as p‐coumarate (orange) and p‐hydroxybenzoate pendent groups (yellow) are elaborated with the structures shown at the side. Proportions for integrated peak volumes are provided as the mean ± SE for three biological replicates.
Derivatisation followed by reductive cleavage reactions were performed next to release intact ester‐linked monolignol conjugates (Fig. 5b). This work revealed that pCA predominantly occurred as conjugates with S‐lignin units (S–DHpCA) since only minor quantities of G conjugates (G–DHpCA) were detected. The activity of HcPMT in transgenic poplar was largely consistent with the native activity in kenaf core tissues, considering that poplar is an S‐lignin‐rich species. Endogenous pHB was conjugated only to S units in poplar, as has been observed previously (Mottiar et al., 2022). As a final confirmation of pCA incorporation, enzyme‐lignin preparations from line 4 and the WT control were examined by NMR (Fig. 5c). HSQC spectra corroborated the thioacidolysis and alkaline hydrolysis results and showed that the incorporation of pCA in poplar slightly altered the S : G lignin monomer ratio and resulted in only a small reduction in pHB groups. Again, no BA groups were observed, indicating that HcPMT was unable to generate monolignol–BA conjugates in poplar likely because the preferred pCA‐CoA substrate was readily available. Volume integrations indicated that pCA groups represented c. 6.6% of the total lignin in transgenic poplar trees expressing HcPMT, much greater than the endogenous levels observed in kenaf core tissues. Inspection of the aliphatics region of the spectra showed that the incorporation of additional γ‐acylated monolignol conjugates also led to a small increase in the proportion of β‐aryl ether linkages at the expense of phenylcoumaran and resinol structures (Fig. S8). Such changes in lignin composition, molecular weight and structure could potentially impact cell wall recalcitrance, thereby affecting the efficiency of industrial biomass utilisation.
Industrial relevance of pCA
The occurrence of pCA in monocot cell walls enhances lignin solubility due to the abundance of free phenolic groups (Lapierre et al., 1989). Similarly, lignin from transgenic Arabidopsis and poplar with novel pCA groups resulting from the expression of BdPMT was significantly more soluble in alkali (Sibout et al., 2016; Lapierre et al., 2021). However, it has been reported that lignin interacts with hemicelluloses in plant cell walls through electrostatic and other non‐covalent forces (Kang et al., 2019). A greater abundance of pCA pendent groups decorating lignin polymers could conceivably promote such interactions, thereby restricting access to structural polysaccharides. As such, the question of whether p‐coumaroylation enhances or impedes cell wall digestibility remains largely unresolved.
To evaluate whether the expression of HcPMT in poplar affected biomass recalcitrance, powdered wood was subjected to various pretreatments (dilute alkali, dilute acid and no pretreatment) followed by 72 h of enzymatic hydrolysis (Fig. S9). Regardless of pretreatment regime, no improvement in saccharification potential was observed among any of the transgenic lines in terms of either glucose or xylose release. In fact, several lines showed slight reductions in digestibility compared with the WT control. This contrasts with the increased sugar release observed for transgenic Arabidopsis expressing BdPMT (Sibout et al., 2016). However, no change in cell wall digestibility was reported for alfalfa lines with elevated pCA levels resulting from the expression of ZmPMT (Marita et al., 2016). In a study of inbred maize lines with varying contents of hydroxycinnamates, pCA was negatively correlated with cell wall degradability (Zhang et al., 2011). A possible explanation for these contrasting observations is that the lignin content was reduced in the transgenic BdPMT Arabidopsis, but unchanged in HcPMT‐expressing poplar and in these other examples. The observed increases in the average molecular weight of lignin in HcPMT transgenic poplar could offer an alternative explanation for reduced digestibility, as lignin polymer size and structure may play an important role in chemical recalcitrance (Yoo et al., 2020). Indeed, recalcitrance is a complex parameter influenced by myriad attributes of the biomass, the severity of the pretreatment regime and the efficacy of enzymatic hydrolysis (McCann & Carpita, 2015).
Whether the introduction of ester‐linked pendent groups impacts biomass recalcitrance remains an open question. In contrast to our findings, a recent report on the expression of BdPMT in transgenic poplar found a significant increase in saccharification due to lignin p‐coumaroylation (Lapierre et al., 2021). Similarly, we also recently reported that transgenic poplar with elevated levels of p‐hydroxybenzoate groups showed reduced recalcitrance (Mottiar et al., 2022). One possible explanation could lie in the timing of lignin acylation. In the current study, we used a constitutive promoter that may have produced acylated monolignols well in advance of lignification. By contrast, those other studies used cell‐wall‐specific promoters, and this could have caused spatiotemporal differences with acylated monomers being incorporated at different stages of lignification and perhaps even in different locations (i.e. in the middle lamella vs in the cell wall). To resolve this question, it would be interesting to analyse poplar biomass from both constructs simultaneously.
Although lignin p‐coumaroylation may or may not improve biomass digestibility, pCA could be a lucrative engineering target in its own right. As pCA groups are ester‐bound to the lignin backbone, treatment with mild alkali can be used to efficiently release these pendent groups. In some applications involving fermentation, such free phenolics could be cytotoxic and detrimental to growth and conversion rates. However, these ‘clip‐offs’ would be commercially valuable if they could be economically separated from the residual biomass (Karlen et al., 2020; Timokhin et al., 2020). Liberated pCA could be used directly as there are well‐established applications in foods, pharmaceuticals and cosmetics (Boz, 2015; Pei et al., 2016; Boo, 2019). Alternatively, pCA could be converted into platform chemicals and then upgraded into various high‐value bioproducts and biomaterials. For example, work is ongoing to develop enhanced strains of Pseudomonas and Rhodococcus capable of efficient catabolism of pCA to produce organic acids that can be readily transformed into an array of valuable chemicals (Otani et al., 2014; Becker et al., 2015; Mohamed et al., 2020). In this way, the discovery and development of new monolignol acyltransferases may provide valuable molecular tools for lignin engineering and the pursuit of designer lignins (Mottiar et al., 2016).
HcPMT differs substantially from monocot PMT sequences
A comparison of PMT amino acid sequences reveals noteworthy differences and similarities (Fig. 6a). Although OsPMT, BdPMT and ZmPMT all share 60% sequence identity, HcPMT is < 30% identical. When amino acid similarities are considered, HcPMT is at most 43% similar to the other PMTs. All four sequences bear the universally conserved HXXXD and DXGWG motifs that are hallmarks of BAHD acyltransferases (St‐Pierre & De Luca, 2000). The conserved histidine in the former sequence is known to deprotonate the hydroxyl group of acyl acceptors, whereas the latter motif is apparently structurally important (Ma et al., 2005). All the PMT sequences also have conserved motifs characteristic of clade Va, namely YPFAGR, QVTXXXCGG and GXYGN (Tuominen et al., 2011).
Fig. 6.
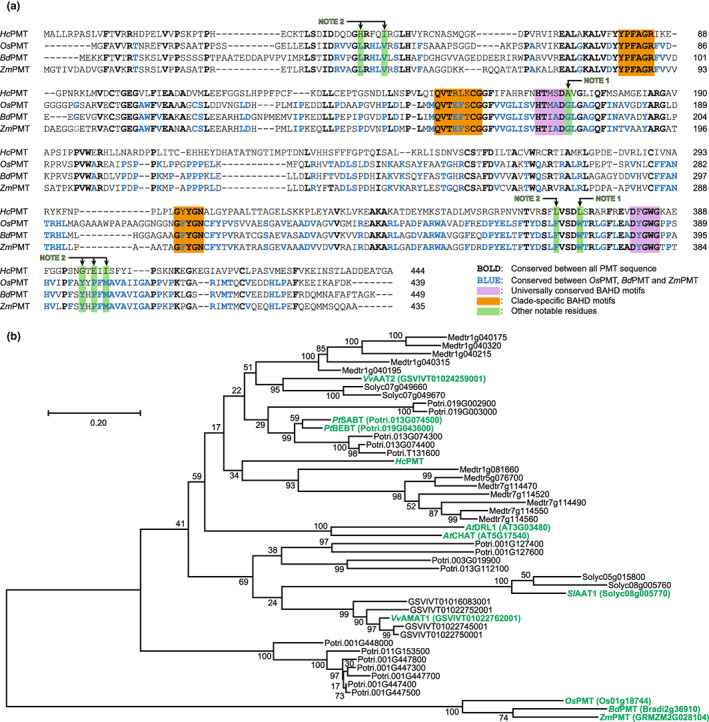
Analysis of PMT sequences. (a) An alignment of the HcPMT amino acid sequence with those of the previously characterised PMTs from rice (OsPMT), Brachypodium (BdPMT) and maize (ZmPMT). Fully conserved residues are shown in bold, and residues conserved only among the commelinid monocot sequences are shown in blue. The universal motifs characteristic of BAHD enzymes are highlighted in purple, clade‐specific motifs are highlighted in orange, and additional residues discussed in the text are highlighted in green (labelled as Note 1 and Note 2). (b) A phylogenetic tree prepared by maximum likelihood analysis showing the relationship of HcPMT with putative homologues identified in Populus trichocarpa (Potri), Arabidopsis thaliana (AT), Vitis vinifera (GSVIV), Medicago truncatula (Medtr) and Solanum lycopersicum (Solyc). Sequences for OsPMT, BdPMT and ZmPMT were included as an outgroup. The values at each node represent statistical confidence based on 1000 bootstrap iterations. The scale bar corresponds to 0.2 substitutions per site. Sequences shown in green are discussed in the text.
Although no crystal structures are currently available for any PMT enzymes, comparisons with other BAHD acyltransferases may be insightful. For example, structure‐functional analysis of the hydroxycinnamoyl‐CoA : shikimate hydroxycinnamoyltransferases (HCT) enzymes from Coffea canephora and Sorghum bicolor revealed that glycine and tryptophan residues guard the entrance to the substrate binding pocket (Lallemand et al., 2012; Walker et al., 2013). Although these residues are conserved in the monocot PMTs, alanine and leucine occur instead in HcPMT, perhaps resulting in a larger active site opening (see Note 1 in Fig. 6a). Similarly, comparison of the HcPMT sequence with the recently reported structure of FMT from Angelica sinensis (Liu et al., 2022) points to a series of residues that comprise the acyl acceptor pocket. Interestingly, none of these are conserved between HcPMT and the other PMT sequences (see Note 2 in Fig. 6a). However, further interpretations, particularly with regard to the binding of the acyl donor, will undoubtedly require a PMT‐specific structural model.
p‐Coumaroylation in kenaf may be the product of convergent evolution
Putative homologues of HcPMT were identified in the genomes of five eudicot species by amino acid homology and then used in the assembly of a phylogenetic tree (Fig. 6b). As has been observed previously with BAHD acyltransferases (Tuominen et al., 2011), these sequences predominantly clustered by taxon, pointing to gene duplication and neofunctionalisation events that occurred after the common ancestors diverged. This pattern of taxon‐specific expansion is typical of genes involved in secondary metabolism (Ober, 2005) and likely reflects the versatility in the active site of BAHD enzymes, which are known to employ a wide range of acyl donors and acceptors.
Included among the putative homologues of HcPMT are several previously‐characterised BAHD enzymes that exhibit diverse activities. For example, AT3G03480 encodes an acetyl‐CoA:(Z)‐3‐hexen‐1‐ol acetyltransferase involved in the biosynthesis of leaf volatiles released upon wounding (D'Auria et al., 2007), and AT5G17540 is involved in the metabolism and homeostasis of brassinosteroids in Arabidopsis (W. Zhu et al., 2013). In grape, GSVIVT01022762001 encodes an anthraniloyl‐CoA : methanol transferase that impacts flavour and aroma (Wang & De Luca, 2005). The alcohol acetyltransferases encoded by Solyc08g005770 and GSVIVT01024259001 in tomato and grape, respectively, accept a range of alcohol substrates (Goulet et al., 2015; Maoz et al., 2018). And finally, Potri.013G074500 and Potri.019G043600 encode BAHD enzymes that act as benzoyl‐CoA : benzyl/salicyl alcohol O‐benzoyltransferases in poplar (Chedgy et al., 2015). Clearly, sequence homology is a poor predictor of activity in BAHD enzymes, as has been noted previously (Luo et al., 2007).
The observation that some of the closest homologues of HcPMT in eudicots have activity on various other acyl donors and acceptors suggests that HcPMT most likely evolved its enzymatic activity independently of the monocot PMTs. Taxon‐specific clustering in the phylogenetic analysis further indicates that HcPMT and the related eudicot BAHD sequences arose following the divergence of their common ancestors (i.e. long after the division between monocots and eudicots). Interestingly, convergent evolution has been documented for BAHD enzymes previously (Luo et al., 2007; Peng et al., 2016).
At present, it remains unclear how broadly pCA occurs within the eudicots beyond kenaf (order: Malvales) and mulberry (order: Rosales; Yamamoto et al., 2020). However, it is also possible that PMT enzymes evolved repeatedly among the eudicots. Only further investigations on lignin acylations in diverse lineages will resolve this question. Of course, it is also possible that p‐coumaroylation, or lignin acylation more generally, could be an ancestral trait that was subsequently lost from many extant genera. However, given that a majority of plant taxa evidently lack lignin acylations altogether, homoplasy appears to offer a more plausible explanation.
Independent evolution of plant cell wall components is hardly atypical. For example, mixed‐linkage glucans evolved separately in horsetails (Equisetum spp.) and in the Poaceae monocots (Fry et al., 2008; Sørensen et al., 2008). Similarly, the biosynthesis of sinapyl alcohol, the monomer that yields S‐lignin units, developed independently in angiosperms, the spikemoss Selaginella moellendorffii and apparently in various basal gymnosperms (Weng et al., 2008; Espiñeira et al., 2011). It has also been postulated that the ability to acylate monolignols with ferulate has arisen at least twice since nonhomologous FMT sequences occur in the eudicot A. sinensis and among the commelinid monocots (Wilkerson et al., 2014; Karlen et al., 2016). Indeed, the same logic may hold for other lignin acylations. For example, p‐hydroxybenzoylation of lignin has been detected in the disparate eudicot families Salicaceae and Araliaceae and in the monocot families Arecaceae and Posidoniaceae (Smith, 1955b; Pearl et al., 1959; Hibino et al., 1994; Rencoret et al., 2020).
The discovery of pCA in a eudicot species elicits new questions on the evolution of lignin. An equally important pursuit for future studies will be to investigate whether lignin acylations have any important biological role(s) in cell walls that could be adaptive for plants. In the meantime, the availability of HcPMT provides a valuable new tool to modify lignin pendent groups and enable further progress in lignin engineering.
Author contributions
YM, RAS and SDK performed the experiments and interpreted the results. JR and SDM conceived the study. YM prepared the manuscript, and all authors contributed to the final version.
Supporting information
Fig. S1 Incorporation of ester‐linked pendent groups into lignin.
Fig. S2 Nucleotide and amino acid sequences of HcPMT.
Fig. S3 SDS‐PAGE of recombinant HcPMT.
Fig. S4 Pairwise enzyme assays with five CoA donors and three monolignol acceptors.
Fig. S5 Expression of HcPMT in transgenic poplar.
Fig. S6 Microscopy of transgenic poplar stems.
Fig. S7 HPLC traces of alkaline hydrolysates from transgenic poplar.
Fig. S8 NMR spectra of transgenic poplar lignin (aliphatics).
Fig. S9 Saccharification potential of transgenic poplar wood.
Table S1 RNA‐seq analysis of candidate BAHDs expressed in kenaf stems.
Table S2 Primers and PCR conditions used in this study.
Table S3 Structural polysaccharides in transgenic poplar.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Acknowledgements
The authors gratefully acknowledge S. A. Peers and C. G. Wilkerson for their help with gene identification. This research was supported by the Great Lakes Bioenergy Research Center, US Department of Energy, Office of Science, Office of Biological and Environmental Research, under award number DE‐SC0018409.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
- Barros J, Serk H, Granlund I, Pesquet E. 2015. The cell biology of lignification in higher plants. Annals of Botany 115: 1053–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Lange A, Fabarius J, Wittmann C. 2015. Top value platform chemicals: bio‐based production of organic acids. Current Opinion in Biotechnology 36: 168–175. [DOI] [PubMed] [Google Scholar]
- Beuerle T, Pichersky E. 2002. Enzymatic synthesis and purification of aromatic coenzyme A esters. Analytical Biochemistry 302: 305–312. [DOI] [PubMed] [Google Scholar]
- Boerjan W, Ralph J, Baucher M. 2003. Lignin biosynthesis. Annual Review of Plant Biology 54: 519–546. [DOI] [PubMed] [Google Scholar]
- Boo YC. 2019. p‐Coumaric acid as an active ingredient in cosmetics: a review focusing on its antimelanogenic effects. Antioxidants 8: 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boz H. 2015. p‐Coumaric acid in cereals: presence, antioxidant and antimicrobial effects. International Journal of Food Science and Technology 50: 2323–2328. [Google Scholar]
- Chedgy RJ, Köllner TG, Constabel CP. 2015. Functional characterization of two acyltransferases from Populus trichocarpa capable of synthesizing benzyl benzoate and salicyl benzoate, potential intermediates in salicinoid phenolic glycoside biosynthesis. Phytochemistry 113: 149–159. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. 2003. A gateway cloning vector set for high‐throughput functional analysis of genes in planta . Plant Physiology 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Auria JC, Pichersky E, Schaub A, Hansel A, Gershenzon J. 2007. Characterization of a BAHD acyltransferase responsible for producing the green leaf volatile (Z)‐3‐hexen‐1‐yl acetate in Arabidopsis thaliana . The Plant Journal 49: 194–207. [DOI] [PubMed] [Google Scholar]
- Espiñeira JM, Novo Uzal E, Gómez Ros LV, Carrión JS, Merino F, Ros Barceló A, Pomar F. 2011. Distribution of lignin monomers and the evolution of lignification among lower plants. Plant Biology 13: 59–68. [DOI] [PubMed] [Google Scholar]
- Fry SC, Nesselrode BHWA, Miller JG, Mewburn BR. 2008. Mixed‐linkage (1→3,1→4)‐β‐d‐glucan is a major hemicellulose of Equisetum (horsetail) cell walls. New Phytologist 179: 104–115. [DOI] [PubMed] [Google Scholar]
- Gani TZH, Orella MJ, Anderson EM, Stone ML, Brushett FR, Beckham GT, Román‐Leshkov Y. 2019. Computational evidence for kinetically controlled radical coupling during lignification. ACS Sustainable Chemistry & Engineering 7: 13270–13277. [Google Scholar]
- Geronikaki AA, Dalimova GN, Abduazimov KA. 1979. Установление сложноэфирной связи в лигнинах кенафа [Establishment of an ester bond in kenaf lignins]. Khimiya Prirodnykh Soedinenii 2: 216–218 [In Russian]. [Google Scholar]
- Goacher RE, Mottiar Y, Mansfield SD. 2021. ToF‐SIMS imaging reveals that p‐hydroxybenzoate groups specifically decorate the lignin of fibres in the xylem of poplar and willow. Holzforschung 75: 452–462. [Google Scholar]
- Goulet C, Kamiyoshihara Y, Lam NB, Richard T, Taylor MG, Tieman DM, Klee HJ. 2015. Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Molecular Plant 8: 153–162. [DOI] [PubMed] [Google Scholar]
- Grabber JH, Quideau S, Ralph J. 1996. p‐Coumaroylated syringyl units in maize lignin: implications for β‐ether cleavage by thioacidolysis. Phytochemistry 43: 1189–1194. [Google Scholar]
- Gutiérrez A, Rodríguez IM, del Río JC. 2004. Chemical characterization of lignin and lipid fractions in kenaf bast fibers used for manufacturing high‐quality papers. Journal of Agricultural and Food Chemistry 52: 4764–4773. [DOI] [PubMed] [Google Scholar]
- Harris PJ, Hartley RD. 1980. Phenolic constituents of the cell walls of monocotyledons. Biochemical Systematics and Ecology 8: 153–160. [Google Scholar]
- Hartley RD, Harris PJ. 1981. Phenolic constituents of the cell walls of dicotyledons. Biochemical Systematics and Ecology 9: 189–203. [Google Scholar]
- Hatfield R, Ralph J, Grabber JH. 2008. A potential role for sinapyl p‐coumarate as a radical transfer mechanism in grass lignin formation. Planta 228: 919–928. [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Marita JM, Frost K. 2008. Characterization of p‐coumarate accumulation, p‐coumaroyl transferase, and cell wall changes during the development of corn stems. Journal of the Science of Food and Agriculture 88: 2529–2537. [Google Scholar]
- Hibino T, Shibata D, Ito T, Tsuchiya D, Higuchi T, Pollet B, Lapierre C. 1994. Chemical properties of lignin from Aralia cordata . Phytochemistry 37: 445–448. [Google Scholar]
- Higuchi T, Ito Y, Shimada M, Kawamura I. 1967. Chemical properties of milled wood lignin of grasses. Phytochemistry 6: 1551–1556. [Google Scholar]
- Huntley SK, Ellis D, Gilbert M, Chapple C, Mansfield SD. 2003. Significant increases in pulping efficiency in C4H‐F5H‐transformed poplars: improved chemical savings and reduced environmental toxins. Journal of Agricultural and Food Chemistry 51: 6178–6183. [DOI] [PubMed] [Google Scholar]
- Kang X, Kirui A, Dickwella Widanage MC, Mentink‐Vigier F, Cosgrove DJ, Wang T. 2019. Lignin‐polysaccharide interactions in plant secondary cell walls revealed by solid‐state NMR. Nature Communications 10: 347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen SD, Zhang C, Peck ML, Smith RA, Padmakshan D, Helmich KE, Free HCA, Lee S, Smith BG, Lu F et al. 2016. Monolignol ferulate conjugates are naturally incorporated into plant lignins. Science Advances 2: e1600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen SD, Smith RA, Kim H, Padmakshan D, Bartuce A, Mobley JK, Free HCA, Smith BG, Harris PJ, Ralph J. 2017. Highly decorated lignins in leaf tissues of the Canary Island date palm Phoenix canariensis . Plant Physiology 175: 1058–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen SD, Free HCA, Padmakshan D, Smith BG, Ralph J, Harris PJ. 2018. Commelinid monocotyledon lignins are acylated by p‐coumarate. Plant Physiology 177: 513–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlen SD, Fasahati P, Mazaheri M, Serate J, Smith RA, Sirobhushanam S, Chen M, Tymokhin VI, Cass CL, Liu S et al. 2020. Assessing the viability of recovery of hydroxycinnamic acids from lignocellulosic biorefinery alkaline pretreatment wastestreams. ChemSusChem 13: 2012–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Li Q, Karlen SD, Smith RA, Shi R, Liu J, Yang C, Tunlaya‐Anukit S, Wang JP, Chang H‐M et al. 2020. Monolignol benzoates incorporate into the lignin of transgenic Populus trichocarpa depleted in C3H and C4H. ACS Sustainable Chemistry & Engineering 8: 3644–3654. [Google Scholar]
- Kim H, Ralph J. 2010. Solution‐state 2D NMR of ball‐milled plant cell wall gels in DMSO‐d 6/pyridine‐d 5 . Organic & Biomolecular Chemistry 8: 576–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Dutta T, Ralph J, Mansfield SD, Simmons BA, Singh S. 2017. Impact of lignin polymer backbone esters on ionic liquid pretreatment of poplar. Biotechnology for Biofuels 10: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolosova N, Miller B, Ralph S, Ellis BE, Douglas C, Ritland K, Bohlmann J. 2004. Isolation of high‐quality RNA from gymnosperm and angiosperm trees. BioTechniques 36: 821–824. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. 2018. Mega X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lallemand LA, Zubieta C, Lee SG, Wang Y, Acajjaoui S, Timmins J, McSweeney S, Jez JM, McCarthy JG, McCarthy AA. 2012. A structural basis for the biosynthesis of the major chlorogenic acids found in coffee. Plant Physiology 160: 249–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapierre C, Jouin D, Monties B. 1989. On the molecular origin of the alkali solubility of Gramineae lignins . Phytochemistry 28: 1401–1403. [Google Scholar]
- Lapierre C, Sibout R, Laurans F, Lesage‐Descauses M‐C, Déjardin A, Pilate G. 2021. p‐Coumaroylation of poplar lignins impacts lignin structure and improves wood saccharification. Plant Physiology 187: 1374–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Dai S, Zhou Y, Liu J, Li D, Zhang J, Zhu Y, Zhao Q, Feng Y, Zhang Y. 2022. Crystal structure of the plant feruloyl–coenzyme A monolignol transferase provides insights into the formation of monolignol ferulate conjugates. Biochemical and Biophysical Research Communications 594: 8–14. [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. 2002. Preliminary evidence for sinapyl acetate as a lignin monomer in kenaf. Chemical Communications 1: 90–91. [DOI] [PubMed] [Google Scholar]
- Lu F, Ralph J. 2008. Novel tetrahydrofuran structures derived from β‐β‐coupling reactions involving sinapyl acetate in Kenaf lignins. Organic & Biomolecular Chemistry 6: 3681–3694. [DOI] [PubMed] [Google Scholar]
- Luo J, Nishiyama Y, Fuell C, Taguchi G, Elliott K, Hill L, Tanaka Y, Kitayama M, Yamazaki M, Bailey P et al. 2007. Convergent evolution in the BAHD family of acyl transferases: identification and characterization of anthocyanin acyl transferases from Arabidopsis thaliana . The Plant Journal 50: 678–695. [DOI] [PubMed] [Google Scholar]
- Ma X, Koepke J, Panjikar S, Fritzsch G, Stöckigt J. 2005. Crystal structure of vinorine synthase, the first representative of the BAHD superfamily. The Journal of Biological Chemistry 280: 13576–13583. [DOI] [PubMed] [Google Scholar]
- Maoz I, Davidovich Rikanati R, Schlesinger D, Bar E, Gonda I, Levin E, Kaplunov T, Sela N, Lichter A, Lewinsohn E. 2018. Concealed ester formation and amino acid metabolism to volatile compounds in table grape (Vitis vinifera L.) berries. Plant Science 274: 223–230. [DOI] [PubMed] [Google Scholar]
- Marita JM, Hatfield RD, Rancour DM, Frost KE. 2014. Identification and suppression of the p‐coumaroyl CoA:hydroxycinnamyl alcohol transferase in Zea mays L. The Plant Journal 78: 850–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marita JM, Rancour D, Hatfield R, Weimer P. 2016. Impact of expressing p‐coumaryl transferase in Medicago sativa L. on cell wall chemistry and digestibility. American Journal of Plant Sciences 7: 2553–2569. [Google Scholar]
- McCann MC, Carpita NC. 2015. Biomass recalcitrance: a multi‐scale, multi‐factor, and conversion‐specific property. Journal of Experimental Botany 66: 4109–4188. [DOI] [PubMed] [Google Scholar]
- Mohamed ET, Werner AZ, Salvachúa D, Singer CA, Szostkiewicz K, Jiménez‐Díaz MF , Eng T, Radi MS, Simmons BA, Mukhopadhyay A et al. 2020. Adaptive laboratory evolution of Pseudomonas putida KT2440 improves p‐coumaric and ferulic acid catabolism and tolerance. Metabolic Engineering Communications 11: e00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison WH III, Akin DE, Archibald DD, Dodd RB, Raymer PL. 1999. Chemical and instrumental characterization of maturing kenaf core and bast. Industrial Crops and Products 10: 21–34. [Google Scholar]
- Mottiar Y, Vanholme R, Boerjan W, Ralph J, Mansfield SD. 2016. Designer lignins: harnessing the plasticity of lignification. Current Opinion in Biotechnology 37: 190–200. [DOI] [PubMed] [Google Scholar]
- Mottiar Y, Gierlinger N, Jeremic D, Master ER, Mansfield SD. 2020. Atypical lignification in eastern leatherwood (Dirca palustris). New Phytologist 226: 704–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Karlen SD, Goacher RE, Ralph J, Mansfield SD. 2022. Metabolic engineering of p‐hydroxybenzoate in poplar lignin. Plant Biotechnology Journal, in press. doi: 10.1111/pbi.13935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mottiar Y, Mansfield SD. 2022. Lignin p‐hydroxybenzoylation is negatively correlated with syringyl units in poplar. Frontiers in Plant Science 13: 938083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novo‐Uzal E, Pomar F, Gómez Ros LV, Espiñeira JM, Ros Barceló A. 2012. Evolutionary history of lignins. Advances in Botanical Research 61: 311–350. [Google Scholar]
- Ober D. 2005. Seeing double: gene duplication and diversification in plant secondary metabolism. Trends in Plant Science 10: 444–449. [DOI] [PubMed] [Google Scholar]
- Otani H, Lee Y‐E, Casabon I, Eltis LD. 2014. Characterization of p‐hydroxycinnamate catabolism in a soil Actinobacterium . Journal of Bacteriology 196: 4293–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearl IA, Beyer DL. 1960. Studies on the chemistry of aspenwood. VI. Products of alkaline hydrolysis of various portions of the aspen tree. TAPPI Journal 43: 611–612. [Google Scholar]
- Pearl IA, Beyer DL, Laskowski D. 1959. Alkaline hydrolysis of representative palms. TAPPI Journal 42: 779–782. [Google Scholar]
- Peers SA. 2012. Identification of a BAHD acyltransferase involved in the acetylation of monolignols in kenaf (Hibiscus cannabinus). MSc thesis, Michigan State University, East Lansing, MI, USA.
- Pei K, Ou J, Huang J, Ou S. 2016. p‐Coumaric acid and its conjugates: dietary sources, pharmacokinetic properties and biological activities. Journal of the Science of Food and Agriculture 96: 2952–2962. [DOI] [PubMed] [Google Scholar]
- Peng M, Gao Y, Chen W, Wang W, Shen S, Shi J, Wang C, Zhang Y, Zou L, Wang S et al. 2016. Evolutionarily distinct BAHD N‐acyltransferases are responsible for natural variation of aromatic amine conjugates in rice. Plant Cell 28: 1533–1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrik DL, Karlen SD, Cass CL, Padmakshan D, Lu F, Liu S, Le Bris P, Antelme S, Santoro N, Wilkerson CG et al. 2014. p‐Coumaroyl‐CoA:monolignol transferase (PMT) acts specifically in the lignin biosynthetic pathway in Brachypodium distachyon . The Plant Journal 77: 713–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H‐JG. 1994. Pathway of p‐coumaric acid incorporation into maize lignin as revealed by NMR. Journal of the American Chemical Society 116: 9448–9456. [Google Scholar]
- Ralph J, Lundquist K, Brunow G, Lu F, Kim H, Schatz PF, Marita JM, Hatfield RD, Ralph SA, Christensen JH et al. 2004. Lignins: natural polymers from oxidative coupling of 4‐hydroxyphenylpropanoids. Phytochemistry Reviews 3: 29–60. [Google Scholar]
- Ralph J. 1996. An unusual lignin from kenaf. Journal of Natural Products 59: 341–342. [Google Scholar]
- Regner M, Bartuce A, Padmakshan D, Ralph J, Karlen SD. 2018. Reductive cleavage method for quantitation of monolignols and low‐abundance monolignol conjugates. ChemSusChem 11: 1600–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rencoret J, Marques G, Serrano O, Kaal J, Martínez AT, del Río JC, Gutiérrez A. 2020. Deciphering the unique structure and acylation pattern of Posidonia oceanica lignin. ACS Sustainable Chemistry & Engineering 8: 12521–12533. [Google Scholar]
- del Río JC, Rencoret J, Gutiérrez A, Kim H, Ralph J. 2022. Unconventional lignin monomers—extension of the lignin paradigm. Advances in Botanical Research 104: 1–39. [Google Scholar]
- Robinson AR, Mansfield SD. 2009. Rapid analysis of poplar lignin monomer composition by a streamlined thioacidolysis procedure and near‐infrared reflectance‐based prediction modeling. The Plant Journal 58: 706–714. [DOI] [PubMed] [Google Scholar]
- Seca AML, Cavaleiro JAS, Domingues FMJ, Silvestre AJD, Evtuguin D, Neto CP. 1998. Structural characterization of the bark and core lignins from kenaf (Hibiscus cannabinus). Journal of Agricultural and Food Chemistry 46: 3100–3108. [Google Scholar]
- Sibout R, Le Bris P, Legée F, Cézard L, Renault H, Lapierre C. 2016. Structural redesigning Arabidopsis lignins into alkali‐soluble lignins through the expression of p‐coumaroyl‐CoA:monolignol transferase PMT. Plant Physiology 170: 1358–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J et al. 2011. Fast, scalable generation of high‐quality protein multiple sequence alignments using Clustal Omega . Molecular Systems Biology 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DCC. 1955a. Ester groups in lignin. Nature 176: 267–268. [Google Scholar]
- Smith DCC. 1955b. p‐Hydroxybenzoate groups in the lignin of aspen (Populus tremula). Journal of the Chemical Society 3: 2347–2351. [Google Scholar]
- Smith RA, Beebe ET, Bingman CA, Vander Meulen K, Eugene A, Steiner AJ, Karlen SD, Ralph J, Fox BG. 2022. Identification and characterization of a set of monocot BAHD monolignol transferases. Plant Physiology 189: 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith RA, Gonzales‐Vigil E, Karlen SD, Park J‐Y, Lu F, Wilkerson CG, Samuels L, Ralph J, Mansfield SD. 2015. Engineering monolignol p‐coumarate conjugates into poplar and Arabidopsis lignins. Plant Physiology 169: 2992–3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen I, Pettolino FA, Wilson SM, Doblin MS, Johansen B, Bacic A, Willats WGT. 2008. Mixed‐linkage (1→3),(1→4)‐β‐d‐glucan is not unique to the Poales and is an abundant component of Equisetum arvense cell walls. The Plant Journal 54: 510–521. [DOI] [PubMed] [Google Scholar]
- St‐Pierre B, De Luca V. 2000. Evolution of acyltransferase genes: origin and diversification of the BAHD superfamily of acyltransferases involved in secondary metabolism. Chapter 9. In: Romeo JT, Ibrahim R, Varin L, De Luca V, eds. Evolution of metabolic pathways. Oxford, UK: Pergamon, Elsevier Science, 285–315. [Google Scholar]
- Stothard P. 2000. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques 28: 1102–1104. [DOI] [PubMed] [Google Scholar]
- Sun R‐C, Sun X‐F, Zhang S‐H. 2001. Quantitative determination of hydroxycinnamic acids in wheat, rice, rye, and barley straws, maize stems, oil palm frond fiber, and fast‐growing poplar wood. Journal of Agricultural and Food Chemistry 49: 5122–5129. [DOI] [PubMed] [Google Scholar]
- Takahama U, Oniki T, Shimokawa H. 1996. A possible mechanism for the oxidation of sinapyl alcohol by peroxidase‐dependent reactions in the apoplast: enhancement of the oxidation by hydroxycinnamic acids and components of the apoplast. Plant & Cell Physiology 37: 499–504. [Google Scholar]
- Timokhin VI, Regner M, Motagamwala AH, Sener C, Karlen SD, Dumesic JA, Ralph J. 2020. Production of p‐coumaric acid from corn GVL‐lignin. ACS Sustainable Chemistry & Engineering 8: 17427–17438. [Google Scholar]
- Tuominen LK, Johnson VE, Tsai C‐J. 2011. Differential phylogenetic expansions in BAHD acyltransferases across five angiosperm taxa and evidence of divergent expression among Populus paralogues. BMC Genomics 12: 236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AM, Hayes RP, Youn B, Vermerris W, Sattler SE, Kang C. 2013. Elucidation of the structure and reaction mechanism of sorghum hydroxycinnamoyltransferase and its structural relationship to other coenzyme A‐dependent transferases and synthases. Plant Physiology 162: 640–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, De Luca V. 2005. The biosynthesis and regulation of biosynthesis of Concord grape fruit esters, including ‘foxy’ methylanthranilate. The Plant Journal 44: 606–619. [DOI] [PubMed] [Google Scholar]
- Weng J‐K, Li X, Stout J, Chapple C. 2008. Independent origins of syringyl lignin in vascular plants. Proceedings of the National Academy of Sciences, USA 105: 7887–7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson C, Ralph J, Withers S, Mansfield SD. 2018. Hibiscus cannabinus feruloyl‐CoA : monolignol transferase. United States Patent US 10,059,955 B2. [WWW document] URL http://www.warf.org/wp‐content/uploads/technologies/ipstatus/P120103US02CON1.pdf [accessed 30 May 2022]
- Wilkerson CG, Mansfield SD, Lu F, Withers S, Park J‐Y, Karlen SD, Gonzales‐Vigil E, Padmakshan D, Unda F, Rencoret J et al. 2014. Monolignol ferulate transferase introduces chemically labile linkages into the lignin backbone. Science 344: 90–93. [DOI] [PubMed] [Google Scholar]
- Withers S, Lu F, Kim H, Zhu Y, Ralph J, Wilkerson CG. 2012. Identification of grass‐specific enzyme that acylates monolignols with p‐coumarate. The Journal of Biological Chemistry 287: 8347–8355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M, Tomiyama H, Koyama A, Okuizumi H, Liu S, Vanholme R, Goeminne G, Hirai Y, Shi H, Nuoendagula et al. 2020. A century‐old mystery unveiled: Sekizaisou is a natural lignin mutant. Plant Physiology 182: 1821–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo CG, Meng X, Pu Y, Ragauskas AJ. 2020. The critical role of lignin in lignocellulosic biomass conversion and recent pretreatment strategies: a comprehensive review. Bioresource Technology 301: 122784. [DOI] [PubMed] [Google Scholar]
- Zhang L, Xu Y, Zhang X, Ma X, Zhang L, Liao Z, Zhang Q, Wan X, Cheng Y, Zhang J et al. 2020. The genome of kenaf (Hibiscus cannabinus L.) provides insights into bast fibre and leaf shape biogenesis. Plant Biotechnology Journal 18: 1796–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Culhaoglu T, Pollet B, Melin C, Denoue D, Barrière Y, Baumberger S, Méchin V. 2011. Impact of lignin structure and cell wall reticulation on maize cell wall degradability. Journal of Agricultural and Food Chemistry 59: 10129–10135. [DOI] [PubMed] [Google Scholar]
- Zhu W, Wang H, Fujioka S, Zhou T, Tian H, Tian W, Wang X. 2013. Homeostasis of brassinosteroids regulated by DRL1, a putative acyltransferase in Arabidopsis. Molecular Plant 6: 546–558. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Regner M, Lu F, Kim H, Mohammadi A, Pearson TJ, Ralph J. 2013. Preparation of monolignol γ‐acetate, γ‐p‐hydroxycinnamate, and γ‐p‐hydroxybenzoate conjugates: selective deacylation of phenolic acetates with hydrazine acetate. RSC Advances 3: 21964–21971. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 Incorporation of ester‐linked pendent groups into lignin.
Fig. S2 Nucleotide and amino acid sequences of HcPMT.
Fig. S3 SDS‐PAGE of recombinant HcPMT.
Fig. S4 Pairwise enzyme assays with five CoA donors and three monolignol acceptors.
Fig. S5 Expression of HcPMT in transgenic poplar.
Fig. S6 Microscopy of transgenic poplar stems.
Fig. S7 HPLC traces of alkaline hydrolysates from transgenic poplar.
Fig. S8 NMR spectra of transgenic poplar lignin (aliphatics).
Fig. S9 Saccharification potential of transgenic poplar wood.
Table S1 RNA‐seq analysis of candidate BAHDs expressed in kenaf stems.
Table S2 Primers and PCR conditions used in this study.
Table S3 Structural polysaccharides in transgenic poplar.
Please note: Wiley is not responsible for the content or functionality of any Supporting Information supplied by the authors. Any queries (other than missing material) should be directed to the New Phytologist Central Office.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request.


