Abstract
Recent years have witnessed a boom of machine learning (ML) applications in chemistry, which reveals the potential of data‐driven prediction of synthesis performance. Digitalization and ML modelling are the key strategies to fully exploit the unique potential within the synergistic interplay between experimental data and the robust prediction of performance and selectivity. A series of exciting studies have demonstrated the importance of chemical knowledge implementation in ML, which improves the model's capability for making predictions that are challenging and often go beyond the abilities of human beings. This Minireview summarizes the cutting‐edge embedding techniques and model designs in synthetic performance prediction, elaborating how chemical knowledge can be incorporated into machine learning until June 2022. By merging organic synthesis tactics and chemical informatics, we hope this Review can provide a guide map and intrigue chemists to revisit the digitalization and computerization of organic chemistry principles.
Keywords: machine learning, molecular embedding, organic synthesis, performance prediction, synthetic dataset
Predicting organic transformations is one of the most critical challenges in molecular syntheses. The synergy between machine learning and chemical knowledge provides a distinctive and powerful strategy for syntheses predictions. This Minireview summarizes the state‐of‐the‐art embedding approaches and model designs for developing synthesis‐sensitive machine learning models to allow for robust yield and selectivity predictions in molecular syntheses.
Introduction
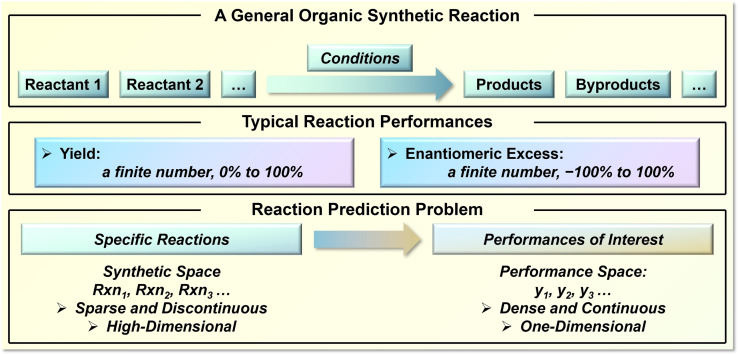
Reaction performance is one of the decisive factors for the success of a synthetic reaction. The ideal yield and selectivity of 100 % is the goal for any synthetic transformation when chemists are making reaction designs, [1] as highlighted in the well‐known concepts of atom‐ and resource‐economy.[ 2 , 3 ] There are nearly infinite combinations of reactive compounds, however, chemists possess a limited experimental efficiency to evaluate reaction performances. Among exciting prediction tasks in organic synthesis (retrosynthesis route,[ 4 , 5 , 6 ] reaction product, [7] reaction condition,[ 8 , 9 ] etc.), the nature of the reaction performance prediction presents its distinctive challenges. The synthetic space, on the one hand, is massive. It not only involves the structural possibilities of involved compounds, but also the choice of reaction conditions as well as the combinations of reactants, which further complicate the issue. [10] On the contrary, the performance space of a defined synthetic reaction is quite simple and usually restricted within a finite range due to detection limits. This requires the desired prediction to connect a sparse and discontinuous high‐dimensional synthetic space to a dense and continuous one‐dimensional performance space (Scheme 1). Therefore, a seemingly trivial displacement in the synthetic space could result in a non‐intuitive and noticeable influence on the reaction performance, such as a subtle structural change of the catalyst[ 11 , 12 ] or switching to a different solvent.[ 13 , 14 ] Consequently, time‐ and resource‐consuming trial‐and‐error attempts continue to be required for an ingenious reaction design.
Scheme 1.
A general definition of the performance prediction problem in organic synthesis. y 1, y 2, and y 3 are reaction performances of interest such as yields, stereoselectivities, etc.
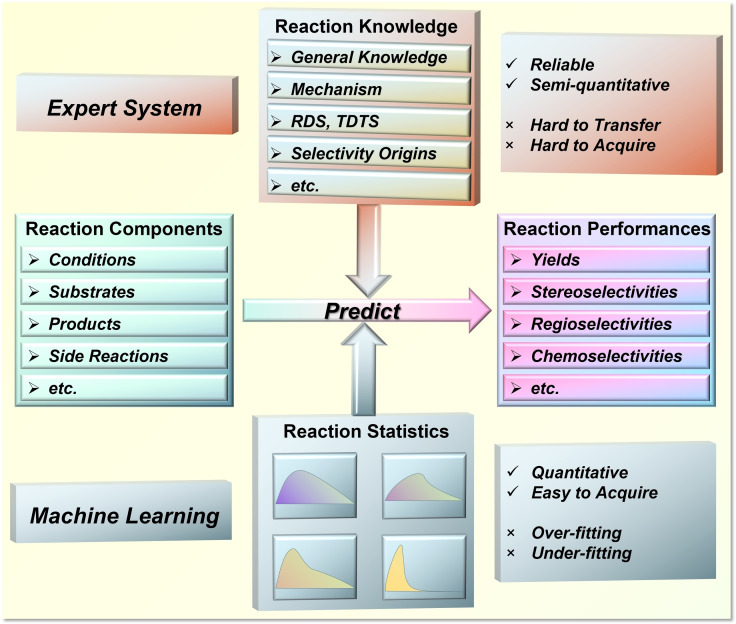
Chemists make predictions of reaction performance based on their domain knowledge. These may include the general information of the reactants (reactivity, stability, solubility, etc.), the molecular level reaction mechanism, the rate‐ and selectivity‐determining elementary steps, and even the quantum chemical origins of target performances (Figure 1). Such knowledge can significantly improve the reliability of their predictions, however, making these domain knowledge‐based predictions is non‐trivial and still often face challenges. Even experienced synthesis and catalysis experts may struggle to provide robust predictions of reaction outcomes, such as yields and selectivities, despite their solid chemical knowledge.[ 10 , 15 ] More importantly, the domain knowledge of organic synthesis cannot be acquired and mastered simply by theoretical deduction like mathematics. The training process of synthetic chemists requires systematic study of chemical theory, but the literature reading and experimental experience are equally important. The resulting knowledge of organic synthesis is difficult to describe in quantitative and programmable expressions, so it is highly challenging to create an “expert system” of organic synthesis.[ 16 , 17 ] An experienced synthetic chemist is of great value, and making an expert prediction of molecular synthesis is precious. The exciting success of SynthiaTM has revealed the remarkable potential of machines in learning sufficient chemical knowledge, becoming experts in organic synthesis once they have been adequately programmed and trained. [15]
Figure 1.
Characteristics of reaction performance prediction by human chemist and ML.
The causal link between synthetic space and reaction performance, as well as the regression nature of the performance prediction task make machine learning (ML) a suitable solution for this challenge.[ 18 , 19 , 20 ] ML is extremely attractive since it is particularly good at determining and locating the hidden connections between intertwined complex factors and targets. [21] Nevertheless, this advantage comes at a cost (Figure 1). Most modern ML techniques require a large dataset for the model training. Insufficient data may lead to an over‐fitting problem, where the model is trained too specifically on the training data and has nearly no predictive ability towards unseen cases, like predicting a new reaction. On the other hand, an under‐fitting problem could emerge due to insufficient data, where the model is incompetent to provide a reliable answer. This results in the paradox between accuracy and data requirement when establishing a regression ML model for reaction performance prediction.
Simply improving the amount of chemical data and applying established regression algorithms cannot provide an all‐purpose artificial intelligence model to make predictions for organic reactions. [22] There are around 10171 possibilities for the Game of Go.[ 23 , 24 ] Synthetic possibilities are beyond this number. For relatively small molecules, the number of possible structures is estimated to be around 1060, [25] which makes any three‐component synthetic reaction to outweigh the complexity of Go. Reaxys® database now contains about 58 million individual transformations. [26] The size of accumulated synthetic data will remain a tiny fraction of synthetic space for an arguably long time to come.
One promising path for artificial intelligence in chemistry is to introduce chemical knowledge when constructing an ML model. [27] Bridging chemical knowledge and ML has the power to harness the benefits of both human intelligence and artificial intelligence, dramatically improving the robustness and predictive ability of ML models given the available data. In addition, the predictor will have improved interpretability,[ 18 , 28 , 29 ] which may offer new chemical inspiration and even knowledge from synthetic statistics. [30]
How can chemical knowledge improve ML? First, chemical knowledge can support the inclusion of fundamental model requisites, such as rotational and translational invariance, [31] which will steer the machine to learn chemically correct models. In addition, chemical knowledge can be beneficial for ML through the implementation of reaction‐specific understandings. Introducing chemically meaningful representations or the inclusion of chemical understandings in model designs will improve the model's differential ability towards the organic transformation as well as its predictive ability towards unseen candidates.[ 27 , 32 , 33 , 34 , 35 , 36 , 37 ] A representative proof of this concept is Sigman's multi‐variant linear regression studies,[ 19 , 38 , 39 ] where powerful models can be built using sophisticated chemical descriptors with primitive regression techniques.
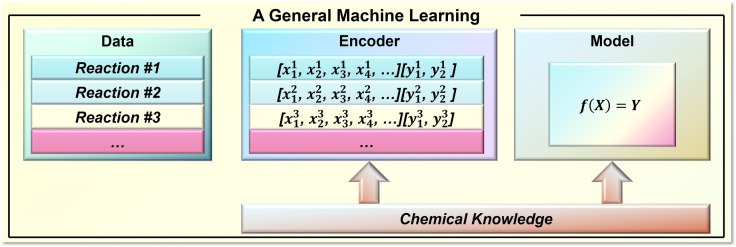
ML prediction generally involves three key components, namely data, encoder and model (Figure 2). It should be noted that encoder is also referred to as molecular representation, which is the more common term. Data provides information on the target transformation, which is the basis for ML training. Particularly for synthetic performance, there is limited availability of open‐source structural databases.[ 40 , 41 , 42 , 43 ] Commercial synthetic databases, such as Reaxys® [26] and SciFinder®, [44] provide access to browse the recorded information upon purchasable licenses, while scalable utilization of the data is unlikely. The available open‐source structural database of organic synthesis mostly stems from research articles on high‐throughput experimentation (HTE) and related ML applications. Prime examples of this strategy include Doyle's database of Ullman–Goldberg/Buchwald–Hartwig cross‐couplings (4140 reaction yields) [18] and Denmark's database of asymmetric imine addition (1075 enantioselectivities), [20] which have now been widely applied as benchmark databases for ML of synthesis/catalysis performance. We recently built a database of asymmetric hydrogenation of olefins (12619 enantioselectivities) based on experimentation literature between the years 2000 and 2020. [45] In addition to the literature data, the reaction data schemes from US patents were extracted as the USPTO reaction database via text‐mining techniques by NextMove.[ 46 , 47 ] However, it is noteworthy that, based on a recent study, there may be some inherent potential problems in the data source. [48] To remove the data barrier of chemistry, Coley and Kearnes proposed an initiative called Open Reaction Database. [49] This initiative provides an open‐access infrastructure for sharing synthetic statistics, which will significantly promote the structural data utilization of organic reactions.
Figure 2.
Key components of ML in reaction performance prediction.
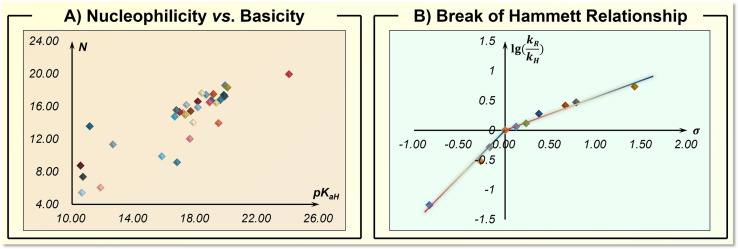
Current implementations of synthesis knowledge in ML are generally realized by the innovative design of encoders or models. The encoder transforms the chemical data into machine‐readable codes. This represents the digital basis for machines to differentiate organic reactions. Through the implementation of chemical knowledge, the desired molecular representation has the ability to distinguish and cluster the relevant synthesis transformations, mostly within a certain specified range. A textbook example constitutes the nucleophilicity vs. basicity, these two reactivity dimensions are closely related, yet not linearly correlated (Figure 3A). [50] Therefore, choosing the wrong parameter will fail to make a reaction prediction, which is a typical out‐of‐distribution (OOD) issue in artificial intelligence. The model treats the encoded data and transforms them into target values. Here, ML discovers the hidden connections and even causalities in synthetic chemistry using mathematic equations and computer algorithms. Chemical knowledge involvement would improve the model's ability to capture and predict the high‐dimensional synthetic relationship. A related case in physical organic chemistry is the well‐known break of the Hammett relationship (Figure 3B). [51] This relationship cannot be described by a linear equation due to the change of the rate‐determining step. If such knowledge is available, projecting these observed statistics in higher dimensional space using algorithms like support vector machines (SVM) would easily provide a predictive regression.
Figure 3.
Typical scenarios where chemical knowledge is critical for the statistical pattern of synthetic performance. A) Nucleophilicity vs. basicity of selected pyrrolidines and imidazolidinones. [50] pK aH are the corresponding Brønsted basicities in acetonitrile and N are the Mayr's nucleophilicity parameters. B) Nonlinear Hammett relationship observed in aminolysis of Y‐substituted‐phenyl 2‐methoxybenzoates in acetonitrile. [51]
In respect to the performance prediction of organic synthesis via ML, a number of exciting studies have emerged with innovative model designs and powerful chemical predictions.[ 18 , 19 , 20 ] The community is witnessing a paradigm‐shifting, and it is exciting to see the dynamic energy from the merging between organic chemistry and artificial intelligence. Representative ML applications[ 27 , 32 , 33 , 34 , 35 , 36 , 37 , 52 , 53 , 54 , 55 , 56 , 57 ] and molecular representations[ 58 , 59 , 60 ] have been summarized in a few excellent reviews, which have elaborated the synthesis targets and the ML performances. In contrast, this Minireview will focus on the strategies of implementing chemical knowledge in ML modelling until June 2022. By critically discussing the key concepts of the highlighted research advances, state‐of‐the‐art chemical knowledge‐based embedding techniques and ML model approaches are analyzed, with diversified applications in ML predictions of reaction yield, chemo‐, regio‐ and stereoselectivity.
Classic Techniques for Molecular Embedding and their Applications in Reaction Performance Prediction
Before diving into the chemical knowledge‐based embedding approaches, we first briefly summarize the classic embedding approaches for molecular structure, which do not require explicit chemical knowledge. These embedding methods have no chemical information about the target reaction, thusly they can only differentiate organic molecules. After the molecular embedding of each reaction component, these digital representations are usually concatenated in a unified order as descriptors for synthetic reactions.
One‐Hot Encoding
The first category of embedding methods for organic molecules is one‐hot encoding (OHE). As the most basic embedding technique in computer science, OHE is a vectorized representation with binary strings, where all the digits of a certain vector are 0 except one digit of 1. The designation of “1” represents the identity of the molecule, usually in the range of a limited selection of compounds.
OHE gives each compound a unique binary vector. The vector itself carries no chemical meaning and has to be transformed into higher dimensions and elusive patterns during model training. Therefore, all the required chemical relationship to precisely predict the reaction performance is learned solely from the training data. These embedding techniques could give predictions of reaction performance when sufficient data are provided.
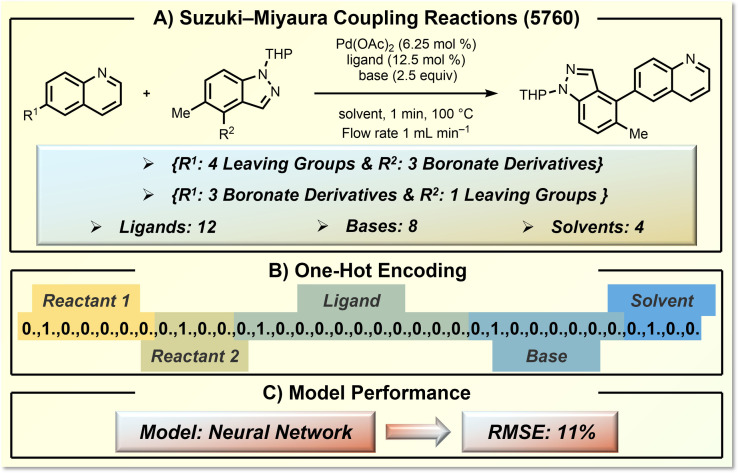
In 2018, Cronin and co‐workers designed an ML‐guided feedback loop to explore synthetic space with a designed liquid‐handling robot. [61] They selected a Suzuki–Miyaura cross‐coupling to examine the power of their ML approach in terms of chemical yield prediction. Figure 4 displays the defined synthesis space of 7 substituted quinolines, 4 substituted 1H‐indazole, 12 ligands, 8 bases and 4 solvents. The entire synthesis space consists of 5760 transformations (Figure 4A). OHE is used to describe this synthetic space. There is a total of 35 different kinds of organic molecules (reactants, ligands, bases, and solvents) in this defined space, thus OHE describes each specific reaction by a unique 35‐digit binary vector (Figure 4B). The concatenation order does not matter in this case but should be held consistent throughout encoding and model training. Based on training with 60 % of the entire synthetic space (3456 reactions of 5760 possibilities), the prediction by the trained neural network model gave a root‐mean‐square error (RMSE) of 11 % for a test set of 1728 transformations. This success of OHE highly relied on the fairly large training set and the fact that the defined synthetic space is already constrained by chemical knowledge, which is a Nobel prize‐winning catalytic reaction with well‐known high functional group tolerance. The trained model is able to capture the statistical patterns of the training data, specifically the presence of which compound or groups of compounds can increase or decrease the reaction yield, enabling the predicting of the reaction yield.
Figure 4.
Application of OHE in yield prediction of Suzuki–Miyaura coupling reactions. A) The target Suzuki–Miyaura coupling reaction and the defined reaction space. B) OHE encoding details. C) The model performance.
Strings
Strings use a naming system to differentiate molecules. There are a few widely applied naming systems, including SMILES, [62] the International Chemical Identifier (InChI), [63] and IUPAC naming. [64] SMILES is perhaps one of the most widely used string representations in modern ML of organic molecules. To overcome certain limitations of the original version of SMILES, such as its non‐uniqueness on a single molecule, innovations of the SMILES syntax rules have led to derivatized versions, such as SMARTS, [65] DEEPSMILES [66] and SELFIES. [67] The string representation does not possess a rich physical chemistry or transformation‐related information, thusly it usually requires a large quantity of data in order to train reliable ML models.
Because string representation is a chemical language that describes the molecular structure, its use for existing compounds or transformations contains the chemical knowledge of why certain compounds or transformations are reasonable and exist in the physical world. This makes it a very useful resource for representation learning. This text‐based representation learning is particularly suitable for the transformer model, [68] which is a trending translation model in computer science with self‐supervised techniques. One representative example is Schwallder's recent study of reaction space mapping; [69] powered by the reaction databases of Pistachio and USPTO, the authors developed a BERT model for reaction classification, which achieved excellent accuracy compared with the rule‐based classification. The learned representation can be used as reaction fingerprints, providing the opportunity for mapping the reaction space without specific synthetic knowledge and can be used for encoding approaches in machine learning predictions of reaction performance, like activation energies [70] and reaction yields. [71] The string representation can also be used for transfer learning involving large datasets, where one highlighted case from Reymond [72] is discussed in section 4 (Figure 12).
Figure 12.
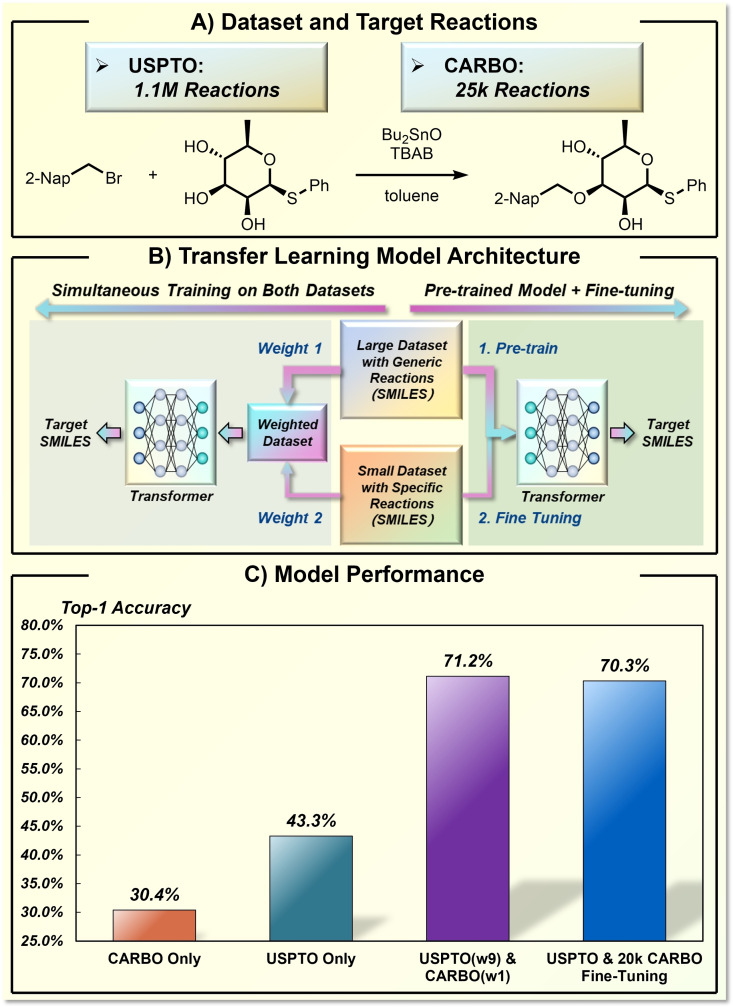
Application of transfer learning strategy in stereoselectivity prediction of carbohydrate transformation. A) The dataset and target reactions. B) The model design for transfer learning. C) The model performance.
Molecular Fingerprints
In addition to OHE and string representation, molecular fingerprint is the third category of molecular embedding methods that applies to any organic molecule and does not require prior chemical knowledge. [73] The idea of molecular fingerprints is to identify the presence of topology, sub‐structure, and/or scaffolds. They were initially designed to determine the similarity of molecules but later have wide applications in ML of organic chemistry. Because the determination rules of MF have large freedom of customized possibilities, there have emerged dozens of diversified MFs, including Morgan fingerprint, [74] Avalon fingerprint [75] and MACCS‐key fingerprint. [76] MFs are usually, albeit not necessarily, binary vectors with hundreds to thousands of digits. Their generation is efficient and structurally sensitive. Though it can be argued that MF has more chemically relevant information than OHE and strings, their utilization does not require any transformation‐related knowledge.
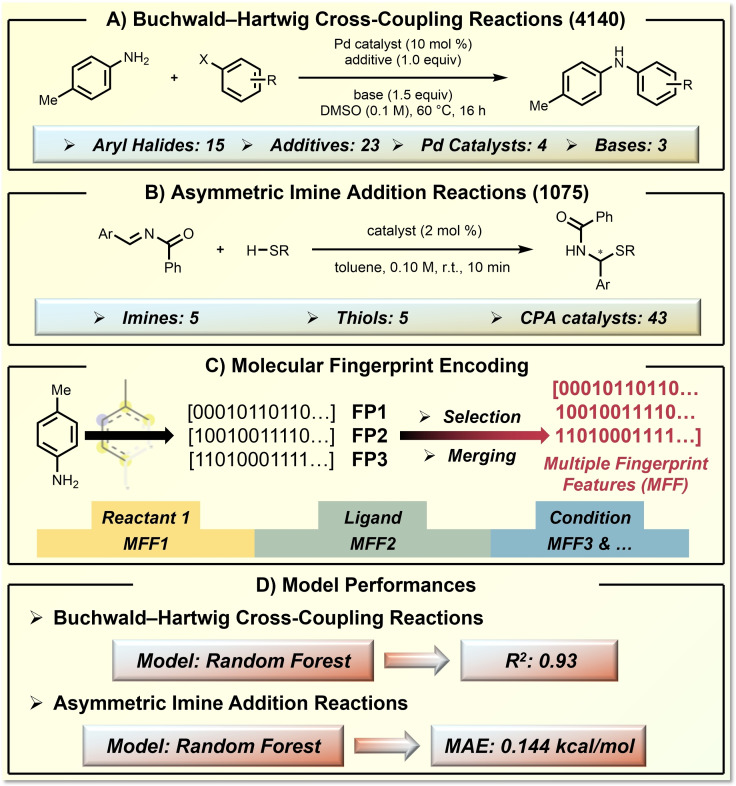
Glorius and co‐workers recently showed that the utilization of multiple fingerprint features can provide useful molecular representations for reaction performance prediction. [77] The effectiveness of their ML approach was demonstrated in both Doyle's Buchwald–Hartwig coupling dataset (Figure 5A) [18] and Denmark's asymmetric imine addition dataset (Figure 5B). [20] The basis of this approach is that different prediction tasks would require different determining molecular properties. Therefore, they combined 24 types of molecular fingerprints (i. e., Morgan fingerprint, [74] Avalon fingerprint [75] and MACCS‐keys fingerprint [76] ) to a universal representation of each reaction component, regardless of the modelling transformation or prediction target (Figure 5C). Through ML, the algorithm is able to capture the determining molecular fingerprints from the synthetic statistics, which builds an adaptive bridge between molecular fingerprints and reaction performance space. The Random Forest (RF) model gave satisfying prediction performances, with a R2 of 0.93 for Doyle's dataset and a mean absolute error (MAE) of 0.144 kcal/mol for Denmark's dataset (Figure 5D).
Figure 5.
Application of molecular fingerprint representation in reaction performance prediction of organic synthesis. A) The target Buchwald–Hartwig coupling reaction and the defined reaction space. B) The target asymmetric imine addition reaction and the defined reaction space. C) The MFF representation technique. D) The model performance.
The above classic molecular embedding techniques are readily available and efficient for ML applications. Their generation does not require any specific chemical understanding of the target transformation, which is user‐friendly for researchers without chemical backgrounds. Nevertheless, it should be noted that these classic embedding techniques can only differentiate the molecules in a relatively primitive fashion. Their predictions are primarily based on the statistics of the training data. If the interested prediction problem is in a well‐defined and limited synthetic space where the statistical pattern can be described by a fairly simple function, the statistics themselves can support a predictive model. When performance prediction tasks involve out‐of‐distribution (i. e., new substrate) and out‐of‐range (i. e., yield optimization) issues, or when the available data size cannot meet the complexity of the target structure‐performance relationship, application of the classic molecular representation approaches may fall into limited success and should be treated with caution.
Chemical Knowledge‐Based Molecular Embedding Approaches and Their Applications in Reaction Performance Prediction
The most straightforward way to introduce chemical knowledge in ML of reaction performance is to design transformation‐related chemical descriptors. One convincing example is Nørskov's discovery of volcano plot which projects the catalytic behavior to a limited number of chemically meaningful dimensions based on scaling relationship, [78] and the concept of scaling relationship and volcano plot was later extended to the realm of homogeneous catalysis by the elegant studies from Corminboeuf[ 35 , 79 , 80 , 81 ] and Nørskov. [82]
Based on the chemical understanding of the target transformation, especially the mechanistic origins of reactivity or selectivity, chemists usually have a solid experience and instinct for determining factors. These factors have strong physical organic relevance, and some are known physical organic parameters (i. e., Sterimol parameters, [83] and Hammett constants [84] ). Through this chemical vectorization, the synthetic space is projected to a descriptor space, which is usually composed of tens of essential chemical factors. Subsequently, the ML model is trained to learn the relationship between the descriptor space and the target reaction performance.
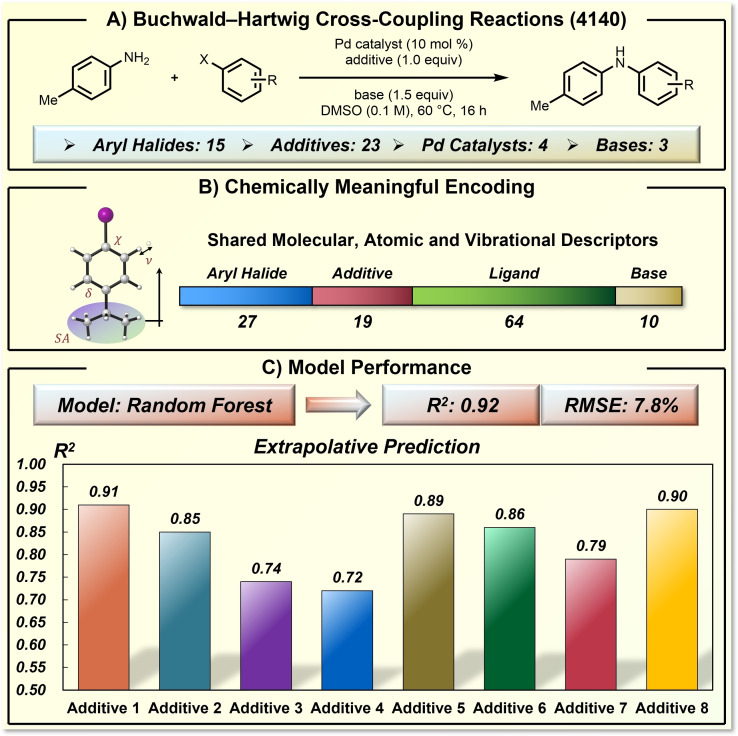
A representative example of chemical knowledge‐based ML prediction of reaction yield is Doyle's study on Buchwald–Hartwig cross‐coupling (Figure 6). [18] They defined a synthetic space with 15 aryl halides, 23 additives, 4 palladium catalysts, and 3 bases, which includes 4140 distinctive transformations. The yields of the entire synthetic space were examined by Merck's mosquito robot system, which provides high parallelism of the experiments and ensures the quality of the generated dataset. Based on the understanding of the coupling mechanism and reactivity‐controlling factors, they used highly compact and chemically meaningful descriptors to encode each reaction component. These descriptors include NMR shifts, electrostatic charges, and others. It is worth noticing that the vibration‐related descriptors of shared substructures were found useful for chemical yield prediction.
Figure 6.
Yield prediction of Buchwald–Hartwig coupling reactions using chemically meaningful descriptors. A) The target Buchwald–Hartwig coupling reaction and the defined reaction space. B) The chemically meaningful descriptors and dimension. C) The model performance.
The influence of chemical vibration on organic reaction performance has also been identified by Sigman et al. [39] The knowledge‐based chemical descriptors and RF algorithm provided a satisfying yield prediction model with a R2 of 0.92 and a RMSE of 7.8 % on a 70/30 split of training and test data (Figure 6C). The authors also examined the challenging extrapolative prediction task. The dataset was split based on additives. The training set was composed of data from 14 additives, while the test set includes the rest data of 8 additives, which were not present in the training set. Even though the additives of the test set were not seen by the model during training, all the R2 values of the predictions are still higher than 0.70. This work by Doyle showed the predictive power of knowledge‐based chemical descriptors and ML models in organic synthesis, which was able to make accurate yield prediction that is challenging for a human chemist.
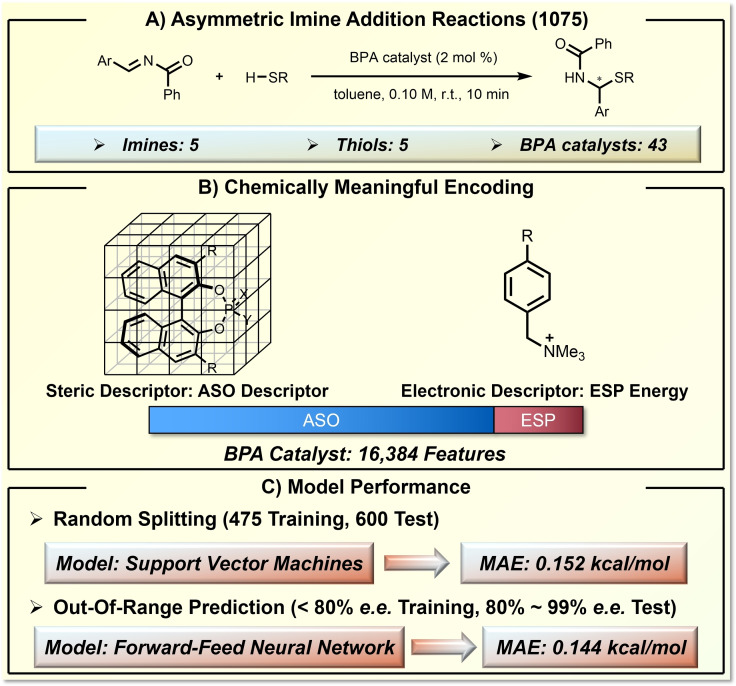
In addition to reaction yield, enantioselectivity can also be predicted in a robust fashion by knowledge‐based descriptor design and ML. Using chiral phosphoric acid‐catalyzed thiol addition to N‐acyl imines as the model reaction, Denmark showed the power of ML prediction in asymmetric catalysis (Figure 7). [20] The reaction space includes 1075 asymmetric transformations, considering the variations of 5 imines, 5 thiols, and 43 BINOL phosphoric acids (BPA) catalysts (Figure 7A). Due to the critical role of the steric environment on chiral induction, the authors developed a novel steric descriptor called average steric occupancy (ASO). By aligning the BPAs with the common scaffold in a grid box (Figure 7B), ASO measures the steric occupancy in each grid (0 for vacant, 1 for occupied). After averaging the steric occupancies of conformers, the generated ASO is a 16384‐dimension vector with float numbers between 0 to 1. In addition to the judicious design of ASO, the authors also applied the electrostatic potential (ESP) as a stereoelectronic descriptor. The vectorization of BPA catalysts was further truncated by removing the redundant features that are identical for the studied BPAs. Using the SVM algorithm, excellent prediction accuracy was achieved; the SVM model based on 475 random training data gave predictions with only 0.152 kcal/mol MAE for the remaining 600 test reactions (Figure 7C). In addition to the prediction in random data splitting, the authors trained a deep feed‐forward neural network (DFNN) model that is able to make out‐of‐range (OOR) predictions. This model, trained with data below 80 % enantiomeric excess (e. e.), can provide a remarkable prediction accuracy of 0.33 kcal/mol MAE for test sets above 80 % e. e. (Figure 7C). This extrapolative ability for OOR prediction is highly desirable in organic synthesis, considering the context of synthetic methodology optimization.
Figure 7.
Enantioselectivity prediction of phosphoric acid‐catalyzed asymmetric imine addition reactions using designed descriptors for chiral environment. A) The target asymmetric imine addition reaction and the defined reaction space. B) The chemically meaningful molecular representation by ASO and ESP. C) The model performance.
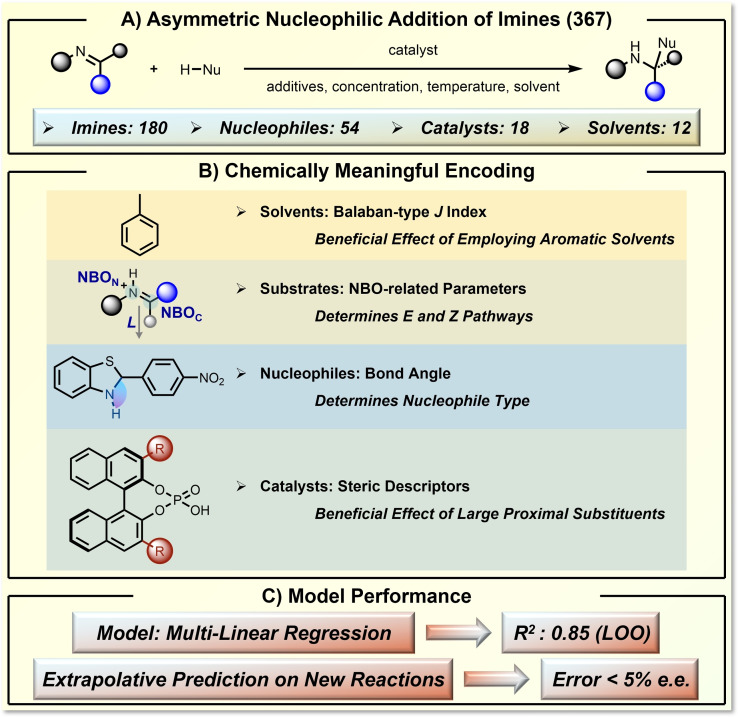
ML can also make reliable predictions without a complete dataset of synthetic space. In 2019, Sigman and co‐workers realized ML prediction of enantioselectivity, focusing on asymmetric phosphoric acid‐catalyzed nucleophilic addition of imines (Figure 8). [19] They curated a dataset of 367 asymmetric transformations from literature reports. This dataset covers a fairly large synthetic space, containing 180 imines, 54 nucleophiles, 18 catalysts, 12 solvents, and additional changes in reaction conditions (Figure 8A). By careful interpretation of the reaction mechanism and evaluation of possible descriptors, the authors found that six important features were enough to encode each reaction component (Figure 8B). These descriptors include the Balaban‐type index, natural bond orbital (NBO) related parameters, nucleophile's bond angle, and steric descriptors of the catalyst. These carefully selected descriptors are highly related to the determination of enantioselectivity. Using the multilinear regression (MLR) algorithm, the ML model is able to achieve a convincing regression performance with a R2 of 0.85 in the leave‐one‐out (LOO) analysis (Figure 8C). The trained coefficients reflected the influence of each chemical descriptor on the overall enantioselectivity prediction. Building on the success of the MLR model, the authors further validated the extrapolative prediction ability where the unseen reactions were correctly predicted within an error of only 5 % e. e. (Figure 8C).
Figure 8.
Multi‐variant‐linear prediction of enantioselectivity of asymmetric nucleophilic addition of imines using designed chemical descriptors. A) The target asymmetric nucleophilic addition of imines reaction and the defined reaction space. B) The chemically meaningful encoding by several selected parameters. C) The model performance.
This highlights the transferability across chemically relevant organic transformations, allowing the desired prediction using existing data of a known reaction to be used for the performance evaluation of a new reaction.
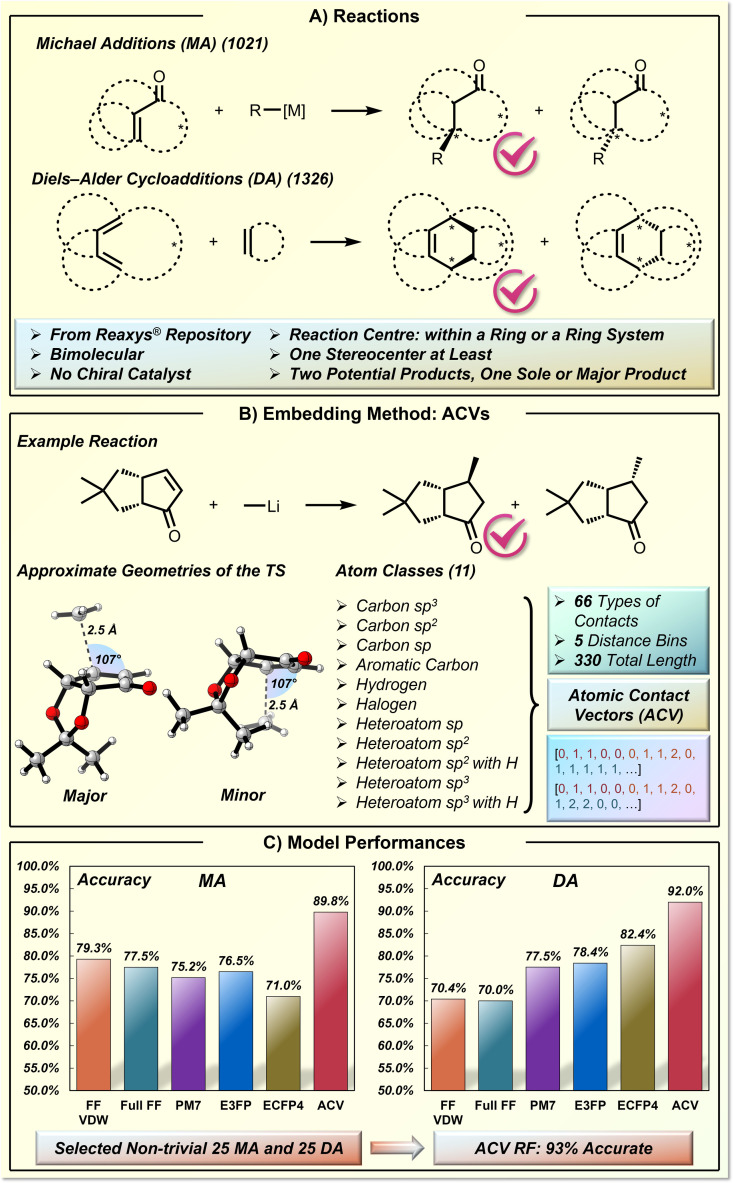
In the above studies,[ 18 , 19 , 20 ] the knowledge‐based vectorization of molecules was achieved by selecting chemically meaningful descriptors based on the local minimum structures (often obtained by DFT‐level optimization). The domain knowledge can also play a role in the determination of the source molecular structure that is responsible for descriptor generation. Grzybowski and co‐workers recently reported an intriguing ML study for stereoselectivity prediction in Michael addition reactions (MA) and Diels‐Alder cycloadditions (DA) (Figure 9). [85] The concave versus convex facial selectivity is challenging to predict due to the involvement of the elusive steric factors and non‐covalent interactions (Figure 9A). To better describe the interacting reaction components in proximity, the authors designed a new type of embedding vector called atomic contact vector (ACV), based on the atom contacts in pre‐assembled transition‐state‐like geometry (Figure 9B). This approach is demonstrated in a selected dataset of 1021 MA and 1326 DA reactions from the Reaxys® repository, which are all bimolecular, ring‐based, and stereospecific without any chiral catalysts. It should be noted that their prediction is not a regression problem for a quantified selectivity target. Instead, the ML model is designed to predict the major product with the correct facial selectivity, which is a binary classification problem.
Figure 9.
Molecular representation based on transition‐state‐like geometry and its application in stereoselectivity prediction of Michael addition and Diels‐Alder cycloaddition. A) The Michael addition and Diels‐Alder cycloaddition reactions. B) The ACV embedding technique. C) The model performance.
The creation of transition‐state‐like geometries requires sophisticated mechanistic knowledge to implement the right geometric constraints. For example, in MA reactions (Figure 9B), the distance of the forming C−C bond is set to be 2.5 Å, and the C(Nu)−C=C angle is set to be the value of Bürgi‐Dunitz angle 107° in order to generate the chemically correct MA transition‐state‐like geometries. Certain flexible fragments in the generated geometries were not considered during the descriptor generation. Subsequently, the authors binned the interatomic distances between each of their defined 11 atom classes into five distance categories, which provides a 330‐dimensional ACV. The authors compared the performances of the ACV representation with other widely applied molecular representations. The latter comprise of reaction energies at various computational levels of theory, extended connectivity fingerprints (ECFP), and extended three‐dimensional fingerprints (E3FP). All these representations showed significantly worse classification performances than ACV (Figure 9C). The final ACV‐based RF model has a classification accuracy of 89.8 % for MA and 92.0 % for DA. Finally, they selected a list of non‐trivial MA and DA reactions, and the ACV‐based model gave a satisfying accuracy of 92 % to predict the correct product, while the human experts accuracy was only 52 %. [85]
Chemical Knowledge‐based ML Model Designs and Their Applications in Reaction Performance Prediction
In addition to the chemical knowledge‐based descriptor design, the design of the ML model itself is equally important for making the correct chemistry prediction. The model designs have generated strong momentum in ML prediction of molecular property.[ 86 , 87 , 88 ] A series of ingenious ML have pushed the performance of molecular property prediction to a remarkable level, which is even comparable to modern quantum mechanical (QM) chemical calculations.[ 89 , 90 , 91 ] For reaction performance prediction, the introduction of chemical knowledge can also improve ML's predictive power and efficiency. In this section, the representative model designs that allow the introduction of chemical knowledge are discussed.
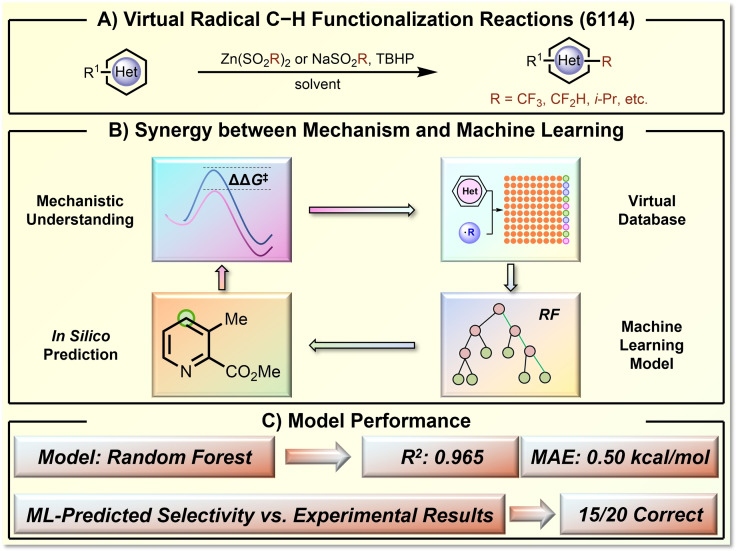
Based on the remarkable advances in computational chemistry, modern quantum chemical calculation has reached an excellent capability in calculating reactivity and selectivity of organic transformation, even with chemical accuracy.[ 92 , 93 , 94 , 95 ] The quantum chemical computational method itself and the generated statistics contain rich chemical knowledge, which can benefit ML purposes if introduced in the right way. In this regard, the Hong group showed that computational structure‐performance statistics could serve as a useful data source to support ML applications where limited experimental results are available.[ 28 , 29 ] An example is represented by the site‐selectivity prediction of radical Minisci‐functionalization of heteroarenes (Figure 10). [28] This transformation is particularly useful when a certain combination of radical and heteroarene can give high regioselectivity. However, only a few experimental regioselectivity results were available.[ 96 , 97 ] Based on the mechanistic understanding that the regioselectivity‐determining step is the radical addition step, the Hong group applied DFT calculations to generate the virtual dataset of 6114 C−H functionalization reactions and 9370 regioisomeric competitions. Relying on this DFT‐computed dataset, the authors selected a set of physical organic descriptors based on chemical knowledge of the regioselectivity‐determining elementary transformation. The selected descriptors include atom‐specific descriptors (bond order, charge and buried volume, among others) and global descriptors (molecular orbital energies and Nucleus‐Independent Chemical Shifts (NICS) values, etc.). The trained RF model provided a satisfying prediction performance with a R2 of 0.965 and a MAE of 0.50 kcal/mol. The DFT statistics‐trained RF model can be directly applied in the predictions of experimental results. For 15 of 20 experimental radical C−H functionalizations, the regioselectivity was correctly predicted despite the fact that none of the experimental data was used in the model training. These findings demonstrated that the computational reaction performance data is a useful resource for ML of synthesis transformation, especially for transformations that are experimentally challenging to access the needed data support for ML purposes.
Figure 10.
ML prediction of the regioselectivity of radical C−H functionalization of heteroarenes based on mechanism‐based computational statistics. A) The virtual radical C−H functionalization reactions. B) The reaction mechanism‐based ML loop. C) The model performance.
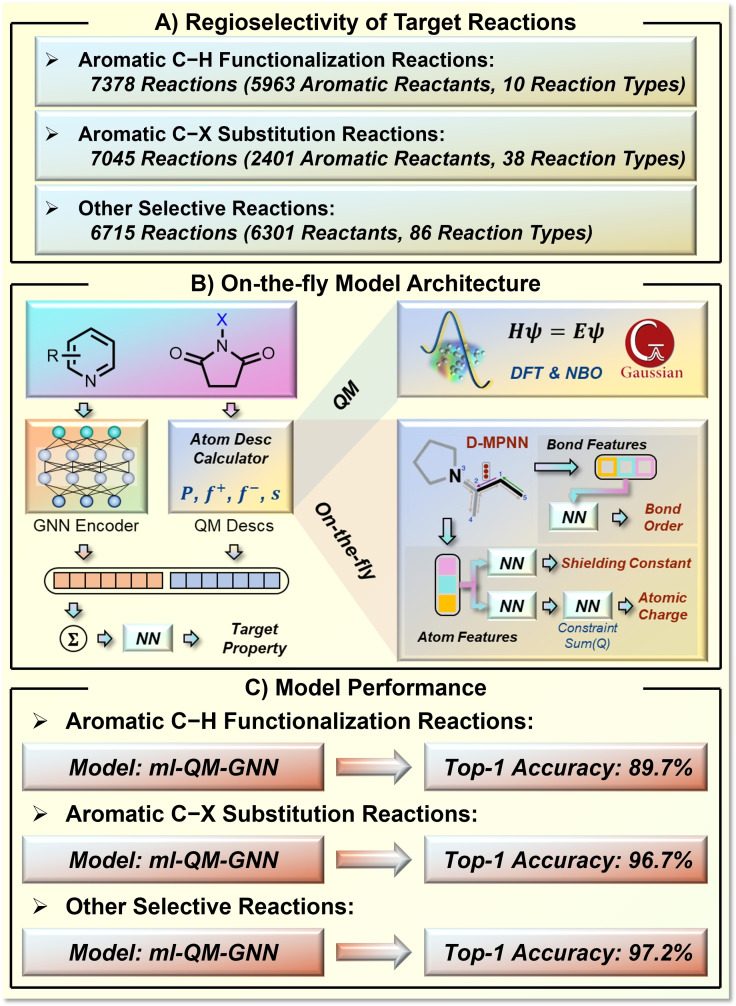
QM computations and statistics can also build a bridge to connect molecular properties and reaction performance prediction. In 2021, Jensen and co‐workers reported a new strategy to synergistically use machine‐learned molecular representation and quantum chemical descriptors (Figure 11). [98] The machine‐learned molecular representation was generated by a graph neural network (GNN) based on the Weisfeiler‐Lehman network (WLN) architecture. This GNN is an annotated graph in which the annotation is based on chemical knowledge. This chemistry‐based annotation in the graph model provides a promising and general strategy to introduce explicit chemical information in molecular representation.[ 99 , 100 ] The selected quantum chemical descriptors include atomic charge, Fukui index, NMR shielding constant, bond length and order. 136,000 organic molecules were computed at the B3LYP/def2‐SVP level of theory using GFN2‐xTB optimized geometries. The authors developed an ML platform to connect the GNN and QM parts by concatenating the GNN‐generated embeddings and QM descriptors, which realizes excellent interpolative and extrapolative regioselectivity predictions for a series of organic transformations (Figure 11A). In addition, the authors showed that it is possible to avoid the expensive DFT calculations of quantum chemical descriptors by using another ML model for QM descriptor prediction. For this purpose, they trained a multitask directed message passing neural network (D‐MPNN), which eventually provided an end‐to‐end fusion ml‐QM‐GNN model for accurate and efficient reactivity performance prediction. This model requires only 70 ms per reaction to predict the selectivity from SMILES. Each using 5000 training reactions from available datasets, this fusion model achieved 89.7 % top‐1 accuracy for aromatic C−H functionalization reactions, 96.7 % top‐1 accuracy for aromatic C−X substitution reactions, and 97.2 % top‐1 accuracy for other substitution reactions in predicting the primary reaction outcome.
Figure 11.
ML prediction of regioselectivity in organic transformations using on‐thy‐fly generated quantum chemical descriptors. A) The target reactions. B) The model design of quantum mechanics descriptors incorporated with GNN. C) The model performance.
Model design can also help the ML model use synthetic data smartly, especially for application scenarios with limited data for target transformation. An emerging artificial intelligence strategy for this problem is transfer learning, which applies the statistics or ML model from an external source to a new related task. In this regard, Reymond and co‐workers reported an insightful framework for reaction performance prediction (Figure 12). [72] The reaction‐related chemical knowledge was learned by machine via a molecular transformer model and then subjected to a specific target reaction via transfer learning. Two datasets were selected to describe the application scenarios. One is a big and all‐purpose dataset, which includes 1.1 million transformations in the USPTO database and represents the general knowledge in synthetic transformations. The other is a small and specialized dataset CARBO, which includes 25 thousand carbohydrate transformations curated from the Reaxys® database and refers to as a target transformation with interesting reaction performances (regio‐ and stereoselectivities in this case). The two datasets can be used for simultaneous training if the big dataset is publicly available. In this multitask scenario, the authors found that the hybrid utilization of the two datasets (weight 9 on the data from USPTO and weight 1 on the data from CARBO) showed excellent performance, with top‐1 accuracy of 71.2 % for prediction in the CARBO test set. The transfer learning strategy is also feasible by fine‐tuning a pre‐trained model even if the big dataset is not available. In the fine‐tuning scenario, the pre‐training was performed on the USPTO dataset and subsequently fine‐tuned based on the data of CARBO. The fine‐tuning model reaches a comparable top‐1 accuracy of 70.3 % without access to USPTO data. This work demonstrated the potential of a transfer learning strategy to connect chemically related synthetic databases as a way to improve the reaction performance prediction.
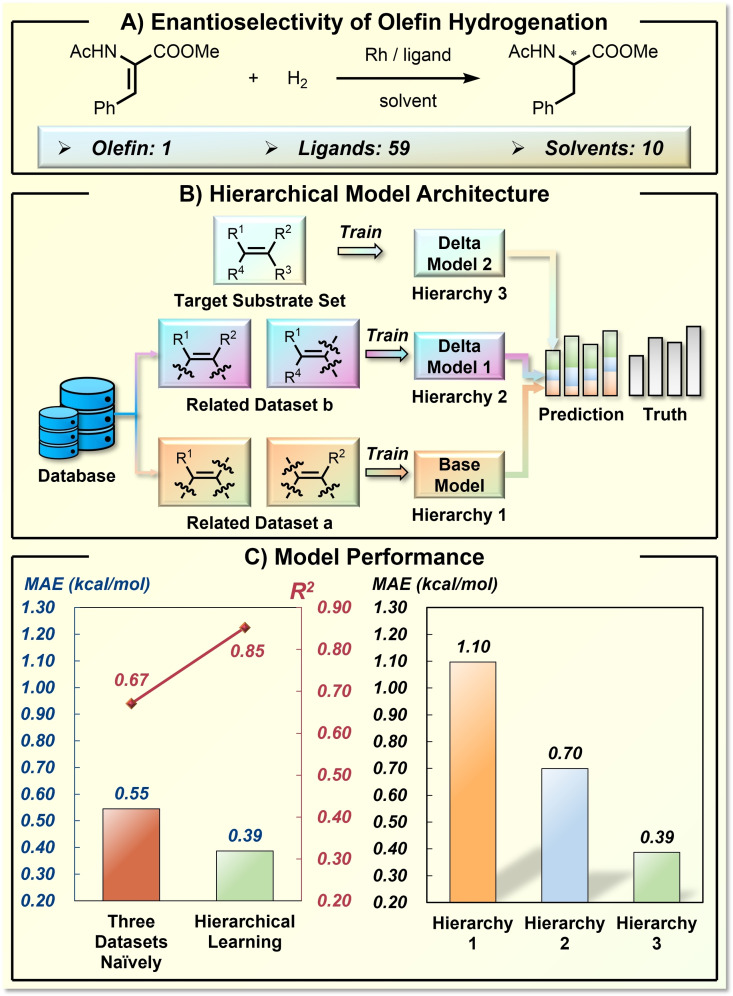
Targeting the few‐shot learning problem in catalysis development, the Hong group recently reported an ML approach to connect synthetically related data in model training. [45] Based on the curated database of asymmetric hydrogenation of olefins (12619 enantioselective transformations from literature), [101] the developed hierarchical learning model can provide satisfying enantioselectivity prediction using only dozens of data with the target olefin. This so‐called hierarchical learning model is essentially an ensemble model that combines individual predictions from different hierarchies (Figure 13). By judging the chemical similarity between the target olefin and the olefins in the database, the data of related asymmetric transformations were split into various hierarchies. With increasing hierarchies, the chemical structure of the involved olefins is closer to that of the target olefin, thusly the data size is also decreasing. The base model, trained by a large amount of data in hierarchy one, provides a general structure‐enantioselectivity relationship prediction, which was further corrected by delta learnings in the subsequent hierarchies. The effectiveness of the hierarchical learning approach is validated by the error reduction with increasing hierarchies, as well as the superior performance compared with the naïve model training (Figure 13C). This work provides a useful ML approach for synthetic method optimization where limited data is available, revealing the critical role of chemical relevance in data utilization.
Figure 13.
Application of transfer learning strategy in stereoselectivity prediction of carbohydrate transformation. A) The general reactions in the curated database of asymmetric hydrogenation of olefins. B) The model design for hierarchical learning. C) The model performance.
Summary and Outlook
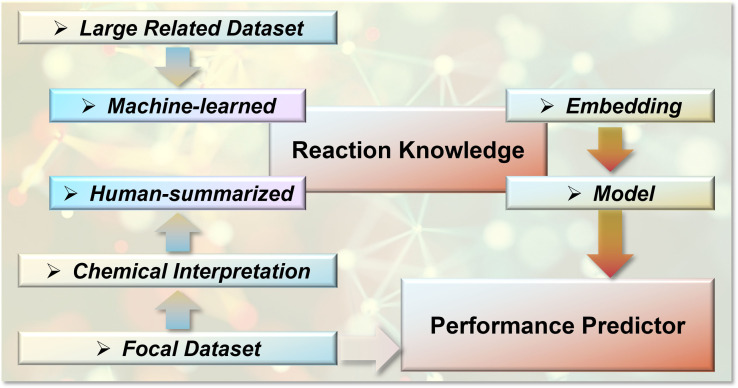
We have discussed representative strategies for implementing chemical knowledge into ML predictions toward reaction performances. The general pipeline for this implementation is presented in Figure 14. The knowledge source of organic synthesis is generally provided by human beings. This allows the installation of explicit rules in ML modelling, such as selecting chemically meaningful descriptors or the assignment of transition‐state‐like geometries. In addition, chemical statistics itself contains rich knowledge, which can be learned by machines to support the performance prediction of a target transformation. The utilization of quantum chemical computations and computed statistics, as well as transfer learning and ensemble learning strategies to connect the sizeable related dataset and the small focal dataset, have been found effective in introducing implicit chemical knowledge from synthetic statistics.
Figure 14.
General pipeline for reaction performance prediction with chemical‐aware ML.
Through the implementation of chemical knowledge, both the molecular embedding and model design can be innovated, which can either improve the prediction performance or mitigate the data requirement, providing powerful predictive tools for molecular syntheses.
Witnessing the exciting advances in ML prediction of organic synthesis, the impact of the data‐driven research paradigm in synthetic chemistry is apparent. The digitalization, computerization, and especially intellectualization of synthetic transformations will provide a strong momentum to push the frontiers of organic synthesis. However, it should be noted that the ML prediction of reaction performance is still in its infancy. There are a few essential but underdeveloped directions. For ML models, synthetic chemists strongly require extrapolative and heuristic predictions. Therefore, there is a strong demand for ML models that can provide predictions to help chemists in the design of new catalysts, reagents or entirely new transformations. Likewise, identifying robust ML tools for highly productive and selective chemical transformations continues to be challenging. These out‐of‐distribution and out‐of‐range prediction challenges require the invention of new ML models, and chemical knowledge is expected to provide critical assistance.
Moreover, the innovation of synthetic methods will play a significant role in facilitating the development of AI‐guided syntheses. For example, If ML provides a catalyst design that requires a twenty‐step synthesis, arguably this prediction will not be followed or valued by the experimentalists. By developing robust and programmable synthetic methods, it can open the gate to a large‐scale and diversified library of molecules with the desired function, this molecule library will provide a critical physical and digital basis to drive the iterative optimization of ML algorithms and synthetic methods. It is without a doubt that the path of AI‐assisted synthesis requires synergistic efforts between chemists and data scientists in order to build the desired bridge that connects chemical knowledge and computer algorithms. We have a strong belief that it will soon become true that artificial intelligence can accelerate, optimize, and even guide the development of organic syntheses.
Conflict of interest
The authors declare no conflict of interest.
1.
Biographical Information
Lutz Ackermann studied Chemistry at the University Kiel. He obtained his PhD in 2001 with Alois Fürstner at the MPI für Kohlenforschung (Mülheim/Ruhr), followed by a postdoctoral stay with Robert G. Bergman at UC Berkeley. In 2003, he initiated his independent career at the Ludwig Maximilians‐University in München. Since 2007, he is Chair and Full Professor at the Georg‐August‐University Göttingen. His awards include an AstraZeneca Excellence in Chemistry Award (2011), an ERC Consolidator Grant (2012), a Gottfried‐Wilhelm‐Leibniz‐Preis (2017) and an ERC Advanced Grant (2021). His current research interests include the development of novel catalysis concepts for sustainable molecular syntheses, with topical foci on strong bond activation, data science, and photo‐ as well as electrocatalysis.

Biographical Information
Xin Hong is a tenured associate professor in Zhejiang University, whose primary research focuses on the reaction mechanism and data‐driven design of organic synthesis. Xin received his B. S. from USTC and Ph.D. from UCLA under the guidance of Prof. K. N. Houk. After postdoc researches with Prof. K. N. Houk in UCLA and Prof. Jens K. Nørskov in Stanford, Xin joined the Zhejiang University in 2016. He has received several awards including CCS Youth Chemist Award (2020) and Thieme Chemistry Journals Award (2022).

Acknowledgments
National Natural Science Foundation of China (21873081 and 22122109, X. H.; 22103070, S.‐Q. Z.), the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (SN‐ZJU‐SIAS‐006, X. H.), Beijing National Laboratory for Molecular Sciences (BNLMS202102, X. H.), CAS Youth Interdisciplinary Team (JCTD‐2021‐11, X. H.), Fundamental Research Funds for the Central Universities (226‐2022‐00140 and 226‐2022‐00224, X. H.), the Center of Chemistry for Frontier Technologies and Key Laboratory of Precise Synthesis of Functional Molecules of Zhejiang Province (PSFM 2021‐01, X. H.) and the State Key Laboratory of Clean Energy Utilization (ZJUCEU2020007, X. H.) are gratefully acknowledged. Generous support by the DFG (SPP1807, SPP2363, and Gottfried‐Wilhelm‐Leibniz award to L. A.) and an ERC Advanced Grant (L. A.) are gratefully acknowledged. Open Access funding enabled and organized by Projekt DEAL.
Zhang S.-Q., Xu L.-C., Li S.-W., Oliveira J. C. A., Li X., Ackermann L., Hong X., Chem. Eur. J. 2023, 29, e202202834.
Contributor Information
Prof. Dr. Lutz Ackermann, Email: lutz.ackermann@chemie.uni-goettingen.de.
Prof. Dr. Xin Hong, Email: hxchem@zju.edu.cn.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.
References
- 1. Noyori R., Nat. Chem. 2009, 1, 5–6. [DOI] [PubMed] [Google Scholar]
- 2.
- 2a. Trost B. M., Angew. Chem. Int. Ed. Engl. 1995, 34, 259–281; [Google Scholar]
- 2b. Trost B. M., Acc. Chem. Res. 2002, 35, 695–705. [DOI] [PubMed] [Google Scholar]
- 3.
- 3a. Meyer T. H., Finger L. H., Gandeepan P., Ackermann L., Trends Chem. 2019, 1, 63–76; [Google Scholar]
- 3b. Ackermann L., You S.-L., Oestreich M., Meng S., MacFarlane D., Yin Y., Trends Chem. 2020, 2, 275–277. [Google Scholar]
- 4. Coley C. W., Green W. H., Jensen K. F., Acc. Chem. Res. 2018, 51, 1281–1289. [DOI] [PubMed] [Google Scholar]
- 5. Liu B., Ramsundar B., Kawthekar P., Shi J., Gomes J., Luu Nguyen Q., Ho S., Sloane J., Wender P., Pande V., ACS Cent. Sci. 2017, 3, 1103–1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Todd M. H., Chem. Soc. Rev. 2005, 34, 247–266. [DOI] [PubMed] [Google Scholar]
- 7. Coley C. W., Barzilay R., Jaakkola T. S., Green W. H., Jensen K. F., ACS Cent. Sci. 2017, 3, 434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gao H., Struble T. J., Coley C. W., Wang Y., Green W. H., Jensen K. F., ACS Cent. Sci. 2018, 4, 1465–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Walker E., Kammeraad J., Goetz J., Robo M. T., Tewari A., Zimmerman P. M., J. Chem. Inf. Model. 2019, 59, 3645–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shields B. J., Stevens J., Li J., Parasram M., Damani F., Alvarado J. I. M., Janey J. M., Adams R. P., Doyle A. G., Nature 2021, 590, 89–96. [DOI] [PubMed] [Google Scholar]
- 11. Knowles R. R., Jacobsen E. N., Proc. Nat. Acad. Sci. 2010, 107, 20678–20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Neel A. J., Milo A., Sigman M. S., Toste F. D., J. Am. Chem. Soc. 2016, 138, 3863–3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aune M., Gogoll A., Matsson O., J. Org. Chem. 1995, 60, 1356–1364. [Google Scholar]
- 14. Burés J., Dingwall P., Armstrong A., Blackmond D. G., Angew. Chem. Int. Ed. 2014, 53, 8700–8704; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2014, 126, 8844–8848. [Google Scholar]
- 15. Mikulak-Klucznik B., Gołębiowska P., Bayly A. A., Popik O., Klucznik T., Szymkuć S., Gajewska E. P., Dittwald P., Staszewska-Krajewska O., Beker W., Badowski T., Scheidt K. A., Molga K., Mlynarski J., Mrksich M., Grzybowski B. A., Nature 2020, 588, 83–88. [DOI] [PubMed] [Google Scholar]
- 16. Corey E. J., Long A. K., Rubenstein S. D., Science 1985, 228, 408–418. [DOI] [PubMed] [Google Scholar]
- 17. Elyashberg M., Williams A. J., Blinov K., Nat. Prod. Rep. 2010, 27, 1296. [DOI] [PubMed] [Google Scholar]
- 18. Ahneman D. T., Estrada J. G., Lin S., Dreher S. D., Doyle A. G., Science 2018, 360, 186–190. [DOI] [PubMed] [Google Scholar]
- 19. Reid J. P., Sigman M. S., Nature 2019, 571, 343–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zahrt A. F., Henle J. J., Rose B. T., Wang Y., Darrow W. T., Denmark S. E., Science. 2019, 363, eaau5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mater A. C., Coote M. L., J. Chem. Inf. Model. 2019, 59, 2545–2559. [DOI] [PubMed] [Google Scholar]
- 22. Strieth-Kalthoff F., Sandfort F., Segler M. H. S., Glorius F., Chem. Soc. Rev. 2020, 49, 6154–6168. [DOI] [PubMed] [Google Scholar]
- 23. Tromp J., Cham 2016, pp. 183–190, in: A. Plaat, W. Kosters, J. van den Herik (Eds.), Springer International Publishing. [Google Scholar]
- 24. Schrittwieser J., Antonoglou I., Hubert T., Simonyan K., Sifre L., Schmitt S., Guez A., Lockhart E., Hassabis D., Graepel T., Lillicrap T., Silver D., Nature 2020, 588, 604–609. [DOI] [PubMed] [Google Scholar]
- 25. Virshup A. M., Contreras-García J., Wipf P., Yang W., Beratan D. N., J. Am. Chem. Soc. 2013, 135, 7296–7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reaxys®, http://www.reaxys.com/, (accessed June 12, 2022).
- 27. Gallegos L. C., Luchini G., John P. C. St., Kim S., Paton R. S., Acc. Chem. Res. 2021, 54, 827–836. [DOI] [PubMed] [Google Scholar]
- 28. Li X., Zhang S.-Q., Xu L.-C., Hong X., Angew. Chem. Int. Ed. 2020, 59, 13253–13259; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 13355–13361. [Google Scholar]
- 29. Yang L.-C., Li X., Zhang S.-Q., Hong X., Org. Chem. Front. 2021, 8, 6187–6195. [Google Scholar]
- 30. Newman-Stonebraker S. H., Smith S. R., Borowski J. E., Peters E., Gensch T., Johnson H. C., Sigman M. S., Doyle A. G., Science. 2021, 374, 301–308. [DOI] [PubMed] [Google Scholar]
- 31. Behler J., J. Chem. Phys. 2011, 134, 74106. [DOI] [PubMed] [Google Scholar]
- 32. Crawford J. M., Kingston C., Toste F. D., Sigman M. S., Acc. Chem. Res. 2021, 54, 3136–3148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Rinehart N. I., Zahrt A. F., Henle J. J., Denmark S. E., Acc. Chem. Res. 2021, 54, 2041–2054. [DOI] [PubMed] [Google Scholar]
- 34. Żurański A. M., Martinez Alvarado J. I., Shields B. J., Doyle A. G., Acc. Chem. Res. 2021, 54, 1856–1865. [DOI] [PubMed] [Google Scholar]
- 35. Wodrich M. D., Sawatlon B., Busch M., Corminboeuf C., Acc. Chem. Res. 2021, 54, 1107–1117. [DOI] [PubMed] [Google Scholar]
- 36. Janet J. P., Duan C., Nandy A., Liu F., Kulik H. J., Acc. Chem. Res. 2021, 54, 532–545. [DOI] [PubMed] [Google Scholar]
- 37. Jorner K., Tomberg A., Bauer C., Sköld C., Norrby P.-O., Nat. Chem. Rev. 2021, 5, 240–255. [DOI] [PubMed] [Google Scholar]
- 38. Niemeyer Z. L., Milo A., Hickey D. P., Sigman M. S., Nat. Chem. 2016, 8, 610–617. [DOI] [PubMed] [Google Scholar]
- 39. Sigman M. S., Harper K. C., Bess E. N., Milo A., Acc. Chem. Res. 2016, 49, 1292–1301. [DOI] [PubMed] [Google Scholar]
- 40. Gensch T., dos Passos Gomes G., Friederich P., Peters E., Gaudin T., Pollice R., Jorner K., Nigam A., Lindner-D'Addario M., Sigman M. S., Aspuru-Guzik A., J. Am. Chem. Soc. 2022, 144, 1205–1217. [DOI] [PubMed] [Google Scholar]
- 41. Sowndarya S. V. S., John P. C. St., Paton R. S., Chem. Sci. 2021, 12, 13158–13166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. John P. C. St., Guan Y., Kim Y., Etz B. D., Kim S., Paton R. S., Sci. Data 2020, 7, 244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Durand D. J., Fey N., Chem. Rev. 2019, 119, 6561–6594. [DOI] [PubMed] [Google Scholar]
- 44.SciFinder®, https://scifinder.cas.org/, (accessed June 12, 2022).
- 45. Xu L.-C., Zhang S.-Q., Li X., Tang M.-J., Xie P.-P., Hong X., Angew. Chem. Int. Ed. 2021, 60, 22804–22811. [DOI] [PubMed] [Google Scholar]
- 46. Schneider N., Lowe D. M., Sayle R. A., Tarselli M. A., Landrum G. A., J. Med. Chem. 2016, 59, 4385–4402. [DOI] [PubMed] [Google Scholar]
- 47.D. Lowe, 2017, DOI 10.6084/m9.figshare.5104873.v1.
- 48. Beker W., Roszak R., Wołos A., Angello N. H., Rathore V., Burke M. D., Grzybowski B. A., J. Am. Chem. Soc. 2022, 144, 4819–4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kearnes S. M., Maser M. R., Wleklinski M., Kast A., Doyle A. G., Dreher S. D., Hawkins J. M., Jensen K. F., Coley C. W., J. Am. Chem. Soc. 2021, 143, 18820–18826. [DOI] [PubMed] [Google Scholar]
- 50. An F., Maji B., Min E., Ofial A. R., Mayr H., J. Am. Chem. Soc. 2020, 142, 1526–1547. [DOI] [PubMed] [Google Scholar]
- 51. Um I.-H., Bae A. R., J. Org. Chem. 2011, 76, 7510–7515. [DOI] [PubMed] [Google Scholar]
- 52. de Almeida A. F., Moreira R., Rodrigues T., Nat. Chem. Rev. 2019, 3, 589–604. [Google Scholar]
- 53. Coley C. W., Eyke N. S., Jensen K. F., Angew. Chem. Int. Ed. 2020, 59, 22858–22893; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23054–23091. [Google Scholar]
- 54. Coley C. W., Eyke N. S., Jensen K. F., Angew. Chem. Int. Ed. 2020, 59, 23414–23436; [DOI] [PubMed] [Google Scholar]; Angew. Chem. 2020, 132, 23620–23643. [Google Scholar]
- 55. Williams W. L., Zeng L., Gensch T., Sigman M. S., Doyle A. G., Anslyn E. V., ACS Cent. Sci. 2021, 7, 1622–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schwaller P., Vaucher A. C., Laplaza R., Bunne C., Krause A., Corminboeuf C., Laino T., WIREs Comput. Mol. Sci. 2022, 12, e1604. [Google Scholar]
- 57. Oliveira J. C. A., Frey J., Zhang S.-Q., Xu L.-C., Li X., Li S.-W., Hong X., Ackermann L., Trends Chem. 2022, 4, 863–885. [Google Scholar]
- 58. David L., Thakkar A., Mercado R., Engkvist O., J. Cheminf. 2020, 12, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Langer M. F., Goeßmann A., Rupp M., npj Comput. Mater. 2022, 8, 41. [Google Scholar]
- 60. Wigh D. S., Goodman J. M., Lapkin A. A., WIREs Comput. Mol. Sci. 2022, 12, e1603. [Google Scholar]
- 61. Granda J. M., Donina L., Dragone V., Long D.-L., Cronin L., Nature 2018, 559, 377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Weininger D., J. Chem. Inf. Model. 1988, 28, 31–36. [Google Scholar]
- 63. Heller S. R., McNaught A., Pletnev I., Stein S., Tchekhovskoi D., J. Cheminf. 2015, 7, 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.IUPAC Naming, https://www.iupacnaming.com/, (accessed June 12, 2022).
- 65.SMARTS - A Language for Describing Molecular Patterns, https://www.daylight.com/dayhtml/doc/theory/theory.smarts.html, (accessed June 12, 2022).
- 66.N. O'Boyle, A. Dalke, ChemRxiv, 2018, DOI 10.26434/chemrxiv.7097960.v1.
- 67. Krenn M., Häse F., Nigam A., Friederich P., Aspuru-Guzik A., Mach. Learn.: Sci. Technol. 2020, 1, 45024. [Google Scholar]
- 68. Schwaller P., Laino T., Gaudin T., Bolgar P., Hunter C. A., Bekas C., Lee A. A., ACS Cent. Sci. 2019, 5, 1572–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Schwaller P., Probst D., Vaucher A. C., Nair V. H., Kreutter D., Laino T., Reymond J.-L., Nat. Mach. Intell. 2021, 3, 144–152. [Google Scholar]
- 70. Jorner K., Brinck T., Norrby P.-O., Buttar D., Chem. Sci. 2021, 12, 1163–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schwaller P., Vaucher A. C., Laino T., Reymond J.-L., Mach. Learn.: Sci. Technol. 2021, 2, 15016. [Google Scholar]
- 72. Pesciullesi G., Schwaller P., Laino T., Reymond J.-L., Nat. Commun. 2020, 11, 4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cereto-Massagué A., Ojeda M. J., Valls C., Mulero M., Garcia-Vallvé S., Pujadas G., Methods 2015, 71, 58–63. [DOI] [PubMed] [Google Scholar]
- 74. Rogers D., Hahn M., J. Chem. Inf. Model. 2010, 50, 742–754. [DOI] [PubMed] [Google Scholar]
- 75. Gedeck P., Rohde B., Bartels C., J. Chem. Inf. Model. 2006, 46, 1924–1936. [DOI] [PubMed] [Google Scholar]
- 76. Durant J. L., Leland B. A., Henry D. R., Nourse J. G., J. Chem. Inf. Comput. Sci. 2002, 42, 1273–1280. [DOI] [PubMed] [Google Scholar]
- 77. Sandfort F., Strieth-Kalthoff F., Kühnemund M., Beecks C., Glorius F., Chem 2020, 6, 1379–1390. [Google Scholar]
- 78. Dahl S., Logadottir A., Jacobsen C. J. H., Nørskov J. K., Appl. Catal. A 2001, 222, 19–29. [Google Scholar]
- 79. Wodrich M. D., Fabrizio A., Meyer B., Corminboeuf C., Chem. Sci. 2020, 11, 12070–12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Cordova M., Wodrich M. D., Meyer B., Sawatlon B., Corminboeuf C., ACS Catal. 2020, 10, 7021–7031. [Google Scholar]
- 81. Yang L.-C., Hong X., Dalton Trans. 2020, 49, 3652–3657. [DOI] [PubMed] [Google Scholar]
- 82. Anand M., Nørskov J. K., ACS Catal. 2020, 10, 336–345. [Google Scholar]
- 83. Verloop A., Hoogenstraaten W., Tipker J., in Drug Des. (Ed.: Ariëns E. J. B. T.-D. D.), Elsevier, Amsterdam, 1976, pp. 165–207. [Google Scholar]
- 84. Hammett L. P., J. Am. Chem. Soc. 1937, 59, 96–103. [Google Scholar]
- 85. Moskal M., Beker W., Szymkuć S., Grzybowski B. A., Angew. Chem. Int. Ed. 2021, 60, 15230–15235. [DOI] [PubMed] [Google Scholar]
- 86. Huang B., von Lilienfeld O. A., Chem. Rev. 2021, 121, 10001–10036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Unke O. T., Chmiela S., Sauceda H. E., Gastegger M., Poltavsky I., Schütt K. T., Tkatchenko A., Müller K.-R., Chem. Rev. 2021, 121, 10142–10186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Westermayr J., Marquetand P., Chem. Rev. 2021, 121, 9873–9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schütt K. T., Sauceda H. E., Kindermans P.-J., Tkatchenko A., Müller K.-R., J. Chem. Phys. 2018, 148, 241722. [DOI] [PubMed] [Google Scholar]
- 90. Unke O. T., Meuwly M., J. Chem. Theory Comput. 2019, 15, 3678–3693. [DOI] [PubMed] [Google Scholar]
- 91. Liu Z., Lin L., Jia Q., Cheng Z., Jiang Y., Guo Y., Ma J., J. Chem. Inf. Model. 2021, 61, 1066–1082. [DOI] [PubMed] [Google Scholar]
- 92. Cheong P. H.-Y., Legault C. Y., Um J. M., Çelebi-Ölçüm N., Houk K. N., Chem. Rev. 2011, 111, 5042–5137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Sperger T., Sanhueza I. A., Kalvet I., Schoenebeck F., Chem. Rev. 2015, 115, 9532–9586. [DOI] [PubMed] [Google Scholar]
- 94. Qi X., Li Y., Bai R., Lan Y., Acc. Chem. Res. 2017, 50, 2799–2808. [DOI] [PubMed] [Google Scholar]
- 95. Zhang S.-Q., Hong X., Acc. Chem. Res. 2021, 54, 2158–2171. [DOI] [PubMed] [Google Scholar]
- 96. Yan M., Lo J. C., Edwards J. T., Baran P. S., J. Am. Chem. Soc. 2016, 138, 12692–12714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Smith J. M., Dixon J. A., DeGruyter J. N., Baran P. S., J. Med. Chem. 2019, 62, 2256–2264. [DOI] [PubMed] [Google Scholar]
- 98. Guan Y., Coley C. W., Wu H., Ranasinghe D., Heid E., Struble T. J., Pattanaik L., Green W. H., Jensen K. F., Chem. Sci. 2021, 12, 2198–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Schütt K. T., Sauceda H. E., Kindermans P.-J., Tkatchenko A., Müller K.-R., J. Chem. Phys. 2018, 148, 241722. [DOI] [PubMed] [Google Scholar]
- 100.N. W. A. Gebauer, M. Gastegger, K. T. Schütt, 2019, 10.48550/arxiv.1906.00957.
- 101.Database of Asymmetric Hydrogenation of Olefins, http://asymcatml.net/, (accessed June 12, 2022).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analyzed in this study.