Abstract
The comprehension of microbial interactions is one of the key challenges in marine microbial ecology. This study focused on exploring chemical interactions between the toxic dinoflagellate Prorocentrum lima and a filamentous fungal species, Aspergillus pseudoglaucus, which has been isolated from the microalgal culture. Such interspecies interactions are expected to occur even though they were rarely studied. Here, a co‐culture system was designed in a dedicated microscale marine‐like condition. This system allowed to explore microalgal–fungal physical and metabolic interactions in presence and absence of the bacterial consortium. Microscopic observation showed an unusual physical contact between the fungal mycelium and dinoflagellate cells. To delineate specialized metabolome alterations during microalgal–fungal co‐culture metabolomes were monitored by high‐performance liquid chromatography coupled to high‐resolution mass spectrometry. In‐depth multivariate statistical analysis using dedicated approaches highlighted (1) the metabolic alterations associated with microalgal–fungal co‐culture, and (2) the impact of associated bacteria in microalgal metabolome response to fungal interaction. Unfortunately, only a very low number of highlighted features were fully characterized. However, an up‐regulation of the dinoflagellate toxins okadaic acid and dinophysistoxin 1 was observed during co‐culture in supernatants. Such results highlight the importance to consider microalgal–fungal interactions in the study of parameters regulating toxin production.
INTRODUCTION
In the marine environment, microalgae are in constant interaction with other microorganisms, at both intra‐ and inter‐specific levels, which span from competition over mutualism, symbiosis and parasitism to predation (Zhang et al., 2018). This large diversity of biotic interactions is governed by nutrient availability and chemical communication mediated by diffusible signalling metabolites (Brown et al., 2019; Puglisi et al., 2014). Among those marine microalgae, toxic dinoflagellates represent particular microorganisms that can drastically impact marine ecosystems, for example, reduction of fish and algal diversity as well as impacting human health through seafood contamination (Grattan et al., 2016). Harmful algal blooms (HABs) are considered detrimental events, and HAB species are increasingly observed in some ecosystems, especially in the proximity of human activities (Stewart et al., 2008). One HAB species involved in such threats is the marine dinoflagellate Prorocentrum lima. This species is able to produce toxins like okadaic acid (OA) and dinophysistoxin‐1 (DTX‐1) associated with diarrheic shellfish poisoning (DSP). Production of such toxins is known to be potentially influenced by nitrogen levels, light intensity, temperature and salinity (Lee et al., 2016; Wang et al., 2015).
Prorocentrum lima has a benthic and typically epiphytic habitat (Nishimura et al., 2020). This microalga was, thus, reported to grow on macroalgae such as Enteromorpha spp., and marine plantae such as Zostera and Ruppia spp. (Foden et al., 2005). Life on such surfaces implies multiple mechanisms to compete for nutrients, light and space (Allen et al., 2016; Wang et al., 2020). Currently, only little information is available on microorganisms co‐occurring with P. lima. Some examples exist related to bacteria found in close association with phytoplankton cells in nature and in culture, such as α‐Proteobacteria (including members of the order Rhodobacterales), γ‐Proteobacteria (Alteromonadales), Bacteroidia (Flavobacteriales), as well as Actinobacteria (Buchan et al., 2014; Tarazona‐Janampa et al., 2020; Traubenberg et al., 1995). However, as an epiphytic organism, P. lima may be part of a complex microbial community frequently associated with algae, involving other microalgae (Wang et al., 2020), bacteria and fungi (Tourneroche et al., 2020). In addition, P. lima can also interact with predators, such as small crustacean (Ajuzie, 2007; Gu et al., 2019). In this context, production of phycotoxins can be considered as a defence mechanism (Ryderheim et al., 2021). Moreover, the complex biosynthesis of such high‐molecular‐weight molecules points out that these compounds should provide competitive advantages to their producers (Chakraborty, 2019). For example, the exposure of P. lima to Artemia salina chemical cues yielded OA over‐production, which impaired survival rates in this predator population (Gu et al., 2019). However, it remains unclear whether OA exerts a broad deterrence effect spectrum, since only a small number of studies directly linked OA exposure to negative predator outcomes (Shaw et al., 1997). Besides predators, P. lima is able to compete with other co‐occurring dinoflagellates (Wang et al., 2020). For instance, P. lima was shown to produce a complex chemical mixture (including OA) exhibiting growth inhibitory activity against co‐occurring dinoflagellate species (Sugg & VanDolah, 1999). Therefore, P. lima seems equipped with various chemical signals suited to its survival or competition.
Various examples of microalgal interactions with bacteria are reported in the literature (Cirri & Pohnert, 2019); however, to our knowledge, very little information is available on P. lima chemicals induced by bacteria and/or fungi. Considering toxins, some bacteria are able to trigger toxin production, by direct competition for nutrients at late culture stages, or alternatively in early stages relying on chemical cues (Wang, Sen, et al., 2018; Wang, Yao, et al., 2018). However, other examples do not highlight such induction, suggesting the occurrence of particular regulation mechanisms depending on bacterial‐microalgal co‐culture (Green et al., 2004, 2010; Ho et al., 2006; Lu et al., 2016; Prol et al., 2009; Uribe & Espejo, 2003).
Fungi are another group of micro‐organisms frequently encountered in the marine environment (Hallegraeff et al., 2014; Li et al., 2017; Wang, Sen, et al., 2018). In the laboratory, fungal genetic material is frequently detected through sequencing using eukaryotic primers during species identification of microalgal cultures (Nicolas Chomérat, Ifremer, personal communication). This prompted us to investigate the possibility of microalgal–fungal interactions. In several initial studies focusing on benthic microalgae (which present similar slow growth as many fungi), we identified several fungal species present in cultures of benthic microalgae (Bagot et al., 2016 unpublished; Chaigne et al., 2017 unpublished). Therefore, we focused in this study on one of the couples of microalgal–fungal associations identified in these initial studies, that is, the microalga P. lima and the fungus Aspergillus pseudoglaucus.
Even though no literature explores P. lima in this context, some examples exist with other microalgae (see above and articles discussed in the studies by Cirri & Pohnert, 2019). A first study described how chytrid zoospores detected diatom preys through their carbohydrate exudates (Scholz et al., 2017). In response, the diatoms adapt their metabolism through the production of polyenal lipids and associated aldehydes, thus altering parasite life cycles (Pohnert, 2000). Beside this example concerning parasitic fungi, to our knowledge, no information is currently available in the literature on the existence of interactions between saprophytic marine fungi and microalgae, even though they are expected to naturally occur, and as both organisms may be present simultaneously. Such ‘interaction’ was only reported artificially during specific bioprocesses and led to microalgal aggregation thus improving biomass harvesting (Wrede et al., 2014).
Microbial community structures are governed by chemical signals that require the production of specialized metabolites (Ianora et al., 2006). However, only a few studies highlight such molecules, mainly due to technical difficulties, which appear in their isolation and subsequent characterization (e.g. quantities) (Ji et al., 2011). To overcome such challenge, microbial interactions are usually studied in vitro in controlled bipartite conditions using metabolomics approaches (Bertrand, Azzollini, et al., 2014; Bertrand, Bohni, et al., 2014). Metabolomics, defined as a comprehensive analytical approach for the identification and quantification of metabolites in biological systems (Wolfender et al., 2015), remains a method of choice to highlight metabolic modifications induced by microbial co‐cultures (Arora et al., 2020; Wolfender et al., 2015). Various examples already showed the induction of specific metabolites in microbial co‐cultures (Arora et al., 2020) through the expression of silent biosynthetic gene clusters (BGCs) (Schroeckh et al., 2009).
To explore the chemical interactions between P. lima and co‐occurring fungi, a fungal strain isolation campaign was initiated using the toxic and non‐axenic P. lima PL4V strain (Bravo et al., 2001). This provided various fungal strains, among which A. pseudoglaucus MMS1589 was selected due to an unusual physical contact with P. lima PL4V cells highlighted by microscopy. A co‐culture experiment in treated (using antibiotics, see Experimental Procedures) and non‐treated conditions was performed between the microalgae and the fungal strain using a high‐performance liquid chromatography coupled to high‐resolution mass spectrometry (HPLC‐HRMS) metabolomics approach. The aims of this study were to explore (1) the presence of fungal strains within the microbiome of this epiphytic dinoflagellate, (2) the chemical mediation between P. lima PL4V and A. pseudoglaucus MMS1589, (3) the impact of co‐occurring bacteria on such chemical mediation and (4) whether the fungus influences microalgal toxin production.
EXPERIMENTAL PROCEDURES
Microalgae
Prorocentrum lima is a benthic dinoflagellate that may be considered a species complex as there is both temperate and tropical strains reported for this species (Lassus et al., 2016). The P. lima strain PL4V was collected in 1985 by colleagues from the Instituto Espanol Oceanografico (IEO, Vigo) from mussel rafts in the Ria de Pontevedra (Galicia, Spain) (Bravo et al., 2001), a temperate area, and maintained since 1985 at the French Research Institute for the Exploitation of the Sea (IFREMER).
Culture media
All media used for fungal strain isolation were prepared using Reef Crystal mixture at 36 g/L (enriched marine salt from Aquarium Systems) to meet standard seawater salt concentrations and autoclaved at 121°C for 20 min. Chloramphenicol (50 mg/ml, Poly Labo, Paul Block & Cie, Strasbourg, France) was only added to the liquid or solid media used for fungal strain isolation, to avoid bacterial growth. Malt extract (ME) liquid medium was prepared with following concentrations: glucose 20 g/L, peptone (Biokar Diagnostics, Beauvais, France) 1 g/L, Malt Extract (Conda pronadisa, Madrid, Spain) 30 g/L, copper sulfate 5 mg/L, zinc sulfate 1 mg/L. The Dextrose Casein (DC) medium was prepared as follows: glucose 40 g/L, peptone 10 g/L. Solid equivalent of these media were also prepared. Dextrose casein agar (DCA) used DCA mixture (Becton, Dickinson and Company, Sparks, NV USA) 65 g/L and the Malt Extract Agar (MEA, Biokar Diagnostics) medium was composed of glucose 20 g/L, peptone 1 g/L, Malt Extract mixture 45 g/L, copper sulfate 5 mg/L, zinc sulfate 1 mg/L.
For the co‐culture experiment, f/2 microalgal growth medium, without silica addition, was used as previously described (Guillard & Ryther, 1962). The solid f/2 glucose agar medium (f/2 GA) was prepared using f/2 liquid medium complemented by 20 g/L of agar and 2 g/L of glucose. To grow microorganisms with limited bacterial development, large spectrum antibiotics cocktail was added to f/2 liquid medium to provide the following final concentration in the culture medium: ampicillin 500 μg/ml, gentamycin 100 μg/ml, and kanamycin 200 μg/ml (Cho et al., 2002). Either condition will further be named as ‘treated condition/sample’ or ‘non‐treated condition/sample’ referring to the use of antibiotic cocktail or not, respectively.
Fungal isolation from microalgal biomass
To isolate fungal strains, P. lima PL4V biomass was inoculated on several liquid and solid culture media (ME, DC, MEA and DCA media). Fungal growth at 27°C was visually evaluated every day. When fungal colonies were observed, each colony was further collected and inoculated on DCA medium for storage and morphological identification based on macroscopic and microscopic features. The MMS1589 strain was further identified based on molecular biology information (ITS and β‐tubulin sequencing) according to previously published protocol (Rédou et al., 2015). The ITS and β‐tubulin sequences for the strain MMS1589 were deposited in GenBank under accession numbers MN134000.1 and MN164633.1, respectively.
Co‐culture growth in miniaturized liquid/solid environment
The miniaturized liquid/solid environment was prepared in tilted 50 ml cell‐culture flasks (75 cm2 surface, CORNING) (Figure S1). First, 10 ml f/2 GA medium were added to the flask. Subsequently, the strain A. pseudoglaucus MMS1589 was inoculated by spiking it into the middle of the f/2 GA medium. After 5 days of fungal growth at 24°C under 12 h/12 h dark–light cycle using artificial light (DENNERLE Nano Light 11 watt, 6000 Kelvin, 900 lumen), 10 ml liquid f/2 medium (with or without antibiotic cocktail), and 15 ml P. lima PL4V inoculum in f/2 medium at 30,600 cells/ml were added to the flask. The co‐culture was then incubated for 6 days under 12 h/12 h dark–light cycle. In parallel, the same design was used for both microorganisms alone (monoculture), where only A. pseudoglaucus MMS1589 or P. lima PL4V was inoculated. In addition, blank samples were obtained by preparing the miniaturized liquid/solid environment (with and without antibiotics) without microorganism inoculation in parallel to the co‐culture growth.
Five replicates of each experiment were performed, leading to 30 samples (with antibiotics: five co‐cultures, five A. pseudoglaucus MMS1589 and five P. lima PL4V monocultures; without antibiotics: five co‐cultures, five A. pseudoglaucus MMS1589 and five P. lima PL4V monocultures). Additional replicates were prepared in parallel for further microscopic observations.
Microscopic observations were performed after sampling the supernatant and/or microalgal/fungal colonies on glass slides. Congo red dye (1% in ethanol) was added to reveal fungal hyphae (Wood, 1980). Binocular and microscopic observations were performed using, respectively, a Zeiss binocular magnifier and a Leica microscope (with a ×10 magnifying eyepiece, combined with a ×10 and ×40 objectives, allowing for ×100 and ×400 magnified observations, respectively).
Biomass extraction
After 6 days, for each growth experiment in liquid/solid environment, the liquid overlaying phase was separated from the solid phase (resulting f/2 GA medium, fungus and/or microalgae biomass) and further centrifuged (5 min at 134g) to remove any cellular debris (P. lima PL4V cells and fungal spores' traces) yielding the supernatant. Each supernatant was extracted three times with 5 ml ethyl acetate (EtOAc), and the three resulting extracts were pooled and dried under vacuum using a SpeedVac™ concentrator (Savant SPD131DDA and Savant RVT400, Thermo Scientific). The solid phase was transferred to glass flasks and extracted twice with 5 ml EtOAc using an ultrasonic bath for 10 min prior to centrifugation and discarding of pellet. This way, the supernatant and solid phase extracts are expected to contain the extra‐ and intra‐cellular content, respectively. All extracts were weighed after evaporation (Figure S2) using a SpeedVac™ concentrator.
LC‐HRMS profiling
Each extract was diluted to a concentration of 1 mg dry weight/ml in HPLC‐grade methanol (Biosolve Chimie, Dieuze, France) and 5 μl were injected. LC‐HRMS profiling was achieved using an ultra‐fast liquid chromatography–electrospray ionization–ion trap–time of flight–mass spectrometer (UFLC‐ESI‐IonTrap‐TOFMS) system (Shimadzu, Marne‐la‐Vallée, France) according to a previously published protocol (Roullier et al., 2016). Briefly, chromatographic separation was achieved using a Kinetex™ C18 column (100 × 2.1 mm, 2.6 μm), at 40°C with a flow rate of 0.3 ml/min. The mobile phase consisted in a gradient starting at 15% acetonitrile + 0.1% formic acid (FA) (B) for 2 min, followed by an increase to 100% over 23 min and further maintained for 5 min, water + FA (A) completing the dual solvent mobile phase. Mass detection was set between m/z 100 and 1000 with fast switching between positive and negative ionization mode (PI and NI), the resolution of the mass spectrometer is 9200 (at m/z 500).
In the sequences, samples and extracts of axenic nutrient medium were injected randomly using a dedicated Excel macro (Bertrand et al., 2013), while MeOH blanks and QC samples were injected every 15 samples. The QC sample was prepared by mixing 30 μl of all samples together. Using the same LC‐HRMS method, an OA standard (Diagnostic Chemicals Limited, Charlottetown, CA USA) was injected at 10 ng/ml and was detected in positive ionization at retention time (t R) 14.6 min as 827.463 [M + Na]+ (main ion). The DTX‐1 was detected at t R 17.1 min as m/z = 841.473 [M + Na]+. All data were deposited in Zenodo (http://doi.org/10.5281/zenodo.7050181).
Data analysis
LC‐HRMS data were exported as CDF files using LC Solution (Shimadzu) to allow automatic peak picking using MZmine 2 as described thereafter (Pluskal et al., 2010). Briefly, mass detection was performed using a centroid algorithm with a noise level of 15,000 arbitrary abundance units (AU), derived from visual inspection of data. Chromatogram building used a minimum time span of 0.03 min, group intensity threshold of 100,000 AU, again a minimal of intensity of 15,000 AU and m/z tolerance of 30 ppm (Myers et al., 2017). Peak deconvolution was performed using the baseline cut‐off algorithm with median m/z centre calculation, and a peak duration range between 0.05 and 10 min. Chromatograms were deisotoped with m/z tolerance of 1 mDa, retention time tolerance of 0.1 min and maximum charge of 3. The peak list was aligned using the Join‐aligner algorithm with an m/z tolerance of 1 mDa or 30 ppm (as used by MZmine), retention time tolerance of 0.2 min, using the same weight for retention time and m/z importance. Subsequently, the gap‐filling step was performed with a peak‐finder algorithm, intensity tolerance of 1.0, m/z tolerance of 1 mDa or 30 ppm, retention time tolerance of 0.2 min. The results were exported as a.csv file containing all peaks observed and referenced by their mass‐to‐charge ratio (m/z) and retention time (t R) together with their respective peak area in each sample. All peaks detected in both the extracts of supernatant and solid phases were concatenated for each of the five biological replicates, generating the final data matrix. The final matrix was therefore composed of peak information from both extract types (with traceability of feature origin). In this way, the metabolic modification of any feature within the biological replicate was highlighted through statistical analysis.
The peaks detected in blanks were withdrawn from the data matrix as background signal. The absence of ions corresponding to antibiotics added for treated conditions (ampicillin, gentamycin, and kanamycin) and their possible anticipated metabolites using Biotransformer (Djoumbou‐Feunang et al., 2019) (with phase I, II CYP450 and microbiota transformation) were confirmed. Analytical consistency during batch analysis was evaluated by principal component analysis (PCA) (Want et al., 2010). As QC samples were grouped within the PCA (Figure S3), no batch correction was applied. As samples were diluted to obtain the same concentration of dry weight of extract, the data matrix containing 6627 features was transformed to reflect actual metabolite content by multiplying each peak area by the corresponding amount of dry extract.
Finally, all data from one culture were concatenated to provide the final data matrix (accessible as supplementary material S1), thus data from replicated samples contained features (m/z at t R with corrected peak areas) obtained from supernatant extract profiles (NI and PI) and from solid‐phase extract profiles (NI and PI). Such concatenation was performed to keep track of feature origin (ionization and compartment). Thus, all 30 samples were characterized by 13,254 features.
The final data matrix was statistically analysed using R 4.0 (CRAN) with the PocheRmon function (Wang & Bertrand, 2019). This function allows analysis of co‐culture data univariately using the Stats package, and multivariately using both the ropls package (Thévenot et al., 2015) and the Projected Orthogonalized CHemical Encounter MONitoring (POChEMon) approach (Jansen et al., 2015) encoded in R (CRAN). The univariate data analysis was performed calculating the fold‐change between the monoculture and the co‐culture and the p‐value for each metabolite according to Student's t‐test. Features of interest were thus selected if they were significant by comparison to both monocultures, according to fold‐changes >2 and p value ≤ 0.05. The PCA and orthogonal projection to latent structure discriminant analysis (OPLS‐DA) were performed after log‐transformation of data and Pareto scaling (van den Berg et al., 2006). The feature selection using OPLS‐DA was performed building two different models comparing co‐culture to each monoculture. Features of interest were selected if their Variable Importance on the Projection (VIP) score was higher than 2 in both models. The POChEMon approach provided two different models (Jansen et al., 2015). First, the mixing model highlighted differences between both monocultures and then provided information about co‐culture similarity to monoculture by the projection of the latter samples within the model. Then, the competition model focussed on analysing the residue of the mixing model, thus highlighting the co‐culture sample specificity. POChEMon model was validated by leave‐one‐out strategy (Figure S4). Finally, the features were selected according to SSrank value (rank product s j in the original publication (Jansen et al., 2015) and the top‐20 features of interest were selected). Features highlighted by the three feature selection approaches were merged (Table 1) and confirmed manually in the raw LC‐HRMS data to remove false peaks detected during the automated peak picking step.
TABLE 1.
Co‐culture associated features using univariate and multivariate data analyses using PocheRmon package
| Detected m/z (ionization mode) | t R (min) | Matrix | Antibiotics | Statistical method | Presence in monoculture (MC) | Molecular formula (error, ppm) | Putative annotations with reported biological sources and adducts a |
|---|---|---|---|---|---|---|---|
| 243.7550 (−) | 1.0 | A | Both | OPLS‐DA | F, M | ||
| 254.9880 (+) | 1.1 | A | Both | OPLS‐DA | F, M | ||
| 433.0163 (−) | 9.0 | Both | No | Ttest, POChEMon | ND | ||
| 435.0291 (+) | 9.0 | L | No | Ttest | ND | ||
| 433.0172 (−) | 8.4 | L | No | Ttest (∞), POChEMon | ND | ||
| 273.0787 (−) | 11.0 | Both | No | Ttest (∞) | ND | ||
| 564.3306 (+) | 11.0 | L | No | OPLS‐DA | ND | ||
| 311.2244 (−) | 12.1 | A | Yes | POChEMon | F | C18H32O4 (5 ppm) | Sporogenic Psi Factor Cα (Mazur et al., 1991) or QMZ37‐I (Qiao et al., 2011) (Aspergillus) [M‐H]‐ |
| 418.0254 (−) | 13.0 | L | No | POChEMon | ND | C15H17N3O5S2 (−12 ppm) | Glionitrin B (Park et al., 2011) (Aspergillus) [M + Cl]− |
| 329.1390 (−) | 14.0 | L | Yes | Ttest | ND | C19H22O5 (−2 ppm) | Arahypin 2 or Arahypin 3 (Sobolev et al., 2011) (Aspergillus) [M‐H]− |
| 415.0061 (−) | 13.2 | L | No | Ttest (∞), POChEMon | ND | ||
| 544.3293 (+) | 11.5 | L | Yes | Ttest (∞) | M | ||
| 442.2601 (+) | 14.2 | L | Yes | Ttest | M | ||
| 606.3650 (−) | 15.2 | L | Yes | Ttest | ND | ||
| 522.5520 (+) | 16.1 | L | No | Ttest | ND b | ||
| 634.3363 (−) | 16.1 | L | No | OPLS‐DA | ND | ||
| 506.3265 (−) | 16.1 | L | No | POChEMon | ND | ||
| 509.2905 (−) | 17.0 | Both | No | OPLS‐DA | M | ||
| 544.4092 (−) | 17.0 | L | No | OPLS‐DA | ND | ||
| 581.3034 (−) | 16.3 | L | No | Ttest | F, M | ||
| 486.2516 (−) | 16.3 | L | No | OPLS‐DA | ND | C28H37NO4 (21 ppm) | Dodecahydro‐11a‐(1H‐indol‐3‐yl)‐1,3a,4,9‐tetramethyl‐8,11c‐methano‐11cH‐benzo[ef]furo[3,2‐c][1]benzoxepin‐3,8a(8H)‐diol (Forseth et al., 2012) (Aspergillus) [M + Cl]− |
| 580.3630 (−) | 17.2 | L | No | Ttest | ND | ||
| 563.3460 (−) | 17.2 | A | No | OPLS‐DA | M | ||
| 514.9216 (−) | 12.8 | L | No | Ttest, POChEMon | ND | ||
| 417.0203 (−) | 13.7 | L | No | Ttest (∞), POChEMon | F | C19H14N2O5S (−27 ppm) | 4‐Methylether‐4′‐sulfate‐xanthocillin X (Antibiotic BU 4704) (Takatsuki et al., 1968) (Aspergillus) [M + Cl]− |
| 419.2678 (−) | 16.4 | A | No | OPLS‐DA | M | ||
| 508.3407 (+) | 16.4 | L | No | OPLS‐DA | ND | ||
| 485.0060 (−) | 12.9 | L | No | Ttest (∞) | ND | ||
| 516.9162 (−) | 12.9 | L | No | Ttest (∞), POChEMon | ND | ||
| 417.0222 (−) | 12.9 | L | No | Ttest, POChEMon | F | C19H14N2O5S (−23 ppm) | 4‐Methylether‐4′‐sulfate‐xanthocillin X (Takatsuki et al., 1968) (Aspergillus) [M + Cl]− |
| 419.0378 (+) | 12.9 | L | No | Ttest, POChEMon | ND | ||
| 464.2782 (−) | 14.8 | L | No | OPLS‐DA | ND | ||
| 507.3061 (+) | 18.4 | L | No | OPLS‐DA | ND | ||
| 566.3484 (−) | 15.8 | L | No | POChEMon | M b | ||
| 509.2915 (−) | 17.6 | L | No | OPLS‐DA | ND | ||
| 542.9179 (+) | 15.9 | Both | No | Ttest (∞), POChEMon | ND | ||
| 542.9179 (−) | 15.9 | L | No | POChEMon | ND | ||
| 523.3067 (−) | 18.6 | A | No | OPLS‐DA | M | ||
| 444.4044 (+) | 22.2 | L | No | Ttest | ND | ||
| 509.9216 (−) | 16.9 | L | No | POChEMon | ND | ||
| 509.7083 (−) | 16.9 | L | No | POChEMon | ND | ||
| 509.2918 (−) | 18.7 | A | No | OPLS‐DA | ND | ||
| 542.4092 (+) | 18.8 | L | No | OPLS‐DA | ND | ||
| 655.5283 (+) | 27.2 | L | No | OPLS‐DA | M | ||
| 545.3470 (−) | 24.8 | A | Both | Ttest | ND | ||
| 802.6100 (+) | 26.7 | L | Yes | POChEMon | M | ||
| 584.4907 (+) | 26.8 | L | No | OPLS‐DA | ND | ||
| 566.4814 (−) | 27.8 | L | No | OPLS‐DA | ND | ||
| 568.4942 (+) | 27.8 | L | No | OPLS‐DA | ND |
Note: Features highlighted using Ttest method correspond to top 20 ones with fold changes higher than 2 with Student's t‐test p value below 5%. The OPLS‐DA method select up‐regulated features with variable importance in the projection (VIP) value higher than 2. Finally, the POChEMon method highlights features with SSrankProduct higher than 0.995. The drastic variable selection yielded low number of features highlighted by different analysis.
Abbreviations: A, detected in Agar; L, detected in Liquid medium; MC, monoculture; ND, not detected in any monoculture replicates; OPLS‐DA, orthogonal projection to latent structure discriminant analysis; POChEMon, Projected Orthogonalised CHemical Encounter MONitoring; ∞, de novo induced features according to Ttest.
Level 3 annotation quality was based on Metabolomics standard initiative (MSI; Sumner et al., 2007)—which correspond to an annotation based on mass and spectral accuracy of the MS1 spectrum and biological source consistency.
Detected in bacterial extracts.
Annotation of statistically relevant features
Highlighted features (Table 1) were further annotated using traditional de‐replication strategy (Wolfender et al., 2019) based on mass and spectral accuracy of the MS spectra. Accurate masses (up to 30 ppm for smaller peaks) were searched taking into consideration adduct mass correction ([M + H]+, [M + Na]+, [M − H]−, [M + Cl]− and [M + formic acid‐H]−; Nielsen et al., 2011) for compounds reported to be produced by either Prorocentrum sp. and Aspergillus sp. (also considering its teleomorph Eurotium sp., Chen, Hubka, et al., 2017) in the Dictionary of Natural Products® (CRC Press, v.29.2) (Chen, de Bruyn Kops, & Kirchmair, 2017). This corresponds to level 3 annotation based on Metabolomics Standard Initiative (MSI) (Sumner et al., 2007). Furthermore, OA standard (Diagnostic Chemicals Limited, Charlottetown, CA USA) was also injected in the LC‐HRMS run sequence and was detected at tR 14.62 min m/z 803.4683 [M‐H]−. This corresponds to level 1 annotation based on MSI (Sumner et al., 2007).
RESULTS
Filamentous fungi were isolated from Prorocentrum lima biomass
To evaluate the presence of fungi in P. lima microbiome, biomass from PL4V strain was used as inoculum for cultivation in a fungal adapted medium (Sallenave‐Namont et al., 2000). Antibiotic (chloramphenicol) was added in the culture to avoid bacterial growth and thus to allow fungal mycelium development. Cultivation was achieved either in liquid or solid conditions (Table S1). After few weeks, fungal colonies were observed and isolated (Figure S5). All strains were identified based on fungal morphology resulting in the isolation of two Penicillium sp. strains (MMS1593, MMS1596) and three Aspergillus sp. strains (MMS1589, MMS1591, MMS1594).
Performing a microbial co‐culture experiment to highlight metabolic modifications according to recent guidelines (Arora et al., 2020) represents a rather complex challenge. As no studies report examples of dinoflagellates grown together with fungi, it was decided to select one fungal strain among the five isolated to perform a microbial co‐culture experiment. Among various isolates, one strain (MMS1589) possessed an unusual morphology with cleistothecia and conidia simultaneously observed (Figure S6), whereas no cleistothecia were observed in the other fungal strains isolated from P. lima. This simultaneous presence of the sexual and asexual reproduction is quite rarely observed in fungi and therefore attracted our attention to further study this strain. After Congo red coloration and microscopic observation, strain MMS1589 revealed a hyaline septate mycelium and conidiophores, bearing uniseriate totally covered vesicles. Conidia were round in shape. Globose cleistothecia were also visible, and sci and ascospores were present next to them. Phylogenetic analyses were performed based on the genes encoding for the fungal small subunit ribosomal RNA (SSU rRNA), internal transcribed spacer 1 (ITS) and β‐tubulin of MMS1589 (all sequences were deposited in GenBank: MN134000.1 and MN164633.1). The MMS1589 ITS partial gene had 99% of identity with the A. pseudoglaucus strain FJAT‐31014 SSU rRNA (partial sequence) and the MMS1589 β‐tubulin partial gene had 99% of identity with the A. pseudoglaucus benA gene for β‐1 and β‐2 tubulin (partial sequence). Finally, this strain was identified as A. pseudoglaucus, formerly known as Eurotium repens (Chaigne, Bertrand, et al., 2017; Chen, Hubka, et al., 2017).
A specific microscale marine‐like environment was designed for co‐culture
The study of chemical interactions between P. lima PL4V and A. pseudoglaucus MMS1589, involved the challenging design of a suitable micro‐environment for microalgal–fungal co‐culture since both species have specific requirements for their growth. On one hand, growth of the benthic dinoflagellate P. lima requires a liquid medium (f/2 medium) without stirring, without organic carbon source and with a light source for night‐and‐day cycle (Moreira‐González et al., 2019). As P. lima is a benthic species, it grows as a biofilm on the bottom of the culture flask. On the other hand, fungal growth requires either liquid or solid media with an organic carbon source. However, A. pseudoglaucus growth in liquid medium without stirring yielded fungal colonies floating over the liquid media, avoiding any contact with the benthic dinoflagellate. Thus, it was decided to culture the fungus on a solid medium (containing a low level of glucose, 2 g/L) prior to the addition of a liquid medium in the culture flask and inoculation of P. lima.
When both organisms were cultivated separately in the composite liquid/solid environment with light (12 h/12 h night‐and‐day cycle), the following observations were made (Figure 1): (1) increase of the fungal colony size at the bottom of the flask (on the surface of the agar layer) and (2) formation of P. lima mucus cellular aggregates. During P. lima culture, bacterial development was observed leading to a cloudy culture medium (Figure 1), as confirmed by microscopic observation. Thus, to study microalgal growth with a very limited bacterial development, a cocktail of antibiotics (ampicillin, gentamycin and kanamycin) was added in parallel (Cho et al., 2002).
FIGURE 1.
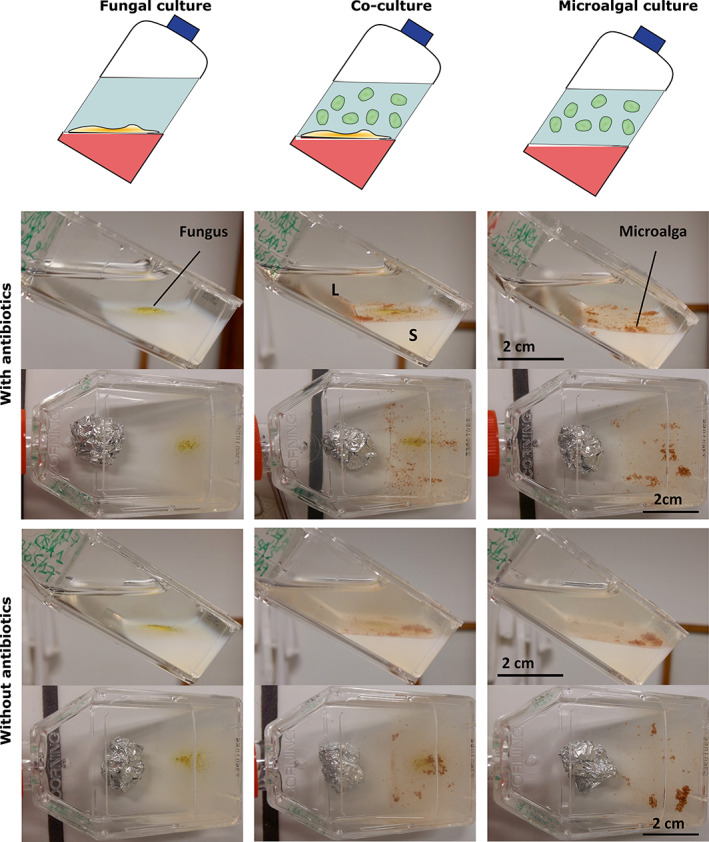
Experimental design of microscale co‐culture experiments of marine benthic chemical interactions (6 incubation days). Notice the clarity of the aqueous supernatant when microalgal inoculum is treated with antibiotics. L, liquid; S, solid
Besides bacterial development in the supernatant, the morphology of both microorganisms in monoculture was similar in absence (non‐treated monocultures) or presence (treated monocultures) of antibiotics. In the case of P. lima PL4V, typical aggregation was observed. Visual examination of co‐cultured samples did not highlight any morphological difference of the microorganisms in comparison to monocultures. However, a biofilm of P. lima PL4V was formed within and around the A. pseudoglaucus MMS1589 colonies. Only a limited number of free P. lima PL4V cells in this biofilm stuck directly to the agar layer while most of the biofilm was found directly in contact with fungal structures (Figure 2A,B), possibly using its filaments as anchor to stick to the solid phase at the bottom of the flask or to the mycelium (Figure 2A,B). The mycelium of A. pseudoglaucus MMS1589 seemed to connect to the dinoflagellates at the flagella location. The red coloration of the connecting organelle indicated its fungal origin, as Congo Red dye is known to stain in red fungal membranes (Figure 2C,D).
FIGURE 2.
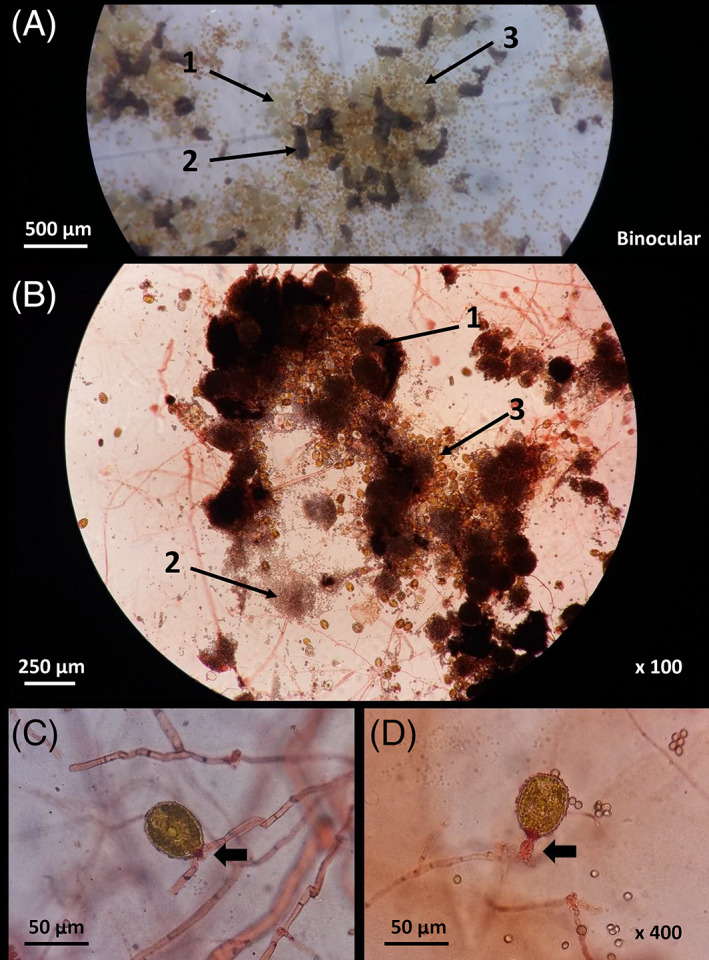
Observation of microalgal–fungal interaction. (A) Binocular observation of co‐culture with aggregated cleistothecia (1) and conidia (2) from Aspergillus pseudoglaucus, and Prorocentrum lima cells (3). (B) Microscopic observation of co‐culture using Congo red staining (×100) highlighting cleistothecia (1) and asexual sporogenic spores (2) of A. pseudoglaucus, and P. lima cells (3). (C, D) Microscopic observation of co‐culture with Congo Red dye (1% in ethanol) (×400). The fungal mycelium is indicated (black arrows) at proximity of the apex of the microalgae where the physical interaction occurs.
Co‐culture of Aspergillus pseudoglaucus MMS1589 with Prorocentrum lima PL4V induces specific metabolites
A co‐culture experiment was carried out with P. lima PL4V and A. pseudoglaucus MMS1589 in the presence and absence of antibiotics (Cho et al., 2002). To unravel early response to co‐culture, the experiment was stopped after 6 days. Then, solid agar phase (bottom part) and supernatant were collected and extracted separately to detect features in each compartment. The solid phase contained fungal mycelium and dinoflagellate cells, while the supernatants contained extracellular compounds, possibly secreted by both organisms. Dry weights of extracts of either compartment (solid or liquid) obtained under different culture conditions did not show any impact of the presence of antibiotics (Figure S2). While the presence of antibiotics in the co‐culture tended to reduce the dry weight of the extract, this effect was not significant, with a p value above 0.05 (Student's t‐test). To obtain a clear view of metabolites present in both types of extracts, they were profiled by LC‐HRMS (Roullier et al., 2016).
To get a global overview of the chemical differences in this metabolomics experiment, a quick look at base peak chromatograms (Figures S7 and S8) was conducted and the number of detected features in each condition was determined (Figure S9). It appeared that monocultures and co‐cultures conditions were not affected by antibiotics presence or absence in the number of detected peaks (statistically insignificant, p value > 0.05 by Student's t‐test), although base peak chromatograms tended to show an effect on fungal monocultures. This might be due to differences in the dry extract weight, while the injected quantities were identical during the HLPC‐HRMS analysis. Co‐culture base peak chromatograms appeared as the combination of both monoculture conditions base peak chromatograms. Therefore, while searching for differences, it should not be neglected that there are only several dozen intense peaks that can be observed in the base peak chromatograms while there are over 6600 features present in the data matrix. Some of these, while minor in intensity, may play an important role in the chemical interaction between P. lima PL4V and A. pseudoglaucus MMS1589. Thus, an in‐depth comparison of the full profiles using a metabolomics strategy was mandatory to highlight chemical variations related to co‐culture. For this in‐depth exploration, the data matrix resulting from MZmine 2 peak picking workflow (see Data Analysis section) was subjected to unsupervised statistical analysis through PCA, to assess the self‐organized metabolome differences between P. lima PL4V and A. pseudoglaucus MMS1589 under monocultures and co‐culture conditions (Figure 3A). The first two components of the resulting PCA model corresponded to 22.1% (PC1) and 11.3% (PC2) of explained variance. This model clearly highlighted the presence of three clusters related to P. lima PL4V, A. pseudoglaucus MMS1589 and their co‐culture. The first component (PC1) was related to the differences between the P. lima PL4V and A. pseudoglaucus MMS1589 monocultures, and the second component clearly differentiated between monocultures and co‐culture conditions.
FIGURE 3.
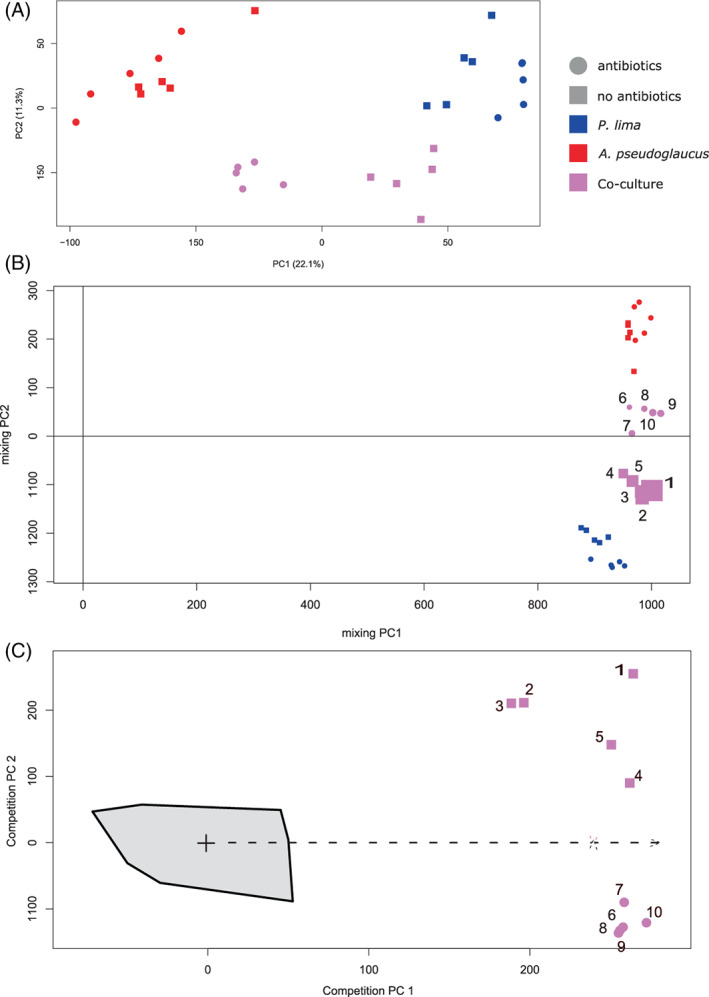
Unsupervised data analysis (n = 5) of LC‐HRMS data showing differences between Prorocentrum lima, Aspergillus pseudoglaucus and their co‐culture samples, in absence or presence of antibiotics. (A) Principal component analysis (log transformed data, pareto scaling); (B) Mixing model using Projected Orthogonalised Encounter Monitoring (POChEMon) data analysis (Jansen et al., 2015). In the mixing model, the size of the co‐culture samples reflects the amount of their unexplained variance within the mixing model (the larger the point, the higher the unexplained variance); (C) Competition model using Projected Orthogonalised Encounter Monitoring (POCheMon) data analysis. The grey zone corresponds to the projected co‐culture domain within the competition model.
The PCA scores plot (Figure 3A) also highlighted a clustering of samples according to the non‐treated xenic and axenic treated status for each culture conditions, with a smaller difference within A. pseudoglaucus MMS1589 monoculture condition (as observed on the PCA with a tight clustering). As signals related to antibiotics were removed from the data, the antibiotic clustering effect was mainly linked to a metabolome alteration due to the influence of bacteria on P. lima PL4V metabolome (chemical and/or physical influences). In the presence of antibiotics, the chemical profiles of the P. lima PL4V monoculture corresponded almost only to compounds produced by the dinoflagellate, while without antibiotics the chemical profiles reflected the metabolome of the microalgae with its associated bacteria as a whole.
Considering addition of antibiotics, greater differences between P. lima PL4V metabolomes under xenic non‐treated and treated axenic conditions were observed in comparison to fungal ones under both conditions (Figure 3A), suggesting that the presence of antibiotics altered the microalgal metabolite profiles (also considering the disappearance of potential bacterial metabolites) with greater significance than the fungal ones (A. pseudoglaucus MMS1589 being already in axenic condition without antibiotics). This difference in the case of microalgal metabolite profiles can also be explained by the relative absence of bacterial metabolites. Furthermore, an even greater impact of the non‐treated condition was observed within co‐cultures. According to the presence or absence of antibiotics, co‐culture replicates appeared even more differentiated across the first component axis. This reflected the impact of each participant of the co‐culture on the metabolome of the other participant. The absence of antibiotics shifted co‐cultured replicates towards the P. lima PL4V monoculture samples (Figure 3A). This highlighted the mutual metabolic changes induced by co‐culture on A. pseudoglaucus MMS1589 and on P. lima PL4V with its associated bacteria. Such conditions, without antibiotics, tended to alter chemical profiles of the co‐culture to resemble more those of the microalgae (and associated bacteria).
This unsupervised analysis did not point out the specific metabolome alteration induced by fungal presence as it encompassed every variation (microbiome‐ or antibiotics‐derived variations). Thus, a more co‐culture centric data mining strategy was used (Bertrand, Azzollini, et al., 2014; Bertrand, Bohni, et al., 2014), called Projected Orthogonalised CHemical Encounter MONitoring (POChEMon) (Jansen et al., 2015). Such an approach uses an unsupervised mixing model, obtained by statistically mixing data from chemical profiles corresponding to strains grown alone whether antibiotics are added or not. This yielded an in silico co‐culture model in absence of co‐culture‐related biotic stress. Then, projection of co‐culture samples within this mixing model allowed, as for the PCA, to compare chemical profiles of the co‐culture to those of monocultures (Figure 3B). This approach further confirmed results from the PCA, showing that in absence of antibiotics the co‐culture metabolome is dominated by the P. lima PL4V one (Figure 3B). Similarly, the presence of antibiotics made the chemical profiles of the co‐culture resemble more to the fungal profiles, the chemical signature of A. pseudoglaucus being more present in the extracts. Additionally, the POChEMon approach provided a global comparison between all co‐culture experiments using the residual information (statistical residue not represented in the mixing model) (Figure 3B). The size of the co‐culture samples (Figure 3B) when represented was directly related to unrepresented variance in the mixing model (Jansen et al., 2015). The size of this residual information (unrepresented variance in the mixing model) was directly related to the amount of information within the dataset related to metabolic alteration during co‐culture for each co‐culture sample. Here, this provided a strategy of unsupervised comparison between the two co‐culture conditions (with or without antibiotics). Clearly, in the non‐treated condition, the size of residual information of the co‐culture sample (statistical residue not represented in the mixing model) was larger than in treated condition (Figure 3B), which means more chemical alteration due to co‐culture in absence of antibiotics.
The co‐culture induction patterns can be explored by interpreting the POChEMon competition model (Figure 3C) (Jansen et al., 2015). This representation corresponded to the unsupervised analysis through PCA of the co‐culture sample residue when co‐culture data were projected in the mixing model. Thus, the POChEMon approach allowed to identify co‐culture related metabolome up‐regulation in every sample (considering de novo induction as a specific case of up‐regulation). In Figure 3C, two co‐culture induction patterns (in presence or absence of antibiotics) were observed and well separated according to the second axis. Interestingly, the overall variability among co‐culture replicates was 10 times lower in presence of antibiotics in comparison to their absence (based on PC1 and PC2 sample distribution).
Identification of co‐culture up‐regulated compounds highlighted Prorocentrum lima toxin induction
As co‐culture induction is a complex mechanism, different complementary supervised statistical approaches were used to highlight induced features (Saccenti et al., 2014). Data were analysed univariately using Student's t‐test and multivariately using both OPLS‐DA and the POChEMon approach (Jansen et al., 2015). The first two methods (univariate and OPLS‐DA) were based on the comparison of the co‐culture to the monoculture independently. Those two approaches selected variables that were significant in both comparisons. Alternatively, the POChEMon approach selected features responsible for the differences between an in silico mixture of both organisms and the co‐culture profiles. This approach has the advantage to highlight features of specific groups of replicates (Jansen et al., 2012, 2015). This is particularly appropriate for analysing simultaneously treated and non‐treated profiles. In all cases, the top 20 most relevant features obtained by each statistical method were selected and non‐redundant ones were reported in Table 1 along with their retention time, m/z value and the statistical approach used to highlight them. Due to the drastic selection of features, low redundancy between statistical approaches was observed (Figure S10). Finally, a set of 49 features was selected as over‐produced (or de novo induced) during co‐culture. Among those features, 12 were detected in the P. lima PL4V extracts (M), 6 in the A. pseudoglaucus MMS1589 extracts (F) and 34 were not detected in any of the monocultures (ND) (Table 1). Considering these 49 features, the following distribution was obtained: 8 and 37 features were observed specifically in agar and liquid media, respectively, the remaining 4 features were common to both media (Figure S10). A majority of features was found in non‐treated conditions (39/49 features). As observed in Table 1, most of those features were highlighted in the supernatant (32 out of the previously selected 39 features). Additionally, six features were only regulated when antibiotics were added, hence confirming a microalgal–fungal interaction‐related chemical induction. Finally, none of the highlighted features were pointed out in three or all four conditions.
Based on high mass and spectral accuracy of the MS1 data, the highlighted features were putatively annotated (Table 1) along with annotation accuracy based on MSI (Sumner et al., 2007). Considering the most significant features, none of the highlighted features was successfully annotated as Prorocentrum spp. metabolites. Furthermore, fungal metabolites annotation was only possible by extending the search to the clade Aspergillus section restrictii. As expected, only a very low number of features were identified (Level 3 annotation based on MS1 spectrum and biological source consistency).
Besides the previously highlighted features (Table 1), the two known toxins OA and DTX‐1 produced by P. lima PL4V were also studied. Both toxins were found induced in co‐culture in the liquid phase using univariate data analysis (Figure 4). OA (main ion detected at tR 14.6 min as m/z = 827.458 [M + Na]+ − 827.456 for C44H68O13Na) was significantly up‐regulated in co‐culture only without antibiotics (not detected in any blanks). In comparison, DTX‐1 (main ion detected at t R 17.1 min as m/z = 841.473 [M + Na]+ − 841.471 for C45H70O13Na) was up‐regulated in co‐culture with or without antibiotic addition to the culture medium (not detected in any blanks). No significant variation was observed in the solid medium profiles (Figure 4) and in between blank samples spiked with OA and DTX‐1 (Figure S11).
FIGURE 4.
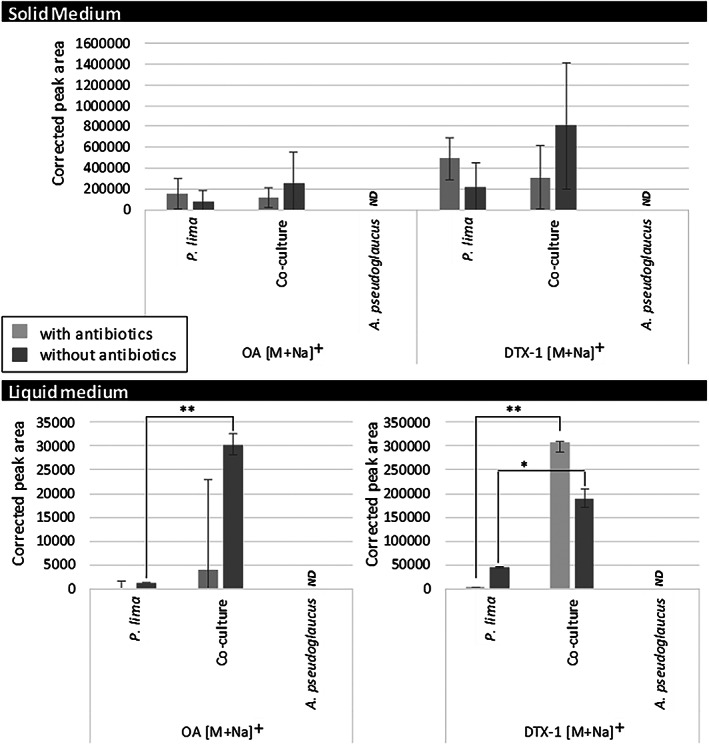
Okadaic acid (OA) [M + Na]+ (m/z = 827.463) and dinophysistoxin 1 (DTX‐1) [M + Na]+ (m/z = 841.473) corrected peak area (peak area corrected by amount of extract) detected within LC‐HRMS data in positive ionization mode according to the type of samples where it was detected. (ND: not detected; variation significance based on student test: *p‐value ≤ 0.05, ** p‐value ≤ 0.01)
DISCUSSION
The functional roles of fungi are relatively underexplored in the marine environment (Amend et al., 2019; Worden et al., 2015). In the ocean, epiphytic fungi were regularly reported associated with algae (Pasqualetti et al., 2020), and can also, more specifically, interact with dinoflagellates (Gémin et al., 2020; Pasqualetti et al., 2020). In such an environment, interactions between microorganisms are highly complex. Beside the presence of microalgae and fungi, bacteria may also play key roles in the algal microbial community (Egan et al., 2013; James et al., 2020).
The present study focused on interactions between the toxic dinoflagellate P. lima PL4V and an associated fungal strain. In fact, various fungal strains from the Penicillium and Aspergillus genera were isolated from the P. lima PL4V strain. Based on the available information, we cannot assert the origin of the fungi. This strain was either isolated as part of the natural microbiome associated with P. lima PL4V since its collection in 1985 (Bravo et al., 2001) or rather the result of a contamination. However, we expect they may co‐occur as such taxa are commonly isolated from marine samples (Amend et al., 2019; Chaigne, Bertrand, et al., 2017; Chen, Hubka, et al., 2017; Nguyen et al., 2020; Sallenave‐Namont et al., 2000). The A. pseudoglaucus strain MMS1589 was selected to further explore chemical mediation between microalga and fungi. Thus, a simple marine microcosm, compatible with fungal and benthic dinoflagellate life style, was designed to study such interaction. Production of specialized metabolites during co‐culture was studied using an LC‐HRMS metabolomics approach after 6 days of growth. Such rather short growth time corresponded to the beginning of the exponential phase of P. lima PL4V (Moreira‐González et al., 2019) and the moment when co‐culture metabolite induction started to be observed (Azzollini et al., 2018; Bertrand, Azzollini, et al., 2014; Bertrand, Bohni, et al., 2014).
The differences in chemical alteration between P. lima PL4V treated and non‐treated conditions clearly emphasized the effect of associated bacteria in the microalgal‐fungal interaction (Figure 3). Interactions between P. lima and bacteria were shown to alter algal metabolites (Tarazona‐Janampa et al., 2020), and it is thus expected that in the present study the bacterial presence (absence of antibiotics) also significantly impacted the metabolite profile. Finally, this showed that, in addition to dinoflagellate signals, the microbiome metabolism was also altered. However, one may wonder if the effect observed here was only related to the addition of antibiotics, which resulted in reduced bacterial abundance. Such a cocktail had already been evaluated for Isochrysis galbana and no growth modification was observed (Cho et al., 2002). In addition, careful examination of toxin production did not show any significant alteration of the toxin production related to the presence of antibiotics (Figure 4 and Figure S11). Thus, the metabolic alteration highlighted in the present study was mostly related to co‐culture alteration in presence and reduced abundance of co‐occurring bacteria. Up to now, chemical induction was mostly reported in fungal–fungal, bacterial–bacterial and fungal–bacterial co‐cultures (Arora et al., 2020).
The chemical signals were tentatively identified considering they were produced by P. lima PL4V and/or A. pseudoglaucus MMS1589. Among the putative annotations for compounds produced in mono‐cultures, Xanthocillin X was reported to possess antiviral activities (Takatsuki et al., 1968), and 5S,8R‐dihydroxy‐9Z,12Z‐octadecadienoic acid (m/z = 311.2244 at t R 12.11 min) was previously reported as psi Cα sporogenic factor of Aspergillus spp. (Mazur et al., 1991), which could reflect a fungal biotic stress adaptation behaviour. Unfortunately, among the features highlighted in co‐cultures only a very limited number of compounds was putatively annotated. The reason may be that (1) frequently, microbial co‐culture induces previously unreported compounds (Arora et al., 2020; Bertrand et al., 2013) and that (2) metabolomes of toxic dinoflagellates are still largely unexplored except toxins (Zendong et al., 2016). Thus, a large effort in studying the metabolome of such organisms will be of great value to better understand their metabolism. Still, two induced compounds were unequivocally identified, OA and DTX‐1, produced by P. lima PL4V. This is particularly interesting, as these toxins are largely involved in seafood contamination (Bauder et al., 2021; Lawrence et al., 2021). In the present study, chemical signals from A. pseudoglaucus MMS1589 and/or physical contact induced the up‐regulation of OA and DTX‐1 in the surrounding liquid environment. Similar results were reported with the domoic acid producer Pseudo‐nitzchia where variation in bacterial community altered toxin levels (Kobayashi et al., 2009; Sison‐Mangus et al., 2014). However, the role of fungi in the modulation of algal toxins remains unknown. Thus, they should be considered to potentially play a significant role in such regulation. Interestingly, OA and its derivatives (DTX‐1 and 7‐deoxyokadaic acid) are known to inhibit Aspergillus niger growth in a paper‐disc assay (Nagai et al., 1990). Thus, OA and DTX‐1 over‐production may reflect that P. lima PL4V may control the fungal growth. Such inhibition was already observed during P. lima interaction with other microalgae (Windust et al., 1996). Thus, toxins produced by P. lima may be part of its global defence mechanism.
During the P. lima PL4V and A. pseudoglaucus MMS1589 interaction, aggregation was also observed (Figure 2A,B). Similar aggregation followed by flocculation was previously reported in biotechnology‐focused microalgal–fungal co‐cultures (Leng et al., 2021). Such aggregation also corresponds well to the epiphytic nature of P. lima which needs to attach to substrate to form a biofilm (Lawrence et al., 2021). This may also suggest the occurrence of nutrient exchange between both organisms, as it could be facilitated by their very close proximity. However, such explanation needs more in‐depth study to be proven. A similar idea was recently explored in the synthetic association between the microalga Chlamydomonas reinhardtii and the yeast Saccharomyces cerevisiae (Hom & Murray, 2014). In this specific association, the two species were forced, under specific conditions, to mutually support each other's growth. In a recent study, during the tripartite interaction between C. reinhardtii, Aspergillus nidulans and Streptomyces iranensis, the fungal partner was demonstrated to physically defend the microalga from the bacterial algicidal compound azalomycin F by forming a protective lipid coat as observed by microscopy (Krespach et al., 2020).
Besides aggregation, and very unexpectedly, the microscopic observation of the interaction between P. lima PL4V and A. pseudoglaucus MMS1589 showed a very unusual physical contact between both organisms. Formerly, the fungal mycelium seemed connected to the dinoflagellate at the flagella location (Figure 2). Such synthetic structured community formation is rather unusual in the literature probably due to the difficulties to observe such type of interaction within the open environment. However, one may consider such contact as a very early stage of lichen formation (Ahmadjian et al., 1978; Trembley et al., 2002). Lichens are classified as a microalgal–fungal symbiosis harbouring various other species to form complex ecosystems (Hawksworth & Grube, 2020). Aspergillus pseudoglaucus was not previously mentioned as a lichen forming fungus (Honegger, 2009); however, some Aspergillus sp. were reported as endolichenic species having antimicrobial properties (Dou et al., 2014; Padhi et al., 2020; Prateeksha et al., 2020).
To conclude, this study raised many questions about microalgal–fungal interactions in the marine environment, especially in the context of toxic microalgae. Further studies are still needed to identify many of the metabolites induced either by A. pseudoglaucus MMS1589 or P. lima PL4V and thus better understand their role in their chemical interaction. For example, it would be of interest to identify the metabolites responsible of OA induction in P. lima PL4V to clarify toxin production regulation. It also would be interesting to understand the function of this unusual physical contact between both partners: is it beneficial for the microalgae as a substrate to fix on/or as a feeding source (i.e. mixotrophy), or beneficial for the fungi through carbon exchange? Thus, exploring this difference of carbon use and flux between the mixotroph dinoflagellate (Johnson, 2015) and the heterotroph fungi during their co‐culture may highlight the still underexplored role of fungi in the marine environment (Worden et al., 2015).
Finally, one may consider the impact of some fungi in the marine environment as a potential risk of microalgal toxicity increase, as demonstrated here, they may act as toxin inducers by chemical mediation or as potential toxin effectors (Ruiz et al., 2010). This should be taken into consideration in regard to recent reports of fungal blooms in the marine environment (Hallegraeff et al., 2014; Hayashi et al., 2016).
AUTHOR CONTRIBUTION
Conceptualisation and methodology: EB, OG, NR, YFP, PH and SB. Investigation: OB, AB, MC, LMC, JW and TRdP. Formal analysis: OB, EB, AB, MC, JJJ, PH and SB. Writing—original draft: OB, EB, LMC and SB. Writing—review and editing: All authors.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
Supporting information
Table S1. Fungal strains isolated from Prorocentrum lima PL4V.
Figure S1. Scheme of the microscale environment designed for this study.
Figure S2. Amount of extract (with standard deviation) obtained in each culture condition.
Figure S3. Principal component analysis (log transformed data with pareto scaling) using peaks detected in the LC‐HRMS profiles in positive (A) and negative (B) ionization modes. QC samples are highlighted in red.
Figure S4. Validation of the POChEMon mixing model, using leave‐one‐out strategy, removing either a mono‐culture sample (A) or a co‐culture sample (B). Sample distribution was represented as a violin plot (in blue: the monoculture sample distribution; in red the co‐culture sample distribution). The removed sample was plotted back (red dot) within the generated model to highlight that no misclassification was observed.
Figure S5. Morphology of fungal strains isolated from Prorocentrum lima PL4V after 3 days (top) after 10 days (bottom). The growth was performed in 5‐cm Petri dishes.
Figure S6. Morphology of Aspergillus pseudoglaucus MMS1589 (binocular view). Cleistothecia (1) and classic mycelium bearing conidiophores (2).
Figure S7. LC‐HRMS profiles in negative ionization of extracts used for metabolomics (only one replicates is presented here). All extracts were profiled at 1 mg/ml concentration and chromatograms were all scaled (y) to the same ion count intensity (inter‐chromatogram y‐step corresponds to 84,000 counts).
Figure S8. LC‐HRMS profiles in positive ionization of extracts used for metabolomics (only one replicates is presented here). All extracts were profiled at 1 mg/ml concentration and chromatograms were all scaled (y) to the same ion count intensity (inter‐chromatogram y‐step corresponds to 140,000 counts).
Figure S9. Number of detected features per conditions, obtained with MZmine 2 processing of LC‐HRMS data. S: supernatant, A: agar solid phase.
Figure S10. Venn Diagram of feature distribution following origin (liquid or solid phase) and antibiotic (ATB) exposure using the selected top features according to the three different statistical treatments applied on the data matrix (highest significance in Student's t‐test, highest VIPs in the OPLS‐DA, best SSrank values) and withdrawing of redundant selected features.
Figure S11. Recovery of okadaic acid (OA) and dinophysistoxin 1 (DTX‐1) after addition to blank samples (with and without antibiotics) showing the absence of extraction bias due to the presence of the antibiotic cocktail.
Additional raw data may be found on Zenodo (http://doi.org/10.5281/zenodo.7050181).
DATA S1: the data matrix as an Excel file.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the technical assistance of Véronique Séchet and Florent Malo for maintaining and providing P. lima cultures. The authors thank the ThalassOMICS Metabolomics Facility—Plateforme Corsaire, Biogenouest, Nantes—for LC‐HRMS analysis and metabolomics expertise. The authors would also like to acknowledge funding received from Ifremer and the regional research federation IUML for funding received for the Master studies of Alizé Bagot and Maud Chaigné.
Berry, O. , Briand, E. , Bagot, A. , Chaigné, M. , Meslet‐Cladière, L. , Wang, J. et al. (2023) Deciphering interactions between the marine dinoflagellate Prorocentrum lima and the fungus Aspergillus pseudoglaucus . Environmental Microbiology, 25(2), 250–267. Available from: 10.1111/1462-2920.16271
Funding information Ifremer; regional research federation IUML
DATA AVAILABILITY STATEMENT
The P. lima PL4V and A. pseudoglaucus MMS1589 strains are stored at IFREMER (PHYTOX, Laboratoire PHYSALG, F‐44311 Nantes, France), and Nantes Université (ISOMer, UR2160, Nantes, France), respectively. They both can be obtained on request.
REFERENCES
- Ahmadjian, V. , Jacobs, J.B. & Russell, L.A. (1978) Scanning electron microscope study of early lichen synthesis. Science, 200, 1062–1064. [DOI] [PubMed] [Google Scholar]
- Ajuzie, C.C. (2007) Palatability and fatality of the dinoflagellate Prorocentrum lima to Artemia salina . Journal of Applied Phycology, 19, 513–519. [Google Scholar]
- Allen, J.L. , Ten‐Hage, L. & Leflaive, J. (2016) Allelopathic interactions involving benthic phototrophic microorganisms: allelopathy in phototrophic biofilms. Environmental Microbiology Reports, 8, 752–762. [DOI] [PubMed] [Google Scholar]
- Amend, A. , Burgaud, G. , Cunliffe, M. , Edgcomb, V.P. , Ettinger, C.L. , Gutiérrez, M.H. et al. (2019) Fungi in the marine environment: open questions and unsolved problems. mBio, 10, e01189–e01118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora, D. , Gupta, P. , Jaglan, S. , Roullier, C. , Grovel, O. & Bertrand, S. (2020) Expanding the chemical diversity through microorganisms co‐culture: current status and outlook. Biotechnology Advances, 40, 107521. [DOI] [PubMed] [Google Scholar]
- Azzollini, A. , Boggia, L. , Boccard, J. , Sgorbini, B. , Lecoultre, N. , Allard, P.‐M. et al. (2018) Dynamics of metabolite induction in fungal co‐cultures by metabolomics at both volatile and non‐volatile levels. Frontiers in Microbiology, 9, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagot, A. , Bertrand, S. & Hess, P. (2016) Mise en évidence d'interactions chimiques entre micro‐algues et champignons filamenteux en milieux marins par profilage métabolique. Nantes, France: ISOMer Nantes Université ‐ PHYC IFREMER: Nantes Université. [Google Scholar]
- Bauder, A.G. , Cembella, A.D. , Bricelj, V.M. & Quilliam, M.A. (2021) Uptake and fate of diarrhetic shellfish poisoning toxins from the dinoflagellate Prorocentrum lima in the bay scallop Argopecten irradians . Marine Ecology Progress Series, 213, 39–52. [Google Scholar]
- Bertrand, S. , Azzollini, A. , Schumpp, O. , Bohni, N. , Schrenzel, J. , Monod, M. et al. (2014) Multi‐well fungal co‐culture for de novo metabolite‐induction in time‐series studies based on untargeted metabolomics. Molecular BioSystems, 10, 2289–2298. [DOI] [PubMed] [Google Scholar]
- Bertrand, S. , Bohni, N. , Schnee, S. , Schumpp, O. , Gindro, K. & Wolfender, J.‐L. (2014) Metabolite induction via microorganism co‐culture: a potential way to enhance chemical diversity for drug discovery. Biotechnology Advances, 32, 1180–1204. [DOI] [PubMed] [Google Scholar]
- Bertrand, S. , Schumpp, O. , Bohni, N. , Bujard, A. , Azzollini, A. , Monod, M. et al. (2013) Detection of metabolite induction in fungal co‐cultures on solid media by high‐throughput differential ultra‐high pressure liquid chromatography–time‐of‐flight mass spectrometry fingerprinting. Journal of Chromatography A, 1292, 219–228. [DOI] [PubMed] [Google Scholar]
- Bravo, I. , Fernández, M.L. , Ramilo, I. & Martínez, A. (2001) Toxin composition of the toxic dinoflagellate Prorocentrum lima isolated from different locations along the Galician coast (NW Spain). Toxicon, 39, 1537–1545. [DOI] [PubMed] [Google Scholar]
- Brown, E.R. , Cepeda, M.R. , Mascuch, S.J. , Poulson‐Ellestad, K.L. & Kubanek, J. (2019) Chemical ecology of the marine plankton. Natural Product Reports, 36, 1093–1116. [DOI] [PubMed] [Google Scholar]
- Buchan, A. , LeCleir, G.R. , Gulvik, C.A. & González, J.M. (2014) Master recyclers: features and functions of bacteria associated with phytoplankton blooms. Nature Reviews Microbiology, 12, 686–698. [DOI] [PubMed] [Google Scholar]
- Chaigne, M. , Bertrand, S. & Hess, P. (2017) Etude des interactions chimiques entre dinoflagellés benthiques et champignons filamenteux en milieu marin par une approche métabolomique. Nantes, France: ISOMer Nantes Université ‐ PHYC IFREMER: Nantes Université. [Google Scholar]
- Chakraborty, S., Pančić, M., Andersen, K.H., & Kiørboe, T. (2019) The cost of toxin production in phytoplankton: the case of PST producing dinoflagellates. The ISME Journal, 13(1), 64–75. 10.1038/s41396-018-0250-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, A. , Hubka, V. , Frisvad, J. , Visagie, C. , Houbraken, J. , Meijer, M. et al. (2017) Polyphasic taxonomy of aspergillus section aspergillus (formerly Eurotium), and its occurrence in indoor environments and food. Studies in Mycology, 88, 37–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Y. , de Bruyn Kops, C. & Kirchmair, J. (2017) Data resources for the computer‐guided discovery of bioactive natural products. Journal of Chemical Information and Modeling, 57, 2099–2111. [DOI] [PubMed] [Google Scholar]
- Cho, J.‐Y. , Choi, J.‐S. , Kong, I.‐S. , Park, S.‐I. & Kerr, R.G. (2002) A procedure for axenic isolation of the marine microalga Isochrysis galbana from heavily contaminated mass cultures. Journal of Applied Phycology, 14, 385–390. [Google Scholar]
- Cirri, E. & Pohnert, G. (2019) Algae−bacteria interactions that balance the planktonic microbiome. New Phytologist, 223, 100–106. [DOI] [PubMed] [Google Scholar]
- Djoumbou‐Feunang, Y. , Fiamoncini, J. , Gil‐de‐la‐Fuente, A. , Greiner, R. , Manach, C. & Wishart, D.S. (2019) BioTransformer: a comprehensive computational tool for small molecule metabolism prediction and metabolite identification. Journal of Cheminformatics, 11, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou, Y. , Wang, X. , Jiang, D. , Wang, H. , Jiao, Y. , Lou, H. et al. (2014) Metabolites from Aspergillus versicolor, an endolichenic fungus from the lichen Lobaria retigera . Drug Discoveries & Therapeutics, 8, 84–88. [DOI] [PubMed] [Google Scholar]
- Egan, S. , Harder, T. , Burke, C. , Steinberg, P. , Kjelleberg, S. & Thomas, T. (2013) The seaweed holobiont: understanding seaweed–bacteria interactions. FEMS Microbiology Reviews, 37, 462–476. [DOI] [PubMed] [Google Scholar]
- Foden, J. , Purdie, D.A. , Morris, S. & Nascimento, S. (2005) Epiphytic abundance and toxicity of Prorocentrum lima populations in the Fleet lagoon, UK. Harmful Algae, 4, 1063–1074. [Google Scholar]
- Forseth, R.R., Amaike, S., Schwenk, D., Affeldt, K.J., Hoffmeister, D., Schroeder, F.C. et al. (2012) Homologous NRPS‐like Gene Clusters Mediate Redundant Small‐Molecule Biosynthesis inAspergillus flavus. Angewandte Chemie International Edition, 52(5), 1590–1594. 10.1002/anie.201207456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grattan, L.M. , Holobaugh, S. & Morris, J.G. (2016) Harmful algal blooms and public health. Harmful Algae, 57, 2–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, D. , Hart, M. , Blackburn, S. & Bolch, C. (2010) Bacterial diversity of Gymnodinium catenatum and its relationship to dinoflagellate toxicity. Aquatic Microbial Ecology, 61, 73–87. [Google Scholar]
- Green, D.H. , Llewellyn, L.E. , Negri, A.P. , Blackburn, S.I. & Bolch, C.J.S. (2004) Phylogenetic and functional diversity of the cultivable bacterial community associated with the paralytic shellfish poisoning dinoflagellate Gymnodinium catenatum . FEMS Microbiology Ecology, 47, 345–357. [DOI] [PubMed] [Google Scholar]
- Gu, S. , Xiao, S.‐W. , Zheng, J.‐W. , Li, H.‐Y. , Liu, J.‐S. & Yang, W.‐D. (2019) ABC transporters in Prorocentrum lima and their expression under different environmental conditions including okadaic acid production. Marine Drugs, 17, 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillard, R.R.L. & Ryther, J.H. (1962) Studies of marine planktonic diatoms. I. Cyclotella nana Hustedt, and Detonula confervacea (Cleve) gran. Canadian Journal of Microbiology, 8, 229–239. [DOI] [PubMed] [Google Scholar]
- Gémin, M.‐P. , Réveillon, D. , Hervé, F. , Pavaux, A.‐S. , Tharaud, M. , Séchet, V. et al. (2020) Toxin content of Ostreopsis cf. ovata depends on bloom phases, depth and macroalgal substrate in the NW Mediterranean Sea. Harmful Algae, 92, 101727. [DOI] [PubMed] [Google Scholar]
- Hallegraeff, G. , Coman, F. , Davies, C. , Hayashi, A. , McLeod, D. , Slotwinski, A. et al. (2014) Australian dust storm associated with extensive Aspergillus sydowii fungal “bloom” in coastal waters. Applied and Environmental Microbiology, 80, 3315–3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth, D.L. & Grube, M. (2020) Lichens redefined as complex ecosystems. New Phytologist, 227, 1281–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi, A. , Crombie, A. , Lacey, E. , Richardson, A. , Vuong, D. , Piggott, A. et al. (2016) Aspergillus sydowii marine fungal bloom in Australian coastal waters, its metabolites and potential impact on Symbiodinium dinoflagellates. Marine Drugs, 14, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho, A. , Hsieh, D. & Qian, P. (2006) Variations in paralytic shellfish toxin and homolog production in two strains of Alexandrium tamarense after antibiotic treatments. Aquatic Microbial Ecology, 42, 41–53. [Google Scholar]
- Hom, E.F.Y. & Murray, A.W. (2014) Niche engineering demonstrates a latent capacity for fungal‐algal mutualism. Science, 345, 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honegger, R. (2009) Lichen‐forming fungi and their photobionts. In: Deising, H.B. (Ed.) The mycota. Berlin, Heidelberg: Springer, pp. 307–333. [Google Scholar]
- Ianora, A. , Boersma, M. , Casotti, R. , Fontana, A. , Harder, J. , Hoffmann, F. et al. (2006) New trends in marine chemical ecology. Estuaries Coast, 29, 531–551. [Google Scholar]
- James, A.K. , English, C.J. , Nidzieko, N.J. , Carlson, C.A. & Wilbanks, E.G. (2020) Giant kelp microbiome altered in the presence of epiphytes. Limnology and Oceanography Letters, 5, 354–362. [Google Scholar]
- Jansen, J.J. , Blanchet, L. , Buydens, L.M.C. , Bertrand, S. & Wolfender, J.‐L. (2015) Projected Orthogonalized CHemical encounter MONitoring (POCHEMON) for microbial interactions in co‐culture. Metabolomics, 11, 908–919. [Google Scholar]
- Jansen, J.J. , Szymańska, E. , Hoefsloot, H.C.J. & Smilde, A.K. (2012) Individual differences in metabolomics: individualised responses and between‐metabolite relationships. Metabolomics, 8, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, X. , Han, X. , Zheng, L. , Yang, B. , Yu, Z. & Zou, J. (2011) Allelopathic interactions between Prorocentrum micans and Skeletonema costatum or Karenia mikimotoi in laboratory cultures. Chinese Journal of Oceanology and Limnology, 29, 840–848. [Google Scholar]
- Johnson, M.D. (2015) Inducible mixotrophy in the dinoflagellate Prorocentrum minimum . Journal of Eukaryotic Microbiolog, 62, 431–443. [DOI] [PubMed] [Google Scholar]
- Kobayashi, K. , Takata, Y. & Kodama, M. (2009) Direct contact between Pseudo‐nitzschia multiseries and bacteria is necessary for the diatom to produce a high level of domoic acid. Fisheries Science, 75, 771–776. [Google Scholar]
- Krespach, M.K.C. , García‐Altares, M. , Flak, M. , Schoeler, H. , Scherlach, K. , Netzker, T. et al. (2020) Lichen‐like association of Chlamydomonas reinhardtii and Aspergillus nidulans protects algal cells from bacteria. ISME Journal, 14, 2794–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassus, P. , Chomérat, N. , Hess, P. & Nézan, E. (2016) Toxic and harmful microalgae of the world ocean. Copenhagen N, Denmark: International Society for the Study of Harmful Algae. [Google Scholar]
- Lawrence, J.E. , Grant, J. , Quilliam, M.A. , Bauder, A.G. & Cembella, A.D. (2021) Colonization and growth of the toxic dinoflagellate Prorocentrum lima and associated fouling macroalgae on mussels in suspended culture. Marine Ecology Progress Series, 201, 147–154. [Google Scholar]
- Lee, T. , Fong, F. , Ho, K.‐C. & Lee, F. (2016) The mechanism of diarrhetic shellfish poisoning toxin production in Prorocentrum spp.: physiological and molecular perspectives. Toxins, 8, 272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng, L. , Li, W. , Chen, J. , Leng, S. , Chen, J. , Wei, L. et al. (2021) Co‐culture of fungi‐microalgae consortium for wastewater treatment: a review. Bioresource Technology, 330, 125008. [DOI] [PubMed] [Google Scholar]
- Li, X. , Xia, Z. , Tang, J. , Wu, J. , Tong, J. , Li, M. et al. (2017) Identification and biological evaluation of secondary metabolites from marine derived fungi‐Aspergillus sp. SCSIOW3, cultivated in the presence of epigenetic modifying agents. Molecules, 22, 1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, Y. , Wohlrab, S. , Groth, M. , Glöckner, G. , Guillou, L. & John, U. (2016) Transcriptomic profiling of Alexandrium fundyense during physical interaction with or exposure to chemical signals from the parasite Amoebophrya . Molecular Ecology, 25, 1294–1307. [DOI] [PubMed] [Google Scholar]
- Mazur, P., Nakanishi, K., El‐Zayat, A.A.E., & Champe, S.P. (1991). Structure and synthesis of sporogenic psi factors from Aspergillus nidulans. Journal of the Chemical Society, Chemical Communications, 20, 1486. 10.1039/c39910001486 [DOI] [Google Scholar]
- Moreira‐González, A.R. , Fernandes, L.F. , Uchida, H. , Uesugi, A. , Suzuki, T. , Chomérat, N. et al. (2019) Variations in morphology, growth, and toxicity among strains of the Prorocentrum lima species complex isolated from Cuba and Brazil. Journal of Applied Phycology, 31, 519–532. [Google Scholar]
- Myers, O.D. , Sumner, S.J. , Li, S. , Barnes, S. & Du, X. (2017) One step forward for reducing false positive and false negative compound identifications from mass spectrometry metabolomics data: new algorithms for constructing extracted ion chromatograms and detecting chromatographic peaks. Analytical Chemistry, 89, 8696–8703. [DOI] [PubMed] [Google Scholar]
- Nagai, H. , Satake, M. & Yasumoto, T. (1990) Antimicrobial activities of polyether compounds of dinoflagellate origins. Journal of Applied Phycology, 2, 305–308. [Google Scholar]
- Nguyen, T.T.T. , Pangging, M. , Bangash, N.K. & Lee, H.B. (2020) Five new records of the family Aspergillaceae in Korea, aspergillus europaeus, A. pragensis, A. tennesseensis, Penicillium fluviserpens, and P. scabrosum . Mycobiology, 48, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen, K.F. , Månsson, M. , Rank, C. , Frisvad, J.C. & Larsen, T.O. (2011) Dereplication of microbial natural products by LC‐DAD‐TOFMS. Journal of Natural Products, 74, 2338–2348. [DOI] [PubMed] [Google Scholar]
- Nishimura, T. , Uchida, H. , Noguchi, R. , Oikawa, H. , Suzuki, T. , Funaki, H. et al. (2020) Abundance of the benthic dinoflagellate Prorocentrum and the diversity, distribution, and diarrhetic shellfish toxin production of Prorocentrum lima complex and P. caipirignum in Japan. Harmful Algae, 96, 101687. [DOI] [PubMed] [Google Scholar]
- Padhi, S. , Masi, M. , Panda, S.K. , Luyten, W. , Cimmino, A. , Tayung, K. et al. (2020) Antimicrobial secondary metabolites of an endolichenic Aspergillus niger isolated from lichen thallus of Parmotrema ravum . Natural Product Research, 34, 2573–2580. [DOI] [PubMed] [Google Scholar]
- Pasqualetti, M. , Giovannini, V. , Barghini, P. , Gorrasi, S. & Fenice, M. (2020) Diversity and ecology of culturable marine fungi associated with Posidonia oceanica leaves and their epiphytic algae Dictyota dichotoma and Sphaerococcus coronopifolius . Fungal Ecology, 44, 100906. [Google Scholar]
- Pluskal, T. , Castillo, S. , Villar‐Briones, A. & Orešič, M. (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry‐based molecular profile data. BMC Bioinformatics, 11, 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohnert, G. (2000) Wound‐activated chemical defense in unicellular planktonic algae. Angewandte Chemie International Edition, 39, 4352–4354. [DOI] [PubMed] [Google Scholar]
- Prateeksha, P. , Bajpai, R. , Yusuf, M.A. , Upreti, D.K. , Gupta, V.K. & Singh, B.N. (2020) Endolichenic fungus, Aspergillus quandricinctus of Usnea longissima inhibits quorum sensing and biofilm formation of Pseudomonas aeruginosa PAO1. Microbial Pathogenesis, 140, 103933. [DOI] [PubMed] [Google Scholar]
- Prol, M.J. , Guisande, C. , Barreiro, A. , Míguez, B. , de la Iglesia, P. , Villar, A. et al. (2009) Evaluation of the production of paralytic shellfish poisoning toxins by extracellular bacteria isolated from the toxic dinoflagellate Alexandrium minutum . Canadian Journal of Microbiology, 55, 943–954. [DOI] [PubMed] [Google Scholar]
- Puglisi, M.P. , Sneed, J.M. , Sharp, K.H. , Ritson‐Williams, R. & Paul, V.J. (2014) Marine chemical ecology in benthic environments. Natural Product Reports, 31, 1510–1553. [DOI] [PubMed] [Google Scholar]
- Qiao, M.‐F., Ji, N.‐Y., Miao, F.‐P., & Yin, X.‐L. (2011) Steroids and an oxylipin from an algicolous isolate of Aspergillus flavus. Magnetic Resonance in Chemistry, 49(6), 366–369. 10.1002/mrc.2748 [DOI] [PubMed] [Google Scholar]
- Roullier, C. , Bertrand, S. , Blanchet, E. , Peigné, M. , Robiou du Pont, T. , Guitton, Y. et al. (2016) Time dependency of chemodiversity and biosynthetic pathways: an LC–MS metabolomic study of marine‐sourced Penicillium . Marine Drugs, 14, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz, N. , Petit, K. , Vansteelandt, M. , Kerzaon, I. , Baudet, J. , Amzil, Z. et al. (2010) Enhancement of domoic acid neurotoxicity on Diptera larvae bioassay by marine fungal metabolites. Toxicon, 55, 805–810. [DOI] [PubMed] [Google Scholar]
- Ryderheim, F. , Selander, E. & Kiørboe, T. (2021) Predator‐induced defence in a dinoflagellate generates benefits without direct costs. ISME J, 15, 2107–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rédou, V. , Navarri, M. , Meslet‐Cladière, L. , Barbier, G. & Burgaud, G. (2015) Species richness and adaptation of marine fungi from deep‐subseafloor sediments. Applied and Environmental Microbiology, 81, 3571–3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saccenti, E. , Hoefsloot, H.C.J. , Smilde, A.K. , Westerhuis, J.A. & Hendriks, M.M.W.B. (2014) Reflections on univariate and multivariate analysis of metabolomics data. Metabolomics, 10, 361–374. [Google Scholar]
- Sallenave‐Namont, C. , Pouchus, Y.F. , Robiou du Pont, T. , Lassus, P. & Verbist, J.‐F. (2000) Toxigenic saprophytic fungi in marine shellfish farming areas. Mycopathologia, 149, 21–25. [DOI] [PubMed] [Google Scholar]
- Scholz, B. , Küpper, F. , Vyverman, W. , Ólafsson, H. & Karsten, U. (2017) Chytridiomycosis of marine diatoms—the role of stress physiology and resistance in parasite–host recognition and accumulation of defense molecules. Marine Drugs, 15, 26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeckh, V. , Scherlach, K. , Nutzmann, H.‐W. , Shelest, E. , Schmidt‐Heck, W. , Schuemann, J. et al. (2009) Intimate bacterial‐fungal interaction triggers biosynthesis of archetypal polyketides in Aspergillus nidulans . Proceedings of the National Academy of Sciences of the United States of America, 106, 14558–14563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw, B.A. , Andersen, R.J. & Harrison, P.J. (1997) Feeding deterrent and toxicity effects of apo‐fucoxanthinoids and phycotoxins on a marine copepod (Tigriopus californicus). Marine Biology, 128, 273–280. [Google Scholar]
- Sison‐Mangus, M.P. , Jiang, S. , Tran, K.N. & Kudela, R.M. (2014) Host‐specific adaptation governs the interaction of the marine diatom, pseudo‐nitzschia and their microbiota. ISME Journal, 8, 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev, V.S., Khan, S.I., Tabanca, N., Wedge, D.E., Manly, S.P., Cutler, S.J. et al. (2011) Biological Activity of Peanut (Arachis hypogaea) Phytoalexins and Selected Natural and Synthetic Stilbenoids. Journal of Agricultural and Food Chemistry, 59(5), 1673–1682. 10.1021/jf104742n [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, I. , Seawright, A.A. & Shaw, G.R. (2008) Chapter 28: cyanobacterial poisoning in livestock, wild mammals and birds – an overview. In: Cyanobacterial harmful algal blooms: state of the science and research needs. Advances in experimental medicine and biology. New York, NY: Springer, p. 25. [DOI] [PubMed] [Google Scholar]
- Sugg, L.M. & VanDolah, F.M. (1999) No evidence for an allelopathic role of okadaic acid among ciguatera‐associated dinoflagellates. Journal of Phycology, 35, 93–103. [Google Scholar]
- Sumner, L.W. , Amberg, A. , Barrett, D. , Beale, M.H. , Beger, R. , Daykin, C.A. et al. (2007) Proposed minimum reporting standards for chemical analysis: chemical analysis working group (CAWG) metabolomics standards initiative (MSI). Metabolomics, 3, 211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuki, A. , Suzuki, S. , Ando, K. , Tamura, G. & Arima, K. (1968) New antiviral antibiotics; Xanthocillin X mono‐ and dimethylether, and methoxy‐xanthocillin X dimethylether. I isolation and characterization (studies on antiviral and antitumor antibiotics V). Journal of Antibiotics, 21, 5. [DOI] [PubMed] [Google Scholar]
- Tarazona‐Janampa, U.I. , Cembella, A.D. , Pelayo‐Zárate, M.C. , Pajares, S. , Márquez‐Valdelamar, L.M. , Okolodkov, Y.B. et al. (2020) Associated bacteria and their effects on growth and toxigenicity of the dinoflagellate Prorocentrum lima species complex from epibenthic substrates along Mexican coasts. Frontiers in Marine Science, 7, 569. [Google Scholar]
- Thévenot, E.A. , Roux, A. , Xu, Y. , Ezan, E. & Junot, C. (2015) Analysis of the human adult urinary metabolome variations with age, body mass index, and gender by implementing a comprehensive workflow for univariate and OPLS statistical analyses. Journal of Proteome Research, 14, 3322–3335. [DOI] [PubMed] [Google Scholar]
- Tourneroche, A. , Lami, R. , Burgaud, G. , Domart‐Coulon, I. , Li, W. , Gachon, C. et al. (2020) The bacterial and fungal microbiota of Saccharina latissima (Laminariales, Phaeophyceae). Frontiers in Marine Science, 7, 587566. [Google Scholar]
- Traubenberg, C.R. , Soyer‐Gobillard, M.‐O. , Géraud, M.‐L. & Albert, M. (1995) The toxic dinoflagellate Prorocentrum lima and its associated bacteria. European Journal of Protistology, 31, 383–388. [Google Scholar]
- Trembley, M.L. , Ringli, C. & Honegger, R. (2002) Morphological and molecular analysis of early stages in the resynthesis of the lichen Baeomyces Rufus . Mycological Research, 106, 768–776. [Google Scholar]
- Uribe, P. & Espejo, R.T. (2003) Effect of associated bacteria on the growth and toxicity of Alexandrium catenella . Applied and Environmental Microbiology, 69, 659–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg, R.A. , Hoefsloot, H.C. , Westerhuis, J.A. , Smilde, A.K. & van der Werf, M.J. (2006) Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics, 7, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, B. , Yao, M. , Zhou, J. , Tan, S. , Jin, H. , Zhang, F. et al. (2018) Growth and toxin production of Gambierdiscus spp. can be regulated by quorum‐sensing bacteria. Toxins, 10, 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, J. & Bertrand, S. (2019) PocheRmon. Available at: https://gitlab.univ-nantes.fr/bertrand-s-1/pochermon [Accessed 09th September 2022]
- Wang, R. , Wu, J. , Zhou, S. , Cao, R. & Chan, L.L. (2020) A preliminary study on the allelopathy and toxicity of the dinoflagellate Karlodinium veneficum . Marine Pollution Bulletin, 158, 111400. [DOI] [PubMed] [Google Scholar]
- Wang, S. , Chen, J. , Li, Z. , Wang, Y. , Fu, B. , Han, X. et al. (2015) Cultivation of the benthic microalga Prorocentrum lima for the production of diarrhetic shellfish poisoning toxins in a vertical flat photobioreactor. Bioresour Technol, 179, 243–248. [DOI] [PubMed] [Google Scholar]
- Wang, Y. , Sen, B. , He, Y. , Xie, N. & Wang, G. (2018) Spatiotemporal distribution and assemblages of planktonic fungi in the coastal waters of the Bohai Sea. Frontiers in Microbiology, 9, 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Want, E.J. , Wilson, I.D. , Gika, H. , Theodoridis, G. , Plumb, R.S. , Shockcor, J. et al. (2010) Global metabolic profiling procedures for urine using UPLC–MS. Nature Protocols, 5, 1005–1018. [DOI] [PubMed] [Google Scholar]
- Windust, A.J. , Wright, J.L.C. & McLachlan, J.L. (1996) The effects of the diarrhetic shellfish poisoning toxins, okadaic acid and dinophysistoxin‐1, on the growth of microalgae. Marine Biology, 126, 19–25. [Google Scholar]
- Wolfender, J.‐L. , Marti, G. , Thomas, A. & Bertrand, S. (2015) Current approaches and challenges for the metabolite profiling of complex natural extracts. Journal of Chromatography A, 1382, 136–164. [DOI] [PubMed] [Google Scholar]
- Wolfender, J.‐L. , Nuzillard, J.‐M. , van der Hooft, J.J.J. , Renault, J.‐H. & Bertrand, S. (2019) Accelerating metabolite identification in natural product research: toward an ideal combination of liquid chromatography–high‐resolution tandem mass spectrometry and NMR profiling, in silico databases, and chemometrics. Analytical Chemistry, 91, 704–742. [DOI] [PubMed] [Google Scholar]
- Wood, P.J. (1980) Specificity in the interaction of direct dyes with polysaccharides. Carbohydr Res, 85, 271–287. [Google Scholar]
- Worden, A.Z. , Follows, M.J. , Giovannoni, S.J. , Wilken, S. , Zimmerman, A.E. & Keeling, P.J. (2015) Rethinking the marine carbon cycle: factoring in the multifarious lifestyles of microbes. Science, 347, 1257594. [DOI] [PubMed] [Google Scholar]
- Wrede, D. , Taha, M. , Miranda, A.F. , Kadali, K. , Stevenson, T. , Ball, A.S. et al. (2014) Co‐cultivation of fungal and microalgal cells as an efficient system for harvesting microalgal cells, lipid production and wastewater treatment. PLoS One, 9, e113497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zendong, Z. , Bertrand, S. , Herrenknecht, C. , Abadie, E. , Jauzein, C. , Lemée, R. et al. (2016) Passive sampling and high resolution mass spectrometry for chemical profiling of French coastal areas with a focus on marine biotoxins. Environmental Science & Technology, 50, 8522–8529. [DOI] [PubMed] [Google Scholar]
- Zhang, S. , Merino, N. , Okamoto, A. & Gedalanga, P. (2018) Interkingdom microbial consortia mechanisms to guide biotechnological applications. Microbial Biotechnology, 11, 833–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Fungal strains isolated from Prorocentrum lima PL4V.
Figure S1. Scheme of the microscale environment designed for this study.
Figure S2. Amount of extract (with standard deviation) obtained in each culture condition.
Figure S3. Principal component analysis (log transformed data with pareto scaling) using peaks detected in the LC‐HRMS profiles in positive (A) and negative (B) ionization modes. QC samples are highlighted in red.
Figure S4. Validation of the POChEMon mixing model, using leave‐one‐out strategy, removing either a mono‐culture sample (A) or a co‐culture sample (B). Sample distribution was represented as a violin plot (in blue: the monoculture sample distribution; in red the co‐culture sample distribution). The removed sample was plotted back (red dot) within the generated model to highlight that no misclassification was observed.
Figure S5. Morphology of fungal strains isolated from Prorocentrum lima PL4V after 3 days (top) after 10 days (bottom). The growth was performed in 5‐cm Petri dishes.
Figure S6. Morphology of Aspergillus pseudoglaucus MMS1589 (binocular view). Cleistothecia (1) and classic mycelium bearing conidiophores (2).
Figure S7. LC‐HRMS profiles in negative ionization of extracts used for metabolomics (only one replicates is presented here). All extracts were profiled at 1 mg/ml concentration and chromatograms were all scaled (y) to the same ion count intensity (inter‐chromatogram y‐step corresponds to 84,000 counts).
Figure S8. LC‐HRMS profiles in positive ionization of extracts used for metabolomics (only one replicates is presented here). All extracts were profiled at 1 mg/ml concentration and chromatograms were all scaled (y) to the same ion count intensity (inter‐chromatogram y‐step corresponds to 140,000 counts).
Figure S9. Number of detected features per conditions, obtained with MZmine 2 processing of LC‐HRMS data. S: supernatant, A: agar solid phase.
Figure S10. Venn Diagram of feature distribution following origin (liquid or solid phase) and antibiotic (ATB) exposure using the selected top features according to the three different statistical treatments applied on the data matrix (highest significance in Student's t‐test, highest VIPs in the OPLS‐DA, best SSrank values) and withdrawing of redundant selected features.
Figure S11. Recovery of okadaic acid (OA) and dinophysistoxin 1 (DTX‐1) after addition to blank samples (with and without antibiotics) showing the absence of extraction bias due to the presence of the antibiotic cocktail.
Additional raw data may be found on Zenodo (http://doi.org/10.5281/zenodo.7050181).
DATA S1: the data matrix as an Excel file.
Data Availability Statement
The P. lima PL4V and A. pseudoglaucus MMS1589 strains are stored at IFREMER (PHYTOX, Laboratoire PHYSALG, F‐44311 Nantes, France), and Nantes Université (ISOMer, UR2160, Nantes, France), respectively. They both can be obtained on request.


