Abstract
Multiple sclerosis (MS) is the most common inflammatory, demyelinating and neurodegenerative disease of the central nervous system in young adults. Chronic‐relapsing experimental autoimmune encephalomyelitis (crEAE) in Biozzi ABH mice is an experimental model of MS. This crEAE model is characterized by an acute phase with severe neurological disability, followed by remission of disease, relapse of neurological disease and remission that eventually results in a chronic progressive phase that mimics the secondary progressive phase (SPEAE) of MS. In both MS and SPEAE, the role of microglia is poorly defined. We used a crEAE model to characterize microglia in the different phases of crEAE phases using morphometric and RNA sequencing analyses. At the initial, acute inflammation phase, microglia acquired a pro‐inflammatory phenotype. At the remission phase, expression of standard immune activation genes was decreased while expression of genes associated with lipid metabolism and tissue remodeling were increased. Chronic phase microglia partially regain inflammatory gene sets and increase expression of genes associated with proliferation. Together, the data presented here indicate that microglia obtain different features at different stages of crEAE and a particularly mixed phenotype in the chronic stage. Understanding the properties of microglia that are present at the chronic phase of EAE will help to understand the role of microglia in secondary progressive MS, to better aid the development of therapies for this phase of the disease.
Keywords: experimental autoimmune encephalitis, microglia, RNA sequencing, secondary progressive MS
Main Points
Microglia switch from a pro‐inflammatory phenotype in acute EAE to a tissue remodeling phenotype in remission.
Chronic EAE microglia resemble remission microglia, but show increased proliferation and regain some features of acute phase microglia.
1. INTRODUCTION
Microglia are the tissue‐resident macrophages of the central nervous system (CNS) and recent genome‐wide association analysis (International Multiple Sclerosis Genetics Consortium, 2019) and animal studies (Ajami et al., 2018; Clark et al., 2021; Heppner et al., 2005; Joost et al., 2019; Masuda et al., 2019; Nissen et al., 2018; Ponomarev et al., 2005; Voß et al., 2012) have increasingly implicated microglia in the progression of multiple sclerosis (MS). In the healthy CNS, microglia are highly ramified and dynamic cells that survey the parenchyma to maintain tissue homeostasis. After injury, microglia acquire an activated, more phagocytic phenotype in response to cues from the environment (Keren‐Shaul et al., 2017; Krasemann et al., 2017; Kreutzberg, 1996). In MS, macrophages, including microglia, phagocytose myelin debris, modulate the extracellular matrix and secrete regenerative factors in order to create a favorable environment for oligodendrocyte progenitor cell (OPC) recruitment and OPC maturation, and therefore are relevant to remyelination (Forbes & Miron, 2021; Kotter et al., 2006; Lampron et al., 2015; Lloyd & Miron, 2019; Robinson & Miller, 1999). However, the extent to which microglia contribute to initiation and repair of MS lesions, especially in the secondary‐progressive phase has been poorly defined (Geladaris et al., 2021). Defining the role of microglia during secondary progressive MS could provide clues to novel therapies. This is relevant as several drugs are effective in reducing neuroinflammation in relapsing remitting MS, but very few effective treatment options are available for progressive MS (Compston & Coles, 2008; Geladaris et al., 2021; Lassmann et al., 2012).
Multiple animal models have been developed to study MS pathology, including demyelination models based on toxins that directly target oligodendrocytes such as cuprizone and lysolecithin (l‐α‐lysophosphatidylcholine or LPC) (Blakemore & Franklin, 2008; Praet et al., 2014), and the widely used experimental autoimmune encephalitis (EAE) model that is induced by eliciting an auto‐immune response against myelin in the periphery (Taylor, 1986). As no animal model fully recapitulates all elements of MS pathology, it often remains difficult to translate the findings to MS (Baker et al., 2011; Baker & Amor, 2014). However, EAE has been instrumental to identify the mechanism of action of interferon‐beta therapy and aided the development of the drug Copaxone (Scheu et al., 2019; Weinstock‐Guttman et al., 2017).
In Biozzi ABH mice, chronic‐relapsing EAE (crEAE) displays a multiphasic progression in contrast to the monophasic EAE models in C57BL/6 mice (Baker et al., 1990; Brown et al., 1982; Cross et al., 1987; Kozlowski et al., 1987; Lassmann & Wisniewski, 1978; Lorentzen et al., 1995). Biozzi ABH crEAE mice develop a reproducible relapsing–remitting disease course due to multiple episodes of demyelination and inflammation (Baker et al., 1990), followed by a progressive increase in disability and a chronic phase (Baker et al., 1990; Pryce et al., 2005). crEAE uniquely mimics the secondary progressive MS phase (SPMS) where patients have accumulating physical disability and neurological damage (Hampton et al., 2008; Jackson et al., 2009; Lassmann et al., 2012). Of note, contrary to other EAE models, pertussis toxin is not used (Baker et al., 1990; Hampton et al., 2008; Jackson et al., 2009). This is relevant in view of the adverse effects of pertussis toxin on microglia and blood brain barrier integrity (Yin et al., 2010). Therefore, crEAE is a highly relevant model to study microglia in the context of SPMS.
Microglia have been mostly studied in the context of acute EAE, where extensive immune infiltration from the periphery is present (Butovsky et al., 2014; Vainchtein et al., 2014; Wlodarczyk et al., 2014; Yamasaki et al., 2014). In view of the more limited involvement of the peripheral immune system in SPMS, microglia have received considerable attention as potential players in SPMS progression (Melief et al., 2013; Peferoen et al., 2015). For this reason, we used the crEAE model to delineate the transcriptional program of microglia during the chronic, secondary progressive phase. We performed immunohistochemistry, microglia morphometrics and RNA sequencing analyses in all stages of the crEAE model to determine the phenotype of microglia at the chronic, secondary progressive phase of EAE.
2. MATERIALS AND METHODS
2.1. Induction of crEAE and progression
A 8–10 weeks old male and female Biozzi ABH mice were housed at the Queen Mary University of London and provided with ad libitum access to food and water (Biozzi et al., 1972). To induce crEAE, Biozzi ABH mice were inoculated as described previously (Al‐Izki et al., 2012; Baker et al., 1990). In brief, a sonicated emulsion of a lyophilized Biozzi ABH mouse spinal cord homogenate with complete Freund's adjuvant (CFA) was injected subcutaneously into the hind flanks on day 0 and day 7. Mice were weighed and monitored for a clinical EAE score using a 6‐point scoring scale daily: 0 = no obvious changes (normal), 1 = limp tail, 2 = limp tail and impaired righting reflex, 3 = limp tail and partial paralysis of hind legs, 4 = limp tail and complete paralysis of hind legs, 5 = moribund, 6 = death. Mice were euthanized in the acute stage when reaching score 4 (n = 19; days 17–20), remission score 0.5 (n = 18; days 27–30) and at the chronic stage (n = 15; days 80–90). Age‐matched, untreated (in other words, did not receive Biozzi ABH mouse spinal cord homogenate with CFA) animals served as controls (n = 22). For all experiments, the number of animals per experimental condition is indicated in the respective figure legend. All experiments were performed according to institutional and international guidelines and approved by the London Animal Welfare Advisory Committee (United Kingdom Animals Act 1986). Aspects of experimental design and use of animals relevant to the ARRIVE guidelines (Amor & Baker, 2012; Baker & Amor, 2012; Kilkenny et al., 2010) have been reported previously (Al‐Izki et al., 2012).
2.2. Acute isolation of microglia
Mice were euthanized using CO2 and perfused with PBS. Subsequently, the spinal cord was isolated and collected in ice‐cold isolation medium (HBSS (PAA Laboratories) supplemented with 15 mM HEPES (PAA) and 0.6% glucose (Sigma‐Aldrich)). A single‐cell suspension was obtained as described previously (de Haas et al., 2008; Vainchtein et al., 2014). In short, the spinal cords were mechanically dissociated using a tissue homogenizer and the suspension was filtered using a 70 μm cell strainer (BD FALCON). Thereafter, cells were pelleted (220 g, 4°C, acc: 9, brake: 9, 10 min), and after supernatant removal the pellet was resuspended in a solution of 22% Percoll (GE Healthcare), 40 mM NaCl and 77% myelin gradient buffer (5.6 mM NaH2PO4˙H20, 20 mM Na2HPO4˙2H20, 140 mM NaCl, 5.4 mM KCl, 11 mM Glucose, pH 7.4). A layer of PBS was placed on top, and this gradient was centrifuged (950 g, 4°C, acc: 4, brake: 0, 20 min) followed by removal of the myelin layer and the remaining supernatant. The pellet was resuspended in Phenol Red deficient isolation medium (HBSS without Phenol Red (PAA) supplemented with 15 mM HEPES (PAA) and 0.6% glucose (Sigma‐Aldrich)). All steps in the isolation procedure were performed on ice.
2.3. Fluorescence activated cell sorting
The cell suspension was Fc receptor blocked with anti‐mouse CD16/CD32 (eBioscience) for 10 min on ice. Subsequently, the cells were incubated with CD11b PE (eBioscience), CD45 FITC (eBioscience), Ly‐6C APC (Biolegend) and CD3 PE/Cy7 (Biolegend) for 30 min. The cell suspension was washed with Phenol Red deficient isolation medium, centrifuged (230 g, 4 °C, acc: 9, brake: 9, 3 min) and after passing through a 35 μm nylon mesh (BD Biosciences) collected in round bottom tubes. Fluorescence activated cell sorting (FACS) was performed on a BD Biosciences FACSAria II cell sorter. After gating for viable cells based on 4′,6‐diamidino‐2‐phenylindole (DAPI; 0.5 μM; Sigma‐Aldrich), microglia were defined as CD11bpos CD45int Ly‐6Cneg and myeloid Ly‐6Cpos infiltrates as CD11bpos CD45high Ly‐6Cpos. The cells were collected in Phenol Red deficient isolation medium and used for further application. FACS plot analysis was performed with Tree Star FlowJo software v10.
2.4. Quantitative real‐time PCR
After FACS the purified cell suspensions were centrifuged (500 g, 4°C, acc: 9, brake: 9, 10 min) and the pellet was lysed in RLT+ buffer (Qiagen). The RNA was extracted using the RNeasy Plus Micro kit (Qiagen) according to the manufacturer's protocol. Subsequently, reverse transcription was performed using a mixture of random hexamers, dNTPs, M‐MLV buffer, Ribolock™RNase Inhibitor and RevertAid™ M‐MuLV Reverse Transcriptase (Fermentas). Quantitative real‐time PCR was performed in 384 well plates (Applied Biosystems) with iQ™ SYBR Green Supermix (Bio‐Rad) on an ABI7900HT machine (Applied Biosystems). All used primers (Table S1) were designed with NCBI Primer‐Blast and ordered from Biolegio (The Netherlands). Data were quantified using the 2–ΔΔCt method where Hmbs (hydroxymethylbilane synthase) was used as a housekeeping gene (Livak & Schmittgen, 2001).
2.5. RNA sequencing and analysis
Sequencing libraries were generated from cDNA using a Nextera XT Sample Prep Kit (Ilumina) according to the manufacturer's protocol. cDNA fragment libraries were sequenced on an Illumina NextSeq500 using default parameters (single read, high output, 75 bp reagents).
Quality control was performed on the raw FASTQ files with FastQC (0.11.4). Reads were trimmed with FastX Trimmer (0.013) where base pairs 1–60 were kept. Alignment of the sequenced reads was done with HiSat2 (Kim et al., 2019) using default parameters (2.0.4) against the GRCm38 genome. Aligned data was sorted with the use of Samtools (1.3.1) and Picard (2.6.4). Quantification was done using HTSeq (0.6.1) and the quality of the processed data was evaluated with RSeQC (2.6.4) (Anders et al., 2015; Wang et al., 2012).
Genes were filtered using a data‐adaptive flag method for RNA‐sequencing (DAFS) to estimate the cut‐off between lowly and highly expressed genes per sample (George & Chang, 2014). Counts were transformed using the vst function as implemented in DESeq2 (v1.26.0). Principal component analysis (PCA) was computed on vst‐transformed counts. Normalization and differential expression analysis were performed with DESeq2 default settings (1.26.0) (Love et al., 2014). Expression changes were adjusted with the function lfcShrink type = apeglm and p‐values were adjusted with the Benjamini‐Hochberg procedure. Genes were considered differentially expressed when Log2 Fold Change >1 or <−1 and p‐adjusted <.01. The crEAE phase‐specific genes were identified by comparing each crEAE profile to control conditions, followed by selection of differential genes unique for each of the crEAE conditions.
GO enrichment analyses were performed on all differentially expressed genes with clusterProfiler (3.14.3) function compareCluster using default settings and the AnnotationDbi mouse genome (org.Mm.eg.db 3.10.0) as background genes (Yu et al., 2012). GO enrichment on signature genes was computed for each signature list using the function enrichGO with identical parameters. In Figure 3b we considered the top five most significantly enriched GO terms per comparison. For GO terms which were semantically highly similar (e.g., “antigen processing and presentation of peptide antigen” and “antigen processing and presentation of exogenous peptide antigen”) the least significant GO term of the two terms was removed. The Venn Diagram was computed by the intersection of three gene lists described in the workflow of Figure S3b and plotted with the R package VennDiagram (1.6.20). The raw count files with cell‐to‐cluster annotations provided by Masuda et al. (2019) were re‐analyzed using Seurat (3.1.5) (Butler et al., 2018). We excluded non‐microglia clusters HuC1 (lymphocytes), HuC9, HuC10 (monocytes). We applied the FindAllMarkers function with parameters only.pos = TRUE and test.use = “negbinom”, to remain close to cluster marker calculation by RaceID2 (Grün et al., 2016) and providing the RaceID2 cluster identities. We excluded HuC4 because it contained <20 single microglia and provided only five cluster marker genes. For mouse microglia from Masuda et al. (2019), we selected microglia isolated from the corpus callosum of cuprizone mice and calculated cluster marker genes with an identical FindAllMarkers procedure. For each set of cluster markers in Table S7 we calculated enrichment of significant single cell cluster markers (p‐adjusted <.05) in the bulk RNAseq samples by calculating the area under curve using the R package AUCell (1.8.0) (Aibar et al., 2017). Genes in each bulk sample were ranked from highest to lowest expression. At each rank, the number of overlapping genes with the single cell gene set was determined. AUC values were z‐transformed across samples.
2.6. Immunohistochemistry
Mice were perfused with saline followed by ice‐cold 4% paraformaldehyde (PFA) and the spinal cord was extracted and post‐fixed overnight in the same solution. Tissues were transferred to 30% sucrose solution for 2 days and subsequently embedded in Tissue Tek optimal cutting temperature (OCT; Sakura) and 50 μm sagittal sections were cut with a cryostat (Jung CM3000, Leica Biosystems). Immunohistochemistry was performed on free‐floating sections. In brief, tissue sections were washed in PBS and blocked for 1 h with 3% bovine albumin serum (BSA) in PBS containing 0.1% TritonX‐100 (PBST). Sections were incubated O/N at 4°C with rabbit anti‐IBA1 (WAKO, 019‐19741, 1:1000) and either goat anti‐IL‐1β (R&D Systems, AF‐401‐NA, 1:40) or goat anti‐AXL (Santa Cruz Biotechnology, sc‐1096, 1:250) in PBST containing 3% BSA. The next day, sections were incubated for 3 h with secondary antibodies (donkey anti‐rabbit Alexa 594, donkey anti‐goat Alexa 488; ThermoFisherScientific, all 1:500).
Triple staining was performed in 24‐well plates and started with an antigen retrieval procedure. Antigen retrieval consisted of 40‐minute incubation of the 50 μm thick free‐floating sections in 10 mM sodium citrate buffer (pH 6.0) at 70°C. Next, sections were incubated 30 min at room temperature (RT) in freshly prepared 0.1% NaBH4. After rinsing with PBS, sections were incubated for 1 h at RT on top of a multipurpose shaker in blocking buffer (0.5% gelatin and 5% normal donkey serum in PBS with 0.01% Triton X‐100). Next, section were incubated overnight at 4°C in blocking buffer containing the primary antibodies. A mixture of Ly‐6C (AbD Serotec, MCA2389GA, 1:250) and IBA1 (WAKO, 019‐19741, 1:1000 or Abcam, ab5076, 1:500) was used in combination with Ki67 (Abcam, ab15580, 1:500), STAT1 (CST, #14995, 1:500) or IL1ß (R&D Systems, AF‐401‐NA, 1:70). The following day, sections were thoroughly washed in PBS and incubated for 1.5 h at RT in blocking buffer containing a mixture of secondary antibodies (donkey anti rat, AF647, JacksonImmunoResearch 712‐605‐153, 1:300; donkey anti rabbit, AF488, ThermoFisherScientific A21206, 1:300; donkey anti goat, AF568, ThermoFisherScientific A11057, 1:300) and Hoechst (f.c. 0.5 μM). After washing with PBS, sections were mounted with Mowiol. Image acquisition was performed using the Leica SP2 AOBS or the Leica TCS SP8 X confocal systems. For immunofluorescence results in Figure S2 (nodules/non‐lesion areas), S6‐8, the images represent maximal projections of a Z‐stack of 9–12 μm thickness with 1 μm intervals along the z‐axis.
2.7. Quantification of morphological parameters
The morphometric parameters were quantified from confocal Z‐stack images of 50 μm thickness with 1 μm intervals along the z‐axis taken from IBA1 and Ly‐6C stained sections at 63x magnification. To determine the process diameter, length, volume, total number, soma volume, terminal endpoints, number of primary branches and total cell volume, microglia were 3D reconstructed and retraced using the Filament Tracer function of Imaris 7.0 software (Bitplane). The number of microglia analyzed varies as only microglia were quantified that were fully captured by the Z‐stack. A ramification index was calculated by dividing the total number of terminal endpoints by the total number of primary branches per cell. For further ramification quantifications, maximal intensity projections were created with ImageJ and converted to binary images. ImageJ was used to perform the Sholl analysis (Sholl analysis plugin) with intersections of a 1‐pixel radius step size (original plugin developed by Ferreira et al., 2014).
2.8. Statistical analysis and graphs
Graphs were created with Prism 9 (Graphpad Prism v9.3.0) and arranged in Adobe Illustrator CC (Adobe). Statistical analysis was performed with Prism 9 (Graphpad Prism v9.3.0) using one‐way ANOVA. Post hoc Tukey's multiple comparison tests were applied to identify differences between groups. Differences were defined as significant when the adjusted p‐value was below .05.
3. RESULTS
3.1. Microglia increase in numbers and acquire a hyper‐ramified morphology in chronic EAE
To experimentally probe the contribution of microglia to SPMS, we induced crEAE in Biozzi ABH mice. crEAE is characterized by an acute phase, where mice develop hind limb paralysis and reach an EAE score of 4, followed by almost complete remission to only minor tail paresis with a score of 0.5 (Figure 1a). Subsequently, paralytic relapses occur with variable degrees of remission and the accumulation of residual deficit. The last phase is characterized by very slow deterioration (Al‐Izki et al., 2012; Pryce et al., 2005), culminating in hind limb paresis with a score 3–3.5, termed here as the chronic phase (Figure 1a). We deployed an optimized FACS‐based protocol to identify and isolate microglia and other immune cells from the indicated EAE stages (Figure S1). Characterization of microglia has always been challenging due to the overlap in cell surface markers between microglia and macrophages. Addition of Ly‐6C to established myeloid markers CD11b and CD45 allowed for a more precise discrimination between microglia and infiltrated monocytes/macrophages (Vainchtein et al., 2014). This allowed us to collect three classes of immune cell types from spinal cords during acute, remission and chronic EAE phases (Figure 1b). Microglia were classified as CD11bpos, CD45pos, and Ly‐6Cneg based on a previously established protocol (Vainchtein et al., 2014) (Figure S1). Peripheral myeloid infiltrates were defined as CD11bpos CD45high Ly‐6Cpos and contained macrophages and granulocytes (Figures 1b and S1). Myeloid cells captured in gate 2 represent a heterogeneous population with varying levels of expression of CD45 and CD11b. Furthermore, the relative abundance of myeloid subpopulations in gate 2 varies over the different EAE stages. The myeloid cells expressing high levels of CD11b are dominating the acute phase, while myeloid cells with lower levels of CD11b are relatively highly abundant in the chronic phase (Figure S1). In addition to myeloid cells, we also quantified CD45high CD3pos T‐lymphocytes (Figures 1b and S1). In line with previous studies, the acute phase of EAE was accompanied by extensive infiltration of innate immune cells and T‐lymphocytes (Allen et al., 1993; Baker et al., 1990; Jackson et al., 2009) that were no longer present at remission (Figures 1b and S1). At the chronic phase, a modest increase in the number of peripheral myeloid infiltrates was observed. These data strongly suggest that peripheral immune cells are actively involved at the acute phase. At later phases, even though the mice display chronic hind leg paresis, the contribution of infiltrated immune cells is limited. In contrast, a persistent increase in microglia numbers was observed at all crEAE phases (Figure 1b).
FIGURE 1.
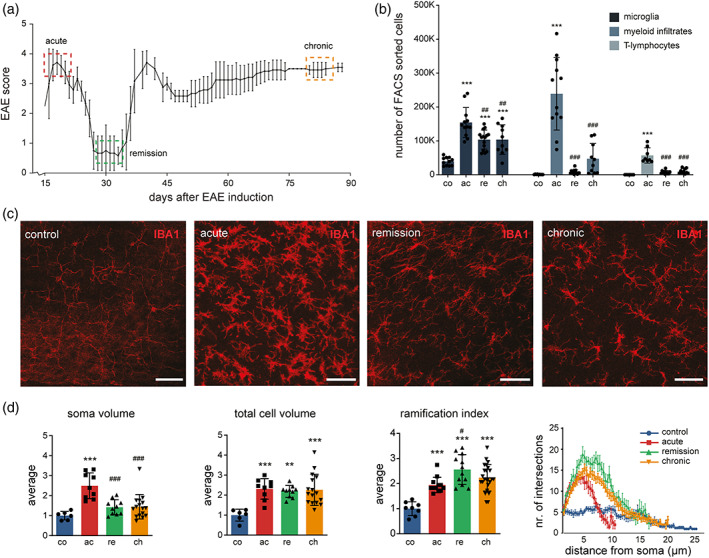
Microglia increase in numbers and acquire a hyper‐ramified morphology in chronic EAE. (a) Mice were sacrificed at the indicated crEAE stages (dashed boxes) and as naïve controls. Controls (n = 22); acute phase (red, n = 19), remission (green, n = 18) and the chronic stage (orange, n = 15). (b) Quantification of the number of sorted cells at crEAE stages. Three distinct cell populations were isolated by FACS gating: microglia as CD11bpos CD45pos Ly‐6Cneg, macrophages/monocytes/granulocytes as CD11bpos CD45high Ly‐6Cpos, T‐lymphocytes as CD45high CD3pos. Significant differences between samples were determined based on one‐way ANOVA and Tukey's multiple comparisons tests. ##p ≤ .01, ***/###p ≤ .001; * represents a significant difference with controls; # indicates a significant difference with acute EAE. Co = controls (n = 15), ac = acute phase (n = 16), re = remission (n = 15), ch = chronic phase (n = 12). Error bars represent the standard deviation. (c) Representative confocal Z‐stack images of 50 μm sagittal spinal cord sections at crEAE stages stained against IBA1. Images represent spinal cord white matter from Biozzi ABH mice. Scale bars indicate 50 μm. (d) Average soma volume, total cell volume, ramification index and Sholl analysis at all crEAE stages. All conditions have been normalized to control microglia. For average soma volume and total cell volume: co = controls (n = 6; n = number of microglia), ac = acute phase (n = 9), re = remission (n = 10), ch = chronic phase (n = 16). Sholl analysis result showing the number of intersections depending on the distance from the soma for each crEAE stage. For ramification index and Sholl analysis: controls (n = 8), acute (n = 12), remission (n = 12) and chronic phase (n = 24). Significant differences were detected based on one‐way ANOVA and Tukey's multiple comparisons tests. #p ≤ .05, **p ≤ .01, ***/###p ≤ .001; * represents a significant difference with controls, # indicates a significant difference with acute EAE. Error bars represent the standard deviation.
Morphological changes of microglia have been associated with functional changes in response to pathology (Dubbelaar et al., 2018; van Olst et al., 2021; Zrzavy et al., 2017). To study whether crEAE induction has a wide‐spread effect on microglia, the morphology of microglia in non‐lesion white matter was analyzed (Figure 1c). Representative examples of non‐lesion white matter are provided in Figure S2. Dramatic changes in microglia morphology were observed. At the acute phase, microglia had shorter, thicker processes, a larger cell body and an increased overall cell volume compared to control microglia (Figures 1d and S3). In the remission and chronic stages, the soma size decreased, while they became more ramified, reaching a hyper‐ramified state when compared to controls (Figure 1d), as exemplified by increased total number of processes and processes terminal endpoints (Figure S3). In brief, these data show that microglia numbers increase at all crEAE stages over control, they retract their processes at the acute phase and become more ameboid like, but acquire a hyper‐ramified morphology at remission and chronic EAE stages.
3.2. Microglia acquire distinct functions throughout crEAE and obtain tissue repair properties in remission and chronic phases
To define the transcriptional changes that occur in microglia during the progression of crEAE, we performed RNA sequencing on FACS‐isolated microglia from acute, remission and chronic phase EAE (Figure 2a and Table S2). Microglia acquire a pro‐inflammatory phenotype at the acute phase (Ajami et al., 2018; Ponomarev et al., 2005) that is defined by an increase in cytokine production, T‐cell activation and cell–cell adhesion, phagocytosis, antigen presentation and interferon signaling (C3, Cd74, Axl, H2‐D1, H2‐Aa, Vim, Stat1, Irf1) (Figures 2b, S4a, and S5a, b, and Tables S3 and S4). This is accompanied by a loss of expression of microglial homeostatic genes (e.g., Fcrls, P2ry12, Hexb, Tmem119) (Figure 2c) as described before (Miedema et al., 2020). Throughout all stages of crEAE, microglia show increased expression of phagocytosis related genes (Figure S5a, b). Representative images of co‐labelling of IBA1 positive microglia and AXL confirmed this notion at the protein level (Figure S5c). In contrast, the pro‐inflammatory signature is lost after the acute phase, and genes related to lipid metabolism (Trem2, Lpl, Plpp1), synapse organization (Plxna4, Cacna1a, Disc1), tissue remodeling (Sparc, Lama3, and Serpine2) and microglial homeostasis are increased in the remission and chronic phase (Figure 2b, c). Interestingly, microglia from remission and chronic EAE are quite similar in terms of gene expression and we only detected 96 differentially expressed genes (DEGs; FDR < 0.01; Figures 2a, S4a and Table S3). Both stages are characterized by minimal infiltration of peripheral immune cells (Figure 1b), but animals in the chronic phase have a high EAE score with obvious paralysis symptoms. Similarities between remission and chronic EAE microglia include interleukin‐1 signaling as part of the overlapping signature genes (Figures 2d, e, S4b,c and Tables S5 and S6). This was confirmed at the protein level, where dense clusters of IL‐1β positive microglia were identified in nodules at the remission and chronic phase (Figure S6). This is reminiscent of previously described hot spots of proliferating microglia that express the IL1 receptor and depend on IL1 signaling for proliferation (Bruttger et al., 2015). A clear proliferative profile was overall most prominent in the chronic phase (cell cycling; Kif14, Kif4, Kif11, Cdca8, and Mki67) (Figure 2b) and chronic EAE microglia also expressed more genes involved in interferon signaling (Stat1, Irf1, Acod1, Iigp1) compared to remission EAE microglia (Figure 2b). Ki67+ proliferating microglia were mostly found in the acute and chronic phase at the protein level, and to a lesser extent in remission (Figure S7). As for interferon signaling, we stained tissues for STAT1, a protein downstream of IFN in the signaling pathway (Regis et al., 2008). Most microglia were STAT1+ in the acute and chronic phase, both in the lesions and surrounding white matter, whereas in remission STAT1 was only found in microglia nodules and lesions (Figure S8). This is relevant to crEAE progression, as interferon signaling has been shown to be important for tissue repair and remodeling (Dorman et al., 2021; Julier et al., 2017). Summarizing, these data suggest that microglia display a tissue supportive phenotype following the acute phase.
FIGURE 2.
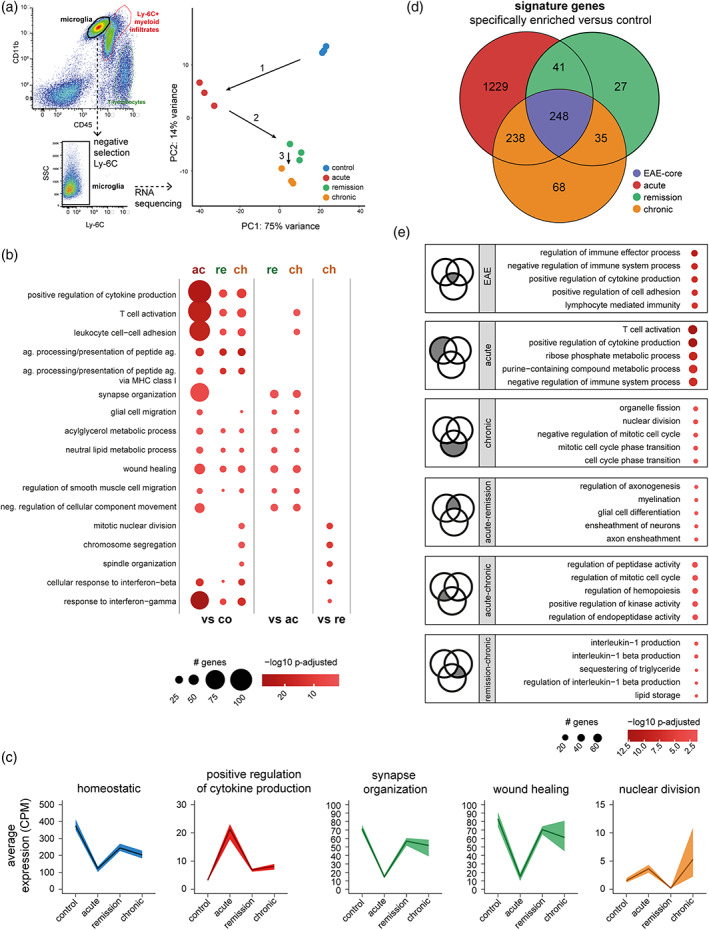
Microglia acquire distinct functions throughout crEAE and obtain tissue repair properties in remission and chronic phases. (a) Schematic overview of the FACS gating strategy and principal component analysis plot representing the variance of microglia transcriptomes at different crEAE stages, n = 3 per condition. Arrows and numbers in the PCA plot indicate the chronological order of crEAE stages. SSC = side‐scatter. (b) Significantly enriched gene ontology (GO) terms associated with differentially enriched genes in the pairwise comparison indicated. (c) Average counts per million (CPM) expression of a gene set across microglia samples (dark line) and minimal and maximal expression (borders) are indicated. In blue: median expression of 25 microglia homeostatic genes published by (Butovsky et al., 2014). In red: median expression of genes enriched in acute vs control and annotated to the GO term “positive regulation of cytokine production”. In green: median expression of genes enriched in remission vs acute and annotated to the GO term “synapse organization” and “wound healing”. In orange: median expression of genes enriched in chronic vs remission and annotated to the GO term “nuclear division”. (d) crEAE stage‐specific genes were determined by comparing gene expression levels of microglia per crEAE stage to microglia from controls. Signature genes (numbers in bold) were defined as enriched with log2 fold change >1 and p‐adjusted <.01 for one stage, but not any other stage. Genes enriched in all crEAE stages compared to control were combined in a core EAE profile (purple). (e) The top five significant GO terms that are associated with signature genes identified in (d) are depicted. Dark gray areas in the Venn diagrams (small insets on the left) indicate which gene set is represented. Significant GO terms were ranked by gene count.
3.3. Chronic EAE microglia likely acquire a mix of phenotypes
In recent years, major efforts have been conducted to characterize the full range of states that microglia can assume during demyelination, MS pathology, development, and aging (Hammond et al., 2019; Maggi et al., 2021; Masuda et al., 2019; Miedema et al., 2022; Safaiyan et al., 2021). Using single‐cell RNA sequencing the cluster markers of different microglia states have been identified (Table S7). To detect the presence of gene expression signatures representing these microglia subsets in chronic EAE, we investigated if cluster markers were enriched in chronic EAE relative to other crEAE stages. Hammond et al. (2019) defined microglia clusters IR2 and IR1 from mice injected with a demyelinating agent LPC or saline, respectively. The injury‐response microglia gene set from cluster 2 (IR2) was expressed in both acute and chronic EAE microglia (Figure 3a). Axon‐tract associated microglia are detected in the corpus callosum at the time of extensive myelination in the early postnatal mouse brain (P4/5) and characterized by expression of genes such as Spp1, Igf1, and Lpl (Figure 3a). The marker genes of the axon‐tract associated microglia are also enriched in the crEAE acute and chronic phase. Similarly, a white matter‐associated microglia expression (WAM) profile (Safaiyan et al., 2021) was observed in both acute and chronic microglia (Figure 3a). The WAM gene set is associated with microglia that form dense nodules and phagocytose damaged myelin in aging wild‐type mice (Safaiyan et al., 2021). Masuda et al. (2019) described microglia clusters from the corpus callosum of mice during cuprizone‐induced toxic demyelination and remyelination (Figure 3b). Cuprizone‐enriched genes are strongly enriched in the acute and chronic phase of EAE, but show a low expression in remission (Figure 3b). Masuda et al. (2019) and Miedema et al. (2022) described human microglia clusters associated with MS. In addition, spatial analysis of white matter lesions identified two microglia subsets, MIMS‐foamy and MIMS‐iron, with an increased abundance at the lesion rim (Absinta et al., 2021). Most of the marker genes representing MS‐associated microglia subsets showed the highest expression in the acute EAE stage, whereas chronic EAE demonstrated a variable enrichment for gene sets representing MS‐associated microglia subsets (Figures 3b and S9a). The human microglia Hu‐C8 cluster markers were most enriched in microglia in the acute and chronic EAE stage (Figure 3b). In remission and chronic EAE microglia the most prominent enrichment in human microglia gene sets was identified for MIMS‐foamy (Figure 3b). This gene set is representing a microglia subset at the rim of chronic‐active MS lesions with an attributed role in myelin clearance (Absinta et al., 2021). Next, to prioritize the most relevant genes in gene sets from single‐cell studies, we intersected the 90 genes enriched in chronic compared to remission EAE microglia (Figure S4a) with gene sets from single‐cell clusters (Table S7). Genes occurring at least three times as single‐cell cluster markers and with enriched expression in chronic EAE microglia were Spp1, Gpnmb, Dab2, and Msr1 (Figures S9b and 3c). Taken together, it seems that chronic EAE microglia are composed of a mixed population, where previously described gene expression profiles overlap partly with acute and injury‐responsive gene expression and partly with remission and homeostatic gene expression profiles.
FIGURE 3.
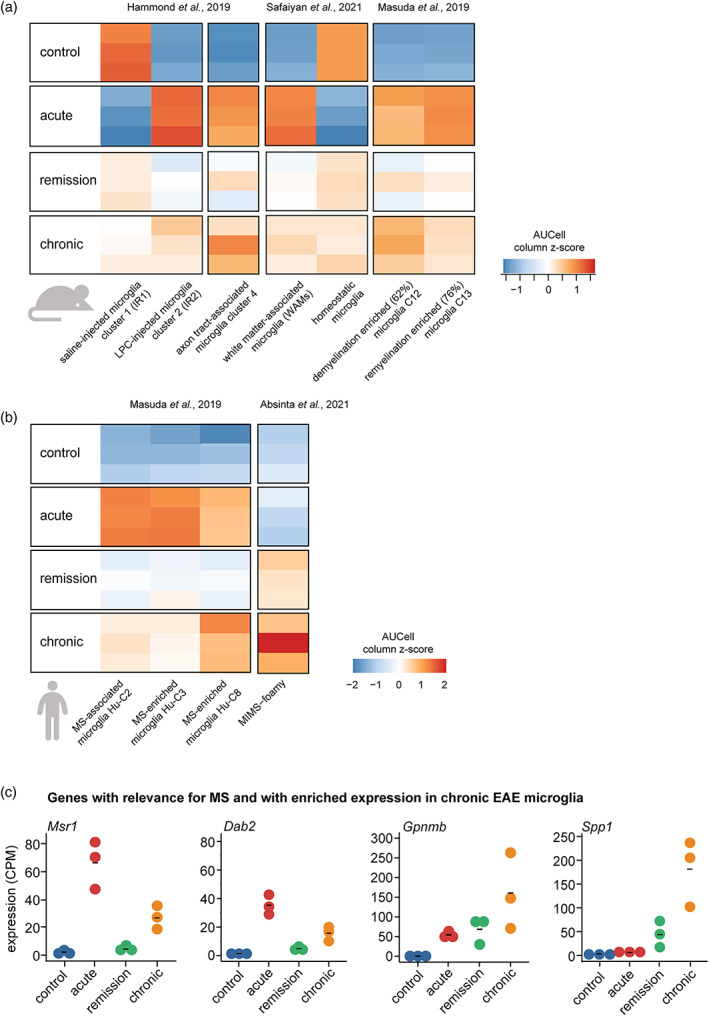
Chronic EAE microglia acquire a mix of phenotypes. (a) Heatmap depicting AUCell enrichment of gene sets representing microglia subsets in different mouse models, including injury‐response microglia cluster markers (IR1 = 43 genes; IR2 = 154 genes), axon tract‐associated microglia cluster markers (140 genes), white matter‐associated microglia cluster marker genes (WAMs = 35 genes; homeostatic cluster = 25 genes) and cuprizone microglia cluster markers from corpus callosum (C12 demyelination‐enriched = 84 genes; C13 remyelination‐enriched = 69 genes) reported by Hammond et al., 2019; Safaiyan et al., 2021 and Masuda et al., 2019 at crEAE stages. Heatmap colors indicate the column z‐scores of AUCell enrichment. LPC = l‐α‐lysophosphatidylcholine. (b) Heatmap depicting AUCell scores of human microglia cluster marker genes, including the single‐cell RNA sequencing clusters reported by Masuda et al., 2019 (Hu‐C2 = 149 genes, Hu‐C3 = 18 genes, Hu‐C8 = 40 genes) and Absinta et al., 2021 (MIMS‐foamy = 100 genes). Heatmap colors indicate the column z‐scores of AUCell enrichment. (c) Gene expression levels in microglia at crEAE stages for selected cluster marker genes. The selected genes occurred at least three times in the investigated single‐cell gene sets and are enriched in chronic EAE microglia compared to remission EAE microglia. C, cluster; CPM; counts per million; Hu, human; MIMS, microglia inflamed in MS; MS, multiple sclerosis
4. DISCUSSION
Here we describe the morphological changes and the transcriptional profile of microglia during crEAE induced in the Biozzi ABH mouse. In this mouse model, disease progresses through an acute phase with severe disability, followed by a remission phase with significant improvement of symptoms, finally resulting in a chronic phase where paralysis is returned (Baker et al., 1990). We found that microglia express genes involved in debris clearance and antigen presentation at all crEAE stages. Looking at distinct disease stages, the acute EAE microglia acquire an immune activated pro‐inflammatory gene expression signature. This has been described previously in multiple neurodegenerative diseases including MS (Jordão et al., 2019; Masuda et al., 2019; Zrzavy et al., 2017). During remission, the inflammatory response in microglia is reduced and there is an increase in some genes involved in lipid metabolism and tissue remodeling. Between remission and chronic phases, we observed a large overlap in gene expression signatures for microglia. However, microglia in chronic EAE partially regain an inflammatory gene signature in conjunction with increased proliferation.
For microglia isolated from the chronic stage of EAE, gene expression signatures and morphological analyses first pointed at an intermediate phenotype between remission and acute microglia. However, when comparing our RNA sequencing data with previously published findings on microglia gene signatures associated with myelination or myelin degradation/regeneration, we observed that the chronic phase has a different enrichment score for each gene signature (Absinta et al., 2021; Hammond et al., 2019; Masuda et al., 2019; Miedema et al., 2022; Safaiyan et al., 2021). This is in contrast to, for example, acute phase microglia that show a consistent enrichment for gene signatures detected in a broad range of contexts (WAMs, development, focal demyelination, and cuprizone‐induced demyelination). Recent single‐cell RNA sequencing studies have demonstrated microglial heterogeneity in both health and disease, with particular microglia subsets arising in disease (Hammond et al., 2019; Masuda et al., 2019; Prinz et al., 2021; Safaiyan et al., 2021). Possibly, the partial or inconsistent overlap of chronic EAE microglia suggests that they are a mix of multiple and partly opposing subsets of microglia: some microglia are involved in repair and remission, some proliferate, while other subsets regain an activated state. A high diversity in myeloid subsets already exists in the context of acute EAE (Jordão et al., 2019), and we can only expect myeloid heterogeneity to expand after a remission and relapse phase.
One transcript, Spp1, showed large increases in chronic EAE microglia compared to remission and was reported multiple times as marker for mouse microglia subpopulations associated with MS pathology (Absinta et al., 2021; Hammond et al., 2019; Masuda et al., 2019; Miedema et al., 2022; Safaiyan et al., 2021). Spp1 transcripts encode the multi‐functional protein osteopontin, which can be cleaved into multiple functional fragments, immobilized in the extracellular matrix to modulate cell‐to‐cell signaling, or can be secreted as soluble cytokine (Rosmus et al., 2022). In EAE, after cleavage of osteopontin by thrombin, osteopontin‐integrin signaling inhibits apoptosis of autoreactive T‐cells (Hur et al., 2007; Steinman, 2009), possibly driving further tissue injury. Osteopontin also stimulates the expression of Th1 and Th17‐type cytokines controlling T‐cell differentiation (Steinman, 2009). In MS patients, three studies reported an increase in osteopontin levels in the CSF and blood during relapses (Chowdhury et al., 2008; Comabella et al., 2005; Vogt et al., 2004). Additionally, the presence of osteopontin‐autoantibodies in a subgroup of MS patients was correlated with a reduced number of relapses (Clemente et al., 2017). Our results indicate that compared to acute EAE microglia, expression of osteopontin is much more pronounced in chronic EAE microglia, the phase when neurodegenerative damage and clinical disability are accumulating. We speculate that osteopontin might not only be associated with glial‐derived inflammation, but may also drive tissue injury. For future studies, it would be highly relevant to investigate possible inhibition of osteopontin in microglia during the chronic, neurodegenerative phase of EAE to develop future therapies against MS.
Transcriptionally, microglia from chronic and remission stages of EAE are quite similar, however, as described above, we did observe some relevant differences. The differences are mainly related to immune activation and proliferation. With regard to immune activation, we hypothesize that a possible cause for this lies in the communication with other (immune) cells. Microglia communicate extensively with other cells through direct cellular interactions, ligand‐receptor interactions and extracellular vesicles. Harmonious intercellular communication involving microglia is important to maintain a healthy state of the tissue environment and to overcome pathology including neuroinflammation (Borst et al., 2021; Forbes & Miron, 2021; Ronzano et al., 2021). Our FACS analysis indicated that, during remission, the majority of myeloid and T cells that infiltrated from the periphery in the acute stage had been cleared from the CNS. However, in the chronic stage there is a certain level of re‐entry of cells in the CNS, in particular of myeloid cells. Studies that distinguish between resident microglia and infiltrating monocyte‐derived macrophages have been complicated by the difficulties in separating these two populations, though recent advances have helped to segregate them, also in case of MS (Masuda et al., 2019). In this study, we have used Ly‐6C to separate microglia from monocyte‐derived macrophages in EAE (Vainchtein et al., 2014). One limitation of the current FACS‐gating strategy is that we cannot exclude that a small population of CNS‐resident perivascular, meningeal and choroid plexus associated macrophages, collectively known as border‐associated macrophages, might be included in the captured cell population (Kierdorf et al., 2019). However, given that CD45 expression varies between microglia and BAMs, the latter showing higher CD45 levels by FACS (Masuda et al., 2020), it is more likely that BAMs are localized to gate 2 and not a major contaminant in gate 1 (Figure S1). More importantly, it has become clear that there is extensive crosstalk between monocytes and microglia that affects their function (Greenhalgh et al., 2018; Plemel et al., 2020). Microglia have been found to modulate the entry of monocytes into the CNS (Plemel et al., 2020) and, at least in vitro, monocyte‐derived macrophages have immuno‐modulatory influence on microglia (Greenhalgh et al., 2018). Therefore, we think it is possible that the signaling‐based interaction between infiltrating cells and microglia results in the induction of a mild inflammatory response in all microglia, or, a more likely scenario, an inflammatory response in a subset of the microglia.
We furthermore observed an increase in the microglial expression of genes related to proliferation in the chronic stage of EAE. In case of CNS pathology, microglia proliferate through a process of selective clonal expansion (Tay et al., 2017). Previous work has demonstrated that also in a classical EAE model, local proliferation of resident macrophages occurred alongside continuous monocytic infiltration up to the peak of disease, followed by apoptosis of resident macrophages (Jordão et al., 2019). Similarly, we observed a significant increase in the number of sorted microglia in the acute stage of EAE that was decreased during remission. Based on Ki67, we detected proliferating microglia, mostly present in the acute phase and chronic phase. Interestingly, we identified the presence of small nodules of microglia expressing IL‐1B in both remission and chronic stages. Similar microglial nodules marked by expression of IL‐1B have been previously observed in animal models for MS and in the CNS of MS patients (Burm et al., 2016;Singh et al., 2013; van Horssen et al., 2012). In MS, axonal degeneration is detected at the sites of microglial nodules and both pro‐ and anti‐inflammatory properties have been ascribed to the associated microglia (Singh et al., 2013; van Horssen et al., 2012).
Considering the hypothesis that distinct microglia subtypes emerge in chronic EAE, in the future, single‐cell RNA‐sequencing for chronic phase EAE microglia and other immune cells could delineate the processes underlying the slow, clinical deterioration during chronic EAE. Analyzing single human microglia from MS donors is complicated due to (a) scarcity of brain tissue, (b) aging bias in control tissue, (c) (anti‐inflammatory) medication, (d) post‐mortem artifacts, (e) the fact that samples are derived from end‐stage disease, and (e) difficulties to obtain high numbers of microglia per donor (Masuda et al., 2019; Miedema et al., 2022). For example, cluster marker re‐analysis of human single microglia by Masuda et al., 2019 was hampered because clusters with limited cell numbers were formed. Hu‐C8 and Hu‐C4 consisted of 34 and 17 single microglia, respectively (the latter was excluded from re‐analysis). The Biozzi ABH mouse model would offer the possibility to investigate pathological events using high numbers of immune cells. Detailed single‐cell RNA sequencing analysis of immune cells at distinct crEAE scores would allow for the correlation of gene sets and immune subtypes to progression in clinical crEAE. The data presented here indicates that microglia obtain various features in different stages of crEAE and in particular in the chronic stage, which might mean that specific targeting of these microglia to modulate disease outcome in the progressive phase might be feasible. Further analysis of microglia and other immune cells in crEAE will contribute to our understanding of the role of microglia in later disease stages. In the future, this helps to determine if subtypes of microglia might be stimulated to perform beneficial roles in repair and to restore CNS homeostasis.
CONFLICT OF INTEREST
The authors declare no conflicts of interest.
Supporting information
Figure S1 FACS gating strategy. Immune cells were isolated from spinal cords at crEAE stages using FACS gating strategies. Three distinct cell populations were isolated: (1) microglia as CD11bpos CD45pos Ly‐6Cneg (first Gate 1 then gated on Ly‐6Cneg cells), (2) macrophages/monocytes/granulocytes as CD11bpos CD45high Ly‐6Cpos (first Gate 2 then gated on Ly‐6Cpos cells), (3) T‐lymphocytes as CD45high CD3pos (first Gate 3 then gated on CD3pos cells). Representative FACS plots are shown.
Figure S2 Peripheral immune infiltrates are not IBA1 positive.
Representative confocal images of sagittal spinal cord sections for DNA (Hoechst, white), IBA1 (magenta) and Ly‐6C (cyan). Images represent spinal cord white matter from Biozzi ABH mice. At all crEAE stages and controls, IBA1 and Ly‐6C did not colocalize. To highlight microglia morphology in nodules (arrow heads) versus non‐lesion areas (asterisks), high magnification images are included. Scale bars in the tile scan overviews (single layer images) indicate 100 μm. Scale bars in magnification (maximal z stack projections) images indicate 25 μm.
Figure S3 Detailed quantification of microglia morphology. The total number of processes, process volume, length, diameter, terminal endpoints and number of primary branches of microglia were quantified using Imaris 3D reconstruction software. Sagittal spinal cord sections of 50 μm were used at all crEAE stages. All conditions were normalized to control microglia (set to 1). Significant differences were determined with one‐way ANOVA and Tukey's multiple comparisons tests. * represents a significant difference with controls, # a significant difference with acute EAE. Error bars represent the standard deviation. */#p ≤ .05, **/##p ≤ .01, ***p ≤ .01, controls (n = 6–7), acute phase (n = 9), remission (n = 10), chronic phase (n = 16). n = number of microglia.
Figure S4 Differentially expressed genes in microglia throughout crEAE stages. (a) Volcano plot showing the fold changes of gene expression (x‐axis log2 scale) and their significance (y‐axis log10 scale) between acute EAE to control microglia (upper panel), remission EAE to acute EAE microglia (middle) and chronic EAE to remission EAE microglia (lower panel). Dotted horizontal lines represent the p‐value threshold of ‐log10 (p‐adjusted = .01). Dotted vertical lines represent the log2 fold change thresholds set for differentially expressed genes at >1 or <−1. (b) Schematic overview of the method to determine signature genes starting from a gene expression matrix. (c) Median expression level of signature genes at crEAE stages. Average CPM (line) and minimal and maximal CPM (borders) are indicated, n = 3 for all conditions.
Figure S5 Phagocytosis‐related gene expression levels in microglia. (a) Gene expression levels in microglia quantified by RNA sequencing at crEAE stages. Genes shown are MHCII components H2‐Aa and Cd74, co‐stimulatory molecule Cd86 and apoptotic cell uptake receptor Axl. Expression levels were normalized for library size using counts per million (CPM). Significant differences were determined with DESeq2 using Wald tests and Benjamini‐Hochberg adjustment. * represent a significant difference with controls, # a significant difference with acute EAE. **/##p‐adjusted ≤.01 and log2 fold change >1 or <−1, n = 3 for all conditions. (b) Relative expression differences in microglia of phagocytosis‐related genes quantified by RT‐qPCR at crEAE stages. RNA expression levels were normalized to Hmbs and controls were set to 1. Significant differences were determined with one‐way ANOVA and Tukey's multiple comparisons tests. * represents a significant difference with controls, # a significant difference with acute EAE. *p ≤ .05, **p ≤ .01, ***p < .001. controls (n = 6), acute phase (n = 7), remission (n = 7), chronic phase (n = 6). co = controls, ac = acute phase, re = remission, ch = chronic phase, CPM = counts per million. (c) Representative confocal images of 50 μm sagittal spinal cord sections stained against AXL and IBA1 at crEAE stages. Images represent spinal cord white matter from Biozzi ABH mice. Scale bars indicate 50 μm.
Figure S6 IL‐1β expressing microglia are detected in nodules. Representative confocal images of sagittal spinal cord sections stained against DNA (white, Hoechst), IBA1 (magenta), Ly6C (cyan) and IL‐1β (yellow) at crEAE stages. Images represent maximal z stack projections of spinal cord white matter from Biozzi ABH mice. Scale bars indicate 25 μm.
Figure S7 Proliferating microglia are detected at acute and chronic EAE stages and decreased in frequency at remission stage. Representative confocal images of sagittal spinal cord sections stained against DNA (white, Hoechst), IBA1 (magenta), Ly6C (cyan) and Ki‐67 (yellow) at crEAE stages. Images represent maximal z stack projections of spinal cord white matter from Biozzi ABH mice. Scale bars indicate 25 μm.
Figure S8 STAT1‐positive microglia are abundantly detected at acute and chronic EAE stages and only a subset of microglia is STAT1‐positive at the remission EAE stage. Representative confocal images of sagittal spinal cord sections stained against DNA (white, Hoechst), IBA1 (magenta), Ly‐6C (cyan) and STAT1 (yellow) at crEAE stages. Images represent maximal z stack projections of spinal cord white matter from Biozzi ABH mice. Scale bars indicate 25 μm.
Figure S9 Enrichment of gene sets representing microglia subsets and the overlap with chronic‐enriched genes. (a) Heatmap depicting AUCell enrichment of gene sets representing a human microglia subset enriched at the rim of chronic‐active white matter lesions expressing ferritin‐related genes (MIMS‐iron = 100 genes) and a microglia subset enriched in white matter lesions expressing phagocytosis‐related genes (Hs7‐microglia = 453 genes). Heatmap colors indicate the column z‐scores of AUCell enrichment. (b) Venn diagrams depicting the overlap between the cluster markers of microglia subsets from the indicated single‐cell RNA sequencing studies (Absinta et al., 2021; Hammond et al., 2019; Masuda et al., 2019; Miedema et al., 2022; Safaiyan et al., 2021) and the significantly enriched genes in chronic EAE microglia compared to remission EAE microglia (chronic‐enriched = 90 genes).
Table S1 qPCR primer information
Table S2. Gene expression data per sample.
Table S3. Differentially expressed genes between crEAE stages.
Table S4. Enriched GO terms for differentially enriched genes at crEAE stages.
Table S5. Signature genes at crEAE stages.
Table S6. Enriched GO terms for signature genes at crEAE stages.
Table S7. Single‐cell RNA sequencing cluster marker genes previously identified for different microglia subsets.
ACKNOWLEDGMENTS
We want to thank Dr. Gary Warnes from the Flow Cytometry Core Facility for his excellent FACS assistance and Dr. Gareth Pryce for his intensive help with the crEAE Biozzi ABH mice from the Blizard Institute, London, United Kingdom. Ilia D. Vainchteinwas supported by an MS Stichting grant (10‐723 MS). Jonathan Vinet was supported by a training fellowship from the Italian MS Foundation (FISM, 2011/B/7). Susanne M. Kooistra was supported by an MS‐fellowship from Stichting MS Research (16‐947). Sequencing was performed with the assistance from Diana Spierings of the ERIBA Research Sequencing Facility.
Vainchtein, I. D. , Alsema, A. M. , Dubbelaar, M. L. , Grit, C. , Vinet, J. , van Weering, H. R. J. , Al‐Izki, S. , Biagini, G. , Brouwer, N. , Amor, S. , Baker, D. , Eggen, B. J. L. , Boddeke, E. W. G. M. , & Kooistra, S. M. (2023). Characterizing microglial gene expression in a model of secondary progressive multiple sclerosis. Glia, 71(3), 588–601. 10.1002/glia.24297
Ilia D. Vainchtein and Astrid M. Alsema contributed equally to this study.
Funding information Italian MS Foundation, Grant/Award Number: 2011/B/7; Stichting MS Research, Grant/Award Numbers: 10‐723, 16‐947
Contributor Information
Ilia D. Vainchtein, Email: ilia.vainchtein@gmail.com.
Susanne M. Kooistra, Email: s.m.kooistra@umcg.nl.
DATA AVAILABILITY STATEMENT
The RNA sequencing datasets generated for this study have been deposited in the NCBI GEO database under accession number GSE194071.
REFERENCES
- Absinta, M. , Maric, D. , Gharagozloo, M. , Garton, T. , Smith, M. D. , Jin, J. , Fitzgerald, K. C. , Song, A. , Liu, P. , Lin, J. , Wu, T. , Johnson, K. R. , Mcgavern, D. B. , Schafer, D. P. , Calabresi, P. A. , & Reich, D. S. (2021). A lymphocyte – microglia – astrocyte axis in chronic active multiple sclerosis. Nature, 597(2020), 709–714. 10.1038/s41586-021-03892-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aibar, S. , González‐Blas, C. B. , Moerman, T. , Huynh‐Thu, V. A. , Imrichova, H. , Hulselmans, G. , Rambow, F. , Marine, J. C. , Geurts, P. , Aerts, J. , Van Den Oord, J. , Atak, Z. K. , Wouters, J. , & Aerts, S. (2017). SCENIC: Single‐cell regulatory network inference and clustering. Nature Methods, 14(11), 1083–1086. 10.1038/NMETH.4463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ajami, B. , Samusik, N. , Wieghofer, P. , Ho, P. P. , Crotti, A. , Bjornson, Z. , Prinz, M. , Fantl, W. J. , Nolan, G. P. , & Steinman, L. (2018). Single‐cell mass cytometry reveals distinct populations of brain myeloid cells in mouse neuroinflammation and neurodegeneration models. Nature Neuroscience, 21(4), 541–551. 10.1038/s41593-018-0100-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al‐Izki, S. , Pryce, G. , O'Neill, J. K. , Butter, C. , Giovannoni, G. , Amor, S. , & Baker, D. (2012). Practical guide to the induction of relapsing progressive experimental autoimmune encephalomyelitis in the Biozzi ABH mouse. Multiple Sclerosis and Related Disorders, 1(1), 29–38. 10.1016/J.MSARD.2011.09.001 [DOI] [PubMed] [Google Scholar]
- Allen, S. J. , Baker, D. , O'Neill, J. K. , Davison, A. N. , & Turk, J. L. (1993). Isolation and characterization of cells infiltrating the spinal cord during the course of chronic relapsing experimental allergic encephalomyelitis in the Biozzi AB/H mouse. Cellular Immunology, 146(2), 335–350. 10.1006/CIMM.1993.1031 [DOI] [PubMed] [Google Scholar]
- Amor, S. & Baker, D . (2012). Checklist for reporting and reviewing studies of experimental animal models of multiple sclerosis and related disorders. Multiple Sclerosis and Related Disorders, 1(3), 111–115. 10.1016/J.MSARD.2012.01.003 [DOI] [PubMed] [Google Scholar]
- Anders, S. , Pyl, P. T. , & Huber, W. (2015). HTSeq–a python framework to work with high‐throughput sequencing data. Bioinformatics, 31(2), 166–169. 10.1093/BIOINFORMATICS/BTU638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, D. & Amor, S . (2012). Publication guidelines for refereeing and reporting on animal use in experimental autoimmune encephalomyelitis. Journal of Neuroimmunology, 242(1–2), 78–83. 10.1016/J.JNEUROIM.2011.11.003 [DOI] [PubMed] [Google Scholar]
- Baker, D. , & Amor, S. (2014). Experimental autoimmune encephalomyelitis is a good model of multiple sclerosis if used wisely. Multiple Sclerosis and Related Disorders, 3(5), 555–564. 10.1016/J.MSARD.2014.05.002 [DOI] [PubMed] [Google Scholar]
- Baker, D. , Gerritsen, W. , Rundle, J. , & Amor, S. (2011). Critical appraisal of animal models of multiple sclerosis. Multiple Sclerosis, 17(6), 647–657. 10.1177/1352458511398885 [DOI] [PubMed] [Google Scholar]
- Baker, D. , O'Neill, J. K., Gschmeissner, S. E., Wilcox, C. E., Butter, C. & Turk, J. L. (1990). Induction of chronic relapsing experimental allergic encephalomyelitis in Biozzi mice. Journal of Neuroimmunology, 28(3), 261–270. 10.1016/0165-5728(90)90019-J [DOI] [PubMed] [Google Scholar]
- Biozzi, G. , Stiffel, C. , Mouton, D. , Bouthillier, Y. , & Decreusefond, C. (1972). Cytodynamics of the immune response in two lines of mice genetically selected for “high” and “low” antibody synthesis. The Journal of Experimental Medicine, 135(5), 1071–1094. 10.1084/JEM.135.5.1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakemore, W. F. , & Franklin, R. J. M. (2008). Remyelination in experimental models of toxin‐induced demyelination. Current Topics in Microbiology and Immunology, 318, 193–212. 10.1007/978-3-540-73677-6_8 [DOI] [PubMed] [Google Scholar]
- Borst, K. , Dumas, A. A. , & Prinz, M. (2021). Microglia: Immune and non‐immune functions. Immunity, 54(10), 2194–2208. 10.1016/J.IMMUNI.2021.09.014 [DOI] [PubMed] [Google Scholar]
- Brown, A. , McFarlin, D. E. , & Raine, C. S. (1982). Chronologic neuropathology of relapsing experimental allergic encephalomyelitis in the mouse. Laboratory Investigation; a Journal of Technical Methods and Pathology, 46(2), 171–185. [PubMed] [Google Scholar]
- Bruttger, J. , Karram, K. , Wörtge, S. , Regen, T. , Marini, F. , Hoppmann, N. , Klein, M. , Blank, T. , Yona, S. , Wolf, Y. , Mack, M. , Pinteaux, E. , Müller, W. , Zipp, F. , Binder, H. , Bopp, T. , Prinz, M. , Jung, S. , & Waisman, A. (2015). Genetic cell ablation reveals clusters of local self‐renewing microglia in the mammalian central nervous system. Immunity, 43(1), 92–106. 10.1016/J.IMMUNI.2015.06.012 [DOI] [PubMed] [Google Scholar]
- Burm, S. M. , Peferoen, L. A. N. , Zuiderwijk‐Sick, E. A. , Haanstra, K. G. , Hart, B. A. , van der Valk, P. , Amor, S. , Bauer, J. , & Bajramovic, J. J. (2016). Expression of IL‐1β in rhesus EAE and MS lesions is mainly induced in the CNS itself. Journal of Neuroinflammation, 13(1), 138. 10.1186/S12974-016-0605-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler, A. , Hoffman, P. , Smibert, P. , Papalexi, E. , & Satija, R. (2018). Integrating single‐cell transcriptomic data across different conditions, technologies, and species. Nature Biotechnology, 36, 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky, O. , Jedrychowski, M. P. , Moore, C. S. , Cialic, R. , Lanser, A. J. , Gabriely, G. , Koeglsperger, T. , Dake, B. , Wu, P. M. , Doykan, C. E. , Fanek, Z. , Liu, L. , Chen, Z. , Rothstein, J. D. , Ransohoff, R. M. , Gygi, S. P. , Antel, J. P. , & Weiner, H. L. (2014). Identification of a unique TGF‐β dependent molecular and functional signature in microglia. Nature Neuroscience, 17(1), 131–143. 10.1038/NN.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, S. A. , Lin, J. , & Sadiq, S. A. (2008). Specificity and correlation with disease activity of cerebrospinal fluid osteopontin levels in patients with multiple sclerosis. Archives of Neurology, 65(2), 232–235. 10.1001/archneurol.2007.33 [DOI] [PubMed] [Google Scholar]
- Clark, I. C. , Gutiérrez‐vázquez, C. , Wheeler, M. A. , Li, Z. , Rothhammer, V. , Linnerbauer, M. , Sanmarco, L. M. , Guo, L. , Blain, M. , Zandee, S. E. J. , Chao, C. , Batterman, K. V. , Schwabenland, M. , Lotfy, P. , Tejeda‐velarde, A. , Hewson, P. , Polonio, C. M. , Shultis, M. W. , Salem, Y. , & Quintana, F. J. (2021). Barcoded viral tracing of single‐cell interactions in central nervous system inflammation. Science, 372(6540), eabf1230. 10.1126/science.abf1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemente, N. , Comi, C. , Raineri, D. , Cappellano, G. , Vecchio, D. , Orilieri, E. , Gigliotti, C. L. , Boggio, E. , Dianzani, C. , Sorosina, M. , Martinelli‐Boneschi, F. , Caldano, M. , Bertolotto, A. , Ambrogio, L. , Sblattero, D. , Cena, T. , Leone, M. , Dianzani, U. , & Chiocchetti, A. (2017). Role of anti‐osteopontin antibodies in multiple sclerosis and experimental autoimmune encephalomyelitis. Frontiers in Immunology, 8(3), 1–12. 10.3389/fimmu.2017.00321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comabella, M. , Pericot, I. , Goertsches, R. , Nos, C. , Castillo, M. , Blas Navarro, J. , Río, J. , & Montalban, X. (2005). Plasma osteopontin levels in multiple sclerosis. Journal of Neuroimmunology, 158(1–2), 231–239. 10.1016/j.jneuroim.2004.09.004 [DOI] [PubMed] [Google Scholar]
- Compston, A. , & Coles, A. (2008). Multiple sclerosis. The Lancet, 372(9648), 1502–1517. 10.1016/S0140-6736(08)61620-7 [DOI] [PubMed] [Google Scholar]
- International Multiple Sclerosis Genetics Consortium . (2019). Multiple sclerosis genomic map implicates peripheral immune cells & microglia in susceptibility. Science, 365(6460), 50. 10.1126/science.aav7188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross, A. H. , McCarron, R. , McFarlin, D. E. , & Raine, C. S. (1987). Adoptively transferred acute and chronic relapsing autoimmune encephalomyelitis in the PL/J mouse and observations on altered pathology by intercurrent virus infection. Laboratory investigation. A Journal of Technical Methods and Pathology, 57(5), 499–512. [PubMed] [Google Scholar]
- de Haas, A. H. , Boddeke, H. W. G. M. , & Biber, K. (2008). Region‐specific expression of immunoregulatory proteins on microglia in the healthy CNS. Glia, 56(8), 888–894. 10.1002/GLIA.20663 [DOI] [PubMed] [Google Scholar]
- Dorman, L. C. , Nguyen, P. T. , Escoubas, C. C. , Vainchtein, I. D. , Xiao, Y. , Lidsky, P. V. , Wang, E. Y. , Taloma, S. E. , Nakao‐Inoue, H. , Rivera, B. M. , Condello, C. , Andino, R. , Nowakowski, T. J. , & Molofsky, A. V. (2021). A type I interferon response defines a conserved microglial state required for effective phagocytosis. BioRxiv, 2021(4), 441889. 10.1101/2021.04.29.441889 [DOI] [Google Scholar]
- Dubbelaar, M. L. , Kracht, L. , Eggen, B. J. L. , & Boddeke, E. W. G. M. (2018). The kaleidoscope of microglial phenotypes. Frontiers in Immunology, 9, 1753. 10.3389/FIMMU.2018.01753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira, T. , Ou, Y. , Li, S. , Giniger, E., & van Meyel, D. (2014). Dendrite architecture organized by transcriptional control of the F‐actin nucleator Spire. Development, 141(3), 650–60. 10.1242/dev.099655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes, L. H. , & Miron, V. E. (2021). Monocytes in central nervous system remyelination. Glia, 70, 797–807. 10.1002/GLIA.24111 [DOI] [PubMed] [Google Scholar]
- Geladaris, A. , Häusler, D. , & Weber, M. S. (2021). Microglia: The missing link to decipher and therapeutically control MS progression? International Journal of Molecular Sciences, 22(7), 3461. 10.3390/ijms22073461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- George, N. I. , & Chang, C. W. (2014). DAFS: A data‐adaptive flag method for RNA‐sequencing data to differentiate genes with low and high expression. BMC Bioinformatics, 15(1), 1–11. 10.1186/1471-2105-15-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh, A. D. , Zarruk, J. G. , Healy, L. M. , Baskar Jesudasan, S. J. , Jhelum, P. , Salmon, C. K. , Formanek, A. , Russo, M. V. , Antel, J. P. , McGavern, D. B. , McColl, B. W. , & David, S. (2018). Peripherally derived macrophages modulate microglial function to reduce inflammation after CNS injury. PLoS Biology, 16(10), e2005264. 10.1371/JOURNAL.PBIO.2005264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grün, D. , Muraro, M. J. , Wiebrands, K. , Lyubimova, A. , Dharmadhikari, G. , Born, M. , Jansen, E. , Clevers, H. , Koning, E. J. P. , & van Oudenaarden, A. (2016). De novo prediction of stem cell identity using single‐cell transcriptome data. Cell Stem Cell, 19, 1–12. 10.1016/j.stem.2016.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond, T. R. , Dufort, C. , Dissing‐Olesen, L. , Giera, S. , Young, A. , Wysoker, A. , Walker, A. J. , Gergits, F. , Segel, M. , Nemesh, J. , Marsh, S. E. , Saunders, A. , Macosko, E. , Ginhoux, F. , Chen, J. , Franklin, R. J. M. , Piao, X. , McCarroll, S. A. , & Stevens, B. (2019). Single‐cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell‐state changes. Immunity, 50(1), 253–271.e6. 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampton, D. W. , Anderson, J. , Pryce, G. , Irvine, K. A. , Giovannoni, G. , Fawcett, J. W. , Compston, A. , Franklin, R. J. M. , Baker, D. , & Chandran, S. (2008). An experimental model of secondary progressive multiple sclerosis that shows regional variation in gliosis, remyelination, axonal and neuronal loss. Journal of Neuroimmunology, 201–202, 200–211. 10.1016/J.JNEUROIM.2008.05.034 [DOI] [PubMed] [Google Scholar]
- Heppner, F. L. , Greter, M. , Marino, D. , Falsig, J. , Raivich, G. , Hövelmeyer, N. , Waisman, A. , Rülicke, T. , Prinz, M. , Priller, J. , Becher, B. , & Aguzzi, A. (2005). Experimental autoimmune encephalomyelitis repressed by microglial paralysis. Nature Medicine, 11(2), 146–152. 10.1038/nm1177 [DOI] [PubMed] [Google Scholar]
- Hur, E. M. , Youssef, S. , Haws, M. E. , Zhang, S. Y. , Sobel, R. A. , & Steinman, L. (2007). Osteopontin‐induced relapse and progression of autoimmune brain disease through enhanced survival of activated T cells. Nature Immunology, 8(1), 74–83. 10.1038/ni1415 [DOI] [PubMed] [Google Scholar]
- Jackson, S. J. , Lee, J. , Nikodemova, M. , Fabry, Z. , & Duncan, I. D. (2009). Quantification of myelin and axon pathology during relapsing progressive experimental autoimmune encephalomyelitis in the Biozzi ABH mouse. Journal of Neuropathology and Experimental Neurology, 68(6), 616–625. 10.1097/NEN.0B013E3181A41D23 [DOI] [PubMed] [Google Scholar]
- Joost, E. , Jordão, M. J. C. , Mages, B. , Prinz, M. , Bechmann, I. , & Krueger, M. (2019). Microglia contribute to the glia limitans around arteries, capillaries and veins under physiological conditions, in a model of neuroinflammation and in human brain tissue. Brain Structure & Function, 224(3), 1301–1314. 10.1007/S00429-019-01834-8 [DOI] [PubMed] [Google Scholar]
- Jordão, M. J. C. , Sankowski, R. , Brendecke, S. M. , Sagar, L. , Tai, Y. H. , Tay, T. L. , Schramm, E. , Armbruster, S. , Hagemeyer, N. , Groß, O. , Mai, D. , Çiçek, Ö. , Falk, T. , Kerschensteiner, M. , Grün, D. , & Prinz, M. (2019). Neuroimmunology: Single‐cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science, 363(6425), eaat7554. 10.1126/science.aat7554 [DOI] [PubMed] [Google Scholar]
- Julier, Z. , Park, A. J. , Briquez, P. S. , & Martino, M. M. (2017). Promoting tissue regeneration by modulating the immune system. Acta Biomaterialia, 53, 13–28. 10.1016/J.ACTBIO.2017.01.056 [DOI] [PubMed] [Google Scholar]
- Keren‐Shaul, H. , Spinrad, A. , Weiner, A. , Matcovitch‐Natan, O. , Dvir‐Szternfeld, R. , Ulland, T. K. , David, E. , Baruch, K. , Lara‐Astaiso, D. , Toth, B. , Itzkovitz, S. , Colonna, M. , Schwartz, M. , & Amit, I. (2017). A unique microglia type associated with restricting development of Alzheimer's disease. Cell, 169(7), 1276–1290. 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- Kierdorf, K. , Masuda, T. , Jordão, M. J. C. , & Prinz, M. (2019). Macrophages at CNS interfaces: Ontogeny and function in health and disease. Nature Reviews Neuroscience, 20(9), 547–562. 10.1038/s41583-019-0201-x [DOI] [PubMed] [Google Scholar]
- Kilkenny, C. , Browne, W. J. , Cuthill, I. C. , Emerson, M. , & Altman, D. G. (2010). Improving bioscience research reporting: The ARRIVE guidelines for reporting animal research. PLoS Biology, 8(6), e1000412. 10.1371/JOURNAL.PBIO.1000412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Paggi, J. M. , Park, C. , Bennett, C. , & Salzberg, S. L. (2019). Graph‐based genome alignment and genotyping with HISAT2 and HISAT‐genotype. Nature Biotechnology, 37(8), 907–915. 10.1038/S41587-019-0201-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotter, M. R. , Li, W. W. , Zhao, C. , & Franklin, R. J. M. (2006). Myelin impairs CNS remyelination by inhibiting oligodendrocyte precursor cell differentiation. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience, 26(1), 328–332. 10.1523/JNEUROSCI.2615-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski, P. B. , Schuller‐Levis, G. B. , & Wisniewski, H. M. (1987). Induction of synchronized relapses in SJL/J mice with chronic relapsing experimental allergic encephalomyelitis. Acta Neuropathologica, 74(2), 163–168. 10.1007/BF00692847 [DOI] [PubMed] [Google Scholar]
- Krasemann, S. , Madore, C. , Cialic, R. , Baufeld, C. , Calcagno, N. , El Fatimy, R. , Beckers, L. , O'Loughlin, E. , Xu, Y. , Fanek, Z. , Greco, D. J. , Smith, S. T. , Tweet, G. , Humulock, Z. , Zrzavy, T. , Conde‐Sanroman, P. , Gacias, M. , Weng, Z. , Chen, H. , & Butovsky, O. (2017). The TREM2‐APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity, 47(3), 566–581. 10.1016/J.IMMUNI.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreutzberg, G. W. (1996). Microglia: A sensor for pathological events in the CNS. Trends in Neurosciences, 19(8), 312–318. 10.1016/0166-2236(96)10049-7 [DOI] [PubMed] [Google Scholar]
- Lampron, A. , Larochelle, A. , Laflamme, N. , Préfontaine, P. , Plante, M. M. , Sánchez, M. G. , Wee Yong, V. , Stys, P. K. , Tremblay, M. È. , & Rivest, S. (2015). Inefficient clearance of myelin debris by microglia impairs remyelinating processes. The Journal of Experimental Medicine, 212(4), 481–495. 10.1084/JEM.20141656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lassmann, H. , Van Horssen, J. , & Mahad, D. (2012). Progressive multiple sclerosis: Pathology and pathogenesis. Nature Reviews. Neurology, 8(11), 647–656. 10.1038/NRNEUROL.2012.168 [DOI] [PubMed] [Google Scholar]
- Lassmann, H. , & Wisniewski, H. M. (1978). Chronic relapsing EAE. Time course of neurological symptoms and pathology. Acta Neuropathologica, 43(1–2), 35–42. 10.1007/BF00684996 [DOI] [PubMed] [Google Scholar]
- Livak, K. J. , & Schmittgen, T. D. (2001). Analysis of relative gene expression data using real‐time quantitative PCR and the 2(‐delta delta C[T]) method. Methods, 25(4), 402–408. 10.1006/METH.2001.1262 [DOI] [PubMed] [Google Scholar]
- Lloyd, A. F. , & Miron, V. E. (2019). The pro‐remyelination properties of microglia in the central nervous system. Nature Reviews. Neurology, 15(8), 447–458. 10.1038/S41582-019-0184-2 [DOI] [PubMed] [Google Scholar]
- Lorentzen, J. C. , Issazadeh, S. , Storch, M. , Mustafa, M. I. , Lassman, H. , Linington, C. , Klareskog, L. , & Olsson, T. (1995). Protracted, relapsing and demyelinating experimental autoimmune encephalomyelitis in DA rats immunized with syngeneic spinal cord and incomplete Freund's adjuvant. Journal of Neuroimmunology, 63(2), 193–205. 10.1016/0165-5728(95)00153-0 [DOI] [PubMed] [Google Scholar]
- Love, M. I. , Huber, W. , & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA‐seq data with DESeq2. Genome Biology, 15(12), 550. 10.1186/S13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi, P. , Kuhle, J. , Schädelin, S. , van der Meer, F. , Weigel, M. , Galbusera, R. , Mathias, A. , Lu, P. J. , Rahmanzadeh, R. , Benkert, P. , La Rosa, F. , Bach Cuadra, M. , Sati, P. , Théaudin, M. , Pot, C. , van Pesch, V. , Leppert, D. , Stadelmann, C. , Kappos, L. , & Granziera, C. (2021). Chronic white matter inflammation and serum neurofilament levels in multiple sclerosis. Neurology, 97(6), e543–e553. 10.1212/WNL.0000000000012326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda, T. , Amann, L. , Sankowski, R. , Staszewski, O. , Lenz, M. , d'Errico, P. , Snaidero, N. , Costa Jordão, M. J. , Böttcher, C. , Kierdorf, K. , Jung, S. , Priller, J. , Misgeld, T. , Vlachos, A. , Luehmann, M. M. , Knobeloch, K. P. , & Prinz, M. (2020). Novel Hexb‐based tools for studying microglia in the CNS. Nature Immunology, 21(7), 802–815. 10.1038/s41590-020-0707-4 [DOI] [PubMed] [Google Scholar]
- Masuda, T. , Sankowski, R. , Staszewski, O. , Böttcher, C. , Amann, L. , Sagar, Scheiwe, C. , Nessler, S. , Kunz, P. , van Loo, G. , Coenen, V. A. , Reinacher, P. C. , Michel, A. , Sure, U. , Gold, R. , Grün, D. , Priller, J. , Stadelmann, C. , & Prinz, M. (2019). Spatial and temporal heterogeneity of mouse and human microglia at single‐cell resolution. Nature, 566(7744), 388–392. 10.1038/s41586-019-0924-x [DOI] [PubMed] [Google Scholar]
- Melief, J. , Schuurman, K. G. , van de Garde, M. D. B. , Smolders, J. , van Eijk, M. , Hamann, J. , & Huitinga, I. (2013). Microglia in normal appearing white matter of multiple sclerosis are alerted but immunosuppressed. Glia, 61(11), 1848–1861. 10.1002/GLIA.22562 [DOI] [PubMed] [Google Scholar]
- Miedema, A. , Gerrits, E. , Brouwer, N. , Jiang, Q. , Kracht, L. , Meijer, M. , Nutma, E. , Baert, R. P. , Pijnacker, A. T. E. , Wesseling, E. M. , Wijering, M. H. C. , Gabius, H. J. , Amor, S. , Eggen, B. J. L. , & Kooistra, S. M. (2022). Brain macrophages acquire distinct transcriptomes in multiple sclerosis lesions and normal appearing white matter. Acta Neuropathologica Communications, 10(8), 1–18. 10.1186/s40478-021-01306-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedema, A. , Wijering, M. H. C. , Eggen, B. J. L. , & Kooistra, S. M. (2020). High‐resolution transcriptomic and proteomic profiling of heterogeneity of brain‐derived microglia in multiple sclerosis. Frontiers in Molecular Neuroscience, 13, 583811. 10.3389/FNMOL.2020.583811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen, J. C. , Thompson, K. K. , West, B. L. , & Tsirka, S. E. (2018). Csf1R inhibition attenuates experimental autoimmune encephalomyelitis and promotes recovery. Experimental Neurology, 307, 24–36. 10.1016/J.EXPNEUROL.2018.05.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peferoen, L. A. N. , Vogel, D. Y. S. , Ummenthum, K. , Breur, M. , Heijnen, P. D. A. M. , Gerritsen, W. H. , Peferoen‐Baert, R. M. B. , van der Valk, P. , Dijkstra, C. D. , & Amor, S. (2015). Activation status of human microglia is dependent on lesion formation stage and remyelination in multiple sclerosis. Journal of Neuropathology and Experimental Neurology, 74(1), 48–63. 10.1097/NEN.0000000000000149 [DOI] [PubMed] [Google Scholar]
- Plemel, J. R. , Stratton, J. A. , Michaels, N. J. , Rawji, K. S. , Zhang, E. , Sinha, S. , Baaklini, C. S. , Dong, Y. , Ho, M. , Thorburn, K. , Friedman, T. N. , Jawad, S. , Silva, C. , Caprariello, A. V. , Hoghooghi, V. , Yue, J. , Jaffer, A. , Lee, K. , Kerr, B. J. , & Wee Yong, V. (2020). Microglia response following acute demyelination is heterogeneous and limits infiltrating macrophage dispersion. Science Advances, 6(3), eaay6324. 10.1126/SCIADV.AAY6324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponomarev, E. D. , Shriver, L. P. , Maresz, K. , & Dittel, B. N. (2005). Microglial cell activation and proliferation precedes the onset of CNS autoimmunity. Journal of Neuroscience Research, 81(3), 374–389. 10.1002/JNR.20488 [DOI] [PubMed] [Google Scholar]
- Praet, J. , Guglielmetti, C. , Berneman, Z. , Van der Linden, A. , & Ponsaerts, P. (2014). Cellular and molecular neuropathology of the cuprizone mouse model: Clinical relevance for multiple sclerosis. Neuroscience and Biobehavioral Reviews, 47, 485–505. 10.1016/J.NEUBIOREV.2014.10.004 [DOI] [PubMed] [Google Scholar]
- Prinz, M. , Masuda, T. , Wheeler, M. A. , & Quintana, F. J. (2021). Microglia and central nervous system‐associated macrophages‐from origin to disease modulation. Annual Review of Immunology, 39, 251–277. 10.1146/ANNUREV-IMMUNOL-093019-110159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryce, G. , O'Neill, J. K. , Croxford, J. L. , Amor, S. , Hankey, D. J. , East, E. , Giovannoni, G. , & Baker, D. (2005). Autoimmune tolerance eliminates relapses but fails to halt progression in a model of multiple sclerosis. Journal of Neuroimmunology, 165(1–2), 41–52. 10.1016/J.JNEUROIM.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Regis, G. , Pensa, S. , Boselli, D. , Novelli, F. , & Poli, V. (2008). Ups and downs: The STAT1:STAT3 seesaw of interferon and gp130 receptor signalling. Seminars in Cell and Developmental Biology, 19(4), 351–359. 10.1016/j.semcdb.2008.06.004 [DOI] [PubMed] [Google Scholar]
- Robinson, S. , & Miller, R. H. (1999). Contact with central nervous system myelin inhibits oligodendrocyte progenitor maturation. Developmental Biology, 216(1), 359–368. 10.1006/DBIO.1999.9466 [DOI] [PubMed] [Google Scholar]
- Ronzano, R. , Roux, T. , Thetiot, M. , Aigrot, M. S. , Richard, L. , Lejeune, F. X. , Mazuir, E. , Vallat, J. M. , Lubetzki, C. , & Desmazières, A. (2021). Microglia‐neuron interaction at nodes of Ranvier depends on neuronal activity through potassium release and contributes to remyelination. Nature Communications, 12(1), 5219. 10.1038/S41467-021-25486-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosmus, D. D. , Lange, C. , Ludwig, F. , Ajami, B. , & Wieghofer, P. (2022). The role of osteopontin in microglia biology: Current concepts and future perspectives. Biomedicine, 10(4), 840. 10.3390/biomedicines10040840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaiyan, S. , Besson‐Girard, S. , Kaya, T. , Cantuti‐Castelvetri, L. , Liu, L. , Ji, H. , Schifferer, M. , Gouna, G. , Usifo, F. , Kannaiyan, N. , Fitzner, D. , Xiang, X. , Rossner, M. J. , Brendel, M. , Gokce, O. , & Simons, M. (2021). White matter aging drives microglial diversity. Neuron, 109(7), 1100–1117. 10.1016/J.NEURON.2021.01.027 [DOI] [PubMed] [Google Scholar]
- Scheu, S. , Ali, S. , Mann‐Nüttel, R. , Richter, L. , Arolt, V. , Dannlowski, U. , Kuhlmann, T. , Klotz, L. , & Alferink, J. (2019). Interferon β‐mediated protective functions of microglia in central nervous system autoimmunity. International Journal of Molecular Sciences, 20, 190. 10.3390/IJMS20010190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, S. , Metz, I. , Amor, S. , van der Valk, P. , Stadelmann, C. , & Brück, W. (2013). Microglial nodules in early multiple sclerosis white matter are associated with degenerating axons. Acta Neuropathologica, 125(4), 595–608. 10.1007/S00401-013-1082-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman, L. (2009). A molecular trio in relapse and remission in multiple sclerosis. Nature Reviews Immunology, 9(6), 440–447. 10.1038/nri2548 [DOI] [PubMed] [Google Scholar]
- Tay, T. L. , Mai, D. , Dautzenberg, J. , Fernández‐Klett, F. , Lin, G. , Sagar, S. , Datta, M. , Drougard, A. , Stempfl, T. , Ardura‐Fabregat, A. , Staszewski, O. , Margineanu, A. , Sporbert, A. , Steinmetz, L. M. , Pospisilik, J. A. , Jung, S. , Priller, J. , Grün, D. , Ronneberger, O. , & Prinz, M. (2017). A new fate mapping system reveals context‐dependent random or clonal expansion of microglia. Nature Neuroscience, 20(6), 793–803. 10.1038/NN.4547 [DOI] [PubMed] [Google Scholar]
- Taylor, W. A. (1986). Experimental allergic encephalomyelitis. A useful model for multiple sclerosis. Progress in clinical and biological research. Journal of Neurology, Neurosurgery, and Psychiatry, 49(3), 339–340. [Google Scholar]
- Vainchtein, I. D. , Vinet, J. , Brouwer, N. , Brendecke, S. , Biagini, G. , Biber, K. , Boddeke, H. W. G. M. , & Eggen, B. J. L. (2014). In acute experimental autoimmune encephalomyelitis, infiltrating macrophages are immune activated, whereas microglia remain immune suppressed. Glia, 62(10), 1724–1735. 10.1002/GLIA.22711 [DOI] [PubMed] [Google Scholar]
- van Horssen, J. , Singh, S. , van der Pol, S. , Kipp, M. , Lim, J. L. , Peferoen, L. , Gerritsen, W. , Kooi, E. J. , Witte, M. E. , Geurts, J. J. G. , de Vries, H. E. , Peferoen‐Baert, R. , van den Elsen, P. J. , van der Valk, P. , & Amor, S. (2012). Clusters of activated microglia in normal‐appearing white matter show signs of innate immune activation. Journal of Neuroinflammation, 9, 156. 10.1186/1742-2094-9-156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Olst, L. , Rodriguez‐Mogeda, C. , Picon, C. , Kiljan, S. , James, R. E. , Kamermans, A. , van der Pol, S. M. A. , Knoop, L. , Michailidou, I. , Drost, E. , Franssen, M. , Schenk, G. J. , Geurts, J. J. G. , Amor, S. , Mazarakis, N. D. , van Horssen, J. , de Vries, H. E. , Reynolds, R. , & Witte, M. E. (2021). Meningeal inflammation in multiple sclerosis induces phenotypic changes in cortical microglia that differentially associate with neurodegeneration. Acta Neuropathologica, 141(6), 881–899. 10.1007/S00401-021-02293-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt, M. H. J. , Floris, S. , Killestein, J. , Knol, D. L. , Smits, M. , Barkhof, F. , Polman, C. H. , & Nagelkerken, L. (2004). Osteopontin levels and increased disease activity in relapsing‐remitting multiple sclerosis patients. Journal of Neuroimmunology, 155(1–2), 155–160. 10.1016/j.jneuroim.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Voß, E. V. , Škuljec, J. , Gudi, V. , Skripuletz, T. , Pul, R. , Trebst, C. , & Stangel, M. (2012). Characterisation of microglia during de‐ and remyelination: Can they create a repair promoting environment? Neurobiology of Disease, 45(1), 519–528. 10.1016/J.NBD.2011.09.008 [DOI] [PubMed] [Google Scholar]
- Wang, L. , Wang, S. , & Li, W. (2012). RSeQC: Quality control of RNA‐seq experiments. Bioinformatics, 28(16), 2184–2185. 10.1093/bioinformatics/bts356 [DOI] [PubMed] [Google Scholar]
- Weinstock‐Guttman, B. , Nair, K. V. , Glajch, J. L. , Ganguly, T. C. , & Kantor, D. (2017). Two decades of glatiramer acetate: From initial discovery to the current development of generics. Journal of the Neurological Sciences, 376, 255–259. 10.1016/J.JNS.2017.03.030 [DOI] [PubMed] [Google Scholar]
- Wlodarczyk, A. , Løbner, M. , Cédile, O. , & Owens, T. (2014). Comparison of microglia and infiltrating CD11c+ cells as antigen presenting cells for T cell proliferation and cytokine response. Journal of Neuroinflammation, 11, 57. 10.1186/1742-2094-11-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki, R. , Lu, H. , Butovsky, O. , Ohno, N. , Rietsch, A. M. , Cialic, R. , Wu, P. M. , Doykan, C. E. , Lin, J. , Cotleur, A. C. , Kidd, G. , Zorlu, M. M. , Sun, N. , Hu, W. , Liu, L. P. , Lee, J. C. , Taylor, S. E. , Uehlein, L. , Dixon, D. , & Ransohoff, R. M. (2014). Differential roles of microglia and monocytes in the inflamed central nervous system. The Journal of Experimental Medicine, 211(8), 1533–1549. 10.1084/JEM.20132477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, J. , Tu, J., Lin, H., Shi, F., Liu, R., Zhao, C., Coons, S. W., Kuniyoshi, S. & Shi, J. , (2010). Centrally administered pertussis toxin inhibits microglia migration to the spinal cord and prevents dissemination of disease in an EAE mouse model. PLoS One, 5(8), e12400. 10.1371/JOURNAL.PONE.0012400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G. , Wang, L.‐G. , Han, Y. , & He, Q.‐Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology, 16(5), 284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrzavy, T. , Hametner, S. , Wimmer, I. , Butovsky, O. , Weiner, H. L. , & Lassmann, H. (2017). Loss of “homeostatic” microglia and patterns of their activation in active multiple sclerosis. Brain: A Journal of Neurology, 140(7), 1900–1913. 10.1093/BRAIN/AWX113 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 FACS gating strategy. Immune cells were isolated from spinal cords at crEAE stages using FACS gating strategies. Three distinct cell populations were isolated: (1) microglia as CD11bpos CD45pos Ly‐6Cneg (first Gate 1 then gated on Ly‐6Cneg cells), (2) macrophages/monocytes/granulocytes as CD11bpos CD45high Ly‐6Cpos (first Gate 2 then gated on Ly‐6Cpos cells), (3) T‐lymphocytes as CD45high CD3pos (first Gate 3 then gated on CD3pos cells). Representative FACS plots are shown.
Figure S2 Peripheral immune infiltrates are not IBA1 positive.
Representative confocal images of sagittal spinal cord sections for DNA (Hoechst, white), IBA1 (magenta) and Ly‐6C (cyan). Images represent spinal cord white matter from Biozzi ABH mice. At all crEAE stages and controls, IBA1 and Ly‐6C did not colocalize. To highlight microglia morphology in nodules (arrow heads) versus non‐lesion areas (asterisks), high magnification images are included. Scale bars in the tile scan overviews (single layer images) indicate 100 μm. Scale bars in magnification (maximal z stack projections) images indicate 25 μm.
Figure S3 Detailed quantification of microglia morphology. The total number of processes, process volume, length, diameter, terminal endpoints and number of primary branches of microglia were quantified using Imaris 3D reconstruction software. Sagittal spinal cord sections of 50 μm were used at all crEAE stages. All conditions were normalized to control microglia (set to 1). Significant differences were determined with one‐way ANOVA and Tukey's multiple comparisons tests. * represents a significant difference with controls, # a significant difference with acute EAE. Error bars represent the standard deviation. */#p ≤ .05, **/##p ≤ .01, ***p ≤ .01, controls (n = 6–7), acute phase (n = 9), remission (n = 10), chronic phase (n = 16). n = number of microglia.
Figure S4 Differentially expressed genes in microglia throughout crEAE stages. (a) Volcano plot showing the fold changes of gene expression (x‐axis log2 scale) and their significance (y‐axis log10 scale) between acute EAE to control microglia (upper panel), remission EAE to acute EAE microglia (middle) and chronic EAE to remission EAE microglia (lower panel). Dotted horizontal lines represent the p‐value threshold of ‐log10 (p‐adjusted = .01). Dotted vertical lines represent the log2 fold change thresholds set for differentially expressed genes at >1 or <−1. (b) Schematic overview of the method to determine signature genes starting from a gene expression matrix. (c) Median expression level of signature genes at crEAE stages. Average CPM (line) and minimal and maximal CPM (borders) are indicated, n = 3 for all conditions.
Figure S5 Phagocytosis‐related gene expression levels in microglia. (a) Gene expression levels in microglia quantified by RNA sequencing at crEAE stages. Genes shown are MHCII components H2‐Aa and Cd74, co‐stimulatory molecule Cd86 and apoptotic cell uptake receptor Axl. Expression levels were normalized for library size using counts per million (CPM). Significant differences were determined with DESeq2 using Wald tests and Benjamini‐Hochberg adjustment. * represent a significant difference with controls, # a significant difference with acute EAE. **/##p‐adjusted ≤.01 and log2 fold change >1 or <−1, n = 3 for all conditions. (b) Relative expression differences in microglia of phagocytosis‐related genes quantified by RT‐qPCR at crEAE stages. RNA expression levels were normalized to Hmbs and controls were set to 1. Significant differences were determined with one‐way ANOVA and Tukey's multiple comparisons tests. * represents a significant difference with controls, # a significant difference with acute EAE. *p ≤ .05, **p ≤ .01, ***p < .001. controls (n = 6), acute phase (n = 7), remission (n = 7), chronic phase (n = 6). co = controls, ac = acute phase, re = remission, ch = chronic phase, CPM = counts per million. (c) Representative confocal images of 50 μm sagittal spinal cord sections stained against AXL and IBA1 at crEAE stages. Images represent spinal cord white matter from Biozzi ABH mice. Scale bars indicate 50 μm.
Figure S6 IL‐1β expressing microglia are detected in nodules. Representative confocal images of sagittal spinal cord sections stained against DNA (white, Hoechst), IBA1 (magenta), Ly6C (cyan) and IL‐1β (yellow) at crEAE stages. Images represent maximal z stack projections of spinal cord white matter from Biozzi ABH mice. Scale bars indicate 25 μm.
Figure S7 Proliferating microglia are detected at acute and chronic EAE stages and decreased in frequency at remission stage. Representative confocal images of sagittal spinal cord sections stained against DNA (white, Hoechst), IBA1 (magenta), Ly6C (cyan) and Ki‐67 (yellow) at crEAE stages. Images represent maximal z stack projections of spinal cord white matter from Biozzi ABH mice. Scale bars indicate 25 μm.
Figure S8 STAT1‐positive microglia are abundantly detected at acute and chronic EAE stages and only a subset of microglia is STAT1‐positive at the remission EAE stage. Representative confocal images of sagittal spinal cord sections stained against DNA (white, Hoechst), IBA1 (magenta), Ly‐6C (cyan) and STAT1 (yellow) at crEAE stages. Images represent maximal z stack projections of spinal cord white matter from Biozzi ABH mice. Scale bars indicate 25 μm.
Figure S9 Enrichment of gene sets representing microglia subsets and the overlap with chronic‐enriched genes. (a) Heatmap depicting AUCell enrichment of gene sets representing a human microglia subset enriched at the rim of chronic‐active white matter lesions expressing ferritin‐related genes (MIMS‐iron = 100 genes) and a microglia subset enriched in white matter lesions expressing phagocytosis‐related genes (Hs7‐microglia = 453 genes). Heatmap colors indicate the column z‐scores of AUCell enrichment. (b) Venn diagrams depicting the overlap between the cluster markers of microglia subsets from the indicated single‐cell RNA sequencing studies (Absinta et al., 2021; Hammond et al., 2019; Masuda et al., 2019; Miedema et al., 2022; Safaiyan et al., 2021) and the significantly enriched genes in chronic EAE microglia compared to remission EAE microglia (chronic‐enriched = 90 genes).
Table S1 qPCR primer information
Table S2. Gene expression data per sample.
Table S3. Differentially expressed genes between crEAE stages.
Table S4. Enriched GO terms for differentially enriched genes at crEAE stages.
Table S5. Signature genes at crEAE stages.
Table S6. Enriched GO terms for signature genes at crEAE stages.
Table S7. Single‐cell RNA sequencing cluster marker genes previously identified for different microglia subsets.
Data Availability Statement
The RNA sequencing datasets generated for this study have been deposited in the NCBI GEO database under accession number GSE194071.


