FIGURE 2.
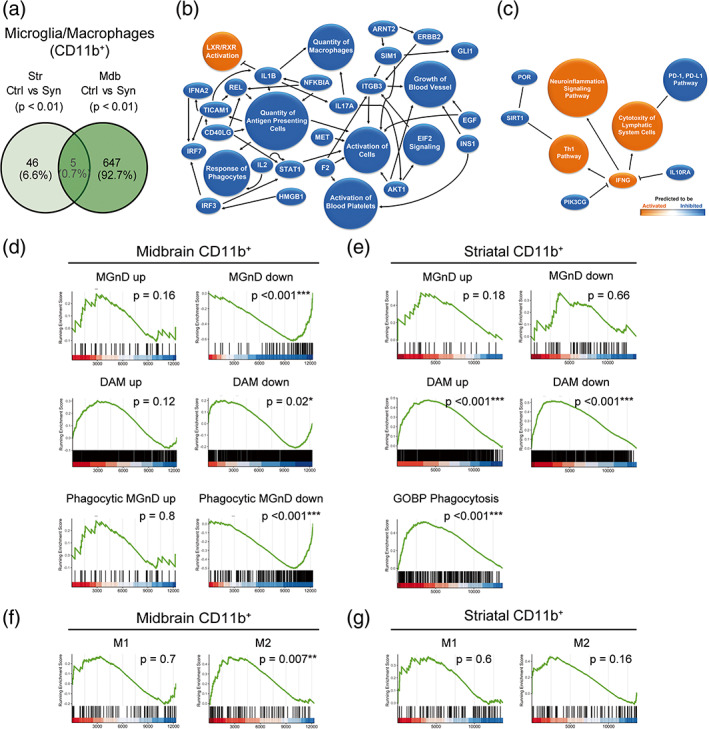
Transcriptomic analysis of myeloid cells purified from the striatum and the midbrain. Animals were sacrificed at 2 weeks after AAV9 administration in the SNpc. The striatum and the midbrain were dissected out and a cell suspension was prepared from these regions. Myeloid cells were separated based on the CD11b expression for RNA sequencing. (a) Venn diagram showing overlap of differentially expressed genes (p < .01) between CD11b+ cells from control and αSyn mice of the two regions. (n = 3 animals/group). (b) Graphical summary of the pathways, upstream regulators, and biological functions predicted to be altered in CD11b+ cells from the midbrain of αSyn mice. (c) Graphical summary of the pathways, upstream regulators, and biological functions predicted to be altered in CD11b+ cells from the striatum of αSyn mice. (d) Gene set enrichment analysis (GSEA) plots of the signatures enriched in midbrain myeloid cells of αSyn mice: Neurodegenerative microglia (MGnD), disease‐associated microglia (DAM) and phagocytic MGnD. GSEA was performed with the upregulated (“signature” up) and downregulated (“signature” down) genes of the signatures separately. (e) GSEA plots of the MGnD, DAM, and GOBP phagocytosis signatures enriched in striatal myeloid cells of αSyn overexpressing mice. GSEA was performed with the upregulated (“signature” up) and downregulated (“signature” down) genes of the MGnD and DAM signatures separately. (f) GSEA plots of the M1/M2 enrichment analysis in midbrain and (g) striatal myeloid cells of αSyn mice.
