ABSTRACT
The seed-borne microbiota and seed metabolites of the grass Achnatherum inebrians, either host to Epichloë gansuensis (endophyte infected [EI]) or endophyte free (EF), were investigated. This study determined the microbial communities both within the seed (endophytic) and on the seed surface (epiphytic) and of the protective glumes by using Illumina sequencing technology. Epichloë gansuensis decreased the richness of the seed-borne microbiota except for the epiphytic fungi of glumes and also decreased the diversity of seed-borne microbiota. In addition, metabolites of seeds and glumes were detected using liquid chromatography-mass spectrometry (LC-MS). Unlike with the seeds of EF plants, the presence of E. gansuensis resulted in significant changes in the content of 108 seed and 31 glume metabolites. A total of 319 significant correlations occurred between seed-borne microbiota and seed metabolites; these correlations comprised 163 (147 bacterial and 16 fungal) positive correlations and 156 (136 bacterial and 20 fungal) negative correlations. Meanwhile, there were 42 significant correlations between glume microbiota and metabolites; these correlations comprised 28 positive (10 bacterial and 18 fungal) and 14 negative (9 bacterial and 5 fungal) correlations. The presence of E. gansuensis endophyte altered the communities and diversities of seed-borne microbes and altered the composition and content of seed metabolites, and there were many close and complex relationships between microbes and metabolites.
IMPORTANCE The present study was to investigate seed-borne microbiota and seed metabolites in Achnatherum inebrians using high-throughput sequencing and LC-MS technology. Epichloë gansuensis decreased the richness of the seed-borne microbiota except for the epiphytic fungi of glumes and also decreased the diversity of seed-borne microbiota. Compared with endophyte-free plants, the content of 108 seed and 31 glume metabolites of endophyte-infected plants was significantly changed. There were 319 significant correlations between seed-borne microbiota and seed metabolites and 42 significant correlations between glume microbiota and metabolites.
KEYWORDS: Achnatherum inebrians, Epichloë gansuensis, microbiome, metabolomics, seed-borne, metabonomics
INTRODUCTION
Achnatherum inebrians is a perennial grass that is becoming increasing widespread in the highly modified arid/semiarid grasslands of northwest China (1–3). A feature of this invasive grass species is that nearly all plants are host to a seed-borne, mutualistic fungal endophyte, either Epichloë gansuensis (2, 4) or Epichloë inebrians (5). The presence of the Epichloë endophyte enhances tolerance to biotic stresses, such as insect pests (6) and pathogenic fungi (7, 8), and abiotic stresses, such as heavy metals (9), low temperature (10), salt (11), drought (12), low N (13), and low P (14). One important advantage provided by the presence of the Epichloë endophyte is deterrence of grazing by livestock due to toxicity from the presence of alkaloids, leading to the increasing prevalence of this grass. This toxicity has led to the common name of drunken horse grass (DHG). Although A. inebrians is commonly regarded as a toxic weed, the presence of these tall plants provides protected space where increasingly rare endemic plants can reestablish and so help to overcome the degradation of these grasslands (3).
The focus of this study was to investigate the fungi and bacteria associated with the seeds of field-grown A. inebrians plants, both when host to an Epichloë endophyte (endophyte infected [EI]) and when endophyte free (EF). Previous studies of seed microbiomes have focused on potentially beneficial (15, 16) and pathogenic (17, 18) microorganisms. Bacteria and fungi on the surfaces of leaves, and other plant tissues, are referred to as epiphytic (19), and those inside leaves are referred to as endophytic (20). Seed-borne microbes are similar to those of leaves. Nelson summarized that the microbes of seeds can be located in internal seed tissues (endophytic species) and on the seed surface (epiphytic species) and that interactions among these microorganisms taking place in, on, and around seeds may have a subsequent impact on plant fitness and productivity (21). To gain additional understanding of the fungi and bacteria associated with EI and EF seeds of A. inebrians, the present study also examined the microbiota of the protective glumes that cover seeds.
In addition to gaining greater understanding of the fungi and bacteria associated with EI and EF seeds, the study examined the metabolites that they contained. In particular, were there differences in the metabolites present in the EI and EF seeds and also in the glumes from EI and EF seeds? Also, were there metabolites present that were known to be secondary metabolites of the Epichloë endophyte and of other fungi and bacteria? Another possibility was that within the seeds were chemicals produced by seed tissues in response to the presence of invading fungi and bacteria.
Plant metabolomics has a wide range of potential applications (22) and has become a key technical method to reveal plant genetic diversity (23) and to better understand plant development (24). It also has a role in investigating the adaptation mechanisms of plants to abiotic stresses, including drought (25, 26), salt stress (27, 28), and temperature (29), and to biotic stresses such as pathogenic fungi (30, 31). This study utilized liquid chromatography-mass spectrometry (LC-MS), as this technology (32) is particularly important for high-sensitivity and nontargeted plant metabolomics and is suitable for the rich diversity of metabolites in plants (33, 34). Secondary metabolites of plants play an important role in the interaction between endophytic fungi and host plants (35). Studies have shown that colonization by endophytic fungi affects the content of metabolites in host plants under different treatments (13, 14).
Previous studies involving DHG found that the presence of E. gansuensis influenced the root-associated fungal community under different growth conditions (36). Based on the morphological identification of spores, research has found that E. gansuensis alters the arbuscular mycorrhizal fungi (AMF) community of DHG rhizosphere soil under different growth conditions (37). Recently, Zhong et al. illustrated that E. gansuensis influences the AMF community of DHG roots and rhizosphere soil under different water conditions, as determined through the application of high-throughput sequencing technology (38). In addition, E. gansuensis and soil moisture also affect the bacterial diversity of roots and rhizosphere soil of DHG by altering rhizosphere soil properties (39). Meanwhile, plant growth-promoting rhizobacteria isolated from the rhizosphere soil of DHG can promote seed germination and seedling growth under salt stress (40). But few studies focused on how the presence of E. gansuensis affects the microbiome diversity and metabolites of DHG seeds.
In order to explore how the presence of the E. gansuensis endophyte affects the microbiome community and metabolites of DHG seeds, we collected EI and EF seeds from a field of the Yuzhong campus (Lanzhou University); these seeds were used to conduct high-throughput sequencing and LC-MS technology analysis to investigate the effects of E. gansuensis on microbiome diversity and metabolism of DHG seeds and glumes.
We hypothesized that (i) E. gansuensis would alter the microbiome communities and diversities of A. inebrians seeds and glumes, (ii) E. gansuensis can influence the content and classes of metabolites in seeds and glumes, and (iii) there are complex and close correlations between seed-borne microbes and metabolites of EI and EF seeds and also between microbes and metabolites of EI and EF glumes.
RESULTS
Bacterial and fungal community compositions of seeds and glumes.
A total of 732,096 bacterial and 1,510,368 fungal sequences were obtained from seed and glume samples by high-throughput sequencing technology (see Table S1 in the supplemental material). All sequences were classified into 1,460 bacterial and 736 fungal operational taxonomy units (OTUs) at a 97% sequence similarity cutoff (Table S1). The 1,460 bacterial OTUs were divided into 11 phyla and 8 genera, and the 736 fungal OTUs were divided into 8 phyla and 10 genera. Among the bacterial communities of seed epiphytic microbes of endophyte-infected plants (SEpI), seed epiphytic microbes of endophyte-free plants (SEpF), seed endophytic microbes of endophyte-infected plants (SEnI), seed endophytic microbes of endophyte-free plants (SEnF), glume microbial community of endophyte-infected plants (GEpI), and glume microbial community of endophyte-free plants (GEpF), 498 OTUs were shared by all eight experimental plot sample repetitions (Fig. S2A; Table S2). In addition, in the fungal communities of SEpI, SEpF, SEnI, SEnF, GEpI, and GEpF, 377 OTUs were shared by all eight sample repetitions (Fig. S2B; Table S3). The bacterial and fungal rarefaction curves suggested that the 16S rRNA and internal transcribed spacer (ITS) gene sequences for all seed and glume samples reach the sequencing depths (Fig. S3A and B).
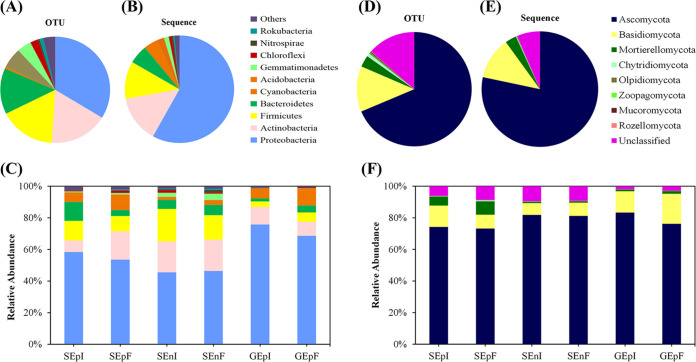
The relative abundances of bacteria and fungi at the phylum level of seed-borne communities were diverse among the three different parts of EI and EF seeds (Fig. 1; Table S1). The seed-borne bacterial communities were dominated by Proteobacteria (493 OTUs, 58% sequences), Actinobacteria (251 OTUs, 14% sequences), Firmicutes (243 OTUs, 11% sequences), and Bacteroidetes (204 OTUs, 5.7% sequences) (Fig. 1A and B; Table S1). The bacterial communities of seeds and glumes, at the genus level, were mostly dominated by Pseudomonas (15 OTUs, 3.6% sequences), Bacillus (10 OTUs, 2.2% sequences) Sphingomonas (9 OTUs, 4.9% sequences), and Allorhizobium (5 OTUs, 13.1% sequences) (Fig. S1A and B; Table S1). In addition, Ascomycota (505 OTUs, 78% sequences) was the most dominant phylum in the fungal community of seeds and glumes (Fig. 1D and E; Table S1); the next most dominant phyla were Basidiomycota (95 OTUs, 12% sequences) and Mortierellomycota (24 OTUs, 3% sequences) (Fig. 1D and E; Table S1). The most common genera of the fungal communities of seeds and glumes were Penicillium (27 OTUs, 2.0% sequences), Mortierella (18 OTUs, 2.8% sequences), and Fusarium (13 OTUs, 4.3% sequences) (Fig. S1D and E; Table S1).
FIG 1.
Taxonomic composition of seed and glume microbial communities. OTUs of bacteria (A) and fungi (D) and sequences of bacteria (B) and fungi (E) in all samples and relative abundances of bacteria (C) and fungi (F) at the phylum level located on or in Achnatherum inebrians seeds and in the endophyte treatments SEpI (seed epiphytic microbes of the seed-borne of endophyte-infected plants), SEpF (seed epiphytic microbes of the seed-borne of endophyte-free plants), SEnI (seed endophytic microbes of the seed-borne of endophyte-infected plants), SEnF (seed endophytic microbes of the seed-borne of endophyte-free plants), GEpI (glume microbial community of endophyte-infected plants), and GEpF (glume microbial community of endophyte-free plants) (n = 8). See also Table S1 in the supplemental material.
Bacterial and fungal community diversity of seeds and glumes.
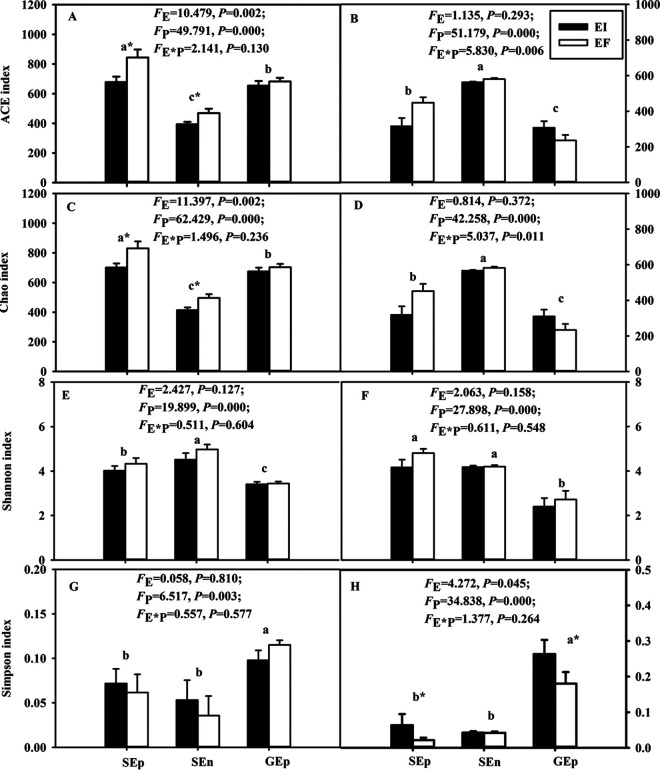
The ACE index and the Chao index of seed-borne bacterial communities were significantly (P < 0.05) influenced by the presence of the Epichloë endophyte (Fig. 2A and C). Meanwhile, the three different parts of EI and EF seeds also had significant (P < 0.05) differences, as illustrated by the ACE index, Chao index, Shannon index, and Simpson index (Fig. 2A, C, E, and G, respectively). For the ACE index and Chao index, epiphytic and endophytic bacterial diversities of seeds were significantly higher than the bacterial diversity of the epiphytic bacteria of glumes (Fig. 2A and C). As the Shannon index illustrated, the diversity of endophytic bacterial communities of A. inebrians seeds was significantly higher than the diversity of the epiphytic bacteria of A. inebrians seeds and of the bacteria of glumes (Fig. 2E). Furthermore, the Simpson index of the epiphytic bacteria of glumes was higher than that of the epiphytic and endophytic bacteria of seeds (Fig. 2G).
FIG 2.
Seed and glume microbial community alpha diversity index. Bacterial (A, C, E, and G) and fungal (B, D, F, and H) alpha diversities in and on seeds and in glumes with and without endophyte. E, Epichloë gansuensis endophyte infection status; P, different parts of EI and EF seeds; EI, endophyte infected; EF, endophyte free; Sep, seed epiphytic; SEn, seed endophytic; GEp, glume epiphytic (n = 8). Values are means ± standard errors of the mean (SEMs), with bars indicating SEs. An asterisk after a lowercase letter indicates significant difference at a P of <0.05 (independent t test) between EI and EF plants at seed and glume at 0.05 level. Different lowercase letters mean a significant difference at a P of <0.05 between seeds and glumes at 0.05 level.
The interaction between the Epichloë endophyte and the three different parts of EI and EF seeds had significant (P < 0.05) effects on the diversity of fungi of seeds and glumes, as summarized by the ACE index and Chao index (Fig. 2B and D). The treatments of different parts of EI and EF seeds had significant (P < 0.05) effects on seed fungal diversities, as summarized by the ACE index, Chao index, Shannon index, and Simpson index. The infection status of the E. gansuensis endophyte had a significant influence on the Simpson index.
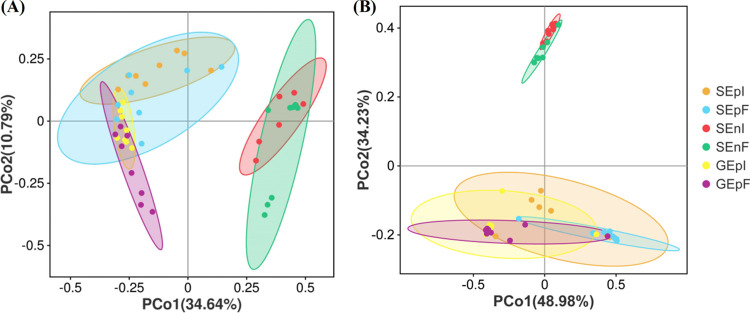
Principal-coordinate analysis (PCoA) showed that the bacterial community compositions of seeds and glumes between EI and EF DHG significantly (P < 0.05) differed among the different parts of EI and EF seed treatments (Fig. 3A; Table 1). In addition, E. gansuensis endophyte infection status, different parts of EI and EF seeds, and their interactions had significant (P < 0.05) effects on the diversity of the fungal communities of seeds and glumes of A. inebrians (Fig. 3B; Table 1).
FIG 3.
Bacteria (A) and fungi (B) of seeds and glumes. PCoA was based on Bray-Curtis dissimilarities at the OTU level (n = 8).
TABLE 1.
ANOSIM and PERMANOVA analysis of differences in bacterial and fungal community compositions in and on seeds and on glumes of EI and EF A. inebrians as calculated by amplicon sequencinga
| Community and treatment | dfb | PERMANOVA (Bray-Curtis) |
ANOSIM (Bray-Curtis) |
||
|---|---|---|---|---|---|
| F | P | R | P | ||
| Bacteria | |||||
| E | 1 | 3.2724 | 0.014 | 0.1835 | 0.001 |
| P | 2 | 14.687 | 0.000 | 0.6539 | 0.000 |
| E × P | 2 | 1.5393 | 0.1396 | ||
| Fungi | |||||
| E | 1 | 2.889 | 0.073 | 0.3160 | 0.000 |
| P | 2 | 35.103 | 0.000 | 0.6627 | 0.000 |
| E × P | 2 | 2.961 | 0.042 | ||
ANOSIM, statistical test of similarity; PERMANOVA, permutational mutlivariate two-way analysis of variance. E, Epichloë gansuensis endophyte infection status; P, different parts of EI and EF seeds. F value is used to measure the influence of a variable on the dependent variable in multivariate analysis of variance. R value represents the degree of linear correlation between the two variables. Boldface values indicate P < 0.05.
df, degrees of freedom.
Effect of Epichloë endophytes on metabolites of seeds and glumes.
In total, 517 and 517 metabolites were successfully detected from EI and EF seeds and glumes of A. inebrians, respectively (Tables S4 and S5). This study calculated the orthogonal projections to latent structures discriminant analysis (OPLS-DA) and principal-component analysis (PCA) on these metabolites of seeds (Fig. S4) and glumes (Fig. S5). The OPLS-DA models of seeds (R2X = 0.609, R2Y = 1, Q2Y = 0.976) and of glumes (R2X = 0.597, R2Y = 0.94, Q2Y = 0.804) are shown in Fig. S4A and S5A, respectively. The PCA results showed that these detected metabolites of seeds (PC1 = 40.67%, PC2 = 28.50%) (Fig. S4C) and glumes (PC1 = 88.52%, PC2 = 7.18%) (Fig. S5C) were significantly diverse between EI and EF seeds and glumes. All showed that there were differences in detected metabolites between EI and EF seeds and glumes (Fig. S4A and S5A).
Based on the fold change (FC) of >2, variable importance in the project (VIP) of >1, and P of 0.05, 108 and 31 different metabolites between EI and EF seeds and glumes, respectively, were separated (Tables S6 and S7). Among metabolites of EI seeds, 69 were upregulated and 39 were downregulated (Fig. S4B; Table S6); among metabolites of EI glumes, 25 were upregulated and 6 were downregulated (Fig. S5B; Table S7).
In seeds, the 108 differential metabolites could be divided into 10 classes; the 7 major classes included alkaloids, lipids, others, phenolic acids, organic acids, amino acids and derivatives, and nucleotides and derivatives (Table 2). In glumes, these 31 differential metabolites could be divided into 6 classes; the 3 major classes were alkaloids, others, and organic acids (Table 2).
TABLE 2.
Classification of 108 and 31 detected differential metabolites in A. inebrians seeds and glumes, respectively
| Class | No. of compounds in: |
|
|---|---|---|
| Seeds | Glumes | |
| Alkaloids | 21 | 9 |
| Lipids | 21 | 3 |
| Others | 16 | 7 |
| Phenolic acids | 15 | |
| Organic acids | 11 | 7 |
| Amino acids and derivatives | 10 | 3 |
| Nucleotides and derivatives | 10 | 2 |
| Terpenoids | 2 | |
| Flavonoids | 1 | |
| Lignans and coumarins | 1 | |
| Total | 108 | 31 |
Compared with EF seeds, a total of 69 upregulated and 39 downregulated differential metabolites were detected in EI seeds (Table S6); the top 10 up- and downregulated metabolites are shown in Table 3. Compared with EF glumes, a total of 25 upregulated and 6 downregulated differential metabolites were detected in EI glumes (Table S7); the top 5 differentially accumulated metabolites are shown in Table 4. In seeds, diethylcarbamazine, 3,4-dimethoxycinnamic acid, methylergonovine, 3-isobutyl-1-methylxanthine (IBMX), hexacosanoic acid, nortriptyline, α-asarone, 6-benzylaminopurine, (−)-tylocrebrine, and d-biotin were significantly (P < 0.05) increased compounds, while the compounds that decreased significantly (P < 0.05) were nicotinamide, linoleoyl ethanolamide, 3-hydroxy-4-methoxycinnamic acid, dl-benzylsuccinic acid, lithocholic acid, eicosapentaenoic acid ethyl ester, alpha-linolenoyl ethanolamide, (+)-alpha-pinene, stearidonic acid, and minoxidil (Table 3). In glumes, hexacosanoic acid, diethylcarbamazine, vitamin D2 (ergocalciferol), dihydrotachysterol, and perillyl alcohol were significantly (P < 0.05) increased compounds, while the compounds that decreased significantly (P < 0.05) were 9-decen-1-ol, 25-hydroxyvitamin D3, 4,6-dioxoheptanoic acid, 2-methylglutaric acid, and pyrocatechol (Table 4).
TABLE 3.
Top 10 differentially accumulated metabolites in EI seeds compared to EF seeds of A. inebriansa
| Compound | Class | No. of SEI | No. of SEF | FC | Log2 FC | Up- or downregulation |
|---|---|---|---|---|---|---|
| Diethylcarbamazine | Alkaloids | 9.59 × 10−4 | 1.76 × 10−2 | 18.32 | 4.49 | Up |
| 3,4-Dimethoxycinnamic acid | Lipids | 2.46 × 10−4 | 1.49 × 10−2 | 60.69 | 6.41 | Up |
| Methylergonovine | Alkaloids | 2.06 × 10−4 | 1.49 × 10−2 | 72.03 | 6.63 | Up |
| IBMX | Nucleotides and derivatives | 1.54 × 10−4 | 7.56 × 10−3 | 48.53 | 6.06 | Up |
| Hexacosanoic acid | Organic acids | 2.03 × 10−4 | 6.80 × 10−3 | 3.34 | 1.73 | Up |
| Nortryptyline | Lipids | 9.16 × 10−5 | 4.80 × 10−3 | 52.31 | 5.86 | Up |
| α-Asarone | Phenolic acids | 8.28 × 10−4 | 4.20 × 10−3 | 5.07 | 2.54 | Up |
| 6-Benzylaminopurine | Nucleotides and derivatives | 1.88 × 10−4 | 3.50 × 10−3 | 18.61 | 4.62 | Up |
| (−)-Tylocrebrine | Alkaloids | 1.75 × 10−4 | 3.06 × 10−3 | 17.27 | 4.09 | Up |
| d-Biotin | Others | 1.15 × 10−4 | 2.9 × 10−3 | 24.82 | 5.73 | Up |
| Nicotinamide | Others | 1.04 × 10−2 | 1.72 × 10−3 | 0.17 | −2.62 | Down |
| Linoleoyl ethanolamide | Lipids | 2.98 × 10−3 | 1.45 × 10−3 | 0.49 | −1.04 | Down |
| 3-Hydroxy-4-methoxycinnamic acid | Phenolic acids | 2.51 × 10−3 | 1.24 × 10−3 | 0.49 | −0.99 | Down |
| dl-Benzylsuccinic acid | Organic acids | 3.393 × 10−3 | 1.18 × 10−3 | 0.35 | −1.52 | Down |
| Lithocholic acid | Organic acids | 2.26 × 10−3 | 9.45 × 10−4 | 0.42 | −1.25 | Down |
| Eicosapentaenoic acid ethyl ester | Lipids | 8.92 × 10−4 | 4.13 × 10−4 | 0.46 | −1.10 | Down |
| Alpha-linolenoyl ethanolamide | Alkaloids | 8.83 × 10−4 | 3.90 × 10−4 | 0.44 | −1.18 | Down |
| (+)-Alpha-pinene | Terpenoids | 8.26 × 10−4 | 2.51 × 10−4 | 0.30 | −1.75 | Down |
| Stearidonic acid | Lipids | 6.15 × 10−4 | 2.49 × 10−4 | 0.40 | −1.31 | Down |
| Minoxidil | Nucleotides and derivatives | 1.57 × 10−3 | 2.47 × 10−4 | 0.16 | −2.23 | Down |
SEI, seed metabolites of endophyte-infected plants; SE, seed metabolites of endophyte-free plants (n = 6).
TABLE 4.
Top five differentially accumulated metabolites in EI glumes compared to EF glumes of A. inebriansa
| Compound | Class | GEI | GEF | FC | Log2 FC | Up- or downregulation |
|---|---|---|---|---|---|---|
| Hexacosanoic acid | Organic acids | 3.72 × 10−3 | 1.59 × 10−2 | 4.27 | 2.05 | Up |
| Diethylcarbamazine | Alkaloids | 7.45 × 10−4 | 7.55 × 10−3 | 10.13 | 3.73 | Up |
| Vitamin D2 (ergocalciferol) | Others | 1.89 × 10−3 | 6.73 × 10−3 | 3.56 | 1.80 | Up |
| Dihydrotachysterol | Others | 6.41 × 10−4 | 5.40 × 10−3 | 8.43 | 3.01 | Up |
| Perillyl alcohol | Others | 1.24 × 10−3 | 4.55 × 10−3 | 3.66 | 1.83 | Up |
| 9-Decen-1-ol | Others | 9.86 × 10−3 | 3.99 × 10−3 | 0.41 | −1.23 | Down |
| 25-Hydroxyvitamin D3 | Others | 2.17 × 10−3 | 1.00 × 10−4 | 0.46 | −1.16 | Down |
| 4,6-Dioxoheptanoic acid | Organic acids | 7.01 × 10−4 | 3.32 × 10−4 | 0.47 | −1.05 | Down |
| 2-Methylglutaric acid | Organic acids | 8.38 × 10−4 | 3.28 × 10−4 | 0.39 | −1.37 | Down |
| Pyrocatechol | Others | 5.29 × 10−4 | 2.43 × 10−4 | 0.46 | −1.14 | Down |
GEI, glume metabolites of EI plants; GEF, glume metabolites of EF plants (n = 6).
The study analyzed different metabolites according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database; the results showed that a total of 34 and 9 metabolites were annotated by KEGG in seeds and glumes, respectively (Tables S6 and S7). There were 20 metabolite pathways significantly enriched (P < 0.05) in EI seeds (Fig. S6A), and 11 metabolites pathways were significantly enriched (P < 0.05) in EI glumes (Fig. S6B). In seeds, KEGG analysis showed that fatty acid biosynthesis and sesquiterpenoid and triterpenoid biosynthesis were major enriched biological pathways (Fig. S6A). In glumes, KEGG analysis showed that the most enriched biological pathways were monoterpenoid biosynthesis, limonene and pinene degradation, d-arginine and d-ornithine metabolism, steroid biosynthesis, phenylalanine tyrosine and tryptophan biosynthesis, nicotinate and nicotinamide metabolism, pyrimidine metabolism, and tryptophan metabolism (Fig. S6B).
Correlation network between metabolites and microbes in seeds and glumes.
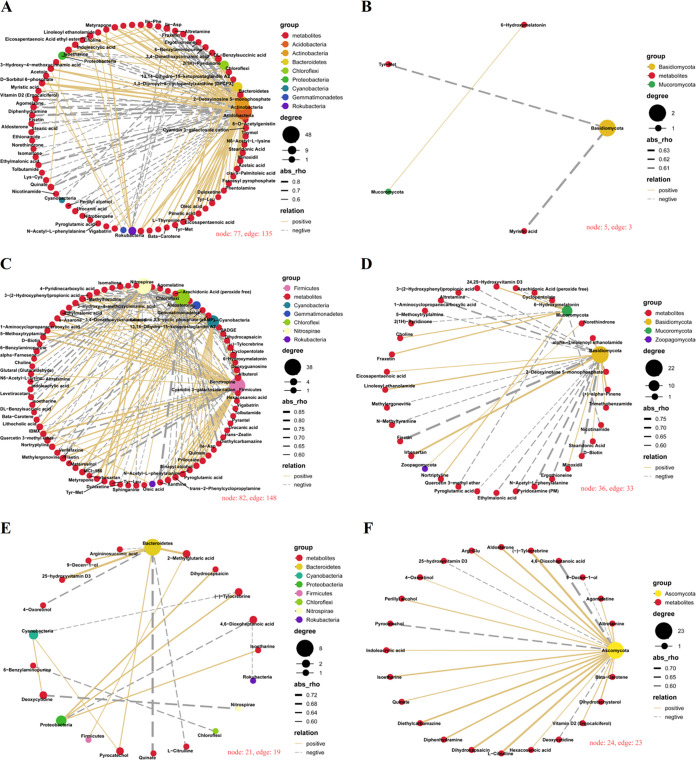
To explore correlations between metabolites and microorganisms, we analyzed the Spearman’s correlation networks between metabolites that were significantly up- or downregulated in seeds and seed-borne microbial phyla (Fig. 4; Tables S8 to S13).
FIG 4.
Spearman's correlation networks between seed differential metabolites and seed-borne microbial phyla. (A) Seed differential metabolites and seed epiphytic bacterial phyla; (B) seed differential metabolites and seed epiphytic fungal phyla; (C) seed differential metabolites and seed endophytic bacterial phyla; (D) seed differential metabolites and seed endophytic fungal phyla; (E) glume differential metabolites and glume epiphytic bacterial phyla; (F) glume differential metabolites and glume epiphytic fungal phyla. node, metabolites or phyla; edge, correlations between metabolites and phyla; group, classification of metabolites and phyla; degree, edge number of one node; abs, extent of correlation; relation, yellow line indicates positive correlation between metabolites and phyla and gray dotted line indicates negative correlation.
There were 135 and 3 significant (P < 0.05) correlations between seed differential metabolites and seed epiphytic bacterial (Fig. 4A; Table S8) and fungal (Fig. 4B; Table S9) phyla, respectively. The main microbial phyla involved with these correlations included Acidobacteria, Actinobacteria, Bacteroidetes, Chloroflexi, Proteobacteria, Basidiomycota, and Mucoromycota. In comparison with SEpF, the relative abundances of Bacteroidetes, Proteobacteria, and Basidiomycota in SEpI were increased but those of other phyla were decreased (Table S1). In the overall network diagram, the bacteria in the Acidobacteria and Actinobacteria were most closely related to the metabolites, with the degrees (degree represents the number of phyla or metabolites connected to other metabolites or phyla) of Acidobacteria and Actinobacteria being 48 and 35, respectively. The phyla next most closely related to metabolites were Bacteroidetes, Chloroflexi, and Proteobacteria, with 16, 14, and 10 correlations with metabolites, respectively, and these correlations between the above phyla and metabolites were all positive (Fig. 4A; Table S8). In addition, the fungi in the Basidiomycota were negatively correlated with Tyr-Met and myristic acid (Fig. 4B; Table S9). In whole bacterial and fungal networks, there were two key metabolites; Tyr-Met was positively correlated with Actinobacteria and Acidobacteria but negatively correlated with Basidiomycota, and myristic acid was positively correlated with Rokubacteria but negatively correlated with Basdiomycota (Fig. 4; Tables S8 and S9).
There were 148 and 33 significant (P < 0.05) correlations between differentially accumulated metabolites of seeds and seed endophytic bacterial (Fig. 4C; Table S10) and fungal (Fig. 4D; Table S11) phyla, respectively. The main microbial phyla involved with these correlations included Chloroflexi, Firmicutes, Nitrospirae, and Basidiomycota. In comparison with SEnF, the relative abundances of Chloroflexi and Firmicutes in SEnI were increased but other phyla were decreased (Table S1). In the network diagram, the bacteria in the Chloroflexi, Firmicutes, and Nitrospirae were mostly correlated with metabolites, with the highest degree being 38; the phylum next most closely related to metabolites was Cyanobacteria, which had 24 correlations with metabolites (Fig. 4C; Table S10). Furthermore, fungi in the Basidiomycota were most closely related to 22 metabolites (Fig. 4D; Table S11). In whole bacterial and fungal networks, 6-hydroxymelatonin was a key metabolite, positively correlated with Firmicutes and negatively correlated with Gemmatimonadetes, Chloroflexi, and Rokubacteria (Fig. 4; Tables S10 and S11).
There were 19 and 23 significant (P < 0.05) correlations between glume differential metabolites and glume bacterial (Fig. 4E; Table S12) and fungal (Fig. 4F; Table S13) phyla, respectively. The main microbial phyla involved with these correlations included Bacteroidetes and Ascomycota. In comparison with GEpF, the relative abundance of Ascomycota was increased and Bacteroidetes was decreased in GEpI (Table S1). In the network diagram, bacteria in the Bacteroidetes were mostly correlated with 8 metabolites (Fig. 4E; Table S12). Moreover, the fungi in the Ascomycota were closely related with 23 glume metabolites, and 18 out of the 23 correlations were positive (Fig. 4F; Table S13).
DISCUSSION
The present study investigated the influence of the E. gansuensis endophyte on seed-borne microbial communities and seed metabolites of DHG. In particular, this study performed correlation analysis between seed-borne microbes and seed metabolites and found that there were many close and complex correlations between them.
Effect of E. gansuensis on seed-borne microbial communities.
Results revealed that seed-borne bacterial communities were mostly dominated by phyla such as Proteobacteria, Actinobacteria, Firmicutes, and Bacteroidetes, and the core genera were Pseudomonas, Bacillus, Sphingomonas, and Allorhizobium. These results were similar to those of the study by Bastías et al., which looked at Lolium multiflorum seeds infected by Epichloë occultans (41). Previous studies reported that seed-borne fungal communities were dominated by Ascomycota and Basidiomycota (42–44), and those results were in line with the present study. Furthermore, Penicillium, Mortierella, and Fusarium were prominent genera of seed-borne fungal communities of DHG.
Bastías et al. not only demonstrated that Epichloë endophytes affected the composition and diversity of the seed microbiota, but they also considered that the bacterial microbes associated with leaf tissues of mother plants are the main source of bacteria for seeds (41). There were many studies that proved that Epichloë endophytes changed the foliar microbiome community of host plants; for example, the presence of Epichloë coenophiala in tall fescue (Festuca arundinacea) modified the leaf-associated fungal communities and the presence of Epichloë festucae var. lolii modified the bacterial communities associated with seedlings of perennial ryegrass (Lolium perenne) plants (45, 46). A recent study found that the presence of Epichloë changed the phyllosphere microbial communities on A. inebrians (47). In accordance with previous studies, the current study’s findings provide strong support for the presence of an Epichloë endophyte modifying the bacterial microbes associated with the foliar tissues and seeds of mother plants.
The presence of E. gansuensis changed the composition and diversity of seed-borne microbes of DHG, which proved the first hypothesis, i.e., that E. gansuensis would modulate the microbial diversity of DHG seeds and glumes. Previous studies also showed that Epichloë endophytes had impacts on the microbial diversity of host plants; for example, the presence of E. occultans in Lolium multiflorum seeds resulted in a higher diversity in the bacterial community than did the presence of endophyte-free seeds (41). Similarly, E. gansusensis increased root-associated AMF diversity under drought conditions of DHG (36). E. gansuensis decreased the Shannon diversity of the root-associated bacterial community but increased the Shannon diversity of the rhizosphere soil bacterial community of A. inebrians (39). Recently, a study found that the Epichloë endophyte increased the diversity of phyllosphere bacterial and fungal communities on A. inebrians leaves (47). Results from the present study showed that E. gansuensis decreased bacterial and fungal diversity and richness except for the GEpI richness. Epichloë endophytes may modulate the fungal and bacterial diversity of seed-borne microbial communities by modifying certain plant physiological responses.
Seed-borne epiphytic, endophytic, and glume microbial community diversity.
This study showed that the decreasing order of the alpha diversity index in DHG seed-borne bacteria was from epiphytic, to endophytic, to glume epiphytic and that the decreasing order of the alpha diversity index in DHG seed-borne fungi was from endophytic, to epiphytic, to glume epiphytic. A previous study showed that microorganisms were more abundant in the seed coat than in the endosperm and the embryo (48). Nelson summarized that it is essential to distinguish between epiphytic and endophytic microbes when work is carried out on the microbiomes of seeds (21). The endophytic microbes of seeds have typically been considered to be composed of commensals or mutualists (16, 43, 49, 50), and the epiphytic microbe communities were dominated by species of well-known plant pathogens of the genera Fusarium, Phoma, Pyrenophora, Alternaria, and Leptosphaeria (21, 51). In studies of the cultivable endophytic bacteria, abundance estimates may range from 101 to 102 CFU/g seed (52, 53) to as high as 106 to 108 CFU/g seed (51, 54). Wang found that the diversity of epiphytic bacteria was higher than that of endophytic bacteria in tomato seeds (55). Most seed-borne fungi were located in the endosperm, with decreasing numbers in the pericarp and embryo in L. perenne seeds, as determined by using a component plating method (56). For the sites of the fungi isolated from Sorghum sudanense seeds, the order of decreasing frequency of fungal species was seed coat, pericarp, endosperm, and embryo (57).
Recently, Bastías et al. reported that the presence of Epichloë endophytes increased the population of Pseudomonas, and these results were in line with the present study (41). Bacteria of the genus Pseudomonas have bene reported to enhance plant growth via production of auxins, phosphate-solubilizing compounds, and other growth-promoting compounds (58). Pseudomonas stutzeri increased Oryza sativa seedling resistance under salt stress via production of exopolysaccharide (59). A Pseudomonas strain protected O. sativa and Poa annua seedlings from fungal pathogens via the production of antimicrobials (60). The present study also showed that E. gansuensis increased the relative abundance of Sphingomonas. A strain of Sphingomonas melonis accumulated and was transmitted across generations in disease-resistant rice seeds, conferring resistance to disease-susceptible phenotypes by producing anthranilic acid (16). The present study also found that the presence of E. gansuensis decreased the abundance of fungi, including Mortierella and Fusarium species. Of importance is that many Fusarium species are pathogens of a wide range of plants (21, 61). Based on the findings of this and other studies, the presence of E. gansuensis seems likely to enhance the resistance of DHG plants to some fungal diseases via increases in the amount of beneficial bacteria and decreases in the presence of pathogenic fungi associated with seeds.
Effect of the E. gansuensis endophyte on seed metabolites.
The present study showed that infection of DHG by E. gansuensis changed the class and content of metabolites of seeds and glumes, which completely supported our second hypothesis. Importantly, metabolomics plays a vital role in tracking temporal changes in metabolites through the entire growth cycle of plants (62–64) and through exposure to stress (27, 28). In many studies, researchers conducted experiments to address the effects of Epichloë endophytes on metabolites of host plants, and these mainly focused on alkaloids (65), acids (organic acids, amino acids, and fatty acids) (13), and amino acids (14).
The absence of alkaloids in EF plants is the biggest difference between EI and EF DHG. Alkaloids, including indol-diterpene, pyrrolapyrazine, pyrrolizidine, and ergot alkaloids, can be detected in Epichloë endophyte-infected plants (66–68). In 1984, the first finding was made of the presence of ergonovine and ergonovinine in DHG plants (69). In 1996, it was reported that the presence of these two alkaloids in DHG plants was associated with the presence of a seed-borne Epichloë endophyte (70). The alkaloid content, and in particular that of ergine and ergonovine, was linked with the growth of DHG plants, and frequent cutting (fortnightly) at a mowing height of 7.5 cm resulted in a significant boost to the content of the bioactive, endophyte-produced alkaloids (71). Further, studies found that the content of alkaloids increased over the DHG growing period under the abiotic stresses of salt and drought, and the content of cytotoxic ergonovine in particular increased under these stress conditions (72, 73). A recent study found that metabolites other than ergonovine and ergine, including epichlicin and cyclosporine T, isolated from host plants with an Epichloë endophyte, had significant antifungal and anti-insect activity (74).
Metabolomics results found that the presence of E. gansuensis influenced a wide range of metabolites produced in seeds or glumes, including purine derivatives, tryptamine derivatives, lignins, and aldehyde in DHG seeds and sterols and phenols in DHG glumes. Other studies on host grasses with an Epichloë endophyte also focused on metabolites in general, rather than just alkaloids. A study found that Neotyphodium uncinatum (=Epichloë uncinata) endophytes in host plants may affect soil insect distribution by altering the presence of root volatiles that affect insect behavior (75). Epichloë coenophiala in tall fescue can affect root exudate composition, including lipids, carbohydrates. and carboxylic acids, affecting plant growth (76). Furthermore, Hou et al. reported that E. gansuensis increased the tolerance of DHG to low N stress by increasing the content of glucose-6-phosphate and organic acids in leaves and increasing the content of fatty acids and amino acids in roots (13). Meanwhile, the presence of Epichloë endophytes improves DHG growth by regulating the metabolic pathway of amino acids, amino acid content, and organic acid content at low P stress (14). Metabolites from alkaloids to nonalkaloids produced by DHG seeds and glumes of E. gansuensis-hosting DHG plants are equally important because the defensive mutualism between grasses and the Epichloë endophytes involves a wider range of metabolites than just the recognized endophyte-produced alkaloids.
Impact factors on the microbes of seeds and glumes.
Soil possesses high microbial diversity, and this directly and indirectly influences seed microbes (75, 76). A recent study showed that many soil bacteria could reach the leaves and flowers of Arabidopsis thaliana (77). Flowers are the origin of seed development and dispersal and also contain a large microbiome (78), and Nelson showed that microbes associated with flowers are an important source of seed-associated microbes (21). Furthermore, the floral nectar, which comprises secondary metabolites, particularly sugars and amino acids, can provide a source of energy and available carbon for seed-borne microbial growth (79).
The present study found that there were complex and close correlations between seed-borne microbes and seed metabolites or correlations between glume microbes and metabolites, which partially supported that there are complex and close correlations between seed-borne microbes and metabolites between EI and EF seeds and also between glume microbes and metabolites between EI and EF glumes. The presence of E. gansuensis changes the seed-borne microbiota and likely increases and decreases the number of beneficial bacteria and pathogenic fungi, respectively. These changes resulting from the presence of E. gansuensis result in the enhanced presence of beneficial antibiotic products, auxin, and alkaloids that promote the growth and persistence of host plants. This study has confirmed that there are close and complex relationships between seed-borne microbes and seed metabolites and so enhances the understanding of how seed-associated microbes interact with each other and play an important role in the growth and development of the following generations.
MATERIALS AND METHODS
Site description and experimental design.
The EI and EF seeds of A. inebrians were collected from an experimental field in Yuzhong campus (104°39′E, 35°89′N; altitude, 1,653 m) of the College of Pastoral Agriculture Science and Technology of Lanzhou University. These EI and EF plots, also used in other studies, were established in 2017 and managed as described by Liu et al. (47). From 2018 to 2019, the leaf sheaths and seeds of all plants were stained with aniline blue (2) to check the endophyte infection status and to ensure that the selected seeds for this study were 100% and 0% infected, respectively.
Sampling.
Eight experimental plots were randomly selected in September 2019 for the collection of seeds for the present study. For each subplot, three mature plants were selected for the collection of seeds, and then these seed samples were mixed into a composite sample. The collected seeds were stored at 4°C in a refrigerator, from which we later randomly selected 50 EI and EF seeds. We wore sterile gloves and separated glumes from seeds on an ultraclean worktable sterilized with UV light for 1 h to ensure the presence of sterility throughout the separation process. The glumes and seeds removed from the seeds were immediately stored in sterile tubes, frozen in liquid nitrogen for 3 h, and then stored in a −80°C freezer for subsequent sequencing.
A total of 16 seed samples (eight EI samples, eight EF samples) were used for the detection of seed epiphytic and endophytic microbes. There were 16 glume samples (eight EI samples, eight EF samples) used for the detection of glume epiphytic microbes.
Seed epiphytic microbes were washed from seed surfaces. Fifty seeds were transferred into 50-mL plastic tubes filled with 30 to 40 mL phosphate-buffered saline (PBS) buffer, along with two blank controls without added seeds, followed by oscillation for 30 to 60 min at 150 to 200 rpm, sonication for 5 min, and further oscillation for 30 to 60 min at 150 to 200 rpm. The seeds were removed, and the suspension was centrifuged at 10,000 × g for 10 min to obtain precipitates containing bacteria, fungal spores, and hyphae dislodged from the surface of the seeds.
For seed endophytic microbes, the above-described harvested seeds were surface sterilized in 1% NaClO (sodium hypochlorite) for 3 min, followed by 70% ethanol for 10 min, and rinsed with sterilized water five times.
Glume epiphytic microbes were washed from the glume surfaces. Fifty glumes were transferred into 50-mL plastic tubes filled with 30 to 40 mL PBS buffer, along with two blank controls without added glumes, followed by oscillation for 30 to 60 min at 150 to 200 rpm, sonication for 5 min, and further oscillation for 30 to 60 min at 150 to 200 rpm. The glumes were removed, and the suspension was centrifuged at 10,000 × g for 10 min to obtain precipitates containing bacteria, fungal spores, and hyphae dislodged from the surface of the glumes. Subsequently, these separated seed and glume samples were stored at −80°C in a freezer before DNA extraction and metabolite detection.
DNA extraction, PCR amplification, and sequencing.
DNA of these seed and glume samples was extracted using an MN NucleoSpin 96 soil DNA kit according to the manufacturer’s instructions. The 16S rRNA genes were amplified using the universal primers 335F (5′-CADACTCCTACGGGAGGC-3′) and 769R (5′-ATCCTGTTTGMTMCCCVCRC-3′). The internal transcribed spacer (ITS1) genes were amplified using the universal primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-GCTGCGTTCTTCATCGATGC-3′). PCR amplification was performed in a total volume of 50 μL, which contained 10 μL buffer, 0.2 μL Q5 high-fidelity DNA polymerase, 10 μL high GC enhancer, 1 μL deoxynucleoside triphosphate, 10 μM each primer, and 60 ng genomic DNA. The PCRs were performed using the following cycling conditions: 98°C for 30 s, followed by 10 cycles of 98°C for 10 s, 65°C for 30 s, and 72°C for 30 s, and a final step of 72°C for 5 min. The reactions were run on 1.8% agarose gels. All PCR products were quantified by Quant-iT double-stranded DNA HS reagent and pooled. High-throughput sequencing analysis of bacterial and fungal rRNA genes was performed on the purified, pooled sample by use of the Illumina NovaSeq 6000 system at Biomarker Technologies Corporation (BMK), Beijing, China.
Bioinformatics analysis.
The raw reads obtained were quality checked and filtered, and reads with a final length of <20 bases were discarded. The remaining sequences without ambiguous bases were assigned to different OTUs at 97% sequence identity by VSEARCH (v10.0). These sequences were determined using Silva (release 128; http://www.arb-silva.de) for bacteria and Unite (release 7.2; http://unite.ut.ee/index.php) for fungi to identity these OTUs. The obtained OTU table was used to determine taxonomic relative abundances and subsequent diversity analysis.
Alpha and beta diversity analysis.
The alpha diversity indices that were calculated using Mothur software (v.1.30) included the Chao index (https://mothur.org/wiki/chao/), the Shannon index (https://mothur.org/wiki/shannon/), the ACE index (https://mothur.org/wiki/ace/), and the Simpson index (https://mothur.org/wiki/simpson/).
Principal-coordinate analysis (PCoA) and the statistically significant differences of bacterial or fungal communities among the three different parts of EI and EF seeds were tested through permutational multivariate analysis of variance (PERMANOVA) and analysis of similarity (ANOSIM) based on the Bray-Curtis dissimilarities using the vegan package in R (v.4.0.3).
Metabolite extraction.
Twelve seed samples (six EI samples, six EF samples) were randomly selected from the 16 microbial sequencing samples for the detection of seed metabolites. Twelve glume samples (six EI samples, six EF samples) were randomly selected from the 16 microbial sequencing samples for the detection of glume metabolites. These EI/EF seed and glume samples were weighed into eppendorf (EP) tubes, respectively, and 500 μL extraction solution (acetonitrile-methanol-water, 2: 2: 1) containing an isotopically labeled internal standard mixture was added. After 30 s of vortexing, these samples were homogenized at 35 Hz for 4 min and sonicated for 5 min in an ice-water bath. The homogenization and sonication cycle was repeated 3 times. These samples were then stored at −40°C for 1 h and then centrifuged at 12,000 rpm for 15 min at 4°C. Two hundred fifty microliters of supernatant was transferred to a fresh tube and dried in a vacuum concentrator at 37°C. The dried samples were reconstituted in 400 μL of 50% acetonitrile by sonication on ice for 10 min and then centrifuged at 13,000 rpm for 15 min at 4°C, and 75 μL of supernatant was transferred to a fresh glass vial for LC-MS analysis.
The ultra-high performance liquid chromatography (UHPLC) separation was carried out using an ExionLC Infinity series UHPLC system (AB Sciex), equipped with a UPLC BEH amide column (2.1 by 100 mm, 1.7 μm; Waters). The mobile phase consisted of 25 mmol/L ammonium acetate and 25 mmol/L ammonia hydroxide in water (pH 9.75) (A) and acetonitrile (B). The analysis was carried with an elution gradient as follows: ~0 to 0.5 min, 95% B; ~0.5 to 7.0 min, ~95% to 65% B; ~7.0 to 8.0 min, ~65% to 40% B; ~8.0 to 9.0 min, 40% B; ~9.0 to 9.1 min, ~40% to 95% B; ~9.1 to 12.0 min, 95% B. The column temperature was 25°C. The autosampler temperature was 4°C, and the injection volume was 2 μL (positive) or 2 μL (negative), respectively.
The TripleTOF 5600 mass spectrometer (AB Sciex) was used for its ability to acquire tandem MS (MS/MS) spectra on an information-dependent basis during an LC-MS experiment. In this mode, the acquisition software (Analyst TF 1.7; AB Sciex) continuously evaluates the full-scan survey MS data as it collects and triggers the acquisition of MS/MS spectra depending on preselected criteria. In each cycle, the most intensive 12 precursor ions with an intensity above 100 were chosen for MS/MS at a collision energy of 30 eV. The cycle time was 0.56 s. Electrospray ionization source conditions were set as follows: gas 1 as 60 lb/in2, gas 2 as 60 lb/in2, curtain gas as 35 lb/in2, source temperature as 600°C, declustering potential as 60 V, and ion spray voltage floating as 5,000 V or −4,000 V in positive or negative mode, respectively. The identification of metabolites of seeds and glumes was done at Biomarker Technologies Corporation (BMK), Beijing, China.
Metabolomics analysis.
The different variables were screened through principal-component analysis (PCA) and orthogonal partial least-squares discriminant analysis (OPLS-DA). Different metabolites were filtered according to variable importance in the project (VIP) of >1, P value of <0.05, and fold change (FC) of >2. The KEGG database was used to analyze the pathway and enzyme for annotation, enrichment, and classification.
Treatments in the study.
In this study, according to the infection status of the E. gansuensis endophyte and different parts of EI and EF seeds, the seed-borne microbial communities were defined as six groups. These were SEpI (seed epiphytic microbes of endophyte-infected plants), SEpF (seed epiphytic microbes of endophyte-free plants), SEnI (seed endophytic microbes of endophyte-infected plants), SEnF (seed endophytic microbes of endophyte-free plants), GEpI (glume microbial community of endophyte-infected plants), and GEpF (glume microbial community of endophyte-free plants).
According to the E. gansuensis infection status and the different parts of EI and EF seeds, the seed metabolites were defined as four groups. These were SEI (seed metabolites of endophyte-infected plants), SEF (seed metabolites of endophyte-free plants), GEI (glume metabolites of endophyte-infected plants), and GEF (glume metabolites of endophyte-free plants).
Statistical analyses.
Visualizations were performed using SigmaPlot (v.12.5) and R software (v.4.0.3) packages (ggplot2 v.3.3.5; igraph v.1.2.6). Differences in seed-borne microbial community diversity under the infection status of the E. gansuensis endophyte and different parts of EI and EF seeds were tested using a two-way analysis of variance (two-way ANOVA) in SPSS 22.0. Significant differences in seed-borne microbial community diversity between EI and EF A. inebrians seeds with treatment of different parts of EI and EF seeds were examined by an independent-sample t test, and significant differences in seed-borne microbial community diversity among the treatment of different parts of EI and EF seeds were examined by one-way analysis of variance (one-way ANOVA). In all tests, a P value of <0.05 was considered a significant difference. For correlation network analysis, if the sample had both metabolite and microbial data, Spearman’s correlation via R software (v.4.0.3) packages (stats v.3.3.5) was used for the sample (80, 81). Metabolite data and microbial data were paired based on individual seed samples to avoid the influence of sample diversity on correlation results.
Compliance with ethics requirements.
This study did not involve any studies with human or animal subjects.
Data availability.
The original sequence data of bacterial and fungal sequences were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/). The BioProject accession numbers of bacterial and fungal raw data are PRJNA865502 and PRJNA865530, respectively.
Supplementary Material
ACKNOWLEDGMENTS
We thank the editor and anonymous reviewers for their valuable comments.
This work was financially supported by the National Nature Science Foundation of China (grant no. 32061123004 and 31772665), the National Basic Research Program of China (grant no. 2014CB138702), and the Fundamental Research Funds for the Central Universities (grant no. jbky-2022-ey21) at Lanzhou University and the Key Scientific Research Platform and Project of Guangdong Education Department (grant no. 2021KCXTD054).
We declare no conflicts of interest.
Footnotes
Supplemental material is available online only.
Contributor Information
Wu Zhang, Email: ldzw1987@163.com.
Xingxu Zhang, Email: xxzhang@lzu.edu.cn.
Patricia Albuquerque, Universidade de Brasilia.
REFERENCES
- 1.Nan ZB, Li CJ. 2000. Neotyphodium in native grasses in China and observations on endophyte/host interactions. In Proceedings of the 4th International Neotyphodium/Grass Interactions Symposium Soest. [Google Scholar]
- 2.Li CJ, Nan ZB, Volker HP, Dapprich PD, Liu Y. 2004. A new Neotyphodium species symbiotic with drunken horse grass (Achnatherum inebrians) in China. Mycotaxon 90:141–147. [Google Scholar]
- 3.Yao X, Fan YB, Chai Q, Johnson RD, Nan ZB, Li CJ. 2016. Modification of susceptible and toxic herbs on grassland disease. Sci Rep 6:30635. doi: 10.1038/srep30635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leuchtmann A, Bacon CW, Schardl CL, White JFJ, Tadych M. 2014. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 106:202–215. doi: 10.3852/13-251. [DOI] [PubMed] [Google Scholar]
- 5.Chen L, Li XZ, Li CJ, Swoboda GA, Young CA, Sugawara K, Leuchtmann A, Schardl CL. 2015. Two distinct Epichloë species symbiotic with Achnatherum inebrians, drunken horse grass. Mycologia 107:863–873. doi: 10.3852/15-019. [DOI] [PubMed] [Google Scholar]
- 6.Zhang XX, Li CJ, Nan ZB, Matthew C. 2012. Neotyphodium endophyte increases Achnatherum inebrians (drunken horse grass) resistance to herbivores and seed predators. Weed Res 52:70–78. doi: 10.1111/j.1365-3180.2011.00887.x. [DOI] [Google Scholar]
- 7.Xia C, Zhang XX, Christensen MJ, Nan ZB, Li CJ. 2015. Epichloë endophyte affects the ability of powdery mildew (Blumeria graminis) to colonise drunken horse grass (Achnatherum inebrians). Fungal Ecol 16:26–33. doi: 10.1016/j.funeco.2015.02.003. [DOI] [Google Scholar]
- 8.Xia C, Li NN, Zhang XX, Feng Y, Christensen MJ, Nan ZB. 2016. An Epichloë endophyte improves photosynthetic ability and dry matter production of its host Achnatherum inebrians infected by Blumeria graminis under various soil water conditions. Fungal Ecol 22:26–34. doi: 10.1016/j.funeco.2016.04.002. [DOI] [Google Scholar]
- 9.Zhang XX, Li CJ, Nan ZB. 2010. Effects of cadmium stress on growth and anti-oxidative systems in Achnatherum inebrians symbiotic with Neotyphodium gansuense. J Hazard Mater 175:703–709. doi: 10.1016/j.jhazmat.2009.10.066. [DOI] [PubMed] [Google Scholar]
- 10.Chen N, He RL, Chai Q, Li CJ, Nan ZB. 2016. Transcriptomic analyses giving insights into molecular regulation mechanisms involved in cold tolerance by Epichloë endophyte in seed germination of Achnatherum inebrians. Plant Growth Regul 80:367–375. doi: 10.1007/s10725-016-0177-8. [DOI] [Google Scholar]
- 11.Wang JF, Tian P, Christensen MJ, Zhang XX, Li CJ, Nan ZB. 2018. Effect of Epichloë gansuensis endophyte on the activity of enzymes of nitrogen metabolism, nitrogen use efficiency and photosynthetic ability of Achnatherum inebrians under various NaCl concentrations. Plant Soil 66:4022–4031. [DOI] [PubMed] [Google Scholar]
- 12.Xia C, Christensen MJ, Zhang XX, Nan ZB. 2018. Effect of Epichloë gansuensis endophyte and transgenerational effects on the water use efficiency, nutrient and biomass accumulation of Achnatherum inebrians under soil water deficit. Plant Soil 424:1–17. [Google Scholar]
- 13.Hou WP, Wang JF, Christensen MJ, Liu J, Zhang YQ, Liu YL, Cheng C. 2021. Metabolomics insights into the mechanism by which Epichloë gansuensis endophyte increased Achnatherum inebrians tolerance to low nitrogen stress. Plant Soil 463:487–508. doi: 10.1007/s11104-021-04930-z. [DOI] [Google Scholar]
- 14.Liu YL, Hou WP, Jin J, Christensen MJ, Gu LJ, Cheng C, Wang JF. 2021. Epichloë gansuensis increases the tolerance of Achnatherum inebrians to low-P stress by modulating amino acids metabolism and phosphorus utilization efficiency. J Fungi (Basel) 7:390. doi: 10.3390/jof7050390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HM, Wu HX, Yan B, Zhao HG, Liu FH, Zhang HH, Sheng Q, Miao F, Liang ZS. 2018. Core microbiome of medicinal plant Salvia miltiorrhiza seed: a rich reservoir of beneficial microbes for secondary metabolism? Int J Mol Sci 19:672. doi: 10.3390/ijms19030672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumoto H, Fan XY, Wang Y, Kusstatscher P, Duan J, Wu SL, Chen SL, Qiao K, Wang YL, Ma B, Zhu GN, Hashidoko Y, Berg G, Cernava T, Wang MC. 2021. Bacterial seed endophyte shapes disease resistance in rice. Nat Plants 7:60–72. doi: 10.1038/s41477-020-00826-5. [DOI] [PubMed] [Google Scholar]
- 17.Bressan W. 2003. Biological control of maize seed pathogenic fungi by use of actinomycetes. Biol Control 48:233–240. [Google Scholar]
- 18.Gitaitis R, Walcott R. 2007. The epidemiology and management of seedborne bacterial diseases. Annu Rev Phytopathol 45:371–397. doi: 10.1146/annurev.phyto.45.062806.094321. [DOI] [PubMed] [Google Scholar]
- 19.Santamaria J, Bayman P. 2005. Fungal epiphytes and endophytes of coffee leaves (Coffea arabica). Microb Ecol 50:1–8. doi: 10.1007/s00248-004-0002-1. [DOI] [PubMed] [Google Scholar]
- 20.Vorholt J. 2012. Microbial life in the phyllosphere. Nat Rev Microbiol 10:828–840. doi: 10.1038/nrmicro2910. [DOI] [PubMed] [Google Scholar]
- 21.Nelson EB. 2018. The seed microbiome: origins, interactions, and impacts. Plant Soil 422:7–34. doi: 10.1007/s11104-017-3289-7. [DOI] [Google Scholar]
- 22.Harrigan GG, Martino-Catt S, Glenn KC. 2007. Metabolomics, metabolic diversity and genetic variation in crops. Metabolomics 3:259–272. doi: 10.1007/s11306-007-0076-0. [DOI] [Google Scholar]
- 23.Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, Willmitzer L, Zamir D, Fernie AR. 2006. Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24:447–454. doi: 10.1038/nbt1192. [DOI] [PubMed] [Google Scholar]
- 24.Tarpley L, Duran AL, Kebrom TH, Sumner LW. 2005. Biomarker metabolites capturing the metabolite variance present in a rice plant developmental period. BMC Plant Biol 5:8. doi: 10.1186/1471-2229-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Catola S, Marino G, Emiliani G, Huseynova T, Musayev M, Akparov Z, Maserti BE. 2016. Physiological and metabolomic analysis of Punica granatum (L.) under drought stress. Planta 243:441–449. doi: 10.1007/s00425-015-2414-1. [DOI] [PubMed] [Google Scholar]
- 26.Meyer E, Aspinwall MJ, Lowry DB, Palacio-Mejía JD, Logan TL, Fay PA, Juenger TE. 2014. Integrating transcriptional, metabolomic, and physiological responses to drought stress and recovery in switchgrass (Panicum virgatum L.). BMC Genomics 15:527. doi: 10.1186/1471-2164-15-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Janz D, Behnke K, Schnitzler JP, Kanawati B, Schmitt-Kopplin P, Polle A. 2010. Pathway analysis of the transcriptome and metabolome of salt sensitive and tolerant poplar species reveals evolutionary adaption of stress tolerance mechanisms. BMC Plant Biol 10:150. doi: 10.1186/1471-2229-10-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sobhanian H, Motamed N, Jazii FR, Nakamura T, Komatsu S. 2010. Salt stress induced differential proteome and metabolome response in the shoots of Aeluropus lagopoides (Poaceae), a halophyte C4 plant. J Proteome Res 9:2882–2897. doi: 10.1021/pr900974k. [DOI] [PubMed] [Google Scholar]
- 29.Zhou J, Wu Y, Long P, Ho CT, Wang Y, Kan ZP, Cao LT, Zhang L, Wan XC. 2019. LC-MS-based metabolomics reveals the chemical changes of polyphenols during high-temperature roasting of large-leaf yellow tea. J Agric Food Chem 67:5405–5412. doi: 10.1021/acs.jafc.8b05062. [DOI] [PubMed] [Google Scholar]
- 30.Lowe RG, Lord M, Rybak K, Trengove RD, Oliver RP, Solomon PS. 2008. A metabolomic approach to dissecting osmotic stress in the wheat pathogen Stagonospora nodorum. Fungal Genet Biol 45:1479–1486. doi: 10.1016/j.fgb.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 31.Sana TR, Fischer S, Wohlgemuth G, Katrekar A, Jung KH, Ronald PC, Fiehn O. 2010. Metabolomic and transcriptomic analysis of the rice response to the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. Metabolomics 6:451–465. doi: 10.1007/s11306-010-0218-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schauer N, Fernie AR. 2006. Plant metabolomics: towards biological function and mechanism. Trends Plant Sci 11:508–516. doi: 10.1016/j.tplants.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 33.Allwood JW, Ellis DI, Goodacre R. 2010. Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol Plantarum 132:117–135. doi: 10.1111/j.1399-3054.2007.01001.x. [DOI] [PubMed] [Google Scholar]
- 34.Allwood JW, Goodacre R. 2010. An introduction to liquid chromatography-mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem Anal 21:33–47. doi: 10.1002/pca.1187. [DOI] [PubMed] [Google Scholar]
- 35.Schulz B, Boyle C. 2005. The endophytic continuum. Mycol Res 109:661–686. doi: 10.1017/s095375620500273x. [DOI] [PubMed] [Google Scholar]
- 36.Zhong R, Xia C, Ju YW, Li NN, Zhang XX, Nan ZB, Christensen MJ. 2018. Effects of Epichloë gansuensis on root-associated fungal communities of Achnatherum inebrians under different growth conditions. Fungal Ecol 31:29–36. doi: 10.1016/j.funeco.2017.10.005. [DOI] [Google Scholar]
- 37.Zhong R, Zhou XR, Zhang ZQ, Xia C, Li NN, Zhang XX. 2017. Effect of Epichloë gansuensis on arbuscular mycorrhizal fungi spore diversity in rhizosphere soli of drunken horse grass under different growth conditions. Pratacultural Science 34:1627–1634. (In Chinese with English abstract.) [Google Scholar]
- 38.Zhong R, Xia C, Ju YW, Zhang XX, Duan TY, Nan ZB, Li CJ. 2021. A foliar Epichloë endophyte and soil moisture modified belowground arbuscular mycorrhizal fungal biodiversity associated with Achnatherum inebrians. Plant Soil 458:105–122. doi: 10.1007/s11104-019-04365-7. [DOI] [Google Scholar]
- 39.Ju YW, Zhong R, Christensen MJ, Zhang XX. 2020. Effects of Epichloë gansuensis endophyte on the root and rhizosphere soil bacteria of Achnatherum inebrians under different moisture conditions. Front Microbiol 11:747. doi: 10.3389/fmicb.2020.00747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ju YW, Kou MZ, Zhong R, Christensen MJ, Zhang XX. 2021. Alleviating salt stress on seedings using plant growth promoting rhizobacteria isolated from the rhizosphere soil of Achnatherum inebrians infected with Epichloë gansuensis endophyte. Plant Soil 465:349–366. doi: 10.1007/s11104-021-05002-y. [DOI] [Google Scholar]
- 41.Bastías DA, Bustos LB, Jáuregui R, Barrera A, Acuña-Rodríguez IS, Molina-Montenegro MA, Gundel PE. 2021. Epichloë fungal endophytes influence seed-associated bacterial communities. Front Microbiol 12:795354. doi: 10.3389/fmicb.2021.795354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guo JX, Bowatte S, Hou FJ. 2021. Diversity of endophytic bacteria and fungi in seeds of Elymus nutans growing in four locations of Qinghai Tibet Plateau, China. Plant Soil 459:49–63. doi: 10.1007/s11104-020-04608-y. [DOI] [Google Scholar]
- 43.Truyens S, Weyens N, Cuypers A, Vangronsveld J. 2015. Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ Microbiol Rep 7:40–50. doi: 10.1111/1758-2229.12181. [DOI] [Google Scholar]
- 44.Kim H, Lee YH. 2021. Spatiotemporal assembly of bacterial and fungal communities of seed-seedling-adult in rice. Front Microbiol 12:708475. doi: 10.3389/fmicb.2021.708475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nissinen R, Helander M, Kumar M, Saikkonen K. 2019. Heritable Epichloë symbiosis shapes fungal but not bacterial communities of plant leaves. Sci Rep 9:5253. doi: 10.1038/s41598-019-41603-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tannenbaum I, Kaur J, Mann R, Sawbridge T, Rodoni B, Spangenberg G. 2020. Profiling the Lolium perenne microbiome: from seed to seed. Phytobiomes J 4:281–289. doi: 10.1094/PBIOMES-03-20-0026-R. [DOI] [Google Scholar]
- 47.Liu BW, Ju YW, Xia C, Zhong R, Christensen MJ, Zhang XX, Nan ZB. 2022. The effect of Epichloë endophyte on phyllosphere microbes and leaf metabolites in Achnatherum inebrians. iScience 25:104144. doi: 10.1016/j.isci.2022.104144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Singh D, Mathur SB. 2004. Histopathology of seed-borne infections. CRC Press, Boca Raton, FL. [Google Scholar]
- 49.Porras-Alfaro A, Bayman P. 2011. Hidden fungi, emergent properties: endophytes and microbiomes. Annu Rev Phytopathol 49:291–315. doi: 10.1146/annurev-phyto-080508-081831. [DOI] [PubMed] [Google Scholar]
- 50.Santoyo G, Moreno-Hagelsieb G, Orozco-Mosqueda MD, Glick BR. 2016. Plant growth-promoting bacterial endophytes. Microbiol Res 183:92–99. doi: 10.1016/j.micres.2015.11.008. [DOI] [PubMed] [Google Scholar]
- 51.Links MG, Demeke T, Gr€afenhan T, Hill JE, Hemmingsen SM, Dumonceaux TJ. 2014. Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and Brassica seeds. New Phytol 202:542–553. doi: 10.1111/nph.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cottyn B, Debode J, Regalado E, Mew TW, Swings J. 2009. Phenotypic and genetic diversity of rice seed-associated bacteria and their role in pathogenicity and biological control. J Appl Microbiol 107:885–897. doi: 10.1111/j.1365-2672.2009.04268.x. [DOI] [PubMed] [Google Scholar]
- 53.Rosenblueth M, Lopez-Lopez A, Martinez J, Rogel MA, Toledo I, Martinez-Romero E. 2012. Seed bacterial endophytes: common genera, seed-to-seed variability and their possible role in plants. Acta Horticulturae 938:39–48. doi: 10.17660/ActaHortic.2012.938.4. [DOI] [Google Scholar]
- 54.Hameed A, Yeh MW, Hsieh YT, Chung WC, Lo CT, Young LS. 2015. Diversity and functional characterization of bacterial endophytes dwelling in various rice (Oryza sativa L.) tissues, and their seed-borne dissemination into rhizosphere under gnotobiotic P-stress. Plant Soil 394:177–197. doi: 10.1007/s11104-015-2506-5. [DOI] [Google Scholar]
- 55.Wang LL. 2018. Study of seed-borne microbial diversity and plasma disinfection method in tomato seeds. Ph.D. dissertation. Chinese Academy of Agricultural Sciences, Beijing, China. [Google Scholar]
- 56.Zhang Y. 2005. Effects of seedborne fungi on the quality of perennial ryegrass (Lolium perenne L.) seeds. China Agricultural University, Beijing, China. [Google Scholar]
- 57.Kong RF. 2009. Seedborne fungi of sudangrass (Sorghum sudanense) and their control. Lanzhou University, Lanzhou, China. [Google Scholar]
- 58.Qessaoui R, Bouharroud R, Furze JN, El Aalaoui M, Akroud H, Amarraque A, Vaerenbergh JV, Tahzima R, Mayad EH, Chebli B. 2019. Applications of new rhizobacteria Pseudomonas isolates in agroecology via fundamental processes complementing plant growth. Sci Rep 9:12832. doi: 10.1038/s41598-019-49216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun XL, Xu ZH, Xie JY, Hesselberg-Thomsen V, Tan T, Zheng DY, Strube ML, Dragoš A, Shen QR, Zhang RF, Kovács ÁT. 2022. Bacillus velezensis stimulates resident rhizosphere Pseudomonas stutzeri for plant health through metabolic interactions. ISME J 16:774–787. doi: 10.1038/s41396-021-01125-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verma SK, Kingsley KL, Bergen MS, Kowalski KP, White JF. 2018. Fungal disease prevention in seedlings of rice (Oryza sativa) and other grasses by growth-promoting seed-associated endophytic bacteria from invasive Phragmites australis. Microorganisms 6:21. doi: 10.3390/microorganisms6010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Diepeningen AD, de Hoog GS. 2016. Challenges in Fusarium, a trans-kingdom pathogen. Mycopathologia 181:161–163. doi: 10.1007/s11046-016-9993-7. [DOI] [PubMed] [Google Scholar]
- 62.Li Y, Chen Y, Zhou L, You SJ, Deng H, Chen Y, Alseekh S, Yuan Y, Fu R, Zhang ZX, Su D, Fernie AR, Bouzayen M, Ma T, Liu MC, Zhang Y. 2020. MicroTom metabolic network: rewiring tomato metabolic regulatory network throughout the growth cycle. Mol Plant 13:1203–1218. doi: 10.1016/j.molp.2020.06.005. [DOI] [PubMed] [Google Scholar]
- 63.He YY, Pan L, Yang T, Wang W, Li C, Chen B, Shen YH. 2021. Metabolomic and confocal laser scanning microscopy (CLSM) analyses reveal the important function of flavonoids in amygdalus pedunculata pall leaves with temporal changes. Front Plant Sci 12:648277. doi: 10.3389/fpls.2021.648277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang CK, Shen SQ, Zhou S, Li YF, Mao YY, Zhou JJ, Shi YH, An LX, Zhou QQ, Peng WJ, Lyu YY, Liu XM, Chen W, Wang SH, Qu LH, Liu XQ, Fernie AR, Luo J. 2021. Rice metabolic regulatory network spanning the entire life cycle. Mol Plant 15:258–275. doi: 10.1016/j.molp.2021.10.005. [DOI] [PubMed] [Google Scholar]
- 65.Franzluebbers AJ, Hill NS. 2005. Soil carbon, nitrogen, and ergot alkaloids with short and long term exposure to endophyte-infected and endophyte-free tall fescue. Soil Sci Soc Am J 69:404–412. doi: 10.2136/sssaj2005.0404. [DOI] [Google Scholar]
- 66.Schardl CL, Young CA, Hesse U, Amyotte SG, Andreeva K, Calie PJ, Fleetwood DJ, Haws DC, Moore N, Oeser B, Panaccione DG, Schweri KK, Voisey CR, Farman ML, Jaromczyk JW, Roe BA, O'Sullivan DM, Scott B, Tudzynski P, An ZQ, Arnaoudova EG, Bullock CT, Charlton ND, Chen L, Cox M, Dinkins RD, Florea S, Glenn AE, Gordon A, Güldener U, Harris DR, Hollin W, Jaromczyk J, Johnson RD, Khan AK, Leistner E, Leuchtmann A, Li CJ, Liu JG, Liu JZ, Liu M, Mace W, Machado C, Nagabhyru P, Pan J, Schmid J, Sugawara K, Steiner U, Takach JE, Tanaka E, et al. 2013. Plant-symbiotic fungi as chemical engineers: multi-genome analysis of the clavicipitaceae reveals dynamics of alkaloid loci. PLoS Genet 9:e1003323. doi: 10.1371/journal.pgen.1003323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roberts E, Lindow S. 2014. Loline alkaloid production by fungal endophytes of Fescue species select for particular epiphytic bacterial microflora. ISME J 8:359–368. doi: 10.1038/ismej.2013.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Young CA, Schardl CL, Panaccione DG, Florea S, Takach JE, Charlton ND, Moore N, Webb JS, Jaromczyk J. 2015. Genetics, genomics and evolution of ergot alkaloid diversity. Toxins 7:1273–1302. doi: 10.3390/toxins7041273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang YJ, Zhu ZQ. 1984. Studies on the chemical compositions of Achnatherum inebrians. Chem Res Chinese U 3:150–152. [Google Scholar]
- 70.Miles CO, Lane GA, di Menna ME, Garthwaite I, Piper EL, Ball OJP, Latch GCM, Allen JM, Hunt MB, Bush LP, Min FK, Fletcher I, Harris PS. 1996. High levels of ergonovine and lysergic acid amide in toxic Achnatherum inebrians accompany infection by an Acremonium-like endophytic fungus. J Agric Food Chem 44:1285–1290. doi: 10.1021/jf950410k. [DOI] [Google Scholar]
- 71.Zhang XX, Li CJ, Nan ZB. 2011a. Effects of cutting frequency and height on alkaloid production in endophyte-infected drunken horse grass (Achnatherum inebrians). Sci China Life Sci 54:567–571. doi: 10.1007/s11427-011-4181-y. [DOI] [PubMed] [Google Scholar]
- 72.Zhang XX, Li CJ, Nan ZB. 2011b. Effects of salt and drought stress on alkaloid production in endophyte-infected drunken horse grass (Achnatherum inebrians). Biochem Syst Ecol 39:471–476. doi: 10.1016/j.bse.2011.06.016. [DOI] [PubMed] [Google Scholar]
- 73.Zhang XX, Nan ZB, Li CJ, Gao K. 2014. Cytotoxic effect of ergot alkaloids in Achnatherum inebrians infected by the Neotyphodium gansuense endophyte. J Agric Food Chem 62:7419–7422. doi: 10.1021/jf502264j. [DOI] [PubMed] [Google Scholar]
- 74.Song QY, Li F, Nan ZB, Coulter JA, Wei WJ. 2020. Do Epichloë endophytes and their grass symbiosis only produce toxic alkaloids to insects and livestock? J Agric Food Chem 68:1169–1185. doi: 10.1021/acs.jafc.9b06614. [DOI] [PubMed] [Google Scholar]
- 75.Rostás M, Cripps MG, Silcock P. 2015. Aboveground endophyte affects root volatile emission and host plant selection of a belowground insect. Oecologia 177:487–497. doi: 10.1007/s00442-014-3104-6. [DOI] [PubMed] [Google Scholar]
- 76.Guo JQ, McCulley RL, McNear DHJ. 2015. Tall fescue cultivar and fungal endophyte combinations influence plant growth and root exudate composition. Front Plant Sci 6:183. doi: 10.3389/fpls.2015.00183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Massoni J, Bortfeld-Miller M, Widmer A, Vorholt JA. 2021. Capacity of soil bacteria to reach the phyllosphere and convergence of floral communities despite soil microbes variation. Proc Natl Acad Sci USA 118:e2100150118. doi: 10.1073/pnas.2100150118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Aleklett K, Hart M, Shade A. 2014. The microbial ecology of flowers: an emerging frontier in phyllosphere research. Botany 92:253–266. doi: 10.1139/cjb-2013-0166. [DOI] [Google Scholar]
- 79.Otano NN, di Pasquo M, Munoz N. 2015. Airborne fungal richness: proxies for floral composition and local climate in three sites at the el palmar National Park (Coln, Entre Rios, Argentina). Aerobiologia 31:537–547. doi: 10.1007/s10453-015-9382-6. [DOI] [Google Scholar]
- 80.Noecker C, Chiu H-C, McNally CP, Borenstein E. 2019. Defining and evaluating microbial contributions to metabolite variation in microbiome-metabolome association studies. mSystems 4:e00579-19. doi: 10.1128/mSystems.00579-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Huang K, Wang YG, Bai Y, Luo QY, Lin XC, Yang QY, Wang SH, Xin HJ. 2022. Gut microbiota and metabolites in atrial fibrillation patients and their changes after catheter ablation. Microbiol Spectr 10:e01077-21. doi: 10.1128/spectrum.01077-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.01350-22-s0001.pdf, PDF file, 1.2 MB (1.3MB, pdf)
Tables S2-S13. Download spectrum.01350-22-s0002.xlsx, XLSX file, 0.9 MB (976.2KB, xlsx)
Data Availability Statement
The original sequence data of bacterial and fungal sequences were submitted to the NCBI database (https://www.ncbi.nlm.nih.gov/). The BioProject accession numbers of bacterial and fungal raw data are PRJNA865502 and PRJNA865530, respectively.