ABSTRACT
Human endogenous retroviruses (HERVs) comprise about 8.3% of the human genome and are capable of producing RNA molecules that can be sensed by pattern recognition receptors, leading to the activation of innate immune response pathways. The HERV-K (HML-2) subgroup is the youngest HERV clade with the highest degree of coding competence. Its expression is associated with inflammation-related diseases. However, the precise HML-2 loci, stimuli, and signaling pathways involved in these associations are not well understood or defined. To elucidate HML-2 expression on a locus-specific level, we used the retroelement sequencing tools TEcount and Telescope to analyze publicly available transcriptome sequencing (RNA-seq) and chromatin immunoprecipitation (ChIP) sequencing data sets of macrophages treated with a wide range of agonists. We found that macrophage polarization significantly correlates with modulation of the expression of specific HML-2 proviral loci. Further analysis demonstrated that the provirus HERV-K102, located in an intergenic region of locus 1q22, constituted the majority of the HML-2 derived transcripts following pro-inflammatory (M1) polarization and was upregulated explicitly in response to interferon gamma (IFN-γ) signaling. We found that signal transducer and activator of transcription 1 and interferon regulatory factor 1 interact with a solo long terminal repeat (LTR) located upstream of HERV-K102, termed LTR12F, following IFN-γ signaling. Using reporter constructs, we demonstrated that LTR12F is critical for HERV-K102 upregulation by IFN-γ. In THP1-derived macrophages, knockdown of HML-2 or knockout of MAVS, an adaptor of RNA-sensing pathways, significantly downregulated genes containing interferon-stimulated response elements (ISREs) in their promoters, suggesting an intermediate role of HERV-K102 in the switch from IFN-γ signaling to the activation of type I interferon expression and, therefore, in a positive feedback loop to enhance pro-inflammatory signaling.
IMPORTANCE The human endogenous retrovirus group K subgroup, HML-2, is known to be elevated in a long list of inflammation-associated diseases. However, a clear mechanism for HML-2 upregulation in response to inflammation has not been defined. In this study, we identify a provirus of the HML-2 subgroup, HERV-K102, which is significantly upregulated and constitutes the majority of the HML-2 derived transcripts in response to pro-inflammatory activation of macrophages. Moreover, we identify the mechanism of HERV-K102 upregulation and demonstrate that HML-2 expression enhances interferon-stimulated response element activation. We also demonstrate that this provirus is elevated in vivo and correlates with interferon gamma signaling activity in cutaneous leishmaniasis patients. This study provides key insights into the HML-2 subgroup and suggests that it may participate in enhancing pro-inflammatory signaling in macrophages and probably other immune cells.
KEYWORDS: HERVs, HERV-K, HML-2, interferon signaling, interferon gamma, type I interferons, macrophage activation, inflammation, human endogenous retroviruses, STAT signaling, Toll-like receptors
INTRODUCTION
Human endogenous retroviruses (HERVs) are the remnants of ancient retroviral insertions in the germ cells of our evolutionary ancestors and comprise approximately 8.3% of the human genome. Among the HERV groups, the HERV-K family subgroup HML-2 contains the most recently integrated proviral loci and the highest degree of coding competence, and it is generally believed to be the most biologically active (1, 2). Although HML-2 expression has not been directly linked to the manifestation of a specific disease, there is a long history of association between high HML-2 expression and the presence of various pathological disease states—typically, those related to chronic inflammation (3 to 7).
Mechanistically, HERV RNA can be sensed by pattern recognition receptors (PRRs) in a manner similar to exogenous viral RNA, termed “viral mimicry,” leading to pro-inflammatory pathway activation (8). Although various PRRs can recognize viral RNA, the retinoic acid-inducible gene I (RIG-I)-like receptors (RLRs) are of interest due to their localization. Unlike the nucleic acid sensing Toll-like receptors (TLRs), which are restricted to surveying endosomal compartments, RLRs are free-floating and respond to viral RNA located in the cytosol (9). The RLR family consists of three members: RIG-I, melanoma differentiation-associated gene 5 (MDA5), and a truncated receptor, the laboratory of genetics and physiology 2 (LGP2). While all three RLRs can bind to viral RNA, only RIG-I and MDA5 contain the tandem caspase recruitment domain (2CARD) which is necessary for downstream signaling through mitochondrial antiviral-signaling protein (MAVS), an adapter protein for RLR signal transduction (9). Activation of MAVS by RIG-I or MDA5 eventually leads to activation and translocation of the transcription factors IRF3, IRF7, and NF-κB (9). As a result, the RIG-I/MDA5/MAVS pathway promotes an antiviral state through the induction of genes which contain interferon-stimulated response elements (ISREs) and NF-κB binding sites and consequently contributes to the inflammatory state through type I interferon (IFN-I) and other inflammatory cytokine upregulation (9–11).
We previously demonstrated that HML-2 is one of the most strongly induced HERV groups in monocyte-derived macrophages (MDMs) in response to ionizing irradiation and that its transcripts interact directly with MDA5 (12). Knockdown of HML-2 significantly decreased the pro-inflammatory response to ionizing irradiation, including IFN-I expression and secretion, suggesting that HML-2 upregulation in macrophages can augment their pro-inflammatory response (12). While the phenomenon of HERV upregulation and their sensing by PRRs is typically documented under abnormal genomic conditions, such as following tumorigenesis, radiation exposure, or treatment with DNA methyltransferase inhibitors (DNMTis), it is possible that specific HML-2 loci are upregulated and sensed in response to clinically relevant antigens and cytokines (13–15). However, the precise HML-2 loci that can be upregulated, the stimuli that can induce HML-2 upregulation, and the signaling pathways involved are not well understood or defined.
In this study, we investigate the dynamics and potential role of HML-2 expression during macrophage polarization and activation. Macrophages were chosen as a model to investigate HML-2 expression dynamics due to their ability to respond to a wide array of agonists and their importance in regulating inflammation during the wound healing process (16–18). We show that pro-inflammatory (M1) polarization significantly correlates with upregulation of the HML-2 provirus located at locus 1q22. This provirus, also known as HERV-K102, is intergenic and previously reported to be elevated in systemic lupus erythematosus (SLE) patients (19). Additional analysis revealed that HERV-K102 upregulation occurred explicitly in response to interferon signaling and relied on an upstream solo long terminal repeat (LTR) that recruits interferon regulatory factor 1 (IRF1) and potentially signal transducer and activator of transcription 1 (STAT1). Induction of HERV-K102 expression by IFN signaling was demonstrated in other cell types, suggesting that this mechanism of HML-2 upregulation may be a universal phenomenon. Using THP1-derived macrophages (TDMs) as a model for MDMs, we show that knockdown of HML-2 or knockout of MAVS leads to significant reduction in the expression of genes that contain ISRE elements and a reporter construct under the control of five tandem ISRE elements. These findings provide a mechanism for HML-2 upregulation in response to inflammation and suggest that this HERV-K subgroup, in particular HERVK-102, may participate in a positive feedback loop to enhance pro-inflammatory signaling.
RESULTS
Macrophage polarization triggers HML-2 upregulation in primary monocyte-derived macrophages and THP1-derived macrophages.
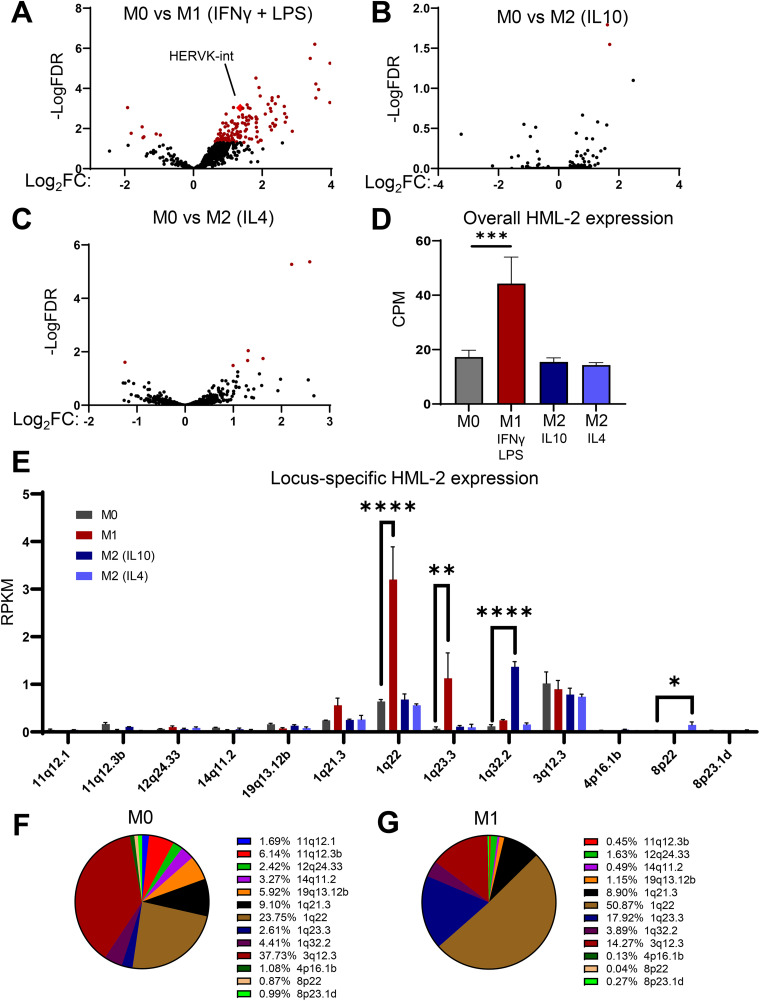
Depending on external signals, macrophages can be polarized toward either pro-inflammatory (M1) or anti-inflammatory (M2) phenotypes. This plasticity and ability to respond to a wide array of agonists makes them an optimal cell type for assessing changes in the retroelement expression landscape in response to pro- versus anti-inflammatory pathway activation. Therefore, we first sought to assess the expression dynamics of all retroelements in response to macrophage polarization. Using the retroelement analysis programs TEcount (20, 21) and Telescope (22), we were able to reanalyze a publicly available RNA-sequencing data set, GSE162698 (23). These software tools use a statistical model to determine the most likely origin of multi-mapped reads, allowing them to accurately measure retroelement expression. In the analyzed data set, primary MDMs were polarized into an M1 phenotype with LPS + IFN-γ and an M2 phenotype with either IL-10 or IL-4 treatment. In response to M1 polarization, over 150 retroelement groups and subgroups were significantly modulated (Fig. 1A, Fig. S1A in the supplemental material) 18 h after treatment. However, fewer than 10 retroelement groups and subgroups were significantly modulated in response to M2 polarization (Fig. 1B and C, Fig. S1A). Notably, among the significantly upregulated retroelement clades following M1 polarization, HERVK-int, which corresponds to HERV-K (HML-2) (24), was the most highly expressed HERV at the basal state (Fig. S1B). This urged us to continue investigating the HML-2 subgroup at a locus-specific level using the retroelement analysis software Telescope (22).
FIG 1.
Pro-inflammatory (M1) polarization of macrophages correlates with retroelement expression. (A to C) Volcano plots showing fold change in retroelement expression measured by TEcount at the subgroup level: M1 polarized, lipopolysaccharide (LPS) + interferon gamma (IFN-γ) (A); M2 polarized, interleukin (IL)-10 (B) and IL-4 (C); versus neutral (M0) monocyte-derived macrophages (MDMs) following 24 h of polarization. Red dots indicate retroelements significantly changed in expression. (D) Expression on RNA level of the HML-2 subgroup (copies per million [CPM]) in M0, M1, M2 (IL-10), and M2 (IL-4) MDMs. (E) Locus-specific expression of the HML-2 subgroup measured by Telescope analysis of transcriptomic sequencing (RNA-seq) data set in M0, M1, M2 (IL-10) and M2 (IL-4) polarized MDMs. RPKM, reads per kilobase per million reads mapped. (F and G) Proportion that each HML-2 provirus constitutes in the total amount of HML-2-derived transcripts in M0 (F) and M1-polarized (G) MDMs, based on analysis shown in panel E. Data are presented as means ± standard error of the mean (SEM) from three donors as biological replicates. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 according to edgeR analysis for panels D and E. Due to the nature of the analysis, only significantly positive fold changes are highlighted in panel E; all fold changes can be found in Table S1. CPM was used for overall HML-2 expression in panel D due to differences in HML-2 proviral length on the locus-specific level.
Although several HML-2 loci are known to be induced following tumorigenesis or chronic inflammatory diseases (25–28), little is known about specific HML-2 loci induction in response to typical inflammatory stimuli in the normal state. Therefore, we repeated our analysis of the data set GSE162698 using Telescope to determine which HML-2 loci displayed a >2-fold increase in expression upon M1 polarization. First, we found that the HML-2 subgroup was upregulated only in response to pro-inflammatory polarization, whereas treatment with M2 polarizing agents did not change the expression of this clade (Fig. 1D). Among the 92 HML-2 proviral loci, only 13 displayed detectable expression. Five loci showed either a high basal expression (3q12.3) or a significant change correlated with the polarization treatment (1q22, 1q23.3, 1q32.2, and 8p22) (Fig. 1E, Table S1). At the basal state, HML-2 expression was dominated by 1q22 and 3q12.3, which constituted 23.75% and 37.3% of the total HML-2-derived transcripts, respectively (Fig. 1F). However, upon M1 polarization, there was significant upregulation of 1q22 and 1q23.3, whereas 3q12.3 expression remained stable (Fig. 1E). This results in 1q22 constituting over 50% of the overall HML-2-derived transcripts (Fig. 1G). In response to M2 polarization, 1q32.2 was significantly upregulated following interleukin (IL)-10 treatment and 8p22 was significantly upregulated following IL-4 treatment (Fig. 1E, Table S1). Of these 5 loci, 3 (1q23.3, 1q32.2, and 8p22) reside within protein-encoding genes. 1q23.3 resides within the intron of CD48 gene, 1q32.2 resides within the intron of CR1, and 8p22 spans the 5′ untranslated region (UTR), the first exon, and the first intron of fibrinogen-like protein 1 (FGL1). The expression patterns of 1q23.3 and 1q32.2 correspond to the expression of the genes they reside within (Fig. S1C). The gene corresponding to 8p22, FGL1, was undetectable in MDMs (Table S2). Previously, FGL1 was found to be expressed in hepatocellular carcinoma and lung cancer tissues (29); thus, in our analysis, its expression could be related to cancer-associated changes that are not present in primary macrophages. The other two proviruses, 1q22 and 3q12.3, are in intergenic loci, and previous literature suggests that transcription likely starts within their own 5′ LTR (30).
Primary macrophages are highly heterogeneous (31–33). To determine if the same trends are consistent in THP1 cells, a monocytic cell line commonly used as a model of a more homogeneous macrophage population, we analyzed another gene expression data set, GSE159112 (34), in which THP1 cells were differentiated to a macrophage phenotype by treatment with phorbol 12-myristate 13-acetate (PMA) (35–37). This data set is similar to the previous one, except that a combination of IL-4 and IL-13 was used for M2 polarization of THP1-derived macrophages (34). Consistent with the primary MDMs, 1q22 and 3q12.3 constitute the majority of the HML-2 derived transcripts, with 1q22 expression significantly upregulated during M1 polarization and 3q12.3 remaining relatively stable (Fig. S1D). However, a notable difference between the primary MDMs and TDMs is the relatively high expression of 8p22 in TDMs compared to that in MDMs, which is likely due to the fact that these cells are cancerous and represent an acute monocytic leukemia cell line with higher expression of FGL1 compared to MDMs (Fig. S1E, Table S2).
The HML-2 provirus at 1q22 is upregulated in response to interferon, but not TLR signaling.
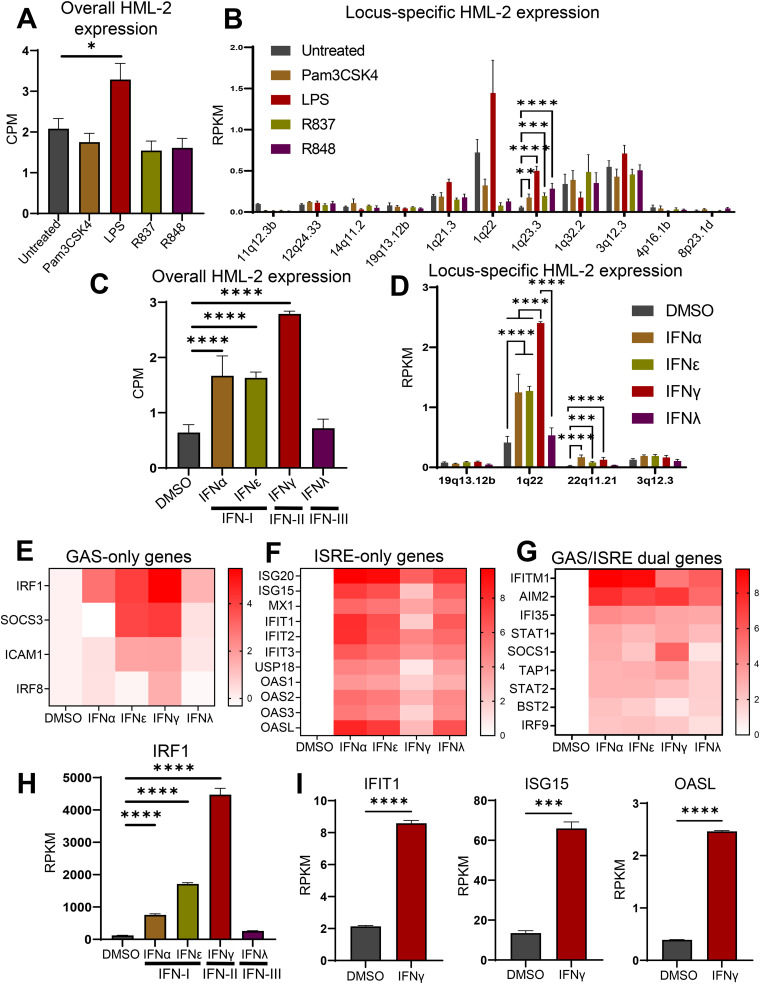
In-vitro M1 polarization involves challenging macrophages with a combination of lipopolysaccharide (LPS) and IFN-γ. Importantly, LPS and IFN-γ activate separate signaling pathways and lead to the activation of different transcription factors (38, 39). Therefore, we sought to determine which of these signaling pathways is directly responsible for the upregulation of 1q22, the most highly upregulated HML-2 provirus following M1 polarization. To determine the impact of TLR signaling on the HML-2 subgroup, we analyzed the RNA-sequencing data set GSE147310 (40). This data set contains transcriptomic data from primary MDMs treated with agonists of Toll-like receptors 1/2 (Pam3csk4), 4 (LPS), 7 (R837), or 7/8 (R848) obtained 18 h post-treatment. When monitoring the overall HML-2 expression using TEcount analysis, we observed a significant upregulation following LPS treatment but not in response to the other agonists (Fig. 2A). In terms of which HML-2 loci were sensitive to the activation of different TLRs, only 1q23.3 was significantly upregulated, and this effect was found in response to each TLR agonist (Fig. 2B, Table S3). Similar to what was shown previously, the expression pattern of 1q23.3 corresponds to the expression of the gene it resides within, CD48 (Fig. S2). While 1q22 was not significantly upregulated by LPS treatment, LPS treatment did result in an upward trend in expression, whereas the other agonists resulted in a downward trend (Fig. 2B).
FIG 2.
Expression ratios of various HML-2 proviruses in response to the activation of Toll-like receptor (TLR) and interferon (IFN) signaling pathways. (A) Overall expression of the HERV-K HML-2 subgroup in MDMs following activation of TLR1/2 with the agonist Pam3CSK4, TLR4 with LPS, TLR7 with R837, and TLR 7/8 with the agonist R848 for 18 h. Analysis of RNA-seq data with TEcount software. (B) Locus-specific expression of the HML-2 subgroup measured by Telescope RNA-seq analysis tool in MDMs following treatment with TLR agonists. The same RNA-seq data as shown in panel A are analyzed. (C) Overall expression of the HML-2 subgroup in MDMs following type I IFN (IFN-α or IFN-ε), type II IFN (IFN-γ), or type III IFN (IFN-λ) treatment for 18 h. Analysis of RNA-seq data with TEcount software. (D) Locus-specific expression of the HML-2 subgroup in MDMs following treatment with dimethyl sulfoxide (DMSO) or type I, II, and III interferons measured by Telescope. The same RNA-seq data as shown in panel C are analyzed. (E to G) Heatmaps of the log2(fold change) in expression for a gamma interferon activation site (GAS)-only containing genes (E), an interferon-stimulated response element (ISRE)-only containing genes (F), and GAS/ISRE dual-containing genes (G) following IFN-α, IFN-ε, IFN-γ, and IFN-λ treatment. Figure based on transcriptomic data analysis using the TEcount pipeline as described in Materials and Methods. (H) Expression of IRF1 gene following IFN-I, IFN-II, and IFN-III treatment: transcriptomic data analysis using the TEcount pipeline as described in Materials and Methods. (I) Expression of selected genes which contain an ISRE but not a GAS element following IFN-γ treatment: transcriptomic data analysis using the TEcount pipeline. Data are presented as means ± SEM from five donors (TLR agonist data set) or three donors (interferon data set) as biological replicates. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 according to edgeR analysis. Due to the nature of the analysis, only significantly positive fold changes are highlighted in panels B and D; all fold changes can be found in Table S2. CPM was used for overall HML-2 expression in panels A and C due to differences in HML-2 proviral length on the locus-specific level. RPKM, reads per kilobase per million reads mapped.
To determine whether interferon signaling could upregulate the HML-2 subgroup, more specifically, 1q22, we analyzed another RNA-sequencing data set, GSE158434 (41). This set contains RNA-seq data from primary MDMs treated with either dimethyl sulfoxide (DMSO), type I IFNs (IFN-α or IFN-ε), type II IFN (IFN-γ), or type III IFN (IFN-λ) for 18 h. We observed that both IFN-I and IFN-II signaling significantly upregulated overall HML-2 expression (Fig. 2C, Table S3). Notably, 1q22 was significantly upregulated in response to both IFN-I and IFN-II but was nearly two times more sensitive to IFN-II signaling than to IFN-I signaling (Fig. 2D).
IFN-I induces gene expression via activation of the transcription factor complex STAT1/STAT2/IRF9, also called ISGF3, which binds to ISREs, whereas IFN-II induces the expression of genes that contain a gamma interferon activation site (GAS) in their promoters via activation of STAT1/STAT1 homodimers (42). This distinction is important for clarifying the phenotypic effect of HML-2 expression, as we suspect that upregulation of this HERV-K subgroup activates the RIG-I/MDA5/MAVS pathway to enhance ISRE-containing gene expression as a secondary effect of IFN-γ signaling (9). Therefore, for downstream experiments and analysis, we were interested in identifying genes which depend on their ISRE site for gene induction following IFN-γ signaling because these genes may be modulated in response to HML-2 knockdown. The ideal targets for this analysis would be genes which are known to contain an ISRE site in their promoter region and are not upregulated abnormally highly or early compared to other ISRE-containing genes, as this may be due to the contribution of additional transcription factor binding sites (TFBSs) outside the ISRE element(s).
To select genes that may be modulated in response to differential HML-2 expression, we examined the induction of genes which contain either ISRE, GAS, or both TFBSs (ISRE-only, GAS-only, and GAS/ISRE dual, respectively). As expected, the genes controlled by GAS-containing promoters, including IRF1, strongly associated with IFN-γ activity (43), were most sensitive to IFN-γ treatment (Fig. 2E and H, Table S4). This is partly consistent with the expression trend of 1q22 following IFN-I versus IFN-II treatment, suggesting that 1q22 may contain IFN-γ-associated transcription factor binding sites.
Treatment with IFN-I or IFN-III remarkably upregulated ISRE-containing genes (Fig. 2F). Among the genes that contained an ISRE site but not a GAS site, we identified three which were upregulated 4- to 6-fold following IFN-γ treatment: IFIT1, ISG15, and OASL (Fig. 2I). These genes were selected as indirect indicators of ISRE activity (demonstrated in later experiments). Although other ISRE-only genes, such as ISG20 and IFIT2, were significantly following IFN-γ treatment, their upregulation was drastically higher than that of the other ISRE-only genes. This led us to exclude them from future analysis. Among the genes containing both GAS and ISRE elements in their promoters, certain genes displayed higher sensitivity to IFN-I (IFITM1, IFI35), whereas other genes were mostly upregulated by IFN-γ treatment (SOCS1, TAP1) (Fig. 2G).
The HML-2 provirus at 1q22 is upregulated in different cell types and pro-inflammatory disease states.
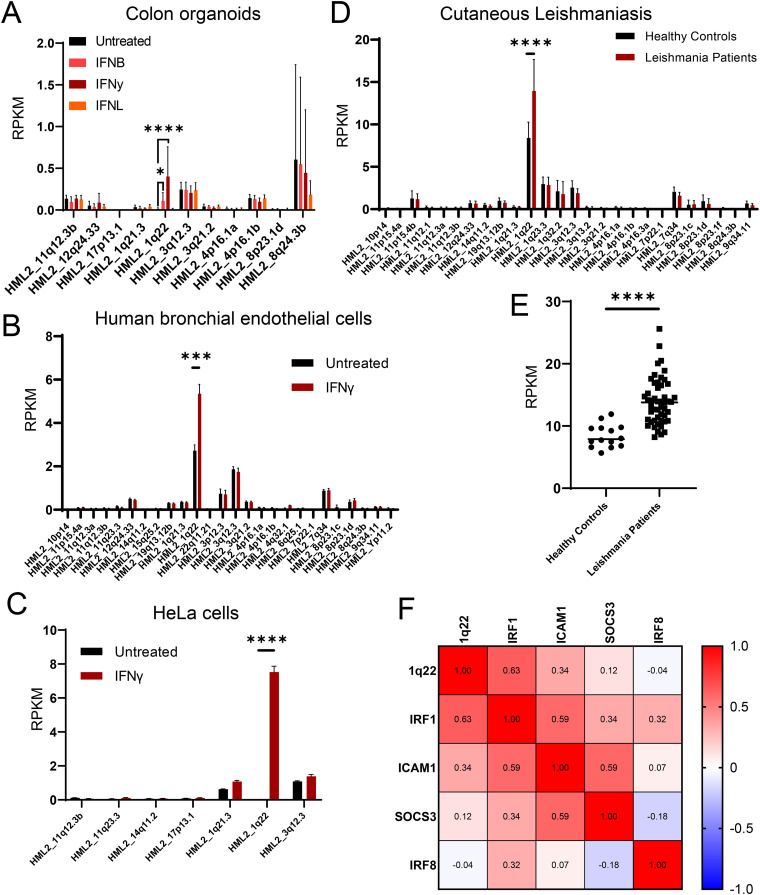
To determine whether 1q22 upregulation by IFN signaling was a universal phenomenon, we examined HML-2 expression on a locus-specific level in response to interferon treatment in primary human colon intestinal epithelial cell organoids (GSE190899), primary human bronchial endothelial cells (GSE185200), and HeLa cells (GSE150196) (Fig. 3A to C). The raw FASTQ files from the indicated RNA-sequencing data sets were obtained from recently published papers (44–46) and analyzed with Telescope, as previously described. In all situations examined, we observed that IFN-γ significantly upregulated expression of 1q22 and that 1q22 constituted the majority of HML-2-derived transcripts following IFN-γ treatment.
FIG 3.
HERV-K102 as an indicator of interferon gamma signaling and inflammation. (A to C) Locus-specific expression of the HML-2 subgroup measured using Telescope software tool in the RNA-seq databases from IFN-β-, IFN-γ-, or IFN-λ-treated colon organoids (A), IFN-γ treated human bronchial endothelial cells (B), and HeLa cells in response to IFN-γ treatment (C). Additional details are provided in Materials and Methods. (D to F) Analysis of transcriptomic data from cutaneous leishmania (CL) patients. (D) Locus-specific expression of the HML-2 subgroup in control and CL patients determined by Telescope software analysis. (E) Individual HERV-K102 expression between control and CL patients, Telescope software analysis. (F) Pearson’s correlation matrix between HML2_1q22, IRF1, ICAM1, SOCS3, and IRF8 expression in CL patients. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 according to edgeR in panels A to D and Pearson’s correlation matrix in panel E. HERV-K, human endogenous retrovirus group K.
Since the sensitivity of 1q22 to IFN-γ appeared to be ubiquitous and the receptors for IFN-γ signaling are nearly ubiquitously expressed (47), we sought to determine whether 1q22 expression in circulating peripheral blood mononuclear cells (PBMCs) was associated with chronic inflammation. Local cutaneous leishmaniasis (CL) is a parasitic disease characterized by chronic inflammation and elevated IFN-γ signaling (19), making it a prime disease for evaluating the relationship between 1q22 and an inflammatory disease state. Therefore, we assessed HML-2 expression on a locus-specific level in healthy control and CL patients (GSE162760), (19). As expected, among the detectably expressed HML-2 loci, only 1q22 was significantly elevated in circulating PBMCs from CL patients (Fig. 3D). Moreover, on an individual-person basis, 1q22 expression had a relatively small overlap between the healthy control and CL patient groups (Fig. 3E) and was significantly correlated with several indicators of IFN-γ signaling, including IRF1 and ICAM1 gene expression (Fig. 3F and Fig. S2).
Interferon gamma signaling induces transcription factor binding and epigenetic modifications upstream of HERV-K102.
Since the rest of our analysis is specific to a single provirus, the HML-2 provirus located at 1q22 is referred to as HERV-K102, a commonly used alias (28, 48, 49). Due to the higher upregulation of HERV-K102 following IFN-γ signaling compared to IFN-I signaling, we assumed that an IFN-γ-associated transcription factor binds to the 5′ LTR of HERV-K102 or an upstream region. As previously described, STAT1 homodimers are directly activated following IFN-γ signaling and mediate the upregulation of genes that contain a GAS site. However, several transcription factors also contain a GAS site and may be preferentially upregulated in response to IFN-γ compared to IFN-I (39, 50), including IRF1 and IRF8 (shown in Fig. 2E), which bind to IRF-binding elements (IRF-Es).
Notably, IRF1 is an IFN-γ-associated transcription factor which contains a GAS site, but not an ISRE site, in its promoter region and is thereby upregulated significantly higher in response to IFN-γ signaling compared to IFN-α or IFN-ε signaling (shown in Fig. 2E and H) (43). Following IFN-γ-mediated upregulation, IRF1 is critical for inducing the expression of various innate immunity genes through binding to IRF-E sites in their promoters, including IFNβ, iNOS, IL-12p35, and many others (43, 51–53).
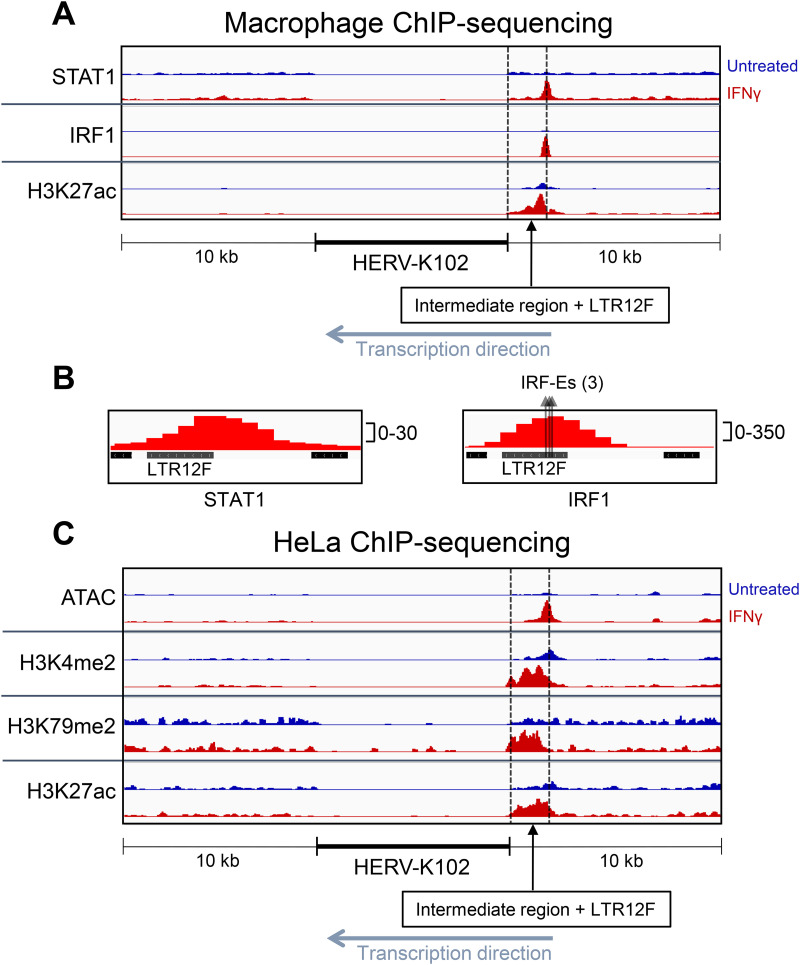
To assess STAT1 and IRF1 occupancy near HERV-K102, we analyzed a publicly available chromatin immunoprecipitation (ChIP)-sequencing data set of primary MDMs treated with IFN-γ for 24 h (GSE43036) (54). Following IFN-γ treatment, STAT1 and IRF1 peaks are significantly enriched in a region upstream (~1.6 kb) of HERV-K102 (Fig. 4A and Fig. S3A). No other STAT1 or IRF1 peaks were identified ±10 kb from HERV-K102. The IRF1 peak is enriched 12-fold (P < 0.0001) compared to the untreated cells and the summit of the peak corresponds to a cluster of three potential IRF-E sites located within a solo LTR, termed LTR12F (Fig. 4B and Fig. S3A). The potential IRF-E sites contain the core consensus binding sequence (AANNGAAA) (55) and match several IRF-1 binding sequence motifs that were enriched in an analysis of IRF-1 bound sequences following IFN-γ treatment (56), highlighted in Fig. S3B. The STAT1 peak is enriched 7.6-fold (P < 0.0001) and its summit is also located within LTR12F. However, the summit of the STAT1 peak is not located at any known GAS or ISRE sequence motifs. It was previously demonstrated that IRF-1 and STAT1 interact and can be co-immunoprecipitated, with speculation that IRF1 (and likely other proteins) can facilitate the recruitment of STAT1 to non-GAS sites (54, 57). Because the distance between the summits of IRF1 and STAT1 is only 40 bp, this could explain the lack of an apparent GAS or ISRE site near the STAT1 summit. The lower peak density of the STAT1 peak compared to the IRF1 peak (30 versus 350 normalized RPKM [reads per kilobase per million reads mapped] at the summit, respectively) may also influence our ability to identify the precise STAT1-binding sequence due to poor resolution. Histone H3 acetylation of lysine 27 (H3K27ac), a histone mark for active enhancers and promoters, was significantly enriched (2.1-fold, P < 0.0001) in the intermediate region between HERV-K102 and LTR12F (Fig. 4A and Fig. S3A) (54, 58, 59).
FIG 4.
Interferon gamma signaling induces transcription factor binding and epigenetic modifications upstream of HERV-K102. (A) Visualization of chromatin immunoprecipitation (ChIP)-sequencing results for STAT1 binding, IRF1 binding, and H3K27ac prior to and following IFN-γ treatment for 24 h in primary MDMs. (B) Close-up visualization of LTR12F and STAT1 or IRF1 binding. Three arrows are depicted for IRF1 binding analysis, with each arrow representing a potential IRF-binding element (IRF-E) sequence motif within LTR12F. (C) Visualization of ATAC-seq (Assay for Transposase-Accessible Chromatin using Sequencing) and ChIP-sequencing for H3K4me2, H3K27ac, and H3K79me2 prior to and following IFN-γ treatment for 24 h in HeLa cells. Peak locations, fold-enrichment values, and P values are shown in Fig. S4.
To more broadly assess the epigenetic landscape of HERV-K102, we analyzed ATAC-seq (Assay for Transposase-Accessible Chromatin using Sequencing) and ChIP sequencing in HeLa cells treated with IFN-γ for 24 h. These are an extension from the RNA-sequencing data set of HeLa cells shown in Fig. 4C and Fig. S3A, which demonstrated that HERV-K102 is upregulated in this cell type following IFN-γ treatment. ATAC-seq is a technique used to assess chromatin accessibility and can be an indicator of active gene regulation (60–62). Following IFN-γ treatment, chromatin accessibility is significantly increased in the region upstream of HERV-K102, encompassing LTR12F and the intermediate region (3-fold enrichment, P < 0.0001) (Fig. 4C). Three histone marks of transcriptional activation were significantly increased in the region between HERV-K102 and LTR12F, including histone H3 dimethylation of lysine 4 (H3K4me2) (two tandem peaks: 6.9-fold, P < 0.0001; 6.6-fold, P < 0.0001) and histone H3 dimethylation of lysine 79 (H3K79me2) (3.8-fold, P < 0.001), and H3K27ac (4.1-fold, P < 0.0001). H3K4me2 marks are often found in actively transcribed or “poised” (regions that are primed for rapid transcriptional activation) promoter regions (46, 63, 64). H3K79me2 is suggested to be an active gene marker that most commonly occurs on the promoter and transcription start site (TSS) region of transcriptionally active genes and is correlated with both high transcription and high transcription elongation rates (65–69). Together, these results suggest that HERV-K102 and the upstream region experience epigenetic modifications which favor or indicate active gene expression.
LTR12F is critical for HERV-K102 upregulation following interferon signaling.
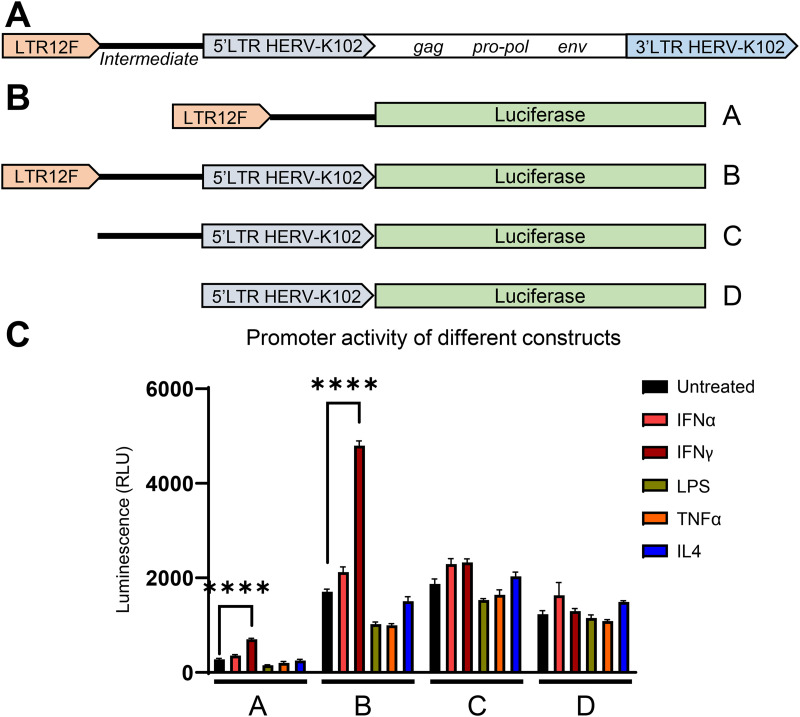
The transcription factor and epigenetic analysis suggests that the region upstream of HERV-K102 may be involved in regulating its expression. A general outline of HERV-K102 and the upstream region is depicted in Fig. 5A. To determine whether the upstream region is critical for HERV-K102 transcriptional upregulation following interferon signaling, we used a promoter-less luciferase reporter lentiviral construct to examine the promoter activity of either the full-size chromosome fragment containing LTR12F + a 1,606-bp intermediate sequence + the 5′ LTR of HERV-K102 or truncated DNA fragments containing the distinct sections of this region (Fig. 5B). None of the constructs experienced increased luciferase activity following treatment with IFN-α, LPS, tumor necrosis factor alpha (TNF-α), or IL-4. However, the constructs containing LTR12F (constructs A and B) saw a nearly 3-fold increase in luciferase activity following IFN-γ treatment (Fig. 5C). Importantly, the increase in reporter activity following IFN-γ treatment was lost when LTR12F was removed from the construct (constructs C and D). As expected, since LTR12F is a truncated LTR and does not contain its own transcription start site, the level of luciferase activity in the construct containing LTR12F without the 5′ LTR of HERV-K102 (construct A) is very low, despite significant upregulation in activity following IFN-γ treatment. When luciferase is expressed under the control of LTR12F followed by the intermediate sequence and the 5′ LTR of HERV-K102 (construct B), which retains its viral transcription start site, luciferase activity is 10-fold higher.
FIG 5.
LTR12F is critical for HERV-K102 upregulation following interferon signaling. (A) General illustration of LTR12F, a downstream intermediate sequence, and 1q22 (HERV-K102 provirus). (B) Representative illustration of the luciferase-expressing constructs used to identify which region spanning the 5′ long terminal repeat (LTR) of HERV-K102 to the upstream solo LTR, LTR12F, was responsible for HERV-K102’s sensitivity to IFN signaling. Lentiviral vectors based on commercial LVR-1048-pLV plasmid (Cellomics Technology) were generated and used for transduction of THP1 monocytes. (C) Comparison of the firefly luciferase reporter activity in the THP1 cells, transduced with the constructs indicated in panel B and differentiated to macrophages using PMA (phorbol 12-myristate 13-acetate) treatment, in response to IFN-α, IFN-γ, LPS, TNF-α, or IL-4 stimulation for 18 h. Data are presented as mean values ± SEM from three biological replicates. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 according to two-way analysis of variance (ANOVA).
HML-2 expression facilitates the MAVS-mediated pathway to enhance IFN-I signaling in response to IFN-γ.
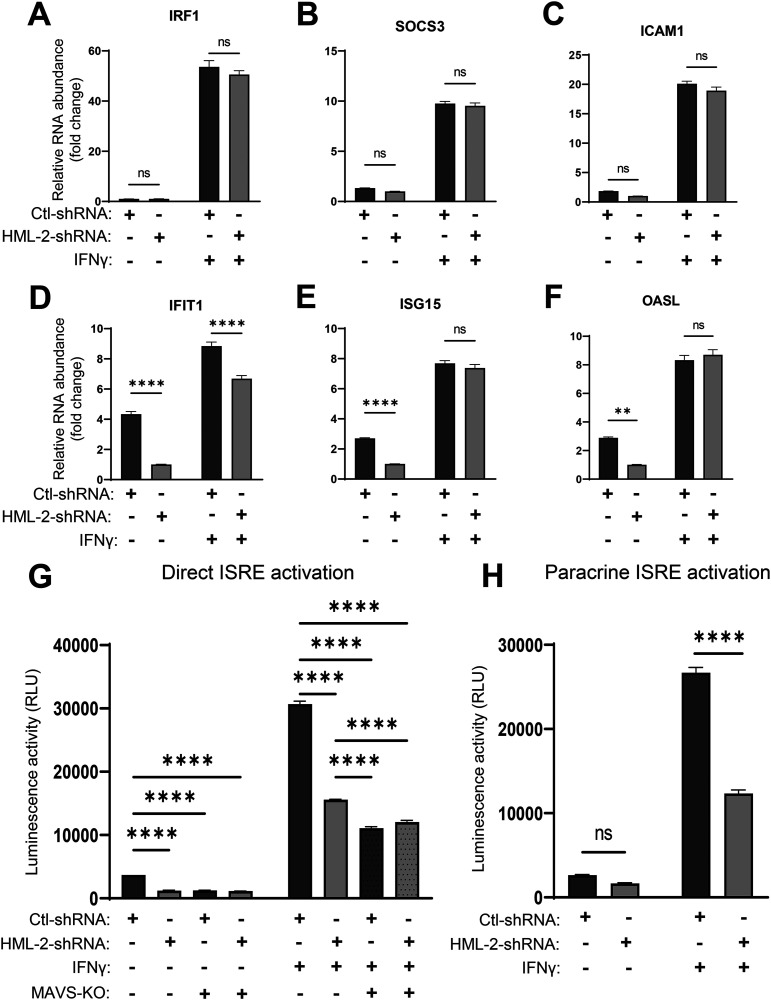
To determine whether the upregulation of genes that contain ISRE sequences shown in Fig. 2F and I was associated with HML-2 expression, we transduced THP-1 cells with either control small hairpin RNA (Ctl-shRNA) or shRNA targeting the env gene of HML-2 (HML-2-shRNA) (described previously [12, 70]; Table S6). Approximately 40% to 50% knockdown in relative HML-2 RNA quantity was achieved in untreated TDMs (Fig. S4). To compare the relative amount of GAS-containing gene induction versus ISRE-containing gene induction, we assessed the expression of three genes which contain a GAS element but not an ISRE element and vice versa by reverse transcriptase quantitative PCR (RT-qPCR). Interestingly, in basal TDMs, all three GAS-containing genes were non-significantly modulated in the shRNA-expressing cells (Fig. 6A to C). However, all three tested ISRE-containing genes displayed significant downregulation in response to HML-2 knockdown (Fig. 6D to F, left two columns). When the cells were treated with IFN-γ, IFIT1 remained significantly downregulated in expression, whereas differences in the expression of the other two genes were leveled, possibly due to the general activation associated with type II interferon and only partial knockdown of HML-2 RNA.
FIG 6.
HML-2 knockdown reduces ISRE activation through direct signaling and distantly via a paracrine mechanism. (A to F) Reverse transcriptase quantitative PCR (RT-qPCR) measured expression (threshold cycle [ΔΔCT] method) of genes IRF1, SOCS3, and ICAM1 which contain a GAS element (A to C) and an ISRE element-containing genes IFIT1, ISG15, and OASL (D to F) in response to IFN-γ treatment for 18 h in TDMs transduced with control shRNA or HML-2-shRNA targeting a conserved region of the HML-2 env gene transcript. (G) Measuring direct activation of ISRE elements in response to IFN-γ treatment using luciferase-reporter THP1-Dual or THP1-Dual MAVS-KO (mitochondrial antiviral-signaling protein-knockout)-derived macrophages transduced with either control-shRNA- or HML-2-shRNA-expressing vectors; 18 h post-IFN-γ treatment. (H) Measuring paracrine activation of ISRE elements in luciferase reporter THP1-Dual-derived macrophages in response to condition medium from TDMs transduced with control shRNA- or HML-2-shRNA-expressing vectors and treated with IFN-γ for 18 h. Data are presented as means ± SEM from three independent measurements in panels A to F and three biological replicates in panels G and H. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 according to two-way ANOVA.
Since the ISRE-containing genes may contain other TFBSs which are indirectly activated following IFN-γ treatment, we engaged reporter THP1 cells, THP1-Dual (InvivoGen), which contain a secreted luciferase gene under the control of ISG54’s promoter region and five upstream ISRE elements. This allows exclusive monitoring of ISRE element activation. Additionally, we used a MAVS-knockout (MAVS-KO) version of these reporter cells to assess the involvement of the MAVS signaling pathway. Both wild-type (WT) and MAVS-KO reporter cells were transduced with control or HML-2 shRNA, differentiated into TDMs, and challenged with IFN-γ. In agreement with the data from our previous experiment, shown above, basal ISRE activation is significantly reduced following HML-2 knockdown (Fig. 6G). Additionally, MAVS-KO reduces ISRE activation to similar levels as HML-2 knockdown in basal TDMs. In response to IFN-γ treatment, ISRE activation is still significantly reduced in both HML-2 knockdown and MAVS-KO cells. MAVS-KO cells have a significantly greater reduction in ISRE activation than the HML-2 knockdown cells. However, addition of HML-2 knockdown did not further decrease ISRE activation in MAVS-KO cells. If MAVS is activated in response to IFN-γ, then type I IFN expression should be induced following MAVS-mediated IRF3/7 activation and secreted to induce ISRE activation in a paracrine manner (71). To determine if HML-2 knockdown influences paracrine ISRE activation in response to IFN-γ treatment, we transferred condition medium from control or HML-2 shRNA-expressing TDMs challenged with IFN-γ to the previously described reporter THP1 cells and assessed ISRE activation. We observed significantly reduced paracrine ISRE activation in response to IFN-γ due to HML-2 knockdown (Fig. 6H).
DISCUSSION
In the present study, we investigated the regulation of HERV-K (HML-2) expression on a locus-specific level in response to inflammatory signaling and examined a potential role of this subgroup in macrophage activation. Several studies have indicated that the HML-2 subgroup is elevated in a variety of inflammation-associated diseases (3–7). However, the precise HML-2 loci which can be induced in response to short-term inflammatory pathway activation have not been identified. One major historical limitation is the highly repetitive nature of HERV proviral loci, making it unfeasible to examine the entire HML-2 subgroup’s expression via RT-qPCR. While a nested PCR followed by sequencing can address this issue, specialized RNA-sequencing analysis tools have proven to be effective and accurate at measuring repetitive element expression on a locus-specific level (20, 22, 72). Briefly, software packages such as TEcount and Telescope assign ambiguously (multi-)mapped reads to the most probable source element through a statistical model that accounts for the number of uniquely mapped reads per element among the list of potential sources for a single ambiguous read (20, 22). Since the standard RNA-seq pipeline ignores repetitive elements, we sought to re-examine publicly available RNA-seq data sets using the retroelement software packages TEcount and Telescope.
Notably, we initially identified that the HML-2 subgroup was the most highly expressed HERV clade that was significantly modulated following M1 polarization in primary MDMs. On a locus-specific level, our HML-2 expression results were consistent with a previous study that implemented single-molecule real-time (SMRT) sequencing and characterized three HML-2 proviral loci (1q22, 1q23.3, and 3q12.3) as constituting over 90% of the HML-2 derived transcripts in circulating human lymphocytes, with these loci constituting the vast majority of our HML-2 derived transcripts across all our data sets (73). SMRT sequencing is advantageous due to its ability to sequence relatively long (~700 bp in this case, up to a maximum of 50,000 bp) reads, allowing for increased sensitivity and accuracy when mapping retroelement reads to the genome (73, 74). Unique to our analysis, we identified that a specific provirus within an intergenic region of locus 1q22, known as HERV-K102, was significantly upregulated and constituted the majority of HML-2 derived transcripts following the pro-inflammatory polarization of primary MDMs with LPS and IFN-γ.
Previous literature on HERV-K102 indicates that its expression is elevated in systemic lupus erythematosus patients and correlates with a higher interferon status (19). The SLE patients also had anti-HERVK-102 Env IgG antibodies and their serum concentration correlated with higher expression of interferon-stimulated genes (ISGs). Although this suggests that there may be a biological consequence of HERV-K102 elevation, the mechanism responsible for HERV-K102 upregulation in these patients was not identified. Interestingly, we found that TLR signaling does not modulate HERV-K102 expression. On the contrary, IFN signaling with either IFN-I or IFN-II significantly upregulated HERV-K102. In agreement with previous literature, we did not find an ISRE or GAS element within the 5′ LTR of HERV-K102 (30, 75, 76), but we did reveal that STAT1 and IRF1 bind to a region upstream of HERV-K102 following IFN-γ signaling. The peaks were co-localized and mapped to a solo LTR, LTR12F, upstream of HERV-K102. Predictably, using a promoter-less luciferase construct, we found that LTR12F was critical for the increase in promoter activity of HERV-K102’s 5′ LTR following IFN-γ signaling. The upregulation of HERV-K102 expression upon IFN-I treatment, which was detected with RNA-sequencing but not in our reporter luciferase constructs that measured promoter activity, may have been a secondary effect of IFN-I signaling which our reporter constructs were not sensitive enough to measure. The hypothetical mechanism of the activation of LTR12F-driven transcription of HERV-K102 by IFN-γ-induced IRF1 is shown in Fig. 7 (steps 1 to 3). Since the promoter of IFN-γ contains a noncanonical ISRE sequence and can be activated by IFN-I signaling (77), it is possible that HERV-K102 upregulation in response to IFN-I may be reliant on IFN-I induced IFN-γ secretion and subsequent autocrine and paracrine IFN-II signaling. However, other possibilities exist, such as the moderate activation of STAT1/STAT1 homodimers (and subsequently, IRF1 upregulation) that can occur following IFN-I signaling, albeit at significantly lower levels than those following IFN-II signaling (42).
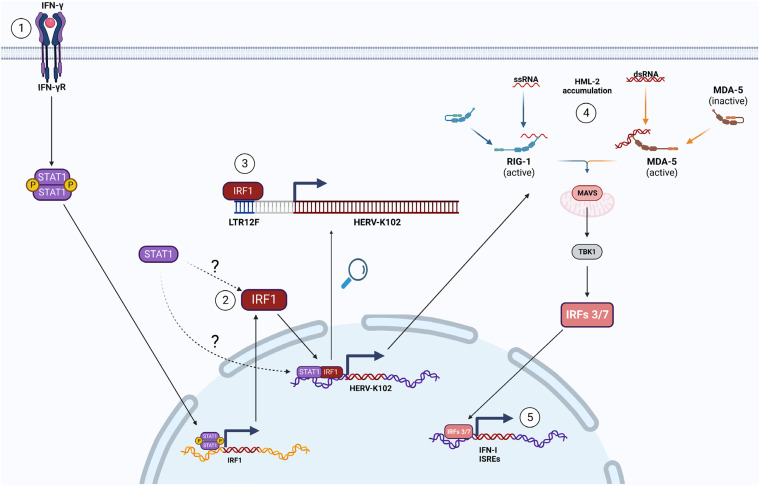
FIG 7.
Hypothetical mechanism of HERV-K102 upregulation and the phenotypic effect. Step 1: IFN-γ binds to and activates IFN-γRs, resulting in STAT1 homodimer formation and translocation into the nucleus. Step 2: STAT1 homodimers bind to the GAS site in IRF1 gene to induce rapid and strong transcription. Following translation, IRF1 translocates into the nucleus. Step 3: IRF1 (and potentially STAT1) bind to LTR12F to induce HERV-K102 upregulation. Step 4: HML-2 transcripts (including HERV-K102) accumulate in the cytoplasm. The RLRs (retinoic acid-inducible gene I-like receptors), RIG-I and MDA-5 (melanoma differentiation-associated gene 5), can respond to increases in HERV RNA, leading to MAVS signaling and subsequently, IRFs 3/7 activation and translocation into the nucleus. Step 5: IRFs 3/7 induce the expression of IFN-I and various ISRE-containing genes to enhance autocrine and paracrine pro-inflammatory signaling.
We and others have previously shown that HERV RNA interacts with the RIG-I/MDA5/MAVS pathway to induce ISRE activation and IFN-I expression in response to strong epigenetic modulators such as ionizing irradiation, UV radiation, and DNMTis (12, 78, 79). However, whether HML-2 RNA mediates ISRE activation in the basal state or in response to IFN-γ signaling is unknown. Surprisingly, we found that HML-2 knockdown significantly reduced ISRE activation in both basal and IFN-γ challenged TDMs. This effect was replicated in MAVS-KO reporter cells and predictably, the reduction in ISRE activation was stronger following MAVS-KO than HML-2 knockdown alone. Importantly, HML-2 knockdown did not have an effect on ISRE activation in MAVS-KO cells, suggesting that HML-2’s activation of ISREs may be mediated through MAVS signaling, as previously reported in response to other conditions (14, 78). We also found that HML-2 knockdown reduced the ability of TDMs to induce ISRE activation following IFN-γ in a paracrine manner, consistent with our previous report on the effect of HML-2 knockdown in response to ionizing irradiation (12). Overall, our phenotypic experiments are in agreement with previous literature on the effects of HERV upregulation and suggest that HML-2 RNA may be sensed and responded to by the RIG-I- or MDA5-MAVS pathway to induce ISRE activity and therefore enhance the type I interferon-related innate immune response (Fig. 7, steps 4 and 5).
Interestingly, a report on HML-2 expression in healthy tissues using the same retroelement analysis software used in our paper, Telescope, found that HERV-K102 is expressed in almost every tissue, suggesting that it is not strongly epigenetically silenced like other HERV loci (30). Since the receptors for IFN-I and IFN-II signaling are nearly ubiquitously expressed (47), we suspected that the IFN sensitivity of HERV-K102 may be a universal phenomenon. Indeed, in every cell type examined, HERV-K102 was significantly upregulated following IFN signaling. Following our findings, we speculated that upregulated HERV-K102 expression alone could be an indicator for the presence of elevated IFN-γ signaling. We chose to investigate this possibility in cutaneous leishmaniasis patients using a publicly available RNA-sequencing data set, provided by Farias Amorim et al. (19), as this disease is characterized by a chronic, systemic IFN-γ signature. Remarkably, HERV-K102 expression in circulating PBMCs was found to be significantly upregulated in CL patients compared to control individuals and was correlated with IRF1 expression.
Taken together, our findings indicate that HERV-K102 is an IFN-sensitive HML-2 provirus which constitutes the majority of HML-2-derived transcripts following IFN signaling. Its upregulation following IFN-γ signaling under inflammatory conditions is likely mediated by an upstream solo LTR, LTR12F, which actively recruits IRF1 to its IRF-E sites following IFN-γ signaling. HML-2 expression in macrophages, even in the basal state, influences ISRE activation and has implications on the paracrine activation of nearby cells following macrophage activation. This suggests that HERV-K102 may play the role of an intracellular enhancer of IFN-γ signaling to increase the secondary IFN-I response and therefore facilitate pro-inflammatory innate immune responses. The impact of HML-2 expression on ISRE activation is most likely mediated through the viral RNA-sensitive MAVS signaling pathway, although the exact molecular mechanism of this effect requires further experimental verification. Future studies will be required to more strongly demonstrate a phenotypic dependence on HML-2 upregulation following IFN signaling and to identify the involvement of the particular RLR sensors of cytoplasmic RNA and the MAVS-mediated pathway in IRF3/7-dependent expression of type I interferons and potentially NF-κB-dependent upregulation of pro-inflammatory modulators.
MATERIALS AND METHODS
Reagents and resources.
All reagents, cell lines, and information resources used are listed in Table S7.
Cells.
Human kidney fibroblast 293T cells were used for lentiviral particle generation. They were maintained at 37°C and 5% CO2 in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% bovine serum albumin (BSA), 1% penicillin/streptomycin, and 1% l-glutamine. The monocytic leukemia cell line THP1 was used for all other in-lab experiments. Wild-type THP1 cells were obtained from the ATCC. The reporter cell variations that measure ISRE activation through a secreted luciferase gene (THP1-Dual cells) were purchased from InvivoGen. All THP1 cells were maintained at 37°C and 5% CO2 in RPMI 1640 culture medium supplemented with 10% BSA, 1% penicillin/streptomycin, and 1% l-glutamine. For culturing of THP1-Dual cells, normocin at a final concentration of 100 μg/mL was added to the culture medium. At 48 h before an experiment, the cells were passaged in regular RPMI 1640 medium.
Design of lentiviral constructs and particle generation.
To generate HML-2-shRNA lentiviral particles, HML-2 env-targeting shRNA was designed using the Invitrogen RNAi Designer tool against a conservative region of the HML-2 subgroup in the env gene. The selected HML-2-shRNA is predicted to interact with 57 loci (Table S6), including the most highly expressed loci, 1q22 and 3q12.3. The control shRNA was also designed using the RNAi Designer tool. The puromycin resistant lentiviral backbone pLKO.1 puro was used for cloning and expressing the control and HML-2-shRNA sequences. The plasmids psPAX2 and pMD2.G were used with the cloned pLKO.1 plasmids to transfect 293T cells with Lipofectamine 3000 and generate control and HML-2-shRNA-expressing lentiviral particles.
To examine the relative promoter activity of different regions between the 5′ LTR of 1q22 and the upstream region, the DNA fragments with appropriate nucleotide sequences were synthesized by GeneScript and then cloned into the puromycin-resistant lentiviral backbone LVR-1048-pLV-Promoterless-Firefly_Luciferase-PGK-puro plasmid vector (Cellomics Technology) using different restriction enzyme digestion sites. Lentiviral particles were generated by transfecting each cloned plasmid with psPAX2 and pMD2.G in 293T cells with Lipofectamine 3000.
Lentiviral transduction of THP1 monocytes.
To transduce THP1 or THP1-Dual cells with the desired lentivirus, 2 × 106 cells were plated in a single well in a 6-well plate with an equal amount of lentiviral particles. Polybrene (Sigma) was added at a final concentration of 8 μg/mL, and spinoculation at 1,000 × g and 25°C for 2 h was performed. After an additional 2 h in the cell incubator at 37°C, the cells were given fresh medium. Two days later, positively transduced cells were selected with puromycin at a concentration of 2 μg/mL (Gibco).
Macrophage differentiation and treatment with IFN-γ.
To obtain THP1-derived macrophages, THP1 cells were seeded in a 24-well plate (0.85 × 106 cells/well) and treated with PMA at a final concentration of 40 nM for 48 h, followed by 48 h in fresh medium prior to subsequent treatment to allow the cells to recover to a neutral state (M0). The resulting TDMs were washed with Dulbecco’s phosphate-buffered saline (DPBS) and given either fresh medium alone or fresh medium containing IFN-γ at a final concentration of 25 ng/mL for 18 h. Depending on the experiment, the TDMs were collected for RNA isolation or the supernatant for downstream experiments.
Measuring direct and paracrine ISRE activation in TDMs.
To measure direct ISRE activation, supernatant was collected from untreated and IFN-γ-treated reporter TDMs, as described above. The reporter cells (THP1-Dual, WT-reporter cells; and THP1-Dual-KO-MAVS, MAVS-knockout cell line) contained a secreted luciferase gene under the control of five upstream ISREs and the promoter region of ISG54. To measure relative ISRE activation, 10 μL of culture supernatant was mixed with 40 μL of substrate and luminescence was measured according to the manufacturer’s protocol. To measure paracrine ISRE activation, culture supernatant from WT-TDMs (transduced to express control shRNA or HML-2-shRNA) was transferred to reporter THP1 cells in a 96-well plate (1 × 105/mL) at a ratio of 1:5 fresh medium to condition medium. Following 18 h, 10 μL of culture supernatant was mixed with 40 μL of substrate and luminescence was measured according to the manufacturer’s protocol.
Measuring relative promoter activity.
To measure the relative promoter activity of the 5′ LTR of HERV-K102 and the upstream region, WT-THP1 cells were transduced with each of the constructs shown in Fig. 4, and positively transduced cells were selected as described previously. Following selection, THP1 cells with each construct were plated in a 96-well plate at a concentration of 1 × 105 and treated with IFN-α (25 ng/mL), IFN-γ (25n/mL), TNF-α (20 ng/mL), LPS (100 ng/mL), or IL-4 (200 ng/mL) for 18 h. To measure luciferase activity, an equal amount of substrate solution was added to the cell suspensions according to the Bright-Glo Luciferase Assay System (Promega) protocol.
RNA isolation, cDNA preparation, and RT-qPCR.
To isolate RNA, cells were collected and washed twice with DPBS. RNA was isolated from cells using RNeasy spin columns (Qiagen) followed by on-column DNase digestion according to the manufacturer’s protocol. Approximately 1,500 ng of RNA was used for cDNA preparation with the High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific), 10 U of RNase Inhibitor (Applied Biosystems), and oligo(dT) primer. Quantitative real-time PCR was performed with the SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) using primers designed for specific genes of interest (primer sequences are provided in Table S7). The optimal PCR program was as follows: 95°C for 3 min, 95°C for 10 sec, and 60°C for 40 sec, running for 41 cycles. Real-time PCRs were carried out at least in triplicate. Relative gene expression was determined by the threshold cycle (ΔΔCT) ratio method using Bio-Rad CFX Manager 3.1 software. The fold-change in gene expression in all experiments was calculated relative to GUSB and GAPDH as the reference genes (80).
RNA-sequencing data acquisition and gene/retroelement expression analysis.
Publicly available raw or pre-processed FASTQ files were downloaded from the NCBI Gene Expression Omnibus repository using the ‘prefetch’ command from the NCBI SRA Toolkit as .sra files and converted into FASTQ files with the ‘fasterq-dump’ command. FASTQ files were adaptor-clipped and quality-trimmed with Trimmomatic. For paired-end sequencing data sets, only paired reads were kept for downstream analysis. The standard Tecount pipeline was followed to map reads to the genome using GRCh38.103 and count the number of mapped reads to normal genes and retroelements (at the family level) (20). Additionally, the standard Telescope pipeline was followed to map reads to the genome and count the number of mapped reads to retroelements (at the locus-specific level) (22). Read counts were imported into R and analyzed with edgeR to normalize the raw data and to determine the CPM, RPKM, and fold change (when applicable) (81). The normalized and raw gene expression data are shown in Supplemental File 2.
ChIP and ATAC sequencing data analysis.
Publicly available raw or pre-processed FASTQ files were downloaded from the NCBI Gene Expression Omnibus repository using the ‘prefetch’ command from the NCBI SRA Toolkit as .sra files and converted into FASTQ files with the ‘fasterq-dump’ command. FASTQ files were adaptor-clipped and quality-trimmed with Trimmomatic. For paired-end sequencing data sets, only paired reads were kept for downstream analysis. Reads were mapped to the human reference genome (GRCh38/hg38) using Bowtie2 with the default parameters. Reads were filtered to remove low-quality reads and reads that mapped to multiple locations (reads with an alignment quality score of <10) were dropped. Bamcoverage was used to create RPKM-normalized bigwig files that were subsequently input into the IGV tool for visualization of the data. MACS2 was used for peak detection, fold-enrichment, and P value calculations.
Quantification and statistical analysis.
The details of the statistical analysis of experiments, including statistical tests used and number of replicates, are provided in the figure legends. Statistical measurements and plotting were performed using GraphPad Prism v9.4.1 software. All values in this study represent means of at least three biological replicates ± standard error of the mean. Two-tailed paired t tests and two-way analysis of variance were used to compare differences between two groups and multiple groups, respectively. Real-time PCR data were quantified and analyzed using Bio-Rad CFX Manager v3.1. The option of Gene Study analysis was performed to assess gene expression data.
ACKNOWLEDGMENTS
We thank the members of the Uniformed Services University of the Health Sciences (USUHS) Department of Pharmacology, Regina Day, Andrew Snow, Brian Cox, and Robert Kortum for productive discussions and helping with reagents and equipment. We are grateful to Clifton Dalgard from the USUHS Department of Anatomy, Physiology and Genetics, and Avindra Nath, Kory Johnson, and Wenxue Li from the National Institute of Neurological Disorders and Stroke, for valuable advice, comments, and help with materials and experimental protocols.
This work was supported in part by start-up funds from the USUHS (to S.I.) and grants from the Congressionally Directed Medical Research Program (JPC-7 2018 BA1-1 to S.I.) and the Armed Forces Radiobiology Research Institute (AFR-11611 to S.I.). E.R. is a recipient of The American Genome Research Center (TAGS) fellowship.
We declare no conflicts of interest. The founding sponsors had no role in the design of the study; the collection, analysis, or interpretation of data; the writing of the manuscript; or the decision to publish the results.
The opinions expressed herein are those of the authors and are not necessarily representative of those of the Uniformed Services University of the Health Sciences, the Department of Defense (DOD), or the United States Army, Navy, or Air Force.
Footnotes
Supplemental material is available online only.
Contributor Information
Sergey Iordanskiy, Email: sergey.iordanskiy@usuhs.edu.
Holly Ramage, Thomas Jefferson University.
REFERENCES
- 1.Bannert N, Kurth R. 2004. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA 101:14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xue B, Zeng T, Jia L, Yang D, Lin SL, Sechi LA, Kelvin DJ. 2020. Identification of the distribution of human endogenous retroviruses K (HML-2) by PCR-based target enrichment sequencing. Retrovirology 17:10. doi: 10.1186/s12977-020-00519-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azebi S, Batsche E, Michel F, Kornobis E, Muchardt C. 2019. Expression of endogenous retroviruses reflects increased usage of atypical enhancers in T cells. EMBO J 38:e101107. doi: 10.15252/embj.2018101107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goering W, Schmitt K, Dostert M, Schaal H, Deenen R, Mayer J, Schulz WA. 2015. Human endogenous retrovirus HERV-K(HML-2) activity in prostate cancer is dominated by a few loci. Prostate 75:1958–1971. doi: 10.1002/pros.23095. [DOI] [PubMed] [Google Scholar]
- 5.Sicat J, Sutkowski N, Huber BT. 2005. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol 32:1821–1831. [PubMed] [Google Scholar]
- 6.Garcia-Montojo M, Simula ER, Fathi S, McMahan C, Ghosal A, Berry JD, Cudkowicz M, Elkahloun A, Johnson K, Norato G, Jensen P, James T, Sechi LA, Nath A. 2022. Antibody response to HML-2 may be protective in amyotrophic lateral sclerosis. Ann Neurol 92:782–792. doi: 10.1002/ana.26466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Büscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. 2005. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res 65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 8.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, Han H, Liang G, Jones PA, Pugh TJ, O’Brien C, De Carvalho DD. 2015. DNA-demethylating agents target colorectal cancer cells by inducing viral mimicry by endogenous transcripts. Cell 162:961–973. doi: 10.1016/j.cell.2015.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rehwinkel J, Gack MU. 2020. RIG-I-like receptors: their regulation and roles in RNA sensing. Nat Rev Immunol 20:537–551. doi: 10.1038/s41577-020-0288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Olagnier D, Lin R. 2016. Host and Viral Modulation of RIG-I-Mediated Antiviral Immunity. Front Immunol 7:662. doi: 10.3389/fimmu.2016.00662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gack MU. 2014. Mechanisms of RIG-I-like receptor activation and manipulation by viral pathogens. J Virol 88:5213–5216. doi: 10.1128/JVI.03370-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikhalkevich N, O’Carroll IP, Tkavc R, Lund K, Sukumar G, Dalgard CL, Johnson KR, Li W, Wang T, Nath A, Iordanskiy S. 2021. Response of human macrophages to gamma radiation is mediated via expression of endogenous retroviruses. PLoS Pathog 17:e1009305. doi: 10.1371/journal.ppat.1009305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petrizzo A, Ragone C, Cavalluzzo B, Mauriello A, Manolio C, Tagliamonte M, Buonaguro L. 2021. Human endogenous retrovirus reactivation: implications for cancer immunotherapy. Cancers (Basel) 13:1999. doi: 10.3390/cancers13091999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee AK, Pan D, Bao X, Hu M, Li F, Li CY. 2020. Endogenous retrovirus activation as a key mechanism of anti-tumor immune response in radiotherapy. Radiat Res 193:305–317. doi: 10.1667/RADE-20-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu M, Thomas SL, DeWitt AK, Zhou W, Madaj ZB, Ohtani H, Baylin SB, Liang G, Jones PA. 2018. Dual inhibition of DNA and histone methyltransferases increases viral mimicry in ovarian cancer cells. Cancer Res 78:5754–5766. doi: 10.1158/0008-5472.CAN-17-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arango Duque G, Descoteaux A. 2014. Macrophage cytokines: involvement in immunity and infectious diseases. Front Immunol 5:491. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon S, Pluddemann A, Martinez Estrada F. 2014. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev 262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosser DM, Hamidzadeh K, Goncalves R. 2021. Macrophages and the maintenance of homeostasis. Cell Mol Immunol 18:579–587. doi: 10.1038/s41423-020-00541-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Farias Amorim C, O Novais F, Nguyen BT, Nascimento MT, Lago J, Lago AS, Carvalho LP, Beiting DP, Scott P. 2021. Localized skin inflammation during cutaneous leishmaniasis drives a chronic, systemic IFN-gamma signature. PLoS Negl Trop Dis 15:e0009321. doi: 10.1371/journal.pntd.0009321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin Y, Tam OH, Paniagua E, Hammell M. 2015. TEtranscripts: a package for including transposable elements in differential expression analysis of RNA-seq datasets. Bioinformatics 31:3593–3599. doi: 10.1093/bioinformatics/btv422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jin Y, Hammell M. 2018. Analysis of RNA-seq data using TEtranscripts. Methods Mol Biol 1751:153–167. doi: 10.1007/978-1-4939-7710-9_11. [DOI] [PubMed] [Google Scholar]
- 22.Bendall ML, de Mulder M, Iniguez LP, Lecanda-Sanchez A, Perez-Losada M, Ostrowski MA, Jones RB, Mulder LCF, Reyes-Teran G, Crandall KA, Ormsby CE, Nixon DF. 2019. Telescope: characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput Biol 15:e1006453. doi: 10.1371/journal.pcbi.1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoppstadter J, Dembek A, Horing M, Schymik HS, Dahlem C, Sultan A, Wirth N, Al-Fityan S, Diesel B, Gasparoni G, Walter J, Helms V, Huwer H, Simon M, Liebisch G, Schulz MH, Kiemer AK. 2021. Dysregulation of cholesterol homeostasis in human lung cancer tissue and tumour-associated macrophages. EBioMedicine 72:103578. doi: 10.1016/j.ebiom.2021.103578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holloway JR, Williams ZH, Freeman MM, Bulow U, Coffin JM. 2019. Gorillas have been infected with the HERV-K (HML-2) endogenous retrovirus much more recently than humans and chimpanzees. Proc Natl Acad Sci USA 116:1337–1346. doi: 10.1073/pnas.1814203116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Goering W, Ribarska T, Schulz WA. 2011. Selective changes of retroelement expression in human prostate cancer. Carcinogenesis 32:1484–1492. doi: 10.1093/carcin/bgr181. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt K, Reichrath J, Roesch A, Meese E, Mayer J. 2013. Transcriptional profiling of human endogenous retrovirus group HERV-K(HML-2) loci in melanoma. Genome Biol Evol 5:307–328. doi: 10.1093/gbe/evt010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grabski DF, Ratan A, Gray LR, Bekiranov S, Rekosh D, Hammarskjold ML, Rasmussen SK. 2021. Upregulation of human endogenous retrovirus-K (HML-2) mRNAs in hepatoblastoma: identification of potential new immunotherapeutic targets and biomarkers. J Pediatr Surg 56:286–292. doi: 10.1016/j.jpedsurg.2020.05.022. [DOI] [PubMed] [Google Scholar]
- 28.Tokuyama M, Gunn BM, Venkataraman A, Kong Y, Kang I, Rakib T, Townsend MJ, Costenbader KH, Alter G, Iwasaki A. 2021. Antibodies against human endogenous retrovirus K102 envelope activate neutrophils in systemic lupus erythematosus. J Exp Med 218:e20191766. doi: 10.1084/jem.20191766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian W, Zhao M, Wang R, Li H. 2021. Fibrinogen-like protein 1 (FGL1): the next immune checkpoint target. J Hematol Oncol 14:147. doi: 10.1186/s13045-021-01161-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burn A, Roy F, Freeman M, Coffin JM. 2022. Widespread expression of the ancient HERV-K (HML-2) provirus group in normal human tissues. PLoS Biol 20:e3001826. doi: 10.1371/journal.pbio.3001826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsang JS, Schwartzberg PL, Kotliarov Y, Biancotto A, Xie Z, Germain RN, Wang E, Olnes MJ, Narayanan M, Golding H, Moir S, Dickler HB, Perl S, Cheung F, Baylor HC, Baylor HIPC Center, CHI Consortium . 2014. Global analyses of human immune variation reveal baseline predictors of postvaccination responses. Cell 157:499–513. doi: 10.1016/j.cell.2014.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buscher K, Ehinger E, Gupta P, Pramod AB, Wolf D, Tweet G, Pan C, Mills CD, Lusis AJ, Ley K. 2017. Natural variation of macrophage activation as disease-relevant phenotype predictive of inflammation and cancer survival. Nat Commun 8:16041. doi: 10.1038/ncomms16041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wynn TA, Chawla A, Pollard JW. 2013. Macrophage biology in development, homeostasis and disease. Nature 496:445–455. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu M, Wang X, Li Y, Geng X, Jia X, Zhang L, Yang H. 2021. Arachidonic acid metabolism controls macrophage alternative activation through regulating oxidative phosphorylation in PPARγ dependent manner. Front Immunol 12:618501. doi: 10.3389/fimmu.2021.618501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Basheeruddin K, Rechtoris C, Mazzone T. 1994. Evaluation of the role of Ap1-like proteins in the enhanced apolipoprotein E gene transcription accompanying phorbol ester induced macrophage differentiation. Biochim Biophys Acta 1218:235–241. doi: 10.1016/0167-4781(94)90021-3. [DOI] [PubMed] [Google Scholar]
- 36.Schwende H, Fitzke E, Ambs P, Dieter P. 1996. Differences in the state of differentiation of THP-1 cells induced by phorbol ester and 1,25-dihydroxyvitamin D3. J Leukoc Biol 59:555–561. doi: 10.1002/jlb.59.4.555. [DOI] [PubMed] [Google Scholar]
- 37.Barilli A, Rotoli BM, Visigalli R, Bussolati O, Gazzola GC, Dall’Asta V. 2011. Arginine transport in human monocytic leukemia THP-1 cells during macrophage differentiation. J Leukoc Biol 90:293–303. doi: 10.1189/jlb.0910510. [DOI] [PubMed] [Google Scholar]
- 38.Palsson-McDermott EM, O'Neill LA. 2004. Signal transduction by the lipopolysaccharide receptor, Toll-like receptor-4. Immunology 113:153–162. doi: 10.1111/j.1365-2567.2004.01976.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bhat MY, Solanki HS, Advani J, Khan AA, Keshava Prasad TS, Gowda H, Thiyagarajan S, Chatterjee A. 2018. Comprehensive network map of interferon gamma signaling. J Cell Commun Signal 12:745–751. doi: 10.1007/s12079-018-0486-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song R, Gao Y, Dozmorov I, Malladi V, Saha I, McDaniel MM, Parameswaran S, Liang C, Arana C, Zhang B, Wakeland B, Zhou J, Weirauch MT, Kottyan LC, Wakeland EK, Pasare C. 2021. IRF1 governs the differential interferon-stimulated gene responses in human monocytes and macrophages by regulating chromatin accessibility. Cell Rep 34:108891. doi: 10.1016/j.celrep.2021.108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schynkel T, Szaniawski MA, Spivak AM, Bosque A, Planelles V, Vandekerckhove L, Trypsteen W. 2020. Interferon-mediated long non-coding RNA response in macrophages in the context of HIV. Int J Mol Sci 21:7741. doi: 10.3390/ijms21207741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Platanias LC. 2005. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol 5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 43.Schroder K, Hertzog PJ, Ravasi T, Hume DA. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J Leukoc Biol 75:163–189. doi: 10.1189/jlb.0603252. [DOI] [PubMed] [Google Scholar]
- 44.Constant DA, Van Winkle JA, VanderHoek E, Dekker SE, Sofia MA, Regner E, Modiano N, Tsikitis VL, Nice TJ. 2022. Transcriptional and cytotoxic responses of human intestinal organoids to IFN types I, II, and III. Immunohorizons 6:416–429. doi: 10.4049/immunohorizons.2200025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bonser LR, Eckalbar WL, Rodriguez L, Shen J, Koh KD, Zlock LT, Christenson S, Woodruff PG, Finkbeiner WE, Erle DJ. 2021. The type 2 asthma mediator IL-13 inhibits SARS-CoV-2 infection of bronchial epithelium. Am J Respir Cell Mol Biol 66:391–401. doi: 10.1165/rcmb.2021-0364OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siwek W, Tehrani SSH, Mata JF, Jansen LET. 2020. Activation of clustered IFN-γ target genes drives cohesin-controlled transcriptional memory. Mol Cell 80:396–409.e6. doi: 10.1016/j.molcel.2020.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.de Weerd NA, Nguyen T. 2012. The interferons and their receptors: distribution and regulation. Immunol Cell Biol 90:483–491. doi: 10.1038/icb.2012.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laderoute MP, Giulivi A, Larocque L, Bellfoy D, Hou Y, Wu HX, Fowke K, Wu J, Diaz-Mitoma F. 2007. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS 21:2417–2424. doi: 10.1097/QAD.0b013e3282f14d64. [DOI] [PubMed] [Google Scholar]
- 49.Khadjinova AI, Wang X, Laine A, Ukadike K, Eckert M, Stevens A, Bengtsson AA, Lood C, Mustelin T. 2022. Autoantibodies against the envelope proteins of endogenous retroviruses K102 and K108 in patients with systemic lupus erythematosus correlate with active disease. Clin Exp Rheumatol 40:1306–1312. doi: 10.55563/clinexprheumatol/2kg1d8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decker T, Kovarik P, Meinke A. 1997. GAS elements: a few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res 17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 51.Salkowski CA, Kopydlowski K, Blanco J, Cody MJ, McNally R, Vogel SN. 1999. IL-12 is dysregulated in macrophages from IRF-1 and IRF-2 knockout mice. J Immunol 163:1529–1536. doi: 10.4049/jimmunol.163.3.1529. [DOI] [PubMed] [Google Scholar]
- 52.Kano S, Sato K, Morishita Y, Vollstedt S, Kim S, Bishop K, Honda K, Kubo M, Taniguchi T. 2008. The contribution of transcription factor IRF1 to the interferon-gamma–interleukin 12 signaling axis and TH1 versus TH-17 differentiation of CD4+ T cells. Nat Immunol 9:34–41. doi: 10.1038/ni1538. [DOI] [PubMed] [Google Scholar]
- 53.Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, Takayanagi H, Ohba Y, Taniguchi T, Honda K. 2006. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in Toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA 103:15136–15141. doi: 10.1073/pnas.0607181103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiao Y, Giannopoulou EG, Chan CH, Park SH, Gong S, Chen J, Hu X, Elemento O, Ivashkiv LB. 2013. Synergistic activation of inflammatory cytokine genes by interferon-γ-induced chromatin remodeling and Toll-like receptor signaling. Immunity 39:454–469. doi: 10.1016/j.immuni.2013.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. 1999. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J 18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rettino A, Clarke NM. 2013. Genome-wide identification of IRF1 binding sites reveals extensive occupancy at cell death associated genes. J Carcinog Mutagen doi: 10.4172/2157-2518.S6-009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chatterjee-Kishore M, Wright KL, Ting JP, Stark GR. 2000. How Stat1 mediates constitutive gene expression: a complex of unphosphorylated Stat1 and IRF1 supports transcription of the LMP2 gene. EMBO J 19:4111–4122. doi: 10.1093/emboj/19.15.4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Buecker C, Wysocka J. 2012. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet 28:276–284. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci USA 107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yan F, Powell DR, Curtis DJ, Wong NC. 2020. From reads to insight: a hitchhiker’s guide to ATAC-seq data analysis. Genome Biol 21:22. doi: 10.1186/s13059-020-1929-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. 2013. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Starks RR, Biswas A, Jain A, Tuteja G. 2019. Combined analysis of dissimilar promoter accessibility and gene expression profiles identifies tissue-specific genes and actively repressed networks. Epigenetics Chromatin 12:16. doi: 10.1186/s13072-019-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Orford K, Kharchenko P, Lai W, Dao MC, Worhunsky DJ, Ferro A, Janzen V, Park PJ, Scadden DT. 2008. Differential H3K4 methylation identifies developmentally poised hematopoietic genes. Dev Cell 14:798–809. doi: 10.1016/j.devcel.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhou VW, Goren A, Bernstein BE. 2011. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet 12:7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 65.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV, Feng Z, Punt N, Daigle A, Bullinger L, Pollock RM, Richon VM, Kung AL, Armstrong SA. 2011. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 20:66–78. doi: 10.1016/j.ccr.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schubeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O’Neill LP, Turner BM, Delrow J, Bell SP, Groudine M. 2004. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev 18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kouskouti A, Talianidis I. 2005. Histone modifications defining active genes persist after transcriptional and mitotic inactivation. EMBO J 24:347–357. doi: 10.1038/sj.emboj.7600516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ooga M, Inoue A, Kageyama S, Akiyama T, Nagata M, Aoki F. 2008. Changes in H3K79 methylation during preimplantation development in mice. Biol Reprod 78:413–424. doi: 10.1095/biolreprod.107.063453. [DOI] [PubMed] [Google Scholar]
- 69.Veloso A, Kirkconnell KS, Magnuson B, Biewen B, Paulsen MT, Wilson TE, Ljungman M. 2014. Rate of elongation by RNA polymerase II is associated with specific gene features and epigenetic modifications. Genome Res 24:896–905. doi: 10.1101/gr.171405.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou F, Li M, Wei Y, Lin K, Lu Y, Shen J, Johanning GL, Wang-Johanning F. 2016. Activation of HERV-K Env protein is essential for tumorigenesis and metastasis of breast cancer cells. Oncotarget 7:84093–84117. doi: 10.18632/oncotarget.11455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bender S, Reuter A, Eberle F, Einhorn E, Binder M, Bartenschlager R. 2015. Activation of type I and III interferon response by mitochondrial and peroxisomal MAVS and inhibition by hepatitis C virus. PLoS Pathog 11:e1005264. doi: 10.1371/journal.ppat.1005264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Agoni L, Guha C, Lenz J. 2013. Detection of human endogenous retrovirus K (HERV-K) transcripts in human prostate cancer cell lines. Front Oncol 3:180. doi: 10.3389/fonc.2013.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brinzevich D, Young GR, Sebra R, Ayllon J, Maio SM, Deikus G, Chen BK, Fernandez-Sesma A, Simon V, Mulder LC. 2014. HIV-1 interacts with human endogenous retrovirus K (HML-2) envelopes derived from human primary lymphocytes. J Virol 88:6213–6223. doi: 10.3389/fonc.2021.658489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amarasinghe SL, Su S, Dong X, Zappia L, Ritchie ME, Gouil Q. 2020. Opportunities and challenges in long-read sequencing data analysis. Genome Biol 21:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dervan E, Bhattacharyya DD, McAuliffe JD, Khan FH, Glynn SA. 2021. Ancient adversary: HERV-K (HML-2) in cancer. Front Oncol 11:658489. doi: 10.3389/fonc.2021.658489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Montesion M, Williams ZH, Subramanian RP, Kuperwasser C, Coffin JM. 2018. Promoter expression of HERV-K (HML-2) provirus-derived sequences is related to LTR sequence variation and polymorphic transcription factor binding sites. Retrovirology 15:57. doi: 10.1186/s12977-018-0441-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Castro F, Cardoso AP, Goncalves RM, Serre K, Oliveira MJ. 2018. Interferon-gamma at the crossroads of tumor immune surveillance or evasion. Front Immunol 9:847. doi: 10.3389/fimmu.2018.00847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, Hein A, Rote NS, Cope LM, Snyder A, Makarov V, Budhu S, Slamon DJ, Wolchok JD, Pardoll DM, Beckmann MW, Zahnow CA, Merghoub T, Chan TA, Baylin SB, Strick R. 2017. Inhibiting DNA methylation causes an interferon response in cancer via dsRNA including endogenous retroviruses. Cell 169:361. doi: 10.1016/j.cell.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 79.Min X, Zheng M, Yu Y, Wu J, Kuang Q, Hu Z, Ouyang L, Lu S, Zhao M. 2022. Ultraviolet light induces HERV expression to activate RIG-I signalling pathway in keratinocytes. Exp Dermatol 31:1165–1176. doi: 10.1111/exd.14568. [DOI] [PubMed] [Google Scholar]
- 80.Zampieri M, Ciccarone F, Guastafierro T, Bacalini MG, Calabrese R, Moreno-Villanueva M, Reale A, Chevanne M, Burkle A, Caiafa P. 2010. Validation of suitable internal control genes for expression studies in aging. Mech Ageing Dev 131:89–95. doi: 10.1016/j.mad.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 81.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download spectrum.04438-22-s0001.pdf, PDF file, 0.5 MB (463.2KB, pdf)
Supplemental material. Download spectrum.04438-22-s0002.xlsx, XLSX file, 0.1 MB (126.3KB, xlsx)