ABSTRACT
India is one of the largest consumers and producers of antibiotics and a hot spot for the emergence and proliferation of antimicrobial resistance genes (ARGs). Indian hospital wastewater (HWW) accumulates ARGs from source hospitals and often merges with urban wastewater, with the potential for environmental and human contamination. Despite its putative clinical importance, there is a lack of high-resolution resistome profiling of Indian hospital wastewater, with most studies either relying on conventional PCR-biased techniques or being limited to one city. In this study, we comprehensively analyzed antibiotic resistomes of wastewater from six Indian hospitals distributed in rural and urban areas of northern India through shotgun metagenomics. Our study revealed the predominance of ARGs against aminoglycoside, macrolide, carbapenem, trimethoprim, and sulfonamide antibiotics in all the samples through both read-based analysis and assembly-based analysis. We detected the mobile colistin resistance gene mcr-5.1 for the first time in Indian hospital sewage. blaNDM-1 was present in 4 out of 6 samples and was carried by Pseudomonas aeruginosa in HWW-2, Klebsiella pneumoniae in HWW-4 and HWW-6, and Acinetobacter baumanii in HWW-5. Most ARGs were plasmid-mediated and hosted by Proteobacteria. We identified virulence factors and transposable elements flanking the ARGs, highlighting the role of horizontal gene transmission of ARGs.
IMPORTANCE There is a paucity of research on detailed antibiotic resistome and microbiome diversity of Indian hospital wastewater. This study reports the predominance of clinically concerning ARGs such as the beta-lactamases blaNDM and blaOXA and the colistin resistance gene mcr and their association with the microbiome in six different Indian hospital wastewaters of both urban and rural origin. The abundance of plasmid-mediated ARGs and virulence factors calls for urgent AMR crisis management. The lack of proper wastewater management strategies meeting international standards and open drainage systems further complicates the problem of containing the ARGs at these hospitals. This metagenomic study presents the current AMR profile propagating in hospital settings in India and can be used as a reference for future surveillance and risk management of ARGs in Indian hospitals.
KEYWORDS: antimicrobial resistance, metagenomics, resistant markers, waste water
INTRODUCTION
Antimicrobial resistance (AMR) is a global public health emergency. In 2019, an estimated 5,000,000 deaths were associated with bacterial AMR, exerting the highest burdens in low- and middle-income countries (LMIC) (1). Exacerbating the problem, a rise in AMR has been accompanied by a reduction in the number of new antibiotics approved for human use (2, 3). The World Health Organization’s “One Health” approach emphasizes the close association of human, animal, and environmental health (4). Reliable and accurate surveillance is critical for characterizing the risk of AMR in a given region, tracking the spread of specific antibiotic resistance genes (ARG) geographically and over time, identifying new ARGs, and supporting preventative measures and interventions against multidrug-resistant (MDR) pathogens.
Hospitals are important reservoirs and vectors of AMR, where the frequent and persistent use of antimicrobials selects for MDR pathogens that can cause hospital-acquired infections (5, 6). Hospital wastewater (HWW) receives liquid medical waste, and the excrement of patients, visitors, and health care professionals, and as such, contains hazardous chemicals, pharmaceutical residues, and human pathogens (7). In turn, it contains a greater diversity and abundance of ARGs than other wastewater systems (8). Additionally, in many settings, particularly in LMICs, HWW is released into municipal sewage systems without treatment (7). This, coupled with open drainage systems and inefficient wastewater treatment plants, can lead to the dissemination of HWW contents, including antimicrobial residues and MDR pathogens, into the local environment and community (9). Metagenomic surveillance of HWW using short-read next-generation sequencing data is an efficient approach for assessing the overall AMR burden in a given hospital (10–12). Unlike traditional culture- and PCR-based approaches, which are laborious and limited to selected taxa and ARGs, metagenomics can quantify thousands of species, ARGs, and virulence factors (VFs) from a single sample at a relatively low cost. Further, surveillance of sewage instead of samples from human patients has the additional advantage of being easily obtained and analyzed without ethical review. However, current research on AMR in HWW has mainly focused on high-income countries, with limited data reported for LMICs (13).
India is one of the largest consumers and producers of antibiotics and is a hot spot for AMR (14). Antibiotic usage in hospitals in Asia is comparatively higher than in European hospitals (15). A study based on data on patient-level antibiotic consumption from 209 surveys and 284,045 children (aged <5 years), collected over 19 years and covering 101 countries, reported a dramatic surge in the median national antibiotic usage in India from 48% in the year 2000 to 67% in 2018. The lack of awareness about judicious antibiotic usage and the detrimental consequences of “over the counter” medication has caused a notable increase in the consumption of fluoroquinolone and third-generation cephalosporin in India from 2000 to 2018 (16). According to the WHO, by the year 2023, 60% of the total antibiotic consumption in a country should be constituted by WHO-Access group antibiotics, the antibiotics that are efficacious against commonly encountered susceptible pathogens, and there are low chances of resistance development against these antibiotics. In 2015, only 30% of the total antibiotic consumption was covered by the WHO-Access group of antibiotics in India (17). Several factors favor the spread of AMR in Indian hospitals such as over-the-counter drug availability, low doctor-to-patient and nurse-to-patient ratios, antibiotics prescription by informal health care providers, and lack of infection prevention and control guidelines (18–20). India is also the hub for the manufacturing and distribution of generic antibiotics for global use, and this industrial-scale manufacturing and its associated waste likely further contribute to the development and spread of antimicrobial resistance (21). The high usage of antimicrobials in Indian hospitals undoubtedly selects for AMR. However, there are limited data on the ARG burden in Indian HWW, with previous reports limited to select ARGs and culturable bacteria and/or to a single city (22, 23).
In this study, we use shotgun metagenome sequencing to characterize the microbiomes and resistomes of HWW from six hospitals in five cities across northern India. To the best of our knowledge, this is the first study from India, comprised of samples from rural and urban hospitals, all of which were equipped with bed facilities and admitted severe infection cases. Our findings suggest that the diversity and abundance of AMR in Indian hospitals are greater than previously thought, which poses an immediate potential risk to patients and the surrounding communities.
RESULTS AND DISCUSSION
Hospital wastewater sample collection.
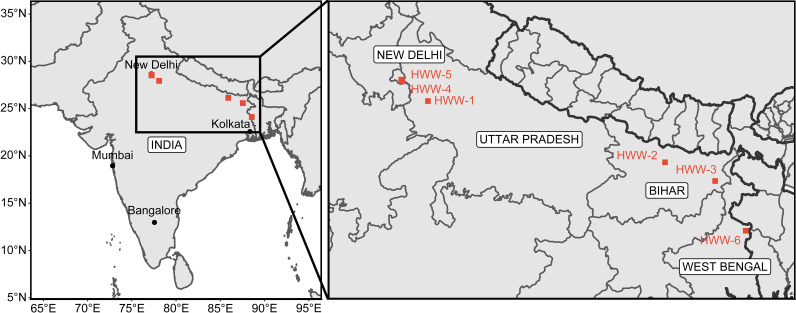
Wastewater samples were collected from six tertiary care hospitals in regions of northern India (Fig. 1), with a range of 300 to 2,400 beds (Table 1). The socioeconomic status of the surrounding population also varied at each site. Sample HWW-1 was collected from Jawaharlal Nehru Medical College and Hospital (J.N.M.C.H.), Aligarh, Uttar Pradesh; HWW-2 was from Darbhanga Medical College and Hospital (D.M.C.H.), Darbhanga, Bihar; HWW-3 was from Katihar Medical College and Hospital (K.M.C.H.), Katihar, Bihar; HWW-4 was from Hamdard Institute of Medical Sciences and Research (H.I.M.S.R.), New Delhi; HWW-5 was from All India Institute of Medical Sciences (A.I.I.M.S.), New Delhi; and HWW-6 was from Domkal Super specialty and subdivisional Hospital, West Bengal. All the hospitals except H.I.M.S.R. are government hospitals with cost-effective treatment. J.N.M.C.H. and D.M.C.H. are located in cities, H.I.M.S.R. and A.I.I.M.S are located in a metropolitan city, whereas K.M.C.H. and D.S.S.H. are located in rural areas. The socio-economic data of patients visiting these hospitals were not retrieved, but according to the 2011 census, the slum population in Aligarh and Darbhanga is approximately 30% and 16%, respectively (24). The population living in villages comprises 91% and 80% population of Katihar and Murshidabad, respectively (24). New Delhi has a predominantly urban population (24). An inexpensive treatment and easier access to these hospitals facilitate a high influx of low-income patients at these hospitals.
FIG 1.
Sampling of hospital wastewater across northern India. HWW-1, HWW-2, HWW-4, and HWW-5 are located in urban areas whereas HWW-3 and HWW-6 are rural hospitals. Due to cost-effective treatment and easier access to HWW-1, HWW-2, HWW-3, and HWW-6, the maximum patient load in these hospitals is from rural areas with below poverty line patients. At HWW-4, both rural and urban populations from rich as well as poor backgrounds come for treatment. HWW-5 has more patients from urban and affluent areas.
TABLE 1.
The information about six hospital wastewater samples collected and analyzed for this study
| Sample collection details | HWW-1 | HWW-2 | HWW-3 | HWW-4 | HWW-5 | HWW-6 |
|---|---|---|---|---|---|---|
| Site of collection | J.N.M.C.H, Aligarh | D.M.C.H., Darbhanga | K.M.C.H., Katihar | H.I.M.S.E.R. | A.I.I.M.S. | Domkal Super Speciality and Subdivisional Hospital |
| State/union territory | Uttar Pradesh | Bihar | Bihar (bordering West Bengal) | New Delhi | New Delhi | West Bengal |
| No. of beds available | 2,400 | 1,050 | 590 | 650 | 710 | 300 |
| Collection date and time | 21 Dec 2019 at 11 a.m. Indian Standard Time (IST). | 10 Jan 2020 at 4 p.m. Indian Standard Time (IST). | 19 Jan 2020 at 2 p.m. Indian Standard Time (IST). | 29 Jan 2020 at 12:40 p.m. Indian Standard Time (IST). | 10 March 2021 at 10:50 a.m. Indian Standard Time (IST). | 21 March 2021 at 5.30 a.m. Indian Standard Time (IST). |
Wastewater samples were collected from the main hospital sewage water pipeline, which receives the effluent of the entire hospital. Samples were processed and submitted for shotgun metagenomic sequencing (Table 2).
TABLE 2.
The classification of reads by Kraken2, visualized in Pavian v1.0
| Sample | No. of raw reads | Classified reads | Unclassified reads | Chordate reads | Artificial reads | Microbial reads | Bacterial reads | Viral reads | Fungal reads | Protozoan reads |
|---|---|---|---|---|---|---|---|---|---|---|
| HWW-1 | 2,67,48,085 | 44.56% | 55.44% | 4.99% | 0.00% | 39.34% | 33.29% | 0.19% | 0.78% | 0.22% |
| HWW-2 | 2,17,73,844 | 49.79% | 50.21% | 13.19% | 0.01% | 29.86% | 22.09% | 0.14% | 0.94% | 0.31% |
| HWW-3 | 2,21,34,911 | 47.24% | 52.76% | 8.93% | 0.00% | 38.08% | 7.53% | 0.25% | 1.77% | 0.77% |
| HWW-4 | 3,72,83,617 | 69.08% | 30.92% | 3.59% | 0.00% | 65.07% | 60% | 0.22% | 0.58% | 0.16% |
| HWW-5 | 2,03,38,684 | 59.81% | 40.19% | 4.38% | 0.00% | 55.24% | 49.83% | 0.12% | 0.77% | 0.15% |
| HWW-6 | 1,97,81,363 | 56.56% | 43.44% | 4.73% | 0.00% | 51.62% | 45.44% | 0.21% | 0.80% | 0.42% |
| Average | 2,46,76,751 |
Dominance of Proteobacteria in HWW microbiomes.
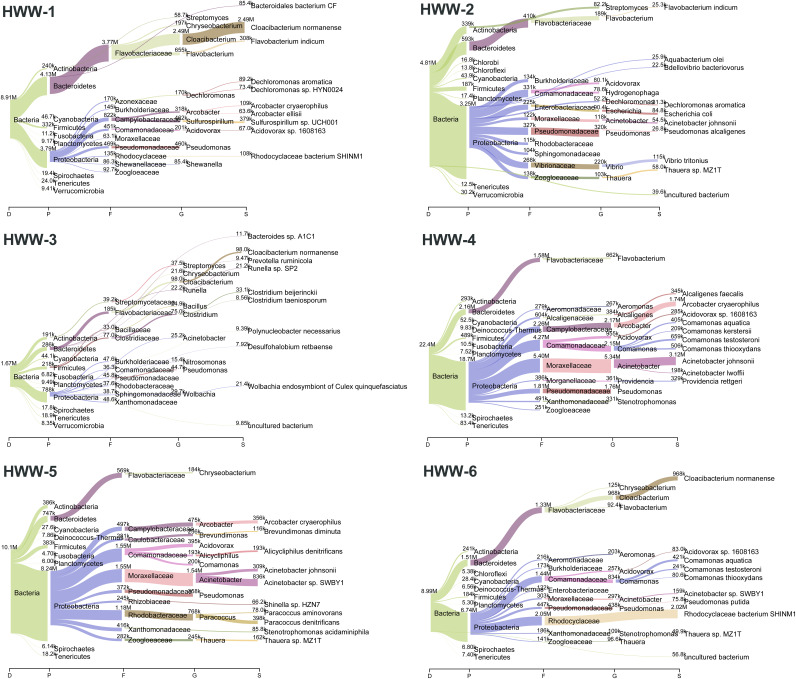
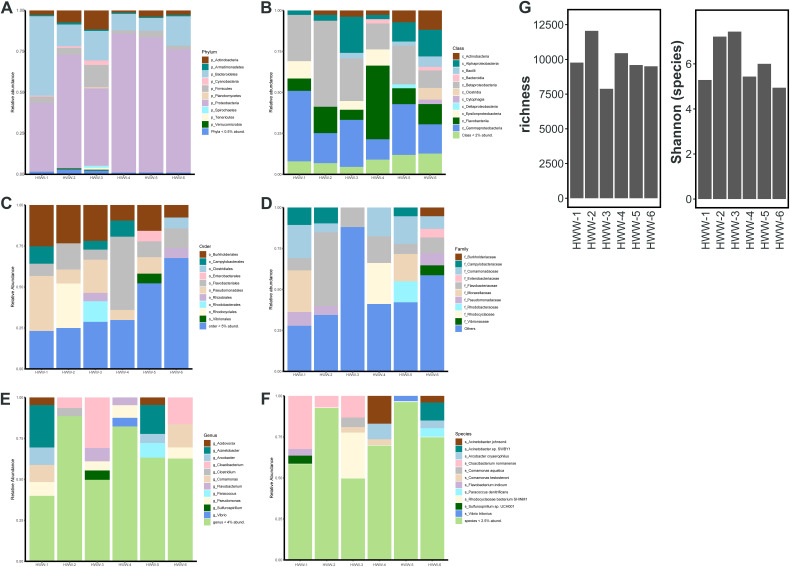
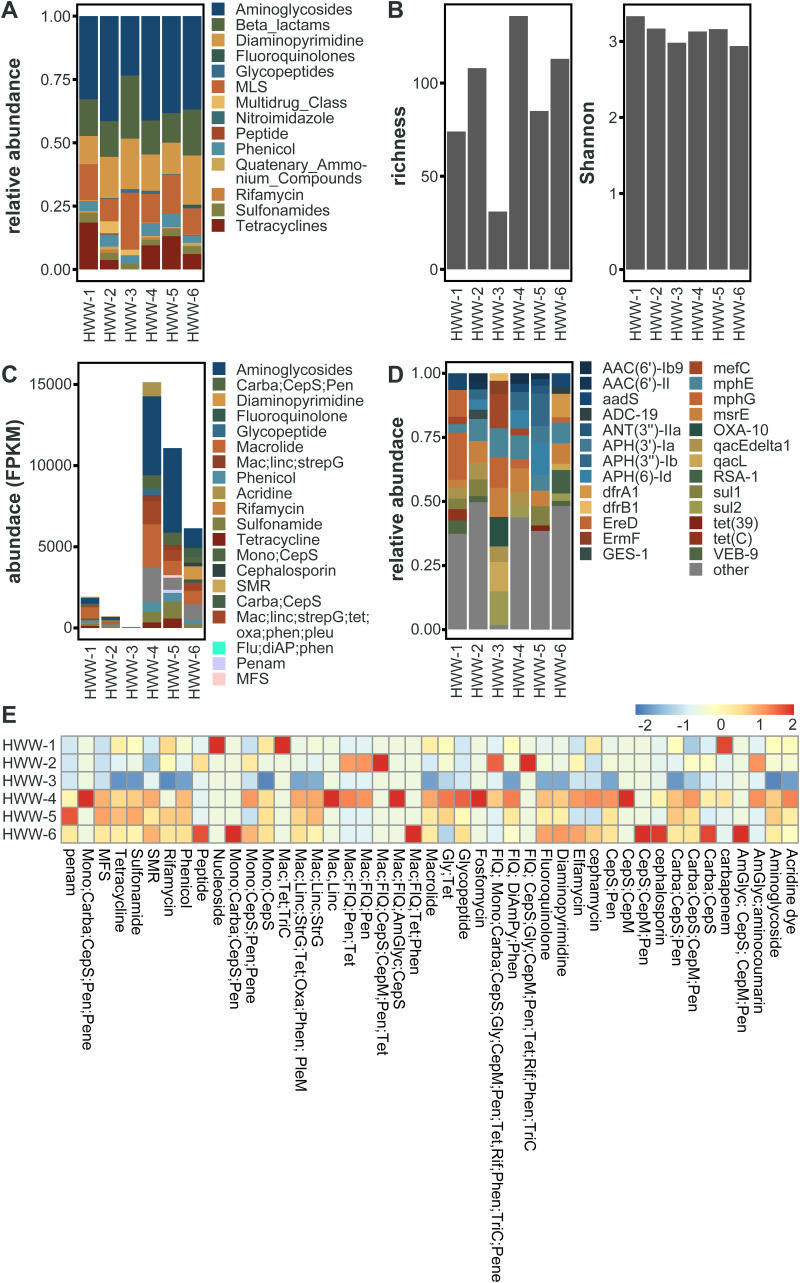
The HWW microbiome is highly complex, with 16 phyla, 39 classes, 108 orders, 247 families, 1,071 genera, and 7,802 species identified. Proteobacteria and Bacteroidetes were the most abundant phyla in all HWW samples (Fig. 2). Proteobacteria constituted approximately 80% of the bacterial population in HWW-2 and HWW-4 to HWW-6 but just half the population was formed in HWW-3 (Fig. 3A to F). HWW-1 had a distinct microbiome from the rest of the samples, with an approximately equal abundance of Bacteroidetes (48%) and Proteobacteria (≈44%). In all the other samples, Bacteroidetes were comparatively low (8% to 17%). Wastewater bacteria Cloacibacterium and Flavobacterium and environmental bacteria Pseudomonas were the predominant genera in HWW-1 to HWW-3, whereas Acinetobacter was dominant in HWW-4 to HWW-6. The Shannon diversity index showed similar microbiome diversity in all the samples (Fig. 3G).
FIG 2.
Taxonomic composition of hospital wastewater microbiomes. Sankey plots representing the abundance of various taxa in terms of reads distribution.
FIG 3.
(A to F) The bacterial diversity in terms of relative abundance at various taxonomic levels in all the samples: Phyla (A), Class (B), Order (C), Family (D), Genera (E), and Species diversity (F). (G) Richness of bacterial content and alp ha diversity of microbiome in all the samples.
Prevalence of ARGs of urgent clinical concern in Indian HWW resistome.
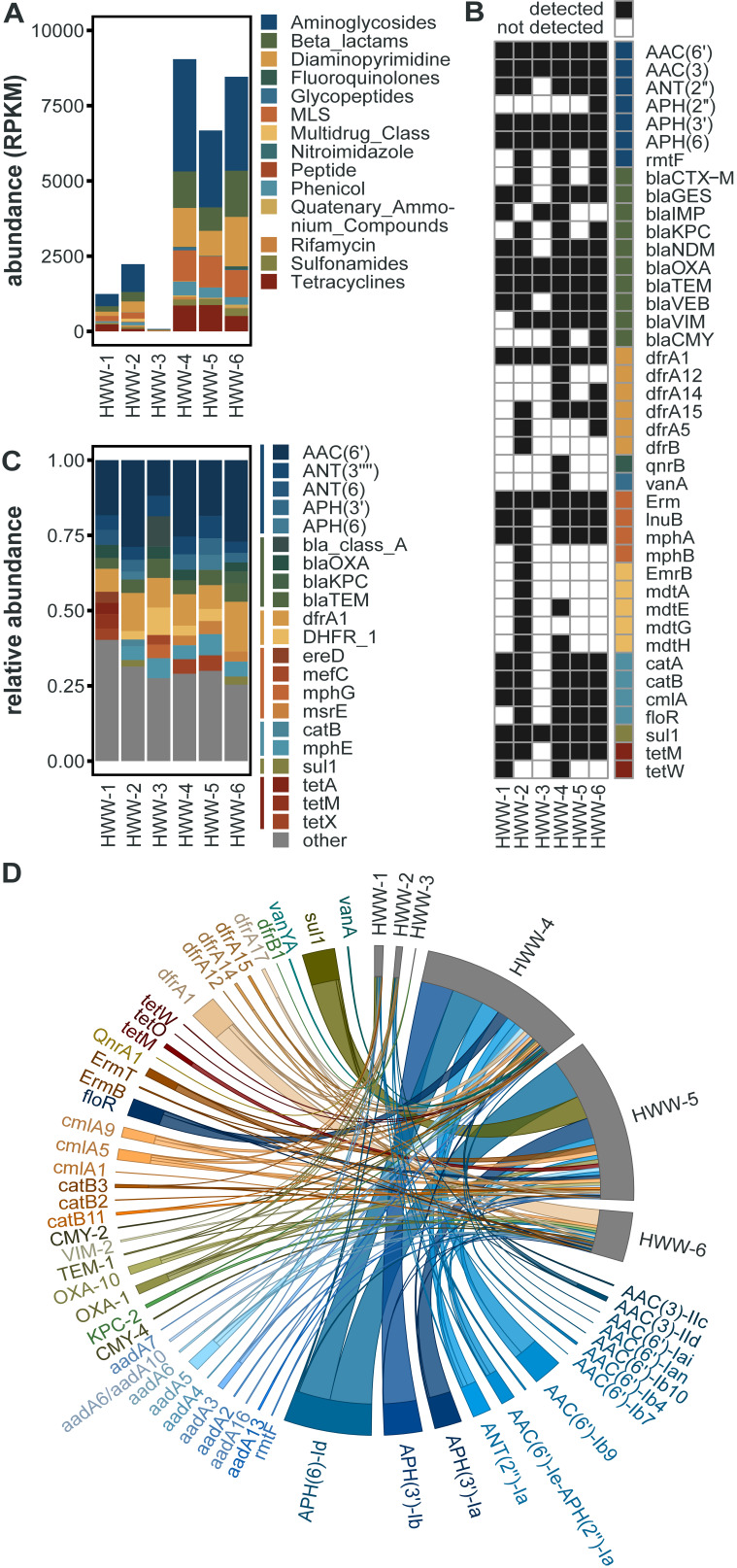
In total, we identified 183 unique AMR determinants using ShortBRED (25). HWW-1 to HWW-3 had lower ARG abundances, quantified as reads per kilobase million (RPKM), compared to HWW-4 to HWW-6 (Fig. 4A). However, the relative abundance of ARGs to specific antibiotic drug classes was comparable across sampling sites (Fig. 5A). We detected the lowest abundance of ARGs in HWW-3, and this is also reflected in as a lower ARG richness, but the Shannon diversity of all HWW samples was comparable (Fig. 5B).
FIG 4.
Identification of ARGs of clinical concern. (A) Abundance of identified ARG markers, quantified as reads per kilobase million (RPKM) and grouped by antibiotic drug target. (B) Presence-absence heatmap of select ARGs of clinical concern. Colored box indicates each ARG’s corresponding antibiotic drug target, colored the same as in Fig. 4A. (C) The relative abundances of the top 10 most abundant ARGs in each sample. ARGs are grouped by antibiotic drug target (thin bar), colored the same as in Fig. 4A. The relative abundances of all other ARGs not in the sample’s top 10 ARGs are grouped as “other.” (D) Abundance of ARGs of clinical concern, quantified as fragments per kilobase million (FPKM) identified through the assembly based method.
FIG 5.
Resistome identification. (A) The relative abundance of ARGs to specific antibiotic drug classes identified by ShortBRED. (B) Richness and Shannon diversity of HWW sample ARGs identified by ShortBRED. (C) Abundance of top 10 drug classes conferring resistance across all the six samples identified by RGI. (D) Relative abundance of the top 10 ARGs in all the HWW samples, identified by RGI. (E) The heatmap of log-transformed abundance of resistance drug classes (AmGlyc, aminoglycoside; CepS, cephalosporin; CepM, cephamycin; Pen, penam; Carba, carbapenem; FlQ, fluoroquinolone; Tet, tetracycline; Rif, rifamycin; TriC, triclosan; DiAmPy, diaminopyrimidine; Phen, phenicol; Gly, glycopeptide; Mono, monobactam; Mac, macrolide; Linc, lincosamide; StrG, streptogramin; Oxa, oxazolidinone; SMR, small multidrug resistance, MFS, Major facilitator superfamily; Pene, penem).
The WHO and other literature have identified several ARGs as being of urgent clinical concern because they have been associated with antibiotic treatment failure in hospitals and/or are widespread on MGEs (26). Many of these were identified in all 6 HWW samples, namely: the aminoglycoside-modifying enzymes aac(6′), aac(3), aph(3′), and aph(6), the carbapenemase blaOXA, the beta-lactamase blaTEM, the trimethoprim resistance gene dfrA1, the macrolide-lincosamide-streptogramin (MLS) resistance gene Erm, and the sulfonamide resistance gene sul1 (Fig. 4B). Further, most of these genes are also among the top 10 most abundant ARGs in each sample (Fig. 4C), indicating that they are not only present but highly prevalent in Indian HWW, and by extension in the hospitals themselves. In addition, numerous other clinically important ARGs were identified in a subset of HWW samples (Fig. 4B), highlighting the severity of AMR propagating in the Indian HWW environment.
Other ARGs of rising concern detected include the tetracycline-inactivating enzyme tet(X), which has been previously identified in one isolate from India (27, 28); however, its presence in five of six HWW samples suggests it may be more prevalent than previously thought. We also detected mobile colistin resistance genes mcr-3 and mcr-5 in HWW-6, which represents the first time mcr-5 was identified in Indian hospital wastewater (29).
To study ARG variants and genetic contexts, we assembled metagenomic reads and identified ARGs using the contig-based tool, RGI (Resistance Gene Identifier) (30). Although homologous ARGs belong to the same gene family, different variants exhibit substantially different risks in terms of host range, mobility potential, and ecological distribution (26). ARG abundances and compositions were consistent among this approach and ShortBRED (Fig. 5C). RGI allowed for the accurate detection of several clinically relevant high-risk ARG variants. Out of 212 unique ARG variants encompassing 43 major antibiotic classes, several were clinically concerning ARGs (53/212), often associated with MDR infections in humans (Fig. 4D). Most of these high-risk ARGs were more prevalent in HWW-4 to HWW-6, as expected owing to high ARG richness in these samples (Fig. 5C). Aminoglycoside resistance (18% to 47%) and macrolide resistance (7.9% to 36.5%) were the top two drug resistance class in all the samples, followed by carbapenemase and sulfonamide resistance. blaOXA was the most prevalent carbapenemase across all the samples (3.6% to 11.5%) except HWW-6. In HWW-6, blaRSA-1 was the dominant carbapenemase (9.1%). Since smaller fragments of ARGs may remain undetected, we used fARGene to validate the prevalence of ARGs by reconstruction of contigs (23). Among the detected ARGs, only four beta-lactamase genes were not identified earlier by RGI, namely, blaCARB-16 and blaVIM-38 in HWW-4 and blaCMY-59 and blaOXA-4 in HWW-6.
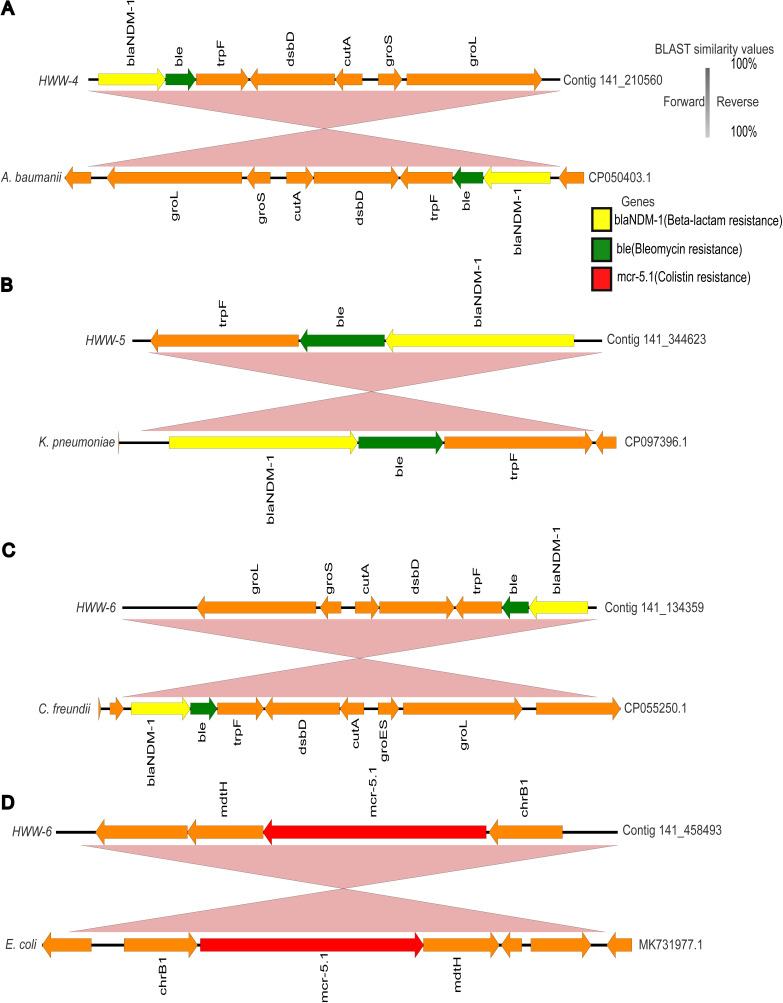
Several ARGs of urgent clinical concern are endemic to India (31, 32). blaNDM, a carbapenemase conferring resistance against most β-lactam antibiotics, was first identified in a Swedish patient returning from New Delhi but in a short span of time was detected in several outbreaks around the world (33). Several studies reported multiple blaNDM variants in J.N.M.C. hospital wastewater (HWW-1), but due to lack of surveillance at other sites of study, this is the first time that we are reporting the presence of blaNDM-1 in HWW-2, HWW-4, HWW-5, and HWW-6 (Fig. 6A to C) (34–36). blaNDM-1 often spreads through plasmid-mediated horizontal gene transfer (HGT) (37), and in this study, blaNDM-1 was predicted to be harbored by plasmid in HWW-2.
FIG 6.
The genetic contexts of blaNDM-1 and mcr-5. (A to C) The genetic context of blaNDM-1 in HWW-4 (A), HWW-5 (B), and HWW-6 (C). (D) The genetic context of identified mcr-5.1 from HWW-6. The genes were annotated through Prokka and BLASTn was performed to find the best hit identity ≥99% and plotted using Easyfig v.2.1. (https://mjsull.github.io/Easyfig). The best hits are represented with accession numbers (CP050403.1, CP097396.1, CP055250.1, and MK731977.1).
Aminoglycosides are typically not included in the first line of antibiotic treatment in many clinical scenarios, yet the high abundance of aminoglycoside resistance genes in HWW-2, HWW-4, HWW-5, and HWW-6 was surprising. As aminoglycosides are used against bacteria already resistant to beta-lactams and fluoroquinolones, the predominance of aminoglycoside resistance genes represents the presence of MDR and pan-drug-resistant bacteria in the hospital environment (38). Colistin is a last resort antibiotic for treating MDR infections, and our detection of the mcr-5.1 gene, likely harbored on an E. coli IncX1 plasmid, is worrisome (Fig. 6D) (29).
ARG co-occurrence and association with MGEs.
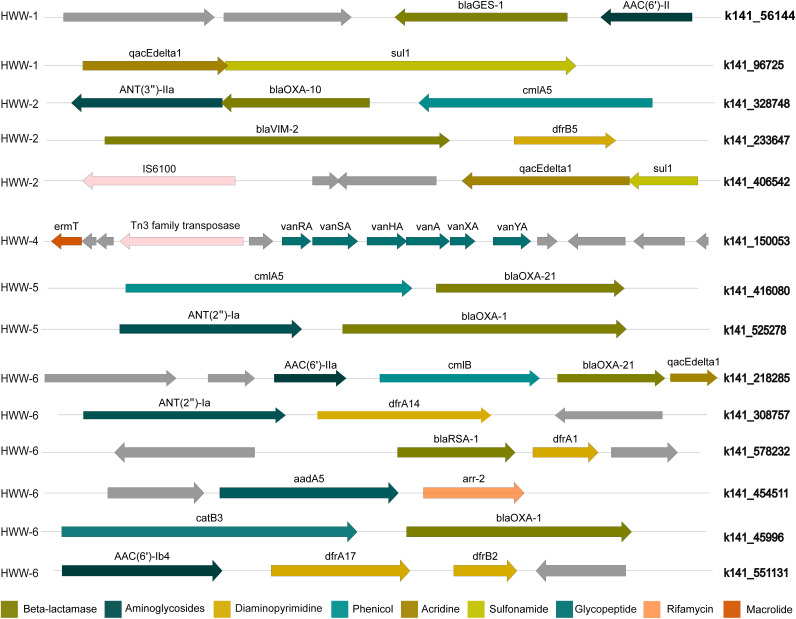
Assembly-based analysis revealed the coexistence of multiple ARGs, which may cause a severe threat to the treatment of clinical infections. We identified ARGs that were present on the same contig (Fig. 7). In HWW-1, five pairs of coexisting ARGs were identified in which three pairs had resistance toward the same drug class. Among the cooccurring resistance genes belonging to different drug classes, blaGES-1, a carbapenemase, was associated with AAC(6’)-II, an aminoglycosidase and qacEdelta1 (acridine dye resistance) with sul1 (sulfonamide resistance gene). The resistance markers belonging to three different drug classes, ANT(3″)-IIa, blaOXA-10, and cmlA5, were encoded by the same contig in HWW-2, whereas in HWW-4 one contig carried seven different glycopeptide resistance genes along with one macrolide resistance gene. blaNDM-1 was coassociated with BRP(MBL) in HWW-4, HWW-5, and HWW-6 (Fig. 6A to C). In HWW-6, blaOXA-21 coexisted with cmlB, AAC(6’)-IIa, and qacEdeltaI.
FIG 7.
ARG co-occurrence. The genetic context of ARGs belonging to different drug classes carried on the same contig. IS element IS6100 in HWW-2 and Tn3 family transposase in HWW-4 is represented in light pink color.
Co-occurrence of ARGs with plasmids and bacterial taxa.
Plasmids play a key role in bacterial ecology and evolution, particularly in regard to the spread of ARGs (39). Therefore, we next sought to identify ARGs associated with plasmids. ARGs in all the HWW samples were predominantly mediated by plasmids, including several important carbapenemases, such as blaOXA variants, blaNDM-1, blaVIM variants, and blaIMP variants. In HWW-5 and HWW-6, a significant proportion of ARGs was carried by plasmids (78% and 72% in HWW-5 and HWW-6, respectively).
Exploring the bacterial host of an ARG is a crucial question to be addressed for understanding their emergence and diversification (40). Hence, we identified the association of ARGs with bacterial taxa. As expected, based on their predominance in the composition of the HWW microbiome, ARGs were more associated with Proteobacteria with an average content of 83.8% (range, 77.9% to 91.9%) followed by Bacteroidetes. The ARGs were mostly carried by the genus Pseudomonas, and P. aeruginosa was the predominant species in all samples except HWW-4 where the genus Acinetobacter and species A. baumannii were most prominent. Both P. aeruginosa and A. baumanii ESKAPE pathogens often cause severe nosocomial infections leading to high mortalities (41, 42). We detected multiple carbapenemase genes such as blaNDM, blaOXA, blaDIM, blaIMP, blaVIM, and blaGES carried by these species. The taxonomic identification of contigs carrying blaNDM-1 showed that it was carried by P. aeruginosa in HWW-2, K. pneumoniae in HWW-4 and HWW-6, and A. baumanii in HWW-5. Colistin resistance gene mcr-5.1 in HWW-6 was detected in E. coli.
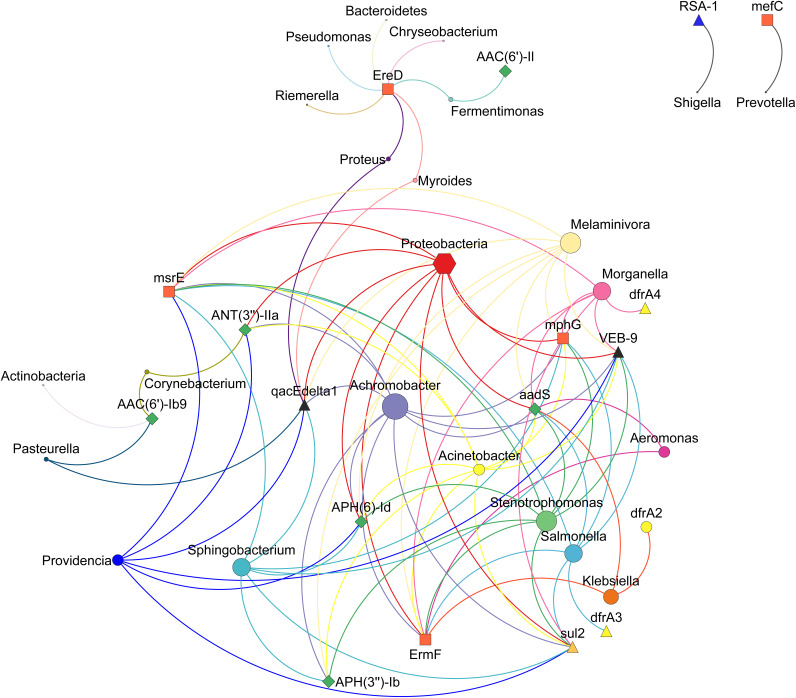
Further, we explored the cooccurrence patterns between the abundance of the top 10 ARGs across all the samples and the associated bacterial abundance at phylum and genus levels. There was a significant Spearman’s rank correlation (Spearman’s ρ = 0.81~1; P < 0.05) between microbial diversity and ARG diversity (Fig. 8). We used network analysis to explore cooccurrence patterns between ARG subtypes and bacterial taxa. Several studies have hypothesized that nonrandom cooccurrence patterns between ARGs and microbial taxa are indicators for possible host information for ARGs if ARGs and coexisting microbial taxa have significantly similar abundance trends (Spearman’s ρ > 0.8; P < 0.05) (43, 44). A total of 20 genera and 3 phyla Proteobacteria, Bacteroidetes, and Actinobacteria were identified as possible hosts of ARGs. Proteobacteria was the host of aminoglycoside resistance genes [aadS, ANT(3′’)-IIa, and APH(6)-Id], macrolide resistance genes (ErmF, mphG, and msrE), sulfonamide resistance gene (sul2), beta-lactamase (blaVEB-9), and qacEdelta1.
FIG 8.
ARG-taxonomy cooccurrence network. The network analysis of cooccurrence of top 10 ARG subtypes in all the samples. The connection pattern represents the Spearman’s correlation with 0.8 cutoff value (P value). The different sizes of nodes are indicative of the number of connections.
Virulence factor distribution.
Virulence factors are the agents of ecological connectivity that play an important role in the delocalization of AMR genes across niches through the formation of biofilms and increased infectivity. Twitching motility proteins mainly belonging to type IV pili and the flagellar proteins were prevalent in most HWW samples (Table 3). Bacteria, especially Pseudomonas spp., with twitching motility proteins have greater cytotoxicity toward the epithelial cells, enhanced transmission to other organs, and higher virulence. The flagellar proteins are responsible for adhesion, biofilm formation, and modulation of the immune system of eukaryotic cells (45). General secretion pathway proteins (gspC, gspF, gspG, gspH, gspI, gspK, and gspM) were most abundant in HWW-2 followed by type IV pili and twitching motility proteins (pilG, pilH, and pilI). In all the HWW samples, most virulence factors were associated with Pseudomonas except in HWW-2. In HWW-2, Escherichia carried most of the virulence factors. HWW-2 represented a distribution of unique VFs compared to other samples. Most of them were E. coli-specific factors like dispersin, type III secretion system effectors, general secretion pathway proteins, and E. coli common pilus chaperones. The presence of alginate biosynthesis, alginate regulation, and alginate biosynthesis virulence factors enhances the capability of bacteria to produce biofilm. The biofilm is highly proficient in transferring AMR and has an innate tolerance toward antibiotics (46). The general secretion pathway proteins have been identified as a major target for the treatment of infections and to curb the growing antibiotic resistance (47).
TABLE 3.
Virulence factors identified through ABRicate v1.0.1 using VFDBa
| Sequence | Gene | Product |
|---|---|---|
| HWW-1 | ||
| k141_344139 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_47463 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_47463 | pilU | (pilU) twitching motility protein PilU [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_359157 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_359157 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_359157 | pilI | (pilI) twitching motility protein PilI [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_359157 | pilJ | (pilJ) twitching motility protein PilJ [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_221870 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_250443 | flgC | (flgC) flagellar basal-body rod protein FlgC [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_272187 | flgC | (flgC) flagellar basal-body rod protein FlgC [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_211659 | flgH | (flgH) flagellar L-ring protein precursor FlgH [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_283651 | flgI | (flgI) flagellar P-ring protein precursor FlgI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_316175 | flgI | (flgI) flagellar P-ring protein precursor FlgI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_132209 | fliE | (fliE) flagellar hook-basal body complex protein FliE [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_446999 | fliG | (fliG) flagellar motor switch protein G [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_61688 | fliI | (fliI) flagellum-specific ATP synthase FliI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_346249 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_346249 | fliN | (fliN) flagellar motor switch protein FliN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_483093 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_483093 | fliQ | (fliQ) flagellar biosynthetic protein FliQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_24580 | flhA | (flhA) flagellar biosynthesis protein FlhA [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_160638 | fleN | (fleN) flagellar synthesis regulator FleN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_153162 | xcpT | (xcpT) general secretion pathway protein G [xcp secretion system (VF0084)] [Pseudomonas aeruginosa PAO1] |
| k141_153162 | xcpS | (xcpS) general secretion pathway protein F [xcp secretion system (VF0084)] [Pseudomonas aeruginosa PAO1] |
| k141_153162 | xcpR | (xcpR) general secretion pathway protein E [xcp secretion system (VF0084)] [Pseudomonas aeruginosa PAO1] |
| k141_250831 | algW | (algW) AlgW protein [Alginate regulation (CVF523)] [Pseudomonas aeruginosa PAO1] |
| k141_438174 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_440675 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_219894 | algC | (algC) phosphomannomutase AlgC [Alginate biosynthesis (CVF522)] [Pseudomonas aeruginosa PAO1] |
| HWW-2 | ||
| k141_340702 | aap/aspU | (aap/aspU) Dispersin [Dispersin (VF0215)] [Escherichia coli O44:H18 042] |
| k141_275495 | aggB | (aggB) fimbrial minor subunit [AAFs (VF0214)] [Escherichia coli 17-2] |
| k141_203453 | csgD | (csgD) DNA-binding transcriptional regulator CsgD [curli fibers/thin aggregative fimbriae (AGF) (AI094)] [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| k141_203453 | csgF | (csgF) curli production assembly/transport protein CsgF [Agf (VF0103)] [Salmonella enterica subsp. enterica serovar Typhimurium str. LT2] |
| k141_200776 | espR4 | (espR4) Type III secretion system effector espR4 [LEE encoded T3SS (SS020)] [Escherichia coli O157:H7 str. EDL933] |
| k141_85666 | espY1 | (espY1) Type III secretion system effector EspY1 [LEE encoded T3SS (SS020)] [Escherichia coli O157:H7 str. EDL933] |
| k141_146443 | fepB | (fepB) ferrienterobactin ABC transporter periplasmic binding protein [Enterobactin (VF0228)] [Escherichia coli CFT073] |
| k141_441654 | fepD | (fepD) ferrienterobactin ABC transporter permease [Enterobactin (VF0228)] [Escherichia coli CFT073] |
| k141_63063 | fimI | (fimI) Fimbrin-like protein fimI precursor [Type 1 fimbriae (VF0221)] [Escherichia coli CFT073] |
| k141_319634 | fliQ | (fliQ) flagellar biosynthetic protein FliQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_272515 | gspC | (gspC) general secretion pathway protein C [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_181603 | gspF | (gspF) general secretion pathway protein F [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_181603 | gspG | (gspG) general secretion pathway protein G [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_234431 | gspH | (gspH) general secretion pathway protein H [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_166675 | gspI | (gspI) general secretion pathway protein I [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_203361 | gspK | (gspK) general secretion pathway protein K [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_799 | gspM | (gspM) general secretion pathway protein M [T2SS (VF0333)] [Shigella dysenteriae Sd197] |
| k141_255827 | hcp1 | (hcp1) type VI secretion system substrate Hcp1 [HSI-I (VF0334)] [Pseudomonas aeruginosa PAO1] |
| k141_295605 | kpsM | (kpsM) KpsM [K1 capsule (VF0239)] [Escherichia coli O18:K1:H7 str. RS218] |
| k141_24329 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_69666 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_356469 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_48103 | pilI | (pilI) twitching motility protein PilI [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_44828 | shuX | (shuX) shu locus protein ShuX [Shu (VF0256)] [Shigella dysenteriae Sd197] |
| k141_466702 | yagV/ecpE | (yagV/ecpE) E. coli common pilus chaperone EcpE [ECP (VF0404)] [Escherichia coli O157:H7 str. EDL933] |
| k141_299262 | yagX/ecpC | (yagX/ecpC) E. coli common pilus usher EcpC [ECP (VF0404)] [Escherichia coli O157:H7 str. EDL933] |
| k141_299262 | yagY/ecpB | (yagY/ecpB) E. coli common pilus chaperone EcpB [ECP (VF0404)] [Escherichia coli O157:H7 str. EDL933] |
| k141_439431 | yagZ/ecpA | (yagZ/ecpA) E. coli common pilus structural subunit EcpA [ECP (VF0404)] [Escherichia coli O157:H7 str. EDL933] |
| k141_439431 | ykgK/ecpR | (ykgK/ecpR) regulator protein EcpR [ECP (VF0404)] [Escherichia coli O157:H7 str. EDL933] |
| HWW-4 | ||
| k141_125532 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_142010 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_298551 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_453859 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_553919 | alg8 | (alg8) alginate-c5-mannuronan-epimerase AlgG [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_446658 | algA | (algA) phosphomannose isomerase / guanosine 5′-diphospho-D-mannose pyrophosphorylase [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_200944 | algB | (algB) two-component response regulator AlgB [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_105172 | algC | (algC) phosphomannomutase AlgC [Alginate biosynthesis (CVF522)] [Pseudomonas aeruginosa PAO1] |
| k141_485400 | algD | (algD) GDP-mannose 6-dehydrogenase AlgD [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_12932 | algI | (algI) alginate o-acetyltransferase AlgI [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_50607 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_185855 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_459309 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_507216 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_568819 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_589694 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_415468 | fimF | (fimF) FimF protein precursor [Type 1 fimbriae (VF0221)] [Escherichia coli CFT073] |
| k141_357125 | fleN | (fleN) flagellar synthesis regulator FleN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_340898 | fleQ | (fleQ) transcriptional regulator FleQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_113633 | flgC | (flgC) flagellar basal-body rod protein FlgC [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_253387 | flgC | (flgC) flagellar basal-body rod protein FlgC [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_428864 | flgC | (flgC) flagellar basal-body rod protein FlgC [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_343807 | flgG | (flgG) flagellar basal-body rod protein FlgG [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_346193 | flgG | (flgG) flagellar basal-body rod protein FlgG [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_274809 | flgH | (flgH) flagellar L-ring protein precursor FlgH [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_591610 | flgH | (flgH) flagellar L-ring protein precursor FlgH [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_274809 | flgI | (flgI) flagellar P-ring protein precursor FlgI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_318595 | flhA | (flhA) flagellar biosynthesis protein FlhA [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_141825 | fliA | (fliA) flagellar biosynthesis sigma factor FliA [Deoxyhexose linking sugar 209 Da capping structure (AI138)] [Pseudomonas aeruginosa PAO1] |
| k141_264415 | fliA | (fliA) flagellar biosynthesis sigma factor FliA [Deoxyhexose linking sugar 209 Da capping structure (AI138)] [Pseudomonas aeruginosa PAO1] |
| k141_397047 | fliE | (fliE) flagellar hook-basal body complex protein FliE [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_106581 | fliG | (fliG) flagellar motor switch protein G [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_3328 | fliG | (fliG) flagellar motor switch protein G [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_266069 | fliI | (fliI) flagellum-specific ATP synthase FliI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_79328 | fliI | (fliI) flagellum-specific ATP synthase FliI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_207084 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_219044 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_433887 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_443952 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_180332 | fliN | (fliN) flagellar motor switch protein FliN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_219044 | fliN | (fliN) flagellar motor switch protein FliN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_186333 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_433887 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_72397 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_88240 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_396457 | fliQ | (fliQ) flagellar biosynthetic protein FliQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_40634 | fliQ | (fliQ) flagellar biosynthetic protein FliQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_127626 | mbtH-like | (mbtH-like) MbtH-like protein from the pyoverdine cluster [pyoverdine (IA001)] [Pseudomonas aeruginosa PAO1] |
| k141_567320 | motA | (motA) flagellar motor protein [Deoxyhexose linking sugar 209 Da capping structure (AI138)] [Pseudomonas aeruginosa PAO1] |
| k141_430957 | motC | (motC) flagellar motor protein [Deoxyhexose linking sugar 209 Da capping structure (AI138)] [Pseudomonas aeruginosa PAO1] |
| k141_14328 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_16339 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_245324 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_545762 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_55210 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_589547 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_597392 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_624550 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_82558 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_119537 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_333795 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_445530 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_592421 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_597392 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_82558 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_314471 | pilM | (pilM) type IV pilus inner membrane platform protein PilM [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_29595 | pilO | (pilO) type IV pilus inner membrane platform protein PilO [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_553621 | pilO | (pilO) type IV pilus inner membrane platform protein PilO [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_44074 | pilR | (pilR) two-component response regulator PilR [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_322422 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_419027 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_113933 | pilU | (pilU) twitching motility protein PilU [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_111109 | waaA | (waaA) lipopolysaccharide core biosynthesis protein WaaP [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_395234 | waaF | (waaF) heptosyltransferase I [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_382804 | waaG | (waaG) B-band O-antigen polymerase [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_220982 | waaP | (waaP) UDP-glucose:(heptosyl) LPS alpha 13-glucosyltransferase WaaG [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_60569 | xcpA/pilD | (xcpA/pilD) type 4 prepilin peptidase PilD [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_222010 | xcpT | (xcpT) general secretion pathway protein G [xcp secretion system (VF0084)] [Pseudomonas aeruginosa PAO1] |
| k141_366811 | xcpT | (xcpT) general secretion pathway protein G [xcp secretion system (VF0084)] [Pseudomonas aeruginosa PAO1] |
| HWW-5 | ||
| k141_107730 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_122640 | waaG | (waaG) B-band O-antigen polymerase [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_122640 | waaP | (waaP) UDP-glucose:(heptosyl) LPS alpha 13-glucosyltransferase WaaG [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_125756 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_149358 | fleN | (fleN) flagellar synthesis regulator FleN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_154646 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_154646 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_154646 | pilJ | (pilJ) twitching motility protein PilJ [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_154979 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_161953 | waaF | (waaF) heptosyltransferase I [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_173408 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_173408 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_214052 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_236530 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_236530 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_236530 | fliG | (fliG) flagellar motor switch protein G [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_237176 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_237176 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_260022 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_275125 | flgG | (flgG) flagellar basal-body rod protein FlgG [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_352540 | algC | (algC) phosphomannomutase AlgC [Alginate biosynthesis (CVF522)] [Pseudomonas aeruginosa PAO1] |
| k141_35606 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_360749 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_38661 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_416308 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_416308 | fliN | (fliN) flagellar motor switch protein FliN [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_416308 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_424073 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_425540 | pilU | (pilU) twitching motility protein PilU [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_427332 | pilU | (pilU) twitching motility protein PilU [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_461073 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_461073 | pilU | (pilU) twitching motility protein PilU [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_465653 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_470086 | algU | (algU) alginate biosynthesis protein AlgZ/FimS [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_476116 | acpXL | (acpXL) acyl carrier protein [LPS (CVF383)] [Brucella melitensis bv. 1 str. 16M] |
| k141_482681 | xcpT | (xcpT) general secretion pathway protein G [xcp secretion system (VF0084)] [Pseudomonas aeruginosa PAO1] |
| k141_488285 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_498478 | fliG | (fliG) flagellar motor switch protein G [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_562960 | pilR | (pilR) two-component response regulator PilR [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_57587 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_60943 | flgI | (flgI) flagellar P-ring protein precursor FlgI [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_60943 | flgG | (flgG) flagellar basal-body rod protein FlgG [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_69665 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| HWW-6 | ||
| k141_14239 | pvdS | (pvdS) extracytoplasmic-function sigma-70 factor [Pyoverdine (VF0094)] [Pseudomonas aeruginosa PAO1] |
| k141_150252 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_191571 | mbtH-like | (mbtH-like) MbtH-like protein from the pyoverdine cluster [pyoverdine (IA001)] [Pseudomonas aeruginosa PAO1] |
| k141_207228 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_218040 | waaG | (waaG) B-band O-antigen polymerase [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_218555 | fliE | (fliE) flagellar hook-basal body complex protein FliE [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_232199 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_232199 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_267491 | waaF | (waaF) heptosyltransferase I [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_270282 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_297256 | hsiB1/vipA | (hsiB1/vipA) type VI secretion system tubule-forming protein VipA [HSI-I (VF0334)] [Pseudomonas aeruginosa PAO1] |
| k141_322579 | algD | (algD) GDP-mannose 6-dehydrogenase AlgD [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_32858 | fliA | (fliA) flagellar biosynthesis sigma factor FliA [Deoxyhexose linking sugar 209 Da capping structure (AI138)] [Pseudomonas aeruginosa PAO1] |
| k141_344288 | waaP | (waaP) UDP-glucose:(heptosyl) LPS alpha 13-glucosyltransferase WaaG [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_354036 | hcp1 | (hcp1) type VI secretion system substrate Hcp1 [HSI-I (VF0334)] [Pseudomonas aeruginosa PAO1] |
| k141_371808 | pilG | (pilG) twitching motility protein PilG [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_378363 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_37950 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_394847 | fleQ | (fleQ) transcriptional regulator FleQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_410255 | algR | (algR) alginate biosynthesis regulatory protein AlgR [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_425127 | pilU | (pilU) twitching motility protein PilU [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_443234 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_443234 | fliQ | (fliQ) flagellar biosynthetic protein FliQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_445882 | algC | (algC) phosphomannomutase AlgC [Alginate biosynthesis (CVF522)] [Pseudomonas aeruginosa PAO1] |
| k141_477016 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_477379 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_492959 | flgH | (flgH) flagellar L-ring protein precursor FlgH [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_494604 | flgC | (flgC) flagellar basal-body rod protein FlgC [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_50664 | algC | (algC) phosphomannomutase AlgC [Alginate biosynthesis (CVF522)] [Pseudomonas aeruginosa PAO1] |
| k141_524666 | fliQ | (fliQ) flagellar biosynthetic protein FliQ [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_524666 | fliP | (fliP) flagellar biosynthetic protein FliP [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_528996 | waaF | (waaF) heptosyltransferase I [LPS (VF0085)] [Pseudomonas aeruginosa PAO1] |
| k141_539669 | fliM | (fliM) flagellar motor switch protein FliM [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
| k141_593211 | algB | (algB) two-component response regulator AlgB [Alginate (VF0091)] [Pseudomonas aeruginosa PAO1] |
| k141_593475 | pilI | (pilI) twitching motility protein PilI [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_599056 | pvdH | (pvdH) diaminobutyrate-2-oxoglutarate aminotransferase PvdH [pyoverdine (IA001)] [Pseudomonas aeruginosa PAO1] |
| k141_64492 | dotU1 | (dotU1) type VI secretion system protein DotU [HSI-I (VF0334)] [Pseudomonas aeruginosa PAO1] |
| k141_64492 | hsiJ1 | (hsiJ1) type VI secretion system hcp secretion island protein HsiJ1 [HSI-I (VF0334)] [Pseudomonas aeruginosa PAO1] |
| k141_73625 | pilH | (pilH) twitching motility protein PilH [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_79819 | pilT | (pilT) twitching motility protein PilT [Type IV pili (VF0082)] [Pseudomonas aeruginosa PAO1] |
| k141_84914 | flgG | (flgG) flagellar basal-body rod protein FlgG [Flagella (VF0273)] [Pseudomonas aeruginosa PAO1] |
VFDB, virulence factor database.
Metagenome-assembled genomes.
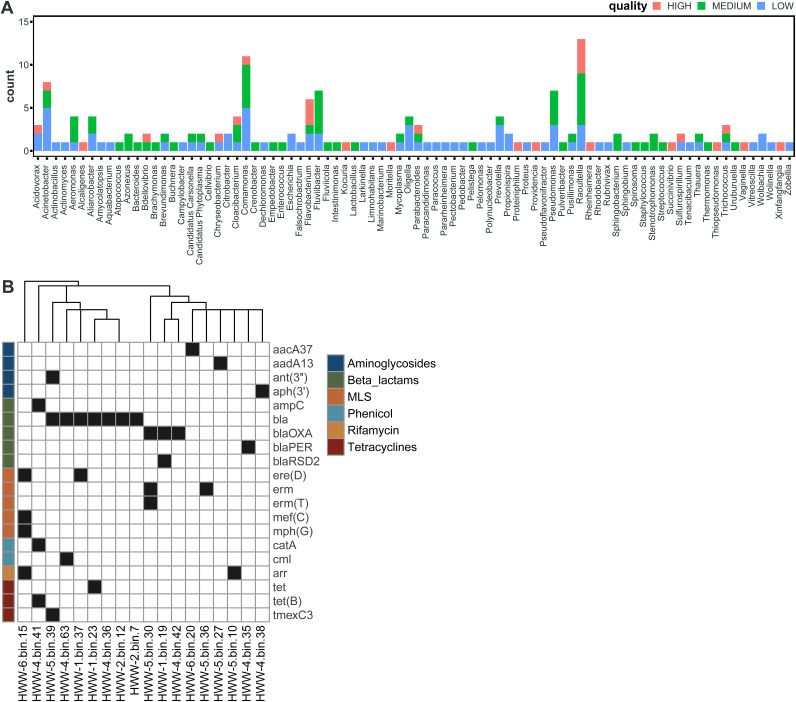
To better study the content of individual genomes and the genetic contexts of ARGs, we constructed metagenome-assembled genomes (MAGs). We recovered a total of 167 draft MAGs of various quality: 26 high-quality, 65 medium-quality, and 76 low-quality drafts (Fig. 9A). The most commonly recovered genera were Raoutella (13/167), Comamonas (11/167), and Acinetobacter (8/167), all Proteobacteria. Of the total 84 unique genera identified, 54 were unique to just 1 MAG. Next, we screened the 26 high-quality MAGs for known ARGs in silico. We identified a total of 20 unique ARGs in 18/26 MAGs (Fig. 9B). Most of these MAGs (12/18) encoded at least one beta-lactamase, including blaOXA carbapenemase and blaPER extended-spectrum β-lactamases (ESBLs).
FIG 9.
Metagenome-assembled genomes. (A) Histogram of MAG species assignment, colored by MAG quality. (B) ARG content of high-quality MAGs identified by AMRFinder.
DISCUSSION
We have shown that Indian HWW samples have a high abundance and diversity of clinically relevant ARGs potentially hosted by high-priority pathogens. We collected samples from six tertiary-care hospitals in north India with various patient loads, located in urban (HWW-1, -2, -4, and -5) and rural areas (HWW-3 and -6) (Fig. 1 and Table 1). Earlier studies have been limited to single hospitals (48). We used shotgun metagenomics to analyze our HWW samples, which has the advantage of quantifying thousands of genes from culturable as well as nonculturable taxa simultaneously. PCR-based approaches may have greater sensitivity to low-abundance ARGs due to targeted amplification (49), but they are limited in the ARGs able to be detected. For example, a study comparing culture-based approaches and metagenomic methods showed that the culture-based technique isolated bacteria from 104 out of 539 clinical samples and therefore, captured resistance against 8 antibiotic types in only 16.17% of the clinical samples. Contrary to the culture method, metagenomic analysis identified 1,573 species and 885 ARG subtypes in the hospital sewage (50).
Hospital wastewater surveillance is useful for monitoring antibiotic-resistant bacteria and the ARG load in the hospital environment (51). In our study, we found high ARG richness (fragments per kilobase million [FPKM]) in hospital wastewater. A study reported a comparably lower abundance of ARGs in urban wastewater compared to our findings (11). In India, hospital wastewater treatment is inadequate and the hospital wastewater mostly gets released into the public sewer network increasing the probability of ARG dissemination to humans, especially to the poor who often reside near the drains (22, 51).
Owing to its high abundance, the phyla Proteobacteria carried most of the ARGs, constituting approximately 85% of the ARG abundance (range, 78% to 84%) in terms of FPKM, followed by Bacteroidetes (HWW-1 to HWW-4), Firmicutes (HWW-5), and Actinobacteria (HWW-6) (Fig. 3 to 5). The taxonomic compositions of our HWW samples are similar to that reported from a single hospital in the city of Mumbai, located in western India (48). They also reported the dominance of Proteobacteria, followed by Bacteroidetes and Firmicutes. Interestingly, they reported Acinetobacter as the dominant genus (30%), which is also true of our HWW samples from New Delhi (HWW-4 = 26% and HWW-5 = 18%); however, Acinetobacter was not among even the top three most abundant genera in the other samples (Fig. 2 and 3). Pseudomonas was among the top four genera (3% to 9%), carrying maximum ARG abundance (23% to 38%) in all the samples except HWW-4. In congruence with the aforementioned study, Acinetobacter predominantly carried 23% ARG abundance in HWW-4 of New Delhi (48). The species carrying most of the ARGs were P. aeruginosa across all the samples, A. baumanii in HWW-4, and P. putida in HWW-6. All these species are high priority, nosocomial pathogens with a high rate of ARG dissemination, mostly involved in hospital-acquired infections (52, 53). The opportunistic pathogens A. baumanii and P. aeruginosa carry natural intrinsic resistance toward multiple drug classes, including beta-lactamases and other carbapenemases. They are not eliminated by wastewater treatment either (54, 55). P. putida is an environmental Gram-negative bacterium. It is rarely a causative agent for human diseases, but there have been reports of serious infections and outbreaks from time to time. Sometimes it functions as an exchange platform for ARGs, spreading ARGs to more pathogenic species like P. aeruginosa (56).
We generally expected HWW from the largest hospitals to have the highest ARG burdens due to increased patient load. Surprisingly, the samples from hospitals with the largest number of beds (HWW-1 = 2,400 beds and HWW-2 = 1,050 beds) had among the lowest ARG abundances. Conversely, HWW-4 and HHW-6 had 4 to 6 times higher ARG abundance but with a fraction of the number of beds (HWW-4 = 650 and HWW-6 = 300). This could be due to improved antibiotic stewardship and waste disposal practices. However, we note that “hospital size” is not definitive because many hospitals in India over admit 3 to 4 times the total bed capacity, and these true numbers may not be reported (22).
Several studies have reported the injudicious use of broad-spectrum antibiotics in Indian hospitals and, consequently, high incidences of beta-lactamase (carbapenemase) resistance (57, 58). We also detected predominant resistance against broad-spectrum antibiotics with a leading abundance of aminoglycoside resistance in all the samples except HWW-1 and HWW-3 where macrolide resistance was most abundant (Fig. 4A and 5) (48). Carbapenem and sulfonamide resistance was among the top five ARG-enriching drug classes in all the samples. Earlier, in Indian HWW, the sul1 sulfonamide resistance gene was reported as the most abundant (11.4%); we also identified sul1 in all HWW samples but it was among the top 10 most abundant ARGs only in HWW-2 and HWW-6 (2.2% and 9.6%, respectively) (Fig. 4) (48). The ARG composition listed in the top 10 ARGs replicated the abundance of resistance evaluated at the drug class level with 8 aminoglycoside resistance genes, 6 macrolide resistance genes, and 3 carbapenemases. Among the top 10 ARGs, the beta-lactamases blaOXA-10, blaGES-1, and blaRSA-1 were dominant. Multiple incidences of blaOXA outbreaks in Indian hospitals have been reported in the past, which correlates with blaOXA being the most abundant carbapenemase in all our samples (59, 60). The blaOXA-10 and GES-type ESBLs have been earlier reported as the most abundant beta-lactamase in one of the Mumbai hospitals (48). The blaNDM-1 is endemic to India and is often flanked by transposable elements. The blaNDM-1 was found prevalent among P. aeruginosa, K. pneumoniae, and A. baumanii (Fig. 6A to C). The use of colistin used as the last-resort antibiotic in extreme clinical cases of MDR and extensively drug-resistant (XDR) infections also leads to the emergence of mcr variants. The detection of plasmid-mediated mcr-5.1 in HWW-6 (Fig. 6D) for the first time in Indian hospital sewage raises concern for future healthcare systems and is an alarming signal for the upcoming antibiotic apocalypse when no antibiotic will work. MAGs construction and analysis also revealed at least one beta-lactamase, including blaOXA carbapenemase and blaPER ESBLs. Most of the ARGs identified in our study can efficiently transmit to different species of bacteria through horizontal gene transfer, and hence, their spread to the local population has become a challenge for healthcare workers. The concentration of several ARGs like blaOXA-1, blaOXA-10, and blaTEM-1 increases with wastewater treatment procedures (61). Plasmid carried the maximum ARGs in all the samples reflecting the high probability of AMR spread through hospital wastewater.
In this study, we identified several virulence factors mostly associated with the general secretion pathway, motility, and alginate biosynthesis often involved in biofilm formation (Table 3). These VFs aid the innate resistance against antibiotics (45, 47, 62).
Our study adds further evidence that India is facing an “AMR pandemic” and needs urgent national surveillance for assessing the ARG risk. Hospital wastewater transports high-risk clinical threats to the public sewer system and facilitates their dissemination to the public through HGT (61). The common practice of open drainage systems and inadequate sanitation measures in most parts of India (63) may lead to infection spread with antibiotic-resistant bacteria and outbreaks in the community as well as a hospital setting. Likely due to our limited sample size, we did not find significant dissimilarities in ARGs and microbial diversity with geographical variations. Overall, the ARG diversity in our samples was in concordance with one of the earlier studies on Indian hospital wastewater samples, carried out in Mumbai (48).
This explorative study shows that Indian HWW contains an abundance of high-risk ARGs that are present in mobile genetic elements and are carried by high-priority pathogens. Antimicrobial risk management strategies should be immediately implemented in hospitals. Efficient wastewater treatment strategies meeting international standards should be implemented in hospitals, as well as areas downstream including the public sewage systems where the HWW are deposited.
MATERIALS AND METHODS
Sample collection and processing.
Wastewater samples were collected from six hospitals located in different regions of northern India from December 21, 2019, to March 21, 2021 (Table 1). From the main sewage pipeline receiving effluents from every other pipeline in the hospital, multiple samples were collected from adjacent points in sterile bottles and pooled into one sample of 2 liters of unfiltered sewage water. Samples were stored on ice without additives or DNA stabilizers. In a sterile environment, samples were vortexed and 50 mL was collected and then centrifuged again at 7,000 × g to separate cell pellets and water. The pellet was stored at –20°C until DNA extraction.
DNA was extracted using the DNeasy PowerSoil kit (Qiagen) as per the manufacturer’s instructions. Extracted DNA was quality checked using NanoDrop 2000 Spectrophotometer (ThermoFisher Scientific), to check for RNA and protein contaminants. Further, to validate the quantitative estimation of the extracted DNA, 50 ng of extracted DNA and Lambda DNA/HindIII Marker (SM0102; ThermoFisher Scientific) was loaded on a 1% agarose gel stained with ultrapure ethidium bromide (ThermoFisher Scientific) and electrophoresed by running the gel at 80 V for 1 h. Finally, the gel was imaged in a SmartView Pro 1100 Imager System (Major Science). Sample DNA concentrations were quantified using Qubit dsDNA HS assay kit (Thermo Fisher Scientific), as per manufacturer’s instructions, using a Qubit 3.0 Fluorometer (Thermo Fisher Scientific) (64).
DNA library preparation and sequencing.
One hundred nanograms of intact DNA was enzymatically fragmented using Covaris targeting the 250-bp fragment size, followed by end repair to convert the overhangs into blunt ends. To the adenylated fragments, loop adapters were ligated and cleaved with a uracil-specific excision reagent (USER) enzyme. The samples were further purified using AMPure beads. DNA was enriched by PCR with six cycles using NEBNext Ultra II Q5 master mix, Illumina universal primer, and sample-specific octamer primers. The amplified products were cleaned by using AM pure beads to remove unused primers. The final DNA libraries were eluted in 15 mL of 0.1× TE buffer, and concentrations were quantified using the Qubit DNA HS assay kit and a Qubit.3 Fluorometer.
Library quality was assessed using DNA 5000 ScreenTape in an Agilent 4150 Tape Station system. Here, 1 mL of the library was mixed with 5 mL sample buffer, vortexed, and then spun to collect the sample to the bottom of the strip. The strip was then loaded into the Agilent 4150 Tape Station instrument. The library qualification criteria were the presence of a broad peak in the range of 200 bp to 1,000 bp, with an average size of 350 bp in the Agilent 4150 TapeStation system, the Qubit concentrations above 2 ng/μL or 10 nmol/L and library devoid of primer, adapter, and larger size peaks.
The quality-passed libraries were diluted to 2 nM and pooled. We then performed shotgun metagenome sequencing on the pooled libraries using Illumina HiSeq, 2 × 150 bp paired-end run. The sequence reads were demultiplexed by barcode using bcl2fastq v2.1.9. Sequence data quality was checked using FastQC v0.11.9 (65) and MultiQC v1.9 (66) for base call quality distribution, percent bases above Q20, Q30, percent GC, and sequencing adapter contamination. The number of reads sequenced (in million) was approximately 54, 45, 45, 78, 43, and 43 for HWW-1, HWW-2, HWW-3, HWW-4, HWW-5, and HWW-6, respectively.
For microbiome composition, and contig-based ARG analyses, these clean reads were assembled using MEGAHIT v1.2.9 (67) with –k-min 35 –k-max 141 –k-step 28 parameters. The contigs shorter than 200 bp were removed from further analysis. Assembly quality was checked using Bowtie2 v2.1.0 (68).
Taxonomic classification.
For taxonomic classification, the reads were quality filtered using fastp v.0.20.1 (69). These clean reads were taxonomically classified using Kraken2 with the NCBI nonredundant nucleotide database as a reference (70). Kraken hits with a relative abundance of <0.1% reads were filtered out.
Identification and quantification of ARG.
For ShortBRED analyses, reads were quality filtered using Trimmomatic v0.38 (71) with the following parameters: ILLUMINACLIP: NexteraPE-PE.fa:2:30:10:1:true SLIDINGWINDOW:4:20 LEADING:10 TRAILING:10 MINLEN:60. Clean read quality was assessed using FastQC v0.11.7 (65) and MultiQC v1.2. (66). ARG abundances were quantified using ShortBRED v0.9.4 (25). We built an ARG-specific markers database from 7,921 antibiotic resistance proteins using ‘shortbred_identify.py’ with the following nondefault parameters: –clustid 0.95 –ref Uniref90 (72). The antibiotic resistance protein sequences include sequences from the CARD database (30), the NCBI-AMR database (73), and antibiotic resistance proteins identified using functional metagenomics in this cohort, and previous studies (74–82). These AMR gene families were then manually curated, and entries with the following criteria were removed because they would not be confidently expected to provide resistance based solely on a short-read marker:
Genes associated with global gene regulators, two-component system proteins, and signaling mediators (e.g., blaZ, vanS-vanR, mecI, mepR, gadW, marR);
Genes encoding subunits that are part of multiple efflux pumps (e.g., tolC, oprM, opmD);
Resistance via mutation in genes (e.g., resistance to antifolate drugs via mutations in dhfr, resistance to rifamycin via mutation in rpoB);
Genes conferring resistance by modifying cell wall charge (e.g., mprF);
Genes that reduce permeability (e.g., omp38, tmrB) or confer resistance through overexpression (e.g., thymidylate synthase); and
General efflux pumps that came through functional selections (e.g., MFS-type, ABC-type).
The relative abundance of AMR gene families was quantified by mapping reads to the filtered set of marker sequences using shortbred_quantify.py. ShortBRED hits were filtered out if they had counts less than 2 or a mean RPKM < 0.001.
For contig-based ARG analyses, the reads were assembled using MEGAHIT v1.2.9 (67) with the following parameters: –k-min 35 –k-max 141 –k-step 28. Contigs less than 200 bp were removed from the downstream analysis. The assembly quality was checked using Bowtie2 v2.1.0 (68). Contigs were assigned taxa using TaxonKit v0.2.3 (83) and the NCBI nucleotide database, and those annotated as Eukaryotic sequences were filtered out. The remainder were annotated using Prokka v1.14.6 –metagenome (84). ARGs were called using the Resistance Gene Identifier (RGI) v5.2.0 and the Comprehensive Antibiotic Resistance Database (CARD) v3.1.1 (https://github.com/arpcard/rgi) (30). Sequences were called as an ARG if they had ≥80% coverage and ≥90% identity to a reference ARG. Sequences with 100% coverage and 100% identity against a reference ARG were classified as “Perfect” hits; those with <95% identity but >65% coverage as putative or potential ARGs. Additionally, fARGene (Fragmented Antibiotic Resistance Gene iENntifiEr) (https://github.com/fannyhb/fargene) was used to identify gene fragments in contigs. ARG abundances were quantified as FPKM (11):
where qi = no. of reads mapped to the contig, li = length of contig, and Q = total no. of mapped reads.
Taxonomic distribution of BLAST hit contigs was identified with TaxonKit v0.2.3. When the ARG containing contig was simultaneously annotated against the CARD database and a microbial taxon in the NCBI NR database, we considered that the associated microbial taxon was the carrier of the corresponding ARG (44). ARGs were identified as located in plasmids or chromosomes using PlasClass and PlasFlow v1.1 (85, 86). Virulence factors were identified using ABRicate v1.0.1 (https://github.com/tseemann/abricate) (Table 3).
Metagenome-assembled genomes.
To extract MAGs, metagenomic assemblies were generated using MEGAHIT v1.1.4 with –min-contig-len 1000. The reads were mapped back to their assembly using Bowtie2 v2.3.5, then converted to the BAM format with SAMtools v1.9 (87). Single-sample metagenomic binning was applied using MetaBAT v2.11.2 with options –minContigLength 1500, producing 215 bins total. MAG quality was assessed using BBMap v38.82, QUAST v4.5 (88), and CheckM v1.0.7 (89). Quality was scored on the basis of the following criteria: high-quality draft: completion >90%, contamination <5%; medium-quality draft: completion ≥50%, contamination <10%; and low-quality draft: completion <50%, contamination <10%. Forty-eight samples did not meet these requirements (i.e., contamination ≥10%) and were excluded. The MAG taxonomy was assigned using Mash v2.2 (90) with a mash sketch of the NCBI RefSeq database (accessed 25 May 2021), reporting the genus of the top-scoring hit. MAGs were screened for known ARGs using AMRFinder v3.9.8 (73).
Statistical analysis.
All the statistical analysis has been done in R 4.1.2 using packages like vegan, cowplot, ggplot, pheatmap, phyloseq, ggpubr, and ggvegan (91). The Spearman’s correlation for ARG-taxa cooccurrence network was calculated in Jamovi v2.2. and was plotted in Cytoscape v3.9.1 (92, 93).
Ethics approval and consent to participate.
Not applicable. Preapproval from the ethical committee is not required in India to work on hospital sewage samples.
Data availability.
All metagenomic sequencing data are available at the National Center for Biotechnology Information (NCBI) database with BioProject accession number PRJNA682952 and SRA accession numbers SRR13227005, SRR13227004, SRR13227003, SRR13227002, SRR15384560, and SRR15384559.
ACKNOWLEDGMENTS
We thank Amina Usmani, Shamaila Tirmeez, Shahid Akhtar, Ali Dawar Mushtaque, and Mohd. Salman for their help in sample collection. The authors also thank the staff at The Edison Family Center for Genome Sciences and Systems Biology at the Washington University School of Medicine in St. Louis, including Eric Martin and Brian Koebbe for computational support, and Bonnie Dee, Kathleen Matheny, and Keith Page for administrative support.
A.U.K. is supported by the DBT, Government of India, grant number BT/PR40148/BTIS/137/20/2021. A.T. is supported by the Council of Scientific and Industrial Research in the form of CSIR-NET/SRF [09/112(0648)/2019-EMR-I]. K.S.B. is supported by the National Institute of Diabetes and Digestive and Kidney Diseases (T32-DK007130). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
We declare no conflict of interest.
A.U.K. conceived the idea and provided all resources. A.T. collected samples, extracted DNA, and generated shotgun sequencing data. A.T. and K.S.B. performed computational analyses and interpreted the results. A.T. and K.S.B. drafted the article with critical revisions from D.G. and A.U.K. All authors reviewed and approved the final manuscript.
Contributor Information
Asad U. Khan, Email: asadukhan72@gmail.com.
Jinxin Liu, Nanjing Agricultural University.
REFERENCES
- 1.Murray CJ, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, de Luca M, Dokova K, et al. 2022. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fischbach MA, Walsh CT. 2009. Antibiotics for emerging pathogens. Science 325:1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper MA, Shlaes D. 2011. Fix the antibiotics pipeline. Nature 472:32–32. doi: 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- 4.Mackenzie JS, Jeggo M. 2019. Tropical medicine and infectious disease the one health approach—why is it so important? TropicalMed 4:88. doi: 10.3390/tropicalmed4020088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blake KS, Choi JH, Dantas G. 2021. Approaches for characterizing and tracking hospital-associated multidrug-resistant bacteria. Cell Mol Life Sci 78:2585–2606. doi: 10.1007/s00018-020-03717-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’souza AW, Potter RF, Wallace M, Shupe A, Patel S, Sun X, Gul D, Kwon JH, Andleeb S, Burnham C-AD, Dantas G. 2019. Spatiotemporal dynamics of multidrug resistant bacteria on intensive care unit surfaces. Nat Commun 10:4569. doi: 10.1038/s41467-019-12563-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Al Aukidy M, Chalabi A, Verlicchi P, Al Aukidy M, Al Chalabi S, Verlicchi P. 2018. Hospital wastewaters-characteristics, management, treatment and environmental risks. Hdb Env Chem 60:171–188. doi: 10.1007/698_2017_5. [DOI] [Google Scholar]
- 8.Durso L, Alduina R, Li B, Zhang S, Huang J, Zhao Z, Cao Y. 2020. Hospital wastewater as a reservoir for antibiotic resistance genes: a meta-analysis. Analysis Front Public Health 8:574968. doi: 10.3389/fpubh.2020.574968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deogratias Ekwanzala M, Lehutso RF, Kasonga TK, Dewar JB, Ndombo M, Momba B. 2020. Environmental dissemination of selected antibiotics from hospital wastewater to the aquatic environment. Antibiotics 9:431. doi: 10.3390/antibiotics9070431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aarestrup FM, Woolhouse ME. 2020. Using sewage for surveillance of antimicrobial resistance. Science 367:630–632. doi: 10.1126/science.aba3432. [DOI] [PubMed] [Google Scholar]
- 11.Hendriksen RS, Munk P, Njage P, van Bunnik B, McNally L, Lukjancenko O, Röder T, Nieuwenhuijse D, Pedersen SK, Kjeldgaard J, Kaas RS, Clausen PTLC, Vogt JK, Leekitcharoenphon P, van de Schans MGM, Zuidema T, de Roda Husman AM, Rasmussen S, Petersen B, Bego A, Rees C, Cassar S, Coventry K, Collignon P, Allerberger F, Rahube TO, Oliveira G, Ivanov I, Vuthy Y, Sopheak T, Yost CK, Ke C, Zheng H, Baisheng L, Jiao X, Donado-Godoy P, Coulibaly KJ, Jergović M, Hrenovic J, Karpíšková R, Villacis JE, Legesse M, Eguale T, Heikinheimo A, Malania L, Nitsche A, Brinkmann A, Saba CKS, Kocsis B, Solymosi N, The Global Sewage Surveillance project consortium , et al. 2019. Global monitoring of antimicrobial resistance based on metagenomics analyses of urban sewage. Nat Commun 10:1124. doi: 10.1038/s41467-019-08853-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perry MR, Lepper HC, McNally L, Wee BA, Munk P, Warr A, Moore B, Kalima P, Philip C, de Roda Husman AM, Aarestrup FM, Woolhouse MEJ, van Bunnik BAD. 2021. Secrets of the hospital underbelly: patterns of abundance of antimicrobial resistance genes in hospital wastewater vary by specific antimicrobial and bacterial family. Front Microbiol 12:703560. doi: 10.3389/fmicb.2021.703560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ekwanzala MD, Dewar JB, Momba MNB. 2020. Environmental resistome risks of wastewaters and aquatic environments deciphered by shotgun metagenomic assembly. Ecotoxicol Environ Saf 197:110612. doi: 10.1016/j.ecoenv.2020.110612. [DOI] [PubMed] [Google Scholar]
- 14.Farooqui HH, Selvaraj S, Mehta A, Heymann DL. 2018. Community level antibiotic utilization in India and its comparison vis-à-vis European countries: evidence from pharmaceutical sales data. PLoS One 13:e0204805. doi: 10.1371/journal.pone.0204805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singh SK, Sengupta S, Antony R, Bhattacharya S, Mukhopadhyay C, Ramasubramanian V, Sharma A, Sahu S, Nirkhiwale S, Gupta S, Rohit A, Sharma S, Raghavan V, Barman P, Sood S, Mamtora D, Rengaswamy S, Arora A, Goossens H, Versporten A. 2019. Variations in antibiotic use across India: multi-centre study through Global Point Prevalence survey. J Hosp Infect 103:280–283. doi: 10.1016/j.jhin.2019.05.014. [DOI] [PubMed] [Google Scholar]
- 16.Browne AJ, Chipeta MG, Haines-Woodhouse G, Kumaran EPA, Hamadani BHK, Zaraa S, Henry NJ, Deshpande A, Reiner RC, Day NPJ, Lopez AD, Dunachie S, Moore CE, Stergachis A, Hay SI, Dolecek C. 2021. Global antibiotic consumption and usage in humans, 2000–18: a spatial modelling study. Lancet Planet Health 5:e893–e904. doi: 10.1016/S2542-5196(21)00280-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein EY, Milkowska-Shibata M, Tseng KK, Sharland M, Gandra S, Pulcini C, Laxminarayan R. 2021. Assessment of WHO antibiotic consumption and access targets in 76 countries, 2000–15: an analysis of pharmaceutical sales data. Lancet Infect Dis 21:107–115. doi: 10.1016/S1473-3099(20)30332-7. [DOI] [PubMed] [Google Scholar]
- 18.Auta A, Hadi MA, Oga E, Adewuyi EO, Abdu-Aguye SN, Adeloye D, Strickland-Hodge B, Morgan DJ. 2019. Global access to antibiotics without prescription in community pharmacies: a systematic review and meta-analysis. J Infect 78:8–18. doi: 10.1016/j.jinf.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 19.Porter G, Kotwani A, Bhullar L, Joshi J. 2021. Over-the-counter sales of antibiotics for human use in India: the challenges and opportunities for regulation. Med Law Int 21:147–173. doi: 10.1177/09685332211020786. [DOI] [Google Scholar]
- 20.Khare S, Purohit M, Sharma M, Tamhankar AJ, Lundborg CS, Diwan V, Pathak A. 2019. Antibiotic prescribing by informal healthcare providers for common illnesses: a repeated cross-sectional study in rural India. Antibiotics (Basel) 8:139. doi: 10.3390/antibiotics8030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pisani E. 2015. Antimicrobial resistance: what does medicine quality have to do with it? Antimicrobial Resistance and Medicine Quality. Pisani for AMR Review. https://amr-review.org/sites/default/files/ElizabethPisaniMedicinesQualitypaper.pdf.
- 22.Lamba M, Graham DW, Ahammad SZ. 2017. Hospital wastewater releases of carbapenem-resistance pathogens and genes in urban India. Environ Sci Technol 51:13906–13912. doi: 10.1021/acs.est.7b03380. [DOI] [PubMed] [Google Scholar]
- 23.Berglund F, Österlund T, Boulund F, Marathe NP, Larsson DGJ, Kristiansson E. 2019. Identification and reconstruction of novel antibiotic resistance genes from metagenomes. Microbiome 1 7:52. doi: 10.1186/s40168-019-0670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Government of India. 2011. Census of India. https://www.census2011.co.in/census/district. Accessed 13 July 2022.
- 25.Kaminski J, Gibson MK, Franzosa EA, Segata N, Dantas G, Huttenhower C. 2015. High-specificity targeted functional profiling in microbial communities with ShortBRED. PLoS Comput Biol 11:e1004557. doi: 10.1371/journal.pcbi.1004557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang A-N, Gaston JM, Dai CL, Zhao S, Poyet M, Groussin M, Yin X, Li L-G, van Loosdrecht MCM, Topp E, Gillings MR, Hanage WP, Tiedje JM, Moniz K, Alm EJ, Zhang T. 2021. An omics-based framework for assessing the health risk of antimicrobial resistance genes. Nat Commun 12:1. doi: 10.1038/s41467-021-25096-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan Y, Awan F, Zhenbao M, Zhang X, Zeng J, Zeng Z, Xiong W. 2020. Preliminary view of the global distribution and spread of the tet(X) family of tigecycline resistance genes. J Antimicrob Chemother 75:2797–2803. doi: 10.1093/jac/dkaa284. [DOI] [PubMed] [Google Scholar]
- 28.Gasparrini AJ, Markley JL, Kumar H, Wang B, Fang L, Irum S, Symister CT, Wallace M, Burnham C-AD, Andleeb S, Tolia NH, Wencewicz TA, Dantas G. 2020. Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance. Commun Biol 3:1–2. doi: 10.1038/s42003-020-0966-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talat A, Usmani A, Khan AU. 2022. Detection of E. coli IncX1 plasmid-mediated mcr-5.1 gene in an Indian hospital sewage water using shotgun metagenomic sequencing: a first report. Microb Drug Resist 28:759–764. doi: 10.1089/MDR.2021.0338. [DOI] [PubMed] [Google Scholar]
- 30.Alcock BP, Raphenya AR, Lau TT, Tsang KK, Bouchard e, Edalatmand A, Huynh W, Nguyen A-Lv, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FS, Hsiao WW, Domselaar G, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. doi: 10.1093/nar/gkz935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chereau F, Opatowski L, Tourdjman M, Vong S. 2017. Risk assessment for antibiotic resistance in South East Asia. BMJ 358:j3393. doi: 10.1136/bmj.j3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian GK, Soundari PG, Ramanathan V, Krishnan P. 2016. Endemic Indian clones of Klebsiella pneumoniae-harbouring New Delhi metallo-beta-lactamase-1 on a hybrid plasmid replicon type: a case of changing New Delhi metallo-beta-lactamase plasmid landscapes in India? Indian J Med Microbiol 34:286–292. doi: 10.4103/0255-0857.188314. [DOI] [PubMed] [Google Scholar]
- 33.Farhat N, Khan AU. 2020. Evolving trends of New Delhi Metallo-betalactamse (NDM) variants: a threat to antimicrobial resistance. Infect Genet Evol 86:104588. doi: 10.1016/j.meegid.2020.104588. [DOI] [PubMed] [Google Scholar]
- 34.Khan AU, Parvez S. 2014. Detection of blaNDM-4 in Escherichia coli from hospital sewage. J Med Microbiol 63:1404–1406. doi: 10.1099/jmm.0.076026-0. [DOI] [PubMed] [Google Scholar]
- 35.Parvez S, Khan AU. 2018. Hospital sewage water: a reservoir for variants of New Delhi metallo-β-lactamase (NDM)- and extended-spectrum β-lactamase (ESBL)-producing Enterobacteriaceae. Int J Antimicrob Agents 51:82–88. doi: 10.1016/j.ijantimicag.2017.08.032. [DOI] [PubMed] [Google Scholar]
- 36.Beg AZ, Khan AU. 2018. Genome analyses of bla NDM-4 carrying ST 315 Escherichia coli isolate from sewage water of one of the Indian hospitals. Gut Pathog 10:17. doi: 10.1186/s13099-018-0247-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Acman M, Wang R, van Dorp L, Shaw LP, Wang Q, Luhmann N, Yin Y, Sun S, Chen H, Wang H, Balloux F. 2022. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene bla NDM. Nat Commun 13:1–3. doi: 10.1038/s41467-022-28819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahmed OB, Asghar AH, Bahwerth FS, Assaggaf HM, Bamaga MA. 2021. The prevalence of aminoglycoside-resistant genes in Gram-negative bacteria in tertiary hospitals. Appl Nanosci (Switzerland) 17:1–7. doi: 10.1007/s13204-021-01887-4. [DOI] [Google Scholar]
- 39.Rodríguez-Beltrán J, DelaFuente J, León-Sampedro R, MacLean RC, San Millán Á. 2021. Beyond horizontal gene transfer: the role of plasmids in bacterial evolution. Nat Rev Microbiol 19:347–359. doi: 10.1038/s41579-020-00497-1. [DOI] [PubMed] [Google Scholar]
- 40.Liu Z, Yao J, Ma H, Rukeya A, Liang Z, Du W, Chen Y. 2022. Bacterial hosts and genetic characteristics of antibiotic resistance genes in wastewater treatment plants of Xinjiang (China) revealed by metagenomics Appl Sci 12:3100. doi: 10.3390/app12063100. [DOI] [Google Scholar]
- 41.Mancuso G, Midiri A, Gerace E, Biondo C. 2021. Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10:1310. doi: 10.3390/pathogens10101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.World Health Organization. 2017. Prioritization of pathogens to guide discovery, research and development of new antibiotics for drug-resistant bacterial infections, including tuberculosis. World Health Organization, Geneva, Switzarlend. [Google Scholar]
- 43.Deng C, Liu X, Li L, Shi J, Guo W, Xue J. 2020. Temporal dynamics of antibiotic resistant genes and their association with the bacterial community in a water-sediment mesocosm under selection by 14 antibiotics. Environ Int 137:105554. doi: 10.1016/j.envint.2020.105554. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y, Fu H, Yang H, Wu J, Chen Z, Jiang H, Liu M, Liu Q, Huang L, Gao J, Chen C. 2022. Extensive metagenomic analysis of the porcine gut resistome to identify indicators reflecting antimicrobial resistance. Microbiome 10:39. doi: 10.1186/s40168-022-01241-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Newman JW, Floyd Rv, Fothergill JL. 2017. The contribution of Pseudomonas aeruginosa virulence factors and host factors in the establishment of urinary tract infections. FEMS Microbiol Lett 364:124. doi: 10.1093/femsle/fnx124. [DOI] [PubMed] [Google Scholar]
- 46.Bowler P, Murphy C, Wolcott R. 2020. Biofilm exacerbates antibiotic resistance: is this a current oversight in antimicrobial stewardship? Antimicrob Resist Infect Control 9:162. doi: 10.1186/s13756-020-00830-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wareham D, Dickey BF, Korotkov KV, Devreese B, Depluverez S, Devos S. 2016. The role of bacterial secretion systems in the virulence of gram-negative airway pathogens associated with cystic fibrosis. Front Microbiol 7:1336. doi: 10.3389/fmicb.2016.01336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marathe NP, Berglund F, Razavi M, Pal C, Dröge J, Samant S, Kristiansson E, Joakim Larsson DG. 2019. Sewage effluent from an Indian hospital harbors novel carbapenemases and integron-borne antibiotic resistance genes. Microbiome 7:97. doi: 10.1186/s40168-019-0710-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Burcham ZM, Schmidt CJ, Pechal JL, Brooks CP, Rosch JW, Benbow ME, Jordan HR. 2019. Detection of critical antibiotic resistance genes through routine microbiome surveillance. PLoS One 14:e0213280. doi: 10.1371/journal.pone.0213280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cai L, Sun J, Yao F, Yuan Y, Zeng M, Zhang Q, Xie Q, Wang S, Wang Z, Jiao X. 2021. Antimicrobial resistance bacteria and genes detected in hospital sewage provide valuable information in predicting clinical antimicrobial resistance. Sci Total Environ 795:148815. doi: 10.1016/j.scitotenv.2021.148815. [DOI] [PubMed] [Google Scholar]
- 51.Mubedi JI, Devarajan N, Le Faucheur S, Mputu JK, Atibu EK, Sivalingam P, Prabakar K, Mpiana PT, Wildi W, Poté J. 2013. Effects of untreated hospital effluents on the accumulation of toxic metals in sediments of receiving system under tropical conditions: case of South India and Democratic Republic of Congo. Chemosphere 93:1070–1076. doi: 10.1016/j.chemosphere.2013.05.080. [DOI] [PubMed] [Google Scholar]
- 52.Ayenew Z, Tigabu E, Syoum E, Ebrahim S, Assefa D, Tsige E. 2021. Multidrug resistance pattern of Acinetobacter species isolated from clinical specimens referred to the Ethiopian Public Health Institute: 2014 to 2018 trend anaylsis. PLoS One 16:e0250896. doi: 10.1371/journal.pone.0250896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Langendonk RF, Neill DR, Fothergill JL. 2021. The building blocks of antimicrobial resistance in pseudomonas aeruginosa: implications for current resistance-breaking therapies. Front Cell Infect Microbiol 11:665759. doi: 10.3389/fcimb.2021.665759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hrenovic J, Goic-Barisic I, Kazazic S, Kovacic A, Ganjto M, Tonkic M. 2016. Carbapenem-resistant isolates of Acinetobacter baumannii in a municipal wastewater treatment plant, Croatia, 2014. Euro Surveill 21:30195. doi: 10.2807/1560-7917.ES.2016.21.15.30195. [DOI] [PubMed] [Google Scholar]
- 55.Slekovec C, Plantin J, Cholley P, Thouverez M, Talon D, Bertrand X, Hocquet D. 2012. Tracking down antibiotic-resistant Pseudomonas aeruginosa isolates in a wastewater network. PLoS One 7:e49300. doi: 10.1371/journal.pone.0049300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Peter S, Oberhettinger P, Schuele L, Dinkelacker A, Vogel W, Dörfel D, Bezdan D, Ossowski S, Marschal M, Liese J, Willmann M. 2017. Genomic characterisation of clinical and environmental Pseudomonas putida group strains and determination of their role in the transfer of antimicrobial resistance genes to Pseudomonas aeruginosa. BMC Genomics 18:859. doi: 10.1186/s12864-017-4216-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hsia Y, Sharland M, Jackson C, Wong ICK, Magrini N, Bielicki JA. 2019. Consumption of oral antibiotic formulations for young children according to the WHO Access, Watch, Reserve (AWaRe) antibiotic groups: an analysis of sales data from 70 middle-income and high-income countries. Lancet Infect Dis 19:67–75. doi: 10.1016/S1473-3099(18)30547-4. [DOI] [PubMed] [Google Scholar]
- 58.Goel N, Wattal C, Oberoi JK, Raveendran R, Datta S, Prasad KJ. 2011. Trend analysis of antimicrobial consumption and development of resistance in non-fermenters in a tertiary care hospital in Delhi, India. J Antimicrob Chemother 66:1625–1630. doi: 10.1093/jac/dkr167. [DOI] [PubMed] [Google Scholar]
- 59.Shankar C, Mathur P, Venkatesan M, Pragasam AK, Anandan S, Khurana S, Veeraraghavan B. 2019. Rapidly disseminating bla OXA-232 carrying Klebsiella pneumoniae belonging to ST231 in India: multiple and varied mobile genetic elements. BMC Microbiol 19:137. doi: 10.1186/s12866-019-1513-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sharma S, Banerjee T, Kumar A, Yadav G, Basu S. 2022. Extensive outbreak of colistin resistant, carbapenemase (bla OXA-48, bla NDM) producing Klebsiella pneumoniae in a large tertiary care hospital, India. Antimicrob Resist Infect Control 11:1. doi: 10.1186/s13756-021-01048-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yao S, Ye J, Yang Q, Hu Y, Zhang T, Jiang L, Munezero S, Lin K, Cui C. 2021. Occurrence and removal of antibiotics, antibiotic resistance genes, and bacterial communities in hospital wastewater. Environ Sci Pollut Res Int 28:57321–57333. doi: 10.1007/s11356-021-14735-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gotkowska-Płachta A. 2021. The prevalence of virulent and multidrug-resistant enterococci in river water and in treated and untreated municipal and hospital wastewater. IJERPH 18:563–519. doi: 10.3390/ijerph18020563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kumar GS, Joseph N. 2012. Drainage and sewerage system in urban India: need for action. Indian J Occup Environ Med 16:150–151. doi: 10.4103/0019-5278.111767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thermo Fisher Scientific. Qubit 3.0 fluorometer catalog number Q33216. https://tools.thermofisher.com/content/sfs/manuals/qubit_3_fluorometer_man.pdf.
- 65.Babraham Bioinformatics. FastQC a quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 66.Ewels P, Ns Magnusson M, Lundin S, Aller MK. 2016. Data and text mining MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32:3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. 2015. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 68.Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen S, Zhou Y, Chen Y, Gu J. 2018. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wood DE, Lu J, Langmead B. 2019. Improved metagenomic analysis with Kraken 2. Genome Biol 20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH, UniProt Consortium . 2015. UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31:926–932. doi: 10.1093/bioinformatics/btu739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MOA, Dantas G. 2012. The shared antibiotic resistome of soil bacteria and human pathogens. Science 337:1107–1111. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Moore AM, Patel S, Forsberg KJ, Wang B, Bentley G, Razia Y, Qin X, Tarr PI, Dantas G. 2013. Pediatric fecal microbiota harbor diverse and novel antibiotic resistance genes. PLoS One 8:e78822. doi: 10.1371/journal.pone.0078822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore AM, Ahmadi S, Patel S, Gibson MK, Wang B, Ndao IM, Deych E, Shannon W, Tarr PI, Warner BB, Dantas G. 2015. Gut resistome development in healthy twin pairs in the first year of life. Microbiome 3:27. doi: 10.1186/s40168-015-0095-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pehrsson EC, Tsukayama P, Patel S, Mejía-Bautista M, Sosa-Soto G, Navarrete KM, Calderon M, Cabrera L, Hoyos-Arango W, Bertoli MT, Berg DE, Gilman RH, Dantas G. 2016. Interconnected microbiomes and resistomes in low-income human habitats. Nature 533:212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, Dantas G. 2014. Bacterial phylogeny structures soil resistomes across habitats. Nature 509:612–616. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clemente JC, Pehrsson EC, Blaser MJ, Sandhu K, Gao Z, Wang B, Magris M, Hidalgo G, Contreras M, Noya-Alarcón Ó, Lander O, McDonald J, Cox M, Walter J, Oh PL, Ruiz JF, Rodriguez S, Shen N, Song SJ, Metcalf J, Knight R, Dantas G, Dominguez-Bello MG. 2015. The microbiome of uncontacted Amerindians. Sci Adv 1:e1500183. doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gibson MK, Wang B, Ahmadi S, Burnham CAD, Tarr PI, Warner BB, Dantas G. 2016. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol 1:16024. doi: 10.1038/nmicrobiol.2016.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsukayama P, Boolchandani M, Patel S, Pehrsson EC, Gibson MK, Chiou KL, Jolly CJ, Rogers J, Phillips-Conroy JE, Dantas G. 2018. Characterization of wild and captive baboon gut microbiota and their antibiotic resistomes. mSystems 3:e00016-18. doi: 10.1128/mSystems.00016-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM, Tarr PI, Warner BB, Dantas G. 2019. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol 4:2285–2297. doi: 10.1038/s41564-019-0550-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Shen W, Xiong J. 2019. TaxonKit: a cross-platform and efficient NCBI taxonomy toolkit. Biorxiv doi: 10.1101/513523. [DOI]
- 84.Seemann T. 2014. Genome analysis Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 85.S Pellow D, Mizrahi I, Shamir R. 2020. PlasClass improves plasmid sequence classification. PLoS Comput Biol 16:e1007781. doi: 10.1371/journal.pcbi.1007781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Krawczyk PS, Lipinski L, Dziembowski A. 2018. PlasFlow: predicting plasmid sequences in metagenomic data using genome signatures. Nucleic Acids Res 46:e35. doi: 10.1093/nar/gkx1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Danecek P, Bonfield JK, Liddle J, Marshall J, Ohan V, Pollard MO, Whitwham A, Keane T, McCarthy SA, Davies RM, Li H. 2021. Twelve years of SAMtools and BCFtools. Gigascience 10:giab008. doi: 10.1093/gigascience/giab008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gurevich A, Saveliev V, Vyahhi N, Tesler G. 2013. QUAST: quality assessment tool for genome assemblies. Bioinformatics 29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ondov BD, Treangen TJ, Melsted P, Mallonee AB, Bergman NH, Koren S, Phillippy AM. 2016. Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 17:132. doi: 10.1186/s13059-016-0997-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.R Core Team. 2021. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 92.The jamovi project. 2021. jamovi (Version 1.6) (computer software). https://www.jamovi.org.
- 93.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All metagenomic sequencing data are available at the National Center for Biotechnology Information (NCBI) database with BioProject accession number PRJNA682952 and SRA accession numbers SRR13227005, SRR13227004, SRR13227003, SRR13227002, SRR15384560, and SRR15384559.