ABSTRACT
This 4-month-long prospective observational study investigated the epidemiological characteristics, genetic composition, transmission pattern, and infection control of carbapenem-resistant Escherichia coli (CREC) colonization in patients at an intensive care unit (ICU) in China. Phenotypic confirmation testing was performed on nonduplicated isolates from patients and their environments. Whole-genome sequencing was performed for all E. coli isolates, followed by multilocus sequence typing (MLST), and antimicrobial resistance genes and single nucleotide polymorphisms (SNPs) were screened. The colonization rates of CREC were 7.29% from the patient specimens and 0.39% from the environmental specimens. Among the 214 E. coli isolates tested, 16 were carbapenem resistant, with the blaNDM-5 gene identified as the dominant carbapenemase-encoding gene. Among the low-homology sporadic strains isolated in this study, the main sequence type (ST) of carbapenem-sensitive Escherichia coli (CSEC) was ST1193, whereas the majority of CREC isolates belonged to ST1656, followed by ST131. CREC isolates were more sensitive to disinfectants than were the carbapenem-resistant Klebsiella pneumoniae (CRKP) isolates obtained in the same period, which may explain the lower separation rate. Therefore, effective interventions and active screening are beneficial to the prevention and control of CREC.
IMPORTANCE CREC represents a public health threat worldwide, and its colonization precedes or occurs simultaneously with infection; once the colonization rate increases, the infection rate rises sharply. In our hospital, the colonization rate of CREC remained low, and almost all of the CREC isolates detected were ICU acquired. Contamination of the surrounding environment by CREC carrier patients shows a very limited spatiotemporal distribution. As the dominant ST of the CSEC isolates found, ST1193 CREC might be considered a strain of notable concern with potential to cause a future outbreak. ST1656 and ST131 also deserve attention, as they comprised the majority of the CREC isolates found, while blaNDM-5 gene screening should play an important role in medication guidance as the main carbapenem resistance gene identified. The disinfectant chlorhexidine, which is used commonly in the hospital, is effective for CREC rather than CRKP, possibly explaining the lower positivity rate for CREC than for CRKP.
KEYWORDS: CREC, carbapenemases, transmission, whole-genome sequencing, disinfectant
INTRODUCTION
Carbapenem-resistant Escherichia coli (CREC) infections are an increasingly severe public health problem, imposing a heavy blow to socioeconomic and health care delivery systems, potentially even causing a global health crisis. CREC is prevalent in developed countries, and its widespread transmission tends to occur in contaminated health care settings rather than in the community. The most common mode of transmission is from one patient to another, primarily via the hands (1). Prior to 2001, the Greek Antimicrobial Drug Resistance Surveillance System reported a carbapenem resistance prevalence of <1%, which increased to 30% in hospital wards and 60% in intensive care units (ICUs) within 7 years (2). Studies in the United States demonstrate that for every 1% increase in colonization pressure, the CREC infection odds increase by 15%. In a study conducted in the Chicago, IL, area, 30% of long-term-hospitalized patients were found to be CREC carriers, and the rate was 3.3% for patients in the ICU. Hospitals have become the major sites of CREC infection transmission (3). E. coli is normally found in the human intestinal tract and can cause diseases under certain conditions.
The causes of carbapenem resistance are complex and heterogeneous. A recent CRACKLE-2 study found that 31% of CREC isolates from the United States contain carbapenemases (4). This rate is as high as 90% in the Middle East and Africa. Geographic heterogeneity is a new challenge in CREC control. The mortality rate of patients with CREC infections in the ICU reaches up to 45.5%, and the infection prolongs hospital stays and increases the economic burden on patients (5).
As a worldwide emphasis on CREC research continues, the World Health Organization has identified CREC as a critical priority pathogen for novel drug development. In addition to the ongoing development of new treatments, strengthening antimicrobial stewardship will be vital to ensuring the continued effectiveness of these treatments in the coming years. The epidemiology and wealth of data generated have demonstrated the urgent need for management of CREC; however, the scientific quality of the evidence supporting the management of CREC infection, especially in assessing antibiotic effectiveness in complex patients, remains low due to the lack of strong randomized controlled trials (6).
Given the lack of data on CREC in the ICU in China, the aim of this study was to explore its relevant epidemiological characteristics. Genomic methods were used to understand the distribution of CREC, genetic pattern, genetic relationship to the corresponding CREC, and relationship with CREC transformation. Toward this end, we conducted a prospective observational study in a 28-bed ICU of a tertiary teaching hospital in China. Genomic sequences were recorded, along with epidemiological and medication administration data, to investigate the diversity of CREC in the ICU and the association between CREC infection, the host, and the environment.
RESULTS
Study design and CREC collection.
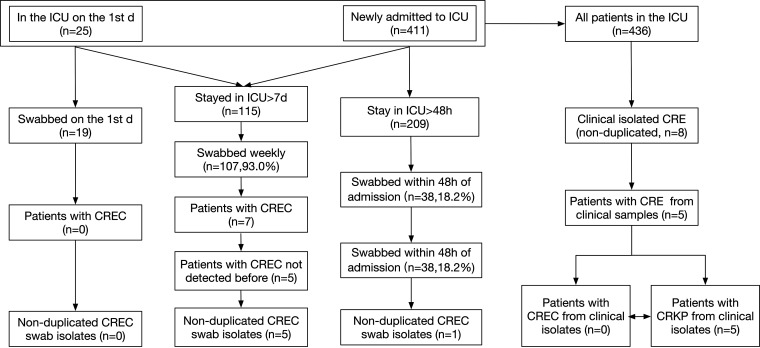
The study duration was 4 months, with 17 weekly sample collections from 436 patients. On day 1 (1 April 2021), 25 patients were admitted to the ICU (Fig. 1). We collected samples, including rectal swabs and oropharyngeal swabs, from 19 of the 25 patients, none of which contained CREC. Of the 411 new ICU admissions during the study period, 209 were admitted to the ICU for longer than 48 h (Fig. 1). Samples (i.e., oropharyngeal and rectal swabs) were collected from 38 of these 209 patients (18.2%) within 48 h of admission. Only 1 of these 38 patients tested positive for CREC. Patient 4 (P4) was admitted to a local hospital with a suspected trauma infection. We considered this patient’s CREC infection to be non-ICU acquired, as CREC was detected on the rectal and gastric tube swabs on the second day of his transfer to the ICU. The carriage rate of CREC at admission was 2.6% (1/38).
FIG 1.
Strain screening method for clinical samples in this study. Strains isolated from oropharyngeal swabs, rectal swabs, nasogastric tubes, nasogastric tubes, tracheal intubation tubes, and tracheotomy tube swabs from the same patient were defined as nonduplicate samples, while strains isolated from the same swab type from the same patient in the same week were defined as duplicate samples. Patients who were already in the intensive care unit (ICU) on the first day of the study without detection of carbapenem-resistant Escherichia coli (CREC) or who were newly admitted to the ICU with negative results of CREC screening within 48 h were defined as patients without CREC.
Five patients were believed to have ICU-acquired CREC. The first patient (P2) had a negative rectal swab on day 5 of ICU admission, followed by two positive swabs thereafter. The second patient (P3) had negative rectal swabs on days 2 and 9 after ICU admission and a positive rectal swab on day 16. The third patient (P6) was identified to have carbapenem-sensitive E. coli (CSEC) in a rectal swab on day 9 after admission, and CREC was detected in a gastric tube sample taken on day 16 of his ICU stay. The fourth patient (P7) had negative rectal, tracheal intubation, and nasogastric tube swabs on day 4 after ICU admission, and CREC was identified in a rectal swab on day 11 after ICU admission. The last patient (P8) was identified to have CSEC in the first two rectal swabs after ICU admission, and CREC was detected in the rectal swab on day 17.
During the study period, eight nonreplicate clinical isolated carbapenem-resistant Enterobacteriaceae (CRE) were found in five different patients: five with carbapenem-resistant Klebsiella pneumoniae (CRKP) and none with CREC (Fig. 1).
Screening for CREC in environmental samples.
We collected a total of 5,824 environmental specimens, 23 of which were positive for E. coli, thus leading to a detection rate of 0.39%. The 23 E. coli-positive environmental samples were from 18 patients, distributed in 13 of 28 beds. E. coli was detected in several environmental samples, including the overflow wall, inner surface of the downspout, nebulizer, bedside table, bed rails, ventilator, micropump, bed linen, hanging cabinet, faucet surface, stethoscope, bed regulator, and locker. Among these 23 samples in which E. coli was detected, 3 were CREC-positive environmental samples from the nebulizer (P4, DY315), bedside cabinet (P4, DY316), and faucet surface (P9, DY317). One patient (P4) had CREC of the same strain, confirmed by genotype sequencing, detected in his rectal swab, nasointestinal tube, and surroundings (nebulizer and bedside table). The other patient (P9) did not carry CREC, but CREC was detected on the faucet surface of his room. Genome sequencing revealed that this strain was homologous (difference of 22 SNPs) to that found in the rectal swab sampled from a patient (P5) in the adjacent room during the same period. The weekly sampling during this patient’s (P4) hospitalization identified two nonduplicate environmental samples of CREC, from the nebulizer (P4, DY315) and bedside table (P4, DY316), only for the duration of 1 week.
Molecular characteristics of E. coli.
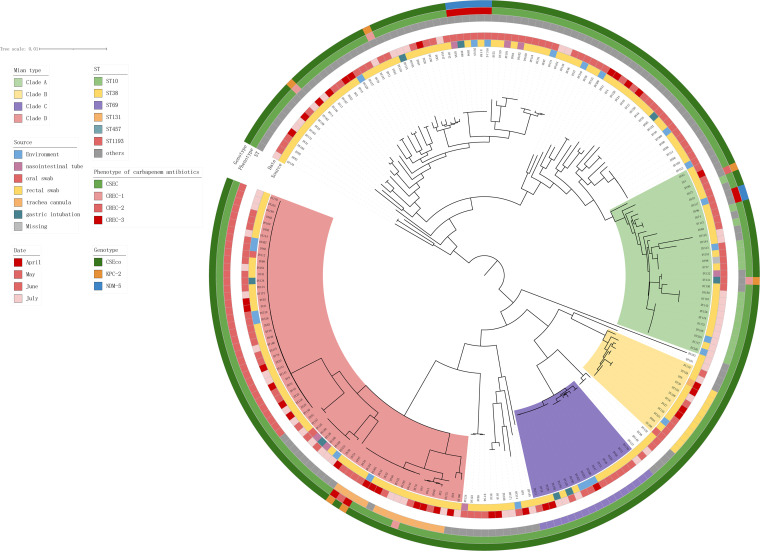
A total of 214 E. coli strains were isolated, of which 191 were isolated from 99 different patients and 23 were isolated from the surrounding environment in the ICU ward. Multilocus sequence typing (MLST) divided the 214 E. coli isolates into four clades: clade A, containing 29 strains, mainly sequence type (ST) 10; clade B, containing 12 strains, mainly ST38; clade C, containing 17 strains, mainly ST69; and clade D, containing 64 strains, mainly ST131 and ST1193 (Fig. 2). Among the strains isolated in this study, ST1193 (n = 36 [16.82%]) was the most prevalent, and all of them clustered in clade D. This was followed by ST10 (n = 18 [8.41%]), ST69 (n = 17 [7.94%]), and ST131 (n = 15 [7.01%]). In clade A, we found four strains of CREC, including two with the blaKPC-2 gene, both with blaNDM-5, and none with ST10. In clade D, we found four strains of CREC, including two with the blaKPC-2 genes and two without carbapenem resistance genes, all belonging to ST131. The remaining strains exhibiting carbapenem resistance did not belong to the above four clades, whereas previous related studies have shown that most CREC strains belong to ST167, followed by ST410 and ST131, and most carry various blaNDM-5 genes (7). ST1193 was the common ST in the E. coli detected in this study (46 strains [21.5%]). In contrast, most of these carbapenem-resistant strains did not belong to the aforementioned STs.
FIG 2.
Phylogenetic tree constructed by the maximum likelihood method reflecting the genetic relationships among all 214 E. coli strains. From inside to outside, the main branches (divided into four branches), sample source, collection time, possible infection site, sequence type (ST), phenotype, and genotype for all E. coli strains are successively listed in each layer. In the phenotype circle, we defined E. coli resistance to one, two, and three kinds of carbapenem drugs as CREC-1, CREC-2, and CREC-3, respectively.
CREC characterization.
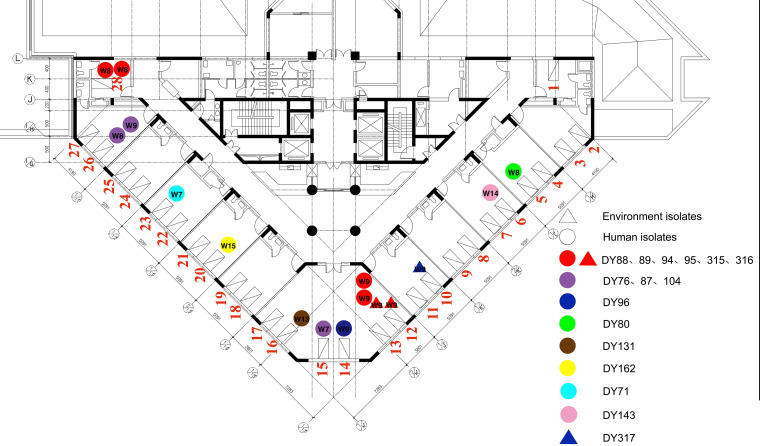
Sixteen carbapenem-resistant E. coli strains, from patients occupying 11 of the 28 beds, were detected in this study (Fig. 3). Two carbapenemases were detected among them: KPC-2 and NDM-5. NDM-5 was the most common (50.00% [8/16]), followed by KPC-2 (37.50% [6/16]), whereas no carbapenemase was detected in two isolates (12.50% [2/16]). Moreover, 15 other β-lactamases were detected (Table 1). According to the antimicrobial susceptibility test results, the rate of resistance to meropenem was 64.70% (11/17), with a MIC range of 0.03 to >256 mg/L. The rate of resistance to imipenem was 64.70% (11/17), with a MIC range of 0.25 to >256 mg/L. The rate of resistance to ertapenem was 100%. The MIC values and ranges for the nine different antibiotics were tested (Table 1).
FIG 3.
ICU floor sketch map and overall distribution of CREC strains. The ICU on the fourth floor of the hospital has a total of 28 beds, including two single rooms (no. 1 and 28), one six-bed room (no. 12, 13, 14, 15, 16, and 17), and 10 two-bed rooms (remaining rooms). Strains from the same bed can be a superposition of positive results collected at different times. Clinical samples are represented by circles, and environmental samples are represented by triangles. The number of the week when the sample was collected is shown on the circle or triangle. Different colors represent CREC strains with different single nucleotide polymorphism (SNP) typing.
TABLE 1.
MIC values of all CREC strains for three carbapenems and other antibiotics, their STs, the genotypes of carbapenemases, the Inc type of carbapenem-carrying plasmid, and other β-lactamase genesa
| Patient ID | Isolated strain | MLST | Carbapenemase | Inc type of carbapenem-carrying plasmid | Other β-lactamase(s) | MIC (mg/L) |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| MEM | IPM | ETP | COL | TGC | CZA | ||||||
| 1 | DY71 | ST101 | KPC-2 | IncFII | CTX-M-14, TEM-1, EC-18 | 1 | 2 | 4 | 0.5 | 0.25 | 0.5 |
| 2 | DY76 | ST131 | CTX-M-27, EC-5 | 4 | 2 | 32 | 1 | 0.125 | 0.5 | ||
| 3 | DY80 | ST3337 | KPC-2 | IncN | CTX-M-3, TEM-1, EC | 1 | 4 | 4 | 1 | 0.5 | 0.25 |
| 2 | DY87 | ST131 | KPC-2 | IncN | CTX-M-27, CTX-M-3, TEM-1, EC-5 | >256 | >256 | >256 | 1 | 0.25 | 1 |
| 4 | DY88 | ST1656 | NDM-5 | IncHI2 | OXA-10, TEM-1, EC-18 | 128 | 256 | 128 | 0.5 | 0.25 | >128 |
| 4 | DY89 | ST1656 | NDM-5 | IncHI2 | OXA-10, TEM-1, EC-18 | 8 | 16 | 32 | 1 | 0.125 | >128 |
| 4 | DY94 | ST1656 | NDM-5 | IncHI2 | OXA-10, TEM-1, EC-18 | 16 | 16 | 32 | 0.5 | 0.25 | >128 |
| 4 | DY95 | ST1656 | NDM-5 | IncHI2 | OXA-10, TEM-1, EC-18 | 4 | 8 | 16 | 0.5 | 0.25 | >128 |
| 5 | DY96 | ST6913 | NDM-5 | IncFIB/IncFIC | EC | 8 | 8 | 32 | 1 | 0.25 | >128 |
| 2 | DY104 | ST131 | KPC-2 | IncN | CTX-M-27, CTX-M-3, TEM-1, EC-5 | >256 | >256 | >256 | 0.5 | 0.125 | 1 |
| 6 | DY131 | ST744 | KPC-2 | IncN | CTX-M-14, CTX-M-3, TEM-1, EC | 2 | 2 | 4 | 0.5 | 0.125 | 0.5 |
| 7 | DY143 | ST131 | CTX-M-14, CTX-M-15, OXA-1, TEM-1, EC-5 | 2 | 1 | 16 | 0.5 | 0.125 | 0.25 | ||
| 8 | DY162 | ST410 | KPC-2 | IncN | CTX-M-3, TEM-1, EC-15 | 1 | 1 | 2 | 1 | 0.125 | <0.0625 |
| 4 | DY315 | ST1656 | NDM-5 | IncHI2 | OXA-10, TEM-1, EC-18 | 8 | 4 | 4 | 0.5 | 0.25 | >128 |
| 4 | DY316 | ST1656 | NDM-5 | IncHI2 | OXA-10, TEM-1, EC-18 | 16 | 16 | 32 | 1 | 0.125 | >128 |
| 9 | DY317 | ST6913 | NDM-5 | IncFIB/IncFIC | EC | 16 | 16 | 16 | 1 | 0.5 | >128 |
CREC, carbapenem-resistant E. coli; ST, sequence type; ID, identifier; MLST, multilocus sequence type; MEM, meropenem; IMP, imipenem; ETP, ertapenem; COL, colistin or polymyxin E; TGC, tigecycline; CZA, ceftazidime-avibactam.
MLST analysis classified the 16 carbapenem-resistant E. coli strains into seven different STs (Table 1). ST1656 was predominant (37.50% [6/16]), followed by ST131 (25.00% [4/16]) and ST6913 (12.50% [2/16]).
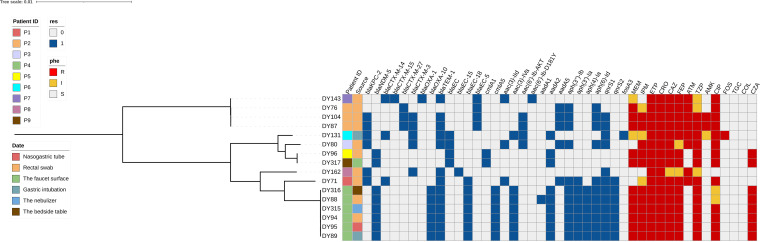
The presence of known resistance genes was screened in 16 carbapenem-resistant isolates. In total, 29 acquired antibiotic resistance genes were identified, conferring resistance to five classes of antibiotics. These included genes encoding extended-spectrum β-lactamases (blaCTX-M-14, blaCTX-M-15, blaCTX-M-27, blaCTX-M-3, blaOXA-1, and blaOXA-10), genes conferring resistance to aminoglycoside antibiotics [aac(3)-IId, aac(3)-Iva, aac(6′)-Ib-AKT, aac(6′)-Ib-D181Y, aadA1, aadA2, aadA5, aph(3″)-Ib, aph(3′)-Ia, aph(4)-Ia, and aph(6)-Id], genes conferring resistance to quinolone antimicrobials (qnrS1, qnrS2, oqxA10, and oqxB17), genes conferring resistance to fosfomycin antibiotics (fosA3), and genes conferring resistance to chloramphenicol antibiotics (cmlA1 and cmlA5) (Fig. 4).
FIG 4.
Resistance genotypes and drug-sensitive phenotypes of 16 nonreplicate samples of CREC detected in 9 patients categorized into four classes of antibiotics (β-lactams, aminoglycosides, quinolones, and fosfomycin) that are mainly used as clinical anti-infection strategies.
Isolates DY96 and DY317, from different patients, were identified to be ST10 and had a close evolutionary relationship, based on a short distance in the phylogenetic tree. Single nucleotide polymorphism (SNP) analysis revealed that they differed by 22 SNPs, indicating that they were of the same origin and that transmission between patient and environment had likely occurred.
A total of five different plasmids carrying carbapenem resistance genes were obtained from 16 strains of CREC isolated from nine patients, three carrying blaKPC-2 and two carrying blaNDM-5. The blaKPC-2-carrying plasmid of DY71 (isolated from patient 1) belonged to the IncFII replicon and was identical to the blaKPC-2-harboring plasmid pC110-KPC (GenBank accession number CP047692.1) isolated from Serratia marcescens strain C110 (see Fig. S1 in the supplemental material). The blaKPC-2 gene in DY71 was flanked by ISKpn27 and ISKpn6 (Fig. S1). Strains DY80, DY87, DY104, and DY131 (isolated from patients 2, 3, and 6) had the same blaKPC-2-carrying IncN-type plasmid that was highly similar to pCRKP-1-KPC (GenBank accession number KX928750.1) carried by a K. pneumoniae strain (Fig. S2). These blaKPC-2 genes were flanked by IS26, ISKpn27, and ISKpn19 (Fig. S2). It is worth noting that the blaKPC-2-harboring plasmid of the DY162 strain (isolated from patient 8) is 36,597 bp and its nucleotide sequence is the same as a part of the pCRKP-1-KPC plasmid mentioned above (Fig. S2). Unfortunately, the blaNDM-5-carrying plasmid in this study could not be assembled completely. Based on the obtained long contig, we found that strains DY88, DY89, DY94, DY95, DY315, and DY316 (isolated from patient 4) had the same blaNDM-5-carrying plasmid that was highly similar to the reported pEC6622-1 isolated from E. coli (GenBank accession number CP096588). This plasmid belonged to the IncHI2 replicon, and the blaNDM-5 gene was flanked by IS3000, ISKpn26, and IS26 (Fig. S3). We also found that strains DY96 and DY317 (isolated from patients 5 and 9) had the same blaNDM-5-carrying plasmid, which was highly similar to the reported plasmid pYUXYEH3783-NDM (GenBank accession number CP110998) (Fig. S4). This plasmid belongs to the IncFIB/IncFIC replicons, and blaNDM-5 is surrounded by IS26, ISKpn26, and ISAba125.
Disinfectant sensitivity of the strains.
To explore why CREC was isolated at a much lower frequency than CRKP, we evaluated the resistance of these strains to disinfectants. We also identified 16 CRKP strains that belong to multiple STs corresponding to the same period of collection as the 16 CREC strains (Table 2). The chlorhexidine MICs for 16 CREC strains ranged from 0.5 to 1 mg/L, while the corresponding MIC range for 16 CRKP strains was 2 to 16 mg/L. The MIC90 for CRKP was eight times higher than that for CREC. However, the MICs of p-chlorophenol and calcium hypochlorite for the 16 CREC strains and 16 CRKP strains presented high-level resistance (MIC>256 mg/L) with no significant difference (Table 3).
TABLE 2.
Information on 16 CREC and CRKP strainsa
| Strain date (mo.day) | CREC |
CRKP |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient ID | Isolate no. | Bed no. | Sample type | ST | Patient ID | Isolate no. | Bed no. | Sample type | ST | |
| 5.18 | 1 | DY71 | 23 | Rectal | 101 | 10 | kp1239 | 8 | Rectal | 15 |
| 5.18 | 2 | DY76 | 15 | Rectal | 131 | 2 | kp1240 | 15 | Rectal | 43 |
| 5.25 | 3 | DY80 | 5 | Rectal | 3337 | 3 | kp1282 | 5 | Rectal | 43 |
| 5.25 | 2 | DY87 | 26 | Rectal | 131 | 2 | kp1306 | 26 | Rectal | 43 |
| 5.25 | 4 | DY88 | 28 | Rectal | 1656 | 11 | kp1283 | 21 | Rectal | 11 |
| 5.25 | 4 | DY89 | 28 | Gastric intubation | 1656 | 12 | kp1292 | 11 | Gastric intubation | 43 |
| 6.01 | 4 | DY94 | 12 | Rectal | 1656 | 13 | kp1326 | 19 | Rectal | 15 |
| 6.01 | 4 | DY95 | 12 | Nasogastric tube | 1656 | 14 | kp1319 | 20 | Nasogastric tube | 11 |
| 6.01 | 5 | DY96 | 14 | Rectal | 6913 | 15 | kp1338 | 4 | Rectal | 11 |
| 6.01 | 2 | DY104 | 26 | Rectal | 131 | 2 | kp1328 | 26 | Rectal | 43 |
| 6.29 | 6 | DY131 | 16 | Gastric intubation | 744 | 6 | kp1442 | 16 | Gastric intubation | 43 |
| 7.06 | 7 | DY143 | 6 | Rectal | 131 | 15 | kp1454 | 23 | Rectal | 11 |
| 7.13 | 8 | DY162 | 20 | Rectal | 410 | 8 | kp1479 | 20 | Rectal | 483 |
| 6.01 | 4 | DY315 | 12 | Nebulizer | 1656 | 16 | kp1315 | 18 | Nebulizer | 43 |
| 6.01 | 4 | DY316 | 12 | Bedside cabinet | 1656 | 17 | kp1332 | 24 | Bedside cabinet | 15 |
| 6.01 | 9 | DY317 | 10 | Faucet surface | 6913 | 18 | kp1342 | 8 | Faucet surface | 43 |
Sampling date, isolate number, bed number, sample type, and ST for 16 CREC and carbapenem-resistant Klebsiella pneumoniae (CRKP) strains collected in the same period.
TABLE 3.
Susceptibility test results of clinically isolated CREC and CRKP isolates for three disinfectants
| Organism | MIC of disinfectant (mg/L) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Chlorhexidine |
p-Chlorophenol |
Calcium hypochlorite |
|||||||
| Range | 50% | 90% | Range | 50% | 90% | Range | 50% | 90% | |
| CREC | 0.5–1 | 0.5 | 1 | 256~512 | 256 | 512 | >256 | >256 | >256 |
| CRKP | 2–16 | 4 | 8 | 256 | 256 | 256 | >256 | >256 | >256 |
| ATCC 25922 | 1 | 512 | >256 | ||||||
| ATCC 700603 | 16 | 256 | >256 | ||||||
DISCUSSION
The emergence of multidrug-resistant Gram-negative bacteria is becoming increasingly serious, causing approximately 700,000 deaths yearly worldwide. This number is expected to reach 10 million by 2050 if no effective solution is found (8). CREC infections are associated with increased mortality, prolonged hospitalization (9, 10), and increased treatment costs compared to CSEC infections (11). An economic prediction model suggested that the increased socioeconomic cost per CREC-infected person ranges from $50,000 to $80,000 (12). First used in the 1980s, carbapenems have become the most effective antibacterial drugs for treating multidrug-resistant Gram-negative bacteria, owing to their advantages of broad-spectrum activity, strong antibacterial effect, stable effect on β-lactamase and cephalosporinase, and low toxicity. Carbapenems have shown good efficacy in the treatment of severe multidrug-resistant bacteria infections (3, 13). However, with extensive clinical use, the detection rate of CREC has gradually increased (14). Asymptomatic CREC colonization precedes or occurs concurrently with infection and is a risk factor for CREC infection, constituting a CREC reservoir (15).
Data from the China Network for Bacterial Resistance Surveillance (CHINET [https://www.chinets.com/Data/AntibioticDrugFast]) showed that the resistance of E. coli to carbapenem antibiotics was relatively stable in China, with rates of 1.1% and 1.4% for imipenem and meropenem, respectively, in 2005, compared with 2.0% and 2.1%, respectively, in 2019. The China Drug Resistance Surveillance Network (http://www.carss.cn/) indicated that the national average rate of resistance of E. coli to carbapenem antibiotics in 2019 was 1.5%, the same as in 2017, with the highest rate in Henan Province at 2.9%, and the overall resistance rate had remained low.
In this study, CREC was detected in only one patient sample taken within 48 h following ICU admission. This patient was admitted to the orthopedic department of a local hospital. Postoperative wound infection was suspected, and carbapenem drugs were administered before ICU admission. He was later transferred to the ICU for continued treatment. Thus, the CREC detected was considered to be hospital acquired. In addition, ICU-acquired CREC from five patients also were hospital-acquired CREC. We can infer that almost all of the CREC cases identified in this study were hospital acquired, and community-acquired CREC is still very rare. Tran et al. (16) also found that the colonization of CREC among patients was mainly hospital acquired, and the rate of CRE colonization increased with the length of ICU stay in a study of 12 hospitals in Vietnam. They also proposed that colonization is a prerequisite for CRE infection.
CREC was detected in the faucet surface sample related to patient P9, the genotype of which was considered similar to that found in a patient (P5) whose rectal swab was taken in the adjacent room during the same period. Since there was no hand-washing sink in the large six-bed ward next door to the patient (P9), it is highly likely that the CREC colonization on the faucet surface was caused by the health care worker or nursing staff managing the patient (P5) in the large ward coming to the hand-washing sink in the next patient’s room (P9). Therefore, we suggest setting up more single isolation rooms, if the hospital’s economic conditions allow, and reducing CREC contamination via the circulation of health care workers by hand hygiene management. Furthermore, because of the low rate of detection of CREC in environmental specimens, we can infer that environmental CREC colonization is limited and that the environmental contamination by CREC carrier patients is highly limited with respect to both the temporal and spatial distribution. This is inextricably linked to our strict hand disinfection management, diligent disinfection of the surrounding environment, and the timely transfer of colonized patients to a single room in our hospital.
Regarding the STs of the strains collected, we found that ST1656 and ST131 were predominant. In NDM-producing E. coli outbreaks globally, ST101, ST405, ST410, ST648, ST156, ST167, and ST131 are the most common clones. ST131 is predominant in India, whereas ST167 is predominant in China. Furthermore, we found that ST410 carrying the blaNDM-5 gene appears to be of greater concern because it is widely disseminated in China and is known to cause infections throughout the country (17). Among the 16 carbapenem-resistant E. coli strains, 2 lacked carbapenemases. It has been previously reported that the mechanisms of carbapenem resistance are closely related to the production of carbapenemases (acquisition of carbapenemase genes), loss of pore proteins in combination with extended-spectrum β-lactamases (ESBLs), and overexpression of efflux pumps. In our study, the two CREC strains lacking carbapenemases might utilize the latter mechanisms, which remains to be investigated (7).
Regarding the MLST results, the genetic analysis for all strains showed low homology among the isolates. The last decade has witnessed a pandemic of ST1193, a new multidrug-resistant clonal group of E. coli. ST1193 has been demonstrated to produce ESBL and is resistant to fluoroquinolones (100%), trimethoprim-sulfamethoxazole, and tetracycline (18–20). Epidemiological studies across developing and developed countries in Asia, Europe, and America recently suggested that ST1193 has become one of the dominant STs of uropathogenic antimicrobial-resistant E. coli, which induces community onset urinary tract infections in children and adults (20–23). In China’s mainland, ST1193 was first reported in a molecular epidemiology study of 590 nonduplicate E. coli isolates from 30 county hospitals in 2015, suggesting that ST1193 is the second most abundant ST among fluoroquinolone-resistant E. coli strains (24). Several studies indicate the possibility of the bidirectional interspecies transmission of E. coli ST1193 among humans, wildlife (such as Australian silver gulls), domestic animals (25), and companion animals (26, 27), which might be one of the causes of the global pandemic of ST1193. Therefore, there is an urgent requirement for increased surveillance to control the rapid worldwide spread of ST1193, which was also the dominant ST identified in this study. In our study, the blaNDM-5-carrying plasmids of DY88, DY315, and DY316 that had the same genetic structure were able to conjugate into E. coli J53. Therefore, once the blaNDM-5-carrying conjugative plasmid is transferred to carbapenem-sensitive E. coli clone ST1193, the epidemic of ST1193 CREC will pose a great threat to public health indeed.
We performed long-read Nanopore sequencing to obtain the complete chromosome and plasmid sequences, which facilitated the analysis of genetic structure and the important mobile elements surrounding resistance genes. In particular, the genes encoding NDM-5 and KPC-2 carbapenemases have attracted attention based on the complete sequence. Three blaKPC-2-carrying and two blaNDM-5-carrying plasmids have been further analyzed. The genetic environments, such as transposon or insertion sequence (IS) surrounding the carbapenemase gene, have been clarified, broadening our understanding of the transfer mechanism around these important antimicrobial resistance genes.
Finally, we found that the CREC was more easily sterilized by commonly used disinfectants. In our study, we chose chlorhexidine, p-chlorophenol, and calcium hypochlorite for investigation. Our results showed that the MIC90 of chlorhexidine for CRKP was 8 mg/L, while that for CREC was 1 mg/L. In addition, we found that calcium hypochlorite and p-chlorophenol had higher MICs for CREC and CRKP. Thus, they are not recommended for use as common disinfectants in clinical practice. Bacteria can acquire disinfectant resistance by expressing relevant resistance genes. Most disinfectant resistance genes are usually carried by the chromosome encoding the bacterial efflux pump system (28). Chen et al. (29) showed that CRE strains collected from patients (including CREC and CRKP) displayed various degrees of resistance to commonly used disinfectants. Continuous monitoring of the decreasing susceptibility of multidrug-resistant bacteria to disinfectants helps to effectively control and prevent the spread of superresistant bacteria. Our previous study revealed that the detected CRKP carried disinfectant resistance genes and virulence clusters and recommended the appropriate use of disinfectants to prevent the development of resistance (30).
Based on these findings, we demonstrate the CREC colonization rate and the relevant genomic information of CREC in Chinese ICUs, including its major multilocus sequence analysis, drug resistance genes, nucleotide polymorphisms, and other important information, which provide a basis for further research. We have also offered recommendations for how physicians and other health care workers can effectively reduce CREC colonization in the ICU and prevent its transmission. Combining this study with previous studies related to CRKP, we found that the transmission of CREC is relatively limited, whereas that of CRKP is widespread (31–33). And we explored the reason why CREC was isolated at a much lower frequency than CRKP.
However, this study is limited by being a single-center observational study. In addition, the sample size was quite small, and the rate of acquisition of samples from relevant patients within 48 h of admission was low, meaning that a few patients carrying CREC were likely missed, thereby reducing the evidence-based strength of the study results.
Conclusion.
In conclusion, the rate of detection of CREC was low in both patients and the environment in this study. Almost all of the CREC strains detected were ICU acquired. Contamination of the surrounding environment by CREC-carrying patients is limited both temporally and spatially. Although all strains showed low and sporadic homology, ST1193 was the primary strain of CSEC detected, indicating that this might be the next potential outbreak strain that requires attention. The next most common strains were ST10 and ST69. ST1656 and ST131 also deserve attention, as they comprise the majority of CREC cases. The blaNDM-5 gene is relevant to empirical antibiotic use as the main carbapenem resistance gene. The difference in clinical transmission between CREC and CRKP is likely due to their different susceptibilities to disinfectants. The results of the susceptibility testing of disinfectants showed that CREC was more susceptible than CRKP. Therefore, effective interventions and active screening are beneficial to the prevention and control of CREC.
MATERIALS AND METHODS
Strains.
This prospective study aimed to explore the colonization, transmission, and distribution of CREC in a tertiary teaching hospital in China. The study period was from 1 April 2021 to 31 July 2021, and screening was performed every Tuesday morning. Screening included 6 sites for each patient and 19 environmental sites around the patient. The study was conducted in the 28-bed ICU of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine. We collected weekly clinical samples such as oropharyngeal, rectal, tracheal intubation, gastric intubation, tracheotomy tube, and nasointestinal tube swabs from all of the patients admitted to the ICU during the study period. Weekly environmental samples were taken from each bed and its surrounding structures, including bed rails, button panels of ventilators, cardiac monitors, microinjection pumps, switch buttons, nebulizers, stethoscopes, hanging towers, bed regulators, bedside tables, computer mice and keyboards, ring hooks of infusion racks, treatment carts, and lockers. We also obtained weekly samples from all ward sinks, including the inside of the downspout, faucet surface, sink countertop, inside surface of the overflow, and the water in the plumbing system.
This study was reviewed and approved by the Ethics Committee of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine (no. 20201217-33).
Strain isolation, strain identification, and drug and disinfectant resistance assays.
Sampling personnel used Copan swabs to obtain samples from the patients, bed units, and sinks. After sampling, the swabs were placed in tryptic soy broth (TSB) and incubated overnight. Twenty microliters of the bacterial solution was then drawn and applied to CHROMagar E. coli screening plates (CHROMagar, Paris, France) for an overnight culture at 37°C. On the second day, a single colony was picked from each plate and inoculated onto a Mueller-Hinton (MH) agar plate. The divisions were marked, and the plates were cultured overnight at 37°C. On the third day, a single colony from each plate was chosen for matrix-assisted laser desorption ionization–time of flight (MALDI-TOF) mass spectrometry (bioMérieux, Marcy-l’Etoile, France) to confirm that the colonies were E. coli strains. The MICs of some drugs, including meropenem, imipenem, ertapenem, ceftriaxone, ceftazidime, cefepime, aztreonam, piperacillin-tazobactam, amikacin, ciprofloxacin, and fosfomycin, were measured using the agar dilution method, while those of ceftazidime-avibactam, colistin, and tigecycline were measured using the broth microdilution method. Both of these methods are recommended by the American Clinical and Laboratory Standards Institute (CLSI) guidelines (2020 edition) (34). All drug sensitivity breakpoints were referenced to the CLSI 2020 implementation criteria except for tigecycline, which was determined according to the FDA 2019 guideline. Furthermore, we applied the broth microdilution method to obtain the MICs of the three frequently used disinfectants in the clinic, including chlorhexidine, p-chlorophenol, and calcium hypochlorite, against the isolated CRKP and CREC strains. The quality control strains were E. coli ATCC 25922 and K. pneumoniae ATCC 700603.
Whole-genome sequencing and analysis.
Genomic DNA was extracted using a Qiagen minikit (Qiagen, Hilden, Germany) and sequenced using the Illumina HiSeq (Illumina, San Diego, CA) and Oxford Nanopore MinION (Nanopore, Oxford, UK) platforms. Illumina read assembly was performed to obtain draft genomes using the shovill pipeline (version 4.4.5 [https://github.com/tseemann/shovill]). The raw data from Nanopore MinION and Illumina sequencing were assembled using Unicycler pipeline to obtain complete genomes under default parameters (35). The assembled genomes were annotated using Prokka (36) and BacAnt (37). A plasmid map was constructed by importing the gbk file into BRIG (version 0.95). The MLST used in the survey was the Achtman scheme, which was performed using mlst (https://github.com/tseemann/mlst) with the PubMLST data set (38). Resistance genes and plasmids were identified using ABRicate v0.8.13 (https://github.com/tseemann/abricate) with the NCBI AMRFinderPlus (39) and PlasmidFinder (40) databases.
Homology analysis.
The core genome alignment file was generated using the panaroo pipeline (version 1.2.7) in strict mode (41). The maximum likelihood tree was generated using the iqtree pipeline (version 2.1.2) (42). The snippy pipeline (version 4.4.5 [https://github.com/tseemann/snippy]) was used to calculate the SNPs of the isolates. An SNP distance of ≤22 bp to the nearest neighbor was set as the criterion for indicating that two strains were homologous (43).
Data availability.
The draft genomes of the isolates in this collection have been deposited in the NCBI database under BioProject accession number PRJNA911045.
ACKNOWLEDGMENTS
This work is supported by the National Natural Science Foundation of China (32141001 and 81830069), and Natural Science Foundation of Zhejiang Province (LY22H190001).
We declare no competing interests.
Footnotes
Supplemental material is available online only.
Contributor Information
Yan Jiang, Email: jiangy@zju.edu.cn.
Yunsong Yu, Email: yvys119@zju.edu.cn.
Katharina Schaufler, Universität Greifswald.
REFERENCES
- 1.Gastmeier P, Vonberg RP. 2014. Klebsiella spp. in endoscopy-associated infections: we may only be seeing the tip of the iceberg. Infection 42:15–21. doi: 10.1007/s15010-013-0544-6. [DOI] [PubMed] [Google Scholar]
- 2.Souli M, Galani I, Antoniadou A, Papadomichelakis E, Poulakou G, Panagea T, Vourli S, Zerva L, Armaganidis A, Kanellakopoulou K, Giamarellou H. 2010. An outbreak of infection due to beta-lactamase Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae in a Greek university hospital: molecular characterization, epidemiology, and outcomes. Clin Infect Dis 50:364–373. doi: 10.1086/649865. [DOI] [PubMed] [Google Scholar]
- 3.Temkin E, Adler A, Lerner A, Carmeli Y. 2014. Carbapenem-resistant Enterobacteriaceae: biology, epidemiology, and management. Ann N Y Acad Sci 1323:22–42. doi: 10.1111/nyas.12537. [DOI] [PubMed] [Google Scholar]
- 4.van Duin D, Arias CA, Komarow L, Chen L, Hanson BM, Weston G, Cober E, Garner OB, Jacob JT, Satlin MJ, Fries BC, Garcia-Diaz J, Doi Y, Dhar S, Kaye KS, Earley M, Hujer AM, Hujer KM, Domitrovic TN, Shropshire WC, Dinh A, Manca C, Luterbach CL, Wang M, Paterson DL, Banerjee R, Patel R, Evans S, Hill C, Arias R, Chambers HF, Fowler VG, Kreiswirth BN, Bonomo RA, Multi-Drug Resistant Organism Network Investigators . 2020. Molecular and clinical epidemiology of carbapenem-resistant Enterobacterales in the USA (CRACKLE-2): a prospective cohort study. Lancet Infect Dis 20:731–741. doi: 10.1016/S1473-3099(19)30755-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Conlan S, Thomas PJ, Deming C, Park M, Lau AF, Dekker JP, Snitkin ES, Clark TA, Luong K, Song Y, Tsai Y-C, Boitano M, Dayal J, Brooks SY, Schmidt B, Young AC, Thomas JW, Bouffard GG, Blakesley RW, Mullikin JC, Korlach J, Henderson DK, Frank KM, Palmore TN, Segre JA, NISC Comparative Sequencing Program . 2014. Single-molecule sequencing to track plasmid diversity of hospital-associated carbapenemase-producing Enterobacteriaceae. Sci Transl Med 6:254ra126. doi: 10.1126/scitranslmed.3009845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Durante-Mangoni E, Andini R, Zampino R. 2019. Management of carbapenem-resistant Enterobacteriaceae infections. Clin Microbiol Infect 25:943–950. doi: 10.1016/j.cmi.2019.04.013. [DOI] [PubMed] [Google Scholar]
- 7.Tian X, Zheng X, Sun Y, Fang R, Zhang S, Zhang X, Lin J, Cao J, Zhou T. 2020. Molecular mechanisms and epidemiology of carbapenem-resistant Escherichia coli isolated from Chinese patients during 2002–2017. Infect Drug Resist 13:501–512. doi: 10.2147/IDR.S232010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shankar PR. 2016. Book review: tackling drug-resistant infections globally. Arch Pharm Pract 7:110–111. doi: 10.4103/2045-080X.186181. [DOI] [Google Scholar]
- 9.Martin A, Fahrbach K, Zhao Q, Lodise T. 2018. Association between carbapenem resistance and mortality among adult, hospitalized patients with serious infections due to Enterobacteriaceae: results of a systematic literature review and meta-analysis. Open Forum Infect Dis 5:ofy150. doi: 10.1093/ofid/ofy150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zilberberg MD, Nathanson BH, Sulham K, Fan W, Shorr AF. 2017. Carbapenem resistance, inappropriate empiric treatment and outcomes among patients hospitalized with Enterobacteriaceae urinary tract infection, pneumonia and sepsis. BMC Infect Dis 17:279. doi: 10.1186/s12879-017-2383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohler PP, Volling C, Green K, Uleryk EM, Shah PS, McGeer A. 2017. Carbapenem resistance, initial antibiotic therapy, and mortality in Klebsiella pneumoniae bacteremia: a systematic review and meta-analysis. Infect Control Hosp Epidemiol 38:1319–1328. doi: 10.1017/ice.2017.197. [DOI] [PubMed] [Google Scholar]
- 12.Bartsch SM, McKinnell JA, Mueller LE, Miller LG, Gohil SK, Huang SS, Lee BY. 2017. Potential economic burden of carbapenem-resistant Enterobacteriaceae (CRE) in the United States. Clin Microbiol Infect 23:48.e9–48.e16. doi: 10.1016/j.cmi.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howatt M, Klompas M, Kalil AC, Metersky ML, Muscedere J. 2021. Carbapenem antibiotics for the empiric treatment of nosocomial pneumonia: a systematic review and meta-analysis. Chest 159:1041–1054. doi: 10.1016/j.chest.2020.10.039. [DOI] [PubMed] [Google Scholar]
- 14.Laxminarayan R, Matsoso P, Pant S, Brower C, Rottingen JA, Klugman K, Davies S. 2016. Access to effective antimicrobials: a worldwide challenge. Lancet 387:168–175. doi: 10.1016/S0140-6736(15)00474-2. [DOI] [PubMed] [Google Scholar]
- 15.Mittal G, Gaind R, Kumar D, Kaushik G, Gupta KB, Verma PK, Deb M. 2016. Risk factors for fecal carriage of carbapenemase producing Enterobacteriaceae among intensive care unit patients from a tertiary care center in India. BMC Microbiol 16:138. doi: 10.1186/s12866-016-0763-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tran DM, Larsson M, Olson L, Hoang NTB, Le NK, Khu DTK, Nguyen HD, Vu TV, Trinh TH, Le TQ, Phan PTT, Nguyen BG, Pham NH, Mai BH, Nguyen TV, Nguyen PTK, Le ND, Huynh TM, Anh Thu LT, Thanh TC, Berglund B, Nilsson LE, Bornefall E, Song LH, Hanberger H. 2019. High prevalence of colonisation with carbapenem-resistant Enterobacteriaceae among patients admitted to Vietnamese hospitals: risk factors and burden of disease. J Infect 79:115–122. doi: 10.1016/j.jinf.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 17.Li F, Ye K, Li X, Ye L, Guo L, Wang L, Yang J. 2021. Genetic characterization of carbapenem-resistant Escherichia coli from China, 2015–2017. BMC Microbiol 21:248. doi: 10.1186/s12866-021-02307-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Peirano G, Pitout JDD. 2019. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79:1529–1541. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen MN, Hoang HTT, Xavier BB, Lammens C, Le HT, Hoang NTB, Nguyen ST, Pham NT, Goossens H, Dang AD, Malhotra-Kumar S. 2021. Prospective One Health genetic surveillance in Vietnam identifies distinct blaCTX-M-harbouring Escherichia coli in food-chain and human-derived samples. Clin Microbiol Infect 27:1515.e1–1515.e8. doi: 10.1016/j.cmi.2021.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Tchesnokova VL, Rechkina E, Larson L, Ferrier K, Weaver JL, Schroeder DW, She R, Butler-Wu SM, Aguero-Rosenfeld ME, Zerr D, Fang FC, Ralston J, Riddell K, Scholes D, Weissman S, Parker K, Spellberg B, Johnson JR, Sokurenko EV. 2019. Rapid and extensive expansion in the United States of a new multidrug-resistant Escherichia coli clonal group, sequence type 1193. Clin Infect Dis 68:334–337. doi: 10.1093/cid/ciy525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holland MS, Nobrega D, Peirano G, Naugler C, Church DL, Pitout JDD. 2020. Molecular epidemiology of Escherichia coli causing bloodstream infections in a centralized Canadian region: a population-based surveillance study. Clin Microbiol Infect 26:1554.e1–1554.e8. doi: 10.1016/j.cmi.2020.02.019. [DOI] [PubMed] [Google Scholar]
- 22.Birgy A, Madhi F, Jung C, Levy C, Cointe A, Bidet P, Hobson CA, Bechet S, Sobral E, Vuthien H, Ferroni A, Aberrane S, Cuzon G, Beraud L, Gajdos V, Launay E, Pinquier D, Haas H, Desmarest M, Dommergues MA, Cohen R, Bonacorsi S, Group of the National Observatory of Urinary tract Infection due to ESBL-producing Enterobacteriaceae in children . 2020. Diversity and trends in population structure of ESBL-producing Enterobacteriaceae in febrile urinary tract infections in children in France from 2014 to 2017. J Antimicrob Chemother 75:96–105. doi: 10.1093/jac/dkz423. [DOI] [PubMed] [Google Scholar]
- 23.Findlay J, Gould VC, North P, Bowker KE, Williams MO, MacGowan AP, Avison MB. 2020. Characterization of cefotaxime-resistant urinary Escherichia coli from primary care in South-West England 2017–18. J Antimicrob Chemother 75:65–71. doi: 10.1093/jac/dkz397. [DOI] [PubMed] [Google Scholar]
- 24.Zhao L, Zhang J, Zheng B, Wei Z, Shen P, Li S, Li L, Xiao Y. 2015. Molecular epidemiology and genetic diversity of fluoroquinolone-resistant Escherichia coli isolates from patients with community-onset infections in 30 Chinese county hospitals. J Clin Microbiol 53:766–770. doi: 10.1128/JCM.02594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukerji S, Stegger M, Truswell AV, Laird T, Jordan D, Abraham RJ, Harb A, Barton M, O’Dea M, Abraham S. 2019. Resistance to critically important antimicrobials in Australian silver gulls (Chroicocephalus novaehollandiae) and evidence of anthropogenic origins. J Antimicrob Chemother 74:2566–2574. doi: 10.1093/jac/dkz242. [DOI] [PubMed] [Google Scholar]
- 26.Kidsley AK, White RT, Beatson SA, Saputra S, Schembri MA, Gordon D, Johnson JR, O’Dea M, Mollinger JL, Abraham S, Trott DJ. 2020. Companion animals are spillover hosts of the multidrug-resistant human extraintestinal Escherichia coli pandemic clones ST131 and ST1193. Front Microbiol 11:1968. doi: 10.3389/fmicb.2020.01968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu FL, Kuan NL, Yeh KS. 2021. Presence of the extended-spectrum-beta-lactamase and plasmid-mediated AmpC-encoding genes in Escherichia coli from companion animals—a study from a university-based veterinary hospital in Taipei, Taiwan. Antibiotics (Basel) 10:1536. doi: 10.3390/antibiotics10121536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong C, Hu H, Chen G, Li Z, Li A, Zhang J. 2021. Disinfectant resistance in bacteria: mechanisms, spread, and resolution strategies. Environ Res 195:110897. doi: 10.1016/j.envres.2021.110897. [DOI] [PubMed] [Google Scholar]
- 29.Chen Y, Liao K, Huang Y, Guo P, Huang H, Wu Z, Liu M. 2020. Determining the susceptibility of carbapenem resistant Klebsiella pneumoniae and Escherichia coli strains against common disinfectants at a tertiary hospital in China. BMC Infect Dis 20:88. doi: 10.1186/s12879-020-4813-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han X, Chen Y, Zhou J, Shi Q, Jiang Y, Wu X, Quan J, Hu H, Wang Q, Yu Y, Fu Y. 2022. Epidemiological characteristics of OXA-232-producing carbapenem-resistant Klebsiella pneumoniae strains isolated during nosocomial clonal spread associated with environmental colonization. Microbiol Spectr 10:e02572-21. doi: 10.1128/spectrum.02572-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y, Wang Q, Yin Y, Chen H, Jin L, Gu B, Xie L, Yang C, Ma X, Li H, Li W, Zhang X, Liao K, Man S, Wang S, Wen H, Li B, Guo Z, Tian J, Pei F, Liu L, Zhang L, Zou C, Hu T, Cai J, Yang H, Huang J, Jia X, Huang W, Cao B, Wang H. 2018. Epidemiology of carbapenem-resistant Enterobacteriaceae infections: report from the China CRE Network Antimicrob Agents Chemother 62:e01882-17. doi: 10.1128/AAC.01882-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiao F, Wei L, Feng Y, Ran S, Zheng L, Zhang Y, Xiang Q, Liu Y, Wu X, Duan X, Zhang W, Li Q, Guo H, Huang W, Zhu S, Wen H, Zong Z. 2020. Handwashing sink contamination and carbapenem-resistant Klebsiella infection in the intensive care unit: a prospective multicenter study. Clin Infect Dis 71:S379–S385. doi: 10.1093/cid/ciaa1515. [DOI] [PubMed] [Google Scholar]
- 33.Hu Y, Liu C, Shen Z, Zhou H, Cao J, Chen S, Lv H, Zhou M, Wang Q, Sun L, Sun Q, Hu F, Wang Y, Zhang R. 2020. Prevalence, risk factors and molecular epidemiology of carbapenem-resistant Klebsiella pneumoniae in patients from Zhejiang, China, 2008–2018. Emerg Microbes Infect 9:1771–1779. doi: 10.1080/22221751.2020.1799721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gary W, Procop PJD, Berkow E, Fuller J, Hanson KE, Holliday NM, Pincus DH, Schuetz AN, Verweij PE, Wiederhold NP, Zelazny AM. 2020. Performance standards for antifungal susceptibility testing of yeasts, 2nd ed. CLSI supplement M60. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 35.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 37.Hua X, Liang Q, Deng M, He J, Wang M, Hong W, Wu J, Lu B, Leptihn S, Yu Y, Chen H. 2021. BacAnt: a combination annotation server for bacterial DNA sequences to identify antibiotic resistance genes, integrons, and transposable elements. Front Microbiol 12:649969. doi: 10.3389/fmicb.2021.649969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feldgarden M, Brover V, Haft DH, Prasad AB, Slotta DJ, Tolstoy I, Tyson GH, Zhao S, Hsu CH, McDermott PF, Tadesse DA, Morales C, Simmons M, Tillman G, Wasilenko J, Folster JP, Klimke W. 2019. Validating the AMRFinder tool and resistance gene database by using antimicrobial resistance genotype-phenotype correlations in a collection of isolates. Antimicrob Agents Chemother 63:e00483-19. doi: 10.1128/AAC.00483-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carattoli A, Zankari E, Garcia-Fernandez A, Voldby Larsen M, Lund O, Villa L, Moller Aarestrup F, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tonkin-Hill G, MacAlasdair N, Ruis C, Weimann A, Horesh G, Lees JA, Gladstone RA, Lo S, Beaudoin C, Floto RA, Frost SDW, Corander J, Bentley SD, Parkhill J. 2020. Producing polished prokaryotic pangenomes with the Panaroo pipeline. Genome Biol 21:180. doi: 10.1186/s13059-020-02090-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.David S, Reuter S, Harris SR, Glasner C, Feltwell T, Argimon S, Abudahab K, Goater R, Giani T, Errico G, Aspbury M, Sjunnebo S, Eu SWG, Group ES, Feil EJ, Rossolini GM, Aanensen DM, Grundmann H, ESGEM Study Group . 2019. Epidemic of carbapenem-resistant Klebsiella pneumoniae in Europe is driven by nosocomial spread. Nat Microbiol 4:1919–1929. doi: 10.1038/s41564-019-0492-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S4. Download spectrum.04010-22-s0001.pdf, PDF file, 1.7 MB (1.7MB, pdf)
Data Availability Statement
The draft genomes of the isolates in this collection have been deposited in the NCBI database under BioProject accession number PRJNA911045.