ABSTRACT
Polymyxin has been the last resort to treat multidrug-resistant Klebsiella pneumonia. However, recent studies have revealed that polymyxin-resistant carbapenem-resistant Klebsiella pneumonia (PR-CRKP) emerged due to the mutations in chromosomal genes or the plasmid-harboring mcr gene, leading to lipopolysaccharide modification or efflux of polymyxin through pumps. Further surveillance was required. In the present study we collected PR-CRKP strains from 8 hospitals in 6 provinces/cities across China to identify the carbapenemase and polymyxin resistance genes and epidemiological features by whole-genome sequencing (WGS). The broth microdilution method (BMD) was performed to determine the MIC of polymyxin. Of 662 nonduplicate CRKP strains, 15.26% (101/662) were defined as PR-CRKP; 10 (9.90%) were confirmed as Klebsiella quasipneumoniae by WGS. The strains were further classified into 21 individual sequence types (STs) by using multilocus sequence typing (MLST), with ST11 being prevalent (68/101, 67.33%). Five carbapenemase types were identified among 92 CR-PRKP, blaKPC-2 (66.67%), blaNDM-1 (16.83%), blaNDM-5 (0.99%), blaIMP-4 (4.95%), and blaIMP-38 (0.99%). Notably, 2 PR-CRKP strains harbored both blaKPC-2 and blaNDM-1. The inactivation of mgrB, associated significantly with high-level polymyxin resistance, was mainly caused by the insertion sequence (IS) insertion (62.96%, 17/27). Furthermore, acrR was inserted coincidently by ISkpn26 (67/101, 66.33%). The deletion or splicing mutations of crrCAB were significantly associated with ST11 and KL47 (capsule locus types), and diverse mutations of the ramR gene were identified. Only one strain carried the mcr gene. In summary, the high IS-inserted mgrB inactivation, the close relationship between ST11 and the deletion or splicing mutations of the crrCAB, and the specific features of PR-K. quasipneumoniae constituted notable features of our PR-CRKP strains in China.
IMPORTANCE Polymyxin-resistant CRKP is a serious public health threat whose resistance mechanisms should be under continuous surveillance. Here, we collected 662 nonduplicate CRKP strains across China to identify the carbapenemase and polymyxin resistance genes and epidemiological features. Polymyxin resistance mechanism in 101 PR-CRKP strains in China were also investigated, 9.8% of which (10/101) were K. quasipneumoniae, as determined via WGS, and inactivation of mgrB remained the most crucial polymyxin resistance mechanism, significantly related to high-level resistance. Deletion or splicing mutations of crrCAB were significantly associated with ST11 and KL47. Diverse mutations of the ramR gene were identified. The plasmid complementation experiment and mRNA expression analysis further confirmed that the mgrB promoter and ramR played a critical role in polymyxin resistance. This multicenter study contributed to the understanding of antibiotic resistance forms in China.
KEYWORDS: polymyxin resistance mechanism, carbapenemase-resistant Klebsiella pneumoniae, antibiotic resistance, mgrB inactivation, K. quasipneumoniae, genetic diversity, multicenter study, polymyxin-resistant Klebsiella pneumoniae
INTRODUCTION
Carbapenem-resistant Klebsiella pneumoniae (CRKP) was highlighted as a notorious pathogen of various infections (1). Polymyxin (polymyxin B and colistin) has been the last-resort treatment for such infections, even with an unfavorable side effect profile, including nephrotoxicity and neurotoxicity (2). However, the emergence of polymyxin resistance has been documented in the setting of increased use and imposed negative impacts on its usage in CRKP treatment (2, 3).
Polymyxin-resistant K. pneumoniae (PRKP) can be conferred by mutations in the two-component system (TCS), mgrB, efflux pumps, repressors, and lipopolysaccharide (LPS)-modifying genes (2). The upregulation of TCS genes, such as phoP/phoQ and pmrA/pmrB, also confers resistance to polymyxins. MgrB is a negative regulator of PhoQ. The insertion sequence (IS)-mediated mgrB gene disruption and inactivation in mgrB and its promoter region constituted the main polymyxin resistance mechanism in K. pneumoniae (4, 5). The addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) and/or phosphoethanolamine (pEtN) to the LPS was mediated by the arnBCADTEF and pmrCAB operons, respectively (2). PmrD, encoded by pmrD, is a connector protein that might activate PmrB/PmrA in response to PhoQ/PhoP stimulation (6). Furthermore, many other mechanisms are involved simultaneously in polymyxin resistance. For example, the ramA, yciM, and lpxM genes, which can mediate LPS alterations, along with the AcrAB-TolC pump and its regulators (acrAB, kpnEF, and SoxSR), contribute to mediating decreased susceptibility of polymyxin in K. pneumoniae (7–11). The plasmid-borne mcr-1 gene and its variant-mediated polymyxin resistance have also been reported in K. pneumoniae in the hospital setting (2, 12).
Moreover, Klebsiella quasipneumoniae was a newly designated species in Klebsiella spp. Few reports described its polymyxin- and carbapenem-resistance molecular features since it was not generally distinguished from K. pneumoniae by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and routine biochemical methods in the clinical laboratory (13).
The increasing rate in clinical isolates of polymyxin- and carbapenem-resistant K. pneumoniae (PR-CRKP) warrants further investigation into their epidemiology and underlying molecular mechanisms. Here, we conducted the study aiming to determine the prevalence of PR-CRKP in six provinces/cities in China, their clonal relationships, and the molecular features via whole-genome sequence (WGS) and bioinformatics analysis.
RESULTS
General characteristics of CRKP and PR-CRKP.
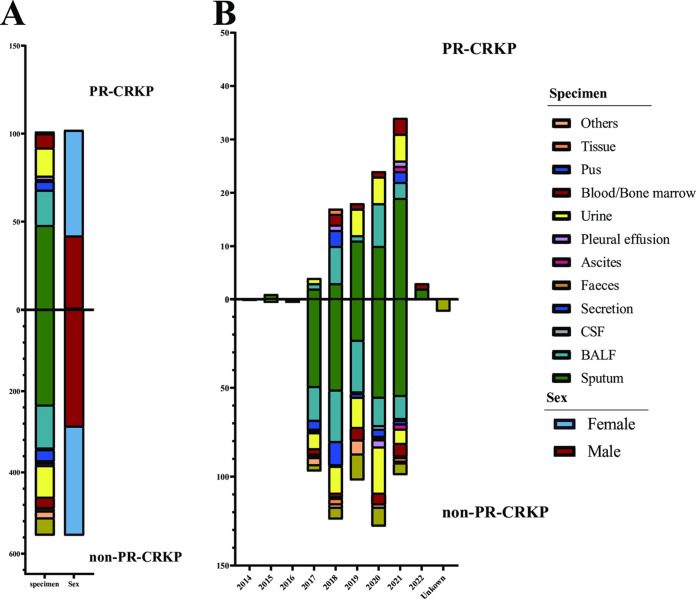
As detailed in Fig. 1, of 662 unduplicated CRKP strains, 15.26% (101/662) were classified as PR-CRKP; 411 (62.08%, 411/662) were recovered from the lower respiratory tract, followed by urine (14.20%, 94/662), secretion (4.83%, 32/662), and blood (4.68%, 31/662). The gender ratio of male/female was approximately 1:1 (333/329). The enrolled patients were aged from 1 day to 102 years, with a median age of 64 years.
FIG 1.
Sample source and distribution. (A) Sample distribution of 101 PR-CRKP strains and 561 non-PR-CRKP strains and the sex of the patients. (B) The number of strains isolated from different specimen types per year among PR-CRKP and non-PR-CRKP.
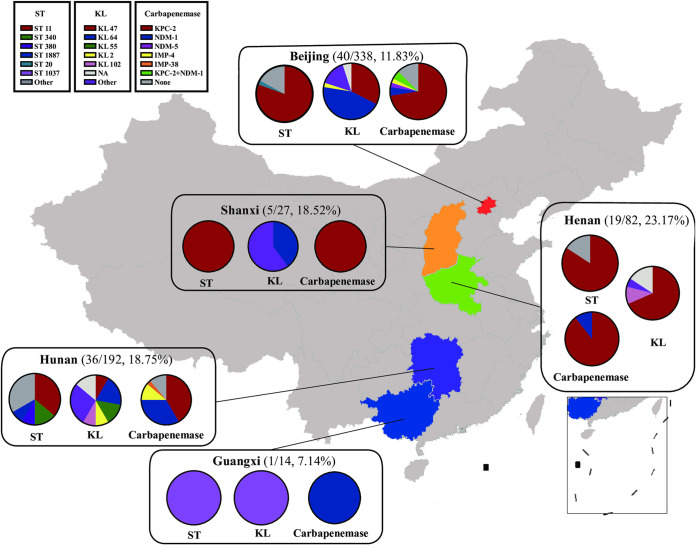
The 101 PR-CRKP strains were isolated from the lower respiratory tract (67.33%, 68, including 48 sputum and 20 bronchoalveolar lavage fluid [BALF] specimens), urine (15.84%, 16), blood (5.94%, 6), wounds (4.95%, 5, including ear, eye, and skin and soft tissue), pleural effusion (1.98%, 2), bone marrow (0.99%, 1), ascites (0.99%, 1), and tissue (0.99%, 1). As shown in Fig. 2 and 3, high-level PRKP (MIC, >16 μg/mL, 58 isolates) were mainly distributed in Beijing (77.50%, 31/40), Hunan (63.89%, 23/36), Shanxi (80.00%, 4/5), and Guangxi (100.00%, 1/1), whereas the low-level ones (MIC, ≤16 μg/mL, 43 isolates) were found mainly in Henan (100.00%, 19/19). No PR-KP isolates were identified from Hebei.
FIG 2.
Geographical locations of polymyxin- and carbapenem-resistant K. pneumoniae (PR-CRKP) strains circulating in China. The color-highlighted provinces/cities are those where PR-CRKP strains were recovered, with the number of strains of PR-CRKP/CRKP shown in parentheses. The distribution of sequence types (ST), capsule locus types (KL types), and carbapenemase genes were also shown. Nine CRKP strains were detected in Hebei, while none were resistant to polymyxin.
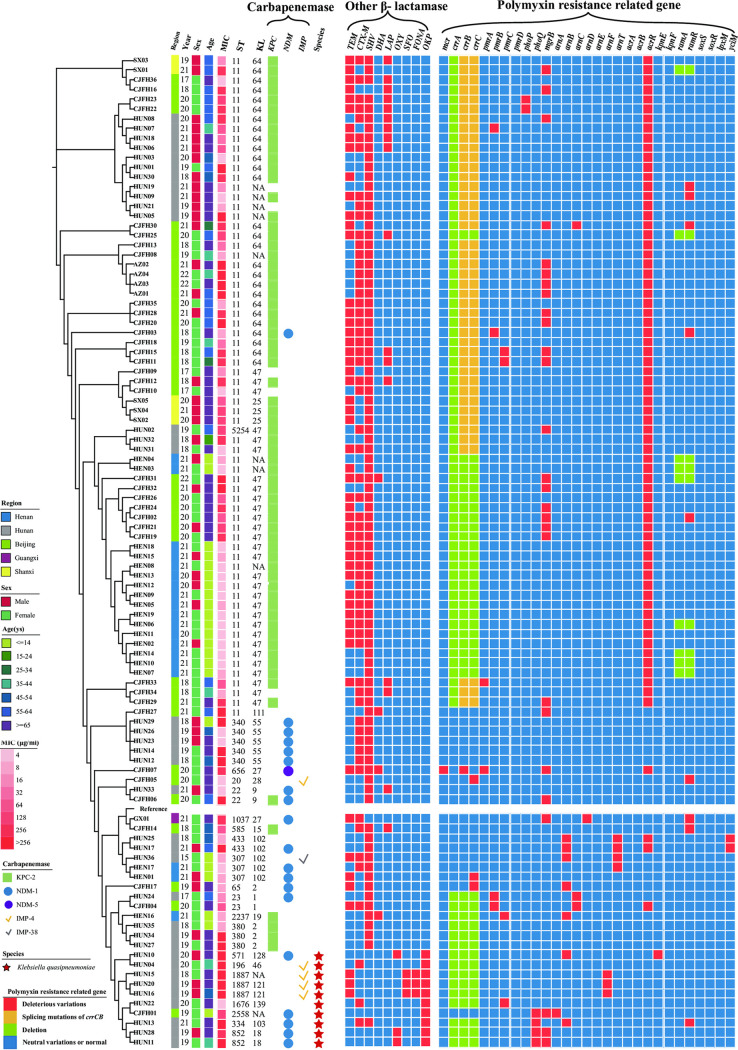
FIG 3.
Phylogenetic tree diagram of 101 polymyxin- and carbapenem-resistant K. pneumoniae (PR-CRKP) strains based on single nucleotide polymorphism (SNP) sequences. From left to right: isolation regions and years, sex, age, MIC, ST, KL, carbapenemase genes, Klebsiella species, other β-lactamases and polymyxin-resistance-related genes.
Antibiotic resistance profiles.
As shown in Table 1, among 101 PR-CRKP strains, more than half of the strains were nonsusceptible to the following antibiotics: aztreonam (90.10%, 91/101), levofloxacin (89.11%, 90/101), ciprofloxacin (88.12%, 89/101), tobramycin (68.67%, 57/101), and amikacin (55.45%, 56/101), followed by trimethoprim-sulfamethoxazole (44.55%, 45/101). Only 14.85% (15/101) were nonsusceptible to tigecycline.
TABLE 1.
Antimicrobial susceptibility profiles and MIC values of 10 antibiotics of the 101 PR-CRKPa
| Antibiotic | Breakpoint (S, I, R) (μg/mL) | Range (μg/mL) | MIC50 (μg/mL) | MIC90 (μg/mL) | No. of strains (%) |
|
|---|---|---|---|---|---|---|
| Susceptible | Nonsusceptibleb | |||||
| IMP | ≤1, 2, ≥4 | 2 to ≥16 | ≥16 | ≥16 | ||
| MEM | ≤1, 2, ≥4 | 2 to ≥16 | ≥16 | ≥16 | ||
| PB | NA, ≤2, >2 | 4 to >256 | 64 | >256 | ||
| ATM | ≤4, NA, ≥16 | 4 to ≥64 | ≥64 | ≥64 | 10 (9.90) | 91 (90.10) |
| AK | ≤16, NA, ≥64 | ≤2 to ≥64 | ≥64 | ≥64 | 45 (44.55) | 56 (55.45) |
| TOB | ≤4, NA, ≥16 | ≤1 to ≥16 | ≥16 | ≥16 | 26 (31.33) | 57 (68.67) |
| CIP | <0.25, 0.5, ≥1 | ≤0.25 to ≥4 | ≥4 | ≥4 | 12 (11.88) | 89 (88.12) |
| LEV | ≤0.25, 1, ≥2 | ≤0.12 to ≥8 | ≥8 | ≥8 | 11 (10.89) | 90 (89.11) |
| TGC | ≤2, 4, ≥8 | 0.25 to 8 | 1 | 4 | 86 (85.15) | 15 (14.85) |
| SXT | ≤40, NA, ≥80 | ≤20 to ≥320 | ≤20 | ≥320 | 56 (55.45) | 45 (44.55) |
NA, not available according to CLSI M100; IMP, imipenem; MEM, meropenem; PB, polymyxin B; ATM, aztreonam; AK, amikacin; TOB, tobramycin; CIP, ciprofloxacin; LEV, levofloxacin; TGC, tigecycline; STX, sulfamethoxazole/trimethoprim; S, susceptible; I, intermediate; R, resistant.
Nonsusceptible contained resistant and intermediary isolates.
Molecular characteristics of PR-CRKP.
Of 101 PR-CRKP strains, 10 (9.90%) were confirmed as K. quasipneumoniae based on WGS. A total of 92 (91.09%) were identified as carbapenemase-producing K. pneumoniae (CPKP) (82.18%, 83/101) and K. quasipneumoniae (8.91%, 9/101). Of 10 K. quasipneumoniae strains, 9 were isolated from two hospitals in Hunan. Further, of 92 CPKP strains, five carbapenemase types were identified, namely, blaKPC-2 (66.67%, 68/101), blaNDM-1 (16.83%, 17/101), blaNDM-5 (0.99%, 1/101), blaIMP-4 (4.95%, 5/101), and blaIMP-38 (0.99%, 1/101). Notably, 2 PR-CRKP (1.98%) strains harbored both blaKPC-2 and blaNDM-1.
As shown in Fig. 3, for other β-lactamases, blaSHV (89.11%, 90/101) was mostly detected, followed by blaCTX-M (60.40%, 61/101, and 44 PR-CRKP strains harbored blaCTX-M-65) and blaTEM (53.47%, 54/101).
A total of 21 individual STs were distinguished, with ST11 dominating (68/101, 67.33%). There was no relationship between ST type and polymyxin resistance level (P = 0.8159). Furthermore, KL47 accounted all the PR-CRKP for 29.70% (30/101), followed by KL64 (27/101, 26.73%), unclassified (10/101, 9.90%), and KL102 (6/101, 5.94%).
Notably, of 10 K. quasipneumoniae strains, 7 ST types were designated, 9 were isolated from Hunan, and 9 harbored metalloenzyme genes, namely, 5 blaNDM-1 and 4 blaIMP-4.
Polymyxin resistance mechanisms.
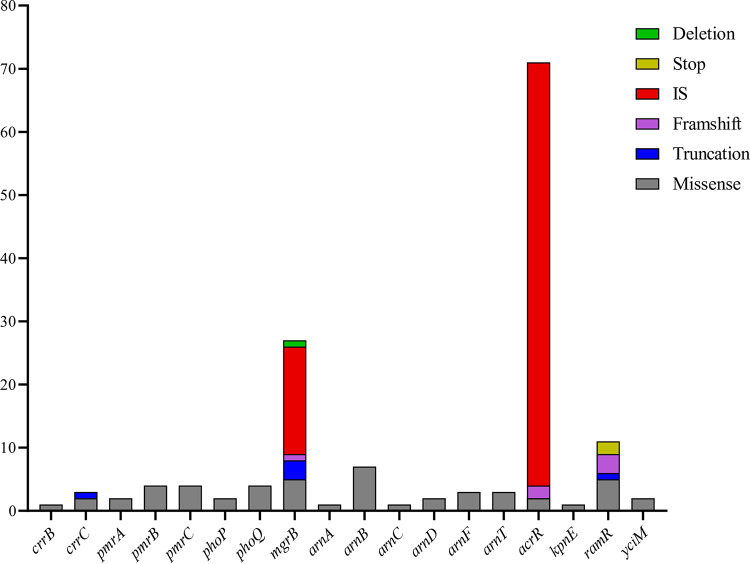
The polymyxin resistance mechanism is shown in Table 2 and Fig. 3 and 4. Of 101 PR-CRKP strains, 168 variations at a variety of sites were identified across all 26 above-mentioned genes possibly involved in polymyxin resistance. The gene variations remaining after removing the neutral variations are listed in Table 2. The acrR gene mutation was the most frequently identified (71, 70.29%), followed by mgrB (27, 26.73%), ramR (11, 10.89%), and arnB (7, 6.93%).
TABLE 2.
Distribution of polymyxin resistance-related gene mutations among 101 polymyxin- and carbapenem-resistant K. pneumoniae isolatesa,b
| Gene (n1) | Gene mutation (+, −/n2) |
|---|---|
| crrA(0) | V167I (−/1), K228R (−/2), D198N (−/2), deletion (−/43) |
| crrB(1) | 1−249 aa del (−/43), C68S (−/20), Q301K (−/2), I27V (−/2), I66V (−/2), Q239H (−/2), T76A (−/2), V193G (+/1), T276A (−/1), Q287K (−/1), M63I (−/1) |
| crrC(2) | 1−22 aa del (−/43), A15V (−/4), V84L (−/3), V82I (−/1), P68T (+/1), 107−129 aa del (+/1) |
| pmrA(2) | A41T (−/4), M66I (−/3), I220N (−/2), D221E (−/2), G53V (+/1), D86E (+/1), E57G (−/1) |
| pmrB(4) | T246A (−/71), R256G (+/51), L213M (−/7), P344L (+/3), G358A (−/3), K220Q (−/3), S363I (−/3), T157P (+/1), M11I (−/1), G345E (−/1) |
| pmrC(4) | I138V (−/92), Q319R (−/81), C27F (−/72), D249E (−/6), L39V (−/3), T224K (−/3), V36I (−/3), P287S (+/2), D75E (−/2), S257L (+/1), A223S (+/1), E408K (−/1), K509T (−/1), I142V (−/1), A162V (−/1) |
| pmrD(0) | T60M (−/8), G80D (−/2) |
| phoP(2) | D191A (+/2), E82D (−/1) |
| phoQ(4) | P424L (+/3), G150D (−/2), D438E (+/1), A145S (−/1), T156S (−/1) |
| mgrB(27) | C28R (+/1), M1V (+/1), I45F (+/1), T-35C (+/2), deletion (+/1), IS903 (+/4), ISkpn14 (+/9), ISkpn18 (+/2), ISkpn26 (+/2), disrupt (+/3), 7Vfs (+/1) |
| arnA(0) | T450N (−/8), L260I (−/5), S18A (−/4), K442N (−/2), A22V (−/1), S061T (−/1), N660H (−/1) |
| arnB(7) | D101A (−/90), T272A (−/5), D274E (−/5), A148V (−/3), A124T (+/1), R269C (+/1), S371F (+/1), G367V (+/1), P264S (+/2), G36D (+/1), I115V (−/1), A261T (−/1), V308A (−/1), I89V (−/1) |
| arnC(1) | T30S (−/2), A36T (−/1), E204V (+/1), I286V (−/1) |
| arnD(2) | L94I (−/20), V300I (−/15), I53V (−/14), S271C (+/2), V187T (−/2), D209E (−/1), V247E (−/1), I300V (−/1), S164P (−/1), S188F (−/1), A225P (−/1) |
| arnE(0) | |
| arnF(3) | A45P (−/8), P100L (+/3), G46S (−/1), G89A (−/1) |
| arnT(3) | H156Q (−/12), I117V (−/11), D441E (−/5), G164S (+/2), S404D (−/2), T153A (−/2), S157R (−/2), K372R (−/2), S404C (+/1), S404S (−/1), K337N (−/1), A153V (−/1), V299M (−/1) |
| acrA(0) | A188T (−/74) |
| acrB(0) | R716L (−/68), E639D (−/4) |
| acrR(71) | ISkpn26 (+/67), L34F (+/2), 81Ffs (+/1), 121Gfs (+/1) |
| kpnE(1) | G67V (+/12), K112Q (−/7), A55V (+/1) |
| kpnF(0) | |
| ramA(0) | Deletion (−/10) |
| ramR(11) | Q122c (+/2), A6V (−/8), L66del (+/2), D77H (−/2), H186N (−/2), V67del (−/2), R3H (+/1), A159T (+/1), 1−38 aa del (+/1), E165K (+/1), G151D (+/1), L111del (+/1), S112R (+/1), A20T (−/1), I141T (−/1), A19V (−/1) |
| LpxM(0) | S253G (−/80), T229S (+/69), N7I (+/32), N6K (−/3), E76D (−/1), deletion (−/10) |
| yciM(2) | N212T (+/83), N212S (−/4), A265V (−/3), V163M (−/3), R350C (+/2), A231T (−/1), V298I (−/1) |
| soxS(0) | I26V (−/10), R29K (−/2) |
| soxR(0) | D142E (−/10), E143D (−/2) |
Gene mutations, involving genes deleted, stopped, truncated, frameshifted, or inserted by an insertion sequence.
n1, the number of deleterious mutations of the corresponding genes in the current study; n2, the number of strains with a gene variation; +, the mutation was predicted as the deleterious mutation; −, the mutation was predicted as the neutral mutation; fs, frameshift mutation.
Stop codon. Del, deletion. The bold variation shows the deleterious variation defined in this study.
FIG 4.
Frequencies of deleterious gene variations conferring polymyxin resistance.
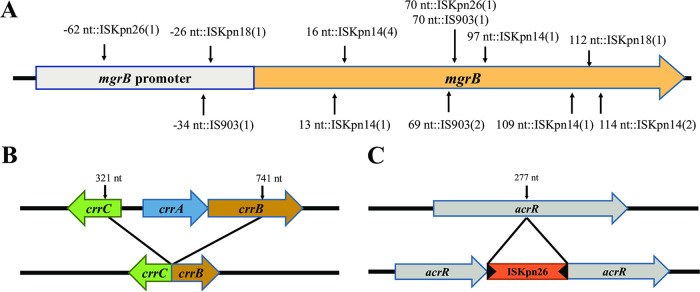
Moreover, the mgrB gene, as the most important polymyxin-related gene, was frequently disrupted and inactivated by insertion sequences (IS; 62.96%, 17/27), namely, ISkpn14 (52.94%, 9/17), IS903 (23.53%, 4/17), ISkpn26 (11.76%, 2/17), and ISkpn18 (11.76%, 2/17), shown in Fig. 5A. No other TCS-related genes were inserted by ISs. Other mutations on mgrB, predicted as deleterious, included missense mutations (3 strains, M1V, C28R, and I45F), a frameshift mutation (1, at the 141st site), overall missing (1), and disruptions (3, all truncated to nucleotide [nt] −30), as shown in Fig. 3. The nucleotide substitution (T-35C) at the promoter region of the mgrB gene (–10 region) was identified in two strains via BPROM. In addition, all 101 PR-CRKP strains were divided into the mgrB-mutation group or not. The Fisher exact test and Mann-Whitney U test showed that the mgrB inactivation was significantly relevant with a higher polymyxin resistance level (P < 0.001).
FIG 5.
(A to C) Schematic illustrations of (A) insertion sequences (IS; ISkpn14, IS903, ISkpn26, and ISkpn18) occurring at different insertion sites of mgrB; (B) the structure of crrCAB with the splicing mutation occurring at the 321st nt of crrC and the 741st nt of crrB, resulting in the crrA deletion; and (C) the IS (ISkpn26) occurring at the 277th nt of acrR.
Missense mutations were observed in other TCS genes, pmrB (4 strains), pmrC (4 strains) phoP (2 strains), and phoQ (4 strains). pmrD was otherwise relatively conserved and had no deleterious variant. The splicing mutations (42.57%, 43/101) in crrB and crrC genes were ubiquitous and caused crrA deletion and crrCAB inactivation; however, this did not increase the MIC of polymyxin. PR-CRKP strains (36.63%, 39/101) harbored no crrCAB system (Fig. 3 and 5B). Furthermore, we divided these strains into two groups, harboring a deletion-splicing mutation or not. Fisher’s exact test revealed that crrCAB was relevant with ST and KL (both P < 0.0001). Then the χ2 test showed a significant relationship between ST11 and crrCAB (P < 0.001), as well as KL47 and crrCAB (P = 0.005).
The efflux pump AcrAB-related genes involved in polymyxin resistance were also evaluated. The ISkpn26-inserted acrR genes, the repressor of the AcrAB, were observed in 67 strains at the same site (+277 nt) (67/101, 66.33%) (Fig. 5C). The PR-CRKP strains were then divided into two groups with or without ISkpn26-mediated insertion of the acrR gene, and it revealed that ST, especially ST11, was significantly associated with this insertion (P < 0.001). Furthermore, one missense mutation (L34F) and two shift mutations (81Ffs, 121Gfs) were also detected in the acrR gene. The ramR gene, the negative regulator of ramA, which can positively regulate efflux pump expression, was the second-most-frequently mutated gene. The amino acid deletions occurred at amino acids 66 and 111 and were overall missing from amino acids 1 to 38. One stop codon was detected at amino acid 122 (Q122*) of the ramR gene in 2 strains. One missense mutation was found in kpnE, an efflux pump-related multidrug resistance gene. No deleterious mutation was identified among the acrA, acrB, kpnF, ramA, soxS, and soxR genes.
In the genes related to LPS synthesis and modification, the missense mutations were identified in the arnB (6 strains) which was one part of arnBCADTEF operon and yciM (1 strain).
The mcr gene was detected only in strain CJFH07 (mcr-8.2), in which the missense mutation in mgrB and crrB and disruption of acrR also played a major role in polymyxin resistance (14).
However, the underlying resistance mechanism of 26 PR-CRKP strains (25.49%) remained elusive.
Complementation experiment and mRNA expression.
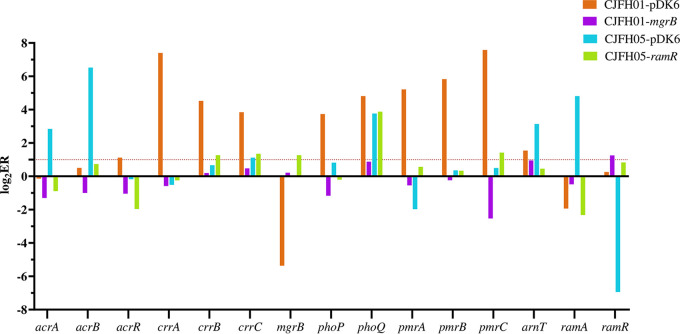
The complementation experiment of the ramR and mgrB genes was performed for strains CJFH05 and CJFH01, whose resistance only resulted from the mutation of ramR and the insertion of the mgrB promoter region, respectively. The polymyxin MIC of CJFH05 fell from 8 μg/mL to 1 μg/mL, whereas the MIC of CJFH01 decreased from 256 μg/mL to 0.5 μg/mL after the complementation experiment with the ramR and mgrB promoter, while the MICs of the control strains were the same as those of the original strains. The mRNA assessment is shown in Fig. 6. The expression of the polymyxin resistance genes (crrA/B/C, pmrA/B/C, and arnT) and efflux pump genes (acrA/B and ramA) reduced to the same level as ATCC 13883 after the plasmid was complemented, suggesting that the mutations in ramR or mgrB played a critical part in polymyxin resistance.
FIG 6.
Assessment of mRNA expression. The x axis represents the detected genes, including the proven and putative polymyxin resistance gene and the efflux pump-related gene. The y axis represents the logarithm of relative expression (RE). RE is the expression ratio (ER) of detected strains divided by the ER of ATCC 13883. ER is the average of the ratio of the detected genes to the rpoB gene for each strain.
DISCUSSION
Polymyxin has become the antibiotic of last resort against the challenge of CRKP infections (15). However, K. pneumoniae isolates that are resistant to both polymyxin and carbapenem have been documented, and the underlying mechanism was under surveillance (10, 16, 17). Predominant molecular characteristics of PR-CRKP strains may differ geographically and over time (10, 16, 18, 19). Therefore, in the study, the genomic survey of clinical PR-CRKP was conducted to investigate the genetic alterations in these isolates circulating in China.
Most PR-CRKP strains were recovered from the lower respiratory tract, similar to a previous survey in China (20). Furthermore, a considerable number of PR-CRKP strains were isolated from children (22.77%) and elderly patients (41.58%).
K. quasipneumoniae, as a novel Klebsiella sp., was commonly misidentified as K. pneumoniae in the clinical laboratory (21). WGS can be used to distinguish the species, and its distribution may be related to the regions and infection sites (13). However, many reports did not distinguish K. quasipneumoniae from K. pneumoniae, and few reports of polymyxin- and carbapenem-resistant K. quasipneumoniae were available online (3, 22, 23). In this study, about 10% of K. pneumoniae were reidentified as K. quasipneumoniae, similar to a previous report from Japan (9.2%) and higher than in Switzerland (4.59%), India (5.23%), and Nigeria (3.73%) (24–27).
Phylogenomic comparison revealed that this national collection of PR-CRKP showed diverse characteristics, with the presence of epidemic clones. Unlike in European countries, where ST258 was prevalent (28), the ST11 blaKPC-2-positive CRKP predominated in China (29). In our study, ST11 accounted for 66.67% of all PR-CRKP strains. The KPC-2-producing strains predominated in our PR-CRKP strains, and the IMP-producing K. pneumoniae strains were clustered in only Hunan, similar to previous reports (1, 30). Coexistence of dual carbapenemases (blaNDM-5 and blaKPC-2) in 2 strains was identified.
For other β-lactamases, blaSHV (89.11%) was the most frequently detected β-lactamase among 101 PR-CRKP strains, while chromosome-harbored blaSHV and blaOKP were intrinsically carried genes were demonstrated earlier (31). The chromosomal blaOKP family, a phylogenetic marker found in group KpII, shared the same ancestry as blaSHV and blaLEN (32, 33). A total of 60.40% (61/101) of PR-CRKP strains harbored blaCTX-M, in which blaCTX-M-65 predominated (44/61, 72.1%). By comparison, blaCTX-M-15 was often documented as the predominant blaCTX-M β-lactamases (34, 35), and this might be explained by geographic differences.
Mutations in mgrB and TCS-related genes were notable in the current study. mgrB gene inactivation was the underlying mechanism of polymyxin resistance of K. pneumoniae reported worldwide (4, 5, 36, 37), as confirmed in our study (26.73%, 27/101). IS-medicated insertion was the main reason for mgrB inactivation (65.38% in this study). Furthermore, the IS types involved ISkpn14 (52.94%), followed by IS903 (23.53%,), ISkpn26 (11.76%), and ISkpn18 (11.76%), consistent with previous reports (16, 18, 38, 39). ISkpn14 was prevalent as the main mgrB-inserted type in our study, which might vary geographically, such as IS5-like in Italy and Switzerland (4, 37) and ISkpn26 in Taiwan (16). The mgrB insertion by ISkpn18 was reported first in 2018 in a Chinese study and rarely elsewhere (40). Furthermore, the ISs were inserted in certain hot spots as previously reported, such as +69 to ~+75 nt (4, 5, 16, 18, 37), as also observed in the present study.
The dysfunction of the crrCAB system might mediate the polymyxin resistance (41, 42). In the present study, if the whole crrCAB system was deleted, as observed in almost all ST11 K. pneumoniae strains, the polymyxin MIC remained unchanged. The regulatory pathway by which crrAB regulated polymyxin resistance was only via crrC (41, 43). Consequently, crrCAB cannot contribute to polymyxin resistance if the whole system is removed. However, the splicing mutations of crrB and crrC may lead to the loss of the crrCAB function. The role of the crrCAB system in the polymyxin resistance of K. pneumoniae requires further in-depth study. In addition, previously unreported novel mutations were identified in our study; crrB (V193G), crrC (P68T), and crrC (107- to 129-amino acid [aa] deletion) were reported.
As for other TCS-related genes, the mutation in pmrB (R256G) was identified in our study, but its role in polymyxin resistance was controversial. It was considered a deleterious mutation according to the PROVEAN result (44). However, Sampaio reported that the substitutions in pmrB (T246A and R256G) were not involved in polymyxin resistance (45). Campos also documented that the expression of the pmrAB system with the above-described mutations had no significant influence on polymyxin resistance (15). Therefore, in our study, we considered it a nondeleterious mutation.
The inactivation in the repressors of efflux pump AcrAB genes has been documented to contribute to polymyxin resistance (10, 11). The efflux function of AcrAB would be hyperactive if acrR, the repressor of the AcrAB, was inactivated (11). In the present study, the inactivated acrR genes inserted coincidently at +277 nt by ISkpn26 were present in about 65.69% of PR-CRKP strains, and all of them belonged to ST11. A total of 25.74% (26/101) strains only harbored this acrR gene mutation and showed low-level polymyxin resistance. Similar to our results, Yang et al. demonstrated that ISkpn26 was always inserted into the acrR gene, resulting in acrR inactivation and then acrAB gene overexpression and multidrug resistance, and the insertion was mainly relevant to the ST11-KPC-2-producing CRKP (46). In their study, several K. pneumoniae isolates presenting an ISKpn26 interrupted the acrR gene and caused only a limited alteration of colistin MIC after exposure with 1-(1-naphthylmethyl)-piperazine (NMP), which was an efflux pump inhibitor. It seemed that the truncated acrR played only a small part in polymyxin resistance. Taken together, ST11 CRKP, as the predominate ST type in China, should be highlighted due to the overactivated efflux pump mediated by the ISkpn26-inserted acrR genes. However, the underlying mechanism remains elusive, and further study should be performed.
The ramR gene might repress the expression of ramA, which can regulate other efflux pumps, such as AcrAB, and then LPS modification and eventually mediate the decreased susceptibility to polymyxin in K. pneumoniae (47, 48), while in our study, the complementation experiment of the ramR gene was employed, indicating that the mutation of ramR will result in polymyxin resistance. Molecular epidemical studies of the ramR gene in PR-CRKP have rarely been reported, and the present study showed that mutations in the ramR gene were diverse, containing deletion, missense mutation, nonsense mutation, and truncation. This deserves more attention.
The arnBCADTEF operon, lpxM and yciM, mediated LPS biosynthesis and modification (3, 15, 22). In the deleterious mutations in the above-mentioned genes, the mutations in arnB were prevalent, similar to the results of a previous study (14). However, in the present study, though predicted as deleterious mutations by PROVEAN, the genes lpxM (T229S), lpxM (N7I), and yciM (N212T) were 100% similar to the reference sequences in GenBank (yciM [N212T]: NCBI reference sequence WP_002901781.1; lpxM [T229S] and lpxM [N7I]: NCBI reference sequence: WP_072353980.1) after alignment using the NCBI BLASTp online tool. Consequently, we defined them as nondeleterious variations. Multiple new mutations were identified in various genes in this study, but the significance of these mutations and their contribution to polymyxin resistance remained unclear.
In the current study, only one isolate carried the mcr gene, showing that polymyxin resistance was most associated with chromosome variation. However, the polymyxin resistance mechanisms of the 26 isolates remained unclear, indicating the presence of an alternative mechanism regulating the expression of these genes, which prompts the demand for exploration of polymyxin resistance.
Our study was limited by several factors. First, the majority of the isolates were collected from a few provinces/cities in China. Therefore, the national surveillance program should be further developed. Second, the underlying resistance mechanism of 26 PR-CRKP strains remained unclear, and we need to investigate this further.
In summary, the current study demonstrated the genetic diversity of the underlying polymyxin resistance mechanism of 101 PR-CRKP strains collected in China. Polymyxin resistance in the study is multifaceted and attributed to mutations in chromosomal genes leading to lipopolysaccharide modification or efflux of polymyxin through pumps. With few transmissible mcr genes identified, the prevalence of polymyxin-resistant strains was low in this unit. Furthermore, the high IS-inserted mgrB inactivation, the close relationship between ST11, the deletion or splicing mutations of crrCAB, and specific features of PR-K. quasipneumoniae constituted notable features of our PR-CRKP strains. The current study will help us to understand the diversity of polymyxin resistance in PR-CRKP circulating in China.
MATERIALS AND METHODS
Ethics approval.
The use of the K. pneumoniae isolates for research purposes was approved by the ethics committee of the China-Japan Friendship Hospital (2022-KY-054). The written informed consent from participants was exempted, and the privacy of involved subjects was not affected.
Strain collection and antibiotic sensitivity test (AST).
A total of 662 unduplicated CRKP strains, confirmed as K. pneumoniae by MALDI-TOF MS (Bruker Daltonics, Germany), were collected from 8 hospitals in 6 provinces/cities across China. Their MIC of carbapenems, polymyxin, and tigecycline were determined by BMD. A MIC of polymyxin of >2 μg/mL was defined as resistance recommended by CLSI M100 guidelines (49), a MIC of tigecycline of >2 μg/mL was defined as nonsusceptible according to the FDA (https://www.fda.gov/). The MIC of other antibiotics, including amikacin, aztreonam, levofloxacin, ciprofloxacin, tobramycin, and sulfamethoxazole/trimethoprim, were determined via the Vitek-2 system with N335 susceptibility cards (bioMérieux, France).
Epidemiological and clinical data.
Medical reports were accessed to collect demographic variables for all patients; 46 (6.95%) non-PR-CRKP cases were lost for follow-up, and their data were unavailable. The following data were retrospectively collected: gender, age, and place of residence.
Definition.
The strains with a MIC of >16 μg/mL were defined as having high-level polymyxin resistance as previously described (50). Deleterious mutations were defined as the mutations of certain genes predicted to affect protein function using PROVEAN.
WGS.
The whole genomes of PR-CRKP strains were sequenced using an Illumina NovaSeq PE150 at the Beijing Novogene Bioinformatics Technology Co., Ltd. Raw reads were filtered to remove low-quality sequences and adaptors. De novo assembly was conducted using SOAP de novo 2.04, SPAdes 3.10.0, and ABySS 1.3.7. The assembly results were integrated with CISA 1.3 software. The gap in preliminary assembly results was filled using Gapcloser 1.12.
Bioinformatics analysis.
Genotyping containing species reidentification, multilocus sequence typing (MLST), and identification of antimicrobial resistance genes, including carbapenemase and other β-lactamases, was conducted using Kleborate 2.0.4 (https://github.com/katholt/Kleborate). Capsular locus typing (KL-typing) was performed with the Kaptive webtool (https://kaptive-web.erc.monash.edu/).
Identification of mutations in proven or putative polymyxin resistance-related genes in K. pneumoniae.
The mutations in the mgrB gene and its promoter region were identified using NCBI-blast+ 2.11.0 with K. pneumoniae MGH 78578 (GenBank accession number CP000647) and K. quasipneumoniae strain ATCC 700603 (GenBank accession number CP014696) as reference strains.
The amino acid mutations encoded by proven or putative polymyxin resistance-related genes, such as TCS-related genes (pmrA, pmrB, pmrC, pmrD, phoP, phoQ, crrA, crrB, crrC), arnBCADTEF operons arnB and yciM, resistance-nodulation-division (RND) multidrug efflux pump AcrAB-involved and -regulating genes (acrA, acrB, acrR, ramA, ramR, soxS, and soxR), small multidrug resistance family (SMR) efflux pump-related genes (kpnE/kpnF), etc., were defined by alignment with the MGH 78578 and ATCC 700603 genomes through blast+ 2.11.0 with the E value set at 1e-50. The ISfinder online tool (https://isfinder.biotoul.fr/blast.php) was used to identify the insertion sequence (IS) type. The PROVEAN platform (http://provean.jcvi.org/index.php) was used to predict alterations in the biological functions of the above-described proteins. The promoter region was predicted using BPROM (http://linux1.softberry.com/berry.phtml).
Phylogenetic reconstruction.
For the phylogenetic analysis, high-quality single-nucleotide polymorphisms (SNPs) were obtained using snippy 3.2-dev against MGH 78578. The alignment file was filtered from variants with elevated densities of base substitutions as a putative recombination event using Gubbins 2.4.1. Then snp-sites 2.5.1 was used to reduce the filtered alignment to the core polymorphic sites. Next, the core alignment output was used to create a randomized accelerated maximum likelihood (RAxML) tree with RAxML 8.2.12 under the general time reversible (GTR)-Gamma model. The tree layout was graphically edited using iTOL 5.6.
Statistical analysis.
The differences between the ST type and crrCAB, the KL type and crrCAB, and the resistance level and related genes were evaluated via the χ2 test or Fisher’s exact test for categorical variables and the Mann-Whitney U test or continuous variables, as appropriate, using the IBM SPSS Statistics software packages 22.0 (SPSS, Inc., Chicago, IL). A P value of <0.05 was considered statistically significant.
Complementation experiment.
Apramycin-resistant pDK6 was used as the vector as previously described, but with modifications (51). The ramR and mgrB sequences were amplified by PCR via PrimeSTAR Max DNA polymerase (TaKaRa, Dalian) with primers ramR-F/R (forward [F]: AGCCAAGCTTGCATGCCTGCAGCCGGGTTCGATACTGCGATAA, reverse [R]: ACAGGAAACAGAATTCGAGCTCCGGCATTAAGAACAAACGGCA) and mgrB-F/R (F: AGCCAAGCTTGCATGCCTGCAGAGCCAGCGATGCCAGATTTA, R: ACAGGAAACAGAATTCGAGCTCCGCCAATCCATAAGATAGCCAC). The plasmid was linearized and PCR products were digested using the double-digestion method. Then, T4 DNA ligase (NEB, USA) was selected for ligating the vector at 16°C overnight, and the plasmid was transformed into CJFH05 and CJFH01 and then inoculated on 30 μg/mL apramycin-containing LB ager. The MICs of polymyxin were determined. The control strains were CJFH05 and CJFH01 with empty vector pDK6 transformed named CJFH01-pDK6 and CJFH05-pDK6, respectively.
mRNA extraction and expression assessment using digital droplet PCR (ddPCR).
Total RNA was extracted from strains CJFH01-pDK6, CJFH01-mgrB, CJFH05-pDK6, and CJFH05-ramR using the RNeasy minikit (Qiagen) according to the manufacturer’s instructions and reverse-transcribed into cDNA via a RevertAid first-strand cDNA synthesis kit with DNase I (Thermo Fisher Scientific, USA). The ddPCR was performed following the manufacturer’s protocol using QX200 ddPCR EvaGreen supermix (Bio-Rad, USA), a QX200 droplet generator (Bio-Rad), a QX200 droplet reader (Bio-Rad), and QuantaSoft software (Bio-Rad). The rpoB gene was used as the internal control. K. pneumonia ATCC 13883 was selected as the control strain. The primers used refer to previous studies (14). The relative expression (RE) of each strain was calculated as follows:
ER is the average of the ratio of the detecting gene to the rpoB gene for each strain. The RE is the ER of detecting strains divided by the ER of ATCC 13883.
Data availability.
All genome sequences have been submitted to the National Center for Biotechnology Information (NCBI) database with BioProject accession number PRJNA839599 and BioSample accession numbers SAMN28546397 to SAMN28546497. The original data presented in the study are included in the article. Further inquiries can be directed to the corresponding author.
ACKNOWLEDGMENT
This work was supported by the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (grant number CIFMS 2021-I2M-1-030).
Contributor Information
Binghuai Lu, Email: zs25041@126.com.
Monica Adriana Garcia-Solache, Brown University.
REFERENCES
- 1.Wang Q, Wang X, Wang J, Ouyang P, Jin C, Wang R, Zhang Y, Jin L, Chen H, Wang Z, Zhang F, Cao B, Xie L, Liao K, Gu B, Yang C, Liu Z, Ma X, Jin L, Zhang X, Man S, Li W, Pei F, Xu X, Jin Y, Ji P, Wang H. 2018. Phenotypic and genotypic characterization of carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012–2016). Clin Infect Dis 67:S196–S205. doi: 10.1093/cid/ciy660. [DOI] [PubMed] [Google Scholar]
- 2.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naha S, Sands K, Mukherjee S, Dutta S, Basu S. 2022. A 12 year experience of colistin resistance in Klebsiella pneumoniae causing neonatal sepsis: two-component systems, efflux pumps, lipopolysaccharide modification and comparative phylogenomics. J Antimicrob Chemother 77:1586–1591. doi: 10.1093/jac/dkac083. [DOI] [PubMed] [Google Scholar]
- 4.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group . 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shankar C, Pragasam AK, Anandan S, Veeraraghavan B. 2019. mgrB as hotspot for insertion sequence integration: change over from multidrug-resistant to extensively drug-resistant Klebsiella pneumoniae? Microb Drug Resist 25:1122–1125. doi: 10.1089/mdr.2018.0415. [DOI] [PubMed] [Google Scholar]
- 6.Chen AI, Albicoro FJ, Zhu J, Goulian M. 2021. Effects of regulatory network organization and environment on PmrD connector activity and polymyxin resistance in Klebsiella pneumoniae and Escherichia coli. Antimicrob Agents Chemother 65:e00889-20. doi: 10.1128/AAC.00889-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clements A, Tull D, Jenney AW, Farn JL, Kim S-H, Bishop RE, McPhee JB, Hancock REW, Hartland EL, Pearse MJ, Wijburg OLC, Jackson DC, McConville MJ, Strugnell RA. 2007. Secondary acylation of Klebsiella pneumoniae lipopolysaccharide contributes to sensitivity to antibacterial peptides. J Biol Chem 282:15569–15577. doi: 10.1074/jbc.M701454200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halaby T, Kucukkose E, Janssen AB, Rogers MR, Doorduijn DJ, van der Zanden AG, Al Naiemi N, Vandenbroucke-Grauls CM, van Schaik W. 2016. Genomic characterization of colistin heteroresistance in Klebsiella pneumoniae during a nosocomial outbreak. Antimicrob Agents Chemother 60:6837–6843. doi: 10.1128/AAC.01344-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Srinivasan VB, Rajamohan G. 2013. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother 57:4449–4462. doi: 10.1128/AAC.02284-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naha S, Sands K, Mukherjee S, Roy C, Rameez MJ, Saha B, Dutta S, Walsh TR, Basu S. 2020. KPC-2-producing Klebsiella pneumoniae ST147 in a neonatal unit: Clonal isolates with differences in colistin susceptibility attributed to AcrAB-TolC pump. Int J Antimicrob Agents 55:105903. doi: 10.1016/j.ijantimicag.2020.105903. [DOI] [PubMed] [Google Scholar]
- 11.Padilla E, Llobet E, Domenech-Sanchez A, Martinez-Martinez L, Bengoechea JA, Albertí S. 2010. Klebsiella pneumoniae AcrAB efflux pump contributes to antimicrobial resistance and virulence. Antimicrob Agents Chemother 54:177–183. doi: 10.1128/AAC.00715-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y-Y, Wang Y, Walsh TR, Yi L-X, Zhang R, Spencer J, Doi Y, Tian G, Dong B, Huang X, Yu L-F, Gu D, Ren H, Chen X, Lv L, He D, Zhou H, Liang Z, Liu J-H, Shen J. 2016. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis 16:161–168. doi: 10.1016/S1473-3099(15)00424-7. [DOI] [PubMed] [Google Scholar]
- 13.Long SW, Linson SE, Ojeda Saavedra M, Cantu C, Davis JJ, Brettin T, Olsen RJ. 2017. Whole-genome sequencing of human clinical Klebsiella pneumoniae isolates reveals misidentification and misunderstandings of Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae. mSphere 2:e00290-17. doi: 10.1128/mSphereDirect.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao J, Li Z, Zhang Y, Liu X, Lu B, Cao B. 2022. Convergence of MCR-8.2 and chromosome-mediated resistance to colistin and tigecycline in an NDM-5-producing ST656 Klebsiella pneumoniae isolate from a lung transplant patient in China. Front Cell Infect Microbiol 12:922031. doi: 10.3389/fcimb.2022.922031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos PA, Fuga B, Ferreira ML, Brigido R, Lincopan N, Gontijo-Filho PP, Ribas RM. 2021. Genetic alterations associated with polymyxin B resistance in nosocomial KPC-2-producing Klebsiella pneumoniae from Brazil. Microb Drug Resist 27:1677–1684. doi: 10.1089/mdr.2020.0531. [DOI] [PubMed] [Google Scholar]
- 16.Yang TY, Wang SF, Lin JE, Griffith BTS, Lian SH, Hong ZD, Lin L, Lu PL, Tseng SP. 2020. Contributions of insertion sequences conferring colistin resistance in Klebsiella pneumoniae. Int J Antimicrob Agents 55:105894. doi: 10.1016/j.ijantimicag.2020.105894. [DOI] [PubMed] [Google Scholar]
- 17.Nang SC, Azad MAK, Velkov T, Zhou Q, Li J, Barker E. 2021. Rescuing the last-line polymyxins: achievements and challenges. Pharmacol Rev 73:679–728. doi: 10.1124/pharmrev.120.000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva DMD, Faria-Junior C, Nery DR, Oliveira PM, Silva LOR, Alves EG, Lima G, Pereira AL. 2021. Insertion sequences disrupting mgrB in carbapenem-resistant Klebsiella pneumoniae strains in Brazil. J Glob Antimicrob Resist 24:53–57. doi: 10.1016/j.jgar.2020.11.003. [DOI] [PubMed] [Google Scholar]
- 19.Nirwan PK, Chatterjee N, Panwar R, Dudeja M, Jaggi N. 2021. Mutations in two component system (PhoPQ and PmrAB) in colistin resistant Klebsiella pneumoniae from North Indian tertiary care hospital. J Antibiot (Tokyo) 74:450–457. doi: 10.1038/s41429-021-00417-2. [DOI] [PubMed] [Google Scholar]
- 20.Jing N, Yan W, Zhang Q, Yuan Y, Wei X, Zhao W, Guo S, Guo L, Gao Y, Zhao L, Shi C, Li Y. 2022. Epidemiology and genotypic characteristics of carbapenem resistant Enterobacterales in Henan, China: a multicentre study. J Glob Antimicrob Resist 29:68–73. doi: 10.1016/j.jgar.2022.01.029. [DOI] [PubMed] [Google Scholar]
- 21.Seki M, Gotoh K, Nakamura S, Akeda Y, Yoshii T, Miyaguchi S, Inohara H, Horii T, Oishi K, Iida T, Tomono K. 2013. Fatal sepsis caused by an unusual Klebsiella species that was misidentified by an automated identification system. J Med Microbiol 62:801–803. doi: 10.1099/jmm.0.051334-0. [DOI] [PubMed] [Google Scholar]
- 22.Pragasam AK, Shankar C, Veeraraghavan B, Biswas I, Nabarro LE, Inbanathan FY, George B, Verghese S. 2016. Molecular mechanisms of colistin resistance in Klebsiella pneumoniae causing bacteremia from India: a first report. Front Microbiol 7:2135. doi: 10.3389/fmicb.2016.02135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He R, Yang Y, Wu Y, Zhong LL, Yang Y, Chen G, Qin M, Liang X, Ahmed M, Lin M, Yan B, Xia Y, Dai M, Chen H, Tian GB. 2021. Characterization of a plasmid-encoded resistance-nodulation-division efflux pump in Klebsiella pneumoniae and Klebsiella quasipneumoniae from patients in China. Antimicrob Agents Chemother 65:e02075-20. doi: 10.1128/AAC.02075-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nagaraj G, Shamanna V, Govindan V, Rose S, Sravani D, Akshata KP, Shincy MR, Venkatesha VT, Abrudan M, Argimón S, Kekre M, Underwood A, Aanensen DM, Ravikumar KL, NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance . 2021. High-resolution genomic profiling of carbapenem-resistant Klebsiella pneumoniae isolates: a multicentric retrospective Indian study. Clin Infect Dis 73:S300–S307. doi: 10.1093/cid/ciab767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afolayan AO, Oaikhena AO, Aboderin AO, Olabisi OF, Amupitan AA, Abiri OV, Ogunleye VO, Odih EE, Adeyemo AT, Adeyemo AT, Obadare TO, Abrudan M, Argimón S, David S, Kekre M, Underwood A, Egwuenu A, Ihekweazu C, Aanensen DM, Okeke IN, NIHR Global Health Research Unit on Genomic Surveillance of Antimicrobial Resistance . 2021. Clones and clusters of antimicrobial-resistant Klebsiella from southwestern Nigeria. Clin Infect Dis 73:S308–S315. doi: 10.1093/cid/ciab769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai K, Ishibashi N, Kodana M, Tarumoto N, Sakai J, Kawamura T, Takeuchi S, Taji Y, Ebihara Y, Ikebuchi K, Murakami T, Maeda T, Mitsutake K, Maesaki S. 2019. Clinical characteristics in blood stream infections caused by Klebsiella pneumoniae, Klebsiella variicola, and Klebsiella quasipneumoniae: a comparative study, Japan, 2014–2017. BMC Infect Dis 19:946. doi: 10.1186/s12879-019-4498-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cuénod A, Wüthrich D, Seth-Smith HMB, Ott C, Gehringer C, Foucault F, Mouchet R, Kassim A, Revathi G, Vogt DR, von Felten S, Bassetti S, Tschudin-Sutter S, Hettich T, Schlotterbeck G, Homberger C, Casanova C, Moran-Gilad J, Sagi O, Rodríguez-Sánchez B, Müller F, Aerni M, Gaia V, van Dessel H, Kampinga GA, Müller C, Daubenberger C, Pflüger V, Egli A. 2021. Whole-genome sequence-informed MALDI-TOF MS diagnostics reveal importance of Klebsiella oxytoca group in invasive infections: a retrospective clinical study. Genome Med 13:150. doi: 10.1186/s13073-021-00960-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macesic N, Nelson B, McConville TH, Giddins MJ, Green DA, Stump S, Gomez-Simmonds A, Annavajhala MK, Uhlemann AC. 2020. Emergence of polymyxin resistance in clinical Klebsiella pneumoniae through diverse genetic adaptations: a genomic, retrospective cohort study. Clin Infect Dis 70:2084–2091. doi: 10.1093/cid/ciz623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu P, Tang Y, Li G, Yu L, Wang Y, Jiang X. 2019. Pandemic spread of blaKPC-2 among Klebsiella pneumoniae ST11 in China is associated with horizontal transfer mediated by IncFII-like plasmids. Int J Antimicrob Agents 54:117–124. doi: 10.1016/j.ijantimicag.2019.03.014. [DOI] [PubMed] [Google Scholar]
- 30.Li J, Huang Z, Tang M, Min C, Xia F, Hu Y, Wang H, Zhou H, Zou M. 2021. Clonal dissemination of multiple carbapenemase genes in carbapenem-resistant Enterobacterales mediated by multiple plasmids in China. Infect Drug Resist 14:3287–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huynh B-T, Passet V, Rakotondrasoa A, Diallo T, Kerleguer A, Hennart M, Lauzanne AD, Herindrainy P, Seck A, Bercion R, Borand L, Pardos de la Gandara M, Delarocque-Astagneau E, Guillemot D, Vray M, Garin B, Collard J-M, Rodrigues C, Brisse S. 2020. Klebsiella pneumoniae carriage in low-income countries: antimicrobial resistance, genomic diversity and risk factors. Gut Microbes 11:1287–1299. doi: 10.1080/19490976.2020.1748257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haeggman S, Löfdahl S, Paauw A, Verhoef J, Brisse S. 2004. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother 48:2400–2408. doi: 10.1128/AAC.48.7.2400-2408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fevre C, Passet V, Weill F-X, Grimont PAD, Brisse S. 2005. Variants of the Klebsiella pneumoniae OKP chromosomal beta-lactamase are divided into two main groups, OKP-A and OKP-B. Antimicrob Agents Chemother 49:5149–5152. doi: 10.1128/AAC.49.12.5149-5152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wyres KL, Nguyen TNT, Lam MMC, Judd LM, van Vinh Chau N, Dance DAB, Ip M, Karkey A, Ling CL, Miliya T, Newton PN, Lan NPH, Sengduangphachanh A, Turner P, Veeraraghavan B, Vinh PV, Vongsouvath M, Thomson NR, Baker S, Holt KE. 2020. Genomic surveillance for hypervirulence and multi-drug resistance in invasive Klebsiella pneumoniae from South and Southeast Asia. Genome Med 12:11. doi: 10.1186/s13073-019-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peirano G, Pitout JDD. 2019. Extended-spectrum β-lactamase-producing Enterobacteriaceae: update on molecular epidemiology and treatment options. Drugs 79:1529–1541. doi: 10.1007/s40265-019-01180-3. [DOI] [PubMed] [Google Scholar]
- 36.Huang PH, Cheng YH, Chen WY, Juan CH, Chou SH, Wang JT, Chuang C, Wang FD, Lin YT. 2021. Risk factors and mechanisms of in vivo emergence of colistin resistance in carbapenem-resistant Klebsiella pneumoniae. Int J Antimicrob Agents 57:106342. doi: 10.1016/j.ijantimicag.2021.106342. [DOI] [PubMed] [Google Scholar]
- 37.Poirel L, Jayol A, Bontron S, Villegas M-V, Ozdamar M, Türkoglu S, Nordmann P. 2015. The mgrB gene as a key target for acquired resistance to colistin in Klebsiella pneumoniae. J Antimicrob Chemother 70:75–80. doi: 10.1093/jac/dku323. [DOI] [PubMed] [Google Scholar]
- 38.Fordham SME, Mantzouratou A, Sheridan E. 2022. Prevalence of insertion sequence elements in plasmids relating to mgrB gene disruption causing colistin resistance in Klebsiella pneumoniae. Microbiologyopen 11:e1262. doi: 10.1002/mbo3.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yan W, Zhang Q, Zhu Y, Jing N, Yuan Y, Zhang Y, Ren S, Hu D, Zhao W, Zhang X, Shi C, Wang M, Li Y. 2021. Molecular mechanism of polymyxin resistance in multidrug-resistant Klebsiella pneumoniae and Escherichia coli isolates from Henan province, China: a multicenter study. Infect Drug Resist 14:2657–2666. doi: 10.2147/IDR.S314490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dong N, Yang X, Zhang R, Chan EW-C, Chen S. 2018. Tracking microevolution events among ST11 carbapenemase-producing hypervirulent Klebsiella pneumoniae outbreak strains. Emerg Microbes Infect 7:146. doi: 10.1038/s41426-018-0146-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng Y-H, Lin T-L, Lin Y-T, Wang J-T. 2016. Amino acid substitutions of CrrB responsible for resistance to colistin through CrrC in Klebsiella pneumoniae. Antimicrob Agents Chemother 60:3709–3716. doi: 10.1128/AAC.00009-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McConville TH, Annavajhala MK, Giddins MJ, Macesic N, Herrera CM, Rozenberg FD, Bhushan GL, Ahn D, Mancia F, Trent MS, Uhlemann A-C. 2020. CrrB positively regulates high-level polymyxin resistance and virulence in Klebsiella pneumoniae. Cell Rep 33:108313. doi: 10.1016/j.celrep.2020.108313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mmatli M, Mbelle NM, Maningi NE, Osei Sekyere J. 2020. Emerging transcriptional and genomic mechanisms mediating carbapenem and polymyxin resistance in Enterobacteriaceae: a systematic review of current reports. mSystems 5:e00783-20. doi: 10.1128/mSystems.00783-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moghnia OH, Al-Sweih NA. 2022. Whole genome sequence analysis of multidrug resistant Escherichia coli and Klebsiella pneumoniae strains in Kuwait. Microorganisms 10:507. doi: 10.3390/microorganisms10030507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sampaio SCF, Bigelli Carvalho R, Mimica MJ, da Silva CB, Mimica LMJ, de Lima AV, Lima K, Rocha D, Sampaio JLM. 2021. Genotyping of paired KPC-producing Klebsiella pneumoniae isolates with and without divergent polymyxin B susceptibility profiles. Braz J Microbiol 52:1981–1989. doi: 10.1007/s42770-021-00600-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang Y, Yang Y, Chen G, Lin M, Chen Y, He R, Galvao KN, El-Gawad El-Sayed Ahmed MA, Roberts AP, Wu Y, Zhong LL, Liang X, Qin M, Ding X, Deng W, Huang S, Li HY, Dai M, Chen DQ, Zhang L, Liao K, Xia Y, Tian GB. 2021. Molecular characterization of carbapenem-resistant and virulent plasmids in Klebsiella pneumoniae from patients with bloodstream infections in China. Emerg Microbes Infect 10:700–709. doi: 10.1080/22221751.2021.1906163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Majumdar S, Yu J, Fookes M, McAteer SP, Llobet E, Finn S, Spence S, Monahan A, Monaghan A, Kissenpfennig A, Ingram RJ, Bengoechea J, Gally DL, Fanning S, Elborn JS, Schneiders T. 2015. Elucidation of the RamA regulon in Klebsiella pneumoniae reveals a role in LPS regulation. PLoS Pathog 11:e1004627. doi: 10.1371/journal.ppat.1004627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu Q, Sheng Z, Hao M, Jiang J, Ye M, Chen Y, Xu X, Guo Q, Wang M. 2021. RamA upregulates multidrug resistance efflux pumps AcrAB and OqxAB in Klebsiella pneumoniae. Int J Antimicrob Agents 57:106251. doi: 10.1016/j.ijantimicag.2020.106251. [DOI] [PubMed] [Google Scholar]
- 49.CLSI. 2020. Performance standards for antimicrobial susceptibility testing, 30th ed. M100. CLSI, Wayne, PA. [Google Scholar]
- 50.Elias R, Spadar A, Phelan J, Melo-Cristino J, Lito L, Pinto M, Goncalves L, Campino S, Clark TG, Duarte A, Perdigao J. 2022. A phylogenomic approach for the analysis of colistin resistance-associated genes in Klebsiella pneumoniae, its mutational diversity and implications for phenotypic resistance. Int J Antimicrob Agents 59:106581. doi: 10.1016/j.ijantimicag.2022.106581. [DOI] [PubMed] [Google Scholar]
- 51.Wei D, Wang M, Shi J, Hao J. 2012. Red recombinase assisted gene replacement in Klebsiella pneumoniae. J Ind Microbiol Biotechnol 39:1219–1226. doi: 10.1007/s10295-012-1117-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All genome sequences have been submitted to the National Center for Biotechnology Information (NCBI) database with BioProject accession number PRJNA839599 and BioSample accession numbers SAMN28546397 to SAMN28546497. The original data presented in the study are included in the article. Further inquiries can be directed to the corresponding author.