ABSTRACT
The dimethyl sulfoxide reductase (or MopB) family is a diverse assemblage of enzymes found throughout Bacteria and Archaea. Many of these enzymes are believed to have been present in the last universal common ancestor (LUCA) of all cellular lineages. However, gaps in knowledge remain about how MopB enzymes evolved and how this diversification of functions impacted global biogeochemical cycles through geologic time. In this study, we perform maximum likelihood phylogenetic analyses on manually curated comparative genomic and metagenomic data sets containing over 47,000 distinct MopB homologs. We demonstrate that these enzymes constitute a catalytically and mechanistically diverse superfamily defined not by the molybdopterin- or tungstopterin-containing [molybdopterin or tungstopterin bis(pyranopterin guanine dinucleotide) (Mo/W-bisPGD)] cofactor but rather by the structural fold that binds it in the protein. Our results suggest that major metabolic innovations were the result of the loss of the metal cofactor or the gain or loss of protein domains. Phylogenetic analyses also demonstrated that formate oxidation and CO2 reduction were the ancestral functions of the superfamily, traits that have been vertically inherited from the LUCA. Nearly all of the other families, which drive all other biogeochemical cycles mediated by this superfamily, originated in the bacterial domain. Thus, organisms from Bacteria have been the key drivers of catalytic and biogeochemical innovations within the superfamily. The relative ordination of MopB families and their associated catalytic activities emphasize fundamental mechanisms of evolution in this superfamily. Furthermore, it underscores the importance of prokaryotic adaptability in response to the transition from an anoxic to an oxidized atmosphere.
IMPORTANCE The MopB superfamily constitutes a repertoire of metalloenzymes that are central to enduring mysteries in microbiology, from the origin of life and how microorganisms and biogeochemical cycles have coevolved over deep time to how anaerobic life adapted to increasing concentrations of O2 during the transition from an anoxic to an oxic world. Our work emphasizes that phylogenetic analyses can reveal how domain gain or loss events, the acquisition of novel partner subunits, and the loss of metal cofactors can stimulate novel radiations of enzymes that dramatically increase the catalytic versatility of superfamilies. We also contend that the superfamily concept in protein evolution can uncover surprising kinships between enzymes that have remarkably different catalytic and physiological functions.
KEYWORDS: DMSO reductase, MopB, biogeochemical cycles, evolution, molybdopterin
INTRODUCTION
A growing body of structural (1–3) and phylogenetic (2, 4, 5) data indicates that the mononuclear molybdenum (Mo)/tungsten (W) enzymes that comprise the “dimethyl sulfoxide reductase” (DMSOR) family were present in the last universal common ancestor (LUCA) of all extant cellular lineages. The essential requirement for Mo/W to mediate crucial bioenergetic reactions for primordial life has already informed hypotheses about the potential mineralogy of the hydrothermal vent fields from which nascent cells may have emerged (6, 7). On the modern Earth, these enzymes mediate central reactions in the global carbon, nitrogen, and sulfur biogeochemical cycles (8) in addition to the biogeochemical cycles of less abundant elements, including arsenic, chlorine, selenium, iodine, and antimony (5, 8–13). Some of these members are implicated in the production of two volatile greenhouse gases, methane and nitrous oxide, that are of central importance to the global climate. These include the formylmethanofuran dehydrogenase B subunit (FwdB/FmdB) of the formylmethanofuran dehydrogenase complex (14) and the cytoplasmic F420-dependent formate dehydrogenase (15), both of which function in hydrogenotrophic methanogenesis, as well as the respiratory nitrate reductase catalytic subunit NarG (16). Methanogens constitute the principal producers of methane in the global carbon biogeochemical cycle (17, 18), whereas denitrifying bacteria constitute a substantial, if unconstrained, source of global nitrous oxide emissions (19, 20).
Despite the centrality of DMSOR members to the emergence of primordial life and modern-day global biogeochemical cycles, a single, unifying characteristic useful for defining this group is lacking in the literature. This assemblage has been variously called the DMSOR family (5, 21), the complex iron-sulfur molybdoenzyme (CISM) family (22), and the molybdopterin or tungstopterin bis(pyranopterin guanine dinucleotide) (Mo/W-bisPGD) enzyme family (8). The name DMSOR follows traditional biochemical convention by having the first enzyme in the group to be extensively characterized, the periplasmic bacterial dimethyl sulfoxide reductase catalytic subunit (DmsA) (23–25), define the group. Yet this name substantially understates the extraordinary catalytic and physiological versatility of known DMSORs. CISM was proposed as most DMSORs contain both a Mo/W-bisPGD cofactor and an N-terminal [4Fe-4S] iron-sulfur cluster cofactor. Additionally, many of the best-characterized DMSORs are associated with distinctive partner subunits that facilitate electron transfer with anaerobic respiratory chains in Bacteria and Archaea. Nonetheless, several DMSORs are known to lack the N-terminal [4Fe-4S] iron-sulfur cluster, and many known DMSORs either work in concert with unrelated partner subunits (8, 14, 26) or do not require additional subunits for catalytic activity (8, 27). Indeed, even the unusual Mo/W-bisPGD cofactor does not seem to define the assemblage, as putative DMSORs have been reported in the literature that lack the cofactor but appear to have the well-conserved structural fold that positions it (28–32). Most of these putative DMSORs are members of bacterial or mitochondrial aerobic respiratory chains.
In our previous study (5), we postulated that this assemblage of metalloenzymes was central to the origin of life and the evolution of global biogeochemical cycles over deep time. It raised questions, however, on how to better define this group and the full breadth of biogeochemical cycles and physiological processes that DMSOR members sustain. In this study, we performed a large-scale phylogenetic analysis of all known and putative DMSORs reported in the literature. We establish that putative DMSORs without Mo/W-bisPGD are indeed true members of this assemblage and show that the loss of this cofactor has occurred three independent times in the evolution of this group. Furthermore, we demonstrate that the radiation of families and novel biogeochemical reactions are frequently stimulated by the gain or loss of specific domains throughout time. Thus, we define DMSORs as a superfamily of enzymes and catalytic subunits united only by the distinctive Mo/W-bisPGD binding fold. We propose naming this assemblage the Mo/W-bisPGD binding (MopB) superfamily based on the name for this specific domain found in the latest version of the NCBI Conserved Domain Database (CDD) (33).
Finally, we generated, for the first time, a data set of over 47,000 putative MopB superfamily members from metagenomic sequence data, confirming that DMSORs are truly ubiquitous across both the bacterial and archaeal domains of life. Phylogenetic analyses using metagenomic and genomic sequence data conclusively establish that formate dehydrogenases were vertically inherited from the LUCA, while nearly all other families clearly evolved first in Bacteria and subsequently were horizontally transferred to hyperthermophilic or halophilic Archaea or, most intriguingly, Asgard Archaea members. Thus, the diversification of most families was driven by Bacteria as the growing oxidation state of surface environments, and the increasing availability of O2, on the Archean Earth (34–36) offered unprecedented redox challenges and novel energy-rich substrates to anaerobic life.
RESULTS
Phylogenetic analyses from cultured genome data sets.
All of the known and putative MopB members included in this analysis are shown in Table 1. Enzymes in boldface type indicate that their membership in the assemblage has yet to be established. We generated phylogenetic trees using data sets containing only canonical MopB members (see Fig. S1 in the supplemental material) and an expanded data set that also contained putative MopB enzymes (Fig. 1). We subjected the data set containing only canonical MopB members to several analyses using different amino acid substitution models and both parametric and nonparametric bootstrap algorithms. These supplemental phylogenies, along with the complete phylogenetic trees of Fig. 1 and Fig. S1 with full bootstrap support, can be accessed via the URL provided in the supplemental material.
TABLE 1.
Enzyme lineages included in the phylogenetic analysis, their function and cellular localization, and the amino acids that coordinate the Mo/W liganda
| Mo/W-bisPGD catalytic subunit(s) (abbreviation[s]) | MopB family lineage(s) | Substrate(s) | Function(s) | Cellular localization | Mo/W ligand | Reference(s) |
|---|---|---|---|---|---|---|
| Formyl-methanofuran dehydrogenase subunit B (FwdB/FmdB) | FwdB/FmdB and FhcB (?) | CO2 | Reduces CO2 to formate in hydrogenotrophic methanogenesis | Cytoplasm | Sec/Cys | 141, 142 |
| Formyltransferase/hydrolase subunit B (FhcB) | FwdB/FmdB and FhcB (?) | None | FhcB serves as a scaffold for the catalytic subunits FhcA and FhcD; the Fhc complex generates formate from formyl-H4MPT during growth on 1-carbon compounds | Cytoplasm | Lacks Mo/W-bisPGD | 32 |
| Formate dehydrogenase N subunit G (FdhG) | FdhG | HCOO−1 | Oxidizes formate to CO2 as an electron donor in anaerobic respiration | Periplasm | Sec/Cys | 143 – 145 |
| NAD-dependent formate dehydrogenase | Cytoplasmic formate dehydrogenases | CO2 | Reduces CO2 to formate during acetogenesis | Cytoplasm | Sec/Cys | 146, 147 |
| F420-dependent formate dehydrogenase | Cytoplasmic formate dehydrogenases | HCOO−1 | Oxidizes formate to CO2 during hydrogenotrophic methanogenesis | Cytoplasm | Sec/Cys | 148, 149 |
| Formate hydrogen lyase (FdhH) | Cytoplasmic formate dehydrogenases | HCOO−1 | Oxidizes excess formate to carbon dioxide during fermentative growth | Cytoplasm | Sec/Cys | 150 |
| NAD+ reducing formate dehydrogenase subunit A | Cytoplasmic formate dehydrogenases | HCOO−1 | Oxidizes excess formate to CO2 during aerobic growth | Cytoplasm | Cys | 151 |
| NADH-quinone oxidoreductase subunit 3 (Nqo3) | NAD- and F420-dependent Fdhs, FdhH, and FdsA (?) | NADH | Transfers electrons from NADH to the quinone pool during aerobic respiration | Cytoplasm | Lacks Mo/W-bisPGD | 28 |
| Assimilatory nitrate reductase catalytic subunits (NasC, NasA, and NarB) | NasC, NasA, and NarB | NO3− | Reduce nitrate to nitrite for assimilation into macromolecules | Cytoplasm | Cys | 152 – 154 |
| Arsenite oxidase catalytic subunit (AioA) | AioA and IdrA (?) | AsO33− | Oxidizes arsenite to arsenate as an electron donor in aerobic respiration and anoxygenic photosynthesis | Periplasm | No amino acid ligand | 155, 156 |
| Iodate reductase catalytic subunit (IdrA) | AioA and IdrA (?) | IO3− | Reduces iodate to iodide as the terminal electron acceptor in anaerobic respiration | Periplasm | No amino acid ligand | 9 |
| Periplasmic nitrate reductase catalytic subunit (NapA) | NapA | NO3− | Reduces nitrate to nitrite and can fulfill various physiological functions, including respiration, redox homeostasis, and assimilation | Periplasm | Cys | 157 |
| Acetylene hydratase (AH) | AH (?) | C2H2 | Hydrates acetylene to acetaldehyde during fermentative growth on acetylene | Cytoplasm | Cys | 27 |
| Haloarchaeal dimethyl sulfoxide reductase catalytic subunit (DmsA) | ? | (CH3)2SO and (CH3)3NO | Reduces DMSO and TMAO to DMS and TMA, respectively, during anaerobic respiration | Periplasm | Asp | 158 |
| Perchlorate reductase catalytic subunit (PcrA) | ? | ClO4− | Reduces perchlorate to chlorite as a terminal electron acceptor during anaerobic respiration | Periplasm | Asp | 159, 160 |
| Steroid C25 dehydrogenase catalytic subunit (S25dA) | DdhA, SerA, and EbdA | Steroid C25 | Hydroxylates the C25 atom of steroid molecules to yield sterol C25 during the anaerobic degradation of cholesterol | Periplasm | Asp | 161 |
| p-Cymene dehydrogenase catalytic subunit (CmdA) | DdhA, SerA, and EbdA | p-Cymene | Hydroxylates p-cymene to dimethyl(4-isopropylbenzyl) succinate during the anaerobic degradation of this hydrocarbon | Periplasm | Asp | 162 |
| Ethylbenzene dehydrogenase catalytic subunit (EbdA) | DdhA, SerA, and EbdA | Ethylbenzene | Hydroxylates ethylbenzene to (S)-1-phenylethanol during the anaerobic degradation of ethylbenzene | Periplasm | Asp | 163 |
| Respiratory selenate reductase catalytic subunit (SerA) | DdhA, SerA, and EbdA | SeO42− | Reduces selenate to selenite (SeO32−) as a terminal electron acceptor during anaerobic respiration | Periplasm | Asp | 164 |
| Respiratory chlorate reductase catalytic subunit (ClrA) | DdhA, SerA, and EbdA | ClO3− | Reduces chlorate to chlorite as a terminal electron acceptor during anaerobic respiration | Periplasm | Asp | 165 |
| Dimethyl sulfide dehydrogenase catalytic subunit (DdhA) | DdhA, SerA, and EbdA | (CH3)2S | Oxidizes DMS to DMSO as an electron donor in either anaerobic respiration or anoxygenic photosynthesis | Periplasm | Asp | 166 |
| Respiratory nitrate reductase catalytic subunit (NarG) | NarG | NO3− | Reduces nitrate to nitrite as a terminal electron acceptor during anaerobic respiration | Periplasm or cytoplasm | Asp | 167 – 169 |
| Bacterial dimethyl sulfoxide reductase catalytic subunit | DmsA | (CH3)2SO, (CH3)3NO, and other S- and N-oxides | Reduces DMSO and TMAO to DMS and TMA, respectively, during anaerobic respiration | Periplasm | Ser | 24 |
| Resorcinol hydroxylase catalytic subunit (RhL) | ? | Resorcinol | Hydroxylates the phenolic compound resorcinol to hydroxyhydroquinone as an electron donor in anaerobic respiration | Cytoplasm | Ser | 170 |
| Pyrogallol-phloroglucinol transhydroxylase catalytic subunit (PgtL) | ? | Pyrogallol | Hydroxylates the polyphenolic compound pyrogallol to phloroglucinol during fermentative growth on pyrogallol | Cytoplasm | Ser | 171 |
| Biotin sulfoxide reductase | ? | Biotin-d-sulfoxide and methionine-S-sulfoxide | Converts biotin-d-sulfoxide to d-biotin and methionine-S-sulfoxide to S-methionine so that d-biotin and S-methionine can be recycled as carbon and sulfur sources, respectively | Cytoplasm | Ser | 172 |
| Dimethyl sulfoxide reductase catalytic subunit (DorA) and trimethylamine N-oxide reductase catalytic subunit (TorA) | DorA and TorA | Various S- and N-oxides, including (CH3)2SO and (CH3)3NO | Reduce DMSO to DMS and TMAO to TMA as terminal electron acceptors in anaerobic respiration | Periplasm or cytoplasm | Ser | 173 – 176 |
| Polysulfide reductase catalytic subunit (PsrA) | PsrA, PhsA, and SrrA | Sn2− | Reduces polysulfides to Sn−12− and S2− as terminal electron acceptors in anaerobic respiration | Periplasm | Cys | 177, 178 |
| Thiosulfate reductase catalytic subunit (PhsA) | PsrA, PhsA, and SrrA | S2O32− | Reduces thiosulfate to sulfite (SO32−) and sulfide (S2−) as terminal electron acceptors in anaerobic respiration | Periplasm | Cys | 179, 180 |
| Respiratory selenite reductase catalytic subunit (SrrA) | PsrA, PhsA, and SrrA | SeO32− | Reduces selenite to elemental selenium (Se0) as a terminal electron acceptor in anaerobic respiration | Periplasm | Cys | 181 |
| Archaeal sulfur reductase catalytic subunit (aSreA) | ? | S0 | Reduces elemental sulfur to S2− as a terminal electron acceptor for anaerobic respiration in hyperthermophilic archaea | Periplasm | Cys | 182 |
| Bacterial sulfur reductase catalytic subunit (bSreA) | ? | S0 | Reduces elemental sulfur to S2− as a terminal electron acceptor for anaerobic respiration in hyperthermophilic bacteria | Cytoplasm | Cys | 183 |
| Sulfite oxidase catalytic subunit (SoeA) | ? | SO32− | Oxidizes sulfite to sulfate as an electron donor in anoxygenic photosynthesis | Cytoplasm | Cys | 184 |
| Tetrathionate reductase catalytic subunit (TtrA) | TtrA, SrdA, and archaeal arsenate reductase | S4O62− | Reduces tetrathionate to thiosulfate as a terminal electron acceptor in anaerobic respiration | Periplasm | Cys | 185 |
| Respiratory selenate reductase catalytic subunit (SrdA) | TtrA, SrdA, and archaeal arsenate reductase | SeO42− | Reduces selenate to selenite as a terminal electron acceptor during anaerobic respiration | Periplasm | Cys | 186 |
| Archaeal arsenate reductase catalytic subunit | TtrA, SrdA, and archaeal arsenate reductase | AsO43− | Reduces arsenate to arsenite (AsO33−) as a terminal electron acceptor during anaerobic respiration in some archaea | Periplasm | Cys | 187 |
| Arsenite oxidase catalytic subunit (ArxA) | ArxA and ArrA | AsO33− | Oxidizes arsenite to arsenate as an electron donor in anaerobic respiration or anoxygenic photosynthesis | Periplasm | Cys | 188 |
| Respiratory arsenate reductase catalytic subunit (ArrA) | ArxA and ArrA | AsO43− | Reduces arsenate to arsenite as a terminal electron acceptor in anaerobic respiration | Periplasm | Cys | 189, 190 |
| Alternative complex III subunit B (ActB) | PsrA, PhsA, and SrrA (?) | Unknown | ActB possibly functions to transfer electrons from the ActA and ActE subunits to the menaquinol-oxidizing ActC subunit during aerobic respiration | Periplasm | Lacks Mo/W-bisPGD | 31 |
Shown are all of the enzyme families and subfamilies utilized in our phylogenetic analyses; their physiological functions, substrates (if known) and cellular localizations; and the amino acid ligands that position the Mo/W-bisPGD cofactor (if present). Boldface type indicates that the family or subfamily has never been subjected to rigorous phylogenetic analysis. Question marks show that the position of the family or subfamily within the superfamily is unknown or hypothetical. TMA is trimethyl amine.
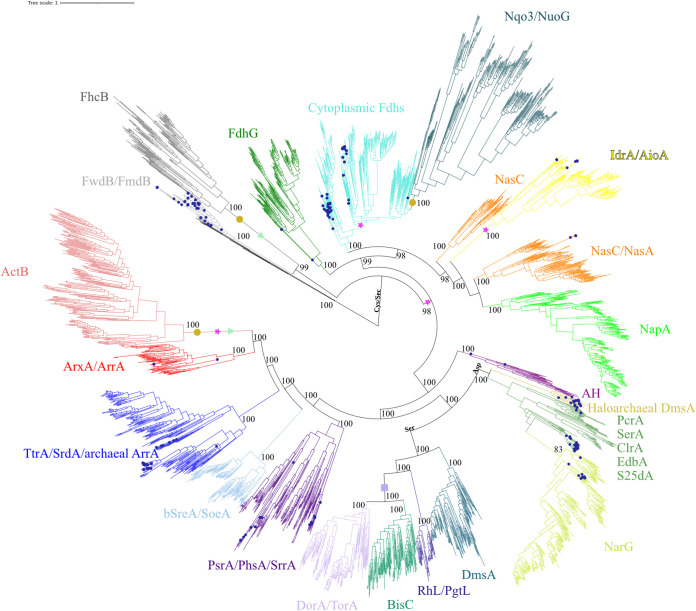
FIG 1.
Maximum likelihood phylogeny of 3,057 MopB domain-containing members constructed using 10,000 ultrafast bootstrap approximations. All sequences came from cultured organisms with sequenced genomes. Branches with blue circles indicate that the MopB homolog was taken from an archaeal genome. The lineages representing MopB families are named in the tree and represented by specific colors. Orange circles at a clade indicate that the lineage has lost the characteristic Mo/W-bisPGD cofactor. Magenta stars indicate that the lineage has acquired novel protein domains not found in other MopB families. Light-green triangles indicate that the MopB homologs in that family lineage no longer have a catalytic function. The light-blue square indicates that the MopB family has lost the N-terminal [4Fe-4S] iron-sulfur cluster.
Nonparametric bootstrapping was the first method used to assess statistical support for tree nodes in maximum likelihood phylogenies (37). Nonparametric bootstraps remain the most rigorous method available, and the parametric bootstraps generated by IQTree (38) and RAxML (39) remain necessary compromises between the need for rigorous statistical assessments of tree nodes and the computational challenges of analyzing ever-larger phylogenetic data sets. Figure S1 represents the largest data set ever subjected to nonparametric bootstrap assessment to our knowledge. All other phylogenetic trees were assessed via the parametric bootstrap offered by IQTree due to the significant computational cost of obtaining nonparametric bootstraps for the larger data sets. All of the tree topologies generated under divergent amino acid substitution models were identical, demonstrating that a genuine phylogenetic signal exists within this assemblage despite their primordial provenance.
Consistent with our previous analyses that used a smaller data set (5), we found robust evidence that the lineage representing the CO2-reducing FwdB subunit was the most ancient representative. Following that, the trees were divided into four large clades that represent major radiations. One clade was comprised of membrane-bound FdhG, the physiologically diverse cytoplasmic formate dehydrogenases, the aerobic arsenite oxidase (AioA), and the assimilatory (variously called NasC, NasA, or NarB) and periplasmic (NapA) nitrate reductase catalytic subunits. FwdB, FdhG, and the various cytoplasmic formate dehydrogenases all coordinate the Mo/W-bisPGD cofactor with either the 21st amino acid selenocysteine (Sec) or Cys. The NasC and NapA catalytic subunits utilize Cys to coordinate Mo/W-bisPGD. The AioA catalytic subunit, unique among MopB domain-containing proteins, does not coordinate Mo/W-bisPGD with an amino acid ligand. The second clade consisted mainly of catalytic subunits that utilize chalcophilic sulfur intermediates and oxyanions of the elements arsenic and selenium as electron donors and terminal electron acceptors. This clade includes enzymes that reduce the sulfur intermediates polysulfide (PsrA) and thiosulfate (PhsA) and the selenium oxyanion selenite (SrrA). Another lineage is specific for arsenic metabolism and encompasses the anaerobic arsenite oxidases (ArxA) and respiratory arsenate reductases (ArrA). The third clade includes tetrathionate reductase (TtrA), a recently discovered selenate reductase (SrdA), and a second alternative respiratory arsenate reductase.
The last major diversification constitutes the MopB domain-containing members that coordinate Mo/W-bisPGD with either a Ser or an Asp residue. The Ser-coordinating members include the DMSO reductase catalytic subunits DmsA (which harbors an N-terminal [4Fe-4S] cluster) and DorA (which lacks this feature). The trimethylamine N-oxide (TMAO) reductase (TorA), pyrogallol-phloroglucinol transhydroxylase (PgtL), resorcinol hydroxylase (RhL), and biotin sulfoxide reductase (BisC) catalytic subunits are also members of the Ser-coordinating group. Both PgtL and RhL mediate hydroxylation reactions. The lineage that utilizes Asp as the Mo/W-bisPGD-coordinating ligand displays the same stunning catalytic versatility as other MopB domain-containing catalytic subunits. The respiratory nitrate reductase catalytic subunit (NarG) is the best known among the members of this group. However, perchlorate reductase (PcrA), chlorate reductase (ClrA), and selenate reductase (SerA) catalytic subunits are also members of this clade, as is a dimethyl sulfide (DMS) dehydrogenase (DdhA) catalytic subunit. There are several catalytic subunits that mediate hydroxylation reactions utilizing aromatic substrates. These include the S25dA (steroid C25 dehydrogenase), CmdA, and EbdA catalytic subunits.
Of particular note is our discovery that the lineages that have lost Mo/W-bisPGD are all distantly related to one another (Fig. 1). Our topology was consistent with speculations drawn from the crystal structure of the Fhc complex that FhcB was closely related to the FwdB subunit of methanogenic Archaea (32). Similarly, the Nqo3 subunit was suggested to be related to the cytoplasmic formate dehydrogenases due to structural homology inferred from the crystal structure (28) and due to the domain architecture shared between Nqo3 and the NAD+-dependent FdsA formate dehydrogenase subunit. FdsA is a cytoplasmic formate dehydrogenase that functions in aerobic bacteria to remove excess formate during the late exponential phase of growth (22). Both share an additional N-terminal [4Fe-4S] cluster and a [2Fe-2S] iron-sulfur cluster, a domain architecture identical to that of iron-only hydrogenases. Therefore, the impetus for the diversification of FdsA away from the other cytoplasmic formate dehydrogenases that participate in acetogenesis, methanogenesis, and carbon fixation was the fusion of a cytoplasmic formate dehydrogenase with an iron-only hydrogenase domain.
Surprisingly, while ActB was inferred from structural homology to be the result of a fusion of a PsrA catalytic subunit with the PsrB electron transfer subunit, we demonstrate conclusively that ActB is more closely related to the ArrAB complex of the respiratory arsenate reductase. However, the impetus for the radiation of ActB subunits from the MopB domain-containing proteins was not merely the fusion of ArrA- and ArrB-like subunits but the loss of Mo/W-bisPGD and 3 [4Fe-4S] clusters and the presence of a novel high-potential [3Fe-4S] cluster (31). This is the first evidence linking the diversification of the ArxA/ArrA lineage, which allowed organisms to exploit the bioenergetic pool of arsenite and arsenate available in the late Archean eon (5, 40), to the ability of anaerobic life to draw energy from the growing reservoir of O2 at the transition between the anoxic and oxic worlds.
Equally as interesting was the positioning of the catalytic subunits of poorly characterized MopB members (Fig. 1). The IdrA subunit of the respiratory iodate reductase clustered with AioA. This is consistent with the sequence analysis of the IdrA subunit performed during the discovery and initial characterization of the enzyme (9). This analysis revealed a distinctive N-terminal high-potential [3Fe-4S] cluster found instead of the typical low-potential [4Fe-4S] cluster of most MopB domain-containing proteins and the lack of an amino acid ligand for Mo/W-bisPGD. Due to the presence of IdrA homologs in the deepest branches of the IdrA/AioA clade, respiratory iodate reduction likely preceded the catalytic function of this lineage to exploit arsenite as an electron donor in aerobic respiration and anoxygenic photosynthesis. While one may presume that the elemental sulfur reductases of hyperthermophilic Archaea and Bacteria share an ancestor, our results demonstrate that they have separate evolutionary histories. Archaeal SreA (aSreA) is phylogenetically indistinguishable from the PsrA/PhsA/SrrA lineage, adding yet another catalytic function to this group. Bacterial SreA (bSreA) (sulfur reductase), however, clearly clustered with SoeA (sulfite oxidase), and the bSreA/SoeA lineage represents a substantial, and heretofore unappreciated, radiation of MopB domain-containing proteins.
Phylogenetic and structural evidence for a MopB superfamily.
The classification of proteins into subfamilies, families, and superfamilies was formally introduced by Dayhoff (41). This concept was integral to the organization of the Protein Sequence Database, established in the 1960s to facilitate early evolutionary studies of enzymes (42). Subfamilies and families both have objective definitions based on percent sequence identity, with families defined as collections of proteins with ~50% sequence identity and subfamilies defined as those with ~80% sequence identity. Superfamilies, in contrast, are assemblages of proteins whose common ancestry can be inferred only from statistical methods (e.g., phylogenetic analyses). We found that there is consistently ~15% sequence identity between different MopB lineages (e.g., between FdhG and NarG and PsrA/PhsA/SrrA). MopB domain-containing proteins thus clearly represent a protein superfamily. The different lineages, with no less than 40% sequence identity between them, constitute distinct families within the superfamily.
The concept of protein superfamilies is analytically useful because it reveals basic mechanisms by which the incredible functional and structural diversity of millions of extant protein families emerged from a limited repertoire of structural folds and catalytic domains that early life exploited for survival. Beyond gene duplication, one of the principal mechanisms by which novel families diversify from superfamilies is the acquisition, loss, and rearrangement of domains around a central domain or structural fold that defines the superfamily (43, 44). In Fig. 1, we highlight major evolutionary domain fusion and loss events, including three independent instances in which Mo/W-bisPGD was lost and two separate events in which catalytic activity itself was lost. As would be expected from a diverse superfamily, these events seemed to drive the diversification of novel families.
In an effort to define what core structural fold or domain may unify this superfamily, we performed structural alignments of all 15 available crystal structures and a single cryogenic electron microscopy (cryo-EM) structure from this superfamily. We observed that the only region of these 16 superfamily members that displays structural homology is a region of 195 amino acids made up of an α-helix and two β-sheets stretching from the N-terminal [4Fe-4S] or [3Fe-4S] cluster, if present, and the pyranopterin guanine dinucleotide (PGD) organic moiety of the Mo/W-bisPGD cofactor proximal to that cluster (Fig. 2). This comprises a single domain that the NCBI Conserved Domain Database refers to as the molybdopterin binding (MopB) domain (accession number cl09928). The Q score for the structural alignment, at 0.0205, demonstrates that the other three to four domains characteristic of MopB superfamily members (22) have not only little conservation of primary structure but also no conservation of tertiary structure. The other domains of different MopB-containing families thus lack any common ancestry. We have provided the strongest evidence to date, therefore, that a single domain is the only feature universally shared by all members. Therefore, this is not a family of Mo- or W-utilizing enzymes and catalytic subunits with broadly shared primary and tertiary structures. These enzymes definitively constitute a catalytically and mechanistically diverse superfamily united only by the single MopB domain and structural fold. This is a fundamentally new insight into the evolution of these enzymes and suggests that the other MopB domains should be more rigorously examined to elucidate how specific families have radiated from the broader superfamily. We propose that this superfamily ought to be named the MopB superfamily.
FIG 2.
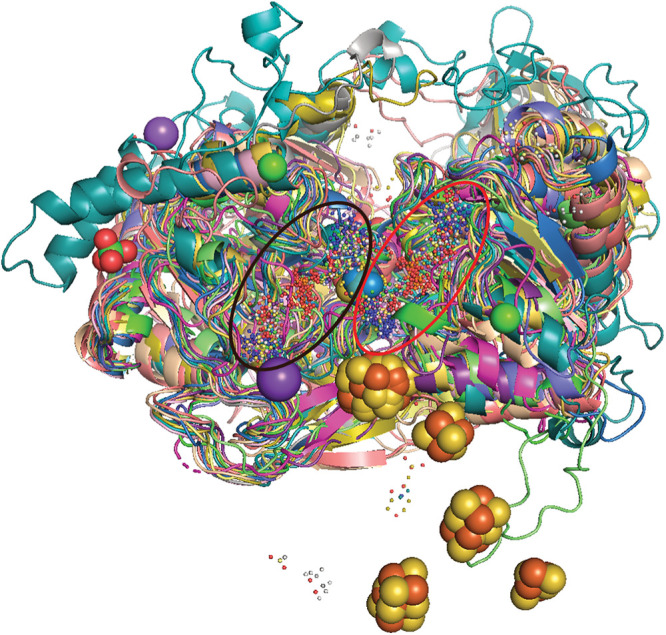
Superposition of all available crystal structures (of which there are 15) of MopB superfamily members and a single cryo-EM structure. The only portion that is retained is the region where the structural alignment of these structures was found to have significant structural homology. This region corresponds to the MopB domain; a stretch between the N-terminal iron-sulfur cluster, if present; and the PGD moiety of Mo/W-bisPGD proximal to that cluster. The proximal PGD is indicated by a black circle, and the distal PGD (the one furthest away) is indicated by a red circle. The cyan atoms at the intersection of these two circles represent Mo atoms, while the gold atoms represent W atoms. The various iron-sulfur clusters were retained in this image to demonstrate that while the orientation and positioning of the Mo/W-bisPGD cofactor of these diverse enzymes and catalytic subunits are conserved, the positionings of the iron-sulfur clusters differ substantially between different superfamily representatives.
Survey for MopB superfamily members across cultured and metagenome-assembled genomes.
We wished to conduct the first evolutionary study of MopB superfamily members across the explosion of new sequence data made available through metagenomics studies (45–47) to better understand the significance of uncultured organisms in global biogeochemical cycles, obtain sequence data from recently discovered archaeal phyla to resolve which MopB families were most likely present in the LUCA, and, finally, determine whether members of the closest known lineage to eukaryotes, Asgardarchaeota, utilize MopB superfamily members to any extent. We were able to obtain a list of 47,011 unique MopB superfamily members (Table 2) across 98 bacterial and 9 archaeal phyla (Table S1). As one would expect, the most prevalent families were those involved in the biogeochemical cycles of the abundant elements carbon, nitrogen, and oxygen. The cytoplasmic formate dehydrogenase family constituted 21.56% of the total sequences that we found. The NasC/NasA family of assimilatory nitrate reductase catalytic subunits similarly represented nearly one-fifth of the MopB superfamily (18.60%). The FdhG family contained 13.10% of the total MopB sequences, and the NarG family contained 9.90%. The ActB family from alternative complex III harbored 7.37%. Note that while the Nqo3 family had only 3.62%, a substantial fraction of the cytoplasmic formate dehydrogenase data set contained members from the Nqo3 family (i.e., they did not have the requisite Cys residue to position Mo/W-bisPGD in sequence alignments). This was also the case for the FhcB and FwdB families.
TABLE 2.
Distribution of MopB family hits from BLASTX searches for MopB superfamily members across genomes from cultured isolates and high-quality metagenome-assembled genomes through GTDB-Tk
| MopB family | No. of hits found | % of hits for each family | No. of archaeal representatives | % from Archaea | No. of bacterial representatives | % from Bacteria |
|---|---|---|---|---|---|---|
| ActB | 3,464 | 7.37 | 0 | 0.00 | 3,381 | 100.00 |
| AH | 452 | 0.96 | 21 | 5.95 | 332 | 94.05 |
| AioA/IdrA | 419 | 0.89 | 5 | 1.21 | 408 | 98.79 |
| ArxA/ArrA | 267 | 0.57 | 11 | 4.21 | 250 | 95.79 |
| Asp-coordinating | 347 | 0.74 | 68 | 25.00 | 204 | 75.00 |
| BisC | 322 | 0.69 | 0 | 0.00 | 305 | 100.00 |
| bSreA/SoeA | 1,354 | 2.88 | 5 | 0.37 | 1,339 | 99.63 |
| DmsA | 1,559 | 3.32 | 13 | 1.07 | 1,204 | 98.93 |
| DorA/TorA | 2,673 | 5.69 | 0 | 0.00 | 2,202 | 100.00 |
| FdhG | 6,147 | 13.08 | 38 | 0.68 | 5,539 | 99.32 |
| Fdhs (cytoplasmic) | 10,134 | 21.56 | 524 | 6.12 | 8,042 | 93.88 |
| FhcB | 94 | 0.20 | 0 | 0.00 | 94 | 100.00 |
| FwdB | 751 | 1.60 | 399 | 72.81 | 149 | 27.19 |
| NapA | 1,800 | 3.83 | 52 | 0.12 | 1,721 | 99.88 |
| NarG | 4,656 | 9.90 | 21 | 0.49 | 4,301 | 99.51 |
| NasC/NasA | 8,746 | 18.60 | 113 | 1.40 | 7,941 | 98.60 |
| Nqo3 | 1,701 | 3.62 | 0 | 0.00 | 1,700 | 100.00 |
| PsrA/PhsA/SrrA | 954 | 2.03 | 50 | 5.82 | 809 | 94.18 |
| RhL/PgtL | 285 | 0.61 | 0 | 0.00 | 196 | 100.00 |
| TtrA/SrdA/alternative ArrA | 886 | 1.89 | 45 | 5.35 | 796 | 94.65 |
| Total superfamily hits | 47,011 | 100 | 1,315 | 40,913 | ||
To gain a better understanding of the extent to which Archaea contribute to various biogeochemical cycles using MopB superfamily members, we generated phylum counts for the full metagenomic data set. We found 42,228 unique MopB family hits in the phylum counts. This means that ~10% of the 47,011 total hits that we retrieved represented multiple copies of homologs from the same family within a single metagenome-assembled genome (MAG) or organism genome. Either the majority of MopB families were confined to the bacterial domain or archaeal representatives constituted ~1.0% or fewer of the total hits (Table 2). However, several families stood out for having significantly higher percentages of archaeal representatives. The most stunning was the FwdB family, where fully 72.81% of FwdB homologs were found in archaeal phyla. This is the only family across the entire superfamily in which archaea constituted the bulk of representatives. One-quarter of Asp-coordinating MopB catalytic subunits were found in archaeal phyla, and nearly all of these were from halophilic Archaea in the phylum Halobacteriota and thus are presumably homologs of the haloarchaeal dimethyl sulfoxide reductase catalytic subunit. Other families that harbored substantial numbers of archaeal homologs were the cytoplasmic formate dehydrogenases (6.12%); acetylene hydratase (AH) (5.95%); and the PsrA/PhsA/SrrA (5.82%), TtrA/SrdA/alternative arsenate reductase (5.35%), and ArxA/ArrA (4.21%) catalytic subunits. These findings demonstrate that members of the domain Archaea comprise a significant fraction of the taxa in MopB families known to be essential drivers of the global sulfur, arsenic, and selenium biogeochemical cycles, even if they acquired the ability to do so through horizontal gene transfer (HGT). Additionally, this analysis further highlights the importance of Archaea in the global carbon biogeochemical cycle.
Phylogenetic analysis of MopB superfamily members using high-quality MAGs.
Using the CD-HIT program (48), we reduced the data set of 47,011 superfamily members to a more computationally feasible 1,570 sequences (Fig. 3) for phylogenetic analyses. The URL where this phylogeny can be accessed, with full node support and branch labels, is provided in the supplemental material. The topology that we obtained was identical to those shown in Fig. 1 and Fig. S1 with respect to the relationship between the chalcophilic oxyanion and sulfur intermediate oxidoreductases and the Asp- and Ser-coordinating MopB superfamily members. The monophyly of these groups was strongly supported. Indeed, the positioning of the AH family ancestral to these three broad groups was also strongly supported (even if the monophyly of this clade was not well supported). The positioning of the ActB family with the ArxA/ArrA family was also robustly supported. Many differences are also apparent. For example, the haloarchaeal dimethyl sulfoxide reductase catalytic subunit was replaced by PcrA as the oldest Asp-coordinating lineage. We also found evidence for novel families within the MopB superfamily in the tree. The multiple branches in black that are basal to the chalcophilic oxyanion and sulfur intermediate oxidoreductases and the Asp- and Ser-coordinating families were all drawn from sequences whose best-supported BLASTX homologs included AH-like, PsrA/PhsA/SrrA-like, and DmsA-like hits and appear to utilize a Cys residue to position Mo/W-bisPGD. A similar seemingly novel lineage was found sister to the TtrA/SrdA/alternative arsenate reductase catalytic subunit family.
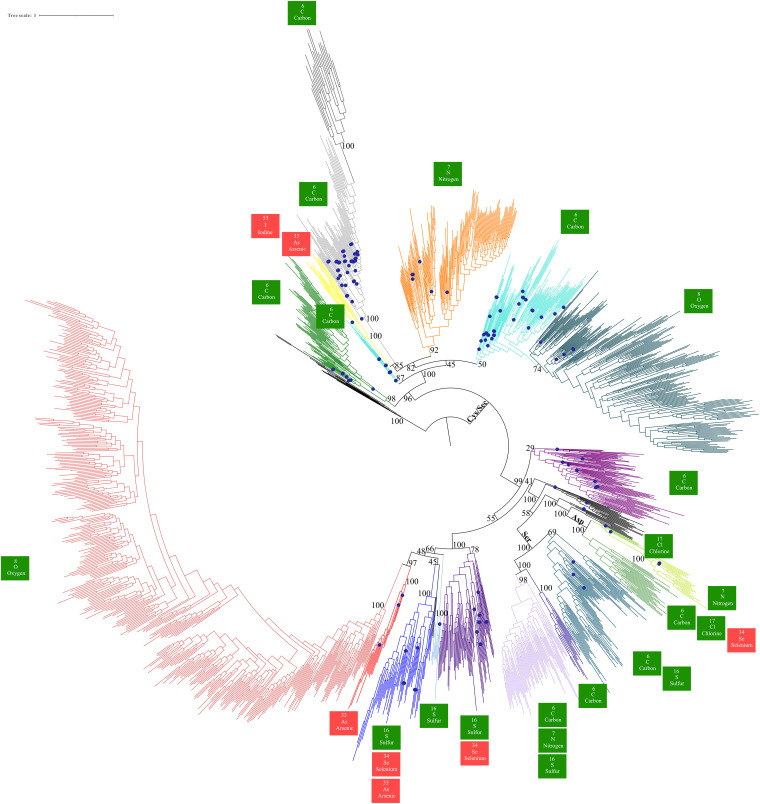
FIG 3.
Maximum likelihood phylogeny of 1,570 MopB domain-containing members constructed using 10,000 ultrafast bootstrap approximations. This phylogeny, unlike the others, contains both genomes from cultured isolates and high-quality MAGs taken from metagenomic studies. Branches with blue circles indicate that the MopB homolog was taken from an archaeal genome or MAG. We have overlaid onto the tree topology bootstrap support at crucial nodes. The color scheme for specific MopB families is identical to the one in Fig. 1. Additionally, to emphasize the catalytic versatility of the MopB superfamily, we have highlighted all known biogeochemical cycles that each MopB family is known to mediate. Elements in green boxes are nonmetals, and elements in red boxes represent metalloids.
The CD-HIT 50% sequence identity setting also removed nearly all of the IdrA/AioA, bSreA/SoeA, and ArxA/ArrA family homologs (approximately 16 sequences from each family were left for the analysis). This could reflect a more recent diversification from the MopB superfamily, strong selective pressure to reduce sequence diversity to remain specialized for a limited array of bioenergetic substrates, or both. Additionally, the phylogenies constructed using this more diverse data set showed that the NasC/NarA and NapA catalytic subunits constituted a single monophyletic family, as did the DorA/TorA and BisC families.
The most substantive differences between the phylogenies constructed solely from MopB representatives from cultured genomes and those that incorporated sequence data from MAGs concern the most ancient radiations from the superfamily. Figure 3 strongly supports an evolutionary scenario in which the FdhG family represents the most ancient lineage within the MopB superfamily. This was followed by the diversification of cytoplasmic formate dehydrogenases, which, in this scenario, no longer form a monophyletic clade. A closeup view of this region of the tree is provided in Fig. 4.
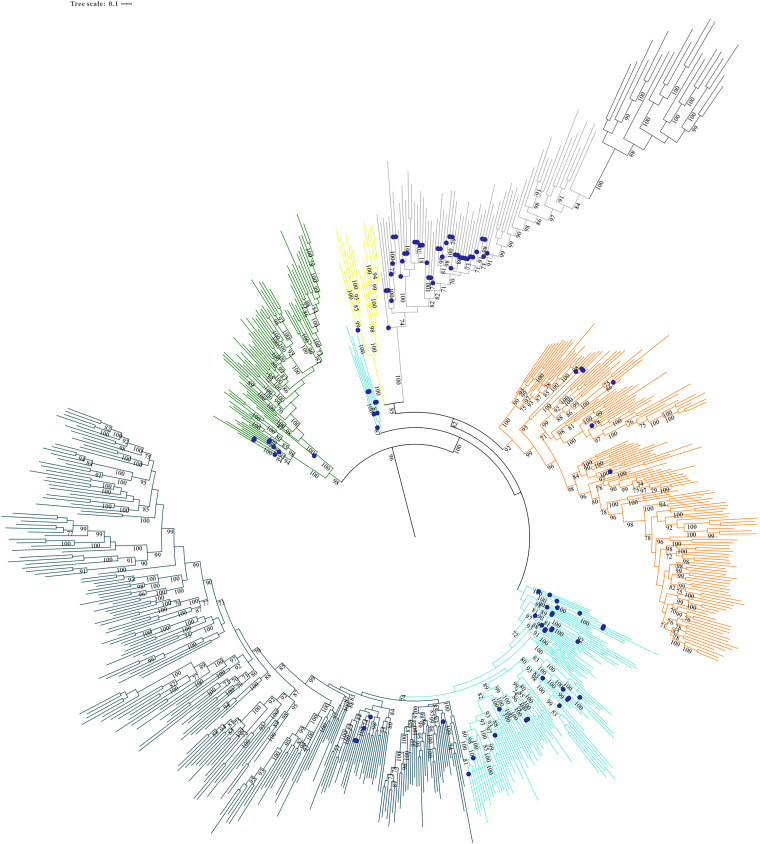
FIG 4.
Subpruned portion of the maximum likelihood phylogeny shown in Fig. 3. This region contains FdhG, the cytoplasmic formate dehydrogenases, and the IdrA/AioA, assimilatory and periplasmic nitrate reductase, and Nqo3 families. Branches with blue circles indicate that the MopB homolog was taken from an archaeal genome or MAG. The color scheme for specific MopB families is identical to the one in Fig. 1. All node supports of ≥70 are provided.
A conserved feature between the topologies in Fig. 1, Fig. 3, and Fig. S1 and previous analysis (5) is the significant uncertainty in the exact positioning of the IdrA/AioA, NasC/NasA, and NapA families. In this tree, the FwdB family is now added to their number. We believe that the most parsimonious explanation for the lack of robust support for the position of these families within the broader superfamily is that they represent diversifications specifically within the cytoplasmic formate dehydrogenases, and the ancestral intermediates for these enzymes that would harbor sequence features consistent with formate dehydrogenase ancestors have been lost through geologic time. Two other features of this region of the tree are notable in Fig. 4. The first concerns the FwdB family.
As described above, the FwdB family is the only family in the entire MopB superfamily that has more archaeal than bacterial representatives. This family has been thought to be involved exclusively in hydrogenotrophic methanogenesis (14), but we show that it does have bacterial representatives. Even among archaeal homologs, 83 of the 548 FwdB homologs came from Asgardarchaeota MAGs, 50 came from the phylum Thermoproteota, and crucially, we found at least 1 FwdB homolog in 8 out of the 9 archaeal phyla that had MopB superfamily representatives in either a genome or a MAG. Thus, the idea that FwdB functions exclusively in methanogenesis is no longer tenable. What is apparent from inspecting the FwdB family in Fig. 4, however, is that it clearly arose in Archaea and was later transferred to the bacterial domain via HGT. This makes the FwdB family the only one in the whole superfamily in which a radiation of enzymes came from Archaea. The second observation is that there is a substantial and varied assemblage of archaeal homologs in the cytoplasmic formate dehydrogenase family and, hence, strong evidence of an origin for this family in the LUCA. Despite this, the propensity of these bioenergetic subunits to be transferred freely between the domains made it difficult for us to confidently assess which MopB families were inherited from the LUCA.
DISCUSSION
Metagenomics confirms the vertical inheritance of formate oxidation and CO2 reduction from the LUCA.
The alkaline hydrothermal vent theory for the origin of life posits that the warm (~70°C), alkaline, and H2-enriched fluids emitted from alkaline hydrothermal vents into the acidic, CO2-enriched waters of the anoxic ocean represented a unique habitat on early Earth in which H+ oxidation is spontaneously coupled to CO2 reduction (49, 50). Such an environment would have been conducive to the evolution of the first chemiosmotic, energy-transducing processes that differentiated biochemically driven oxidation-reduction reactions from geochemically driven ones. A major consequence of such a theory is that CO2 reduction must have been the earliest electron acceptor in anaerobic respiration and the source of carbon for the synthesis of organic macromolecules. The results of phylogenomic analyses of genomes from cultured Bacteria and Archaea seemed to be consistent with this hypothesis in that components of the Wood-Ljungdahl pathway of carbon assimilation appear to have been vertically inherited between these two domains (4). Unlike that previous study, however, we conducted phylogenetic analyses of the formate dehydrogenases themselves, and thus, we obtained a phylogenetic signal from the periplasmic and cytoplasmic formate dehydrogenase families that is independent of other components of bacterial and archaeal genomes. Additionally, we have taken advantage of sequence data from archaeal phyla that researchers utilizing genomic resources do not include.
Cultured isolates of the domain Archaea are comprised mainly of methanogens, hyperthermophiles, and halophiles. A principal finding of our analyses is that Archaea acquired all MopB families involved in acetylenotrophy and the nitrogen, sulfur, arsenic, and, possibly, selenium biogeochemical cycles via HGT. Two major archaeal groups that frequently inherited these bioenergetic traits via HGT were hyperthermophiles and halophiles. It seems likely that these two extremophilic archaeal groups were able to acquire MopB family members and compete effectively with bacterial communities for access to electron donors and terminal electron acceptors because they inhabit rare environmental niches in which Archaea can predominate over Bacteria, as demonstrated by observations of Archaea dominating hypersaline and soda lake environments (51) and high-temperature settings (52–55). Thus, metagenomic data are essential for identifying genuine vertical inheritance in the domain Archaea.
Our study provides crucial new insights into carbon metabolism in the LUCA. First, it is surprising that both the periplasmic and cytoplasmic formate dehydrogenase families were broadly distributed throughout the bacterial and archaeal domains. This suggests that both families had begun to diversify from the MopB superfamily within the LUCA. That is, our analyses indicate that the LUCA had developed distinct enzymes for periplasmic formate oxidation and cytoplasmic CO2 reduction before the domains diverged. There is significant discordance between the seemingly deep antiquity of FwdB and cytoplasmic formate dehydrogenases in our comparative genomics analyses and the results of comparative metagenomic analyses that supported a deep ancestry for periplasmic FdhG and cytoplasmic formate dehydrogenases. The most parsimonious interpretation of these conflicting phylogenetic signals is that both formate dehydrogenase families were likely inherited from the LUCA.
Our metagenomic studies have added additional context to the alkaline hydrothermal vent hypothesis by demonstrating that FwdB was an evolutionary innovation of Archaea that was transferred by HGT to a limited array of bacterial taxa. The hypothesis posits that CO2 served as an ancestral electron acceptor in acetogenesis in Bacteria and hydrogenotrophic methanogenesis in Archaea (49, 50). FwdB is the subunit in the formylmethanofuran dehydrogenase complex that catalyzes the reduction of CO2, and our metagenomic phylogenetic analyses strongly suggest that FwdB was not present in the LUCA. Indeed, most archaeal phyla do not conserve energy via methanogenesis. However, cytoplasmic formate dehydrogenases are central to acetogenesis (56), and recent studies have shown that several uncultured Archaea are capable of energy conservation via acetogenesis (57, 58). Therefore, we contend that Bacteria and Archaea inherited a single, primordial pathway (i.e., acetogenesis) for energy conservation.
The antiquity of acetylenotrophy shows that early life was adapted to an organic haze atmosphere.
A wealth of geochemical data indicates that a dense organic haze atmosphere, redolent of Titan, was present throughout the Archean Earth (59–63). Acetylene, while a rare trace gas on the modern Earth (64, 65), would have been comparatively enriched in such an atmosphere (66–69). We provide the first evidence from the molecular evolutionary record that acetylene was a crucial source of energy for Bacteria as they moved beyond deep-sea hydrothermal vent fields to colonize surface environments on the Archean Earth. All of our tree topologies robustly support an evolutionary scenario in which acetylenotrophy evolved before polysulfide, thiosulfate, tetrathionate, dimethyl sulfoxide (DMSO), and nitrate respiration, making AH one of the most ancient diversifications from the MopB superfamily. These phylogenetic data are the strongest support to date for previous hypotheses that acetylenotrophy was an early metabolic adaptation supporting ancient bacterial communities (70–72).
A relative ordination of major catalytic expansions in biogeochemical cycles through deep time.
Geochemical data clearly indicate that the Earth at the dawn of the Archean eon was a rocky world with a reducing surface environment and a thick organic haze atmosphere enriched in hydrocarbons (63, 73). By the dawn of the Proterozoic eon (2.5 to 0.541 Gya [billion years ago]), the Earth was characterized by oxidized surface environments and stable concentrations of O2 produced by oxygenic photosynthesis (34, 74). A range of geochemical data (75, 76) and some molecular data (36), however, suggest that transient pools of O2 or some other oxidant formed ~3.1 Gya and steadily increased until ~2.4 Gya. The exact beginning of the Earth’s surface environment oxidation, the nature of the oxidants that drove it, and the rate at which the Archean Earth became oxidized remain fiercely contested questions in the geobiological literature. The MopB superfamily catalyzes a wealth of geochemically relevant redox reactions on a bevy of substrates that span an enormous gradient of redox potentials. A thorough understanding of when specific catalytic activities diversified from the superfamily could help resolve these debates.
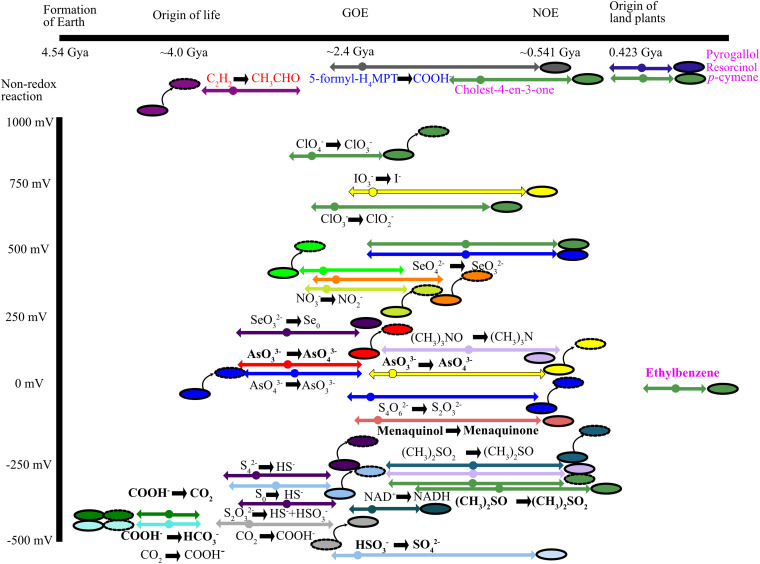
As we noted previously, a crucial challenge in providing an absolute ordination for when specific families diversified from the superfamily is the lack of reliable geochemical proxies for the presence of most MopB superfamily substrates in the geological record (5). By superimposing the reactions catalyzed by each enzyme family onto Fig. 3, however, we were able to relatively ordinate when MopB families diversified from the superfamily over geologic time (Fig. 5). We ordinated these diversification events over both geologic time and the known redox potential of the substrate and product of the oxidation-reduction reaction, if known. We used several events as crucial benchmarks when establishing this relative ordination. These include the origin of life, the Great Oxygenation Event (GOE) ~2.4 Gya, the Neoproterozoic Oxygenation Event (NOE) (~0.541 Gya), and the origin and diversification of land plants (dates for when these events occurred [in billions of years ago] were taken from a previous study by Knoll and Nowak [74]). The FdhG and cytoplasmic formate dehydrogenase families, given the robust evidence that we have for inheritance from the LUCA, should have diversified from the superfamily at around the time of the origin of life (~4.0 Gya). The midpoint potential for the formate (HCOO−1)/CO2 couple is −432 mV.
FIG 5.
Relative ordination of when MopB superfamily substrates became available over geologic time against the midpoint potential of the conversion of the substrate to the product. This is possible only for oxidation-reduction reactions for which the midpoint potential is known. Nonredox reactions are placed above the y axis (midpoint potential in millivolts). The x axis corresponds to billions of years ago (Gya). Major evolutionary events are also highlighted on the axis. Each dot is accompanied by error bars, indicating the rough estimates for when the substrate might conceivably have been available to life. The colors of both the dots and error bars match the family with which each reaction is associated. When the same reaction evolved in multiple families, we tried to put them in as close spatial proximity as possible. For the conversion of the substrate to the product, normal text indicates that the reaction is a reduction. Boldface type represents oxidase or dehydrogenase reactions, blue text indicates transferases, red text indicates hydration, and pink text indicates hydroxylation reactions. Beside each dot, we also include graphical depictions of cells, again colored by the family from which the reaction evolved. Rod-shaped cells with undotted black borders represent bacterial cells. Rod-shaped cells with dotted black borders represent archaeal cells. If bacterial and archaeal cells are positioned side by side, this indicates that the catalytic function was most likely present in the LUCA. If only one rod-shaped cell is present, this catalytic function is known in only one domain of life. The acquisition of a catalytic subunit by one domain from the other via HGT is depicted using an arrow, indicating the direction of the HGT event. For example, an arrow from a bacterial cell to an archaeal cell indicates that Archaea within this family acquired it from the bacterial domain.
It is also reasonably simple to estimate when in geologic time the bacterial Nqo3 and ActB families diversified from the superfamily. Both families are known to participate exclusively in aerobic respiration, and multiple robust geochemical proxies exist to trace the concentrations of O2 through geologic time (35, 74). The Nqo3 and ActB subunits should have diversified from the superfamily sometime between the initial “whiffs of oxygen” in the hundreds of millions of years prior to the GOE all the way to the NOE, a secondary burst of oxygenation in which O2 concentrations first approached modern-day levels and the deep oceans became permanently oxygenated. It is likely that the Nqo3 family diversified from the cytoplasmic formate dehydrogenases closer to the GOE given the ubiquity of bacterial respiratory complex I among aerobes (77, 78). The more limited phylogenetic distribution of ActB (79) suggests that the complex may have evolved at a later date when O2 concentrations were higher. Another family whose physiological function is associated with aerobic respiration is the FhcB family. The Fhc complex exists exclusively in aerobic methylotrophs to convert formyl-tetrahydromethanopterin to formate (32) and so should have diversified from the superfamily at around the same time as both Nqo3 and ActB.
Similarly, the most recent diversifications of catalytic functions in the MopB superfamily are easy to ordinate. These include CmdA and EdbA from the Asp-coordinating family and RhL and PgtL from the RhL/PgtL family. The substrate for CmdA, p-cymene, is a terpene found in many diverse plant species (80). Ethylbenzene, the substrate for EbdA, is an aromatic hydrocarbon component of many fossil fuels (81). Pyrogallol, the substrate for PgtL, and resorcinol, the substrate for RhL, are phenolic compounds produced during the degradation of lignin (82, 83). The association of each of these substrates specifically with plant matter demonstrates that these various hydroxylation reactions could have evolved only once land plants first occupied terrestrial environments (~0.423 Gya). S25dA, another Asp-coordinating family member, allows various bacteria to utilize various cholesterols as growth substrates (also through a hydroxylation reaction). Assuming that this catalytic subunit cannot exploit the bacterial sterol analogs hopanoids (84), this function could have evolved only once eukaryotes emerged (~1.5 Gya) given that eukaryotes are the only organisms known to produce cholesterol (85).
The remainder of the catalytic functions within the MopB superfamily are substantially more challenging to ordinate given that there are no robust geochemical proxies for the presence of the remaining substrates through geologic time. If hydrogenotrophic methanogenesis was not a feature of the LUCA’s physiology, it is unquestionably an ancient adaptation for energy conservation in the archaeal domain (86–88). Thus, the radiation of the FwdB family likely occurred shortly after the divergence of Archaea from the LUCA as we note above for the AH family in Bacteria. Other bioenergetic substrates that could have stimulated diversifications in the MopB superfamily early in the Archean eon include polysulfide (S42−), elemental sulfur (S0), and thiosulfate (S2O32−). Neither S42− nor S0 requires any oxygen atoms (although S0 is thermodynamically stable only in high-temperature settings [89]), and S2O32− can form abiotically from the oxidation of HS− by Fe(III) under anoxic conditions (90). All three terminal electron acceptors also have low redox potentials of −260 mV for S42−/HS−, −270 mV for S0/HS−, and −402 mV for S2O32−/HS−+HSO3−.
As for the diversification of catalytic functions to exploit arsenic, chlorine, nitrogen, and selenium oxyanions, ordinating these is challenging given how little is known concerning the redox state of surface environments on the Archean Earth. Arsenite would have been widely available in reducing surface environments (91, 92), protected from photooxidation by a thick organic haze atmosphere. However, if the organic haze atmosphere dissipated toward the late Archean, both perchlorate (ClO4−) (93) and selenite (SeO32−) (94) could have formed from UV photooxidation. If the organic haze atmosphere remained stable, then most of the diversifications involved in chlorine, nitrogen, and selenium biogeochemical cycling would have been formed only in oases of oxygen (or, at least, oxidants) at localized environments from ~2.7 to 2.4 Gya (35, 74, 95, 96).
A suite of catalytic diversifications also followed between the GOE and the origin of land plants. The sulfur intermediate tetrathionate (S4O62−) can be formed abiotically only when pyrites are oxidized by substantial concentrations of O2 or Mn(IV) oxides (97). Selenate (SeO42−) would have had thermodynamic stability only when O2 concentrations began to approach modern-day levels (i.e., around the NOE) (94, 98). The various diversifications in the MopB superfamily related to DMSO respiration (and, in the case of DdhA, a DMSO-producing dehydrogenase) are challenging to ordinate. The precursor of DMSO, dimethyl sulfide (DMS), is produced from dimethylsulfoniopropionate (DMSP), and DMSP production is thought to have evolved in marine algae, which marine bacteria subsequently evolved mechanisms to metabolize (99). That would correspond to an origin for DMSO reduction to, at the earliest, the origin of eukaryotes ~1.5 Gya. Similarly, the methylamine compound trimethylamine N-oxide (TMAO) is produced in marine eukaryotes as an osmolyte (100, 101). Caution is warranted in assuming that DMSO and TMAO represent the ancestral substrates of the Ser-coordinating families. Both bacterial (102) and archaeal (103) DMSO reductases from DmsA, DorA, and the haloarchaeal Asp-coordinating DMSO reductase catalytic subunits can efficiently reduce a bevy of N- and S-oxides (and in the case of Archaea, it has been shown these substrates can also be utilized as terminal electron acceptors). The TMAO reductase TorA catalytic subunit, in contrast, can efficiently reduce only an array of N-oxide molecules (102). Thus, the ancestral substrates of both Ser- and Asp-coordinating families within the MopB superfamily may be N- or S-oxide compounds that are as yet unknown.
A model for how evolutionary diversifications were stimulated in the MopB superfamily.
Consideration of the various arsenite oxidase, arsenate reductase, and nitrate reductase catalytic subunits suggests important mechanisms for the diversification of families within the MopB superfamily. It is curious to note that there are two chalcophilic oxyanion and sulfur intermediate oxidoreductase families that mediate arsenic oxyanion transformations. The first, ArxA/ArrA, is highly specific for arsenic oxyanions (no other physiological functions have been identified). The second, the TtrA/SrdA/alternative arsenate reductase family, shows substantial catalytic versatility. Given that ArxA preceded the evolution of ArrA (5) and that arsenite (AsO33−) would have been abundant through the Archean eon, it is likely that Bacteria first began exploiting arsenic as a bioenergetic substrate through the use of AsO33− as an electron donor in anaerobic respiration and anoxygenic photosynthesis. For most of the Archean eon, the pool of arsenate (AsO43−) produced by this reaction would have been transient and unstable (as sulfur intermediates are under any thermodynamic conditions). The midpoint potential of the AsO33−/AsO43− redox couple is 60 mV. Thus, there would be a powerful selective advantage for organisms that evolved the capacity to exploit AsO43−. This could initially be accomplished through the alternate arsenate reductase catalytic subunit, which clearly has little catalytic specificity. It seems likely that as the Earth’s surface environments became more oxidizing and, thus, AsO43− became more stable, selection for an enzyme more specific to AsO43−, ArrA, would strongly be favored. In contrast, AioA, the predominantly aerobic arsenite oxidase, could have evolved only at around the time of the GOE given that respiratory iodate reduction (IdrA) appears to be the ancestral function of this family.
In a similar manner, our phylogeny illustrates how the availability of nitrate stimulated multiple diversification events in the MopB superfamily, with the first radiation comprising an enzyme with multiple physiological functions to two lineages with more specialized physiological functions. In our search for NapA and NasC/NasA homologs in the genomes of cultured isolates, we observed that NapA alone is present in anaerobic taxa. Our phylogeny in Fig. 3, however, did not differentiate these families, and MopB superfamily representatives from anaerobic taxa with best matches to NasC/NasA homologs clustered in the basal branches with representatives from anaerobic taxa that had best matches to NapA homologs. NasC/NasA homologs function only in the presence of oxygen (104) and function exclusively in nitrate (NO3−) assimilation, whereas NapA reduces NO3− for a variety of physiological functions, including anaerobic respiration and redox homeostasis (26). Therefore, when NO3− first became available in Archean environments, NapA most likely was the first diversification in the MopB superfamily associated with nitrogen metabolism. NapA would allow organisms to exploit NO3− (a rich source of energy given that the NO3−/NO2− couple has a midpoint potential of 433 mV) for a variety of physiological functions. Sometime later, an enzyme that is specialized for nitrate respiration, NarG, would have diversified from the superfamily. Only once a substantial quantity of O2 had formed in the atmosphere would a NasC/NasA/NapA-like subunit become associated with partner subunits characteristic of NO3− assimilation. This scenario is consistent with a recent molecular clock study of nitrogen metabolism genes that found an origin for NapA and NarG of ~2.8 Gya and an origin for NasC/NasA of ~2.5 Gya (105). Note that if PcrA is indeed the ancestral substrate of all Asp-coordinating MopB representatives, then NarG could have diversified from the superfamily only after a sizable pool of energy-rich perchlorate (ClO4−) had formed.
HGT between Asgard Archaea and Bacteria has implications for models of eukaryogenesis.
It is remarkable that we did not find evidence for biogeochemical innovations in the MopB superfamily driven by the archaeal domain of life, besides the FwdB family. Archaea acquired all MopB families involved in acetylenotrophy and the nitrogen, sulfur, arsenic, and, possibly, selenium biogeochemical cycles via HGT. The finding that members of Asgardarchaeota acquired MopB family members through HGT at least partially supports the current model for eukaryogenesis (106). We found 7 homologs of the AH family in Asgardarchaeota MAGs, but the remainder of the homologs acquired through HGT represent substrates with a relatively high redox potential (i.e., they would be available primarily at oxic-anoxic interfaces in modern environments). These include 4 Asp-coordinating members (the potential function of these homologs cannot be inferred without operon context), 11 DmsA-like members, 2 NarG-like members, and 2 NasC/NasA/NapA-like members. The phylogeny in Fig. 3 includes Asgard archaeal representatives from the AH-like, Asp-like, DmsA-like, and NasC/NasA/NapA-like family homologs, and these robustly cluster with homologs from the phyla Firmicutes and Desulfobacterota. This robust evidence for HGT events between these Archaea and organisms of Firmicutes and Desulfobacterota is consistent with the current model that posits that the first syntrophic association between these organisms and anaerobic Bacteria, such as sulfur-reducing bacteria, was stimulated by the need of Asgard Archaea for a ready supply of H2 (106, 107). Given that Asgard archaeal MAGs are found exclusively in anoxic environments (e.g., anoxic marine sediments), it is likely that only a single precursor lineage to eukaryotes evolved adaptations to environments with trace concentrations of O2 sometime around the GOE, leaving no trace of such events in extant Archaea.
Conclusions.
Our phylogenetic analysis has defined the MopB superfamily with unprecedented rigor, demonstrating that what is frequently regarded as a limited repertoire of metalloenzymes that mediate obscure biogeochemical reactions is in actuality an incredibly diverse superfamily scattered throughout the prokaryotic domains of life and is present in eukaryotic mitochondrial respiratory chains. We have provided independent support for the origin of the superfamily in formate and CO2 metabolism, consistent with the most experimentally rigorous hypothesis for the origin of life. We have added new depth to this hypothesis by providing molecular evidence that the LUCA did not generate energy from a precursor pathway that would later evolve into acetogenesis and hydrogenotrophic methanogenesis in Bacteria and Archaea but that the LUCA specifically likely utilized CO2 as an electron acceptor in acetogenesis. We have found molecular evidence that supports the idea that an organic haze atmosphere existed on the Archean Earth and that one of the constituents of that atmosphere, acetylene, became a substrate for energy conservation early in the evolution of the domain Bacteria. Finally, our relative ordination of catalytic activities in the superfamily demonstrates that Bacteria repeatedly exploited MopB superfamily members to gain access to new electron donors and terminal electron acceptors for energy conservation made available by increasing pools of O2 on Earth’s surface environments and to utilize a suite of macromolecules generated by multicellular life.
We also reveal that the primary drivers of catalytic innovations within this superfamily are Bacteria. The sheer variety of substrates that these enzymes and catalytic subunits have evolved to exploit increasingly taxes the ability of human recollection. Yet we demonstrate here that incorporating all of that biochemical and physiological detail into an evolutionary study yields vital insights into how this superfamily has diversified through geologic time. MopB superfamily catalytic radiations, and the associated innovations in anaerobic energy conservation, did not cease with the GOE. Bacteria and Archaea have actively altered the Earth’s atmosphere, and global climate, from the early Archean Earth, when methanogens fed methane into an organic haze atmosphere, to the slow, inevitable oxidation of the Earth’s atmosphere from the GOE to the NOE driven by oxygenic photosynthesis. As we grapple with the substantial changes in global biogeochemical cycles and atmospheric composition wrought by industrialization in what is now often referred to as the Anthropocene epoch (108–110), it is likely that this superfamily will continue to evolve to allow prokaryotic life to survive yet another environmental and climatic transition.
MATERIALS AND METHODS
Strategies for automated searches of genomic and metagenomic databases and manual curation.
Representative sequences for each MopB family were selected from each of the studies included in Table 1 as BLAST queries to construct comprehensive libraries of MopB homologs. For the comparative genomic analyses, DELTA-BLAST searches (111) were performed, and MopB homologs were selected only if the sequence came from an organism that had been isolated in a pure culture or a defined coculture, the sequence aligned over at least 95% of the query with an amino acid identify of at least 30%, the sequence length was consistent with the sequence length of the query, and the primary sequence contained motifs considered characteristic of the enzyme family (e.g., a twin-arginine translocation motif, a [4Fe-4S] or [3Fe-4S] cluster binding motif, and a Mo/W-bisPGD binding motif). Candidates were additionally screened using the Integrated Microbial Genomics (IMG) platform (112) to view the genomic context of the putative homolog. Sequences were retained only if the primary sequences were conserved between the NCBI database and the IMG database and the operon contained other subunits consistent with the operon structures described in model organisms previously (e.g., a four-[4Fe-4S] cluster-containing protein, a [2Fe-2S] Rieske protein, or a membrane anchor).
The resources of GTDB-Tk (113) were utilized to expand our phylogenetic analyses to MopB homologs from high-quality MAGs. Further Asgard archaeal MAGs were taken from a previous study by Liu et al. (114). These sequence libraries were downloaded, and the nucleotide sequences were searched against a database containing our protein query sequences using BLASTX (115). This approach was necessitated by the frequent use of Sec in the FwdB, FdhG, and cytoplasmic formate dehydrogenases. Genome annotation programs nearly always interpret the UGA codon as a stop codon in proteins that exploit UGA as the selenocysteine codon (94), resulting in truncated protein fragments. The BLASTX searches for all query proteins in the database were executed simultaneously so that each possible MopB superfamily member could match only one of the protein queries, avoiding sequence redundancy in the data set. The search results were then filtered to remove any sequences that did not have ≥70% query coverage and ≥30% sequence identity. The use of BLASTX, however, introduced an additional complication. The DNA coordinates are included in the output files, but the protein sequences (or even the corresponding nucleotide sequences) are not included. We were able to retrieve the nucleotide sequences encoding putative MopB superfamily homologs using BEDTools (116). These nucleotide sequences were subsequently translated back into protein sequences using the transeq program from the EMBOSS open software suite (117). The standard codon table was used to translate nucleotide sequences.
Manual inspection of each of the MopB superfamily hits that we obtained through automated searches involved several filtering steps. The first priority was to remove any sequences that had a large number of undetermined amino acids (designated with the letter X), which suggested that the organism from which the sequence data were obtained did not use the standard codon table. Following this, each sequence was inspected to ensure that there were no BLASTX returns that contained essentially the same protein from the same genome or MAG but with slightly different DNA start and end coordinates. Finally, all sequences were manually inspected to ensure that the region between the N-terminal iron-sulfur cluster(s) (if present) and the Mo/W-coordinating amino acid ligand was fully present. Any sequence with missing amino acid data for this region was excised, as this region defines the MopB domain that characterizes the superfamily. Each sequence was also inspected to ensure that the correct amino acid ligand was present for the family to which the sequence was assigned based on its best BLASTX match.
Selection of outgroups for phylogenetic analyses.
All outgroups used in these analyses were molybdo- or tungstoenzyme families that lack the distinctive Mo/W-bisPGD found in many MopB superfamily members. The MopB superfamily, molybdenum hydroxylase, and aldehyde ferredoxin oxidoreductase lineages constitute the only known assemblages of mononuclear Mo/W-containing enzymes (21). Molybdenum hydroxylases utilize a molybdopterin cytosine dinucleotide cofactor, while aldehyde ferredoxin oxidoreductases are known to exploit W only in the form of a tungstopterin cofactor without any nucleotide moieties. Crucially, these three broad groups of metalloenzymes do not harbor similar structural folds or domains and therefore represent evolutionarily distinct superfamilies or families. The tungstopterin aldehyde ferredoxin oxidoreductases were used to generate Fig. 1 and Fig. S1 in the supplemental material, whereas the molybdenum hydroxylase family was used to generate Fig. 3. Phylogenies using tungstopterin aldehyde ferredoxin oxidoreductases as outgroups for the MAG-derived sequence data set failed to resolve.
Tungstoperin aldehyde ferredoxin oxidoreductase outgroups included the aldehyde oxidoreductases from Moorella thermoacetica (118) and Pyrococcus furiosus (119), formaldehyde oxidoreductases from P. furiosus (120) and Thermococcus litoralis (121), and glyceraldehyde-3-phosphate ferredoxin oxidoreductases from P. furiosus (122), Methanococcus maripaludis (123), and Pyrobaculum aerophilum (124). Molybdenum hydroxylase outgroup representatives included characterized xanthine dehydrogenase catalytic subunits from Eubacterium barkeri (125), Gottschalkia acidurici (126), and Rhodobacter capsulatus (127) as well as putative xanthine dehydrogenase catalytic subunits in the archaeal domain, including BAN90858.1 from Aeropyrum camini, GGM71171.1 from Thermogymnomonas acidicola, HIQ30525.1 from “Candidatus Caldiarchaeum subterraneum,” and MCD6514100.1 from an Asgard archaeon. As of now, no xanthine dehydrogenases have been characterized in this domain of life. Aldehyde oxidoreductases from Desulfovibrio gigas (128) and Escherichia coli (129) were also included as outgroups. Finally, carbon monoxide large subunits from Afipia carboxidovorans (130) and Hydrogenophaga pseudoflava (131) were also included.
Structural alignments.
Structural alignments of the 15 X-ray crystal structures and the single cryo-EM structure from the MopB superfamily were generated with the Secondary Structure Matching program of PDBeFold (132), using default parameters. The overlapping structures were subsequently viewed in Swiss-PdbViewer (133), and the alignment was manually trimmed to a single region with shared structural homology among all 16 structures. This region corresponds to the MopB domain; a stretch between the N-terminal iron-sulfur cluster, if present; and the PGD moiety of Mo/W-bisPGD proximal to that cluster.
Phylogenetic analyses.
Sequences were aligned using the online platform of MAFFT for large-scale sequence alignments (134). Untrimmed alignments were analyzed directly. A single trimmed alignment (described above) was generated for the well-characterized MopB superfamily members using the trimAl tool (135). The alignment was trimmed such that all columns with gaps in more than 20% of MopB sequences or with a similarity score of below 0.001 were omitted, with the caveat that 60% of the columns be conserved for the analysis. The amino acid selection models that best fit our data were chosen using the ModelFinder program (136). The amino acid substitution model used for each tree is described in the legend of each figure and supplemental figure. Maximum likelihood phylogenies were generated using IQTREE (137). All phylogenies generated using the ultrafast bootstrap approximation were run for 10,000 replicates. The tree generated with nonparametric bootstraps (Fig. S1) was run for 200 replicates. The model selection and maximum likelihood analyses for the genomic data sets were performed using the CIPRES gateway portal (138). The automated searches and the subsequent phylogenetic analyses of the metagenomic data set were performed using RMACC Summit (139). All phylogenies were visualized using the Interactive Tree of Life (iTOL) program (140). Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Footnotes
Supplemental material is available online only.
Contributor Information
John F. Stolz, Email: stolz@duq.edu.
Noha H. Youssef, Oklahoma State University
REFERENCES
- 1.Baymann F, Lebrun E, Brugna M, Schoepp-Cothenet B, Giudici-Orticoni M-T, Nitschke W. 2003. The redox protein construction kit: pre-last universal common ancestor evolution of energy-conserving enzymes. Philos Trans R Soc Lond B Biol Sci 358:267–274. doi: 10.1098/rstb.2002.1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schoepp-Cothenet B, van Lis R, Philippot P, Magalon A, Russell MJ, Nitschke W. 2012. The ineluctable requirement for the trans-iron elements molybdenum and/or tungsten in the origin of life. Sci Rep 2:263. doi: 10.1038/srep00263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schoepp-Cothenet B, van Lis R, Atteia A, Baymann F, Capowiez L, Ducluzeau A-L, Duval S, ten Brink F, Russell MJ, Nitschke W. 2013. On the universal core of bioenergetics. Biochim Biophys Acta 1827:79–93. doi: 10.1016/j.bbabio.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 4.Weiss MC, Sousa FL, Mrnjavac N, Neukirchen S, Roettger M, Nelson-Sathi S, Martin WF. 2016. The physiology and habitat of the last universal common ancestor. Nat Microbiol 1:16116. doi: 10.1038/nmicrobiol.2016.116. [DOI] [PubMed] [Google Scholar]
- 5.Wells M, Kanmanii NJ, Al Zadjali AM, Janecka JE, Basu P, Oremland RS, Stolz JF. 2020. Methane, arsenic, selenium and the origins of the DMSO reductase family. Sci Rep 10:10946. doi: 10.1038/s41598-020-67892-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nitschke W, McGlynn SE, Milner-White EJ, Russell MJ. 2013. On the antiquity of metalloenzymes and their substrates in bioenergetics. Biochim Biophys Acta 1827:871–881. doi: 10.1016/j.bbabio.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Duval S, Baymann F, Schoepp-Cothenet B, Trolard F, Bourrié G, Grauby O, Branscomb E, Russell MJ, Nitschke W. 2019. Fougerite: the not so simple progenitor of the first cells. Interface Focus 9:20190063. doi: 10.1098/rsfs.2019.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. 2013. The prokaryotic Mo/W-bisPGD enzymes family: a catalytic workhorse in bioenergetic. Biochim Biophys Acta 1827:1048–1085. doi: 10.1016/j.bbabio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Yamazaki C, Kashiwa S, Horiuchi A, Kasahara Y, Yamamura S, Amachi S. 2020. A novel dimethylsulfoxide reductase family of molybdenum enzyme, Idr, is involved in iodate respiration by Pseudomonas sp. SCT. Environ Microbiol 22:2196–2212. doi: 10.1111/1462-2920.14988. [DOI] [PubMed] [Google Scholar]
- 10.Abin CA, Hollibaugh JT. 2019. Transcriptional response of the obligate anaerobe Desulfuribacillus stibiiarsenatis MLFW-2T to growth on antimonate and other terminal electron acceptors. Environ Microbiol 21:618–630. doi: 10.1111/1462-2920.14503. [DOI] [PubMed] [Google Scholar]
- 11.Shi L-D, Wang M, Han Y-L, Lai C-Y, Shapleigh JP, Zhao H-P. 2019. Multi-omics reveal various potential antimonate reductases from phylogenetically diverse microorganisms. Appl Microbiol Biotechnol 103:9119–9129. doi: 10.1007/s00253-019-10111-x. [DOI] [PubMed] [Google Scholar]
- 12.Wang Q, Warelow TP, Kang Y-S, Romano C, Osborne TH, Lehr CR, Bothner B, McDermott TR, Santini JM, Wang G. 2015. Arsenite oxidase also functions as an antimonite oxidase. Appl Environ Microbiol 81:1959–1965. doi: 10.1128/AEM.02981-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, Ye L, Jing C. 2020. Genetic identification of antimonate respiratory reductase in Shewanella sp. ANA-3. Environ Sci Technol 54:14107–14113. doi: 10.1021/acs.est.0c03875. [DOI] [PubMed] [Google Scholar]
- 14.Wagner T, Ermler U, Shima S. 2016. The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters. Science 354:114–117. doi: 10.1126/science.aaf9284. [DOI] [PubMed] [Google Scholar]
- 15.Stock T, Rother M. 2009. Selenoproteins in archaea and Gram-positive bacteria. Biochim Biophys Acta 1790:1520–1532. doi: 10.1016/j.bbagen.2009.03.022. [DOI] [PubMed] [Google Scholar]
- 16.González PJ, Correia C, Moura I, Brondino CD, Moura JJG. 2006. Bacterial nitrate reductases: molecular and biological aspects of nitrate reduction. J Inorg Biochem 100:1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 17.Thauer RK, Kaster A-K, Seedorf H, Buckel W, Hedderich R. 2008. Methanogenic archaea: ecologically relevant differences in energy conservation. Nat Rev Microbiol 6:579–591. doi: 10.1038/nrmicro1931. [DOI] [PubMed] [Google Scholar]
- 18.Mayumi D, Mochimaru H, Tamaki H, Yamamoto K, Yoshioka H, Suzuki Y, Kamagata Y, Sakata S. 2016. Methane production from coal by a single methanogen. Science 354:222–225. doi: 10.1126/science.aaf8821. [DOI] [PubMed] [Google Scholar]
- 19.Beaulieu JJ, Tank JL, Hamilton SK, Wollheim WM, Hall RO, Mulholland PJ, Peterson BJ, Ashkenas LR, Cooper LW, Dahm CN, Dodds WK, Grimm NB, Johnson SL, McDowell WH, Poole GC, Valett HM, Arango CP, Bernot MJ, Burgin AJ, Crenshaw CL, Helton AM, Johnson LT, O’Brien JM, Potter JD, Sheibley RW, Sobota DJ, Thomas SM. 2011. Nitrous oxide emission from denitrification in stream and river networks. Proc Natl Acad Sci USA 108:214–219. doi: 10.1073/pnas.1011464108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bakken LR, Bergaust L, Liu B, Frostegård A. 2012. Regulation of denitrification at the cellular level: a clue to the understanding of N2O emissions from soils. Philos Trans R Soc Lond B Biol Sci 367:1226–1234. doi: 10.1098/rstb.2011.0321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hille R, Hall J, Basu P. 2014. The mononuclear molybdenum enzymes. Chem Rev 114:3963–4038. doi: 10.1021/cr400443z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothery RA, Workun GJ, Weiner JH. 2008. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim Biophys Acta 1778:1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Bilous PT, Cole ST, Anderson WF, Weiner JH. 1988. Nucleotide sequence of the dmsABC operon encoding the anaerobic dimethylsulphoxide reductase of Escherichia coli. Mol Microbiol 2:785–795. doi: 10.1111/j.1365-2958.1988.tb00090.x. [DOI] [PubMed] [Google Scholar]
- 24.Weiner JH, MacIsaac DP, Bishop RE, Bilous PT. 1988. Purification and properties of Escherichia coli dimethyl sulfoxide reductase, an iron-sulfur molybdoenzyme with broad substrate specificity. J Bacteriol 170:1505–1510. doi: 10.1128/jb.170.4.1505-1510.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cammack R, Weiner JH. 1990. Electron paramagnetic resonance spectroscopic characterization of dimethyl sulfoxide reductase of Escherichia coli. Biochemistry 29:8410–8416. doi: 10.1021/bi00488a030. [DOI] [PubMed] [Google Scholar]
- 26.Sparacino-Watkins C, Stolz JF, Basu P. 2014. Nitrate and periplasmic nitrate reductases. Chem Soc Rev 43:676–706. doi: 10.1039/c3cs60249d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seiffert GB, Ullmann GM, Messerschmidt A, Schink B, Kroneck PMH, Einsle O. 2007. Structure of the non-redox-active tungsten/[4Fe:4S] enzyme acetylene hydratase. Proc Natl Acad Sci USA 104:3073–3077. doi: 10.1073/pnas.0610407104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sazanov LA, Hinchliffe P. 2006. Structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus. Science 311:1430–1436. doi: 10.1126/science.1123809. [DOI] [PubMed] [Google Scholar]
- 29.Wirth C, Brandt U, Hunte C, Zickermann V. 2016. Structure and function of mitochondrial complex I. Biochim Biophys Acta 1857:902–914. doi: 10.1016/j.bbabio.2016.02.013. [DOI] [PubMed] [Google Scholar]
- 30.Yanyushin MF, del Rosario MC, Brune DC, Blankenship RE. 2005. New class of bacterial membrane oxidoreductases. Biochemistry 44:10037–10045. doi: 10.1021/bi047267l. [DOI] [PubMed] [Google Scholar]
- 31.Sun C, Benlekbir S, Venkatakrishnan P, Wang Y, Hong S, Hosler J, Tajkhorshid E, Rubinstein JL, Gennis RB. 2018. Structure of the alternative complex III in a supercomplex with cytochrome oxidase. Nature 557:123–126. doi: 10.1038/s41586-018-0061-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hemmann JL, Wagner T, Shima S, Vorholt JA. 2019. Methylofuran is a prosthetic group of the formyltransferase/hydrolase complex and shuttles one-carbon units between two active sites. Proc Natl Acad Sci USA 116:25583–25590. doi: 10.1073/pnas.1911595116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A. 2020. CDD/SPARCLE: the Conserved Domain Database in 2020. Nucleic Acids Res 48:D265–D268. doi: 10.1093/nar/gkz991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knoll AH, Bergmann KD, Strauss JV. 2016. Life: the first two billion years. Philos Trans R Soc Lond B Biol Sci 371:20150493. doi: 10.1098/rstb.2015.0493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer WW, Hemp J, Valentine JS. 2016. How did life survive Earth’s great oxygenation? Curr Opin Chem Biol 31:166–178. doi: 10.1016/j.cbpa.2016.03.013. [DOI] [PubMed] [Google Scholar]
- 36.Jabłońska J, Tawfik DS. 2021. The evolution of oxygen-utilizing enzymes suggests early biosphere oxygenation. Nat Ecol Evol 5:442–448. doi: 10.1038/s41559-020-01386-9. [DOI] [PubMed] [Google Scholar]
- 37.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 38.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sforna MC, Philippot P, Somogyi A, van Zuilen MA, Medjoubi K, Schoepp-Cothenet B, Nitschke W, Visscher PT. 2014. Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nat Geosci 7:811–815. doi: 10.1038/ngeo2276. [DOI] [Google Scholar]
- 41.Dayhoff MO. 1976. The origin and evolution of protein superfamilies. Fed Proc 35:2132–2138. [PubMed] [Google Scholar]
- 42.Barker WC, George DG, Mewes HW, Tsugita A. 1992. The PIR-International Protein Sequence Database. Nucleic Acids Res 20(Suppl):2023–2026. doi: 10.1093/nar/20.suppl.2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Todd AE, Orengo CA, Thornton JM. 2001. Evolution of function in protein superfamilies, from a structural perspective. J Mol Biol 307:1113–1143. doi: 10.1006/jmbi.2001.4513. [DOI] [PubMed] [Google Scholar]
- 44.Baier F, Copp JN, Tokuriki N. 2016. Evolution of enzyme superfamilies: comprehensive exploration of sequence-function relationships. Biochemistry 55:6375–6388. doi: 10.1021/acs.biochem.6b00723. [DOI] [PubMed] [Google Scholar]
- 45.Rinke C, Schwientek P, Sczyrba A, Ivanova NN, Anderson IJ, Cheng J-F, Darling A, Malfatti S, Swan BK, Gies EA, Dodsworth JA, Hedlund BP, Tsiamis G, Sievert SM, Liu W-T, Eisen JA, Hallam SJ, Kyrpides NC, Stepanauskas R, Rubin EM, Hugenholtz P, Woyke T. 2013. Insights into the phylogeny and coding potential of microbial dark matter. Nature 499:431–437. doi: 10.1038/nature12352. [DOI] [PubMed] [Google Scholar]
- 46.Hug LA, Baker BJ, Anantharaman K, Brown CT, Probst AJ, Castelle CJ, Butterfield CN, Hernsdorf AW, Amano Y, Ise K, Suzuki Y, Dudek N, Relman DA, Finstad KM, Amundson R, Thomas BC, Banfield JF. 2016. A new view of the tree of life. Nat Microbiol 1:16048. doi: 10.1038/nmicrobiol.2016.48. [DOI] [PubMed] [Google Scholar]
- 47.Zaremba-Niedzwiedzka K, Caceres EF, Saw JH, Bäckström D, Juzokaite L, Vancaester E, Seitz KW, Anantharaman K, Starnawski P, Kjeldsen KU, Stott MB, Nunoura T, Banfield JF, Schramm A, Baker BJ, Spang A, Ettema TJG. 2017. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature 541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 48.Fu L, Niu B, Zhu Z, Wu S, Li W. 2012. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin W, Russell MJ. 2007. On the origin of biochemistry at an alkaline hydrothermal vent. Philos Trans R Soc Lond B Biol Sci 362:1887–1925. doi: 10.1098/rstb.2006.1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sousa FL, Thiergart T, Landan G, Nelson-Sathi S, Pereira IAC, Allen JF, Lane N, Martin WF. 2013. Early bioenergetic evolution. Philos Trans R Soc Lond B Biol Sci 368:20130088. doi: 10.1098/rstb.2013.0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oren A. 2002. Molecular ecology of extremely halophilic Archaea and Bacteria. FEMS Microbiol Ecol 39:1–7. doi: 10.1111/j.1574-6941.2002.tb00900.x. [DOI] [PubMed] [Google Scholar]
- 52.Takai K, Gamo T, Tsunogai U, Nakayama N, Hirayama H, Nealson KH, Horikoshi K. 2004. Geochemical and microbiological evidence for a hydrogen-based, hyperthermophilic subsurface lithoautotrophic microbial ecosystem (HyperSLiME) beneath an active deep-sea hydrothermal field. Extremophiles 8:269–282. doi: 10.1007/s00792-004-0386-3. [DOI] [PubMed] [Google Scholar]
- 53.Niederberger TD, Ronimus RS, Morgan HW. 2008. The microbial ecology of a high-temperature near-neutral spring situated in Rotorua, New Zealand. Microbiol Res 163:594–603. doi: 10.1016/j.micres.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 54.Mardanov AV, Gumerov VM, Beletsky AV, Perevalova AA, Karpov GA, Bonch-Osmolovskaya EA, Ravin NV. 2011. Uncultured archaea dominate in the thermal groundwater of Uzon Caldera, Kamchatka. Extremophiles 15:365–372. doi: 10.1007/s00792-011-0368-1. [DOI] [PubMed] [Google Scholar]
- 55.Antranikian G, Suleiman M, Schäfers C, Adams MWW, Bartolucci S, Blamey JM, Birkeland N-K, Bonch-Osmolovskaya E, da Costa MS, Cowan D, Danson M, Forterre P, Kelly R, Ishino Y, Littlechild J, Moracci M, Noll K, Oshima T, Robb F, Rossi M, Santos H, Schönheit P, Sterner R, Thauer R, Thomm M, Wiegel J, Stetter KO. 2017. Diversity of bacteria and archaea from two shallow marine hydrothermal vents from Vulcano Island. Extremophiles 21:733–742. doi: 10.1007/s00792-017-0938-y. [DOI] [PubMed] [Google Scholar]
- 56.Ragsdale SW, Pierce E. 2008. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta 1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He Y, Li M, Perumal V, Feng X, Fang J, Xie J, Sievert SM, Wang F. 2016. Genomic and enzymatic evidence for acetogenesis among multiple lineages of the archaeal phylum Bathyarchaeota widespread in marine sediments. Nat Microbiol 1:16035. doi: 10.1038/nmicrobiol.2016.35. [DOI] [PubMed] [Google Scholar]
- 58.Seitz KW, Lazar CS, Hinrichs K-U, Teske AP, Baker BJ. 2016. Genomic reconstruction of a novel, deeply branched sediment archaeal phylum with pathways for acetogenesis and sulfur reduction. ISME J 10:1696–1705. doi: 10.1038/ismej.2015.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Domagal-Goldman SD, Kasting JF, Johnston DT, Farquhar J. 2008. Organic haze, glaciations and multiple sulfur isotopes in the mid-Archean era. Earth Planet Sci Lett 269:29–40. doi: 10.1016/j.epsl.2008.01.040. [DOI] [Google Scholar]
- 60.Zerkle AL, Claire MW, Domagal-Goldman SD, Farquhar J, Poulton SW. 2012. A bistable organic-rich atmosphere on the Neoarchaean Earth. Nat Geosci 5:359–363. doi: 10.1038/ngeo1425. [DOI] [Google Scholar]
- 61.Izon G, Zerkle AL, Zhelezinskaia I, Farquhar J, Newton RJ, Poulton SW, Eigenbrode JL, Claire MW. 2015. Multiple oscillations in Neoarchaean atmospheric chemistry. Earth Planet Sci Lett 431:264–273. doi: 10.1016/j.epsl.2015.09.018. [DOI] [Google Scholar]
- 62.Arney G, Domagal-Goldman SD, Meadows VS, Wolf ET, Schwieterman E, Charnay B, Claire M, Hébrard E, Trainer MG. 2016. The pale orange dot: the spectrum and habitability of hazy Archean Earth. Astrobiology 16:873–899. doi: 10.1089/ast.2015.1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Catling DC, Zahnle KJ. 2020. The Archean atmosphere. Sci Adv 6:eaax1420. doi: 10.1126/sciadv.aax1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Goldman A, Murcray FJ, Blatherwick RD, Gillis JR, Bonomo FS, Murcray FH, Murcray DG, Cicerone RJ. 1981. Identification of acetylene (C2H2) in infrared atmospheric absorbtion [sic] spectra. J Geophys Res 86:12143–12146. doi: 10.1029/JC086iC12p12143. [DOI] [Google Scholar]
- 65.Rudolph J, Ehhalt DH, Khedim A. 1984. Vertical profiles of acetylene in the troposphere and stratosphere. J Atmos Chem 2:117–124. doi: 10.1007/BF00114125. [DOI] [Google Scholar]
- 66.Zahnle KJ. 1986. Photochemistry of methane and the formation of hydrocyanic acid (HCN) in the Earth’s early atmosphere. J Geophys Res 91:2819–2834. doi: 10.1029/JD091iD02p02819. [DOI] [Google Scholar]
- 67.Kasting JF. 1993. Earth’s early atmosphere. Science 259:920–926. doi: 10.1126/science.11536547. [DOI] [PubMed] [Google Scholar]
- 68.Kasting JF. 2004. When methane made climate. Sci Am 291:78–85. doi: 10.1038/scientificamerican0704-78. [DOI] [PubMed] [Google Scholar]
- 69.Trainer MG, Pavlov AA, Curtis DB, McKay CP, Worsnop DR, Delia AE, Toohey DW, Toon OB, Tolbert MA. 2004. Haze aerosols in the atmosphere of early Earth: manna from heaven. Astrobiology 4:409–419. doi: 10.1089/ast.2004.4.409. [DOI] [PubMed] [Google Scholar]
- 70.Culbertson CW, Strohmaier FE, Oremland RS. 1988. Acetylene as a substrate in the development of primordial bacterial communities. Orig Life Evol Biosph 18:397–407. doi: 10.1007/BF01808218. [DOI] [PubMed] [Google Scholar]
- 71.Oremland RS, Voytek MA. 2008. Acetylene as fast food: implications for development of life on anoxic primordial Earth and in the outer solar system. Astrobiology 8:45–58. doi: 10.1089/ast.2007.0183. [DOI] [PubMed] [Google Scholar]
- 72.Akob DM, Sutton JM, Fierst JL, Haase KB, Baesman S, Luther GW, III, Miller LG, Oremland RS. 2018. Acetylenotrophy: a hidden but ubiquitous microbial metabolism? FEMS Microbiol Ecol 94:fiy103. doi: 10.1093/femsec/fiy103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sleep NH. 2010. The Hadean-Archaean environment. Cold Spring Harb Perspect Biol 2:a002527. doi: 10.1101/cshperspect.a002527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Knoll AH, Nowak MA. 2017. The timetable of evolution. Sci Adv 3:e1603076. doi: 10.1126/sciadv.1603076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ostrander CM, Johnson AC, Anbar AD. 2021. Earth’s first redox revolution. Annu Rev Earth Planet Sci 49:337–366. doi: 10.1146/annurev-earth-072020-055249. [DOI] [Google Scholar]
- 76.Johnson AC, Ostrander CM, Romaniello SJ, Reinhard CT, Greaney AT, Lyons TW, Anbar AD. 2021. Reconciling evidence of oxidative weathering and atmospheric anoxia on Archean Earth. Sci Adv 7:eabj0108. doi: 10.1126/sciadv.abj0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Berrisford JM, Baradaran R, Sazanov LA. 2016. Structure of bacterial respiratory complex I. Biochim Biophys Acta 1857:892–901. doi: 10.1016/j.bbabio.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 78.Kaila VRI, Wikström M. 2021. Architecture of bacterial respiratory chains. Nat Rev Microbiol 19:319–330. doi: 10.1038/s41579-020-00486-4. [DOI] [PubMed] [Google Scholar]
- 79.Majumder ELW, King JD, Blankenship RE. 2013. Alternative complex III from phototrophic bacteria and its electron acceptor auracyanin. Biochim Biophys Acta 1827:1383–1391. doi: 10.1016/j.bbabio.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 80.Eaton RW. 1997. p-Cymene catabolic pathway in Pseudomonas putida F1: cloning and characterization of DNA encoding conversion of p-cymene to p-cumate. J Bacteriol 179:3171–3180. doi: 10.1128/jb.179.10.3171-3180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ball HA, Johnson HA, Reinhard M, Spormann AM. 1996. Initial reactions in anaerobic ethylbenzene oxidation by a denitrifying bacterium, strain EB1. J Bacteriol 178:5755–5761. doi: 10.1128/jb.178.19.5755-5761.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schink B, Pfennig N. 1982. Fermentation of trihydroxybenzenes by Pelobacter acidigallici gen. nov. sp. nov., a new strictly anaerobic, non-sporeforming bacterium. Arch Microbiol 133:195–201. doi: 10.1007/BF00415000. [DOI] [Google Scholar]
- 83.Gallus C, Gorny N, Ludwig W, Schink B. 1997. Anaerobic degradation of α-resorcylate by a nitrate-reducing bacterium, Thauera aromatica strain AR-1. Syst Appl Microbiol 20:540–544. doi: 10.1016/S0723-2020(97)80023-9. [DOI] [Google Scholar]
- 84.Sáenz JP, Grosser D, Bradley AS, Lagny TJ, Lavrynenko O, Broda M, Simons K. 2015. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc Natl Acad Sci USA 112:11971–11976. doi: 10.1073/pnas.1515607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mouritsen OG, Zuckermann MJ. 2004. What’s so special about cholesterol? Lipids 39:1101–1113. doi: 10.1007/s11745-004-1336-x. [DOI] [PubMed] [Google Scholar]
- 86.Ueno Y, Yamada K, Yoshida N, Maruyama S, Isozaki Y. 2006. Evidence from fluid inclusions for microbial methanogenesis in the early Archaean era. Nature 440:516–519. doi: 10.1038/nature04584. [DOI] [PubMed] [Google Scholar]
- 87.Wolfe JM, Fournier GP. 2018. Horizontal gene transfer constrains the timing of methanogen evolution. Nat Ecol Evol 2:897–903. doi: 10.1038/s41559-018-0513-7. [DOI] [PubMed] [Google Scholar]
- 88.Berghuis BA, Yu FB, Schulz F, Blainey PC, Woyke T, Quake SR. 2019. Hydrogenotrophic methanogenesis in archaeal phylum Verstraetearchaeota reveals the shared ancestry of all methanogens. Proc Natl Acad Sci USA 116:5037–5044. doi: 10.1073/pnas.1815631116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter KO. 1998. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev 22:353–381. doi: 10.1111/j.1574-6976.1998.tb00376.x. [DOI] [Google Scholar]
- 90.Pyzik AJ, Sommer SE. 1981. Sedimentary iron monosulfides: kinetics and mechanism of formation. Geochim Cosmochim Acta 45:687–698. doi: 10.1016/0016-7037(81)90042-9. [DOI] [Google Scholar]
- 91.Oremland RS, Saltikov CW, Wolfe-Simon F, Stolz JF. 2009. Arsenic in the evolution of Earth and extraterrestrial ecosystems. Geomicrobiol J 26:522–536. doi: 10.1080/01490450903102525. [DOI] [Google Scholar]
- 92.van Lis R, Nitschke W, Duval S, Schoepp-Cothenet B. 2013. Arsenics as bioenergetic substrates. Biochim Biophys Acta 1827:176–188. doi: 10.1016/j.bbabio.2012.08.007. [DOI] [PubMed] [Google Scholar]
- 93.Catling DC, Claire MW, Zahnle KJ, Quinn RC, Clark BC, Hecht MH, Kounaves S. 2010. Atmospheric origins of perchlorate on Mars and in the Atacama. J Geophys Res 115:E00E11. doi: 10.1029/2009JE003425. [DOI] [Google Scholar]
- 94.Wells M, Stolz JF. 2020. Microbial selenium metabolism: a brief history, biogeochemistry and ecophysiology. FEMS Microbiol Ecol 96:fiaa209. doi: 10.1093/femsec/fiaa209. [DOI] [PubMed] [Google Scholar]
- 95.Anbar AD, Duan Y, Lyons TW, Arnold GL, Kendall B, Creaser RA, Kaufman AJ, Gordon GW, Scott C, Garvin J, Buick R. 2007. A whiff of oxygen before the Great Oxidation Event? Science 317:1903–1906. doi: 10.1126/science.1140325. [DOI] [PubMed] [Google Scholar]
- 96.Stüeken EE, Buick R, Anbar AD. 2015. Selenium isotopes support free O2 in the latest Archean. Geology 43:259–262. doi: 10.1130/G36218.1. [DOI] [Google Scholar]
- 97.Schippers A, Jørgensen BB. 2001. Oxidation of pyrite and iron sulfide by manganese dioxide in marine sediments. Geochim Cosmochim Acta 65:915–922. doi: 10.1016/S0016-7037(00)00589-5. [DOI] [Google Scholar]
- 98.Stüeken EE, Buick R, Bekker A, Catling D, Foriel J, Guy BM, Kah LC, Machel HG, Montañez IP, Poulton SW. 2015. The evolution of the global selenium cycle: secular trends in Se isotopes and abundances. Geochim Cosmochim Acta 162:109–125. doi: 10.1016/j.gca.2015.04.033. [DOI] [Google Scholar]
- 99.Bullock HA, Luo H, Whitman WB. 2017. Evolution of dimethylsulfoniopropionate metabolism in marine phytoplankton and bacteria. Front Microbiol 8:637. doi: 10.3389/fmicb.2017.00637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Yancey PH, Clark ME, Hand SC, Bowlus RD, Somero GN. 1982. Living with water stress: evolution of osmolyte systems. Science 217:1214–1222. doi: 10.1126/science.7112124. [DOI] [PubMed] [Google Scholar]
- 101.Lidbury ID, Murrell JC, Chen Y. 2015. Trimethylamine and trimethylamine N-oxide are supplementary energy sources for a marine heterotrophic bacterium: implications for marine carbon and nitrogen cycling. ISME J 9:760–769. doi: 10.1038/ismej.2014.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.McCrindle SL, Kappler U, McEwan AG. 2005. Microbial dimethylsulfoxide and trimethylamine-N-oxide respiration. Adv Microb Physiol 50:147–198. doi: 10.1016/S0065-2911(05)50004-3. [DOI] [PubMed] [Google Scholar]
- 103.Sorokin DY, Roman P, Kolganova TV. 2021. Halo(natrono)archaea from hypersaline lakes can utilize sulfoxides other than DMSO as electron acceptors for anaerobic respiration. Extremophiles 25:173–180. doi: 10.1007/s00792-021-01219-y. [DOI] [PubMed] [Google Scholar]
- 104.Ruiz B, Le Scornet A, Sauviac L, Rémy A, Bruand C, Meilhoc E. 2019. The nitrate assimilatory pathway in Sinorhizobium meliloti: contribution to NO production. Front Microbiol 10:1526. doi: 10.3389/fmicb.2019.01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Parsons C, Stüeken EE, Rosen CJ, Mateos K, Anderson RE. 2021. Radiation of nitrogen-metabolizing enzymes across the tree of life tracks environmental transitions in Earth history. Geobiology 19:18–34. doi: 10.1111/gbi.12419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Imachi H, Nobu MK, Nakahara N, Morono Y, Ogawara M, Takaki Y, Takano Y, Uematsu K, Ikuta T, Ito M, Matsui Y, Miyazaki M, Murata K, Saito Y, Sakai S, Song C, Tasumi E, Yamanaka Y, Yamaguchi T, Kamagata Y, Tamaki H, Takai K. 2020. Isolation of an archaeon at the prokaryote-eukaryote interface. Nature 577:519–525. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Spang A, Stairs CW, Dombrowski N, Eme L, Lombard J, Caceres EF, Greening C, Baker BJ, Ettema TJG. 2019. Proposal of the reverse flow model for the origin of the eukaryotic cell based on comparative analyses of Asgard archaeal metabolism. Nat Microbiol 4:1138–1148. doi: 10.1038/s41564-019-0406-9. [DOI] [PubMed] [Google Scholar]
- 108.Ruddiman WF. 2013. The Anthropocene. Annu Rev Earth Planet Sci 41:45–68. doi: 10.1146/annurev-earth-050212-123944. [DOI] [Google Scholar]
- 109.Lewis SL, Maslin MA. 2015. Defining the Anthropocene. Nature 519:171–180. doi: 10.1038/nature14258. [DOI] [PubMed] [Google Scholar]
- 110.Ruddiman WF, Ellis EC, Kaplan JO, Fuller DQ. 2015. Defining the epoch we live in. Science 348:38–39. doi: 10.1126/science.aaa7297. [DOI] [PubMed] [Google Scholar]
- 111.Boratyn GM, Schäffer AA, Agarwala R, Altschul SF, Lipman DJ, Madden TL. 2012. Domain enhanced lookup time accelerated BLAST. Biol Direct 7:12. doi: 10.1186/1745-6150-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, Lykidis A, Mavromatis K, Ivanova N, Kyrpides NC. 2006. The Integrated Microbial Genomes (IMG) system. Nucleic Acids Res 34:D344–D348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH. 2019. GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36:1925–1927. doi: 10.1093/bioinformatics/btz848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Liu Y, Makarova KS, Huang W-C, Wolf YI, Nikolskaya AN, Zhang X, Cai M, Zhang C-J, Xu W, Luo Z, Cheng L, Koonin EV, Li M. 2021. Expanded diversity of Asgard archaea and their relationships with eukaryotes. Nature 593:553–557. doi: 10.1038/s41586-021-03494-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 118.White H, Feicht R, Huber C, Lottspeich F, Simon H. 1991. Purification and some properties of the tungsten-containing carboxylic acid reductase from Clostridium formicoaceticum. Biol Chem Hoppe Seyler 372:999–1005. doi: 10.1515/bchm3.1991.372.2.999. [DOI] [PubMed] [Google Scholar]
- 119.Mukund S, Adams MW. 1991. The novel tungsten-iron-sulfur protein of the hyperthermophilic archaebacterium, Pyrococcus furiosus, is an aldehyde ferredoxin oxidoreductase. Evidence for its participation in a unique glycolytic pathway. J Biol Chem 266:14208–14216. doi: 10.1016/S0021-9258(18)98669-2. [DOI] [PubMed] [Google Scholar]
- 120.Hu Y, Faham S, Roy R, Adams MWW, Rees DC. 1999. Formaldehyde ferredoxin oxidoreductase from Pyrococcus furiosus: the 1.85 Å resolution crystal structure and its mechanistic implications. J Mol Biol 286:899–914. doi: 10.1006/jmbi.1998.2488. [DOI] [PubMed] [Google Scholar]
- 121.Mukund S, Adams MW. 1993. Characterization of a novel tungsten-containing formaldehyde ferredoxin oxidoreductase from the hyperthermophilic archaeon, Thermococcus litoralis. A role for tungsten in peptide catabolism. J Biol Chem 268:13592–13600. doi: 10.1016/S0021-9258(19)38690-9. [DOI] [PubMed] [Google Scholar]
- 122.Mukund S, Adams MW. 1995. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase, a novel tungsten-containing enzyme with a potential glycolytic role in the hyperthermophilic archaeon Pyrococcus furiosus. J Biol Chem 270:8389–8392. doi: 10.1074/jbc.270.15.8389. [DOI] [PubMed] [Google Scholar]
- 123.Park M-O, Mizutani T, Jones PR. 2007. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase from Methanococcus maripaludis. J Bacteriol 189:7281–7289. doi: 10.1128/JB.00828-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Reher M, Gebhard S, Schönheit P. 2007. Glyceraldehyde-3-phosphate ferredoxin oxidoreductase (GAPOR) and nonphosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN), key enzymes of the respective modified Embden-Meyerhof pathways in the hyperthermophilic crenarchaeota Pyrobaculum aerophilum and Aeropyrum pernix. FEMS Microbiol Lett 273:196–205. doi: 10.1111/j.1574-6968.2007.00787.x. [DOI] [PubMed] [Google Scholar]
- 125.Schräder T, Rienhöfer A, Andreesen JR. 1999. Selenium-containing xanthine dehydrogenase from Eubacterium barkeri. Eur J Biochem 264:862–871. doi: 10.1046/j.1432-1327.1999.00678.x. [DOI] [PubMed] [Google Scholar]
- 126.Wager R, Cammack R, Andreesen JR. 1984. Purification and characterization of xanthine dehydrogenase from Clostridium acidiurici grown in the presence of selenium. Biochim Biophys Acta 791:63–74. doi: 10.1016/0167-4838(84)90282-6. [DOI] [Google Scholar]
- 127.Leimkühler S, Kern M, Solomon PS, McEwan AG, Schwarz G, Mendel RR, Klipp W. 1998. Xanthine dehydrogenase from the phototrophic purple bacterium Rhodobacter capsulatus is more similar to its eukaryotic counterparts than to prokaryotic molybdenum enzymes. Mol Microbiol 27:853–869. doi: 10.1046/j.1365-2958.1998.00733.x. [DOI] [PubMed] [Google Scholar]
- 128.Huber R, Hof P, Duarte RO, Moura JJ, Moura I, Liu MY, LeGall J, Hille R, Archer M, Romão MJ. 1996. A structure-based catalytic mechanism for the xanthine oxidase family of molybdenum enzymes. Proc Natl Acad Sci USA 93:8846–8851. doi: 10.1073/pnas.93.17.8846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Correia MAS, Otrelo-Cardoso AR, Schwuchow V, Sigfridsson Clauss KGV, Haumann M, Romão MJ, Leimkühler S, Santos-Silva T. 2016. The Escherichia coli periplasmic aldehyde oxidoreductase is an exceptional member of the xanthine oxidase family of molybdoenzymes. ACS Chem Biol 11:2923–2935. doi: 10.1021/acschembio.6b00572. [DOI] [PubMed] [Google Scholar]
- 130.Dobbek H, Gremer L, Kiefersauer R, Huber R, Meyer O. 2002. Catalysis at a dinuclear [CuSMo(O)OH] cluster in a CO dehydrogenase resolved at 1.1-Å resolution. Proc Natl Acad Sci USA 99:15971–15976. doi: 10.1073/pnas.212640899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hänzelmann P, Dobbek H, Gremer L, Huber R, Meyer O. 2000. The effect of intracellular molybdenum in Hydrogenophaga pseudoflava on the crystallographic structure of the seleno-molybdo-iron-sulfur flavoenzyme carbon monoxide dehydrogenase. J Mol Biol 301:1221–1235. doi: 10.1006/jmbi.2000.4023. [DOI] [PubMed] [Google Scholar]
- 132.Krissinel E, Henrick K. 2004. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 133.Guex N, Peitsch MC. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 134.Nakamura T, Yamada KD, Tomii K, Katoh K. 2018. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 34:2490–2492. doi: 10.1093/bioinformatics/bty121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Miller MA, Pfeiffer W, Schwartz T. 2010. Creating the CIPRES Science Gateway for inference of large phylogenetic trees, p 1–8. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE). Institute of Electrical and Electronics Engineers, New York, NY. doi: 10.1109/GCE.2010.5676129. [DOI] [Google Scholar]
- 139.Anderson J, Burns PJ, Milroy D, Ruprecht P, Hauser T, Siegel HJ. 2017. Deploying RMACC Summit: an HPC resource for the Rocky Mountain region, p 1–7. In Proceedings of the Practice and Experience in Advanced Research Computing 2017 on Sustainability, Success and Impact. Association for Computing Machinery, New York, NY. [Google Scholar]
- 140.Letunic I, Bork P. 2019. Interactive Tree of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res 47:W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Karrasch M, Börner G, Thauer RK. 1990. The molybdenum cofactor of formylmethanofuran dehydrogenase from Methanosarcina barkeri is a molybdopterin guanine dinucleotide. FEBS Lett 274:48–52. doi: 10.1016/0014-5793(90)81326-j. [DOI] [PubMed] [Google Scholar]
- 142.Schmitz RA, Richter M, Linder D, Thauer RK. 1992. A tungsten-containing active formylmethanofuran dehydrogenase in the thermophilic archaeon Methanobacterium wolfei. Eur J Biochem 207:559–565. doi: 10.1111/j.1432-1033.1992.tb17082.x. [DOI] [PubMed] [Google Scholar]
- 143.Jormakka M, Törnroth S, Byrne B, Iwata S. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 144.Raaijmakers H, Macieira S, Dias JM, Teixeira S, Bursakov S, Huber R, Moura JJG, Moura I, Romão MJ. 2002. Gene sequence and the 1.8 Å crystal structure of the tungsten-containing formate dehydrogenase from Desulfovibrio gigas. Structure 10:1261–1272. doi: 10.1016/s0969-2126(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 145.Costa C, Teixeira M, LeGall J, Moura JJG, Moura I. 1997. Formate dehydrogenase from Desulfovibrio desulfuricans ATCC 27774: isolation and spectroscopic characterization of the active sites (heme, iron-sulfur centers and molybdenum). J Biol Inorg Chem 2:198–208. doi: 10.1007/s007750050125. [DOI] [Google Scholar]
- 146.Yamamoto I, Saiki T, Liu SM, Ljungdahl LG. 1983. Purification and properties of NADP-dependent formate dehydrogenase from Clostridium thermoaceticum, a tungsten-selenium-iron protein. J Biol Chem 258:1826–1832. doi: 10.1016/S0021-9258(18)33062-X. [DOI] [PubMed] [Google Scholar]
- 147.Graentzdoerffer A, Rauh D, Pich A, Andreesen JR. 2003. Molecular and biochemical characterization of two tungsten- and selenium-containing formate dehydrogenases from Eubacterium acidaminophilum that are associated with components of an iron-only hydrogenase. Arch Microbiol 179:116–130. doi: 10.1007/s00203-002-0508-1. [DOI] [PubMed] [Google Scholar]
- 148.Wood GE, Haydock AK, Leigh JA. 2003. Function and regulation of the formate dehydrogenase genes of the methanogenic archaeon Methanococcus maripaludis. J Bacteriol 185:2548–2554. doi: 10.1128/JB.185.8.2548-2554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Jones JB, Stadtman TC. 1981. Selenium-dependent and selenium-independent formate dehydrogenases of Methanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem 256:656–663. doi: 10.1016/S0021-9258(19)70024-6. [DOI] [PubMed] [Google Scholar]
- 150.Boyington JC, Gladyshev VN, Khangulov SV, Stadtman TC, Sun PD. 1997. Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine, and an Fe4S4 cluster. Science 275:1305–1308. doi: 10.1126/science.275.5304.1305. [DOI] [PubMed] [Google Scholar]
- 151.Niks D, Duvvuru J, Escalona M, Hille R. 2016. Spectroscopic and kinetic properties of the molybdenum-containing, NAD+-dependent formate dehydrogenase from Ralstonia eutropha. J Biol Chem 291:1162–1174. doi: 10.1074/jbc.M115.688457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ramos F, Blanco G, Gutiérrez JC, Luque F, Tortolero M. 1993. Identification of an operon involved in the assimilatory nitrate-reducing system of Azotobacter vinelandii. Mol Microbiol 8:1145–1153. doi: 10.1111/j.1365-2958.1993.tb01659.x. [DOI] [PubMed] [Google Scholar]
- 153.Ogawa K, Akagawa E, Yamane K, Sun ZW, LaCelle M, Zuber P, Nakano MM. 1995. The nasB operon and nasA gene are required for nitrate/nitrite assimilation in Bacillus subtilis. J Bacteriol 177:1409–1413. doi: 10.1128/jb.177.5.1409-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Kuhlemeier CJ, Logtenberg T, Stoorvogel W, van Heugten HA, Borrias WE, van Arkel GA. 1984. Cloning of nitrate reductase genes from the cyanobacterium Anacystis nidulans. J Bacteriol 159:36–41. doi: 10.1128/jb.159.1.36-41.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Ellis PJ, Conrads T, Hille R, Kuhn P. 2001. Crystal structure of the 100 kDa arsenite oxidase from Alcaligenes faecalis in two crystal forms at 1.64 Å and 2.03 Å. Structure 9:125–132. doi: 10.1016/s0969-2126(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 156.Warelow TP, Pushie MJ, Cotelesage JJH, Santini JM, George GN. 2017. The active site structure and catalytic mechanism of arsenite oxidase. Sci Rep 7:1757. doi: 10.1038/s41598-017-01840-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Dias JM, Than ME, Humm A, Huber R, Bourenkov GP, Bartunik HD, Bursakov S, Calvete J, Caldeira J, Carneiro C, Moura JJ, Moura I, Romão MJ. 1999. Crystal structure of the first dissimilatory nitrate reductase at 1.9 Å solved by MAD methods. Structure 7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 158.Müller JA, DasSarma S. 2005. Genomic analysis of anaerobic respiration in the archaeon Halobacterium sp. strain NRC-1: dimethyl sulfoxide and trimethylamine N-oxide as terminal electron acceptors. J Bacteriol 187:1659–1667. doi: 10.1128/JB.187.5.1659-1667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Liebensteiner MG, Pinkse MWH, Schaap PJ, Stams AJM, Lomans BP. 2013. Archaeal (per)chlorate reduction at high temperature: an interplay of biotic and abiotic reactions. Science 340:85–87. doi: 10.1126/science.1233957. [DOI] [PubMed] [Google Scholar]
- 160.Bender KS, Shang C, Chakraborty R, Belchik SM, Coates JD, Achenbach LA. 2005. Identification, characterization, and classification of genes encoding perchlorate reductase. J Bacteriol 187:5090–5096. doi: 10.1128/JB.187.15.5090-5096.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dermer J, Fuchs G. 2012. Molybdoenzyme that catalyzes the anaerobic hydroxylation of a tertiary carbon atom in the side chain of cholesterol. J Biol Chem 287:36905–36916. doi: 10.1074/jbc.M112.407304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Strijkstra A, Trautwein K, Jarling R, Wöhlbrand L, Dörries M, Reinhardt R, Drozdowska M, Golding BT, Wilkes H, Rabus R. 2014. Anaerobic activation of p-cymene in denitrifying Betaproteobacteria: methyl group hydroxylation versus addition to fumarate. Appl Environ Microbiol 80:7592–7603. doi: 10.1128/AEM.02385-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Kniemeyer O, Heider J. 2001. Ethylbenzene dehydrogenase, a novel hydrocarbon-oxidizing molybdenum/iron-sulfur/heme enzyme. J Biol Chem 276:21381–21386. doi: 10.1074/jbc.M101679200. [DOI] [PubMed] [Google Scholar]
- 164.Schröder I, Rech S, Krafft T, Macy JM. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J Biol Chem 272:23765–23768. doi: 10.1074/jbc.272.38.23765. [DOI] [PubMed] [Google Scholar]
- 165.Thorell HD, Stenklo K, Karlsson J, Nilsson T. 2003. A gene cluster for chlorate metabolism in Ideonella dechloratans. Appl Environ Microbiol 69:5585–5592. doi: 10.1128/AEM.69.9.5585-5592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.McDevitt CA, Hanson GR, Noble CJ, Cheesman MR, McEwan AG. 2002. Characterization of the redox centers in dimethyl sulfide dehydrogenase from Rhodovulum sulfidophilum. Biochemistry 41:15234–15244. doi: 10.1021/bi026221u. [DOI] [PubMed] [Google Scholar]
- 167.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NCJ. 2003. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol 10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 168.Afshar S, Johnson E, de Vries S, Schröder I. 2001. Properties of a thermostable nitrate reductase from the hyperthermophilic archaeon Pyrobaculum aerophilum. J Bacteriol 183:5491–5495. doi: 10.1128/JB.183.19.5491-5495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ramírez-Arcos S, Fernández-Herrero LA, Berenguer J. 1998. A thermophilic nitrate reductase is responsible for the strain specific anaerobic growth of Thermus thermophilus HB8. Biochim Biophys Acta 1396:215–227. doi: 10.1016/s0167-4781(97)00183-8. [DOI] [PubMed] [Google Scholar]
- 170.Darley PI, Hellstern JA, Medina-Bellver JI, Marqués S, Schink B, Philipp B. 2007. Heterologous expression and identification of the genes involved in anaerobic degradation of 1,3-dihydroxybenzene (resorcinol) in Azoarcus anaerobius. J Bacteriol 189:3824–3833. doi: 10.1128/JB.01729-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Messerschmidt A, Niessen H, Abt D, Einsle O, Schink B, Kroneck PMH. 2004. Crystal structure of pyrogallol-phloroglucinol transhydroxylase, an Mo enzyme capable of intermolecular hydroxyl transfer between phenols. Proc Natl Acad Sci USA 101:11571–11576. doi: 10.1073/pnas.0404378101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Pierson DE, Campbell A. 1990. Cloning and nucleotide sequence of bisC, the structural gene for biotin sulfoxide reductase in Escherichia coli. J Bacteriol 172:2194–2198. doi: 10.1128/jb.172.4.2194-2198.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Schneider F, Löwe J, Huber R, Schindelin H, Kisker C, Knäblein J. 1996. Crystal structure of dimethyl sulfoxide reductase from Rhodobacter capsulatus at 1.88 Å resolution. J Mol Biol 263:53–69. doi: 10.1006/jmbi.1996.0555. [DOI] [PubMed] [Google Scholar]
- 174.Mouncey NJ, Choudhary M, Kaplan S. 1997. Characterization of genes encoding dimethyl sulfoxide reductase of Rhodobacter sphaeroides 2.4.1T: an essential metabolic gene function encoded on chromosome II. J Bacteriol 179:7617–7624. doi: 10.1128/jb.179.24.7617-7624.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Méjean V, Iobbi-Nivol C, Lepelletier M, Giordano G, Chippaux M, Pascal MC. 1994. TMAO anaerobic respiration in Escherichia coli: involvement of the tor operon. Mol Microbiol 11:1169–1179. doi: 10.1111/j.1365-2958.1994.tb00393.x. [DOI] [PubMed] [Google Scholar]
- 176.Czjzek M, Dos Santos JP, Pommier J, Giordano G, Méjean V, Haser R. 1998. Crystal structure of oxidized trimethylamine N-oxide reductase from Shewanella massilia at 2.5 Å resolution. J Mol Biol 284:435–447. doi: 10.1006/jmbi.1998.2156. [DOI] [PubMed] [Google Scholar]
- 177.Krafft T, Bokranz M, Klimmek O, Schröder I, Fahrenholz F, Kojro E, Kröger A. 1992. Cloning and nucleotide sequence of the psrA gene of Wolinella succinogenes polysulphide reductase. Eur J Biochem 206:503–510. doi: 10.1111/j.1432-1033.1992.tb16953.x. [DOI] [PubMed] [Google Scholar]
- 178.Sorokin DY, Kublanov IV, Gavrilov SN, Rojo D, Roman P, Golyshin PN, Slepak VZ, Smedile F, Ferrer M, Messina E, La Cono V, Yakimov MM. 2016. Elemental sulfur and acetate can support life of a novel strictly anaerobic haloarchaeon. ISME J 10:240–252. doi: 10.1038/ismej.2015.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Heinzinger NK, Fujimoto SY, Clark MA, Moreno MS, Barrett EL. 1995. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J Bacteriol 177:2813–2820. doi: 10.1128/jb.177.10.2813-2820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Haja DK, Wu C-H, Poole FL, Sugar J, Williams SG, Jones AK, Adams MWW. 2020. Characterization of thiosulfate reductase from Pyrobaculum aerophilum heterologously produced in Pyrococcus furiosus. Extremophiles 24:53–62. doi: 10.1007/s00792-019-01112-9. [DOI] [PubMed] [Google Scholar]
- 181.Wells M, McGarry J, Gaye MM, Basu P, Oremland RS, Stolz JF. 2019. Respiratory selenite reductase from Bacillus selenitireducens strain MLS10. J Bacteriol 201:e00614-18. doi: 10.1128/JB.00614-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Laska S, Lottspeich F, Kletzin A. 2003. Membrane-bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens. Microbiology (Reading) 149:2357–2371. doi: 10.1099/mic.0.26455-0. [DOI] [PubMed] [Google Scholar]
- 183.Guiral M, Tron P, Aubert C, Gloter A, Iobbi-Nivol C, Giudici-Orticoni M-T. 2005. A membrane-bound multienzyme, hydrogen-oxidizing, and sulfur-reducing complex from the hyperthermophilic bacterium Aquifex aeolicus. J Biol Chem 280:42004–42015. doi: 10.1074/jbc.M508034200. [DOI] [PubMed] [Google Scholar]
- 184.Dahl C, Franz B, Hensen D, Kesselheim A, Zigann R. 2013. Sulfite oxidation in the purple sulfur bacterium Allochromatium vinosum: identification of SoeABC as a major player and relevance of SoxYZ in the process. Microbiology (Reading) 159:2626–2638. doi: 10.1099/mic.0.071019-0. [DOI] [PubMed] [Google Scholar]
- 185.Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. 1999. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Mol Microbiol 32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- 186.Kuroda M, Yamashita M, Miwa E, Imao K, Fujimoto N, Ono H, Nagano K, Sei K, Ike M. 2011. Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J Bacteriol 193:2141–2148. doi: 10.1128/JB.01197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Haja DK, Wu C-H, Ponomarenko O, Poole FL, George GN, Adams MWW. 2020. Improving arsenic tolerance of Pyrococcus furiosus by heterologous expression of a respiratory arsenate reductase. Appl Environ Microbiol 86:e01728-20. doi: 10.1128/AEM.01728-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Zargar K, Hoeft S, Oremland R, Saltikov CW. 2010. Identification of a novel arsenite oxidase gene, arxA, in the haloalkaliphilic, arsenite-oxidizing bacterium Alkalilimnicola ehrlichii strain MLHE-1. J Bacteriol 192:3755–3762. doi: 10.1128/JB.00244-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Krafft T, Macy JM. 1998. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur J Biochem 255:647–653. doi: 10.1046/j.1432-1327.1998.2550647.x. [DOI] [PubMed] [Google Scholar]
- 190.Afkar E, Lisak J, Saltikov C, Basu P, Oremland RS, Stolz JF. 2003. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol Lett 226:107–112. doi: 10.1016/S0378-1097(03)00609-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 and links to phylogenies and data set. Download spectrum.04145-22-s0001.docx, DOCX file, 1.9 MB (2MB, docx)