Abstract
Background & objectives:
Atypical El Tor strains of Vibrio cholerae are frequently implicated in outbreaks of cholera. It is important to understand genetic variations of such strains which impact clinical and epidemiological outcomes. The present study was carried out to characterize an outbreak of cholera which occurred between July 8 and 13, 2018, in a remote settlement in Nashik district, Maharashtra.
Methods:
A large number of acute diarrhoea cases were reported in Rahude village, Nashik, Maharashtra since July 8, 2018. Molecular characterization of the isolated strains of V. cholerae was done.
Results:
195 cases of cholera were detected from a population of 850 (attack rate 22.9%) with two deaths (Case Fatality Ratio of 1.03). A non-haemolytic polymyxin B sensitive strain of V. cholerae O1 Ogawa was isolated from 5/14 fecal samples. Molecular characterization of the isolates indicated that this strain was an altered El Tor (AET) strain. Deletion of the trinucleotide ‘GTA’ in the rstB gene, a unique feature of classical strains, was observed.
Interpretation & conclusions:
A cholera outbreak caused by a non-haemolytic polymixin B sensitive AET strain, occurred from July 8 to 13, 2018, in a remote settlement in western India. The molecular characterization of the outbreak strains highlighted an assortment of genetic determinants, stressing the need to monitor the genetic attributes of V. cholerae O1 in outbreaks for better understanding and mapping of clinical and epidemiological changes.
Keywords: Altered El Tor, cholera, India, outbreak
Cholera poses a serious public health threat in India, particularly in the background of unsafe water and poor sanitation1. Earlier, the classical and El Tor biotypes could be readily distinguished by a group of phenotypic tests which began in 19612. However, in the recent past, this categorization has been confounded by the emergence of El Tor strains known as altered El tor (AET) strains, which possess genetic determinants of the classical strains3. The seventh cholera pandemic (1961), spread from the Bay of Bengal area in three waves. Of these, wave 3 strains predominate presently4. The emergence of such strains is alarming for the public health system, as these strains show survival and transmission advantages in the natural habitats of the organism2. After the Haiti outbreak in 2010, outbreaks caused by AET strains with several genetic modifications throughout the genome of Vibrio cholerae, are observed. There is therefore an urgent need to understand the genetic character of V. cholerae isolates causing outbreaks1.
In India, more than 200 alerts/outbreaks of cholera were cumulatively reported in the three years (2015-2017) preceding this outbreak5. However, due to lack of appropriate infrastructure, laboratory confirmation of the aetiology is often not available, particularly if the outbreaks occur in remote locations. This in turn also hampers molecular surveillance.
The present study was carried out to characterize an outbreak of cholera which occurred between July 8 and 13, 2018, in a remote settlement in Nashik district, Maharashtra. The study focused on the laboratory confirmation and molecular characterization of the isolated strains of V. cholerae. Such an approach may help track the emergence and dissemination of new variant strains in real-time, and foster a better understanding of the clinical and epidemiological consequences.
Material & Methods
This was a retrospective study of a district level outbreak investigation of cholera in Rahude village, Nashik, Pune between July 8 to 13, 2018. The stool samples were collected and sent to bacteriology laboratory, ICMR-National Institute of Virology, Pune, India, for evaluation. The Institutional Ethics Committee was informed about the outbreak and approval taken for conducting post outbreak. However, due to the setting of common practices and exposures of the study population, this was not possible. A household survey was carried out instead to study the affected cohort, in accordance with the ethical standards as laid down in the ICMR Guidelines 2017.
Setting: Rahude village is located in a notified area in Surgana taluka, Nashik district of western Maharashtra with latitude and longitude of 20.01 and 73.78, respectively. The total population of the affected village is 850 with adaptation to agricultural practices in recent years. Male members of the village migrate to nearby cities in summer to work as contract labourers.
Outbreak response: Following an alert about a large number of cases of acute diarrhoea in the village on July 8, 2018, the district rapid response team started investigation within 24 h. The district integrated disease surveillance programme (IDSP) records for the previous three years (2015-2017), were reviewed to confirm the occurrence of an outbreak.
Case identification and management: A case of acute diarrhoea was defined as more than three loose stools occurring within a day, among the residents of Rahude village since July 5, 2018. Case management was based on the level of dehydration. Antibiotics (tetracycline and ciprofloxacin) were sparingly used. House-to-house search was conducted by trained health workers for active case finding. The cases identified were managed on the basis of disease severity. Health education was provided to the villagers by the local health workers.
Prevention measures: Safe water was provided through water tankers and quality was monitored at source and household by ortho toluidine test (informed by IDSP, Nashik, personal communication). Chlorination of the only source of drinking water - an open well, repair of the well wall and construction of the well apron was undertaken. Vaccination of at-risk population was not undertaken.
Identification of source of infection: A sanitary survey was conducted to understand the possible source of infection for the outbreak. Local weather conditions including rainfall and flooding were documented.
Laboratory investigations: Stool samples of 14 representative hospitalized patients, collected on the day of onset of illness, were referred to the ICMR-NIV, Pune for detection of viral aetiology. The samples were transported in Cary Blair transport medium (Hi media Laboratories, Mumbai) to the bacteriology laboratory.
Viral diagnostics: Faecal specimens were tested for known viral causative agents responsible for diarrhoeal outbreaks namely, Rotavirus (RVA, RVB and RVC), Norovirus, Enteric adenovirus, Astrovirus and Enterovirus. All specimens were tested for the presence of RVA using antigen capture ELISA (Premier Rotaclone, Meridian Bioscience, Inc. USA) as per the manufacturer’s instructions. Reverse transcription (RT) - PCR for different viral agents was carried out using the primers and methods described earlier6–11.
Bacterial diagnostics: Isolation and characterization of bacterial diarrhoeal pathogens were carried out as per standard protocols12. Briefly, swabs from the Cary-Blair tube were inoculated on MacConkey (MAC) and Sorbitol MAC agar plates (BD Difco, Sparks, MD, USA) and into alkaline peptone water (APW; Hi media Laboratories, Mumbai, India) and selenite F broth (BD Difco) for enrichment. A loopful of APW and selenite F were plated on selective media, thiosulfate citrate bile salt sucrose (TCBS) agar and xylose lysine deoxycholate agar (BD Difco) respectively, after incubation for 8 h at 37°C. All plates were incubated overnight at 37°C and further, if needed, for 18-24 h. Suspicious colonies were selected and processed by standard biochemical tests, for preliminary identification12. For identification of V. cholerae, standard procedures were used13. Organisms which produced characteristic yellow colonies on TCBS, were oxidase-positive and showed growth at zero and six per cent but not eight per cent NaCl, were selected for serological confirmation by agglutination with specific V. cholerae polyvalent O1 antisera, followed by Ogawa and Inaba serotype-specific antisera (Denka Seiken, Tokyo, Japan). Biotyping was carried out by the recommended phenotypic tests [chicken red blood cell agglutination, Voges-Proskauer reaction, sheep erythrocyte lysis and polymyxin B (50U) sensitivity]12. Antimicrobial susceptibility testing was performed by Kirby Bauer disk diffusion method14 using CLSI (Clinical Laboratory Standards Institute) guidelines15.
Water samples (n=5) of 50 ml each, from the well, collected on July 9, before chlorination, were tested at the district public health laboratory, Nashik for evidence of faecal contamination using methodology reported earlier12.
Molecular characterization: Molecular characterization of the V. cholerae isolates was performed for ctxB, rstA, rstB and tcpA genes. The genomic DNA was extracted from the five V. cholerae isolates using QIAamp DNA kit (Qiagen, USA) and PCR assays were performed for specified targets using AccuPrime Taq DNA Polymerase System (Thermo Fisher Scientific, CA, USA), primers and PCR conditions as described earlier16–19. The PCR products were cloned in TOPO-TA cloning kit (Thermo Fisher Scientific, CA, USA) and the ligated products transformed using Escherichia coli JM109 competent cells (Thermo Fisher Scientific). The sequencing of both the strands was performed with M13 forward and reverse primers using Big-Dye terminator cycle sequencing kit (Applied Biosystems, USA) followed by sequencing using automated nucleotide sequencer (ABI PRISM 3100 Genetic Analyzer, Applied Biosystems, USA).
The results were confirmed by an alignment search using NCBI BLAST. Multiple sequence alignment was performed using the MUltiple Sequence Comparison by Log-Expectation (MUSCLE, EMBL-EBI, UK) tool of the Molecular Evolutionary Genetic Analysis (MEGA) programme version 7 and the phylogenetic tree of the trimmed alignment was prepared using the Neighbour-joining method with 2000 bootstraps19. Diarrhoeagenic E. coli were detected using procedures described earlier20.
Results
Characterization of the outbreak: The first cases of cholera were reported on July 8, 2018. According to the available data, the peak (75 cases) was observed on the same day. No specific index case could be identified. The outbreak continued till July 13, 2018 with no new cases detected on or after July 14, 2018 (Figure).
Figure.
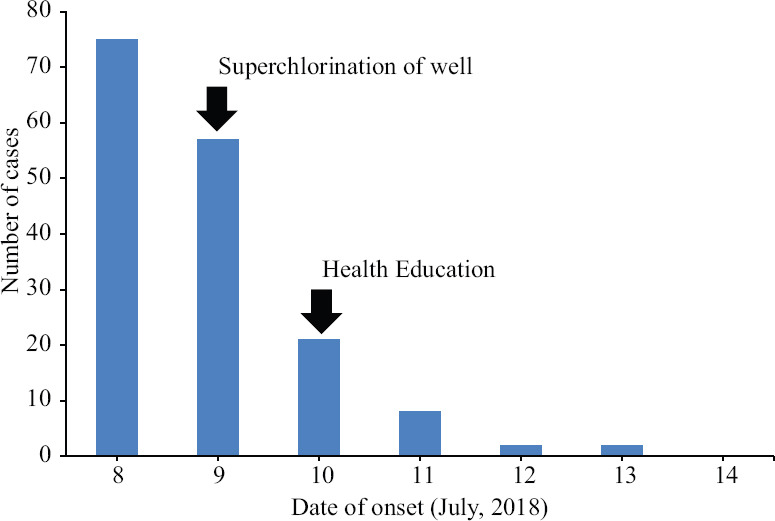
Epidemic curve.
A total of 195 case patients were identified from among the population of 850, with an attack rate of 22.9 per cent. Sex ratio (Male: Female) of cases was 0.7:1 as compared to the population sex ratio of 1.1:1. Majority of the cases (n=142; 72.8%) occurred among the adult population. Out of 130 households, 46 (35%) reported one case-patient, 49 (38%) reported more than one case-patient and 35 (27%) reported none.
All the 195 cases presented with diarrhoea, 80 per cent (n=156) presented with vomiting, 30 per cent (n=59) with weakness and 19 per cent (n=37) with abdominal pain. Fever was noted in six per cent (n=12) of the cases. The severity of illness as indicated by hospitalization was observed in 61 patients (31.3%) with two deaths (CFR: 1.03%). Both the deaths occurred in elderly hospitalized male patients with other co-morbidities (renal failure and chronic obstructive pulmonary disease, respectively). The remaining 134 patients were treated on an outpatient basis.
Environmental conditions: The general hygiene and living conditions in the village were poor. A single open well was the only source of water for drinking and all other domestic purposes. There were no records of regular water chlorination practice available at the village administrative office. The sanitary survey showed that the drinking water well was located at a lower level than the entire village, and the nearby fields, which were used for open defecation. A drainage channel was situated within 10 m of the well. The well had no apron and a pipe had been inserted through the wall for obtaining water. There was no fencing or facility for covering the well, when not in use.
The outbreak occurred in the month of July during the rainy season. In the days before the outbreak, there was heavy rainfall leading to flooding of the area. There was overflow of water from the surrounding village and fields used for open defecation into the well.
Laboratory: All the faecal samples were negative for the viral agents tested by ELISA and RT-PCR. The samples were culture negative for Shigella spp., Salmonella spp. and E. coli O157. V. cholerae O1 Ogawa was isolated from five of 14 referred fecal samples. The isolates agglutinated chick red blood cells and produced acetoin. The strains were also non-haemolytic on sheep blood agar and sensitive to polymyxin B showing a zone of inhibition of 14 mm. Based on these findings, the isolates were phenotypically identified as a polymyxin-sensitive El Tor variant21. All the isolates of V. cholerae were susceptible to azithromycin, tetracycline and resistant to nalidixic acid, ampicillin and cefotaxime. All the water samples from the well showed >18 coliform counts per 50 ml of water, and were also positive for thermotolerant coliforms22.
Molecular characterization of V. cholerae isolates: The sequences of virulence genes from all the representative V. cholerae isolates were identical, indicating that one strain was prevalent in the outbreak. ctxB gene analysis confirmed an AET strain of CT- genotype 7, with three SNPs- A58, C115 and C203. The A58 SNP which confirms the strain contains the 7th allele (ctbB7) of the ctxB caused mutation of histidine to asparagine at the 20th amino acid of the signal region of cholera toxin subunit B. tcpA CIRS with the SNP A266G was detected, causing N64S mutation of the tcpA protein. The rstA partial sequence (from 435 to 1083 nucleotides) gene showed three SNPs C927, T933 and T942. This confirmed the occurrence of a Wave 3 strain. Deletion of the trinucleotide ‘GTA’ at position 74-76 nt in the rstB gene was observed.
The sequences of genes were submitted to GenBank® database and their respective IDs are as follows - MK873008 (ctxB), MK873009 (rstA), MK873010 (rstB) and MK873011(tcpA).
Two E. coli isolates from fecal samples negative for V. cholerae were found to be positive for virulence eae (intimin) gene, a marker for enteropathogenic E. coli (EPEC) strain but negative for another EPEC gene bfpA (bundle forming pili).
Post outbreak epidemiological study: Out of the 130 households, 107 with a population of 765 villagers, participated in the survey. All the households were overcrowded with an average of seven members per household. The entire population studied were of low socio-economic status. Literacy was low with 67/107 (63%) of heads of the surveyed households, never attending school. Toilets were available in 39/107 (36%) of the households surveyed. However, these were in use for other purposes like storage, bathing, etc. and not used for defecation. In all the surveyed households, open defecation was practiced in nearby fields.
Discussion
Cholera is still an important public health concern in several parts of India. This study focused on the molecular characterization of V. cholerae in the setting of an outbreak24. Such an approach might help to track the emergence and dissemination of new variant strains in real-time, which fosters a better understanding of the clinical and epidemiological consequences.
The district level outbreak response towards the outbreak was in line with the WHO’s recommended strategy25, with the exception of non-vaccination and non-availability of laboratory support at a peripheral level. While the samples were sent to a referral laboratory for confirmation, the use of rapid diagnostic tests at primary care level, would have been helpful to quickly screen outbreak samples locally, with referral of positive samples for confirmation.
Since the beginning of the 21st century, AET strains have been reported to cause almost all cholera outbreaks with molecular analysis showing that these strains originated in the early 1990s in South Asia.
V. cholerae O1 serogroup strains are categorized based on the CTX prophage that they harbour and the SNPs in the CTX prophage genes26. The CTX phage consists of 10 genes (rstR, rstA, rstB, psh, cep, orfU, ace, zot, ctxA and ctxB). The rstR is phage type-specific, other genes differ between phages by several SNPs, except for ctxA3. The molecular characterization of the strain in the present outbreak gave several key messages based on the variations in the ctxB, tcpA, rstA and rstB genes. Our strain belonged to Wave 3, isolates of which are known to harbour the CT-genotype 74. The clade containing ctxB7 allele, to which our strain belongs, emerged in Kolkata in 200627 and has since been reported to have disseminated widely28. The tcpA gene codes for the major structural protein of the toxin-co-regulated pilus, which is the second major virulence factor of V. cholerae20. We detected variant tcpA (tcpACIRS) in our strain. This variant came into prominence after the devastating Haiti outbreak, but has since been reported in outbreaks in Africa, Asia and subsequently identified as having originated in Kolkata in 200329.
The emergence of polymyxin B sensitive El Tor strains in 2012 is considered ‘a major event in the history of cholera after 1961’4. A recent retrospective study has detected such strains in 12 States across India, including Maharashtra in 2015-201621. Our study reaffirms the spread of this strain by detection of an ongoing outbreak from a remote location. Hemolysis mediated by the HlyA gene is traditionally used for differentiation of classical and El Tor strains. Haemolysin is also considered as one of the virulence factors produced by V. cholerae30. The phenotypic differentiation of classical and El Tor strains is confounded by the occurrence of such non-haemolytic polymyxin-sensitive strains.
A novel finding of our study was the deletion of GTA at 74-76 nucleotide position of rstB. This unique feature of classical strains is detected in CTX phage type 2, 5, 6 and 6b4 and also reported in the Haitian strain13. However, this deletion has not yet been reported from the Indian subcontinent so far24.
The occurrence of (eae+, bfpA-) strain is suggestive of an atypical EPEC strain31. Deletion of eae gene reduces the pathogenicity of EPEC, whereas no significant effects happen in absence of deletion of bfpA gene32. The positivity of atypical-EPEC (eae+, bfpA-) in samples collected during the cholera outbreak is intriguing, as bfpA and tcpA both belong to the same class of type IVb pilins33.
The occurrence of cholera is well documented in rural areas of India which have traditionally had limited access to safe water and good sanitation practices. Despite tremendous progress in recent years, these areas still lag behind urban centres. The inadequate supply of safe water and persistent open defecation were important triggers for this outbreak.
The population had several risk factors for cholera including poverty, overcrowding, lack of good sanitary practices and limited access to safe drinking water, and illiteracy. The recently constructed toilets were not used for defecation, emphasizing the need to move beyond physical infrastructure and focus on measures for behavioural change. Despite the remote location and adverse environmental conditions, swift action and efficient management ensured rapid control within a short period of five days with relatively low case fatality. Although the exact age of all cases was not recorded due to the sudden influx of patients and limitations of the setting, adults were more affected with 72.8 per cent of cases occurring among adult population, suggesting that the population was naive for cholera.
Women were more affected than men in the investigated outbreak. While cholera is considered an ‘equal opportunity infection’, it is not considered as gender-neutral. Women are disproportionately reported to be affected by a cholera outbreak as they are also burdened with a disproportionate amount of domestic work such as caring for sick family members, fetching and carrying water and preparing food with contaminated water34. This increased exposure to the contaminated water, however, can be an additional risk factor for acquiring the infection.
The question which remains is regarding the origin of the outbreak. Although Maharashtra and Nashik district have reported cholera cases35,36, the affected area itself was naive with no reports of cholera or acute watery diarrhoea in the past several years. V. cholerae is an indigenous bacterium in water37. The organism is capable of surviving in the aquatic environment which can act as a source for outbreaks. It is possible that such an unidentified reservoir triggered the present outbreak. Another possibility is that members of the population who migrate to the cities for work, may have introduced the infection to the village, through the practice of open defecation leading to contamination of the source of water. The supply well in the village, was located downstream from the site of open defecation and due to the heavy rain and flooding, faecal contamination was probably washed down into the well. The epidemic curve with a large number of cases on the very first day suggests a common point source outbreak. As 27 per cent of the households did not report a case, it is possible that these households were using water collected and stored before the contamination occurred.
Our study was not without some limitations. The sudden influx of a large number of cases in this remote and inaccessible location resulted in the index case not being traced. Also, the source of contamination could not be confirmed. The detection of V. cholerae could not be attempted from the well water, which had been chlorinated on the July 9, 2018. Also, AET strains are known to possess mutations in several virulence genes, all of which were not studied in the present outbreak.
In conclusion, a cholera outbreak occurred in a remote village of western Maharashtra. The molecular characterization of the outbreak highlighted an assortment of genetic determinants, stressing the need to monitor the genetic attributes of V. cholerae O1 in outbreaks for better understanding and mapping of clinical and epidemiological changes.
Acknowledgment:
The authors acknowledge Drs Pradip Awate and Vijay Dekate for facilitating the field investigation, Shrimati Nutan Chavan and Shri Manohar Shinde for technical support, Ms Sonali Zute, Servshree Godse and RN Khedkar for logistic support and all the health workers for contributing to the household survey.
Footnotes
Financial support & sponsorship: None.
Conflicts of Interest: None.
References
- 1.Saha GK, Ganguly NK. Spread and endemicity of cholera in India: Factors beyond the numbers. J Infec Dis. 2021;224((12 Suppl 2)):S710–6. doi: 10.1093/infdis/jiab436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vicente ACP. On the emergence of atypical Vibrio cholerae O1 El Tor &cholera epidemic. Indian J Med Res. 2011;133:366–7. [PMC free article] [PubMed] [Google Scholar]
- 3.Kim EJ, Yu HJ, Lee JH, Kim JO, Han SH, Yun CH, et al. Replication of Vibrio cholerae classical CTX phage. Proc Natl Acad Sci U S A. 2017;114:2343–8. doi: 10.1073/pnas.1701335114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramamurthy T, Mutreja A, Weill FX, Das B, Ghosh A, Nair GB. Corrigendum: Revisiting the global epidemiology of cholera in conjunction with the genomics of Vibrio cholerae. Front Public Health. 2019;7:237. doi: 10.3389/fpubh.2019.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.National Rural Health Mission. Disease alerts/outbreaks reported and responded to by states/UTs through integrated disease surveillance programme (IDSP) [accessed on April 16, 2018]. Available from: http://idsp.nic.in/writereaddata/l892s/522017.pdf .
- 6.Iturriza Gómara M, Wong C, Blome S, Desselberger U, Gray J. Molecular characterization of VP6 genes of human rotavirus isolates: Correlation of genogroups with subgroups and evidence of independent segregation. J Virol. 2002;76:6596–601. doi: 10.1128/JVI.76.13.6596-6601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joshi MS, Jare VM, Gopalkrishna V. Group C rotavirus infection in patients with acute gastroenteritis in outbreaks in western India between 2006 and 2014. Epidemiol Infect. 2017;145:310–5. doi: 10.1017/S0950268816002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joshi MS, Ganorkar NN, Ranshing SS, Basu A, Chavan NA, Gopalkrishna V. Identification of group B rotavirus as an etiological agent in the gastroenteritis outbreak in Maharashtra, India. J Med Virol. 2017;89:2244–8. doi: 10.1002/jmv.24901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girish R, Broor S, Dar L, Ghosh D. Foodborne outbreak caused by a Norwalk-like virus in India. J Med Virol. 2002;67:603–7. doi: 10.1002/jmv.10145. [DOI] [PubMed] [Google Scholar]
- 10.Ando T, Monroe SS, Gentsch JR, Jin Q, Lewis DC, Glass RI. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and southern hybridization. J Clin Microbiol. 1995;33:64–71. doi: 10.1128/jcm.33.1.64-71.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allard A, Girones R, Juto P, Wadell G. Polymerase chain reaction for detection of adenoviruses in stool samples. J Clin Microbiol. 1990;28:2659–67. doi: 10.1128/jcm.28.12.2659-2667.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murray PR, Rosenthal KS, Pfaller MA. Manual of Medical Microbiology. 8th ed. Washington: Elsevier; 2016. [Google Scholar]
- 13.Garg P, Aydanian A, Smith D, Glenn MJ, Jr, Nair GB, Stine OC. Molecular epidemiology of O139 Vibrio cholerae: Mutation, lateral gene transfer, and founder flush. Emerg Infect Dis. 2003;9:810–4. doi: 10.3201/eid0907.020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bauer AW, Kirby WM, Sherris JC, Turck M. Antibiotic susceptibility testing by a standardized single disk method. Am J Clin Pathol. 1966;45:493–6. [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. 27th ed. CLSI supplement M100. Wayne, PA: CLSI; 2017. [Google Scholar]
- 16.Chatterjee S, Ghosh K, Raychoudhuri A, Chowdhury G, Bhattacharya MK, Mukhopadhyay AK, et al. Incidence, virulence factors, and clonality among clinical strains of non-O1, non-O139 Vibrio cholerae isolates from hospitalized diarrheal patients in Kolkata, India. J Clin Microbiol. 2009;47:1087–95. doi: 10.1128/JCM.02026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim EJ, Lee D, Moon SH, Lee CH, Kim DW. CTX prophages in Vibrio cholerae O1 strains. J Microbiol Biotechnol. 2014;24:725–31. doi: 10.4014/jmb.1403.03063. [DOI] [PubMed] [Google Scholar]
- 18.Liao F, Mo Z, Chen M, Pang B, Fu X, Xu W, et al. Comparison and evaluation of the molecular typing methods for toxigenic Vibrio cholerae in Southwest China. Front Microbiol. 2018;9:905. doi: 10.3389/fmicb.2018.00905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Son MS, Megli CJ, Kovacikova G, Qadri F, Taylor RK. Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes. J Clin Microbiol. 2011;49:3739–49. doi: 10.1128/JCM.01286-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lavania M, Viswanathan R, Bhardwaj SD, Oswal JS, Chavan N, Shinde M, et al. Detection of Echovirus-18 in children suspected with SARS-CoV-2 infection with multisystem inflammatory syndrome: A case report from India. Front Public Health. 2022;10:897662. doi: 10.3389/fpubh.2022.897662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samanta P, Saha RN, Chowdhury G, Naha A, Sarkar S, Dutta S, et al. Dissemination of newly emerged polymyxin B sensitive Vibrio cholerae O1 containing Haitian-like genetic traits in different parts of India. J Med Microbiol. 2018;67:1326–33. doi: 10.1099/jmm.0.000783. [DOI] [PubMed] [Google Scholar]
- 22.Park K. Parks textbook of preventive and social medicine. 24th ed. Jabalpur: Banarsidas Bhanot Publishers; 2017. [Google Scholar]
- 23.Ghosh P, Kumar D, Chowdhury G, Singh P, Samanta P, Dutta S, et al. Characterization of Vibrio cholerae O1 strains that trace the origin of Haitian-like genetic traits. Infect Genet Evol. 2017;54:47–53. doi: 10.1016/j.meegid.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 24.Kim EJ, Lee D, Moon SH, Lee CH, Kim SJ, Lee JH, et al. Molecular insights into the evolutionary pathway of Vibrio cholerae O1 atypical El Tor variants. PLoS Pathog. 2014;10:e1004384. doi: 10.1371/journal.ppat.1004384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Global Task Force on Cholera Control. Ending cholera a global roadmap To 2030. [accessed on November 27, 2017]. Available from:http://www.who.int/cholera/publications/global-roadmap/en/
- 26.Hu D, Liu B, Feng L, Ding P, Guo X, Wang M, et al. Origins of the current seventh cholera pandemic. Proc Natl Acad Sci U S A. 2016;113:E7730–9. doi: 10.1073/pnas.1608732113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ghosh P, Naha A, Pazhani GP, Ramamurthy T, Mukhopadhyay AK. Genetic traits of Vibrio cholerae O1 Haitian isolates that are absent in contemporary strains from Kolkata, India. PLoS One. 2014;9:e112973. doi: 10.1371/journal.pone.0112973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smirnova NI, Krasnov YM, Agafonova EY, Shchelkanova EY, Alkhova ZV, Kutyrev VV. Whole-genome sequencing of Vibrio cholerae O1 El Tor Strains Isolated in Ukraine (2011) and Russia (2014) Genome Announc. 2017;5:e01640–16. doi: 10.1128/genomeA.01640-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ghosh P, Naha A, Basak S, Ghosh S, Ramamurthy T, Koley H, et al. Haitian variant tcpA in Vibrio cholerae O1 El Tor strains in Kolkata, India. J Clin Microbiol. 2014;52:1020–1. doi: 10.1128/JCM.03042-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan Y, Li Z, Li Z, Li X, Sun H, Li J, et al. Nonhemolysis of epidemic El Tor biotype strains of Vibrio cholerae is related to multiple functional deficiencies of hemolysin A. Gut Pathog. 2019;11:38. doi: 10.1186/s13099-019-0316-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouzari S, Aslani MM, Oloomi M, Jafari A, Dashti A. Comparison of multiplex PCR with serogrouping and PCR-RFLP of fliC gene for the detection of enteropathogenic Escherichia coli (EPEC) Braz J Infect Dis. 2011;15:365–9. doi: 10.1016/s1413-8670(11)70206-9. [DOI] [PubMed] [Google Scholar]
- 32.Donnenberg MS, Tacket CO, James SP, Losonsky G, Nataro JP, Wasserman SS, et al. Role of the eaeA gene in experimental enteropathogenic Escherichia coli infection. J Clin Invest. 1993;92:1412–7. doi: 10.1172/JCI116717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Donnenberg MS, Kaper JB. Enteropathogenic Escherichia coli . Infect Immun. 1992;60:3953–61. doi: 10.1128/iai.60.10.3953-3961.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.United Nations Children's Fund. Briefing Note:Strategy for integrating a gendered response in Haiti's cholera epidemic. UNICEF Haiti child protection section/GBV program;2010. [accessed on January 3, 2021]. Available from: https://gbvguidelines.org/wp/wp-content/ uploads/2020/03/29_Haiti_UNICEF_Briefing_Note_Gender_Cholera.pdf .
- 35.Duthade MM, Bhakre JB, Gaikwad AA, Khaparkuntikar MN, Damle AS. An outbreak of cholera in and around Aurangabad. J Commun Dis. 2010;42:165–7. [PubMed] [Google Scholar]
- 36.The Times of India. 70 cases of acute diarrhoea in Peth: ZP. [accessed on January 3, 2021]. Available from: https://timesofindia.indiatimes.com/city/nashik/70-cases-of-acute-diarrhoea-in-Peth-ZP/articleshow /45002156.cms .
- 37.Colwell RR. Infectious disease and environment:Cholera as a paradigm for waterborne disease. Int Microbiol. 2004;7:285–9. [PubMed] [Google Scholar]
- 38.Svenungsson B, Lagergren A, Ekwall E, Evengård B, Hedlund KO, Kärnell A, et al. Enteropathogens in adult patients with diarrhea and healthy control subjects:A 1-year prospective study in a Swedish clinic for infectious diseases. Clin Infect Dis. 2000;30:770–8. doi: 10.1086/313770. [DOI] [PubMed] [Google Scholar]
- 39.Houng HSH, Sethabutr O, Echeverria P. A simple polymerase chain reaction technique to detect and differentiate Shigella and enteroinvasive Escherichia coli in human feces. Diagn Microbiol Infect Dis. 1997;28:19–25. doi: 10.1016/s0732-8893(97)89154-7. [DOI] [PubMed] [Google Scholar]
- 40.Gómez-Duarte OG, Bai J, Newell E. Detection of Escherichia coli, Salmonella spp., Shigella spp., Yersinia enterocolitica, Vibrio cholerae, and Campylobacter spp. enteropathogens by 3-reaction multiplex polymerase chain reaction. Diagn Microbiol Infect Dis. 2009;63:1–9. doi: 10.1016/j.diagmicrobio.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]


