Abstract
Background & objectives:
BK virus (BKV) is a polyomavirus and cause of a common infection after renal transplantation which could be preceded to BKV-associated nephropathy. It has four main subtypes (I–IV). BKV subtypes II and III are rare, whereas subtype I shows a ubiquitous distribution. The objective of the present study was to investigate the prevailing BKV subtypes and subgroups in renal transplant patients in Sri Lanka.
Methods:
The presence of BKV in urine was tested through virus load quantification by real-time PCR from 227 renal transplant patients who were suspected to have BKV infection. Of these patients only 41 were found to be BKV infected (>103copies/ml) and those were subjected to conventional PCR amplification of VP1 gene followed by BKV genotyping via phylogenetic analysis based on DNA sequencing data.
Results:
Persistent BK viral loads varied from 1×103 to 3×108 copies/ml. Of the 41 patient samples, 25 gave positive results for PCR amplification of subtyping region of VP1 gene of BKV. BKV genotyping resulted in detecting subtype I in 18 (72%) and subtype II in seven (28%) patients. BKV subgroups of Ia, Ib-1 and Ib-11, and Ic were identified with frequencies of 6/18 (33.3%), 6/18 (33.3%), 5/18 (27.8%), and 1/18 (5.6%), respectively.
Interpretation & conclusions:
Findings from this preliminary study showed a high occurrence of subtype I, while the presence of subtype II, which is rare and less prevalent, was a novel finding for this Asian region. This emphasizes the need for further molecular and serological studies to determine the prevalence of different BKV subtypes in Sri Lanka.
Keywords: BK virus, renal transplant patients, subtypes and subgroups, Sri Lanka, VP1 gene
The name of the BK virus (BKV) was derived from the initials of the patient from whom the virus was first isolated in 19711. BKV could exist quiescent in the epithelial cells of urogenital tract or either could be transferred from donor to recipient while the renal transplantation and may get reactivated and replicate in recipient later during immunosuppressive medication. Therefore, BKV is a cause of a common infection after renal transplantation and initially starts as viraemia (presence of viruses in the urine) followed by viraemia (presence of a virus in the blood) before developing into tubulointerstitial nephritis known as BKV-associated nephropathy (BKVAN) which causes serious inflammatory complications. BKV associated nephropathy occurs in up to 10 per cent of renal transplant recipients and can result in graft loss2,3. In most instances BKV infection is asymptomatic and associated with gradual rise in serum creatinine level, and BK viral DNA could be detected in urine prior to plasma4. If the viral load in urine is greater than 3.2×104 copies/ml, it may indicate a risk for BKVAN5.
A genotyping scheme for BKV has been introduced based on either restriction enzyme analysis or nucleotide sequence data of virus capsid protein (VP) 1. All subsisting BKVs were classified into four subtypes (I–IV) and subtyping region of the VP1 gene was based on a very short segment of the VP1 gene consisting of nucleotides 1744 and 1812 (amino acids 61 to 83)6. Four subgroups of subtype I (Ia, Ib-1, Ib-II, and Ic) and six subgroups of subtype IV (IVa-1, IVa-2, IVb-1 and IVb-2, and IVc-1 and IVc-2) have been identified through DNA sequencing followed by phylogenetic analyses7.
Prolonged diabetes8, hypertension9 and various forms of glomerulonephritis conditions10 can cause chronic kidney disease (CKD) eventually. Over the past two decades, CKD have become more prevalent in rural agricultural areas of Sri Lanka. This is known as CKD of unknown aetiology. The consumption of water with contaminants11 has been found to be a cause for the increase in cases of kidney failure significantly and at present approximately 300 kidney transplants take place annually in Sri Lanka12. However, the information on BK viral infection followed by BKV-associated nephropathy which could be led to allograft rejection during post transplantation period is limited in Sri Lanka13,14. Therefore, this preliminary study was conducted to investigate prevailing BKV subtypes and subgroups among renal transplant patients through DNA sequencing followed by genotyping of polymorphic subtyping region of VP1 gene of each BKV present in renal transplant patient.
Material & Methods
A total of 227 renal transplant patients who were admitted to the National Hospital of Sri Lanka-Colombo; National Institute of Nephrology Dialysis and Transplantation, Maligawatta; Teaching Hospital, Karapitiya and Sri Lanka Army Hospital, Narahenpita, from July to December 2017, were tested for the presence of BKV in urine at the department of Virology of Medical Research Institute (MRI) through its routine laboratory BKV test of viral load quantification by RT-PCR method (RealStar® BKV PCR Kit 1.0, Altona DIAGNOSTICS, Germany). The majority of patients were suspected for BKV infection during their post-transplantation period due to rising levels of serum creatinine and obstructive uropathies. Samples with a BK viral load of more than 1×103 copies/ml were selected and 41 were confirmed for urinary presence of BKV out of 227 samples. Of these 41 patients, both urine and purified DNA samples were available for 13 patients, whereas either serum or urine or purified DNA samples were available for 2, 12 and 14 patients, respectively. Scientific and ethical approvals were obtained from the Research Committee and the Ethics Review Committee at MRI, respectively, under the project number 12/2017.
Processing of urine samples: If urine processing was done within 24 h after collection, it was stored at 4°C whereas if processing could not be done at the same day of collection, urine samples were stored in a sterile container at −20°C for further usage. To extract the BK viral DNA from urine samples, processing of urine was carried out at 4°C in three different ways using two samples containing 12 ml in each to identify which method will give a high yield: (i) processing of whole urine in which urine samples were homogenized (vortexed), and 200 µl was used for the viral DNA extraction; (ii) processing of urine after centrifugation in which the homogenized urine sample was centrifuged at 2000 g for 10 min (Eppendorf - Centrifuge 5810R, Thomas Scientific, USA), and the pellet containing 200 µl of urine was used for the viral DNA extraction15 and (iii) processing of urine after aliquoting and centrifugation in which the homogenized urine was aliquoted into three 4 ml samples and centrifuged at 2800 g for 15 min (SORVALL ST16R, Thermo Fisher Scientific, USA). Then pellets in all three aliquots were pooled into 200 µl of urine and used for the viral DNA extraction. DNA extraction was carried out using QIAamp® DNA Mini Kit (Qiagen, Hilden, Germany)16,17.
When available urine volume was not sufficient to extract DNA using the above-mentioned methods of precipitating pellets containing cell-associated virus particles (12 ml) or when extracted DNA (using QIAamp® DNA mini kit) was shown unresponsive to conventional PCR of VP1 gene amplification, already purified DNA which was extracted using QIAamp® Viral RNA Mini Kit (Qiagen, Hilden, Germany) was obtained from the department of Virology, MRI which had been stored at −20°C. BKV DNA elutes were concentrated to 20 µl using the ethanol and sodium acetate precipitation method to enhance the sensitivity in conventional PCR18.
PCR amplification and DNA sequencing: PCR was performed using previously described primers and given conditions18. The forward primer (5′-AGTGCCAAAACTACTAATAAAAG-3′, nt 1632-1654) and reverse primer (5′-CTGGGCTGTTGGG TTTTTAG-3′, nt 2121-2102) were used to amplify a region with 489 bp of the BKV VP1 gene. PCR was carried out in a total volume of 20 µl containing 5.25 µl of PCR water, 0.5 µl of forward and reverse primer (10 pmoles/µl), 0.5 µl of 10 mM dNTPs, 2 µl of 25 mM MgCl2, and 0.25 µl of 5 U of GoTaq® DNA Polymerase in the presence of 5 µl of 5X GoTaq® Reaction Buffer (Promega, USA). Five µl of viral DNA (10–100 ng/µl) extracted from urine were added to the PCR mixture. PCR amplification was performed in a b960 thermal cycler (Heal Force Advance, China). Cleaning the cycle sequencing PCR products was performed using a commercially available PCR purification kit, Wizard® SV gel and PCR clean-up system (Promega, USA) according to the manufacturer’s instructions. Nucleotide sequences of both DNA strands were determined by direct double-strand DNA cycle sequencing with the same forward and reverse primers using the BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, USA). After purification of sequencing PCR products, samples were denatured for three minutes at 95°C, immediately chilled on ice, and loaded on an Applied Biosystems® 3500Dx Capillary Sequencer (Applied Biosystems). DNA sequences were analyzed using the BioEdit7.0.5 sequence alignment editor software19.
Identification of BK virus (BKV) subtypes and subgroups: The resulting sequence data of subtyping region of VP1 gene (nucleotides 1744-1812) of the BKV were aligned with five reference sequences subtyping region of VP1 gene from NCBI (Subtype I- Z_19534, Z_19537, Subtype II - Z_19536, Subtype III - M_23122, Subtype IV- Z_19535) to categorize them into known BKV subtypes6. Since PCR amplification yields a product of 489 bp in size that lies in-between nucleotides 1,632 to 2,121 in the VP1 gene, sequencing results in this region were uploaded into NCBI GenBank database (MH930957 – MH930981) and used to construct a Neighbor Joining (NJ) phylogenetic tree20 after aligning DNA sequences using Clustal W21 available in the Molecular Evolutionary Genetics Analysis (MEGA) version 6.022 with known positive sequences retrieved from BKV subgroups (Subgroup Ia - NC_001538, AB_263926, Subgroup Ib-1- AB_211369, AB_211371, Subgroup Ib-11 - AB_211370, AB_263918, Subgroup Ic- AB_211372, AB_211377, AB_211381, Subtype II - AB_263916, AB_263920, Subtype III - M_23122, AB_211386, Subtype IV - AB_211389, AB_211390, AB_211391, AB_211387)7,23. NJ analysis was performed as described previously20,24 and bootstrap probabilities (BPs) were assessed with 1000 bootstrap replicates to evaluate the confidence level for the each clades in phylogenetic tree25. Baboon Polyomavirus SA12 (GenBank accession number of AY614708) was used as the out group26. References sequences and out group were downloaded from the NCBI GenBank database, and their accession numbers were included within the taxon labels in each phylogenetic tree.
Statistical analysis: The one-way analysis of variance test was used to assess the mean differences between viral loads related to BKV subtypes and subgroups. All statistical analyses were performed using SPSS software version 20.0 for Windows (SPSS, Chicago, IL, USA) and significance level was set at the 95 per cent confidence interval.
Results
The age of the 25 renal transplant recipients for whom BKV genotyping was achieved varried between 22 and 59 years. The highest occurrence of BKV infection was identified in those who are between 34 and 52 yr (mean±SD; 43.28±9.98 yr). BK viral loads varied from 1.06 × 103 to 3.016 × 108 copies/ml, and the median value of the viral load was 3.7 × 105 copies/ml (mean±SD; 23597012.4±67223861.79). Their serum creatinine levels varied in between 68.97 and 605 (mean±SD; 208.32±130.55) μmol/l (Table I).
Table I.
Baseline characteristics of BK virus (BKV) infected renal transplant patients (n=25)
| Patient (sample) ID | NCBI-GenBank accession number | Clinical and demographic data of BKV infected patients | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| Age (yr) | Gender | Post-transplantation period (wk) | BK viral load (copies/ml) | Serum creatinine level (µmol/l) | ||
| 17-32975 | MH930964 | 53 | Female | 24 | 1.69×106 | 138.82 |
| 17-32248 | MH930965 | 59 | Male | 4 | 1.10×103 | 457 |
| 17-33804 | MH930963 | 50 | Male | 8 | 1.59×108 | 166 |
| 17-31031 | MH930970 | 54 | Female | 24 | 4.93×106 | 123.79 |
| 17-32121 | MH930966 | 41 | Male | 20 | 4.75×106 | 137 |
| 17-30655 | MH930972 | 56 | Male | 28 | 3.016×108 | 185 |
| 17-30657 | MH930971 | 35 | Male | 104 | 7.29×106 | 159.16 |
| 17-31620 | MH930967 | 33 | Male | 20 | 5.04×104 | 135 |
| 17-31264 | MH930969 | 39 | Female | 144 | 1.71×104 | 273 |
| 17-28871 | MH930973 | 22 | Male | 28 | 7.94×106 | - |
| 17-22696 | MH930977 | 41 | Female | 78 | 2.05×103 | 167.11 |
| 17-23102 | MH930976 | 54 | Male | 120 | 1.06×103 | 128 |
| 17-34763 | MH930962 | 44 | Male | 36 | 1.69×105 | 158 |
| 17-38510 | MH930959 | 50 | Male | 24 | 1.21×107 | 127.2 |
| 17-37771 | MH930960 | 46 | Female | 128 | 2.63×103 | 133 |
| 17-5744 | MH930981 | 41 | Male | 52 | 1.00×106 | 605 |
| 17-26714 | MH930974 | 27 | Male | 232 | 1.83×103 | 155.62 |
| 17-23149 | MH930975 | 29 | Male | 68 | 3.51×104 | 496.04 |
| 17-39770 | MH930957 | 41 | Male | 31 | 3.67×105 | 178 |
| 17-38590 | MH930958 | 44 | Male | 16 | 7.57×107 | 185.68 |
| 17-35477 | MH930961 | 41 | Male | 24 | 8.31×106 | 178 |
| 17-21792 | MH930980 | 30 | Male | 13 | 1.64×103 | 300.67 |
| 17-21848 | MH930979 | 58 | Male | 16 | 2.64×104 | 171.78 |
| 17-22281 | MH930978 | 48 | Male | 47 | 3.70×105 | 172 |
| 17-31275 | MH930968 | 46 | Female | 4 | 4.57×106 | 68.97 |
The two serum samples (0.4 - 0.6 ml) with a viral load of >1 × 104 copies/ml (from which DNA was extracted followed by conventional PCR amplification of VP1 gene), showed no PCR amplicons. Therefore, obtaining serum samples for BK viral DNA extraction was discontinued for this study, and it was decided to carry out DNA extraction using the available urine samples from rest of the patients’ samples pool with adequate minimum volume of 12 ml of urine.
Among the three different methods of processing urine prior to viral DNA extraction, the third method resulted in the highest positivity rate for conventional PCR (>75%) followed by second method (<50%). Urine samples processed using the first method did not give positive results for the conventional PCR. Thus, 18 of the 25 (72%) urine samples, which were processed after centrifugation for generating pellets containing cell-associated viruses, followed by BKV DNA extraction, were given positive amplification for conventional PCR. Only seven of the 27 of BKV DNA elute samples, (25.92%) which were received from MRI, were shown to yield positive PCR amplifications after DNA concentration using the ethanol and sodium acetate precipitation method. Therefore, due to the unresponsiveness to conventional PCR, BKV subtyping could be achieved with only 25 patient samples out of the total 41 BKV-infected renal transplant patients.
In this study, BKV subtypes I and II were identified after aligning at the subtyping region of VP1 gene (in between the nucleotides of 1744 and 1812) with known positive BKV sequences retrieved from NCBI with frequencies of 18/25 (72%) and 7/25 (28%), respectively (Fig. 1A and B). Subgroups Ia, Ib-1, Ib-11 and Ic were identified with frequencies of 6/18 (33.3%), 6/18 (33.3%), 5/18 (27.8%), and 1/18 (5.6%) respectively (Fig. 2). The subgroups of Ia, Ib-1, Ib-11, and Ic were aligned with subtype I and rest of the subgroups resulting from phylogenetic analysis aligned well with subtype II. Mean viral load (104 copies/ml) of BKV subgroups Ia, Ib-1, Ib-11, Ic and subtype II were 1.226±1.405, 354.267±373.524, 4590.250±7546.121, 0.205±0 and 5499.592±11221.212, respectively (Table II). However, the mean differences of persistent viral loads between each subgroup representing subtypes I and II were not significant at the 95 per cent confidence level, suggesting that BKV subtype or subgroup did not affect the amount of BKV present in these patients.
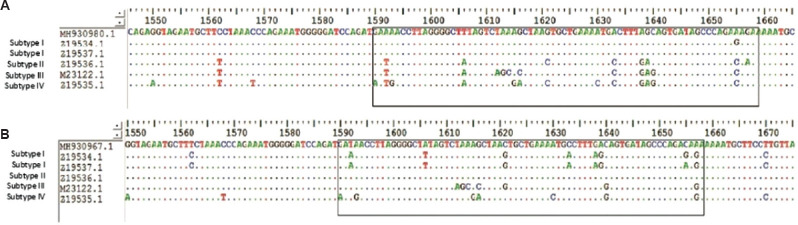
Fig. 1.
(A) Alignment of forward sequence of MH930980 with BKV reference sequence of subtype I at the subtyping region of VP1 gene. (B) Alignment of forward sequence of MH930967 with BKV reference sequence of subtype II at the subtyping region of VP1 gene.
Fig. 2.
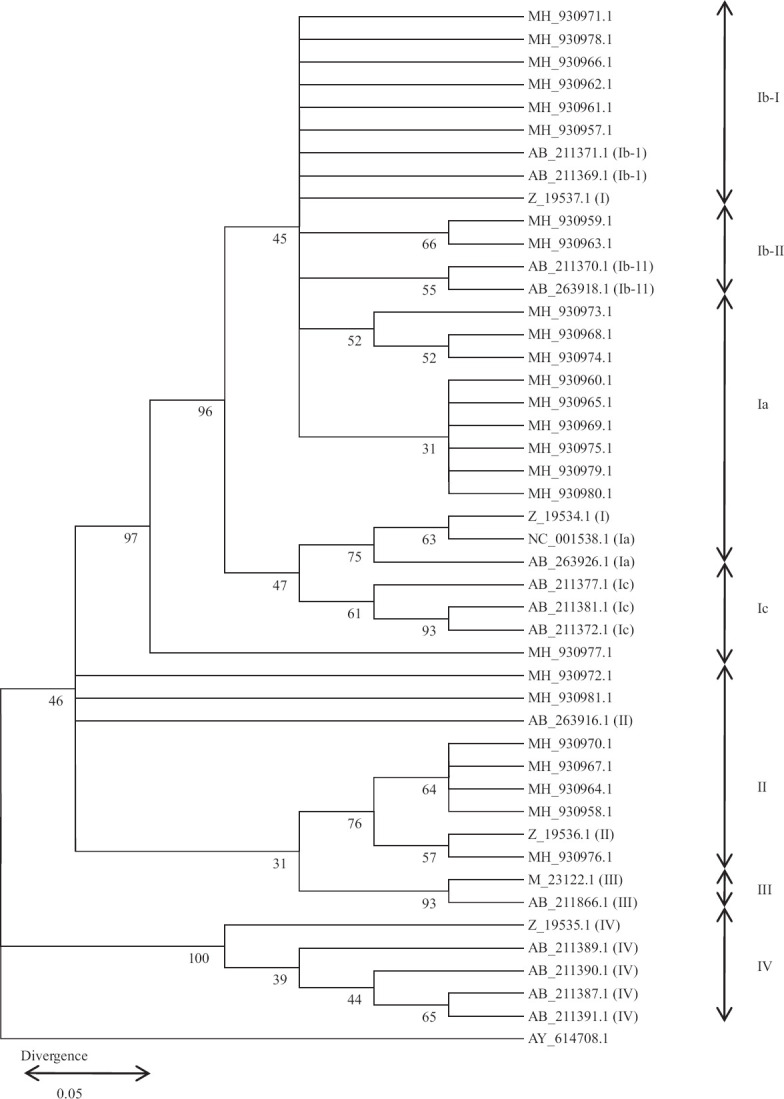
NJ phylogenetic tree classifying 41 BKV into subtypes and subgroups. The 430 bp of subtyping sequences detected around the Sri Lanka including 11 reference sequences7,23 were used to reconstruct an NJ phylogenetic tree23,24. The corresponding sequence of the baboon polyomavirus SA12 (GenBank accession number, AY614708) was used as the outgroup26. BKV subtypes II and subgroups belonging to subtype I are indicated at the right side of the tree and GenBank accession number of retrieved BKV isolates in this study are shown (MH930957 – MH930981). The numbers at the nodes are BPs (%) obtained for 1000 replicates25. (BKV Subgroup Ia - Z_19534, NC_001538, AB_263926, MH_930960, MH_930965, MH_930969, MH_930975, MH_930979, MH_930980; Subgroup Ib-1- Z_19537, AB_211369, AB_211371, MH_930971, MH_930978, MH_930966, MH_930962, MH_930961, MH930957: Subgroup Ib-11 – AB_211370, AB_263918, MH_930959, MH_930963, MH_930973, MH_930968, MH_930974; Subgroup Ic – AB_211372, AB_211377, MH_930977 and Subtype II – Z_19536, AB_263916, AB_263920, MH_930972, MH_930981, MH_930976, MH_930958, MH_930964, MH_930967, MH_930970, Subtype III - M_23122, AB_211386, Subtype IV - Z_19535, AB_211389, AB_211390, AB_211391, AB_211387).
Table II.
Mean viral load pertaining to BK virus (BKV) subtypes and subgroups
| BKV subtype and subgroup | Relevant NCBI-GenBank accession numbers | BK viral load (copies/ml) | Mean±SD value of BKV load (×104 copies/ml) |
|---|---|---|---|
| Ia | MH930960 | 2.63×103 | 1.226±1.405 |
| MH930965 | 1.10×103 | ||
| MH930969 | 1.71×104 | ||
| MH930975 | 3.51×104 | ||
| MH930979 | 2.64×104 | ||
| MH930980 | 1.64×103 | ||
| Ib-1 | MH930957 | 3.67×105 | 354.267±373.524 |
| MH930961 | 8.31×106 | ||
| MH930962 | 1.69×105 | ||
| MH930966 | 4.75×106 | ||
| MH930971 | 7.29×106 | ||
| MH930978 | 3.70×105 | ||
| Ib-11 | MH930959 | 1.21×107 | 4590.250±7546.121 |
| MH930963 | 1.59×108 | ||
| MH930968 | 4.57×106 | ||
| MH930973 | 7.94×106 | ||
| MH930974 | 1.83×103 | ||
| Ic | MH930977 | 2.05×103 | 0.205±0 |
| II | MH930958 | 7.57×107 | 5499.592±11221.212 |
| MH930964 | 1.69×106 | ||
| MH930967 | 5.04×104 | ||
| MH930970 | 4.93×106 | ||
| MH930972 | 3.016×108 | ||
| MH930976 | 1.06×103 | ||
| MH930981 | 1.00×106 |
Discussion
Molecular characterization of BKV is important for tracing multiple BK viral strains as an epidemiologic investigation, identification of the role of a particular subtype or a variant of BKV toward the development or the pathogenesis of BKV nephropathy. In this study, majority of renal transplant patients belonged to BKV subtype I (72%). The rest of the patients (28%) were found to be infected by subtype II which was previously known as a rare subtype and it was a novel finding for Asian region. The presence of BKV subtype I observed in this study was consistent with the worldwide BKV prevalence studies where subtype I was found as the most frequent in normal adults in the general population worldwide (80%)24 as well as in Asian countries such as in India (50%)27 and Japan (70-80%)24,28. BKV subtype IV has been presented as the second most frequent subtype worldwide (15%) and also in India (33.3%) whereas subtypes II and III are infrequently detected in normal adults24,27. In contrast, other Asian countries such as Mongolia, China and Vietnam have reported higher prevalence of subtype IV followed by subtype I28. The first BKV genotyping scheme introduced by Jin6 showed that the BKV subtype I was the most frequent genotype (63.4%) followed by type II (21.9%), IV (9.8%) and III (4.9%) in England. Furthermore, the second highest prevalence of BKV subtype II was identified among Brazilian, Dutch, Tunisian and Spanish renal transplant patients29–32.
With respect to global distribution of BKV subgroups, Ia was the most prevalent in Africans, Ib1 in Southeast Asians, Ib2 in Europeans, American populations and West Asians and Ic in Northeast Asians33,34. In the current study, subgroups of Ia, Ib-1 were identified equally and considerably, followed by Ib-11 and Ic in Sri Lankan renal transplant patients.
One of the major limitations of PCR-based tests is inhibition of the nucleic acid amplification process by substances present in samples35. In urine, substances such as urea can act as PCR inhibitors, which can lead to inhibition of DNA polymerase or reverse transcriptase activity36,37. Another limitation was samples with infectious agent levels that were below the assay’s sensitivity. Isolation and culturing of BKV from each sample could not be performed due to resource and time limitations. The use of alternative methodology for BKV genotyping such as quantitative PCR /real time reverse transcription-PCR is shown to have many advantages over conventional PCR such as high sensitivity and capability of determining the subtype of patient samples with lower viral loads38,39.
In conclusion, this preliminary study showed a high occurrence of BKV subtype I in renal transplant recipients in Sri lanka. The BKV subtype II, which is considered rare worldwide, was also observed in this study. This emphasizes the need for further molecular and serological studies to determine the prevalence of different BKV subtypes in Sri Lanka.
Footnotes
Financial support & sponsorship: This study was funded by the Institute of Biochemistry, Molecular Biology and Biotechnology (IBMBB), University of Colombo and Department of Virology, MRI, Colombo.
Conflicts of Interest: None.
References
- 1.Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B. K.) isolated from urine after renal transplantation. Lancet. 1971;297:1253–7. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- 2.Babel N, Volk HD, Reinke P. BK polyomavirus infection and nephropathy:The virus-immune system interplay. Nat Rev Nephrol. 2011;7:399–406. doi: 10.1038/nrneph.2011.59. [DOI] [PubMed] [Google Scholar]
- 3.Kuypers DR. Management of polyomavirus-associated nephropathy in renal transplant recipients. Nat Rev Nephrol. 2012;8:390–402. doi: 10.1038/nrneph.2012.64. [DOI] [PubMed] [Google Scholar]
- 4.Hirsch HH, Randhawa P AST Infectious Diseases Community of Practice. BK polyomavirus in solid organ transplantation. Am J Transplant. 2013;13((Suppl 4)):179–88. doi: 10.1111/ajt.12110. [DOI] [PubMed] [Google Scholar]
- 5.BK Virus, Molecular Detection, Quantitative, PCR, Urine. Mayo Clinic Laboratories. 2020. [accessed on October 31, 2020]. Available from: https://www.mayocliniclabs.com/test-catalog/Clinical+and+Interpretive/87859 .
- 6.Jin L. Rapid genomic typing of BK virus directly from clinical specimens. Mol Cell Probes. 1993;7:331–4. doi: 10.1006/mcpr.1993.1047. [DOI] [PubMed] [Google Scholar]
- 7.Zhong S, Yogo Y, Ogawa Y, Oshiro Y, Fujimoto K, Kunitake T, et al. Even distribution of BK polyomavirus subtypes and subgroups in the Japanese archipelago. Arch Virol. 2007;152:1613–21. doi: 10.1007/s00705-007-0997-y. [DOI] [PubMed] [Google Scholar]
- 8.ADVANCE Collaborative Group. Patel A, MacMahon S, Chalmers J, Neal B, Billot L, et al. Intensive blood glucose control and vascular outcomes in patients with type 2 diabetes. N Engl J Med. 2008;358:2560–72. doi: 10.1056/NEJMoa0802987. [DOI] [PubMed] [Google Scholar]
- 9.Roehm B, Weiner DE. Blood pressure targets and kidney and cardiovascular disease:same data but discordant guidelines. Curr Opin Nephrol Hypertens. 2019;28:245–50. doi: 10.1097/MNH.0000000000000492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chronic Kidney Disease Prognosis Consortium. Matsushita K, Van der Velde M, Astor BC, Woodward M, Levey AS, et al. Association of estimated glomerular filtration rate and albuminuria with all-cause and cardiovascular mortality in general population cohorts:A collaborative meta-analysis. Lancet. 2010;375:2073–81. doi: 10.1016/S0140-6736(10)60674-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jayasumana C, Gunatilake S, Senanayake P. Glyphosate, hard water and nephrotoxic metals:Are they the culprits behind the epidemic of chronic kidney disease of unknown etiology in Sri Lanka? Int J Environ Res Public Health. 2014;11:2125–47. doi: 10.3390/ijerph110202125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jha V, Ur-Rashid H, Agarwal SK, Akhtar SF, Kafle RK, Sheriff R. The state of nephrology in South Asia. Kidney Intl. 2019;95:31–7. doi: 10.1016/j.kint.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Premathilake MIP, Jayamaha CJS. Comparison of BK virus viruria and viremia in post renal transplant patients –A single centre study. Sri Lanka Coll Microbiol Bull. 2016;14:25. [Google Scholar]
- 14.Gunawardena KW, Jayamaha JS, Wijewickrama ES, Lanerolle RD. BK virus vireamia and viruria among a group of post kidney transplant patients in Sri Lanka. Ceylon Med J. 2017;62:114. doi: 10.4038/cmj.v62i2.8480. [DOI] [PubMed] [Google Scholar]
- 15.Pinto GG, Poloni JA, Carneiro LC, Baethgen LF, Barth AL, Pasqualotto AC. Evaluation of different urine protocols and DNA extraction methods for quantitative detection of BK viruria in kidney transplant patients. J Virol Methods. 2013;188:94–6. doi: 10.1016/j.jviromet.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 16.Bergallo M, Galliano I, Loiacono E, Ferro F, Montanari P, Ravanini P. Impact of two different commercial DNA extraction methods on BK virus viral load. Microbiologia Medica. 2016;4825;31 [Google Scholar]
- 17.Taheri S, Kafilzadeh F, Shafa M, Yaran M, Mortazavi M, Seirafian S, et al. Comparison of polyomavirus (BK virus and JC viruses) viruria in renal transplant recipients with and without kidney dysfunction. J Res Med Sci. 2011;16:916–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Tremolada S, Delbue S, Castagnoli L, Allegrini S, Miglio U, Boldorini R, et al. Mutations in the external loops of BK virus VP1 and urine viral load in renal transplant recipients. J Cell Physiol. 2010;222:195–9. doi: 10.1002/jcp.21937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Department of Microbiology, North Carolina State University. BioEdit. Program and documentation, Ver. 7.2.5. North Carolina, USA: North Carolina State University: 2005. [Google Scholar]
- 20.Saitou N, Nei M. The neighbor-joining method:A new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–25. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 21.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W:Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6:Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nishimoto Y, Takasaka T, Hasegawa M, Zheng HY, Chen Q, Sugimoto C, et al. Evolution of BK virus based on complete genome data. J Mol Evol. 2006;63:341–52. doi: 10.1007/s00239-005-0092-5. [DOI] [PubMed] [Google Scholar]
- 24.Takasaka T, Goya N, Tokumoto T, Tanabe K, Toma H, Ogawa Y, et al. Subtypes of BK virus prevalent in Japan and variation in their transcriptional control region. J Gen Virol. 2004;85:2821–7. doi: 10.1099/vir.0.80363-0. [DOI] [PubMed] [Google Scholar]
- 25.Felsenstein J. Confidence limits on phylogenies:An approach using the bootstrap. Evolution. 1985;39:783–91. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 26.Cantalupo P, Doering A, Sullivan CS, Pal A, Peden KW, Lewis AM, et al. Complete nucleotide sequence of polyomavirus SA12. J Virol. 2005;79:13094–104. doi: 10.1128/JVI.79.20.13094-13104.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Madhavan HN, Bagyalakshmi R, Revathy M, Aarthi P, Malathi J. Optimisation and analysis of polymerase chain reaction based DNA sequencing for genotyping polyoma virus in renal transplant patients:A report from South India. Indian J Med Microbiol. 2015;33((Suppl)):37–42. doi: 10.4103/0255-0857.150878. [DOI] [PubMed] [Google Scholar]
- 28.Chen Q, Zheng HY, Zhong S, Ikegaya H, He HX, Wei W, et al. Subtype IV of the BK polyomavirus is prevalent in East Asia. Arch Virol. 2006;151:2419–29. doi: 10.1007/s00705-006-0814-z. [DOI] [PubMed] [Google Scholar]
- 29.Zalona AC, Lopes GS, Schrago CG, Gonçalves RT, Zalis MG, Varella RB. Molecular characterization of BK polyomavirus subtypes in renal transplant recipients in Brazil. J Med Virol. 2011;83:1401–5. doi: 10.1002/jmv.22117. [DOI] [PubMed] [Google Scholar]
- 30.Wunderink HF, De Brouwer CS, Gard L, De Fijter JW, Kroes ACM, Rotmans JI, et al. Source and relevance of the BK polyomavirus genotype for infection after kidney transplantation. Open Forum Infect Dis. 2019;6:ofz078. doi: 10.1093/ofid/ofz078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boukoum H, Nahdi I, Foulongne V, Zallema D, Aloui S, Achour A, et al. Distribution of BK polyomavirus genotypes in Tunisian renal transplant recipients. J Med Virol. 2011;83:725–30. doi: 10.1002/jmv.22035. [DOI] [PubMed] [Google Scholar]
- 32.Ledesma J, Bouza E, González-Nicolás MA, García de Viedma D, Rodríguez-Sánchez B, Muñoz P. BK polyomavirus genotyping at inter- and intra-patient level in Spain. J Med Virol. 2013;85:1402–8. doi: 10.1002/jmv.23612. [DOI] [PubMed] [Google Scholar]
- 33.Zheng HY, Nishimoto Y, Chen Q, Hasegawa M, Zhong S, Ikegaya H, et al. Relationships between BK virus lineages and human populations. Microbes Infect. 2007;9:204–13. doi: 10.1016/j.micinf.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 34.Yogo Y, Sugimoto C, Zhong S, Homma Y. Evolution of the BK polyomavirus:Epidemiological, anthropological and clinical implications. Rev Med Virol. 2009;19:185–99. doi: 10.1002/rmv.613. [DOI] [PubMed] [Google Scholar]
- 35.Bergallo M, Costa C, Gribaudo G, Tarallo S, Baro S, Negro Ponzi A, et al. Evaluation of six methods for extraction and purification of viral DNA from urine and serum samples. New Microbiol. 2006;29:111–9. [PubMed] [Google Scholar]
- 36.Al-Soud WA, Jönsson LJ, Râdström P. Identification and characterization of immunoglobulin G in blood as a major inhibitor of diagnostic PCR. J Clin Microbiol. 2000;38:345–50. doi: 10.1128/jcm.38.1.345-350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Abu Al-Soud W, Rådström P. Effects of amplification facilitators on diagnostic PCR in the presence of blood, feces, and meat. J Clin Microbiol. 2000;38:4463–70. doi: 10.1128/jcm.38.12.4463-4470.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gambarino S, Costa C, Astegiano S, Piasentin EA, Segoloni GP, Cavallo R, et al. Genotyping of polyomavirus BK by real time PCR for VP1 gene. Mol Biotechnol. 2011;49:151–8. doi: 10.1007/s12033-011-9386-6. [DOI] [PubMed] [Google Scholar]
- 39.Gard L, Niesters HG, Riezebos-Brilman A. A real time genotyping PCR assay for polyomavirus BK. J Virol Methods. 2015;221:51–6. doi: 10.1016/j.jviromet.2015.04.024. [DOI] [PubMed] [Google Scholar]