Abstract
Background
Some human polymorphisms of ACE1, ACE2, IFITM3, TMPRSS2 and TNFα genes may have an effect on the susceptibility to SARS-CoV-2 infection and increase the risk to develop severe COVID-19. We conducted a systematic review of current evidence to investigate the association of genetic variants of these genes with the susceptibility to virus infection and patient prognosis.
Methods
We systematically searched Medline, Embase and The Cochrane Library for articles published until May 2022, and included observational studies covering genetic association of ACE1, ACE2, IFITM3, TMPRSS2 and TNFα genes with COVID-19 susceptibility or prognosis. We evaluated the methodological quality of included studies, and pooled data as convenient in meta-analysis (MA). Odds ratio (OR) values and 95% confidence intervals were calculated.
Results
We included 35 studies (20 on ACE, 5 each on IFITM3, TMPRSS2, TNFα), enrolling 21,452 participants, of them 9401 were COVID-19 confirmed cases. ACE1 rs4646994 and rs1799752, ACE2 rs2285666, TMPRSS2 rs12329760, IFITM3 rs12252 and TNFα rs1800629 were identifies as common polymorphisms. Our MA showed an association between genetic polymorphisms and susceptibility to SARS-CoV-2 infection for IFITM3 rs12252 CC (OR 5.67) and CT (OR 1.64) genotypes. Furthermore, MA uncovered that both ACE DD (OR 1.27) and IFITM3 CC (OR 2.26) genotypes carriers had a significantly increased risk of developing severe COVID‐19.
Discussion
These results provide a critical evaluation of genetic polymorphisms as predictors in SARS-CoV-2 infection. ACE1 DD and IFITM3 CC polymorphisms would lead to a genetic predisposition for severe lung injury in patients with COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10238-023-01038-9.
Keywords: SARS-CoV-2, Polymorphism, Meta-analysis
Background
The infection by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), causing the coronavirus disease 2019 (COVID-19), has emerged as a global health problem. The mechanism underlying the infection was studies by several authors to identify main cause of susceptibility, and responsible factors of severe form of COVID-19. Polymorphism in genes mediating virus entry in target cells has been at the centre of attention. To entry into cells, the virus uses angiotensin-converting enzyme 2 (ACE2) as the major receptor for viral entry in humans. SARS-CoV-2 spike glycoprotein binds via its receptor-binding domain (RDB) with a high affinity to human ACE2 and mediates virus internalization [1]. This phenomenon suggests that this gene as a factor for increasing susceptibility to disease [2]. Likewise, the presence of polymorphism in ACE1 has been shown to be associated with COVID-19 [3]. Indeed, several studies have also demonstrated an association between the frequency of ACE D/D polymorphism and both prevalence and mortality rates of COVID-19 [4, 5].
Single-nucleotide polymorphisms (SNPs) in the ACE and ACE2 genes have been described, and their association with the risk of various diseases, included COVID-19 has been indicated [6]. In addition to ACE, several other molecules, such as the transmembrane protease serine 2 (TMPRSS2), are also involved in the process of SARS-CoV-2 virus entry [1]. TMPRSS2 facilitates the cleavage of the S protein, enabling membrane fusion and endocytic entry of the virus particles. This has suggested the hypothesis that genetic variability within the TMPRSS2 gene may play a role in determining SARS-CoV-2 infection [7, 8]. The Interferon-induced transmembrane proteins (IFITMs) play an important role in the antiviral defence in the adaptive and innate immune response [9], blocking the fusion of enveloped-viruses with the cell membranes. IFITMs seem to play a role also in the response to coronavirus as inhibitors of infection. In particular, polymorphisms in the IFITM3 genes would affect the susceptibility to viral infection [10]. Furthermore, the SARS‐CoV‐2 infection induces pathogenic T helper 1 (Th1) cells to secrete proinflammatory cytokines such interleukin‐1 (IL-1) and IL‐6, which, in turn, trigger CD14 + CD16 + inflammatory monocytes to generate large amounts of IL‐6, TNF‐α, and other cytokines. Genetic variations within some inflammatory cytokines, including TNFα, have been already associated with the increased risk of severe COVID-19 [11].
Thus, we conducted a comprehensive systematic review with meta‐analysis aimed to evaluate the association of genetic polymorphisms of ACE1, ACE2, IFITM3, TMPRSS2 and TNFα genes with the susceptibility to SARS-CoV-2 infection and risk to develop severe COVID-19.
Methods
Protocol
The review protocol was registered in PROSPERO, the International Prospective Register of Systematic Reviews (CRD42022356627). We followed the Preferred Reporting Items for Systematic Reviews and Meta-analyses guideline for reporting systematic review [12].
Literature search
A systematic literature search was conducted in Medline (PubMed), EMBASE and Cochrane Central Register of Controlled Trials (CENTRAL). We scanned also reference lists of articles for additional records. Search strategy adopted was similar across the databases and it was developed using applying the following keywords: COVID-19, genetic polymorphisms, mutation, ACE1, ACE2, IFITM3, TMPRSS2 and TNFα. We limited the search to studies in humans and published in English, Italian or Spanish. The search was performed on May 2022.
Inclusion and exclusion criteria
We included studies meet the following inclusion criteria: (i) examined the association between genetic polymorphisms of genes of interest and susceptibility and severity to SARS-CoV-2 infection; (ii) enrolled human subjects with infection of SARS-CoV-2; (iii) reported the COVID-19-related SNPs and genes.
We excluded editorial, abstracts, conference proceedings, unpublished reports, review articles, meta-analyses, comments, editorials and repeated literature, animal studies and studies with human subjects involving other coronaviruses, studies that did not provide enough information or were performed on paediatric patients. Our approach was ‘inclusive’ so as to obtain a pragmatic overall picture of research in this field.
Selection of studies
Two investigators (VP and MC) independently screened title and abstract of each citation included in reference list of potentially eligible studies. After examining the entire text of the retrieved documents, only those articles satisfying the inclusion criteria were included. Any disagreements were resolved by discussion and consensus.
Data extraction
We collected information about characteristic of: (i) the publication (author, year of publication, and country), (ii) included study (study design and total number of patients included), (iii) the study population (age and gender), and (iv) outcomes of interest (prevalence of each genotype, and association between SNP and susceptibility and severity of SARS-CoV-2 infection).
Quality assessment
The Newcastle–Ottawa Scale (NOS) [13] was used to evaluate quality of eligible cohort and case‐control studies included in this systematic review with meta‐analysis. Two authors (VP and MC) independently evaluated each included study considering the following domains: selection, comparability, and exposure. The maximum NOS scores of each domain were 4, 2, and 3 stars, respectively. The study was rated as high quality if it received a total score of 7–9, moderate quality with a total score of 4–6, or low quality with a total score of 0–3 stars.
Statistical analysis
We stratified studies by genes and carried out meta-analyses for each polymorphism. Pooled odds ratio (OR) with 95% confidence intervals (CIs) was calculated. We assessed the presence of heterogeneity utilising the I-squared statistics (I2), which estimates the percentage of variation between study results that is due to heterogeneity rather than sampling error. The I2 statistics indicates the percentage of the overall variability that is due to between-study (or inter-study) variability, as opposed to within-study (or intra-study) variability. An I2 value smaller than 50% reveals low heterogeneity, I2 included between 50 and 75% moderate heterogeneity, and I2 greater than 75% substantial heterogeneity. In the absence of heterogeneity between studies, we pooled data using Mantel–Haenszel methods for a fixed-effects model [14], otherwise we combined the studies using the random-effects model [15]. Meta-analysis was performed when at least three articles studying the same subgroup were available. A p value < 0.05 was considered as statistically significant. Analyses were performed with the REVMAN 5.4 (The Cochrane Collaboration, The Nordic Cochrane Centre, Copenhagen, Denmark) software.
Results
Studies identification and selection
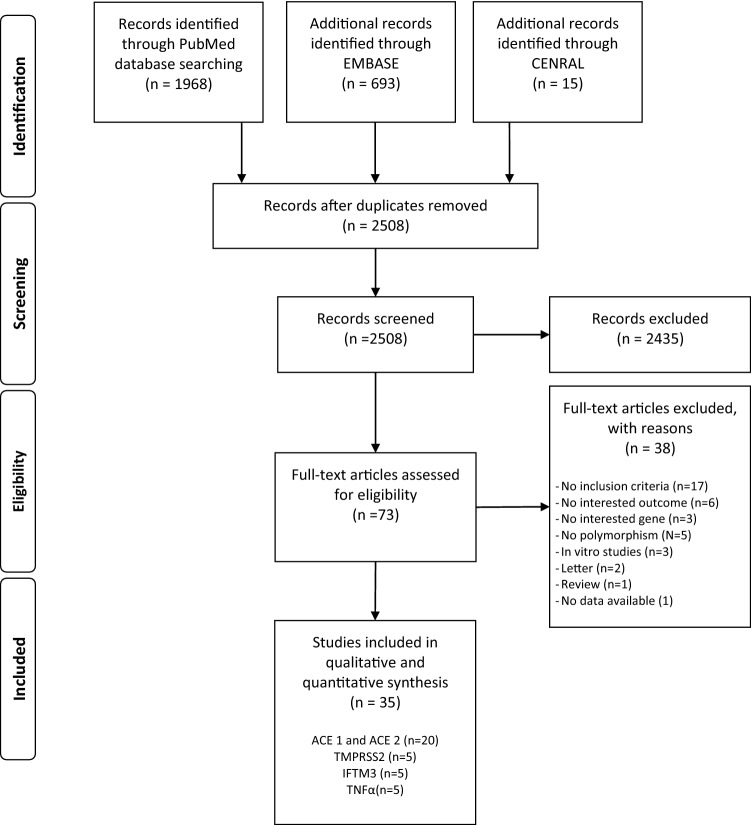
The literature search, after the exclusion of duplicates and irrelevant records, identified 2508 references. Of these, 2435 were excluded because they did not meet the inclusion criteria. There were 73 studies considered eligible for inclusion and details were obtained from full texts. From full-text analysis, further 38 texts were excluded, leaving a total of 35 studies [6, 11, 16–48] included in this systematic review (Fig. 1).
Fig. 1.
PRISMA flow diagram of the studies selection process
Characteristics of included studies
We included 35 studies (enrolling 21,452 participants, of them 9401 COVID-19 confirmed cases), 20 on ACE, 5 on TMPRSS2, 5 on IFITM3, and 5 on TNFα. We included ten cohort studies and 25 case–control studies. Overall, the number of participants ranged from 39 to 4759. ACE1 rs4646994 and rs1799752, ACE2 rs2285666, TMPRSS2 rs12329760 and IFTM3 rs12252 were identifies as common polymorphisms. Details are reported in Table 1.
Table 1.
Characteristics of included studies
| Author | Study design | Country | Genotyping method | Gene | Polymorphism | N participants | Male (%) | N patients with COVID 19 | NOS SCORE |
|---|---|---|---|---|---|---|---|---|---|
| Akbari et al. [16] | Case–control | Iran | PCR | ACE 1 | rs1799752 | 182 | 105 (56) | 91 | 7 |
| Aladag e al. [17] | Cross-sectional | Turkey | PCR | ACE 1 | rs4646994 | 412 | – | 112 | 6 |
| Alimorandi et al. [18] | Case–control | Iran | PCR | ACE 1 | rs4343 | 129 | 67 (52) | 79 | 9 |
| ACE 2 | rs2285666 | ||||||||
| Annunziata et al. [19] | Case–control | Italy | RT-PCR | ACE 1 | I/D polymorphism | 39 | – | 20 | 6 |
| Bastug et al. [20] | Cohort | Turkey | RT-PCR | ACE 1 | rs1799752 | 100 | 59 (59) | 100 | 8 |
| Cafiero et al. [21] | Cross-sectional | Italy | PCR | ACE 1 | rs1799752 | 104 | 58 (56) | 104 | 6 |
| ACE 2 | rs2074192 | ||||||||
| ACE 2 | rs2106809 | ||||||||
| Calabrese et al. [22] | Case–control | Italy | Not Reported | ACE 1 | rs1799752 | 290 | – | 68 | 7 |
| Celik et al. [23] | Cohort | Turkey | PCR | ACE 1 | ACE I/D | 155 | 78 (50) | 155 | 5 |
| ACE 2 | rs2106809 | ||||||||
| Gomez et al. [6] | Case–control | Spain | PCR | ACE 1 | rs4646994 | 740 | 373 (50) | 204 | 8 |
| ACE2 | rs2285666 | ||||||||
| Gong et al. [24] | Case–control | China | PCR | ACE 1 | I/D polymorphism | 862 | – | 419 | 8 |
| Gunal et al. [25] | Cohort | Turkey | (RT)-qPCR | ACE 1 | I/D polymorphism | 90 | 59 (65) | 90 | 8 |
| Hubacek et al. [26] | Case–control | Czech Republic | PCR | ACE 1 | rs4646994 I/D polymorphism | 2969 | 1388 (47) | 410 | 8 |
| Kouhpayeh et al. [27] | Case–control | Iran | RT-PCR | ACE 1 | rs4646994 | 504 | 276 (55) | 258 | 8 |
| Mahmood et al. [28] | Cohort | Iraq | PCR | ACE 1 | rs4646994 | 195 | 98 (50) | 99 | 8 |
| ACE 2 | rs2285666 G/A | ||||||||
| Martınez-Gomez et al. [29] | Cross-sectional | Mexico | RT-PCR | ACE 1 | I/D polymorphism | 481 | 290 (60) | 481 | 7 |
| ACE 2 | rs2285666 | ||||||||
| ACE 2 | rs2074192 | ||||||||
| Mir et al. [30] | Case–control | Saudi Arabia | RT-qPCR | ACE 1 | rs4646994 I/D | 267 | 185 (69) | 117 | 8 |
| Mohlendick et al. [31] | Cohort | Germany | RT-PCR | ACE 1 | rs1799752 | 550 | 323 (59) | 297 | 8 |
| ACE 2 | rs2285666 | ||||||||
| Papadopoulou et al. [32] | Case–control | Greece | PCR | ACE 1 | I/D polymorphism | 397 | - | 81 | 8 |
| Saad et al. [33] | Case–control | Lebanon | PCR | ACE 1 | rs1799752 | 358 | 195 (54) | 232 | 9 |
| Verma et al. [34] | Cohort | India | PCR- AFLP | ACE 1 | rs4646994 | 269 | 170 (63) | 269 | 6 |
| Alghamdi et al. [35] | Cohort | Saudi Arabia | PCR | IFTM3 | rs12252 | 880 | – | 825 | 6 |
| Cuesta-Llavona et al. [36] | Case–control | Spain | RT-PCR | IFTM3 | rs34481144 C/T | 666 | 369 (55) | 484 | 2 |
| rs12252 A/G | |||||||||
| Gomez et al. [37] | Case–control | Spain | RT-PCR | IFTM3 | rs12252 | 751 | 374 (50) | 311 | 7 |
| Schonfelder et al. [38] | Case–control | Germany | RT-PCR | IFTM3 | rs12252 | 492 | 288 (59) | 239 | 8 |
| rs34481144 | |||||||||
| Zhang et al. [39] | Cohort | China | Not Reported | IFTM3 | rs12252 | 80 | 33 (41) | 80 | 8 |
| Andolfo et al. [40] | Cohort | Italy | TaqMan, WES | TMPRSS2 | rs12329760 | 4759 | – | 996 | 7 |
| Ravikanth et al. [41] | Cohort | India | WES | TMPRSS2 | rs12329760 | 1030 | 809 (79) | 510 | 8 |
| Rokni et al. [42] | Case–control | Iran | RTqPCR | TMPRSS2 | s12329760 C/T | 576 | 325 (56) | 288 | 9 |
| rs75603675 C/A | |||||||||
| rs17854725 A/G | |||||||||
| rs4303795 A/G | |||||||||
| Schonfelder et al. [43] | Case–control | Germany | RT-PCR | TMPRSS2 | rs2070788 G/A | 492 | 288 (59) | 239 | 7 |
| rs12329760 C/T | |||||||||
| rs383510 T/C | |||||||||
| Wulandari et al. [44] | Cohort | Indonesia | PCR | TMPRSS2 | rs12329760 | 95 | 60 (63) | 95 | 7 |
| Ali et al. [45] | Case–control | Iraq | rRT PCR | TNFα | rs1800629 | 239 | 104 (44) | 125 | 6 |
| Fishchuk et al. [46] | Cohort | Ukraine | PCR–RFLP | TNFα | rs1800629 | 31 | 16 (52) | 31 | 6 |
| Heidari Nia et al. [47] | Case–control | Iran | RT-PCR | TNFα | rs1800629 | 550 | 316 (57) | 275 | 8 |
| Rokni et al. [48] | Case–control | Iran | PCR–RFLP | TNFα | rs1800629 | 634 | 359 (57) | 317 | 9 |
| Saleh et al. [11] | Case–control | Egypt | RT-qPCR | TNFα | rs1800629 | 1084 | 600 (55) | 900 | 9 |
Methodological quality of included studies
Following the NOS, the most of included studies (n = 26, 74%) were of high methodological quality (7–9 stars), while eight studies (23%) were of moderate quality (4–6 stars) and only one study was of scarce quality (Table1).
Susceptibility to SARS-CoV-2 infection
Thirty-five studies reported data about allele and genotype frequencies of ACE1 rs4646994, ACE1 rs1799752, ACE2 rs2285666, IFITM3 rs12252, TMPRSS2 rs12329760 and TNFα (Table 1).
For ACE1, ACE2, TMPRSS2, and TNFα, meta-analyses showed not significant association between genetic polymorphisms and SARS-CoV-2 infection in patients tested positive respected to negative, with high heterogeneity among included studies (Table 2).
Table 2.
Meta-analyses on susceptibility considering different genotypes
| ACE1_rs4646994_rs1799752Gene | N studies | Cases | Controls | OR (95% CI) | p | I2 | ||
|---|---|---|---|---|---|---|---|---|
| Events | Total | Events | Total | |||||
| ACE1_rs4646994_rs1799752 | ||||||||
| DD | 10 | 750 | 20,664 | 1405 | 4827 | 1.41 (0.97–2.05) | 0.07 | 87% |
| DI | 10 | 917 | 2398 | 0.8 (0.51–1.26) | 0.34 | 92% | ||
| II | 10 | 399 | 1034 | 0.69 (0.48–1) | 0.05 | 80% | ||
| ACE2_rs2285666 | ||||||||
| GG | 4 | 368 | 679 | 409 | 935 | 1.27 (0.58–2.82) | 0.55 | 90% |
| AG | 4 | 96 | 149 | 0.77 (0.46–1.29) | 0.33 | 64% | ||
| AA | 4 | 44 | 54 | 1.12 (0.26–4.82) | 0.88 | 84% | ||
| IFTM3_rs12252 | ||||||||
| CC | 3 | 9 | 1034 | 0 | 875 | 5.67 (1.01–31.77) | 0.05 | 0% |
| CT | 3 | 101 | 55 | 1.64 (1.15–2.33) | 0.007 | 0% | ||
| TT | 3 | 924 | 820 | 0.56 (0.39–0.79) | 0.001 | 0% | ||
| TMPRSS2_rs12329760 | ||||||||
| CC | 4 | 1165 | 2033 | 3094 | 4824 | 0.87 (0.68–1.11) | 0.27 | 72% |
| CT | 4 | 718 | 1456 | 1.10 (0.94–1.3) | 0.24 | 37% | ||
| TT | 4 | 150 | 254 | 1.01 (0.54–1.91) | 0.97 | 84% | ||
| TNFα_rs1800629 | ||||||||
| AA | 4 | 543 | 1617 | 170 | 890 | 1.11 (0.51–2.40) | 0.79 | 89% |
| GA | 4 | 601 | 366 | 1.22 (0.74–2.01) | 0.44 | 85% | ||
| GG | 4 | 473 | 360 | 0.63 (0.29–1.38) | 0.25 | 94% | ||
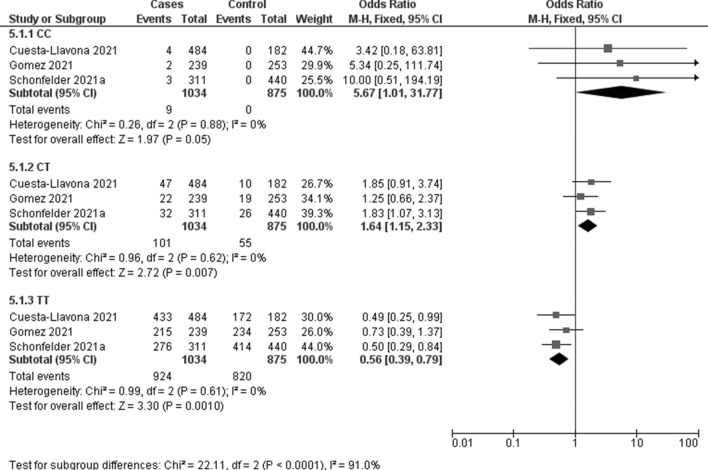
The association between IFITM3 rs12252 and COVID-19 susceptibility was evaluated in three studies including 1034 COVID-19 positive patients and 875 controls. Meta-analysis showed a significant association with C recessive (OR 5.67, 95% CI 1.01–31.77; p = 0.05; I2 = 0%, Fig. 2) and CT heterozygous models (OR 1.64, 95% CI 1.15–2.33; p = 0.007; I2 = 0%, Fig. 2).
Fig. 2.
Forest plot on IFTM3 rs12252 association with COVID-19 susceptibility
Severity of SARS-CoV-2 infection
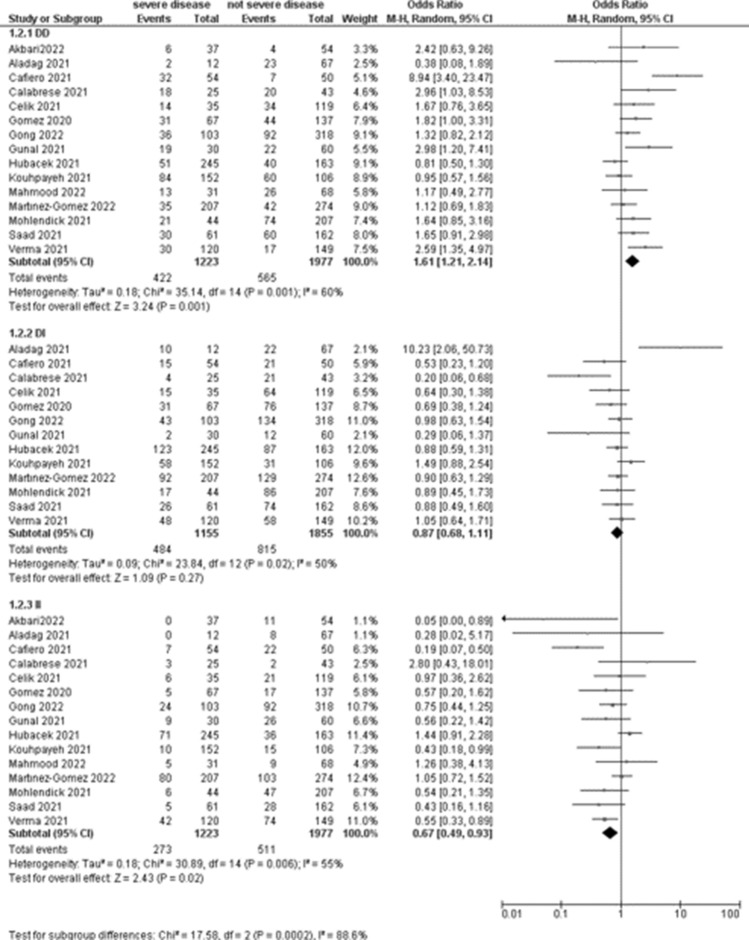
The association between COVID-19 severity and ACE1 rs4646994 and ACE1 rs1799752 was evaluated in 15 studies (1223 patients with severe disease). Meta-analyses showed that the DD genotype was associated with an increased risk of severe disease (OR 1.61, 95% CI 1.21–2.14; p = 0.001; Table 3, Fig. 3) respect to patients with not severe disease, with high heterogeneity among included studies (I2 = 60%).
Table 3.
Meta-analyses on severity considering different genotypes
| Gene | N studies | Severe disease | Not severe disease | OR (95% CI) | p | I2 | ||
|---|---|---|---|---|---|---|---|---|
| Events | Total | Events | Total | |||||
| ACE1_rs4646994_rs1799752 | ||||||||
| DD | 15 | 422 | 1223 | 565 | 1977 | 1.61 (1.21–2.14) | 0.001 | 60% |
| DI | 13 | 484 | 1155 | 815 | 1855 | 0.87 (0.68–1.11) | 0.27 | 50% |
| II | 15 | 273 | 1223 | 511 | 1977 | 0.67 (0.49–0.93) | 0.02 | 55% |
| ACE2_rs2285666 | ||||||||
| GG | 4 | 290 | 480 | 284 | 454 | 1.47 (0.77–2.80) | 0.24 | 62% |
| AG | 4 | 64 | 105 | 0.51 (0.35–0.74) | 0.0005 | 0% | ||
| AA | 4 | 126 | 65 | 0.89 (0.31–2.54) | 0.83 | 61% | ||
| IFTM3_rs12252 | ||||||||
| CC | 4 | 16 | 332 | 21 | 782 | 2.26 (1.05–4.89) | 0.04 | 0% |
| CT | 4 | 41 | 97 | 1.00 (0.67–1.49) | 0.98 | 39% | ||
| TT | 4 | 275 | 664 | 0.82 (0.55–1.21) | 0.32 | 0% | ||
| TMPRSS2_rs12329760 | ||||||||
| CC | 4 | 145 | 384 | 367 | 749 | 0.92 (0.45–1.91) | 0.83 | 83% |
| CT | 4 | 172 | 302 | 1.04 (0.68–1.59) | 0.86 | 54% | ||
| TT | 4 | 67 | 80 | 0.93 (0.46–1.87) | 0.84 | 41% | ||
| TNFα_rs1800629 | ||||||||
| AA | 5 | 420 | 896 | 125 | 752 | 1.91 (0.44–8.32) | 0.39 | 94% |
| GA | 5 | 316 | 292 | 0.87 (0.58–1.3) | 0.50 | 61% | ||
| GG | 5 | 160 | 335 | 0.36 (0.07–1.75) | 0.20 | 95% | ||
Meta-analyses with p < 0.005 are in bold
Fig. 3.
Forest plot on ACE association with COVID-19 severity
A significant association between ACE1 polymorphism with an increased risk to develop severe disease was observed in dominant (OR 1.50), homozygous (OR 1.53) and additive (OR 1.4) models (Table 4, Supplemental Figure I), while there was not association in recessive model (Table 4).
Table 4.
Meta-analyses on severity considering different genetic models
| Gene | N studies | N participants | OR (95% CI) | p | I2 |
|---|---|---|---|---|---|
| ACE1_rs4646994_rs1799752 | |||||
| Dominant (DD vs II + DI) | 13 | 3042 | 1.50 (1.10–2.06) | 0.01 | 67% |
| Recessive (DD + ID vs II) | 13 | 3105 | 1.31 (0.94–1.81) | 0.11 | 59% |
| Homozygous (DD vs II) | 13 | 1720 | 1.53 (1.23–1.9) | 0.0002 | 63% |
| Additive () | 13 | 2275 | 1.41 (1.02–1.94) | 0.04 | 62% |
| ACE2_rs2285666 | |||||
| Dominant (GG + GA vs AA) | 3 | 454 | 2.24 (0.90- 5.61) | 0.08 | 0% |
| Recessive (GG vs GA + AA) | 3 | 454 | 2.18 (1.28- 3.72) | 0.04 | 16% |
| Homozygous (GG vs AA) | 3 | 370 | 2.52 (1.00—6.33) | 0.05 | 0% |
| Additive (GG vs GA) | 3 | 418 | 2.03 (1.10- 3.76) | 0.02 | 0% |
| IFTM3_rs12252 | |||||
| Dominant (CC + CT vs TT) | 3 | 630 | 1.14 (0.68- 1.94) | 0.61 | 0% |
| Recessive (CC vs CT + TT) | 3 | 630 | 2.27 (0.98- 5.25) | 0.05 | 0% |
| Homozygous (CC vs TT) | 3 | 539 | 1.60 (0.58- 4.42) | 0.37 | 0% |
| Additive (CC vs CT) | 3 | 124 | 2.60 (1.04- 6.52) | 0.04 | 0% |
| TMPRSS2_rs12329760 | |||||
| Dominant (CC + CT vs TTI) | 4 | 2492 | 0.72 (0.04- 12.18) | 0.82 | 96% |
| Recessive (CC vs CT + TT) | 4 | 1132 | 0.92 (0.44- 1.89) | 0.82 | 83% |
| Homozygous (CC vs TT) | 4 | 659 | 1.08 (0.33- 3.47) | 0.90 | 73% |
| Additive (CC vs CT) | 4 | 986 | 0.89 (0.44- 1.80) | 0.74 | 80% |
| TNFα_rs1800629 | |||||
| Dominant (AA + AG vs GG) | 4 | 748 | 1.17 (0.85–1.60) | 0.34 | 38% |
| Recessive (AA vs AG + GG) | 4 | 440 | 1.26 (0.82–1.92) | 0.29 | 0% |
| Homozygous (AA vs GG) | 4 | 428 | 1.14 (0.96–1.36) | 0.14 | 0% |
| Additive (AA vs GA) | 4 | 445 | 1.13 (0.72–1.77) | 0.61 | 0% |
The association between COVID-19 severity and ACE2 rs2285666 polymorphism was evaluated in four studies enrolling 480 patients with severe disease. Meta-analysis showed that this polymorphism was not associated with an increased risk to develop severe disease respect to patients with not severe disease (Table 3). Likewise, this polymorphism was not associated with an increased risk to develop severe disease in any genetic model, but meta-analyses showed high heterogeneity among included studies. After the exclusion of the Martinez-Gomez study [29] in the sensitivity analysis, meta-analyses showed a significant association in recessive, homozygous and additive models without heterogeneity among included studies (Table 4, Supplemental Figure II).
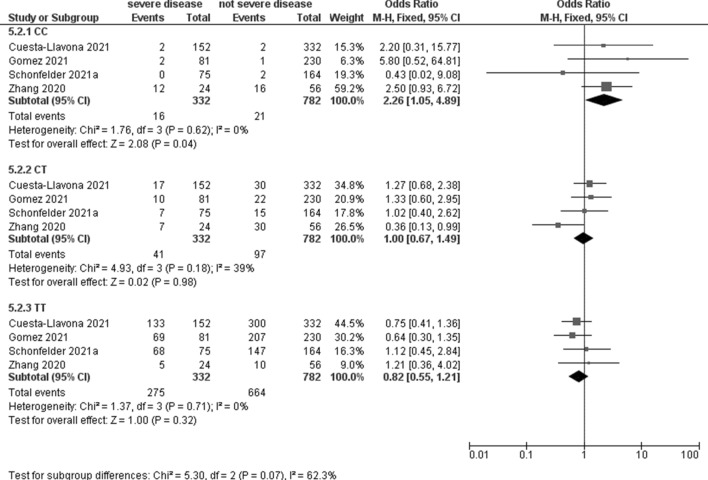
Four studies, including 332 patients with severe disease and 782 with not severe disease, evaluated the association between IFTM3 rs12252 and COVID-19 severity with a significant association for the C recessive model (OR 2.26, 95% CI 1.05–4.89; p = 0.04; I2 = 0%, Table 3, Fig. 4). No significant association was observed under any genetic model (Table 4).
Fig. 4.
Forest plot on IFTM3 rs12252 association with COVID-19 severity
The association between COVID-19 severity and TMPRSS2 rs12329760 polymorphism was evaluated in four studies enrolling 384 patients with severe disease. Meta-analysis showed that this polymorphism was not associated with an increased risk to develop severe disease respect to patients with not severe disease (Table 3). Likewise, no association between polymorphism and a higher risk to develop severe disease was observed under any genetic model (Table 4).
The association between COVID-19 severity and TNFα rs1800629 polymorphism was evaluated in five studies enrolling 896 patients with severe disease. Meta-analysis showed that this polymorphism was not associated with an increased risk to develop severe disease respect to patients with not severe disease (Table 3). Likewise, no association between polymorphism and a higher risk to develop severe disease was observed under any genetic model, even after the exclusion of the Saleh study [11] in the sensitivity analysis (Table 4).
Mortality
The association with death was analysed in three studies for ACE1 [25, 30, 31] including 98 patients who died and 406 survivors, three for IFITM3 [35, 36, 39] including 121 subjects who died and 991 survivors, two for TMPRSS2 [42, 44] including 30 subjects who died and 290 survivors, and three for TNFα [11, 47, 48] including 112 subjects who died and 1380 survivors.
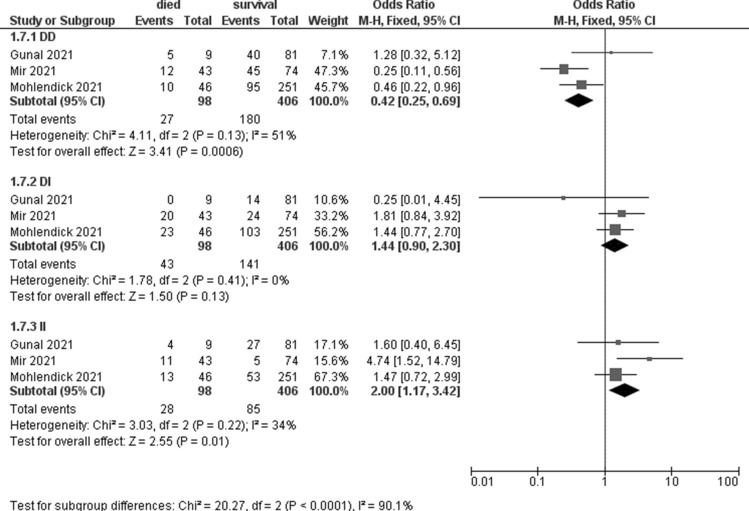
Meta-analyses showed that the ACE 1 II genotype seem to be associated with an increased risk of death (OR 2; 95% CI 1.17–3.42, p = 0.01, I2 = 34%, Table 5, Fig. 5). No significant association was observed for TMPRSS2 and TNFα (Table 5).
Table 5.
Meta-analyses on mortality considering different genotypes
| Gene | N studies | Died | Survivors | OR (95% CI) | P | I2 | ||
|---|---|---|---|---|---|---|---|---|
| Events | Total | Events | Total | |||||
| ACE1_rs4646994_rs1799752 | ||||||||
| DD | 3 | 27 | 98 | 180 | 406 | 0.42 (0.25–0.69) | 0.0006 | 51% |
| DI | 3 | 43 | 141 | 1.44 (0.90–2.30) | 0.13 | 0% | ||
| II | 3 | 28 | 85 | 2.00 (1.17–3.42) | 0.01 | 34% | ||
| ACE2_rs2285666 | ||||||||
| GG | 1 | 41 | 46 | 189 | 251 | 2.69 (1.02–7.11) | 0.05 | n.a |
| AG | 1 | 4 | 36 | 0.57 (0.19–1.68) | 0.31 | n.a | ||
| AA | 1 | 1 | 26 | 0.19 (0.03–1.45) | 0.11 | n.a | ||
| IFTM3_rs12252 | ||||||||
| CC | 3 | 3 | 121 | 31 | 991 | 2.52 (0.59–10.84) | 0.21 | 0% |
| CT | 3 | 27 | 183 | 1.58 (0.99–2.54) | 0.06 | 0% | ||
| TT | 3 | 88 | 762 | 0.58 (0.37–0.91) | 0.02 | 0% | ||
| TMPRSS2_rs12329760 | ||||||||
| CC | 2 | 3 | 30 | 74 | 290 | 0.30 (0.08–1.07) | 0.06 | 0% |
| CT | 2 | 19 | 128 | 2.29 (1.02–5.16) | 0.04 | 25% | ||
| TT | 2 | 7 | 79 | 0.81 (0.34–1.96) | 0.65 | 37% | ||
| TNFα_rs1800629 | ||||||||
| AA | 3 | 70 | 112 | 533 | 1380 | 2.4 (0.08–70.13) | 0.61 | 93% |
| GA | 3 | 25 | 539 | 0.39 (0.06–2.43) | 0.31 | 87% | ||
| GG | 3 | 10 | 308 | 0.35 (0.05–2.75) | 0.32 | 81% | ||
n.a. not applicable
Fig. 5.
Forest plot on ACE1 association with mortality
Discussion
This systematic review with meta-analysis includes all relevant studies providing evidence about the association of genetic variation in some genes of interest and SARS-CoV-2 infection susceptibility or risk to develop severe COVID-19.
Selected genes included ACE1, ACE2, IFITM3, TMPRSS2 and TNFα based on their involvement in SARS-CoV-2 tropism to the human cells. Several studies have found that SARS-CoV-2, to enter into host cells, utilizes ACE2 to attach the receptor-binding domain (RBD) and TMPRSS2 to cleave the spike (S) protein and also helps the virus escape the immune system [49]. Hence, genetic variations among some molecules responsible for cellular entry might alter the observed responses to virus infection among different individuals [1]. Given the involvement of these proteins in the entry of SARS-CoV-2 into host cells, as well as host-immune response to the virus, the relationship with disease severity may be due to single-nucleotide polymorphisms (SNPs) in the corresponding genes.
For ACE1, ACE2, TMPRSS2 and TNFα, our meta-analysis showed no significant association in test positive respect to negative subjects. For IFITM3 was a higher susceptibility for patients with C allele. Although the evaluated SNPs have been reportedly associated with viral pathogenesis, the results on host susceptibility indicated no connections between genetic polymorphisms of those genes and COVID‐19 susceptibility, probably due to limited availability of studies. Furthermore, it is important to consider that numerous factors could influence vulnerability of a population to SARS‐CoV‐2 infection, such as age, gender, ethnicity, and co‐morbidities, in addition to genetic factors, and these factors are not considered in our work. [50–52]
Furthermore, our results showed that ACE1 DD and IFITM3 CC polymorphisms could lead to a genetic predisposition for severe lung injury in patients infected by SARS-CoV-2. Notably, a significant association between ACE1 polymorphism and a higher risk to develop severe disease was observed for dominant, homozygous and additive models. Accordingly, also for ACE2 polymorphism, our meta-analyses, after the sensitivity analysis, showed a significant association with developing severe disease for recessive, homozygous and additive models. The inclusion of Martinez-Gomez study in the meta-analysis reversed results and increased heterogeneity from 0 to 97%. Patients with IFITM3 CC genotype presented higher risk to develop severe COVID-19. Finally, meta-analyses showed that the ACE 1 II genotype seem to be associated with an increased risk of death, instead no significant association was observed for TMPRSS2 and TNFα.
Despite this systematic review with meta-analysis contributes to our current understanding of host genetic susceptibility to SARS-CoV-2 infection, the following limitation should be considered. First, small number of studies was included, reducing the statistical power of the analysis. Second, included studies enrolled patients came from Europe and Asia, limiting our conclusions to a narrow ethnic group. Thirty, the analysis not considered co-founding factors, including age, gender and comorbidity that may influence the infection prognosis.
The course of SARS-CoV-2 infection can differ greatly among individuals, ranging from asymptomatic to severe disease and death. The factors that underlying these clinical manifestations are still under debate. Several studies showed that multiple viral factors such as the number of viral particles and mutations in the virus genome can influence the disease severity [53]. However, our meta-analyses showed that the genetic background of the host could influence the severity of the infection and disease outcome. Similarly, host factors such as race, age, gender, immune status, diabetes, hypertension, cardiovascular disease, chronic respiratory disease or cancer, might influence the symptoms and outcome of disease. Unfortunately, the studies included in our work not reported these information. Other studies are needed to confirm their role on susceptibility and severity of SARS-CoV-2 infection. Finding the factors that affect the virulence of SARS-CoV-2 will contribute to the development of appropriate treatment strategies and better infection control.
As in our review, studies included in previous works contributed with few data about severity. In fact, our results are aligned with finding reported by de Araújo [54] about ACE1 association with COVID-19 severity. In addition, our meta-analysis showed a higher risk to develop severe disease for patients with ACE1 dominant genotype, as reported in a previous systematic review [55].
In virology, is well known that the host genetic background plays a pivotal role in determining susceptibility to viral infections. The genetic characteristics of the host influence the recognition of viral particles, presentation of viral peptides to the host-immune system and neutralization of the viral infection. Likewise, host genetic variants of genes and innate immunity might alter susceptibility, and prognosis of COVID-19. Evidences suggest that genetic variants contribute to individual variability in human immunity, and these may affect innate and adaptive responses to SARS-CoV-2 infection.
As has been described for the other coronaviruses, host genetic variation may influence the susceptibility, severity, and overall clinical outcomes of COVID-19. Due to the emergence of the SARS‐CoV‐2, few studies evaluating the genetic characteristics of the host cell on susceptibility to the COVID‐19 has been conducted.
To identify genetic determinants of COVID-19 susceptibility, severity, and outcomes, an international COVID-19 host genetics initiative (https://www.covid19hg.org/) has been launched. This project aims to analyse genetic information for millions of individuals in order to identify genetic variants associated with SARS-CoV-2 infection as well as COVID-19 hospitalization and disease severity. Recently published meta-analyses conducted whiting the COVID-19 HGI project, identified 13 genomic loci that are significantly associated with SARS-CoV-2 infection and/or COVID-19 severity confirming that this disease has a strong underlying genetic component [56].
We now know that some host cell molecule, such as ACE and TMPRSS2, are used by SARS-CoV-2 for cell entry and spike protein cleavage, and their polymorphisms gave an impact on COVID-19 susceptibility. In addition, also individual biological characteristics such as ethnicity, age and gender, carry specificity variants of genes directly involved in viral infection, and differential expression of these genes may have different susceptibility to COVID-19, which may explain the broad spectrum of symptoms and severity of disease. Currently, the physiological basis of this heterogeneous predisposition is unknown, and population studies integrating analysis of genetic variant and immunogenetic are need to understand the inter-individual variability of COVID-19 severity [57].
Furthermore, understanding the interaction between SARS-CoV-2 and host antiviral defence mechanism could be fundamental to create effective vaccines. But many questions about the genetic variants and immunity mechanisms remain without answer. Main gaps which should be filled to fully understand the disease prevention, pathogenesis and treatment are about: [1] the role of host genes (such as ACE and TMPRSS2) on SARS-CoV-2 infection; (2) the association between individual characteristics (ethnicity, gender, and age) and clinical outcomes; (3) the effects of viral mutations and recombination on infectivity, transmissibility and disease severity in consideration of host factors which influence host gene expression.
In conclusion, genetic polymorphism of ACE1 and IFITM3 is associated with higher risk of severe COVID-19, but further studies considering ethnicity and comorbidities of patients are need to corroborate our results.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contributions
All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission. VP conceived and designed the systematic review. VP designed and implemented the search strategy. VP and MC extracted data and performed the quality assessment of included studies. VP analysed data. VP wrote the paper. All authors were involved in the critical revision of the intellectual content of the manuscript.
Funding
The authors received no financial support for the research, authorship, and/or publication of this article.
Declarations
Conflict of interests
The authors declared no potential conflict of interest with respect to the research, authorship, and/or publication of this article.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Valentina Pecoraro, Email: v.pecoraro@ausl.mo.it.
Michela Cuccorese, Email: m.cuccorese@ausl.mo.it.
Tommaso Trenti, Email: t.trenti@ausl.mo.it.
References
- 1.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wan Y, Shang J, Graham R, et al. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J Virol. 2020;94:e00127–e220. doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rigat B, Hubert C, Alhencgelas F, et al. An insertion deletion polymorphismin the angiotensin I-converting enzyme gene accounting for half the variance of serum enzyme levels. J Clin Invest. 1990;86:1343–1346. doi: 10.1172/JCI114844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zheng H, Cao JJ. Angiotensin-converting enzyme gene polymorphism and severe lung injury in patients with coronavirus disease 2019. Am J Pathol. 2020;190:2013–2017. doi: 10.1016/j.ajpath.2020.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemmati D, Tisato V. Genetic hypothesis and pharmacogenetics side of renin-angiotensin-system in COVID-19. Genes. 2020;11:1044. doi: 10.3390/genes11091044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gomez J, Albaiceta GM, García-Clemente M, et al. Angiotensin-converting enzymes (ACE, ACE2) gene variants and COVID-19 outcome. Gene. 2020;762:145102. doi: 10.1016/j.gene.2020.145102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pozzi G, Masselli E, Gobbi G, Mirandola P, et al. Hydrogen sulfide inhibits TMPRSS2 in human airway epithelial cells: Implications for SARS-CoV-2 infection. Biomedicines. 2021;9(9):1273. doi: 10.3390/biomedicines9091273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hou Y, Zhao J, Martin W, et al. New insights into genetic susceptibility of COVID-19: an ACE2 and TMPRSS2 polymorphism analysis. BMC Med. 2020;18(1):1–8. doi: 10.1186/s12916-020-01673-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bailey CC, Zhong G, Huang IC, Farzan M. IFITM-family proteins: the cell's first line of antiviral defense. Annu Rev Virol. 2014;1:261–283. doi: 10.1146/annurev-virology-031413-085537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao X, Li J, Winkler CA, et al. IFITM genes, variants, and their roles in the control and pathogenesis of viral infections. Front Microbiol. 2019;9:3228. doi: 10.3389/fmicb.2018.03228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saleh A, Sultan A, Elashry MA, et al. Association of TNF-α G-308 a promoter polymorphism with the course and outcome of COVID-19 patients. Immunol Invest. 2022;51(3):546–557. doi: 10.1080/08820139.2020.1851709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liberati A, Altman DG, Tetzlaff J, et al. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;21(339):b2700. doi: 10.1136/bmj.b2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wells GA, Shea B, O'Connell D, et al. The Newcastle-Ottawa Scale (NOS) for Assessing the Quality of Nonrandomised Studies in Meta-analyses. Ottawa: Ottawa Hospital Research Institute. Available at http://www.ohri.ca/programs/clinical_epidemiology/oxford.asp. Accessed 22 Aug 2022
- 14.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 15.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 16.Akbari M, Taheri M, Mehrpoor G, et al. Assessment of ACE1 variants and ACE1/ACE2 expression in COVID-19 patients. Vascul Pharmacol. 2022;142:106934. doi: 10.1016/j.vph.2021.106934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aladag E, Tas Z, Ozdemir BS, et al. Human ace D/I polymorphism could affect the clinicobiological course of COVID-19. J Renin Angiotensin Aldosterone Syst. 2021;2021:5509280. doi: 10.1155/2021/5509280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alimoradi N, Sharqi M, Firouzabadi D, et al. SNPs of ACE1 (rs4343) and ACE2 (rs2285666) genes are linked to SARS-CoV-2 infection but not with the severity of disease. Virol J. 2022;19(1):48. doi: 10.1186/s12985-022-01782-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Annunziata A, Coppola A, Di Spirito V, et al. The angiotensin converting enzyme deletion/deletion genotype is a risk factor for severe COVID-19: implication and utility for patients admitted to emergency department. Medicina (Kaunas) 2021;57(8):844. doi: 10.3390/medicina57080844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baştuğ S, Çavdarlı B, Baştuğ A, et al. Are angiotensin converting enzyme (ACE1/ACE2) gene variants associated with the clinical severity of COVID-19 pneumonia? A single-center cohort study. Anatol J Cardiol. 2022;26(2):133–140. doi: 10.5152/AnatolJCardiol.2021.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cafiero C, Rosapepe F, Palmirotta R, et al. Angiotensin system polymorphisms' in SARS-CoV-2 positive patients: assessment between symptomatic and asymptomatic patients: a pilot study. Pharmgenomics Pers Med. 2021;14:621–629. doi: 10.2147/PGPM.S303666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calabrese C, Annunziata A, Coppola A, et al. ACE gene I/D polymorphism and acute pulmonary embolism in COVID19 pneumonia: a potential predisposing role. Front Med (Lausanne) 2021;7:631148. doi: 10.3389/fmed.2020.631148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Çelik KS, Çakmak Genç G, Pişkin N, et al. Polymorphisms of ACE (I/D) and ACE2 receptor gene (Rs2106809, Rs2285666) are not related to the clinical course of COVID-19: a case study. J Med Virol. 2021;93(10):5947–5952. doi: 10.1002/jmv.27160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong P, Mei F, Li R, et al. Angiotensin-converting enzyme genotype-specific immune response contributes to the susceptibility of COVID-19: a nested case-control study. Front Pharmacol. 2022;12:759587. doi: 10.3389/fphar.2021.759587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunal O, Sezer O, Ustun GU, et al. Angiotensin-converting enzyme-1 gene insertion/deletion polymorphism may be associated with COVID-19 clinical severity: a prospective cohort study. Ann Saudi Med. 2021;41(3):141–146. doi: 10.5144/0256-4947.2021.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hubacek JA, Dusek L, Majek O, et al. ACE I/D polymorphism in Czech first-wave SARS-CoV-2-positive survivors. Clin Chim Acta. 2021;519:206–209. doi: 10.1016/j.cca.2021.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouhpayeh HR, Tabasi F, Dehvari M, et al. Association between angiotensinogen (AGT), angiotensin-converting enzyme (ACE) and angiotensin-II receptor 1 (AGTR1) polymorphisms and COVID-19 infection in the southeast of Iran: a preliminary case-control study. Transl Med Commun. 2021;6(1):26. doi: 10.1186/s41231-021-00106-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahmood ZS, Fadhil HY, Abdul Hussein TA, Ad'hiah AH. Severity of coronavirus disease 19: profile of inflammatory markers and ACE (rs4646994) and ACE2 (rs2285666) gene polymorphisms in Iraqi patients. Meta Gene. 2022;31:101014. doi: 10.1016/j.mgene.2022.101014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martínez-Gómez LE, Herrera-López B, Martinez-Armenta C, et al. ACE and ACE2 gene variants are associated with severe outcomes of COVID-19 in men. Front Immunol. 2022;13:812940. doi: 10.3389/fimmu.2022.812940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mir MM, Mir R, Alghamdi MAA, et al. Strong association of angiotensin converting enzyme-2 gene insertion/deletion polymorphism with susceptibility to SARS-CoV-2, Hypertension, coronary artery disease and COVID-19 disease mortality. J Pers Med. 2021;11(11):1098. doi: 10.3390/jpm11111098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Möhlendick B, Schönfelder K, Breuckmann K, et al. ACE2 polymorphism and susceptibility for SARS-CoV-2 infection and severity of COVID-19. Pharmacogenet Genomics. 2021;31(8):165–171. doi: 10.1097/FPC.0000000000000436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Papadopoulou A, Fragkou PC, Maratou E, et al. Angiotensin-converting-enzyme insertion/deletion polymorphism, ACE activity, and COVID-19: a rather controversial hypothesis. A case-control study. J Med Virol. 2022;94(3):1050–1059. doi: 10.1002/jmv.27417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saad H, Jabotian K, Sakr C, et al. The role of angiotensin converting enzyme 1 insertion/deletion genetic polymorphism in the risk and severity of COVID-19 infection. Front Med (Lausanne) 2021;8:798571. doi: 10.3389/fmed.2021.798571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verma S, Abbas M, Verma S, et al. Impact of I/D polymorphism of angiotensin-converting enzyme 1 (ACE1) gene on the severity of COVID-19 patients. Infect Genet Evol. 2021;91:104801. doi: 10.1016/j.meegid.2021.104801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alghamdi J, Alaamery M, Barhoumi T, et al. Interferon-induced transmembrane protein-3 genetic variant rs12252 is associated with COVID-19 mortality. Genomics. 2021;113(4):1733–1741. doi: 10.1016/j.ygeno.2021.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cuesta-Llavona E, Albaiceta GM, García-Clemente M, et al. Association between the interferon-induced transmembrane protein 3 gene (IFITM3) rs34481144/rs12252 haplotypes and COVID-19. Curr Res Virol Sci. 2021;2:100016. doi: 10.1016/j.crviro.2021.100016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gómez J, Albaiceta GM, Cuesta-Llavona E, et al. The Interferon-induced transmembrane protein 3 gene (IFITM3) rs12252 C variant is associated with COVID-19. Cytokine. 2021;137:155354. doi: 10.1016/j.cyto.2020.155354. [DOI] [PubMed] [Google Scholar]
- 38.Schönfelder K, Breuckmann K, Elsner C, et al. The influence of IFITM3 polymorphisms on susceptibility to SARS-CoV-2 infection and severity of COVID-19. Cytokine. 2021;142:155492. doi: 10.1016/j.cyto.2021.155492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang Y, Qin L, Zhao Y, et al. Interferon-induced transmembrane protein 3 genetic variant rs12252-C associated with disease severity in coronavirus disease 2019. J Infect Dis. 2020;222(1):34–37. doi: 10.1093/infdis/jiaa224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andolfo I, Russo R, Lasorsa VA, et al. Common variants at 21q22.3 locus influence MX1 and TMPRSS2 gene expression and susceptibility to severe COVID-19. Iscience. 2021;24(4):102322. doi: 10.1016/j.isci.2021.102322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravikanth V, Sasikala M, Naveen V, et al. A variant in TMPRSS2 is associated with decreased disease severity in COVID-19. Meta Gene. 2021;29:100930. doi: 10.1016/j.mgene.2021.100930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rokni M, Heidari Nia M, Sarhadi M, et al. Association of TMPRSS2 gene polymorphisms with COVID-19 severity and mortality: a case-control study with computational analyses. Appl Biochem Biotechnol. 2022;194(8):3507–3526. doi: 10.1007/s12010-022-03885-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schönfelder K, Breuckmann K, Elsner C, et al. Transmembrane serine protease 2 polymorphisms and Susceptibility to severe acute respiratory syndrome coronavirus type 2 infection: a German case-control study. Front Genet. 2021;12:667231. doi: 10.3389/fgene.2021.667231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wulandari L, Hamidah B, Pakpahan C, et al. Initial study on TMPRSS2 p.Val160Met genetic variant in COVID-19 patients. Hum Genomics. 2021;15(1):29. doi: 10.1186/s40246-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ali HN, Niranji SS, Al-Jaf SMA. Association of tumor necrosis factor alpha -308 single nucleotide polymorphism with SARS CoV-2 infection in an Iraqi Kurdish population. J Clin Lab Anal. 2022;36(5):e24400. doi: 10.1002/jcla.24400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fishchuk L, Rossokha Z, Pokhylko V, et al. Modifying effects of TNF-α, IL-6 and VDR genes on the development risk and the course of COVID-19. Pilot study. Drug Metab Pers Ther. 2021;37(2):133–139. doi: 10.1515/dmpt-2021-0127. [DOI] [PubMed] [Google Scholar]
- 47.Heidari Nia M, Rokni M, Mirinejad S, et al. Association of polymorphisms in tumor necrosis factors with SARS-CoV-2 infection and mortality rate: a case-control study and in silico analyses. J Med Virol. 2022;94(4):1502–1512. doi: 10.1002/jmv.27477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rokni M, Sarhadi M, Heidari Nia M, et al. Single nucleotide polymorphisms located in TNFA, IL1RN, IL6R, and IL6 genes are associated with COVID-19 risk and severity in an Iranian population. Cell Biol Int. 2022;46(7):1109–1127. doi: 10.1002/cbin.11807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shang J, Wan Y, Luo C, et al. Cell entry mechanisms of SARS-CoV-2. Proc Natl Acad Sci. 2020;117(21):11727. doi: 10.1073/pnas.2003138117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benetti E, Tita R, Spiga O, et al. ACE2 gene variants may underlie interindividual variability and susceptibility to COVID-19 in the Italian population. Eur J Hum Genet. 2020;28(11):1602–1614. doi: 10.1038/s41431-020-0691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Novelli A, Biancolella M, Borgiani P, et al. Analysis of ACE2 genetic variants in 131 Italian SARS-CoV-2-positive patients. Hum Genomics. 2020;14:10–15. doi: 10.1186/s40246-020-00279-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Torre-Fuentes L, Matías-Guiu J, Hern´andez-Lorenzo L, et al. ACE2, TMPRSS2, and Furin variants and SARS-CoV-2 infection in Madrid, Spain. J Med Virol. 2020 doi: 10.1002/jmv.26319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.de Araújo JLF, Menezes D, de Aguiar RS, de Souza RP. IFITM3, FURIN, ACE1, and TNF-α genetic association with COVID-19 outcomes: systematic review and meta-analysis. Front Genet. 2022;13:775246. doi: 10.3389/fgene.2022.775246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saengsiwaritt W, Jittikoon J, Chaikledkaew U, Udomsinprasert W. Genetic polymorphisms of ACE1, ACE2, and TMPRSS2 associated with COVID-19 severity: a systematic review with meta-analysis. Rev Med Virol. 2022;32(4):e2323. doi: 10.1002/rmv.2323. [DOI] [PubMed] [Google Scholar]
- 55.Debnath M, Banerjee M, Berk M. Genetic gateways to COVID-19 infection: implications for risk, severity, and outcomes. FASEB J. 2020;34(7):8787–8795. doi: 10.1096/fj.202001115R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ovsyannikova IG, Haralambieva IH, Crooke SN, et al. The role of host genetics in the immune response to SARS-CoV-2 and COVID-19 susceptibility and severity. Immunol Rev. 2020;296(1):205–219. doi: 10.1111/imr.12897. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.