Abstract
Background: The Environmental Determinants of Islet Autoimmunity (ENDIA) pregnancy-birth cohort investigates the developmental origins of type 1 diabetes (T1D), with recruitment between 2013 and 2019. ENDIA is the first study in the world with comprehensive data and biospecimen collection during pregnancy, at birth and through childhood from at-risk children who have a first-degree relative with T1D. Environmental exposures are thought to drive the progression to clinical T1D, with pancreatic islet autoimmunity (IA) developing in genetically susceptible individuals. The exposures and key molecular mechanisms driving this progression are unknown. Persistent IA is the primary outcome of ENDIA; defined as a positive antibody for at least one of IAA, GAD, ZnT8 or IA2 on two consecutive occasions and signifies high risk of clinical T1D.
Method: A nested case-control (NCC) study design with 54 cases and 161 matched controls aims to investigate associations between persistent IA and longitudinal omics exposures in ENDIA. The NCC study will analyse samples obtained from ENDIA children who have either developed persistent IA or progressed to clinical T1D (cases) and matched control children at risk of developing persistent IA. Control children were matched on sex and age, with all four autoantibodies absent within a defined window of the case’s onset date. Cases seroconverted at a median of 1.37 years (IQR 0.95, 2.56). Longitudinal omics data generated from approximately 16,000 samples of different biospecimen types, will enable evaluation of changes from pregnancy through childhood.
Conclusions: This paper describes the ENDIA NCC study, omics platform design considerations and planned univariate and multivariate analyses for its longitudinal data. Methodologies for multivariate omics analysis with longitudinal data are discovery-focused and data driven. There is currently no single multivariate method tailored specifically for the longitudinal omics data that the ENDIA NCC study will generate and therefore omics analysis results will require either cross validation or independent validation.
KEY MESSAGES
The ENDIA nested case-control study will utilize longitudinal omics data on approximately 16,000 samples from 190 unique children at risk of type 1 diabetes (T1D), including 54 who have developed islet autoimmunity (IA), followed during pregnancy, at birth and during early childhood, enabling the developmental origins of T1D to be explored.
Introduction
Background and rationale
The clinical presentation of type 1 diabetes (T1D) follows an extended period of months to years during which the immune system attacks and destroys insulin-producing cells in the islets of the pancreas. The presence of autoantibodies targeting beta cell antigens, referred to as islet autoimmunity (IA), signals a high risk of clinical T1D.
The Environmental Determinants of Islet Autoimmunity (ENDIA) cohort study is the first and largest study in the world with meta-data and prospective sample collection from women commencing in early pregnancy, at the delivery and immediate post-partum period and from at-risk children from birth and continuing throughout childhood [1]. ENDIA addresses the hypothesis that modifiable environmental exposures in the prenatal and early postnatal period increase the penetrance of T1D risk genes and drive the development of IA progressing to clinical T1D in children. As the onset of islet autoimmunity at a young age [2], predicts progression to T1D, there is high interest in the gestational environment exposures and maternal – child transmission at birth. ENDIA is positioned amongst other large at-risk cohorts [3–6] that have recruited from three months onwards after birth, to provide a unique perspective on the significance of early-life antecedent factors in childhood T1D, by investigating exposures during pregnancy and the perinatal period in accordance with the Developmental Origins of Health and Disease (DOHaD) [7] paradigm.
Many of the environmental exposures investigated in ENDIA will involve characterization of various omes, that is, the totality of certain classes of biological molecules or analytes in a range of samples and cell types and their association with the development of IA. Omics studies take a holistic approach to the identification and quantification of analytes that characterize the structure and function of biological systems. Omics technologies hold great potential for elucidating disease mechanisms and identifying new early-stage biomarkers of risk. While several international cohorts have reported on omics outcomes [8–10], the ENDIA NCC has the capacity to track the expression of analytes, and their precursors, across the antenatal and postnatal environments.
Here we describe a case-control study design nested within the ENDIA cohort study, also known as a nested case-control (NCC) study design [11], that will enable elucidation of how omics change over time and relate to progression to persistent IA. A NCC study design is a retrospective observational design that includes all individuals who have experienced an outcome of interest (known as cases) and matches them using incidence density sampling [12] to individuals at-risk at the time of the case event, but who have not yet experienced the defined event (known as controls) [13]. The ENDIA NCC study was planned when the number of children who developed persistent IA (the primary outcome) reached approximately half the projected number of seroconverters in the total cohort. The NCC study analysis of omics data represents an interim analysis within the context of the long-term ENDIA cohort study.
NCC studies with correctly matched representative controls can yield efficient and unbiased estimates of the odds ratio that would be obtained from a full cohort analysis. Their benefits include savings in time and cost, particularly with regard to omic investigations that may be infeasible in the entire cohort [14–16]. Using high-throughput platforms, the ENDIA study will comprehensively generate omics data from samples collected during pregnancy and from children since birth. Omics investigations undertaken in the ENDIA study will characterize the epigenome for modifications of gene activity, transcriptome for gene expression, proteome for proteins, lipidome for lipids, glycome for glycans and glycoconjugates, virome for viruses, metabolome for small molecules and microbiome metagenome for bacteria and fungi.
Profiling individuals across different omics platforms and sample types in the ENDIA NCC study will compare hundreds to thousands of analytes in parallel, allowing the identification of those analytes that underlie and interact in the development of persistent IA and thus contribute to T1D pathogenesis. This discovery-focused, hypothesis generating approach will complement targeted analyses and will be refined in validation studies conducted on the completed ENDIA cohort and other international cohorts.
In addition, because ENDIA has prospectively collected longitudinal samples it provides an opportunity to explore how omics analytes change over time, how they interact at different stages of the life-course and how this contributes to the development of persistent IA.
Objectives and research question
The purpose of this paper is to: (1) describe the selection of participants in the ENDIA NCC design; (2) describe baseline characteristics of the NCC cohort; (3) discuss the design of omics studies within the NCC; (4) discuss issues and potential solutions when using the NCC design for single time and longitudinal omics data and (5) outline principles for the statistical analysis to be used for single time and longitudinal omics data when generated in accordance with the NCC design.
Hypotheses and aims
The main hypotheses associated with omics investigations utilizing the ENDIA NCC design are that exposures to environmental factors result in perturbations in the omes and are associated with the outcome of persistent IA, reflecting critical aspects of biology that underpin T1D risk.
The aim is to identify any differences in analytes between cases and controls at the time of development of persistent IA in the child, across the pregnancy, birth and early childhood time trajectory. Individual analytes will be considered independently as well as looking at interactions between analytes.
Methods
ENDIA study
Recruitment of 1214 women during pregnancy or early postpartum to the Australia-wide ENDIA study [1], commenced in February 2013 and concluded in December 2019 [17]. Informed written consent was obtained from study participants. The eligibility criteria were unborn children, or children less than six months of age, who had at least one first-degree relative (mother, father or sibling) with T1D (known as a proband). Children are followed every three months until aged 2 years, then six-monthly thereafter until aged 10 years.
The primary outcome of ENDIA is the development of persistent IA defined as positivity to one or more islet autoantibodies on consecutive tests at least three months apart (i.e. the case definition). Autoantibodies to insulin (IAA), glutamic acid decarboxylase (GAD), insulinoma antigen-2 (IA2) and the Zinc transporter 8 (ZnT8) were measured.
Rationale and participants for the NCC study
The ENDIA study commenced with intention to recruit 1400 eligible mother-child dyads. By the end of 2017, 813 ENDIA children had been born with 14 children having seroconverted to persistent IA. Based on those data, the future seroconversion rate was simulated (Supplementary Material A). Given the estimate that close to 100 ENDIA children would develop persistent IA by the end of 2024, it was determined that the NCC study would commence at the end of 2019 when approximately half the projected number of cases (i.e. 50) had emerged. ENDIA children developing persistent IA from 1 January 2020 will eventually form a ‘validation’ set for discoveries from the NCC study.
There were 1307 children eligible for this NCC study, who were born before 30 September 2019 (inclusive) and eligible for their first islet autoantibody testing by the cut-off date of 31 December 2019 (inclusive).
Power considerations
Odds ratio (OR) estimations were based on 50 children and 150 matched controls. Given a Type I error of 0.05, power of 0.8 and a two-sided test, the detectable odds ratio can be calculated [18] for the ENDIA NCC study. Assumptions for the correlation (φ) between matched case and control children and the probability of exposure of (p0) to a particular omics analyte amongst the control children, are required for OR estimation. Dupont [18] suggests when no estimate of the correlation is available, a value of 0.2 can be used. ORs have been calculated for the ENDIA NCC study assuming that the correlation can vary between 0 and 0.3 and the probability of exposure is between 0.01 and 0.3 (Supplementary Material A, Table A3). The function epi.sscc from the R software [19] package epiR (v 2.0.38) [20] was used for this calculation.
As expected, the detectable OR increases as the correlation increases and decreases as the probability of exposure in the controls increases. For the ENDIA NCC study, with a sample size of 50 cases plus 150 matched controls (i.e. 3 controls for each case), the study is estimated to be powered to detect an OR of 2.9 (p0=0.2 and φ = 0.2) or higher. This is a conservative estimate based on a cross-sectional analysis at a single time point; that is undertaking the primary analysis for the NCC study at the time of IA onset using a conditional logistic regression model. It does not take into consideration the increased power that would be obtained from multiple samples from the same child across time [21].
Cases
In the ENDIA NCC study, cases were defined as children who had either developed persistent IA or had progressed to clinical T1D. The development of persistent islet autoantibodies was defined as having at least one positive antibody result for any one of IAA, GAD, IA2 and ZnT8 on two consecutive occasions. To be included in the NCC study, the first positive must have occurred by 31 December 2019 (inclusive), with the second positive antibody result confirmed by 30 June 2020 (inclusive). Confirmation with a second positive result was required at least three months from the initial visit. While not recorded as such, when determining seroconversion missing samples are by default assumed not positive. It is possible that once defined, a case may serorevert, that is, have a single or persistently negative antibody results after being defined as a case.
Autoantibodies to IAA were measured by a radiobinding assay. Prior to October 2017, autoantibodies to GAD, IA2 and ZnT8 were measured by immunoprecipitation of 35S-methionine-labelled recombinant human proteins. From October 2017, autoantibodies to GAD, IA2 and ZnT8 were initially assayed by the ELISA RSR 3-screen ICA kit; samples exceeding the positive threshold were re-tested in separate ELISA RSR kits for autoantibodies to GAD, IA2 and ZnT8.
Results were expressed in arbitrary units (U) in comparison with positive and negative controls. The threshold for autoantibody positivity was IAA > =0.7 U. Prior to October 2017, autoantibody positivity was defined as GAD ≥ 5 U, IA2 ≥ 13 U and ZnT8 ≥ 3.1 U. From October 2017, autoantibody positivity was defined as ELISA RSR 3-screen ICA > 20 U, ELISA RSR GAD ≥ 5 U, IA2 ≥ 7.5 U and ZnT8 ≥ 15 U. Positive results prior to October 2017 were confirmed by repeat testing in ELISA assays. The assays had 100%, 98%, 100%, 94% specificity and 36%, 84%, 72%, 76% sensitivity for IAA, GAD, IA2 and ZnT8 autoantibodies respectively, in the 2020 Islet Autoantibody Standardization Program (University of Florida).
Once confirmed persistent, the date of the first positive was established as the onset date, or event time. For children whose autoimmune status was determined around the time of clinical diagnosis of T1D with no prior testing for persistent islet autoantibodies, the time of case onset corresponded to the earlier of either the date of the first blood draw where one or more islet autoantibodies were detected, or the diagnosis date of T1D. To be included in the NCC, clinical T1D status had to be identified by 31 December 2019 (inclusive).
There were 54 individuals who met the definition of a case. Of these, 35/54 children were positive for a single islet autoantibody and 19/54 children were positive for two or more islet autoantibodies at the time of onset. There were seven children whose autoimmune status was determined at the clinical diagnosis of type 1 diabetes (median age years 1.4, IQR 0.79, 1.93) as no prior testing for persistent antibodies had been undertaken, with 13 individuals progressing to T1D before the NCC study cut-off date. It is noteworthy that the case median age at IA seroconversion in the ENDIA NCC study was close to 17 months of age (median age years 1.37, IQR 0.95, 2.56). While the persistent IA outcome includes single IA and multiple IA, children as young as this would be expected to progress to T1D more frequently than older children with single IA [2] as reported in analyses of rates of progression according to the number of islet autoantibodies [22,23].
Controls
Children eligible to be controls were initially matched on sex and age, as these are known risk factors for T1D [24,25]. An eligible child’s date of birth had to be within 45 days (inclusive) of the case’s. Eligible children matched on sex and age, also had to have a negative autoantibody test for all four autoantibodies within a defined window of the case’s onset date. For cases aged ≤2 years, negative tests were required to be within 45 days (inclusive) of case onset and within 90 days (inclusive) of case onset for cases aged >2 years. This corresponded with the ENDIA study visit timetable of three-monthly visits until age two years and six-monthly visits thereafter. The target ratio of matching was three controls to one case.
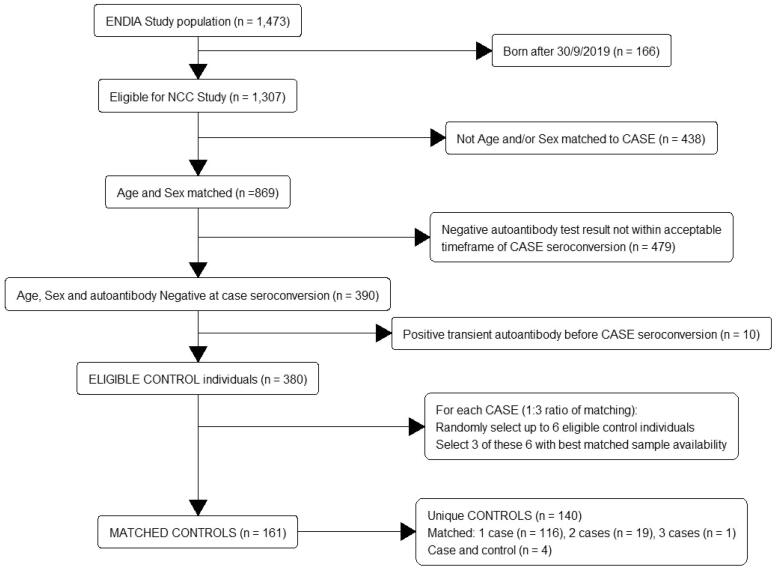
Matching of eligible controls to cases was performed (Figure 1) using incidence density sampling [12].
Figure 1.
Flow chart for inclusion of children in the nested case control study: matching of controls to cases. Note: Cases are eligible to be controls up until their date of onset of persistent IA. Fifty-three cases had three matched controls, one case had two matched controls.
Eligible controls also required consistently negative tests in all islet autoantibody tests prior to the case event time. Children who had one or more autoantibodies identified at a single visit that did not persist at the subsequent visit were defined as children with transient autoantibodies. While children with transient antibodies were excluded as eligible controls from the date of their first transient autoantibody (inclusive), they were eligible to be controls to cases if the case event time was prior to their transient autoantibody visit. This ensured that all samples from the same eligible child had negative tests, enabling longitudinal comparisons between the control and their case. Eligible controls could be matched to multiple cases as long as they were at-risk at the date of case onset and met matching criteria. Cases were eligible to be controls up until their event time.
Eligible control children with autoantibodies that had been transferred across the placenta from their mothers (i.e. not host derived), were included. Transplacental transfer was defined as a positive result at the first autoantibody test during the first year of life with regressing titres at subsequent visits until testing negative. Any child who tested positive during the first year of life after a previously negative result, or with an increasing titre relative to the previous visit, was deemed to have developed independent IA. There was a pool of 380 age, sex and autoantibody negative matched children that were eligible to be controls.
The numbers of eligible matched controls for cases ranged from two to 19. For cases with six or less eligible controls, all are initially chosen. For cases with more than six eligible controls, six were randomly selected. A hierarchical stepwise selection strategy was then applied based on optimal sample availability (i.e. a non-random selection) until three matched controls per case could be definitively selected. This strategy was based on the NCC design utilized by the TEDDY study [26] as follows:
most stool samples matched between the control and case up to and including IA onset,
largest number of stool samples overall until IA onset,
most serum samples matched between the control and case up to and including IA onset,
largest number of serum samples overall until IA onset.
Of the 54 cases, 53 had three matched eligible controls and one had only two matched controls, so that in total there were 161 matched controls. There were 190 unique children included in the ENDIA NCC study: 50 were included only as cases; 116 were controls matched to a single case; 19 were controls to two different cases; 1 was a control to three different cases and 4 were included as both controls and cases because of their time-varying outcome status (i.e. were matched as a control to a different case, who had an earlier onset date). Each case and their matched controls formed a sampled risk-set.
Matching was done with reproducible sample code written in the R software environment [19] (Supplementary Material B, C, D and E).
Notably, two of the cases of the same sex not only had a date of birth but also an event time within two days of each other. For these cases, the controls sampled at the date of onset are identical and the initial random selection had five of the six controls overlapping for these two cases. Therefore, these two cases and selected controls will form a single risk-set in the analysis. There were 15 sets of siblings in the NCC as no restriction was imposed on the inclusion of siblings. However, siblings were not from the same pregnancy (i.e. not a twin or triplet) and none were in the same risk-set. There was one individual in the NCC who was a twin, but the sibling of the twin was not chosen as a control.
Baseline participant characteristics
The characteristics of the NCC study participants are outlined in Table 1 and compared to the eligible cohort.
Table 1.
Characteristics of the NCC study participants.
| Cases | Matched controls | Eligible cohort | |
|---|---|---|---|
| (N = 54) | (N = 161)a | (N = 1307)b | |
| Child sex | |||
| Female | 28 (51.9%) | 84 (52.2%) | 633 (48.4%) |
| Male | 26 (48.1%) | 77 (47.8%) | 673 (51.5%) |
| Missing | 0 (0%) | 0 (0%) | 1 (0.1%) |
| Proband relationship | |||
| Mother | 17 (31.5%) | 87 (54.0%) | 787 (60.2%) |
| Father | 19 (35.2%) | 45 (28.0%) | 342 (26.2%) |
| Sibling | 9 (16.7%) | 21 (13.0%) | 137 (10.5%) |
| Multiple | 9 (16.7%) | 8 (5.0%) | 36 (2.8%) |
| Other | 0 (0%) | 0 (0%) | 5 (0.4%) |
| Child HLAc | |||
| DR3,4 | 17 (31.5%) | 14 (8.7%) | 140 (12.0%) |
| Not DR3,4 | 36 (66.7%) | 142 (88.2%) | 1023 (88.0%) |
| Indeterminant | 0 (0%) | 4 (2.5%) | 4 (0.3%) |
| Missing | 1 (1.9%) | 1 (0.6%) | 140 (10.7%) |
| Gestational age at birth (weeks) | |||
| Median (Q1–Q3) | 38.5 (37.2–39.4) | 38.0 (36.9–39.4) | 37.9 (36.6–38.7) |
| Missing | 0 (0%) | 0 (0%) | 4 (0.3%) |
| Mode of birth | |||
| Caesarean | 33 (61.1%) | 89 (55.3%) | 766 (58.6%) |
| Vaginal | 21 (38.9%) | 72 (44.7%) | 533 (40.8%) |
| Missing | 0 (0%) | 0 (0%) | 8 (0.6%) |
| Birth weight (grams) | |||
| Mean (SD) | 3540 (480) | 3580 (535) | 3500 (656) |
| Missing | 0 (0%) | 0 (0%) | 8 (0.6%) |
| Plurality of pregnancy | |||
| Singleton | 53 (98.1%) | 161 (100%) | 1269 (97.1%) |
| Twins | 1 (1.9%) | 0 (0%) | 35 (2.7%) |
| Triplets | 0 (0%) | 0 (0%) | 3 (0.2%) |
| Any breastfeedingc | |||
| Yes | 53 (98.1%) | 156 (96.9%) | 1206 (98.2%) |
| No | 0 (0%) | 5 (3.1%) | 22 (1.8%) |
| Missing | 1 (1.9%) | 0 (0%) | 79 (6.0%) |
| Apgar score at 5 min | |||
| Median (Q1–Q3) | 9.00 (9.00–9.00) | 9.00 (9.00–9.00) | 9.00 (9.00–9.00) |
| Missing | 0 (0%) | 0 (0%) | 14 (1.1%) |
aThere are 161 matched controls (Figure 1), includes duplicated information from individuals that are controls to more than one case and controls that later become cases.
bEligible children (Figure 1).
cFor those count variables with >5% missing, the % of Yes and No are based on those children with data (i.e. exclude missing).
Samples
Childhood samples are collected at scheduled study visits:-birth, three monthly until two years of age and six monthly thereafter. Pregnancy samples can include samples from Trimester 1, 2 and 3, depending on recruitment.
Details of total sample numbers for the different sample types, as well as the mean and median numbers of samples per individual that are available for the omics studies, during childhood and for pregnancy are shown in Tables 2 and 3, respectively.
Table 2.
Childhood (including birth) total numbers of samples, mean (standard deviation) and median (interquartile range) number of samples per child, by sample type for cases and controls of the ENDIA nested-case control study.
| Sample type | Case | Control |
|---|---|---|
| (N = 54) | (N = 161) | |
| Serum | ||
| Total samples | 210 | 863 |
| Mean (SD) | 3.89 (2.81) | 5.36 (3.15) |
| Median (Q1–Q3) | 3.00 (2.00–5.00) | 4.00 (3.00–7.00) |
| Plasma | ||
| Total samples | 189 | 794 |
| Mean (SD) | 3.50 (2.56) | 4.93 (3.03) |
| Median (Q1–Q3) | 3.00 (2.00–4.00) | 4.00 (3.00–6.00) |
| Stool | ||
| Total samples | 252 | 1000 |
| Mean (SD) | 4.67 (2.44) | 6.22 (2.84) |
| Median (Q1–Q3) | 4.00 (3.00–6.00) | 6.00 (4.00–8.00) |
| Urine | ||
| Total samples | 202 | 782 |
| Mean (SD) | 3.74 (2.84) | 4.86 (3.05) |
| Median (Q1–Q3) | 3.00 (2.00–4.00) | 4.00 (3.00–6.00) |
| Throat swab | ||
| Total samples | 265 | 1050 |
| Mean (SD) | 4.91 (2.86) | 6.53 (3.21) |
| Median (Q1–Q3) | 4.00 (3.00–6.00) | 6.00 (4.00–9.00) |
| Tongue swab | ||
| Total samples | 273 | 1070 |
| Mean (SD) | 5.06 (2.82) | 6.62 (3.21) |
| Median (Q1–Q3) | 4.00 (3.25–6.00) | 6.00 (4.00–9.00) |
| Buccal swab | ||
| Total samples | 270 | 1060 |
| Mean (SD) | 5.00 (2.87) | 6.60 (3.22) |
| Median (Q1–Q3) | 4.00 (3.25–6.00) | 6.00 (4.00–9.00) |
| Nasal swab | ||
| Total samples | 271 | 1070 |
| Mean (SD) | 5.02 (2.82) | 6.65 (3.21) |
| Median (Q1–Q3) | 4.00 (3.25–5.75) | 6.00 (4.00–9.00) |
| Skin swab | ||
| Total samples | 301 | 1180 |
| Mean (SD) | 5.57 (2.90) | 7.30 (3.24) |
| Median (Q1–Q3) | 5.00 (4.00–6.00) | 6.00 (5.00–10.0) |
| Breastmilk | ||
| Total samples | 137 | 533 |
| Mean (SD) | 2.54 (1.92) | 3.31 (2.04) |
| Median (Q1–Q3) | 2.00 (1.00–4.00) | 3.00 (2.00–5.00) |
| Total | ||
| Total samples | 2370 | 9402 |
| Mean (SD) | 43.9 (24.9) | 58.4 (27.1) |
| Median (Q1–Q3) | 37.5 (28.3–48.8) | 52.0 (38.0–73.0) |
Note: Case:control ratio was 1:3 (Figure 1). Childhood samples are collected at scheduled study visits:-birth, three monthly until two years of age, then six monthly. Samples from a case included those up until the date of onset of persistent IA (inclusive). Samples from a control included those available until it’s matched case onset time.
Table 3.
Pregnancy total numbers of samples, mean (standard deviation) and median (interquartile range) number of samples per mother, by sample type for cases and controls of the ENDIA nested-case control study.
| Sample type | Case | Control |
|---|---|---|
| (N = 39) | (N = 135) | |
| Serum | ||
| Total samples | 66 | 266 |
| Mean (SD) | 1.69 (0.800) | 1.97 (0.872) |
| Median (Q1–Q3) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Plasma | ||
| Total samples | 66 | 265 |
| Mean (SD) | 1.69 (0.800) | 1.96 (0.867) |
| Median (Q1–Q3) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Stool | ||
| Total samples | 51 | 235 |
| Mean (SD) | 1.31 (0.800) | 1.74 (0.969) |
| Median (Q1–Q3) | 1.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Urine | ||
| Total samples | 65 | 271 |
| Mean (SD) | 1.67 (0.838) | 2.01 (0.806) |
| Median (Q1–Q3) | 1.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Throat swab | ||
| Total samples | 68 | 272 |
| Mean (SD) | 1.74 (0.818) | 2.01 (0.810) |
| Median (Q1–Q3) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Tongue swab | ||
| Total samples | 69 | 275 |
| Mean (SD) | 1.77 (0.777) | 2.04 (0.796) |
| Median (Q1–Q3) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Buccal swab | ||
| Total samples | 69 | 275 |
| Mean (SD) | 1.77 (0.810) | 2.04 (0.796) |
| Median (Q1–Q3) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Nasal swab | ||
| Total samples | 70 | 275 |
| Mean (SD) | 1.79 (0.767) | 2.04 (0.796) |
| Median (Q1–Q3) | 2.00 (1.00–2.00) | 2.00 (1.00–3.00) |
| Skin swab | ||
| Total samples | 37 | 128 |
| Mean (SD) | 0.949 (0.223) | 0.948 (0.223) |
| Median (Q1–Q3) | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) |
| Vaginal swab | ||
| Total samples | 36 | 118 |
| Mean (SD) | 0.923 (0.270) | 0.874 (0.333) |
| Median (Q1–Q3) | 1.00 (1.00–1.00) | 1.00 (1.00–1.00) |
| Total | ||
| Total samples | 597 | 2380 |
| Mean (SD) | 15.3 (5.97) | 17.6 (6.54) |
| Median (Q1–Q3) | 16.0 (10.0–18.0) | 18.0 (10.0–25.0) |
Note: Case:control ratio was 1:3. Mothers of siblings will appear in the table multiple times. Pregnancy samples can include samples from Trimester 1, 2 and 3. There are 26 control children with no maternal samples, 25 of these were postnatal recruits and 1 was recruited during pregnancy but had no maternal samples. There are 15 case children with no maternal samples, 13 of these were postnatal recruits and 2 were recruited during pregnancy but had no maternal samples.
Longitudinal availability of matched serum samples for cases and controls by risk-set is shown in Supplementary Material F. The longitudinal distribution of other sample types will be similar, as collection of all samples from an individual generally occurs at the same scheduled visit. Standard operating procedures for quality control of sample collection, processing and biobanking are briefly described in Supplementary Material G.
Omics laboratory design
The aim of laboratory design in omics investigations is to ensure that laboratory variation is not confounded with sample variation. A general discussion of appropriate designs for omics studies is included in Supplementary Material H.
In ENDIA, processing of samples in the omics studies will be undertaken in batches. Where possible, longitudinal samples from individuals in the same risk-set will be analysed in the same batch to minimize variation within risk-set and maximize the potential to detect biologically meaningful differences. Related risk-sets, which have individuals in common, because individuals are controls for more than one case or are both a control and case, will be processed in the same batch where possible. This ensures unique samples from those individual cases and controls that appear more than once in the NCC design in different risk-sets, will only be processed once and creates linkages between different risk-sets, inducing confounding of risk-sets and batches.
Limitations
The NCC study will be used for omics analyte discovery and the cases that emerge for persistent IA between 2020 and 2024/5 for validation. The estimated proportion of exposure to analytes in control children of 20% is of a reasonable size that will enable common exposures to be found. Related ORs greater than 2.9 have been reported for analytes, for example cytokines in children with IA versus persistently negative children [27] as well as in recent viral metagenomics analyses performed in the TEDDY study [28]. The opportunity for finding rarer exposures will be limited as indicated by the larger ORs under proportions of exposures of 1% to 5% (Supplementary Material A, Table A3).
The time of onset of persistent IA has been based on the first antibody positive sample. Seroconversion is therefore assumed to develop between the last negative and first positive serum sample, with the exact time of onset of autoantibodies between these dates unknown. The time between these samples, will also vary depending on the age of the child (due to the sampling regime − three monthly before two years; six monthly thereafter), and if samples are unavailable for testing due to missed visits. Bias may be introduced as the recorded age at onset is the last possible time and therefore potentially inflated. These limitations however, apply to all at – risk cohorts with interval testing of islet autoimmunity.
Limitations relating to the design of omics studies are discussed (Supplementary Material H), including the potentially prohibitive cost of sample processing, the quantity of sample available given these samples represent a precious limited resource and the sample quality.
The persistent IA outcome includes children with single and multiple IA, and children that have progressed to T1D, however, there is insufficient power in the ENDIA NCC study to separate these groups.
Analysis approaches
Univariate and multivariate approaches to the analysis of omics data are considered (Table 4). Here, a univariate omics analysis is used to refer to a model in which a single omics analyte is considered in isolation and therefore assumes that analytes are acting independently. Multivariate approaches refer to methods that consider omics analytes simultaneously and therefore take a holistic approach. Both approaches consider analytes from a single omics platform only. Multi-omics approaches that consider the complexity of the entire ‘biomolecular system’ (i.e. interactions of analytes from multiple omes) are not considered in Table 4.
Table 4.
Summary of univariate and multivariate analysis approaches considered and those to be implemented for a single time and longitudinal omic data in the ENDIA study.
| Univariatea | Multivariatee | |
|---|---|---|
| Single time |
Unweighted conditional logistic regression
b
Weighted logistic regression b Weighted conditional logistic regression c |
Principal component analysis (PCA)
Partial least square-discriminant analysis (PLS-DA) |
| Longitudinal |
Linear or generalized linear mixed modelsd IA status as response IA status as explanatory factor |
N-way PLS-DA
Clustering Network analysis methods |
aUnivariate methods will require false discovery rate (FDR)29 adjustment with <0.1 cut-off.
bDiscussed in Supplementary Material I.
cThis is the primary analysis, weight calculations are in Supplementary Material J and example of R code to fit models in Supplementary Material K.
dThe hierarchy of models fitted are discussed in Supplementary Material N.
eMultivariate approaches will use adjusted analyte values (Table 3) and are discussed in Supplementary material O.
Adjustment will be made for design factors and confounders in all approaches (Table 5). Models will be fitted with and without confounders; while design variables will be kept in all models. Where appropriate, robust variance estimators will be calculated [29].
Table 5.
Summary of potential variables included in the univariate and multivariate models.
| Explanatory variables | Design variables | Confounders | |
|---|---|---|---|
| Fixed effects | Analytea IA statusb (factor) Timec
IA status and time interactionc Sample IDd (factor) |
Risk-sete (factor) Matching variablese
|
Confounder variables or propensity scoreeh |
| Random effects | Laboratory design (factors)e Motheref (factor) |
||
| Log offset | Weightseg |
aPrimary analysis only, may be either continuous or a factor (e.g. presence/absence).
bSecondary analyses only.
cLongitudinal analyses only.
dMultivariate approach only, where adjusted sample analyte values are to be used which are determined based on a model with an explanatory variable relating to sample ID, discussed in Supplementary Material O.
eDiscussed in Supplementary Material K.
fInduces a correlation between NCC siblings who have the same mother.
gCalculation of weights is shown in Supplementary Material J.
hDiscussed in Supplementary Material L.
For univariate (including longitudinal) models, false discovery rate (FDR) [30] adjustment with a cut-off of less than 0.1 will be used to reduce the likelihood of type I errors when examining multiple models (i.e. one for each analyte in a omics platform).
Primary analyses
For the ENDIA study, the primary analysis is a weighted conditional logistic regression analysis [31] for each measured analyte independently (Supplementary Material I).
For samples other than serum, matching control samples may not be available at the case time of onset (i.e. within the 45 or 90 days cut-off). In this situation, the closest control sample within 12 months will be used.
Secondary analyses
Longitudinal univariate data
The matching of controls to cases in the ENDIA NCC design is based on a single time point – the onset of persistent IA. In a longitudinal study such as ENDIA, where testing children for the primary outcome of IA occurs over time, with controls matched to cases on age, this incidentally also ensures matching longitudinally of case and control samples (i.e. a sample-level matching) [31]. Longitudinal data in an NCC study have some general considerations for analysis that are not applicable to single time point analysis (Supplementary Material M). These include centring time at the case date of onset; sample allocation to risk-sets for children that are represented in more than one risk-set and mothers with children that are siblings participating in the NCC study and whether to include IA status as the response or explanatory variable in the analysis.
For children’s univariate longitudinal data, for each analyte (response variable), a series of linear mixed models or generalized linear mixed models will be fitted. A hierarchical approach will be taken, with the base model including fixed main effects of time and IA status and their interaction as explanatory variables so that independent trajectories can be fitted for cases and controls (Supplementary Material N).
The preferred sample allocation for risk-sets samples from the same child who is represented in more than one risk set, will duplicate samples within each risk-set, so that each risk-set will have the full trajectory of relevant samples from that child. As there are four children included as cases and controls, sensitivity analyses will be undertaken that will (a) exclude the samples of these children as cases (remove the risk-sets), but leave the samples of them as controls, (b) vice versa and (c) exclude all data of these children.
When only maternal longitudinal data is considered, there are three time points. The model fitted here will include main effects and an interaction between time and IA status, with time as a factor with levels corresponding to trimesters.
For the mothers who have children from different pregnancies (siblings) participating in the NCC, due to the age matching, the siblings will be allocated to different risk-sets. For these maternal samples, allocation will be to both siblings’ risk-sets. In the majority of cases, both the siblings are controls or both are cases. However, for four mothers, one of the siblings is a case and the other a control. A sensitivity analysis will be conducted that (a) excludes the samples of the mother of the case sibling, but leaves the samples of the control sibling and (b) vice versa and (c) excludes all data of these mothers (i.e. both as cases and controls).
Depending on the omics platform, a transformation of the response variable (omics analyte) may need to be considered for linear mixed models assuming a normal error distribution to ensure that model assumptions are met. Generalized linear mixed models with appropriate distributions including a negative binomial model, quasi-Poisson model or zero inflated models [10] (e.g. for count data with many zero values in microbiome data) will be considered.
Multivariate omics analysis
Detailed discussion of multivariate analysis approaches are in Supplementary Material O. Sample values adjusted for design factors and confounders (Table 5) will be used in multivariate analyses. For a single time point data, partial least square-discriminant analysis PLS-DA is the preferred method for the ENDIA study with cross-validation to evaluate the predictive performance of the method [32].
Currently methods and software for omics longitudinal data with disease groups (i.e. case versus control) are critically lacking. Some methods that may be suitable for exploring data from the ENDIA NCC study in the context of analysing multivariate longitudinal data are presented (Table 4). This is underpinned by the proviso that this is a rapidly expanding field with methodology and software continually being updated and developed. New approaches may emerge to be more suitable for the ENDIA NCC study at the actual time of analysis, thus taking advantage of the contemporary tools available [33]. Cross-validation and/or validation using an independent data set, of all results determined from multivariate approaches will be undertaken. Finally, multi-omics methods that combine data from different omics platforms (e.g. microbiome and proteome) with clinical and demographic data, may also be appropriate exploratory approaches. These could be undertaken as developments in methodology emerge and will make better use of the data generated from the individual omic studies.
Conclusions
We have outlined a framework for the analysis of longitudinal omics data with disease data within an NCC study, discussing appropriate design and considerations for analysis. Omics data can deliver benefits for determining disease aetiology, potentially leading to translation to personalized medicine solutions and multi-level clinical trials.
For univariate analysis of omics data, standard methods can be used. Multivariate approaches are exploratory and predictive rather than employing causal inference. Cross-validation and/or independent validation cohorts are required to confirm the associations found. For multivariate and multi-omics analysis, methods and software for longitudinal data with disease groups (i.e. case versus control) are critically lacking.
Supplementary Material
Acknowledgements
The ENDIA Study Group would like to thank all those institutions and individuals for their contribution to ENDIA recruitment and follow-up.
Lead Clinical Recruitment/Follow-up Sites: The Women’s and Children’s Hospital, Royal Melbourne Hospital, Barwon Health, Monash Health, Children’s Hospital at Westmead, Royal Hospital for Women, St George Hospital, Princess Margaret Hospital/Perth Children’s Hospital, Mater Mother’s Hospital/Queensland Children’s Hospital. Lead Academic Sites: The University of Adelaide, Walter and Eliza Hall Institute, University of New South Wales, University of Sydney, University of Western Australia/Telethon Kids Institute/Harry Perkins Institute, University of Melbourne and University of Queensland. We also gratefully acknowledge the participants and their families who contribute to the ENDIA study.
Members of The ENDIA Study Group (December 2022): Simon C Barry, Maria E Craig, Peter G Colman, Jennifer J Couper, Elizabeth A Davis, Emma E Hamilton-Williams, Mark Harris, Leonard C Harrison, Aveni Haynes, Ki Wook Kim, Grant Morahan, Helena Oakey, Megan A S Penno, William D Rawlinson, Richard O Sinnott, Georgia Soldatos, Rebecca L Thomson, Peter J Vuillermin, John M Wentworth. Amanda J Anderson, Pat Ashwood, James D Brown, William Hu, Dao Huynh, Kelly J McGorm. Kelly Watson. Jason Tye-Din, Tony Huynh, Claire Morbey, Prudence Lopez. Sarah Beresford, Samantha Bertram, Debra Bezuidenhout, Susan Brandrick, Carlie Butterworth, Jacki Catteau, Nakita Clements, Kyana Gartrell, Abbey Gilbert, Helen Griffiths, Alison Gwiazdzinski, Candice Hall, Gail Harper, Amanda Hulley, Mikayla Hoffman, Renee Kludas, Belinda Moore, Benjamin Ramoso, Alison Roberts, Alexandra Tully, Isabelle Vicary, Rosemary Wood. Rachel Battersby. Chris Hope, Tim Sadlon, Alexandra Roth-Schulze, Ying-Ying Wong, Enrique Zozaya-Valdes. Sabrina Binkowski, Bek Brittain, Minh Bui, Dexing Huang, Asma Minhaj, Gaetano Naselli, Katrina Ngui, Trung Nguyen, Natalie Stone, Emily Ward, Yan Xu, Cynthia Yau. Thank you to Jennie Louise (University of Adelaide) for her comments and suggestions on an early draft of the manuscript.
Funding Statement
This research was supported by JDRF Australia, the recipient of the Commonwealth of Australia grant for Accelerated Research under the Medical Research Future Fund and with funding from The Leona M. and Harry B. Helmsley Charitable Trust (grant keys 3-SRA-2020-966-M-N, 3-SRA-2019-899-M-N, 2-SRA-2021-1083-M-B, 2-SRA-2022-1094-M-B, 2-SRA-2021-1085-M-B). In addition, support was provided by The National Health and Medical Research Council of Australia (APP1078106), JDRF Australia Pilot and Innovation Award scheme (JDRF 2-SRA-2022-1100-M-B), JDRF International Strategic Research Award scheme (JDRF 3-SRA-2017-417-A-N, 3-SRA-2019-730-S-B, 17-2013-526), The Leona M. and Harry B. Helmsley Charitable Trust (G-2112-04908, G-2103-05117) and Diabetes SA. KWK and AH are the recipients of JDRF International Postdoctoral Fellowships (JDRF 3-PDF-2020-940-A-N and 3-PDF-2020-939-A-N). LCH is the recipient of an NHMRC Leadership Investigator Grant (APP1173945). MEC is the recipient of an NHMRC Practitioner fellowship (APP1136735).
Ethical approval
The ENDIA study was reviewed and approved at each clinical site, with the Women’s and Children’s Hospital Network Human Research Ethics Committee in Adelaide acting as the lead under the Australian National Mutual Acceptance Scheme (reference number HREC/16/WCHN/066). Conduct in Western Australia was approved by the Women and Newborn Health Service Ethics Committee (reference number RGS0000002639).
Registration details
ENDIA is registered on the Australia New Zealand Clinical Trials Registry (ACTRN12613000794707), https://www.anzctr.org.au/Trial/Registration/TrialReview.aspx?id=364573&isReview=true.
Author contributions
HO, RLT, LCG, EJK, MEC, PGC, EEHW, AH, KWK, WDR, JJC and MASP made substantial contributions to the conception or design of the work. HO, RLT, LCG, KALC, PA, JDB, EJK, SCB, MEC, PGC, LCH, KWK, KAM, KM, GM, WDR, ROS, GS, JJC and MASP were involved in the acquisition, management, analysis, or interpretation of data for the work. HO, RLT, LCG, MEC, PGC, EAD, EEHW, LCH, AH, KWK, KM, GM, WDR, ROS, GS, JMW, JJC and MASP were involved in the management of ENDIA staff and/or sites. MEC, PGC, LCH, KM, GS, JMW, JJC and MASP were involved in recruitment of participants to the ENDIA study. MEC, LCH, ROS, JJC and MASP were involved in sample collection. HO and MASP wrote the paper. HO wrote the R code. All authors read, critically revised and approved the final manuscript. All authors agree to be accountable for all aspects of the work.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
Data sharing is not applicable to this article as no new data were created or analysed in this study. Data dictionaries and study protocols are available here: https://www.endia.org.au/for-researchers/. Data and samples can be requested by researchers who provide a methodologically sound proposal that aligns with the goals of the ENDIA Study Group as described here: https://www.endia.org.au/for-researchers/.
References
- 1.Penno MA, Couper JJ, Craig ME, et al. . Environmental Determinants of Islet Autoimmunity (ENDIA): a pregnancy to early life cohort study in children at-risk of type 1 diabetes. BMC Pediatr. 2013;13(1):1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krischer JP, Liu X, Lernmark Å, et al. . Predictors of the initiation of islet autoimmunity and progression to multiple autoantibodies and clinical diabetes: the TEDDY study. Diabetes Care. 2022;45(10):2271–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rewers M, Bugawan T, Norris J, et al. . Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia. 1996;39(7):807–812. [DOI] [PubMed] [Google Scholar]
- 4.Group TS. The environmental determinants of diabetes in the young (TEDDY) study. Ann N Y Acad Sci. 2008;1150(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peet A, Kool P, Ilonen J, et al. . Birth weight in newborn infants with different diabetes-associated HLA genotypes in three neighbouring countries: Finland, Estonia and Russian Karelia. Diabetes Metab Res Rev. 2012;28(5):455–461. [DOI] [PubMed] [Google Scholar]
- 6.Ziegler A-G, Hummel M, Schenker M, et al. . Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: the 2-year analysis of the German BABYDIAB study. Diabetes. 1999;48(3):460–468. [DOI] [PubMed] [Google Scholar]
- 7.Gillman MW. Developmental origins of health and disease. N Engl J Med. 2005;353(17):1848–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcazar O, Hernandez LF, Nakayasu ES, et al. . Parallel multi-omics in high-risk subjects for the identification of integrated biomarker signatures of type 1 diabetes. Biomolecules. 2021;11(3):383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Parikh H, Butterworth MD, et al. . Longitudinal metabolome-wide signals prior to the appearance of a first islet autoantibody in children participating in the TEDDY study. Diabetes. 2020;69(3):465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lamichhane S, Ahonen L, Dyrlund TS, et al. . Dynamics of plasma lipidome in progression to islet autoimmunity and type 1 diabetes–type 1 diabetes prediction and prevention study (DIPP). Sci Rep. 2018;8(1):10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Prentice RL. On the design of synthetic case-control studies. Biometrics. 1986;42(2):301–310. [PubMed] [Google Scholar]
- 12.Richardson D. An incidence density sampling program for nested case-control analyses. Occup Environ Med. 2004;61(12):e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borgan Ø, Keogh R.. Nested case–control studies: should one break the matching? Lifetime Data Anal. 2015;21(4):517–541. [DOI] [PubMed] [Google Scholar]
- 14.Breslow NE. Statistics in epidemiology: the case-control study. J Am Stat Assoc. 1996;91(433):14–28. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein L, Langholz B.. Asymptotic theory for nested case-control sampling in the cox regression model. Ann Statist. 1992;20(4):1903–1928. [Google Scholar]
- 16.Ngo LH, Inouye SK, Jones RN, et al. . Methodologic considerations in the design and analysis of nested case-control studies: association between cytokines and postoperative delirium. BMC Med Res Methodol. 2017;17(1):1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGorm KJ, Brown JD, Thomson RL, et al. . A long-term evaluation of Facebook for recruitment and retention in the ENDIA type 1 diabetes pregnancy-birth cohort study. J Diabet Sci Technol. 2022:19322968221079867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dupont WD. Power calculations for matched case-control studies. Biometrics. 1988;44(4):1157–1168. [PubMed] [Google Scholar]
- 19.R Core Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2020. [Google Scholar]
- 20.Stevenson M, Sergeant E. epiR: tools for the analysis of epidemiological data. 2022. R package version 2.0.48. https://CRAN.R-project.org/package=epiR
- 21.Guo Y, Logan HL, Glueck DH, et al. . Selecting a sample size for studies with repeated measures. BMC Med Res Methodol. 2013;13(1):1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ziegler AG, Rewers M, Simell O, et al. . Seroconversion to multiple islet autoantibodies and risk of progression to diabetes in children. J Am Med Assoc. 2013;309(23):2473–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orban T, Sosenko JM, Cuthbertson D, et al. . Pancreatic islet autoantibodies as predictors of type 1 diabetes in the diabetes prevention trial–type 1. Diabetes Care. 2009;32(12):2269–2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dahlquist GG, Nyström L, Patterson CC.. Incidence of type 1 diabetes in Sweden among individuals aged 0–34 years, 1983–2007: an analysis of time trends. Diabetes Care. 2011;34(8):1754–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maahs DM, West NA, Lawrence JM, et al. . Epidemiology of type 1 diabetes. Endocrinol Metab Clin North Am. 2010;39(3):481–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee HS, Burkhardt BR, McLeod W, et al. . Biomarker discovery study design for type 1 diabetes in the environmental determinants of diabetes in the young (TEDDY) study. Diabetes Metab Res Rev. 2014;30(5):424–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeung W-CG, Al-Shabeeb A, Pang CNI, et al. . Children with islet autoimmunity and enterovirus infection demonstrate a distinct cytokine profile. Diabetes. 2012;61(6):1500–1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vehik K, Lynch KF, Wong MC, et al. . Prospective virome analyses in young children at increased genetic risk for type 1 diabetes. Nat Med. 2019;25(12):1865–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin IF, Paik MC.. Matched case—control data analysis with selection bias. Biometrics. 2001;57(4):1106–1112. [DOI] [PubMed] [Google Scholar]
- 30.Storey JD, Tibshirani R.. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee HS, Lynch KF, Krischer JP.. Nested case-control data analysis using weighted conditional logistic regression in the environmental determinants of diabetes in the young (TEDDY) study: a novel approach. Diabetes Metab Res Rev. 2020;36(1):e3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruiz-Perez D, Guan H, Madhivanan P, et al. . So you think you can PLS-DA? BMC Bioinf. 2020;21(Suppl 1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Balzano-Nogueira L, Ramirez R, Zamkovaya T, et al. . Integrative analyses of TEDDY omics data reveal lipid metabolism abnormalities, increased intracellular ROS and heightened inflammation prior to autoimmunity for type 1 diabetes. Genome Biol. 2021;22(1):1–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing is not applicable to this article as no new data were created or analysed in this study. Data dictionaries and study protocols are available here: https://www.endia.org.au/for-researchers/. Data and samples can be requested by researchers who provide a methodologically sound proposal that aligns with the goals of the ENDIA Study Group as described here: https://www.endia.org.au/for-researchers/.