Abstract
International airports can have a key role in screening, detecting, and mitigating cross-border transmission of SARS-CoV-2 and potentially other infectious diseases. With aircraft passengers representing a subpopulation of a country or region, aircraft-based wastewater surveillance can be a promising approach to effectively identifying emerging viruses, tracing their evolution, and mapping global spread with international flights. Therefore, we propose the development of a global aircraft-based wastewater genomic surveillance network, with the busiest international airports as central nodes and continuing air travel journeys as vectors. This surveillance programme requires routinely collecting aircraft wastewater samples for microbiological analysis and sequencing and linking the resulting data with associated international air traffic information. With the creation of a strong international alliance between the airline industry and health authorities, this surveillance network will potentially complement public health systems with a true early warning ability to inform decision making for new variants and future global health risks.
Introduction
International air traffic strongly influenced the onward global transmission of SARS-CoV-2.1, 2 Many countries and regions that were initially and most heavily affected by SARS-CoV-2 and its variants were highly connected international hubs with many commercial flights and overseas travellers. The first detection of SARS-CoV-2 and novel variants at a new location was usually associated with inbound air travellers from known or suspected hotspots. At the onset of the COVID-19 pandemic, the cross-border transmission of SARS-CoV-2 associated with air travellers leaving China was assessed using epidemiological models based on air travel data, human mobility data, and case reports.3, 4 The travel-related transmission pattern continued in the subsequent global spread of new SARS-CoV-2 variants.5 For instance, the first cases of the omicron (B.1.1.529 and BA.1) variants in many countries were detected at local international airports or in returning overseas travellers.6, 7
International travel as the vector
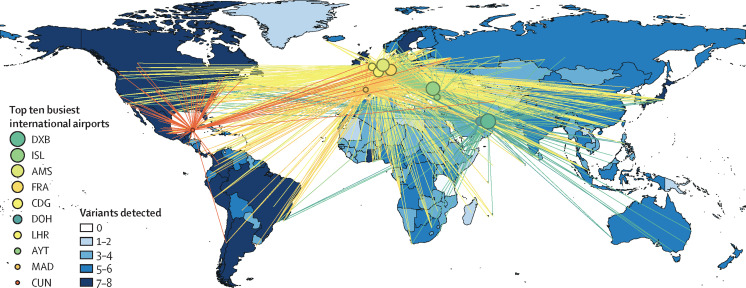
The notable travel-associated spread pattern of SARS-CoV-2 (figure 1 ) indicates that international airport hubs can be used as the key nodes across the global transportation network for first-line screening, detection, and mitigation of emerging health risks. Experience gained from the previous Ebola virus epidemic that severely impacted west Africa in 2013–16 and, most recently, the COVID-19 pandemic has emphasised the need for continuous and effective health surveillance for the rapid detection and real-time tracking of emerging infectious viruses and mutations among representative communities, such as air travellers.10, 11, 12 We therefore propose the formation of a global aircraft-based surveillance network to assess emergence, diversity, and trends in the global transmission of SARS-CoV-2 variants and other pathogens with pandemic and epidemic potential, linking with real-time international air traffic information. This surveillance network would specify strategic international airports across five continents as central nodes, arriving aircrafts as monitoring targets, and continuing air travel journeys as vectors. Selection of aircrafts for monitoring can be based on the prevalence of health risks in the place of origin, with onboard passengers being considered a representative subpopulation of local communities. A real-time picture of the spread of monitored infectious diseases worldwide can be created by combining multiple data sources, including sample testing, air travel history, and available clinical data. This network will allow the introduction of potential health risks to be identified at the point of entry and traced back to the most likely places of origin. The identification of an emerging variant or pathogen at an early stage will further drive public health efforts towards the specific geographical origin and inform enhanced testing, sequencing, public health measures, and social measures.
Figure 1.
Spread of SARS-CoV-2 variants across five continents and flight routes from the busiest ten international airports in 2021
The top ten busiest airports worldwide in 2021 were published by the Airports Council International. The ranking was based on international passenger traffic.8 Flight route data were obtained from OpenFlights. Data for SARS-CoV-2 variants were obtained from GISAID,9 including variants of concern (alpha [B.1.1.7], beta [B.1.351], gamma [P.1], delta [B.1.617.2 and AY], and omicron [B.1.1.529 and BA.]), previous variants of interest (lambda [C.37] and mu [B.1.621 and B.1.621.1]), and variants under monitoring (GH/490R [B.1.640]). DXB=Dubai International Airport, Dubai, United Arab Emirates. ISL=Atatürk International Airport, Istanbul, Türkiye. AMS=Amsterdam Schipol Airport, Amsterdam, Netherlands. FRA=Frankfurt Airport, Frankfurt, Germany. CDG=Paris Charles de Gaulle Airport, Paris, France. DOH=Hamad International Airport, Doha, Qatar. LHR=London Heathrow, London, UK. AYT=Antalya Airport, Antalya, Türkiye. MAD=Adolfo Suárez Madrid–Barajas Airport, Madrid, Spain. CUN=Cancun International Airport, Cancun, Mexico.
The establishment of this aircraft-based global genomic surveillance network will build on existing strong links between public health authorities and the aviation industry, which is of national and international importance. The Future of the Airline Industry 2035 report, commissioned by the International Air Transport Association in 2018, had the foresight to identify the airport as “a strategic asset supporting governments’ public health objectives for both detection and containment of diseases”, taking increased responsibility for supporting “a global approach to managing infectious diseases”.13 Compared with mass clinical testing during high-prevalence periods of the COVID-19 pandemic, collecting representative samples regularly from specific regions or communities for continued and sustainable genomic surveillance is more important in the current and post-pandemic era. The proposed aircraft-based surveillance will allow the centralised monitoring of representative samples of aircraft carrying many thousands of overseas travellers to a destination airport every day. Furthermore, this system can exploit new next-generation sequencing (NGS) technologies to identify known and novel viruses and pathogens and evaluate their diversity and circulation in relevant communities in areas with emerging infectious diseases of public health concern. The formation of this global network also aligns with the new WHO 10-year strategy (2022–32) to scale up and strengthen genomic surveillance of pathogens worldwide.14
Aircraft wastewater surveillance
Wastewater testing is the only current environmental surveillance technique that allows for understanding excreted human health indicators at the population level using a non-invasive sample, following the wastewater-based epidemiological method. Aircraft wastewater samples can be used to effectively monitor SARS-CoV-2 and new variants carried by onboard passengers, allowing interpretation of virus diversity and circulation in departure countries and regions.6, 11, 15, 16, 17, 18 The feasibility of aircraft wastewater testing depends on a reasonable proportion of passengers on long-haul flights using lavatories and depositing oronasal fluids or faeces that contain sufficient gene fragments of SARS-CoV-2.19 Results of aircraft wastewater samples and clinical nasopharyngeal swab samples collected from arriving travellers were congruent; however, some samples were positive for SARS-CoV-2 despite all passengers testing negative on an RT-PCR before departure.11, 15, 18 NGS technologies, such as whole-genome sequencing, amplicon-based sequencing, and metagenomic sequencing, have been applied to detect individual and more than one SARS-CoV-2 variants in wastewater, including variants that were not yet present in clinical sequence databases.10, 17, 20, 21, 22 Specifically, omicron variants have been successfully detected in aircraft wastewater, with quantification cycle (Cq) values ranging from 29·8 to 35·5 in collected samples.6, 11, 17 Karthikeyan and colleagues developed improved virus concentration and deconvolution methods to obtain high-quality sequence data for alpha, beta, delta, epsilon (B.1.427 and B.1.429), and gamma SARS-CoV-2 variants, allowing high genome coverage in wastewater samples with Cq values ranging from 23·9 to 39·9.10 Other studies reported similarly high coverage of SARS-CoV-2 variants from wastewater, with a wide range of Cq values—ie, 29·5–36·2 (nucleocapsid gene) and 26·8–36 (envelope gene)—using metatranscriptomic, whole-genome, or high-throughput SARS-CoV-2 amplicon sequencing.21, 22, 23
With the surge of new SARS-CoV-2 subvariants (eg, the XBB lineage) and China's border reopening for international travellers from January, 2023, testing aircraft wastewater of international flights is being pushed for and done in many countries and regions as a complementary tool to ceasing clinical testing. An aircraft surveillance programme is being developed and implemented in Europe, coordinating EU Member States to test wastewater from aircraft coming from countries outside of the EU as part of the EU Sewage Sentinel System for SARS-CoV-2. Ad-hoc guidance was published by the European Commission in January, 2023 to address the absence of standardised operating procedures for aircraft wastewater sampling and testing in Europe.24 Although potentially feasible approaches were proposed, this guidance emphasised the necessity of harmonising the knowledge and experience gained through the expansion of surveillance activities to establish best practice methods. In the meantime, the US Centers for Disease Control and Prevention, Public Health Agency of Canada, and health authorities and an airline company in Australia are initiating aircraft wastewater sampling programmes to detect emerging variants of concern entering the country, track variants of international origin, and strengthen the global surveillance capacity. Aircraft wastewater testing, especially in combination with NGS, is a potentially cost-effective, efficient, and robust surveillance approach to support public health decision making. Beyond the COVID-19 pandemic period, the capacity of wastewater surveillance for infectious disease surveillance could be further enhanced, including to track influenza virus, monkeypox virus, and antimicrobial resistance, and NGS could be used to discover new microbes in untreated sewage, as shown in previous studies.25, 26
True early warning
Building an aircraft-based wastewater genomic surveillance network will importantly complement public health surveillance with a distinctive early warning ability, addressing key challenges that are associated with sampling bias and the global inequality of resources used for genomic surveillance. Wastewater surveillance will show its true early warning potential when the background level of a disease is low and clinical testing of populations is infrequent or deficient.27 Mass testing and individual traveller testing is being phased out worldwide, reducing the information that is available for the real-time monitoring of new SARS-CoV-2 variant transmission within and across borders. The widespread adoption of rapid antigen testing at home has resulted in a marked reduction in the proportion of positive samples that reach the laboratory for sequencing. The sampling strategy for genomic surveillance primarily relies on the routine random sampling of a subset of a given target population, such as weekly monitoring of people testing positive for (or with suspected) SARS-CoV-2, which can result in a long turnaround time (ie, the time in days between sample collection and genome submission to GISAID, ranging from <21 days in high-income countries to >71–109 days in low-income countries) and a low sequencing intensity (ie, <0·5% of total confirmed COVID-19 cases were sequenced).28 Sampling bias can further affect the interpretation of variant transmission routes and rates when genomes from a small number of countries (eg, the UK and the USA) dominate the global dataset. This bias reflects another challenge for the expansion of global genomic sequencing, namely the socioeconomic inequalities in terms of resources and experience across different countries. According to WHO, one in every three countries does not have the capacity to use genomic surveillance technology, especially low-income and middle-income countries.14 In these under-resourced areas, local transmission of new variants was typically not recognised until weeks or even months after substantial international spread, which was normally associated with outbound air travellers.29, 30
Key components
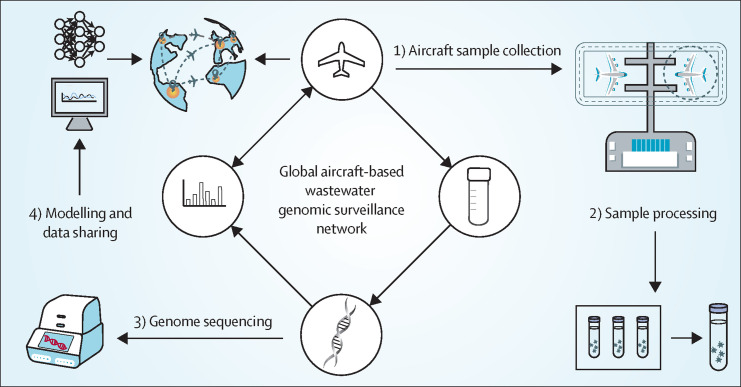
This proposed global aircraft-based wastewater surveillance network will tackle conventional surveillance challenges by prioritising the sampling and sequencing effort at well resourced international hubs that are highly connected to regions across the world and can gather diverse genomic information, including from areas with less sequencing capacity. We describe four key components of the entire surveillance approach (figure 2 ).
Figure 2.
Flow diagram of the proposed global aircraft-based wastewater genomic surveillance network
The first component is aircraft sample collection. Establishing this surveillance network relies on airline companies permitting sample collection from arriving aircrafts at the airports selected for genomic surveillance. Interception wastewater sampling can be conducted alongside normal offloading operations,31 during which the aircraft wastewater tank is drained immediately to the lavatory service truck on arrival.18 A standardised sampling protocol is important to efficiently coordinate ground crew for effective sample collection and transport and should be formulated jointly by members of the surveillance network. For example, the routine sample collection can be based on one aircraft from a specific country per day, perhaps departing from one of its largest cities. A potentially more practical approach is a responsive mode that can be activated to enact intensive sampling for individual flights from departure areas with increasing likelihood of, say, a high number of SARS-CoV-2 cases. When the initial intent of a local health authority is to detect the entry of new variants into the community, wastewater sampling can be conducted at airports to provide a composite sample (eg, 24-h time-proportional or flow-proportional sample) that contains integrated genetic information derived from disposed wastewater of all landing flights during a period of time, with simpler logistics than aircraft sampling. The ad-hoc guidance published by the European Commission specified the advantages and disadvantages of different sampling approaches by considering the resolution (eg, from single aircraft to airport scales) and reliability of information and the logistics of the approach.24
The second component is sample processing. The collected samples are transported to the laboratory for sample processing and archiving, with sample information uploaded to the local laboratory information management system. Sample pooling is an approach to improving process efficiency by reducing the number of samples to be processed and increasing the genome abundance in a pooled sample. For instance, for aircrafts departing from the same country or region during a defined period (eg, within 1 day or over a few days), small samples of individual wastewater can be combined to produce a pooled sample. Moreover, the archived samples (ie, both pooled samples and original samples of individual aircrafts) provide the capacity to perform retrospective analyses of specific sample pools or individual samples of a given pool, when the reference genome of a new virus or mutation is confirmed. These kinds of analyses will allow tracking of new variants or pathogens back to the departing dates and geographical locations. The pooled samples will be processed through DNA or RNA concentration and extraction and submitted for sequencing on a regular basis. For countries or regions with an increasing likelihood of health risks, aircraft wastewater samples will be prioritised for genomic sequencing.
The third component is genome sequencing. Targeted sequencing can be used to detect known specific variants or pathogens, such as through targeted amplification to increase sensitivity and achieve high sequencing coverage and depth, particularly for the samples with low amounts of nucleic acids that are collected during the early stages of outbreaks.20, 26 Considering the primary use of NGS, however, aircraft wastewater sequencing should not be limited to known viral sequences and should be expanded to discover new pathogens through non-targeted metagenomic and metatranscriptomic shotgun sequencing. These types of sequencing require experienced scientists to undertake bioinformatic analysis and submit new metadata to open-access genomic databases, such as GISAID, reporting the detection of new variants or pathogens with the related air travel information. Challenges and uncertainties in genome sequencing might exist due to the complex nature of aircraft wastewater, low abundance and recovery of targeted genome, and the detection of so-called novel mutations, which might be detected due to technical errors.10, 20 Further improvements in sample extraction, NGS, and bioinformatic approaches can be applied to address relevant problems, such as assignment and misclassification. The turnaround time for sequencing (eg, metagenomics) is longer than for RT-quantitative-PCR analysis of wastewater. Nevertheless, compared with clinical testing of many thousands of arriving travellers, the collection and analysis of pooled wastewater samples requires less labour and resources and costs less, with a reasonable anticipated turnaround time (ie, 72–96 h17 or within 1–2 weeks10).
The fourth component is modelling and data sharing. The purpose of this global surveillance network is to link aircraft-based genomic sequencing data with air travel history and facilitate virus tracking, even back to the potential primary outbreak location. Timely data, such as the frequency of flights and the volume of arriving and transfer passengers (ie, passengers landing at and departing from the airport within 24 h to their final destination) from various countries at local airports, should be collected from airline companies and airports.24 Data-driven models that incorporate aircraft sequencing data and a dynamic flight network, building on epidemiological models (eg, Flight-SEIR32 and the integrated GLEAM-SIR model33) and artificial intelligence algorithms,3 will enhance the ability to predict global virus spread with travel. Challenges in this step include the possibility of transfer passengers, which might confound virus tracking; however, data from these passengers also indicate the introduction of emerging viruses to the transiting locations. Another potential challenge is the availability of sufficient population mobility or clinical data for aircraft passengers at the time of flight, which will be important to interpret and validate the routes of virus transmission.
The capacity for rapid data sharing across many sectors and disciplines is important for successful global health surveillance. The leadership role of international health agencies, such as WHO, and national health authorities in convening and coordinating capacities at country and regional levels is crucial to motivate partnerships with key stakeholders, coordinate resource allocation, and inform decision making to address potential health threats. To ensure effective and transparent data exchange, the core of this network will be creating a united information platform that gathers data from many sources and disseminates real-time virus identification and distribution results to a broad community. Overall, we propose the creation of a strong international alliance of the airline industry, airports, public health authorities, biotechnology industry, laboratories, research institutes, and other relevant industries and stakeholders. The development of a global aircraft-based wastewater genomic surveillance network will support the ability of the public health system to address the threat of novel SARS-CoV-2 variants and future global health risks.
Declaration of interests
IH is an employee of Qantas Airways. DP is a consultant adviser to the International Air Transport Association. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This work received funding from JFM's Australian Research Council Laureate Fellowship (FL200100028). We thank Warish Ahmed from the Commonwealth Scientific and Industrial Research Organisation Environment Business Unit, Australia, for providing helpful comments and suggestions to this manuscript.
Editorial note: The Lancet Group takes a neutral position with respect to territorial claims in published maps and institutional affiliations.
Contributors
JLi is responsible for conceptualisation, visualisation, writing of the original draft, and revising of the final manuscript. IH, DP, BT, JLa, KVT, and JFM contributed to conceptualisation, review, and editing of the manuscript. All authors have read and approved the final version of the manuscript. All authors had final responsibility for the decision to submit for publication.
References
- 1.du Plessis L, McCrone JT, Zarebski AE, et al. Establishment and lineage dynamics of the SARS-CoV-2 epidemic in the UK. Science. 2021;371:708–712. doi: 10.1126/science.abf2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakamura H, Managi S. Airport risk of importation and exportation of the COVID-19 pandemic. Transp Policy. 2020;96:40–47. doi: 10.1016/j.tranpol.2020.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu JT, Leung K, Leung GM. Nowcasting and forecasting the potential domestic and international spread of the 2019-nCoV outbreak originating in Wuhan, China: a modelling study. Lancet. 2020;395:689–697. doi: 10.1016/S0140-6736(20)30260-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chinazzi M, Davis JT, Ajelli M, et al. The effect of travel restrictions on the spread of the 2019 novel coronavirus (COVID-19) outbreak. Science. 2020;368:395–400. doi: 10.1126/science.aba9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Lai S, Gao GF, Shi W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature. 2021;600:408–418. doi: 10.1038/s41586-021-04188-6. [DOI] [PubMed] [Google Scholar]
- 6.Agrawal S, Orschler L, Tavazzi S, Greither R, Gawlik BM, Lackner S. Genome sequencing of wastewater confirms the arrival of the SARS-CoV-2 omicron variant at Frankfurt Airport but limited spread in the city of Frankfurt, Germany, in November 2021. Microbiol Resour Announc. 2022;11 doi: 10.1128/MRA.01229-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Viana R, Moyo S, Amoako DG, et al. Rapid epidemic expansion of the SARS-CoV-2 omicron variant in southern Africa. Nature. 2022;603:679–686. doi: 10.1038/s41586-022-04411-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Airports Council International The top 10 busiest airports in the world revealed. April 11, 2022. https://aci.aero/2022/04/11/the-top-10-busiest-airports-in-the-world-revealed
- 9.GISAID Tracking of hCoV-19 variants. https://www.gisaid.org/hcov19-variants
- 10.Karthikeyan S, Levy JI, De Hoff P, et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609:101–108. doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Targa L, Wurtz N, Lacoste A, et al. SARS-CoV-2 testing of aircraft wastewater shows that mandatory tests and vaccination pass before boarding did not prevent massive importation of omicron variant into Europe. Viruses. 2022;14 doi: 10.3390/v14071511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardy JL, Loman NJ. Towards a genomics-informed, real-time, global pathogen surveillance system. Nat Rev Genet. 2018;19:9–20. doi: 10.1038/nrg.2017.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.International Air Transport Association Future of the airline industry 2035. Sept 20, 2018. https://www.iata.org/contentassets/086e8361b2f4423e88166845afdd2f03/iata-future-airline-industry.pdf
- 14.WHO Global genomic surveillance strategy for pathogens with pandemic and epidemic potential, 2022–2032. March 28, 2022. https://www.who.int/publications/i/item/9789240046979 [DOI] [PMC free article] [PubMed]
- 15.Ahmed W, Bertsch PM, Angel N, et al. Detection of SARS-CoV-2 RNA in commercial passenger aircraft and cruise ship wastewater: a surveillance tool for assessing the presence of COVID-19 infected travellers. J Travel Med. 2020;27 doi: 10.1093/jtm/taaa116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Albastaki A, Naji M, Lootah R, et al. First confirmed detection of SARS-COV-2 in untreated municipal and aircraft wastewater in Dubai, UAE: the use of wastewater based epidemiology as an early warning tool to monitor the prevalence of COVID-19. Sci Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.143350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ahmed W, Bivins A, Smith WJM, et al. Detection of the omicron (B.1.1.529) variant of SARS-CoV-2 in aircraft wastewater. Sci Total Environ. 2022;820 doi: 10.1016/j.scitotenv.2022.153171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahmed W, Bivins A, Simpson SL, et al. Wastewater surveillance demonstrates high predictive value for COVID-19 infection on board repatriation flights to Australia. Environ Int. 2022;158 doi: 10.1016/j.envint.2021.106938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones DL, Rhymes JM, Wade MJ, et al. Suitability of aircraft wastewater for pathogen detection and public health surveillance. Sci Total Environ. 2023;856 doi: 10.1016/j.scitotenv.2022.159162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Izquierdo-Lara R, Elsinga G, Heijnen L, et al. Monitoring SARS-CoV-2 circulation and diversity through community wastewater sequencing, the Netherlands and Belgium. Emerg Infect Dis. 2021;27:1405–1415. doi: 10.3201/eid2705.204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fontenele RS, Kraberger S, Hadfield J, et al. High-throughput sequencing of SARS-CoV-2 in wastewater provides insights into circulating variants. Water Res. 2021;205 doi: 10.1016/j.watres.2021.117710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crits-Christoph A, Kantor RS, Olm MR, et al. Genome sequencing of sewage detects regionally prevalent SARS-CoV-2 variants. mBio. 2021;12:e02703–e02720. doi: 10.1128/mBio.02703-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bar-Or I, Weil M, Indenbaum V, et al. Detection of SARS-CoV-2 variants by genomic analysis of wastewater samples in Israel. Sci Total Environ. 2021;789 doi: 10.1016/j.scitotenv.2021.148002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Deere DA, Jones DL, Ahmed W, et al. Ad-hoc guidance: wastewater sampling of aircrafts for SARS-CoV-2 surveillance. Jan 9, 2023. https://wastewater-observatory.jrc.ec.europa.eu/static/pdf/Sampling%20Aircrafts_FINAL_Version%209%20Jan%202023.pdf
- 25.Ng TF, Marine R, Wang C, et al. High variety of known and new RNA and DNA viruses of diverse origins in untreated sewage. J Virol. 2012;86:12161–12175. doi: 10.1128/JVI.00869-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy JI, Andersen KG, Knight R, Karthikeyan S. Wastewater surveillance for public health. Science. 2023;379:26–27. doi: 10.1126/science.ade2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Safford HR, Shapiro K, Bischel HN. Opinion: wastewater analysis can be a powerful public health tool—if it's done sensibly. Proc Natl Acad Sci USA. 2022;119 doi: 10.1073/pnas.2119600119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brito AF, Semenova E, Dudas G, et al. Global disparities in SARS-CoV-2 genomic surveillance. Nat Commun. 2022;13 doi: 10.1038/s41467-022-33713-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grubaugh ND, Hodcroft EB, Fauver JR, Phelan AL, Cevik M. Public health actions to control new SARS-CoV-2 variants. Cell. 2021;184:1127–1132. doi: 10.1016/j.cell.2021.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hodcroft EB, Zuber M, Nadeau S, et al. Spread of a SARS-CoV-2 variant through Europe in the summer of 2020. Nature. 2021;595:707–712. doi: 10.1038/s41586-021-03677-y. [DOI] [PubMed] [Google Scholar]
- 31.Ehret J, Cook J, King T, Hosegood I, Mueller J. Aircraft lavatory wastewater sampling utilising the Qantas Sample Trap (QST) MK III. May 20, 2022. https://zenodo.org/record/6565202#.Y5aAzuxBxUd
- 32.Ding X, Huang S, Leung A, Rabbany R. Incorporating dynamic flight network in SEIR to model mobility between populations. Appl Netw Sci. 2021;6:42. doi: 10.1007/s41109-021-00378-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nikolaou P, Dimitriou L. Identification of critical airports for controlling global infectious disease outbreaks: stress-tests focusing in Europe. J Air Transp Manage. 2020;85 doi: 10.1016/j.jairtraman.2020.101819. [DOI] [PMC free article] [PubMed] [Google Scholar]