Summary
Background
Nirmatrelvir-ritonavir (Paxlovid) and ensitrelvir are 3-chymotrypsin-like cysteine protease (3CLpro) inhibitors which have been approved for the treatment of COVID-19 in 2021 and 2022, respectively. Previous studies have identified 3CLpro mutations that are associated with reduced susceptibility to these antivirals. The aim of the current study was to estimate the global prevalence of 3CLpro inhibitor-resistant SARS-CoV-2 strains.
Methods
We compiled a list of 3CLpro mutations which have been associated with nirmatrelvir or ensitrelvir resistance based on either viral replication or 3CLpro activity assays, and determined their prevalence among 13.4 million sequences deposited in GISAID as of December 14, 2022, about 1 year after the approval of nirmatrelvir-ritonavir. We analyzed the prevalence for different time periods, SARS-CoV-2 lineages and geographical locations.
Findings
Overall, 0.5% (67,095/13,446,588) of the sequences contained at least one mutation that was shown to affect the inhibitory activity of nirmatrelvir or ensitrelvir on viral replication or 3CLpro activity. We did not observe any increasing trend of resistance after the widespread clinical use of nirmatrelvir-ritonavir. G15S (2070 per million) and T21I (1386 per million) were the most prevalent mutations, and these mutations were dominant in some SARS-CoV-2 lineages. E166V and S144E, previously shown to affect the inhibitory activity of nirmatrelvir on viral replication or protease activity by > 100-folds, were found in <1 per million sequences.
Interpretation
Our data suggest that 3CLpro inhibitor resistance is currently rare. However, continuous global genotypic and phenotypic surveillance would be crucial in the early detection of resistant mutants.
Funding
Richard and Carol Yu, May Tam Mak Mei Yin, The Shaw Foundation Hong Kong, Michael Tong, Marina Lee, Government Consultancy Service, the Emergency Key Program of Guangzhou Laboratory (See acknowledgements for full list).
Keywords: COVID-19, SARS-CoV-2, Antiviral resistance, Nirmatrelvir, Ensitrelvir, Protease inhibitor
Research in context.
Evidence before this study
Nirmatrelvir-ritonavir (Paxlovid), which inhibits SARS-CoV-2 3CL protease (3CLpro), has been approved in late 2021 for the treatment of COVID-19. Ensitrelvir, another 3CLpro inhibitor, has been approved in Japan. Several studies have identified mutations in the 3CLpro which confer resistance to nirmatrelvir or ensitrelvir. Since these 3CLpro mutations may lead to treatment failure, it is important to know the prevalence of these mutations among circulating strains. We searched PubMed without language restrictions on 11th January 2023 for articles using the terms “COVID-19” or “SARS-CoV-2” and the terms “antiviral resistance”, “nirmatrelvir”, or “ensitrelvir”. Several studies have isolated nirmatrelvir or ensitrelvir-resistant strains by serially passaging SARS-CoV-2 strains in the presence of these drugs, and have identified the 3CLpro mutations that were associated with the resistance. Some studies have reported the prevalence of some mutations up to June 2022. However, the prevalence of most of these mutations were not reported.
Added value of this study
By analyzing over 13 million SARS-CoV-2 sequences deposited into a public database up to December 2022, we found that about 0.5% of the viral sequences contain at least one mutation that has been associated with resistance against nirmatrelvir or ensitrelvir. We did not observe any significant increase in resistance rate after the approval of Paxlovid. G15S and T21I were the most frequently identified mutations associated with 3CLpro inhibitor resistance, and these mutations were fixed in some lineages.
Implications of all the available evidence
The low prevalence of 3CLpro inhibitor resistance suggest that nirmatrelvir and ensitrelvir remain clinically useful. However, since some of these mutations were fixed in certain lineages, there is a potential for 3CLpro inhibitor-resistant viruses to spread in the future. Continuous genotypic and phenotypic surveillance is important for early detection of 3CLpro inhibitor-resistant viruses.
Introduction
Coronavirus Disease 2019 (COVID-19) pandemic contributed to an estimated 14.83 million excess deaths globally between 2020 and 2021.1 The case-fatality rate is higher among older adults, individuals with comorbidities and those who have not received COVID-19 vaccines.2 Pharmacological therapies, including antivirals and immunomodulatory agents, have shortened the duration of hospitalization or reduced death rates.3,4
Among the virus-targeting antivirals, remdesivir and monoclonal antibodies were the mainstay of treatment in 2020 and 2021. However, these antivirals are given via the intravenous route and are therefore only suitable for treating hospitalized patients. Furthermore, most monoclonal antibodies are now ineffective due to the emergence of immune escape mutants. In late 2021, two oral antivirals, nirmatrelvir-ritonavir (Paxlovid) and monulpiravir, were approved for clinical use, which have made out-patient treatment possible. Since then, several other oral antivirals have been found to be clinically effective and have been approved in different countries, including entrelsivir furamic acid (S-217622; approved in Japan in November 2022), azvudine (approved in China in August 2022) 5and deuremidevir hydrobromide (VV116; approved in China in January 2023).6
Both nirmatrelvir-ritonavir and ensitrelvir furamic acid inhibit the 3-chymotrypsin-like cysteine protease (3CLpro), also known as main protease (Mpro) and non-structural protein 5 (nsp5), cleaves the polyproteins pp1a and pp1ab, a critical step in SARS-CoV-2 replication.7 In a phase 3 clinical trial, nirmatrelvir-ritonavir was demonstrated to reduce the risk of severe disease.8 Ensitrelvir furamic acid was associated with lower viral load in a hamster model and in a phase 2/3 clinical trial.9,10
Antiviral resistance can result in treatment failure. Although naturally-occurring nirmatrelvir-resistant SARS-CoV-2 strains have not been reported, nirmatrelvir-resistant mutants have been found after serial passage in cell lines.11, 12, 13 Several studies have demonstrated the effect of individual mutations on the inhibitory activity of 3CLpro.11,13, 14, 15, 16, 17, 18, 19, 20 Previous studies assessing sequences deposited into public databases up to June 2022 showed that mutations that affect nirmatrelvir antiviral activity or mutations located at the 3CLpro inhibitor binding site were rarely found.11,16,21 In the current study, we determined the prevalence of SARS-CoV-2 mutants with reduced susceptibility to nirmatrelvir or ensitrelvir based on 3CLpro genetic mutations.
Methods
Compilation of a list of 3CLpro inhibitor mutation
First, based on studies or FDA reports published on or before January 11, 2023, we compiled a list of mutations which showed reduced susceptibility to nirmatrelvir or ensitrelvir in terms of viral replication or 3CLpro activity (Tables 1 and 2).11,13, 14, 15, 16, 17, 18, 19, 20 We excluded mutations that were only shown to affect binding or occurred in resistant mutants but had not been experimentally verified to affect viral replication or 3CLpro activity.
Table 1.
3CLpro mutations that were associated with reduced nirmatrelvir inhibitory activity of SARS-CoV-2 replication or 3CLpro activity.
| Mutation | Reduction in susceptibility of SARS-CoV-2 replication (fold change) | Reduction in inhibition of 3CLpro activity (fold change) | References |
|---|---|---|---|
| G15S | N/A | 4.4 | 14 |
| T21I | 1.1–4.6 | N/A | 11,13, 14 |
| L50F | 1.4–4.2 | N/A | 11,13, 14 |
| Y54A | N/A | 24.0 | 14 |
| Y54C | 2.6–8.3 | N/A | 16 |
| T135I | N/A | 3.2 | 14 |
| F140A | N/A | 39.0 | 14 |
| F140L | N/A | 5.4 | 14 |
| S144A | 2.2–12.2 | 20.5–92 | 11,14,18,19 |
| S144E | N/A | 470.0 | 14 |
| S144F/G/M/Y | N/A | 19.2–38.0 | 18 |
| S144T | N/A | 160.0 | 14 |
| H164N | N/A | 6.4 | 14 |
| M165T | N/A | 29.9 | 18 |
| E166A | 3.3 | 10.0–33.0 | 14,17 |
| E166G | N/A | 16.0–16.4 | 14,18 |
| E166V | 25.0–288 | N/A | 13, 14, 15 |
| L167F | 3.6–16.5 | N/A | 16,17 |
| Del P168 | 5.1 | N/A | 19 |
| H172Q/F | N/A | >42.0 | 18 |
| H172Y | 24 | >133.7 | 14,18,20 |
| A173T | 4.1 | N/A | 19 |
| A173V | 0.9–11.6 | 26.0 | 11,14,19 |
| V186G | N/A | 13.0 | 14 |
| Q189K | N/A | 65.0 | 14 |
| Q192L | 4.3 | 28.0 | 14,18 |
| Q192P | 7.6 | 33.0 | 14,18 |
| Q192T/S/A/I/H/V/W/C/F | N/A | >22.2 | 18 |
| D248E | N/A | 3.7 | 14 |
| P252L | 5.9 | N/A | 11,14 |
| T304I | 2.1–5.5 | N/A | 11,13,14 |
IC50: 50% inhibitory concentration; N/A, data not available.
This table only shows the effect of single mutations.
SARS-CoV-2 sequence analysis
We downloaded the metadata of SARS-CoV-2 sequences from GISAID on 14th December 2022.22 A SARS-CoV-2 sequence was included if it had been marked as complete in the metadata file, and was directly obtained from a human clinical specimen without in vitro passage. A sequence was excluded if collection date information was not complete. All processing was performed on Python v3.9.1223 (pandas v1.4.2,24 numpy v1.22.4,25 openpyxl v3.0.10,26 json v2.0.9) running on Anaconda Software Distribution v 4.11.0. The script started by loading the metadata into the RAM in chunks of 10,000 sequences. The sequences in each chunk were selected for further analysis based on the inclusion and exclusion criteria. The date strings were then converted into pandas datetime objects, and each strain was then assigned with the respective quarter that it was collected in.
To calculate the prevalence of each mutation, the strains containing the mutation identifier in their “AA Substitutions” column were extracted. Then, the number of strains belonging to each continent was counted using the value_counts () function. If we were to search for the prevalence of a set of mutations, we would use a pipe symbol ‘|’ to separate all the possible mutations, as pandas.series.str.contains default supports regular expression (regex) searching. The number of mutations was then normalized to strain per million.
To extract the metadata of strains with 3CLpro E166V or both E166V and L50F, each strain was searched for the target mutation(s) within their “AA Substitutions” column. If a target was found, the GISAID accession ID and lineage was displayed on the screen. The number of strains containing at least one 3CLpro inhibitor affecting mutation was calculated using the same method by counting the number of targets found.
Ethics
Ethical approval is not required as anonymised data retrieved from GISAID were used in this study.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report.
Results
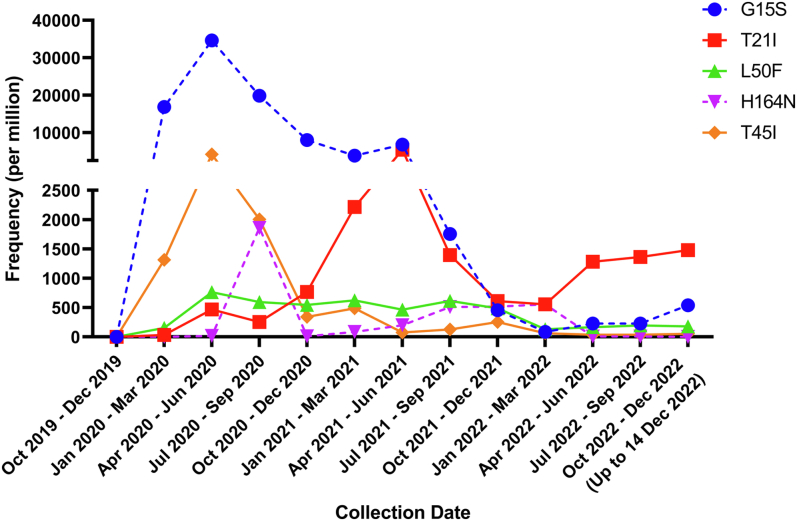
A total of 14,157,266 SARS-CoV-2 sequences were obtained from GISAID on 14th December 2022, of which 13,446,588 satisfied our selection criteria. 67,095 (4990/million) sequences contained at least one amino acid mutation that has been shown to affect the inhibitory activity of nirmatrelvir or ensitrelvir on viral replication or 3CLpro activity, including 36,072 (2683/million) sequences containing mutations that affect the inhibitory activity of 3CLpro activity, 31,065 (2310/million) sequences with mutations that have been shown to affect the inhibitory activity on viral replication, and 279 (21/million) sequences with mutations that have been shown to affect the inhibitory activity on both viral replication and 3CLpro activity. Nirmatrelvir-ritonavir (Paxlovid) was first approved on 22nd December 2021. We analyzed the prevalence of the five most frequently identified mutations since 2020. However, we did not find any evidence of increasing frequency of resistant mutations after nirmatrelvir-ritonavir was clinically approved (Fig. 1). There are also differences in the geographical distribution of the prevalence of mutations. South America has the highest proportion of sequences carrying mutations associated with nirmatrelvir or ensitrelvir resistance (29,032 per million), followed by Africa (24,437 per million) and North America (4809 per million) (Supplementary Table S1).
Fig. 1.
Global temporal trend of 3CLpro mutations that have been shown to affect the inhibitory activity of nirmatrelvir or ensitrelvir on viral replication or 3CLpro activity. Only the 5 most frequently found mutations are shown. Solid line indicates 3CLpro mutations which have been demonstrated to affect inhibitory activity of nirmatrelvir and/or ensitrelvir on viral replication. Dotted line indicates 3CLpro mutations which have been demonstrated to affect inhibitory activity of nirmatrelvir or ensitrelvir on protease activity only, but without data on viral replication.
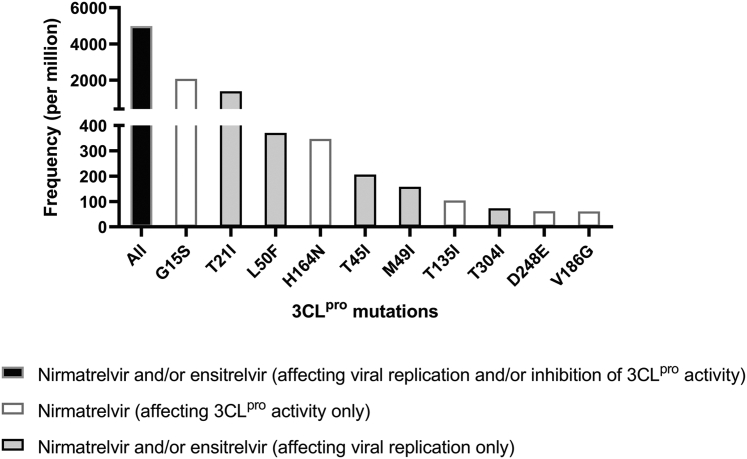
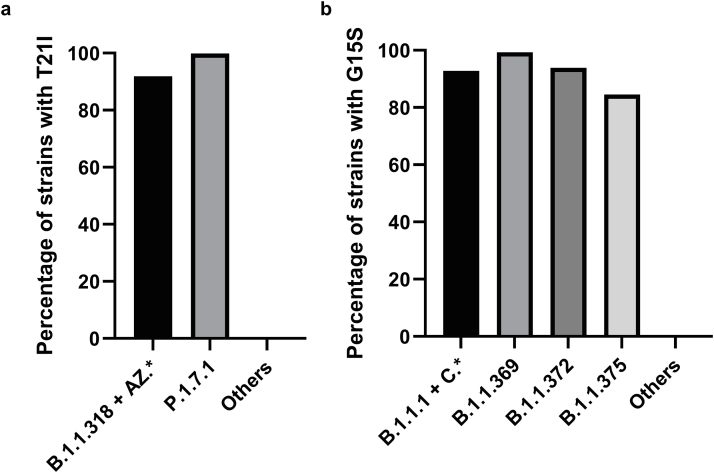
Overall, the most frequent mutation was G15S (2070/million [27,828/13,446,588]) (Fig. 2 and Supplementary Fig. S1), which conferred reduced susceptibility to nirmatrelvir 3CLpro inhibitory activity.14 G15S was present in almost all strains in the lineages B.1.1.1, B.1.1.369, B.1.1.372, B.1.1.375, and lineage C, including the Variant of Interest (VOI) Lambda [C.37]) (Supplementary Fig. S2a). Currently, there is no published data on the effect of G15S mutation on the inhibitory effect of 3CLpro inhibitor on viral replication.
Fig. 2.
Frequency of 3CLpro mutations (sequences per million) among sequences deposited onto GISAID up to 14th December 2022. Only the 10 most frequently found mutations affecting the inhibitory activity of nirmatrelvir or ensitrelvir are shown.
The second most frequently found mutation was T21I (1386/million [18,637/13,446,588]), which was shown to affect the inhibitory effect of nirmatrelvir on viral replication.11 T21I was present in almost all strains in the B.1.1.318 lineage, a VOI prevalent in West Africa in early 2021,27,28 and the Gamma variant sublineage P.1.7.1, which was prevalent in South America (Supplementary Fig. S2b). In a study assessing the emergence of nirmatrelvir-resistant strains during serial passage, T21I was shown to be one of the five precursor mutations that were present in all resistant strains.11
Among mutations that have been shown to affect the inhibitory activity of nirmatrelvir or ensitrelvir on viral replication, E166V conferred the greatest reduction in susceptibility (25-288-fold reduction for nirmatrelvir; 23-fold reduction for ensitrelvir) (Tables 1 and 2). E166V was found in 13 sequences (0.97/million) (Supplementary Table S2 and S3). Four were collected from a 65-year-old man from Japan at 1-week intervals between 30 May and 17 June 2022, and belonged to BA.1.1 lineage. All other strains with E166V mutations belonged to different lineages and were collected from different places. Amino acid 166 is a key residue that interacts with nirmatrelvir, including polar contact with the pyrrolidone group and hydrogen bonds between the tert-butyl moiety of nirmatrelvir.29,30 Previous studies showed that E166V confers a loss of viral fitness, but can be compensated by L50F mutation.11,13 Among the 13 sequences with E166V mutations, one (7.7%) sequence also contained L50F mutation (0.07/million). Among mutations that have been shown to affect the inhibitory activity of nirmatrelvir or ensitrelvir on 3CLpro activity, S144E confers the greatest reduction in the inhibition of nirmatrelvir on 3CLpro activity (470-fold reduction in 3CLpro activity) (Table 1). However, it occurs in less than 1 per million (Supplementary Figure S1).
Table 2.
3CLpro mutations that were associated with reduced ensitrelvir inhibitory activity of SARS-CoV-2 replication or 3CLpro activity.
| Mutation | Reduction in susceptibility of SARS-CoV-2 replication (fold change) | Reduction in inhibition of 3CLpro activity (fold change) | References |
|---|---|---|---|
| T21I | 1.7 | N/A | 11 |
| T45I | 4.1 | N/A | 19 |
| D48Y | 5.0 | N/A | 19 |
| M49I/L/T/V | 2.6–25.4 | N/A | 19 |
| L50F | 2.8 | N/A | 11 |
| S144A | 13–16.9 | N/A | 11,19 |
| E166V | 23 | N/A | 11 |
| Del P168 | 6.8 | N/A | 19 |
| A173V | 1.7 | N/A | 11 |
| P252L | 1.9 | N/A | 11 |
| T304I | 1.6 | N/A | 11 |
This table only shows the effect of single mutations.
Discussion
3CLpro inhibitors represent a major breakthrough in the treatment of COVID-19, especially for outpatient treatment since these antivirals can be taken orally. Our analysis indicates that 3CLpro inhibitor resistant mutations remain rare and mostly sporadic up to one year after the approval of Paxlovid. Only 0.5% of analyzed sequences had mutations associated with 3CLpro inhibitor resistance.
There are several unique features in our study. First, we have examined mutations or deletions at 23 amino acid positions of the 3CLpro protein which have been experimentally verified to affect either the viral replication or 3CLpro activity. Second, we have assessed the prevalence of mutations up to January 2023, which is more than 1 year after the approval of nirmatrelvir-ritonavir. Third, we have performed subgroup analysis on the prevalence of mutations in different sublineages.
While it is reassuring that 3CLpro inhibitors will likely remain clinically effective, there are several reasons why we should continue to monitor for 3CLpro inhibitor resistance. First, the most frequent mutations, G15S and T21I, were the dominant mutations within some lineages, suggesting that these mutations do not affect viral fitness and can be widely disseminated. These mutations first appeared before the clinical use of 3CLpro inhibitors, suggesting that they can arise spontaneously and are not selected by the usage of protease inhibitors. However, with higher selective pressure after widespread use, these mutations may spread and become fixed in a population. Second, the low prevalence of 3CLpro resistant mutations may be related to the low consumption of nirmatrelvir-ritonavir after clinical approval.31 Possible reasons for low consumption include the fear of viral rebound and side effects. With increasing use of 3CLpro inhibitors, resistance may emerge in the future. Third, antiviral resistance can occur suddenly. For example, oseltamivir was first introduced in late 1990s as an antiviral for influenza, but a major surge in oseltamivir resistance only occurred about 10 years later, when oseltamivir resistance for seasonal influenza A (H1N1) in 2008 suddenly jumped from <10% before the 2006–2007 season to almost 100% in the 2008–2009 season.32 Fourth, there is a growing concern that widespread use of molnupiravir, another widely used oral antiviral for the treatment of SARS-CoV-2 infection that acts by introducing viral genome mutations, may accelerate the mutation rate of circulating viruses.33,34
We found a difference in the geographical distribution of SARS-CoV-2 3CLpro mutations, with South America and Africa having the highest prevalence. The high prevalence in these two continents is because some mutations were present in widely circulating lineage in those continents. Specifically, G15S was present in Lambda variant which circulates widely in South America in 2021, while T21I was present in the B.1.1.318 which circulated widely in West Africa in early 2021.
There are several limitations in this study. First, there could be other mutations that confer 3CLpro inhibitor resistance which have not been described previously. In the study, we compiled a list of mutations that influence viral replication or 3CLpro activity, which are associated with reduced susceptibility to nirmatrelvir or ensitrelvir from previous studies and FDA reports published on or before January 11, 2023 (Tables 1 and 2). We only included the mutations that were previously experimentally investigated with fold change in viral replication and inhibition of 3CLpro activity. Some mutations, such as substitutions at G143,21 were not experimentally tested, therefore fold change results were not available. The effects of multiple mutations were also not considered in our study. Second, as the genomic surveillance is poorer in some countries, emergence of resistance in these places could be missed if the sequences and metadata are not published online. Third, as GISAID is an open public database, it is possible that the sequences retrieved can be duplicated or from the same person. Fourth, the 3CLpro sequences obtained from clinical specimens available on GISAID do not contain information regarding the viability of the viral strains. Our analysis would not be able to estimate the proportion of strains with mutations that are from infectious virus particles.
In conclusion, our study shows that the prevalence of protease inhibitor resistance remain low one year after the approval of nirmatrelvir-ritonavir. Early detection of the emergence of 3CLpro inhibitor resistance would require a global effort in genomic and phenotypic characterization with prompt dissemination of information.
Contributors
JDI and KKWT contributed to the conception and design of the study. JDI, AWHC, WMC, SMUA, RCYL contributed to data acquisition. JDI, SMUA, YS, RCYL and KKWT contributed to data analysis and interpretation. All authors critically revised the manuscript for important intellectual content. KKWT obtained the funding. KKWT supervised the research. JDI and KKWT have accessed and verified the data. All authors read and approved the final version of the manuscript.
Data sharing statement
The Python source code used in this study have been deposited at https://github.com/Jonathan-D-Ip/Global-prevalence-of-SARS-CoV-2-3CL-protease-mutations-associated-with-nirmatrelvir-or-ensitrelvir.
Declaration of interests
KKWT report collaboration with Sinovac, Sinopharm and Wantai BioPharm. Other authors declare no competing interests.
Acknowledgments
We gratefully acknowledge the authors originating and submitting laboratories of the SARS-CoV-2 genetic sequences and metadata made available through GISAID on which this research is based. This work was partly supported by the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Diseases and Research Capability on Antimicrobial Resistance for Department of Health of the Hong Kong, and the Emergency Key Program of Guangzhou Laboratory (EKPG22-01), and the Hong Kong Innovation and Technology Fund MPR/071/20X. We acknowledge funding received from private donors including the Shaw Foundation Hong Kong, Richard Yu and Carol Yu, May Tam Mak Mei Yin, Michael Seak-Kan Tong, Respiratory Viral Research Foundation Limited, Hui Ming, Hui Hoy and Chow Sin Lan Charity Fund Limited, Chan Yin Chuen Memorial Charitable Foundation, and Marina Man-Wai Lee.
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2023.104559.
Appendix A. Supplementary data
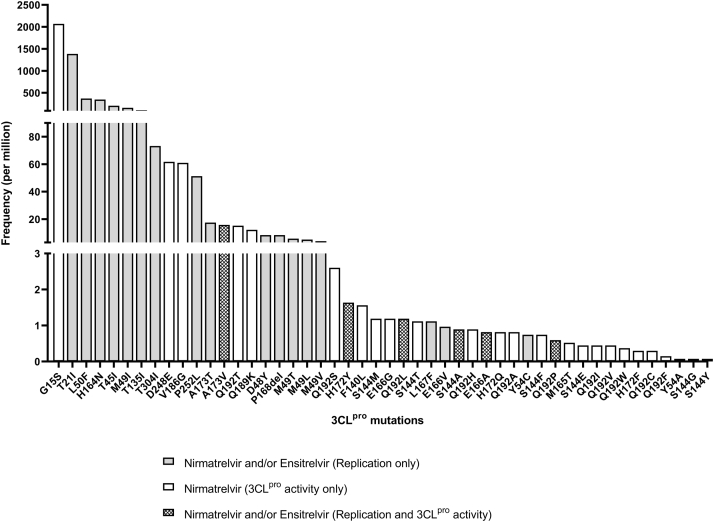
Supplementary Fig. S1.
Frequency of 3CLpro mutations (sequences per million) among sequences deposited onto GISAID up to 14th December 2022. All mutations listed in Tables 1 and 2 are shown.
Supplementary Fig. S2.
Frequency of (a) G15S and (b) T21I in selected SARS-CoV-2 lineages. It should be noted that G15S and T21I are present in the Variant of Interests B.1.1.318 and Lambda variant (C.37), respectively.
References
- 1.Msemburi W., Karlinsky A., Knutson V., Aleshin-Guendel S., Chatterji S., Wakefield J. The WHO estimates of excess mortality associated with the COVID-19 pandemic. Nature. 2023;613(7942):130–137. doi: 10.1038/s41586-022-05522-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen L.L., Abdullah S.M.U., Chan W.M., et al. Contribution of low population immunity to the severe Omicron BA.2 outbreak in Hong Kong. Nat Commun. 2022;13(1):3618. doi: 10.1038/s41467-022-31395-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hung I.F., Lung K.C., Tso E.Y., et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet. 2020;395(10238):1695–1704. doi: 10.1016/S0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Consortium W.H.O.S.T. Remdesivir and three other drugs for hospitalised patients with COVID-19: final results of the WHO Solidarity randomised trial and updated meta-analyses. Lancet. 2022;399(10339):1941–1953. doi: 10.1016/S0140-6736(22)00519-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J.L., Li Y.H., Wang L.L., et al. Azvudine is a thymus-homing anti-SARS-CoV-2 drug effective in treating COVID-19 patients. Signal Transduct Target Ther. 2021;6(1):414. doi: 10.1038/s41392-021-00835-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cao Z., Gao W., Bao H., et al. VV116 versus nirmatrelvir-ritonavir for oral treatment of Covid-19. N Engl J Med. 2023;388(5):406–417. doi: 10.1056/NEJMoa2208822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.To K.K., Sridhar S., Chiu K.H., et al. Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic. Emerg Microb Infect. 2021;10(1):507–535. doi: 10.1080/22221751.2021.1898291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammond J., Leister-Tebbe H., Gardner A., et al. Oral nirmatrelvir for high-risk, nonhospitalized adults with Covid-19. N Engl J Med. 2022;386(15):1397–1408. doi: 10.1056/NEJMoa2118542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sasaki M., Tabata K., Kishimoto M., et al. S-217622, A SARS-CoV-2 main protease inhibitor, decreases viral load and ameliorates COVID-19 severity in hamsters. Sci Transl Med. 2022 doi: 10.1126/scitranslmed.abq4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mukae H., Yotsuyanagi H., Ohmagari N., et al. Efficacy and safety of ensitrelvir in patients with mild-to-moderate COVID-19: the phase 2b part of a randomized, placebo-controlled, phase 2/3 study. Clin Infect Dis. 2022 doi: 10.1093/cid/ciac933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iketani S., Mohri H., Culbertson B., et al. Multiple pathways for SARS-CoV-2 resistance to nirmatrelvir. Nature. 2022 doi: 10.1038/s41586-022-05514-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelnabi R., Jochmans D., Donckers K., et al. Nirmatrelvir-resistant SARS-CoV-2 is efficiently transmitted in Syrian hamsters. bioRxiv. 2022 doi: 10.1101/2022.09.28.509903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Y., Gammeltoft K.A., Ryberg L.A., et al. Nirmatrelvir-resistant SARS-CoV-2 variants with high fitness in an infectious cell culture system. Sci Adv. 2022;8(51) doi: 10.1126/sciadv.add7197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anonymous Fact sheet for healthcare providers: emergency use authorization for Paxlovid (revised in 09/2022) 2022. https://www.fda.gov/media/155050/download Available at:
- 15.Iketani S., Hong S.J., Sheng J., et al. Functional map of SARS-CoV-2 3CL protease reveals tolerant and immutable sites. Cell Host Microbe. 2022;30(10):1354–13562.e6. doi: 10.1016/j.chom.2022.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heilmann E., Costacurta F., Moghadasi S.A., et al. SARS-CoV-2 3CL(pro) mutations selected in a VSV-based system confer resistance to nirmatrelvir, ensitrelvir, and GC376. Sci Transl Med. 2023;15(678) doi: 10.1126/scitranslmed.abq7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jochmans D., Liu C., Donckers K., et al. The substitutions L50F, E166A, and L167F in SARS-CoV-2 3CLpro are selected by a protease inhibitor in vitro and confer resistance to nirmatrelvir. mBio. 2023 doi: 10.1128/mbio.02815-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Y., Lewandowski E.M., Tan H., et al. Naturally occurring mutations of SARS-CoV-2 main protease confer drug resistance to nirmatrelvir. bioRxiv. 2022 doi: 10.1101/2022.06.28.497978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moghadasi S.A., Heilmann E., Khalil A., et al. Transmissible SARS-CoV-2 variants with resistance to clinical protease inhibitors. bioRxiv. 2022 doi: 10.1101/2022.08.07.503099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.de Oliveira V.M., Ibrahim M.F., Sun X., Hilgenfeld R., Shen J. H172Y mutation perturbs the S1 pocket and nirmatrelvir binding of SARS-CoV-2 main protease through a nonnative hydrogen bond. bioRxiv. 2022 doi: 10.1101/2022.07.31.502215. [DOI] [Google Scholar]
- 21.Lee J.T., Yang Q., Gribenko A., et al. Genetic surveillance of SARS-CoV-2 M(pro) reveals high sequence and structural conservation prior to the introduction of protease inhibitor Paxlovid. mBio. 2022;13(4) doi: 10.1128/mbio.00869-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shu Y., McCauley J. GISAID: global initiative on sharing all influenza data - from vision to reality. Euro Surveill. 2017;22(13) doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Rosuum G., Drake F.L., Jr. Python v3.9.12. 2023. https://www.python.org/ Available at:
- 24.McKinney W. Proceedings of the 9th Python in Science Conference. Vol. 445. 2010. Data structures for statistical computing in Python; pp. 51–56. [Google Scholar]
- 25.Harris C.R., Millman K.J., van der Walt S.J., et al. Array programming with NumPy. Nature. 2020;585(7825):357–362. doi: 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gazoni E., Clark C. Openpyxl - a Python library to read/write Excel 2010 xlsx/xlsm files. 2023. https://openpyxl.readthedocs.io/en/stable/
- 27.Manouana G.P., Nzamba Maloum M., Bikangui R., et al. Emergence of B.1.1.318 SARS-CoV-2 viral lineage and high incidence of alpha B.1.1.7 variant of concern in the Republic of Gabon. Int J Infect Dis. 2022;114:151–154. doi: 10.1016/j.ijid.2021.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morang'a C.M., Ngoi J.M., Gyamfi J., et al. Genetic diversity of SARS-CoV-2 infections in Ghana from 2020-2021. Nat Commun. 2022;13(1):2494. doi: 10.1038/s41467-022-30219-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Owen D.R., Allerton C.M.N., Anderson A.S., et al. An oral SARS-CoV-2 M(pro) inhibitor clinical candidate for the treatment of COVID-19. Science. 2021;374(6575):1586–1593. doi: 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- 30.Noske G.D., de Souza Silva E., de Godoy M.O., et al. Structural basis of nirmatrelvir and ensitrelvir activity against naturally occurring polymorphisms of the SARS-CoV-2 main protease. J Biol Chem. 2023;299(3) doi: 10.1016/j.jbc.2023.103004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kozlov M. COVID drug Paxlovid was hailed as a game-changer. What happened? Nature. 2023 doi: 10.1038/d41586-022-04576-6. [DOI] [PubMed] [Google Scholar]
- 32.Weinstock D.M., Zuccotti G. The evolution of influenza resistance and treatment. JAMA. 2009;301(10):1066–1069. doi: 10.1001/jama.2009.324. [DOI] [PubMed] [Google Scholar]
- 33.Fountain-Jones N.M., Vanhaeften R., Williamson J., et al. Antiviral treatments lead to the rapid accrual of hundreds of SARS-CoV-2 mutations in immunocompromised patients. medRxiv. 2022 doi: 10.1101/2022.12.21.22283811. [DOI] [Google Scholar]
- 34.Service R.F. Could a popular antiviral supercharge the pandemic? Science. 2023;379(6632):526. doi: 10.1126/science.adh0582. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.