Abstract
The timing of parturition is crucial for neonatal survival and infant health. Yet, its genetic basis remains largely unresolved. We present a maternal genome-wide meta-analysis of gestational duration (n = 195,555), identifying 22 associated loci (24 independent variants) and an enrichment in genes differentially expressed during labor. A meta-analysis of preterm delivery (18,797 cases, 260,246 controls) revealed seven associated loci and large genetic similarities with gestational duration. Analysis of the parental transmitted and nontransmitted alleles (n = 136,833) shows that 15 of the gestational duration genetic variants act through the maternal genome, whereas 7 act both through the maternal and fetal genomes and 2 act only via the fetal genome. Finally, the maternal effects on gestational duration show signs of antagonistic pleiotropy with the fetal effects on birth weight: maternal alleles that increase gestational duration have negative fetal effects on birth weight. The present study provides insights into the genetic effects on the timing of parturition and the complex maternal–fetal relationship between gestational duration and birth weight.
Subject terms: Genetics research, Genome-wide association studies
Maternal genome-wide analyses identify variants associated with gestational duration and preterm delivery. Maternal alleles positively associated with gestational duration exhibit negative fetal effects on birth weight, likely reflecting antagonistic pleiotropy.
Main
In humans, similar to mammals broadly, the timing of delivery is crucial for neonatal survival and health. Preterm delivery is the world-leading direct cause of death in neonates and children under five years of age1. Although the rate of neonatal mortality has substantially decreased in recent years, the reduction attributable to preterm delivery is one of the lowest among the major causes of mortality2. This fact partly reflects the relatively poor knowledge of the processes governing the timing of delivery in humans. Parturition may be initiated by a diversity of biological and mechanical pathways. Some of these are part of the physiological timing process, whereas others may override pregnancy maintenance with fail-safe mechanisms (for example, in the case of uterine infection)3. The diversity of the mechanisms has led to the conceptualization of preterm delivery as a syndrome4, with various pathophysiological processes contributing to its etiology. Both maternal and fetal genomes are involved in these mechanisms. Yet, genetic studies have identified only a handful of loci associated with the timing of parturition5,6.
Gestational duration is the major determinant of birth weight (that is, the longer the gestation, the heavier the newborn). At the same time, uterine load is one of the known triggers of parturition7, evidenced by half of twin pregnancies delivering preterm8. Both the maternal and fetal genomes contribute to birth weight as well, as revealed in recent genome-wide association studies (GWAS)9,10, and over evolutionary time may have even conflicted on gestational duration and birth weight, as proposed in the hypothesis of the genetic conflicts of pregnancy11. This hypothesis suggests that the maternal genome favors slightly shorter gestations and lower birth weight, whereas the fetal genome favors the opposite. Coadaptation theory, instead, suggests that maternal and fetal genomes may invest resources to achieve an optimal gestational duration or birth weight that increases fitness12. These known contributions, potential conflicts and coadaptation of gestational duration and birth weight may ultimately create a complex relationship between the two.
What and how distinct are the maternal genetic effects on gestational duration and preterm delivery? What is the relationship between fetal growth and gestational duration? Is there evidence suggesting maternal–fetal coadaptation on these traits? To address these questions, we conducted a GWAS meta-analysis of gestational duration and preterm and post-term delivery in >190,000 maternal samples with spontaneous onset of delivery. We further analyzed these results using the parental transmitted and nontransmitted alleles in >135,000 parent-offsprings.
Results
Genome-wide association analyses
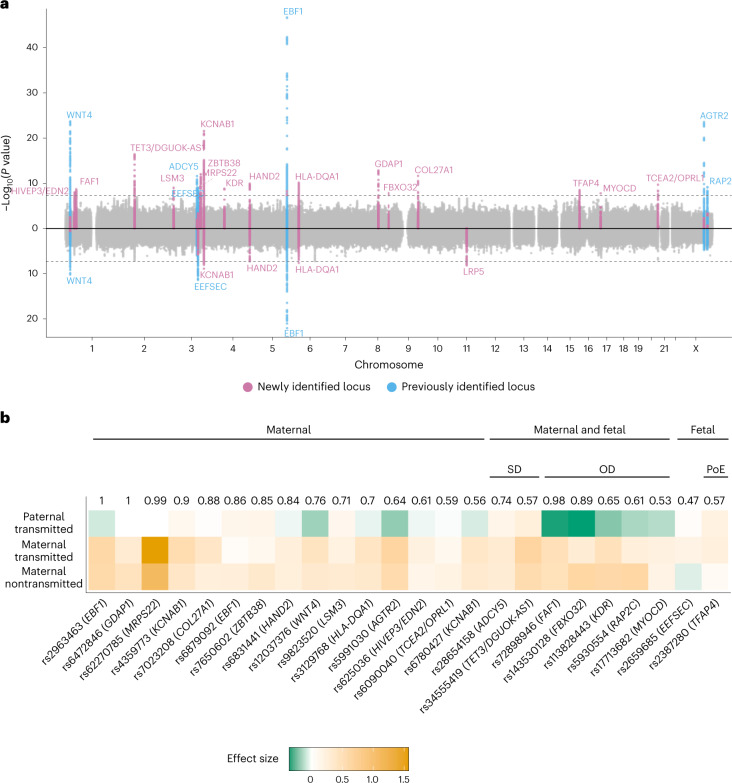
We conducted a GWAS meta-analysis of gestational duration in 195,555 women of recent European ancestry (Supplementary Table 1), a fourfold increase in sample size compared to the largest published maternal GWAS of gestational duration to date5. After quality control (QC), genetic variants at 22 loci were associated with gestational duration at genome-wide significance (Fig. 1, Supplementary Table 2 and Supplementary Fig. 1). Approximate conditional and joint (COJO) analysis revealed two conditionally independent signals at EBF1 and KCNAB1 gene regions. Sixteen of the loci did not overlap with any previously reported gestational duration-associated locus5. Effect sizes were relatively small, ranging from 7 (HIVEP3/EDN2) to 27 (MRPS22) hours of gestation per allele (average duration of gestation = 282 days, 40.3 weeks). Heterogeneity in the effect estimates was limited to loci previously identified (EBF1, WNT4, ADCY5, EEFSEC and AGTR2), likely due to winner’s curse13 (Supplementary Table 2 and Supplementary Fig. 2). Out-of-sample reanalysis of previously reported gestational duration-associated lead single-nucleotide polymorphisms (SNPs) (n = 6) showed that all four that were available after QC replicate at nominal significance (Supplementary Table 3). In addition, all six loci (±250 kb from lead SNP) replicated at suggestive evidence.
Fig. 1. GWAS of the timing of parturition and dissection of maternal–fetal effects.
a, Miami plot illustrating the GWAS for gestational duration (top) and preterm delivery (bottom). The x-axis shows the chromosome position and the y-axis the two-sided P-value of the fixed-effect inverse-variance weighted meta-analysis. The dashed line represents the genome-wide significance threshold (P = 5 × 10−8). Each genome-wide significant locus is labeled by its nearest protein-coding gene. b, Clustering of the effect origin for the index SNPs for gestational duration using transmitted and nontransmitted parental alleles (n = 136,833). Numbers depicted above the heatmap are the highest probability observed for that SNP, and group names define the cluster to which the highest probability refers to. The probabilities were estimated using model-based clustering. Heatmap represents effect size and effect direction for the parental transmitted and nontransmitted alleles. For comparison purposes, the maternal alleles with positive effects were chosen as reference alleles. Three major groups were identified according to the highest probability: maternal-only effect, fetal-only effect and maternal and fetal effect. Within variants with both maternal and fetal effects, two clusters were observed: same (SD) or opposite (OD) effect direction from maternal and fetal genomes. One of the fetal effects was further clustered as having a parent-of-origin effect (PoE), specifically, an effect from the maternal transmitted allele.
To prioritize candidate genes, we performed colocalization analysis14 with cis-expression quantitative trait loci (cis-eQTLs) in induced pluripotent stem cells15, endometrium16, uterus, vagina and ovary17 (Supplementary Table 4). cis-eQTLs for seven protein-coding (OPRL1, ZBTB38, RGS19, TET3, COL27A1, CRISPLD1 and ADCY5) and four non-coding genes colocalized with gestational duration. Furthermore, colocalization analysis with blood protein QTLs18 showed several trans associations: ZBTB38 with three proteins, and TCEA2/OPRL1 and WNT4 with one each. Particularly interesting are the associations with OPRL1 and POMC, which play a role in modulating nociception and pain perception; in vitro studies in tissues from pregnant rats and humans suggest that the administration of nociceptin inhibits uterine contractions, mediated by the OPRL1 receptors19,20.
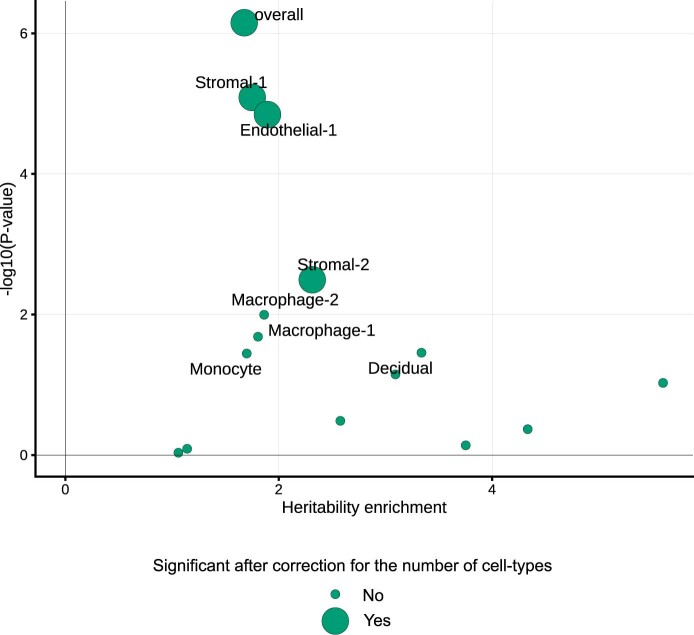
RNA tissue-specific enrichment of top genes highlighted the endometrium and other female reproductive and smooth muscle tissues (Supplementary Fig. 3), results further supported at the genome-wide scale using stratified linkage disequilibrium (LD)-score regression (Supplementary Fig. 4). Previous genetic studies have suggested a critical role of the decidua (endometrium) in the timing of parturition, indicating an effect early in pregnancy21. Using stratified LD-score regression, we show that the heritability of gestational duration is enriched in regions harboring genes differentially expressed during labor (enrichment = 1.7, P = 7.1 × 10−7; Extended Data Fig. 1)22, suggesting the SNPs associated with gestational duration may as well act during labor.
Extended Data Fig. 1. SNP-heritability enrichment of gestational duration for genes differentially expressed during labor in different cell types of the myometrium and overall.
LD-score regression was used to partition heritability and estimate the heritability enrichment for each cell type and overall. We calculated LD scores (European individuals from phase 3 of the 1000 Genomes project) for sets of genes differentially expressed at labor (± 100 kb) for each cell type separately and for the overall set of genes differentially expressed in the myometrium. Each dot represents a cell type, the x-axis shows the heritability enrichment, and the y-axis the -log10(P-value) of a two-sided test. Larger dots denote significant heritability enrichment after Bonferroni correction for multiple comparisons (that is, number of cell types; P-value < 0.05/15). See Online Methods for a cautionary note regarding the comparison of different cell-type enrichment P-values.
Stratified LD-score regression (Supplementary Fig. 5) revealed an enrichment in background selection, superenhancers, CpG content, H3K23ac and DNA methylation. Using the mosaic pipeline23, we confirm that gestational duration loci have diverse evolutionary histories, including evolutionary conservation, excess population differentiation and negative selection (Supplementary Fig. 6).
We also performed a GWAS meta-analyses of preterm delivery (controls, delivery between 39 and 42 gestational weeks, n = 260,246; cases, delivery <37 completed weeks, n = 18,797) and post-term delivery (controls, delivery between 39 and 42 gestational weeks = 115,307, cases >42 completed weeks, n = 15,972) (Fig. 1a, Supplementary Table 2 and Supplementary Figs. 7 and 8). We observed a lower number of associated loci: seven and one for preterm and post-term delivery, respectively. COJO analysis identified a secondary conditionally independent SNP associated with preterm delivery at the EBF1 gene region. We identified only one locus associated with preterm delivery (rs312777, P = 6.6 × 10−9) that showed weak evidence of association with gestational duration (P = 3.9 × 10−3).
We observed a modest genetic correlation (rg = −0.62; 95% confidence interval (CI) = −0.72, −0.51) between gestational duration and preterm delivery, suggesting similarities between the two phenotypes (Supplementary Figs. 9 and 10). Post-term delivery, instead, showed a perfect genetic correlation with gestational duration (rg = 1.17; 95% CI = 0.93, 1.41), suggesting no differences in the maternal genetic effects on such traits.
Resolving maternal–fetal effect origin
The genetic effects on pregnancy traits may be driven by two correlated genomes: the maternal and the fetal. To investigate whether the gestational duration signals originate in either or both genomes, we used phased genotype data to estimate the effects of the parental transmitted and nontransmitted alleles from 136,833 parent-offspring trios or mother-child duos (Fig. 1b, Supplementary Table 5 and Extended Data Fig. 2; the maternal samples of these duos/trios were part of the GWAS meta-analysis). Based on pattern similarity using Gaussian mixture model-based clustering10, SNPs were assigned to three large groups. Of the 24 index variants, 15 had the highest probability of a maternal effect, seven of both maternal and fetal effects (five with opposite effect directions, and the remaining two with the same direction), and two were grouped as having a fetal-only effect: the first, independent of the parent of origin (TFAP4, probability = 0.57), and the second limited to the maternal transmitted allele (EEFSEC). Caution should be taken when interpreting the latter considering the low probability (0.47).
Extended Data Fig. 2. Ternary plot representing the probabilities of having maternal, fetal or maternal, and fetal effect for each index SNP.
The sum of all probabilities for each index SNP is 1. Lines are colored according to the axis they belong to. All points in a horizontal line (green) have the same probability of ‘fetal-only effect’, points on a line (yellow) parallel to the right side of the triangle have the same probability of a ‘Maternal-only effect’, and lines (black) parallel to the left side of the triangle have the same probability of a ‘Maternal and fetal effect’. Probabilities were obtained using Gaussian Mixture models clustering using the effect size and standard error estimates of the parental transmitted and nontransmitted alleles (n = 136,833 parent-offsprings). While five different clusters were identified, the fetal effect was broken down into two groups (parent-of-origin and independent of parent-of-origin), and the maternal and fetal effects also into two groups (same or opposite maternal and fetal direction). For this figure, probability of a ‘Fetal only effect’ is the sum of the two groups with fetal effect, and ‘Maternal and fetal effect’ is the sum of the probabilities of the two clusters with maternal and fetal effects.
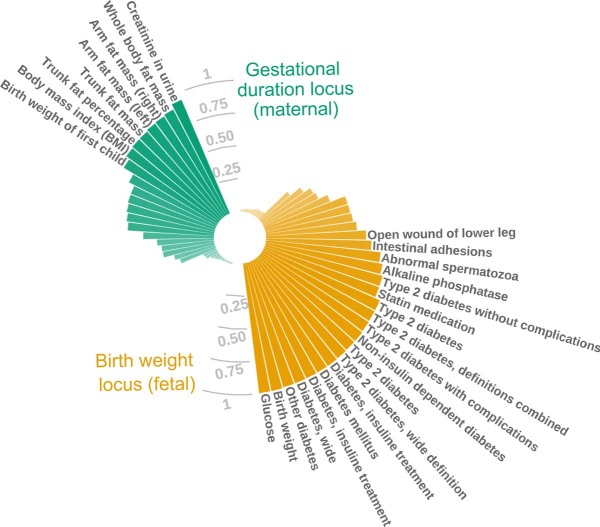
The index SNP at the ADCY5 locus (rs28654158) had both maternal and fetal effects on gestational duration with the same effect direction. Interestingly, a SNP also located in the first intron of ADCY5 harbors maternal and fetal effects on birth weight, but in opposite directions, attributed to the fetal insulin hypothesis9,10. The two index SNPs for gestational duration (rs28654158) and birth weight (rs11708067) are located 50 kb apart from each other and are in low LD (r2 < 0.2). The birth weight SNP, also implicated in diabetes, likely acts through ADCY5 (ref. 24), but it is unknown whether the gestational duration variant also acts through the same gene, although it colocalizes with ADCY5 gene expression in the uterus (Supplementary Table 4). Despite being physically close to each other, differences between the two loci are evident in the traits they colocalize with. The gestational duration locus also affects fat-mass-related traits, whereas the birth weight locus affects glucose-related ones (Extended Data Fig. 3).
Extended Data Fig. 3. Colocalization between the maternal effects on gestational duration (green) and fetal effects on birth weight (yellow) and other phenotypes from UK Biobank and FinnGen at the ADCY5 locus.
Posterior probability of colocalization between the maternal effect on gestational duration (rs28654158) and the fetal-only effect on birth weight (rs11708067) with traits from UK Biobank and FinnGen. Only traits with a posterior probability of colocalization ≥ 0.01 are plotted, and names are only shown if the posterior probability is > 0.5. Maternal locus on gestational duration was centered around rs28654158 (± 1.5 Mb), and the fetal locus on birth weight around rs11708067 (± 1.5 Mb).
The only fetal index SNP identified to date in a GWAS (rs7594852; minor allele frequency = 0.49; beta = 0.37 days; 95% CI = 0.22, 0.51)6 clustered as having a fetal-only effect (Supplementary Table 5, probability = 1), independent of the parent of origin (beta paternal transmitted allele = −0.42, P = 2.7 × 10−6).
Polygenic score of gestational duration and preterm delivery
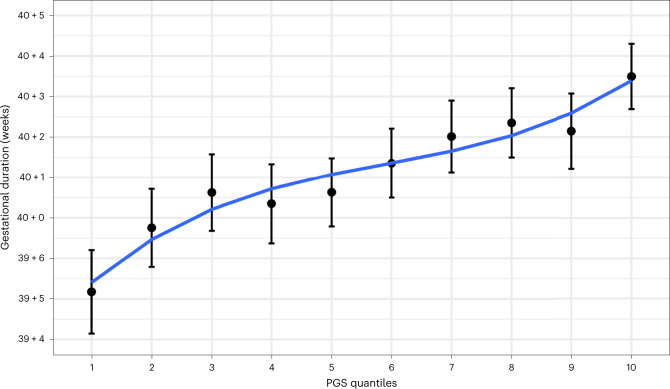
We built polygenic scores for gestational duration and preterm delivery using the corresponding GWAS results in the MoBa cohort (including the X chromosome) using LDpred2 (ref. 25) and estimated their effect on both traits. The polygenic score for gestational duration explains 2.2% of its variance (beta = 0.22 days per z-score; 95% CI = 0.02, 0.03; n = 3,943). The lowest decile had a mean gestational duration of 278 days (95% CI = 278, 279), whereas the highest decile had a mean of 283 days (95% CI = 282, 284) (Fig. 2). The polygenic score was also statistically significantly associated with preterm delivery (Supplementary Table 6 and Supplementary Fig. 11; odds ratio = 0.994; 95% CI = 0.990, 0.997) with an area under the curve of 0.61 (95% CI = 0.55, 0.67). For comparison, a polygenic score for preterm delivery was built using the same samples as above. This polygenic score was also significantly associated with preterm delivery (Supplementary Table 6 and Supplementary Fig. 11; odds ratio = 1.005, 95% CI = 1.001, 1.009), with effect estimate similar to that obtained for the gestational duration polygenic score (after matching the direction). This reflects the genetic similarity between gestational duration and preterm delivery.
Fig. 2. Polygenic prediction of gestational duration.
Mean (95% CI) gestational duration for each decile of the gestational duration polygenic score (n = 3,943). Only spontaneous deliveries were considered. PGS, polygenic score.
Pleiotropy between sex hormones and the timing of parturition
To examine the potential shared genetic basis between the timing of parturition and other traits, we estimated the genetic correlations between 14 female reproductive traits and the maternal effects on gestational duration and preterm delivery (Fig. 3). These estimates were generally comparable, with the latter being consistently higher. Calculated bioavailable testosterone (CBAT; rg = 0.40; 95% CI = 0.26, 0.54), testosterone (rg = 0.35; 95% CI = 0.19, 0.51) and sex hormone binding globulin (SHBG; rg = −0.16; 95% CI = −0.27, −0.06) in women were modestly genetically correlated with preterm delivery, whereas there was little genetic correlation with levels of the same hormones in men (Supplementary Table 7). We observed a positive genetic correlation between preterm delivery and the number of live births, and although this finding may be counterintuitive, it is in line with a positive genetic correlation reported between miscarriage and the number of live births26. The genetic correlation between preterm delivery and the number of live births was twice as high in cohorts where the women’s whole reproductive history was available (rg = 0.27; 95% CI = 0.11, 0.43) compared to cohorts based on a random pregnancy (rg = 0.13; 95% CI = 0.00, 0.26), indicating an increased probability of preterm delivery with an increasing number of live births. We also detected a negative genetic correlation with age at first birth and age at menopause.
Fig. 3. Genetic correlations between gestational duration and preterm delivery and other female reproductive traits.
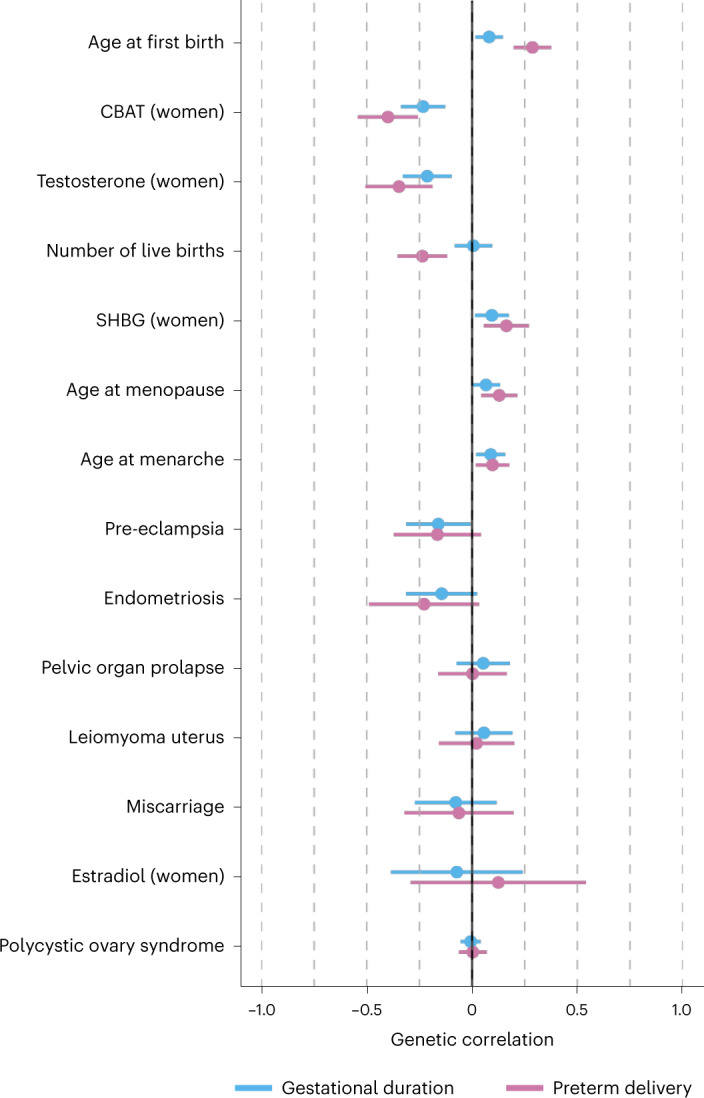
a, Genetic correlations between gestational duration (n = 195,555) and preterm delivery (18,797 cases, 260,246 controls) and other female reproductive traits were estimated using LD-score regression. Dots are the genetic correlation estimate, and error bars are the 95% CI. The direction of the genetic correlations with preterm delivery was flipped so that term deliveries were considered as cases and preterm deliveries as controls. Hence, the direction of the genetic correlations of preterm delivery matches that of gestational duration, providing a clear comparison of the 95% CI.
Genetic correlations can arise due to pleiotropy or due to a trait being causally upstream of the other. To distinguish between these situations, we used a latent causal variable (LCV)27 model between sex hormones and preterm delivery and gestational duration (Supplementary Table 8). We observed evidence for full or nearly full genetic causality of CBAT, testosterone and SHBG on preterm delivery (0.7 < GCP ≤ 0.8), but not on gestational duration (0.4 ≤ GCP < 0.5). In a two-sample Mendelian randomization analysis, the concentrations of these sex hormones (Supplementary Tables 9 and 10), including a set of variants that have consistent effects on testosterone, but no aggregate effects on SHBG28, were associated with gestational duration and preterm delivery. Although the MR-Egger intercept was not significantly different from 0 (Supplementary Table 10 and Extended Data Fig. 4), colocalization analyses across the genome confirmed that distinct variants underlie the associations for sex hormones and the timing of parturition (Supplementary Fig. 12).
Extended Data Fig. 4. Association between testosterone levels (women) and maternal effect on gestational duration.
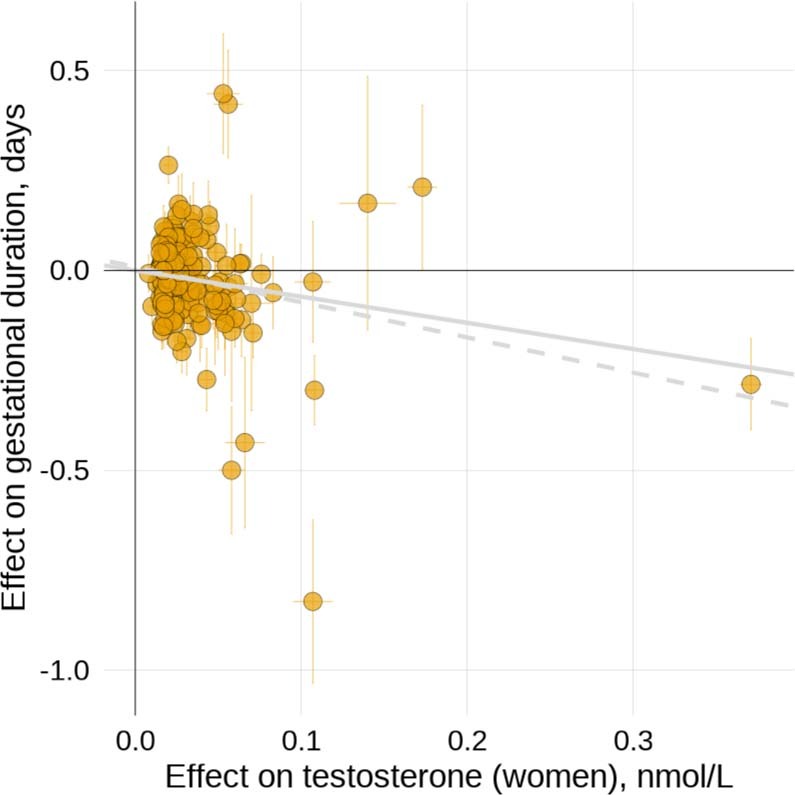
Scatterplot for two-sample Mendelian randomization analysis for the effect of testosterone in nmol/L (x-axis, independent of SHBG, n = 230,454) on gestational duration in days (y-axis, maternal effect, n = 195,555). Each dot represents one of the testosterone associated SNPs. Horizontal and vertical error bars represent the 95% CI. The gray line depicts the inverse-variance weighted method estimate, and the gray-dashed line the MR-Egger estimate.
Using the parental transmitted and nontransmitted alleles in individual-level parent-offspring data from Iceland and Norway (deCODE, MoBa and HUNT; n = 46,105 parent-offsprings; Supplementary Table 11), we observed a nominally significant association between the maternal nontransmitted alleles polygenic scores for CBAT and testosterone and gestational duration.
Testosterone and SHBG levels have a complex genetic link with the timing of parturition, likely explained by partial causality, as pointed out by the LCV analysis on gestational duration.
Gestational duration partially mediates maternal effects on birth weight
We sought to understand the genetic relationship between gestational duration and birth weight and how the interplay between the maternal and fetal genomes affect this relationship. We used published summary statistics of birth weight (<15% of samples adjusted for gestational duration) derived from two different models9: maternal-only effect (adjusted by fetal effects) and fetal-only effect (adjusted by maternal effects). These models were obtained using weighted linear modeling and provide unbiased estimates for the maternal and fetal effects, respectively. The fetal effects on gestational duration were obtained from a previously published GWAS6. The more recent GWAS meta-analysis of fetal growth10 had >40% of samples adjusted for gestational duration, which is the reason why we did not use it in this section.
The maternal effects on gestational duration are strongly correlated with those on birth weight (Supplementary Fig. 13; rg = 0.65; 95% CI = 0.54, 0.75). Conversely, neither the maternal (rg = −0.05; −0.15, 0.04) nor the fetal (rg = −0.02; 95% CI = −0.15, 0.11) effects on gestational duration were genetically correlated with the fetal-only effects on birth weight. We suggest the maternal effects on birth weight are at least partially mediated by gestational duration, whereas the effects of the fetus on birth weight are not.
We then tested the extent of this mediation. Using multitrait COJO analysis29, we conditioned the genetic effects on birth weight on the maternal effects on gestational duration. After conditioning, the maternal effects on birth weight changed substantially: the SNP heritability was reduced by 53% (P = 9.4 × 10−7; Supplementary Table 12), and the effect size of 87 suggestive SNPs decreased (Fig. 4a; median relative difference = −11%, Wilcoxon rank-sum test P = 1.3 × 10−8). Applying the same method on genome-wide significant variants classified with a maternal-only effect on birth weight9 provided very similar results (Supplementary Table 13 and Supplementary Fig. 14). This finding was further replicated using individual-level data by directly adjusting for gestational duration in the linear model on birth weight (using genotypes in Icelandic data and the maternal nontransmitted alleles in MoBa, Norway; Supplementary Table 13 and Supplementary Fig. 14). In contrast, for fetal effects on birth weight, conditioning on gestational duration did not change the effect estimates or the heritability (Fig. 4a and Supplementary Table 12 for results with 108 suggestive SNPs, and Supplementary Table 13 and Supplementary Fig. 14 with genome-wide significant variants classified as having a fetal-only effect9).
Fig. 4. Genetic relationship between gestational duration and birth weight.
a, Distribution of the relative difference in effect size before and after conditioning the effect on birth weight by the maternal effect on gestational duration using approximate multitrait COJO analysis. After conditioning, we split the genome into approximately LD-independent regions and selected the SNPs with the lowest P value on birth weight (P < 5 × 10−6) from each region (n SNPs maternal effect = 87; n SNPs fetal effect = 108). Fetal, pink; maternal, blue. b,c, Scatterplot for two-sample Mendelian randomization analysis for the maternal effect of gestational duration on birth weight (b, maternal effects; c, fetal effects). Each dot represents one of the gestational duration index SNPs. Effect sizes and standard errors (horizontal or vertical error bars) from the index SNPs for gestational duration derived from the maternal nontransmitted alleles were obtained from the meta-analysis of parent-offspring data (n = 136,833). The maternal-only and the fetal-only effects on birth weight were extracted from a previous GWAS meta-analysis (n = 210,248 and 297,356, respectively). The x-axis shows the SNP effect of the maternal nontransmitted alleles on gestational duration (days), and the y-axis the effect on birth weight (z-scores). Horizontal and vertical error bars represent the standard error. The solid line depicts the inverse-variance weighted method estimate, and the dashed line the MR-Egger estimate. Colors represent the clustering of the SNP effects on gestational duration, performed using model-based clustering.
In summary, although the maternal effects on birth weight are partially driven by gestational duration, we found no evidence for this for the fetal effects on birth weight.
The maternal genome drives the association between gestational duration and birth weight
It is widely accepted that longer gestations lead to heavier newborns. Here, we sought to obtain causal estimates of the effect of gestational duration on birth weight.
We used the index SNPs from our discovery GWAS and the effect estimates from the maternal nontransmitted alleles as genetic instruments in a two-sample Mendelian randomization analysis (Fig. 4b and Supplementary Fig. 15) on the maternal-only effects on birth weight (derived using a weighted linear model9). The maternal nontransmitted gestational duration-increasing alleles were associated with higher birth weight (beta = 0.06 z-scores per day; 95% CI = 0.05, 0.08; P = 1.7 × 10−16). The estimated effect (approximately 23 g per day) is concordant with the phenotypic association between gestational duration and birth weight (25 g per day in 18,452 samples from the MoBa cohort). We observed no effect from the paternal transmitted gestational duration-increasing alleles on birth weight. The LCV model confirmed a full or nearly full causal (GCP = 0.6, P = 0.002; Supplementary Table 8) effect of gestational duration on birth weight.
Maternal effects on gestational duration and fetal effects on birth weight exhibit signs of antagonistic pleiotropy
First, we evaluated the impact of fetal growth on gestational duration by instrumenting fetal growth using 68 SNPs with fetal-only effect on birth weight (n = 35,280 and 48,741 parent-offsprings; Supplementary Table 14)9. Higher paternally transmitted birth weight score was associated with shorter duration of gestation, and the estimated effect was larger when estimated using the last menstrual period (beta = −1.9 days per z-score, P = 4.0 × 10−4) than ultrasound. This result supports previous evidence showing faster fetal growth is associated with shorter duration of gestation30. To investigate whether this was due to antagonistic pleiotropy between the fetal effects on birth weight and the maternal effects on gestational duration, we assessed the relation between birth weight-increasing alleles and maternal effects on gestational duration. The fetal birth weight-increasing alleles were not associated with maternal effects on gestational duration (Supplementary Table 15), suggesting that the results presented above are likely not due to antagonistic pleiotropic effects.
Next, we used summary statistics to investigate potential pleiotropy between the genetic effects on gestational duration and fetal birth weight. Using methods borrowed from Mendelian randomization analysis, we evaluated the association between the maternal gestational duration-increasing alleles and the fetal effects on birth weight. We observe that the alleles that increase gestational duration through a maternal effect tend to reduce birth weight through a fetal effect (Fig. 4c and Supplementary Table 15). Interestingly, this effect was not limited to the maternal transmitted alleles (beta = −0.02 z-scores per day; 95% CI = −0.03, −0.01; P = 3.4 × 10−4) but was also observed for the maternal nontransmitted gestational duration-increasing alleles (beta = −0.01 z-scores per day; 95% CI = −0.02, −0.01; P = 6.2 × 10−3). The paternal transmitted gestational duration-increasing alleles were not associated with fetal-only effects on birth weight (Supplementary Table 15).
Discussion
The timing of parturition is crucial for neonatal survival and health. Yet, discovery of maternal and fetal genetic effects lags behind that of other pregnancy traits such as birth weight9 and fetal growth10. In this GWAS meta-analysis of parturition timing, we identified 17 loci not previously reported, one of which was more strongly linked to preterm delivery than to gestational duration. The results support large similarities in the maternal genetic effects on gestational duration and preterm delivery. By including parent-offspring data with a similar sample size to that of the discovery GWAS, we were able to discern maternal from fetal effects with high certainty for most index SNPs. Finally, the results show a complex genetic relationship between the maternal and fetal genomes on gestational duration and birth weight.
Our understanding of the molecular signals governing the timing of parturition in humans has not advanced significantly. Previous genomic evidence suggests a critical role of the decidua21, denoting an effect on the timing of parturition as early as implantation. We report that the SNP heritability of gestational duration is enriched in genes differentially expressed during labor in the myometrium. We suggest the maternal effects on the duration of gestation may as well act during labor, for instance, by inhibiting uterine contractions. Genetic studies of gestational duration may prove useful in the discovery of drug targets as tocolytic agents or for labor induction. At the same time, the genetic effects on gestational duration and preterm delivery are largely similar; this is opposed to the heterogeneity observed at the phenotypic and transcriptomic levels31,32. As an example, although the polygenic score of gestational duration is still inadequate for clinical use, it had a similar effect on preterm delivery as a polygenic score of preterm delivery itself.
Gestational duration is the major determinant of birth weight. Although the maternal genome affects offspring birth weight through many different causal pathways (for example, maternal glucose levels9,10), the effects are partly mediated by gestational duration. This has implications for the interpretation of GWAS of birth weight and downstream analyses, such as Mendelian randomization. In contrast, the fetal genetic effects on birth weight are not mediated by gestational duration, suggesting the fetal genome mainly acts on birth weight by modulating fetal growth. Interestingly, the maternal gestational duration-increasing alleles have negative fetal effects on birth weight, likely reflecting antagonistic pleiotropy. The opposite was not true; fetal birth weight-increasing alleles were not associated with maternal effects on gestational duration. We speculate that the fetal effects on birth weight have likely co-adapted to increase the fitness of the fetus in pregnancies genetically predisposed to a shorter duration. It has been suggested that both gestational duration and birth weight are under balancing selection, with intermediate values of these traits having highest fitness3,33. As exemplified here, this could lead to antagonistic pleiotropy favoring the coadaptation of maternal and fetal effects to attain optimal gestational duration and birth weight12.
The presented results have several limitations. First, we analyzed data from participants of European ancestry. Over 70% of the samples were obtained from Nordic countries, with genotype data linked to the Medical Birth Registers; in these countries, the preterm delivery rate is one of the lowest in the world1. Studying diverse ancestries would propel the identification of novel loci associated with gestational duration and aid in fine-mapping efforts, as has been previously shown for other traits34. Second, to understand the relationship between gestational duration and birth weight, we used summary statistics from a previously published birth weight GWAS that was partially adjusted for gestational duration (<15% of samples) and excluded preterm deliveries, which is likely to affect our analyses by reducing their power. Third, we assumed a causal association between gestational duration and birth weight. Although this is known to be true to some extent (that is, longer gestations are linked to heavier newborns), pleiotropy between gestational duration and birth weight could be very well at play. Fourth, phenotypic heterogeneity between cohorts (for example, gestational duration estimation method) may have hindered the identification of additional signals.
In conclusion, the present results provide evidence of large genetic similarities between gestational duration and preterm delivery and further our understanding of the complex relationship between gestational duration and birth weight. Particularly, we show that the maternal effects on birth weight are largely driven by gestational duration and that the maternal and fetal genomes have antagonistic pleiotropic effects on gestational duration and birth weight.
Methods
Phenotype definition
In this study, we included pregnancies with a singleton live birth and a spontaneous onset of delivery: medically initiated deliveries (either by induction or planned cesarean section) were excluded or part of controls for preterm delivery. Gestational duration in days was estimated using either the last menstrual period date or ultrasound. We excluded pregnancies lasting <140 days (20 completed weeks) or >310 days (44 completed weeks), as well as women with health complications prior to or during pregnancy and congenital fetal malformations. Spontaneous preterm delivery was defined as a spontaneous delivery <259 days (37 completed gestational weeks) or by using the ICD-10 O60 code, and controls as a delivery occurring between 273 and 294 days (39 and 42 gestational weeks). Post-term delivery was defined as a delivery occurring >294 days (42 completed weeks) or ICD-10 O48 code, and controls as a spontaneous delivery between 273 and 294 days (39 and 42 gestational weeks). Given the perfect genetic correlation between gestational duration and post-term delivery GWAS, and the small power of the latter, all downstream analyses are focused on gestational duration and preterm delivery.
Study cohorts and individual-level GWAS
This study consists of cohorts participating in the Early Growth Genetics (EGG) Consortium and the Norwegian Mother, Father and Child Cohort study (MoBa)35, deCODE genetics10, Trøndelag Health Study (HUNT)36, Danish Blood Donor Study (DBDS)37, the Estonian Genome Center of the University of Tartu (EGCUT)38 and summary statistics from FinnGen39 and from a previous GWAS of gestational duration and preterm delivery performed using 23andMe data5. A total of 18 different cohorts (Supplementary Table 1) provided GWAS data under an additive model for meta-analysis for the maternal genome, resulting in 195,555 samples for gestational duration, 276,218 samples for preterm delivery (n cases = 18,797) and 131,279 samples for post-term delivery (n cases = 15,972) of recent European ancestries (indicated by principal component analysis). For binary outcomes (preterm and post-term deliveries), only cohorts with an effective sample size >100 were included. Detailed description of the cohorts included can be found in the Supplementary Note. All study participants provided a signed informed consent, and all research studies were approved by the relevant institutional ethics review boards (Supplementary Note).
Each individual cohort applied specific QC procedures, data imputation and analysis independently following the consortium recommendations. Unless more stringent, samples were excluded if genotype call rate <95%, autosomal mean heterozygosity >3 standard deviations from the cohort mean, sex mismatch or major recent ancestry was other than European (HapMap central European). Genetic variants were excluded if genotype call rate <98%, Hardy-Weinberg equilibrium P value < 1 × 10−6 or minor allele frequency <1%. Reference panels for imputation were either 1000 Genomes Project40, Haplotype Reference Consortium41, 10KUK or a combination of one of the mentioned reference panels and own whole-genome sequencing data (deCODE, HUNT, DBDS and FinnGen). Each individual cohort performed a GWAS using an additive linear regression model adjusted for, at least, genetic principal components or relationship matrix on autosomal chromosomes and chromosome X. Summary statistics for each individual cohort were stored centrally and underwent QC procedures before meta-analysis (Supplementary Note).
Meta-analysis of GWASs
After QC, individual-cohort GWAS summary statistics were pooled using fixed-effects inverse-variance weighted meta-analysis with METAL42 without genomic control correction. We also performed an analysis of heterogeneity of effects (Supplementary Table 2; I2 statistic). After meta-analysis, we removed genetic variants reported in less than half the number of available samples for each phenotype, resulting in 9-10 million genetic variants. For example, the variant observed in the largest number of samples for gestational duration was available in 195,555 individuals; only variants reported in at least 97,778 were kept. Genomic inflation factors were low for all three phenotypes (Supplementary Table 16; gestational duration λ = 1.14, preterm delivery λ = 1.08 and post-term delivery λ = 1.05). LD-score regression intercepts were substantially lower than genomic inflation factors, suggesting that the inflation in test statistics was mostly due to polygenicity (Supplementary Table 16). Test statistics were not further adjusted for genomic control for any of the phenotypes. If not otherwise stated, all analyses presented in this study are two-sided tests.
Initially, we naively defined independent loci based on physical distance, where SNPs within 250 kb from the index SNP were considered to be at the same locus. Novel loci were defined as loci not overlapping previously reported gestational duration loci in the largest GWAS performed to date5. Finally, we used conditional analysis to resolve independent loci (see below).
Conditional analysis
We looked for conditionally independent associations within each locus using approximate conditional and joint (COJO) analysis43 implemented in Genome-wide Complex Trait Analysis (GCTA) software44. We ran a stepwise model selection (-cojo-slct) to identify conditionally independent genetic variants at P < 5 × 10−8 for each of the genome-wide significant loci (using a radius of 1.5 Mb from the index SNP). Overlapping loci were merged into a single locus (only two loci overlapped, at 3q23). LD between genetic variants was estimated from 19,092 maternal samples from the Norwegian Mother, Father and Child Cohort, after excluding variants with imputation INFO score <0.4. We converted the reference panel from BGEN files to hard-called PLINK binary format (.bed). As per default in COJO, genetic variants >10 Mb apart were assumed to be in complete linkage equilibrium.
Gene prioritization
To prioritize genes at the gestational duration loci identified, we set the baseline as the nearest protein-coding gene to the index SNP at each independent locus. Although naive, this approach has been consistently shown to outperform other single metrics for locus-to-gene mapping45,46. Next, we performed colocalization analysis for cis-eQTLs in 1,367 human induced pluripotent stem cell lines from the i2QTL resource (±250 kb from gene start and stop position)15, endometrium (± 250 kb from gene start and stop position)16 and uterus, vagina and ovary from GTEx (±1 Mb around transcription start site)17. None of the variants we identified were in LD (r2 > 0.6) with missense variants. To complement the prioritization of genes, we queried each of the index SNPs for blood protein QTLs18 (both in cis and trans). For all index SNPs that were protein QTLs (P < 5 × 10−6), we performed colocalization analyses (±1.5 Mb around the index SNP). We excluded the HLA region due to its large pleiotropic effects.
Colocalization
We utilized genetic colocalization to identify pleiotropic effects between gestational duration and expression and protein quantitative trait locus (see Gene prioritization) and with other female and reproductive traits. To this end, we applied COLOC14, which evaluates, in a Bayesian statistical framework, whether a single locus from two different phenotypes best fits a model where the associations are due to a single shared variant or distinct variants in close LD (Supplementary Note).
Prior probabilities for each for the non-null hypotheses were set as suggested by Wallace (prior probabilities that a random SNP in the loci is associated with phenotype A, phenotype B, or both phenotypes, 1 × 10−4, 1 × 10−4, and 5 × 10−6, respectively), which are considered more conservative than the ones set by default47. Strong evidence of colocalization was defined as a posterior probability of colocalization >0.9.
Enrichment analysis
We tested for enrichment based on top loci and genome-wide using partitioned LD-score regression. To test for overrepresentation in tissue-specific RNA expression (Human Protein Atlas, RNA consensus tissue gene data)48, a Wilcoxon rank-sum test was performed on normalized RNA for genes within our set (above-mentioned) and all other genes. Significance for this test was set at Bonferroni correction for the number of tissues (P < 0.05/61), and suggestive evidence at P < 0.1/61. At the genome-wide level, we performed partitioned heritability using LD-score regression to test for enrichment in 97 different annotations49,50, tissue-specific RNA expression using 205 different tissues/cell types51, using precomputed partitioned LD-scores for subjects of recent European ancestry (baseline-LD model v2.2) and for enrichment in regions harboring genes differentially expressed during labor in single cells from myometrium22.
Genetic correlations
We estimated genetic correlations by performing LD-score regression52 locally using precomputed LD-scores from 1000 Genomes Project samples of recent European ancestry. The MHC region (chr6:28477797-33448354) was removed prior to running LD-score regression.
Resolving effect origin
To classify the identified index SNPs for gestational duration as having maternal, fetal, or maternal and fetal origin, we performed an association analysis using the parental transmitted and nontransmitted alleles on gestational duration. We used phased genotype data (that is estimated haplotypes) in parent-offsprings or mother-child duos to infer the parent-of-origin of the genotyped/imputed alleles as previously described30. Once the transmitted allele was identified, the nontransmitted maternal allele was extracted. Briefly, parental origin of each allele was inferred using genotypes of relatives, reference cohort data, or distributions of genotypes within the cohort and LD measurements. Different methods were used for phasing in each of the cohorts providing data for this analysis10,53–56 (Supplementary Table 17). Details of the phasing strategy used by each cohort are described in Supplementary Note.
For each index SNP, we fit the following linear regression model:
where MnT and MT refer to the maternal nontransmitted and transmitted alleles respectively, and PT refers to the paternal transmitted alleles. The latter is interpreted as a fetal-only genetic effect, whereas the effect of the maternal nontransmitted allele is a maternal-only genetic effect. We first estimated the effects of the index SNPs in each birth cohort separately; effect sizes were then combined through fixed-effect meta-analysis, totaling a sample size of 136,833 (Supplementary Note and Supplementary Table 17). To classify the identified genetic variants into classes with similar patterns of effect, we used model-based clustering10. Variants were clustered based on estimated effects of the transmitted and nontransmitted parental alleles into five clusters. Two clusters assume fetal effect only, one with effect independent of parent of origin, and one where the effect is limited to the maternally transmitted allele; a cluster with maternal effect only; and two clusters with both maternal and fetal effects, either in opposite or same direction.
Locus pleiotropy at 3q21
After identifying locus pleiotropy between the maternal effect on gestational duration and the fetal-only effect on birth weight at the ADCY5 gene region, we set out to investigate differences between the two top SNPs in their colocalization with other traits. Phenome-wide colocalization for the two regions was performed using summary statistics from FinnGen (data freeze 5) and Pan UK Biobank data (https://pan.ukbb.broadinstitute.org, in participants of recent European ancestry; Supplementary Note).
Female reproductive traits
We obtained summary statistics for several female reproductive traits from different sources (minimum sample size 10,000). We included summary statistics from the following traits: miscarriage26, gestational duration (fetal genome)6, age at first birth, age at menarche (Neale lab, http://www.nealelab.is), age at menopause57, number of live births (Neale lab, http://www.nealelab.is), testosterone58, CBAT58, SHBG58, estradiol (women, Neale lab, http://www.nealelab.is), pelvic organ prolapse (FinnGen), polycystic ovary syndrome (59 and FinnGen), endometriosis (Neale lab, http://www.nealelab.is), leiomyoma uterus (FinnGen) and pre-eclampsia60. For polycystic ovary syndrome, we meta-analyzed summary statistics from the largest published GWAS59 and FinnGen. We estimated genetic correlations between gestational duration and preterm delivery and these traits, and latent causal variable analysis between sex hormones (testosterone, CBAT and SHBG) and gestational duration and preterm delivery. We further explored causality using two-sample Mendelian randomization and inspected whether the effects originated in the maternal or the fetal genome (see below, ‘Mendelian randomization’). Finally, when one trait is causally upstream of the other, it is expected that the two traits would share a causal variant at some of the trait-associated loci. To test for this at the genome-wide scale, we performed colocalization analysis between sex hormones and gestational duration and preterm delivery using approximately LD-independent regions61.
Gestational duration and preterm delivery polygenic scores
To obtain an independent sample for training and validation of a polygenic score, the meta-analyses for gestational duration were rerun, excluding the MoBa cohort. These new meta-analysis results were used as the base data sets to calculate the polygenic scores. After applying the same exclusion criteria as used for the study samples in the meta-analysis, and removing duplicated samples and those with a kinship of greater than 0.125, the MoBa cohort was randomly split, using 80% (n = 15,768) as the training cohort and the remaining 20% (n = 3,942) as the validation cohort. LDpred2 was used for the calculation of the polygenic scores25. A description of polygenic score training can be found in Supplementary Note.
Polygenic score validation
We constructed polygenic scores converted to z-scores to enable comparison of the gestational duration and the preterm delivery polygenic scores. To test the performance of the polygenic score, a linear regression was conducted for gestational duration by the polygenic score. A second model was used that adjusted for five principal components and genotyped batch. R2 was calculated for the models to quantify variance explained.
The utility of the polygenic score for the prediction of preterm delivery was also assessed. Gestational duration was dichotomized into preterm delivery (<37 weeks) or full term (≥39 weeks and <41 weeks). Two models were analyzed, one assessing just the polygenic score and a second adjusting for five principal components and genotype batch. Receiver operating characteristic, area under the curve were calculated for each model and used as assessment of diagnostic accuracy.
Mendelian randomization
We performed Mendelian randomization to study the effects of gestational duration (maternal) on birth weight (maternal) and the effects of fetal growth (fetal effect on birth weight) and sex hormones on gestational duration.
To study the effect of gestational duration on birth weight, we employed two-sample Mendelian randomization. The 24 index SNPs (22 autosomal SNPs) from the present gestational duration meta-analysis and the effect sizes from the parental transmitted and nontransmitted alleles analysis were used to instrument gestational duration. Birth weight was instrumented using summary statistics from a previous GWAS of offspring’s birth weight with minimal adjustment by gestational duration (<15% of samples)9.
We assessed the effect of sex hormones (testosterone, SHBG and CBAT) on gestational duration using two-sample Mendelian randomization and instrumenting the hormones using a polygenic score for the parental transmitted and nontransmitted alleles. For each sex hormone, we obtained a list of independent SNPs genome-wide associated with these traits (Supplementary Table 9) by performing GWAS clumping (r2 > 0.001) using the following PLINK command:
plink–bfile <1000 Genomes > –clump {GWAS summary statistics}–clump-r2 0.001–clump-kb 1000–clump-p1 5e-8–clump-p2 1e-5.
We also used a set of SNPs associated with testosterone, but with no aggregated effects on SHBG, as clustered in28. Such variants were used as instrumental variables in the two-sample Mendelian randomization analysis and to construct the polygenic score for the parental transmitted and nontransmitted alleles. The current meta-analysis results were employed as outcome for the two-sample Mendelian randomization analysis (inverse-variance weighted and MR-Egger). We subsequently constructed the polygenic score for the maternal transmitted and nontransmitted alleles and the paternal transmitted alleles in 46,105 parent-offsprings from Iceland and Norway. We estimated the effects of the maternal nontransmitted (MnTPGS) and transmitted (MTPGS) and paternal transmitted (PTPGS) alleles polygenic score using the following linear model:
Again, effects from each of the three data sets (Iceland, MoBa and HUNT) were combined using fixed-effect inverse-variance weighted meta-analysis.
To understand the impact of fetal growth on gestational duration, we used individual genetic data from 35,280 (ultrasound-gestational duration) and 48,741 (last menstrual period-gestational duration) parent-offsprings from Iceland, the MoBa cohort and HUNT. To instrument fetal growth, we used 68 SNPs with fetal-only effect on birth weight as classified in Warrington et al. using Structural Equation Modeling9. Based on these 68 SNPs, we constructed a fetal growth polygenic score for the parental transmitted and nontransmitted alleles and regressed these on gestational duration (estimated by ultrasound or last menstrual period, separately). We estimated the effects of the maternal nontransmitted (MnTPGS) and transmitted (MTPGS) and paternal transmitted (PTPGS) alleles polygenic scores as above.
Effect estimates from each of the three data sets (Iceland, MoBa and HUNT) were pooled using fixed-effects inverse-variance weighted meta-analysis.
Multitrait conditional analysis
GCTA was used to perform bi-directional multitrait COJO (mtCOJO)29 analysis using summary statistics. The gestational duration GWAS was conditioned on the birth weight GWAS and vice versa (Supplementary Note), using birth weight summary statistics from the largest GWAS meta-analysis of birth weight9. We did not condition on the fetal effects on gestational duration due to a lack of power in the fetal GWAS6.
Maternal–fetal pleiotropy on gestational duration and birth weight
We further investigated what are the fetal effects on birth weight for the maternal gestational duration-increasing alleles, and the maternal effects on gestational duration for the fetal birth weight-increasing alleles. To study this, we borrowed the inverse-variance weighted analysis from Mendelian randomization, but using the effects of two distinct genomes, the maternal and fetal. We caution that this should not be interpreted under a causal framework.
To understand what the maternal gestational duration-raising alleles do to birth weight when present in the fetus, we used the effect sizes and standard errors of the parental transmitted and nontransmitted alleles for the 22 autosomal index SNPs on gestational duration and assessed its effects on the same SNPs with a fetal-only effect on birth weight. To understand what the fetal birth weight-raising alleles do to gestational duration when present in the mother, we used the effect sizes and standard errors of 68 autosomal SNPs associated with fetal effects on birth weight and the effect sizes and standard errors from the current maternal GWAS of gestational duration.
Evolutionary analysis
To examine the evolutionary history of the regions identified in the GWAS meta-analysis, we ran the significant variants through the MOSAIc pipeline23. This pipeline is designed to detect enrichment in evolutionary signals using a variety of sequence-based metrics of selection (Supplementary Note).
Variant annotation
Variants were annotated using Ensembl’s Variant Effect Predictor (hg19) command line tool62. Physical coordinates of protein-coding genes were obtained from the UCSC Table Browser63, and were matched to the index SNPs using bedtools v2.29.2 (ref. 64).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Online content
Any methods, additional references, Nature Portfolio reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at 10.1038/s41588-023-01343-9.
Supplementary information
Supplementary Note and Supplementary Figures 1–16.
Supplementary Tables 1–17.
Acknowledgements
B.J. received funding from The Swedish Research Council, Stockholm, Sweden (2015-02559 and 2019-01004), The Research Council of Norway, Oslo, Norway (FRIMEDBIO #547711, #273291) and March of Dimes (#21-FY16-121). Research reported in this publication (B.J., G.Z. and R.M.F.) was supported by the Eunice Kennedy Shriver National Institute Of Child Health & Human Development of the National Institutes of Health under Award Number R01HD101669. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. G.-H.M. has received funding from the Norwegian Diabetes Association and Nils Normans minnegave. G.-H.M. is supported by the Norwegian Research Council (postdoctoral mobility research grant 287198). M.C.B.’s contribution to this work was supported by a UK Medical Research Council (MRC) Skills Development Fellowship (MR/P014054/1) and a University of Bristol Vice-Chancellor’s fellowship. M.C.B. and D.A.L. are supported by the British Heart Foundation (AA/18/7/34219) and work in a Unit that receives funding from the University of Bristol and UK Medical Research Council (MRC) (MC_UU_00011/6). M.V. is supported by the Research Council of Norway (project #301178). D.A.L. is supported by a British Heart Foundation Chair (CH/F/20/90003). S.F.A.G. is supported by Daniel B. Burke Chair for Diabetes Research and NIH Grant R01 HD056465, IDF to CAG center from CHOP; CHOP’s Endowed Chair in Genomic Research. Funding for T.L. was provided by the European Regional Development Fund and the programme Mobilitas Pluss (MOBTP155). B.M.S. is a core member of the NIHR Exeter Clinical Research Facility. R.M.F. and R.N.B. were funded by a Wellcome Trust and Royal Society Sir Henry Dale Fellowship (WT104150). R.M.F. is funded by a Wellcome Trust Senior Research Fellowship (WT220390). B.F. was supported by the Oak Foundation. L.B. is a senior research scholar from the Fonds de la recherche du Québec en santé (FRQS) and member of the CR-CHUS, a FRQS-funded Research Center. E.O. has received funding from the US National Institutes of Health. D.W. is funded by the Novo Nordisk Foundation (NNF18SA0034956, NNF14CC0001, NNF17OC0027594). Additional funding statements for each cohort are available in the Supplementary Note.
Extended data
Author contributions
The core working group comprised P.S.-N., C.F., V.S., M.V., J.C., P.N., L.J.M., R.M.F., S.J., G.Z., B.J. The following authors performed analyses in their respective cohorts: P.S.-N., C.F., M.V., J.C., T.L., A.L.L., A.A., D.W., B.B., L.S., M.C.B., A.M., M.W., F.L., C.B., C.A.W., G.-H.M., R.N.B., J.P.B., G.T., O.T., G.S., H.X., D.F.G., S.L.R.-S., D.S. B.F. Individual cohort designers and principal investigators included V.S., J.B., J.J., A.M., M.E.G., E.A.N., E.H., S.M.W., R. Menon, M. Melbye, W.L., L.B., E.O., A.R., R.T.L., K.T., M.H., T.J., H.H., B.M.S., L.S., M. Merialdi, D.S., H.U., C.P., M.N., J.A.C., A.H.S., P.M., O.A.A., U.T., S.F.A.G., E.Q., C.E.P., M.-F.H., G.M.H., M.I.M., D.A.L., H.S.N., R. Mägi, K.H., K.S., B.F., L.J.M., R.M.F., S.J., G.Z. and B.J. Sample collection, phenotyping and/or genotyping was performed by V.S., J.B., J.J., Ø.H., C.A.W., G.T., M.E.G., S.R.O., D.M., E.A.N., E.H., A.S., C.A., A.T.H., M. Melbye, W.L., L.B., E.O., O.B.P., C.E., E.S., K.T., H.U., M.H., T.J., H.H., B.M.S., L.S., T.S., C.P., J.W., A.H.S., P.M., O.A.A., U.T., S.F.A.G., E.Q., C.E.P., M.-F.H., G.M.H., M.-R.J., M.I.M., D.A.L., R. Mägi, K.H., B.F., P.N., L.J.M., G.Z. and B.J.
Peer review
Peer review information
Nature Genetics thanks John Perry, Fasil Tekola-Ayele and Heping Zhang for their contribution to the peer review of this work. Peer reviewer reports are available.
Funding
Open access funding provided by University of Gothenburg.
Data availability
Cohorts should be contacted individually for access to raw genotype and phenotype data, as each cohort has different data access policies. Summary statistics from the meta-analysis, excluding 23andMe, are available at the EGG website (https://egg-consortium.org/), and access to the weights for constructing the polygenic score of gestational duration excluding 23andMe are available at the PGS Catalog (https://www.pgscatalog.org/, score ID: PGS002806). Access to the full set, including 23andMe results, can be obtained after approval from 23andMe is presented to the corresponding author or by completion of a Data Transfer Agreement (https://research.23andme.com/dataset-access/), which exists to protect the privacy of 23andMe participants. Access to the Danish National Birth Cohort (phs000103.v1.p1), Hyperglycemia and Adverse Pregnancy Outcome (phs000096.v4.p1) and Genomic and Proteomic Network (phs000714.v1.p1) individual-level phenotype and genetic data can be obtained through dbGaP Authorized Access portal (https://dbgap.ncbi.nlm.nih.gov/dbgap/aa/wga.cgi?page=login). The informed consent under which the data or samples were collected is the basis for determining the appropriateness of sharing data through unrestricted-access databases or NIH-designated controlled-access data repositories. The summary statistics used in this publication other than the one generated are available at the following links: fetal GWAS of gestational duration (http://egg-consortium.org/gestational-duration-2019.html), fetal and maternal GWAS of birth weight (http://egg-consortium.org/birth-weight-2019.html), miscarriage (http://www.geenivaramu.ee/tools/misc_sumstats.zip), age at first birth, estradiol (women), endometriosis, number of live births and age at menarche (http://www.nealelab.is), age at menopause (https://www.reprogen.org), testosterone (women)58, SHBG, testosterone and CBAT (10.6084/m9.figshare.c.5304500.v1), pelvic organ prolapse and leiomyoma of the uterus (https://www.finngen.fi/fi), polycystic ovary syndrome (https://www.repository.cam.ac.uk/handle/1810/283491 and https://www.finngen.fi/fi) and pre-eclampsia (European Genome-phenome Archive, https://ega-archive.org, EGAD00010001984). Pan-UK Biobank data are available at https://pan.ukbb.broadinstitute.org/. Precomputed LD scores for European populations (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2) and multi-tissue gene expression precomputed stratified LD scores (https://alkesgroup.broadinstitute.org/LDSCORE/LDSC_SEG_ldscores/Multi_tissue_gene_expr_1000Gv3_ldscores.tgz) are available. eQTL data from GTEx are available at https://gtexportal.org/home/ and from endometrium at
http://reproductivegenomics.com.au/shiny/endo_eqtl_rna/. Protein QTL data were obtained from https://www.omicscience.org/apps/pgwas/. Genome Reference Consortium Human Build 37 (hg19) available at https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_000001405.13/.
Code availability
Code for this project has been structured using a Snakemake workflow65 and is available at https://github.com/PerinatalLab/metaGWAS. A public release of it has been deposited in Zenodo (10.5281/zenodo.7311977).
Competing interests
As of January 2020, A.M. is an employee of Genentech and a holder of Roche stock. The views expressed in this article are those of the author(s) and not necessarily those of the NHS, NIHR or Department of Health. M.I.M. has served on advisory panels for Pfizer, Novo Nordisk and Zoe Global; has received honoraria from Merck, Pfizer, Novo Nordisk and Eli Lilly; and has received research funding from Abbvie, AstraZeneca, Boehringer Ingelheim, Eli Lilly, Janssen, Merck, Novo Nordisk, Pfizer, Roche, Sanofi Aventis, Servier and Takeda. As of June 2019, M.I.M. is an employee of Genentech and a holder of Roche stock. D.A.L. receives support from several national and international government and charitable research funders, as well as from Medtronic and Roche Diagnostics for research unrelated to that presented here. H.S. obtained speaker fees from Ferring Pharmaceuticals, Merck A/S, AstraZeneca and Cook Medical. V.S., G.T., G.S., D.F.G., U.T. and K.S. are employees of deCODE genetics/Amgen. The remaining authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ge Zhang, Bo Jacobsson.
Lists of authors and their affiliations appear at the end of the paper.
Change history
5/10/2023
A Correction to this paper has been published: 10.1038/s41588-023-01412-z
Contributor Information
Pol Solé-Navais, Email: pol.sole.navais@gu.se.
Bo Jacobsson, Email: bo.jacobsson@obgyn.gu.se.
Early Growth Genetics Consortium:
Extended data
is available for this paper at 10.1038/s41588-023-01343-9.
Supplementary information
The online version contains supplementary material available at 10.1038/s41588-023-01343-9.
References
- 1.Chawanpaiboon S, et al. Global, regional, and national estimates of levels of preterm birth in 2014: a systematic review and modelling analysis. Lancet Glob. Health. 2019;7:e37–e46. doi: 10.1016/S2214-109X(18)30451-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Perin J, et al. Global, regional, and national causes of under-5 mortality in 2000-19: an updated systematic analysis with implications for the Sustainable Development Goals. Lancet Child Adolesc. Health. 2022;6:106–115. doi: 10.1016/S2352-4642(21)00311-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rokas A, et al. Developing a theoretical evolutionary framework to solve the mystery of parturition initiation. eLife. 2020;9:e58343. doi: 10.7554/eLife.58343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science. 2014;345:760–765. doi: 10.1126/science.1251816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang G, et al. Genetic associations with gestational duration and spontaneous preterm birth. N. Engl. J. Med. 2017;377:1156–1167. doi: 10.1056/NEJMoa1612665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu X, et al. Variants in the fetal genome near pro-inflammatory cytokine genes on 2q13 associate with gestational duration. Nat. Commun. 2019;10:3927. doi: 10.1038/s41467-019-11881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bacelis J, et al. Uterine distention as a factor in birth timing: retrospective nationwide cohort study in Sweden. BMJ Open. 2018;8:e022929. doi: 10.1136/bmjopen-2018-022929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldenberg RL, et al. The preterm prediction study: risk factors in twin gestations. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. Am. J. Obstet. Gynecol. 1996;175:1047–1053. doi: 10.1016/S0002-9378(96)80051-2. [DOI] [PubMed] [Google Scholar]
- 9.Warrington NM, et al. Maternal and fetal genetic effects on birth weight and their relevance to cardio-metabolic risk factors. Nat. Genet. 2019;51:804–814. doi: 10.1038/s41588-019-0403-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juliusdottir T, et al. Distinction between the effects of parental and fetal genomes on fetal growth. Nat. Genet. 2021;53:1135–1142. doi: 10.1038/s41588-021-00896-x. [DOI] [PubMed] [Google Scholar]
- 11.Haig D. Genetic conflicts in human pregnancy. Q. Rev. Biol. 1993;68:495–532. doi: 10.1086/418300. [DOI] [PubMed] [Google Scholar]
- 12.Jb W, Ed B. The coadaptation of parental and offspring characters. Evol. Int. J. Org. Evol. 1998;52:299–308. doi: 10.2307/2411068. [DOI] [PubMed] [Google Scholar]
- 13.Kraft P. Curses—winner’s and otherwise—in genetic epidemiology. Epidemiol. Camb. Mass. 2008;19:649–651. doi: 10.1097/EDE.0b013e318181b865. [DOI] [PubMed] [Google Scholar]
- 14.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonder MJ, et al. Identification of rare and common regulatory variants in pluripotent cells using population-scale transcriptomics. Nat. Genet. 2021;53:313–321. doi: 10.1038/s41588-021-00800-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mortlock S, et al. Tissue specific regulation of transcription in endometrium and association with disease. Hum. Reprod. 2020;35:377–393. doi: 10.1093/humrep/dez279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pietzner M, et al. Mapping the proteo-genomic convergence of human diseases. Science. 2021;374:eabj1541. doi: 10.1126/science.abj1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deak BH, et al. Uterus-relaxing effects of nociceptin and nocistatin: studies on preterm and term-pregnant human myometrium in vitro. Reprod. Syst. Sex. Disord. 2013;2:117. [Google Scholar]
- 20.Klukovits A, et al. Nociceptin inhibits uterine contractions in term-pregnant rats by signaling through multiple pathways. Biol. Reprod. 2010;83:36–41. doi: 10.1095/biolreprod.109.082222. [DOI] [PubMed] [Google Scholar]
- 21.Sakabe NJ, et al. Transcriptome and regulatory maps of decidua-derived stromal cells inform gene discovery in preterm birth. Sci. Adv. 2020;6:eabc8696. doi: 10.1126/sciadv.abc8696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pique-Regi R, et al. A single-cell atlas of the myometrium in human parturition. JCI Insight. 2022;7:e153921. doi: 10.1172/jci.insight.153921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.LaBella AL, et al. Accounting for diverse evolutionary forces reveals mosaic patterns of selection on human preterm birth loci. Nat. Commun. 2020;11:3731. doi: 10.1038/s41467-020-17258-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sinnott-Armstrong N, et al. A regulatory variant at 3q21.1 confers an increased pleiotropic risk for hyperglycemia and altered bone mineral density. Cell Metab. 2021;33:615–628.e13. doi: 10.1016/j.cmet.2021.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Privé F, Arbel J, Vilhjálmsson BJ. LDpred2: better, faster, stronger. Bioinformatics. 2020;36:5424–5431. doi: 10.1093/bioinformatics/btaa1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Laisk T, et al. The genetic architecture of sporadic and multiple consecutive miscarriage. Nat. Commun. 2020;11:5980. doi: 10.1038/s41467-020-19742-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Connor LJ, Price AL. Distinguishing genetic correlation from causation across 52 diseases and complex traits. Nat. Genet. 2018;50:1728–1734. doi: 10.1038/s41588-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruth KS, et al. Using human genetics to understand the disease impacts of testosterone in men and women. Nat. Med. 2020;26:252–258. doi: 10.1038/s41591-020-0751-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu Z, et al. Causal associations between risk factors and common diseases inferred from GWAS summary data. Nat. Commun. 2018;9:224. doi: 10.1038/s41467-017-02317-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, et al. Dissecting maternal and fetal genetic effects underlying the associations between maternal phenotypes, birth outcomes, and adult phenotypes: a mendelian-randomization and haplotype-based genetic score analysis in 10,734 mother-infant pairs. PLoS Med. 2020;17:e1003305. doi: 10.1371/journal.pmed.1003305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Romero R, et al. The preterm parturition syndrome. BJOG. 2006;113:17–42. doi: 10.1111/j.1471-0528.2006.01120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eidem HR, Ackerman WE, McGary KL, Abbot P, Rokas A. Gestational tissue transcriptomics in term and preterm human pregnancies: a systematic review and meta-analysis. BMC Med. Genom. 2015;8:27. doi: 10.1186/s12920-015-0099-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karn MN, Penrose LS. Birth weight and gestation time in relation to maternal age, parity and infant survival. Ann. Eugen. 1951;16:147–164. doi: 10.1111/j.1469-1809.1951.tb02469.x. [DOI] [PubMed] [Google Scholar]
- 34.Wojcik GL, et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Magnus P, et al. Cohort profile: the norwegian mother and child cohort study (MoBa) Int. J. Epidemiol. 2006;35:1146–1150. doi: 10.1093/ije/dyl170. [DOI] [PubMed] [Google Scholar]
- 36.Brumpton BM, et al. The HUNT study: A population-based cohort for genetic research. Cell Genom. 2022;2:100193. doi: 10.1016/j.xgen.2022.100193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hansen TF, et al. DBDS Genomic Cohort, a prospective and comprehensive resource for integrative and temporal analysis of genetic, environmental and lifestyle factors affecting health of blood donors. BMJ Open. 2019;9:e028401. doi: 10.1136/bmjopen-2018-028401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leitsalu L, et al. Cohort Profile: Estonian Biobank of the Estonian Genome Center, University of Tartu. Int. J. Epidemiol. 2015;44:1137–1147. doi: 10.1093/ije/dyt268. [DOI] [PubMed] [Google Scholar]
- 39.Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature613, 508–518 (2023). [DOI] [PMC free article] [PubMed]
- 40.1000 Genomes Project Consortium. et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCarthy S, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat. Genet. 2016;48:1279–1283. doi: 10.1038/ng.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat. Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Backman JD, et al. Exome sequencing and analysis of 454,787 UK Biobank participants. Nature. 2021;559:628–634. doi: 10.1038/s41586-021-04103-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mountjoy E, et al. An open approach to systematically prioritize causal variants and genes at all published human GWAS trait-associated loci. Nat. Genet. 2021;53:1527–1533. doi: 10.1038/s41588-021-00945-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallace C. Eliciting priors and relaxing the single causal variant assumption in colocalisation analyses. PLoS Genet. 2020;16:e1008720. doi: 10.1371/journal.pgen.1008720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlén M, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 49.Finucane HK, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat. Genet. 2015;47:1228–1235. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gazal S, et al. Linkage disequilibrium–dependent architecture of human complex traits shows action of negative selection. Nat. Genet. 2017;49:1421–1427. doi: 10.1038/ng.3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Finucane HK, et al. Heritability enrichment of specifically expressed genes identifies disease-relevant tissues and cell types. Nat. Genet. 2018;50:621–629. doi: 10.1038/s41588-018-0081-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat. Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong A, et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462:868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.O’Connell J, et al. A general approach for haplotype phasing across the full spectrum of relatedness. PLoS Genet. 2014;10:e1004234. doi: 10.1371/journal.pgen.1004234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loh P-R, et al. Reference-based phasing using the Haplotype Reference Consortium panel. Nat. Genet. 2016;48:1443–1448. doi: 10.1038/ng.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ruth KS, et al. Genetic insights into biological mechanisms governing human ovarian ageing. Nature. 2021;596:393–397. doi: 10.1038/s41586-021-03779-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sinnott-Armstrong N, Naqvi S, Rivas M, Pritchard JK. GWAS of three molecular traits highlights core genes and pathways alongside a highly polygenic background. eLife. 2021;10:e58615. doi: 10.7554/eLife.58615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Day F, et al. Large-scale genome-wide meta-analysis of polycystic ovary syndrome suggests shared genetic architecture for different diagnosis criteria. PLoS Genet. 2018;14:e1007813. doi: 10.1371/journal.pgen.1007813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Steinthorsdottir V, et al. Genetic predisposition to hypertension is associated with preeclampsia in european and central asian women. Nat. Commun. 2020;11:5976. doi: 10.1038/s41467-020-19733-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Berisa T, Pickrell JK. Approximately independent linkage disequilibrium blocks in human populations. Bioinformatics. 2016;32:283–285. doi: 10.1093/bioinformatics/btv546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McLaren W, et al. The ensembl variant effect predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Quinlan AR. BEDTools: The Swiss-Army tool for genome feature analysis. Curr. Protoc. Bioinforma. 2014;47:11.12.1–34. doi: 10.1002/0471250953.bi1112s47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mölder F, et al. Sustainable data analysis with Snakemake. F1000Research. 2021;10:33. doi: 10.12688/f1000research.29032.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Note and Supplementary Figures 1–16.
Supplementary Tables 1–17.
Data Availability Statement
Cohorts should be contacted individually for access to raw genotype and phenotype data, as each cohort has different data access policies. Summary statistics from the meta-analysis, excluding 23andMe, are available at the EGG website (https://egg-consortium.org/), and access to the weights for constructing the polygenic score of gestational duration excluding 23andMe are available at the PGS Catalog (https://www.pgscatalog.org/, score ID: PGS002806). Access to the full set, including 23andMe results, can be obtained after approval from 23andMe is presented to the corresponding author or by completion of a Data Transfer Agreement (https://research.23andme.com/dataset-access/), which exists to protect the privacy of 23andMe participants. Access to the Danish National Birth Cohort (phs000103.v1.p1), Hyperglycemia and Adverse Pregnancy Outcome (phs000096.v4.p1) and Genomic and Proteomic Network (phs000714.v1.p1) individual-level phenotype and genetic data can be obtained through dbGaP Authorized Access portal (https://dbgap.ncbi.nlm.nih.gov/dbgap/aa/wga.cgi?page=login). The informed consent under which the data or samples were collected is the basis for determining the appropriateness of sharing data through unrestricted-access databases or NIH-designated controlled-access data repositories. The summary statistics used in this publication other than the one generated are available at the following links: fetal GWAS of gestational duration (http://egg-consortium.org/gestational-duration-2019.html), fetal and maternal GWAS of birth weight (http://egg-consortium.org/birth-weight-2019.html), miscarriage (http://www.geenivaramu.ee/tools/misc_sumstats.zip), age at first birth, estradiol (women), endometriosis, number of live births and age at menarche (http://www.nealelab.is), age at menopause (https://www.reprogen.org), testosterone (women)58, SHBG, testosterone and CBAT (10.6084/m9.figshare.c.5304500.v1), pelvic organ prolapse and leiomyoma of the uterus (https://www.finngen.fi/fi), polycystic ovary syndrome (https://www.repository.cam.ac.uk/handle/1810/283491 and https://www.finngen.fi/fi) and pre-eclampsia (European Genome-phenome Archive, https://ega-archive.org, EGAD00010001984). Pan-UK Biobank data are available at https://pan.ukbb.broadinstitute.org/. Precomputed LD scores for European populations (https://data.broadinstitute.org/alkesgroup/LDSCORE/eur_w_ld_chr.tar.bz2) and multi-tissue gene expression precomputed stratified LD scores (https://alkesgroup.broadinstitute.org/LDSCORE/LDSC_SEG_ldscores/Multi_tissue_gene_expr_1000Gv3_ldscores.tgz) are available. eQTL data from GTEx are available at https://gtexportal.org/home/ and from endometrium at
http://reproductivegenomics.com.au/shiny/endo_eqtl_rna/. Protein QTL data were obtained from https://www.omicscience.org/apps/pgwas/. Genome Reference Consortium Human Build 37 (hg19) available at https://www.ncbi.nlm.nih.gov/data-hub/genome/GCF_000001405.13/.
Code for this project has been structured using a Snakemake workflow65 and is available at https://github.com/PerinatalLab/metaGWAS. A public release of it has been deposited in Zenodo (10.5281/zenodo.7311977).