Abstract
Viruses have evolved a multitude of mechanisms to combat barriers to productive infection in the host cell. Virally-encoded miRNAs are one such means to regulate host gene expression in ways that benefit the virus lifecycle. miRNAs are small non-coding RNAs that regulate protein expression but do not trigger the adaptive immune response, making them powerful tools encoded by viruses to regulate cellular processes. Diverse viruses encode for miRNAs but little sequence homology exists between miRNAs of different viral species. Despite this, common cellular pathways are targeted for regulation, including apoptosis, immune evasion, cell growth and differentiation. Herein we will highlight the viruses that encode miRNAs and provide mechanistic insight into how viral miRNAs aid in lytic and latent infection by targeting common cellular processes. We also highlight how viral miRNAs can mimic host cell miRNAs as well as how viral miRNAs have evolved to regulate host miRNA expression. These studies dispel the myth that viral miRNAs are subtle regulators of gene expression, and highlight the critical importance of viral miRNAs to the virus lifecycle.
Keywords: Virus, miRNA, EBV, KSHV, HSV, HCMV, MDV
1. Introduction
A successful viral infection necessitates that viral gene products re-engineer the infected cell to support the viral lifecycle. This includes preventing apoptosis and other antiviral responses as well as altering host metabolism to support viral gene expression, DNA replication and the production of new virions. While viruses encode proteins that have a myriad of functions to subvert and alter host cell responses, until recently the importance and functions of viral non-coding RNAs has not been as well appreciated. Virus-encoded long non-coding RNAs (lncRNA), circular RNAs (circRNAs) and microRNAs (miRNAs) as well as many other small non-coding RNAs have critical roles in modulating host responses to infection and supporting the viral lifecycle.
In this review we focus on the role of virally-encoded miRNAs in regulating host gene expression and describe how viral miRNAs play a powerful role in supporting the different phases of viral replication. We describe the viruses that encode miRNAs as well as how viral miRNAs regulate (i) apoptosis, (ii) innate and adaptive immunity and (iii) cellular signaling pathways regulating cell growth and differentiation. Furthermore, we discuss how viral miRNAs regulate transcription factor and cytokine expression, mimic cellular miRNAs and regulate cellular miRNA expression and function. These studies highlight the common pathways and processes regulated by viral miRNAs and provide insights into the mechanistic details of viral infection. Finally, we discuss viral miRNA packaging into exosomes and their use as biomarkers of infection and disease states and how viral miRNA targeting of viral genes plays an essential role in latency and reactivation.
2. Virus-encoded miRNAs
2.1. miRNA biogenesis and function
miRNAs are 22 nucleotide (nt) RNAs that are formed via multiple cleavage events of stem loop-containing transcripts that destabilize and degrade targeted transcripts. The biogenesis of viral miRNAs follows the same general rules as host-encoded miRNAs (with several exceptions – see below) and is outlined briefly here. miRNA-encoding transcripts are commonly transcribed by RNA polymerase II as primary miRNAs (pri-miRNAs), can be many kilobases long, and contain one or several stem-loop structures. The stem-loop structures are recognized in the nucleus by the Microprocessor complex consisting of the RNAse III-type protein Drosha and DiGeorge syndrome critical region gene 8 (DGCR8). This complex cleaves the pri-miRNA and releases an ~65 nt hairpin, termed the pre-miRNA, which is recognized by exportin 5 and translocated to the cytoplasm. In the cytoplasm the pre-miRNA is cleaved near the terminal loop into a ~22 nt miRNA duplex by the action of TAR RNA binding protein (TRBP) and the RNAse III enzyme Dicer. In association with an Argonaute protein, the miRNA duplex is unwound with the guide strand remaining associated with the RNA-induced silencing complex (RISC). The guide strand is typically the strand with the weaker base pairing at the 5’ end. The passenger strand (sometimes referred as the star strand) is often degraded, but in some cases can also associate with RISC to mediate effects on gene expression. When the RISC complex is associated with a region of complementarity on an mRNA it recruits decapping factors to inhibit translation initiation and/or deadenylation factors to mediate mRNA degradation, thereby post-transcriptionally limiting expression of the encoded protein [1, 2]. One exception to these rules of miRNA biogenesis occurs with murine gammaherpesvirus 68 (MHV-68), where the miRNA stem loops are encoded within viral tRNA structures (TMERs) transcribed by RNA polymerase III and are processed by tRNAse Z and not Drosha [3, 4]. Likewise, the miRNAs of Bovine Leukemia virus-1(BLV-1) are also transcribed by RNA polymerase III [5, 6]. Finally, Herpesvirus saimiri (HVS) uses the host small nucleolar RNA processing pathway to produce pre-miRNAs from small RNAs termed Sm class U RNAs (HSURs) [7].
Within the 22 nt mature miRNA, the seed region—nucleotides 2 through 8—generally defines the specificity of the miRNA for its targets. In most cases, complete or near-complete complementarity between the seed region and the target mRNA are required for the RISC complex to exert its effects. Additional sequences at the 3’ end of the miRNA can also contribute to specificity in an undefined manner. miRNAs generally recognize sequences within the 3’ untranslated region (UTR) of an mRNA, but other non-canonical modes of interaction have also been identified, including within 5’ UTRs and open reading frames (ORFs); however, the mechanism of post-transcriptional regulation for these modes of recognition has not been elucidated [8, 9]. While an individual miRNA rarely reduces protein expression greater than 50% [10, 11], multiple sites for the same miRNA, or sites for additional miRNAs within the transcript can have both additive and cooperative effects [12, 13]. Furthermore, miRNAs are very stable, with a half-life of days [14], and complex regulation of gene expression can be achieved both by the cell type and developmentally timed expression of specific miRNAs as well as the number of molecules of a given miRNA in a cell, adding an additional layer of control to regulatory networks. Virally-encoded miRNAs hack into these complex networks to tip the balance of gene expression in favor of the virus.
Host and virus-encoded miRNAs have substantial regulatory potential, with some estimates suggesting that individual miRNAs could target upwards of 100 different transcripts [15]. This implies that miRNAs could regulate essentially all cellular processes and disease states. In mice, deletion of individual miRNA families has revealed defects in development of solid organs, vasculature, skin, skeletal and hematopoietic compartments along with physiological and behavioral defects [16]. Furthermore, miRNAs are involved in regulating many disease states including tumorigenesis, epilepsy, immune disorders and responses to infectious diseases [17]. High-throughput approaches such as HITS-CLIP, PAR-CLIP and qCLASH have identified many thousands of cellular targets of viral miRNAs and revealed extensive gene networks regulated by herpesvirus miRNAs [9, 18–20]. In light of these findings, it is essential to consider the phenotype of a miRNA mutant virus as the sum of the effects of the miRNA on many different transcripts. As discussed below, mutations in individual or combinations of viral miRNAs have profound effects on the virus lifecycle. These findings are contrary to the idea that miRNAs have redundant or only subtle effects on gene expression.
2.2. Viruses that encode miRNAs
Given the power of miRNAs to regulate gene expression networks, it is perhaps unsurprising that viruses have evolved to encode their own miRNAs. miRNAs take up relatively little genetic space, have the potential to regulate expression of numerous genes and are not recognized by the adaptive immune system and therefore represent a stealth means of altering host responses during persistent infection. The first viruses shown to encode miRNAs were members of the Herpesviridae. Epstein Barr virus (EBV) is a gammaherpesvirus that can transform B cells to form lymphoblastoid cell lines (LCLs) that are amenable to studying EBV latency and replication. 5 EBV-encoded miRNAs were first cloned from an EBV-infected LCL in seminal work by Pfeffer et al [21]. Subsequently, miRNAs were also found encoded by members of the alpha-and betaherpesviridae [1, 2]. To date, Varicella zoster virus and HHV-7 are the only human herpesviruses for which miRNAs have not been identified [22, 23] , although several recent reports identified small non-coding RNAs in neurons both latently and productively infected with VZV, these small RNAs await validation as bone fide miRNAs [24, 25]. In general, miRNA sequences are highly conserved between viruses infecting humans and Old World non-human primates, but little sequence conservation occurs between more distinct species. We refer the reader to Table 1 for a list of viruses that encode miRNAs.
Table 1.
List of viruses encoding miRNAs.
| Virus Family/Subfamily | Virus | Host | # pre-miRNAs | # mature miRNAs |
|---|---|---|---|---|
| Alphaherpesvirinae | Herpes Simplex Virus 1 | Human | 9a/18b | 27 |
| Herpes Simplex Virus 2 | Human | 18 | 24 | |
| Varicella Zoster Virus | Human | 1c | 1c | |
| Herpes B Virus | Rhesus macaque | 10 | 12 | |
| Rhesus macaque rhadinovirus | Rhesus macaque | 7 | 15 | |
| Japanese macaque rhadinovirus | Rhesus macaque | 15 | 23 | |
| Herpesvirus saimiri | New World Primates | 3 | 6 | |
| Pseudorabiesvirus | Swine | 13 | 13 | |
| Bovine Herpesvirus 1 | Cattle | 10 | 12 | |
| Marek’s Disease Virus type 1/2 | Chicken | 14 | 26/36 | |
| Betaherpesvirinae | Human Cytomegalovirus | Human | 12 | 22 |
| Human Herpesvirus 6A/B | Human | 4 | 8 | |
| Rhesus Cytomegalovirus | Rhesus | 17 | 17 | |
| Mouse Cytomegalovirus | Mouse | 18 | 21 | |
| Rat Cytomegalovirus | Rat | 14 | 24 | |
| Gammaherpesvirinae | Kaposi’s Sarcoma-Associated Herpesvirus | Human | 12 | 25 |
| Epstein-Barr Virus | Human | 25 | 44 | |
| Rhesus Lymphocryptovirus | Rhesus | 35 | 68 | |
| Herpesvirus papio | Baboon | 4 | 8 | |
| Polyomaviridae | Merkel Cell Polyomavirus | Human | 1 | 2 |
| JC Polyomavirus | Human | 1 | 2 | |
| BK Polyomavirus | Human | 1 | 2 | |
| Simian Virus 40 | Rhesus macaque | 1 | 2 | |
| Murine Polyomavirus | Mouse | 1 | 2 | |
| Papillomaviridae | HPV17 | Human | 1 | 1 |
| HPV47 | Human | 1 | 1 | |
| HPV41 | Human | 1 | 1 | |
| FCPV | Avian | 2 | 2 | |
| Retroviridae | Bovine Leukemia Virus 1 | Cattle | 5 | 10 |
| Ascoviridae | Heliothis virescens ascovirus | Insect | 1 | 1 |
| Baculoviridae | Bombyx mori nucleopolyhedrosis virus | Insect | 4d | 4 |
| Autographa californica nucleopolyhedrovirus | Insect | 5d | 5 |
Virally-encoded miRNAs are likely limited to viruses that infect eukaryotes. While plant viruses readily utilize host miRNAs as an antiviral defense mechanism [26], miRNA-like small RNAs have only been detected in two plant viruses, Sugarcane streak mosaic virus, and Hibiscus chlorotic ringspot virus, and no functions have been attributed to these small RNAs [27, 28]. Moreover, because bacteria are not known to express the machinery required for miRNA biogenesis, bacteriophages are unlikely to express miRNAs. The majority of virally-encoded miRNAs have been identified from the herpesvirus family, but a number of other nuclear replicating DNA viruses also encode miRNAs, including polyomaviruses and papillomaviruses (Table 1) [29–33]. Furthermore, several DNA viruses that infect insects, including ascoviruses and baculoviruses also encode miRNAs [34, 35]. Adenoviruses express two miRNA-like molecules from the VA RNA which fold into a structure that can be cleaved by Dicer [36, 37]. The VA-RNA-encoded miRNA 3’mivaRNAI is incorporated into RISC [38, 39] but appears to play no role in the adenovirus lifecycle and may be a byproduct of the large amount of VA-RNA that is produced during infection [40, 41]. However, more recent studies suggest that a second VA-RNA-associated miRNA enhances viral replication [42]. Finally, the ability of RNA viruses to encode their own miRNAs has been controversial due to the potential for cleavage of the RNA virus genome as well as the cytoplasmic replication cycle of many RNA viruses. It was recently proposed that the coronavirus SARS-CoV-2 expresses miRNAs [43], although this finding awaits further validation. One retrovirus, BLV-1, encodes a cluster of miRNAs which are transcribed by RNA pol III [5, 6], Furthermore, RNA viruses including Sindbis virus, tick-borne encephalitic virus and influenza virus can be engineered to express artificial miRNAs [44], although how these are processed remains unclear. Repeated attempts to identify miRNAs encoded by HIV and other RNA viruses [45, 46] has not conclusively shown the association of virus-encoded miRNAs with RISC in the context of viral infection. The remainder of this review will focus mainly on gene expression regulated by herpesvirus miRNAs and highlight common targets among viruses that encode miRNAs.
Why herpesviruses have evolved to express miRNAs is likely due to the life-long nature of herpesvirus infection. The hallmark of herpesviruses is their ability to establish a latent infection, where the viral genome is maintained in the infected cell but no new virus is produced until specific cellular signals trigger reactivation of viral DNA replication and production of new virions. During latency, expression from the viral genome is reduced, but not completely silenced, as the virus retains a need to manipulate the cell to maintain the viral genome and dampen extracellular signals. This leads to some exposure to the host immune system, necessitating an immune evasion strategy which is at least partially achieved by expression of viral miRNAs. Alpha- and gammaherpesvirus family miRNAs are found grouped in regions of the genome expressed during lytic infection but also specifically during latency. In contrast, betaherpesvirus miRNAs are scattered across the viral genome. Work by several groups has shown that at least a subset of HCMV miRNAs are expressed in experimental models of latency in myeloid lineage cells [47–49]. While all herpesviruses encode a core set of relatively highly conserved genes that are important for the production of new virions, miRNA sequences across herpesviruses are highly divergent. Despite this lack of sequence conservation, many similar proteins and pathways are targeted by herpesvirus miRNAs, indicating that viral miRNAs have undergone convergent evolution to overcome common obstacles during infection regardless of the different cellular environments that each family of herpesviruses encounters. Here we will highlight the common cellular processes and pathways that are regulated by viral miRNAs in order to appreciate how virally-encoded miRNAs have evolved to alter host gene expression to aid in latency and replication in diverse cell types.
3. Cellular processes targeted by viral miRNAs
3.1. Viral miRNAs modulate apoptosis during infection
Apoptosis is a carefully controlled cell death that is an essential component of development and hematopoiesis as well as an important mechanism to remove infected, damaged or aberrantly growing cells. The ability to control the death of cells is a powerful tool to respond to physiological and pathological insults and is regulated by sophisticated signaling pathways. The extrinsic apoptosis pathway involves the activation of death receptors at the cell surface by extracellular signals, while the intrinsic pathway of apoptosis is triggered by intracellular injuries and cell stress [50]. Additionally, cells can be triggered to undergo apoptosis when recognized by activated CD8+ T cells which results in perforin/granzyme-dependent killing [51, 52]. Activation of each of these pathways converges at the cleavage of caspase-3 which leads to the hallmarks of apoptosis including DNA fragmentation, protein degradation and expression of phagocytic receptor ligands [53]. Unsurprisingly, in the arms race of virus-host evolution, viruses encode numerous methods to block apoptosis. In the context of latency, miRNAs with redundant functions have proven to be an effective means to regulate apoptotic responses.
Oncogenic viruses like EBV and Kaposi’s Sarcoma-associated herpesvirus (KSHV) must overcome apoptotic responses not only to maintain the virally-infected cells, but also to drive tumorigenesis. EBV miRNAs are involved in regulating proteins involved in the intrinsic apoptotic response at the mitochondria including Bim, Bid, Bad and PUMA as well as directly targeting caspase-3 [54–57]. Moreover, through regulating p53 expression with viral miRNAs, EBV can also exert control over the expression of pro-apoptotic proteins [58]. Further work established that the BHRF1 miRNAs protect primary B cells from spontaneous apoptosis, but the miRNA targets involved have not been identified [59]. Paradoxically, expression of EBV miR-BART-15-3p induces apoptosis through targeting the inhibitor of apoptosis BRUCE and likely other still undiscovered anti-apoptotic genes, although the relevance of this to EBV infection is still unclear [60].
Similar to EBV, KSHV encodes multiple miRNAs that directly target caspase-3 [61] and Bim [62], indicating the importance of regulating these pro-apoptotic proteins in gammaherpesvirus infection. KSHV also encodes miRNAs to target additional specific components of the apoptosis pathways, including the extrinsic apoptosis receptor TWEAKR by miR-K10a, which limits TWEAK-induced caspase activation [63]. Furthermore, KSHV miR-K9 inhibits apoptosis induced by the p53 activator Nutlin-3 by downregulation of GADD45 [64]. miR-K12-3 downregulates cathepsin D which regulates cleavage of components of the intrinsic apoptosis pathway in endothelial cells [65], while several KSHV miRNAs target the pro-apoptotic protein BCLAF1 [66]. Finally, regulation of the NFκB signaling pathway by KSHV miRNAs, including targeting IκBα, also acts to limit apoptosis [67].
The betaherpesvirus Human cytomegalovirus (HCMV) establishes latency in CD34+ hematopoietic progenitor cells (HPCs), a cell type that is highly sensitive to apoptotic stimuli. As such, HCMV uses virally-encoded miRNAs to target the pro-apoptotic transcription factor FOXO3 in these cells [68]. Other viral miRNAs target components of the extrinsic (FAS and FADD), the intrinsic (CASP7) apoptosis pathways and like the gammaherpesviruses, targets caspase-3 directly [20].
Less is known about the role of the alphaherpesvirus Herpes Simplex virus (HSV) miRNAs in regulating apoptosis. HSV establishes latency in long-lived neuronal cells which are susceptible to apoptosis. The HSV miRNAs are encoded within the Latency-Associated Transcript (LAT) which itself is known to prevent apoptosis by inhibiting the activation of pro-apoptotic caspases [69]. The contribution of the miRNAs encoded within LAT to this phenotype is unclear. The only evidence that HSV miRNAs regulate apoptosis is a recent study that showed that HSV-2 miR-H4-5p blocks actinomycin D-induced apoptosis in HeLa cell lines [70]. The oncogenic alphaherpesvirus, MDV-1 also encodes miRNAs that block apoptosis including the cellular miR-155 homolog miR-M4-5p which targets the tumor suppressor WWOX [71]. Moreover, miR-M2-5p blocks MYOD1 to limit caspase-3 cleavage [72]. Finally, miR-M3 regulates TGFβ-induced apoptosis through targeting SMAD2 [73]. Interestingly, the miRNA encoded by mouse polyomavirus (MPyV) also targets SMAD2 which contributes to its ability to suppress apoptosis in vivo [74].
In summary, it is clear that suppression of apoptosis is an important function of viral miRNAs, especially during latency. Given the multiple signaling pathways that contribute to the induction of apoptosis, viruses have evolved redundant methods to inhibit apoptosis using viral proteins and non-coding RNAs (Figure 1). Even within the spectrum of viral miRNAs, redundant functions exist, likely due to the modulatory nature of miRNA function. In this case, viruses use multiple miRNAs and target multiple components of the pathway to exert a strong phenotypic effect. Caspase-3 sits at the convergence point of all apoptosis pathways and is key in regulating the downstream effects of apoptosis and as such is highly targeted by many herpesviruses using multiple miRNAs. Viral miRNA regulation of other types of cell death, including necroptosis and pyroptosis have not been investigated, but it would be unsurprising to find viral miRNAs are involved in regulating these processes as well.
Figure 1. A model of viral miRNA regulation of apoptosis.
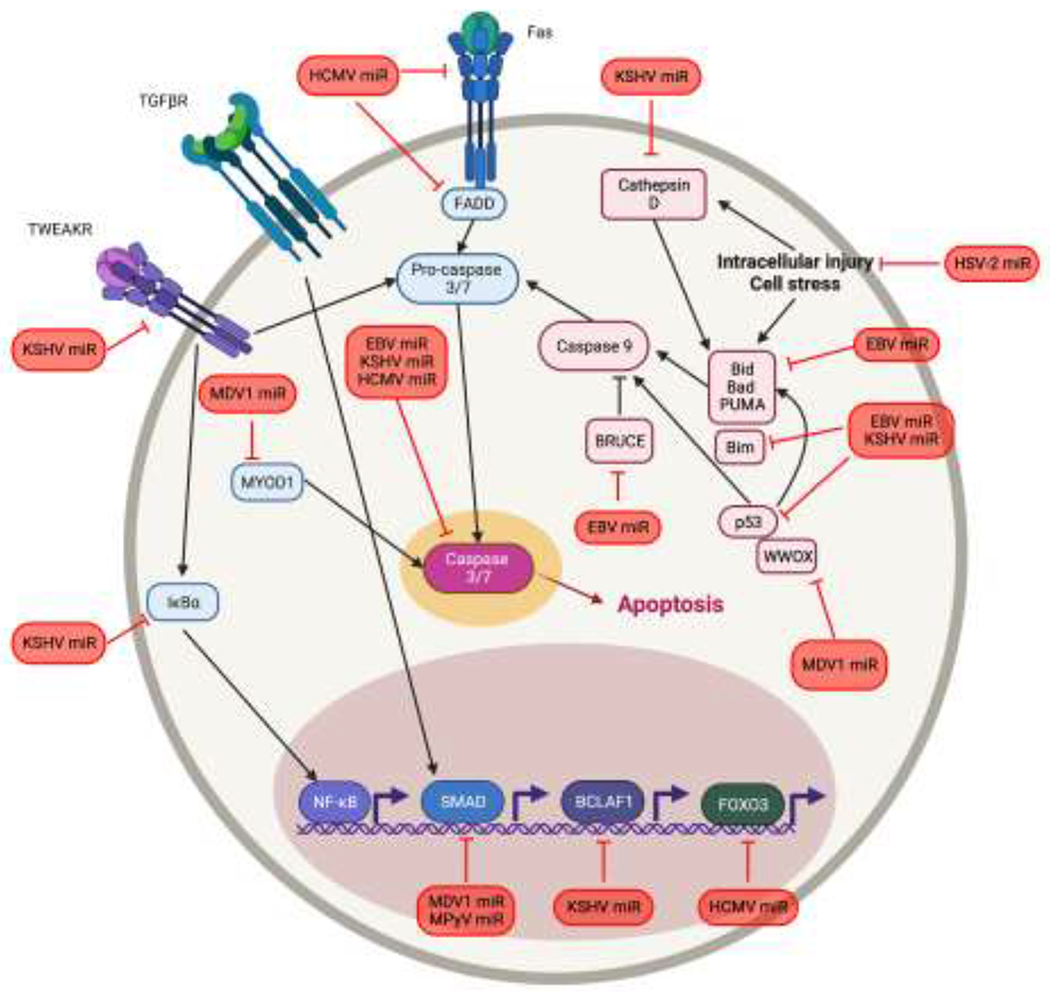
Viruses modulate cell death stimulated by physiological insults as well as infection by targeting both the intrinsic and extrinsic apoptosis pathways. Extrinsic pathway proteins are shown in blue, intrinsic pathways are shown in pink, and these pathways converge on Caspase-3. Viral miRNAs are shown in red.
3.2. Viral miRNA regulation of the host immune response
As a means of antiviral defense, viruses are recognized by cellular sensors which act to signal the infection to neighboring cells using the production and secretion of type I IFNs and proinflammatory cytokines. Innate immune cells, such as natural killer (NK) cells as well as virus-specific lymphocytes direct killing of infected cells and ultimately provide long-term immunity against pathogens. Thus, long-term persisting viruses devote significant effort to evading the intrinsic, innate and adaptive immune responses elicited by the host, including encoding miRNAs that target components of these pathways. Below, we highlight mechanisms utilized by viral miRNAs to attenuate antiviral responses (summarized in Figure 2).
Figure 2. A model of viral miRNA regulation of the host immune response.
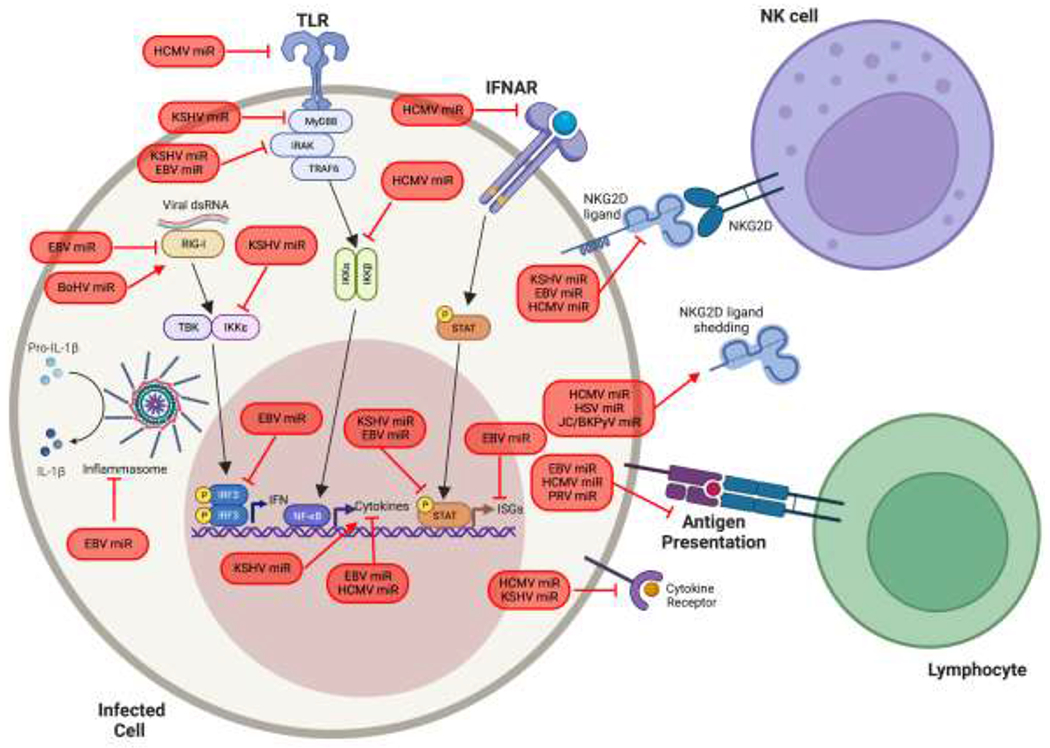
Viruses must combat the intrinsic, innate, and adaptive immune responses employed by cells to detect and control viral infection. Viruses that encode miRNAs have evolved myriad ways to target these host immune response. Viral miRNAs are shown in red.
Intrinsic immunity
Host cells constitutively express antiviral factors to limit replication of the incoming virus as an early defense against infection, before the innate and adaptive immune responses are activated. Unsurprisingly, viruses have evolved numerous mechanisms to target the intrinsic antiviral defenses of the cell. HSV-1 miR-H1 and HSV-2 miR-H6 target ATRX [75], a component of promyelocytic leukemia nuclear bodies (PML-NBs), also known as nuclear domains 10 (ND10). PML-NBs can sequester viral DNA/protein complexes [76] and this results in epigenetic silencing of viral genomes [77]. Given that miRNAs must first be expressed from the viral genome, they do not exert an effect at the earliest stages of infection. Instead, viral miRNAs may contribute to sustained inhibition PML-NBs late in lytic infection or during latent infection in order to maintain the genome is a state poised for reactivation. In addition to HSV, Merkel cell polyomavirus (MCPyV) miR-M1 targets the ND10 component SP100. miRNA-mediated regulation of SP100 results in decreased CXCL8 secretion, and reduced neutrophil chemotaxis during MCPyV infection [78]. Therefore, targeting PML-NB components may represent a general mechanism employed by virus-encoded miRNAs to escape intrinsic epigenetic silencing.
Innate immunity
Recognizing and responding to invading pathogens is a critical function of all cells. This is accomplished by pattern recognition receptors (PRRs) which recognize pathogen-associated molecular patterns (PAMPs) such as viral proteins and nucleic acids [79]. One such group of PRRs is the nucleotide-binding oligomerization domain and leucine-rich repeat-containing receptors (NLRs) that recognize numerous PAMPs and form inflammasomes, multiprotein complexes containing NLRs, adapter proteins, and caspase-1, that cleave pro-IL-1β and pro-IL18 [80]. EBV limits IL-1β secretion using miR-BART15 which targets NLRP3 to inhibit formation of the NLRP3 inflammasome [81]. Another critical PRR is RIG-I, which detects viral dsRNA in the cytoplasm [82]. Viral miRNAs themselves can be recognized by RIG-I since miRNAs are exported to the cytoplasm as double-stranded pre-miRNAs. Indeed, EBV-encoded small noncoding RNAs are recognized by RIG-I and induce expression of type I interferon (IFN) as well as IFN stimulated gene (ISG) induction [83]. EBV miR-BART6-3p directly targets RIG-I and modulates IFN signaling during lytic infection in BJAB cells, suggesting that knockdown of RIG-I may be important for immune evasion during EBV latency [84]. Another group confirmed EBV miRNA-mediated targeting of RIG-I, but implicated miR-BART3 and miR-BART19 as the miRNAs responsible for this effect [85]. Like EBV, miRNAs encoded by Bovine herpesvirus-1 (BoHV-1) directly stimulate RIG-I and induce IFNβ and ISG production. Surprisingly, infection of calves with a mutant virus lacking these miRNAs impaired the establishment of latency in TG neurons [86]. The authors hypothesized that stimulation of the IFN response downstream of RIG-I is important for dampening viral gene expression in order to establish latency. It is important to note that the BoHV-1 miRNA mutant is also lacking expression of ORF2, and it is unclear what role ORF2 plays in latency establishment of BoHV-1. Further study is needed to elucidate the intriguing role of viral miRNA triggering of RIG-I in the establishment of latency.
TOLL-like receptors (TLRs) are transmembrane PRRs expressed on plasma or endocytic membranes that recognize viruses during entry and trafficking to the nucleus [79]. Binding of PAMPs to TLRs stimulates activation of MYD88, an adaptor protein that recruits IRAK1 and results in activation of NFκB and MAP kinase signaling cascades to promote the production of proinflammatory cytokines [87]. TLR2 recognizes PAMPs produced by herpesviruses [88–91]. HCMV miR-UL112-3p directly targets TLR2 during infection, and ectopic expression of this miRNA decreased NFκB activity similar to knockdown of TLR2. Moreover, stimulation with a TLR2 agonist induced expression of proinflammatory cytokines IL-1β, IL-6, and IL-8, but expression of miR-UL112-3p reduced this effect [92]. Interestingly, KSHV also disrupts the TLR pathway but instead directly targets IRAK and MYD88 via miR-K9 and miR-K5, respectively. Similar to HCMV miR-UL112-3p, the KSHV miRNAs inhibit NFκB activity and production of IL-6 and IL-8 [93]. In addition, EBV targets IRAK2, indicating that many herpesviruses target TLR signaling using their miRNAs [85]. HCMV also inhibits NFκB signaling downstream of TLRs. HCMV miR-US5-1 and miR-UL112-3p target IKKα and IKKβ to inhibit NFκB induction of proinflammatory cytokines IL-6 and CCL5. HCMV lacking these miRNAs showed increased levels of IKKα and IKKβ as well as IL-6 and CCL5 secretion during infection. Expression of shRNAs directed to target IKKα and IKKβ from the miRNA mutant virus genome restored cytokine production to WT levels, providing further evidence that miRNA targeting of IKKα and IKKβ is directly responsible for this effect on cytokine production [94]. The effect of HCMV miRNAs on proinflammatory cytokine production was investigated only in the context of lytic infection, and so it is currently unclear whether HCMV targeting of the TLR/IKK/NFκB pathway is important for HCMV latency. Interestingly, KSHV miR-K12-11 targets the noncanonical IKK protein IKKε to inhibit the IFN response, including induction of IFNα, IFNβ, and CXCL-10. Overexpression of IKKε or inhibition of miR-K12-11 with a miRNA sponge enhances KSHV reactivation [95]. Collectively, herpesvirus miRNAs commonly target the TLR/IKK/NFκB signaling pathways, albeit by interfering with different parts of the pathway, with the ultimate goal of supporting virus infection.
IFNs are widely expressed cytokines that can be induced by PRR signaling cascades. IFNs produced by the infected cell bind IFN receptors in an autocrine or paracrine manner to trigger downstream signaling, primarily through the JAK-STAT pathway. STAT activation and translocation to the nucleus induces transcription of numerous ISGs which perform a variety of roles in resisting and controlling infection [96]. Viruses devote considerable efforts to subverting the production and signaling of IFNs using proteins and non-coding RNAs. Type I IFNs include IFNα and IFNβ, that bind the interferon-α/β receptor (IFNAR). HCMV miR-US29 targets IFNAR1 and inhibits STAT1 translocation to the nucleus and ISG induction [97]. EBV targets multiple components of the type I IFN pathway: (1) those involved in the production of type I IFN, including RIG-I and IRAK2; (2) proteins involved in the signaling downstream of type I IFN, including JAK1, JAK2, IKKβ, and IRF9; and (3) ISGs themselves, including RSAD2/viperin and OAS2 [85]. EBV miR-BART16 targets the transcriptional coactivator CREBBP to inhibit ISG expression in response to IFNα stimulation, and this blocks the antiproliferative effect of IFNα on latently infected Burkitt’s lymphoma (BL) cells [98]. KSHV miRNAs target multiple proteins involved in STAT3 activation downstream of IL-6 and IFNα, and knockdown of STAT3 expression or treatment with a STAT3 inhibitor in latently infected cells increased lytic reactivation [99]. In addition, KSHV miR-K12-10a-3p directly targets STAT3 and indirectly downregulates ICAM1 expression, which could affect recruitment of leukocytes; whether KSHV miRNA targeting of STAT3 affects IFN signaling remains untested [100]. The type II IFN IFNγ signals through the interferon-gamma receptor (IFNGR) and activates cells of the immune system, including NK cells, macrophages, and B cells, making this cytokine a critical link between the innate and adaptive immune responses [96]. STAT1 and IFNγ are both direct targets of EBV miR-BART20-5p and miR-BART8. In EBV+ cell lines, expression of these miRNAs correlated with a decrease in IFNγ/STAT1 levels which the authors hypothesize promotes viral replication [101]. Thus, viral miRNAs participate in an essential function of virus replication by contributing to the inhibition of innate immune signaling.
NK cells are potent cytotoxic lymphocytes that can recognize and kill pathogen-infected cells. Patients with NK cell deficiencies are more susceptible to herpesvirus infections, highlighting these cells an important contributor to controlling infections [102, 103]. Upon infection or other stresses, cells can express so-called ‘kill me’ ligands, which are recognized by activating receptors such as NKG2D, expressed on NK cells. miRNA targeting of one of these ligands, MICB, is a general mechanism of immune evasion by herpesviruses. Although these miRNAs are not similar in sequence, EBV miR-BART2-5p, KSHV miR-K12-7, and HCMV miR-UL112-3p all bind MICB mRNA via different target sites in the 3’UTR [104, 105]. Ectopic expression of any of these miRNAs reduces NK cell cytotoxicity, and infection with an HCMV miR-UL112 mutant resulted in increased NK cell killing [105]. Another way viral miRNAs contribute to evasion of NK cell killing is to promote shedding of NKG2D ligands from infected cells; if the ligand is no longer attached to the cell, NK cells will be unable to recognize the cell as infected. HCMV miR-US25-2-3p targets the matrix metalloprotease inhibitor TIMP3 leading to increased shedding of MICA during infection. Moreover, CMV disease in transplant patients correlated with elevated MICA serum levels [106], suggesting that HCMV miRNAs could contribute to MICA shedding from infected cells into the blood. Likewise, HSV-1 miR-H8 targets PIGT, a component of the glycosylphosphatidylinositol (GPI) lipid anchor pathway, which is responsible for anchoring ligands recognized by NK cells to the cell surface [107]. miRNA-mediated targeting of NKG2D ligands is not limited to herpesviruses: human JC polyomavirus (JCPyV) and human BK polyomavirus (BKPyV) each express a miRNA, identical in sequence, that targets the NKG2D ligand ULBP3. Inhibition of miR-J1-3p with a miRNA sponge restored ULBP3 levels and enhanced NK cell killing of JCPyV-infected cells [108]. NK cell killing is a powerful tool used by the host to respond to viral infection, and viruses that encode miRNAs have evolved ways to target proteins that contribute to recognition of the infected cell as a potent means of immune evasion.
Adaptive immunity
The adaptive arm of the immune response involves specialized cells that recognize specific pathogen infections and provide long-lasting protection. Somatic rearrangement of T cell receptors and immunoglobulin (Ig) genes results in T and B cells capable of recognizing and responding to infection by specific pathogens. Virus-specific antigens can be expressed on infected cells themselves or taken up by antigen presenting cells that are then recognized by antigen-specific receptors expressed on T- and B-lymphocytes [109].
In order to prevent the recognition of virally infected cells, many viruses affect the processing and presentation of antigens on the cell surface using a variety of methods. EBV miRNAs have been implicated in preventing antigen processing and presentation through downregulation of the peptide transport protein TAP2 by miR-BHRF1-3 and miR-BART17, thus reducing the ability of virus-specific CD8+ T cell to recognize infected cells [110]. Surface levels of HLA class I and II proteins and co-receptors were also downregulated as a result of miRNA targeting of the EBV latent membrane protein 1 (LMP1) [111]. HCMV miR-US4 and miR-UL112-5p target ERAP1, an ER protein that is important for trimming peptides for antigen presentation, which prevents pp65 presentation to CTLs [112, 113]. Swine leukocyte antigen-1 (SLA-1; equivalent to MHC in human cells) and TAP1 are targeted by Pseudorabies virus (PRV) miR-LLT11a [114]. Collectively, this suggests that herpesvirus utilize miRNAs to regulate the cellular pathways necessary for antigen presentation to limit detection of infected cells.
Due to the complexity of the adaptive immune response, investigation into the role of virus-specific miRNAs often requires in vivo approaches. miRNA mutants of MCMV were the first demonstrated to have a functional phenotype in vivo. Knockout of miR-M23-2 and miR-m21-1 results in reduced virus production in the salivary glands, although the miRNA target(s) responsible for this phenotype have not been identified. Depletion of NK cells and CD4+ T cells restored virus production in the salivary glands, suggesting that these miRNAs act to evade the host immune response and promote virus replication and transmission [115]. Similarly, infection with an MPyV miRNA mutant also results in reduced shedding, and this defect can be rescued in immunodeficient mice [116]. All polyomaviruses encode a single pre-miRNA opposite to the large T antigen (TAg) gene [117]. Therefore, the phenotype of the MPyV miRNA mutant may represent the in vivo phenotype of other polyomaviruses. Studies utilizing humanized mouse models have been useful for studying the role of human virus-encoded miRNAs during infection in vivo. An EBV miRNA deficient virus has been examined in a humanized mouse system and shown to replicate to lower titers and induce fewer EBV-infected B cells compared to WT infected mice. Interestingly, if the mice were depleted of CD8+ T cells, mutant virus replication could be restored to WT levels, indicating that EBV miRNAs are important for CD8+ T cell evasion [118], which correlates with the identification of miRNAs that target the antigen processing pathways during EBV infection [110, 111]. While in vivo studies are beneficial to expand our understanding of the importance of viral miRNAs during infection, more work is needed to understand the miRNA targets that are responsible for these effects.
3.3. Viral miRNA regulation of cell proliferation, differentiation and tumorigenesis
The processes of cell proliferation and differentiation are critical to the health of tissue and organ systems as well as the development and maintenance of the hematopoietic compartment. These processes are carefully regulated by cellular signaling pathways that are triggered by both extracellular signaling molecules binding to cell surface receptors as well as intracellular cues driven by different physiological states. Intricate and interconnecting signaling pathways work together to cause cells to grow and change with changing conditions. Dysregulation of these pathways alters the delicate balance that controls the health and function of the cells and can lead to effects on apoptosis, cell proliferation and the capacity of cells to differentiate. Keeping these processes in check is essential to preventing the uncontrolled growth associated with tumorigenesis. Oncogenic viruses, including the gammaherpesviruses EBV, KSHV and MHV68 and the alphaherpesvirus MDV, transform their host cells partly through the actions of virally-encoded miRNAs by regulating host signaling networks to promote cell growth and transformation. These viruses cause a variety of different cancers based on their cell tropism. EBV is associated with lymphomas, nasopharyngeal carcinomas (NPCs) and gastric cancer (GC) [59, 119]. EBV miRNAs are directly involved in the transformation process [119, 120]. KSHV causes Kaposi’s sarcoma, a cancer of the lymphatic endothelium, primary effusion lymphoma (PEL), an aggressive non-Hodgkin’s lymphoma caused by KSHV infection of B cells, and multicentric Castleman disease, a systemic inflammatory lymphoproliferative disease. KSHV miRNAs have also been implicated in cellular transformation and oncogenesis [67]. MHV68, genetically related to EBV and KSHV, also causes lymphoproliferative disorders [121]. In contrast, MDV transforms CD4+ T cells in vivo where it integrates into host telomeres and causes T cell lymphomas in chickens [122], with a viral miRNA playing an important role in this process. The non-oncogenic betaherpesviruses also directly affect cell proliferation and differentiation as a means to enhance their replication, and in the case of HCMV, to regulate reactivation from latency.
Several major signaling hubs regulate cell proliferation and differentiation including PI3K/AKT, MAPK, Wnt and TGFβ signaling (summarized in Figure 3). Below we outline how viral miRNAs interfere with these signaling pathways to alter host cell proliferation and differentiation and how, in the case of oncogenic herpesviruses, this can lead to tumorigenesis.
Figure 3. A model of viral miRNA regulation of signaling pathways involved in proliferation, differentiation, and tumorigenesis.
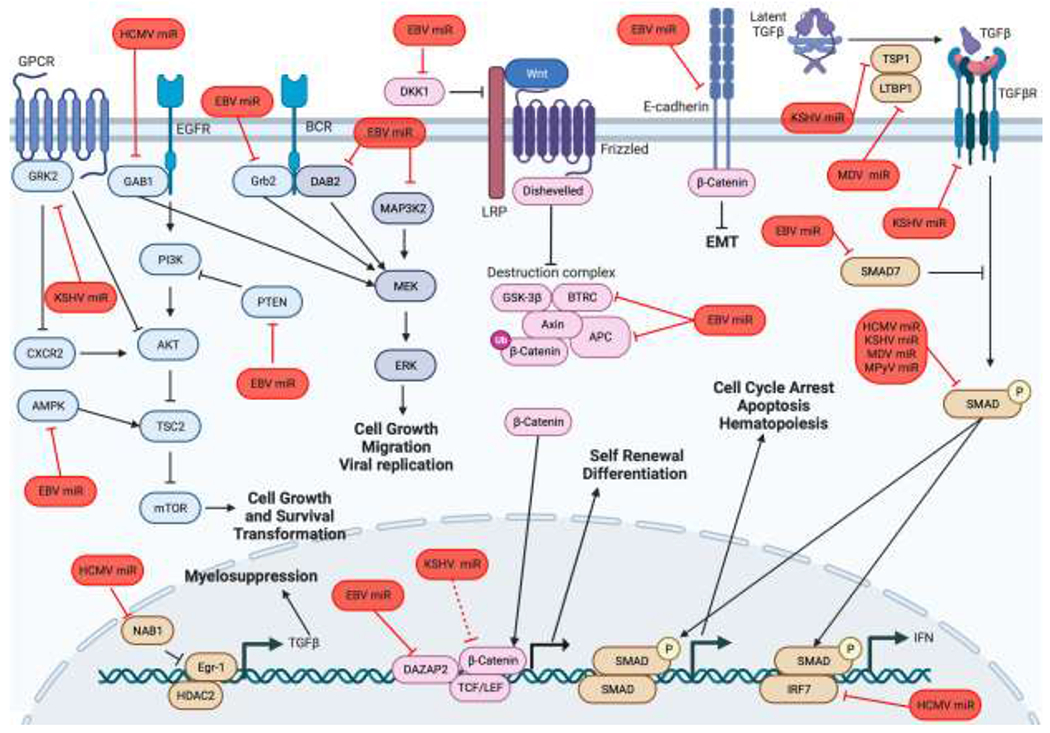
Viral miRNAs alter host cell signaling depending on the type of cell infected to control lytic and latent infection. The major signaling hubs are highlighted here (Akt signaling in blue, MAPK signaling in dark blue, Wnt signaling in pink, and TGFβ signaling in brown). Viral miRNAs are shown in red.
Viral miRNA interference with PI3K/AKT signaling
Signaling to the serine/threonine kinase AKT leads to activation of many downstream pathways regulating cell survival, growth, protein synthesis, glucose metabolism and migration. AKT is activated by phosphorylation downstream of phosphoinositide 3-kinase (PI3K) and further regulated via dephosphorylation by phosphatase and tensin homolog (PTEN). The PI3K-AKT signaling network is the most commonly dysregulated pathway in human cancers [123]. Activation of this pathway in tumorigenesis uncouples cell growth and metabolic pathways from external stimuli leading to uncontrolled cell proliferation. Activating this pathway during viral replication is important for limiting the induction of apoptosis and supporting the metabolic needs of the virus during replication, and sustained PI3K/AKT signaling caused by oncogenic viruses is essential to their pathogenesis. EBV miRNAs play an important role in activating AKT in both NPCs and GCs by targeting the AKT inhibitor PTEN using miR-BART7-3p, miR-BART1 and miR-BHRF1-3 [124, 125]. Additionally, miR-BART1-5p targets the alpha subunit of AMPK to affect AKT/mTOR signaling for enhanced glycolysis and angiogenesis [126]. PI3K signaling is also important for growth of LCLs transformed by EBV and through targeting the adaptor protein Grb2, miR-BHRF1-2 participates in the transformation process [127]. KSHV induces the activation of the AKT signaling pathway through miR-K3 targeting the G protein-coupled receptor kinase GRK2 whose downregulation promotes the migration and invasion of endothelial cells and angiogenesis in PEL [128]. Additionally, reducing GRK2 protein levels increases the expression of CXCR2 which in turn directly activates AKT [128, 129], thus miR-K3 acts to enforce AKT activation in multiple ways. Using qCLASH to identify targets of MHV68 miRNAs, mTOR and PI3K signaling pathway members were highly enriched during infection of both fibroblasts and B cells, although the implications of this targeting to viral infection remains to be determined [130]. Finally, unlike the oncogenic herpesviruses, HCMV inhibits AKT activity during infection which is important for lytic replication and may also be an essential component of reactivation from latency [131–133]. HCMV miR-US5-2 targets the adaptor protein GAB1, which is required for epidermal growth factor (EGF)-mediated activation of AKT signaling [134].
Viral miRNA interference with MAPK signaling
MAPK pathways are also major signaling hubs in the cell that links extracellular signals with cell growth, proliferation, differentiation and apoptosis. Thus, when these signaling pathways are disrupted such that extracellular signals are no longer required to activate the pathways, uncontrolled cell growth can be the result [135]. During EBV infection, targeting the receptor tyrosine kinase adaptor Grb2 affects MAPK signaling and LCL growth [127], while targeting the mitogen-responsive adaptor protein DAB2 can affect the migration of GC cells [136]. Furthermore, EBV miR-BART-18-5p targets MAP3K2, which plays a role in regulating viral replication [137]. HCMV miRNA targeting of GAB1 also inhibits downstream MAPK/ERK signaling which may be important in the latent-to-lytic switch [134]. KSHV miRNAs are predicted to redundantly target components of MAPK signaling pathways [67], although the relevance of this to viral infection has not been elucidated. Furthermore, crosstalk between MAPK and other signaling pathways likely means that viral miRNAs regulate these signaling pathways in ways that have yet to be examined.
Viral miRNA interference with Wnt signaling
The Wnt signaling pathways play important roles in development, including regulating cell fate determination, motility, polarity, organogenesis and stem cell renewal via signaling through cell surface receptors that results in the stabilization of the transcription factor β-catenin and expression of β-catenin-regulated genes [138]. Like with the PI3K/AKT and MAPK pathways, mutations that uncouple responses to extracellular Wnt ligands results in unregulated expression of genes that lead to oncogenesis. This pathway is frequently dysregulated in cancers and is involved in the epithelial-to-mesenchymal transition (EMT) process by causing changes in cell adhesion properties and motility [139]. EBV-associated gastric cancer and NPCs are associated with dysregulated EMT and viral miRNAs play a role in stimulating this pathway. miR-BART9 targeting of E-cadherin contributes to EMT [140] while miR-BART10-3p targeting of BTRC, which ubiquitinates β-catenin, does the same [141]. In gastric carcinoma cells, the Wnt signaling inhibitor DKK1 is targeted by miR-BART10-3p [142]. Furthermore, WIF1, NLK and APC are inhibitory molecules of the Wnt pathway regulated by EBV miRNAs in NPCs [143]. Therefore, it appears that EBV utilizes its miRNAs to induce signaling through the Wnt pathway as a means to promote tumorigenesis. However, one target of miR-BART3, DAZAP2, interacts with Tcf/Lef family members to activate Wnt-responsive genes [19]. Thus, regulation of this pathway during EBV infection is complex and not completely understood. Gene expression analysis of cells infected with WT or a KSHV miRNA mutant virus revealed that Wnt pathway components are specifically downregulated in the presence of KSHV miRNAs, although the relevance of this to viral infection has not been further investigated [67]. In fact, in silico analysis suggests that the Wnt signaling pathway is targeted by miRNAs from multiple herpesviruses [144] and high throughput sequencing methods have also identified Wnt components as targets of gammaherpesvirus miRNAs [18, 19, 145]. Given that there is still much to learn about how Wnt signaling is regulated, it is perhaps unsurprising that the relevance of this pathway to herpesvirus infections is still unclear, but given that components of the pathway are redundantly regulated by viral miRNAs, the pathway is clearly important.
Viral miRNA interference in TGFβ signaling
TGFβ signaling regulates multiple different cellular processes including apoptosis, proliferation, EMT and differentiation. TGFβ signaling is normally anti-proliferative and can suppress growth of premalignant cells. However, inactivation of this pathway results in abnormal expression of cell cycle regulators and ultimately tumor progression. In B cells, TGFβ can induce cell cycle arrest and apoptosis, and hence EBV and KSHV invest heavily in viral proteins and miRNAs to affect this pathway. KSHV miRNAs target the TGFβ type II receptor [146] and SMAD5 in order to become resistant to the growth inhibitory effects induced by TGFβ [147]. Thrombospondin-1 is involved in the release of latent TGFβ from the extracellular matrix and is a target of multiple KSHV miRNAs [148]. The TGFβ signaling inhibitor SMAD7 is a target of EBV miR-BART7-3p resulting in enhanced stemness of NPC cells and resistance to chemotherapeutic drugs [149]. This is in contrast to the role of numerous EBV proteins in inhibiting TGFβ signaling [150, 151], suggesting context-specific roles for TGFβ signaling in EBV infection. HCMV establishes latency in hematopoietic progenitor cells that are exquisitely sensitive to the effects of TGFβ. Intriguingly, HCMV both induces the expression of TGFβ and blocks the TGFβ signaling cascade using viral miRNAs. miR-US5-2 targeting of the TGFβ transcriptional repressor NAB1 enhances TGFβ production and secretion from the infected cell, resulting in myelosuppression of the uninfected HPCs in the microenvironment [152] which likely contributes to the myelosuppression observed upon CMV reactivation in transplant patients [153]. At the same time the virus utilizes miR-UL22A to directly target SMAD3 and block TGFβ signaling within the infected cell, protecting it from the negative effects of TGFβ signaling in order to maintain the viral genome during latent infection [152]. During lytic HCMV infection, viral miRNA targeting of SMAD3 contributes to the block in IFN induction mediated by the interaction between SMAD3 and IRF7 [154]. MDV also uses viral miRNAs to limit TGFβ signaling, including targeting SMAD2 [73] and the latent TGFβ-binding protein LTBP1 [155]. The polyomavirus MPyV also targets SMAD2 to limit apoptosis [74]. Thus, like with the other signaling pathways highlighted here, TGFβ signaling is carefully regulated by DNA virus miRNAs through targeting multiple components of the signaling pathway to limit the detrimental effects on viral replication.
Viral miRNA interference with cell cycle regulation
While herpesviruses encode for many of the proteins necessary for replicating their DNA, they also regulate the cell cycle in order to enrich for host factors and metabolites necessary for their replication. Herpesvirus infection most often cause a block at the G1/S transition and selective regulation of components of the cell cycle pathways aid in lytic viral replication [156]. During latency, however, different herpesviruses encounter very different environments depending on the cell type in which they reside. HSV establishes latency in long-lived neuronal cells and likely requires very little regulation of cell cycle components. In contrast, the oncogenic herpesviruses promote cell cycling and transformation while HCMV establishes latency in hematopoietic progenitor cells and limits their proliferation.
HCMV regulates expression of a number of cell cycle genes using virally-encoded miRNAs including miR-US25-1 regulation cyclin E2, CCND1, CDK1 and Geminin [8, 157], however the importance of targeting these proteins during lytic or latent infection is unclear. miR-US25-1 also targets the Rho GTPase RhoA, which is important for limiting the proliferation of latently infected HPCs [158]. The oncogenic herpesviruses also utilize viral miRNAs to regulate cell cycle progression and contribute to oncogenesis. When B cells are newly infected with EBV lacking the BHRF1 miRNAs, the cells progress inefficiently through the cell cycle and show reduced proliferation [120], however this is not the case in established LCLs [119]. PRDM1 has been identified as one target of miR-BHRF1-2 that could contribute to this phenotype [159]. High throughput sequencing techniques identified components involved in control of the cell cycle as highly enriched as EBV miRNA targets [145]. Furthermore, by targeting the tumor suppressor p53, miR-BART5-3p regulates expression cell cycle genes like the cyclin dependent kinase inhibitor CDKN1A (p21) [58]. WT KSHV also promotes cell cycle progression and stimulates proliferation but the role of viral miRNAs in this process is unclear. miR-K1 targeting of IκBα and the cyclin dependent kinase inhibitor p21 may be means of regulating cell cycle progression, although this requires further investigation [160, 161]. Thus, KSHV miRNAs may play an important role in preventing cell cycle arrest and along with viral oncoproteins participate in the tumorigenesis of KSHV. Hence, as has been found for cellular miRNAs [162], viral miRNAs can exert significant control over the cell cycle during lytic and latent infection (Figure 4).
Figure 4. A model of viral miRNA regulation of the cell cycle.
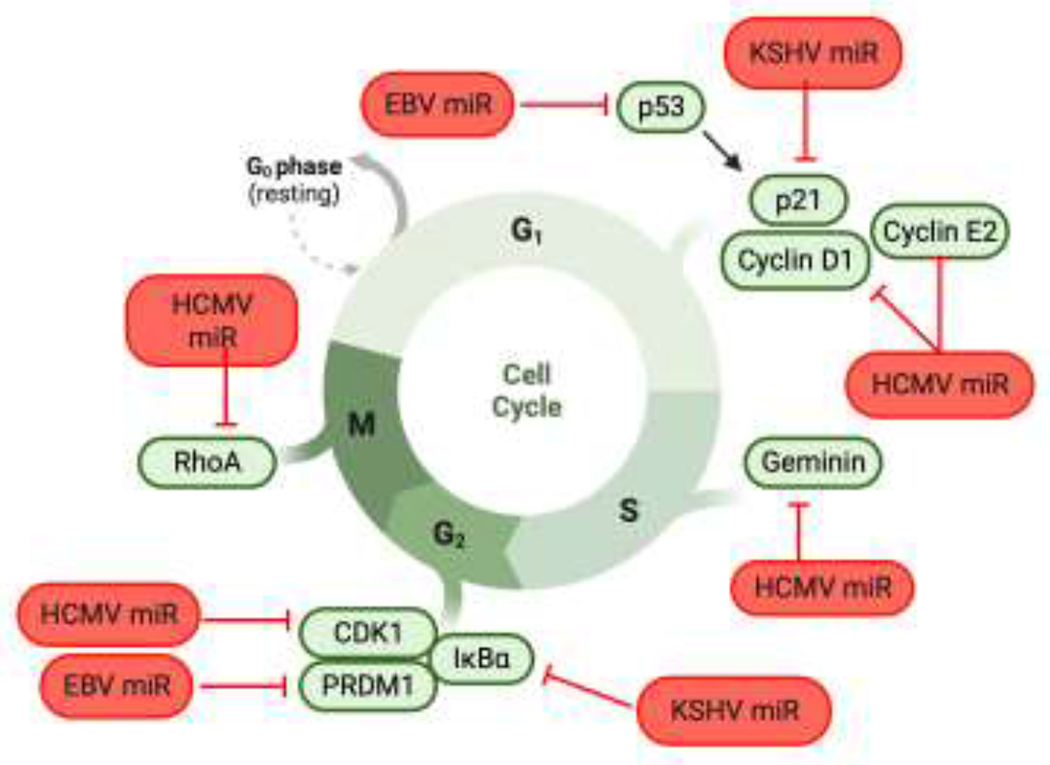
Viral miRNAs are employed to control different stages of the cell cycle, depending on the host cell type. Oncogenic viruses such as EBV and KSHV promote cell cycling, while HCMV arrests the cell cycle in hemopoietic progenitor cells to maintain latency. Indicated proteins that promote progression through G1, S, G2, and M are shown in green, and viral miRNAs targeting these proteins are shown in red.
3.4. Viral miRNAs directly target host transcription factors
miRNAs and transcription factors are both components of the complicated regulatory networks that mediate gene expression in the cell. Through modulating expression of host transcription factors, miRNAs add an additional layer of regulation to these networks, altering the output of signal transduction pathways and modulating viral gene expression. Below are examples of how viruses use miRNAs to support both lytic and latent infection by directly targeting general and cell-type-specific transcription factors.
Since EBV establishes latency in and transforms B cells, it is unsurprising that it uses viral miRNAs to attenuate expression of genes regulated by cell-type specific transcription factors. For example, both BCL6 [163], a transcriptional repressor associated with B cell differentiation, and EBF1, involved in germinal center formation [164], are regulated by a number of EBV miRNAs. Regulation of cellular transcription factors has also been implicated in EBV-mediated transformation, where miRNA targeting of KLF2 in gastric carcinoma cells is associated with enhanced anchorage-independent growth and migration [165]. In another intriguing example, two EBV miRNAs have been implicated in regulating PD-L1 expression in NPC and GC cell lines. These miRNAs target two transcriptional repressors, FOXP1 and PBRM1 that normally inhibit PD-L1 expression. By reducing expression of these negative regulators of PD-L1 expression, the EBV miRNAs cause apoptosis of infiltrating T cells and promote tumor immune escape [166].
Like EBV, KSHV also targets transcription factors that play a role in development and lymphomagenesis. The miR-155 mimic miR-K-12-11 targets C/EBPβ [167] which plays a role in B cell expansion. KSHV infection of lymphatic endothelial cells reprograms these cells to express markers of blood vessel endothelial cells. 3 KSHV miRNAs directly reduce the expression of MAF, a transcriptional repressor of blood vessel endothelial cell markers which contributes to KSHV-induced endothelial cell reprogramming [168]. KSHV miRNAs also target transcription factors that regulate expression of viral proteins that promote lytic reactivation, reiterating the role of KSHV miRNAs in enforcing latency. miR-K12-3 and miR-K12-11 suppress reactivation by targeting the transcription factors MYB and Ets-1, which regulate the lytic transactivator RTA [169, 170].
In contrast, HCMV regulates expression of transcription factors to both enforce latency in CD34+ HPCs as well as to re-launch the lytic cycle of infection. HCMV miRNAs target the transcription factor FOXO3, which regulates the expression of genes involved in induction of apoptosis [68]. Furthermore, HCMV miR-US5-2 targets the TGFβ transcriptional repressor NAB1 in order to induce the expression of TGFβ and mediate myelosuppression [152]. Additionally, EGFR signaling to the transcription factor Egr-1 is required to sustain HCMV latency by stimulating the expression of the viral gene UL138 [133]. HCMV miR-US22 plays a role in virus reactivation by directly targeting Egr-1 and reducing UL138 expression [47]. Thus, viral miRNA targeting of transcriptional activators and repressors is a powerful means to reprogram cells and prevent expression of viral proteins that affect replication.
3.5. Viral miRNAs affect expression and secretion of cytokines and chemokines
Expression and secretion of cytokines and chemokines is an important means for the infected cell to communicate to neighboring uninfected and immune cells in order to limit the spread of infection. One way that viral miRNAs inhibit proinflammatory cytokine secretion is to target immunogenic viral genes that stimulate the innate immune response, as discussed below. For example, HSV-1 miR-H6 indirectly inhibits IL-6 secretion from infected cells by targeting ICP4 [171]. Viral miRNAs also target cellular proteins at various points in signaling cascades with the ultimate objective of inhibiting secretion of cytokines, chemokines, and growth factors.
Viral infection triggers signaling cascades that result in the production and secretion of proinflammatory cytokines, and so viruses have evolved ways to block all aspects of cytokine signaling. First, miRNAs can target the receptors responsible for triggering signaling cascades resulting in cytokine production. For example, KSHV miR-K10a targets the cell surface receptor TWEAKR to limit TWEAK-induced IL-8 and MCP-1 production [63]. HCMV miR-UL148D targets the activin A receptor, ACVR1B. Monocytes infected with HCMV lacking miR-UL148D exhibited increased ACVR1B levels and were more sensitive to activin A stimulation resulting in increased production of IL-6 [49]. Moreover, many viral miRNAs directly target cytokines and chemokines in order to reduce their expression. Several EBV miRNAs reduce IL-12 and IL-23 secretion by directly targeting the IL-12p40 subunit encoded by the IL12B gene. IL-12 secretion is important for Th1 cell differentiation; when B cells infected with WT virus were co-cultured with autologous CD4+ T cells, Th1 differentiation was repressed compared to B cells infected with EBV lacking the miRNAs [111]. In addition, EBV miR-BHRF1-3 inhibits both basal levels and IFNγ induction of the T cell chemoattractant CXCL-11 either directly, or indirectly [172]. HCMV miRNAs have also been implicated in targeting multiple cytokines and chemokines, including IL-32 and RANTES [173, 174], although the functional consequences of this targeting have not been tested.
Blocking the secretion of cytokines can also be accomplished by altering the vesicular trafficking pathways needed for cytokine release. Three HCMV miRNAs—miR-US5-1, miR-US5-2, and miR-UL112-3p—target multiple endocytic proteins during infection to promote formation of the virion assembly compartment (VAC) and block the secretion of cytokines. Strikingly, ectopic expression of these three miRNAs alone was sufficient to reorganize endocytic membranes into a VAC-like structure and inhibit IL-6 release. Mutation of the miRNAs in HCMV resulted in increased export of noninfectious particles as well as proinflammatory cytokines, suggesting that the reorganization of vesicular structures by the virus is important for virion and cytokine release [175].
Taken together, viral miRNAs play a significant role in modulating cytokine and chemokine secretion—through decreasing signaling receptors, directly targeting cytokines and chemokines or their regulators, or rewiring trafficking pathways—to escape immune detection.
4. Viral miRNAs affect cellular miRNA biogenesis and function
Another mechanism used by viruses to alter the host gene expression landscape is to alter the production or function of cellular miRNAs. Disrupting the cellular miRNA regulatory networks can have profound impacts both on a cellular and organism level [15] and plays an important role in viral infection. Infection with most viruses results in changes in the expression levels of specific cellular miRNAs through direct effects on their transcription [176]. Moreover, interaction between miRNAs and RNA transcripts that exhibit high complementarity can induce target-mediated miRNA degradation (TDMD) by recruiting a ubiquitin ligase that causes degradation of the Argonaute complex and cleavage of the exposed miRNA [177, 178]. Here we highlight some interesting ways used by viruses to alter cellular miRNA expression.
The betaherpesvirus human herpesvirus 6 (HHV-6) is the causative agent of roseola in children and establishes latency by integration into host telomeric regions [179]. Recent work shows that the HHV-6A-encoded miRNA miR-aU14 inhibits the processing of multiple miR-30 family members through direct interaction with miR-30 hairpin loops within the pri-miRNA. By affecting the cellular miR-30 regulatory network, miR-aU14 affects mitrochondrial architecture and type I IFN responses as well as playing a role in reactivation from latency, indicating that the viral miRNA and its regulatory network is an essential component of the HHV-6A latent-lytic switch. This is an intriguing example of a new method of regulation by a viral miRNA, although the precise mechanism of how miR-aU14 regulates miR-30 processing through binding the pri-miRNA hairpin loop remains to be determined [180].
Other herpesviruses use different strategies to affect cellular miRNA stability which may be examples of TDMD, although this has not been formally demonstrated. MCMV and HCMV as well as HVS encode transcripts which contain cellular miRNA binding sites that functionally reduce the steady-state level of cellular miRNAs in the infected cell. miR-27 is rapidly degraded during MCMV infection [181] and several groups determined that the ORF m169 contains binding sites for miR-27 [182, 183]. Mutating m169 to be complementary to a different miRNA resulted in efficient retargeting [182, 183] indicating the generalizability of the phenomenon. Mutation of the miR-27 target site resulted in a virus that is attenuated for growth in vivo [183], suggesting that regulation of miR-27 is important for MCMV infection, although mechanistic details are still lacking. HCMV uses a similar strategy to regulate specific miRNAs within the cellular miR-17-92 cluster using the UL144-145 transcript. Despite inducing expression of pri-miR-17-92, of the six miRNAs within the cluster, only miR-17 and miR-20a were downregulated during infection. The UL144-145 transcript of HCMV contains binding sites for miR-17 and miR-20a, termed the miRNA decay element (miRDE). The relevance of degradation of miR-17 and miR-20a during HCMV infection remains to be fully explored, but a miRDE mutant replicates with slightly delayed kinetics in human fibroblasts [184]. It may be that regulation of miR-17 and miR-20a is more relevant to latent infection of HCMV. HVS encodes miRNA binding sites for cellular miR-142-3p, miR-27a and miR-16 within the HSUR RNAs [7]. While miR-27a interaction with HSUR1 mediates its degradation, miR-142-3p and miR-16 levels are stable throughout infection. Recent work determined that HSUR2 acts as an adaptor to bring the miRNAs to target mRNAs involved in the induction of apoptosis [185, 186], highlighting another novel means of regulating cellular miRNA function. Working in a potentially similar manner, virus-encoded circular RNAs (circRNAs) can act as miRNA ‘sponges’ to reduce the levels of available miRNA in the infected cell. CircRNAs are formed by back-splicing in which the 5’ and 3’ ends of the transcript are covalently joined resulting in very stable non-coding RNA structures. In the context of EBV and KSHV infection, virus-encoded circRNAs modulate the levels of cellular miRNAs and alter gene regulation networks [187–192].
In a final example, the baculovirus Bombyx mori nucleopolyhedrosis virus (BmNPV) miRNA miR-1 downregulates the expression of the GTP-binding protein Ran involved in exportin-5-mediated small RNA transport to the cytoplasm. Targeting Ran and blocking the export of small RNAs resulted in enhanced BmNPV loads, possibly through preventing the export of host pre-miRNAs involved in limiting BmNPV IE protein expression [34]. This approach is conceptually paradoxical, since the viral pre-miRNA would also undergo reduced transport to the cytoplasm, but the sheer amount of viral pre-miRNA made during infection may result in sufficient viral RNA export. Collectively, the findings discussed here indicate that viruses interfere with cellular miRNA processing and functions in unique and interesting ways in order to benefit the virus lifecycle (Figure 5).
Figure 5. A model of viral miRNAs that affect cellular miRNA biogenesis and function.
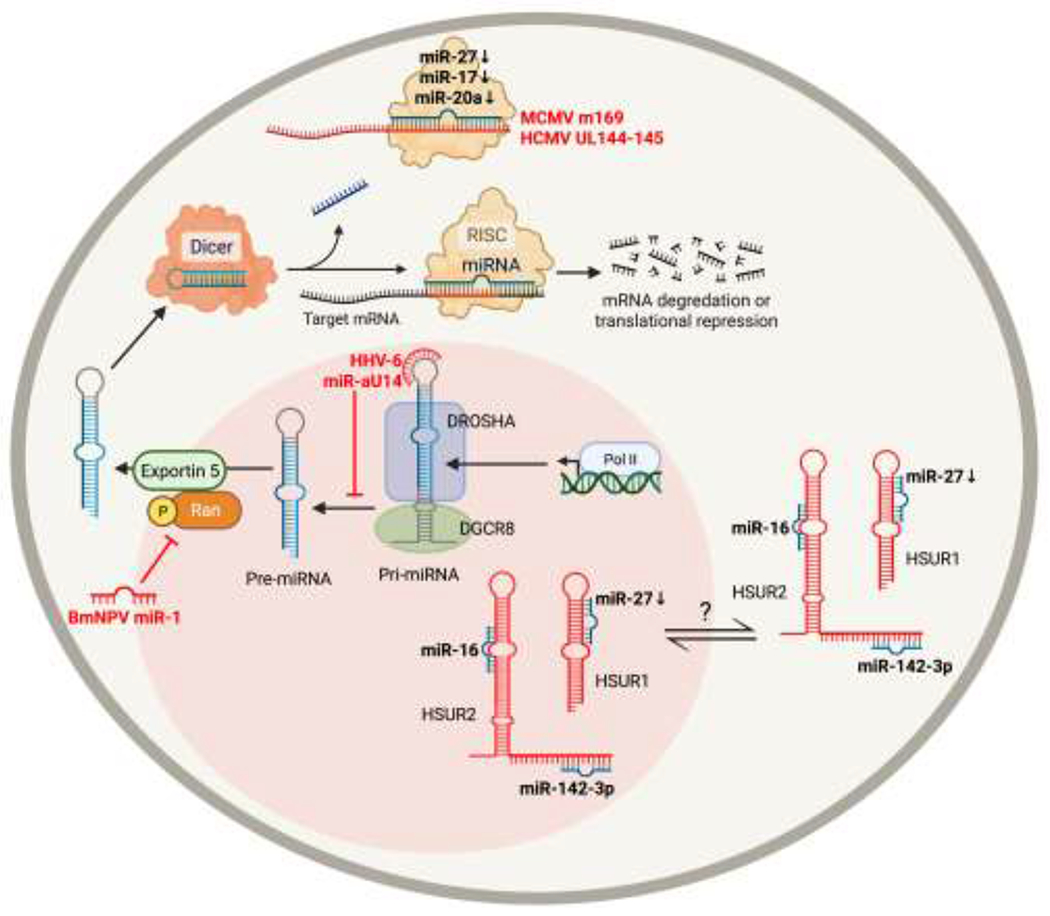
miRNA-encoding transcripts are transcribed by RNA polymerase II as a pri-miRNAs, which is cleaved by Drosha and DGCR8 to generate pre-miRNAs. pre-miRNAs are transported to the cytoplasm by exportin-5, which is dependent on Ran-GTP. In the cytoplasm, the pre-miRNA is further processed by TRBP and Dicer to generate a miRNA duplex. Argonaute unwinds this duplex, and the resulting single strand mature miRNA associates with RISC and a complementary region of mRNA to inhibit translation. Viruses have evolved ways to directly affect biogenesis and/or function of cellular miRNAs. BmNPV miR-1 directly targets the GTP-binding protein Ran to inhibit transport of pre-miRNAs to the cytoplasm. HHV-6 miR-aU14 inhibits processing of miR-30 family miRNAs through direct interaction with the stem-loops in the pri-miRNA. MCMV and HCMV encode transcripts (UL145 and m169, respectively) that bind cellular miRNAs to reduce levels of available miRNAs. HVS HSUR RNAs contain binding sites for cellular miRNAs miR-142-3p, miR-27, and miR-16, which either mediates miRNA degradation or stabilizes the miRNA.
5. Viral mimicry of host miRNAs
The life-long nature of most herpesvirus infections and the limited coding capacity of viral genomes has resulted in numerous examples of convergent evolution of viral molecules that are functionally similar to host molecules. Within the context of viral miRNAs, numerous examples exist of viral miRNAs which share identical or highly similar seed sequences to host miRNAs, thus allowing the virus to tap into finely tuned and extensive host regulatory networks. One estimate suggests that up to 15% of all human virus-encoded miRNAs are complete or partial seed sequence mimics for host miRNAs [193]. One of the first virally-encoded cellular miRNA mimics was identified in KSHV, where miR-K12-11 is a seed sequence homolog of cellular miR-155 [62, 194]. This results in common mRNA targets involved in regulating the cell cycle and suggests that the viral miRNA, like its host-encoded counterpart [195], could contribute to B cell transformation seen in PELs [62, 194]. In fact, ectopic expression of miR-K12-11 or miR-155 in various mouse models resulted in increased detection of CD19+ B cells in lymphoid organs [167, 196]. Conversely, Sin et al. showed that expression of the KSHV latency locus, including miR-K12-11, could complement B cell deficiencies when crossed into miR-155 knockout mice [197]. Furthermore, the oncogenic alphaherpesvirus MDV-1 also encodes a seed sequence homolog to miR-155, miR-M4, which is critical for the formation of lymphomas in chickens [198]. Deletion or mutation of miR-M4 results in a virus no longer capable of inducing lymphomas, whereas revertant virus, or miR-M4 deletion viruses that express cellular gga-miR-155 are able to rescue to oncogenic effects of the virus [199]. Further work determined that while miR-M4 plays an important role in tumor formation, it is not essential for the proliferation or maintenance of the transformed phenotype [200, 201]. The oncogenic gammaherpesvirus EBV does not encode its own miR-155 homolog, instead inducing the expression of miR-155 through LMP1-induced activation of NFκB signaling [202, 203].
KSHV encodes miR-K6-5p which shares sequence homology with the miR-15/16 family and regulates a subset of the same targets. Intriguingly, miR-K6-5p acts as a tumor suppressor miRNA, as inhibition of the miRNA in transformed B cells confers a significant growth advantage. Thus, this KSHV miRNA mimic paradoxically acts to limit the oncogenic potential of the virus [204].
Finally, KSHV miR-K10a_+1_5 is a seed homolog of the hematopoietic miRNA miR-142-3p and regulates a number of the same targets. Since miR-142-3p expression is high in B cells, KSHV may use its viral miRNA to regulate miR-142-3p targets instead in endothelial cells [18]. This mimicry goes even further, as 5’ isomiRs of miR-142-3p also exist and partially overlap with miR-K10a, significantly enlarging the potential pool of cellular miRNA targets of the viral miRNAs [205].
Herpesviruses have also evolved to encode mimics of the miR-17-92 family involved in proliferation and apoptosis amongst other functions, including EBV miR-BART5, rLCV miR-rL1-8 and MHV68 miR-M1-7-5p sharing seed homology with miR-18a/b [206–208]. Likewise, Japanese macaque rhadinovirus (JMRV) miR-J8, rhesus macaque rhadinovirus (RRV) miR-rR1-8-3p and Macaca nemestrina rhadinovirus (MneRv2) miRc-N1-3p are seed homologs of miR-17 [209]. The functional relevance of these viruses mimicking the miR-17-92 family has not been fully elucidated. Additional viral mimics of cellular miRNAs have been identified by various sequencing methods (Table 2), but their functions during viral infection have not been extensively investigated. One important consideration of viral miRNA mimicry is that the viral and cellular miRNAs are often only seed sequence homologs, with the remainder of the 22 nts having very little similarity between virus and host, which could have implications in the efficiency of miRNA targeting. Thus, while a viral and cellular miRNA may share common targets, the targetome of the two miRNAs may not be completely overlapping.
Table 2.
List of viral miRNAs known to mimic host miRNA.
| Viral miRNA | Cellular miRNA | Homology | Effect | Reference |
|---|---|---|---|---|
| KSHV miR-K12-11 | miR-155 | Seed | Cell cycle targets, B cell transformation | [62, 194] |
| Seed | Evade detection of CD19+ B cells in lymphoid organs | [167, 196] | ||
| Seed | Complements B cell deficiency in miR-155 KO mice | [197] | ||
| KSHV miR-K12-3, miR-K12-3+1 | miR-23 | Seed | Confer miR-23 targeting to B cells | [250] |
| KSHV miR-K10a_+1_5 | miR-142-3p | Seed | Shares targets | [18] |
| EBV miR-BART5 | miR-18a | Seed | Shares targets | [206] |
| MDV-1 miR-M4 | gga-miR-155 | Seed | Shares targets Contributes to formation of lymphomas | [198, 199] |
| rLCV miR-rL1-8 | miR-18a | Seed | Shares targets | [207] |
| MHV68 miR-M1-7-5p | miR-18a/b? miR-17-92 family | Seed | ? | [208] |
| JMRV miR-J8 | miR-17 | Seed | Shares targets | [209] |
| RRV miR-rR1-8-3p | miR-17 | Seed | Shares targets | [209] |
| MneRv2 miRc-N1-3P | miR-17 | Seed | Shares Targets | [209] |
| HCMV miR-UL112 | hsa-miR-373 | Overlapping binding sites | Acts synergistically with hsa-miR376a to decrease MICB levels | [251] |
| HHV6B mir_Ro6-2 | miR-582-5p | Seed ortholog | Targets SMAD3 | [252] |
| BLV miR-B4 | miR-29 | Seed | Shared targets HBP1, PXDN possibly contributes to tumorigenesis | [5] |
| MHV68 miR-M1-4 | Mmu-miR-151-5p | Seed | ? | [253] |
6. Viral miRNA packaging into exosomes
Exosomes are secreted 30-150 nm vesicles that can transfer cargos—including lipids, proteins, DNA, mRNA, and miRNAs—to neighboring or distant cells. Exosomes are derived from multivesiclar bodies (MVBs), also known as late endosomes, and their main biological function is cell-to-cell communication, whereby the cell releasing exosomes can modulate the behavior of recipient cells [210]. Viral infection is known to influence the composition of host factors in exosomes to create a pro-viral microenvironment [211], and emerging evidence suggests that viral miRNAs are themselves loaded into exosomes as a way of influencing bystander cells without direct infection. Incorporation of viral miRNAs into extracellular vesicles and delivery to host cells have to be, however, carefully considered. For human miRNAs, the majority of circulating miRNAs are associated with proteins, not exosomes [212]. Furthermore, copy numbers of a given miRNA are less than 1 per 100 exosomes [213], bringing into question the functional relevance of viral miRNAs packaged in exosomes. Nevertheless, we will discuss current evidence for viral miRNA packaging into exosomes and delivery to bystander cells.
Pegtel et al. [214] showed that most mature EBV miRNAs, but not their corresponding pre-miRs, are selectively packaged into exosomes secreted from LCLs. Likewise, exosomes derived from EBV-infected NPCs were enriched for specific viral miRNAs compared to intracellular levels [215], suggesting that exosome composition during viral infection can be cell-type dependent.
Components of RISC have been detected in exosomes [216], suggesting that exosomes are poised to knock down targets of viral miRNAs in recipient cells. In agreement with this, co-culture experiments showed the transfer of fluorescently labelled exosomes between EBV-infected LCLs and uninfected monocyte-derived dendritic cells (MoDC) and corresponding downregulation of CXCL11 [214], presumably by EBV BHRF1-3 [172]. Moreover, EBV miR-BART15, which targets NLRP3, is also packaged into exosomes from infected B cells. Treatment of THP-1 cells with purified exosomes from an EBV-transformed B cell line resulted in reduced NLRP3 levels compared to treatment with purified exosomes from an EBV-naive B cell line [81]. Therefore, viral miRNAs can be functionally transferred between cells via exosomes. However, it is unclear how many exosomes are needed to functionally modulate the behavior of recipient cells, and whether this transfer can occur in vivo. The most direct evidence for this comes from Pegtel et al. [214], who showed that EBV miRNAs could be detected in non-B cell fractions of PBMCs, while EBV DNA was detected only in B cells. This suggests that viral miRNAs are transferred to uninfected cells within the host.
Our understanding of how exosomal transfer of viral miRNAs to uninfected cells can benefit the viral life cycle is still poorly understood. Recent work from HSV-1 has demonstrated that miR-H28 and miR-H29 are secreted in exosomes [217] and can induce IFNγ production in uninfected cells [218], although the target(s) of the miRNAs responsible for modulating IFNγ levels and the effects on viral replication are unknown.
Exosome-associated viral miRNAs have also been investigated as potential biomarkers of various disease states. Exosomes can be isolated from blood, and this method was used to show that MDV-encoded miRNAs are present in blood of infected chickens [219]. Additionally, miRNAs encoded by EBV, KSHV, and HCMV have been detected in plasma samples from human patients [220–222] as has BLV miRNAs in the serum of seropositive cattle [223]. The presence of viral miRNAs in blood suggests some mechanism to protect the miRNA from degradation, although it is currently unclear whether viral miRNAs in the blood are contained in exosomes. Cellular miRNAs can also be stabilized in circulation through RNA-binding protein complexes and high-density lipoproteins [224–226], and so studies reporting the presence of viral miRNAs in blood, serum, or plasma does not necessarily denote the presence of exosomal miRNAs. Nevertheless, numerous studies have linked circulating viral miRNAs with various disease states. EBV BART-13-3p levels are elevated in blood from NPC patients compared to asymptomatic patients [227], suggesting this miRNA can serve as a biomarker for specific disease states. Moreover, elevated plasma levels of several EBV miRNAs correlated with disease severity [228]. Plasma levels of KSHV miR-K12-10b and miR-K12-12 were elevated in septic patients compared to non-septic controls [229]. There is a higher prevalence of HCMV miR-UL112-3p detection in patients with hypertension, Diabetes Mellitus, or Glioblastoma multiforme compared to healthy controls [230, 231]. Collectively, these data suggest that viral miRNAs can be used as biomarkers for prognosis of disease states.
7. Viral miRNA regulation of viral gene expression
Extensive research has established that most viruses encode miRNAs that target at least one viral gene [117, 193, 232–238]. During lytic infection, viral miRNAs are often expressed with early or late kinetics, when many viral genes are already highly expressed, suggesting that viral miRNAs act to regulate viral gene expression only later in infection. Below, we will briefly describe two main themes that have emerged from the study of miRNA-mediate targeting of viral proteins: (1) evasion of the host immune response and (2) regulation of viral latency and reactivation.
Viral proteins are inherently immunogenic, and one means to limit immune recognition is to regulate viral gene expression. SV40 miR-S1 targets TAg during late viral replication, when TAg is no longer needed but can cause recognition of the infected cell by cytotoxic T lymphocytes (CTLs) [239]. A mutant virus lacking this miRNA replicates similar to WT virus, but is more sensitive to CTL lysis and induces higher cytokine expression levels than WT virus [30]. Subsequently, miRNA-mediated targeting of TAg has emerged as a universal phenomenon observed in polyomaviruses [117]. Evolutionary conservation of miRNA-mediated targeting of early genes has also been demonstrated for papillomaviruses; human papillomavirus HPV41 and avian papillomavirus FcPV1 target early viral genes E1 and E2, although the functional consequence of this targeting has not been elucidated [29]. While it was initially theorized that herpesviruses targeted viral genes to promote the establishment of latency, there is evidence to suggest that herpesviruses also utilize miRNAs to target highly expressed viral genes that evoke an immune response. HCMV miR-UL112-3p was predicted to regulate latency as it targets the immediate early lytic protein IE72 [238, 240]. Interestingly, infection with an HCMV mutant lacking miR-UL112 did not affect latency establishment in CD14+ monocytes. However, increased levels of IE72 in mutant infected cells resulted in increased CTL killing [241]. Thus, multiple viruses target their own proteins using viral miRNAs in order to limit immune cell recognition. Whether this is generalizable to all miRNA-encoding viruses remains to be determined.
There are numerous accounts of viral miRNAs that target viral proteins important for lytic replication, presumably to promote latency. Herpesviruses with tractable latency models have provided evidence for this well-accepted hypothesis. HSV-1 miR-H2 and its HSV-2 homolog targets the viral transactivator ICP0, and these miRNAs are important for maintaining latency in trigeminal neurons [242–245]. Moreover, HSV-1 miR-H6 targets the immediate early transcriptional activator ICP4 [171], which is required for early and late viral gene expression and represses the latency-associate transcript (LAT) [246, 247], suggesting that HSV-1 miRNA targeting of ICP4 is important for the establishment of latency. KSHV miR-K12-7 and miR-K12-9 target the immediate early transcriptional activator RTA, and miRNA-mediated knockdown of this protein prevents reactivation [248, 249]. Herpesvirus miRNAs also target cellular proteins that regulate viral gene expression. HCMV miR-UL148D targets IER5 to modulate the IER5-CDC25B-CDK1 signaling pathway, controlling IE1 expression. Knockout of miR-UL148D in the NR-1 strain of HCMV resulted in a failure to establish latency [48]. Likewise, EBV miR-BART18-5p targets MAP3K2 to prevent viral replication in latency [137]. Collectively, these data suggest that herpesvirus miRNAs contribute to regulating viral gene expression in order to regulate entrance into and exit from latency.
8. Conclusions
Here we have highlighted the extensive ways in which viral miRNAs regulate host gene expression to influence the outcome of infection and how they have been useful tools to uncover novel processes regulated by virus infection. Most viral miRNAs come from the Herpesvirus family, and despite very little sequence homology between the miRNAs of the various subfamilies, general themes and targeting of common signaling pathways has clearly emerged from nearly two decades of research and have helped to uncover the cellular processes requiring regulation during viral infection. Unsurprisingly, key cellular signaling pathways related to apoptosis, immune evasion, cell growth and differentiation are carefully regulated by viral miRNAs since modulating these processes are key to successful viral infection, despite the different cellular environments encountered by each virus. The breadth of cellular targets identified for viral miRNAs by high throughput techniques highlights the difficulty in attributing a phenotype of a viral miRNA to a single target. It is more likely that the effects of a viral miRNA during infection is the result of the cumulative effect of regulation of numerous cellular and viral genes. This complexity is underscored by work demonstrating that a viral miRNA, or several miRNAs, can target multiple genes within the same pathway [8, 85, 127, 152, 175], suggesting that while a single target may not be responsible for an effect of a viral miRNA, the phenotype of the miRNA may nevertheless result from regulation of specific signaling pathways. Thus, it will be critical in future studies to take on a holistic approach when considering host targets of viral miRNAs.
It is apparent that viral miRNAs act most powerfully during latent infection and can replace the functions of viral proteins in order to maintain the latent infection and/or stimulate reactivation in the appropriate context. Given that latency and reactivation are key to the pathology of herpesvirus infection, understanding how viral miRNAs regulate these processes is essential to developing tools for interventions. Despite the wealth of knowledge summarized here, there are still many viral miRNAs whose functions have not been explored and uncovering their roles during infection will continue to add to our understanding of the molecular determinants of virus-host interactions.
Acknowledgements
This work was supported by grant P01 AI127335 from the National Institute of Allergy and Infectious Diseases, NIH. The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Figures were generated using BioRender.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References:
- [1].O'Brien J, Hayder H, Zayed Y, Peng C, Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation, Front Endocrinol (Lausanne) 9 (2018) 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ha M, Kim VN, Regulation of microRNA biogenesis, Nat Rev Mol Cell Biol 15(8) (2014) 509–24. [DOI] [PubMed] [Google Scholar]
- [3].Bogerd HP, Karnowski HW, Cai X, Shin J, Pohlers M, Cullen BR, A mammalian herpesvirus uses noncanonical expression and processing mechanisms to generate viral MicroRNAs, Mol Cell 37(1) (2010) 135–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Diebel KW, Smith AL, van Dyk LF, Mature and functional viral miRNAs transcribed from novel RNA polymerase III promoters, RNA 16(1) (2010) 170–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kincaid RP, Burke JM, Sullivan CS, RNA virus microRNA that mimics a B-cell oncomiR, Proc Natl Acad Sci U S A 109(8) (2012) 3077–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rosewick N, Momont M, Durkin K, Takeda H, Caiment F, Cleuter Y, Vernin C, Mortreux F, Wattel E, Burny A, Georges M, Van den Broeke A, Deep sequencing reveals abundant noncanonical retroviral microRNAs in B-cell leukemia/lymphoma, Proc Natl Acad Sci U S A 110(6) (2013) 2306–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Cazalla D, Xie M, Steitz JA, A primate herpesvirus uses the integrator complex to generate viral microRNAs, Mol Cell 43(6) (2011) 982–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Grey F, Tirabassi R, Meyers H, Wu G, McWeeney S, Hook L, Nelson JA, A Viral microRNA Down-Regulates Multiple Cell Cycle Genes through mRNA 5′UTRs, PLOS Pathogens 6(6) (2010) e1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Gay LA, Sethuraman S, Thomas M, Turner PC, Renne R, Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) Identifies Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Targets in Endothelial Cells, J Virol 92(8) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Baek D, Villén J, Shin C, Camargo FD, Gygi SP, Bartel DP, The impact of microRNAs on protein output, Nature 455(7209) (2008) 64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N, Widespread changes in protein synthesis induced by microRNAs, Nature 455(7209) (2008) 58–63. [DOI] [PubMed] [Google Scholar]
- [12].Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP, MicroRNA targeting specificity in mammals: determinants beyond seed pairing, Mol Cell 27(1) (2007) 91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Saetrom P, Heale BS, Snøve O Jr., Aagaard L, Alluin J, Rossi JJ, Distance constraints between microRNA target sites dictate efficacy and cooperativity, Nucleic Acids Res 35(7) (2007) 2333–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Zlotorynski E, Insights into the kinetics of microRNA biogenesis and turnover, Nature Reviews Molecular Cell Biology 20(9) (2019) 511–511. [DOI] [PubMed] [Google Scholar]
- [15].Bartel DP, MicroRNAs: Target Recognition and Regulatory Functions, Cell 136(2) (2009) 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Park CY, Choi YS, McManus MT, Analysis of microRNA knockouts in mice, Hum Mol Genet 19(R2) (2010) R169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Li Y, Kowdley KV, MicroRNAs in common human diseases, Genomics Proteomics Bioinformatics 10(5) (2012) 246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Gottwein E, Corcoran DL, Mukherjee N, Skalsky RL, Hafner M, Nusbaum JD, Shamulailatpam P, Love CL, Dave SS, Tuschl T, Ohler U, Cullen BR, Viral microRNA targetome of KSHV-infected primary effusion lymphoma cell lines, Cell Host Microbe 10(5) (2011) 515–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Skalsky RL, Corcoran DL, Gottwein E, Frank CL, Kang D, Hafner M, Nusbaum JD, Feederle R, Delecluse HJ, Luftig MA, Tuschl T, Ohler U, Cullen BR, The viral and cellular microRNA targetome in lymphoblastoid cell lines, PLoS Pathog 8(1) (2012) e1002484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Kim S, Seo D, Kim D, Hong Y, Chang H, Baek D, Kim VN, Lee S, Ahn K, Temporal Landscape of MicroRNA-Mediated Host-Virus Crosstalk during Productive Human Cytomegalovirus Infection, Cell Host Microbe 17(6) (2015) 838–51. [DOI] [PubMed] [Google Scholar]
- [21].Pfeffer S, Zavolan M, Grasser FA, Chien M, Russo JJ, Ju J, John B, Enright AJ, Marks D, Sander C, Tuschl T, Identification of virus-encoded microRNAs, Science 304(5671) (2004) 734–6. [DOI] [PubMed] [Google Scholar]
- [22].Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR, Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia, J Virol 83(20) (2009) 10677–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Depledge DP, Ouwendijk WJD, Sadaoka T, Braspenning SE, Mori Y, Cohrs RJ, Verjans G, Breuer J, A spliced latency-associated VZV transcript maps antisense to the viral transactivator gene 61, Nat Commun 9(1) (2018) 1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Markus A, Golani L, Ojha NK, Borodiansky-Shteinberg T, Kinchington PR, Goldstein RS, Varicella-Zoster Virus Expresses Multiple Small Noncoding RNAs, J Virol 91(24) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Golani-Zaidie L, Borodianskiy-Shteinberg T, Bisht P, Das B, Kinchington PR, Goldstein RS, Bioinformatically-predicted varicella zoster virus small non-coding RNAs are expressed in lytically-infected epithelial cells and neurons, Virus Res 274 (2019) 197773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Liu SR, Zhou JJ, Hu CG, Wei CL, Zhang JZ, MicroRNA-Mediated Gene Silencing in Plant Defense and Viral Counter-Defense, Front Microbiol 8 (2017) 1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Gao R, Liu P, Wong SM, Identification of a plant viral RNA genome in the nucleus, PLoS One 7(11) (2012) e48736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Viswanathan C, Anburaj J, Prabu G, Identification and validation of sugarcane streak mosaic virus-encoded microRNAs and their targets in sugarcane, Plant Cell Rep 33(2) (2014) 265–76. [DOI] [PubMed] [Google Scholar]
- [29].Chirayil R, Kincaid RP, Dahlke C, Kuny CV, Dälken N, Spohn M, Lawson B, Grundhoff A, Sullivan CS, Identification of virus-encoded microRNAs in divergent Papillomaviruses, PLoS Pathog 14(7) (2018) e1007156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Sullivan CS, Grundhoff AT, Tevethia S, Pipas JM, Ganem D, SV40-encoded microRNAs regulate viral gene expression and reduce susceptibility to cytotoxic T cells, Nature 435(7042) (2005) 682–6. [DOI] [PubMed] [Google Scholar]
- [31].Sullivan CS, Sung CK, Pack CD, Grundhoff A, Lukacher AE, Benjamin TL, Ganem D, Murine Polyomavirus encodes a microRNA that cleaves early RNA transcripts but is not essential for experimental infection, Virology 387(1) (2009) 157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Seo GJ, Chen CJ, Sullivan CS, Merkel cell polyomavirus encodes a microRNA with the ability to autoregulate viral gene expression, Virology 383(2) (2009) 183–7. [DOI] [PubMed] [Google Scholar]
- [33].Seo GJ, Fink LH, O'Hara B, Atwood WJ, Sullivan CS, Evolutionarily conserved function of a viral microRNA, J Virol 82(20) (2008) 9823–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Singh CP, Singh J, Nagaraju J, A baculovirus-encoded MicroRNA (miRNA) suppresses its host miRNA biogenesis by regulating the exportin-5 cofactor Ran, J Virol 86(15) (2012) 7867–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hussain M, Taft RJ, Asgari S, An insect virus-encoded microRNA regulates viral replication, J Virol 82(18) (2008) 9164–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Aparicio O, Razquin N, Zaratiegui M, Narvaiza I, Fortes P, Adenovirus virus-associated RNA is processed to functional interfering RNAs involved in virus production, J Virol 80(3) (2006) 1376–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao H, Chen M, Pettersson U, Identification of adenovirus-encoded small RNAs by deep RNA sequencing, Virology 442(2) (2013) 148–155. [DOI] [PubMed] [Google Scholar]
- [38].Xu N, Segerman B, Zhou X, Akusjärvi G, Adenovirus virus-associated RNAII-derived small RNAs are efficiently incorporated into the rna-induced silencing complex and associate with polyribosomes, J Virol 81(19) (2007) 10540–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Bellutti F, Kauer M, Kneidinger D, Lion T, Klein R, Identification of RISC-associated adenoviral microRNAs, a subset of their direct targets, and global changes in the targetome upon lytic adenovirus 5 infection, J Virol 89(3) (2015) 1608–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kamel W, Segerman B, Öberg D, Punga T, Akusjärvi G, The adenovirus VA RNA-derived miRNAs are not essential for lytic virus growth in tissue culture cells, Nucleic Acids Res 41(9) (2013) 4802–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Machitani M, Sakurai F, Wakabayashi K, Tomita K, Tachibana M, Mizuguchi H, Dicer functions as an antiviral system against human adenoviruses via cleavage of adenovirus-encoded noncoding RNA, Sci Rep 6 (2016) 27598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wakabayashi K, Machitani M, Tachibana M, Sakurai F, Mizuguchi H, A MicroRNA Derived from Adenovirus Virus-Associated RNAII Promotes Virus Infection via Posttranscriptional Gene Silencing, J Virol 93(2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Singh M, Chazal M, Quarato P, Bourdon L, Malabat C, Vallet T, Vignuzzi M, van der Werf S, Behillil S, Donati F, Sauvonnet N, Nigro G, Bourgine M, Jouvenet N, Cecere G, A virus-derived microRNA targets immune response genes during SARS-CoV-2 infection, EMBO Rep 23(2) (2022) e54341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Zhan S, Wang Y, Chen X, RNA virus-encoded microRNAs: biogenesis, functions and perspectives on application, ExRNA 2(1) (2020) 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lin J, Cullen BR, Analysis of the interaction of primate retroviruses with the human RNA interference machinery, J Virol 81(22) (2007) 12218–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Skalsky RL, Olson KE, Blair CD, Garcia-Blanco MA, Cullen BR, A “microRNA-like” small RNA expressed by Dengue virus?, Proceedings of the National Academy of Sciences 111(23) (2014) E2359–E2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Mikell I, Crawford LB, Hancock MH, Mitchell J, Buehler J, Goodrum F, Nelson JA, HCMV miR-US22 down-regulation of EGR-1 regulates CD34+ hematopoietic progenitor cell proliferation and viral reactivation, PLoS Pathog 15(11) (2019) e1007854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pan C, Zhu D, Wang Y, Li L, Li D, Liu F, Zhang CY, Zen K, Human Cytomegalovirus miR-UL148D Facilitates Latent Viral Infection by Targeting Host Cell Immediate Early Response Gene 5, PLoS Pathog 12(11) (2016) e1006007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lau B, Poole E, Krishna B, Sellart I, Wills MR, Murphy E, Sinclair J, The Expression of Human Cytomegalovirus MicroRNA MiR-UL148D during Latent Infection in Primary Myeloid Cells Inhibits Activin A-triggered Secretion of IL-6, Sci Rep 6 (2016) 31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lossi L, The concept of intrinsic versus extrinsic apoptosis, Biochemical Journal 479(3) (2022) 357–384. [DOI] [PubMed] [Google Scholar]
- [51].Salti SM, Hammelev EM, Grewal JL, Reddy ST, Zemple SJ, Grossman WJ, Grayson MH, Verbsky JW, Granzyme B regulates antiviral CD8+ T cell responses, J Immunol 187(12) (2011) 6301–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Chattopadhyay PK, Betts MR, Price DA, Gostick E, Horton H, Roederer M, De Rosa SC, The cytolytic enzymes granyzme A, granzyme B, and perforin: expression patterns, cell distribution, and their relationship to cell maturity and bright CD57 expression, J Leukoc Biol 85(1) (2009) 88–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Cullen SP, Martin SJ, Mechanisms of granule-dependent killing, Cell Death & Differentiation 15(2) (2008) 251–262. [DOI] [PubMed] [Google Scholar]
- [54].Marquitz AR, Mathur A, Nam CS, Raab-Traub N, The Epstein-Barr Virus BART microRNAs target the pro-apoptotic protein Bim, Virology 412(2) (2011) 392–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Kim H, Choi H, Lee SK, Epstein-Barr Virus MicroRNA miR-BART20-5p Suppresses Lytic Induction by Inhibiting BAD-Mediated caspase-3-Dependent Apoptosis, Journal of Virology 90(3) (2016) 1359–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Choy EY, Siu KL, Kok KH, Lung RW, Tsang CM, To KF, Kwong DL, Tsao SW, Jin DY, An Epstein-Barr virus-encoded microRNA targets PUMA to promote host cell survival, J Exp Med 205(11) (2008) 2551–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Shinozaki-Ushiku A, Kunita A, Isogai M, Hibiya T, Ushiku T, Takada K, Fukayama M, Profiling of Virus-Encoded MicroRNAs in Epstein-Barr Virus-Associated Gastric Carcinoma and Their Roles in Gastric Carcinogenesis, J Virol 89(10) (2015) 5581–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Zheng X, Wang J, Wei L, Peng Q, Gao Y, Fu Y, Lu Y, Qin Z, Zhang X, Lu J, Ou C, Li Z, Zhang X, Liu P, Xiong W, Li G, Yan Q, Ma J, Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression, J Virol 92(23) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Seto E, Moosmann A, Grömminger S, Walz N, Grundhoff A, Hammerschmidt W, Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells, PLoS Pathog 6(8) (2010) e1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Choi H, Lee H, Kim SR, Gho YS, Lee SK, Epstein-Barr Virus-Encoded MicroRNA BART15-3p Promotes Cell Apoptosis Partially by Targeting BRUCE, Journal of Virology 87(14) (2013) 8135–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Suffert G, Malterer G, Hausser J, Viiliäinen J, Fender A, Contrant M, Ivacevic T, Benes V, Gros F, Voinnet O, Zavolan M, Ojala PM, Haas JG, Pfeffer S, Kaposi's sarcoma herpesvirus microRNAs target caspase 3 and regulate apoptosis, PLoS Pathog 7(12) (2011) e1002405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Skalsky RL, Samols MA, Plaisance KB, Boss IW, Riva A, Lopez MC, Baker HV, Renne R, Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155, J Virol 81(23) (2007) 12836–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Abend JR, Uldrick T, Ziegelbauer JM, Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi's sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression, J Virol 84(23) (2010) 12139–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Liu X, Happel C, Ziegelbauer JM, Kaposi's Sarcoma-Associated Herpesvirus MicroRNAs Target GADD45B To Protect Infected Cells from Cell Cycle Arrest and Apoptosis, J Virol 91(3) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Gay LA, Stribling D, Turner PC, Renne R, Kaposi's Sarcoma-associated Herpesvirus microRNA mutants modulate cancer hallmark phenotypic differences in human endothelial cells, J Virol 95(7) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Ziegelbauer JM, Sullivan CS, Ganem D, Tandem array-based expression screens identify host mRNA targets of virus-encoded microRNAs, Nat Genet 41(1) (2009) 130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Moody R, Zhu Y, Huang Y, Cui X, Jones T, Bedolla R, Lei X, Bai Z, Gao SJ, KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways, PLoS Pathog 9(12) (2013) e1003857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Hancock MH, Crawford LB, Perez W, Struthers HM, Mitchell J, Caposio P, Human Cytomegalovirus UL7, miR-US5-1, and miR-UL112-3p Inactivation of FOXO3a Protects CD34(+) Hematopoietic Progenitor Cells from Apoptosis, mSphere 6(1) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Perng GC, Jones C, Ciacci-Zanella J, Stone M, Henderson G, Yukht A, Slanina SM, Hofman FM, Ghiasi H, Nesburn AB, Wechsler SL, Virus-induced neuronal apoptosis blocked by the herpes simplex virus latency-associated transcript, Science 287(5457) (2000) 1500–3. [DOI] [PubMed] [Google Scholar]
- [70].Zhao Y, Yang J, Liu Y, Fan J, Yang H, HSV-2-encoded miRNA-H4 Regulates Cell Cycle Progression and Act-D-induced Apoptosis in HeLa Cells by Targeting CDKL2 and CDKN2A, Virol Sin 34(3) (2019) 278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Zhu Z-J, Teng M, Li H-Z, Zheng L-P, Liu J-L, Yao Y, Nair V, Zhang G-P, Luo J, Virus-encoded miR-155 ortholog in Marek's disease virus promotes cell proliferation via suppressing apoptosis by targeting tumor suppressor WWOX, Veterinary microbiology 252 (2021) 108919. [DOI] [PubMed] [Google Scholar]
- [72].Zhu Z-J, Teng M, Li H-Z, Zheng L-P, Liu J-L, Chai S-J, Yao Y-X, Nair V, Zhang G-P, Luo J, Marek's Disease Virus ( Gallid alphaherpesvirus 2)-Encoded miR-M2-5p Simultaneously Promotes Cell Proliferation and Suppresses Apoptosis Through RBM24 and MYOD1-Mediated Signaling Pathways, Frontiers in microbiology, 2020, p. 596422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Xu S, Xue C, Li J, Bi Y, Cao Y, Marek's Disease Virus Type 1 MicroRNA miR-M3 Suppresses Cisplatin-Induced Apoptosis by Targeting SMAD2 of the Transforming Growth Factor Beta Signal Pathway, Journal of Virology 85(1) (2011) 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Sung CK, Yim H, Andrews E, Benjamin TL, A mouse polyomavirus-encoded microRNA targets the cellular apoptosis pathway through Smad2 inhibition, Virology 468-470 (2014) 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Jurak I, Silverstein LB, Sharma M, Coen DM, Herpes simplex virus is equipped with RNA- and protein-based mechanisms to repress expression of ATRX, an effector of intrinsic immunity, J Virol 86(18) (2012) 10093–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Rivera-Molina YA, Rojas BR, Tang Q, Nuclear domain 10-associated proteins recognize and segregate intranuclear DNA/protein complexes to negate gene expression, Virol J 9 (2012) 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Scherer M, Otto V, Stump JD, Klingl S, Muller R, Reuter N, Muller YA, Sticht H, Stamminger T, Characterization of Recombinant Human Cytomegaloviruses Encoding IE1 Mutants L174P and 1-382 Reveals that Viral Targeting of PML Bodies Perturbs both Intrinsic and Innate Immune Responses, J Virol 90(3) (2016) 1190–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Akhbari P, Tobin D, Poterlowicz K, Roberts W, Boyne JR, MCV-miR-M1 Targets the Host-Cell Immune Response Resulting in the Attenuation of Neutrophil Chemotaxis, J Invest Dermatol 138(11) (2018) 2343–2354. [DOI] [PubMed] [Google Scholar]
- [79].Li D, Wu M, Pattern recognition receptors in health and diseases, Signal Transduct Target Ther 6(1) (2021) 291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Swanson KV, Deng M, Ting JP, The NLRP3 inflammasome: molecular activation and regulation to therapeutics, Nat Rev Immunol 19(8) (2019) 477–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Haneklaus M, Gerlic M, Kurowska-Stolarska M, Rainey AA, Pich D, McInnes IB, Hammerschmidt W, O'Neill LA, Masters SL, Cutting edge: miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production, J Immunol 189(8) (2012) 3795–9. [DOI] [PubMed] [Google Scholar]
- [82].Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C, RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates, Science 314(5801) (2006) 997–1001. [DOI] [PubMed] [Google Scholar]
- [83].Samanta M, Iwakiri D, Kanda T, Imaizumi T, Takada K, EB virus-encoded RNAs are recognized by RIG-I and activate signaling to induce type I IFN, EMBO J 25(18) (2006) 4207–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Lu Y, Qin Z, Wang J, Zheng X, Lu J, Zhang X, Wei L, Peng Q, Zheng Y, Ou C, Ye Q, Xiong W, Li G, Fu Y, Yan Q, Ma J, Epstein-Barr Virus miR-BART6-3p Inhibits the RIG-I Pathway, J Innate Immun 9(6) (2017) 574–586. [DOI] [PubMed] [Google Scholar]
- [85].Bouvet M, Voigt S, Tagawa T, Albanese M, Chen YA, Chen Y, Fachko DN, Pich D, Gobel C, Skalsky RL, Hammerschmidt W, Multiple Viral microRNAs Regulate Interferon Release and Signaling Early during Infection with Epstein-Barr Virus, mBio 12(2) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Silva LF, Jones C, Two microRNAs encoded within the bovine herpesvirus 1 latency-related gene promote cell survival by interacting with RIG-I and stimulating NF-kappaB-dependent transcription and beta interferon signaling pathways, J Virol 86(3) (2012) 1670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].El-Zayat SR, Sibaii H, Mannaa FA, Toll-like receptors activation, signaling, and targeting: an overview, Bulletin of the National Research Centre 43(1) (2019) 187. [Google Scholar]
- [88].Boehme KW, Guerrero M, Compton T, Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells, J Immunol 177(10) (2006) 7094–102. [DOI] [PubMed] [Google Scholar]
- [89].Compton T, Kurt-Jones EA, Boehme KW, Belko J, Latz E, Golenbock DT, Finberg RW, Human cytomegalovirus activates inflammatory cytokine responses via CD14 and Toll-like receptor 2, J Virol 77(8) (2003) 4588–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Fiola S, Gosselin D, Takada K, Gosselin J, TLR9 contributes to the recognition of EBV by primary monocytes and plasmacytoid dendritic cells, J Immunol 185(6) (2010) 3620–31. [DOI] [PubMed] [Google Scholar]
- [91].Kurt-Jones EA, Chan M, Zhou S, Wang J, Reed G, Bronson R, Arnold MM, Knipe DM, Finberg RW, Herpes simplex virus 1 interaction with Toll-like receptor 2 contributes to lethal encephalitis, Proc Natl Acad Sci U S A 101(5) (2004) 1315–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA, Human Cytomegalovirus miR-UL112-3p Targets TLR2 and Modulates the TLR2/IRAK1/NFkappaB Signaling Pathway, PLoS Pathog 11(5) (2015) e1004881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM, Kaposi's sarcoma-associated herpesvirus microRNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression, J Virol 86(21) (2012) 11663–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Hancock MH, Hook LM, Mitchell J, Nelson JA, Human Cytomegalovirus MicroRNAs miR-US5-1 and miR-UL112-3p Block Proinflammatory Cytokine Production in Response to NF-kappaB-Activating Factors through Direct Downregulation of IKKalpha and IKKbeta, mBio 8(2) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Liang D, Gao Y, Lin X, He Z, Zhao Q, Deng Q, Lan K, A human herpesvirus miRNA attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKepsilon, Cell Res 21(5) (2011) 793–806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Platanias LC, Mechanisms of type-I- and type-II-interferon-mediated signalling, Nat Rev Immunol 5(5) (2005) 375–86. [DOI] [PubMed] [Google Scholar]
- [97].Zhang Q, Song X, Ma P, Lv L, Zhang Y, Deng J, Zhang Y, Human Cytomegalovirus miR-US33as-5p Targets IFNAR1 to Achieve Immune Evasion During Both Lytic and Latent Infection, Front Immunol 12 (2021) 628364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hooykaas MJG, van Gent M, Soppe JA, Kruse E, Boer IGJ, van Leenen D, Groot Koerkamp MJA, Holstege FCP, Ressing ME, Wiertz E, Lebbink RJ, EBV MicroRNA BART16 Suppresses Type I IFN Signaling, J Immunol 198(10) (2017) 4062–4073. [DOI] [PubMed] [Google Scholar]
- [99].Ramalingam D, Ziegelbauer JM, Viral microRNAs Target a Gene Network, Inhibit STAT Activation, and Suppress Interferon Responses, Sci Rep 7 (2017) 40813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Gallaher AM, Das S, Xiao Z, Andresson T, Kieffer-Kwon P, Happel C, Ziegelbauer J, Proteomic screening of human targets of viral microRNAs reveals functions associated with immune evasion and angiogenesis, PLoS Pathog 9(9) (2013) e1003584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Huang WT, Lin CW, EBV-encoded miR-BART20-5p and miR-BART8 inhibit the IFN-gamma-STAT1 pathway associated with disease progression in nasal NK-cell lymphoma, Am J Pathol 184(4) (2014) 1185–97. [DOI] [PubMed] [Google Scholar]
- [102].Münz C, Chijioke O, Natural killer cells in herpesvirus infections, F1000Res 6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Biron CA, Byron KS, Sullivan JL, Severe herpesvirus infections in an adolescent without natural killer cells, N Engl J Med 320(26) (1989) 1731–5. [DOI] [PubMed] [Google Scholar]
- [104].Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O, Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells, Cell Host Microbe 5(4) (2009) 376–85. [DOI] [PubMed] [Google Scholar]
- [105].Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, Goldman-Wohl D, Greenfield C, Yagel S, Hengel H, Altuvia Y, Margalit H, Mandelboim O, Host immune system gene targeting by a viral miRNA, Science 317(5836) (2007) 376–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Esteso G, Luzon E, Sarmiento E, Gomez-Caro R, Steinle A, Murphy G, Carbone J, Vales-Gomez M, Reyburn HT, Altered microRNA expression after infection with human cytomegalovirus leads to TIMP3 downregulation and increased shedding of metalloprotease substrates, including MICA, J Immunol 193(3) (2014) 1344–52. [DOI] [PubMed] [Google Scholar]
- [107].Enk J, Levi A, Weisblum Y, Yamin R, Charpak-Amikam Y, Wolf DG, Mandelboim O, HSV1 MicroRNA Modulation of GPI Anchoring and Downstream Immune Evasion, Cell Rep 17(4) (2016) 949–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Bauman Y, Mandelboim O, MicroRNA based immunoevasion mechanism of human polyomaviruses, RNA Biol 8(4) (2011) 591–4. [DOI] [PubMed] [Google Scholar]
- [109].Chaplin DD, Overview of the immune response, J Allergy Clin Immunol 125(2 Suppl 2) (2010) S3–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Albanese M, Tagawa T, Bouvet M, Maliqi L, Lutter D, Hoser J, Hastreiter M, Hayes M, Sugden B, Martin L, Moosmann A, Hammerschmidt W, Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8+ T cells, Proc Natl Acad Sci U S A 113(42) (2016) E6467–E6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Tagawa T, Albanese M, Bouvet M, Moosmann A, Mautner J, Heissmeyer V, Zielinski C, Lutter D, Hoser J, Hastreiter M, Hayes M, Sugden B, Hammerschmidt W, Epstein-Barr viral miRNAs inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing, J Exp Med 213(10) (2016) 2065–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Kim S, Lee S, Shin J, Kim Y, Evnouchidou I, Kim D, Kim YK, Kim YE, Ahn JH, Riddell SR, Stratikos E, Kim VN, Ahn K, Human cytomegalovirus microRNA miR-US4-1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1, Nat Immunol 12(10) (2011) 984–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Romania P, Cifaldi L, Pignoloni B, Starc N, D'Alicandro V, Melaiu O, Li Pira G, Giorda E, Carrozzo R, Bergvall M, Bergstrom T, Alfredsson L, Olsson T, Kockum I, Seppala I, Lehtimaki T, Hurme MA, Hengel H, Santoni A, Cerboni C, Locatelli F, D'Amato M, Fruci D, Identification of a Genetic Variation in ERAP1 Aminopeptidase that Prevents Human Cytomegalovirus miR-UL112-5p-Mediated Immunoevasion, Cell Rep 20(4) (2017) 846–853. [DOI] [PubMed] [Google Scholar]
- [114].Liu H, Yang L, Shi Z, Lv R, Yang X, Wang C, Chen L, Chang H, Functional analysis of prv-miR-LLT11a encoded by pseudorabies virus, J Vet Sci 20(6) (2019) e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Dolken L, Krmpotic A, Kothe S, Tuddenham L, Tanguy M, Marcinowski L, Ruzsics Z, Elefant N, Altuvia Y, Margalit H, Koszinowski UH, Jonjic S, Pfeffer S, Cytomegalovirus microRNAs facilitate persistent virus infection in salivary glands, PLoS Pathog 6(10) (2010) e1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Burke JM, Bass CR, Kincaid RP, Ulug ET, Sullivan CS, The Murine Polyomavirus MicroRNA Locus Is Required To Promote Viruria during the Acute Phase of Infection, J Virol 92(16) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zou W, Imperiale MJ, Biology of Polyomavirus miRNA, Front Microbiol 12 (2021) 662892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Murer A, Ruhl J, Zbinden A, Capaul R, Hammerschmidt W, Chijioke O, Munz C, MicroRNAs of Epstein-Barr Virus Attenuate T-Cell-Mediated Immune Control In Vivo, mBio 10(1) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Feederle R, Haar J, Bernhardt K, Linnstaedt SD, Bannert H, Lips H, Cullen BR, Delecluse HJ, The members of an Epstein-Barr virus microRNA cluster cooperate to transform B lymphocytes, J Virol 85(19) (2011) 9801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Seto E, Moosmann A, Gromminger S, Walz N, Grundhoff A, Hammerschmidt W, Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells, PLoS Pathog 6(8) (2010) e1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Wang Y, Tibbetts SA, Krug LT, Conquering the Host: Determinants of Pathogenesis Learned from Murine Gammaherpesvirus 68, Annual Review of Virology 8(1) (2021) 349–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Bertzbach LD, Kheimar A, Ali FAZ, Kaufer BB, Viral Factors Involved in Marek’s Disease Virus (MDV) Pathogenesis, Current Clinical Microbiology Reports 5(4) (2018) 238–244. [Google Scholar]
- [123].Hoxhaj G, Manning BD, The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism, Nat Rev Cancer 20(2) (2020) 74–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Bernhardt K, Haar J, Tsai MH, Poirey R, Feederle R, Delecluse HJ, A Viral microRNA Cluster Regulates the Expression of PTEN, p27 and of a bcl-2 Homolog, PLoS Pathog 12(1) (2016) e1005405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Cai L, Ye Y, Jiang Q, Chen Y, Lyu X, Li J, Wang S, Liu T, Cai H, Yao K, Li JL, Li X, Epstein-Barr virus-encoded microRNA BART1 induces tumour metastasis by regulating PTEN-dependent pathways in nasopharyngeal carcinoma, Nat Commun 6 (2015) 7353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Lyu X, Wang J, Guo X, Wu G, Jiao Y, Faleti OD, Liu P, Liu T, Long Y, Chong T, Yang X, Huang J, He M, Tsang CM, Tsao SW, Wang Q, Jiang Q, Li X, EBV-miR-BART1-5P activates AMPK/mTOR/HIF1 pathway via a PTEN independent manner to promote glycolysis and angiogenesis in nasopharyngeal carcinoma, PLoS Pathog 14(12) (2018) e1007484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Chen Y, Fachko D, Ivanov NS, Skinner CM, Skalsky RL, Epstein-Barr virus microRNAs regulate B cell receptor signal transduction and lytic reactivation, PLoS Pathog 15(1) (2019) e1007535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Li W, Jia X, Shen C, Zhang M, Xu J, Shang Y, Zhu K, Hu M, Yan Q, Qin D, Lee MS, Zhu J, Lu H, Krueger BJ, Renne R, Gao SJ, Lu C, A KSHV microRNA enhances viral latency and induces angiogenesis by targeting GRK2 to activate the CXCR2/AKT pathway, Oncotarget 7(22) (2016) 32286–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Hu M, Wang C, Li W, Lu W, Bai Z, Qin D, Yan Q, Zhu J, Krueger BJ, Renne R, Gao S-J, Lu C, A KSHV microRNA Directly Targets G Protein-Coupled Receptor Kinase 2 to Promote the Migration and Invasion of Endothelial Cells by Inducing CXCR2 and Activating AKT Signaling, PLOS Pathogens 11(9) (2015) e1005171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Bullard WL, Kara M, Gay LA, Sethuraman S, Wang Y, Nirmalan S, Esemenli A, Feswick A, Hoffman BA, Renne R, Tibbetts SA, Identification of murine gammaherpesvirus 68 miRNA-mRNA hybrids reveals miRNA target conservation among gammaherpesviruses including host translation and protein modification machinery, PLOS Pathogens 15(8) (2019) e1007843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Zhang H, Domma AJ, Goodrum FD, Moorman NJ, Kamil JP, The Akt Forkhead Box O Transcription Factor Axis Regulates Human Cytomegalovirus Replication, mBio 0(0) e01042–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].McKinney C, Perez C, Mohr I, Poly(A) binding protein abundance regulates eukaryotic translation initiation factor 4F assembly in human cytomegalovirus-infected cells, Proceedings of the National Academy of Sciences 109(15) (2012) 5627–5632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Buehler J, Carpenter E, Zeltzer S, Igarashi S, Rak M, Mikell I, Nelson JA, Goodrum F, Host signaling and EGR1 transcriptional control of human cytomegalovirus replication and latency, PLOS Pathogens 15(11) (2019) e1008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Hancock MH, Mitchell J, Goodrum FD, Nelson JA, Human Cytomegalovirus miR-US5-2 Downregulation of GAB1 Regulates Cellular Proliferation and UL138 Expression through Modulation of Epidermal Growth Factor Receptor Signaling Pathways, mSphere 5(4) (2020) e00582–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Dhillon AS, Hagan S, Rath O, Kolch W, MAP kinase signalling pathways in cancer, Oncogene 26(22) (2007) 3279–3290. [DOI] [PubMed] [Google Scholar]
- [136].Min K, Kim JY, Lee SK, Epstein-Barr virus miR-BART1-3p suppresses apoptosis and promotes migration of gastric carcinoma cells by targeting DAB2, Int J Biol Sci 16(4) (2020) 694–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Qiu J, Thorley-Lawson DA, EBV microRNA BART 18-5p targets MAP3K2 to facilitate persistence in vivo by inhibiting viral replication in B cells, Proc Natl Acad Sci U S A 111(30) (2014) 11157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].MacDonald BT, Tamai K, He X, Wnt/beta-catenin signaling: components, mechanisms, and diseases, Dev Cell 17(1) (2009) 9–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Basu S, Cheriyamundath S, Ben-Ze'ev A, Cell-cell adhesion: linking Wnt/β-catenin signaling with partial EMT and stemness traits in tumorigenesis, F1000Res 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Hsu CY, Yi YH, Chang KP, Chang YS, Chen SJ, Chen HC, The Epstein-Barr virus-encoded microRNA MiR-BART9 promotes tumor metastasis by targeting E-cadherin in nasopharyngeal carcinoma, PLoS Pathog 10(2) (2014) e1003974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Yan Q, Zeng Z, Gong Z, Zhang W, Li X, He B, Song Y, Li Q, Zeng Y, Liao Q, Chen P, Shi L, Fan S, Xiang B, Ma J, Zhou M, Li X, Yang J, Xiong W, Li G.-y., EBV-miR-BART10-3p facilitates epithelial-mesenchymal transition and promotes metastasis of nasopharyngeal carcinoma by targeting BTRC, Oncotarget 6 (2015) 41766–41782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Min K, Lee SK, EBV miR-BART10-3p Promotes Cell Proliferation and Migration by Targeting DKK1, Int J Biol Sci 15(3) (2019) 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Zhao Z, Liu W, Liu J, Wang J, Luo B, The effect of EBV on WIF1, NLK, and APC gene methylation and expression in gastric carcinoma and nasopharyngeal cancer, Journal of Medical Virology 89(10) (2017) 1844–1851. [DOI] [PubMed] [Google Scholar]
- [144].Gao G, Li J-T, Kong L, Tao L, Wei L, Human herpesvirus miRNAs statistically preferentially target host genes involved in cell signaling and adhesion/junction pathways, Cell Research 19(5) (2009) 665–667. [DOI] [PubMed] [Google Scholar]
- [145].Riley KJ, Rabinowitz GS, Yario TA, Luna JM, Darnell RB, Steitz JA, EBV and human microRNAs co-target oncogenic and apoptotic viral and human genes during latency, Embo j 31(9) (2012) 2207–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Lei X, Zhu Y, Jones T, Bai Z, Huang Y, Gao SJ, A Kaposi's sarcoma-associated herpesvirus microRNA and its variants target the transforming growth factor β pathway to promote cell survival, J Virol 86(21) (2012) 11698–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Liu Y, Sun R, Lin X, Liang D, Deng Q, Lan K, Kaposi's sarcoma-associated herpesvirus-encoded microRNA miR-K12-11 attenuates transforming growth factor beta signaling through suppression of SMAD5, J Virol 86(3) (2012) 1372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Samols MA, Skalsky RL, Maldonado AM, Riva A, Lopez MC, Baker HV, Renne R, Identification of cellular genes targeted by KSHV-encoded microRNAs, PLoS Pathog 3(5) (2007) e65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Cai L, Long Y, Chong T, Cai W, Tsang CM, Zhou X, Lin Y, Ding T, Zhou W, Zhao H, Chen Y, Wang J, Lyu X, Cho WC, Li X, EBV-miR-BART7-3p Imposes Stemness in Nasopharyngeal Carcinoma Cells by Suppressing SMAD7, Front Genet 10 (2019) 939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Wan XX, Yi H, Qu JQ, He QY, Xiao ZQ, Integrated analysis of the differential cellular and EBV miRNA expression profiles in microdissected nasopharyngeal carcinoma and non-cancerous nasopharyngeal tissues, Oncol Rep 34(5) (2015) 2585–601. [DOI] [PubMed] [Google Scholar]
- [151].Chan JY, Gao W, Ho WK, Wei WI, Wong TS, Overexpression of Epstein-Barr virus-encoded microRNA-BART7 in undifferentiated nasopharyngeal carcinoma, Anticancer Res 32(8) (2012) 3201–10. [PubMed] [Google Scholar]
- [152].Hancock MH, Crawford LB, Pham AH, Mitchell J, Struthers HM, Yurochko AD, Caposio P, Nelson JA, Human Cytomegalovirus miRNAs Regulate TGF-β to Mediate Myelosuppression while Maintaining Viral Latency in CD34(+) Hematopoietic Progenitor Cells, Cell Host Microbe 27(1) (2020) 104–114.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Randolph-Habecker J, Iwata M, Torok-Storb B, Cytomegalovirus mediated myelosuppression, J Clin Virol 25 Suppl 2 (2002) S51–6. [DOI] [PubMed] [Google Scholar]
- [154].Pham AH, Mitchell J, Botto S, Pryke KM, DeFilippis VR, Hancock MH, Human cytomegalovirus blocks canonical TGFβ signaling during lytic infection to limit induction of type I interferons, PLoS Pathog 17(8) (2021) e1009380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [155].Chi JQ, Teng M, Yu ZH, Xu H, Su JW, Zhao P, Xing GX, Liang HD, Deng RG, Qu LH, Zhang GP, Luo J, Marek's disease virus-encoded analog of microRNA-155 activates the oncogene c-Myc by targeting LTBP1 and suppressing the TGF-β signaling pathway, Virology 476 (2015) 72–84. [DOI] [PubMed] [Google Scholar]
- [156].Flemington EK, Herpesvirus lytic replication and the cell cycle: arresting new developments, J Virol 75(10) (2001) 4475–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Jiang S, Huang Y, Qi Y, He R, Liu Z, Ma Y, Guo X, Shao Y, Sun Z, Ruan Q, Human cytomegalovirus miR-US5-1 inhibits viral replication by targeting Geminin mRNA, Virol Sin 32(5) (2017) 431–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Diggins NL, Crawford LB, Hancock MH, Mitchell J, Nelson JA, Human Cytomegalovirus miR-US25-1 Targets the GTPase RhoA To Inhibit CD34(+) Hematopoietic Progenitor Cell Proliferation To Maintain the Latent Viral Genome, mBio 12(2) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Ma J, Nie K, Redmond D, Liu Y, Elemento O, Knowles DM, Tam W, EBV-miR-BHRF1-2 targets PRDM1/Blimp1: potential role in EBV lymphomagenesis, Leukemia 30(3) (2016) 594–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Lei X, Bai Z, Ye F, Xie J, Kim CG, Huang Y, Gao SJ, Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA, Nat Cell Biol 12(2) (2010) 193–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Gottwein E, Cullen BR, A human herpesvirus microRNA inhibits p21 expression and attenuates p21-mediated cell cycle arrest, J Virol 84(10) (2010) 5229–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Chivukula RR, Mendell JT, Circular reasoning: microRNAs and cell-cycle control, Trends Biochem Sci 33(10) (2008) 474–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Martín-Pérez D, Vargiu P, Montes-Moreno S, León EA, Rodríguez-Pinilla SM, Lisio LD, Martínez N, Rodríguez R, Mollejo M, Castellvi J, Pisano DG, Sánchez-Beato M, Piris MA, Epstein-Barr virus microRNAs repress BCL6 expression in diffuse large B-cell lymphoma, Leukemia 26(1) (2012) 180–183. [DOI] [PubMed] [Google Scholar]
- [164].Ross N, Gandhi MK, Nourse JP, The Epstein-Barr virus microRNA BART11-5p targets the early B-cell transcription factor EBF1, Am J Blood Res 3(3) (2013) 210–24. [PMC free article] [PubMed] [Google Scholar]
- [165].Yoon JH, Min K, Lee SK, Epstein-Barr Virus miR-BART17-5p Promotes Migration and Anchorage-Independent Growth by Targeting Kruppel-Like Factor 2 in Gastric Cancer, Microorganisms 8(2) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Wang J, Ge J, Wang Y, Xiong F, Guo J, Jiang X, Zhang L, Deng X, Gong Z, Zhang S, Yan Q, He Y, Li X, Shi L, Guo C, Wang F, Li Z, Zhou M, Xiang B, Li Y, Xiong W, Zeng Z, EBV miRNAs BART11 and BART17-3p promote immune escape through the enhancer-mediated transcription of PD-L1, Nature Communications 13(1) (2022) 866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Boss IW, Nadeau PE, Abbott JR, Yang Y, Mergia A, Renne R, A Kaposi's sarcoma-associated herpesvirus-encoded ortholog of microRNA miR-155 induces human splenic B-cell expansion in NOD/LtSz-scid IL2Rgammanull mice, J Virol 85(19) (2011) 9877–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Hansen A, Henderson S, Lagos D, Nikitenko L, Coulter E, Roberts S, Gratrix F, Plaisance K, Renne R, Bower M, Kellam P, Boshoff C, KSHV-encoded miRNAs target MAF to induce endothelial cell reprogramming, Genes Dev 24(2) (2010) 195–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Plaisance-Bonstaff K, Choi HS, Beals T, Krueger BJ, Boss IW, Gay LA, Haecker I, Hu J, Renne R, KSHV miRNAs decrease expression of lytic genes in latently infected PEL and endothelial cells by targeting host transcription factors, Viruses 6(10) (2014) 4005–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Yang Y, Boss IW, McIntyre LM, Renne R, A systems biology approach identified different regulatory networks targeted by KSHV miR-K12-11 in B cells and endothelial cells, BMC Genomics 15(1) (2014) 668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Duan F, Liao J, Huang Q, Nie Y, Wu K, HSV-1 miR-H6 inhibits HSV-1 replication and IL-6 expression in human corneal epithelial cells in vitro, Clin Dev Immunol 2012 (2012) 192791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Xia T, O'Hara A, Araujo I, Barreto J, Carvalho E, Sapucaia JB, Ramos JC, Luz E, Pedroso C, Manrique M, Toomey NL, Brites C, Dittmer DP, Harrington WJ Jr., EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1-3, Cancer Res 68(5) (2008) 1436–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Kim Y, Lee S, Kim S, Kim D, Ahn JH, Ahn K, Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection, PLoS Pathog 8(3) (2012) e1002577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Huang Y, Qi Y, Ma Y, He R, Ji Y, Sun Z, Ruan Q, The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112-1, Virol J 10 (2013) 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA, Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion, Cell Host Microbe 15(3) (2014) 363–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Skalsky RL, Cullen BR, Viruses, microRNAs, and host interactions, Annu Rev Microbiol 64 (2010) 123–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Han J, LaVigne CA, Jones BT, Zhang H, Gillett F, Mendell JT, A ubiquitin ligase mediates target-directed microRNA decay independently of tailing and trimming, Science 370(6523) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Shi CY, Kingston ER, Kleaveland B, Lin DH, Stubna MW, Bartel DP, The ZSWIM8 ubiquitin ligase mediates target-directed microRNA degradation, Science 370(6523) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Agut H, Bonnafous P, Gautheret-Dejean A, Laboratory and Clinical Aspects of Human Herpesvirus 6 Infections, Clinical Microbiology Reviews 28(2) (2015) 313–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Prusty BK, Gulve N, Chowdhury SR, Schuster M, Strempel S, Descamps V, Rudel T, HHV-6 encoded small non-coding RNAs define an intermediate and early stage in viral reactivation, npj Genomic Medicine 3(1) (2018) 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Buck AH, Perot J, Chisholm MA, Kumar DS, Tuddenham L, Cognat V, Marcinowski L, Dolken L, Pfeffer S, Post-transcriptional regulation of miR-27 in murine cytomegalovirus infection, RNA 16(2) (2010) 307–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Libri V, Helwak A, Miesen P, Santhakumar D, Borger JG, Kudla G, Grey F, Tollervey D, Buck AH, Murine cytomegalovirus encodes a miR-27 inhibitor disguised as a target, Proceedings of the National Academy of Sciences 109(1) (2012) 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Marcinowski L, Tanguy M, Krmpotic A, Rädle B, Lisnić VJ, Tuddenham L, Chane-Woon-Ming B, Ruzsics Z, Erhard F, Benkartek C, Babic M, Zimmer R, Trgovcich J, Koszinowski UH, Jonjic S, Pfeffer S, Dölken L, Degradation of cellular mir-27 by a novel, highly abundant viral transcript is important for efficient virus replication in vivo, PLoS Pathog 8(2) (2012) e1002510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Lee S, Song J, Kim S, Kim J, Hong Y, Kim Y, Kim D, Baek D, Ahn K, Selective degradation of host MicroRNAs by an intergenic HCMV noncoding RNA accelerates virus production, Cell Host Microbe 13(6) (2013) 678–90. [DOI] [PubMed] [Google Scholar]
- [185].Gorbea C, Mosbruger T, Cazalla D, A viral Sm-class RNA base-pairs with mRNAs and recruits microRNAs to inhibit apoptosis, Nature 550(7675) (2017) 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [186].Gorbea C, Mosbruger T, Nix DA, Cazalla D, Viral miRNA adaptor differentially recruits miRNAs to target mRNAs through alternative base-pairing, Elife 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Ungerleider N, Concha M, Lin Z, Roberts C, Wang X, Cao S, Baddoo M, Moss WN, Yu Y, Seddon M, Lehman T, Tibbetts S, Renne R, Dong Y, Flemington EK, The Epstein Barr virus circRNAome, PLoS Pathog 14(8) (2018) e1007206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Toptan T, Abere B, Nalesnik MA, Swerdlow SH, Ranganathan S, Lee N, Shair KH, Moore PS, Chang Y, Circular DNA tumor viruses make circular RNAs, Proc Natl Acad Sci U S A 115(37) (2018) E8737–e8745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Tagawa T, Gao S, Koparde VN, Gonzalez M, Spouge JL, Serquiña AP, Lurain K, Ramaswami R, Uldrick TS, Yarchoan R, Ziegelbauer JM, Discovery of Kaposi's sarcoma herpesvirus-encoded circular RNAs and a human antiviral circular RNA, Proc Natl Acad Sci U S A 115(50) (2018) 12805–12810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Liu Q, Shuai M, Xia Y, Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs, Cancer Manag Res 11 (2019) 8023–8031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Huang JT, Chen JN, Gong LP, Bi YH, Liang J, Zhou L, He D, Shao CK, Identification of virus-encoded circular RNA, Virology 529 (2019) 144–151. [DOI] [PubMed] [Google Scholar]
- [192].Gong LP, Chen JN, Dong M, Xiao ZD, Feng ZY, Pan YH, Zhang Y, Du Y, Zhang JY, Bi YH, Huang JT, Liang J, Shao CK, Epstein-Barr virus-derived circular RNA LMP2A induces stemness in EBV-associated gastric cancer, EMBO Rep 21(10) (2020) e49689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Kincaid RP, Sullivan CS, Virus-encoded microRNAs: an overview and a look to the future, PLoS Pathog 8(12) (2012) e1003018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Gottwein E, Mukherjee N, Sachse C, Frenzel C, Majoros WH, Chi JT, Braich R, Manoharan M, Soutschek J, Ohler U, Cullen BR, A viral microRNA functions as an orthologue of cellular miR-155, Nature 450(7172) (2007) 1096–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K, Regulation of the germinal center response by microRNA-155, Science 316(5824) (2007) 604–8. [DOI] [PubMed] [Google Scholar]
- [196].Dahlke C, Maul K, Christalla T, Walz N, Schult P, Stocking C, Grundhoff A, A microRNA encoded by Kaposi sarcoma-associated herpesvirus promotes B-cell expansion in vivo, PLoS One 7(11) (2012) e49435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Sin SH, Kim YB, Dittmer DP, Latency locus complements MicroRNA 155 deficiency in vivo, J Virol 87(21) (2013) 11908–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Zhao Y, Yao Y, Xu H, Lambeth L, Smith LP, Kgosana L, Wang X, Nair V, A functional MicroRNA-155 ortholog encoded by the oncogenic Marek's disease virus, J Virol 83(1) (2009) 489–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Zhao Y, Xu H, Yao Y, Smith LP, Kgosana L, Green J, Petherbridge L, Baigent SJ, Nair V, Critical role of the virus-encoded microRNA-155 ortholog in the induction of Marek's disease lymphomas, PLoS Pathog 7(2) (2011) e1001305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Yu ZH, Teng M, Sun AJ, Yu LL, Hu B, Qu LH, Ding K, Cheng XC, Liu JX, Cui ZZ, Zhang GP, Luo J, Virus-encoded miR-155 ortholog is an important potential regulator but not essential for the development of lymphomas induced by very virulent Marek's disease virus, Virology 448 (2014) 55–64. [DOI] [PubMed] [Google Scholar]
- [201].Zhang Y, Tang N, Luo J, Teng M, Moffat K, Shen Z, Watson M, Nair V, Yao Y, Marek's Disease Virus-Encoded MicroRNA 155 Ortholog Critical for the Induction of Lymphomas Is Not Essential for the Proliferation of Transformed Cell Lines, J Virol 93(17) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Yin Q, Wang X, Roberts C, Flemington EK, Lasky JA, Methylation status and AP1 elements are involved in EBV-mediated miR-155 expression in EBV positive lymphoma cells, Virology 494 (2016) 158–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Gatto G, Rossi A, Rossi D, Kroening S, Bonatti S, Mallardo M, Epstein – Barr virus latent membrane protein 1 trans-activates miR-155 transcription through the NFi B pathway, 2008. [DOI] [PMC free article] [PubMed]
- [204].Morrison K, Manzano M, Chung K, Schipma MJ, Bartom ET, Gottwein E, The Oncogenic Kaposi's Sarcoma-Associated Herpesvirus Encodes a Mimic of the Tumor-Suppressive miR-15/16 miRNA Family, Cell Rep 29(10) (2019) 2961–2969.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Manzano M, Forte E, Raja AN, Schipma MJ, Gottwein E, Divergent target recognition by coexpressed 5'-isomiRs of miR-142-3p and selective viral mimicry, Rna 21(9) (2015) 1606–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Yoon CJ, Chang MS, Kim DH, Kim W, Koo BK, Yun SC, Kim SH, Kim YS, Woo JH, Epstein-Barr virus-encoded miR-BART5-5p upregulates PD-L1 through PIAS3/pSTAT3 modulation, worsening clinical outcomes of PD-L1-positive gastric carcinomas, Gastric Cancer 23(5) (2020) 780–795. [DOI] [PubMed] [Google Scholar]
- [207].Skalsky RL, Kang D, Linnstaedt SD, Cullen BR, Evolutionary conservation of primate lymphocryptovirus microRNA targets, J Virol 88(3) (2014) 1617–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [208].Gottwein E, Cullen BR, Viral and cellular microRNAs as determinants of viral pathogenesis and immunity, Cell Host Microbe 3(6) (2008) 375–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Skalsky RL, Barr SA, Jeffery AJ, Blair T, Estep R, Wong SW, Japanese Macaque Rhadinovirus Encodes a Viral MicroRNA Mimic of the miR-17 Family, J Virol 90(20) (2016) 9350–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Kalluri R, LeBleu VS, The biology, function, and biomedical applications of exosomes, Science 367(6478) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Nahand JS, Mahjoubin-Tehran M, Moghoofei M, Pourhanifeh MH, Mirzaei HR, Asemi Z, Khatami A, Bokharaei-Salim F, Mirzaei H, Hamblin MR, Exosomal miRNAs: novel players in viral infection, Epigenomics 12(4) (2020) 353–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [212].Turchinovich A, Weiz L, Langheinz A, Burwinkel B, Characterization of extracellular circulating microRNA, Nucleic Acids Res 39(16) (2011) 7223–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Chevillet JR, Kang Q, Ruf IK, Briggs HA, Vojtech LN, Hughes SM, Cheng HH, Arroyo JD, Meredith EK, Gallichotte EN, Pogosova-Agadjanyan EL, Morrissey C, Stirewalt DL, Hladik F, Yu EY, Higano CS, Tewari M, Quantitative and stoichiometric analysis of the microRNA content of exosomes, Proc Natl Acad Sci U S A 111(41) (2014) 14888–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM, Functional delivery of viral miRNAs via exosomes, Proc Natl Acad Sci U S A 107(14) (2010) 6328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].Meckes DG Jr., Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N, Human tumor virus utilizes exosomes for intercellular communication, Proc Natl Acad Sci U S A 107(47) (2010) 20370–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O, Multivesicular bodies associate with components of miRNA effector complexes and modulate miRNA activity, Nat Cell Biol 11(9) (2009) 1143–9. [DOI] [PubMed] [Google Scholar]
- [217].Han Z, Liu X, Chen X, Zhou X, Du T, Roizman B, Zhou G, miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells, Proc Natl Acad Sci U S A 113(7) (2016) E894–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [218].Huang R, Wu J, Zhou X, Jiang H, Guoying Zhou G, Roizman B, Herpes Simplex Virus 1 MicroRNA miR-H28 Exported to Uninfected Cells in Exosomes Restricts Cell-to-Cell Virus Spread by Inducing Gamma Interferon mRNA, J Virol 93(21) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Neerukonda SN, Tavlarides-Hontz P, McCarthy F, Pendarvis K, Parcells MS, Comparison of the Transcriptomes and Proteomes of Serum Exosomes from Marek's Disease Virus-Vaccinated and Protected and Lymphoma-Bearing Chickens, Genes (Basel) 10(2) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Gourzones C, Ferrand FR, Amiel C, Verillaud B, Barat A, Guerin M, Gattolliat CH, Gelin A, Klibi J, Chaaben AB, Schneider V, Guemira F, Guigay J, Lang P, Jimenez-Pailhes AS, Busson P, Consistent high concentration of the viral microRNA BART17 in plasma samples from nasopharyngeal carcinoma patients--evidence of non-exosomal transport, Virol J 10 (2013) 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [221].Komabayashi Y, Kishibe K, Nagato T, Ueda S, Takahara M, Harabuchi Y, Circulating Epstein-Barr virus-encoded micro-RNAs as potential biomarkers for nasal natural killer/T-cell lymphoma, Hematol Oncol 35(4) (2017) 655–663. [DOI] [PubMed] [Google Scholar]
- [222].Koupenova M, Mick E, Corkrey HA, Huan T, Clancy L, Shah R, Benjamin EJ, Levy D, Kurt-Jones EA, Tanriverdi K, Freedman JE, Micro RNAs from DNA Viruses are Found Widely in Plasma in a Large Observational Human Population, Sci Rep 8(1) (2018) 6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Casas E, Ma H, Lippolis JD, Expression of Viral microRNAs in Serum and White Blood Cells of Cows Exposed to Bovine Leukemia Virus, Front Vet Sci 7 (2020) 536390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [224].Wang K, Zhang S, Weber J, Baxter D, Galas DJ, Export of microRNAs and microRNA-protective protein by mammalian cells, Nucleic Acids Res 38(20) (2010) 7248–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M, Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma, Proc Natl Acad Sci U S A 108(12) (2011) 5003–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT, MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins, Nat Cell Biol 13(4) (2011)423–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Ramayanti O, Verkuijlen S, Novianti P, Scheepbouwer C, Misovic B, Koppers-Lalic D, van Weering J, Beckers L, Adham M, Martorelli D, Middeldorp JM, Pegtel DM, Vesicle-bound EBV-BART13-3p miRNA in circulation distinguishes nasopharyngeal from other head and neck cancer and asymptomatic EBV-infections, Int J Cancer 144(10) (2019) 2555–2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Kawano Y, Iwata S, Kawada J, Gotoh K, Suzuki M, Torii Y, Kojima S, Kimura H, Ito Y, Plasma viral microRNA profiles reveal potential biomarkers for chronic active Epstein-Barr virus infection, J Infect Dis 208(5) (2013) 771–9. [DOI] [PubMed] [Google Scholar]
- [229].Tudor S, Giza DE, Lin HY, Fabris L, Yoshiaki K, D'Abundo L, Toale KM, Shimizu M, Ferracin M, Challagundla KB, Cortez MA, Fuentes-Mattei E, Tulbure D, Gonzalez C, Henderson J, Row M, Rice TW, Ivan C, Negrini M, Fabbri M, Morris JS, Yeung SC, Vasilescu C, Calin GA, Cellular and Kaposi's sarcoma-associated herpes virus microRNAs in sepsis and surgical trauma, Cell Death Dis 5 (2014) e1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [230].Li S, Zhu J, Zhang W, Chen Y, Zhang K, Popescu LM, Ma X, Lau WB, Rong R, Yu X, Wang B, Li Y, Xiao C, Zhang M, Wang S, Yu L, Chen AF, Yang X, Cai J, Signature microRNA expression profile of essential hypertension and its novel link to human cytomegalovirus infection, Circulation 124(2) (2011) 175–84. [DOI] [PubMed] [Google Scholar]
- [231].Mohammad AA, Rahbar A, Lui WO, Davoudi B, Catrina A, Stragliotto G, Mellbin L, Hamsten A, Ryden L, Yaiw KC, Soderberg-Naucler C, Detection of circulating hcmv-miR-UL112-3p in patients with glioblastoma, rheumatoid arthritis, diabetes mellitus and healthy controls, PLoS One 9(12) (2014) e113740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Diggins NL, Hancock MH, HCMV miRNA Targets Reveal Important Cellular Pathways for Viral Replication, Latency, and Reactivation, Noncoding RNA 4(4) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Diggins NL, Skalsky RL, Hancock MH, Regulation of Latency and Reactivation by Human Cytomegalovirus miRNAs, Pathogens 10(2) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].De Re V, Caggiari L, De Zorzi M, Fanotto V, Miolo G, Puglisi F, Cannizzaro R, Canzonieri V, Steffan A, Farruggia P, Lopci E, d’Amore ESG, Burnelli R, Mussolin L, Mascarin M, Epstein-Barr virus BART microRNAs in EBV-associated Hodgkin lymphoma and gastric cancer, Infectious Agents and Cancer 15(1) (2020) 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Gottwein E, Kaposi's Sarcoma-Associated Herpesvirus microRNAs, Front Microbiol 3 (2012) 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [236].Jurak I, Griffiths A, Coen DM, Mammalian alphaherpesvirus miRNAs, Biochim Biophys Acta 1809(11-12) (2011) 641–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Hancock MH, Skalsky RL, Roles of Non-coding RNAs During Herpesvirus Infection, Curr Top Microbiol Immunol 419 (2018) 243–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Murphy E, Vanicek J, Robins H, Shenk T, Levine AJ, Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: Implications for latency, Proceedings of the National Academy of Sciences 105(14) (2008) 5453–5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Sullivan CS, Pipas JM, T antigens of simian virus 40: molecular chaperones for viral replication and tumorigenesis, Microbiol Mol Biol Rev 66(2) (2002) 179–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Grey F, Meyers H, White EA, Spector DH, Nelson J, A human cytomegalovirus-encoded microRNA regulates expression of multiple viral genes involved in replication, PLoS Pathog 3(11) (2007) e163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Lau B, Poole E, Van Damme E, Bunkens L, Sowash M, King H, Murphy E, Wills M, Van Loock M, Sinclair J, Human cytomegalovirus miR-UL112-1 promotes the down-regulation of viral immediate early-gene expression during latency to prevent T-cell recognition of latently infected cells, J Gen Virol 97(9) (2016) 2387–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242].Tang S, Patel A, Krause PR, Novel less-abundant viral microRNAs encoded by herpes simplex virus 2 latency-associated transcript and their roles in regulating ICP34.5 and ICP0 mRNAs, J Virol 83(3) (2009) 1433–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [243].Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR, An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor, Proc Natl Acad Sci U S A 105(31) (2008) 10931–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [244].Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR, MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs, Nature 454(7205) (2008) 780–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Umbach JL, Wang K, Tang S, Krause PR, Mont EK, Cohen JI, Cullen BR, Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2, J Virol 84(2) (2010) 1189–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].DeLuca NA, McCarthy AM, Schaffer PA, Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4, J Virol 56(2) (1985) 558–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Smith CA, Bates P, Rivera-Gonzalez R, Gu B, DeLuca NA, ICP4, the major transcriptional regulatory protein of herpes simplex virus type 1, forms a tripartite complex with TATA-binding protein and TFIIB, J Virol 67(8) (1993) 4676–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Bellare P, Ganem D, Regulation of KSHV lytic switch protein expression by a virus-encoded microRNA: an evolutionary adaptation that fine-tunes lytic reactivation, Cell Host Microbe 6(6) (2009) 570–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [249].Lin X, Liang D, He Z, Deng Q, Robertson ES, Lan K, miR-K12-7-5p encoded by Kaposi's sarcoma-associated herpesvirus stabilizes the latent state by targeting viral ORF50/RTA, PLoS One 6(1) (2011) e16224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Manzano M, Shamulailatpam P, Raja AN, Gottwein E, Kaposi's sarcoma-associated herpesvirus encodes a mimic of cellular miR-23, J Virol 87(21) (2013) 11821–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Nachmani D, Lankry D, Wolf DG, Mandelboim O, The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination, Nat Immunol 11(9) (2010) 806–13. [DOI] [PubMed] [Google Scholar]
- [252].Tuddenham L, Jung JS, Chane-Woon-Ming B, Dolken L, Pfeffer S, Small RNA deep sequencing identifies microRNAs and other small noncoding RNAs from human herpesvirus 6B, J Virol 86(3) (2012) 1638–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Zhu JY, Strehle M, Frohn A, Kremmer E, Hofig KP, Meister G, Adler H, Identification and Analysis of Expression of Novel MicroRNAs of Murine Gammaherpesvirus 68, Journal of Virology 84(19) (2010) 10266–10275. [DOI] [PMC free article] [PubMed] [Google Scholar]


