Abstract
Membrane disruption using Bulk Electroporation (BEP) is a widely used non-viral method for delivering biomolecules into cells. Recently, its microfluidic counterpart, Localized Electroporation (LEP), has been successfully used for several applications ranging from reprogramming and engineering cells for therapeutic purposes to non-destructive sampling from live cells for temporal analysis. However, the side effects of these processes on gene expression, that can affect the physiology of sensitive stem cells are not well understood. Here, we use single cell RNA sequencing (scRNA-seq) to investigate the effects of BEP and LEP on murine neural stem cell (NSC) gene expression. Our results indicate that unlike BEP, LEP does not lead to extensive cell death or activation of cell stress response pathways that may affect their long-term physiology. Additionally, our demonstrations show that LEP is suitable for multi-day delivery protocols as it enables better preservation of cell viability and integrity as compared to BEP.
Keywords: Localized electroporation, Bulk electroporation, Intracellular delivery, Stem cell, Single cell RNA sequencing, Cell stress response
Graphical abstract
Highlights
-
•
Localized electroporation emerges as an efficient and precise alternative to bulk electroporation for cell transfection.
-
•
Adverse effects of electroporation on stem cell physiology are poorly understood.
-
•
Bulk electroporation induces extensive stress response gene expression in stem cells.
-
•
Minimal stress in localized electroporation is beneficial for multi-day transfections.
1. Introduction
Intracellular delivery of functional molecular cargo is a critical step in cell engineering and manipulation tasks within a broad range of applications such as studying the mechanisms of development or diseases, generating desirable cell phenotypes in vitro, and manufacturing novel cell based therapeutics [1,2]. Traditionally, viral vectors and bulk electroporation (BEP) are the commonly used methods to accomplish these cell engineering tasks. Viral vectors are efficient delivery vehicles for a wide range of cell types [1,2] and have been used to engineer therapeutic cells in pre-clinical studies as well as clinical trials [3,4]. However, viral vectors have limited payloads, can elicit an immune response, and require specialized facilities for manufacturing [5,6]. On the other hand, BEP has been a popular non-viral delivery method of choice but leads to massive losses in cell viability due to the high voltages applied, especially in the case of primary immune and stem cells [7,8]. More recently, it has also been shown that BEP leads to non-specific activation and loss of function in primary T-cells and Hematopoietic Stem and Progenitor Cells (HSPCs) [9,10].
To address these limitations, several microfluidic methods have been developed that provide promising new alternatives for intracellular delivery. For instance, flow-based microfluidic systems that mechanically perturb cells in micro-channels have been successfully used to engineer cells, particularly those of the hematopoietic lineage [11,12]. Although, these systems provide very high throughputs, they are restricted by cell-size dependent device design, clogging issues and the requirement to dissociate cells before flowing them through the micro-channels. This may not be ideal for sensitive adherent cell types that can undergo detachment induced apoptosis [13]. Probe-based technologies that use hollow nanopipettes [[14], [15], [16], [17]] or AFM cantilevers [18,19] for targeted single cell manipulation, alleviate this issue by delivering materials into cells in their adherent state. However, their serial nature limits their throughput to a few hundred cells in a single run. Substrate-based methods on the other hand employ arrays of high aspect-ratio nanostructures such as nanochannels [[20], [21], [22], [23]], nanoneedles [24,25] and nanostraws [10,26] for intracellular delivery. In many of these methods, localized electroporation (LEP) which is based on the application of a spatially controlled electric potential, is employed to disrupt the cell membrane, at regions where it interfaces with the nanostructures, to facilitate exogenous material delivery. These methods provide intermediate throughputs and have been proven to enable highly efficient intracellular delivery with superior dosage control, while preserving cell viability, in a variety of adherent and suspended cell types [27,28]. Consequently, they have been utilized for non-destructive extraction and temporal analysis of intracellular contents from live cells [29,30]. Thus, they present versatile platforms for cell manipulation and analysis. Although LEP using high-aspect ratio nanostructures offers a versatile method for cell manipulation, the potential off target effects of electroporation on cells are not completely understood. Consequently, its suitability for intracellular delivery or non-destructive extraction at multiple time points without disrupting normal cell health and function is also unknown. In this work and as a case study, we use single-cell RNA sequencing (scRNA-seq) to compare the effects of LEP and BEP treatment on murine neural stem cell (NSC) gene expression. We employ a 24 well-plate version of our previously demonstrated LEP system called the localized electroporation device (LEPD). The LEPD is a microfluidic platform that uses transparent polycarbonate (PC) membranes with nanochannels for transfection, long-term culture (∼7 days), and on-chip differentiation of murine NSCs in their adherent state [22]. Our results show that compared to BEP, LEP reduces the adverse effects of applied electric fields on the NSCs and does not lead to widespread stress response gene expression. It is worth noting here that previous studies have shown BEP to enhance cell stress as compared to microfluidic delivery methods [9,10]. However, those studies were performed using bulk gene expression assays that do not capture the heterogeneity of cellular response at the single cell level and its implications in normal health and function of the cell population. For example, in this case study of differentiating NSCs, use of bulk assays would not allow deciphering disparate effects of electroporation-induced cell stress in various sub-populations (such as glial progenitors, astrocytes and oligodendrocytes) and their implications on lineage commitment and cell fate. Furthermore, we report the use of LEP (using the LEPD) to deliver an siRNA into NSCs on consecutive days, to knockdown the expression of the transcription factor SOX9, which promotes glial lineage commitment. The experiments revealed high siRNA delivery efficiency, successful SOX9 knockdown, and preserved cell viability after a multi-day delivery protocol. Analysis of lineage specific markers in electroporated NSCs revealed that SOX9 knockdown effectively restricted the differentiation of NSCs into astrocytes. These results suggest that LEP can be used for multi-timepoint electroporation of cells without compromising their integrity and functionality. Overall, our study demonstrates that LEP is a promising method for minimally disruptive multi-timepoint intracellular delivery, which may find utility in developing efficient protocols for in vitro differentiation and control of cell fate.
2. Results
2.1. LEPD architecture and delivery process
Previous LEP platforms employing high-aspect ratio nanostructures have usually involved complex fabrication procedures, thus hindering their widescale adoption in research. Moreover, the substrates often lack optical transparency and are not suitable for long-term cell culture. As a result, treated cells require dissociation and re-plating onto compatible substrates for subsequent culture and monitoring, which again limits their application for sensitive adherent cell types. In this study we used a 24 well-plate format LEPD [23] that overcomes these limitations, allowing for multi-timepoint electroporation mediated delivery and investigation of the impact of LEP on differentiating NSCs. The 24 well-plate LEPD design enables the execution of multiple electroporation experiments in parallel (Fig. 1A and B and Supplementary Fig. 1E). Each LEPD unit consists of a glass cloning cylinder bonded to a track-etched PC membrane having nanochannels. To perform a delivery experiment, cells (∼50,000 per well) are first plated in the LEPDs and allowed to adhere on the surface of the PC membranes. Usually, the membrane surface is coated with an extracellular matrix to promote cell adhesion. Here, the membranes were coated with poly-D-lysine for NSC culture and differentiation. Once the cells adhere, an electric field is applied across the LEPD to permeabilize the cells and introduce the molecular cargo of interest. The applied electric field is localized only at the interface of the cell membrane and the nanochannels, which makes the process gentle, reduces the perturbation on the cells, and enhances electrophoretic cargo delivery [20,21]. Critically, the far field voltage applied in this process (20 V - 40 V) is much lower than that used in BEP (100–1000 V), which minimizes issues of joule heating, bubble formation, and changes in pH that are detrimental to cell health [1]. It is important to note that at the operating voltage range of LEP, the BEP systems cannot produce sufficiently strong electric fields across the cell membrane for permeabilization and cargo delivery. The PC membranes used in the LEPD are also biocompatible and optically transparent, allowing for the long-term culture and imaging of cells. To apply the electric field, the LEPD is placed between two electrodes. For the 24 well-plate configuration, the bottom electrode consists of an array of gold pads on a printed circuit board (PCB). This bottom electrode PCB is bonded to a bottomless 24 well-plate. Similarly, the top electrode is an array of gold coated electrode pins projecting from a PCB. The bottom electrode pads, the LEPDs, and the top electrode pins are designed to be concentric with the individual wells of the 24 well-plate. Once the 24 well-plate system is assembled, a train of electroporation pulses (see Methods) is applied to deliver the molecular cargo into the cells within the LEPDs. Importantly, each row in the 24 well-plate LEPD can be independently addressed allowing for the application of multiple pulse parameters within a single run. This allows for quick protocol optimization or execution of multiple experimental conditions simultaneously. Post electroporation, the LEPDs are transferred to a regular well plate for subsequent culture and analysis.
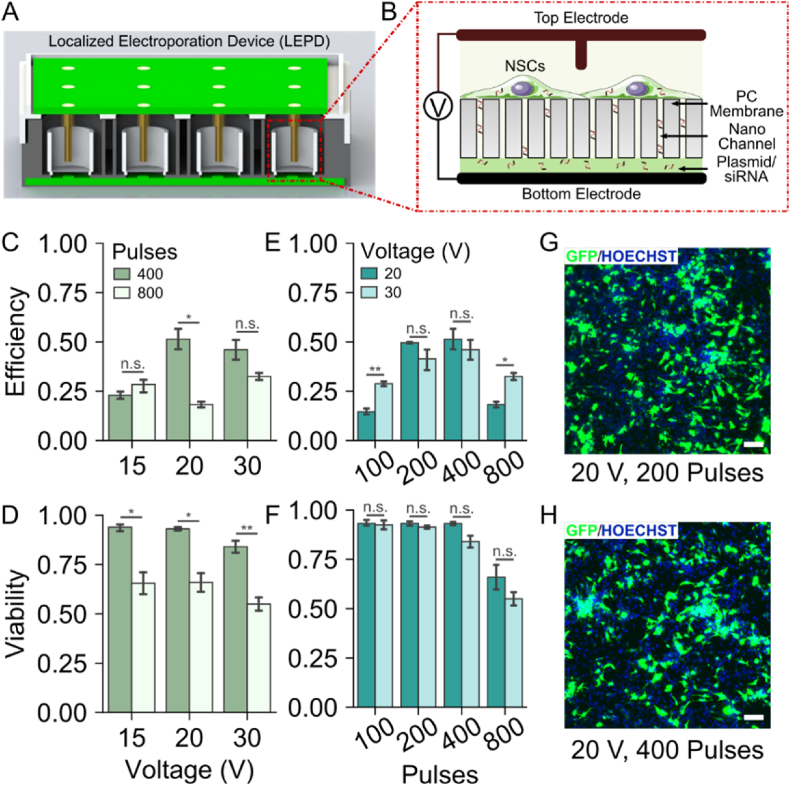
Fig. 1.
LEPD Design and Optimization. A) Cross section of the 24 well-plate LEPD CAD model. B) Schematic of the localized electroporation process using polycarbonate (PC) membranes with nanochannels. C) Transfection efficiency of GFP expressing plasmid in NSCs using the LEPD platform for different voltages, 24 h post-delivery. D) Viability of NSCs 24 h post electroporation with the LEPD for different voltages. E) Transfection efficiency of GFP expressing plasmid in NSCs using the LEPD platform for different pulse numbers, 24 h post-delivery. F) Viability of NSCs 24 h post electroporation with the LEPD for different pulse numbers. (Error Bars indicate S. E. M., n = 3. Statistical significance using Student's t-test; n.s. = no significance, ∗p ≤ 0.05, ∗∗p ≤ 0.01) G-H) Composite fluorescence micrographs showing successfully transfected NSCs (green) and all cell nuclei (blue) for the two optimal conditions (20 V, 200 Pulses and 20 V 400 Pulses). Images were acquired 24 h post electroporation with the LEPD. (Scale Bars: 50 μm).
2.2. Optimization of electroporation pulse parameters for NSC transfection
To effectively compare the downstream effects of LEP and BEP mediated intracellular delivery on NSCs, we first optimized the electroporation parameters to maximize performance of the two systems. We delivered a plasmid encoding green fluorescent protein (GFP), for expression into NSCs, and evaluated the transfection efficiency and cell viability 24 h later. For the LEPD we used a bilevel pulse (Supplementary Figs. 1A–B) for electroporation. We varied the voltage of the first level (15–40 V) and the total number of pulses applied (100–800 pulses). The pulse frequency (20 Hz), voltage of the second level (10 V) and the duration of the first and the second level of the pulse (0.25 ms and 2.0 ms respectively) were kept constant. The experiments were performed using a commercially available electroporation buffer. We observed that the 20 V, 200 pulse and 20 V, 400 pulse conditions yielded the best performance (Fig. 1C–H and Supplementary Fig. 2) both in terms of transfection efficiency (49.62 ± 0.63% and 51.40 ± 5.27%, respectively) and cell viability (93.17 ± 1.67% and 93.12 ± 1.42%, respectively). Intermediate voltages and pulse numbers were most effective, which corroborates with previous computational models explaining LEP mediated intracellular transport and experimental studies using LEP platforms [14,20,21,31]. At lower voltages and pulse numbers, the transfection efficiency was lower possibly due to low membrane permeabilization and insufficient material delivery. On the other hand, at higher voltages and pulse numbers the viability suffered likely due to electroporation induced cell death and DNA toxicity [20,21]. Additionally, similar transfection efficiencies were obtained for electroporation using cell culture media (Supplementary Fig. 1C) with the highest efficiency obtained for the 20 V condition (50.47 ± 5.30%). However, normal cell morphology was best preserved using the 20 V, 200 pulse condition in the electroporation buffer (Supplementary Fig. 1D). In comparison to LEPD, the optimized conditions of BEP yielded a transfection efficiency of 46.5 ± 5.37% after 24 h s and viability of 64.46 ± 8.92%, which is consistent with reported values of 4D nucleofector mediated electroporation of neural stem cells [32]. A comparison of transfection efficiency and viability for the optimal LEPD and BEP conditions is provided in Supplementary Table 1. Considering these findings, subsequent experiments with the LEPD were carried out using the 20 V, 400 pulse and 20 V, 200 pulse conditions.
2.3. Single cell RNA sequencing
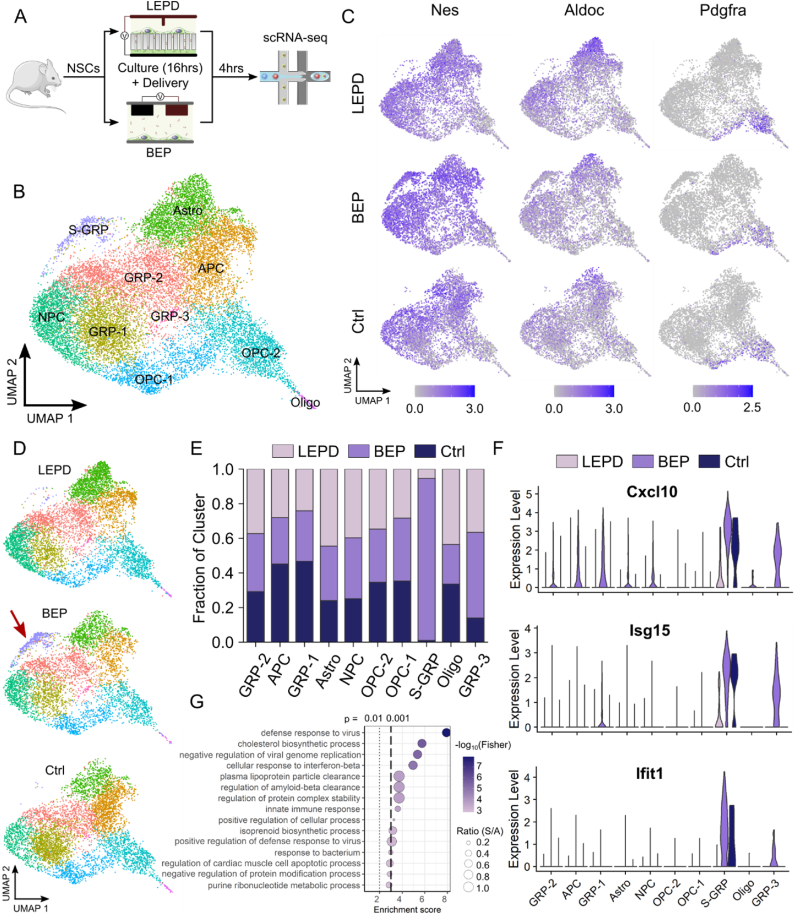
Recent reports have demonstrated that BEP can lead to transcriptomic level changes in HSPC and T-cell gene expression, while inhibiting their proliferative capacity [9,10]. This presents a concern, as genetic mis-regulation can compromise normal cell function. As a result, we were prompted to investigate the impact of LEP and BEP treatments on NSC gene expression, which may adversely affect their expected differentiation and lineage commitment trajectories. To this end, we performed a scRNA-seq experiment on NSCs electroporated using BEP or our LEPD system, 4 h post treatment (Fig. 2A). We note that for the previous optimization study, it was necessary to wait 24 h to maximize GFP expression and allow for cell apoptosis to obtain accurate transfection efficiency and cell viability metrics. On the other hand, our initial qPCR experiments showed that stress marker transcripts in BEP treated NSCs are not significantly different from no electroporation control at 24 h post electroporation (see Supplementary Fig. 3). This is likely because the stressed cells had already undergone apoptosis, and the expression of stress markers in the remaining cells had returned to baseline levels at 24 h. Hence, to capture the transient cell stress response due to electroporation, the sequencing analysis was performed at the 4 h time point. The integrated dataset of BEP treated, LEPD treated, and control (CTRL) NSC populations yielded 17,314 cells with over 5000 cells and an average of 3196 genes detected per cell in each of the three conditions, after filtering out the low-quality cells. Unsupervised clustering identified 10 clusters in the integrated dataset. We classified the clusters based on the expression of canonical markers expressed in neural cell types at similar developmental stages in mouse and human cortex [[33], [34], [35]], cell cycling phases, similarities/differences in gene expression across clusters and pseudo-time trajectories (Supplementary Figs. 4–7 and Supplementary Table 4). The populations we identified included neural progenitor cells (NPC), glial restricted progenitors (GRP), astrocyte progenitors (APC), oligodendrocyte progenitors (OPC), astrocytes (Astro) and oligodendrocytes (Oligo) (Fig. 2B). The expression pattern of stem cell and lineage-specific markers were found to be similar across all the three conditions (Fig. 2C and Supplementary Fig. 8). It is important to note that the ubiquitous expression of the NPC markers (e.g., Nes, Sox3 and Mki67) in all clusters indicates that the cell population is still in an early stage of development and all of them express progenitor-like characteristics. We labeled the cluster in the earliest stage of differentiation as NPCs. We labeled the other clusters as astrocytes, oligodendrocytes or their lineage restricted progenitors based on specific markers of those lineages and their cell cycling phases. Specifically, OPC-1 and OPC-2 clusters share expression of OPC markers but can be distinguished by cell cycle gene expression in OPC-1 and differentiation-associated gene expression in OPC-2, suggesting the two clusters may represent the same cell type in different cell cycle states or different points along the differentiation trajectory (OPC-1 = cycling, OPC-2 = more differentiated). Similarly, all GRP clusters share expression of glial progenitor associated genes but exhibit differences in cell cycle (GRP-1 = G1/S, GRP-2 = G2/M) or metabolic states (GRP-3). We found that the cells within individual clusters were evenly distributed across LEPD, BEP and CTRL conditions for the OPC, Oligo and GRP-2 populations (Fig. 2E and Supplementary Fig. 9). However, both the electroporated conditions (LEPD and BEP) constituted a larger proportion of the NPC and Astro clusters, whereas the CTRL condition constituted a larger proportion of the APC and GRP-1 clusters (Fig. 2E). Both electroporated conditions also induced enrichment of a small glial progenitor population that shares transcriptomic similarities with the GRP-2/OPC clusters, which we designated as GRP-3. Gene Ontology (GO) analysis revealed that GRP-3 shows elevated expression of genes regulating oxidative phosphorylation, protein instability, response to metal ions, pH imbalance and regulation of apoptosis (Supplementary Fig. 10) suggesting that GRP-3 is a population of stressed cells responding to known detrimental effects of BEP [36,37]. The GRP-3 cluster is predominantly represented by cells from the BEP condition (49.47% for BEP, 36.57% for LEPD and 13.96% for CTRL), consistent with the idea that cells under the BEP condition incur more stress (Fig. 2E and Supplementary Fig. 9).
Fig. 2.
Effects of bulk and localized electroporation on NSCs as revealed by scRNA-seq. A) Design of scRNA-seq experiment. B) UMAP plot of the clusters in the integrated dataset with cells from all conditions (LEPD, BEP and CTRL). Clusters were identified and labeled based on the expression of known markers. NPC – Neural Progenitor Cells, GRP – Glial Restricted Progenitors, S-GRP – Stressed Glial Restricted Progenitors, APC – Astrocyte Progenitor Cells, OPC – Oligodendrocyte Progenitor Cells, Astro – Astrocytes, Oligo – Oligodendrocytes. C) Gene expression distribution of a canonical marker for NSC (Nes), Astro (Aldoc) and Oligo (Pdgfra) populations respectively for each condition visualized in a UMAP feature plot. D) UMAP visualization of clusters in each condition separately, showing the overrepresentation of S-GRP (arrow) population in the BEP condition. E) Composition of clusters by condition. Cells were normalized to the total number of cells per sample prior to comparison of conditions. F) Expression of upregulated genes in S-GRP cluster across all clusters and conditions. G) Gene Ontology analysis using differentially expressed genes (DEGs) showing the top biological processes assigned to the S-GRP cluster. In Ratio (S/A), ‘S’ indicates the number of significant genes from the dataset (DEGs) found in a pathway and ‘A’ indicates the total number of genes annotated in that pathway.
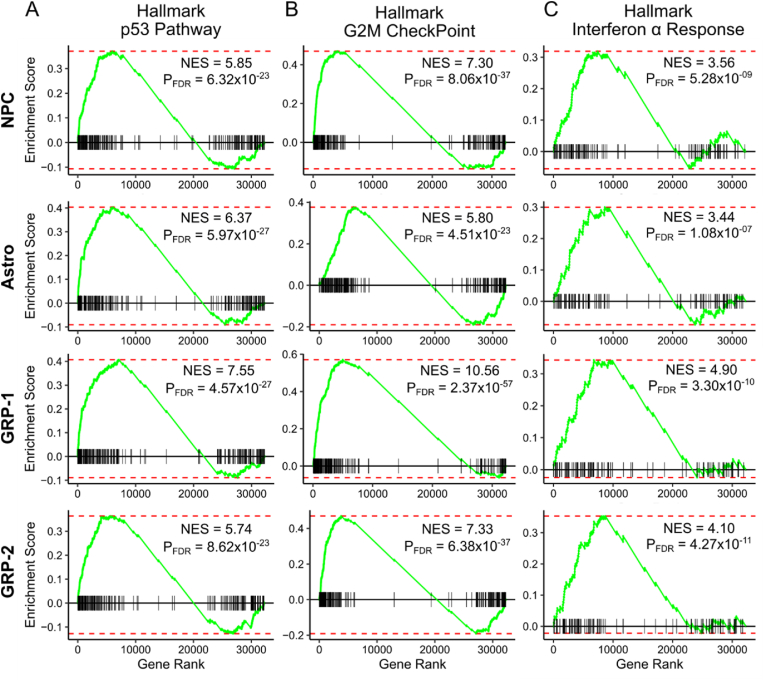
Interestingly, a distinct population of GRPs with upregulation of genes related to an immune response (Fig. 2F and Supplementary Fig. 8) was also present in the dataset, which we classified as stressed GRP (S-GRP). This population of S-GRP cells was prominent in the BEP condition with 92.50% of the cluster comprised of BEP treated cells (Fig. 2D and E). In comparison, LEPD treated, and control cells comprised 6.36% and 1.14% of this population respectively (Fig. 2E). To further investigate the characteristics of the S-GRP cells, we identified the differentially expressed genes (DEGs) for this cluster. Then, we used the DEGs to perform GO Analysis, which revealed the activation of interferon signaling and induction of apoptotic pathways in this population (Fig. 2G). These results indicate that electroporation mediated cargo delivery, especially with BEP, elicits an elevated cell stress response within a small subgroup of cells (S-GRP). Although clustering revealed a stressed cell population, additional adverse impact on the normal cell populations due to LEP and BEP treatment could still not be ruled out. Notably, some of the immune response markers present in the S-GRP cluster (e.g., Cxcl10 and Isg15) were also upregulated in the other clusters, for the BEP condition (Fig. 2F). This motivated us to further examine subtle gene expression changes in cell cluster-specific manner because of BEP and LEP treatments. For this purpose, we used Gene Set Enrichment Analysis (GSEA) with Hallmark gene-sets and performed a cluster wise comparison of the BEP and LEPD treated NSCs. From our analysis we observed an enrichment of p53 mediated stress, G2M checkpoint and interferon signaling pathways for most of the regular clusters (7 out of 9) in the BEP condition (Fig. 3A–C and Supplementary Fig. 12). The enrichment of these pathways indicate that BEP treatment induces a greater degree of cell stress, leading to an inhibition of cell cycle progression and possibly apoptosis. Some of these results also corroborate with prior studies where delayed cell proliferation was observed in BEP treated cells [9,10]. Here, it must be mentioned that these cell stress and apoptotic pathways are enriched in both BEP and LEPD populations when compared to the control population. However, as the analysis shows, the perturbation is more pronounced in the BEP treated cells. Overall, these results indicate that unlike BEP, the LEP offers a gentler mechanism of intracellular delivery and minimally perturbs cell function. This makes LEP well suited for multi-timepoint delivery experiments where minimizing cell death and preserving normal cell phenotype is of utmost importance.
Fig. 3.
Gene Set Enrichment Analysis of BEP treated NSCs in comparison to LEPD treated NSCs. Enrichment plots and normalized enrichment scores (NES) of p53 signaling, G2M checkpoint, and interferon-α signaling pathways using Broad Institute's Hallmark gene sets in different cell clusters (NPC, Astro, GRP-1 and GRP-2) for BEP treatment as compared to LEPD treatment (A–C). Enrichment of these pathways indicate elevated cell stress, impaired cell cycle progression, and potential apoptosis in the BEP condition. Genes were sorted in descending order based on the area under the receiver operating characteristics curve (AUROC) parameter obtained from a differential expression analysis using a Wilcoxon Rank Sum Test. Individual clusters from the BEP condition were compared to the LEPD condition.
2.4. Multi-timepoint cargo delivery
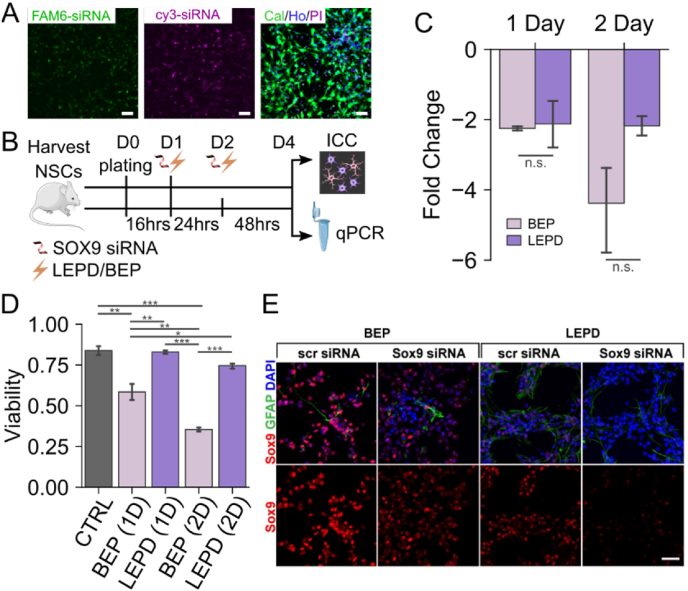
We hypothesized that since LEP is a gentle process and cell detachment/re-plating is not required in the LEPD, it should be possible to perform multiple rounds of intracellular delivery on NSCs using the LEPD to elicit a functional outcome, without any significant loss of cell viability. Directing stem cell fate by gene silencing via RNA interference has been reported previously [38]. Hence, to test our hypothesis of functional multi-timepoint delivery, we performed electroporation of small-interfering RNA (siRNA) targeted against SOX9, a transcription factor necessary for the differentiation of astrocytes from neural progenitors [[39], [40], [41]]. First, we confirmed successful delivery on consecutive days with the LEPD by using fluorophore tagged scrambled siRNAs (Fig. 4A). High cell viability was maintained (83.78 ± 3.09%) after the two-day delivery process (Fig. 4A). Next, we introduced SOX9 siRNA in adherent NSCs with BEP or LEPD for 1 day and examined at 72 hs post electroporation, or electroporated twice 24 hs apart and examined at the same 72 hs end point (Fig. 4B). Sox9 transcript level was examined via quantitative PCR, and cell differentiation was assessed by immunocytochemistry. We found that 1 day electroporation using either BEP or LEPD lead to approximately 50% reduction in SOX9 transcript level at the 72 hs time point (Fig. 4C). Although two-day electroporation with BEP increased the average SOX9 transcript reduction to 75% and SOX9 transcript in the LEPD samples remained at 50% knockdown, no statistical significance was observed between the two conditions (Fig. 4C). However, we found that two-day BEP leads to greater than 60% cell death while cell death with LEPD was less than 30% (Fig. 4D). At the protein level, electroporation of Sox9 siRNA with either BEP or LEPD led to a significant reduction in Sox9 expression compared to scrambled control siRNA as detected by immunocytochemistry at the 72 hs time point (Fig. 4E). To confirm that SOX9 siRNA electroporation is effective in altering astrocyte lineage commitment, we examined neural stem cell differentiation into the astrocytic lineage via immunocytochemistry at 72 hs post electroporation. We found that both BEP and LEPD electroporation of SOX9 siRNA reduce the number of GFAP-expressing cells and increase the number of Nestin-expressing cells when compared with scrambled siRNA electroporated controls, substantiating that SOX9 siRNA electroporation via both BEP and LEPD inhibited neural progenitor differentiation (Supplementary Fig. 13). These finding confirmed our previous assessment that while multi-timepoint BEP may prove advantageous in delivering cargo at a higher efficiency, LEPD is preferred in scenarios where cell survival and physiological stability is imperative to the study.
Fig. 4.
Multi-Timepoint Functional Cargo Delivery into NSCs using LEPD and BEP. A) Representative fluorescence micrographs showing successful delivery of Scrambled siRNA into NSCs using LEPD over two days while preserving cell viability. A FAM6 tagged scrambled siRNA was delivered to the NSCs on Day 1 (Left). A Cy3 tagged scrambled siRNA was delivered to the same cells on Day 2 (Middle). High cell viability was observed on Day 3. Cal: Calcein staining for live cells, Ho: Hoechst staining for cell nuclei, PI: Propidium Iodide staining for dead cells. Scale Bar: 50 μm. B) Schematic showing the experimental design, timeline, and subsequent validation for multi-day delivery of siRNA targeting SOX9. D0 – D4: Day 0 to Day 4, ICC: Immunocytochemistry. C) Expression of SOX9 in NSCs electroporated using BEP or LEPD with SOX9 targeting siRNA over multiple days. SOX9 expression analyzed by qRT-PCR 72 hs post the first round of electroporation. Expression levels are presented as fold change relative to control groups delivered with scrambled siRNA (Error Bars indicate S. E. M, n = 3 per condition. Statistical significance using Student's t-test; n.s. = no significance). D) Quantification of cell viability following electroporation of Scrambled siRNA using BEP or LEPD (Error Bars indicate S. E. M., n = 3 per condition. Statistical significance using analysis of variance (ANOVA) and post hoc Tukey test; n.s. = no significance, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). E) Representative images of immunocytochemical analysis of Sox9 (red) expression in cells 72 hs after BEP or LEPD mediated delivery of Scrambled siRNA (scr siRNA) or Sox9 siRNA. Reduced Sox9 were detected in both BEP and LEPD samples. (Scale bar, 50 μm).
3. Discussions
In this study we explore the implications of electroporation in general and LEP in particular, on cell physiology and function. First, we demonstrated that LEP (using our LEPD platform) can efficiently transfect NSCs (∼50%) while maintaining high cell viability (∼90%). Importantly, the LEPD design and electroporation pulse parameters were optimized for this study (see Methods) to enable successful transfection of stem cells with plasmids. Although previous platforms employing LEP have demonstrated efficient transfection of stem cells, these studies we primarily restricted to the use of smaller cargo such as mRNA that do not require nuclear delivery for gene expression [10,21]. Moreover, our 24 well-plate LEPD design enables the execution of multiple experiments in parallel along with long-term cell culture and imaging. In comparison to the LEPD, a commercially available gold standard BEP system provides similar transfection efficiencies (∼47%) but leads to considerable losses in cell viability (∼65%). This observation is consistent with previous reports of poor cell survival in stem cells due to a combination of detachment, BEP and delivery cargo induced stress [8]. Importantly, such significant losses are not observed for the LEPD, as cell detachment is not necessary, and the electroporation process is less stressful to the cells. Although the losses in BEP can be offset by starting with a larger cell population, the LEPD may be advantageous in situations where the starting sample is small or cannot be proliferated in vitro indefinitely (e.g., primary or rare cell populations).
Next, we compared the effects of BEP and LEP treatment on NSC gene expression using scRNA-seq. Previous studies have compared BEP with microfluidic methods using bulk transcriptomic methods, which preclude the resolution of a heterogeneous cellular response and its potential implications. In our scRNA-seq data, we observed the presence of a cell population (GRP-3) that exhibited glial progenitor expression profiles but with upregulation of pathways related to metal ion toxicity, oxidative stress, protein instability, intracellular pH regulation, and apoptosis. Generation of toxic metal ions, reactive oxygen species, and pH changes at the electrode surface has been reported to induce mitochondria mediated stress response and apoptosis in electroporated cells [36]. Moreover, joule heating can destabilize intracellular lipids and denature proteins. The presence of the GRP-3 population primarily in the electroporated samples and particularly in the BEP condition, indicates elevated cell stress caused due to the drastic changes in the cellular micro-environment brought about by the high applied voltages. Analysis of putative transcription factor (TF) binding sites shared among the GRP-3 enriched DEGs revealed E2F1, ELK3, and TAL1 as the top three TF regulators of GRP-3 enriched DEGs [42]. Future studies examining whether functional inhibition of these transcription factors alter expression of GRP-3 associated DEG may elucidate the molecular mechanisms underlying electroporation associated stress responses. In addition to the GRP-3 population, we identified a distinct stressed cluster (S-GRP) comprised of cells predominantly from the BEP condition. This cluster expressed elevated interferon and apoptotic signaling likely due to electroporation induced stress, which is consistent with previous studies that demonstrated that oxidative stress could induce interferon signaling, hinder neurogenesis and increase apoptosis in NSCs [43,44]. Involvement of the interferon signaling pathway is also supported by the analysis of putative TF binding sites in S-GRP enriched DEGs, which identified interferon regulatory factors (IRF1 and IRF8) as potential TF regulators of S-GRP enriched DEGs [42]. Moreover, the presence of this cluster could have two implications. First, it could indicate the heterogenous effect of BEP on different sub-populations or that cells are experiencing non-uniform and disparate electric fields, leading to a varied stress response with the small S-GRP sub-group exhibiting heightened stress response. Alternatively, it could indicate a temporal variation in stress response, where the cells in the normal clusters may exhibit heightened stress levels at a later timepoint. The heterogeneity of the stress response could have important implications in cases where cell types of interest could undergo selective apoptosis due to differential responses to EP-induced stress. Alternatively, activation of cellular stress signals in subpopulations of cells may lead to changes in cell-cell signaling, resulting in altered differentiation trajectories and affect terminal cell fate. Further, a comparison of cell clusters between BEP and LEP conditions showed that BEP treatment activates the p53 and G2M checkpoint pathways to a greater extent, indicating increased cell stress levels due to possible DNA damage that results in cell cycle arrest, loss of proliferative capacity, and cellular senescence [45,46]. Loss of cell proliferation due to BEP has also been reported for HSPCs in recent studies [9,10]. Although the scRNA-seq results suggest that BEP induces greater cell stress as compared to LEP, we did not observe any drastic deviations in lineage composition for either electroporation conditions as compared to the control sample. However, effects on lineage in the long-term cannot still be ruled out. Moreover, we did not observe any significant mis-regulation of signaling pathways other than stress related ones due to BEP as reported in prior comparative studies between BEP and other micro/nano technology-based delivery methods. This could be due to two reasons. First, human cells of the hematopoietic lineage were used in these studies [9,10], which could be more prone stress induced disruptions as compared to murine NSCs. Second, in the previously reported studies, earlier versions of commercial BEP technologies were used. The electroporation pulse protocol and buffer provided by the manufacturer may have been further optimized to reduce cell death and stress. This highlights the fact that bulk delivery methods are continuously improving and benchmarking new microfluidic methods against the latest bulk technologies is important to evaluate their potential. In summary, comparison of other electroporation and microfluidic intracellular delivery methods to the latest BEP technologies to determine their effects on overall cellular physiology could be an interesting future study.
Finally, we demonstrated a functional multi-day siRNA delivery protocol using both BEP and LEP to knock down the transcription factor SOX9. We observed that although for a single day delivery protocol both BEP and the LEPD platform exhibited similar knockdown efficiencies, BEP led to greater SOX9 knockdown compared to the LEPD over two days of delivery. However, the viability of cells was much lower for the BEP case. The lower knockdown efficiency using the LEPD over the two-day delivery protocol could result from nanochannel clogging that hinders cargo transport. Additionally, the local electric field experienced by the cells can be reduced after the first electroporation cycle, as cell death and proliferation alter the cell confluency. The impact of cell confluency on the electric field strength and the efficacy of LEP mediated delivery has been reported previously [20]. Further investigation and optimization of the electroporation pulse parameters and cargo concentration for the LEPD is necessary to improve its performance over multi-timepoint delivery, which is left for future studies. Overall, knockdown of SOX9 was effective in restricting astrocytic lineage commitment and enrich the cultures for NSC populations in both electroporation formats. This again highlights the scenarios where each technology may be beneficial – if highly efficient delivery is required, BEP is more suitable, while if cell survival is important, then LEP is preferable. Here, we must also mention that it may be possible to enhance the knockdown efficiency of the LEPD over the two-day delivery protocol to match BEP by applying a harsher pulse condition at the expense of a slight decrease in viability (Fig. 1). In general, the LEPD is likely to find applications in cases where the starting cell population is small and precious or multi-timepoint delivery is necessary such as for fibroblast reprogramming [47] or stem cell differentiation [48]. Not surprisingly, several recent studies have used LEP for non-destructive extraction and analysis of live cell contents due to its non-perturbative nature [29,30,49]. However, caution must be exercised in drawing inferences from multi-timepoint extraction data, as electroporation induced cell stress at the first timepoint can affect gene expression and confound subsequent extraction and analysis steps if the electroporation timepoints are not well spaced out to allow for sufficient cell recovery.
4. Conclusion
In summary, we present a comparative study of LEP and BEP, highlighting the advantages and disadvantages of each system and discuss possible use case scenarios. We report the first scRNA-seq study on cell-induced stress, which has implications not only in applications involving biomolecular delivery but also for temporal cell analysis using localized electroporation mediated sampling [29,30,50]. We note that while BEP is a mature technology, LEP is still in its infancy with great scope for improvement. For example, the nanostructure geometry, distribution, and density used for LEP, the role of electroporation buffer in LEP, and various electroporation pulse designs for different cell types or molecular cargo require systematic exploration to arrive at optimal solutions. Additionally, ways of scaling up LEP based platforms to handle several millions of cells must also be explored. With further developments, we expect LEP based platforms to be useful for both research and clinical applications, where cell survival, recovery, and maintenance of normal function is of primary importance.
5. Methods
5.1. Device fabrication
The PCBs that housed the top and bottom electrodes were designed using electronic-automation design software (EAGLE: AutoDesk) and fabricated in a PCB foundry. The bottom electrode PCB was bonded to a bottom-less 24 well-plate (Greiner Bio-One) using a silicone pressure adhesive (Adhesives Research). The gold-coated electrode pin heads were mounted on the top PCB using push-fit receptacles (MillMax). The LEPDs were assembled using sterilized Pyrex-glass cloning cylinders (Millipore Sigma) bonded to a track-etched polycarbonate (PC) membrane (it4ip) using silicon pressure adhesive. For this study, membranes with a nanochannel density of 2e6/cm2 and diameter of 400 nm were used.
5.2. Cell seeding and culture on LEPD
For NSC culture, the nanoporous PC membranes of the LEPDs were first coated with poly-D-Lysine (PDL, Sigma) and incubated for 4 h. Following this, the devices were washed with DPBS (Gibco). Neural progenitors were isolated and cultured as neurospheres from postnatal day 1 mice following established protocol [51]. Electroporation studies were performed using dissociated neurospheres within 4 passages. Neurospheres were dissociated by treating the spheres with 0.05% trypsin (Thermo Fisher Scientific) for 5 min which was then neutralized using trypsin inhibitor (Millipore Sigma). The spheres were then pipetted multiple times to obtain a suspension of single NSCs. In each LEPD, 50,000 NSCs were seeded and 200 μL of NSC media was added (see Supplementary Table 1 for media formulation). The NSCs were then cultured on the PC surface overnight in the incubator (at 37 °C with 5% CO2) to promote cell adhesion and tight nanopore-cell membrane contact before electroporation the next day. Post electroporation, the LEPD arrays were filled with fresh NSC media, transferred to 24 well-plates in an incubator and cultured for downstream imaging or molecular assays.
5.3. LEPD experimental protocol
To deliver the molecular cargo of interest into cells, a solution at the required concentration (plasmid – 200 ng/μL, siRNA – 10 μM) was prepared in the electroporation buffer. Following this, a 4 μL droplet of the solution was pipetted at the center of each well of the 24 well-plate LEPD system, on top of the bottom gold electrodes. Then, an array of LEPDs with cultured NSCs were placed over these cargo droplets. The LEPDs were filled with the appropriate electroporation buffer. The droplets formed a thin film between the bottom electrode and the nanoporous PC membrane of the LEPDs having the cells. Finally, the lid of the well plate, housing the top gold coated electrode pins, was placed over the LEPDs. These electrodes were immersed in the buffer within the LEPD chambers and formed a closed electrical circuit. A function generator (Agilent) connected to a voltage amplifier (OPA445, Texas Instruments) was used to apply the electroporation pulses (bilevel pulses (V1 = 10–40 V; T1 = 0.25 ms; V2 = 10 V; T2 = 2.0 ms), 100–800 pulses, 20 Hz). Resistance was measured for each LEPD in a well using a multimeter (Agilent) to ensure good electrical connection. The voltage traces were verified on an oscilloscope (Agilent). The pulse application, resistance measurement and voltage trace verification were controlled from a PC using a custom software written in C++.
5.4. Bulk electroporation
BEP was performed using the P3 primary cell kit (Cat#V4XP-3012, Lonza Bioscience) with the 4D-Nucleofector (Core Unit: AAF-1002 B, X Unit: AAF-1002X, Y Unit: AAF-1002Y, Lonza Bioscience) following the manufacturer's instructions. Both X and Y units were used to electroporate NSCs cultured in suspension or adherent condition for comparison with the LEPD. For suspension cultures, 2.5 × 106 NPCs were used per electroporation using program DS137 in the X unit, followed by plating 100,000 cells per well in 24 well-plates. Efficiency and viability were tested 4 and 24 hs post electroporation. For adherent cultures, 50,000 cells were plated 16 h s prior to electroporation using program DS137 with the Y unit, followed by single cell RNA sequencing analysis at 4 h s or viability/efficiency assessment at 4 hs and 24 hs post electroporation. In both methods, 5 μg of pMAX-GFP was used as the cargo to introduce into the cells.
5.5. Single cell RNA sequencing
Postnatal neural progenitors plated on poly-D-lysine (PDL)-coated polystyrene dishes (NUNC) or PDL-coated LEPD were dissociated using 0.05% Trypsin-EDTA 4 hs after electroporation. Cells from multiple wells (N = 6) were collected for sequencing for both BEP and LEP. Single cell gel bead generation and barcoding was performed with 10,000 cells from each experimental condition using the NextGEM single cell 3’ v3 expression chip on the Chromium platform (10x Genomics), followed by RNA sequencing using the HiSeq 4000 system (Illumina). Cell Ranger (version 6) was used to align the FASTQ files for each sample to the mouse reference genome (GRCm38) and generate the matrix files for subsequent analyses. The sequencing library summary metrics are provided in Supplementary Table 3.
5.6. scRNA-seq data preprocessing and quality control
ScRNA-seq data was processed using the R package Seurat [52] unless specified otherwise. First, a seurat object was created from the matrix files using the CreateSeuratObject function with samples (cells) that had at least 150 features (genes). Then, quality control steps were performed using the subset routine to remove cells with unique molecular identifier counts below 1500 (low quality cells or empty droplets) and above 6000 (multiple cells). Cells with more than 10% mitochondrial genes (low quality or dying cells) were also removed. The data was then normalized and scaled using the NormalizeData routine. For this purpose, the LogNormalize method was used that normalizes the gene expression measurements from each cell with the total expression, multiplies it with a scale factor (10,000 in this case) and then log transforms the resulting value. Following this, the principal components (top 30) were found using the top 2000 variable features. The FindVariableFeatures routine using the VST selection method was used for this purpose. Then the three datasets (BEP, LEPD and CTRL) were integrated using matched biological states (anchors). This was done using the FindIntegrationAnchors and IntegrateData functions of Seurat. The data was then scaled and centered using the ScaleData function to ensure that the mean is 0 and variance is 1 for all the features. Finally, the top 30 principal components were computed using the RunPCA routine which was used for the subsequent clustering and visualization steps.
5.7. Clustering and visualization
The high dimensional data was visualized in two dimensions using UMAP. Differences in cell cycle scores between G2M and S phase cells were regressed out using Seurat's CellCycleScoring function. Clustering was performed using shared-nearest neighbor graph construction and the Louvain algorithm. Differential expression testing for the clusters was performed using the MAST algorithm [53] with a log fold change threshold of 0.25 and a threshold of 0.1 for the minimum fraction of cells expressing the gene using the FindAllMarkers function (see Supplementary Table 5). The DEGs for each cluster were cross-referenced with multiple single cell RNA sequencing studies of neural cell types at similar developmental stages in mouse and human cortex [[33], [34], [35]]. Heatmaps and dot plots of the DEGs were constructed using the DoHeatmap and DotPlot functions respectively to correlate gene expression and cluster identity. Additionally, a pseudo-temporal trajectory was constructed using the Monocle3 package in Seurat to inform cluster identification. Cluster-specific expression and cluster identity were further validated with a small list of known neural cell type markers using the Featureplot function in Seurat to represent marker expressing cells in the UMAP plot.
5.8. Gene Ontology analysis
The differentially expressed genes from a cluster were used for Gene Ontology (GO) analysis. The analysis was performed using the TopGO Bioconductor package. Statistical significance was determined using Fisher's exact test.
5.9. Gene Set Enrichment Analysis
For each cluster, cells from BEP treated condition were compared to the cells from the LEPD treated condition. Cells from the BEP and LEPD conditions were also individually compared to the CTRL condition. Genes were ranked using the Wilcoxon rank sum test. The Area Under the Receiver Operating Characteristics Curve (AUROC) metric, arranged in descending order was used as the ranking statistic in all the cases. The AUROC metric was calculated using the presto package. This pre-ranked gene list was used for Gene Set Enrichment Analysis using the fgsea [54] package and Broad Institute's Hallmark gene sets.
5.10. Wide field fluorescence imaging
Fluorescence images were acquired on a Nikon Eclipse TE 2000 Microscope equipped with an Andor Zyla 5.5 sCMOS camera. Image acquisition was controlled using Micro-Manager software. A custom python script interfacing with Micro-Manager was used to acquire multi-channel images from the 24 well-plate format LEPDs.
5.11. Quantitative real-time PCR
RNA from cultured cells was collected using the RNAqueous micro kit (Thermo Fisher Scientific) at designated experimental time points, followed by cDNA generation using the Superscript IV VILO Mastermix kit (Thermo Fisher Scientific). qPCR was performed with specified primers using the SYBR green master mix (Applied Biosystems) in a CFX connect thermocycler (BioRad). Gene expression is analyzed using GAPDH as the standard and compared among experimental groups. At least three independent experiments were performed for all qPCR studies.
5.12. Immunocytochemistry and imaging
To examine the phenotypic changes of sequential siRNA electroporation via BEP or LEPD, electroporated cells were fixed at 72 h s after electroporation with 4% paraformaldehyde for 20mins at 4 °C and washed twice with phosphate buffer saline (PBS) before proceeding to immunostaining. Immunocytochemistry was performed following the general guidelines of previously described protocols [55]. Specific to this study is the use of antibody against Sox9 (Abcam ab185230, 1:1000). Stained coverslips were mounted in Prolong Gold antifade reagent (Thermo Fisher Scientific). Images of immunocytochemically stained cells were obtained on a SP5 confocal microscope (Leica Microsystem) using the 20x objective and 10 μm z-series with 1um optical thickness. 5 images from random fields of the stained coverslip from each experimental condition were taken for qualitative analysis of Sox9, GFAP, and Nestin expression.
5.13. Transfection efficiency and viability analysis
For estimating transfection efficiency and viability, the NSCs were first stained with Hoechst 33,342 (0.1 mg/mL, Life Technologies) to label all cell nuclei and propidium iodide (PI, 0.01 mg/mL, Life Technologies) to label dead cell nuclei. The stained NSCs were imaged using florescence microscopy. Transfection efficiency was calculated as the ratio of the number of GFP positive cells to the total number of cell nuclei (stained by Hoechst 33,342) in the field of view. Viability was calculated as the number of live cells (nuclei not stained by PI) to the total number of cells (nuclei stained by Hoechst 33,342) in the field of view.
5.14. Image analysis
Image analysis procedures was performed using the open-source image processing package, FIJI [56].
5.15. Statistical analysis
Statistical comparisons were performed using two-tailed Student's t-tests and ANOVA with post hoc Tukey test.
Credit author statement
P.M. and C.P. contributed equally to the mansucript. H.D.E. and J.A.K. conceived the project and provided overall guidance. C.A.P., P.M., N.P. and H.D.E. designed and validated the mutli-well LEPD system. P.M., C.P. and T.M. designed and conducted all the biological experiments, imaging and associated data processing. P.M., C.P., J.W.H. and C.H.P. analyzed the scRNA-seq data. R.B. provided guidance on scRNA-seq experimental design and data analysis. All authors discussed the results obtained and contributed towards writing the mansucript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Research reported in this publication was supported by the National Cancer Institute of the National Institutes of Health under award number U54CA19909 and NIH R21 award number 1R21GM132709-01.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2023.100601.
Contributor Information
John A. Kessler, Email: jakessler@northwestern.edu.
Horacio D. Espinosa, Email: espinosa@northwestern.edu.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
The scRNA-seq dataset discussed in this publication has been deposited in NCBI's Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE205507. Additional data will be made available on request.
References
- 1.Stewart M.P., et al. Vitro and ex vivo strategies for intracellular delivery. Nature. 2016;538:183–192. doi: 10.1038/nature19764. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee P., et al. Integrating micro and nano technologies for cell engineering and analysis: toward the next generation of cell therapy workflows. ACS Nano. 2022;16:15653–15680. doi: 10.1021/acsnano.2c05494. [DOI] [PubMed] [Google Scholar]
- 3.Vakulskas C.A., et al. A high-fidelity Cas9 mutant delivered as a ribonucleoprotein complex enables efficient gene editing in human hematopoietic stem and progenitor cells. Nat. Med. 2018;24:1216–1224. doi: 10.1038/s41591-018-0137-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stadtmauer E.A., et al. CRISPR-engineered T cells in patients with refractory cancer. Science. 2020;367:eaba7365. doi: 10.1126/science.aba7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colella P., Ronzitti G., Mingozzi F. Emerging issues in AAV-mediated in vivo gene therapy. Molecular Therapy - Methods & Clinical Development. 2018;8:87–104. doi: 10.1016/j.omtm.2017.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas C.E., Ehrhardt A., Kay M.A. Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 2003;4:346–358. doi: 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- 7.Roth T.L., et al. Reprogramming human T cell function and specificity with non-viral genome targeting. Nature. 2018;559:405–409. doi: 10.1038/s41586-018-0326-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li X.-L., et al. Highly efficient genome editing via CRISPR–Cas9 in human pluripotent stem cells is achieved by transient BCL-XL overexpression. Nucleic Acids Res. 2018;46:10195–10215. doi: 10.1093/nar/gky804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DiTommaso T., et al. Cell engineering with microfluidic squeezing preserves functionality of primary immune cells in vivo. Proc. Natl. Acad. Sci. USA. 2018;115 doi: 10.1073/pnas.1809671115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmiderer L., et al. Efficient and nontoxic biomolecule delivery to primary human hematopoietic stem cells using nanostraws. Proc. Natl. Acad. Sci. USA. 2020;117 doi: 10.1073/pnas.2001367117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharei A., et al. A vector-free microfluidic platform for intracellular delivery. Proc. Natl. Acad. Sci. USA. 2013;110:2082. doi: 10.1073/pnas.1218705110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deng Y., et al. Intracellular delivery of nanomaterials via an inertial microfluidic cell hydroporator. Nano Lett. 2018;18:2705–2710. doi: 10.1021/acs.nanolett.8b00704. [DOI] [PubMed] [Google Scholar]
- 13.Ohgushi M., et al. Molecular pathway and cell state responsible for dissociation-induced apoptosis in human pluripotent stem cells. Cell Stem Cell. 2010;7:225–239. doi: 10.1016/j.stem.2010.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Nathamgari S.S.P., et al. Nanofountain probe electroporation enables versatile single-cell intracellular delivery and investigation of postpulse electropore dynamics. Small. 2020;16 doi: 10.1002/smll.202002616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babakinejad B., et al. Local delivery of molecules from a nanopipette for quantitative receptor mapping on live cells. Anal. Chem. 2013;85:9333–9342. doi: 10.1021/ac4021769. [DOI] [PubMed] [Google Scholar]
- 16.Yang R., et al. Monoclonal cell line generation and CRISPR/Cas9 manipulation via single-cell electroporation. Small. 2018;14 doi: 10.1002/smll.201702495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee P., Patino C.A., Pathak N., Lemaitre V., Espinosa H.D. Deep learning-assisted automated single cell electroporation platform for effective genetic manipulation of hard-to-transfect cells. Small. 2022;18 doi: 10.1002/smll.202107795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kang W., et al. Nanofountain probe electroporation (NFP-E) of single cells. Nano Lett. 2013;13:2448–2457. doi: 10.1021/nl400423c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meister A., et al. FluidFM: combining atomic force microscopy and nanofluidics in a universal liquid delivery system for single cell applications and beyond. Nano Lett. 2009;9:2501–2507. doi: 10.1021/nl901384x. [DOI] [PubMed] [Google Scholar]
- 20.Mukherjee P., Nathamgari S.S.P., Kessler J.A., Espinosa H.D. Combined numerical and experimental investigation of localized electroporation-based cell transfection and sampling. ACS Nano. 2018;12:12118–12128. doi: 10.1021/acsnano.8b05473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Y., et al. Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. Proc. Natl. Acad. Sci. USA. 2019;116:7899. doi: 10.1073/pnas.1818553116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang W., et al. Microfluidic device for stem cell differentiation and localized electroporation of postmitotic neurons. Lab Chip. 2014;14:4486–4495. doi: 10.1039/C4LC00721B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patino, C. A. et al. Multiplexed high-throughput localized electroporation workflow with deep learning–based analysis for cell engineering. Sci. Adv. 8, eabn7637, doi:10.1126/sciadv.abn7637. [DOI] [PMC free article] [PubMed]
- 24.Chiappini, C. et al. Tutorial: using nanoneedles for intracellular delivery. Nat. Protoc. 16, 4539-4563, doi:10.1038/s41596-021-00600-7.(2021). [DOI] [PubMed]
- 25.Yoh H.Z., et al. Polymeric nanoneedle arrays mediate stiffness-independent intracellular delivery. Adv. Funct. Mater. 2022;32 doi: 10.1002/adfm.202104828. [DOI] [Google Scholar]
- 26.Cao, Y. et al. Universal intracellular biomolecule delivery with precise dosage control. Sci. Adv. 4, eaat8131, doi:10.1126/sciadv.aat8131. [DOI] [PMC free article] [PubMed]
- 27.Brooks J., et al. High throughput and highly controllable methods for in vitro intracellular delivery. Small. 2020;16 doi: 10.1002/smll.202004917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang W., McNaughton R.L., Espinosa H.D. Micro- and nanoscale technologies for delivery into adherent cells. Trends Biotechnol. 2016;34:665–678. doi: 10.1016/j.tibtech.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao Y., et al. Nondestructive nanostraw intracellular sampling for longitudinal cell monitoring. Proc. Natl. Acad. Sci. USA. 2017;114:E1866. doi: 10.1073/pnas.1615375114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukherjee P., et al. Temporal sampling of enzymes from live cells by localized electroporation and quantification of activity by SAMDI mass spectrometry. Small. 2020;16 doi: 10.1002/smll.202000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nathamgari S.S.P., Mukherjee P., Kessler J.A., Espinosa H.D. Localized electroporation with track-etched membranes. Proc. Natl. Acad. Sci. USA. 2019;116:22909–22910. doi: 10.1073/pnas.1911718116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bertram B., Wiese S., von Holst A. High-efficiency transfection and survival rates of embryonic and adult mouse neural stem cells achieved by electroporation. J. Neurosci. Methods. 2012;209:420–427. doi: 10.1016/j.jneumeth.2012.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Borrett M.J., et al. Single-cell profiling shows murine forebrain neural stem cells reacquire a developmental state when activated for adult neurogenesis. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.108022. [DOI] [PubMed] [Google Scholar]
- 34.Fu Y., et al. Heterogeneity of glial progenitor cells during the neurogenesis-to-gliogenesis switch in the developing human cerebral cortex. Cell Rep. 2021;34 doi: 10.1016/j.celrep.2021.108788. [DOI] [PubMed] [Google Scholar]
- 35.Weng Q., et al. Single-cell transcriptomics uncovers glial progenitor diversity and cell fate determinants during development and gliomagenesis. Cell Stem Cell. 2019;24:707–723.e708. doi: 10.1016/j.stem.2019.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart M.P., Langer R., Jensen K.F. Intracellular delivery by membrane disruption: mechanisms, strategies, and concepts. Chem. Rev. 2018;118:7409–7531. doi: 10.1021/acs.chemrev.7b00678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perl A., Gergely P., Puskas F., Banki K. Metabolic switches of T-cell activation and apoptosis. Antioxidants Redox Signal. 2002;4:427–443. doi: 10.1089/15230860260196227. [DOI] [PubMed] [Google Scholar]
- 38.Yau W.W.Y., Rujitanaroj P.-o., Lam L., Chew S.Y. Directing stem cell fate by controlled RNA interference. Biomaterials. 2012;33:2608–2628. doi: 10.1016/j.biomaterials.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 39.Kang P., et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74:79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stolt C.C., et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Canals I., et al. Rapid and efficient induction of functional astrocytes from human pluripotent stem cells. Nat. Methods. 2018;15:693–696. doi: 10.1038/s41592-018-0103-2. [DOI] [PubMed] [Google Scholar]
- 42.Lachmann A., et al. ChEA: transcription factor regulation inferred from integrating genome-wide ChIP-X experiments. Bioinformatics. 2010;26:2438–2444. doi: 10.1093/bioinformatics/btq466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hwang I., et al. Cellular stress signaling activates type-I IFN response through FOXO3-regulated lamin posttranslational modification. Nat. Commun. 2021;12:640. doi: 10.1038/s41467-020-20839-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Borsini A., et al. Interferon-alpha reduces human hippocampal neurogenesis and increases apoptosis via activation of distinct STAT1-dependent mechanisms. Int. J. Neuropsychopharmacol. 2018;21:187–200. doi: 10.1093/ijnp/pyx083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Harris S.L., Levine A.J. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 46.Taylor W.R., Stark G.R. Regulation of the G2/M transition by p53. Oncogene. 2001;20:1803–1815. doi: 10.1038/sj.onc.1204252. [DOI] [PubMed] [Google Scholar]
- 47.Kogut I., et al. High-efficiency RNA-based reprogramming of human primary fibroblasts. Nat. Commun. 2018;9:745. doi: 10.1038/s41467-018-03190-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoldan J., et al. Directing human embryonic stem cell differentiation by non-viral delivery of siRNA in 3D culture. Biomaterials. 2011;32:7793–7800. doi: 10.1016/j.biomaterials.2011.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.He G., et al. Hollow nanoneedle-electroporation system to extract intracellular protein repetitively and nondestructively. ACS Sens. 2018;3:1675–1682. doi: 10.1021/acssensors.8b00367. [DOI] [PubMed] [Google Scholar]
- 50.Patino C.A., et al. High-throughput microfluidics platform for intracellular delivery and sampling of biomolecules from live cells. ACS Nano. 2022;16:7937–7946. doi: 10.1021/acsnano.2c00698. [DOI] [PubMed] [Google Scholar]
- 51.Bonaguidi M.A., et al. LIF and BMP signaling generate separate and discrete types of GFAP-expressing cells. Development. 2005;132:5503–5514. doi: 10.1242/dev.02166. [DOI] [PubMed] [Google Scholar]
- 52.Stuart T., et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finak G., et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol. 2015;16 doi: 10.1186/s13059-015-0844-5. 278-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sergushichev A.A. An algorithm for fast preranked gene set enrichment analysis using cumulative statistic calculation. bioRxiv. 2016 doi: 10.1101/060012. [DOI] [Google Scholar]
- 55.Chen J., et al. BMP-responsive protease HtrA1 is differentially expressed in astrocytes and regulates astrocytic development and injury response. J. Neurosci. 2018;38:3840. doi: 10.1523/JNEUROSCI.2031-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schindelin J., et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq dataset discussed in this publication has been deposited in NCBI's Gene Expression Omnibus (GEO) and is accessible through GEO Series accession number GSE205507. Additional data will be made available on request.