Abstract
Background
The genus Induratia is based on Induratia apiospora, a xylarialean pyrenomycete from New Zealand with clypeate uniperitheciate stromata, hyaline apiospores and a nodulisporium-like anamorph. However, because of the lack of DNA data from the generic type, its phylogenetic affinities have remained unresolved. Recently, two fungal species with teleomorphs strikingly similar to Induratia were discovered in Thailand. However, they did not produce an anamorph and were found to be phylogenetically close to the species classified within the hyphomycete genus Muscodor, which was described after Induratia. Therefore, in 2020 the species of Muscodor were transferred to Induratia, and a new family Induratiaceae was proposed.
Results
We have encountered an unpublished ex-holotype strain of Induratia apiospora among the holdings of the ATCC collection, enabling detailed morphological and molecular phylogenetic investigations. We observed the characteristic nodulisporium-like anamorph described in the original publication. Phylogenetic analyses of multigene sequence data revealed a close relationship of Induratia apiospora to the Barrmaeliaceae, while a close relationship to the Induratia species formerly classified within Muscodor was rejected.
Conclusions
We here classify Induratia apiospora within the Barrmaeliaceae and consider Induratiaceae to be synonymous with the former. As the holotype specimen of Induratia apiospora is apparently lost, an isotype specimen from WSP is selected as lectotype. We also propose that the genus Muscodor is resurrected within the Xylariaceae, and formally transfer several Induratia species to Muscodor.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40529-023-00372-1.
Keywords: Fungi, Lectotypification, Sordariomycetes, Phylogeny, New combination
Background
The taxonomy of the Sordariomycetes and other Ascomycota has changed drastically in the past decade, owing to the advent of multi-locus phylogenies, which were often combined in polyphasic studies, using morphological and chemotaxonomic data as additional evidence. The currently proposed classification of genera and higher taxa (cf. Hyde et al. 2020; Wijayawardene et al. 2022) is steadily changing as new evidence becomes available, in particular when the species that have been first described in the pre-molecular era are cultured and sequenced for the first time. A fair example are genera of the Xylariales, and especially so the Xylariaceae s. lat. where it has become evident due to the availability of molecular data that the classical discrimination of higher taxa based on ascus structure (uni-, bitunicate), fruiting bodies, anamorph-teleomorph connections and ascospore morphology alone is not feasible. This has been reflected by the recent re-organization of the families (Wendt et al. 2018), where the results of a four-locus genealogy were better in agreement with chemotaxonomy and anamorphic morphology than with the classical concept based on ascospore shape. Interestingly, the aforementioned phylogeny was even backed up at genus level by a concurrent phylogenomic study using the amino acid sequences of 4912 orthologue genes for the core genera of the Hypoxylaceae (Wibberg et al. 2021). Other families and genera of Xylariales, for which by far not that many data are available, are in bad need of further studies. It is to be expected that it will take several years more to generate sufficient amounts of data to attain a stable phylogeny. Numerous taxa are still only known from old morphological descriptions (often restricted to the teleomorphs), or have only recently been recollected and cultured to generate molecular data and study their anamorphs for the first time. These studies sometimes revealed rather unexpected phylogenetic affinities but also showed the limits of a morphocentric approach for phylogenetic assessments (Jaklitsch et al. 2014; 2016; Voglmayr et al. 2022).
Aside from attempts to re-discover fresh specimens corresponding to the old fungal taxa, it may at times also be feasible to screen the inventories of the large culture collections, since those may contain valuable reference or ex-type strains that have not been reported in the original literature. The current paper describes such a case.
The genus Induratia was originally described by Samuels et al. (1987) based on a single specimen from New Zealand that featured uniperitheciate stromata, asci with an amyloid ascal apparatus and apiosporous ascospores. The authors also observed a nodulisporium-like conidial stage in the mycelial culture they obtained, and this new combination had at that time given rise to the erection of a new monotypic genus. In the following decades the genus almost remained forgotten, except that Miller and Huhndorf (2005) included a specimen they referred to as “Induratia sp. SMH 1255” originating from Puerto Rico in their phylogenetic study of Sordariales and other Sordariomycetes. However, Miller and Huhndorf (2005) neither included any morphological data of the specimen (which is housed in the fungarium of the University of Illinois as ILLS 82598), nor cultured it nor studied the anamorph. Samarakoon et al. (2020) later included the DNA sequences derived from this collection and provided microscopic details of the stromata and ascogenic structures. These morphological features resembled the drawings of Induratia apiospora by Samuels et al. (1987), even though the holotype specimen of this species has apparently been lost and could neither be located at the ZT nor the PDD herbarium. The molecular data of Induratia sp. SMH 1255 resembled those derived from two specimens that were freshly collected from Thailand. Moreover, they also clustered with the sequences of all hitherto described species of the genus Muscodor. This gave rise to the synonymisation of Induratia and Muscodor, with the former, older name taking priority over the younger Muscodor, and the erection of the new family Induratiaceae, which also included the similar genus Emarcea (Samarakoon et al. 2020).
However, we have recently encountered a culture labeled Induratia apiospora in the catalogue of the ATCC (Manassas, USA) via a random Google search on information on the genus on the Internet and decided to order and study it for comparison. The current paper is dedicated to the description of its characteristics and the necessary changes in the taxonomy of the Xylariales.
Methods
Morphological studies on strain ATCC 60639
A Google search for Induratia apiospora revealed a culture deposited as ATCC 60639 by G.J. Samuels not mentioned by Samuels et al. (1987). From discussions with two of the authors of the original paper, it was confirmed that this strain was indeed derived from the holotype specimen (O. Petrini and G.J. Samuels, personal communications). The strain Induratia apiospora (Samuels et al. 1987) was thus purchased from the American Type Culture Collection (ATCC, Mannassas, Virginia) under the accession ATCC 60639 and cultured on Yeast-Malt agar (10 g/L malt extract, 4 g/L D-glucose, 4 g/L yeast extract, supplemented with 20 g/L agar and adjusted to pH 6.3 prior to sterilization). The mycelia were transferred to a new plate once the strain had covered the medium by excision of a 5 mm2 square overgrown with mycelium on a regular basis (2–4 weeks). Plates were frequently monitored for sporulation.
For the morphological analysis of strain ATCC 60639, a small piece of sporulating mycelium was extracted and the dimensions of conidiogenous structures measured in distilled water and lactic acid. To observe the macro-morphology of the cultures, the strains were grown on Yeast-Malt agar (YM6.3; malt extract 10 g/L, yeast extract 4 g/L, D-glucose 4 g/L, agar 20 g/L, pH 6.3 before autoclaving), 2% Malt Extract Agar (MEA), Oatmeal Agar (OA, Sigma-Aldrich, Steinheim, Germany), and potato dextrose agar (PDA, Himedia, Mumbai, India) and the cultures checked at four weeks after inoculation. Photomicrographs were obtained using a DS-Fi3 camera connected to a Nikon eclipse Ni-U microscope (Nikon Europe BV, Amsterdam, Netherlands).
DNA extraction, PCR amplification and sequencing
The DNA extraction protocol and the solutions used for PCR amplification followed the description of Wendt et al. (2018). PCR programs followed Samarakoon et al. (2020), with the exemption of using the primer pair ITS1f and ITS4 (White et al. 1990) instead of ITS5 and ITS4. Briefly, the following settings were used: ITS: 94 °C for 30 s, 56 °C for 50 s, 72 °C for 60 s; LSU: LR0R/LR5: 94 °C for 30 s, 55 °C for 50 s, 72 °C for 60 s (Vilgalys and Hester 1990); SSU: NS1/NS4: 94 °C for 30 s, 54 °C for 50 s, 72 °C for 60 s (White et al. 1990); rpb2: fRPB2-5F/fRPB2-7cR: 95 °C for 45 s, 57 °C for 50 s, 72 °C for 90 s (Liu et al. 1999); tub2: T1/T22: 95 °C for 60 s, 54 °C for 110 s, 72 °C for 120 s (O’Donnell and Cigelnik 1997). PCR amplicons were purified as described in Wendt et al. (2018) and sequences generated with the Sanger sequencing method at the Microsynth sequencing company, using respective forward and reverse primers (Microsynth SeqLab GmbH, Göttingen, Germany). Sequences were assembled as a consensus sequence from both reads by using the de-novo assembly program contained in Geneious® R7.1.9 (Kearse et al. 2012). The generated and further used sequences in this study are listed with their respective GenBank Acc. No. in Table 1.
Table 1.
Feature table listing all sequences used for molecular phylogenetic inference. GenBank sequence accession numbers, type status of taxa, country of origin and specimen/strain numbers are given in the respective columns
| Species | Specimen or strain number | Origin | Status | GenBank accession numbers | References | |||
|---|---|---|---|---|---|---|---|---|
| ITS | LSU | rpb2 | tub2 | |||||
| Albicollum longisporum | CBS 147283 | Spain | HT | ON869286 | ON869286 | ON808465 | ON808509 | Voglmayr et al. (2022) |
| Albicollum vincensii | CBS 147286 | Austria | ET | ON869297 | ON869297 | ON808475 | ON808519 | Voglmayr et al. (2022) |
| Amphirosellinia nigrospora | HAST 91092308 | Taiwan | HT | GU322457 | N/A | GQ848340 | GQ495951 | Hsieh et al. (2010) |
| Annulohypoxylon truncatum | CBS 140778 | Texas | ET | KY610419 | KY610419 | KY624277 | KX376352 | Kuhnert et al. (2017), Wendt et al. (2018) |
| Anthostomelloides krabiensis | MFLUCC 15–0678 | Thailand | HT | KX305927 | KX305928 | KX305929 | N/A | Tibpromma et al. (2017) |
| Astrocystis concavispora | MFLUCC 14–0174 | Italy | KP297404 | KP340545 | KP340532 | KP406615 | Daranagama et al. (2015) | |
| Barrmaelia macrospora | CBS 142768 | Austria | ET | KC774566 | KC774566 | MF488995 | MF489014 | Voglmayr et al. (2018) |
| Barrmaelia moravica | CBS 142769 | Austria | ET | MF488987 | MF488987 | MF488996 | MF489015 | Voglmayr et al. (2018) |
| Barrmaelia oxyacanthae | CBS 142770 | Austria | MF488988 | MF488988 | MF488997 | MF489016 | Voglmayr et al. (2018) | |
| Barrmaelia rappazii | CBS 142771 | Norway | HT | MF488989 | MF488989 | MF488998 | MF489017 | Voglmayr et al. (2018) |
| Barrmaelia rhamnicola | CBS 142772 | France | ET | MF488990 | MF488990 | MF488999 | MF489018 | Voglmayr et al. (2018) |
| Biscogniauxia marginata | MFLUCC 12–0740 | France | KJ958407 | KJ958408 | KJ958409 | KJ958406 | Daranagama et al. (2015) | |
| Camillea obularia | ATCC 28093 | Puerto Rico | KY610384 | KY610429 | KY624238 | KX271243 | Wendt et al. (2018) | |
| Clypeosphaeria mamillana | CBS 140735 | France | ET | KT949897 | KT949897 | MF489001 | MH704637 | Jaklitsch et al. (2016), Voglmayr et al. (2018), Liu et al. (2019) |
| Collodiscula japonica | CBS 124266 | China | JF440974 | JF440974 | KY624273 | KY624316 | Jaklitsch and Voglmayr (2011), Wendt et al. (2018) | |
| Creosphaeria sassafras | STMA 14087 | Argentina | KY610411 | KY610468 | KY624265 | KX271258 | Wendt et al. (2018) | |
| Daldinia concentrica | CBS 113277 | Germany | AY616683 | KY610434 | KY624243 | KC977274 | Triebel et al. (2005), Kuhnert et al. (2014), Wendt et al. (2018) | |
| Dematophora necatrix | CBS 349.36 | Argentina | AY909001 | KF719204 | KY624275 | KY624310 | Pelaez et al. (2008), Wendt et al. (2018) | |
| Diatrype disciformis | CBS 197.49 | Netherlands | N/A | DQ470964 | DQ470915 | N/A | Zhang et al. (2006) | |
| Digitodochium amoenum | CBS 147285 | Austria | ET | ON869303 | ON869303 | ON808481 | ON808525 | Voglmayr et al. (2022) |
| Emarcea castanopsidicola | CBS 117105 | Thailand | HT | AY603496 | MK762717 | MK791285 | MK776962 | Duong et al. (2004), Samarakoon et al. (2020) |
| Emarcea eucalyptigena | CBS 139908 | Malaysia | HT | KR476733 | MK762718 | MK791286 | MK776963 | Crous et al. (2015), Samarakoon et al. (2020) |
| Entalbostroma erumpens | ICMP 21152 | New Zealand | HT | KX258206 | N/A | KX258204 | KX258205 | Johnston et al. (2016) |
| Entoleuca mammata | 100 J.D.R | France | GU300072 | N/A | GQ844782 | GQ470230 | Hsieh et al. (2010) | |
| Entonaema liquescens | ATCC 46302 | USA | KY610389 | KY610443 | KY624253 | KX271248 | Wendt et al. (2018) | |
| Entosordaria perfidiosa | CBS 142773 | Austria | ET | MF488993 | MF488993 | MF489003 | MF489021 | Voglmayr et al. (2018) |
| Entosordaria quercina | CBS 142774 | Greece | HT | MF488994 | MF488994 | MF489004 | MF489022 | Voglmayr et al. (2018) |
| Eutypa lata | UCR-EL1 | USA | JGI | JGI | JGI | JGI | ||
| Graphostroma platystomum | CBS 270.87 | France | JX658535 | DQ836906 | KY624296 | HG934108 | Zhang et al. (2006), Stadler et al. (2014), Koukol et al. (2015), Wendt et al. (2018) | |
| Hypocreodendron sanguineum | J.D.R. 169 | Mexico | GU322433 | N/A | GQ844819 | GQ487710 | Hsieh et al. (2010) | |
| Hypomontagnella monticulosa | MUCL 54604 | French Guiana | ET | KY610404 | KY610487 | KY624305 | KX271273 | Wendt et al. (2018) |
| Hypoxylon fragiforme | MUCL 51264 | Germany | ET | KC477229 | KM186295 | KM186296 | KX271282 | Stadler et al. (2013), Daranagama et al. (2015), Wendt et al. (2018) |
| Induratia apiospora | ATCC 60639 | New Zealand | HT | OP862879 | OP862881 | OP879469 | OP879468 | This study |
| Jackrogersella multiformis | CBS 119016 | Germany | ET | KC477234 | KY610473 | KY624290 | KX271262 | Kuhnert et al. (2014), Kuhnert et al. (2017), Wendt et al. (2018) |
| Kretzschmaria deusta | CBS 163.93 | Germany | KC477237 | KY610458 | KY624227 | KX271251 | Stadler et al. (2013), Wendt et al. (2018) | |
| Leptomassaria simplex | CBS 147282 | Austria | ET | ON869305 | ON869305 | ON808483 | ON808527 | Voglmayr et al. (2022) |
| Linosporopsis ischnotheca | CBS 145761 | Switzerland | ET | MN818952 | MN818952 | MN820708 | MN820715 | Voglmayr and Beenken (2020) |
| Linosporopsis ochracea | CBS 145999 | Germany | ET | MN818958 | MN818958 | MN820714 | MN820721 | Voglmayr et al. (2022) |
| Lopadostoma turgidum | CBS 133207 | Austria | ET | KC774618 | KC774618 | KC774563 | MF489024 | Jaklitsch et al. (2014), Voglmayr et al. (2018) |
| Muscodor albus | 9-6 | N/A | HM034857 | HM034865 | N/A | HM034844 | Zhang et al. (2010) | |
| Muscodor albus | MONT 620 | HT | AF324336 | N/A | N/A | N/A | Worapong et al. (2001) | |
| Muscodor brasiliensis | LGMF 1256 | HT | KY924494 | N/A | MF510171 | N/A | Pena et al. (2019) | |
| Muscodor camphorae | NFCCI 3236 | HT | KC481681 | N/A | N/A | N/A | Meshram et al. (2017) | |
| Muscodor cinnanomi | BCC 38842 | HT | GQ848369 | N/A | N/A | N/A | Suwannarach et al. (2010) | |
| Muscodor coffeanum | COAD 1842 | Brazil | HT | KM514680 | N/A | KP862881 | N/A | Hongsanan et al. (2015) |
| Muscodor crispans | MONT 2347 | HT | EU195297 | N/A | N/A | N/A | Mitchell et al. (2008) | |
| Muscodor darjeelingensis | NFCCI 3095 | HT | JQ409997 | N/A | N/A | N/A | Saxena et al. (2014) | |
| Muscodor equiseti | JCM 18233 | HT | JX089322 | N/A | N/A | N/A | Suwannarach et al. (2013) | |
| Muscodor fengyangensis | CGMCC 2862 | China | HT | HM034856 | HM034859 | HM034849 | HM034843 | Zhang et al. (2010) |
| Muscodor ghoomensis | NFCCI 3234 | HT | KF537625 | N/A | N/A | N/A | Meshram et al. (2015) | |
| Muscodor indicus | NFCCI 3235 | HT | KF537626 | N/A | N/A | N/A | Meshram et al. (2015) | |
| Muscodor kashayum | NFCCI 2947 | HT | KC481680 | N/A | N/A | N/A | Meshram et al. (2013) | |
| Muscodor musae | JCM 18230 | HT | JX089323 | N/A | N/A | N/A | Suwannarach et al. (2013) | |
| Muscodor oryzae | JCM 18231 | HT | JX089321 | N/A | N/A | N/A | Suwannarach et al. (2013) | |
| Muscodor roseus | MONT 2098 | HT | AH010859 | N/A | N/A | N/A | Worapong et al. (2002) | |
| Muscodor sp. | SMH 1255 | MN250031 | AY780069 | N/A | AY780119 | Miller and Huhndorf (2005), Samarakoon et al. (2020) | ||
| Muscodor strobelii | NFCCI 2907 | HT | JQ409999 | N/A | N/A | N/A | Meshram et al. (2014) | |
| Muscodor suthepensis | JCM 18232 | HT | JN558830 | N/A | N/A | N/A | Suwannarach et al. (2013) | |
| Muscodor suturae | MSUB 2380 | HT | JF938595 | N/A | N/A | N/A | Kudalkar et al. (2012) | |
| Muscodor thailandica | MFLUCC 17-2669 | Thailand | HT | MK762707 | MK762714 | MK791283 | MK776960 | Samarakoon et al. (2020) |
| Muscodor tigerensis | NFCCI 3172 | HT | JQ409998 | N/A | N/A | N/A | Saxena et al. (2015) | |
| Muscodor vitigenus | MONT P-15 | HT | AY100022 | N/A | N/A | N/A | Daisy et al. (2002) | |
| Muscodor yucatanensis | MEXU 25511 | HT | FJ917287 | N/A | N/A | N/A | González et al. (2009) | |
| Muscodor yunnanensis | CGMCC 3.18908 | China | HT | MG866046 | MG866038 | MG866059 | MG866066 | Chen et al. (2019) |
| Muscodor ziziphi | MFLUCC 17-2662 | Thailand | HT | MK762705 | MK762712 | MK791281 | MK776958 | Samarakoon et al. (2020) |
| Magnostiolata mucida | MFLU 19-2133 | Thailand | HT | MW240673 | MW240603 | MW658652 | MW775618 | Samarakoon et al. (2020) |
| Nemania ethancrensonii | CBS 148337 | USA | HT | ON869311 | ON869311 | ON808489 | ON808533 | Voglmayr et al. (2022) |
| Nemania primolutea | HAST 91102001 | Taiwan | HT | EF026121 | N/A | GQ844767 | EF025607 | Hsieh et al. (2010) |
| Nemania uda | CBS 148422 | Austria | ON869312 | ON869312 | ON808488 | ON808532 | Voglmayr et al. (2022) | |
| Obolarina dryophila | MUCL 49882 | France | GQ428316 | GQ428316 | KY624284 | GQ428322 | Pažoutová et al. (2010), Wendt et al. (2018) | |
| Occultitheca rosae | HKAS 102393 | China | HT | MW240672 | MW240602 | MW658651 | MW775617 | Samarakoon et al. (2020) |
| Oligostoma insidiosum | CBS 147288 | Switzerland | ET | ON869314 | ON869314 | ON808491 | ON808535 | Voglmayr et al. (2022) |
| Podosordaria mexicana | WSP 176 | Mexico | GU324762 | N/A | GQ853039 | GQ844840 | Hsieh et al. (2010) | |
| Poronia punctata | CBS 656.78 | Australia | HT | KT281904 | KY610496 | KY624278 | KX271281 | Senanayake et al. (2015), Wendt et al. (2018) |
| Pyrenopolyporus hunteri | MUCL 52673 | Ivory Coast | ET | KY610421 | KY610472 | KY624309 | KU159530 | Kuhnert et al. (2017), Wendt et al. (2018) |
| Rhopalostroma angolense | CBS 126414 | Ivory Coast | KY610420 | KY610459 | KY624228 | KX271277 | Wendt et al. (2018) | |
| Rosellinia corticium | MUCL 51693 | France | KY610393 | KY610461 | KY624229 | KX271254 | Wendt et al. (2018) | |
| Rostrohypoxylon terebratum | CBS 119137 | Thailand | HT | DQ631943 | DQ840069 | DQ631954 | DQ840097 | Tang et al. (2007), Fournier et al. (2011) |
| Ruwenzoria pseudoannulata | MUCL 51394 | D. R. Congo | HT | KY610406 | KY610494 | KY624286 | KX271278 | Wendt et al. (2018) |
| Sarcoxylon compunctum | CBS 359.61 | South Africa | KT281903 | KY610462 | KY624230 | KX271255 | Senanayake et al. (2015), Wendt et al. (2018) | |
| Spiririma gaudefroyi | CBS 147284 | Spain | ET | ON869320 | ON869320 | ON808497 | ON808541 | Voglmayr et al. (2022) |
| Stilbohypoxylon elaeicola | Y.M.J. 173 | French Guiana | EF026148 | N/A | GQ844826 | EF025616 | Hsieh et al. (2010) | |
| Stromatoneurospora phoenix | BCC 82040 | Thailand | MT735133 | MT735133 | MT742605 | MT700438 | Becker et al. (2020) | |
| Thamnomyces dendroideeus | CBS 123578 | French Guiana | FN428831 | KY610467 | KY624232 | KY624313 | Stadler et al. (2010), Wendt et al. (2018) | |
| Xylaria apoda | HAST 90080804 | Taiwan | GU322437 | N/A | GQ844823 | GQ495930 | Hsieh et al. (2010) | |
| Xylaria arbuscula | CBS 126415 | Germany | KY610394 | KY610463 | KY624287 | KX271257 | Fournier et al. (2011), Wendt et al. (2018) | |
| Xylaria atrosphaerica | HAST 91111214 | Taiwan | GU322459 | N/A | GQ848342 | GQ495953 | Hsieh et al. (2010) | |
| Xylaria digitata | HAST 919 | Ukraine | GU322456 | N/A | GQ848338 | GQ495949 | Hsieh et al. (2010) | |
| Xylaria hypoxylon | CBS 122620 | Sweden | ET | KY610407 | KY610495 | KY624231 | KX271279 | Sir et al. (2016b), Wendt et al. (2018) |
| Xylaria ianthinovelutina | HAST 553 | French West Indies | GU322441 | N/A | GQ844828 | GQ495934 | Hsieh et al. (2010) | |
| Xylaria laevis | HAST 419 | French West Indies | GU324746 | N/A | GQ848359 | GQ502695 | Hsieh et al. (2010) | |
| Xylaria longipes | CBS 148.73 | Germany | MH860649 | MH872351 | KU684280 | KU684204 | Vu et al. (2019), U'Ren et al. (2016) | |
| Xylaria oxyacanthae | J.D.R. 859 | USA | GU322434 | N/A | GQ844820 | GQ495927 | Hsieh et al. (2010) | |
| Xylaria polymorpha | MUCL 49884 | France | KY610408 | KY610464 | KY624288 | KX271280 | Wendt et al. (2018) | |
Taxon selection and molecular phylogenetic inference
To assess the affinities of the newly generated sequences within the order Xylariales, the manually curated alignment and taxon set recently presented by Voglmayr et al. 2022 was used, further restricting sequences derived from Xylariaceae from 154 to 94 taxa. The newly generated sequences of the ITS, LSU, rpb2 and tub2 loci were inserted into the data matrix and manually checked for consistency. The different loci were subjected to IQTree2 (Minh et al. 2020) for molecular phylogenetic inference using Maximum-Likelihood criterion with options for a partitioned analysis (Chernomor et al. 2016) of the supermatrix with prior testing for the optimal nucleotide substitution model by using ModelFinder (Kalyaanamoorthy et al. 2017) following BIC criterion and 1000 non-parametric bootstrap (Felsenstein 1985) replicates. Concurrently, the optimal nucleotide substitution models for each locus were calculated with PartitionFinder2 (Lanfear et al. 2016) as implemented in the PhyloSuite V.1.2.2 (Zhang et al. 2020) program package and to-be-tested models restricted to the ones available in MrBayes 3.2.7a (Ronquist et al. 2012). Partitions were regarded as unlinked and testing options set to BIC optimality criterion and test strategy set to test all. A phylogenetic inference using MrBayes 3.2.7a followed, with settings used and described by Kemkuignou et al. (2022). Briefly, a random starting tree was used to calculate 200.000.000 generations with convergence controlled to arrive at an average split frequency of 0.01. Tree sampling was done every 1000 generations, of which the first 25% were discarded as “burn-in”. Four incrementally heated chains were used for the Markov Chain Monte Carlo (MCMC), with temperature set to 0.15. The BEAGLE library (Ayres et al. 2012) and a parallel Metropolis coupling for the MCMC (Altekar et al. 2004) were used to calculate in total four chains in parallel. The resulting bootstrap (bs) ≥ 70% and posterior probabilities ≥ 0.95 were mapped on the respective bipartition on the found maximum-likelihood tree.
Results
Morphological studies on strain ATCC 60639
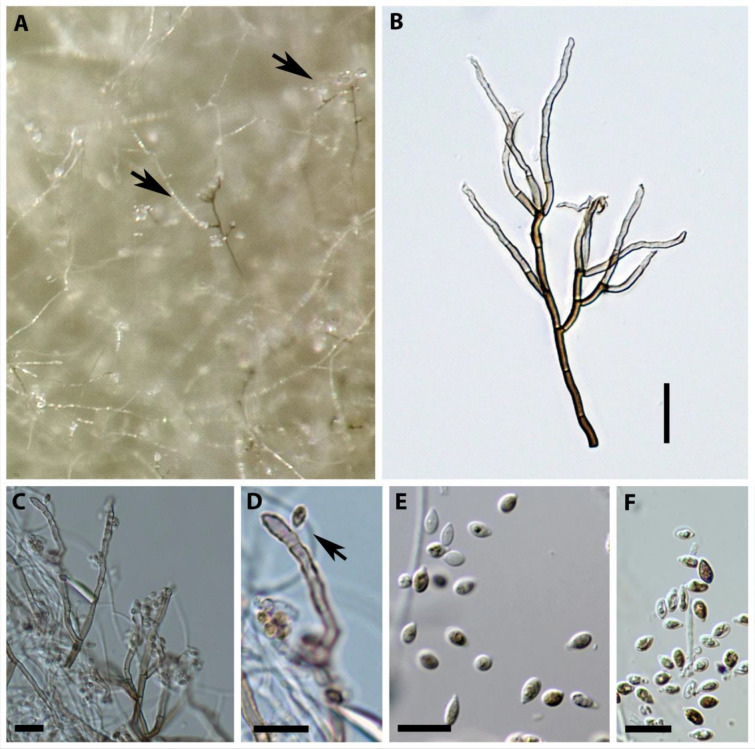
The culture we obtained from ATCC showed the following characteristics (Fig. 1):
Fig. 1.
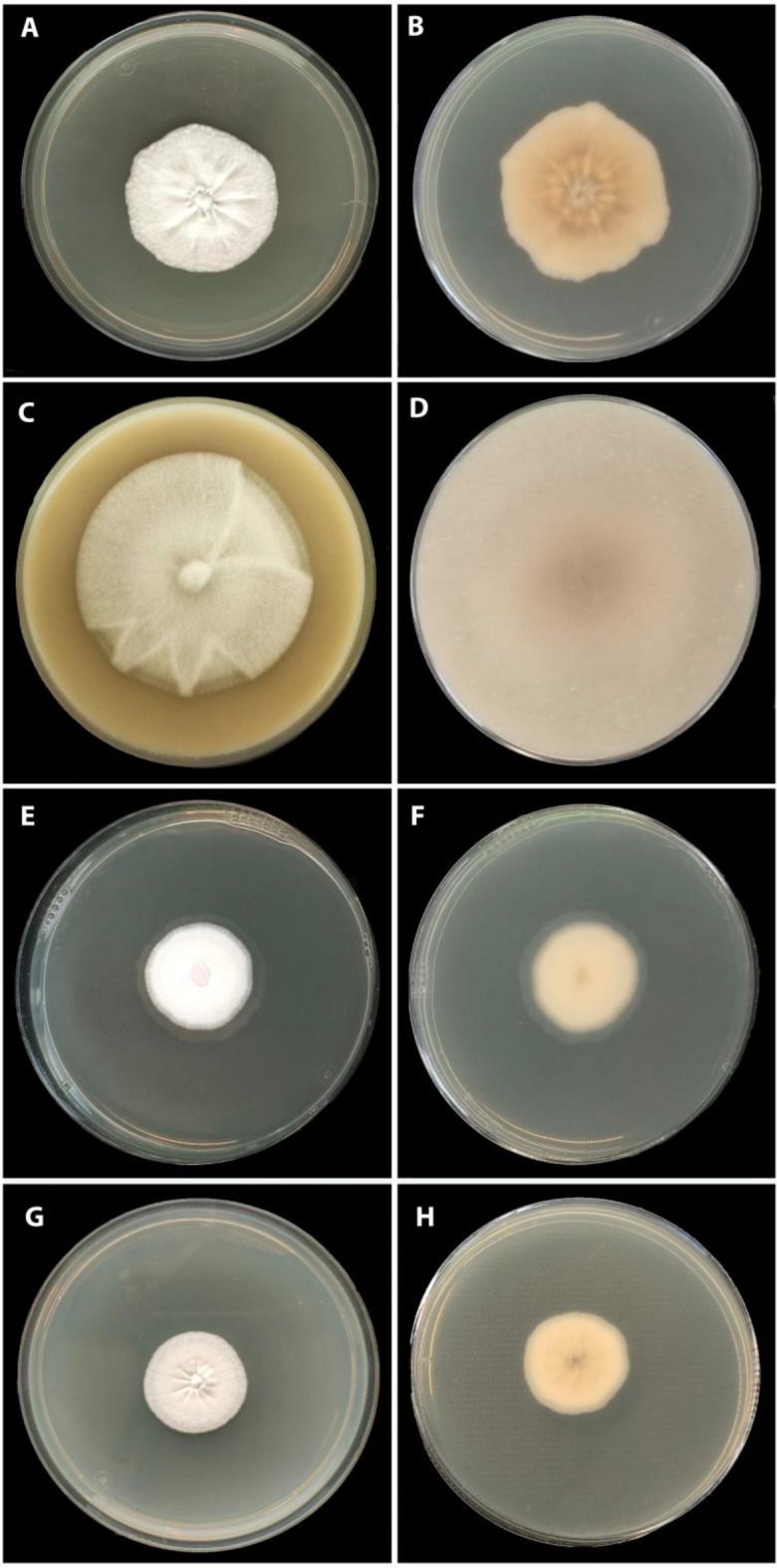
Colonies of Induratia apiospora (ATCC 60639) after four weeks A, B on MEA; C, D on OA; E, F on PDA; G, H on YM6.3
Culture characteristics: On YM6.3 and PDA: 2.6 cm in four weeks. Mycelium white, wavy or cottony hyphal growth, margin filiform to slightly undulate, flat to slightly elevate. Reverse white to pale. On MEA: 4.2 cm in four weeks, differs by having a mycelium with a margin slightly undulate, flat. On OA: 5.8 cm in four weeks, differing to have a margin entire. The information is summarized in Table 2. Sporulating regions: in patches, citrine (13) olivaceous (48). Conidiogenous structure nodulisporium-like, abundant, smooth to slightly roughened. Conidiogenous cells: melanized, smooth to slightly roughened, 34.5–66.5 × 2–3 µm (n = 23). Conidia: hyaline with cytoplasm content melanized, smooth, ellipsoidal to obovoid, 4–6 × 2–4 µm (n = 41).
Table 2.
Rates of growth and culture characteristics of Induratia apiospora ATCC60639 in four different media at four weeks of incubation
| Media | area (cm2) | diameter (cm) | Temperature | Margin | Elevation | Mycelium color |
|---|---|---|---|---|---|---|
| YM6.3 | 5.5 | 2.6 | 25 °C | filiform to slightly undulate | slightly elevate | White |
| MEA | 14.0 | 4.2 | slightly undulate | flat | White | |
| OA | 26.5 | 5.8 | Entire | slightly elevate | White | |
| PDA | 5.4 | 2.6 | filiform to slightly undulate | slightly elevate | White |
These characteristics (see also Fig. 2) are indeed well in accordance with what Samuels et al. (1987) had reported for Induratia apiospora. According to the description by Samuels et al. (1987), the anamorph is nodulisporium-like, conidiogenous cells are light brown, slightly roughened, conidia are hyaline, ellipsoidal to obovoid, 4–6 × 2–4 µm.
Fig. 2.
Morphology of the anamorph of the ex-type strain of Induratia apiospora (ATCC 60639) on YM6.3. A Conidiophore on surface mycelium. B Single nodulisporium-like conidiophore. C, D Conidiogenous cells. E, F Conidia. Scale bars: B 20 µm, E, F 10 µm
Molecular phylogenetic inference
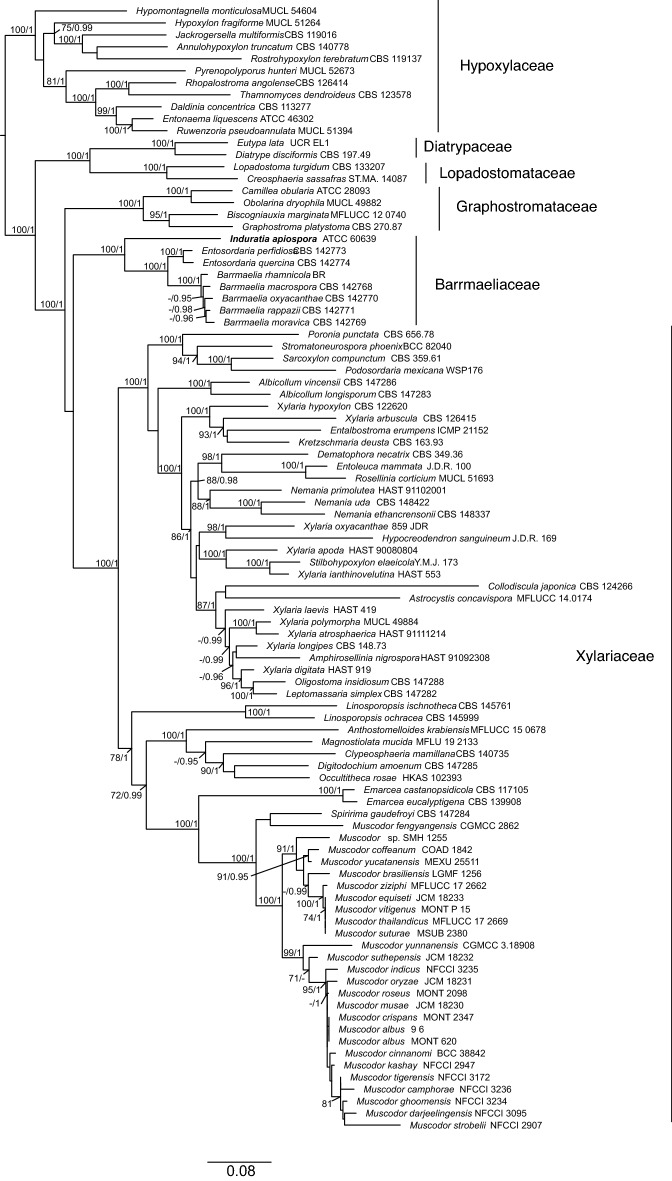
The alignment subjected to IQTree and MrBayes consisted of 4679 sites in total, distributed among loci corresponding to the ITS (584 sites), LSU (1329 sites), rpb2 (1235 sites) and tub2 (1531 sites). The generated sequence of the SSU for Induratia apiospora ATCC 60639 has not been used for phylogenetic reconstruction, but has been deposited under the GenBank Acc. No. OQ748100 for the scientific community to use. A detailed list of alignment features and the tested models per partition, as well as the alignments can be found in the (Additional file 1: Tables S1–S3). The final inferred tree following maximum-likelihood analysis had an lLn of − 89939.9947 (Fig. 3). The topology of trees generated from the ML and Bayesian approach were identical, showing a similar arrangement of the taxa as previously presented by Voglmayr et al. (2022). Briefly, the Hypoxylaceae, Diatrypaceae, Lopadostomataceae, Graphostromataceae, Xylariaceae and Barrmaeliaceae received maximum support. The position of the latter was not resolved. Sequences derived from Induratia sensu Samarakoon et al. (2020; given as Muscodor in Fig. 3), Spiririma gaudefroyi and Emarcea received maximum support in a clade nested inside the Xylariaceae. Surprisingly, the sequences of Induratia apiospora ATCC 60639 clustered in a basal position to a clade formed by Entosordaria and Barrmaelia representing the Barrmaeliaceae, each clade receiving maximum support.
Fig. 3.
Maximum Likelihood Tree (lLn = − 89939.9947) inferred from a manually edited alignment of ITS, LSU, rpb2 and tub2 sequences featuring sequences derived from Hypoxylaceae, Diatrypaceae, Lopadostomataceae, Graphostromataceae, Barrmaeliaceae and Xylariaceae. The position of the newly generated and concatenated sequences of Induratia apiospora are marked in bold. Bootstrap and Bayesian posterior probabilities ≥ 70% and ≥ 0.95, respectively, are given at bipartitions
Taxonomy
Our phylogenetic study showed that the ex-type strain of Induratia resolves within the Barrmaeliaceae. The Induratiaceae are thus regarded as a synonym of Barrmaeliaceae and Muscodor is resurrected. The holotype specimen of Induratia could still not be recovered, hence the isotype (WSP73242; MyCoPortal 2022) located at Shaw Mycology Herbarium (WSP; Washington State University) is chosen as lectotype. The genera Emarcea and Muscodor, which were previously classified within Induratiaceae, are now formally accommodated in the Xylariaceae.
Barrmaeliaceae Voglmayr & Jaklitsch, Mycol. Progr. 17(1–2): 162 (2017) [2018], emend. Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert.
MycoBank MB 822042
Type genus: Barrmaelia Rappaz
≡ Induratiaceae Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Diversity 101: 188 (2020) [MB833443], syn. nov.
Other genera in the family: Entosordaria Höhn. (see Voglmayr et al. 2018), Induratia Samuels, E. Müll. & Petrini (see below).
Saprobic on wood or bark. Stroma if present mostly in wood and blackening the surface in wide areas or in elongate bands, sometimes darker or carbonized around the ostioles; entostroma prosenchymatous, poorly developed, sometimes delimited by a black carbonized line (Induratia), without KOH-extractable pigments; ectostroma variable, from virtually absent, poorly developed to strongly carbonized and clypeus-like. Ascomata (perithecia) globose, sometimes raising the substrate, singly, in small groups or gregarious. Peridium melanized, pseudoparenchymatous to prosenchymatous. Hamathecium of numerous persistent, hyaline, septate paraphyses. Asci eight-spored, cylindrical, persistent, with inamyloid or amyloid apical ascus apparatus. Ascospores hyaline, yellow to dark brown; unicellular with or without germ slit (Barrmaelia), or two-celled with septum near one end, the small cell hyaline, the large cell dark brown and with an apical germ apparatus consisting of radial slits (Entosordaria), or hyaline, two-celled, apiosporous without germ slit (Induratia); allantoid, ellipsoid or fusoid, inequilateral, slightly inequilateral or nearly equilateral, with narrowly or broadly rounded ends. Anamorph, where known, libertella-like (Barrmaelia; Rappaz 1995), or nodulisporium-like (Induratia).
Key to the genera of Barrmaeliaceae
1. Ascospores one-celled, asymmetrically ellipsoid to allantoid, uniformly light to dark brown, with or without a longitudinal germ slit, without appendages; anamorph libertella-like……………….……………Barrmaelia.
1. Ascospores two-celled, apiosporous, germ locus absent or consisting of apical radial slits, with or without appendages…………………………………2
2. Ascospores with submedian septum, entirely hyaline, without germ locus, with hyaline cellular appendages at each end while still in the ascus; anamorph nodulisporium-like………………………………………Induratia.
2. Ascospores with septum near one end, small ascospore cell hyaline, large ascospore cell dark brown and with an apical germ apparatus consisting of radial slits, without appendages; anamorph unknown………………………………………………Entosordaria.
Induratia Samuels, E. Müll. & Petrini, Mycotaxon 28(2): 484 (1987).
Type species: Induratia apiospora Samuels, E. Müll. & Petrini, Mycotaxon 28(2): 484 and Fig. 5 (1987).
MycoBank MB 130900
Holotype: New Zealand, North Island, Hokianga Co., Waipoua State Forest, near Yakas Track, on decorticated wood, 30 May 1982, G.J. Samuels and P. Johnston (PDD 44399, lost). Lectotype: (designated here, MBT 10010382) New Zealand North Island, Hokianga Co., Waipoua State Forest, near Yakas Track, on decorticated wood, 30 May 1982, G.J. Samuels and P. Johnston (WSP73242).
Ex-type culture: ATCC 60639, deposited by G.J. Samuels; duplicates sent to CBS under accession CBS 149733 and ICMP 24754.
Resurrection of Muscodor
The genus Muscodor is now placed in the Xylariaceae, where it had originally been accommodated based on phylogenetic relationship, however anamorph morphology, a traditionally important character for the characterization of Xylariales, is still lacking. Below we list all species that were formerly accepted in Muscodor, including those that have been invalidly described in the original publications. The typifications have already been corrected and changed by Samarakoon et al. (2020) according to the rules of the International Code of Nomenclature for Algae, Fungi, and Plants (ICN), when those designations were validated in Induratia. We here introduce new combinations for these Induratia names in Muscodor.
Muscodor Worapong, Strobel & W.M. Hess, Mycotaxon 79: 71 (2001).
Type species: Muscodor albus Worapong, Strobel & W.M. Hess, Mycotaxon 79: 71 (2001).
≡ Induratia alba (Worapong, Strobel & W.M. Hess) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Other accepted species
Muscodor brasiliensis (L.C. Pena, Servienski & V. Kava ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846432
Basionym: Induratia brasiliensis L.C. Pena, Servienski & V. Kava ex Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 196 (2020).
[originally described as: Muscodor brasiliensis L.C. Pena, Servienski & V. Kava, Microbiol. Res. 221: 32 (2019), nom. inval., Art. 40.7 (Shenzhen)]
Muscodor camphorae Meshram, N. Kapoor, G. Chopra & S. Saxena [as ‘camphora’], Mycosphere 8(4): 571 (2017).
≡ Induratia camphorae (Meshram, N. Kapoor, G. Chopra & S. Saxena) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor cinnamomi Suwannar., Bussaban, K.D. Hyde & Lumyong, Mycotaxon 114: 19 (2011) [2010].
≡ Induratia cinnamomi (Suwannar., Bussaban, K.D. Hyde & Lumyong) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor coffeanus A.A.M. Gomes, Pinho & O.L. Pereira [as ‘coffeanum’], in Hongsanan et al., Cryptog. Mycol. 36(3): 368 (2015).
≡ Induratia coffeana (A.A.M. Gomes, Pinho & O.L. Pereira) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor crispans A.M. Mitch., Strobel, W.M. Hess, Pérez-Vargas & Ezra, Fungal Divers. 31: 41 (2008).
≡ Induratia crispans (A.M. Mitch., Strobel, W.M. Hess, Pérez-Vargas & Ezra) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor darjeelingensis (Meshram, N. Kapoor & S. Saxena ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846443
Basionym: Induratia darjeelingensis Meshram, N. Kapoor & S. Saxena ex Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 198 (2020).
[originally described as: Muscodor darjeelingensis Meshram, N. Kapoor & S. Saxena, Sydowia 66(1): 61 (2014), nom. inval., Art. 40.7 (Shenzhen)]
Muscodor equiseti Suwannar. & Lumyong, in Suwannarach, Kumla, Bussaban, Hyde, Matsui & Lumyong, Ann. Microbiol. 63(4): 1350 (2013).
≡ Induratia equiseti (Suwannar. & Lumyong) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor fengyangensis (Chu L. Zhang ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846442
Basionym: Induratia fengyangensis Chu L. Zhang ex Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 198 (2020).
[originally described as: Muscodor fengyangensis Chu L. Zhang, Fungal Biol. 114(10): 801 (2010), nom. inval., Art. 40.1 (Shenzhen)]
Muscodor ghoomensis S. Saxena, M. Gupta & Meshram, Sydowia 67: 136 (2015).
≡ Induratia ghoomensis (S. Saxena, M. Gupta & Meshram) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor heveae (Siri-Udom & Lumyong ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846436
Basionym: Induratia heveae Siri-Udom & Lumyong ex Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 199 (2020).
[originally described as: Muscodor heveae Siri-Udom & Lumyong, Ann. Microbiol. 66(1): 442 (2015), nom. inval., Art. 40.7 (Shenzhen)]
Muscodor indicus S. Saxena, M. Gupta & Meshram [as ‘indica’], Sydowia 67: 136 (2015).
≡ Induratia indica (S. Saxena, M. Gupta & Meshram) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor kashay (Meshram, N. Kapoor & S. Saxena ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846437
Basionym: Induratia kashay Meshram, N. Kapoor & S. Saxena ex Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 199 (2020).
[originally described as: Muscodor kashayum Meshram, N. Kapoor & S. Saxena, Mycology 4(4): 198 (2013), nom. inval., Art. 40.7 (Shenzhen)]
Muscodor musae Suwannar. & Lumyong, in Suwannarach, Kumla, Bussaban, Hyde, Matsui & Lumyong, Ann. Microbiol. 63(4): 1347 (2013).
≡ Induratia musae (Suwannar. & Lumyong) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 193 (2020).
Muscodor oryzae Suwannar. & Lumyong, in Suwannarach, Kumla, Bussaban, Hyde, Matsui & Lumyong, Ann. Microbiol. 63(4): 1349 (2013).
≡ Induratia oryzae (Suwannar. & Lumyong) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 194 (2020).
Muscodor roseus Worapong, Strobel & W.M. Hess, Mycotaxon 81: 467 (2002).
≡ Induratia rosea (Worapong, Strobel & W.M. Hess) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 194 (2020).
Muscodor strobelii Meshram, S. Saxena & N. Kapoor, Mycotaxon 128: 96 (2014).
≡ Induratia strobelii (Meshram, S. Saxena & N. Kapoor) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 194 (2020).
Muscodor suthepensis Suwannar. & Lumyong, in Suwannarach, Kumla, Bussaban, Hyde, Matsui & Lumyong, Ann. Microbiol. 63(4): 1349 (2013).
≡ Induratia suthepensis (Suwannar. & Lumyong) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 194 (2020).
Muscodor suturae Kudalkar, Strobel & Riy.-Ul-Hass. [as ‘sutura’], Mycoscience 53(4): 322 (2012).
≡ Induratia suturae (Kudalkar, Strobel & Riy.-Ul-Hass.) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 194 (2020).
Muscodor thailandicus (Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846434
Basionym: Induratia thailandica Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 194 (2020).
Muscodor tigerensis (S. Saxena, Meshram & N. Kapoor ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846438
Basionym: Induratia tigerensis S. Saxena, Meshram & N. Kapoor ex Samarak., Thongbai, K.D. Hyde & M. Stadler Fungal Divers. 101(1): 199 (2020).
[originally described as: Muscodor tigerensis S. Saxena, Meshram & N. Kapoor, Ann. Microbiol. 65(1): 53 (2014) [2015], [as ‘tigerii’], nom. inval., Art. 40.7 (Shenzhen)].
Muscodor vitigenus Daisy, Strobel, Ezra & W.M. Hess, in Daisy, Strobel, Ezra, Castillo, Baird & Hess, Mycotaxon 84: 45 (2002).
≡ Induratia vitigena (Daisy, Strobel, Ezra & W.M. Hess,) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 196 (2020).
Muscodor yucatanensis M.C. González, A.L. Anaya, Glenn & Hanlin, Mycotaxon 110: 365 (2009).
≡ Induratia yucatanensis (M.C. González, A.L. Anaya, Glenn & Hanlin) Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 196 (2020).
Muscodor yunnanensis (C.L. Zhang ex Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846435
Basionym: Induratia yunnanensis C.L. Zhang ex Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 199 (2020).
[originally described as: Muscodor yunnanensis C.L. Zhang, in Chen et al., Mycosphere 10(1): 193 (2019), nom. inval., Art. 40.8 (Shenzhen)]
Muscodor ziziphi (Samarak., Thongbai, K.D. Hyde & M. Stadler) Cedeño-Sanchez, M. Stadler, Voglmayr & C. Lambert, comb. nov.
MycoBank MB 846433
Basionym: Induratia ziziphi Samarak., Thongbai, K.D. Hyde & M. Stadler, Fungal Divers. 101(1): 196 (2020).
Notes
The specimen SMH 1255 from Puerto Rico, originally studied by Miller and Huhndorf (2005) and later by Samarakoon et al. (2020), has so far been treated as a member of the genus Induratia. It should henceforth be treated as a species of Muscodor, as its phylogenetic affinities are with the latter genus. Even though the species has not been formally described, the taxonomic name associated with the GenBank Acc. Nos for its DNA sequences ought to be changed.
Emarcea Duong, Jeewon & K.D. Hyde, Stud. Mycol. 50(1): 255 (2004).
MycoBank MB 500070.
Type species: Emarcea castanopsidicola Duong, Jeewon & K.D. Hyde, Stud. Mycol. 50(1): 255 (2004).
Notes
Since the genus Emarcea is phylogenetically closely related to the species in the Muscodor clade, it is again classified within the Xylariaceae, together with the Muscodor species discussed above.
Discussion
Multi-locus molecular phylogenetic analysis proved to serve as a powerful tool to emend placements of taxonomic groups in the Xylariales over the last years, exemplified by the erection of the Barrmaeliaceae to accommodate a phylogenetically distinct and well-supported clade of Barrmaelia and Entosordaria (Voglmayr et al. 2018). Other examples include the emendation of the Hypoxylaceae in concordance with chemotaxonomic information (Wendt et al. 2018) or the resurrection of Dematophora, distinguished from Rosellinia species by its synnematal geniculosporium-like anamorph and often a phytopathogenic lifestyle (Wittstein et al. 2020). Regarding the recent synonymisation of Muscodor with Induratia (Samarakoon et al. 2020), some doubts remained on the validity of that approach, e.g., because the anamorph described by Samuels et al. (1987) was hitherto not observed in the Muscodor cultures, which seem to be unable to sporulate. The proposed taxonomy of Samarakoon et al. (2020) was based on the rather similar teleomorphs of M. thailandicus (≡ I. thailandica) and M. ziziphi (≡ I. ziziphi) and the Muscodor sp. (≡ Induratia sp.) specimen reported by Miller and Huhndorf (2005) and characterized by Samarakoon et al. (2020), featuring typical xylariaceous asci and apiospores. The results of the corresponding phylogenies unfortunately did not include any sequences of Induratia before erection of its eponymous family, due to a lack of access to molecular data to compare (Samarakoon et al. 2020). Only two years later, Voglmayr et al. (2022) found that DNA sequences of Spiririma (≡ Helicogermslita) gaudefroyi, clustered within the Induratiaceae clade. Since S. gaudefroyi features dark ascospores, this gave reason to challenge the concept of restricting apiosporous Xylariales to the Induratiaceae. In our study, the ex-holotype culture of Induratia apiospora isolated by Samuels et al. (1987) clustered in a basal position with Barrmaelia and Entosordaria, while the position of the Barrmaeliaceae remained unchanged. As the Induratia spp. transferred from Muscodor showed no relationship with the ex-holotype culture of I. apiospora, the genus Muscodor is resurrected. Furthermore, this implied that the Induratiaceae cannot be retained as an own family, from a phylogenetic perspective, but should be merged with Barrmaeliaceae (Voglmayr et al. 2022). Morphological characters formerly used to segregate the Induratiaceae from other members of the Xylariaceae, i.e. the presence of apiosporous ascospores, can apparently not serve beyond species discrimination. The resurrected Muscodor still showed a paraphyletic structure, indicated by a clade formed by Spiririma and Muscodor fengyangensis, which may result in further re-arrangements in the future, e.g., when a larger number of anamorph/teleomorph relationships has been established.
Serious degrees of synapomorphies are well documented within the Xylariaceae, illustrated by the paraphyly of the genus Xylaria, and so far it has not been possible to resolve this problem even by extensive taxon sampling (Hsieh et al. 2010; Voglmayr et al. 2018, 2022; Voglmayr and Beenken 2020). A similar situation was previously encountered for another family in the Xylariales, the Hypoxylaceae. An important feature of the Hypoxylaceae is the prolific secondary metabolism of its members, which has long been exploited for chemotaxonomic purposes. Recently, it was possible to match chemotaxonomic data by performing genome mining for secondary metabolite-encoding biosynthesis gene clusters (Wibberg et al. 2021; Kuhnert et al. 2021) after sequencing of the full genomes of some representative strains using 3rd generation techniques with phylogenomic approaches. This example indicates the feasibility to sample for additional phenotypic data, i.e. data about the secondary metabolism, and attempting to link it with phylogenetic data on a larger scale to explore taxonomic affinities. Almost all Xylariales are well-known to be very prolific secondary metabolite producers, and in particular Muscodor species are known to produce volatile antibiotics. Therefore, following the previously presented example of the Hypoxylaceae, we suggest the inclusion of chemotaxonomic data and in particular studies on the non-volatile metabolites, combined with robust synthetic chemistry enabling the identification of the produced metabolites for the future.
Supplementary Information
Additional file 1. Images of the isotype located in the Shaw Mycological Herbarium (WSP), provided by Monique H. Slipher, are shown in Fig. S1. Auxilliary information and characteristics covering the molecular phylogenetic analysis as well as the alignment are given in the Tables S1–S3.
Acknowledgements
We thank Anke Skiba for expert technical assistance and, Adrienne Stanton, Peter Johnston, Gary J. Samuels, Orlando Petrini, for valuable correspondence in the course of our search for the type of Induratia apiospora. Monique H. Slipher is thanked for providing information and images about the state of the isotype of I. apiospora located in WSP.
This article is dedicated to the memory of Prof. Jack D. Rogers
Author contributions
CL: conceptualization, supervision, analysis, writing—original draft preparation; HV: analysis; MCS: methods and analysis; RS: methods and analysis; MS: analysis, writing—original draft preparation, resources; KB: analysis, writing—review and editing. All authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL. MCS gratefully acknowledges a PhD stipend from The National Secretariat of Science, Technology and Innovation of the Republic of Panama (SENACYT) and the Institute for the Development of Human Resources (IFARHU). We are also grateful to the LifeScience Foundation (Munich) for a PhD stipend for CL. MS would like to thank the Deutsche Forschungsgemeinschaft for a Research Unit grant in the SPP 1991 (TaxonOmics).
Data availability
All data except for the DNA sequences, which are deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) are available in the manuscript or the Additional file 1.
Declarations
Competing interests
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. Parallel metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinformatics. 2004;20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- Ayres DL, Darling A, Zwickl DJ, Beerli P, Holder MT, Lewis PO, Huelsenbeck JP, Ronquist F, Swofford DL, Cummings MP, Rambaut A, Suchard MA. BEAGLE: an application programming interface and high-performance computing library for statistical phylogenetics. Syst Biol. 2012;61:170–173. doi: 10.1093/sysbio/syr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Wongkanoun S, Wessel AC, Bills GF, Stadler M, et al. Phylogenetic and chemotaxonomic studies confirm the affinities of Stromatoneurospora phoenix to the coprophilous Xylariaceae. J Fungi. 2020;6:144. doi: 10.3390/jof6030144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Feng XX, Xia CY, Kong DD, Qi ZY, Liu F, Chen D, Lin FC, Zhang CL. Confirming the phylogenetic position of the genus Muscodor and the description of a new Muscodor species. Mycosphere. 2019;10:187–201. doi: 10.5943/mycosphere/10/1/2. [DOI] [Google Scholar]
- Chernomor O, von Haeseler A, Minh BQ. Terrace aware data structure for phylogenomic inference from supermatrices. Syst Biol. 2016;65:997–1008. doi: 10.1093/sysbio/syw037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous PW, Wingfield MJ, Guarro J, Sotton DA, Acharya K, et al. Fungal Planet description sheets: 320–370. Persoonia. 2015;34:167–266. doi: 10.3767/003158515X688433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daisy B, Strobel G, Ezra D, Castillo U, Baird G, Hess WM. Muscodor vitigenus anam. sp. nov., an endophyte from Paullinia paullinioides. Mycotaxon. 2002;84:39–50. [Google Scholar]
- Daranagama DA, Camporesi E, Tian Q, Liu X, Chamyuang S, Stadler M, Hyde KD. Anthostomella is polyphyletic comprising several genera in Xylariaceae. Fungal Divers. 2015;73:203–238. doi: 10.1007/s13225-015-0329-6. [DOI] [Google Scholar]
- Duong LM, Lumyong S, Hyde KD, Jeewon R. Emarcea castanopsidicola gen. et sp. nov. from Thailand, a new xylariaceous taxon based on morphology and DNA sequences. Stud Mycol. 2004;50:253–260. [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- Fournier J, Flessa F, Peršoh D, Stadler M. Three new Xylaria species from southwestern Europe. Mycol Prog. 2011;10:33–52. doi: 10.5248/113.209. [DOI] [Google Scholar]
- González MC, Anaya AL, Glenn AE, Macías-Rubalcava ML, Hernández-Bautista BE, Hanlin RT. Muscodor yucatanensis, a new endophytic ascomycete from Mexican chakah, Bursera simaruba. Mycotaxon. 2009;110:363–372. doi: 10.5248/110.363. [DOI] [Google Scholar]
- Hongsanan S, Hyde KD, Bahkali AH, Camporesi E, Chomnunti P, Ekanayaka H, Gomes AAM, Hofstetter V, Jones EBG, Pinho DB, Pereira OL, Tian Q, Wanasinghe DN, Xu JC, Buyck B. Fungal biodiversity profiles 11–20. Cryptog Mycol. 2015;36:355–380. doi: 10.7872/crym/v36.iss3.2015.355. [DOI] [Google Scholar]
- Hsieh HM, Lin CR, Fang MJ, Rogers JD, Fournier J, Lechat C, Ju YM. Phylogenetic status of Xylaria subgenus Pseudoxylaria among taxa of the subfamily Xylarioideae (Xylariaceae) and phylogeny of the taxa involved in the subfamily. Mol Phylogen Evol. 2010;54:957–969. doi: 10.1016/j.ympev.2009.12.015. [DOI] [PubMed] [Google Scholar]
- Hyde KD, Norphanphoun C, Maharachchikumbura SSN, et al. Refined families of Sordariomycetes. Mycosphere. 2020;11:305–1059. doi: 10.5943/mycosphere/11/1/7. [DOI] [Google Scholar]
- Jaklitsch WM, Voglmayr H. Phylogenetic relationships of five genera of Xylariales and Rosasphaeria gen. nov. (Hypocreales) Fungal Divers. 2011;52:75–98. doi: 10.1007/s13225-011-0104-2. [DOI] [Google Scholar]
- Jaklitsch WM, Fournier J, Rogers JD, Voglmayr H. Phylogenetic and taxonomic revision of Lopadostoma. Persoonia. 2014;32:52–82. doi: 10.3767/003158514X679272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaklitsch WM, Gardiennet A, Voglmayr H. Resolution of morphology-based taxonomic delusions: Acrocordiella, Basiseptospora, Blogiascospora, Clypeosphaeria, Hymenopleella, Lepteutypa, Pseudapiospora, Requienella, Seiridium and Strickeria. Persoonia. 2016;37:82–105. doi: 10.3767/003158516X690475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston PR, Rogers JD, Park D, Martin NA. Entalbostroma erumpens gen. et sp. nov. (Xylariaceae) from Phormium in New Zealand. Mycotaxon. 2016;131:765–771. doi: 10.5248/131.765. [DOI] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemkuignou BM, Schweizer L, Lambert C, Anoumedem EGM, Kouam SF, Stadler M, Marin-Felix Y. New polyketides from the liquid culture of Diaporthe breyniae sp. nov. MycoKeys. 2022;90:85–118. doi: 10.3897/mycokeys.90.82871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koukol O, Kelnarová I, Černý K. Recent observations of sooty bark disease of sycamore maple in Prague (Czech Republic) and the phylogenetic placement of Cryptostroma corticale. Forest Pathol. 2015;45:21–27. doi: 10.1111/efp.12129. [DOI] [Google Scholar]
- Kudalkar P, Strobel G, Riyaz-Ul-Hassan S, Geary B, Sears J. Muscodor sutura, a novel endophytic fungus with volatile antibiotic activities. Mycoscience. 2012;53:319–325. doi: 10.47371/S10267-011-0165-9. [DOI] [Google Scholar]
- Kuhnert E, Fournier J, Peršoh D, Luangsa-ard JJD, Stadler M. New Hypoxylon species from Martinique and new evidence on the molecular phylogeny of Hypoxylon based on ITS rDNA and β-tubulin data. Fungal Divers. 2014;64:181–203. doi: 10.1007/s13225-013-0264-3. [DOI] [Google Scholar]
- Kuhnert E, Sir EB, Lambert C, Hyde KD, Hladki AI, Romero AI, Rohde M, Stadler M. Phylogenetic and chemotaxonomic resolution of the genus Annulohypoxylon (Xylariaceae) including four new species. Fungal Divers. 2017;85:1–43. doi: 10.1007/s13225-016-0377-6. [DOI] [Google Scholar]
- Kuhnert E, Navarro-Muñoz JC, Becker K, Stadler M, Collemare J, Cox RJ. Secondary metabolite biosynthetic diversity in the fungal family Hypoxylaceae and Xylaria hypoxylon. Stud Mycol. 2021;99:100118. doi: 10.1016/j.simyco.2021.100118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2016;4(3):772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Whelen S, Hall BD. Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol Biol Evol. 1999;16:1799–1808. doi: 10.1093/oxfordjournals.molbev.a026092. [DOI] [PubMed] [Google Scholar]
- Liu F, Bonthond G, Groenewald JZ, Cai L, Crous PW. Sporocadaceae, a family of coelomycetous fungi with appendage-bearing conidia. Stud Mycol. 2019;92:287–415. doi: 10.1016/j.simyco.2018.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshram V, Kapoor N, Saxena S. Muscodor kashayum sp. nov.—a new volatile anti-microbial producing endophytic fungus. Mycology. 2013;4:196–204. doi: 10.1080/21501203.2013.877990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meshram V, Saxena S, Kapoor N. Muscodor strobelii, a new endophytic species from south India. Mycotaxon. 2014;128:93–104. doi: 10.5248/128.93. [DOI] [Google Scholar]
- Meshram V, Gupta M, Saxena S. Muscodor ghoomensis and Muscodor indica: new endophytic species based on morphological features, molecular and volatile organic analysis from northeast India. Sydowia. 2015;67:133–146. [Google Scholar]
- Meshram V, Kapoor N, Chopra G, Saxena S. Muscodor camphora, a new record from Cinnamomum camphora. Mycosphere. 2017;8:568–582. doi: 10.5943/mycosphere/8/4/6. [DOI] [Google Scholar]
- Miller AN, Huhndorf SM. Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi) Mol Phylogen Evol. 2005;35:60–75. doi: 10.1016/j.ympev.2005.01.007. [DOI] [PubMed] [Google Scholar]
- Minh BQ, Schmidt HA, Chernomor O, Schrempf D, Woodhams MD, von Haeseler A, Lanfear R. IQ-TREE 2: New models and efficient methods for phylogenetic inference in the genomic era. Mol Biol Evol. 2020;37(5):1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A, Strobel G, Hess W, Vargas P, Ezra D. Muscodor crispans, a novel endophyte from Ananas ananassoides in the Bolivian Amazon. Fungal Divers. 2008;31:37–43. [Google Scholar]
- MyCoPortal. 2022. https://www.mycoportal.org/portal/collections/individual/index.php?occid=6595195. Accessed 09 Dec 2022.
- O’Donnell K, Cigelnik E. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol Phylogenet Evol. 1997;7:103–116. doi: 10.1006/mpev.1996.0376. [DOI] [PubMed] [Google Scholar]
- Pažoutová S, Šrůtka P, Holuša J, Chudíčková KM. The phylogenetic position of Obolarina dryophila (Xylariales) Mycol Prog. 2010;9:501–507. doi: 10.1007/s11557-010-0658-5. [DOI] [Google Scholar]
- Peláez F, González V, Platas G, Sánchez-Ballesteros J, Rubio V. Molecular phylogenetic studies within the Xylariaceae based on ribosomal DNA sequences. Fungal Divers. 2008;31:111–134. [Google Scholar]
- Pena LC, Jungklaus GH, Savi DC, Ferreira-Maba L, Servienski A, Maia BHLNS, Annies V, Galli-Terasawa LV, Glienke C, Kava V. Muscodor brasiliensis sp. nov. produces volatile organic compounds with activity against Penicillium digitatum. Microbiol Res. 2019;221:28–35. doi: 10.1016/j.micres.2019.01.002. [DOI] [PubMed] [Google Scholar]
- Rappaz F. Anthostomella and related xylariaceous fungi on hard wood from Europe and North America. Mycol Helv. 1995;7:99–168. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarakoon MC, Thongbai B, Hyde KD, Broenstrup M, Beutling U, Lambert C, Stadler M. Elucidation of the life cycle of the endophytic genus Muscodor and its transfer to Induratia in Induratiaceae fam. nov., based on a polyphasic taxonomic approach. Fungal Divers. 2020;101:177–210. doi: 10.1007/s13225-020-00443-9. [DOI] [Google Scholar]
- Samuels GJ, Müller E, Petrini O. Studies in the Amphisphaeriaceae (sensu lato). III: new species of Monographella and Pestalosphaeria, and two new genera. Mycotaxon. 1987;28:473–499. [Google Scholar]
- Saxena S, Meshram V, Kapoor N. Muscodor darjeelingensis, a new endophytic fungus of Cinnamomum camphora collected from northeastern Himalayas. Sydowia. 2014;66:55–67. doi: 10.12905/0380.sydowia66(1)2014-0055. [DOI] [Google Scholar]
- Saxena S, Meshram V, Kapoor N. Muscodor tigerii sp. nov. Volatile antibiotic producing endophytic fungus from the northeastern Himalayas. Ann Microbiol. 2015;65:47–57. doi: 10.1007/s13213-014-0834-y. [DOI] [Google Scholar]
- Senanayake IC, Maharachchikumbura SSN, Hyde KD, Bhat JD, Jones EBG, McKenzie EHC, et al. Towards unraveling relationships in Xylariomycetidae (Sordariomycetes) Fungal Divers. 2015;73:73–144. doi: 10.1007/s13225-015-0340-y. [DOI] [Google Scholar]
- Sir EB, Lambert C, Wendt L, Hladki AI, Romero AI, Stadler M. A new species of Daldinia (Xylariaceae) from the Argentine subtropical montane forest. Mycosphere. 2016;7(9):1378–1388. doi: 10.5943/mycosphere/7/9/11. [DOI] [Google Scholar]
- Siri-udom S, Suwannarach N, Lumyong S. Existence of Muscodor vitigenus, M. equiseti and M. heveae sp. nov. in leaved of the rubber tree (Hevea brasiliensis Müll. Arg.), and their biocontrol potential. Ann Microbiol. 2015;66:437–448. doi: 10.1007/s13213-015-1126-x. [DOI] [Google Scholar]
- Stadler M, Fournier J, Læssøe T, Chlebicki A, Lechat C, Flessa F, Rambold G, Peršoh D. Chemotaxonomic and phylogenetic studies of Thamnomyces (Xylariaceae) Mycoscience. 2010;51:189–207. doi: 10.1007/S10267-009-0028-9. [DOI] [Google Scholar]
- Stadler M, Kuhnert E, Peršoh D, Fournier J. The Xylariaceae as model example for a unified nomenclature following the “one fungus-one name” (1F1N) concept. Mycology. 2013;4:5–21. doi: 10.1080/21501203.2013.782478. [DOI] [Google Scholar]
- Stadler M, Laessoe T, Fournier J, Decock C, Schmieschek B, Tichy HV, Persoh DA. A polyphasic taxonomy of Daldinia (Xylariaceae) Stud Mycol. 2014;77:1–143. doi: 10.3114/sim0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwannarach N, Bussaban B, Hyde KD, Lumyong S. Muscodor cinnamomi, a new endophytic species from Cinnamomum bejolghota. Mycotaxon. 2010;114:15–23. doi: 10.5248/114.15. [DOI] [Google Scholar]
- Suwannarach N, Kumla J, Bussaban B, Hyde KD, Matsui K. Molecular and morphological evidence support four new species in the genus Muscodor from northern Thailand. Ann Microbiol. 2013;63:1341–1351. doi: 10.1007/s13213-012-0593-6. [DOI] [Google Scholar]
- Tang AMC, Jeewon R, Hyde KD. Phylogenetic relationships of Nemania plumbea sp. nov. and related taxa based on ribosomal ITS and RPB2 sequences. Mycol Res. 2007;111:392–402. doi: 10.1016/j.mycres.2007.01.009. [DOI] [PubMed] [Google Scholar]
- Tibpromma S, Daranagama DA, Boonmee S, Promputtha I, Nontachaiyapoom S, Hyde KD. Anthostomelloides krabiensis gen. et sp. nov. (Xylariaceae) from Pandanus odorifer (Pandanaceae) Turkish J Bot. 2017;40:107–116. doi: 10.3906/bot-1606-45. [DOI] [Google Scholar]
- Triebel D, Peršoh D, Wollweber H, Stadler M. Phylogenetic relationships among Daldinia, Entonaema and Hypoxylon as inferred from ITS nrDNA sequences. Nova Hedw. 2005;80:25–43. doi: 10.1127/0029-5035/2005/0080-0025. [DOI] [Google Scholar]
- U’Ren JM, Miadlikowska J, Zimmerman NB, Lutzoni F, Stajich JE, Arnold AE. Contributions of North American endophytes to the phylogeny, ecology, and taxonomy of Xylariaceae (Sordariomycetes, Ascomycota) Mol Phylogen Evol. 2016;98:210–232. doi: 10.1016/j.ympev.2016.02.010. [DOI] [PubMed] [Google Scholar]
- Vilgalys R, Hester M. Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol. 1990;172:4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Beenken L. Linosporopsis, a new leaf-inhabiting scolecosporous genus in Xylariaceae. Mycol Progr. 2020;19:205–222. doi: 10.1007/s11557-020-01559-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Friebes G, Gardiennet A, Jaklitsch WM. Barrmaelia and Entosordaria in Barrmaeliaceae (fam. nov., Xylariales), and critical notes on Anthostomella-like genera based on multi-gene phylogenies. Mycol Prog. 2018;17:155–177. doi: 10.1007/s11557-017-1329-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H, Tello S, Jaklitsch WM, Friebes G, Baral HO, Fournier J. About spirals and pores: Xylariaceae with remarkable germ loci. Persoonia. 2022;49:58–98. doi: 10.3767/persoonia.2022.49.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu D, Groenewald M, De Vries M, et al. Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud Mycol. 2019;92:135–154. doi: 10.1016/j.simyco.2018.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt L, Sir EB, Kuhnert E, Heitkämper S, Lambert C, Hladki AI, Romero AI, Luangsa-ard JJ, Srikitikulchai P, Peršoh D, Stadler M. Resurrection and emendation of the Hypoxylaceae, recognized from a multigene phylogeny of the Xylariales. Mycol Prog. 2018;17:115–154. doi: 10.1007/s11557-017-1311-3. [DOI] [Google Scholar]
- White TJ, Bruns T, Lee S, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York: Academic; 1990. pp. 315–322. [Google Scholar]
- Wibberg D, Stadler M, Lambert C, Bunk B, Spröer C, Rückert C, Kalinowski J, Cox RJ, Kuhnert E. High quality genome sequences of thirteen Hypoxylaceae (Ascomycota) strengthen the phylogenetic family backbone and enable the discovery of new taxa. Fungal Divers. 2021;106:7–28. doi: 10.1007/s13225-020-00447-5. [DOI] [Google Scholar]
- Wijayawardene NN, Hyde KD, Dai DQ, et al. Outline of Fungi and fungus-like taxa-2021. Mycosphere. 2022;13:53–453. doi: 10.5943/mycosphere/13/1/2. [DOI] [Google Scholar]
- Wittstein K, Cordsmeier A, Lambert C, Wendt L, Sir EB, Weber J, Wurzler N, Petrini LE, Stadler M. Identification of Rosellinia species as producers of cyclodepsipeptide PF1022A and resurrection of the genus Dematophora as inferred from polythetic taxonomy. Stud Mycol. 2020;96:1–16. doi: 10.1016/j.simyco.2020.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worapong J, Strobel G, Ford EJ, Li JY, Baird G, Hess WM. Muscodor albus anam. gen. et sp nov., an endophyte from Cinnamomum zeylanicum. Mycotaxon. 2001;79:67–79. [Google Scholar]
- Worapong J, Strobel GA, Daisy B, Castillo UF, Baird G, Hess WM. Muscodor roseus anam. sp. nov., an endophyte from Grevillea pteridifolia. Mycotaxon. 2002;81:463–475. [Google Scholar]
- Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, et al. An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia. 2006;98:1076–1087. doi: 10.1080/15572536.2006.11832635. [DOI] [PubMed] [Google Scholar]
- Zhang CL, Wang GP, Mao LJ, Komon-Zelazowska M. Muscodor fengyangensis sp. nov. from southeast China: morphology, physiology and production of volatile compounds. Fungal Biol. 2010;114:797–808. doi: 10.1016/j.funbio.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Zhang D, Gao F, Jakovlić I, Zou H, Zhang J, Li WX, Wang GT. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol Ecol Res. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Images of the isotype located in the Shaw Mycological Herbarium (WSP), provided by Monique H. Slipher, are shown in Fig. S1. Auxilliary information and characteristics covering the molecular phylogenetic analysis as well as the alignment are given in the Tables S1–S3.
Data Availability Statement
All data except for the DNA sequences, which are deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) are available in the manuscript or the Additional file 1.