Abstract
Bacteriophages (phages) are nanostructured viruses with highly selective antibacterial properties that have gained attention beyond eliminating bacteria. Specifically, M13 phages are filamentous phages that have recently been studied in various aspects of nanomedicine due to their biological advantages and more compliant engineering capabilities over other phages. Having nanofiber-like morphology, M13 phages can reach varied target sites and self-assemble into multidimensional scaffolds in a relatively safe and stable way. In addition, genetic modification of the coat proteins enables specific display of peptides and antibodies on the phages, allowing for precise and individualized medicine. M13 phages have also been subjected to novel engineering approaches, including phage-based bionanomaterial engineering and phage-directed nanomaterial combinations that enhance the bionanomaterial properties of M13 phages. In view of these features, researchers have been able to utilize M13 phages for therapeutic applications such as drug delivery, biodetection, tissue regeneration, and targeted cancer therapy. In particular, M13 phages have been utilized as a novel bionanomaterial for precisely mimicking natural tissue environment in order to overcome the shortage in tissue and organ donors. Hence, in this review, we address the recent studies and advances of using M13 phages in the field of nanomedicine as therapeutic agents based upon their characteristics as novel bionanomaterial with biomolecules displayed. This paper also emphasizes the novel engineering approach that enhances M13 phage's bionanomaterial capabilities. Current limitations and future approaches are also discussed to provide insight in further progress for M13 phage-based clinical applications.
Keywords: Bacteriophages, M13, Phage display, Tissue regeneration, Cancer therapy
Graphical abstract
1. Introduction
Bacteriophages, or phages, are primarily nano-structured (one geometric dimension less than 100 nm) viruses that naturally infect bacteria [1]. Phages have recently made a comeback to the forefront of medicine following a rise in antibiotic-resistant bacteria that pose a serious health threat to individuals and place a heavy burden on the healthcare system [[2], [3], [4]]. While phages are well known for their role as antibiotic alternatives, their use as bionanomaterials has many potential applications. Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), a recent pandemic virus, has become one of the largest public health and economic crises in present time due to its rapid spread. Along with vaccines and medicines, researchers have suggested the use of multivalent nanomaterials, such as multivalent phage-based motif displaying viral mimotopes of SARS-CoV-2 spike proteins. These materials can bind with receptors of human cells and provide better understanding of the mechanism of viral infection, including potential algorithms for ongoing virus evolution [5].
The advancement of phage-based bionanomaterials is made possible by the convergence of biological and material science enabled by phage display technology, a Nobel-prize winning discovery first introduced by George Smith in 1985 [6]. Phage display involves insertion of foreign peptide sequences into the phage coat protein gene, which results in external display of the peptide for the corresponding sequences. By using phages' mutability and replication, billions of phage clones each displaying unique peptides could be created for peptide libraries, allowing affinity selection of receptors and enzymes for therapeutic use [[7], [8], [9]]. Among different types of phages, filamentous phage, especially M13 phage, is the most commonly used vector for display and bionanomaterial production [10,11]. Originally discovered and isolated from Escherichia coli (E. coli) in 1963, M13 phage has been discovered to offer a desirable capsid protein site for peptide display due to their wide range of compatibility and ability to display multiple peptides on one single phage for multifunctional nanomaterials, making them attractive for present-day biomedical applications [9,12].
Unlike the lytic tailed phages of the Caudovirales (T1, T4, T7, etc.) seen often in phage therapy, M13 phage is a lysogenic phage that establishes chronic infection in its host and generates new phages without lytic disruption. M13 phage, alongside other filamentous phages such as fd and f1 phage, belongs to the Inoviridae family and is characterized by its semi-flexible nanofiber-like shape capable of self-assembling into different ordered structures according to its environment [[13], [14], [15]]. While nuclear magnetic resonance spectroscopy revealed that both M13 and fd phage possess very conserved and stable structures suitable for withstanding harsh conditions, M13 phage shows slightly higher rigidity [16]. Considering these characteristics, M13 phage has been exploited as a preferred resource for bionanomaterial across different fields from therapeutic to energy production [7,17,18].
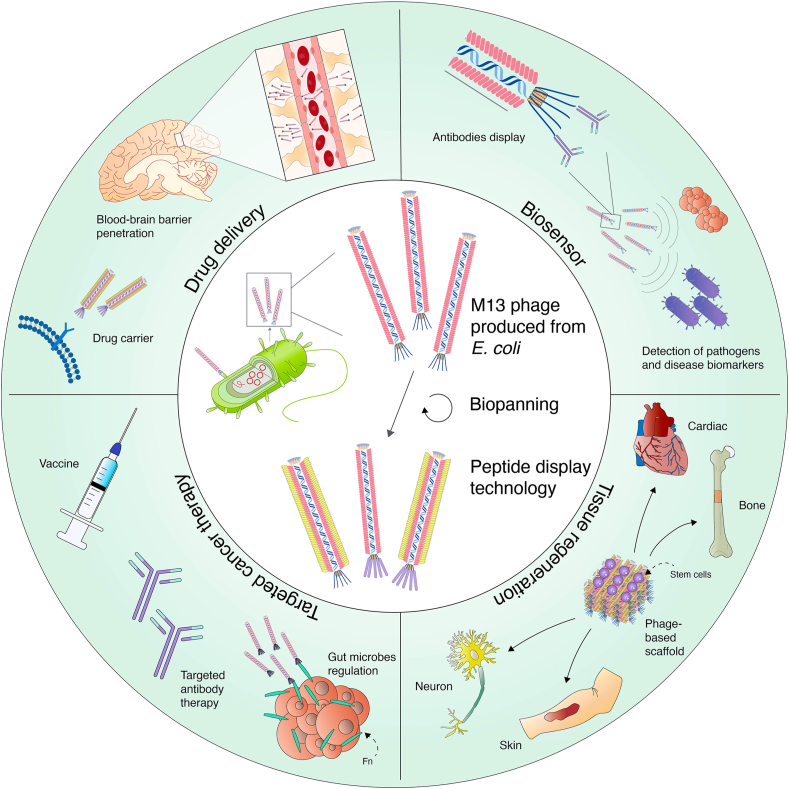
Notably, as depicted in Fig. 1, recent advances in phage display and bionanomaterial fabrication have allowed M13 phage to be used in a variety of clinical applications, including: 1) drug and gene delivery system. 2) biomedical sensor. 3) tissue regeneration, and 4) targeted cancer therapy [14,[19], [20], [21], [22]]. This review focuses on collecting current research on such medical applications and discussing in depth how bionanomaterial using M13 phage may aid in improving clinical diagnosis and treatment going forward. As previous insightful reviews have addressed the use of phages as genetically modifiable biomaterial for nanomedicine [9,[23], [24], [25]], this review gives an updated summary of the use of M13 phage, covers a broader range of diseases, and highlights recent advances in tissue regeneration using M13 phage and associated stem cell therapy for improving the recovery of tissue damage. Moreover, this paper emphasizes the novel engineering approach, particularly the phage-based bionanomaterial engineering and phage-directed nanomaterial combination that enhances M13 phage's bionanomaterial capabilities. Ultimately, this review provides a comprehensive overview of M13 phage research in clinical settings and discusses its significance and future potential in nanomedicine research.
Fig. 1.
Overview of M13 phage for clinical applications in drug delivery, biosensor, tissue regeneration, and targeted cancer therapy.
2. Biology of M13 phage as bionanomaterials
This section presents the advantages of M13 phage as a template for nanomaterial synthesis by describing its structure and phage life cycle.
2.1. Structure and feature of M13 phage
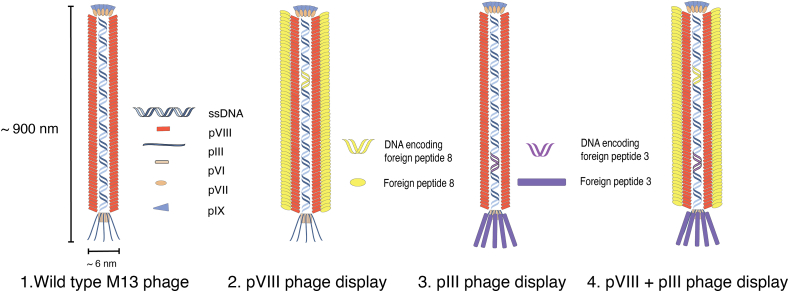
M13 phage is a filamentous virus with inherent structural advantages for forming complex nanostructures. Approximately 900 nm long and 6 nm wide, M13 phage is a linear nanostructure with a long-rod shape and monodispersed arrangement that could self-assemble in response to external stimuli [15,26] (Fig. 2). When rod-shaped structures collectively self-assemble, they form unique hierarchical structures that reveal their liquid crystalline characteristics. Such trait allows the fabrication of various self-aligned two- and three-dimensional structures on the nanometer to centimeter scales [27,28]. The liquid-crystal like properties also enhance phage delivery into targeted cells due to increased ligand-receptor interactions [14]. Interestingly, the linear shape and structure of the M13 phage allows the particle to penetrate the blood-brain barrier (BBB), which has rigorous permeability restrictions but is vital to overcome for delivering drugs to the nervous system in an effective manner [22].
Fig. 2.
Schematic representation of the nanofibrous structure of M13 phage displaying distinct peptides on coat proteins for different display system.
The M13 phage contains a single-stranded DNA genome (ssDNA) with a length of 6407 bp comprising nine genes that encode 11 proteins [29]. Among 11 different proteins, five of them are coat proteins (proteins located on the capsid), including ∼ 2700 copies of the major coat protein pVIII located on the side wall of the phage body, and five copies each of the pIII, pIV, pVII, and pIX minor coat proteins located on two ends (pIII and pVI on one end, pVII and pIX on the other end), as seen in Fig. 2 [30,31]. pVIII (g8p) and pIII (g3p) are the most commonly exploited and modified coat protein for peptide display due to wide range of compatibility [8]. With regard to interacting with foreign molecules, the pIII proteins are primarily responsible for biorecognition, while pVIII proteins are involved in the structural assembly of phage [32]. These different protein groups on a viral particle can be modified independently through genetic engineering to display biochemical ligands precisely down to the single amino acid level (see section 3.1) [33]. The ability to finely program viral protein functions add to the flexibility of assembling hierarchical nanostructures. Moreover, researchers reported a low chemical shift deviation over the entire capsid assembly, suggesting that the capsid is a conserved and robust protective layer vital for resistance to environmental stresses (pH, temperature, salinity condition, etc.) [16,34].
2.2. M13 phage life cycle
Filamentous phage's primary host is Gram-negative bacteria [35]. In particular, M13 phages has a natural affinity for bacteria that express F-pilus (sex pilus on the F+ male cells), especially E. coli, from which the virus can produce up to more than 1012 phage particles per milliliter overnight [36,37]. In contrast to lytic phages, M13 phages infect bacteria and produce progeny virions without killing the host cells [1]. The infection begins when the minor coat protein pIII binds to the F-pilus of a bacterial cell [31]. The circular ssDNA then enters the host's cytoplasm, where it begins to integrate into the bacterial genome through site-specific recombination and synthesize double stranded replicative form (RF) for further replication. RF produces linear single stranded DNA (M13 genome) via rolling circle replication [31,38]. The proteins translated from the genome assemble into progeny phages and are extruded from the bacterium with the 11 coat proteins neatly assembled along the virion [30,38,39]. Although it is reported that lysogenic phages can occasionally enter the lytic cycle upon stimulation by certain environmental factors, M13 phages are generally benign and cause mild immune reactions as they do not disrupt cell walls and membranes [35,40]. Furthermore, the error-free replication and the specificity of infection further suggest M13 phages' safe use in humans [40,41].
2.3. Biological advantages
Briefly, M13 phage nanomaterials offer the following biological advantages suitable for therapeutic applications: 1) The nanoscale fiber-like structures allow phage to self-assemble into multidimensional scaffolds, while thin structures also provide increased ligand-receptor interactions and versatile mobility for phage to reach target sites [14,22,41]. 2) The ability to precisely and easily modify surface proteins, as well as display multiple peptides at once, makes this an ideal phage display vector [42]. 3) The virion capsids provide robust protection for viruses in unpredictable human body environments to maintain concentration and stability during drug delivery or biopanning [16]. 4) Being one of the most abundant biological entities, M13 phage can generate large numbers of progeny phages upon infecting bacteria, making it an economical and accessible source of biomedical material [13,37]. 5) By virtue of the lysogenic cycle and error-free replication, M13 phage can be safely used in vivo without damaging mammalian cells [14].
3. Engineering of M13 phage for nano-applications
3.1. Genetic engineering
3.1.1. Phage display technology
The phage display system is a genetic engineering process that involves inserting foreign peptides into phage capsid proteins to enable corresponding peptides to display during the phage assembly process [8]. M13 phages and related phages fd and f1 are the most widely used vectors in phage display [1]. In terms of phage display locations, as mentioned earlier, the M13 phage typically display peptides at pIII and pVIII coat proteins, despite all five coat proteins capable of displaying material specific peptides [8,43]. The pIII protein has only five copies, but it can insert more than 100 amino acids. The major coat protein pVIII, on the other hand, displays thousands of copies of peptides, though shorter in amino acid length (up to 10 amino acids) [8].
3.1.2. Construction of peptide library
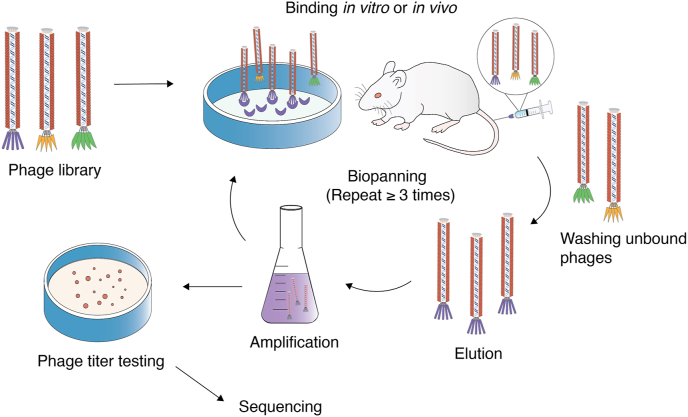
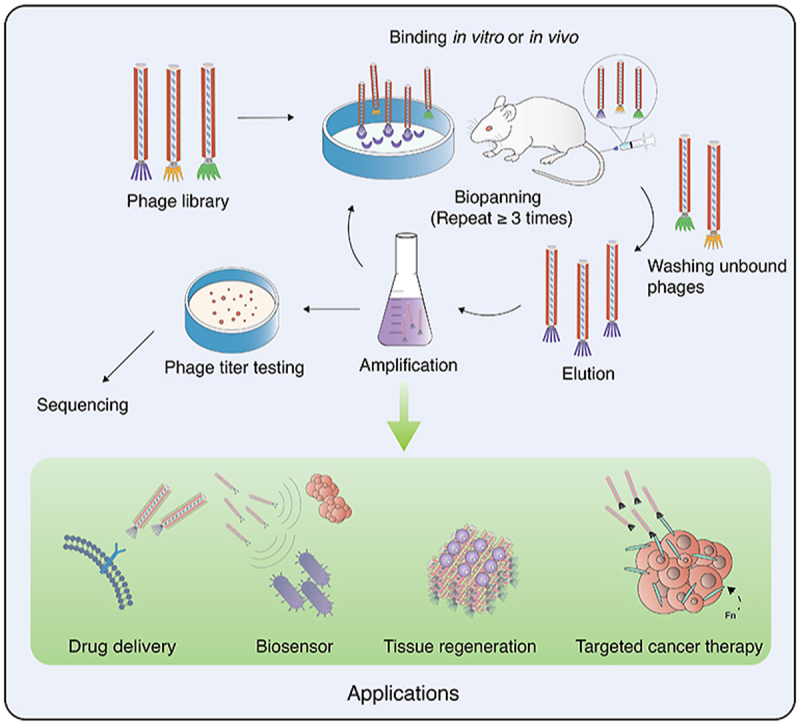
In phage display technology, the first step is to assemble a phage display library, also known as peptide library, which consists of billions of phage clones that display unique peptide sequences based on genotype to phenotype association [14]. This library is prepared by creating random oligonucleotide sequences through codon degeneracy and polymerase chain reaction amplification [9]. By inserting randomized sequences into specific sites of the phage coat protein DNA, diverse peptides are expressed on the phage nanoparticle surface for construction of random peptide library (Fig. 3). A peptide library typically contains 109 independent phage clones [44]. The pIII and pVIII phage libraries were designed to provide broad compatibility under a variety of situations due to their distinct properties. As part of New England Biolabs' Ph. D series of phage displayed peptide libraries, the commonly adopted Ph. D-7 and Ph. D-12 libraries display random sequences of heptapeptides and dodecapeptides at the N-terminus of the M13 bacteriophage minor coat protein pIII [45]. This system is constructed from the M13-based vector M13KE, a M13 derivative that replicates easily using standard M13 techniques without antibiotic selection or helper phage superinfection [[44], [45], [46]]. With the application of phage display technology becoming more prevalent, phage libraries have become more diverse, incorporating sequences of antibodies and receptor-selective proteins to the phage DNA for the preparation of corresponding display library [11,[47], [48], [49]]. The aim of all phage display libraries is to select a specific variant of interest in a library through screening, which is governed by the affinity of peptides displayed and their associated ligands [50]. Such affinity-based screening of the phage display libraries is also referred to as phage biopanning.
Fig. 3.
Phage biopanning process.
3.1.3. Phage biopanning
Phage biopanning technique is the process of capturing target-binding phages that display the desired trait from a pool containing billions of variants [30]. The ligand of interest is mounted on a solid support, and the phage display library in solution is applied to selectively bind variants through affinity selection. Based on the binding targets, biopanning can be conducted at different settings [9]. For binding with the cell targets in vitro, phage biopanning is conducted on suspension cells in which phage libraries are incubated. Similarly, material surfaces can also be biopanned to identify peptides that bind to natural or synthetic materials. Biopanning in vivo is a process that entails injecting a phage library into the mice tail vein to screen out therapeutic peptides from the target tissue [[51], [52], [53], [54]]. As seen in Fig. 3, during any biopanning process, non-binding virions in the libraries are removed by washing buffers, and bound virions are collected by elution buffers. The remaining phages with high affinity for the ligands are propagated by infecting E. coli cells. The selected phage clones are used as an input for the subsequent screening to identify the best targeting phages. Repeating the process several times increases the percentage of phage that recognizes the target [8]. Through phage DNA gene sequencing, the desired target sequence can be identified and applied for fabrication of phage-based nanoparticles with targeting peptide (or antibodies, proteins, enzymes, etc.) attached. Furthermore, by means of phage display technology and biopanning, the M13 phage can display specific targeting peptides to attach to particular ligands or biomarkers, fulfilling the need for personalized treatment. Patients even with similar diseases have unique protein profiles, therefore it is crucial to ensure personalized treatment with target-specific binding, which can be accomplished particularly with M13 phage display due to its ease of display and other biological advantages (refer section 2.3) [9].
3.2. M13 phage-based bionanomaterial engineering
3.2.1. Electrospinning
Along with exploiting genetic engineering of M13 phage for phage display technology, researchers have also engineered M13 phage into distinctive nanomaterials that modify the template's morphologies and physiological characteristics to enhance its functions [14,33,55]. One of the conventional methods is electrospinning. The electrospinning technique fabricates fibers in the nanometer to micrometer range by passing polymer solutions through high voltage to create electrically charged jets that are then used to generate fibrous membranes [56,57]. M13 phage employs the electrospinning process to tailor fibrous properties and fabricate bionanofiber for biomedical applications, especially in tissue engineering [55,58,59]. For instance, Shin et al. produced hybrid nanofiber sheets by electrospinning mixed solution of poly (lactic-co-glycolic acid) (PLGA) and peptide-displaying M13 phages to create an ideal cell-adhesive substrate [12]. Electrospun bionanofibers have several advantages, particularly in tissue regeneration, including: 1) High level of versatility. Through adjusting polymer parameters such as the molecular weight, solubility, viscosity and conductivity, or processing variables such as degradation rate, voltage and feed rate, the morphological and functional characteristics of the nanofibers could be customized, as well as the release kinetics of phage particles [56,[60], [61], [62]]. 2) High surface area, and enhanced permeability. The complex network of porous structure and high surface-to-volume ratio of the electrospun nanofibers resemble the extracellular matrix (ECM) of the native tissues, as well as provide moisture and nutrients to the wound site, necessary for activating angiogenesis, fibroblast proliferation and migration, epithelialization, and cell differentiation [56,58,63]. 3) Mixture with bioactive molecules. Electrospun nanofibers can incorporate many bioactive molecules, such as antibiotics, anti-inflammatories, anti-cancer drugs, genomic DNA, enzymes, vitamins, proteins, probiotics, etc., into the polymer solution during spinning, thereby offering a high efficient drug delivery system while synthesizing nanomaterials [60,64]. The bioactive surface could also facilitate the detection and identification of pathogens and disease biomarkers as desired biomedical sensors [65,66]. 4) Other attractive features include high rigidity and cost effectiveness [58,64].
3.2.2. Hierarchical nano-to microstructures
Building a nano-to-micro hierarchical structure is an innovative strategy for assembling M13 phage into unique patterns, which can be used to develop alternative forms of virus-based materials for therapeutic purposes [67]. Synthetic building blocks with varying degrees of anisotropy (irregular patterns such as rough gratings and ridges) provide topography cues as well as a supportive environment for cell differentiation [68,69]. In particular, the disordered structures were demonstrated to instigate neurite growth, as well as promote stem cells to differentiate along the mesodermal lineage, such as neuronal cell differentiation, by strengthening the adhesion between adjacent cells [70]. In addition, astrocyte differentiation has been studied using nanofibrous structures [[71], [72], [73]]. In this regard, Zhou et al. developed a form of hierarchical structure, referred to as nanoridge-in-microridge (NiM) structure, as a mimicking matrix to induce bidirectional neural differentiation of stem cells into neurons and astrocytes [69]. The NiM structure was constructed by operating dip-pulling method, which involves dipping a glass slide precoated with positively charged polylysine into a monodisperse phage solution and then vertically pulling out of the solution at stable speed, for self-assembly of M13 phages. As the polylysine coated glass slide was pulled through the dip-pulling process, negatively charged phages were attracted to its surface and aligned into a hierarchical NiM structure [69]. Such assembly of phages into NiM was possible due to the electrostatic interaction between phages and the polylysine, as well as the air–fluid–solid interface meniscus evaporation caused by the pulling force [69]. The phage films show the presence of nanoridges (assembled from parallel-aligned phages) organized into microridges separated by the microvalleys. The topography of such phage films was a combination of anisotropic gratings and nanofibers, resulting in neuronal and astrocyte differentiation without additional neural differentiation inducers [69].
Hierarchical nano-to microstructures composed of highly aligned nanofibers were also observed in the study by Tanaka et al. [74]. Researchers have demonstrated that the M13 phage can be self-assembled into single-layer, well-packed and aligned microstructures at the liquid/liquid interface after mixing M13 phage solution with organic solvents, which results in exceptionally stable emulsions that are characterized by a decrease in surface free energy [74]. Such hierarchical microstructure showed long-term stability which is essential for further phage-directed nanomaterial synthesis.
3.3. M13 phage-directed nanomaterial combinations
3.3.1. Carbon nanotube
Multivalent structures such as the M13 phages can be further modified by integrating different compounds to synthesize phage-directed fusion substances [14]. The nano-assembly process is often carried out using components with their own chemical and physical properties suited to biosensing, scaffold building, microbiome mediating, etc. [75]. Over the past few years, carbon nanotubes have been assembled along the surface of M13 phage for the detection of bacteria and tumor through fluorescence imaging [[76], [77], [78]]. Carbon nanotubes are often constructed as single-walled carbon nanotubes (SWNTs) [76]. SWNTs offer increased signal amplification and decreased area spread in deep tissues due to their higher Stokes' shift (high Stokes' shift indicates reduced self-quenching from light reabsorption, which makes them more efficient at harvesting light), low-autofluorescence background, and high photoluminescence spectrum at 900–1400 nm. These characteristics make them more valuable for detecting specific infections in deep living tissues with enhanced contrast imaging [76,79]. Studies by Bardhan et al. Yi et al. and Ghosh et al. employed similar approaches in constructing M13-SWNT conjugates and showed comparable effects on monitoring specific bacteria, prostate cancer and ovarian cancer, respectively, especially in hard-to-reach areas [[76], [77], [78]].
3.3.2. Metal nanoparticles
In addition to the fluorescent imaging with M13-carbon nanotubes, the combination of M13 phage with metal nanoparticles has also been demonstrated to substantially improve fluorescence in bioimaging [80]. Among combination with metals, M13 phage has shown to form complexes commonly with silver nanoparticles (AgNP) [79,80,80,81] and gold nanoparticles (AuNP) [82,83]. Metal nanoparticles amplify the fluorescence excitation of the nearby fluorophores by increasing the local electric field, as well as prolonging the fluorescence duration by modulating the decay rate [80]. Metal nanoparticle conjugation with M13 phage could be used to detect bacteria [80,83] and cancer cells [84]. Moreover, AgNPs on M13 phage can also serve as cell targeting agents [24,81]. By deconstructing bacterial cell walls and intracellular biomolecules, as well as by causing oxidative stress, the metal nanoparticles on the viral structure, designed by Dong et al. selectively killed protumoral bacteria for gut microbiota regulation [81,85].
3.3.3. Magnetic nanoparticles
Immunomagnetic approaches have been exploited for detection and capturing of cancer cells [86,87]. Though commonly used, the traditional immunomagnetic approaches usually results in low delivery and capture efficiency [42,88]. Utilizing the multivalent properties of M13 phage, Ghosh et al. designed a phage vector containing a tumor-targeting peptide and a peptide motif that directs the assembly of magnetic iron oxide nanoparticles (MNPs) along the major coat protein pVIII, thereby providing a higher concentration of MNPs [42]. The greater magnetic resonance potential and the higher fluorescence uptake of M13-MNPs over MNPs indicate that the former exhibits better contrast properties, making it ideal for dark contrast imaging for selective cancer detection [42]. In contrast to attaching MNPs to the major coat protein of M13 phages, Sm et al. assembled M13 phages on magnetic particles through a “tentacle-like action”, in which the ends of M13 phages were bound to magnetic particles [89]. Compared to conventional magnetic particles, the nanotentacle-structured particles showed vastly improved binding affinity for cancer cells that captured the cells more efficiently and effectively [89].
4. Theranostic application of M13 phage
4.1. Phage-based delivery system for drugs
Target specificity is an important feature in drug delivery. Nontargeted drugs release active substances and transport throughout the human body without accumulating in any specific organ, resulting in ineffective drug action, and requiring higher doses that cause more side effects [90]. Drug delivery systems without controlled release may cause difficulty in preserving drug efficacy and precise delivery at the desired site of action [91]. Drugs may also be restricted in their delivery to certain organs by physiological barriers, instead reaching and affecting peripheral tissues [92,93]. To address these issues, recent studies have focused on nanotechnology-based approaches [90]. Among them is the M13 phage, which serves as multivalent nanocarriers to enhance drug delivery in two ways: 1) Displaying targeting peptides or antibodies to enhance targeted delivery. 2) Making use of the nanofiber-like thin structure to penetrate barriers, especially in the brain. As several targeting compounds are capable of inhibiting cancer cell growth, the multivalent M13 phage could simultaneously display therapeutic agents for targeted therapy [94]. In addition, M13 phage system has shown to be a safe and stable process due to its lysogenic properties and robust structure [14,16].
4.1.1. Improved drug delivery with phage display technique
Through biopanning, researchers have identified cancer targeting peptides that could be displayed on the coat proteins of M13 phage to assist drug delivery and generate targeted cancer therapy (see section 4.4). Wang et al. and Ghosh et al. have both displayed cancer cell targeting ligands while loading therapeutic agents on different phage display sites to target colorectal cancer and prostate cancer, respectively [95,96]. Moreover, other studies have investigated the use of overexpressed extracellular proteins in cancer as peptide-binding targets for precise drug delivery. For instance, as denatured collagens are upregulated in various cancers, Jin et al. designed M13 phage that displayed collagen mimetic peptide (7 GPP), which exhibited strong affinity to TGF-β1 induced abnormal collagens in human lung adenocarcinoma cells for potential targeted drug delivery [97].
4.1.2. Improved drug delivery in the brain
Neurodegenerative diseases are on the rise, but remain unsolved [98]. While accurate targeting of pathogenic factors in the cerebral region could allow for proactive treatment and effective monitoring of drugs, the crossing of BBB is a major obstacle [99]. This section explores the possibility of applying nanofiber shaped M13 phage as a novel delivery vector to target the protein dysregulation of these diseases, such as Alzheimer's disease (AD) and Parkinson's diseases (PD), through effective penetration of the barriers in the brain.
The most prevalent explanation for the pathogenesis of AD is the increased accumulation of amyloid-β polypeptides (Aβ), which are proteolytic products from amyloid precursor proteins (APP) [100]. With M13 phage's linear structure and high permeability, researchers have used genetically modified M13 phages displaying single-chain variable fragment (ScFv) of antibodies against the Aβ fragment to deliver the antibodies to the central nervous system (CNS) [101]. To first demonstrate the filamentous phage's ability to reach the CNS, Solomon injected M13 phage displaying anti-Aβ ScFv into Alzheimer's APP transgenic mice through intranasal route, and showed positive Aβ (detected both by Thioflavin-S staining and fluorescent labeled anti-phage antibodies) in the olfactory and hippocampal regions, followed by 72-h clearance of the phage [101]. A six-month Intranasal administration of antibody-displaying phages to transgenic mice overexpressing human amyloid-β protein precursor (hAPP) led to a significant reduction in brain inflammation and a reduction in plaque load by 50%, as well as improved cognitive functions evaluated by spatial and temporal navigation assay [101]. Hence, the delivery of M13 phage displayed with anti-Aβ ScFv could be used not only to dissolve Aβ plaques, but also as a highly specific diagnostic tool to monitor the formation of amyloid plaques in AD patients. Notably, De Plano et al. used biopanning screening of the pVIII M13 phage display library to construct structural peptide segments that form amyloid-like structures. These structures serve as a target for serum IgG of patients with AD, which could aid in the diagnosis of the disease's state and stage [102].
Through infiltration of the CNS, moreover, M13 phage itself has proven to be a potential treatment for PD, which is associated with amyloidosis characterized by deposits of amyloid fibrils of α-synuclein (AS) [103,104]. Upon one-week incubation of phage with fibril solutions, significant disaggregation of fibrils occurred, presumably due to the interaction between phage and the central hydrophobic domain of AS [104]. To conclude, M13 phages, with or without displayed antibodies, proved to be effective and safe viral delivery vectors to the brain, demonstrating the ability to penetrate into the CNS while binding to and disrupting protein aggregation associated with neurodegenerative diseases.
4.2. Biosensors
With M13 phage's natural affinity for F-pilus bacteria and its programmable phage display, the phage-based bionanomaterial, often combined with other imaging moieties, can be used as a biosensor to detect a wide range of substances, such as pathogens, biomolecules, and cancer cells [7,36,78,80].
4.2.1. Detection of pathogens
According to recent systematic reviews, bacterial antimicrobial resistance was associated with 4.95 million deaths in 2019 globally with an estimated financial impact of $1.0 to $3.5 trillion per year by 2030 [[105], [106], [107]]. A global burden and large increase in antibiotic-resistant bacterial infections call for more sophisticated imaging modalities to detect and treat infections early and precisely, ensuring better chances of successful treatment while maintaining antibiotic efficacy [108,109]. Among the current biodetective methods are conventional bacterial culture and cell counting, polymerase chain reaction (PCR), as well as antibody-based detection [110,111]; More recent approaches include polymer-based detection (e.g., polydiacetylenes) [112], microelectronic devices [113], metal nanoprobes [114], etc. However, the current methods have disadvantages in their long detection times (e.g., bacterial culture), high cost of equipment, tendency to show false positives (e.g., PCR tests), low tolerance to extreme environments (e.g., antibody), and lack of ability to detect in vivo (e.g., polymer-based detection) [7,82,112,115].
The challenges could be overcome by incorporating above-mentioned biodetective methods with M13 phage. M13 phage-based biosensors for pathogen detection could capture bacteria using M13 phage and occasionally bind antibodies to enhance or specify target recognition, while using integrated sensing probes to generate detection signals. The M13 phage allows for a more accurate pathogen detection within living tissues. Bardhan et al. engineered a multifaceted M13 phage vector capable of dispersing and stabilizing SWNT, a tissue specific, highly sensitive, and aqueous-dispersed nanoprobe for non-invasive detection, imaging, and monitoring deep buried specific F′-negative bacteria in vivo [76]. The M13 phage was developed as a biological scaffold with SWNTs attached via major coat protein pVIII and anti-bacterial antibodies attached via minor coat protein pIII. The anti-Staphylococcus aureus (S. aureus) antibody provides greater specificity towards the relevant strains of bacteria, showing a 3.8-fold increase in fluorescence signal enhancement of the deep-tissue S. aureus endocarditis mouse model for the antibody-integrated model compared to the non-integrated model [76]. Furthermore, in the same study, the SWNTs exhibited a higher signal amplification at greater depths than traditional fluorescent dyes in chicken model, as the SWNT group had a 1.4-fold higher amplification signal. By stabilizing and dispersing SWNTs in an aqueous suspension, the M13 phage allows for in vivo bioimaging with high specificity and high photoluminescence properties sustained throughout tissue depths [76]. The stability of the M13 phage-based biosensor was demonstrated by Sedki et al. who conjugated M13 phage with AuNP on a glassy carbon electrode for rapid screening of fecal coliform bacteria [82]. The biosensors showed no significant difference in their sensitivity at 45 °C for up to 16 days, and their sensitivity remained unaffected at pH ranging from 4.5 to 8, with slight reductions at pH 3 and 10 [82].
In light of the global burden of antibiotic resistance, researchers have taken a broader approach than merely detecting the causing pathogens [107,108,115,116]. To ensure effective antibiotic stewardship, rapid and accurate characterization of antibiotic susceptibility profiles (ASP) of infectious bacterial strains is also essential as it helps clinicians identify appropriate antibiotics and their dosages at early stage without having to prescribe broad-spectrum antibiotics [117]. Unlike the classic characterization methods, such as disk diffusion and broth dilution that are time-consuming and laborious, Peng et al.‘s M13-AuNP assay could rapidly detect bacteria's antibiotic susceptibilities via phenotypic analysis [115,117]. The tetracycline-resistant E. coli strain ER2738, which is sensitive to ampicillin and kanamycin, was tested for its ASP by diluting into tubes loaded with one of the three antibiotics. By taking advantage of M13-AuNP assay's rapid detection of bacteria even at low concentrations, the phage-based technique shortened the time needed for ASP characterization by reducing the time required to produce detectable growth and assess growth. The improved assay duration is approximately 2.5 h (2 h bacterial growth and 30 min determination), which is considerably shorter than the 16 – 24-h test duration of both disk diffusion and broth dilution methods [115,117].
4.2.2. Detection of disease biomarkers
The M13 phage can be used to detect proteins as biomarker molecules to screen diseases including cancer. Protein-coupled receptors and integrins are overexpressed in certain diseases such as glioblastoma multiforme, in which heterodimer αvβ3 integrin cell surface receptors are highly expressed [118]. In addition, the integrin binding motif Arg-Gly-Asp (RGD) is identified to recognize integrins for tumor targeting [119]. As RGD can be displayed on M13 phage coat proteins to recognize overexpressed integrins in cancer cells, such binding could aid in disease diagnosis and imaging [24].
Detection of antigens is relatively common and is made possible by the structural property of M13 phages. The pIII minor protein located at the tip of the phage structure could be used to display antibodies that specifically bind target antigens. In the study by Hou et al. the M13 phage was genetically engineered at pIII protein site to display ScFv of antibody molecules that targets colorectal cancer cells with overexpressed carcinoembryonic antigen (CEA) [120]. The phage conjugation could specifically attach to Caco-2 and HT29 cells with CEA expressed, but hardly bound to HEK293T cells that didn't express CEA. Additionally, the signal-to-noise ratio of the M13 phage conjugated probe (S/N = 36.7) was substantially higher than that of the negative control (M13KO7 helper phage) (S/N = 8.3), indicating that the former with anti-CEA ScFv offers higher specificity when detecting the targeted cancer cell [120]. While pIII minor protein is involved in biological recognition, the pVIII major protein located at the bilateral sides of the phage could be simultaneously modified to attach for protein sensing moieties [14]. Lee et al. engineered M13 phages conjugated with multiple DNA oligonucleotides, antigoat immunoglobulin (IgG) and AuNPs to generate cost-effective protein sensors that rapidly detect the presence of target antigen [121]. The pVIII bound AuNPs act as signal-producing materials that enhance its interaction with M13 phages as antigen concentration in the solution increases, resulting in immediate color changes. Moreover, the conjugated M13 phages also allow identification of the detected antigen by eluding the AuNPs from the phage and transferring the sequence to DNA microarrays for analysis [121].
Upregulation of matricellular protein, specifically secreted protein acidic and rich in cysteine (SPARC), is often observed in certain tumor progressions such as cervical cancer, renal cancer, ovarian cancer and prostate cancer [[122], [123], [124]]. Ghosh et al. isolated a SPARC-binding peptide (SBP) using phage biopanning and genetically engineered to display SBP on the pIII minor protein of M13 phage, while the MNPs were displayed along the PVIII coat protein (see section 3.3.3) [42]. The M13-SBP-MNP conjugation was injected intravenously in mice model with either C4–2 B (prostate cancer cell line with high SPARC expression) or DU145 control (prostate cancer cell line with low SPARC expression) prostate cancer cell line inoculated for imaging of prostate cancer. After staining the tumor section using anti-human SPARC antibody, the results show colored stains on the tumor cells with high expression of SPARC in comparison to the control group with no staining observed, thereby suggesting M13 phage-derived biosensor as a promising tool for selectively screening SPARC expressed tumors [42]. A similar study was conducted by Ceppi et al. who designed SBP-M13 phage-based probe with SWNTs as near-infrared fluorophores assembled along the phage to accurately detect microscopic tumors in ovarian cancer mouse model [122].
Leading research has been conducted by Bhasin et al. who demonstrated the use of virus bioresistor (VBR) as a platform for incorporating M13 phage into an electoral apparatus to detect DJ-1 protein, a bladder cancer biomarker [125]. The M13 phages are embedded on the 3,4-ethylenedioxythiophene to create bioaffinity layers that could rapidly bind antibodies. The channel resistance (RVBR) reflects the information on the targeted protein [126]. The author demonstrated that VBR with M13 phage incorporated could respond to the presence of DJ-1 protein with a high signal-to-noise ratio (S/N > 100) and excellent reproducibility between sensors [125]. To further illustrate the versatility of M13 phage-based biosensors for detecting a variety of biomarkers, Cho et al. used M13 phage for biopanning and displaying targeted peptide for identification of neutrophil gelatinase-associated lipocalin (NGAL), a highly selective biomarker for acute kidney injury diagnosis [127]. To assess the binding affinity and specificity between the peptide displayed and NGAL, enzyme-linked immunosorbent assay (ELISA) and electrochemical impedance spectroscopy (EIS) were conducted, with the results showing a high binding affinity at certain concentrations and an increase in impedance, respectively, indicating high specificity and sensitivity in NGAL detection using M13-derived sensor system [127].
4.3. Tissue regeneration
4.3.1. What is tissue regeneration?
Tissue regeneration is a complex cellular process that involves the restoration of normal morphology and functionality to injured tissues [128,129] Tissue and organ lesions are among the most common challenges in medicine, and the shortage of organ donors, increasing transplantation rates, ineffective compatibility of transplants with the host, and the possible rejection of the foreign tissue implant necessitate the development of biocompatible materials based on novel techniques [56,130]. In order to understand how M13 phage is applied to tissue engineering, a comprehensive understanding of normal tissue regeneration mechanisms is necessary as well as a review of current biomimicking approaches.
Tissues are comprised of specialized cells and their supporting matrix, called extra cellular matrix (ECM). The ECM is a nanofibrous protein network (diameters ranging from 3 to 300 nm) that carries biochemical ligands and bioactive molecules (e.g., growth factors and cytokines) for orchestrating nearby cell fates, including those of seed cells (e.g., differentiated cells and stem cells). The mechanically rigid self-organized structure also provides physical supports and instructive cues through topographical presentation for regulating cell activities [20,131]. During regular tissue rejuvenation or minor injury healing, undamaged tissue cells, including stem cells or differentiated normal cells, can replace the damaged cells in injured tissue to restore its normal structure and function. By contrast, when significant traumatic injuries take place, as in the case of military patients or those with chronic diseases wounds, the routine regenerative process is impaired due to various reasons such as ongoing infections, vein obstructions, and peripheral neuropathy [132,133]. For such irreparable extensive damages, the design of a biodegradable, 3D network similar to the ECM holds promise for not only restoring tissue growth and function, but also improving tissue compatibility during transplantation when needed [1]. To further promote the interactions between cells and tissues, autologous cellular elements are incorporated into the nanostructure [132]. Hence, the biomimetic materials serve as structural scaffolds (in shapes of nanoparticles, nanofibrous, 3D printed) with bioactive molecules and seed cells attached to regulate cell behavior using chemical and physical cues [20,134].
Currently, treatments using ECM-mimicking scaffolds are limited by biomolecule inclusion, such as the inability to economically produce bioactive molecules, and the difficulty in integrating them into scaffolds to sustain their functionality [134]. Moreover, a lack of precise control over chemically conjugated peptides may also limit the ability to effectively manipulate stem cell fate with currently available peptide-coated biomaterial substrates [23]. Conversely, peptides can be genetically introduced onto M13 phage coat proteins in an efficient and precise way, which further facilitates more effective integration of biomolecules [20]. M13 phage is therefore a potential option for tissue regeneration materials due to its genetic control and modularity properties.
4.3.2. M13 phages as novel bionanomaterials for mimicking natural tissue environment
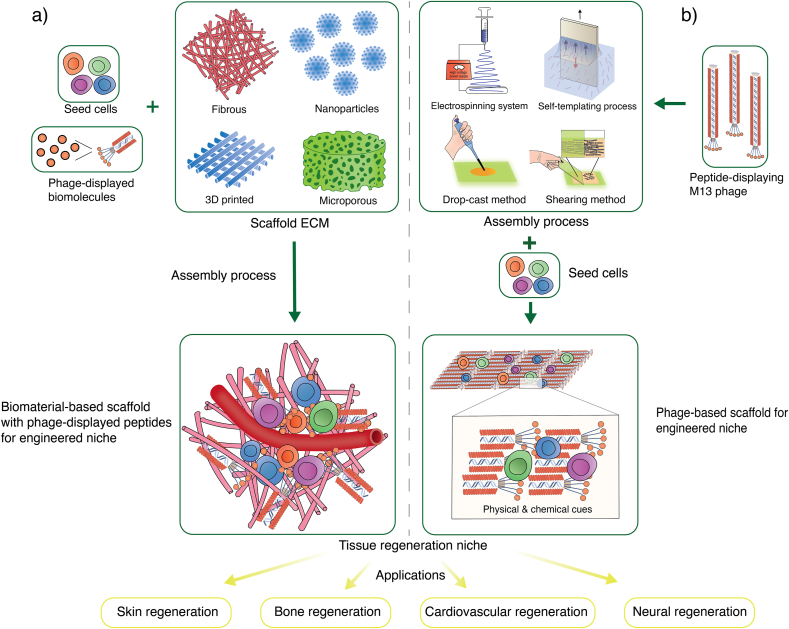
M13 phage is a highly effective tissue regenerating materials due to its biological advantages (see section 2.3). The ability to precisely express functional peptides as signaling molecules on the surface [41], to regulate the behavior of macroscopic cells [18], to self-assemble into different order structures for diverse topological effects [27], to be biocompatible and biodegradable [135], to provide high surface area and adaptability for cell interaction and material integration [20] make M13 phage ideal bionanomaterial for creating artificial cell “niche” (Fig. 4) [41]. Such multifunctional niche provides both chemical and physical cues to create a conducive environment for cellular growth. Furthermore, by mixing with bioactive molecules, therapeutic cells and polymeric solutions during the electrospinning process, M13 phage can transform into ECM-mimicking electrospun nanofibers with integration of molecular components in high efficacy (see section 3.2.1) [64]. Nevertheless, some studies use M13 phage for displaying functional peptides and integrating into complementary scaffolds instead of using it as a component for building tissue scaffolds, as seen in Fig. 4a [136].
Fig. 4.
Demonstration of two distinct artificial cell niches using M13 phage as bionanomaterial. (a) Conventional biomaterial-based scaffold with phage-displayed peptides and seeded stem cells for engineered niche. (b) Biomimetic phage-based scaffold with seeded stem cells for engineered niche.
M13 phages are genetically engineered for multivalent display of various peptide ligands, such as RGD, the cell adhesion integrin binding motif, at their N-termini of pVIII major coat protein to regulate intercellular interactions [18]. By binding to integrins on activated endothelial cells (ECs) as well as other cell types, the RGD peptide regulates the cell behavior and stimulates angiogenesis, neurogenesis, and osteogenesis within the ECM [[137], [138], [139]].
4.3.3. Skin regeneration and wound healing
Based on a retrospective analysis conducted in the U.S in 2014, approximately 8.2 million patients suffered from chronic wounds [140]. In order to reduce chronic wound burdens on health systems, more research is needed on the mechanisms of wound healing and novel approaches to healing chronic wounds. Wound healing consists of four phases: hemostasis, inflammation, proliferation, and remodeling [141,142]. Among four phases, poor proliferation, or impaired regeneration of tissue, significantly hinders chronic wound healing and is one of the most challenging obstacles to overcome, due to the difficulty in designing tissue regeneration materials with precise structural arrangements while exhibiting signaling motifs to direct cell behavior [18,142]. Proliferation stage mainly involves reconstruction of vascular channels (angiogenesis) and growth of fibroblasts (fibroplasia) to form granulation tissues. As fibroplasia occurs, the surrounding ECs proliferate, assisting in further blood vessel healing within the granulation tissue for ensuring proper blood flow and providing wound healing factors [141]. By inducing angiogenesis and fibroplasia, M13 phage nanofibers can be used to promote skin cell proliferation.
Normally, the balance between vessel proliferation and stagnation determines angiogenesis, a hallmark for healing wounds [143]. Angiogenesis fails as a result of loss of proangiogenic mediators when factors such as apoptosis of ECs and macrophage deficits affect the balance [132]. In accordance, angiogenic and/or integrin-binding peptides (e.g., RGD) could be introduced on to M13 phage, allowing the nanofiber to function as an angiogenic factor as well as an ECM component [144]. Yoo et al. developed a vascular-like niche, with angiogenic peptides (Ser-Asp-Lys-Pro; SDKP) and RGD peptides displayed on pVIII and pIII coat proteins of M13 phage, respectively [144]. RGD peptide promotes angiogenesis by binding to EC integrin and thereby promoting the migration of quiescent endothelial cells to form new blood vessels, whereas the SDKP peptide stimulates EC proliferation. Interestingly, through topographical cues of the phage patterns, M13 phage itself could promote EC-specific activity, as shown by elongated EC morphologies. The phage nanofiber's own angiogenic effects were further enhanced by the expression of SDKP and RGD peptides as biochemical cues [144].
Fibroblast proliferation and migration are activated upon fibroblast growth factor (FGF), a family of growth factors involved in cellular signaling pathways for tissue regeneration [145]. The expression of FGF, however, is reduced during chronic wounds due to downregulated activity of FGF receptors (FGFR) and low sensitivity to external environmental cues [132]. Though topically administered FGFs have been introduced as therapeutic agents, they are easily degradable in vivo and poorly absorbed through the skin, resulting in a lack of bioactivity in the wound bed [146,147]. Instead, functional peptides could be displayed on M13 phage to activate FGF and FGFR-associated signaling pathways. Through biopanning with Ph.D.-7 library, Zhao et al. identified phage clones displaying novel peptide H1 that strongly binds to FGFR2IIIc (recombinant receptor protein responsible for skin tissue repair), which activates intracellular signals and regulates cellular behavior [146]. The H1 peptide, which was topically applied to the wound site, accelerated rat full-thickness wound healing in vivo, stimulated proliferation and migration of fibroblasts and human umbilical vein endothelial cells (HUVEC) in accordance with its angiogenic property, and improved skin wound healing by simultaneously activating MAPKERK1/2, PI3K-AKT, and vascular endothelial growth factor (VEGF) signals (cell proliferation and angiogenesis inducers) [146].
It is important to note that wound healing is a multi-stage process that involves both tissue damage repair and infection prevention. FGF-based therapies that neglect bacterial complications could result in compromised treatment outcomes [148]. Thus, to achieve optimum wound healing, antibacterial treatment must also be used for combating bacterial infection and biofilm formation at the wound site [149]. Peng et al. engineered M13 phage nanofiber conjugates, called phanorod-Zn, that inhibited the growth of Gram-negative pathogens and provided faster healing results in vivo compared to conventional antibiotics or antibodies treatment [150]. The phanorod was based on the conjugation of gold nanorods (AuNRs), an effective photothermal agent against bacteria, and M13-g3p (Pf1), a genetically modified M13 phage with the receptor-binding domain of phage Pf1 swapped at pIII to attach to type IV pili of Pseudomonas aeruginosa (P.aeruginosa). In addition, phanorods displayed zinc-binding peptides designed to release zinc ions upon photothermal heating, which promoted collagen deposition and wound healing. With phanorod-Zn treatment, infected wounds of mice at day 4 had a size under 20% of an untreated wound, and they were completely healed after 8 days. Phanorod-Zn approach also showed a 50% reduction in wound size by day 2, versus ciprofloxacin at day 6, demonstrating a three-fold faster healing rate than standard-of-care antibiotics. According to the bacterial load measurements, the decrease in wound size corresponded to a decrease in cfu at wound site by > 10-fold on day 2 and > 100-fold on day 4 [150]. Overall, since nanofiber M13 and its antimicrobial peptides and materials displayed can effectively control bacterial infection and skin tissue proliferation, such phage-based approaches could be highly desirable in novel topical formulations to treat chronic wounds.
4.3.4. Bone regeneration
Bone is made up of bone cells surrounded by the ECM, which consists of organic compounds (40%) including collagen type 1 and noncollagenous proteins such as proteoglycans, glycoproteins, and glycophosphoproteins, along with inorganic compounds (60%) mainly composed of calcium phosphate hydroxyapatite (HAP) [151]. Bone cells consist of multipotent mesenchymal stem cells (MSCs), osteoblasts, osteocytes, and osteoclasts (hematopoietic progenitor-derived cells that are involved in bone degradation) [152]. During osteogenic differentiation, osteoblasts are differentiated from MSCs and secrete collagen matrix, as well as mineralize bone matrix. Osteocytes are formed when immature osteoblasts are surrounded by the secreted matrix that activates their morphological transformation [152]. The cellular components, therefore, dynamically interact with bone ECM to facilitate bone regeneration. Moreover, to maintain structural stability and health of the skeleton, bone relies on well-developed blood vessels to supply blood, nutrients and hormones consistently [153]. Accordingly, current M13 phage-based bone tissue regeneration materials include elements of ECM-mimicking scaffolds, biomolecules, along with or without additional seed cells to promote osteoinductivity, angiogenesis, or both.
Yoo et al. studied the effects of collagen-derived peptides displayed on the collagen-like structure of the M13 phage on early osteogenic differentiation of bone stem cells [154]. Researchers constructed nanofibrous tissues-like matrices by genetically introducing DGEA-peptides, natural compounds derived from collagen type 1 with osteodifferentiation properties, onto pVIII of M13 phage with high density, in order to examine its effects on mouse preosteoblast cells (MC3T3). As a result, the DGEA-phage matrices showed 70% higher alkaline phosphatase (ALP) activity than the control group, as did other osteogenic differentiation markers. Shin et al. also reported the enhanced proliferation of mouse preosteoblastic cells (MC3T3-E1) when seeded onto the 3D porous scaffold composed of electrospun RGD-M13 phage-PLGA nanofiber sheet, which served to promote adhesion of the cells [12]. Based on Lee's research, mechanical cues can modulate osteogenic differentiation of MC3T3 cells by shifting the stiffness of the ECM-like matrix [155]. The M13 phage was genetically engineered to express RGD and HPQ, a biotin-like peptide that binds to streptavidin for phage-to-phage crosslinking in a controlled manner. The increase in scaffold stiffness resulted in greater osteogenic gene expression in preosteoblastic cells, indicating that mechanical cues play a key role in osteogenic differentiation, but only when cell-matrix interaction is present, as with RGD peptide display [155].
Moreover, HAP-containing bone regeneration scaffolds have demonstrated enhanced osteoconductivity and osteoinductivity, for which phage biopanning effectively identifies HAP binding peptides [156,157]. Chung et al. used Ph.D.-12, −7, and -C7C phage library to screen out the best HAP-binding peptide NPYHPTIPQSVH (CLP12 peptide) after four rounds of screening [156]. Isolated CLP12 peptide could induce HAP nucleation and growth in a sequence- and composition-selective manner compared with the control group, suggesting its potential display on M13 phage for synthesis of bone-mimicking biochemical scaffold. It is important to note that Li et al. by displaying HAP-binding peptide (DSSTPSST, termed DT peptide) on the pVIII coat protein of fd phage to form phage-HAP scaffold, showed such bone-mimicking biochemical scaffold to support the proliferation of MSCs in high stabilization and strong surface interactions with the host cell [157]. Further studies, therefore, should focus on producing scaffolds with nanoscale alignment of the M13 phage nanofibers displaying HAP-binding peptides to compare bone repair outcomes. Nevertheless, studies by Wang et al. and He et al. both demonstrated the formation of M13 phage-based nanocrystalline structure with oriented nucleation of HAP accumulated along phage bundles via anionic and cationic reaction [158,159]. The M13 phage, genetically engineered to become anionic by fusing negatively charged peptide to pVIII, self-assembled in the presence of cationic precursor (Ca + ions) and anionic precursor (PO43- ions), leading to the oriented nucleation of HAP to form hybrid nanostructures of HAP and M13 phage, which resembled the hierarchical structure of bone [159]. Future research should investigate the in vivo application of such nanostructures.
Several recent bone regeneration strategies, such as the targeting of type H vascular ECs, focus on improving vasculature in the skeletal system for promoting tissue development [160,161]. Meanwhile, M13 phage nanofibers displaying RGD peptides are also employed to facilitate activated EC migration and adhesion to generate bone tissue with high density of blood vessels [136]. Wang et al. designed a virus-activated matrix (VAM) with M13 phage-RGD electrostatically integrated into the pores of chitosan-decorated 3D printed biomimetic bone scaffold consisting of biphasic calcium phosphate, HAP, and β-tricalcium phosphate. The VAM was then seeded with MSCs and implanted into the damaged rat bone. As a result, VAM led to enhanced bone formation following the induction of vascularized bone tissue, as evidenced by the increased expression of the EC markers and the formation of new blood vessels [136].
4.3.5. Cardiovascular regeneration
Cardiovascular disease (CVD) remains the leading cause of death worldwide, including in China, where the disease accounts for 40% of all deaths in the Chinese population [162]. Atherosclerotic CVD (ASCVD), including ischemic heart disease and stroke, results in the majority of CVD deaths and the burden is steadily increasing [162]. Among the promising therapeutic options is stem cell transplantation that aims to restore cardiovascular function using progenitor cells such as endothelial progenitor cells (EPCs) and human cardiac progenitor cells (hCPCs), which can differentiate into cardiomyocytes, endothelial cells, and smooth muscle cells [163,164]. However, limitations lie in the low survival rate of stem cells implanted into the ischemic area and decreased bioactivity of implanted cells, possibly due to impairment or absence of the vascular “niche” [165,166]. In this regard, M13 phage has been exploited both as nano-carrier and as biochemical scaffold to enhance engraftment ability and stem cell functions.
Lee et al. constructed an artificial stem cell niche comprising of engineered M13 phage for enhanced EPC function [165]. The researchers first reported the dual functional motif of M13 phage displaying RGD peptide, a cell-adhesive motif, and thymosin β4 (Ser-Asp-Lys-Pro; SDKP), a pro-angiogenic and anti-fibrotic motif, for constructing peptide-displaying scaffold. EPCs were then treated with engineered M13 phage. Based on the expression levels of cyclin D, scratched wound healing assay, and Matrigel tube formation assay, the M13 phage displaying both RGD and SDKP promoted proliferation, migration, and tube formation in EPCs, respectively, suggesting a synergistic effect between the two displayed peptides. Engineered M13 phage also exhibited anti-oxidative and anti-inflammatory properties in EPCs. Accordingly, by transplanting M13 phage-treated EPCs into a murine hindlimb ischemia model, researchers observed increased EPC survival, neovascularization, and functional recovery [165]. Genetically engineered M13 phages displaying RGD and SDKP peptides at pIII and pVIII, respectively, were also designed by Jang et al. in order to investigate the efficacy and viability of transplanting hCPCs in ischemic heart regions [166]. Despite no morphological changes observed in hCPC after pre-treatment with engineered M13 phage, the engineered M13 carrier significantly increased hCPC retention in the myocardial infarct zone of mice after three days. Moreover, hCPCs treated with engineered M13 phage exhibited enhanced angiogenic activity, as demonstrated by their increased ability to migrate and form tubes in vitro, as well as the increased number of smooth muscle cell marker in vivo [166].
4.3.6. Neural regeneration
Neural regeneration in the CNS involves regrowth of axons from newborn neural cells following traumatic brain injury [167]. Moreover, in neurological injuries such as brain stroke, autophagy and degradation of injured tissue cause impairment of the ECM, which hampers regeneration of brain tissue [139]. Hence, research has turned to constructing ECM-mimicking scaffolds with therapeutic cells, such as neural progenitor cells (NPCs), neural stem cells (NSCs) and human induced pluripotent stem cells (hiPSCs) for neurite regeneration, as well as biomolecules that guide neuron network restoration [18,69,139,168]. Cell delivery vehicles such as M13 phages could carry therapeutic cells while functional peptides with biochemical cues could be displayed along the nanostructured virion that assembles into an ECM-like scaffold or integrates with complementary scaffolds.
Merzlyak et al. designed M13 phages that displayed either RGD or IKVAV peptides (motifs known to promote neural cell adhesion and neurite extension) at pVIII and arranged them into three-dimensional nanofiber matrices with NPCs embedded for cellular proliferation and differentiation [18]. Within seven-day period, the adult rats' NPCs aligned along either phage fiber were able to differentiate neural cells and extend long neurites parallel to the fiber's long axis [18]. Similar study was conducted by Chung et al. who genetically engineered M13 phages to display RGD peptides and induced virion self-assembly into matrices featuring anisotropic topography capable of guiding NPC growth. Due to the biochemical and topographical cues, the seeded NPCs showed considerably higher proliferation rate in RGD-phage films compared to that of the control group with wild-type-phage films. Compared to the control group, differentiated NPC neurites grew at a much faster rate, and their extension length was four to seven times longer on day 4 [169]. In addition to neurons, astrocytes, also differentiated from NPCs, are equally essential for the building of the brain and may stimulate growth-promoting ECM components like fibronectin and laminin [68,155]. Hence, research conducted by Zhou et al. explored the bidirectional differentiation of hiPSC-derived NPCs into both neurons and astrocytes using NiM structure composed of M13 phage [68]. The topographical cues, in conjunction with the RGD peptides, greatly induced NPCs to express neuron markers, as well as to generate astrocytes within 8 days, as opposed to 30 days required with astrocyte inducer medium [69].
Similar to other tissue repair, restoring optimum function in damaged brain tissue also depends on the formation of abundant vascular networks. Liu et al. explored the application option of using M13 phages to display functional peptides and integrate them to microparticles (MPs) as templates to seed NSCs for achieving both neurogenesis and angiogenesis after stroke [139]. To achieve this, Liu et al. genetically introduced RGD peptide to display on pVIII of M13 phage in high density to form “R-phage”. Silk fibroin MPs were prepared from silkworm cocoons for biocompatibility, then coated with polyethyleneimine (PEI) to allow R-phage attachment to the MPs electrostatically [139]. As the NSC-seeded R-phage-MPs were injected into the stroke sites in the rat brains, the phage conjugation improved stroke-damaged brain tissue repair by stimulating NSC differentiation, thereby regenerating densely connected axon-rich neurons in the brain and improving animal limb motor function significantly. In addition, the compounds stimulated revascularization and angiogenesis by activating endothelial cells, which further promoted the delivery of nutrients and oxygen for neurogenesis [139].
4.4. Targeted cancer therapy
4.4.1. Breast cancer
Breast cancer (BC) is the most prevalent type of cancer in women [170]. In clinical practice, conventional treatment methods for BC include surgery, radiotherapy (RT), chemotherapy, endocrinotherapy and targeted therapy. However, these methods could induce resistance and strong side effects, especially in the case of triple negative BC (TNBC) [171]. The alarming numbers in BC prevalence and deficiency in conventional treatments demand for extensive research in novel tools for BC therapy.
The utilization of M13 phage display in BC therapy is a promising method in identifying tumor-targeting agents as well as carriers and potential vaccines. In 2004, An et al. managed to isolate an anticancer peptide, F56, through the screening of M13 phage library. F56 was found to be both antiangiogenic and antimetastatic when tested in severe combined immunodeficient mice implanted with human BC cells (BICR-H1) [172]. Similarly, a human antibody, scFv 12H7, was recognized with tumor inhibitory effects on TNBC cells both in vitro and in vivo [173]. The use of this antibody significantly reduced tumor size when compared to the isotype control. BC undergoes epithelial to mesenchymal transition (EMT) prior to metastasis, hence recognition of EMT could be promising in BC diagnostics. Jones and colleagues identified LGLRGSL, a phage peptide that binds specifically to EMT BC cells, possibly providing early identification of metastatic BC [174].
By displaying exogenous linear dodecapeptides to the minor coat protein pIII of M13 phage, peptides with high affinity to BC cell line MCF-7 were isolated and then evaluated for efficacy in cancer drug delivery [175]. A MCF-7 xenograft model on female nude mice was used, demonstrating results of improved antitumor effect of targeted drug delivery. In a study by Bartolacci et al. M13 phage was engineered to display Δ16HER2 (splice variant of HER2 that accelerates BC progression) epitopes on their coat protein pIII to serve as an anticancer vaccine [19]. The vaccine was able to avoid immune tolerance as well as induce a protective response against Δ16HER2. Moreover, a recent study discussed recombinant M13 phage particles displaying a total of eight vaccine immunogens, all of which carrying special proteins and multiepitope regions [176]. Using three of the eight vaccines studied, vaccination of 4T1 BC mouse models showed clear tumor growth inhibition.
Interestingly, Li et al. demonstrated genetic engineering of the fd phage to display both an angiogenin-binding peptide on its side surface and MCF-7 homing peptides on its tip [177]. This could enable significant tumor suppression, as phages not only home in breast tumors, but also bind to angiogenin which prevents angiogenesis. Considering the similarities between M13 and fd phage, this study suggests that the M13 phage may be effective at inhibiting angiogenesis in BC therapy, a research area that could be explored in future studies.
4.4.2. Brain tumor
Glioblastoma, also known as glioblastoma multiforme (GBM), is an incurable, malignant, and most common primary cerebral tumor in adults with a less than 2 years median survival time and a 5-year survival rate of only 5.4% in the United States [178]. Current therapy consisting of radiation, chemotherapy, and surgery have not produced effective results in prognosis or mortality [179,180]. In a novel attempt to increase survival and potentially open routes to cure the disease, it was proposed that a M13 phage was used to increase the efficacy of current chemotherapy drugs [118].
In order to accomplish this, the M13 phage will have to cross the BBB as mentioned previously. The BBB normally blocks out 98% of small molecules, thus presenting a large obstacle for current drug delivery options for glioblastoma [181,182]. Current phage-based drug delivery has been credited with thermodynamic stability, biocompatibility, homogeneity, high carrying capacity, self-assembly, scalability, and low toxicity [183]. Przystal et al. proposed in 2019 that Temozolomide (TMZ), a chemotherapy drug able to cross the BBB, can be combined with gene therapy and delivered via the M13 phage directly to the glioblastoma site as a result of a synergistic action [118]. Their research was conducted in vitro in common human cellular models of glioblastoma (LN229, U87, and SNB19) and determined that tumor cells treated with TMZ resulted in effective glioblastoma cell killing in vitro as well as glioblastoma growth inhibition in vivo. Moreover, their research determined that the M13 phages not only targeted existing cancer cells, but also cells that help new blood vessels form, and cancer stem cells which possess the ability to evade initial treatment because of its undifferentiated state [118].
Another factor affecting this form of treatment is the method of delivery. A novel approach known as convection-enhanced delivery (CED) was used for chemotherapy drug administration in pontine glioma brain tumor patients aged 3–21 years old [184]. This route was also determined to be a safe and effective method for M13 phage distribution to the brain [185]. Further research in method of delivery, recombinant adeno-associated virus genome therapy, and clinical viability are still needed.
4.4.3. Colorectal cancer
Colorectal cancer (CRC) is one of the leading causes of cancer death worldwide, often being diagnosed in its later stages with poor response to conventional chemotherapy and cancer immunotherapy [186]. Several underlying factors in the tumor microenvironment (TME) could account for the suppression of anti-tumor immunity in cancer immunotherapy. According to current research, the gut microbiota, notably the overgrowth of Fusobacterium nucleatum (Fn) in CRC tissue, triggers the recruitment of immunosuppressive myeloid cells, creating an immunosuppressive TME that inhibits T cell responses [187,188]. To regulate the microbiota, the use of antibiotics has been proposed, but discouraged due to its broad target spectrum and its negative effects on immunotherapy [81,189]. In this case, the M13 phage could be applied to rebuild tumor-immune microenvironment by precise manipulation of gut microbiota as well as activating innate immune cells [81,190].
Phage-directed nanomaterial combination therapy was proposed by Dong et al. to accurately manipulate gut microbiota for CRC suppression [81]. While M13 phage has shown its capability of screening various targets, researchers identified a specific Fn-binding M13 phage strain that demonstrated optimal targeting of bacteria based on three rounds of biopanning. In order for M13 phage to effectively eliminate Fn, AgNPs were electrostatically attached to antibacterial phage surfaces to create a hybridization system called M13@Ag. Results after intravenous injection of M13@Ag in vivo revealed that AgNPs selectively eliminated protumoral Fn following precise phage recognition system and further inhibited myeloid-derived suppressor cell expansion, thereby suppressing tumor cells and eliminating immunosuppressive TME. Interestingly, M13@Ag also promoted antigen-presenting cells (APC) activation in vivo and contributed to antitumor immune response by activating innate immune cells such as dendritic cells and macrophages [81]. M13 phage induces innate immune cell inflammation to alleviate tumor-associated immunosuppression through its immunogenic nature [191].
An additional study demonstrated that M13 phage could be used to actively promote tumor infiltration of innate immune cells by specifically targeting highly expressed antigens on the surface of CRC cells [190]. By designing M13 phage displayed with CEA-specific ScFv at the minor coat protein, Murgas et al. generated a CEA-targeting phage termed M13 αCEA. Both intratumoral and systemic administration of M13 αCEA diminished tumor growth and prolonged survival in CRC mouse models, with a mechanism involving activation of macrophages, neutrophils, and dendritic cells, along with activation of CD8+ T-cell responses [190]. With existing strategies focused on overcoming immunosuppressive TME to protect against CRC, M13 phage offers the possibility of restoring tumor immunity. Nonetheless, future studies should explore the combination of novel and distinctive nanomaterials to maximize the therapeutic potential against CRC.
4.4.4. Other cancer
Various other types of cancers have been treated with tumor specific M13 phages using phage display technology. To exploit the efficacy of tumor-targeting phages against melanoma, Eriksson et al. designed two genetically modified M13 phages for selective targeting of specific cancer cells [192]. One (WDC-2 phage) displaying specific cell-binding peptide identified through in vivo biopanning, another (HLA-A2 specific Fab-phage) expressing tumor specific antibody fragments on the phage. While both treatments resulted in complete tumor regression and prolonged survival of the mice, the tumor suppression was likely accomplished through engineered phage's pro-inflammatory process via activation of toll-like receptor 9 on innate immune cells that further induced adaptive immune responses [192].
Additionally, with the M13 phage, targeted photodynamic therapy (PDT) can be performed in epithelial growth factor receptor (EFGR)-overexpressing cancer cell to effectively destroy tumors by selectively accumulating photosensitizer (PS) in desired target [33,193]. As EFGR gene mutations are often linked to cancer progression, Ulfo et al. first demonstrated the use of EGFR peptide ligands (SYPIPDT) fused to M13 phages along with Rose Bengal (RB) as effective PS for promising EFGR targeted PDT [33]. The same EGFR-targeting peptide was coupled to chlorin e6 (Ce6), one of the most widely used PS molecules, for targeting ovarian cancer cells. Due to the selective targeting of mitochondria-located EFGR that modulated cell death pathway, the phage-based phototheranostic platform demonstrated enhanced efficacy in eliminating two ovarian cancer cell lines by promoting mitochondrial localization and autophagy induction [193]. Moreover, EGFR has also been investigated as a potential therapeutic target in other cancer types, including cervical cancer, bladder cancer, and head and neck cancer [194]. M13 phage-based bionanomaterials could thus be a promising novel approach to cancer therapy.
5. Current limitations and future outlooks
With all possibilities come limitations. Phage application including the use of phage display continues to raise ethical concerns and challenges half a century after its discovery. Despite being widely used in animal experiments, M13 phages as bionanomaterials have been limited in human trials due to the following reasons: 1) Restrictions in peptides displayed. In order for M13 phages to function as peptide display particles, they must first be expressed and assembled within a bacterial host, then secreted without disrupting the host cells, therefore limiting the types, lengths, and amounts of peptides displayed. In addition, the large aspect ratio of nanofiber-like phage inhibits the display of large peptides or antibodies to the pIII protein due to its small tail proportion. Having a limited range of peptides to display decreases the efficiency of phage-based applications such as biosensors. A number of studies have proposed ways to reduce the length of M13 phage as much as possible while retaining the minor coat protein [195,196]. In future studies, shorter M13 phages could be studied to compare their efficacy in therapeutic applications. 2) Issues with stability. Despite their structural robustness, M13 phages are still susceptible to losing activity under various physicochemical conditions. As various encapsulation approaches have shown to retain bioactivity and stability in other phages, future studies may explore such strategies for M13 phage [197]. 3) The safety concerns. While M13 phages do not cause cellular lysis, they are viruses that exhibit immunogenicity. The immunogenic properties are likely due to repeated structures of coat proteins which have shown to potentially stimulate humoral and cellular immune responses [198,199]. In patients at high risk, therefore, use of phages might be restricted due to their impaired immunity and the immune-inducing properties of the particles. Additionally, since M13 phages are constructed in bacteria, they may contain different levels of lipopolysaccharide (LPS), making it necessary to develop a more effective purification method for future studies [14]. 4) Lack of funding. As Henein et al. stated, research on phage-based therapeutic applications might be hindered by the lack of funding from large pharmaceutical companies and require massive funds for clinical trials [200]. Current clinical trials have yet to show breakthrough progress in efficacy and agencies including the FDA have yet to approve widespread use due to this fact [201]. We believe both public and clinical awareness regarding phage-based applications remain a target of improvement, especially in safety and basic knowledge regarding the phage concept. Nevertheless, the use of M13 phages as bionanomaterials has demonstrated impressive results that demonstrate their potential for use in a wide variety of clinical settings.
6. Conclusions
This overview demonstrates that phage application goes beyond merely bacteria elimination but also in various aspects of nanoscience. M13 phages, in particular, are naturally occurring nanomaterials with filamentous shapes and monodispersed properties that are versatile in applications. Through the advancement of phage display technology and material engineering, the M13 phage has been developed as a virus-based bionanomaterial that is useful in drug delivery, biodetection, tissue regeneration, and targeted cancer therapy. In order to bridge the gap between animal and human clinical trials, moreover, future relevant research should overcome limitations related to peptide display, stability, safety concerns, and funding shortages. Importantly, a thorough understanding of how the M13 phage will behave once it enters the human body through different pathways is essential. Overall, the use of M13 phage as bionanomaterial, based on phage display technology, will become increasingly valuable for precision and individualized medicine as ongoing biotechnology innovations allow for novel phage derived nanotherapeutic applications to be discovered.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the National Natural Science Foundation of China (Grant No. 32170141), China Medical Board (no. 20–365) and Shanghai Jiao Tong University Integrated Innovation Fund (no. 2020–01).
Contributor Information
Ru-Lin Huang, Email: huangrulin@sjtu.edu.cn.
Qingtian Li, Email: qingtianli@sjtu.edu.cn.
Yongzhang Zhu, Email: yzhzhu@sjtu.edu.cn, yzhzhu@hotmail.com.
Data availability
No data was used for the research described in the article.
References
- 1.Paczesny J., Bielec K. Application of bacteriophages in nanotechnology. Nanomaterials. 2020;10:1944. doi: 10.3390/nano10101944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hatfull G.F., Dedrick R.M., Schooley R.T. Phage therapy for antibiotic-resistant bacterial infections. Annu. Rev. Med. 2022;73:197–211. doi: 10.1146/annurev-med-080219-122208. [DOI] [PubMed] [Google Scholar]
- 3.Murray C.J., Ikuta K.S., Sharara F., et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Naylor N.R., Atun R., Zhu N., Kulasabanathan K., Silva S., Chatterjee A., Knight G.M., Robotham J.V. Estimating the burden of antimicrobial resistance: a systematic literature review. Antimicrob. Resist. Infect. Control. 2018;7:58. doi: 10.1186/s13756-018-0336-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrenko V.A., Gillespie J.W., De Plano L.M., Shokhen M.A. Phage-displayed mimotopes of SARS-CoV-2 spike protein targeted to authentic and alternative cellular receptors. Viruses. 2022;14:384. doi: 10.3390/v14020384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith G.P. Filamentous fusion phage: novel expression vectors that display cloned antigens on the virion surface. Science. 1985;228:1315–1317. doi: 10.1126/science.4001944. [DOI] [PubMed] [Google Scholar]
- 7.Jiang H., Li Y., Cosnier S., Yang M., Sun W., Mao C. Exploring phage engineering to advance nanobiotechnology. Mater. Today Nano. 2022;19 [Google Scholar]
- 8.Smith G.P., Petrenko V.A. Phage display. Chem. Rev. 1997;97:391–410. doi: 10.1021/cr960065d. [DOI] [PubMed] [Google Scholar]
- 9.Xu H., Cao B., Li Y., Mao C. Phage nanofibers in nanomedicine: biopanning for early diagnosis, targeted therapy, and proteomics analysis. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2020;12:e1623. doi: 10.1002/wnan.1623. [DOI] [PubMed] [Google Scholar]
- 10.Hertveldt K., Beliën T., Volckaert G. General M13 phage display: M13 phage display in identification and characterization of protein-protein interactions. Methods Mol. Biol. Clifton NJ. 2009;502:321–339. doi: 10.1007/978-1-60327-565-1_19. [DOI] [PubMed] [Google Scholar]
- 11.Ledsgaard L., Kilstrup M., Karatt-Vellatt A., McCafferty J., Laustsen A.H. Basics of antibody phage display technology. Toxins. 2018;10:236. doi: 10.3390/toxins10060236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shin Y.C., Lee J.H., Kim M.J., Park J.H., Kim S.E., Kim J.S., Oh J.-W., Han D.-W. Biomimetic hybrid nanofiber sheets composed of RGD peptide-decorated PLGA as cell-adhesive substrates. J. Funct. Biomater. 2015;6:367–378. doi: 10.3390/jfb6020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dion M.B., Oechslin F., Moineau S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020;18:125–138. doi: 10.1038/s41579-019-0311-5. [DOI] [PubMed] [Google Scholar]
- 14.Karimi M., Mirshekari H., MoosaviBasri M., Bahrami S., Moghoofei M., Hamblin M.R. Bacteriophages and phage-inspired nanocarriers for targeted delivery of therapeutic cargos. Adv. Drug Deliv. Rev. 2016;106:45–62. doi: 10.1016/j.addr.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee S.-W., Mao C., Flynn C.E., Belcher A.M. Ordering of quantum dots using genetically engineered viruses. Science. 2002;296:892–895. doi: 10.1126/science.1068054. [DOI] [PubMed] [Google Scholar]
- 16.Morag O., Abramov G., Goldbourt A. Similarities and differences within members of the ff family of filamentous bacteriophage viruses. J. Phys. Chem. B. 2011;115:15370–15379. doi: 10.1021/jp2079742. [DOI] [PubMed] [Google Scholar]
- 17.Kim H., Lee J.-H., Lee J.H., et al. M13 virus triboelectricity and energy harvesting. Nano Lett. 2021;21:6851–6858. doi: 10.1021/acs.nanolett.1c01881. [DOI] [PubMed] [Google Scholar]
- 18.Merzlyak A., Indrakanti S., Lee S.-W. Genetically engineered nanofiber-like viruses for tissue regenerating materials. Nano Lett. 2009;9:846–852. doi: 10.1021/nl8036728. [DOI] [PubMed] [Google Scholar]
- 19.Bartolacci C., Andreani C., Curcio C., et al. Phage-based anti-HER2 vaccination can circumvent immune tolerance against breast cancer. Cancer Immunol. Res. 2018;6:1486–1498. doi: 10.1158/2326-6066.CIR-18-0179. [DOI] [PubMed] [Google Scholar]
- 20.Jin H.-E., Lee S.-W. Engineering of M13 bacteriophage for development of tissue engineering materials. Methods Mol. Biol. Clifton NJ. 2018;1776:487–502. doi: 10.1007/978-1-4939-7808-3_32. [DOI] [PubMed] [Google Scholar]
- 21.Peng H., Chen I.A. Rapid colorimetric detection of bacterial species through the capture of gold nanoparticles by chimeric phages. ACS Nano. 2019;13:1244–1252. doi: 10.1021/acsnano.8b06395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Podlacha M., Grabowski Ł., Kosznik-Kawśnicka K., Zdrojewska K., Stasiłojć M., Węgrzyn G., Węgrzyn A. Interactions of bacteriophages with animal and human organisms—safety issues in the light of phage therapy. Int. J. Mol. Sci. 2021;22:8937. doi: 10.3390/ijms22168937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cao B., Yang M., Mao C. Phage as a genetically modifiable supramacromolecule in chemistry, materials and medicine. Acc. Chem. Res. 2016;49:1111–1120. doi: 10.1021/acs.accounts.5b00557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sunderland K.S., Yang M., Mao C. Phage-enabled nanomedicine: from probes to therapeutics in precision medicine. Angew. Chem., Int. Ed. Engl. 2017;56:1964–1992. doi: 10.1002/anie.201606181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue H., Li Y., Yang M., Mao C. T7 phage as an emerging nanobiomaterial with genetically tunable target specificity. Adv. Sci. 2021;9 doi: 10.1002/advs.202103645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Berkowitz S.A., Day L.A. Mass, length, composition and structure of the filamentous bacterial virus fd. J. Mol. Biol. 1976;102:531–547. doi: 10.1016/0022-2836(76)90332-6. [DOI] [PubMed] [Google Scholar]
- 27.Park S.M., Kim W.-G., Kim J., Choi E.-J., Kim H., Oh J.-W., Yoon D.K. Fabrication of chiral M13 bacteriophage film by evaporation-induced self-assembly. Small Weinh. Bergstr. Ger. 2021;17 doi: 10.1002/smll.202008097. [DOI] [PubMed] [Google Scholar]
- 28.Lee J.H., Fan B., Samdin T.D. Phage-based structural color sensors and their pattern recognition sensing system. ACS Nano. 2017;11:3632–3641. doi: 10.1021/acsnano.6b07942. [DOI] [PubMed] [Google Scholar]
- 29.van Wezenbeek P.M.G.F., Hulsebos T.J.M., Schoenmakers J.G.G. Nucleotide sequence of the filamentous bacteriophage M13 DNA genome: comparison with phage fd. Gene. 1980;11:129–148. doi: 10.1016/0378-1119(80)90093-1. [DOI] [PubMed] [Google Scholar]
- 30.Kehoe J.W., Kay B.K. Filamentous phage display in the new millennium. Chem. Rev. 2005;105:4056–4072. doi: 10.1021/cr000261r. [DOI] [PubMed] [Google Scholar]
- 31.Stassen A.P., Folmer R.H., Hilbers C.W., Konings R.N. Single-stranded DNA binding protein encoded by the filamentous bacteriophage M13: structural and functional characteristics. Mol. Biol. Rep. 1994;20:109–127. doi: 10.1007/BF00990543. [DOI] [PubMed] [Google Scholar]
- 32.Straus S.K., Bo H.E. Filamentous bacteriophage proteins and assembly. Subcell. Biochem. 2018;88:261–279. doi: 10.1007/978-981-10-8456-0_12. [DOI] [PubMed] [Google Scholar]
- 33.Ulfo L., Cantelli A., Petrosino A. Orthogonal nanoarchitectonics of M13 phage for receptor targeted anticancer photodynamic therapy. Nanoscale. 2022;14:632–641. doi: 10.1039/d1nr06053h. [DOI] [PubMed] [Google Scholar]
- 34.Plano L.M.D., Franco D., Rizzo M.G., Zammuto V., Gugliandolo C., Silipigni L., Torrisi L., Guglielmino S.P.P. Role of phage capsid in the resistance to UV-C radiations. Int. J. Mol. Sci. 2021;22:3408. doi: 10.3390/ijms22073408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hay I.D., Lithgow T. Filamentous phages: masters of a microbial sharing economy. EMBO Rep. 2019;20 doi: 10.15252/embr.201847427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Murray P., Rosenthal K., Pfaller M. Medical Microbiology. Elsevier/Saunders; 2013. Bacterial metabolism and genetics. [Google Scholar]
- 37.Reddy P., McKenney K. Improved method for the production of M13 phage and single-stranded DNA for DNA sequencing. Biotechniques. 1996;20:854–856. doi: 10.2144/96205st05. 858, 860. [DOI] [PubMed] [Google Scholar]
- 38.Rakonjac J., Feng J. n, Model P. Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J. Mol. Biol. 1999;289:1253–1265. doi: 10.1006/jmbi.1999.2851. [DOI] [PubMed] [Google Scholar]
- 39.Loh B., Kuhn A., Leptihn S. The fascinating biology behind phage display: filamentous phage assembly. Mol. Microbiol. 2019;111:1132–1138. doi: 10.1111/mmi.14187. [DOI] [PubMed] [Google Scholar]
- 40.Merril C.R., Scholl D., Adhya S.L. The prospect for bacteriophage therapy in Western medicine. Nat. Rev. Drug Discov. 2003;2:489–497. doi: 10.1038/nrd1111. [DOI] [PubMed] [Google Scholar]
- 41.Shrestha K.R., Yoo S.Y. Phage-based artificial niche: the recent progress and future opportunities in stem cell therapy. Stem Cell. Int. 2019;2019 doi: 10.1155/2019/4038560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh D., Lee Y., Thomas S., Kohli A.G., Yun D.S., Belcher A.M., Kelly K.A. M13-templated magnetic nanoparticles for targeted in vivo imaging of prostate cancer. Nat. Nanotechnol. 2012;7:677–682. doi: 10.1038/nnano.2012.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Georgieva Y., Konthur Z. Design and screening of M13 phage display cDNA libraries. Mol. Basel Switz. 2011;16:1667–1681. doi: 10.3390/molecules16021667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sloth A.B., Bakhshinejad B., Jensen M. Analysis of compositional bias in a commercial phage display peptide library by next-generation sequencing. Viruses. 2022;14:2402. doi: 10.3390/v14112402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen K.T.H., Adamkiewicz M.A., Hebert L.E., et al. Identification and characterization of mutant clones with enhanced propagation rates from phage-displayed peptide libraries. Anal. Biochem. 2014;462:35–43. doi: 10.1016/j.ab.2014.06.007. [DOI] [PubMed] [Google Scholar]
- 46.Zygiel E.M., Noren K.A., Adamkiewicz M.A., et al. Various mutations compensate for a deleterious lacZα insert in the replication enhancer of M13 bacteriophage. PLoS One. 2017;12 doi: 10.1371/journal.pone.0176421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Levisson M., Spruijt R.B., Winkel I.N., Kengen S.W.M., van der Oost J. Phage display of engineered binding proteins. Methods Mol. Biol. Clifton NJ. 2014;1129:211–229. doi: 10.1007/978-1-62703-977-2_19. [DOI] [PubMed] [Google Scholar]
- 48.Roscow O., Zhang W. Using phage display to develop ubiquitin variant modulators for E3 ligases. J. Vis. Exp. 2021 doi: 10.3791/62950. Epub ahead of print 27 August. [DOI] [PubMed] [Google Scholar]
- 49.Vetter S.W. Phage display selection of peptides that target calcium-binding proteins. Methods Mol. Biol. Clifton NJ. 2013;963:215–235. doi: 10.1007/978-1-62703-230-8_14. [DOI] [PubMed] [Google Scholar]
- 50.Rakonjac J., Bennett N.J., Spagnuolo J., Gagic D., Russel M. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011;13:51–76. [PubMed] [Google Scholar]
- 51.Zahid M., Phillips B.E., Albers S.M., Giannoukakis N., Watkins S.C., Robbins P.D. Identification of a cardiac specific protein transduction domain by in vivo biopanning using a M13 phage peptide display library in mice. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee K.J., Lee J.H., Chung H.K., Ju E.J., Song S.Y., Jeong S.-Y., Choi E.K. Application of peptide displaying phage as a novel diagnostic probe for human lung adenocarcinoma. Amino Acids. 2016;48:1079–1086. doi: 10.1007/s00726-015-2153-4. [DOI] [PubMed] [Google Scholar]
- 53.Lee K.J., Lee J.H., Chung H.K., et al. Novel peptides functionally targeting in vivo human lung cancer discovered by in vivo peptide displayed phage screening. Amino Acids. 2015;47:281–289. doi: 10.1007/s00726-014-1852-6. [DOI] [PubMed] [Google Scholar]
- 54.Pleiko K., Põšnograjeva K., Haugas M. In vivo phage display: identification of organ-specific peptides using deep sequencing and differential profiling across tissues. Nucleic Acids Res. 2021;49:e38. doi: 10.1093/nar/gkaa1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shin Y.C., Lee J.H., Jin L., Kim M.J., Oh J.-W., Kim T.W., Han D.-W. Cell-adhesive RGD peptide-displaying M13 bacteriophage/PLGA nanofiber matrices for growth of fibroblasts. Biomater. Res. 2014;18:14. doi: 10.1186/2055-7124-18-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dos Santos D.M., Correa D.S., Medeiros E.S., Oliveira J.E., Mattoso L.H.C. Advances in functional polymer nanofibers: from spinning fabrication techniques to recent biomedical applications. ACS Appl. Mater. Interfaces. 2020;12:45673–45701. doi: 10.1021/acsami.0c12410. [DOI] [PubMed] [Google Scholar]
- 57.Xie J., MacEwan M.R., Schwartz A.G., Xia Y. Electrospun nanofibers for neural tissue engineering. Nanoscale. 2010;2:35–44. doi: 10.1039/b9nr00243j. [DOI] [PubMed] [Google Scholar]
- 58.Arida I.A., Ali I.H., Nasr M., El-Sherbiny I.M. Electrospun polymer-based nanofiber scaffolds for skin regeneration. J. Drug Deliv. Sci. Technol. 2021;64 [Google Scholar]
- 59.Shin Y.C., Kim C., Song S.J. Ternary aligned nanofibers of RGD peptide-displaying M13 bacteriophage/PLGA/graphene oxide for facilitated myogenesis. Nanotheranostics. 2018;2:144–156. doi: 10.7150/ntno.22433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ling H., Lou X., Luo Q., He Z., Sun M., Sun J. Recent advances in bacteriophage-based therapeutics: insight into the post-antibiotic era. Acta Pharm. Sin. B. 2022 doi: 10.1016/j.apsb.2022.05.007. Epub ahead of print 13 May. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miguel S.P., Figueira D.R., Simões D., Ribeiro M.P., Coutinho P., Ferreira P., Correia I.J. Electrospun polymeric nanofibres as wound dressings: a review. Colloids Surf. B Biointerfaces. 2018;169:60–71. doi: 10.1016/j.colsurfb.2018.05.011. [DOI] [PubMed] [Google Scholar]
- 62.Wang J., Planz V., Vukosavljevic B., Windbergs M. Multifunctional electrospun nanofibers for wound application – novel insights into the control of drug release and antimicrobial activity. Eur. J. Pharm. Biopharm. 2018;129:175–183. doi: 10.1016/j.ejpb.2018.05.035. [DOI] [PubMed] [Google Scholar]
- 63.Nazarnezhad S., Baino F., Kim H.-W., Webster T.J., Kargozar S. Electrospun nanofibers for improved angiogenesis: promises for tissue engineering applications. Nanomater. Basel Switz. 2020;10:E1609. doi: 10.3390/nano10081609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kaur S., Kumari A., Kumari Negi A., Galav V., Thakur S., Agrawal M., Sharma V. Nanotechnology based approaches in phage therapy: overcoming the pharmacological barriers. Front. Pharmacol. 2021;12 doi: 10.3389/fphar.2021.699054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen S.Y., Harrison M., Ng E.K., Sauvageau D., Elias A. Immobilized reporter phage on electrospun polymer fibers for improved capture and detection of Escherichia coli O157:H7. ACS Food Sci. Technol. 2021;1:1085–1094. [Google Scholar]
- 66.Sugimoto R., Lee J.H., Lee J.-H., Jin H.-E., Yoo S.Y., Lee S.-W. Bacteriophage nanofiber fabrication using near field electrospinning. RSC Adv. 2019;9:39111–39118. doi: 10.1039/c9ra07510k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lee J.H., Warner C.M., Jin H.-E., Barnes E., Poda A.R., Perkins E.J., Lee S.-W. Production of tunable nanomaterials using hierarchically assembled bacteriophages. Nat. Protoc. 2017;12:1999–2013. doi: 10.1038/nprot.2017.085. [DOI] [PubMed] [Google Scholar]
- 68.Yang K., Yu S.J., Lee J.S., et al. Electroconductive nanoscale topography for enhanced neuronal differentiation and electrophysiological maturation of human neural stem cells. Nanoscale. 2017;9:18737–18752. doi: 10.1039/c7nr05446g. [DOI] [PubMed] [Google Scholar]
- 69.Zhou N., Li Y., Loveland C.H., Wilson M.J., Cao B., Qiu P., Yang M., Mao C. Hierarchical ordered assembly of genetically modifiable viruses into nanoridge-in-microridge structures. Adv. Mater. Deerfield Beach Fla. 2019;31 doi: 10.1002/adma.201905577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dalby M.J., Gadegaard N., Oreffo R.O.C. Harnessing nanotopography and integrin-matrix interactions to influence stem cell fate. Nat. Mater. 2014;13:558–569. doi: 10.1038/nmat3980. [DOI] [PubMed] [Google Scholar]
- 71.Nisbet D.R., Yu L.M.Y., Zahir T., Forsythe J.S., Shoichet M.S. Characterization of neural stem cells on electrospun poly(epsilon-caprolactone) submicron scaffolds: evaluating their potential in neural tissue engineering. J. Biomater. Sci. Polym. Ed. 2008;19:623–634. doi: 10.1163/156856208784089652. [DOI] [PubMed] [Google Scholar]
- 72.Rasti Boroojeni F., Mashayekhan S., Abbaszadeh H.-A., Ansarizadeh M., Khoramgah M.-S., Rahimi Movaghar V. Bioinspired nanofiber scaffold for differentiating bone marrow-derived neural stem cells to oligodendrocyte-like cells: design, fabrication, and characterization. Int. J. Nanomed. 2020;15:3903–3920. doi: 10.2147/IJN.S248509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang T.-Y., Forsythe J.S., Nisbet D.R., Parish C.L. Promoting engraftment of transplanted neural stem cells/progenitors using biofunctionalised electrospun scaffolds. Biomaterials. 2012;33:9188–9197. doi: 10.1016/j.biomaterials.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 74.Tanaka M., Sawada T., Li X., Serizawa T. Controlled assembly of filamentous viruses into hierarchical nano- to microstructures at liquid/liquid interfaces. RSC Adv. 2020;10:26313–26318. doi: 10.1039/d0ra04529b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Passaretti P., Sun Y., Dafforn T.R., Oppenheimer P.G. Determination and characterisation of the surface charge properties of the bacteriophage M13 to assist bio-nanoengineering. RSC Adv. 2020;10:25385–25392. doi: 10.1039/d0ra04086j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bardhan N.M., Ghosh D., Belcher A.M. Carbon nanotubes as in vivo bacterial probes. Nat. Commun. 2014;5:4918. doi: 10.1038/ncomms5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghosh D., Bagley A.F., Na Y.J., Birrer M.J., Bhatia S.N., Belcher A.M. Deep, noninvasive imaging and surgical guidance of submillimeter tumors using targeted M13-stabilized single-walled carbon nanotubes. Proc. Natl. Acad. Sci. U. S. A. 2014;111:13948–13953. doi: 10.1073/pnas.1400821111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yi H., Ghosh D., Ham M.-H., Qi J., Barone P.W., Strano M.S., Belcher A.M. M13 phage-functionalized single-walled carbon nanotubes as nanoprobes for second near-infrared window fluorescence imaging of targeted tumors. Nano Lett. 2012;12:1176–1183. doi: 10.1021/nl2031663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Meinardi F., McDaniel H., Carulli F., Colombo A., Velizhanin K.A., Makarov N.S., Simonutti R., Klimov V.I., Brovelli S. Highly efficient large-area colourless luminescent solar concentrators using heavy-metal-free colloidal quantum dots. Nat. Nanotechnol. 2015;10:878–885. doi: 10.1038/nnano.2015.178. [DOI] [PubMed] [Google Scholar]
- 80.Huang S., Qi J., deQuilettes D.W., Huang M., Lin C.-W., Bardhan N.M., Dang X., Bulović V., Belcher A.M. M13 virus-based framework for high fluorescence enhancement. Small Weinh. Bergstr. Ger. 2019;15 doi: 10.1002/smll.201901233. [DOI] [PubMed] [Google Scholar]
- 81.Dong X., Pan P., Zheng D.-W., Bao P., Zeng X., Zhang X.-Z. Bioinorganic hybrid bacteriophage for modulation of intestinal microbiota to remodel tumor-immune microenvironment against colorectal cancer. Sci. Adv. 2020;6 doi: 10.1126/sciadv.aba1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sedki M., Chen X., Chen C., Ge X., Mulchandani A. Non-lytic M13 phage-based highly sensitive impedimetric cytosensor for detection of coliforms. Biosens. Bioelectron. 2020;148 doi: 10.1016/j.bios.2019.111794. [DOI] [PubMed] [Google Scholar]
- 83.Wang X.-Y., Yang J.-Y., Wang Y.-T., Zhang H.-C., Chen M.-L., Yang T., Wang J.-H. M13 phage-based nanoprobe for SERS detection and inactivation of Staphylococcus aureus. Talanta. 2021;221 doi: 10.1016/j.talanta.2020.121668. [DOI] [PubMed] [Google Scholar]
- 84.Lentini G., Fazio E., Calabrese F., et al. Phage-AgNPs complex as SERS probe for U937 cell identification. Biosens. Bioelectron. 2015;74:398–405. doi: 10.1016/j.bios.2015.05.073. [DOI] [PubMed] [Google Scholar]
- 85.Tang S., Zheng J. Antibacterial activity of silver nanoparticles: structural effects. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201701503. [DOI] [PubMed] [Google Scholar]
- 86.Wilson R.E., O'Connor R., Gallops C.E. Immunomagnetic capture and multiplexed surface marker detection of circulating tumor cells with magnetic multicolor surface-enhanced Raman scattering nanotags. ACS Appl. Mater. Interfaces. 2020;12:47220–47232. doi: 10.1021/acsami.0c12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tian X., Sha X., Feng Y. A magnetic dynamic microbiointerface with biofeedback mechanism for cancer cell capture and release. ACS Appl. Mater. Interfaces. 2019;11:41019–41029. doi: 10.1021/acsami.9b13140. [DOI] [PubMed] [Google Scholar]
- 88.Deng Z., Wu S., Wang Y., Shi D. Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine. 2022;83 doi: 10.1016/j.ebiom.2022.104237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sm J., Jj L., W H., Hs K. Nanotentacle-structured magnetic particles for efficient capture of circulating tumor cells. Small Weinh. Bergstr. Ger. 2015;11 doi: 10.1002/smll.201402619. Epub ahead of print 24 April. [DOI] [PubMed] [Google Scholar]
- 90.Jain K.K. An overview of drug delivery systems. Methods Mol. Biol. Clifton NJ. 2020;2059:1–54. doi: 10.1007/978-1-4939-9798-5_1. [DOI] [PubMed] [Google Scholar]
- 91.Pinto A.M., Silva M.D., Pastrana L.M., Bañobre-López M., Sillankorva S. The clinical path to deliver encapsulated phages and lysins. FEMS Microbiol. Rev. 2021;45 doi: 10.1093/femsre/fuab019. fuab019. [DOI] [PubMed] [Google Scholar]
- 92.Hammond S.M., Aartsma-Rus A., Alves S. Delivery of oligonucleotide-based therapeutics: challenges and opportunities. EMBO Mol. Med. 2021;13 doi: 10.15252/emmm.202013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang J., Ni Q., Wang Y., Zhang Y., He H., Gao D., Ma X., Liang X.-J. Nanoscale drug delivery systems for controllable drug behaviors by multi-stage barrier penetration. J. Control. Release Off. J. Control. Release Soc. 2021;331:282–295. doi: 10.1016/j.jconrel.2020.08.045. [DOI] [PubMed] [Google Scholar]
- 94.Ngweniform P., Abbineni G., Cao B., Mao C. Self-assembly of drug-loaded liposomes on genetically engineered target-recognizing M13 phage: a novel nanocarrier for targeted drug delivery. Small Weinh. Bergstr. Ger. 2009;5:1963–1969. doi: 10.1002/smll.200801902. [DOI] [PubMed] [Google Scholar]
- 95.Wang H.Y., Chang Y.-C., Hu C.-W. Development of a novel cytokine vehicle using filamentous phage display for colorectal cancer treatment. ACS Synth. Biol. 2021;10:2087–2095. doi: 10.1021/acssynbio.1c00266. [DOI] [PubMed] [Google Scholar]
- 96.Ghosh D., Kohli A.G., Moser F., Endy D., Belcher A.M. Refactored M13 bacteriophage as a platform for tumor cell imaging and drug delivery. ACS Synth. Biol. 2012;1:576–582. doi: 10.1021/sb300052u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Jin H.-E., Farr R., Lee S.-W. Collagen mimetic peptide engineered M13 bacteriophage for collagen targeting and imaging in cancer. Biomaterials. 2014;35:9236–9245. doi: 10.1016/j.biomaterials.2014.07.044. [DOI] [PubMed] [Google Scholar]
- 98.Hansson O. Biomarkers for neurodegenerative diseases. Nat. Med. 2021;27:954–963. doi: 10.1038/s41591-021-01382-x. [DOI] [PubMed] [Google Scholar]
- 99.Sweeney M.D., Sagare A.P., Zlokovic B.V. Blood-brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018;14:133–150. doi: 10.1038/nrneurol.2017.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Dugger B.N., Dickson D.W. Pathology of neurodegenerative diseases. Cold Spring Harbor Perspect. Biol. 2017;9:a028035. doi: 10.1101/cshperspect.a028035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Solomon B. Filamentous bacteriophage as a novel therapeutic tool for Alzheimer's disease treatment. J Alzheimers Dis. 2008;15:193–198. doi: 10.3233/jad-2008-15205. [DOI] [PubMed] [Google Scholar]
- 102.De Plano L.M., Carnazza S., Franco D. Innovative IgG biomarkers based on phage display microbial amyloid mimotope for state and stage diagnosis in Alzheimer's disease. ACS Chem. Neurosci. 2020;11:1013–1026. doi: 10.1021/acschemneuro.9b00549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Meade R.M., Fairlie D.P., Mason J.M. Alpha-synuclein structure and Parkinson's disease – lessons and emerging principles. Mol. Neurodegener. 2019;14:29. doi: 10.1186/s13024-019-0329-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dimant H., Sharon N., Solomon B. Modulation effect of filamentous phage on alpha-synuclein aggregation. Biochem. Biophys. Res. Commun. 2009;383:491–496. doi: 10.1016/j.bbrc.2009.04.048. [DOI] [PubMed] [Google Scholar]
- 105.Antimicrobial Resistance Collaborators Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet Lond. Engl. 2022;399:629–655. doi: 10.1016/S0140-6736(21)02724-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Drug-Resistant Infections. World Bank; 2017. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 107.Jit M., Ng D.H.L., Luangasanatip N., Sandmann F., Atkins K.E., Robotham J.V., Pouwels K.B. Quantifying the economic cost of antibiotic resistance and the impact of related interventions: rapid methodological review, conceptual framework and recommendations for future studies. BMC Med. 2020;18:38. doi: 10.1186/s12916-020-1507-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jin P., Wang L., Sha R. A blood circulation-prolonging peptide anchored biomimetic phage-platelet hybrid nanoparticle system for prolonged blood circulation and optimized anti-bacterial performance. Theranostics. 2021;11:2278–2296. doi: 10.7150/thno.49781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Buckel W.R., Veillette J.J., Vento T.J., Stenehjem E. Antimicrobial stewardship in community hospitals. Med. Clin. 2018;102:913–928. doi: 10.1016/j.mcna.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 110.Hillman Y., Gershberg J., Lustiger D., et al. Monoclonal antibody-based biosensor for point-of-care detection of type III secretion system expressing pathogens. Anal. Chem. 2021;93:928–935. doi: 10.1021/acs.analchem.0c03621. 2021. [DOI] [PubMed] [Google Scholar]
- 111.Zhao X., Hilliard L.R., Mechery S.J., Wang Y., Bagwe R.P., Jin S., Tan W. A rapid bioassay for single bacterial cell quantitation using bioconjugated nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2004;101:15027–15032. doi: 10.1073/pnas.0404806101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Huang Q., Wu W., Ai K., Liu J. Highly sensitive polydiacetylene ensembles for biosensing and bioimaging. Front. Chem. 2020;8 doi: 10.3389/fchem.2020.565782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ashoori S., Naderpour M., Ghezelayagh M.M., Zadeh R.M., Raissi F. Ultrasensitive bio-detection using single-electron effect. Talanta. 2021;224 doi: 10.1016/j.talanta.2020.121769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Pardehkhorram R., Alshawawreh F., Gonçales V.R., Lee N.A., Tilley R.D., Gooding J.J. Functionalized gold nanorod probes: a sophisticated design of sers immunoassay for biodetection in complex media. Anal. Chem. 2021;93:12954–12965. doi: 10.1021/acs.analchem.1c02557. [DOI] [PubMed] [Google Scholar]
- 115.Peng H., Borg R.E., Nguyen A.B.N., Chen I.A. Chimeric phage nanoparticles for rapid characterization of bacterial pathogens: detection in complex biological samples and determination of antibiotic sensitivity. ACS Sens. 2020;5:1491–1499. doi: 10.1021/acssensors.0c00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rentschler S., Kaiser L., Deigner H.-P. Emerging options for the diagnosis of bacterial infections and the characterization of antimicrobial resistance. Int. J. Mol. Sci. 2021;22:E456. doi: 10.3390/ijms22010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Behera B., Anil Vishnu G.K., Chatterjee S., Sitaramgupta V, V. S. N. Sreekumar N., Nagabhushan A., Rajendran N., Prathik B.H., Pandya H.J. Emerging technologies for antibiotic susceptibility testing. Biosens. Bioelectron. 2019;142 doi: 10.1016/j.bios.2019.111552. [DOI] [PubMed] [Google Scholar]
- 118.Przystal J.M., Waramit S., Pranjol M.Z.I., et al. Efficacy of systemic temozolomide-activated phage-targeted gene therapy in human glioblastoma. EMBO Mol. Med. 2019;11 doi: 10.15252/emmm.201708492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hafsi M., Preveral S., Hoog C. RGD-functionalized magnetosomes are efficient tumor radioenhancers for X-rays and protons. Nanomed. Nanotechnol. Biol. Med. 2020;23 doi: 10.1016/j.nano.2019.102084. [DOI] [PubMed] [Google Scholar]
- 120.Hou J., Shen J., Zhao N., Yang C.-T., Thierry B., Zhou X., Zhu J., Mao C. Detection of a single circulating tumor cell using a genetically engineered antibody-like phage nanofiber probe. Mater. Today Adv. 2021;12 [Google Scholar]
- 121.Lee J.H., Domaille D.W., Cha J.N. Amplified protein detection and identification through DNA-conjugated M13 bacteriophage. ACS Nano. 2012;6:5621–5626. doi: 10.1021/nn301565e. [DOI] [PubMed] [Google Scholar]
- 122.Ceppi L., Bardhan N.M., Na Y., Siegel A., Rajan N., Fruscio R., Del Carmen M.G., Belcher A.M., Birrer M.J. Real-time single-walled carbon nanotube-based fluorescence imaging improves survival after debulking surgery in an ovarian cancer model. ACS Nano. 2019;13:5356–5365. doi: 10.1021/acsnano.8b09829. [DOI] [PubMed] [Google Scholar]
- 123.Ghanemi A., Yoshioka M., St-Amand J. Secreted protein acidic and rich in cysteine as a molecular physiological and pathological biomarker. Biomolecules. 2021;11:1689. doi: 10.3390/biom11111689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bao J.-M., Dang Q., Lin C.-J. SPARC is a key mediator of TGF-β-induced renal cancer metastasis. J. Cell. Physiol. 2021;236:1926–1938. doi: 10.1002/jcp.29975. [DOI] [PubMed] [Google Scholar]
- 125.Bhasin A., Drago N.P., Majumdar S., Sanders E.C., Weiss G.A., Penner R.M. Viruses masquerading as antibodies in biosensors: the development of the virus BioResistor. Acc. Chem. Res. 2020;53:2384–2394. doi: 10.1021/acs.accounts.0c00474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bhasin A., Ogata A.F., Briggs J.S., Tam P.Y., Tan M.X., Weiss G.A., Penner R.M. The virus bioresistor: wiring virus particles for the direct, label-free detection of target proteins. Nano Lett. 2018;18:3623–3629. doi: 10.1021/acs.nanolett.8b00723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Cho C.H., Kim J.H., Song D.-K., Park T.J., Park J.P. An affinity peptide-incorporated electrochemical biosensor for the detection of neutrophil gelatinase-associated lipocalin. Biosens. Bioelectron. 2019;142 doi: 10.1016/j.bios.2019.111482. [DOI] [PubMed] [Google Scholar]
- 128.Eming S.A., Wynn T.A., Martin P. Inflammation and metabolism in tissue repair and regeneration. Science. 2017;356:1026–1030. doi: 10.1126/science.aam7928. [DOI] [PubMed] [Google Scholar]
- 129.Lee J.-H., Parthiban P., Jin G.-Z. Materials roles for promoting angiogenesis in tissue regeneration. Prog. Mater. Sci. 2021;117 [Google Scholar]
- 130.Shafiee A., Atala A. Tissue engineering: toward a new era of medicine. Annu. Rev. Med. 2017;68:29–40. doi: 10.1146/annurev-med-102715-092331. [DOI] [PubMed] [Google Scholar]
- 131.Ma Z., Mao C., Jia Y. Extracellular matrix dynamics in vascular remodeling. Am. J. Physiol. Cell Physiol. 2020;319:C481–C499. doi: 10.1152/ajpcell.00147.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Blanco-Fernandez B., Castaño O., Mateos-Timoneda M.Á., Engel E., Pérez-Amodio S. Nanotechnology approaches in chronic wound healing. Adv. Wound Care. 2021;10:234–256. doi: 10.1089/wound.2019.1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Fleming J.W., Capel A.J., Rimington R.P., Player D.J., Stolzing A., Lewis M.P. Functional regeneration of tissue engineered skeletal muscle in vitro is dependent on the inclusion of basement membrane proteins. Cytoskelet. Hoboken NJ. 2019;76:371–382. doi: 10.1002/cm.21553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Cao B., Li Y., Yang T., Bao Q., Yang M., Mao C. Bacteriophage-based biomaterials for tissue regeneration. Adv. Drug Deliv. Rev. 2019;145:73–95. doi: 10.1016/j.addr.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jj B. A bacteriophages journey through the human body. Immunol. Rev. 2017;279 doi: 10.1111/imr.12565. Epub ahead of print September. [DOI] [PubMed] [Google Scholar]
- 136.Wang J., Yang M., Zhu Y., Wang L., Tomsia A.P., Mao C. Phage nanofibers induce vascularized osteogenesis in 3D printed bone scaffolds. Adv. Mater. Deerfield Beach Fla. 2014;26:4961–4966. doi: 10.1002/adma.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Safari Z., Soudi S., Jafarzadeh N. Promotion of angiogenesis by M13 phage and RGD peptide in vitro and in vivo. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47413-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lee D.-Y., Lee H., Kim Y. Phage as versatile nanoink for printing 3-D cell-laden scaffolds. Acta Biomater. 2016;29:112–124. doi: 10.1016/j.actbio.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 139.Liu X., Yang M., Lei F., Wang Y., Yang M., Mao C. Highly effective stroke therapy enabled by genetically engineered viral nanofibers. Adv. Mater. Deerfield Beach Fla. 2022;34 doi: 10.1002/adma.202201210. [DOI] [PubMed] [Google Scholar]
- 140.Nussbaum S.R., Carter M.J., Fife C.E., DaVanzo J., Haught R., Nusgart M., Cartwright D. An economic evaluation of the impact, cost, and medicare policy implications of chronic nonhealing wounds. Value Health J. Int. Soc. Pharmacoeconomics Outcomes Res. 2018;21:27–32. doi: 10.1016/j.jval.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 141.Pinto A.M., Cerqueira M.A., Bañobre-Lópes M., Pastrana L.M., Sillankorva S. Bacteriophages for chronic wound treatment: from traditional to novel delivery systems. Viruses. 2020;12:E235. doi: 10.3390/v12020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Girija A.R., Balasubramanian S., Cowin A.J. Nanomaterials-based drug delivery approaches for wound healing. Curr. Pharmaceut. Des. 2022;28:711–726. doi: 10.2174/1381612828666220328121211. [DOI] [PubMed] [Google Scholar]
- 143.Han C., Barakat M., DiPietro L.A. Angiogenesis in wound repair: too much of a good thing? Cold Spring Harbor Perspect. Biol. 2022:a041225. doi: 10.1101/cshperspect.a041225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Yoo S.Y., Shrestha K.R., Jeong S.-N. Engineered phage nanofibers induce angiogenesis. Nanoscale. 2017;9:17109–17117. doi: 10.1039/c7nr03332j. [DOI] [PubMed] [Google Scholar]
- 145.Farooq M., Khan A.W., Kim M.S., Choi S. The role of fibroblast growth factor (FGF) signaling in tissue repair and regeneration. Cells. 2021;10:3242. doi: 10.3390/cells10113242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Zhao Y., Wang Q., Jin Y., et al. Discovery and characterization of a high-affinity small peptide ligand, H1, targeting FGFR2IIIc for skin wound healing. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018;49:1033–1048. doi: 10.1159/000493287. [DOI] [PubMed] [Google Scholar]
- 147.Liu Y., Liu Y., Deng J. Fibroblast growth factor in diabetic foot ulcer: progress and therapeutic prospects. Front. Endocrinol. 2021;12 doi: 10.3389/fendo.2021.744868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhang J., Ge J., Xu Y., et al. Bioactive multi-engineered hydrogel offers simultaneous promise against antibiotic resistance and wound damage. Int. J. Biol. Macromol. 2020;164:4466–4474. doi: 10.1016/j.ijbiomac.2020.08.247. [DOI] [PubMed] [Google Scholar]
- 149.Gupta P., Singh H.S., Shukla V.K., Nath G., Bhartiya S.K. Bacteriophage therapy of chronic nonhealing wound: clinical study. Int. J. Low. Extrem. Wounds. 2019;18:171–175. doi: 10.1177/1534734619835115. [DOI] [PubMed] [Google Scholar]
- 150.Peng H., Rossetto D., Mansy S.S., Jordan M.C., Roos K.P., Chen I.A. Treatment of wound infections in a mouse model using Zn2+-releasing phage bound to gold nanorods. ACS Nano. 2022;16:4756–4774. doi: 10.1021/acsnano.2c00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lin X., Patil S., Gao Y.-G., Qian A. The bone extracellular matrix in bone formation and regeneration. Front. Pharmacol. 2020:11. doi: 10.3389/fphar.2020.00757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alcorta-Sevillano N., Macías I., Infante A., Rodríguez C.I. Deciphering the relevance of bone ECM signaling. Cells. 2020;9:E2630. doi: 10.3390/cells9122630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tuckermann J., Adams R.H. Crosstalk between endothelial cells and bone in development, homeostasis and inflammatory disease. Nat. Rev. Rheumatol. 2021;17:608–620. doi: 10.1038/s41584-021-00682-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yoo S.Y., Kobayashi M., Lee P.P., Lee S.-W. Early osteogenic differentiation of mouse preosteoblasts induced by collagen-derived DGEA-peptide on nanofibrous phage tissue matrices. Biomacromolecules. 2011;12:987–996. doi: 10.1021/bm1013475. [DOI] [PubMed] [Google Scholar]
- 155.Lee H.-S., Kang J.-I., Chung W.-J., Lee D.H., Lee B.Y., Lee S.-W., Yoo S.Y. Engineered phage matrix stiffness-modulating osteogenic differentiation. ACS Appl. Mater. Interfaces. 2018;10:4349–4358. doi: 10.1021/acsami.7b17871. [DOI] [PubMed] [Google Scholar]
- 156.Chung W.-J., Kwon K.-Y., Song J., Lee S.-W. Evolutionary screening of collagen-like peptides that nucleate hydroxyapatite crystals. Langmuir. 2011;27:7620–7628. doi: 10.1021/la104757g. [DOI] [PubMed] [Google Scholar]
- 157.Li Y., Cao B., Modali S., Lee E.M.Y., Xu H., Petrenko V., Gray J.J., Yang M., Mao C. Understanding the interactions between bone mineral crystals and their binding peptides derived from filamentous phage. Mater. Today Adv. 2022;15 [Google Scholar]
- 158.He T., Abbineni G., Cao B., Mao C. Nanofibrous bio-inorganic hybrid structures formed through self-assembly and oriented mineralization of genetically engineered phage nanofibers. Small. 2010;6:2230–2235. doi: 10.1002/smll.201001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wang F., Cao B., Mao C. Bacteriophage bundles with pre-aligned Ca initiate the oriented nucleation and growth of hydroxylapatite. Chem. Mater. Publ. Am. Chem. Soc. 2010;22:3630–3636. doi: 10.1021/cm902727s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Filipowska J., Tomaszewski K.A., Niedźwiedzki Ł., Walocha J.A., Niedźwiedzki T. The role of vasculature in bone development, regeneration and proper systemic functioning. Angiogenesis. 2017;20:291–302. doi: 10.1007/s10456-017-9541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Peng Y., Wu S., Li Y., Crane J.L. Type H blood vessels in bone modeling and remodeling. Theranostics. 2020;10:426–436. doi: 10.7150/thno.34126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zhao D., Liu J., Wang M., Zhang X., Zhou M. Epidemiology of cardiovascular disease in China: current features and implications. Nat. Rev. Cardiol. 2019;16:203–212. doi: 10.1038/s41569-018-0119-4. [DOI] [PubMed] [Google Scholar]
- 163.Sebastião M.J., Serra M., Pereira R. Human cardiac progenitor cell activation and regeneration mechanisms: exploring a novel myocardial ischemia/reperfusion in vitro model. Stem Cell Res. Ther. 2019;10:77. doi: 10.1186/s13287-019-1174-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Huang H., Huang W. Regulation of endothelial progenitor cell functions in ischemic heart disease: new therapeutic targets for cardiac remodeling and repair. Front Cardiovasc Med. 2022;9 doi: 10.3389/fcvm.2022.896782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Lee J.H., Kim S.W., Ji S.T., et al. Engineered M13 nanofiber accelerates ischemic neovascularization by enhancing endothelial progenitor cells. Tissue Eng. Regen. Med. 2017;14:787–802. doi: 10.1007/s13770-017-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jang W.B., Ji S.T., Park J.H., et al. Engineered M13 peptide carrier promotes angiogenic potential of patient-derived human cardiac progenitor cells and in vivo engraftment. Tissue Eng. Regen. Med. 2020;17:323–333. doi: 10.1007/s13770-020-00244-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cooke P., Janowitz H., Dougherty S.E. Neuronal redevelopment and the regeneration of neuromodulatory axons in the adult mammalian central nervous system. Front. Cell. Neurosci. 2022:16. doi: 10.3389/fncel.2022.872501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Cao H., Liu T., Chew S.Y. The application of nanofibrous scaffolds in neural tissue engineering. Adv. Drug Deliv. Rev. 2009;61:1055–1064. doi: 10.1016/j.addr.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 169.Chung W.-J., Merzlyak A., Yoo S.Y., Lee S.-W. Genetically engineered liquid-crystalline viral films for directing neural cell growth. Langmuir ACS J. Surf. Colloids. 2010;26:9885–9890. doi: 10.1021/la100226u. [DOI] [PubMed] [Google Scholar]
- 170.Siegel R.L., Miller K.D., Fuchs H.E., Jemal A. Cancer statistics, 2022. CA. Cancer J. Clin. 2022;72:7–33. doi: 10.3322/caac.21708. [DOI] [PubMed] [Google Scholar]
- 171.Wein L., Loi S. Mechanisms of resistance of chemotherapy in early-stage triple negative breast cancer (TNBC) Breast Edinb. Scotl. 2017;34(Suppl 1) doi: 10.1016/j.breast.2017.06.023. S27–S30. [DOI] [PubMed] [Google Scholar]
- 172.An P., Lei H., Zhang J., et al. Suppression of tumor growth and metastasis by a VEGFR-1 antagonizing peptide identified from a phage display library. Int. J. Cancer. 2004;111:165–173. doi: 10.1002/ijc.20214. [DOI] [PubMed] [Google Scholar]
- 173.Zheng P., Zhang Y., Zhang B., Wang Y., Wang Y., Yang L. Synthetic human monoclonal antibody targets hIL1 receptor accessory protein chain with therapeutic potential in triple-negative breast cancer. Biomed. Pharmacother. Biomedecine Pharmacother. 2018;107:1064–1073. doi: 10.1016/j.biopha.2018.07.099. [DOI] [PubMed] [Google Scholar]
- 174.Jones K.M., Karanam B., Jones-Triche J., Sandey M., Henderson H.J., Samant R.S., Temesgen S., Yates C., Bedi D. Phage ligands for identification of mesenchymal-like breast cancer cells and cancer-associated fibroblasts. Front. Oncol. 2018;8:625. doi: 10.3389/fonc.2018.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Zhang H., Guo Z., He B., Dai W., Zhang H., Wang X., Zhang Q. The improved delivery to breast cancer based on a novel nanocarrier modified with high-affinity peptides discovered by phage display. Adv. Healthc. Mater. 2018;7 doi: 10.1002/adhm.201800269. [DOI] [PubMed] [Google Scholar]
- 176.Domínguez-Romero A.N., Martínez-Cortés F., Munguía M.E., Odales J., Gevorkian G., Manoutcharian K. Generation of multiepitope cancer vaccines based on large combinatorial libraries of survivin-derived mutant epitopes. Immunology. 2020;161:123–138. doi: 10.1111/imm.13233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Li Y., Qu X., Cao B., et al. Selectively suppressing tumor angiogenesis for targeted breast cancer therapy by genetically engineered phage. Adv. Mater. 2020;32 doi: 10.1002/adma.202001260. [DOI] [PubMed] [Google Scholar]
- 178.Tan A.C., Ashley D.M., López G.Y. Management of glioblastoma: state of the art and future directions. Ca - Cancer J. Clin. 2020;70:299–312. doi: 10.3322/caac.21613. [DOI] [PubMed] [Google Scholar]
- 179.Roshdy E., ElNaggar M., Atta H., Kandeel A., Abdel-Wanis M., Abd Elbadee O.M., Abdelhafez Y.G., Mohamed Y. Role of post-therapy 99mTc-MIBI single-photon emission computed tomography/computed tomography scan in predicting survival in patients with high-grade glioma. Nucl. Med. Commun. 2021;42:625–632. doi: 10.1097/MNM.0000000000001385. [DOI] [PubMed] [Google Scholar]
- 180.Zhang J., Chen H., Chen C., Liu H., He Y., Zhao J., Yang P., Mao Q., Xia H. Systemic administration of mesenchymal stem cells loaded with a novel oncolytic adenovirus carrying IL-24/endostatin enhances glioma therapy. Cancer Lett. 2021;509:26–38. doi: 10.1016/j.canlet.2021.03.027. [DOI] [PubMed] [Google Scholar]
- 181.Lo Y.-L., Lin H.-C., Hong S.-T., Chang C.-H., Wang C.-S., Lin A.M.-Y. Lipid polymeric nanoparticles modified with tight junction-modulating peptides promote afatinib delivery across a blood–brain barrier model. Cancer Nanotechnol. 2021;12:13. [Google Scholar]
- 182.Pandit R., Chen L., Götz J. The blood-brain barrier: physiology and strategies for drug delivery. Adv. Drug Deliv. Rev. 2020;165–166:1–14. doi: 10.1016/j.addr.2019.11.009. [DOI] [PubMed] [Google Scholar]
- 183.Wang Y., Sheng J., Chai J., Zhu C., Li X., Yang W., Cui R., Ge T. Filamentous bacteriophage-A powerful carrier for glioma therapy. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.729336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Souweidane M.M., Kramer K., Pandit-Taskar N., et al. Convection-enhanced delivery for diffuse intrinsic pontine glioma: a single-centre, dose-escalation, phase 1 trial. Lancet Oncol. 2018;19:1040–1050. doi: 10.1016/S1470-2045(18)30322-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ksendzovsky A., Walbridge S., Saunders R.C., Asthagiri A.R., Heiss J.D., Lonser R.R. Convection-enhanced delivery of M13 bacteriophage to the brain. J. Neurosurg. 2012;117:197–203. doi: 10.3171/2012.4.JNS111528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Xi Y., Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Transl. Oncol. 2021;14 doi: 10.1016/j.tranon.2021.101174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Sepich-Poore G.D., Zitvogel L., Straussman R., Hasty J., Wargo J.A., Knight R. The microbiome and human cancer. Science. 2021;371 doi: 10.1126/science.abc4552. eabc4552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.V M., Cs C., Tf G. vol. 160. 2021. Cancer and the Microbiome-Influence of the Commensal Microbiota on Cancer, Immune Responses, and Immunotherapy. Gastroenterology. Epub ahead of print January. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Xie Y.-H., Chen Y.-X., Fang J.-Y. Comprehensive review of targeted therapy for colorectal cancer. Signal Transduct. Targeted Ther. 2020;5:22. doi: 10.1038/s41392-020-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Murgas P., Bustamante N., Araya N., et al. A filamentous bacteriophage targeted to carcinoembryonic antigen induces tumor regression in mouse models of colorectal cancer. Cancer Immunol. Immunother. CII. 2018;67:183–193. doi: 10.1007/s00262-017-2076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Manivannan A.C., Dhandapani R., Velmurugan P., Thangavelu S., Paramasivam R., Ragunathan L., Saravanan M. Phage in cancer treatment – biology of therapeutic phage and screening of tumor targeting peptide. Expet Opin. Drug Deliv. 2022;0:1–10. doi: 10.1080/17425247.2022.2094363. [DOI] [PubMed] [Google Scholar]
- 192.Eriksson F., Culp W.D., Massey R., Egevad L., Garland D., Persson M.A.A., Pisa P. Tumor specific phage particles promote tumor regression in a mouse melanoma model. Cancer Immunol. Immunother. CII. 2007;56:677–687. doi: 10.1007/s00262-006-0227-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bortot B., Apollonio M., Baj G., et al. Advanced photodynamic therapy with an engineered M13 phage targeting EGFR: mitochondrial localization and autophagy induction in ovarian cancer cell lines. Free Radic. Biol. Med. 2022;179:242–251. doi: 10.1016/j.freeradbiomed.2021.11.019. [DOI] [PubMed] [Google Scholar]
- 194.Shi K., Wang G., Pei J. Emerging strategies to overcome resistance to third-generation EGFR inhibitors. J Hematol OncolJ Hematol Oncol. 2022;15:94. doi: 10.1186/s13045-022-01311-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Sattar S., Bennett N.J., Wen W.X., Guthrie J.M., Blackwell L.F., Conway J.F., Rakonjac J. Ff-nano, short functionalized nanorods derived from Ff (f1, fd, or M13) filamentous bacteriophage. Front. Microbiol. 2015;6 doi: 10.3389/fmicb.2015.00316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Tsedev U., Lin C.-W., Hess G.T., Sarkaria J.N., Lam F.C., Belcher A.M. Phage particles of controlled length and genome for in vivo targeted glioblastoma imaging and therapeutic delivery. ACS Nano. 2022;16:11676–11691. doi: 10.1021/acsnano.1c08720. [DOI] [PubMed] [Google Scholar]
- 197.Dj M. Approaches for manufacture, formulation, targeted delivery and controlled release of phage-based therapeutics. Curr. Opin. Biotechnol. 2021;68 doi: 10.1016/j.copbio.2021.02.009. Epub ahead of print April. [DOI] [PubMed] [Google Scholar]
- 198.Venturini C., Petrovic Fabijan A., Fajardo Lubian A. Biological foundations of successful bacteriophage therapy. EMBO Mol. Med. 2022;14 doi: 10.15252/emmm.202012435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.González-Mora A., Calvillo-Rodríguez K.M., Hernández-Pérez J. Evaluation of the immune response of a candidate phage-based vaccine against rhipicephalus microplus (cattle tick) Pharmaceutics. 2021;13:2018. doi: 10.3390/pharmaceutics13122018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Henein A. What are the limitations on the wider therapeutic use of phage? Bacteriophage. 2013;3 doi: 10.4161/bact.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Nilsson A.S. Pharmacological limitations of phage therapy. Ups. J. Med. Sci. 2019;124:218–227. doi: 10.1080/03009734.2019.1688433. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No data was used for the research described in the article.