Abstract
Soft rot is a widespread, catastrophic disease caused by Pectobacterium carotovorum subsp. carotovorum (Pcc) that severely damages the production of Amorphophallus spp. This study evaluated the rhizosphere bacterial and fungal communities in Pcc-infected and uninfected plants of two species of Amorphophallus, A. muelleri and A. konjac. Principal component analysis showed that the samples formed different clusters according to the Pcc infection status, indicating that Pcc infection can cause a large number of changes in the bacterial and fungal communities in the Amorphophallus spp. rhizosphere soil. However, the response mechanisms of A. muelleri and A. konjac are different. There was little difference in the overall microbial species composition among the four treatments, but the relative abundances of core microbiome members were significantly different. The relative abundances of Actinobacteria, Chloroflexi, Acidobacteria, Firmicutes, Bacillus, and Lysobacter were lower in infected A. konjac plants than in healthy plants; in contrast, those of infected A. muelleri plants were higher than those in healthy plants. For fungi, the relative abundances of Ascomycota and Fusarium in the rhizosphere of infected A. konjac plants were significantly higher than those of healthy plants, but those of infected A. muelleri plants were lower than those of healthy plants. The relative abundance of beneficial Penicillium fungi was lower in infected A. konjac plants than in healthy plants, and that of infected A. muelleri plants was higher than that of healthy plants. These findings can provide theoretical references for further functional research and utilization of Amorphophallus spp. rhizosphere microbial communities in the future.
Keywords: Amorphophallus konjac, Amorphophallus muelleri, Pectobacterium carotovorum subsp. carotovorum, rhizosphere microbial community, soft rot
Cultivated Amorphophallus spp. are perennial herb of the Araceae family and can accumulate a large amount of glucomannan (KGM), which is used in the food, chemical, and pharmaceutical industries (Behera and Ray, 2016; Yang et al., 2022). China is the world’s largest producer of konjac, accounting for 60% of the world’s total output. However, bacterial soft rot caused by Pectobacterium carotovorum subsp. carotovorum (Pcc) severely restricts konjac cultivation and KGM production in China. Pcc infects all parts of Amorphophallus spp. plants (leaves, petioles, and tubers), usually causing leaf yellowing and wilting, soft rot of internal petiole tissues and underground bulbs, and ultimately death of the whole plant. The average incidence of Pcc infection in the field is more than 35%, which can reduce the Amorphophallus spp. yield by 30–70% or even result in a failed harvest across a large area (Wu et al., 2018). In addition to Amorphophallus spp., Pcc can cause a wide range of diseases on a variety of plant hosts around the world (Xu et al., 2021), including soft rot in bananas (Basim et al., 2019), potato (Anajjar et al., 2014), artichoke (Gallelli et al., 2009), lettuce (Cariddi and Sanzani, 2013), and Chinese cabbage (Popović et al., 2019). However, its prevention and control have always been difficult problems during production.
At present, the control of Amorphophallus spp. soft rot is mainly based on chemical and agricultural control, including land exchange, crop rotation, seed disinfection, and soil disinfection (Cui and Li, 2009). These methods doubtlessly all have certain limitations and are incapable of eradicating soft rot. Therefore, the utilization of disease-resistant varieties is considered one of the most promising strategies for preventing soft rot. A. muelleri is a species that can be propagated by seeding (triploid apomixis 2n = 39) and has a high reproduction factor, high yield, and high resistance to soft rot compared to the traditionally cultivated species A. konjac and A. albus (Xue et al., 2022). Previous studies have elaborated the molecular mechanisms of defense against soft rot in Amorphophallus spp. in terms of physiology, disease resistance genes, etc. (Wei et al., 2022), but the mechanism of Amorphophallus spp. resistance to soft rot remains incompletely elucidated.
The occurrence of soil-borne diseases is closely related to the imbalance of the soil microecosystem (Kim and Anderson, 2018; Raaijmakers et al., 2009; She et al., 2017), and soil physical and chemical properties, climatic conditions, geographical location, and rhizosphere microbial diversity all play important roles in the occurrence of soil-borne diseases (Ahmed et al., 2022a; Cai et al., 2021). Among them, rhizosphere soil microorganisms have attracted considerable attention in recent years as the most physiologically active part of the rhizosphere soil microecological environment (Xiaolong et al., 2022; Zheng et al., 2022). Mendes et al. (2013) and Dong et al. (2019) indicated that most disease resistance and developmental mechanisms in plants are directly related to rhizosphere microorganism diversity. Furthermore, plant genotype and soil type are two important factors for a healthy rhizosphere microbiota (Cai et al., 2021). In the present study, we conducted an in-depth evaluation of the rhizosphere bacterial and fungal communities of Pcc-infected Amorphophallus spp. with different levels of resistance and healthy plants using 16S rRNA and 18S rRNA high-throughput sequencing analysis and predicted the bacterial community function by PICRUSt. The results may provide a theoretical reference for further functional research and utilization of Amorphophallus spp. rhizosphere microbial communities in the future.
Materials and Methods
Microorganisms and plant resources
The soft rot pathogen P. carotovorum subsp. carotovorum strain EccK-23B (accession no. MN653919) used in this study has been preserved in our laboratory, the Yunnan Urban Agricultural Engineering and Technological Research Center. After the strain was activated, it was inoculated into Luria-Bertani broth (10 g/l trypsin, 5 g/l yeast extract, 5 g/l NaCl, pH 7.0 ± 0.2), shaken overnight, and then diluted with sterile distilled water to a seeding concentration of 1 × 108 cfu/ml to inoculate the plants.
In this study, two species of Amorphophallus spp. with different levels of resistance were selected: A. konjac and A. muelleri; A. muelleri is highly resistant to Pcc, whereas A. konjac is highly susceptible. The disease resistance of A. konjac and A. muelleri against Pcc was described in our previous study (Wei et al., 2022). We selected one-year-old underground bulbs of the two species of Amorphophallus spp. and planted them in the same greenhouse at the Yunnan Urban Agricultural Engineering and Technological Research Center in Kunming City, Yunnan Province, China. A. konjac and A. muelleri plants grown under the same light, temperature, watering (three times per week), and fertilization (compound fertilizer once per month) conditions were used as test materials.
Pathogen inoculation and soil sample collection
Healthy A. konjac and A. muelleri plants growing in the greenhouse for after five months were selected; 100 μl of Pcc bacterial solution (1 × 108 cfu/ml) was inoculated at 1–2 cm from the base of the petiole, and at the same time, the same amount of sterile water was inoculated as a control. Four treatments were implemented, with three replicates for each treatment containing three inoculated plants each.
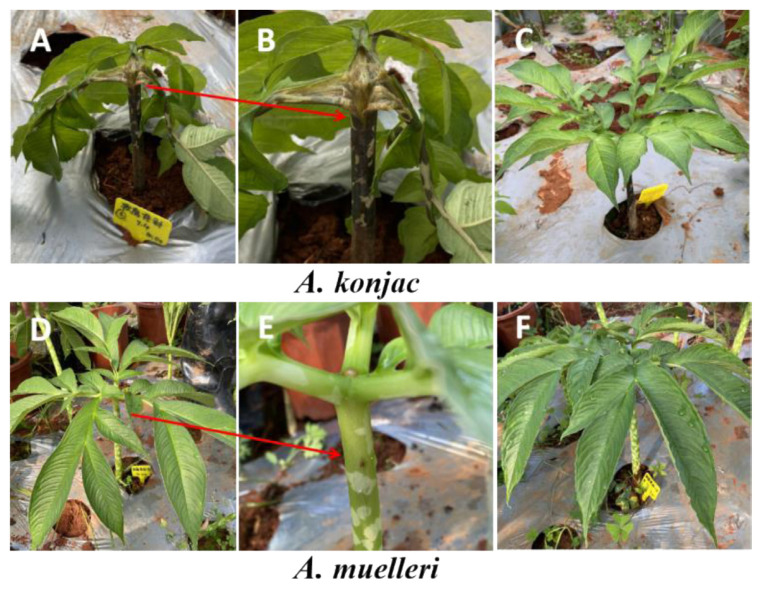
When A. konjac plants infected with Pcc showed obvious disease symptoms (soft rot symptoms on petioles and stems) (Fig. 1), all treatment groups were sampled. After removing the topsoil, the whole plants of each treatment groups were removed, and the loose soil was shaken off. After the 0–4 mm layer of soil attached to the roots was collected as the rhizosphere soil, the rhizosphere soil of the three konjac plants in each replicate was mixed together as a sample, for a total of 12 samples, and the treatment groups were named AK_CK (A. konjac inoculated with sterile water), AK_PCC (A. konjac inoculated with Pcc), AM_CK (A. muelleri inoculated with sterile water), and AM_PCC (A. muelleri inoculated with Pcc).
Fig. 1.
Symptoms of Amorphophallus konjac and A. muelleri plants infected by Pectobacterium carotovorum subsp. carotovorum (Pcc). (A, B) A. konjac stems and petioles showed obvious soft rot symptoms after injection with Pcc. (C) Healthy A. konjac plants injected with sterile water (CK). (D, E) A. muelleri stems and petioles had no soft rot symptoms after injection with Pcc. (F) Healthy A. muelleri plants injected with sterile water (CK). The arrow points to a clearer phenotype at the inoculation site.
DNA extraction, polymerase chain reaction amplification, and sequencing
The DNA of each rhizosphere soil sample was extracted by referring to the instructions of the FastDNA Spin Kit for Soil (MP Biomedicals, Solon, OH, USA) soil genomic DNA extraction kit. DNA concentration and purity were examined using a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington, DE, USA), and DNA extraction quality was examined using 1% agarose gel electrophoresis. Diversity analysis was performed by amplifying the V3–V4 region of the 16S rRNA gene with the universal primers 338F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′). Fungal diversity was analyzed using the universal primers TS1 F (5′-ACTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 R (5′-BGCTGCGTTCTTCATCGATGC-3′). The polymerase chain reaction (PCR) amplification procedure was described by Xiaolong et al. (2022) and Zheng et al. (2022). The PCR product was recovered using a 2% agarose gel, further purified using the AxyPrep DNA Gel Extraction Kit (Axygen, Union City, CA, USA) according to the manufacturer’s instructions, and sent to Shanghai Majorbio Bio-pharm Technology Co., Ltd. (Shanghai, China) for sequencing on the Illumina MiSeq PE300 platform (Illumina, San Diego, CA, USA).
Bioinformatics analysis
Trimmomatic software was used for the quality control of the original sequence, and FLASH software (Caporaso et al., 2011) was used for sequence splicing; the obtained sequences were filtered by Usearch software (Edgar, 2013), and chimeric sequences were removed to obtain valid sequences; UPARSE software was used to classify the operational taxonomic units (OTUs) at 97% similarity. The representative sequences were annotated with the RDP classifier software (Wang et al., 2007) and the SILVA database (Altschul et al., 1990), and Mothur software was used to draw the dilution curve. The library coverage (Coverage), Shannon, Simpson, ACE, and Chao1 indices were calculated to evaluate the species diversity and abundance index, and the Bray-Curtis distance algorithm established by QIIME software was used for principal coordinate analysis (PCoA). Circos diagrams were built using Circos-0.67-7 software (Yan et al., 2020). Linear discriminant analysis (LDA) combined with linear discriminant analysis effect size (LEfSe) was used to search for significantly different biomarkers between treatments using LEfSe software. Functional prediction of bacterial populations in each sample was performed using PICRUSt software.
Results
Analysis of OTUs and assessment of community structure
After 16S rRNA and 18S rRNA high-throughput sequencing, 668,630 effective bacterial sequences and 645,801 effective fungal sequences were obtained from the 12 rhizosphere soil samples, with average lengths of 415 bp and 240 bp, respectively. The abundance of bacterial and fungal communities in the different treatment groups was analyzed using sparsity curves (Supplementary Fig. 1), and the results indicated that the sparsity of bacterial and fungal communities reached reasonable reads in all samples.
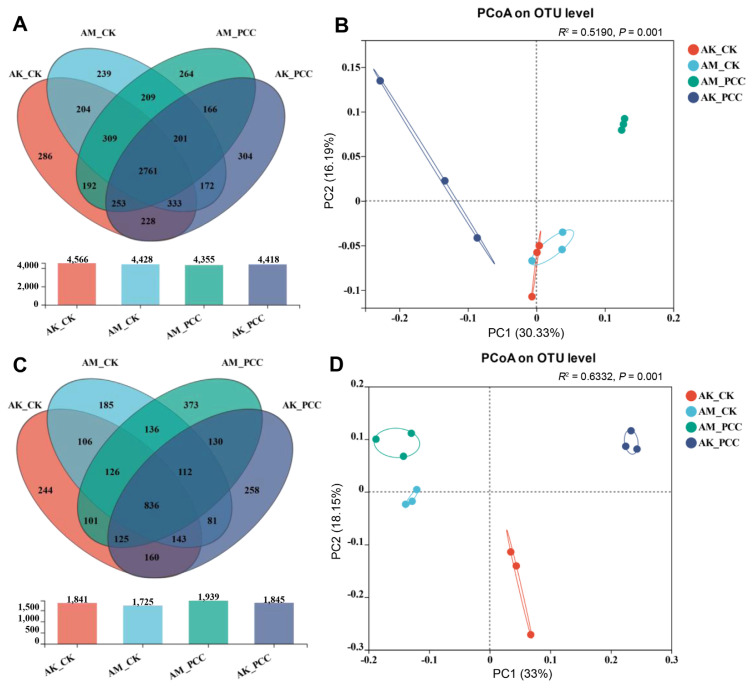
The number of OTUs in the different samples (97% clustering level) is shown in Fig. 2. A total of 6,121 bacterial OTUs and 3,116 fungal OTUs were isolated from the different treatment groups, of which 2,761 bacterial OTUs and 836 fungal OTUs were common to all treatments (Fig. 2A and C). The Pcc treatment groups for two species of Amorphophallus had a higher number of unique bacterial and fungal OTUs than their respective control group: AM_PCC (bacteria, 264; fungi, 373) had more OTUs than AM_CK (bacteria, 239; fungi, 185), and AK_PCC (bacteria, 304; fungi, 258) had more OTUs than AK_CK (bacteria, 286; fungi, 244). These results indicated that soft rot pathogens had effects on both bacterial and fungal communities.
Fig. 2.
Diversity and structure of Amorphophallus konjac and A. muelleri rhizosphere microbial communities under different treatments. A Venn diagram showing the number of unique, shared, and common bacterial (A) and fungal (C) operational taxonomic units (OTUs). Principal coordinate analysis (PCoA) plots based on the Bray-Curtis dissimilarity matrix showing the changes in the structure of bacterial (B) and fungal (D) communities. AK_CK, A. konjac inoculated with sterile water; AM_CK, A. muelleri inoculated with sterile water; AM_PCC, A. muelleri inoculated with Pectobacterium carotovorum subsp. carotovorum (Pcc); AK_PCC, A. konjac inoculated with Pcc.
Using the Bray-Curtis distance algorithm for PCoA principal coordinate analysis, the explanation degrees of principal component 1 (PC1) and principal component 2 (PC2) for sample variability were 30.33%, 16.19% (bacteria), and 33.00%, and 18.15% (fungi), respectively, which together can explain 46.52% and 51.15% of the samples. Fig. 2B and D reveals that the bacterial and fungal community compositions of the different treatments were significantly different. When the two species of Amorphophallus were compared, the AM_CK and AK_CK bacterial communities were closer in distance, and the community composition was more similar; however, the AM_CK fungal community was mainly distributed on the left side of the PC1 axis, AK_CK was mainly distributed on the right side of the PC1 axis, and the community composition of the two species was significantly different, indicating that the species had a greater impact on the composition of the fungal community. Interestingly, the bacterial and fungal communities of the different species in the Pcc treatment groups were significantly separated from CK, AM_PCC and AK_PCC were mainly distributed on the upper side of the PC2 axis, and AM_CK and AK_CK were mainly distributed on the lower side of the PC2 axis, indicating that the microbial communities of the two treatments varied widely. Overall, samples from the two species of Amorphophallus formed distinct clusters according to Pcc infection status, suggesting that the largest source of microbial community variability may be caused by Pcc.
Microbial community diversity analysis
We assessed the within-sample diversity of bacterial and fungal communities at the 97% threshold level (alpha diversity index: Shannon, Simpson, ACE, and Chao 1), and the results are shown in Supplementary Fig. 2. The Shannon, ACE, and Chao 1 indices of the bacterial communities were ranked AM_PCC < AM_CK and AK_PCC < AK_CK, and the Simpson indices were ranked AM_PCC > AM_CK and AK_PCC > AK_CK, indicating that the diversity and abundance of bacterial communities decreased after Pcc treatment compared with CK. The Shannon indices of the fungal community were ranked AM_PCC < AM_CK and AK_PCC > AK_CK, the ACE and Chao 1 indices were ranked AM_PCC > AM_CK and AK_PCC < AK_CK, and the Simpson indices were ranked AM_PCC > AM_CK and AK_PCC < AK_CK.
Microbial community composition and relative abundance
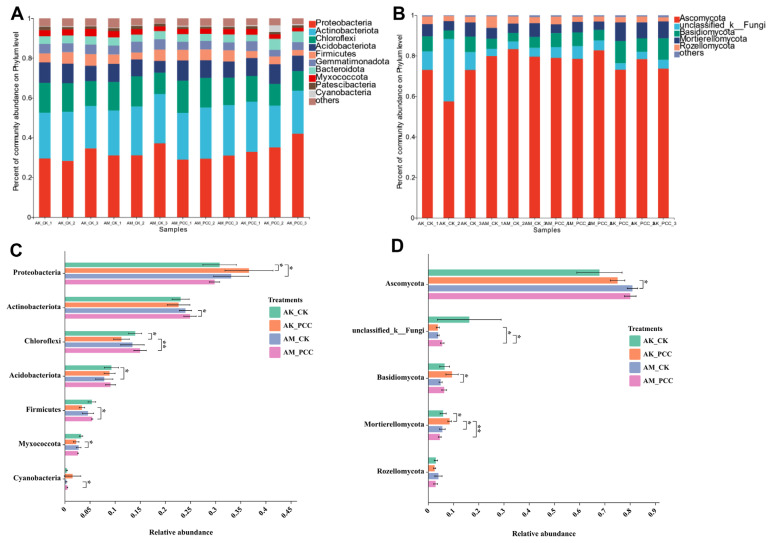
At the phylum level, the bacterial flora of Amorphophallus spp. rhizosphere soil in the different treatments was mainly composed of Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Firmicutes, Gemmatimonadetes, and Bacteroidota (Fig. 3A), which together accounted for more than 91.0% of the total soil bacteria. The results of significant difference analysis (Fig. 3C) revealed that the relative abundance of Proteobacteria in AK_PCC (36.63%) was significantly higher than that in AK_CK (30.79%) in the A. konjac treatment group (t test, P < 0.05), but the relative abundance of Chloroflexi in AK_PCC (11.26%) was significantly lower than that in AK_CK (13.99%) (t test, P < 0.05); in addition, the relative abundances of Actinobacteria, Acidobacteria, Firmicutes, and Gemmatimonadota was lower in AK_PCC than in AK_CK, but the difference was not significant. In the A. muelleri treatment group, the relative abundance of Proteobacteria in AM_CK (33.09%) was higher than that of AM_PCC (29.78%), but the difference was not significant; in contrast, the relative abundances of Actinobacteria and Cyanobacteria in AM_CK (23.99%, 0.29%) were significantly lower than those in AM_PCC (24.89%, 0.50%) (t test, P < 0.05). Meanwhile, the relative abundances of Chloroflexi, Acidobacteria, and Firmicutes were also ranked AM_CK < AM_PCC, but the difference was not significant.
Fig. 3.
Distribution of the most dominant bacterial (A) and fungal (B) communities at the phylum level under the different treatments. The bar plots for some bacterial (C) and fungal (D) phyla showed significant differences among treatments according to a t test (P < 0.05). Asterisks represent significant differences at *P < 0.05 and **P < 0.01. AK_CK, A. konjac inoculated with sterile water; AM_CK, A. muelleri inoculated with sterile water; AM_PCC, A. muelleri inoculated with Pectobacterium carotovorum subsp. carotovorum (Pcc); AK_PCC, A. konjac inoculated with Pcc.
The fungal communities of the different treatments were dominated by Ascomycota, Basidiomycota and Mortierellomycota, accounting for more than 88.8% of the total soil fungal community (Fig. 3B). The relative abundance of Ascomycota in AK_CK was lower than that in AK_PCC and higher in AM_CK than in AM_PCC, but the difference was not significant. The relative abundance of Basidiomycota in the two species rhizosphere soils showed the same ranking AK_CK < AK_PCC and AM_CK < AM_PCC. The relative abundance of Mortierellomycota in AK_PCC (8.44%) was significantly higher than that of AK_CK (5.86%) (t test, P < 0.05); in contrast, AM_PCC (4.54%) was lower than AM_CK (5.58%), but the difference was not significant (Fig. 3D).
Genus-level microbial community composition and relative abundance
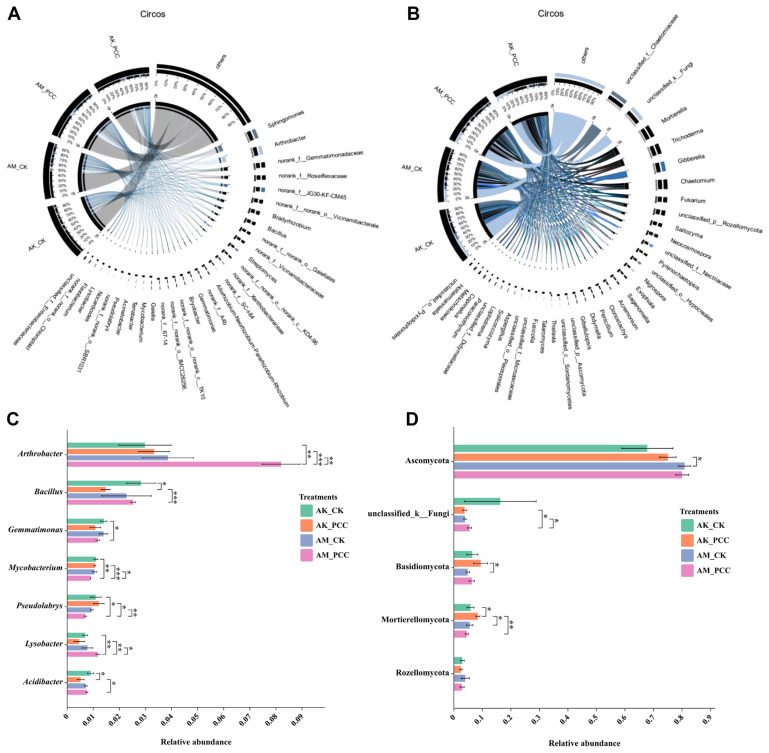
The dominant bacterial genera of Amorphophallus spp. rhizosphere soil bacteria in different treatments were Sphingomonas, Arthrobacter, norank_f__Gemmatimonadaceae, norank_f__Roseiflexaceae, norank_f__JG30-KF-CM45, norank_f__norank_o__Vicinamibacterales, Bradyrhizobium, and Bacillus (Fig. 4A). The relative abundance of Sphingomonas was higher in AM_CK (5.99%) than in AK_CK (4.37%); after Pcc infection, AM_PCC (4.64%) was lower in abundance, while AK_PCC (4.78%) abundance was higher compared with that of AM_CK and AK_CK. The change in Arthrobacter abundance in the two species of Amorphophallus rhizosphere soils was the same, with the Pcc infection treatment producing higher abundance than CK. The differences between AK_CK (2.98%) and AK_PCC (3.33%) were not statistically significant (P > 0.05); however, abundance in AM_PCC (8.20%) was significantly higher than that in AM_CK (3.86%) (t test, P < 0.01). The relative abundance of Bacillus was significantly lower in AK_PCC (1.48%) than in AK_CK (2.83%) (t test, P < 0.05) and was higher in AM_PCC (2.53%) than in AM_CK (2.28%), but the difference was not significant (Fig. 4C). The relative abundances of Mycobacterium and Pseudolabrys were both significantly higher in AM_CK (1.05%, 0.94%) than in AM_PCC (0.90%, 0.70%), and the relative abundance of Lysobacter was significantly lower in AM_CK (0.78%) than in AM_PCC (1.18%) (t test, P < 0.05). In addition, after Pcc infection, the relative abundances of Streptomyces and Bryobacter in two species of Amorphophallus rhizosphere soils was lower.
Fig. 4.
Distribution of the most dominant bacterial (A) and fungal (B) communities at the genus level under the different treatments. The bar plots for some bacterial (C) and fungal (D) genera showed significant differences among treatments according to a t test (P < 0.05). Asterisks represent significant differences at *P < 0.05, **P < 0.01, and ***P < 0.001. AK_CK, A. konjac inoculated with sterile water; AM_CK, A. muelleri inoculated with sterile water; AM_PCC, A. muelleri inoculated with Pectobacterium carotovorum subsp. carotovorum (Pcc); AK_PCC, A. konjac inoculated with Pcc.
For the fungal community, the main dominant genera included unclassified_f__Chaetomiaceae, unclassified_k__Fungi, Mortierella, Trichoderma, Gibberella, Chaetomium, Fusarium, unclassified_p__Rozellomycota, Saitozyma, and Neocosmospora (Fig. 4B). Among them, Mortierella had the highest abundance in AK_PCC (8.42%), and its relative abundance in AK_PCC was significantly higher than that in AK_CK (5.83%), AM_CK (5.54%), and AM_PCC (4.53%) (t test, P < 0.05) (Fig. 4D). The relative abundance of Gibberella was also significantly different among treatments, with AK_PCC (2.68%) and AM_PCC (4.10%) showing significantly lower abundance than AK_CK (6.53%) and AM_CK (7.10%), respectively (t test, P < 0.05). The relative abundance of Fusarium was higher in AK_PCC (2.83%) than in AK_CK (2.67%) but lower in AM_PCC (3.36%) than in AM_CK (3.87%). The relative abundance of Penicillium was lower in AK_PCC (1.24%) than in AK_CK (1.52%) and higher in AM_PCC (1.43%) than in AM_CK (1.32%). These results indicated that Pcc infection had an effect on the distribution and composition of the bacterial and fungal communities in the Amorphophallus spp. rhizosphere soil.
Multilevel species difference analysis based on LEfSe
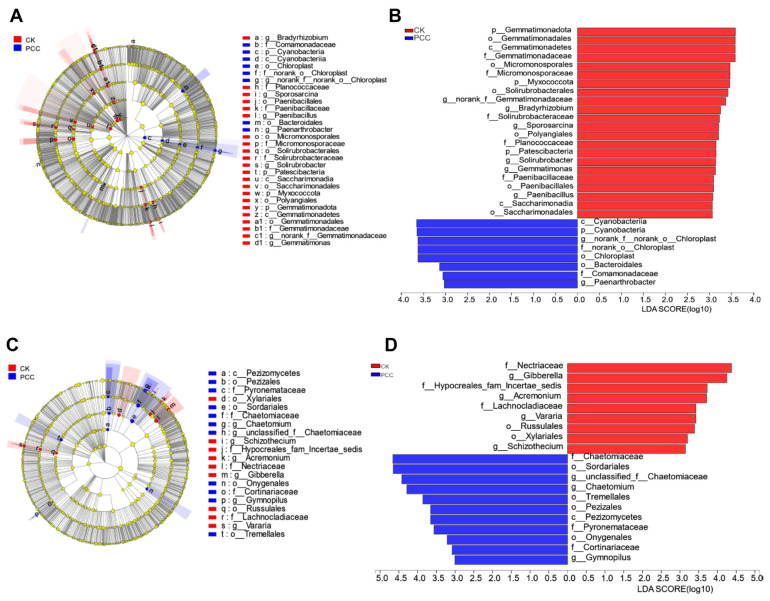
To further determine the source of differences in the rhizosphere soil microbial community of Amorphophallus spp. under Pcc infection, we divided the A. muelleri and A. konjac samples into Pcc infection and CK groups. Using LEfSe multilevel species difference analysis, the samples were subjected to LDA according to different grouping conditions based on taxonomic composition to determine the communities or species that had significantly different effects on the sample division. Fig. 5 indicates that in bacterial communities with LDA values greater than 3, Cyanobacteria (phylum to class), norank_f__norank_o__Chloroplast (order to genus), Bacteroidales (order), Comamonadaceae (family), and Paenarthrobacter (genus) were mainly enriched in Pcc-infected konjac rhizosphere soils. Gemmatimonadota (phylum to genus), Micromonosporales (order to family), Myxococcota (phylum), Solirubrobacterales (order to genus), Bradyrhizobium (genus), and others were mainly enriched in the healthy konjac rhizosphere. For fungal communities, Chaetomiaceae (family to genus), Sordariales (order), Tremellales (order), Pezizales (class to order), and others were enriched in Pcc-infected konjac rhizosphere soil; Nectriaceae (family), Gibberella (genus), Hypocreales_fam_Incertae_sedis (family), Acremonium (genus), and others were mainly enriched in the healthy konjac rhizosphere. These microorganisms enriched in the different samples may be important groups that cause differences in the community structure.
Fig. 5.
Linear discriminant analysis effect size analyses of the aggregated groups of microbial communities in the rhizosphere soil of Pectobacterium carotovorum subsp. carotovorum (Pcc)-infected plants and healthy plants (taxa from the phylum to the genus level). Cladogram representing the abundance of bacterial (A) and fungal (C) taxa. Histogram of the bacterial (B) and fungal (D) microbiota with linear discriminant analysis = 3. CK, inoculated with sterile water; PCC, inoculated with Pectobacterium carotovorum subsp. carotovorum; LDA, linear discriminant analysis.
Predictive analysis of microbial community function
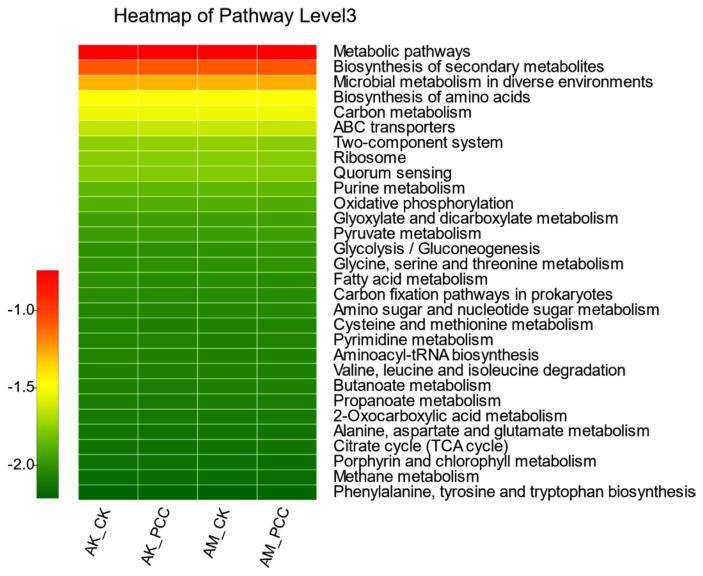
The functional characteristics of the Amorphophallus spp. rhizosphere soil bacterial community were predicted using PICRUSt software. PICRUSt identifies 16S rRNA gene sequences based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway database and infers possible gene content. Most of the predicted functional genes of the Amorphophallus spp. rhizosphere soil bacterial community were assigned to functional groups such as metabolism, environmental information processing, genetic information processing, and cellular processes. Among the top 30 functions with relative abundance at the pathway level 3 (Fig. 6), 83.33% were metabolism-related functions, 6.67% were environmental information processing-related functions, 6.67% were genetic information processing-related functions, and 3.33% were cellular process-related functions. Among them, some important metabolism-related functions were significantly different between treatments. First, the relative abundances of amino acid synthesis, pyruvate metabolism, glycolysis/gluconeogenesis, amino sugar and nucleotide sugar metabolism, phenylalanine/tyrosine, and metabolite and tryptophan metabolism in the AM_PCC treatment were the highest and were significantly higher than those in AK_CK, AK_PCC, and AM_CK (P < 0.05). Second, carbon metabolism, glycine, serine and threonine metabolism, cysteine and methionine metabolism, pyrimidine metabolism, tricarboxylic acid cycle, and methane metabolism all showed the same differences; that is, the difference between AM_PCC and AM_CK did not reach a significant level but was significantly higher than AK_CK and AK_PCC. Oxidative phosphorylation was significantly higher in AM_CK than in AM_PCC, but there was no significant difference between AK_CK and AK_PCC. The above results indicate that when Pcc infects Amorphophallus spp., it affects the basic life functions of the rhizosphere soil bacterial community and the information transfer process with the surrounding environment and then affects the ecological effect of the bacterial community in the soil environment.
Fig. 6.
Heatmap of the relative abundances of the top 30 PICRUSt-predicted genes in the different treatments (n = 3/treatment). The abscissa is the treatment, and the ordinate is the function name of pathway level 3. The color gradients of the different color blocks represent the changes in different functional abundances in the treatments. The legend is the value represented by the color gradient. AK_CK, A. konjac inoculated with sterile water; AM_CK, A. muelleri inoculated with sterile water; AM_PCC, A. muelleri inoculated with Pectobacterium carotovorum subsp. carotovorum (Pcc); AK_PCC, A. konjac inoculated with Pcc.
Discussion
Soft rot caused by P. carotovorum subsp. carotovorum is one of the most common and most damaging diseases in Amorphophallus spp. production and is referred to as a “cancer” of konjac. The disease can occur throughout the Amorphophallus spp. growth period, and the yield loss caused by severe outbreaks can reach more than 80%. The rhizosphere microbiota is considered the first line of defense against soil pathogen infection and abiotic stress and is closely related to plant health (Ahmed et al., 2022a; Bulgarelli et al., 2013; Mendes et al., 2013). In this study, by 16S rRNA and 18S rRNA high-throughput sequencing, we analyzed that different response mechanisms of the rhizosphere microbial communities of A. muelleri and A. konjac to soft rot stress.
After Pcc infection, the number of unique bacterial and fungal OTUs in A. konjac and A. muelleri was higher than that in CK. Alpha diversity results indicate that in the A. konjac and A. muelleri rhizospheres after Pcc infection, the diversity and abundance of the bacterial community were lower than that of CK; for fungi, the rhizosphere fungal community diversity of Pcc-infected A. muelleri was lower but abundance was higher than in CK, while in the A. konjac rhizosphere, the differences in the fungal community were the opposite, suggesting that the changes in the rhizosphere microbial community composition and diversity of Amorphophallus spp. may be driven by the Pcc infection status, and the responses of Amorphophallus spp. with different levels of resistance may be different. We also analyzed the distribution of the rhizosphere bacterial and fungal communities of the two species of Amorphophallus under different infection states. PCoA revealed that the bacterial and fungal communities were significantly separated between Pcc-infected and uninfected plants (CK), and the samples formed different clusters according to the state of Pcc infection, indicating that the microbial communities in the infected and uninfected states were different, further revealing that the largest source of microbial community variability may be Pcc.
To further clarify the differences in the responses of A. konjac and A. muelleri rhizosphere microbial communities to Pcc infection, we analyzed differences in bacterial and fungal communities at the phylum and genus levels among treatments. In this study, the dominant bacterial phyla in the Amorphophallus spp. rhizosphere soil were Proteobacteria, Actinobacteria, Chloroflexi, Acidobacteria, Firmicutes, Gemmatimonadetes, and Bacteroidota, and the dominant fungal phyla were Ascomycota, Basidiomycota, and Mortierellomycota. Additionally, we detected the bacterial genera Sphingomonas, Arthrobacter, Bradyrhizobium, Bacillus, Mycobacterium, Pseudolabrys, Lysobacter, Streptomyces, and Bryobacter identified from various plant rhizosphere soils along with the fungal genera Mortierella, Trichoderma, Gibberella, Chaetomium, Fusarium, Saitozyma, and Neocosmospora. The overall microbial species composition of the four treatments was not significantly different because the microbial communities attached to these phyla and genera were considered the universal core soil microbiome, and the composition of the soil microbial communities did not appreciably differ (Greening et al., 2015; Huang et al., 2020). However, we found that the relative abundances of core microbiome members differed significantly between treatments. In terms of bacterial communities, Actinomycetes and Firmicutes can degrade cellulose, lignin, and lignocellulose by secreting hydrolases, thereby utilizing recalcitrant C sources, and Actinomycetes can also produce antibiotics that inhibit the growth and development of a variety of soil plant pathogens (Wang et al., 2020; Wei et al., 2018); The phylum Chloroflexi included anaerobic thermophiles and anaerobic organohalide respirers, which was used to utilize organohalide compounds (Wang et al., 2020); Acidobacteria are mainly involved in iron cycling and single-carbon metabolism and are important in degrading cellulose and lignin from plant residues (Wang et al., 2016). After Pcc infection, the relative abundances of the above bacteria in A. konjac and A. muelleri were completely opposite. The relative abundances of Actinobacteria, Chloroflexi, Acidobacteria, and Firmicutes in the rhizosphere soil of Pcc-infected A. konjac were lower than those of healthy plants, but the relative abundances of these types of bacteria in Pcc-infected A. muelleri was higher, to different degrees, than in healthy plants. The high abundance of these bacteria in the rhizosphere soil of A. muelleri can enhance the cycling of essential nutrients, thereby improving soil fertility and sustainable utilization.
At the genus level, many previous studies have reported that beneficial microorganisms (such as Bacillus, Streptomyces, Sphingomonas, Bradyrhizobium, Arthrobacter, Bryobacter, Micromonospora, and Lysobacter) are abundant in healthy soils, which can improve soil nutrients and promote plant growth and the control of soil-borne diseases (Wang et al., 2017). Bradyrhizobium inhibits fungal pathogens and root-knot nematodes (Siddiqui and Shaukat, 2002). Streptomyces is known worldwide for its powerful ability to produce antibiotics (Schlatter et al., 2009). Our results also indicated that their relative abundances were significantly higher in the rhizosphere soil of healthy A. konjac and A. muelleri. Some strains of Bacillus can inhibit the occurrence of disease through mechanisms that induce systemic disease resistance, direct antagonism, and colonization in host plants (Ahmed et al., 2022b). Lysobacter produces antimicrobial compounds that protect plants from pathogens (Ji et al., 2008). Some strains of Sphingomonas are closely related to nitrogen fixation, which can enhance the survivability of plants under environmental stress by improving the soil environment and degrading toxic substances (Xie and Yokota, 2006). Our results revealed that in infected A. konjac plants, the relative abundance of Bacillus and Lysobacter in the rhizosphere was lower than that in healthy plants, but the relative abundance in infected A. muelleri plants was higher than that in healthy plants. The relative abundance of Sphingomonas was higher in infected A. konjac plants than in healthy plants and lower in infected A. muelleri plants than in healthy plants.
With respect to the fungal community, Ascomycota contains a large number of pathogenic bacteria, but these pathogens are not pathogenic to crops; they usually gather in the roots of plants to cause damage to the root surface, creating conditions for infection by some pathogenic bacteria (Zheng et al., 2022). The relative abundance of Ascomycota in the rhizosphere of infected A. konjac plants was higher than that of healthy plants, but there was little difference between infected and healthy A. muelleri plants. At the genus level, Fusarium is the most harmful filamentous fungus to plants, and its toxins can cause more than 100 animal and plant diseases (Schlatter et al., 2009). Our results indicate that in infected A. konjac plants, the relative abundance of Fusarium in the rhizosphere was higher than that in healthy plants, but the abundance in infected A. muelleri was significantly lower than that in healthy plants. Penicillium can participate in the decomposition of organic matter, promote the cycles of C, N, P, and other elements, and can degrade a variety of environmentally harmful substances (Xiaolong et al., 2022). The relative abundance of Penicillium was lower in infected A. konjac than in healthy plants but higher in infected A. muelleri plants than in healthy plants. These results further illustrate that A. konjac and the rhizosphere microbial community of A. muelleri respond differently to Pcc infection, and A. muelleri tends to recruit some beneficial microorganisms, which may contribute to a healthier rhizosphere microecological environment.
PICRUSt function prediction indicated that among the top 30 functions in relative abundance, several important metabolism-related functions were significantly more active in the rhizosphere bacterial community of infected A. muelleri plants than in the remaining three treatments, but there were no significant differences between infected A. konjac and healthy plants. Soil microorganisms mainly participate in the material cycle and transformation of soil through their metabolic activities and regulate the metabolic processes of organisms (Wu et al., 2016). For example, amino acids are precursors of many secondary metabolites and play critical roles in many physiological processes, such as plant signaling, stress defense, and interactions with other organisms (Florencio-Ortiz et al., 2018). Glycolysis is a key pathway of carbohydrate metabolism that can provide energy and metabolites for plant and bacterial growth and development (Megguer et al., 2017). During Pcc infection, the higher metabolic function of the rhizosphere bacterial community in A. muelleri may be closely related to the stress response and plant stress tolerance. The results revealed that the basal life functions of rhizobacterial communities of A. konjac and A. muelleri also responded differently to Pcc infection. However, at present, the PICRUSt preliminary prediction of the functions of related bacteria is still greatly limited. In the future, we will combine traditional isolation and culture methods with metagenomics and other research methods to comprehensively analyze the correlative functions of rhizosphere bacterial communities during the resistance of Amorphophallus spp. to Pcc stress.
In this study, we concluded that Pcc infection caused substantial changes in the Amorphophallus spp. rhizosphere soil bacterial and fungal communities, but highly resistant A. muelleri and highly susceptible A. konjac responded differently. A. muelleri tended to recruit more beneficial microbes (e.g., Actinobacteria, Chloroflexi, Acidobacteria, Firmicutes, Bacillus, Lysobacter, Penicillium), these beneficial microorganisms may contribute to a healthier rhizosphere microecological environment for plants to resist infection. Taken together, our results indicate that the response mechanisms of the rhizosphere microbial communities of A. muelleri and A. konjac to Pcc stress are different. These findings can provide a theoretical basis for a comprehensive understanding of the assembly process of Amorphophallus spp. rhizosphere microbial communities and the potential function of the community under Pcc stress.
Acknowledgments
This study was supported by the Yunnan Provincial Science and Technology Department (Nos. 2019FH001-051, 2019FH001-008, 2021530000242017) and MOE (2019-NYZD-25-9 and 2019J0574).
Footnotes
Conflicts of Interest
No potential conflict of interest relevant to this article was reported.
Electronic Supplementary Material
Supplementary materials are available at The Plant Pathology Journal website (http://www.ppjonline.org/).
References
- Ahmed W., Dai Z., Liu Q., Munir S., Yang J., Karunarathna S. C., Li S., Zhang J., Ji G., Zhao Z. Microbial cross-talk: dissecting the core microbiota associated with flue-cured tobacco (Nicotiana tabacum) plants under healthy and diseased state. Front. Microbiol. 2022a;13:845310. doi: 10.3389/fmicb.2022.845310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Dai Z., Zhang J., Li S., Ahmed A., Munir S., Liu Q., Tan Y., Ji G., Zhao Z. Plant-microbe interaction: mining the impact of native Bacillus amyloliquefaciens WS-10 on tobacco bacterial wilt disease and rhizosphere microbial communities. Microbiol. Spectr. 2022b;10:e0147122. doi: 10.1128/spectrum.01471-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Anajjar B., Azelmat S., Terta M., Ennaji M. M. Evaluation of phytopathogenic effect of Pectobacterium carotovorum subsp. carotovorum isolated from symptomless potato tuber and soil. Curr. J. Appl. Sci. Technol. 2014;4:67–78. [Google Scholar]
- Basim H., Basim E., Bakİ D., Turgut A. Wet rot disease of banana (Musa Sp.) caused by Pectobacterium carotovorum subsp. carotovorum in Turkey. Can J. Plant Pathol. 2019;41:174–187. [Google Scholar]
- Behera S. S., Ray R. C. Konjac glucomannan, a promising polysaccharide of Amorphophallus konjac K. Koch in health care. Int. J. Biol. Macromol. 2016;92:942–956. doi: 10.1016/j.ijbiomac.2016.07.098. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren van Themaat E., Schulze-Lefert P. Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 2013;64:807–838. doi: 10.1146/annurev-arplant-050312-120106. [DOI] [PubMed] [Google Scholar]
- Cai Q., Zhou G., Ahmed W., Cao Y., Zhao M., Li Z., Zhao Z. Study on the relationship between bacterial wilt and rhizospheric microbial diversity of flue-cured tobacco cultivars. Eur. J. Plant Pathol. 2021;160:265–276. [Google Scholar]
- Caporaso J. G., Lauber C. L., Walters W. A., Berg-Lyons D., Lozupone C. A., Turnbaugh P. J., Fierer N., Knight R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U. S. A. 2011;108(Suppl 1):4516–4522. doi: 10.1073/pnas.1000080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cariddi C., Sanzani S. M. A severe outbreak of bacterial lettuce soft rot caused by Pectobacterium carotovorum subsp. carotovorum in Apulia (Italy) J. Plant Pathol. 2013;95:441–446. [Google Scholar]
- Cui M., Li C. Research progress on occurrence regularity and control technology of soft rot of Amorphophallus spp. China Plant Prot. 2009;6:33–35. (in Chinese) [Google Scholar]
- Dong C.-J., Wang L.-L., Li Q., Shang Q.-M. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS ONE. 2019;14:e0223847. doi: 10.1371/journal.pone.0223847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat. Methods. 2013;10:996–998. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- Florencio-Ortiz V., Sellés-Marchart S., Zubcoff-Vallejo J., Jander G., Casas J. L. Changes in the free amino acid composition of Capsicum annuum (pepper) leaves in response to Myzus persicae (green peach aphid) infestation. A comparison with water stress. PLoS ONE. 2018;13:e0198093. doi: 10.1371/journal.pone.0198093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallelli A., Galli M., De Simone D., Zaccardelli M., Loreti S. Phenotypic and genetic variability of Pectobacterium carotovorum isolated from artichoke in the sele valley. J. Plant Pathol. 2009;91:757–761. [Google Scholar]
- Greening C., Carere C. R., Rushton-Green R., Harold L. K., Hards K., Taylor M. C., Morales S. E., Stott M. B., Cook G. M. Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging. Proc. Natl. Acad. Sci. U. S. A. 2015;112:10497–10502. doi: 10.1073/pnas.1508385112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang K., Jiang Q., Liu L., Zhang S., Liu C., Chen H., Ding W., Zhang Y. Exploring the key microbial changes in the rhizosphere that affect the occurrence of tobacco root-knot nematodes. AMB Express. 2020;10:72. doi: 10.1186/s13568-020-01006-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji G.-H., Wei L.-F., He Y.-Q., Wu Y.-P., Bai X.-H. Biological control of rice bacterial blight by Lysobacter antibioticus strain 13–1. Biol. Control. 2008;45:288–296. [Google Scholar]
- Kim Y. C., Anderson A. J. Rhizosphere pseudomonads as probiotics improving plant health. Mol. Plant Pathol. 2018;19:2349–2359. doi: 10.1111/mpp.12693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megguer C. A., Fugate K. K., Lafta A. M., Ferrareze J. P., Deckard E. L., Campbell L. G., Lulai E. C., Finger F. L. Glycolysis is dynamic and relates closely to respiration rate in stored sugarbeet roots. Front. Plant Sci. 2017;8:861. doi: 10.3389/fpls.2017.00861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R., Garbeva P., Raaijmakers J. M. The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013;37:634–663. doi: 10.1111/1574-6976.12028. [DOI] [PubMed] [Google Scholar]
- Popović T., Jelušić A., Marković S., Iličić R. Characterization of Pectobacterium carotovorum subsp. carotovorum isolates from a recent outbreak on cabbage in Bosnia and Herzegovina. Pestic. Phytomed. (Belgrade) 2019;34:211–222. [Google Scholar]
- Raaijmakers J. M., Paulitz T. C., Steinberg C., Alabouvette C., Moënne-Loccoz Y. The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2009;321:341–361. [Google Scholar]
- Schlatter D., Fubuh A., Xiao K., Hernandez D., Hobbie S., Kinkel L. Resource amendments influence density and competitive phenotypes of Streptomyces in soil. Microb. Ecol. 2009;57:413–420. doi: 10.1007/s00248-008-9433-4. [DOI] [PubMed] [Google Scholar]
- She S., Niu J., Zhang C., Xiao Y., Chen W., Dai L., Liu X., Yin H. Significant relationship between soil bacterial community structure and incidence of bacterial wilt disease under continuous cropping system. Arch. Microbiol. 2017;199:267–275. doi: 10.1007/s00203-016-1301-x. [DOI] [PubMed] [Google Scholar]
- Siddiqui I. A., Shaukat S. S. Mixtures of plant disease suppressive bacteria enhance biological control of multiple tomato pathogens. Biol. Fertil. Soils. 2002;36:260–268. [Google Scholar]
- Wang G.-H., Liu J.-J., Yu Z.-H., Wang X.-Z., Jin J., Liu X.-B. Research progress of Acidobacteria ecology in soils. Biotechnol. Bull. 2016;32:14–20. (in Chinese) [Google Scholar]
- Wang Q., Garrity G. M., Tiedje J. M., Cole J. R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Zhang H., Sun L., Qi G., Chen S., Zhao X. Microbial community composition is related to soil biological and chemical properties and bacterial wilt outbreak. Sci. Rep. 2017;7:343. doi: 10.1038/s41598-017-00472-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Liu L., Yang J., Duan Y., Luo Y., Taherzadeh M. J., Li Y., Li H., Awasthi M. K., Zhao Z. The diversity of microbial community and function varied in response to different agricultural residues composting. Sci. Total Environ. 2020;715:136983. doi: 10.1016/j.scitotenv.2020.136983. [DOI] [PubMed] [Google Scholar]
- Wei H., Wang L., Hassan M., Xie B. Succession of the functional microbial communities and the metabolic functions in maize straw composting process. Bioresour. Technol. 2018;256:333–341. doi: 10.1016/j.biortech.2018.02.050. [DOI] [PubMed] [Google Scholar]
- Wei H., Yang M., Ke Y., Liu J., Chen Z., Zhao J., Zhao Y., Huang F., Yu L. Comparative physiological and transcriptomic profiles reveal regulatory mechanisms of soft rot disease resistance in Amorphophallus spp. Physiol. Mol. Plant Pathol. 2022;118:101807. [Google Scholar]
- Wu X., Yang M., Liu J.-N., Chen Z.-B., Wang D.-K., Zhao J.-R., Zhong Y., Wu D.-X., Yu L. Identification and evaluation of resistance of Amorphophallus spp. to Erwinia carotovora subsp. carotovora. Subtrop. Plant Sci. 2018;47:176–180. (in Chinese) [Google Scholar]
- Wu Z., Hao Z., Sun Y., Guo L., Huang L., Zeng Y., Wang Y., Yang L., Chen B. Comparison on the structure and function of the rhizosphere microbial community between healthy and root-rot Panax notoginseng. Appl. Soil Ecol. 2016;107:99–107. [Google Scholar]
- Xiaolong C., Lingling G., Xiaopeng D., Yongfeng Y., Jianwei W., Zhan Z., Yongzhan C., Feiyan H., Min Y., Wenjie T., Lei Y. Effects of Meloidogyne incognita on the fungal community in tobacco rhizosphere. Rev. Bras. Cienc. Solo. 2022;46:e0210127. [Google Scholar]
- Xie C.-H., Yokota A. Sphingomonas azotifigens sp. nov., a nitrogen-fixing bacterium isolated from the roots of Oryza sativa. Int. J. Syst. Evol. Microbiol. 2006;56:889–893. doi: 10.1099/ijs.0.64056-0. [DOI] [PubMed] [Google Scholar]
- Xu P., Wang H., Qin C., Li Z., Lin C., Liu W., Miao W. Analysis of the taxonomy and pathogenic factors of Pectobacterium aroidearum L6 using whole-genome sequencing and comparative genomics. Front. Microbiol. 2021;12:679102. doi: 10.3389/fmicb.2021.679102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Z., Huang F., Liu J., Ke Y., Wei H., Gao P., Qi Y., Yu L. A high trans-zeatin nucleoside concentration in corms may promote the multileaf growth of Amorphophallus muelleri. Front. Plant Sci. 2022;13:964003. doi: 10.3389/fpls.2022.964003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan M., Chen S., Huang T., Li B., Li N., Liu K., Zong R., Miao Y., Huang X. Community compositions of phytoplankton and eukaryotes during the mixing periods of a drinking water reservoir: dynamics and interactions. Int. J. Environ. Res. Public Health. 2020;17:1128. doi: 10.3390/ijerph17041128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M., Qi Y., Liu J., Wu Z., Gao P., Chen Z., Huang F., Yu L. Dynamic changes in the endophytic bacterial community during maturation of Amorphophallus muelleri seeds. Front. Microbiol. 2022;13:996854. doi: 10.3389/fmicb.2022.996854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y., Wang J., Zhao W., Cai X., Xu Y., Chen X., Yang M., Huang F., Yu L., He Y. Effect of bacterial wilt on fungal community composition in rhizosphere soil of tobaccos in tropical Yunnan. Plant Pathol J. 2022;38:203–211. doi: 10.5423/PPJ.OA.03.2022.0035. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.