DNA methylation analysis reveals the molecular characteristics of PCBM and may serve as a starting point for efforts to identify and target susceptibilities of these rare metastases.
Abstract
Metastases from primary prostate cancers to rare locations, such as the brain, are becoming more common due to longer life expectancy resulting from improved treatments. Epigenetic dysregulation is a feature of primary prostate cancer, and distinct DNA methylation profiles have been shown to be associated with the mutually exclusive SPOP-mutant or TMPRSS2-ERG fusion genetic backgrounds. Using a cohort of prostate cancer brain metastases (PCBM) from 42 patients, with matched primary tumors for 17 patients, we carried out a DNA methylation analysis to examine the epigenetic distinction between primary prostate cancer and PCBM, the association between epigenetic alterations and mutational background, and particular epigenetic alterations that may be associated with PCBM. Multiregion sampling of PCBM revealed epigenetic stability within metastases. Aberrant methylation in PCBM was associated with mutational background and PRC2 complex activity, an effect that is particularly pronounced in SPOP-mutant PCBM. While PCBM displayed a CpG island hypermethylator phenotype, hypomethylation at the promoters of genes involved in neuroactive ligand–receptor interaction and cell adhesion molecules such as GABRB3, CLDN8, and CLDN4 was also observed, suggesting that cells from primary tumors may require specific reprogramming to form brain metastasis. This study revealed the DNA methylation landscapes of PCBM and the potential mechanisms and effects of PCBM-associated aberrant DNA methylation.
Significance:
DNA methylation analysis reveals the molecular characteristics of PCBM and may serve as a starting point for efforts to identify and target susceptibilities of these rare metastases.
Introduction
Clinical outcomes for prostate cancer are highly variable, ranging from indolent tumors requiring no intervention to highly aggressive, metastatic disease. Concerted efforts to characterize somatic features of prostate cancer have identified early and mutually exclusive events such as TMPRSS2-ETS gene fusions (most frequently TMPRSS2-ERG) and SPOP somatic mutations in therapy-naïve localized tumors (1–3) and androgen receptor alterations in the metastatic setting under androgen deprivation therapy (4–7), among other frequent molecular alterations.
Beyond their genomic profiles, numerous studies have compared gene expression and DNA-methylation profiles of primary prostate cancers and benign tissue (8–10), and the distinction between primary tumors harboring SPOP mutations or TMPRSS2-ERG fusions is detectable at the epigenetic level (11). Tumors with these two common genetic alterations show distinct methylomes (11), with the activity of EZH2 in non–TMPRSS2-ERG cases implicated in driving this distinction (12). A hypermethylator phenotype has been described in common sites of prostate cancer metastasis, such as bone, lymph node, and liver, along with regulation of oncogenes by intergenic DNA methylation (13, 14). More recently, epigenetic stability was reported between metastases from different sites within the same patient (15). However, while improvements in treatment for prostate cancer have led to increased survival, it has also brought an increase in metastasis to rare locations such as the brain (16), and a deep molecular understanding of these is lacking.
We, therefore, analyzed DNA methylation data from a cohort of 42 patients with metastasis from primary prostate cancer to the brain, with both primary and metastatic samples available for 17 of these. Multiregion sampling permitted the evaluation of intratumoural epigenetic heterogeneity at the primary and metastatic sites. Furthermore, we examined whether the established DNA methylation features of prostate cancer are retained in this setting and sought insights into mechanisms driving metastasis to the brain.
Materials and Methods
Patient selection and tumor procurement
Tumor samples were collected from Pathology Departments in University and Cantonal Hospitals across Switzerland (Institute of Pathology, Bern; Institute of Neuropathology, Zurich; Institute of Medical Genetics and Pathology, Basel; Institute of Pathology, Aarau; Institute of Pathology, Münsterlingen Institute of Pathology, Liestal; Institute of Pathology, St. Gallen) and from the Departments of Pathology and Laboratory Medicine, and Urology, Emory University School of Medicine, Atlanta, GA. Inclusion criteria were defined as patients having available formalin-fixed paraffin-embedded (FFPE) blocks from confirmed central nervous system (CNS) or meningeal metastases of prostate carcinoma and, if available, from the matched primary tumor and normal tissue (Supplementary Table S1). All analyses were carried out following protocols approved by the Ethical Committee Bern (project ID: 2019-00328). No participant compensation was applied for the current study.
Study population
Prostate cancer is a cancer that only involves men, thus all subjects are male. There was no exclusion of subjects due to age, although prostate cancer is most often diagnosed in the sixth or seventh decade of life. Our cohort included samples from 43 male patients, with prostate cancer brain metastases (PCBM) tissue from 42. Only the primary tumor was available for patient P41 because the brain metastasis tissue did not pass quality controls for methylation analysis. Patients qualified for inclusion in this study if written consent or no documented refusal was available (Human Research Act, HRA, Swiss Confederation; Art. 34). We collected archived FFPE tissue from CNS (brain/spinal cord) and meningeal metastases. Most tumor samples corresponded to diagnostic biopsies (from prostate or CNS/dura), transurethral resections, or prostatectomy specimens. Primary tumors and metastases from patients P1, P32, P43, P44, P48, and P49 were taken from autopsy tissues. Additional diagnostic biopsies were available for patient P1. At least one metastatic sample was included from 42 patients (i.e., except P41). For patient P43, metastatic samples at several timepoints were collected. In addition, from 17 of 42 patients with metastatic tissue, primary tumor tissue was also available, including 5 patients (P1, P6, P9, P29, and P44) with primary tumor samples at multiple timepoints. In total, we analyzed 155 areas, which included 57 from primary tumors and 95 from metastases, two from normal prostates, and one from normal brain. All 155 selected areas underwent DNA methylation analysis and were integrated with whole-exome sequencing and targeted RNA sequencing (RNA-seq) data on the same samples, produced as described previously (17).
DNA extraction and Illumina EPIC methylation array
After deparaffinization, DNA was extracted from selected FFPE core biopsies (1 mm diameter) of matched tumor and normal tissue using the QIAamp DNA micro kit (Qiagen). Quality and quantity were determined by real-time PCR (Agilent NGS FFPE QC Kit, catalog no. G9700B) and Qubit Fluorometer (Invitrogen; Thermo Fisher Scientific Qubit fluorimeter, RRID:SCR_018095). Methylation profiling was performed using Infinium MethylationEPIC BeadChips (850K; catalog no. 20087706) as described previously (18). The Epigenomic Services from Diagenode (RRID:SCR_014807) carried out this assay.
EPIC methylation array processing
After whole-genome amplification and enzymatic fragmentation, the samples were hybridized to the BeadChip, and scanning was conducted with Illumina iScan (RRID:SCR_016388). Idat files were exported and analyzed using the minfi package in R (RRID:SCR_012830; 19). Probes associated with SNPs or with a detection P value > 0.01 in any sample were removed before analysis. Data were normalized using the functional normalization algorithm from the minfi package (19). Probes were annotated using the IlluminaHumanMethylationEPICanno.ilm10b2.hg19 package in Bioconductor (RRID:SCR_006442). Estimating tumor purity was done using the R package InfiniumPurify v2.0, and beta values were purity corrected using the R function adjustBetas (https://github.com/StaafLab/adjustBetas, latest commit 19a5bc9). Purity-corrected M values were derived from the corrected beta values using the B2M function from the R package ENmix v.1.31.02. Principal component analysis (PCA) was performed using the top 1% (8,038) most variable CpG sites in each set of samples being analyzed. Comparisons of the intragroup variation, defined as the within-group pairwise Euclidean distance based on their principal components (PC), were performed using Wilcoxon tests. Comparisons of the intergroup variation, as measured by pairwise Euclidean distance based on their PCs between samples of different groups, were performed using Wilcoxon tests.
Probe-level differential methylation analysis was performed for 803,841 CpG sites using limma (RRID:SCR_010943; ref. 20). Probes with |logFC| > 1.5 and FDR < 0.05 were considered differentially methylated (DM). DM regions (DMR) were called using DMRcate using the parameters “lambda = 500, C = 5” (21). Regions with ≥ 5 CpGs and a mean change in B value > |20%| and FDR < 0.05 were considered differentially methylated. DMRs were annotated using the annotatePeak function from ChIPSeeker (RRID:SCR_021322) and the TxDb.Hsapiens.UCSC.hg19.knownGene packages from Bioconductor (22). Enrichment for DM at different genomic regions was performed using Fisher exact test with Benjamini–Hochberg correction for multiple testing. Gene set enrichment analysis on the CpG level was performed using the ebGSEA algorithm implemented in the ChAMP package v.2.24.0 (RRID:SCR_012891; refs. 21, 23). Gene ontology analysis on promoter DMRs was performed using goregion from the missMethyl package v.1.28.0 (24).
Unsupervised hierarchical consensus clustering was performed using the ConsensusClusterPlus package (RRID:SCR_016954), 1,000 subsamplings, and ward.D2 linkage. The optimal number of clusters was selected using the elbow method applied to the plot of change in area under the cumulative density function curve (25).
Integration with published datasets
Data from 100 nonbrain prostate cancer metastases was kindly provided by Prof. Felix Feng (UCSF Helen Diller Family Comprehensive Cancer Center; ref. 13) and analyzed using the bsseq package (RRID:SCR_001072). Data were filtered to sites covered to at least 10X and present in the postfiltering EPIC array. CpGs were then annotated using the IlluminaHumanMethylationEPICanno.ilm10b2.hg19 package to analyze CpG island methylation.
Illumina 450k methylation data for 502 primary prostate cancers from The Cancer Genome Atlas (TCGA) cohort (RRID:SCR_003193; ref. 26) were downloaded as TCGA Prostate Adenocarcinoma (PRAD) cohort using TCGABiolinks on May 18, 2022 (RRID:SCR_017683; refs. 27, 28).
Before the analysis of promoter DMR methylation levels, the three datasets were merged using the CpG IDs and batch corrected using the removeBatchEffect function from the limma package. The batch-corrected data were then reduced to the CpGs included in the GABRB3, CLDN8, and CLDN4 promoter DMRs using subsetByOverlaps from the GenomicRanges package (RRID:SCR_000025; ref. 29). Levels of methylation at the CpG level, and after averaging the level of methylation at each DMR, were compared between datasets using the Wilcoxon test.
Data availability
The data generated in this study are publicly available in The European Genome-phenome Archive (EGA; EGAD00010002372).
Results
Interpatient and intrapatient epigenetic heterogeneity in PCBM
Using the Illumina Infinium EPIC array, we acquired DNA methylation data from 155 samples from 43 patients (17 patients with PCBM with matched primary tumor tissue, 25 with PCBM only, and 1 with primary tumor only). The 155 samples comprised 95 regions from brain metastases, 57 regions from primary tumors, two normal prostate, and one from normal brain tissue (Supplementary Table S1).
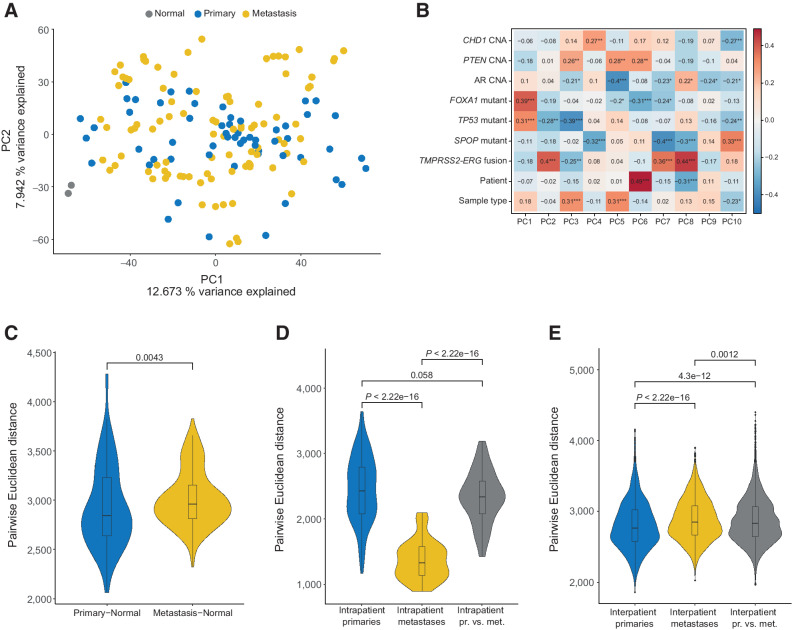
We performed PCA using the 1% of most variably methylated CpG sites (8,038 CpGs) across all primary and metastatic samples (Supplementary Table S2). No obvious separation of primary and metastatic samples was revealed by PC1 and PC2 (Fig. 1A), suggesting other factors contributing to the observed variance. Correlating the variation with the mutation status of prostate cancer driver genes, the presence of mutations in TP53 and/or FOXA1 and/or TMPRSS2-ERG fusions showed significant correlations with the calculated eigenvectors for PCs 1 and 2 (Fig. 1B). PC components 3 to 10 correlated with other features such as SPOP mutations, sample type (metastasis or primary tumor), and other genomic aberrations (Fig. 1B). To assess whether metastatic samples were more dissimilar to normal prostate tissues than primary samples, we calculated the pairwise distance between the tumor samples and the normal prostate reference tissues. Indeed, we found that the metastatic samples were slightly farther from the normal prostate samples than the primary samples were (P = 0.0043, Wilcoxon test; Fig. 1C). Interestingly, the intrapatient variation between metastases was significantly lower compared with the variation between primary tumors or between metastases and primary tumors (P < 2.22 × 10−16, Wilcoxon test; Fig. 1D). The opposite was true for interpatient variation, where metastases showed modestly higher variation compared with primary tumors (P < 2.22 × 10−16, Wilcoxon test; Fig. 1E).
Figure 1.
The methylome of brain metastases from prostate cancer is largely inherited from primary tumors and is driven by genomic background. A, PCA using the 1% most variably methylated CpG sites from the Illumina EPIC array (8,038 sites). B, Spearman correlation between eigenvectors of PCs 1–10, with sample type (i.e., primary tumor vs. metastasis), patient, and mutational status. *, P < 0.05; **, P < 0.01; ***, P < 0.001. C, Euclidean distance between primary samples and normal prostate samples and between metastatic and normal samples. D, Euclidean distance between primary samples within each patient, metastatic samples within each patient, and between primary and metastatic samples within each patient. E, As in D, but comparison done between patients. PCBM primary, n = 57; PCBM metastasis, n = 95; normal prostate, n = 2.
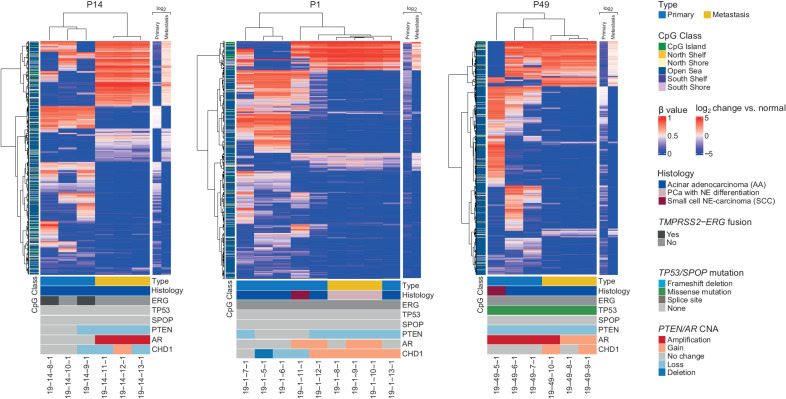
To obtain a focused view of intrapatient heterogeneity, we performed unsupervised hierarchical clustering, using the top 1% most variably methylated CpGs, on a per-patient basis for the 17 patients for which primary and metastatic samples were available. This highlighted the separation between primary and metastatic samples within each patient, as these clustered separately in 12 of 16 cases with three or more samples (Fig. 2; Supplementary Fig. S1). We further observed that the top 1% of most variably methylated CpGs tended to be hypomethylated in the metastases compared with normal prostate. In some cases (such as P4 and P32), the metastatic samples harbored large blocks of hypomethylated CpG sites. However, the opposite (P9) or mixed patterns were observed in some cases (Supplementary Fig. S1). Interestingly, the only two samples annotated as small cell neuroendocrine (NE) carcinoma, in P1 and P49, clustered separately from the other samples from the same patient (Fig. 2), even though they are genetically related as they shared a large fraction of clonal somatic mutations (17). We further examined the top 1% most variably methylated CpG sites in the 18 cases with more than one metastatic sample and again observed the tendency of global hypomethylation among these samples (Supplementary Fig. S2). Some of these cases, for example, P40, P45, and P47, had larger blocks of hypermethylated CpGs than the normal samples.
Figure 2.
Intrapatient variation between selected primary and metastatic samples. The samples from each patient were clustered using the 1% most variably methylated CpG sites from the Illumina EPIC array (8,038 sites). Heatmap shows β values for primary and metastatic samples. Class of CpG (in relation to CpG island) is shown in blue/green heatmap on the left. Average log2-fold change of the primary tumors and metastases compared with normal prostate tissue is shown on the right. Sample type (primary or metastatic), histology, and genetic alterations from whole-exome sequencing are annotated below.
These data suggest that, while primary prostate cancer samples have epigenomes that distinguish them from normal prostate, metastatic samples acquire additional DNA methylation changes, further separating them epigenetically from normal tissue and primary cancer. The smaller distance between metastatic samples suggests reduced epigenetic heterogeneity within PCBM compared with primary prostate cancer.
DNA methylation changes associated with early driver mutations distinguish PCBM
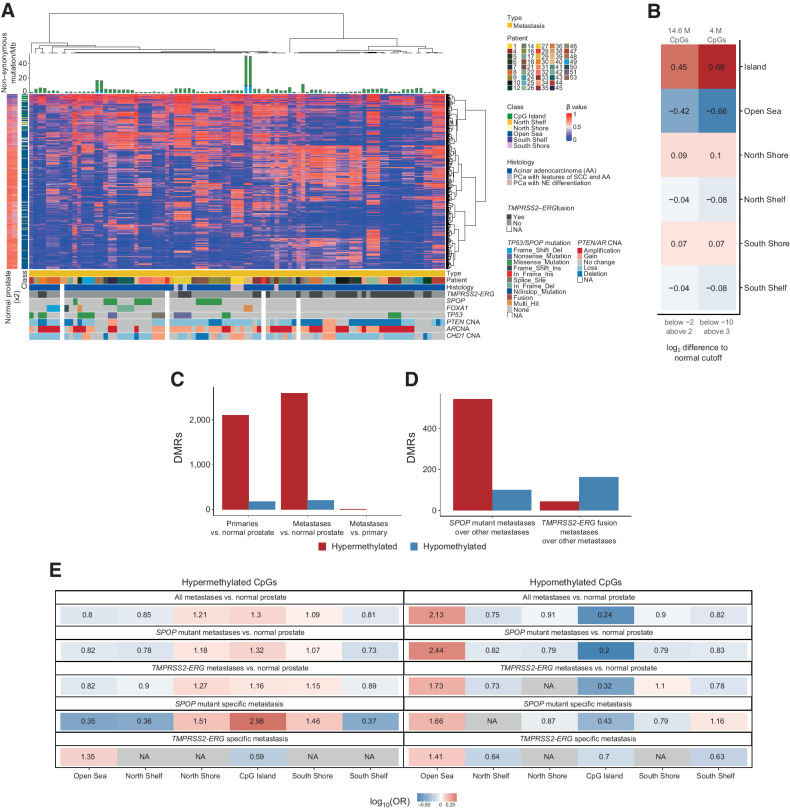
It has previously been reported that prostate cancer with and without the TMPRSS2-ERG fusion show distinct methylomes (13), and TMPRSS2-ERG fusion-negative prostate cancers acquire DNA hypermethylation dependent on EZH2 (12). However, whether the epigenetic distinction resulting from these mutually exclusive driver events persists following metastasis has not been addressed. Using the 95 metastatic cases annotated with genotypic information in our previously published study (17), we performed unsupervised hierarchical consensus clustering using the top 1% of most variably methylated CpG sites across the metastatic samples. This separated most of the TMPRSS2-ERG fused samples from the nonfused samples, most of which harbored mutations in SPOP, FOXA1, or TP53 (Fig. 3A). Rather than separating by sample site, this separation by mutational background was also observed when clustering all metastatic and primary samples (Supplementary Fig. S3). We calculated the mean methylation change of the SPOP-mutant, TP53-mutant, or TMPRSS2-ERG fusion samples compared with normal prostate tissue. While the TP53 mutants had overall more CpG sites that were significantly different from the normal tissue (either hypomethylated or hypermethylated), we observed the previously reported hypermethylator phenotype in the SPOP-mutant samples (Supplementary Fig. S4; ref. 13). We observed that strongly hypermethylated/hypomethylated CpG sites in tumors (compared with normal tissue) were more strongly associated with CpG islands than weakly hypermethylated/hypomethylated CpG sites, suggesting the potential to regulate gene expression (Fig. 3B). Strongly hypomethylated CpGs were more frequent than strongly hypermethylated CpGs in all our tumor samples (Supplementary Fig. S5), indicating global hypomethylation, which was also observed in other cancer types (30).
Figure 3.
SPOP-mutant and TMPRSS2-ERG fusion PCBM have distinct methylomes. A, Unsupervised hierarchical consensus clustering of metastatic samples from 42 patients. Samples from each patient were clustered using 1% most variably methylated CpG sites from the Illumina EPIC array (8,038 sites). The heatmap shows the β values. Class of CpG (in relation to CpG island) is shown in blue/green heatmap on the left, along with methylation status of CpG sites in normal prostate tissue. Mutational burden (mutations/Mb) is shown in the barplot on top. Sample type (primary or metastatic), histology, and genetic alterations from whole-exome sequencing are annotated below. B, Enrichment in CpG types among the variably methylated CpGs, showing CpGs with log2-fold difference to mean β value of normal prostates below −2 and above 2 (left column), and below −10 and above 3 (right column). Values and colors indicate Pearson correlation coefficients. P values were < 2.22 × 10−16 in all cases. C, Number of DMRs in primary cancers compared with normal prostate and metastases compared with normal prostate (DMR ≥ 5 CpGs, >|20%| change in methylation, q < 0.05). D, Number of DMRs in metastases with SPOP mutation versus samples with neither SPOP mutation nor TMPRSS2-ERG fusion and metastases with TMPRSS2-ERG fusion versus samples with neither SPOP mutation nor TMPRSS2-ERG fusion (DMR ≥ 5 CpGs, >|20%| change in methylation). E, Enrichment for differentially methylated CpGs between all metastases and normal prostate tissue, SPOP-mutant metastases and normal prostate tissue, TMPRSS2-ERG fusion metastases, and normal prostate tissue, SPOP-mutant metastases compared with non–TMPRSS2-ERG/SPOP-mutant metastases, and TMPRSS2-ERG fusion metastases compared with non–TMPRSS2-ERG/SPOP-mutant metastases. Values indicate the log-OR from Fisher exact tests.
We used limma to call DM CpG sites (|log2FC| > 1.5 and q < 0.05) and DMRcate to call DMRs (|mean difference in β value| > 20%, q < 0.05, ≥ 5 CpGs) in all primary prostate cancers against normal prostate tissue, and in metastases against normal prostate tissue. In contrast to the global hypomethylation of CpG sites, we detected 2,102 DMRs hypermethylated in the primaries compared with the normals and 180 hypomethylated DMRs (Supplementary Table S3). The metastases showed a greater acquisition of DMRs compared with normals (2,596 hypermethylated and 211 hypomethylated; Supplementary Table S4), but only a further 15 hypermethylated DMRs in comparison with the primary prostate cancer tissue (Fig. 3C; Supplementary Table S5). Extending this analysis to look specifically at methylation changes occurring in the SPOP-mutant or TMPRSS2-ERG fusion metastases, in comparison with normal prostate tissue, beyond those seen in metastases with neither alteration, reflected the observations from the global methylation analysis. While the SPOP-mutant metastases showed hypermethylation at 542 regions and hypomethylation at 101 regions, the TMPRSS2-ERG fusion metastases had only 44 hypermethylated and 162 hypomethylated regions in comparison with metastases lacking either mutation (Fig. 3D; Supplementary Tables S6 and S7).
Considering the distribution of the DM CpGs in relation to CpG islands, compared with the overall distribution of analyzed CpGs, we found enrichment for hypermethylated CpGs at CpG islands in the comparison of metastases with normal prostate samples [OR = 1.30, 95% confidence interval (CI) = 1.26–1.33, q < 2.22 × 10−16, Fisher exact test; Fig. 3E]. This enrichment of hypermethylated CpG islands was very similar when comparing SPOP-mutant metastases and normal prostate (OR = 1.32, 95% CI = 1.30–1.34, q < 2.22×10−16, Fisher exact test), but was weaker in the TMPRSS2-ERG fusion metastases compared with normal prostate (OR = 1.16, 95% CI = 1.13–1.19, q < 2.22×10−16, Fisher exact test). However, the enrichment was more pronounced when examining the hypermethylated CpGs detected in SPOP mutant–specific metastases (i.e., metastases with SPOP mutations vs. metastases without SPOP mutations nor TMPRSS2-ERG fusions; OR = 2.98, 95% CI = 2.89–3.08, q < 2.22 × 10−16, Fisher exact test). The same increase in enrichment for hypermethylation at CpG islands was not observed in the TMPRSS2-ERG fusion–specific methylation changes (OR = 0.59, 96% CI = 0.54–0.65, q < 2.22 × 10−16, Fisher exact test; Fig. 3E). Similarly, when compared with normal prostate, while we found a depletion of hypomethylated CpGs at CpG islands in the metastases (OR = 0.24, 95% CI = 0.22–0.26, q < 2.22 × 10−16, Fisher exact test; Fig. 3E), SPOP-mutant metastases showed a stronger depletion of hypomethylated CpG island CpGs than TMPRSS2-ERG fusion metastases (SPOP OR = 0.20, 95% CI = 0.18–0.21, q < 2.22 × 10−16, TMPRSS2-ERG, OR = 0.32, 95% CI = 0.30–0.34, q < 2.22 × 10−16, Fisher exact tests).
Our results showed that early driver mutations in PCBM are associated with distinct methylation changes. In particular, the enrichment of hypermethylation and depletion of hypomethylation in CpGs at CpG islands in SPOP-mutant metastases suggest a role in transcriptional regulation.
Activity of the PRC2 complex drives PCBM-associated DNA methylation changes
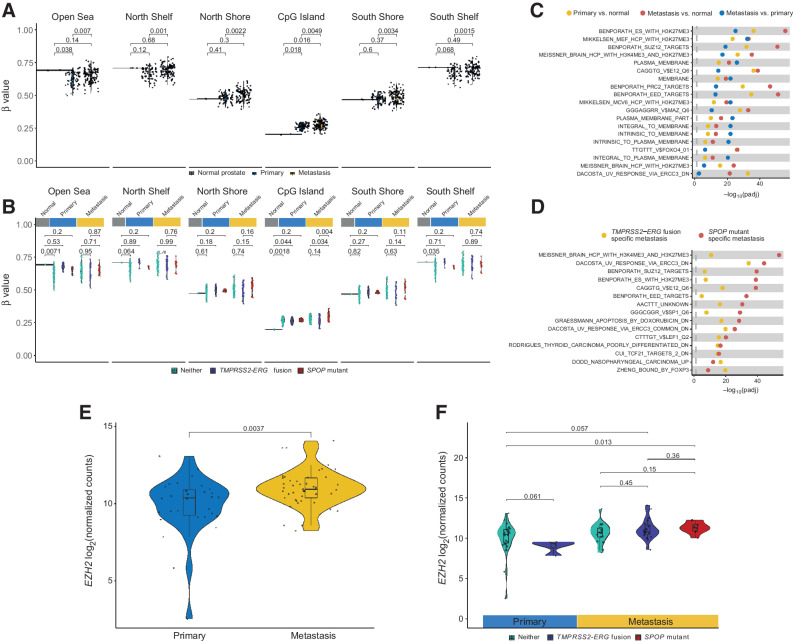
Examining the results of the DMR analysis, we found hypermethylation of the promoters of genes such as SMARCA2 in the metastases, suggesting possible transcriptional downregulation (Supplementary Fig. S6), in keeping with reports of its role in the progression of metastatic castration-resistant prostate cancer (mCRPC; refs. 31, 32). This hypermethylation was strongest in SPOP-mutant metastases (Supplementary Fig. S6). We, therefore, examined genome-wide methylation occurring at and around CpG islands and in open sea regions (Fig. 4A). We found a significant reduction in global methylation at open sea regions in the primary samples, in keeping with the well-described phenomenon of global hypomethylation in cancer (P = 0.038, Wilcoxon test; ref. 33). However, this difference did not persist in the metastatic samples (P = 0.14, Wilcoxon test), suggesting the level of methylation at open sea regions may have slightly increased in the PCBM. Indeed, when examining CpG island–associated CpGs, we observed a significant increase in overall methylation in the primary and PCBM samples compared with the normal prostate tissues (P = 0.018 and P = 0.016, respectively, Wilcoxon tests). However, there was also a marked increase in CpG island methylation in the PCBM compared with the primary samples (P = 0.0049, Wilcoxon test). Interestingly, at CpG island shores and shelves (2 and 4 kb from the center of a CpG island, respectively), no significant difference was found between the tumor and normal samples, while PCBM were hypermethylated compared with the primaries (North shelf P = 0.001, North shore P = 0.0022, South shore P = 0.0034, South shelf P = 0.0015, Wilcoxon tests). These data suggest hypermethylation of CpG islands and surrounding regions is a characteristic of the metastatic samples in our cohort.
Figure 4.
Global CpG island hypermethylation in PCBM is associated with the PRC2 complex. A, Mean methylation of CpG sites in relation to CpG islands in normal prostate, primary, and metastatic samples. P values computed from Wilcoxon tests. B, Mean methylation of CpG sites in relation to CpG islands in normal prostate, primary, and metastatic samples, stratified by TMPRSS2-ERG fusion, SPOP mutation, or neither. P values computed from Wilcoxon tests. C, Gene set enrichment analysis on DM CpG sites between primary tumors and normal prostates, metastases and normal prostates, and metastases and primary tumors, using curated gene sets from MSigDB. Pathways shown are from the union of the top 10 DM pathways from each of the three comparisons. D, Same as C but showing the union of the 10 most DM pathways between SPOP-mutant or TMPRSS2-ERG fusion metastases against metastases with neither alteration. E, Expression of EZH2 in primary tumors and metastases from targeted RNA-seq. P values computed from Wilcoxon tests. F, Same as E, with samples stratified by TMPRSS2-ERG fusion, SPOP mutation, or neither.
As we had identified distinct methylomes associated with the SPOP-mutant and TMPRSS2-ERG fusion metastases, we asked whether this global hypermethylation of CpG islands was more strongly associated with one of these genetic backgrounds. We found that CpG island methylation was significantly higher in the SPOP-mutant metastases than TMPRSS2-ERG fusion PCBM or PCBM with neither an SPOP mutation nor the TMPRSS2-ERG fusion (P = 0.004 and P = 0.034, respectively, Wilcoxon tests; Fig. 4B). The TMPRSS2-ERG fusion PCBM showed no significant difference in CpG island methylation compared with PCBM with neither the fusion nor SPOP mutation (P = 0.14, Wilcoxon test). Together, these findings suggest that CpG island hypermethylation is, in fact, predominantly a feature of the SPOP-mutant PCBMs.
We asked whether this SPOP mutant–specific CpG island hypermethylation was unique to PCBM or whether it was also observed in metastases to other locations (i.e., nonbrain metastases) that harbored SPOP mutations. Using a previously published dataset (13), we examined the methylation status of the same CpG islands in TMPRSS2-ERG fusion or SPOP-mutant metastases. While this dataset only included five SPOP-mutant metastases, we found no significant difference in the level of CpG island methylation between these and the TMPRSS2-ERG fusion nonbrain metastases (P = 0.29, Wilcoxon test; Supplementary Fig. S7). This may suggest that the SPOP mutant–specific CpG island hypermethylation is a specific characteristic of PCBM.
We performed gene set enrichment analysis using the CpG-level DM data to understand which processes might drive the aberrant methylation. Comparing primaries against normal, metastases against normal, and metastases against primaries, highlighted regions of known histone 3 lysine 27 trimethylation (H3K27me3), as well as targets of the PRC2 complex, which catalyzes the formation of this histone modification complex, and its subunits (EZH1/2, SUZ12, and EED; Fig. 4C). While the enrichment for H3K27me3 regions and PRC2 targets (e.g., Benporath_ES_with_H3K27me3 and Benporath_PRC2_targets) was significant for all three comparisons, it was most pronounced in the metastasis versus normal comparison, and this pattern persists for all examined gene sets related to its subunits examined. Likewise, when examining the methylation changes specifically associated with either the TMPRSS2-ERG fusion or SPOP-mutant backgrounds, in comparison to metastases with neither alteration, the same PRC2-associated pathways were enriched in the SPOP-mutant metastases and not enriched, or enriched to a far lesser extent, in the TMPRSS2-ERG fusion metastases (Fig. 4D). Using targeted RNA-seq data from these samples, we observed a significant increase in the expression of EZH2, a component of PRC2, in the metastases compared with primary samples (P = 0.0037, Wilcoxon test; Fig. 4E). Furthermore, when stratifying the samples by TMPRSS2-ERG fusion or SPOP mutation, we found that only the SPOP-mutant metastases showed a slight numerical increase in EZH2 expression compared with metastases with neither TMPRSS2-ERG fusion nor SPOP mutation (Fig. 4F).
Together, these findings demonstrate that CpG island hypermethylation is prevalent in PCBM and even higher in SPOP-mutant metastases. Our results suggest that aberrant methylation in PCBM is driven by PRC2 activity and may be associated with H3K27me3.
PCBM DNA methylation changes may suggest mechanisms driving PCBM
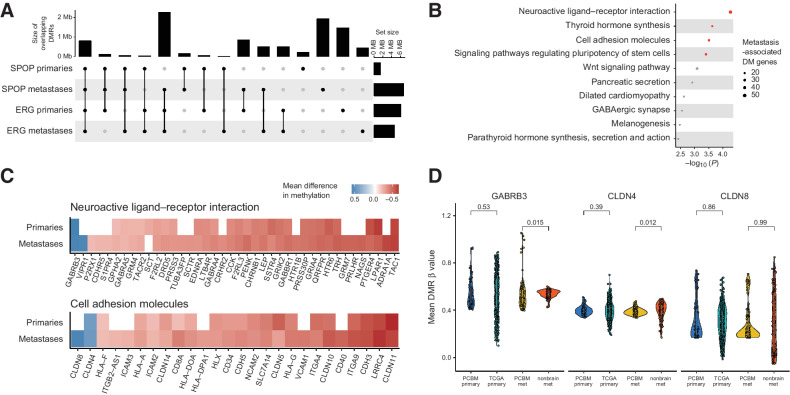
While we have, so far, focused on the different DNA methylation landscapes of PCBM with different mutational drivers, it is clear that these samples share the crucial characteristic of having metastasized to the brain and so may share epigenetic alterations associated with this. Indeed, overlapping the DMRs called in the SPOP-mutant or TMPRSS2-ERG fusion primary and metastatic samples confirmed a substantial overlap of DMRs (Fig. 5A). We, therefore, selected DMRs from the comparison of all metastases against the normal prostate samples falling within 1,500 bp of transcription start site, identified their corresponding genes, and performed a gene ontology analysis using the Kyoto Encyclopedia of Genes and Genomes pathways, to identify the aberrantly methylated biological pathways that may permit, or drive, metastasis to the brain. This highlighted the neuroactive ligand–receptor interaction, thyroid hormone synthesis, cell adhesion molecules, and pluripotency of stem cells gene sets as the significantly enriched pathways (q < 0.05, overrepresentation test, Wallenius’ noncentral hypergeometric distribution; Fig. 5B). Interestingly, the neuroactive ligand–receptor interaction gene set was identified in a transcriptomic analysis of brain-specific breast cancer metastases (34), while cell adhesion molecules play an important role in cancer migration, immune response, and metabolism (35). Moreover, thyroid hormones (36, 37) and the pluripotency of stem cells (38) may play a role in the progression of prostate cancer. We further examined the genes in these sets. While their associated promoter DMRs were frequently hypermethylated, we found hypomethylation of key genes in these pathways, such as GABRB3, VIPR1, CLDN8, and CLDN4 (Fig. 5C). As promoter hypomethylation may be associated with increased transcription, this is in line with studies showing upregulation of these genes in prostate cancer, other primary cancers, and metastases (34, 39, 40). Notably, the methylation status of the CpGs associated with these genes was similar in normal brain tissue to normal prostate tissue (Supplementary Fig. S8), suggesting that these hypomethylation events may be specific to prostate cancer and PCBM. To address this, we examined the CpGs failing in the hypomethylated DMRs at the promoters of GABRB3, CLDN8, and CLDN4 in the dataset of nonbrain metastases (13) and TCGA cohort of primary prostate cancers (CpGs associated with the promoter of VIPR1 were not present in the external datasets; ref. 28). In PCBM, the promoters of GABRB3 and CLDN4 were significantly hypomethylated compared with the nonbrain metastases (P = 0.015 and P = 0.012, respectively, Wilcoxon test; Fig. 5D. While methylation of the promoter of CLDN8 varied in all samples, there was a noticeable subpopulation of PCBM with hypomethylation of this promoter (Fig. 5D). We observed some variation at the CpG level in these comparisons (Supplementary Fig. S9). However, most CpGs (present in all three datasets) supported the above results, and some showed even more striking differences between PCBM and nonbrain metastases (Supplementary Fig. S9). Interestingly, there was no difference in methylation status at these loci when comparing primary samples between cohorts (P < 0.05).
Figure 5.
PCBM DNA methylation changes may suggest mechanisms driving PCBM. A, Overlap between DMRs in SPOP-mutant primaries, SPOP-mutant metastases, TMPRSS2-ERG fusion primaries, and TMPRSS2-ERG fusion metastases, compared with normal prostates. Dots and lines show sets being intersected, bar plots on top show the intersection size (Mb), and bar plots on the right show the size of set (Mb). B, Gene ontology analysis on genes with promoter-associated DMRs in PCBM compared with normal prostate tissue. C, Heatmap showing mean difference in methylation level at DM promoters of genes in the neuroactive ligand–receptor interaction and cell adhesion molecules gene sets for primary tumors and metastases, compared with normal prostates. White, the absence of a DMR at a given promoter. D, Mean level of methylation at promoter DMRs of GABRB3, CLDN4, and CLDN8 in primary samples from the PCBM cohort, TCGA primary prostate cancers, metastatic samples from the PCBM cohort, and nonbrain metastases from Zhao and colleagues (13). P values computed from Wilcoxon tests. PCBM primary, n = 57; TCGA primary, n = 502; PCBM metastasis, n = 95; nonbrain metastasis, n = 100.
These data suggest that there are consistencies in the DNA methylation patterns of PCBM. These are related to the methylation status of genes in the neuroactive ligand–receptor and cell adhesion molecules pathways implicated in other cancers. Our findings suggest that methylation changes affecting these genes may also be specific to brain metastasis in prostate cancer.
Discussion
Here, we present the DNA methylation landscapes of a cohort of 42 patients with brain metastases from prostate cancer, complementing our recent study on the genetic landscapes of these patients (17). Our findings highlight the interpatient and intrapatient epigenetic heterogeneity present in PCBM, demonstrate that DNA methylation landscapes associated with early driver genetic events persist in PCBM, and provide potential explanations for the mechanisms underlying PCBM-associated DNA methylation patterns.
Epigenetic subtypes of advanced prostate cancers have been identified (13, 41), and epigenetic stability is observed between metastases at different sites within a patient (15). With this, we have shown that epigenetic patterns linked to genetic driver alterations persist through the process of metastasis to the brain, supporting the idea that epigenetic stability may be exploited in the effective, systematic treatment of mCRPC, even when involving rare metastatic sites such as the brain. Furthermore, the multiregion sampling employed in this study allowed us to examine intrametastatic epigenetic heterogeneity at fine resolution. Interestingly, the methylation landscape of intrapatient brain metastases was relatively homogeneous compared with intrapatient primary tumors, potentially suggesting clonal selection in the metastatic process.
We identified salient characteristics of PCBM, particularly global hypermethylation affecting CpG islands, which was most prominent in PCBM with SPOP mutations, and appears to be linked to the activity of the PRC2 complex. PRC2-associated aberrant DNA methylation is well-studied characteristic of advanced prostate cancer. They have been linked to prostate cancer progression using single-cell transcriptomics (42) and are particularly prevalent in TMPRSS2-ERG fusion-negative prostate cancer (12). However, the relevance of PRC2-associated aberrant DNA methylation events in metastatic prostate cancers is less well studied. Although the number of normal prostate samples used in this study was limited, our findings related to PRC2 activity in PCBM suggest that findings from studies on primary prostate cancers hold true in this specific metastatic setting and again reinforce the importance of early driver mutations in shaping the DNA methylation landscape of metastatic prostate cancer.
PRC2-associated aberrant DNA methylation was a characteristic of this dataset and was associated with the two different genetic backgrounds on which we focused. However, examining the DNA methylation changes consistent across the metastatic samples provided some interesting insight into how DNA methylation changes, and hypomethylation, in particular, might promote brain metastases, as these changes appeared to be specific to PCBM when compared with nonbrain metastases. While transcriptomic data were lacking from this study, overexpression of neuroactive ligand receptors (GABRB3) has previously been linked to brain metastases from other cancers (34), possibly suggesting that metastasizing cells from disparate primary tumors may require similar reprogramming to establish brain metastases.
The study represents an initial characterization of the DNA methylation features of PCBM. Future work will focus on elucidating whether CpG island hypermethylation, driven by the activity of PRC2 or hypomethylation of genes such as GABRB3, CLDN4, and CLDN8, confer brain-specific metastatic potential to prostate cancer cells (39, 40, 43). Integration of these data with transcriptomics from PCBM will improve understanding of how epigenetically driven transcriptional changes may drive PCBM and provide insights into therapeutic strategies for the systematic treatment of mCPRC, encompassing less common metastatic sites such as the brain.
Supplementary Material
Supplementary Figures
Supplementary Tables
Acknowledgments
This study was supported by an NCI (NIH) grant P50 CA211024 (M.A. Rubin), Swiss Personalized Health Network grant SOCIBP (H. Moch, M.A. Rubin), and the Swiss Cancer Research Foundation grants KFS-4982-02-2020 (M.A. Rubin), KFS-4988-02-2020-R (S. Piscuoglio), and KFS-4543-08-2018 (C.K.Y. Ng). The Prof. Dr. Max Cloëtta Foundation supports S. Piscuoglio. A. Rodriguez-Calero was supported by a Prostate Cancer Foundation (PCF) Young Investigator award.
The authors would like to thank Mariana Ricca from the Department of Biomedical Research (University of Bern) for expert assistance in editing and preparing the article for submission and the Translational Research Unit (TRU) of the Institute of Pathology (University of Bern) for technical support.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
L. Bubendorf reports grants and personal fees from Janssen, AstraZeneca, Bayer, Takeda, Novartis, Thermo Fisher Scientific, and Systems Oncology outside the submitted work. H. Moch reports grants from Roche and personal fees from AstraZeneca, Merck, Janssen, Tecan, and Bayer outside the submitted work. F.Y. Feng reports personal fees from Bluestar Genomics, Astellas, Foundation Medicine, Exact Sciences, Tempus, POINT Biopharma, Janssen, Bayer, Myovant, Roivant, Bristol Meyers Squibb, Novartis, and POINT Biopharma; personal fees and other support from SerImmune and Artera outside the submitted work. S. Gillessen reports personal fees from ProteoMedix, Amgen, MSD, AstraZeneca, and other support from MSD Merck Sharp&Dome, Bayer, Roche, Pfizer, Pfizer, Telixpharma, Amgen, Amgen, Bristol-Myers Squibb, AAA International SA, Orion, Silvio Grasso Consulting, Tolremo, WebMD-Medscape, AstraZeneca, Myriad Genetic, MSD, and PeerVoice outside the submitted work; in addition, S. Gillessen has a patent for WO2009138392 issued. C.K.Y. Ng reports grants from Swiss Cancer League during the conduct of the study and personal fees from Repare Therapeutics USA outside the submitted work. S. Piscuoglio reports grants from Swiss Cancer Research Foundation and The Prof. Dr. Max Cloëtta Foundation during the conduct of the study. No disclosures were reported by the other authors.
Authors' Contributions
J. Gallon: Conceptualization, data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. A. Rodriguez-Calero: Conceptualization, data curation, investigation, visualization, methodology, writing–original draft, project administration, writing–review and editing. A. Benjak: Data curation, software, formal analysis, visualization, writing–review and editing. D. Akhoundova: Resources, funding acquisition. S. Maletti: Data curation, formal analysis. U. Amstutz: Resources, funding acquisition. E. Hewer: Resources, funding acquisition. V. Genitsch: Resources, funding acquisition. A. Fleischmann: Resources, validation. E.J. Rushing: Resources, validation. R. Grobholz: Resources, validation. I. Fischer: Resources. W. Jochum: Resources. G. Cathomas: Resources. A.O. Osunkoya: Resources. L. Bubendorf: Resources. H. Moch: Resources. G. Thalmann: Resources. F.Y. Feng: Resources, data curation, supervision. S. Gillessen: Resources. C.K.Y. Ng: Conceptualization, data curation, software, formal analysis, supervision, investigation, methodology, writing–original draft, project administration, writing–review and editing. M.A. Rubin: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing. S. Piscuoglio: Conceptualization, resources, supervision, funding acquisition, investigation, writing–original draft, project administration, writing–review and editing.
References
- 1. Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun X-W, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644–8. [DOI] [PubMed] [Google Scholar]
- 2. Barbieri CE, Baca SC, Lawrence MS, Demichelis F, Blattner M, Theurillat J-P, et al. Exome sequencing identifies recurrent SPOP, FOXA1, and MED12 mutations in prostate cancer. Nat Genet 2012;44:685–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fraser M, Rouette A. Prostate cancer genomic subtypes. Adv Exp Med Biol 2019;1210:87–110. [DOI] [PubMed] [Google Scholar]
- 4. Robinson D, Van Allen EM, Wu Y-M, Schultz N, Lonigro RJ, Mosquera J-M, et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015;162:454. [DOI] [PubMed] [Google Scholar]
- 5. Quigley DA, Dang HX, Zhao SG, Lloyd P, Aggarwal R, Alumkal JJ, et al. Genomic hallmarks and structural variation in metastatic prostate cancer. Cell 2018;175:889. [DOI] [PubMed] [Google Scholar]
- 6. van Dessel LF, van Riet J, Smits M, Zhu Y, Hamberg P, van der Heijden MS, et al. The genomic landscape of metastatic castration-resistant prostate cancers reveals multiple distinct genotypes with potential clinical impact. Nat Commun 2019;10:5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceder Y, Bjartell A, Culig Z, Rubin MA, Tomlins S, Visakorpi T. The molecular evolution of castration-resistant prostate cancer. Eur Urol Focus 2016;2:506–13. [DOI] [PubMed] [Google Scholar]
- 8. Skvortsova K, Masle-Farquhar E, Luu P-L, Song JZ, Qu W, Zotenko E, et al. DNA hypermethylation encroachment at CpG island borders in cancer is predisposed by H3K4 monomethylation patterns. Cancer Cell 2019;35:297–314. [DOI] [PubMed] [Google Scholar]
- 9. Saghafinia S, Mina M, Riggi N, Hanahan D, Ciriello G. Pan-cancer landscape of aberrant DNA methylation across human tumors. Cell Rep 2018;25:1066–80. [DOI] [PubMed] [Google Scholar]
- 10. Yu YP, Ding Y, Chen R, Liao SG, Ren B-G, Michalopoulos A, et al. Whole-genome methylation sequencing reveals distinct impact of differential methylations on gene transcription in prostate cancer. Am J Pathol 2013;183:1960–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. The Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell 2015;163:1011–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Börno ST, Fischer A, Kerick M, Fälth M, Laible M, Brase JC, et al. Genome-wide DNA methylation events in TMPRSS2-ERG fusion-negative prostate cancers implicate an EZH2-dependent mechanism with miR-26a hypermethylation Cancer Discov 2012;2:1024–35. [DOI] [PubMed] [Google Scholar]
- 13. Zhao SG, Chen WS, Li H, Foye A, Zhang M, Sjöström M, et al. The DNA methylation landscape of advanced prostate cancer. Nat Genet 2020;52:778–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wu A, Cremaschi P, Wetterskog D, Conteduca V, Franceschini GM, Kleftogiannis D, et al. Genome-wide plasma DNA methylation features of metastatic prostate cancer. J Clin Invest 2020;130:1991–2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, et al. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med 2013;5:169ra10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sacks P, Rahman M. Epidemiology of brain metastases. Neurosurg Clin N Am 2020;31:481–8. [DOI] [PubMed] [Google Scholar]
- 17. Rodriguez-Calero A, Gallon J, Akhoundova D, Maletti S, Ferguson A, Cyrta J, et al. Alterations in homologous recombination repair genes in prostate cancer brain metastases. Nat Commun 2022;13:2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gallon J, Coto-Llerena M, Ercan C, Bianco G, Paradiso V, Nuciforo S, et al. Epigenetic priming in chronic liver disease impacts the transcriptional and genetic landscapes of hepatocellular carcinoma. Mol Oncol 2021;16:665–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, et al. Minfi: a flexible and comprehensive bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 2014;30:1363–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peters TJ, Buckley MJ, Chen Y, Smyth GK, Goodnow CC, Clark SJ. Calling differentially methylated regions from whole genome bisulphite sequencing with DMRcate. Nucleic Acids Res 2021;49:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Yu G, Wang L-G, He Q-Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 2015;31:2382–3. [DOI] [PubMed] [Google Scholar]
- 23. Tian Y, Morris TJ, Webster AP, Yang Z, Beck S, Feber A, et al. ChAMP: updated methylation analysis pipeline for illumina BeadChips. Bioinformatics 2017;33:3982–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from illumina's HumanMethylation450 platform. Bioinformatics 2016;32:286–8. [DOI] [PubMed] [Google Scholar]
- 25. Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics 2010;26:1572–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-origin patterns dominate the molecular classification of 10,000 tumors from 33 types of cancer. Cell 2018;173:291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Colaprico A, Silva TC, Olsen C, Garofano L, Cava C, Garolini D, et al. TCGAbiolinks: an R/Bioconductor package for integrative analysis of TCGA data. Nucleic Acids Res 2016;44:e71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Consortium TIP-CA of WG, The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature 2020;578:82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lawrence M, Huber W, Pagès H, Aboyoun P, Carlson M, Gentleman R, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol 2013;9:e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nishiyama A, Nakanishi M. Navigating the DNA methylation landscape of cancer. Trends Genet 2021;37:1012–27. [DOI] [PubMed] [Google Scholar]
- 31. Shen H, Powers N, Saini N, Comstock CES, Sharma A, Weaver K, et al. The SWI/SNF ATPase Brm is a gatekeeper of proliferative control in prostate cancer. Cancer Res 2008;68:10154–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cyrta J, Augspach A, De Filippo MR, Prandi D, Thienger P, Benelli M, et al. Role of specialized composition of SWI/SNF complexes in prostate cancer lineage plasticity. Nat Commun 2020;11:5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zelic R, Fiano V, Grasso C, Zugna D, Pettersson A, Gillio-Tos A, et al. Global DNA hypomethylation in prostate cancer development and progression: a systematic review. Prostate Cancer Prostatic Dis 2014;18:1–12. [DOI] [PubMed] [Google Scholar]
- 34. Zhang L, Fan M, Napolitano F, Gao X, Xu Y, Li L. Transcriptomic analysis identifies organ-specific metastasis genes and pathways across different primary sites. J Transl Med 2021;19:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang K, Dong S, Higazy D, Jin L, Zou Q, Chen H, et al. Inflammatory environment promotes the adhesion of tumor cells to brain microvascular endothelial cells. Front Oncol 2021;11:691771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aksoy O, Pencik J, Hartenbach M, Moazzami AA, Schlederer M, Balber T, et al. Thyroid and androgen receptor signaling are antagonized by μ-Crystallin in prostate cancer. Int J Cancer 2021;148:731–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Delgado-González E, Sánchez-Tusie AA, Morales G, Aceves C, Anguiano B. Triiodothyronine attenuates prostate cancer progression mediated by β-adrenergic stimulation. Mol Med 2016;22:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu Y, Jiang W. Pluripotent stem cell differentiation as an emerging model to study human prostate development. Stem Cell Res Ther 2020;11:285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ashikari D, Takayama K-I, Obinata D, Takahashi S, Inoue S. CLDN8, an androgen-regulated gene, promotes prostate cancer cell proliferation and migration. Cancer Sci 2017;108:1386–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng B, Rong A, Zhou Q, Li W. CLDN8 promotes colorectal cancer cell proliferation, migration, and invasion by activating MAPK/ERK signaling. Cancer Manag Res 2019;11:3741–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X, Jordahl KM, Zhu C, Livingstone J, Rhie SK, Wright JL, et al. Methylation subtypes of primary prostate cancer predict poor prognosis. Cancer Epidemiol Biomarkers Prev 2022;31:1473–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bolis M, Bossi D, Vallerga A, Ceserani V, Cavalli M, Impellizzieri D, et al. Dynamic prostate cancer transcriptome analysis delineates the trajectory to disease progression. Nat Commun 2021;12:7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Luo J, Wang H, Chen H, Gan G, Zheng Y. CLDN4 silencing promotes proliferation and reduces chemotherapy sensitivity of gastric cancer cells through activation of the PI3K/Akt signalling pathway. Exp Physiol 2020;105:979–88. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures
Supplementary Tables
Data Availability Statement
The data generated in this study are publicly available in The European Genome-phenome Archive (EGA; EGAD00010002372).