Single-cell transcriptomic profiling and multiomic cross-validation to uncover therapeutic targets identifies 38 myeloma marker genes, including 11 transcribing surface proteins with previously uncharacterized potential for targeted antitumor therapy.
Abstract
Multiple myeloma (MM) is a highly refractory hematologic cancer. Targeted immunotherapy has shown promise in MM but remains hindered by the challenge of identifying specific yet broadly representative tumor markers. We analyzed 53 bone marrow (BM) aspirates from 41 MM patients using an unbiased, high-throughput pipeline for therapeutic target discovery via single-cell transcriptomic profiling, yielding 38 MM marker genes encoding cell-surface proteins and 15 encoding intracellular proteins. Of these, 20 candidate genes were highlighted that are not yet under clinical study, 11 of which were previously uncharacterized as therapeutic targets. The findings were cross-validated using bulk RNA sequencing, flow cytometry, and proteomic mass spectrometry of MM cell lines and patient BM, demonstrating high overall concordance across data types. Independent discovery using bulk RNA sequencing reiterated top candidates, further affirming the ability of single-cell transcriptomics to accurately capture marker expression despite limitations in sample size or sequencing depth. Target dynamics and heterogeneity were further examined using both transcriptomic and immuno-imaging methods. In summary, this study presents a robust and broadly applicable strategy for identifying tumor markers to better inform the development of targeted cancer therapy.
Significance:
Single-cell transcriptomic profiling and multiomic cross-validation to uncover therapeutic targets identifies 38 myeloma marker genes, including 11 transcribing surface proteins with previously uncharacterized potential for targeted antitumor therapy.
Introduction
Multiple myeloma (MM) is an adult hematologic malignancy, characterized by the uncontrolled proliferation of bone marrow (BM) plasma cells (PC), that is highly prone to drug-resistant relapse. Targeted immunotherapies (1), which may reduce toxicity toward healthy tissues, have shown promise in treating MM (2). Immunotherapies currently in development include monoclonal antibodies, bispecific T-cell engagers, antibody–drug conjugates, and adoptive cellular therapies such as CAR-T, CAR-natural killer (NK), and TCR-T (2), which have demonstrated substantial efficacy in combating liquid tumors, including MM (3, 4, 5).
Despite these advances, major limitations in the development of targeted immunotherapies lie both in the discovery of optimal candidate targets with sufficient tissue specificity and tumor coverage and in the risk of evolutionary downregulation of target proteins once subjected to engineered immune surveillance (6). Recent single-cell studies characterize the inherent subclonal heterogeneity and longitudinal evolution of MM tumors (7, 8), further underlining the need for a nuanced understanding of myeloma tumor epitopes. Unlike bulk strategies that have been used to date as the rationale for immunotherapeutic strategies against MM (9), single-cell approaches minimally obscure tumor architecture and do not rely on selection strategies that may bias resulting tumor profiles. Applications of single-cell RNA-sequencing (scRNA-seq) in diseases have demonstrated the utility of single-cell data in overcoming tumor heterogeneity when optimizing treatment regimens (10). In this study, we build upon these established principles by directly profiling the MM patient population and tailoring our approach to inform targeted immunotherapy against MM.
To systematically identify myeloma markers of potential therapeutic relevance, we used scRNA-seq to perform an unbiased search for genes with specific expression in PCs and/or B cells (relative to other detected cell types) on 53 BM samples taken from 41 patients. We collectively analyzed 146,725 single cells from these patients, including 40,177 PCs, 7,050 B cells, and 99,498 other immune cells. We then used publicly available databases to annotate plasma/B-cell–specific genes with known tissue expression and the predicted cellular localization of their protein products. After filtering candidates' expression, their prevalence across our cohort, and tissue specificities, we examined their expression correlation and coexpression patterns across patients. Consistent with previous studies (7), we found PCs from each patient to be transcriptionally distinct, with unique sets of highly expressed genes driving patient-specific cluster resolution in ways not seen in other BM cell types. Additionally, we observed that certain targets were expressed dynamically during disease development within the same patient and that the preferential expression of certain targets may be related to disease progression. Finally, we cross-validated the expression of candidate targets using bulk RNA-seq and protein quantification. This study represents the first effort using high-throughput scRNA-seq for the systematic discovery of myeloma-associated antigens with the potential for therapeutic applications, paving the way for expanding targeted immunotherapy options in MM.
Materials and Methods
Ethics approval and consent to participate
All procedures performed in studies involving human participants were in accordance with ICH guidelines, applicable regulations and guidelines governing clinical study conduct, and the ethical principles that have their origin in the Declaration of Helsinki. Written informed patient consent was obtained from all patients for the collection and analysis of their samples. The CoMMpass study was conducted in accordance with recognized ethical guidelines in the US and EU and the Institutional Review Board (IRB) at each participating center provided protocol oversight. All work conducted at Washington University (WashU) was performed under the oversight of the IRB at Washington University in St. Louis.
Processing of BMMCs prior to library preparation
WashU Cohort 1, WashU Cohort 2, and Multiple Myeloma Research Foundation (MMRF) Immune Atlas Pilot Cohort BM aspirates were collected at varying disease time points. BM mononuclear cells (BMMC) were isolated using Ficoll-Paque purification and cryopreserved in a 1:10 mixture of dimethyl sulfoxide and fetal bovine serum. Upon thawing in 37°C water baths, whole BMMCs from WashU Cohort 1 were loaded onto the 10X Genomics Chromium Controller using 10X Genomics Chromium Single-Cell 3′v2 Library Kits.
BMMC aliquots from WashU Cohort 2, MMRF Pilot, and healthy donors were centrifuged upon thawing at 300 × g for 5 minutes to pellet cells. All supernatant was removed. To prepare cells for the Miltenyi Dead Cell Removal Kit, cells were resuspended in 100 μL of beads and incubated at room temperature for 15 minutes. Cells were then run through the DepeleS selection using the autoMACSPro Separator. The negative fraction (live cells) was pelleted by centrifugation at 450 × g for 5 minutes. Cells were finally resuspended in ice-cold phosphate buffer saline (PBS) and 0.5% BSA and loaded onto the 10X Genomics Chromium Controller. WashU Cohort 2 and healthy donor samples were loaded using 10X Genomics Chromium Next GEM Single-Cell 3′ GEM, Library and Gel Bead Kit v3.2. MMRF Pilot samples were loaded using 10X Genomics Chromium Controller and using the Chromium Next GEM Single-Cell 3′ GEM, Library and Gel Bead Kit v3.3.
Single-cell library prep and sequencing
Using the 10X Genomics Chromium Single-Cell and Chromium instrument, approximately 16,500 to 20,000 cells were partitioned into nanoliter droplets to achieve single-cell resolution for a maximum of 10,000 individual cells per sample. The resulting cDNA was tagged with a common 16nt cell barcode and 10nt Unique Molecular Identifier during reverse transcription. Full-length cDNA from poly-A mRNA transcripts was enzymatically fragmented and size selected for ∼400 bp cDNA amplicons optimal for library construction (10X Genomics). The concentration of the 10X single-cell library was determined through qPCR (Kapa Biosystems) or automated electrophoresis (Agilent TapeStation) to produce cluster counts appropriate for the HiSeq 4000 or NovaSeq 6000 platform (Illumina). 26 × 98 bp sequence data were generated targeting between 25K and 50K read pairs/cell, which provided digital gene-expression profiles for each individual cell.
scRNA-seq data quantification preprocessing
For scRNA-seq, data were demultiplexed to FASTQ, aligned to the human reference genome (GRCh38), and transposed into gene-by-cell UMI count matrices using the proprietary software tool Cell Ranger (v3.0.0) from 10X Genomics.
Seurat v3.0.0 (11) was used for all subsequent analyses. First, all data underwent quality filtering (as recommended by Seurat) to remove barcodes with: too few total transcript counts (<300); too few genes expressed (<200) or too few UMIs (<1,000), indicating possible debris; too many genes expressed (>50,000) and too many UMIs (>10,000), indicating the presence of multiplet; or too high proportion of mitochondrial gene expression over the total transcript counts (>20%), indicating a dying cell. The median number of genes detected per plasma cell is about 1,500 in our data sets.
Seurat objects were constructed for each sample using its unfiltered feature-barcode matrix. Each sample was scaled and normalized using Seurat's “SCTransform” function to correct for batch effects (with parameters: vars.to.regress = c(“nCount_RNA,” “percent.mito”), return.only.var.genes = F).
scRNA-seq cell type annotation
Cell types were assigned to each cluster by manually reviewing the expression of marker genes. The marker genes used were CD79A, CD79B, MS4A1 (B cells); CD8A, CD8B, CD7, CD3E (CD8+ T cells); CD4, IL7R, CD7, CD3E (CD4+ T cells); NKG7, GNLY (NK cells); MZB1, SDC1, IGHG1 (PCs); FCGR3A (macrophages); CD14, LYZ (monocytes); FCER1A, CLEC10A (dendritic cells); AHSP1, HBA, HBB (erythrocytes); CLEC4C, LILRA4 (pDC); and AZU1, MPO, ELANE (neutrophils).
Differential expression analysis
Differential expression analysis was performed using the default test (Wilcoxon rank sum test) of function FindMarkers (from the Seurat package) with the specified parameters: min.pct = 0.25, logfc.threshold = 0.25, and only.pos = T.
scRNA-seq–driven tumor cell–associated marker discovery
Potential tumor-specific marker discovery was done in Seurat by comparing gene expression between tumor cells and nontumor cells in patient samples. A gene is determined as tumor cell specific if both the following criteria are satisfied: (i) the average expression of the gene is higher in tumor cells compared with any other cell type, respectively, for at least one sample, and that all the differences are of statistical significance (log-fold change >0; adjusted P < 0.05); (ii) the average expression of the gene is higher in tumor cells compared with nontumor cells (as a combined population) for 90% of the samples and that such differences are found to be statistically significant in at least 75% of the samples. Here, all P values were adjusted stringently by Bonferroni correction.
Tumor cell–associated marker subcellular location annotation
To find potential antigens, we further annotated tumor cell–specific genes by their subcellular location and tissue specificity. We used three databases to curate the subcellular location information: (i) Gene Ontology (GO) Term 0005886; (ii) Mass Spectrometric-Derived Cell-Surface Protein Atlas (CSPA; ref. 12); (iii) The Human Protein Atlas (HPA) subcellular location data based on HPA version 19.3 and Ensembl version 92.38.
GTEx tissue expression specificity analysis
Expression data and metadata were downloaded from the Genotype-Tissue Expression (GTEx) portal (gtexportal.org). We then designed a more focused analysis than the ∼3,500 hypothesis tests implied by a would-be round-robin approach for the 53 GTEx tissue type/subtype categories and plasma and/or B-cell–specific genes in Fig. 1. First, we distilled GTEx categorizations into 31 major tissue types, generally corresponding to the first word in the GTEx SMTSD tissue type designation. For each gene, we then identified good hypothesis test candidates, i.e., those showing a high average value of transcripts per million as evaluated over the major tissue types, using Peirce's criterion (13). Each candidate combination of tissue and gene was then tested against the null hypothesis that its average expression did not exceed the grand average of the remaining tissue types in that gene, the specific test being a standard difference-of-means t test for unequal variances. Then, P values were corrected for multiple tests using the standard Benjamini–Hochberg false discovery rate (FDR).
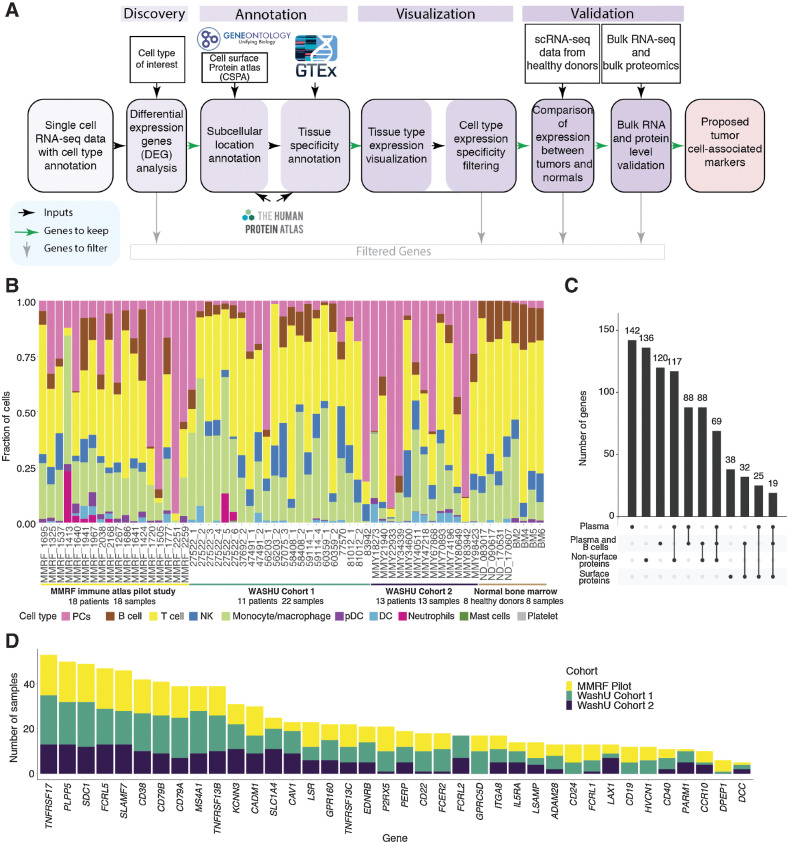
Figure 1.
Myeloma cell–associated therapeutic protein discovery workflow, data sets, and overview. A, Tumor cell–associated therapeutic protein discovery pipeline. B, Stacked bar plot of cell type fractions for each sample, including both tumor BM samples for discovery and normal BM samples for validation. Bar segments are colored by cell types, grouped by data sets. C, Overview of plasma-specific genes and combined plasma and B-specific genes, intersections of the two as well as their cellular localization. D, Stacked bar charts showing number of samples with differentially expressed surface proteins in plasma and/or B cells compared with other cell types, colored by data sets.
HPA tissue expression specificity analysis
Normal tissue protein expression data were downloaded from the HPA portal (proteinatlas.org). These data are based on the HPA version 19.3 and Ensembl version 92.38. We sought to identify proteins that are statistically significantly highly expressed in BM and lymphoid tissues. For each gene, we tested expression for each tissue against the remaining tissue expressions as a background (“take one out” approach) using standard t testing. The resulting P values were corrected for multiple tests using the standard Benjamini–Hochberg FDR. In addition, tissue expression specificity analysis was also performed based on RNA consensus tissue gene data downloaded from HPA.
Manual revision to refine tissue specificity annotation and protein localization assignment
To remove false positives from GTEx tissue expression specificity statistical analysis and HPA tissue expression specificity statistical analysis, we manually reviewed the expression of candidate targets across tissue on the GTEx webpage (https://www.gtexportal.org/home/) and HPA webpage (https://www.proteinatlas.org/). Candidate targets with high and specific expression in lymphocytes (according to GTEx) as well as BM and lymphoid tissues (according to HPA) are defined as tissue-specific targets. In terms of protein localization assignment, in addition to subcellular location annotated by three databases, namely, GO, CSPA, and HPA, we refined the protein cellular location annotation by reviewing literature support.
Comparison of gene expression between tumors and normals
Single-cell-level normalized expression was used to compare expression of myeloma cell–associated genes between 53 tumor BMs and 8 normal bone marrows, again using standard t-testing and FDR correction.
Pathway analysis
We performed pathway overrepresentation analysis on the gene list output from our filtering pipeline using ConsensusPathDB (14) against pathways defined by the Kyoto Encyclopedia of Genes and Genomes (KEGG; ref. 15) and Reactome databases (16), with a 0.05 P value cutoff. Forty-two pathways were identified as significantly overrepresented by our gene list; we then categorized each pathway under a broad biological function using descriptive information provided by the database sources. Gene Ontology enrichment analysis was also performed using ConsensusPathDB, using the “Biological Function” (B) level 3 search filter with enrichment cutoff FDR (q) < 0.05.
Essential genes analysis
Functional dependency scores of proposed myeloma markers were obtained from the DepMap database via the depmap R package (https://depmap.org/portal/).
Flow validation
Antibodies used (clone and source in parenthesis): CD38-BUV395 (HB7; BD Biosciences), CD8-BUV496 (RPA-T8; BD Biosciences), HLA-DR-BUV661 (G46-6; BD Biosciences), CD56-BUV737 (NCAM16.2; BD Biosciences), CD4-BV510 (SK3; BD Biosciences), CD3-BV650 (UCHT1; BD Biosciences), CD20-BV785 (2H7; BioLegend), CD138-VioBrightB515 (44F9; Miltenyi Biotec), gamma/delta TCR-BB700 (11F2; BD Biosciences), CD33-PECF594 (WM53; BD Biosciences), CD19-PE/Cyanine7 (HIB19; BioLegend), CD279-APC (REA315; Miltenyi Biotec), CD45-R718 (HI30; BD Biosciences), and CD14-APC-Vio770 (REA599; Miltenyi Biotec). Sytox AADvanced (Invitrogen) was used for viability. The FCRL5 antibody (clone 952) was generated for us by Alloy Therapeutics by immunizing humanized Gx mice with human domain 9 protein. We conjugated the FCRL5 antibody to PE using a lightning-link kit (Innova Biosciences Ltd.). Purified CS1 antibody (Luc90) was obtained from Creative Biolabs and used with a BV421 IgG secondary antibody. Cryopreserved unsorted bone marrow aspirates from myeloma patients (WashU) and healthy peripheral blood mononuclear cell (PBMC) obtained from leftover platelet apheresis products were thawed and run on a ZE5 Cell Analyzer (Bio-Rad), and data were analyzed using FCS Express flow cytometry software (De Novo Software).
Clustering plasma cells by candidate target expression profiles
We used customized features, which are candidate targets, to compute principal component analysis on seurat_object ← RunPCA(seurat_object, npcs = 30, features = candidate_targets).
Major histocompatibility complex peptide binding prediction
Major histocompatibility complex (MHC) binding prediction of protein sequences was performed using NetMHC (v4.0). Fasta sequences of myeloma-associated intracellular proteins were parsed into 8–11mer peptides and predicted binding affinity for frequently occurring MHC class-I HLA-A, -B, and -C alleles in the US population were calculated and compared with a set of 400,000 random natural peptides to determine strong binding (default top 0.5% of affinity values used as strong binder threshold).
Bulk RNA-seq data generation for WashU internal samples
RNA extraction for bulk sequencing is done using the RNeasy Mini Kit (Qiagen). Following library construction, samples are sequenced to a targeted depth of 80 million reads/sample using NovaSeq 6000 (Illumina).
MMRF whole-exome sequencing and bulk RNA-seq data downloading
We obtained whole-exome sequencing (WES) and bulk RNA-seq data from the MMRF CoMMpass (Relating Clinical Outcomes in MM to Personal Assessment of Genetic Profile) Study (NCT01454297). dbGaP Study Accession: phs000748.
Expression analysis and deconvolution of bulk RNA-seq data
Gene expression was estimated using Kallisto (v0.43.1; ref. 17). The abundance of each cell type was inferred by xCell on sample-by-gene transcript per million (TPM) matrices for each cohort (18), which performed the cell type enrichment analysis from gene-expression data for 64 immune and stromal cell types (default xCell signature). xCell is a gene signatures-based method learned from thousands of pure cell types from various sources. We input the FPKM-UQ expression matrix of this study in xCell using the expression levels ranking.
Bulk RNA-seq-driven plasma cell–associated marker discovery
First, we selected 1,261 genes with expression above the 95 percentile across genes in at least 75% samples of 892 CD138+ sorted samples from the MMRF CoMMpass Study. We then identified genes with significantly high expression in MM cell lines compared with other cancer cell lines in Cancer Cell Line Encyclopedia (CCLE; n = 1,061; CCLE accessed March 23, 2021). For each gene, we identified good hypothesis test candidates, i.e., those showing a high average value of transcripts per million as evaluated over the major malignant cell lines, using Peirce's criterion (13). Each candidate combination of cell line and gene was then tested against the null hypothesis that its average expression did not exceed the grand average of the remaining cell lines in that gene, the specific test being a standard difference-of-means t test for unequal variances. Then, P values were corrected for multiple tests using the standard Benjamini–Hochberg FDR.
Proteomics data generation and analysis
Protein digestion
Viable frozen BMMC aliquots were thawed and incubated with Miltenyi CD138 microbeads prior to PosselD selection using the autoMACSPro Separator. Retained positive fractions were washed by ice-cold PBS and resuspended in cell lysis buffer (50 mmol/L tetraethylammonium bicarbonate, TEABC, pH 8.0, 8 M urea, 1% protease, and phosphatase inhibitor, pH 8.0). Protein concentrations were measured with a Pierce BCA protein assay (Thermo Fisher Scientific). Proteins were reduced with 10 mmol/L dithiothreitol for 30 minutes at 25°C and subsequently alkylated with 50 mmol/L iodoacetamide for 30 minutes at 25°C in the dark. The concentration of urea was then diluted to 2 M by 50 mmol/L TEABC for enzymatic digestion. Proteins in the sorted cells were digested with 0.1 mg Lys-C (Wako) and 0.1 mg sequencing-grade modified trypsin (Promega, V5117) at 25°C for 4 and 16 hours, respectively. For boosting samples, cell pellets from 3 cell lines (OPM, MM1S, and Jurkat) were washed and digested similarly except for a 1:10 enzyme-to-substrate ratio for both Lys-C and trypsin digestion. Digested peptides were desalted by C18 solid-phase extraction (SPE) extraction. Resulting peptides from sorted cells were dissolved in 5 mL 200 mmol/L 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES, pH = 8.5) and further digested by 0.1 mg Lys-C and 0.1 mg sequencing-grade modified trypsin at 25°C to reduce the percentage of missed cleavage.
Tandem mass tag labeling
Digested peptides (in 200 mmol/L HEPES) were mixed with 2 mL tandem mass tag (TMT)-16 reagents (20 mg/mL) dissolved in 100% acetonitrile (ACN) and allowed to react for 1 hour. An optimized ratio of TMT to peptide amount of 1:1 (w/w) was used (i.e., 100 mg of peptides labeled by 100 mg of TMT reagent). The labeling reactions were stopped by adding 5% hydroxylamine (final concentration is 0.5%) for 15 minutes and then acidified with trifluoroacetic acid (TFA; final concentration is 0.5%). Peptides labeled with different TMT tags were mixed in the same tube, after which, the ACN concentration was adjusted to below 5% (v/v) and the samples were desalted by C18 SPE.
Peptide fractionation by basic reversed-phase liquid chromatography
TMT-labeled peptide (50 μg) was dissolved in 0.1% TFA with 2% ACN (1 mg/mL) and fractionated by the high-pressure, high-resolution separations coupled with intelligent selection and multiplexing (PRISM) fractionation method (19). A nanoACQUITY UPLC system (Waters Corporation) equipped with a reversed-phase capillary liquid chromatography (LC) column (3 mm Jupiter C18-bonded particles packed in 200 mm i.d. × 50 cm) and an autosampler were used for fraction collection. Separations were performed by basic reversed-phase liquid chromatography fractionation at a flow rate of 2.2 mL/minute on the binary pump systems using 10 mmol/L ammonium formate (pH 10) in water as mobile phase A and 10 mmol/L ammonium formate (pH 10) in 90% ACN as mobile phase B. 45 mL of sample was loaded onto the reversed-phase capillary LC column and separated using a 190-minute gradient of (minutes:%B): 35:1, 37:10, 52:15, 87:25, 112:35, 125:45, 150:90, and 156:1. A total of 96 fractions was collected at equal time intervals and subsequently concatenated into 24 fractions for global proteome analysis.
LC-MS/MS analysis and protein identification
The LC-MS/MS setting was similar to a previous study (20). Lyophilized peptides were reconstituted in 12 mL (TMT labeling) or 30 mL (label-free) of 0.1% formic acid (FA) with 2% ACN and separated by a nanoACQUITY UPLC system (Waters; buffer A: 0.1% FA with 3% ACN and buffer B: 0.1% FA in 90% ACN) as previously described. Peptides were separated by a gradient mixture with an analytical column (75 mm i.d. × 20 cm) packed using 1.9-mm ReproSil C18 and with a column heater set at 50°C. Peptides were separated by an LC gradient: 2% to 6% buffer B in 1 minute, 6% to 30% buffer B in 84 minutes, 30% to 60% buffer B in 9 minutes, 60% to 90% buffer B in 1 minute, and finally 90% buffer B for 5 minutes at 200 nL/minute. For TMT-labeled samples, data were acquired by Orbitrap Eclipse Tribrid mass spectrometer (Thermo Scientific) in a data-dependent mode with a full MS scan (m/z 400–1,600) at a resolution of 120K with automatic gain control (AGC) setting set to 1×106 and maximum ion injection period set to 246 ms. The isolation window for MS/MS was set at 0.7 m/z and optimal higher-energy C-trap dissociation (HCD) fragmentation was performed at a normalized collision energy of 32% with AGC set as 1×106 and a maximum ion injection time of 246 ms. The MS/MS spectra were acquired at a resolution of 120K. For label-free analysis, data were acquired by Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Scientific) in a data-dependent mode with a full MS scan (m/z 350–1,800) at a resolution of 60K with AGC setting set to 4×105 and maximum ion injection period set to 50 ms. The isolation window for MS/MS was set at 2 m/z and optimal HCD fragmentation was performed at a normalized collision energy of 30% with AGC set at 1×105 and a maximum ion injection time of 105 ms. The MS/MS spectra were acquired at a resolution of 50K.
Data analysis
MaxQuant (21, 22) was used to process the raw MS/MS data. MS/MS spectra were searched against a human UniProt database (fasta file dated April 12, 2017, with 20,198 sequences) or specific fasta file generated by UniProt search for genes of interest. The default setting (4.5 ppm for peptide tolerance and 20 ppm for MS/MS match tolerance) was used for mass tolerance for precursor ions and fragment ions. The search type was set to “Reporter ion MS2” for isobaric label measurements. A peptide search was performed with trypsin/P and allowed a maximum of two missed cleavages. Carbamidomethyl (C) was set as a fixed modification; acetylation (protein N-term) and oxidation (M) were set as variable modifications for global proteome analysis. The FDR was set to 1% at the level of proteins, peptides, and modifications; no additional filtering was performed. The intensities of all 10 TMT reporter ions and label-free quantitation (LFQ) values were extracted from MaxQuant outputs and analyzed by Perseus (23) for statistical analyses.
scRNA-seq data integration
Cells from multiple samples were merged in Seurat, followed by scaling and normalization. All cells were then clustered using the original Louvain algorithm and top 30 principal component analysis dimensions via “FindNeighbors” and “FindClusters” (with parameters: resolution = 0.5) functions. The resulting merged and normalized matrix was used for the subsequent analysis.
Coexpression and mutually exclusive expression analysis using scRNA-seq data
Sample-level average normalized expression in all sample merged Seurat object was used to perform Pearson correlation analysis. To determine the threshold of correlation coefficient (r) for coexpressed gene pairs and mutually exclusively expressed gene pairs, we sampled 2,000 random genes and plotted the distribution of their correlation coefficients. Also, we manually reviewed the expression of some gene pairs in Uniform Manifold Approximation and Projection (UMAP) and decided to use 0.05 and 0.30 as cutoffs for mutually exclusive expression and coexpression of a gene pair, respectively.
Coexpression and mutually exclusive expression analysis using bulk RNA-seq data
Transcript quantification was performed using Kallisto v0.46.2 (17), against the Ensembl transcript reference (release 95, GRCh38). Subsequent analysis was performed using a python script to aggregate transcript-level data to the gene level. Then, Pearson correlation analysis was performed on TPM estimates of target gene candidates.
Immunofluorescent imaging of BM biopsies
5-mm sections of formalin-fixed paraffin-embedded (FFPE) core biopsies of patient BM were incubated at 55°C to prevent lifting and rehydrated using xylene, followed by successive washes in ethanol at decreasing concentrations. Antigen retrieval was done using 10 mmol/L sodium citrate (prewarmed to 80°C–90°C) for 25 minutes, then blocked using 100 mmol/L glycine (2 rounds of 10-minute incubations) once cooled to room temperature and washed with 1× PBS. Sections were then incubated for 1 hour in blocking solution (10% normal donkey serum and 1% BSA in PBS) and rinsed twice in 1× PBS. Primary antibodies (CD138: Akoya #4450008; BCMA: BioLegend #357502; CS1: BioLegend #331802; FCRL5: BioLegend #340302) were diluted in blocking serum (all at 1:50) and incubated on sections overnight at 4°C. Sections were then washed using 1× PBS and incubated for 1 hour with secondary antibodies (Alexa Fluor 647 AffiniPure F(ab')₂ Fragment Donkey Anti-Mouse IgG (H+L) and green second antibody (1:1,000): Alexa Fluor 488 AffiniPure Donkey Anti-Rabbit IgG, both from Jackson ImmunoResearch) diluted to 1:1,000 in blocking serum. Following secondary staining, sections were incubated with Hoescht 33342 staining (Thermo Fisher; 1:2,000 in PBS), then washed in 1× PBS. Slides were mounted with VECTASHIELD (Vector Labs) mounting medium and analyzed on a Leica DMi8.
Somatic mutation detection from WES
Somatic variants were called using our SomaticWrapper pipeline, which integrates the established bioinformatic tools Strelka, Mutect, VarScan2 (2.3.83), and Pindel (0.2.54; refs. 24–27). SNVs identified by any 2 callers among Mutect, VarScan, and Strelka and INDELs identified by any 2 callers among VarScan, Strelka, and Pindel were retained. We then applied coverage cutoffs for these merged SNVs and INDELs of 14× and 8× for tumor and normal, respectively. We also filtered SNVs and INDELs with a high-pass variant allele fraction (VAF) of 0.05 in tumor and a low-pass VAF of 0.02 in normal. The SomaticWrapper pipeline is freely available from GitHub at https://github.com/ding-lab/somaticwrapper. We also applied this pipeline to MMRF samples.
Neoantigen prediction
The wild-type protein sequences are obtained from Ensembl Database. We constructed different epitope lengths (8mer, 9mer, 10mer, and 11mer) from the translated protein sequence. Each sample's HLA type was predicted by OptiType (28) based on bulk RNA-seq data. We predicted the binding affinity between epitopes and the MHC using NetMHC4 (29). Novel epitopes with a binding affinity 500 nmol/L, which are also not present in the wild-type transcript, are reported as neoantigens.
Data availability statement
Single-cell RNA-seq and bulk RNA-seq expression matrices are available at NCBI GEO under accession numbers GSE223060 and GSE223061, respectively.
Results
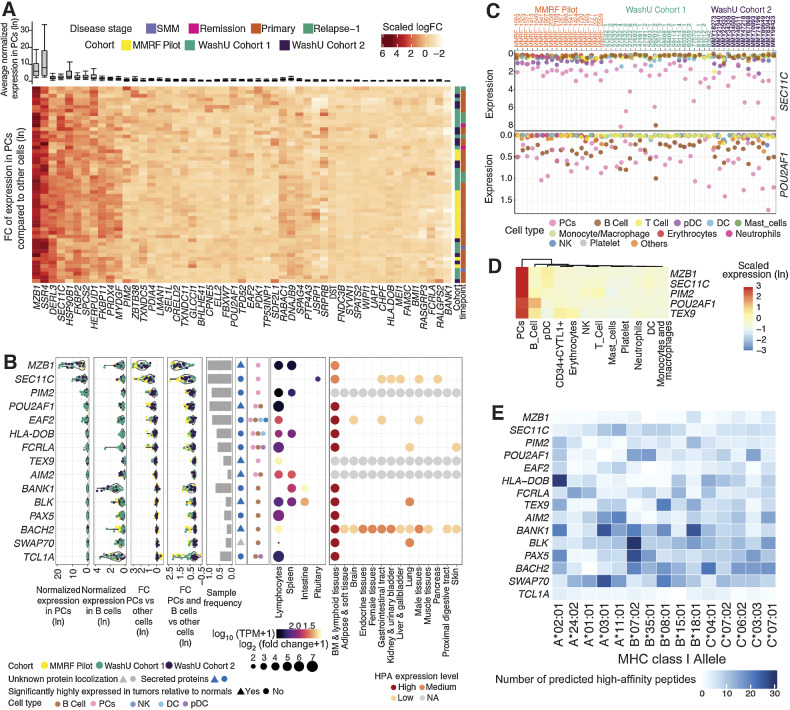
Identification of myeloma targets using scRNA-seq–driven strategies with three independent data sets
We developed an scRNA-seq–based strategy for identifying MM markers (Fig. 1A) using samples collected from 3 independent studies: the Immune Atlas Pilot study led by the MMRF and 2 internal studies at WashU (Cohort 1 and Cohort 2). In total, 53 BM samples were analyzed from 41 patients (patient 83942 was included in both WashU Cohort 1 and WashU Cohort 2; Supplementary Table S1) using 3′ scRNA-seq (Materials and Methods). Of these, 18 samples belonging to the MMRF Immune Atlas Pilot (30) were CD138-depleted fractions taken at diagnosis from patients in the MMRF CoMMpass study (CD138+ PC fractions having been used for earlier bulk sequencing studies by the same name). Despite prior sorting, PCs were still detectable in remaining fractions. The remaining 35 samples were sourced from internal studies and represent 23 individuals with varied disease stages and treatment histories (Supplementary Table S1). The proportions of captured PC and immune cell types varied across patients (Fig. 1B), with PC proportions ranging from 0.5% to 94.3% and B-cell proportions ranging from 0.03% to 29.6%. scRNA-seq of 8 BM samples taken from healthy donors were included as a control group (Fig. 1B).
To identify MM markers, we first defined PCs, which make up the myeloma neoplasm, and B cells, which developmentally precede PCs and may harbor premalignant subsets (31), as our lineages of interest. We began by comparing patient PCs against all other cells detected in each scRNA sample. From this analysis, we identified 142 genes with a positive fold change in PCs of at least 90% of samples and reaching significance in at least 75% of samples (adjusted P < 0.05; Fig. 1C). We then compared combined PCs and B cells against other lineages (including T, NK, and myeloid cells), identifying 120 genes that are overexpressed by this compound group (subject to the same prevalence and significance criteria). We then annotated these genes and their encoded proteins with predicted cellular localization and known tissue specificity using information from GO (32), CSPA (12), HPA (33), and GTEx (Materials and Methods; Fig. 1A; ref. 34). Following these first-pass statistical filtering, we undertook extensive manual revision to eliminate tissue-specificity false positives and refine the protein localization assignment (Materials and Methods). Lastly, because relative (rather than absolute) expression values were used to identify our genes of interest, we subsequently compared patient PC/B cells to healthy donor PC/B cells, eliminating any genes whose absolute expressions were in fact lower than normal (by 2-sided t test, FDR < 0.05; Fig. 1A). These filtering and annotation steps ultimately led to the retention of the following: 136 genes encoding intracellular or secreted proteins and 38 encoding proteins that localize to the cell-surface (Fig. 1C); 15 surface-protein–encoding genes specific to PC/B cells but not expressed by tissues outside of the lymphoid system; 23 surface-protein–encoding genes specific to PC/B cells within the BM but expressed in other tissues; and 15 genes encoding intracellular or secreted products that are highly specific to both lymphoid tissues and the PC/B cells within them. Bulk RNA-seq and bulk global proteomics were then utilized to cross-validate the expression of potential targets.
Of the 38 MM-associated surface-protein–encoding genes, 32 (84%) were discovered in all three cohorts, suggesting high concordance in differential expression among the three data sets (Fig. 1D). This gene set included the top 20 candidate genes as ranked by magnitude of positive fold change in the lineages of interest. Importantly, our strategy recapitulated several of the most promising MM therapeutic targets currently under preclinical and clinical evaluation, including TNFRSF17, SLAMF7, CD38, FCRL5, GPRC5D, and SDC1 (35, 36, 4, 5), which we use as benchmarks of accuracy. We also identified less well-known targets (Fig. 1D) that might have the potential for CAR targeting.
Candidate targets are enriched in immune signaling and protein-processing–related pathways
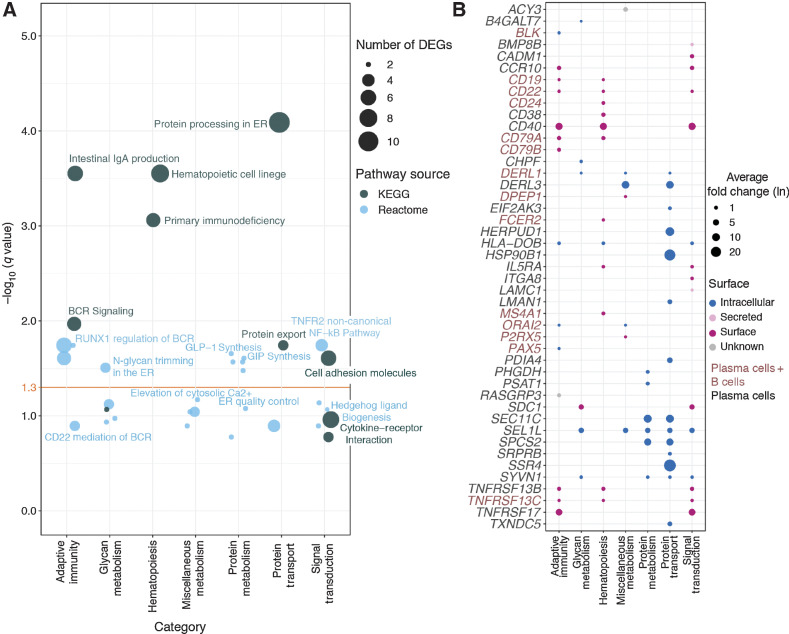
To understand the biological functions of myeloma-specific genes, we used pathway enrichment analysis, finding a significant overrepresentation of cellular processes related to hematopoiesis and protein synthesis, modification, and secretion. In particular, protein processing in the endoplasmic reticulum (ER; q = 8.13×10−5) and protein export (q = 1.81×10−2) are highly enriched, which is consistent with current understanding of active antibody production in malignant PCs (Fig. 2A). Enrichment of N-glycan trimming (q = 0.03) and glucagon-like peptide-1 (GLP-1) synthesis (q = 0.02) may reflect changes in glycoprotein-mediated immune signaling in the myeloma microenvironment, as well as serum glycosylation changes in MM (Fig. 2A; ref. 37). Several genes, including CD19, MS4A1 (CD20), CD22, CD38, CD40, and IL5RA, also enrich for hematopoietic cell lineage (q = 2.81×10−4; Fig. 2B), as they represent canonical B-cell markers with important lineage-determining functions. Consistent with observations that B-cell receptor signaling is enriched in monoclonal gammopathy of undetermined significance (MGUS; ref. 38), B-cell receptor (BCR) signaling (q = 0.01) is also represented in our findings by high expression of RASGRP3, CD79A/B, CD19, and CD22. Notably, we see evidence that the NF-κB pathway, which has been shown in multiple studies to play a critical role in the proliferation and drug resistance of myeloma cells (39, 40), may be affected in myeloma cells due to enrichment of noncanonical TNF-mediated NF-κB signaling (q = 1.81×10−2). Overall, we find that many myeloma-associated intracellular markers, including DERL3, HERPUD1, HSP90B1, SEC11C, MZB1, SPCS2, and SSR4, coordinate protein transport and metabolism. Compared with intracellular markers, which have high expression fold change relative to other cells, genes encoding myeloma-associated surface proteins often have relatively smaller fold differences, and are often related to signal transduction and hematopoiesis. Such members include CD19, CD22, CD24, CD38, CD40, SDC1, TNFRSF17 (BCMA), TNFRSF13B (TACI), and TNFRSF13C (Fig. 2B). GO enrichment of myeloma-associated genes likewise shows overrepresentation of protein metabolism, cell proliferation, and molecular signaling processes (Supplementary Fig. S1A).
Figure 2.
Pathway overrepresentation analysis of plasma and B-cell marker genes using the KEGG and Reactome databases. A, Bubble plot of significantly enriched pathways identified via overrepresentation analysis. Pathways are binned into functional categories and plotted by significance (−log10 of FDR, q); bubble size indicates the number of genes called in the enrichment result. Yellow horizontal line indicates a 5% significance threshold. DEG, differentially expressed genes; ER, endoplasmic reticulum; GIP, gastric inhibitory polypeptide; GLP, glucagon-like peptide; BCR, B-cell receptor. B, Bubble plot of fold change and cellular localization of differentially expressed plasma and B-cell genes identified as functionally relevant by pathway enrichment. Bubble size indicates fold change (ln) of gene expression in either plasma cells or combined plasma and B cells (as indicated by gene name color) relative to other BM cells; color denotes predicted cellular localization of the encoded protein.
To assess the functional importance of proposed myeloma markers, we compared reported dependency scores from the DepMap CRISPR-Cas9 gene knockout data set (41). Roughly half of our marker genes were consistently found to have a detrimental loss-of-function effect across 21 plasma cell lines (Supplementary Fig. S1B). Most strikingly, PIM2 and POU2AF1, which encode intracellular proteins, reached dependency scores similar to or exceeding those of pan-essential genes in nearly all plasma cell lines.
In considering how upregulated PC/B functions may be disrupted for antitumor treatment, we next examined the utility of targeting chemokine systems to enhance tumor access by CAR-T or other immune cell modalities. Cancers often rely on altered chemokine signaling channels for growth and metastasis, and receptors that modulate bone architecture or immune surveillance play important roles in MM pathogenesis (42). For cell-based immunotherapies, taking advantage of existing communication patterns between tumor and the immune microenvironment may improve tumor penetration. In our cohort, we found that CCL3, CXCL10, and CXCL12 are upregulated in PCs in a few patients (Supplementary Fig. S1B). Interestingly, CXCL12 is strikingly highly expressed in 27522_2, the only remission sample in our data sets (Supplementary Table S1). Some chemokine receptors (CXCR6 and CCR4) are found to be specifically expressed in T cells in most patients (Supplementary Fig. S1C). Although our findings are inconclusive regarding the role of chemokines or their receptors in this cohort, this analysis may provide insights toward enhancing tumor homing by taking advantage of chemotaxis gradients in the diseased BM.
Characterization of surface proteins’ potentials as antigenic targets of CAR-T cells
Ideal target antigens for CAR-T therapy are plasma membrane-localized proteins with accessible extracellular domains that have both unique and abundant expression in tumor cells of most patients (43). We further examined the 38 surface-protein–encoding genes identified from our discovery pipeline to assess expression specificity and magnitude across individual samples. Although certain genes, including CD79A, EDNRB, CD40, and MS4A1 (CD20), show sample-specific overexpression, most have relatively uniform overexpression across samples (Fig. 3A). As expected, known myeloma markers, including TNFRSF17 (BCMA), SDC1 (CD138), FCRL5, CD38, SLAMF7 (CS1), and CD79A, were identified as top-ranking genes upregulated in PCs relative to other immune populations, with high average expression in PCs as well (Fig. 3A).
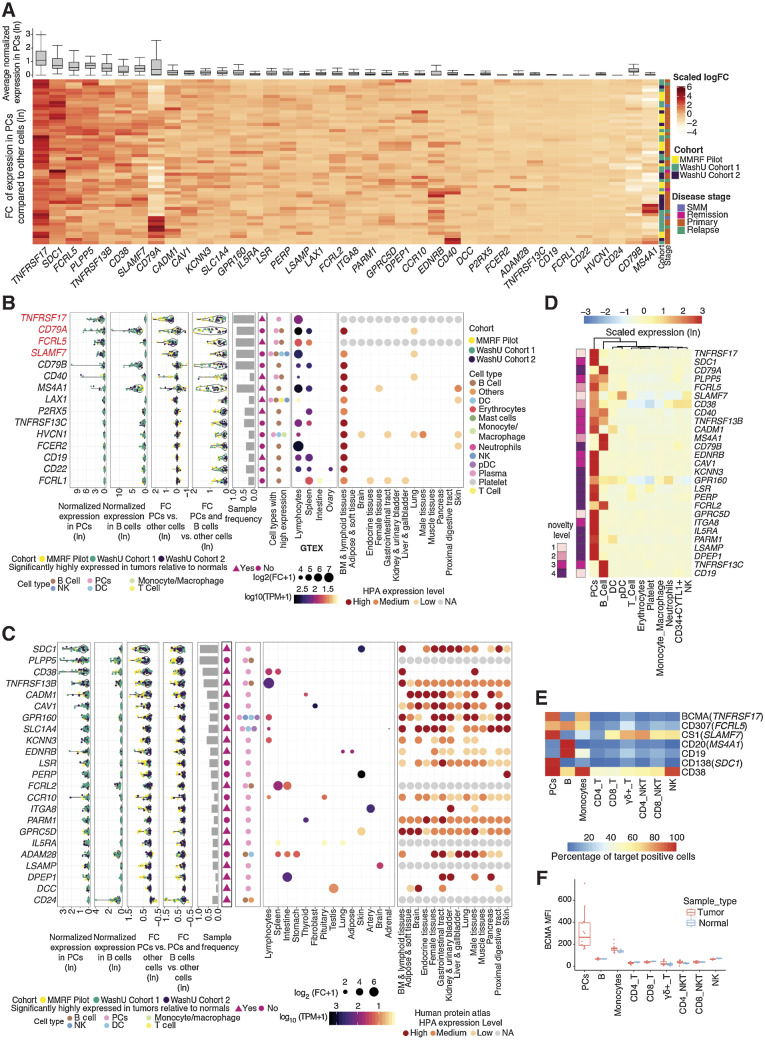
Figure 3.
Characterization of myeloma cell–associated surface proteins. A, Average gene expression in plasma cells (top, box and whisker plot) and hierarchical clustering of z-score scaled log-fold change between plasma and nonplasma cells across samples (bottom, heat map). B, Summarized characterization of primary candidate targets expressed on the cell surface with specific expression in lymphoid tissues. Genes are ordered by average expression in PCs. Top four genes are top-tier primary targets, highlighted in red. Average normalized expression in plasma and B cells are shown in the first two sections, respectively. Average expression log-fold change between PCs and other cells is shown in the third section, followed by expression log-fold change of combined plasma and B cells compared with other cells shown in the fourth section (FC, fold change). Each dot represents a sample, colored by its corresponding cohort. Sample frequency is the proportion of samples having genes differentially expressed in PCs or in combined plasma and B-cell population if the gene is only expressed in B cells. In the sixth section, triangle and circle denote whether genes are significantly highly expressed in tumors relative to normal BMs. Cell types with target expression are annotated in the seventh section. In the GTEx tissue expression specificity analysis section, only gene-tissue type relationships with a significant FDR are plotted. Size corresponds to fold difference and is colored by expression. HPA protein expression was shown in the last section, with color indicating expression level. NA, not available. C, Summarized characterization of secondary candidate targets expressed on the cell surface without specific expression in lymphoid tissues. Figure layout is the same as B. D, Heat map showing the z-score scaled average expression of known and novel targets across cell populations in scRNA-seq. Columns are ordered based on the hierarchical clustering of target expressions; rows are ordered by average normalized expression in plasma cells. Left annotation indicates novelty level: 1, currently under clinical study as CAR-based therapy; 2, currently under clinical study as antibody-based therapy; 3, potential therapeutic utility is supported by existing literature; 4, previously uncharacterized in its capacity as a myeloma marker. Further details for novelty ranking are included in Supplementary Table S2. E, Heat map showing the average percentage of cells with positive expression of surface antigens across cell populations from 12 patient samples (11 MM and 1 SMM patients) in flow cytometry. F, Box plot showing mean fluorescence intensity (MFI) of BCMA in patient BMMC (n = 12, including 11 MM and 1 SMM) or PBMC from healthy donors (n = 3) across cell populations in flow cytometry.
To avoid nonspecific toxicity, ideal target antigens must have a limited expression on critical normal tissues. We stratified surface proteins into primary (specific) and secondary targets (nonspecific or unknown) based on tissue expression as reported in GTEx (44) and HPA (Fig. 3B and C; ref. 45). We found further support for TNFRSF17 as being exclusively high in lymphocytes (GTEx; fold change ∼100; Fig. 3B). CD79A, a receptor involved in adaptive immunity that is restricted to lymphoid tissues, has the second-highest average expression in PCs among primary targets and is overexpressed in the combined PC/Bs of 46/53 samples (Figs. 2B and 3B). However, CD79A is also highly expressed in normal PCs and B cells due to its critical role in B-cell development and activation (46). FCRL5, a member of immunoglobulin receptor superfamily, ranks third in PC-specific overexpression and, with the exception of the spleen, appears exclusively expressed in lymphocytes according to GTEx [fold change (FC) ∼50; Fig. 3B; Supplementary Fig. S2]. SLAMF7, a primary target already in clinical application (5, 47, 48), ranks fourth; however, its expression in other immune cells, such as dendritic cells (DC), NK cells, and T cells, is relatively high (Fig. 3B; Supplementary Fig. S2). Other primary targets, exhibiting lower average expression in PCs than the aforementioned candidates but still meeting our specificity criteria, include CD79B, CD40, MS4A1, LAX1, P2RX5, TNFRSF13C (BAFF-R), HVCN1, FCER2, CD19, and FCRL1. (Figs. 2B and 3B). Of these, CD79B, TNFRSF13C, and FCER2 are strikingly overexpressed in B cells and may be therapeutically relevant for eradicating the B developmental reservoir of MM PCs (Fig. 3B). The preclinical efficacy of CD19/CD20 bispecific CARs in limiting MM antigen escape (49), the success of BAFF-inhibitory drugs (50), and the evidence that FCER2, which canonically encodes a B-cell-specific antigen, is strongly associated with chromosomal abnormalities in myeloma (51) all indicate B-cell involvement in MM. CD79B, which encodes part of the BCR signaling complex, has previously been studied as a therapeutic target in B-cell lymphoma (52), but its role in myeloma is not yet extensively studied. Although LAX1 and HVCN1 are expressed in multiple BM cell types, they may still be considered MM targets in their comparability to SLAMF7, which is also expressed in non-PC/B immune cells (Fig. 3B; Supplementary Fig. S2). Taken together, CD79A, P2RX5, FCRL1, LAX1, and HVCN1, for which there are not yet any published preclinical studies in the context of MM, merit further investigation.
Secondary targets, which have either unknown protein-level specificity or have been reported in other tissues, remain relevant considerations for dual-targeted CAR-T-cell immunotherapy approaches currently under development (53). It must also be noted that database-reported expression in nonlymphoid compartments does not necessarily indicate toxicity, as MM PC expression levels may far exceed those of normal tissues. For example, SDC1 (CD138) and CD38 have been extensively investigated as targets for anti-MM monoclonal antibodies (54, 55) despite showing abundant expression in nonhematopoietic tissues in HPA (Fig. 3C). Like the aforementioned TNFRSF17, TNFRSF13B (TACI) belongs to the tumor necrosis factor (TNF) superfamily and is expressed primarily in lymphocytes (GTEx; FC ∼200). In our data, 46/53 samples exhibited a logFC > 1.5 of TNFRSF17 in PCs versus non-PCs, whereas TNFRSF13B showed similar PC fold elevation in only 14/53 samples. In preclinical MM models, dual targeting of BCMA and TACI successfully maintained tumor control in the absence of BCMA due antigen escape (56). GPRC5D, a member of G protein-coupled receptor family, is another secondary target currently in clinical trials as both an antitumor CAR-T target and as a GPRC5D/CD3 bispecific antibody (57). In addition to these known targets, our study identified PLPP5, CADM1, CAV1, GPR160, KCNN3, EDNRB, LSR, FCRL2, and several other genes as novel secondary targets with potential therapeutic utility (Fig. 3C and D).
Taking into consideration the fact that protein expression data reported in GTEx and HPA may be incomplete or misleading, we present 20 candidate genes not yet under clinical investigation whose PC and/or B-cell–specific overexpression can at least be confirmed in BM populations by our scRNA-seq data set. Of these genes, 9 are supported by existing publications as myeloma markers of possible clinical interest, and 11 are novel targets whose expression in myeloma has to date been largely uncharacterized. Representative studies describing known targets are provided in Supplementary Table S2. Novel candidates include KCNN3, LSR, PERP, FCRL2, GPR160, and IL5RA. Our candidate genes, highlighted in Fig. 3D, may provide therapeutic opportunities for patients refractory to current therapies. Although further investigation is of course necessary to determine protein abundance and off-target toxicity, our analyses greatly narrow the search space in identifying tumor markers. Flow cytometry of an independent cohort including 11 MM patients and one Smoldering multiple myeloma (SMM) patient (clinical information in Supplementary Table S1) indicated that protein-level expression of TNFRSF17 (BCMA), FCRL5 (CD307), MS4A1 (CD20), CD19, SDC1 (CD138), and CD38 are concordant with our RNA-level findings and even suggest underestimation of sample prevalence in scRNA-seq (Fig. 3E and F; Supplementary Fig. S3), further supporting the utility and robustness of our discovery approach.
Potential combinatorial targeting partners revealed by correlation analysis
Simultaneously targeting multiple tumor markers may mitigate antigen escape and enhance efficacy. To evaluate target coexpression, we aggregated samples using an integration method that corrects for batch-effect-induced biases (Materials and Methods; ref. 58). We observed that most immune lineages (e.g., T cells and monocytes) from different samples clustered together by cell type, whereas PCs formed 49 clusters with unique patient origin, indicating intrinsic transcriptomic diversity of tumors (Supplementary Fig. S4A). Samples taken from the same individual tended to cluster together, as in the cases of 81012, 58408, and 47491 (Fig. 4A). To reduce noise from potentially irrelevant differences in gene expression, we reclustered PCs using candidate target expression instead of global expression (Materials and Methods). We then observed that most samples cluster together, whereas cells from 19 patients remained distinct. This suggests that most patients may in fact share common target profiles, whereas others will require more personalized targeted therapy (Supplementary Fig. S4B).
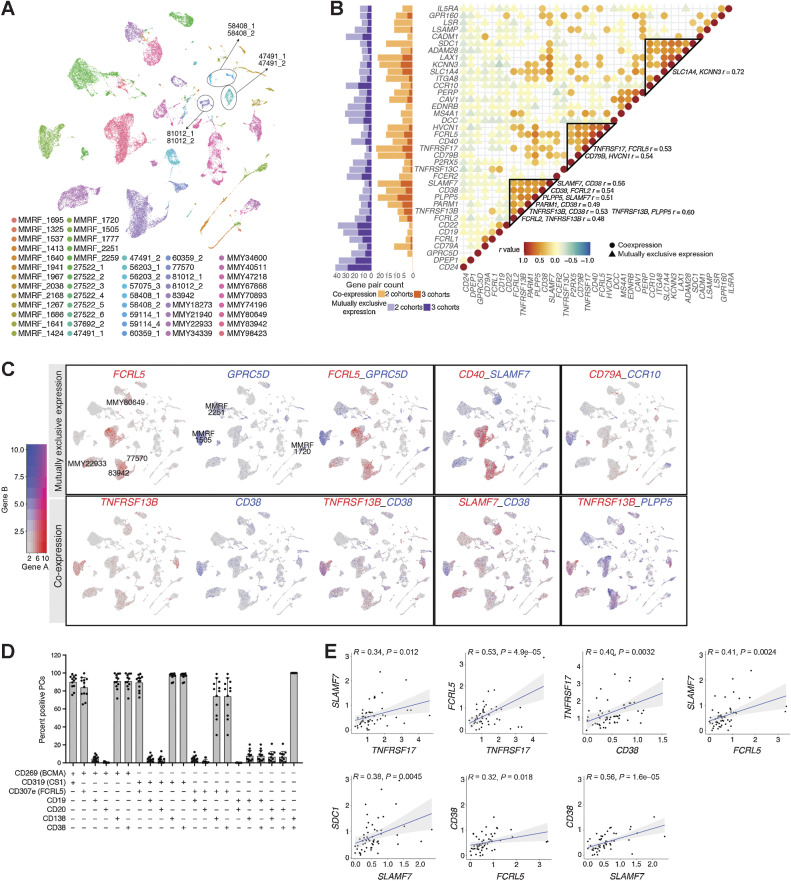
Figure 4.
Expression correlation of myeloma cell–associated genes encoding surface proteins. A, UMAP showing plasma cells from 53 patients reveals tumor-specific clusters, colored by patient. B, Dot plots showing correlation of gene expression averaged by sample in all 53 samples, colored by Pearson correlation coefficient (r value). Only predicted coexpressed gene pairs (r > 0.30) and mutually exclusively expressed gene pairs (r < 0.05) are shown. Coexpressed gene groups are highlighted in triangles, with visually confirmed ones listed on the right. Bar charts showing the number of predicted gene pairs (r < 0.05 or r > 0.30) in two data sets or three data sets. C, Gene pairs showing mutually exclusive expressions (top row) or coexpressions (bottom row). The first box of each row shows an example of expression of gene A (first UMAP), expression of gene B (second UMAP), and dual expression of gene A and gene B (third UMAP). D, Bar chart showing the percentage of plasma cells from 12 patient samples (11 MM and 1 SMM), with coexpression of surface proteins detected by flow cytometry. E, Pearson correlation plots of gene pair (same genes shown in D) expression in scRNA-seq.
Using Pearson correlation analysis for sample-level average expression in PCs, we identified coexpressed (r > 0.30) and mutually exclusive target pairs (r < 0.05; Fig. 4B; Supplementary Table S3). The r value thresholds were determined based on the distribution of correlation of 2000 random genes, and by visualizing the expression of gene pairs in UMAP (Supplementary Fig. S4C; Materials and Methods). To account for potential cohort-related biases, we performed this correlation analysis within each cohort, finding that 66% (25/38) of genes have coexpression partners and 97% (37/38) of genes have at least one mutually exclusive partner in all three cohorts (Fig. 4B; Supplementary Fig. S4D; Supplementary Table S3). We observed generally high concordance between the cohort-specific and pooled correlation analyses, meaning that genes with the greatest number of coexpression or mutual-exclusion partners in the pooled analyses show similar trends in each individual cohort.
Although many candidate genes were simultaneously expressed at the sample level, we saw evidence for distinct groups of closely-correlated target genes (Fig. 4B; indicated by triangular outlines). One major group includes SDC1, LAX1, SLC1A4, and KCNN3; another includes HVCN1, FCRL5, CD40, and TNFRSF17; and a third includes SLAMF7, CD38, PLPP5, and TNFRSF13B. Consistent with sample-level analysis, SLAMF7 and CD38 expression are largely concordant in UMAP visualization of individual cells (Fig. 4C; “Co-expression” row), as are TNFRSF13B, CD38, and PLPP5. GPRC5D, MS4A1, and CD79A are coexpressed as well (Supplementary Fig. S5A). Conversely, CD40 and SLAMF7 are very rarely simultaneously expressed by individual tumors (Fig. 4C; “Mutually exclusive expression” row). Other mutually exclusive pairs include FCRL5/GPRC5D, CD79A/CCR10, SLC1A4/KCNN3, and HVCN1/CD79B (Fig. 4C; Supplementary Fig. S5A). These findings suggest that certain tumor phenotypes may respond best to targeting against specific markers: for example, FCRL5 may be a more broadly applicable alternative than CS1 (SLAMF7) for patients who also qualify for anti-BCMA (TNFRSF17) therapies but may not be appropriate for tumors expressing GPRC5D. As clinical reagents are in development for several of these targets, our data indicate that further study is warranted to characterize their interchangeability. As yet, however, our interpretation is limited by the low expression of some targets and the possibly insufficient coverage or sample size for thoroughly interrogating these relationships in our scRNA cohort.
To address these weaknesses of scale and depth, we next examined coexpression across the much larger CD138+ sorted CoMMpass bulk RNA-seq data set, wherein we see two distinct groups of highly correlated surface targets (Supplementary Fig. S4E). The first group includes the well-characterized myeloma markers SDC1, SLAMF7, TNFRSF17, and CD38, along with LAX1, CAV1, and SLC1A4. The distinct correlation patterns between these targets, which we see in scRNA-based coexpression analysis, are largely obscured in this data set. This may be due to the limitations discussed above, but the CD138 selection of tumors for the CoMMpass data set may also contribute to this discrepancy. The second highly correlated gene group contains certain known B-cell-associated genes such as MS4A1, CD19, TNFRSF13C, and CD79A, which are also highly expressed in PCs from our scRNA-seq data set. Interestingly, these two gene groups are negatively correlated with each other, suggesting that most samples preferentially express either one set or the other, and that there may be a distinct “B-cell-like” phenotype among myeloma tumors (7). However, we do not see the resolution of a distinct B-like coexpression group in our scRNA data, and due to the imperfect isolation of PCs and the aggregate nature of bulk studies, it remains inconclusive whether this apparent mutual exclusivity is actually an artifact of B-cell contamination.
Given potential differences in mRNA and protein half-life, as well as possible dropout error in scRNA-seq, we next assessed coexpression of well-characterized myeloma markers using flow cytometry of 12 unsorted patient samples (Fig. 4D; clinical demographics in Supplementary Table S1). PCs were identified by CD138+/CD38+ gating. Flow cytometry confirmed overall CS1 (SLAMF7), BCMA (TNFRSF17), FCRL5, CD38, and CD138 coexpression as observed in scRNA-seq (Fig. 4D and E); among these markers, FCRL5 expression was the least prevalent. In our flow cytometry results, CD19 and CD20 (MS4A1) expression appeared restricted to CD138−/BCMA− cells, which we categorized as nonplasma B cells for lack of better evidence. Whether a CD19+/CD20+ subset of PCs exists in a subset of patients, as suggested by our scRNA-seq findings, remains to be further investigated.
Finally, we note the observed subcluster heterogeneity of marker expression, particularly the single-positive expression of several gene pairs including the canonical MM markers (59) TNFRSF17 (BCMA) and SDC1 (CD138), SLAMF7 (CS1/CD319) and SDC1, and FCRL5 (CD307e) and SDC1 (Supplementary Fig. S5B). The relative abundance of single positives varies between individuals. For example, PCs from patient MMY83942 exhibit a slight preference for either TNFRSF17 or SDC1 with low–mid expression of both genes, whereas PCs from MMY22933 highly express one or the other. Several PC subpopulations exhibit high SLAMF7 or FCRL5 expression and low or no SDC1 expression (and vice-versa). When comparing RNA expression of SLAMF7 and FCRL5 to TNFRSF17 instead of to SDC1, we again see subpopulations that skew toward one over the other, with relative proportions differing between patients (Supplementary Fig. S5B). The proportion of FCRL5+/SDC1−/TNFRSF17− PCs in MMY83942, MMY80649, and 77570 are 45%, 48%, 33%, respectively. As CD138 selection is frequently used to isolate MM PCs for study, our results suggest that current MM cell isolation techniques are likely not sufficient to capture all MM clones within a sample and that additional techniques should be considered. To see whether this may be explained by technical dropout or differential mRNA metabolism irrespective of protein expression, we next performed immunofluorescence (IF) costaining of these pairs (Supplementary Fig. S5C). The presence of both coexpressing and single-positive cells (expressing either CD138 or one of the other three genes) is corroborated by IF in patient s10-10686 (newly diagnosed sample; BCMA vs. CD138, FCRL5 vs. CD138, and SLAMF7 vs. CD138 in Supplementary Fig. S5C). A mounting body of evidence that CD138 may be rapidly shedding from the PC surface (60, 61) renders inconclusive whether BCMA+, CS1+, or FCRL5+ PCs are truly distinct from CD138+ PCs, but an alternative explanation may lie in the stochastic fluctuation of either marker in individual cells, which scRNA-seq and imaging of fixed tissues cannot detect. These findings demonstrate both the potential difficulty in capturing MM tumors with any single marker and the importance of selecting the right set of markers when designing combinatorial strategies.
Characterization of intracellular or secreted proteins with potential relevance for MHC-based therapies
As the majority of our detected PC/B-cell–specific genes encode nonsurface proteins, we next evaluated these genes for potential utility as therapeutic targets. Cytosolic proteins are presented as peptides on the cell surface by the MHC, and intracellular tumor markers may therefore be targeted via T-cell receptor (TCR) recognition of specific MHC-bound peptides (62). Of the 136 genes encoding nonsurface targets highly expressed in PC and/or B-cell populations, 11 were specifically upregulated in PCs relative to other BM immune populations in nearly all samples, namely, MZB1, SSR4, DERL3, SEC11C, HSP90B1, FKBP2, SPCS2, HERPUD1, FKBP11, PRDX4, and MYDGF (Fig. 5A). In contrast to the signaling and developmental pathways enriched by surface protein targets, 6 of these 11 genes (SSR4, DERL3, SEC11C, HSP90B1, SPCS2, and HERPUD1) are involved in protein transport and metabolism (Fig. 2B). Of note, MZB1 (marginal zone B and B1 cell-specific protein) and SSR4 (signal sequence receptor subunit delta, an ER-targeted protein) are extremely highly expressed in PCs. These observations are consistent with previous reports characterizing MZB1 as a candidate marker of MM in a functional proteomics study (63) and reaffirm the important role of ER stress in MM (64).
Figure 5.
Characterization of myeloma cell–associated intracellular proteins. A, Average gene expression in PCs shown in box and whisker plot (top) and hierarchical clustering of z-score scaled log-fold change (bottom) between plasma and nonplasma cells across samples shown in heat map. B, Summarized characterization of intracellular targets specifically expressed in lymphoid tissues. Genes are ordered by average expression in PCs. Average normalized expression in plasma and B cells is shown in the first two sections, respectively. Average expression log-fold change between PCs and other cells is shown in the third section, followed by expression log-fold change of combined plasma and B cells compared with other cells shown in the fourth section. FC, fold change. Each dot represents a sample, colored by its corresponding cohort. Sample frequency is the proportion of samples having genes differentially expressed in PCs or in combined plasma and B-cell population if the gene is only expressed in B cells. In the sixth section, triangle and circle denote whether genes are significantly highly expressed in tumors relative to normal BMs. Blue, secreted proteins. Gray, unknown protein localization. Cell types with target expression are annotated in the seventh section. In the GTEx tissue expression specificity analysis section, only gene–tissue type relationships with a significant FDR are plotted. Size corresponds to fold difference and is colored by expression. HPA protein expression is shown in the last section, with color indicating expression level. NA, not available. C, Bubble plots showing the normalized expression of SEC11C and POU2AF1 averaged by sample and cell type. D, Heat map showing the z-score scaled average expression of promising intracellular targets across cell populations in scRNA-seq. Columns are ordered based on the hierarchical clustering of target expressions. E, Number of peptides for each candidate gene product predicted to have high-affinity binding to common MHC class I alleles in the US population as determined by NetMHC.
Considering reported gene expression in non-BM tissues, we retained 15/136 genes showing specific expression in lymphocytes or lymphoid tissues only (Fig. 5B). We then compared expression in patient PC/B cells to those from normal BM, as true tumor antigens have more therapeutic potential than proteins native to healthy tissue in a TCR-based approach. MZB1 meets key criteria for a good target antigen, namely, significant high expression in patient PC/Bs and lymphocyte/lymphoid tissue-restricted expression. SEC11C, which encodes another ER-related protein restricted to lymphoid tissues, has the second-highest average normalized expression in PCs among the 15 genes (Fig. 5B–D, note the scale). Similarly, PIM2, which encodes a serine/threonine kinase, is differentially expressed in PCs in 83% of samples and specific to lymphocytes in GTEx (FDR ∼10−10; Fig. 5B). Studies have shown that PIM2 is upregulated and required for MM proliferation, suggesting the potential clinical efficacy of targeting PIM2 despite this limitation (65). POU2AF1 (POU class 2 homeobox associating factor 1) may be another promising target, as it is highly expressed in PCs in our cohort in 49/53 of samples (Fig. 5C) and has been found to promote MM cell growth by direct transactivation of TNFRSF17 (66). As with MZB1, the expression levels of POU2AF1, TEX9, BLK, BACH2, and SWAP70 were significantly elevated in patient PCs and/or B cells compared with those from normal BM. Given their specific and abundant expression in PCs, MZB1, SEC11C, PIM2, POU2AF1, and TEX9 are highlighted as priority tissue-specific intracellular targets (Fig. 5D).
We next analyzed candidate target protein sequences using NetMHC (29) to predict high-affinity peptide binding for the five most frequently occurring MHC class-I HLA-A, -B, and -C alleles in the US population (Fig. 5E; ref. 67). Predicted binding affinity of 8–11mer peptides were compared with a set of 400,000 random natural peptides to determine strong binding (Supplementary Table S4). Overall, 4,921 peptides obtained from these 15 genes (Fig. 5E) were predicted to bind with high affinity (defined as the top 0.5% of predicted affinity values; see Materials and Methods) to the 15 HLA alleles (Supplementary Table S4). At least one potentially high-affinity binding peptide was obtained from each gene for 14 out of the 15 HLA alleles examined (Fig. 5E). Fifty-seven predicted high-affinity peptides from MZB1 cover alleles including A*01:01, A*02:01, A*03:01, A*11:01, and A*24:02, each of which represents significant (10%–40%) fractions of the US population (68). With respect to its high prevalence and tumor specificity, MZB1 may be a model candidate for TCR-based targeting. Certain genes (POU2AF1, HLA-DOB, TEX9, and AIM2) appear to have distinct preferential binding for a subset of HLA alleles, whereas others (BACH2, SWAP70, BLK, and BANK1) are predicted to have high affinity for several common HLA alleles. As MHC presentation is required for TCR-based adoptive cell therapy, HLA compatibility is an important factor in choosing targets. However, high native immunogenicity may present a potential complication as tumors may be under selective pressure to downregulate highly expressed and highly presented proteins (69). Further study is necessary to fully evaluate the utility of our proposed candidates in a TCR-based approach in MM.
Expression of candidate targets in relation to clinical characteristics
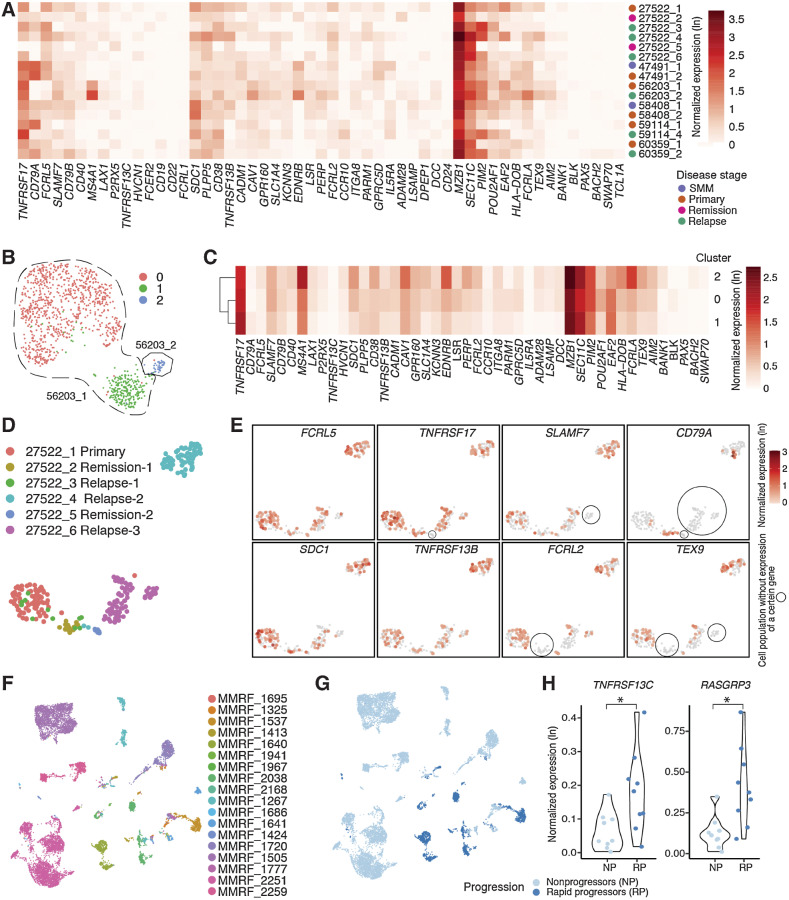
To better understand how target persistence may be affected by cycles of treatment and relapse, we next compared PCs sampled longitudinally from 6 individuals (Fig. 6A; Supplementary Table S1). Notably, certain genes appear more stably expressed across time points than others. For example, TNFRSF17, CD79A, MZB1, and SEC11C expression fluctuate minimally, whereas SLAMF7, LAX1, PLPP5, and PIM2 show evidence of increased expression in later time points. In patient 27522, PIM2 is significantly downregulated at remission (27522_2 and 27522_5). This evidence aligns well with previous studies identifying Pim2 as a key modulator of MMPC proliferation (70). Baseline heterogeneity between individuals was also evident; for instance, CD79A and GPRC5D are highly expressed in patient 47491, and MS4A1 in 56203 compared with other patients at all time points (Fig. 6A).
Figure 6.
Expression of myeloma cell–associated therapeutic targets in relation to clinical features. A, Heat map showing normalized expression of candidate targets across longitudinal samples. Different disease stages are annotated in different colors on the right of the heat map. B, UMAP showing plasma cells from samples taken at two different stages of patient 56203, colored by seurat clusters. C, Heat map showing normalized expression of candidate targets across three plasma populations in patient 56203. Genes detected in fewer than five cells were not considered during normalization and thus were not included in the heat map. D, UMAP showing plasma cells from samples taken at six different time points of patient 27522, colored by sample. E, Expression of typical myeloma cell–associated therapeutic targets reveals diverse and dynamic change of gene expression along the disease progression in the same patient. F, UMAP showing plasma cells from samples in MMRF Immune Atlas Pilot study, colored by patient. G, UMAP showing plasma cells from samples in MMRF Immune Atlas Pilot study, colored by patients’ progression features. H, Violin plots showing expression of TNFRSF13C and RASGRP3 is significantly elevated in rapid progressors compared with nonprogressors (P < 0.05).
Upon integrating longitudinal samples from patient 56203, we see that the primary tumor (56203_1) forms two distinct clusters (0 and 1 in Fig. 6B), whereas the remission tumor (56203_2) forms a third. Several markers appear upregulated in cluster 0 relative to cluster 1, including SLAMF7, SDC1, EDNRB, LSR, POU2AF1, and FCRLA; conversely, TNFRSF17 and SEC11C are lower (Fig. 6C). In the relapse cluster 2, upregulation of marker genes, including MS4A1, SDC1, CAV1, EDNRB, and FCRLA, become increasingly pronounced. This pattern of overall higher marker expression at relapse can be found in patients 59114 and 60329 as well, though to lesser degrees (Fig. 6A). Conversely, cluster 2 expression of TNFRSF13B and POU2AF1 are lower than cluster 0 and similar to cluster 1 levels. These observations suggest that cluster 0 may be an intermediate tumor phenotype for 56203 and that escalating levels of tumor marker expression may in general be indicative of advancing disease.
Our most complete longitudinal profile was of patient 27522, for whom we sampled from six time points representing primary tumor (timepoint 1), first remission (2), first and second relapses (3, 4), second remission (5), and third relapse (timepoint 6; Fig. 6D; clinical details in Supplementary Table S1). TNFRSF17 is lost at the second remission (5) and recovered at the third relapse (6); CD79A is absent from the primary tumor but appears in the first remission (2) as well as in relapse time points 3 and 4; FCRL2 and TEX9 are absent from the first remission (2) but reappear in later time points (Fig. 6E). SLAMF7 is lost in an isolated subcluster at the third relapse (timepoint 6), which also lacks TEX9 expression. Even within a given time point, we see transcriptomic heterogeneity. In contrast, FCRL5, SDC1, and TNFRSF13B are expressed fairly uniformly at all time points. Further studies may elucidate whether these fluctuations reflect physiologic changes of the tumor in response to specific treatment regimens.
We further investigated the evolution of immunogenic mutations in patient 27522 by comparing predicted immunogenic peptides across three time points using bulk WES and RNA-seq, which were derived from a previous study (Supplementary Fig. S6A; ref. 7). Twenty putative neoantigens were detected in the primary MM sample and retained during MM progression, including potentially immunogenic peptides associated with the driver mutation NRAS G13R. Seven putative neoantigens (BRD2-P243Q, XRN1-W1305C, SOCS6-S106F, EVPL-T1677S, TP63-F181Y, PHLPP1-N666S, and NIPBL-A1250S) were lost during MM progression, suggesting potential immune-mediated clearance of subclones harboring these mutations in the primary sample. We then expanded our analysis to 34 patient samples from the CoMMpass bulk RNA-seq data set and WES data, which confirmed that the landscape of immunogenic peptides evolves appreciably as the disease progresses. Only an average of 40% of predicted immunogenic mutations overlapped across successive time points (Supplementary Fig. S6B). In MMRF patient 1931, new immunogenic mutations, such as KRAS-G12R, SAMD1-G309V, and CDK13-L1011F, arose at a later time point, whereas others, such as UTRN-M394I and CABIN1-N1252K, had receded (Supplementary Fig. S6C). These evolutionary dynamics suggest that a tumor's ability to initiate antigenic immune responses is time dependent. Interestingly, we found four recurrent mutations with commonly predicted neoantigen across patients (Supplementary Table S5). They are from four genes, including KRAS, PABPC1, KMT2C, and RHPN2. As there are a limited number of predicted neoantigens overall, this strategy may be challenging for MM.
We next sought to shed light on expression patterns associated with specific clinical features. Given the diverse treatment background in WashU cohorts compared with the relatively uniform MMRF cohort, we restricted this analysis to MMRF samples and integrated all PCs from these 18 patients (Fig. 6F). Nine subjects were rapid progressors [progression-free survival (PFS) < 18 months], and nine subjects were nonprogressors (PFS not reached; Fig. 6G). TNFRSF13C (BAFF-R) and RASGRP3 are significantly highly expressed in rapid progressors compared with nonprogressors (Fig. 6H; logistic regression test; P = 0.02 and 0.03, respectively). RASGRP3 is involved in BCR signaling and is critical in B-cell receptor-mediated Ras activation (71, 72). Although the role of RASGRP3 in MM is not well understood, our observations suggest that TNFRSF13C and RASGRP3 may serve as indicators of MM progression.
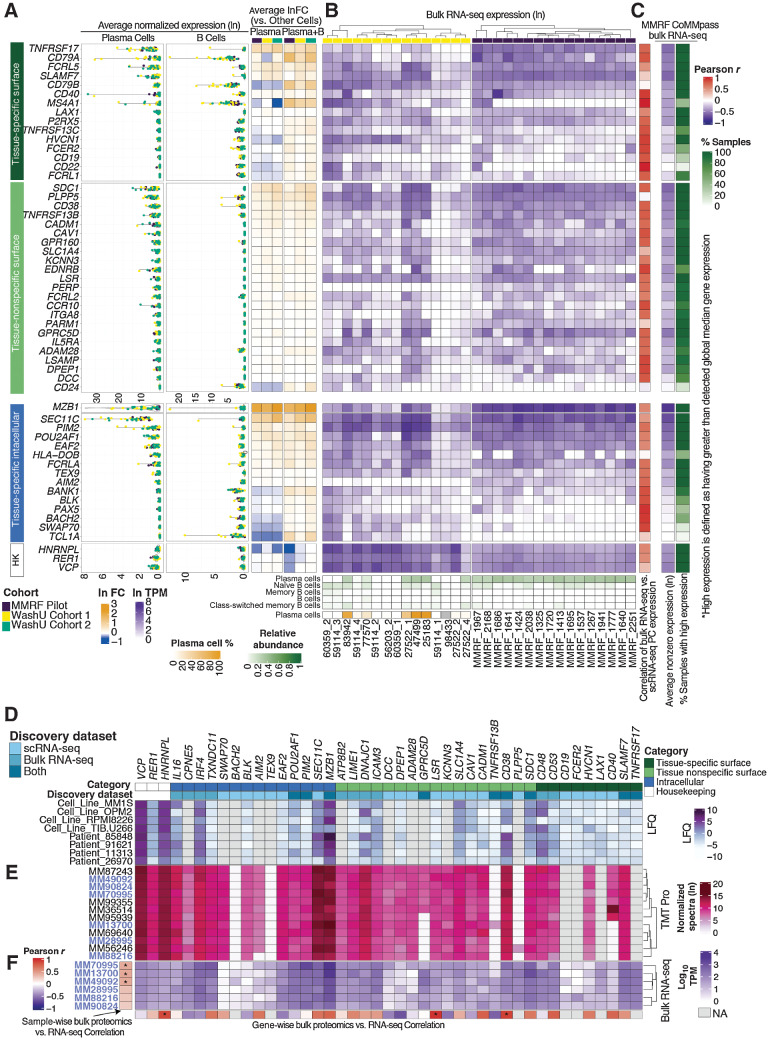
Cross-referencing bulk RNA-seq and bulk protein expression for multifaceted characterization of myeloma-associated markers
To better assess expression patterns across larger sample sizes, we first analyzed average normalized expression and logFC of candidate genes in integrated PCs from our combined discovery cohorts (Fig. 7A; integrated PCs shown in Fig. 4A). We then cross-referenced gene-expression data with 907 bulk RNA-sequencing samples, including 892 CD138+ sorted samples from the MMRF CoMMpass Study and 15 unsorted samples with matching single-cell data from WashU Cohort 1. When comparing PC gene expression from scRNA-seq to matching bulk CD138+ RNA-seq across the MMRF Immune Atlas Pilot cohort, we see a high positive correlation for a majority of genes (Pearson r ranging between 0.60 and 0.99; Fig. 7B). Notably, TNFRSF13C (r = 0.80), CD79A (r = 0.80), TNFRSF13B (r = 0.76), CD38 (r = 0.74), and TNFRSF17 (r = 0.72) were among the genes with highest positive correlation, whereas CD24 (r = −0.08), PLPP5 (r = −0.05), CD79B (r = 0.05), and SLAMF7 (r = 0.23) were among genes with the lowest (Supplementary Table S6). When comparing PCs in scRNA-seq to bulk, unsorted RNA-seq from WashU Cohort 1, we see that bulk expression of proposed target genes is highest in samples predicted by in silico deconvolution to have high plasma cell content, such as 83942, 27522_1, 47499, and 25183 (Fig. 7B).
Figure 7.
Comparison of scRNA-seq, bulk RNA-seq, and bulk proteomic expression of candidate targets. A, Average normalized expression (ln) and average fold change (ln) of target genes across scRNA-seq discovery cohorts. HK, housekeeping genes. B, Unsorted bulk RNA-seq expression of WashU Cohort 1 and CD138+-sorted bulk RNA-seq of the MMRF Immune Atlas Pilot Cohort. XCell deconvolution of plasma and B-cell relative abundance is shown along the bottom. Plasma cell percentages from matching scRNA-seq data is shown for WashU Cohort 1. Correlations between plasma cell expression (scRNA) and bulk CD138+ expression of each gene for MMRF Pilot samples are shown in the rightmost column. C, Average nonzero expression and expression prevalence across MMRF CoMMpass bulk RNA-seq. Expression prevalence is defined as a percentage of all samples wherein target expression is higher than global median nonzero gene expression. D, Bulk protein expression of target gene products assayed via label-free proteome; LFQ values were analyzed by MaxQuant, Log2 transformed, and subsequently internally normalized with missing values imputed. E, Bulk protein expression of target gene products assayed via TMT Pro; TMT intensities were analyzed by MaxQuant, Ln transformed, and internally normalized. F, Bulk RNA expression of 6 samples (names identified in purple) concurrently processed via TMT Pro. Sample-wise RNA-vs.-protein correlations across all target genes shown is displayed along the y-axis; gene-wise RNA vs. protein correlations across samples is displayed along the x-axis.
Looking at overall trends in MMRF CoMMpass data, we see that average nonzero bulk expression levels of target genes largely agree with normalized expression in PCs from our scRNA-seq cohort. Twenty-nine out of 38 genes encoding candidate surface targets are detected at levels higher than global median nonzero gene expression in >80% of the entire MMRF bulk RNA-seq cohort (Fig. 7C). These genes may represent potentially effective targets for large portions of the myeloma patient population. Among them, TNFRSF17, SLAMF7, FCRL5, CD79B, P2RX5, LAX1, CD40, HVCN1, and CD79A are primary candidates with high tissue specificity. Primary surface candidates with lesser prevalence include TNFRSF13C (higher expression than global median in 67% of samples), FCER2 (52%), MS4A1 (42%), and CD19 (30%). Nine of 15 tissue-specific intracellular targets also have higher-than-median expression with >80% prevalence, including EAF2, PIM2, FCRLA, AIM2, and BANK1.
Due to the limitations of scRNA-seq in both cohort size and sequencing depth, and having established the general concordance of both data types, we next utilized the CoMMpass bulk RNA-seq data set as a supplementary cohort for target discovery. We selected 1,261 genes with expression above the 95 percentile in at least 75% of 892 CD138+ sorted samples. Next, we stratified candidate genes based on their tissue specificity (evaluated via GTEx and HPA) into categories previously described: primary and secondary surface targets, and tissue-specific intracellular targets. We also examined the expression of these genes across over 1,000 cancer cell lines in the CCLE, retaining 21 genes with significantly higher average expression across 25 MM cell lines compared with those of other tissues (FDR < 0.1; Supplementary Fig. S7A). Top candidate genes identified by this method (including SLAMF7, TNFRSF17, FCRL5, CD79B, and GPRC5D) had been identified in scRNA-seq cohorts as well, reaffirming the validity of our scRNA-seq–based target discovery strategy. Other targets, such as CD53 (a possible growth regulator of hematopoietic cells; ref. 73), CD48 (which encodes a protein involved in lymphocyte adhesion and activation; ref. 74), DNAJC1, TXNDC11, IRF4, and CPNE5, were not captured by our scRNA-seq discovery. Among these, however, only TXNDC11, a thioredoxin that localizes to the ER (75), is both restricted to MM cell lines (CCLE; Supplementary Fig. S7B) and specifically overexpressed in PCs/B cells in our scRNA-seq data (Supplementary Fig. S7C). Its omission from our scRNA-seq–driven discovery may be due to sampling variation, as it failed to meet our criteria for the prevalence of overexpression. Other targets discovered only via bulk RNA-seq were not specifically expressed by PCs and/or B cells in our scRNA data. Although sampling variation may affect these results, our findings demonstrate the imprecision of sort-based lineage isolation upon which bulk sequencing strategies rely.
To assess the protein-level expression of candidate genes, we performed global proteomic analysis via LFQ and TMT mass spectrometry. Four MM cell lines (TIB.U266, RPMI8226, OPM2, and MM1ST) and four patient samples (Fig. 7D; clinical details in Supplementary Table S1) were analyzed by LFQ. The subsequent TMT-Pro assay was comprised of 12 patient samples (Fig. 7E; Supplementary Table S1), 6 of which were also divided for concurrent bulk RNA-seq (Fig. 7F). For each sample with paired bulk RNA and proteomic data, we see overall positive correlation across 40 target and 3 housekeeping genes (Fig. 7F). Across these 6 samples, 23 target genes had positive protein-vs.-RNA correlation, 14 with r > 0.5 and 2 (CD38, LSR) reaching significance. Other genes with high positive correlation include CD53 (r = 0.75), CADM1(r = 0.74), MZB1(r = 0.73), and CD40 (r = 0.72). Overall, trends in protein expression appear consistent across samples and between batches; however, we see clear differential expression between patients for CD40 and GPRC5D in the second batch (Fig. 7E). Genes with highly negative protein-vs.-RNA correlation include DPEP1 (r = −0.65), TNFRSF13B (r = −0.59), and KCNN3 (r = −0.52). As low protein detection may be due to protease incompatibility during peptide extraction or other technical artifacts, these negative correlations remain inconclusive. Taken together, in our two proteomic data sets 57% (30/53) of targets discovered via scRNA-seq and 90% (19/21) of targets discovered via bulk RNA had detectable protein expression (Fig. 7D and E). Five genes (CD38, DNAJC1, MZB1, SEC11C, and IRF4) were expressed at levels surpassing the detected global median in nearly all 20 samples profiled by proteomic quantitation, and 8 more (SLAMF7, CD53, CD48, SDC1, SLC1A4, ICAM3, POU2AF1, and IL16; Supplementary Fig. S7D) surpassed global medians in a majority of samples in one of the two batches. Although further functional studies are required, our proteomics data serve as an important starting reference that demonstrates the overall concordance between RNA and protein levels for candidate targets, as well as the heterogeneity between patients in both RNA and protein expression.
Discussion
We have described a strategy for the high-throughput discovery of MM therapeutic targets that captures known CAR-T targets and identifies previously understudied MM markers. Our analysis strategy has three key advantages: (i) systematic search for genes specifically overexpressed in PC/B cells without relying on prior sorting; (ii) direct comparison of cell types simultaneously captured by scRNA-seq that minimizes technical biases, (iii) versatility and ease of application toward identifying tumor markers in other cancer types.
Through pathway enrichment analysis, we offered a “broad-strokes” view of how upregulated genes in PC/B lineages may affect protein synthesis and immune signaling. The power of database-driven functional analysis lies in its ability to narrow the search space for relevant biological processes. However, further study is needed to elucidate how individual genes promote or inhibit these processes before therapeutic strategies can be built around disrupting their function.
We proceeded to examine tumor heterogeneity at both interpatient and subtumor levels using multiple data types profiling multiple patient cohorts. Taken together, our data reinforce the notion that patient heterogeneity restricts target selection when designing treatment strategies. Consistent coexpression may constitute a particular disease profile, wherein intratumor subpopulations exhibit heterogeneous expression of a limited group of markers, but are unlikely to express an altogether different set of markers. Alternatively, pairing targets from consistently exclusive marker groups may be more effective in “catching” tumor cells that undergo antigen escape as they transition into different disease phenotypes.
Our findings suggest the existence of discrete target coexpression phenotypes, as well as the potential stochastic variation in consistently co-occurring targets within otherwise highly uniform tumor clusters. This latter finding requires further study, as distinguishing consistent phenotypical differences from dynamic expression changes is critical to solving the challenges of antigen escape and antigen coverage. To cross-validate our proposed targets, we examined marker gene expression in bulk RNA-seq from a large cohort of CD138+ sorted MM PCs, and quantified encoded protein levels in MM cell lines and primary samples using immunophenotyping and mass spectrometry. Although limited by technical incompatibilities, we demonstrated overall concordance between data types for most targets.
Our strategy can be improved in several ways. Ideal discovery data sets would include patients exposed to uniform treatment regimens and controlled for clinical demographics. The three independent cohorts in our study were sequenced using two different library preparation chemistries, and although we were largely able to remove batch effects, technical biases may persist. Due to our limited cohort of normal BM, we were unable to deeply investigate therapeutic opportunities in MM precursors, and due to the low coverage of scRNA-seq data, we are yet unable to perform expression correlation at single-cell resolution. Our analysis has revealed intratumor heterogeneity of tumor markers (Fig. 6B–E), and further investigation may elucidate the clinical or pathologic significance of MM subpopulations. We were unable to confirm cellular localization of all proposed targets due to limited antibody availability for conclusive immunophenotyping, and further experimental validation is therefore required. Future studies functionally characterizing our proposed MM markers, and assessing the toxicity and efficacy of targeting them, will ultimately decide their clinical utility.
We are mindful that the full collection of MM-associated antigens may still be incomplete; however, this study represents the most systematic search of myeloma antigens to date, and our findings provide researchers with a reference list of candidate targets to aid the development of immunotherapy and other targeted therapies against MM.
Supplementary Material
supplementary figures
Clinical information of patients in scRNA-seq discovery cohort, flow cytometry validation cohort and proteomics validation cohort, including age, gender, race, ISS stage, time to disease progression (TTPD). Disease stage for WashU cohort 1.
Novelty description of candidate tumor markers. Each gene is ranked according to existing literature and/or clinical trials investigating its role in multiple myeloma. Representative studies are listed.
Batch specific co-expression analysis.
NetMHC analysis output for tissue-specific intracellular genes. Only peptides identified as strong binders (% rank <0.5) are shown. Detailed explanation of each column can be found here: http://www.cbs.dtu.dk/services/NetMHC/output.php.
List of recurrent mutations with potential utility as neoantigens
Pearson R correlation values between bulk RNA expression and scRNA expression in PCs for target genes.
Acknowledgments
The authors thank the multiple myeloma patients, families, and professionals who have contributed to this study and the Multiple Myeloma Research Foundation CoMMpass Study and Immune Atlas Pilot study. This study has also been supported by the Paula C. and Rodger O. Riney Blood Cancer Research Fund to J.F. DiPersio, R. Vij, and L. Ding, NCI U24CA211006, U2CCA233303, and PJ000021702 funds to L. Ding, NCI R35 CA210084 to J.F. DiPersio, and NCI R50CA211466 to M.P. Rettig.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
R.G. Jayasinghe reports other support from Multiple Myeloma Research Foundation outside the submitted work. J. O'Neal receives royalties from Wugen. S.M. Foltz reports grants from Paula C. and Rodger O. Riney Blood Cancer Research Initiative Fund during the conduct of the study. E. Storrs reports grants from NIH during the conduct of the study. J.F. DiPersio is a cofounder of and has equity in Magenta and Wugen. No disclosures were reported by the other authors.
Authors' Contributions
L. Yao: Data curation, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. J.T. Wang: Software, formal analysis, validation, investigation, methodology, writing–original draft. R.G. Jayasinghe: Supervision, investigation, methodology, writing–review and editing. J. O'Neal: Formal analysis, validation, methodology. C.-F. Tsai: Validation, investigation, methodology. M.P. Rettig: Conceptualization, data curation. Y. Song: Formal analysis. R. Liu: Formal analysis. Y. Zhao: Validation. O.M. Ibrahim: Formal analysis. M.A. Fiala: Conceptualization, data curation, methodology. J.M. Fortier: Resources, data curation. S. Chen: Validation. L. Gehrs: Data curation. F.M. Rodrigues: Investigation. M.C. Wendl: Writing–review and editing. D. Kohnen: Data curation. A. Shinkle: Validation. S. Cao: Formal analysis, methodology. S.M. Foltz: Investigation. D.C. Zhou: Conceptualization. E. Storrs: Conceptualization. M.A. Wyczalkowski: Visualization. S. Mani: Data curation. S.R. Goldsmith: Data curation. Y. Zhu: Formal analysis. M. Hamilton: Resources. T. Liu: Supervision. F. Chen: Supervision. R. Vij: Conceptualization, supervision. L. Ding: Conceptualization, supervision, writing–review and editing. J.F. DiPersio: Conceptualization, supervision.
References
- 1. June CH, Sadelain M. Chimeric antigen receptor therapy. N Engl J Med 2018;379:64–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Franssen LE, Mutis T, Lokhorst HM, van de Donk NWCJ. Immunotherapy in myeloma: how far have we come? Ther Adv Hematol 2019;10:2040620718822660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Filley AC, Henriquez M, Dey M. CART immunotherapy: development, success, and translation to malignant gliomas and other solid tumors. Front Oncol 2018;8:453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Drent E, Poels R, Mulders MJ, van de Donk NWCJ, Themeli M, Lokhorst HM, et al. Feasibility of controlling CD38-CAR T cell activity with a Tet-on inducible CAR design. PLoS One 2018;13:e0197349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gogishvili T, Danhof S, Prommersberger S, Rydzek J, Schreder M, Brede C, et al. SLAMF7-CAR T cells eliminate myeloma and confer selective fratricide of SLAMF7+ normal lymphocytes. Blood 2017;130:2838–47. [DOI] [PubMed] [Google Scholar]
- 6. Majzner RG, Mackall CL. Tumor antigen escape from CAR T-cell therapy. Cancer Discov 2018;8:1219–26. [DOI] [PubMed] [Google Scholar]
- 7. Liu R, Gao Q, Foltz SM, Fowles JS, Yao L, Wang JT, et al. Co-evolution of tumor and immune cells during progression of multiple myeloma. Nat Commun 2021;12:2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ledergor G, Weiner A, Zada M, Wang S-Y, Cohen YC, Gatt ME, et al. Single cell dissection of plasma cell heterogeneity in symptomatic and asymptomatic myeloma. Nat Med 2018;24:1867–76. [DOI] [PubMed] [Google Scholar]
- 9. Shah N, Chari A, Scott E, Mezzi K, Usmani SZ. B-cell maturation antigen (BCMA) in multiple myeloma: rationale for targeting and current therapeutic approaches. Leukemia 2020;34:985–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim K-T, Lee HW, Lee H-O, Song HJ, Jeong DE, Shin S, et al. Application of single-cell RNA sequencing in optimizing a combinatorial therapeutic strategy in metastatic renal cell carcinoma. Genome Biol 2016;17:80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol 2019;20:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bausch-Fluck D, Hofmann A, Bock T, Frei AP, Cerciello F, Jacobs A, et al. A mass spectrometric-derived cell surface protein atlas. PLoS One 2015;10:e0121314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ross SM. Peirce's criterion for the elimination of suspect experimental data. J Eng Technol 2003;20:38–41. [Google Scholar]
- 14. Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB–a database for integrating human functional interaction networks. Nucleic Acids Res 2009;37:D623–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res 2020;48:D498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 2016;34:525–7. [DOI] [PubMed] [Google Scholar]
- 18. Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol 2017;18:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi T, Fillmore TL, Sun X, Zhao R, Schepmoes AA, Hossain M, et al. Antibody-free, targeted mass-spectrometric approach for quantification of proteins at low picogram per milliliter levels in human plasma/serum. Proc Natl Acad Sci U S A 2012;109:15395–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tsai C-F, Zhao R, Williams SM, Moore RJ, Schultz K, Chrisler WB, et al. An improved boosting to amplify signal with isobaric labeling (iBASIL) strategy for precise quantitative single-cell proteomics. Mol Cell Proteomics 2020;19:828–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 2008;26:1367–72. [DOI] [PubMed] [Google Scholar]
- 22. Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc 2016;11:2301–19. [DOI] [PubMed] [Google Scholar]
- 23. Tyanova S, Temu T, Sinitcyn P, Carlson A, Hein MY, Geiger T, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 2016;13:731–40. [DOI] [PubMed] [Google Scholar]
- 24. Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Saunders CT, Wong WSW, Swamy S, Becq J, Murray LJ, Cheetham RK. Strelka: accurate somatic small-variant calling from sequenced tumor–normal sample pairs. Bioinformatics 2012;28:1811–7. [DOI] [PubMed] [Google Scholar]
- 27. Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009;25:2865–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Szolek A, Schubert B, Mohr C, Sturm M, Feldhahn M, Kohlbacher O. OptiType: precision HLA typing from next-generation sequencing data. Bioinformatics 2014;30:3310–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Andreatta M, Nielsen M. Gapped sequence alignment using artificial neural networks: application to the MHC class I system. Bioinformatics 2016;32:511–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao L, Jayasinghe RG, Lee BH, Bhasin SS, Pilcher W, Doxie DB, et al. Comprehensive characterization of the multiple myeloma immune microenvironment using integrated scRNA-seq, CyTOF, and CITE-seq analysis. Cancer Res Commun 2022;2:1255–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Johnsen HE, Bøgsted M, Schmitz A, Bødker JS, El-Galaly TC, Johansen P, et al. The myeloma stem cell concept, revisited: from phenomenology to operational terms. Haematologica 2016;101:1451–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, et al. The gene ontology (GO) database and informatics resource. Nucleic Acids Res 2004;32:D258–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science 2015;347:1260419. [DOI] [PubMed] [Google Scholar]
- 34. GTEx Consortium. The genotype-tissue expression (GTEx) project. Nat Genet 2013;45:580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brudno JN, Maric I, Hartman SD, Rose JJ, Wang M, Lam N, et al. T cells genetically modified to express an anti-B-cell maturation antigen chimeric antigen receptor cause remissions of poor-prognosis relapsed multiple myeloma. J Clin Oncol 2018;36:2267–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chu J, He S, Deng Y, Zhang J, Peng Y, Hughes T, et al. Genetic modification of T cells redirected toward CS1 enhances eradication of myeloma cells. Clin Cancer Res 2014;20:3989–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Zhang Z, Westhrin M, Bondt A, Wuhrer M, Standal T, Holst S. Serum protein N-glycosylation changes in multiple myeloma. Biochim Biophys Acta Gen Subj 2019;1863:960–70. [DOI] [PubMed] [Google Scholar]
- 38. Chattopadhyay S, Thomsen H, da Silva Filho MI, Weinhold N, Hoffmann P, Nöthen MM, et al. Enrichment of B cell receptor signaling and epidermal growth factor receptor pathways in monoclonal gammopathy of undetermined significance: a genome-wide genetic interaction study. Mol Med 2018;24:30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Demchenko YN, Kuehl WM. A critical role for the NFkB pathway in multiple myeloma. Oncotarget 2010;1:59–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roy P, Sarkar UA, Basak S. The NF-κB activating pathways in multiple myeloma. Biomedicines 2018;6:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, et al. Defining a cancer dependency map. Cell 2017;170:564–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Aggarwal R, Ghobrial IM, Roodman GD. Chemokines in multiple myeloma. Exp Hematol 2006;34:1289–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu B, Yan L, Zhou M. Target selection of CAR T cell therapy in accordance with the TME for solid tumors. Am J Cancer Res 2019;9:228–41. [PMC free article] [PubMed] [Google Scholar]
- 44. Carithers LJ, Moore HM. The genotype-tissue expression (GTEx) project. Biopreserv Biobank 2015;13:307–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Thul PJ, Lindskog C. The human protein atlas: a spatial map of the human proteome. Protein Sci 2018;27:233–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Luger D, Yang Y-A, Raviv A, Weinberg D, Banerjee S, Lee M-J, et al. Expression of the B-cell receptor component CD79a on immature myeloid cells contributes to their tumor promoting effects. PLoS One 2013;8:e76115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palumbo A, Sonneveld P. Preclinical and clinical evaluation of elotuzumab, a SLAMF7-targeted humanized monoclonal antibody in development for multiple myeloma. Expert Rev Hematol 2015;8:481–91. [DOI] [PubMed] [Google Scholar]
- 48. Prommersberger S, Reiser M, Beckmann J, Danhof S, Amberger M, Quade-Lyssy P, et al. CARAMBA: a first-in-human clinical trial with SLAMF7 CAR-T cells prepared by virus-free Sleeping Beauty gene transfer to treat multiple myeloma. Gene Ther 2021;28:560–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sidana S, Shah N. CAR T-cell therapy: is it prime time in myeloma? Blood Adv 2019;3:3473–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hengeveld PJ, Kersten MJ. B-cell activating factor in the pathophysiology of multiple myeloma: a target for therapy? Blood Cancer J 2015;5:e282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Walters M, Olteanu H, Van Tuinen P, Kroft SH. CD23 expression in plasma cell myeloma is specific for abnormalities of chromosome 11, and is associated with primary plasma cell leukaemia in this cytogenetic sub-group. Br J Haematol 2010;149:292–3. [DOI] [PubMed] [Google Scholar]
- 52. Ding S, Mao X, Cao Y, Wang N, Xu H, Zhou J. Targeting CD79b for chimeric antigen receptor T-cell therapy of B-cell lymphomas. Target Oncol 2020;15:365–75. [DOI] [PubMed] [Google Scholar]
- 53. Spiegel JY, Patel S, Muffly L, Hossain NM, Oak J, Baird JH, et al. CAR T cells with dual targeting of CD19 and CD22 in adult patients with recurrent or refractory B cell malignancies: a phase 1 trial. Nat Med 2021;27:1419–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yu T, Chaganty B, Lin L, Xing L, Ramakrishnan B, Wen K, et al. VIS832, a novel CD138-targeting monoclonal antibody, potently induces killing of human multiple myeloma and further synergizes with IMiDs or bortezomib in vitro and in vivo. Blood Cancer J 2020;10:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Morandi F, Horenstein AL, Costa F, Giuliani N, Pistoia V, Malavasi F. CD38: a target for immunotherapeutic approaches in multiple myeloma. Front Immunol 2018;9:2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee L, Draper B, Chaplin N, Philip B, Chin M, Galas-Filipowicz D, et al. An APRIL-based chimeric antigen receptor for dual targeting of BCMA and TACI in multiple myeloma. Blood 2018;131:746–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pillarisetti K, Edavettal S, Mendonça M, Li Y, Tornetta M, Babich A, et al. A T-cell-redirecting bispecific G-protein-coupled receptor class 5 member D x CD3 antibody to treat multiple myeloma. Blood 2020;135:1232–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, et al. Comprehensive integration of single-cell data. Cell 2019;177:1888–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Stewart AK, Krishnan AY, Singhal S, Boccia RV, Patel MR, Niesvizky R, et al. Phase I study of the anti-FcRH5 antibody-drug conjugate DFRF4539A in relapsed or refractory multiple myeloma. Blood Cancer J 2019;9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Frigyesi I, Adolfsson J, Ali M, Christophersen MK, Johnsson E, Turesson I, et al. Robust isolation of malignant plasma cells in multiple myeloma. Blood 2014;123:1336–40. [DOI] [PubMed] [Google Scholar]
- 61. Grigoriadis G, Whitehead S. CD138 shedding in plasma cell myeloma. Br J Haematol 2010;150:249. [DOI] [PubMed] [Google Scholar]
- 62. He Q, Jiang X, Zhou X, Weng J. Targeting cancers through TCR-peptide/MHC interactions. J Hematol Oncol 2019;12:139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Chanukuppa V, Paul D, Taunk K, Chatterjee T, Sharma S, Shirolkar A, et al. Proteomics and functional study reveal marginal zone B and B1 cell specific protein as a candidate marker of multiple myeloma. Int J Oncol 2020;57:325–37. [DOI] [PubMed] [Google Scholar]
- 64. Nikesitch N, Lee JM, Ling S, Roberts TL. Endoplasmic reticulum stress in the development of multiple myeloma and drug resistance. Clin Transl Immunology 2018;7:e1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Lu J, Zavorotinskaya T, Dai Y, Niu X-H, Castillo J, Sim J, et al. Pim2 is required for maintaining multiple myeloma cell growth through modulating TSC2 phosphorylation. Blood 2013;122:1610–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zhao C, Inoue J, Imoto I, Otsuki T, Iida S, Ueda R, et al. POU2AF1, an amplification target at 11q23, promotes growth of multiple myeloma cells by directly regulating expression of a B-cell maturation factor, TNFRSF17. Oncogene 2008;27:63–75. [DOI] [PubMed] [Google Scholar]
- 67. Maiers M, Gragert L, Klitz W. High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 2007;68:779–88. [DOI] [PubMed] [Google Scholar]
- 68. Gonzalez-Galarza FF, McCabe A, Santos EJMD, Jones J, Takeshita L, Ortega-Rivera ND, et al. Allele frequency net database (AFND) 2020 update: gold-standard data classification, open access genotype data and new query tools. Nucleic Acids Res 2020;48:D783–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang S, He Z, Wang X, Li H, Liu X-S. Antigen presentation and tumor immunogenicity in cancer immunotherapy response prediction. Elife 2019;8:e49020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramachandran J, Santo L, Siu KT, Panaroni C, Raje N. Pim2 is important for regulating DNA damage response in multiple myeloma cells. Blood Cancer J 2016;6:e462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yan H, Zheng G, Qu J, Liu Y, Huang X, Zhang E, et al. Identification of key candidate genes and pathways in multiple myeloma by integrated bioinformatics analysis. J Cell Physiol 2019;234:23785–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Aiba Y, Oh-hora M, Kiyonaka S, Kimura Y, Hijikata A, Mori Y, et al. Activation of RasGRP3 by phosphorylation of Thr-133 is required for B cell receptor-mediated Ras activation. Proc Natl Acad Sci U S A 2004;101:16612–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Dunlock VE. Tetraspanin CD53: an overlooked regulator of immune cell function. Med Microbiol Immunol 2020;209:545–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. McArdel SL, Terhorst C, Sharpe AH. Roles of CD48 in regulating immunity and tolerance. Clin Immunol 2016;164:10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wang D, De Deken X, Milenkovic M, Song Y, Pirson I, Dumont JE, et al. Identification of a novel partner of duox: EFP1, a thioredoxin-related protein. J Biol Chem 2005;280:3096–103. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary figures
Clinical information of patients in scRNA-seq discovery cohort, flow cytometry validation cohort and proteomics validation cohort, including age, gender, race, ISS stage, time to disease progression (TTPD). Disease stage for WashU cohort 1.
Novelty description of candidate tumor markers. Each gene is ranked according to existing literature and/or clinical trials investigating its role in multiple myeloma. Representative studies are listed.
Batch specific co-expression analysis.
NetMHC analysis output for tissue-specific intracellular genes. Only peptides identified as strong binders (% rank <0.5) are shown. Detailed explanation of each column can be found here: http://www.cbs.dtu.dk/services/NetMHC/output.php.
List of recurrent mutations with potential utility as neoantigens
Pearson R correlation values between bulk RNA expression and scRNA expression in PCs for target genes.
Data Availability Statement
Single-cell RNA-seq and bulk RNA-seq expression matrices are available at NCBI GEO under accession numbers GSE223060 and GSE223061, respectively.