Abstract
The genus Vibrio includes bacteria with different morphological and metabolic characteristics responsible for different human and animal diseases. An accurate identification is essential to assess the risks in regard to aquatic organisms and consequently to public health. The Multilocus Sequence Analysis (MLSA) scheme developed on the basis of 4 housekeeping genes (gyrB, pyrH, recA and atpA) was applied to identify 92 Vibrio strains isolated from crustaceans in 2011. Concatenated sequences were used for the phylogenetic and population analyses and the results were compared with those from biochemical identification tests. From the phylogenetic analysis, 10 clusters and 4 singletons emerged, whereas the population analysis highlighted 12 subpopulations that were well supported by phylogeny with few exceptions. The retrospective analysis allowed correct re-attribution of isolated species, indicating how, for some pathogens, there may be an overestimation of phenotypic identification (e.g. V. parahaemolyticus). Use of the PubMLST Vibrio database highlighted a possible genetic link between Sequence Type (ST) 529 and ST195 (V. alginolyticus) isolated from a human case in Norway during 2018. In addition to the identification of major risk groups of V. cholerae, V. vulnificus and V. parahaemolyticus, MLSA could be a valid support for species considered a minor risk, such as V. alginolyticus, V. mimicus and V. fluvialis. Due to the increased incidence of vibriosis in Europe, the application of different tools will also have to be considered to investigate the possible epidemiological links of the various species in the perspective of Open Science to protect the consumer.
Key words: Vibrio spp, crustacean, MLSA, emerging risks
Introduction
At present, few data are available about vibriosis incidence and the related outbreaks both, foodborne and extra-intestinal infections (Onohuean et al., 2022). The underestimation of this foodborne illness is due to mild gastrointestinal symptoms that do not require any medical treatment; therefore, foodborne vibriosis is not a notifiable disease in most European countries (Amato et al., 2022). However, due to climate change and extreme meteorological events (heatwave, flood, changes in water salinity), the number of cases related to Vibrio species causing vibriosis has dramatically increased in Europe in the last decade (Brehm et al., 2021; ECDC, 2021; Amato et al., 2022). Moreover, eating habits play an important role in the transmission of vibriosis. Italian consumers enjoy raw or slightly cooked seafood such as crustaceans or shellfish, that may be accidentally contaminated by foodborne pathogens, including Vibrio spp. This behaviour can increase safety concerns and the spread of outbreaks.
Vibrio is one of the most studied genera found in aquatic ecosystems and includes the major culturable bacteria in marine and estuarine environments; indeed, many species of Vibrio are part of the indigenous aquatic microbiota. According to recent species updates, there are around 147 species of Vibrio and 4 subspecies (Sampaio et al., 2022), but the description of new species has led to a constantly changing taxonomy.
Austin (2010) suggested a classification of zoonotic Vibrio spp. in 2 groups named higher risk vibrios (V. cholerae, V. parahaemolyticus and V. vulnificus, the main species causing serious foodborne gastroenteritis in humans) and lower risk vibrios (V. alginolyticus, V. fluvialis, V. furnissii, V. harveyi, V. metschnikovii and V. mimicus).
Currently, the European legislation still lacks microbiological criteria on the punctual monitoring of Vibrio contamination in fishery products. However, the Italian guidelines related to EC Regulations 882/2004 (European Commission, 2004) and 854/2004 (European Commission, 2004) have indicated Vibrio cholerae (V. cholerae O1, V. cholerae 0139, V. cholerae non-O1, V. cholerae non-O139) and V. parahaemolyticus as hazards to detect in fishery products during the official controls. Moreover, the guidelines mentioned the methods suitable for the detection of the potentially enteropathogenic Vibrio species. Although the Regulation 625/2017 (European Commission, 2017) repeals the previous regulations (EC Reg. 882/2004 and 854/2004), the guidelines are still valid in Italy according to the note No. 0069887/2019 (Italian Health Ministry, 2019).
Numerous phenotypic schemes and biomolecular methods have been developed to characterize and classify Vibrio species. Classical biochemical tests are usually applied to identify the Vibrio genus, but the great phenotypic diversity among strains makes their application difficult. Moreover, traditional analyses are time and resourceconsuming. For this reason, researchers are focused on the simplification of the classical identification protocols to implement the specific ISO (ISO, 2017) and to test the efficacy of molecular methods for the detection of the most important Vibrio human pathogens and their putative virulence markers (Hartnell et al., 2019). 16S rRNA gene sequencing can give an accurate identification of vibrios at the family and genus levels but identification at the species and strain levels requires the application of genomic analysis.
The Multilocus Sequence Analysis (MLSA) approach is a valid alternative to biochemical as well as fingerprint patternbased methods for species identification. MLSA has proved to be a very practical and reliable method and one of the main advantages of it is the reproducibility among different laboratories. Several molecular markers, e.g. recA, pyrH, gyrB and atpA in single or concatenated sequences, have been used to identify Vibrionaceae species and many different specific schemes are available for Vibrio spp. or Vibrionaceae (Rahman et al., 2014).
The aim of this retrospective study was to apply the MLSA scheme previously developed by Rahman et al. (2014) to identify and characterize Vibrio spp. isolated from crustaceans of the northeast Italian market. The data were analysed using different approaches in order to define the Vibrio species associated with different commercialized crustaceans and the possible genetic relationships among the strains. The direct comparison of sequences and allelic profiles deposited on the public database Vibrio spp. PubMLST (https://pubmlst.org/ organisms/vibrio-spp) allowed the definition of additional links between strains collected from shellfish and others implicated in human cases of vibriosis. A comparison between MLSA attribution and the biochemical identification highlighted some limits of the phenotypic methods. In the frame of open science, this study aimed to represent a first step of the hazard identification to characterize Vibrio spp. associated with crustacean marketing and consumption in northeast Italy.
Materials and Methods
The MLSA scheme was applied to identify Vibrio species isolated from fresh and defrosted samples of various crustacean species (Palaemon spp., Crangon crangon, Squilla mantis, Hymenopenaeus muelleri, Carcinus aestuarii). Samples were collected during a market survey in Venice from July to December 2011 (see the PubMLST Vibrio spp. website for sampling details: https://pubmlst.org/organisms/vibrio-spp; ID isolate collection from 1147 to 1244; Supplementary Table 1 and the paper of Caburlotto et al. (2016) for additional information. The shellfish originated mainly from the North Adriatic (Chioggia area, Venice Lagoon, Po Delta – Goro) and South Adriatic Sea.
Isolation of Vibrio strains and species identification by biochemical methods
In collaboration with the National Reference Laboratory for Fish, Crustacean and Mollusc Pathologies, IZSVe (Adria, Italy), the samples were prepared according to Caburlotto et al. (2016). In brief, for the first enrichment, 25 g of sample (crustacean pulp and a portion of the carapace) was homogenized in 225 mL of common alkaline peptone water (APW) and APW with 2% NaCl and incubated at 37°C for 18–24 hours. The Vibrio spp. were enumerated by the Most Probable Number (MPN) procedure and pure colonies were obtained from each enrichment medium streaked on thiosulphate citrate bile salt sucrose agar (TCBS) and on ChromAgar plates (37°C for 18–24 hours). The Vibrio presumptive colonies (6–8 per sample) were then subjected to Gram staining, resistance to vibriostatic O129, oxidase test, and O/F test and growth at different salt concentrations. Gram-negative, oxidase-positive and facultative anaerobic (+/+ for O/F test) isolates were identified with miniaturized biochemical tests (API20NE, bioMérieux, Florence, Italy).
Figure 1.

Neighbour-joining phylogenetic tree (compressed) with concatenated sequences of 4 housekeeping genes for the Vibrio strains isolated from crustacean samples in 2011. Numbers in brackets describe the number of strains included in the reference species group (black triangle).
MLSA approach - DNA extraction, polymerase chain reaction amplification and sequencing
The MLSA scheme followed in this study was from Rahman et al. (2014). In brief, four housekeeping genes (gyrB, pyrH, recA and atpA) were chosen for the MLSA.
DNA were extracted by boiling from 107 pure colonies classified as Vibrionaceae by API20NE (102 Vibrio species and 5 other genera). The Polymerase chain reaction (PCR) amplification was performed in a Euroclone One Advanced thermal cycler (Celbio, Milan, Italy) following different amplification conditions described in detail in Rahman et al. (2014) and on Vibrio spp. PubMLST (https://pubmlst.org/static/organisms/vibriospp/ Vibrio_primers.pdf). The amplicon of each gene was verified by BLAST search for an initial species attribution.
Phylogenetic analysis of MLSA data
The concatenated sequences were aligned for phylogenetic analysis by using MEGA v5.04 (Tamura et al., 2011) according to the Kimura two-parameter model and the phylogenetic tree was constructed using the neighbour-joining method.
In order to better describe the phylogenetic relatedness among isolates, we also sequenced 16 Vibrio reference strains and included the sequences downloaded from the NCBI database (Supplementary Table 2). The taxon name of each cluster was attributed according to the available reference/NCBI strains clustered in the same group. When the isolates were considered related but clearly distinct, the strain name representative for the cluster was used (e.g. Vibrio sp. Vi20). All strains were also screened for virulence genes markers by specific PCR protocols (genes: ToxR, tlh, tdh and trh; (Bej et al., 1999; Kim et al., 1999).
Structure analysis and genetic relationship
The linkage model was used to identify groups with distinct allele frequencies in STRUCTURE software (Falush et al., 2003). This procedure assigns a probability of ancestry for each polymorphic nucleotide for a given number of groups, K, and it estimates q, the combined probability of ancestry from each of the K groups for each individual isolate (Rahman et al., 2017). This analysis was conducted in order to verify the phylogenetic species attribution and to compare MLSA and API20NE classifications.
All the new sequences, allelic profiles and new sequence types were submitted to the public database Vibrio spp. (https://pubmlst.org/organisms/vibrio-spp). The PHYLOViZ (www.phyloviz.net/) program was applied to verify the possible relationships between the epidemiological information (geographic area of isolation, Vibrio species, year of isolation and source of isolation such as clinical/environment) provided by the public database Vibrio spp. and the genotypic profiles of Italian strains. On the date of analysis (2022-07-05), the database included 969 allele sequences, 1184 isolates and 272 genomes of vibrios.
Results and Discussion
Among 107 putative Vibrio strains isolated from crustacean samples (Supplementary Table 1), 7 strains amplified only with the atpA gene and were identified as Shewanella spp. by BLAST search. Another 8 strains did not amplify with one or more genes of the MLSA scheme and were excluded by the subsequent analyses (Supplementary Table 1). However, it was possible to define the genus of these strains, where 7 are Vibrio and 1 Photobacterium (Supplementary Table 1).
Phylogenetic analysis with a neighbourjoining tree showed 10 clusters and 4 singletons (Figure 1). In particular, the database was mainly formed by V. parahaemolyticus (43% of total Vibrio strains), V. diabolicus (9%), V. alginolyticus (17%), V. harveyi group (7.6%) and V. splendidus (5.4%). The only higher risk Vibrio species identified in crustacean samples was V. parahaemolyticus, while there were no V. cholerae or V. vulnificus. The most represented lower risk vibrios were V. alginolyticus followed by V. diabolicus and V. splendidus. In one study by Traoré et al. (2012) to assess the risk of Vibrio spp. transmission from crustaceans to humans, they identified 40% of the isolates as V. alginolyticus, 36% as V. parahaemolyticus and 24% as the nontoxigenic V. cholerae. Koralage et al. (2012) in their investigation on the prevalence of Vibrio spp. in shrimp farms, found V. parahaemolyticus was the most common (91.2%) followed by V. alginolyticus (18.8%), V. cholerae non- O1/non-O139 (4.1%) and V. vulnificus (2.4%). At the market level, the prevalence of many Vibrio species (such as V. mimicus, V. vulnificus, V. alginolyticus and V. parahaemolyticus) was found in around 20% of crustacean samples (Álvarez-Contreras et al., 2021).
Figure 2.
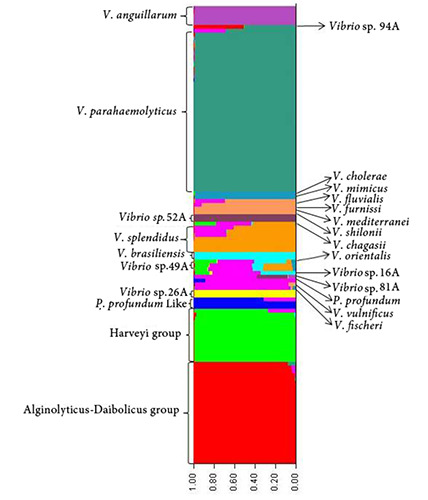
Population clustering (strains isolated from crustacean samples in 2011) identified by STRUCTURE software on the concatenated sequences of 4 genes. A single colour corresponds to a single population, while columns with mixed colours include strains carrying DNA from different populations. The analysis showed 12 ancestral groups. Groups with more than one isolate are indicated on the left side and single strains are showed on the right side (V. cholerae, V. mimicus, V. fluvialis, V. furnissii, V. mediterranei, V. shilonii, V. chagasii, V. orientalis, V. vulnificus, V. fischeri and P. profundum only represent the reference strains, not isolates in our study).
The Harveyi clade contains 4 species (V. harveyi, V. campbellii, V. rotiferianus and V. owensii) that are pathogens for marine animals (Pretto, 2020; Harrison et al., 2022). The BLAST analysis highlighted the putative attribution of only two species of this group, V. harveyi and V. rotiferianus.
Structure analysis recognized 12 subpopulations with the highest delta K value of 31.136 (Figure 2). The population structure confirmed the phylogenetic analysis, while V. alginolyticus-diabolicus were included in the same genetic population. Both analyses agreed with the previous findings on the MLSA application, with a similar Vibrio species definition as described by Rahman et al. (2014).
Finally, 92 strains were analysed using the MLSA approach, of which 52 (56.5%) strains had the same identification as for the biochemical method. The Sankey diagram (Figure 3) showed the cases of misclassification between methods. In V. parahaemolyticus, 11 false positive and 4 false negative strains were identified as compared to the biochemical approach. In total, 40 V. parahaemolyticus were identified by MLSA, whereas 51 were identified using the biochemical method. 10% of V. parahaemolyticus were finally assigned to the genus Shewanella according to the atpA sequence. The V. parahaemolyticus strains were also checked using species-specific toxR and tlh genes; moreover, the tdh and trh genes were tested to assess the virulence properties. No virulence factors were detected; moreover, the identification of strains by MLSA totally agreed with the toxR results. The application of these genetic markers is strongly recommended to identify V. parahaemolyticus and to detect putative enteropathogenic isolates (CSR 212/2016, 2016). Moreover, for the three major Vibrio species (V. cholerae, V. parahaemolyticus and V. vulnificus), the application of standardized protocols for biochemical identification is considered as an important prerequisite for diagnostic laboratories, especially for environmental strains as in the case of shellfish and crustacean samples (Hartnell et al., 2019). No probable enteropathogenic strains were detected in the present Vibrio database; however, the number of samples/strains should be enlarged. The V. parahaemolyticus strains included in this retrospective analysis are only a part of those described by Caburlotto et al. (2016). However, the main focus of this retrospective work was to elucidate the feasibility of the MLSA approach on strains collected from this matrix and to define a first detailed evaluation of all the Vibrio species from crustaceans. In particular, the API20NE identification tended to underestimate several minor species such as L. anguillarum, V. harveyi group and V. splendidus. Moreover, the MLSA allowed the classification of undefined Vibrio sp. strains such as V. alginolyticus-diabolicus genetic clusters.
MLSA/Multi-locus Sequence Typing (MLST) is also a suitable tool to highlight links between isolates from different sources. In this regard, the comparison of sequence types (STs) available on public databases allowed fast and easy detection of epidemiological relatedness. Moreover, the possibility of comparing data from whole genome analysis could increase the resolution and detail of these comparisons.
The analysis of allelic profiles showed the presence of 72 STs with the definition of 67 new STs that were deposited in the Vibrio spp. PubMLST website. Interestingly, from the same crustacean sample, not only the copresence of different Vibrio species detected, but also different STs for each species (see isolate details) were found. The sequence analysis of a large number of Vibrio isolates per sample allowed the definition of a complex picture of the Vibrio strains associated with crustaceans, as in the case of Crangon crangon (sample 269/ITT) where four different V. parahaemolyticus STs were detected, or Palaemon elegans (sample 234/TT) with the presence of many different Vibrio species and different STs. These observations suggest screening many isolates per sample to better define the genetic variability of each Vibrio species.
Figure 3.
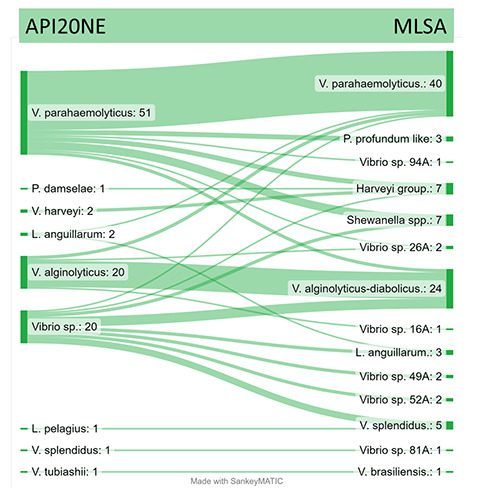
Sankey diagram of the whole Vibrio dataset (99 strains). The two blocks of nodes are related to the different identification methods (phenotypic API20NE vs Multilocus Sequence Analysis). Each node is the taxonomic attribution according to each classification method; the stream fields between the blocks represent the different attribution of these clusters in relation to each Vibrio species.
Strain relationships were analysed using the PHYLOViZ program to identify potential clonal complexes (CCs) and founders. First, a recognition of the most represented species in the Vibrio spp. PubMLST database was performed by full MLST analysis (Figure 4A). The Minimum Spanning Tree-like structure formed with all available isolates showed two major branches represented by V. alginolyticusdiabolicus and V. parahaemolyticus. Only four species are displayed in Figure 4A, while others were not included in the analysis; moreover, the species attribution is not reported for all isolates of the database.
Clonal relationships among the STs collected from Italian samples and worldwide isolates at the triple-locus-variant level are reported in Figure 4B. The analysis evidenced 65 CCs, the biggest of which included 480 STs (a core cluster formed by V. alginolyticus-diabolicus). This CC also included many STs derived from strains collected from different cases of vibriosis. Several STs from crustaceans and molluscs are inside this CC; moreover, the few additional clonal complexes are formed mainly by Vibrio isolates from shellfish.
A clear relation was highlighted from ST529 and ST195 originating from human vibriosis (Figure 4B). ST195 was defined for the strain NO_VA_18_16, a V. alginolyticus isolated in Norway during 2018 (Amato et al., 2022). In many countries of the Nordic- Baltic region, a dramatic increase of vibriosis cases associated with heatwaves was reported (ECDC, 2021; Amato et al., 2022). V. alginolyticus was the cause of 34% of vibriosis infections during the period 2014–2017; 90% of cases were ear- or wound-related while only a few strains were isolated from faeces (2%). Despite the lesser importance of V. alginolyticus species as a foodborne pathogen, the manipulation, preparation and processing of crustaceans could be a risk for fishers and operators due to the probability of skin and soft tissue lesions (Neill et al., 2020). The application of MLSA/MLST schemes and public websites can help researchers to postulate new epidemiological links and sources of infections also for low-risk species and to discover new threats.
Figure 4.
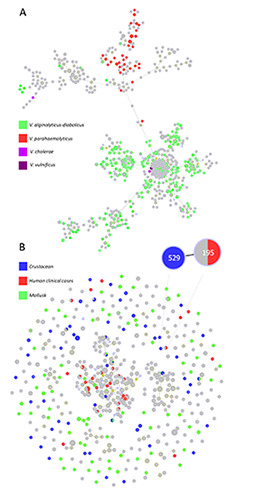
A) Full MST of all PubMLST database isolates; the sequence types are coloured according to the species attribution (4 species were considered); B) goeBURST analysis at the triple-locus-variant level to define clonal complexes. The proportion of each node is related to the frequency of each sequence type. The close-up shows a specific link between sequence types from crustaceans and human cases.
Conclusions
This retrospective analysis demonstrated that MLSA is a very fast and accurate analytic method to discriminate Vibrio species. The distribution and clustering of the analysed species achieved a high supported degree of discrimination that confirmed the results of previous analyses conducted on Vibrio spp. The 4 genes used in this study are sufficient to give suitable results and represent, of course, a faster way to analyse the genus Vibrio.
Sharing the data on public databases can deepen the understanding of seafood, such as crustaceans, as a vehicle of Vibrio spread and their distribution in the final product and can provide detailed information on their potential pathogenicity.
Funding Statement
Funding: This paper is linked to the project IZS VE RC16/10 entitled Occurrence of Vibrio spp. in crustaceans and their potential pathogenicity for seafood consumers, that was funded by the Italian Ministry of Health in the framework of the Ricerca Corrente 2010.
References
- Álvarez-Contreras AK, Quiñones-Ramírez EI, Vázquez-Salinas C, 2021. Prevalence, detection of virulence genes and antimicrobial susceptibility of pathogen Vibrio species isolated from different types of seafood samples at “La Nueva Viga” market in Mexico City. Antonie van Leeuwenhoek 114:1417–29. [DOI] [PubMed] [Google Scholar]
- Amato E, Riess M, Thomas-lopez D, Linkevicius M, Pitkänen T, Wołkowicz T, Rjabinina J, Jernberg C, Hjertqvist M, macdonald E, Antony-Samy JK, Bjerre KD, Saara Salmenlinna S, Fuursted K, Hansen A, Naseer U, 2022. Epidemiological and microbiological investigation of a large increase in vibriosis, northern Europe, 2018. Euro Surveill 27:2101088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austin B, 2010. Vibrios as causal agents of zoonoses. Vet Microbiol 140:310–7. [DOI] [PubMed] [Google Scholar]
- Bej AK, Patterson DP, Brasher CW, Vickery MCL, Jones DD, Kaysner CA, 1999. Detection of total and hemolysinproducing Vibrio parahaemolyticus in shellfish using multiplex PCR amplification of tl, tdh and trh. J Microbiol Methods 36:215–25. [DOI] [PubMed] [Google Scholar]
- Brehm TT, Berneking L, Martins MS, Dupke S, Jacob D, Drechsel O, Bohnert J, Becker K, Kramer A, Christner M, Aepfelbacher M, Schmiedel S, Rohde H, Balau V, Baufeld E, Brechmann S, Briedigkeit L, Diedrich S, Ebert U, Fickenscher H, Grgic B, Heidecke CD, Hinz P, Hoffmann A, Holbe M, Ignatius R, Kaup O, Kern M, Kerwat M, Klempien I, Lamprecht G, Meerbach A, Mischnik A, Podbielski A, Schaefer S, Schwarz R, Strauch E, Warnke P, Weikert-Asbec S, Witte M, Zaki W, 2021. Heatwave-associated Vibrio infections in Germany, 2018 and 2019. Euro Surveill 26:2002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caburlotto G, Suffredini E, Toson M, Fasolato L, Antonetti P, Zambon M, Manfrin A, 2016. Occurrence and molecular characterisation of Vibrio parahaemolyticus in crustaceans commercialised in Venice area, Italy. Int J Food Microbiol 220:39–49. [DOI] [PubMed] [Google Scholar]
- Conferenza stato regioni Intesa Stato Regioni 212/2016, 2016. 854. 882 212/16. Linee guida per il controllo ufficiale ai sensi dei Regolamenti (CE) 882-854. [Google Scholar]
- ECDC E.C. for D.P. and C., 2021. Communicable disease threats. [Google Scholar]
- European Commission, 2004. Regulation of European parliament and of the Council of 29 April 2004 (EC) on official controls performed to ensure the verification of compliance with feed and food law, animal health and animal welfare rules 882/2004/CE. In: Official Journal. L165/1. 30/4/2004 No longer in force, Date of end of validity: 13/12/2019. [Google Scholar]
- European Commission, 2004. Regulation of European parliament and of the Council of 29 April 2004 (EC) laying down specific rules for the organisation of official controls on products of animal origin intended for human consumption 854/2004 /CE. In: Official Journal. L155/206. 30/4/2004 No longer in force, Date of end of validity: 13/12/2019. [Google Scholar]
- European Commission, 2017. Regulation of European parliament and of the Council of 15 March 2017 (EC) on official controls and other official activities performed to ensure the application of food and feed law, rules on animal health and welfare, plant health and plant protection products, amending Regulations (EC) No 999/2001, (EC) No 396/2005, (EC) No 1069/2009, (EC) No 1107/2009, (EU) No 1151/2012, (EU) No 652/2014, (EU) 2016/429 and (EU) 2016/2031 of the European Parliament and of the Council, Council Regulations (EC) No 1/2005 and (EC) No 1099/2009 and Council Directives 98/58/EC, 1999/74/EC, 2007/43/EC, 2008/119/EC and 2008/120/EC, and repealing Regulations (EC) No 854/2004 and (EC) No 882/2004 of the European Parliament and of the Council, Council Directives 89/608/EEC, 89/662/EEC, 90/425/EEC, 91/496/EEC, 96/23/EC, 96/93/EC and 97/78/EC and Council Decision 92/438/EEC (Official Controls Regulation) 625/2017/CE. In: Official Journal. L95/1. 7/4/2017. [Google Scholar]
- Falush D, Stephens M, Pritchard JK, 2003. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J, Nelson K, Morcrette H, Morcrette C, Preston J, Helmer L, Titball RW, Butler CS, Wagley S, 2022. The increased prevalence of Vibrio species and the first reporting of Vibrio jasicida and Vibrio rotiferianus at UK shellfish sites. Water Res 211:117942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartnell RE, Stockley L, Keay W, Rosec JP, Hervio-Heath D, Van den Berg H, Leoni F, Ottaviani D, Henigman U, Denayer S, Serbruyns B, Georgsson F, Krumova- Valcheva G, Gyurova E, Blanco C, Copin S, Strauch E, Wieczorek K, Lopatek M, Britova A, Hardouin G, Lombard B, in’t Veld P, Leclercq A, Baker-Austin C, 2019. A pan-European ring trial to validate an International Standard for detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus in seafoods. Int J Food Microbiol 288:58–65. [DOI] [PubMed] [Google Scholar]
- ISO, 2017. Microbiology of the food chain — Horizontal method for the determination of Vibrio spp. — Part 1: Detection of potentially enteropathogenic Vibrio parahaemolyticus, Vibrio cholerae and Vibrio vulnificus. Norm 21872-1. International Standardization Organization ed., Geneva Switzerland. [Google Scholar]
- Kim YB, Okuda J, Matsumoto C, Takahashi N, Hashimoto S, Nishibuchi M, 1999. Identification of Vibrio parahaemolyticus strains at the species level by PCR targeted to the toxR gene. J Clin Microbiol 37:1173–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralage MSG, Alter T, Pichpol D, Strauch E, Zessin K-H, Huehn S, 2012. Prevalence and Molecular Characteristics of Vibrio spp. Isolated from Preharvest Shrimp of the North Western Province of Sri Lanka. J Food Prot 75:1846–50. [DOI] [PubMed] [Google Scholar]
- Neill MA, 2020. Other Pathogenic Vibrios. In Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. 9th ed. Philadelphia (PA): Saunders. Pp.2645-9. [Google Scholar]
- Onohuean H, Agwu E, Nwodo UU, 2022. A Global Perspective of Vibrio species and associated diseases: three-decade metasynthesis of research advancement. Environ Health Insights 16:11786302221099406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretto T, 2020. Vibriosis due to Vibrio harveyi: etiopathogenesis and vaccine efficacy studies in sea bass (Dicentrarchus labrax). Degree Diss., Alma Mater Stud. Università di Bologna, Italy. [Google Scholar]
- Rahman MS, Carraro R, Cardazzo B, Carraro L, Boscolo Meneguolo D, Martino ME, Andreani NA, Bordin P, Mioni R, Barco L, Novelli E, Fasolato L, 2017. Molecular typing of Vibrio parahaemolyticus strains isolated from mollusks in the North Adriatic Sea. Foodborne Pathog Dis 14:454-64. [DOI] [PubMed] [Google Scholar]
- Rahman MS, Martino ME, Cardazzo B, Facco P, Bordin P, Mioni R, Novelli E, Fasolato L, 2014. Vibrio trends in the ecology of the Venice lagoon. Appl Environ Microbiol 80:2372-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampaio A, Silva V, Poeta P, Aonofriesei F, 2022. Vibrio spp.: Life strategies, ecology, and risks in a changing environment. Diversity 14:1–26. [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S, 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traoré SG, Bonfoh B, Krabi R, Odermatt P, Utzinger J, Rose KN, Tanner M, Frey J, Quilici ML, Koussémon M, 2012. Risk of Vibrio transmission linked to the consumption of crustaceans in coastal towns of Côte d’Ivoire. J Food Prot 75:1004-11. [DOI] [PMC free article] [PubMed] [Google Scholar]


