SUMMARY
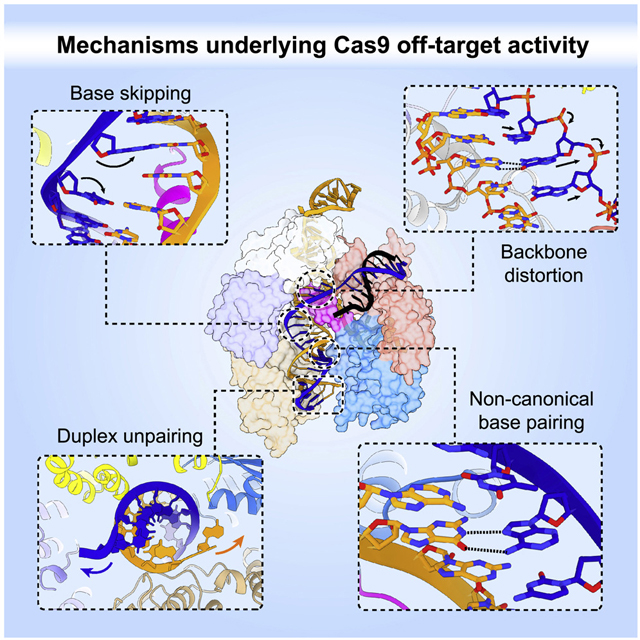
The target DNA specificity of the CRISPR-associated genome editor nuclease Cas9 is determined by complementarity to a 20-nucleotide segment in its guide RNA. However, Cas9 can bind and cleave partially complementary off-target sequences, which raises safety concerns for its use in clinical applications. Here we report crystallographic structures of Cas9 bound to bona fide off-target substrates, revealing that off-target binding is enabled by a range of non-canonical base pairing interactions within the guide–off-target heteroduplex. Off-target sites containing single-nucleotide deletions relative to the guide RNA are accommodated by base skipping or multiple non-canonical base pairs rather than RNA bulge formation. Additionally, PAM-distal mismatches result in duplex unpairing and induce a conformational change of the Cas9 REC lobe that perturbs its conformational activation. Together, these insights provide a structural rationale for the off-target activity of Cas9 and contribute to the improved rational design of guide RNAs and off-target prediction algorithms.
Graphical Abstract
INTRODUCTION
Cas9, the effector nuclease of prokaryotic Type II CRISPR adaptive immune systems (Makarova et al., 2020), cleaves double-stranded DNA substrates complementary to a guide CRISPR RNA (crRNA) (Jinek et al., 2012). By changing the sequence of the guide RNA, the target DNA specificity of the CRISPR-Cas9 system is readily programmable (Jinek et al., 2012), a feature that has been widely exploited for genome engineering applications (Anzalone et al., 2020). Cas9 functions in conjunction with a trans-activating crRNA (tracrRNA), which is required both for crRNA loading and subsequent DNA binding and cleavage (Deltcheva et al., 2011; Jinek et al., 2012). Target DNA binding and cleavage is further dependent on the presence of a protospacer-adjacent motif (PAM) flanking the target sequence (Anders et al., 2014; Jinek et al., 2012). Due to its high activity and 5’-NGG-3’ PAM specificity, Streptococcus pyogenes Cas9 (SpCas9) remains the most widely used CRISPR-Cas nuclease for gene editing applications. However, despite a high intrinsic accuracy in generating targeted DNA breaks, SpCas9 can nevertheless cleave genomic DNA sequences with imperfect complementarity to the guide RNA, resulting in off-target editing (Cameron et al., 2017; Hsu et al., 2013; Pattanayak et al., 2013; Tsai et al., 2015). The off-target activity of SpCas9, as well as other Cas9 enzymes, thus presents a safety concern for their therapeutic applications.
Off-target sites typically contain one or several nucleobase mismatches relative to the guide RNA (Cameron et al., 2017; Tsai et al., 2017; Tsai et al., 2015). Recent studies have established that the type of mismatch, its positioning, and the total number of mismatches are important determinants of off-target DNA binding and cleavage (Boyle et al., 2017; Boyle et al., 2021; Doench et al., 2016; Jones et al., 2021; Zhang et al., 2020). PAM-proximal mismatches within the seed region of the guide RNA (gRNA)-target strand (TS) DNA heteroduplex typically have a dramatic impact on substrate DNA binding and R-loop formation (Boyle et al., 2021; Ivanov et al., 2020; Singh et al., 2016; Zhang et al., 2020). In contrast, PAM-distal mismatches are compatible with stable binding; however, their presence often results in the formation of a catalytically incompetent complex (Boyle et al., 2021; Dagdas et al., 2017; Ivanov et al., 2020; Jones et al., 2021; Sternberg et al., 2015; Yang et al., 2018; Zhang et al., 2020). In addition, Cas9 has been shown to cleave off-target substrates containing insertions or deletions relative to the guide RNA sequence, which have been proposed to be recognized through the formation of nucleotide “bulges” in the gRNA-TS DNA heteroduplex (Boyle et al., 2021; Cameron et al., 2017; Doench et al., 2016; Jones et al., 2021; Lin et al., 2014; Tsai et al., 2015).
Numerous computational tools have been developed to predict possible genomic off-target sites based on sequence similarity (Bae et al., 2014; Stemmer et al., 2015). However, the majority of actual off-target cleavage events remain unpredicted (Cameron et al., 2017; Tsai et al., 2015). Furthermore, although Cas9 is able to bind genomic sites harbouring as few as five complementary nucleotides, only a relatively small number of off-target sites are actually cleaved and result in detectable off-target editing in cells (Kuscu et al., 2014; O’Geen et al., 2015; Wu et al., 2014). Several structures of target-bound Cas9 complexes have been determined to date (Anders et al., 2014; Jiang et al., 2016; Nishimasu et al., 2014; Zhu et al., 2019) that have shed light on the mechanism of on-target binding and cleavage. However, the same processes for off-target sites remain poorly understood.
To elucidate the mechanism of mismatch tolerance of Cas9, we determined crystal structures of a comprehensive set of bona fide off-target–bound complexes. These structures reveal that the formation of non-canonical base pairs and preservation of heteroduplex shape underpin the off-target tolerance of Cas9. We also observe that multiple consecutive mismatches can be accommodated by base skipping of a guide RNA nucleotide, as opposed to nucleotide bulging. Finally, the structure of an off-target complex containing three PAM-distal mismatches exhibits REC2/3 domain rearrangements, which likely perturbs conformational activation of Cas9 and thus modulates cleavage efficiency. Taken together, our structural data reveal the diversity of mechanisms enabling off-target recognition and lay the foundation for improved off-target prediction and engineering optimized CRISPR-Cas9 complex designs for gene editing.
RESULTS
In vitro profiling reveals diversity of Cas9 off-targets
Multiple studies have investigated the off-target activity of Cas9, suggesting context-dependent tolerance of nucleobase mismatches between the guide RNA and off-target DNA sequences (Boyle et al., 2021; Cameron et al., 2017; Lazzarotto et al., 2020; Tsai et al., 2015; Zhang et al., 2020). To investigate the effect of mismatches on Cas9 binding and cleavage, we performed the SITE-Seq® assay (Cameron et al., 2017) to define the off-target landscapes of 12 well-studied guide RNAs to select suitable off-targets for further evaluation (Table S1, Table S2) (Figure S1A, Table 1). The SITE-Seq assay analysis revealed a total of 3,848 detectable off-target sites at the highest Cas9 ribonucleoprotein (RNP) concentration, with a median of 5 mismatches per off-target site (Figure S1C). The detected mismatches covered all possible base mismatch combinations and were distributed throughout the length of the gRNA-TS DNA heteroduplex (Figure S1D–E).
Table 1.
Biochemical analysis of the binding and cleavage efficiency of selected off-target sites.
| Gene | Target | 24 h cleavage (%) | kobs (min−1) | kon (M−1 . s−1) | koff (s−1) | Kd (pM) | 24 h cleavage (%) PAMme r | kobs (min−1) PAMmer |
|---|---|---|---|---|---|---|---|---|
| AAVS1 | on-target | 92.2 | 1.624 ± 0.159 | 3.95 ± 0.26 × 106 | 5.74 ± 0.73 × 10−5 | 14.5 ± 2.1 | 95.0 | 0.562 ± 0.073 |
| AAVS1 | off-target #1 | 92.7 | 0.071 ± 0.005 | 4.73 ± 0.71 × 106 | 6.29 ± 0.29 × 105 | 13.3 ± 2.1 | 87.5 | 0.239 ± 0.031 |
| AAVS1 | off-target #2 | 95.1 | 0.034 ± 0.002 | 8.75 ± 0.31 × 106 | 3.38 ± 0.08 × 10−3 | 386 ± 16 | 85.5 | 0.004 ± 0.0001 |
| AAVS1 | off-target #3 | 96.7 | 0.051 ± 0.002 | 3.30 ± 0.11 × 106 | 2.51 ± 0.15 × 10−3 | 761 ± 51 | 82.7 | 0.081 ± 0.010 |
| AAVS1 | off-target #4 | 94.4 | 0.004 ± 0.0002 | 1.09 ± 0.03 × 107 | 3.28 ± 0.06 × 10−3 | 301 ± 9 | 89.6 | 0.065 ± 0.007 |
| AAVS1 | off-target #5 | 18.9 | 0.00013 ± 0.0003 |
ND | ND | ND | 70.1 | 0.007 ± 0.0001 |
| FANCF | on-target | 97.5 | 0.238 ± 0.013 | 3.45 ± 0.19 × 106 | 7.46 ± 0.97 × 10−5 | 21.6 ± 3.1 | 98.3 | 0.565 ± 0.054 |
| FANCF | off-target #1 | 35.1 | 0.001 ± 0.0001 | 3.97 ± 0.06 × 106 | 2.09 ± 0.06 × 10−3 | 528 ± 17 | 97.4 | 0.069 ± 0.006 |
| FANCF | off-target #2 | 62.4 | 0.001 ± 0.0002 | 1.42 ± 0.03 × 106 | 2.45 ± 0.06 × 10−3 | 1730 ± 60 | 92.9 | 0.233 ± 0.018 |
| FANCF | off-target #3 | 0.0 | 0 | 1.22 ± 0.05 × 107 | 2.37 ± 0.11 × 10−3 | 193 ± 13 | 4.2 | 0.00003 ± 0.0003 |
| FANCF | off-target #4 | 53.0 | 0.0005 ± 0.0002 | 3.35 ± 0.05 × 106 | 1.91 ± 0.07 × 10−3 | 571 ± 24 | 38.9 | 0.001 ± 0.0001 |
| FANCF | off-target #5 | 80.4 | 0.001 ± 0.0001 | 1.27 ± 0.05 × 106 | 2.55 ± 0.10 × 10−3 | 2010 ± 110 | 92.9 | 0.058 ± 0.005 |
| FANCF | off-target #6 | 8.2 | 0.00006 ± 0.0003 |
1.50 ± 0.09 × 106 | 2.03 ± 0.15 × 10−3 | 1350 ± 130 | 66.6 | 0.001 ± 0.00005 |
| FANCF | off-target #7 | 5.2 | 0.004 ± 0.0003 | 2.95 ± 0.20 × 106 | 3.21 ± 0.23 × 10−3 | 1090 ± 110 | 94.5 | 0.013 ± 0.001 |
| PTPRC- tgt2 |
on-target | 96.8 | 0.459 ± 0.022 | 6.08 ± 0.39 × 106 | 2.19 ± 0.21 × 10−4 | 36.0± 4.2 | 95.5 | 0.074 ± 0.015 |
| PTPRC- tgt2 |
off-target #1 | 0.0 | 0 | 1.22 ± 0.07 × 107 | 2.39 ± 0.12 × 10−3 | 196 ± 15 | 91.4 | 0.001 ± 0.0003 |
| TRAC | on-target | 97.7 | 0.381 ± 0.015 | 1.02 ± 0.21 × 106 | 3.23 ± 0.53 × 10−4 | 31.8 ± 8.5 | 93.5 | 0.181 ± 0.013 |
| TRAC | off-target #1 | 95.8 | 0.019 ± 0.001 | 1.37 ± 0.45 × 106 | 1.77 ± 0.1 × 10−4 | 130 ± 44 | 90.7 | 0.081 ± 0.006 |
| TRAC | off-target #2 | 65.0 | 0.001 ± 0.00006 | 9.43 ± 1.12 × 106 | 3.27 ± 0.27 × 10−4 | 34.6 ± 5.0 | 88.4 | 0.026 ± 0.002 |
Kinetic and thermodynamic parameters of off-target substrates. The cleavage rate constants (kobs) were derived from single-exponential function fitting of measured cleavage rates of the target-strand labelled DNA substrates. The binding and dissociation rate constants (kon and koff) and the equilibrium dissociation constant (Kd) were determined using a DNA nanolever binding (switchSENSE) assay. Intervals indicate standard error of mean.
To probe the thermodynamics of on- and off-target substrate DNA binding and the kinetics of DNA cleavage, we focused on a subset of four guide RNAs (AAVS1, FANCF, PTPRC-tgt2, and TRAC) and a total of 15 bona fide off-target sites detectable in vivo (Cameron et al., 2017; Donohoue et al., 2021; Tsai et al., 2017; Tsai et al., 2015) (Figure 1A) that covered all 12 possible base mispair types. To benchmark the editing efficiency of the selected off-target substrates in vivo, we transfected human primary T cells with recombinant Cas9 RNPs at multiple concentrations and quantified indel frequencies. We observed efficient cellular editing at the on-target site for each guide RNA at all RNP concentrations, and detected editing at 7 of the 15 off-target sites in our set, including the deletion-containing FANCF off-target #3 (Figure S3, Table S3), which contained a mismatch-reverting single-nucleotide polymorphism (SNP) relative to the consensus human genome build.
Figure 1. Biochemical and structural analysis of Cas9 off-targets.

(A) Guide RNA and (off-)target DNA sequences selected for biochemical and structural analysis. Matching bases in off-targets are denoted by a dot; nucleotide mismatches and deletions (–) are highlighted. (B) Top: Schematic representation of the guide RNA (orange), TS (blue), and NTS (black) sequences used for crystallisation. The PAM sequence in the DNA is highlighted in yellow. Bottom: Structure of the Cas9 FANCF on-target complex. Individual Cas9 domains are coloured according to the legend; substrate DNA target strand (TS) is coloured blue, non-target strand (NTS) black, and the guide RNA orange.
In vitro nuclease activity assays revealed that all selected off-target sequences were cleaved slower than the corresponding on-target substrates, with 20–500-fold reductions in the calculated rate constants (Table 1, Figure S2A). To distinguish whether the cleavage defects were due to slow R-loop formation or perturbations in downstream steps, we also quantified cleavage kinetics using partially single-stranded PAMmer DNA substrates (Anders et al., 2014; O’Connell et al., 2014). These experiments revealed that the slower cleavage kinetics of most off-target substrates was due to perturbed R-loop formation (Table 1, Figure S2B). However, for some off-targets, notably AAVS1 off-targets #2 and #5, FANCF off-targets #3, #4, and #6, and PTPRC-tgt2 off-target #1, the rate of PAMmer substrate cleavage was more than 100-fold slower as compared to their respective on-target sequences (Table 1, Figure S2B), indicating perturbations in the conformational activation checkpoint downstream of guide-target hybridization or inhibition of cleavage by direct steric hindrance of the Cas9 HNH domain (Chen et al., 2017; Dagdas et al., 2017).
Complementary quantification of substrate DNA binding using DNA nanolever (switchSENSE) methodology revealed perturbations in the binding affinities of most off-targets (Table 1, Data S1). Notably, the reductions in binding affinities were almost entirely due to increased dissociation rates (koff), while on-binding rates (kon) were largely unperturbed (Table 1), indicating that most of the tested off-targets promote DNA dissociation, likely due to R-loop collapse. However, there was little correlation between the observed reductions in cleavage rates and binding constants (Figure S2C), confirming that the molecular basis for off-target discrimination by Cas9 is not based on substrate binding alone, in agreement with prior studies (Boyle et al., 2021; Chen et al., 2017; Dagdas et al., 2017; Jones et al., 2021; Yang et al., 2018; Zhang et al., 2020). The dissociation rate (koff) correlated significantly only with the number of mismatches located in the seed region (R2=0.46, p=0.001) (Figure S2C), suggesting that seed mismatches promote R-loop collapse and non-target strand (NTS) rehybridization (Boyle et al., 2017; Gong et al., 2018; Singh et al., 2016; Sternberg et al., 2014).
Finally, we compared the measured cleavage rate constants (kobs) with predicted data from a leading biophysical model of Cas9 off-target cleavage that accounts for mismatch number and position but utilises position-independent weights for mismatch type (Jones et al., 2021). Despite good overall correlation between the model and our data (R2=0.46, p=0.004) (Figure S2C), there were several prominent outliers (AAVS1 off-target #2 and off-target #3, and FANCF off-target #4), suggesting that accurate modelling of off-target interactions requires accounting for position-specific effects of individual mismatch types (Figure S2D).
Crystallographic analysis of off-target interactions
To obtain insights into the structural basis of off-target recognition and mismatch tolerance, we employed a previously described approach (Anders et al., 2014) to co-crystallize Cas9 with sgRNA guides and partially duplexed off-target DNA substrates (Figure 1C). Focusing on our set of AAVS1, FANCF, PTPRC-tgt2 and TRAC off-targets (Figure 1A), covering all 12 possible mismatch types, we determined a total of 15 off-target complex structures at resolutions of 2.25–3.30 Å (Figure 1C, Table S4). Overall, the off-target complex structures have very similar conformations, with the Cas9 polypeptide backbone superimposing with a mean root mean square deviation of 0.41Å over 1330 Cα atoms (as referenced to the FANCF on-target complex structure, excluding FANCF off-target #4, as discussed below). Of note, the AAVS1 on-target complex structure reveals substantial repositioning of the REC2 domain, where it undergoes a 12o rotation (relative to FANCF and TRAC on-target complexes) (Figure S4A, Video S1), with concomitant shortening of the α-helix comprising residues 301–305 and restructuring of the loop comprising residues 175–179 (Figure S4B), enabled by the absence of crystal contacts involving the REC2 and REC3 domains.
However, the structures display considerable local variation of the gRNA-TS DNA heteroduplex conformation (Table S5). Base pairing and base stacking are mostly preserved throughout the heteroduplexes (Table S6), with the exception of positions 1–3 within the PAM-distal end of the guide-TS duplex, where the presence of mismatches results in duplex unpairing (Figure S5). Interestingly, such unpairing results in the base stacking of the terminal residue of the sgRNA on top of the unpaired DNA base, as observed in a recent cryo-EM structure of a PAM-distal mismatch off-target complex (Bravo et al., 2022). Despite the observed conformational variation, the off-target structures preserve almost all intermolecular contacts between the Cas9 protein and the bound nucleic acids (Figure S6), further underscoring the structural plasticity of Cas9 in accommodating mismatch-induced heteroduplex distortions.
Non-canonical base-pairing interactions facilitate off-target recognition
A substantial fraction of the base pair mismatches observed in the determined off-target complex structures (34 out of 49) is accommodated by non-canonical base pairing interactions that preserve at least one hydrogen bond between the mispaired bases. The most common off-target mismatches, both in our data set (Table S1, Table S2) and as reported by other studies (Boyle et al., 2017; Doench et al., 2016; Hsu et al., 2013; Jones et al., 2021; Pattanayak et al., 2013; Zhang et al., 2020), are rG-dT (Figure S7) and rU-dG (Figure 2A, Figure S8), which have the potential to form wobble base pairs (Kimsey et al., 2015). Indeed, all rG-dT mismatches in the determined structures are accommodated by wobble base pairing. Observed at duplex positions 4, 13 and 15, the dT base undergoes a ~1 Å shear displacement into the major groove of the gRNA–TS DNA heteroduplex to form the wobble base pair (Figure S7A–E), whereas at duplex position 2, wobble base pairing is enabled by a minor groove displacement of the rG base (Figure S7F-G). In contrast, rU-dG base pairs in the determined structures exhibit considerable structural variation. At duplex position 10, the rU base is able to undergo the major groove displacement required for wobble base-pairing (Figure 2A, Figure S8A). In contrast, at duplex position 5, the backbone of the RNA strand makes extensive contacts with Cas9 (Figure S8B–D, Figure S6) and as a result, the rU-dG base pairs are instead accommodated by compensatory shifts of the dG base to maintain hydrogen-bonding interactions (Figure S8B–D). At duplex position 9 in the TRAC off-target #1 complex, the rU-dG mismatch is accommodated by wobble base-pairing enabled by a minor groove displacement of the dG base (Figure S8E). At the same duplex position in the AAVS1 off-target #1 and #2 complexes, however, this mismatch occurs next to rC-dA and rC-dT mismatches, respectively, and adopts the sterically prohibited Watson-Crick geometry (Figure S8F–G), implying a tautomeric shift or base deprotonation to accommodate this otherwise unfavorable base pairing mode (Figure S8H). Collectively, these observations suggest that the ability of rU-dG (and likely rG-dT) mismatches to form wobble base pairing interactions is determined not only by backbone constraints at the specific position within the gRNA-TS DNA heteroduplex (Figure S6), but also by local sequence context and/or the presence of neighboring mismatches (Figure S8C–G).
Figure 2. Cas9 off-target binding is enabled by non-canonical base pairing.
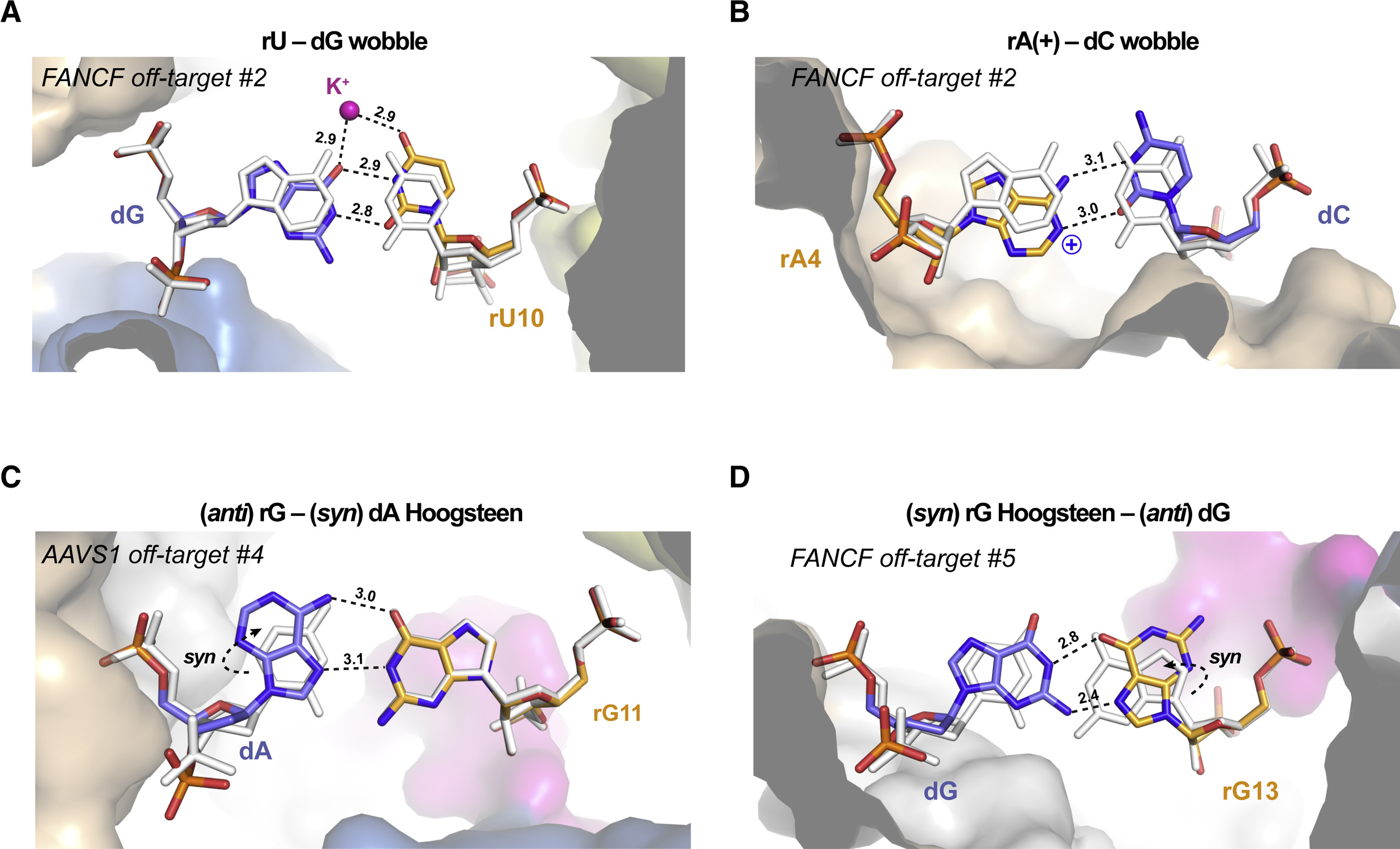
Close-up views of (A) rU-dG wobble base pair at duplex position 10 in FANCF off-target #2 complex, (B) rA-dC wobble base pair at position 4 in FANCF off-target #2 complex, (C) rG-dA Hoogsteen base pair at duplex position 11 in AAVS1 off-target #4 complex and (D) rG-dG Hoogsteen base pair at duplex position 13 in FANCF off-target #5 complex. Hydrogen bonding interactions are indicated with dashed lines. Numbers indicate interatomic distances in Å. Corresponding on-target Watson-Crick base pairs are shown in white. Dashed arrows indicate anti-syn isomerization of the dA and rG bases to enable Hoogsteen-edge base pairing. A bound monovalent ion, modelled as K+, is depicted as a purple sphere. See also Figures S7, S8, S9.
rA-dC or rC-dA mismatches can also form wobble-like base pairs when the adenine base is protonated at the N1 position (Garg and Heinemann, 2018; Wang et al., 2011). In the rA-dC mispair found at duplex position 4, the dC base undergoes a wobble displacement compatible with the formation of two hydrogen bonds with the adenine base, indicative of adenine protonation (Figure 2B, Figure S9A). At other duplex positions in our data set, the rA-dC or rC-dA mispairs are instead accommodated by slight displacements of the adenine base within the base pair plane resulting in the formation of a single hydrogen bond in each case (Figure S9B–D).
Accommodating purine-purine mismatches by Watson-Crick-like interactions would normally require severe distortion of the guide–off-target duplex to increase its width by more than 2 Å (Leontis et al., 2002). At positions where the duplex width is constrained by Cas9 interactions (Figure S6), rG-dA and rA-dG mispairs are accommodated by anti-to-syn isomerization of the adenine base to form two hydrogen-bonding interactions via its Hoogsteen base edge. This is observed at duplex position 11 in the AAVS1 off-target #4 complex (rG-dA mispair) (Figure 2C) and at position 7 in the AAVS1 off-target #2 complex (rA-dG mispair) (Figure S9E). Similarly, the rG-dG mispair at duplex position 13 in the FANCF off-target #5 complex is accommodated by Hoogsteen base-pairing as a result by anti-to-syn isomerization of the guide RNA base (Figure 2D). Overall, the observed Hoogsteen base pairing interactions are near-isosteric with Watson-Crick base pairs and maintain duplex width without excessive backbone distortion (Table S6).
Duplex backbone rearrangements accommodate otherwise non-productive mismatches
Whereas wobble (G-U/T or A-C) and Hoogsteen (A-G or G-G) base pairs are generally compatible with the canonical A-form geometry of an RNA-DNA duplex, other nucleotide mismatches only form non-isosteric base pairs that require considerable distortion of the (deoxy)ribose-phosphate backbone. The formation of pyrimidine-pyrimidine base pairs is expected to occur by a substantial reduction in duplex width (Leontis et al., 2002). This is observed at the rU-dC mismatch at duplex position 9 in the FANCF off-target #1 complex (Figure 3A). Here, the guide RNA backbone is able to shift towards the target DNA strand, resulting in a reduction of the C1’–C1’ distance to 8.65 Å as compared to 10.0 Å in the FANCF on-target complex. This facilitates the formation of two hydrogen bonding interactions within the rU-dC base pair, which is further enabled by a substantial increase in base propeller twist (Figure 3A). In contrast, rC-dT mismatches remain unpaired at duplex positions 6 and 7 in (Figure S10A-B), or form only a single hydrogen bond at position 8 (Figure S10C), likely due to backbone steric constraints at these positions imposed by Cas9 interactions (Figure S6). Of note, the FANCF off-target #7 rC-dT mismatch is bridged by hydrogen bonding interactions with the side chain of Arg895 inserted into the minor groove of the heteroduplex (Figure S10B); however, the interaction is not essential for the tolerance of rC-dT mismatches at this position (Figure S11, Table S7). Backbone steric constraints also likely influence the formation of rU-dT base pairs (Figure S6). At duplex position 7, the mismatch remains unpaired (Figure S10D), whereas productive pairing is seen at duplex positions 8 and 9, facilitated by distortions of the guide RNA and TS backbone, respectively (Figure 3B, Figure S10E–F).
Figure 3. Duplex backbone distortions facilitate formation of non-canonical base pairs.
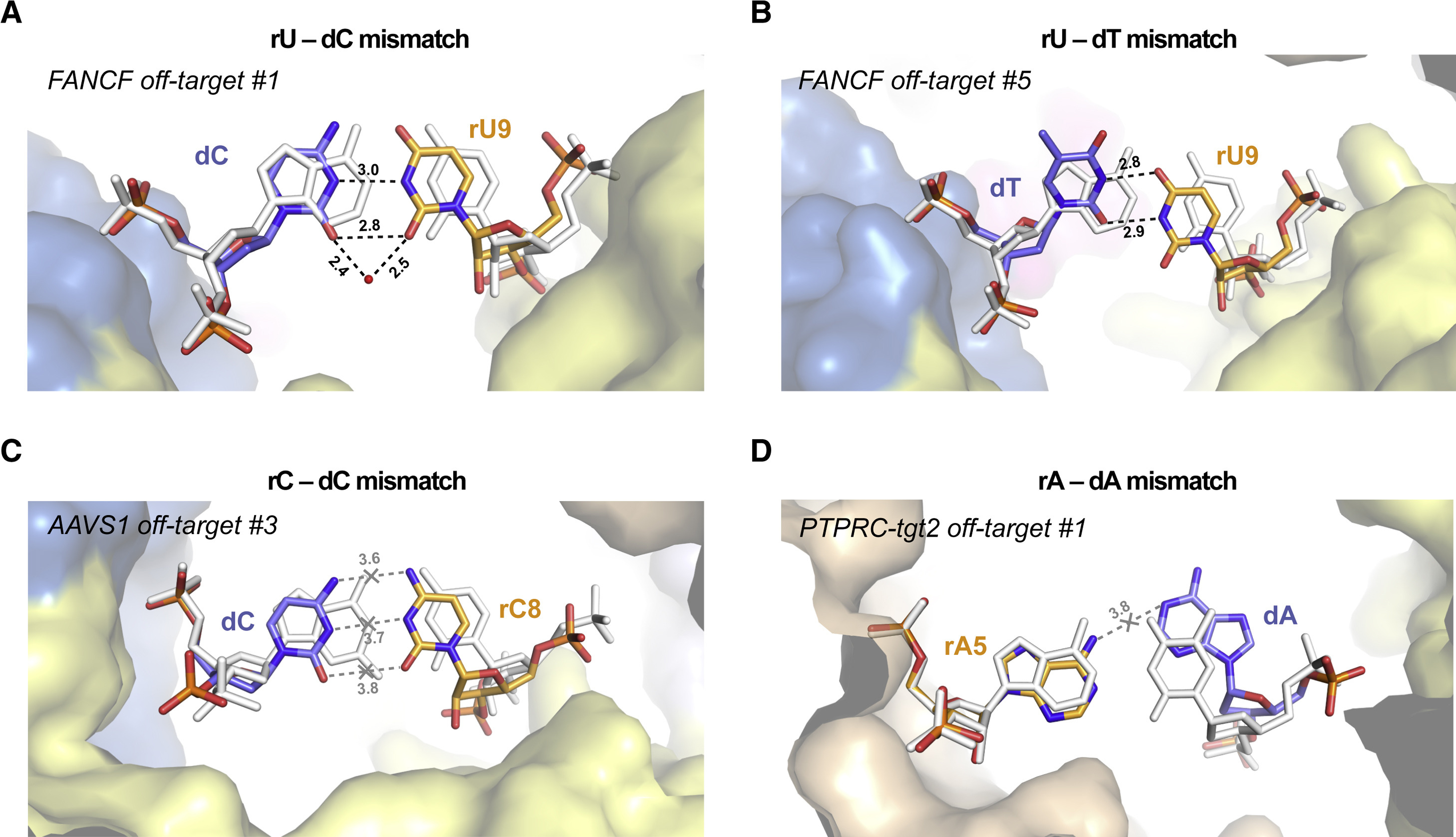
(A) Close-up view of the rU-dC base pair at duplex position 9 in FANCF off-target #1 complex, facilitated by lateral displacement of the guide RNA backbone. (B) Zoomed-in view of the rU-dT base pair at position 9 in FANCF off-target #5 complex. (C) Zoomed-in view of the rC-dC mismatch at duplex position 8 in AAVS1 off-target #3 complex. The distances between the cytosine bases indicate lack of hydrogen bonding. (D) Zoomed-in view of the rA-dA mismatch at duplex position 5 in PTPRC-tgt2 off-target #1 complex. Hydrogen bonding interactions are indicated with dashed lines. Corresponding on-target Watson-Crick base pairs are shown in white (for PTPRC-tgt2, the FANCF on-target structure was used and bases were mutated in silico). Numbers indicate interatomic distances in Å. Bound water molecule is depicted as red sphere. See also Figures S10, S11, S12, S13.
rC-dC mismatches only form productive hydrogen-bonding interactions if bridged by a water molecule or when one of the cytosine bases is protonated (Leontis et al., 2002). Only the former is observed in the determined structures, at duplex position 5 in the AAVS1 off-target #2 complex (Figure S10G). In contrast, at duplex position 8 and 15, the bases remain unpaired while maintaining intra-strand base stacking interactions (Figure 3C, Figure S10H). Similarly, rA-dA mismatches are unable to form productive hydrogen-bonding interactions within the constraints of an A-form duplex (Leontis et al., 2002). Accordingly, the rA-dA mismatch at duplex position 5, where duplex width is constrained by Cas9 (Figure S6), is accommodated by extrusion of the dA nucleobase out of the base stack into the major groove of the duplex (Figure 3D) which is enabled by local distortion of the TS backbone (Figure S12A). Analysis of our SITE-Seq assay data set revealed that off-target rA-dA mismatches occur at all positions within the gRNA–TS DNA heteroduplex (Figure S12B), in agreement with previous studies (Boyle et al., 2017; Boyle et al., 2021; Doench et al., 2016; Jones et al., 2021; Zhang et al., 2020). This suggests that rA-dA mismatches do not encounter steric barriers within Cas9 that would disfavour their presence, which is consistent with the absence of specific contacts with Cas9 along the length of the major groove of the heteroduplex (Figure S6).
To gain further insights into the mechanism of rA-dA mismatch accommodation and extrapolate our structural observations to other heteroduplex positions, we performed molecular dynamics (MD) simulations by introducing single rA-dA mismatches at all 20 positions along the heteroduplex in the context of the catalytically active state of Cas9 (PDB: 6O0Y) and compared the resulting trajectories to the corresponding on-target system. rA-dA mismatch simulations at positions 5, 18, and 19 showed very good agreement with the determined crystal structures (PTPRC-tgt2 off-target #1, FANCF off-target #6, AAVS1 off-target #2), predicting both dA base extrusion and phosphate backbone distortions (Figure S12C–D). In-depth analysis of the rA-dA conformational dynamics was performed by computing the geometrical descriptors defining the base pair complementarity along the heteroduplex (Figure S12D, Figure S13) (Lavery et al., 2009). These reveal that at all positions, the rA-dA mismatch is primarily accommodated by base extrusion from the duplex, with positions 3, 5 ,15, 16, 18, 19 and 20 undergoing dA base extrusion, and positions 10 and 17 rA base extrusion. Notably, at duplex positions 2, 4, 6, 8, 11 and 12, the broad or bimodal distributions of base pair shear and opening parameters are indicative of considerable conformational dynamics, allowing the rA-dA mismatch to be accommodated by either rA or dA base extrusion (Figure S13A).
PAM-proximal mismatches are accommodated by TS distortion due to seed sequence rigidity
The seed sequence of the guide RNA (nucleotides 11–20) makes extensive interactions with Cas9, both in the absence and presence of bound DNA (Anders et al., 2014; Jiang et al., 2015; Nishimasu et al., 2014; Zhu et al., 2019). Structural pre-ordering of the seed sequence by Cas9 facilitates target DNA binding and contributes to the specificity of on-target DNA recognition (Jiang et al., 2015; O’Geen et al., 2015; Wu et al., 2014). Conversely, binding of off-target DNAs containing PAM-proximal mismatches is inhibited and results in accelerated off-target dissociation (Boyle et al., 2017; Boyle et al., 2021; Ivanov et al., 2020; Jones et al., 2021; Singh et al., 2016; Zhang et al., 2020). Nevertheless, Cas9 does tolerate most base mismatch types within the seed region of the guide RNA, leading to detectable off-target DNA cleavage (Boyle et al., 2021; Doench et al., 2016; Jones et al., 2021; Zhang et al., 2020). In particular, the first two PAM-proximal positions display a markedly higher tolerance for mismatches than the rest of the seed region (Cofsky et al., 2022; Doench et al., 2016; Hsu et al., 2013; Mekler et al., 2017; Zeng et al., 2018); this is supported by our SITE-Seq assay data as the frequency of mismatches at PAM-proximal positions 18–20 is 1.4–2.0x higher than at seed positions 14–17 (Figure S1D–E).
Unlike the seed region of the guide RNA, the complementary PAM-proximal TS nucleotides are not directly contacted by Cas9 in the pre-cleavage state and are thus under fewer steric constraints (Figure S6), with the exception of duplex position 20 in which the phosphodiester group of the TS nucleotide makes extensive interactions with the phosphate lock loop of Cas9 (Anders et al., 2014) (Figure S14A). In agreement with this, four of our off-target complex structures reveal that PAM-proximal base mismatches are accommodated solely by structural distortions of the TS backbone (Figure 4), while the conformation of the guide RNA backbone and base stacking within the seed region remain unperturbed (Figure S14B-C). The presence of an rA-dA mismatch in the PAM-proximal position 18 results in the extrusion of the TS nucleobase into the major groove (Figure 4A), likely due to steric constraints on duplex width at this position. In contrast, the rA-dA mismatch at duplex position 19 is instead accommodated by a marked distortion in the TS backbone that results in increased duplex width, which preserves base stacking within the duplex in the absence of productive pairing between the adenine bases (Figure 4B, Figure S14B). Similarly, the rA-dG mismatch at position 19 is accommodated by a ~2 Å displacement of the TS backbone, increasing duplex width. This not only preserves base stacking but also facilitates rA-dG base paring by two hydrogen bonding interactions via their Watson-Crick edges (Figure 4C, Figure S14C). This off-target complex also contains a rU-dG mismatch at duplex position 20 which does not undergo wobble base pairing as the rU20 nucleotide is extensively contacted by Cas9 (Figure S6) and unable to shift towards the major groove and is instead accommodated by a slight shift in the dG nucleotide (Figure 4C). Finally, the rU-dT base mismatch at duplex position 20 in the AAVS1 off-target #4 complex remains unpaired and the dT base lacks ordered electron density (Figure 4D). This is likely a result of the dT nucleotide maintaining contact with the phosphate-lock loop of Cas9 (Figure S14A), which prevents a reduction in the duplex width and precludes productive base pairing. Overall, these observations indicate that off-target DNAs containing mismatches to the seed sequence of the guide RNA can be accommodated by Cas9 due to limited interactions with the TS DNA in the seed-binding region
Figure 4. TS distortion facilitates mismatch accommodation in the seed region of the guide–off-target heteroduplex.
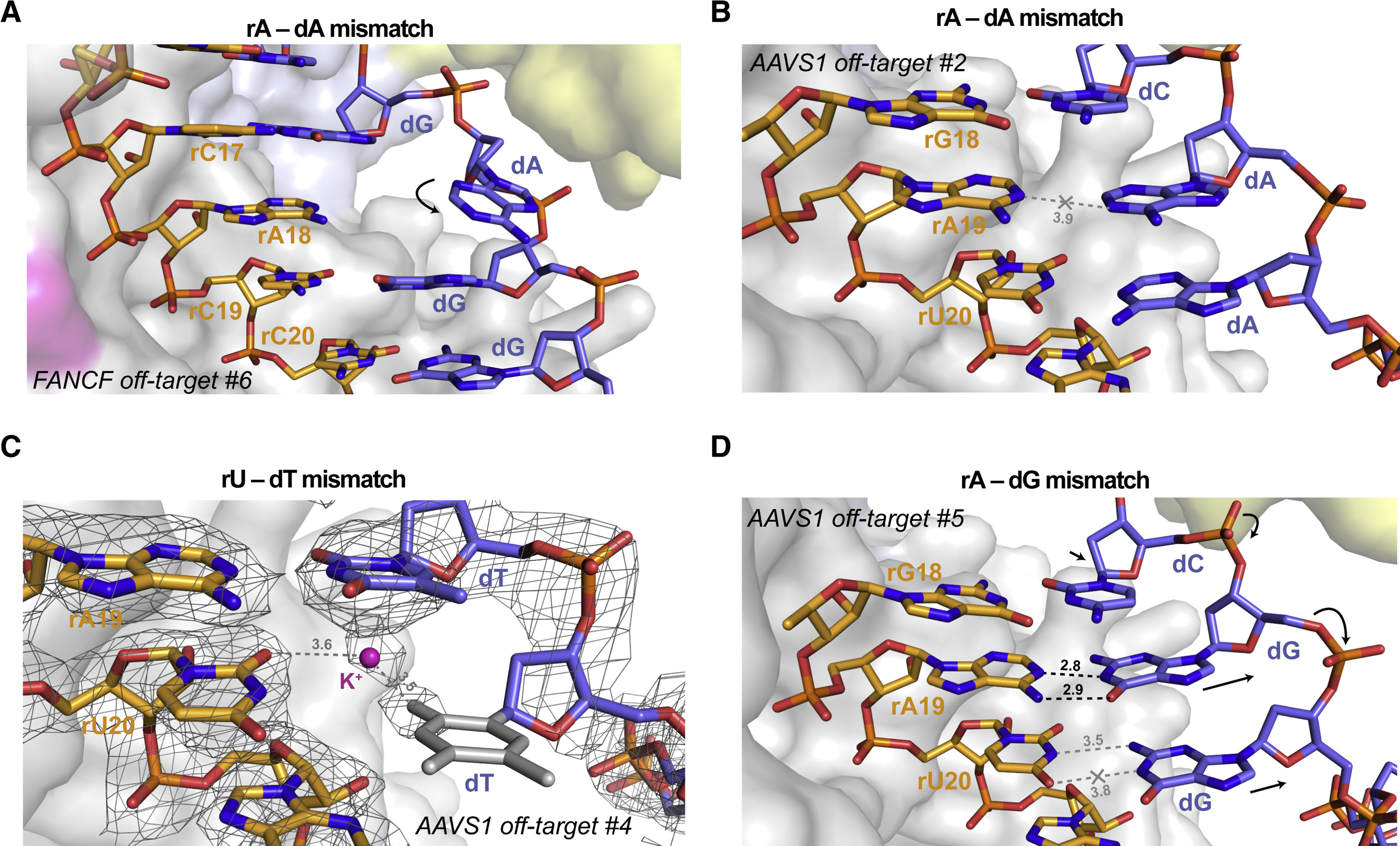
(A) Close-up view of the rA-dA mismatch at position 18 in FANCF off-target #6 complex, showing major groove extrusion of the dA base. (B) Close-up view of the rA-dA mismatch at position 19 in AAVS1 off-target #2 complex, showing retention of the dA base in the duplex stack. (C) Zoomed-in view of the rA-dG base pair at position 19 and the unpaired rU-dG mismatch at position 20 in the AAVS1 off-target #5 complex. (D) Close-up rU-dT mismatch at the PAM-proximal position 20 in AAVS1 off-target #4 complex. Residual electron density indicates the presence of an ion or solvent molecule. Refined 2mFo−DFc electron density map of the heteroduplex, contoured at 1.5σ, is rendered as a grey mesh. Structurally disordered thymine nucleobase for which no unambiguous density is present is in grey. Arrows indicate conformational changes in the TS backbone relative to the on-target complex. See also Figure S14.
Cas9 recognizes off-targets with single-nucleotide deletions by base skipping or via multiple mismatches
A substantial fraction of off-target sites recovered in our SITE-Seq assay analysis (46.4%, when not considering the possibility of nucleotide insertions or deletions) contained six or more mismatched bases to the guide RNA (Figure S1C, Table S1, Table S2). Such off-target sequences have previously been proposed to be accommodated by bulging out or skipping of nucleotides (Boyle et al., 2021; Cameron et al., 2017; Doench et al., 2016; Jones et al., 2021; Lin et al., 2014; Tsai et al., 2015), which would result in a shift of the nucleotide register to re-establish correct base pairing downstream of the initially encountered mismatch. The PTPRC-tgt2 off-target #1 and FANCF off-target #3 sites are predicted to contain single nucleotide deletions at duplex positions 15 and 17, respectively (Figure 1A). The structures of the corresponding off-target complexes reveal that the single nucleotide deletions in these off-target substrates are not accommodated by bulging out the unpaired guide RNA nucleotide. Instead, the conformations of the guide RNAs remain largely unperturbed and the off-target TS DNAs “skip over” the unpaired gRNA bases to resume productive base-pairing downstream (Figure 5A–B). The seed regions of the gRNAs make extensive interactions with the bridge helix and the REC1 domain, whereas the TS DNA phosphate backbones are displaced by almost 3 Å (Figure S17A–B). The base pair skips are accommodated by considerable buckling and tilting of the base pairs immediately downstream of the skip site (Table S6). An additional consequence of the base pairing register shift is the formation of non-canonical base pairs between the off-target DNA and the extra 5’-terminal guanine nucleotides present in the guide RNA as a consequence of in vitro transcription by T7 RNA polymerase (Figure S17C–D). This potentially explains the impact of the 5’-guanines on both R-loop stability and in vitro cleavage activity (Kulcsar et al., 2020; Mullally et al., 2020; Okafor et al., 2019).
Figure 5. Off-targets with single-nucleotide deletions are accommodated by base skipping or multiple consecutive mismatches.
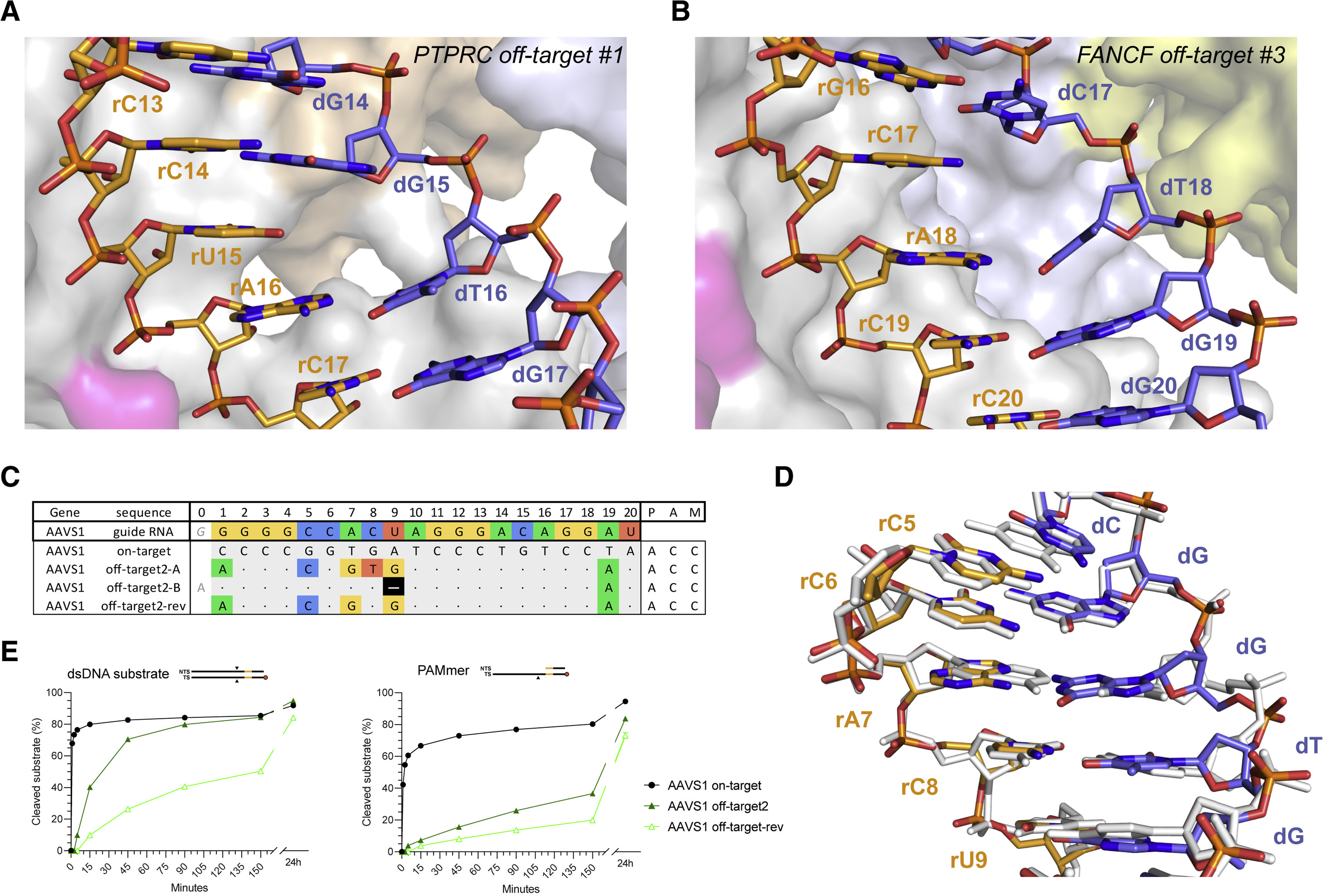
(A) Zoomed-in view of the base skip at duplex position 15 in the PTPRC-tgt2 off-target #1 complex. (B) Zoomed-in view of the base skip at duplex position 17 in the FANCF off-target #3 complex. (C) Schematic depiction of alternative base pairing interactions in the AAVS1 off-target #2 complex. AAVS1 off-target #2-rev substrate was designed based on the AAVS1 off-target #2, with the reversal of a single mismatch in the consecutive region back to the corresponding canonical base pair. (D) Structural overlay of the AAVS1 off-target #2 (coloured) and AAVS1 on-target (white) heteroduplexes. (E) Cleavage DNA kinetics of AAVS1 on-target, off-target #2 and off-target #2-rev substrates. See also Figures S15, S16, S17, S18, S19.
Originally, our SITE-Seq assay analysis classified the AAVS1 off-target #2 as a single-nucleotide deletion at duplex position 9 (Figure 5C). Unexpectedly, the structure of the AAVS1 off-target #2 complex instead reveals that the off-target substrate is bound in the unshifted register, resulting in the formation of five base mismatches in the PAM-distal half of the guide RNA–TS duplex (Figure 5D), including a partially paired rC-dC mismatch at position 5, an rA-dG Hoogsteen pair at position 7, a partially paired rC-dT mismatch at position 8, and a tautomeric rU-dG pair at position 9. The backbone conformations of the gRNA and the off-target TS exhibit minimal distortions and are nearly identical with the corresponding on-target heteroduplex (Figure 5D). This implies that some mismatch combinations might synergistically result in guide RNA and TS backbone geometries that mimic the on-target conformation and result in off-target tolerance. To test this hypothesis, we reverted the rC-dT mismatch at position 8 to the on-target rC-dG pair, thereby reducing the total amount of off-target mismatches from 6 to 5 (Figure 5C). The resulting off-target substrate (AAVS1 off-target #2-rev) exhibited substantially reduced cleavage rates in both dsDNA and PAMmer formats, as well as a significantly increased dissociation rate (Figure 5E, Table S8). We replicated these results for two other AAVS1 off-target substrates with different mismatch patterns that were initially predicted to contain PAM-distal deletions (Figure S18, Table S8). Together, these results suggest that some bona fide off-target substrates containing multiple consecutive mismatches, the reversal of one mismatch may affect the structural integrity of the guide RNA–TS DNA heteroduplex and interfere with DNA binding and/or conformational activation of Cas9, despite a reduction in the total number of mismatches.
To further investigate the accommodation of PAM-distal deletions in Cas9 off-target sites, we selected an additional efficiently cleaved off-target substrate (CD34 off-target #9, Figure S19A,B), containing a dT substitution at position 17 (resulting in a rU-dT mismatch with the CD34 sgRNA) and a predicted single-nucleotide deletion at position 6. Crystal structure of the resulting complex revealed that the rU-dT mismatch involves the syn conformer of the rU17 nucleotide (Figure S19C). The deletion at position 6 is accommodated by base skipping, leaving the base of rA6 unpaired within the guide-TS heteroduplex stack (Figure S19E) and the register shift results enables the 5’-terminal guanine nucleotide of the gRNA (introduced during in vitro transcription) to base pair with 3’-terminal TS nucleotide (Figure S19F). The backbone distortions in the TS are accompanied by compensatory rearrangements of the REC2 and REC3 domains of Cas9 (Figure S19G), which preserve all contacts with the heteroduplex (Figure S6).
Collectively, these results indicate that deletion-containing off-target complexes are accommodated either by RNA base skipping, as opposed to RNA nucleotide bulging, or by the formation of multiple base mismatches, with the precise mechanism dependent in part on the position of the deletion.
PAM-distal mismatches perturb the Cas9 conformational checkpoint
FANCF off-target #4, which contains three PAM-distal mismatches at positions 1–3 and a G-U mismatch in position 10 (Figure 1A), is reproducibly the top ranking off-target site for the FANCF guide RNA, as detected by SITE-Seq assay analysis at the lowest Cas9 RNP concentrations (Table S1, Table S2). The off-target substrate exhibits slow cleavage kinetics in vitro with both dsDNA and PAMmer substrates (Figure 2B, Figure S2A–B), indicating a perturbation of the conformational activation checkpoint of Cas9. The structure of the FANCF off-target #4 complex reveals that the RNA–DNA heteroduplex is unpaired at positions 1–3 as a result of the PAM-distal mismatches, with nucleotides 1–2 of the guide RNA and 19–20 of the TS disordered (Figure 6A). Furthermore, Cas9 undergoes structural rearrangements of its REC lobe and the HNH domain (Figure 6B, Video S2), resulting in a root mean square displacement of the REC2 and REC3 domains of 3.7 Å (1,315 Cα atoms) relative to the FANCF on-target complex structure. The REC3 domain undergoes a 19-degree rotation (Figure 6B), facilitated by extending the helix comprising residues 703–712 through restructuring of loop residues 713–716 (Figure S20A), to accommodate the altered guide RNA conformation. The REC2 domain rotates 32o away from the REC3 domain (Figure 6B). This is accompanied by restructuring of the hinge loop residues 174–180 and disordering of loops 258–264, 284–285, and 307–309. Concomitantly, the HNH domain rotates 11o away from the heteroduplex, as compared to the FANCF on-target structure, to accommodate distortion of the TS DNA (Figure 6B).
Figure 6. FANCF off-target #4 exhibits conformational changes in the REC2/3 and HNH domains due to PAM-distal duplex unpairing.

(A) Close-up view of the unpairing of mismatched bases at the PAM-distal end of the FANCF off-target #4 heteroduplex. The last two nucleotides on each strand could not be modelled due to structural disorder. (B) Overlay of the FANCF off-target #4 and FANCF on-target complex structures. The FANCF off-target #4 complex is coloured according to the domain legend in Figure 1A, FANCF on-target complex is shown in white. The REC1, RuvC, and PAM-interaction domains have been omitted for clarity, as no structural differences are observed. See also Figures S20, S21.
The unpaired 5’ end of the sgRNA is located at the interface between the REC3 and the RuvC domain and maintains interactions with heteroduplex-sensing residues Lys510, Tyr515, and Arg661 of the REC3 domain (Figure S20B). In contrast to the corresponding on-target complex structure, the unpaired 3’ end of the off-target TS breaks away from the REC3 lobe and instead points towards the REC2 domain, forming unique interactions with Arg895, Asn899, Arg905, Arg919 and His930 in the HNH domain (Figure S20C). These interactions (Figure S20D) could be responsible for the observed repositioning of the REC lobe and HNH domain, and they may impede the formation of a cleavage-competent complex.
The conformation of the FANCF off-target #4 complex is distinct from the conformations observed in cryo-EM reconstructions of the pre- and post-cleavage states of the Cas9 complex (Zhu et al., 2019) (Figure S21A-B). Instead, the off-target complex structure most closely resembles that of a high-fidelity variant xCas9 3.7 containing amino acid substitutions that disrupt interactions with the TS DNA (Guo et al., 2019). Although the xCas9 3.7 complex adopts a slightly different REC lobe conformation (Figure S21C), the PAM-distal duplex also undergoes unpairing at positions 1–3 and displays a comparable degree of structural disorder (Figure S21D). These structural observations thus suggest that the presence of multiple mismatches in the PAM-distal region of a guide RNA–off-target DNA duplex leads to conformational perturbations in the DNA-bound complex that resemble the structural consequences of specificity-enhancing mutations in high-fidelity Cas9 variants.
DISCUSSION
The off-target activity of Cas9 has been extensively documented in prior genome editing, biochemical and biophysical studies (Boyle et al., 2017; Boyle et al., 2021; Doench et al., 2016; Jones et al., 2021; Lazzarotto et al., 2020; Zhang et al., 2020). Although numerous methods have been devised for computational prediction of genomic off-target sites and their experimental validation, these have reported highly variable mismatch tolerance profiles depending on the screening method and the target sequence. Thus, a comprehensive understanding of this phenomenon is still lacking, particularly as to whether off-target tolerance has an underlying structural basis. In this study, we used the SITE-Seq assay to examine the off-target landscape of 12 well-studied guide RNAs, observing a broad variation of cleavage activities associated with individual off-target substrates. To shed light on the molecular mechanisms underpinning off-target activity, we determined atomic structures of a representative set of 16 off-target complexes, thus providing fundamental insights into the structural aspects of off-target recognition.
Role of non-canonical base pairing in off-target recognition
The principal, and largely unexpected, finding of our structural analysis is that the majority of nucleotide mismatches in bona fide off-target substrates are accommodated by non-canonical base pairing interactions. These range from simple rG-dT/rU-dG wobble or rA-dG/rG-dA/rG-dG Hoogsteen base pairing interactions, to pyrimidine-pyrimidine pairs that rely on (deoxy)ribose-phosphate backbone distortions that reduce duplex width. With the notable exception of rA-dA mismatches, which are accommodated by extrahelical base extrusion, the structural rearrangements associated with base mismatch accommodation largely preserve base stacking, which is the primary determinant of nucleic acid duplex stability (Yakovchuk et al., 2006). For some off-target sequences, our structures are suggestive of base protonation or tautomerization facilitating hydrogen bonding interactions in otherwise non-permissive mismatches, such as rA-dC. These rare base pair forms have been previously observed in both RNA and DNA duplexes and are thought to be important contributors to DNA replication and translation errors (Kimsey et al., 2015; Kimsey et al., 2018). Future studies employing complementary structural methods, such as nuclear magnetic resonance, will help confirm the occurrence of non-canonical base states in off-target complexes.
The mismatch tolerance of Cas9 can be explained primarily by two factors. Firstly, Cas9 does not directly contact the major- or minor-groove edges of the guide RNA–TS DNA heteroduplex base pairs at any of the duplex positions and thus lacks a steric mechanism to enforce Watson-Crick base pairing (Figure S6). This is further underscored by Cas9’s tolerance of base modifications in target DNA, including cytosine 5-hydroxymethylation and, at least at some duplex positions, glucosyl-5-hydroxymethylation (Vlot et al., 2018). In this respect, Cas9 differs from other molecular systems, notably the ribosome and replicative DNA polymerases, which enhance the specificity of base-pairing by direct readout of base-pair shape and steric rejection of mismatches (Kunkel and Bebenek, 2000; Rodnina and Wintermeyer, 2001; Timsit, 1999).
Secondly, Cas9 is a multidomain protein that displays considerable conformational plasticity and is therefore able to accommodate local distortions in the guide–TS duplex geometry by compensatory rearrangements of the REC2, REC3 and HNH domains (Chen et al., 2017; Donohoue et al., 2021). Indeed, in most off-target structures reported in this study, almost all atomic contacts between Cas9 and the guide–TS heteroduplex are preserved (Figure S6). Thus, Cas9 only detects guide-target mismatches by indirect readout of the guide RNA–TS DNA heteroduplex width, except at the PAM-distal end of the heteroduplex where base mismatches result in duplex unpairing, as discussed below. Our observations are consistent with recent molecular dynamics simulation studies showing that internally positioned mismatches within the guide RNA–TS DNA heteroduplex are readily incorporated within the heteroduplex and have only minor effects on Cas9 interactions (Mitchell et al., 2020). The lack of a steric base-pair enforcement mechanism and the resulting off-target promiscuity likely reflects the biological function of Cas9 in CRISPR immunity, where mismatch tolerance enables targeting of closely related viruses and hindering immune evasion by mutations or covalent base modifications (Deveau et al., 2008; Semenova et al., 2011; van Houte et al., 2016; Yaung et al., 2014). On the other hand, such conformational plasticity also enables the incorporation of various chemical modifications in the guide RNA that are compatible with the A-form geometry of the heteroduplex (Cromwell et al., 2018; Donohoue et al., 2021; Hendel et al., 2015; O’Reilly et al., 2019; Yin et al., 2018; Yin et al., 2017). These could potentially disfavour the formation of certain non-canonical base pairs or reduce the backbone flexibility of the guide RNA, thereby enhancing off-target discrimination. However, the effect of specific guide RNA modifications on particular types of mismatches is yet to be closely examined mechanistically (Donohoue et al., 2021).
Structural rigidity of the guide RNA seed region and implications for off-target recognition
The seed sequence of the Cas9 guide RNA (nucleotides 11–20) is the primary determinant of target DNA binding, a consequence of its structural pre-ordering in an A-like conformation by extensive interactions with Cas9 (Anders et al., 2014; Jiang et al., 2015; Nishimasu et al., 2014; Zhu et al., 2019). Our data indicate that structural rigidity of the guide RNA seed sequence also affects off-target recognition, as base mismatches in the seed region of the heteroduplex can only be accommodated by conformational distortions of TS DNA, which is subject to only a few steric constraints, notably at position 20 due to interactions with the phosphate lock loop (Anders et al., 2014). This increases the energetic penalty of base mispairing in the seed region of the heteroduplex, and thus contributes to mismatch sensitivity of Cas9 within the seed region. Although structural distortions of TS DNA facilitate binding of off-target substrates containing seed mismatches, they may nevertheless inhibit off-target cleavage by steric hindrance of the HNH domain, thereby further contributing to off-target discrimination. The contrasting structural plasticities of the guide RNA and TS DNA strands are manifested in the differential activities of Cas9 against off-targets containing rU-dG and rG-dT mismatches within the seed region (Boyle et al., 2021; Doench et al., 2016; Hsu et al., 2013; Jones et al., 2021; Zhang et al., 2020). Whereas rG-dT mismatches can be readily accommodated by wobble base pairing, seed sequence rigidity is expected to hinder rU-dG wobble base pairing. Combined with a lower energetic penalty associated with rG-dT mismatch binding (binding an off-target with an rG-dT mismatch requires unpairing a dT-dA base pair in the off-target DNA, while rU-dG off-target recognition requires dC-dG unpairing), these effects thus help Cas9 discriminate against rU-dG mismatches in the seed region.
Recognition of off-targets containing insertions and deletions
Bona fide off-target sites containing insertions or deletions have been detected in a number of studies (Boyle et al., 2021; Cameron et al., 2017; Doench et al., 2016; Jones et al., 2021; Tsai et al., 2015). Nucleotide “bulging” has been proposed as a mechanism to recognize such off-targets, which would otherwise result in large numbers of consecutive base mismatches. However, as Cas9 encloses the guide RNA–TS DNA heteroduplex in a central channel and makes extensive interactions along the entire length of the guide RNA strand, the formation of RNA bulges is precluded by steric clashes, pointing to a different mechanism.
Indeed, our structural analysis provides no evidence for RNA bulging. Instead, the structures of PTPRC-tgt2 off-target #1 and FANCF off-target #3 complexes reveal that off-target sequences predicted to contain single-nucleotide deletions in the seed region of the heteroduplex are recognized by base skipping, resulting in an unpaired guide RNA base within the duplex stack. This is enabled by the lack of extensive contacts of Cas9 with the TS, while rigid coordination of the guide RNA in the seed region disfavours extrahelical guide RNA bulging. However, base skipping in the seed region of the heteroduplex likely incurs a large energetic penalty. As the seed region of the TS DNA is devoid of Cas9 contacts in the pre-cleavage state (Zhu et al., 2019), off-targets containing single-nucleotide insertions in the seed region of the heteroduplex are likely to be recognized by DNA nucleotide bulging, likewise incurring a large energetic penalty as unwinding an off-target DNA sequence containing an insertion requires breaking an extra base pair. Additionally, TS DNA distortion might inhibit cleavage by steric hindrance of the HNH domain. These observations thus explain why Cas9 appears to tolerate mismatches better than insertions or deletions and why deletions and insertions within the seed region are particularly deleterious (Boyle et al., 2021; Cameron et al., 2017; Doench et al., 2016; Jones et al., 2021).
In contrast, off-target sequences containing insertions or deletions in the PAM-distal region of the heteroduplex (positions 1–10) can be recognized either by base skipping, as in the case of CD34 off-target #9, or bound in the unshifted register, with multiple base mismatches accommodated by non-canonical base pairing interactions, as seen for AAVS1 off-target #2, which was previously predicted to contain an RNA bulge or skip (Cameron et al., 2017; Lazzarotto et al., 2020). Which of the two mechanisms is used likely depends on the off-target sequence and the position of the deletion, which in turn dictate the number of mismatches between the guide and the off-target sequence in the unshifted register. Off-target accommodation by multiple non-canonical base pairs likely relies on their synergistic effect to mimic the on-target heteroduplex geometry, which enables unperturbed binding, as supported by our observations that mismatch reversal in several off-target substrates reduces their rates of cleavage. Although our structural analysis does not examine off-target substrates containing insertions, we posit that PAM-distal insertions are recognized as multiple mismatches due to steric hindrance of extrahelical bulging. Thus, our structural findings suggest that a substantial fraction of off-target sites predicted to contain insertions or deletions may be bound via multiple mismatches instead (Boyle et al., 2021; Doench et al., 2016; Jones et al., 2021).
PAM-distal base pairing and the conformational checkpoint of Cas9
Upon substrate DNA hybridization and R-loop formation, Cas9 undergoes conformational activation of its nuclease domains (Bravo et al., 2022; Pacesa et al., 2022; Zhu et al., 2019). The Cas9 REC3 domain plays a key role in the process, as it senses the integrity of the PAM-distal region of the guide RNA–TS DNA heteroduplex and allosterically regulates the REC2 and HNH domains, providing a conformational checkpoint that traps Cas9 in a conformationally inactive state in the absence of PAM-distal hybridization (Chen et al., 2017; Dagdas et al., 2017; Palermo et al., 2018; Zhu et al., 2019). Our structural data confirm that mismatches at the PAM-distal end of the heteroduplex (positions 1–3) result in heteroduplex unpairing, incomplete R-loop formation and structural repositioning of the REC3 domain (Figure 6), indicating a perturbation of the Cas9 conformational checkpoint. We envision that the observed conformational state mimics the structural effect of 5’-truncated guide RNAs, which have been shown to improve targeting specificity (Fu et al., 2014). Furthermore, similarities with the structure of a high-fidelity Cas9 variant (Guo et al., 2019) suggest a shared underlying mechanism for increased specificity.
Implications for off-target prediction
Our structural data reveal that Cas9 plays a limited steric role in off-target discrimination insofar as only sensing the integrity and general shape of the guide–target heteroduplex. Off-target activity is thus largely determined by the kinetics and energetics of R-loop formation, that is off-target DNA strand separation and concomitant guide RNA–TS DNA hybridization, and subsequent Cas9 conformational activation. We observe on multiple occasions that a given mismatch adopts different conformational arrangements depending on its position along the guide RNA–TS DNA heteroduplex, as further supported by MD simulations of rA-dA mismatches. This poses a challenge for ab initio modelling of off-target activity, as biophysical models of off-target binding and cleavage are bound to be of limited accuracy unless they incorporate position-dependent energetic penalties for each base mismatch type and for deletions, as well as position- and base-specific penalties for insertions (Boyle et al., 2021; Jones et al., 2021; Zhang et al., 2020). As illustrated by our study, MD simulations can complement experimental data to provide structural information on specific mismatches in remaining positions within the heteroduplex. Thus, ongoing structural and computational studies, combined with machine learning approaches, will assist in generating complete models for off-target prediction.
Furthermore, as certain off-target sequences that are incompatible with dsDNA cleavage can undergo NTS nicking (Fu et al., 2019; Jones et al., 2021; Murugan et al., 2020; Zeng et al., 2018), future bioinformatic models need to be able to predict off-target nicking activity as well. Furthermore, accurate modelling of off-target interactions remains difficult due to context-dependent effects, as documented in previous studies showing that the binding and cleavage defects of consecutive mismatches deviate from additivity (Boyle et al., 2021; Cameron et al., 2017; Lazzarotto et al., 2020; Zhang et al., 2020). Indeed, our structural data rationalize this by showing that the conformation of a given base mismatch is highly sensitive to the presence of neighbouring mismatches. As seen in the case of AAVS1 off-target #2 complex, multiple mismatched bases can synergistically combine to preserve an on-target-like heteroduplex conformation that passes the REC3 conformational checkpoint, supporting nearly on-target efficiencies of cleavage (Zhang et al., 2020). This is in line with recent cryo-EM structural studies suggesting that indirect readout of heteroduplex conformation is coupled to nuclease activation, while mismatches disrupt this coupling (Bravo et al., 2022; Pacesa et al., 2022). Critically, reversion of one of the mismatches in this off-target substrate impairs cleavage activity. Similar effects have been described for other DNA binding proteins such as transcription factors, where mismatches modulate transcription factor binding activity by affecting the conformation of the DNA duplex (Afek et al., 2020). In an analogy with Cas9, these proteins check for correct binding sites through indirect sequence readout by sampling for the correct duplex shape rather than base sequence (Abe et al., 2015; Kitayner et al., 2010; Rohs et al., 2009a; Rohs et al., 2009b).
In conclusion, structural insights presented in this study establish an initial framework for understanding the molecular basis for the off-target activity of Cas9. In conjunction with ongoing computational studies, these findings will help achieve improved energetic parametrization of off-target mismatches and deletions/insertions, thus contributing to the development of more accurate off-target prediction algorithms and more specific guide RNA designs. In doing so, these studies will contribute towards increasing the precision of CRISPR-Cas9 genome editing and the safety of its therapeutic applications.
Limitations of the study
The structural dataset presented in this study is necessarily restricted in scope. Although all mismatch types are covered, a much larger collection of off-target complex structures would be required to cover all mismatches at all positions to achieve a complete structural overview of the mismatch tolerance underpinning intrinsic off-target activity of Cas9. Although MD simulations can help fill in the gaps in the experimental data, our findings suggest that mismatch accommodation appears to be sequence context-dependent, thus limiting their predictive power. Although we provide insights into off-target substrates containing single-nucleotide deletions, it is presently difficult to predict which mechanism occurs at a given heteroduplex position. Moreover, we currently lack structural data for off-target substrates containing insertions, which will be addressed in future studies. Finally, not all structurally characterized off-targets (~50%) are efficiently cleaved in cells, despite detectable cleavage in vitro, implying that other factors affect Cas9 off-targeting in the context of eukaryotic genomes. Thus, further work combining structural and computational approaches will be needed to accurately predict the off-target activity of Cas9.
STAR★METHODS
METHODS
DNA oligonucleotides and substrates
Sequences of DNA oligonucleotides used in this study are summarised in Table S9. Crystallisation substrates were synthesised by Sigma Aldrich without further purification, sgRNA transcription templates and ATTO-532-labelled cleavage substrates were synthesised by Integrated DNA Technologies, Inc., with PAGE and HPLC purification, respectively. Partially double stranded crystallisation substrates were prepared by mixing complementary oligonucleotides in a 1:1 molar ratio (as determined by 260 nm absorption), heating to 95 °C for 5 minutes and slow cooling to room-temperature. Cleavage substrates were prepared similarly, except that a 2-fold molar excess of the non-target strand was used.
Cas9 protein expression and purification
Streptococcus pyogenes Cas9 wild type protein and the nuclease dead mutant (D10A, H840A) were both recombinantly expressed for 16 hours at 18 °C in Escherichia coli Rosetta 2 (DE3) (Novagen) N-terminally fused to a hexahistidine affinity tag, the maltose binding protein (MBP) polypeptide, and the tobacco etch virus (TEV) protease cleavage site. Cells were resuspended and lysed in 20 mM HEPES-KOH pH 7.5, 500 mM KCl, 5 mM imidazole, and supplemented with added protease inhibitors. Clarified lysate was loaded on a 10 ml Ni-NTA Superflow column (QIAGEN), washed with 7 column volumes of 20 mM HEPES-KOH pH 7.5, 500 mM KCl, 5 mM imidazole, and eluted with 10 column volumes of 20 mM HEPES-KOH pH 7.5, 250 mM KCl, 200 mM imidazole. Salt concentration is adjusted and protein is loaded on a 10 ml HiTrap Heparin HP column (GE Healthcare) equilibrated in 20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM DTT. The column is washed with 5 column volumes of 20 mM HEPES-KOH pH 7.5, 250 mM KCl, 1 mM DTT, and Cas9 is eluted with 17 column volumes of 20 mM HEPES-KOH pH 7.5, 1.5 M KCl, 1 mM DTT, in a 0–32% gradient (peak elution around 500 mM KCl). His6-MBP tag was removed by TEV protease cleavage overnight with gentle shaking. The untagged Cas9 was concentrated and applied to a Superdex 200 16/600 (GE Healthcare) and eluted with 20 mM HEPES-KOH pH 7.5, 500 mM KCl, 1 mM DTT. Purified protein was concentrated to 10 mg/ml, flash frozen in liquid nitrogen and store at -80 °C. DTT was omitted in the size-exclusion step of the purification when protein was used for switchSENSE measurements.
sgRNA transcription and purification
sgRNAs are transcribed from a double stranded PCR product template amplified from a plasmid in a 5 ml transcription reaction (30 mM Tris-HCl pH 8.1, 25 mM MgCl2, 2 mM spermidine, 0.01% Triton X-100, 5 mM CTP, 5 mM ATP, 5 mM GTP, 5 mM UTP, 10 mM DTT, 1 μM DNA transcription template, 0.5 units inorganic pyrophosphatase (Thermo Fischer), 250 μg homemade T7 RNA polymerase. The reaction is incubated at 37 °C for 5 hours, and then treated for 30 minutes with 15 units of RQ1 DNAse (Promega). The transcribed sgRNAs are subsequently PAGE purified on an 8% denaturing (7 M urea) polyacrylamide gel, and lastly ethanol precipitated and resuspended in DEPC treated water.
Crystallisation of Cas9 ternary complexes and structure determination
To assemble the Cas9 on-/off-target ternary complexes, the Cas9 protein is first mixed with the sgRNA in a 1:1.5 molar ratio and incubated at room temperature for 10 minutes. Next, the binary complex is diluted to 2 mg/ml with 20 mM HEPES-KOH 7.5, 250 mM KCl, 1 mM DTT, 2 mM MgCl2 buffer, pre-annealed 100 μM DNA substrate is added in a 1:1.8 molar ratio and the complex is incubated another 10 minutes at room temperature. For crystallisation, 1 μl of the ternary complex (1–2 mg/ml) is mixed with 1 μl of the reservoir solution (0.1 M Tris-acetate pH 8.5, 0.3–0.5 M KSCN, 17–19% PEG3350) and crystals are grown at 20 °C using the hanging drop vapour diffusion method. In some cases, microseeding was be used to improve crystal morphology. Crystals are typically harvested after 2–3 weeks, cryoprotected in 0.1 M Tris-acetate pH 8.5, 0.4 M KSCN, 30% PEG3350, 15% ethylene glycol, 1 mM MgCl2, and flash-cooled in liquid nitrogen. Diffraction data was obtained at beamlines PXI and PXIII of the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) and were processed using the XDS package (Kabsch, 2010). Structures were solved by molecular replacement through the Phaser module of the Phenix package (Adams et al., 2010) using the PDB ID: 5FQ5 model omitting the RNA-DNA target duplex from the search. Model adjustment and duplex building was completed using COOT software (Emsley et al., 2010). Atomic model refinement was performed using Phenix.refine (Adams et al., 2010). Protein-nucleic acid interactions were analysed using the PISA web server (Krissinel and Henrick, 2007). Characterisation of the guide-protospacer duplex was performed using the 3DNA 2.0 web server (Li et al., 2019). Structural figures were generated using PyMOL and ChimeraX (Pettersen et al., 2021).
In vitro nuclease activity assays
Cleavage reactions were performed at 37 °C in reaction buffer, containing 20 mM HEPES pH 7.5, 250 mM KCl, 5 mM MgCl2 and 1 mM DTT. First, Cas9 protein was pre-incubated with sgRNA in 1:1.25 ratio for 10 minutes at room temperature. The protein-RNA complex was rapidly mixed with the target strand-ATTO-532-labelled dsDNA, to yield final concentrations of 1.67 μM protein and 66.67 nM substrate in a 7.5 μl reaction. Time points were harvested at 1, 2.5, 5, 15, 45, 90, 150 minutes, and 24 hours. Cleavage was stopped by addition of 2 μl of 250 mM EDTA, 0.5% SDS and 20 μg of Proteinase K. Formamide was added to the reactions with final concentration of 50%, samples were incubated at 95 °C for 10 minutes, and resolved on a 15% denaturing PAGE gel containing 7M urea and imaged using a Typhoon FLA 9500 gel imager. Depicted error bars correspond to the standard deviation from four independent cleavage reactions. Rate constants (kobs) were extracted from single exponential fits: [Product] = A*(1-exp(−kobs*t))
switchSENSE analysis
The target strands (TS) containing a 3’ flanking sequence complementary to the ssDNA covalently bound to the chip electrode, and the non-target strands (NTS) (Table S9) were resuspended in a buffer containing 10 mM Tris-HCl pH 7.4, 40 mM NaCl, and 0.05% Tween 20. The matching TS:NTS duplex is pre-annealed and hybridised to the chip anode. The Cas9 protein was mixed with the sgRNAs at a 1:2 protein:RNA molar ratio, and the complex was incubated for 30 min at 37 °C in association buffer containing 20 mM HEPES-KOH pH 7.5, 150 mM KCl, 2 mM MgCl2, 0.01% Tween 20. All switchSENSE experiments were performed on a DRX analyser using CAS-48–1-R1-S chips (Dynamic Biosensors GmbH, Martinsried, Germany). Kinetics experiments were performed at 25 °C in association buffer, with an association time of 5 min, dissociation time of 20 min, and a flow rate of 50 μl/min.
SITE-Seq assay
SITE-Seq assay reaction conditions were performed as described previously (Cameron et al., 2017). Briefly, high molecular weight genomic DNA (gDNA) was purified from human primary T cells using the Blood & Cell Culture DNA Maxi Kit (Qiagen) according to the manufacturer’s instructions. RNPs comprising the guides were biochemically assembled for gDNA digestion. Specifically, equal molar amounts of crRNA and tracrRNA were mixed and heated to 95 °C for 2 min then allowed to cool at room temperature for ~5 min. Three-fold molar excess of the guides were incubated with Streptococcus pyogenes Cas9 (SpCas9) in cleavage reaction buffer (20 mM HEPES pH 7.4, 150 mM KCl, 10 mM MgCl2, 5% glycerol) at 37 °C for 10 min. In a 96-well plate format, 10 μg of gDNA was treated with 0.2 pmol (4 nM), 0.8 pmol (16 nM), 3.2 pmol (64 nM), and 12.8 pmol (256 nM) of each RNP in 50 μL total volume in cleavage reaction buffer. Each cleavage reaction was performed in triplicate. Negative control reactions were assembled in parallel and did not include RNP. gDNA was treated with RNPs for 4 hours at 37 °C. SITE-Seq assay library preparation and sequencing was performed as described previously and the final library was loaded onto the Illumina NextSeq platform (Illumina, San Diego, CA), and ~1–3 M reads were obtained for each sample.
SITE-Seq assay analysis and selection for cellular validation
SITE-Seq assay recovered off-targets were filtered for sites that had read-pileups proximal to the expected cut site, a PAM comprising at least one guanine base, fewer than 12 mismatches (reasoning that sites with 12 or more mismatches are likely spurious peaks not resulting from Cas9-induced double-strand breaks), and all sites with 11 mismatches were visually inspected and included in analysis if a putative deletion or insertion would result in a reduction of at least 4 mismatches relative to the spacer sequence.
In silico mismatch, deletion, and insertion classification
Predictive classification of SITE-Seq assay recovered off-target sites as pure mismatches, deletions, or insertions was executed using a scoring algorithm which consisted of the following sequential steps (Figure S15): (i) For each off-target, a gap library was generated where a single nucleotide gap was introduced between each nucleotide in the off-target sequence. (ii) The off-target gap library was then aligned to the spacer sequence and each alignment was scored based on the number of matched bases between the spacer and gapped off-target pair. If the gapped off-target with the highest alignment score improved alignment by at least 4 nucleotides relative to the non-gapped spacer–off-target alignment, the off-target sequence was marked as a single-nucleotide deletion and removed from subsequent analysis. (iii) The remaining pool of off-targets were then aligned to a spacer gapped library where a single nucleotide gap was introduced at each positing in the spacer. (iv) The spacer gap library was then aligned to each off-target sequence and each alignment was scored based on the number of matched bases between the off-target and the gapped spacer pair. If the gapped spacer with the highest alignment score improved alignment by at least 4 nucleotides relative to the non-gapped spacer–off-target alignment, the off-target sequence was marked as a single-nucleotide insertion and removed from subsequent analysis. (v) The remaining off-targets for which the spacer–off-target alignment was not improved by single-nucleotide deletions or insertions were annotated as a mismatched off-targets.
Generation of ribonucleoprotein complexes (RNP) for electroporation
Synthetic crRNA and tracrRNA oligonucleotides were ordered from Integrated DNA Technologies (Coralville, IA) as AltR™ reagents. Ribonucleoprotein complexes (RNPs) were formulated by incubating 480 pmols of each crRNA and tracrRNA and heated to 95 °C for 2 min then allowed to cool at room temperature for ~5 min. Annealed guides were then mixed with 160 pmols of Cas9 protein (1:3 molar ratio of Cas9 to guides) in RNP assembly buffer (20 mM HEPES pH 7.4, 150 mM KCl, 10 mM MgCl2, 5% glycerol) and incubated at 37 °C for 10 min. RNP were then serial diluted two-fold by mixing with equal volume of RNP assembly buffer across seven concentrations down to 2.5 pmol of Cas9 and 7.5 pmols of crRNA-tracrRNA. RNPs were kept at 4 °C or on ice until transfection.
T cell handling and transfections
T cells were isolated from peripheral blood mononuclear cells (PBMCs) using the RoboSep-S cell isolation platform (STEMCELL Technologies) with EasySep Human T cell Isolation Kit (STEMCELL Technologies). Cells were then activated for 3 days in “Complete media” (Immunocult-XF T cell expansion media (STEMCELL Technologies), CTS Immune Cell serum replacement (Fisher Scientific), 1x antibiotic-antimycotic (Fisher Scientific), and recombinant human Interleukin-2 (rhIL-2, 100 units/mL)) in the presence of anti-CD3/CD28 Dynabeads (Fisher Scientific) at a bead:cell ratio of 1:1. On the third day, beads were removed and cells expanded in Complete media for 24 hours. Cells were then harvested, washed once with phosphate-buffered saline (PBS) and resuspended at 107 cells/mL in P4 Primary Cell Nucleofector Solution (Lonza). 20 μL of resuspended cells (corresponding to ~ 200,000 cells) were then combined with 2.5 μL of RNP and electroporated using the Lonza 4D 96-well Nucleofector electroporation system using pulse code CA137. Transfected cells were then recovered in 180 μL/sample of Complete media. After 48 hours, the electroporated T cells were pelleted and gDNA was isolated by adding 50 μL/well QuickExtract DNA extraction solution (Epicentre), followed by incubation at 37 °C for 10 minutes, 65 °C for 6 minutes, and 95 °C for 3 minutes. The isolated gDNA was diluted with 50 μL sterile water to achieve ~ 2,000 genome equivalents/μL and samples were stored at −20 °C until NGS analysis.
Targeted amplicon sequencing
Cas9 on- and off-target sites were amplified in a two-step PCR reaction. In brief, 3.75 μl (corresponding to ~7,500 cells) of lysate was used as a template for PCR amplification with Q5 Hot-Start High Fidelity DNA Polymerase (NEB) and unique primer pairs containing an internal locus-specific region and an outer Illumina-compatible adapter sequence. A second PCR reaction targeting the outer-adapter sequence was performed to append unique indices to each amplicon. Sites were sequenced on a MiSeq with 2 × 151 paired-end reads and v2 chemistry, or NextSeq with 2 × 151 paired- end reads and v2 or v2.5 chemistry (Illumina), and aligned to the hg38 genomic assembly. For each site, indels were tallied if they occurred within 3 nucleotides of the putative Cas9 cut site, and editing efficiencies were calculated by subtracting the percentage of indels in untransfected cells from the percentage of indels in RNP-transfected cells. Depth of coverage was ~5,000–20,000 reads per amplicon, and all samples with <500 reads aligning to the predicted amplicon were discarded. Lower limit of detection (e.g., ≥0.1%) was determined by titration of NIST genomic standards and assessment of expected versus measured values (Data not shown).
Molecular Dynamics (MD) simulations
MD simulations were performed using a well-established protocol, previously employed in several computational studies of CRISPR-Cas9 (Mitchell et al., 2020; Ricci et al., 2019). The Amber ff14SB force field (Maier et al., 2015) was adopted, including the ff99bsc1 corrections for DNA (Ivani et al., 2016) and the ff99bsc0+χOL3 corrections for RNA (Banas et al., 2010; Zgarbova et al., 2011). Explicit water molecules were described using the TIP3P model (Jorgensen et al., 1983), while the Li & Merz model was used for Mg2+ ions (Li and Merz, 2014). An integration time step of 2 fs was applied. All bond lengths involving hydrogen atoms were constrained using the SHAKE algorithm. Temperature control (300 K) was performed via Langevin dynamics (Turq et al., 1977), with a collision frequency γ = 1. Pressure control was accomplished by coupling the system to a Berendsen barostat (Berendsen et al., 1984), at a reference pressure of 1 atm and with a relaxation time of 2 ps. The systems were subjected to energy minimization to relax water molecules and counter ions, keeping the protein, the RNA, DNA and Mg2+ ions fixed with harmonic position restraints of 300 kcal/mol · Å2. Then, the systems were heated up from 0 to 100 K in the canonical ensemble (NVT), by running two simulations of 5 ps each, imposing position restraints of 100 kcal/mol · Å2 on the above-mentioned elements of the system. The temperature was further increased up to 200 K in ~100 ps of MD in the isothermal-isobaric ensemble (NPT), reducing the restraint to 25 kcal/mol · Å2. Subsequently, all restraints were released, and the temperature of the systems was raised up to 300 K in a single NPT simulation of 500 ps. After ~ 1.1 ns of equilibration, ~10 ns of NPT runs were carried out allowing the density of the systems to stabilize around 1.01 g cm-3. Finally, production runs were carried out in the NVT ensemble, collecting ~500 ns in three replicates for each of the systems. These simulations have been performed using the GPU-empowered version of AMBER 20 (D.A. Case, 2021; Salomon-Ferrer et al., 2013). Analysis of the RNA:DNA conformational dynamics has been done using the CURVES+ code (Lavery et al., 2009). Molecular simulations were based on three X-ray structures of CRISPR-Cas9: TRAC on-target (PDB: 7OX8), AAVS1 on-target #2 and FANCF on-target, displaying 20 base pair long RNA:TS DNA hybrid and a cleaved NTS. To study the accommodation of rA-dA mismatches at various positions along the heteroduplex, we systematically mutated the base pairs to rA-dA mismatches (TRAC on-target for positions 1, 11, 15, and 20; FANCF on-target for positions 3–5, 9, and 18; AAVS1 on-target for positions 2, 6–8, 10, 12–14, 16, 17, and 19), using the LEaP tool in the AMBER 20 code (Lavery et al., 2009). Additionally, the pre-cleavage state crystal structure of CRISPR-Cas9 with no mismatches (PDB: 4UN3) was also simulated for comparison. The obtained model systems were embedded in explicit waters, and counterions were added to neutralize the total charge at physiological conditions, leading to periodic simulation cells of ~145*115*150 Å3 and a total of ~220,000 atoms for each system.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Prof. Martin Jinek (jinek@bioc.uzh.ch)
Material availability
This study did not generate new unique reagents.
Data and code availability
Coordinates have been deposited to the Protein Data Bank under the PDB accession numbers: 7QQO, 7QR7, 7QQP, 7QQQ, 7QQR, 7QQS, 7QQT, 7QQU, 7QQV, 7QQW, 7QQX, 7QR5, 7QQZ, 7QR8, 7QR0, 7QR1 and 7ZO1. Raw sequencing files for the SITE-Seq assay have been deposited to BioProject. Accession numbers are listed in the key resource table. All deposited data is publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Alt-R S.p. Cas9 Nuclease V3 | IDT | Cat# 1081058 |
| RoboSep Filter Tip Racks (8) | StemCell Technologies | Cat# 20125 |
| Lonza Nucleofection Kits (10 96-well plates and buffer) P3 | Lonza Walkersville, Inc. | Cat# V4SP-3960 |
| Recombinant Human IL-2 | PeproTech, Inc | Cat# 200–02 |
| QuickExtract DNA Extraction Solution (10 mL Aliquots) | Lucigen | Cat# QE09050-PQ1329 |
| RoboSep Buffer (5X Concentrate) | StemCell Technologies | Cat# 20124 |
| RoboSep Kit # 17951 (Human T cell Isolation Kit) | StemCell Technologies | Cat# 17951 |
| Dynabeads Human T-Activator CD3/CD28 (5 × 2 mL) | Fisher Scientific Co LLC | Cat# 11132D |
| DNAse I Solution (1 mg/mL, 1 mL) | StemCell Technologies | Cat# 7900 |
| Antibiotic/Antmycotic (6 × 100 mL) | Fisher Scientific Co LLC | Cat# MT30004CI |
| CTS Immune Cell Serum Replacement (50 mL) | ThermoFisher Scientific | Cat# A2596101 |
| Immunocult-XF T Cell Expansion Media | StemCell Technologies | Cat# 10981 |
| SPRIselect Reagent Kit | Beckman Coulter | Cat# B233318 |
| Dynabeads M-280 Streptavidin | Invitrogen | Cat# 11206D |
| Q5^ Hot Start High-Fidelty 2x Master Mix | New England Biolabs | Cat# M0494L |
| NEBNext double-stranded DNA (dsDNA) Fragmentase | New England Biolabs | Cat# M0348L |
| T4 DNA Ligase | New England Biolabs | Cat# M0202S |
| 10x T4 DNA Ligase Reaction Buffer | New England Biolabs | Cat# B0202S |
| Proteinase K | ThermoFisher Scientific | Cat# EO0491 |
| HEPES | Roth | Cat# HN78.1 |
| Magnesium chloride | Fluka | Cat# 63064–500G |
| Potassium chloride | Roth | Cat# 6781.2 |
| Potassium thiocyanate | Roth | Cat# P753.1 |
| Poly(ethylene glycol) 3,350 | Sigma-Aldrich | Cat# 202444–500G |
| Ethylenediaminetetraacetic acid | Roth | Cat# 8040.3 |
| Ni Sepharose 6 Fast Flow resin (500 mL) | Cytiva | CAT# 17–5318-03 |
| HiPrep 26/10 desalting columns (pack of 4) | Cytiva | CAT# 17–5087-02 |
| 5 mL HiTrap SP HP | Cytiva | CAT# 17–1152-01 |
| HiLoad Superdex 200 26/600 | Cytiva | CAT# 28–9893-36 |
|
| ||
| Critical commercial assays | ||
|
| ||
| NextSeq 500/550 High Output V2.5 (150 cycles) | Illumina | Cat# 20024907 |
| MiSeq Reagent Kit v2 (300- cycles) | Illumina, Inc | Cat# MS-102– 2002 |
| NextSeq PhiX Control Kit | Illumina, Inc | Cat# FC-110– 3002 |
| Blood & Cell Culture DNA Maxi Kit | Qiagen | Cat# 13323 |
| High Sensitivity DNA Bioanalyzer Kit | Agilent | Cat# 5067–4626 |
| NEBNext^ dA-tailing Kit | NEB | Cat# E6053L |
| NEBNext^ Ultra End Repair / dA-tailing module | NEB | Cat# E7442L |
| NEBNext^ Ultra Ligation Kit | NEB | Cat# E7445L |
| NextSeq 500/550 High Output V2.5 (150 cycles) | Illumina | Cat# 20024907 |
|
| ||
| Deposited data | ||
|
| ||
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 on-target DNA substrate | Donohoue et al., 2021 | PDB: 7OX9 |
| Atomic coordinates and structure factors of the ternary | This study | PDB: 7QQO |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #2 DNA substrate |
This study | PDB: 7QR7 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #3 DNA substrate |
This study | PDB: 7QQP |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #4 DNA substrate |
This study | PDB: 7QQQ |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #5 DNA substrate |
This study | PDB: 7QQR |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF on-target DNA substrate |
This study | PDB: 7QQS |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #1 DNA substrate |
This study | PDB: 7QQT |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #2 DNA substrate |
This study | PDB: 7QQU |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #3 DNA substrate |
This study | PDB: 7QQV |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #4 DNA substrate |
This study | PDB: 7QQW |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #5 DNA substrate |
This study | PDB: 7QQX |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #6 DNA substrate |
This study | PDB: 7QR5 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #7 DNA substrate |
This study | PDB: 7QQZ |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the PTPRC off-target #1 DNA substrate |
This study | PDB: 7QR8 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the TRAC on-target DNA substrate |
Donohoue et al., 2021 | PDB: 7OX8 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the TRAC off-target #1 DNA substrate |
This study | PDB: 7QR0 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the TRAC off-target #2 DNA substrate |
This study | PDB: 7QR1 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the CD34 off-target #9 DNA substrate |
This study | PDB: 7ZO1 |
| Raw SITE-Seq data | This study | BioProject ID: PRJNA862989 |
| Raw SITE-Seq data – TRAC target | Donohoue et al., 2021 | BioProject ID: PRJNA744493 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli Rosetta 2 (DE3) | Novagen | Cat# 71400–3 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Transcription templates, cleavage substrates, crystallization substrates | This study | Table S9 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pMJ806 (WT SpCas9) | AddGene | #39312 |
| pMJ841 (SpCas9 D10A, H840A) | AddGene | #39318 |
|
| ||
| Software and algorithms | ||
|
| ||
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.0 Schrö dinger, LLC | https://www.pymol.org/ |
| DynDom | Poornam et al., 2009 | http://fizz.cmp.uea.ac.uk/dyndom/ |
| PDBePISA | Krissinel and Henrick, 2007 | http://www.ebi.ac.uk/pdbe/pisa/ |
| ChimeraX | Pettersen et al., 2021 | https://www.rbvi.ucsf.edu/chimerax/ |
| GraphPad Prism 9 | GraphPad Prism 9 for Windows, Version 9.1.2. | https://www.graphpad.com |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
Experimental model and subject details
The E. coli strains Rosetta 2 (DE3) was used for recombinant protein expression for structural studies and biochemical in vitro experiments. T cells used for cellular editing experiments were isolated from human primary blood mononuclear cells were purchased from STEMCELL Technologies.
Supplementary Material
Figure S1. Off-target profiling of selected genomic sites using SITE-Seq.
(A) Selected genomic targets and the corresponding guide RNA sequences selected for the SITE-Seq assay off-target profiling. Heatmap indicates frequency of nucleotide identity across each position for the selected targets. (B) SITE-seq assay analysis for RNPs assembled with indicated crRNAs. The numbers of detected off-target sites are shown as a function of RNP concentration. Checked boxes indicate recovery of the on-target site. n=3 replicates per sample. (C) Number of off-target sites recovered by the SITE-Seq assay are shown as a function of the number of mismatches between the guide RNA and the off-target sequence. (D) Frequency of nucleotide mismatches at each guide RNA–off-target DNA heteroduplex position for all off-target sites identified in (B). (E) Number of total identified mismatches per heteroduplex position.
Figure S2. in vitro cleavage of a subset of Cas9 off-target substrates.
(A) In vitro clevage kinetics of fully double stranded on- and off-target synthetic DNA substrates labelled on the target-strand for each guide RNA used in the study. Black triangles in the substrate schematic (top) indicate position of cleavage sites. Each data point represents a mean of four independent replicates. Error bars represent standard deviation for each time point. (B) In vitro clevage kinetics of partially single stranded (PAMmer) on- and off-target substrates. (C) Heatmap representation of mutual correlations between measured kinetic and thermodymamic parameters including cleavage (kobs), substrate DNA binding (kon), substrate dissociation (koff) rate constants, equilibrium dissociation constant (Kd) with numbers of nucleotide mismatches in the off- target sites (total and within seed), the GC content of the spacer (%GC) and cleavage rate predicted using the NucleaSeq algorithm (NucleaSeq kobs). The values were calculated across all off-targets for both dsDNA (lower left half, in blue), and partially single stranded (PAMmer) substrates (upper right half, in red). ns, no significant correlation. (D) Correlation between measured and NucleaSeq-predicted kobs rate constants. Off-target sites with significant deviations are highlighted in yellow.
Figure S3. In vivo editing at on- and off-target sites.
(A) Editing activity of crystalized on- and off-targets in primary human T cells. Matching bases in off-targets are denoted by a dot, whereas base pair mismatches and deletions (–) are highlighted. Asterisk indicated single-nucleotide polymorphism in PBMC donor sequence different from hg38 genomic reference. Editing rates are from highest Cas9 concentration shown in (B), n=3 replicates per sample, limit of detection = 0.1%. (B) Dose titration of Cas9 in human T cells for selected on and off-target sites. Off-target numbering correspond to sample shown in (A), and/or reference number (“ref”) as indicated in Table S1. Cas9 concentration shown below x-axis (1:3 molar ratio of Cas9 to guides), N=3 replicates per sample.
Figure S4. Alternative REC2 conformation in AAVS1 on-target complex.
(A) Overlay of REC2 domain conformations in the AAVS1 (pink), FANCF (purple) and TRAC (light blue) on-target complexes (B) Close-up view of helix REC2 helix spanning Cas9 residues 174–180. Linear and angular displacements of the helix in the AAVS1 on-target complex relative to the FANCF and TRAC on-target complexes are indicated.
Figure S5. PAM-distal mismatches result in unpairing and disordering of guide RNA nucleobase in position 1.
Close-up views of the PAM-distal end of the guide RNA-TS heteroduplex in (A) AAVS1 off-target #2, (B) AAVS1 off-target #4, (C) FANCF off-target #5 and (D) TRAC off-target #1 complexes. Arrowheads indicate nucleotides with disordered bases. Refined 2mFo−DFc electron density maps of the heteroduplexes are rendered as a grey mesh and contoured at 1.2σ for (A) and 1.0σ for (B)-(D).
Figure S6. Cas9-nucleic acid interactions in on-target complexes.
Schematic diagram depicting Cas9 residues interacting with the guide RNA-target DNA heteroduplex. Dotted lines represent hydrogen bonding interactions; dashed lines represent salt bridges; solid lines represent stacking/hydrophobic interactions. Target strand is coloured in blue, guide RNA in orange. Phosphates are represented by circles, ribose moieties by pentagons, and nucleobases by rectangles.
Figure S7. Wobble base pairing of rG-dT mismatches.
Close-up views of rG-dT mismatches at (A) heteroduplex position 15 in the TRAC off-target #1 complex, (B) position 13 in FANCF off-target #7 complex, (C) position 13 in FANCF off-target #2 complex, (D) position 4 in AAVS1 off-target #5 complex, (E) position 4 in AAVS1 off-target #1 complex, (F) position 2 in TRAC off-target #1 complex and (G) position 2 in TRAC off-target #2 complex. Corresponding on-target Watson-Crick base pairs are shown in white. Monovalent ions, modeled as K+, are depicted as purple spheres. In (A)-(E), the dT base is displaced into the major groove and forms a canonical wobble base pair with the rG base. In (F)-(G), the the rG base instead shifts towards the minor groove to facilitate wobble pairing.
Figure S8. rU-dG wobble base pairs adopt duplex position-specific conformations.
Close-up views of of rU-dG mismatches at (A) heteroduplex position 10 in FANCF off-target #4 complex, (B) position 5 in FANCF off-target #1 complex, (C) position 5 in FANCF off-target #3 complex, (D) position 5 in FANCF off-target #6 complex, (E) TRAC off-target #1 complex, and (F) AAVS1 off-target #2, and (G) AAVS1 off-target #1 complex. Corresponding on-target Watson-Crick base pairs are shown in white. Bound potassium ions are depicted as purple spheres. In (A), rU-dG wobble base pairing is achieved by minor groove displacement of the guanine base. In (B)-(D), the rU-dG mismatches adopt atypical conformations. In (E), the guanine base is shifted into the minor groove to form a wobble base pair, whereas at the identical heteroduplex position in (F) and (G), the rU-dG base pairs do not engage in wobble pairing, instead adopting alternative tautomeric forms. (H) Schematic depicting hydrogen bonding interactions between rU and dG bases in (F) and (G).
Figure S9. Non-canonical pyrimidine-purine base pairs within Cas9 off-target complexes.
(A) Close-up view of rA-dC wobble base pairing at position 4 in FANCF off-target #3 complex. The base pair geometry is consistent with base protonation or tautomerism to enable productive hydrogen bonding between the bases. (B) Close-up view of rC-dA mismatch at position 11 of FANCF off-target #7 complex, facilitated by base tilting at positions 11 and 12. (C) Close-up view of partially paired rC-dA mismatch at position 8 in AAVS1 off-target #1 complex. (D) Close-up view of rA-dC mismatch at position 10 in AAVS1 off-target #5 complex. (E) Close-up view of Hoogsteen-edge rA-dG base pair at position 7 in AAVS1 off-target #2 complex. Corresponding on-target Watson-Crick base pairs are shown in white.
Video S1. Morph of REC2 domain between AAVS1 on-target and FANCF on-target structures, Related to Figure S4
Video S2. Morph between FANCF off-target #4 and FANCF on-target showing conformational changes in the REC2, REC3, and HNH domains, Related to Figure S20
Table S1. SITE-Seq assay analysis of Cas9 off-target sites for 12 selected genomic targets. Related to Figure S1
Columns indicate the recovered off-target sequence; motif location; number of substitutions in recovered target sequence compared to the on-target (substitutions); strand designation of PAM; the lowest recovery concentration of each target; and whether the off-target is predicted to contain inserts or deletions based. Off-target sites recovered at lower concentrations were also recovered at higher concentrations (e.g., all 4 nM sites were also recovered at 16 nM, 64 nM, and 256 nM).
Table S2. List of recovered off-target sites by SITE-Seq. Related to Figure S1 and S191
Off-target alignments classified by genomic target and by the presence of insertions, deletions or purely mismatched targets. Indexes correspond to off-target sequence numbering in Table S1.
Table S3. Cellular editing data, Related to Figure S3
Table S4. Crystallographic data collection and refinement statistics of Cas9 on-target and off-target complexes, Related to STAR methods
Table S5. Summary of non-canonical base pairing and nucleobase distortions observed in structural data, Related to Figure 1
Data S1. SwitchSENSE association and dissociation curves, Related to Table 1
Acknowledgements
We thank members of the Jinek laboratory and Caribou Biosciences, Inc. for discussion and critical reading of the manuscript. We thank Christelle Chanez and Michael Schmitz for their assistance with preparing reagents. We thank Kenny B. Jungfer for assistance with crystallographic data collection. We thank John Hawkins and Ilya Finkelstein for providing access to the NucleaSEQ prediction pipeline. We thank Vincent Olieric, Meitian Wang, and Takashi Tomizaki (Swiss Light Source, Paul Scherrer Institute) for assistance with crystallographic data collection, and Nena Matscheko from Dynamic Biosensors for support with switchSENSE experiments. We thank the NCCR RNA and Disease for providing infrastructural support. This work was supported by the Swiss National Science Foundation Grant 31003A_182567 (to M.J.), the National Institutes of Health (Grant No. R01GM141329 to G.P.) and National Science Foundation (Grant No. CHE-1905374 to G.P.). M.J. is an International Research Scholar of the Howard Hughes Medical Institute and Vallee Scholar of the Bert L & N Kuggie Vallee Foundation.
Footnotes
Conflict of interest statement
P.D.D. is a current employee of Caribou Biosciences, Inc., and C-H.L., M.J.I, and P.C. are former employees of Caribou Biosciences, Inc. M.J. is a co-founder of Caribou Biosciences, Inc. M.J., M.J.I., P.C., and P.D.D. are named inventors on patents and patent applications related to CRISPR-Cas technologies. SITE-Seq is a registered trademark of Caribou Biosciences, Inc.
References
- Abe N, Dror I, Yang L, Slattery M, Zhou T, Bussemaker HJ, Rohs R, and Mann RS (2015). Deconvolving the recognition of DNA shape from sequence. Cell 161, 307–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afek A, Shi H, Rangadurai A, Sahay H, Senitzki A, Xhani S, Fang M, Salinas R, Mielko Z, Pufall MA, et al. (2020). DNA mismatches reveal conformational penalties in protein-DNA recognition. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders C, Niewoehner O, Duerst A, and Jinek M. (2014). Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature 513, 569–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzalone AV, Koblan LW, and Liu DR (2020). Genome editing with CRISPR-Cas nucleases, base editors, transposases and prime editors. Nat Biotechnol 38, 824–844. [DOI] [PubMed] [Google Scholar]
- Bae S, Park J, and Kim JS (2014). Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 30, 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banas P, Hollas D, Zgarbova M, Jurecka P, Orozco M, Cheatham TE 3rd, Sponer J, and Otyepka M. (2010). Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins. J Chem Theory Comput 6, 3836–3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendsen HJC, Postma JPM, Gunsteren W.F.v., DiNola A, and Haak JR (1984). Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics 81, 3684–3690. [Google Scholar]
- Boyle EA, Andreasson JOL, Chircus LM, Sternberg SH, Wu MJ, Guegler CK, Doudna JA, and Greenleaf WJ (2017). High-throughput biochemical profiling reveals sequence determinants of dCas9 off-target binding and unbinding. Proc Natl Acad Sci U S A 114, 5461–5466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Becker WR, Bai HB, Chen JS, Doudna JA, and Greenleaf WJ (2021). Quantification of Cas9 binding and cleavage across diverse guide sequences maps landscapes of target engagement. Sci Adv 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo JPK, Liu MS, Hibshman GN, Dangerfield TL, Jung K, McCool RS, Johnson KA, and Taylor DW (2022). Structural basis for mismatch surveillance by CRISPR-Cas9. Nature 603, 343–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron P, Fuller CK, Donohoue PD, Jones BN, Thompson MS, Carter MM, Gradia S, Vidal B, Garner E, Slorach EM, et al. (2017). Mapping the genomic landscape of CRISPR-Cas9 cleavage. Nat Methods 14, 600–606. [DOI] [PubMed] [Google Scholar]
- Chen JS, Dagdas YS, Kleinstiver BP, Welch MM, Sousa AA, Harrington LB, Sternberg SH, Joung JK, Yildiz A, and Doudna JA (2017). Enhanced proofreading governs CRISPR-Cas9 targeting accuracy. Nature 550, 407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cofsky JC, Soczek KM, Knott GJ, Nogales E, and Doudna JA (2022). CRISPR-Cas9 bends and twists DNA to read its sequence. Nat Struct Mol Biol 29, 395–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromwell CR, Sung K, Park J, Krysler AR, Jovel J, Kim SK, and Hubbard BP (2018). Incorporation of bridged nucleic acids into CRISPR RNAs improves Cas9 endonuclease specificity. Nat Commun 9, 1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Case DA, M.A. H, Belfon K, Ben-Shalom IY, Brozell SR, Cerutti DS, Cheatham TE III, Cisneros GA, Cruzeiro VWD, Darden TA, Duke RE, Giambasu G, Gilson MK, Gohlke H, Goetz AW, Harris R, Izadi S, Izmailov SA, Jin C, Kasavajhala K, Kaymak MC, King E, Kovalenko A, Kurtzman T, Lee TS, LeGrand S, Li P, Lin C, Liu J, Luchko T, Luo R, Machado M, Man V, Manathunga M, Merz KM, Miao Y, Mikhailovskii O, Monard G, Nguyen H, O’Hearn KA, Onufriev A, Pan F, Pantano S, Qi R, Rahnamoun A, Roe DR, Roitberg A, Sagui C, Schott-Verdugo S, Shen J, Simmerling CL, Skrynnikov NR, Smith J, Swails J, Walker RC, Wang J, Wei H, Wolf RM, Wu X, Xue Y, York DM, Zhao S, and Kollman PA (2021). Amber 2021. University of California, San Francisco. [Google Scholar]
- Dagdas YS, Chen JS, Sternberg SH, Doudna JA, and Yildiz A. (2017). A conformational checkpoint between DNA binding and cleavage by CRISPR-Cas9. Sci Adv 3, eaao0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, and Charpentier E. (2011). CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471, 602–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deveau H, Barrangou R, Garneau JE, Labonte J, Fremaux C, Boyaval P, Romero DA, Horvath P, and Moineau S. (2008). Phage response to CRISPR-encoded resistance in Streptococcus thermophilus. J Bacteriol 190, 1390–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench JG, Fusi N, Sullender M, Hegde M, Vaimberg EW, Donovan KF, Smith I, Tothova Z, Wilen C, Orchard R, et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohoue PD, Pacesa M, Lau E, Vidal B, Irby MJ, Nyer DB, Rotstein T, Banh L, Toh MS, Gibson J, et al. (2021). Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells. Mol Cell 81, 3637–3649 e3635. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, and Cowtan K. (2010). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu BXH, Smith JD, Fuchs RT, Mabuchi M, Curcuru J, Robb GB, and Fire AZ (2019). Target-dependent nickase activities of the CRISPR-Cas nucleases Cpf1 and Cas9. Nat Microbiol 4, 888–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sander JD, Reyon D, Cascio VM, and Joung JK (2014). Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32, 279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg A, and Heinemann U. (2018). A novel form of RNA double helix based on G.U and C.A(+) wobble base pairing. RNA 24, 209–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Yu HH, Johnson KA, and Taylor DW (2018). DNA Unwinding Is the Primary Determinant of CRISPR-Cas9 Activity. Cell Rep 22, 359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ren K, Zhu Y, Tang Z, Wang Y, Zhang B, and Huang Z. (2019). Structural insights into a high fidelity variant of SpCas9. Cell Res 29, 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendel A, Bak RO, Clark JT, Kennedy AB, Ryan DE, Roy S, Steinfeld I, Lunstad BD, Kaiser RJ, Wilkens AB, et al. (2015). Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol 33, 985–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivani I, Dans PD, Noy A, Perez A, Faustino I, Hospital A, Walther J, Andrio P, Goni R, Balaceanu A, et al. (2016). Parmbsc1: a refined force field for DNA simulations. Nat Methods 13, 55–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IE, Wright AV, Cofsky JC, Aris KDP, Doudna JA, and Bryant Z. (2020). Cas9 interrogates DNA in discrete steps modulated by mismatches and supercoiling. Proc Natl Acad Sci U S A 117, 5853–5860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Taylor DW, Chen JS, Kornfeld JE, Zhou K, Thompson AJ, Nogales E, and Doudna JA (2016). Structures of a CRISPR-Cas9 R-loop complex primed for DNA cleavage. Science 351, 867–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang F, Zhou K, Ma L, Gressel S, and Doudna JA (2015). STRUCTURAL BIOLOGY. A Cas9-guide RNA complex preorganized for target DNA recognition. Science 348, 1477–1481. [DOI] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, and Charpentier E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SK Jr., Hawkins JA, Johnson NV, Jung C, Hu K, Rybarski JR, Chen JS, Doudna JA, Press WH, and Finkelstein IJ (2021). Massively parallel kinetic profiling of natural and engineered CRISPR nucleases. Nat Biotechnol 39, 84–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics 79, 926–935. [Google Scholar]
- Kabsch W. (2010). Xds. Acta Crystallogr D Biol Crystallogr 66, 125–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey IJ, Petzold K, Sathyamoorthy B, Stein ZW, and Al-Hashimi HM (2015). Visualizing transient Watson-Crick-like mispairs in DNA and RNA duplexes. Nature 519, 315–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimsey IJ, Szymanski ES, Zahurancik WJ, Shakya A, Xue Y, Chu CC, Sathyamoorthy B, Suo Z, and Al-Hashimi HM (2018). Dynamic basis for dG*dT misincorporation via tautomerization and ionization. Nature 554, 195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayner M, Rozenberg H, Rohs R, Suad O, Rabinovich D, Honig B, and Shakked Z. (2010). Diversity in DNA recognition by p53 revealed by crystal structures with Hoogsteen base pairs. Nat Struct Mol Biol 17, 423–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K. (2007). Inference of macromolecular assemblies from crystalline state. J Mol Biol 372, 774–797. [DOI] [PubMed] [Google Scholar]
- Kulcsar PI, Talas A, Toth E, Nyeste A, Ligeti Z, Welker Z, and Welker E. (2020). Blackjack mutations improve the on-target activities of increased fidelity variants of SpCas9 with 5’G-extended sgRNAs. Nat Commun 11, 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, and Bebenek K. (2000). DNA replication fidelity. Annu Rev Biochem 69, 497–529. [DOI] [PubMed] [Google Scholar]
- Kuscu C, Arslan S, Singh R, Thorpe J, and Adli M. (2014). Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 32, 677–683. [DOI] [PubMed] [Google Scholar]
- Lavery R, Moakher M, Maddocks JH, Petkeviciute D, and Zakrzewska K. (2009). Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res 37, 5917–5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazzarotto CR, Malinin NL, Li Y, Zhang R, Yang Y, Lee G, Cowley E, He Y, Lan X, Jividen K, et al. (2020). CHANGE-seq reveals genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, and Westhof E. (2002). The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res 30, 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, and Merz KM (2014). Taking into Account the Ion-Induced Dipole Interaction in the Nonbonded Model of Ions. Journal of Chemical Theory and Computation 10, 289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Olson WK, and Lu XJ (2019). Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res 47, W26–W34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, Deshmukh H, Ranjan P, Sarode N, Wile BM, Vertino PM, Stewart FJ, and Bao G. (2014). CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res 42, 7473–7485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, and Simmerling C. (2015). ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J Chem Theory Comput 11, 3696–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, Charpentier E, Cheng D, Haft DH, Horvath P, et al. (2020). Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 18, 67–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekler V, Minakhin L, and Severinov K. (2017). Mechanism of duplex DNA destabilization by RNA-guided Cas9 nuclease during target interrogation. Proc Natl Acad Sci U S A 114, 5443–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell BP, Hsu RV, Medrano MA, Zewde NT, Narkhede YB, and Palermo G. (2020). Spontaneous Embedding of DNA Mismatches Within the RNA:DNA Hybrid of CRISPR-Cas9. Frontiers in Molecular Biosciences 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullally G, van Aelst K, Naqvi MM, Diffin FM, Karvelis T, Gasiunas G, Siksnys V, and Szczelkun MD (2020). 5’ modifications to CRISPR-Cas9 gRNA can change the dynamics and size of R-loops and inhibit DNA cleavage. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murugan K, Seetharam AS, Severin AJ, and Sashital DG (2020). CRISPR-Cas12a has widespread off-target and dsDNA-nicking effects. J Biol Chem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimasu H, Ran FA, Hsu PD, Konermann S, Shehata SI, Dohmae N, Ishitani R, Zhang F, and Nureki O. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell MR, Oakes BL, Sternberg SH, East-Seletsky A, Kaplan M, and Doudna JA (2014). Programmable RNA recognition and cleavage by CRISPR/Cas9. Nature 516, 263–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Geen H, Henry IM, Bhakta MS, Meckler JF, and Segal DJ (2015). A genome-wide analysis of Cas9 binding specificity using ChIP-seq and targeted sequence capture. Nucleic Acids Res 43, 3389–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly D, Kartje ZJ, Ageely EA, Malek-Adamian E, Habibian M, Schofield A, Barkau CL, Rohilla KJ, DeRossett LB, Weigle AT, et al. (2019). Extensive CRISPR RNA modification reveals chemical compatibility and structure-activity relationships for Cas9 biochemical activity. Nucleic Acids Res 47, 546–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okafor IC, Singh D, Wang Y, Jung M, Wang H, Mallon J, Bailey S, Lee JK, and Ha T. (2019). Single molecule analysis of effects of non-canonical guide RNAs and specificity-enhancing mutations on Cas9-induced DNA unwinding. Nucleic Acids Res 47, 11880–11888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacesa M, and Jinek M. (2021). Mechanism of R-loop formation and conformational activation of Cas9. bioRxiv, 2021.2009.2016.460614. [Google Scholar]
- Pacesa M, Loeff L, Querques I, Muckenfuss LM, Sawicka M, and Jinek M. (2022). Mechanism of R-loop formation and conformational activation of Cas9. bioRxiv, 2021.2009.2016.460614. [Google Scholar]
- Palermo G, Chen JS, Ricci CG, Rivalta I, Jinek M, Batista VS, Doudna JA, and McCammon JA (2018). Key role of the REC lobe during CRISPR-Cas9 activation by ‘sensing’, ‘regulating’, and ‘locking’ the catalytic HNH domain. Q Rev Biophys 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, and Liu DR (2013). High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nat Biotechnol 31, 839–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021). UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30, 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricci CG, Chen JS, Miao Y, Jinek M, Doudna JA, McCammon JA, and Palermo G. (2019). Deciphering Off-Target Effects in CRISPR-Cas9 through Accelerated Molecular Dynamics. ACS Cent Sci 5, 651–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodnina MV, and Wintermeyer W. (2001). Fidelity of aminoacyl-tRNA selection on the ribosome: kinetic and structural mechanisms. Annu Rev Biochem 70, 415–435. [DOI] [PubMed] [Google Scholar]
- Rohs R, West SM, Liu P, and Honig B. (2009a). Nuance in the double-helix and its role in protein-DNA recognition. Curr Opin Struct Biol 19, 171–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohs R, West SM, Sosinsky A, Liu P, Mann RS, and Honig B. (2009b). The role of DNA shape in protein-DNA recognition. Nature 461, 1248–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon-Ferrer R, Gotz AW, Poole D, Le Grand S, and Walker RC (2013). Routine Microsecond Molecular Dynamics Simulations with AMBER on GPUs. 2. Explicit Solvent Particle Mesh Ewald. J Chem Theory Comput 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, and Severinov K. (2011). Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A 108, 10098–10103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh D, Sternberg SH, Fei J, Doudna JA, and Ha T. (2016). Real-time observation of DNA recognition and rejection by the RNA-guided endonuclease Cas9. Nat Commun 7, 12778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmer M, Thumberger T, Del Sol Keyer M, Wittbrodt J, and Mateo JL (2015). CCTop: An Intuitive, Flexible and Reliable CRISPR/Cas9 Target Prediction Tool. PLoS One 10, e0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, LaFrance B, Kaplan M, and Doudna JA (2015). Conformational control of DNA target cleavage by CRISPR-Cas9. Nature 527, 110–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg SH, Redding S, Jinek M, Greene EC, and Doudna JA (2014). DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 507, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timsit Y. (1999). DNA structure and polymerase fidelity. J Mol Biol 293, 835–853. [DOI] [PubMed] [Google Scholar]
- Tsai SQ, Nguyen NT, Malagon-Lopez J, Topkar VV, Aryee MJ, and Joung JK (2017). CIRCLE-seq: a highly sensitive in vitro screen for genome-wide CRISPR-Cas9 nuclease off-targets. Nat Methods 14, 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SQ, Zheng Z, Nguyen NT, Liebers M, Topkar VV, Thapar V, Wyvekens N, Khayter C, Iafrate AJ, Le LP, et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol 33, 187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turq P, Lantelme F, and Friedman HL (1977). Brownian dynamics: Its application to ionic solutions. The Journal of Chemical Physics 66, 3039–3044. [Google Scholar]
- van Houte S, Ekroth AK, Broniewski JM, Chabas H, Ashby B, Bondy-Denomy J, Gandon S, Boots M, Paterson S, Buckling A, et al. (2016). The diversity-generating benefits of a prokaryotic adaptive immune system. Nature 532, 385–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot M, Houkes J, Lochs SJA, Swarts DC, Zheng P, Kunne T, Mohanraju P, Anders C, Jinek M, van der Oost J, et al. (2018). Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes. Nucleic Acids Res 46, 873–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Hellinga HW, and Beese LS (2011). Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci U S A 108, 17644–17648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S, et al. (2014). Genome-wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32, 670–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yakovchuk P, Protozanova E, and Frank-Kamenetskii MD (2006). Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res 34, 564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Peng S, Sun R, Lin J, Wang N, and Chen C. (2018). The Conformational Dynamics of Cas9 Governing DNA Cleavage Are Revealed by Single-Molecule FRET. Cell Rep 22, 372–382. [DOI] [PubMed] [Google Scholar]
- Yaung SJ, Esvelt KM, and Church GM (2014). CRISPR/Cas9-mediated phage resistance is not impeded by the DNA modifications of phage T4. PLoS One 9, e98811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Suresh S, Kwan SY, Wu Q, Walsh S, Ding J, Bogorad RL, Zhu LJ, Wolfe SA, et al. (2018). Partial DNA-guided Cas9 enables genome editing with reduced off-target activity. Nat Chem Biol 14, 311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H, Song CQ, Suresh S, Wu Q, Walsh S, Rhym LH, Mintzer E, Bolukbasi MF, Zhu LJ, Kauffman K, et al. (2017). Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nat Biotechnol 35, 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Cui Y, Zhang Y, Zhang Y, Liang M, Chen H, Lan J, Song G, and Lou J. (2018). The initiation, propagation and dynamics of CRISPR-SpyCas9 R-loop complex. Nucleic Acids Res 46, 350–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE 3rd, and Jurecka P. (2011). Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J Chem Theory Comput 7, 2886–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Rube HT, Vakulskas CA, Behlke MA, Bussemaker HJ, and Pufall MA (2020). Systematic in vitro profiling of off-target affinity, cleavage and efficiency for CRISPR enzymes. Nucleic Acids Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Clarke R, Puppala AK, Chittori S, Merk A, Merrill BJ, Simonovic M, and Subramaniam S. (2019). Cryo-EM structures reveal coordinated domain motions that govern DNA cleavage by Cas9. Nat Struct Mol Biol 26, 679–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Off-target profiling of selected genomic sites using SITE-Seq.
(A) Selected genomic targets and the corresponding guide RNA sequences selected for the SITE-Seq assay off-target profiling. Heatmap indicates frequency of nucleotide identity across each position for the selected targets. (B) SITE-seq assay analysis for RNPs assembled with indicated crRNAs. The numbers of detected off-target sites are shown as a function of RNP concentration. Checked boxes indicate recovery of the on-target site. n=3 replicates per sample. (C) Number of off-target sites recovered by the SITE-Seq assay are shown as a function of the number of mismatches between the guide RNA and the off-target sequence. (D) Frequency of nucleotide mismatches at each guide RNA–off-target DNA heteroduplex position for all off-target sites identified in (B). (E) Number of total identified mismatches per heteroduplex position.
Figure S2. in vitro cleavage of a subset of Cas9 off-target substrates.
(A) In vitro clevage kinetics of fully double stranded on- and off-target synthetic DNA substrates labelled on the target-strand for each guide RNA used in the study. Black triangles in the substrate schematic (top) indicate position of cleavage sites. Each data point represents a mean of four independent replicates. Error bars represent standard deviation for each time point. (B) In vitro clevage kinetics of partially single stranded (PAMmer) on- and off-target substrates. (C) Heatmap representation of mutual correlations between measured kinetic and thermodymamic parameters including cleavage (kobs), substrate DNA binding (kon), substrate dissociation (koff) rate constants, equilibrium dissociation constant (Kd) with numbers of nucleotide mismatches in the off- target sites (total and within seed), the GC content of the spacer (%GC) and cleavage rate predicted using the NucleaSeq algorithm (NucleaSeq kobs). The values were calculated across all off-targets for both dsDNA (lower left half, in blue), and partially single stranded (PAMmer) substrates (upper right half, in red). ns, no significant correlation. (D) Correlation between measured and NucleaSeq-predicted kobs rate constants. Off-target sites with significant deviations are highlighted in yellow.
Figure S3. In vivo editing at on- and off-target sites.
(A) Editing activity of crystalized on- and off-targets in primary human T cells. Matching bases in off-targets are denoted by a dot, whereas base pair mismatches and deletions (–) are highlighted. Asterisk indicated single-nucleotide polymorphism in PBMC donor sequence different from hg38 genomic reference. Editing rates are from highest Cas9 concentration shown in (B), n=3 replicates per sample, limit of detection = 0.1%. (B) Dose titration of Cas9 in human T cells for selected on and off-target sites. Off-target numbering correspond to sample shown in (A), and/or reference number (“ref”) as indicated in Table S1. Cas9 concentration shown below x-axis (1:3 molar ratio of Cas9 to guides), N=3 replicates per sample.
Figure S4. Alternative REC2 conformation in AAVS1 on-target complex.
(A) Overlay of REC2 domain conformations in the AAVS1 (pink), FANCF (purple) and TRAC (light blue) on-target complexes (B) Close-up view of helix REC2 helix spanning Cas9 residues 174–180. Linear and angular displacements of the helix in the AAVS1 on-target complex relative to the FANCF and TRAC on-target complexes are indicated.
Figure S5. PAM-distal mismatches result in unpairing and disordering of guide RNA nucleobase in position 1.
Close-up views of the PAM-distal end of the guide RNA-TS heteroduplex in (A) AAVS1 off-target #2, (B) AAVS1 off-target #4, (C) FANCF off-target #5 and (D) TRAC off-target #1 complexes. Arrowheads indicate nucleotides with disordered bases. Refined 2mFo−DFc electron density maps of the heteroduplexes are rendered as a grey mesh and contoured at 1.2σ for (A) and 1.0σ for (B)-(D).
Figure S6. Cas9-nucleic acid interactions in on-target complexes.
Schematic diagram depicting Cas9 residues interacting with the guide RNA-target DNA heteroduplex. Dotted lines represent hydrogen bonding interactions; dashed lines represent salt bridges; solid lines represent stacking/hydrophobic interactions. Target strand is coloured in blue, guide RNA in orange. Phosphates are represented by circles, ribose moieties by pentagons, and nucleobases by rectangles.
Figure S7. Wobble base pairing of rG-dT mismatches.
Close-up views of rG-dT mismatches at (A) heteroduplex position 15 in the TRAC off-target #1 complex, (B) position 13 in FANCF off-target #7 complex, (C) position 13 in FANCF off-target #2 complex, (D) position 4 in AAVS1 off-target #5 complex, (E) position 4 in AAVS1 off-target #1 complex, (F) position 2 in TRAC off-target #1 complex and (G) position 2 in TRAC off-target #2 complex. Corresponding on-target Watson-Crick base pairs are shown in white. Monovalent ions, modeled as K+, are depicted as purple spheres. In (A)-(E), the dT base is displaced into the major groove and forms a canonical wobble base pair with the rG base. In (F)-(G), the the rG base instead shifts towards the minor groove to facilitate wobble pairing.
Figure S8. rU-dG wobble base pairs adopt duplex position-specific conformations.
Close-up views of of rU-dG mismatches at (A) heteroduplex position 10 in FANCF off-target #4 complex, (B) position 5 in FANCF off-target #1 complex, (C) position 5 in FANCF off-target #3 complex, (D) position 5 in FANCF off-target #6 complex, (E) TRAC off-target #1 complex, and (F) AAVS1 off-target #2, and (G) AAVS1 off-target #1 complex. Corresponding on-target Watson-Crick base pairs are shown in white. Bound potassium ions are depicted as purple spheres. In (A), rU-dG wobble base pairing is achieved by minor groove displacement of the guanine base. In (B)-(D), the rU-dG mismatches adopt atypical conformations. In (E), the guanine base is shifted into the minor groove to form a wobble base pair, whereas at the identical heteroduplex position in (F) and (G), the rU-dG base pairs do not engage in wobble pairing, instead adopting alternative tautomeric forms. (H) Schematic depicting hydrogen bonding interactions between rU and dG bases in (F) and (G).
Figure S9. Non-canonical pyrimidine-purine base pairs within Cas9 off-target complexes.
(A) Close-up view of rA-dC wobble base pairing at position 4 in FANCF off-target #3 complex. The base pair geometry is consistent with base protonation or tautomerism to enable productive hydrogen bonding between the bases. (B) Close-up view of rC-dA mismatch at position 11 of FANCF off-target #7 complex, facilitated by base tilting at positions 11 and 12. (C) Close-up view of partially paired rC-dA mismatch at position 8 in AAVS1 off-target #1 complex. (D) Close-up view of rA-dC mismatch at position 10 in AAVS1 off-target #5 complex. (E) Close-up view of Hoogsteen-edge rA-dG base pair at position 7 in AAVS1 off-target #2 complex. Corresponding on-target Watson-Crick base pairs are shown in white.
Video S1. Morph of REC2 domain between AAVS1 on-target and FANCF on-target structures, Related to Figure S4
Video S2. Morph between FANCF off-target #4 and FANCF on-target showing conformational changes in the REC2, REC3, and HNH domains, Related to Figure S20
Table S1. SITE-Seq assay analysis of Cas9 off-target sites for 12 selected genomic targets. Related to Figure S1
Columns indicate the recovered off-target sequence; motif location; number of substitutions in recovered target sequence compared to the on-target (substitutions); strand designation of PAM; the lowest recovery concentration of each target; and whether the off-target is predicted to contain inserts or deletions based. Off-target sites recovered at lower concentrations were also recovered at higher concentrations (e.g., all 4 nM sites were also recovered at 16 nM, 64 nM, and 256 nM).
Table S2. List of recovered off-target sites by SITE-Seq. Related to Figure S1 and S191
Off-target alignments classified by genomic target and by the presence of insertions, deletions or purely mismatched targets. Indexes correspond to off-target sequence numbering in Table S1.
Table S3. Cellular editing data, Related to Figure S3
Table S4. Crystallographic data collection and refinement statistics of Cas9 on-target and off-target complexes, Related to STAR methods
Table S5. Summary of non-canonical base pairing and nucleobase distortions observed in structural data, Related to Figure 1
Data S1. SwitchSENSE association and dissociation curves, Related to Table 1
Data Availability Statement
Coordinates have been deposited to the Protein Data Bank under the PDB accession numbers: 7QQO, 7QR7, 7QQP, 7QQQ, 7QQR, 7QQS, 7QQT, 7QQU, 7QQV, 7QQW, 7QQX, 7QR5, 7QQZ, 7QR8, 7QR0, 7QR1 and 7ZO1. Raw sequencing files for the SITE-Seq assay have been deposited to BioProject. Accession numbers are listed in the key resource table. All deposited data is publicly available as of the date of publication.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Chemicals, peptides, and recombinant proteins | ||
|
| ||
| Alt-R S.p. Cas9 Nuclease V3 | IDT | Cat# 1081058 |
| RoboSep Filter Tip Racks (8) | StemCell Technologies | Cat# 20125 |
| Lonza Nucleofection Kits (10 96-well plates and buffer) P3 | Lonza Walkersville, Inc. | Cat# V4SP-3960 |
| Recombinant Human IL-2 | PeproTech, Inc | Cat# 200–02 |
| QuickExtract DNA Extraction Solution (10 mL Aliquots) | Lucigen | Cat# QE09050-PQ1329 |
| RoboSep Buffer (5X Concentrate) | StemCell Technologies | Cat# 20124 |
| RoboSep Kit # 17951 (Human T cell Isolation Kit) | StemCell Technologies | Cat# 17951 |
| Dynabeads Human T-Activator CD3/CD28 (5 × 2 mL) | Fisher Scientific Co LLC | Cat# 11132D |
| DNAse I Solution (1 mg/mL, 1 mL) | StemCell Technologies | Cat# 7900 |
| Antibiotic/Antmycotic (6 × 100 mL) | Fisher Scientific Co LLC | Cat# MT30004CI |
| CTS Immune Cell Serum Replacement (50 mL) | ThermoFisher Scientific | Cat# A2596101 |
| Immunocult-XF T Cell Expansion Media | StemCell Technologies | Cat# 10981 |
| SPRIselect Reagent Kit | Beckman Coulter | Cat# B233318 |
| Dynabeads M-280 Streptavidin | Invitrogen | Cat# 11206D |
| Q5^ Hot Start High-Fidelty 2x Master Mix | New England Biolabs | Cat# M0494L |
| NEBNext double-stranded DNA (dsDNA) Fragmentase | New England Biolabs | Cat# M0348L |
| T4 DNA Ligase | New England Biolabs | Cat# M0202S |
| 10x T4 DNA Ligase Reaction Buffer | New England Biolabs | Cat# B0202S |
| Proteinase K | ThermoFisher Scientific | Cat# EO0491 |
| HEPES | Roth | Cat# HN78.1 |
| Magnesium chloride | Fluka | Cat# 63064–500G |
| Potassium chloride | Roth | Cat# 6781.2 |
| Potassium thiocyanate | Roth | Cat# P753.1 |
| Poly(ethylene glycol) 3,350 | Sigma-Aldrich | Cat# 202444–500G |
| Ethylenediaminetetraacetic acid | Roth | Cat# 8040.3 |
| Ni Sepharose 6 Fast Flow resin (500 mL) | Cytiva | CAT# 17–5318-03 |
| HiPrep 26/10 desalting columns (pack of 4) | Cytiva | CAT# 17–5087-02 |
| 5 mL HiTrap SP HP | Cytiva | CAT# 17–1152-01 |
| HiLoad Superdex 200 26/600 | Cytiva | CAT# 28–9893-36 |
|
| ||
| Critical commercial assays | ||
|
| ||
| NextSeq 500/550 High Output V2.5 (150 cycles) | Illumina | Cat# 20024907 |
| MiSeq Reagent Kit v2 (300- cycles) | Illumina, Inc | Cat# MS-102– 2002 |
| NextSeq PhiX Control Kit | Illumina, Inc | Cat# FC-110– 3002 |
| Blood & Cell Culture DNA Maxi Kit | Qiagen | Cat# 13323 |
| High Sensitivity DNA Bioanalyzer Kit | Agilent | Cat# 5067–4626 |
| NEBNext^ dA-tailing Kit | NEB | Cat# E6053L |
| NEBNext^ Ultra End Repair / dA-tailing module | NEB | Cat# E7442L |
| NEBNext^ Ultra Ligation Kit | NEB | Cat# E7445L |
| NextSeq 500/550 High Output V2.5 (150 cycles) | Illumina | Cat# 20024907 |
|
| ||
| Deposited data | ||
|
| ||
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 on-target DNA substrate | Donohoue et al., 2021 | PDB: 7OX9 |
| Atomic coordinates and structure factors of the ternary | This study | PDB: 7QQO |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #2 DNA substrate |
This study | PDB: 7QR7 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #3 DNA substrate |
This study | PDB: 7QQP |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #4 DNA substrate |
This study | PDB: 7QQQ |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the AAVS1 off-target #5 DNA substrate |
This study | PDB: 7QQR |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF on-target DNA substrate |
This study | PDB: 7QQS |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #1 DNA substrate |
This study | PDB: 7QQT |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #2 DNA substrate |
This study | PDB: 7QQU |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #3 DNA substrate |
This study | PDB: 7QQV |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #4 DNA substrate |
This study | PDB: 7QQW |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #5 DNA substrate |
This study | PDB: 7QQX |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #6 DNA substrate |
This study | PDB: 7QR5 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the FANCF off-target #7 DNA substrate |
This study | PDB: 7QQZ |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the PTPRC off-target #1 DNA substrate |
This study | PDB: 7QR8 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the TRAC on-target DNA substrate |
Donohoue et al., 2021 | PDB: 7OX8 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the TRAC off-target #1 DNA substrate |
This study | PDB: 7QR0 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the TRAC off-target #2 DNA substrate |
This study | PDB: 7QR1 |
| Atomic coordinates and structure factors of the ternary SpCas9 complex bound to the CD34 off-target #9 DNA substrate |
This study | PDB: 7ZO1 |
| Raw SITE-Seq data | This study | BioProject ID: PRJNA862989 |
| Raw SITE-Seq data – TRAC target | Donohoue et al., 2021 | BioProject ID: PRJNA744493 |
|
| ||
| Bacterial and virus strains | ||
|
| ||
| E. coli Rosetta 2 (DE3) | Novagen | Cat# 71400–3 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Transcription templates, cleavage substrates, crystallization substrates | This study | Table S9 |
|
| ||
| Recombinant DNA | ||
|
| ||
| pMJ806 (WT SpCas9) | AddGene | #39312 |
| pMJ841 (SpCas9 D10A, H840A) | AddGene | #39318 |
|
| ||
| Software and algorithms | ||
|
| ||
| XDS | Kabsch, 2010 | http://xds.mpimf-heidelberg.mpg.de/ |
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| PyMOL | The PyMOL Molecular Graphics System, Version 2.0 Schrö dinger, LLC | https://www.pymol.org/ |
| DynDom | Poornam et al., 2009 | http://fizz.cmp.uea.ac.uk/dyndom/ |
| PDBePISA | Krissinel and Henrick, 2007 | http://www.ebi.ac.uk/pdbe/pisa/ |
| ChimeraX | Pettersen et al., 2021 | https://www.rbvi.ucsf.edu/chimerax/ |
| GraphPad Prism 9 | GraphPad Prism 9 for Windows, Version 9.1.2. | https://www.graphpad.com |
| MolProbity | Chen et al., 2010 | http://molprobity.biochem.duke.edu |
| COOT | Emsley et al., 2010 | https://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |


