Abstract
Native ion mobility-mass spectrometry (IM-MS) has emerged as an information-rich technique for gas phase protein structure characterization; however, IM resolution is currently insufficient for the detection of subtle structural differences in large biomolecules. This challenge has spurred the development of collision-induced unfolding (CIU) which utilizes incremental gas phase activation to unfold a protein in order to expand the number of measurable descriptors available for native protein ions. Although CIU is now routinely used in native mass spectrometry studies, the interlaboratory reproducibility of CIU has not been established. Here we evaluate the reproducibility of the CIU data produced across three laboratories (University of Michigan, Texas A&M University, and Vanderbilt University). CIU data were collected for a variety of protein ions ranging from 8.6-66 kDa. Within the same laboratory, the CIU fingerprints were found to be repeatable with root mean square deviation (RMSD) values of less than 5%. Collision cross section (CCS) values of the CIU intermediates were consistent across the laboratories, with most features exhibiting an interlaboratory reproducibility of better than 1%. In contrast, the activation potentials required to induce protein CIU transitions varied between the three laboratories. To address these differences, three source assemblies were constructed with an updated ion activation hardware design utilizing higher mechanical tolerance specifications. The production-grade assemblies were found to produce highly consistent CIU data for intact antibodies, exhibiting high precision ion CCS and CIU transition values, thus opening the door to establishing databases of CIU fingerprints to support future biomolecular classification efforts.
Native mass spectrometry (MS) has rapidly grown as a robust technique for making measurements of proteins and their complexes.1 Samples are prepared in aqueous, pH-adjusted electrolyte solutions of volatile salt (usually ammonium acetate) and ionized gently, preserving transient, non-covalent interactions from solution to the gas phase. Improvements in ionization2,3 and instrumentation4–11 have expanded the accessible mass range, enabling routine analysis of larger proteins such as intact antibodies,12–15 membrane protein complexes,16–22 protein chaperones,9,10,23–25 and complete viral particles.26. The coupling of ion mobility (IM) with native MS has spurred the field of native ion mobility-mass spectrometry (IM-MS) wherein ions are separated by size, shape, and charge prior to MS analysis. For uniform field drift tubes, ion arrival times can be converted to a rotationally averaged collision cross section (CCS) via the low-field IM relationship prescribed by the Mason-Schamp equation.27 Such CCS values can facilitate structural comparisons with other experimentally measured or otherwise estimated CCS values corresponding to available 3-dimensional structures.28–38
Previous drift tube IM-MS (DTIM-MS) studies have shown that CCS measurements can be obtained within 0.3% RSD for small and medium sized molecules,39 and within ~0.4% for larger native-like protein ions.9 While these results established the reproducibility of such IM measurements, in the context of protein structure, IM-MS alone remains unable to resolve many key conformational states critical for biomolecular function. At its core, the native IM-MS experiment provides two key descriptors for differentiating protein states: the mass-to-charge (m/z) ratio of the ion and its CCS (Figure 1A). The inherent complexity of proteins results in broad structural ensembles, and the IM resolution of IM-MS instruments is often insufficient to detect subtle but biologically relevant structural variations based on CCS alone. To overcome these limitations, the gradual and controlled collisional activation of gas-phase protein ions can be used to induce structural changes via collision-induced unfolding (CIU). CIU acts to populate a wide range of non-native intermediate states corresponding to unfolded gas-phase protein ions (Figure 1B).40 By correlating the CCS distributions of ions against the instrument potentials applied, activation-correlated CCS plots (CIU fingerprints) can be generated. Most native-like globular protein ions exhibit a single, monomodal CCS distribution at low activation energy across all charge states observed. However, CIU fingerprints contain information regarding additional non-native, unfolded, and collisionally activated conformer populations and the accelerating potentials necessary to induce each transition (CIU50) detected, thus expanding the pool of structural descriptors 2-5 fold (Figure 1). Using this expanded set of structural descriptors, CIU can be used to detect subtle changes in protein structure that are otherwise unresolvable by standard IM techniques alone. To date, CIU has been deployed in the analysis of various protein classes including kinases,41 membrane proteins,19–21 metalloproteins,42,43 and biotherapeutics.12,13,44
Figure 1: Expanding Gas Phase Ion Structure Descriptors.
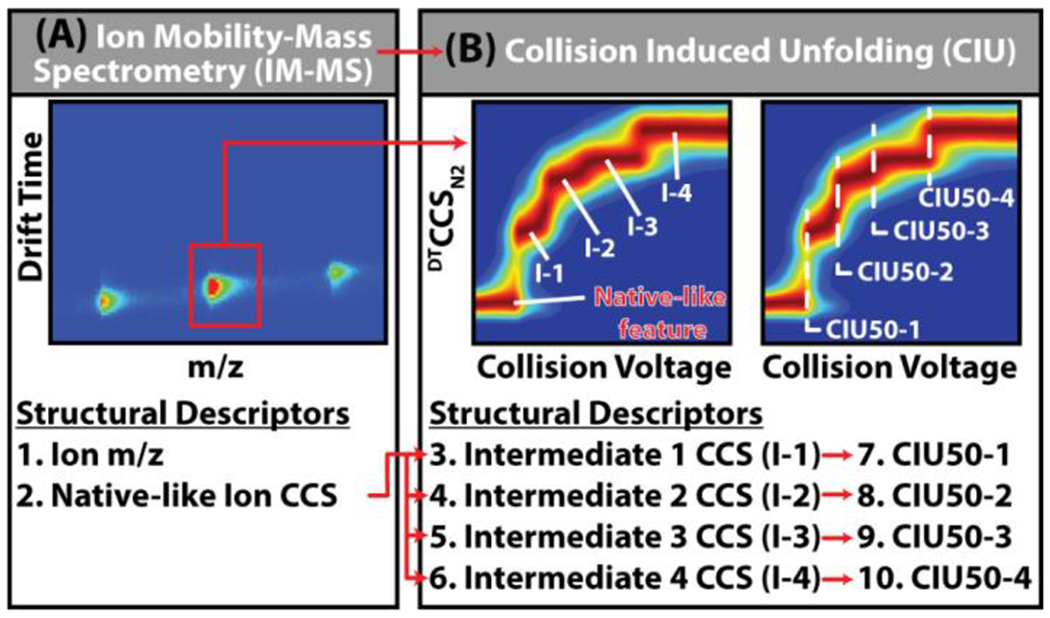
(A) Typical IM-MS analysis of proteins yields two structural descriptors per ion, the m/z and collision cross sections recorded for native-like ions, however the resolution of these measurements alone is not currently sufficient to differentiate critically important structural microstates that dictate much of protein function. (B) CIU expands the structural descriptors of protein ions to include unfolding intermediates (I-1,−2,−3,−4 etc.) in addition to the native like CCS (N), and the activation voltages necessary to achieve these unfolding events (CIU50-1,−2,−3,−4 etc.), scaling the number of available structural descriptors 2-to 5-fold. Using this expanded set of structural descriptors, analytes can be differentiated based on subtle structural differences which are not captured by IM-MS alone.
In support of the rapidly growing applications of CIU, we have previously described our development of several modified DTIM-MS instruments equipped with prototype high-energy source hardware capable of increased in-source activation necessary for CIU experiments. To date, our work has demonstrated the ability of this IM-MS platform to produce CIU data similar to those reported previously.9,15,45,46 In this study, we present the reproducibility of CIU fingerprints on three geographically distinct DTIM-MS instruments each equipped with prototype high-energy source hardware of the same design. Furthermore, we improved upon previously described methods, achieving higher-energy CIU of large proteins and protein complexes using a standard ESI source without the requisite addition of heavier dopant gases (i.e. sulfur hexafluoride).9 We demonstrate that CCS measurements of CIU features observed across all laboratories are highly reproducible (<1% RSD), although CIU50 values vary significantly between datasets. Finally, we compare our interlaboratory CIU results with CIU data collected across multiple production-grade high-energy source hardware designed using higher-tolerance specifications and observe excellent reproducibility across both CIU features and CIU50 values. Establishing the reproducibility of CIU data obtained across multiple production-grade hardware assemblies alludes to the possibility of comparing CIU data acquired in different laboratories. The successful benchmarking of CIU reproducibility on different instruments of the same platform (Agilent 6560c DTIM-QTOF), is essential to enabling future evaluations of CIU reproducibility across different instrument platforms. Ultimately, this, and future work support the curation of a CIU fingerprint database with potential applications in proteomics, structural biology, and the pharmaceutical sciences.
Experimental Section
Sample Preparation
Ammonium acetate, triethylammonium acetate, and lyophilized protein standards of bovine erythrocyte ubiquitin, equine heart myoglobin, Streptomyces avidinii streptavidin, bovine serum albumin (BSA), and universal monoclonal antibody standard (IgG1), were obtained from MilliporeSigma (St. Louis, MO). Product numbers for these standards are included in Table S1. Low Concentration Tune Mix was obtained from Agilent Technologies (Santa Clara, CA). The protein standards for the interlaboratory investigation were reconstituted to 5 μM in 200 mM ammonium acetate at pH ~7.2. Aliquots (150 μL) of each protein solution were flash frozen prior to being distributed to each laboratory. Myoglobin, BSA and IgG1 samples were desalted in 200 mM ammonium acetate by Micro BioSpin P-6 columns (Bio-Rad, Hercules, CA) immediately prior to IM-MS analysis. Streptavidin samples were prepared by desalting into 160 mM ammonium acetate supplemented with 40 mM triethylammonium acetate to facilitate charge reduction. Ubiquitin samples were not desalted to avoid sample loss in the desalting columns. From each sample, the highest intensity ion signals, exhibiting both unimodality and native-like CCS values were chosen for subsequent collision-induced unfolding experiments.
Ion Mobility-Mass Spectrometry
Instruments at all sites were tuned to optimize transmission of native-like, compact ions using parameters compiled from several previous studies utilizing this instrument platform for intact protein analyses.9,47–49 We collaboratively cross-examined native IM-MS spectra acquired at all sites and determined the optimal tuning conditions to ensure similar native-like ions were generated and measured at all sites. Samples were introduced via direct infusion into an electrospray ionization (ESI) source (Agilent Jet Stream) of a DTIM-MS (6560 IM-QTOF, Agilent Technologies) equipped with a prototype desolvation assembly consisting of high-energy in-source ion activation hardware (Figure S1). The sources were equipped with a micronebulizer assembly which supports low sample flow rates (2-5 μL/min). The following ESI settings were used: Ion transfer capillary voltage, 2.5-3.5 kV; ion focusing nozzle voltage, 1-2 kV; drying gas flow, 5 L/min; drying gas temperature, 140 °C; sheath gas flow, 11 L/min; sheath gas temperature 140 °C. The use of lower sheath gas temperature compared to those employed in under standard Agilent Jet Stream ESI operation (325 °C) is likely due to the lower flow rates enabled by the microflow nebulizer. In addition, for BSA and IgG1 samples, drying gas temperature was maintained at 250 °C. The source and ion transfer conditions were optimized for each protein to minimize activation and best preserve native MS conditions. Similar tuning conditions were used for myoglobin, streptavidin, and BSA. Ubiquitin, the smallest protein studied here, required lowered radio frequency voltages (RF) and electric fields in the pre-IM region to prevent unintentional ion activation. In contrast, SigmaMAb the largest protein in this study required higher pre-IM RFs and electric fields to improve ion transmission. Detailed tuning conditions can be found in Table S2. The high-pressure funnel, ion trap funnel, and drift tube were operated with high purity N2 at 4.80 ± 0.10 Torr, 3.800 ± 0.025 Torr, and 3.950 ± 0.005 Torr (autoregulated by a gas flow controller) respectively, unless otherwise noted. The drift tube was operated at ambient temperature at an electric field of ~18 V/cm. The maximum drift time was set to 90 ms for all analytes, and the trap fill and release times were set to 80 ms and 1 ms, respectively. All post-IM tune settings used default values determined by performing a “System Tune” in the MassHunter Acquisition software. The post-IM settings used on the UM 6560 platform varied slightly due to the presence of a linear ExD cell (eMSion, Corvalis, OR); however, the ability of this instrument to perform native protein measurements has been extensively characterized previously.9 Representative native mass spectra of all proteins used in the interlaboratory evaluation are available in Figure S2.
All collision cross sections were measured using the single-field calibration method (DTCCSN2), which is a previously described linear calibration approach derived from the Mason-Schamp equation.39 This approach incorporates instrument specific coefficients (β and tfix) that are obtained via linear regression analysis of arrival time measurements from Agilent tune mix ions (m/z 622-2722). Previous studies established that the single-field method produces CCS measurements within ~1.6% of the standard stepped-field method for a range of small molecules, metabolites, and proteins up to ~800 kDa.9,39
Collision-Induced Unfolding
CIU has been previously demonstrated on the 6560 DTIM-MS platform previously. Data contained in this report was acquired on three such instruments located in laboratories at University of Michigan (UM), Texas A&M University (TAMU), and Vanderbilt University (VU). Each instrument was equipped with prototype high-energy source hardware to enable ion activation prior to IM-MS analysis. The modified source includes the addition of an ion lens element (termed the fragmentor lens) positioned at the exit of the ion transfer capillary and the entrance to first ion funnel (Figure 2A). Ramping the potential difference between the ion transfer capillary exit and the fragmentor lens up to 450 V (depending on specific hardware) when operating in high purity N2, enables sufficient activation to achieve protein unfolding prior to IM separation (Figure 2B). A fourth instrument at the Agilent Technologies Research & Development Laboratory was used to evaluate new in-source ion activation hardware assemblies built to final commercial specifications (production-grade). The three identical production-grade hardware assemblies were evaluated to assess the CIU experiment reproducibility and performance. All instruments were also upgraded with QTOF firmware to enable time-of-flight mass spectrometer tuning and operation up to m/z 20,000.
Figure 2: Source Modifications Enabling Collision Induced Unfolding.
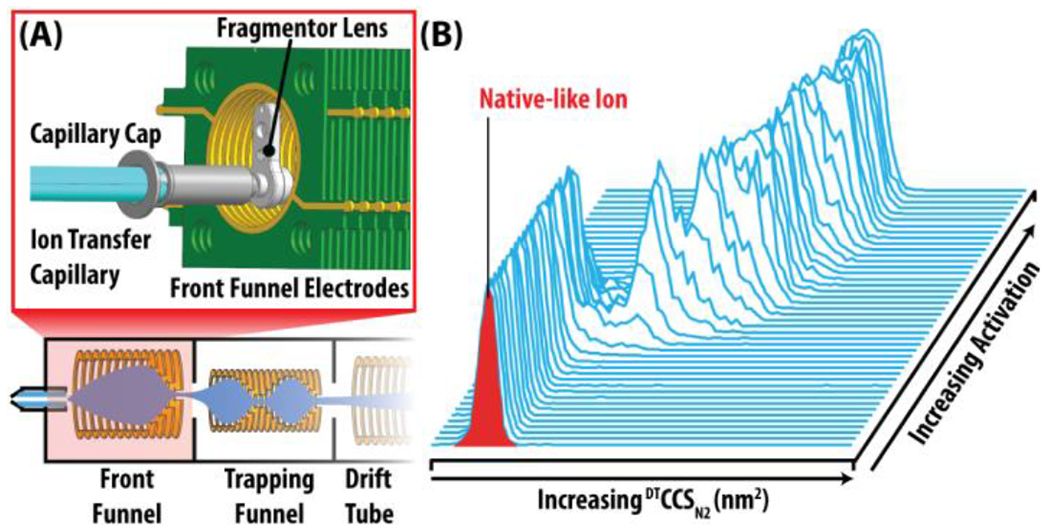
(A) Schematic of the modified Agilent 6560 Source Region (Full instrument diagram in SI). The expanded red box depicts a CAD rendition of the high energy source optics. (B) Collision induced unfolding (CIU) occurs when native-like ions are gradually activated by increasing amounts of in-source activation, resulting in ion unfolding. Ion unfolding is monitored by an increase in CCS relative to the initial CCS. The ion CCS is plotted versus in-source activation to visualize gas phase unfolding.
Our CIU data acquisition methods were designed using the time segment feature in MassHunter Acquisition software 10.0 (Agilent Technologies), enabling the collection of multiple activation steps in a single data file. All IM-MS data were analyzed and calibrated for CCS in IM-MS Browser 10.0 (Agilent Technologies), and the activation-resolved IM data were extracted and analyzed using CIUSuite2.50 CIU fingerprints were generated by plotting DTCCSN2 distributions as a function of increasing applied collision voltage, referred to as “in-source collision energy” (In-source CE) in MassHunter Acquisition (Agilent Technologies). Additional CIUSuite2 fitting parameters are included in Table S3. To assess reproducibility, all CIU fingerprint data were obtained in triplicate from each laboratory, and averaged fingerprints and corresponding RMSD values were obtained using software features currently available in CIUSuite2.
Results and Discussion
Interlaboratory CIU Results for Small Proteins
Figure 3 presents the CIU fingerprints for the three lowest molecular weight proteins investigated in this study: ubiquitin (+6, [M+6H]+6), myoglobin (+8, [M+8H]+8), and streptavidin (+11, [M+11H]+11), panels (A) - (C), respectively. Each CIU fingerprint is an average of three intralaboratory repeats (technical replicates) and the corresponding RMSDs are provided at the upper left corner of each fingerprint as well as summarized in Table S4. In general, the Intralaboratory CIU reproducibility was excellent, with all proteins analyzed producing CIU data with RMSD <4.5% in all laboratories. Importantly, the CIU fingerprints obtained for each protein are qualitatively similar across the different laboratories, in that all proteins sample similar intermediate CIU features, supporting the use of CIU fingerprints to support proteoform identification.51
Figure 3. Cross-Laboratory CIU Fingerprints and Feature CCS Reproducibility.
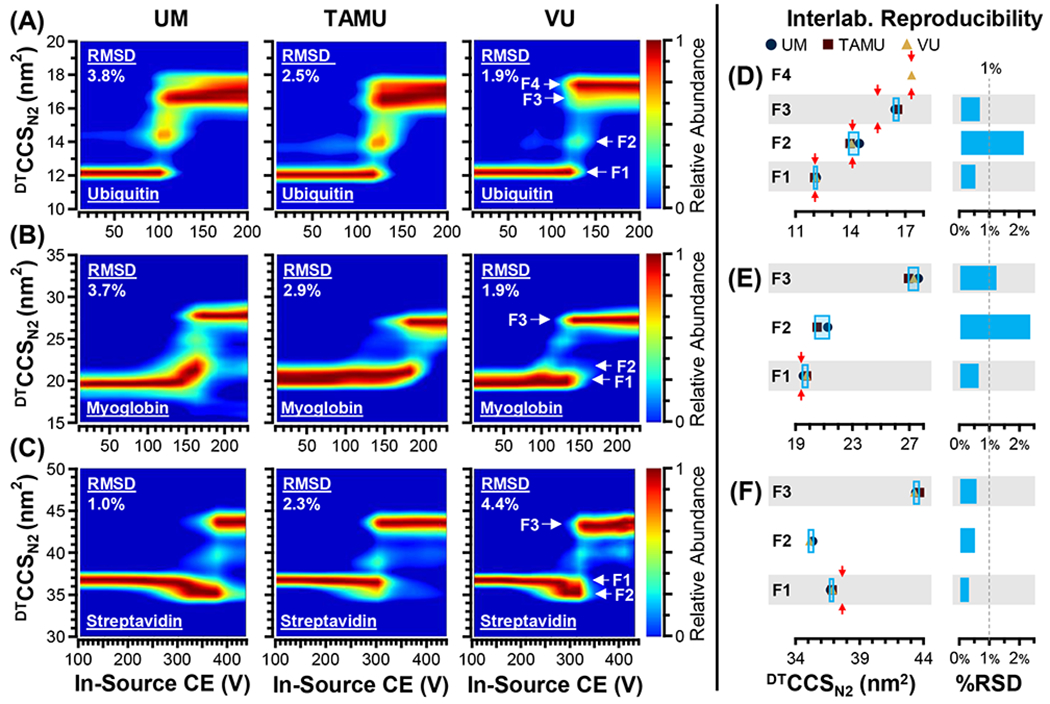
(A) ubiquitin +6, (B) myoglobin +8, and (C) streptavidin +11. Significant spectral features identified by CIUSuite2 (F1, F2, etc.) are annotated in the VU fingerprints. Interlaboratory reproducibility of feature CCS measurements for each protein are summarized, delineated into distinct spectral features (F1, F2, etc.). (D-F) The interlaboratory standard deviations are indicated with the light blue boxes, and previous literature values (DTCCSN2), when available, are indicated with red arrows. The interlaboratory relative standard deviations (RSDs) for most (67%) CCS measurements, are within 1%. (D) Ubiquitin, (E) Myoglobin, (F) Streptavidin.
While these CIU fingerprints were found to be highly reproducible within each laboratory, there are interlaboratory differences observed in the CIU fingerprints we recorded, particularly with respect to various stable intermediate structural families, referred to as CIU “features” (F). For example, ubiquitin (+6) exhibits a clear population of intermediate conformers (~14 nm2 CCS) which appear with different degrees of prominence across all three laboratory datasets (Figure 3A). Ubiquitin (+6) also exhibits two unfolded features (F3 and F4) that vary in abundance in the CIU fingerprints. For myoglobin (+8), two low-abundance intermediate features can be observed: (1) a feature exhibiting slightly larger CCS than native-like ions (F2, ~21 nm2), observed in two out of three fingerprints (UM & TAMU), and (2) a set of intermediate CIU features (~25 nm2) observed in all three fingerprints, but not in sufficient abundance to be labeled as a feature in these data (Figure 3B). Likewise, for streptavidin (+11), at least two intermediate features are observed: (1) a feature with a CCS likely corresponding to a collapsed state of the tetramer adopting a smaller CCS than the value measured for the ion population observed at lowest activation energies (F2, ~35 nm2), observed in two out of three fingerprints (UM & VU), and (2) a low-abundance feature between the compact and fully-extended states, with a CCS value of ~40 nm2 (Figure 3C). The transient nature of these intermediate features ultimately limits the cross-laboratory reproducibility of CIU fingerprints using prototype in-source activation hardware.
The CCS measurements obtained for all CIU features observed are summarized in Figure 3D–F, Figure S3 and Table S5, with criteria used for feature identification provided in Table S3. For those protein features which appear in sufficient abundance across all laboratories, the CCS measurement reproducibility was found to be excellent, with the majority of features (6/9, 67%) exhibiting an interlaboratory RSD of less than 1%. In addition, one feature exhibits a reproducibility just above this arbitrary 1% threshold (myoglobin F3, 1.3%). The remaining features have an interlaboratory RSD of ~2%. Overall, this work presents the remarkable reproducibility of CIU feature CCS’s, especially when considering that these features correspond to transient gas-phase protein unfolding intermediates (Figure 3D–F). Previously reported drift tube CCS measurements are available for the native-like states of some of the CIU features studied here (Table S5) and the average literature DTCCSN2 values are indicated in Figure 3D–F. The CCS measurements presented in this study generally agree with previous reports, with significant deviations noted for one extended state of ubiquitin (F3, our value is 6.2% larger), as well as the starting native-like state sampled for streptavidin (F1, our value is 2.1% smaller). The four other CCS measurements for which literature values were available exhibited a relative bias of less than 2%, and in two cases (ubiquitin F1, F4) the interlaboratory CCS measurements were within 0.5% of the averaged literature values. While the interlaboratory reproducibility of native-like protein ion CCS has been evaluated before, this work is the first to report the interlaboratory reproducibility of transient, gas-phase protein unfolding intermediates, establishing that the CCS of CIU features can be reliably used as reliable structural descriptors when differentiating gas-phase protein ions.
Despite the high degree of reproducibility observed for CIU feature CCS, larger differences were observed in the levels of activation required to achieve CIU, commonly referred to as “CIU50” values. CIU50 voltages varied significantly between laboratories for the small proteins measured. Despite the excellent intralaboratory RMSDs (<4.5%) for all proteins measured by CIU, the interlaboratory RMSDs were ~18-40% (Table S4) driven primarily by CIU50 variation found when using the prototype hardware assemblies located at UM, TAMU, and VU.
Interlaboratory CIU Results for BSA
Interlaboratory CIU comparisons of bovine serum albumin (BSA), a protein previously established as a CIU standard,9 produced similar results as those observed for ubiquitin, myoglobin, and streptavidin. Comparisons of the CIU fingerprints from all laboratories demonstrate that similar unfolding pathways were observed for the BSA +16 ion, with the protein starting in a natively structured form (F1) with a CCS of ~45 nm2. As the in-source CE is increased, the +16 ion gradually unfolds and populates several stable intermediate structures (F2, F3, F4) en route to a fully unfolded state (F5) (Figure 4A–C). The three hardware assemblies were able to reproduce the fingerprints with an intralaboratory reproducibility of <3% RMSD (3 replicates). The interlaboratory reproducibility for the CCSs of each of the observable features of BSA +16 was also found to be excellent (≤1.3% RSD). The most notable difference detected in our data was the pronounced variation in CIU feature intensity, resulting in a “missing” second feature (F2) in the BSA CIU fingerprint produced at TAMU, wherein F2 never achieved sufficient signal for feature detection (Figure 4B, Figure S3, Table S5). Our analysis instead detected the first feature at ~45 nm2 (F1) and the third feature at ~58 nm2 (F3), only populating F2 transiently enroute during the F1-to-F3 transition (Figure S3D). While our interlaboratory measurements of BSA +16 were found to be consistent, the observed CIU50 values once again varied. For example, F1 unfolds into F2 at ~180 V in the UM fingerprint (Figure 4A), however, F2 is undetected in the TAMU data (Figure 4B) and the same transition occurs at a lower volage (~140 V) in the VU fingerprint (Figure 4C). These differences appeared to be systematic within a given CIU dataset. This effect is particularly apparent when comparing feature F5 in our BSA CIU experiments. This final unfolded feature appears at ~440 V in UM data, while the TAMU and VU fingerprints show F5 appearing at ~370 V, suggesting that the TAMU and VU prototype sources are more activating than the UM source. As was noted in our analysis of Figure 3, CIU50 differences are the primary contributor to the high interlaboratory CIU differences detected in our BSA CIU data (Figure 4D, Table S4).
Figure 4: Interlaboratory CIU Fingerprints of BSA +16 and Development of Production-grade Source Hardware.
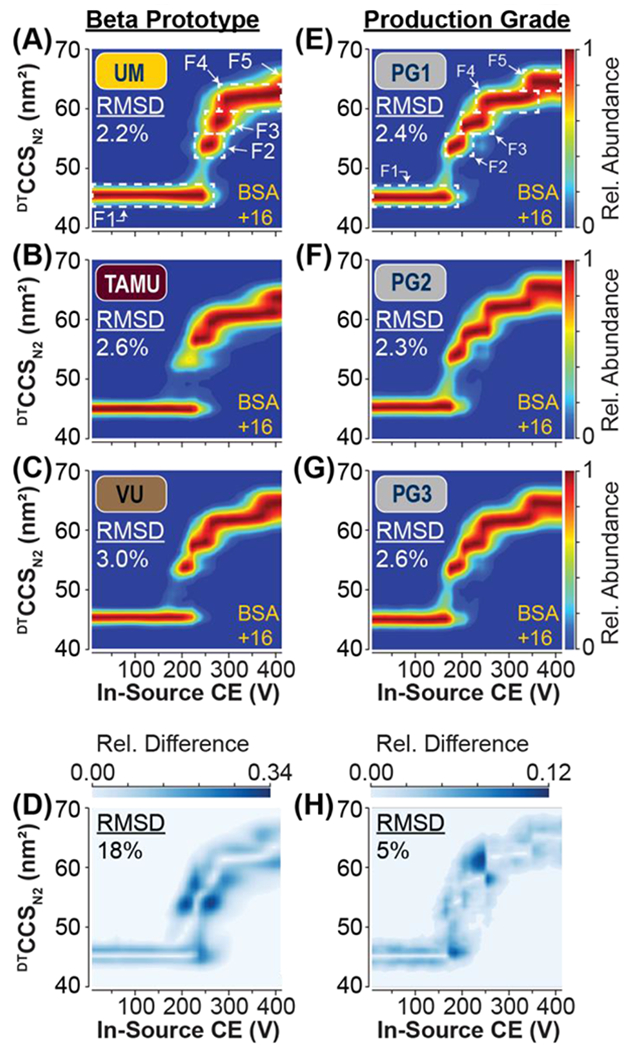
Average CIU fingerprint of BSA +16 ions acquired on three independent beta prototype DTIM-MS instruments located at (A) UM, (B) TAMU, and (C) VU. Intralaboratory CIU reproducibility is indicated by RMSD in the top left corner of each fingerprint. (D) CIU difference plot indicating the areas of highest difference between the CIU fingerprints from the three laboratories (A-C), resulting in an interlaboratory RMSD of 18%. CIU of BSA +16 ions acquired on three independent production-grade hardware assemblies (E-G). CIU difference plot indicating the areas of highest difference between the CIU fingerprints from PG1-3 (E-G), resulting in an interhardware RMSD of 5%.
Based on the interlaboratory evaluation of CIU reproducibility for 4 proteins (8-66 kDa), we concluded that CIU experiments obtained from different laboratories sample similar CIU features; however, the prototype source assemblies used in our experiments presented challenges associated with carrying out a rigorous interlaboratory comparison of CIU50 values. Limiting the interlaboratory CIU comparisons to unfolding features alone, eliminates 50% of the structural descriptors typically extracted from CIU data (Figure 1), and recovering this information content thus motivates our development of improved ion activation hardware capable of higher degrees of CIU reproducibility.
Evaluation of Production-grade Hardware
Bovine Serum Albumin (BSA)
To overcome the lack of CIU50 reproducibility observed in our interlaboratory study, three updated production-grade source assemblies (PG1, PG2, PG3) were constructed using high-precision tolerances to define the dimensions and inter-lens distances within the source assemblies constructed. In addition, the Fragmentor counter electrode was repositioned, improving both the effective activation capabilities of the production-grade source assemblies, as well as the reproducibility of CIU experiments. To test the reproducibility of these new source assemblies independent of other instrument variables, the same DTIM-MS instrument, in Santa Clara, CA was used for all measurements across all three production-grade source assemblies. The instrument was vented completely after completing all measurements with each production-grade source, and the source assemblies were exchanged. This decision was also justified by the findings of the interlaboratory evaluation which established that the CIU feature CCS is highly reproducible across multiple DTIM-MS instruments and geographic locations. The production-grade sources were evaluated in terms of their CIU reproducibility using BSA and SigmaMAb IgG1 standard samples.
CIU collected for BSA +16 ions on PG1, PG2, and PG3 was highly reproducible with intrahardware replicates achieving an RMSDs ≤2.6% (Figure 4E–G). Furthermore, an interhardware RMSD of 4.7% was obtained by comparing CIU data collected across all three production-grade sources (PG1 vs. PG2 vs. PG3), which represents a ~4-fold improvement over the 18.3% RMSD measured for interlaboratory comparisons of BSA +16 CIU fingerprints using prototype hardware (Figure 4H, Table S4, Table S6). Features F1-F5 were detected in all three production-grade hardware tests, addressing the previous inconsistencies in CIU feature detection during our prototype source evaluation (Figure 4B, Figure S3D). Each CIU feature CCS had an interhardware replicate RSD of ≤0.3%, indicating that the features associated with low-energy structures and those related to gas phase unfolding intermediates were highly reproducible across the hardware assemblies tested. The greatest improvement was observed in the reproducibility of CIU50 voltages, which resulted in an RSD of ≤3% in our interhardware evaluation (Table S6). CIU50-1, and CIU50-3 displayed some variation; however, producing RSDs of ~3%. In contrast, the CIU50-2 and CIU50-4 values were highly reproducible across our production-grade hardware tests, leading to RSDs of 0.2% and 1%, respectively. The slight differences in CIU50s for CIU50-1 and CIU50-3 are likely a result of the PG2 source requiring ~10V more in-source CE to induce feature transitions compared to PG1 and PG3. CIU50-1 and CIU50-3 are the two highest intensity regions in the CIU difference plot (Figure 4H), indicating that they are the main contributors to the slightly higher RSMD calculated for the interhardware tests compared to our intrahardware data.
In addition to and improvement in CIU-50 reproducibility, a modest improvement was observed in CIU feature reproducibility from 0.9% (interlaboratory) to 0.2% (interhardware). While this is of note, the overall RMSD improvement is primarily a function of the CIU-50 reproducibility. Overall, the production-grade source assemblies outperformed prototype source hardware in terms of CIU reproducibility improving upon the interlaboratory reproducibility of BSA CIU by approximately ~4-fold.
Immunoglobulin G1 (IgG1)
Throughout the pharmaceutical industry, stability measurements act as critical elements in the development of biotherapeutic monoclonal antibodies (mAbs). CIU has long been proposed as an ideal approach for inclusion in biotherapeutic pipelines, with a substantial body of work supporting its ability to characterize relevant mAb structures.12,14,15,46,52,53 A high level of technical reproducibility is required to conduct comparative analyses across mAb subtypes. Although the BSA results discussed above can be used to positively project the reproducibility of such CIU experiments for protein-based pharmaceuticals, mAbs are over two times larger (~150 kDa), and thus pose unique challenges for high-precision IM-MS and CIU.
CIU of SigmaMAb was conducted across each of the three production prototype hardware assemblies (PG1, PG2, PG3), and replicate RMSDs were calculated for each hardware assembly independently, as well as for all three assemblies. Representative mass spectra from our CIU experiments were plotted against the in-source CE potential values demonstrating an increase in signal intensity as the activation level increased (Figure 5A). The increase in signal intensity is due to both an associated improvement in ion transmission efficiency and improved desolvation of the large IgG1 ions at elevated source potentials. Seven mAb charge states (+25 to +31) of intact IgG1 were observed in these mass spectra, and the 5 highest intensity charge states (+26 to +30) were extracted for CIU analysis. Three technical replicates were acquired from each hardware assembly, resulting in nine CIU fingerprints which were averaged to produce the interhardware CIU fingerprint displayed here (Figure 5B–F). Corresponding CIU difference plots were also produced for each charge state, indicating that most of the differences between individual CIU fingerprints are the result of variations in CIU50 values (Figure 5G–K). The intrahardware RMSDs ranged from 1.8-5.0% (Table S7), with interhardware RMSDs of 2.6-4.4% (Figure 5G–K). In both instances the magnitude of the RMSDs recorded was inversely correlated with the intensities of the charge states selected for CIU analysis. The slightly higher interhardware RMSD observed for the CIU data extracted from +26 and +30 mAb ions is likely attributable to the relatively low abundances of these (Table S7, Figure 5A), however such RMSD values are <5%.
Figure 5: IgG1 CIU using Production-grade CIU Hardware.
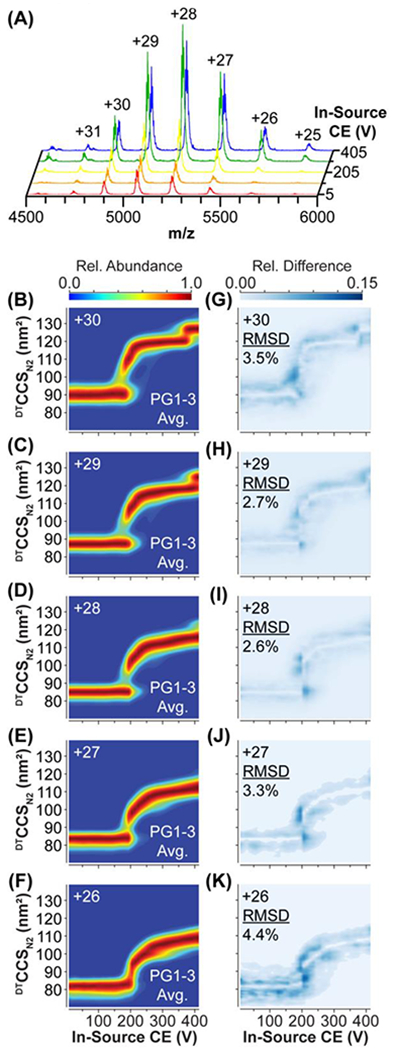
(A) IgG1 mass spectra during a CIU experiment at 5 V (red), 105 V (orange), 205 V (yellow), 305 V (green), and 405 V (blue). Interhardware average CIU fingerprints (B-F) and CIU Difference plots (G-K) For +30 to +26 charge states, indicating areas of greatest difference between CIU fingerprints from the 3 production prototypes, and the interhardware RMSD.
Our CIU data for SigmaMAb yields fingerprints similar to those reported previously,12 with ions across all charge states producing a similarly gradual transition from the CIU first feature to the second. All fingerprints were fit to two (+27, +29), or three (+26, +28, +30) features (Figure S4). In CIU data acquired for +29 mAb ions, a third feature is apparent at ~124 nm2; however, it was not included in our fits due to lack of sufficient sampling of the associated voltage slices where these structures are observed in our CIU data. We record an average interhardware feature CCS reproducibility value for SigmaMAb of 0.2%, similar to the ~0.3% feature RSD measured for BSA (Table S7). Continuing with this trend, we observe an average CIU50 RSD 1.5%, which is comparable to the ~2% measured for BSA (Table S7). If we filter our data to search for the most reproducible CIU fingerprints within our mAb dataset, we obtain interhardware RSDs across CIU features and CIU50s of 0.03%, and 1.2% respectively (28+ and 29+ data only), representing exceptional interhardware CIU reproducibility for such a large, structurally dynamic protein ion.
Conclusions
We evaluated the interlaboratory reproducibility of CIU data acquired using prototype source hardware for a variety of small proteins (8-66 kDa). These measurements were performed at 3 independent sites to rigorously assess the interlaboratory reproducibility of CIU data. Our analysis of this prototype hardware revealed the CIU experiments were generally reproducible, with all three laboratories reporting similar CCS measurements (RSD<3%) for gas-phase unfolding intermediates observed during CIU experiments. However, the results also indicated that prototype source construction tolerances were insufficient to produce high-precision CIU50 measurements across the different test sites.
These results spurred the development of the production-grade CIU hardware which performs ion activation equally across DTIM-MS platforms. Three production prototype hardware assemblies were built to final commercial specifications and were evaluated for their ability to reproduce CIU experiments for larger proteins such as BSA and SigmaMAb (66-150 kDa). We found that the production prototype hardware assemblies were capable of conducting CIU experiments with a high level of overall interhardware CIU reproducibility (≤4.4% RMSD). Furthermore, we observed improved feature reproducibility, to a value of <0.5%, and critically collected CIU50 measurements with a reproducibility of <2% RSD using our calibrated activation source optics.
With CIU becoming a more commonly utilized technology for applications in structural biology and the pharmaceutical sciences, the reproducibility of such data is paramount to achieving its full potential as a laboratory-independent comparative technique. Presently, most CIU practitioners only compare CIU data with other datasets acquired in the same laboratory. The reproducibility of CIU data reported here opens the door to broader interlaboratory comparisons of CIU fingerprints, including the creation of CIU databases, potentially enabling broader uses of such data extending to protein identification and protein biomarker tracking.51
Supplementary Material
Acknowledgements
V.V.G., B.R.J., B.T.R. acknowledge support from Agilent Technologies Thought Leader Award and University Relations grant programs. CIU method development in the Ruotolo laboratory is supported through an Agilent Technologies Applications and Core Technology University Research (ACT-UR) Grant. C.S.M. and D.H.R acknowledge support from NIH grants R01GM121751 and P41GM1285771. J.C.M. and J.A.M. acknowledge the National Institutes of Health, National Institute of Child Health and Human Development (NIH HHS, R01HD102752) for funding support as well as the Center for Innovative Technology (CIT) at Vanderbilt for providing resource support for the experiments conducted at Vanderbilt University.
Conflicts of Interest
V.V.G., B.R.J, and B.T.R. acknowledge that the Ruotolo laboratory at University of Michigan is designated as an Agilent Thought Leader Laboratory and certify that their contributions are scientifically objective. J.C.M and J.A.M acknowledge the Vanderbilt University Center for Innovative Technology is designated as an Agilent Thought Leader Laboratory and certify that their contributions are scientifically objective. R.T.K. and J.C.F. are employees of Agilent Technologies, the manufacturer of the IM-MS platforms evaluated in this report.
Footnotes
Supporting Information
The supporting information (SI) can be found at (insert link). The SI includes sample purchasing numbers, instrument tuning settings, data analysis settings for CIUSuite2, tables summarizing IM-MS and CIU measurements, and figures detailing the instrument, representative mass spectra, and CIU Feature fitting.
References
- (1).Leney AC; Heck AJR Native Mass Spectrometry: What Is in the Name? J Am Soc Mass Spectrom 2017, 28 (1), 5–13. 10.1007/s13361-016-1545-3. [DOI] [PubMed] [Google Scholar]
- (2).Wilm M; Mann M Analytical Properties of the Nanoelectrospray Ion Source. Analytical Chemistry 1996, 68 (1), 1–8. 10.1021/ac9509519. [DOI] [PubMed] [Google Scholar]
- (3).Tahallah N; Pinkse M; Maier CS; Heck AJR The Effect of the Source Pressure on the Abundance of Ions of Noncovalent Protein Assemblies in an Electrospray Ionization Orthogonal Time-of-Flight Instrument. Rapid Communications in Mass Spectrometry 2001, 15 (8), 596–601. 10.1002/rcm.275. [DOI] [PubMed] [Google Scholar]
- (4).Sobott F; Hernández H; McCammon MG; Tito MA; Robinson CV A Tandem Mass Spectrometer for Improved Transmission and Analysis of Large Macromolecular Assemblies. Analytical Chemistry 2002, 74 (6), 1402–1407. 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- (5).Li H; Wolff JJ; Van Orden SL; Loo JA Native Top-down Electrospray Ionization-Mass Spectrometry of 158 KDa Protein Complex by High-Resolution Fourier Transform Ion Cyclotron Resonance Mass Spectrometry. Analytical Chemistry 2014, 86 (1), 317–320. 10.1021/ac4033214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).May JC; Goodwin CR; Lareau NM; Leaptrot KL; Morris CB; Kurulugama RT; Mordehai A; Klein C; Barry W; Darland E; Overney G; Imatani K; Stafford GC; Fjeldsted JC; McLean JA Conformational Ordering of Biomolecules in the Gas Phase: Nitrogen Collision Cross Sections Measured on a Prototype High Resolution Drift Tube Ion Mobility-Mass Spectrometer. Analytical Chemistry 2014, 86 (4), 2107–2116. 10.1021/ac4038448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).May JC; Dodds JN; Kurulugama RT; Stafford GC; Fjeldsted JC; Mclean JA Broadscale Resolving Power Performance of a High Precision Uniform Field Ion Mobility-Mass Spectrometer †. Analyst 2015, 140, 6824. 10.1039/c5an00923e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Dyachenko A; Wang G; Belov M; Makarov A; De Jong RN; Van Den Bremer ETJ; Parren PWHI; Heck AJR Tandem Native Mass-Spectrometry on Antibody-Drug Conjugates and Submillion Da Antibody-Antigen Protein Assemblies on an Orbitrap EMR Equipped with a High-Mass Quadrupole Mass Selector. Analytical Chemistry 2015, 87 (12), 6095–6102. 10.1021/acs.analchem.5b00788. [DOI] [PubMed] [Google Scholar]
- (9).Gadkari V. v.; Ramírez CR; Vallejo DD; Kurulugama RT; Fjeldsted JC; Ruotolo BT Enhanced Collision Induced Unfolding and Electron Capture Dissociation of Native-like Protein Ions. Analytical Chemistry 2020, 92 (23), 15489–15496. 10.1021/acs.analchem.0c03372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Mallis CS; Zheng X; Qiu X; McCabe JW; Shirzadeh M; Lyu J; Laganowsky A; Russell DH Development of Native MS Capabilities on an Extended Mass Range Q-TOF MS. International Journal of Mass Spectrometry 2020, 458, 116451. 10.1016/j.ijms.2020.116451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Giles K; Ujma J; Wildgoose J; Pringle S; Richardson K; Langridge D; Green M A Cyclic Ion Mobility-Mass Spectrometry System. Analytical Chemistry 2019, 91 (13), 8564–8573. 10.1021/acs.analchem.9b01838. [DOI] [PubMed] [Google Scholar]
- (12).Tian Y; Han L; Buckner AC; Ruotolo BT Collision Induced Unfolding of Intact Antibodies: Rapid Characterization of Disulfide Bonding Patterns, Glycosylation, and Structures. Analytical Chemistry 2015, 87 (22), 11509–11515. 10.1021/acs.analchem.5b03291. [DOI] [PubMed] [Google Scholar]
- (13).Tian Y; Lippens JL; Netirojjanakul C; Campuzano IDG; Ruotolo BT Quantitative Collision-Induced Unfolding Differentiates Model Antibody–Drug Conjugates. Protein Science 2019, 28 (3), 598–608. 10.1002/pro.3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Hernandez-Alba O; Wagner-Rousset E; Beck A; Cianférani S Native Mass Spectrometry, Ion Mobility, and Collision-Induced Unfolding for Conformational Characterization of IgG4 Monoclonal Antibodies. Analytical Chemistry 2018, 90 (15), 8865–8872. 10.1021/acs.analchem.8b00912. [DOI] [PubMed] [Google Scholar]
- (15).Vallejo DD; Polasky DA; Kurulugama RT; Eschweiler JD; Fjeldsted JC; Ruotolo BT A Modified Drift Tube Ion Mobility-Mass Spectrometer for Charge-Multiplexed Collision-Induced Unfolding. Analytical Chemistry 2019, 91 (13), 8137–8146. 10.1021/acs.analchem.9b00427. [DOI] [PubMed] [Google Scholar]
- (16).Laganowsky A; Reading E; Allison TM; Ulmschneider MB; Degiacomi MT; Baldwin AJ; Robinson C. v. Membrane Proteins Bind Lipids Selectively to Modulate Their Structure and Function. Nature 2014, 510 (7503), 172–175. 10.1038/nature13419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Hopper JTS; Yu YTC; Li D; Raymond A; Bostock M; Liko I; Mikhailov V; Laganowsky A; Benesch JLP; Caffrey M; Nietlispach D; Robinson CV Detergent-Free Mass Spectrometry of Membrane Protein Complexes. Nature Methods 2013, 10 (12), 1206–1208. 10.1038/nmeth.2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Laganowsky A; Reading E; Hopper JTS; Robinson CV Mass Spectrometry of Intact Membrane Protein Complexes. Nature Protocols 2013, 8 (4), 639–651. 10.1038/nprot.2013.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Fantin SM; Parson KF; Niu S; Liu J; Polasky DA; Dixit SM; Ferguson-Miller SM; Ruotolo BT Collision Induced Unfolding Classifies Ligands Bound to the Integral Membrane Translocator Protein. Anal. Chem 2019, 91, 38. 10.1021/acs.analchem.9b03208. [DOI] [PubMed] [Google Scholar]
- (20).Fantin SM; Huang H; Sanders CR; Ruotolo BT Collision-Induced Unfolding Differentiates Functional Variants of the KCNQ1 Voltage Sensor Domain. J Am Soc Mass Spectrom 2020, 01, 13. 10.1021/jasms.0c00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Hutchison JM; Shih KC; Scheidt HA; Fantin SM; Parson KF; Pantelopulos GA; Harrington HR; Mittendorf KF; Qian S; Stein RA; Collier SE; Chambers MG; Katsaras J; Voehler MW; Ruotolo BT; Huster D; McFeeters RL; Straub JE; Nieh MP; Sanders CR Bicelles Rich in Both Sphingolipids and Cholesterol and Their Use in Studies of Membrane Proteins. J Am Chem Soc 2020, 142 (29), 12715–12729. 10.1021/jacs.0c04669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Fantin SM; Parson KF; Yadav P; Juliano B; Li GC; Sanders CR; Ohi MD; Ruotolo BT Ion Mobility–Mass Spectrometry Reveals the Role of Peripheral Myelin Protein Dimers in Peripheral Neuropathy. Proceedings of the National Academy of Sciences 2021, 118 (17). 10.1073/PNAS.2015331118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Sobott F; Benesch JLP; Vierling E; Robinson C. v. Subunit Exchange of Multimeric Protein Complexes: REAL-TIME MONITORING OF SUBUNIT EXCHANGE BETWEEN SMALL HEAT SHOCK PROTEINS BY USING ELECTROSPRAY MASS SPECTROMETRY. Journal of Biological Chemistry 2002, 277 (41), 38921–38929. 10.1074/JBC.M206060200. [DOI] [PubMed] [Google Scholar]
- (24).Aquilina JA; Benesch JLP; Bateman OA; Slingsby C; Robinson CV Polydispersity of a Mammalian Chaperone: Mass Spectrometry Reveals the Population of Oligomers in AB-Crystallin. Proceedings of the National Academy of Sciences 2003, 100 (19), 10611–10616. 10.1073/PNAS.1932958100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Mitra R; Gadkari V. v.; Meinen BA; van Mierlo CPM; Ruotolo BT; Bardwell JCA Mechanism of the Small ATP-Independent Chaperone Spy Is Substrate Specific. Nature Communications 2021, 12 (1), 1–13. 10.1038/s41467-021-21120-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Uetrecht C; Barbu IM; Shoemaker GK; van Duijn E; Heck AJR Interrogating Viral Capsid Assembly with Ion Mobility-Mass Spectrometry. Nature Chemistry 2011, 3 (2), 126–132. 10.1038/nchem.947. [DOI] [PubMed] [Google Scholar]
- (27).Mason EA; McDaniel EW Transport Properties of Ions in Gases; 1988. 10.1107/97809553602060000907. [DOI] [Google Scholar]
- (28).Marklund EG; Degiacomi MT; Robinson CV; Baldwin AJ; Benesch JLP Collision Cross Sections for Structural Proteomics. Structure 2015, 23 (4), 791–799. 10.1016/j.str.2015.02.010. [DOI] [PubMed] [Google Scholar]
- (29).Ewing SA; Donor MT; Wilson JW; Prell JS Collidoscope: An Improved Tool for Computing Collisional Cross-Sections with the Trajectory Method. J Am Soc Mass Spectrom 2017, 28 (4), 587–596. 10.1007/S13361-017-1594-2/SUPPL_FILE/JS8B05514_SI_001.PDF. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Bleiholder C; Contreras S; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (IV). Application to Polypeptides. International Journal of Mass Spectrometry 2013, 354–355, 275–280. 10.1016/J.IJMS.2013.06.011. [DOI] [Google Scholar]
- (31).Anderson SE; Bleiholder C; Brocker ER; Stang PJ; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (III): Application to Supramolecular Coordination-Driven Assemblies with Complex Shapes. International Journal of Mass Spectrometry 2012, 330–332, 78–84. 10.1016/J.IJMS.2012.08.024. [DOI] [Google Scholar]
- (32).Bleiholder C; Contreras S; Do TD; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (II). Model Parameterization and Definition of Empirical Shape Factors for Proteins. International Journal of Mass Spectrometry 2013, 345–347, 89–96. 10.1016/J.IJMS.2012.08.027. [DOI] [Google Scholar]
- (33).Bleiholder C; Wyttenbach T; Bowers MT A Novel Projection Approximation Algorithm for the Fast and Accurate Computation of Molecular Collision Cross Sections (I). Method. International Journal of Mass Spectrometry 2011, 308 (1), 1–10. 10.1016/J.IJMS.2011.06.014. [DOI] [Google Scholar]
- (34).Shvartsburg AA; Jarrold MF An Exact Hard-Spheres Scattering Model for the Mobilities of Polyatomic Ions. Chemical Physics Letters 1996, 261 (1–2), 86–91. 10.1016/0009-2614(96)00941-4. [DOI] [Google Scholar]
- (35).Mesleh MF; Hunter JM; Shvartsburg AA; Schatz GC; Jarrold MF Structural Information from Ion Mobility Measurements: Effects of the Long-Range Potential. Journal of Physical Chemistry 1996, 100 (40), 16082–16086. 10.1021/JP961623V. [DOI] [Google Scholar]
- (36).Shvartsburg AA; Hudgins RR; Dugourd P; Jarrold MF Structural Information from Ion Mobility Measurements: Applications to Semiconductor Clusters. Chemical Society Reviews 2001, 30 (1), 26–35. 10.1039/A802099J. [DOI] [Google Scholar]
- (37).Shvartsburg AA; Jarrold MF An Exact Hard-Spheres Scattering Model for the Mobilities of Polyatomic Ions. Chemical Physics Letters 1996, 261 (1–2), 86–91. 10.1016/0009-2614(96)00941-4. [DOI] [Google Scholar]
- (38).Davidson KL; Oberreit DR; Hogan CJ; Bush MF Nonspecific Aggregation in Native Electrokinetic Nanoelectrospray Ionization. International Journal of Mass Spectrometry 2017, 420, 35–42. 10.1016/J.IJMS.2016.09.013. [DOI] [Google Scholar]
- (39).Stow SM; Causon TJ; Zheng X; Kurulugama RT; Mairinger T; May JC; Rennie EE; Baker ES; Smith RD; McLean JA; Hann S; Fjeldsted JC An Interlaboratory Evaluation of Drift Tube Ion Mobility-Mass Spectrometry Collision Cross Section Measurements. Analytical Chemistry 2017, 89 (17), 9048–9055. 10.1021/acs.analchem.7b01729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (40).Dixit SM; Polasky DA; Ruotolo BT Collision Induced Unfolding of Isolated Proteins in the Gas Phase: Past, Present, and Future. Current Opinion in Chemical Biology 2018, 42, 93–100. 10.1016/j.cbpa.2017.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Rabuck JN; Hyung SJ; Ko KS; Fox CC; Soellner MB; Ruotolo BT Activation State-Selective Kinase Inhibitor Assay Based on Ion Mobility-Mass Spectrometry. Analytical Chemistry 2013, 85 (15), 6995–7002. 10.1021/ac4012655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Dong S; Wagner ND; Russell DH Collision-Induced Unfolding of Partially Metalated Metallothionein-2A: Tracking Unfolding Reactions of Gas-Phase Ions. Analytical Chemistry 2018, 90 (20), 11856–11862. 10.1021/ACS.ANALCHEM.8B01622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Dong S; Shirzadeh M; Fan L; Laganowsky A; Russell DH Ag+ Ion Binding to Human Metallothionein-2A Is Cooperative and Domain Specific. Analytical Chemistry 2020, 92 (13), 8923–8932. 10.1021/ACS.ANALCHEM.0C00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (44).Hernandez-Alba O; Wagner-Rousset E; Beck A; Cianférani S Native Mass Spectrometry, Ion Mobility, and Collision-Induced Unfolding for Conformational Characterization of IgG4 Monoclonal Antibodies. Analytical Chemistry 2018, 90 (15), 8865–8872. 10.1021/acs.analchem.8b00912. [DOI] [PubMed] [Google Scholar]
- (45).Zheng X; Kurulugama RT; Laganowsky A; Russell DH Collision-Induced Unfolding Studies of Proteins and Protein Complexes Using Drift Tube Ion Mobility-Mass Spectrometer. Anal. Chem 2020, 92, 23. 10.1021/acs.analchem.0c00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Vallejo DD; Kang J; Coghlan J; Ramírez CR; Polasky DA; Kurulugama RT; Fjeldsted JC; Schwendeman AA; Ruotolo BT Collision-Induced Unfolding Reveals Stability Differences in Infliximab Therapeutics under Native and Heat Stress Conditions. Analytical Chemistry 2021, 93 (48), 16166–16174. 10.1021/ACS.ANALCHEM.1C03946/SUPPL_FILE/AC1C03946_SI_001.PDF. [DOI] [PubMed] [Google Scholar]
- (47).Harrison JA; Kelso C; Pukala TL; Beck JL Conditions for Analysis of Native Protein Structures Using Uniform Field Drift Tube Ion Mobility Mass Spectrometry and Characterization of Stable Calibrants for TWIM-MS. J. Am. Soc. Mass Spectrom 2019, 30 (2), 256–267. 10.1007/s13361-018-2074-z. [DOI] [PubMed] [Google Scholar]
- (48).May JC; Jurneczko E; Stow SM; Kratochvil I; Kalkhof S; McLean JA Conformational Landscapes of Ubiquitin, Cytochrome c, and Myoglobin: Uniform Field Ion Mobility Measurements in Helium and Nitrogen Drift Gas. International Journal of Mass Spectrometry 2018, 427, 79–90. 10.1016/j.ijms.2017.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Gabelica V; Livet S; Rosu F Optimizing Native Ion Mobility Q-TOF in Helium and Nitrogen for Very Fragile Noncovalent Structures. J Am Soc Mass Spectrom 2018, 29 (11), 2189–2198. 10.1007/s13361-018-2029-4. [DOI] [PubMed] [Google Scholar]
- (50).Polasky DA; Dixit SM; Fantin SM; Ruotolo BT CIUSuite 2: Next-Generation Software for the Analysis of Gas-Phase Protein Unfolding Data. Anal. Chem 2019. 10.1021/acs.analchem.8b05762. [DOI] [PubMed] [Google Scholar]
- (51).Ruotolo BT Collision Cross Sections for Native Proteomics: Challenges and Opportunities. Journal of Proteome Research 2021, 21 (1), 2–8. 10.1021/ACS.JPROTEOME.1C00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Pisupati K; Tian Y; Okbazghi S; Benet A; Ackermann R; Ford M; Saveliev S; Hosfield CM; Urh M; Carlson E; Becker C; Tolbert TJ; Schwendeman SP; Ruotolo BT; Schwendeman A A Multidimensional Analytical Comparison of Remicade and the Biosimilar Remsima. Analytical Chemistry 2017, 89 (9), 4838–4846. 10.1021/ACS.ANALCHEM.6B04436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Botzanowski T; Hernandez-Alba O; Malissard M; Wagner-Rousset E; Deslignière E; Colas O; Haeuw J-F; Beck A; Cianférani S Middle Level IM–MS and CIU Experiments for Improved Therapeutic Immunoglobulin Subclass Fingerprinting. Analytical Chemistry 2020, 92 (13), 8827–8835. 10.1021/ACS.ANALCHEM.0C00293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


