Abstract
We previously reported that breast milk from women with (W) or without (WO) vaginal yeast infection during pregnancy differs in its immunological and antimicrobial properties, especially against pathogenic vaginal Candida sp.. Here, we investigated the differences in microbiota profiles of breast milk from these groups. Seventy-two breast milk samples were collected from lactating mothers (W, n=37; WO, n=35). The DNA of bacteria was extracted from each breast milk sample for microbiota profiling by 16S rRNA gene sequencing. Breast milk from the W-group exhibited higher alpha diversity than that from the WO-group across different taxonomic levels of class (P=0.015), order (P=0.011), family (P=0.020), and genus (P=0.030). Compositional differences between groups as determined via beta diversity showed marginal differences at taxonomic levels of phylum (P=0.087), family (P=0.064), and genus (P=0.067). The W-group showed higher abundances of families Moraxellaceae (P=0.010) and Xanthomonadaceae (P=0.008), and their genera Acinetobacter (P=0.015), Enhydrobacter (P=0.015), and Stenotrophomonas (P=0.007). Meanwhile, the WO-group showed higher abundances of genus Staphylococcus (P=0.046) and species Streptococcus infantis (P=0.025). This study shows that, although breast milk composition is affected by vaginal infection during pregnancy, this may not pose a threat to infant growth and development.
Keywords: breast milk, microbial community composition, microbiota, vaginal yeast infection
INTRODUCTION
Human breast milk is acknowledged as a crucial source of first food to newborns, with many documented benefits for the health and development of infants, including for their immunity, and brain and behavioral development (Martin et al., 2016). While the main components of breast milk have always been considered to be its chemical constituents, such as human milk oligosaccharides, which have attracted substantial attention, information on its biological components, especially on the highly diverse microbial population, is also now accumulating. This population is responsible for the production of myriad metabolites, which ultimately affect infant health status. In addition, milk microbiota has been proven to exert probiotic effects with myriad health benefits, ranging from ameliorating metabolic disorders (Fung and Liong, 2010; Lye et al., 2017) and enhancing immunity (Chong et al., 2019) to antimicrobial effects (Tham et al., 2012) and modulating the gut microenvironment (Hor et al., 2019). Notably, these microbial populations are also among the first to form the microbiota in infants’ gut, which may have a long-term effect on health. Infant gut microbiota has long been associated with the ability to acquire new immunity, develop and strengthen innate immunity, directly inhibit gut pathogenesis, and possibly develop autoimmune disorders later in life (Turroni et al., 2020).
The microbiota profile of breast milk is known to vary depending on various factors, particularly geographical location, diet, and maternal health status (Zimmermann and Curtis, 2020). While some distinct diseases and disorders such as inflammatory bowel disease, colorectal cancer, liver cirrhosis, gestational diabetes, and mastitis during the lactation period have been reported to alter the populations of bacteria in breast milk (Patel et al., 2017), less serious diseases have less pronounced effects on the microbiota profile of breast milk, which has been attributed either to limited information about the effects being available or a lack of assessment of factors causing biological changes of breast milk.
Vaginal infections are among the most common gynecological conditions affecting women globally, varying in prevalence from 5% to 50% among different study populations across major regions such as the United States, Europe, and South Asia (Begum et al., 2011). The most common causes of vaginal infections are vaginal candidiasis and bacteriosis. Centers for Disease Control and Prevention (CDC) reported that there was a higher prevalence of such infections among pregnant women, causing increased risks of systemic infection in neonates and prematurity after delivery (CDC, 2022). Although rarely fatal, vaginal infections often cause various other discomforts, such as vaginal redness, swelling, itching and/or soreness, and pain during sexual intercourse, leading to a reduced quality of life emotionally, socially, and sexually.
We previously reported that breast milk from women with (W) or without (WO) vaginal yeast infections during pregnancy share similar levels of biological components such as interleukin-10, immunoglobulins (IgA, IgM, and IgG), and growth factors (epidermal growth factor and transforming growth factor-α) (Nisaa et al., 2022). However, breast milk from the WO-group exhibited higher antimicrobial potential against vaginal pathogenic Candida sp. than that from the W-group, via both growth inhibition and aggregation of yeast cells. We postulate that all of these biological and psychological changes inflicted by vaginal infections during pregnancy may eventually affect other biological profiles of breast milk, typically the microbiota populations within it. It remains unclear whether breast milk from women in the W- and WO-groups differs in its microbiota composition and whether vaginal pathogens are carried forward into breast milk after pregnancy. To the best of our knowledge, no reports of studies assessing this issue have yet been published. Thus, the present study was established to resolve some of these questions.
MATERIALS AND METHODS
Recruitment of subjects
The design of population included in this study has been described in our previous report (Nisaa et al., 2022). Written informed consent was obtained from all subjects prior to study initiation. Subjects were recruited from Universiti Sains Malaysia (USM) campuses of Kelantan and Penang, Malaysia. Briefly, lactating mothers were screened based on the inclusion and exclusion criteria. For inclusion in this study, subjects had to be lactating women. Exclusion criteria included long-term medication due to certain illnesses for over 3 months during pregnancy, gestational diabetes during pregnancy, history of diabetes, coronary heart disease, and hypertension at any age, body mass index before pregnancy >24.9, consumed probiotics during pregnancy, and consumed probiotics during lactation period.
Study protocol
The lactating women were separated into those W and WO vaginal infection during pregnancy. Vaginal infections were confirmed by taking their previous medical history during the period of pregnancy. This study was conducted in accordance with the Declaration of Helsinki (World Medical Association, 2001). All procedures involving human subjects were approved by the JEPeM-USM Review Panel on Clinical Studies (approval no. USM/JEPeM/20090500) and the study was registered at ClinicalTrials.gov (identifier no. NCT05005286).
Breast milk was collected via hand expression. Gloves were put on prior to washing with sterilized and individually packed hand detergent liquid and sterilized water, and swabbing with a 70% isopropyl alcohol pad prior to the expression of breast milk. In addition, nipples were cleaned with an alcohol swab. The first ten drops of breast milk were discarded. Fifteen milliliters of fresh breast milk was collected in a sterile tube and stored at −20°C until further analyzes.
Microbiota analyses
DNA extraction and purification from all samples were performed as previously described (Liu et al., 2020). Purified DNA was analyzed using a NanoDrop 2000 ultraviolet-visible Spectrophotometer (Thermo Scientific). The V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified with the primers 341F (5’-CCTAYGGGRBGCASCAG-3’) and 806R (5’-GGACTACHVGGGTWTCTAAT-3’) by a thermocycler PCR system (Gene-Amp 9700, Applied Biosystems). The PCR reactions were performed in triplicate with a reaction volume of 20 μL containing 4 μL of 5× FastPfu Buffer (TransStart), 2 μL of 2.5 mM dNTPs, 0.8 μL of each primer (5 μM), 0.4 μL of FastPfu Polymerase (TransStart), and 10 ng of template DNA, in the following sequence: 3 min of denaturation at 95°C; 27 cycles of 30 s at 95°C, 30 s for annealing at 55°C, and 45 s for elongation at 72°C; and final extension at 72°C for 10 min. The PCR products were then extracted from a 2% agarose gel and further purified using the AxyPrep DNA Gel Extraction Kit (Axygen Bio-sciences) and quantified using QuantiFluor-ST (Promega), in accordance with the manufacturer’s protocol.
The purified amplicons were pooled in equimolar samples and paired-end sequenced (2×300) on an Illumina MiSeq platform (Illumina). The 16S rRNA gene sequences were processed using QIIME v.1.9.1 (Caporaso et al., 2010) and USEARCH v.10.0 (Edgar, 2010). Raw FASTQ files were quality-filtered by Trimmomatic and merged by USEARCH (Edgar, 2010) with the following criteria: removal of barcodes and primers, filtering of low-quality reads, and finding non-redundant reads. The merged raw reads numbered at least 50,000 per sample. Operational taxonomic units (OTUs) were clustered with 97% similarity cutoff using UPARSE. The taxonomy for each 16S rRNA gene sequence was analyzed by the Ribosomal Database Project Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva 132 16S rRNA database using a 60% confidence threshold.
Alpha (within-sample richness) and beta diversity (be-tween-sample dissimilarity) estimates were computed using MicrobiomeAnalyst phyloseq-R package version 3.6.1 (https://www.microbiomeanalyst.ca/MicrobiomeAnalyst/home.xhtml) for different taxonomic levels.
Statistical analyses
Data were analyzed using SPSS version 20.0 (IBM Corp.). The primary hypothesis tested in this study was that the microbiota profiles differed between groups. Considering the skewed distribution and nonparametric nature of our data, differences in OTU relative abundance between groups were analyzed using the Mann-Whitney U-test, whereas the correlation analyzes were evaluated using Spearman’s rank correlations with rho (r) as the correlation coefficient. Alpha diversity of gut microbiota was measured by Chao1 and observed richness indexes, then compared using Mann-Whitney U-test, whereas beta diversity was calculated by principal coordinates analysis on Bray-Curtis dissimilarity and compared using permutational analysis of variance (PERMANOVA) and analysis of similarities (ANOSIM). As a continuation of our previous study, data on abundance and coaggregation of Candida species used in the present correlation analyzes were obtained from our previous study (Nisaa et al., 2022). All tests were two-sided with P<0.05 considered to indicate statistically signifi-cance.
RESULTS
Baseline characteristics
Based on the 85 subjects recruited in our previous study (Nisaa et al., 2022), only 72 breast milk samples qualified for microbiota analyzes based on the quantity and quality of their DNA (WO-group, n=35; W-group, n=37). No significant differences were observed between the groups in all of the general characteristics (Table 1).
Table 1.
Baseline characteristics of 72 lactating women with or without vaginal yeast infections during pregnancy
| Characteristic | W-group (n=37) | WO-group (n=35) | P-value1) |
|---|---|---|---|
| Occurrence of vaginal infection (pregnancy wk) | 24.19±6.88 | − | − |
| Age (yr) | 31.59±5.75 | 31.43±3.99 | 0.821 |
| Lactating period (mon) | 5.03±4.30 | 4.87±2.97 | 0.534 |
| n (%) | n (%) | P-value2) | |
| Body mass index | |||
| Normal (18.5∼24.9) | 24 (64.86) | 29 (82.86) | 0.083 |
| Overweight (25∼29.9) | 8 (21.62) | 5 (14.29) | 0.419 |
| Obesity (≥30) | 5 (13.51) | 1 (2.85) | 0.102 |
| Family income (per month) | |||
| ≤RM 5,999 | 24 (64.86) | 24 (68.57) | 0.739 |
| >RM 6,000 | 13 (35.14) | 11 (31.43) | 0.739 |
| Occupation | |||
| Housewife | 16 (43.24) | 8 (22.86) | 0.067 |
| Self-employed | 3 (8.11) | 5 (14.29) | 0.404 |
| Student | 2 (5.41) | 1 (2.86) | 0.589 |
| Waged worker | 16 (43.24) | 21 (60.00) | 0.155 |
| Level of education | |||
| Secondary | 7 (18.92) | 4 (11.43) | 0.377 |
| Tertiary | 20 (54.05) | 21 (60.00) | 0.611 |
| Postgraduate | 10 (27.03) | 10 (28.57) | 0.884 |
| Number of people living together in the household | |||
| ≤4 | 17 (45.95) | 15 (42.86) | 0.792 |
| >4 | 20 (54.05) | 20 (57.14) | 0.792 |
| Residential location | |||
| Suburban | 10 (27.03) | 7 (20.00) | 0.483 |
| Urban | 27 (72.97) | 28 (80.00) | 0.483 |
Data are presented as mean±standard error or number (%).
1)P-value obtained via Mann-Whitney U-test.
2)P-value obtained via chi-squared test.
W-group, with vaginal infection; WO-group, without vaginal infection; RM, ringgit Malaysia.
Alpha and beta diversity
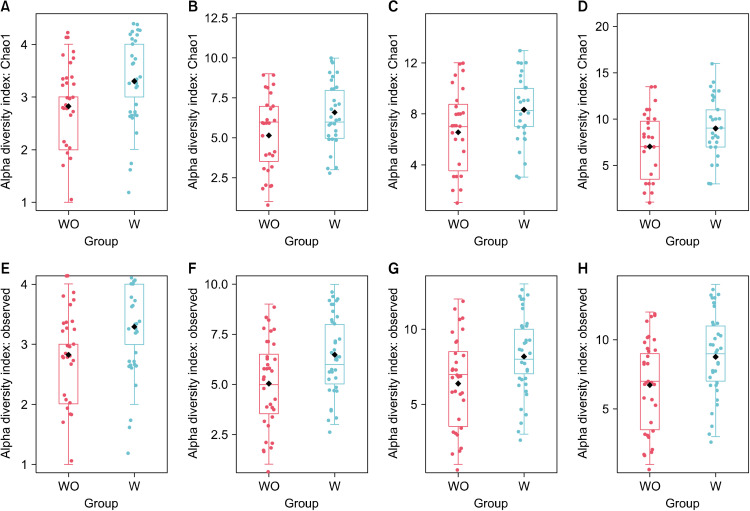
Alpha diversity measures differences within samples. While both the observed and Chao1 indexes provide a measure of alpha diversity in terms of OTU “richness”, only Chao1 equally takes into account frequent and rare OTUs, while the observed index only takes into account observed OTUs. Comparable findings were made for the breast milk from the two groups in terms of richness and evenness at the phylum level, but they differed at the class, order, family, and genus levels for both observed and Chao1 indexes (P<0.05; Fig. 1), where women with vaginal infections during pregnancy (W-group) showed higher alpha diversity than those without such infection (WO-group). Meanwhile, differences of alpha diversity in terms of OTU diversity, as measured by the Shannon and Simpson indexes, were not significant. While richness represents the number of species in a community, diversity represents not only the number of species in a community but also the abundance of each species.
Fig. 1.
Alpha diversity plots for breast milk from women without (WO) or with (W) vaginal infection during pregnancy. Diversity measured by Chao1 richness index for class (A, P=0.015), order (B, P=0.011), family (C, P=0.020), and genus (D, P=0.030). Diversity measured by observed richness index for class (E, P=0.015), order (F, P=0.009), family (G, P=0.014), and genus (H, P=0.014). The line inside the box represents the median, whereas the whiskers represent the lowest and highest values within the interquartile range. Outliers, as well as individual sample values, are shown as dots. Statistical significance was analyzed using the Mann-Whitney U-test (Total=72; W, n=37; WO, n=35).
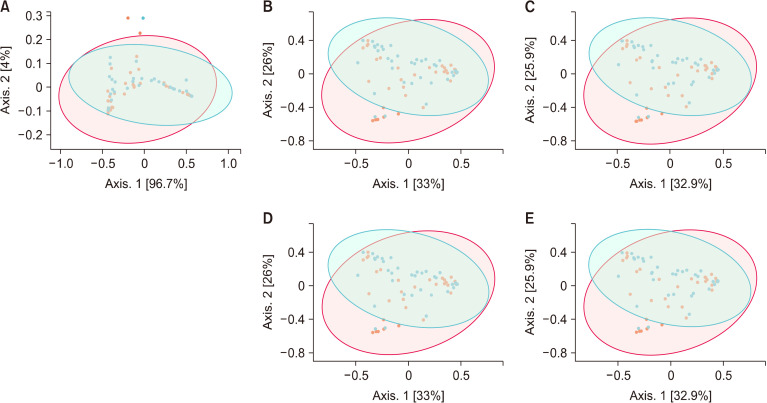
Beta diversity measures differences between samples. The Bray-Curtis index considers both the co-occurrence and differential abundance of OTUs. While both PERMANOVA and ANOSIM are suitable for analyzing nonparametric data, they do not measure the same parameters. PERMANOVA tests whether distance differs between groups, while ANOSIM tests whether the difference between groups is greater than that within groups. The difference in beta diversity between groups was only observed to reach marginal levels of significance (P<0.10) and only at the taxonomic levels of phylum (P=0.087), family (P=0.064), and genus (P=0.067; Fig. 2), as tested by PERMANOVA, and only reached significance at the levels of family (P=0.045) and genus (P=0.041; Fig. 2), as tested by ANOSIM. These findings indicated that vaginal infections during pregnancy contributed to a mild shift in composition of the microbial community, with a more prevalent effect at the lower taxonomic levels of family and genus.
Fig. 2.
Beta diversity as measured by Bray-Curtis dissimilarity and principal coordinates analysis for different taxonomic levels in breast milk from women without (WO; red) or with (W; blue) vaginal infections during pregnancy. Diversity in microbial community composition was evaluated by permutational analysis of variance for phylum (A, P=0.087), family (B, P=0.064), and genus (C, P=0.067), and by analysis of similarities for family (D, P=0.045) and genus (E, P=0.041) with the total of 72 (W, n=37; WO, n=35).
Compositional differences
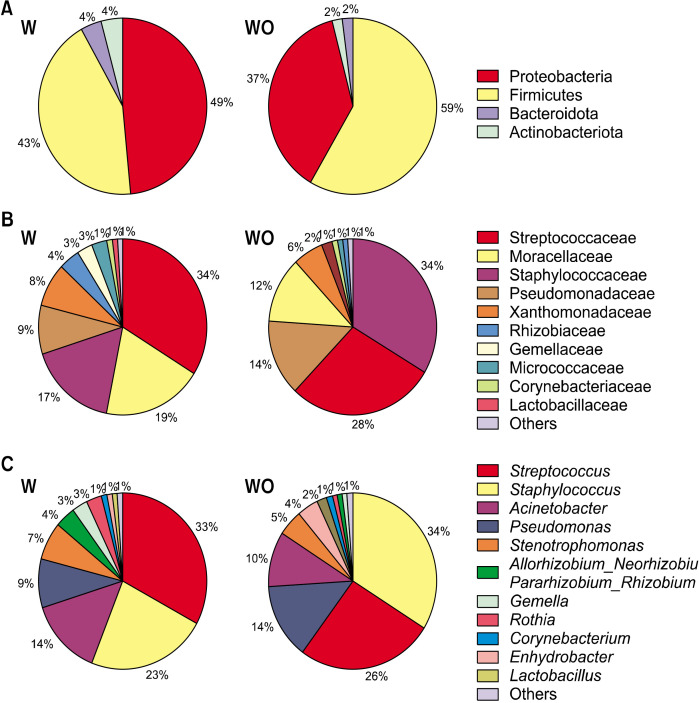
As alpha and beta diversity analyzes revealed differences between W- and WO-groups, we further analyzed the differences in microbiota of breast milk at different taxonomic levels. The dominant phyla of breast milk were consistently Firmicutes and Proteobacteria, with there being no significant differences in their levels between the W- and WO-groups (Fig. 3A). Meanwhile, regarding the less dominant phyla, the W-group had higher abundances of Actinobacteriota (P=0.006) and Bacteroidota (P=0.001) than the WO-group. These findings were attributable mainly to higher abundance at lower taxonomic levels in the W-group, such as for Micrococcaceae (P=0.001), Prevotellaceae (P=0.005), and Weeksellaceae (P=0.002) at the family level (Fig. 3B), and their genera Prevotella (P=0.009) and Chryseobacterium (P=0.003; Fig. 3C), compared with the levels in the WO-group.
Fig. 3.
Relative abundance of breast milk microbiota at taxonomic levels of phylum (A), family (B), and genus (C) from women with (W) or without (WO) vaginal infections during pregnancy (total=72; W, n=37; WO, n=35).
Although the abundance of the phylum Proteobacteria in breast milk remained similar between the groups (n=72), at lower taxonomic levels there were differences between the groups. Specifically, the W-group showed higher abundance of the family Moraxellaceae (P=0.01; Fig. 3B) and its genera Acinetobacter (P=0.015) and Enhydrobacter (P=0.015; Fig. 3C), along with the species Acinetobacter baumannii (P=0.046), compared with the levels in the WO-group.
Similarly, the abundances of the family Xanthomonadaceae (P=0.008), its genus Stenotrophomonas (P=0.007), and its species Stenotrophomonas pavanii (P=0.002) were also higher in breast milk of the W-group than in that of the WO-group. Resembling the findings for the phylum Proteobacteria, the phylum Firmicutes also showed similar abundances between groups, but differences between the groups emerged at lower taxonomic levels. Breast milk from the WO-group showed higher abundances of the genus Staphylococcus (P=0.046; Fig. 3C) and the species Streptococcus infantis (P=0.025) than the W-group.
Correlation analyzes
We previously reported that breast milk of the W-group had lower potential to inhibit Candida species commonly found in vaginal regions of women with vaginal candidiasis than that from the WO-group (Nisaa et al., 2022). This may be an effect of various microbiota in breast milk that are present at higher levels in the W-group as observed here. Chryseobacterium had positive correlations with the growth of Candida albicans (P=0.018, r=0.279), Candida glabrata (P=0.039, r=0.243), Candida krusei (P=0.002, r=0.363), and Candida tropicalis (P=0.002, r=0.359), along with negative correlations with the co-aggregation potential of breast milk against C. albicans (P=0.038, r=−0.245), C. glabrata (P=0.004, r=−0.338), C. krusei (P=0.002, r=−0.363), and C. tropicalis (P=0.010, r=−0.301). A similar observation was obtained for S. pavanii, which had positive correlations with the growth of C. albicans (P=0.038, r=0.245), C. krusei (P=0.010, r=0.302), and Candida parasilopsis (P=0.013, r=0.293), along with negative correlations with the co-aggregation potential of breast milk against C. albicans (P=0.031, r=−0.255), C. krusei (P=0.001, r=−0.368), and C. parasilopsis (P=0.005, r=−0.330). To a lesser degree, Prevotella showed a positive correlation with the growth of C. krusei (P=0.001, r=0.380), along with a negative correlation with the co-aggregation potential against C. krusei (P=0.002, r=−0.366), while Staphylococcus showed a negative correlation with the growth of the same pathogen (P=0.019, r=−0.276), along with a positive correlation with the co-aggregation potential (P=0.004, r=0.334). As we previously reported, antimicrobial effects of breast milk may be exhibited via cellular aggregation potential, where the ability of aggregated pathogenic cells to attach to the hosts’ urogenital epithelial surface is reduced, leading to greater elimination from the hosts’ systems (Nisaa et al., 2022).
The abundance of Enhydrobacter in breast milk showed positive correlations with the growth of C. glabrata (P=0.003, r=0.346), C. krusei (P=0.044, r=0.238), C. parasilopsis (P=0.019, r=0.276), C. tropicalis (P=0.034, r=0.251), and A. baumannii (P=0.030, r=0.256). The present data led us to believe that a higher abundance of Enhydrobacter in breast milk of the W-group may be crucial as an immune defense against vaginal Candida sp., which may be transferred to newborns during natural birth, and also against any potential harm associated with the higher abundance of A. baumannii in breast milk of the W-group. Breast milk as a natural first food for newborns may have its own protective and balancing mechanisms to sustain the general wellbeing of infants.
DISCUSSION
The diversity of microbial communities in breast milk often affects the profiles of gut microbiota in infants, with human breast milk having been reported to reduce the richness of infant gut microbiota (Haddad et al., 2021). This was mainly attributed to the presence of human milk oligosaccharides that favor the growth of specific microbial species. The introduction of formula feeding eventually diversifies the gut microbiota of infants to include taxa with a higher degree of proteolytic fermentation (Laursen, 2021). It is thus important to evaluate microbiota communities in breast milk. Maternal health conditions during pregnancy have been reported to affect the bacterial diversity of breast milk, potentially leading to microbial dysbiosis. This includes diseases such as inflammatory bowel disease, rheumatoid arthritis, colorectal cancer, liver cirrhosis, and type 2 diabetes during pregnancy, and mastitis during lactation, where microbial diversity and species richness were lower than in healthy women (Patel et al., 2017). However, this remains controversial as some other health conditions had little or opposite impacts on the microbial diversity profiles of breast milk. Regardless of the immunity status, HIV-positive women as determined by high or low cluster of differentiation count did not show a difference in the levels of microbiota diversity of breast milk (Maqsood et al., 2021). Meanwhile, gestational weight gain or intrapartum antibiotic use (penicillin, cephalothin, or cephalexin) has been shown to contribute to increased levels of microbial diversity in breast milk samples compared with that in normal controls (Hermansson et al., 2019; Lundgren et al., 2019). Our present study indicated that vaginal infections during pregnancy contributed to a shift in the richness and evenness of breast milk samples, which were consistent across different taxonomic levels.
The compositional differences in the W-group may have contributed to a higher microbiota diversity within breast milk samples of the W-group, as supported by the higher alpha diversity that we identified. While maternal gut Prevotella during pregnancy has been shown to be associated with protection against food allergy in infants (Vuillermin et al., 2020), the vaginal microbiota in cases of bacterial vaginosis is also often dominated by Prevotella, compared with the more lactobacillus-dominated profile in healthy women (Superti and De Seta, 2020). Similarly, while Chryseobacterium is found in common pairs between breast milk and infant stool samples (Duale et al., 2022), this genus is also found in the high vaginal regions (Bernardet et al., 2005) and in the urobiomes of women with urinary tract infections (Omar et al., 2014). Based on the higher abundance of Prevotella and Chryseobacterium in breast milk of the W-group compared with the levels in the WO-group, we have reason to believe that these populations may be passed on during vaginal infections via breast milk. This is supported by the positive correlations of Chryseobacterium with the growth of several pathogenic yeasts, namely, C. albicans, C. glabrata, C. krusei, and C. tropicalis, during inhibitory assays. However, considering that Chryseobacterium is rarely virulent (Duale et al., 2022), while Prevotella aids in the metabolism of gut carbohydrates (Prasoodanan et al., 2021), we postulate that the higher abundance of these genera in breast milk may not pose a health risk for infants.
The family Moraxellaceae includes species that colonize mucosal membranes or the skin of humans and other animals and can occasionally cause a variety of infections. This is notable in species such as A. baumannii, which has emerged as an important agent of opportunistic infections in healthcare-associated infections that are difficult to treat (Rosenberg et al., 2014). It primarily affects those who are immunocompromised, so it is increasingly being reported as a hospital-derived infection isolate (Howard et al., 2012). The human milk oligosaccharides have been found to inhibit biofilm activity, and have been employed against multidrug-resistant pathogens and susceptible isolates of A. baumannii (Spicer et al., 2021). While Enhydrobacter is a major taxon of the human skin and within the ocular microbiome (Leung et al., 2020), it may be opportunistic and has been detected in individuals with blepharitis (Fredricks, 2001). In breast milk, Enhydrobacter has been identified in sample pairs of breast milk and infant feces (Zhu et al., 2021), and also found at high levels in human colostrum, where it is associated with the infant metabolism of nutrients and immune development against diseases (Klein-Jöbstl et al., 2019).
The Xanthomonadaceae family consists of opportunistic human pathogens such as those from the genus Stenotrophomonas (Pak et al., 2015). The species S. pavanii has been identified to be multidrug-resistant against carbapenems and aminoglycosides, harboring many resistance genes against various antibiotics in the genome, with the potential to transmit antibiotic resistance in the environment (Kenzaka and Tani, 2018). The abundance of Stenotrophomonas was reportedly increased in breast milk in women undergoing chemotherapy for the treatment of Hodgkin’s lymphoma compared with the level in women not undergoing this treatment. This was accompanied by reduced concentrations of inositol and docosahexaenoic acid in breast milk, which are crucial components that reduce the risks of preterm birth and infant brain maldevelopment, respectively (Fernández et al., 2020). In the present study, the abundance of Stenotrophomonas was higher in breast milk of the W-group than in the WO-group, suggesting that Stenotrophomonas may be passed on during vaginal infections via breast milk. This is supported by the positive correlations of S. pavanii with the growth of C. albicans, C. krusei, and C. parasilopsis during inhibitory assays.
Staphylococcus is a common genus in breast milk (Togo et al., 2019), where many species of staphylococci play inhibitory roles against the growth and virulence of S. aureus that are present in breast milk (Heikkilä and Saris, 2003). Considering that the abundance of S. aureus specifically remains similar between the groups, we postulate that the higher abundance of Staphylococcus in breast milk in the WO-group does not pose a health risk to infants. This is supported by a potential protective role of Staphylococcus given its negative correlation with the growth of C. krusei in inhibitory assays.
S. infantis is a non-inflammatory and commensal species commonly reported in breast milk samples, and in paired samples of breast milk and breastfed infants’ oral swabs (Biagi et al., 2017), breast milk and infant feces (Biagi et al., 2017), and mother and infant feces (Kageyama et al., 2022). During vaginal infection which vaginal pH increases, the abundance of S. infantis was reported to decrease along with its replacement by Prevotella species, compared with the findings in women with normal vaginal pH (Xu et al., 2020). Hormone therapy also decreases the abundance of S. infantis in urobiomes of women compared with that of women without hormone therapy (Neugent et al., 2021). While S. infantis is a common oral pathogen, this species often acts as a mediator in pathways to eliminate gut pathogens such as Escherichia coli (He et al., 2014). We postulate that the higher abundance of S. infantis in breast milk may be beneficial for infant immunity, growth, and development.
Taken together, our findings show that the composition of breast milk differs between groups, as an effect of vaginal infection during pregnancy. While some vaginal pathogens may have been passed on via breast milk, the current differences in composition between women with or without vaginal infections during pregnancy may not pose a health threat to infant growth and development, which is mainly attributed to the presence of beneficial and/or protective microbiota to counter potential detrimental impacts. Breast milk is clearly a complex ecosystem, where microbiota is merely a fraction of the entirety that makes it beneficial as a newborn’s first food.
ACKNOWLEDGEMENTS
We thank Probionic Corp., Korea, and Universiti Sains Malaysia (USM) for funding this project.
Footnotes
FUNDING
This work is supported by a USM-Probionic Grant (grant no. 304/PTEKIND/6501096) and a USM-Industry Re-search Matching Grant (grant no. 1001/PTEKIND/8070012).
AUTHOR DISCLOSURE STATEMENT
The authors declare no conflict of interest.
AUTHOR CONTRIBUTIONS
Concept and design: GL, MTL, CEO. Analysis and interpretation: AAN, GL, CEO, FZ, DR, FFR, WSJ, MTL. Data collection: AAN, SS, VB, JJT, NSMN, ZZD. Writing the article: AAN, MTL. Critical revision of the article: HZ, SDT, MTL, YHP. Final approval of the article: all authors. Statistical analysis: AAN, MTL. Overall responsibility: MTL.
REFERENCES
- Begum N, Muazzam N, Shamsuzzaman SM, Islam MDU, Chowdhury AK, Begum SA. Prevalence of bacterial vaginosis among the PID patients in Bangladesh. Faridpur Med Coll J. 2011;6:10–13. doi: 10.3329/fmcj.v6i1.7403. [DOI] [Google Scholar]
- Bernardet JF, Vancanneyt M, Matte-Tailliez O, Grisez L, Tailliez P, Bizet C, et al. Polyphasic study of Chryseobacterium strains isolated from diseased aquatic animals. Syst Appl Microbiol. 2005;28:640–660. doi: 10.1016/j.syapm.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Biagi E, Quercia S, Aceti A, Beghetti I, Rampelli S, Turroni S, et al. The bacterial ecosystem of mother's milk and infant's mouth and gut. Front Microbiol. 2017;8:1214. doi: 10.3389/fmicb.2017.01214. https://doi.org/10.3389/fmicb.2017.01214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention, author. STDs during preg-nancy-CDC detailed fact sheet. 2022. [cited 2022 Oct 30]. Available from: https://www.cdc.gov/std/pregnancy/stdfact-pregnancy-detailed.htm .
- Chong HX, Yusoff NAA, Hor YY, Lew LC, Jaafar MH, Choi SB, et al. Lactobacillus plantarum DR7 improved upper respiratory tract infections via enhancing immune and inflammatory parameters: A randomized, double-blind, placebo-controlled study. J Dairy Sci. 2019;102:4783–4797. doi: 10.3168/jds.2018-16103. [DOI] [PubMed] [Google Scholar]
- Duale A, Singh P, Al Khodor S. Breast milk: a meal worth having. Front Nutr. 2022;8:800927. doi: 10.3389/fnut.2021.800927. https://doi.org/10.3389/fnut.2021.800927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Fredricks DN. Microbial ecology of human skin in health and disease. J Investig Dermatol Symp Proc. 2001;6:167–169. doi: 10.1046/j.0022-202x.2001.00039.x. [DOI] [PubMed] [Google Scholar]
- Fernández L, Pannaraj PS, Rautava S, Rodríguez JM. The microbiota of the human mammary ecosystem. Front Cell Infect Microbiol. 2020;10:586667. doi: 10.3389/fcimb.2020.586667. https://doi.org/10.3389/fcimb.2020.586667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung WY, Liong MT. Evaluation of proteolytic and ACE-inhibitory activity of Lactobacillus acidophilus in soy whey growth medium via response surface methodology. LWT. 2010;43:563–567. doi: 10.1016/j.lwt.2009.10.004. [DOI] [Google Scholar]
- Haddad EN, Sugino KY, Kerver JM, Paneth N, Comstock SS. The infant gut microbiota at 12 months of age is associated with human milk exposure but not with maternal pre-pregnancy body mass index or infant BMI-for-age z-scores. Curr Res Physiol. 2021;4:94–102. doi: 10.1016/j.crphys.2021.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, McLean JS, Guo L, Lux R, Shi W. The social structure of microbial community involved in colonization resistance. ISME J. 2014;8:564–574. doi: 10.1038/ismej.2013.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikkilä MP, Saris PE. Inhibition of Staphylococcus aureus by the commensal bacteria of human milk. J Appl Microbiol. 2003;95:471–478. doi: 10.1046/j.1365-2672.2003.02002.x. [DOI] [PubMed] [Google Scholar]
- Hermansson H, Kumar H, Collado MC, Salminen S, Isolauri E, Rautava S. Breast milk microbiota is shaped by mode of delivery and intrapartum antibiotic exposure. Front Nutr. 2019;6:4. doi: 10.3389/fnut.2019.00004. https://doi.org/10.3389/fnut.2019.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hor YY, Lew LC, Jaafar MH, Lau AS, Ong JS, Kato T, et al. Lactobacillus sp. improved microbiota and metabolite profiles of aging rats. Pharmacol Res. 2019;146:104312. doi: 10.1016/j.phrs.2019.104312. https://doi.org/10.1016/j.phrs.2019.104312. [DOI] [PubMed] [Google Scholar]
- Howard A, O'Donoghue M, Feeney A, Sleator RD. Acinetobacter baumannii: an emerging opportunistic pathogen. Virulence. 2012;3:243–250. doi: 10.4161/viru.19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kageyama S, Furuta M, Takeshita T, Ma J, Asakawa M, Yamashita Y. High-level acquisition of maternal oral bacteria in formula-fed infant oral microbiota. mBio. 2022;13:e0345221. doi: 10.1128/mbio.03452-21. https://doi.org/10.1128/mbio.03452-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzaka T, Tani K. Draft genome sequence of multidrug-resistant Stenotrophomonas pavanii BWK1, isolated from Mareca penelope feces. Genome Announc. 2018;6:e00187–18. doi: 10.1128/genomeA.00187-18. https://doi.org/10.1128/genomeA.00187-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein-Jöbstl D, Quijada NM, Dzieciol M, Feldbacher B, Wagner M, Drillich M, et al. Microbiota of newborn calves and their mothers reveals possible transfer routes for newborn calves' gastrointestinal microbiota. PLoS One. 2019;14:e0220554. doi: 10.1371/journal.pone.0220554. https://doi.org/10.1371/journal.pone.0220554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen MF. Gut microbiota development: influence of diet from infancy to toddlerhood. Ann Nutr Metab. 2021;77:21–34. doi: 10.1159/000517912. [DOI] [PubMed] [Google Scholar]
- Leung MHY, Tong X, Bastien P, Guinot F, Tenenhaus A, Appenzeller BMR, et al. Changes of the human skin microbiota upon chronic exposure to polycyclic aromatic hydrocarbon pollutants. Microbiome. 2020;8:100. doi: 10.1186/s40168-020-00874-1. https://doi.org/10.1186/s40168-020-00874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Chong HX, Chung FY, Li Y, Liong MT. Lactobacillus plantarum DR7 modulated bowel movement and gut microbiota associated with dopamine and serotonin pathways in stressed adults. Int J Mol Sci. 2020;21:4608. doi: 10.3390/ijms21134608. https://doi.org/10.3390/ijms21134608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundgren SN, Madan JC, Karagas MR, Morrison HG, Hoen AG, Christensen BC. Microbial communities in human milk relate to measures of maternal weight. Front Microbiol. 2019;10:2886. doi: 10.3389/fmicb.2019.02886. https://doi.org/10.3389/fmicb.2019.02886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lye HS, Kato T, Low WY, Taylor TD, Prakash T, Lew LC, et al. Lactobacillus fermentum FTDC 8312 combats hypercholesterolemia via alteration of gut microbiota. J Biotechnol. 2017;262:75–83. doi: 10.1016/j.jbiotec.2017.09.007. [DOI] [PubMed] [Google Scholar]
- Maqsood R, Reus JB, Wu LI, Holland LA, Nduati R, Mbori-Ngacha D, et al. Breast milk virome and bacterial microbiome resilience in Kenyan women living with HIV. mSystems. 2021;6:e01079–20. doi: 10.1128/mSystems.01079-20. https://doi.org/10.1128/mSystems.01079-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin CR, Ling PR, Blackburn GL. Review of infant feeding: key features of breast milk and infant formula. Nutrients. 2016;8:279. doi: 10.3390/nu8050279. https://doi.org/10.3390/nu8050279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugent ML, Kumar A, Hulyalkar NV, Lutz KC, Nguyen VH, Fuentes JL, et al. Recurrent urinary tract infection and estrogen shape the taxonomic ecology and functional potential of the postmenopausal urobiome. bioRxiv. 2021 doi: 10.1101/2021.11.06.467345. https://doi.org/10.1101/2021.11.06.467345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisaa AA, Oon CE, Sreenivasan S, Balakrishnan V, Tan JJ, Teh CS, et al. Breast milk from healthy women has higher anti-Candida properties than women with vaginal infections during pregnancy. Food Sci Biotechnol. 2022 doi: 10.1007/s10068-022-01088-x. https://doi.org/10.1007/s10068-022-01088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar A, Camara M, Fall S, Ngom-Cisse S, Fall B, Ba-Diallo A, et al. Chryseobacterium indologenes in a woman with acute leukemia in Senegal: a case report. J Med Case Rep. 2014;8:138. doi: 10.1186/1752-1947-8-138. https://doi.org/10.1186/1752-1947-8-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pak TR, Altman DR, Attie O, Sebra R, Hamula CL, Lewis M, et al. Whole-genome sequencing identifies emergence of a quinolone resistance mutation in a case of Stenotrophomonas maltophilia bacteremia. Antimicrob Agents Chemother. 2015;59:7117–7120. doi: 10.1128/AAC.01723-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SH, Vaidya YH, Patel RJ, Pandit RJ, Joshi CG, Kunjadiya AP. Culture independent assessment of human milk microbial community in lactational mastitis. Sci Rep. 2017;7:7804. doi: 10.1038/s41598-017-08451-7. https://doi.org/10.1038/s41598-017-08451-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasoodanan PKV, Sharma AK, Mahajan S, Dhakan DB, Maji A, Scaria J, et al. Western and non-western gut microbiomes reveal new roles of Prevotella in carbohydrate metabolism and mouth-gut axis. NPJ Biofilms Microbiomes. 2021;7:77. doi: 10.1038/s41522-021-00248-x. https://doi.org/10.1038/s41522-021-00248-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F. The prokaryotes: gammaproteobacteria. 4th ed. Springer; Berlin, Germany: 2014. pp. 443–476. [Google Scholar]
- Spicer SK, Moore RE, Lu J, Guevara MA, Marshall DR, Manning SD, et al. Antibiofilm activity of human milk oligosaccharides against multidrug resistant and susceptible isolates of Acinetobacter baumannii. ACS Infect Dis. 2021;7:3254–3263. doi: 10.1021/acsinfecdis.1c00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Superti F, De Seta F. Warding off recurrent yeast and bacterial vaginal infections: lactoferrin and lactobacilli. Microorganisms. 2020;8:130. doi: 10.3390/microorganisms8010130. https://doi.org/10.3390/microorganisms8010130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tham CSC, Peh KK, Bhat R, Liong MT. Probiotic properties of bifidobacteria and lactobacilli isolated from local dairy products. Ann Microbiol. 2012;62:1079–1087. doi: 10.1007/s13213-011-0349-8. [DOI] [Google Scholar]
- Togo A, Dufour JC, Lagier JC, Dubourg G, Raoult D, Million M. Repertoire of human breast and milk microbiota: a systematic review. Future Microbiol. 2019;14:623–641. doi: 10.2217/fmb-2018-0317. [DOI] [PubMed] [Google Scholar]
- Turroni F, Milani C, Duranti S, Lugli GA, Bernasconi S, Margolles A, et al. The infant gut microbiome as a microbial organ influencing host well-being. Ital J Pediatr. 2020;46:16. doi: 10.1186/s13052-020-0781-0. https://doi.org/10.1186/s13052-020-0781-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vuillermin PJ, O'Hely M, Collier F, Allen KJ, Tang MLK, Harrison LC, et al. Maternal carriage of Prevotella during pregnancy associates with protection against food allergy in the offspring. Nat Commun. 2020;11:1452. doi: 10.1038/s41467-020-14552-1. https://doi.org/10.1038/s41467-020-14552-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Medical Association, author. World Medical Association Declaration of Helsinki-Ethical principles for medical research involving human subjects. Bull World Health Organ. 2001;79:373–374. [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bian G, Zheng M, Lu G, Chan WY, Li W, et al. Fertility factors affect the vaginal microbiome in women of reproductive age. Am J Reprod Immunol. 2020;83:e13220. doi: 10.1111/aji.13220. https://doi.org/10.1111/aji.13220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng S, Lin K, Xu X, Lv L, Zhao Z, et al. Effects of infant formula supplemented with prebiotics and OPO on infancy fecal microbiota: a pilot randomized clinical trial. Front Cell Infect Microbiol. 2021;11:650407. doi: 10.3389/fcimb.2021.650407. https://doi.org/10.3389/fcimb.2021.650407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann P, Curtis N. Breast milk microbiota: A review of the factors that influence composition. J Infect. 2020;81:17–47. doi: 10.1016/j.jinf.2020.01.023. [DOI] [PubMed] [Google Scholar]