Abstract
Since the early 1940s, androgen ablation has been the cornerstone of treatment for prostate cancer (PC). Importantly, androgen receptor (AR) signaling is vital not only for the initiation of PC, which is initially androgen-dependent, but also for castration-resistant disease. Recent studies demonstrated clear promise of the poly(ADP-ribose) Polymerase 1 (PARP-1) inhibitors for targeting prostate cancer cells harboring mutations in DNA damage-repair genes. In addition, it has been established that PARP-1 inhibition suppresses growth of AR-positive prostate cancer cells in cell and animal models. Thus, prostate cancer represents a particularly promising disease site for targeting PARP-1, given that both DNA repair and AR-mediated transcription depend on PARP-1 function. Here, we describe the development and use of cell based assay to evaluate the impact of PARP-1 inhibitors on the AR signaling in prostate cancer cells.
Keywords: Prostate, cancer, PARP-1, androgen receptor
1. INTRODUCTION
The burden of PC is tremendous. It is estimated that about 268,000 men will be diagnosed with PC, and more than 34,000 men will die from this disease in the United States alone in 2022 [1]. Therefore, development of curative therapeutic agents for the treatment of prostate cancer represents an urgent clinical need. AR signaling is regarded as the main oncogenic driver in prostate carcinogenesis [2–6]. Thus, modulation of AR activation and/or expression represents a logical target for PC prevention and treatment. Contemporary therapeutic agents, such as abiraterone acetate (Zytiga) and enzalutamide (Xtandi) have shown impressive results in pre- and post-chemotherapy settings, prolonging the survival of patients with castration-resistant PC (CRPC) [7–10]. However, nearly all patients ultimately develop a hormone-resistant form of PC [11–14]. Also, AR-mediated functions are not completely abrogated by existing and emerging hormone therapies. Therapeutic failure is accompanied by various molecular alterations including increased expression of AR and/or androgens, stimulation of ligand-independent AR activity and the expression of constitutively active AR splice variants (AR-Vs) lacking the ligand-binding domain [2,15,16]. PC cells expressing AR-Vs are ultimately resistant to conventional antiandrogens. Current literature and some clinical trials already concluded now show clear promise of the poly(ADP-ribose) Polymerase 1 (PARP-1) inhibitors for the treatment of PC tumors harboring mutations in DNA damage-repair genes [17,18]. In addition, recent studies have established that PARP-1 activity is required for AR function, as well as generation and maintenance of the castration resistant phenotype in PC cells [19]. Indeed, PARP-1 inhibition suppresses growth of AR-positive prostate cancer cells in both in vitro and in vivo [19]. Thus, PC represents a particularly promising disease site for targeting PARP-1, given that both DNA repair and AR-mediated transcription depend on PARP-1 function.
2. MATERIALS
-
2.1
Androgen-dependent LNCaP, castration-resistant C4–2 PC cell lines (ATCC) or any other AR- positive PC cell lines.
-
2.2
Culture flasks (T25 or T75)
-
2.3
96-well white opaque and 6- or 12 well clear cell culture treated plates
-
2.4
Plastic pipettes (1, 5,10, and 25 ml)
-
2.5
Pipette tips (10, 200, and 1000 ul)
-
2.6
Centrifuge tubes (15 or 50ml)
-
2.7
Single and multichannel pipettes
-
2.8
Motorized Pipette
-
2.9
RPMI 1640 cell culture medium supplemented with penicillin/streptomycin (100 μg/ml), sodium pyruvate (1 mM) and non-essential amino acids (0.1 mM)
-
2.10
Fetal Bovine Serum (FBS) and charcoal-stripped FBS
-
2.11
Phosphate buffered saline (PBS)
-
2.12
AR agonist R1881 (MilliporeSigma)
-
2.13
Firefly AR Response Element Luciferase Reporter and Control Renilla Luciferase Reporter Lentiviral particles (G&P Biosciences, GeneTarget)
-
2.14
Dual-Glo Luciferase Assay System (Promega)
-
2.15
Trypsin-EDTA: 0.25% w/v trypsin and 0.02% w/v EDTA in PBS without Ca2+ and Mg2+
-
2.16
CO2 source
-
2.17
CO2 chamber
-
2.18
Centrifuge
3. METHODS
3.1. Cell preparation
-
3.1.1
Grow the cells in culture flasks (T25 or T75) in RPMI 1640 cell culture medium supplemented with 10% FBS, penicillin/streptomycin (100 μg/ml), sodium pyruvate (1 mM) and non-essential amino acids (0.1 mM).
-
3.1.2
When cells are 70–90% confluent, remove culture medium and wash cells with PBS. Add trypsin-EDTA solution (T25 flask - ~ 0.75 – 1.0 ml; T75 flask - ~ 2.5 ml). Incubate for 2–3 min at 37°C, then shake or tap the flask to detach the cells.
-
3.1.3
Re-suspend the cells in 10 mL of culture medium supplemented with 10% FBS and transfer to a centrifuge tube. Centrifuge at 200g for 10 min, and resuspend the pellet in RPMI 1640 cell culture medium supplemented with 10% charcoal-stripped FBS, penicillin/streptomycin (100 μg/ml), sodium pyruvate (1 mM) and non-essential amino acids (0.1 mM).
-
3.1.4
Quantitate the number of cells using a hemocytometer or any other method.
-
3.1.5
Plate the empirically established number of cells, depending on their doubling time, in 12- or 6-well plates. By the time of transduction, the cells should be 50–60% confluent.
3.2. Cell transduction
-
3.2.1
Establish the minimum concentration of the specific antibiotic that results in complete death of naïve parental cells by adding increasing amounts of the appropriate antibiotic to duplicate wells. Include a no-antibiotic control wells. Incubate the cells for 8–10 days replacing selective medium with or without the specific antibiotic every 3–4 days. Visually examine the culture for signs of toxicity on a daily basis. Establish the minimal concentration of the specific antibiotic that results in complete death of the cells. Choose the antibiotic concentration, which is just 10–15% above the one, which shows complete cell death. This is the optimal antibiotic concentration to use for stable selection.
-
3.2.2
To prepare cells for lentiviral transduction, repeat steps 1 to 5 described in Section 3.1 (Cell preparation). Replace the medium with fresh medium when the cells reach 50–60% confluency. Add polybrene (PB) at final concentration 4–8 μg/ml and AR response element luciferase reporter lentiviral stock (the specific amount depends on the manufacture’s recommendations).
-
3.2.3
Replace the medium with fresh medium after 24 hours. Culture the cells for the additional 24 hours and add the appropriate antibiotic at concentrations established in step 6 for the selection of clones carrying antibiotic resistance gene. Use non-transduced PC cells as a control to monitor antibiotic-mediated cell death. As population of resistant cells becomes confluent, expand the cells into larger plates or flasks. The antibiotic selection should last for as long as it takes the control untransduced cells to completely die.
-
3.2.4
The Renilla luciferase reporter is commonly used as a control in luciferase-based reporter gene assays to normalize the values of the experimental reporter gene for variations that could be caused by transfection efficiency and sample handling. To establish cells stably expressing both firefly and Renilla luciferase reporters, use Renilla luciferase reporter lentivirus carrying resistance gene to a different antibiotic. Repeat steps 1–3 described in Section 3.2 (Cell transduction). As a control use the cells with stable expression of AR response element reporter.
3.3. Testing the effect of PARP-1 inhibitors on the AR transcriptional activity in PC cells
-
3.3.1
Plate 5,000–10,000 PC cells concomitantly expressing AR response element and Renilla luciferase reporters per well in 96-well white opaque plates (total volume 0.1 ml) in complete RPMI 1640 medium supplemented with 10% FBS. Next day, replace medium with RPMI 1640 medium supplemented with 10% charcoal-stripped FBS.
-
3.3.2
Culture the cells for the additional 24–48 hours. Pre-incubate the cells with PARP-1 inhibitor(s) for 30–60 minutes following by stimulation with the synthetic AR agonist R1881 (0.1–2.5nM) for an additional 6–24 hours. The optimal time should be determined empirically.
-
3.3.3
Assay the cells for firefly and Renilla luciferase activities using Dual-Glo Luciferase Assay System and plate reader capable of measuring luminescence intensity.
4. NOTES
-
4.1
Even in the absence of cell death during the antibiotic selection, the cell media should be changed every 2–3 days.
-
4.2
AR reporter activity may be normalized using cell viability assays such as CellTiter-Blue or CellTiter-Glo (Promega) assays. The cells should be treated in parallel under the same conditions.
-
4.3
Cells can be transduced simultaneously with both, firefly (AR reporter) and Renilla (control) containing lentiviruses. If so, the selection should be performed using two antibiotics simultaneously to the transduced cells. A separate control should be used for each antibiotic. The selection can be considered as complete when untransduced cells are completely dead in both controls.
-
4.4
Some cells may be sensitive to the recommended PB concentrations. If cell viability is affected during transduction, PB concentration should be reduced to 2–4 ug/ml.
-
4.5
If all transduced cells die during antibiotic selection, it can indicate a low efficacy of transduction. In this case, the volume (amount) of lentiviral stock used for transduction may be increased as well as a time of transduction.
Figure 1. PARP-1 activity is required for AR function.
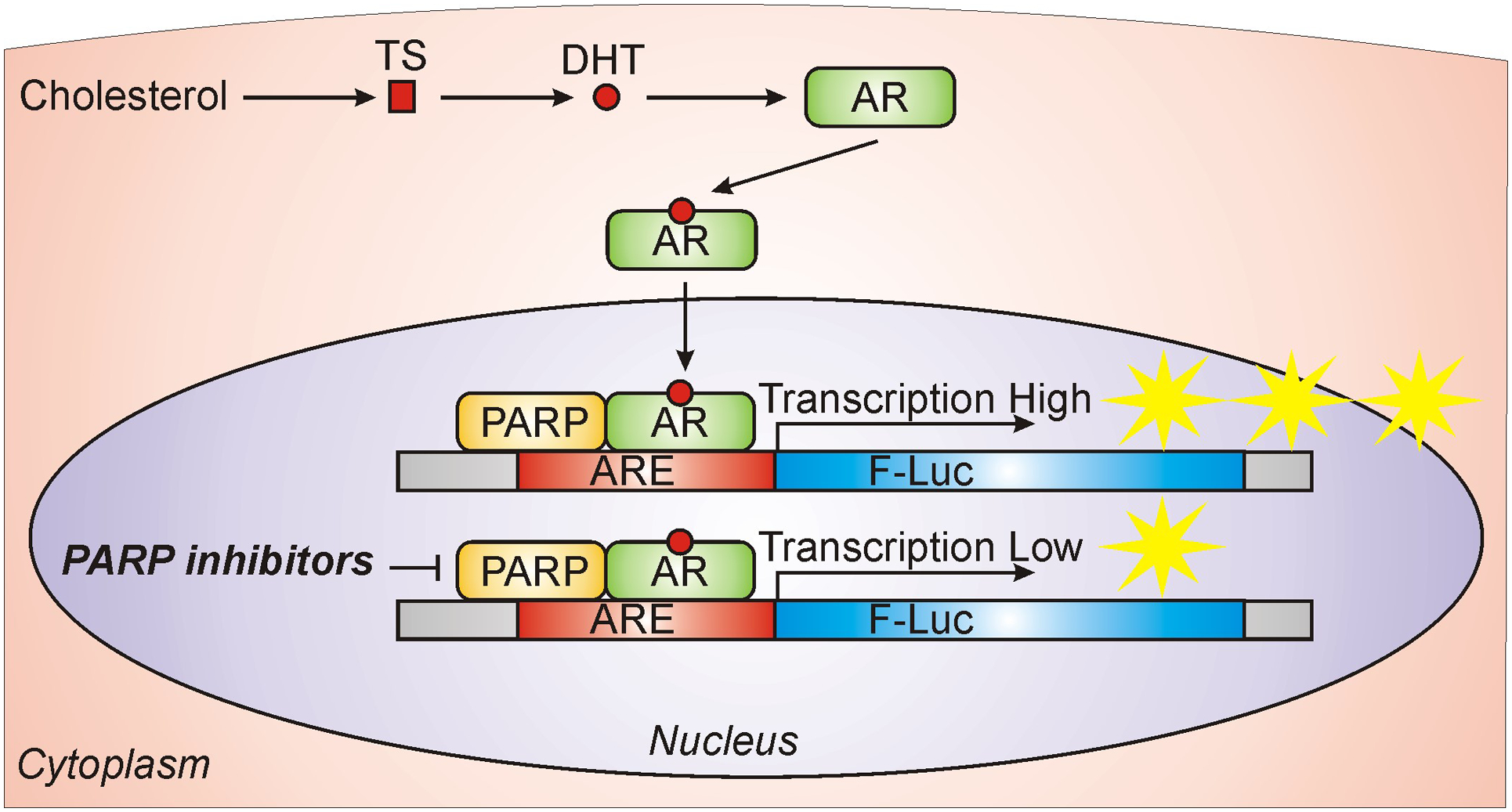
PC is a hormone-sensitive cancer that is influenced by androgens produced from cholesterol such as testosterone (TS) or its 5α-reduced metabolite, dihydrotestosterone (DHT). Pharmacological inhibition of PARP-1 results in diminished AR activity in PC cells.
Figure 2. Schematic representation of generation of PC cells stably expressing ARE driven Firefly luciferase (Fluc) and TATA driven Renilla luciferase (RLuc).
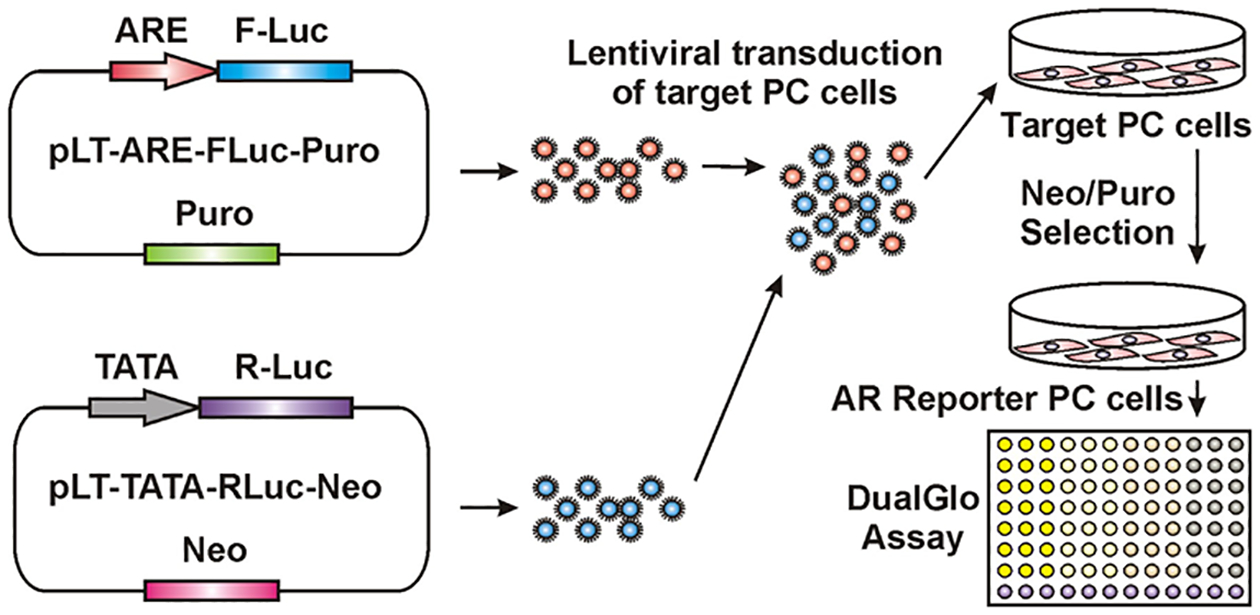
pLT lentiviral vectors contain indicated selection markers, Puromycin (Puro) or Neomycin (Neo).
REFERENCES:
- 1.Siegel RL, Miller KD, Fuchs HE, Jemal A (2022) Cancer statistics, 2022. CA Cancer J Clin 72 (1):7–33. doi: 10.3322/caac.21708 [DOI] [PubMed] [Google Scholar]
- 2.Aragon-Ching JB (2014) The evolution of prostate cancer therapy: targeting the androgen receptor. Front Oncol 4:295. doi: 10.3389/fonc.2014.00295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cai C, Chen S, Ng P, Bubley GJ, Nelson PS, Mostaghel EA, Marck B, Matsumoto AM, Simon NI, Wang H, Chen S, Balk SP (2011) Intratumoral de novo steroid synthesis activates androgen receptor in castration-resistant prostate cancer and is upregulated by treatment with CYP17A1 inhibitors. Cancer Res 71 (20):6503–6513. doi: 10.1158/0008-5472.CAN-11-0532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dehm SM, Tindall DJ (2007) Androgen receptor structural and functional elements: role and regulation in prostate cancer. Mol Endocrinol 21 (12):2855–2863. doi:me.2007–0223 [pii] 10.1210/me.2007-0223 [DOI] [PubMed] [Google Scholar]
- 5.Lonergan PE, Tindall DJ (2011) Androgen receptor signaling in prostate cancer development and progression. J Carcinog 10:20. doi: 10.4103/1477-3163.83937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pisano C, Tucci M, Di Stefano RF, Turco F, Scagliotti GV, Di Maio M, Buttigliero C (2021) Interactions between androgen receptor signaling and other molecular pathways in prostate cancer progression: Current and future clinical implications. Crit Rev Oncol Hematol 157:103185. doi: 10.1016/j.critrevonc.2020.103185 [DOI] [PubMed] [Google Scholar]
- 7.Mori K, Miura N, Mostafaei H, Quhal F, Sari Motlagh R, Pradere B, Kimura S, Kimura T, Egawa S, Briganti A, Karakiewicz PI, Shariat SF (2020) Sequential therapy of abiraterone and enzalutamide in castration-resistant prostate cancer: a systematic review and meta-analysis. Prostate Cancer Prostatic Dis. doi: 10.1038/s41391-020-0222-6 [DOI] [PubMed] [Google Scholar]
- 8.Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Flechon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS, Investigators A (2012) Increased Survival with Enzalutamide in Prostate Cancer after Chemotherapy. New Engl J Med 367 (13):1187–1197. doi: 10.1056/Nejmoa1207506 [DOI] [PubMed] [Google Scholar]
- 9.Attard G, Reid AH, A’Hern R, Parker C, Oommen NB, Folkerd E, Messiou C, Molife LR, Maier G, Thompson E, Olmos D, Sinha R, Lee G, Dowsett M, Kaye SB, Dearnaley D, Kheoh T, Molina A, de Bono JS (2009) Selective inhibition of CYP17 with abiraterone acetate is highly active in the treatment of castration-resistant prostate cancer. J Clin Oncol 27 (23):3742–3748. doi:JCO.2008.20.0642 [pii] 10.1200/JCO.2008.20.0642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Bono JS, Logothetis CJ, Molina A, Fizazi K, North S, Chu L, Chi KN, Jones RJ, Goodman OB Jr., Saad F, Staffurth JN, Mainwaring P, Harland S, Flaig TW, Hutson TE, Cheng T, Patterson H, Hainsworth JD, Ryan CJ, Sternberg CN, Ellard SL, Flechon A, Saleh M, Scholz M, Efstathiou E, Zivi A, Bianchini D, Loriot Y, Chieffo N, Kheoh T, Haqq CM, Scher HI (2011) Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med 364 (21):1995–2005. doi: 10.1056/NEJMoa1014618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buttigliero C, Tucci M, Bertaglia V, Vignani F, Bironzo P, Di Maio M, Scagliotti GV (2015) Understanding and overcoming the mechanisms of primary and acquired resistance to abiraterone and enzalutamide in castration resistant prostate cancer. Cancer Treat Rev 41 (10):884–892. doi: 10.1016/j.ctrv.2015.08.002 [DOI] [PubMed] [Google Scholar]
- 12.Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM (2013) Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res 73 (2):483–489. doi: 10.1158/0008-5472.CAN-12-3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vellky JE, Ricke WA (2020) Development and prevalence of castration-resistant prostate cancer subtypes. Neoplasia 22 (11):566–575. doi: 10.1016/j.neo.2020.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galletti G, Leach BI, Lam L, Tagawa ST (2017) Mechanisms of resistance to systemic therapy in metastatic castration-resistant prostate cancer. Cancer Treatment Reviews 57:16–27. doi: 10.1016/j.ctrv.2017.04.008 [DOI] [PubMed] [Google Scholar]
- 15.Mitsiades N (2013) A road map to comprehensive androgen receptor axis targeting for castration-resistant prostate cancer. Cancer Res 73 (15):4599–4605. doi: 10.1158/0008-5472.CAN-12-4414 0008–5472.CAN-12–4414 [pii] [DOI] [PubMed] [Google Scholar]
- 16.Siegel RL, Miller KD, Jemal A (2016) Cancer statistics, 2016. CA Cancer J Clin 66 (1):7–30. doi: 10.3322/caac.21332 [DOI] [PubMed] [Google Scholar]
- 17.Deshmukh D, Qiu Y (2015) Role of PARP-1 in prostate cancer. American journal of clinical and experimental urology 3 (1):1–12 [PMC free article] [PubMed] [Google Scholar]
- 18.Mateo J, Carreira S, Sandhu S, Miranda S, Mossop H, Perez-Lopez R, Nava Rodrigues D, Robinson D, Omlin A, Tunariu N, Boysen G, Porta N, Flohr P, Gillman A, Figueiredo I, Paulding C, Seed G, Jain S, Ralph C, Protheroe A, Hussain S, Jones R, Elliott T, McGovern U, Bianchini D, Goodall J, Zafeiriou Z, Williamson CT, Ferraldeschi R, Riisnaes R, Ebbs B, Fowler G, Roda D, Yuan W, Wu YM, Cao X, Brough R, Pemberton H, A’Hern R, Swain A, Kunju LP, Eeles R, Attard G, Lord CJ, Ashworth A, Rubin MA, Knudsen KE, Feng FY, Chinnaiyan AM, Hall E, de Bono JS (2015) DNA-Repair Defects and Olaparib in Metastatic Prostate Cancer. N Engl J Med 373 (18):1697–1708. doi: 10.1056/NEJMoa1506859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schiewer MJ, Goodwin JF, Han S, Brenner JC, Augello MA, Dean JL, Liu F, Planck JL, Ravindranathan P, Chinnaiyan AM, McCue P, Gomella LG, Raj GV, Dicker AP, Brody JR, Pascal JM, Centenera MM, Butler LM, Tilley WD, Feng FY, Knudsen KE (2012) Dual roles of PARP-1 promote cancer growth and progression. Cancer Discov 2 (12):1134–1149. doi: 10.1158/2159-8290.CD-12-0120 2159–8290.CD-12–0120 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]


