Abstract
Precise modulation of neuronal activity by neuroactive molecules is essential for understanding brain circuits and behavior. However, tools for highly controllable molecular release are lacking. Here, we developed a photoswitchable nanovesicle with azobenzene-containing phosphatidylcholine (azo-PC), coined ‘azosome’, for neuromodulation. Irradiation with 365 nm light triggers the trans-to-cis isomerization of azo-PC, resulting in a disordered lipid bilayer with decreased thickness and cargo release. Irradiation with 455 nm light induces reverse isomerization and switches the release off. Real-time fluorescence imaging shows controllable and repeatable cargo release within seconds (< 3 s). Importantly, we demonstrate that SKF-81297, a dopamine D1-receptor agonist, can be repeatedly released from the azosome to activate cultures of primary striatal neurons. Azosome shows promise for precise optical control over the molecular release and can be a valuable tool for molecular neuroscience studies.
Keywords: azobenzene, photoswitch, liposome, controlled release, neuromodulation
1. Introduction
Understanding the role of neuroactive compounds in brain circuits and behavior is fundamentally essential in neuroscience. The delivery of neuroactive compounds via permanently implanted cannulae within the brain allows studying the effect of molecules on neural activity and behavior [1], but it may cause neuroinflammation and limit the activities of animals [2]. The remotely controlled release of neuroactive compounds using external stimuli, such as a magnetic field [3], focused ultrasound [4, 5] or light [6], offers new possibilities. Among these methods, light has the benefit of high temporal and spatial resolution. One of the main methods using light is to chemically attach a ‘cage’ to a molecule of interest to block its activity until photoremoval of the protecting group [7, 8]. Caged compounds, such as caged glutamate or γ-aminobutyric acid (GABA), are widely used by neurophysiologists to alter cell signaling in neurons and glia, and have contributed significantly to our understanding of the nervous system [9]. However, caged compounds have several limitations, such as challenges in ensuring biological inertness, solubility, uncaging efficiency [10], and potential off-target activity [11, 12]. Another approach for neuromodulation is to photorelease from nanocarriers based on the photothermal [13–15], photochemical [16–18], or photomechanical effects [19, 20]. However, the photorelease is not temporally controlled and cannot switch off. It remains challenging to develop a fast and photoswitchable nanosystem that responds to a low light intensity without associated side effects (such as heating and generation of reactive oxygen species).
Azobenzene groups have been extensively used to build various bioactive photoswitches, and many have been applied in neuroscience using one-photon and two-photon stimulation [21–28]. Recently, this approach has been expanded to lipids, and a series of photoswitchable lipids have been developed, including a photoswitchable phosphatidylcholine derivative, azo-PC. Azo-PC has enabled optical control of membrane organization and permeability [29–33]. Chander et al. demonstrated light-induced release of the anticancer drug doxorubicin from azo-PC containing lipid nanoparticles in vitro and in vivo [34]. However, it remains unclear whether azo-PC would support highly controllable and photoswitchable release of compounds, which is critical to perform in vivo neuromodulation.
Here we have developed a new class of photoswitchable nanovesicles, coined ‘azosomes’, by incorporating azo-PC into liposome formulations to deliver neuroactive compounds. Azo-PC undergoes reversible isomerization upon irradiation with ultraviolet (U.V.) and blue light, respectively, which rapidly changes the thickness of the lipid bilayer. The light-induced change of the lipid bilayer leads to increased permeability for encapsulated molecules and allows for switching the release of molecules on and off. Short light pulses enable temporally controlled release within seconds (< 3 s) from the photoswitchable azosome, demonstrating that the azosome is a promising tool for neuromodulation.
2. Results and Discussions
2.1. Photophysical properties of azosomes
First, we studied the reversible photoswitching property of azosomes. Figure 1(a) shows that one of the two lipid tails of phosphatidylcholine was modified to incorporate an azobenzene group, which can be isomerized between its cis- and trans-isomers upon the irradiation with U.V. and blue light, respectively. To increase the liposome stability, we mixed azo-PC with 1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[methoxyl(polyethylene glycol)-2000] (DSPE-PEG2000), 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), and cholesterol to prepare photoswitchable liposomes (azosomes) by the thin-film hydration method. Varying the percentage of azo-PC in the formulation leads to a series of azosomes after extruding through membranes with the pore size of 400 nm and 200 nm. The hydrodynamic diameters of these azosomes were around 160 nm (Fig. S1(a) in the Electronic Supplementary Material (ESM)). Cryogenic transmission electron microscopy (Cryo-TEM) images show that azosomes are mostly spherical and unilamellar and had a narrow size distribution (Fig. 1(b) and Fig. S1(b) in the ESM). To investigate the photoisomerization of azo-PC in the azosome, we monitored the ultraviolet-visible (UV-Vis) spectra of the azosome before and after the irradiation of 365 nm and 455 nm light. Trans-azo-PC was a thermally stable isomer, as confirmed by the strong absorption at 326 nm before light irradiation (Fig. 1(c)). Fig. 1(c) also shows that the absorption peak of the azosome at 326 nm decreased, and the peak at 440 nm increased in an irradiation time-dependent manner under 365 nm light. In contrast, the absorption peak at 326 nm increased, and the peak at 440 nm decreased with irradiation time under 455 nm light (Fig. 1(d)). These results indicate that the irradiation of 365 nm light triggered the isomerization of trans-azo-PC to cis-azo-PC. The process was reversed under the irradiation of 455 nm light. The absolute absorbance changes at 326 nm under the irradiation of 365 nm and 455 nm overlapped well (Fig. 1(e)), suggesting that the photoisomerization is reversible. The spontaneous cis-to-trans isomerization in the dark was measured at 22, 37, and 45 °C (Fig. 1(f)–1(h) and Fig. S2 in the ESM). The half-lives (t1/2) of cis-azo-PC in the dark increase with temperature due to the intrinsically exothermic process [35, 36] and were much longer than that (seconds) with the 455 nm irradiation. These results demonstrate that isomerization with 365 nm light should induce the cis state with little reversal to the trans state, unless photostimulated with 455 nm light, providing good temporal control of isomerization. Next, we investigated the effect of photoisomerization of azo-PC on the azosome size. Figure 1(i) and Fig. S1(c) in the ESM show that the azosomes swelled by around 5 nm after the 365 nm light irradiation and can be switched back by the 455 nm light irradiation, suggesting that the reversible isomerization of azo-PC leads to reversible changes in azosome size.
Figure 1.
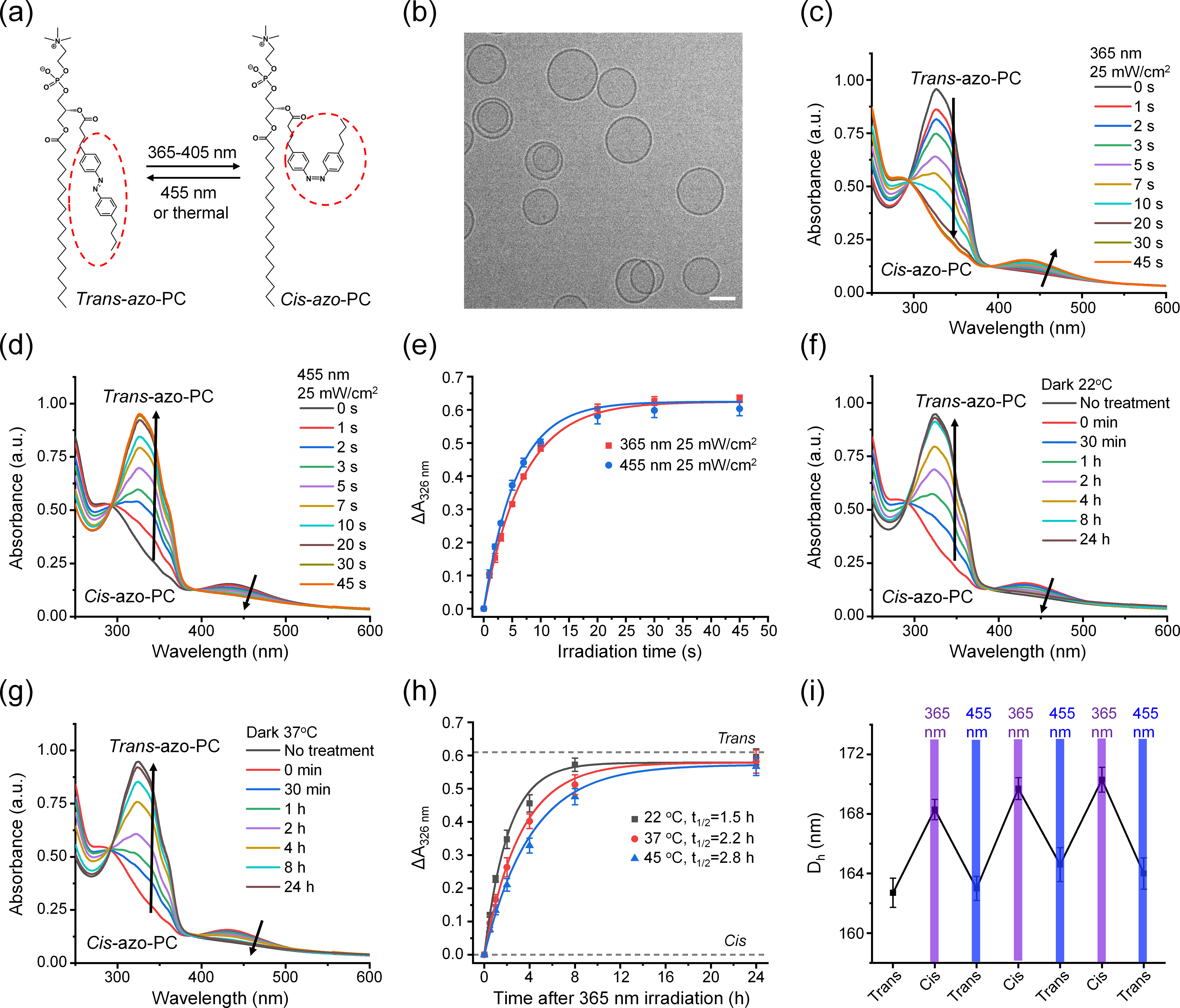
Photophysical properties of asozomes. (a) Schematic of the photoisomerization of azo-PC. (b) Cryo-TEM image of azosomes. Scale bar: 100 nm. (c) and (d) UV–Vis spectra change of azosome upon the irradiation of (c) 365 nm light and (d) 455 nm light. (e) The absorbance change of azosome at 326 and 455 nm light as a function of irradiation time. (f) and (g) UV–Vis spectra change of azosome in the dark at (f) 22 °C and (g) 37 °C. 365 nm light irradiation (25 mW/cm2, 60 s) was performed on the azosome to switch trans-azo-PC to cis-azo-PC at 0 min. (h) The absorbance change of azosome at 326 nm over time in the dark at 22, 37 and 45 °C. The data were fit based on a first-order reaction to give the half-life of cis-azo-PC. (i) The Z-average hydrodynamic diameter of azosome (azo-PC: 25%) measured by dynamic light scattering (DLS) under photoisomerization of azo-PC. 365 or 455 nm light was irradiated on the azosome for 30 s at 25 mW/cm2.
2.2. Computational modeling of azo-PC bilayer
Second, we performed all-atom molecular dynamics simulations to study the azosome bilayer at the molecular level. Simulations of pure azo-PC bilayers revealed distinct structural differences between trans-azo-PC and cis-azo-PC. Figure 2(a) shows that the azobenzene moieties aligned with each other in the trans isomer, resulting in an ordered bilayer structure. In contrast, azobenzenes in the cis isomer were disordered and spanned the bilayer hydrophobic core (Fig. 2(a)). The density profiles across the bilayer confirmed these differences in the molecular arrangements (Fig. 2(b)). In the cis-azo-PC bilayer, the azo functional groups spanned the bilayer hydrophobic core and significantly overlapped with the head group region. Notably, the bilayer thickness, defined as the peak-to-peak distance of the lipid headgroups, was smaller in the cis-azo-PC bilayer than in the trans-azo-PC bilayer. We hypothesized that the disordered distribution of cis-azobenzene groups in the bilayer, the decreased bilayer thickness, and the lack of a methyl trough separating the upper and lower leaflets led to the increased permeability across the cis-azo-PC bilayer. To demonstrate the robustness of our computational approach, we changed the torsional force field parameters of azobenzene to impose either the cis or trans-state every 100 ns, resulting in reversible and reproducible bilayer properties (area per lipid shown in Fig. 2(c)). These reversible bilayer changes are consistent with the experimental findings (Fig. 1(i)). From the pure (100%) azo-PC area per lipid increase upon isomerization, we simply scaled by the azo-PC lipid composition to predict the azosome size increase as a function of azo-PC content. This linear scaling is justified by the linear nature of the experimental data (Fig. 2(d)), and the comparison demonstrates that the simulation and experimental data are in reasonable agreement.
Figure 2.
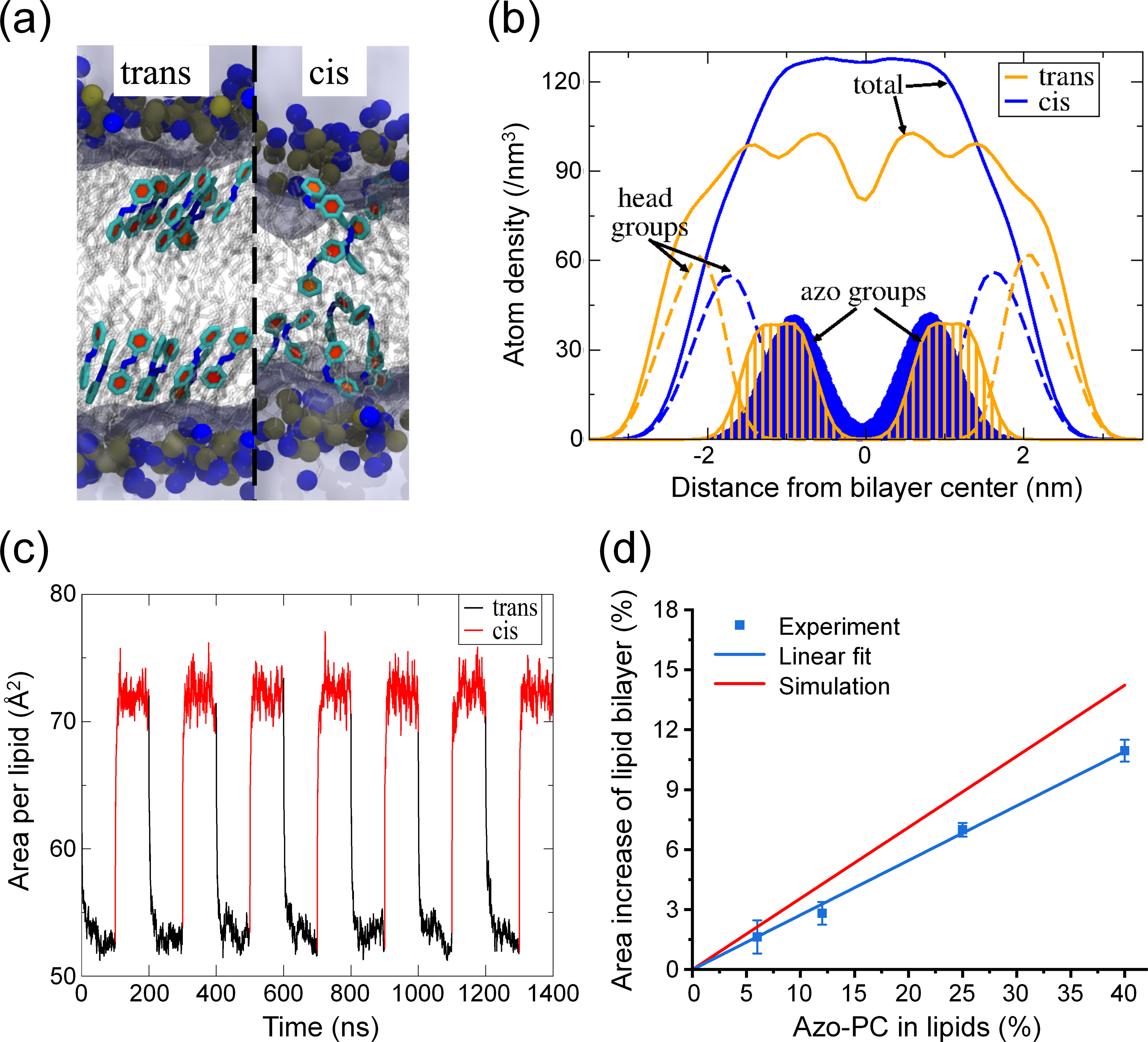
Molecular dynamics simulation for azo-PC lipid bilayer. (a) Snapshots of the trans and cis bilayers. Selected azobenzene groups are highlighted in light cyan with the ring planes in red. Lipid tails are shown in transparent gray. The blue and brown spheres represent nitrogen and phosphorous atoms on the head group, respectively. Water is represented as light purple regions. (b) The lipid atom density profile of the azo-PC bilayer as a function of the distance from the bilayer center. The orange and blue lines represent the trans-azo-PC and cis-azo-PC bilayers. Solid lines: the overall lipid density; dashed lines: lipid head groups; shaded regions: azobenzene groups. (c) Area per lipid in the tensionless ensemble of the azo-PC bilayer when the isomeric state of every lipid is altered between cis and trans every 100 ns. (d) Experimental and simulated azosome area increase with different percentages of azo-PC after the irradiation with 365 nm light (25 mW/cm2, 30 s). Data are expressed as mean ± standard deviation (S.D.) Simulation data were extrapolated to 0% azo-PC from the data in (c).
2.3. Photoswitchable fluorophore release from azosomes
Next, we investigated the photoswitchable release from azosome by alternating 365 and 455 nm light. We hypothesized that the trans-to-cis isomerization under 365 nm light irradiation increased the permeability of the liposome membrane and caused cargo release, while the reversed isomerization by 455 nm light restored the membrane integrity impeding the cargo escape from the azosome and switched off the cargo release (Fig. 3(a)). To test this hypothesis, a self-quenching polyanionic dye, calcein, was encapsulated in the azosome (calcein-azosome). We predicted the fluorescence would increase upon release due to the dequenching of calcein. Minimal calcein dye leakage (< 5%) was observed with azosomes with a percentage of azo-PC up to 25% for 24 h in 0.01 M phosphate saline buffer at room and physiological temperatures, while the 40% azo-PC formulation leads to uncontrolled leakage (Fig. S3 in the ESM). This observation may be due to the low phase transition temperature with a high percentage of azo-PC. The photorelease efficiency increased with the higher azo-PC composition under 365 nm light irradiation (Fig. 3(b)). We selected the calcein-azosome with 12% azo-PC for the following tests due to its high stability and photosensitivity. The photorelease efficiency was highly controllable by the intensity and duration of 365 nm light (Fig. 3(c) and Fig. S4 in the ESM). To explore the kinetics of the calcein release with a high temporal resolution, we recorded the real-time fluorescence of calcein using a high-speed camera. The results show that short irradiation (0.1–0.5 s) at 365 nm triggered a fast release of calcein from azosome within 1–2 s, followed by a slow-phase release that lasted 10–20 s (Fig. 3(d) and Fig. S5 in the ESM). To interrupt the slow phase of release, we applied a second irradiation at 455 nm, which terminated the release immediately (Fig. 3(d)). Furthermore, the amount of released calcein was controllable by the pattern of the sequential irradiations, as shown by the higher amount of release with a 0.9 s delay between 365 and 455 nm light (Fig. 3(d)). Interestingly, 1 s of 455 nm irradiation was sufficient to switch off the release triggered by 0.1 s of 365 nm light (Fig. 3(d)), while 2 s of 455 nm irradiation was necessary to give a complete switch-off under longer 365 nm irradiation (0.2–0.5 s, Fig. S5 in the ESM). The longer irradiation time at 455 nm compared to 365 nm could be due to the strong absorption of calcein at 455 nm (Fig. S6 in the ESM). These results suggest the ability to temporally control the release within seconds (< 3 s). In addition, the photoswitchable property allows for step-wise release by multiple pulses of sequential irradiations of 365 nm and 455 nm light (Fig. 3(e)). These results demonstrate that the photoswitchable azosome offers a platform for on-demand and repeatable release with both “on” and “off” switches.
Figure 3.
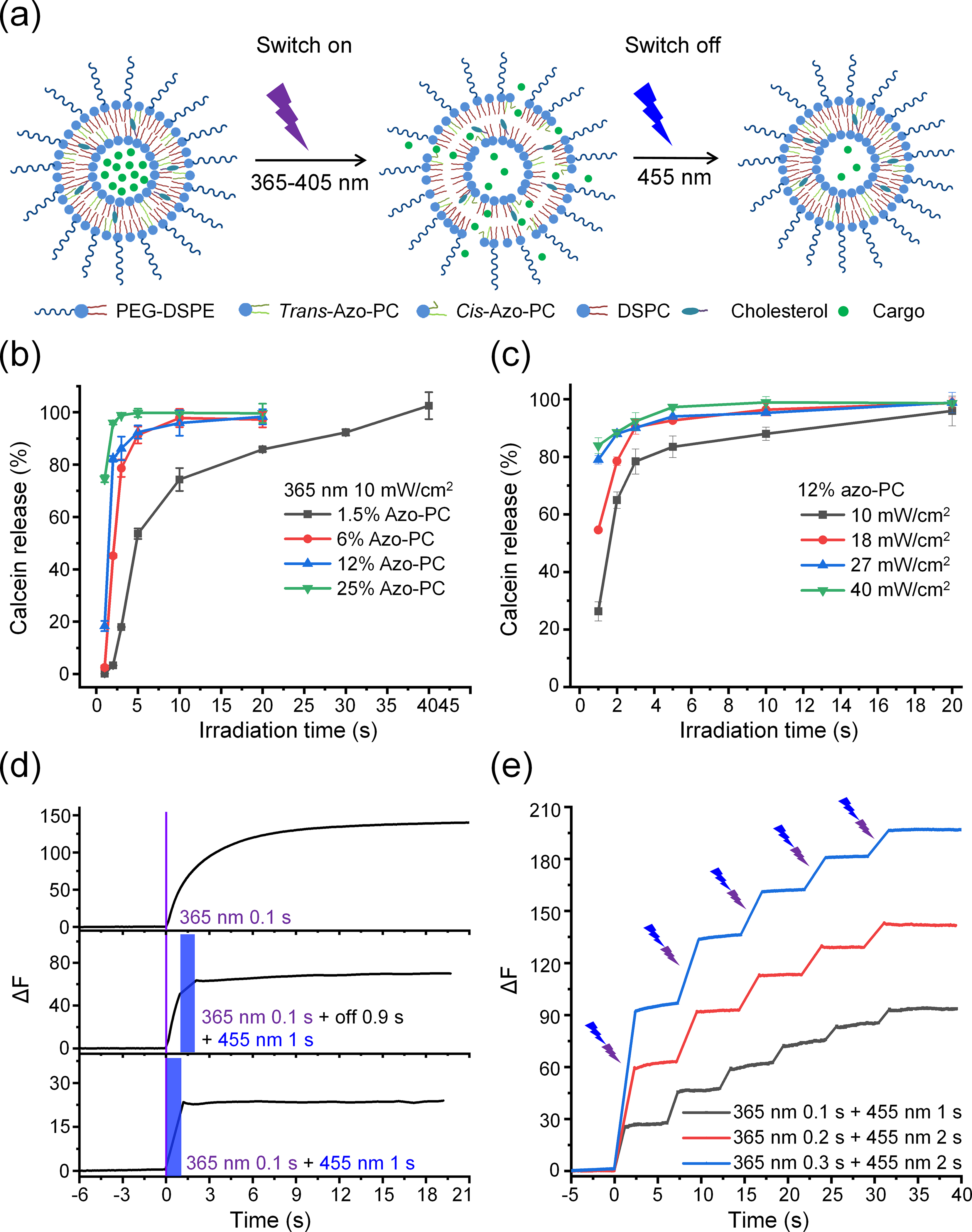
Photoswitchable fluorophore release from asozomes. (a) Schematic of the photoswitchable release from azosome. (b) Plot of calcein release efficiency of azosomes with different percentages of azo-PC upon 365 nm light irradiation (10 mW/cm2). (c) Plot of calcein release efficiency from azosomes at different 365 nm light intensities and durations. Calcein fluorescence was measured 30 s after irradiation in (b) and (c). (d) Real-time fluorescence intensity of calcein (ΔF) plotted over time under the sequential irradiations of 365 and 455 nm light (40 mW/cm2). The purple and blue rectangles indicate 365 and 455 nm light irradiation, respectively. (e) Real-time fluorescence intensity of calcein (ΔF) plotted over time under multiple cycles of irradiations. The purple and blue arrows indicate the sequential irradiations of 365 and 455 nm light every 5 s (40 mW/cm2). Data are expressed as mean ± S.D.
2.4. Photoswitchable neuromodulator release from azosomes
Lastly, we tested the application of azosome for neuromodulation. Here we encapsulated SKF-81297, a dopamine D1-like receptor agonist, in the azosome (SKF-azosome, Fig. 4(a)). Approximately half of the striatal neuron population expresses the D1 receptor [37]. We predicted that the release of SKF-81297 would activate the D1 receptor on striatal neurons and increase intracellular Ca2+ and neuronal excitability [38, 39]. Primary striatal neurons were cultured on glass-bottom dishes and used for Ca2+ imaging after the neurons had matured (14 days). We first stained neurons with Ca2+-sensitive dye Fluo-4 as a proxy for neural activity and applied Dil-labeled azosomes to the extracellular medium. After 1 h, there was no evidence of significant endocytosis of azosome by neurons, suggesting that azosomes remain predominantly extracellular (Fig. S7 in the ESM). We confirmed that the same irradiation time of 455 as 365 nm light could switch cis-azo-PC back to trans-azo-PC in the SKF-azosome (Fig. S8 in the ESM). After the first sequential irradiations at 365 and 455 nm, 29% of the neurons incubated with SKF-azosomes exhibited an increase in fluorescence, indicating an increase in intracellular Ca2+ (as identified by the average ΔF/F0 > (mean + 3δ) of baseline, Figs. 4(b) and (c)). The second and third irradiations activated 20% and 30% of neurons, respectively, with increased irradiation time. For control, we examined the effect of irradiating SKF-liposomes lacking azo-PC and observed no change in fluorescence (Fig. 4(d)). We also investigated any potential cytotoxicity; a live/dead cell viability assay indicated no significant toxicity of azosomes or the irradiation paradigm (Fig. S9 in the ESM). These results demonstrate that azosome is a useful platform for the controlled and repeated release of neurochemicals for neuromodulation.
Figure 4.
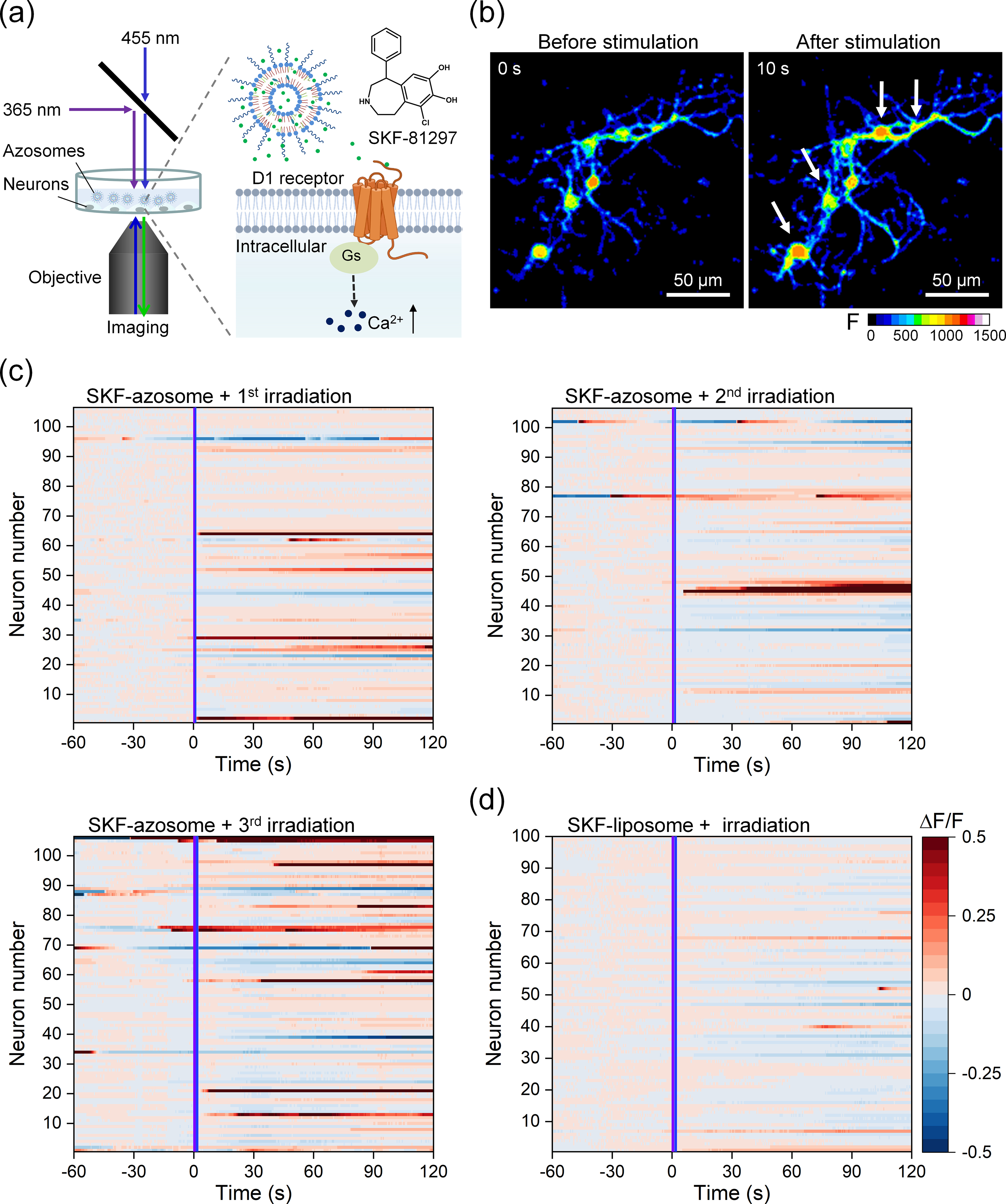
Controlled release of a D1 receptor agonist for neuromodulation. (a) Schematic photorelease of SKF-81297 from the azosome to induce Ca2+ elevation in primary mouse striatal neurons. (b) Real-time fluorescent images of primary mouse striatal neurons before and after the irradiation of sequential 365 nm light and 455 nm light. Fluo-4 was used as the Ca2+ indicator. Scale bar: 50 μm. (c) and (d) The fluorescence change (ΔF/F) plotted as a function of time from individual neurons incubated with (c) SKF-azosome or (d) SKF-liposome. The irradiation of 365 and 455 nm light was performed during the imaging at 0 s. Three repeated irradiations (25 mW/cm2) were performed on SKF-azosome with an interval of 10 min (1st, 365 nm 0.3 s + 455 nm 0.3 s; 2nd, 365 nm 0.5 s + 455 nm 0.5 s; 3rd, 365 nm 1 s + 455 nm 1 s). The irradiation condition (25 mW/cm2) in (d) was 365 nm 1 s + 455 nm 1 s. The activated neuron was counted if the mean fluorescence change (ΔF/F) after stimulation was larger than the mean + 3δ (δ: standard deviation) of baseline. The neurons were labeled in the same order in (c).s
Near-infrared (NIR) light is a particularly attractive option for uncaging or photorelease in the brain due to the deep tissue penetration and reduced phototoxicity [15, 19]. Nanotransducers with NIR light have enabled remote neuromodulation in freely moving mice [40–43]. With the emerging red-azo-PC that can undergo photoisomerization at a longer wavelength (≥ 630 nm) [30], the azosome is expected to be photoswitchable under the irradiation of tissue-penetrating red light. Combined with upconversion nanoparticles which can convert NIR light into emissions in the UV/Vis region [44, 45], the azosome will be able to perform the on-off release by NIR irradiation. These possibilities are being explored to broaden the application of azosomes.
3. Conclusion
In summary, we developed a photoswitchable nanovesicle, referred to as ‘azosome’, based on the reversible photoisomerization of azo-PC under the irradiation of 365 nm and 455 nm light. The trans-to-cis isomerization triggered by 365 nm light decreases the thickness of the liposome bilayer, increases the azosome diameter, and leads to efficient cargo release. In contrast, the reversed cis-to-trans isomerization induced by 455 nm light increases the thickness of the liposome bilayer, decreases the azosome diameter, and switches off the release. We demonstrate that photorelease is controllable within seconds (< 3 s), is repeatable, and can specifically activate targeted neurons by releasing the dopamine D1-like receptor agonist SKF-81297. The azosome formulation shows promise for precise optical control over the release of neuroactive compounds for neuromodulation and can be a valuable tool for molecular neuroscience studies.
4. Experimental methods
4.1. Materials
DSPC (> 99%), DSPE- PEG2000 (> 99%) and cholesterol (ovine wool, >98%) were purchased from Avanti Polar lipids, Inc. Calcein sodium salt (644.5 g/mol, 108750-13-6) was purchased from Alfa Aesar. SKF-81297 hydrobromide (370.67 g/mol, ≥ 98%) was purchased from Tocris Bioscience. 1,1’-Dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine perchlorate (DilC18(3)), ≥ 90%) was purchased from Fisher Scientific. All other chemicals were analytical grade.
4.2. Preparation and characterization of azosome
Preparation of different azo-PC-containing liposomes was performed by the thin-film hydration method reported previously [19, 20]. DSPC, PEG-DSPE, cholesterol, and azo-PC were mixed in chloroform at the molar ratio of 57: 1: 30: 12. The lipid film was prepared with N2 blow for several minutes and dried under vacuum overnight. 10 mM phosphate-buffered saline (PBS) containing either calcein (75 mM) or sulfate ammonium (300 mM) was added to the film for 1 h at 65 °C. After 5 freeze-thaw cycles (1 min in liquid N2 and 2 min in a 65 °C water bath, respectively), the liposome solution was extruded through 400 and 200 nm polycarbonate membranes (Whatman, USA) for 11 passages each using a Mini Extruder (Avanti Polar Lipids, USA). Afterward, free calcein or sulfate ammonium was removed by size exclusion chromatography with a Sephacryl® S-500 HR column (Cat. # GE17-0613-10, Sigma-Aldrich). The calcein-loaded azosome was used directly. To load the SKF-81297, the sulfate ammonium-loaded azosome was incubated with 1 mM SKF-81297 at 70 °C for 30 min. SKF-81287 loaded azosome was further purified by dialysis. The molar percentage of azo-PC in the azosome was adjusted to 1.5%, 6%, 12%, 25%, and 40% to investigate its effect on release efficiency, and the percentage of DSPC was changed accordingly. As a control group, SKF-81297 loaded liposome (without azo-PC) was prepared by similar methods. 0.5% of DilC18(3) was mixed with lipids to label azosome (Dil-azosome).
The hydrodynamic size, size distribution, and zeta-potential of the liposomes were determined by dynamic light scattering measurement (Malvern Zetasizer Nano ZS) at room temperature. A DU800 spectrophotometer (Beckman Coulter) was used to track the UV-Vis absorption spectrum change of azo-PC before and after the photoisomerization. The morphology of liposomes was observed by a transmission electron microscope with a FEI Tecnai 300kV field emission gun and a Gatan Summit K2 direct electron detector camera at the Characterization Facility, University of Minnesota.
4.3. In vitro photoisomerization and release
To investigate the photoisomerization of azo-PC, 100 μL of empty azosome was added to a 96-well plate and irradiated by 365 and 455 nm light (25 mW/cm2). Afterward, the UV-Vis absorption spectra of azosome before and after irradiation were recorded immediately. To investigate the release efficiency of azosome, calcein-loaded azosome was added to a 96-well plate and irradiated by 365 and 455 nm light at a series of intensities or durations. The fluorescence intensity (485 nm excitation and 520 nm emission) was measured using a microplate reader (Synergy H2). For 100% release, azosomes were treated with 0.5% Triton X-100. The percentage release of calcein was calculated by the following formula:
| (1) |
where F0, Ft, and Ftotal represent the initial fluorescence signal, fluorescence signal after light irradiation, and after being treated by Triton, respectively.
The release kinetics of azosome was investigated following our previously reported method [20]. Briefly, an aliquot of well-dispersed calcein-loaded azosome was placed on a glass slide, covered by a cover slide, and sealed by nail polish. The samples were then immobilized onto a microscope (Olympus IX73) stage and irradiated by 365 and 455 nm light. A high-speed digital camera (Hamamatsu Photonics, ORCA-Flash 4.0) was used to record the real-time fluorescent intensity profile every 50 ms. A series of fluorescent images were obtained, and the fluorescence intensity was analyzed by Image J.
4.4. Primary neuron culture and neuromodulation
Cryopreserved primary mouse neurons derived from the striatum of day 18 embryonic CD1 mouse brain (Cat. # M8812N-10) and the culture kit (Cat. # M8812NK-10) were purchased from Cell Applications, Inc. The glass-bottom dish (diameter of glass: 10 mm) was treated with a neuron coating solution in the kit overnight and rinsed with PBS before seeding. After thawing, the cryopreserved primary mouse neurons were transferred to a 50 mL tube containing a 5 mL mouse neuron plating medium. The neurons were then aliquoted into 8 dishes (only on the glass) to ensure the seeding density is 100,000 cells per cm2 or above and cultured in a cell culture incubator (37 °C, 5% CO2, humidified). Half of the mouse neuron culture medium was changed every three days. After 2 weeks, the neurons were incubated with Fluo-4 AM (Cat. # F14201, Molecular ProbesTM) at 3 μM for 30 min and then washed with PBS three times. 300 uL of artificial cerebrospinal fluid (ACSF, 124 mM NaCl, 5 mM KCl, 26 mM NaHCO3, 1.25 mM NaH2PO4, 10 mM D-glucose, 1.3 mM MgCl2 and 1.5 mM CaCl2) containing SKF-81297 loaded azosome (12% azo-PC, total lipids: 2 mM) was added into the glass area in the dish. The fluorescence of Fluo-4 in the neurons was recorded by a spinning disk microscope (Olympus, SD-OSR) at the excitation of 488 nm. During the recording, the whole glass area was irradiated by sequential 365 nm light and 455 nm light to control the release of SKF-81297. Three repeated irradiations were performed on SKF-azosome (25 mW/cm2; 1st, 365 nm 0.3 s + 455 nm 0.3 s; 2nd, 365 nm 0.5 s + 455 nm 0.5 s; 3rd, 365 nm 1 s + 455 nm 1 s). The fluorescence intensity change (ΔF/F0) from individual neurons was analyzed by Image J. F0 value of each neuron was measured by averaging its fluorescent intensity during the 60 s before the irradiation (baseline). The activated neuron was counted if the average fluorescence change (ΔF/F) after stimulation was larger than the mean+3δ (δ: standard deviation) of baseline. To investigate the distribution of azosome, Dil-azosome (total lipids: 20 mM) was incubated with neurons for 1 h and imaged by the microscope at the excitation of 561 nm. After washing, the neurons were imaged again under the same condition.
4.5. In vitro toxicity
The cytotoxicity of azosomes on neurons was evaluated by the live/dead staining method. The primary striatal neurons were cultured in the 96-well plate. After 2 weeks, the neurons were incubated with azosome (12% azo-PC, total lipids: 2 mM) for 1 and 24 h. The light irradiation paradigm used in the neuromodulation experiments was treated on the neurons. After the treatment, the live and dead cells were stained with 3 μM calcein-AM (ThermoFisher, Cat. # C3100MP) and 5 μM propidium iodide (PI, ThermoFisher, Cat. # P1304MP) for 20 min, respectively. After wash, the fluorescence of calcein and PI was measured by the plate reader. The fluorescent images of stained cells were taken by a confocal microscope (Olympus, FV3000RS).
4.6. All-atom modeling of the lipid bilayer
All-atom simulations of azo-PC were performed using the CHARMM36 lipid [46] force field and the NAMD simulation software (version 2.13) [47]. The initial azo-PC structure was created in Avogadro (version 1.2.0) [48] and minimized using the built-in steepest descent algorithm. The topology entry for azo-PC was created from the existing entry of DSPC and manually altered to incorporate the azobenzene moiety. Atomistic force field parameters of azo-PC were created from a mixture of existing DSPC parameters and previously published parameters for the azobenzene moiety [49], and missing parameters were obtained through CGenFF [50–54]. Packmol was used to create the initial bilayer structures [55], and the VMD Solvate Plugin was used to solvate the bilayer with the TIP3P water model [56]. All simulations were performed in the tensionless ensemble at 1 atm and 323 K. Analysis was performed using custom Python scripts utilizing the MD Analysis package [57, 58].
Torsional force field parameters to control the azobenzene isomeric state were used following our previous method [49]. To mimic photoisomerization in a simulation, the torsional parameters corresponding to the desired photoisomeric state were enabled while simultaneously disabling the torsional parameters for the opposite state. This parameter change was made between simulation runs, using a restart or checkpoint file to continue with the new parameter set. Thus, for a given simulation run the parameters for only a single photoisomeric state were enabled at a time.
The following equation (2) was used to calculate the change of the azosome surface area during isomerization from the experimental diameter data.
| (2) |
where Dcis and Dtrans are the azosome diameters for azo-PC in the cis and trans state, respectively.
The areas per lipid (APL) from the simulations were calculated using the following equation (3):
| (3) |
where Xcell and Ycell are the cell dimensions in the X- and Y-directions, respectively, and Nlipids is the number of lipids per bilayer leaflet.
The change of bilayer area in the simulation was calculated by the following equation (4):
| (4) |
Supplementary Material
Acknowledgements
This work was partially supported by National Science Foundation under award number 2123971 (Z.Q., P.S., S.N.), National Institute of Neurological Disorders and Stroke of the National Institutes of Health under award number RF1NS110499 (Z.Q., P.S.), and a postdoc research grant from the Phospholipid Research Center (Heidelberg, Germany) to H.X.
Footnotes
Electronic Supplementary Material: Supplementary material (further characterizations of azosomes and in vitro toxicity) is available in the online version of this article at https://doi.org/10.1007/s12274-022-4853-x.
References
- [1].Dagdeviren C; Ramadi KB; Joe P; Spencer K; Schwerdt HN; Shimazu H; Delcasso S; Amemori K.-i.; Nunez-Lopez C; Graybiel AM Miniaturized neural system for chronic, local intracerebral drug delivery. Sci. Transl. Med. 2018, 10, eaan2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Feiner R; Dvir T Tissue-electronics interfaces: from implantable devices to engineered tissues. Nat. Rev. Mater. 2017, 3, 1–16. [Google Scholar]
- [3].Rao S; Chen R; LaRocca AA; Christiansen MG; Senko AW; Shi CH; Chiang P-H; Varnavides G; Xue J; Zhou Y Remotely controlled chemomagnetic modulation of targeted neural circuits. Nat. Nanotechnol. 2019, 14, 967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Airan RD; Meyer RA; Ellens NP; Rhodes KR; Farahani K; Pomper MG; Kadam SD; Green JJ Noninvasive targeted transcranial neuromodulation via focused ultrasound gated drug release from nanoemulsions. Nano Lett. 2017, 17, 652–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Wang JB; Aryal M; Zhong Q; Vyas DB; Airan RD Noninvasive ultrasonic drug uncaging maps whole-brain functional networks. Neuron 2018, 100, 728–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Rapp TL; DeForest CA Targeting drug delivery with light: A highly focused approach. Adv. Drug Deliv. Rev. 2021, 171, 94–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Ellis-Davies GC Caged compounds: photorelease technology for control of cellular chemistry and physiology. Nat. Methods 2007, 4, 619–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Taura J; Nolen EG; Cabré G; Hernando J; Squarcialupi L; López-Cano M; Jacobson KA; Fernández-Dueñas V; Ciruela F Remote control of movement disorders using a photoactive adenosine A2A receptor antagonist. J. Control. Release 2018, 283, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ellis-Davies GC Useful caged compounds for cell physiology. Acc. Chem. Res. 2020, 53, 1593–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Silva JM; Silva E; Reis RL Light-triggered release of photocaged therapeutics - Where are we now? J. Control. Release 2019, 298, 154–176. [DOI] [PubMed] [Google Scholar]
- [11].Maier W; Corrie JET; Papageorgiou G; Laube B; Grewer C Comparative analysis of inhibitory effects of caged ligands for the NMDA receptor. J. Neurosci. Methods 2005, 142, 1–9. [DOI] [PubMed] [Google Scholar]
- [12].Noguchi J; Nagaoka A; Watanabe S; Ellis-Davies GC; Kitamura K; Kano M; Matsuzaki M; Kasai H In vivo two-photon uncaging of glutamate revealing the structure-function relationships of dendritic spines in the neocortex of adult mice. J. Physiol. 2011, 589, 2447–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Li B; Wang Y; Gao D; Ren S; Li L; Li N; An H; Zhu T; Yang Y; Zhang H; Xing C Photothermal modulation of depression-related ion channel function through conjugated polymer nanoparticles. Adv. Funct. Mater. 2021, 31, 2010757. [Google Scholar]
- [14].Nakano T; Mackay SM; Wui Tan E; Dani KM; Wickens J Interfacing with neural activity via femtosecond laser stimulation of drug-encapsulating liposomal nanostructures. eNeuro 2016, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Li W; Luo R; Lin X; Jadhav AD; Zhang Z; Yan L; Chan C-Y; Chen X; He J; Chen C-H; Shi P Remote modulation of neural activities via near-infrared triggered release of biomolecules. Biomaterials 2015, 65, 76–85. [DOI] [PubMed] [Google Scholar]
- [16].Huu VAN; Luo J; Zhu J; Zhu J; Patel S; Boone A; Mahmoud E; McFearin C; Olejniczak J; de Gracia Lux C; Lux J; Fomina N; Huynh M; Zhang K; Almutairi A Light-responsive nanoparticle depot to control release of a small molecule angiogenesis inhibitor in the posterior segment of the eye. J. Control. Release 2015, 200, 71–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Kohman RE; Cha SS; Man HY; Han X Light-triggered release of bioactive molecules from DNA nanostructures. Nano Lett. 2016, 16, 2781–2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Veetil AT; Chakraborty K; Xiao K; Minter MR; Sisodia SS; Krishnan Y Cell-targetable DNA nanocapsules for spatiotemporal release of caged bioactive small molecules. Nat. Nanotechnol. 2017, 12, 1183–1189. [DOI] [PubMed] [Google Scholar]
- [19].Xiong H; Li X; Kang P; Perish J; Neuhaus F; Ploski JE; Kroener S; Ogunyankin MO; Shin JE; Zasadzinski JA; Wang H; Slesinger PA; Zumbuehl A; Qin Z Near-infrared light triggered-release in deep brain regions using ultra-photosensitive nanovesicles. Angew. Chem. 2020, 132, 8686–8693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Li X; Che Z; Mazhar K; Price TJ; Qin Z Ultrafast near-infrared light-triggered intracellular uncaging to probe cell signaling. Adv. Funct. Mater. 2017, 27, 1605778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Cabré G; Garrido-Charles A; Moreno M; Bosch M; Porta-de-la-Riva M; Krieg M; Gascón-Moya M; Camarero N; Gelabert R; Lluch JM; Busqué F; Hernando J; Gorostiza P; Alibés R Rationally designed azobenzene photoswitches for efficient two-photon neuronal excitation. Nat. Commun. 2019, 10, 907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].DiFrancesco ML; Lodola F; Colombo E; Maragliano L; Bramini M; Paternò GM; Baldelli P; Serra MD; Lunelli L; Marchioretto M; Grasselli G; Cimò S; Colella L; Fazzi D; Ortica F; Vurro V; Eleftheriou CG; Shmal D; Maya-Vetencourt JF; Bertarelli C; Lanzani G; Benfenati F Neuronal firing modulation by a membrane-targeted photoswitch. Nat. Nanotechnol. 2020, 15, 296–306. [DOI] [PubMed] [Google Scholar]
- [23].Kellner S; Berlin S Two-photon excitation of azobenzene photoswitches for synthetic optogenetics. Appl. Sci. 2020, 10. [Google Scholar]
- [24].Morstein J; Dacheux MA; Norman DD; Shemet A; Donthamsetti PC; Citir M; Frank JA; Schultz C; Isacoff EY; Parrill AL; Tigyi GJ; Trauner D Optical control of lysophosphatidic acid signaling. J. Am. Chem. Soc. 2020, 142, 10612–10616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Morstein J; Hill RZ; Novak AJE; Feng S; Norman DD; Donthamsetti PC; Frank JA; Harayama T; Williams BM; Parrill AL; Tigyi GJ; Riezman H; Isacoff EY; Bautista DM; Trauner D Optical control of sphingosine-1-phosphate formation and function. Nat. Chem. Biol. 2019, 15, 623–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Morstein J; Romano G; Hetzler BE; Plante A; Haake C; Levitz J; Trauner D Photoswitchable serotonins for optical control of the 5-HT2A receptor. Angew. Chem. Int. Ed. 2022, 61, e202117094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mukhopadhyay TK; Morstein J; Trauner D Photopharmacological control of cell signaling with photoswitchable lipids. Curr. Opin. Pharmacol. 2022, 63, 102202. [DOI] [PubMed] [Google Scholar]
- [28].Bahamonde MI; Taura J; Paoletta S; Gakh AA; Chakraborty S; Hernando J; Fernández-Dueñas V; Jacobson KA; Gorostiza P; Ciruela F Photomodulation of G protein-coupled adenosine receptors by a novel light-switchable ligand. Bioconjug. Chem. 2014, 25, 1847–1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Pernpeintner C; Frank JA; Urban P; Roeske CR; Pritzl SD; Trauner D; Lohmüller T Light-controlled membrane mechanics and shape transitions of photoswitchable lipid vesicles. Langmuir 2017, 33, 4083–4089. [DOI] [PubMed] [Google Scholar]
- [30].Pritzl SD; Konrad DB; Ober MF; Richter AF; Frank JA; Nickel B; Trauner D; Lohmüller T Optical membrane control with red light enabled by red-shifted photolipids. Langmuir 2021, 38, 385–393. [DOI] [PubMed] [Google Scholar]
- [31].Pritzl SD; Urban P; Prasselsperger A; Konrad DB; Frank JA; Trauner D; Lohmüller T Photolipid bilayer permeability is controlled by transient pore formation. Langmuir 2020, 36, 13509–13515. [DOI] [PubMed] [Google Scholar]
- [32].Urban P; Pritzl SD; Konrad DB; Frank JA; Pernpeintner C; Roeske CR; Trauner D; Lohmüller T Light-controlled lipid interaction and membrane organization in photolipid bilayer vesicles. Langmuir 2018, 34, 13368–13374. [DOI] [PubMed] [Google Scholar]
- [33].Urban P; Pritzl SD; Ober MF; Dirscherl CF; Pernpeintner C; Konrad DB; Frank JA; Trauner D; Nickel B; Lohmueller T A lipid photoswitch controls fluidity in supported bilayer membranes. Langmuir 2020, 36, 2629–2634. [DOI] [PubMed] [Google Scholar]
- [34].Chander N; Morstein J; Bolten JS; Shemet A; Cullis PR; Trauner D; Witzigmann D Optimized photoactivatable lipid nanoparticles enable red light triggered drug release. Small 2021, 17, 2008198. [DOI] [PubMed] [Google Scholar]
- [35].Ishiba K; Morikawa M.-a.; Chikara C; Yamada T; Iwase K; Kawakita M; Kimizuka N Photoliquefiable ionic crystals: a phase crossover approach for photon energy storage materials with functional multiplicity. Angew. Chem. Int. Ed. 2015, 54, 1532–1536. [DOI] [PubMed] [Google Scholar]
- [36].Zhou H; Xue C; Weis P; Suzuki Y; Huang S; Koynov K; Auernhammer GK; Berger R; Butt H-J; Wu S Photoswitching of glass transition temperatures of azobenzene-containing polymers induces reversible solid-to-liquid transitions. Nat. Chem. 2017, 9, 145–151. [DOI] [PubMed] [Google Scholar]
- [37].Gagnon D; Petryszyn S; Sanchez M; Bories C; Beaulieu J; De Koninck Y; Parent A; Parent M Striatal neurons expressing D1 and D2 receptors are morphologically distinct and differently affected by dopamine denervation in mice. Sci. Rep. 2017, 7, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Dai R; Ali MK; Lezcano N; Bergson C A crucial role for cAMP and protein kinase a in D1 dopamine receptor regulated intracellular calcium transients. NeuroSignals 2008, 16, 112–123. [DOI] [PubMed] [Google Scholar]
- [39].Jeroen Vermeulen R; Drukarch B; Rob Sahadat MC; Goosen C; Wolters EC; Stoof JC The selective dopamine D1 receptor agonist, SKF 81297, stimulates motor behaviour of MPTP-lesioned monkeys. Eur. J. Pharmacol. 1993, 235, 143–147. [DOI] [PubMed] [Google Scholar]
- [40].Wu X; Jiang Y; Rommelfanger NJ; Yang F; Zhou Q; Yin R; Liu J; Cai S; Ren W; Shin A; Ong KS; Pu K; Hong G Tether-free photothermal deep-brain stimulation in freely behaving mice via wide-field illumination in the near-infrared-II window. Nat. Biomed. Eng. 2022, 6, 754–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Li J; Duan H; Pu K Nanotransducers for near-infrared photoregulation in biomedicine. Adv. Mater. 2019, 31, 1901607. [DOI] [PubMed] [Google Scholar]
- [42].Li X; Xiong H; Rommelfanger N; Xu X; Youn J; Slesinger PA; Hong G; Qin Z Nanotransducers for wireless neuromodulation. Matter 2021, 4, 1484–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Lyu Y; Xie C; Chechetka SA; Miyako E; Pu K Semiconducting polymer nanobioconjugates for targeted photothermal activation of neurons. J. Am. Chem. Soc. 2016, 138, 9049–9052. [DOI] [PubMed] [Google Scholar]
- [44].Yao C; Wang P; Li X; Hu X; Hou J; Wang L; Zhang F Near-infrared-triggered azobenzene-liposome/upconversion nanoparticle hybrid vesicles for remotely controlled drug delivery to overcome cancer multidrug resistance. Ad. Mater. 2016, 28, 9341–9348. [DOI] [PubMed] [Google Scholar]
- [45].Zhang Y; Zhang Y; Song G; He Y; Zhang X; Liu Y; Ju H, A DNA–azobenzene nanopump fueled by upconversion luminescence for controllable intracellular drug release. Angew. Chem. Int. Ed. 2019, 58, 18207–18211. [DOI] [PubMed] [Google Scholar]
- [46].Klauda JB; Venable RM; Freites JA; O’Connor JW; Tobias DJ; Mondragon-Ramirez C; Vorobyov I; MacKerell AD Jr; Pastor RW Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J. Phys. Chem. B 2010, 114, 7830–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Phillips JC; Hardy DJ; Maia JD; Stone JE; Ribeiro JV; Bernardi RC; Buch R; Fiorin G; Hénin J; Jiang W Scalable molecular dynamics on CPU and GPU architectures with NAMD. J. Chem. Phys. 2020, 153, 044130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hanwell MD; Curtis DE; Lonie DC; Vandermeersch T; Zurek E; Hutchison GR Avogadro: an advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Siriwardane DA; Kulikov O; Batchelor BL; Liu Z; Cue JM; Nielsen SO; Novak BM UV-and thermo-controllable azobenzene-decorated polycarbodiimide molecular springs. Macromolecules 2018, 51, 3722–3730. [Google Scholar]
- [50].Gutiérrez IS; Lin F-Y; Vanommeslaeghe K; Lemkul JA; Armacost KA; Brooks III CL; MacKerell AD Jr Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand–protein interactions. Bioorg. Med. Chem. 2016, 24, 4812–4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Vanommeslaeghe K; Hatcher E; Acharya C; Kundu S; Zhong S; Shim J; Darian E; Guvench O; Lopes P; Vorobyov I CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2010, 31, 671–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Vanommeslaeghe K; MacKerell AD Jr Automation of the CHARMM General Force Field (CGenFF) I: bond perception and atom typing. J. Chem. Inf. Model. 2012, 52, 3144–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Vanommeslaeghe K; Raman EP; MacKerell AD Jr Automation of the CHARMM General Force Field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 2012, 52, 3155–3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Yu W; He X; Vanommeslaeghe K; MacKerell AD Jr Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J. Comput. Chem. 2012, 33, 2451–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Martínez L; Andrade R; Birgin EG; Martínez JM, PACKMOL: a package for building initial configurations for molecular dynamics simulations. J. Comput. Chem. 2009, 30, 2157–2164. [DOI] [PubMed] [Google Scholar]
- [56].Humphrey W; Dalke A; Schulten K VMD: visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [DOI] [PubMed] [Google Scholar]
- [57].Gowers RJ; Linke M; Barnoud J; Reddy TJE; Melo MN; Seyler SL; Domanski J; Dotson DL; Buchoux S; Kenney IM MDAnalysis: a Python package for the rapid analysis of molecular dynamics simulations. In Proceedings of the 15th Python in Science Conference, Austin, Texas, United States, 2016. [Google Scholar]
- [58].Michaud-Agrawal N; Denning EJ; Woolf TB; Beckstein O MDAnalysis: a toolkit for the analysis of molecular dynamics simulations. J. Comput. Chem. 2011, 32, 2319–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


