Summary
Cerebral ischemia triggers a powerful inflammatory reaction involving both peripheral leukocytes and brain resident cells. Recent evidence indicates that their differentiation into a variety of functional phenotypes contributes to both tissue injury and repair. However, the temporal dynamics and diversity of post-stroke immune cell subsets remain poorly understood. To address these limitations, we performed a longitudinal single-cell transcriptomic study of both brain and mouse blood to obtain a composite picture of brain-infiltrating leukocytes, circulating leukocytes, microglia and endothelium diversity over the ischemic/reperfusion time. Brain cells and blood leukocytes isolated from mice 2 or 14 days after transient middle cerebral artery occlusion or sham surgery were purified by FACS sorting and processed for droplet-based single-cell transcriptomics. The analysis revealed a strong divergence of post-ischemic microglia, macrophages, and neutrophils over time, while such diversity was less evident in dendritic cells, B, T and NK cells. Conversely, brain endothelial cells and brain associated-macrophages showed altered transcriptomic signatures at 2 days post-stroke, but low divergence from sham at day 14. Pseudotime trajectory inference predicted the in-situ longitudinal progression of monocyte-derived macrophages from their blood precursors into day 2 and day 14 phenotypes, while microglia phenotypes at these two time points were not connected. In contrast to monocyte-derived macrophages, neutrophils were predicted to be continuously de-novo recruited from the blood. Brain single-cell transcriptomics from both female and male aged mice did not show major changes in respect to young mice, but aged and young brains differed in their immune cell composition. Furthermore, blood leukocyte analysis also revealed altered transcriptomes after stroke. However, brain-infiltrating leukocytes displayed higher transcriptomic divergence than their circulating counterparts, indicating that phenotypic diversification into cellular subsets occurs within the brain in the early and the recovery phase of ischemic stroke. In addition, this resource report contains a searchable database https://anratherlab.shinyapps.io/strokevis/ to allow user-friendly access to our data. The StrokeVis tool constitutes a comprehensive gene expression atlas that can be interrogated at the gene and cell type level to explore the transcriptional changes of endothelial and immune cell subsets from mouse brain and blood after stroke.
Introduction
Ischemic stroke (IS) accounts for about 87% of all strokes and occurs when a vessel supplying blood to the brain is obstructed. Approaches for treatment of acute stroke are targeted at rapid recanalizing of the blocked artery by endovascular thrombectomy and/or administration of thrombolytic agents 1,2. Despite recent successes in both prevention and treatment, stroke is yet the second leading cause of death and third leading cause of disability worldwide 3,4.
The immune system actively participates in the acute and chronic pathogenesis of IS. Damaged neurons lead to a secondary inflammatory reaction that aggravates brain injury, increasing neurologic deficits 5–7. This response progresses for days to weeks and involves glial and brain endothelium activation, recruitment of peripheral immune cells, and release of cytokines. Whereas there is evidence that the acute inflammatory response contributes to the progression of ischemic brain injury, more recent research points at a more multifaceted role of immune cells in brain ischemia, where they participate in repair processes during the sub-acute and chronic stages 8–11. In addition, post-stroke chronic inflammation and adaptive immunity have been implicated in the long-term sequelae such as depression and dementia 12.
Emerging single-cell RNA sequencing (scRNA-seq) studies reveal a highly transcriptional cellular heterogeneity in response to IS 13, supporting the previously reported functional plasticity of immune cells after stroke 14. Although the precise mechanisms inducing cellular diversity have not yet been determined, signals released by damaged brain cells as well as other environmental cues like metabolic alterations after ischemia, are most likely to induce phenotypic differentiation of the brain cells. Furthermore, the ischemic brain and systemic immunity interact in a bidirectional fashion. While the immune system supplies the brain with immune cells that participate in the local inflammatory response, neural and humoral factors generated by the ischemic brain communicate to peripheral organs 7. Thus, activation of the immune system through brain-derived molecules or via autonomic nervous system, might lead to transcriptional differentiation of immune cells before entering the brain 7,15
Some studies using scRNA-seq technology have begun to decipher the cellular heterogeneity of brain cells after IS 16–20. Here we used single-cell RNA sequencing to gain deeper insights into the impact of IS on transcriptional diversity of brain immune cells, brain endothelial cells (EC) and peripheral blood leukocytes. Specifically, this study focuses on assessing differences of transcriptomic signatures related to cell origin (brain resident vs. recruited cells), cellular localization (periphery vs. brain), time post-stroke (acute versus sub-acute) and age (young vs. aged mice).
Results
Dynamics of mouse brain and blood cell heterogeneity over the ischemic-reperfusion time.
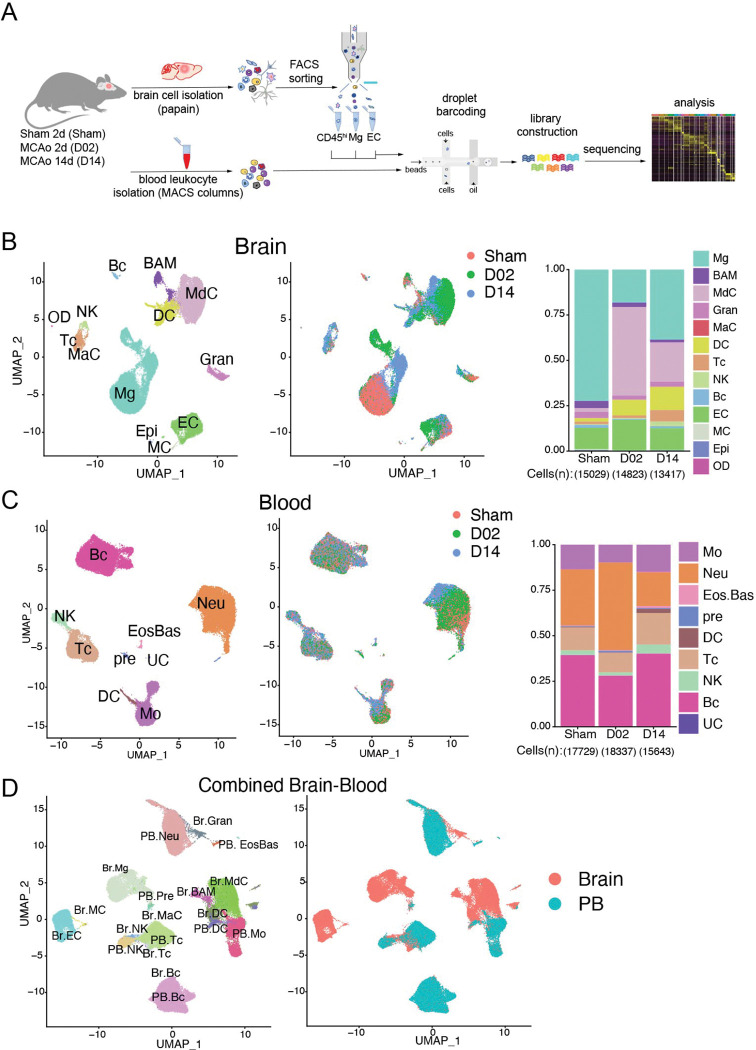
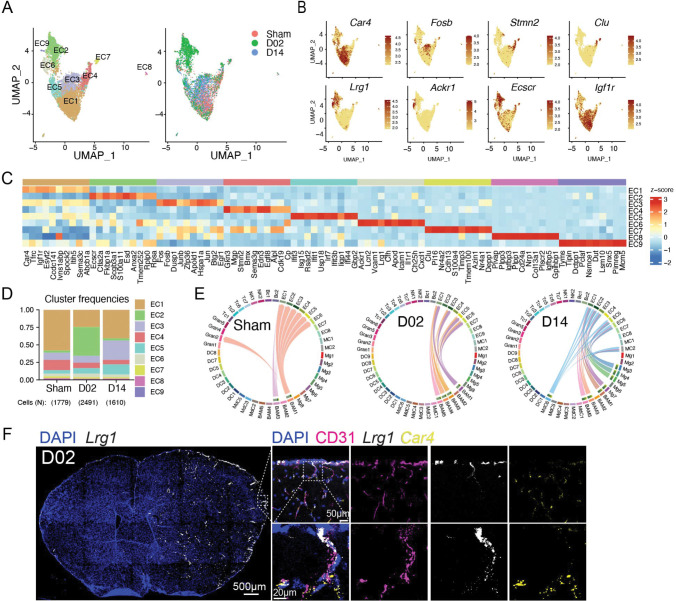
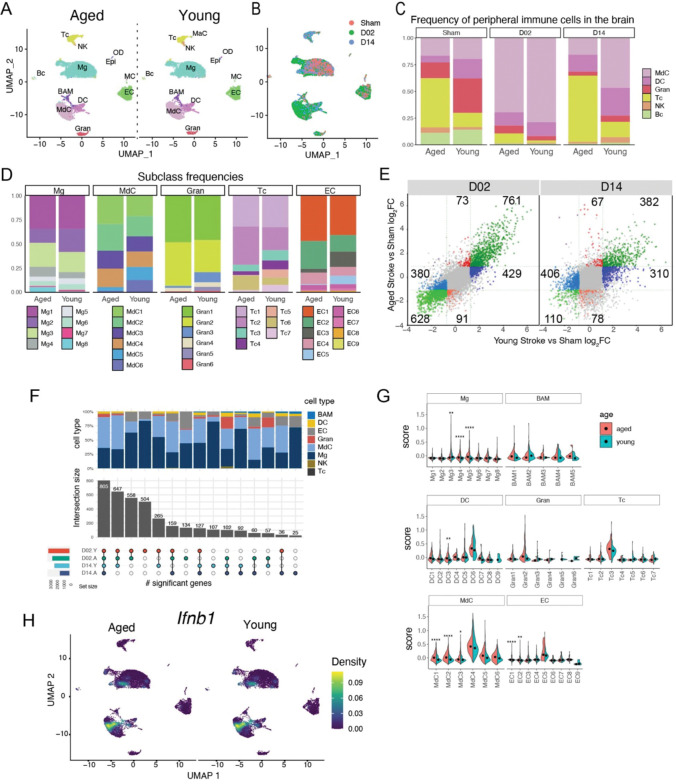
To obtain a comprehensive picture of the immune cell diversity and temporal dynamics over the acute and subacute phases after cerebral ischemia, we isolated immune cells from the brains of stroke mice and performed droplet-based scRNA-seq. We also isolated endothelial cells (EC), the first cells to interact with leukocytes migrating into the ischemic tissue. We prepared brain single cell suspensions from mice 2 days after sham surgery (Sham) or 2 and 14 days after stroke (D02 and D14, respectively, Fig.1A). Subsequently, we flow sorted peripheral immune cells and border-associated macrophages (BAM; CD45hi), microglia (CD45intCX3CR1+), and EC (CD45lowLy6Chi)21,22. Cells were mixed in a 6:1:1 ratio (CD45hi:microglia:EC) and processed for droplet-based RNA library preparation (Fig.1A). We enriched the samples for CD45hi cells because we anticipated higher cellular heterogeneity of CD45hi population than in either microglia or EC. We detected 13 distinct major cell type clusters in 43269 cells after performing unsupervised clustering and Uniform Manifold Approximation and Projection (UMAP). On the basis of the expression of established marker genes and unsupervised cell type annotation, clusters were identified as microglia (Mg), BAM, monocyte-derived cells (MdC), granulocytes (Gran), mast cells (MaC), dendritic cells (DC), T cells (Tc), NK cells (NK), B cells (Bc), endothelial cells (EC), vascular mural cells (MC), epithelial-like cells (Epi), and oligodendrocytes (OD) (Fig.1B; Fig. S3). As anticipated, we observed that the relative abundance of each cell cluster changed notably with ischemia-reperfusion. For instance, MdC increased at D02, whereas the increase in lymphocytes (Tc and NK) was more pronounced at D14, as previously reported 6,23,24. Amid brain resident cells, we observed divergent clustering of microglia across Sham, D02 and D14, indicating that ischemic injury markedly modified microglia transcriptomes. On the other hand, the UMAP representation indicated that BAM and EC clusters were less heterogeneous across groups (Fig. 1B).
Figure 1. Single-cell transcriptomic profiling of mouse brain and blood cells after transient focal cerebral ischemia.
(A) Schematic representation of Drop-Seq scRNA-seq pipeline used to analyze brain and blood cells isolated from either control surgery (Sham) or stroke mice 2 and 14 days (D02, D14) after injury. Brain cells were dissociated by enzymatic digestion with papain. Infiltrating leukocytes (CD45hi), microglia (Mg) and endothelial cells (EC) were isolated by flow cytometry sorting. Blood leukocytes were purified after erythrocyte removal. Brain and blood single cell suspensions were subjected to Drop-Seq, sequencing and analysis.
(B, C) Left: Uniform Manifold Approximation and Projection (UMAP) plot representing color-coded cell clusters identified in merged brain (B) or blood (C) single-cell transcriptomes; Middle: UMAP of 3 color-coded time point overlay of brain (B) or blood (C) single-cell transcriptomes; Right: bar graph showing relative frequencies of each cell type across Sham, D02 and D14 groups of either brain (B) or blood (C) identified cell type clusters.
(D) Left: UMAP plot of the combined brain (Br) and blood (PB) dataset showing cell clustering similarities between brain and blood Gran, Tc, Bc and brain myeloid cells (BAM, MdC, DC) with blood monocytes (left). Left: UMAP plot shows the distribution of brain and blood cells overlying the identified clusters. Border-associated macrophages (BAM), monocyte-derived cells (MdC), granulocytes (Gran), mast cells (MaC), dendritic cells (DC), T cells (Tc), NK cells (NK), B cells (Bc), vascular mural cells (MC), epithelial-like cells (Epi), oligodendrocytes (OD); Eosinophils-Basophils (EosBas); Monocytes (Mo); hematopoietic precursors (pre); unclassified (UC).
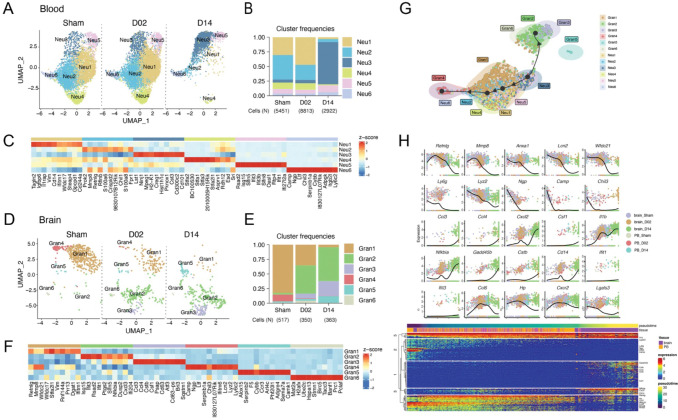
Because brain ischemia elicits a systemic immune response that ultimately affects stroke outcome 7, we next sought to explore the transcriptomes of peripheral blood cells from stroke mice. We performed scRNA-seq of purified blood leukocytes from Sham, D02 and D14 mice. Leukocytes grouped into 8 distinct clusters that were annotated by the expression of commonly used marker genes, as follows: monocytes (Mo), granulocytes (Gran), eosinophils/basophils (Eos/Bas), dendritic cells (DC), T cells (Tc), NK cells (NK), B cells (Bc), various precursors (pre), and one unclassified cluster (UC) (Fig.1C; Fig. S4). Overall, the major peripheral blood immune cell types showed conserved positioning in the UMAP space across Sham, D02 and D14 groups. Only Gran and Mo clusters were slightly divergent at D14 from their respective Sham and D02 clusters. Thus, these results point to lesser phenotypic differentiation in circulating leukocytes than in brain recruited leukocytes after stroke.
To assess transcriptional similarities between leukocytes from both tissues we analyzed a combined brain and blood dataset. We observed that non-brain resident immune cells clustered together independently of their tissue origin. On the other hand, whereas BAM, MdC, and brain DC clustered with blood monocytes and DC, the microglia cluster remained spatially separated from blood myeloid cells reflecting their unique transcriptome (Fig. 1D).
Cerebral ischemia induces different transcriptomic states in microglia at early and late phases.
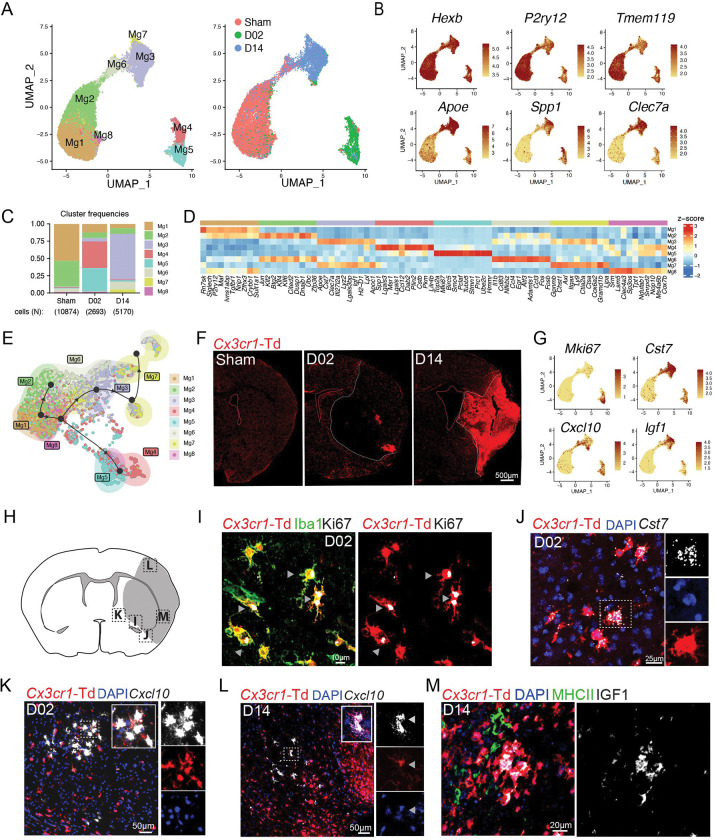
To determine whether distinct transcriptional programs of microglia develop over the ischemia-reperfusion time, we re-clustered microglia which resulted in 8 clusters (Mg1-8) all showing high expression of canonical microglial markers Hexb, Olfml3, and Fcrls (Fig. 2A–D; Fig. S5A). Sham microglia mostly fell into clusters Mg1 and Mg2, which exhibited high expression of genes found in homeostatic microglia including P2ry12, Tmem119, and Sall1. Whereas Mg1 was mainly characterized by expression of homeostatic genes (Siglech, P2ry12, Tmem119), Mg2 showed upregulation of immediate early genes (Jun, Fos, Erg1, Klf2, Klf4 and Atf3), a group of transcription factors typically expressed in adult microglia and involved in establishing microglia surveilling functions 25–28. In line with recent reports, we observed that microglial homeostatic genes, such as P2ry12 and Tmem119, were downregulated after cerebral ischemia 16,29 (Fig. 2B). The expression of Apoe, Lpl, Spp1, Clec7a or Cst7 genes, which have been linked to microglial responses to demyelination 30, Alzheimer’s disease 31, as well as stroke 16,29, were upregulated in both D02 and D14 microglia (Fig. 2A–D). Furthermore, cerebral ischemia led to distinct D02 and D14 microglia clusters. D02 microglia was mainly composed of Mg4 and Mg5 clusters, while the most prominent clusters at D14 were Mg3, Mg6, and Mg7 (Fig. 2C). Among D02 microglia clusters, Mg4 showed high expression of genes related to the clearance of damaged cells and tissue repair (Spp1, Msr1 and Lgals3) 29,32–34, and high expression of chemokine genes (Ccl2, Ccl12) 35. The Mg5 cluster was characterized by the expression of mitotic genes including Top2A, Mki67 and Stmn1 20, indicating the presence of proliferative microglia during the acute phase of stroke. Mg3, the predominant cluster at D14, was mainly defined by expression of genes associated with disease-associated microglia (DAM) including Apoe, Cst7, Clec7a, Lyz2, Lgals3bp, Igf1, and Lpl 31 (Fig. S5B). Similar to Mg3, Mg7 was characterized by upregulation of DAM genes (Spp1, Apoe, Cst7, Igf1, Lgals3bp, Apoc1, Lpl, Gpnmb, Itgax)31 and by expression of genes observed in the response of microglia to neurodegeneration (Gpnmb, Axl, Itgax, Spp1, Apoe)36. Mg6 showed upregulation of immune genes (Il1b, Nfkbiz, Cd83, Ccl4) and downregulation of Spp1 and Lpl, which suggests a pro-inflammatory phenotype, previously described in ischemic stroke and subarachnoid hemorrhage 29,37. In addition, Mg6 showed enriched gene expression signature of disease inflammatory macrophages (DIM; Il1b, Cd83, Nfkbiz, Atf3, Ccl4, Egr1, Fosb), indicating Mg6 cluster as DIM-like microglial cells (Fig. S5B)38. Pseudo-time trajectory predicted the progression of Sham microglia (Mg1) to either D02 or D14 microglia, whereas we could not observe a trajectory from D02 to D14 microglia (Fig. 2E).
Figure 2. Subclustering analysis of microglia reveals altered transcriptional states through the acute and subacute stroke phases.
(A) Left: UMAP analysis of merged Sham, D02 and D14 microglial single cell transcriptomes reveals 8 subclusters; Right: UMAP of overlaid time points reveals high segregation of microglial clusters among Sham, D02 and D14 groups.
(B) Feature plots depicting single-cell gene expression of individual genes characterizing homeostatic and activated microglia. Scale bar represents log of normalized gene expression.
(C) Bar graph showing relative frequencies of each microglial subcluster across Sham, D02 and D14 groups.
(D) Heatmap displaying expression of the top 10 upregulated genes in each microglial cluster. Scale bar represents Z-score of average gene expression (log).
(D) Heatmap displaying expression of the top 10 upregulated genes in each microglial cluster. Scale bar represents Z-score of average gene expression (log).
(E) Pseudotime analysis of microglial subclusters showing transitional trajectories from Sham-Mg1 cluster to either Sham-Mg2, or D02 Mg4-5 clusters or D14 Mg3-7 clusters.
(F) Representative fluorescence images of the cerebral hemisphere after Sham surgery or after 2 (D02) or 14 (D14) days MCA occlusion in Cx3cr1CreERT2:R26tdTomato mice. Microglia were identified as Td+ cells (red). Images show that the number of microglia decreased in the center of the lesion (white dashed outline) at D02 whereas its number increased at D14.
(G) UMAP plots depicting single-cell gene expression of individual marker genes that characterize ischemic microglial clusters. Scale bar represents log of normalized gene expression.
(H) Graphical representation of brain coronal section indicating anatomical regions where images I-L were acquired.
(I) Immunofluorescence images showing Ki67 (white) expression by Cx3cr1-Td+(red) Iba1+(green) microglia 2 days after MCA occlusion. Arrowheads indicate Ki67 staining.
(J) RNAscope in situ hybridization fluorescence (FISH) and immunofluorescence (IF) images validating Cst7 (white) expression in D02 Cx3cr1-Td+ microglia (Td+, red). Nuclei are stained with DAPI (blue).
(K, L) FISH images showing Cxcl10 (white) expression in D02 (K) and D14 (L) Cx3cr1-Td+ microglia (Td+, red). Nuclei are stained with DAPI (blue).
(M) IF images showing IGF1 (white) expression by Cx3cr1-Td+(red) MHCII− (green) microglia 14 days after MCA occlusion.
Microglia undergo significant morphological and functional differentiation after cerebral ischemia, which could confound discrimination from infiltrating MdC 39,40. To verify microglial identity we used tamoxifen-inducible Cx3cr1CreERT2:R26tdTomato (Cx3cr1-Td) mice, a model that distinguishes Td+ microglia and BAM from hematopoietic myeloid cells 41–43. We subjected Cx3cr1-Td mice that had been injected with tamoxifen 6–8 weeks prior to Sham, D02 or D14 stroke and performed scRNA-seq on Td+ brain cells sorted by flow cytometry. Cells were annotated using the WT brain dataset as a reference. Clustering analysis of tdTomato expressing cells revealed that microglia constituted the majority of Td+ cells (92.7%) followed by BAM (2.9%), MdC (2.0%) and DC (1.3%). Considering that the number of MdC is nine times higher than the number of BAM, and that the number of DC is three times higher, we infer that the majority of BAM were Td+ while only a small proportion of MdC and DC retained Td expression (Fig. S5C).
To investigate microglia spatial distribution, we examined brains of Cx3cr1-Td stroke mice by histology and used FISH-IF to spatially map major microglia clusters (Fig. 2F, H–M). We observed that Cx3cr1-Td+ cells were significantly reduced in the ischemic core at D02, whereas they accumulated in the core at D14 (Fig. 2F). We observed colocalization of Cx3cr1-Td+Iba1+ microglia with the mitotic marker Ki67 at the infarct border at D02 (Fig. 2I; Fig. S6A), suggesting that proliferative microglia contribute to the repopulation of the ischemic territory seen at D14.
We found the DAM marker Cst7 31,44 in both D02 and D14 microglia. Cst7 colocalized with ramified microglial cells at the infarct border and with ameboid cells in the ischemic core (Fig. 2J; Fig. S6B) suggesting that cell morphology was not associated with a DAM signature. We also identified Cxcl10 and Igf1 among the DEGs that were upregulated in microglia after ischemia (Fig. 2G). Microglial induction of the inflammatory chemokine Cxcl10, a IFN type I stimulated gene, has previously been reported in models of traumatic and ischemic brain injury, EAE and AD 29,45–48, while IGF-1 expressing microglia has been reported to be associated with a pro-neurogenic phenotype after stroke in rats 49. We observed that Cxcl10 was upregulated at D02 and D14, whereas Igf1 was mainly induced at D14 (Fig. 2G). By histology, we also found Cxcl10 expressing microglia in the infarct border at D02 and in the lesioned tissue at D14, while IGF1+ microglia were found in the ischemic core (Fig. K–M; Fig. S6C–D). In some instances, Td+Cxcl10+ microglia were found near Td−Cxcl10+ cells, suggesting microglia and non-microglial cells expressing Cxcl10 organize into discrete cell clusters (Fig. 2K).
Border-associated macrophages exhibit distinct transcriptional states early after stroke.
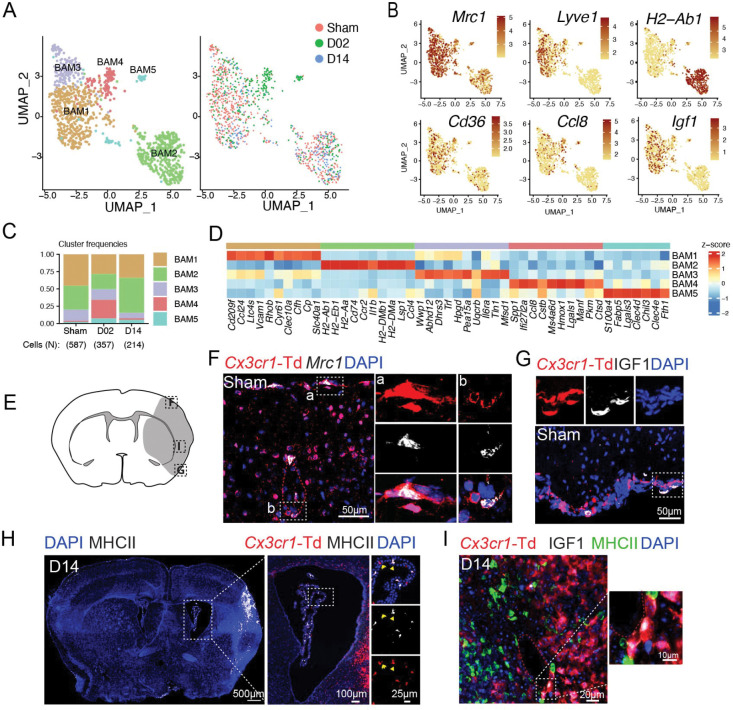
We identified five BAM clusters (Fig. 3A). BAM1 and BAM2 were the most abundant clusters in all groups (Fig. 3C). BAM1 showed transcriptional signature of subdural BAM (Cd209f, Ccl24, Clec10a, Slc40a1, Stab1) 50. Some of these genes have been found in homeostatic BAM and are related to leukocyte recruitment (Ccl24), phagocytosis (Cd209f) and iron metabolism (Slc40a1, Cp) 51,52. BAM2 showed the highest separation from all other BAM clusters and displayed upregulation of canonical marker genes of choroid plexus macrophages (ChMp) such MHC class II genes (H2-Ab1, H2-Eb1, H2-Aa, H2-DMb1, H2-DMa), the MHCII-associated gene Cd74, Ccr2, and low level of the perivascular macrophages (PVM) marker Lyve1 (Fig. 3B,D) 52–55. BAM3, which was less frequent at D14, featured high expression of genes coding for enzymes Wwp1, Abhd12, Dhrs3, Hpgd, and transferrin (Trf). BAM4 was largely confined to D02 and was characterized by the upregulation of genes linked to macrophage/microglia activation in either ischemic stroke or other neurodegenerative diseases (Spp1, Ccl8, Cstb, Lgals1) 50,56,57. BAM5 was associated with induction of genes related to lipid metabolism (Fabp5, Lgals3) 58 and damage-associated molecular patterns (DAMPs)-recognition molecules (S100A4, Lgals3, Clec4d, Clec4e) 59.
Figure 3. Transcriptional changes of border-associated macrophages (BAM) after stroke.
(A) Left: UMAP analysis of merged Sham, D02 and D14 BAM transcriptomes discloses 5 subclusters; Right: UMAP of overlaid time points shows overlap of clusters among Sham, D02 and D14 groups, except for BAM4, which is confined to D02.
(B) UMAP plots depicting expression of individual marker genes for BAM (Mrc1, Cd36), meningeal and perivascular BAM (Lyve1), choroid plexus BAM (H2-Ab1) and activated meningeal and perivascular BAM (Ccl8, Igf1). Scale bar represents log of normalized gene expression.
(C) Bar graph showing relative frequencies of BAM subclusters across Sham, D02 and D14 groups.
(D) Heatmap displaying expression of the top 10 upregulated genes in each BAM cluster. Scale bar represents Z-score of average gene expression (log).
(E) Graphical representation of brain coronal section indicating anatomical regions where images F-I were acquired.
(F) RNAscope in situ hybridization fluorescence (FISH) combined with immunofluorescence (IF) images of brain cortical areas showing Mrc1 expression (white) in resident macrophages (Td+, red) on the brain surface and around vessels of Cx3cr1-Td+ mice 2 days after Sham surgery (Sham). Nuclei are stained with DAPI (blue).
(G, I) IF images validating IGF1 (white) expression by pial BAM (Td+, red) (G) in Sham Cx3cr1-Td+mice and in perivascular macrophages (PVM) (I) of the ischemic brain at D14 (Middle and left).
(H) Left: Representative IF image of a whole brain section from a Cx3cr1-Td+ mouse subjected to 14 days of MCA occlusion (D14), showing MHCII+ cells (white, binary mask) localization and nuclear DAPI staining (blue); Right: IF images of magnified areas of the choroid plexus (ChP) showing MHCII expression (white) by ChP macrophages (Td+, red).
Utilizing Cx3cr1-Td mice, we mapped BAM on the surface of the brain or lining parenchymal blood vessels by Mrc1 (coding for CD206) and Td co-detection. Consistent with the scRNA-seq data, MHC class II antigen was only detected in ChMp, while PVM and pial macrophages (pMp), but not ChMp, expressed IGF1 (Fig. 3B, E–I).
Brain monocyte-derived cells arise from circulating inflammatory monocytes and diversify over time in the brain after stroke.
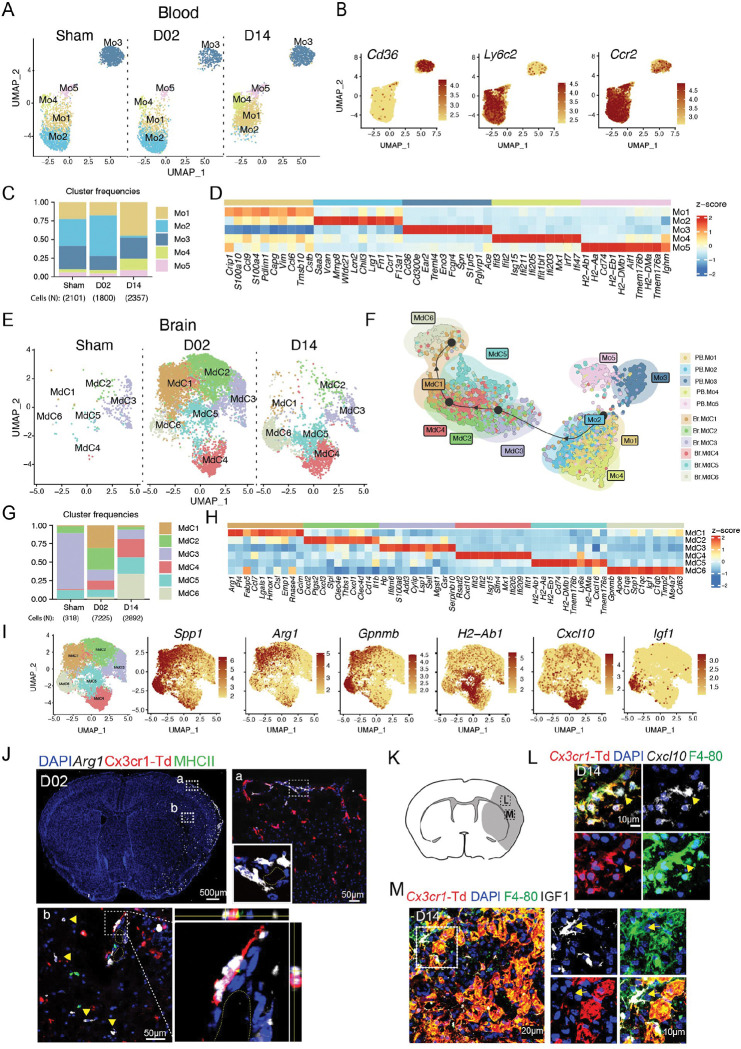
After sub-setting and re-clustering, we identified 5 clusters of blood monocytes (Mo1-5). All the clusters except for Mo2, which was almost absent at D14, were present at all studied time points (Fig. 4A). Based on Ly6c2 and Cd36 expression, Mo clusters were annotated as inflammatory (Ly6c2 high, Cd36 low) or patrolling (Ly6c2 low, Cd36 high) monocytes (Geissmann et al., 2003; Marcovecchio et al., 2017). Inflammatory monocytes separated into 4 clusters (Mo1,2,4 and 5), whereas patrolling monocytes constituted a single cluster (Mo3) (Fig. 4B). Mo1 was most abundant at D14 and was characterized by increased expression of S100 proteins and chemokines (S100a10, S100a4, Ccl9, Ccl6). On the other hand, Mo2 was highest at D02 and showed transcriptional profile of ‘neutrophil-like’ Ly6Chi monocytes (Saa3, Mmp8, Lcn2, Wfdc21, Lrg1, Chil3) 60. In addition to Cd36, Mo3 expressed several genes characteristic of patrolling monocytes (Ear2, Treml4, Eno3, Aceas) 60.
Figure 4. Inflammatory blood monocytes give rise to infiltrating brain macrophages (MdC) after stroke.
(A) UMAP plot of peripheral blood monocyte transcriptomes (Mo) in control surgery (Sham) or stroke mice 2 or 14 days (D02, D14) after injury, showing 5 monocyte clusters (Mo1-5).
(B) UMAP plots depicting single-cell gene expression of individual marker genes for monocytes (Ccr2) and for inflammatory vs. patrolling monocyte identification (Cd36, Ly6c2). Scale bar represents log of normalized gene expression.
(C) Bar graph showing relative frequencies of monocytes subclusters across Sham, D02 and D14 groups.
(D) Heatmap displaying expression of the top 10 upregulated genes in each monocyte cluster. Scale bar represents Z-score of average gene expression (log).
(E) UMAP plot of brain monocyte derived cell (MdC) transcriptomes for each studied time point shows 6 clusters.
(F) Slingshot trajectory analysis of peripheral blood monocytes (Mo) and brain macrophages (Mdc) subclusters. Each point is a cell and is colored according to its cluster identity shown in (A) and (E). The inferred trajectory shows transition from blood inflammatory monocyte clusters Mo1 and Mo2 to fully differentiated brain macrophages MdC1 and MdC6.
(G) Bar graph showing relative frequencies of MdC subclusters across Sham, D02 and D14 groups.
(H) Heatmap displaying expression of the top 10 upregulated genes in each MdC subcluster. Scale bar represents Z-score of average gene expression (log).
(I) UMAP plots displaying expression of marker genes for each identified MdC brain clusters. Scale bar represents Z-score of average gene expression (log).
(J) RNAscope fluorescent in situ hybridization (FISH) for Arg1 (white) combined with immunofluorescence (IF) brain images from Cx3cr1-Td+ mice 2 days after stroke. Top left: Overview of Arg1 expression (binary mask) in a whole brain section co-stained for DAPI (blue), showing high upregulation of Arg1 in cortical areas of the ischemic hemisphere. Top right and bottom: images of magnified areas (a) and (b) showing localization of Arg1 in Cx3cr1-Td− and MHCII− cells. Bottom right: orthogonal view of magnified area showing Arg1 expression in an adjacent cell to a PVM (Td+, red).
(K) Graphical representation of brain coronal section indicating anatomical regions where L and M images were acquired.
(L) FISH-IF images of the ischemic cortical region in D14-Cx3cr1-Td+ mice showing Cxcl10 expression (white) by MdC (Td−, F4-80+ cells, yellow arrow) near microglia (Td+, red; F4/80+, green) expressing Cxcl10. Nuclei are stained with DAPI (blue).
(M) IF images of the ischemic cortical region in D14-Cx3cr1-Td+ mice validating IGF1 expression (white) by MdC. Left: Image shows IGF1 expression by MdC (Td−, F4/80+, green) on brain surface. Right: Image shows clusters of mixed microglia (Td+, red) and MdC (Td−, F4/80+, green) expressing IGF1.
Furthermore, Mo4 was characterized by an interferon-stimulated gene (ISG) signature (Ifit3, Ifit2, Isg15, Ifi211, Ifi205, Ifit1bl1, Ifi203) and Mo5 by high expression of MHC class II genes (H2-Ab1, H2-Eb1, H2-Aa, H2-DMb1, H2-DMa) and Cd74, resembling monocyte-derived dendritic cells 61 (Fig. 4C–D).
Sub-clustering of brain MdC identified six populations. Sham brain displayed low numbers of MdC and most of them (~75%) clustered as MdC3, a cluster closely related to blood monocytes (Serpinb10, Plac8, Sell) 62(Fig. 4E and 4G, Fig. S7A–B). MdC1 and MdC2 were the predominant clusters at D02 and MdC6 was the major cluster at D14. MdC1 expressed a gene signature (Fabp5, Spp1, Gpnmb, Ctsl, Cd63, Ctsb, Ctsd, Arg1) that has been found in stroke-associated macrophages 16 and which is reminiscent of the gene signature found in either foamy macrophages in atherosclerotic plaques or lipid-associated macrophages in myocardial infarct (Fig. 4H, Fig. S7B) 63,64. MdC2 showed upregulation of neutrophil chemoattractants (Cxcl1, Cxcl2, Cxcl3) and pro-inflammatory genes (Ptgs2, Il1b, Clec4e), suggesting a role in driving post-stroke inflammation. Like MdC1, MdC6 showed increased expression of genes characteristic of stroke-associated macrophages and foamy macrophages (Gpnmb, Spp1, Cd63, Trem2 and Fabp5), but differ from MdC1 in that they were enriched for the repair growth factor Igf1 65. MdC4 cells showed an ISG signature (Rsad2, Cxcl10, Ifi205, Ifit2, Ifit1, Ifit3, Ifit1bl1, Isg15) (Fig. S7B), while MdC5 exhibited upregulation of MHC class II associated genes (H2-Ab1, H2-Aa, H2-Eb1, Cd74, H2-DMb1), a signature which identifies this cluster as monocyte derived-DC 61.
Slingshot trajectory analysis showed sequential transition of inflammatory peripheral blood Mo1 and Mo2 clusters to brain MdC3 cluster, followed by MdC2, MdC4 and MdC1 transition and final differentiation into MdC6. MdC5 appeared as the only MdC cluster dissociated from the trajectory (Fig. 4F). Neither patrolling Mo3 nor Mo5 were identified as MdC sources. Spearman correlation analysis between Mo and MdC clusters also showed proximity of blood monocytes to MdC3 and, consistent with the trajectory analysis, patrolling Mo3 cluster showed the lowest correlation with all brain MdC clusters (Fig. S7A). Thus, these analyses suggest that MdC in the inflamed brain derive from inflammatory monocytes, as recently described 60 and that both MdC1 and MdC6 constituted bona-fide tissue macrophages, while MdC2, MdC3, and MdC4 were clusters associated with transitional phenotypes.
Spp1 (osteopontin) and Gpnmb (osteoactivin) were preferentially upregulated in fully differentiated MdC1 and MdC6 clusters (Fig. 4H–I). Upregulation of these two genes in brain macrophages has been linked to tissue regeneration and neuroprotection after stroke 66,67. MdC1 was characterized by Arg1 expression, a gene associated with efferocytosis 68. Detection of Arg1 by combined FISH-IF revealed Arg1 increased at D02 in infiltrating macrophages (Cx3cr1-Td− cells) located on the brain surface or structures resembling parenchymal blood vessel as previously reported 69 (Fig4. J). In addition, we found upregulation of Cxcl10 at both D02 and D14 (MdC4) and of Igf1 at D14 (MdC6), similar to the one observed in microglia (Fig. 4K–M, Fig. S7C–D). Many of the Cxcl10 expressing MdC were organized in cell clusters together with Cxcl10 producing microglia indicating that local environmental cues in selected brain regions might drive the ISG phenotype in MdC and microglia.
Endothelial cells subclusters display a reactive signature early after stroke identified by Lrg1 expression.
EC segregated into nine subclusters (EC1-9), which by marker gene expression could be attributed to four arteriovenous segments: EC1 (venous capillaries; Car4, Tfrc), EC6 (large veins; Vcam1, Cfh, Scl38a5), EC3 (arterial capillaries; Fos, Fosb), EC4 (arteries; Gkn3, Mgp, Stmn2, Bmx), and EC7 (arteries; Clu, Cdh13, Mgp, Stmn2, Bmx) 70–72. EC2, EC6 and EC9, which clustered together in the UMAP space, showed upregulation of the endothelial venule marker Lrg1, a gene associated with angiogenesis after ischemic stroke 73. In addition, EC6 was defined by high expression of the venular atypical chemokine receptor Ackr1 74, lipocalin-2 (Lcn2), which has been implicated in stroke damage 75, and, consistent with a venous phenotype, adhesion molecules Icam1 and Vcam1. Attesting to their reactive state, EC2, which was the predominant cluster at D02, was defined by expression of Ecscr, a gene related to EC migration and vessel formation 76, Anxa2 and S100a11 implicated in endothelial fibrinolysis 77,78, and Scgb3a1, a endothelial secreted protein crucial for metastasis 79. EC9 was characterized by genes involved in EC metabolic reprogramming (Tyms, Dut, Dctpp1) 80 and proliferation (Pimreg, Pclaf). EC8 showed signature of fenestrated brain vascular ECs, likely stemming from choroid plexus EC (Plvap, Plpp3, Igfbp3, Plpp1, Cd24a, Ldb2) 81. Moreover, EC5 displayed increased ISG gene expression (Ifit3, Isg15, Rsad2, Ifit1, Usp18, Irf7, Ifit3b, Iigp1, Ifi44, Gbp2, Ifit2, Gbp3) (Fig. 5A–D). It has been reported that brain EC express IFN type I inducible genes at homeostasis, and that after stroke, EC show reduced ISG expression 82. Despite our data showing significant ISG scores in the Sham group, they were further increased at 2 or 14 days after stroke (Fig. S8). We also found that ECs, particularly EC1, EC3 and EC5, exhibited high Igf1r expression (Fig. 5B). Igf1r levels are elevated in brain endothelial cells compared to peripheral tissues 83. Because we found Igf1 expression in microglia, MdC, and BAM, we performed CellChat analysis to investigate interactions between ECs and IGF1 producing cells. Whereas Igf1-Igf1r communication occurred between BAM and EC in Sham and D02 groups, both microglia and MdC6 contributed to Igf1-Igf1r signaling network at D14 (Fig. 5E).
Figure 5. Endothelial cell (ECs) transcriptional changes and Igf1r signaling after stroke.
(A) Left: UMAP analysis of merged Sham, D02 and D14 EC transcriptomes discloses 9 subclusters; Right: UMAP of overlaid time points shows dissociation of EC2 cluster at D02 and general overlap of all the other clusters across Sham, D02 and D14 groups.
(B) UMAP plots depicting single-cell gene expression of individual marker genes for capillary (Car4, Fosb), arterial (Stmn2, Clu) and venular (Lrg1, Ackr1) ECs and for the endothelial receptors Ecscr and Igf1r. Scale bar represents log of normalized gene expression.
(C) Heatmap displaying scaled differential expression of the top 10 upregulated genes in each EC cluster. Scale bar represents Z-score of average gene expression (log).
(D) Bar graph showing relative frequencies of EC subclusters across Sham, D02 and D14 groups.
(E) Chord plots showing CellChat inferred ligand-receptor interactions between Igfr1 and Igf1 in Sham, D02 and D14 stroke mice. The strength of the interaction is indicated by the edge thickness. The color of the chord matches the cell cluster color sending the signal (Igf1). The number of cell cluster receptors (Igfr1), and their weight in the interactions, is indicated by the color-matched stacked bar next to each sender cluster.
(F) RNAscope in situ hybridization fluorescence (FISH) combined with immunofluorescence (IF) of Lrg1. Left: Overview of Lrg1 expression (binary mask, white) in a whole brain section co-stained for DAPI (blue), showing high upregulation of Lrg1 in the ischemic hemisphere Right: magnified images of brain cortical areas showing Lrg1 (white) colocalization with the EC marker CD31 (magenta) but not with the capillary marker Car4 (yellow). Nuclei are stained with DAPI (blue).
FISH-IF analysis for the venule marker Lrg1 70 revealed strong upregulation of vascular Lrg1 in the ischemic hemisphere (Fig. 5F). Co-detection of Lrg1 and Car4, a marker for venule capillaries, showed low overlap between both markers, indicating that Lrg1 was mainly induced in larger veins. LRG1, is a secreted glycoprotein primarily synthesized by neutrophils and hepatocytes, however, in conditions of altered homeostasis its transcription is upregulated in endothelial cells 84. Indeed, LRG1 expression increased in the mouse brain 1–3 days after MCA occlusion 85, although its localization in the endothelium was not demonstrated. Among its described vascular effects, LRG1 has been linked to pathological angiogenesis through modulation of endothelial cell TGFβ signaling 86 and it has also been described as a predictor for arterial stiffness and endothelial dysfunction 84,87. On the other hand, LRG1, produced from myeloid cells, contributed to cardiac repair after myocardial infarction in mice 88. Although we did not explore LRG1 activity in our stroke model, our data identify Lrg1 as a powerful marker of a specific subset of venular brain EC that respond early after ischemia.
Transcriptional changes of brain granulocytes indicate continuous recruitment from the circulation.
We performed subclustering and cell trajectory analyses to investigate whether granulocytes states evolve following a transcriptional continuum defined by either their migration from periphery to the injured brain or by their transition over the ischemic-reperfusion time. We detected 6 clusters of blood neutrophils, which relative frequencies were similar between Sham and D02 groups whereas at D14, blood Neu1, 2 and 4 clusters were practically absent and Neu3 became the predominant cluster (Fig. 6A–C). Neu1 showed increased expression of Cd14, Cebpb, Il1b and Lrg1. Neu2 was characterized by upregulation of genes described in immature neutrophils (Mmp8, Retnlg, Lcn2, Ly6g) 89, and Neu4 cluster was enriched in stefinA family genes (Stfa1, Stfa2, Stfa3, Stfa2l1, BC100530), which are cytoplasmic inhibitors of proteases such as cathepsins 90. On the other hand, Neu3, the predominant cluster at D14, was characterized by genes associated with neutrophil infiltration (Ninj1, Cd300c2) 91, cell growth inhibition (Lst1, Creg1) or defining a mature neutrophil state (Cd101) 92.
Figure 6. Granulocyte transcriptional changes through ischemia-reperfusion.
(A, D) UMAP plots of peripheral blood neutrophil (Neu) (A) and brain granulocyte (Gran)(C) transcriptomes by studied time point: control surgery or stroke mice 2 or 14 days after injury (Sham, D02, D14). Six clusters were identified in each data set (Neu1-6; Gran1-6).
(B, E) Bar graph showing relative frequencies of peripheral blood Neu (B) and brain Gran (D) clusters across Sham, D02 and D14 groups.
(C, F) Heatmap displaying scaled differential expression of the top 10 upregulated genes in each peripheral blood Neu (E) and brain Gran (F) clusters. Scale bar represents Z-score of average gene expression (log).
(G) Slingshot trajectory of combined peripheral blood neutrophils (Neu) and brain granulocytes (Gran) showing predicted cluster transition. Each point is a cell and is colored according to its cluster identity. The analyses point out transcriptional states changes of blood and brain granulocyte through the ischemic-reperfusion time.
(H) Top: Expression of single gene plotted as a function of pseudotime. Dot plots show expression levels of top cluster genes of combined blood Neu and brain Gran data sets along ischemic-reperfusion pseudotime. Each dot represents the expression levels (log) for each gene in a cell and is colored according to the group. Lines show average expression. Bottom: The top 100 genes that specifically covary with pseudotime were identified using generalized additive models and the log normalized expression values were plotted along the pseudotime axis. The location of genes plotted above is indicated.
Regarding the brain, granulocyte subclustering also identified 6 subsets. Most of the sham brain granulocytes fell into Gran1 and 4 clusters, which, similar to blood Neu2, were characterized by the upregulation of the immature neutrophil marker genes Retnlg, Mmp8, and Wfdc21 89. In addition, Gran4 was specifically characterized by induction of genes associated with early stage neutrophil development (Camp, Ltf, Chil3, Lcn2) 89. We observed that Gran1 and 4 frequencies gradually decreased over the ischemia-reperfusion time, whereas the frequencies of Gran2, which showed upregulation of ISG genes (Isg15, Ifit3, Rsad2, Ifit, Ifi204) and Gran 3, characterized by the cytokine genes (Ccl3, Ccl4, Csf1) augmented progressively over D02 and D14. Furthermore, we detected two small clusters, Gran5, which also contained eosionophil markers (Ccr3, Alox15), and Gran6, which showed upregulation of the cell cycle genes Pclaf, Banf1 and Prc1 (Fig. 6D–F).
Slingshot trajectory analysis of combined blood (Neu) and brain (Gran) granulocytes, indicated similarities between Gran1, which is the major Sham brain granulocyte cluster, and Neu1, 2, and 4, which were the main blood clusters at Sham and D02. On the contrary, Gran2 and 3 were disconnected from sham brain granulocytes (Gran1 and 4) and closer to blood Neu3, the predominat blood granulocyte cluster at D14 (Fig. 6G). Thus, this analysis suggests that, in contrary to MdC, granulocyte transcriptomic states are not developing within the tissue but suggests recruitment of brain infiltrating granulocytes from the circulating pool at early and late phases after ischemia-reperfusion. To further investigate granulocyte transcriptomic patterns over the ischemia-reperfusion time, we plotted gene expression as a function of pseudotime of selected genes that showed the strongest correlations with granulocyte transcriptomic shifts (Fig. 6H). We found that the pseudotime trajectory was characterized by the early expression of Retnlg, Mmp8, Ly6g, Anxa1 and Lcn2 genes, top marker genes describing Sham brain granulocytes and Sham and D02 blood neutrophils. Conversely, late expressed genes included Ccl3, Ccl4, Csf1, Gadd45b in D14 brain granulocytes, suggesting different functions of brain neutrophils under homeostatic conditions, the early and late phases of ischemic injury.
Transcriptional changes in dendritic cells (DC).
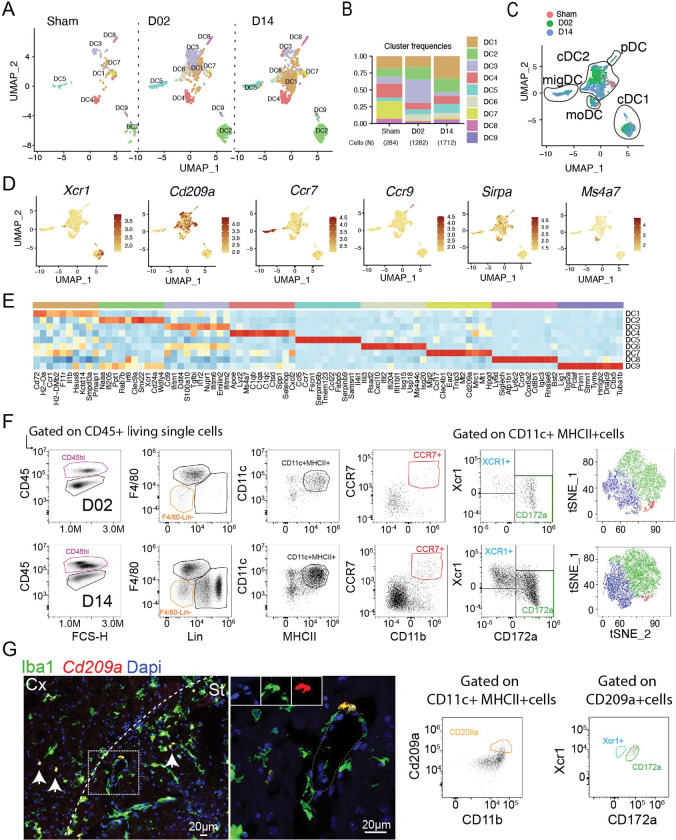
We identified nine DC subclusters in the stroke mouse brain. Low numbers of DC were found in brains of Sham mice, which gradually increased over the ischemia-reperfusion time (Fig. 7A–B). Expression signatures of the clusters were related to the five main DC populations based on canonical markers: conventional cDC1 (Xrc1, Clec9a) and cDC2 (CD209a, Sirpa (CD172a)), the plasmacytoid DC (pDC; Siglech, Ccr9, Bst2), a subpopulation with high Ccr7 expression representing migratory DC (migDC), and monocyte-derived DC (moDC; Ms4a7, Lyz2) 20,55,93–97 (Fig. 7C–E). We confirmed by flow cytometry the presence of DC (CD45hiF480−Lin-CD11c+MHCII+) subtypes cDC1 (XCR1+), cDC2 (CD172a+ and CD209a+) and migDC (CCR7+) in the ischemic brain (Fig. 7F). cDC1 were comprised by clusters DC2 and DC9. DC2 showed upregulation of Irf8 transcription factor, which is required for the full development of cDC198,99, whereas DC9 additionally showed transcriptional signature of cDC1 dividing cells (Lig1, Top2a, Mki67, Pcna) 100. cDC2, the largest population of brain DC, comprised DC1, DC3, DC6 and DC7 clusters. DC1, the cDC2 cluster displaying the highest frequency at D14, showed prominent upregularion of genes associated with antigen presentation (Cd72, H2-Oa, H2-DMb2), while DC7 showed expression of scavenger receptors (Clec4b1, Mrc1, Cd209a). DC4 exhibited high expression of monocyte/macrophage marker genes (Lyz2, Csf1r, Apoe, genes encoding complement C1q chains C1qa, C1qb, C1qc, Ms4a7, Trem2 and Cd14) identifying them as moDC 101 and DC6 was characterized by the upregulation of ISG genes (Ifit3, Rsad2, Cxcl10, Ifit2, Ifi204, Ifit1bl1, Isg15, Usp18). migDC were composed of a single subcluster (DC5) which, in addition to Ccr7, expressed other genes characteristic for migDC including Fscn1, Tmem123, Ccl22, and Socs2 55,102. DC8 classified as plasmacytoid DC based on the expression of Ccr9, Bst1, Il3ra, Siglech, Irf7, Ly6d 103 (Fig. 7E).
Figure 7. Transcriptional changes in brain dendritic cells.
(A) UMAP plots of brain dendritic cells (DC) transcriptomes for each studied time point identifies 9 clusters ((DC1-9).
(B) Bar graph showing relative frequencies of DC subclusters across Sham, D02 and D14 groups.
(C) UMAP of 3 color-coded time point overlay of brain DC. Classification of the clusters in DC subtypes is -DC (moDC) and plasmacytoid DC (pDC).
(D) UMAP plots displaying expression of marker genes for each identified DC cluster in the brain. Scale bar represents log of normalized gene expression.
(E) Heatmap displaying scaled differential expression of the top 10 upregulated genes in each DC subcluster. Scale bar represents Z-score of average gene expression (log).
(F) Flow cytometry analysis validating brain cDC1 (XCR1+), cDC2 (CD172a+) and migDC (CCR7+) subtypes identified by scRNA-seq after stroke.
(G) Left: FISH of Cd209a (red) expression in the brain, combined with IF for Iba1 (green) and nuclear satining with DAPI, showing Cd209a+Iba1+ cells around the vessels. Cx: cortex; St: striatum. Right: Flow cytometry analysis showing double positive CD209a+CD172a+ cells.
Regardig peripheral blood, we detected few DC as compared to other leukocyte types (Fig.1B). Subclustering analysis and automated cell type annotation identified 5 clusters which corresponded to cDC2 (DC1, DC4), moDC (DC2), and pDC (DC5) (Fig. S9). Whereas the number of blood cDC2 clusters DC1, DC2 and DC3 increased over the ischemic-reperfusion time, the number of DC5 (pDC) remained unchanged (Fig. S9A)
Transcriptional changes in lymphoid cells.
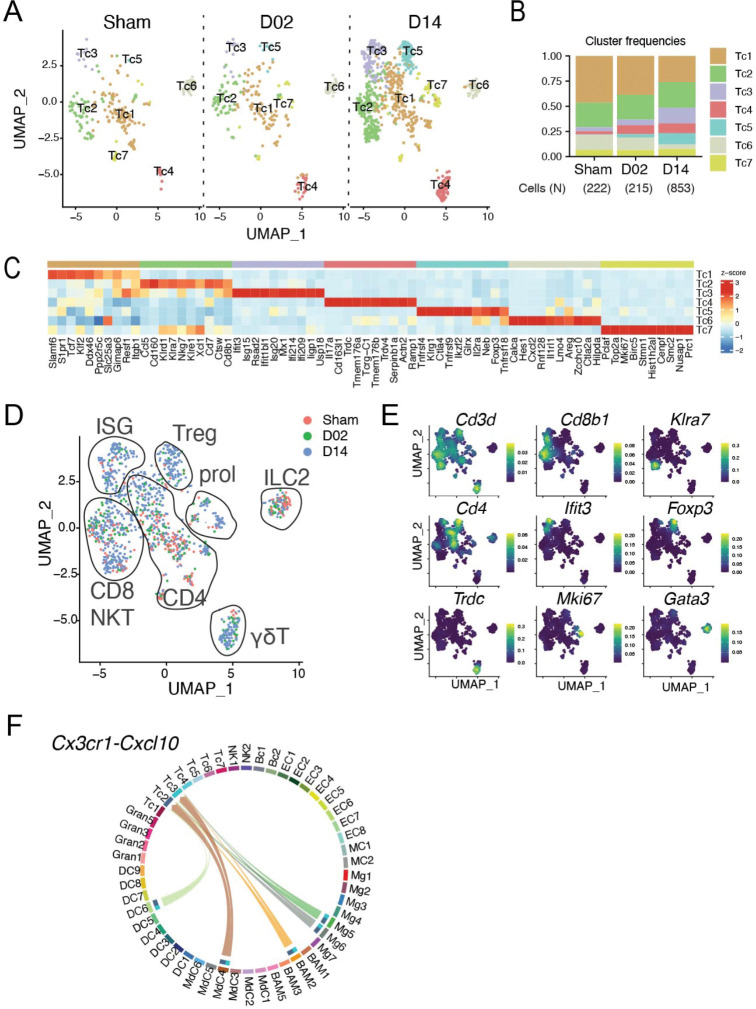
Brain-associated T cells split into seven clusters (Fig. 8A). T cells were more numerous at D14 as compared to Sham and D02 (Fig. 8B). Expression of Cd3d identified all but one cluster (Tc6) as bona fide T cells (Fig. 8E). Tc6 was classified as type 2 innate lymphoid cells (ILC2) based on the expression of Il1rl1, Gata3, Areg, and Calca 104 (Fig. S2A–B). Tc1 expressed Cd4 (Fig. 8E) and showed high S1pr1 and Klf2 expression (Fig. 8C) consistent with a low activation and differentiation state 105,106. Tc2 expressed features of cytotoxic T cells including CD8b1 and several killer cell lectin-like receptors (Klrd1, Klra7, Klrg7, Klre1) suggesting that the cluster was composed of conventional CD8 and natural killer T (NKT) cells. Cluster Tc3 was defined by a ISG signature (Ifit3, Isg15, Rsad2, Usp18 among others) and included CD8 as well as CD4 T cells (Fig. 8C–E). This cluster was expanded at D14 after stroke indicating increased IFN signaling during the subacute phase of ischemic injury (Fig. 8B). Of note, CellChat analysis showed Tc2 and Tc3 cells interacting with Mg5, Mg6, BAM2, MdC4 and DC6 through Cxcr3-Cxcl10 pathway at D14, indicating that myeloid Cxcl10 expressing cells might have a role in the recruitment of CD8+ T cells into the brain 107 (Fig. 8F). Tc4 expressed T cell receptors of the γδT cell lineage (Trdc, Trdv4, Tcrg-C1). Expression of Il17a, a cytokine that has been implicated in aggrevating stroke pathology 108–110, was confined to the Tc4 cluster suggesting that γδT and not Th17 are the major IL-17 producing T cells after ischemic brain injury. Cluster Tc5 showed features of regulatory T cells (Treg) including the expression of the canonical transcription factors Foxp3 and Ikzf2 (also known as HELIOS). Interestingly, cells in this cluster expressed TNF receptors Tnfrsf4, Tnfrsf9 and Tnfrsf18, which have been found in non-lymphoid tissue Treg but not in lymphoid-tissue associated Treg 111, possibly indicating that brain associated Treg don’t originate from lymphoid organs such as lymph nodes or spleen, but rather are recruited from non-lymphoid tissues or develop locally as previously suggested 11. Similar to Tc3 and consistent with previous studies addressing Treg kinetics after ischemic brain injury 11,112, Tc5 was expanded at D14 after stroke (Fig. 8A–B). Cluster Tc7 was characterized by expression of cell proliferation markers including Mki67, Top2a and Birc5 consistent with the presence of in situ T cell proliferation which was however not changed by stroke (Fig. 8B–C). No major changes were observed in the longitudinal composition of blood Tc (Fig. S10) which could be divided into five clusters constituting naïve CD8 (Tc1), CD4 (Tc2), memory CD8/NKT (Tc3) that showed a similar gene expression pattern as the brain Tc2 cluster, ISG T cells (Tc4), and a cluster with high expression of Lgals1, Rora, and several S100 genes (Tc5).
Figure 8. Transcriptional changes in brain lymphoid cells.
(A) UMAP plots of brain lymphoid cells (Tc) transcriptomes for each studied time point identifies 7 clusters (Tc1-7).
(B) Bar graph showing relative frequencies of Tc subclusters across Sham, D02 and D14 groups.
(C) Heatmap displaying expression of the top 10 upregulated genes in each Tc subcluster. Scale bar represents Z-score of average gene expression (log).
(D) UMAP of merged Sham, D02 and D14 Tc transcriptomes depicting identified Tc subtypes in diagrams: CD4, CD8, NKT, ILC2, ISG signature, γδT, Treg and proliferating T cells (prol).
(E) Density plots in the UMAP space showing the expression of selected marker genes used for lymphoid cell type identification. Scale bar represents densities based on kernel density estimation of gene expression.
(F) Chord plot showing cell-cell interactions between Cxcr3 and Cxcl10 in grouped Sham, D02 and D14 stroke mice. The strength of the interaction is indicated by the edge thickness. The color of the chord matches the cell cluster color sending the signal (Cxcl10). The number of cell cluster receptors (Cxcr3), and their weight in the interactions, is indicated by the color-matched stacked bar next to each cluster sender.
Similar to T cells, brain NK cells were increased at the D14 time point (Fig. S11B) and could be separated into two clusters (Fig. S11A). Brain NK1 was more similar to blood NK cells than brain NK2 as evidenced by higher Spearman correlation coefficient (Fig. S11D). CellChat analysis showed two major secretory axes confined to NK cells and some T cell, Il18 and Xcl1 signaling (Fig S11E–F). MdC4 were the main subset interacting with NK cells expressing Il18r1 receptors, a known activator of NK cells 113, whereas clusters DC2 and DC9 interacted with NK cells via Xcl1-Xcr1 pathway, possibly indicating a role of in cDC1 recruitment as previously reported 114.
Numbers and transcriptomes of brain associated B cells were not changed by stroke (Fig S11G) suggesting a minor role of B cells during the acute and subacute phases of ischemic brain injury.
Brain transcriptomic changes after stroke in aged mice.
The inflammatory response to ischemic brain injury differs between young and aged mice both in the transcriptomic response and the cellular composition of brain infiltrating immune cells 115,116. Therefore, we explored if aging alters cell signatures of immune and endothelial cells after stroke by preparing single cell transcriptomes from aged (17–20 months) Sham, D02, and D14 mice (Fig. S12A–E). The aged dataset was merged with the young brain dataset which was randomly down-sampled to match cell numbers of the aged dataset (aged = 11,999 cells). The combined data set showed distinct clusters for the major brain cell types including Mg, MdC, DC, Gran, Tc, BC, EC, and MC. The clusters of young and aged brains showed largely overlapping positioning in the UMAP space indicating the retention of the core transcriptome between the two groups (Fig. 9A). However, the frequency of infiltrating peripheral immune cell types differed between young and aged mice (Fig. 9C–D). Aged brains of Sham mice showed higher frequency of T cells while granulocytes and DC were more prominent in Sham brains of young mice indicating increased brain-associated T cells in brains of aged mice as previously reported 117. Overall, the cellular profile at D02 after stroke was comparable between groups, with a modest increase in neutrophils and decreased MdC in aged mice as previously reported 118 (Fig. 9C). At day 14, we observed increased Tc and reduced MdC and DC participation in aged as compared to young mice. At the subclass level we found that the distribution of Mg subclusters was similar between the age groups (Fig 9D). MdC clusters MdC5 and MdC6 were reduced in aged brains possibly reflecting the overall decreased brain MdC content at D14. Granulocytes showed a reduction in clusters Gran3 and Gran5 without overt changes in gene expression (Fig. 9D). T cells showed an expansion of CD8/NKT (Tc2) and ILC2 (Tc6) clusters whereas the frequency of γδT cells (Tc4) and Treg (Tc5) was reduced in aged brains 119,120. We analyzed differential gene expression of all cell clusters between aged and young mice and found that using a 2-fold change in expression level as a cutoff, overall, the number of genes that were exclusively either up- or downregulated in young stroke mice over young sham mice (D02 = 429 up, 380 down; D14 = 310 up, 406 down) was higher than in aged mice over aged sham mice (D02 = 73 up, 91 down; D14 = 67 up, 78 down) (Fig. 9E), whereas the majority of genes were regulated in both aged and young mice (Fig. 9E). We observed that young mice show higher number of regulated genes than aged mice, and that acute stroke (D02) led to higher exclusively regulated genes than subacute stroke (D14) in both young and aged mice (Fig. 9F). In addition, most of the exclusive DEG were detected in Mg, MdC, EC and to a lesser extend in Gran cell types (Fig. 9F). Among specifically upregulated genes in aged as compared to young mice, some belonged to the family of interferon inducible genes (Ifi204, Cxcl10, Ifit3, Nfkbiz) 82, which was reflected by a higher ISG score in several Mg, MdC, DC, and EC clusters in aged mice (Fig. 9G, Fig. S12F). Although it has been suggested that IFNβ signaling attenuates post-ischemic inflammation 121, the cellular sources of type I interferons in the post-ischemic brain have not been elucidated. In this study we found that Ifnb1 was upregulated in some subsets of MdC and microglia in both young and aged mice (Fig. 9H).
Figure 9. Comparison of the cellular composition and transcriptome signatures of brain and blood cells in aged and young stroke mice.
(A) UMAP plot representing color-coded brain cell clusters identified in aged and young single-cell transcriptomes.
(B) UMAP of 3 color-coded time point overlay of combined aged and young brain single-cell transcriptomes.
(C) Bar graph of relative frequencies of infiltrating peripheral immune cells in the brain across Sham, D02 and D14 groups.
(D) Bar graph showing relative frequencies of microglia (Mg), myeloid derived cells (MdC), granulocytes (Gran), T cells (Tc) and endothelial cells (EC) subclusters in either aged or young stroke mice.
(E) Scatterplot comparing stroke-induced differential gene expression (DGE) versus Sham in young and aged mice at D02 and D14. Genes with log2FC > 1 are highlighted in color. Green color indicates differentially regulated genes in both aged and young mice. Blue color indicates differentially regulated genes in only young mice. Red color indicates differentially regulated genes in only aged mice. The number of differentially regulated genes in each subgroups is indicated in the corresponding subquadrants.
(F) Upset plot of DGE results showing overlapping and age-specific (A, aged; Y, young) differentially expressed genes by cell type in response to either acute (D02) or subacute (D14) stroke.
(G) Module-score analysis for interferon response gene sets in aged and young brain cell clusters.
(H) Density plots in the UMAP space showing the expression of Ifnb1 in the brain of young and age mice. Scale bar represents densities based on kernel density estimation of gene expression.
Together, the data provide a high-resolution landscape of the inflammatory response to ischemic brain injury in aged mice, which, compared to the response of young mice, is characterized by a different cellular composition but remarkable stability of the transcriptomic response at the single-cell level.
Discussion
We sought to investigate the cellular immune landscape in the brain after transient cerebral ischemia during the early and late phases of the injury, and relate cellular signatures found in the brain to the cellular states of blood immune cells. In addition, we compared the cellular response between young and aged mice.
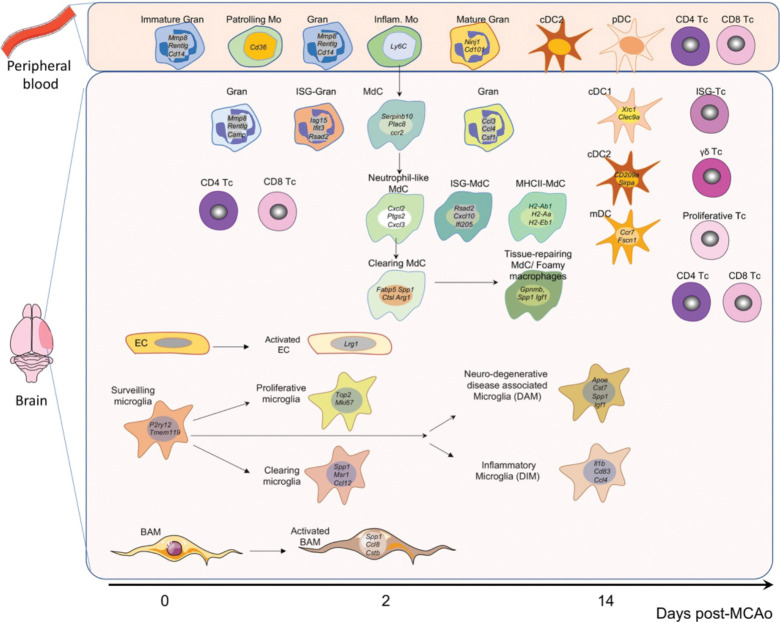
A major finding of this study is that there are distinct cellular transcriptomic responses of brain resident immune cells, infiltrating immune cells, and endothelial cells in the early and late phases of the tissue damage. Importantly, trajectory analysis showed that the transcriptome of blood-borne myeloid cells found in the brain after stroke remained distinct from their counterparts in the blood. These findings indicate that the local tissue milieu rather than peripheral immune priming determines the cellular state of monocyte-derived cells and neutrophils (Fig. 10).
Figure 10. Summary of the main cell clusters identified in the blood and brain after experimental stroke.
Among the brain resident cells analyzed in this study, microglia showed the largest diversification in their transcriptional response across time points, whereas BAM and EC showed the strongest deviation from Sham states at D02 while the transcriptomes at D14 were more similar to Sham condition.
Another key finding of this study is that microglia showed a strong proliferative response at D02 possibly triggered by the pronounced loss of microglia in the ischemic core. Similar proliferative responses have been observed after pharmacologic depletion of microglia 122, suggesting that depleting the microglial niche rather than a selective response to ischemic injury is inducing microglia proliferation. We found that proliferating microglia where primarily localized in the periinfarct region and that, at the D14 time point, the ischemic territory including the core areas was repopulated by microglia at a higher density than the one observed in non-injured tissue. Microglia acquired transcriptional states that were specific for the time points analyzed while there was no clear trajectory of microglia transcriptional states that would indicate a transition from the proliferative state observed at D02 to the inflammatory state that is prominent at D14.
Upregulation of interferon-stimulated genes has been observed in a variety of brain disease models including stroke 47,48,123. Similar to a previous study that identified ISG signatures in Mg, MdC, BAM, neutrophils, EC, and vascular smooth muscle cells one day after stroke 17, our study found ISG signatures in the same cell types but also in DC and Tc, suggesting a generalized IFN type I response independent of cell types and post-stroke time points (Fig. 10). Consistent with previous studies utilizing bulk RNA sequencing 124, we found a marginal but significant increase of ISG expression in aged animals after stroke, although the cell types and clusters showing this response were not changed. The IFN type I response is initiated by IFNα or IFNβ signaling, and we found Ifnb1 expression primarily in MdC and to a lesser extend in Mg with a similar pattern in young and aged mice whereas we could not detect the expression of any of the Ifna gene subtypes (14 genes in the mouse).
MdC showed a continuous pseudotime trajectory originating from Ccr2hi/Ly6c2hi blood monocytes, gradually loosing monocytic marker genes and acquiring transcriptomic characteristics of tissue macrophages (Fig. 10). This is consistent with a longitudinal in situ transformation of macrophage phenotypes that is independent of de novo recruitment of blood monocytes as previously suggested 125,126. In contrast, and consistent with their short life span, brain granulocytes did not exhibit a pseudotime trajectory that was independent of their blood origins resulting in trajectories that suggest continuous recruitment from the circulating pool.
Another finding of this study is that cellular states of different myeloid populations can converge into similar transcriptomic phenotypes. For example, “monocyte derived-DC” (MdC5) showed high resemblance to DC clusters in their expression of MHC class II and co-stimulatory molecules while retaining an overall monocyte/MdC phenotype (expression of Ly6c2, Lyz2, Cd14, Ms4a7). Similarly, Mg clusters Mg3 and Mg7 share marker genes (Spp1, Gpnmb, Apoe, Igf1) with MdC6. In addition, we observed that microglia and MdC expressing the ISG marker gene Cxcl10 were arranged into discrete clusters possibly indicating that myeloid cells organize into functional units to execute specific immune programs independently of their ontogeny.
A major limitation of this study is the restriction to two time points after cerebral ischemia. Our analysis at D02 and D14 identified high plasticity in the immune response and the transition of cell states can be only partially modeled on the provided data. For example, we have no information on the fate of proliferating microglia at D02 as we could not detect gene trajectories that would indicate progression to Mg clusters at D14. This is most likely due to the fact that transitional states might be present at time points not analyzed in this study. Similarly, endothelial cells showed a response at D02 that indicated initiation of migratory and proliferative response, whereas their transcriptomic profile at D14 was similar to Sham EC. Given that endothelial cells surrounding the infarcted brain area start to proliferate as early as 12–24 hours following occlusion and, that the angiogenic response and formation of new vessels continued more than 21 days post-stroke 127,128, it is likely that this response is not covered by our data.
Although our scRNA-seq study is one of the first to incorporate young and aged animals, we were not able to include aged male mice for the 14 day time point due to the high mortality rate observed in this group after transient focal ischemia, a drawback that has also been reported by others 120
Finally, because our investigation only included immune and endothelial cells, the response of other cell types including neurons, astrocytes, oligodendroglia, and pericytes were not assessed. This limitation is pertinent to the overall study design where we prioritized in depth resolution of cellular states of low abundance cell types such as BAM over the coverage of all brain cell populations. As more single cell RNA-seq datasets from experimental stroke studies become available it might be possible to integrate data from studies that used untargeted and targeted approaches to deliver a more comprehensive landscape of the immune response after stroke.
Taken together, by analyzing the immune response in the brain and peripheral blood at single cell resolution during acute and sub-acute stages after stroke, this study identifies cell type and time point specific immune programs and contributes to the ongoing efforts to compile a longitudinal cellular map of the immune response after stroke.
Methods
Mice
All procedures were approved by the institutional animal care and use committee of Weill Cornell Medicine and were conducted in accordance with the ARRIVE guidelines 129,130. Experiments were performed in young (8–12 week-old) male wild type mice obtained from Jackson Laboratory (IMSR_JAX:000664; Bar Harbor, ME) and aged (17–18 month-old) male and female wild type mice obtained from the NIA-NIH. All in house bred mice were on a C57Bl/6J background and included B6.Cg-Gt(ROSA)26Sortm14(CAG-tdTomato)Hze/J (IMSR_JAX:007914), B6.129P2(C)-Cx3cr1tm2.1(cre/ERT2)Jung/J (IMSR_JAX:020940) and C57BL/6-Tg(CAG-EGFP)131Osb/LeySopJ (IMSR_JAX:006567).
Middle Cerebral Artery occlusion
Transient focal cerebral ischemia was induced using the intraluminal filament model of middle cerebral artery (MCA) occlusion as described previously 131. Briefly, under isoflurane anesthesia (maintenance 1.5–2%), the MCA was occluded for 35 minutes using a 6–0 Doccol monofilament (#L12; Sharon, MA). Reperfusion was confirmed by measuring the cerebral blood flow (CBF) in the MCA territory by transcranial laser Doppler flowmetry (Periflux System 5010, Perimed, King Park, NY). Only animals with CBF reduction of >85% during MCA occlusion and CBF recovered by >80% after 10 minutes of reperfusion were included in the study (Jackman et al., 2011). Rectal temperature was monitored and kept constant (37.0±0.5°C) during the surgical procedure and in the recovery period until the animals regained full consciousness. After, mice were housed in a single chamber environmental incubator (Darwin Chambers, Saint Louis, MO) to hold mice at 29–32°C for up to 4 days. Topical lidocaine and bupivacaine (0.25%, 0.1 ml, transdermal) were used for pre-operative analgesia and buprenorphine (0.5 mg/kg q12h; s.c.) as post-operative analgesia for 72 hours.
Tamoxifen treatment
Four-week old male Cx3cr1CreER-Td-tomato mice were treated with tamoxifen (T5648, Sigma; 80mg/Kg) dissolved in corn oil (C8267, Sigma) by intraperitoneal injection during 5 consecutive days. The mice were used for experiments 6 to 8 week after treatment.
Cell isolation
Isolation of brain and blood cells from sham and stroke mice were performed at the same time of the day (8:00–9:00 am) for all experiments to limit the effects of circadian gene expression variation 132. Mice were anesthetized with pentobarbital (100 mg/Kg, i.p.) and transcardially perfused with heparinized PBS (2U/ml). Briefly, either control-sham or ischemic hemispheres were separated from the cerebellum and olfactory bulb and gently triturated using a Gentle MACS dissociator (Miltenyi Biotec, Auburn, CA). Single cell suspensions were obtained by enzymatic digestion with papain (Neural Tissue Dissociation Kit (P), #130-092-628, Miltenyi Biotec) following the manufacturer’s instructions. Four to five hemispheres were pooled for each experiment. In order to increase cell viability and to preserve the transcriptional state during the generation of single-cell suspensions, Brilliant Blue G (BBG, P2X7 receptor antagonist, 1μM, Sigma), AP-5 (25 nM, NMDA receptor blocker, Tocris), and actinomycin D (RNA polymerase inhibitor, 5μg/ml, Sigma) were added to the dissociation solution 133 134. Next, cell suspension was filtered through a 70 μm filter, resuspended in 30% Percoll (GE Healthcare)-HBSS containing 1μM BBG, and spun for 10 minutes at 700 g. After gradient centrifugation, the myelin layer was removed, and the cell pellet was resuspended in 2%FBS-PBS buffer and filtered through a 40 μm filter. Isolated cells were washed and resuspended in 100 μl of blocking buffer per hemisphere to proceed for FACS staining and cell sorting.
For isolation of peripheral leukocytes, mice were anesthetized with pentobarbital (100 mg/Kg, i.p.) and 0.5 ml of blood was collected by cardiac puncture into heparinized tubes. For each experiment the blood from two mice (1 ml total blood) was pooled and erythrocytes were lysed. BBG (1μM) and actinomycin D (5μg/ml) inhibitors were added during the isolation procedure. After erythrolysis, blood cells were resuspended in MACS buffer (PBS supplemented with 2% FBS, 2 mM EDTA; 300 μl/107 cells) and incubated with a biotinylated Ter-119 antibody (Table S1) and remaining erythrocytes were depleted with anti-biotin microbeads according to the manufacturer’s instructions (Miltenyi Biotec). Afterward, cells were washed and resuspended in 0.01% BSA-PBS at a concentration of 105 leukocytes/ml for Drop-seq processing.
Flow Cytometry and Cell Sorting
For Drop-seq experiments, brain single cell suspensions were incubated with anti-CD16/CD32 antibody for 10 min at 4°C to block Fc receptors, followed by staining with CD45-BV510, Ly6C-FITC, CX3CR1-PE, Ly6G-PercP-Cy5.5 and CD11b-APC-Cy7 antibodies for 15 min at 4°C (Table S1). CD45hi cells, microglia (CD45hiCD11b+CX3CR1+) and endothelial cells (CD45−Ly6C+) were sorted on an Aria II cytometer (BD Bioscience) and collected in 0.5 ml of 0.01% BSA-PBS for Drop-seq. Flow cytometry gating strategy is described in Fig. S1. Analytical flow cytometry was performed on a NovoCyte Flow Cytometer (Agilent, Santa Clara, CA). The antibodies used are described in Table S1. Appropriate isotype controls, ‘fluorescence minus one’ staining, and staining of negative populations were used to establish gating parameters.
Generation of single cell RNA libraries by Drop-seq
Single-cell transcriptomes of sorted brain cells and purified blood leukocytes were prepared by drop-seq as described 135 with modifications. Cells were resuspended in PBS-0.01%BSA to a final concentration of 100 cells/μl. Barcoded capture beads (ChemGenes Corporation, Wilmington, MA) were resuspended in 1.8 ml lysis buffer consisting of 4M Guanidine HCL (Thermofisher Scientific, Waltham, MA), 6% Ficoll PM-400 (Sigma-Aldrich), 0.2% Sarkosyl (Sigma-Aldrich, St. Louis, MO), 20mM EDTA (Thermofisher Scientific), 200mM Tris pH 7.5 (Sigma-Aldrich), 50mM DTT (Sigma-Aldrich) at a concentration of 120 beads/μl. A 5mm diameter, 1.7mm thick PVDF encapsulated magnetic stir disc and rotary magnetic tumble stirrer (V&P Scientific, San Diego, CA) was used along with a 3 ml syringe that contained the beads in lysis buffer to keep the beads in suspension. Thereafter, single cells and beads were encapsulated in nanoliter-scale droplets using a Drop-seq microfluidic device coated with Aquapel (FlowJEM, Toronto Canada), droplet generation oil (BioRad, Hercules, CA) using flow rates of 4 ml/h for cells and beads and 15 ml/h for oil. Each run typically lasted about 18 minutes. After removing the oil, droplets were resuspended in 30 ml of room temperature 6X SSC (Promega, Madison, WI) and 1 ml perfluorooctanol (Sigma-Aldrich, St. Louis, MO) and shaken vigorously 6 times vertically to break the droplets. The beads were captured by loading them into a 20 ml syringe with an attached 0.22μm Millex-Gv syringe filter (Millipore Sigma, Burlington, MA) as previously described 136. Beads were washed with 2 × 20 ml of ice cold 6X SCC. The syringe filter was then inverted, and a 10 ml syringe was used to flush the beads out with 10 ml ice cold 6X SSC repeated for a total of 3 times. Beads were collected by centrifugation at 1,250 × g for 2 min at 4°C with low brake setting. The remainder of the Drop-seq protocol followed the one published 135. cDNA was amplified by PCR using the following parameters: 95°C (3 min); 4 cycles of 98°C (20 s), 65°C (45 s), 72°C (3 min); 11 cycles of 98°C (20 s), 67°C (20 s), 72°C (3 min). Libraries were quantified by quantitative PCR and checked for quality and size distribution on a Bioanalyzer (Agilent) before sequencing on an Illumina NextSeq500 instrument using the 75 cycle High Output v2 kit (Genomics Core Facility, Cornell University, Ithaca, NY). Three to four libraries were multiplexed into a single run. We loaded 1.8 pM library and provided Drop-seq Custom Read1 Primer at 0.3 μM in position 7 of the reagent cartridge without PhiX spike-in using a read configuration of 20 bases (Read1), 8 bases (Index1), and 64 bases (Read2). Details regarding each Drop-seq run and characteristics of the strain, sex, age, and number of mice used for DropSeq can be found in Table S2.
Data pre-processing
Demultiplexed fastq files were cleaned of reads not passing the Illumina Passing Filter with fastq_illumina_filter (version 0.1) and processed with the Drop-seq Tools (version 2.3.0) pipeline 137. Briefly, each transcriptome Read2 was tagged with the cell barcode (bases 1 to 12) and unique molecular identifier (UMI) barcode (bases 13 to 20) obtained from Read1, trimmed for sequencing adapters and poly-A sequences, and aligned using STAR v2.7.3a 138 to the mouse reference genome assembly (Ensembl GRCm38.94 release). Reads aligning to exons were tagged with the respective gene symbol and counts of UMI-deduplicated reads per gene within each singular cell barcode were used to build a digital gene expression (DGE) matrix. The DGE matrix contained 40,000 cell barcodes associated with the highest numbers of UMIs. We used the DecontX method from the R package celda 139, a Bayesian hierarchical model to estimate and remove cross-contamination from ambient RNA, to construct a corrected DGE matrix. Cells with fewer than 200 UMIs, more than 10,000 UMIs, or more than 20% mitochondrial genes were excluded. We used Doubletfinder (RRID:SCR_018771) 140 to computationally detect cell doublets with an expected doublet rate of 5% 135 as input parameter. Cells tagged with a “Doublet” call were removed. Finally, the corrected DGE matrices were merged into a single matrix.
Bioinformatic analysis and statistics
We used Seurat (version 4.1.0; RRID:SCR_016341) 141 for down-stream analysis.
Counts were log-normalized for each cell using the natural logarithm of 1 + counts per ten thousand.
The 3000 most variable genes were identified by calling FindVariableFeatures.
We next standardized expression values for each gene across all cells by Z-score transformation (ScaleData).
Principle Component Analysis (PCA) was performed on the scaled variable gene matrix. The R package Harmony (RRID:SCR_022206) 142 was used to correct the matrix for batch effects. The data from replicate experiments were combined into four sets so that each set included a Sham, D02, and D14 experiment. We run Harmony on the first 40 PCA dimensions with a maximum of 20 iterations.
We used the Uniform Manifold Approximation and Projection (UMAP) 143 for dimensional reduction and visualization of Harmony-derived embeddings in a two-dimensional space with preset parameters by invoking the RunUMAP function in Seurat utilizing the 40 first components of the Harmony reduction.
We used the Louvain algorithm as implemented in FindClusters with a resolution setting of 1.2 for the brain and 0.7 for the peripheral blood dataset to perform graph-based clustering on the neighbor graph that was constructed with the FindNeighbors function call on Harmony-derived embeddings.
After clustering, we used the model-based analysis of single cell transcriptomics (MAST) algorithm 144 in the FindAllMarkers function to find differentially expressed genes in each cluster based on the log-normalized expression matrix with parameters: only.pos = T, min.pct = 0.1, logfc.threshold = log2(1.5), max.cells.per.ident=2000.
We performed unsupervised cell type annotation using the SingleR package (RRID:SCR_023120) 145 with ImmGen 146, BrainImmuneAtlas 55, and Tabula Muris 147 as reference datasets (see Fig. S2). Assignments were further manually validated by scoring the 10 most differentially expressed genes for the presence of canonical marker genes for each cell type. On these bases, we assigned the metacells to microglia (Mg), border-associated macrophages (BAM), monocyte-derived cells (MdC), granulocytes (Gran), mast cells (MaC), dendritic cells (DC), T cells (Tc), NK cells (NK), B cells (Bc), endothelial cells (EC), vascular mural cells (MC), epithelial-like cells (Epi), and oligodendrocytes (OD) clusters for the brain dataset, and monocytes (Mo), granulocytes (Gran), eosinophils/basophils (Eos/Bas), dendritic cells (DC), T cells (Tc), NK cells (NK), B cells (Bc), various precursors (pre), and one unclassified cluster (UC) for the peripheral blood dataset.
To achieve further resolution of cell states, individual count matrices were generated based on the initial cluster designation and steps 1–7 were repeated with the following modifications: Mitochondrial, ribosomal, and gene model (Gm) annotated genes were removed; Step 2, 2000 variable features were selected; Step 4, Harmony was run on the first 15 PCA dimensions; Step 5, The “min.dist” parameter in the RunUMAP function was set to 0.1 for clusters with more than 2500 cells; Step 6, FindClusters was performed at a resolution of 0.4. Differentially expressed genes with an FDR < 0.05 were ranked by their log2 fold change and z-scores were computed on the average gene expression across clusters for visualization in heatmaps.
Cluster pruning and metacell exclusion
After subclustering we detected several clusters within the brain dataset with high expression of microglial (Hexb, Siglech, P2ry12) marker genes together with either granulocyte (S100a8, S100a9, Cxcr2), DC (Cd209a, Xcr1, Ccr7), T cells (Trac, Trbc2, Cd3d), NK cells (Nkg7, Gzma), and macrophage (Mrc1, Lyve1) genes. Although we cannot exclude that this is due to biological processes such as transcriptomic changes or engulfment of living cells by microglia as previously reported for neutrophils and lymphocytes 148, we opted to exclude these cells as potential cell doublets. Therefore, we manually removed 1730 Mg, 287 DC, 145 BAM, 70 Gran, 65 Tc, and 41 NK metacells. After exclusion of these cells, we rerun steps 1–7 on the main dataset and performed de novo subclustering as described above.
Analysis of combined young and aged brain datasets
The dataset from aged (17–20 month) mice brains were preprocessed as described above. Cell identities in the aged mice brain dataset were assigned using the FindTransferAnchors and TransferData functions in Seurat using the young brain dataset as reference. Seurat objects from young and aged mice were merged into a single object by retaining all cells of the aged brains and randomly down-sampling the young brain dataset. This resulted in balanced cell numbers among cell type classes in both datasets (Table S3). Raw count data were processed as described above. The Harmony algorithm was run on the first 40 principal components of the PCA. UMAP was computed on the first 40 Harmony dimensions. Differential gene expression (Fig 9E) was computed with limma-voom with default parameters 149 after “pseudobulk” conversion of individual experiments using the aggregateAcrossCells function of the scuttle package 150 following the removal of mitochondrial, ribosomal, hemoglobin, gene model (Gm) annotated genes, and the sex-specific genes (Tsix, Xist).
Cell trajectory inference
We conducted the trajectory inference (TI) analysis using the Dyno package 151. Brain and peripheral blood datasets were merged and randomly downsampled to a maximum of 1000 cells per cluster and treatment. The merged dataset was subset to contain either Mg, blood Mo and brain MdC, or blood Neu and brain Gran clusters. The TI was performed on the 2000 most variable genes selected with Seurat FindVariableFeatures function. For Gran and Mo/MdC, blood granulocytes and monocytes were designated as starting population, respectively. For Mg, the starting population consisted of all Mg from Sham brains. The most appropriate TI method (Slingshot) 152 was selected based on Dyno (guidelines_shiny function) recommendations. UMAP was calculated on the first 15 PCA dimensions and the 2-dimensional space and trajectories were visualized by the plot_dimred function of the Dyno package.
CellChat analysis
Cell-cell interaction networks were constructed using the CellChat R package (RRID:SCR_021946) 153 with a custom mouse ligand-receptor interaction database that contained combined entries of the curated RNAMagnet 154 and CellChat databases. Briefly, the processed Seurat object was split by treatment into Sham, D02, and D14 datasets. Then, cell clusters were randomly downsampled to contain no more than 1000 cells. The matrices were used as input for the createCellChat function and processed using its standard pipeline. Differentially expressed genes and interactions were identified in the CellChat object via identifyOverExpressedGenes and identifyOverExpressedInteractions, respectively. The CellChat algorithm was then run to calculate the probable interactions at the cell-to-cell level via computeCommunProb with “truncatedMean” as the method for computing the average gene expression after removing 15% of observations from each end of the gene expression vector (parameter: trim = 0.15). The filterCommunication function was used to filter out interactions with less than 20 cells in each cluster. Communication probabilities on the signaling pathway level were then calculated by invoking the computeCommunProbPathway function. Ligand-receptor interaction probabilities were visualized via netVisual_aggregate and netVisual_individual. For joint analysis of different group datasets (Sham, D02, D14), CellChat objects were merged and compareInteractions was used to compare the total number and strength of interactions between treatments followed by the rankNet function for visualization.
Modular score calculation and visualization
AddModuleScore function in the Seurat package was used to calculate the functional signatures of each cell cluster. The type I interferon-response score was calculated using interferon-stimulated genes (ISG) as previously reported 155. Disease-associated microglia (DAM) and disease inflammatory macrophages (DIM) scores were calculated using the gene list reported by Silvin et al.38. Additional module scores were calculated for Foamy Macrophages 63, Stroke-Associated macrophages (SAM) 16, and monocytes 156. All marker genes used for score calculation can be found in Supplementary Table S4. Module scores were visualized in R using the ggplot2 (V3.3.6) package (RRID:SCR_014601) 157.
Correlation plots
Cluster-wise average gene expression was calculated with the AverageExpression function of the Seurat package. The cor function in R were used to construct a Spearman correlation matrix of gene expression between blood monocyte and brain MdC clusters. The correlation matrix was visualized with the corrplot function of the corrplot (V0.92) package (RRID:SCR_023081) 158.
Combined analysis of young and aged brain scRNA-seq data
Cell identities were assigned by aligning both datasets in the high-dimensional space and projecting cluster annotations from the young dataset onto the aged dataset essentially as previously described 141 and by using SingleR to adjudicate cell identities using the young dataset as a reference. For each cell the maximal correlation score obtained by the two methods was used to assign the cell identity. The young dataset was down-sampled to contain no more than 5000 cells per treatment and both datasets were merged and processed using steps 1–6 as outlined above (Bioinformatics analysis). Differential gene expression between aged and young mice was calculated for each cell cluster using the MAST algorithm of the FindMarkers function in Seurat with preset parameters after excluding mitochondrial, ribosomal, hemoglobin, sex-specific (Tsix, Xist), and gene model (Gm) annotated genes. Genes were considered differentially regulated if they showed higher than two-fold change in expression level and an FDR < 0.05.
Brain histology
Mice were deeply anesthetized with sodium pentobarbital and transcardially perfused with ice cold PBS (30ml) followed by 4% paraformaldehyde (PFA) in PBS (100ml). Brains were dissected, post-fixed in 4% PFA overnight, dehydrated in 30% PBS-sucrose solution for 1–2 days and then frozen using dry ice. Frozen brains were then embedded in Epredia™ M-1 Embedding Matrix (Thermo Ficher Scientific) and cut in coronal sections using a cryostat (Leica CM3050S, Mannheim, Germany), and mounted on slides for either fluorescence immunocytochemistry (IF) downstream applications or RNA-fluorescence in situ hybridization (FISH).
For IF, coronal 18 μm-thick sections were permeabilized with 0.5% Triton X-100 (Sigma) in PBS (PBST), blocked with 5% normal donkey serum (NDS) in 0.1% PSBT for 1 hour and incubated overnight at 4°C with primary antibodies (table 2) in 1%NDS-0.1% PBST. After ON incubation, sections were washed 3×5min with 0.1%PBST and incubated with secondary antibodies in 1%NDS-0.1%PBST for 1 hour at RT. Sections were washed with 0.1% PBST 2×5min, followed by a 1×5min wash in 0.1%PBST-DAPI (1:100, 12.5ng/ul), mounted with FluorSave Reagent (Millipore) and visualized using either the Olympus IX83 or Leica TCS SP8 confocal microscopes. Primary and secondary antibodies used are described in Table S5.
RNA-FISH was performed using RNAscope Multiplex Fluorescent Kit v2 (ACD-Bio-Techne, Newark, CA) following the manufacturer’s instructions. Briefly, 10-μm thickness sections were processed using RNAscope hydrogen peroxide, followed by boiling Target Retrieval solution, dehydrated by 100% ethanol, and incubated in Protease III solution. Afterwards, tissue sections were hybridized with the target probes (Table S6) for 2 h at 40 °C, followed by a series of signal amplification and washing steps. Hybridization signals were detected by fluorescent signal using peroxidase-based Tyramide Signal Amplification-Plus Fluorescein or TSA Plus Cyanine 5 (Perkinelmer, Shelton, CT). Finally, the sections were counterstained with DAPI and coverslipped using ProLong Gold Antifade Mountant. Images were acquired using either an epi-fluorescent (Olympus IX83, Waltham, MA) or a confocal (Leica) microscope. Images were analyzed using Fiji (RRID:SCR_002285) 159. When FISH and IF staining were combined, the slides were washed in RNAscope wash buffer after the development of Tyramide Signal and then, IF was sequentially performed as described above but skipping the permeabilization step.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Mason for helpful discussions. The generous support of the Feil Family Foundation is gratefully acknowledged.
Funding
This work was supported by NIH grants R01NS081179 (JA), R01NS34179 (CI), the Leducq Foundation (StrokeIMPaCT Network; JA), and the Sackler Brain and Spine Institute Research Grant (LGB)
Footnotes
Declaration of interests
The authors declare no competing interests.
Data availability
The raw and processed data and metadata of all scRNA-seq datasets included in this study are available in the GEO repository (GSE225948). A public accessible interactive web portal for exploring the scRNA-seq data included in this study has been developed (https://anratherlab.shinyapps.io/strokevis/).
References
- 1.Nogueira R. G. et al. Thrombectomy 6 to 24 Hours after Stroke with a Mismatch between Deficit and Infarct. N Engl J Med 378, 11–21 (2018). 10.1056/NEJMoa1706442 [DOI] [PubMed] [Google Scholar]
- 2.Albers G. W. et al. Thrombectomy for Stroke at 6 to 16 Hours with Selection by Perfusion Imaging. N Engl J Med 378, 708–718 (2018). 10.1056/NEJMoa1713973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Collaborators, G. B. D. S. Global, regional, and national burden of stroke and its risk factors, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Neurol 20, 795–820 (2021). 10.1016/S1474-4422(21)00252-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mensah G. A. et al. Decline in Cardiovascular Mortality: Possible Causes and Implications. Circ Res 120, 366–380 (2017). 10.1161/CIRCRESAHA.116.309115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shi K. et al. Global brain inflammation in stroke. Lancet Neurol 18, 1058–1066 (2019). 10.1016/S1474-4422(19)30078-X [DOI] [PubMed] [Google Scholar]
- 6.Garcia-Bonilla L., Iadecola C. & Anrather J. Cerebral Ischemia and Inflammation. 7th edn, 117–128.e5 (Elsevier, 2021). [Google Scholar]
- 7.Iadecola C., Buckwalter M. S. & Anrather J. Immune responses to stroke: mechanisms, modulation, and therapeutic potential. J Clin Invest 130, 2777–2788 (2020). 10.1172/JCI135530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sakai S. & Shichita T. Inflammation and neural repair after ischemic brain injury. Neurochem Int 130, 104316 (2019). 10.1016/j.neuint.2018.10.013 [DOI] [PubMed] [Google Scholar]
- 9.Pedragosa J. et al. CCR2 deficiency in monocytes impairs angiogenesis and functional recovery after ischemic stroke in mice. J Cereb Blood Flow Metab 40, S98–S116 (2020). 10.1177/0271678X20909055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cuartero M. I. et al. N2 neutrophils, novel players in brain inflammation after stroke: modulation by the PPARgamma agonist rosiglitazone. Stroke 44, 3498–3508 (2013). 10.1161/STROKEAHA.113.002470 [DOI] [PubMed] [Google Scholar]
- 11.Ito M. et al. Brain regulatory T cells suppress astrogliosis and potentiate neurological recovery. Nature 34, 609 (2019). 10.1038/s41586-018-0824-5 [DOI] [PubMed] [Google Scholar]
- 12.Endres M. et al. Immune Pathways in Etiology, Acute Phase, and Chronic Sequelae of Ischemic Stroke. Circ Res 130, 1167–1186 (2022). 10.1161/CIRCRESAHA.121.319994 [DOI] [PubMed] [Google Scholar]
- 13.Qiu M. et al. Cell Heterogeneity Uncovered by Single-Cell RNA Sequencing Offers Potential Therapeutic Targets for Ischemic Stroke. Aging Dis 13, 1436–1454 (2022). 10.14336/AD.2022.0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iadecola C. & Anrather J. The immunology of stroke: from mechanisms to translation. Nat Med 17, 796–808 (2011). 10.1038/nm.2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu L. et al. Interactions between the Autonomic Nervous System and the Immune System after Stroke. Compr Physiol 12, 3665–3704 (2022). 10.1002/cphy.c210047 [DOI] [PubMed] [Google Scholar]
- 16.Beuker C. et al. Stroke induces disease-specific myeloid cells in the brain parenchyma and pia. Nat Commun 13, 945 (2022). 10.1038/s41467-022-28593-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zheng K. et al. Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J Cereb Blood Flow Metab, 0271678X211026770 (2021). 10.1177/0271678X211026770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakahashi-Oda C. et al. CD300a blockade enhances efferocytosis by infiltrating myeloid cells and ameliorates neuronal deficit after ischemic stroke. Sci Immunol 6, eabe7915 (2021). 10.1126/sciimmunol.abe7915 [DOI] [PubMed] [Google Scholar]
- 19.Kim S. et al. The antioxidant enzyme Peroxiredoxin-1 controls stroke-associated microglia against acute ischemic stroke. Redox Biol 54, 102347 (2022). 10.1016/j.redox.2022.102347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li X. et al. Single-cell transcriptomic analysis of the immune cell landscape in the aged mouse brain after ischemic stroke. J Neuroinflammation 19, 83 (2022). 10.1186/s12974-022-02447-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Bonilla L. et al. Inducible nitric oxide synthase in neutrophils and endothelium contributes to ischemic brain injury in mice. J Immunol 193, 2531–2537 (2014). 10.4049/jimmunol.1400918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garcia-Bonilla L. et al. Role of microglial and endothelial CD36 in post-ischemic inflammasome activation and interleukin-1beta-induced endothelial activation. Brain Behav Immun 95, 489–501 (2021). 10.1016/j.bbi.2021.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gelderblom M. et al. Temporal and spatial dynamics of cerebral immune cell accumulation in stroke. Stroke 40, 1849–1857 (2009). 10.1161/STROKEAHA.108.534503 [DOI] [PubMed] [Google Scholar]
- 24.Jin R., Yang G. & Li G. Inflammatory mechanisms in ischemic stroke: role of inflammatory cells. J Leukoc Biol 87, 779–789 (2010). 10.1189/jlb.1109766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Butovsky O. et al. Identification of a unique TGF-beta-dependent molecular and functional signature in microglia. Nat Neurosci 17, 131–143 (2014). 10.1038/nn.3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matcovitch-Natan O. et al. Microglia development follows a stepwise program to regulate brain homeostasis. Science 353, aad8670 (2016). 10.1126/science.aad8670 [DOI] [PubMed] [Google Scholar]
- 27.Holtman I. R., Skola D. & Glass C. K. Transcriptional control of microglia phenotypes in health and disease. J Clin Invest 127, 3220–3229 (2017). 10.1172/JCI90604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ochocka N. et al. Single-cell RNA sequencing reveals functional heterogeneity of glioma-associated brain macrophages. Nat Commun 12, 1151 (2021). 10.1038/s41467-021-21407-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zheng K. et al. Single-cell RNA-seq reveals the transcriptional landscape in ischemic stroke. J Cereb Blood Flow Metab 42, 56–73 (2022). 10.1177/0271678X211026770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammond T. R. et al. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity 50, 253–271 e256 (2019). 10.1016/j.immuni.2018.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keren-Shaul H. et al. A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217 (2017). 10.1016/j.cell.2017.05.018 [DOI] [PubMed] [Google Scholar]
- 32.Lalancette-Hebert M. et al. Galectin-3 is required for resident microglia activation and proliferation in response to ischemic injury. J Neurosci 32, 10383–10395 (2012). 10.1523/JNEUROSCI.1498-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Venkatraman A. et al. Galectin-3: an emerging biomarker in stroke and cerebrovascular diseases. Eur J Neurol 25, 238–246 (2018). 10.1111/ene.13496 [DOI] [PubMed] [Google Scholar]
- 34.Shichita T. et al. MAFB prevents excess inflammation after ischemic stroke by accelerating clearance of damage signals through MSR1. Nat Med 23, 723–732 (2017). 10.1038/nm.4312 [DOI] [PubMed] [Google Scholar]
- 35.Llovera G. et al. The choroid plexus is a key cerebral invasion route for T cells after stroke. Acta Neuropathol 134, 851–868 (2017). 10.1007/s00401-017-1758-y [DOI] [PubMed] [Google Scholar]
- 36.Krasemann S. et al. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity 47, 566–581 e569 (2017). 10.1016/j.immuni.2017.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zheng J. et al. Single-cell RNA-seq analysis reveals compartment-specific heterogeneity and plasticity of microglia. iScience 24, 102186 (2021). 10.1016/j.isci.2021.102186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Silvin A. et al. Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55, 1448–1465 e1446 (2022). 10.1016/j.immuni.2022.07.004 [DOI] [PubMed] [Google Scholar]
- 39.Zanier E. R., Fumagalli S., Perego C., Pischiutta F. & De Simoni M. G. Shape descriptors of the “never resting” microglia in three different acute brain injury models in mice. Intensive Care Med Exp 3, 39 (2015). 10.1186/s40635-015-0039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang C. T., Wu W. F., Deng Y. H. & Ge J. W. Modulators of microglia activation and polarization in ischemic stroke (Review). Mol Med Rep 21, 2006–2018 (2020). 10.3892/mmr.2020.11003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jordao M. J. C. et al. Single-cell profiling identifies myeloid cell subsets with distinct fates during neuroinflammation. Science 363 (2019). 10.1126/science.aat7554 [DOI] [PubMed] [Google Scholar]
- 42.Yona S. et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 38, 79–91 (2013). 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda T. et al. Specification of CNS macrophage subsets occurs postnatally in defined niches. Nature 604, 740–748 (2022). 10.1038/s41586-022-04596-2 [DOI] [PubMed] [Google Scholar]
- 44.Ofengeim D. et al. RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc Natl Acad Sci U S A 114, E8788–E8797 (2017). 10.1073/pnas.1714175114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Somebang K. et al. CCR2 deficiency alters activation of microglia subsets in traumatic brain injury. Cell Rep 36, 109727 (2021). 10.1016/j.celrep.2021.109727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khorooshi R. et al. Induction of endogenous Type I interferon within the central nervous system plays a protective role in experimental autoimmune encephalomyelitis. Acta Neuropathol 130, 107–118 (2015). 10.1007/s00401-015-1418-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McDonough A. et al. Ischemia/Reperfusion Induces Interferon-Stimulated Gene Expression in Microglia. J Neurosci 37, 8292–8308 (2017). 10.1523/JNEUROSCI.0725-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roy E. R. et al. Type I interferon response drives neuroinflammation and synapse loss in Alzheimer disease. J Clin Invest 130, 1912–1930 (2020). 10.1172/JCI133737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thored P. et al. Long-term accumulation of microglia with proneurogenic phenotype concomitant with persistent neurogenesis in adult subventricular zone after stroke. Glia 57, 835–849 (2009). 10.1002/glia.20810 [DOI] [PubMed] [Google Scholar]
- 50.Van Hove H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 22, 1021–1035 (2019). 10.1038/s41593-019-0393-4 [DOI] [PubMed] [Google Scholar]
- 51.Dani N. et al. A cellular and spatial map of the choroid plexus across brain ventricles and ages. Cell 184, 3056–3074 e3021 (2021). 10.1016/j.cell.2021.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerganova G., Riddell A. & Miller A. A. CNS border-associated macrophages in the homeostatic and ischaemic brain. Pharmacol Ther 240, 108220 (2022). 10.1016/j.pharmthera.2022.108220 [DOI] [PubMed] [Google Scholar]
- 53.Mildenberger W., Stifter S. A. & Greter M. Diversity and function of brain-associated macrophages. Curr Opin Immunol 76, 102181 (2022). 10.1016/j.coi.2022.102181 [DOI] [PubMed] [Google Scholar]
- 54.Goldmann T. et al. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol 17, 797–805 (2016). 10.1038/ni.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Van Hove H. et al. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci 13, 206 (2019). 10.1038/s41593-019-0393-4 [DOI] [PubMed] [Google Scholar]
- 56.Rajan W. D. et al. Defining molecular identity and fates of CNS-border associated macrophages after ischemic stroke in rodents and humans. Neurobiol Dis 137, 104722 (2020). 10.1016/j.nbd.2019.104722 [DOI] [PubMed] [Google Scholar]
- 57.Miedema A. et al. Brain macrophages acquire distinct transcriptomes in multiple sclerosis lesions and normal appearing white matter. Acta Neuropathol Commun 10, 8 (2022). 10.1186/s40478-021-01306-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaitin D. A. et al. Lipid-Associated Macrophages Control Metabolic Homeostasis in a Trem2-Dependent Manner. Cell 178, 686–698 e614 (2019). 10.1016/j.cell.2019.05.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamaguchi A. et al. Temporal expression profiling of DAMPs-related genes revealed the biphasic post-ischemic inflammation in the experimental stroke model. Mol Brain 13, 57 (2020). 10.1186/s13041-020-00598-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Amorim A. et al. IFNgamma and GM-CSF control complementary differentiation programs in the monocyte-to-phagocyte transition during neuroinflammation. Nat Immunol 23, 217–228 (2022). 10.1038/s41590-021-01117-7 [DOI] [PubMed] [Google Scholar]
- 61.Menezes S. et al. The Heterogeneity of Ly6C(hi) Monocytes Controls Their Differentiation into iNOS(+) Macrophages or Monocyte-Derived Dendritic Cells. Immunity 45, 1205–1218 (2016). 10.1016/j.immuni.2016.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patsalos A. et al. A growth factor-expressing macrophage subpopulation orchestrates regenerative inflammation via GDF-15. J Exp Med 219 (2022). 10.1084/jem.20210420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams J. W. et al. Limited proliferation capacity of aortic intima resident macrophages requires monocyte recruitment for atherosclerotic plaque progression. Nat Immunol 21, 1194–1204 (2020). 10.1038/s41590-020-0768-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rizzo G. et al. Dynamics of monocyte-derived macrophage diversity in experimental myocardial infarction. Cardiovasc Res (2022). 10.1093/cvr/cvac113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mangale V. et al. Microglia influence host defense, disease, and repair following murine coronavirus infection of the central nervous system. Glia 68, 2345–2360 (2020). 10.1002/glia.23844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gliem M. et al. Macrophage-derived osteopontin induces reactive astrocyte polarization and promotes re-establishment of the blood brain barrier after ischemic stroke. Glia 63, 2198–2207 (2015). 10.1002/glia.22885 [DOI] [PubMed] [Google Scholar]
- 67.Nakano Y. et al. Glycoprotein nonmetastatic melanoma protein B (GPNMB) as a novel neuroprotective factor in cerebral ischemia-reperfusion injury. Neuroscience 277, 123–131 (2014). 10.1016/j.neuroscience.2014.06.065 [DOI] [PubMed] [Google Scholar]
- 68.Cai W. et al. STAT6/Arg1 promotes microglia/macrophage efferocytosis and inflammation resolution in stroke mice. JCI Insight 4 (2019). 10.1172/jci.insight.131355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rajan W. D. et al. Dissecting functional phenotypes of microglia and macrophages in the rat brain after transient cerebral ischemia. Glia 67, 232–245 (2019). 10.1002/glia.23536 [DOI] [PubMed] [Google Scholar]
- 70.Kalucka J. et al. Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764–779 e720 (2020). 10.1016/j.cell.2020.01.015 [DOI] [PubMed] [Google Scholar]
- 71.Vanlandewijck M. et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature 554, 475–480 (2018). 10.1038/nature25739 [DOI] [PubMed] [Google Scholar]
- 72.Chen M. B. et al. Brain Endothelial Cells Are Exquisite Sensors of Age-Related Circulatory Cues. Cell Rep 30, 4418–4432 e4414 (2020). 10.1016/j.celrep.2020.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meng H. et al. LRG1 promotes angiogenesis through upregulating the TGFbeta1 pathway in ischemic rat brain. Mol Med Rep 14, 5535–5543 (2016). 10.3892/mmr.2016.5925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Minten C. et al. DARC shuttles inflammatory chemokines across the blood-brain barrier during autoimmune central nervous system inflammation. Brain 137, 1454–1469 (2014). 10.1093/brain/awu045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang G. et al. Neutralization of Lipocalin-2 Diminishes Stroke-Reperfusion Injury. Int J Mol Sci 21 (2020). 10.3390/ijms21176253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhuang X., Cross D., Heath V. L. & Bicknell R. Shear stress, tip cells and regulators of endothelial migration. Biochemical Society Transactions 39, 1571–1575 (2011). 10.1042/BST20110746 [DOI] [PubMed] [Google Scholar]
- 77.Madureira P. A. et al. The role of the annexin A2 heterotetramer in vascular fibrinolysis. Blood 118, 4789–4797 (2011). 10.1182/blood-2011-06-334672 [DOI] [PubMed] [Google Scholar]
- 78.Tezuka K. et al. Activation of Annexin A2 signaling at the blood-brain barrier in a mouse model of multiple sclerosis. J Neurochem 160, 662–674 (2022). 10.1111/jnc.15578 [DOI] [PubMed] [Google Scholar]
- 79.Hongu T. et al. Perivascular tenascin C triggers sequential activation of macrophages and endothelial cells to generate a pro-metastatic vascular niche in the lungs. Nature Cancer 3, 486–504 (2022). 10.1038/s43018-022-00353-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rohlenova K. et al. Single-Cell RNA Sequencing Maps Endothelial Metabolic Plasticity in Pathological Angiogenesis. Cell Metabolism 31, 862–877.e814 (2020). 10.1016/j.cmet.2020.03.009 [DOI] [PubMed] [Google Scholar]
- 81.Matsuoka R. L., Buck L. D., Vajrala K. P., Quick R. E. & Card O. A. Historical and current perspectives on blood endothelial cell heterogeneity in the brain. Cell Mol Life Sci 79, 372 (2022). 10.1007/s00018-022-04403-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Androvic P. et al. Decoding the Transcriptional Response to Ischemic Stroke in Young and Aged Mouse Brain. Cell Rep 31, 107777 (2020). 10.1016/j.celrep.2020.107777 [DOI] [PubMed] [Google Scholar]
- 83.Zhang W. et al. Differential expression of receptors mediating receptor-mediated transcytosis (RMT) in brain microvessels, brain parenchyma and peripheral tissues of the mouse and the human. Fluids Barriers CNS 17, 47 (2020). 10.1186/s12987-020-00209-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Camilli C., Hoeh A. E., De Rossi G., Moss S. E. & Greenwood J. LRG1: an emerging player in disease pathogenesis. J Biomed Sci 29, 6 (2022). 10.1186/s12929-022-00790-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jin J. et al. LRG1 Promotes Apoptosis and Autophagy through the TGFbeta-smad1/5 Signaling Pathway to Exacerbate Ischemia/Reperfusion Injury. Neuroscience 413, 123–134 (2019). 10.1016/j.neuroscience.2019.06.008 [DOI] [PubMed] [Google Scholar]
- 86.Wang X. et al. LRG1 promotes angiogenesis by modulating endothelial TGF-beta signalling. Nature 499, 306–311 (2013). 10.1038/nature12345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pek S. L. et al. Elevation of a novel angiogenic factor, leucine-rich-alpha2-glycoprotein (LRG1), is associated with arterial stiffness, endothelial dysfunction, and peripheral arterial disease in patients with type 2 diabetes. J Clin Endocrinol Metab 100, 1586–1593 (2015). 10.1210/jc.2014-3855 [DOI] [PubMed] [Google Scholar]
- 88.Kumagai S. et al. Myeloid cell-derived LRG attenuates adverse cardiac remodelling after myocardial infarction. Cardiovasc Res 109, 272–282 (2016). 10.1093/cvr/cvv273 [DOI] [PubMed] [Google Scholar]
- 89.Grieshaber-Bouyer R. et al. The neutrotime transcriptional signature defines a single continuum of neutrophils across biological compartments. Nat Commun 12, 2856 (2021). 10.1038/s41467-021-22973-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Turk V. et al. Cysteine cathepsins: from structure, function and regulation to new frontiers. Biochim Biophys Acta 1824, 68–88 (2012). 10.1016/j.bbapap.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lee H. K. et al. Neuroprotective and Anti-inflammatory Effects of a Dodecamer Peptide Harboring Ninjurin 1 Cell Adhesion Motif in the Postischemic Brain. Mol Neurobiol 55, 6094–6111 (2018). 10.1007/s12035-017-0810-1 [DOI] [PubMed] [Google Scholar]
- 92.Evrard M. et al. Developmental Analysis of Bone Marrow Neutrophils Reveals Populations Specialized in Expansion, Trafficking, and Effector Functions. Immunity 48, 364–379 e368 (2018). 10.1016/j.immuni.2018.02.002 [DOI] [PubMed] [Google Scholar]
- 93.Gelderblom M. et al. IL-23 (Interleukin-23)-Producing Conventional Dendritic Cells Control the Detrimental IL-17 (Interleukin-17) Response in Stroke. Stroke 49, 155–164 (2018). 10.1161/STROKEAHA.117.019101 [DOI] [PubMed] [Google Scholar]
- 94.Posel C. et al. Flow cytometric characterization of brain dendritic cell subsets after murine stroke. Exp Transl Stroke Med 6, 11 (2014). 10.1186/2040-7378-6-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Clarkson B. D. et al. CCR7 deficient inflammatory Dendritic Cells are retained in the Central Nervous System. Sci Rep 7, 42856 (2017). 10.1038/srep42856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ludewig P. et al. Dendritic cells in brain diseases. Biochim Biophys Acta 1862, 352–367 (2016). 10.1016/j.bbadis.2015.11.003 [DOI] [PubMed] [Google Scholar]
- 97.Miller J. C. et al. Deciphering the transcriptional network of the dendritic cell lineage. Nat Immunol 13, 888–899 (2012). 10.1038/ni.2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Durai V. et al. Cryptic activation of an Irf8 enhancer governs cDC1 fate specification. Nat Immunol 20, 1161–1173 (2019). 10.1038/s41590-019-0450-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grajales-Reyes G. E. et al. Batf3 maintains autoactivation of Irf8 for commitment of a CD8alpha(+) conventional DC clonogenic progenitor. Nat Immunol 16, 708–717 (2015). 10.1038/ni.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brown C. C. et al. Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity. Cell 179, 846–863 e824 (2019). 10.1016/j.cell.2019.09.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cochain C. et al. Single-Cell RNA-Seq Reveals the Transcriptional Landscape and Heterogeneity of Aortic Macrophages in Murine Atherosclerosis. Circ Res 122, 1661–1674 (2018). 10.1161/CIRCRESAHA.117.312509 [DOI] [PubMed] [Google Scholar]
- 102.Ngoi S. et al. Migrating Type 2 Dendritic Cells Prime Mucosal Th17 Cells Specific to Small Intestinal Commensal Bacteria. J Immunol 209, 1200–1211 (2022). 10.4049/jimmunol.2200204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dress R. J. et al. Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat Immunol 20, 852–864 (2019). 10.1038/s41590-019-0420-3 [DOI] [PubMed] [Google Scholar]
- 104.Xu H. et al. Transcriptional Atlas of Intestinal Immune Cells Reveals that Neuropeptide α-CGRP Modulates Group 2 Innate Lymphoid Cell Responses. Immunity 51, 696–708.e699 (2019). 10.1016/j.immuni.2019.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kiner E. et al. Gut CD4+ T cell phenotypes are a continuum molded by microbes, not by TH archetypes. Nat. Immunol. 22, 216–228 (2021). 10.1038/s41590-020-00836-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lee J.-Y. et al. The Transcription Factor KLF2 Restrains CD4+ T Follicular Helper Cell Differentiation. Immunity 42, 252–264 (2015). 10.1016/j.immuni.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cai W. et al. Neuroprotection against ischemic stroke requires a specific class of early responder T cells in mice. J Clin Invest 132 (2022). 10.1172/JCI157678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Benakis C. et al. Commensal microbiota affects ischemic stroke outcome by regulating intestinal γδ T cells. Nat. Med. 22, 516–523 (2016). 10.1038/nm.4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gelderblom M. et al. Neutralization of the IL-17 axis diminishes neutrophil invasion and protects from ischemic stroke. Blood 120, 3793–3802 (2012). 10.1182/blood-2012-02-412726 [DOI] [PubMed] [Google Scholar]
- 110.Shichita T. et al. Pivotal role of cerebral interleukin-17-producing gammadeltaT cells in the delayed phase of ischemic brain injury. Nat. Med. 15, 946–950 (2009). 10.1038/nm.1999 [DOI] [PubMed] [Google Scholar]
- 111.Miragaia R. J. et al. Single-Cell Transcriptomics of Regulatory T Cells Reveals Trajectories of Tissue Adaptation. Immunity 50, 493–504.e497 (2019). 10.1016/j.immuni.2019.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Shi L. et al. Treg cell-derived osteopontin promotes microglia-mediated white matter repair after ischemic stroke. Immunity (2021). 10.1016/j.immuni.2021.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bjorkstrom N. K., Strunz B. & Ljunggren H. G. Natural killer cells in antiviral immunity. Nat Rev Immunol 22, 112–123 (2022). 10.1038/s41577-021-00558-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Bottcher J. P. et al. NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037 e1014 (2018). 10.1016/j.cell.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ritzel R. M. et al. Aging alters the immunological response to ischemic stroke. Acta Neuropathol 136, 89–110 (2018). 10.1007/s00401-018-1859-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shi L. et al. Genome-wide transcriptomic analysis of microglia reveals impaired responses in aged mice after cerebral ischemia. J Cereb Blood Flow Metab 40, S49–S66 (2020). 10.1177/0271678X20925655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Ritzel R. M. et al. Age-Associated Resident Memory CD8 T Cells in the Central Nervous System Are Primed To Potentiate Inflammation after Ischemic Brain Injury. J Immunol 196, 3318–3330 (2016). 10.4049/jimmunol.1502021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ritzel R. M. et al. Aging alters the immunological response to ischemic stroke. Acta Neuropathol. (Berl). 136, 89–110 (2018). 10.1007/s00401-018-1859-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ritzel R. M. et al. Age-Associated Resident Memory CD8 T Cells in the Central Nervous System Are Primed To Potentiate Inflammation after Ischemic Brain Injury. The Journal of Immunology 196, 3318–3330 (2016). 10.4049/jimmunol.1502021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ahnstedt H. et al. Sex differences in T cell immune responses, gut permeability and outcome after ischemic stroke in aged mice. Brain, Behavior, and Immunity 87, 556–567 (2020). 10.1016/j.bbi.2020.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Inacio A. R. et al. Endogenous IFN-beta signaling exerts anti-inflammatory actions in experimentally induced focal cerebral ischemia. J Neuroinflammation 12, 211 (2015). 10.1186/s12974-015-0427-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Huang Y. et al. Repopulated microglia are solely derived from the proliferation of residual microglia after acute depletion. Nat Neurosci 21, 530–540 (2018). 10.1038/s41593-018-0090-8 [DOI] [PubMed] [Google Scholar]
- 123.Todd B. P. et al. Traumatic brain injury results in unique microglial and astrocyte transcriptomes enriched for type I interferon response. J Neuroinflammation 18, 151 (2021). 10.1186/s12974-021-02197-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Androvic P. et al. Decoding the Transcriptional Response to Ischemic Stroke in Young and Aged Mouse Brain. Cell Reports 31, 107777 (2020). 10.1016/j.celrep.2020.107777 [DOI] [PubMed] [Google Scholar]
- 125.Garcia-Bonilla L. et al. Spatio-temporal profile, phenotypic diversity, and fate of recruited monocytes into the post-ischemic brain. J Neuroinflammation 13, 285 (2016). 10.1186/s12974-016-0750-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miro-Mur F. et al. Immature monocytes recruited to the ischemic mouse brain differentiate into macrophages with features of alternative activation. Brain Behav Immun 53, 18–33 (2016). 10.1016/j.bbi.2015.08.010 [DOI] [PubMed] [Google Scholar]
- 127.Hayashi T., Noshita N., Sugawara T. & Chan P. H. Temporal profile of angiogenesis and expression of related genes in the brain after ischemia. J Cereb Blood Flow Metab 23, 166–180 (2003). 10.1097/01.WCB.0000041283.53351.CB [DOI] [PubMed] [Google Scholar]
- 128.Beck H. & Plate K. H. Angiogenesis after cerebral ischemia. Acta Neuropathol 117, 481–496 (2009). 10.1007/s00401-009-0483-6 [DOI] [PubMed] [Google Scholar]
- 129.Kilkenny C. et al. Animal research: reporting in vivo experiments--the ARRIVE guidelines. J Cereb Blood Flow Metab 31, 991–993 (2011). 10.1038/jcbfm.2010.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Percie du Sert N. et al. The ARRIVE guidelines 2.0: updated guidelines for reporting animal research. J Physiol (2020). 10.1113/JP280389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Jackman K., Kunz A. & Iadecola C. Modeling focal cerebral ischemia in vivo. Methods Mol Biol 793, 195–209 (2011). 10.1007/978-1-61779-328-8_13 [DOI] [PubMed] [Google Scholar]
- 132.Ximerakis M. et al. Single-cell transcriptomic profiling of the aging mouse brain. Nat Neurosci 22, 1696–1708 (2019). 10.1038/s41593-019-0491-3 [DOI] [PubMed] [Google Scholar]
- 133.Soltoff S. P., McMillian M. K. & Talamo B. R. Coomassie Brilliant Blue G is a more potent antagonist of P2 purinergic responses than Reactive Blue 2 (Cibacron Blue 3GA) in rat parotid acinar cells. Biochem Biophys Res Commun 165, 1279–1285 (1989). 10.1016/0006-291x(89)92741-1 [DOI] [PubMed] [Google Scholar]
- 134.Hrvatin S. et al. Single-cell analysis of experience-dependent transcriptomic states in the mouse visual cortex. Nat Neurosci 21, 120–129 (2018). 10.1038/s41593-017-0029-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Macosko E. Z. et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161, 1202–1214 (2015). 10.1016/j.cell.2015.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Datlinger P. et al. Pooled CRISPR screening with single-cell transcriptome readout. Nat Meth 14, 297–301 (2017). 10.1038/nmeth.4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Saunders A. et al. Molecular Diversity and Specializations among the Cells of the Adult Mouse Brain. Cell 174, 1015–1030.e1016 (2018). 10.1016/j.cell.2018.07.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Campbell J., Yang S., Wang Z., Corbett S. & Koga Y. celda: CEllular Latent Dirichlet Allocation. (2022).
- 140.McGinnis C. S., Murrow L. M. & Gartner Z. J. DoubletFinder: Doublet Detection in Single-Cell RNA Sequencing Data Using Artificial Nearest Neighbors. Cell Systems 8, 329–337.e324 (2019). 10.1016/j.cels.2019.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Stuart T. et al. Comprehensive Integration of Single-Cell Data. Cell 177, 1888–1902.e1821 (2019). 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Korsunsky I. et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Meth 16, 1289–1296 (2019). 10.1038/s41592-019-0619-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.McInnes L., Healy J. & Melville J. UMAP: Uniform Manifold Approximation and Projection for Dimension Reduction. arXiv:1802.03426 [cs, stat] (2020). [Google Scholar]
- 144.Finak G. et al. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biology 16, 278 (2015). 10.1186/s13059-015-0844-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Aran D. et al. Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat. Immunol. 20, 163–172 (2019). 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Heng T. S. P. et al. The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). 10.1038/ni1008-1091 [DOI] [PubMed] [Google Scholar]
- 147.Tabula Muris C. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018). 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Neumann J. et al. Microglia cells protect neurons by direct engulfment of invading neutrophil granulocytes: a new mechanism of CNS immune privilege. J Neurosci 28, 5965–5975 (2008). 10.1523/JNEUROSCI.0060-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Ritchie M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43, e47 (2015). 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.McCarthy D. J., Campbell K. R., Lun A. T. & Wills Q. F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics 33, 1179–1186 (2017). 10.1093/bioinformatics/btw777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Saelens W., Cannoodt R., Todorov H. & Saeys Y. A comparison of single-cell trajectory inference methods. Nat Biotechnol 37, 547–554 (2019). 10.1038/s41587-019-0071-9 [DOI] [PubMed] [Google Scholar]
- 152.Street K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018). 10.1186/s12864-018-4772-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jin S. et al. Inference and analysis of cell-cell communication using CellChat. Nat Commun 12, 1088 (2021). 10.1038/s41467-021-21246-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Baccin C. et al. Combined single-cell and spatial transcriptomics reveal the molecular, cellular and spatial bone marrow niche organization. Nat Cell Biol 22, 38–48 (2020). 10.1038/s41556-019-0439-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kim H. et al. Development of a Validated Interferon Score Using NanoString Technology. Journal of Interferon & Cytokine Research 38, 171–185 (2018). 10.1089/jir.2017.0127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ballesteros I. et al. Co-option of Neutrophil Fates by Tissue Environments. Cell 183, 1282–1297 e1218 (2020). 10.1016/j.cell.2020.10.003 [DOI] [PubMed] [Google Scholar]
- 157.Wickham H. ggplot2: Elegant graphics for data analysis. (Springer-Verlag; New York, 2016). [Google Scholar]
- 158.Wei T. & Simko V. R package ‘corrplot’: Visualization of a correlation matrix. (2021). [Google Scholar]
- 159.Schindelin J. et al. Fiji: an open-source platform for biological-image analysis. Nat Methods 9, 676–682 (2012). 10.1038/nmeth.2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw and processed data and metadata of all scRNA-seq datasets included in this study are available in the GEO repository (GSE225948). A public accessible interactive web portal for exploring the scRNA-seq data included in this study has been developed (https://anratherlab.shinyapps.io/strokevis/).